
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 September 2018
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01303
The MYB transcription factors (TFs) is a plant TF families, which involves in hormone signal transduction, and abiotic stress tolerance, etc. However, there are few studies on the MYB TFs family and its regulatory mechanism in Tamarix hispida. In this study, 14 MYB genes (named ThMYB1 - ThMYB14) were cloned and characterized from T. hispida. The transcription profiles of ThMYBs in T. hispida under different abiotic stress conditions were monitored using qRT-PCR. Most of studied ThMYBs were significantly downregulated and/or upregulated by salt and osmotic stress, ABA, GA3 and JA treatments in at least one organ. Especially, ThMYB13 was induced in the leaves and roots of T. hispida when exposed to NaCl treatment at all study periods, indicating that it may involve in salt stress. To further study ThMYB13 function, ThMYB13 overexpression and knock-down plants and control plants transformed with an empty pROKII were obtained using a transient transformation system. Overexpression of ThMYB13 in T. hispida displayed the lowest O2-, H2O2 and MDA accumulation, minimal cell death, the most stable K+/Na+ ratio and the lowest electrolyte leakage rate among the three kinds of transient expression in T. hispida. Conversely, the RNAi-silencing, transiently transformed plants displayed the opposite physiological changes. Therefore, ThMYB13 might play a role in salt stress tolerance in transgenic T. hispida plants.
The MYB TFs is most abundant in plants (Peng et al., 2016), which contains the MYB domain serving as DNA binding. MYB TFs is classified according to the repeats present that varying from 1 to 4 in their sequences. Therefore, the MYB is divided with 4 groups, 1R-, R2R3-, 3R- and 4RMYB (Roy, 2016).
In previous studies, the MYB family genes, R2R3 MYBs, were involved in diverse processes, including cell cycle control, hormone signaling, secondary metabolism, meristem formation and cellular morphogenesis. Additionally, some MYB genes were found to regulate responses to abiotic stress (Cao et al., 2013). For, AtMYB96 induced pathogen resistance through the pathway of ABA signaling that regulating stomatal movement, increased tolerance to drought and disease (Seo et al., 2009; Seo and Park, 2010). Moreover, Arabidopsis AtMYB15 and AtMYB44, were found to involve in stomatal closure to improve drought tolerance (Jung et al., 2008; Ding et al., 2009). The silencing of GbMYB5 reduced antioxidant enzyme activities and proline content, leading to decreased the drought tolerance. Furthermore, overexpression of GbMYB5 in tobacco improved tolerance to drought accompanied with decreased water loss, elevated the proline level and ROS scanvenging activities; meanwhile, the expression of SOD, CAT, GST, SAMDC and ADC1 were significantly induced (Chen et al., 2015).
The MYB gene had been found to involve in plant salt stress responses. The plants overexpression of AtMYB20 increased salt tolerance, however, the plants repressing AtMYB20-SRDX showed decreased tolerance to salt stress (Cui et al., 2013). AtMYB73 could be induced by salt stress. The peak expression of AtMYB73 occurred at NaCl treatment for 6 h. In addition, AtMYB73 played a negative role in SOS induction in Arabidopsis (Kim et al., 2013). Arabidopsis plants expressing TaMYB3R1 produced more rosette leaves during the stage of vegetative growth; whereas they produced more inflorescences at the reproductive stage. The transformed lines showed improved tolerance to salt and drought treatments (Cai et al., 2015).
Additionally, some MYB TFs also participate in light, low-temperature, and osmotic stress induction responses. AtMYB18/LAF1 and AtMYB38 control hypocotyl elongation responding to far-red light (Yang et al., 2009) and blue (Hong et al., 2008) in seedlings, respectively. The OsMYB4 gene from rice enhanced frost tolerance and improved germination in transgenic barley plants under unfavorable conditions (Soltész et al., 2012). Arabidopsis expressing the apple MdMYB10 gene displayed enhanced tolerance to osmotic stress (Gao et al., 2011).
Tamarix hispida is a woody halophyte. This kind of perennial shrub or small tree is highly resistant to drought and soil salinity. This plant can absorb a high amount of salt from the soil and can accumulate it in the cells, with the transfer of salt achieved upon harvest. This resistance of T. hispida makes it an ideal material for cloning salt-tolerant genes. Therefore, in order to screen for genes with excellent resistance to abiotic tolerance, 14 T. hispida MYBs were cloned and their expression under salt or osmotic stress was analyzed by qRT-PCR. Further, ThMYB13 was selected for transiently transformed into T. hispida. The results showed that ThMYB13 could significantly improve salt tolerance of transgenic T. hispida, and it was an excellent salt tolerance gene. This investigate will provide new insights into the function of ThMYBs in tolerance to salt.
Tamarix hispida seeds were collected from Turpan Desert Botanical Garden of Chinese Academy Sciences (Xinjiang province, China) and were planted in a mixture of sand and turf peat (1:2, v/v) with the conditions of 14 h light/10 h darkness photocycle and 70–75% relative humidity under the temperature of 24°C. The seedlings (approximately 5–6 cm in height, 2-month old) with good growth and uniform size were used for stress treatment. The seedlings were irrigated every 24 h on roots with the solution of 20% (w/v) PEG6000, 0.4 M NaCl, 100 μM ABA, 50 μM GA3 or 100 μM JA solution and were harvested at 6, 12, 24, 48, and 72 h. Meanwhile, the T. hispida plants irrigated with fresh water were served as the controls. After these treatments, roots or leaves from about 20 seedlings were pooled together at each time point, and stored at -80°C.
Seven transcriptome libraries had been constructed with T. hispida plants with 2-month-old, including 4 root transcriptomes treated respectively with NaHCO3 for 0, 12, 24, and 48 h, and 3 leaf transcriptomes treated with NaHCO3 for 0, 12, and 24 h (Wang et al., 2014). The unigenes were analyzed using BLASTX program for searching the Swiss-Prot and NR databases. The phrase “MYB transcription factor (TFs)” was searched against the unigenes with functional annotation to identify MYB family genes. Then, the ThMYB genes with complete open reading frames (ORFs) were selected by ORF finder1. The Mol.Wt and theoretical pI of the proteins encoded by ThMYBs were studied by ProtParam2.
Arabidopsis MYB proteins were retrieved from TAIR database3. The MEGA5.0 software (Tamura et al., 2011) was used to systematically analyze the MYB proteins of Arabidopsis and T. hispida. Neighbour-joining (NJ) was employed to predict the phylogenetic tree, which was subjected to a Bootstrap method with 1000 replications (Tamura et al., 2011). Multiple sequence alignments was performed by Clustal X to search the conserved regions of the ThMYBs with the gap extension penalties and a gap open of 10 and 0.1, respectively (Thompson et al., 1997). ExPASy-PROSITE4 was used to predict the conserved domains of ThMYBs protein sequences.
Total RNA was extracted from T. hispida samples using an RNA extraction kit (BioTeke Corporation, Beijing, China). About 1 μg of total RNA was transcribed into cDNA. The synthesized cDNA was diluted to 10-fold with sterile water to act as the template for qRT-PCR. In addition, actin (FJ618517), α-tubulin (FJ618518), and β-tubulin (FJ618519) were used as the internal reference genes in T. hispida to normalize the templates in the PCR reactions. All primer sequences used were shown as Supplementary Table S1. qRT-PCR was conducted on an Opticon 2 system (Bio-Rad). The PCR reaction with a volume of 20 μl contains the followings: 2 μl of template cDNA, 0.5 μM of each reverse and forward primer, 10 μl of SYBR Green Real-time PCR Master Mix (Toyobo). PCR program was set as following conditions: 95°C for 30 s, followed by 45 cycles of 95°C for 15 s, 58°C for 15 s, 72°C for 30 s and 1 s at 80°C for reading plate. All the reactions were performed in triplicate. Expression levels were determined according to the 2-ΔΔCt (Livak and Schmittgen, 2001).
To verify the salt tolerance function of ThMYB genes, the overexpression and RNAi vectors of ThMYB13 were constructed. The CDS of ThMYB13 was inserted into pROKII driving by 35S CaMV promoter to overexpress ThMYB13 (35S:MYB). An inverted repeat truncated CDS of ThMYB13 with the length of 246 bp was inserted into pFGC5941 (Kerschen et al., 2004) at the two sides of the CHSA intron (pFGC:MYB) for silencing ThMYB13 expression. All primer used are shown in Supplementary Table S2. The genetic transformation of T. hispida plants was carried out following Ji et al. (2014). In brief, 40 day tissue cultures of T. hispida seedlings with similar sizes were used for genetic transformation. The seedlings were incubated in a solution for transformation [1/2 MS + 150 μM acetosyringone + 3% (w/v) sucrose + 0.01% (w/v) Tween 20 + 0.6 OD600 Agrobacterium tumefaciens, pH 5.6] shaking with 120 rpm at 25°C for 6 h. The seedlings were washed twice using distilled water and planted vertically on 1/2 MS solid medium. In total, 3 types of transiently transformed plantlets were cultured, including plants transiently transformed with 35S:MYB to overexpress ThMYB13 (OE), with pFGC:MYB for silencing ThMYB13 (RNAi) expression, or control (Con) plants transformed with the empty pROKII plasmid.
After culturing for 36 h on 1/2 MS agar medium, the seedlings from transient transgenic T. hispida (OE, Con and RNAi) were transferred to new 1/2 MS medium (as control) or 1/2 MS with 150 mM NaCl stress 2 h for biochemical staining, and then these seedlings were incubated with NBT, DAB or Evans blue solutions. The procedures for NBT and DAB staining were according to Zhang et al. (2011). Evans blue staining was performed as described by Yang et al. (2014). H2O2 was measured following the protocol of Dal Santo et al. (2012).
In addition, after culturing for 36 h on 1/2 MS solid medium, the plants were transferred to 1/2 MS medium with 150 mM NaCl for stress for 12, 24, or 36 h. The plants were harvested for qRT-PCR and physiological analyses. The expression of ThMYB13 in the transgenic plants were measured with qRT-PCR as described above. MDA measurements were conducted descried by Wang et al. (2010), while the electrolyte leakage measurement was conducted as described by Ji et al. (2014). The determination of the Na+ and K+ content was described in Chen et al. (2005) and Ma et al. (2011). All experiments were conducted with triplicate.
The data were analyzed with the Statistical Software Package for Social Science (SPSS) version 17.0. Using Student’s t-test to compare the data, if P < 0.05, the data were considered significantly different. ∗Indicates significant difference (∗P < 0.05).
A total of 238 unigenes were found to be the MYB family genes from the seven transcriptomes. BLASTX analysis was performed, and 14 unigenes of the ThMYBs with full ORF were finally obtained and searched in the NR protein database. In addition, these 14 ThMYB genes were named ThMYB1 to ThMYB14. The proteins of these 14 ThMYBs ranged from 284 to 554 amino acids residues, their pI were between 5.05 and 9. 66 and the Mol.Wt ranging from 30.6 to 61.8 kDa (Table 1).
The conserved domains of 14 ThMYB protein sequences were analyzed using ExPASy-PROSITE. The results showed that the structural domain of each ThMYB protein had at least one MYB conserved domain. To further analyze the similarities and characterization of the protein sequences of 14 ThMYB, their deduced amino acid sequences of were sequenced by Clustal X software. As shown in Supplementary Figure S1, 14 ThMYB protein sequences have a conserved MYB domain, and the conserved domains have high similarity. The N-terminus of ThMYB2, 3, 4, 8, 9, and 13 protein sequences contain two conserved MYB domains. The conserved domain of ThMYB12 exists at the end of the peptide chain. The conserved domains of ThMYB5, 6, 7, 10, and 11 were located in the middle of the peptide chain. ThMYB1 and ThMYB14 contain a conserved domain of MYB-type in the N-terminus.
Further analysis of the sequences of these conserved domains revealed that ThMYB2, 3, 4, 8, 9, and 13 all contained the R2R3 conserved domain (Supplementary Figure S2). It was suggested that these 6 ThMYB genes belong to R2R3-MYB family, in which the first tryptophan in the R3 domain of ThMYB8 is replaced by leucine (L) and the first tryptophan in the R3 domain of the other five proteins is replaced by phenylalanine (F). In addition, the peptide chains of ThMYB5, 6, 7, 10, and 11 contain a highly conserved domain of MYB-type HTH DNA in the middle (Supplementary Figure S3).
A NJ phylogenetic tree of 14 T. hispida ThMYB proteins and 125 Arabidopsis MYB proteins were constructed (Figure 1). The phylogenetic distribution indicated that MYB proteins can be classified into 13 groups, while the 14 ThMYB TFs were divided into 5 subgroups: class II, IV, V, VI, and X. Among them, class X was the largest subgroup and contained 8 ThMYB proteins, ThMYB1, 5, 6, 7, 10, 11, 12, and 14. All of these proteins contained only one conserved domain. The other ThMYB proteins (including ThMYB 2, 3, 4, 8, 9, and 13) that contained two conserved MYB domains were clustered into four individual clades.
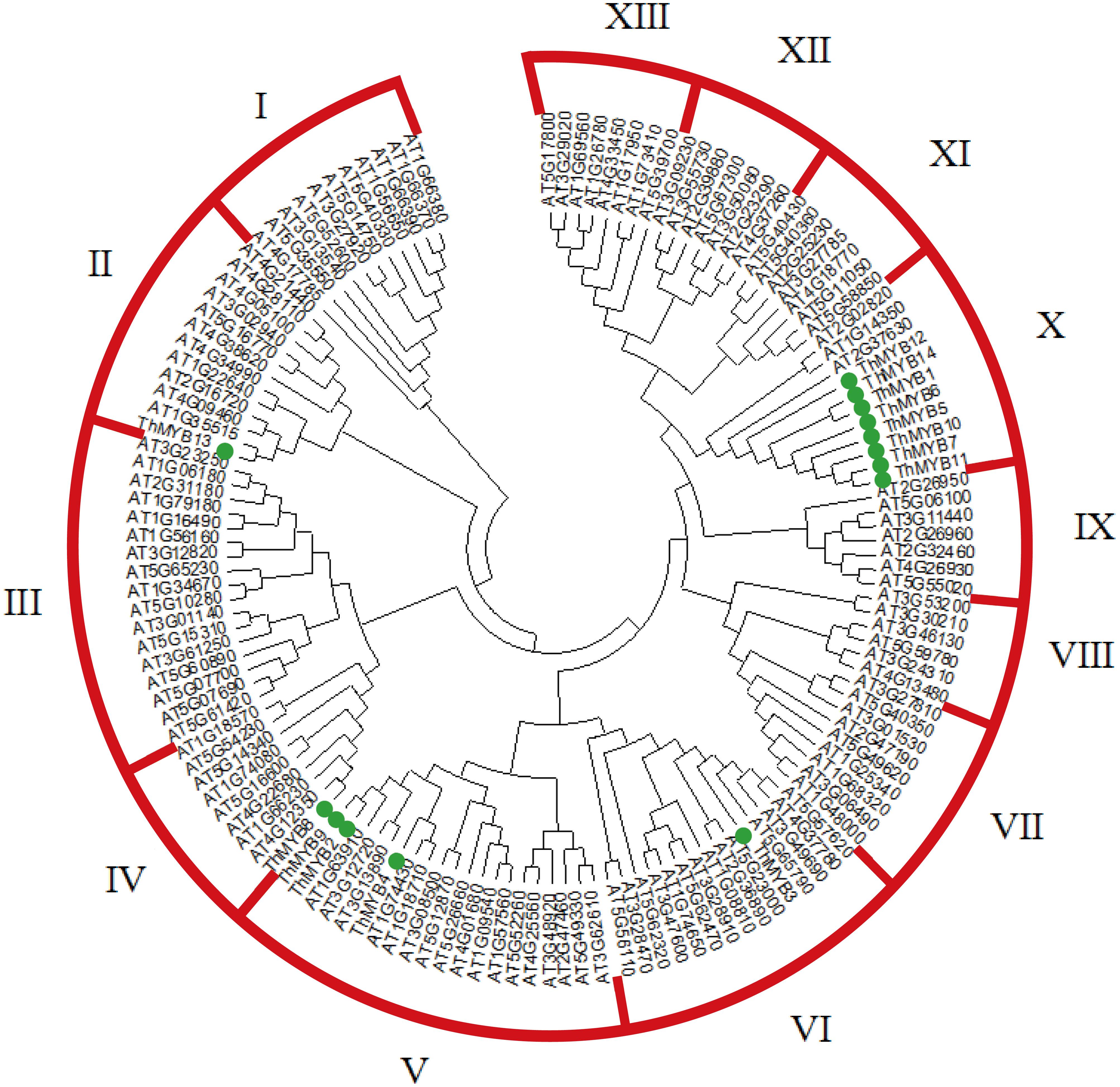
FIGURE 1. The phylogenetic tree of MYB proteins from T. hispida and Arabidopsis. In total, 14 MYB proteins in T. hispida and 125 in Arabidopsis were analyzed using Clustal X 2.0, and the neighbor-joining tree was constructed using MEGA 5.0. The bootstrap value was 1000 replicates.
To study the expression patterns of these 14 ThMYB genes in T. hispida leaves and roots without stress, their expressions were measured by qRT-PCR. The lowest transcript level genes (i.e., the highest delta Ct value) were set as 1 to normalize the transcript levels of the other ThMYBs (Figure 2). The relative expression levels of the 14 ThMYBs exhibited marked differences in leaves and roots. Interestingly, ThMYB9 was the least abundant in both the leaves and roots. However, ThMYB6 was the most abundant gene in leaves, and ThMYB12 was the most abundant gene in roots. The abundance of ThMYB6 was 2939 times higher than the lowest abundance (ThMYB9) in leaves, and the abundance of ThMYB12 was 2391 times greater than ThMYB9 in the roots, indicating that ThMYB6 in the leaves and ThMYB12 in the roots might play important functions than the other ThMYBs.
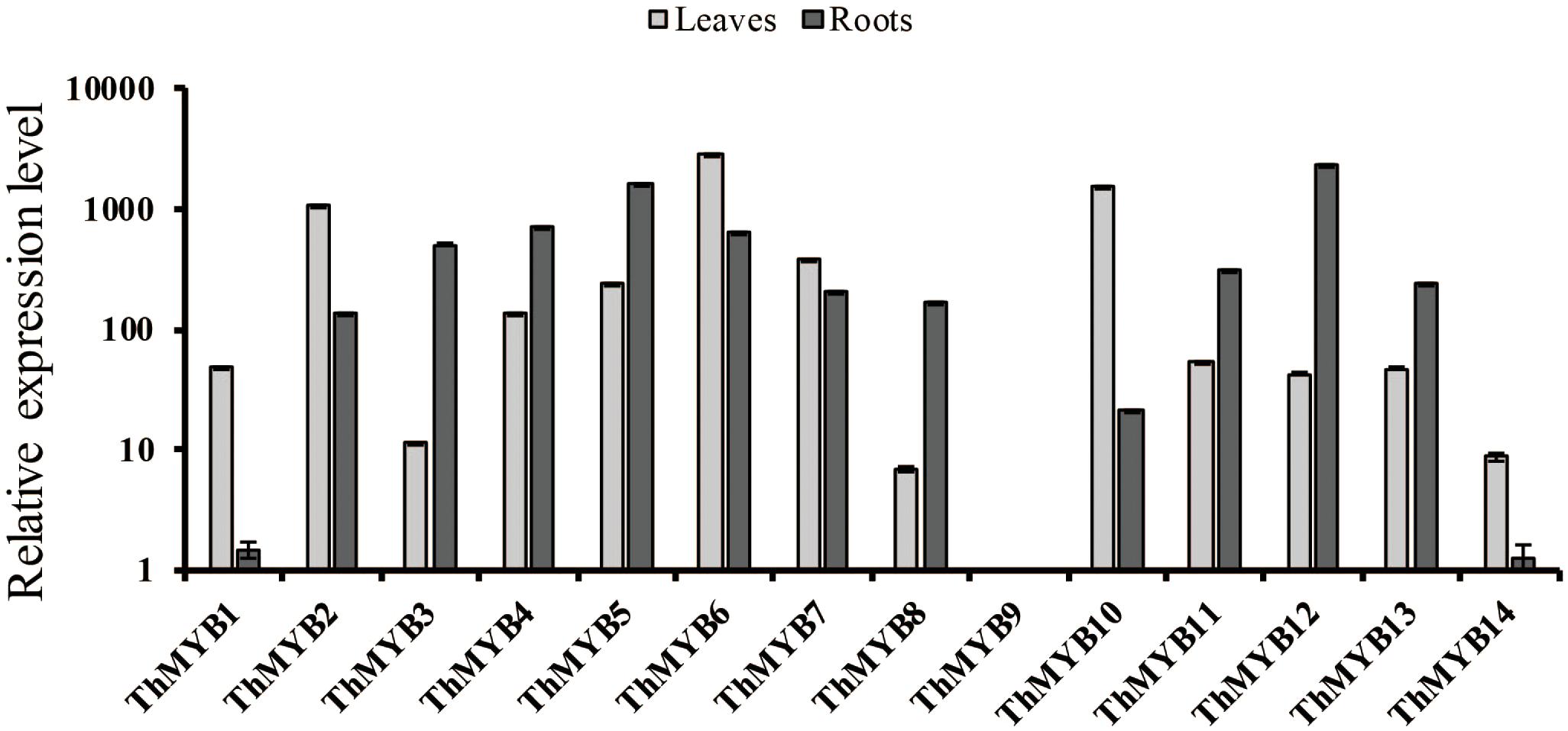
FIGURE 2. Relative expression levels of the 14 ThMYBs in leaf and root tissues of T. hispida under normal conditions. The ThMYB9 gene, which had the lowest expression levels (the highest delta Ct value) in both of the tissues of leaf and root, were assigned as 1. The expression levels of other ThMYB genes were plotted relative to the expression of ThMYB9. The y-axis shows the relative expression level of ThMYB genes, and the x-axis shows the ThMYB genes. The relative expression levels of different ThMYBs were log10-transformed.
To analyze the stress response function of ThMYBs, the expression profiles of 14 ThMYBs under different stress, including high salt, osmotic stress, and hormones treatments (ABA, GA3, JA) were analyzed by qRT-PCR.
Under NaCl stress, most of ThMYB genes had upregulated expression in the leaves. Especially, ThMYB1, 3, 4, 11, 13 and 14 had all induced expression at all stress points. The expression of ThMYB9 was significantly upregulated at most stress times (6, 24, and 48 h). The relative expression levels of ThMYB5 and ThMYB12 in leaves were upregulated at 12–72 h but downregulated rapidly at 6 h after salt stress. In contrast, ThMYB6 and ThMYB10 were induced at 6 h and inhibited with the other studied stress times. In the roots, the expression patterns of ThMYB1, 3, 4, 11, 13, and 14 were the same as those in leaves, and they were upregulated at all stress points. However, ThMYB6, 10 and ThMYB12 were significantly different from those in the leaves. The expressions of ThMYB6 and 10 were mainly upregulated at all stress points, and ThMYB12 was downregulated at all studied time points. In addition, the transcript of ThMYB8 was significantly downregulated at 6 h, and the lowest expression was only 1.65% of the control (Figure 3A).
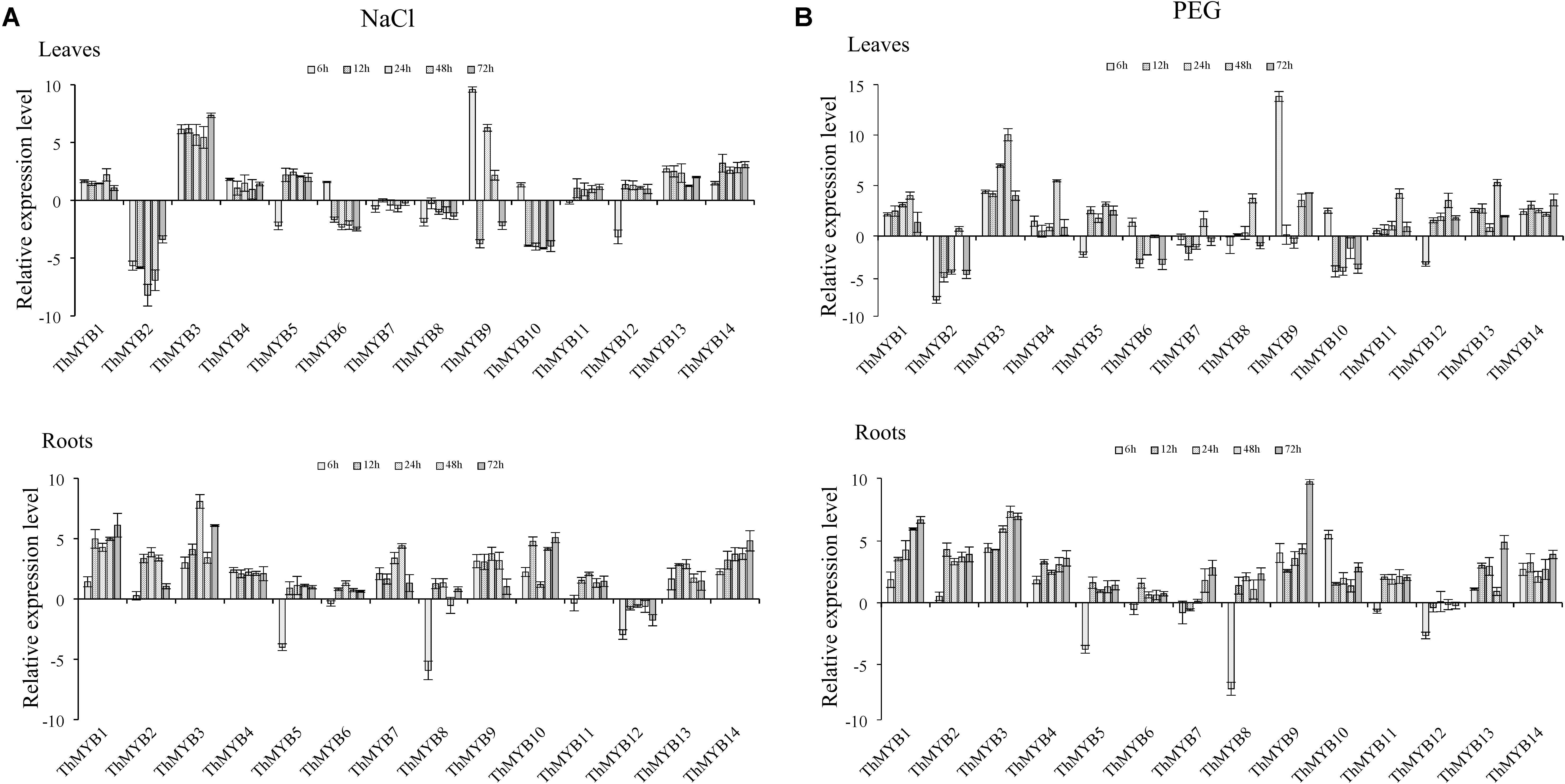
FIGURE 3. Expression analysis of the 14 ThMYBs responding to abiotic stresses (NaCl, PEG) in leaves and roots. (A) NaCl. (B) PEG. The relative expression level = transcription level under stress treatment/transcription level under control conditions. All relative transcription levels were log2-transformed. The error bars were obtained from multiple replicates of qRT-PCR.
Under PEG stress, the expressions of ThMYB1, 3, 13, and 14 were significantly upregulated at all stress points in the leaves. In addition, the most induced gene was ThMYB9, in which the peak expression level was 14,563-fold (6 h) that of the control. The relative expressions of ThMYB5 and ThMYB12 in leaves were also upregulated after stress at 12 h. In contrast, some ThMYB genes showed downregulated expression after PEG stress. Especially, the expression of ThMYB2 was significantly downregulated at most stress times (except 48 h), which was only 1.24% of the control at PEG stress at 6 h. Similarly, ThMYB6 and ThMYB10 were downregulated with most of the studied stress times (except 6 h). In the roots, almost all of the genes were significantly upregulated at all stress points, except for ThMYB5, 7, 8, and 12. Especially, with the prolonged stress time, the relative expression level of ThMYB1 gradually increased. The expressions of ThMYB5, 8, and 12 were significantly downregulated at 6 h. However, at 12–72 h, the relative expressions of ThMYB5 and 8 were upregulated, while the expression of ThMYB12 did not change significantly (Figure 3B).
Under ABA treatment, the transcripts of ThMYB11 were not significantly differentially regulated at all stress points in the leaves. However, most the ThMYB genes expressed were significantly different after ABA stress. Especially, ThMYB1, 3, and 14 showed significantly upregulated expression at all stress points, while ThMYB2, 7, and 8 were downregulated at all studied stress times. The relative expression of ThMYB5 and ThMYB12 in leaves was upregulated at 12-72 h but downregulated rapidly at 6 h after ABA stress. In contrast, ThMYB6 and ThMYB10 were induced at 6 h and inhibited with the other studied stress times. In the roots, the transcripts of all ThMYB were classified into three groups. One group was included by ThMYB1, 3, 4, 6, 9, 10, 13, and 14 and was clearly upregulated at all stress points. The second ThMYB group composed of ThMYB2, 5, 7, 8, and 11 was upregulated at 12–72 h but downregulated at 6 h after treatment of ABA. The expression patterns of ThMYB12 were different from those of the two groups, which was down regulated at all stress time points (Figure 4A).
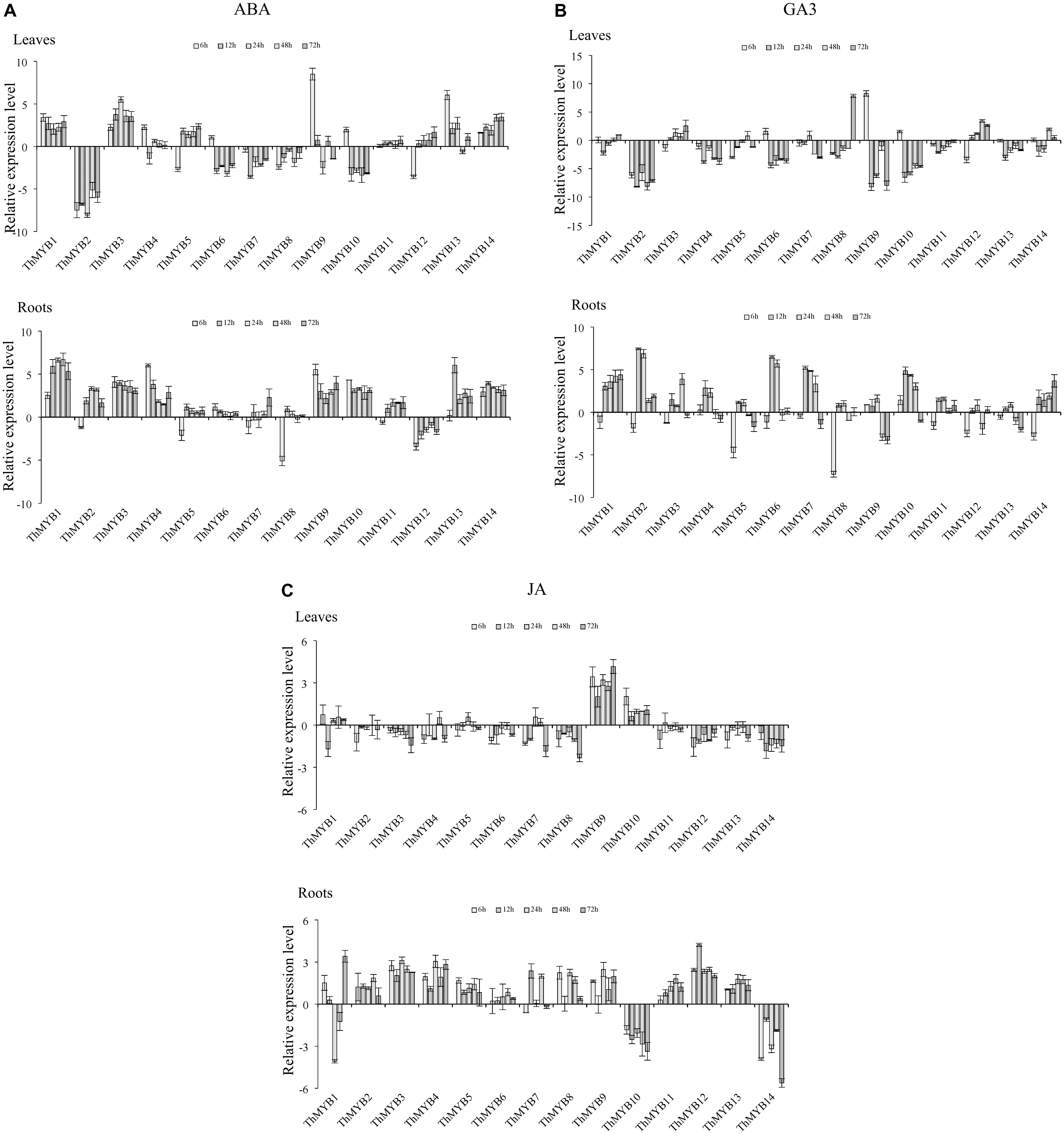
FIGURE 4. Expression analysis of the 14 ThMYBs responding to hormones treatments (ABA, GA3, JA) in leaves and roots. (A) ABA. (B) GA3. (C) JA. The relative expression level = transcription level under stress treatment/transcription level under control conditions. All relative transcription levels were log2-transformed. The error bars were obtained from multiple replicates of the qRT-PCR.
Under GA3 treatment, most of ThMYB genes had downregulated expression after GA3 stress in the leaves. Especially, ThMYB2, 4, 11, and 13 were downregulated at all stress points. In addition, the relative expression of ThMYB6, 9 and 10 was significantly downregulated at most stress times (12–72 h). In contrast, ThMYB3 and ThMYB12 were mainly upregulated at most of the studied stress times. In the roots, most of the ThMYB genes expressed were significantly inhibited at 6 h and induced at the other studied stress time points. ThMYB10 was significantly different than the other ThMYBs, which was significantly upregulated at the early stress times (6–48 h) and downregulated rapidly at 72 h after GA3 treatment (Figure 4B).
Under JA stress, except ThMYB1, 9, and 10, almost all of the ThMYB genes transcribed showed downregulation at all stress points in the leaves. In addition, ThMYB9 was highly upregulated at all of the studied stress times. The relative expression reached its peak level at 72 h (17.8-fold higher than the control). Interestingly, most of the ThMYBs expressed (except ThMYB9 and 14) showed distinct expression patterns between the leaves and roots. In the roots, except for ThMYB1, ThMYB10 and ThMYB14, the other genes showed significant upregulation at all stress points. However, ThMYB10 was significantly downregulated at all stress points (Figure 4C).
In general, the expression of 14 ThMYBs in leaf and root tissues were changed at least at one stress time point, which indicated that these ThMYBs might be one or several stress response genes, which might be involved in the stress tolerance of T. hispida. Especially, ThMYB2, 6, and 7 were mainly downregulated in leaves and upregulated in roots under five stress conditions. However, ThMYB1, 3, 13, and 14 genes were mainly upregulated in both leaves and roots in response to NaCl, ABA and PEG treatment conditions, suggesting that they might play an important role in stress tolerance. Therefore, the ThMYB13 gene was selected for further study of stress resistance.
To determine whether transient overexpression and suppression of the ThMYB13 gene in T. hispida were successful, the expression of ThMYB13 in Con (transformed with empty pROKII), OE and RNAi plants were investigated by qRT-PCR. The transcripts of ThMYB13 in OE or RNAi plants was normalized by the expression of Con plants at 0 h. The results suggested that the transcript of ThMYB13 in the OE plants was the highest among the three types of transiently transformed seedlings. At 36 h, the expression of ThMYB13 in OE was 33.94 times that of Con. While the expression of ThMYB13 in RNAi plants was significantly lower than those in Con, it was only 5.37% of control at 36 h (Figure 5). These results indicated that we successfully obtained transient overexpression and inhibition of ThMYB13 plants.
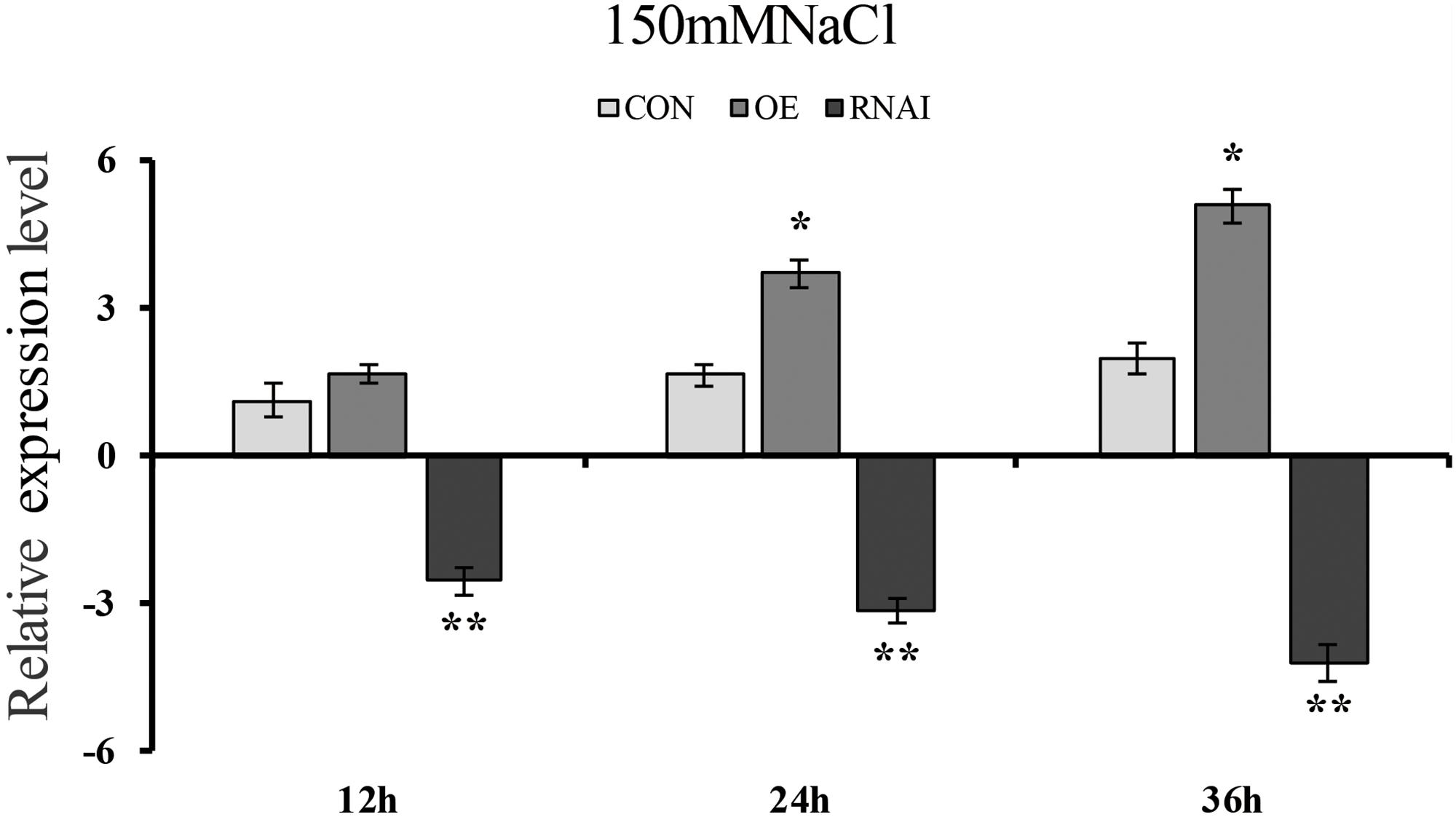
FIGURE 5. ThMYB13 transcript levels in T. hispida plants with transient overexpression or knockdown of ThMYB13. The expression data were log2 transformed. Two-month-old T. hispida plants were transiently transformed with empty pROKII, 35S:MYB or pFGC:MYB. After transformation for 36 h, T. hispida plants were treated with 150 mM NaCl for 12, 24, or 36 h, and the expression of ThMYB13 was determined. Con, plants transformed with empty pROKII; OE, plants transformed with 35S:MYB for overexpression of ThMYB13; RNAi, plants transformed with pFGC:MYB for silence the expression of ThMYB13. ∗Represents a significant difference (P < 0.05). ∗∗Represents a very significant difference (0.01 < P < 0.05).
To preliminarily identify the function of ThMYB13 gene, we analyzed and compared the biochemical staining and related physiological indexes of three kinds of transient transformed T. hispida. DAB and NBT in situ staining were carried out to study H2O2 and O2- accumulation, respectively. In the Con, OE and RNAi plants. Oxygen ions released by H2O2 in cells can oxidize DAB to form brown precipitates, and the amount of H2O2 released from cells can be displayed according to the depth of staining. Similarly, NBT can be oxidized by superoxide ion O2- to blue formazan, which can reflect the content of O2- through the blue shade of formazan. The results of both NBT and DAB staining showed that the OE plants showed greatly reduced levels while RNAi plants accumulated high levels compared with Con plants (Figures 6A,B). The content of H2O2 was further determined. The results showed that the content of H2O2 in the three types of plants was significantly different after 24 h of salt stress, and the level of H2O2 in RNAi plants was the highest, which was 1.22 times that of Con plants. While H2O2 content in the OE plants was the lowest, it was only 82.41% of the content of Con plants (Figure 6D). These results indicated that in the OE plants, ROS was highly reduced, whereas in RNAi plants, ROS was accumulated highly under salt conditions. Increasing the expression of ThMYB13 can improve ROS clearance.
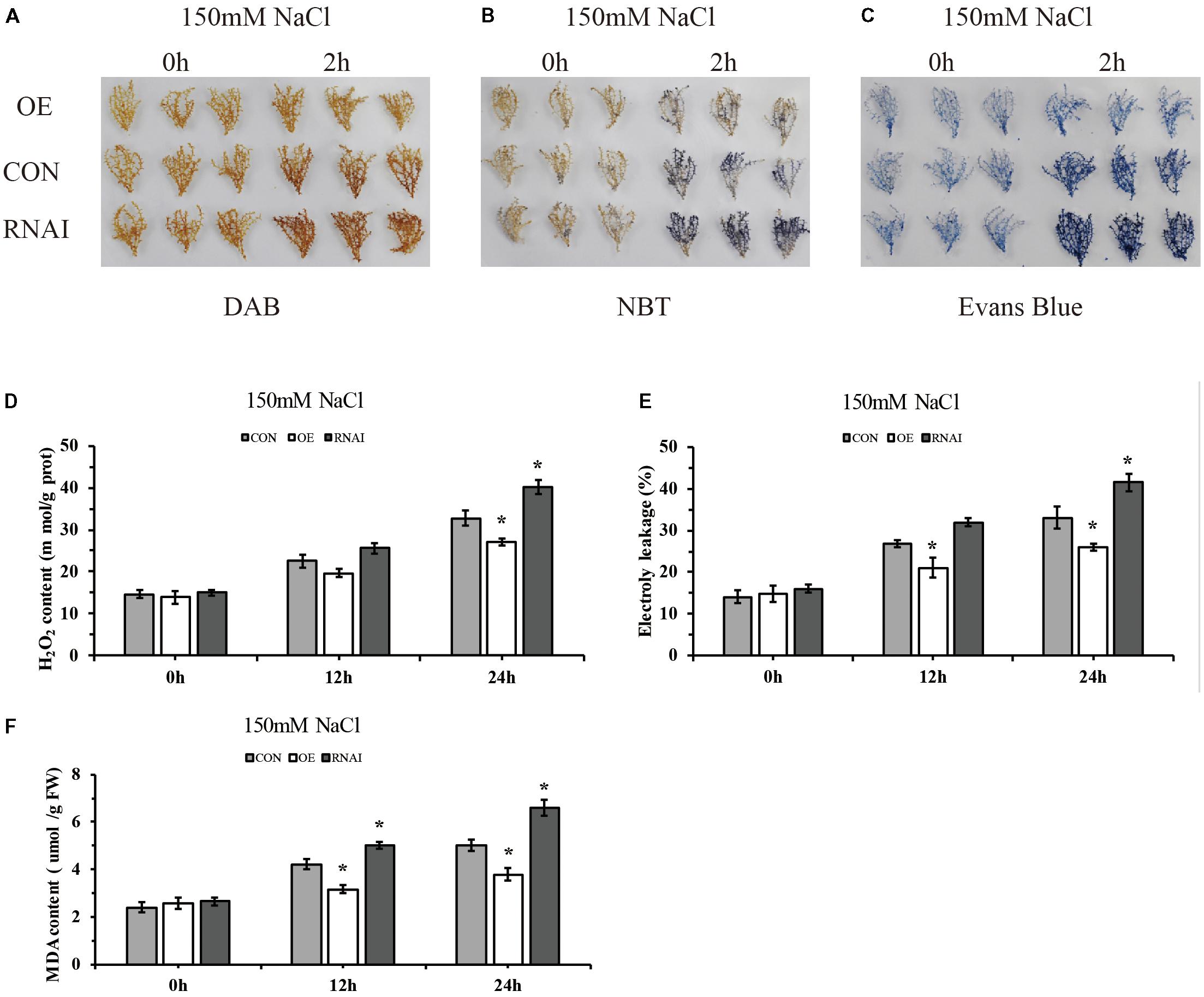
FIGURE 6. Histochemical staining and related physiological changes analyses of transformed T. hispida. (A,B) The plants were stained with DAB (A) and NBT (B) to reveal the accumulation of O2- and H2O2, respectively. (C) Analysis of cell death by Evans blue staining. (D) Determination of H2O2 contents. (E) Analysis of cell death by measurement of electrolyte leakage. (F) MDA content analysis of Con, OE and RNAi plants. The experiments were conducted with three independent biological replications. ∗The significant difference (t-test, P < 0.05) compared with Con plants.
To monitor the level of cell death, electrolyte leakage and evans blue staining were analyzed. Evans blue in situ staining indicated that in OE plants, cell death was reduced, but it was increased in that of RNAi plants when compared with Con plants exposed to salt stress (Figure 6C). Electrolyte leakage results confirmed these results. The relative electrical conductivities of RNAi plants were the highest at 24 h, 1.26 times that of Con plants, meanwhile those of OE were 0.78 folds those of Con plants (Figure 6E). At the same time, there was no difference among the three types of plants in MDA level under normal conditions. Under salt conditions, OE plants showed significantly lower MDA level than the Con plants, and the content of MDA in RNAi plants was 1.32 folds higher than MDA in the Con plants at 24 h after salt stress (Figure 6F). These results indicated that membrane lipid peroxidation was highly increased in RNAi plants, but was decreased in OE plants.
We further determined the contents of K+ and Na+ in transformed plant roots and leaves. Under normal conditions, the sodium and potassium ion contents in the leaves and roots of three types of plants were not significantly different. Under salt conditions, all three types of plants showed higher Na+ content in roots than leaves. However, the RNAi plants accumulated higher Na+ content than the OE and Con plants, and the OE plants showed significantly lower accumulation of Na+ than Con plants in leaves and roots (Figures 7A,B). Conversely, all three types of plants showed lower K+ content in roots than leaves under salt stress conditions, the RNAi plants still had the lowest K+ content, whereas the OE plants showed the highest K+ contents in the leaves or roots, followed by that in the Con plants (Figures 7C,D). Consistently, all three types of plants displayed a higher K+/Na+ ratio in leaves than roots. At the same time, the OE plants showed the highest K+/Na+ ratio in both roots and leaves, followed by the Con plants, with the RNAi plants having the lowest K+/Na+ ratio. After salt stress for 24 h, the ratio of K+/Na+ in OE plants in leaves and roots was 1.48 and 2.13 times higher than that in Con plants, while the K+/Na+ ratio of RNAi plants was only 45.67 and 41.81% of Con plants (Figures 7E,F).
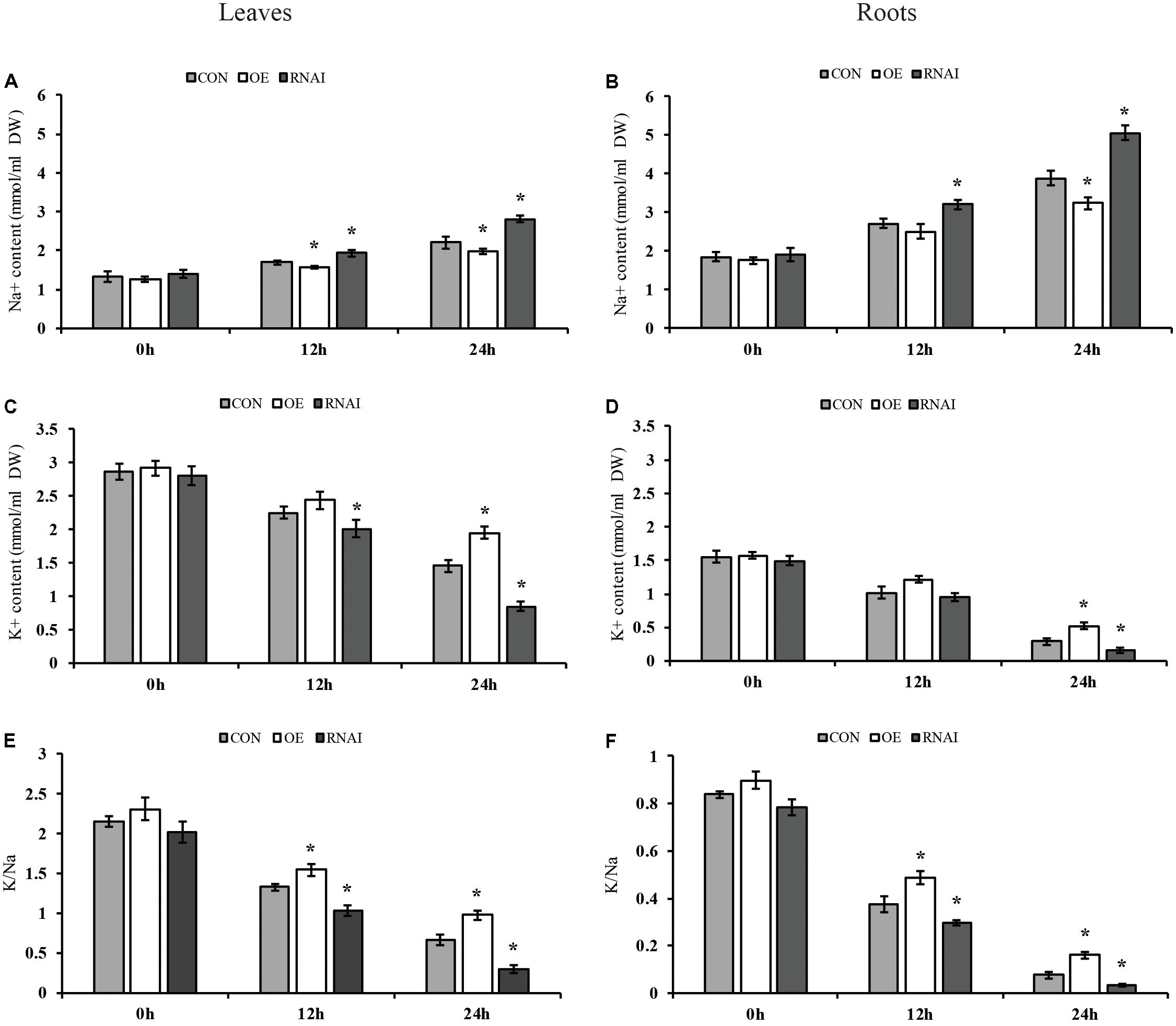
FIGURE 7. Analysis of Na+ and K+ content. Three transiently transfected plants were treated with 150 mM NaCl for 12, 24 or 36 h, in which ThMYB13 was overexpressed at 35S: MYB (OE), the expression of ThMYB13 was silenced with pFGC: ThMYB13 (RNAi), or the empty pROKII plasmid was used as a control. (A,B) Na+ content in leaves (A) and roots (B). (C,D) K+ content in leaves (C) and roots (D). (E,F) K+/Na+ ratio in leaves (E) and roots (F); ∗indicates significant differences between OE plants and Con, or between Con and the RNAi plants under the same conditions (P < 0.05).
The MYB family plays an important role in plants. The MYBs function was systematically researched in Arabidopsis (Dubos et al., 2010; Mondal and Roy, 2017), Setaria italica (Muthamilarasan et al., 2014), Vitis vinifera (Wong et al., 2016), Zea mays (Du et al., 2012), Populus trichocarpa (Wilkins et al., 2009), Gossypium raimondii (He et al., 2016) and other plants, especially Arabidopsis and Oryza sativa (Katiyar et al., 2012). However, these plants are found mostly in sweet soils, and there are few studies on the ThMYB TF family and its regulatory mechanism in T. hispida.
In a recent study, a TaODORANT1 (R2R3-type MYB TF) from wheat, whose overexpression in tobacco was reported to enhance salt and drought tolerance by enhancing RWC and reducing H2O2, MDA, and NaCl accumulation, as well as decreasing water loss (Wei et al., 2017). Compared with the control, transgenic BplMYB46 overexpressed birch plants increased salt and osmotic tolerance, maintained high lignin and cellulose content and reduced hemicellulose content (Guo et al., 2017). In addition, our study also confirmed the ability of the ThMYB13 gene to increase salt tolerance through transient transformation techniques from homologous overexpression and suppression of expression, providing the basis for further investigation of the salt tolerance mechanism of ThMYB13 gene.
In this study, 14 monomorphic and intact ORFs of ThMYBs were cloned. The 14 ThMYB genes all had the distinct characteristics of the MYB family gene and contained MYB at different positions in the protein sequence domain. In addition, the 14 ThMYB TFs were divided into 5 subgroups: class II, IV, V, VI, and X.
The MYB TFs in subgroup II were mainly involved in stress response. AT4G38620, AT4G34990, AT1G22640, AT2G16720 and AT1G35515 were involved in Arabidopsis in responding to salt stress, osmotic stress, cold acclimation, salicylic acid, ABA and JA treatments (Mondal and Roy, 2017), suggesting that ThMYB13 TF (the member of class II MYB TFs) may also respond to abiotic stress.
The ThMYB8 and Arabidopsis class IV members (AT4G12350, AT1G66230, AT4G22680 and AT5G16600) are closely related. In a previous study, the Arabidopsis class IV members were reported to play a role in the secondary cell wall biosynthesis regulation in Arabidopsis (Zhong et al., 2008), suggesting that ThMYB8 may also participate in biosynthesis of secondary cell wall. The TF of subgroup VI is related to the formation of axillary meristems (Dubos et al., 2010); ThMYB3 belongs to subgroup VI, so it is speculated that ThMYB3 may be involved in the formation of axillary meristems.
Further, in order to analyze the abiotic stress response function, the expressions patterns of 14 ThMYBs in the leaves and roots of T. hispida in response to different abiotic stresses (salt, osmotic stress) and hormone treatments (ABA, GA3, JA) were analyzed. The expressions of most of the ThMYBs were significantly changed by salt and osmotic stress, ABA, GA3 and JA treatments in at least one organ. Especially, ThMYB1, 3, 13, and 14 genes were mainly induced in the leaves and roots of T. hispida under NaCl, PEG and ABA treatment conditions. ThMYB13 was induced in the leaves and roots exposed to salt stress during the study periods. Consistently, these results further showed that ThMYB13 might play a role in response to salt stress.
To further study the salt stress response function of ThMYB13, transgenic T. hispida plants overexpressing or knocking down ThMYB13 and empty pROKII (control) were generated using a transient genetic transformation system. Under salt stress conditions, overexpression of ThMYB13 displayed the lowest O2-, H2O2 and MDA accumulation, minimal cell death and the lowest electrolyte leakage rate among the three kinds of transiently expressed T. hispida. Conversely, the RNAi-silencing transiently transformed plants displayed the opposite physiological changes. In addition, our results showed that the ratio of K+/Na+ in the overexpression of ThMYB13 was higher than that of the Con and RNAi plants when exposed to salt stress conditions. This indicated that the expression of the ThMYB13 gene increased the uptake of K+ in plants and reduced the accumulation of Na+. It has been increasingly recognized that plant salt tolerance is directly reflected by the ratio of K+/Na+ in the cytoplasm; the higher the ratio of K+/Na+, the higher ability of salt tolerance of the plant (Ma et al., 2011; Mishra et al., 2014; Alkhateeb et al., 2015; Zang et al., 2016). These results indicated that ThMYB13 might play an important role in tolerance to salt in transgenic T. hispida plants. In future studies, the salt stress regulatory mechanism of ThMYB13 will be further studied.
In this study, 14 ThMYBs with full ORFs were cloned and identified. The expression patterns of 14 ThMYBs in response to different abiotic stresses (salt and osmotic) and hormones (ABA, GA3, JA) were analyzed using qRT-PCR. The results indicated that ThMYB13 was induced in the leaves and roots of T. hispida treated with NaCl at all study periods, indicating that it may involved in salt stress in plants. Further, T. hispida plants knockdown or overexpression of ThMYB13 and the control transformed with empty pROKII vector were generated using a transient transformation system. Overexpression of ThMYB13 in T. hispida plants displayed the lowest O2-, H2O2 and MDA level, minimal cell death, the most stable K+/Na+ ratio and the lowest electrolyte leakage under salt stress conditions. Conversely, the RNAi-silencing transiently transformed plants displayed the opposite physiological changes. These results suggested that ThMYB13 might play an important physiological role in salt tolerance in transgenic T. hispida plants. In addition, this study may provide new insights into the function of ThMYBs in tolerance to abiotic stress.
TZ wrote the manuscript and performed some of the assays (data shown in Figures 2, 5–7). YZ and ZL performed the assays (cloned and identified the 14 ThMYBs with full ORFs) and analyzed the expression profiles of 14 ThMYB genes under different abiotic stresses and hormones by qRT-PCR; data shown in Figures 1, 3, 4). YW performed the data analysis and also revised the manuscript. CG provided funds for the current study, designed the study and revised the manuscript.
This work was supported by Heilongjiang Province Outstanding Youth Science Foundation (JC2017004) and the National Natural Science Foundation of China (No. 31370676).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01303/full#supplementary-material
FIGURE S1 | Multiple-sequence alignment of the 14 ThMYB proteins from T. hispida. Names of all the14 members were listed on the left side of the figure. Conserved amino acid residues were indicated by black shading. Conserved domains of each ThMYB protein sequence corresponds to a different position of the peptide chain.
FIGURE S2 | ThMYB2, ThMYB3, ThMYB4, ThMYB8, ThMYB9 and ThMYB13 proteins conserved domain alignment. The N-terminus of its protein sequence contains two conserved MYB domains (R2, R3).
FIGURE S3 | ThMYB5, ThMYB6, ThMYB7, ThMYB10 and ThMYB11 proteins conserved domain alignment. The conserved domain was present in the middle of the peptide chain.
TABLE S1 | qRT-PCR primers of 14 ThMYBs. α-tubulin (FJ618518), Actin (FJ618517) and β-tubulin (FJ618519) were the internal reference genes of T. hispida.
TABLE S2 |ThMYB13 gene of pROKII and pFGC5941 vector primers.
ABA, abscisic acid; DAB, 3,3′-Diaminobenzidine tetrahydrochloride; GA3, Gibberellin A3; JA, jasmonic acid; MDA, malondialdehyde; Mol.Wt, molecular weight; NBT, nitroblue tetrazolium; pI, isoelectric point; qRT-PCR, real-time quantitative reverse-transcribed polymerase chain reaction; ROS, reactive oxygen species.
Alkhateeb, S. A., Alkhateeb, A. A., and Solliman, M. E. (2015). In vitro response of date palm (Phoenix dactylifera L.) to K/Na ratio under saline conditions. Biol. Res. 48:63. doi: 10.1186/s40659-015-0055-2
Cai, H., Tian, S., Dong, H., and Guo, C. (2015). Pleiotropic effects of TaMYB3R1 on plant development and response to osmotic stress in transgenic Arabidopsis. Gene 558, 227–234. doi: 10.1016/j.gene.2014.12.066
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., and Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS One 8:e69955. doi: 10.1371/journal.pone.0069955
Chen, T., Li, W., Hu, X., Guo, J., Liu, A., and Zhang, B. (2015). A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 56, 917–929. doi: 10.1093/pcp/pcv019
Chen, Z., Newman, I., Zhou, M., Mendham, N., Zhang, G., and Shabala, S. (2005). Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant Cell Environ. 28, 1230–1246. doi: 10.1111/j.1365-3040.2005.01364.x
Cui, M. H., Yoo, K. S., Hyoung, S., Nguyen, H. T., Kim, Y. Y., Kim, H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587, 1773–1778. doi: 10.1016/j.febslet.2013.04.028
Dal Santo, S., Stampfl, H., Krasensky, J., Kempa, S., Gibon, Y., Petutschnig, E., et al. (2012). Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell 24, 3380–3392. doi: 10.1105/tpc.112.101279
Ding, Z. H., Li, S. M., An, X. L., Liu, X., Qin, H. J., and Wang, D. W. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. doi: 10.1016/S1673-8527(09)60003-5
Du, H., Feng, B. R., Yang, S. S., Huang, Y. B., and Tang, Y. X. (2012). The R2R3-MYB transcription factor gene family in maize. PLoS One 7:e37463. doi: 10.1371/journal.pone.0037463
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Gao, J. J., Zhang, Z., Peng, R. H., Xiong, A. S., Xu, J., Zhu, B., et al. (2011). Forced expression of MdMYB10, a MYB transcription factor from apple, enhances tolerance to osmotic stress in transgenic Arabidopsis. Mol. Biol. Rep. 38, 205–211. doi: 10.1007/s11033-010-0096-0
Guo, H., Wang, Y., Wang, L., Hu, P., Wang, Y., Jia, Y., et al. (2017). Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 15, 107–121. doi: 10.1111/pbi.12595
He, Q., Jones, D. C., Wei, L., Xie, F., Ma, J., Sun, R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 6:22980. doi: 10.1038/srep22980
Hong, S. H., Kim, H. J., Ryu, J. S., Choi, H., Jeong, S., Shin, J., et al. (2008). CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J. 55, 361–371. doi: 10.1111/j.1365-313X.2008.03508.x
Ji, X., Zheng, L., Liu, Y., Nie, X., Liu, S., and Wang, Y. (2014). A transient transformation system for the functional characterization of genes involved in stress response. Plant Mol. Biol. Rep. 32, 732–739. doi: 10.1007/s11105-013-0683-z
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kerschen, A., Napoli, C. A., Jorgensen, R. A., and Müller, A. E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566, 223–228. doi: 10.1016/j.febslet.2004.04.043
Kim, J. H., Nguyen, N. H., Jeong, C. Y., Nguyen, N. T., Hong, S. W., and Lee, H. (2013). Loss of the R2R3 MYB, AtMYB73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170, 1461–1465. doi: 10.1016/j.jplph.2013.05.011
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2011). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63, 305–317. doi: 10.1093/jxb/err280
Mishra, S., Alavilli, H., Lee, B. H., Panda, S. K., and Sahoo, L. (2014). Cloning and functional characterization of a vacuolar Na+/H+ antiporter gene from mungbean (VrNHX1) and its ectopic expression enhanced salt tolerance in Arabidopsis thaliana. PLoS One 9:e106678. doi: 10.1371/journal.pone.0106678
Mondal, S. K., and Roy, S. (2017). Genome-wide sequential, evolutionary, organizational and expression analyses of phenylpropanoid biosynthesis associated MYB domain transcription factors in Arabidopsis. J. Biomol. Struct. Dyn. 2017, 1–25. doi: 10.1080/07391102.2017.1329099
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Identification and molecular characterization of MYB Transcription Factor Superfamily in C4 model plant foxtail millet (Setaria italica L.). PLoS One 9:e109920. doi: 10.1371/journal.pone.0109920
Peng, X. J., Liu, H., Wang, D., and Shen, S. H. (2016). Genome-wide identification of the Jatropha curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genomics 17:251. doi: 10.1186/s12864-016-2576-7
Roy, S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. doi: 10.1080/15592324.2015.1117723
Seo, P. J., and Park, C. M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. doi: 10.1111/j.1469-8137.2010.03183.x
Seo, P. J., Xiang, F., Qiao, M., Park, J.-Y., Lee, Y. N., Kim, S.-G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Soltész, A., Vágújfalvi, A., Rizza, F., Kerepesi, I., Galiba, G., Cattivelli, L., et al. (2012). The rice OsMYB4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. J. Appl. Genet. 53, 133–143. doi: 10.1007/s13353-011-0081-x
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang, L., Wang, C., Wang, D., and Wang, Y. (2014). Molecular characterization and transcript profiling of NAC genes in response to abiotic stress in Tamarix hispida. Tree Genet. Genomes 10, 157–171. doi: 10.1007/s11295-013-0672-2
Wang, Y., Gao, C., Liang, Y., Wang, C., Yang, C., and Liu, G. (2010). A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol. 167, 222–230. doi: 10.1016/j.jplph.2009.09.008
Wei, Q., Luo, Q., Wang, R., Zhang, F., He, Y., Zhang, Y., et al. (2017). A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 8:1374. doi: 10.3389/fpls.2017.01374
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Wong, D. C. J., Schlechter, R., Vannozzi, A., Höll, J., Hmmam, I., Bogs, J., et al. (2016). A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 23, 451–466. doi: 10.1093/dnares/dsw028
Yang, G., Wang, Y., Xia, D., Gao, C., Wang, C., and Yang, C. (2014). Overexpression of a GST gene (ThGSTZ1) from Tamarix hispida improves drought and salinity tolerance by enhancing the ability to scavenge reactive oxygen species. Plant Cell Tissue Organ Cult. 117, 99–112. doi: 10.1007/s11240-014-0424-5
Yang, S. W., Jang, I. C., Henriques, R., and Chua, N. H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21, 1341–1359. doi: 10.1105/tpc.109.067215
Zang, D., Li, H., Xu, H., Zhang, W., Zhang, Y., Shi, X., et al. (2016). An Arabidopsis zinc finger protein increases abiotic stress tolerance by regulating sodium and potassium homeostasis, reactive oxygen species scavenging and osmotic potential. Front. Plant Sci. 7:1272. doi: 10.3389/fpls.2016.01272
Zhang, X., Wang, L., Meng, H., Wen, H., Fan, Y., and Zhao, J. (2011). Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol. Biol. 75, 365–378. doi: 10.1007/s11103-011-9732-x
Keywords: abiotic stress, MYB transcription factor, gene expression, Tamarix hispida, Na+ and K+ content
Citation: Zhang T, Zhao Y, Wang Y, Liu Z and Gao C (2018) Comprehensive Analysis of MYB Gene Family and Their Expressions Under Abiotic Stresses and Hormone Treatments in Tamarix hispida. Front. Plant Sci. 9:1303. doi: 10.3389/fpls.2018.01303
Received: 08 January 2018; Accepted: 17 August 2018;
Published: 19 September 2018.
Edited by:
Vicent Arbona, Universitat Jaume I, SpainReviewed by:
Ramesh Katam, Florida A&M University, United StatesCopyright © 2018 Zhang, Zhao, Wang, Liu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caiqiu Gao, Z2FvY2FpcWl1QG5lZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.