- 1Department of Molecular Biology, Sejong University, Seoul, South Korea
- 2Department of Bioindustry and Bioresource Engineering, Sejong University, Seoul, South Korea
- 3Plant Engineering Research Institute, Sejong University, Seoul, South Korea
Hemoglobin (Hb) proteins are ubiquitous in plants, and non-symbiotic class 1 hemoglobin (Hb1) is involved in various biotic and abiotic stress responses. Here, the expression of the tobacco (Nicotiana tabacum) hemoglobin gene NtHb1 in Arabidopsis (Arabidopsis thaliana) showed higher cadmium (Cd) tolerance and lower accumulations of Cd, nitric oxide (NO), and reactive oxygen species (ROS) like hydrogen peroxide (H2O2). NtHb1-expressing Arabidopsis exhibited a reduced induction of NO levels in response to Cd, suggesting scavenging of NO by Hb1. In addition, transgenic plants had reduced accumulation of ROS and increased activities of antioxidative enzymes (catalase, superoxide dismutase, and glutathione reductase) in response to Cd. While the expression of the Cd exporters ABC transporter (PDR8) and Ca2+/H+ exchangers (CAXs) was increased, that of the Cd importers iron responsive transporter 1 (IRT1) and P-type 2B Ca2+ ATPase (ACA10) was reduced in response to Cd. When Col-0 plants were treated with the NO donor sodium nitroprusside (SNP) and H2O2, the expression pattern of Cd transporters (PDR8, CAX3, IRT1, and ACA10) was reversed, suggesting that NtHb1 expression decreased the Cd level by regulating the expression of Cd transporters via decreased NO and ROS. Correspondingly, NtHb1-expressing Arabidopsis showed increased Cd export. In summary, the expression of NtHb1 reduces Cd levels by regulating Cd transporter expression via decreased NO and ROS levels in Arabidopsis.
Introduction
Cadmium is a non-essential metal and a major hazardous environmental pollutant because it has toxic effects even at low concentrations. Cadmium negatively influences several aspects of plant metabolism and development, such as growth, transpiration, photosynthesis, respiration, and nutrient distribution (Das et al., 1997; Sanita et al., 1999; Zanella et al., 2016; Ronzan et al., 2017). Moreover, Cd toxicity causes various alterations in plants at genetic, biochemical, and physiological levels and results in phytotoxicity (Wahid et al., 2010). Due to similar oxidation states, Cd can be exchanged with Fe2+, Zn2+, and Ca2+ in some protein structures, leading to protein malfunction (Goyer, 1997). In addition, at excess concentrations, Cd induces the formation of ROS, leading to lipid peroxidation and DNA damage, as well as calcium homeostasis modification (Stohs and Bagchi, 1995). The mechanism of Cd uptake by plants is significantly affected by various factors, such as pH, temperature, aeration, Cd concentration in the environment, and concentration of other micro and macro elements (McLaughlin et al., 1996). Uptake of Cd by plant roots occurs through divalent cation transport systems, including those for iron, zinc, and calcium (Verbruggen et al., 2009).
Several Cd transporters involved in the uptake, efflux, and sequestration of Cd have been identified. Among these proteins, plasma membrane-localized transporters, such as AtIRT1 (Connolly et al., 2002), OsIRT1 (Lee and An, 2009), AhIRT1 (Chen C. et al., 2017), AtNRAMP1 (Thomine et al., 2000), AhNRAMP1 (Chen C. et al., 2017), NtNRAMP1 (Sano et al., 2012), OsNRAMP5 (Sasaki et al., 2012), HvNramp5 (Wu et al., 2016), OsMTP1 (Yuan et al., 2012), OsZIP1 (Ramesh et al., 2003), and SaNramp6 (Chen S. S. et al., 2017), have been implicated in Cd uptake. Furthermore, members of the HMA family, including AtHMA2 (Mills et al., 2003), AtHMA4 (Hussain et al., 2004; Verret et al., 2004), and OsHMA2 (Takahashi et al., 2012), as well as the ATP-binding cassette transporter AtPDR8 (Kim et al., 2007), are located at the plasma membrane and are involved in Cd efflux. In addition, vacuolar transporters, including AtCAX1 (Wu et al., 2011), AtCAX2 (Hirschi et al., 2000), AtCAX4 (Korenkov et al., 2007), AtHMA3 (Morel et al., 2009), TcHMA3 (Ueno et al., 2011), FlHMA3 (Guo et al., 2017), AtABCC1/AtABCC2 (Park et al., 2012), and AtABCC3 (Brunetti et al., 2015), play considerable roles in Cd transport into the vacuole.
Calcium (Ca2+) is a vital nutrient and signaling molecule that is implicated in various metabolic and signal transduction pathways (Yang and Poovaiah, 2003; Demidchik et al., 2018). It has been reported that Ca interferes with Cd uptake and transport by regulating Cd transporter expressions (Kim et al., 2002; Zeng et al., 2017). Moreover, due to their similar ionic radii, Ca and Cd may compete with each other for uptake and transport into plant cells. It was shown that Cd treatment repressed the activity of Ca2+ channels (Li S. et al., 2012). So far, several genes have been found to be involved in Ca2+ transport as well as Cd tolerance. Over-expression of OsACA6 encoding P-type 2B Ca2+ ATPase improved Cd tolerance by mediating Cd distribution (enhanced and decreased Cd levels in roots and shoots, respectively) and lowering oxidative stress in tobacco (Shukla et al., 2014). In addition, plant cadmium resistance 1 (PCR1) protein is able to transport Ca2+ (Song et al., 2011) and promotes Cd tolerance by reducing Cd accumulation in yeasts and Arabidopsis protoplasts (Song et al., 2004).
Nitric oxide is a short-lived free-radical reactive gas that functions in a wide range of physiological processes in plants, such as growth and development, iron homeostasis, and responses to biotic and abiotic stresses (Ramirez et al., 2011). Production of NO in plants is affected by biotic and abiotic stresses (Lamattina et al., 2003; Leitner et al., 2009). NO production in plants under Cd stress conditions can act as either an enhancer or reducer of Cd toxicity. Cd-induced NO production primarily contributes to Cd toxicity through elevating oxidative stress by enhancing ROS, RNS and lipid peroxidation, as well as by repressing the activity of antioxidant enzymes (Groppa et al., 2008; De Michele et al., 2009; Arasimowicz-Jelonek et al., 2012; Kulik et al., 2012). By contrast, NO protects plants against Cd-induced oxidative stress by enhancing antioxidant enzyme activity and reducing ROS accumulation (Verma et al., 2013; Perez-Chaca et al., 2014; Yang et al., 2016). Therefore, a decrease in NO by Cd results in higher ROS levels and toxicity (Rodriguez-Serrano et al., 2009; Ortega-Galisteo et al., 2012; Gupta et al., 2017). Moreover, treatment with exogenous NO donors, such as SNP, mitigates Cd-induced oxidative stress by enhancing the activities of antioxidant enzymes (Kopyra and Gwozdz, 2003; Hsu and Kao, 2004; Li L. et al., 2012; He et al., 2014). In addition, NO accumulation is involved in programmed cell death under Cd toxicity conditions (Ye et al., 2013). NO can also mediate the induction or inhibition of Cd toxicity by increasing Cd accumulation (Ma et al., 2010; Arasimowicz-Jelonek et al., 2012; Chmielowska-Bąk and Deckert, 2013; Han et al., 2014), potentially by enhancing Cd uptake (Besson-Bard et al., 2009; Luo et al., 2012; Zhu et al., 2012) or by decreasing Cd accumulation (Li L. et al., 2012; Zhang et al., 2012; He et al., 2014).
Hemoglobin (Hb), an ubiquitous protein in plants, was first identified in the root nodules of soybean (Glycine max) plants and implicated in oxygen binding and transport (Appleby, 1992). There are two classes of plant non-symbiotic Hb genes (class 1 and 2), which have over 50% sequence identity, but are distinct in terms of their phylogenetic characteristics, gene expression, and oxygen binding features (Kundu et al., 2003; Igamberdiev et al., 2011). Several lines of evidence indicate a significant role for non-symbiotic class 1 hemoglobin (Hb1) in NO detoxification (Dordas et al., 2003; Igamberdiev et al., 2004; Perazzolli et al., 2004; Hebelstrup et al., 2006). Therefore, Hb1 can participate in plant responses to biotic and abiotic stress by modulating the level of NO. Transgenic Arabidopsis plants that overexpress AtHB1 have higher tolerance to hypoxia stress (Hunt et al., 2002). In addition, tolerance to submergence, salinity, and osmotic stresses are increased by ZmHb expression in tobacco (Zhao et al., 2008). Furthermore, heterologous expression of MsHb1 in tobacco and Arabidopsis (Seregelyes et al., 2004; Maassen and Hennig, 2011), GhHb1 in Arabidopsis (Qu et al., 2006), and AtHb1 in barley (Hebelstrup et al., 2014) improves defense responses against pathogen attack. Down-regulation of cold-responsive genes, as well as mitigated oxidative stress, has also been observed in transgenic Arabidopsis expressing AtHb1 (Cantrel et al., 2011; Thiel et al., 2011). Previous studies have revealed that the growth responses and development of plants are mediated by Hb1, as its expression delays bolting in Arabidopsis (Hebelstrup and Jensen, 2008) and reduces plant growth and development in barley (Hebelstrup et al., 2014). Recently, we also showed that over-expression of NtHb1 enhances Cd tolerance by reducing the NO and Cd levels in transgenic tobacco (Lee and Hwang, 2015). Interestingly, over-expression of spinach SoHb in Arabidopsis reduces the NO content and tolerance to nitrate, NaCl, and osmotic stresses (Bai et al., 2016). Moreover, AtHb1-overexpressing Arabidopsis plants fumigated with NO showed improved growth parameters and NO fixation ability compared with the WT plants (Kuruthukulangarakoola et al., 2017). In this study, to understand a mechanism for Hb1-induced Cd tolerance in Arabidopsis, we over-expressed NtHb1 in Arabidopsis and examined concentrations of Cd, NO, and ROS and expression levels of the Cd transporters Ca2+/H+ exchangers (CAXs), ABC transporter (PDR8), and iron responsive transporter 1 (IRT1).
Materials and Methods
Plasmid Construction and Transformation of Arabidopsis
The coding sequence of NtHb1 (GenBank Access No. KJ808726.1) was amplified by RT-PCR using gene specific primers flanked by XbaI and BamHI restriction sites and sub-cloned into the pBS vector. After sequencing the insert, the XbaI-BamHI fragment was cloned into the binary vector pBI121. The pBI121 vector harboring NtHb1 was transformed into Agrobacterium tumefaciens strain GV3103 by the freeze–thaw method, and plant transformation was achieved by the floral dipping method (Clough and Bent, 1998). To generate pAtHb1::GUS-expressing Arabidopsis, the promoter region of AtHb1 (1-kb upstream of the start codon) was amplified using primers with HindIII and XbaI restriction sites, sequenced, and cloned into the binary vector pBI121. The primer pairs used for these constructs are shown in Supplementary Table S1.
Quantitative RT-PCR (Real-Time PCR)
Total RNA was isolated from transgenic Arabidopsis seedlings (T3 homozygous) grown on 1/2 MS agar plates for 3 weeks using the Plant RNA extraction Kit (Intron, South Korea). After quantification of the RNA using a NanoDrop (BioSpec-nano, Shimadzu, Japan), first-strand cDNA was synthesized from 2 μg of RNA in RNase-free water using PrimeScriptTM RT reagent kit (Takara, Otsu, Japan) according to the manufacturer’s instructions. Semi-quantitative PCR was employed for the analysis of NtHb1 expression by using gene-specific primers. Actin was used as an internal control. The expression levels of cadmium transporter genes in Arabidopsis were evaluated by using qRT-PCR. Isolation of total RNA was performed by using transgenic Arabidopsis seedlings (T3 homozygous) grown on 1/2 MS agar plates supplemented with 0 and 50 μM CdSO4 for 3 weeks, and cDNA was synthesized as described above. Quantitative RT-PCR was accomplished using a CFX ConnectTM Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, United States). The reaction mixture consisted of 10 μl of SYBER Supermix (SsoAdvanced®Universal SYBRTM Green Supermix), 1 μl of cDNA, 7 μl of nuclease-free water, and 1 μl of each primer to a final volume of 20 μl. The following qRT-PCR reaction thermal conditions were used: 95°C for 30 s, followed by 40 cycles at 95°C for 15 s and 57°C for 20 s. The Arabidopsis ACTIN2 gene was used as an internal quantitative control, and the relative expression level of each gene was calculated based on the 2−ΔΔCt method (Pfaffl, 2001). Each qRT-PCR experiment was performed three times with different cDNA sets obtained from three independent biological replicates. The gene-specific primers are shown in Supplementary Table S1.
Western Blot Analysis
Arabidopsis wild-type plants (Col-0) were grown on 1/2 MS agar plates containing 0 and 50 μM CdSO4 for 3 weeks, and subsequently, the roots and shoots were separated and homogenized in extraction buffer (20 mM Tris–Hcl, pH 8.0, 1 mM DTT (dithiothreitol), 0.3 mM EDTA and protease inhibitor cocktail). Protein samples (40 μg) were loaded and separated on SDS-PAGE gels and then transferred to nitrocellulose membranes (Hybond-C Extra, Amersham Biosciences) and probed with AtHb1 primary antibodies (Abmart Co.,) and the binding of secondary antibody rabbit anti-mouse horseradish peroxidase-conjugated IgG (Amersham Biosciences) was detected by enhanced chemiluminescence (Amersham Biosciences). Rubisco Ponceau S-stained large subunit was used as a loading control. Three independent biological experiments were performed.
Analysis of Cadmium Tolerance
To measure cadmium tolerance, seeds of transgenic Arabidopsis lines (T3 homozygous) were surface-sterilized, germinated and incubated on 1/2 MS agar supplemented with 0 and 50 μM of cadmium sulfate for 3 weeks. This cadmium concentration reduced the fresh weight of wild-type seedlings by 50%. The Arabidopsis seedlings were photographed after 3 weeks. These experiments were performed in three independent biological replicates for each line. To calculate the cadmium tolerance, the fresh weight of each seedling (n = 90) on cadmium plates was divided by the fresh weight of seedlings grown on control plates (1/2 MS), and finally expressed as the relative fresh weight (%).
Measurement of Cadmium Concentration in Plants
For the Cd concentration measurement, control and transgenic Arabidopsis seeds (T3 homozygous) were surface-sterilized, germinated and incubated on 1/2 MS agar supplemented with 50 μM of cadmium sulfate for 3 weeks. The seedlings on each plate were harvested after 21 days, washed with ice-cold 5 mM CaCl2 (three times) and dried for 72 h at 60 °C. Then, the dried sample (0.5 g) was digested by using concentrated HNO3 and HClO4 in a Teflon Digestion Vessel (Savillex, United States). ICP Mass Spectroscopy (Varian 720-MS, United States) was employed to specify the concentration of Cd at a wavelength of 214.44 nm at the National Instrumentation Center for Environmental Management (NICEM) at Seoul National University, South Korea. Three independent biological experiments were performed.
Localization of Cadmium in Plants
LeadmiumTM Green AM (Invitrogen, United States) was used to monitor the localization of Cd in Arabidopsis roots following the manufacturer’s instructions. Briefly, control and transgenic Arabidopsis seeds were germinated and grown on 1/2 MS medium containing 0 and 50 μM of cadmium. After 21 days, the root samples were harvested and rinsed in distilled water. The working solution of LeadmiumTM Green AM was obtained through dilution (1:10) of its stock solution [1 μg/μl in DMSO in 0.85% NaCl. Next, the samples were immersed in staining solution [0.04% (v/v) of LeadmiumTM Green AM in 0.85% NaCl] and kept in the dark. After 1 h, the samples were briefly washed with saline solution (0.85% NaCl) (Du et al., 2015) and visualized using a fluorescence microscope (Leica, MZ10F, Germany) with 490 nm excitation and 510–520 nm emission filters under proper magnification (12.5-25X). At least 10 plants were examined per line, and three independent biological experiments were performed.
ROS (Hydrogen Peroxide) Detection in Roots Using a Fluorescent Probe (CM-H2DCFDA)
Visualization of ROS in root was performed by using the CM-H2DCFDA [5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate] fluorescent probe (Molecular Probes1) as previously described (Wang et al., 2014), with some modifications. The roots of three-week-old plants grown in 1/2 MS medium supplemented with 0 and 50 μM of cadmium were harvested and briefly washed with deionized water. After immersing the roots in CM-H2DCFDA solution (10 μM) for 15 min in the dark, samples were rinsed with water and photographed using a fluorescence microscope (Leica, MZ10F, Germany) with 485 nm excitation and 510–530 nm emission filters under proper magnification (12.5-20X). Image J software (NIH, United States) was employed for quantification of the fluorescence signal. At least 10 plants were examined per line, and three independent biological experiments were performed.
Visualization of NO Production in Roots
The NO-specific fluorescent probe DAF-2DA (Sigma Co.) was used to detect NO production as previously described (Besson-Bard et al., 2009), with some modifications. Briefly, the roots of three-week-old plants grown in 1/2 MS medium supplemented with 0 and 50 μM of cadmium were incubated with 10 mM of DAF-2DA (prepared in 20 mM HEPES-NaOH, pH 7.5) under dark conditions for 1 h. Subsequently, the samples were washed with the same buffer and photographed using a fluorescence microscope (Zeiss Axioplan2) with 488 nm excitation and 515–565 nm emission filters. For treatment of NO donor SNP (sodium prusside, Sigma Co.) and NO scavenger cPTIO (Sigma), 500 μM of each chemical was prepared in 20 mM of HEPES-NaOH, pH 7.5 and used for the pre-treatment of the root samples prior to staining with DAF-2DA and visualization. Image J software (NIH, United States) was used for Quantification of the NO signal intensity. At least 10 plants were examined per line, and the experiment was repeated independently three times.
Measurement of Anti-oxidative Enzyme Activity
Antioxidant enzymes assays were performed using the crude extract of the leaves as the enzyme source. Briefly, leaf samples (0.2 g) of 3-week-old transgenic Arabidopsis plants grown on 1/2 MS medium supplemented with or without 50 μM of cadmium sulfate were ground to a fine powder in liquid nitrogen and then homogenized in 1.2 ml of 0.2 M potassium phosphate buffer (pH 7.8 containing 0.1 mM EDTA) and centrifuged at 15,000 × g at 4°C for 20 min to obtain a crude enzyme extract.
Catalase (1.11.1.6) activity, was measured by monitoring the decrease in absorbance at 240 nm as previously described (Aebi, 1984). To start the reaction, leaf crude extract was added to 2.5 μl of H2O2 (30% solution) prepared in 50 mM of potassium phosphate buffer (pH 7.5) at a final volume of 1 ml. The enzyme activity was calculated according to the extinction coefficient of H2O2 (40 M−1 cm−1 at 240 nm) and expressed in terms of millimoles of H2O2 per minute per gram fresh weight.
To determine the SOD activity (SOD; EC 1.15.1.1), the modified NBT method of Bayer and Fridovich (Beyer and Fridovich, 1987) was employed. Briefly, the assay solution consisted of 50 mM phosphate buffer (pH 7.8) containing 2 mM EDTA, 9.9 mM L-methionine, 55 μM NBT, 0.025% Triton X-100 and 1 mM riboflavin. Following the addition of the leaf extract, the reaction was initiated after exposing the samples to a fluorescent lamp (15 W) for 10 min. The absorbance of the reaction mixture was measured at 560 nm against a similar mixture lacking the leaf extract as a control. The SOD standard curve was used to evaluate the enzyme activity (per gram fresh weight) of the samples.
Glutathione reductase activity (GR; EC 1.8.1.7) was measured as previously described (Smith et al., 1918). The assay solution (1 ml) was prepared in 100 mM potassium phosphate buffer (pH 7.5) and 0.75 mM DTNB, 0.1 mM NADPH, 1 mM GSSG (glutathione disulfide), and 10 μl of leaf extract. GSSG was added to start the reaction, and the absorbance increment at 412 nm was measured when DTNB was reduced to TNB by GSH in the reaction. The activity of GR was determined based on the extinction coefficient of TNB (14.15 M−1 cm−1) and expressed in terms of millimoles of TNB minute per gram fresh weight. Three independent biological experiments were performed, in which each measurement was repeated three times.
Measurement of Cd Efflux
Seeds of control (pBI121) and NtHb1-expressing Arabidopsis were germinated and grown on 1/2 MS media plate for 3 weeks, and subsequently, all plants were transferred to 50 μM Cd (cadmium sulfate) agar plates, incubated for 24 h, washed with distilled water, and placed in a Cd-free liquid medium (DW). After 6 and 24 h, the Cd concentrations in the liquid media were determined as described in the above section. Three independent biological experiments were performed.
GUS Staining Analysis
Detection of AtHb1::GUS activity was performed as previously described (Prasinos et al., 2005). Briefly, 7-day-old seedlings grown vertically on 1/2 MS agar medium containing 0 and 50 μM of cadmium were incubated in GUS staining solution (50 mM NaPO4 buffer, pH 6.8, 0.5 M EDTA, pH 8.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 20% Triton X-100 and 2 mM X-gluc), for 2 h at 37°C. For short-term experiments, 7-day-old seedlings grown vertically on 1/2 MS agar medium were treated with 50 μM of cadmium for 2, 6, 12, and 24 h. To observe GUS expression in the leaves, the samples were washed with ethanol (70%) to remove green chlorophyll pigments. Images were captured with a microscope (Zeiss Axioplan2). This experiment was repeated 3 times, and 10 seedlings were observed each time.
Statistical Analysis
All data were subjected to analysis of variance (Two-way ANOVA) by using SAS software (version 9.1), and comparisons of the means were performed according to Tukey’s test at P ≤ 0.05 or P ≤ 0.01.
Results
Over-Expression of NtHb1 Enhances Cd Tolerance but Decreases Cd Accumulation in Transgenic Arabidopsis
To examine whether NtHb1 increases Cd tolerance in Arabidopsis similar to that in tobacco (Lee and Hwang, 2015), NtHb1 was over-expressed in Arabidopsis using the binary pBI121 vector. As shown in Figure 1A, two lines (4 and 5) of transgenic Arabidopsis expressing NtHb1 exhibited higher levels of NtHb1 transcript compared with that of the vector only-expressing control Arabidopsis, which had no NtHb1 transcript. As expected, NtHb1-expressing Arabidopsis displayed higher tolerance to Cd than the control plants (Figures 1B,C). In contrast, the accumulation of Cd in NtHb1-Arabidopsis was lower than that in control plants (Figure 1D–F). Additionally, Cd was highly accumulated in the meristematic zone and stele (likely in the xylem).
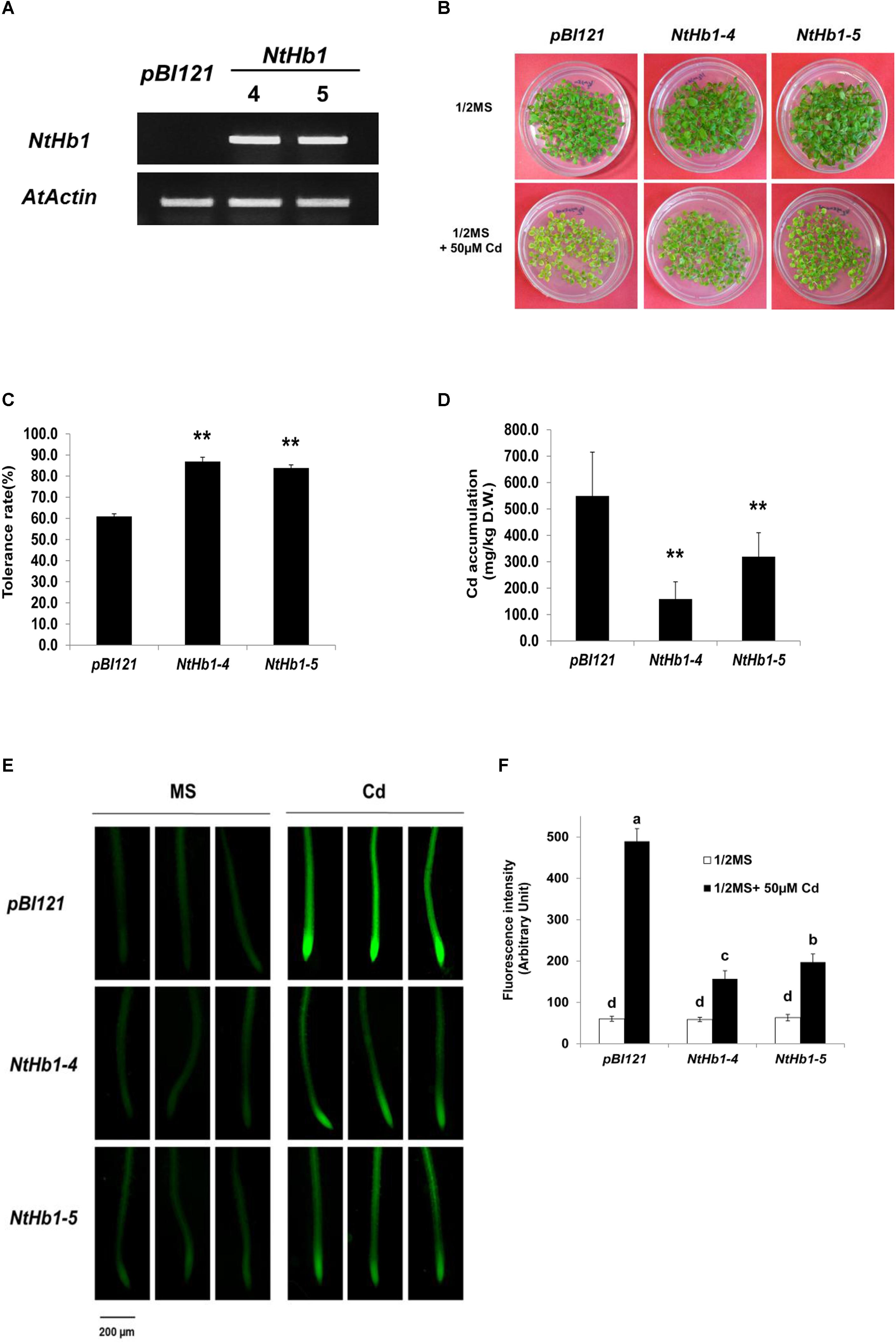
Figure 1. Cd tolerance and accumulation in the control and NtHb1-expressing Arabidopsis. (A) qRT-PCR results showing higher levels of NtHb1 transcripts in transgenic Arabidopsis. Control plants contain the empty vector pBI121, and NtHb1-4 and NtHb1-5 refer to two different lines of NtHb1-expressing transgenic Arabidopsis. EtBr-stained qRT-PCR product of the actin gene (AtActin) was used as a loading control. (B) Cadmium tolerance of transgenic Arabidopsis expressing pBI121 (control) and NtHb1 (NtHb1-4 and NtHb1-5). Plants were germinated and grown for 3 weeks on 1/2 MS agar plates without (upper) and with (lower) 50 μM CdSO4 and then photographed. (C) Graph showing the Cd tolerance rates of transgenic Arabidopsis shown in (B). Each value corresponds to the means ± standard error (SE) (n = 3). (D) Accumulation level of Cd in transgenic Arabidopsis. Values correspond to the means ± SE (n = 3). (E) Visualization of Cd in Arabidopsis roots using LeadmiumTM Green AM. Plants were germinated and grown for 3 weeks on 1/2 MS plates without (left) and with (right) 50 μM Cd. The experiments were performed 3 times, and 10 plants were examined per line for each experiment. (F) The intensity of fluorescence (five plants from each line) was measured using Image J software (NIH, United States). Each value corresponds to the means ± SE (n = 5). Asterisks indicate significant differences between control and transgenic Arabidopsis (p ≤ 0.01). Different letters over the bars indicate significant differences according to Tukey’s multiple comparison test (p ≤ 0.05).
NO and ROS Are Less Induced by Cd in NtHb1-Expressing Arabidopsis
Because NO production is enhanced in response to Cd (Besson-Bard et al., 2009) and Hb scavenges NO in Arabidopsis (Dordas et al., 2003; Perazzolli et al., 2004; Hebelstrup et al., 2006), the NO level was examined in control and transgenic Arabidopsis germinated and grown with 50 μM CdSO4 for 21 days. As shown in Figures 2A,B, the NO level was highly enhanced (6.0-fold) by Cd in control Arabidopsis expressing the pBI121 vector only. In contrast, the NO level was less increased (2-fold) by Cd in NtHb1-expressing Arabidopsis, indicating that Cd-induced NO accumulation was decreased by increased Hb in NtHb1-expressing Arabidopsis, leading to increased Cd tolerance. In addition, the increased accumulation of NO in the meristem and stele of Cd-treated plants (Figure 2A) corresponded to the pattern of Cd accumulation (Figure 1E).
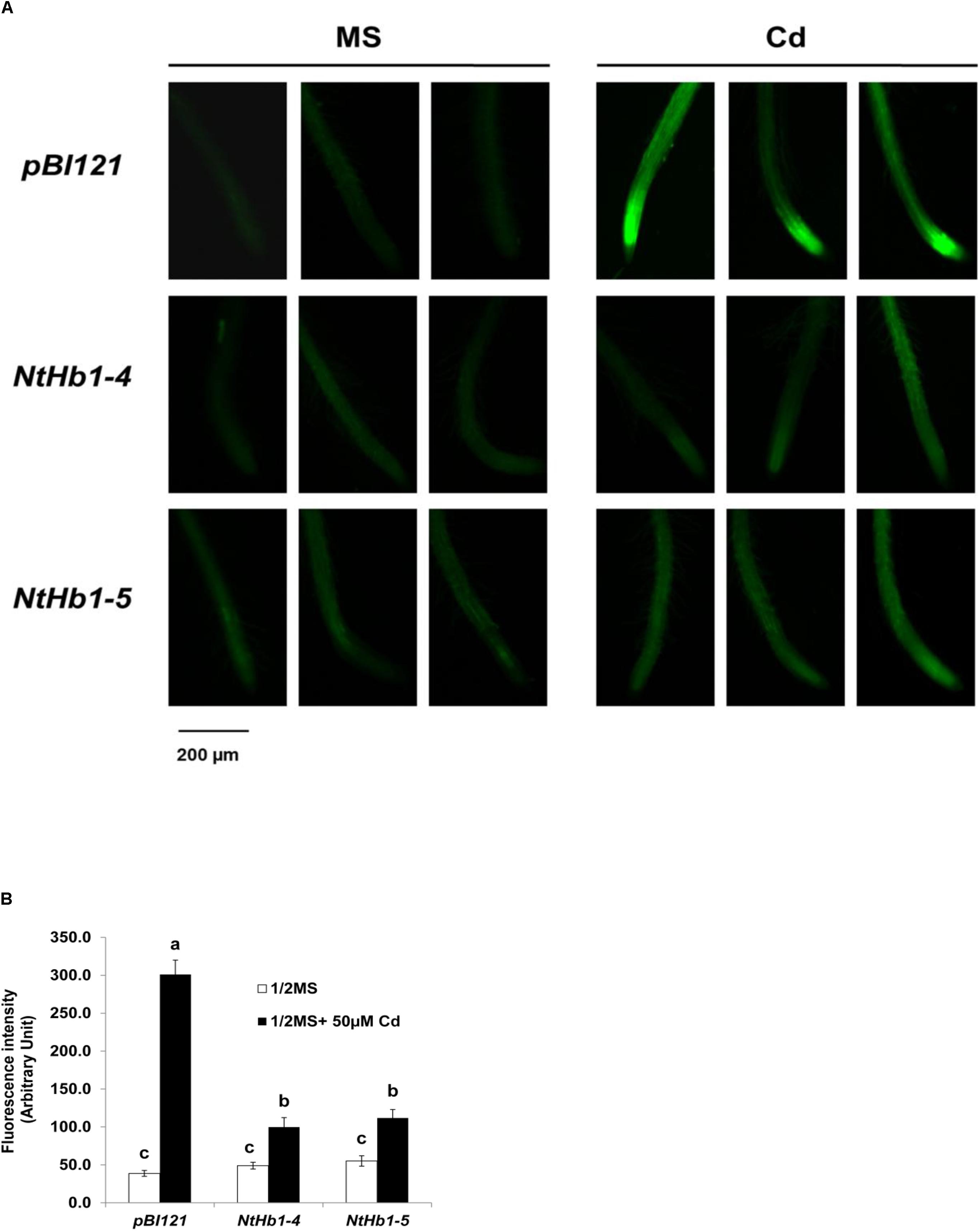
Figure 2. NO production in response to Cd in the control and NtHb1-expressing Arabidopsis. (A) NO production was visualized using DAF-2DA in the control (pBI121) and NtHb1-expressing Arabidopsis (NtHb1-4 and NtHb1-5), which were germinated and grown for 3 weeks on 1/2 MS agar media without (left) and with (right) 50 μM CdSO4. (B) The intensity of NO fluorescence (five plants from each line) was measured using Image J software. Each value corresponds to the means ± SE (n = 5). Different letters over the bars indicate significant differences according to Tukey’s multiple comparison test (p ≤ 0.05).
Because ROS are also generated by Cd in Arabidopsis (Bi et al., 2009), the ROS level was measured using a fluorescent probe (CM-H2DCFDA). As shown in Figure 3, ROS accumulation was greatly increased (14-fold) by Cd in control plants, while the ROS content was less enhanced (3∼4-fold) in NtHb1-expressing Arabidopsis. Interestingly, ROS evenly accumulated in the roots in response to Cd (Figure 3), while the Cd and NO levels were higher in the meristem and stele (Figure 2). To examine the involvement of anti-oxidative enzymes in decreasing ROS levels, the activities of CAT, SOD, and GR were measured in control and transgenic plants. As shown in Figure 4, all three enzymes were activated by Cd in both control and NtHb1-expressing Arabidopsis plants, but were highly induced in transgenic plants compared with levels in control plants.
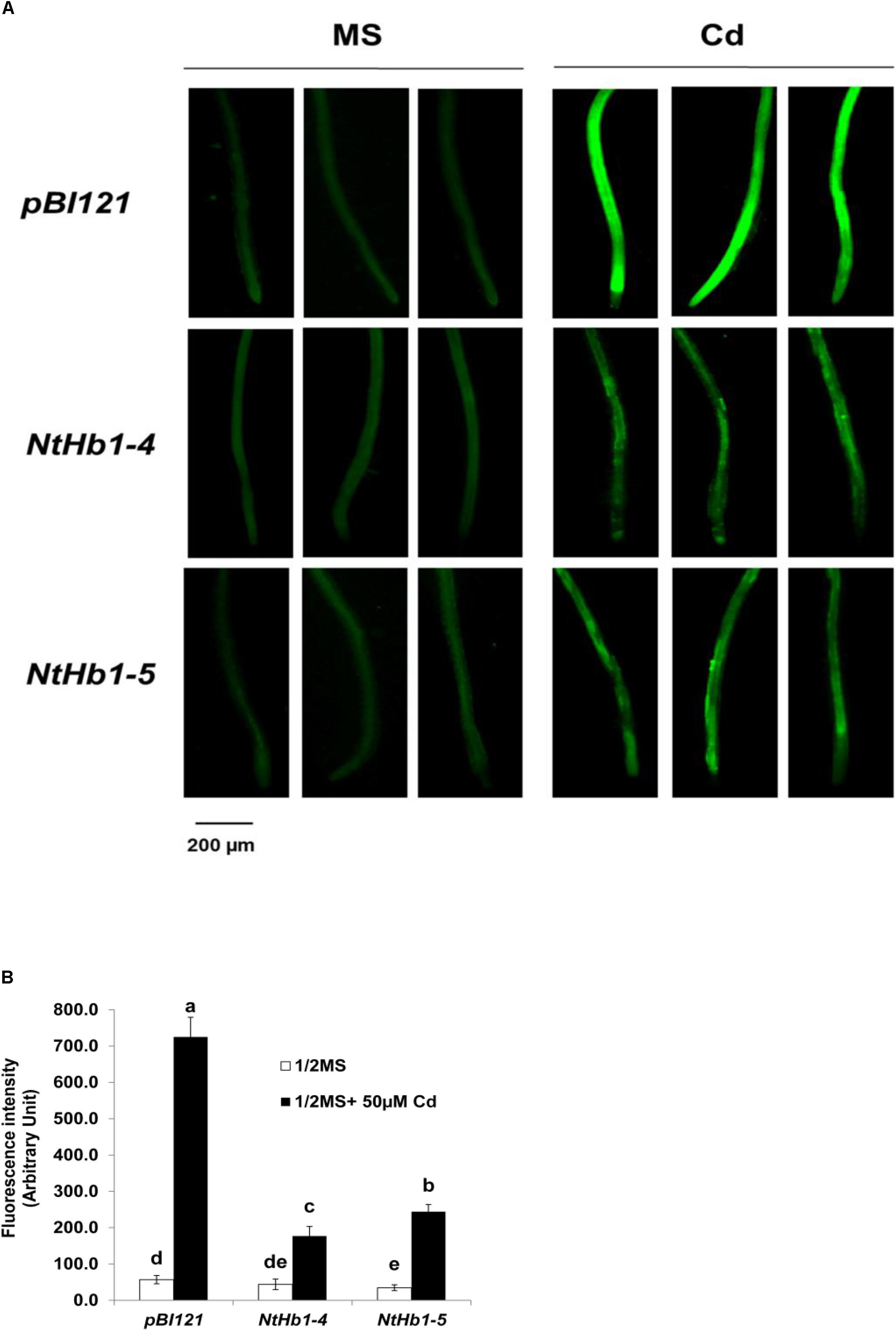
Figure 3. ROS levels in response to Cd in the control and NtHb1-Arabidopsis. (A) ROS staining with a fluorescent probe (CM-H2DCFDA). Plants were germinated and grown for 3 weeks on 1/2 MS agar media without and with 50 μM Cd. Experiments were performed 3 times, and 10 plants from each line were examined in each experiment. (B) Quantification of the ROS shown in (A). The fluorescence signal was quantified using Image J software. Data are presented as the means of three independent experiments, and the error bars indicate SE. Different letters over the bars indicate significant differences according to Tukey’s multiple comparison test (p ≤ 0.05).
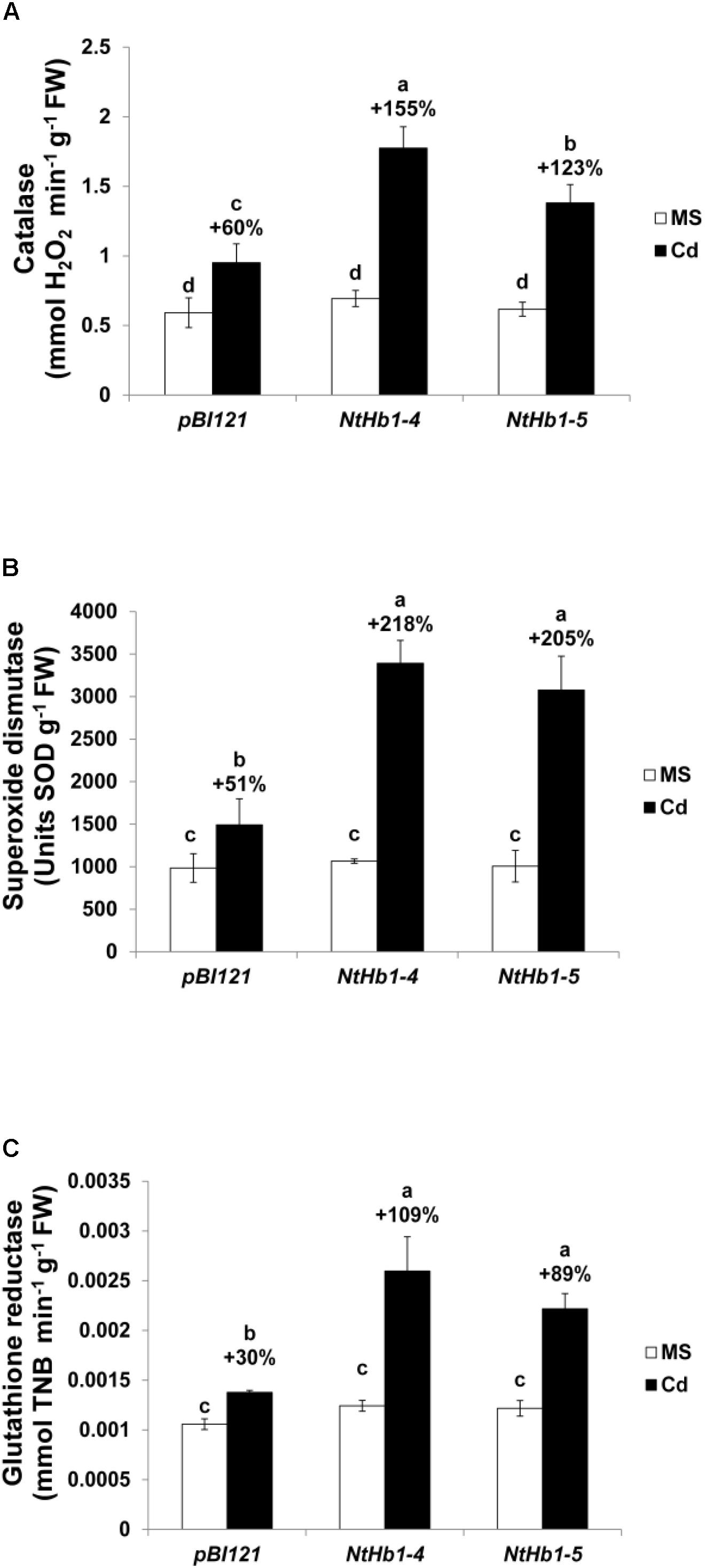
Figure 4. Activity of anti-oxidative enzymes in response to Cd in the control and NtHb1-expressing Arabidopsis. Activity of catalase (A), superoxide dismutase (B), and glutathione reductase (C) in control (pBI121) and NtHb1 Arabidopsis. Plants were germinated and grown for 3 weeks on 1/2 MS agar media without (white) and with (black) 50 μM Cd. Data correspond to the means of three independent experiments, and the error bars indicate SE. Different letters over the bars indicate significant differences according to Tukey’s multiple comparison test (p ≤ 0.05).
Expression of CAXs and PDR8 Is Enhanced While That of IRT1 Is Decreased in Response to Cd, Leading to Higher Cd Export in NtHb1-Expressing Arabidopsis
To elucidate the mechanism by which Cd accumulation is decreased in NtHb1-Arabidopsis, qualitative reverse transcription–polymerase chain reaction (qRT-PCR) was performed to examine the expression levels of various Cd transporters, including AtCAX1 (vacuolar transporter, AT2G38170), AtCAX2 (vacuolar transporter, AT3G13320), AtCAX3 (vacuolar transporter, AT3G51860), AtCAX4 (vacuolar transporter, AT5G01490), AtPDR8 (efflux transporter, AT1G59870), AtIRT1 (uptake transporter, AT4G19690), AtHMA2 (efflux transporter, AT4G30110), AtHMA4 (xylem loading transporter, AT2G19110), AtNRAMP3 (vacuolar transporter, AT2G23150), AtABCC1 (vacuolar transporter, AT1G30400), AtACA10 (putative uptake transporter, OsACA6 homolog, AT4g29900.1), and AtPCR1 (efflux transporter, AT1G14880.1). As shown in Figure 5A, the expression of AtCAX3 and AtPDR8 was increased by Cd in both control and NtHb1-expressing Arabidopsis plants, but transgenic plants showed greater increases and thus higher levels than those in control plants. In contrast, the expression of AtIRT1 and ACA10 was induced by Cd in control plants but reduced in NtHb1-Arabidopsis, resulting in a lower level of IRT1 and ACA10 in transgenic plants than in control plants. The expression of CAX1 and CAX2 in response to Cd was also higher in transgenic plants because the expression of these genes was already increased in transgenic plants in the absence of Cd. The expression of AtHMA2, AtHMA4, AtNRAMP3, AtABCC1, and AtPCR1 did not show significant differences between control and transgenic plants in the absence and presence of Cd. Moreover, phytochelatins have been shown to play a significant role in Cd tolerance by sequestrating Cd in vacuoles (Pomponi et al., 2006; Brunetti et al., 2011). To examine whether phytochelatins are involved in Cd tolerance of NtHb1-expressing plants, we measured the expression level of AtPCS1 (phytochelatin synthase, AT5G44070) in control and NtHb1-expressing plants. As shown in Figure 5A, the transcript level of AtPCS1 did not change significantly in control and transgenic plants in the absence and presence of Cd, indicating that phytochelatins may not participate in enhancing the Cd tolerance observed in NtHb1-expressing transgenic plants. Furthermore, the Cd export rates of plants were examined. As shown in Figure 5B, NtHb1-expressing Arabidopsis showed higher Cd efflux rates than those of control plants, suggesting the involvement of CAX1∼3 and PDR8 in reducing Cd levels in transgenic plants.
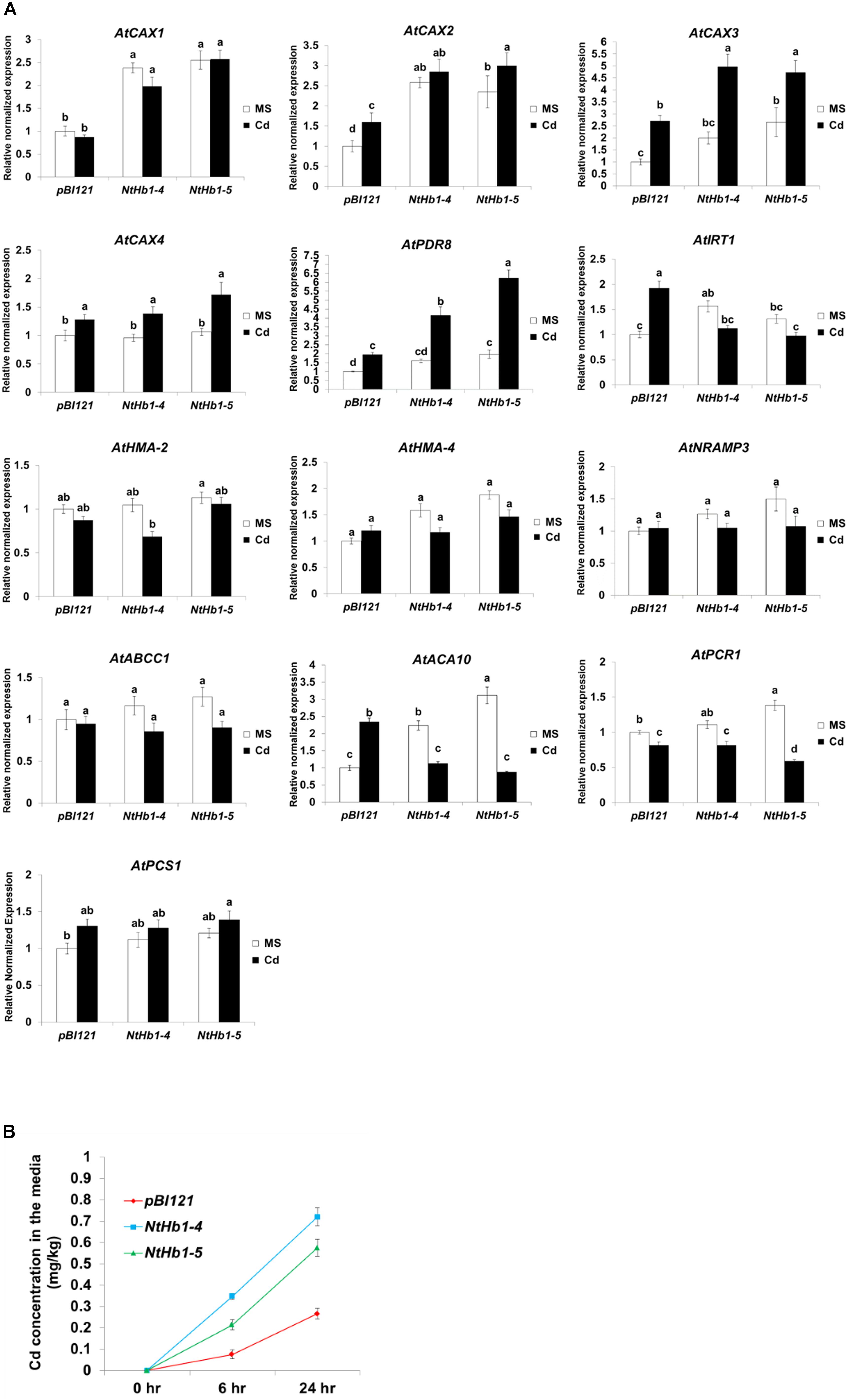
Figure 5. Expression of Cd transporter genes in response to Cd and Cd efflux in the control and NtHb1-Arabidopsis. (A) qRT-PCR results showing the transcript levels of various Cd transporter genes in transgenic Arabidopsis.Notably, pBI121 refers to control plants containing the empty vector pBI121, and NtHb1-4 and NtHb1-5 refer to two different lines of NtHb1-expressing transgenic Arabidopsis. qRT-PCR was performed using total RNA isolated from plants that were germinated and grown for 3 weeks on 1/2 MS plates without or with 50 μM Cd. The relative transcript levels were assessed by normalization to actin transcript abundance as an endogenous control. (B) Cd efflux in plants. Control and transgenic plants were germinated and grown for 3 weeks on 1/2 MS agar plates, transferred to agar plates with 50 μM Cd, incubated for 24 h, and then placed in Cd-free liquid media for 6 h and 24 h. Subsequently, the Cd concentrations in liquid media were determined. All samples were run in triplicate (three biological repeats) for cDNA and each primer set, and the error bars indicate SE. Different letters over the bars indicate significant differences according to Tukey’s multiple comparison test (p ≤ 0.05).
Taken together, these results suggest that the increased expression of CAX1∼3 and PDR8 and the decreased expression of IRT1 and ACA10 are involved in reducing Cd accumulation by enhancing the Cd efflux and decreasing the Cd influx in NtHb1-expressing Arabidopsis.
Expression of CAX3, PDR8, IRT1, and ACA10 Is Regulated by NO and ROS in Arabidopsis
Because NtHb1-Arabidopsis showed a lower level of NO, it is likely that the expression of Cd transporters was regulated by the decreased NO content. To examine this hypothesis, we treated Col-0 Arabidopsis with SNP (an NO donor) for 24 h and subsequently examined the expression of CAX3, PDR8, IRT1, and ACA10 using qRT-PCR. As shown in Figure 6, when plants were germinated and grown for 3 weeks and thereafter treated with SNP for 24 h, the transcript levels of CAX3 and PDR8 were decreased, while those of IRT1 and ACA10 were increased, and these expression patterns were reversed in NtHb1-expressing Arabidopsis (Figure 5). In addition, the transcript levels of CAX3 and PDR8 were decreased, while those of IRT1 and ACA10 were increased in plants germinated and grown for 3 weeks and thereafter treated with H2O2 for 24 h (Figure 7). These data indicate that the enhanced expression of CAX3 and PDR8 and decreased expression level of IRT1 and ACA10 in NtHb1-Arabidopsis are attributed to the reduced levels of NO and ROS.
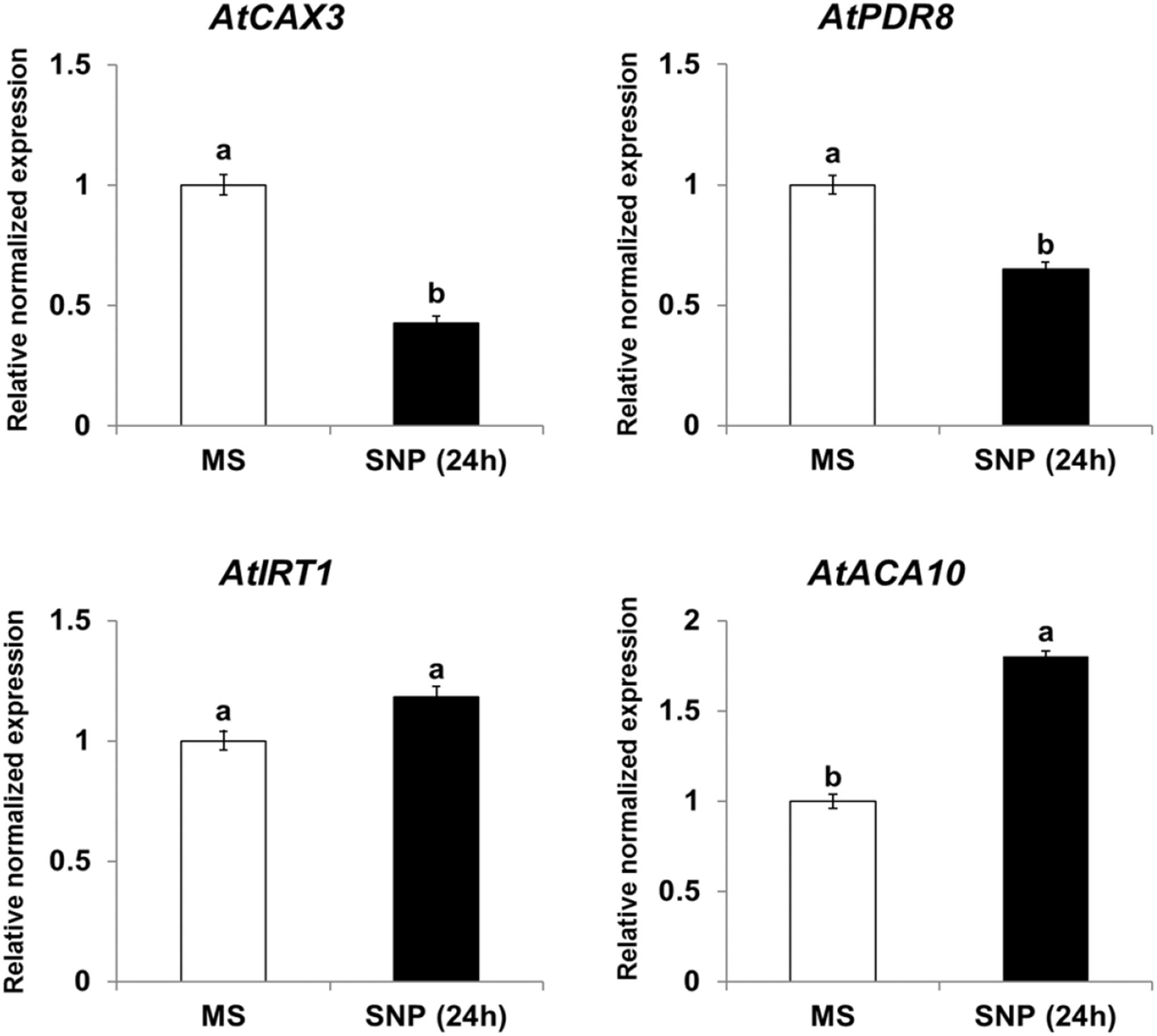
Figure 6. Short-term effect of SNP (NO donor) on the phenotype and gene expression of Cd transporters in Col-0 Arabidopsis. qRT-PCR results showing the transcript levels of various Cd-transporter genes in Col-0 Arabidopsis after short-term (24-h) exposure to SNP. qRT-PCR was performed using total RNA isolated from Col-0 plants, which were germinated and grown for 3 weeks on 1/2 MS agar plate and subsequently treated with 1/2 MS liquid (labeled as MS) and 500 μM SNP (labeled as SNP) for 24 h. All experiments were performed independently in triplicate, and the error bars indicate SE.
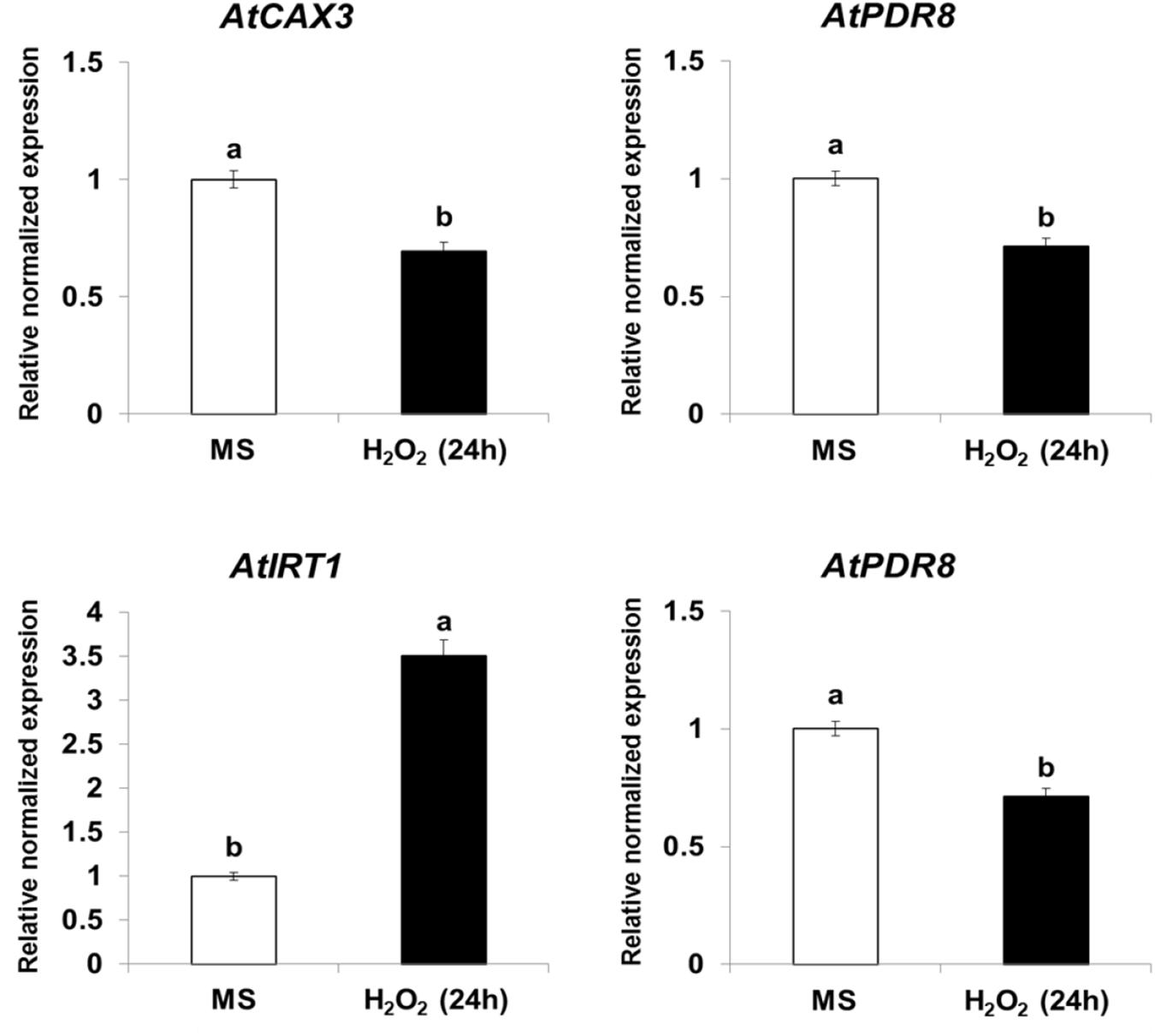
Figure 7. Short-term effect of H2O2 on the gene expression of Cd transporters in Col-0 Arabidopsis. qRT-PCR results showing the transcript levels of various Cd-transporter genes in Col-0 Arabidopsis after short-term (24-h) exposure to H2O2. qRT-PCR was performed using total RNA isolated from Col-0 plants, which were germinated and grown for 3 weeks on 1/2 MS agar plate and subsequently treated with 1/2 MS liquid (labeled as MS) and 100 μM H2O2 for 24 h.
AtHb1 Expression Is Enhanced Greatly in the Roots and Slightly in the Shoots in Response to Cd
To examine whether AtHb1 is naturally involved in decreasing the NO level, which is induced by Cd in plants, the transcript level of AtHb1 in response to Cd was measured (Figure 8). The expression of AtHb1 was increased by 78% in the shoots and 428% in the roots in Col-0 Arabidopsis germinated and grown for 3 weeks on Cd media (Figure 8A). The AtHb1 expression level in the roots was 32 and 317% higher, respectively, than that in the shoots in the absence and presence of Cd. The AtHb1 transcript level was elevated by up to 14-fold in the roots within 24 h after Cd treatment (Figure 8B). Furthermore, the AtHb1 protein level was increased by 83% in the shoots and 305% in the roots in response to Cd, while the AtHb1 level was 47 and 246% higher, respectively, in the roots than in the shoots in the absence and presence of Cd (Figures 8F,G). In addition, transgenic Arabidopsis expressing the pAtHb1-GUS construct was generated to study the tissue localization of AtHb1. As shown in Figures 8C,E, AtHb1 was highly expressed in the roots but slightly expressed in cauline leaves. In the absence of Cd, AtHb1 was highly expressed in the root tips (meristems) and slightly expressed around the stele in the upper part of the roots. In contrast, AtHb1 expression was highly induced by Cd in every part of the roots, with the highest level of expression in the meristem, which corresponds to the NO pattern (Figures 2A, 8D), but AtHb1 induction was not clear in the shoots (Figures 8C,E). Regarding the 78% induction of the AtHb1 transcript levels by Cd in the shoots, the non-induction of GUS by Cd in pAtHb1-GUS plants may be ascribed to the shorter Cd treatment (7 days) compared with the 3-week Cd treatment in the AtHb1 transcript measurement, while GUS expression in the roots was enhanced by the 7-day Cd treatment. The time-dependent slight increase in Hb1 expression without Cd challenge (Figure 8D) was likely due to a buffer-induced hypoxia condition. These data suggest that Hb1 is naturally involved in removing NO, which is induced by Cd in the roots. AtHb1 expression was observed in cauline leaf tips and rosette leaf hydathodes in Arabidopsis Col-0 (Hebelstrup et al., 2006). Furthermore, AtHb1 expression was also observed in the shoot apical meristem of 16-day-old Arabidopsis plants (Hebelstrup and Jensen, 2008). By contrast, AtHb1 was only expressed in the roots of the plant (C-24), and it was absent in the shoots (Hunt et al., 2001).
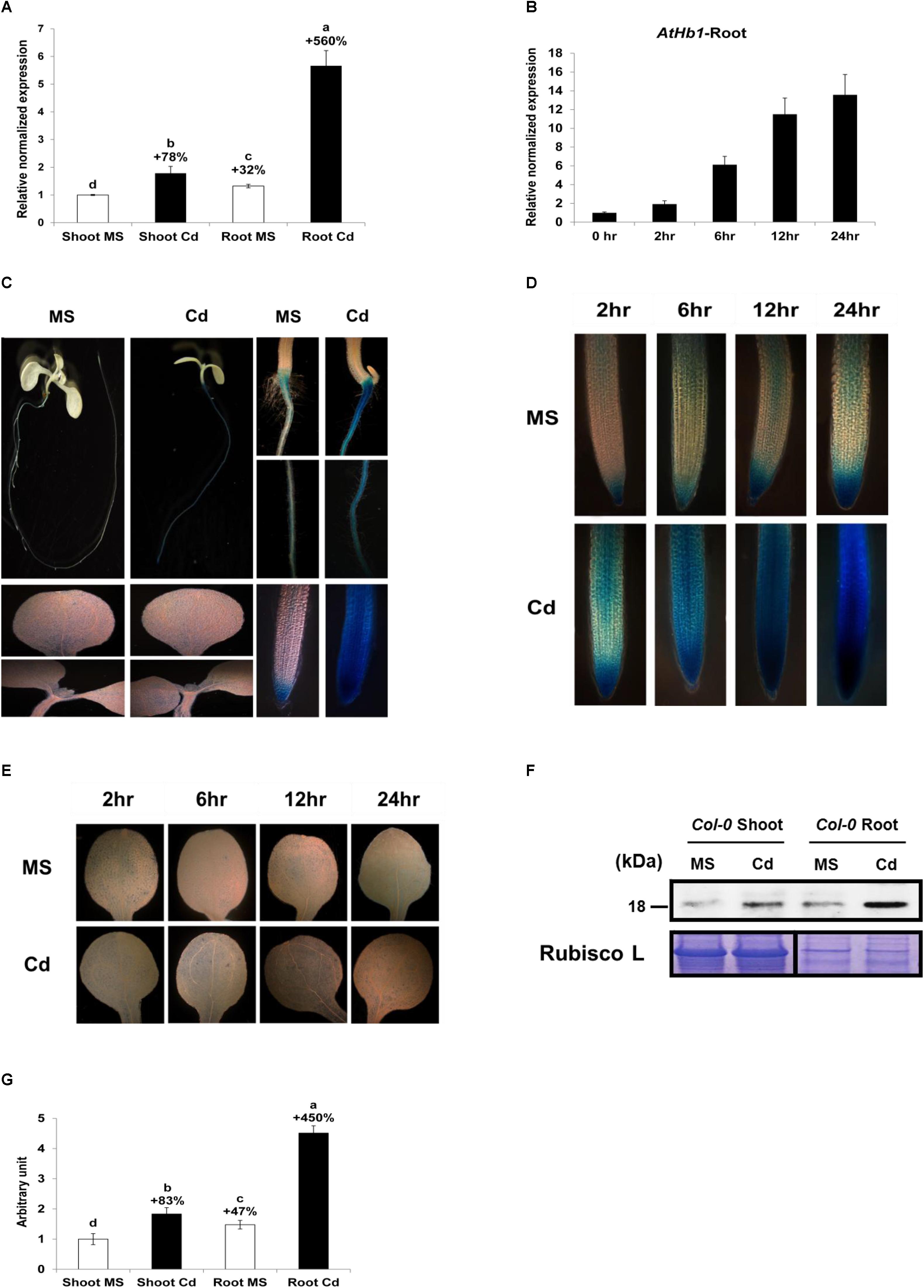
Figure 8. Long-term and short-term effect of Cd on AtHb1 expression in Col-0 Arabidopsis. (A) Long-term (3-week) effect of Cd on the expression level (qRT-PCR) of AtHb1 in Arabidopsis. Plants were germinated and grown for 3 weeks on 1/2 MS agar media without and with 50 μM Cd. The values over the bars indicate the increment compared with the shoot MS. (B) Short-term (24-h) effect of Cd on AtHb1 expression. Plants were germinated and grown for 2 weeks on 1/2 MS agar media and then treated with 50 μM Cd for 2, 6, 12, and 24 h. (C) GUS staining of pAtHB1::GUS plants after long-term (7-day) exposure to Cd. Plants were germinated and grown for 7 days on 1/2 MS agar media with 50 μM Cd and stained. (D,E) GUS staining of pAtHB1::GUS plants after short-term (24-h) exposure to Cd. Plants were germinated and grown for 7 days on 1/2 MS agar media and then treated with Cd for 2, 6, 12, and 24 h, followed by staining. (F) Western blot of AtHb1 protein in Col-0 Arabidopsis. Plants were germinated and grown on 1/2 MS and 1/2 MS with 50 μM Cd for 3 weeks, and protein was extracted from the roots and shoots. Rubisco L, used as a loading control for shoots, refers to the large subunit of RuBP carboxylase/oxygenase. (G) Quantification of the AtHb1 protein shown in (F) by densitometry. The values over the bars indicate the increment compared with the shoot MS. Data correspond to the means of three independent experiments. Data of pAtHB1::GUS plants were produced from one transgenic line used as the representative of three transgenic lines.
Discussion
Transgenic Arabidopsis expressing the hemoglobin gene NtHb1 showed an enhancement in Cd tolerance and reductions in Cd, NO, and ROS levels. The increase in Cd tolerance could be ascribed to the decreased Cd accumulation, as well as the reduction in oxidative stress, including NO and ROS. The reduction in the Cd level might be due to the enhanced expression of CAX3 and PDR8 and reduced expression of IRT1 and ACA10, which was attributed to the diminished levels of NO and ROS.
NO Accumulation Is Increased by Cd and Inhibited by Hemoglobin in Arabidopsis
Nitric oxide levels were enhanced by Cd in both control and NtHb1-expressing Arabidopsis, but NO was less increased (103%) in transgenic plants than in control plants (679%), resulting in lower levels of NO in transgenic plants (Figure 2). This finding suggests that the enhanced Hb1 diminishes the NO level, likely by scavenging more NO in NtHb1-expressing Arabidopsis.
The NO level was enhanced by Cd treatment in diverse plant species, including wheat (Groppa et al., 2008), Arabidopsis (Besson-Bard et al., 2009; De Michele et al., 2009; Zhu et al., 2012; Ye et al., 2013), white poplar (Balestrazzi et al., 2011), barley (Valentovicova et al., 2010), tobacco (Ma et al., 2010; Kulik et al., 2012; Lee and Hwang, 2015), pea (Lehotai et al., 2011), yellow lupine (Arasimowicz-Jelonek et al., 2012), Indian mustard (Verma et al., 2013), and soybean (Perez-Chaca et al., 2014). In contrast, some studies reported that NO content was decreased by Cd (Rodriguez-Serrano et al., 2009; Xiong et al., 2009; Ortega-Galisteo et al., 2012; Gupta et al., 2017).
Decreased NO and Cd Levels Reduce the ROS Level, Contributing to Cd Tolerance
In addition to the lower accumulation of NO in response to Cd, NtHb1-expressing Arabidopsis also had a lower ROS level (Figure 3). The lower levels of NO and ROS contribute to Cd tolerance higher in transgenic plants than that in control plants. Cd toxicity is mediated by NO by increasing ROS (De Michele et al., 2009; Arasimowicz-Jelonek et al., 2012; Kulik et al., 2012), RNS (Arasimowicz-Jelonek et al., 2012), and lipid peroxidation (Groppa et al., 2008; Zhu et al., 2012) as well as by inhibiting the anti-oxidative enzymes CAT and ascorbate peroxidase (De Michele et al., 2009). These reports suggest that Cd toxicity may not be enhanced if NO is not elevated in response to Cd. Consistently, in transgenic Arabidopsis expressing NtHb1, the NO level was less elevated in response to Cd, while Cd tolerance was more promoted than it was in control plants (Figures 1B,C, 2). Therefore, we concluded that the enhancement of Cd tolerance in NtHb1-Arabidopsis is attributed to the reduced accumulation of NO, which is responsible for Cd toxicity by promoting oxidative stress.
Furthermore, the lower ROS content in transgenic plants suggests that the ROS level is also decreased by Hb and that NO functions as ROS or produces ROS in Arabidopsis. In support of this result, it was reported that Hb was involved in decreasing ROS in Arabidopsis (Yang et al., 2005), and NO increased the ROS level (De Michele et al., 2009; Arasimowicz-Jelonek et al., 2012; Kulik et al., 2012). In addition, the highly enhanced activities of the anti-oxidative enzymes CAT, SOD, and GR by Cd in transgenic plants are responsible for the smaller increase of ROS (Figure 4). The higher activities of anti-oxidative enzymes in transgenic plants may be ascribed to the lower levels of NO and Cd. Interestingly, ROS accumulated evenly in roots in response to Cd (Figure 3), whereas the NO level was higher in the meristem and stele, indicating that the pattern of ROS production/accumulation is different from that of NO synthesis/accumulation in response to Cd in Arabidopsis.
Decreased NO and ROS Levels Reduce Cd Accumulation by Enhancing Cd Efflux via Regulation of the Expression of Cd Transporters
Cd-tolerant NtHb1-expressing Arabidopsis displayed lower levels of Cd and NO than those of control plants (Figures 1D–F, 2). This result is supported by reports that NO mediates Cd toxicity by enhancing Cd levels (Ma et al., 2010; Arasimowicz-Jelonek et al., 2012; Chmielowska-Bąk and Deckert, 2013; Han et al., 2014). Among the putative Cd transporters examined, the expression of AtCAX1∼3 (vacuolar transporter) and AtPDR8 (efflux transporter) was higher in NtHb1-Arabidopsis than in control plants in the presence of Cd, while the AtIRT1 (uptake transporter) and AtACA10 (putative uptake transporter) transcript levels were lower. This finding suggests that the altered expressions of these transporters are involved in decreasing Cd accumulation in NtHb1-Arabidopsis. This is supported by the enhanced Cd efflux in transgenic plants (Figure 5B). IRT1 expression is induced by Cd exposure in tobacco (Yoshihara et al., 2006), and the over-expression of IRT1 reduces Cd tolerance in Arabidopsis and rice, which is accompanied by increased accumulation of Cd (Connolly et al., 2002; Lee and An, 2009). The proposed roles for PDR8 and IRT1 in NtHb1-expressing Arabidopsis are supported by previous reports that PDR8 functions in pumping out Cd from the cytoplasm (Kim et al., 2007) and IRT1 plays a role in Cd absorption (Korshunova et al., 1999; Connolly et al., 2002). Like IRT1, ACA10 expression was induced by Cd in control plants but was reduced in transgenic plants, leading to a lower level than that in control plants (Figure 5A). In support of our result, the transcript level of OsACA6 (homologous to Arabidopsis ACA10) was increased by Cd treatment in rice, and tobacco expressing OsACA6 exhibited higher Cd tolerance and accumulation in the roots (Shukla et al., 2014). It has been reported that Cd uptake can be mediated by the transporters involved in uptake and transport of other divalent cations, such as Ca2+, Fe2+, and Mn2+ (Vert et al., 2002; Sasaki et al., 2012; Chen et al., 2018). However, it was not elucidated how OsACA6 expression enhanced Cd levels in roots.
In contrast, it is not clear how CAXs are involved in reducing the Cd levels in transgenic plants because CAXs have been reported as tonoplast transporters that sequester Ca, Cd, and Mg into vacuoles in Arabidopsis (Pittman and Hirschi, 2016). Therefore, enhanced CAXs may indirectly participate in decreasing Cd accumulation in transgenic Arabidopsis through modulation of the cytosol Ca concentration. Furthermore, other studies suggest that CAXs may not contribute to reducing Cd accumulation in plants. AtCAX1-expressing petunia showed higher Cd tolerance and accumulation (Wu et al., 2011), and the ectopic expression of Arabidopsis CAX2 and CAX4 improved Cd tolerance along with higher Cd accumulation (Korenkov et al., 2007). In addition, CAXs can promote Cd tolerance by regulating ROS signaling and accumulation in plants. Knockout Arabidopsis of CAX1 showed less tolerance to Cd at low concentration of Ca and a higher ROS level compared with the WT plant (Baliardini et al., 2015). Very recently, CAX1 has been shown to increase the Cd tolerance and diminish ROS accumulation under Cd stress in Arabidopsis halleri (Ahmadi et al., 2018). Therefore, it is very possible that the higher expression level of CAXs in NtHb1-Arabidopsis also lowered ROS accumulation compared with the control plants, resulting in a higher Cd tolerance.
Sodium nitroprusside (NO donor) treatment experiments (Figure 6) demonstrated that expression of CAX3 and PDR8 was decreased by NO, but IRT1 expression was enhanced, emphasizing the correctness of the reversed expression pattern in NtHb1-expressing Arabidopsis. Expression of IRT1 is enhanced by NO, which leads to an increase in Cd uptake (Besson-Bard et al., 2009; Luo et al., 2012; Zhu et al., 2012). However, the repression of CAX3 and PDR8 expression, as well as induction of ACA10 expression, by NO was first reported in this research. Regarding the increased expression of CAX1∼3, PDR8, and ACA10 in NtHb1-Arabidopsis in the absence of Cd (Figure 5), the higher level of Hb1 in transgenic plants is likely involved in enhancing the expression of CAX1∼3, PDR8, and ACA10 by decreasing NO, even without a Cd challenge. In summary, the reduced accumulation of Cd in NtHb1-Arabidopsis was attributed to the enhanced expression of CAX1∼3 and PDR8, and to the reduced expression of IRT1 and ACA10, which was caused by the decreased level of NO.
It seems that ROS, in addition to NO, is able to regulate the expression of Cd transporters. In response to H2O2 treatment, the transcript levels of CAX3 and PDR8 were decreased, while those of IRT1 and ACA10 were increased (Figure 7). NtCAX3 expression was increased and NtIRT1 expression was reduced in NtUBC1-expressing tobacco, which accumulated a lower level of ROS under Cd stress conditions (Bahmani et al., 2017). Moreover, NRAMP3 and NRAMP4 were up-regulated by H2O2 treatment (Molins et al., 2013), and ABCC3, ABCC4, and ABCC6 expression was also enhanced in wheat plants exposed to H2O2 (Bhati et al., 2015). The next step is to elucidate how NO or ROS regulates gene expression of Cd transporters.
In summary, we propose a model (Figure 9) showing that the over-expression of NtHb1 reduces NO and ROS levels, by which Cd efflux is enhanced and Cd influx is decreased.
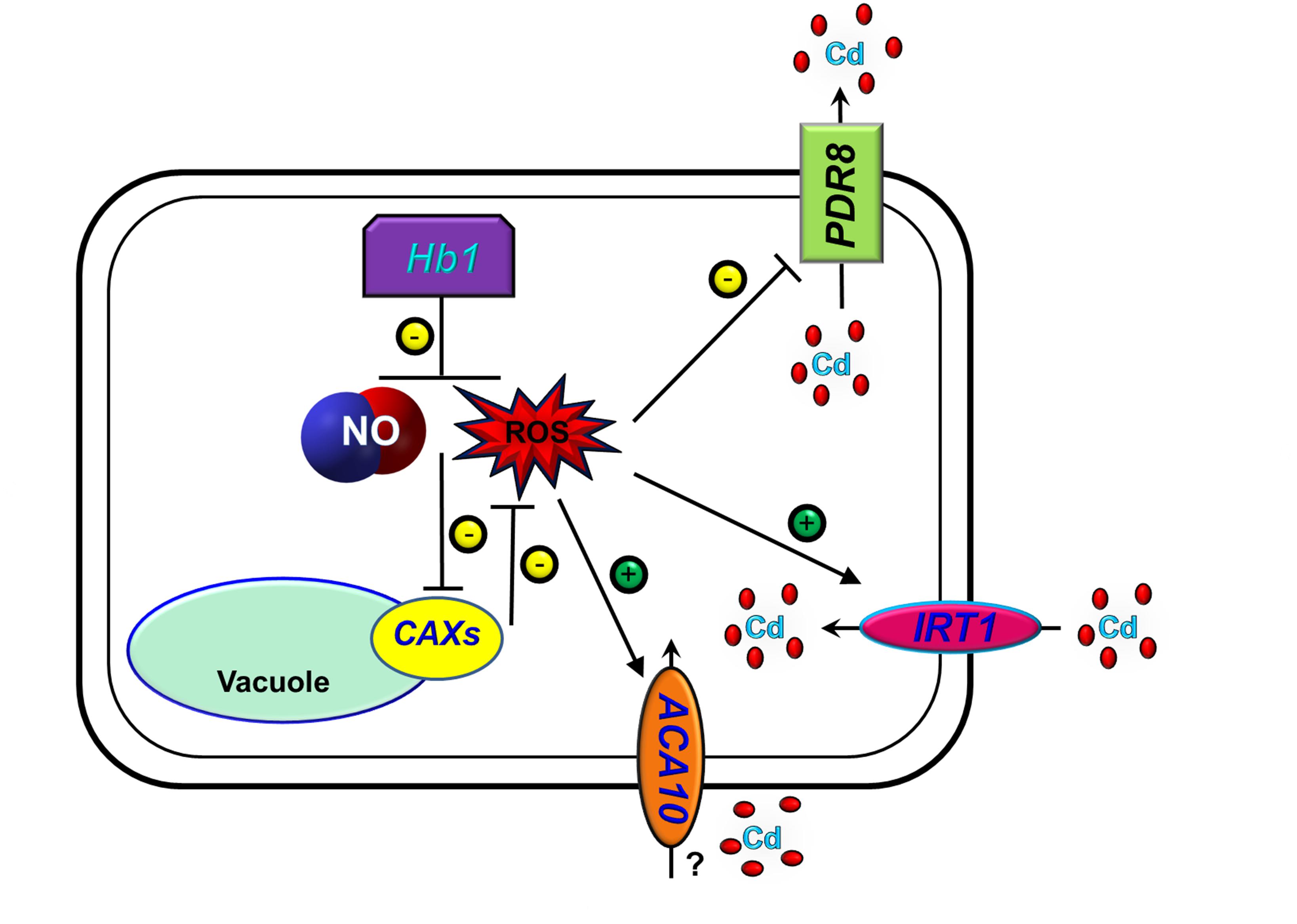
Figure 9. A proposed model for the function of Hb1 in response to cadmium. The arrows show positive regulation, whereas the blunt lines show negative regulation.
Conclusion
The expression of NtHb1 in Arabidopsis reduces Cd levels and increases Cd tolerance by inhibiting expression of IRT1 and ACA10 and enhancing expression of PDR8 and CAXs via decreased NO and ROS (H2O2) levels. In addition, AtHb1 expression is greatly enhanced in the roots and slightly enhanced in the shoots in response to Cd.
Author Contributions
SH acquired a research fund, designed and supervised whole research, and wrote the article with contributions of all authors. RB performed most experiments. DK and JN performed the minor parts of experiments.
Funding
This work was financially supported through a grant from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Agri-Bio industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (IPET 316087-4), South Korea.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00201/full#supplementary-material
Abbreviations
ANOVA, analysis of variance; CaCl2, calcium chloride; CAT, catalase; CM-H2DCFDA, 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate; cPTIO, 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxid; DAF-2DA, 4,5-diaminofluorescein; DMSO, Dimethyl sulfoxide; DTNB, 2-nitrobenzoic acid; DTT, dithiothreitol; EDTA, ethylene diamine tetraacetic acid; EGTA, ethylene glycol tetraacetic acid; GR, glutathione reductase; GSSG, glutathione disulphide; H2O2, hydrogen peroxide; ICP, inductively coupled plasma, MS, Murashige and Skoog; NBT, nitroblue tetrazolium; NO, nitric oxide; qRT-PCR, quantitative real-time PCR; RNS, reactive Nitrogen Species; ROS, reactive oxygen species; SNP, sodium nitroprusside; SOD, superoxide dismutase; TNB, 2-nitro-5-thiobenzoate; WT, Wild type.
Footnotes
References
Aebi, H. (1984). Catalase in vitro. Methods Enzymol. 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Ahmadi, H., Corso, M., Weber, M., Verbruggen, N., and Clemens, S. (2018). CAX1 suppresses Cd-induced generation of reactive oxygen species in Arabidopsis halleri. Plant Cell Environ. 41, 2435–2448. doi: 10.1111/pce.13362
Arasimowicz-Jelonek, M., Floryszak-Wieczorek, J., Deckert, J., Rucinska-Sobkowiak, R., Gzyl, J., Pawlak-Sprada, S., et al. (2012). Nitric oxide implication in cadmium-induced programmed cell death in roots and signaling response of yellow lupine plants. Plant Physiol. Biochem. 58, 124–134. doi: 10.1016/j.plaphy.2012.06.018
Bahmani, R., Kim, D., Lee, B. D., and Hwang, S. (2017). Over-expression of tobacco UBC1 encoding a ubiquitin-conjugating enzyme increases cadmium tolerance by activating the 20S/26S proteasome and by decreasing Cd accumulation and oxidative stress in tobacco (Nicotiana tabacum). Plant Mol. Biol. 94, 433–451. doi: 10.1007/s11103-017-0616-6
Bai, X. G., Long, J., He, X. Z., Yan, J. P., Chen, X. Q., Tan, Y., et al. (2016). Overexpression of spinach non-symbiotic hemoglobin in Arabidopsis resulted in decreased NO content and lowered nitrate and other abiotic stresses tolerance. Sci. Rep. 6:26400. doi: 10.1038/srep26400
Balestrazzi, A., Agoni, V., Tava, A., Avato, P., Biazzi, E., Raimondi, E., et al. (2011). Cell death induction and nitric oxide biosynthesis in white poplar (Populus alba) suspension cultures exposed to alfalfa saponins. Physiol. Plant. 141, 227–238. doi: 10.1111/j.1399-3054.2010.01436.x
Baliardini, C., Meyer, C. L., Salis, P., Saumitou-Laprade, P., and Verbruggen, N. (2015). CATION EXCHANGER1 cosegregates with cadmium tolerance in the metal hyperaccumulator Arabidopsis halleri and plays a role in limiting oxidative stress in arabidopsis Spp. Plant Physiol. 169, 549–559. doi: 10.1104/pp.15.01037
Besson-Bard, A., Gravot, A., Richaud, P., Auroy, P., Duc, C., Gaymard, F., et al. (2009). Nitric oxide contributes to cadmium toxicity in arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 149, 1302–1315. doi: 10.1104/pp.108.133348
Beyer, W. F., and Fridovich, I. (1987). Assaying for superoxide-dismutase activity - some large consequences of minor changes in conditions. Anal. Biochem. 161, 559–566. doi: 10.1016/0003-2697(87)90489-1
Bhati, K. K., Sharma, S., Aggarwal, S., Kaur, M., Shukla, V., Kaur, J., et al. (2015). Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Front. Plant Sci. 6:488. doi: 10.3389/fpls.2015.00488
Bi, Y. H., Chen, W. L., Zhang, W. N., Zhou, Q., Yun, L. J., and Xing, D. (2009). Production of reactive oxygen species, impairment of photosynthetic function and dynamic changes in mitochondria are early events in cadmium-induced cell death in Arabidopsis thaliana. Biol. Cell 101, 629–643. doi: 10.1042/Bc20090015
Brunetti, P., Zanella, L., De Paolis, A., Di Litta, D., Cecchetti, V., Falasca, G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829. doi: 10.1093/jxb/erv185
Brunetti, P., Zanella, L., Proia, A., De Paolis, A., Falasca, G., Altamura, M. M., et al. (2011). Cadmium tolerance and phytochelatin content of Arabidopsis seedlings over-expressing the phytochelatin synthase gene AtPCS1. J. Exp. Bot. 62, 5509–5519. doi: 10.1093/jxb/err228
Cantrel, C., Vazquez, T., Puyaubert, J., Reze, N., Lesch, M., Kaiser, W. M., et al. (2011). Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 189, 415–427. doi: 10.1111/j.1469-8137.2010.03500.x
Chen, C., Xia, S. L., Deng, R. B., Liu, C. F., and Shi, G. R. (2017). AhIRT1 and AhNRAMP1 metal transporter expression correlates with Cd uptake in peanuts under iron deficiency. PLoS One 12:e0185144. doi: 10.1371/journal.pone.0185144
Chen, S. S., Han, X. J., Fang, J., Lu, Z. C., Qiu, W. M., Liu, M. Y., et al. (2017). Sedum alfredii SaNramp6 metal transporter contributes to cadmium accumulation in transgenic Arabidopsis thaliana. Sci. Rep. 7:e0185144. doi: 10.1038/s41598-017-13463-4
Chen, X. H., Ouyang, Y. N., Fan, Y. C., Qiu, B. Y., Zhang, G. P., and Zeng, F. R. (2018). The pathway of transmembrane cadmium influx via calcium-permeable channels and its spatial characteristics along rice root. J. Exp. Bot. 69, 5279–5291. doi: 10.1093/jxb/ery293
Chmielowska-Bąk, J., and Deckert, J. (2013). Nitric oxide mediates Cd-dependent induction of signaling- associated genes. Plant Signal. Behav. 8:e26664. doi: 10.4161/psb.26664
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Connolly, E. L., Fett, J. P., and Guerinot, M. L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14, 1347–1357. doi: 10.1105/tpc.001263
Das, P., Samantaray, S., and Rout, G. R. (1997). Studies on cadmium toxicity in plants: a review. Environ. Pollut. 98, 29–36. doi: 10.1016/S0269-7491(97)00110-3
De Michele, R., Vurro, E., Rigo, C., Costa, A., Elviri, L., Di Valentin, M., et al. (2009). Nitric oxide is involved in cadmium-induced programmed cell death in arabidopsis suspension cultures. Plant Physiol. 150, 217–228. doi: 10.1104/pp.108.133397
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A., and Pottosin, I. (2018). Calcium transport across plant membranes: mechanisms and functions. New Phytol. 220, 49–69. doi: 10.1111/nph.15266
Dordas, C., Hasinoff, B. B., Igamberdiev, A. U., Manac’h, N., Rivoal, J., and Hill, R. D. (2003). Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. Plant J. 35, 763–770. doi: 10.1046/j.1365-313X.2003.01846.x
Du, Z. Y., Chen, M. X., Chen, Q. F., Gu, J. D., and Chye, M. L. (2015). Expression of Arabidopsis acyl-CoA-binding proteins AtACBP1 and AtACBP4 confers Pb(II) accumulation in Brassica juncea roots. Plant Cell Environ. 38, 101–117. doi: 10.1111/pce.12382
Goyer, R. A. (1997). Toxic and essential metal interactions. Ann. Rev. Nutr. 17, 37–50. doi: 10.1146/annurev.nutr.17.1.37
Groppa, M. D., Rosales, E. P., Lannone, M. F., and Benavides, M. P. (2008). Nitric oxide, polyamines and Cd-induced phytotoxicity in wheat roots. Phytochemistry 69, 2609–2615. doi: 10.1016/j.phytochem.2008.07.016
Guo, Q., Meng, L., Humphreys, M. W., Scullion, J., and Mur, L. A. J. (2017). Expression of FIHMA3, a P-1B2-ATPase from Festulolium loliaceum, correlates with response to cadmium stress. Plant Physiol. Biochem. 112, 270–277. doi: 10.1016/j.plaphy.2017.01.013
Gupta, D. K., Pena, L. B., Romero-Puertas, M. C., Hernández, A., Inouhe, M., and Sandalio, L. M. (2017). NADPH oxidases differentially regulate ROS metabolism and nutrient uptake under cadmium toxicity. Plant Cell Environ. 40, 509–526. doi: 10.1111/pce.12711
Han, B., Yang, Z., Xie, Y. J., Nie, L., Cui, J., and Shen, W. B. (2014). Arabidopsis HY1 confers cadmium tolerance by decreasing nitric oxide production and improving iron homeostasis. Mol. Plant 7, 388–403. doi: 10.1093/mp/sst122
He, J. Y., Ren, Y. F., Chen, X. L., and Chen, H. (2014). Protective roles of nitric oxide on seed germination and seedling growth of rice (Oryza sativa L.) under cadmium stress. Ecotoxicol. Environ. Safety 108, 114–119. doi: 10.1016/j.ecoenv.2014.05.021
Hebelstrup, K. H., Hunt, P., Dennis, E., Jensen, S. B., and Jensen, E. O. (2006). Hemoglobin is essential for normal growth of Arabidopsis organs. Physiol. Plant. 127, 157–166. doi: 10.1111/j.1399-3054.2006.00653.x
Hebelstrup, K. H., and Jensen, E. O. (2008). Expression of NO scavenging hemoglobin is involved in the timing of bolting in Arabidopsis thaliana. Planta 227, 917–927. doi: 10.1007/s00425-007-0667-z
Hebelstrup, K. H., Shah, J. K., Simpson, C., Schjoerring, J. K., Mandon, J., Cristescu, S. M., et al. (2014). An assessment of the biotechnological use of hemoglobin modulation in cereals. Physiol. Plant. 150, 593–603. doi: 10.1111/ppl.12115
Hirschi, K. D., Korenkov, V. D., Wilganowski, N. L., and Wagner, G. J. (2000). Expression of arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124, 125–133. doi: 10.1104/pp.124.1.125
Hsu, Y. T., and Kao, C. H. (2004). Cadmium toxicity is reduced by nitric oxide in rice leaves. Plant Growth Regul. 42, 227–238. doi: 10.1023/B:Grow.0000026514.98385.5c
Hunt, P. W., Klok, E. J., Trevaskis, B., Watts, R. A., Ellis, M. H., Peacock, W. J., et al. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 17197–17202. doi: 10.1073/pnas.212648799
Hunt, P. W., Watts, R. A., Trevaskis, B., Llewelyn, D. J., Burnell, J., Dennis, E. S., et al. (2001). Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 47, 677–692. doi: 10.1023/A:1012440926982
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16, 1327–1339. doi: 10.1105/Tpc.020487
Igamberdiev, A. U., Bykova, N. V., and Hill, R. D. (2011). Structural and functional properties of class 1 plant hemoglobins. Iubmb Life 63, 146–152. doi: 10.1002/iub.439
Igamberdiev, A. U., Seregelyes, C., Manac’h, N., and Hill, R. D. (2004). NADH-dependent metabolism of nitric oxide in alfalfa root cultures expressing barley hemoglobin. Planta 219, 95–102. doi: 10.1007/s00425-003-1192-3
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Kim, Y. Y., Yang, Y. Y., and Lee, Y. (2002). Pb and Cd uptake in rice roots. Physiol. Plant. 116, 368–372. doi: 10.1034/j.1399-3054.2002.1160312.x
Kopyra, M., and Gwozdz, E. A. (2003). Nitric oxide stimulates seed germination and counteracts the inhibitory effect of heavy metals and salinity on root growth of Lupinus luteus. Plant Physiol. Biochem. 41, 1011–1017. doi: 10.1016/j.plaphy.2003.09.003
Korenkov, V., Hirschi, K., Crutchfield, J. D., and Wagner, G. J. (2007). Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 226, 1379–1387. doi: 10.1007/s00425-007-0577-0
Korshunova, Y. O., Eide, D., Clark, W. G., Guerinot, M. L., and Pakrasi, H. B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40, 37–44. doi: 10.1023/A:1026438615520
Kulik, A., Anielska-Mazur, A., Bucholc, M., Koen, E., Szymanska, K., Zmienko, A., et al. (2012). SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883. doi: 10.1104/pp.112.194472
Kundu, S., Trent, J. T., and Hargrove, M. S. (2003). Plants, humans and hemoglobins. Trends Plant Sci. 8, 387–393. doi: 10.1016/S1360-1385(03)00163-8
Kuruthukulangarakoola, G. T., Zhang, J. L., Albert, A., Winkler, B., Lang, H., Buegger, F., et al. (2017). Nitric oxide-fixation by non-symbiotic haemoglobin proteins in Arabidopsis thaliana under N-limited conditions. Plant Cell and Environ. 40, 36–50. doi: 10.1111/pce.12773
Lamattina, L., Garcia-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Ann. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Lee, B. R., and Hwang, S. (2015). Over-expression of NtHb1 encoding a non-symbiotic class 1 hemoglobin of tobacco enhances a tolerance to cadmium by decreasing NO (nitric oxide) and Cd levels in Nicotiana tabacum. Environ. Exp. Bot. 113, 18–27. doi: 10.1016/j.envexpbot.2015.01.003
Lee, S., and An, G. (2009). Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ 32, 408–416. doi: 10.1111/j.1365-3040.2009.01935.x
Lehotai, N., Peto, A., Bajkan, S., Erdei, L., Tari, I., and Kolbert, Z. (2011). In vivo and in situ visualization of early physiological events induced by heavy metals in pea root meristem. Acta Physiol. Plant. 33, 2199–2207. doi: 10.1007/s11738-011-0759-z
Leitner, M., Vandelle, E., Gaupels, F., Bellin, D., and Delledonne, M. (2009). NO signals in the haze Nitric oxide signalling in plant defence. Curr. Opin. Plant Biol. 12, 451–458. doi: 10.1016/j.pbi.2009.05.012
Li, L., Wang, Y. Q., and Shen, W. B. (2012). Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots. Biometals 25, 617–631. doi: 10.1007/s10534-012-9551-9
Li, S., Yu, J. L., Zhu, M. J., Zhao, F. G., and Luan, S. (2012). Cadmium impairs ion homeostasis by altering K+ and Ca2+channel activities in rice root hair cells. Plant Cell Environ. 35, 1998–2013. doi: 10.1111/j.1365-3040.2012.02532.x
Luo, B. F., Du, S. T., Lu, K. X., Liu, W. J., Lin, X. Y., and Jin, C. W. (2012). Iron uptake system mediates nitrate-facilitated cadmium accumulation in tomato (Solanum lycopersicum) plants. J. Exp. Bot. 63, 3127–3136. doi: 10.1093/jxb/ers036
Ma, W. W., Xu, W. Z., Xu, H., Chen, Y. S., He, Z. Y., and Ma, M. (2010). Nitric oxide modulates cadmium influx during cadmium-induced programmed cell death in tobacco BY-2 cells. Planta 232, 325–335. doi: 10.1007/s00425-010-1177-y
Maassen, A., and Hennig, J. (2011). Effect of Medicago sativa Mhb1gene expression on defense response of Arabidopsis thaliana plants. Acta Biochim. Pol. 58, 427–432.
McLaughlin, M. J., Tiller, K. G., Naidu, R., and Stevens, D. P. (1996). Review: the behaviour and environmental impact of contaminants in fertilizers. Aust. J. Soil Res. 34, 1–54. doi: 10.1071/Sr9960001
Mills, R. F., Krijger, G. C., Baccarini, P. J., Hall, J. L., and Williams, L. E. (2003). Functional expression of AtHMA4, a P-1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J. 35, 164–176. doi: 10.1046/j.1365-313X.2003.01790.x
Molins, H., Michelet, L., Lanquar, V., Agorio, A., Giraudat, J., Roach, T., et al. (2013). Mutants impaired in vacuolar metal mobilization identify chloroplasts as a target for cadmium hypersensitivity in Arabidopsis thaliana. Plant Cell Environ. 36, 804–817. doi: 10.1111/pce.12016
Morel, M., Crouzet, J., Gravot, A., Auroy, P., Leonhardt, N., Vavasseur, A., et al. (2009). AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 149, 894–904. doi: 10.1104/pp.108.130294
Ortega-Galisteo, A. P., Rodriguez-Serrano, M., Pazmino, D. M., Gupta, D. K., Sandalio, L. M., and Romero-Puertas, M. C. (2012). S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103. doi: 10.1093/jxb/err414
Park, J., Song, W. Y., Ko, D., Eom, Y., Hansen, T. H., Schiller, M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Perazzolli, M., Dominici, P., Romero-Puertas, M. C., Zago, E., Zeier, A., Sonoda, M., et al. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16, 2785–2794. doi: 10.1105/tpc.104.025379
Perez-Chaca, M. V., Rodriguez-Serrano, M., Molina, A. S., Pedranzani, H. E., Zirulnik, F., Sandalio, L. M., et al. (2014). Cadmium induces two waves of reactive oxygen species in Glycine max (L.) roots. Plant Cell Environ. 37, 1672–1687. doi: 10.1111/pce.12280
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pittman, J. K., and Hirschi, K. D. (2016). CAX-ing a wide net: Cation/H+ transporters in metal remediation and abiotic stress signalling. Plant Biol. 18, 741–749. doi: 10.1111/plb.12460
Pomponi, M., Censi, V., Di Girolamo, V., De Paolis, A., Sanita, di Toppi, L., et al. (2006). Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd2+ stop tolerance and accumulation but not translocation to the shoot. Planta 223, 180–190. doi: 10.1007/s00425-005-0073-3
Prasinos, C., Krampis, K., Samakovli, D., and Hatzopoulos, P. (2005). Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J. Exp. Bot. 56, 633–644. doi: 10.1093/jxb/eri035
Qu, Z. L., Zhong, N. Q., Wang, H. Y., Chen, A. P., Jian, G. L., and Xia, G. X. (2006). Ectopic expression of the cotton non-symbiotic hemoglobin gene GhHb1 triggers defense responses and increases disease tolerance in Arabidopsis. Plant Cell Physiol. 47, 1058–1068. doi: 10.1093/pcp/pcj076
Ramesh, S. A., Shin, R., Eide, D. J., and Schachtman, D. P. (2003). Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 133, 126–134. doi: 10.1104/pp.103.026815
Ramirez, L., Simontacchi, M., Murgia, I., Zabaleta, E., and Lamattina, L. (2011). Nitric oxide, nitrosyl iron complexes, ferritin and frataxin: a well equipped team to preserve plant iron homeostasis. Plant Sci. 181, 582–592. doi: 10.1016/j.plantsci.2011.04.006
Rodriguez-Serrano, M., Romero-Puertas, M. C., Pazmino, D. M., Testillano, P. S., Risueno, M. C., del Rio, L. A., et al. (2009). Cellular response of pea plants to cadmium toxicity: cross talk between reactive oxygen species. Nitric Oxide, and Calcium. Plant Physiol. 150, 229–243. doi: 10.1104/pp.108.131524
Ronzan, M., Zanella, L., Fattorini, L., Della Rovere, F., Urgast, D., Cantamessa, S., et al. (2017). The morphogenic responses and phytochelatin complexes induced by arsenic in Pteris vittata change in the presence of cadmium. Environ. Exp. Bot. 133, 176–187. doi: 10.1016/j.envexpbot.2016.10.011
Sanita, di Toppi, L., and Gabbrielli, R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41, 105–130. doi: 10.1016/S0098-8472(98)00058-6
Sano, T., Yoshihara, T., Handa, K., Sato, M. H., Nagata, T., and Hasezawa, S. (2012). “Metal ion homeostasis mediated by NRAMP transporters in plant cells-focused on increased resistance to iron and cadmiumion,” in Crosstalk and Integration of Membrane Trafficking Pathways, ed. R. Weigert (London: IntechOpen Limited), 213–228.
Sasaki, A., Yamaji, N., Yokosho, K., and Ma, J. F. (2012). Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167. doi: 10.1105/tpc.112.096925
Seregelyes, C., Igamberdiev, A. U., Maassen, A., Hennig, J., Dudits, D., and Hill, R. D. (2004). NO-degradation by alfalfa class 1 hemoglobin (Mhb 1): a possible link to PR-1a gene expression in Mhb1-overproducing tobacco plants. Febs Lett. 571, 61–66. doi: 10.1016/j.febslet.2004.06.055
Shukla, D., Huda, K. M. K., Banu, M. S. A., Gill, S. S., Tuteja, R., and Tuteja, N. (2014). OsACA6, a P-type 2B Ca2+ ATPase functions in cadmium stress tolerance in tobacco by reducing the oxidative stress load (vol 240, pg 809, 2014). Planta 240, 825–825. doi: 10.1007/s00425-014-2145-8
Smith, I. K., Vierheller, T. L., and Thorne, C. A. (1988). Assay of glutathione-reductase in crude tissue-homogenates using 5,5’-Dithiobis(2-Nitrobenzoic Acid). Anal. Biochem. 175, 408–413. doi: 10.1016/0003-2697(88)90564-7
Song, W. Y., Choi, K. S., Alexis, D., Martinoia, E., and Lee, Y. (2011). Brassica juncea plant cadmium resistance 1 protein (BjPCR1) facilitates the radial transport of calcium in the root. Proc. Natl. Acad. Sci. U.S.A. 108, 19808–19813. doi: 10.1073/pnas.1104905108
Song, W. Y., Martinoia, E., Lee, J., Kim, D., Kim, D. Y., Vogt, E., et al. (2004). A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 135, 1027–1039. doi: 10.1104/pp.103.037739
Stohs, S. J., and Bagchi, D. (1995). Oxidative mechanisms in the toxicity of metal-ions. Free Radic. Biol. Med. 18, 321–336. doi: 10.1016/0891-5849(94)00159-H
Takahashi, R., Ishimaru, Y., Shimo, H., Ogo, Y., Senoura, T., Nishizawa, N. K., et al. (2012). The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 35, 1948–1957. doi: 10.1111/j.1365-3040.2012.02527.x
Thiel, J., Rolletschek, H., Friedel, S., Lunn, J. E., Nguyen, T. H., Feil, R., et al. (2011). Seed-specific elevation of non-symbiotic hemoglobin AtHb1: beneficial effects and underlying molecular networks in Arabidopsis thaliana. BMC Plant Biol. 11:48. doi: 10.1186/1471-2229-11-48
Thomine, S., Wang, R., Ward, J. M., Crawford, N. M., and Schroeder, J. I. (2000). Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. U.S.A. 97, 4991–4996. doi: 10.1073/pnas.97.9.4991
Ueno, D., Milner, M. J., Yamaji, N., Yokosho, K., Koyama, E., Clemencia Zambrano, M., et al. (2011). Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 66, 852–862. doi: 10.1111/j.1365-313X.2011.04548.x
Valentovicova, K., Haluskova, L., Huttova, J., Mistrik, I., and Tamas, L. (2010). Effect of cadmium on diaphorase activity and nitric oxide production in barley root tips. J. Plant Physiol. 167, 10–14. doi: 10.1016/j.jplph.2009.06.018
Verbruggen, N., Hermans, C., and Schat, H. (2009). Mechanisms to cope with arsenic or cadmium excess in plants. Curr. Opin. Plant Biol. 12, 364–372. doi: 10.1016/j.pbi.2009.05.001
Verma, K., Mehta, S. K., and Shekhawat, G. S. (2013). Nitric oxide (NO) counteracts cadmium induced cytotoxic processes mediated by reactive oxygen species (ROS) in Brassica juncea: cross-talk between ROS, NO and antioxidant responses. Biometals 26, 255–269. doi: 10.1007/s10534-013-9608-4
Verret, F., Gravot, A., Auroy, P., Leonhardt, N., David, P., Nussaume, L., et al. (2004). Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. Febs Lett. 576, 306–312. doi: 10.1016/j.febslet.2004.09.023
Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Wahid, A., Arshad, M., and Farooq, M. (2010). “Cadmium phytotoxicity: responses, mechanisms and mitigation strategies: a review,” in Organic Farming, Pest Control and Remediation of Soil Pollutants: Organic Farming, Pest Control and Remediation of Soil Pollutants, ed. E. Lichtfouse (Dordrecht: Springer), 371–403.
Wang, L., Guo, Y. J., Jia, L. X., Chu, H. Y., Zhou, S., Chen, K. M., et al. (2014). Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in arabidopsis seedlings1[C][W]. Plant Physiol. 164, 2184–2196. doi: 10.1104/pp.113.229369
Wu, D., Yamaji, N., Yamane, M., Kashino-Fujii, M., Sato, K., Feng, et al. (2016). The HvNramp5 transporter mediates uptake of cadmium and manganese, but not iron. Plant Physiol. 172, 1899–1910. doi: 10.1104/pp.16.01189
Wu, Q., Shigaki, T., Williams, K. A., Han, J. S., Kim, C. K., Hirschi, K. D., et al. (2011). Expression of an Arabidopsis Ca2+/H+ antiporter CAX1 variant in petunia enhances cadmium tolerance and accumulation. J. Plant Physiol. 168, 167–173. doi: 10.1016/j.jplph.2010.06.005
Xiong, J., Lu, H., Lu, K. X., Duan, Y. X., An, L. Y., and Zhu, C. (2009). Cadmium decreases crown root number by decreasing endogenous nitric oxide, which is indispensable for crown root primordia initiation in rice seedlings. Planta 230, 599–610. doi: 10.1007/s00425-009-0970-y
Yang, L. M., Ji, J. H., Harris-Shultz, K. R., Wang, H., Wang, H. L., Abd-Allah, E. F., et al. (2016). The dynamic changes of the plasma membrane proteins and the protective roles of nitric oxide in rice subjected to heavy metal cadmium stress. Front. Plant Sci. 7:190. doi: 10.3389/fpls.2016.00190
Yang, L. X., Wang, R. Y., Ren, F., Liu, J., Cheng, J., and Lu, Y. T. (2005). AtGLB1 enhances the tolerance of Arabidopsis to hydrogen peroxide stress. Plant Cell Physiol. 46, 1309–1316. doi: 10.1093/pcp/pci40
Yang, T. B., and Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8, 505–512. doi: 10.1016/j.tplants.2003.09.004
Ye, Y., Li, Z., and Xing, D. (2013). Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium-induced Arabidopsis thaliana programmed cell death. Plant Cell Environ. 36, 1–15. doi: 10.1111/j.1365-3040.2012.02543.x
Yoshihara, T., Hodoshima, H., Miyano, Y., Shoji, K., Shimada, H., and Goto, F. (2006). Cadmium inducible Fe deficiency responses observed from macro and molecular views in tobacco plants. Plant Cell Rep. 25, 365–373. doi: 10.1007/s00299-005-0092-3
Yuan, L. Y., Yang, S. G., Liu, B. X., Zhang, M., and Wu, K. Q. (2012). Molecular characterization of a rice metal tolerance protein, OsMTP1. Plant Cell Rep. 31, 67–79. doi: 10.1007/s00299-011-1140-9
Zanella, L., Fattorini, L., Brunetti, P., Roccotiello, E., Cornara, L., D’Angeli, S., et al. (2016). Overexpression of AtPCS1 in tobacco increases arsenic and arsenic plus cadmium accumulation and detoxification. Planta 243, 605–622. doi: 10.1007/s00425-015-2428-8
Zeng, L. H., Zhu, T., Gao, Y., Wang, Y. T., Ning, C. J., Bjorn, L. O., et al. (2017). Effects of Ca addition on the uptake, translocation, and distribution of Cd in Arabidopsis thaliana. Ecotoxicol. Environ. Safety 139, 228–237. doi: 10.1016/j.ecoenv.2017.01.023
Zhang, L., Chen, Z., and Zhu, C. (2012). Endogenous nitric oxide mediates alleviation of cadmium toxicity induced by calcium in rice seedlings. J. Environ. Sci. 24, 940–948. doi: 10.1016/S1001-0742(11)60978-9
Zhao, L., Gu, R. L., Gao, P., and Wang, G. Y. (2008). A nonsymbiotic hemoglobin gene from maize, ZmHb, is involved in response to submergence, high-salt and osmotic stresses. Plant Cell Tissue Organ Cult. 95, 227–237. doi: 10.1007/s11240-008-9436-3
Keywords: ACA10, cadmium, CAX, hemoglobin, IRT1, nitric oxide, PDR8, ROS
Citation: Bahmani R, Kim DG, Na JD and Hwang S (2019) Expression of the Tobacco Non-symbiotic Class 1 Hemoglobin Gene Hb1 Reduces Cadmium Levels by Modulating Cd Transporter Expression Through Decreasing Nitric Oxide and ROS Level in Arabidopsis. Front. Plant Sci. 10:201. doi: 10.3389/fpls.2019.00201
Received: 01 October 2018; Accepted: 06 February 2019;
Published: 22 February 2019.
Edited by:
Luisa M. Sandalio, Spanish National Research Council (CSIC), SpainReviewed by:
Toshiro Shigaki, The University of Tokyo, JapanLiliana Beatriz Pena, Universidad de Buenos Aires, Argentina
Copyright © 2019 Bahmani, Kim, Na and Hwang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seongbin Hwang, c2Jod2FuZ0BzZWpvbmcuYWMua3I=
 Ramin Bahmani
Ramin Bahmani DongGwan Kim
DongGwan Kim JongDuk Na
JongDuk Na Seongbin Hwang
Seongbin Hwang