
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 October 2018
Sec. Plant Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01503
This article is part of the Research TopicNew Insights into Mechanisms of Epigenetic Modifiers in Plant Growth and DevelopmentView all 15 articles
Single-stranded DNA-binding proteins (SSBs) are assumed to involve in DNA replication, DNA repairmen, and gene transcription. Here, we provide the direct evidence on the functionality of an Arabidopsis SSB, WHIRLY1, by using loss- or gain-of-function lines. We show that WHIRLY1 binding to the promoter of WRKY53 represses the enrichment of H3K4me3, but enhances the enrichment of H3K9ac at the region contained WHIRLY1-binding sequences and TATA box or the translation start region of WRKY53, coincided with a recruitment of RNAPII. In vitro ChIP assays confirm that WHIRLY1 inhibits H3K4me3 enrichment at the preinitiation complex formation stage, while promotes H3K9ac enrichment and RNAPII recruitment at the elongation stage, consequently affecting the transcription of WRKY53. These results further explore the molecular actions underlying SSB-mediated gene transcription through epigenetic regulation in plant senescence.
Plant senescence is the last stage of plant development. It is a controlled process that plants response to internal factors (age and hormone) and external factors (abiotic stress and biotic stress), in which plants remobilize nutrients from source leaves to developing tissues. In the past decades, molecular components underlying the onset of senescence have been studied, however, the age-dependent mechanisms that control the onset of senescence remains opening.
WRKY is a major transcription factor (TF) family of plants, in which many of them are the central players in gene regulation during leaf senescence. WRKY6 was found to activate several senescence-associated genes during leaf senescence (Robatzek and Somssich, 2002). Mutation and overexpression of WRKY6 retarded and accelerated both developmentally and dark-induced senescence (Robatzek and Somssich, 2002; Zhang et al., 2018). Similarly, overexpression or knockout of WRKY22 also showed accelerated and delayed senescence phenotype in dark condition (Zhou et al., 2011). WRKY53 and WRKY70 were reported to be positive and negative regulators of senescence, respectively. Loss of WRKY53 delayed the leaf senescence (Miao et al., 2004), while wrky70 mutant showed aggravated senescent phenotype during development and dark treatment (Ulker et al., 2007). WRKY54 can co-operate with WRKY70 to repress leaf senescence (Besseau et al., 2012). Additionally, WRKY57 function as a repressor in JA-induced leaf senescence (Jiang et al., 2014) and WRKY45 positively regulate age-triggered leaf senescence via gibberellin (GA)-mediated signaling pathway (Chen et al., 2017). Mutual regulation exists between WRKYs and WRKYs can be regulated at different levels (Phukan et al., 2016).
WRKY53, a well-known regulator to plant early leaf senescence, is a convergence node tightly regulated by various process (Zentgraf et al., 2010). WRKY53 binds to more than 60 target genes directly (Miao et al., 2004). It has been reported that WRKY53 protein is regulated by MEKK1 kinase, epithiospecifying senescence regulator (Miao and Zentgraf, 2007; Miao et al., 2007), and UPL5 in the protein level (Miao and Zentgraf, 2010). Moreover, the expression of WRKY53 gene is activated by GATA4 (Zentgraf et al., 2010), activation domain protein (AD protein) (Miao et al., 2008), and REVOLUTA protein (Xie et al., 2014) in the transcriptional level in leaf senescence, while it is repressed by ssDNA binding protein WHIRLY1 (Miao et al., 2013). The histone modification at WRKY53 locus seemed to associate with SUVH2 histone methylase (Ay et al., 2009). Current research showed WRKY53 can interact with histone deacetylase 9 (HDA9) affecting downstream gene expression in leaf senescence (Chen et al., 2016). The AT-rich motif in WHIRLY1 binding domain of WRKY53 has been shown to relate to chromatin structure and epigenetic mark modification (Lim et al., 2007). However, up to now, there is no further evidence about the transcriptional regulation of WRKY53 by single stranded DNA binding WHIRLY1 protein with chromatin modification.
Single-stranded DNA binding proteins are ubiquitous in organisms and essential in recognition and processing of ssDNA during various cellular processes (Dickey et al., 2013). They bind to ssDNA with high sequence specificity or independent of sequence, stabilizing the ssDNA intermediates during several cellular processes, such as DNA replication, recombination, and repair as well as telomere maintenance (Dickey et al., 2013; Ribeiro et al., 2016; Hedglin and Benkovic, 2017; Huang S.H. et al., 2017). SSB proteins is also given a role in the regulation of gene expression (MacDonald et al., 1995; Liu et al., 2000; Kim et al., 2005; Choi et al., 2009; Miao et al., 2013) and regulation of the activity of many other DNA metabolic proteins (Lohman and Ferrari, 1994; Roy et al., 2007; Shereda et al., 2008; Roy et al., 2009). As a single-stranded DNA binding protein, WHIRLY domains are four structural topologies that have been structurally characterized with ssDNA (Dickey et al., 2013). WHIRLY domains are approximately 180-amino-acid-long domains characterized by two roughly parallel four-stranded β sheets with interspersed helical elements (Desveaux et al., 2005; Cappadocia et al., 2013).
As a member of WHIRLY family, WHIRLY1 has been proven to be a plastid nucleoid-associated protein affecting DNA replication (Krupinska et al., 2014b) and have a function in repair of organelle DNA (Etminan et al., 2010; Maréchal and Zou, 2014) and in maintenance of plastid genome stability (Lepage et al., 2013; Zampini et al., 2015). WHIRLY1 has been likewise implicated in telomere maintenance through binding to four copies of the telomere repeat (Yoo et al., 2007). However, WHIRLY1 protein first has been reported to bind to the inverted repeat sequence of the elicitor response element (ERE) on the promoter of PR-10a gene in potato, acting as a transcription activator (Desveaux et al., 2000). Then other studies have shown WHIRLY1 together with WHIRLY3 can bind to the AT-rich region of kinesin gene promoter to activate kinesin gene expression in Arabidopsis (Xiong et al., 2009). Recently, WHIRLY1 has been reported to bind to the GTCAAT motif of S40 promoter in barley by Nanoelectrospray Mass Spectroscopy (Krupinska et al., 2014a). In our previous study, we have found that WHIRLY1 can bind to combination motif of GTNNNAAT and AT-rich motif of downstream target genes, such as WRKY53, WRKY33, SPO11, and PR1 by in vivo chromatin immunoprecipitation sequencing (ChIP-seq). It has been verified that WHIRLY1 bind to the promoter of WRKY53 to repress expression of WRKY53 and WRKY33 in Arabidopsis leaf senescence (Miao et al., 2013; Ren et al., 2017). Nevertheless, how WHIRLY1 as a single-stranded protein mediate the transcription of downstream genes in a fine-tune case remains elusive.
In this study, we first address WHIRLY1 occupancy on WRKY53 promoter, enrichment of several histone modifications and recruitment of RNAPII at promoter and translation start regions of WRKY53, as well as gene expression of WRKY53 in chronological during leaf aging. Biochemical and genetically evidences demonstrate that WHIRLY1 occupancy at the WRKY53 promoter affects not only the enrichment of H3K4me3 and H3K9ac but also RNAPII recruitment at WHIRLY1 bind region and translation start region of WRKY53 in vivo and in vitro, which influences the transcription of WRKY53 in the development manner. This study provides an implication for exploration of SSB-mediated transcription in epigenetically modification level in plants.
All the plants used were Arabidopsis thaliana (L.) Heynold ecotype Columbia 0 background, WHIRLY1 T-DNA insertion line Salk_023713 (why1) were provided by the European Arabidopsis Stock Centre, while WHIRLY1 overexpression mutants (oepnWHY1 and oenWHY1), WHIRLY1 complementary line (PWHY1) which harbors its own promoter, and WHIRLY1 CDS plus HA target were constructed at previous research (Miao et al., 2013). Seeds were germinated on wet filter paper after 48 h of vernalization. Then they were transplanted in pots in vermiculite in a climatic chamber with a 13-h light (100 μE/h) /11-h dark photoperiod, 22°C/18°C day–night temperature regime, and 60% relative humidity. Rosette leaves were labeled with colored threads after emergence, as described previously (Hinderhofer and Zentgraf, 2001). 5–8th rosette leaves from PWHY1 mutants at 6th week were collected for treatment with 3-Deazaneplanocin A (Dzenp) to detect the influence of WHY1 binding affinity and enrichment of H3K4me3, H3K9ac.
Chlorophyll fluorescence of leaf 5 from different developmental plants was measured using a Pocket PEA Chlorophyll Fluorimeter (Hansatech) after 15-minimum dark incubation. The average Fv/Fm value of leaf 5 from at least 12 individual plants was calculated. Chlorophyll concentrations of leaf 5 from 12 different developmental plants were measured by Dualex 4. Three points of each leaf were detected.
Total RNA from 5th to 8th rosette leaves was isolated according to the manufacturer’s protocol of TransZol UP (TRANSGEN), and was then treated with RNase-free DNase I (EN0521, Thermo Scientific). First-strand cDNA was generated from 1 μg portion of total RNA using RevertAid First-Strand cDNA Synthesis Kit (K1622, Thermo Fisher), following the instruction. PCR was performed to analyze the expression of genes. To determine the relative expression rate, data were normalized to the expression level of wild-type or of 5-week-old plants (which were set to 1) after normalized to the internal control of GAPC2. Additionally, three technical replicates of three biological replicates and the determination of a melting curve of the amplified PCR products were carried out.
The ChIP assay was performed with a modified method (Gendrel et al., 2005). About 0.75 g leaves (fifth to eighth leaves of WT and WHIRLY1 mutants during different developmental stages and whole rosettes of darkness-treated plants) were used. The detailed procedure is shown in Supplementary Material. The immunoprecipitated DNA was isolated by Universal DNA purification kit (TIANGEN, DP214-03). Purified DNA was analyzed by real-time PCR with specific primers (see Supplementary Tables). Relative histone modification levels in WT during leaf development were therefore normalized to the input, while the enrichment in WHIRLY1 mutants was normalized to WT again, referring to ΔΔCt method1.
The recombinant WHY1 protein was expressed in E. coli as described by Miao et al. (2013).
Chromatin was assembled and transcription assays were performed as described by Active & Motif Chromatin Assembly Manuel2. pG5ML with promoter of WRKY53 including mutated WRKY53II [mutant (GTNNNGGT) m1 or mutant (CTNNNNAAAT) m2 WHIRLY1 binding motif] (Miao et al., 2013) or mutated TATA-box or wild-type fragment was used as DNA template. The detail showed in Supplementary Material.
In vitro ChIP assays were performed as described (Lauberth et al., 2013) (the detail in Supplementary Material). The levels of H3K4me3, H3K9ac are given relative to the total H3 levels. In vitro ChIP to detect the effect of WHIRLY1 on transcription stage was performed in the presence or absence of 0.01% sarkosyl, which inhibits PIC assembly but does not affect elongation by pre-formed complexes (Cai and Luse, 1987; Hawley and Roeder, 1987) or 800 nM B2 RNA, which inhibits transcription prior to PIC formation (Espinoza et al., 2004).
In previous work, we performed ChIP-Seq and WHIRLY1 was found binding on the promoter of WRKY53 as a repressor in the developmental manner (Miao et al., 2013). Leaves of 5–8 from plants grown under 80 μmolm-2s-1 radiation were chosen to analyze the relationship of WHIRLY1 and downstream gene expression at the fine-turn level (Supplementary Figure S1). The expression of RBCS, a gene encoding the small subunit of ribulose 1,5-bisphosphate carboxylase, was downregulated at the 6th week while the expression of SAG12 gene encoding a cysteine protease which expressed only in senescent tissues, was drastically induced at 8th week in current case (Supplementary Figures S1A–C). Therefore, we inferred that 5th to 7th week was the initiation and early stage of leaf senescence. WHIRLY1 was phosphorylated by calcineurin B-like-interacting protein kinase14 (CIPK14) and accumulated steady in the nucleus after 5W (Ren et al., 2017) (Supplementary Figure S1E). Although occupancy of WHIRLY1 on the promoter of WRKY53 was conversed with the accumulation of nuclear form WHIRLY1, the expression level of WRKY53 strikingly increased from 5th week to 6th week while the occupancy of WHIRLY1 on the promoter of WRKY53 declined (Supplementary Figures S1D,E). Taken together, these results demonstrate that WHIRLY1 chronologically repressed WRKY53 transcription and expression via binding to its promoter at senescence initiation stage from 5th week to 7th week.
To investigate whether WHIRLY1 binding on the WRKY53 promoter affect the enrichment of selected histone methylation at senescence initiation stage, chromatin immunoprecipitation experiments were carried out with antibodies against H3K4me2, H3K4me3 (associated with transcriptional active), and H3K27me2 (associated with transcriptional inactive) (Supplementary Figure S2) using 5- to 8-week-old wild-type plants. The ChIP assay showed that enrichments of H3K4me2 and H3K4me3 at WRKY53P region which was downstream of WHIRLY1 binding domain at the promoter of WRKY53 and at WRKY53II region which contained WHIRLY1 binding domain with TATA-box (Figure 1A), were at high level in 5-week-old plants, then significantly declined from 5th to 6th, and started to increase from 6th to 8th week, and significantly climbed up to 20% high level at 8th week (Figures 1B,C and Supplementary Figure S3). The enrichment of H3K4me2, H3K4me3 at the residual detected regions of WRKY53 promoter were at low level compared to that of WRKY53P. H3K27me2 was the lowest at all the detected regions of WRKY53 gene (Supplementary Figure S4A). Although the enrichment of WHIRLY1 at WRKY53 promoter decreased gradually, still maintained at 40% level at 7th week (Supplementary Figure S1F), and WRKY53 expression level increased rapidly from 5th to 6th week then declined at 7th week (Supplementary Figure S1D). Taken together, these results suggest that the occupancy of WHIRLY1 protein at WRKY53 promoter may be associated with the enrichment of H3K4me2 and H3K4me3 at WRKY53. However, WHIRLY1 occupancy did not repress WRKY53 expression via directly correlated with H3K4me2 and H3K4me3.
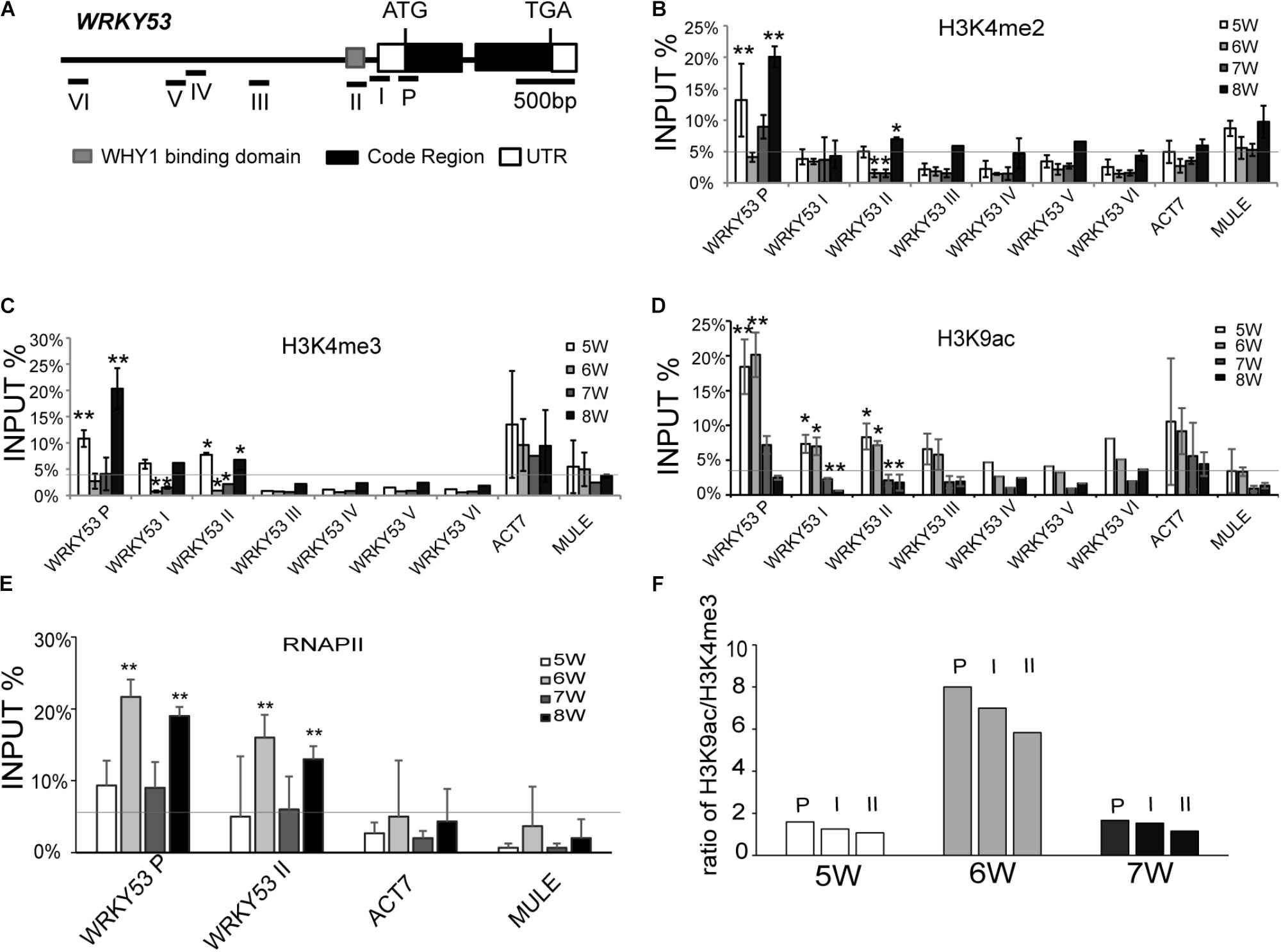
FIGURE 1. Histone modification and RNAPII occupancy at the promoter regions and translation start region of WRKY53. (A) Schematic diagram of the genomic structure of the WRKY53 gene. The lines with number represent qPCR amplicons in different regions of WRKY53 gene. Gray box, black box, and blank box represent WHIRLY1 binding domain (gray), exon (black), and untranslated regions (blank). (B–E) ChIP analyses of changes in H3K4me2 (B), H3K4me3 (C), H3K9ac (D) levels, and RNAPII (E) occupancy at different regions of WRKY53 from 5th to 8th week. The antibodies used for ChIP were optimized (Supplementary Figure S2). The quantitative PCR was performed with primers (I–VI) around the predicted WHIRLY1 binding sites on the promoter of WRKY53 and primer (P) covered the 5′ untranslated region and translational start region of WRKY53 (Figure 2) (Ay et al., 2009). ACT7 (Actin-related gene 7, AT5G09810) and MULE (Mutator-like transposable element, AT2G15810) were analyzed as controls. The relative level was normalized to INPUT DNA. Three biological replicates and three technique replicates were used to analyze. Error bar show the SD (n = 3×3). Asterisk indicates significant differences (∗P < 0.05 and ∗∗P < 0.01) based on Student’s t-test. (F) Alteration of ratio of H3K9ac/H3K4me2-3 at different regions of WRKY53 from 5th to 7th week compared to occupancy of WHIRLY1 and transcripts level of WRKY53.
We also detected the pattern of H3K9ac, H4ac, and RNAPII occupancy at WRKY53 in wild-type plants during leaf aging. The H3K9ac at promoter and P regions of WRKY53 maintained high level at 5th and 6th week but decreased sharply from 7th week (Figure 1D and Supplementary Figure S5). H3K9ac has been reported to positively correlate with the recruitment of RNAPII (Zhang et al., 2017). Expectedly, the recruitment of RNAPII at WRKY53P and WRKY53II regions significantly increased at 6th week, which is consistent with the transcription of WRKY53 (Supplementary Figure S6 and Figure 1E). The enrichment of H4ac at promoter and P region of WRKY53 showed a constant low level during leaf senescence in our study (Supplementary Figure S4B). Taken together, these results indicate that histone modifications at the promoter and translational start region of WRKY53 are dynamic from 5th to 8th week and H3K9ac seems more closely related to the transcription of WRKY53.
A combinatorial interplay between posttranslational modifications on the same histone was proposed based on the patterns of H3 methylation and acetylation at promoters of specific target genes (Taverna et al., 2007). We calculated the ratio of H3K9ac and H3K4me2-3 at the WRKY53 promoter during the initiation period of plant senescence from 5th to 7th week. The results showed the ratio pattern of H3K9ac/H3K4me2-3 enrichment was chronologically associated with the occupancy of WHIRLY1 on the WRKY53 promoter (Supplementary Figure S1F) and the transcription level of WRKY53 Supplementary Figures S1D,E and Figure 1F) during the initiation period of plant senescence, which suggests that H3K9ac/H3K4me2-3 synergistically control WRKY53 transcription in plant developmental manner.
To further investigate whether the binding of WHIRLY1 affects the enrichment of H3K4me2 and H3K4me3 at promoter and P regions of WRKY53, we detected the pattern of H3K4me2, H3K4me3, and RNAPII recruitment at WRKY53II and WRKY53P regions of WRKY53 in WHIRLY1 knockout line (why1), overexpression nucleus-located WHIRLY1 line (oenWHY1), WHIRLY1 complementary line (PWHY1) as well as WT plants (Miao et al., 2013). The ChIP–qPCR results showed that the enrichments of H3K4me2 and H2K4me3 at WRKY53P region of WRKY53 as well as H3K4me2 at WRKY53II region of WRKY53 increased significantly at 6th week in why1 mutant compared to WT plants (Figure 2A and Supplementary Figure S3), but RNAPII recruitment did not change (Supplementary Figure S6). Interestingly, overexpression nuclear isoform WHIRLY1 (Miao et al., 2013) did not have effect on H3K4me2, H3K4me3, and RNAPII recruitment at WRKY53P region, but the enrichment of H3K4me2 at WRKY53II region which contained the WHIRLY1 binding domain decreased (Figures 2A,B). The results suggest that loss of WHIRLY1 enhances the H3K4me2 and H3K4me3 enrichment at the 5′ untranslated region and translational start region of WRKY53 (WRKY53P), while the occupancy of WHIRLY1 seems to inhibit the enrichment of H3K4me2 and H3K4me3 at its binding site on WRKY53 at senescence initiation stage.
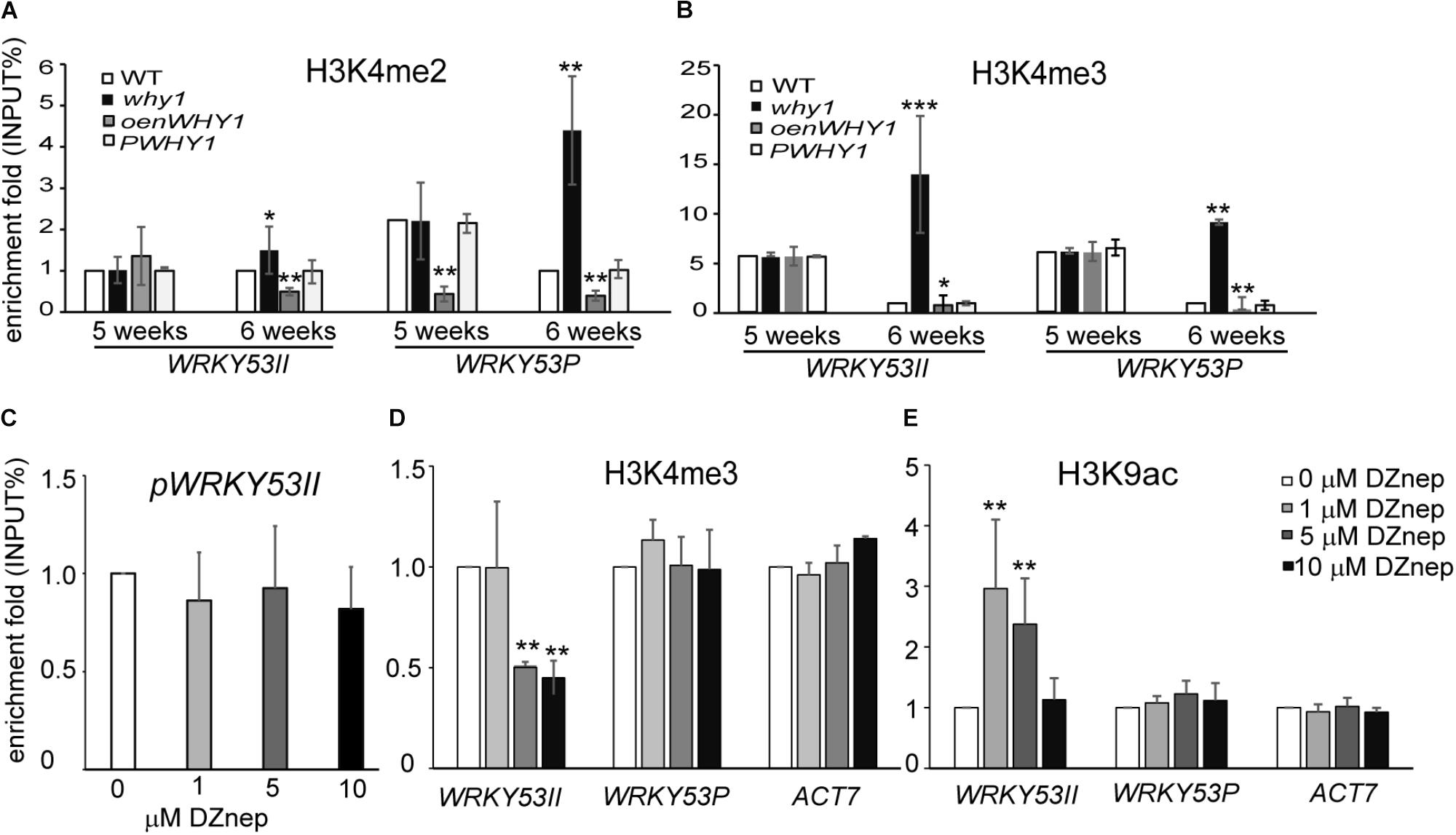
FIGURE 2. WHIRLY1 regulates the H3K4me3 enrichment at WRKY53 at senescence initiation stage. (A,B) ChIP experiments were performed to assess H3K4me2 (A) and H3K4me3 (B) levels at WHIRLY1 binding region (WRKY53II) and translation start region (WRKY53P) of WRKY53 using rosette leaves of why1, oenWHY1, wild-type (WT), and PWHY1 plants. why1, whirly1; oenWHY1, oenWHIRLY1; and PWHY1, Pwhirly1:WHIRLY1. The relative level (input %) was normalized to that of 5-week-old WT. Three biological replicates and three technique replicates were used to analyze. Error bar shows the SD (n = 3×3). Asterisk indicate significant differences (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001) based on Student’s t-test. (C–E) The binding affinity of WHIRLY1 at promoter of WRKY53 (pWRKY53II) (C), H3K4me3 (D), and H3K9ac (E) levels at WRKY53II and WRKY53P regions of WRKY53 using 6-week-old rosette leaves of PWHY1 plants after 24 h induction with 1, 5, and 10 μM 3-Deazaneplanocin A, dimethylsulfoxide (Dznep) was used as a control in treatment. Actin7 was used as an inner control for ChIP–qPCR. ChIP–qPCR were carried out with antibody against HA, H3K4me3, and H3K9ac. Three biological replicates and three technique replicates were used to analyze. Asterisk indicate significant differences (∗P < 0.05 and ∗∗P < 0.01) based on Student’s t-test.
H3K4me3 at promoters and 5′-end regions have been reported to significantly correlate with active gene expression (Santos-Rosa et al., 2002; Zhang et al., 2009; Songa et al., 2014), and the H3K9 acetylation has been supposed to serve as a template for the gain of H3K4me3 marks during leaf senescence (Brusslan et al., 2015). We next investigate whether the enrichment of H3K4me3 affects the WHIRLY1 occupancy and H3K9ac enrichment at WRKY53. We treated 6-week-old PWHY1 plants with different concentrations(0, 1, 5, and 10 μM) of 3-Deazaneplanocin A (DZnep), which is a S-adenosyl homocysteine hydrolase inhibitor and has been reported to affect H3K4me3 enrichment at the promoters of 3% genes in zebrafish (Ostrup et al., 2014). The results showed that DZnep deduce the enrichment of H3K4me3 at WRKY53II region of WRKY53 in a dose-dependent manner (Figure 2D). However, DZnep had no effect on the occupancy of WHIRLY1 at WRKY53 promoter (Figure 2C). Interestingly, DZnep treatment resulted in increase of H3K9ac at WRKY53II region of WRKY53 (Figure 2E). Therefore, H3K4me3 marks at WRKY53 promoter did not directly affect the occupancy of WHIRLY1 at WRKY53 promoter, but H3K4me3 marks correlate with H3K9ac.
To determine whether WHIRLY1 occupancy affects H3K9ac level and RNAPII recruitment at WRKY53, we performed ChIP experiment in why1, oenWHY1, PWHY1, and WT plants with anti-H3K9ac and anti-RNAPII antibody (Supplementary Figure S2). As showed in Figure 3 and Supplementary Figure S5, the loss of WHIRLY1 led to an increase in H3K9ac at WRKY53II region of WRKY53 at 6-week-old and 7-week-old wild-type plants, and enhanced the H3K9ac enrichment at WRKY53P region of WRKY53 at 7-week-old wild-type plants. H3K9ac enrichment at WRKY53P region of WRKY53 was inhibited at 6-week-old and 7-week-old oenWHY1 mutant, while H3K9ac enrichment at WRKY53II region of WRKY53 was only inhibited at 7-week-old oenWHY1 mutant (Figure 3A). Interestingly, significant enrichment of RNAPII at WRKY53II and WRKY53P regions of WRKY53 were also detected in the 7-week-old why1 mutants. Overexpression of WHIRLY1 reduced the RNAPII recruitment at WRKY53II and WRKY53P regions of WRKY53 at 7-week-old PWHY1 (Figure 3B). As shown in the Figures 3C,D, higher transcript level of WRKY53 and SAG12 in the why1 mutant and lower transcript level of WRKY53 and SAG12 in the oenWHY1 were detected, showing high proportion of yellow leaves and less chlorophyll content in the why, reversely, less proportion of yellow leaves and high chlorophyll content in the oenWHY1 line. These results demonstrated that WHIRLY1 binding on WRKY53 accelerated the deacetylation of H3K9ac and repressed RNAPII recruitment at promoter and translational start region of WRKY53 to determine WRKY53 transcript level and senescence-related parameter in a developmental manner.
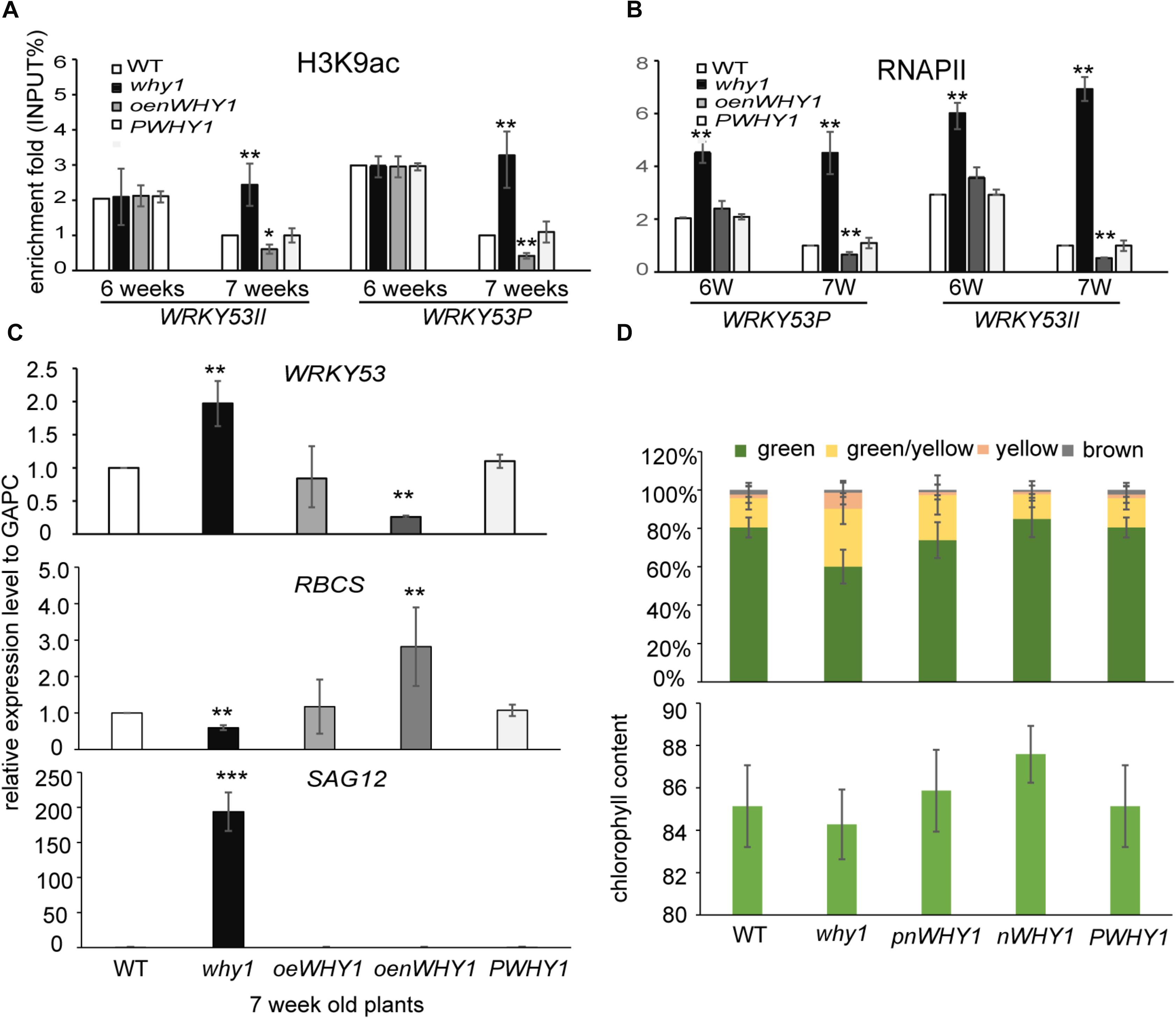
FIGURE 3. WHIRLY1 enhances H3K9 deacetylation and represses RNAPII recruitment at WRKY53 at leaf early senescence stage. (A,B) ChIP experiments were performed to assess H3K9ac (A) and RNAPII occupancy (B) at WHIRLY1 binding region (WRKY53II) and translation start region (WRKY53P) of WRKY53 using rosette leaves of why1, oenWHY1, wild-type (WT), and PWHY1 plants. The relative level (input %) was normalized to that of 5-week-old WT. Three biological replicates and three technique replicates were used to analyze. Error bar shows the SD (n = 3×3). Asterisk indicate significant differences (∗P < 0.05 and ∗∗P < 0.01) based on Student’s t-test. (C) Expression of senescence-associated genes in 7-week-old why1, oenWHY1, WT, and PWHY1 plants. The transcript level in each case was normalized to that of GAPC2 as a reference gene and the expression level of WT was set as 1. Three biological replicates and three technique replicates were used to analyze. Asterisk indicate significant differences (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001) based on Student’s t-test. (D) Senescent leaf fraction and chlorophyll content in 7-week-old why1, oenWHY1, WT, and PWHY1 plants. Mean and SD of at least 12 independent measurements are shown. Error bars represent SE.
To confirm occupancy of WHIRLY1 protein at the WRKY53 promoter directly impacts H3K4 methylation, H3K9ac and WRKY53 transcription initiation, we recruited a cell-free transcription system (Figure 4A) (An and Roeder, 2004). Chromatin was assembled by recombinant factors using Hela core histone and pG5ML template with the promoter of WRKY53 including mutated WRKY53II or mutated TATA-box or wild-type fragment (Figure 4B). Micrococcal nuclease digestion showed that all templates are chromatinized equivalently (Supplementary Figure S7). In vitro ChIP was performed with assembled chromatin. Chromatin was incubated with or without recombinant WHIRLY1 protein expressed in E. coli (Miao et al., 2013) and immunoprecipitated with antibodies against WHIRLY1, H3, H3K4me3, or H3K9ac or RNAPII. The ChIP-quantitative PCR was carried out using primer containing WHIRLY1 binding sites (WRKY53II) and TATA box. The enrichment of WHIRLY1 on chromatin templates with wild-type WRKY53II and mutant TATA was obviously higher than that on chromatin templates with mutant WRKY53II (WRKY53IIm1 and WRKY53IIm2) (Figure 4C) (Miao et al., 2013). Interestingly, on chromatin templates with wild-type WRKY53II and mutant TATA, in the presence of WHIRLY1, H3K4me3 enrichment was inhibited while H3K9ac enrichment and RNAPII recruitment were enhanced (Figure 4C). Further the transcript run-on assay and quantitative RT-PCR were used to detect the transcript elongation and accumulation of TBP report genes. Surprisingly, the results showed that report gene TBP transcript elongation and accumulation were increased with the time course in the presence of WHIRLY1, while TBP transcription did not process when pWRKY53II were mutated or in the deficiency of WHIRLY1 (Figures 4D,E). Together, these results indicate that WHIRLY1 binding on WRKY53II in vitro represses the enrichment of H3K4me3 and enhance the enrichment of H3K9ac and RNAPII recruitment, and activate the TBP transcription.
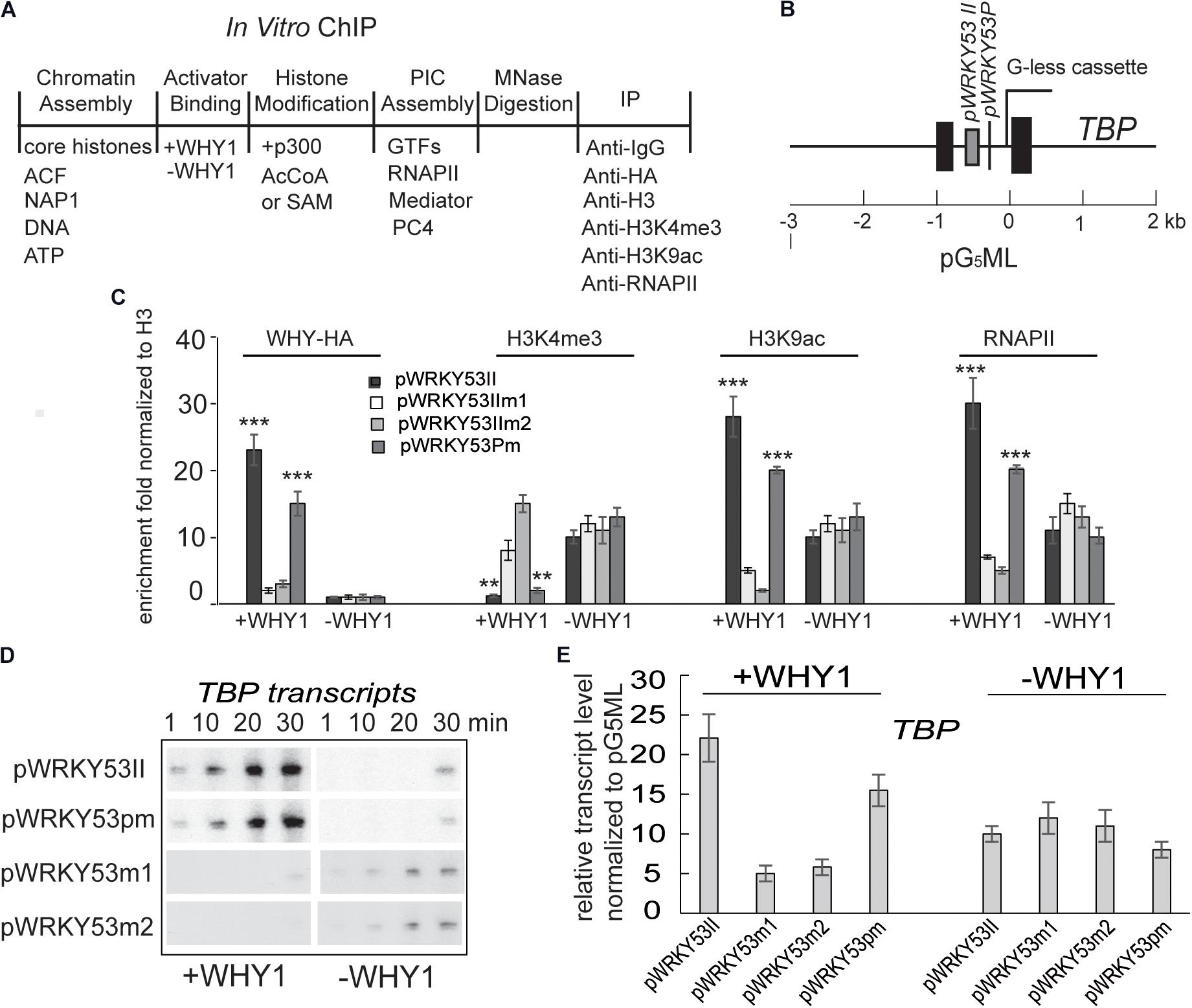
FIGURE 4. WHIRLY1 impacts H3K4me3, H3K9ac levels on WRKY53 promoter in vitro. (A) Schematic of the in vitro ChIP assays. (B) Schematic of the pG5ML template indicating the amplicons used for qRT-PCR. (C) In vitro ChIP was performed with assembled chromatin. Chromatin was incubated with or without recombinant WHIRLY1 protein expressed in E. coli (Miao et al., 2013) acetylated by p300 or methylated by S-adenosyl-l-methionine (SAM), digested with MNase, and immunoprecipitated with antibodies against WHIRLY1, H3, H3K4me3 or H3K9ac, or RNAPII (A). The ChIP-quantitative PCR was carried out using primer containing WHIRLY1 binding sites (WRKY53II) and TATA box. H3K4me3 and H3K9ac levels were relative to H3 levels. Three biological replicates and three technique replicates were used to analyze. Asterisk indicate significant differences (∗∗P < 0.01 and ∗∗∗P < 0.001) based on Student’s t-test. Error bars represent SE. (D) The report gene transcription by run-on assay. (E) The report gene transcription level by qRT-PCR. Three biological replicates and three technique replicates were used to analyze. Error bars represent SE.
H3K4me3 has been reported to involve in the preinitiation complex (PIC) assembly during transcription (Lauberth et al., 2013; Songa et al., 2014). We wondered WHIRLY1 occupancy on the WRKY53 promoter impact H3K4me3 before or after PIC formation, therefore, 0.01% sarkosyl, which inhibits PIC assembly but does not affect elongation by pre-formed complexes (Cai and Luse, 1987; Hawley and Roeder, 1987), or B2 RNA, which binds to RNA polymerase II and inhibits transcription before PIC formation (Espinoza et al., 2004), was used to treat assembled chromatin in presence or absence of WHIRLY1 (Figure 5A). ChIP–qPCR results showed that neither sarkosyl nor B2 RNA affected the enrichment of WHIRLY1 at the WRKY53 promoter. However, sarkosyl treatment decreased the enrichment of H3K4me3 at WRKY53 promoter while B2 RNA treatment inhibited enrichment of H3K9ac and RNAPII recruitment at WRKY53 promoter (Figure 5B). Occupancy of WHIRLY1 on WRKY53 promoter also inhibited the enrichment of H3K4me3 in the presence of B2 RNA and enhanced the enrichment of H3K9ac and RNAPII recruitment in the presence of sarkosyl (Figure 5B). The accumulation of TBP transcript was shown in the presence of sarkosyl (Figure 5C). These results suggest that WHIRLY1 binding to the WRKY53 promoter represses the enrichment of H3K4me3 at the preinitiation stage of PIC formation, while promotes the enrichment of H3K9ac and RNAPII recruitment during the elongation by pre-formed complexes, consequently promotes the TBP transcription
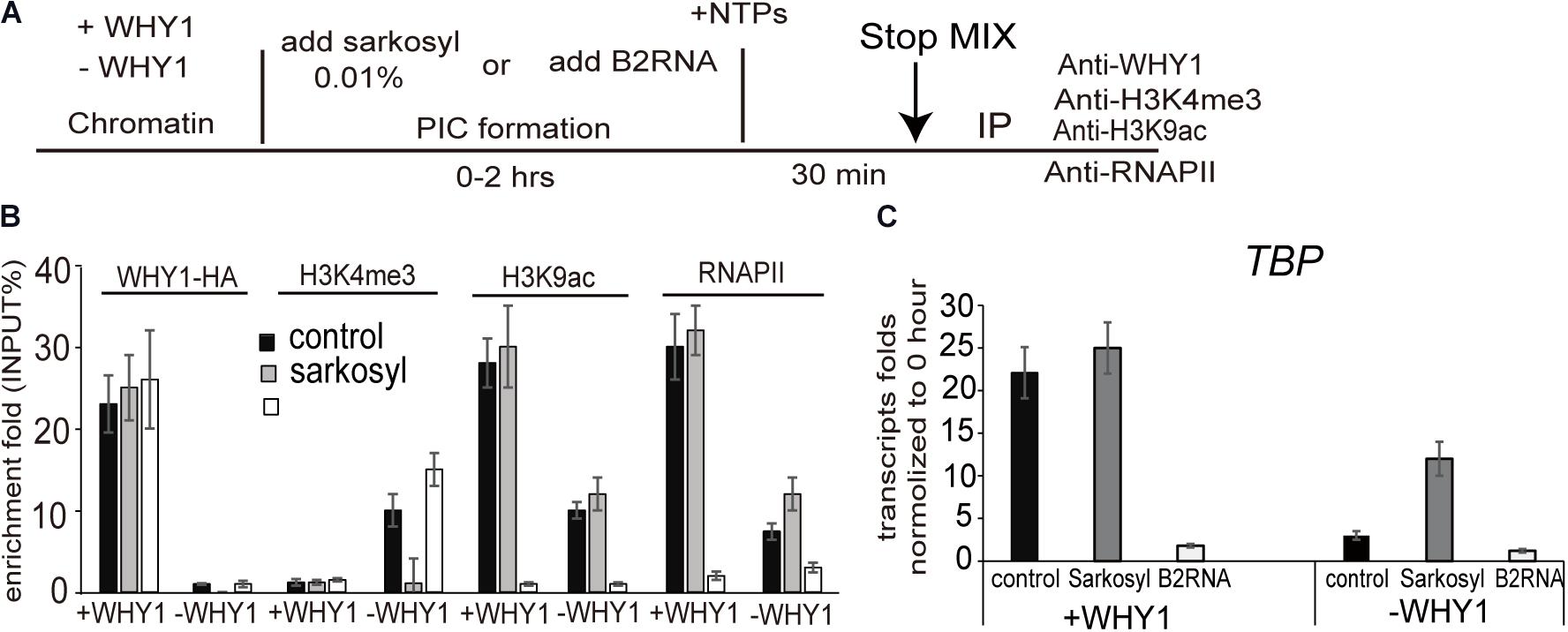
FIGURE 5. WHIRLY1 affects H3K4me3 and H3K9ac at the preinitiation conformation stage. (A) Schematic of the in vitro transcription assay. (B) ChIP experiments were performed on chromatin assembled in vitro after transcription in presence or absence of 0.01% sarkosyl or 800 nM B2RNA with indicated antibodies. H3K4me3 and H3K9ac levels were relative to H3 levels. Three biological replicates and three technique replicates were used to analyze. Error bars represent SE. (C) The report gene transcription level by qRT-PCR. Three biological replicates and three technique replicates were used to analyze. Error bars represent SE.
To investigate whether WHIRLY1 regulates H3K4me3, H3K9ac, and RNAPII recruitment at the WRKY53 promoter in transcriptional level, differentially expression genes related to histone modifications were picked from the dataset of RNA-seq between whirly1 and wild-type plants (Lin et al., 2018, unpublished data; Supplementary Figure S8). SWI3D, HD2D, JMJ22, NFA2, and HTA4 were selected to be further analyzed in 6-week-old why1, oenWHY1 mutants as well as wild-type plants by qRT-PCR. Surprisingly, our qPCR results showed the expression of HD2D and JMJ22 in both why1 mutants and oenWHY1 mutants was lower than that of WT (Figure 6), which contradicts with the previous RNA-seq data. Overexpressing or knocking out WHIRLY1 has no effect on gene expression of SWI3D, NFA2, and HTA4. Moreover, the expression levels of histone methyltransferase ATX1 (Arabidopsis Trithorax-like protein 1), ATX2, SUVH2, and histone deacetylases HDA15, HDA6, HDA2, HDA5, and HDA9 were also detected by qPCR. ATX1 and ATX2 play roles for trimethylating and dimethylating K4 of histone H3, respectively (Saleh et al., 2008), and SUVH2 involved in regulating histone methylation marks at WRKY53 (Ay et al., 2009). The expression of ATX1 was slightly increased in the why1 and decreased in the oenWHY1 mutants, while ATX2 and SUVH2 showed no difference in expression between WHIRLY1 mutants and wild-type plants (Figure 6). Hence, WHIRLY1 is likely mediated histone methylation by regulating expression of histone methyltransferases ATX1. HDA2, HDA5, HDA6, HDA9, and HDA15 which has been shown highly expression in leaves based on Arabidopsis eFP database3 were also selected for qPCR. Interestingly, the loss of WHIRLY1 decreased the transcription level of HDA2, HDA5, HDA6, and HDA9, while overexpressing WHIRLY1 only increased HDA6 and HDA9 expression. These results suggest that WHIRLY1 eFP database might involve in regulating gene expression of histone deacetylases to remove the H3K9ac marks at the promoter of WRKY53.
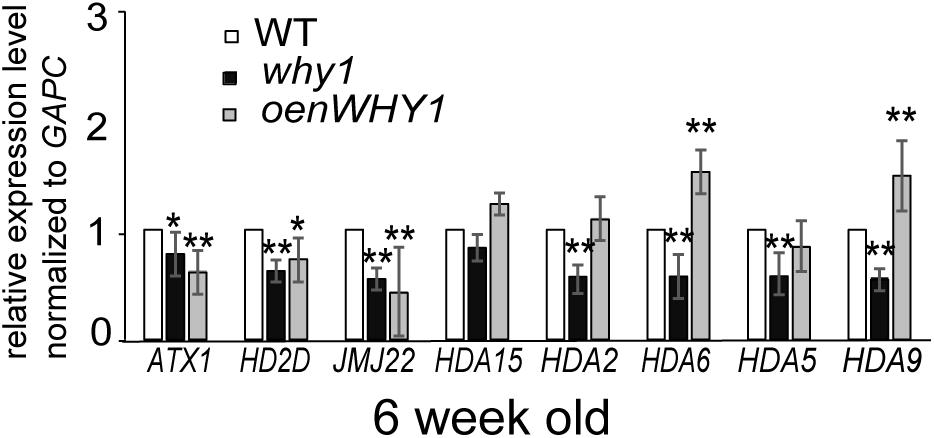
FIGURE 6. WHIRLY1 affects the HDAs gene expression. The expression of histone modification-related genes in 6-week-old why1, oenWHY1, and wild-type (WT) plants by qRT-PCR. The transcript level in each case was normalized to that of GAPC2 as a reference gene, and the expression level at WT was set as 1. Three biological replicates and three technique replicates were used to analyze. Error bars represent SE. Asterisk indicate significant differences (∗P < 0.05 and ∗∗P < 0.01) based on Student’s t-test.
WHIRLY1 was reported to be a repressor binding on the GNNNAAATT, plus an AT-rich telomeric repeat-like sequence in the WRKY53 promoter (Miao et al., 2013). In this study, we have found that enrichment of H3K4me3 and H3K9ac at promoter region contained WHIRLY1 binding domain and TATA box and translation start region of WRKY53, and recruitment of RNAPII, as well as transcription of WRKY53 are coordinated by WHRLY1 protein in a developmental manner. It demonstrates that the occupancy of WHIRLY1 represses the enrichment of H3K4me3 before senescence initiation and enhances the enrichment of H3K9ac and the recruitment of RNAPII at senescence initiation stage, and high ratio of H3K9ac/H3K4me2-3 determines the transcription level of WRKY53 and leaf senescence initiation. It illuminates that WHIRLY1 works as repressor of WRKY53 transcription associated with H3K4me2-3/H3K9ac balance in the developmental manner.
H3K4me2/3 is high at promoter or around the transcription start site (TSS) regions of active or poised genes in animals, while H3K4me2/3 is enriched at the proximal promoter and TSS site of genes in plants (Xiao et al., 2016). Both in animals and plants, H3K4me2/3 as well as H3K9ac which is also enriched at 5′-end and ATG site of genes were reported to correlate with active gene expression (Zhou et al., 2010; Songa et al., 2014). In Arabidopsis, H3K4me2/3 mark at 5′untranslated region and translational start region (from -155 bp to +27 bp) as well as gene body (from +367 bp to +549 bp) of WRKY53 locus increased in senescent leaves which consisted with the expression of WRKY53 (Ay et al., 2009). However, currently, genome-wide data of ChIP-Seq and RNA-seq showing a constant high level of H3K4me3 and H3K9ac marks in WRKY53 gene did not directly relate to WRKY53 expression (Brusslan et al., 2015). WRKY53 is one example of the many genes that are marked before significant up-regulation of mRNA levels during plant aging. It was explained that this inconsistencies between H3K4me3 marks and gene expression might be down-regulated by WHIRLY1 (Miao et al., 2013; Brusslan et al., 2015), or which may explain the coincidence of low transcript levels and high levels of H3K4me3 marks, as well as the examples of posttranscriptional regulation mediated by small RNAs have been identified during leaf senescence (Kim et al., 2009; Thatcher et al., 2015; Swida-Barteczka et al., 2018). In this study, we clearly showed only some specific regions of WRKY53 locus showed significantly change in H3K4me2-3 and H3K9ac at specific time point (Figure 1). The observation that the level of both H3K4me2/3 and H3K9ac at selected regions (WRKY53II and WRKY53P) of WRKY53 from 5th to 7th week correlated with the enrichment of WHIRLY1 protein binding on WRKY53 promoter. It indicates that WHIRLY1 spatio-temporally affect H3K4 methylation and H3K9 acetylation at WRKY53 during leaf aging. In vitro assay further confirms WHIRLY1 binding on specified motifs (GNNNAAATT) of WRKY53 promoter repressed enrichment of H3K4me3 and potentiated enrichment of H3K9ac, RNAPII recruitment, and TBP report gene transcription (Figure 4), implicating that WHIRLY1 involves in modification of local chromatin states.
Both H3K4me3 and H3K9ac are closely associated with active genes and play an important role in transcription. In human cells, H3K9ac potentiates the interaction of H3K4me3 and basal TF TFIID (Vermeulen et al., 2007). There are numerous studies demonstrated a strong relationship between TF occupancy and chromatin features (Liu et al., 2016). In this study, WHIRLY1 binding on WRKY53 promoter affected H3K4me3 and H3K9ac in vivo and in vitro. We utilized two transcription inhibitor sarkosyl and B2 RNA to address this question (Lauberth et al., 2013). Sarkosyl inhibits PIC assembly but does not affect elongation by pre-formed complexes (Cai and Luse, 1987; Hawley and Roeder, 1987), while B2 RNA binds to RNA polymerase II and inhibits transcription before PIC formation (Espinoza et al., 2004). Results of in vitro cell-free experiments indicated that WHIRLY1 occupied on WRKY53 promoter inhibit enrichment of H3K4me3 at the preinitiation stage of PIC formation, in contrary, WHIRLY1 enhances the enrichment of H3K9ac and RNAPII recruitment during the elongation by pre-formed complexes (Figure 5).
Compass-like complex is known to facilitate PIC assembly and generate H3K4me3 (Songa et al., 2014), our results showed WHIRLY1 which did not interact with core components of compass-like complex and H3K4 methyltransferases ATX1 then affected the enrichment of H3K4me3. WHIRLY1 binding motif at WRKY53 promoter was upstream and close to TATA box, in addition, the structure analysis of transcription initiation by RNA polymerase II showed PIC form around TATA box (Grunberg and Hahn, 2013), WHIRLY1 binding on promoter of WRKY53 may block the site for TATA-box binding protein and repress H3K4me3 enrichment by inhibiting the PIC assembly.
A possible combinatorial interplay between posttranslational modifications on the same histone was proposed based on the patterns of H3 methylation and acetylation at promoters of specific target genes (Taverna et al., 2007). Interestingly, our study suggested a relationship between the enrichment of H3K4me3 and H3K9ac at WRKY53 promoter region (WRKY53II). Enrichment of H3K4me3 at WRKY53II was inhibited by histone methylation inhibitor DZnep, while the enrichment of H3K9ac was improved by Dznep treatment, in turn, the enrichment of H3K9ac at WRKY53II region decreased in oehada15 mutant plants while H3K4me3 level at WRKY53II region increased in them. Many protein complexes and epigenetic modifications in the local chromatin environment are necessary for the progress of RNAPII-mediated transcription (Lauberth et al., 2013; Stasevich et al., 2014; Gates et al., 2017). H3K4me3 and H3K9ac were reported to involve in transcription initiation and elongation (Santos-Rosa et al., 2002; Ding et al., 2012; Gates et al., 2017; Zhang et al., 2017) and the interplay between H3K4me3 and H3K9ac at gene promoter was mediated by several factors such as HATs general control nonderepressible 5 (GCN5) (Foulds et al., 2013). WHIRLY1 was involved in the transcription of WRKY53 by interplay between H3K4me3 and H3K9ac.
WRKY53 is a well-known transcription factor, plays remarkable role in leaf senescence and plant senescence initiation (Hinderhofer and Zentgraf, 2001; Miao et al., 2004, 2007). It regulated more than 60 senescence-related gene expressions and their related signaling networks, showing delaying senescence phenotype in the wrky53 line, early senescence phenotype in the oeWRKY53 lines (Miao et al., 2004, 2007). The expression of WRKY53 is tightly controlled by multiple layers of regulation, including at the level of chromatin and transcription, as well as by ubiquitination and phosphorylation regulation. (Ay et al., 2009; Miao and Zentgraf, 2010; Zentgraf et al., 2010; Xie et al., 2014; Chen et al., 2016). In this study, WHIRLY1 had been found to bind on the promoter of WRKY53 and regulate the enrichment of H3K4me3 and H3K9ac in a developmental manner. It may seem paradoxical that WHIRLY1 itself binding on WRKY53 promoter enhances the enrichment of H3K9ac at leaf senescence initiation while it affects HDACs gene expression to promote the deacetylation of H3K9 at early senescence stage (Figure 7). Occupancy of WHIRLY1 on WRKY53 promoter mainly occurs as WRKY53 is being shutdown and decreases followed by aging. However, in vitro ChIP assay WHIRLY1 increased H3K9ac enrichment and RNAPII recruitment and upregulate downstream target gene transcription. It is a possibility that WHIRLY1 protein not only acts as a block of WRKY53 transcription before senescence initiation, inhibits activated transcription, but also acts as an activator of other downstream genes such as PR10a in potato or S40 in barley (Desveaux et al., 2000; Krupinska et al., 2014a), which might be coordinated with chromatin remodeling factor BRM or HDA6, HDA19 (Efroni et al., 2013; Chen et al., 2016). Additionally, the occupancy of WHIRLY1 may affect the accessibility of DNA by adjusting themselves to be more complicated complex under specific signals (Cappadocia et al., 2012). This may be the reason why the expression of HD2D and JMJ decreased in both why1 mutants and oenWHY1 mutants.
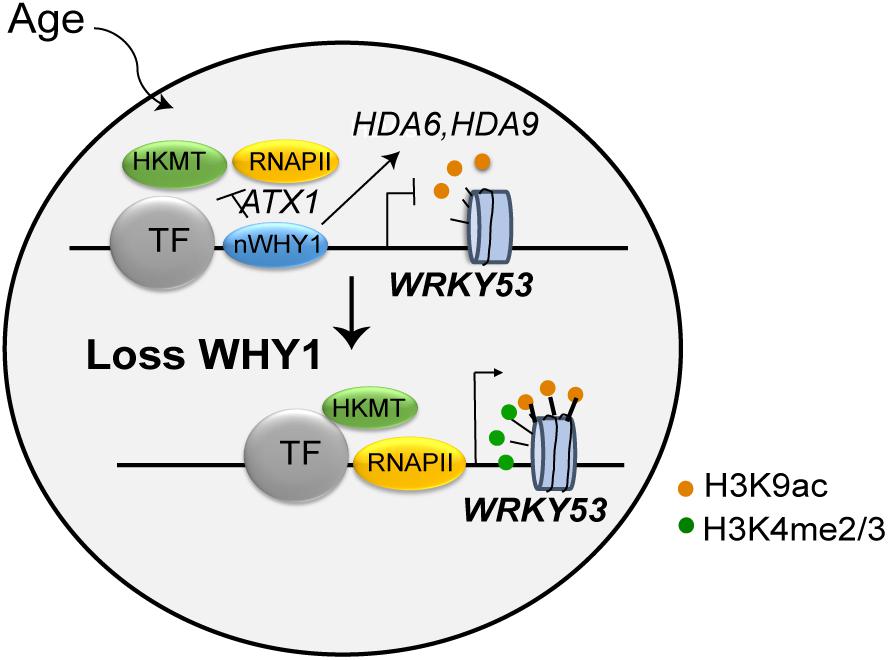
FIGURE 7. Working model of WHIRLY1 regulates WRKY53 transcription. WHIRLY1 binding on WRKY53 promoter represses enrichment of H3K4 methylation by ATX1 inhibiting the PIC formation in leaf senescence initiation and modulates the enrichment of H3K9ac by association with HDACs and high-level transcription at early senescence stage. In absence of WHIRLY1, other transcription activators recruit histone lysine methyltransferase and HDACs is dissociated from WRKY53 resulting in elevated H3K4me3, H3K9ac levels, and higher occupancy of RNAPII in WRKY53 loci, thus increasing the transcription of WRKY53. TF, transcription factor; HDACs, histone deacetylases; RNAPII, RNA polymerase II; HKMT, histone lysine methyltransferase; nWHY1, nuclear WHY1 protein.
Conclusively, we propose a model that WHIRLY1 binding on WRKY53 promoter represses enrichment of H3K4 methylation by ATX1 inhibits the PIC formation in leaf senescence initiation and modulates the enrichment of H3K9ac by association with HDAs and high-level transcription at early senescence stage (Figure 7). Further, what signals or factors promoted and controlled the action of WHIRLY1 on WRKY53 promoter during leaf senescence are still speculated. The accumulation of WHIRLY1 in nucleus is mediated by CIPK14 kinase which was promoted by light conditions, sugar, cytokinin, and calcium–calmodulin signal (Gan and Amasino, 1995; Lee et al., 2005; Akaboshi et al., 2008; Qin et al., 2010; Yan et al., 2014; Ren et al., 2017). Indeed, WHIRLY1 involved in response to different light conditions (Huang D. et al., 2017; Kucharewicz et al., 2017), further revealing that their relationships might provide insights into the upstream signals of WHIRLY1 function on SSB binding on target genes.
YM designed the study and performed in vitro ChIP assay. DH and BD performed ChIP–qPCR, Western blots, and phenotyping. WL performed qRT-PCR. DL crossed double mutant and screen them, phenotyping. WfL and YR for qRT-PCR and RNA-seq data analyses. DH and YM analyzed the data and wrote the paper.
This work was supported by a grant from the Natural Science Foundation of China (NSFC, Nos. 31470383 and 31770318) and Fujian “Hundred Talents Program” and German research Grant (DFG MI1392/1-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Professor Keqiang Wu (University of Taiwan) for providing the oeHDA15-GFP, hda15 seeds, Professor Zoya. Avramova (University of Nebraska-Lincoln) for providing the atx1 and atx2 seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01503/full#supplementary-material
Akaboshi, M., Hashimoto, H., Ishida, H., Saijo, S., Koizumi, N., Sato, M., et al. (2008). The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 377, 246–257. doi: 10.1016/j.jmb.2008.01.006
An, W., and Roeder, R. G. (2004). Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 377, 460–474. doi: 10.1016/s0076-6879(03)77030-x
Ay, N., Irmler, K., Fischer, A., Uhlemann, R., Reuter, G., and Humbeck, K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58, 333–346. doi: 10.1111/j.1365-3139.2008.03782.x
Besseau, S., Li, J., and Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. doi: 10.1093/jxb/err450
Brusslan, J. A., Bonora, G., Rus-Canterbury, A. M., Tariq, F., Jaroszewicz, A., and Pellegrini, M. (2015). A genome-wide chronological study of gene expression and two histone modifications, H3K4me3 and H3K9ac, during developmental leaf senescence. Plant Physiol. 168, 1246. doi: 10.1104/pp.114.252999
Cai, H., and Luse, D. S. (1987). Transcription initiation by RNA polymerase II in vitro, properties of preinitiation, initiation, and elongation complexes. J. Biol. Chem. 262, 298–304.
Cappadocia, L., Parent, J. S., Sygusch, J., and Brisson, N. (2013). A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69(Pt 11), 1207–1211. doi: 10.1107/S1744309113028698
Cappadocia, L., Parent, J. S., Zampini, E., Lepage, E., Sygusch, J., and Brisson, N. (2012). A conserved lysine residue of plant Whirly proteins is necessary for higher order protein assembly and protection against DNA damage. Nucleic Acids Res. 40, 258–269. doi: 10.1093/nar/gkr740
Chen, L., Xiang, S., Chen, Y., Li, D., and Yu, D. (2017). Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 10, 1174–1189. doi: 10.1016/j.molp.2017.07.008
Chen, X., Lu, L., Mayer, K. S., Scalf, M., Qian, S., Lomax, A., et al. (2016). Powerdress interacts with histone deacetylase 9 to promote aging in Arabidopsis. Elife 5:e17214. doi: 10.7554/eLife.17214.001-10.7554/eLife.17214.002
Choi, H. S., Hwang, C. K., Song, K. Y., Law, P. Y., Wei, L. N., and Loh, H. H. (2009). Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun. 380, 431–436. doi: 10.1016/j.bbrc.2009.01.136
Desveaux, D., Despres, C., Joyeux, A., Subramaniam, R., and Brisson, N. (2000). PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12, 1477–1489.
Desveaux, D., Maréchal, A., and Brisson, N. (2005). Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 10, 95–102. doi: 10.1016/j.tplants.2004.12.008
Dickey, T. H., Altschuler, S. E., and Wuttke, D. S. (2013). Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure 21, 1074–1084. doi: 10.1016/j.str.2013.05.013
Ding, Y., Ndamukong, I., Xu, Z., Lapko, H., Fromm, M., and Avramova, Z. (2012). ATX1-generated H3K4me3 is required for efficient elongation of transcription, not initiation, at ATX1-regulated genes. PLoS Genet. 8:e1003111. doi: 10.1371/journal.pgen.1003111
Efroni, I., Han, S. K., Kim, H. J., Wu, M. F., Steiner, E., Birnbaum, K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24, 438–445. doi: 10.1016/j.devcel.2013.01.019
Espinoza, C. A., Allen, T. A., Hieb, A. R., Kugel, J. F., and Goodrich, J. A. (2004). B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 11, 822–829. doi: 10.1038/nsmb812
Etminan, M., Fitzgerald, J. M., Gleave, M., and Chambers, K. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186, 299–317. doi: 10.1007/s10552-005-0334-2
Foulds, C. E., Feng, Q., Ding, C., Bailey, S., Hunsaker, T. L., Malovannaya, A., et al. (2013). Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Mol. Cell 51, 185–199. doi: 10.1016/j.molcel.2013.06.007
Gan, S., and Amasino, R. M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. doi: 10.1126/science.270.5244.1986
Gates, L. A., Shi, J., Rohira, A. D., Feng, Q., Zhu, B., Bedford, M. T., et al. (2017). Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 292, 14456. doi: 10.1074/jbc.M117.802074
Gendrel, A. V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2, 213–218. doi: 10.1038/nmeth0305-213
Grunberg, S., and Hahn, S. (2013). Structural insights into transcription initiation by RNA polymerase II. Trends Biochem. Sci. 38, 603–611. doi: 10.1016/j.tibs.2013.09.002
Hawley, D. K., and Roeder, R. G. (1987). Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 262, 3452–3461.
Hedglin, M., and Benkovic, S. J. (2017). Replication protein A prohibits diffusion of the PCNA sliding clamp along single-stranded DNA. Biochemistry 56, 1824–1835. doi: 10.1021/acs.biochem.6b01213
Hinderhofer, K., and Zentgraf, U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213, 469–473. doi: 10.1007/s004250000512
Huang, D., Lin, W., Deng, B., Ren, Y., and Miao, Y. (2017). Dual-located WHIRLY1 interacting with LHCA1 alters photochemical activities of photosystem I and is involved in light adaptation in Arabidopsis. Int. J. Mol. Sci. 18:2352. doi: 10.3390/ijms18112352
Huang, S. H., Hart, M. A., Wade, M., Cozart, M. R., McGrath, S. L., and Kobryn, K. (2017). Biochemical characterization of Borrelia burgdorferi’s RecA protein. PLoS One 12:e0187382. doi: 10.1371/journal.pone.0187382
Jiang, Y., Liang, G., Yang, S., and Yu, D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26, 230–245. doi: 10.1105/tpc.113.117838
Kim, J. H., Woo, H. R., Kim, J., Lim, P. O., Lee, I. C., Choi, S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. doi: 10.1126/science.1166386
Kim, S. S., Pandey, K. K., Choi, H. S., Kim, S. Y., Law, P. Y., Wei, L. N., et al. (2005). Poly(C) binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol. Pharmacol. 68, 729–736. doi: 10.1124/mol.105.012245
Krupinska, K., Dähnhardt, D., Fischer-Kilbienski, I., Kucharewicz, W., Scharrenberg, C., Trösch, M., et al. (2014a). Identification of WHIRLY1 as a factor binding to the promoter of the stress- and senescence-associated gene HvS40. J. Plant Growth Regul. 33, 91–105. doi: 10.1007/s00344-013-9378-9
Krupinska, K., Oetke, S., Desel, C., Mulisch, M., Schafer, A., Hollmann, J., et al. (2014b). WHIRLY1 is a major organizer of chloroplast nucleoids. Front. Plant Sci. 5:432. doi: 10.3389/fpls.2014.00432
Kucharewicz, W., Distelfeld, A., Bilger, W., Muller, M., Munne-Bosch, S., Hensel, G., et al. (2017). Acceleration of leaf senescence is slowed down in transgenic barley plants deficient in the DNA/RNA-binding protein WHIRLY1. J. Exp. Bot. 68, 983–996. doi: 10.1093/jxb/erw501
Lauberth, S. M., Nakayama, T., Wu, X., Ferris, A. L., Tang, Z., Hughes, S. H., et al. (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036. doi: 10.1016/j.cell.2013.01.052
Lee, E.-J., Iai, H., Sano, H., and Koizumi, N. (2005). Sugar responsible and tissue specific expression of a gene encoding AtCIPK14, an Arabidopsis CBL-interacting protein kinase. Biosci. Biotechnol. Biochem. 69, 242–245. doi: 10.1271/bbb.69.242
Lepage, E., Zampini, E., and Brisson, N. (2013). Plastid genome instability leads to reactive oxygen species production and plastid-to-nucleus retrograde signaling in Arabidopsis. Plant Physiol. 163, 867–881. doi: 10.1104/pp.113.223560
Lim, P. O., Kim, Y., Breeze, E., Koo, J. C., Woo, H. R., Ryu, J. S., et al. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52, 1140–1153. doi: 10.1111/j.1365-313X.2007.03317.x
Liu, J., He, L., Collins, I., Ge, H., Libutti, D., Li, J., et al. (2000). The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol. Cell 5, 331–341. doi: 10.1016/S1097-2765(00)80428-1
Liu, L., Zhao, W., and Zhou, X. (2016). Modeling co-occupancy of transcription factors using chromatin features. Nucleic Acids Res. 44:e49. doi: 10.1093/nar/gkv1281
Lohman, T. M., and Ferrari, M. E. (1994). Escherichia Coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570. doi: 10.1146/annurev.bi.63.070194.002523
MacDonald, G. H., Itoh-Lindstrom, Y., and Ting, J. P. (1995). The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J. Biol. Chem. 270, 3527–3533.
Maréchal, A., and Zou, L. (2014). RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 25:9. doi: 10.1038/cr.2014.147
Miao, Y., Jiang, J., Ren, Y., and Zhao, Z. (2013). The single-stranded DNA binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis thaliana. Plant Physiol. 163, 746–756. doi: 10.1104/pp.113.223412
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-004-2142-6
Miao, Y., Laun, T. M., Smykowski, A., and Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. doi: 10.1007/s11103-007-9198-z
Miao, Y., Smykowski, A., and Zentgraf, U. (2008). A novel upstream regulator of WRKY53 transcription during leaf senescence in Arabidopsis thaliana. Plant Biol. 10, 110–120. doi: 10.1111/j.1438-8677.2008.00083.x
Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. doi: 10.1105/tpc.106.042705
Miao, Y., and Zentgraf, U. (2010). A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 63, 179–188. doi: 10.1111/j.1365-313X.2010.04233.x
Ostrup, O., Reiner, A. H., Alestrom, P., and Collas, P. (2014). The specific alteration of histone methylation profiles by DZNep during early zebrafish development. Biochim. Biophys. Acta 1839, 1307–1315. doi: 10.1016/j.bbagrm.2014.09.013
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qin, Y., Guo, M., Li, X., Xiong, X., He, C., Nie, X., et al. (2010). Stress responsive gene CIPK14 is involved in phytochrome A-mediated far-red light inhibition of greening in Arabidopsis. Sci. China Life Sci. 53, 1307–1314. doi: 10.1007/s11427-010-4078-1
Ren, Y., Li, Y., Jiang, Y., Wu, B., and Miao, Y. (2017). Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual dunctions in Arabidopsis. Mol. Plant 10, 749–763. doi: 10.1016/j.molp.2017.03.011
Ribeiro, J., Abby, E., Livera, G., and Martini, E. (2016). RPA homologs and ssDNA processing during meiotic recombination. Chromosoma 125, 265–276. doi: 10.1007/s00412-015-0552-7
Robatzek, S., and Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Roy, R., Kozlov, A. G., Lohman, T. M., and Ha, T. (2007). Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 369, 1244–1257. doi: 10.1016/j.jmb.2007.03.079
Roy, R., Kozlov, A. G., Lohman, T. M., and Ha, T. (2009). SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature 461:1092. doi: 10.1038/nature08442
Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20, 568–579. doi: 10.1105/tpc.107.056614
Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., et al. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. doi: 10.1038/nature01080
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M., and Keck, J. L. (2008). SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. 43:289. doi: 10.1080/10409230802341296
Songa, Z.-T., Suna, L., Lua, S.-J., Tianb, Y., Dingb, Y., and Liu, J.-X. (2014). Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. U.S.A. 112, 2900–2905. doi: 10.1073/pnas.1419703112
Stasevich, T. J., Hayashi-Takanaka, Y., Sato, Y., Maehara, K., Ohkawa, Y., Sakata-Sogawa, K., et al. (2014). Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516, 272–275. doi: 10.1038/nature13714
Swida-Barteczka, A., Krieger-Liszkay, A., Bilger, W., Voigt, U., Hensel, G., Szweykowska-Kulinska, Z., et al. (2018). The plastid-nucleus located DNA/RNA binding protein WHIRLY1 regulates microRNA-levels during stress in barley (Hordeum vulgare L.). RNA Biol. 15, 886–891. doi: 10.1101/197202
Taverna, S. D., Ueberheide, B. M., Liu, Y., Tackett, A. J., Diaz, R. L., Shabanowitz, J., et al. (2007). Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc. Natl. Acad. Sci. U.S.A. 104, 2086–2091. doi: 10.1073/pnas.0610993104
Thatcher, S. R., Burd, S., Wright, C., Lers, A., and Green, P. J. (2015). Differential expression of miRNAs and their target genes in senescing leaves and siliques: insights from deep sequencing of small RNAs and cleaved target RNAs. Plant Cell Environ. 38, 188–200. doi: 10.1111/pce.12393
Ulker, B., Shahid Mukhtar, M., and Somssich, I. E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226, 125–137. doi: 10.1007/s00425-006-0474-y
Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W., van Schaik, F. M. A., Varier, R. A., et al. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69. doi: 10.1016/j.cell.2007.08.016
Xiao, J., Lee, U. S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xie, Y., Huhn, K., Brandt, R., Potschin, M., Bieker, S., Straub, D., et al. (2014). REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141, 4772–4783. doi: 10.1242/dev.117689
Xiong, J. Y., Lai, C. X., Qu, Z., Yang, X. Y., Qin, X. H., and Liu, G. Q. (2009). Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol. Biol. 71, 437–449. doi: 10.1007/s11103-009-9533-7
Yan, J., Niu, F., Liu, W.-Z., Zhang, H., Wang, B., Yang, B., et al. (2014). Arabidopsis CIPK14 positively regulates glucose response. Biochem. Biophys. Res. Commun. 450, 1679–1683. doi: 10.1016/j.bbrc.2014.07.064
Yoo, H. H., Kwon, C., Lee, M. M., and Chung, I. K. (2007). Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 49, 442–451. doi: 10.1111/j.1365-313X.2006.02974.x
Zampini, E., Lepage, E., Tremblay-Belzile, S., Truche, S., and Brisson, N. (2015). Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 25, 645–654. doi: 10.1101/gr.188573.114
Zentgraf, U., Laun, T., and Miao, Y. (2010). The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 89, 133–137. doi: 10.1016/j.ejcb.2009.10.014
Zhang, B. L., Guo, T. W., Gao, L. L., Ji, G. Q., Gu, X. H., Shao, Y. Q., et al. (2017). Egr-1 and RNA POL II facilitate glioma cell GDNF transcription induced by histone hyperacetylation in promoter II. Oncotarget 2017, 45105–45116. doi: 10.18632/oncotarget.15126
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Zhang, Y., Liu, Z., Wang, X., Wang, J., Fan, K., Li, Z., et al. (2018). DELLA proteins negatively regulate dark-induced senescence and chlorophyll degradation in Arabidopsis through interaction with the transcription factor WRKY6. Plant Cell Rep. 37, 981–992. doi: 10.1007/s00299-018-2282-9
Zhou, J., Wang, X., He, K., Charron, J. B., Elling, A. A., and Deng, X. W. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Keywords: ssDNA binding protein, transcription, histone modification, leaf senescence, Arabidopsis
Citation: Huang D, Lan W, Li D, Deng B, Lin W, Ren Y and Miao Y (2018) WHIRLY1 Occupancy Affects Histone Lysine Modification and WRKY53 Transcription in Arabidopsis Developmental Manner. Front. Plant Sci. 9:1503. doi: 10.3389/fpls.2018.01503
Received: 15 July 2018; Accepted: 26 September 2018;
Published: 19 October 2018.
Edited by:
Ming Luo, South China Botanical Garden (CAS), ChinaReviewed by:
Diqiu Yu, Xishuangbanna Tropical Botanical Garden (CAS), ChinaCopyright © 2018 Huang, Lan, Li, Deng, Lin, Ren and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Miao, eW1pYW9AZmFmdS5lZHUuY24=; eW1pYW8yMDEzQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.