
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 September 2018
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01304
The transgenic technology using a single gene has been widely used for crop improvement. But the transgenic pyramiding of multiple genes, a promising alternative especially for enhancing complexly inherited abiotic stress tolerance, has received little attention. Here, we developed and evaluated transgenic rice lines with a single Salt Responsive Protein 3-1 (SaSRP3-1) gene as well as pyramids with two-genes SaSRP3-1 and Vacuolar H+-ATPase subunit c1 (SaVHAc1) derived from a halophyte grass Spartina alterniflora L. for salt tolerance at seedling, vegetative, and reproductive stages. The overexpression of this novel gene SaSRP3-1 resulted in significantly better growth of E. coli with the recombinant plasmid under 600 mM NaCl stress condition compared with the control. During early seedling and vegetative stages, the single gene and pyramided transgenic rice plants showed enhanced tolerance to salt stress with minimal wilting and drying symptoms, improved shoot and root growth, and significantly higher chlorophyll content, relative water content, and K+/Na+ ratio than the control plants. The salt stress screening during reproductive stage revealed that the transgenic plants with single gene and pyramids had better grain filling, whereas the pyramided plants showed significantly higher grain yield and higher grain weight compared to control plants. Our study demonstrated transgenic pyramiding as a viable approach to achieve higher level of salt tolerance in crop plants.
The productivity of rice (Oryza sativa L.) and other food crops is severely affected by abiotic stresses such as drought, salinity, flooding, temperature extremes, and nutrient deficiency or toxicity worldwide. Among these, soil salinity is a major abiotic stress negatively affecting the growth and productivity of rice and other major agricultural crops. Improving salt tolerance of crop plants and thereby minimizing yield loss is of paramount importance for any crop improvement program.
During last few decades, considerable efforts have been made through classical breeding to introgress genes from salt tolerant donor parents as well as wild relatives. As a result, commercial rice varieties have been improved through introgression of salt tolerance genes from promising landraces like Nona Bokra and Pokkali (Gregorio et al., 2002). However, major obstacles faced by plant breeders are the transfer of many undesirable attributes linked to the targeted salt tolerance genes (Thomson et al., 2010) and narrow genetic variation for salt tolerance in rice germplasm (Pearson and Bernstein, 1959; Kaddah, 1963; Lutts et al., 1995). Likewise, quantitative trait loci (QTL) mapping and modern genomics tools have been exploited for a better understanding of salt tolerance mechanisms as well as the development of new varieties. Several QTLs associated with salt tolerance traits have been identified in rice using several mapping populations derived from salt tolerant genotypes (Zhang et al., 1995; Koyama et al., 2001; Lin et al., 2003; Thomson et al., 2010). Similarly, dissection of salt tolerance through genome-wide association study (GWAS) revealed candidate salt tolerance-related genes in rice (Yu et al., 2017) and provided the platform for their functional analysis of salt tolerance. Since salt tolerant traits are controlled by many genes and influenced by environmental interactions (Flowers et al., 2000; Koyama et al., 2001; Lin et al., 2003), the development of salt tolerant varieties through marker assisted selection (MAS) requires extensive genotyping efforts and longer time to introgress many desirable QTLs into elite cultivars. Therefore, transgenic approach using novel salt tolerant genes from halophytes offers a promising alternative to augment the limited natural variation for salt tolerance attributes available in the rice germplasm. Smooth cordgrass is a halophytic grass species widely grown in coastal salt marsh areas of the United States. It can grow well in areas, where salinity is a major obstacle for plant growth. It can tolerate salt concentration up to 0.6 M NaCl (Vasquez et al., 2006). Since halophytes have evolved physiological mechanisms to survive under natural saline conditions, these species could be considered as a potential reservoir for isolating salt tolerance genes. During the last decade, extensive efforts have been made to isolate and characterize salt tolerant genes from halophytes like smooth cordgrass (Baisakh et al., 2008), Thellungiella salsuginea, a close relative of Arabidopsis (Wu et al., 2012; Ellouzi et al., 2014), Salicornia europaea L. (Lv et al., 2017), and Brachypodium distachyon (Takahashi et al., 2017). Recently, transgenic rice plants overexpressing a soluble inorganic pyrophosphatase gene ThPP1 of Thellungiella halophila showed enhanced tolerance to alkaline stress compared to the wild type (He et al., 2017).
Generally, the plasma membrane is a primary structure which helps to maintain the intracellular ion homeostasis (Lv et al., 2017), and it selectively allows the exchange of other organic compounds. Salt stress is known to alter the structure and composition of membrane proteome, and thereby affecting the stability and solubility of essential components of membrane proteins (Katz et al., 2007). The PMP3 protein from Saccharomyces cerevisiae was first reported to be involved in salt tolerance (Navarre and Goffeau, 2000) followed by the discovery of its homolog RCI2A in Arabidopsis (Mitsuya et al., 2005) and AcPMP3-1 in sheep grass (Aneurolepidium chinense). These genes helped to improve salt tolerance by restricting excessive uptake of Na+ (Inada et al., 2005).
Previous studies from the analysis of expressed sequence tags (ESTs) from salt stressed tissues demonstrated that Spartina alterniflora L. is a rich repository of salt stress-related genes (Baisakh et al., 2008) and indicated enhanced expression of plasma membrane protein3 (SaPMP3) and other unknown plasma membrane proteins under salt stress. In the present study, we evaluated the role of a novel salt responsive protein, SaSRP3-1 in enhancing salt tolerance in rice through transgenic approach. The SaSRP3-1 protein has a single transmembrane domain which is a typical characteristic of plasma membrane proteins (Singh and Mittal, 2016). In addition to single gene transgenic lines, pyramided transgenic rice lines with both SaSRP3-1 and vacuolar ATPase Subunit c1 gene (SaVHAc1) were generated and evaluated for salt tolerance at various stages of crop growth.
Cocodrie, a high yielding long grain rice variety released by the Louisiana State University Agricultural Experiment Station (Linscombe et al., 2000), was used for Agrobacterium transformation.
An EST sequence from S. alterniflora L. (GenBank Acc. No. EH277327) corresponding to Salt Responsive Protein3-1 (SaSRP3-1) gene, highly expressed under salt stress condition (Baisakh et al., 2008), was used to develop transgenic rice plants. The complete open reading frame (ORF) of SaSRP3-1 from the S. alterniflora root cDNA library was amplified with primers SRP3-1F and SRP3-1R (Supplementary Table S1) for cloning into plant expression vector pCAMBIA 1305.2. In addition to single gene transgenic lines, pyramided transgenic rice lines were generated by crossing the homozygous SaSRP3-1 plants with the previously developed transgenic plants overexpressing SaVHAc1 from S. alterniflora L. (Baisakh et al., 2012).
For Escherichia coli overexpression, blunt-end PCR products from the complete ORF of SaSRP3-1 was directionally cloned by adding four bases to the forward primer (CACC) into pET101/D-TOPO® vector (Life Technologies, Grand Island, NY, United States). The gene was cloned after T7 promoter, which was induced in response to isopropyl β-D-1-thiogalactopyranoside (IPTG). The resulting recombinant vector was referred to as pET-SaSRP3-1 (Figure 1A). All primers used in this study for cloning, reverse transcription PCR, and confirmation of transgenic plants are listed in Supplementary Table S1. The recombinant plasmid pET-SaSRP3-1 as well as the plasmid pET-C, without the target gene, was transformed into competent E. coli strain BL21 (DE3). Transformed cells were inoculated in 3 mL of LB medium containing kanamycin (50 mg/L) and allowed to grow overnight in a shaker. The secondary culture was then initiated in 20 mL LB medium containing kanamycin (50 mg/L) with an equal concentration of each transformed cells at 37°C with constant shaking (250 rpm). In order to induce the protein synthesis in BL21 (DE3) cells, IPTG (100 mM) was added to the media when the OD600 value reached 0.5. For salt tolerance screening, BL21 (DE3) cells transformed with SaSRP3-1 and expression control were streaked on selection media plates with varying salt concentration of 200, 400, 600, 800, and 1000 mM NaCl and incubated at 37°C. Based on the relative growth pattern, threshold NaCl concentration was determined to differentiate the growth of expression between control and recombinant cells.
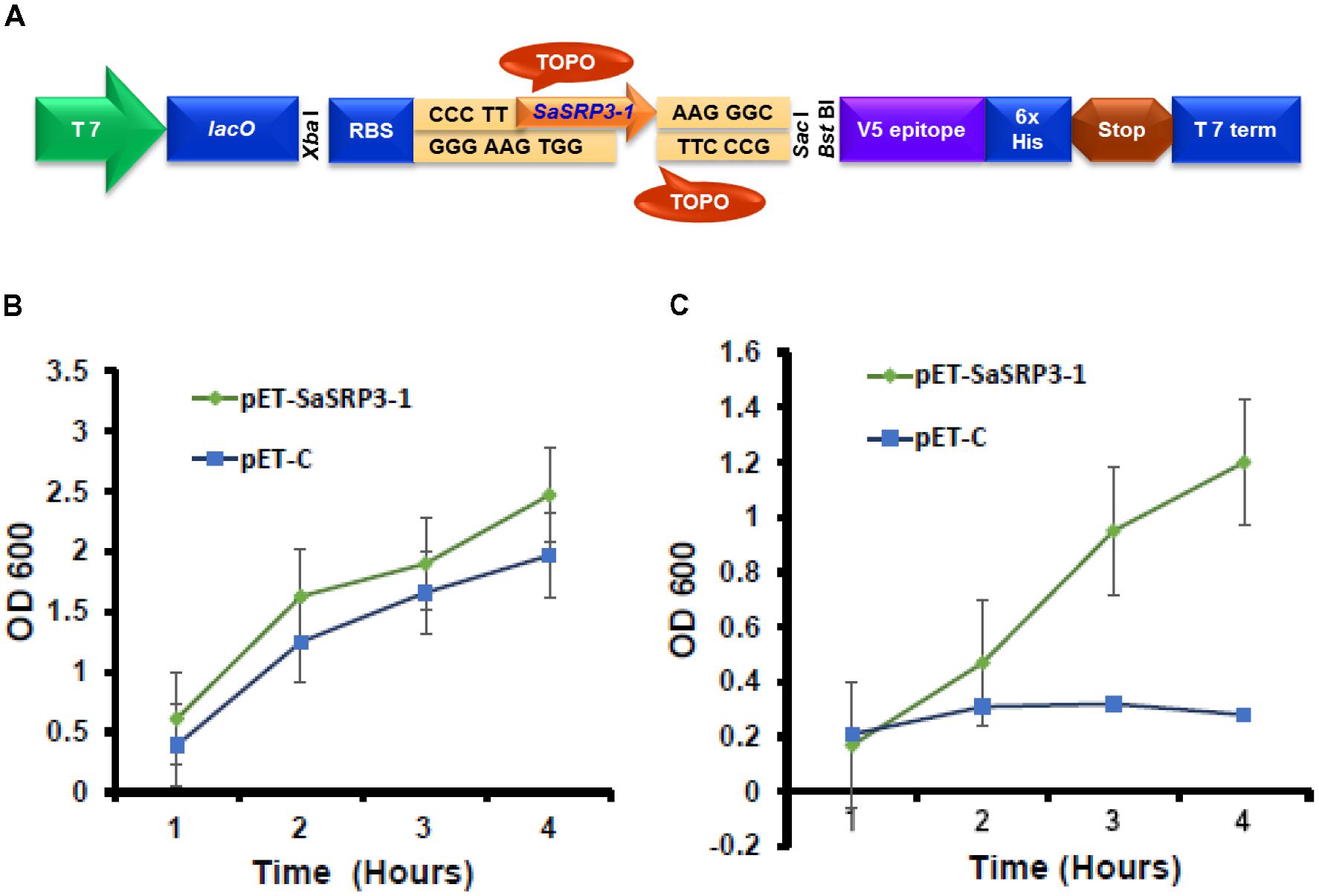
FIGURE 1. Cloning of SaSRP3-1 into E. coli expression vector pET101/D-TOPO®. (A) Schematic representation of pET101/D-TOPO vector with cloning site, (B) Differences in the growth response curve of E. coli cells grown in the absence of NaCl in liquid LB culture. (C) Growth of E. coli cells after adding 600 mM NaCl to liquid LB media SaSRP3-1: pET-SaSRP3-1, ExCt1: pET-C. The mean of three independent measurements (OD600) are indicated at different time intervals.
For liquid growth assay, a secondary culture was initiated in 50 mL LB medium following the same procedure as described above and after induction of protein expression, 600 mM NaCl was added to LB broth in one flask but not to the control flask. The experiment was conducted in three replications, and the optical density of the culture was noted after a regular time interval (1, 4, 8, and 16 h) to construct the growth curve.
The pCAMBIA 1305.2 (CAMBIA, Australia), a binary plasmid vector showing stable replication in E. coli and Agrobacterium tumefaciens, was used to deliver the SaSRP3-1 gene into embryonic calli. The plasmid contains the hygromycin B phosphotransferase (hptII) antibiotic resistance gene to screen the positive plants and kanamycin (nptIII) resistance gene for E. coli and A. tumefaciens selection (Broothaerts et al., 2005). The A. tumefaciens strain harboring pTOK233 plasmid and the virulence loci necessary for T-DNA transfer was used for callus infection (Hiei et al., 1994, 1997). Full-length cDNA clone of SaSRP3-1 was cloned under the control of CaMV 35 promoter in pCAMBIA 1305.2 vector at BglII and PmlI restriction endonucleases sites (Figure 2A). The plant expression vector pC-SaSRP3-1 was confirmed by both restriction analysis and DNA sequencing and then transformed into A. tumefaciens strain LBA4404 (Hoekema et al., 1983) for rice calli infection.
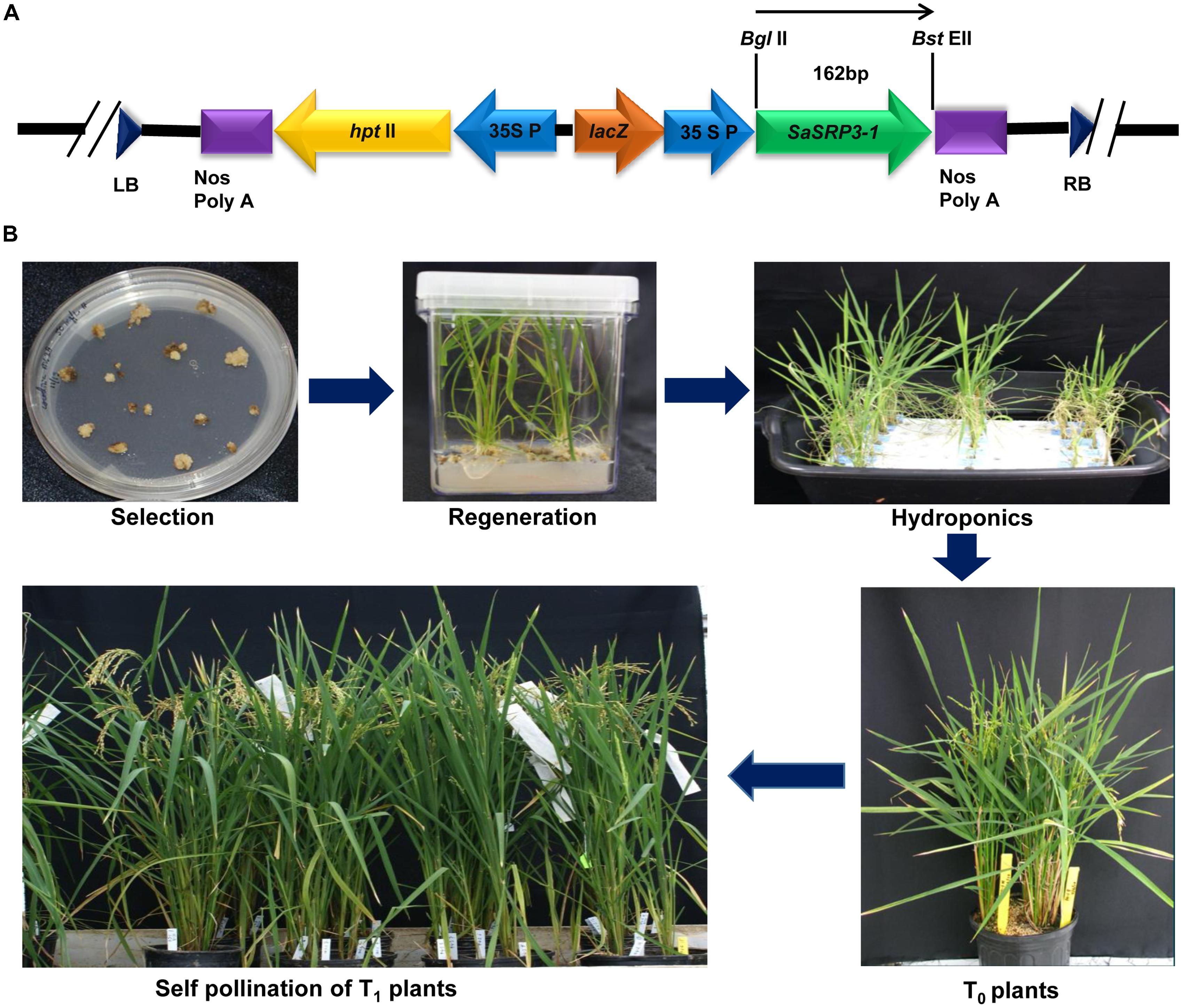
FIGURE 2. Plasmid construction for the gene SaSRP3-1 and development of transgenic plants. (A) The schematic diagram of the partial linear plasmid vector and position of the gene SaSRP3-1. The diagram depicts the details of the pCAMBIA1305.2 vector containing right and left borders, SaSRP3-1 gene between BglII and BstEII sites, 35S CaMV promoter, plant selection marker (hptII), multiple cloning sites, and termination sites; (B) Transformation and regeneration of Cocodrie-SaSRP3-1 transgenic rice plants showing selection, regeneration, and self-pollination of transgenic plants after Agrobacterium mediated transformation.
Dehusked rice seeds from Cocodrie variety were used for callus induction following the method of Hiei et al. (1994). Four to six weeks old light yellowish and compact embryogenic calli were used for Agrobacterium transformation. After Agrobacterium co-cultivation, calli were placed on selection media with hygromycin (50 mg/L) and 3–4 cycles of selection were performed. Subsequently, these calli were transferred to regeneration media and then into the rooting medium (Figure 2B and Supplementary Table S5). After 5 days of root regeneration, small seedlings of 3 to 8 cm height were initially grown in hydroponics under control conditions of light with six 40W Sylvania cool white fluorescent bulbs (∼100 μE m−2 s−1) at 26°C in 1/4th strength Yoshida media (Yoshida et al., 1976). The hydroponic nutrient media was replaced with fresh media after every 7 days. Transgenic plants were identified by PCR amplification of the target gene as well as hygromycin B phosphotransferase gene. The positive seedlings were grown in the greenhouse and self-pollinated to advance the generation (Figure 2B). The greenhouse growing condition was set at 13:11 h (day: night) photoperiod with the temperature setting of at 28°C/25°C (day/night) and a light intensity of 350 μE m−2 s−1. Five seedlings were allowed to grow per 1-gallon pot of 6.25-inch depth × 6.5-inch diameter size.
Regenerated individual plants from hydroponic media at 4–5 leaf stage were analyzed to confirm the successful transformation. Transgenic plants were identified by PCR amplification of SaSRP3-1 and hptII genes by gene specific primers in T0 generation. The positive transgenic plants were then transferred to greenhouse and selfed for two successive generations to get homozygous plants. All PCR amplification reactions were performed using the protocol of Baisakh et al. (2008).
Transgenic rice plants selected by PCR analysis were further confirmed through Southern hybridization (Sambrook and Russell, 2001). Ten micrograms of genomic DNA from independent positive plants as well as untransformed control plants was digested with HindIII (Fermentas, Waltham, MA, United States), which cuts at a single restriction site within the T-DNA region (pCAMBIA 1305.2). The separated DNA on agarose gel was transferred to HybondTM N+ membrane (GE Healthcare Biosciences, Piscataway, NJ, United States) following standard procedure and DNA on blot was fixed by UV cross-linking.
Southern hybridization was done using the ECL direct nucleic acid labeling and detection system (GE Healthcare Biosciences, Piscataway, NJ, United States). Transformed SaSRP3-1 gene present on T-DNA was used as a probe to detect the transgenic plants The blots were exposed to autoradiography film (Hyperfilm ECL, GE Healthcare, Piscataway, NJ, United States) for 5 h to visualize the gene specific bands (Supplementary Figure S3).
Total RNA was isolated from T2 homozygous rice seedlings using the RNeasy Plant Mini Kit (QIAGEN, Valencia, CA, United States) and was eluted in RNase-free water for gel analysis and RT-PCR. First strand complementary DNA (cDNA) was synthesized using the qScriptTM Flex cDNA Synthesis Kit (Quanta BioSciences, Gaithersburg, MD, United States). The cDNA was subsequently used as a template for RT-PCR with gene specific primers. The amplification of elongation factor 1-α gene (ef1α) was used as an internal control for comparing gene expression in wild type and transgenic rice plants. The amplified products were separated on 1.2% agarose gel for comparison of gene expression.
For phenotypic characterization T2 homozygous SaSRP3-1, T3 homozygous SaVHAc1, and homozygous pyramided plants with two-genes SaSRP3-1 and SaVHAc1 were used for salt stress screening at seedling, vegetative, and reproductive stages. For seedling screening, rice seeds were germinated in Petri plates and 5-day-old seedlings were transferred to Yoshida nutrient media (Yoshida et al., 1976) under a 14 h light and 10 h dark at 25°C. The seedlings were supported on a styrofoam sheet fabricated with supporting sponge or nylon mesh at the bottom. Seedlings were allowed to grow under normal condition for 10 days. Salt stress was imposed to 12-day-old seedlings by adding 100 and 150 mM NaCl to the nutrient solution and pH 5.2 was maintained. The control plants were not subjected to salt stress. Plant growth related traits such as shoot and root length were measured on five seedlings both before and 12 days after stress treatment for comparison. For vegetative stage screening, plants were exposed to 60 and 80 mM NaCl 45 days after planting. Physiological observations (chlorophyll content, relative water content [RWC], and proline content) were recorded 12 days after salt stress under greenhouse condition. The unstressed leaves at the vegetative stage were used for leaf disk assay.
The wild type and transgenic plants were used for salinity tolerance screening at reproductive stage under greenhouse conditions by adding 60 mM NaCl at the time of boot leaf emergence. During both vegetative and reproductive stage screening, fresh salt solution was added regularly at 3 days intervals to maintain the desired salt stress and pH 5.2. For reproductive stage screening, salt stress and pH were maintained until harvesting. Phenotypic traits such as grain yield per plant, 100-grain weight, and seed sterility (%) were recorded. The 100-grain weight was measured on 100 seeds after drying at 45°C in an oven for 5 days after harvesting. The number of filled and chaffy spikelets was counted at the time of harvesting and seed sterility percentage was computed as follows.
Leaf injury due to leakage of ion and damage to the photosynthetic unit was visually evaluated by floating leaf disk assay. During vegetative stage, fully expanded leaf sections of 2 cm length from one leaf below the flag leaf were used for comparison. Leaf disks were floated in control and NaCl concentrations of 100 mM and 150 mM. After 72 h, the phenotypic difference in chlorophyll retention was visually compared (Munns and James, 2003; Baisakh et al., 2012).
The chlorophyll content was measured to see the possible differences between transgenic plants and control plants. Leaf samples from vegetative stage screening were used for comparing the variation in total chlorophyll content. Fresh leaf weight was measured, and chlorophyll was extracted with 80% acetone from individual transgenic and control plants. The absorbance of the supernatant was measured at 645 and 663 nm. The concentration of chlorophyll a (Chl-a), chlorophyll b (Chl-b), and total chlorophyll (Chl-t) were estimated using the extinction coefficient equations (Arnon, 1949). Total chlorophyll content in leaf sample was expressed as milligram per gram of fresh leaf weight (mg/g).
A leaf section of 2 cm length from one leaf below the flag leaf was used to determine the RWC during vegetative stage. Ten leaf sections of 2 cm length were collected 5 cm below the leaf tip from each plant and fresh weights were taken. The leaf sections were then transferred to fresh Yoshida nutrient solution (Yoshida et al., 1976) for about 3 h at room temperature under dark condition and turgid weight was taken. Leaf samples were dried in oven at 70°C overnight and dry weights were taken. The RWC was calculated using the following formula.
where FW is fresh weight, DW is dry weight, and TW is turgid weight.
Proline accumulation in response to hyperosmotic stress was well documented in Arabidopsis (Toka et al., 2010) and other plants. The amount of accumulated proline in transgenic and control plants was quantified during the vegetative stage after exposure to salt stress (60 and 80 mM NaCl) and control conditions following the procedure of Bates et al. (1973). Known quantity of leaf samples were homogenized in 3% 5-sulfosalicylic acid. The amount of proline content in each sample was quantified by ninhydrin labeling. Total amount of proline content in the leaf sample was expressed as microgram per gram of fresh leaf weight (μg/g).
Total Na+ and K+ content in shoots were measured under stress and control conditions during seedling stage salt stress. Leaf samples were collected and rinsed thoroughly in distilled water to ensure the exclusion of external salt deposition. Samples were completely dried at 80°C for 2 days and powdered. One hundred micrograms of powdered samples were incubated with 5 mL of 4 M HCl at 37°C overnight to extract Na+ and K+ (Zhao et al., 2007). The supernatant was filtered, and diluted samples were used to determine the total Na+ and K+ by a Flame photometer (Jenway Model PFP7, Bibby Scientific Limited, United Kingdom). Total concentration of Na+ and K+ in plant samples were estimated based on the concentration determined in NaCl and KCl standard curve and, K+/Na+ ratio was used for comparing the transgenic and control plants.
The effect of salt stress on various morphological and physiological traits was analyzed by one-way ANOVA using the mean observations from replicated experiments separately for salt stress and control condition. Differences in mean values of traits for transgenic and control plants were analyzed using paired t-test. Post hoc comparison was made using Dunnett’s test, when one-way ANOVA results showed the difference in genotypic mean values at 0.05 probability level. All statistical analyses were performed using SAS version 9.3 for Windows (SAS Institute, 2011).
The ORF of the EST SaSRP3-1 (Baisakh et al., 2008) was 162 bp long and the protein was made up of 54 amino acids (Supplementary Figures S1A,B). The secondary structure prediction indicated the presence of one prominent α helix structure and very small size β strand as well (Supplementary Figure S1C). Amino acid analysis indicated that 44 amino acids were hydrophobic in SaSRP3-1 and 14 to 28th amino acids formed a single hydrophobic transmembrane domain (TMD) (Supplementary Figure S1D). Moreover, comparison of its nucleotide sequence to accessions in the non-redundant sequence database using BLASTN interface indicated 94–95 percent similarity to stress induced hydrophobic protein PMP from other plant species (Supplementary Table S2). The presence of a single TMD indicated the possibility of unique PMP evolution in S. alterniflora to form an uncharacterized but crucial protein to survive under abiotic stress environments.
The results from the SaSRP3-1 overexpression experiment in E. coli indicated normal bacterial colony growth under control, 200, and 400 mM NaCl concentrations and there was no significant difference in growth between the pET-SaSRP3-1 transformed and control E. coli cells (Supplementary Figure S2). Since control condition, 200 and 400 mM NaCl concentrations did not differentiate the growth of SaSRP3-1 transformed E. coli from E. coli cells without transgene, we observed similar pattern of colony growth at these NaCl treatments. Interestingly, more colony growth was observed at 600 mM NaCl only with pET-SaSRP3-1 transformed E. coli cells compared to expression control E. coli cells (pET-C). The critical difference between the growth of transformed and control E. coli was found only at 600 mM NaCl, where the transformed E. coli cells showed a gradual increase in growth with maximum growth difference 4 h after salt stress. Therefore, above results implies that SaSRP3-1 contributed toward salt tolerance in E. coli at 600 mM NaCl and at low salt stress, up to 400 mM NaCl, intrinsic mechanism of E. coli cells supported normal growth, which was not observed at 600 mM NaCl and higher level of salt stress. However, the growth of both transformed and control cells ceased at NaCl concentrations beyond 600 mM. This scenario implies that SaSRP3-1 was contributing for salt tolerance at 600 mM, whereas higher salt concentration seems to be detrimental.
To validate how this above observation is translated into realistic growth pattern, the quantitative growth difference at 600 mM NaCl was measured using liquid growth curve assay, which is more accurate. The growth of bacterial cells in terms of optical density (OD 600 nm) was plotted at different time intervals to examine the relationship between protein expression and salt tolerance. Optical density (OD 600 nm) observations revealed that even under control condition, there were differences in the growth of E. coli cells transformed with expression vector control and cells transformed with SaSRP3-1 (Figure 1B). Moreover, the optical density (OD) of the cells transformed with SaSRP3-1 was more than the expression control and the magnitude of this effect increased with incubation time. This observation indicated that SaSRP3-1 protein has little influence on the growth of E. coli cells under control condition. But when these cells were grown under 600 mM NaCl and their OD600 were compared, there was a significant difference between the growth of E. coli cells transformed with SaSRP3-1 and cells transformed with expression control vector at several time points (Figure 1C). Moreover, cells transformed with SaSRP3-1 showed normal growth curve pattern at 600 mM NaCl, whereas E. coli cells transformed with expression control vector did not grow normally. We also observed faster growth of transformed E. coli cells immediately after approximately 1 h of salt exposure, which is the characteristic of a log phase of growth. Based on this E. coli expression results, it can be inferred that SaSRP3-1 plays an important role in the adaptation of transformed E. coli cells under salt stress.
Seven independent transgenic rice lines were obtained using the Agrobacterium-mediated transformation method, and the transformation efficiency was 29.2% (Figures 3A,B). Progenies from three independent transgenic plants were used for RT-PCR, and four independent transgenic plants were used for both Southern confirmation and phenotypic screening at different levels of salt stress. Transgenic rice lines generated after selection and regeneration were confirmed by PCR, RT-PCR, and Southern hybridization. As an initial step of molecular screening, T0 generation transgenic plants were confirmed by PCR amplification of SaSRP3-1 and hptII genes by gene specific primers. PCR analysis confirmed the presence of SaSRP3-1 and hptII genes in seven regenerated lines (Figures 3A,B). Subsequently, homozygous SaSRP3-1 plants were obtained by selfing and then crossed with SaVHAc1 transgenic plants to obtain pyramided plants carrying both genes (Figures 4A,B). The resulting two gene combination pyramided rice lines were confirmed by PCR amplification of both SaSRP3-1 and SaVHAc1 genes. Twenty-one plants were confirmed for the presence of both transgenes out of thirty-three progenies (Figures 4C,D). Further, southern hybridization results revealed the presence of multiple copies of SaSRP3-1 in the selected transgenic lines (Supplementary Figure S3). The RT-PCR analysis showed expression of SaSRP3-1 in the selected transgenic lines (Figure 3C). However, plant #A22 did not show amplification in RT-PCR despite being confirmed for the presence of SaSRP3-1 based on regular PCR. It implies that plant #A22 did not actively express the SaSRP3-1 transgenic gene. Post-transcriptional gene silencing influenced by various environmental factors could be the possible reason for non-detection of the transcript in RT-PCR.
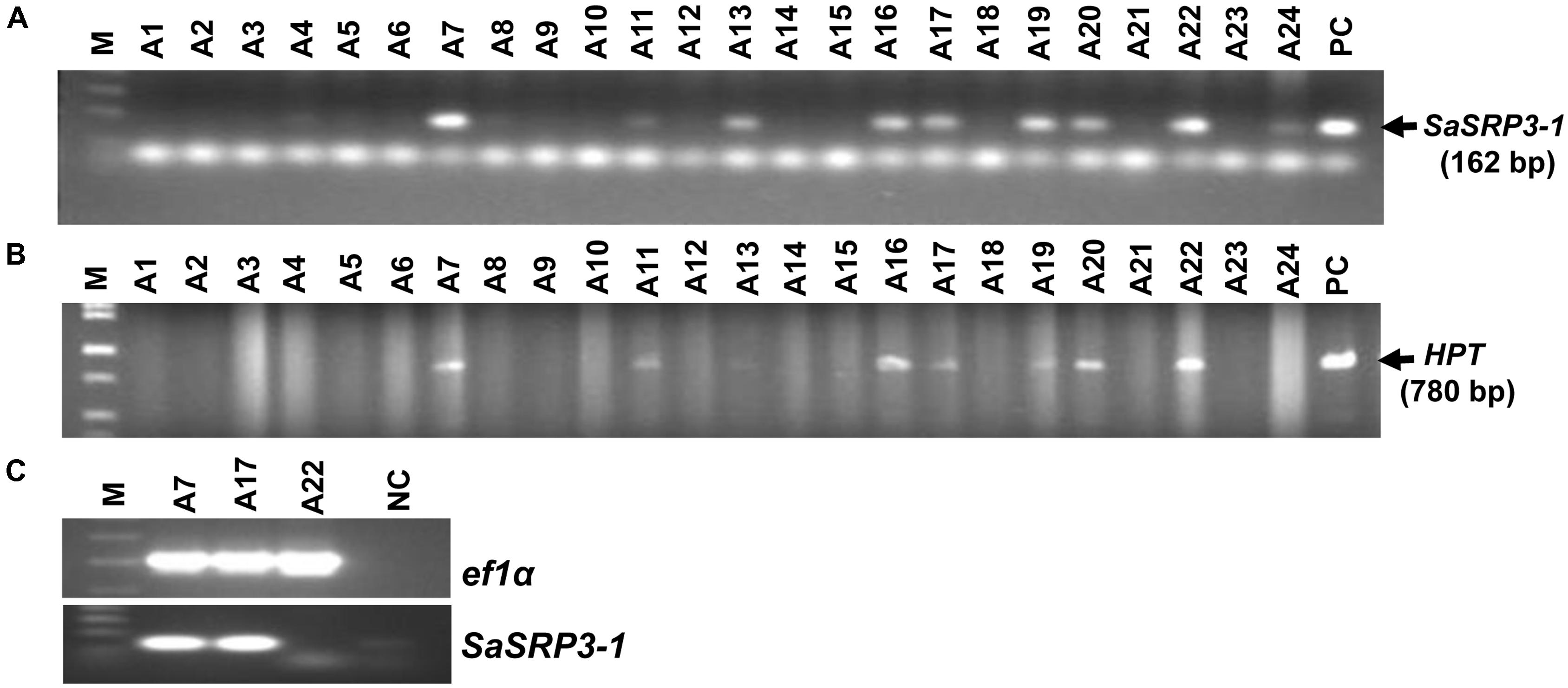
FIGURE 3. Confirmation of SaSRP3-1 containing T0 transgenic rice plants by PCR and detection of SaSRP3-1 mRNA by semi-quantitative RT-PCR analysis of transgenic seedlings grown in greenhouse condition. (A) PCR confirmation using SaSRP3-1 specific primers; (B) PCR confirmation of T0 plants using hygromycin phosphotransferase (hptII) specific primers. M: 1 Kb DNA ladder, A1 to A24: DNA isolated from individual T0 transgenic plants, PC: Positive control; (C) a representative RT-PCR experiment for detection of expression of elongation factor 1-α (ef1α) and SaSRP3-1 mRNA in seedling stage leaf samples of transgenic and Cocodrie (WT) plants. M: 1 Kb ladder, A7, A17, and A22: transgenic plants with SaSRP3-1 gene, NC: Negative control without any sample.
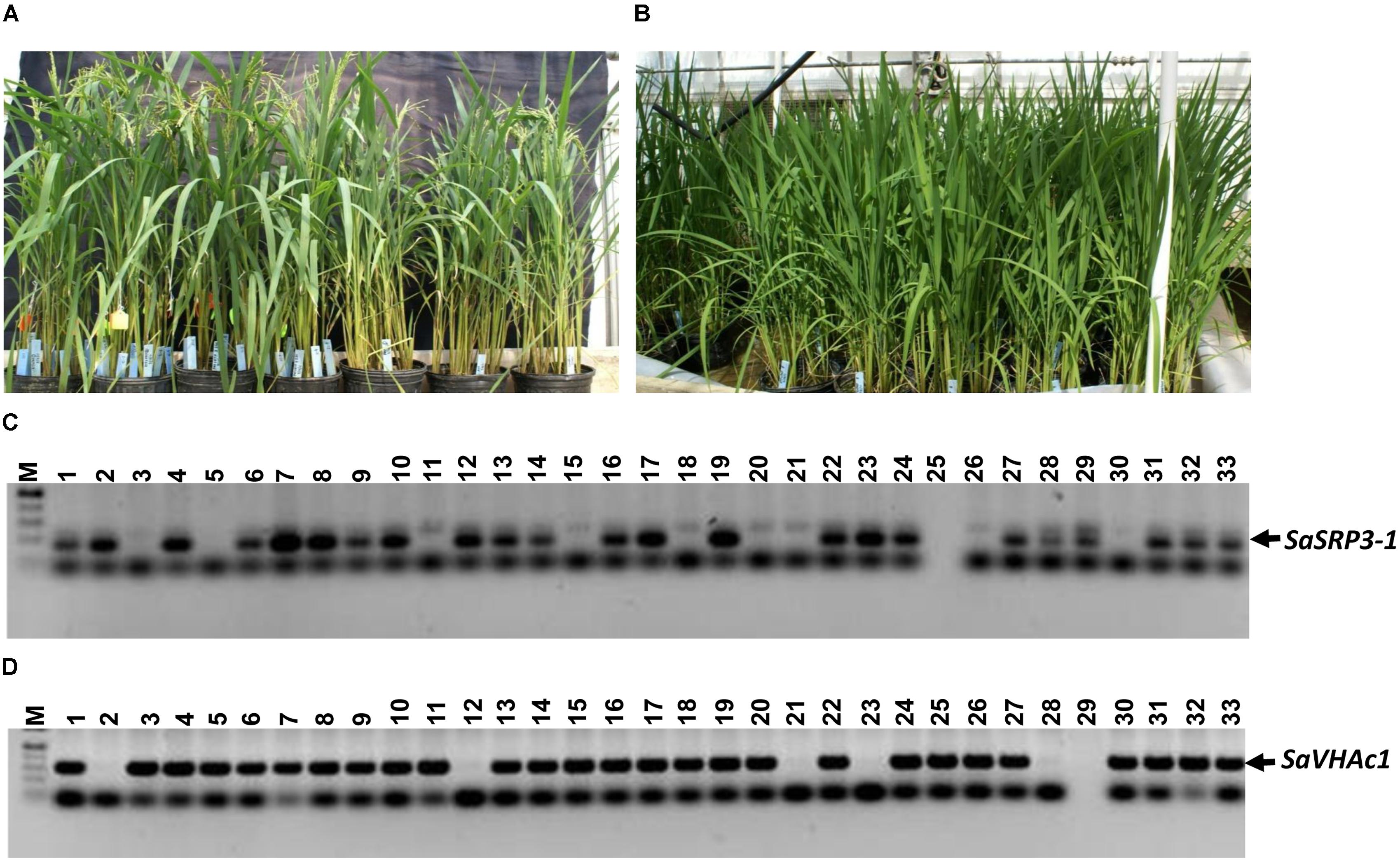
FIGURE 4. Transgenic plants grown in green house and PCR analysis to confirm the integration of SaSRP3-1 and SaVHAc1 genes in pyramided progenies. (A) Pyramiding of SaSRP3-1 (T1) and SaVHAc1 (T2) genes by crossing homozygous plants during flowering stage; (B) Growth of pyramided progenies with both genes; (C) PCR amplification showing the presence of SaSRP3-1 gene; (D) Pyramided transgenic rice plants showing the amplification of SaVHAc1 gene using gene specific primers.
Five-day old seedlings with uniform growth were transferred to hydroponic culture, and after 7 days of growth under the normal hydroponic condition, seedlings were transferred to different salt treatments (Supplementary Figure S4). Shoot and root lengths were compared after imposing different salt stress for 12 days. Notable changes in plant height were observed at 100 mM NaCl stress compared to the control condition (Figure 5A). All transgenic plants showed increased shoot growth compared to Cocodrie (WT) under 100 mM NaCl salt stress (Figure 5B). As expected, the growth of transgenic and WT seedlings was normal under control condition, and all plants had similar shoot length. But there was increase in the shoot length of transgenic plants compared to the WT under salt stress (Supplementary Table S3).
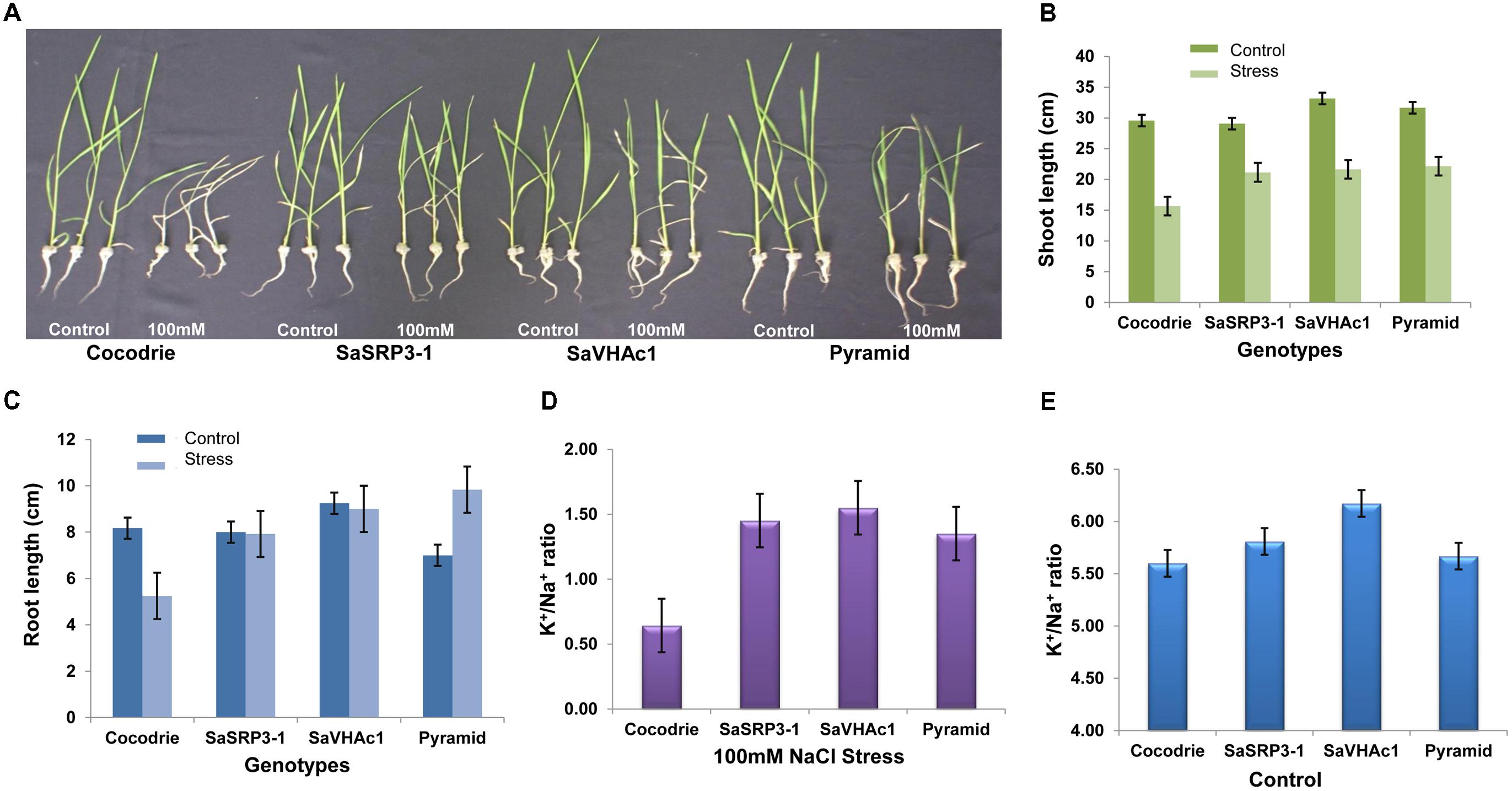
FIGURE 5. Salt tolerance evaluation of transgenic and wild type plants. (A) Responses of transgenic and wild type rice plants to 100 mM NaCl stress during seedling stage; (B) Comparison of shoot length; (C) Comparison of root length; (D) K+/Na+ ratio in shoot 12 days after exposure to 100 mM salt stress; (E) K+/Na+ ratio in shoot under control condition; Values represent means ± SD of five independent replicates. Error bars represent standard error.
The root length of Cocodrie plants ceased to grow at 100 mM NaCl compared to their root growth under the control condition. In contrast, 100 mM NaCl did not negatively affect the root growth in transgenic plants carrying either SaSRP3-1 or SaVHAc1 gene. We observed the non-significant effect on root growth pattern between control and 100 mM NaCl stress condition in single gene transgenic plants. However, the growth of root was significantly higher in pyramided transgenic plants carrying both SaSRP3-1 and SaVHAc1 genes at 100 mM NaCl salt stress than the control condition (Figure 5C). These results showed that the root growth was not affected by salt stress in transgenic plants with either single or two transgenes, which accounts for the relative ineffectiveness of salt stress on root growth of transgenic plants compared to WT plants (Supplementary Table S4). This normal growth of roots observed in transgenic plants is a clear indication of salt tolerance level of SaSRP3-1 and SaVHAc1 genes. Moreover, the higher root growth found in pyramided plants could be a complementary effect of these two genes. However, analyses of seedling growth at 150 mM salt stress did not generate any valuable data as plants showed severe wilting symptoms immediately after 2 days of salt stress.
Our results on Na+ and K+ content in shoot tissue indicated that SaSRP3-1 overexpressing transgenic plants had significantly higher K+/Na+ ratio than non-transformed plants under salt stress condition (Figure 5D). Likewise, the transgenic plants with SaVHAc1 and pyramided transgenic plants with both genes had significantly higher K+/Na+ ratio in shoot tissues than WT plants under 100 mM NaCl salt stress. The shoot tissue had more available K+ in all transgenic plants compared to control plants. In contrast, leaf K+ and Na+ ratio under control condition did not show any significant difference between single gene transgenic lines and WT. However, only SaVHAc1 transgenic plants had significantly higher leaf K+/Na+ ratio than others under control condition (Figure 5E).
The leaf disk assay is a sensitive and rapid technique for preliminary identification of salt tolerant plants. In the present investigation, it was used to assess the extent of chlorophyll retention under varying salt stress conditions. Based on visual observation, there was no visible difference between transgenic and WT plants under control condition (Supplementary Figure S5). However, transgenic plants over-expressing SaSRP3-1, SaVHAc1, and pyramided line with two genes retained more chlorophyll compared to Cocodrie (WT) at 100 mM and 150 mM NaCl salt stress (Figure 6A). This implies that loss of chlorophyll was less in transgenic plants under salt stress. Since this assay is still a qualitative approach, the actual quantity of total chlorophyll content in leaves was measured to confirm the normal functioning of photosynthetic activity at a physiological level. The total chlorophyll content of transgenic plants was least affected under salt stress compared with the control plants.
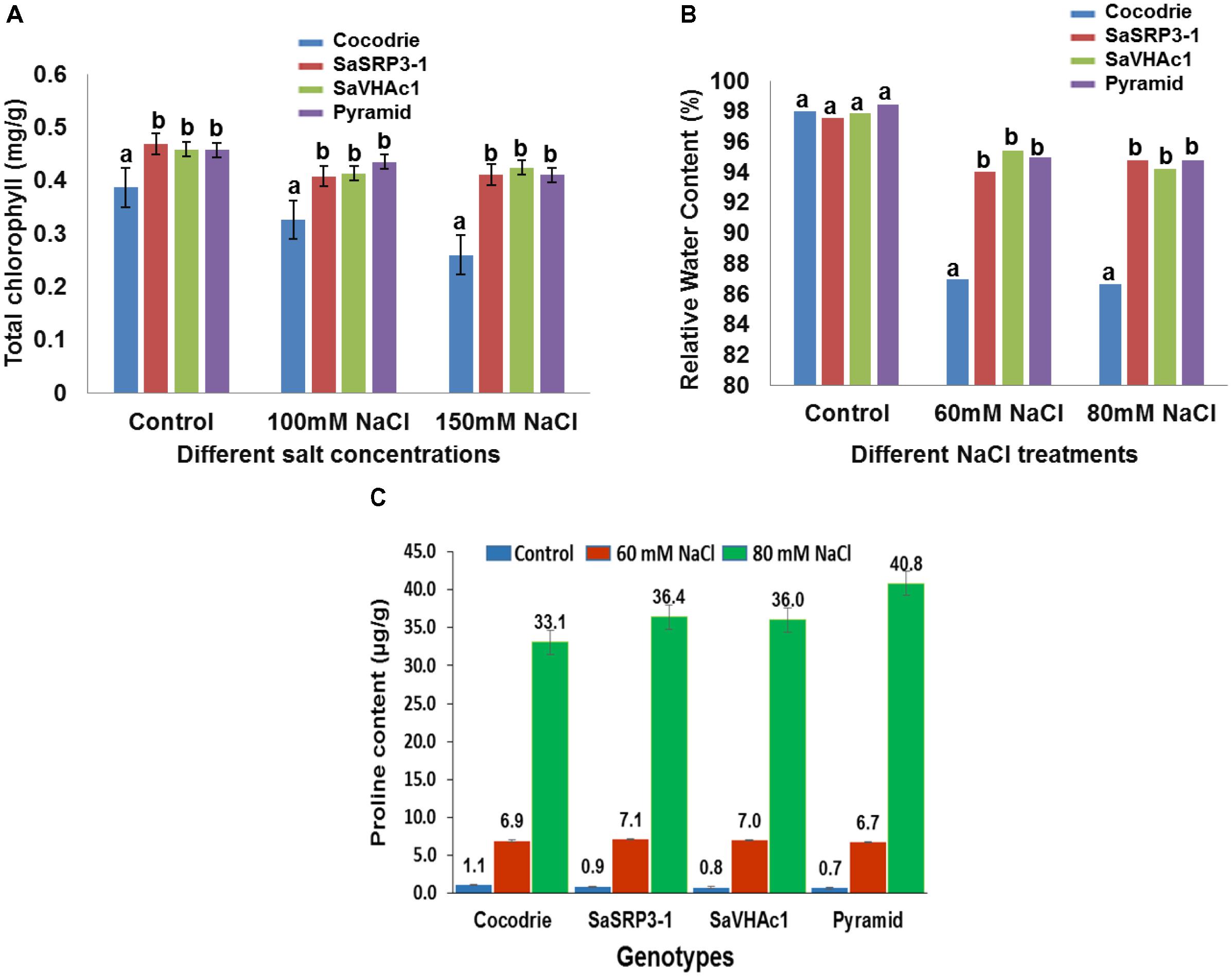
FIGURE 6. Physiological analysis of Cocodrie, single gene (SaSRP3-1 and SaVHAc1) and pyramided transgenic rice plants. (A) Total chlorophyll content in leaf samples from different transgenic and Cocodrie (WT) plants 4 days after salt stress (100 and 150 mM NaCl); (B) Comparison of relative water content (RWC) under control and salt stress (60 and 80 mM NaCl) conditions; (C) Proline accumulation in transgenic and WT plants under salt stress (60 and 80 mM NaCl) 10 days after salt stress. Error bars represent standard error. Different letters on bar diagram indicates statistical significance at 5% level based on Dunnett’s test.
Relative water content has proven to be a valuable index for measuring leaf turgidity, which is known to be affected by salt stress at initial stages of salt stress. There was clearly retention of more water in transgenic rice plants compared to Cocodrie (Figure 6B). There was no significant difference in RWC between transgenic and WT plants under control conditions. However, RWC was significantly higher in transgenic plants with single and two gene combinations at 60 and 80 mM NaCl compared to WT. These observations suggested that the transgenic plants maintained normal leaf turgidity and photosynthetic function under salt stress condition. Although we evaluated the plants at 60, 80, 100, and 150 mM NaCl conditions, critical difference was found only at 60 and 80 mM NaCl concentrations. As the higher salt stress of 100 and 150 mM NaCl treatments showed the severe loss in RWC in both WT and transgenic plants, the results were not presented.
Accumulation of proline, an organic compatible osmolyte, in response to salt stress was measured 10 days after exposure to NaCl stress using ninhydrin method. All plants showed an average of 7-fold increase in proline accumulation at 60 mM NaCl compared to the control condition, and there was no difference between Cocodrie and transgenic plants. Further, when salt stress was increased to 80 mM NaCl, there was 33-fold, 36-fold, and a 40-fold increase in free proline accumulation in leaf compared to the control condition in Cocodrie, single gene transgenic plants, and pyramided plants, respectively (Figure 6C). Transgenic plants accumulated more proline content at 80 mM NaCl stress than control plants. The single gene and pyramided transgenic plants accrued 9 and 21% more proline at 80mM salt stress compared to the control condition, respectively. But the increased accumulation was more in pyramided plants at 80 mM NaCl stress compared to their single gene transgenic and WT plants. The significant increase of proline accumulation in pyramided plants was most likely conferred by the presence of two salt tolerance genes.
The transgenic plants were evaluated at the reproductive stage to determine the effect of salt stress on traits influencing yield. The percent seed sterility was calculated to explore the effect of salt stress on grain filling. As expected, there was no significant difference in seed sterility among the genotypes under the control condition (Figure 7A). In contrast, the seed sterility was significantly lower in all the plants carrying single transgene as well as pyramided plants with two gene combination than Cocodrie (WT) plants under 60 mM salt stress condition. The results indicated that plants carrying single transgene as well as pyramided plants could enhance salt tolerance at the reproductive stage.
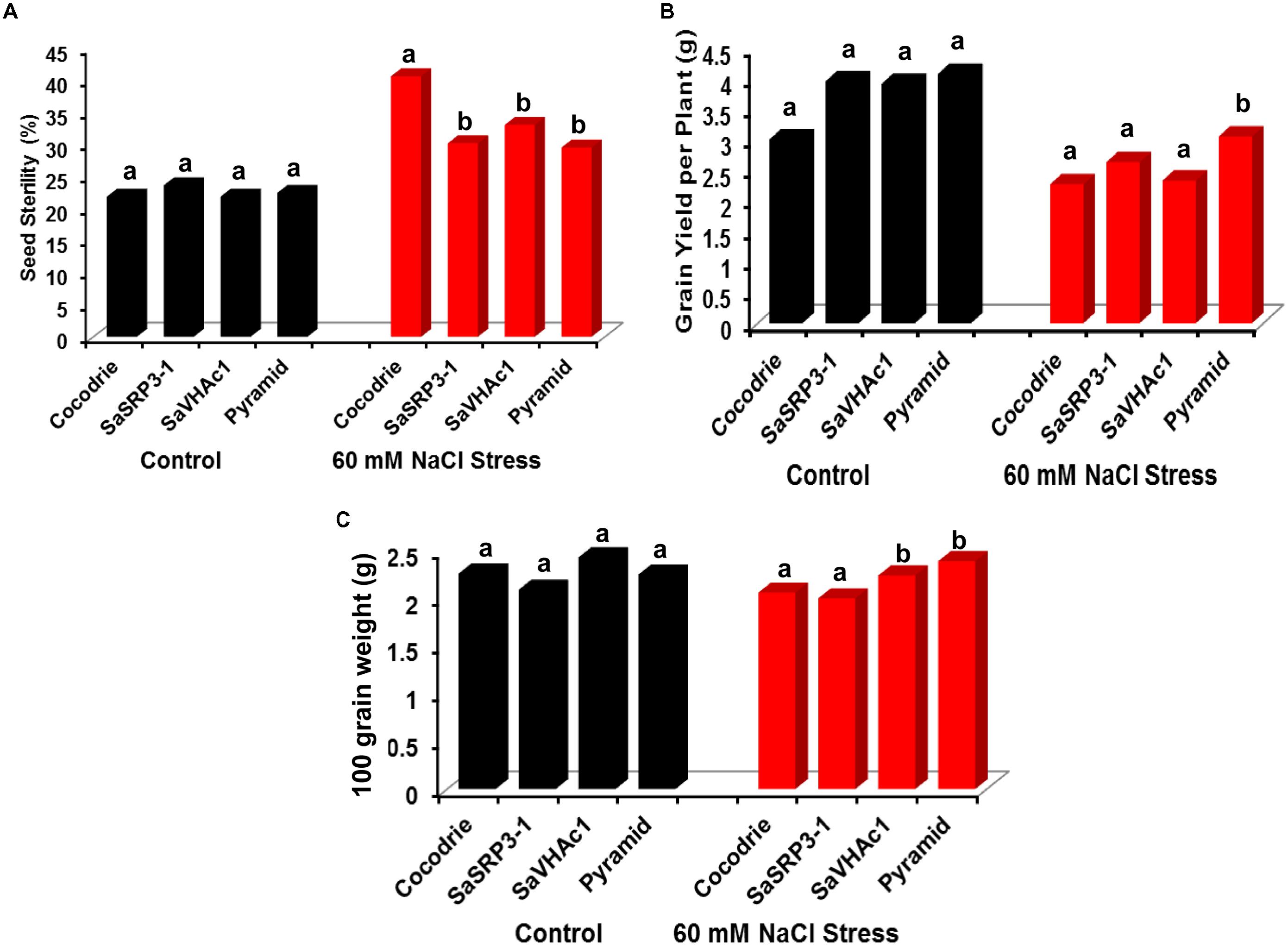
FIGURE 7. Analysis of yield parameters in transgenic and control plants under salt stress at the reproductive stage in greenhouse. (A) Impact of salt stress on grain filling and seed sterility; (B) comparison of grain yield per plant under salt stress; (C) variation in 100-grain weight due to salt stress. Different letters on bar diagram indicate the statistical significance at 5% level based on Dunnett’s test.
There was no significant difference in grain yield per plant between transgenic and WT plants under control conditions (Figure 7B). But the grain yield was affected in WT as well as transgenic plants with single genes under 60 mM NaCl stress. However, only pyramided transgenic plants showed significantly higher grain yield per plant than others. Further, we found no significant difference in 100-grain weight between transgenic lines and Cocodrie under control condition (Figure 7C). However, upon imposing 60 mM salt stress, rice plants transformed with SaVHAc1 and pyramided plants exhibited significantly higher 100-grain weight than WT and transgenic plants with SaSRP3-1.
Salt tolerance of plants is a complex trait due to the involvement of multiple genes associated with numerous mechanisms. The physiological responses to salinity depend on the phenology of the plants (Munns, 2002) and environmental influence (Flowers et al., 2000; Koyama et al., 2001; Lin et al., 2003). Although considerable efforts have been made to improve salt tolerance of field crops through conventional breeding as well as QTL approach using salt tolerant donors and wild relatives (Gregorio et al., 2002; Thomson et al., 2010), the progress in improving growth and yield parameters were not satisfactory under salt stress conditions. In this study, we demonstrated that overexpression of a halophytic gene SaSRP3-1 as well as its combination with SaVHAc1 from S. alterniflora L., in pyramided transgenic plants, could improve the salt tolerance at seedling, vegetative, and reproductive stages.
The amino acid sequence of SaSRP3-1 showed no homology to any of the functionally characterized protein in the NCBI non-redundant protein database. It has a hydrophobic transmembrane helix from 14th to 28th amino acid sequences. The presence of uncharacterized single transmembrane domain having a high homology of 94–95 percent to uncharacterized hydrophobic proteins (Supplementary Table S2) indicated that SaSRP3-1 is a novel salt-responsive protein. In addition, the presence of a transmembrane domain (TMD) based on the secondary protein structure suggests that SaSRP3-1 may be a subunit of big plasma membrane protein 3 (PMP 3) protein due to similarity of its TMD to the TMD2 of the PMP3. However, the exact function of the protein is not known. It is possible that SaSRP3-1 may be the product of alternative splicing regulated either by histone modifications or non-coding RNAs. Recent advances in the next generation sequencing technologies have revealed the prevalence of alternative splicing across eukaryotic genomes leading to the production of multiple distinct functional transcripts from a single gene (Luco and Misteli, 2011; Walters et al., 2013).
Functional characterization of genes in microbial model organisms such as E. coli and yeast has been widely used for screening salt-sensitive mutations (Yamada et al., 2003). The liquid growth curve assay of E. coli cells transformed with SaSRP3-1 showed that E. coli cells transformed with SaSRP3-1 grew normally at 600 mM NaCl concentration, while expression control cells ceased to grow (Figure 1C). Therefore, SaSRP3-1 may be involved in some cellular activity, which is crucial for survival under salt stress. This is the first line of evidence indicating the involvement of SaSRP3-1 gene in salt tolerance mechanism. Previous studies proved the suitability of a similar E. coli overexpression study to test salt tolerance of mangrove allene oxide cyclase (AOC) and characterized the salt tolerance role of AOC in E. coli, yeast, and tobacco cells (Yamada et al., 2002). Apart from overexpression, another advantage of E. coli system is the feasibility of protein purification and subsequent enzymatic characterization. Liu et al. (2011) overexpressed ascorbate peroxidase (APX) gene from Brassica napus in E. coli BL21 (DE3) and demonstrated that enzymatic assay conducted from purified protein increased the activity of APX decompose H2O2 produced during salt stress.
Since some physiological traits are adversely affected under salt stress, it is necessary to understand how overexpression of SaSRP3-1 influences vegetative growth and grain yield in rice. Transgenic lines selected in this study had multiple copies of SaSRP3-1. Interestingly, one out of three transgenic rice lines failed to show SaSRP3-1 expression, which might be due to SaSRP3-1 transgene silencing or epigenetic modification (Rajeevkumar et al., 2015). It was clearly demonstrated that the SaSRP3-1 and pyramided transgenic plants retained significantly higher chlorophyll content and RWC under salt stress (Figures 6A,B). Additional evidence for enhancing salt tolerance in transgenic rice was substantiated by the maintenance of higher K+/Na+ during salt stress at the vegetative stage (Figure 5D). In contrast, control plants showed low RWC, increased bleaching of chlorophyll as well as the low K+/Na+ in leaves. Based on the research findings in yeast (Navarre and Goffeau, 2000), tomato (Žižková et al., 2015), Arabidopsis (Mitsuya et al., 2005; Karan and Subudhi, 2012a), and rice (Song et al., 2011), the decrease in total chlorophyll content and low RWC in Cocodrie can be attributed to toxicity caused by accumulation of more Na+ in WT plants. Consequently, these results infer that higher chlorophyll and higher RWC observed in transgenic plants could be due to the prevention of excess Na+ accumulation in root and shoot, which in turn helps to maintain ion homeostasis and normal cellular functions.
Excess accumulation of Na+ was toxic to cellular activities, and it may directly affect the photosynthetic activity through the reduction in chlorophyll content at cellular level. The bleaching of chlorophyll under salt stress has been reported by many researchers (Shrivastava et al., 2010; Karan and Subudhi, 2012b; Žižková et al., 2015; Srivastava et al., 2016). In addition, improvement in seedling growth and high K+/Na+ has been demonstrated in rice plants overexpressing SaVHAc1 (Baisakh et al., 2012), OsNAC5 (Song et al., 2011) and Arabidopsis plants overexpressing of SaSce9 (Karan and Subudhi, 2012a). Likewise, the significant reduction of RWC in control plants suggests osmotic stress caused by salt stress, whereas transgenic plants showed tolerance to water loss during salt stress. The tolerance to water loss could be due to reduced stomatal openings in transgenic plants. Evidences for such higher RWC under salt stress has been reported in Arabidopsis plants overexpressing AVP1-1 and AVP1-2 (Gaxiola et al., 2001) and SaARF1 (Karan and Subudhi, 2014) and rice plants overexpressing SaVHAc1 (Baisakh et al., 2012) and transcription factor HYR (Ambavaram et al., 2014).
Proline accumulation has been one of the strategy to maintain the normal functioning of cells under different environmental stresses, such as drought, high salinity, and low temperature in plants (Sanchez et al., 2008; Usadel et al., 2008; Lugan et al., 2010; Karan and Subudhi, 2012b; Jaarsma et al., 2013). The results indicated a marked difference in the amount of proline accumulation between the transgenic and WT plants at 80 mM salt stress than the control condition. The magnitude of difference was more and distinct at 80 mM stress between transgenic and control plants than at 60 mM NaCl stress (Figure 6C) suggesting the role of proline as osmoprotectant during salt stress as well as in maintaining normal cellular functions (Karan and Subudhi, 2012b; Jaarsma et al., 2013). Furthermore, the higher proline accumulation by pyramided plants indicates that SaSRP3-1 and SaVHAc1 genes have a different mechanism of salt tolerance and together they are acting in a synergistic manner to increase the level of salt tolerance in pyramided plants. Alternatively, this result also implies the possibility of a decrease in protein synthesis and protein degradation during vegetative stage salt stress. The general increase in proline accumulation at 60 mM NaCl stress by all plants, including Cocodrie along with transgenic plants, supports the hypothesis that proline accumulation is an indication of the onset of salt stress as well as plants’ tolerance level.
Furthermore, both single and two-gene transgenic plants displayed significantly more shoot and root growth in response to salt stress during the early seedling stage. These transgenic plants not only survived from wilting and drying symptoms but also showed significantly enhanced shoot and root growth under salt stress (Figures 5B,C). The enhanced salt tolerance of pyramided lines could be attributed to V-ATPase, which is known to participate in the vacuolar Na+ compartmentation by regulating both vacuolar H+-ATPase (V-ATPase) and vacuolar H+-PPase (V-PPase) activity as demonstrated in the euhalophyte S. europaea (Lv et al., 2017). In contrast, we noticed a significant reduction in both shoot and root growth in wild type. The reduction in shoot and root length might be due to osmotic stress imposed by salt stress or due to the direct effect of salinity affecting the functioning of photosynthetic apparatus. Several earlier studies reporting significant reduction in vegetative growth and development observed under salt stress screening conditions in Arabidopsis (Karan and Subudhi, 2012b, 2014; Jia et al., 2015), rice (Jain et al., 2006; Shrivastava et al., 2010; Baisakh et al., 2012), maize (Henry et al., 2015), tomato (Žižková et al., 2015), and faba bean (Tavakkoli et al., 2010), were supportive of our findings.
Generally, salt stress during the reproductive stage negatively affect grain yield parameters. The WT plants showed 40 percent seed sterility, which was significantly higher than all transgenic plants under salt stress (Figure 7A). Such high seed sterility under abiotic stress conditions could be due to the extreme vulnerability of the pollen development stage during early reproductive and seed maturation stages (Dolferus et al., 2011). Similarly, differences were also observed for grain yield per plant and 100-grain weight. There was a significant increase in grain yield per plant and 100-grain weight only in transgenic pyramided plants than others (Figures 7B,C). The lower seed sterility and subsequent improvement in yield parameters observed in transgenic plants under salt stress implied possible mechanism of salt tolerance by preventing excess accumulation of Na+ to the developing panicle. The SaSRP3-1 plants had less seed sterility and more grain yield than SaVHAc1 plants and likewise SaVHAc1 plants showed higher 100-grain weight and root length compared to SaSRP3-1 plants. Interestingly, the results of salt stress revealed a decrease in seed sterility, increased root length, grain yield, and 100-grain weight in pyramided plants compared to the single transgenic plants. The probable explanation for this improved performance of pyramided plants under salt stress condition could be due to the complementary effect of SaSRP3-1 and SaVHAc1 genes. Thus, these results support the hypothesis that SaSRP3-1 and SaVHAc1 genes function synergistically to impart higher level of salt tolerance. The poor seed setting, low grain weight, and low grain yield observed in control rice plants compared to transgenic plants could be due to alteration of proteins associated with photosynthesis and carbon metabolism under salt stress (Li et al., 2015) and reduced carbon dioxide fixation to stomatal closure (Zhu, 2001; Wang et al., 2015).
In conclusion, our study demonstrated that SaSRP3-1 and SaVHAc1 genes function synergistically under salt stress and pyramiding of multiple genes involved in different salt tolerance mechanisms may be a viable approach to achieve higher level of salt tolerance in crop plants. The novel gene SaSRP3-1 was involved in preventing excess accumulation of Na+ and maintaining the structural integrity of plasma membrane under salt stress.
PS conceived and supervised the complete study and revised the manuscript. HB and RK conducted the experiments, analyzed the data, and wrote the manuscript. All authors read and approved the manuscript.
The work presented here was funded by the Economic Development Assistantship (EDA) program of the Graduate School of Louisiana State University, Baton Rouge, LA, United States.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Norimoto Murai, Louisiana State University, Baton Rouge, LA, United States for kindly sharing A. tumefaciens strain, EHA105. The manuscript is approved for publication by the Director of Louisiana Agricultural Experiment Station, United States as manuscript number 2018-306-32283.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01304/full#supplementary-material
cDNA, complementary DNA; EST, expressed sequence tag; IPTG, isopropyl β-D-1-thiogalactopyranoside; NCBI, national center for biotechnology information; ORF, open reading frame; PMP, plasma membrane protein; RWC, relative water content; RT-PCR, reverse transcription PCR; TMD, transmembrane domain; WT, wild type.
Ambavaram, M. M. R., Basu, S., Krishnan, A., Ramegowda, V., Batlang, U., Rahman, L., et al. (2014). Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 5:5302. doi: 10.1038/ncomms6302
Arnon, D. (1949). Copper enzymes in isolated chloroplasts, polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Baisakh, N., RamanaRao, M. V., Rajasekaran, K., Söbudhi, P., Janda, J., Galbraith, D., et al. (2012). Enhanced salt stress tolerance of rice plants expressing a vacuolar H+-ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora Löisel. Plant Biotechnol. J. 10, 453–464. doi: 10.1111/j.1467-7652.2012.00678.x
Baisakh, N., Subudhi, P. K., and Varadwaj, P. (2008). Primary responses to salt stress in a halophyte, smooth cordgrass (Spartina alterniflora Loisel.). Funct. Integr. Genomics 8, 287–300. doi: 10.1007/s10142-008-0075-x
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Broothaerts, W., Mitchell, H. J., Weir, B., Kaines, S., Smith, L. M. A., Yang, W., et al. (2005). Gene transfer to plants by diverse species of bacteria. Nature 433, 629–633. doi: 10.1038/nature03309
Dolferus, R., Ji, X., and Richards, R. A. (2011). Abiotic stress and control of grain number in cereals. Plant Sci. 181, 331–341. doi: 10.1016/j.plantsci.2011.05.015
Ellouzi, H., Hamed, K. B., Hernández, I., Cela, J., Müller, M., Magné, C., et al. (2014). A comparative study of the early osmotic, ionic, redox, and hormonal signaling response in leaves and roots of two halophytes and a glycophyte to salinity. Planta 240, 1299–1317. doi: 10.1007/s00425-014-2154-7
Flowers, T. J., Koyama, M. L., Flowers, S. A., Sudhakar, C., Singh, K. P., and Yeo, A. R. (2000). QTL: their place in engineering tolerance of rice to salinity. J. Exp. Bot. 51, 99–106. doi: 10.1093/jexbot/51.342.99
Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., Alper, S. L., et al. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc. Natl. Acad. Sci. U.S.A. 98, 11444–11449. doi: 10.1073/pnas.191389398
Gregorio, G. B., Senadhira, D., Mendoza, R. D., Manigbas, N. L., Roxas, J. P., and Cuerta, C. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
He, R., Yu, G., Han, X., Han, J., Li, W., Wang, B., et al. (2017). ThPP1 gene, encodes an inorganic pyrophosphatase in Thellungiella halophila, enhanced the tolerance of the transgenic rice to alkali stress. Plant Cell Rep. 36, 1929–1942. doi: 10.1007/s00299-017-2208-y
Henry, C., Bledsoe, S. W., Griffiths, C. A., Kollman, A., Paul, M. J., Sakr, S., et al. (2015). Differential role for trehalose metabolism in salt-stressed maize. Plant Physiol. 169, 1072–1089. doi: 10.1104/pp.15.00729
Hiei, Y., Komari, T., and Kubo, T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218. doi: 10.1023/A:1005847615493
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hoekema, A., Hirsch, P. R., Hooykaas, P. J. J., and Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Inada, M., Ueda, A., Shi, W., and Takabe, T. (2005). A stress-inducible plasma membrane protein 3 (AcPMP3) in a monocotyledonous halophyte, Aneurolepidium chinense, regulates cellular Na(+) and K(+) accumulation under salt stress. Planta 220, 395–402. doi: 10.1007/s00425-004-1358-7
Jaarsma, R., de Vries, R. S. M., and de Boer, A. H. (2013). Effect of salt stress on growth, Na+ accumulation and proline metabolism in potato (Solanum tuberosum) cultivars. PLoS One 8:e60183. doi: 10.1371/journal.pone.0060183
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa). BMC Plant Biol. 6:1. doi: 10.1186/1471-2229-6-1
Jia, F., Wang, C., Huang, J., Yang, G., Wu, C., and Zheng, C. (2015). SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J. Exp. Bot. 66, 4683–4697. doi: 10.1093/jxb/erv245
Kaddah, M. T. (1963). Salinity effects on growth of rice at the seedling and inflorescence stages of development. Soil Sci. 96, 105–111. doi: 10.1097/00010694-196308000-00006
Karan, R., and Subudhi, P. K. (2012a). A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol. 12:187. doi: 10.1186/1471-2229-12-187
Karan, R., and Subudhi, P. K. (2012b). Overexpression of a nascent polypeptide associated complex gene (SaβNAC) of Spartina alterniflora improves tolerance to salinity and drought in transgenic Arabidopsis. Biochem. Biophy. Res. Commun. 424, 747–752. doi: 10.1016/j.bbrc.2012.07.023
Karan, R., and Subudhi, P. K. (2014). Overexpression of an adenosine diphosphate-ribosylation factor gene from the halophytic grass Spartina alterniflora confers salinity and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 33, 373–384. doi: 10.1007/s00299-013-1537-8
Katz, A., Waridel, P., Shevchenko, A., and Pick, U. (2007). Salt-induced changes in the plasma membrane proteome of the halotolerant alga Dunaliella salina as revealed by blue native gel electrophoresis and Nano-LC-MS/MS Analysis. Mol. Cell. Proteomics 6, 1459–1472. doi: 10.1074/mcp.M700002-MCP200
Koyama, M. L., Levesley, A., Koebner, R. M. D., Flowers, T. J., and Yeo, A. R. (2001). Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol. 125, 406–422. doi: 10.1104/pp.125.1.406
Li, W., Zhao, F., Fang, W., Xie, D., Hou, J., Yang, X., et al. (2015). Identification of early salt stress responsive proteins in seedling roots of upland cotton (Gossypium hirsutum L.) employing iTRAQ-based proteomic technique. Front. Plant Sci. 6:732. doi: 10.3389/fpls.2015.00732
Lin, H. X., Zhu, M. Z., Yano, M., Gao, J. P., Liang, Z. W., Su, W. A., et al. (2003). QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 108, 253–260. doi: 10.1007/s00122-003-1421-y
Linscombe, S. D., Jodari, F., Bollich, P. K., Groth, D. E., White, L. M., Chu, Q. R., et al. (2000). Registration of ‘Cocodrie’ rice. Crop Sci. 40:294. doi: 10.2135/cropsci2000.0007rcv
Liu, Z. B., Wan, Q., Yue, Y. F., Yang, H. W., Wang, J. M., Li, X. F., et al. (2011). Expression in Escherichia coli of the gene encoding ascorbate peroxidase from Brassica napus enhances salt tolerance of bacterial cells. Russ. J. Plant Physiol. 58, 478–483. doi: 10.1134/S1021443711030113
Luco, R. F., and Misteli, T. (2011). More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 21, 366–372. doi: 10.1016/j.gde.2011.03.004
Lugan, R., Niogret, M. F., Leport, L., Guegan, J. P., Larher, F. R., Savoure, A., et al. (2010). Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J. 64, 215–229. doi: 10.1111/j.1365-313X.2010.04323.x
Lutts, S., Kinet, J. M., and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J. Exp. Bot. 46, 1843–1852. doi: 10.1093/jxb/46.12.1843
Lv, S., Jiang, P., Tai, F., Wang, D., Feng, J., Fan, P., et al. (2017). The V-ATPase subunit A is essential for salt tolerance through participating in vacuolar Na+ compartmentalization in Salicornia europaea. Planta 246, 1177–1187. doi: 10.1007/s00425-017-2762-0
Mitsuya, S., Taniguchi, M., Miyake, H., and Takabe, T. (2005). Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta 222, 1001–1009. doi: 10.1007/s00425-005-0043-9
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R., and James, R. A. (2003). Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253, 201–218. doi: 10.1023/A:1024553303144
Navarre, C., and Goffeau, A. (2000). Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 19, 2515–2524. doi: 10.1093/emboj/19.11.2515
Pearson, G. A., and Bernstein, L. (1959). Salinity effects at several growth stages of rice. Agron. J. 51, 654–657.
Rajeevkumar, S., Anunanthini, P., and Sathishkumar, R. (2015). Epigenetic silencing in transgenic plants. Front. Plant Sci. 6:693. doi: 10.3389/fpls.2015.00693
Sambrook, J., and Russell, R. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold spring harbor laboratory press.
Sanchez, D. H., Siahpoosh, M. R., Roessner, U., Udvardi, M., and Kopka, J. (2008). Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol. Plant 132, 209–219.
Shrivastava, V., Pekar, M., Grosser, E., Im, J., and Vigodner, M. (2010). SUMO proteins are involved in the stress response during spermatogenesis and are localized to DNA double-strand breaks in germ cells. Reproduction 139, 999–1010. doi: 10.1530/REP-09-0492
Singh, S., and Mittal, A. (2016). Transmembrane domain lengths serve as signatures of organismal complexity and viral transport mechanisms. Sci. Rep. 6:22352. doi: 10.1038/srep22352
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345. doi: 10.1007/s00425-011-1403-2
Srivastava, A. K., Zhang, C., Yates, G., Bailey, M., Brown, A., and Sadanandom, A. (2016). SUMO is a critical regulator of salt stress responses in rice. Plant Physiol. 170, 2378–2391. doi: 10.1104/pp.15.01530
Takahashi, Y., Tahara, M., Yamada, Y., Mitsudomi, Y., and Koga, K. (2017). Characterization of the polyamine biosynthetic pathways and salt stress response in Brachypodium distachyon. J. Plant Growth Regul. 37, 625–634. doi: 10.1007/s00344-017-9761-z
Tavakkoli, E., Rengasamy, P., and McDonald, G. K. (2010). High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 61, 4449–4459. doi: 10.1093/jxb/erq251
Thomson, M. J., Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3, 148–160. doi: 10.1007/s12284-010-9053-8
Toka, I., Planchais, S., Cabassa, C., Justin, A. M., De Vos, D., Richard, L., et al. (2010). Mutations in the hyperosmotic stress-responsive mitochondrial BASIC AMINO ACID CARRIER2 enhance proline accumulation in Arabidopsis. Plant Physiol. 152, 1851–1862. doi: 10.1104/pp.109.152371
Usadel, B., Blasing, O. E., Gibon, Y., Poree, F., Hohne, M., Gunter, M., et al. (2008). Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ. 31, 518–547. doi: 10.1111/j.1365-3040.2007.01763.x
Vasquez, E. A., Glenn, E. P., Guntenspergen, G. R., Brown, J. J., and Nelson, S. G. (2006). Salt tolerance and osmotic adjustment of Spartina alterniflora (Poaceae) and the invasive M haplotype of Phragmites australis (Poaceae) along a salinity gradient. Am. J. Bot. 93, 1784–1790. doi: 10.3732/ajb.93.12.1784
Walters, B., Lum, G., Sablok, G., and Min, X. J. (2013). Genome-wide landscape of alternative splicing events in Brachypodium distachyon. DNA Res. 20, 163–171. doi: 10.1093/dnares/dss041
Wang, J., Meng, Y., Li, B., Ma, X., Lai, Y., Si, E., et al. (2015). Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ. 38, 655–669. doi: 10.1111/pce.12428
Wu, H. J., Zhang, Z., Wang, J. Y., Oh, D. H., Dassanayake, M., Liu, B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224. doi: 10.1073/pnas.1209954109
Yamada, A., Saitoh, T., Mimura, T., and Ozeki, Y. (2002). Expression of mangrove allene oxide cyclase enhances salt tolerance in Escherichia coli, yeast, and tobacco cells. Plant Cell Physiol. 43, 903–910. doi: 10.1093/pcp/pcf108
Yamada, A., Tsutsumi, K., Tanimoto, S., and Ozeki, Y. (2003). Plant RelA/SpoT homolog confers salt tolerance in Escherichia coli and Saccharomyces cerevisiae. Plant Cell Physiol. 44, 3–9. doi: 10.1093/pcp/pcg001
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1976). Laboratory Manual for Physiological Studies of Rice, 3rd Edn. Manila: International Rice Research Institute.
Yu, J., Zao, W., He, Q., Kim, T. S., and Park, Y. J. (2017). Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage. Mol. Genet. Genom. 292, 1391–1403. doi: 10.1007/s00438-017-1354-9
Zhang, G. Y., Guo, Y., Chen, S. L., and Chen, S. Y. (1995). RFLP tagging of a salt tolerance gene in rice. Plant Sci. 110, 227–234. doi: 10.1016/0168-9452(95)04219-K
Zhao, F., Song, C. P., He, J., and Zhu, H. (2007). Polyamines improve K+/Na+ homeostasis in barley seedlings by regulating root ion channel activities. Plant Physiol. 145, 1061–1072. doi: 10.1104/pp.107.105882
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Keywords: abiotic stress, halophyte, Oryza sativa, salt responsive protein 3-1 gene, smooth cordgrass, transgene stacking, vacuolar H+-ATPase subunit c1 gene
Citation: Biradar H, Karan R and Subudhi PK (2018) Transgene Pyramiding of Salt Responsive Protein 3-1 (SaSRP3-1) and SaVHAc1 From Spartina alterniflora L. Enhances Salt Tolerance in Rice. Front. Plant Sci. 9:1304. doi: 10.3389/fpls.2018.01304
Received: 25 April 2018; Accepted: 17 August 2018;
Published: 12 September 2018.
Edited by:
Lam-Son Tran, RIKEN, JapanReviewed by:
Sitakanta Pattanaik, University of Kentucky, United StatesCopyright © 2018 Biradar, Karan and Subudhi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasanta K. Subudhi, cHN1YnVkaGlAYWdjZW50ZXIubHN1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.