
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 February 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00213
 Ning Jia1,2,3
Ning Jia1,2,3 Jiqin Liu4
Jiqin Liu4 Penghui Tan5
Penghui Tan5 Yufeng Sun1,2
Yufeng Sun1,2 Yuemeng Lv3
Yuemeng Lv3 Jiameng Liu1,2
Jiameng Liu1,2 Jing Sun1,2
Jing Sun1,2 Yatao Huang1,2
Yatao Huang1,2 Jia Lu1,2
Jia Lu1,2 Nuo Jin1,2
Nuo Jin1,2 Minmin Li1,2
Minmin Li1,2 Khandaker Md Sharif Uddin Imam3
Khandaker Md Sharif Uddin Imam3 Fengjiao Xin3*
Fengjiao Xin3* Bei Fan1,2*
Bei Fan1,2*Varieties of Citrus are commercially important fruits that are cultivated worldwide and are valued for being highly nutritious and having an appealing flavor. Lignification of citrus fruit juice sacs is a serious physiological disorder that occurs during postharvest storage, for which the underlying transcriptional regulatory mechanisms remain unclear. In this study, we identified and isolated a candidate MYB transcription factor, CsMYB85, that is involved in the regulation of lignin biosynthesis in Citrus sinensis, which has homologs in Arabidopsis and other plants. We found that during juice sac lignification, CsMYB85 expression levels increase significantly, and therefore, suspected that this gene may control lignin biosynthesis during the lignification process. Our results indicated that CsMYB85 binds the CsMYB330 promoter, regulates its expression, and interacts with CsMYB308 in transgenic yeast and tobacco. A transient expression assay indicated that Cs4CL1 expression levels and lignin content significantly increased in fruit juice sacs overexpressing CsMYB85. At4CL1 expression levels and lignin content were also significantly increased in Arabidopsis overexpressing CsMYB85. We accordingly present convincing evidence for the participation of the CsMYB85 transcription factor in fruit juice sac lignification, and thereby provide new insights into the transcriptional regulation of this process in citrus fruits.
Varieties of Citrus are commercially important fruits cultivated worldwide that are valued for being highly nutritious and having an appealing flavor. Citrus fruit juice sac granulation (lignification) is a serious physiological disorder that occurs during the postharvest storage of these fruits (Shomer et al., 1989). In the granulation process, the pulp tissue becomes hard and lignified, which diminishes fruit nutritional and commercial value (Pan et al., 1999). As fruit juice sac granulation increases, the cell walls thicken by lignification. Consequently, the juice sacs become deformed and atrophied, and the number of normal juice cells gradually decreases (Shomer et al., 1989; Pan et al., 1999). Previous studies have demonstrated that juice sac lignin content increases during granulation and plays a vital role in this process (Zhang et al., 2016; Jia et al., 2018). Lignin accumulation is significantly correlated with the juice sac granulation index (Zhang et al., 2016). During the granulation process, transcription levels of the CmMYB330 and CsMYB308 transcription factor genes are either significantly upregulated or downregulated and homologs identified in Arabidopsis and other plant species have also been identified as being involved in lignin biosynthesis regulation (Yang et al., 2017a; Jia et al., 2018). Accordingly, an in-depth functional study of lignin biosynthesis-related genes may enhance our understanding of juice sac granulation.
The biosynthesis of lignin involves a series of sequential enzymatic reactions, in the first step of which, cinnamic acid hydroxylase (C4H), phenylalanine ammonia-lyase (PAL), hydroxystyrene CoA, and ligase (4CL) participate in the phenylpropylene ring pathway. Thereafter, caffeic acid-O-methyltransferase (COMT) and ferulate 5-hydroxylase (F5H) methylate lignin monomers, which are further modified via cinnamyl alcohol dehydrogenase (CAD) and cinnamate-CoA reductase (CCR) (Kawaoka et al., 2010; Vanholme et al., 2010), followed by polymerization catalyzed by PER/LAC.
A number of MYB and NAC transcription factors play important roles in the transcription and regulation of lignin synthesis-related genes (Zhong et al., 2006; Golfier et al., 2017; Koshiba et al., 2017), as both transcriptional activators and repressors. Previously, it has been demonstrated that overexpression of AtMYB46 or AtMYB83 promotes the upregulation of lignin synthesis-related genes, resulting in abnormal secondary cell wall accumulation (Ko et al., 2009). In the Arabidopsis myb46myb83 double mutant, plants are unable to form secondary walls and die during the seedling stage (Wang and Dixon, 2012). In other plant species, several MYB transcription factors are associated with lignin synthesis, including PtrMYB3, PtrMYB20, EgMYB2, PvMYB4, PtMYB1, PtMYB4, PtMYB8, OsMYB46, and ZmMYB (Patzlaff et al., 2003; Bomal et al., 2008; McCarthy et al., 2010; Rahantamalala et al., 2010; Zhong et al., 2011; Shen et al., 2012). In tomato, SlMYB1 and SlMYB2 have been found to regulate cellulose metabolism (Shi et al., 2017), whereas in chrysanthemum, overexpression of the CmMYB19 transcription enhances lignin content and aphid tolerance in transgenic plants (Wang et al., 2017). Numerous MYB transcription factors, including AtMYB58, AtMYB63, and AtMYB85, recognize and bind lignin synthesis gene promoters, thereby regulating their expression (Zhong et al., 2007; Zhou et al., 2009).
Transcriptional repressors that have been implicated in lignin synthesis include Arabidopsis AtMYB32 and AtMYB4 (Jin et al., 2000; Preston et al., 2004). Overexpression of AtMYB4 in tobacco results in a significant downregulation of lignin synthesis-related gene expression and retards growth (Jin et al., 2000), whereas AtMYB32 binds to the COMT promoter and upregulates COMT expression in the myb32 mutant (Preston et al., 2004). Several transcriptional repressors have also been identified in other plants, including eucalyptus (EgMYB1) (Legay, 2005), Antirrhinum (AmMYB330 and AmMYB308) (Tamagnone et al., 1998), Eriobotrya japonica (EjMYB2) (Xu et al., 2014), and maize (ZmMYB31 and ZmMYB42) (Sonbol et al., 2009; Fornalé et al., 2010). EgMYB1 downregulates EgCCR and EgCAD expression (Legay et al., 2007), whereas PtoMYB156 downregulates secondary wall formation during xylogenesis in Populus tomentosa via the phenylpropanoid pathway (Yang et al., 2017b), and overexpression of Maize ZmMYB31 in Arabidopsis thaliana reduces lignin content of transgenic plants, which show a dwarf phenotype (Fornalé et al., 2010).
Aspects of the regulatory mechanisms underlying fruit lignification have been reported previously. In Citrus sinensis, the CsMYB330 and CsMYB308 transcription factors have been found to regulate fruit juice sac lignification by regulating the expression of Cs4CL1 (Jia et al., 2018). Loquat fruit lignin content significantly increases following cold exposure, and in this context, and it has been shown that EjMYB1 and EjMYB2 recognize and bind to the AC element of the Ej4CL1 promoter, thereby regulating Ej4CL1 expression and lignin biosynthesis (Xu et al., 2014). EjMYB8 has also been found to regulate lignin biosynthesis in loquat fruits (Wang et al., 2016) and EjNAC3 regulates EjCAD-like expression and influences lignin content (Ge et al., 2017). Although EjAP2-1 is known to regulate lignin biosynthesis in loquat fruits, it does not directly influence lignin synthesis gene expression, but instead controls interactions between the EjMYB1 and EjMYB2 proteins (Zeng et al., 2015).
Nevertheless, although a number of MYB transcription factors related to lignin metabolism have been widely studied, their functions in citrus fruit lignification have seldom been analyzed. In this study, we isolated and identified the R2R3 MYB transcription factor CsMYB85 and examined its roles in citrus fruits juice sac lignification. Our results indicate that this CsMYB transcription factor indirectly regulates citrus fruit juice sac lignification.
In June 2017, fresh naturally ripened Citrus sinensis fruits of similar size and maturity were harvested and selected in Yichang, Hubei Province, China. They were maintained in ventilated cold storage in which the climate was controlled at 8.0 ± 2.0°C and 75.5% ± 5.0% relative humidity (RH). The fruit juice sacs showed different levels of granulation according to the method described by Zhang et al. (2016).
Wild-type (WT) Arabidopsis thaliana (Columbia) and Nicotiana benthamiana were grown either in perlite/vermiculite/peat soil (1:1:1 v/v/v) or on Murashige and Skoog (MS) media plates in a greenhouse under 16 h light and 8 h darkness at 22°C and 40–60% RH.
Total RNA and genomic DNA of C sinensis fruit juice sacs and Arabidopsis plants were extracted using RNA and DNA extraction kits (Aidlab, Beijing, China). The cDNA was synthesized from 2.5 μg of total RNA using a RevertAid premium first-strand cDNA synthesis kit (Fermentas, Thermo Fisher Scientific, Rochester, NY, United States), and used as a template for quantitative real-time polymerase chain reaction (qRT-PCR) and reverse-transcription PCR (RT-PCR) analyses. For qRT-PCR, we used a SYBR Green PCR MasterMix (TaKaRa Bio, Inc., Kusatsu, Shiga, Japan). Amplification was performed using a 7500 fast quantitative PCR system (Applied Biosystems, Foster City, CA, United States). CsActin and AtActin were used as reference genes to normalize expression levels. All qRT-PCR analyses were repeated on four biological replicates. The primers used for amplification are listed in Supplementary Table 1.
On the basis of the sweet orange genome sequence (Xu et al., 2013), the CsMYB85 coding region was cloned via RT-PCR using gene-specific primers (Supplementary Table 2). The promoters were isolated by PCR using the promoter-specific primers listed in Supplementary Table 2. The RT-PCR products were cloned in the SK vector. The protein sequences were blasted in the NCBI database to identify homologous sequences in other plant species. DNAMAN v. 6.0 (Lynnon LLC, San Ramon, CA, United States) was used to align and construct a phylogenetic tree of MYB proteins.
The subcellular localization of CsMYB85 was determined by cloning the full-length coding region lacking a stop codon into the 3302GFP vector fused with the green fluorescent protein (GFP) gene. Pro-35S::CsMYB85-GFP and control 3302GFP (Pro-35S::GFP) vectors were transfected into Agrobacterium (GV3101) and subsequently transiently expressed in N. benthamiana leaf cells (Voinnet et al., 2003). After infiltration for 48 h, fluorescence images were observed under a confocal laser scanning microscope (SP8; Leica Microsystems, Wetzlar, Germany). Cell nuclei were stained with 4’,6-diamidino-2- phenylindole (DAPI). The primers used for cloning are listed in Supplementary Table 1.
Lignin content was determined using the method described by Zhang et al. (2016). All lignin content analyses were performed with three biological replicates.
To assess CsMYB85 transcriptional activation in yeast cells, CsMYB85 was cloned into the pGBKT7 (BD) vector (Clontech Laboratories, Mountain View, CA, United States). BD-CsMYB85, pGADT7 (AD), and a negative control (AD + BD vector) were transformed into the yeast strain Y2H Gold (Clontech Laboratories, Mountain View, CA, United States) using PEG/LiAc method. The yeast cells were grown on SD medium lacking Leu and Trp (SD/-Leu-Trp or SD/-L-T) or Leu, Trp, and His (SD/-Leu-Trp-His or SD/-L-T-H). Transcriptional activation by CsMYB85 was evaluated according to yeast growth status.
CsMYB85 coding sequences were ligated into to the AD vector, whereas the CsMYB330 and CsMYB308 promoters were inserted into the pHIS2 vector. The primers used for vector construction are listed in Supplementary Table 2. Other vectors used have been described previously (Jia et al., 2018). Recombinant pHIS2 and empty AD (control) vectors were transformed into yeast strain Y187 (Clontech Laboratories, Mountain View, CA, United States), which was grown on SD screening medium (SD/-Leu-Trp-His) containing different concentrations of 3-amino-1,2,4-triazole (3-AT), an appropriate concentration of which suppresses background histidine leakiness in pHIS2 vectors. Interactions between CsMYB85 and its promoters were assessed on SD screening medium (SD/-Leu-Trp-His) containing the optimal 3-AT concentration.
The coding sequences of CsMYB85 and CsMYB308 were inserted into the pGADT7 (AD) and pGBKT7 (BD) vectors. BD-53 and AD-T were used as positive controls and empty BD and AD were used as negative controls. The plasmids were co-transfected into yeast strain AH109 (Clontech Laboratories, Mountain View, CA, United States) via PEG-mediated transformation, and the transformed yeast cells were grown on SD screening medium (SD/-Leu-Trp and SD/-Leu-Trp-His) for 3 days at 30°C.
On the basis of the results of the one-hybrid yeast assay, transcriptional activity was analyzed according to Liu et al. (2016) and Jia et al. (2018). The CsMYB330 promoter was cloned into the 35S-LUC-GUS vector to drive GUS reporter gene expression, with LUC being used as an internal standard. The coding sequence of the CsMYB85 transcription factor was cloned into the 35S-LUC-GUS vector driven by the cauliflower mosaic virus 35S (CAM35S) promoter. The plasmids were transformed into Agrobacterium GV3101, which was cultured in infiltration buffer containing 10 mM MgCl2, 10 mM MES, and 100 μM acetosyringone until OD600 = 0.5. For transcriptional activity analysis, N. benthamiana plants were infiltrated with Agrobacterium GV3101 containing recombinant plasmids using needleless syringes. GUS and LUC activities were measured after 3 days. The primers used for vector construction are listed in Supplementary Table 2.
BiFC assays were performed using N. benthamiana leaves as described previously (Voinnet et al., 2003). CsMYB85 and CsMYB308 sequences were inserted into 35S-SPYNE and 35S-SPYCE vectors and fused with N-terminal yellow fluorescent protein (YFPN) and/or C-terminal YFP (YFPC). The BiFC vectors were introduced into Agrobacterium GV3101, which was infiltrated into N. benthamiana leaves using a needleless syringe. YFP fluorescence signals were visualized under a confocal laser scanning microscope (SP8; Leica Microsystems, Wetzlar, Germany) 3 days after infiltration. The primers used for BiFC vector construction are listed in Supplementary Table 2.
Juice sacs were separated from sweet orange fruits and placed into a Petri dish containing MS medium supplemented with 0.2% gelrite (Endo et al., 2007). The coding sequence of CsMYB85 sequence was inserted into the SK vector under control of the CAM35S promoter. Empty SK vector was used as a control. The PDS-1000/He particle delivery system (Bio-Rad Laboratories, Hercules, CA, United States) was used for particle gun bombardment according to manufacturer instructions. Gold particles (1.0 μm in diameter) were packed with SK-CsMYB85 or SK plasmids. The target distance between the Petri dish and the stop screen was 9 cm and helium pressure was 9.3 MPa. After bombardment, the juice sacs were incubated on MS medium at 25°C and subsequently collected at 24 and 120 h for gene expression analysis and lignin content assay, respectively.
CsMYB85 isolated from sweet orange (C. sinensis) and its orthologs in other plants play important roles in lignin biosynthesis. The amino acid sequence of this transcription factor is similar to that of AtMYB85 from Arabidopsis (60%), and was accordingly named CsMYB85. CsMYB85 encodes a protein comprising 267 amino acids and has an isoelectric point and molecular weight of 5.08 and 30.14 kDa, respectively. CsMYB85 has a conserved R2R3 domain and is thus classified as an R2R3 MYB transcription factor (Figure 1A). In a phylogenetic tree constructed for MYB transcription factors, we found that CsMYB85 was clustered with the lignin-related AtMYB85 and AtMYB42 (Figure 1B), which may also regulate lignin biosynthesis (Zhong et al., 2008). We accordingly speculate that CsMYB85 is involved in the regulation of lignin biosynthesis.
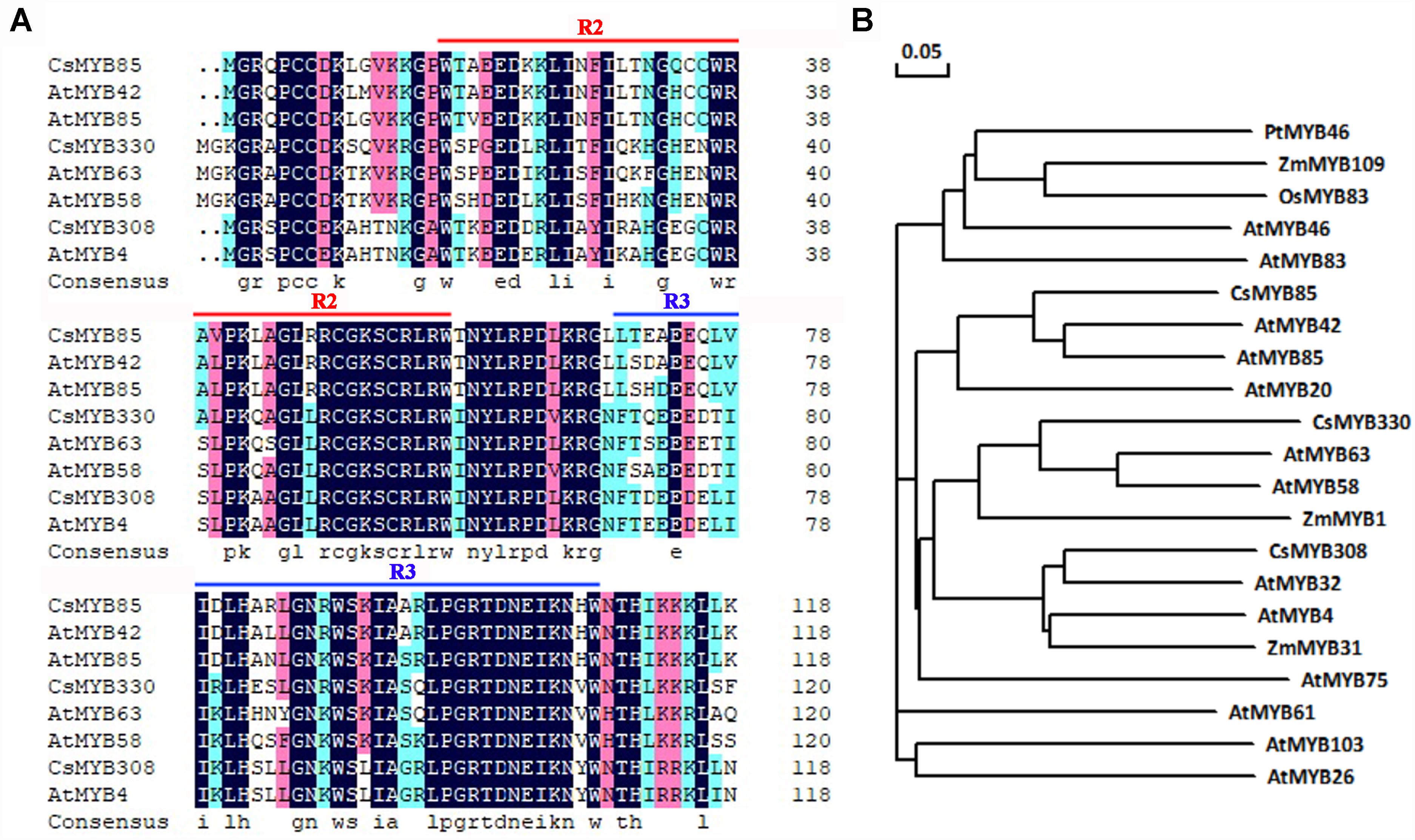
Figure 1. CsMYB85 sequence and phylogenetic analysis. (A) Alignment of the conserved R2R3 domains in CsMYB85 with those in AtMYB42, AtMYB85, AtMYB58, AtMYB63, AtMYB4, CsMYB330, and CsMYB308. (B) Phylogenetic tree for CsMYB85 and MYB transcription factors from other plants: Populus trichocarpa, Arabidopsis thaliana, Zea mays, and Oryza sativa. The accession numbers of the transcription factor proteins are as follows: PtMYB46 (XP_024465368.1), ZmMYB109 (NP_001241859.1), OsMYB83 (XP_015619488.1), AtMYB46 (AT5G12870), AtMYB83 (At3g08500), CsMYB85 (XP_006477265.1), AtMYB42 (AT4G12350), AtMYB85 (AT4G22680), AtMYB20 (AT4G66230), CsMYB330 (NC_023049), AtMYB63 (AT1G79180), AtMYB58 (AT1G16490), ZmMYB1 (P20024.1), CsMYB308 (NC_023053), AtMYB32 (AT4G34990), AtMYB4 (AT4G38620), ZmMYB31 (NP_001105949), AtMYB75 (AT1G56650), AtMYB61 (AT1G09540), AtMYB103 (AT1G63910), and AtMYB26 (AT3G13890) The phylogenetic tree was constructed using DNAMAN v. 6.0.
In previous studies, it has been observed that lignin content increases significantly with the progression of citrus fruit granulation (Zhang et al., 2016; Jia et al., 2018). On the basis of CsMYB85 expression during citrus fruit granulation, we performed qRT-PCR analysis to determine its association with lignin biosynthesis. We accordingly found that CsMYB85 transcription levels increased with the advance of citrus fruit granulation, being relatively low at granulation level 0, but showing an approximate 13-fold increase by level 4 (Figure 2). This increase in the expression level CsMYB85 is consistent with the increase in lignin content during citrus fruit granulation (Figure 2).
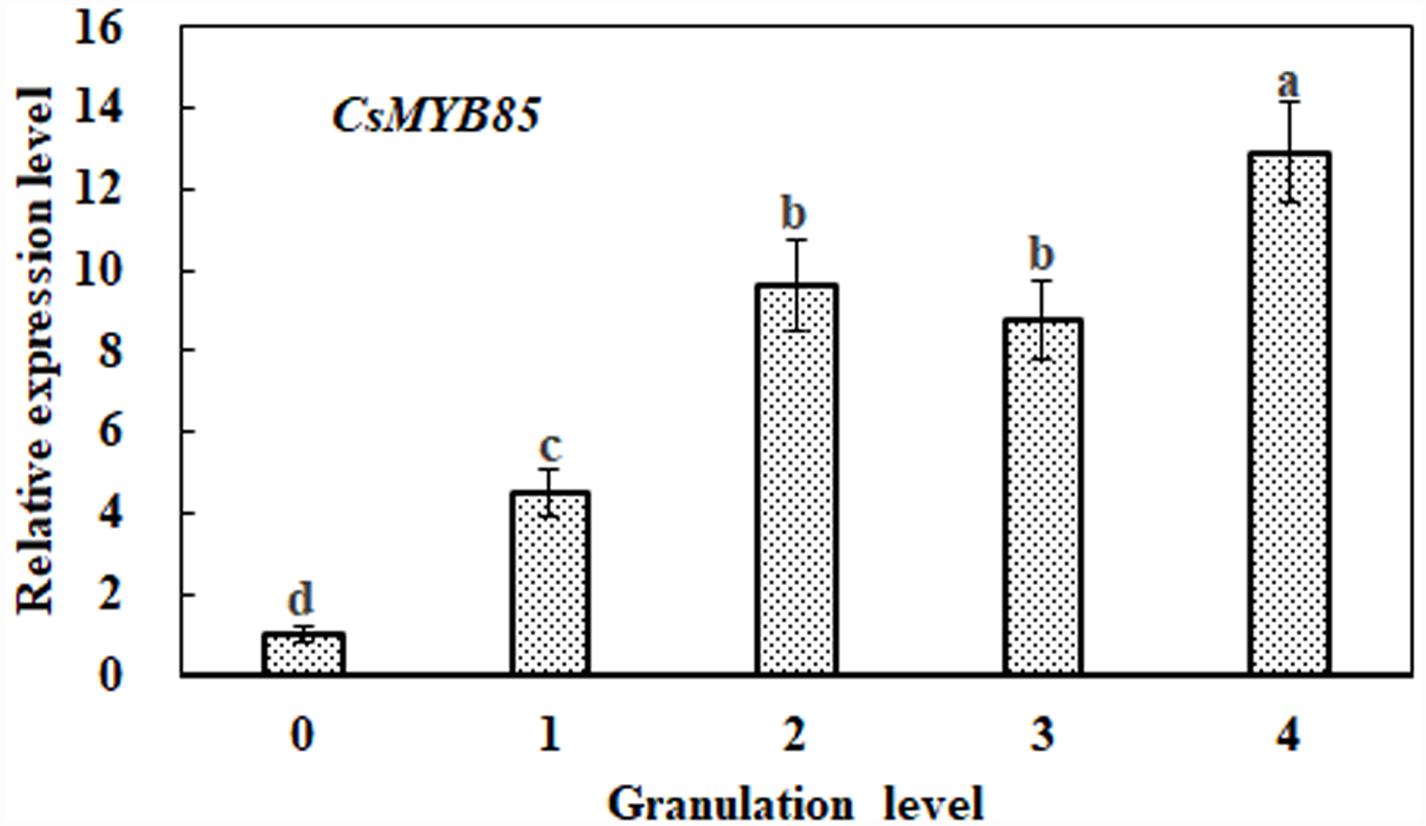
Figure 2. Relative expression of the CsMYB85 gene at different stages of citrus fruit granulation. CsMYB85 expression was determined by real-time qRT-PCR and normalized to the CsActin reference gene. Means were obtained from four biological replicates. Error bars represent standard errors and were determined using Duncan’s multiple range test (α = 0.05) in SAS software (SAS Institute, Cary, NC, United States). The ‘a–d’ above each column indicates P < 0.05 and the same letter indicates that the difference is not significant.
MYBs transcription factors are typically nuclear proteins (Zhong et al., 2007; Xu et al., 2014; Jia et al., 2018). Consistently, we found that when a vector harboring CsMYB85 and GFP under control of the CaMV 35S promoter was expressed in tobacco leaves, CsMYB85-GFP signals were detected in the nucleus, whereas positive control GFP signals were observed in both nucleus and cytoplasm (Figure 3A). To examine CsMYB85 transcriptional activity, a pGBKT7 (BD)-CsMYB85 recombinant vector was constructed and transformed into the Y2HGold strain of yeast along with pGADT7 (AD). We observed that the pGADT7 (AD) vector could activate Leu synthase and the pGBKT7 (BD) vector containing CsMYB85 could activate Trp and His synthase, and thus defective Y2HGold yeast containing the BD-CsMYB85 and AD vectors grew well on SD medium lacking Leu/Trp/His (–L/–T/–H) medium for 3 days at 30°C (Figure 3B), thereby indicating that CsMYB85 is transcriptionally active in yeast.
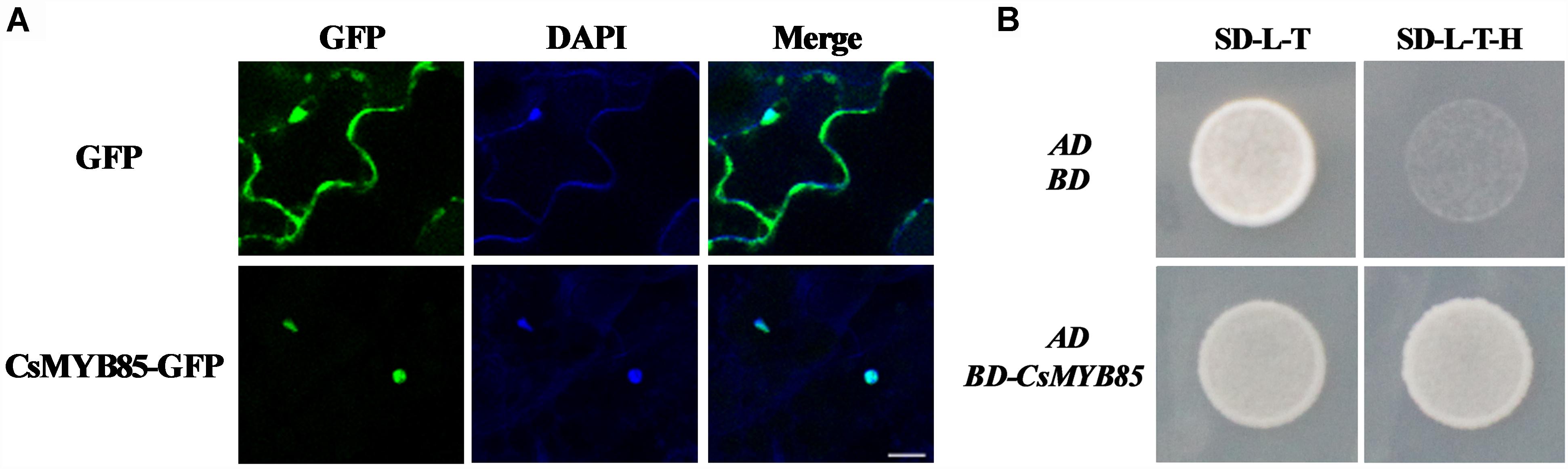
Figure 3. Subcellular localization and transcriptional activation of CsMYB85. (A) Subcellular localization of CsMYB85 in tobacco leaf cells. 35S:GFP was localized in both the nucleus and cytoplasm of tobacco leaf cells. In contrast, CsMYB330:GFP was observed only in the nucleus. 4’,6-Diamidino-2-phenylindole (DAPI) signals were localized only in the nucleus. Merged images show GFP and DAPI colocalization. Bar = 20 μm. (B) Transcriptional activation of CsMYB85 in yeast cells. The full-length coding sequence of CsMYB85 was inserted into the pGBKT7 (BD) vector. The pGADT7 (AD) vector and either BD-CsMYB85 or BD were transformed into cells of the Y2HGold yeast strain. Yeast cells containing AD and BD vectors were used as negative controls. The yeast was grown on SD media lacking –Leu/–Trp (–L/–T) or –Leu/–Trp/–His (–L/–T/–H) for 3 days at 30°C. Results were obtained from three independent transformation experiments.
To confirm the interaction between CsMYB85 and lignin synthesis-related genes, their promoter fragments were cloned from citrus fruit genomic DNA. To establish whether CsMYB85 binds directly to the promoters of these genes, we used the yeast one-hybrid (Y1H) system. The constructed pHIS2 and empty AD vectors were transformed into yeast strain Y187, which was subsequently cultured on screening medium (SD/-Leu-Trp-His) containing different concentrations of 3-AT ranging from 0 to 250 mM. The Y1H assay indicated that CsMYB85 interacts directly and exclusively with the CsMYB330 promoter to activate its expression (Figure 4A), and therefore the CsMYB330 promoter was used in subsequent experiments.
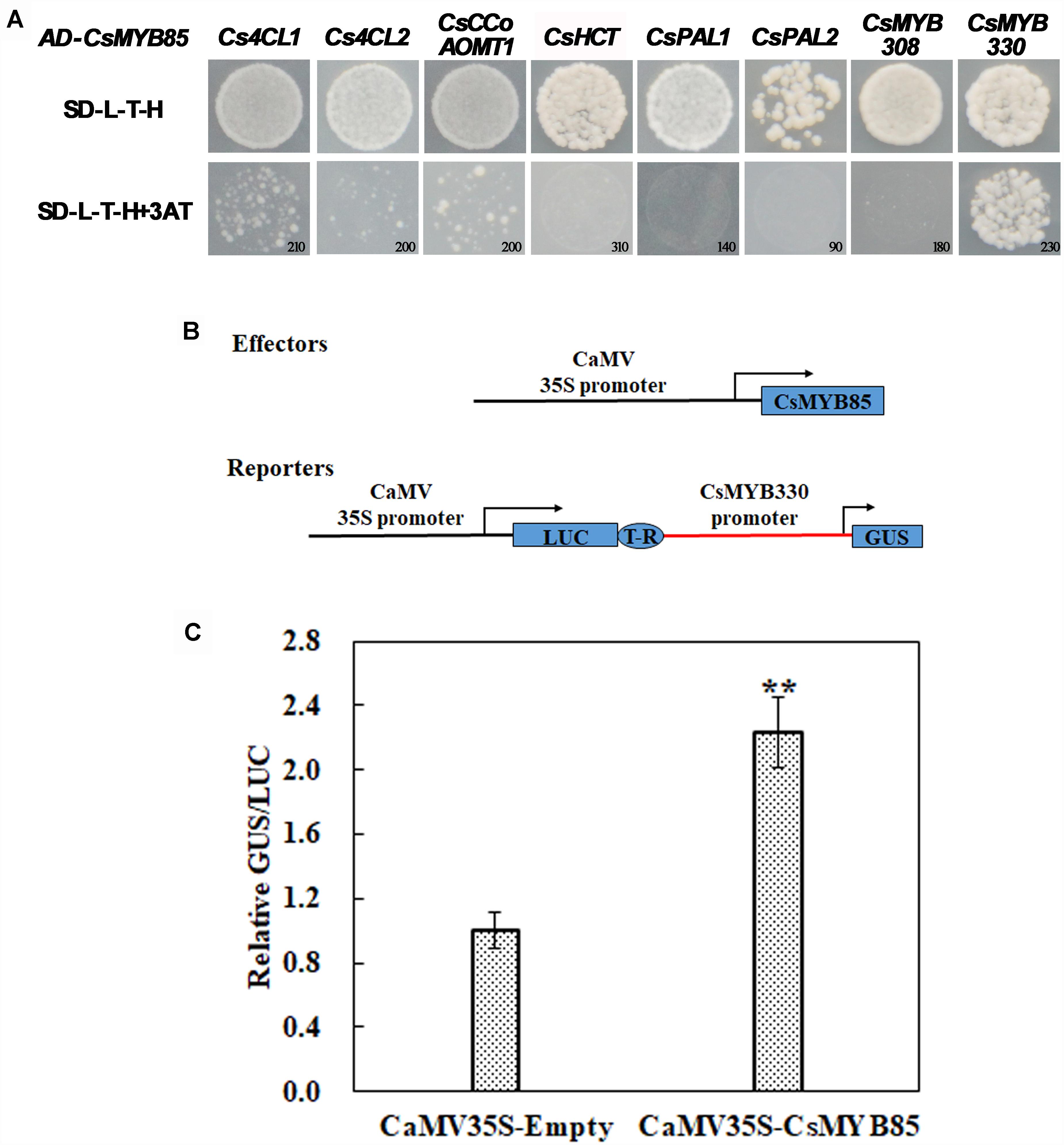
Figure 4. Interactions between CsMYB85 and the promoters of lignin biosynthesis-related genes. (A) Interactions of CsMYB85 with the promoters of CsMYB330, CsMYB308, Cs4CL1, Cs4CL2, Cs4CCoAOMT1, CsHCT, CsPAL1, and CsPAL2 were identified using Y1H assays. The promoters were inserted into pHIS2 vectors. Empty AD and constructed pHIS2 vectors were transfected into yeast Y187 cells, which were grown on SD/–L/–T/–H dropout medium containing various concentrations of 3-amino-1,2,4-triazole (3-AT) to suppress background histidine leakiness. Numbers in the lower-right corner of the images indicate the optimal 3-AT concentrations. Results were obtained from three independent transformation experiments. (B) Activation of the CsMYB330 promoter by CsMYB85 was assayed transiently in Nicotiana benthamiana leaves using an effector and reporter system. The effector and reporter constructs are shown in the schematic diagram. The reporter vector contained LUC normalization and GUS reporter genes driven by the CaMV35S and CsMYB330 promoters, respectively. The effector vector contained a CsMYB85 gene under control of the CaMV35S promoter. T-R, terminator; Boxes, various DNA sequences. (C) Transcriptional activity of CsMYB85 was analyzed using a GUS reporter gene driven by the CsMYB330 promoter. The GUS/LUC ratio in leaves transformed with the empty vector (control) harboring the CsMYB330 promoter was set to 1. Error bars represent standard errors. Means were obtained from five biological replicates. Student’s t-test: ∗∗P < 0.01.
We cloned the CsMYB330 promoter, which we used to drive the GUS reporter gene, and CsMYB85 was driven by the CaMV35S promoter (Figure 4B). Agrobacterium (GV3101) cells containing 35S::CsMYB85 and ProCsMYB330::GUS plasmids were used to transform tobacco plants for transient expression analysis, and the results showed that CsMYB85 activated expression of the GUS reporter gene driven by the CsMYB330 promoter in tobacco leaves (Figure 4C). Thus, we deduced that CsMYB85 activates CsMYB330 expression.
The results of our previous study indicated that the CsMYB308 transcription factor binds directly binds to the Cs4CL1 promoter in yeast cells and participates in juice sac lignification (Jia et al., 2018). However, CsMYB85 does not bind directly bind to lignin biosynthesis-related gene promoters. Therefore, we assumed that CsMYB85 may interact with other CsMYB transcription factors in lignin biosynthesis. To examine protein–protein interactions between CsMYB85 and CsMYB308, we used the yeast two-hybrid system. The coding sequences of CsMYB308 and CsMYB85 were inserted into the AD and BD vectors, respectively, and we found that whereas BD-CsMYB308 was not auto-activated in yeast (Jia et al., 2018), auto-activation was observed with BD-CsMYB85 (Figure 3B). Co-transfection of cells of the yeast strain AH109 with the BD-CsMYB308 and AD-CsMYB85 vectors indicates that CsMYB85 interacts with CsMYB308 (Figure 5).
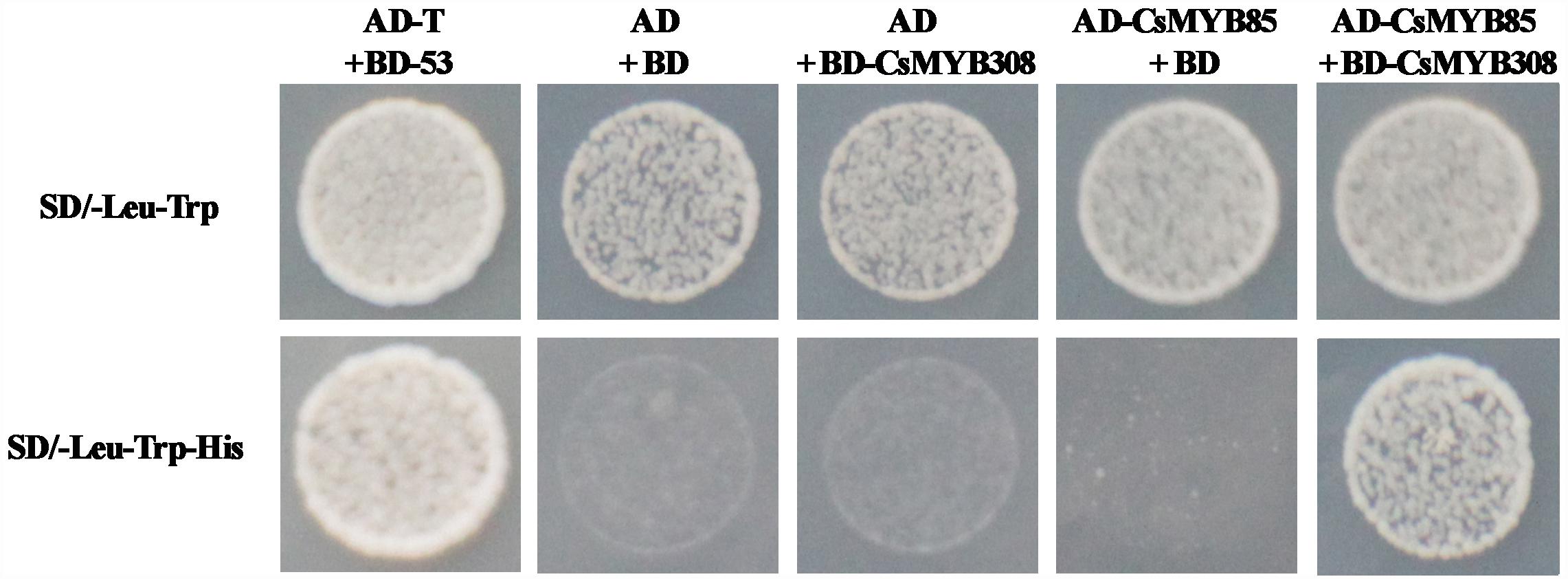
Figure 5. Interaction between CsMYB85 and CsMYB308 determined using a yeast two-hybrid assay. Analyses of CsMYB85 and CsMYB308 interactions in yeast cells. The CsMYB85 and CsMYB308 coding regions were cloned into the AD and BD vectors, respectively. AD-T containing BD-53 and AD containing BD were used as positive and negative controls, respectively. Yeast clones were grown on SD media lacking –Leu/–Trp or –Leu/–Trp/–His for 3 days at 30°C. Results were obtained from three independent transformation experiments.
To validate the results of the yeast two-hybrid assays, BiFC was used to analyze the interaction between CsMYB85 and CsMYB308. These two transcription factors were fused with N-terminal YFP (YFPN) and/or C-terminal YFP (YFPC) driven by the CaMV35S promoter, and the fusion proteins were expressed in N. benthamiana leaves via Agrobacterium (GV3101)-mediated infiltration. A YFP fluorescent signal was detected in the epidermal cell nuclei of plants transformed with 35S:CsMYB85-YFPN, 35S:CsMYB308-YFPC/35S:CsMYB85-YFPC, and 35S:CsMYB308-YFPN constructs, indicating that the CsMYB85 fusion proteins interact with CsMYB308 (Figure 6), thereby confirming the results obtained using the yeast two-hybrid assay.
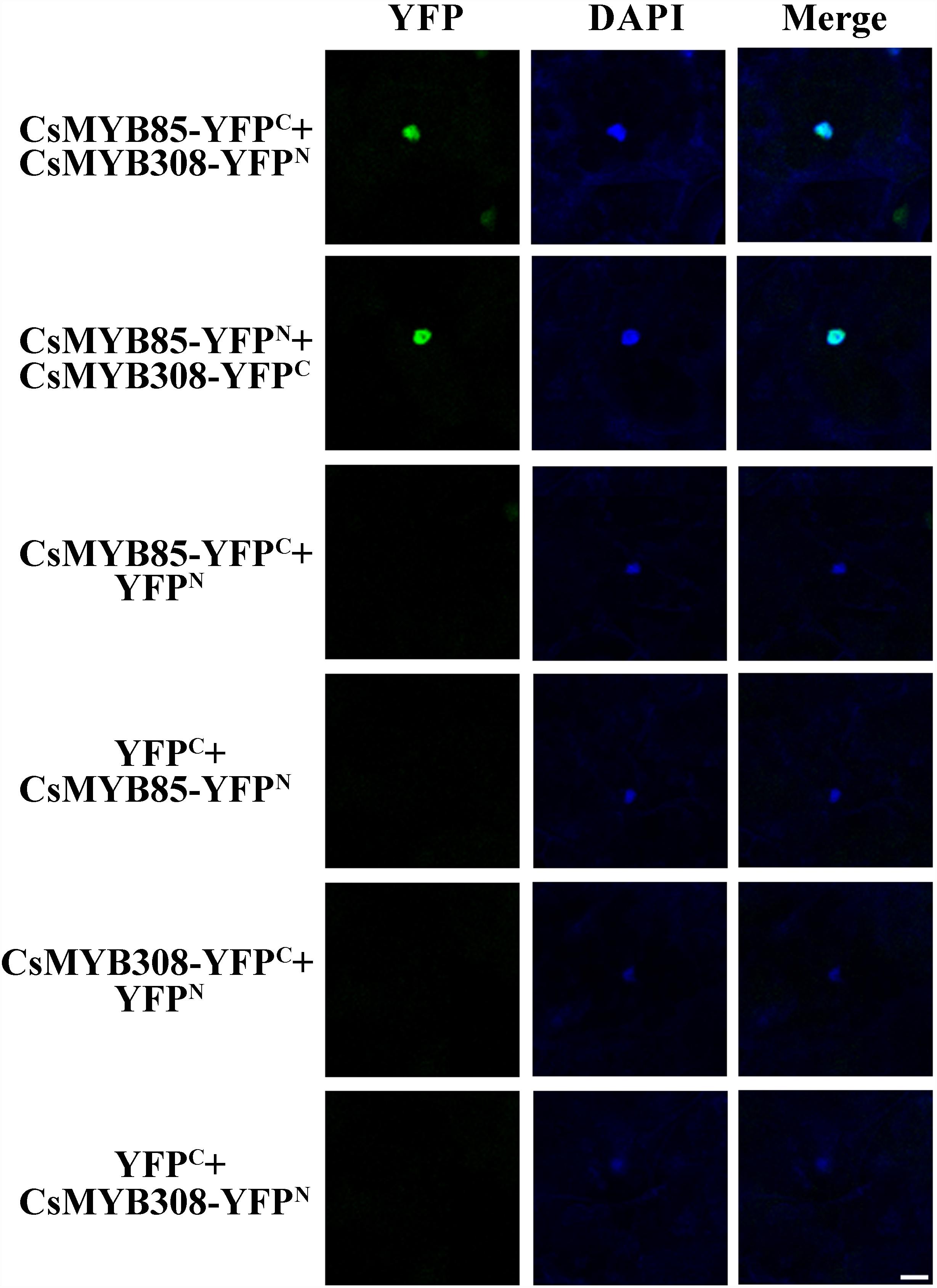
Figure 6. Interaction between CsMYB85 and CsMYB308 determined using a BiFC assay. CsMYB85 and CsMYB308 were fused with C-terminal yellow fluorescent protein (YFPC) and N-terminal YFP (YFPN), respectively, and driven by the CaMV35S promoter. The vectors were introduced into Nicotiana benthamiana leaves by agroinfiltration. YFP fluorescent signals were imaged under a confocal microscope 72 h after infiltration. Panels from left to right: YFP signal (YFP), 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining signal, and merged YFP and DAPI signals. Empty vectors (YFPC and YFPN) and the aforementioned vectors were co-expressed in epidermal cells as a control. Bar = 50 μm.
Obtaining transgenic C. sinensis lines is typically laborious and time-consuming, and therefore we investigated the function of the CsMYB85 transcription factor in lignin biosynthesis regulation via a transient juice sac assay based on particle gun bombardment. We transiently overexpressed CsMYB85 driven by the CaMV35S promoter in juice sac tissue and subsequently analyzed gene expression and lignin content. As shown in Figure 7A, CsMYB85 expression was significantly higher in plants overexpressing CsMYB85 than in those transformed with the empty SK control. Figures 4, 5 indicate that CsMYB85 binds the CsMYB330 promoter and regulates its expression in yeast and tobacco leaf cells. We performed a real-time qRT-PCR assay to evaluate the expression levels of CsMYB330, CsMYB308, and Cs4CL1 in cells containing the SK and SK-CsMYB85 constructs, and the results indicated that the expression levels of all three genes were upregulated in transiently overexpressing juice sac tissue (Figure 7B). We found that the lignin content of juice sac tissue transiently overexpressing SK-CsMYB85 was approximately 1.4-fold higher than in tissue harboring the empty SK vector (Figure 7C). These results accordingly indicate that CsMYB85 indirectly regulates lignin biosynthesis.
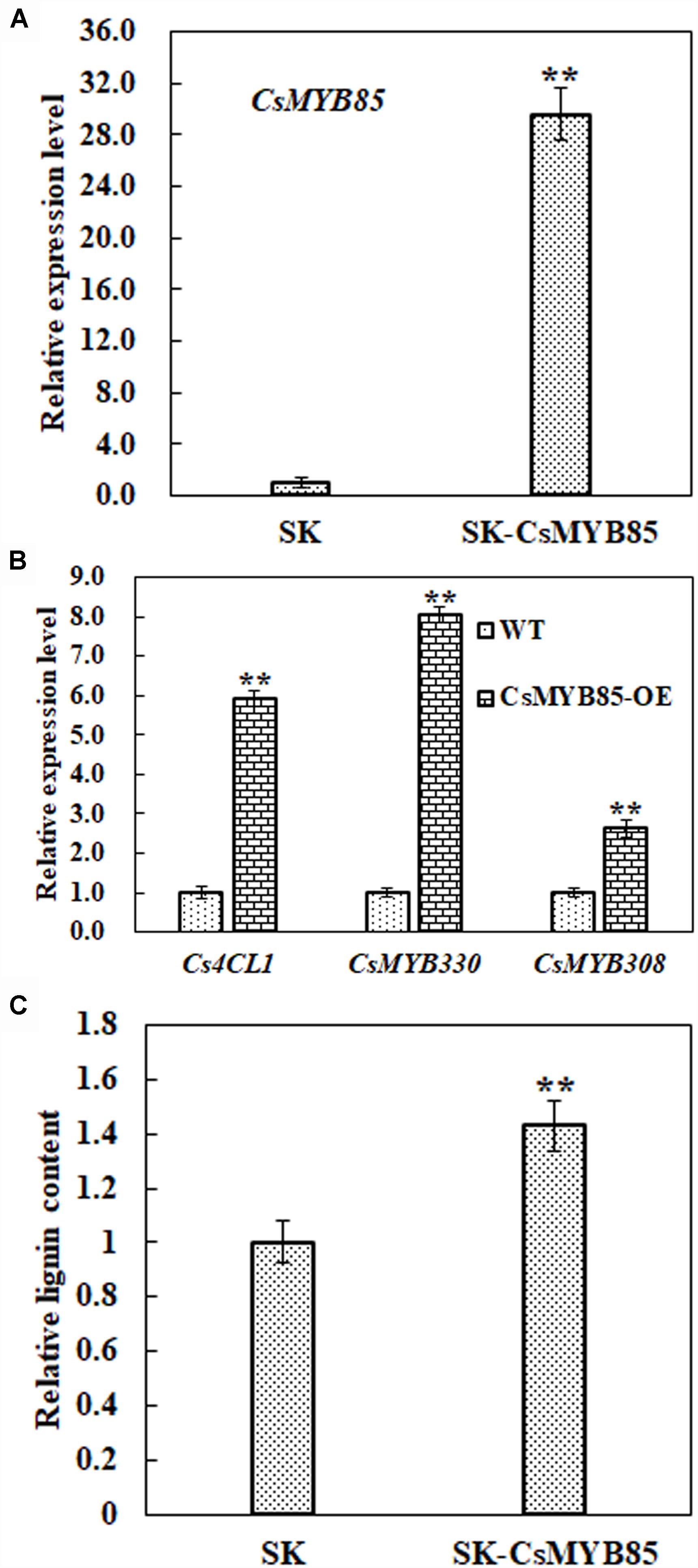
Figure 7. Lignin content and gene expression analyses in transiently overexpressing juice sacs. (A) Expression levels of the CsMYB85 gene in transiently overexpressing juice sac tissues were determined using real-time qRT-PCR. Empty SK vector was used as a control. The CsMYB85 expression level was set to 1 in the cintrol. Expression of the CsActin reference gene (NW_006256915.1) was used to normalize the CsMYB85 expression level. Error bars represent the standard error of four biological replicates. Student’s t-test: ∗∗P < 0.01. (B) Transcription levels of CsMYB330, CsMYB308, and Cs4CL1 (NW_006257196.1) in transiently overexpressing juice sac tissues determined by real-time qRT-PCR and normalized to the CsActin reference gene. Expression levels of CsMYB330, CsMYB308, and Cs4CL1 in the SK control were set to 1. Error bars represent the standard error of four biological replicates. Student’s t-test: ∗∗P < 0.01. (C) Lignin content in juice sac tissues. Means represent the averages of three replicate lignin content determinations in transiently overexpressing juice sac tissues. Error bars represent standard errors. Student’s t-test: ∗∗P < 0.01.
We overexpressed CsMYB85 under the control of the CaMV35S promoter in wild-type (WT) Arabidopsis (Figure 8) and evaluated gene expression and lignin content in the transgenic plants. We accordingly obtained nine CsMYB85 overexpression (CsMYB85-OE) lines and subsequently analyzed overexpression lines 1 and 2, which were characterized by smaller rosettes than those of WT plants (Figure 8A). The Arabidopsis genes AtMYB58 and AtMYB4 are orthologous to the CsMYB330 and CsMYB308 genes, respectively, in C. sinensis (Jia et al., 2018). We performed qRT-PCR to determine the transcription levels of AtMYB58, AtMYB4, and At4CL1 in WT Arabidopsis and CsMYB85-OE transgenic plants, and accordingly observed that AtMYB58, AtMYB4, and At4CL1 expression levels were upregulated in CsMYB85-OE transgenic plants (Figure 8B) and that the lignin content in these transgenic plants was 188% higher than that in the WT plants (Figure 8C).
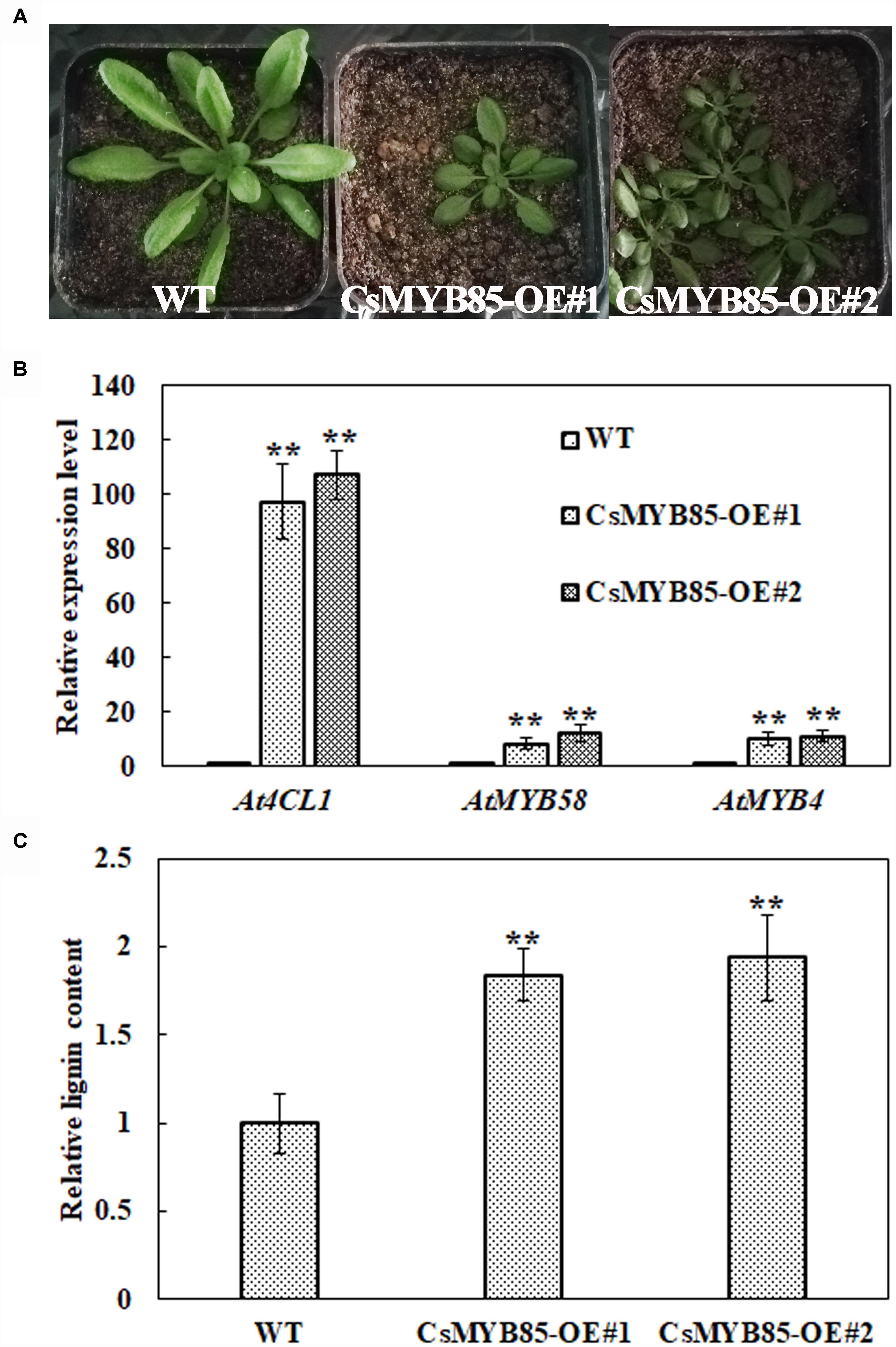
Figure 8. CsMYB85 overexpression increased lignin content in Arabidopsis thaliana. (A) Six-week-old seedlings of wild-type A. thaliana (WT, left), MYB85-OE#1 (CsMYB85 overexpression line 1, center), and MYB85-OE#2 (CsMYB85 overexpression line 2, right). Note the smaller leaves of MYB85-OE#1 and MYB85-OE#2 plants relative to those of the WT. (B) Expression levels of At4CL1 (AT1G51680), AtMYB58, and AtMYB4 were increased in CsMYB85-OE#1 and MYB85-OE#2 plants compared with those in WT plants. At4CL1, AtMYB58, and AtMYB4 expression levels in the WT were set to 1. AtActin was used as an internal reference gene. Error bars represent the standard errors of four biological replicates. Student’s t-test: ∗∗P < 0.01. (C) Lignin content in WT, CsMYB85-OE#1, and MYB85-OE#2 plants. Data are the averages of four biological replicates in WT, CsMYB85-OE#1, and MYB85-OE#2 plants. Error bars represent standard errors. Student’s t-test: ∗∗P < 0.01.
In this study, we identified and isolated the MYB transcription factor CsMYB85 from Citrus sinensis. Sequence analysis based on alignment and phylogenetic tree construction indicated that the function of CsMYB85 is conserved with its orthologs in other plant species (Figure 1). CsMYB85 expression was found to increase significantly during the progression of juice sac granulation (Figure 2), and we determined that CsMYB85 localizes in the nuclei of transformed N. benthamiana cells and has transcriptional activity in yeast (Figure 3). Furthermore, yeast one-hybrid and transcriptional activation assays indicated that CsMYB85 regulates CsMYB330 expression by binding the CsMYB330 promoter (Figure 4). In addition, CsMYB85 overexpression in juice sacs and A. thaliana revealed that CsMYB85 regulates lignin biosynthesis (Figures 5, 6), and therefore we surmise that CsMYB85 may regulate lignin biosynthesis during fruit juice sac granulation in C. sinensis.
Previous studies have shown that cell wall thickening and lignin deposition occur during juice sac granulation (Burns and Achor, 1989; Shomer et al., 1989), and that the levels of lignin, pectin, hemicellulose, and cellulose all increase in the cell walls of granulating fruit juice sacs (Shomer et al., 1989; Zhang et al., 2016). To date, however, little attention has been focused on the identity and role of the transcription factors involved in this process. In a previous paper, we reported that the transcription factors CsMYB330 and CsMYB308 finely regulate Cs4CL1 expression and participate in citrus fruit juice sac lignification (Jia et al., 2018). We also established that CsMYB85 does not bind to the promoters of lignin synthesis-related genes, but does bind to the promoter of the CsMYB330 transcription factor, via which it is assumed to indirectly regulate citrus fruit juice sac lignification.
Transcription factors play prominent roles in regulating gene expression (Taylorteeples et al., 2015). In Arabidopsis and other plants, MYB and NAC family members, including AtMYB85, AtMYB58, AtMYB63, NST1, and NST2, regulate secondary cell wall biosynthesis (Zhou et al., 2009; Zhong et al., 2011; Taylorteeples et al., 2015). Lignin has been found to be ectopically deposited in stem epidermal and cortical cells of plants overexpressing AtMYB85, whereas no corresponding ectopic deposition of cellulose or xylan deposition was observed in these cells. Therefore, it is assumed that overexpression of AtMYB85 promotes the exclusive induction of the lignin biosynthetic pathway. Previously, it was found that AtMYB85 specifically induces GUS expression under control of the At4CL1 promoter (Zhong et al., 2008), and in the present study, we found that lignin content was increased and At4CL1 expression was significantly upregulated in Arabidopsis plants overexpressing CsMYB85 (Figure 8). Given that yeast one-hybrid assays indicated that CsMYB85 does not bind to the Cs4CL1 promoter, we propose that CsMYB85 regulates Cs4CL1 expression via other transcription factors such as CsMYB330 or CsMYB308.
Analysis of the promoters of lignin synthesis-related genes revealed that the AC cis-acting elements of these promoters are recognized by MYB transcription factors and have widespread occurrence (Rogers and Campbell, 2004). Numerous studies have found that Arabidopsis AtMYB4, AtMYB58, and AtMYB63, C. sinensis CsMYB330 and CsMYB308, E. japonica EjMYB1 and EjMYB2, and maize ZmMYB31 transcription factors can bind to AC elements, and thereby regulate gene expression (Zhao et al., 2007; Zhou et al., 2009; Fornalé et al., 2010; Xu et al., 2014; Jia et al., 2018). In the present study, we determined that the CsMYB85 transcription factor binds to the promotor of CsMYB330 in vitro (Figure 4). However, that fact that the CsMYB330 promoter lacks AC elements indicates that some MYB transcription factors recognize sequences other than AC elements and are thus functionally differentiated within their family. Although the Cs4CL1 promoter contains AC elements (Jia et al., 2018) and CsMYB85 induces Cs4CL1 expression (Figure 7B), CsMYB85 cannot physically interact with the Cs4CL1 promoter (Figure 4), and thus it must be assumed that CsMYB85 indirectly regulate Cs4CL1 expression. Similar findings have been reported for the AP2/ERF transcription factor family. For example, EjAP2-1 has been shown to induce E. japonica fruit lignification by interacting with EjMYB transcription factors (Zeng et al., 2015). In Arabidopsis, the AP2 transcription factor AtSHINE (AtSHN) is associated with increased cellulose levels and decreases lignin content by interacting with the promoters of putative rice NAC and MYB transcription factor genes, thereby regulating their expression. Certain transcription factors, including NST1/2/SND1, MYB20/43, MYB58/63, and VND4/5/6, are known to be secondary wall regulators (Ambavaram et al., 2011), and thus EjAP2-1 and AtSHINE are assumed to regulate lignification via different mechanisms. Similarly, our findings revealing that CsMYB85 binds to the CsMYB330 promoter and interacts with CsMYB308 indicate that CsMYB85 regulates citrus fruit lignification through different routes.
Figures 7, 8 show that the expression levels of CsMYB308 and AtMYB4 were significantly upregulated in response to overexpression of CsMYB85. However, Figure 4 indicates that CsMYB85 does not bind to the CsMYB308 promoter, although Figures 5, 6 reveal that CsMYB85 does interact with CsMYB308 in yeast and tobacco leaves. In a previous study, we found that CsMYB308 expression levels were downregulated during juice sac lignification and identified CsMYB308 as a transcriptional repressor in citrus fruit juice sac lignification (Jia et al., 2018). Given that, in the present study, we found that CsMYB308 expression levels were significantly upregulated in response to overexpression of CsMYB85 (Figure 7), we speculate that citrus may inhibit fruit juice sac lignification in response to stress. In Citrus maxima “Hongrou-miyou,” CmMYB330 expression levels were highest 208 days after flowering and thus CmMYB330 may have strongly inhibited fruit juice sac lignification during this period. It is therefore conceivable that CmMYB330 inhibits the onset of granulation and impede its progress (Yang et al., 2017a).
In the present study, we identified and isolated the transcription factor CsMYB85, a novel regulator of Citrus fruit juice sac lignification. The results showed that CsMYB85 regulates Citrus fruit juice sac lignification via different mechanisms. CsMYB85 binds the promoter of CsMYB330, which is a lignin biosynthetic-related regulator. In this way, it induces both CsMYB330 and its downstream genes, including Cs4CL1. These observations suggest that the CsMYB85 has regulatory functions similar to those of its transcription factor homolog in Arabidopsis. Together with the findings from previous studies have revealed a regulatory network is involved in the transcriptional activation of Citrus sinensis fruit juice sacs lignification (Figure 9). We believe that the findings of the present study contribute to establishing the functional classification and elucidating the regulatory mechanisms of MYB transcription factors in postharvest citrus fruit lignification.
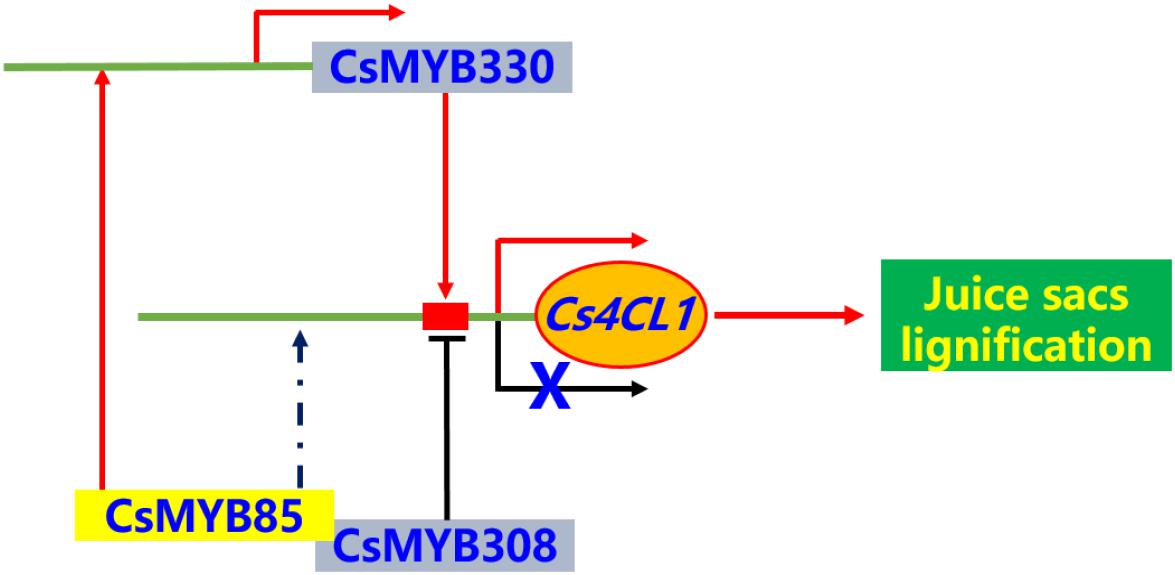
Figure 9. The brief model for regulatory network of Citrus sinensis fruit juice sacs lignification. The MYB transcription factors can activate or inhibit the expression of lignin biosynthesis related genes in Citrus sinensis fruit juice sacs. The Cs4CL1 gene directly involved in lignin biosynthesis. The transcription factors, CsMYB330 and CsMYB330, regulated expression of Cs4CL1 gene, directly. The CsMYB85 transcription factor can interactive with CsMYB308 and regulated expression of CsMYB330 gene, and indirectly regulated expression of Cs4CL1 gene.
NiJ, FX, and BF designed the study. NiJ, JiqL, PT, YS, YL, JiaL, JS, YH, JL, NuJ, ML, and KM performed the experiments. NiJ, FX, and BF wrote the manuscript. All authors have read and approved the manuscript.
This work was supported by the Special Fund for Supervision of Quality and Safety of Agricultural Products (Risk Assessment) from the Chinese Ministry of Agriculture (Grant No. GJFP2018011), the National Key Research and Development Plan (Grant No. 2017YFD0400204), and the Natural Science Foundation of China (Grant No. 31571963).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Drs. Zejian Guo and Xujun Chen for providing the 35S-LUC-GUS vector and for technical assistance with the promoter activation analysis, and Drs. Ke Teng and Zhi Li for helpful discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00213/full#supplementary-material
Ambavaram, M. M. R., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi: 10.1104/pp.110.168641
Bomal, C., Bedon, F., Caron, S., Mansfield, S. D., Levasseur, C., Cooke, J. E., et al. (2008). Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J. Exp. Bot. 59, 3925–3939. doi: 10.1093/jxb/ern234
Burns, J. K., and Achor, D. S. (1989). Cell wall changes in juice vesicles associated with “section drying” in stored late-harvested grapefruit. J. Am. Soc. Hortic. Sci. 114, 283–287.
Endo, T., Shimada, T., Fujii, H., Moriguchi, T., and Omura, M. (2007). Promoter analysis of a type 3 metallothionein-like gene abundant in Satsuma mandarin (Citrus unshiu Marc.) fruit. Sci. Hortic. 112, 207–214. doi: 10.1016/j.scienta.2006.12.042
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644. doi: 10.1111/j.1365-313X.2010.04363.x
Ge, H., Zhang, J., Zhang, Y. J., Li, X., Yin, X. R., Grierson, D., et al. (2017). EjNAC3 transcriptionally regulates chilling-induced lignification of loquat fruit via physical interaction with an atypical CAD-like gene. J. Exp. Bot. 68, 5129–5136. doi: 10.1093/jxb/erx330
Golfier, P., Volkert, C., He, F., Rausch, T., and Wolf, S. (2017). Regulation of secondary cell wall biosynthesis by a NAC transcription factor from Miscanthus. Plant Direct 1, 1–13. doi: 10.1002/pld3.24
Jia, N., Liu, J., Sun, Y., Tan, P., Cao, H., Xie, Y., et al. (2018). Citrus sinensis MYB transcription factors CsMYB330 and CsMYB308 regulate fruit juice sac lignification through fine-tuning expression of the Cs4CL1 gene. Plant Sci. 277, 334–343. doi: 10.1016/j.plantsci.2018.10.006
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Kawaoka, A., Kaothien, P., Yoshida, K., Endo, S., Yamada, K., and Ebinuma, H. (2010). Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J. 22, 289–301. doi: 10.1046/j.1365-313x.2000.00737.x
Ko, J. H., Kim, W. C., and Han, K. H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665. doi: 10.1111/j.1365-313X.2009.03989.x
Koshiba, T., Yamamoto, N., Tobimatsu, Y., Yamamura, M., Suzuki, S., Hattori, T., et al. (2017). MYB-mediated upregulation of lignin biosynthesis in Oryza sativa towards biomass refinery. Plant Biotechnol. 34, 7–15. doi: 10.5511/plantbiotechnology.16.1201a
Legay, S. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. Cell Mol. Biol. 43:553.
Legay, S., Lacombe, E., Goicoechea, M., Brière, C., Séguin, A., Mackay, J., et al. (2007). Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 173, 542–549. doi: 10.1016/j.plantsci.2007.08.007
Liu, J., Chen, X., Liang, X., Zhou, X., Yang, F., Liu, J., et al. (2016). Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 171, 1427–1442. doi: 10.1104/pp.15.01921
McCarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. doi: 10.1093/pcp/pcq064
Pan, D., Zheng, G., Chen, G., She, W., Guo, Z., Shi, M., et al. (1999). Analysis of the reasons caused granulation of juice sacs in guanximiyou pomelo variety. J. Fruit Sci. 16, 202–209.
Patzlaff, A., Newman, L. J., Dubos, C., Whetten, R. W., Smith, C., McInnis, S., et al. (2003). Characterisation of Pt MYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 53, 597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Rahantamalala, A., Rech, P., Martinez, Y., Chaubetgigot, N., and Grimapettenati, J. (2010). Coordinated transcriptional regulation of two key genes in the lignin branch pathway–CAD and CCR–is mediated through MYB-binding sites. BMC Plant Biol. 10:130. doi: 10.1186/1471-2229-10-130
Rogers, L. A., and Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Shen, H., He, X. Z., Poovaiah, C. R., Wuddineh, W. A., Ma, J. Y., Mann, D. G. J., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136. doi: 10.1111/j.1469-8137.2011.03922.x
Shi, Y. N., Liu, X. F., Li, X., Dong, W. C., Grierson, D., Yin, X. R., et al. (2017). SIMYB1 and SIMYB2, two new MYB genes from tomato, transcriptionally regulate cellulose biosynthesis in tobacco. J. Integr. Agric. 16, 65–75. doi: 10.1016/S2095-3119(16)61389-8
Shomer, I., Chalutz, E., Vasiliver, R., Lomaniec, E., and Berman, M. (1989). Sclerification of juice sacs in pummelo (Citrus grandis) fruit. Can. J. Bot. 67, 625–632. doi: 10.1139/b89-084
Sonbol, F. M., Fornalé, S., Capellades, M., Encina, A., Touriño, S., Torres, J. L., et al. (2009). The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 70, 283. doi: 10.1007/s11103-009-9473-2
Tamagnone, L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F. A., Roberts, K., et al. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10, 135–154. doi: 10.1105/tpc.10.2.135
Taylorteeples, M., Lin, L., Lucas, M., De, Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol 153, 895–905. doi: 10.1104/pp.110.155119
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. doi: 10.1046/j.1365-313X.2003.01676.x
Wang, H. Z., and Dixon, R. A. (2012). On-off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, W. Q., Jing, Z., Hang, G., Li, S. J., Xian, L., Yin, X. R., et al. (2016). EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS One 11:e0154399. doi: 10.1371/journal.pone.0154399
Wang, Y., Sheng, L., Zhang, H., Du, X., An, C., Xia, X., et al. (2017). CmMYB19 over-expression improves aphid tolerance in chrysanthemum by promoting lignin synthesis. Int. J. Mol. Sci. 18:619. doi: 10.3390/ijms18030619
Xu, Q., Chen, L., Ruan, X., Chen, D., Zhu, A., Chen, C., et al. (2013). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–U92. doi: 10.1038/ng.2472
Xu, Q., Yin, X., Zeng, J., Ge, H., Song, M., Xu, C., et al. (2014). Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 65, 4349–4359. doi: 10.1093/jxb/eru208
Yang, H., Cai, W., Lv, S., Xu, S., Yu, L., Pan, T., et al. (2017a). Expression analysis of CmMYB330 gene from Citrus maxima “Hongroumiyou.”. J. Fruit Sci. 34, 268–278.
Yang, L., Zhao, X., Ran, L., Li, C., Fan, D., and Luo, K. (2017b). PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 7:41209. doi: 10.1038/srep41209
Zeng, J., Li, X., Xu, Q., Chen, J., Yin, X., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zhang, J., Wang, M., Cheng, F., Dai, C., Sun, Y., Lu, J., et al. (2016). Identification of microRNAs correlated with citrus granulation based on bioinformatics and molecular biology analysis. Postharvest. Biol. Technol. 118, 59–67. doi: 10.1016/j.postharvbio.2016.03.010
Zhao, J., Zhang, W., Zhao, Y., Gong, X., Guo, L., Zhu, G., et al. (2007). SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19:3805. doi: 10.1105/tpc.106.048900
Zhong, R., and Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, R., Lee, C., Zhou, J., Mccarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., Richardson, E. A., and Ye, Z. H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. doi: 10.1105/tpc.107.053678
Keywords: Citrus sinensis, CsMYB85, juice sacs, lignification, postharvest
Citation: Jia N, Liu J, Tan P, Sun Y, Lv Y, Liu J, Sun J, Huang Y, Lu J, Jin N, Li M, Md Sharif Uddin Imam K, Xin F and Fan B (2019) Citrus sinensis MYB Transcription Factor CsMYB85 Induce Fruit Juice Sac Lignification Through Interaction With Other CsMYB Transcription Factors. Front. Plant Sci. 10:213. doi: 10.3389/fpls.2019.00213
Received: 03 November 2018; Accepted: 07 February 2019;
Published: 25 February 2019.
Edited by:
Benoit Schoefs, University of Maine, FranceReviewed by:
Stanislas Thiriet-Rupert, University of Maine, FranceCopyright © 2019 Jia, Liu, Tan, Sun, Lv, Liu, Sun, Huang, Lu, Jin, Li, Md Sharif Uddin Imam, Xin and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengjiao Xin, MjAwMmhvbmd6aGkzMEAxNjMuY29t Bei Fan, ZmFuYmVpNTE3QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.