- 1Rice Research Institute, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Southwest Crop Genetic Resources and Genetic Improvement, Ministry of Education, Chengdu, China
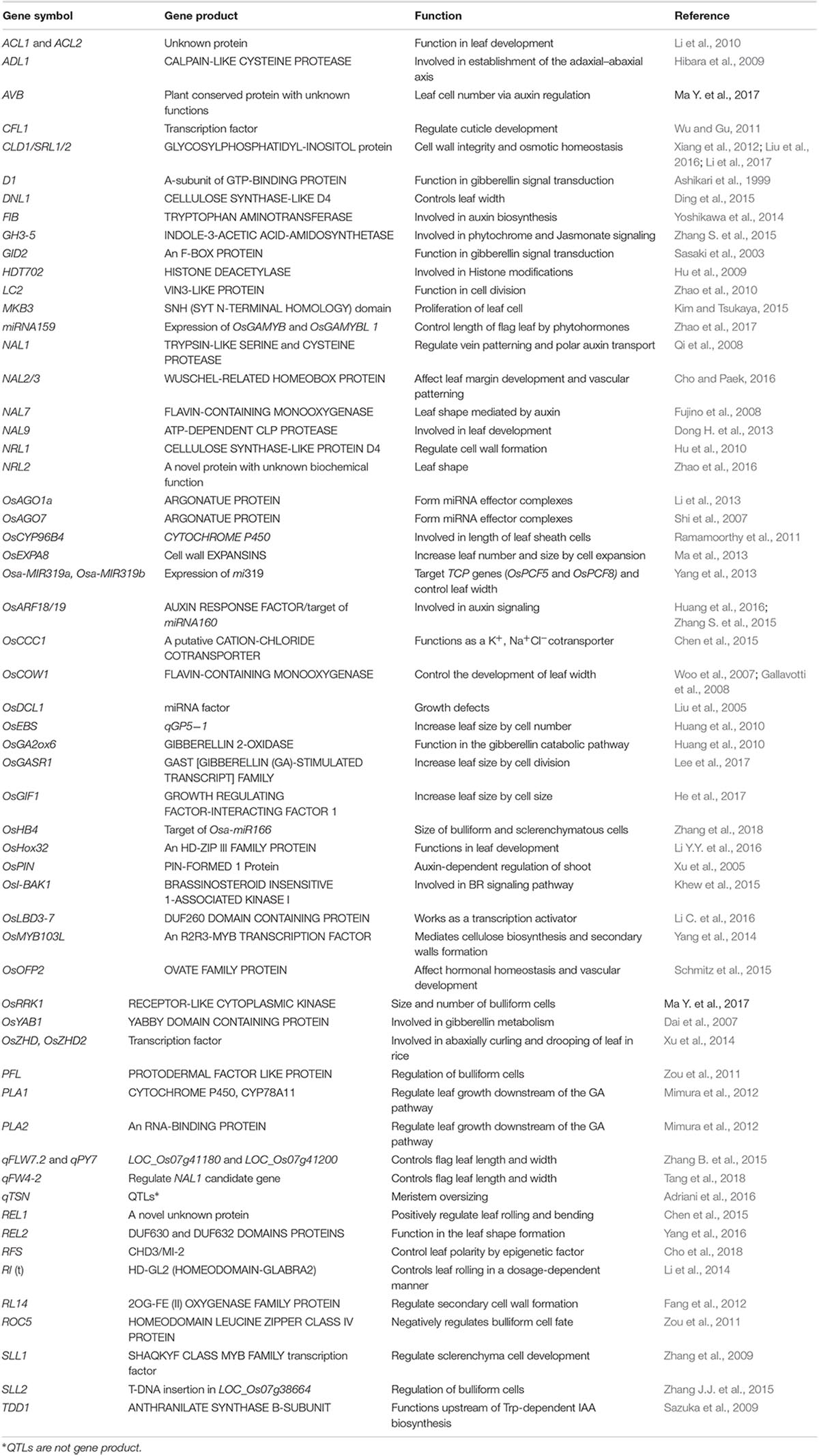
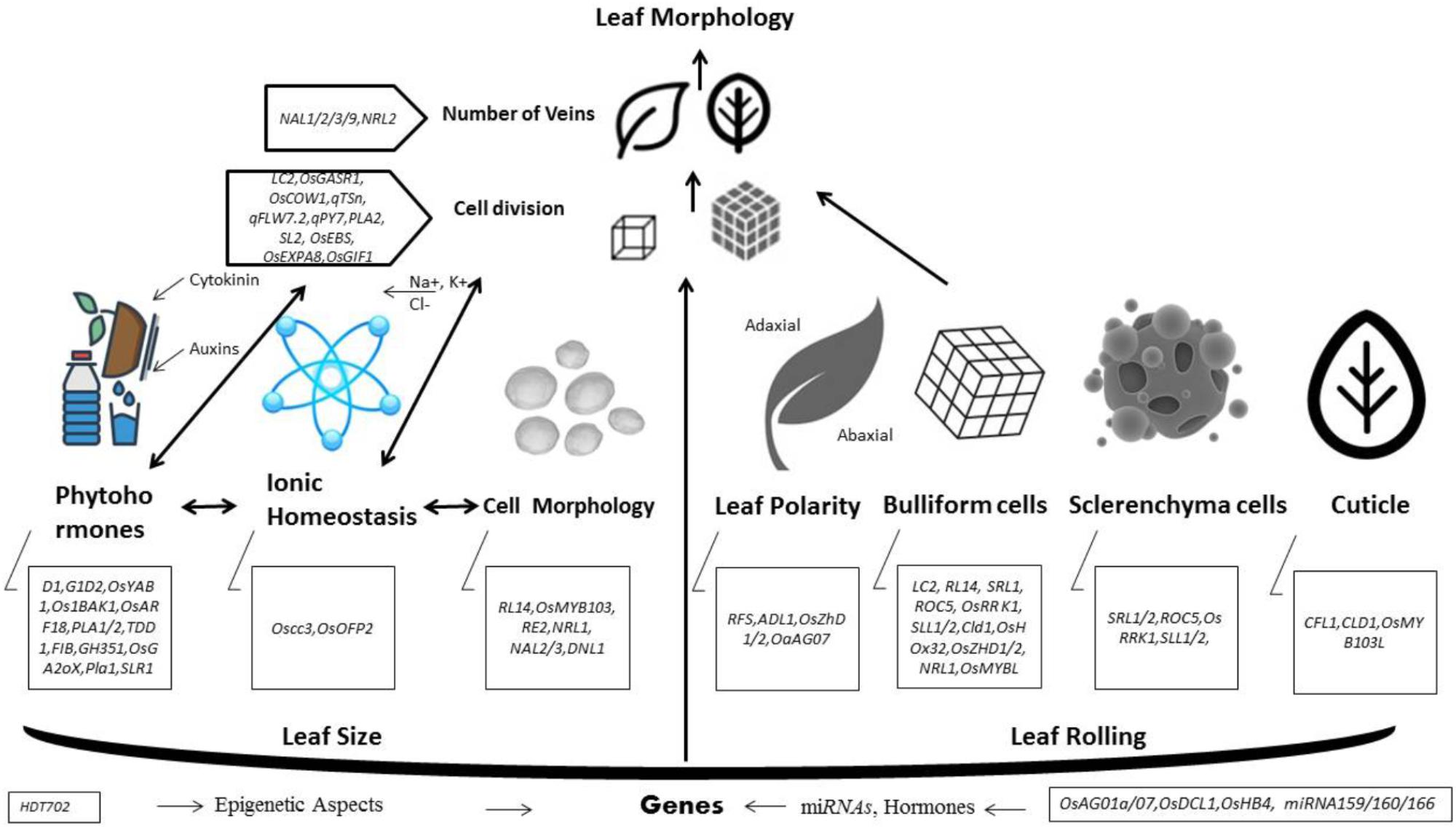
Yield is majorly affected by photosynthetic efficiency. Leaves are essential structure for photosynthesis and their morphology especially size and shape in a plant canopy can affect the rate of transpiration, carbon fixation and photosynthesis. Leaf rolling and size are considered key agronomic traits in plant architecture that can subsidize yield parameters. In last era, a number of genes controlling leaf morphology have been molecularly characterized. Despite of several findings, our understanding toward molecular mechanism of leaf rolling and size are under-developed. Here, we proposed a model to apprehend the physiological basis of different genes organized in a complex fashion and govern the final phenotype of leaf morphology. According to this leaf rolling is mainly controlled by regulation of bulliform cells by SRL1, ROC5, OsRRK1, SLL2, CLD1, OsZHD1/2, and NRL1, structure and processes of sclerenchyma cells by SLL1 and SRL2, leaf polarity by ADL1, RFS and cuticle formation by CFL1, and CLD1. Many of above mentioned and several other genes interact in a complex manner in order to sustain cellular integrity and homeostasis for optimum leaf rolling. While, leaf size is synchronized by multifarious interaction of PLA1, PLA2, OsGASR1, and OsEXPA8 in cell division, NAL1, NAL9, NRL1, NRL2 in regulation of number of veins, OsCOW1, OsPIN1, OsARF19, OsOFP2, D1 and GID in regulation of phytohormones and HDT702 in epigenetic aspects. In this review, we curtailed recent advances engrossing regulation and functions of those genes that directly or indirectly can distress leaf rolling or size by encoding different types of proteins and genic expression. Moreover, this effort could be used further to develop comprehensive learning and directing our molecular breeding of rice.
Introduction
Rice is a model plant of monocots and an important crop that feeds more than half of the population around the globe. According to estimates, food supply will be insufficient as growth rate of population and increase in yield is not harmonious (Zhu et al., 2010). Yield is a complex quantitative trait determined by various agronomic factors of plant morphology and environment. With recent advances made in the field of molecular biology, genes and QTLs for grain yield have been identified and characterized functionally. Incorporation of those genes could into local cultivars can assist molecular breeding for improvement of yield in rice (Lafitte et al., 2004; Liu et al., 2008).
The leaf is a major photosynthetic organ in plants and its morphology such as size and rolling are key components in plant architecture that significantly affect crop yield (Walter et al., 2009). Leaf rolling is normally caused in plant species due to water scarcity (Chandra and Dubey, 2009). However, other abiotic stress factors such as temperature, salt, UV radiation and heavy metals also affect leaf morphology and yield (Subashri et al., 2009). Morphology and physiology of leaf can affect light interception, carbon fixation, photosynthetic rate, transpiration and storage assimilation (Yuan et al., 2015). Leaf rolling is a protective mechanism to avoid photo damage in plants (Nar et al., 2009; Kadioglu et al., 2012). The leaf rolling reduces the exposed leaf area to sunlight, and it is regulated by complex mechanism of several interacting genes (Kadioglu et al., 2012).
The ideotype breeding of ‘Super rice’ suggests that uppermost three leaves should be erect, long, rolled (V-shaped), narrow, and dark green in color (Zhang et al., 2009). Engineering of molecular breeding based on ideotype could be a helpful approach to increase yield (Yang et al., 2014). A long and thick leaf contains more abundant photosynthetic pigment and content of nitrogen assimilates (Ghiglione et al., 2008). Moreover, thick leaf has low tendency to bend down as it has more potential to stay erect (Nelson et al., 2002). Appropriate leaf rolling is beneficial in rice to enhance its erectness and optimum leaf angle helps in proper light interception (Yuan et al., 2015). Morphological diversity in leaf folding (inward or outward) also allows adjustments for efficient photosynthetic activity per unit leaf area (Yuan et al., 2015). Optimum leaf rolling not only enhances the accumulation of dry matter but also slows down transpiration rate by reducing absorbance of solar radiation on leaves (Lang et al., 2004). Previous studies revealed low water potential and decreased turgidity in bulliform cells cause leaf rolling (Price et al., 1997). Partial leaf rolling is effective for better water use efficiency than fully flattened or extremely curved leaf on adaxial or abaxial side (Juarez et al., 2004). To date, a large number of genes are reported that regulate leaf morphology affecting factors especially leaf rolling and size. Our review will present a comprehensive understanding about the function of these genes.
Role of Genes in Optimum Leaf Rolling
Advancements in molecular methodologies lead to discovery of several leaf rolling genes that have been cloned and functionally characterized in rice. We have categorized those genes based on their part, which they play in final appearance of leaf.
Leaf Polarity (Adaxial/Abaxial)
Adaxial–abaxial determinants control leaf polarity in developing leaves of plant (Yamaguchi et al., 2012). Leaf morphogenesis is an important feature of plants, as they need to establish proximodistal, mediolateral, and adaxial–abaxial axes during leaf development (Hibara et al., 2009). Manipulation in molecular breeding can modify the adaxial–abaxial axis for adjustment in organogenesis. Leaf polarity is characterized in two stages, i.e., meristem-dependent and -independent stage (Toriba et al., 2011). Adaxial–abaxial polarity is considered to have an important effect on leaf rolling. Rice ADL1 (ADAXIALIZED LEAF 1) gene was isolated from adl1 mutant using positional cloning, and it encodes a CALPAIN-LIKE CYSTEINE PROTEINASE. Deficiency of CALPAIN-LIKE CYSTEINE PROTEINASE leads to rolling of leaves abaxially. Morphological analysis revealed bulliform cells specifically appear only on the adaxial side of leaf normally were also found on the abaxial side in adl1 mutant (Hibara et al., 2009). The faulty establishment of adaxial–abaxial polarity might be responsible for the rolling of leaf. Additionally, loss of a transcription factor, SLL1 (SHALLOT-LIKE 1) and overexpression of OsAGO7 (ARGONATUE) caused leaf rolling (Shi et al., 2007; Zhang et al., 2009). A crucial gene ROLLED FINE STRIPED (RFS) also showed leaf rolling phenotypes in rice due to poor development of vascular bundles on adaxial side of leaf. Knock down of key elements of CHD3/Mi-2 (chromatin remodeling factor of RFS) caused severe leaf rolling in rfs-1 mutant (Cho et al., 2018).
Bulliform Cells (Number and Size)
Many genes regulate leaf rolling by affecting cytological architecture of leaf cells, e.g., bulliforms, cuticle of leaves, and sclerenchymatous cells. Bulliform cells are located in ridges of vascular bundle near midrib on the adaxial epidermis of leaf (Li et al., 2010). These are highly vacuolated and play an important role in leaf rolling by regulating their number and size1. Thus far, dozens of genes modulating bulliform cell number and size have been functionally characterized. SRL1 (SEMI ROLLED LEAF 1) encodes a putative GPI (GLYCOSYLPHOSPHATIDYLINOSITOL) anchored protein in rice that is located in plasma membrane. The loss-of-function mutant of SRL1 exhibits rolling of leaves adaxially due to an augmented number of bulliform cells on adaxial surface of leaf. Further studies demonstrated SRL1 negatively normalizes the expression of genes encoding vacuolar H+-PYROPHOSPHATASE and H+-ATPASE that usually impede the development of bulliform cells (Xiang et al., 2012). Transgenic plants overexpressing OsHox32 that belongs to HD-ZIPIII (HOMEODOMAIN LEUCINE ZIPPER) gene family had a similar phenotype to that of loss-of-function mutant of SRL1. However, besides increasing, reduced number of bulliform cells are also responsible for leaf rolling (Li Y.Y. et al., 2016).
Curled leaf and dwarf 1 (cld1) mutant displayed defects of leaf rolling and is allelic to SRL1. CLD1/SRL1 encodes a GPI anchored protein that plays its role in formation of cell wall. The loss-of-function mutant of CLD1/SRL1 showed lesser contents of lignin and cellulose in epidermis of bulliform cells. Defects in cell wall formation cause more rapid water loss and reduce water retaining capacity of leaves (Li et al., 2017).
Compared to adaxial rolling of leaves, suppression of YAB1, a member of YABBY gene family leads to a higher number of bulliform cells and showed abaxial rolling of leaf (Dai et al., 2007). Likewise, overexpression of OsZHD1 (ZINC FINGER HOMEODOMAIN 1) or its homolog OsZHD2 and a defect in LC2 (LEAF INCLINATION 2) induced more number of bulliform cells that causes abaxial rolling of leaves (Zhao et al., 2010; Xu et al., 2014). Some genes control leaf rolling via regulating size of bulliform cells. Characterization of a hal1(half-pipe-like leaf 1) mutant in rice exhibited adaxial curling of leaves phenotype due to underdeveloped size of bulliform cells. Mutation in hal1 mutant also affected size of leaf blade and spikelet (Matsumoto et al., 2017). RL14 (ROLLING LEAF 14) encodes a 2OG-Fe (OXYGENASE PROTEIN) that is convoluted in formation of secondary cell wall of leaf (Fang et al., 2012). NRL1 (NARROW AND ROLLED LEAF 1) (Hu et al., 2010), encodes a CELLULOSE SYNTHASE-LIKE PROTEIN that is essential for normal biosynthesis of cell-wall (Fang et al., 2012). OsMYB103L, a R2R3MYB transcription factor usually targets CESA genes that are involved in regulation of cellulose synthesis (Fang et al., 2012; Yafeng et al., 2015). Transgenic plants with reduced expression level of RL14 or NRL1, or overexpression of OsMYBL103L displayed adaxial rolling of leaves due to reduced size of bulliform cells. Moreover, all of these genes were involved in cell-wall formation, suggesting these genes may contribute to cell division and ultimately regulate size of bulliform cells.
In rice a series of genes, such as ROC5 (RICE OUTER CELL SPECIFIC 5) and its downstream gene (PFL) PROTODERMAL FACTOR LIKE (Zou et al., 2011), ACL1 (ABAXIALLY CURLED LEAF 1), its homologous gene ACL2 (Li et al., 2010), ROLLED AND ERECT LEAF 1 (REL1) (Chen et al., 2015), REL2 (Yang et al., 2016), (LATERAL ORGAN BOUNDARIES DOMAIN 3-7) OsLBD 3-7 (Li C. et al., 2016), (NARROW LEAF 7) NAL7 (Fujino et al., 2008), NAL2/3 (Cho et al., 2013), (BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1) OsI-BAK1 (Khew et al., 2015), and (AUXIN RESPONSE FACTOR) OsARF18 (Huang et al., 2016) regulate leaf rolling by affecting both number and area of bulliform cells. For example, defects of ROC5, a member of HDZIP CLASS IV gene family displayed abaxial rolling of leaf. Studies have also reported that reduced or increased expression level of ROC5 can affect leaf rolling, it serves as a negative regulator leaf rolling by mediating number and size of bulliform cells. Further studies also disclosed ROC5 probably regulates leaf rolling by binding to L1 motif box in promoter region of PFL, as loss-of-function of PFL also displayed phenotypes of abaxial leaf rolling (Zou et al., 2011). Higher expression of ROC5 in rl (t) mutant revealed positive correlation of bulliform cell area and LRI (leaf rolling index) in rice (Li et al., 2014). OsARF18 is involved in auxin signaling pathway and is a major target of miRNA160. The transgenic plants expressing miR160-resistance for OsARF18 displayed adaxial curling of leaves, due to decreased number and size of bulliform cells (Huang et al., 2016). Similarly, SLL2 (SHALLOT-LIKE 2) showed more adaxial curling of leaves in order to increase the photosynthetic activity through regulation of bulliform cells and other related genes (Zhang J.J. et al., 2015). OsRRK1 (RECEPTOR-LIKE CYTOPLASMIC KINASE 1) also showed an erect morphology with decreased number and size of bulliform cells in order to get high seed setting (Ma Y. et al., 2017). Keeping in view the functions of above mentioned genes it can be suggested that bulliform cells regulate leaf rolling by changing their size and number through complex interaction of several genes.
Sclerenchymatous Cells
Rice SLL1 encodes a transcription factor of KANADI family and its loss-of-function mutant showed extreme rolling of leaves phenotype caused by defective development of sclerenchymatous cells on the abaxial side. Further studies exposed SLL1 regulates leaf rolling by mediating programmed death of sclerenchymatous cells and inhibiting development of abaxial features (Zhang et al., 2009). SRL2 deficiency also brings about abnormal development of sclerenchymatous cells on the abaxial surface of leaf blade leading to adaxial rolling of leaves. Although, SLL1 and SRL2 both were involved in the sclerenchymatous cells development, the analysis of sll1:srl2 double mutant revealed that these two genes play role in different pathways to regulate leaf rolling (Liu et al., 2016). Moreover, NRL2 protein interacted with RL14 and distresses contents of cellulose, lignin and differentiation of sclerenchymatous cells (Zhao et al., 2016). These findings suggested that sclerenchymatous cells also play their part in regulation of leaf rolling.
Cuticle Development
Histological studies indicated that defective cuticle development is responsible for the rolling of leaves (Wu and Gu, 2011). Molecular mechanism of CFL1 (CURLY FLAG LEAF 1) in cuticle development exhibited its involvement in leaf rolling. CFL1 encodes a WW DOMAIN PROTEIN that was isolated from cfl1 mutant with curling leaf phenotype. Moreover, overexpression of OsMYB103L encodes a transcription factor R2R3MYB that regulates contents of cellulose and mechanical strength of leaves (Yang et al., 2014). It signifies role of cuticle development in developing normal leaf rolling.
miRNAs
There is a consensus that miRNAs play an important role in leaf development by negatively regulating expression of genes at post-transcriptional level (Moon and Hake, 2011). For example, AGO protein characterized by PAZ and PIWI domains is a core component of RNA-induced silence complex (RISCs) that play crucial role in rice leaf development. Overexpression of OsAGO7 leads to upward rolling of leaf blade (Shi et al., 2007). Meanwhile, transgenic plants that were knocked down against AGO1s via an RNAi silencing approach exhibited pleiotropic defects in phenotypic development, e.g., low height, narrow and rolled leaves (Wu et al., 2009). Additionally, miRNA160 has been found to be involved in leaf rolling by targeting OsARF18 (Huang et al., 2016). HOMEODOMAIN CONTAINING PROTEIN 4 (OsHB4) is major target of miRNA166 and play role in development of xylem and transpiration rate. Knockdown of miRNA166 in short tandem target mimic (sttm166) line resulted in rolling of leaf, which had smaller bulliform cells and reduced stomatal conductance (Zhang et al., 2018). The RNaseIII enzyme DICER-LIKE 1 (DCL1) is required for the miRNA biogenesis and has pivotal influences on plant growth and development. Down-regulation of OsDCL1 in RNAi lines leads to a phenotype of narrow leaf blade (Liu et al., 2005). Thus, some miRNAs indirectly can affect the leaf rolling via targeting other genes such as OsAGO7 and OsHB4.
Role of Genes in Controlling Leaf Size
Leaf size has long been regarded as central agronomic trait in rice and received tremendous attention. A range of QTLs for leaf size have been identified on several chromosomes in rice and most of genes governing leaf size were mainly identified through recessive mutations. Leaf size is regulated by different genes through various internal and external features of plant and environment, which are described below.
Number of Veins
Grasses leaves are of mostly strap shape, in which veins run in parallel fashion from base to tip and distance among the veins create the differentiation of C3 from C4 plants (Wang et al., 2017). The vascular system found in veins is thought to be directly associated with leaf size. NAL1 encodes a plant specific protein and preferentially expressed in vascular tissues. Reduced expression of NAL1 leads to decreased number of leaf veins that might be responsible for the narrow leaf phenotype (Qi et al., 2008). Besides of reduced expression, recessive mutation in NRL1 resulted in narrow leaves with reduced number of veins (Jiang et al., 2010). NAL2 and NAL3 (NAL2/3) encode WUSCHEL-RELATED HOMEOBOX PROTEIN in rice that are duplicate orthologs of genes NS1 (NARROW SHEATH 1) and NS2 in maize, respectively. The nal2:nal3 double mutant leaves contains decreased number of veins with extremely narrow leaves (Cho et al., 2013). Similarly, NAL9 and NRL2 also known as ClpP and SRL2, respectively, regulate leaf size through affecting the number of veins (Dong H. et al., 2013; Li et al., 2013). OsaMIR319a or Osa-MIR319b overexpression, both results in wider leaves, owing to an increased number of longitudinal small veins, suggesting miRNAs also regulate leaf size in rice (Yang et al., 2013).
Fourteen QTLs for flag leaf length and nine for breadth were detected in chromosomal substitution line (CSSL) in rice and candidate gene for qFW4-2 was NAL1 (Tang et al., 2018). Yield per plant and flag leaf width have positive association found in qFLW7.2 and qPY7 and both of them were positioned on chromosome 7 (Zhang B. et al., 2015). These evidences support the idea that leaf size had an obvious positive correlation with number of leaf veins.
Cell Division
NAL1, NRL1, and AVB (ABNORMAL VASCULAR BUNDLES) all are reported to be involved in cell division, implying these genes probably affect cell division to regulate leaf size (Jiang et al., 2015; Ma L. et al., 2017). OsCCC1, a member of CATION-CHLORIDE CO-TRANSPORTER family localized in plasma membrane played its role in ionic transportation. Recent finding demonstrates that OsCCC1 participates in elongation of cells by mediating ionic (K+, Cl-, and Na+) homeostasis to sustain osmoregulation, and loss-of- function of OsCCC1 leads to narrow leaves illustrating cell division also accounts for narrow leaves phenotype (Kong et al., 2011; Zhi et al., 2016). Additionally, PLA1 (PLASTOCHRON 1) and PLA2 encode cytochrome P450 and an RNA-binding protein, respectively, and both of them were reported to regulate leaf size mainly due to increase in cell size (Miyoshi et al., 2004). Two QTLs, qTSN4 and qTSN12 found in their NILs revealed, additional leaf area was produced by oversizing of meristems (Adriani et al., 2016). OsGASR1 (GA-STIMULATED RICE 1) that belongs to family of GAST [GIBBERELLIN (GA)-STIMULATED TRANSCRIPT] showed higher expression in regions of cell proliferation and increase leaf blade size due to increase in cell length. Its mRNA expression could be triggered by an exogenous application of gibberellins (Lee et al., 2017). OsGIF1 (GRF-INTERACTING FACTOR 1) influenced grain production and size of leaves in rice by regulating leaf cell size (He et al., 2017). OsGIF1 positively regulates cell proliferation and revealed conserved functional control of MAKIBA3 (MKB3) and ANGUSTIFOLIA3 (AN3) in rice and Arabidopsis, respectively. A loss-of-function mutant MKB3 exhibited narrowed- and rolled-leaf phenotype (Shimano et al., 2018). Overexpression of an α-expansin gene OsEXPA8 (EXPANSIN 8) produced improved root system, enhanced leaf number and enlarged leaf size in rice. Further analysis of OsEXPA8 line showed increased lignin content in cell wall and enhanced length of leaf cells (Ma et al., 2013). Expression of OsEBS (ENHANCING BIOMASS AND SPIKELET NUMBER) in rice caused increase in plant height, leaf size and spikelet number per panicle due to increase in cell number (Dong X. et al., 2013). A semi-dwarf mutant in which a single copy of transposon dissociator (Ds) was inserted into gene OsCYP96B4 (CYTOCHROME P450 96B4). It showed defects in plant height and length of leaf sheath cells (Ramamoorthy et al., 2011). So, keeping in view function of these genes in cell division (i.e., size and number), their use in breeding can provide an effective tool to engineer plants with more leaf area for better agronomic yield.
Phytohormones
Plant hormones such as auxin and gibberellins are necessary for plant development and play a substantial role in regulation of leaf size. Several genes have been characterized and involved in biosynthesis, transport and signal transduction pathways of some phytohormones. OsCOW1 (CONSTITUTIVELY WILTED 1) gene is identical to NAL7 that encodes a FLAVIN-CONTAINING MONO-OXYGENASE protein and indicated resemblance with YUCCA in Arabidopsis and FLOOZY in petunia, which encode for auxin biosynthesis (Woo et al., 2007). Overexpression and knock-down of OsCOW1 formed wide and narrow leaves, respectively. Further studies have also disclosed function of some members of YUCCA gene family that were involved in a tryptophan-dependent IAA biosynthetic pathway of rice and Arabidopsis (Gallavotti et al., 2008). Physiological and quantitative real-time PCR analysis of OsPIN1 (PIN-FORMED 1) showed that it can serve as an auxin efflux facilitator (Xu et al., 2005). NAL1 regulates polar transport of auxin in rice by OsPIN1 (Jiang et al., 2015). Another, leaf size gene NAL2/3 also found to be involved in distribution of auxin. In addition, TDD1 (TRYPTOPHAN DEFICIENT DWARF MUTANT 1) and FIB (FISH BONE) both were involved in auxin biosynthesis and mutants with reduced expression of TDD1 or FIB showed narrow leaves phenotype (Sazuka et al., 2009; Yoshikawa et al., 2014). Overexpression of OsARF19 (Zhang S. et al., 2015) or its downstream gene OsGH3-5, also revealed narrow leaves phenotype due to changes in level of phytohormones (Zhang S. et al., 2015).
Another class of phytohormones, gibberellin has proved to be involved in leaf expansion. A recent study showed that NAL2/3 not only regulate distribution of auxin but also provide negative feedback to gibberellin biosynthesis for gibberellin homeostasis in rice. It is indicating that NAL2/3 probably regulate leaf size via the crosstalk of GA and auxin (Cho and Paek, 2016). Rice PLA1 and PLA2 genes act downstream in gibberellin signal transduction pathway and their loss-of-function caused shortening of leaves (Mimura et al., 2012). In contrast, the mutant disrupted in SLR1 that is a negative regulatory factor for the gibberellin signal transduction, displayed elongated leaves in rice. Phenotypes of pla1 and pla2 mutants displayed rapid leaf emergence and small organs, and found gibberellin is the major phytohormone associated with PLA1 and PLA2 functions. Both, PLA1 and PLA2 act downstream of GA signal transduction pathway to regulate leaf development (Mimura et al., 2012). Catabolism of double mutants indicated that PLA1 and PLA2 were partially necessary for leaf elongation depending on the gibberellin contents. Moreover, altered expression of (OVATE FAMILY PROTEIN 2) OsOFP2 (Schmitz et al., 2015), OsGA2ox6 (Huang et al., 2010), (DWARF 1) D1 (Ashikari et al., 1999), and (GIBBERELLIN-INSENSITIVE DWARF 2) GID2 (Sasaki et al., 2003) all were involved in regulation of gibberellin pathway and causes changes in leaf size. Besides auxin and gibberellin altered leaf size was also observed in mutants that were driven by different genes involved in regulation of other phytohormones. However, little is known at present whether or how extent the leaf size is associated with other phytohormones.
Interestingly, in most cases the genes controlling leaf size also have an effect on plant height. For example NAL1 regulates both plant height and leaf size. DNL1 (DWARF AND NARROW LEAF 1) allelic to NRL1, is a QTL for leaf size and plant height (Ding et al., 2015). Compared to wild type, mutant showing loss-of-function of SLL1 gene displayed narrow rolled leaves and reduced plant height (Zhang et al., 2009). Although, number of other changes were also observed in plants showing reduced activity of NAL2/3. Overexpression of NAL2/3 leads to dwarf phenotype in rice (Cho et al., 2013). However, the leaf size doesn’t have positive correlation with plant height, for example, dwarf 1 (d1) mutant have wider leaves phenotype and overexpression of SG1 (SHORT GRAIN 1) results in reduced plant height and increased leaf size (Nakagawa et al., 2012). Suppression of miR159 controls plant height and leaf length in sttm159 transgenic plants by expression of those genes that were involved in phytohormones homeostasis (Zhao et al., 2017). Study of osgasr1 mutant suggested that OsGASR1 played important roles in expression of α-amylase gene and regulate growth of seedling by increasing cell length (Lee et al., 2017). This anomalous relationship between leaf size and plant height needs further investigations.
Epigenetic Aspects
Histone modifications are important part of epigenetic mechanisms and have determinant role in controlling leaf size. Different stresses act as stimulus and change the genic expression level by various epigenetic mechanisms, e.g., DNA methylation, histone modifications and miRNA (Saraswat et al., 2017). HDT702 is a member of the HISTONE DEACETYLASE (HDAC) gene family that plays an important function in histone modifications and plant gene expression. Down-regulation of HDT702 results in narrow leaves, indicating that histone modifications are also involved in regulation of leaf size (Hu et al., 2009).
Conclusion
In past several years, molecular and genomic studies disclosed important advancements in identification of genes or QTLs controlling leaf morphology (Table 1). Innovative cloning and sequencing technologies has made possible to identify new genes much easier and faster than before. In order to conclude our understanding, we built a hypothetical working model (Figure 1) that is showing a logical mechanistic control of leaf morphology. According to this model, different genes involved in leaf polarity, bulliform cells, sclerenchyma cells and cuticle development exhibit the final phenotype of leaf rolling. While, leaf size is mainly controlled by numerous genes encoding phytohormones, cell division, number of veins, ionic homeostasis and epigenetic aspects. Defective development of cell wall formation, impaired cell division, abnormal contents of cellulose and lignin, reduced water retaining capacity, failure of developing homeostasis in cellular structures of leaf, and poor development of sclerenchyma cells on either side of leaf blade are main causes of leaf rolling. Leaf size genes are partially obsessed or affected by epigenetic mechanisms, ionic homeostasis, number of veins in leaf, cell division and regulation of phytohormones.
Biotic and a-biotic stresses such as high temperature, drought, fungus and insects cause severe yield losses to plants (Kadioglu et al., 2012). Plant makes adjustments in leaf morphology to avoid or compensate the effect of those stresses by gene regulation and plant metabolism. Genes that are controlling the ideal morphology of leaf, and enable the plant to cope challenges of changing climate and environment will be an important area of future research. Finding of physiological and molecular mechanisms underlying genes that are governing desired characteristics would be beneficial in wheat, rice, maize and other crops. Knowledge of cause of changes in morphology and mechanism determining these changes may lead in way to develop better strategies in molecular breeding. Keeping in view of this need, this review can provide an umbrella of genes function to understand leaf morphogenesis of monocot especially in rice. We can utilize these gene in molecular breeding of rice in order to improve grain yield by gaining advantages of ideal leaf morphology. In spite of great progress, some areas still need further research, e.g., which leaf morphology associated genes are under epigenetic control? Which other factors are playing role in upstream of signaling pathways that are regulating leaf morphology. Is the regulation of leaf morphology is conserved in all plant species?
Author Contributions
PX and AA wrote the review. XW read and approved the contents. BH helped in literature and reference digestion.
Funding
We acknowledge grants-in-aid from Science and Technology Program of Sichuan Province (2017NZZJ005) and NSFC (Natural Science Foundation of China) Grant No. 31771763.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors are thankful to reviewers for their critics and suggestions.
Footnotes
References
Adriani, D. E., Lafarge, T., Dardou, A., Fabro, A., Clément-Vidal, A., Yahya, S., et al. (2016). The qTSN positive effect on panicle and flag leaf size of rice is associated with an early down-regulation of tillering. Front. Plant Sci. 6:1197. doi: 10.3389/fpls.2015.01197
Ashikari, M., Wu, J., Yano, M., Sasaki, T., and Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. 96, 10284–10289. doi: 10.1073/pnas.96.18.10284
Chandra, A., and Dubey, A. (2009). Assessment of ploidy level on stress tolerance of Cenchrus species based on leaf photosynthetic characteristics. Acta Physiol. Plant. 31, 1003–1013. doi: 10.1007/s11738-009-0317-0
Chen, Q., Xie, Q., Gao, J., Wang, W., Sun, B., Liu, B., et al. (2015). Characterization of Rolled and Erect Leaf 1 in regulating leave morphology in rice. J. Exp. Bot. 66, 6047–6058. doi: 10.1093/jxb/erv319
Cho, S.-H., Lee, C.-H., Gi, E., Yim, Y., Koh, H.-J., Kang, K., et al. (2018). The Rice Rolled Fine Striped (RFS) CHD3/Mi-2 chromatin remodeling factor epigenetically regulates genes involved in oxidative stress responses during leaf development. Front. Plant Sci. 9:364. doi: 10.3389/fpls.2018.00364
Cho, S.-H., and Paek, N.-C. (2016). Regulatory role of the OsWOX3A transcription factor in rice root development. Plant Signal. Behav. 11:e1184807. doi: 10.1080/15592324.2016.1184807
Cho, S. H., Yoo, S. C., Zhang, H., Pandeya, D., Koh, H. J., Hwang, J. Y., et al. (2013). The rice NARROW LEAF 2 and NARROW LEAF 3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 198, 1071–1084. doi: 10.1111/nph.12231
Dai, M., Zhao, Y., Ma, Q., Hu, Y., Hedden, P., Zhang, Q., et al. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133. doi: 10.1104/pp.107.096586
Ding, Z., Lin, Z., Li, Q., Wu, H., Xiang, C., and Wang, J. (2015). DNL1, encodes cellulose synthase-like D4, is a major QTL for plant height and leaf width in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 457, 133–140. doi: 10.1016/j.bbrc.2014.12.034
Dong, H., Fei, G. L., Wu, C. Y., Wu, F. Q., Sun, Y. Y., Chen, M. J., et al. (2013). A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 162, 1867–1880. doi: 10.1104/pp.113.217604
Dong, X., Wang, X., Zhang, L., Yang, Z., Xin, X., Wu, S., et al. (2013). Identification and characterization of OsEBS, a gene involved in enhanced plant biomass and spikelet number in rice. Plant Biotechnol. J. 11, 1044–1057. doi: 10.1111/pbi.12097
Fang, L., Zhao, F., Cong, Y., Sang, X., Du, Q., Wang, D., et al. (2012). Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Fujino, K., Matsuda, Y., Ozawa, K., Nishimura, T., Koshiba, T., Fraaije, M. W., et al. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genomics. 279, 499–507. doi: 10.1007/s00438-008-0328-3
Gallavotti, A., Barazesh, S., Malcomber, S. T., Hall, D. H., Jackson, D., Schmidt, R. J., et al. (2008). SPARSE INFLORESCENCE1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. U.S.A. 105, 15196–15201. doi: 10.1073/pnas.0805596105
Ghiglione, H., Gonzalez, F. G., Serrago, R. A., Maldonado, S., Chilcott, C., Cura, J. A., et al. (2008). Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 55, 1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x
He, Z., Zeng, J., Ren, Y., Chen, D., Li, W., Gao, F., et al. (2017). OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice. Front. Plant Sci. 8:1730. doi: 10.3389/fpls.2017.01730
Hibara, K., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF 1 gene functions in leaf and embryonic pattern formation in rice. Develop. Biol. 334, 345–354. doi: 10.1016/j.ydbio.2009.07.042
Hu, J., Zhu, L., Zeng, D., Gao, Z., Guo, L., Fang, Y., et al. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. doi: 10.1007/s11103-010-9614-7
Hu, Y., Qin, F., Huang, L., Sun, Q., Li, C., Zhao, Y., et al. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388, 266–271. doi: 10.1016/j.bbrc.2009.07.162
Huang, J., Li, Z., and Zhao, D. (2016). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 6:29938. doi: 10.1038/srep29938
Huang, J., Tang, D., Shen, Y., Qin, B., Hong, L., You, A., et al. (2010). Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genomics 37, 23–36. doi: 10.1016/S1673-8527(09)60022-9
Jiang, D., Fang, J., Lou, L., Zhao, J., Yuan, S., Yin, L., et al. (2015). Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS One 10:e0118169. doi: 10.1371/journal.pone.0118169
Jiang, H., Li, Z., Zeng, D., Gao, Z., Guo, L., Fang, Y., et al. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. doi: 10.1007/s11103-010-9614-7
Juarez, M. T., Twigg, R. W., and Timmermans, M. C. P. (2004). Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544. doi: 10.1242/dev.01328
Kadioglu, A., Terzi, R., Saruhan, N., and Saglam, A. (2012). Current advances in the investigation of leaf rolling caused by biotic and abiotic stress factors. Plant Sci. 182, 42–48. doi: 10.1016/j.plantsci.2011.01.013
Khew, C. Y., Teo, C. J., Chan, W. S., Wong, H. L., Namasivayam, P., and Ho, C. L. (2015). Brassinosteroid insensitive 1-associated kinase 1 (OsI-BAK1) is associated with grain filling and leaf development in rice. J. Plant Physiol. 182, 23–32. doi: 10.1016/j.jplph.2015.05.003
Kim, J. H., and Tsukaya, H. (2015). Regulation of plant growth and development by the growth-regulating factor and GRF-interacting factor duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Kong, X. Q., Gao, X. H., Sun, W., An, J., Zhao, Y. X., and Zhang, H. (2011). Cloning and functional characterization of a cation–chloride cotransporter gene OsCCC1. Plant Mol.Biol. 75, 567–578. doi: 10.1007/s11103-011-9744-6
Lafitte, H. R., Price, A. H., and Courtois, B. (2004). Yield response to water deficit in an upland rice mapping population: associations among traits and genetic markers. Theor. Appl. Genet. 109, 1237–1246. doi: 10.1007/s00122-004-1731-8
Lang, Y., Zhang, Z., Gu, X., Yang, J., and Zhu, Q. (2004). Physiological and ecological effects of crimpy leaf character in rice (Oryza sativa L.) I. Leaf orientation, canopy structure and light distribution. Acta Agron. Sin. 30, 806–810.
Lee, S.-C., Kim, S.-J., Han, S.-K., An, G., and Kim, S.-R. (2017). A gibberellin-stimulated transcript, OsGASR1, controls seedling growth and α-amylase expression in rice. J. Plant Physiol. 214, 116–122. doi: 10.1007/s00122-004-1731-8
Li, C., Zou, X., Zhang, C., Shao, Q., Liu, J., Liu, B., et al. (2016). OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS One 11:e0156413. doi: 10.1371/journal.pone.0156413
Li, L., Shi, Z. Y., Li, L., Shen, G. Z., Wang, X. Q., An, L. S., et al. (2010). Overexpression of ACL1 (ABAXIALLY CURLED LEAF 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant 3, 807–817. doi: 10.1093/mp/ssq022
Li, L., Xue, X., Chen, Z., Zhang, Y., Ma, Y., Pan, C., et al. (2014). Isolation and characterization of rl (t), a gene that controls leaf rolling in rice. Chin. Sci. Bull. 59, 3142–3152. doi: 10.1007/s11434-014-0357-8
Li, W., Wu, C., Hu, G., Xing, L., Qian, W., Si, H., et al. (2013). Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J. Integ. Plant Biol. 55, 1016–1025. doi: 10.1111/jipb.12098
Li, W.-Q., Zhang, M.-J., Gan, P.-F., Qiao, L., Yang, S.-Q., Miao, H., et al. (2017). CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923. doi: 10.1111/tpj.13728
Li, Y. Y., Shen, A., Xiong, W., Sun, Q. L., Luo, Q., Song, T., et al. (2016). Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice 9:46. doi: 10.1186/s12284-016-0118-1
Liu, B., Li, P., Li, X., Liu, C., Cao, S., Chu, C., et al. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139, 296–305. doi: 10.1104/pp.105.063420
Liu, G., Yang, J., Xu, H., Hayat, Y., and Zhu, J. (2008). Genetic analysis of grain yield conditioned on its component traits in rice (Oryza sativa L.). Crop Pasture Sci. 59, 189–195. doi: 10.1071/AR07163
Liu, X., Ming, L., Kai, L., Ding, T., Sun, M., Li, Y., et al. (2016). SEMI-ROLLED LEAF 2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 67:erw029. doi: 10.1093/jxb/erw029
Ma, L., Sang, X., Zhang, T., Yu, Z., Li, Y., Zhao, F., et al. (2017). ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 213, 275–286. doi: 10.1111/nph.14142
Ma, N., Wang, Y., Qiu, S., Kang, Z., Che, S., Wang, G., et al. (2013). Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS One 8:e75997. doi: 10.1371/journal.pone.0075997
Ma, Y., Zhao, Y., Shangguan, X., Shi, S., Zeng, Y., Wu, Y., et al. (2017). Overexpression of OsRRK1 changes leaf morphology and defense to insect in rice. Front. Plant Sci. 8:1783. doi: 10.3389/fpls.2017.01783
Matsumoto, H., Yasui, Y., Kumamaru, T., and Hirano, H.-Y. (2017). Characterization of a half-pipe-like leaf1 mutant that exhibits a curled leaf phenotype. Genes Genet. Syst. 92, 287–291. doi: 10.1266/ggs.17-00013
Mimura, M., Nagato, Y., and Itoh, J.-I. (2012). Rice PLASTOCHRON genes regulate leaf maturation downstream of the gibberellin signal transduction pathway. Planta 235, 1081–1089. doi: 10.1007/s00425-012-1639-5
Miyoshi, K., Ahn, B.-O., Kawakatsu, T., Ito, Y., Itoh, J.-I., Nagato, Y., et al. (2004). PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. U.S.A. 101, 875–880. doi: 10.1073/pnas.2636936100
Moon, J., and Hake, S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14, 24–30. doi: 10.1016/j.pbi.2010.08.012
Nakagawa, H., Tanaka, A., Tanabata, T., Ohtake, M., Fujioka, S., Nakamura, H., et al. (2012). SHORT GRAIN 1 decreases organ elongation and brassinosteroid response in rice. Plant Physiol. 158, 1208–1219. doi: 10.1104/pp.111.187567
Nar, H., Saglam, A., Terzi, R., Varkonyi, Z., and Kadioglu, A. (2009). Leaf rolling and photosystem II efficiency in Ctenanthe setosa exposed to drought stress. Photosynthetica 47, 429–436. doi: 10.1007/s11099-009-0066-8
Nelson, J. M., Lane, B., and Freeling, M. (2002). Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development 129, 4581–4589.
Price, A. H., Young, E., and Tomos, A. D. (1997). Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa). New Phytol. 137, 83–91. doi: 10.1046/j.1469-8137.1997.00818.x
Qi, J., Bu, Q., Li, S., Chen, Q., Sun, J., Liang, W., et al. (2008). Mutation of the Rice NARROW LEAF 1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Ramamoorthy, R., Jiang, S.-Y., and Ramachandran, S. (2011). Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PLoS One 6:e28069. doi: 10.1371/journal.pone.0028069
Saraswat, S., Yadav, A. K., Sirohi, P., and Singh, N. K. (2017). Role of epigenetics in crop improvement: water and heat stress. J. Plant Biol. 60, 231–240. doi: 10.1007/s12374-017-0053-8
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. doi: 10.1126/science.1081077
Sazuka, T., Kamiya, N., Nishimura, T., Ohmae, K., Sato, Y., Imamura, K., et al. (2009). A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 60, 227–241. doi: 10.1111/j.1365-313X.2009.03952.x
Schmitz, A. J., Begcy, K., Sarath, G., and Walia, H. (2015). Rice OVATE FAMILY PROTEIN 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 241, 177–188. doi: 10.1016/j.plantsci.2015.10.011
Shi, Z., Wang, J., Wan, X., Shen, G., Wang, X., and Zhang, J. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. doi: 10.1007/s00425-006-0472-0
Shimano, S., Hibara, K.-I., Furuya, T., Arimura, S.-I., Tsukaya, H., and Itoh, J.-I. (2018). Conserved functional control, but distinct regulation of cell proliferation in rice and Arabidopsis leaves revealed by comparative analysis of GRF-INTERACTING FACTOR 1 orthologs. Development 145:dev159624. doi: 10.1242/dev.159624
Subashri, M., Robin, S., Vinod, K. K., Rajeswari, S., Mohanasundaram, K., and Raveendran, T. S. (2009). Trait identification and QTL validation for reproductive stage drought resistance in rice using selective genotyping of near flowering RILs. Euphytica 166, 291–305. doi: 10.1007/s10681-008-9847-6
Tang, X., Gong, R., Sun, W., Zhang, C., and Yu, S. (2018). Genetic dissection and validation of candidate genes for flag leaf size in rice (Oryza sativa L.). Theor. Appl. Genet. 131, 801–815. doi: 10.1007/s00122-017-3036-8
Toriba, T., Ohmori, Y., and Hirano, H. (2011). Common and distinct mechanisms underlying the establishment of adaxial and abaxial polarity in stamen and leaf development. Plant Signal. Behav. 6, 430–433. doi: 10.4161/psb.6.3.14494
Walter, A., Silk, W. K., and Schurr, U. (2009). Environmental effects on spatial and temporal patterns of leaf and root growth. Annu. Rev. Plant Biol. 60, 279–304. doi: 10.1146/annurev.arplant.59.032607.092819
Wang, P., Karki, S., Biswal, A. K., Lin, H.-C., Dionora, M. J., Rizal, G., et al. (2017). Candidate regulators of early leaf development in maize perturb hormone signalling and secondary cell wall formation when constitutively expressed in rice. Sci. Rep. 7:4535. doi: 10.1038/s41598-017-04361-w
Woo, Y. M., Park, H. J., Su’Udi, M., Yang, J. I., Park, J. J., Back, K., et al. (2007). CONSTITUTIVELY WILTED 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136. doi: 10.1007/s11103-007-9203-6
Wu, L., Zhang, Q., Zhou, H., Ni, F., Wu, X., and Qi, Y. (2009). Rice microRNA effector complexes and targets. Plant Cell 21, 3421–3435. doi: 10.1105/tpc.109.070938
Wu, R., and Gu, H. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23, 3392–3411. doi: 10.1105/tpc.111.088625
Xiang, J. J., Zhang, G. H., Qian, Q., and Xue, H. W. (2012). SEMI-ROLLED LEAF1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. doi: 10.1104/pp.112.199968
Xu, M., Zhu, L., Shou, H., and Wu, P. (2005). A PIN1 Family Gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 46, 1674–1681. doi: 10.1093/pcp/pci183
Xu, Y., Wang, Y., Long, Q., Huang, J., Wang, Y., Zhou, K., et al. (2014). Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239, 803–816. doi: 10.1093/pcp/pci183
Yafeng, Y., Binmei, L., Meng, Z., Kun, W., Weimin, C., Xiangbin, C., et al. (2015). CEF1/OsMYB103L is involved in GA-mediated regulation of secondary wall biosynthesis in rice. Plant Mol. Biol. 89, 385–401. doi: 10.1007/s11103-015-0376-0
Yamaguchi, T., Nukazuka, A., and Tsukaya, H. (2012). Leaf adaxial-abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53, 1180–1194. doi: 10.1093/pcp/pcs074
Yang, C., Li, D., Liu, X., Ji, C., Hao, L., Zhao, X., et al. (2014). OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 14:158. doi: 10.1186/1471-2229-14-158
Yang, C., Li, D., Mao, D., Liu, X., Ji, C., Li, X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218. doi: 10.1111/pce.12130
Yang, S. Q., Li, W. Q., Miao, H., Gan, P. F., Qiao, L., Chang, Y. L., et al. (2016). REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 9:37. doi: 10.1186/s12284-016-0105-6
Yoshikawa, T., Ito, M., Sumikura, T., Nakayama, A., Nishimura, T., Kitano, H., et al. (2014). The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. Cell Mol. Biol. 78, 927–936. doi: 10.1111/tpj.12517
Yuan, S., Li, Y., and Peng, S. (2015). Leaf lateral asymmetry in morphological and physiological traits of rice plant. PLoS One 10:e0129832. doi: 10.1371/journal.pone.0129832
Zhang, B., Ye, W., Ren, D., Tian, P., Peng, Y., Gao, Y., et al. (2015). Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice 8:2. doi: 10.1186/s12284-014-0039-9
Zhang, G., Xu, Q., Zhu, X., Qian, Q., and Xue, H. (2009). SHALLOT-LIKE 1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhang, J. J., Wu, S. Y., Jiang, L., Wang, J. L., Zhang, X., Guo, X. P., et al. (2015). A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biol. 17, 437–448. doi: 10.1104/pp.17.01432
Zhang, J., Zhang, H., Srivastava, A. K., Pan, Y., Bai, J., Fang, J., et al. (2018). Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 176, 2082–2094. doi: 10.1104/pp.17.01432
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhao, S., Zhao, L., Liu, F., Wu, Y., Zhu, Z., Sun, C., et al. (2016). NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integ. Plant Biol. 58, 983–996. doi: 10.1111/jipb.12503
Zhao, S. Q., Hu, J., Guo, L. B., Qian, Q., and Xue, H. W. (2010). RICE LEAF INCLINATION 2, a VIN3-1ike protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20, 935–947. doi: 10.1038/cr.2010.109
Zhao, Y., Wen, H., Teotia, S., Du, Y., Zhang, J., Li, J., et al. (2017). Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 17:215. doi: 10.1186/s12870-017-1171-7
Zhi, C. C., Yamaji, N., Kashino-Fujii, M., and Jian, F. M. (2016). A cation-chloride cotransporter gene is required for cell elongation and osmoregulation in rice. Plant Physiol. 17, 494–507. doi: 10.1104/pp.16.00017
Zhu, X.-G., Long, S. P., and Ort, D. R. (2010). Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 61, 235–261. doi: 10.1146/annurev-arplant-042809-112206
Keywords: bulliform cells, cell proliferation, cell wall integrity, homeostasis, phytohormones
Citation: Xu P, Ali A, Han B and Wu X (2018) Current Advances in Molecular Basis and Mechanisms Regulating Leaf Morphology in Rice. Front. Plant Sci. 9:1528. doi: 10.3389/fpls.2018.01528
Received: 26 June 2018; Accepted: 28 September 2018;
Published: 23 October 2018.
Edited by:
Taku Takahashi, Okayama University, JapanCopyright © 2018 Xu, Ali, Han and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianjun Wu, d3V4anNhdUAxMjYuY29t
†These authors have contributed equally to this work
 Peizhou Xu
Peizhou Xu Asif Ali
Asif Ali Baolin Han
Baolin Han Xianjun Wu
Xianjun Wu