- 1The State Key Laboratory of Subtropical Silviculture, Zhejiang Agriculture and Forestry University, Lin’an, China
- 2Shenzhen Key Laboratory of Marine Bioresource & Eco-Environmental Science, Guangdong Engineering Research Center for Marine Algal Biotechnology, College of Life Science and Oceanography, Shenzhen University, Shenzhen, China
- 3Key Laboratory of Saline-alkali Vegetation Ecology Restoration in Oil Field (SAVER), Ministry of Education, Alkali Soil Natural Environmental Science Center (ASNESC), Northeast Forestry University, Harbin, China
- 4Asian Natural Environment Science Center (ANESC), University of Tokyo, Tokyo, Japan
A large proportion of the world’s arable land is saline-alkali land, and this is becoming an urgent environmental problem for agriculture. One approach to address this problem is to develop new varieties of stress-resistant plants through genetic engineering. The algae (Chlorella sp.) JB6, which was previously isolated from saline-alkali land, was found to exhibit strong NaHCO3 tolerance. Here, we explored saline-alkali-tolerance genes in this alga that might be useful for producing abiotic stress-resistant transgenic plants. We identified a gene encoding acyl-CoA-binding protein 1 (ACBP1) from JB6 by screening a full-length cDNA library in yeast under NaHCO3 stress. Northern blot analyses showed that the ChACBP1 mRNA levels were significantly up-regulated under abiotic stresses such as salinity, oxidation, heavy metals, and low temperature stresses. The recombinant ChACBP1 protein was found to bind phosphatidylcholine in vitro. Green fluorescent protein-labeled ChACBP1 was localized to the cytosol. Overexpression of ChACBP1 in yeast and Arabidopsis increased their resistance to high salinity, oxidation, heavy metals, and low temperature stresses. These results suggested that ChACBP1 may mediate plant abiotic stress adaptation through phospholipid metabolism. Thus, ChACBP1 may be useful to genetically improve the tolerance of plants to saline-alkali soil.
Introduction
Saline-alkali soil can contain different types of salts (e.g., Na2SO4, NaCl, NaHCO3). Neutral salts (NaCl and Na2SO4) and alkaline salts (NaHCO3 and Na2CO3) are highly destructive to plants (Guo et al., 2016; Song et al., 2017). Soils rich in NaHCO3 and Na2CO3 are characterized by a high pH, a high exchangeable sodium percentage, a very low water-permeability range, and saturated hydraulic conductivity (Shi et al., 2012; Qiao et al., 2015). Such environments, which are known as “carbonate stressed” environments, are usually barren with only a few scattered plants (Qiao et al., 2015). Carbonate stress threatens the development of agriculture and livestock husbandry. Carbonate stress impairs plants in several ways by imposing ionic stress, drought stress, and oxidative stress, resulting in nutritional deficiency, metabolic disorders, and membrane damage. These changes affect plant growth and development and agricultural yield. The induction of proteins associated with lipid metabolism and lipid signaling has been proposed to be an important factor in plants’ acclimation to stress conditions (Xiao and Chye, 2011).
In plants, lipids are one of the main components of biological membranes and play important roles in diverse biological processes, such as the provision of energy for cell metabolism and the maintenance of organelle integrity and composition. During lipid metabolism in plant cells, lipids, and their derivatives are transferred within or across subcellular compartments with the aid of lipid-transfer proteins or acyl-CoA-binding proteins (ACBPs) (Xiao and Chye, 2009). Arabidopsis thaliana has six ACBPs (AtACBP1–AtACBP6) with a conserved acyl-CoA-binding domain that range in size from 10 to 73 kDa (Xiao and Chye, 2011). In vitro, ACBPs bind phospholipids such as phosphatidylcholine (PC) (Chen et al., 2008), phosphatidic acid (PA) (Du et al., 2010), phosphatidylethanolamine (PE) (Xiao et al., 2011), and lysophosphatidylcholine (lysoPC) (Gao et al., 2010). Plant ACBPs participate in several biological processes, including early embryogenesis (Chen et al., 2010) and leaf senescence (Xiao et al., 2011), and in responses to heavy metals (Xiao et al., 2008; Gao et al., 2010; Du et al., 2015), drought (Du et al., 2013), and freezing temperatures (Chen et al., 2008). A small 10-kDa ACBP has been identified in several plants including Brassica napus (Hills et al., 1994; Brown et al., 1998), A. thaliana (Engeseth et al., 1996), and agricultural crops including cotton (Gossypium hirsutum) (Reddy et al., 1996) and rice (Oryza sativa) (Suzui et al., 2006). The small ACBP has a housekeeping function in the intracellular transport of acyl-CoAs (Mandrupl et al., 1992) and has been well-characterized in many eukaryotic organisms (Metzner et al., 2000). In vitro, the small ACBPs of A. thaliana (AtACBP6) and Helianthus annuus (HaACBP) have been observed to bind only PCs (Chen et al., 2008; Aznar-Moreno et al., 2016). In addition to its involvement in lipid metabolism, AtACBP6 has also been reported to enhance freezing tolerance and affect jasmonate composition (Chen et al., 2008; Ye et al., 2016). Although several studies have examined the role of ACBP in abiotic stresses in higher plants, little is known about its role in algae.
Previously, we found a species of Chlorella, JB6, in the alkaline-saline soil of northeastern China. This alga was found to show extremely high tolerance to NaHCO3 and NaCl (Wang et al., 2011; Qiao et al., 2015). Therefore, it is of great significance as a genetic resource to generate transgenic stress-tolerant plants, and for studies on the responses to abiotic stresses.
In this study, we identified a gene encoding a small ACBP (ChACBP) from Chlorella (JB6) by screening its full-length cDNA library expressed in yeast under NaHCO3 stress. We investigated ChACBP expression in response to several abiotic stresses (high salinity, oxidation, heavy metals, and low temperature). Transgenic yeast and Arabidopsis overexpressing ChACBP showed enhanced tolerance to salinity, oxidation, heavy metals, and low-temperature stresses. Filter-binding assays revealed that ChACBP interacted with lipids. These results have clarified the molecular mechanism of ChACBP and indicate that the gene encoding this protein may be useful for generating transgenic plants resistant to saline-alkali soil.
Materials and Methods
Materials
The saline-alkali-tolerant (SAT) microalga Chlorella JB6 was isolated and screened from extremely alkaline-saline soil (pH > 10) from the Songnen Plain (46°27′N, 125°22′E, Heilongjiang Province, China) (Wang et al., 2011; Qiao et al., 2015) and was cultivated in liquid Bold’s basal medium (BBM) (Bold and Wynne, 1978). The Chlorella cells were cultured at 23 ± 1°C under white light (40 μmol photons m-1 s-1) and an 16 h light/8 h dark photoperiod. Total RNA was isolated from Chlorella after 100 mM NaHCO3 treatment. An In-Fusion®SMARTer®Directional cDNA Library Construction Kit (Cat. 634933, Clontech, Palo Alto, CA, United States) was used to construct the full-length cDNA library. Mixed plasmids were expressed in Saccharomyces cerevisiae InVSCI using the PEG/LiAC method. The full-length cDNA yeast library was provided by the Alkali Soil Natural Environmental Science Center (ASNESC) at Northeast Forestry University (Harbin, China).
Gene Cloning and Analysis
The Chlorella acyl-CoA-binding protein gene (ChACBP) was amplified from a cDNA library expressed in yeast under 25 mM NaHCO3 stress. The full-length sequence of ChACBP was obtained from the NCBI website1. The open reading frame (ORF) and the predicted protein sequence were determined and analyzed using DNASTAR Lasergene v7.1 software2. Multiple sequences were aligned with Genedoc 3.0. A phylogenetic tree was constructed using the neighbor-joining (NJ) method with Mega 3.1 software.
Yeast Transformation and Stress Tolerance Assays
The cDNA fragment of ChACBP was ligated into the pYES2 vector (Invitrogen, Carlsbad, CA, United States) digested with BamHI and NotI to construct the plasmid pYES2-ChACBP. This construct and the pYES2 empty vector (control) were transformed into S. cerevisiae (InVSCI) using the PEG/LiAC method. To check the stress tolerance of transgenic lines, yeast cells expressing the pYES2-ChACBP vectors were incubated in liquid uracil-minus medium overnight at 30°C (FunGenome, Beijing, China), then adjusted to an OD600 of 0.5, and further diluted to 10-1, 10-2, 10-3, and 10-4 with sterile H2O. Then, 4.5 μL of each dilution series was spotted onto solid medium (yeast extract 20 g/L; peptone 20 g/L; galactose 20%) supplemented with 35 mM NaHCO3, 0.8 M NaCl, 3 mM H2O2, or 8 mM CuCl2. The cultures were grown for 3–7 days at 30°C. For the low temperature treatment, diluted yeast cells spotted onto solid medium were cultured for 20 days at 10°C.
Purification of Recombinant His-Tagged ChACBP for Filter-Binding Assays
To construct the pQE30-ChACBP expression plasmid, the ChACBP cDNA fragment was amplified from the plasmid pYES2-ChACBP using the following primers: forward: 5′-GGAT CCATGGGCCTCAAGGAAGAC-3′ (BamHI site underlined); and reverse 5′-GTCGACTCAAGCGTACTTCGCCTTC-3′ (SalI site underlined). The amplified fragment was inserted into the pQE30 vector (Qiagen, Hilden, Germany), which was then transformed into Escherichia coli M15 cells. The ChACBP protein was induced, ultrasonicated, and loaded onto Ni–NTA Sefinose (Qiagen). The bound fusion protein was eluted using 250 mM imidazole (Sigma-Aldrich, St. Louis, MO, United States). Protein samples were denatured at 100°C for 8 min and then separated by SDS-PAGE. After electrophoresis, the protein products were transferred to a nylon membrane (Amersham, Little Chalfont, United Kingdom) using a HoeferTM TE 70 semi-dry transfer unit (Amersham) for 90 min, then blocked for 1 h in 1% blocking buffer (QIAexpress Anti-His HRP Conjugate kit), probed with Penta anti-(His)6 horseradish peroxidase (HRP)-conjugate antibody (Qiagen) for 90 min at 25°C, and then detected with an ECL Western Blotting Substrate kit (Abcam, Cambridge, United Kingdom) using a Luminescent Image Analyzer LAS-4000 (Fujifilm, Tokyo, Japan). Binding of ChACBP to various lipids on a nitrocellulose filter membrane was detected as described previously with minor modifications (Chen et al., 2008). Briefly, various lipids were spotted onto the membrane and incubated overnight at 25°C in the dark. The lipids PC, PA, 16:0-PC, 18:0-PC, and 18:1-PC (total acyl carbon: double bonds) were purchased from Sigma; and phosphatidylglycerol (PG), phosphatidyl-serine (PS), PE, and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) were purchased from Echelon Biosciences (Salt Lake City, UT, United States). The lipid-bound membrane was blocked with 1% (w/v) nonfat milk for 1 h, and then with 2 μg/mL ChACBP protein for 2 h. The membrane was incubated with the Penta anti-(His)6 HRP antibody for 2.5 h at 25°C, detected with ECL reagent, and analyzed using the LAS-4000 imager.
Expression Analysis of ChACBP Under Stress Treatments
To check the expression level of ChACBP under various abiotic stresses including high salinity, oxidation, and heavy metal stresses, Chlorella cells were grown in liquid medium supplemented with 200 mM NaHCO3, 200 mM NaCl, 2 mM H2O2, or 100 μM CuCl2, respectively. Low temperature stress was applied by incubating the microalga at 4°C. Samples were collected at 0, 3, 6, 12, 24, and 48 h for analysis. The concentration gradients in the stress treatments were as follows: 0, 50, 100, 200, 300, and 400 mM NaHCO3; 0, 50, 100, 150, 200, and 300 mM NaCl; 0, 1, 2, 3, 4, and 5 mM H2O2; 0, 50, 100, 200, 300, and 500 μM CuCl2; and temperatures of 24°C, 18°C, 16°C, 14°C, 10°C, and 4°C. Samples were collected at 6 h of these treatments, and ground in liquid nitrogen with a mortar and pestle.
Total RNA was extracted using RNAiso plus (TaKaRa, Kyoto, Japan). Northern blot analysis was performed using the Digoxigenin Nucleic Acid Detection kit (Roche, Basel, Switzerland). Total RNA (3 μg) was separated on a 1.5% agarose gel and transferred to a Hybond-N membrane (Amersham). The ChACBP cDNA probes were obtained using the PCR Digoxigenin Probe Synthesis kit according to the manufacturer’s instructions (Roche). Hybridization was performed according to standard procedures recommended by the manufacturer (Roche), and then detected using the CDP Star system with the LAS-4000 imager.
Localization of ChACBP Protein in Plant Cells
To construct the expression plasmid pBI121-ChACBP-GFP, ChACBP was amplified from the pYES2-ChACBP plasmid by PCR using the primers ChACBP-GFP-F (5′-GGATCCATGGGC CTCAAGGAAGACTTTG-3′; BamHI site underlined) and ChACBP-GFP-R (5′-GGTACCGGAGCGTACTTCGCCTTCAG CG-3′; KpnI site underlined). The PCR product was introduced into the pEGFP plasmid (Clontech) at the BamH1 and KpnI sites. Then, ChACBP-pEGFP was digested with BamHI and NotI and ligated into the pYES2 vector (Invitrogen). The plasmid pYES2-ChACBP-GFP was inserted into the pBI121 vector via the BamHI and XhoI digestion sites to construct the plasmid pBI121-ChACBP-GFP. Agrobacterium tumefaciens strain EHA105 was transformed by electroporation with either pBI121-GFP or pBI121-ChACBP-GFP. Transgenic A. tumefaciens cells were transformed into wild-type (WT) Arabidopsis (Columbia-0) by the floral dip method (Clough and Bent, 1998). Transgenic lines overexpressing pBI121-ChACBP-GFP were selected on 1/2 MS medium containing kanamycin (40 μg/mL) and confirmed by northern blot analysis using a ChACBP cDNA probe. Arabidopsis protoplasts were prepared by Sheen’s method (Sheen, 2002). All GFP signals were detected using a laser-scanning confocal imaging system (Olympus, Tokyo, Japan).
Generation of ChACBP-Overexpressing Plants
To construct the plasmid pBI121-ChACBP, the full-length ChACBP was amplified from pYES2-ChACBP by PCR using the primers pBI-F (5′-GGATCCATGG GCCTCAAGGAAGACTTTG-3′; BamHI site underlined) and pBI-R (5′- GTCGACTCAAGCGTACTTCGCCTTCAG-3′; SalI site underlined). The PCR product was ligated into the pBI121 plasmid (Clontech) via the BamHI and SalI sites. The construct was amplified in E. coli and then used to transform A. tumefaciens. The positive transformants were confirmed by Northern blot and Southern blot analyses using a ChACBP probe. Genomic DNA was extracted from the leaves of 12-day-old seedlings (WT, OX-2, OX-3, and OX-4) using the CTAB method. The genomic DNA was digested with BamHI, and then separated by electrophoresis on a 1.5% agarose gel and blotted onto a nylon membrane. Southern blotting was conducted with the CDP Star system and analyzed using the LAS-4000 imager.
Stress Tolerance of ChACBP Transgenic Plants
The WT and transgenic Arabidopsis seeds were sterilized as described by Zhang et al. (2008). To measure root growth and fresh weight, the WT and transgenic Arabidopsis seeds were sown on 1/2 MS solid agar plates supplemented with 2 and 3 mM NaHCO3, 125, and 150 mM NaCl, 2, and 3 mM H2O2, or 50 and 80 μM CuCl2. The seedlings were grown for 7–14 days at 23 ± 1°C under a 8-h light/16-h dark photoperiod. For the low temperature stress, the plates were incubated at 16°C and 14°C for 30 days. Root length and fresh weight were measured, and the mean ± standard error were calculated from three independent experiments. Statistical analysis was performed using SPSS 13.0 software. An LSD t-test was used to compare the mean values among different groups (P < 0.05).
Electrolyte Leakage
Two-week-old seedlings of WT and three overexpressing lines were grown under NaHCO3 (0, 1, 3, 5, 7, and 10 mM), NaCl (0, 100, 125, 150, 175, and 200 mM), H2O2 (0, 1, 3, 5, 7, and 9 mM), CuCl2 (0, 20, 50, 80, 100, and 120 μM), and temperature (24°C, 18°C, 16°C, 14°C, 12°C, and 10°C) stress conditions for 24 h. Seedling samples were immersed in deionized water. The solution was gently agitated at 25°C for 1 h before measuring its conductivity. The total ion content was determined after heating samples to 100°C for 10 min and then cooling to 25°C. Ionic leakage was detected using a conductivity meter (DDS-307, Leici Co., Ltd., Shanghai, China).
Results
Identification and Characterization of ChACBP
The SAT Chlorella was isolated from extremely alkaline-saline soil (pH > 10). The Chlorella cells were previously shown to tolerate 1 M NaHCO3 and 600 mM NaCl (Qiao et al., 2015). A cDNA library of Chlorella was expressed and screened in the InVSC1 strain of S. cerevisiae to identify potential genes involved in the strong stress tolerance of this microalga. Potential overexpression candidates were further verified under high NaHCO3 conditions (Supplementary Figure S1), and genes were isolated and sequenced (Supplementary Figure S2). The acyl-CoA-binding protein gene (ACBP) was isolated from SAT Chlorella for the first time (white arrow), and was named ChACBP1. Yeast tolerance analyses showed that ChACBP-expressing transformants grew better than the controls (YPD) under 35 mM NaHCO3, 0.8 M NaCl, 3 mM H2O2, 8 mM CuCl2, and at 10°C (Figure 1).
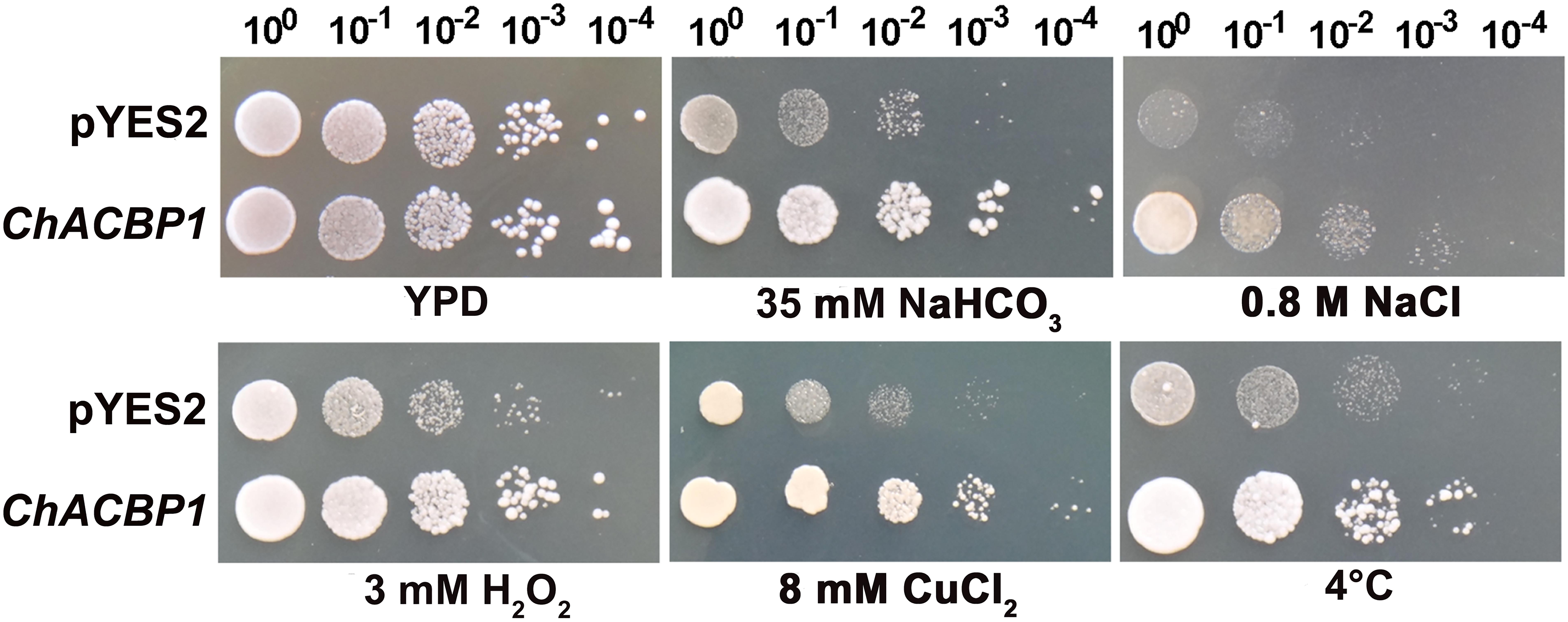
FIGURE 1. Tolerance analysis of yeast expressing ChACBP1 to various abiotic stresses. Yeast cells containing either pYES2 empty vector or pYES2-ChACBP1 were incubated as described in Materials and Methods. Serial dilutions were spotted onto yeast extract/peptone/galactose solid plates supplemented with NaHCO3, NaCl, H2O2, or CuCl2 at indicated concentrations. Solid yeast extract/peptone/glucose media (YPD) was the control. Growth was monitored for 3–7 days at 30°C. In the cold stress treatment, yeast cells were grown for 15 days at 10°C.
Analysis of the ChACBP1 sequence indicated that the ORF was 264 nucleotides long, and encoded a predicted protein of 87 amino acids. The cDNA sequence was found to contain a 57-bp 5’-untranslated region (UTR) and a 204-bp 3’-UTR. The amino acid sequence of ChACBP1 was found to share high identity with previously published ACBP sequences from other algae, plants, and animals. A comparison of the ACB domains of these ACBPs suggested conservation of the YKQA and KWDAW motifs (Figure 2A). In a neighbor-joining (NJ) tree of the ACBPs, ChACBP1 was closer to the ACBP of Chlorella variabilis (XP_005848979.1) than to the ACBPs of other species. In the tree, the SAT Chlorella first clustered with unicellular microalgae, then with plants and animals (Figure 2B), in agreement with the expected taxonomy. In a circular NJ tree, ChACBP1 was closer to AtACBP6 and OsACBP4 than to the ACBPs of Arabidopsis and rice (Figure 2C).
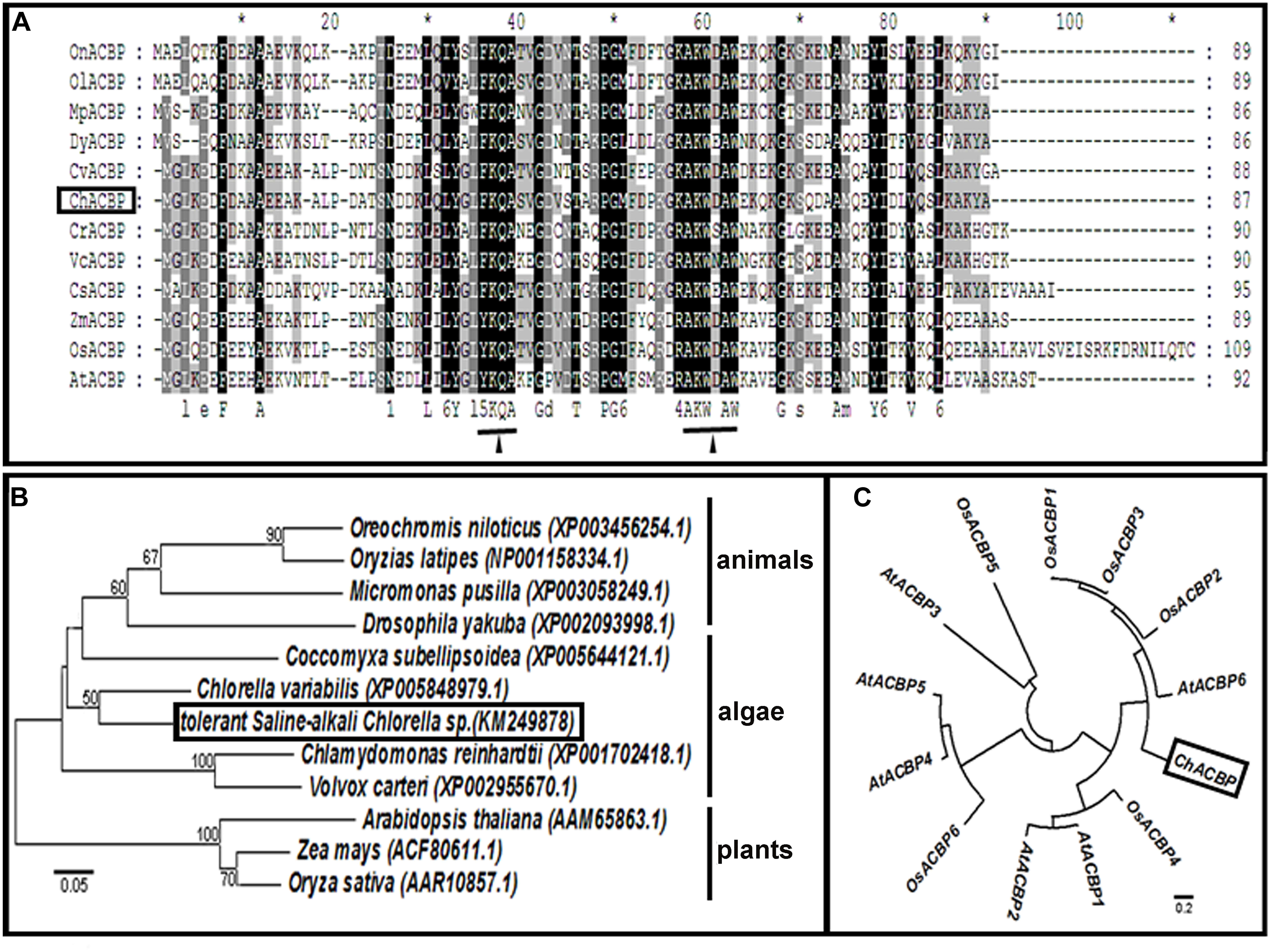
FIGURE 2. Phylogenetic and sequence analyses of acyl-CoA-binding protein 1 (ChACBP1) from saline-alkali-tolerant Chlorella. (A) Sequence alignment of ACB domain from ChACBP1 with those of other species. YKQA and KWDAW motifs are underlined; capital letters indicate identical residues in all ACBPs; small letters indicate residues identical in most ACBPs (arrowheads). Black background indicates conserved residues. On, Oreochromis niloticus; Ol, Oryzias latipes; Mp, Micromonas pusilla; Dy, Drosophila yakuba; Cv, Chlorella variabilis; Ch, Saline-alkali-tolerant Chlorella sp.; Cr, Chlamydomonas reinhardtii; Vc, Volvox carteri; Cs, Coccomyxa subellipsoidea; Zm, Zea mays; Os, Oryza sativa; At, Arabidopsis thaliana; ChACBP1 is indicated in boldface in box. (B) Neighbor-joining (NJ) phylogenetic relationships among acyl-CoA-binding protein (ChACBP) from saline-alkali-tolerant Chlorella and various ACBPs from other species. Bootstrap values were calculated 1,000 times; values < 50% are not shown. Saline-alkali-tolerant Chlorella is indicated in boldface in box. GenBank accession numbers are as follows: OnACBP (XP_003456254.1), OlACBP (NP_001158334.1), MpACBP (XP_003058249.1), DyACBP1 (XP_002093998.1), CvACBP1 (XP_005848979.1), ChACBP (KM249878), CrACBP (XP_001702418.1), VcACBP (XP_002955670.1), CsACBP (XP_005644121.1), ZmACBP (ACF80611.1), OsACBP (AAR10857.1), and AtACBP (AAM65863.1). (C) Neighbor-joining phylogenetic relationships among ChACBP and members of ACBP families in Arabidopsis thaliana and rice. Box indicates ChACBP.
ChACBP Interacted With Phospholipid PC in vitro
The E. coli M15 cells transformed with the pQE30-ChACBP1 plasmid produced His-ChACBP1 fusion protein (9.5 kDa) (Figure 3A, lane 1). Elution of the His-ChACBP1 fusion protein with 250 mM imidazole gave a pure protein of the expected size (Figure 3A, lane 2). Western blot analysis showed that the purified ChACBP1 protein was not degraded (Figure 3B). To detect the interaction between ChACBP1 and phospholipids (PC, PA, PS, PG, PE, DMPC), which influences abiotic stress tolerance, we tested the binding of purified ChACBP1 to various lipids. In the binding assays, ChACBP1 specifically bound PC, but not other lipids (Figure 3C). Further analyses showed that ChACBP1 was able to bind several species of PC (16:0-PC, 18:0-PC, and 18:1-PC) (Figure 3D).
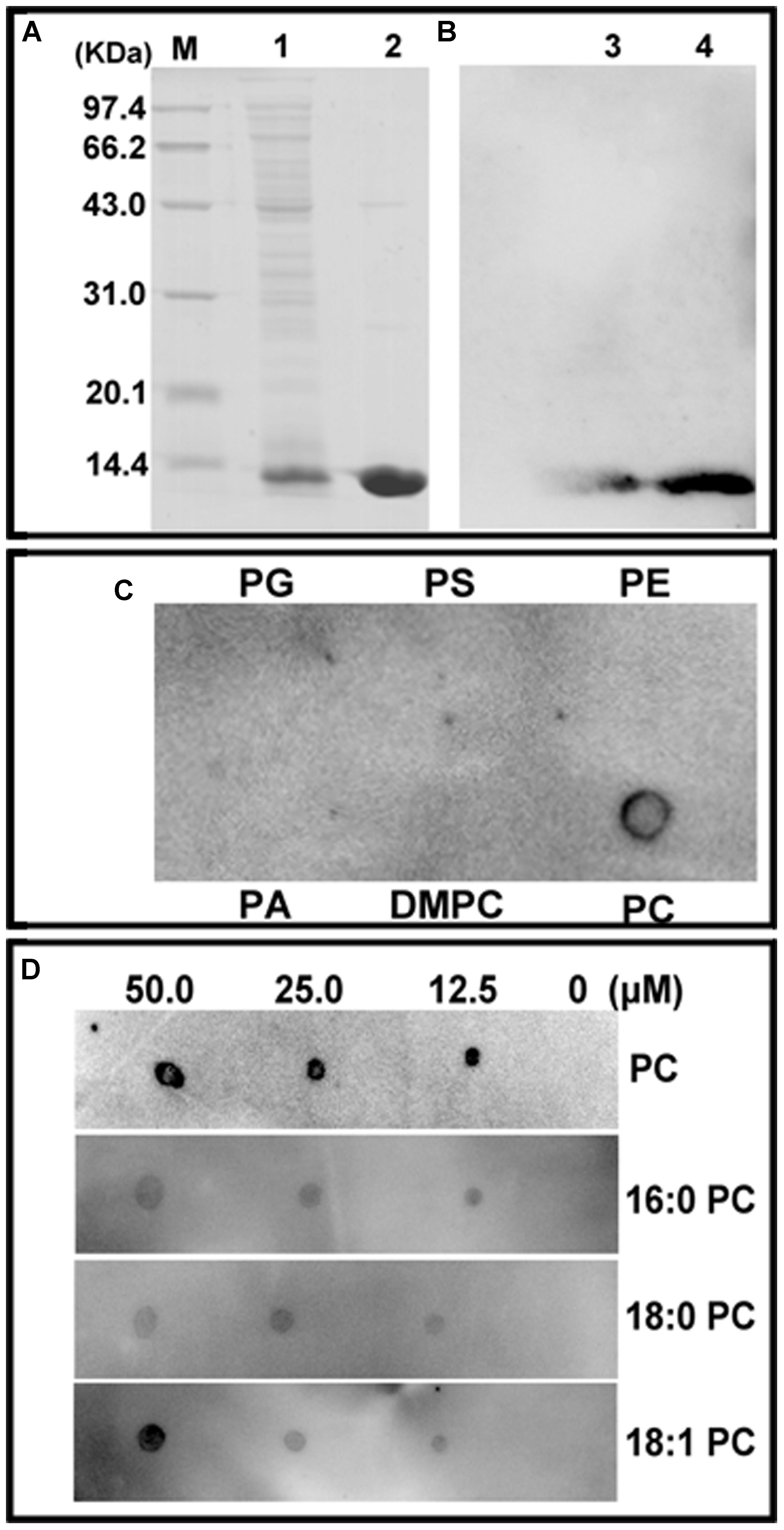
FIGURE 3. Purification of (His)6-ChACBP1 recombinant protein and its interaction with phosphatidylcholine (PC). (A) Purification of ChACBP1 protein: 9.5-kDa ChACBP1 protein was purified by Ni–NTA agarose and analyzed by SDS-PAGE. M, Marker; Lane 1, pQE30-ChACBP bacterial lysate; lane 2, purified ChACBP1 protein. (B) Western blot analysis of ChACBP1: protein was transferred to nylon membrane, then probed with HRP-conjugated anti-(His)6 antibodies. Lanes 3–4, immunoblot of (His)6-ChACBP1 fusion protein. (C) Binding of (His)6-ChACBP1 and lipid on filter membrane. Lipids (50.0 μM of PG, PS, PE, PA, DMPC, and PC) were spotted onto nitrocellulose and incubated with ChACBP1 protein. Binding of ChACBP1 to lipids was detected by ECL reagent with HRP-conjugated anti-(His)6 antibodies. (D) Binding of (His)6-ChACBP1 to various PC acyl species. Different concentrations (0, 10, 25.0, and 50.0 μM) of 16:0 PC, 18:0 PC, and 18:1 PC were spotted onto nitrocellulose and incubated with ChACBP1. Binding of ChACBP1 to lipids was detected by ECL reagent.
Induction of ChACBP1 Expression by Multiple Abiotic Stresses
Northern blot analyses were used to examine the response of ChACBP1 expression to various abiotic stresses. Analyses of total RNA extracted from SAT Chlorella exposed to NaHCO3, NaCl, H2O2, CuCl2, and 4°C for 0, 3, 6, 12, 24, and 48 h indicated that the ChACBP1 mRNA levels increased in the 200 mM NaHCO3 treatment from 3 to 48 h, compared with the control (0 h). ChACBP1 mRNA began to increase after 3 h in the 200 mM NaCl treatment and peaked after 24 h of treatment. ChACBP gene expression was induced at 12 h of the 2 mM H2O2 treatment and its expression level peaked at 48 h. The expression level of ChACBP1 was higher after 3 h of the 100 μM CuCl2 treatment than in the control. Subsequently, its expression level increased gradually at 6, 12, and 24 h of the 100 μM CuCl2 treatment, but was similar to the control after 48 h. The expression level of ChACBP1 began to increase after 3 h at 4°C and peaked at 48 h (Figure 4A). The level of ChACBP1 expression was affected by the concentration as well as the duration of the stress elicitors.
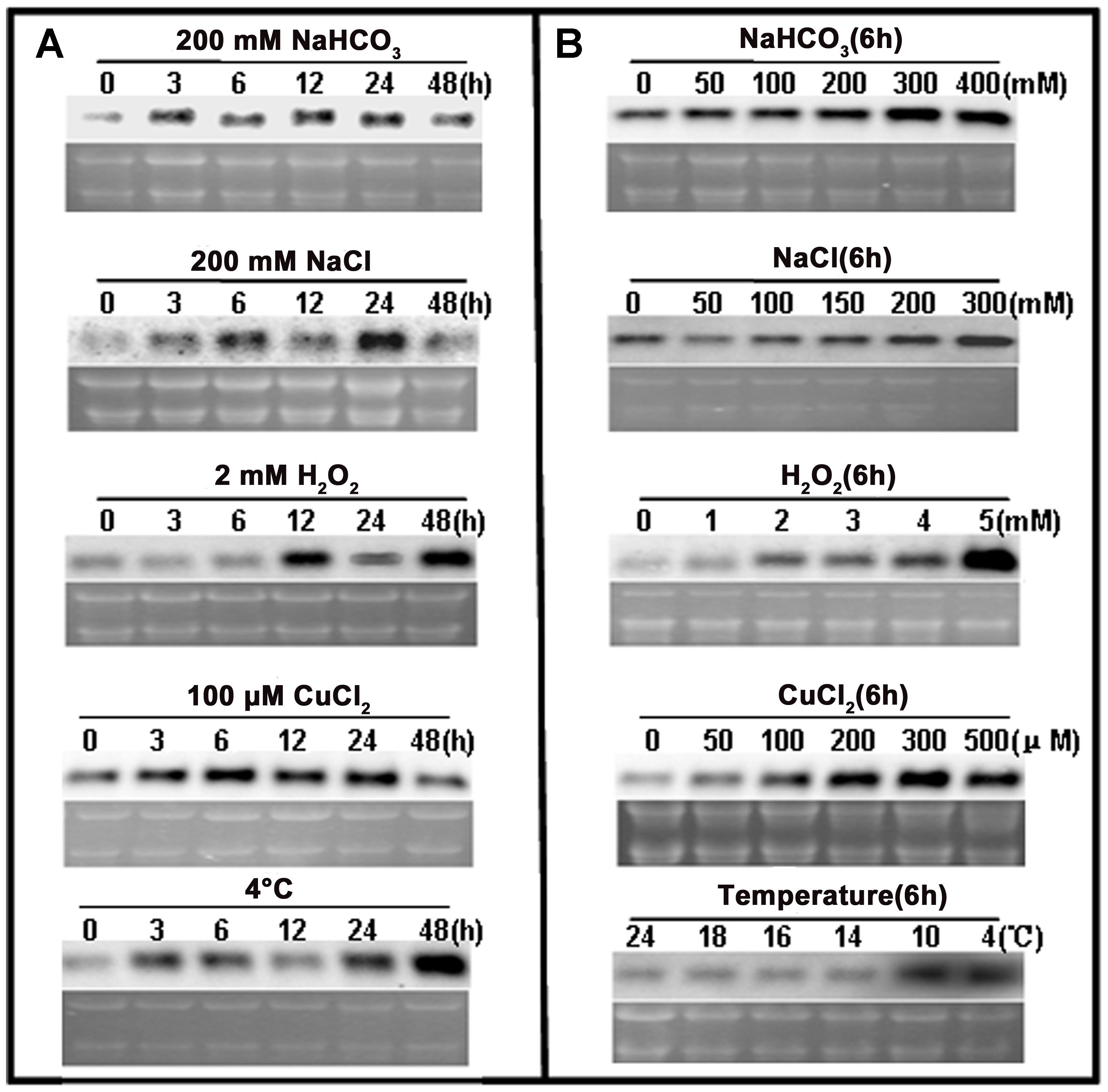
FIGURE 4. ChACBP1 expression under abiotic stresses as detected by Northern blot analysis with digoxigenin-labeled ChACBP1 cDNA probe. (A) Northern blot analysis using total RNA (3 μg) extracted from Chlorella cells treated with 200 mM NaHCO3, 200 mM NaCl, 2 mM H2O2, 100 μM CuCl2, or low temperature (4°C) at indicated time points. (B) Northern blot analysis using total RNA (3 μg) extracted from Chlorella cells treated with different concentrations of NaHCO3 (0, 50, 100, 200, 300, and 400 mM); NaCl (0, 50, 100, 150, 200, and 300 mM); H2O2 (0, 1, 2, 3, 4, and 5 mM); CuCl2 (0, 50, 100, 200, 300, and 500 μM); or different temperatures (24°C, 18°C, 16°C, 14°C, 10°C, and 4°C). All samples were collected at 6 h of treatment.
Northern blot analyses were conducted using total RNA extracted from SAT Chlorella exposed to different concentrations of NaHCO3, NaCl, H2O2, and CuCl2, and different temperatures for 6 h. At this time point, ChACBP1 mRNA levels were slightly higher in the 50 mM NaHCO3 treatment than in the control and were higher in the treatments with higher NaHCO3 concentrations. The peak expression level of ChACBP1 among the NaHCO3 treatments was in the 300 mM NaHCO3 treatment. The highest ChACBP1 expression level among the NaCl treatments was in the 300 mM NaCl treatment. The expression level of ChACBP1 was slightly increased in the 1 mM H2O2 treatment, compared with the control, and its highest expression level among the H2O2 treatments was in the 5 mM H2O2 treatment. The expression of ChACBP1 was induced by 50 μM CuCl2 and its expression level gradually increased as the concentration increased to 300 μM CuCl2. The expression levels of ChACBP1 in the 18°C, 16°C, and 14°C treatments were not significantly different from that in the control (24°C), but were increased in the 10°C treatment to a level similar to that measured at 4°C (Figure 4B). These data suggested that the expression of ChACBP1 was induced by NaHCO3, NaCl, H2O2, CuCl2, and low temperature stresses.
ChACBP1 Localizes to the Cytosol
The expression of ChACBP1-GFP mRNA in three independent pBI121-ChACBP1-GFP transgenic Arabidopsis lines was detected by Northern blot analysis using a ChACBP1 cDNA probe (Figure 5A). Analyses of the shoots of line 2 expressing pBI121-ChACBP1-GFP revealed that the GFP signal was localized in the cytosol. The GFP control was also expressed in the cytosol (Figure 5B).
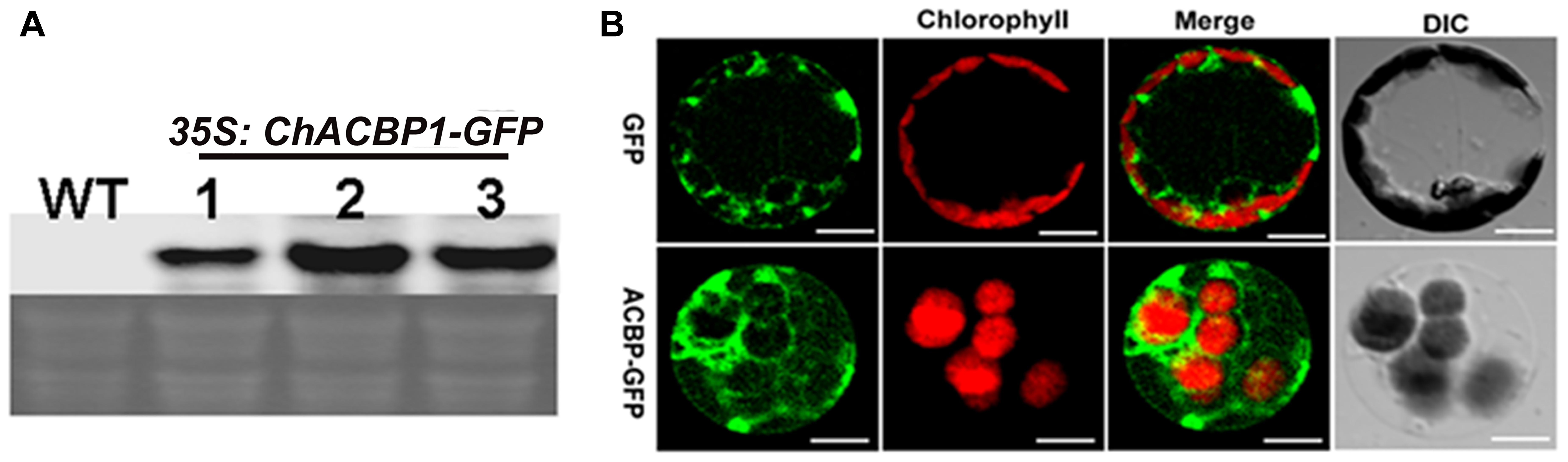
FIGURE 5. Subcellular localization of ChACBP1 in Arabidopsis protoplasts. (A) Northern blot analysis with ChACBP1 cDNA probe in three independent pBI121-ChACBP1-GFP transgenic lines (lanes 1–3). WT, Wild type. (B) Confocal microscope images of protoplasts of Arabidopsis pBI121-ChACBP1-GFP line 2 indicating localization of ChACBP1-GFP in the cytosol. GFP empty vector is shown at the top. Bars = 20 μm.
Generation of ChACBP1 Transgenic Arabidopsis
To test whether ChACBP1 overexpression enhanced abiotic tolerance, the ChACBP1 full-length cDNA was expressed in the Arabidopsis by an Agrobacterium-mediated method. Four independent ChACBP1-overexpressing lines (OX-1, OX-2, OX-3, and OX-4) were identified by Northern blot analysis using ChACBP1 mRNA. A Southern blot analysis indicated that OX-2, OX-3, and OX-4 had one or two gene copies (Supplementary Figure S3).
Overexpression of ChACBP1 in Arabidopsis Enhanced Tolerance to Multiple Abiotic Stresses
When grown on 1/2 MS medium, the three transgenic lines (OX-2, OX-3, and OX-4) over-expressing ChACBP1 were not noticeably different from WT (Figures 6, 7). Under 2 and 3 mM NaHCO3 treatment, root growth was significantly enhanced in ChACBP1-overexpressing Arabidopsis lines, compared with WT (P < 0.05). The entire seedling fresh weight was greater in the overexpressing lines than in the WT only in the 3 mM NaHCO3 treatment (P < 0.05; Figure 7). In the presence of 125 and 150 mM NaCl, the root length and fresh weight were higher in the overexpressing lines than in WT (P < 0.05; Figures 6, 7). When grown under oxidative stress (H2O2), root length was not notably different between the overexpressing lines and WT. However, the leaves of the overexpressing lines grew better than did those of WT (P < 0.05; Figures 6, 7). The fresh weights of the overexpressing lines were higher than those of WT in the 2 and 3 mM H2O2 treatments (P < 0.05; Figure 7). When the plants were exposed to CuCl2 and low temperature stresses, root lengths and fresh weights were higher in the overexpressing lines than in the WT (P < 0.05; Figures 6, 7).
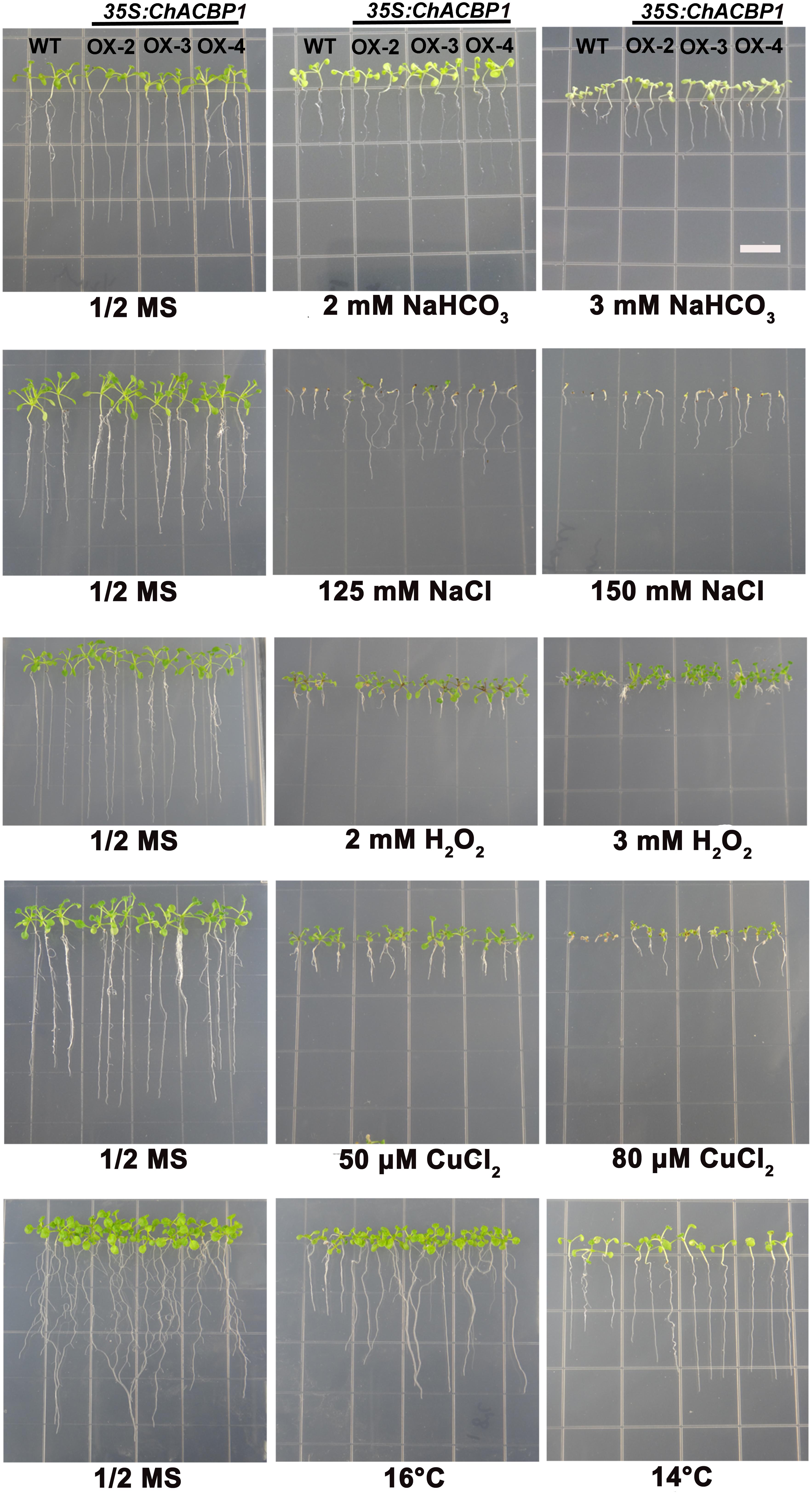
FIGURE 6. Effects of abiotic stresses on root length in wild-type (WT) and ChACBP1-overexpressing transgenic plants. Seeds of WT and transgenic lines were grown on 1/2 MS medium supplemented with 2 and 3 mM NaHCO3, 125 and 150 mM NaCl, 2 and 3 mM H2O2, or 50 and 80 μM CuCl2. Seedlings were grown for 7–14 days. For low temperature stress treatment, plants were grown at 16°C and 14°C for 30 days. Bars = 1 cm.
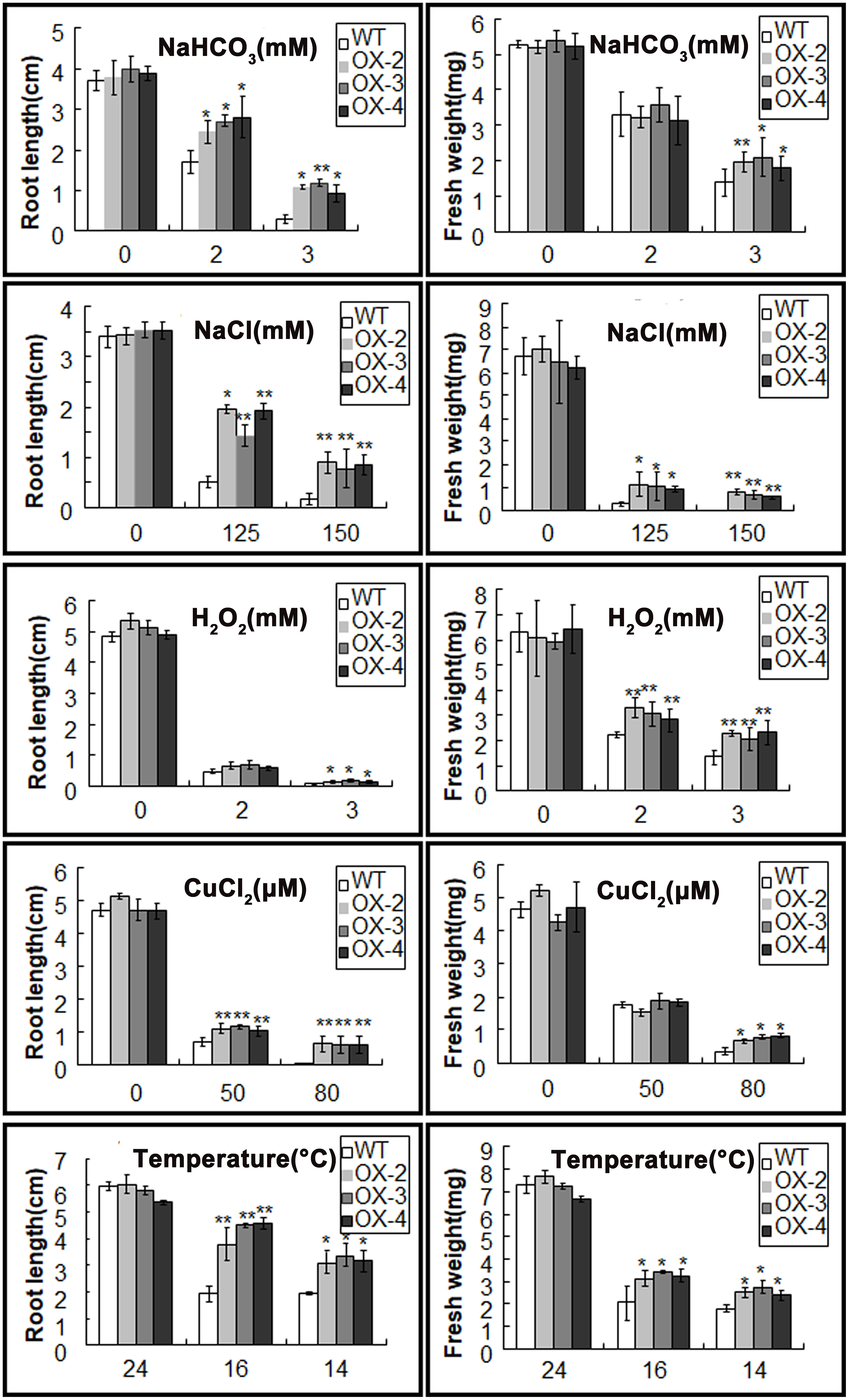
FIGURE 7. Root length and fresh weight of wild-type (WT) and ChACBP1-overexpressing transgenic plants after indicated stress treatments. Values are mean ± SE of four plants. Significant differences (P < 0.05) were determined by t-test. Asterisks indicate significant differences from WT (∗∗P < 0.01, ∗0.01 < P < 0.05).
To evaluate membrane injury after different stress treatments, electrolyte leakage was measured in 2-week-old seedlings from WT and ChACBP1-overexpressing plants treated with different concentrations of NaHCO3, NaCl, H2O2, and CuCl2, and different temperatures. The results showed that ionic leakage was not obviously different between WT and overexpressing lines in the 1 mM NaHCO3 treatment, but was significantly higher in overexpressing lines than in WT plants in the 3 to 10 mM NaHCO3 treatments (P < 0.05; Figure 8A). In comparison, the ionic leakage in ChACBP1-overexpressing seedlings was significantly lower (P < 0.05) than that in WT in the 100, 125, 150, and 175 mM NaCl treatments. There was no obvious difference in ionic leakage between WT and overexpression lines in the 200 mM NaCl treatment (Figure 8B). The ionic leakage was significantly lower (P < 0.05) in ChACBP1-overexpressing plants than in WT plants in all the temperature treatments (18°C, 16°C, 14°C, 12°C, and 10°C) (P < 0.05; Figure 8E). In the presence H2O2 and CuCl2, ionic leakage was lower in the overexpressing plants than in the WT only at the higher concentrations (9 mM H2O2 and 120 μM CuCl2, respectively) (P < 0.05; Figures 8C,D).
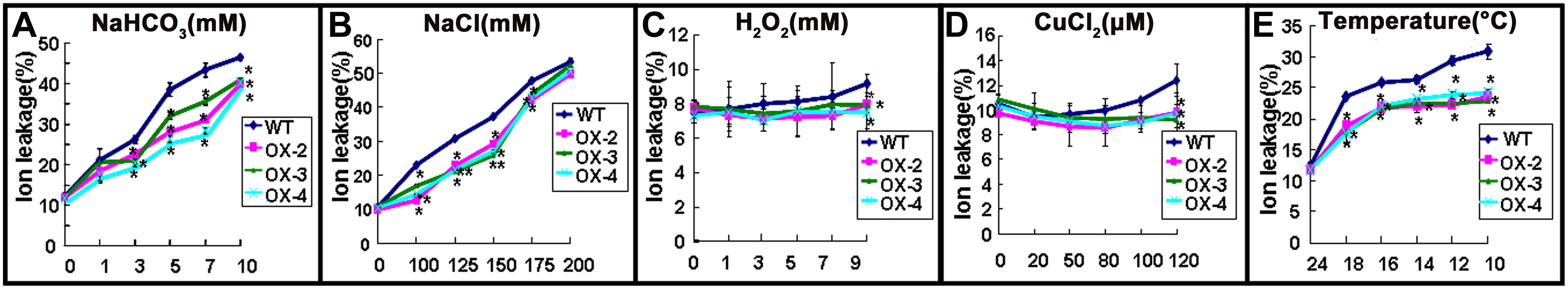
FIGURE 8. Electrolyte leakage of wild-type (WT) and ChACBP1-overexpressing (OX-2, OX-3, and OX-4) plants after 24 h of treatment with NaHCO3 (0, 1, 3, 5, 7, and 10 mM) (A), NaCl (0, 100, 125, 150, 175, and 200 mM) (B), H2O2 (0, 1, 3, 5, 7, and 9 mM) (C), CuCl2 (0, 20, 50, 80, 100, and 120 μM) (D), or low temperature (24°C, 18°C, 16°C, 14°C, 12°C, and 10°C) (E). Values are means ± SD (n = 3) from three independent experiments. Asterisks indicate significant differences from WT (∗∗P < 0.01, ∗0.01 < P < 0.05).
Discussion
China has 99.13 million hectares of saline-alkali soil, which accounts for 10% of the world’s saline-alkali land. The alkalinity in these regions is generally high, resulting in oxidative and salt stress. Drought stress is also common in these environments. As a result, few plants can survive under such conditions. In recent years, the area of saline-alkali soil has gradually increased as a result of environmental changes and human activities (Wang et al., 2011; Qiao et al., 2015). These changes have resulted in severe losses in agricultural production. We previously reported that Chlorella JB6 can survive in extreme saline-alkali soil with pH > 9.5 (Qiao et al., 2015). Therefore, this species may be a useful resource for plant genetic engineering.
We isolated ChACBP1 by screening a cDNA library of SAT Chlorella JB6. The full-length protein sequence was found to be homologous with the amino acid sequences of the small ACBPs from other species used to construct the NJ tree. Some functionally critical motifs in ACBPs have been relatively well conserved throughout evolution. However, there were some differences between the N-terminal or C-terminal regions of ChACBP1 and those of ACBPs in other species (Figure 2), suggesting that the function of ChACBP1 may be slightly different from that of ACBPs in other species. The YKQA and KWDAW motifs in the ACB conserved domain are considered to be essential for binding PC (Zhang et al., 2008). An evolutionary analysis of the Chlorella, Arabidopsis, and rice ACBP families indicated that ChACBP1 might be close to AtACBP6 and OsACBP4 in its structure and function. Therefore, the ACBP gene in Chlorella was considered to encode an acyl-CoA-binding protein. We tested the binding of ChACBP1 to phospholipids in filter-binding assays and observed that ChACBP1 bound PC (Figure 3), similar to the lipid-binding properties of AtACBP6 (Chen et al., 2008). It was hinted that ChACBP1 might transport multiple PCs associated with phospholipid metabolism. Phospholipids are the main components of cell membranes. In this study, the ion leakage (membrane stability index) of ChACBP1-overexpressing Arabidopsis was lower than that of WT under different stresses (Figure 8). This result suggested that ACBP may play a role in protecting the plasma membrane.
Our results showed that the Chlorella ACBP1 gene was induced by NaHCO3, NaCl, H2O2, CuCl2, and low temperatures, and that overexpression of ChACBP1 in yeast and Arabidopsis improved stress tolerance compared with that of their WT controls (Figures 1, 6). In cotton, GhACBP3 and GhACBP6 were found to be up-regulated under high salinity stress (Qin et al., 2016). In Arabidopsis, ACBP2-overexpressing lines showed enhanced tolerance to H2O2 (Gao et al., 2009), and both AtACBP1- and AtACBP2-overexpressors displayed enhanced tolerance to heavy metals (Pb or Cd) (Xiao et al., 2008; Gao et al., 2009). The expression level of AtACBP6 was found to be up-regulated by cold treatment (Chen et al., 2008). In the present study, ChACBP1 was isolated from a cDNA library expressed in yeast grown under NaHCO3 stress, and overexpression of ChACBP1 in yeast and Arabidopsis improved their tolerance to NaHCO3 (Figures 1, 7). Because NaHCO3 treatment likely leads to other stresses, such as salinity, alkalinity, oxidative, and drought stresses, it is not surprising that ChACBP1 can confer tolerance to multiple abiotic stresses. However, the mechanism of ChACBP1 tolerance is still unclear.
Overexpression of ACBP6 in Arabidopsis was shown to enhance the activities of phospholipase Dδ (PLDδ), which in turn enhanced cold tolerance by stabilizing the membrane skeleton (Chen et al., 2008). PLDδ can convert PC into PG, PS, and PE, all of which stabilize the cell membrane. Therefore, we propose a model in which exposure of the plasma membrane to abiotic stresses induces the expression of ChACBP1, and the subsequent increase in ChACBP1 protein leads to increased binding and transport of PC to the plasma membrane from within the cytosol. Hence, the increase in PG, PS, and PE contents increases the stability of the cell membrane (Figure 9). However, this model is still hypothetical. Like ACBPs from higher plants, ChACBP1 conferred tolerance to various abiotic stresses. The proposal that ChACBP1 mediates plant abiotic stress adaptation through phospholipid metabolism should be tested experimentally in further studies.
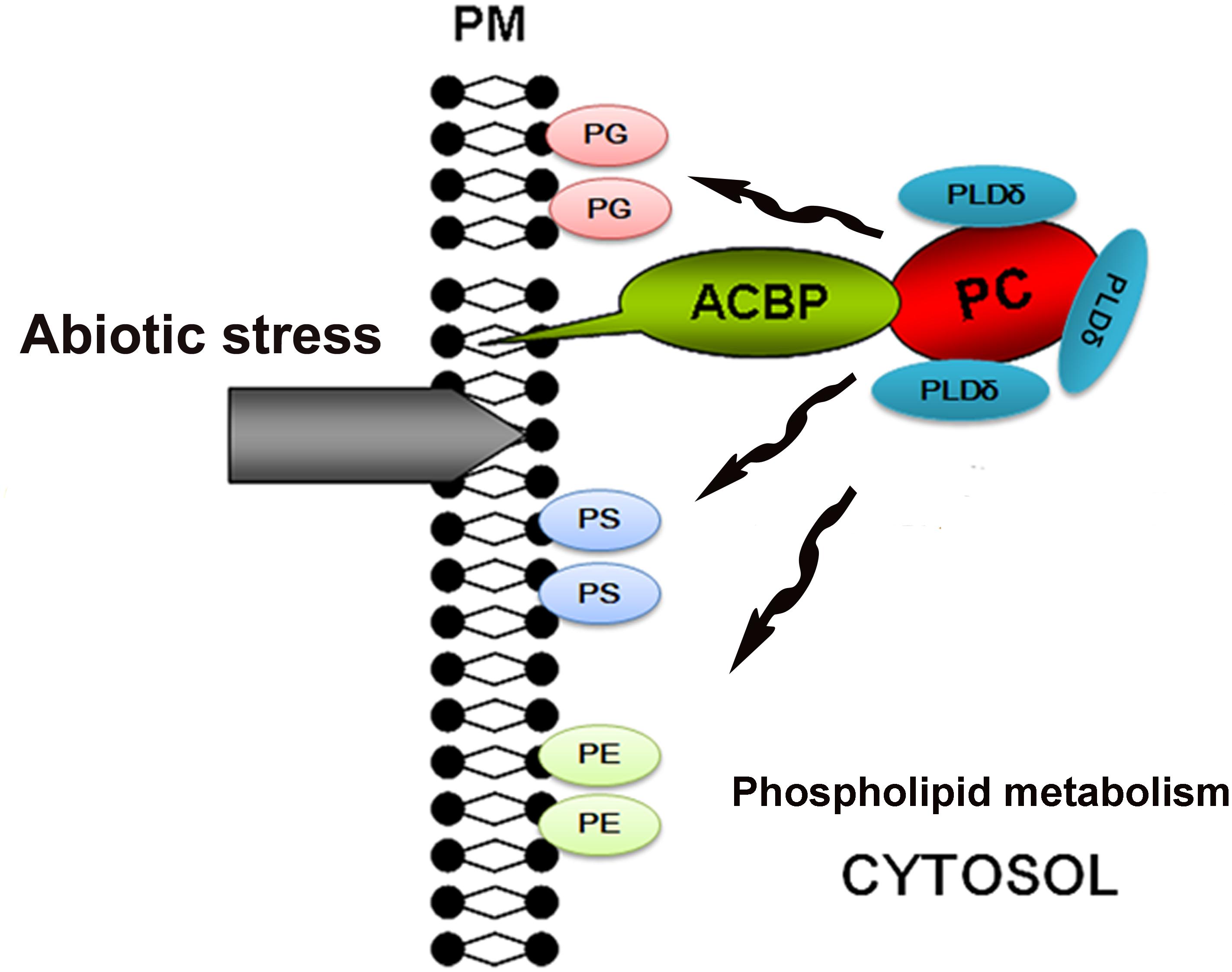
FIGURE 9. Hypothetical model for stress tolerance function of Chlorella ACBP. ACBP, acyl-CoA-binding protein; PC, phosphatidylcholine; PG, phosphatidylglycerol; PS, phosphatidyl-serine; PE, phosphatidylethanolamine; PLDδ, phospholipases Dδ; PM, plasma membrane.
Ethics Statement
This study was approved by the ethics committee of Zhejiang Agriculture and Forestry University.
Author Contributions
SL and TT conceived and designed the research. KQ and MW conducted the experiments. KQ analyzed the data and wrote the manuscript. All authors revised and approved the manuscript.
Funding
This work was supported by Special Fund for Forest Scientific Research in the Public Welfare (201404220), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) (IRT_17R99), and Fundamental Research Funds for the Central Universities (252016AA56).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01772/full#supplementary-material
Footnotes
References
Aznar-Moreno, J. A., Venegas-Caleron, M., Du, Z. Y., Garces, R., Tanner, J. A., Chye, M. L., et al. (2016). Characterization of a small acyl-CoA-binding protein (ACBP) from Helianthus annuus L. and its binding affinities. Plant Physiol. Biochem. 102, 141–150. doi: 10.1016/j.plaphy.2016.02.025
Brown, A. P., Johnson, P., Rawsthorne, S., and Hills, M. J. (1998). Expression and properties of acyl-CoA binding protein from Brassica napus. Plant Physiol. Biochem. 36, 629–635. doi: 10.1016/S0981-9428(98)80011-9
Chen, Q. F., Xiao, S., and Chye, M. L. (2008). Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme a-binding protein ACBP6 enhances freezing tolerance. Plant Physiol. 148, 304–315. doi: 10.1104/pp.108.123331
Chen, Q. F., Xiao, S., Qi, W. Q., Mishra, G., Ma, J. Y., Wang, M. F., et al. (2010). The Arabidopsis acbp1acbp2 double mutant lacking the acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytol. 186, 843–855. doi: 10.1111/j.1469-8137.2010.03231.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Du, Z. Y., Chen, M. X., Chen, Q. F., Gu, J. D., and Chye, M. L. (2015). Expression of Arabidopsis acyl-CoA-binding proteins AtACBP1 and AtACBP4 confers Pb(II) accumulation in Brassica juncea roots. Plant Cell Environ. 38, 101–117. doi: 10.1111/pce.12382
Du, Z. Y., Chen, M. X., Chen, Q. F., Xiao, S., and Chye, M. L. (2013). Overexpression of Arabidopsis acyl-CoA-binding protein ACBP2 enhances drought tolerance. Plant Cell Environ. 36, 300–314. doi: 10.1111/j.1365-3040.2012.02574.x
Du, Z. Y., Xiao, S., Chen, Q. F., and Chye, M. L. (2010). Depletion of the membrane-associated acyl-Coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiol. 152, 1585–1597. doi: 10.1104/pp.109.147066
Engeseth, N. J., Pacovsky, R. S., Newman, T., and Ohlrogge, J. B. (1996). Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch. Biochem. Biophys. 331, 55–62. doi: 10.1006/abbi.1996.0282
Gao, W., Li, H. Y., Xiao, S., and Chye, M. L. (2010). Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. Plant J. 62, 989–1003. doi: 10.1111/j.1365-313X.2010.04209.x
Gao, W., Xiao, S., Li, H. Y., Tsao, S. W., and Chye, M. L. (2009). Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein ATFP6. New Phytol. 181, 89–102. doi: 10.1111/j.1469-8137.2008.02631.x
Guo, R., Shi, L. X., Yang, C. W., Yan, C. R., Zhong, X. L., Liu, Q., et al. (2016). Comparison of ionomic and metabolites response under alkali stress in old and young leaves of cotton (Gossypium hirsutum l.) seedlings. Front. Plant Sci. 7:1785. doi: 10.3389/fpls.2016.01785
Hills, M. J., Dann, R., Lydiate, D., and Sharpe, A. (1994). Molecular cloning of a cDNA from Brassica napus L. for a homologue of acyl-CoA-binding protein. Plant Mol. Biol. 25, 917–920. doi: 10.1007/BF00028886
Mandrupl, S., Hummell, R., Ravn, S., Jensen, G., Andreasen, P. H., Gregersen, N., et al. (1992). Acyl-CoA-binding protein/diazepam-binding inhibitor gene and pseudogenes: a typical housekeeping gene family. J. Mol. Biol. 228, 1011–1022. doi: 10.1016/0022-2836(92)90888-Q
Metzner, M., Ruecknagel, K. P., Knudsen, J., Kuellertz, G., Mueller-Uri, F., and Diettrich, B. (2000). Isolation and characterization of two acyl-CoA-binding proteins from proembryogenic masses of Digitalis lanata Ehrh. Planta 210, 683–685. doi: 10.1007/s004250050060
Qiao, K., Takano, T., and Liu, S. K. (2015). Discovery of two novel highly tolerant NaHCO3 Trebouxiophytes: identification and characterization of microalgae from extreme saline-alkali soil. Algal Res. 9, 245–253. doi: 10.1016/j.algal.2015.03.023
Qin, P. F., Shang, X. G., Song, J., and Guo, W. Z. (2016). Genome-wide identification of Acyl-CoA-binding protein (ACBP) gene family and their functional analysis in abiotic stress tolerance in cotton. Acta Agron. Sin. 42, 1577–1591. doi: 10.3724/SP.J.1006.2016.01577
Reddy, A. S., Ranganathan, B., Haisler, R. M., and Swize, M. A. (1996). A cDNA encoding acyl-CoA-binding protein from cotton. Plant Physiol.111:348.
Sheen, J. (2002). A Transient Expression Assay Using Arabidopsis MesophyII Protoplasts. Available at: http://genetics.mgh.harvard.edu/sheenweb/
Shi, W., Takano, T., and Liu, S. K. (2012). Isolation and characterization of novel bacterial taxa from extreme alkali–saline soil. World J. Microb. Biotechnol. 28, 2147–2157. doi: 10.1007/s11274-012-1020-7
Song, T. T., Xu, H. H., Sun, N., Jiang, L., Tian, P., Yong, Y. Y., et al. (2017). Metabolomic analysis of alfalfa (Medicago sativa L.) root-symbiotic rhizobia responses under alkali stress. Front. Plant Sci. 8:1208. doi: 10.3389/fpls.2017.01208
Suzui, N., Nakamura, S., Fujiwara, T., Hayashi, H., and Yoneyama, T. (2006). A putative acyl-CoA-binding protein is a major phloem sap protein in rice (Oryza sativa L.). J. Exp. Bot. 57, 2571–2576. doi: 10.1093/jxb/erl021
Wang, J., Shi, W., Takano, T., and Liu, S. K. (2011). Purification and resistance analysis of algae of soda soil in northeast China. Mol. Soil Biol.3, 1–4.
Xiao, S., and Chye, M. L. (2009). An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 47, 479–484. doi: 10.1016/j.plaphy.2008.12.002
Xiao, S., and Chye, M. L. (2011). New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 50, 141–151. doi: 10.1016/j.plipres.2010.11.002
Xiao, S., Gao, W., Chen, Q. F., Chan, S. W., Zheng, S. X., Ma, J. Y., et al. (2011). Overexpression of Arabidopsis acyl-CoA-binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22, 1463–1482. doi: 10.1105/tpc.110.075333
Xiao, S., Gao, W., Chen, Q. F., Ramalingam, S., and Chye, M. L. (2008). Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant J. 54, 141–151. doi: 10.1111/j.1365-313X.2008.03402.x
Ye, Z. W., Lung, S. C., Hu, T. H., Chen, Q. F., Suen, Y. L., Wang, M. F., et al. (2016). Arabidopsis acyl-CoA-binding protein ACBP6 localizes in the phloem and affects jasmonate composition. Plant Mol. Biol. 92, 717–730. doi: 10.1007/s11103-016-0541-0
Keywords: saline-alkali-tolerant (SAT) Chlorella, acyl-CoA-binding protein 1, gene expression, stress tolerance, in vitro filter-binding, phosphatidylcholine
Citation: Qiao K, Wang M, Takano T and Liu S (2018) Overexpression of Acyl-CoA-Binding Protein 1 (ChACBP1) From Saline-Alkali-Tolerant Chlorella sp. Enhances Stress Tolerance in Arabidopsis. Front. Plant Sci. 9:1772. doi: 10.3389/fpls.2018.01772
Received: 26 June 2018; Accepted: 14 November 2018;
Published: 28 November 2018.
Edited by:
Alejandra A. Covarrubias, National Autonomous University of Mexico, MexicoReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences, JapanWricha Tyagi, Central Agricultural University, India
Copyright © 2018 Qiao, Wang, Takano and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenkui Liu, c2hlbmt1aWxpdUBuZWZ1LmVkdS5jbg==
 Kun Qiao
Kun Qiao Min Wang3
Min Wang3 Tetsuo Takano
Tetsuo Takano Shenkui Liu
Shenkui Liu