
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 January 2019
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01979
 Qibin Ma1,2,3,4†
Qibin Ma1,2,3,4† Zhenglin Xia1,2,3,4†
Zhenglin Xia1,2,3,4† Zhandong Cai1,2,3,4†
Zhandong Cai1,2,3,4† Lu Li1,2,3,4
Lu Li1,2,3,4 Yanbo Cheng1,2,3,4
Yanbo Cheng1,2,3,4 Jia Liu1,2,3,4
Jia Liu1,2,3,4 Hai Nian1,2,3,4*
Hai Nian1,2,3,4*The WRKY transcription factors (TFs) are one of the largest families of TFs in plants and play multiple roles in plant development and stress response. In the present study, GmWRKY16 encoding a WRKY transcription factor in soybean was functionally characterized in Arabidopsis. GmWRKY16 is a nuclear protein that contains a highly conserved WRKY domain and a C2H2 zinc-finger structure, and has the characteristics of transcriptional activation ability, presenting a constitutive expression pattern with relative expression levels of over fourfold in the old leaves, flowers, seeds and roots of soybean. The results of quantitative real time polymerase chain reaction (qRT-PCR) showed that GmWRKY16 could be induced by salt, alkali, ABA, drought and PEG-6000. As compared with the control, overexpression of GmWRKY16 in Arabidopsis increased the seed germination rate and root growth of seedlings in transgenic lines under higher concentrations of mannitol, NaCl and ABA. In the meantime, GmWRKY16 transgenic lines showed over 75% survival rate after rehydration and enhanced Arabidopsis tolerance to salt and drought with higher proline and lower MDA accumulation, less water loss of the detached leaves, and accumulated more endogenous ABA than the control under stress conditions. Further studies showed that AtWRKY8, KIN1, and RD29A were induced in GmWRKY16 transgenic plants under NaCl treatment. The expressions of the ABA biosynthesis gene (NCED3), signaling genes (ABI1, ABI2, ABI4, and ABI5), responsive genes (RD29A, COR15A, COR15B, and RD22) and stress-related marker genes (KIN1, LEA14, LEA76, and CER3) were regulated in transgenic lines under drought stress. In summary, these results suggest that GmWRKY16 as a WRKY TF may promote tolerance to drought and salt stresses through an ABA-mediated pathway.
The growth and yield of plants are seriously affected and inhibited by various biotic and abiotic stresses such as high salinity, drought and extreme temperatures, which are the main limiting factors (Hennig, 2012; Liang et al., 2017; He et al., 2018). Plants have evolved some effective mechanisms to face environmental challenges (Munns, 2002; He et al., 2018). Among all the stress-resistance systems in plants, many transcription factors (TFs) related to plant tolerance to stresses have been identified as playing important roles in abiotic stresses. Under stressful conditions, transcriptome changes are the earliest responses in plants. The TFs from the bZIP, NAC, AP2, WRKY, PHD, DREB, or MYB families are essential for plant stress response by binding specific cis-acting elements to form a complex regulatory network. Overexpression of these TF genes usually increases the adaptability of plants to drought and salt stresses (Liu et al., 2012; Ying et al., 2012; Zong et al., 2016; Ullah et al., 2018).
The WRKY TFs, dominating the genetic transcription, have become one of the largest TF families in plants (Eulgem et al., 2000; Rushton et al., 2010; Bencke-Malato et al., 2016; Goel et al., 2016; Meng et al., 2016; Yu Y. et al., 2016; Wu J. et al., 2017; Xiao et al., 2017; Bai et al., 2018; Finatto et al., 2018; Song et al., 2018; Xie et al., 2018). All the WRKY proteins have one or two conserved WRKY domains of approximately 60 amino acids with a highly conserved sequence motif WRKYGQK and a Zn-finger motif at N and C termini, respectively (Eulgem et al., 2000). The WRKY TFs can be classified into three groups (I, II, and III) according to the number of WRKY domains and the Zn-finger motif type. The WRKY proteins of group I have two domains of WRKY and a C2H2-type zinc-finger structure, while the WRKY proteins of groups II and III have a WRKY domain with C2H2 and/or C2HC-type Zn-finger motifs, respectively. Most TF proteins of group II with one WRKY domain are divided into five subgroups (IIa–IIe) on their phylogenetic clades (Eulgem et al., 2000; Zhang and Wang, 2005; Rushton et al., 2010; Zhang et al., 2017; Song et al., 2018; Xie et al., 2018; Yang et al., 2018).
Since the first WRKY gene SPF1 was isolated from sweet potato (Ishiguro and Nakamura, 1994), more WRKY family genes with multiple roles in plants were discovered and widely reported to be particularly involved in response to biotic and abiotic stresses (Dai et al., 2016; Kiranmai et al., 2018) as well as regulatory roles such as leaf senescence (Miao and Zentgraf, 2007; Rinerson et al., 2015), trichome development (Bakshi and Oelmüller, 2014), root growth (Grunewald et al., 2013), seed development (Schluttenhofer and Yuan, 2015), dormancy (Ding et al., 2014), and secondary metabolites biosynthesis (Eulgem et al., 2000).
Recently, the functional characterization of WRKY proteins in plants was focused on their response to abiotic stresses. An individual WRKY gene can enhance plant tolerance to abiotic stress (Wang et al., 2013; Bakshi and Oelmüller, 2014; Dai et al., 2016; Kiranmai et al., 2018). In rice, overexpression of OsWRKY11 and OsWRKY42 improved heat and/or salt tolerance in transgenic rice seedlings (Wu et al., 2009; Pillai et al., 2018), and overexpression of OsWRKY11 and OsWRKY30 enhanced drought tolerance in transgenic rice (Wu et al., 2009; Shen et al., 2012), whereas OsWRKY76 played opposite roles in cold stress tolerance and blast disease resistance (Yokotani et al., 2013). In Arabidopsis, overexpression of AtWRKY25, AtWRKY26 and AtWRKY30 enhanced resistance to heat and salt stresses, respectively (Li et al., 2011; Scarpeci et al., 2013); overexpression of AtWRKY46, AtWRKY54, AtWRKY70, and ABO3 enhanced resistance to drought stress (Ren et al., 2010; Chen et al., 2017). In contrast to rice and Arabidopsis, many WRKY genes in other plants have been identified specifically in relation to abiotic stress. For example, GhWRKY41/SpWRKY1 enhances salt and drought tolerance in transgenic tobacco by regulating stomatal conductance and ROS levels (Li et al., 2015; Chu et al., 2016). Other WRKY genes such as TaWRKY2 and TaWRKY19 (wheat), TaWRKY146 (wheat), TaWRKY1 and TaWRKY33 (wheat), VaWRKY14 (grapevine), ZmWRKY17 and ZmWRKY40 (maize), and PeWRKY83 (moso bamboo) are overexpressed in Arabidopsis, and the heterologous transgenic plants of the WRKY genes conferred tolerance to drought and/or salt stresses, respectively (Niu et al., 2012; He et al., 2016; Cai et al., 2017; Ma et al., 2017; Wu M. et al., 2017; Wang et al., 2018; Zhang et al., 2018). Among the WRKY genes in soybean, the overexpression of GmWRKY21 enhanced the tolerance to cold stress in Arabidopsis, whereas GmWRKY54 conferred salt and drought tolerance. By contrast, overexpression of GmWRKY13 resulted in more sensitivity to salt and mannitol stress (Zhou et al., 2008). GmWRKY27 overexpression improved salt and drought tolerance by interacting with GmMYB174 to reduce expression of GmNAC29 (Wang et al., 2015).
The plant hormone abscisic acid (ABA) plays important roles in the regulation of plant responses to adverse environmental stresses including drought, extreme temperatures and osmotic stress partly mediated by ABA-independent and -dependent pathways (Shinozaki and Yamaguchi-Shinozaki, 1997; Fujii and Zhu, 2009; Umezawa et al., 2010; Rushton et al., 2012; Shang et al., 2012; Yoshida et al., 2014; Song et al., 2016). Recently, some WRKY genes responding to abiotic stresses have been reported within the ABA signaling pathways. GhWRKY6-like from cotton improved Arabidopsis tolerance to salt stress by activating the ABA signaling pathway and eliminating reactive oxygen species (Ullah et al., 2018). GhWRKY17 in transgenic nicotiana enhanced the sensitivity of plants to salt and drought stresses through the ABA signaling pathway by reducing the level of ABA and transcript levels of some ABA-inducible genes (Yan et al., 2014). The overexpression of ZmWRKY17 in Arabidopsis resulted in the enhanced sensitivity of transgenic plants to salt stress and decreased ABA sensitivity by regulating the expression of some ABA- and stress-responsive genes (Cai et al., 2017). GsWRKY20 and ABO3 in Arabidopsis increased drought tolerance of transgenic plants by regulating gene expression of the ABA signaling pathway (Ren et al., 2010; Luo et al., 2013). CmWRKY1 and CmWRKY10 enhanced drought and dehydration tolerance of chrysanthemum by an ABA-mediated pathway through the regulation of ABA-associated genes (Fan et al., 2016; Jaffar et al., 2016).
In this study, GmWRKY16, a stress responsive gene induced by drought, salt, alkali, PEG, and ABA, was analyzed for its functional characterization of stress tolerance in Arabidopsis. Transgenic Arabidopsis plants of GmWRKY16 enhanced drought and salt tolerance as compared to wild type. Furthermore, the regulated genes responding to stresses were investigated in Arabidopsis for potential mechanisms through an ABA-mediated pathway.
A spring-sowing soybean cultivar, “Huachun 2” (HC2), bred by the Guangdong Subcenter of National Center for Soybean Improvement in South China Agricultural University, China, was used to investigate the tissue expression pattern of GmWRKY16 and its responses to various stresses including mannitol, salt, drought, and ABA. Soybean seeds of HC2 were germinated, and its seedlings grew in growth chamber under a 12 h light/12 h dark cycle with 100 μM photons m-2 s-1 at 25–30°C for 3 weeks as described previously by Zeng et al. (2012). The soybean roots of 3-week-old seedlings were immersed in Hoagland solution saturated with 200 mM NaCl, 50 mM NaHCO3, 20% (w/v) PEG-6000 or 100 μM ABA, respectively (Luo et al., 2013; Chen et al., 2015). Root samples were harvested after time intervals of 0, 1, 3, 6, 9, 12, and 24 h, respectively. Samples were frozen immediately in liquid nitrogen and subsequently stored for RNA extraction at -80°C.
Ecotype Col-0 of Arabidopsis was used as the receptor for genetic transformation of GmWRKY16 gene. All the Arabidopsis seeds of wild-type and transgenic lines were grown on the agar medium of 1/2 Murashige and Skoog (MS) in darkness for 2 days at 4°C after the seeds were sterilized with ddH2o. The seedlings were then cultured for several days under long-day conditions (16 h light/8 h dark cycle) at 22°C in new 1/2 MS medium. The chemical compounds of mannitol, NaCl, and ABA were dissolved in MS medium, respectively. The taken samples of Arabidopsis were frozen immediately in liquid nitrogen and stored at -80°C (Chen et al., 2015).
According to previous reports, the sequence information of the GmWRKY16 gene involved in abiotic stress was obtained from the database of the National Center for Biotechnology Information (NCBI) with the accession number XP_003518509. The full-length sequence of GmWRKY16 was amplified by RT-PCR using specific primers (Supplementary Table S1) and Super-Fidelity DNA polymerase (Phanta Max, Vazyme Biotech Co., Ltd.; Nanjing, China). The seedlings of HC2 under the treatment of 100 mM NaCl were used to extract total RNA by TRIzol reagent (Invitrogen). The methods of generated cDNA, RT-PCR reaction and agarose gel electrophoresis were described in detail previously (Lü et al., 2015). The purified PCR product was then inserted into the multiple cloning sites of the pLB vector (Tiangen Rapid DNA Ligation Kit, Beijing, China). The positive clones in Escherichia coli were used to obtain the full cDNA sequence of GmWRKY16 identified by PCR, enzyme digestion and sequencing [Sangon Biotech (Shanghai) Co., Ltd., China] (Lü et al., 2015; Ma et al., 2018).
The nucleotide sequence of GmWRKY16 and the amino acid sequence of GmWRKY16 protein were used to search its homologous genes and proteins using the database of NCBI1. The alignment of nucleotide sequences was performed with the software of DNAMAN. The structure prediction of the GmWRKY16 protein was carried out using the NCBI blast results. Phylogenetic analysis was enforced by using the software of MEGA 7 (Kumar et al., 2016). The alignment was adjusted manually, while the unrooted phylogenetic trees were constructed by the Neighbor-Joining method (Sievers et al., 2011).
The full-length coding sequence of GmWRKY16 (Supplementary Table S1) amplified from the GmWRKY16-pLB vector was inserted into the BamHI and KpnI sites of a pTF-101 vector using specific primers (Supplementary Table S1). The pTF-101-GmWRKY16 fusion construct with a β-glucuronidase (GUS) reporter was generated under the control of the cauliflower mosaic virus 35S (CaMV 35S) promoter. The pTF-101-GmWRKY16 plasmid was then transformed into Agrobacterium tumefaciens GV3101 competent cells by the electroporation method. The full-flowering Arabidopsis plants were used for genetic transformation by the floral dip method (Clough and Bent, 1998). All the GmWRKY16 transgenic seedlings were screened by an herbicide to obtain the positive plants.
Localization of the GmWRKY16 protein was carried out using the method described previously (Lü et al., 2015). The complete coding sequence of GmWRKY16 without a stop codon amplified by PCR using the specific primers was inserted into the NcoI and SpeI restriction sites to generate a fusion construct under the control of CaMV 35S promoter. The plasmids of pCAMBIA1302 and pCAMBIA1302-GmWRKY16 were transformed into Agrobacterium tumefaciens strain GV3101 by the heat shock method. The young leaves of 4-week-old tobacco plants were contaminated by the recombinant Agrobacterium tumefaciens using the method according to Kokkirala et al. (2010). The leaves after 48 h agro-infiltration were photographed with a confocal laser scanning microscope (Leica, Germany).
The full-length cDNA of GmWRKY16 amplified by PCR using specific primers was inserted into the EcoRI and SacI sites to create a fusion construct of pGBKT7-GmWRKY16 (Supplementary Table S1). The pGBKT7-GmWRKY16 plasmid was transformed into yeast strain Y2H. An empty pGBKT7 vector was used as a negative control. Transcriptional activation was analyzed according to the methods described previously (Yang et al., 2010).
Total RNA was extracted from the seedlings of soybean or Arabidopsis using the plant total RNA kit (GeneMark, Taiwan, China). Reversing transcription for the first strand cDNA synthesis was performed with 2 μg of total RNA using a Prime ScriptTM RT reagent Kit (Takara, Beijing, China). The qRT-PCR analyses were performed on a CFX96TM Real-Time system (United States) with SYBR Premix ExTaq II (Takara, Beijing, China). The inner reference gene β-tubulin was used to normalize the data. The quantitative variations of gene expression between the examined replicates were evaluated by the 2-ΔΔCt method described previously (Willems et al., 2008; Lü et al., 2015). Three independent biological repeats were performed to ensure accurate statistical analysis. The specific primers were listed in Supplementary Table S1.
To detect seed germination of GmWRKY16 transgenic lines under the condition of mannitol, NaCl or ABA stresses, the surface-sterilized Arabidopsis seeds of wild type and transgenic lines were planted on solid 1/2 MS media with various concentrations of mannitol (0, 100, 200, and 300 mM), NaCl (0, 100, 150, and 200 mM), and ABA (0, 0.5, 1, and 1.5 μM). The rates of seed germination were recorded every day with images taken on the 4th and 7th day after seed was sown. For the root length assay, the sterilized seeds of transgenic lines and WT were grown on 1/2 MS media for 3 days, and then the seedlings were transplanted to fresh media supplemented with mannitol (0, 150, and 300 mM), NaCl (0, 75, and 150 mM), and ABA (0, 0.5, and 1.0 μM) for another 7 days to examine the root length (Rushton et al., 2012; Seo et al., 2012; Shang et al., 2012; Chen et al., 2015). The root elongation of Arabidopsis plants was monitored and analyzed using Image J software2.
For drought treatments, Arabidopsis seeds of the T3 generation of WT and GmWRKY16 overexpression lines were sown in mixed soil (vermiculite and flower nutrient soil, 1:1) for 3 weeks under optimal irrigation conditions. The following drought stress treatments were set as 5, 7, and 9 days without watering, respectively. All the Arabidopsis plants were watered again for 5 days after their photographs were taken and their survival rates were determined (Seo et al., 2012; Ying et al., 2012; Babitha et al., 2013). For salt treatments, Arabidopsis seeds of the WT and GmWRKY16 overexpression lines of the T3 generation were sown in mixed soil (vermiculite and flower nutrient soil, 1:1) in the chambers. The 3-week-old Arabidopsis plants were then irrigated with a solution of 200 mM NaCl for 15 days (Niu et al., 2012; Ying et al., 2012; Babitha et al., 2013).
The content of free proline in plants was determined following methods described previously (Bates et al., 1973). Briefly, 0.5 g of plant samples were homogenized in 5 ml of 3% sulfosalicylic acid using a mortar. The 2 ml supernatant was transferred to a new tube, and then 2 ml of acid ninhydrin and 2 ml of glacial acetic acid were added to form the extraction solution. The reaction mixture was boiled in a water bath at 100°C for 1 h and then stored at 4°C for 30 min. After the mixtures were centrifuged at 3,000 g for 5 min, the proline content for each sample was measured at 520 nm absorbance. The 3-week-old Arabidopsis seedlings were prepared in accordance with the above description. After treatment of 200 mM NaCl for 1 week, the leaf samples were taken to measure the MDA contents using the method of thiobarbituric acid (TBA) described in detail previously (Heath and Packer, 1968). For measuring dehydration, the leaves of rosettes were cut from the 3-week-old plants and immediately placed on a laboratory bench (40% relative humidity). Water loss of fresh leaves was weighed at treatment intervals set as 0, 0.5, 1, 1.5, 2, 3, 4, 5, and 6 h, respectively. All of the experiments comprised three replicates. ABA content was measured using an enzyme-linked immunosorbent assay (ELISA), as previously described (Yang et al., 2001).
All data were presented as the mean of three biological replicates ± SD. Student’s test at P = 0.01 or P = 0.05 was used to identify the difference between observation values (Lü et al., 2015).
The WRKY superfamily is one of the biggest transcription factor (TF) families in soybean and plays multiple roles in plant growth and development, abiotic stresses, etc. (Bencke-Malato et al., 2014; Dias et al., 2016; Yu Y. et al., 2016; Yang et al., 2017). Based on recent reports on the functions of WRKY TFs in soybean (Yu Y. et al., 2016), a salt and/or drought stress-induced gene encoding a transcription factor of WRKY proteins was obtained from the database under gene locus LOC100790175 and protein accession number of XP_003518509. The WRKY gene, located on chromosome 2 of soybean and designed as GmWRKY16, was cloned using the specific primers (Supplementary Table S1) from the cultivar HC2. The full-length genome sequence of GmWRKY16 included 3 exons and 2 introns with a full-length cDNA of 1014 bp (data not shown). The predicted GmWRKY16 protein comprises 337 amino acids (AA) with 38.113 kDa of molecular weight (data not shown).
The NCBI BLAST analysis of the full amino acids sequence indicated that the GmWRKY16 protein had a WRKY DNA-binding domain at the location of the peptide chain between 190 AA and 250 AA (data not shown). The results of multiple alignment using the Phytozome data showed that the GmWRKY16 protein contained two small domains with a motif of WRKYGQK and a C2H2 zinc-finger motif similar to those of five other WRKY proteins from Arabidopsis and soybean (Figure 1A). To investigate the relationship between GmWRKY16 and other WRKY proteins from soybean, rice and Arabidopsis, a phylogenetic tree was constructed by the neighbor-joining method using the MEGA 7.0 software (Figure 1B). The results indicated that all the 80 WRKY proteins were divided into three major groups: Group I (15 proteins), Group II (59 proteins), and Group III (6 proteins). In addition, 59 WRKY proteins in Group II were further divided into five subgroups: Group II-a, Group II-b, Group II-c, Group II-d, and Group II-e. GmWRKY16 protein belongs to the WRKY members of Group II-c with 93% similarity to GmWRKY129 protein (Figure 1B). Therefore, the bioinformatics analysis suggest that GmWRKY16 protein may have a function in abiotic stresses.
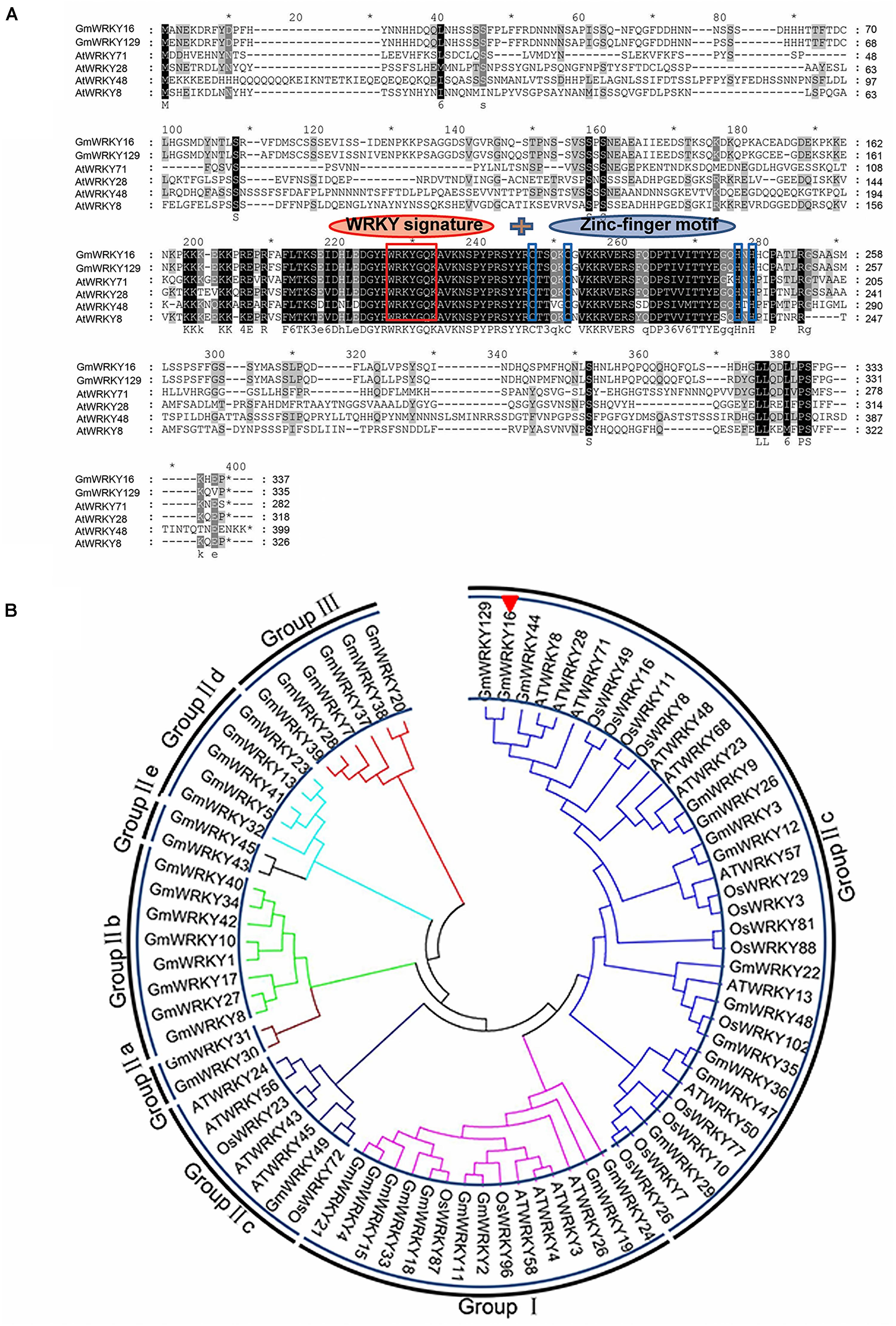
Figure 1. Homology analysis of GmWRKY16 and other WRKY transcription factors. (A) Multiple sequence alignment of GmWRKY16 and WRKY family members from Arabidopsis and soybean. (B) Phylogenetic tree analysis of GmWRKY16 and WRKY proteins. The comparison of amino acid sequences was conducted by the software of DNAMAN6.0. The phylogenetic tree was constructed by the neighbor-joining method using the software of MEGA 7.0. All the amino acid sequences and the accession numbers of WRKY proteins from soybean, rice and Arabidopsis were obtained from the databases of NCBI (https://www.ncbi.nlm.nih.gov/). The detailed information of WRKY proteins was available from the Supplementary Table S2.
To determine the transcriptional activity of GmWRKY16 protein, the ORF sequence of GmWRKY16 was inserted into the EcoRI and SacI sites of pGBKT7 to form a GmWRKY16-pGBKT7 construct (data not shown). The fusion plasmid of GmWRKY16-pGBKT7 and pGBKT7 alone (negative control) were transformed into the cells of yeast strain Y2H, respectively. As shown in Figure 2A, yeast cells transformed by the GmWRKY16-pGBKT7 and pGBKT7 vectors could grew well on the SD/-Trp medium without any differences. However, the cell deposits transformed by the GmWRKY16-pGBKT7 construct turned blue on the SD/-Trp/X-α-Gal medium colored with chromogenic substrate of X-gal (Figure 2A). These results indicated that GmWRKY16 protein had transcriptional activity in yeast cell.
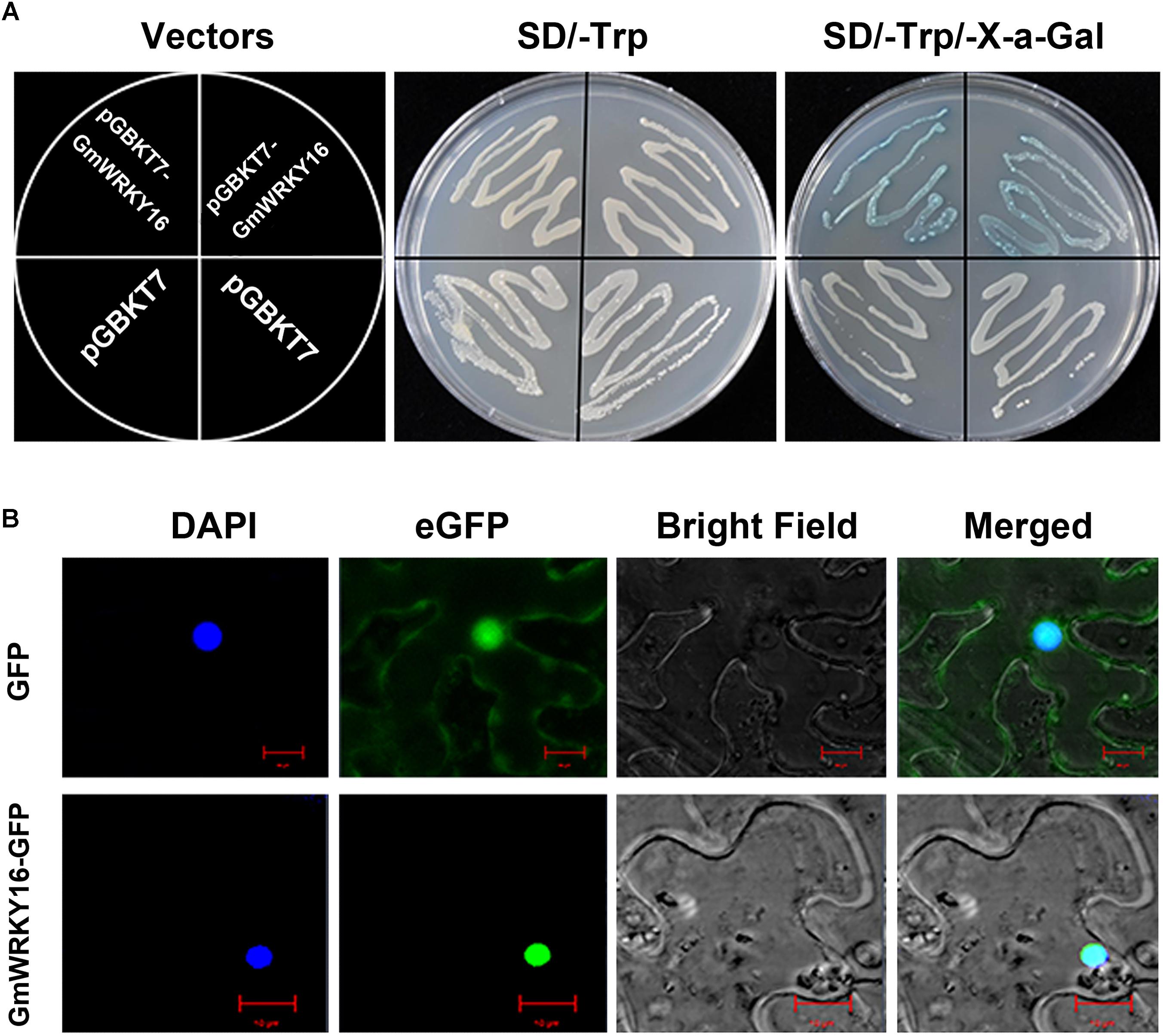
Figure 2. Transcriptional activation and subcellular localization of GmWRKY16 protein. (A) Transcriptional activation of GmWRKY16 protein in yeast cells. (B) Subcellular localization of GmWRKY16 protein in leaf epidermal cells of tobacco. The ORF sequence of GmWRKY16 was inserted into the sites of EcoRI and SacI of pGBKT7 vector to form the fusion carrier of GmWRKY16-pGBKT7. The competent yeast cells were transformed using the heat-shock method. After being cultured at 30°C for 48 h, the transcriptional activation of GmWRKY16 protein was detected by the method of color reaction in yeast cells using X-α-Gal as a substrate. The full coding sequence of GmWRKY16 (without TAA) was inserted into the NcoI and SpeI sites of pCAMBIA1302 vector to obtain the GmWRKY16-GFP fusion construct. The vectors of pCAMBIA1302 and pCAMBIA1302-GmWRKY16 were transformed into the young leaves of 4-week-old tobacco plants using the method of Kokkirala mediated by Agrobacterium tumefaciens (Kokkirala et al., 2010). The protein expression of GFP or GmWRKY16-GFP from the leaves after agro-infiltration for 48 h was visualized using a confocal laser scanning microscope (Leica, Germany) (Zhang et al., 2011).
To verify the subcellular localization of GmWRKY16 protein, the full-length GmWRKY16 cDNA without the termination codon was fused in-frame to the 5′ end of GFP gene at the NcoI and SpeI sites of the pCAMBIA1302 vector to obtain the GmWRKY16-GFP fusion construct (data not shown). The fusion plasmid of pCAMBIA1302-GmWRKY16 and pCAMBIA1302 alone were transformed into the young leaves of 4-week-old tobacco plants, respectively. As shown in Figure 2B, the GmWRKY16-GFP protein accumulated mainly in the nucleus with strong signals of green fluorescence. In contrast, GFP alone was distributed evenly throughout all parts of the cell including the nucleus and cytoplasm (Figure 2B). The result of confocal microscopic analysis suggests that the GmWRKY16 protein may function as a transcription factor.
To investigate the expression patterns of GmWRKY16 response to abiotic stresses, quantitative real-time PCR (qRT-PCR) was performed to analyze the transcript abundance of GmWRKY16 under salt, alkali, PEG-6000, ABA and drought treatments. As shown in Figure 3A, GmWRKY16 was quickly induced by 200 mM NaCl with increasing expression levels. Total RNA of GmWRKY16 was increased up to the highest level of 3.5-fold at the 6 h treatment in roots compared to that of the control, and then declined to a lower level under the 9 h to 24 h treatments. Similarly, under treatment of 50 mM NaHCO3, GmWRKY16 increased slowly under the treatments from 1 to 9 h up to the highest expression level of 5.8-fold (Figure 3B). Meanwhile, GmWRKY16 was induced rapidly by PEG-6000 up to the highest expression level of 6.8-fold (Figure 3C). For ABA treatment, GmWRKY16 was also induced rapidly up to its highest expression level of 2.5-fold, and then the expression of GmWRKY16 maintained at a relatively high level (Figure 3D). Furthermore, the expression of GmWRKY16 was increased along with the treatments of dehydration with its highest expression level of 4.6-fold (Figure 3E). These findings suggest that GmWRKY16 might play a role in multiple abiotic stresses.
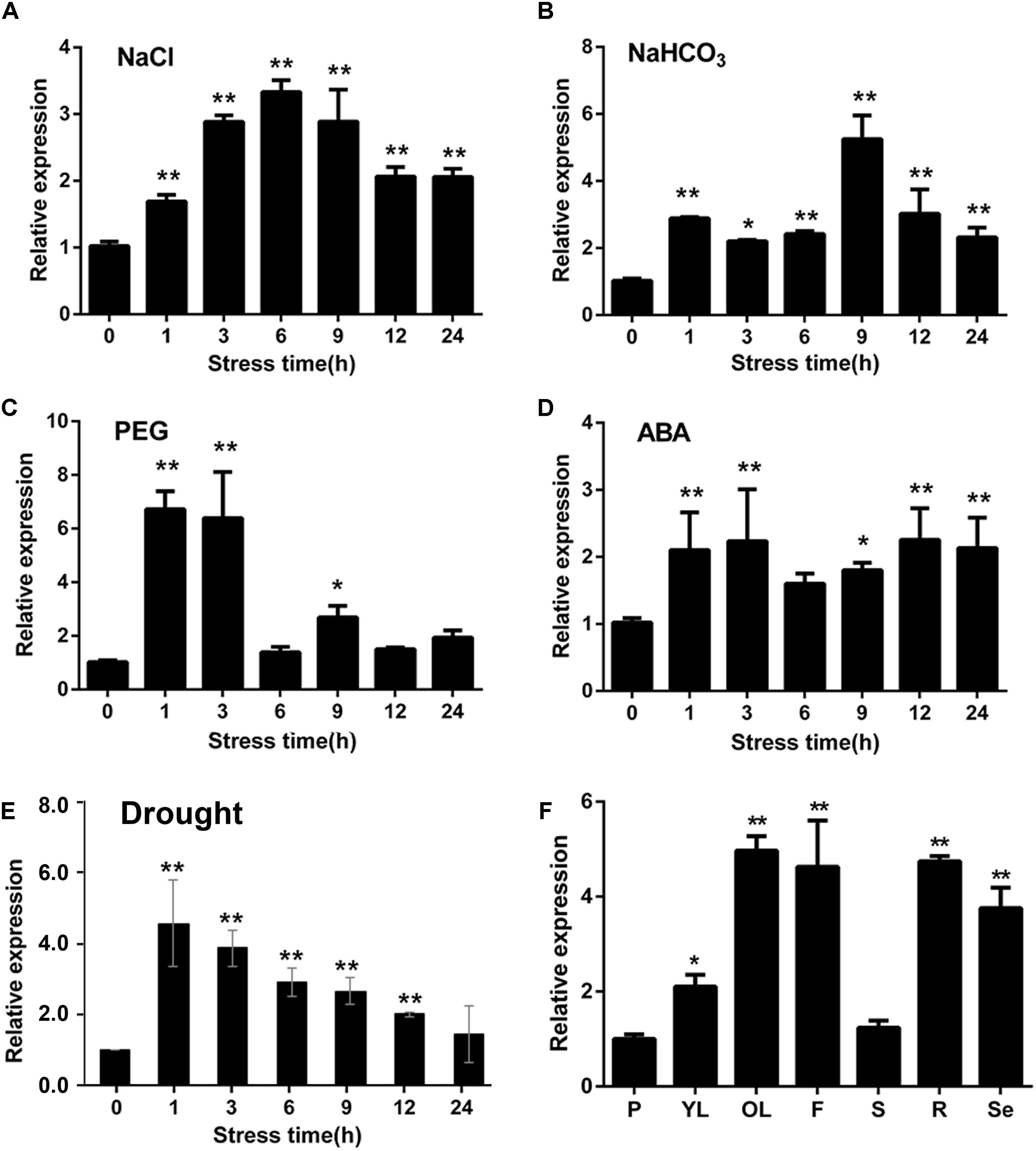
Figure 3. Expression patterns of GmWRKY16 under different abiotic stress conditions and in soybean tissues. (A–E) Patterns of GmWRKY16 expression under the different conditions of abiotic stresses. (B) Tissue expression pattern of GmWRKY16 in soybean. The soybean roots of 3-week-old seedlings were immersed in Hoagland solution saturated with 200 mM NaCl (A), 50 mM NaHCO3 (B), 20% PEG-6000 (C), and 100 μm ABA (D), respectively. Root samples were harvested at 0, 1, 3, 6, 9, 12, and 24 h after different treatments, respectively. The 1-week-old seedlings were dehydrated at room temperature (E), while the seedling samples were taken at treatment times of 0, 1, 3, 6, 9, 12, and 24 h, respectively. (F) Tissue samples were taken at different stages of soybean growth and development. GmWRKY16 transcript abundance was assessed by qRT-PCR using the 2-ΔΔCt method with the actin ACT3 gene as an internal control (Lü et al., 2015). The data are represented as the averages of three independent biological experiments ± SD, and asterisks indicate a significant difference (∗P = 0.05; ∗∗P = 0.01) compared with the corresponding controls. P, young pod; YL, young leaf; OL, old leaf; F, flower; S, stem; R, root; Se, seed.
To detect the tissue expression pattern of GmWRKY16, the leaf, stem, pod, flower, root, and seed samples were taken from the soybean cultivar HC2. The analysis of qRT-PCR indicated that GmWRKY16 was expressed constitutively in soybean with a more than fivefold expression level in old leaves compared to those in the stem and young pod (Figure 3F). In addition, GmWRKY16 has higher expression of more than 3.8-fold levels in soybean organs including flower, root, and seed (Figure 3F). These results suggest that GmWRKY16 may have some functions in soybean tissues and/or organs.
To investigate the tolerance to osmotic stress of GmWRKY16 transgenic lines after molecular identification (Supplementary Figure S1), different treatments of mannitol and NaCl were performed on 1/2 MS medium to determine the germination rates and root elongation of Arabidopsis (Figures 4, 5). Statistical results showed that seed germination of Arabidopsis was inhibited by mannitol stress (Figures 4A,B). As compared with the control (0 mM mannitol), the transgenic Arabidopsis lines of GmWRKY16 overexpression enhanced seed germination rates at 100, 200, and 300 mM of mannitol, respectively. The germination rates of GmWRKY16 transgenic lines were almost up to 100% under the mannitol treatments at the 3rd, 4th, and 5th days, respectively. The wild-type germination was severely suppressed, with lower germination rates compared to those of GmWRKY16 transgenic lines under treatments of 200 mM and 300 mM mannitol, respectively (Figures 4A,B). When Arabidopsis seeds were treated with NaCl, GmWRKY16 transgenic plants germinated faster than those of WT in the first 3 days. However, no significant differences in germination rate were found between WT and GmWRKY16 transgenic lines after 3 days under the treatments of 75, 100, and 150 mM NaCl, respectively (Figures 4C,D).
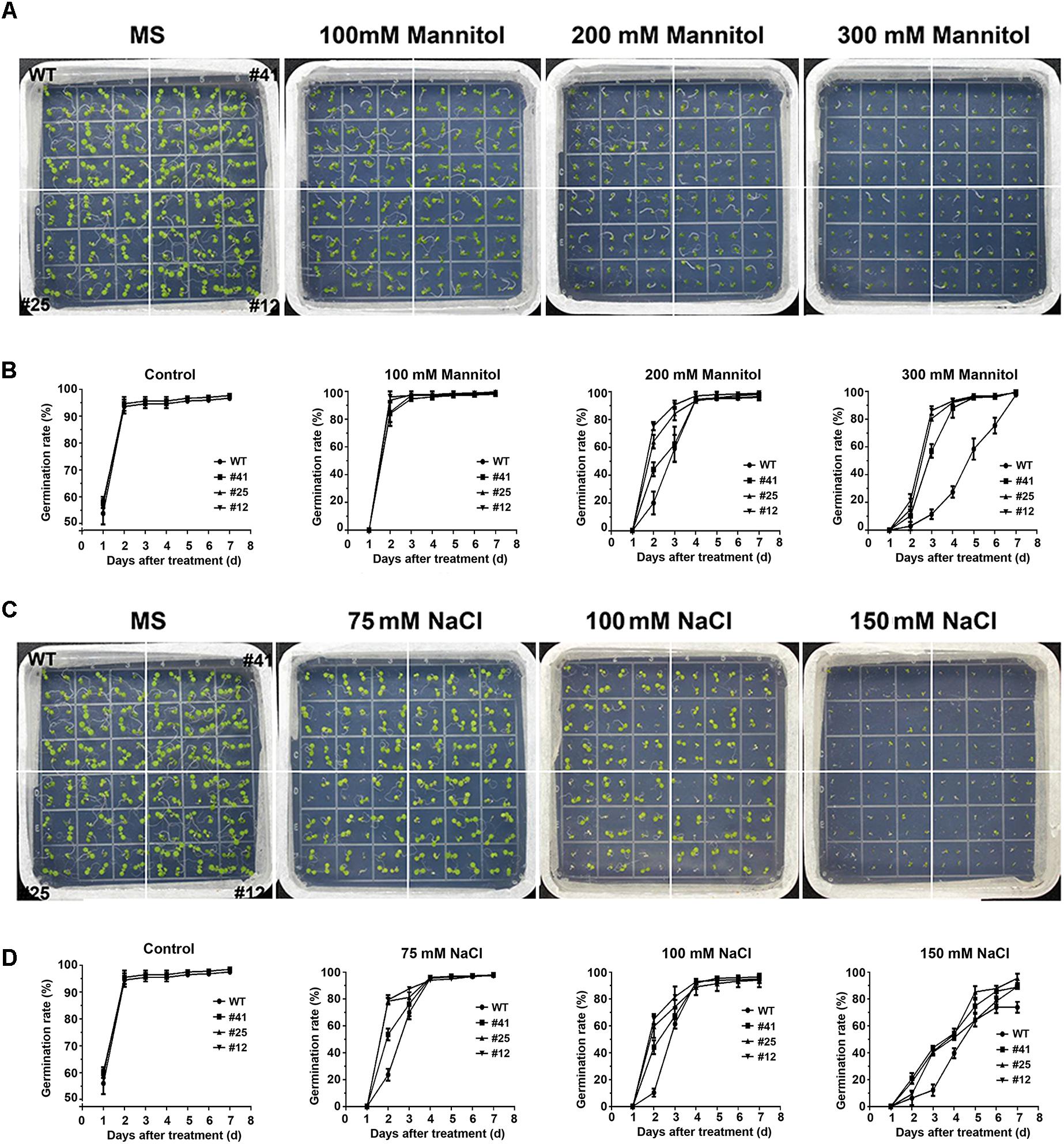
Figure 4. Overexpression of GmWRKY16 increased the germination rates of Arabidopsis under the conditions of osmotic stress. (A,B) Seed germination under the treatment of mannitol. (C,D) Seed germination under the treatment of salt. The surface-sterilized Arabidopsis seeds of wild-type and transgenic lines were sown on solid media of 1/2 MS containing mannitol (0, 100, 200, and 300 mM) and NaCl (0, 100, 150, and 200 mM). The status of seed germination was taken by photos after the treatment of osmotic stress for 4 and 7 days, respectively, while the germination rate of seeds was counted every day. Error bars represent ± SD. The observation values were the averages of three repetitions (n = 3). Three independent biological experiments were carried out to investigate the seed germination of WT and GmWRKY16 transgenic lines under osmotic stress. WT, wild type; #12, 25, 41: GmWRKY16 Arabidopsis transgenic lines of T3 generations.
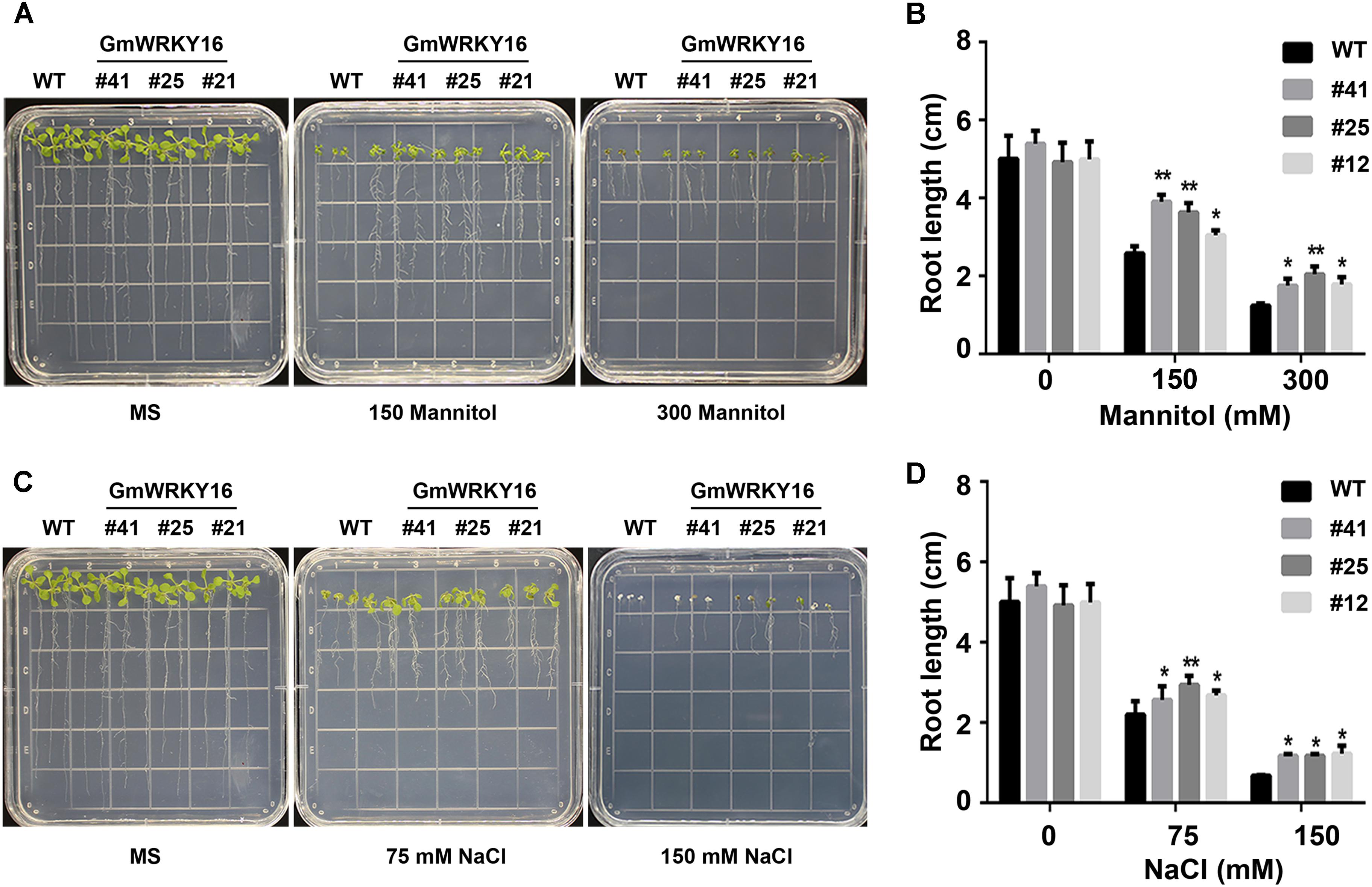
Figure 5. Overexpression of GmWRKY16 improved the tolerance of Arabidopsis seedlings to osmotic stress. (A,B) Seedlings under the treatment of mannitol. (C,D) Seedlings under the treatment of salt. The surface-sterilized Arabidopsis seeds of wild type and transgenic lines were sown on the solid media of 1/2 MS without mannitol and NaCl. The 3-day-old seedlings from 1/2 MS medium were transferred to the plates of 1/2 MS containing mannitol (0, 150, and 300 mM) and NaCl (0, 75, and 200 mM). The status of seedlings was taken by photos and root lengths were measured when Arabidopsis plants were treated under osmotic stress for 7 days. The vertical columns for the average observation value of the three repetitions represent the means ± SD. Three independent biological experiments were carried out to investigate the status of seedlings and root growth of WT and GmWRKY16 transgenic lines under osmotic stress. Asterisks indicate significant differences between WT and GmWRKY16 transgenic lines (∗P = 0.05; ∗∗P = 0.01). WT, wild type; #12, 25, 41: GmWRKY16 Arabidopsis transgenic lines of T3 generations.
The root lengths of Arabidopsis were inhibited by mannitol and NaCl stresses with the increase of the treatment concentrations (Figure 5). The root lengths of WT plants were 2.5 cm long under treatment of 150 mM mannitol, which was almost half the length of the control, while the root lengths of GmWRKY16 transgenic lines were more than 3 cm up to 4.1 cm long under treatment of 150 mM mannitol (Figures 5A,B). Under treatment of 300 mM mannitol, the root lengths of WT and of GmWRKY16 transgenic lines were inhibited to 1.5 cm and more than 2 cm long, respectively (Figures 5A,B). In the meantime, the inhibition of root elongation was also found under salt stress (Figures 5C,D). Compared to the control, the root lengths of WT were 2.2 and 0.7 cm long under the treatments of 75 and 150 mM NaCl, respectively, while the root lengths of GmWRKY16 transgenic lines were over 2.5 and 1.1 cm long under the corresponding treatments (Figures 5C,D). These results suggest that overexpression of GmWRKY16 enhances tolerance to osmotic stress.
To test the performance of the adult transgenic plants under NaCl stress, 21-day-old Arabidopsis plants were irrigated with a solution of 200 mM NaCl for 15 days. We observed that the leaves of the wild type were severely affected by salinity, whereas the leaves of GmWRKY16 transgenic lines were less affected (Figure 6A). It was found that 200 mM NaCl treatment induced proline accumulation both in wild-type plants and GmWRKY16 transgenic lines, with 65 and over 135 mg.g-1 of fresh weight, respectively (Figure 6B). In contrast, a greater MDA accumulation of 0.019 μmol.g-1 of fresh weight was found in WT compared to those of GmWRKY16 transgenic lines, with less than 0.013 μmol.g-1 of fresh weight under salt stress (Figure 6C). Meanwhile, ABA accumulation of over 130 μg.g-1 of fresh weight was determined in GmWRKY16 transgenic plants compared to those of WT, with less than 100 μg.g-1 of fresh weight (Figure 6D).
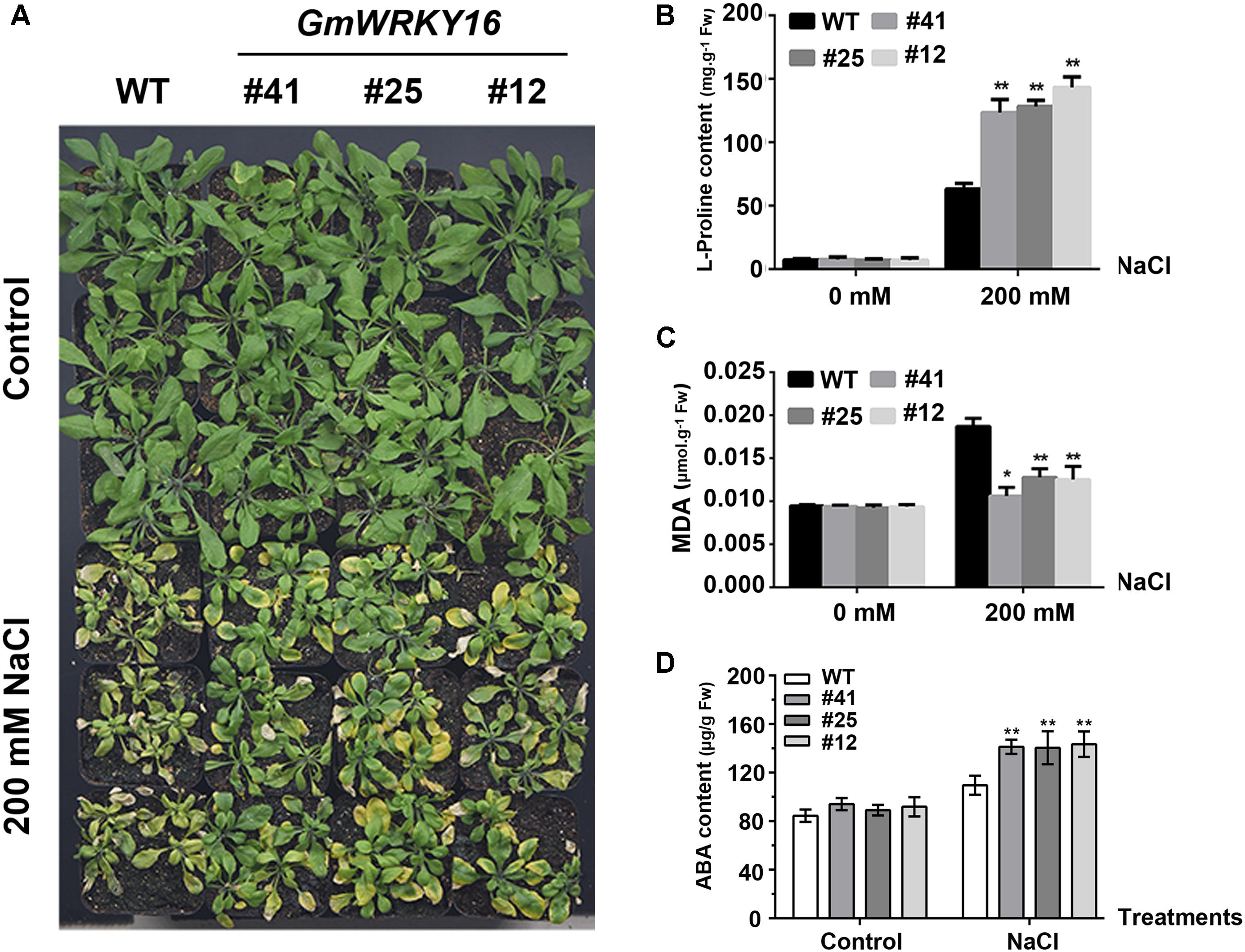
Figure 6. Overexpression of GmWRKY16 in Arabidopsis enhanced salt tolerance. (A) The phenotypes of GmWRKY16 transgenic lines tolerant to salt stress. (B) The determination of free proline content. (C) The determination of MDA content. (D) The determination of ABA content. Arabidopsis seeds of WT and GmWRKY16 transgenic lines of T3 generation were sown in mixed soil (vermiculite and flower nutrient soil, 1:1) and cultured in the chamber room. The 3-week-old Arabidopsis plants were then irrigated with a solution of 200 mM NaCl for 15 days to determinate the contents of free proline, MDA and ABA. The vertical columns for the average observation value of the three repetitions represent the means ± SD. Three independent biological experiments were carried out to investigate the status of seedlings and accumulations of proline, MDA and ABA in plants of WT and GmWRKY16 transgenic lines under salt stress. Asterisks indicate significant differences between WT and GmWRKY16 transgenic lines (∗P = 0.05; ∗∗P = 0.01). WT, wild type; #12, 25, 41: GmWRKY16 Arabidopsis transgenic lines of T3 generations.
To test plant response to drought stress, 21-day-old Arabidopsis plants under optimal irrigation conditions were treated without watering for 6, 8, or 10 days, respectively. The observation results showed that Arabidopsis plants were damaged by water shortage. As shown in Figure 7A, WT plants were severely damaged, with symptoms of leaf yellowing and crimping as well as a 60% survival rate after 10 days of drought treatment (Figure 7B), while GmWRKY16 transgenic plants grew well with an over 80% survival rate (Figure 7B). Furthermore, detached leaves of WT lost water much faster than those of GmWRKY16 transgenic lines under the dehydration treatment (Figure 7C). Meanwhile, ABA accumulation of over 150 μg.g-1 of fresh weight was determined in GmWRKY16 transgenic plants compared to those of WT, with less than 120 μg.g-1 of fresh weight (Figure 7D). Therefore, the data indicated that overexpression of GmWRKY16 enhanced Arabidopsis tolerance to drought stress.
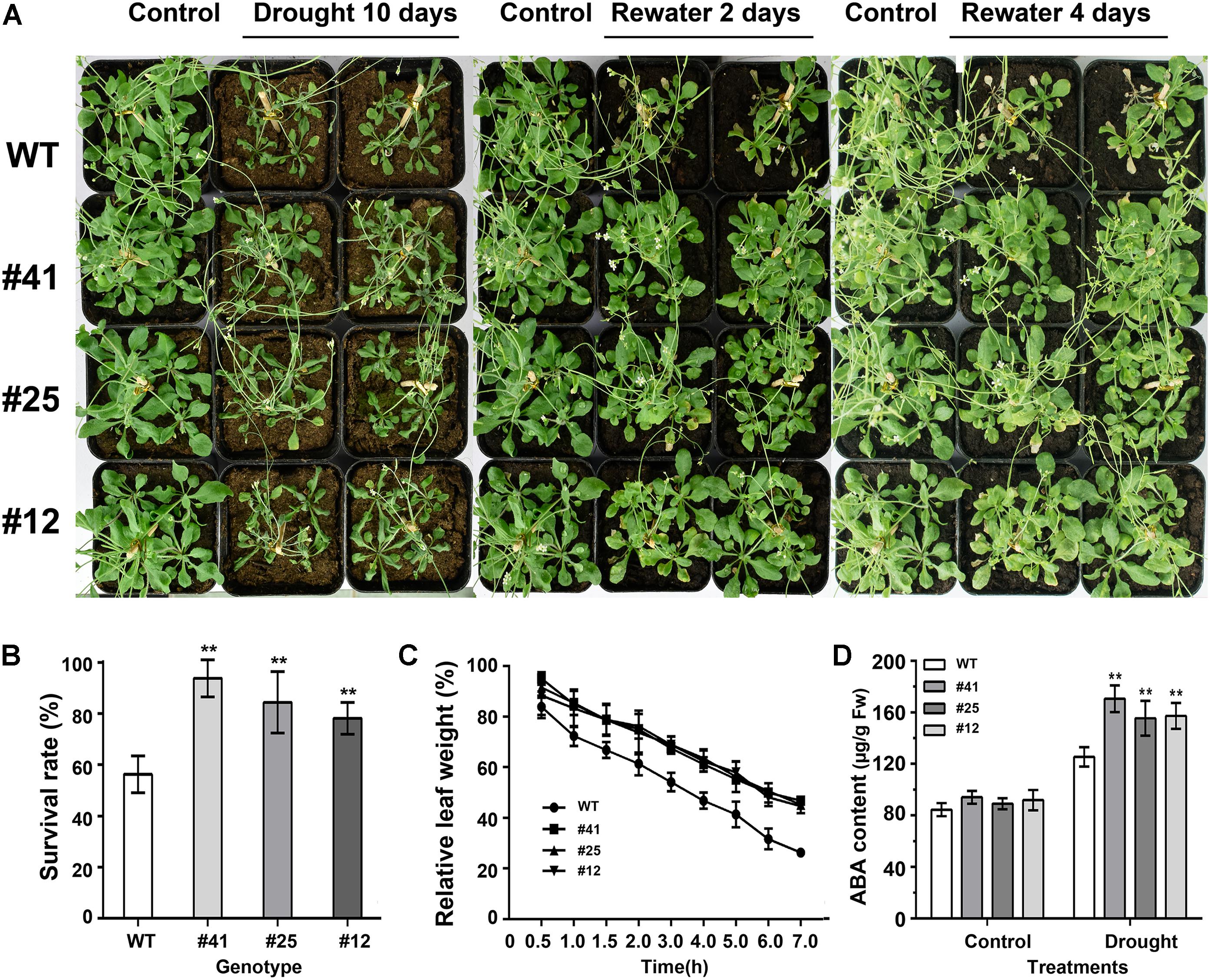
Figure 7. Overexpression of GmWRKY16 Arabidopsis conferred drought tolerance. (A) The phenotypes of GmWRKY16 transgenic lines tolerant to drought stress. (B) The survival rates of Arabidopsis plants. (C) The water loss of detached leaves. (D) The determination of ABA content. Arabidopsis seeds of WT and GmWRKY16 transgenic lines of T3 generation were sown in mixed soil (vermiculite and flower nutrient soil, 1:1) and cultured in the chamber room. The 3-week-old Arabidopsis plants were subjected to water shortage for 5, 7, and 9 days, respectively. All the drought-treated plants were then rehydrated after they were dried for 5 days. The fresh weight was measured at the set intervals of drought stress. The vertical columns for the average observation value of the three repetitions represent the means ± SD. Three independent biological experiments were carried out to investigate the status of seedlings, the survival rate and water loss of the detached leaves of WT and GmWRKY16 transgenic lines under drought stress. Asterisks indicate the significant differences between WT and GmWRKY16 transgenic lines (∗P = 0.05; ∗∗P = 0.01). WT: wild type; #12, 25, 41: GmWRKY16 Arabidopsis transgenic lines of T3 generations.
To study GmWRKY16 sensitivity to ABA, different treatments of ABA were performed on 1/2 MS medium to determine the germination rates and root elongation of Arabidopsis (Figure 8). The statistical results showed that seed germination of Arabidopsis was inhibited by ABA stress with increasing concentrations (Figures 8A,B). When compared to the control (0 μM ABA), the GmWRKY16 transgenic lines enhanced seed germination rates under ABA treatments of 0.5, 1.0, and 1.5 mM, respectively. The germination rates of GmWRKY16 transgenic lines were almost up to 100% under the ABA treatments at the 6th day. Meanwhile, the germination rates of the wild type were less than 90, 80, and 70% with increasing concentrations under ABA treatments (Figures 8A,B).
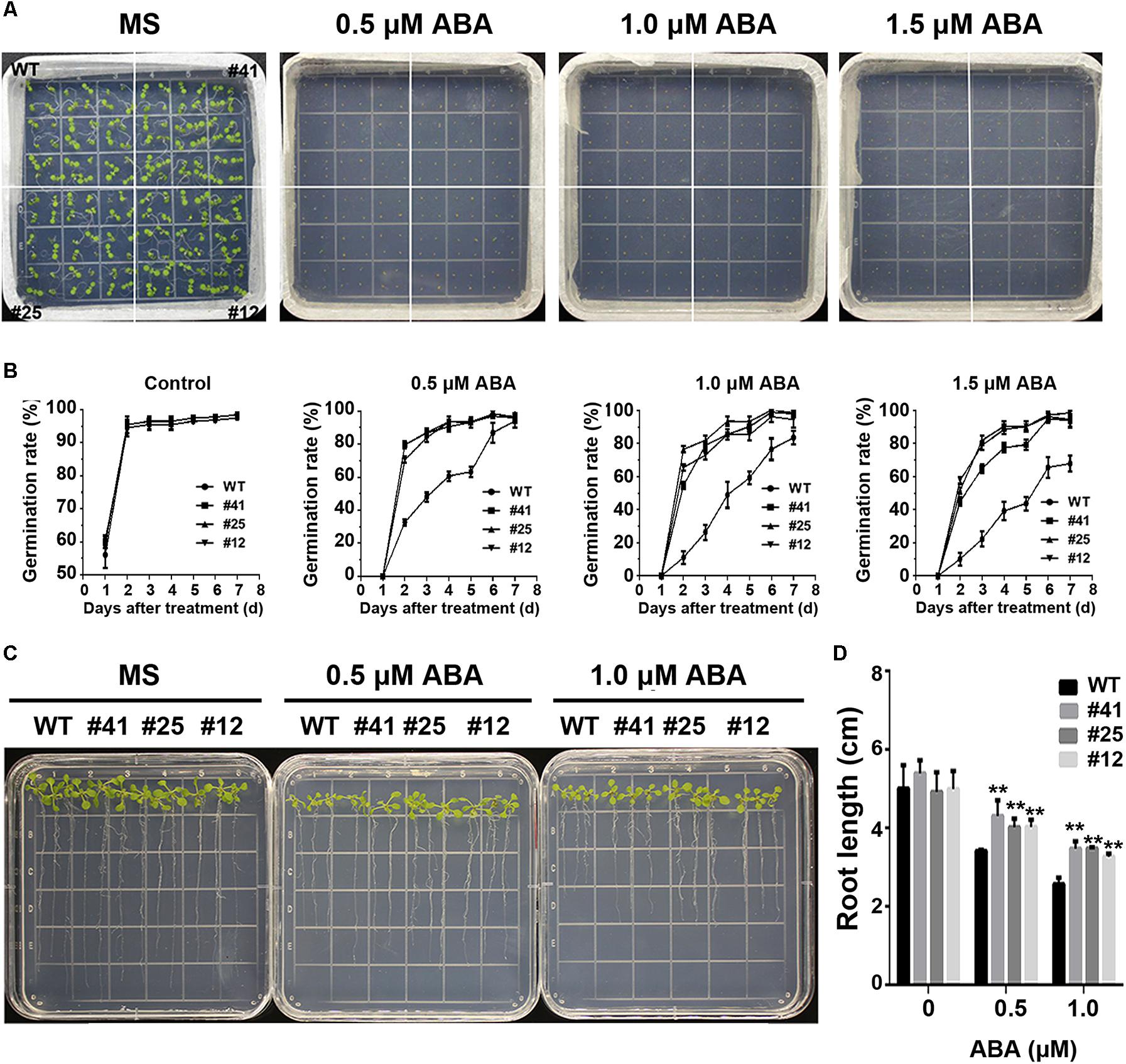
Figure 8. Overexpression of GmWRKY16 in Arabidopsis confers the tolerance to ABA. (A,B) Seed germination under ABA treatment. (C,D) Seedlings under ABA treatment. The surface-sterilized Arabidopsis seeds of wild-type and transgenic lines were sown on the solid media of 1/2 MS containing ABA (0, 0.5, 1.0, and 1.5 μM). The photos were taken for the status of seed germination after the ABA treatment for 4 and 7 days, respectively. The 3-day-old seedlings from 1/2 MS medium were transferred to the plates of 1/2 MS containing ABA (0, 0.5, and 1.0 μM). The root lengths were measured when Arabidopsis plants were treated under the ABA stress for 7 days. Error bars represent ± SD. The observation values were the averages of three repetitions. Three independent biological experiments were carried out to investigate the seed germination and root growth of WT and GmWRKY16 transgenic lines under ABA stress. WT, wild type; #12, 25, 41: GmWRKY16 Arabidopsis transgenic lines of T3 generations.
The root lengths of Arabidopsis were also inhibited by ABA stress (Figures 8C,D). The root lengths of WT plants were 3.5 cm long under the treatment of 0.5 μM ABA, or around 1.8 cm shorter than that under the control, while the root lengths of GmWRKY16 transgenic lines were over 4.1 cm long under the treatment of 0.5 μM ABA (Figures 8C,D). Under the treatment of 1 μM ABA, the root elongations of WT and GmWRKY16 transgenic lines were also inhibited with 2.8 cm and over 3.2 cm long, respectively (Figures 8C,D). These results suggest that overexpression of GmWRKY16 enhances the Arabidopsis tolerance to ABA stress.
To further investigate the pathway regulated by GmWRKY16 under abiotic stress, expression patterns of stress/ABA responsive genes were performed by qRT-PCR. Two genes of KIN1 and RD29A were induced with much higher expression levels, over 220- and 130-fold greater under the NaCl treatment compared with those of the control, respectively (Figure 9A). Meanwhile, AtWRKY8 was induced with an expression level of over 12-fold greater under the NaCl treatment compared to that of the control (Figure 9A).
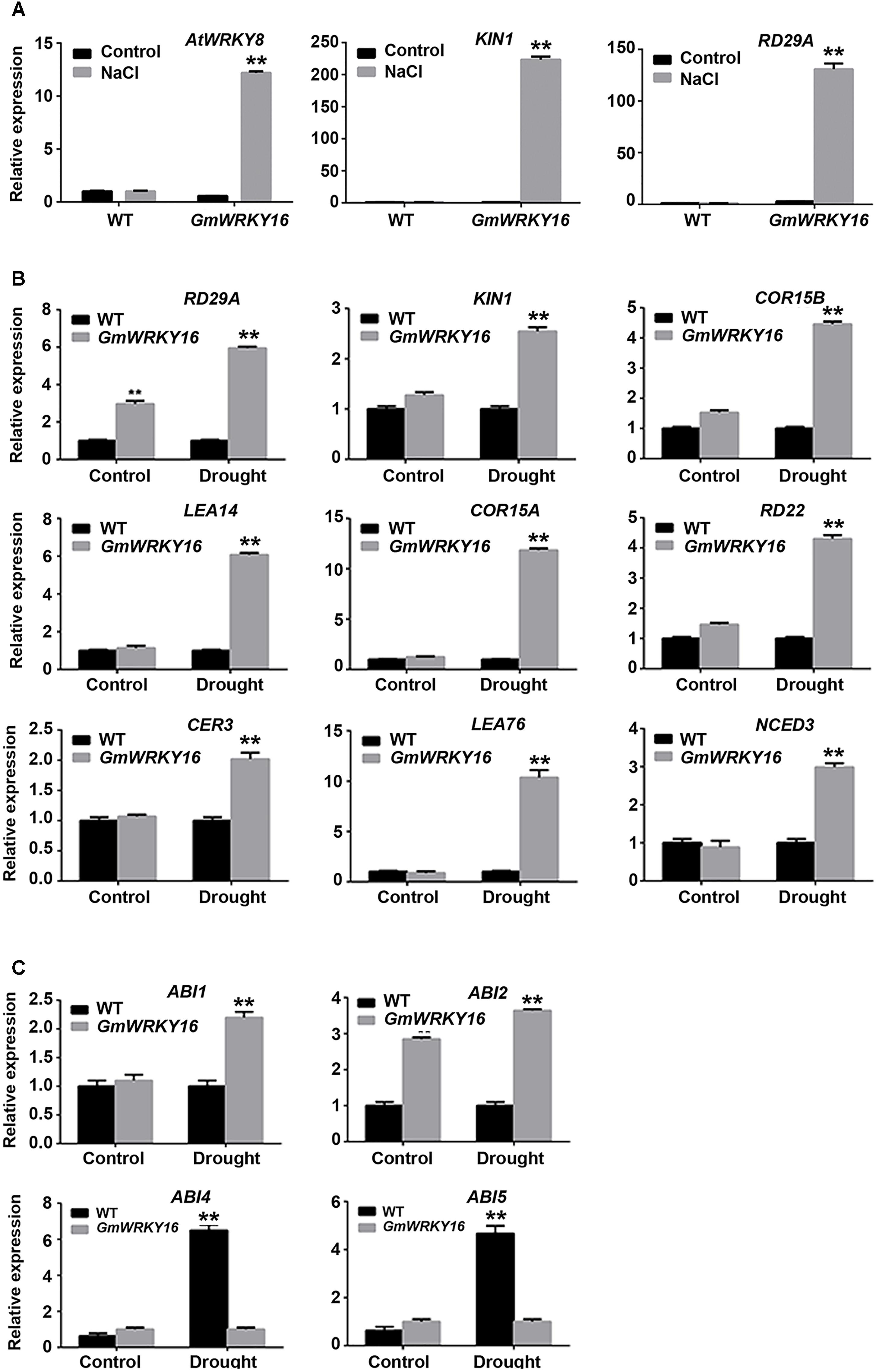
Figure 9. Expression patterns of stress/ABA responsive genes regulated by GmWRKY16. (A) Expression patterns of genes responsive to salt stress. (B) Expression patterns of ABA and/or stress-responsive genes under drought stress. (C) Expression patterns of ABA-responsive genes under drought stress. Arabidopsis seeds of WT and GmWRKY16 transgenic line #12 of T3 generation were sown in mixed soil (vermiculite and flower nutrient soil, 1:1) and cultured in the chamber room. The 2-week-old seedlings were subjected to drought treatment (withholding water) and 200 mM NaCl treatment for 10 days. The samples from the aerial parts of Arabidopsis plants were taken for total RNA extraction. The relative expression of drought- and/or salt stress-responsive genes was quantified by qRT-PCR using ACT3 as the reference gene to normalize the data (∗∗P = 0.01). The 2-ΔΔCt method was used to evaluate the quantitative variation between the examined replicates (Lü et al., 2015). The details for the specific primers of the GmWRKY16, ACT3 and stress-responsive genes are listed in Supplementary Table S1.
Under drought stress, the expressions of COR15A and LEA76 were upregulated in GmWRKY16 transgenic lines that were increased over 10-fold as compared to those in the wild type (Figure 9B). The expressions of LEA14, RD29A, KINI, COR15B, RD22, CER3, and NCED3 were upregulated in GmWRKY16 transgenic lines by less than sixfold as compared to those of wild type (Figure 9B). Among all the detected genes, RD29A was induced by GmWRKY16 in transgenic lines with about a sixfold increased expression level in comparison to that of the wild type (Figure 9B). ABI2 was induced by GmWRKY16 in transgenic lines, with 3.6- and 2.8-fold increased expression levels under the drought treatment and the control, respectively. However, ABI1 was induced by GmWRKY16 in transgenic lines with a 2.2-fold increased expression level under the drought treatment (Figure 9C). In contrast, two drought-induced genes of ABI4 and ABI5 were inhibited with much lower expression levels under the treatment of drought stress (Figure 9C). All the results suggest that GmWRKY16 may enhance Arabidopsis resistance to salt and drought stresses through ABA and/or other pathways.
The WRKY TFs constitute one of the biggest transcription factor families, which play multiple roles in plants. Since the SPF1 gene was firstly discovered in sweet potato (Ishiguro and Nakamura, 1994), some WRKY family genes have been investigated for their functional roles and mechanisms involved particularly in biotic and abiotic stresses (Dai et al., 2016; Kiranmai et al., 2018). However, little is known about the WRKYs functional roles and their mechanisms in soybean. In this study, GmWRKY16 encoding a TF protein of the soybean WRKY family was cloned to determine its potential role in soybean resistance to salt and drought stresses (Figures 6, 7). The phylogenetic tree analysis of GmWRKY16 and other WRKY proteins from soybean, rice and Arabidopsis showed that GmWRKY16 protein in subgroup IIc had more similarity to GmWRKY129 and other WRKY proteins such as AtWRKY8, AtWRKY28, AtWRKY48, AtWRKY71 (Figure 1B). Previous reports indicated that AtWRKY8 enhanced the tolerance of Arabidopsis to salinity stress through modulating its interacting partner VQ9 protein (Hu et al., 2013), that AtWRKY28 conferred resistance to abiotic stress in Arabidopsis by co-expression of AtbHLH17 (Babitha et al., 2013), and that AtWRKY71 hastened flowering in Arabidopsis thaliana in the presence of salt stress (Yu et al., 2018). Furthermore, the multiple alignment of amino acid sequence indicated that GmWRKY16 protein had a conserved WRKY motif and a zinc-finger motif at the central region with characteristics similar to other five WRKY proteins (Figure 1A). The analysis of the biochemistry characteristics of GmWRKY16 protein indicated that GmWRKY16 localizing to the nucleus could activate the expression of GFP in leaf epidermal cells of tobacco and held the transcriptional activation in yeast cells (Figure 2). Therefore, the results above suggest that GmWRKY16 protein may regulate various functions as a WRKY transcription factor involved in abiotic stress and/or development in plants.
Previous studies revealed that WRKY TFs were widely investigated for key roles in abiotic stress responses to drought and salt stresses (Zhou et al., 2008; Wang et al., 2015; Liu et al., 2016). In the present study, the WRKY gene GmWRKY16 in soybean was quickly induced to the highest expression levels at 3–6 h after treatments of NaCl, PEG, drought, and ABA, respectively (Figure 3). Overexpression of GmWRKY16 in Arabidopsis promoted seed germination and root elongation of seedlings under NaCl and mannitol treatments (Figures 4, 5), and enhanced the tolerance of transgenic plants to salt and drought stresses (Figures 6, 7). Similar resistant phenotypes to abiotic stress were also found in other WRKY TF genes in soybean. For example, GmWRKY54 enhanced the tolerance of transgenic Arabidopsis plants to salt and drought stresses through the regulation of DREB2A and STZ/Zat10 (Zhou et al., 2008). Furthermore, GmWRKY27 was found to improve tolerance to salt and drought stress in soybean plants by interacting with GmMYB174 (Wang et al., 2015). Recent research indicates that abiotic stresses of drought and salt impose osmotic stresses, which result in the enhanced production of ROS that cause lipid peroxidation, protein oxidation, nucleic acid damage, activation of programmed cell death and then cell death. Meanwhile, it was found that transgenic plants enhanced tolerance to drought as well as osmotic and salt stress through the scavenging of ROS (Foyer and Noctor, 2005; Sharma et al., 2012). The MDA level was an indicator of ROS destructive effects under stress conditions (Sharma et al., 2012), and the content of H2O2 and MDA in overexpressing transgenic plants were significantly reduced under salt and osmotic stresses (Sairam et al., 1998; Ullah et al., 2018). Proline as an osmolyte plays important roles in stabilizing macromolecules and membranes in cells by a higher accumulation in transgenetic plants under salt and osmotic stress (Wang et al., 2003; Mahajan and Tuteja, 2005; Couée et al., 2006; Cao et al., 2009; Ullah et al., 2018). The contents of proline and MDA in GmWRKY16 transgenic lines and the wild type were also determined to detect the response of Arabidopsis to drought and salt stresses (Figures 6, 7), and the results were consistent with those of previous reports (Ding et al., 2014; Wang et al., 2015; Yu L. et al., 2016; Ullah et al., 2018), suggesting that overexpression of GmWRKY16 may enhance the tolerance of drought and salinity by the accumulation of proline and the reduction of MDA in transgenic plants.
Recently, mores studies have indicated that WRKY TFs play vital roles in various responses to abiotic stresses in plants with the potential tolerance mechanisms related to ABA signaling (Zhou et al., 2008; Jiang et al., 2012; Luo et al., 2013; Sun et al., 2015; Cai et al., 2017). Some physiological processes such as seed germination, seedling growth and plant development under various stresses may be inhibited by the accumulation of ABA induced through the ABA-dependent signaling pathway (Xiong et al., 2006; Wang et al., 2014; Wu M. et al., 2017). Overexpression of GmWRKY16 resulted in better seed germination and longer roots of seedlings under the stress treatments of NaCl, mannitol and exogenous ABA compared to those of the wild type (Figures 4, 5, 8). Moreover, the transgenic Arabidopsis plants exhibited a distinct resistance to salt and drought stresses with higher accumulations of ABA in comparison with the WT plants (Figures 6, 7). Furthermore, overexpression of GmWRKY16 caused a reduction of stomatal aperture under ABA treatments (Supplementary Figure S2) and decreased water loss rate under drought treatment (Figure 7). There are some reports about ABA role in tolerance to stress. For example, it was found that stoma controlling transpiration water loss in plants is an important factor related to plant resistance to drought stress through changes in ABA level (Holbrook et al., 2002); Luo et al. (2013) found that overexpression of GsWRKY20 exhibited great tolerance of transgenic Arabidopsis plants to drought stress by decreasing stomatal density under ABA treatment (Luo et al., 2013); and the cotton WRKY transcription factor GhWRKY17 regulated the increased sensitivity of transgenic nicotiana plants to drought stress by reducing the ABA level (Yan et al., 2014). All these results suggest that GmWRKY16 may enhance plant tolerance to salt, osmotic and drought stresses through positive regulation of ABA pathways. Similar observations were found in other plant species (Zhou et al., 2008; Wu M. et al., 2017; Ullah et al., 2018; Wang et al., 2018).
Some studies have demonstrated that several WRKY TF genes enhanced tolerance to salt and drought stresses through expression regulation of stress/ABA-responsive genes (Yoshida et al., 2014; He et al., 2016; Jaffar et al., 2016; Ullah et al., 2018). To gain insight into the molecular function basis of GsWRKY16 in salt and drought stress responses, we detected several well-characterized marker genes in transgenic lines (Figure 9). The qRT-PCR results of gene expression showed that AtWRKY8 and two stress-related marker genes of KIN1 and RD29A were significantly upregulated in transgenic Arabidopsis plants under salt treatment (Figure 9A). Previous reports indicated that AtWRKY8 directly bound the promoter of RD29A to modulate salinity stress tolerance (Hu et al., 2013), suggesting that GmWRKY16 may function in an ABA-independent pathway. NCED3 (9-cis epoxycarotenoid dioxygenase 3) encoding a key and rate-limiting enzyme was supposed to participate in ABA biosynthesis to regulate the drought-stress response (Qin and Jan, 1999; Thompson et al., 2000; Seki et al., 2003). Under drought conditions, NCED3 was determined in GmWRKY16 transgenic plants with higher levels than those in the WT plants as a result of ABA accumulation (Figures 7D, 9B). Similar results were also found in other WRKY genes, such as AtWRKY57, which conferred drought tolerance of Arabidopsis by elevating ABA levels induced by NCED3 (Jiang et al., 2012). These results suggest that GmWRKY16 may play a role in stress-induced ABA-biosynthesis along with additional factors. The ABI1 and ABI2 genes encode two serine/threonine phosphatases 2C (PP2C) proteins that acted as negative regulators of ABA response (Merlot et al., 2001), and the ABI4 and ABI5 genes are the best characterized positive regulators of ABA signaling (Reeves et al., 2011; Rushton et al., 2012). In this study, ABI1 and ABI2 were upregulated by GmWRKY16, whereas ABI4 and ABI5 were downregulated in transgenic lines under drought stress (Figure 9C). Previous reports indicated that there was no positive correlation between the gene expression of the ABA signaling pathway and the stress tolerance of transgenic plants to abiotic stresses. Several reports showed that some transgenic plants increased stress tolerance with downregulation of ABI1/2, while other transgenic plants decreased stress tolerance with upregulation of ABI1/2 (Jung et al., 2007; Seo et al., 2010). However, contrary phenotypes of drought tolerance by regulating ABI1/2 have also been reported over the past several years. For example, overexpression of GmbZIP44, GmbZIP62, or GmbZIP78 enhanced more tolerance of transgenic Arabidopsis plants to abiotic stress, which may play an important role in ABA signaling by upregulation of ABI1 and ABI2 (Liao et al., 2008). Overexpression of GsWRKY20 in Arabidopsis increased the tolerance of transgenic plants to drought stress by regulating the expression of some key ABA signaling regulators with upregulation of ABI1/2 and downregulation of ABI4/5 (Luo et al., 2013). However, ZmPP2C transgenic plants decrease tolerance to salt and drought stresses with lower expression of ABI1 and ABI2 (Liu et al., 2009). Furthermore, ABREs were detected in a number of ABA-responsive genes (RD29, COR15, and RD22) in several plant species (Kim, 2005). A group of LEA genes, which play important roles in dehydration tolerance (Ingram and Bartels, 1996), was also investigated. In the present study, overexpression of GmWRKY16 promoted drought tolerance of transgenic plants through regulating expression levels of these stress-related marker genes including RD29A, KINI, COR15A, COR15B, RD22, CER3, LEA14, and LEA76 (Figure 9B). All of these results suggest that overexpression of GmWRKY16 in Arabidopsis improves the tolerance of transgenic plants to drought and salt stresses through inducing ABA synthesis and regulating the expression levels of ABA signaling and/or stress-related genes.
In summary, we identified a soybean WRKY transcription factor, GmWRKY16, which is a multiple stress-inducible gene induced by salt drought and ABA stresses and enriched in the old leaves, flowers, seeds, and roots of soybean. GmWRKY16 overexpression enhanced the resistance of transgenic plants to drought and salt stresses, and less sensitivity to ABA during the processes of seed germination and seedling root growth. The analysis of molecular mechanisms revealed that enhanced tolerance to drought and salt might result in comprehensive roles in upregulating and/or downregulating transcripts of the stress- and/or ABA-responsive genes with ABA and proline accumulation, and MDA decrease. Therefore, these results suggest that GmWRKY16 may increase the resistance to drought and salt stresses through ABA-mediated and/or independent pathways.
QM and HN conceived of and designed the study. ZX, QM, ZC, LL, and JL conducted the experiments. ZX, QM, and YC performed data as well as statistical analysis. QM and ZX prepared the manuscript. All authors read and approved the final manuscript.
This work was supported by the grants from the Projects of Science and Technology of Guangzhou (201804020015), the National Natural Science Foundation of China (31771816 and 31371642), the National Key R&D Program of China (2017YFD0101500), the New Varieties Cultivation of Genetically Modified Organisms (2016ZX08004002-007), the China Agricultural Research System (CARS-04-PS09), and the Research Project of the State Key Laboratory of Agricultural and Biological Resources Protection and Utilization in Subtropics (4100-31331).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Bo Zhang (Assistant Professor, Virginia Tech, United States) for her critical comments and language improvement of the present manuscript. We are very grateful to Prof. Yaoguang Liu (South China Agricultural University, China) for kindly providing us with the pCAMBIA1302 vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01979/full#supplementary-material
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8
Bai, Y., Sunarti, S., Kissoudis, C., Visser, R. G. F., and van der Linden, C. G. (2018). The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9:801. doi: 10.3389/fpls.2018.00801
Bakshi, M., and Oelmüller, R. (2014). WRKY transcription factors. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Bencke-Malato, M., Cabreira, C., Wiebke-Strohm, B., Bucker-Neto, L., Mancini, E., and Osorio, M. B. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. doi: 10.1186/s12870-014-0236-0
Bencke-Malato, M., Cabreira, C., Wiebke-Strohm, B., Bücker-Neto, L., Mancini, E., and Osorio, M. B. (2016). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. Phys. Chem. Chem. Phys. 18, 6607–6617. doi: 10.1186/s12870-014-0236-0
Cai, R., Dai, W., Zhang, C., Wang, Y., Wu, M., Zhao, Y., et al. (2017). The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 246, 1215–1231. doi: 10.1007/s00425-017-2766-9
Cao, Y., Wei, Q., Liao, Y., Song, H., Li, X., Xiang, C., et al. (2009). Ectopic overexpression of AtHDG11 in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep. 28, 579–588. doi: 10.1007/s00299-008-0659-x
Chen, C., Sun, X., Duanmu, H., Zhu, D., Yu, Y., Cao, L., et al. (2015). GsCML27, a gene encoding a calcium-binding EF-hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses. PLoS One 10:e141888. doi: 10.1371/journal.pone.0141888
Chen, J., Nolan, T., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell 29, 425–1439. doi: 10.1105/tpc.17.00364
Chu, X., Wang, C., Chen, X., Lu, W., Li, H., Wang, X., et al. (2016). Correction: the cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 11:e157026. doi: 10.1371/journal.pone.0157026
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Couée, I., Sulmon, C., Gouesbet, G., and El Amrani, A. (2006). Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57, 449–459. doi: 10.1093/jxb/erj027
Dai, X., Wang, Y., and Zhang, W. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Dias, L. P., de Oliveira-Busatto, L. A., and Bodanese-Zanettini, M. H. (2016). The differential expression of soybean [Glycine max (L.) Merrill] WRKY genes in response to water deficit. Plant Physiol. Biochem. 107, 288–300. doi: 10.1016/j.plaphy.2016.06.018
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fan, Q., Song, A., Jiang, J., Zhang, T., Sun, H., Wang, Y., et al. (2016). CmWRKY1 enhances the dehydration tolerance of chrysanthemum through the regulation of ABA-associated genes. PLoS One 11:e150572. doi: 10.1371/journal.pone.0150572
Finatto, T., Viana, V. E., Woyann, L. G., Busanello, C., Maia, L. C. D., and Oliveira, A. C. D. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 41, 533–544. doi: 10.1590/1678-4685-GMB-2017-0232
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Fujii, H., and Zhu, J. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8335. doi: 10.1073/pnas.0903144106
Goel, R., Pandey, A., Trivedi, P. K., and Asif, M. H. (2016). Genome-wide analysis of the musa WRKY gene family: evolution and differential expression during development and stress. Front. Plant Sci. 7:299. doi: 10.3389/fpls.2016.00299
Grunewald, W., Smet, D. I., Rybel, D. B., Robert, H. S., Cotte, V. D. B., Willemsen, V. A., et al. (2013). Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep. 14, 1136–1142. doi: 10.1038/embor.2013.169
He, G., Xu, J., Wang, Y., Liu, J., Li, P., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16:116. doi: 10.1186/s12870-016-0806-4
He, L., Wu, Y., Zhao, Q., Wang, B., Liu, Q., and Zhang, L. (2018). Chrysanthemum DgWRKY2 gene enhances tolerance to salt stress in transgenic Chrysanthemum. Int. J. Mol. Sci. 19:2062. doi: 10.3390/ijms19072062
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hennig, L. (2012). Plant gene regulation in response to abiotic stress. Biochim. Biophys. Acta 1819:85. doi: 10.1016/j.bbagrm.2012.01.005
Holbrook, N. M., Shashidhar, V., James, R. A., and Rana, M. (2002). Stomatal control in tomato with ABA-deficient roots: response of grafted plants to soil drying. J. Exp. Bot. 373, 1503–1514.
Hu, Y., Chen, L., Wang, H., Zhang, L., Wang, F., and Yu, D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47:377. doi: 10.1146/annurev.arplant.47.1.377
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jaffar, M., Song, A., Faheem, M., Chen, S., Jiang, J., Liu, C., et al. (2016). Involvement of CmWRKY10 in drought tolerance of Chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 17:693. doi: 10.3390/ijms17050693
Jiang, Y., Liang, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Molecular Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2007). Overexpression of AtMYB44 enhances tomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kim, S. Y. (2005). The role of ABF family bZIP class transcription factors in stress response. Physiol. Plantarum 126, 519–527. doi: 10.1111/j.1399-3054.2005.00601.x
Kiranmai, K., Lokanadha Rao, G., Pandurangaiah, M., Nareshkumar, A., Amaranatha Reddy, V., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9:346. doi: 10.3389/fpls.2018.00346
Kokkirala, V. R., Yonggang, P., Abbagani, S., Zhu, Z., and Umate, P. (2010). Subcellular localization of proteins of Oryza sativa L. in the model tobacco and tomato plants. Plant Signal. Behav. 5, 1336–1341. doi: 10.4161/psb.5.11.13318
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, J. B., Luan, Y. S., and Liu, Z. (2015). Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 155, 248–266. doi: 10.1111/ppl.12315
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Liang, Q., Wu, Y., Wang, K., Bai, Z., Liu, Q., Pan, Y., et al. (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 7:4799. doi: 10.1038/s41598-017-05170-x
Liao, Y., Zou, H., Wei, W., Hao, Y., Tian, A., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240. doi: 10.1007/s00425-008-0731-3
Liu, C., Wu, Y., and Wang, X. (2012). bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235, 1157–1169. doi: 10.1007/s00425-011-1564-z
Liu, L., Hu, X., Song, J., Zong, X., Li, D., and Li, D. (2009). Over-expression of a Zea mays L. protein phosphatase 2C gene (ZmPP2C) in Arabidopsis thaliana decreases tolerance to salt and drought. J. Plant Physiol. 166, 531–542. doi: 10.1016/j.jplph.2008.07.008
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., and Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253, 1265–1281. doi: 10.1007/s00709-015-0885-3
Lü, J., Suo, H., Yi, R., Ma, Q., and Nian, H. (2015). Glyma11g13220, a homolog of the vernalization pathway gene VERNALIZATION 1 from soybean [Glycine max (L.) Merr.], promotes flowering in Arabidopsis thaliana. BMC Plant Biol. 15:232. doi: 10.1186/s12870-015-0602-6
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Ma, J., Gao, X., Liu, Q., Shao, Y., Zhang, D., Jiang, L., et al. (2017). Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 8:2036. doi: 10.3389/fpls.2017.02036
Ma, Q., Yi, R., Li, L., Liang, Z., Zeng, T., Zhang, Y., et al. (2018). GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in Arabidopsis thaliana. BMC Plant Biol. 18:212. doi: 10.1186/s12870-018-1397-z
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Meng, D., Li, Y., Bai, Y., Li, M., and Cheng, L. (2016). Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 103, 71–83. doi: 10.1016/j.plaphy.2016.02.006
Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25, 295–303. doi: 10.1046/j.1365-313x.2001.00965.x
Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. doi: 10.1105/tpc.106.042705
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Niu, C., Wei, W., Zhou, Q., Tian, A., Hao, Y., Zhang, W., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Pillai, S. E., Kumar, C., Patel, H. K., and Sonti, R. V. (2018). Overexpression of a cell wall damage induced transcription factor, OsWRKY42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BMC Plant Biol. 18:177. doi: 10.1186/s12870-018-1391-5
Qin, X., and Jan, A. D. Z. (1999). The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. U.S.A. 96, 15354–15361. doi: 10.1073/pnas.96.26.15354
Reeves, W. M., Lynch, T. J., Mobin, R., and Finkelstein, R. R. (2011). Direct targets of the transcription factors ABA-insensitive (ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 75, 347–363. doi: 10.1007/s11103-011-9733-9
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Rinerson, C. I., Scully, E. D., Palmer, N. A., Donze-Reiner, T., Rabara, R. C., Tripathi, P., et al. (2015). The WRKY transcription factor family and senescence in switchgrass. BMC Genomics 16:912. doi: 10.1186/s12864-015-2057-4
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sairam, R., Deshmukh, P., and Saxena, D. (1998). Role of antioxidant systems in wheat genotypes tolerance to water stress. Biol. Plant. 3, 387–394. doi: 10.1007/s11356-015-5452-0
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., and Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Schluttenhofer, C., and Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Seki, M., Kamei, A., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2003). Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr. Opin. Biotechnol. 14, 194–199. doi: 10.1016/S0958-1669(03)00030-2
Seo, D. H., Ryu, M. Y., Jammes, F., Hwang, J. H., Turek, M., Kang, B. G., et al. (2012). Roles of four Arabidopsis U-Box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160, 556–568. doi: 10.1104/pp.112.202143
Seo, Y., Park, J., Cho, Y., Jung, C., Seo, H., Park, S., et al. (2010). Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cells 3, 271–277. doi: 10.1007/s10059-010-0114-z
Shang, Y., Yan, L., Liu, Z., Cao, Z., Mei, C., Xin, Q., et al. (2012). The Mg-Chelatase H subunit ofarabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:217037. doi: 10.1155/2012/217037
Shen, H., Liu, C., Zhang, Y., Meng, X., Zhou, X., Chu, C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80, 241–253. doi: 10.1007/s11103-012-9941-y
Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115, 327–334. doi: 10.1104/pp.115.2.327
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Song, H., Sun, W., Yang, G., and Sun, J. (2018). WRKY transcription factors in legumes. BMC Plant Biol. 18:243. doi: 10.1186/s12870-018-1467-2
Song, L., Huang, S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354:g1550. doi: 10.1126/science.aag1550
Sun, J., Hu, W., Zhou, R., Wang, L., Wang, X., Wang, Q., et al. (2015). The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34, 23–35. doi: 10.1007/s00299-014-1684-6
Thompson, A., Jackson, A., Symonds, R., Mulholland, B., Dadswell, A., Blake, P., et al. (2000). Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23, 363–374. doi: 10.1046/j.1365-313x.2000.00789.x
Ullah, A., Sun, H., Hakim, Yang, X., and Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 162, 439–454. doi: 10.1111/ppl.12651
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Wang, C., Deng, P., Liulin, C., Xiatian, W., Hui, M., Wei, H., et al. (2013). A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One 6:e65120. doi: 10.1371/journal.pone.0065120
Wang, C., Ru, J., Liu, Y., Yang, J., Li, M., Xu, Z., et al. (2018). The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 19:2580. doi: 10.3390/ijms19092580
Wang, F., Chen, H., Li, Q., Wei, W., Li, W., Zhang, W., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14:103. doi: 10.1186/1471-2229-14-103
Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Willems, E., Leyns, L., and Vandesompele, J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379, 127–129. doi: 10.1016/j.ab.2008.04.036
Wu, J., Chen, J., Wang, L., and Wang, S. (2017). Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 8:380. doi: 10.3389/fpls.2017.00380
Wu, M., Liu, H., Han, G., Cai, R., Pan, F., and Xiang, Y. (2017). A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci. Rep. 7:11721. doi: 10.1038/s41598-017-10795-z
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xiao, Y., Zhou, L., Lei, X., Cao, H., Wang, Y., Dou, Y., et al. (2017). Genome-wide identification of WRKY genes and their expression profiles under different abiotic stresses in Elaeis guineensis. PLoS One 12:e189224. doi: 10.1371/journal.pone.0189224
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., and He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 19:490. doi: 10.1186/s12864-018-4880-x
Xiong, L., Wang, R., Mao, G., and Koczan, J. (2006). Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol. 142, 1065–1074. doi: 10.1104/pp.106.084632
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yang, J., Zhang, J., Wang, Z., Zhu, Q., and Wang, W. (2001). Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 127, 315–323. doi: 10.1104/pp.127.1.315
Yang, L., Ji, W., Zhu, Y., Gao, P., Li, Y., Cai, H., et al. (2010). GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 61, 2519–2533. doi: 10.1093/jxb/erq084
Yang, X., Li, H., Yang, Y., Wang, Y., Mo, Y., Zhang, R., et al. (2018). Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS One 13:e191308. doi: 10.1371/journal.pone.0191308
Yang, Y., Zhou, Y., Chi, Y., Fan, B., and Chen, Z. (2017). Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7:17804. doi: 10.1038/s41598-017-18235-8
Ying, S., Zhang, D., Fu, J., Shi, Y., Song, Y., Wang, T., et al. (2012). Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235, 253–266. doi: 10.1007/s00425-011-1496-7
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Yoshida, T., Mogami, J., and Kazuko, Y. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, L., Wu, S., Peng, Y., Liu, R., Chen, X., Zhao, P., et al. (2016). Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. doi: 10.1111/pbi.12358
Yu, Y., Wang, L., Chen, J., Liu, Z., Park, C., and Xiang, F. (2018). WRKY71 Acts Antagonistically Against Salt-Delayed Flowering in Arabidopsis thaliana. Plant Cell Physiol. 59, 414–422. doi: 10.1093/pcp/pcx201
Yu, Y., Wang, N., Hu, R., and Xiang, F. (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. SpringerPlus 5:920. doi: 10.1186/s40064-016-2647-x
Zeng, Q., Yang, C., Ma, Q., Li, X., Dong, W., and Nian, H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12:182. doi: 10.1186/1471-2229-12-182
Zhang, C., Wang, D., Yang, C., Kong, N., Shi, Z., Zhao, P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12:e181573. doi: 10.1371/journal.pone.0181573
Zhang, L., Cheng, J., Sun, X., Zhao, T., Li, M., Wang, Q., et al. (2018). Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 37, 1159–1172. doi: 10.1007/s00299-018-2302-9
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol.Biol. 5:1.
Zhou, Q., Tian, A., Zou, H., Xie, Z., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: GmWRKY16, soybean, salt, drought, ABA, Arabidopsis thaliana
Citation: Ma Q, Xia Z, Cai Z, Li L, Cheng Y, Liu J and Nian H (2019) GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Front. Plant Sci. 9:1979. doi: 10.3389/fpls.2018.01979
Received: 07 June 2018; Accepted: 20 December 2018;
Published: 21 January 2019.
Edited by:
Ruth Grene, Virginia Tech, United StatesReviewed by:
Yang Zhao, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2019 Ma, Xia, Cai, Li, Cheng, Liu and Nian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Nian, aG5pYW5Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.