- 1Laboratory of Chromosome Structure and Function, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany
- 2Laboratory of Genetic Basis of Plant Identification, Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia
- 3Laboratory of Molecular Karyology, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia
Five diploid Aegilops species of the Sitopsis section: Ae. speltoides, Ae. longissima, Ae. sharonensis, Ae. searsii, and Ae. bicornis, two tetraploid species Ae. peregrina (= Ae. variabilis) and Ae. kotschyi (Aegilops section) and hexaploid Ae. vavilovii (Vertebrata section) carry the S-genomes. The B- and G-genomes of polyploid wheat are also the derivatives of the S-genome. Evolution of the S-genome species was studied using Giemsa C-banding and fluorescence in situ hybridization (FISH) with DNA probes representing 5S (pTa794) and 18S-5.8S-26S (pTa71) rDNAs as well as nine tandem repeats: pSc119.2, pAesp_SAT86, Spelt-1, Spelt-52, pAs1, pTa-535, and pTa-s53. To correlate the C-banding and FISH patterns we used the microsatellites (CTT)10 and (GTT)9, which are major components of the C-banding positive heterochromatin in wheat. According to the results obtained, diploid species split into two groups corresponding to Emarginata and Truncata sub-sections, which differ in the C-banding patterns, distribution of rDNA and other repeats. The B- and G-genomes of polyploid wheat are most closely related to the S-genome of Ae. speltoides. The genomes of allopolyploid wheat have been evolved as a result of different species-specific chromosome translocations, sequence amplification, elimination and re-patterning of repetitive DNA sequences. These events occurred independently in different wheat species and in Ae. speltoides. The 5S rDNA locus of chromosome 1S was probably lost in ancient Ae. speltoides prior to formation of Timopheevii wheat, but after the emergence of ancient emmer. Evolution of Emarginata species was associated with an increase of C-banding and (CTT)10-positive heterochromatin, amplification of Spelt-52, re-pattering of the pAesp_SAT86, and a gradual decrease in the amount of the D-genome-specific repeats pAs1, pTa-535, and pTa-s53. The emergence of Ae. peregrina and Ae. kotschyi did not lead to significant changes of the S*-genomes. However, partial elimination of 45S rDNA repeats from 5S* and 6S* chromosomes and alterations of C-banding and FISH-patterns have been detected. Similarity of the Sv-genome of Ae. vavilovii with the Ss genome of diploid Ae. searsii confirmed the origin of this hexaploid. A model of the S-genome evolution is suggested.
Introduction
Evolutionary goat grasses, or Aegilops are closely related to wheat and contributed two of the three subgenomes of hexaploid bread wheat (Sears, 1969; Kihara, 1975; Feldman, 2001). The natural distribution area of the genus Aegilops L. covers the Mediterranean basin, southwestern and central Asia (Witcombe, 1983; Kimber and Feldman, 1987; Van Slageren, 1994; Kilian et al., 2011). Their center of origin is thought to be located in Transcaucasia (Hammer, 1980; Van Slageren, 1994), or in the Fertile Crescent (Kimber and Feldman, 1987). These regions contain the highest concentration of Aegilops species. Goat grasses inhabit a broad range of environments and are characterized by very wide adaptation. Owing to this, many goat grasses exhibit good resistance to fungal diseases and pests (Hammer, 1980; Gill et al., 1985; Makkouk et al., 1994; El Bouhssini et al., 1998; Monneveux et al., 2000; Schneider et al., 2008; Zhao et al., 2016), heat, drought or frost tolerance and cold hardiness (Limin and Fowler, 1985; Damania et al., 1992; Monneveux et al., 2000; Pradhan et al., 2012). Some Aegilops accessions are characterized by high grain quality and increased micronutrient content (Rawat et al., 2011; Farkas et al., 2014; Rakszegi et al., 2017) that can be used for wheat improvement. Although many agronomically useful genes have already been transferred from Aegilops to common wheat varieties or breeding lines (Knott and Dvorák, 1976; Schneider et al., 2008; Rawat et al., 2011; McIntosh et al., 2013; Zhang et al., 2015), their genetic potential in broadening genetic diversity of wheat is not fully exploited. Utilization of gene pool of Aegilops requires good knowledge of genetics and genomics of these species, including their karyotypes and chromosomal structures.
In addition to the great potential for wheat breeding, goat grasses can also be an attractive model for studying mechanisms of reticulate evolution. Depending on taxonomical system, the genus Aegilops is classified into 20 (Kihara, 1954), 22 (Zhukovsky, 1928; Eig, 1929; Van Slageren, 1994), 24 (Kimber and Feldman, 1987), 25 (Chennaveeraiah, 1960), or 26 species (Witcombe, 1983). These species are split into sections based on morphological criteria or genome composition. At present, the system suggested by Van Slageren (1994) is commonly accepted; therefore, we will follow this nomenclature. According to it, 10 Aegilops species are diploid and 12 – polyploid, that were formed as a result of hybridization of different diploid progenitors.
Based on a series of pioneering works of the famous Japanese geneticist (Kihara, 1937, 1949, 1954, 1957, 1963; Lilienfeld, 1951; Kihara et al., 1959), diploid Aegilops were divided into three major genomic groups, C, D, and S. The C-genome group included two species; the D-genome group included four species; and the S-genome group consisted of three species of the Sitopsis (Jaub. & Spach) Zhuk. section: Ae. longissima Schweinf. & Muschl. (including Ae. sharonensis Eig), Ae. bicornis (Forssk.) Jaub. & Spach, and Ae. speltoides Tausch (Kihara, 1937, 1949; Lilienfeld, 1951). A new diploid species of the Sitopsis section—Ae. searsii Feldman and Kislev ex Hammer, has been discovered later by Feldman and Kislev (1977). Analysis of the karyotype, meiotic chromosome pairing, pollen fertility and seed set in Ae. longissima x Ae. searsii hybrids showed that Ae. searsii possesses the S*-genome (Feldman et al., 1979; Yen and Kimber, 1990a).
Thus, current taxonomy recognizes five diploid species carrying the S-genome: Ae. speltoides including ssp. ligustica (Savign.) Fiori (SS) and ssp. speltoides Boiss., Ae. bicornis (SbSb), Ae. searsii (SsSs), Ae. sharonensis (SshSsh), and Ae. longissima (SlSl) (Van Slageren, 1994; Kilian et al., 2011; Feldman and Levy, 2015). These species are morphologically similar, but can be easily distinguished by their habitat, climatic adaptation, and distribution areas. Based on differences in spike morphology, Eig (1929) divided the Sitopsis group into two sub-sections, Truncata and Emarginata. Subsection Truncata includes only one species–Ae. speltoides (SS), which grows in central, eastern, and northern part of the Sitopsis area. This species consists of two forms, ligustica and auscheri, which differ in their fruiting spike and the mode of seed dispersal (Eig, 1929; Zohary and Imber, 1963), but are similar in karyotype structure (Chennaveeraiah, 1960). Their hybrids are fully fertile and show complete meiotic chromosome pairing (Zohary and Imber, 1963). Ae. speltoides has the lowest nuclear DNA content (1C = 5.81 ± 0.123 pg) within the Sitosis group (Eilam et al., 2007) and differs significantly from Emarginata species in its chromosome morphology (Chennaveeraiah, 1960), Giemsa C-banding (Teoh and Hutchinson, 1983; Friebe and Gill, 1996; Friebe et al., 2000) and FISH patterns (Yamamoto, 1992a,b; Jiang and Gill, 1994b; Badaeva et al., 1996a,b; Salina et al., 2006b; Raskina et al., 2011; Belyayev and Raskina, 2013).
The subsection Emarginata includes four species: Ae. bicornis, Ae. searsii, Ae. sharonensis, and Ae. longissima, which grow in the central and southern part of the Sitopsis section habitat (Feldman and Kislev, 1977). Study of the chromosome pairing of intraspecific hybrids (Kihara, 1954, 1963; Feldman et al., 1979; Yen and Kimber, 1989, 1990a,b,c), similarity of karyotype structure (Riley et al., 1958; Chennaveeraiah, 1960), the number and distribution of 5S and 45S rDNA loci (Yamamoto, 1992a,b; Badaeva et al., 1996b), and the distribution of pSc119.2 sequence (Badaeva et al., 1996a) suggest a close relationship of Emarginata species, although they differ from each other in genome size (Eilam et al., 2007) and C-banding patterns (Friebe and Gill, 1996).
Morphologically, Ae. bicornis is the most primitive species in this group (Eig, 1929). It is more difficult to produce hybrids with Ae. bicornis than with other Aegilops of the S-genome group (Kimber and Feldman, 1987). Genome size of Ae. bicornis (1C = 6.84 ± 0.097 pg) is only little larger than that of Ae. searsii (1C = 6.65 ± 0.091 pg), and is lower than of Ae. longissima (1C = 7.48 ± 0.082 pg) or Ae. sharonensis (1C = 7.52 ± 1.000 pg) (Eilam et al., 2007). Morphologically Ae. searsii resembles Ae. longissima, but differs from it in a number of morphological traits which are considered as evolutionary advanced (Feldman and Kislev, 1977). Ae. longissima x Ae. searsii hybrids exhibit meiotic irregularities and are highly sterile (Feldman et al., 1979). By contrast, the F1 hybrids Ae. longissima x Ae. sharonensis are fertile and show complete chromosome pairing in meiosis. Isolation of these species is caused by different ecological requirements (Feldman and Levy, 2015). According to other hypothesis (Waines and Johnson, 1972), Ae. sharonensis could be a hybrid between Ae. longissima and Ae. bicornis. Ae. longissima carries a species-specific 4S*/7S* translocation (Tanaka, 1955; Yen and Kimber, 1990b; Friebe et al., 1993; Naranjo, 1995), while no structural rearrangements have been identified in other species of this group (Yen and Kimber, 1989, 1990a,b,c; Maestra and Naranjo, 1997, 1998; Luo et al., 2005; Dobrovolskaya et al., 2011).
The similarity of Emarginata species and separate position of Ae. speltoides within the Sitopsis section was confirmed by molecular analyses of nuclear and cytoplasmic DNA. Based on the variation of repeated nucleotide sequences (RNS) Dvorák and Zhang (1992) showed that the Sitopsis species are phylogenetically similar, but Ae. speltoides is clearly separated from species of the Emarginata group. RAPD- and AFLP analyses revealed that Ae. speltoides forms a cluster with polyploid wheats, which is separated from other Sitopsis species (Kilian et al., 2007, 2011; Goryunova et al., 2008). Study of organellar DNAs by PCR-single-strand conformational polymorphism (PCR-SSCP) revealed high similarity of Ae. bicornis - Ae. sharonensis - Ae. longissima plasmons and their distinctness from plasmon of Ae. speltoides (Wang et al., 1997).
Comparative sequence analysis provided further insights into the evolution of Triticum and Aegilops and allowed the estimation of divergence time of different genomic groups. Comparison of chloroplast (Yamane and Kawahara, 2005; Golovnina et al., 2007; Gornicki et al., 2014; Middleton et al., 2014; Bernhardt et al., 2017) and nuclear DNA sequences (Petersen et al., 2006; Salse et al., 2008; Marcussen et al., 2014) strongly suggest that Ae. speltoides occupies a basal position on the phylogenetic tree of Aegilops/Triticum (Petersen et al., 2006; Kawahara, 2009). Probably Ae. speltoides diverged from the progenitor of the Triticeae much earlier than diploid wheat and Aegilops species (Yamane and Kawahara, 2005; Salse et al., 2008; Gornicki et al., 2014; Middleton et al., 2014; Bernhardt et al., 2017). Estimates obtained from the analyses of nuclear DNA sequences placed the possible divergence time within the period from ~7 MYA (Marcussen et al., 2014) to 3.5–2.7 MYA (Dvorák and Akhunov, 2005; Salse et al., 2008). Estimates obtained from chloroplast DNA favored a more recent origin of Ae. speltoides – 4.1–3.6 MYA (Bernhardt et al., 2017) to 2.67 ± 1.1. MYA (Middleton et al., 2014). Marcussen et al. (2014) supposed that the D-genome lineage (which indeed included D, M, and S* genome species, Sandve et al., 2015) emerged through ancient homoploid hybridization between A and S genomes. The members of Emarginata group are thought to radiate from common ancestor approximately 1.0–2.0 MYA (Ae. searsii) – 1.4 MYA (Ae. bicornis) – to 0.4 MYA (Ae. sharonensis) (Marcussen et al., 2014; Feldman and Levy, 2015).
Hypothesis that the B and G genomes of polyploid wheats originated from a diploid S-genome Aegilops species was put forward in the middle XXth (Sears, 1956; Riley et al., 1958). Different taxa were suggested as potential progenitors of polyploid wheat (Haider, 2013). All species of the Sitopsis section have been considered as the B-genome donors: Ae. speltoides (Sarkar and Stebbins, 1956; Tanaka et al., 1979; Bahrman et al., 1988; Kerby et al., 1990; Daud and Gustafson, 1996; Maestra and Naranjo, 1998; Yan et al., 1998; Blake et al., 1999; Rodríguez et al., 2000a; Haider, 2013), Ae. bicornis (Sears, 1956), Ae. longissima (Tanaka, 1956; Konarev et al., 1976; Konarev, 1980; Peacock et al., 1981), Ae. searsii (Feldman and Kislev, 1977; Nath et al., 1983, 1984; Kerby et al., 1990; Liu et al., 2003), Ae. sharonensis (Kushnir and Halloran, 1981) or yet unknown species of the Emarginata group (Kerby et al., 1990). Molecular analyses of common wheat genome and genomes of related species confirmed the ancestry of wheat B- genome from Ae. speltoides or the species close to it (Talbert et al., 1991; Petersen et al., 2006; Goryunova et al., 2008; Salse et al., 2008; Marcussen et al., 2014). Based on the analysis of nuclear or plastid DNA, ancient tetraploid emmer could emerge 0.4–0.8 MYA (Huang et al., 2002; Dvorák and Akhunov, 2005; Yamane and Kawahara, 2005; Golovnina et al., 2007; Gornicki et al., 2014; Marcussen et al., 2014; Middleton et al., 2014; Bernhardt et al., 2017).
The origin of the G-genome of Triticum timopheevii Zhuk. from the S-genome of Ae. speltoides was first hypothesized by Giorgi and Bozzini (1969) based on comparison of chromosome morphologies and was later confirmed by numerous studies including chromosome pairing analysis of intraspecific hybrids (Shands and Kimber, 1973; Tanaka et al., 1979; Maestra and Naranjo, 1999; Rodríguez et al., 2000a), comparison of C-banding (Badaeva et al., 1996a) and ISH patterns (Jiang and Gill, 1994a,b; Salina et al., 2006b), isozyme profiles (Konarev et al., 1976; Nakai, 1978; Jaaska, 1980), AFLP- (Kilian et al., 2007, 2011) and RFLP-analyses (Dvorák and Zhang, 1990; Talbert et al., 1991; Dvorák, 1998), sequencing of nuclear (Huang et al., 2002) and cytoplasmic DNA (Sasanuma et al., 1996; Yamane and Kawahara, 2005; Golovnina et al., 2007; Gornicki et al., 2014). These studies revealed that Ae. speltoides is more closely related to the G genome of T. timopheevii than to the B-genome of common wheat and suggested that ancient T. timopheevii could emerge approximately 0.4 MYA (Huang et al., 2002; Gornicki et al., 2014).
The S*-genome is identified in two tetraploid Aegilops species belonging to the section Aegilops L.: Ae. peregrina (Hach. in Fraser) Maire & Weiller (= Ae. variabilis Eig, UpUpSpSp) and Ae. kotschyi Boiss. (UkUkSkSk). Based on the “analyzer” method H. Kihara (1954) proposed that Ae. peregrina is a hybrid between Ae. umbellulata Zhuk. and a diploid species of the Sitopsis group (Lilienfeld, 1951), although conventional chromosome staining did not reveal the S*-genome in these species (Chennaveeraiah, 1960). Cytoplasmic genomes of Ae. peregrina and Ae. kotschyi are most closely related to the cytoplasmic genome of Ae. searsii (Ogihara and Tsunewaki, 1988; Siregar et al., 1988). However, meiotic analysis of the F1 hybrids between Ae. kotschyi and induced autotetraploid of three Sitopsis species showed that Ae. kotschyi shared the S* genome with Ae. longissima (Yen and Kimber, 1990d). Yu and Jahier (1992) come to the same conclusion based on chromosome pairing analysis in hybrids of Ae. variabilis (= Ae. peregrina) with different Sitopsis species. RFLP profiles of RNS suggested that the S* genome of Ae. peregrina and Ae. kotschyi could have originated from Ae. longissima or Ae. sharonensis or the species immediately preceding the divergence of these diploids (Zhang et al., 1992). C-banding and FISH analyses confirmed highest similarity of the S*-genome of these tetraploids with Ae. longissima or Ae. sharonensis (Jewell, 1979; Jewell and Driscoll, 1983; Friebe et al., 1996; Badaeva et al., 2004; Zhao et al., 2016).
Ae. vavilovii (Zhuk.) Chennav. (D1D1XcrXcrSvSv) is a hexaploid taxa belonging to section Vertebrata Zhuk. emend Kihara, complex Crassa. Ae. vavilovii originated from hybridization of tetraploid Ae. crassa Boiss. with a species of Emarginata group, possibly Ae. longissima (Kihara, 1963; Kihara and Tanaka, 1970). Originally Ae. vavilovii was treated as a subspecies of hexaploid Ae. crassa, and its taxonomic rank was raised to independent biological species by Chennaveeraiah (1960). Although this author was unable to determine genome constitution of Ae. vavilovii, he noticed a pairwise similarity of the satellite chromosomes in karyotype of this species.
Yen and Kimber (1992) failed to identify the exact donor of the Sv-genome of Ae. vavilovii based on analysis of chromosome pairing in the F1 hybrids of Ae. vavilovii with induced autotetraploids of the Sitopsis species and proposed that the Sv-genome is substantially modified. By using molecular markers (Talbert et al., 1991) showed that the Sv-genome of Ae. vavilovii is related to the S*-genome of Emarginata group. Data collected by molecular methods (Zhang and Dvorák, 1992), C-banding and FISH analyses (Badaeva et al., 2002; Zhang et al., 2002) confirmed, that Ae. vavilovii contains the Sv-genome that could probably derive from Ae. searsii (Badaeva et al., 2002).
Because of the genetic relatedness of the S-genome Aegilops species and polyploid wheats as well as of their potential for wheat improvement, they have been attracting the attention of researchers over the past century. Numerous intraspecific hybrids have been created to transfer desired genes from Aegilops to wheat (Schneider et al., 2008). Sets of addition, substitution or translocation wheat-Aegilops lines, including Ae. speltoides (Friebe et al., 2000; Liu et al., 2016), Ae. searsii (Pietro et al., 1988; Friebe et al., 1995), Ae. sharonensis (Olivera et al., 2013), Ae. longissima (Friebe et al., 1993), and polyploid Ae. peregrina, (Jewell and Driscoll, 1983; Friebe et al., 1996; Yang et al., 1996) and Ae. kotschyi (Rawat et al., 2011) were obtained and characterized using a combination of C-banding and analyses with the group-specific molecular or isozyme markers. As a result of these studies, the genetic classifications were developed for C-banded chromosomes of several S-genome species (Friebe and Gill, 1996).
From another side, the S-genomes were extensively examined by FISH with various DNA probes (Yamamoto, 1992a; Badaeva et al., 1996a,b, 2002, 2004; Belyayev et al., 2001; Zhang et al., 2002; Giorgi et al., 2003; Salina et al., 2006b, 2009; Raskina et al., 2011; Ruban et al., 2014; Molnár et al., 2016; Zhao et al., 2016). Probe pSc119.2 was used most frequently (Badaeva et al., 1996a, 2002, 2004; Molnár et al., 2016; Zhao et al., 2016), however, the pSc119.2 signals are located predominantly in subtelomeric chromosome regions, thus hindering unequivocal chromosome identification. Probe pAs1, which proves to be highly informative for many Aegilops species, is not very useful for the S-genome analysis owing to a small number of detected sites (Badaeva et al., 1996a). In most papers FISH-labeled Aegilops chromosomes were classified based on their morphology, which is not sufficient to determine their correspondence to the genetic nomenclature of C-banded chromosomes. Owing to this, it was necessary to find FISH markers for the precise identification of all S-genome chromosomes and coordination of classification systems.
Recently, Komuro et al. (2013) isolated and characterized a number of repetitive DNAs from the wheat genome, which can potentially be used for molecular-cytogenetic analysis of wheat and Aegilops species. Several new sequences have been described in other papers (Salina et al., 1998, 2009; Adonina et al., 2015; Badaeva et al., 2015; Zhao et al., 2016). In this study we characterized the S genomes of diploid and polyploid Triticum and Aegilops species using C-banding and FISH with a set of “classical” [pSc119.2, pAs1, pTa71, pTa794, Spelt-1, Spelt-52] and novel [pAesp_SAT86, (CTT)n, (GTT)n, pTa-535, pTa-s53] probes in order to assess evolutionary changes in the Triticum-Aegilops alliance.
Materials and Methods
Plant Material
Five diploid (Aegilops speltoides, Ae. longissima, Ae. sharonensis, Ae. searsii, Ae. bicornis), two tetraploid (Ae. peregrina and Ae. kotschyi) and one hexaploid (Ae. vavilovii) Aegilops species carrying the S-genome have been examined in comparison with two tetraploid wheats, T. timopheevii and T. dicoccoides. The list of accessions, their ploidy level, genome constitution and the origin are given in Table S1.
DNA Probes
Following probes were used for FISH:
Plasmid clones pTa71 - a 9 kb long sequence of common wheat encoding 18S, 5.8S and 26S rRNA genes including spacers (Gerlach and Bedbrook, 1979), pTa794 – a 420 bp long sequence of wheat containing the 5S rRNA gene and intergenic spacer (Gerlach and Dyer, 1980), pAs1 - a 1 kb fragment derived from Ae. tauschii and belonging to Afa family (Rayburn and Gill, 1986), pSc119.2 – a 120 bp long sequence isolated from rye (Bedbrook et al., 1980), pTa-s53 – a 587 bp DNA fragment isolated from common wheat (Komuro et al., 2013), Spelt-1 – a 150 bp fragment isolated from Ae. speltoides (Salina et al., 1997), Spelt-52 (homolog of pAesKB52) – a 276 bp long DNA fragment isolated from Ae. speltoides (Salina et al., 2004a), and pAesp_SAT86 - a new satellite family with a monomer length of 86 bp isolated from Ae. speltoides genomic DNA (Badaeva et al., 2015) and showing 91-94% similarity to wheat repeat pTa-713 described in Komuro et al. (2013) were labeled with dUTP-ATTO-488, dUTP-ATTO-550, dUTP-ATTO-647N by nick-translation using an Atto NT Labeling Kit (Jena Bioscience, Germany) or with FITC (fluorescein-12-dUTP, Roche, Germany) or biotin (biotin-16-dUTP, Roche, Germany) by nick-translation using the Nick Translation Mix (Roche, Germany) according to manufacturers' instruction.
Probe pTa535-1 was used as 5′ 6-carboxyfluorescein (6-FAM) or 6-carboxytetramethylrhodamine (TAMRA) end-labeled (MWG, Germany) oligo probe (5′-AAA AAC TTG ACG CAC GTC ACG TAC AAA TTG GAC AAA CTC TTT CGG AGT ATC AGG GTT TC-3′) (Komuro et al., 2013; Tang et al., 2014).
The oligo-(CTT)10 or complementary oligo-(GAA)10 probes [thereafter (CTT)n] were labeled with 5/6-Sulforhodamine 101-PEG3-Azide or 6-Carboxyfluorescein Azide by click chemistry (Baseclick, Germany).
The oligo-(GTT)9 probe labeled at the 3′-end with fluorescein-12-dUTP was synthesized in the Laboratory of Biological Microchips at the Engelhardt Institute of Molecular Biology, Moscow, Russia.
Giemsa C-Banding Method
The Giemsa C-banding method described in Badaeva et al. (1994) was used for analysis. Seeds were soaked in water for 24 h at room temperature and then kept at 4°C overnight on wet filter paper in Petri dishes. For the next 24 h Petri dishes were placed at 24°C. Roots were cut and treated with 0.05% colchicine for 3 h. Further, roots were fixed in 45% acetic acid for 4 h, washed with distilled water and treated with 0.2 N HCl for 15 min at 4°C and for 5 min at 60°C. After overnight treatment with a 4 mg/ml Cellulysine (Fluka, Switzerland) solution at 24°C root meristems were squashed in drop of 45% acetic acid. Slides were frozen in liquid nitrogen and coverslips were removed. After that slides were placed into 96% ethanol at room temperature. Chromosomes of wheat were classified according to nomenclature suggested in Gill et al. (1991), Badaeva et al. (2016); chromosomes of Aegilops species were classified according to the nomenclature of Friebe et al. (1993, 1995, 1996, 2000), Friebe and Gill (1996). Karyotype of one typical accession per each species was taken as standard for alignment of C-banding and FISH patterns.
Fluorescence in situ Hybridization
Detailed protocols of the pretreatment of the materials, fixation and chromosomal preparation are given in Badaeva et al. (2017). Briefly, seeds were germinated in Petri dishes on wet filter paper at 24°C in dark. Roots were excised when 2 cm long, treated with ice-cold water for 24 h, and fixed with ethanol:acetic acid (3:1) fixative for at least 4 days at room temperature. Before slide preparation roots were stained in 2% acetocarmine for 15 min. Meristems were cut off and squashed in a drop of 45% acetic acid. Slides were frozen in liquid nitrogen and coverslips were removed with a razor blade. The slides were kept in 96% ethanol in a freezer.
Hybridization mixture contained 1 g dextran sulfate dissolved in 1 ml of distilled water, 5 ml deionized formamide, 1 ml of 20x SSC, 1 ml Herring sperm DNA (10 mg/ml, Promega, USA). Per slide 40–60 ng of each labeled probe were added to 18 μl hybridization mixture. Post hybridization washes were carried out as follows: for probes labeled with biotin or fluorescein the slides were washed in 0.1x SSC 2 × 10 min, then in 2x SSC 2 × 10 min at 42°C. Slides hybridized with directly labeled probes were washed at 58°C in 2x SSC for 20 min. The probes labeled with fluorescein were detected using anti-fluorescein/Oregon green®, rabbit IgG fraction, Alexa Fluor® 488 conjugated antibody (Molecular Probes, USA). Biotin was detected with sptreptavidin-Cy3 (Amersham Pharmacia Biotech, USA). The slides were counter-stained with DAPI (4′,6-diamidino-2-phenylindole) in Vectashield mounting media (Vector laboratories, Peterborough, UK) and examined with a Zeiss Imager D-1 microscope. Selected metaphase cells were captured with an AxioCam HRm digital camera using software AxioVision, version 4.6. Images were processed in Adobe PhotoshopR, version CS5 (Adobe Systems, Edinburgh, UK). For classification, chromosomes were aligned with the C-banding patterns based on the hybridization patterns of labeled CTT- and GTT-satellite sequences.
Results
Analysis of Diploid Species
According to the C-banding and FISH patterns of nine probes, five diploid species of the Sitopsis section split into two groups corresponding to taxonomically recognized sub-sections Truncata (Ae. speltoides) and Emarginata (Ae. longissima, Ae. sharonensis, Ae. searsii, Ae. bicornis).
Sub-section Truncata: Ae. speltoides
The karyotype of Ae. speltoides consists of metacentric or submetacentric chromosomes; the chromosome pairs 1S and 6S carry large satellites in their short arms (Figure S1). All chromosomes contain large Giemsa-positive pericentromic heterochromatin, prominent subtelomeric C-bands, and some small or medium sized interstitial bands. Giemsa-patterns allowed the identification of all Ae. speltoides chromosomes. We observed significant variations of Giemsa bands between plants within and between accessions. Heteromorphism of homologous chromosome has been recorded in all studied genotypes (Figure S1).
The (CTT)10 clusters (Figure 1, CTT) are located in proximal and interstitial chromosome regions, overlapping with Giemsa N-bands (Jiang and Gill, 1994b). No (CTT)10 signals were found in the sub-telomeric parts of the chromosomes possessing C-bands. The (GTT)9 probe forms prominent proximal clusters (Figure 1, GTT, Figures 2, 3f), often exceeding the size of (CTT)n-signals. The abundance of the GTT-microsatellite is an important diagnostic feature of Ae. speltoides chromosomes.
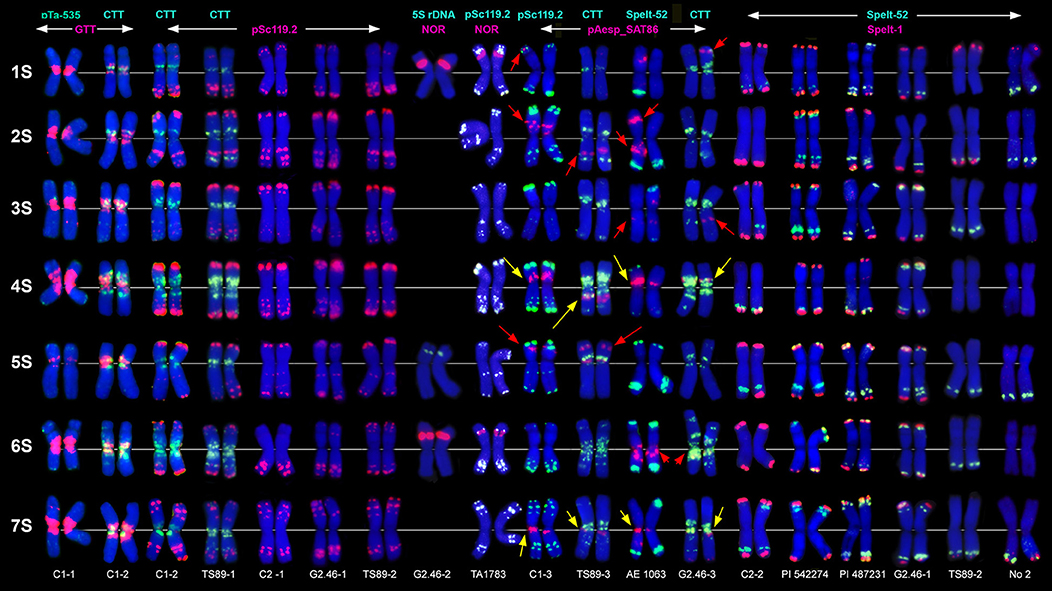
Figure 1. Localization of different DNA sequences on chromosomes of Ae. speltoides. Probe combinations are shown on the top; signal color corresponds to probe name. Accessions numbers are indicated in the bottom: C1-1–C1-3 genotypes from Technion park, Haifa, Israel; TS89-1–TS89-3–genotypes from Katzir, Israel; C2-1–C2-2–genotypes from Nahal Mearot, Israel; G2.46-1–G2.46-3–genotypes from Ramat haNadiv, Israel. Permanent pAesp_SAT86 loci are indicated with yellow arrows; polymorphic sites are shown with red arrows.
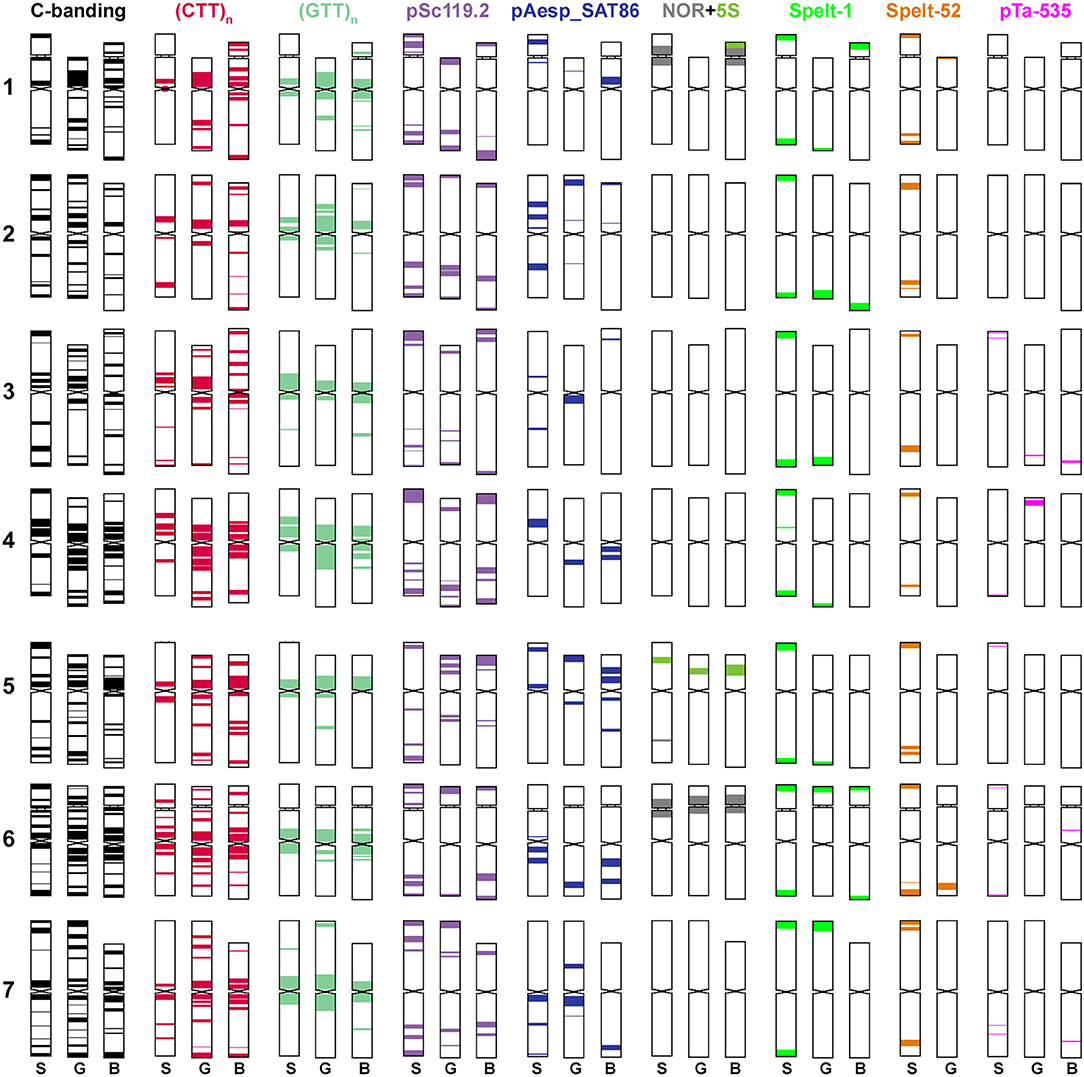
Figure 2. Idiogram showing relative positions of C-bands and nine DNA probes (probe names are given on the top) on chromosomes of Ae. speltoides (S), the B-genome (B) of common wheat and G-genome (G) of T. timopheevii.
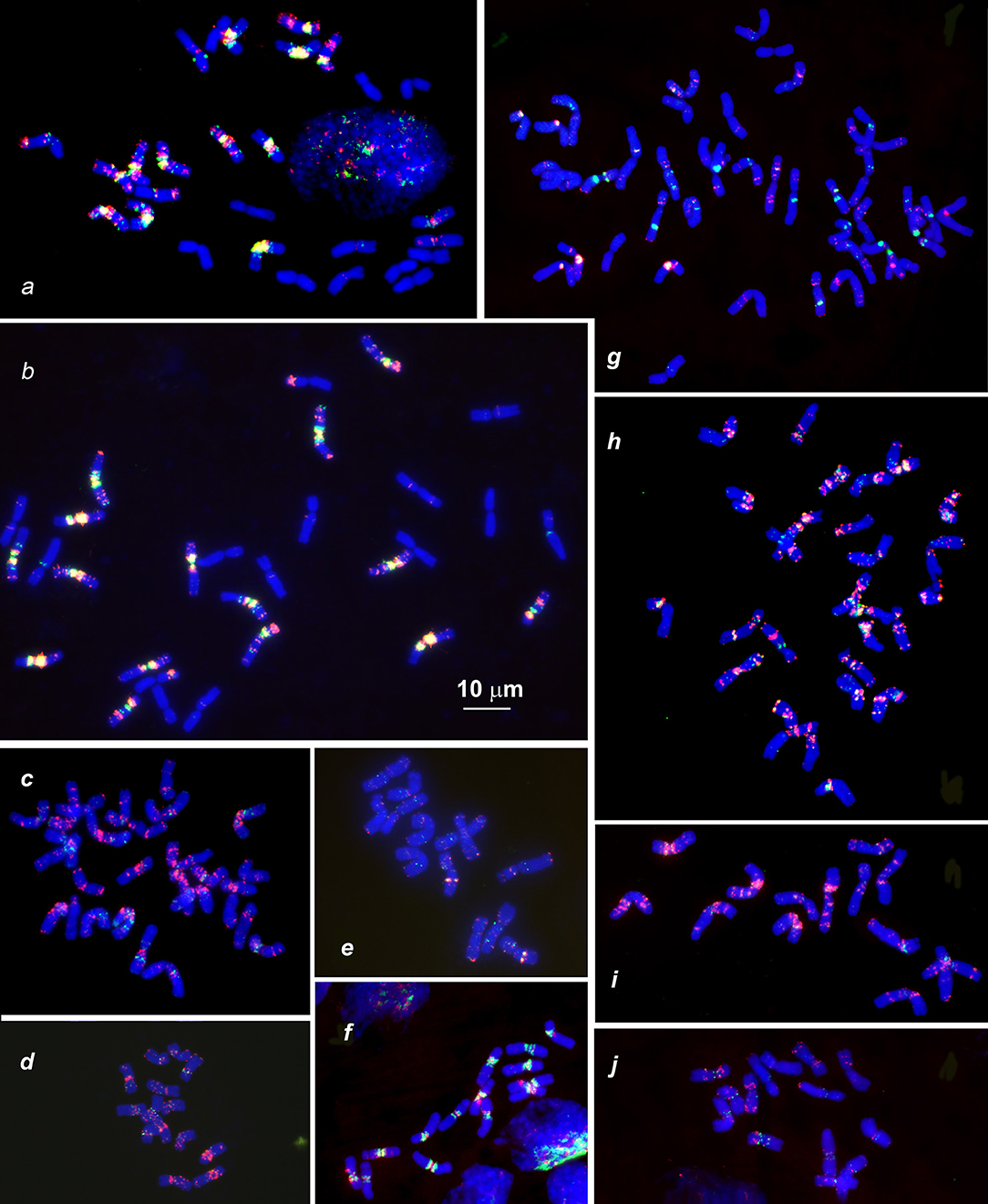
Figure 3. Distribution of (CTT)10 and (GTT)9 probes (red and green colors respectively) on chromosomes of wheat and Aegilops species: (a), Triticum dicoccoides (IG 46396); (b), T. araraticum (IG 116164); (c), Ae. kotschyi (TA2206); (d), Ae. longissmia (AE 904); (e), Ae. searsii (AE 1071); (f), Ae. speltoides (C1, Technion park, Israel); (g), Ae. vavilovii (K-3637); (h), Ae. peregrina (C11, Nahal Mearot, Israel); (i), Ae. sharonensis (C6, Keshon, Israel); (j), Ae. bicornis (K-666). Scale bar, 10 μm.
The pSc119.2 labeling patterns are represented by subtelomeric and interstitial signals allowing the discrimination of all Ae. speltoides chromosomes. Some hybridization sites are found in all genotypes, whereas other vary in the presence and signal size (Figure 1). Based on dual-color FISH with (CTT)10 and pSc119.2 probes we corrected previously published classification of pSc119.2-labeled chromosomes (Badaeva et al., 1996a) according to the genetic nomenclature (Friebe et al., 2000). In particular, the chromosomes 2S and 3S have been renamed.
Major NORs are detected on chromosomes 1S and 6S, and one pair of 5S rDNA loci are mapped on the chromosome 5S (Figures 1, 2). In addition, accession TA1873 shows one minor site on the long arm of one 5S chromosome.
Repeat pAesp_SAT86 exhibits significant variation of labeling patterns between Ae. speltoides genotypes (Figure 1). Two sites located in the short arm of 4S and pericentromeric region of 7SL are permanent (Figure 1, yellow arrows). In genotypeTS89 this repeat is transferred to the long arm of 4S, probably due to a pericentric inversion. Several facultative pAesp_SAT86 sites were found in more than one genotype (Figure 1, red arrows), while some signals were detected in single genotypes on either one or both homologous chromosomes.
The labeling patterns of Spelt-1 and Spelt-52 probes are highly polymorphic (Figure 1). The Spelt-1 sequence is located in subtelomeric regions of either one or two chromosome arms. The number of loci per diploid genome varied from six (TS89 Katzir and No2 from Turkey) to 27 (PI 542274 from Turkey). Genotypes differ from each other in the size and chromosome location of the Spelt-1 clusters. The Spelt-52 signals of variable size are located in distal chromosome regions, proximally to Spelt-1. The number of Spelt-52 clusters per diploid genome varied from eight to 22 (Figure 1), the size and chromosomal distribution are highly polymorphic. Genotypes differ from each other in a ratio of Spelt-1/Spelt-52 repeats. Thus, the Spelt-1 could significantly prevail over Spelt-52, or the Spelt-52 could be more abundant (Figure 1).
Only few inconsistent, dot-like pTa-535 signals have been detected in Ae. speltoides (Figure 1). No hybridization was found with pAs1 and pTa-s53 probes.
Sub-section Emarginata
Four species of the Emarginata sub-section have a similar karyotype, which is distinct from that of Ae. speltoides (Figures S1, S2). Chromosome pairs 5S* and 6S* carry unequal satellites: large on 6S* and small on 5S* chromosomes (Figure S2). Most Ae. sharonensis genotypes collected in Keshon (Israel) are heterozygotes (Figures S2c1,c2) indicating that open pollination is common in this population.
The karyotypes of Emarginata species differ in heterochromatin content detected by Giemsa staining. Ae. bicornis and Ae. searsii showed small-to-medium C-bands located in interstitial chromosome regions (Figures S2a1–b4). Ae. sharonensis and Ae. longissima exhibit prominent pericentromeric and subtelomeric and many interstitial C-bands (Figures S2c1–d10). C-banding patterns allowed the chromosome identification in all Emarginata species. A species-specific translocation between 4S* and 7S* is found in all Ae. longissima accessions.
The (CTT)10-hybridization pattern (Figures 3d,e,i,j, 4, 5) corresponds to the C-banding pattern. As expected, Ae. bicornis and Ae. searsii carry predominantly small CTT-signals (Figures 3e,j), while Ae. sharonensis and Ae. longissima possess prominent pericentromeric and distinct interstitial CTT-clusters. In contrast to Ae. speltoides, the (GTT)9 probe hybridizes poorly on the chromosomes of Emarginata species. Probably, accumulation of heterochromatin in this evolutionary lineage was mainly due to amplification of CTT-repeat, contributing to an increase of nuclear DNA content in Ae. sharonensis/Ae. longissima genomes as compared to Ae. bicornis/Ae. searsii (Eilam et al., 2007).
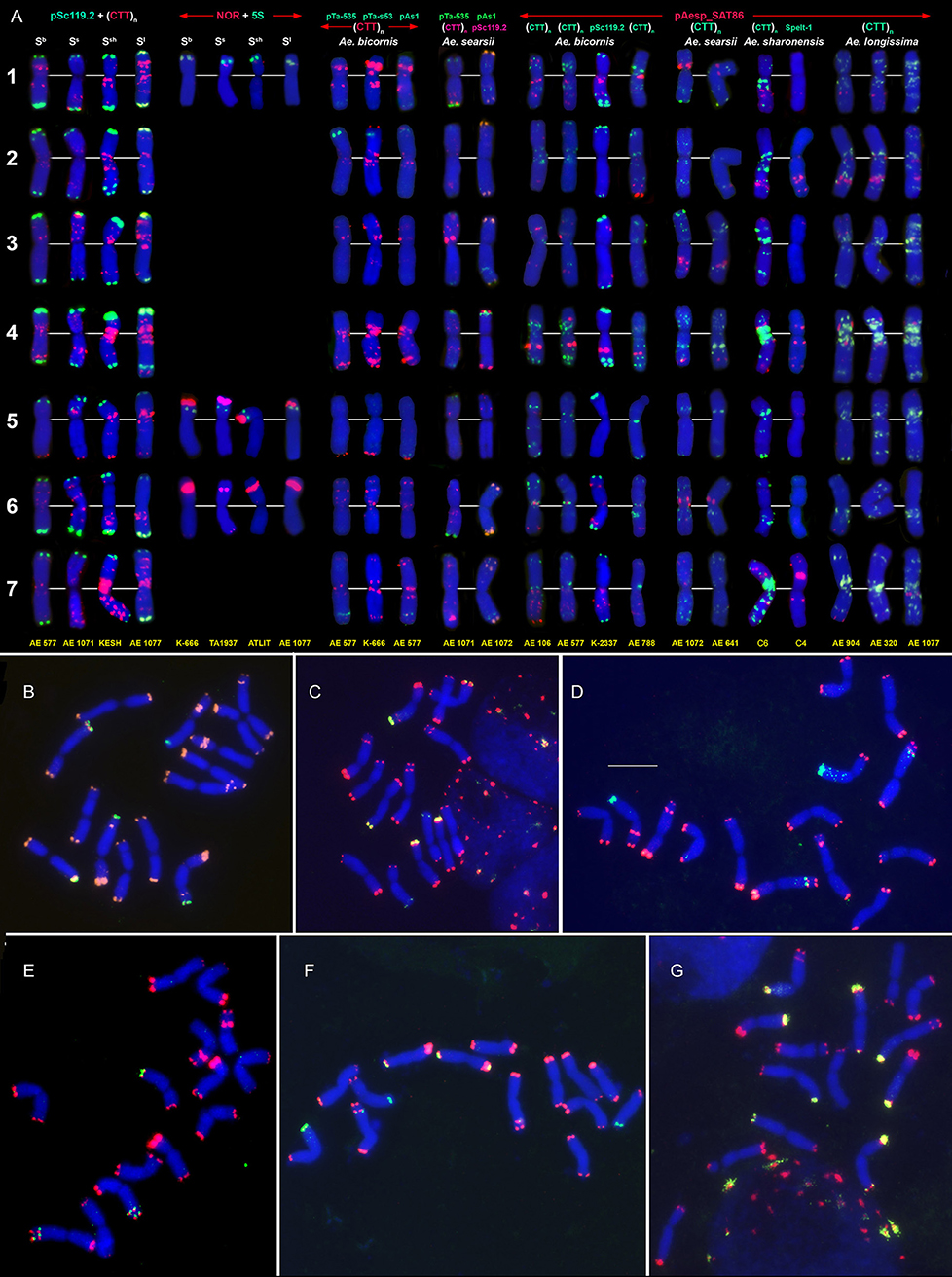
Figure 4. Distribution of repeated DNA sequences on chromosomes of four species of Emarginata group (A), Ae. bicornis (Sb), Ae. searsii (Ss), Ae. sharonensis (Ssh), and Ae. longissima (Sl). (A) Probe combinations are given on the top, accession names are shown below karyograms. Signal color corresponds to probe name. 1–7 – homoeologous groups. Polymorphisms of Spelt-52 patterns on Ae. longissima (B–D) and Ae. sharonensis (E–G) chromosomes: (B), K-905; (C), K-907; (D), C3 (HaBonim); (E), C6 (Keshon); (F), C7 (HaBonim); (G), i-570030. The pSc119.2 signals are shown in red, Spelt-52–in green. Scale bar, 10 μm.
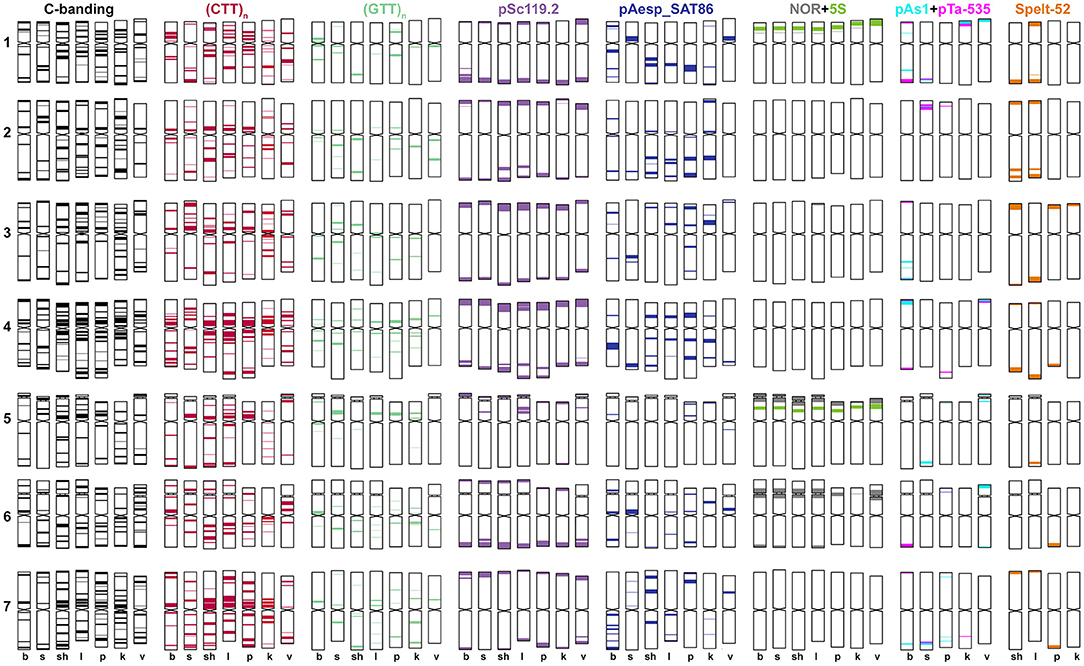
Figure 5. Idiogram showing relative positions of C-bands and nine DNA probes (probe names are given on the top) on the S*genome chromosomes of Ae. bicornis (b), Ae. searsii (s), Ae. sharonensis (sh), Ae. longissima (l), Ae. peregrina (p), Ae. kotschyi (k), and Ae. vavilovii (v).
Emarginata species display similar pSc119.2 hybridization patterns consisting of subtelomeric signals of variable size in one or both chromosome arms. Interstitial loci were rarely found (Figures 4, 5). Permanent interstitial sites are found on 2S*S (Ae. sharonensis and Ae. bicornis), 4SlL (Ae. longissima), and 7Ss (Ae. searsii) only. The pSc119.2 cluster in the middle of 5S*S is present in all Ae. searsii accessions and some Ae. longissima and Ae. sharonensis lines (Figure 4). One or two polymorphic pSc119.2 sites were rarely observed on 1SbL and 4SbL of Ae. bicornis.
The number and location of 5S and 45S rDNA loci in Emarginata species is similar and differ from that in Ae. speltoides (Figures 1, 4). Major NORs are located on 5S*S and 6S*S and permanent minor NORs are found on 1S*S (Figure 4). Additional minor sites were detected in the terminal region of 6S*L of all Ae. searsii accessions and some lines of Ae. bicornis and Ae. longissima. All species possess two 5S rDNA loci located in the short arms of chromosome 1S* and 5S*, distally (1S*) or proximally (5S*) to the 45S rDNA loci.
The distribution of pAesp_SAT86 signals is species- and chromosome-specific. An intraspecific polymorphism was detected in Ae. bicornis and Ae. longissima, labeling patterns are virtually invariable in Ae. searsii (Figures 4, 5). Distribution of pAesp_SAT86 clusters on Ae. sharonensis and Ae. longissima chromosomes is similar and differs from Ae. bicornis and Ae. searsii, which, in turn, are clearly distinct from each other. No similarity between homoeologous chromosomes of different species has been observed, though the chromosome 3Ss (Ae. searsii) shows almost the same distribution of pAesp_SAT86 sequence as the chromosomes 2S* of Ae. sharonensis and Ae. longissima.
The Spelt-1 repeat was not found in any Emarginata species, and Spelt-52 is detected in Ae. sharonensis and Ae. longissima only (Figures 4B–G). Signals of variable size are located in terminal regions of either one or both arms of all chromosomes except 6S*. Only two interstitial loci are found in the long arms of 2S* and 4S*. Distribution of Spelt-52 is highly diverse and polymorphisms are often observed even between homologous chromosomes. Depending on genotype, the number of signals ranges from 0 to 14. Most Ae. longissima accessions carry a Spelt-52 site in the long arm of 5S*, while it is absent in six out of 8 Ae. sharonensis accessions (Table S2). No other differences in labeling patterns were found between these species.
Distinct signals of the D-genome specific probes pAs1, pTa-535 or pTa-s53 are revealed in Ae. bicornis and Ae. searsii only (Figures 4A, 5). Two small pTa-535 sites are found in the distal parts of 2SbS and 7SbL chromosomes of Ae. bicornis; the first one overlaps with pTa-s53, and the second—with pAs1 sites. The pTa-535 probe hybridizes to subterminal regions of five pairs of Ae. searsii chromosomes, 1SsL and 6SsL exhibiting the largest signals. A relatively intense pAs1 signal is detected in a terminus of 4SsS and few very weak interstitial signals are observed on 1SsL, 3SsL, and 7SsL. Faint, dispersed, non-specific pAs1 signals are distributed in distal halves of Ae. sharonensis and Ae. longissima chromosomes, while pTa-s53 and pTa-535 did not hybridize to the chromosomes of these species.
Analysis of Polyploid Species: Wheats
Differences between emmer and Timopheevii wheat are mainly due to species-specific translocations identified in both evolutionary lineages (Naranjo et al., 1987; Liu et al., 1992; Jiang and Gill, 1994a; Maestra and Naranjo, 1999; Salina et al., 2006a). The (CTT)10 signals on T. araraticum and T. dicoccoides chromosomes (Figures 3a,b, 6a,c,h) mainly correspond to the C-bands, whereas (GTT)9 forms large clusters in proximal regions of all B- and G-genome chromosomes (Figure 2, G,B; Figures 3a,b); their positions mainly overlapped with the location of (CTT)10 clusters. A similar pattern is also observed in Ae. speltoides (Figure 3f).
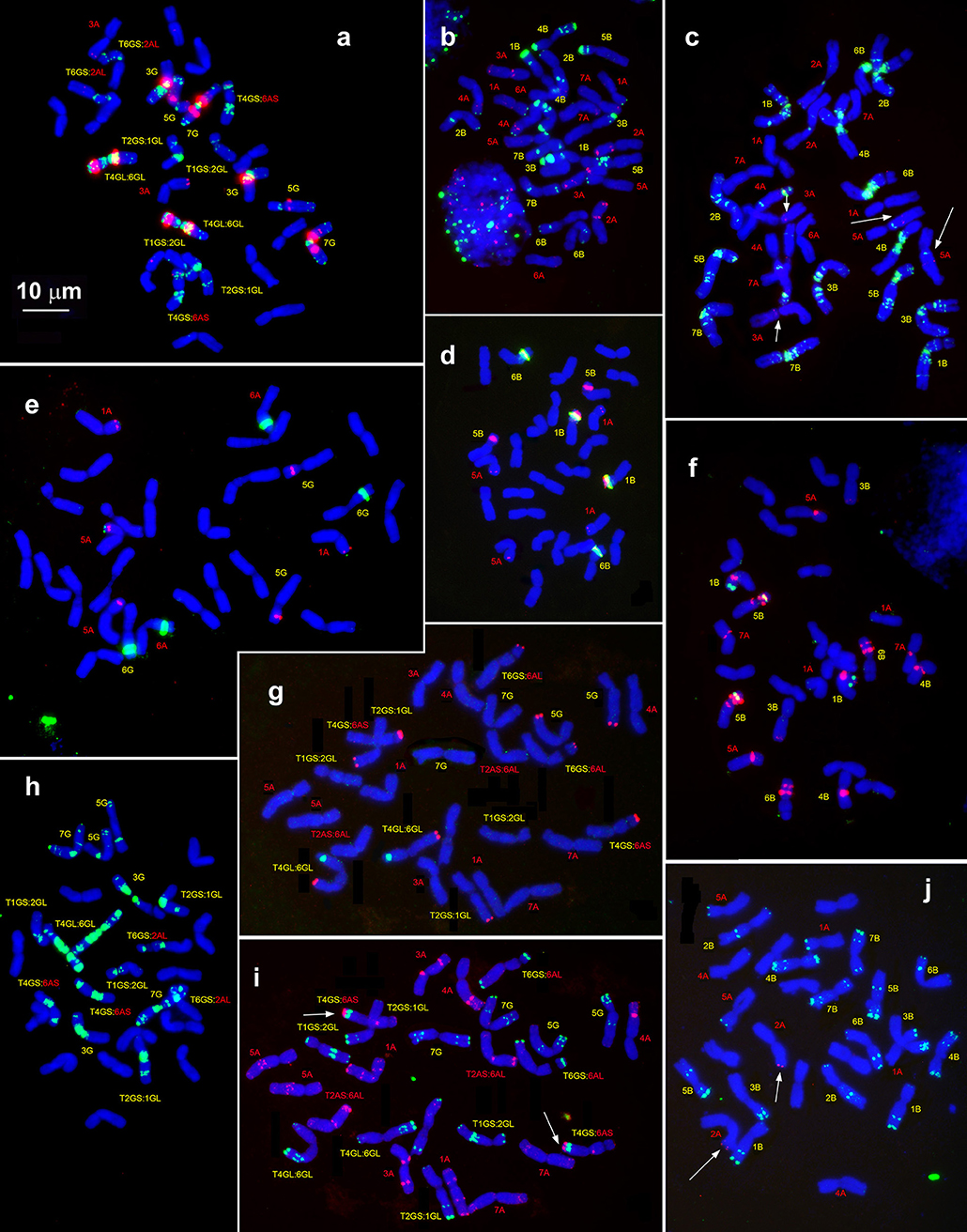
Figure 6. Hybridization patterns of (CTT)10 (a,c,h, green), pSc119.2 (b,i,j, green), pTa-535 (c,i, red), pTa-s53 (b,h, red), pAesp_SAT86 (f,h, red), NORs (d,e, green), and 5S rDNA (d,e, red), 5S rDNA (f, green); Spelt-1 (j,g, red) and Spelt-52 (g, green) on metaphase chromosomes of T. dicoccoides, IG 46396 (b–d,f,j) and T. araraticum, K-59940 (a,e,h,g,i). Position of pTa-s53 hybridization sites on T. dicoccoides chromosomes (c), huge cluster of pTa-535 sequence on the chromosome 4GS (i) and Spelt-1 site on T. dicoccoides chromosome 2AL (j) are indicated with arrows. Scale bar, 10 μm.
Although the pSc119.2 hybridization patterns in these two wheat species are distinct and species-specific, they share some similar features. As in Ae. speltoides, pSc119.2 signals are located in interstitial and subtelomeric regions of orthologous chromosome allowing a complete chromosome identification.
Two chromosome pairs of T. araraticum and T. dicoccoides carry major NORs (Figures 6d,e). These are 1B and 6B in T. dicoccoides and 6G and 6At in T. araraticum (transfer of NORs from 1G to 6At is due to species-specific translocation 1G-4G-6At in Timopheevii lineage, Jiang and Gill, 1993). Group 1 and 5 chromosomes of T. dicoccoides display eight 5S rDNA signals (Figure 6d), but only six - in T. araraticum (chromosomes 1At, 5At, and 5G, see Figure 6e). Chromosome 5S of Ae. speltoides shows one 5S rDNA locus, therefore 1S likely lost the 5S rDNA locus in the progenitor of Ae. speltoides after the formation of ancient emmer, but prior to the divergence of Timopheevii wheat.
The pAesp_SAT86 clusters are found on both A and B/G genome chromosomes (Figures 6a,f), T. dicoccoides and T. araraticum show different labeling patterns and both exhibit broad intraspecific polymorphisms (Badaeva, unpublished). A large pAesp_SAT86 signal is found on 1BS of all emmer (Figure 6f) and common wheat (Komuro et al., 2013), but it is absent on 1G of T. timopheevii (Figure 6a). By contrast, huge 3GL- and 7GL-located pericentromeric pAesp_SAT86 clusters are missing on the homoeologous chromosomes of emmer wheat. At the same time, similar labeling patterns were observed on 4B/4G, 5B/5G, and 6B/6G of these species.
Very weak Spelt-52 signals were seen on 1GS and large on 6GL of T. araraticum. The same sequence was not detectable in wild emmer. Two faint Spelt-1 signals were revealed on the chromosome pair 2A of T. dicoccoides (Figure 6j), whereas ten clear signals were observed on chromosomes 6AtS, 1GL, 4GL, 5GL, and 6GS of T. araraticum (Figure 6g).
Probe pTa-535 hybridized predominantly on the A-genome chromosomes of both wheat species (Figures 6b,i, red color). A large pTa-535 cluster was found on the short arm of 4G of T. araraticum (Figure 6i, indicated with arrows). Overlapping, small pAs1/ pTa-535 signals are detected in distal halves of 3GL and 3BL. In addition, faint pAs1 signals were found in the satellite of 1B, in the middle of 6BS and 7BL of wild emmer. T. araraticum carries small pAs1 loci on 5GL and 7GL and in the satellite of 6At (data not shown). Only weak pTa-s53 signals were observed on chromosomes 3AS and 5AL of T. dicoccoides (Figure 6c), and no hybridization was found on T. araraticum (Figure 6h).
Polyploid Aegilops: Ae. peregrina and Ae. kotschyi
Ae. peregrina and Ae. kotschyi are both tetraploids with the same genome constitutions UUS*S*. Their C-banding patterns are generally similar, however, some differences in morphology and heterochromatin distribution on chromosomes 2S*, 4S*, and 7S* are observed (Figure S3). According to C-banding patterns, Ae. peregrina carries 4S-7S* translocation and therefore the Sp-genome is originated from Ae. longissima. The Sk genome of Ae. kotschyi is more diverged from the S*-genomes of Emarginata species, but shares similar structure and C-banding pattern of chromosome 4S* with Ae. sharonensis (Figures S2, S3).
FISH with (CTT)10 and (GTT)9 probes reveals large CTT-clusters on all chromosomes, but only few weak GTT-signals on some U and S*-genome chromosomes of both species (Figures 3c,h). Distribution of (CTT)10 probe corresponds to the C-banding patterns (Figure 7, Figure S3), and dual-color FISH allows aligning of the CTT/C-banding and pSc119.2-FISH patterns (Figure 5). Positions of pSc119.2 clusters on chromosomes of the two species are similar except for 4S*, which carries two prominent subtelomeric signals in Ae. kotschyi, but one huge cluster in the short and two smaller sites in the long arm in Ae. peregrina (Figure 7). Labeling patterns varies between the accessions. Owing to subterminal location of pSc119.2 sites and polymorphism of labeling patterns, applicability of the pSc119.2 probe for chromosome identification of Ae. peregrina and Ae. kotschyi is limited.
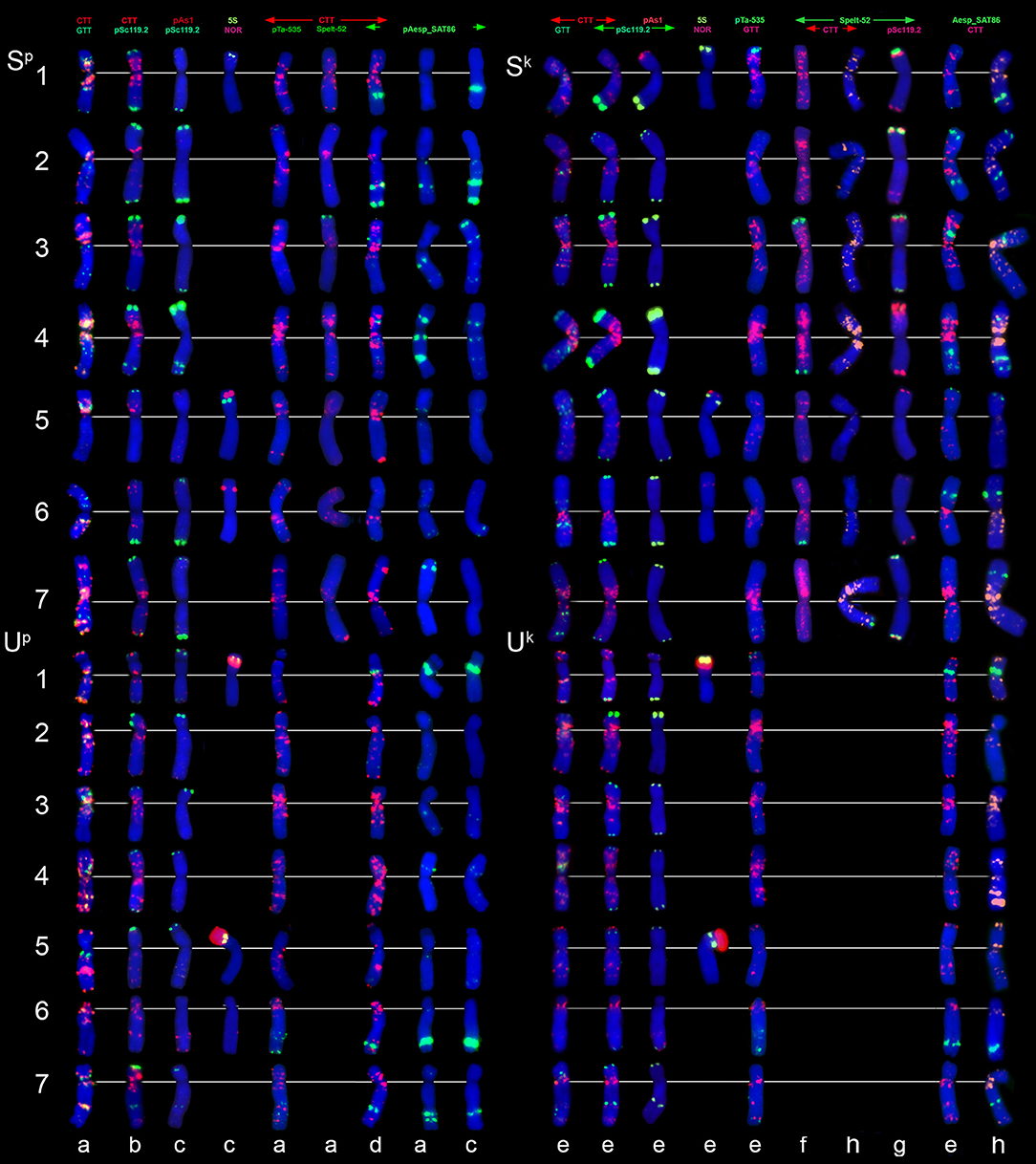
Figure 7. Distribution of repetitive DNA families on chromosomes of Ae. peregrina left side and Ae. kotschyi (right side of the figure): (a), C11 (Nahal Mearot, Israel); (b), C8 (Haifa, Carmel, Israel); (c), K-61; (d), C9 (Keshon, Israel); (e), TA2206; (f), K-91; (g), hybrid Ae. umbellulata TU04 × Ae. sharonensis TH02; (h), K-2905. Probe combinations are given on the top; signal color corresponds to probe name. The S-genome chromosomes are shown on the top, the U-genome–on the bottom part of the figure.
Distribution of 45S and 5S rDNA loci on Ae. peregrina and Ae. kotschyi chromosomes is similar Figure 7, (NOR+5S) and corresponds to that in the parental species. Signal size of pTa71 probe (45S rDNA) on 5Sk and especially 6Sk chromosomes of Ae. kotschyi is significantly smaller than on the orthologous chromosomes of Ae. peregrina, which can be an indicative of more extensive gene loss at the respective loci.
FISH reveals similar hybridization patterns of pAesp_SAT86 probe on chromosomes of Ae. peregrina and Ae. kotschyi. According to dual-color FISH, the largest pAesp_SAT86 signals are located on chromosomes 1S*L (polymorphic), 2S*L, 1US, 6UL, and 7UL. Chromosomes 3S* and 4S* carry medium and 5S*S, 6S*L, and 4US – faint signals (Figure 7). Labeling patterns of chromosomes 3S*, 4S*, 6S*, and 7S* are polymorphic. In contrast to Ae. peregrina and diploid Emarginata species, the chromosome 6Sk of Ae. kotschyi carries large pAesp_SAT86 cluster in the short arm.
The Spelt-1 repeat is not found in these tetraploid species, while Spelt-52 is revealed only in few accessions of Ae. kotschyi and Ae. peregrina. Small Spelt-52 clusters are observed on four out of seven S*-genome chromosome: 3S*S, 4S*L, 6S*L, and 7S*L. Number of signals varies from two to six (Figure 7), nearly half of genotypes we examined do not exhibit any hybridization. This is strictly different from what is observed in a newly synthesized hybrid Ae. umbellulata x Ae. sharonensis, in which 12 distinct Spelt-52 signals are observed in either one or both arms of chromosome pairs 1S*, 2S*, 3S*, and 7S* (Figure 7g ).
Very few weak pAs1 and pTa-535 signals are located predominantly on the U-genome chromosomes of Ae. kotschyi and Ae. peregrina (Figure 7), while the pTa-s53 sequence is totally absent.
Polyploid Aegilops: Ae. vavilovii
The hexaploid species Ae. vavilovii with the genome constitution D1D1XcrXcrSvSv is characterized by a medium amount of Giemsa bands. Small and medium sized bands are distributed predominantly in interstitial chromosome regions (Figure 8). Two chromosome pairs are submetacentrics with small satellites, which morphologically correspond to 5S*. Two other pairs are metacentrics with large satellites, which is typical for chromosome 6S*. The C-banding pattern of Ae. vavilovii is similar to the parental species: Ae. crassa (Badaeva et al., 1998, 2002) and Ae. searsii (Friebe et al., 1995; Friebe and Gill, 1996). Intraspecific variations due to chromosomal rearrangements were identified in two of the three accessions of Ae. vavilovii.
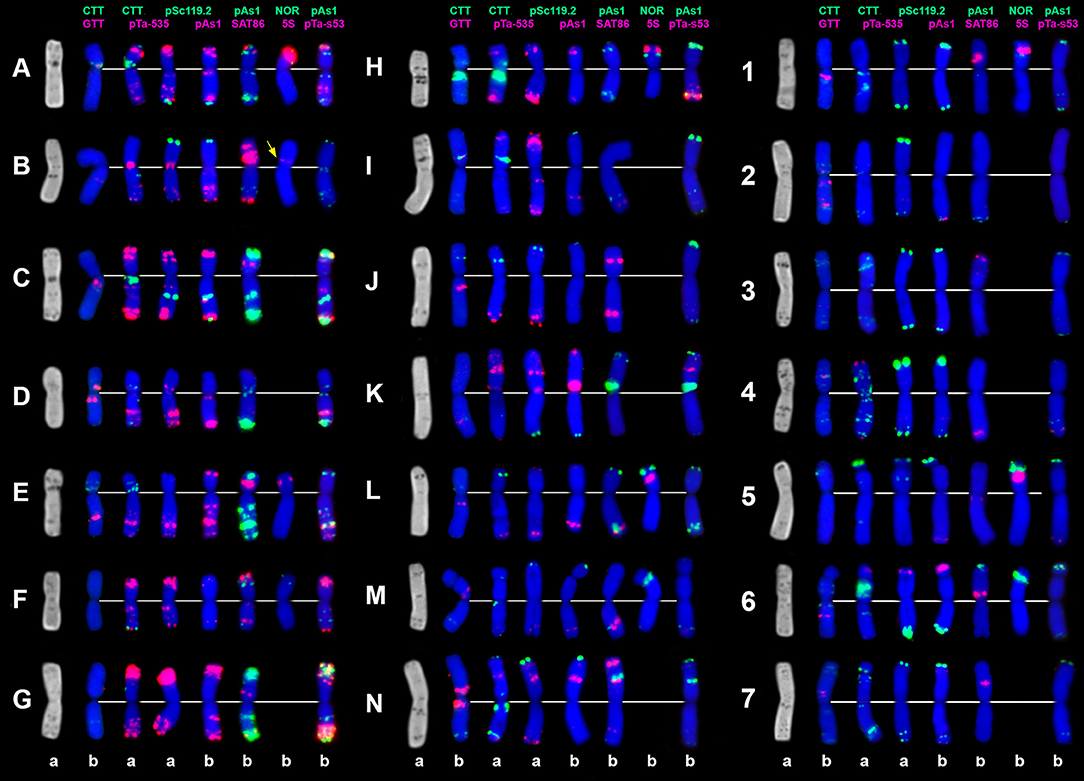
Figure 8. C-banding (left) and FISH patterns of Ae. vavilovii chromosomes: a – K-3635; b – K-3637. Chromosomes are assigned to genome D1 (left), Xcr (middle), and to the Sv genome (right) according to similarity with chromosomes of Ae. crassa (Badaeva et al., 1998) and Ae. searsii (Friebe et al., 1995). Probe combinations are shown on the top, signal color corresponds to probe name. Chromosomes derived from Ae. crassa are designated with letters (A–N); the Sv genome chromosomes are numbered 1–7 according to genetic nomenclature (Friebe et al., 1995).
The distribution of (CTT)10 signals is generally similar to the observed C-banding patterns (Figure 8). The (GTT)9 probe results in distinct signals on five pairs of the Sv genome chromosomes and small to medium clusters on seven pairs of the D1 and Xcr genome chromosome. (GTT)9 signals only partially overlap with (CTT)10 loci (Figure 3g).
Probe pSc119.2 hybridized with all Sv and some Xcr genome chromosomes. Signals are located in subterminal chromosome regions; interstitial sites were found in the middle of 5SvS and in the distal region of 4SvL and 7SvL (Figure 8).
Probe pTa71 revealed eight major and eight minor NOR sites in Ae. vavilovii. The major NORs are located on group 5 and 6 chromosomes belonging to Sv and Xcr genomes. Minor NORs mapped on all three pairs of group 1 chromosomes and, surprisingly on 6D1S. Six of 5S rDNA sites are located on group 1 and 5 chromosomes. An additional, minor 5S rDNA locus is detected in the proximal region of an unknown small metacentric chromosome (Figure 8, shown with arrow).
The pAesp_SAT86 signals of different sizes were detected on many Ae. vavilovii chromosomes; the number of loci varies from one to three per chromosome (Figure 8). Distribution of pAesp_SAT86 sites on 2Sv, 3Sv, and 7Sv is different from Ae. searsii, while the remaining chromosomes of these genomes show similar labeling patters.
The A/D-genome-specific probes pAs1, pTa-535, and pTa-s53 hybridize mainly to the D1-genome and partially to Xcr genome chromosomes of Ae. vavilovii (Figure 8). The Sv genome possesses the lowest amount of these sequences. Small pAs1 signals were observed only in terminal regions of 4SvS and 6SvS. Neither Spelt-1, nor Spelt-52 hybridization sites were detected in Ae. vavilovii.
Discussion
Karyotype analysis as a tool for studying evolutionary processes must be based on an unified chromosome nomenclature. The first classification of chromosomes according to their homoeologous relationships and genome affinities was developed for common wheat by Sears (1954), and since then it is used as standard in genetic and cytogenetic studies of the Triticeae. Although the nomenclature of Giemsa C-banded chromosomes is now available for many Aegilops species, including Ae. speltoides, Ae. searsii, Ae. longissima, and Ae. peregrina (Friebe and Gill, 1996), their correspondence to the distribution of FISH probes is not known.
In order to link C-banding and FISH patterns (Jiang and Gill, 1993) developed a method of sequential C-banding and in situ hybridization analysis. An alternative approach was suggested by Pedersen and Langridge (1997), who used the barley probe pHvG38 containing a GAA-satellite sequence for the identification of wheat chromosomes. Later this sequence was successfully used for the FISH analyses of wheat, barley, rye and some other cereal chromosomes (Pedersen et al., 1996; Cuadrado et al., 2000; Vrána et al., 2000; Cuadrado and Jouve, 2002; Kubaláková et al., 2005; Kato, 2011; Komuro et al., 2013; Adonina et al., 2015; Badaeva et al., 2016), however it was rarely applied for Aegilops species (Molnár et al., 2005, 2016; Mirzaghaderi et al., 2014).
The CTT–labeling patterns of Aegilops chromosomes obtained in our study basically correspond to their C-banding patterns. Therefore, we used the CTT-signals as landmark to identify chromosomes according to the genetic nomenclature. This allowed us to compare karyotypes based on chromosome homoeology and to trace chromosomal changes that could have occurred over the course of species evolution.
Ae. speltoides and Polyploid Wheats Are Cytogenetically Distinct From the S*-Genome of Other Diploid and Polyploid Aegilops Species
Based on C-banding and FISH patterns it is possible to divide the S-genome chromosomes of diploid and polyploid wheat and Aegilops species into two distinct groups. The first one includes Ae. speltoides and polyploid wheat. The second contains four diploid species of the Emarginata sub-section and three polyploid Aegilops, in agreement with molecular phylogenetic analyses (Yamane and Kawahara, 2005; Golovnina et al., 2007; Goryunova et al., 2008; Salse et al., 2008; Gornicki et al., 2014; Marcussen et al., 2014; Middleton et al., 2014; Feldman and Levy, 2015). The main diagnostic features of these groups can be described as follows.
1. The satellite chromosomes of the S-genome of Ae. speltoides and B/G-genomes of polyploid wheats belong to homoeologous groups 1 and 6 (Dvorák et al., 1984; Friebe et al., 2000). The satellites are large and nearly equal in size (Chennaveeraiah, 1960). The satellite of T. timopheevii chromosome 1G is transferred to 6At as a result of a species-specific translocation (Jiang and Gill, 1994a; Rodríguez et al., 2000b; Dobrovolskaya et al., 2011). Major 45S rDNA sites are located on the short arms of group 1 and 6 chromosomes (Figure 2) (Yamamoto, 1992a,b; Jiang and Gill, 1994b; Badaeva et al., 1996b; Raskina et al., 2011; Belyayev and Raskina, 2013; Molnár et al., 2016). In addition to major NORs, Jiang and Gill (1994b) revealed minor 45S rDNA loci in the long arm of chromosome 1B of common and durum wheat, 1G of T. timopheevii and 1S of Ae. speltoides, which were never observed in other S*-genome Aegilops species. Diploid Emarginata species possess two pairs of satellite chromosomes assigned to genetic groups 5 and 6 (Friebe et al., 1993, 1995; Friebe and Gill, 1996); satellites significantly differ in size (Chennaveeraiah, 1960). The secondary constrictions of 5S* and 6S* are suppressed in polyploid Ae. peregrina and Ae. kotschyi, but are extended in hexaploid Ae. vavilovii. FISH with the probe pTa71 revealed major 45S rDNA sites on 5S* and 6S* chromosomes of diploid and polyploid Aegilops species, but signal sizes were significantly reduced in tetraploid Ae. peregrina and Ae. kotschyi (Figures 4, 7). Permanent minor 45S rDNA loci were present on chromosome 1S*, and additional minor site was detected in the terminus of 6S*L of all Ae. searsii and some Ae. bicornis and Ae. longissima accessions (Figure 5). Earlier we also found minor 45S rDNA locus in a terminus of the short arm of an unknown chromosome, probably 3Sl, of Ae. longissima, accession TA1912 (Badaeva et al., 1996b). These observations are in agreement with previously published results (Yamamoto, 1992a,b; Friebe et al., 1993; Badaeva et al., 1996b, 2002, 2004).
2. The S, B, and G genomes are enriched in GTT-repeats (Figures 1–3). This microsatellite is especially abundant in proximal chromosome regions, but rarely appears in interstitial locations. The GTT-sites do not always overlap with the CTT-clusters, and proximal GTT-signals could be observed in chromosome regions lacking Giemsa C-bands. By contrast, the S*-genome chromosomes of Aegilops species show poor labeling with the (GTT)9 probe (Figures 3, 5). The GTT-interstitial signals mainly overlap with the (CTT)n clusters (Figures 3c–e, h–j).
3. The distribution of pSc119.2 repeat in Ae. speltoides and the B/G genomes of wheat observed in our study (Figures 1, 2) is similar to what was reported before (Jiang and Gill, 1994a; Badaeva et al., 1996a; Schneider et al., 2003; Kubaláková et al., 2005; Salina et al., 2006b; Komuro et al., 2013) and is distinct from the S*-genome chromosomes of other Aegilops species in preferentially interstitial signal location.
4. The Spelt-1 sequence is present in the S-genome of Ae. speltoides (Salina et al., 1997, 2006b; Raskina et al., 2011; Belyayev and Raskina, 2013) and the B/G genomes of polyploid wheats (Salina, 2006; Salina et al., 2006b; Zoshchuk et al., 2007, 2009), but it is absent from the S*-genome of other diploid and polyploid Aegilops species.
Different Families of Repetitive DNA Show Different Evolutionary Rates
Our data and previous findings imply that the evolutionary rate varies between different families of repetitive DNAs. Despite distinct differences between Ae. speltoides/polyploid wheats and other S*-genome Aegilops species in the distribution of rDNA probes, the patterns of 45S and 5S rDNA loci was highly conserved within each group. Only minor intra-and inter-specific variations were observed,
(1) Regarding the appearance of minor NORs, which occur at similar positions on the orthologous chromosomes (Yamamoto, 1992a,b; Badaeva et al., 1996b), and
(2) The decrease of signal size on the S*-genome chromosomes of tetraploid Aegilops species (Yamamoto, 1992a,b; Badaeva et al., 2004). Such signal reduction could be explained by uniparental elimination of genes (Shcherban et al., 2008).
The distribution of the rye-derived pSc119.2 repeat is also found to be relatively conserved within each of the two S-genome groups. This sequence with a 120 bp-long repeat unit is broadly distributed in the Triticaea and some Aveneae species and constitutes large and evolutionary old component of their genomes (Contento et al., 2005). The repeat units isolated from wheat, rye, barley and Aegilops species showed 70-100% similarity to each other. Nucleotide sequences of pSc119.2 repeat units are not species-specific, and one site may contain diverse members of the family (Contento et al., 2005). The authors proposed that these individual pSc119.2 sites are transferred as blocks and can be translocated within the genome resulting in position variation and site numbers. Similar was observed in our material. Most cereals, including barley (Taketa et al., 2000; Zhao et al., 2018), Aegilops (Badaeva et al., 1996a, 2002, 2004; Linc et al., 1999; Molnár et al., 2005, 2016), Agropyron (Brasileiro-Vidal et al., 2003; Li et al., 2018; Said et al., 2018), Elytrigia (Linc et al., 2012), Haynaldia (Zhang et al., 2013), possess predominantly subtelomeric pSc119.2 clusters. Therefore, a terminal location of pSc119.2 satellite family is probably a more primitive character compared to interstitial locations. Intercalary pSc119.2 sites are typical for Ae. speltoides (Badaeva et al., 1996a; Molnár et al., 2016), B- and G-genomes of polyploid wheats (Jiang and Gill, 1994a; Schneider et al., 2003), and rye (Cuadrado and Jouve, 2002); the rye genome being highly rearranged relative to wheat (Liu et al., 1992). Strong differences in the distribution of pSc119.2 sites in the R and S genome chromosomes suggest that transposition of this repeat proceeded in genomes of rye and Ae. speltoides independently, likely, after their radiation from the ancestral form.
Comparison of C-banding patterns with the distribution CTT+ GTT-microsatellite sequences shows that heterochromatin blocks detected by Giemsa staining in different Triticum and Aegilops species could have different sequence composition. Thus, Ae. speltoides chromosomes carry prominent proximal and telomeric C-bands and only few intercalary bands, which is considered as primitive karyotype structure (Stebbins, 1971). Only proximal bands overlap with both (CTT)10 and (GTT)9 clusters. The GTT-repeat is more abundant in these chromosomal regions. Intercalary C-bands correspond to CTT-signals, and probably they are composed by this microsatellite mainly. Neither (CTT)10, nor (GTT)9 signals were detected in telomeric heterochromatin, which is enriched in Spelt-1 and Spelt-2 repeats.
The C-banding patterns of the S*-genome Emarginata species and their polyploid derivatives are very similar to their CTT-hybridization patterns indicating that this sequence is a major component of Giemsa-positive heterochromatin. The GTT-microsatellite is present in much lower quantities, and only few C-bands contain this sequence solely. Species of this genomic group exhibit drastic differences in the content of C-positive heterochromatin. Diploid Ae. bicornis, Ae. searsii, and hexaploid Ae. vavilovii are low heterochromatic; the (CTT)10-signals are small and located mainly in the intercalary chromosome regions. Karyotypically Ae. searsii is distinct from other diploid species and its divergence was accompanied mainly by heterochromatin re-pattering visualized by Giemsa-staining and FISH with the CTT-microsatellite probe. The genomes of Ae. sharonensis, Ae. longissima, Ae. kotschyi, and Ae. peregrina are highly heterochromatic; prominent C-bands and CTT-signals are distributed in proximal and intercalary chromosome regions (Figure 5). Thus, massive amplification of the CTT-repeat occurred at the stage of radiation of Ae. sharonensis and Ae. longissima, resulting in an increase of nuclear DNA (Eilam et al., 2007) and the amount of heterochomatin.
Three tandemly repeated DNA families, pAesp_SAT86, Spelt-1, and Spelt-52 show the highest rate of evolution in the Triticum-Aegilops group. pAesp_SAT86 sequence is detected in all S-genome species (Figures 1, 3, 7, 8) and the B/G genomes of polyploid wheat (Figure 6). The labeling patterns are extremely variable in Ae. speltoides (Figure 1) and differ from polyploid wheat species which, in turn, are distinct from each other (Komuro et al., 2013; Badaeva et al., 2016). Diploid Emarginata species and their polyploid derivatives display species-specific patterns of pAesp_SAT86 probe (Figures 4–8). Ae. bicornis shows the highest degree of intraspecific pAesp_SAT86-polymorphism, while little variation has been observed in Ae. searsii, Ae. sharonensis (Figure 4), Ae. kotschyi (Figure 7), and Ae. vavilovii (data not shown). Ae. bicornis and Ae. searsii differ from each other and from other species of this group (Figure 5). Ae. sharonensis is more similar with Ae. longissima and Ae. peregrina in the distribution of pAesp_SAT86 clusters and only slightly different from Ae. kotschyi. The pTa-713 (homolog of pAesp_SAT86) hybridization patterns of Ae. peregrina reported by Zhao et al. (2016) is consistent with our results, though there are some discrepancies in chromosome designations.
The Spelt-1 repeat is found in Ae. speltoides and the B/G genomes of polyploid wheats. In Ae. speltoides it comprises nearly 2% of the nuclear genome (105–106 copies). The copy number of constituent sequence related to Spelt-1 is ~40–60 reduced in genomes of tetraploid wheats, and ~1200–2400 times reduced in genomes of other Triticeae (Pestsova et al., 1998; Salina et al., 1998; Salina, 2006). Minor amounts of Spelt-1 exist in genomes of rye, cultivated barley, most diploid and polyploid wheat as well as Aegilops species indicates that this sequence was already present in minor quantities in the common ancestor of the Triticeae (Salina et al., 1998). High homology (97–100%) of individual repetitive units implies that massive amplification of Spelt-1 repeat occurred in ancient Ae. speltoides after radiation from the common ancestor of the Triticeae (Salina et al., 1998; Salina, 2006). Spelt52 is homologous to the pAesKB52 repeat isolated earlier from Ae. speltoides by Anamthawat-Jonsson and Heslop-Harrison (1993). This repeat consists of monomers of two types, Spelt52.1 and Spelt52.2, which share a homologous stretch of 280 bp and have two regions without sequence similarity of 96 and 110 bp, respectively. Ae. speltoides displays intraspecific variation in the occurrence of Spelt52 monomer types, whereas Ae. longissima, Ae. sharonensis, and Ae. bicornis showed no interspecific variation (Salina et al., 2004a). The Spelt-52 is abundant in Ae. speltoides accounting for approximately 1% of nuclear genome (Anamthawat-Jonsson and Heslop-Harrison, 1993; Salina, 2006) and is also highly represented (1.0 × 104 – 2.5 × 105 copies) in Ae. longissima and Ae. sharonensis, but it present in minor quantities in Ae. bicornis and Ae. searsii (Salina, 2006).
Coincidently with previous findings (Salina et al., 2006b; Raskina et al., 2011; Belyayev and Raskina, 2013), we observed significant intraspecific variation of Spelt-1 and Spelt-52 labeling patterns (Figure 2). Strict differences in a ratio of Spelt-1/ Spelt-52 repeats detected between genotypes can be due to geographical origin of the material. Earlier, Raskina et al. (2011) found that the amount of Spelt-1 and, in lower extent, the Spelt-52 repeat decreases in marginal populations of Ae. speltoides.
Although pAs1 and pTa-535 repeats are abundant in some cereal genomes (Rayburn and Gill, 1986; Badaeva et al., 1996a; Taketa et al., 2000; Komuro et al., 2013), they are poorly represented in the S genomes of Triticum and Aegilops. Thus, we failed to detect any pAs1signals in Ae. speltoides, but Molnár et al. (2016) revealed small pAs1 signals on the chromosome 3S. Wheat chromosomes 3BL - 3GL and 7BL - 7GL possess pAs1 and pTa-535 clusters in similar positions (Schneider et al., 2003; Badaeva et al., 2016), although they are not detected in Ae. speltoides. Probably, these loci were present in the genome of ancient Ae. speltoides, but they were eliminated after radiation of polyploid wheats. The pAs1 and pTa-535 repeats are also poorly represented in genomes of Emarginata species. Two distinct interstitial pAs1 sites overlapping with either pTa-535, or with pTa-s53 loci are found in Ae. bicornis. pAs1 and pTa-535 are less abundant in Ae. searsii, Ae. sharonensis and Ae. longissima. Only chromosome 1Sk of tetraploid Ae. kotschyi contains a distinct pAs1/pTa-535 cluster, and these sequences are absent from the S* genomes of Ae. peregrina and Ae. vavilovii.
Evolution of the S-Genome
Summarizing our data and the results of other authors (Kihara, 1954; Chennaveeraiah, 1960; Kihara and Tanaka, 1970; Yen and Kimber, 1990b; Zhang and Dvorák, 1992; Zhang et al., 1992; Dvorák, 1998; Feldman and Levy, 2015), the following scenario of the S-genome evolution can be suggested (Figure 9).
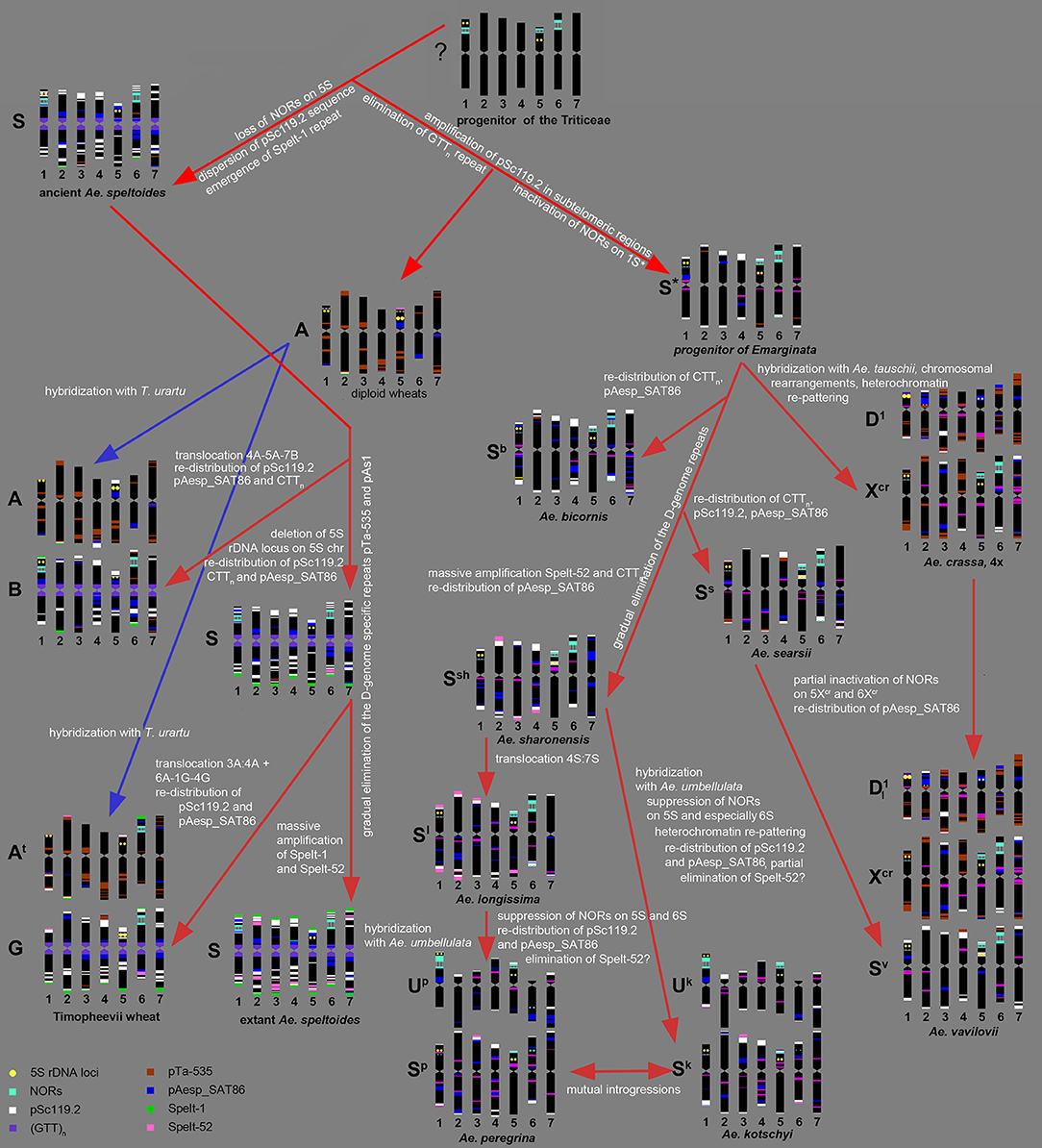
Figure 9. Hypothetic scheme of the S-genome evolution deduced from the results of molecular-cytogenetic analysis.
According to molecular phylogeny, Ae. speltoides is the most distinct diploid Aegilops, which diverged from the common ancestor very early, prior to the split of diploid wheat and Aegilops species (Salse et al., 2008; Gornicki et al., 2014; Marcussen et al., 2014; Feldman and Levy, 2015). Divergence of Ae. speltoides from an ancestral form was not associated with major translocations, because neither meiotic analysis (Rodríguez et al., 2000a), nor microsatellite mapping (Dobrovolskaya et al., 2011) detected structural chromosmal rearrangements in the S-genome. However some genomic changes not causing linkage group perturbations did probably occur at the early stages of Ae. speltoides speciation. As was shown earlier, major NORs in Triticum and Aegilops species are located on group 1, 5, and 6 chromosomes (Appels et al., 1980; Appels and Honeucutt, 1986), while 5S rDNA loci are located separately from NORs in the short arms of group 1 and 5 chromosomes (Appels et al., 1980; Dvorák et al., 1989). The chromosome 5S of Ae. speltoides and B/G genome of polyploid wheats does not contain 45S rDNA loci, therefore the loss of respective NOR probably occurred prior to formation of ancient emmer. Other early genomic changes of ancient Ae. speltoides included the transposition of the pSc119.2 repeat from subtelomeric to interstitial chromosome regions and also the amplification of Spelt-1 repeat.
As mentioned above, Ae. speltoides and the B/G genomes of polyploid wheats are characterized by the abundance of GTT-microsatellite (Cuadrado et al., 2000; Badaeva et al., 2016), which is poorly represented in diploid wheats (Badaeva et al., 2015) and most Aegilops species (Figure 3). This difference can be caused by massive amplification of GTT-repeat in the ancient Ae. speltoides. Alternatively, this repeat could be eliminated from the progenitor of wheat and Aegilops species. Taking into consideration the abundance of GTT-repeat in rye and Hordeum (Cuadrado and Jouve, 2002, 2007; Dou et al., 2016), the second scenario seems to be more likely. The progenitor of Ae. speltoides probably possessed minor amounts of Spelt-52 and the D-genome specific repeats pTa-535, or pAs1, as they are still present in Ae. speltoides and the B/G genomes of polyploid wheats (Schneider et al., 2003; Badaeva et al., 2016; Molnár et al., 2016). However, these sequences could be of the A-genome origin, which spread to the S-genome following allopolyploidization.
The emergence of tetraploid emmer was accompanied by the species-specific translocation involving the chromosomes 4A-5A-7B (Naranjo et al., 1987; Liu et al., 1992; Maestra and Naranjo, 1999). In addition to structural chromosome rearrangements, other genetic and epigenetic changes occurred in a newly formed polyploid, including inactivation of the 45S rDNA loci on the A-genome chromosomes, re-distribution of Giemsa C-bands and repetitive DNA families on both A and B-genome chromosomes. Evolution of polyploid wheat resulted in polymorphisms of various DNA sequences and heterochromatin patterns that were described in many publications (Friebe and Gill, 1996; Schneider et al., 2003; Badaeva et al., 2016).
Subsequent evolution of Ae. speltoides occurred independently of polyploid emmer and was accompanied by several transposon insertions (Salse et al., 2008) and the loss of the 5S rDNA locus on the chromosome 1S, which is present in emmer and common wheat (Mukai et al., 1990), but absent in T. timopheevii (Badaeva et al., 2016) and modern Ae. speltoides. Although T. timopheevii derived from the same parental species as emmer, different parental genotypes were involved in the origin of these two lineages (Golovnina et al., 2007). Timopheevii wheat emerged much later, than ancient emmer - nearly 0.4 MYA (Gornicki et al., 2014) and its formation was accompanied by different species-specific translocation involving the chromosomes 1G-4G-6At + 3At-4At (Jiang and Gill, 1994a; Maestra and Naranjo, 1999; Rodríguez et al., 2000b; Dobrovolskaya et al., 2011). As a result, a major NOR was translocated from chromosome 1G to 6At, and a massive cluster of the A/ D-genome specific repeat pTa-535 appeared on the short arm of chromosome 4G (Figure 6g). Existence of Spelt-52 sites and a spread of Spelt-1 to most T. timopheevii chromosomes (Salina et al., 2006b; Zoshchuk et al., 2007; Badaeva et al., 2016) suggests a massive amplification of these sequences in Ae. speltoides prior to emergence of ancient T. timopheevii.
Results of molecular cytogenetic analysis suggest that genome re-structuring process is still ongoing in natural populations of Ae. speltoides. This is exemplified by intraspecific C-banding polymorphisms and diversity of labeling patterns of pAesp_SAT86, Spelt-1 and Spelt-52 probes observed in this and other studies (Belyayev and Raskina, 2011, 2013; Raskina et al., 2011), fluctuation of copy number of retrotransposons and tandem repeats, and high number of chromosomal rearrangements (Belyayev and Raskina, 2013; Shams and Raskina, 2018).
The species of the Emarginata group are closely related to each other (Eig, 1929; Kihara, 1954; Friebe and Gill, 1996; Kilian et al., 2011; Gornicki et al., 2014; Feldman and Levy, 2015), which is supported by their similar karyotypes (Chennaveeraiah, 1960), C-banding and pSc119.2-labeling patterns (Badaeva et al., 1996a), distribution of rDNA probes (Yamamoto, 1992a,b; Badaeva et al., 1996b). Separation of Emarginata species from a common ancestor was associated with inactivation of major NORs on chromosome 1S* accompanied with the significant loss of 45S rDNA repeat copies. Despite similarity of pSc119.2 labeling patterns, there are obvious, but discontinuous changes in the patterns of other sequences. Our data show that most drastic changes occurred probably at the stage of radiation of Ae. sharonensis-Ae. longissima. These are massive amplification of Spelt-52 and CTT-repeats resulting in the gain of heterochromatin in Ae. sharonensis and Ae. longissima, leading to an approximately 12% increase of nuclear DNA content in Ae. sharonensis/ Ae. longissima as compared to Ae. searsii/ Ae. bicornis (Eilam et al., 2007). By contrast, the amount of the D-genome repeats pTa-535, pAs1 and especially pTa-s53 gradually decreased, and these sequences nearly disappeared in genomes of Ae. sharonensis and Ae. longissima. Spelt-52 patterns of Ae. sharonensis and Ae. longissima chromosomes are highly polymorphic. The similar distribution of all analyzed DNA sequences on chromosomes of Ae. sharonensis and Ae. longissima (Figures 3, 4) point to a rather recent divergence of these species, which was accompanied by the species-specific translocation 4S*-7S* in Ae. longissima.
Formation of tetraploid Ae. peregrina and Ae. kotschyi did not cause significant alterations of the parental genomes. Considering the structure of chromosome 4S*, the Sp-genome of Ae. peregrina was donated by Ae. longissima, while Ae. sharonensis or the form preceding the split of these diploids could be the source of the S*-genome of Ae. kotschyi. These data are consistent with observations of other authors (Yu and Jahier, 1992; Zhang et al., 1992; Friebe et al., 1996), however they contradict the hypothesis about the possible ancestry of Ae. searsii in the origin of Ae. peregrina (Siregar et al., 1988). Merging of U and S* genomes in the tetraploid Ae. peregrina and Ae. kotschyi led to inactivation of 45S rDNA loci on the S*-genome chromosomes (Figure 7). Similar was also recorded in the artificial allopolyploid Ae. umbellulata × Ae. sharonensis (Shcherban et al., 2008). Significantly smaller 45S rDNA sites on Ae. kotschyi chromosome 6Sk compared to the 6Sp of Ae. peregrina evidences in favor of a higher extent of gene loss at the respective locus, which can be due to earlier origin of Ae. kotschyi. The assumption that Ae. kotschyi is an older species is also supported by higher divergence of C-banding patterns relative to the parental species.
Interestingly, Ae. peregrina and Ae. kotschyi both possess only minor quantities of the Spelt-52 repeat, which is abundant in their diploid parents. According to the analyses of artificial wheat-Aegilops or Aegilops-Aegilops hybrids, the Spelt-52 was either amplified or retained at the same level upon polyploidization (Salina et al., 2004b). Considering these results we can expect massive amplification of the Spelt-52 sequence in Ae. peregrina and Ae. kotschyi genomes. However, this is not the case. Low amount of Spelt-52 in these species can be caused by the so-called “originator effect,” if they obtained their S*genomes from genotype depleted with this repeat, or it can be caused by sequence elimination after formation of tetraploids.
The Sv-genome chromosomes of Ae. vavilovii are very similar to the Ss-genome chromosomes of Ae. searsii, which further supports their close relationships (Zhang and Dvorák, 1992; Dubkovsky and Dvorák, 1995). Our results strongly suggest that the Xcr genome of Ae. vavilovii is also the derivative of the S* genome of an unknown Emarginata species, but not of Ae. speltoides as proposed by Dubkovsky and Dvorák (1995); Edet et al. (2018). Significant differences between the Xcr and Ss genomes, as well between Xcr and S*-genomes of all diploid Emarginata species in the C-banding and labeling patterns demonstrate that the Xcr genome was significantly modified during speciation.
Conclusions
Analysis of the S-genomes of diploid and polyploid Triticum and Aegilops species using FISH with nine DNA probes, including 5S and 45S rDNA, two microsatellites and five tandem repeats showed an isolated position of Ae. speltoides among other Aegilops species. In addition, close relationships with the B and G genomes of polyploid wheats were observed, thus confirming previous molecular-phylogenetic data (Yamane and Kawahara, 2005; Petersen et al., 2006; Golovnina et al., 2007; Salse et al., 2008; Gornicki et al., 2014; Marcussen et al., 2014; Middleton et al., 2014; Bernhardt et al., 2017). The evolution of polyploid wheats was associated with different species-specific chromosome translocations and the amplification/ elimination of repeats, re-pattering or, possibly with an exchange of repetitive DNA families with the A-genome chromosomes. Evolutionary changes in the Ae. speltoides genome occurred independently from polyploid wheats.
Diploid Aegilops species of Emarginata group are similar, but are substantially different from Ae. speltoides based on C-banding and FISH patterns. The genome evolution in this group was mainly associated with an increase of high copy DNA fraction due to amplification of CTT-repeat, re-distribution of C-bands, (CTT)n-, (GTT)n-, and pAesp_SAT86-clusters, massive amplification of Spelt-52 and gradual elimination of the D-genome-specific sequences pAs1, pTa-535 and pTa-s53. These changes were more profound at the stage of divergence of Ae. sharonensis/Ae. longissima. Tetraploid Ae. peregrina and Ae. kotschyi originated independently from hybridization of Ae. umbellulata with Ae. longissima (Ae. peregrina) or Ae. sharonensis or its immediate precursor (Ae. kotschyi). The S*-genomes of both tetraploids show little differences to the parental species. The Sk-genome is characterized by more modifications than the Sp-genome, suggesting that Ae. kotschyi is older than Ae. peregrina. Chromosome introgressions recorded in some accessions of both species (Badaeva et al., 2004) can be explained by gene flow between Ae. peregrina and Ae. kotschyi.
Our study confirmed that Ae. vavilovii is a natural hybrid between tetraploid Ae. crassa and Ae. searsii. The similarity of C-banding and FISH patterns of Ae. vavilovii and corresponding parental species points to rather recent origin of this hexaploid. The assumption that the Xcr genome is an additional derivative of the S*genome obtained from an unknown or extinct species of the Emarginata group, which was substantially modified over the course of evolution is supported.
Author Contributions
EB planned the research, performed and coordinate the analysis. EB and AR performed the research and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Drs. O.M. Raskina and A.A. Belyayev (Institute of Evolution, University of Haifa, Israel) for providing seeds and for the opportunity to collect seed materials from their natural habitat. Additional material was obtained from gene banks of the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK, Gatersleben, Germany); VIR Collection of Plant Genetic Resources (Saint Petersburg, Russia); Institute of Evolution, University of Haifa (Haifa, Israel); Weizmann Institute of Science (Rehovot, Israel); USDA-ARS (Aberdeen, Idaho, USA); Wheat Genetics and Genomic Resource Center, Kansas State University (Manhattan, Kansas, USA); ICARDA (Aleppo, Syria). We thank Dr. A. Houben [Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany] for critical reading and valuable comments. We thank Dr. S. Surzhikov (Engehardt Institute of Molecular Biology, Russian Academy of Sciences) for the synthesis of oligo-probes for analysis. This work was supported by grant from Russian State Foundation of Basic Research (project No 17-04-00087). The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01756/full#supplementary-material
Figure S1. Diversity of the C-banding patterns in Aegilops speltoides accessions: (a), No1, from Turkey (provided by Dr. N. Aminov); (b–f), genotypes collected from Israeli populations; (b,c), C1, Technion park, Haifa; (d), C2, Nahal Mearot; (e), G2.46, Ramat haNadiv; (f), C14, Keshon; (g), no 2734 (unknown provided by Dr. B. Kilian); (h), No 2 from Iran (provided by Dr. N. Aminov); (i), i-570060; (j), TS89, Katzir, Israel; (k), PI 487233, and (l), PI 487231 (from Syria); (m), PI 542269 (Turkey). Arrows show unbalanced chromosome modifications.
Figure S2. Interspecific and intraspecific variation of the C-banding patterns in Ae. bicornis (a1–a3), Ae. searsii (b1–b4), Ae. sharonensis (c1–c8), and Ae. longissima (d1–d10). Accession codes: (a1), TA1942; (a2), TB04-3; (a3), TB10-2; (b1), G.7.15; (b2), TE01-1; (b3), G7.12; (b4), IG 47619; (c1,c2), C6, Keshon; (c3), C5, Caesaria; (c4), TH04; (c5), TH01; (c6), TH02; (c7), C4, Atlit; (c8), C7, HaBonim; (d1), TL06; (d2), TL01; (d3), G6.77 (Sa'ad); (d4), G6.58 (Tel Akko); (d5), TL03; (d6), C3 (HaBonim); (d7), G6.32 (Nizzanim); (d8), G6.55 (Zomet Shoked); (d9), G17-3; (d10), TL05.
Figure S3. C-banding polymorphism of Ae. peregrina (a–f) and Ae. kotschyi (g–j) chromosomes: (a), TA1888; (b), C11 (Nahal Mearot, Israel); (c), K-61; (d), C12 (Caesaria, Israel); (e), C13 (Natufia, Israel); (f), i-570632; (g), PI 487279; (h), K-91; (i), TA2206; (j), K-201; (k) K-2905. 1–7, homoeologous groups.
Table S1. List of material and their origins.
Table S2. Distribution of Spelt-52 probe on chromosomes of Ae. longissima and Ae. sharonensis.
References
Adonina, I. G., Goncharov, N. P., Badaeva, E. D., Sergeeva, E. M., Petrash, N. V., and Salina, E. A. (2015). (GAA)n microsatellite as an indicator of the A genome reorganization during wheat evolution and domestication. Comp. Cytogenet. 9, 533–547. doi: 10.3897/CompCytogen.v9i4.5120
Anamthawat-Jonsson, K., and Heslop-Harrison, J. S. (1993). Isolation and characterisation of genome-specific DNA sequences in Triticeae species. Mol. Gen. Genet. 240, 151–158. doi: 10.1007/BF00277052
Appels, R., Gerlach, W. L., Dennis, E. S., Swift, H., and Peacock, W. J. (1980). Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 78, 293–311. doi: 10.1007/BF00327389
Appels, R., and Honeucutt, R. L. (1986). “rDNA evolution over a billion years,” in DNA Systematics, ed S. K. Dutta (FL: CRC Press), 81–125.
Badaeva, E. D., Amosova, A. V., Goncharov, N. P., Macas, J., Ruban, A. S., Grechishnikova, I. V., et al. (2015). A set of cytogenetic markers allows the precise identification of all A-genome chromosomes in diploid and polyploid wheat. Cytogenet. Genome Res. 146, 71–79. doi: 10.1159/000433458
Badaeva, E. D., Amosova, A. V., Muravenko, O. V., Samatadze, T. E., Chikida, N. N., Zelenin, A. V., et al. (2002). Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Syst. Evol. 231, 163–190. doi: 10.1007/s006060200018
Badaeva, E. D., Amosova, A. V., Samatadze, T. E., Zoshchuk, S. A., Shostak, N. G., Chikida, N. N., et al. (2004). Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst. Evol. 246, 45–76. doi: 10.1128/AEM.02052-08
Badaeva, E. D., Badaev, N. S., Gill, B. S., and Filatenko, A. A. (1994). Intraspecific karyotype divergence in Triticum araraticum (Poaceae). Plant Syst. Evol. 192, 117–145. doi: 10.1007/BF00985912
Badaeva, E. D., Friebe, B., and Gill, B. S. (1996a). Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 39, 293–306. doi: 10.1139/g96-040
Badaeva, E. D., Friebe, B., and Gill, B. S. (1996b). Genome differentiation in Aegilops. 2. Physical mapping of 5S and 18S-26S ribosomal RNA gene families in diploid species. Genome 39, 1150–1158. doi: 10.1139/g96-145
Badaeva, E. D., Friebe, B., Zoshchuk, S. A., Zelenin, A. V., and Gill, B. S. (1998). Molecular cytogenetic analysis of tetraploid and hexaploid Aegilops crassa. Chrom. Res. 6, 629–637. doi: 10.1023/A:1009257527391
Badaeva, E. D., Ruban, A. S., Aliyeva-Schnorr, L., Municio, C., Hesse, S., and Houben, A. (2017). “In situ hybridization to plant chromosomes,” in Fluorescence in situ Hybridization (FISH) Application Guide, ed T. Liehr. (Berlin; Heidelberg: Springer), 477-494. doi: 10.1007/978-3-662-52959-1
Badaeva, E. D., Ruban, A. S., Zoshchuk, S. A., Surzhikov, S. A., Knüpffer, H., and Kilian, B. (2016). Molecular cytogenetic characterization of Triticum timopheevii chromosomes provides new insight on genome evolution of T. zhukovskyi. Plant Syst. Evol. 302, 943–956. doi: 10.1007/s00606-016-1309-3
Bahrman, N., Zivi, M., and Thiellement, H. (1988). Genetic relationships in the Sitopsis section of Triticum and the origin of the B genome of polyploid wheats. Heredity 61, 473–480. doi: 10.1038/hdy.1988.141
Bedbrook, R. J., Jones, J., O'Dell, M., Thompson, R. J., and Flavell, R. B. (1980). A molecular description of telomeric heterochromatin in Secale species. Cell 19, 545–560. doi: 10.1016/0092-8674(80)90529-2
Belyayev, A., and Raskina, O. (2011). “Transposable elements in a marginal population of Aegilops speltoides: temporal fluctuations provide new Insights into genome evolution of wild diploid wheat,” in Evolutionary Biology – Concepts, Biodiversity, Macroevolution and Genome Evolution, ed P. Pontarotti (Berlin; Heidelberg: Springer), 313–324. doi: 10.1007/978-3-642-20763-1_18
Belyayev, A., and Raskina, O. (2013). Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences. Ann. Bot. 111, 531–538. doi: 10.1093/aob/mct023
Belyayev, A., Raskina, O., and Nevo, E. (2001). Evolutionary dynamics and chromosomal distribution of repetitive sequences on chromosomes of Aegilops speltoides revealed by genomic in situ hybridization. Heredity 86, 738–742. doi: 10.1046/j.1365-2540.2001.t01-1-00891.x
Bernhardt, N., Brassac, J., Kilian, B., and Blattner, F. R. (2017). Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evol. Biol. 17:141. doi: 10.1186/s12862-017-0989-9
Blake, N. K., Lehfeldt, B. R., Lavin, M., and Talbert, L. E. (1999). Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: the B genome of wheat. Genome 42, 351–360. doi: 10.1139/g98-136
Brasileiro-Vidal, A. C., Cuadrado, A., Brammer, S. P., Zanatta, A. C. A., Prestes, A. M., Moraes-Fernandes, M. I. B., et al. (2003). Chromosome characterization in Thinopyrum ponticum (Triticeae, Poaceae) using in situ hybridization with different DNA sequences. Genet. Mol. Biol. 26, 505–510. doi: 10.1590/S1415-47572003000400014
Chennaveeraiah, M. S. (1960). Karyomorphologic and cytotaxonomic studies in Aegilops. Acta Horti Gotoburgensis 23, 85–186.
Contento, A., Heslop-Harrison, J. S., and Schwarzacher, T. (2005). Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenet. Genome Res. 109, 34–42. doi: 10.1159/000082379
Cuadrado, A., and Jouve, N. (2002). Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J. Hered. 93, 339–345. doi: 10.1093/jhered/93.5.339
Cuadrado, A., and Jouve, N. (2007). The nonrandom distribution of long clusters of all possible classes of trinucleotide repeats in barley chromosomes. Chrom. Res. 15, 711–720. doi: 10.1007/s10577-007-1156-8
Cuadrado, A., Schwarzacher, T., and Jouve, N. (2000). Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor. Appl. Genet. 101, 711–717. doi: 10.1007/s001220051535
Damania, A. B., Altunji, H., and Dhaliwal, H. S. (1992). Evaluation of Aegilops spp. for Drought and Frost Tolerance. Genetic Resources Unit Annual Report, ICARDA, 45–46.
Daud, H. M., and Gustafson, J. P. (1996). Molecular evidence for Triticum speltoides as a B-genome progenitor of wheat (Triticum aestivum). Genome 39, 543–548. doi: 10.1139/g96-069
Dobrovolskaya, O., Boeuf, C., Salse, J., Pont, C., Sourdille, P., Bernard, M., et al. (2011). Microsatellite mapping of Ae. speltoides and map-based comparative analysis of the S, G, and B genomes of Triticeae species. Theor. Appl. Genet. 123, 1145–1157. doi: 10.1007/s00122-011-1655-z
Dou, Q., Liu, R., and Yu, F. (2016). Chromosomal organization of repetitive DNAs in Hordeum bogdanii and H. brevisubulatum (Poaceae). Compar. Cytogenet. 10, 465–481. doi: 10.3897/CompCytogen.v10i4.9666
Dubkovsky, J., and Dvorák, J. (1995). Genome identification of the Triticum crassum complex (Poaceae) with the restriction patterns of repeated nucleotide sequences. Am. J. Bot. 182, 131–140. doi: 10.1002/j.1537-2197.1995.tb15657.x
Dvorák, J. (1998). “Genome analysis in the Triticum-Aegilops alliance,” in Proceedings of the 9th International Wheat Genetics Symposium, ed A. E. Slinkard (Saskatoon, SK: Printcrafters Inc.), 8–11.
Dvorák, J., and Akhunov, E. D. (2005). Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171, 323–332. doi: 10.1534/genetics.105.041632
Dvorák, J., Lassner, M. W., Kota, R. S., and Chen, K. C. (1984). The distribution of the ribosomal RNA genes in the Triticum speltoides and Elytrigia elongata genomes. Can. J. Genet. Cytol. 62, 628–632. doi: 10.1139/g84-097
Dvorák, J., and Zhang, H.-B. (1990). Variation in repeated nucleotide sequences sheds light on the phylogeny of the wheat B and G genomes. Proc Natl Acad Sci USA 87, 9640–9644. doi: 10.1073/pnas.87.24.9640
Dvorák, J., and Zhang, H.-B. (1992). Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide sequences. Theor. Appl. Genet. 84, 419–429. doi: 10.1007/BF00229502
Dvorák, J., Zhang, H.-B., Kota, R. S., and Lassner, M. (1989). Organization and evolution of the 5S ribosomal RNA gene family in wheat and related species. Genome 32, 1003–1016. doi: 10.1139/g89-545
Edet, O. U., Gorafi, Y. S. A., Nasuda, S., and Tsujimoto, H. (2018). DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 8:16397. doi: 10.1038/s41598-018-34811-y
Eilam, T., Anikster, Y., Millet, E., Manisterski, J., Sagi-Assif, O., and Feldman, M. (2007). Genome size and genome evolution in diploid Triticeae species. Genome 50, 1029–1037. doi: 10.1139/G07-083
El Bouhssini, M., Benlhabib, O., Nachit, M. M., Houari, A., Bentika, A., Nsarellah, N., et al. (1998). Identification in Aegilops species of resistant sources to Hessian fly (Diptera: Cecidomyiidae) in Morocco. Genet Res. Crop Evol. 45, 343–345. doi: 10.1023/A:1008675029389
Farkas, A., Molnár, I., Dulai, S., Rapi, S., Oldal, V., Cseh, A., et al. (2014). Increased micronutrient content (Zn, Mn) in the 3Mb(4B) wheat – Aegilops biuncialis substitution and 3Mb.4BS translocation identified by GISH and FISH. Genome 57, 61–67. doi: 10.1139/gen-2013-0204
Feldman, M. (2001). “Origin of cultivated wheat,” in The World Wheat Book A History of Wheat Breeding, eds A. P. Bonjean and W. J. Angus (Londres; Paris; New York, NY: Tec & Doc/Intersept Ltd.), 3–56.
Feldman, M., and Kislev, M. E. (1977). Aegilops searsii, a new species of section Sitopsis (Platystachys). Israel J. Bot. 28, 190–201.
Feldman, M., and Levy, A. A. (2015). “Origin and evolution of wheat and related Triticeae species,” in Alien Introgression in Wheat: Cytogenetics, Molecular Biology, and Genomics, eds M. Molnár-Láng, C. Ceoloni, and J. Doležel (Cham: Springer International Publishing), 21–76.
Feldman, M., Strauss, I., and Vardi, A. (1979). Chromosome pairing and fertility of F1 hybrids of Aegilops longissima and Ae. searsii. Can. J. Genet. Cytol. 21, 261–272. doi: 10.1139/g79-031
Friebe, B., and Gill, B. S. (1996). “Chromosome banding and genome analysis in diploid and cultivated polyploid wheats,” in Methods in Genome Analysis in Plants: Their Merits and Pitfals, ed P. P. Jauhar (Boca Ration, FL; New York, NY; London; Tokyo: CRC Press), 39–60.
Friebe, B., Qi, L. L., Nasuda, S., Zhang, P., Tuleen, N. A., and Gill, B. S. (2000). Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theor. Appl. Genet. 101, 51–58. doi: 10.1007/s001220051448
Friebe, B., Tuleen, N., Jiang, J., and Gill, B. S. (1993). Standard karyotype of Triticum longissimum and its cytogenetic relationship with T. aestivum. Genome 36, 731–742. doi: 10.1139/g93-098
Friebe, B., Tuleen, N. A., Badaeva, E. D., and Gill, B. S. (1996). Cytogenetic identification of Triticum peregrinum chromosomes added to common wheat. Genome 39, 272–276. doi: 10.1139/g96-037
Friebe, B., Tuleen, N. A., and Gill, B. S. (1995). Standard karyotype of Triticum searsii and its relationship with other S-genome species and common wheat. Theor. Appl. Genet. 91, 248–254. doi: 10.1007/BF00220885
Gerlach, W. L., and Bedbrook, J. R. (1979). Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acid Res. 7, 1869–1885. doi: 10.1093/nar/7.7.1869
Gerlach, W. L., and Dyer, T. A. (1980). Sequence organization of the repeated units in the nucleus of wheat which contains 5S-rRNA genes. Nucleic Acid Res. 8, 4851–4865
Gill, B. S., Friebe, B., and Endo, T. R. (1991). Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34, 830–839. doi: 10.1139/g95-030
Gill, B. S., Sharma, C., Raupp, W. J., Browder, L. E., Heachett, J. H., Harvey, T. L., et al. (1985). Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, Hessian fly, and greenbug. Plant Dis. 69, 314–316.
Giorgi, B., and Bozzini, A. (1969). Karyotype analysis in Triticum. IV. Analysis of (Aegilops speltoides × Triticum boeoticum) amphiploid and a hypothesis on the evolution of tetraploid wheats. Caryologia 22, 289–306. doi: 10.1080/00087114.1969.10796348
Giorgi, D., D'Ovidio, R., Tanzarella, O. A., Ceoloni, C., and Porceddu, E. (2003). Isolation and characterization of S genome specific sequences from Aegilops sect. Sitopsis species. Genome 46, 478–489. doi: 10.1139/g03-022
Golovnina, K., Glushkov, S., Blinov, A., Mayorov, V., Adkison, L., and Goncharov, N. (2007). Molecular phylogeny of the genus Triticum, L. Plant Syst. Evol. 264, 195–216. doi: 10.1007/s00606-006-0478-x
Gornicki, P., Zhu, H., Wang, J., Challa, G. S., Zhang, Z., Gill, B. S., et al. (2014). The chloroplast view of the evolution of polyploid wheat. New Phytol. 204, 704–714. doi: 10.1111/nph.12931
Goryunova, S. V., Chikida, N. N., and Kochieva, E. Z. (2008). Molecular analysis of the phylogenetic relationships among the diploid Aegilops species of the section Sitopsis. Russ. J. Genet. 44, 115–118. doi: 10.1134/s1022795408010146
Haider, N. (2013). The origin of the B-genome of bread wheat (Triticum aestivum L.). Russ. J. Genet. 49, 263–274. doi: 10.1134/s1022795413030071
Hammer, K. (1980). Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops, L. Kulturpflanze 28, 33–180. doi: 10.1007/BF01997266
Huang, S., Sirikhachornkit, A., Su, X., Faris, J., Gill, B., Haselkorn, R., et al. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. U.S.A. 99, 8133–8138. doi: 10.1073/pnas.072223799
Jaaska, V. (1980). Electrophoretic survey of seedling esterases in wheats in relation to their phylogeny. Theor. Appl. Genet. 56, 273–284. doi: 10.1007/BF00282570
Jewell, D. C. (1979). Chromosome banding in Triticum aestivum cv. Chinese spring and Aegilops variabilis Chromosoma 71, 129–134. doi: 10.1007/BF00426370
Jewell, D. C., and Driscoll, C. J. (1983). The addition of Aegilops variabilis chromosomes to Triticum aestivum and their identification. Can. J. Genet. Cytol. 25, 76–84. doi: 10.1139/g83-014
Jiang, J., and Gill, B. S. (1993). Sequential chromosome banding and in situ hybridization analysis. Genome 36, 792–795. doi: 10.1139/g93-104
Jiang, J., and Gill, B. S. (1994a). Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheats. Chrom. Res. 2, 59–64. doi: 10.1007/BF01539455
Jiang, J., and Gill, B. S. (1994b). New 18S-26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 103, 179–185. doi: 10.1007/BF00368010
Kato, A. (2011). High-density fluorescence in situ hybridization signal detection on barley (Hordeum vulgare L.) chromosomes with improved probe screening and reprobing procedures. Genome 54, 151–159. doi: 10.1139/G10-098
Kawahara, T. (2009). Molecular phylogeny among Triticum-Aegilops species and of the tribe Triticeae. Breed. Sci. 59, 499–504. doi: 10.1270/jsbbs.59.499
Kerby, K., Kuspira, J., Jones, B. L., and Lookhart, G. L. (1990). Biochemical data bearing on the origin of the B genome in the polyploid wheats. Genome 33, 360–368. doi: 10.1139/g90-055
Kihara, H. (1937). Genomanalyse bei Triticum und Aegilops. VII. Kurze Uebersicht über die Ergebnisse der Jahre 1934-1936. Mem. Coll. Agric. Kyoto Imp. Univ. 41, 1–61.
Kihara, H. (1949). Genomanalyse bei Triticum und Aegilops. IX. Systematischer Aufbau der Gattung Aegilops auf genomanalytischer Grundlage. Cytologia 14, 135–144.
Kihara, H. (1954). Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 19, 336–357. doi: 10.1508/cytologia.19.336
Kihara, H. (1957). Completion of genome-analysis of three 6x species of Aegilops. Wheat Inf. Serv. 6:11.
Kihara, H. (1975). Origin of cultivated plants with special reference to wheat. Seiken Ziho 25-26, 1–24.
Kihara, H., and Tanaka, M. (1970). Attendum to the classification of the genus Aegilops by means of genome analysis. Wheat Inf. Serv. 30, 1–2.
Kihara, H., Yamashita, K., and Tanaka, M. (1959). Genomes of 6x species of Aegilops. Wheat Inf. Serv. 8, 3–5.
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al. (2011). “Aegilops.” in Wild crop relatives: Genomics and breeding resources Cereals, ed C. Kole (Berlin; Heidelberg: Springer), 1–76.
Kilian, B., Ozkan, H., Deusch, O., Effgen, S., Brandolini, A., Kohl, J., et al. (2007). Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 24, 217–227. doi: 10.1093/molbev/msl151
Kimber, G., and Feldman, M. (1987). Wild wheat, an Introduction. College of Agriculture University of Missouri, Columbia, Mo.
Knott, D. R., and Dvorák, J. (1976). Alien germ plasm as a source of resistance to disease. Ann. Rev. Phytopatol. 14, 211–235. doi: 10.1146/annurev.py.14.090176.001235
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 131–137. doi: 10.1139/gen-2013-0003
Konarev, V. G., Gavriliuk, E. P., Penyova, T. I., Kovnarev, A. V., Khakimova, A. G., and Migushova, E. F. (1976). On the nature and origin of wheat genomes by the data of biochemistry and immunochemistry of seed proteins. Agric. Biol. 11, 656–665.
Kubaláková, M., Kovárová, P., Suchánková, P., Číhalíková, J., Bartoš, J., Lucretti, S., et al. (2005). Chromosome sorting in tetraploid wheat and its potential for genome analysis. Genetics 170, 823–829. doi: 10.1534/genetics.104.039180
Kushnir, U., and Halloran, G. M. (1981). Evidence for Aegilops sharonensis Eig as the donor of the B genome in wheat. Genetics 99, 495–512.
Li, D., Li, T., Wu, Y., Zhang, X., Zhu, W., Wang, Y., et al. (2018). FISH-based markers enable identification of chromosomes derived from tetraploid Thinopyrum elongatum in hybrid lines. Front. Plant Sci. 9:526. doi: 10.3389/fpls.2018.00526
Lilienfeld, F. A. (1951). H. Kihara: Genome-analysis in Triticum and Aegilops. X. Concluding review. Cytologia 16, 101–123.
Limin, A. E., and Fowler, D. B. (1985). Cold hardiness in Triticum and Aegilops species. Can. J. Plant Sci. 65, 71–77. doi: 10.4141/cjps85-010
Linc, G., Friebe, B.R., Kynast, R.G., Molnar-Lang, M., Köszegi, B., Sutka, J., et al. (1999). Molecular cytogenetic analysis of Aegilops cylindrica host. Genome 42, 497–503. doi: 10.1139/gen-42-3-497
Linc, G., Sepsi, A., and Molnár-Láng, L. (2012). A FISH karyotype to study chromosome polymorphisms for the Elytrigia elongata E genome. Cytogenet. Genome Res. 136, 138–144. doi: 10.1159/000334835
Liu, B., Segal, G., Rong, J. K., and Feldman, M. (2003). A chromosome-specific sequence common to the B genome of polyploid wheat and Aegilops searsii. Plant Syst. Evol. 241, 55–66. doi: 10.1007/s00606-003-0015-0
Liu, C. J., Atkinson, M. D., Chinoy, C. N., Devos, K. M., and Gale, M. D. (1992). Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor. Appl. Genet. 83, 305–312. doi: 10.1007/BF00224276
Liu, W., Koo, D.-H., Friebe, B., and Gill, B. S. (2016). A set of Triticum aestivum-Aegilops speltoides Robertsonian translocation lines. Theor. Appl. Genet. 129, 2359–2368. doi: 10.1007/s00122-016-2774-3
Luo, M.-C., Deal, K. R., Yang, Z.-L., and Dvorak, J. (2005). Comparative genetic maps reveal extreme crossover localization in the Aegilops speltoides chromosomes. Theor. Appl. Genet. 111, 1098–1106. doi: 10.1007/s00122-005-0035-y
Maestra, B., and Naranjo, T. (1997). Homoeologous relationships of Triticum sharonense chromosomes to T. aestivum. Theor. Appl. Genet. 94, 657–663. doi: 10.1007/s001220050463
Maestra, B., and Naranjo, T. (1998). Homoeologous relationships of Aegilops speltoides chromosomes to bread wheat. Theor. Appl. Genet. 97, 181–186. doi: 10.1007/s001220050883
Maestra, B., and Naranjo, T. (1999). Structural chromosome differentiation between Triticum timopheevii and T. turgidum and T. aestivum. Theor. Appl. Genet. 98, 744–750. doi: 10.1007/s001220051130
Makkouk, K., Comeau, A., and Ghulam, W. (1994). Resistance to barley yellow dwarf luteovirus in Aegilops species. Can J Plant Sci 74, 631–634. doi: 10.4141/cjps94-113
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., Jakobsen, K. S., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092. doi: 10.1126/science.1250092
McIntosh, R. A., Yamazaki, Y., Dubkovsky, G., Rogers, J., Morris, C. F., Appels, R., et al. (2013). “Catalogue of Gene Symbols for Wheat,” in The 12th International Wheat Genetics Symposium (Yokohama), 395.
Middleton, C. P., Senerchia, N., Stein, N., Akhunov, E. D., Keller, B., Wicker, T., et al. (2014). Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe. PLoS ONE 9:e85761. doi: 10.1371/journal.pone.0085761
Mirzaghaderi, G., Houben, A., and Badaeva, E. (2014). Molecular-cytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenet. 7:91. doi: 10.1186/s13039-014-0091-6
Molnár, I., Schneider, A., and Molnar-Lang, M. (2005). Demonstration of Aegilops biuncialis chromosomes in wheat background by genomic in situ hybridization (GISH) and identification of U chromosomes by FISH using GAA sequences. Cereal Res. Commun. 33, 673–680. doi: 10.1556/CRC.33.2005.2-3.134
Molnár, I., Vrána, J., Burešová, V., Cápal, P., Farkas, A., Darkó, É., et al. (2016). Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J. 88, 452–467. doi: 10.1111/tpj.13266
Monneveux, P., Zaharieva, M., and Rekika, D. (2000). “The utilisation of Triticum and Aegilops species for the improvement of durum wheat,” in Durum Wheat Improvement in the Mediterranean Region: New challenges, eds C. Royo, M. Nachit, and N. Di Fonzo (Zaragoza: CIHEAM), 71–81.
Mukai, Y., Endo, T. R., and Gill, B. S. (1990). Physical mapping of the 5S rRNA multigene family in common wheat. J. Hered. 81, 290–295. doi: 10.1093/oxfordjournals.jhered.a110991
Nakai, Y. (1978). “The origin of the tetraploid wheats revealed from the study on esterase isozymes,” in Proceedings of the 5th International Wheat Genetics Symposium, ed S. Ramanujan (New Dehli: Indian Society of Genetics and Plant Breeding), 108–119.
Naranjo, T. (1995). Chromosome structure of Triticum longissimum relative to wheat. Theor. Appl. Genet. 91, 105–109. doi: 10.1007/BF00220865
Naranjo, T., Roca, A., Goicoecha, P. G., and Giraldez, R. (1987). Arm homoeology of wheat and rye chromosomes. Genome 29, 873–882. doi: 10.1139/g87-149
Nath, J., Hanzel, J. J., Thompson, J. P., and McNay, J. W. (1984). Additional evidence implicating Triticum searsii as the B genome donor to wheat. Biochem. Genet. 22, 37–50. doi: 10.1007/BF00499285
Nath, J., McNay, J. W., Paroda, C. M., and Gulati, S. C. (1983). Implication of Triticum searsii as the B-genome donor to wheat using DNA hybridization. Biochem. Genet. 21, 745–759. doi: 10.1007/BF00498921
Ogihara, Y., and Tsunewaki, K. (1988). Diversity and evolution of chloroplast DNA in Triticum and Aegilops as revealed by restriction fragment analysis. Theor. Appl. Genet. 76, 321–332. doi: 10.1007/BF00265331
Olivera, P. D., Kilian, A., Wenzl, P., and Steffenson, B. J. (2013). Development of a genetic linkage map for Sharon goatgrass (Aegilops sharonensis) and mapping of a leaf rust resistance gene. Genome 56, 367–376. doi: 10.1139/gen-2013-0065
Peacock, W. J., Dennis, E. S., and Gerlach, W. L. (1981). “Molecular aspects of wheat evolution: repeated DNA sequences,” in Wheat Science - Today and Tomorrow, eds T. Evans and W. J. Peacock (Cambridge: Cambridge University Press), 41–60.
Pedersen, C., and Langridge, P. (1997). Identification of the entire chromosome complement of bread wheat by two-color FISH. Genome 40, 589–593. doi: 10.1139/gen-40-5-589
Pedersen, C., Rasmussen, S. K., and Linde-Laursen, I. (1996). Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39, 93–104. doi: 10.1139/g96-013
Pestsova, E. G., Goncharov, N. P., and Salina, E. A. (1998). Elimination of a tandem repeat of telomeric heterochromatin during the evolution of wheat. Theor. Appl. Genet. 97, 1380–1386. doi: 10.1007/s001220051032
Petersen, G., Seberg, O., Yde, M., and Berthelsen, K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. doi: 10.1016/j.ympev.2006.01.023
Pietro, M. E., Tuleen, N. A., and Hart, G. E. (1988). “Development of wheat-Triticum searsii disomic chromosome addition lines,” in Proceedings of the 7th International Wheat Genetics Symposium, eds T. E. Miller and R. M. D. Koebner (Cambridge: Bath Press; Avon), 409–414.
Pradhan, G. P., Prasad, P. V. V., Fritz, A. K., Kirkham, M. B., and Gill, B. S. (2012). High temperature tolerance in Aegilops species and Its potential transfer to wheat. Crop Sci. 52, 292–304. doi: 10.2135/cropsci2011.04.0186
Rakszegi, M., Molnár, I., Lovegrove, A., Darkó, É., Farkas, A., Láng, L., et al. (2017). Addition of Aegilops U and M chromosomes affects protein and dietary fiber content of wholemeal wheat flour. Front. Plant Sci. 8:1529. doi: 10.3389/fpls.2017.01529
Raskina, O., Brodsky, L., and Belyayev, A. (2011). Tandem repeats on an eco-geographical scale: outcomes from the genome of Aegilops speltoides. Chrom. Res. 19, 607–623. doi: 10.1007/s10577-011-9220-9
Rawat, N., Neelam, K., Tiwari, V. K., Randhawa, G. S., Friebe, B., Gill, B. S., et al. (2011). Development and molecular characterization of wheat-Aegilops kotschyi addition and substitution lines with high grain protein, iron, and zinc. Genome 54, 943–953. doi: 10.1139/g11-059
Rayburn, A. L., and Gill, B. S. (1986). Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep. 4, 102–109. doi: 10.1007/BF02732107
Riley, R., Unrau, J., and Chapman, V. (1958). Evidence on the origin of the B genome of wheat. J. Hered. 49, 91–98. doi: 10.1093/oxfordjournals.jhered.a106784
Rodríguez, S., Maestra, B., Perera, E., Díez, M., and Naranjo, T. (2000a). Pairing affinities of the B- and G-genome chromosomes of polyploid wheats with those of Aegilops speltoides. Genome 43, 814–819. doi: 10.1139/g00-055
Rodríguez, S., Perera, E., Maestra, B., Díez, M., and Naranjo, T. (2000b). Chromosome structure of Triticum timopheevii relative to T. turgidum. Genome 43, 923–930. doi: 10.1139/g00-062
Ruban, A., Fuchs, J., Marques, A., Schubert, V., Soloviev, A., Raskina, O., et al. (2014). B Chromosomes of Aegilops speltoides are enriched in organelle genome-derived sequences. PLoS ONE 9:e90214. doi: 10.1371/journal.pone.0090214
Said, M., Hribová, E., Danilova, T. V., Karafiátová, M., Čížkov, á J., Friebe, B., et al. (2018). The Agropyron cristatum karyotype, chromosome structure and cross-genome homoeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor. Appl. Genet. 131, 2213–2227. doi: 10.1007/s00122-018-3148-9
Salina, E. A. (2006). The Structure and Evolution of Genomes of Polyploid Wheats and Their Wild Relatives: Analyses Using Macro- and Microsatellites. Siberian Branch of the Russian Academy of Sciences, Institute of Cytology and Genetics, Novosibirsk.
Salina, E. A., Adonina, I. G., Vatolina, T. Y., and Kurata, N. (2004a). A comparative analysis of the composition and organization of two subtelomeric repeat families in Aegilops speltoides Tausch. and related species. Genetica 122, 227–237. doi: 10.1007/s10709-004-5602-7
Salina, E. A., Leonova, I. N., Efremova, T. T., and Roder, M. S. (2006a). Wheat genome structure: translocations during the course of polyploidization. Funct. Integr. Genomics 6, 71–80. doi: 10.1007/s10142-005-0001-4
Salina, E. A., Lim, Y. K., Badaeva, E. D., Scherban, A. B., Adonina, I. G., Amosova, A. A., et al. (2006b). Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids. Genome 49, 1023–1035. doi: 10.1139/g06-050
Salina, E. A., Numerova, O. M., Ozkan, H., and Feldman, M. (2004b). Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat. Genome 47, 860–867. doi: 10.1139/g04-044
Salina, E. A., Pestsova, E. G., Adonina, I. G., and Vershinin, A. V. (1998). Identification of a new family of tandem repeats in Triticeae genomes. Euphytica 100, 231–237. doi: 10.1023/A:1018360324242
Salina, E. A., Pestsova, E. G., and Vershinin, A. V. (1997). “Spelt-1” - a new family of tandem repeats. Rus. J. Genet. 33, 437–442,
Salina, E. A., Sergeeva, E. M., Adonina, I. G., Shcherban, A. B., Afonnikov, D. A., Belcram, H., et al. (2009). Isolation and sequence analysis of the wheat B genome subtelomeric DNA. BMC Genomics 10:141. doi: 10.1186/1471-2164-10-414
Salse, J., Chagué, V., Bolot, S., Magdelenat, G., Huneau, C., Pont, C., et al. (2008). New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics 9, 1–12. doi: 10.1186/1471-2164-9-555
Sandve, S. R., Marcussen, T., Mayer, K., Jakobsen, K. S., Heier, L., Steuernagel, B., et al. (2015). Chloroplast phylogeny of Triticum/Aegilops species is not incongruent with an ancient homoploid hybrid origin of the ancestor of the bread wheat D-genome. New Phytol. 208, 9–10. doi: 10.1111/nph.13487
Sarkar, P., and Stebbins, G. L. (1956). Morphological evidence concerning the origin of the B genome in wheat. Am. J. Bot. 43, 297–304. doi: 10.1002/j.1537-2197.1956.tb10494.x
Sasanuma, T., Miyashita, N. T., and Tsunewaki, K. (1996). Wheat phylogeny determined by RFLP analysis of nuclear DNA. 3. Intra- and interspecific variations of five Aegilops Sitopsis species. Theor. Appl. Genet. 92, 928–934. doi: 10.1007/BF00224032
Schneider, A., Linc, G., and Molnar-Lang, M. (2003). Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed 122, 396–400. doi: 10.1046/j.1439-0523.2003.00891.x
Schneider, A., Molnár, I., and Molnár-Láng, M. (2008). Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Sears, E. R. (1954). The aneuploids of common wheat. Columbia, MO: Missouri Agricultural Experiment Station and U.S. Department of Agriculture cooperating.
Sears, E. R. (1969). Wheat Cytogenetics. Ann. Rev. Genet 3, 451–468. doi: 10.1146/annurev.ge.03.120169.002315
Shams, I., and Raskina, O. (2018). Intraspecific and intraorganismal copy number dynamics of retrotransposons and tandem repeat in Aegilops speltoides Tausch (Poaceae, Triticeae). Protoplasma 255, 1023–1038. doi: 10.1007/s00709-018-1212-6
Shands, H., and Kimber, G. (1973). “Reallocation of the genomes of Triticum timopheevii Zhuk,” in Proceedings of the 4th International Wheat Genetics Symposium, eds E. R. Sears and L. M. S. Sears (Columbia, MO: Missouri Agricultural Experiment Station), 101–108.
Shcherban, A. B., Badaeva, E. D., Amosova, A. V., Adonina, I. G., and Salina, E. A. (2008). Genetic and epigenetic changes of rDNA in a synthetic allotetraploid, Aegilops sharonensis × Ae. umbellulata. Genome 51, 261–271. doi: 10.1139/G08-006
Siregar, U. J., Ishii, T., and Tsunewaki, K. (1988). “Aegilops searsii is a possible donor to Ae. kotschyi and Ae. variabilis,” in Proceedings of the 7th International Wheat Genetics Symposium, eds T. E. Miller and R. M. D. Koebner (Cambridge: Bath Press; Avon), 145–151.
Taketa, S., Ando, H., Takeda, K., Harrison, G. E., and Heslop-Harrison, J. S. (2000). The distribution, organization and evolution of two abundant and widespread repetitive DNA sequences in the genus Hordeum. Theor. Appl. Genet. 100, 169–176. doi: 10.1007/s001220050023
Talbert, L. E., Magyar, G. M., Lavin, M., Moylan, S. L., and Blake, T. K. (1991). Molecular evidence for the origin of S-derived genomes of polyploid Triticum species. Am. J. Bot. 78, 340–349. doi: 10.2307/2444956
Tanaka, M. (1955). Chromosome pairing in hybrids between Aegilops sharonensis and some species of Aegilops and Triticum. Wheat Inf. Serv. 2, 7–8.
Tanaka, M. (1956). Chromosome pairing and fertility in the hybrid between the new amphiploid SlSl-AA and emmer wheat. Wheat Inf. Serv. 3, 21–22.
Tanaka, M., Kawahara, T., and Sano, J.i. (1979). The origin and the evolution of wild tetraploid wheats. Wheat Inf. Serv. 47–48, 7–11.
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Teoh, S. B., and Hutchinson, J. (1983). Interspecific variation in C-banded chromosomes of diploid Aegilops species. Theor. Appl. Genet. 65, 31–40. doi: 10.1007/BF00276259
Van Slageren, M. W. (1994). Wild Wheats: A Monograph of Aegilops, L. and Amblyopyrum (Jaub. et Spach) Eig (Poaceae). Wageningen Agricultural University, Wageningen and ICARDA, Aleppo; Wageningen.
Vrána, J., Kubaláková, M., Simková, H., Cíhalíková, J., Lysák Martin, A., and Dolezel, J. (2000). Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 156, 2033–2041. http://www.genetics.org/content/156/4/2033.abstract
Waines, J. G., and Johnson, B. L. (1972). Genetic differences between Aegilops longissima, A. sharonensis, and A. bicornis. Can. J. Geneti. Cytol. 14, 411–415. doi: 10.1139/g72-051
Wang, G.-Z., Miyashita, N. T., and Tsunewaki, K. (1997). Plasmon analyses of Triticum (wheat) and Aegilops: PCR-single-strand conformational polymorphism (PCR-SSCP) analyses of organellar DNAs. Proc. Natl. Acad. Sci. U.S.A. 94, 14570–14577. doi: 10.1073/pnas.94.26.14570
Witcombe, J. R. (1983). A Guide to the Species of Aegilops, L. Their Taxonomy, Morphology and Distribution. Rome: IBPGR Secretariat.
Yamamoto, M. (1992a). Detection of ribosomal RNA genes in Aegilops by in situ hybridization. Bull. Osaka Priv. Coll. Assoc. 29, 77–82.
Yamamoto, M. (1992b). Distribution of ribosomal RNA genes in Aegilops and Triticum chromosomes. Bull Kansai Women Coll. 2, 25–37.
Yamane, K., and Kawahara, T. (2005). Intra- and interspecific phylogenetic relationships among diploid Triticum-Aegilops species (Poaceae) based on base-pair substitutions, indels, and microsatellites in chloroplast noncoding sequences. Am. J. Bot. 92, 1887–1898. doi: 10.3732/ajb.92.11.1887
Yan, L., Fairclough, R., and Bhave, M. (1998). “Molecular evidence supporting the origin of the B genome of Triticum turgidum from T. speltoides,” in Proceedings of the 9th International Wheat Genetics Symposium, ed A. E. Slinkard (Saskatoon, SK: Printcrafters Inc.) 119–121.
Yang, Y. C., Tuleen, N. A., and Hart, G. E. (1996). Isolation and identification of Triticum aestivum L. em. Thell. cv Chinese Spring-T. peregrinum Hackel disomic chromosome addition lines. Theor. Appl. Genet. 92, 591–598. doi: 10.1007/BF00224563
Yen, Y., and Kimber, G. (1989). Triploid hybrid between autotetraploid Triticum longissimum and Triticum speltoides. Cereal Res. Commun. 17, 259–264.
Yen, Y., and Kimber, G. (1990a). Genomic relationships of Triticum searsii to other S-genome diploid Triticum species. Genome 33, 369–373. doi: 10.1139/g90-056
Yen, Y., and Kimber, G. (1990b). Genomic relationships of Triticum sharonense with other S-genome diploid Triticum species. Plant Breed. 104, 53–57. doi: 10.1111/j.1439-0523.1990.tb00402.x
Yen, Y., and Kimber, G. (1990c). Production and meiotic analysis of autotriploid Triticum speltoides and T. bicorne. Theor. Appl. Genet. 79, 525–528. doi: 10.1007/BF00226163
Yen, Y., and Kimber, G. (1990d). Reinvestigation of the S-genome in Triticum kotschyi. Genome 33, 521–524. doi: 10.1139/g90-077
Yen, Y., and Kimber, G. (1992). The S genome in Triticum syriacum. Genome 35, 709–713. doi: 10.1139/g92-109
Yu, M. Q., and Jahier, J. (1992). Origin of Sv genome of Aegilops variabilis and utilization of the Sv as analyser of the S genome of the Aegilops species in the Sitopsis section. Plant Breed. 108, 290–295. doi: 10.1111/j.1439-0523.1992.tb00133.x
Zhang, H.-B., Dvorák, J., and Waines, J. G. (1992). Diploid ancestry and evolution of Triticum kotschyi and T. peregrinum examined using variation in repeated nucleotide sequences. Genome 35, 182–191. doi: 10.1139/g92-029
Zhang, H. B., and Dvorák, J. (1992). The genome origin and evolution of hexaploid Triticum crassum and Triticum syriacum determined from variation in repeated nucleotide sequences. Genome 35, 806–814. doi: 10.1139/g92-123
Zhang, P., Dundas, I. S., McIntosh, R. A., Xu, S. S., Park, R. F., Gill, B. S., et al. (2015). “Wheat– Aegilops introgressions,” in Alien Introgression in Wheat Cytogenetics, Molecular Biology, and Genomics, eds M. Molnár-Láng, C. Ceoloni, and J. Doležel (Heidelberg; New York, NY; Dodrecht; London: Springer International Publishing AG Switzerland), 221–244.
Zhang, P., Friebe, B., and Gill, B. S. (2002). Variation in the distribution of a genome-specific DNA sequence on chromosomes reveals evolutionary relationships in the Triticum and Aegilops complex. Plant Syst. Evol. 235, 169–179. doi: 10.1007/s00606-002-0224-y
Zhang, W., Zhang, R., Feng, Y., Bie, T., and Chen, P. (2013). Distribution of highly repeated DNA sequences in Haynaldia villosa and its application in the identification of alien chromatin. Chin. Sci. Bull. 58, 890–897. doi: 10.1007/s11434-012-5598-9
Zhao, L., Ning, S., Yi, Y., Zhang, L., Yuan, Z., Wang, J., et al. (2018). Fluorescence in situ hybridization karyotyping reveals the presence of two distinct genomes in the taxon Aegilops tauschii. BMC Genomics 19:3. doi: 10.1186/s12864-017-4384-0
Zhao, L., Ning, S., Yu, J., Hao, M., Zhang, L., Yuan, Z., et al. (2016). Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed. Sci. 66, 522–529. doi: 10.1270/jsbbs.16011
Zhukovsky, P. M. (1928). A critical-systematical survey of the species of the genus Aegilops, L. Bul. Appl. Bot. Genet Pl Breed. 18, 417–609.
Zohary, D., and Imber, D. (1963). Genetic dimorphism in fruit types in Aegilops speltoides. Heredity 18, 223–231. doi: 10.1038/hdy.1963.24
Zoshchuk, S. A., Badaeva, E. D., Zoshchuk, N. V., Adonina, I. G., Shcherban,', A. B, and Salina, E. A. (2007). Intraspecific divergence in wheats of the Timopheevi group as revealed by in situ hybridization with tandem repeats of the Spelt1 and Spelt52 families. Rus. J. Genet. 43, 636–645. doi: 10.1134/S1022795407060063
Keywords: wheat, Aegilops, S-genome of Ae. speltoides, S*-genome of other Aegilops species, chromosome, karyotype evolution, C-banding, FISH
Citation: Ruban AS and Badaeva ED (2018) Evolution of the S-Genomes in Triticum-Aegilops Alliance: Evidences From Chromosome Analysis. Front. Plant Sci. 9:1756. doi: 10.3389/fpls.2018.01756
Received: 30 July 2018; Accepted: 12 November 2018;
Published: 04 December 2018.
Edited by:
István Molnár, Centre for Agricultural Research (MTA), HungaryReviewed by:
Elena Khlestkina, Institute of Cytology and Genetics, Russian Academy of Sciences, RussiaShuhei Nasuda, Kyoto University, Japan
Copyright © 2018 Ruban and Badaeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ekaterina D. Badaeva, a2F0ZXJpbmFiYWRhZXZhQGdtYWlsLmNvbQ==
 Alevtina S. Ruban
Alevtina S. Ruban Ekaterina D. Badaeva
Ekaterina D. Badaeva