
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 November 2018
Sec. Plant Metabolism and Chemodiversity
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01599
 Congxi Cai1,2,3
Congxi Cai1,2,3 Wenxin Yuan1
Wenxin Yuan1 Huiying Miao1,2
Huiying Miao1,2 Mingdan Deng1
Mingdan Deng1 Mengyu Wang1
Mengyu Wang1 Jiayao Lin1
Jiayao Lin1 Wei Zeng1
Wei Zeng1 Qiaomei Wang1,2*
Qiaomei Wang1,2*R2R3-MYB transcription factor MYB51 is known to control indole glucosinolate (indole GSL) biosynthesis in Arabidopsis. Here, two copies of BoaMYB51 have been isolated in Chinese kale (Brassica oleracea var. alboglabra Bailey), designated BoaMYB51.1 and BoaMYB51.2, which exhibit overlapping but distinct expression levels among different organs and respond to signaling molecules in a similar pattern. It has been demonstrated a structural and functional conservation between BoaMYB51s and AtMYB51 by phylogenetic analysis, complementation studies and transient expression assay. To further investigate the transcriptional mechanism, we identified the transcriptional activation domain (TAD) and putative interacting proteins of BoaMYB51s by means of yeast (Saccharomyces cerevisiae) two hybrid. Using tobacco (Nicotiana benthamiana) transient expression assay, we confirmed that the carboxy-end is required for transcriptional activation activity of BoaMYB51s. In addition, several BoaMYB51-interacting proteins have been identified by yeast two-hybrid screening. These results provide important insights into the molecular mechanisms by which MYB51 transcriptionally regulates indole GSL biosynthesis.
Chinese kale (Brassica oleracea var. alboglabra Bailey), a biennial vegetable belonging to the family of Brassicaceae, is widespread in southern China and Southeast Asia, with a large growing area and a marketable supply in these regions. Generally, Chinese kale is consumed for its bolting stems as common edible parts, and the tender rosette leaves and sprouts are also widely consumed (Wang et al., 2017). In addition to good flavor, numerous studies indicate that Chinese kale has abundant glucosinolates (GSLs) (Sun et al., 2011; Qian et al., 2016).
GSLs are a class of nitrogen- and sulfur-containing amino acid-derived secondary metabolites, which are common in members of the Brassicaceae family including the model plant Arabidopsis thaliana and agriculturally important Brassica crops such as Chinese cabbage (Brassica rapa ssp. pekinensis), broccoli (Brassica oleracea var. italica), and cauliflower (Brassica oleracea var. botrytis) (Halkier and Gershenzon, 2006; Mithen et al., 2010; Cai et al., 2016; Seo et al., 2016; Capriotti et al., 2018). In recent years, GSL have evolved as a model system for study of secondary metabolite in plants. Depending on the origin of the core amino acid, GSLs are generally classified into three groups: aliphatic GSL derived from methionine, leucine, isoleucine or valine; indole GSL derived from tryptophan; and benzolic GSL derived from tyrosine or phenylalanine (Fahey et al., 2001; Kliebenstein et al., 2001; Wittstock and Halkier, 2002), of which more than 200 different GSLs have been identified (Clarke, 2010). It is known that GSLs and their breakdown products contribute to the protective effects against cancer (Gross et al., 2000; Mithen et al., 2003; Grubb and Abel, 2006). Besides the benefits to the human health, these compounds also play an active role in plant defense against microbial pathogens (Clay et al., 2009) as well as herbivorous insects (Kliebenstein et al., 2002; Levy et al., 2005), and therefore, it will be a potential strategy for improvement of Brassica vegetables by regulating GSL.
Over the past two decades, indole GSL biosynthetic pathway has been well elucidated, with the almost complete characterization of enzymes involved (Halkier and Gershenzon, 2006). The composition and content of GSLs vary drastically in response to environmental stimuli, and GSL biosynthesis can be controlled by multiple signals. An understudied aspect of GSL research is to illustrate this regulatory machinery. Recent observations have begun to provide evidence that transcriptional regulation plays a central role in biosynthesis of secondary metabolites (Stracke et al., 2007; Gonzalez et al., 2008). R2R3-MYB transcription factors have been shown to function in a variety of plant-specific processes, which control development, primary and secondary metabolism and response to biotic and abiotic stresses (Dubos et al., 2010). In A. thaliana, six R2R3-MYB transcription factors having a characteristic “[L/F]LN[K/R]VA” motif (Stracke et al., 2001) are known to be involved in transcriptional regulation of aliphatic GSL (MYB28, MYB29, and MYB76), and indole GSL (MYB34, MYB51, and MYB122) (Gigolashvili et al., 2007a,b, 2008; Malitsky et al., 2008; Sønderby et al., 2010). Among them, MYB51 controls indole GSL biosynthesis mainly in the shoots and have been demonstrated to exclusively trans-activates the expression of indole GSL biosynthetic genes and differentially respond to phytohormone signaling molecules such as abscisic acid (ABA), jasmonate (JA), ethylene (ETH), and salicylate (SA) (Gigolashvili et al., 2007a; Frerigmann and Gigolashvili, 2014), but the underlying transcriptional mechanism remains relatively poorly defined.
A class of JA-signaled bHLH transcription factors (MYC2, MYC3, and MYC4) has been shown to interact with the six GSL-related MYB transcription factors, respectively (Schweizer et al., 2013; Frerigmann et al., 2014). These MYB-bHLH protein complexes have been linked to GSL biosynthesis regulating, and similar scenarios can also be found in other plant secondary metabolites (Gonzalez et al., 2008; Xu et al., 2014). Since previous analyses of the reference genome sequence of B. oleracea identified a whole-genome triplication (WGT) event after its divergence from a common ancestor of A. thaliana in the Brassica ancestor, most genes in Chinese kale were found to be duplicated or triplicated (Cheng et al., 2014). Thus, construction of Chinese kale cDNA prey library for yeast two-hybrid screening will be enriched for cDNAs of naturally co-occurring proteins, which will increase the possibility to identify novel GSL-related MYB interactors and enable us to seek other types of MYB-bHLH protein complex regulating GSL biosynthesis.
Despite the identification of aliphatic GSL biosynthesis regulator MYB28 and MYB29 in Brassica rapa ssp. pekinensis (Kim et al., 2013), B. juncea (Augustine et al., 2013), and B. oleracea (Araki et al., 2013; Yin et al., 2017), none of the transcription factors regulating indole GSL in Brassica crops have been well investigated to date. Previous study only identified one BoMYB51 from broccoli (Yu et al., 2018). Here, we report that both two BoaMYB51 genes have been isolated with identification of their transcriptional activation domain (TAD) and novel protein interactors. Our results highlight the importance of BoaMYB51s in regulating indole GSL biosynthesis in Chinese kale, which could be potential in improvement of pest and pathogen resistance as well as health benefits of B. oleracea by metabolic engineering.
Chinese kale cultivar “DSCH” germinated and grown in Zhejiang University (Hangzhou, China) was used as material in this study. Chinese kale seeds were sterilized for 30 s in 75% ethanol and washed with sterile water twice, and then immersed in 10% bleach for 2 min, followed by washing with sterile water five times. They were then soaked in sterile-distilled water for 48 h at 25°C in dark. Chinese kale sprouts were cultured in petri dishes with three pieces of wet filter paper in a plant growth chamber at 25°C with a 16-h-light/8-h-dark photoperiod. Five-day-old sprouts and six-month-old Chinese kale plants were used to collect different organs to analyze BoaMYB51 expression patterns. For epi-brassinolide (eBL) (Sigma), methyl-jasmonate (MeJA) (Sigma), SA (Sigma), and flagelin22 (flg22) (Chinese Peptide, Hangzhou, China) treatment, 5-day-old Chinese kale sprouts were grown in sterile half-strength Murashige and Skoog (MS) media at 25°C with a 16-h-light/8-h-dark photoperiod.
Arabidopsis mutant myb34myb51 (Frerigmann and Gigolashvili, 2014) was used for functional complementation studies. Arabidopsis seeds were sterilized for 12 min in 10% bleach and washed with sterile water 5 times, and then were stratified for 3 days at 4°C. Arabidopsis plants were grown in sterile half-strength Murashige and Skoog (MS) media at 22°C with a 16-h-light/8-h-dark photoperiod.
Multiple protein sequence alignments were performed using Clustal W. The phylogenetic tree was produced based on the amino acid sequences of MYB34, MYB51, and MYB122 proteins from B. rapa, B. oleraceae, and A. thaliana by applying the NJ (Neighbor-joining) method with MEGA v.7.0 software (Kumar et al., 2016), and bootstrap values with 1,000 replicates were calculated.
Plant samples were ground using Tissuelyser-24 (Jingxin, Shanghai, China). Total RNA was isolated as described previously with minor modifications (Guo et al., 2013). Reverse transcription was performed using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara). qRT-PCR was performed on ABI StepOne Real-time PCR System (Thermo Fisher). BoaACTIN2 gene was used as an endogenous control and the expression of other genes was computed using the 2−ΔΔCT method. Primers used are listed in Supplementary Table 1. Data were analyzed from three independent sets of biological replicates.
Constructs for plant transformation were generated using Gateway system (Invitrogen). The full-length coding DNA sequences (CDS) of BoaMYB51.1 and BoaMYB51.2 were amplified with Gateway-compatible primers (see Supplementary Table 2). The PCR product was cloned using pENTR/D-TOPO CLONING KIT (Invitrogen) and then recombined with the binary vector pGWB2 (Nakagawa et al., 2007) to generate the 35Spro:BoaMYB51.1 and 35Spro:BoaMYB51.2 constructs. For the generation of BoaMYB51pro:GUS constructs, the promoter regions (~2 kb) of the two BoaMYB51 genes were isolated from genomic DNA of and cloned into the gateway binary vector pGWB3 (Nakagawa et al., 2007). Histochemical analysis was performed as previously described (Sun et al., 2009). Primers are tabulated in Supplementary Table 2. The above constructs were then transformed into Agrobacterium tumefaciens strain GV3101, which was used for transformation of Arabidopsis plants by floral dip method.
GSL were extracted and analyzed as previously described (Guo et al., 2013). Extract was applied to a DEAE-Sephadex A-25 (35 mg) column (Pyridine acetate form) (Sigma). ONPG (Sigma) was used as an internal standard for HPLC analysis. The GSL concentration was expressed as μmol g−1fresh weight (FW).
The transient expression assays were carried out in tobacco leaves as previously described (Zhai et al., 2015). The promoters of BoaCYP79B2.1, BoaCYP83B1, BoaSOT16.1, BoaCYP79F1, and AtCYP83B1 were amplified and cloned into entry vector using the pENTR/D-TOPO CLONING KIT (Thermo Fisher). Then promoters were fused with the luciferase reporter gene LUC into pGWB35 and pGreenII 0800-LUC to generate the reporter constructs BoaCYP79B2.1pro:LUC, BoaCYP83B1pro:LUC, BoaSOT16.1pro:LUC, BoaCYP79F1pro:LUC, and AtCYP83B1pro:LUC. Effector constructs 35Spro:BoaMYB51.1-YFP, 35Spro:BoaMYB51.2-YFP, and 35Spro:BoaMYB28.1-YFP were generated using ClonExpress II One Step Cloning Kit (Vazyme Biotech). We used a low-light cooled CCD imaging apparatus (NightOWL II LB983) to capture the LUC image and to count luminescence intensity. The leaves were sprayed with 100 mM luciferin (Promega) and were placed in darkness for 3 min before luminescence detection.
Full-length CDS of BoaMYB51.1 and BoaMYB51.2 and their derivatives were amplified with listed primers (see Supplementary Table 2). PCR products were recombined with the pGBKT7 vector using ClonExpress II One Step Cloning Kit (Vazyme Biotech) for fusion with the BD domain at their N terminal. The resulting constructs were then transformed into the yeast strain Saccharomyces cerevisiae AH109, and the presence of the transgenes was verified by PCR and growth on an SD/-Trp plate. The Matchmaker GAL4 two-hybrid systems (Clontech) was used for the transactivation activity assay. Each yeast liquid culture was serially diluted to OD600 = 1.0, and 5 μl of each dilution was spread on the plates containing SD/-Ade/-His/-Trp synthetic dropout medium.
The TAD deletion constructs of BoaMYB51.1 and BoaMYB51.2 were used for the Y2H screening of the pGADT7-based Chinese kale cDNA library, which was generated with mRNAs isolated from Chinese kale sprouts, leaves, roots, stems and flowers. Y2H assays were based on Matchmaker GAL4 two-hybrid systems (Clontech). Yeast transformants were exhaustively selected on SD/-Ade/-His/-Leu/-Trp/X-α-Gal medium. Putative BoaMYB51 interacting clones were characterized and sequenced.
Statistical analysis was performed using the SPSS package program version 11.5 (SPSS inc. Chicago, IL, USA). For Figure 7, Data was analyzed by one-way ANOVA, followed by Turkey's HSD multiple comparison test. The values are reported as means with their standard error for all results. Differences were considered significant at p < 0.05.
Chinese kale sequence information can be found in the Brassica database (BRAD, http://brassicadb.org) under the following accession numbers: BoaMYB51.1 (Bol013207), BoaMYB51.2 (Bol030761), BoaMYB28.1 (Bol017019), BoaCYP79B2.1 (Bol018585), BoaCYP83B1 (Bol033477), BoaSOT16.1 (Bol026200), BoaCYP79F1 (Bol038222), BoaBIM1.1 (Bol043819), BoaBIM1.2 (Bol008832), and BoaACTIN2 (Bol030974). All Arabidopsis genes used in this article are referenced in the Arabidopsis Genome Initiative under the following accession numbers: AtMYB51 (At1G18570), AtCYP83B1 (At4G31500) and AtSOT16 (At1G74100).
According to Brassica database (BRAD, http://brassicadb.org), in B. oleracea genome, there are two MYB51 homologs distributed in different chromosomes, namely BoaMYB51.1 (Bol013207, C08) and BoaMYB51.2 (Bol030761, C05). First, primers based on reference sequences from BRAD were used to isolate the two BoaMYB51 genes from Chinese kale cultivar “DSCH.” Next, the full-length coding sequences of two BoaMYB51 genes were confirmed with multiple amplifications in other Chinese kale cultivars. As a result, the length of CDS of BoaMYB51.1 is 1002 bp encoding a protein of 333 aa, whereas the CDS of BoaMYB51.2 is 990 bp encoding a protein of 329 aa (Table 1). Compared with reference sequences from BRAD, the coding sequence of BoaMYB51.1 isolated from Chinese kale has three different nucleotides and three different amino acids (Supplementary Figure 1), while BoaMYB51.2 has eight different nucleotides and six different amino acids (Supplementary Figure 2). Comparison of the coding sequence with their corresponding genomic sequences revealed that both BoaMYB51 genes consisted of three exons and two introns (Table 1).
The amino acid sequences of two BoaMYB51 proteins are 76% identical and share 74 and 70% similarity with AtMYB51, respectively (Supplementary Table 3). We also found that BoaMYB51.1 protein sequence shares maximum identity (97%) with BrMYB51.2 (Bra016553), and BoaMYB51.2 protein sequence showed the highest level of identity (94%) with BrMYB51.3 (Bra025666) (Supplementary Table 3). Furthermore, amino acid sequence alignment of two BoaMYB51s with AtMYB51 demonstrated a conserved N-terminal region (Figure 1), containing R2R3-MYB domain which was predicated to serve as DNA binding domain (Dubos et al., 2010). Nuclear localized signal (NLS) and characteristic “[L/F]LN[K/R]VA” motif belonging to R2R3-type MYB subgroup 12 were also found in BoaMYB51s (Figure 1). Compared with the highly conserved N-terminal domain, the C-terminal region of BoaMYB51 exhibited conservation in patches (Figure 1).
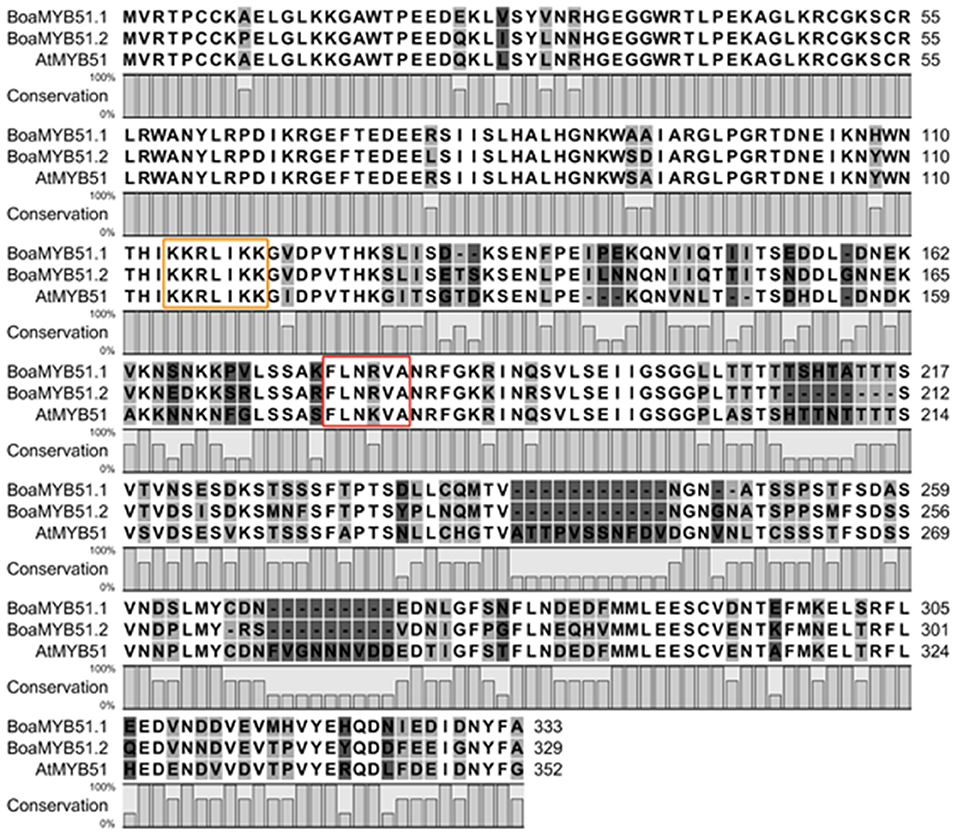
Figure 1. Multiple alignments of the BoaMYB51 and AtMYB51 protein sequences. Multiple alignments were performed using CLC Main Workbench software (QIAGEN). Nuclear localized signal (NLS) and characteristic “[L/F]LN[K/R]VA” motif were marked with yellow and red rectangle, respectively.
To investigate the evolutionary relationship, we identified all indole GSL-related MYB genes (MYB34, MYB51, and MYB122) in B. rapa, B. oleracea and A. thaliana. We constructed a phylogenetic tree of all deduced MYB amino acid sequences using the neighbor-joining method (Figure 2). Phylogenetic analyses revealed that in B. rapa genome there are three MYB51 orthologs, four MYB34 orthologs, and two MYB122 orthologs, while in B. oleracea genome, there are two MYB51 orthologs, one MYB34 ortholog, and one MYB122 ortholog (Figure 2).
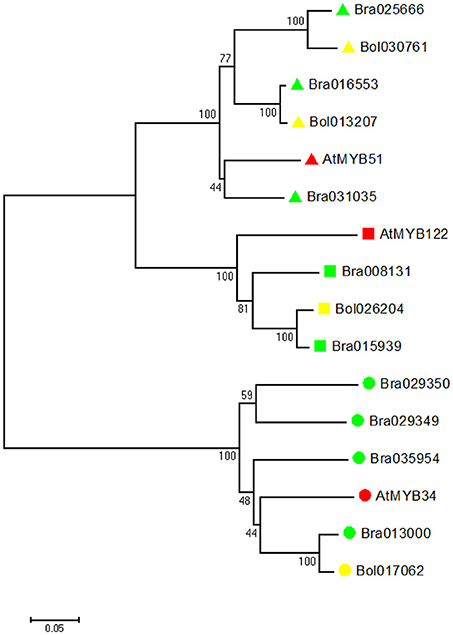
Figure 2. Evolutionary relationships of MYB34, MYB51, and MYB122 proteins in A. thaliana, B. rapa and B. oleracea. Phylogenetic analysis was performed using the neighbor-joining method (bootstrap test with 1000 replicates). Triangle indicates MYB51, square indicates MYB122, and circle indicates MYB34. A. thaliana proteins are labeled with red, B. rapa proteins are labeled with green, and B. oleracea proteins are labeled with yellow.
Distinct expression profiles among homologous genes in Brassica crops have been observed in many studies (Augustine et al., 2013; Zhang et al., 2015; Nour-Eldin et al., 2017). To test if two BoaMYB51 genes are differentially expressed, qRT-PCR analysis was performed to measure the expression levels of the two genes during different developmental stages and among different tissues in Chinese kale. In general, two BoaMYB51 genes showed overlapping expression profiles in Chinese kale and BoaMYB51.1 was more highly expressed (Figures 3A,B). At seedling stage, BoaMYB51.1 as well as BoaMYB51.2 was expressed in both shoots and roots, and BoaMYB51.1 exhibited a higher expression level than BoaMYB51.2 (Figure 3A). At reproductive stage, BoaMYB51 gene expression levels differed among different tissues. BoaMYB51.1 was highly expressed in roots, leaves and flowers with the highest level in leaves while lowest level in siliques. However, BoaMYB51.2 was expressed abundantly only in roots and flowers, and a trace accumulation of BoaMYB51.2 transcript was detected in leaves and siliques (Figure 3B). In order to confirm the result of qRT-PCR analysis, we performed the GUS histochemical analysis of BoaMYB51pro:GUS transgenic lines in Arabidopsis. As shown in Supplementary Figure 3, both two BoaMYB51 genes were expressed in aerial part and underground part, and a higher GUS staining was observed in BoaMYB51.1pro:GUS.
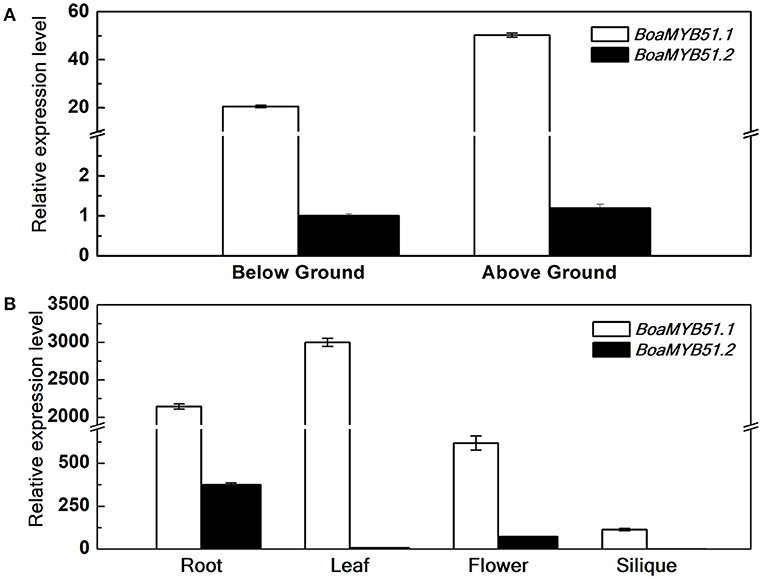
Figure 3. Relative gene expression of BoaMYB51s in Chinese kale organs at the seedling stage (5 day) (A) and reproductive stage (6 month) (B). Each data point represents the mean of three independent biological replicates (mean ± SE). (A) Values are shown compared with expression level of BoaMYB51.2 in below ground tissues. (B) Values are shown compared with expression level of BoaMYB51.2 in siliques.
The transcript level of MYB51 varies in response to phytohormones (Frerigmann and Gigolashvili, 2014). To address whether two BoaMYB51s respond to signaling molecules such as eBL, MeJA, SA, and flg22, we examined the expression patterns of both two genes in Chinese kale sprouts, which had been treated with corresponding signaling molecules. Generally, eBL and MeJA treatment caused a repression of BoaMYB51s, whereas SA and flg22 treatment induced the expression of BoaMYB51 genes (Figures 4A–H). eBL and MeJA repressed the expression of BoaMYB51.1 in a similar mode, which differed from the expression pattern of BoaMYB51.2. The expression level of BoaMYB51.2 was reduced at 6 h under eBL treatment while at 1h with MeJA treatment (Figures 4A–D). In contrast, SA and flg22 treatment caused a significant induction of both BoaMYB51.1 and BoaMYB51.2 in a similar pattern. BoaMYB51s reached the highest level at 3 h with SA treatment while the peak time was 1 h after treatment with flg22 (Figures 4E–H).
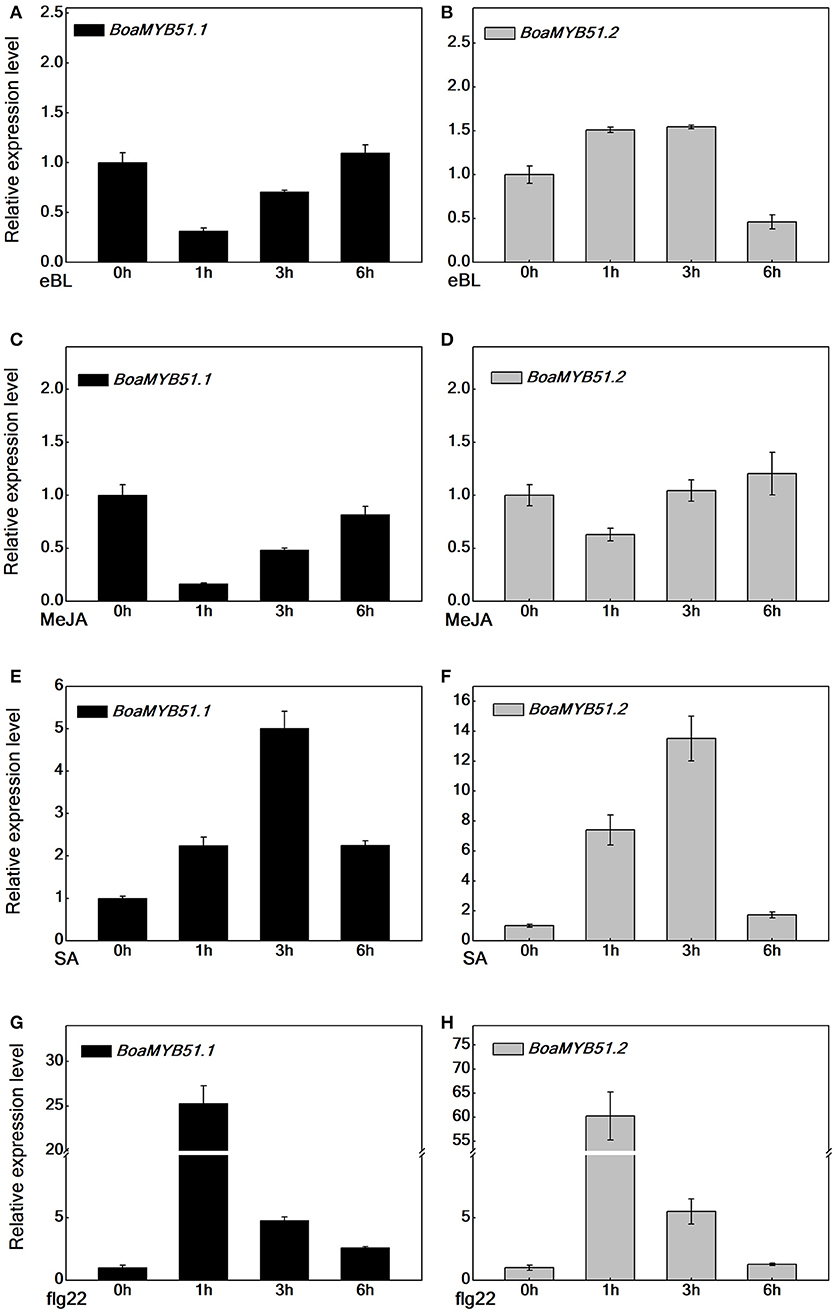
Figure 4. Time-course expression of BoaMYB51s in response to eBL, MeJA, SA, and flg22 treatments. Five-day-old Chinese kale sprouts were treated with 1 μM eBL (A,B), 100 μM MeJA (C,D), 100 μM SA (E,F), and 100 nM flg22 (G,H) for indicated times before total RNAs were extracted for real time-qPCR assays. Values are shown compared with gene expression level at 0 h. Each data point represents the mean of three independent biological replicates (mean ± SE).
To investigate the biological function of BoaMYB51s in Chinese kale, we analyzed the transcriptional activation ability of the two transcription factors using transient assays. N. benthamiana leaves were infiltrated with Agrobacteria strain GV3101 carrying the 35Spro:BoaMYB51 or 35Spro:BoaMYB28.1 construct as effector expression and reporter constructs containing the promoter of indole GSL biosynthetic genes (BoaCYP79B2.1, BoaCYP83B1, and BoaSOT16.1) or aliphatic GSL biosynthetic gene (BoaCYP79F1) fused with LUC. We showed that all tested promoters of indole GSL biosynthetic genes were activated by two BoaMYB51s but not affected by BoaMYB28.1 (Figures 5A–L and Supplementary Figures 4A–D). On the contrary, co-transformation of 35Spro:BoaMYB51 with BoaCYP79F1pro:LUC failed to cause an obvious increase of luminescence intensity (Figures 6A–D and Supplementary Figures 5A,B). These results implicated that BoaMYB51s are specifically involved in the transcriptional regulation of indole GSL biosynthesis in Chinese kale.
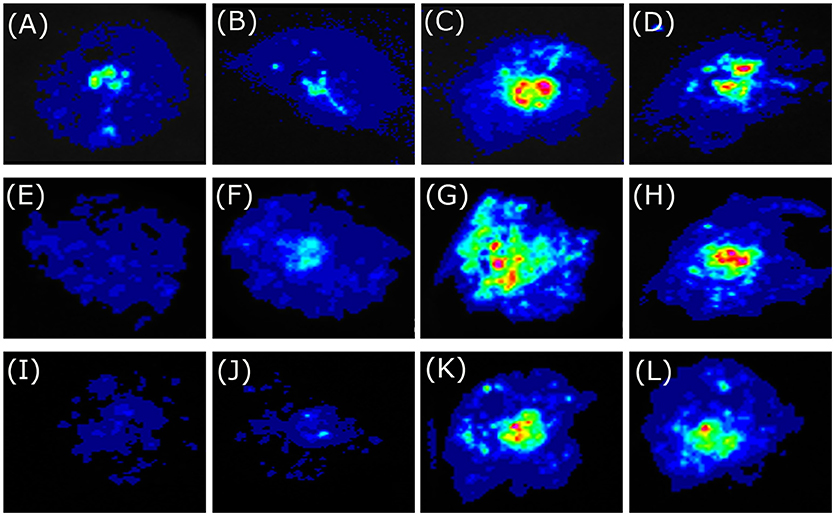
Figure 5. Tobacco transient expression assays showing that BoaMYB51 trans-activates the expressions of Chinese kale indole glucosinolate biosynthetic genes. (A–D) Co-infiltration of BoaCYP79B2.1pro:LUC and 35Spro:YFP, 35Spro:BoaMYB28.1, 35Spro:BoaMYB51.1 or 35Spro:BoaMYB51.2, respectively. (E–H) Co-infiltration of BoaCYP83B1pro:LUC and 35Spro:YFP, 35Spro:BoaMYB28.1, 35Spro:BoaMYB51.1 or 35Spro:BoaMYB51.2, respectively. (I–L) Co-infiltration of BoaSOT16.1pro:LUC and 35Spro:YFP, 35Spro:BoaMYB28.1, 35Spro:BoaMYB51.1 or 35Spro:BoaMYB51.2, respectively. Representative images of N. benthamiana leaves 72 h after infiltration are shown.
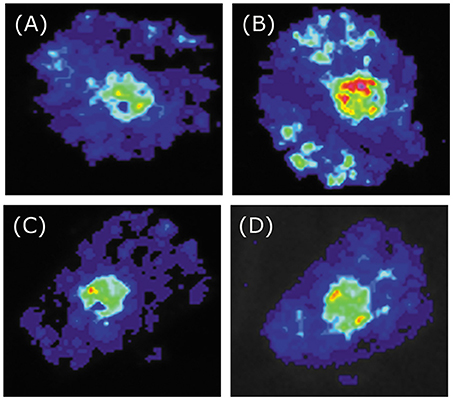
Figure 6. Tobacco transient expression assays showing that BoaMYB51 fail to trans-activates the expressions of BoaCYP79F1. (A–D) Co-infiltration of BoaCYP79F1pro:LUC and 35Spro:YFP, 35Spro:BoaMYB28.1, 35Spro:BoaMYB51.1 or 35Spro:BoaMYB51.2, respectively. Representative images of N. benthamiana leaves 72 h after infiltration are shown.
To substantiate the functional contribution of each BoaMYB51 to the regulation on indole GSL biosynthesis, two BoaMYB51s were over-expressed in the Arabidopsis mutant myb34myb51. Three independent homozygous lines of each BoaMYB51 were analyzed for total as well as individual indole GSL contents in 6-week old rosette leaves. We showed that both two BoaMYB51 genes complemented not only the accumulation of total indole GSL contents but also individual indole GSL fractions (Figure 7). The increased indole GSL accumulation in transgenic lines correlated with increased mRNA levels of the indole GSL pathway genes of Arabidopsis. As shown in Supplementary Figure 6, the gene expression levels of CYP79B2, CYP79B3, CYP83B1, and SOT16 were considerably increased in complementation lines compared with mutants. Moreover, transient assay revealed that both two BoaMYB51 transcription factors were able to activate the expression of AtCYP83B1pro:LUC (Supplementary Figures 7A–D). Collectively, these results clearly suggested that both two BoaMYB51 genes are active and positively regulate the indole GSL biosynthesis.
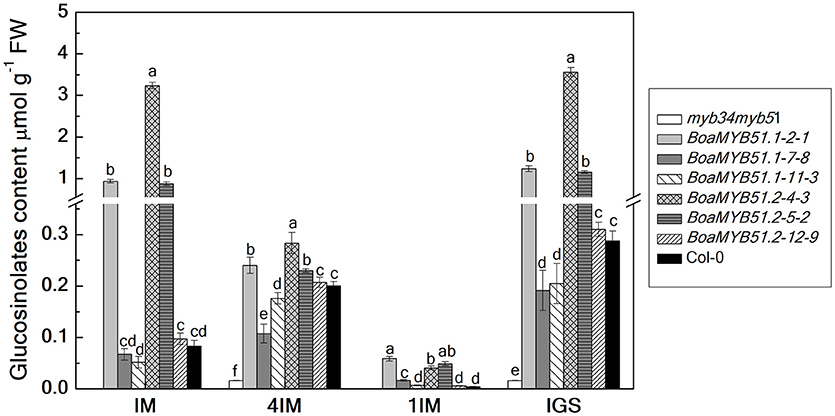
Figure 7. Functional complementation analyses of BoaMYB51 genes in Arabidopsis thaliana myb34myb51. The glucosinolate content and profile were determined in 6-week-old rosette leaves. Three independent mutant-complemented lines for each BoaMYB51 gene were analyzed. Each data point represents the mean of three independent biological replicates (mean ± SE).
Next, to better understand the transcriptional activation mechanism of BoaMYB51, we defined the transcriptional activation domain (TAD) of two BoaMYB51s using the MATCHMAKER GAL4-based Two-Hybrid System 3 (Clontech). In these assays, we found that both two BoaMYB51s had strong transcriptional activation activity whereas BoaMYB51.1 derivative containing amino acids from 1 to 274, and BoaMYB51.2 derivative containing amino acids from 1 to 270, do not, which suggested that the carboxy-end domain of both BoaMYB51.1 and BoaMYB51.2 could be the TAD (Figure 8A). To validate this in planta, we co-expressed construct encoding full-length BoaMYB51s or TAD-deleted BoaMYB51 derivatives with reporter BoSOT16.1pro:LUC in N. benthamiana leaves. Our results showed that BoaMYB51sΔTAD led to a substantial decrease of luminescence intensity when compared with full-length BoaMYB51s, suggesting that deletion of C-end strongly weakened the transcriptional activation activity of BoaMYB51s (Figure 8B and Supplementary Figures 8A–D). Taken together, these data indicated that C-end is required for transcriptional activation activity of BoaMYB51s.
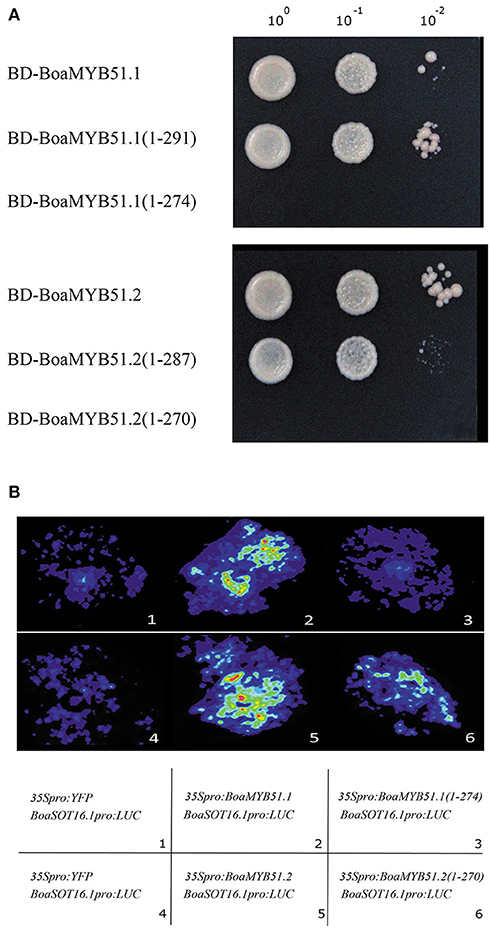
Figure 8. C-end is the TAD of BoaMYB51. (A) Yeast assay shows the deletion of C-end abolishes the transcriptional activity of BoaMYB51s. Yeast clones were grown on yeast synthetic dropout lacking Trp or on selective media lacking Ade, His, and Trp. one-tenth and one-hundred dilution yeast growth in media were also shown. (B) Tobacco transient expression assays show C-end is required for the transcriptional activity of BoaMYB51s. Representative images of N. benthamiana leaves 72 h after infiltration are shown. Each experiment was repeated at least three times with similar results.
To identify proteins that function in cooperation with BoaMYB51, a yeast two-hybrid assay with a Chinese kale complementary DNA (cDNA) library as prey was performed. To circumvent the problem that full-length BoaMYB51s have autoactivation activity, we used BoaMYB51ΔTAD as bait. In total, we analyzed plasmids from 52 (BoaMYB51.1ΔTAD as bait) and 82 (BoaMYB51.2ΔTAD as bait) yeast colonies by sequencing. Using the basic local alignment search tool (BLAST) of BRAD, we identified a total of 34 unique inserts, most of which represent full length or nearly full-length coding sequences. Duplicates and sequences unambiguously belonging to proteins of the photosynthetic and the ribosomal machinery were excluded from further analysis, leaving a curated list of 30 candidate interactors (Table 2).
Among the proteins interacting with MYB51 were transcription factors (Bol014189, Bol030761, and Bol043819), other nuclear-localized regulatory proteins (Bol044700, and Bol024700, and Bol026467), protein regulatory proteins (Bol037760, Bol030585, and Bol044051), and a protein with unassigned functions (Bol022070). In the current study, we further analyzed the interaction between BoaMYB51 and BoaBIM1.1 (Bol043819), because previous studies demonstrated that MYB-bHLH protein complex is important for transcriptional regulation of GSL biosynthesis (Schweizer et al., 2013; Frerigmann et al., 2014).
BES1-interacting Myc-like1 (BIM1), a basic helix-loop-helix (bHLH) transcription factor, was first identified by yeast two-hybrid assay using BES1 as bait in Arabidopsis. BIM1 was reported to be involved in brassinosteroid responses such as hypocotyl elongation (Yin et al., 2005). There are two BIM1 paralogs in Chinese kale, namely BoaBIM1.1 (Bol043819) and BoaBIM1.2 (Bol008832), and BoaBIM1.2 showed a higher expression level than BoaBIM1.1 (Supplementary Figure 9). Screened by yeast two-hybrid assay, BoaBIM1.1 was identified to be a protein interactor of BoaMYB51s. To verify this new identified MYB-bHLH complex, we cloned full-length BoaBIM1.1 and BoaBIM1.2 from Chinese kale and fused them to the GAL4-activation domain. We found that both BoaBIM1.1 and BoaBIM1.2 showed interaction with two BoaMYB51s (Figure 9A). Next, we mapped the interaction domain of BoaBIM1.1 with BoaMYB51s using yeast two-hybrid assays. We found that C-end but not N-end of BoaBIM1.1 was required for its interaction with BoaMYB51s (Figure 9B).
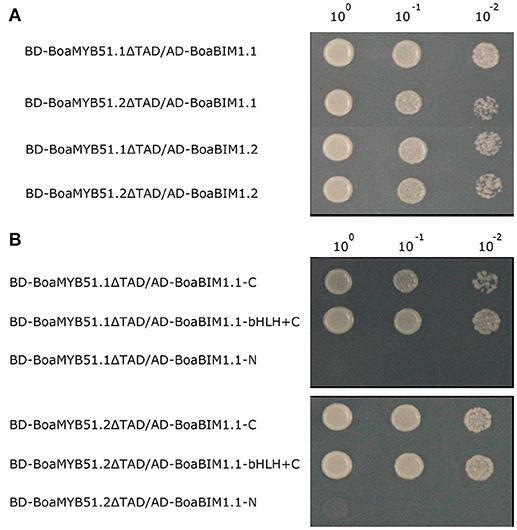
Figure 9. BoaMYB51s interact with BoaBIMs. (A) A yeast two-hybrid assay shows the interaction between BoaMYB51s and BoaBIMs. (B) C-end of BoaBIM1.1 was sufficient for its interaction with BoaMYB51s. BoaBIM1.1-C, C-terminal domain of BoaBIM1.1 (332–545), BoaBIM1.1-bHLH+C, bHLH and C-terminal domain of BoaBIM1.1 (278–545), and BoaBIM1.1-N, N-terminal domain of BoaBIM1.1 (1–277). Yeast clones were grown on yeast synthetic dropout lacking Trp and Leu or on selective media lacking Ade, His, Trp, and Leu. One-tenth and one-hundred dilution yeast growth in media were also shown. Each experiment was repeated at least three times with similar results.
Chinese kale is abundant in GSL accumulation and MYB51 acts as an important regulator of indole GSL biosynthesis. Here, we isolated both two MYB51 homologs for the first time in Brassica crops. BoaMYB51s exhibited overlapping but distinct expression profiles in Chinese kale and BoaMYB51.1 exhibited a higher expression level than BoaMYB51.2 (Figure 3). Previous study in Arabidopsis indicated that MYB51 seems to be the major regulator of indole GLS biosynthesis in shoots, while MYB34 is decisive for the production of indole GLS in roots (Frerigmann and Gigolashvili, 2014). In our current research, BoaMYB51.1 has a higher transcript level in shoots when compared to roots (Figure 3A), which was in line with the study in Arabidopsis. Next, we found that two BoaMYB51 genes responded to signaling molecules in a similar pattern, which were repressed by MeJA while induced by SA and flg22 (Figure 4). In Arabidopsis, the expression level of MYB51 has also been demonstrated to be activated by SA and flg22 (Millet et al., 2010; Frerigmann and Gigolashvili, 2014), while was decreased upon MeJA and eBL treatment (Guo et al., 2013; Frerigmann and Gigolashvili, 2014). However, MYB34 gene expression was repressed by SA but triggered by MeJA (Frerigmann and Gigolashvili, 2014). To analyze whether these signaling molecules affect indole GSL accumulation, we measure the GSL content of Chinese kale sprouts after signaling molecules treatment. Our results revealed that flg22 and SA treatment elevated the content of indole GSL, while eBL treatment reduced the accumulation of indole GSL, thus demonstrating the importance of BoaMYB51.1 for signal-mediated indole GSL regulation (Supplementary Figure 10). However, MeJA treatment also increased the content of indole GSL, substantiating the importance of MYB34 upon JA signaling (Supplementary Figure 10). Collectively, it seems that BoaMYB51.1 act as a major regulator of indole GSL biosynthesis while BoaMYB51.2 perform as an accessory role. To obtain insights into BoaMYB51s, we analyzed their transcriptional activation mechanisms, defined their transcriptional activation domains, and identified BoaMYB51-interacting proteins.
In the present study, complementation experiments in A. thaliana myb34myb51 double mutants and tobacco transient expression assay demonstrated that both two BoaMYB51 proteins are involved in controlling indole GSL biosynthesis in Chinese kale. Two BoaMYB51 proteins were shown to exclusively trans-activate the indole GSL biosynthetic genes including BoaCYP79B2, BoaCYP83B1 and BoaSOT16 (Figure 5) while they failed to induce the aliphatic GSL biosynthetic gene BoaCYP79F1 (Figure 6), which is in consistency with previous report of AtMYB51 (Gigolashvili et al., 2007b). Moreover, it has been revealed that two BoaMYB51s also activates AtCYP83B1 (Supplementary Figures 7A–D), implying that MYB51-mediated transcription regulation of indole GSL biosynthesis is conserved. Amino acid sequence alignment of two BoaMYB51 and AtMYB51 proteins showed conserved N-terminal domain among them (Figure 1). It is known that N-terminal domain of R2R3-MYB is responsible for its DNA binding function (Dubos et al., 2010), which explains the conservation of MYB51-mediated regulation on GSL biosynthesis. Besides, when promoter sequences of BoaSOT16.1 and AtSOT16 genes were aligned, a conserved cis-element was observed (Supplementary Figure 11), which is a putative binding site of MYB51. In addition to indole GSL regulator, we also found that BoaMYB28 positively regulates aliphatic GSL biosynthetic gene without directly affecting indole GSL biosynthesis in Chinese kale, which is in line with the results in A. thaliana and other Brassica crops (Gigolashvili et al., 2007a; Augustine et al., 2013), suggesting that the MYB-mediated regulatory mechanism of aliphatic GSL and indole GSL biosynthesis is conserved in Brassicaceae. To further study the MYB-mediated specific regulation on GSL biosynthesis, we swapped the N-terminal fragments of BoaMYB28 for the N-terminal domain of BoaMYB51.1 and co-expressed this chimeric protein with BoaSOT16.1pro:LUC and BoaCYP79F1pro:LUC, respectively in tobacco leaves (Supplementary Figure 12). Our results revealed that BoaMYB51.1N−SWAP protein failed to activate the promoter of BoaSOT16.1, whereas co-transfomation with BoaCYP79F1pro:LUC led to a significant increase of luminescence intensity (Supplementary Figure 12). We also generated another chimeric protein with N-terminal fragments of BoaMYB51.1 and C-terminal fragments of BoaMYB28 and this BoaMYB51.1C−SWAP protein showed an opposite pattern in tobacco transient assay when compared with BoaMYB51.1N−SWAP (Supplementary Figure 12). Collectively, these results suggest that the N-terminal domain is critical for the specific regulation of GSL biosynthesis.
Typically, a TF consists of a DNA-binding domain (DBD) and a transcription regulatory domain. It is generally considered that plant R2R3-MYB TFs share a conserved N-terminal MYB DBD, and C-terminal part of the protein usually harbors an activation or repression domain (Dubos et al., 2010). However, the experimentally proven details about TAD in R2R3-MYB TFs is limited. From studies by Goff et al. (1992), the TAD of the ZmC1 is located in a carboxy-terminal acidic region. Moreover, the C-terminal acidic region of AtMYB2 and AtMYB12 was found to be able to activate transcription (Urao et al., 1996; Stracke et al., 2017). Here, we identified the TAD of BoaMYB51, which is also located in the C-terminal region, by using yeast two-hybrid system and tobacco transient assay (Figures 8A,B). According to previous reports, the TAD of a TF often contains acidic, glutamine-rich, or proline-rich stretches of amino acids (Stracke et al., 2017). The TAD of BoaMYB51 defined in this study is enriched in acidic amino acids, but proline-rich or glutamine-rich amino acid stretches were not identified, which is in good accordance with the features of the TADs of above-mentioned R2R3-MYB TFs. It is a general phenomenon that the acidic TADs of TFs overlap closely with the destruction elements (Salghetti et al., 2000). Whether the acidic TAD of BoaMYB51 also functions as a degron which may account for the unstable characteristic of MYB51 protein needs further analysis. Since the amino sequences between AtMYB51 and BoaMYB51 are highly conserved, we also identified the TAD of AtMYB51 based on sequence alignment which is located in the amino acid region 294 to 352, and the result was validated by yeast two-hybrid system (Supplementary Figure 13). Furthermore, we also found that the TAD of AtMYB28 and BoaMYB28.1 are localized to C-end (data not shown), indicating that the C-terminally localized TAD is shared as a conserved feature by GSL-related MYBs.
The bHLH protein BIM1 is a brassinosteroid signaling component involved in mediating BR-responsive gene expression. It has been demonstrated that BIM1 regulates hypocotyl elongation, embryo patterning, and UVR8-mediated photomorphogenesis via interacting with the atypical bHLH protein BES1, AP2 protein DRN and UV-B light photoreceptor UVR8, respectively (Yin et al., 2005; Chandler et al., 2009; Liang et al., 2018). Here, BoaBIM1.1 was identified from a two-hybrid screen using BoaMYB51.1ΔTAD as bait (Table 2), and protein-protein interaction between BoaBIM1s and BoaMYB51s has been further confirmed. Gene expression analysis revealed that both BoaBIM1.1 and BoaBIM1.2 are expressed in Chinese kale leaves at a relatively high level (Supplementary Figure 9), in which BoaMYB51s are also highly expressed (Figure 3). To study the role of BIM1 on GSL biosynthesis, Arabidopsis triple mutant bim123 was used for GSL and qRT-PCR analysis. We found that indole GSL content and expression levels of indole GSL biosynthetic genes AtCYP79B2, AtCYP79B3, and AtCYP83B1 were decreased (Supplementary Figures 14A,B), implicating that BIM1 is a putative regulator of indole GSL biosynthesis. Besides, it has also been showed that the transcript level of AtMYB51 was not affected in bim123, suggesting that BIM1 interact with MYB51 and regulate the transcription of indole GSL biosynthetic genes.
Protein-protein interaction between MYB and bHLH transcription factors is a well-known paradigm of combinatorial gene regulation in plants and are instrumental in various biological processes (Feller et al., 2011). MYB-bHLH protein complex has been revealed to regulate anthocyanin biosynthesis (Gonzalez et al., 2008) and trichome morphogenesis (Zhao et al., 2008). Besides, the bHLH protein MYC2, known as a component of JA signaling pathway, and its close homologs MYC3 and MYC4 were shown to interact with GSL-related MYBs and affect thereby the GSL biosynthesis (Schweizer et al., 2013; Frerigmann et al., 2014). In this study, we identified another MYB-bHLH protein complex involved in the GSL biosynthesis regulation. It is known that the three MYCs bind to GSL-related MYBs via an N-terminal domain, the so-called JID domain (JAZ interaction domain). However, the C-terminal domain of BoaBIM1 show interaction with BoaMYB51 in yeast two-hybrid assays (Figure 9B), suggesting that the interaction mechanism between these two MYB-bHLH protein complexes is different.
CC and QW designed the research, CC and WY conducted the experiments. WY, HM, MD, MW, JL, and WZ analyzed the data. CC and QW wrote the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a grant from National Science Foundation of China (NO. 31270343, 31470385, 31801864) and China Postdoctoral Science Foundation (NO. 2017M621943).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01599/full#supplementary-material
Araki, R., Hasumi, A., Nishizawa, O. I., Sasaki, K., Kuwahara, A., Sawada, Y., et al. (2013). Novel bioresources for studies of Brassica oleracea: identification of a kale MYB transcription factor responsible for glucosinolate production. Plant Biotechnol. J. 11, 1017–1027. doi: 10.1111/pbi.12095
Augustine, R., Majee, M., Gershenzon, J., and Bisht, N. C. (2013). Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 64, 4907–4921. doi: 10.1093/jxb/ert280
Cai, C. X., Miao, H. Y., Qian, H. M., Yao, L. S., Wang, B. L., and Wang, Q. M. (2016). Effects of industrial pre-freezing processing and freezing handling on glucosinolates and antioxidant attributes in broccoli florets. Food Chem. 210, 451–456. doi: 10.1016/j.foodchem.2016.04.140
Capriotti, A. L., Cavaliere, C., La Barbera, G., Montone, C. M., Piovesana, S., Zenezini Chiozzi, R., et al. (2018). Chromatographic column evaluation for the untargeted profiling of glucosinolates in cauliflower by means of ultra-high performance liquid chromatography coupled to high resolution mass spectrometry. Talanta 179, 792–802. doi: 10.1016/j.talanta.2017.12.019
Chandler, J. W., Cole, M., Flier, A., and Werr, W. (2009). BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol. Biol. 69, 57–68. doi: 10.1007/s11103-008-9405-6
Cheng, F., Wu, J., and Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Hortic. Res. 1:14024. doi: 10.1038/hortres.2014.24
Clarke, D. B. (2010). Glucosinolates, structures and analysis in food. Anal Methods UK. 2, 310–325. doi: 10.1039/b9ay00280d
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fahey, J. W., Zalcmann, A. T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51. doi: 10.1016/S0031-9422(00)00316-2
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Frerigmann, H., Berger, B., and Gigolashvili, T. (2014). bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166, 349–369. doi: 10.1104/pp.114.240887
Frerigmann, H., and Gigolashvili, T. (2014). MYB34, MYB51, and MYB122 distinctly regulate indole glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7, 814–828. doi: 10.1093/mp/ssu004
Gigolashvili, T., Berger, B., Mock, H. P., Muller, C., Weisshaar, B., and Flugge, U. I. (2007a). The transcription factor HIG1/MYB51 regulates indole glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50, 886–901. doi: 10.1111/j.1365-313X.2007.03099.x
Gigolashvili, T., Engqvist, M., Yatusevich, R., Muller, C., and Flugge, U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177, 627–642. doi: 10.1111/j.1469-8137.2007.02295.x
Gigolashvili, T., Yatusevich, R., Berger, B., Muller, C., and Flugge, U. I. (2007b). The R2R3–MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261. doi: 10.1111/j.1365-313X.2007.03133.x
Goff, S. A., Cone, K. C., and Chandler, V. L. (1992). Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Gene Dev. 6, 864–875. doi: 10.1101/gad.6.5.864
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Gross, H. B., Dalebout, T., Grubb, C. D., and Abel, S. (2000). Functional detection of chemopreventive glucosinolates in Arabidopsis thaliana. Plant Sci. 159, 265–272. doi: 10.1016/S0168-9452(00)00354-X
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Guo, R. F., Qian, H. M., Shen, W. S., Liu, L. H., Zhang, M., Cai, C. X., et al. (2013). BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 64, 2401–2412. doi: 10.1093/jxb/ert094
Halkier, B. A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Bio. 57, 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Kim, Y. B., Li, X., Kim, S. J., Kim, H. H., Lee, J., Kim, H., et al. (2013). MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis). Molecules 18, 8682–8695. doi: 10.3390/molecules18078682
Kliebenstein, D. J., Lambrix, V. M., Reichelt, M., Gershenzon, J., and Mitchell-Olds, T. (2001). Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell. 13, 681–693. doi: 10.1105/tpc.13.3.681
Kliebenstein, D. J., Pedersen, D., Barker, B., and Mitchell-Olds, T. (2002). Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161, 325–332.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Levy, M., Wang, Q., Kaspi, R., Parrella, M. P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96. doi: 10.1111/j.1365-313X.2005.02435.x
Liang, T., Mei, S., Shi, C., Yang, Y., Peng, Y., Ma, L., et al. (2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell. 44, 512–523. doi: 10.1016/j.devcel.2017.12.028
Malitsky, S., Blum, E., Less, H., Venger, I., Elbaz, M., Morin, S., et al. (2008). The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 148, 2021–2049. doi: 10.1104/pp.108.124784
Millet, Y., Danna, C., Clay, N., Songnuan, W., Simon, M., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Mithen, R., Bennett, R., and Marquez, J. (2010). Glucosinolate biochemical diversity and innovation in the Brassicales. Phytochemistry 71, 2074–2086. doi: 10.1016/j.phytochem.2010.09.017
Mithen, R., Faulkner, K., Magrath, R., Rose, P., Williamson, G., and Marquez, J. (2003). Development of isothiocyanate-enriched broccoli, and its enhanced ability to induce phase 2 detoxification enzymes in mammalian cells. Theor. Appl. Genet. 106, 727–734. doi: 10.1007/s00122-002-1123-x
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Nour-Eldin, H. H., Madsen, S. R., Engelen, S., Jørgensen, M. E., Olsen, C. E., Andersen, J. S., et al. (2017). Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 35, 377–382. doi: 10.1038/nbt.3823
Qian, H. M., Liu, T. Y., Deng, M. D., Miao, H. Y., Cai, C. X., Shen, W. S., et al. (2016). Effects of light quality on main health-promoting compounds and antioxidant capacity of Chinese kale sprouts. Food Chem. 196, 1232–1238. doi: 10.1016/j.foodchem.2015.10.055
Salghetti, S. E., Muratani, M., Wijnen, H., Futcher, B., and Tansey, W. P. (2000). Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. U.S.A. 97, 3118–3123. doi: 10.1073/pnas.97.7.3118
Schweizer, F., Fernández-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 25, 3117–3732. doi: 10.1105/tpc.113.115139
Seo, M. S., Jin, M., Chun, J. H., Kim, S. J., Park, B. S., Shon, S. H., et al. (2016). Functional analysis of three BrMYB28 transcription factors controlling the biosynthesis of glucosinolates in Brassica rapa. Plant Mol. Biol. 90, 503–516. doi: 10.1007/s11103-016-0437-z
Sønderby, I. E., Burow, M., Rowe, H. C., Kliebenstein, D. J., and Halkier, B. A. (2010). A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol. 153, 348–363. doi: 10.1104/pp.109.149286
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3–MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Turgut-Kara, N., and Weisshaar, B. (2017). The AtMYB12 activation domain maps to a short C-terminal region of the transcription factor. Z. Naturforsch. C 72, 251–257. doi: 10.1515/znc-2016-0221
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sun, B., Liu, N., Zhao, Y. T., Yan, H. Z., and Wang, Q. M. (2011). Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem. 124, 941–947. doi: 10.1016/j.foodchem.2010.07.031
Sun, J. Q., Xu, Y. X., Ye, S. Q., Jiang, H. L., Chen, Q., Liu, F., et al. (2009). Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21, 1495–1511. doi: 10.1105/tpc.108.064303
Urao, T., Noji, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1996). A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. 10, 1145–1148. doi: 10.1046/j.1365-313X.1996.10061145.x
Wang, Y. Q., Hu, L. P., Liu, G. M., Zhang, D. S., and He, H. J. (2017). Evaluation of the nutritional quality of Chinese kale (Brassica alboglabra Bailey) using UHPLC-Quadrupole-Orbitrap MS/MS-based metabolomics. Molecules 22:1262. doi: 10.3390/molecules22081262
Wittstock, U., and Halkier, B. A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 6, 263–270. doi: 10.1016/S1360-1385(02)02273-2
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Yin, L., Chen, H., Cao, B., Lei, J., and Chen, G. (2017). Molecular characterization of MYB28 involved in aliphatic glucosinolate biosynthesis in Chinese kale (Brassica oleracea var. alboglabra Bailey). Front. Plant Sci. 8:1083. doi: 10.3389/fpls.2017.01083
Yin, Y., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Yu, Q., Hao, G., Zhou, J., Wang, J., Evivie, E. R., and Li, J. (2018). Identification and expression pattern analysis of BoMYB51 involved in indole glucosinolate biosynthesis from broccoli (Brassica oleracea var. italica). Biochem. Biophys. Res. Commun. 501, 598–604. doi: 10.1016/j.bbrc.2018.05.058
Zhai, Q. Z., Zhang, X., Wu, F. M., Feng, H. L., Deng, L., Xu, L., et al. (2015). Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27, 2814–2828. doi: 10.1105/tpc.15.00619
Zhang, J., Liu, Z., Liang, J., Wu, J., Cheng, F., and Wang, X. W. (2015). Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J. Exp. Bot. 66, 6205–6218. doi: 10.1093/jxb/erv331
Keywords: BoaMYB51, Chinese kale, indole glucosinolate, protein-protein interaction, transcription regulation
Citation: Cai C, Yuan W, Miao H, Deng M, Wang M, Lin J, Zeng W and Wang Q (2018) Functional Characterization of BoaMYB51s as Central Regulators of Indole Glucosinolate Biosynthesis in Brassica oleracea var. alboglabra Bailey. Front. Plant Sci. 9:1599. doi: 10.3389/fpls.2018.01599
Received: 21 June 2018; Accepted: 17 October 2018;
Published: 06 November 2018.
Edited by:
Xiaoya Chen, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, ChinaReviewed by:
Gaojie Hong, ZheJiang Academy of Agricultural Sciences, ChinaCopyright © 2018 Cai, Yuan, Miao, Deng, Wang, Lin, Zeng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaomei Wang, cW13YW5nQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.