
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 September 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01274
 Dongryung Lee1
Dongryung Lee1 Gileung Lee1
Gileung Lee1 Backki Kim1
Backki Kim1 Su Jang1
Su Jang1 Yunjoo Lee1
Yunjoo Lee1 Yoye Yu1
Yoye Yu1 Jeonghwan Seo1
Jeonghwan Seo1 Seongbeom Kim2
Seongbeom Kim2 Yong-Hwan Lee2
Yong-Hwan Lee2 Joohyun Lee3
Joohyun Lee3 Sunghan Kim4
Sunghan Kim4 Hee-Jong Koh1*
Hee-Jong Koh1*Lesion mimic mutants (LMMs) commonly exhibit spontaneous cell death similar to the hypersensitive defense response that occurs in plants in response to pathogen infection. Several lesion mimic mutants have been isolated and characterized, but their molecular mechanisms remain largely unknown. Here, a spotted leaf sheath (sles) mutant derived from japonica cultivar Koshihikari is described. The sles phenotype differed from that of other LMMs in that lesion mimic spots were observed on the leaf sheath rather than on leaves. The sles mutant displayed early senescence, as shown, by color loss in the mesophyll cells, a decrease in chlorophyll content, and upregulation of chlorophyll degradation-related and senescence-associated genes. ROS content was also elevated, corresponding to increased expression of genes encoding ROS-generating enzymes. Pathogenesis-related genes were also activated and showed improved resistance to pathogen infection on the leaf sheath. Genetic analysis revealed that the mutant phenotype was controlled by a single recessive nuclear gene. Genetic mapping and sequence analysis showed that a single nucleotide substitution in the sixth exon of LOC_Os07g25680 was responsible for the sles mutant phenotype and this was confirmed by T-DNA insertion line. Taken together, our results revealed that SLES was associated with the formation of lesion mimic spots on the leaf sheath resulting early senescence and defense responses. Further examination of SLES will facilitate a better understanding of the molecular mechanisms involved in ROS homeostasis and may also provide opportunities to improve pathogen resistance in rice.
Leaf senescence, the final stage of leaf development, is primarily governed by leaf age. However, leaf senescence is also influenced by various internal and environmental signals that are integrated with the age information (Lim et al., 2007). Lesion mimic mutants (LMMs) are often associated with early leaf senescence. An spl5 mutant continuously developed small reddish-brown necrotic lesions on leaves, leading to early senescence (Chen et al., 2012). An lmes1 mutant also exhibited early senescence, with tiny brown spots developing initially at the leaf tip and spreading to the entire leaf surface (Li et al., 2014). LMMs can be divided into two classes according to the mechanisms involved in controlling cell death: (1) initiation mutants and (2) feedback or propagation mutants (Lorrain et al., 2003). Initiation mutants, such as acd5, cpn1, and cpr5, form localized necrotic spots of determinate size whereas formation rate and lesion extent are not controlled in propagation mutants, such as acd2, lsd1, and svn1 (Dietrich et al., 1997; Boch et al., 1998; Lin and De, 1999; Greenberg et al., 2000; Jambunathan et al., 2001; Mach et al., 2001).
The necrotic spots formed in LMMs resemble those formed during the pathogen infection-induced hypersensitive response (HR). Reactive oxygen species (ROS) are thought to prime the orchestration of the HR (Zurbriggen et al., 2010). During HR, rapid production of ROS, such as hydrogen peroxide (H2O2), superoxide (O), is stimulated in mitochondria and chloroplast as well as in the cytoplasmic level (Zurbriggen et al., 2010). Plants use two types of defense mechanisms to combat oxidative stress. These mechanisms involve non-enzymatic antioxidants such as ascorbate and glutathione, or enzymatic antioxidants such as catalase, superoxide dismutase, and ascorbate peroxidase (Navabpour et al., 2003). Either over-accumulation of ROS or the failure of these oxidative stress defense mechanisms will result in cell death. HR is an important resistance mechanism that prevents pathogen spread to adjacent cells by inducing cell death in infected regions (Lam et al., 2001). HR is usually accompanied by the activation of pathogenesis-related (PR) genes. Expression of PR1a and PR1b, which encodes acidic and basic proteins, respectively, was induced upon infection with rice blast fungus (Agrawal et al., 2000a,b). Expression of PR5 family genes, which encodes thaumatin-like proteins, was induced in plants in response to infection by plant pathogens, elicitors, stresses, and developmental signals (Bryngelsson and Green, 1989). Proteins of the PR10 family were involved in multiple anti-pathogen processes, and are generally localized in the intracellular spaces, in contrast to the extracellular nature of most PR proteins (Jwa et al., 2001).
LMM genes encode wide range of functional protein types, such as heat stress transcription factor (Yamanouchi et al., 2002), U-Box/Armadillo repeat protein (Zeng et al., 2004), zinc finger proteins (Wang et al., 2005), membrane-associated proteins (Noutoshi et al., 2006), ion channel family member (Mosher et al., 2010), clathrin-associated adaptor protein (Qiao et al., 2010), and splicing factor 3b subunit 3 (Chen et al., 2012), indicating the involvement of complex and diverse molecular mechanisms in lesion mimic spot formation. Recently, Wang et al. (2015) showed that Mitogen-Activated Protein Kinase Kinase Kinase (MAPKKK) was also involved in the formation of lesion mimic spots. Several studies revealed that MAPKKK plays an important role in regulating a range of biological processes, such as ethylene signaling (Kieber et al., 1993), plant cytokinesis (Krysan et al., 2002), innate immunity (Asai et al., 2002), defense responses (Suarez-Rodriguez et al., 2007), responses to various stresses (Gao and Xiang, 2008), stomatal development (Kim et al., 2012), and ABA signaling (Wang et al., 2015). However, the relationship between lesion mimic spot formation and MAPK cascades remains poorly understood.
In this study, a new lesion mimic mutant (sles) was identified. In contrast to other LMMs, which exhibited necrotic spots on leaves, lesion mimic spots in the sles mutant covered the leaf sheath resulting in early senescence. Fine-mapping and sequence analysis revealed that the sles locus encoded a kinase domain (KD) containing protein of the Raf MAPKKK family. Greenness and chlorophyll content were adversely affected in mesophyll cells in the sles mutant. Expression of genes encoding ROS-generating enzymes was induced in the sles mutant and ROS accumulation increased accordingly. Defense response genes were also activated and showed enhanced resistance to pathogen infection in sles mutant. These results are relevant to future research into the mechanisms involved in the formation of lesion mimic spots, ROS homeostasis, and resistance to diseases in plants.
The sles mutant was isolated through EMS treatment of the japonica cultivar Koshihikari. The sles mutant was crossed with Koshihikari (japonica) and Milyang 23 (M.23). The M.23 genetic background is similar to that of indica. For phenotypic characterization and genetic mapping, plants were grown by conventional culture at the Experimental Farm of Seoul National University, Suwon, Korea. F2 populations and parents were seeded in a plastic tunnel seedbed. Forty-day-old seedlings were then transplanted, one plant per hill, into a paddy field. The two-tailed Student t-test was used to compare the agronomic traits of sles mutant and wild type plants. 10 independent plants were measured to calculate the means values.
For light microscopic study, thin sections of 100-day-old wild-type penultimate leaf sheath and non-spotted and spotted regions of the sles mutant leaf sheath were cut using a sharp blade and observed using an Olympus CX31 dissecting microscope (Olympus, Japan) under white light. Photographs were taken using an Olympus eXcope T500 digital camera (Olympus, Japan). Three independent plants were used for the anatomical characterization.
Chlorophyll (Chl) a, Chlb, and carotenoid (Car) contents were assessed in the penultimate leaf sheath from 100-day-old wild-type and sles mutant plants. Absorption values were measured as described by Arnon (1949) using UV/Vis spectrophotometer (Biochrom Libra S22, USA). The two-tailed Student t-test was used to compare the pigment intensity between sles mutant and wild type plants. Three biological replicates were used for the experiment.
For O determination, penultimate leaves and leaf sheath samples from 100-day old plants were vacuum-infiltrated (three cycles of 10 min) in 0.5 mg ml−1 nitro blue tetrazolium (NBT) in 10 mM potassium phosphate buffer (pH 7.8) for 16 h. For H2O2 detection, samples were vacuum-infiltrated (three cycles of 10 min) in 1 mg ml−1 3,3′-diaminobenzidine (DAB) containing 10 mM MES (pH 6.5) for 16 h. Reactions were stopped by transferring tissue to 90% ethanol and incubating at 70°C until chlorophyll was completely removed. The cleared leaves and leaf sheaths were examined and photographed after a 2–4 h incubation period. Trypan blue staining was performed on fresh leaf and leaf sheath as previously described by Qiao et al. (2010). Samples were submerged in lactic acid-phenol-trypan blue solution (LPTB; 2.5 mg ml−1 trypan blue, 25% (w/v) lactic acid, 23% water-saturated phenol, and 25% glycerol in H2O) at 70°C, infiltrated by slow-release vacuum for 4 min, and then re-infiltrated. Samples in LPTB were heated in boiling water for 2 min and then cooled for 1.5 h before LPTB solution was replaced with visikol for destaining. The cleared leaves and leaf sheaths were examined and photographed after 3 d incubation period. All the experiments were performed in 10 biological replicates.
The Magnaporthe oryzae strain KJ201 was provided by Department of Agricultural Biotechnology, College of Agriculture and Life Sciences, Seoul National University, Seoul, Korea. The sles mutant and wild-type plants were grown in the greenhouse at 28/25°C, day/night, and were inoculated with KJ201 suspension into the leaf sheath of 50-day-old seedlings in a procedure described by Koga et al. (2004). The blast resistance evaluation was determined 2 days after inoculation by the invasive hyphae on inoculated leaf sheath.
Total RNA was extracted from the penultimate leaf sheath of 100-day-old sles mutant and wild-type plants using Iso-Plus reagent (Takara Bio, Japan) according to the manufacturer's instructions. RNA was then treated with RNA-free DNase I (Promega, USA) to remove any remaining genomic DNA. DNase-treated RNA was reverse transcribed to first-strand cDNA using M-MLV reverse transcriptase (Promega, USA). Real-time PCR was performed using a CFX96 Real-time PCR detection system with SYBR Premix Ex Taq (Takara Bio, Japan). Primer3web (http://bioinfo.ut.ee/primer3/) was used to design primers that spanned an intron to enhance specific amplification of target fragments. Primers used for gene-specific PCR are listed in Supplementary Table 1. Data were analyzed using the comparative Ct method. The two-tailed Student t-test was used to compare the expression level between sles mutant and wild type plants.
For genetic analysis, F2 populations were developed from two crosses: sles mutant × M.23 and sles mutant × Koshihikari. Bulked segregant analysis (BSA) was performed for preliminary genetic mapping using sequence-tagged site (STS) markers designed at the Crop Molecular Breeding Lab, Seoul National University (Chin et al., 2007). Two molecular markers flanking the primary candidate region were used to screen recombination events from 628 F2 individuals. To fine map sles, new STS markers between the two flanking markers were designed based on the sequence difference between japonica variety Nipponbare and the indica variety 93-11. Primers used for genetic mapping are listed in Supplementary Table 1.
Gene prediction analysis was performed using the Gramene database (http://www.gramene.org) and sles candidate genes were analyzed further. The sequence of the AP005101 BAC clone was used to design 28 specific primers for sequence analysis of sles candidate genes. PCR-amplified products were purified using IncloneTM Gel & PCR purification kit (Inclone Biotec, Republic of Korea), TA-cloned into the pGEM-T Easy Vector (Promega, USA), and transformed into E. coli strain DH5α for sequencing.
Twenty dehulled T1 seeds were surfaced-sterilized, placed on 1/2 MS media containing 50 mg/ml hygromycin, and allowed to germinate in the dark at 37°C. To genotype T-DNA insertion lines, three primers were designed based on sequence information for T-DNA insertion positions available at RiceGE (http://signal.salk.edu/cgi-bin/RiceGE). Primers used for PCR are listed in Supplementary Table 1. PCR was used to test co-segregation between flanking sequences and the mutation phenotype.
Homologs of SLES were identified in other species using search functions at the NCBI website. Multiple sequence alignments were conducted using Clustal X (http://www.clustal.org/) and edited with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). NCBI web-based searches were used for conserved domain prediction of the SLES protein (https://www.ncbi.nlm.nih.gov/cdd).
Wild-type and mutant plants were phenotypically and agronomically compared. The mutant exhibited lesion mimic spots on the leaf sheath (Figure 1A). Sparse spots appeared initially at the two-leaf stage and later expanded to cover the entire leaf sheath, resulting in earlier senescence than in wild type (Figure 1B). Lesion mimic spots were restricted to the leaf sheath and were not observed on leaves. Based on these observations, the mutant was designated spotted leaf sheath (sles). Lesion mimic spots also appeared on sles mutant roots (Figure 1C), initiating at the same growth stage as spot appearance on the leaf sheath.

Figure 1. Morphological comparisons between wild-type and sles mutant plants. (A) Wild type (left) and sles mutant (right) at 60 day after germination. (B) 90-day-old and 120-day-old wild type (left) and sles mutant (right). (C) Root color in wild type (left) and sles mutant (right).
Wild-type and sles mutant plants were significantly different with respect to agronomic traits (Table 1). Seed germination rate was substantially lower in the mutant compared to wild-type. Seedling vigor at the 3-leaf-seedling stage, as determined by shoot length, root length, fresh weight, and dry weight, was significantly reduced in the sles mutant compared to wild type. Leaf emergence was also slower in the sles mutant than in wild type. In the reproductive stage, plant height and number of tillers were significantly reduced in the sles mutant compared to wild type. Grains were smaller and thinner in the sles mutant than in wild-type. However, grain shape, which was determined by the grain length/width ratio, was similar between wild type and the sles mutant. Heading was delayed by a week in the sles mutant compared to wild type. Nevertheless, the sles mutant senesced more rapidly than wild type and sles leaves yellowed 4 weeks after heading while wild-type plants remained green (Figure 1B). Yield-related agronomic traits such as spikelet number per panicle, seed-setting rate, and 1,000-grain weight were all significantly adversely impacted compared to wild type.
Differences in mesophyll greenness level were apparent between wild type and sles mutant tissues (Figure 2). Palisade parenchyma of mesophyll cells were green and filled with chloroplast in leaf sheath sections from wild type and non-spotted regions close to spots in the sles mutant (Figures 2A,B,D,E). However, although the palisade parenchyma of mesophyll cells from non-spotted sles regions were green, dark brown areas were sometimes observed in the spongy parenchyma of the mesophyll cells (Figures 2B,E). In the spotted region of sles mutant leaf sheath, however, mesophyll cells were completely dark brown (Figures 2C,F), indicating their death.
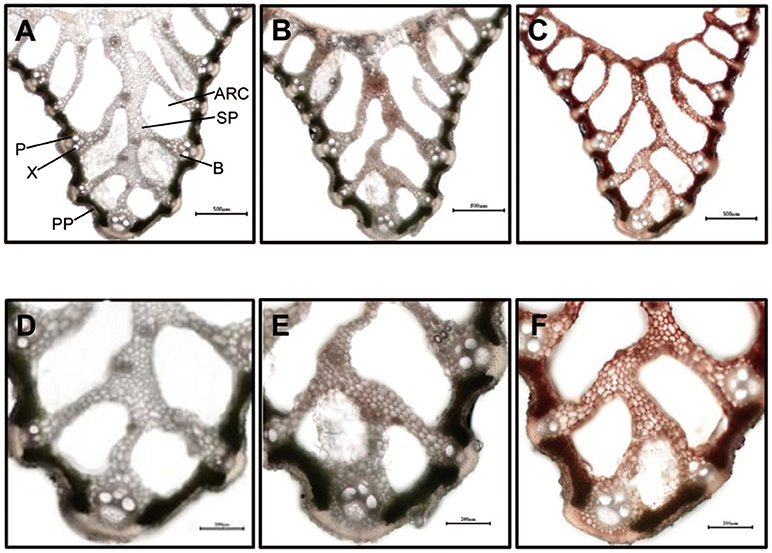
Figure 2. Light microscopic analysis of spotted and non-spotted leaf sheath from sles mutant and wild type plants. Transverse sections of penultimate leaf sheaths were observed under white light. (D–F) are magnified view of (A–C), respectively. (A,D) are wild type leaf sheath sections. (B,E) are non-spotted, and (C,F) are spotted leaf sheath sections from the sles mutant. Indications in (A) are ARC, aerenchyma; B, bundle sheath; P, phloem; PP, palisade parenchyma; SP, spongy parenchyma; X, xylem.
Contents of chlorophyll and carotenoid, the two most important pigments in rice, were compared between sles mutant and wild type (Figure 3A). Total chlorophyll content and Chla and Chlb levels were significantly lower in sles spotted regions than in wild type. The Chla/Chlb ratio was also significantly lower in the sles mutant than in wild type, indicating that Chla content had decreased to a greater extent than Chlb content in the sles mutant. The Chla/Chlb ratio was also significantly different between the non-spotted region of the sles mutant and the wild type, but there were no apparent differences in total chlorophyll content or Chla and Chlb levels. Carotenoid content was also significantly lower in the spotted region of the sles mutant leaf sheath than in wild type. Overall, the dark brown spotted regions of the sles mutant leaf sheath correlated with reductions in chlorophyll content.
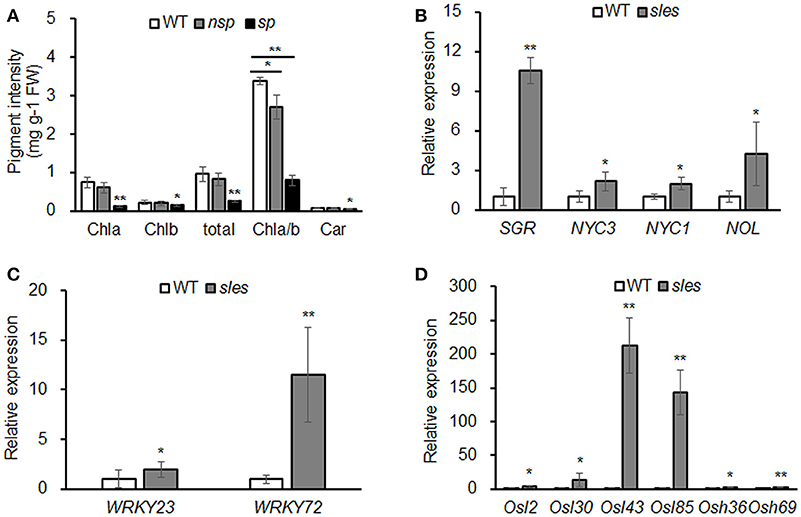
Figure 3. Early senescence in the sles mutant leaf sheath. (A) Abundance of major plant pigments in non-spotted (nsp) and spotted (sp) leaf sheaths from the sles mutant and wild-type (WT) leaf sheaths. (B) Expression of chlorophyll degradation-related genes. (C) Expression of senescence transcription factors. (D) Expression of senescence-associated genes. Real-time PCR (three biological replicates and three technical replicates) was performed with WT leaf sheath samples and sles leaf sheath samples from regions with legion mimic spots. Asterisks indicate the statistical significance levels according to Student's t-test: **P < 0.01 and *P < 0.05.
We examined the expression of chlorophyll degradation-related genes such as STAY GREEN (SGR), NON-YELLOW COLORING (NYC1), NON-YELLOW COLORING3 (NYC3), and NYC1-like (NOL) (Kusaba et al., 2007; Park et al., 2007; Morita et al., 2009; Sato et al., 2009). Expression analysis showed that the chlorophyll degradation-related genes, particularly SGR, were dramatically upregulated in the sles mutant compared to wild type (Figure 3B). To confirm that senescence occurred in the leaf sheath of the sles mutant, expression of senescence transcription factors (OsWRKY23 and OsWRKY72) and senescence-associated genes (Osl2, Osl30, Osl43, Osl85, Osh36, and Osh69) were examined using real-time PCR (Figures 3C,D). All eight genes, particularly Osl43 and Osl85, exhibited elevated expression in the leaf sheath of the sles mutant, compared to wild type, consistent with the early senescence phenotype.
Histochemical markers were examined to investigate putative mechanisms underlying the development of lesion mimic spots in the sles mutant (Figure 4A). The leaf sheath of sles mutant exhibited strong blue color of cells compared to that of wild type after staining with trypan blue, which is a histochemical indicator of irreversible membrane damage or cell death. There was no evidence of ROS production in the wild-type leaf sheath, but the pattern of NBT staining, an indicator of O accumulation, correlated strongly with lesion formation on the sles mutant leaf sheath. In leaves, there were negligible differences in ROS production between sles mutant and wild type. Similar results were obtained with 3,3′-diaminobenzidine (DAB) staining, which indicated H2O2 accumulation. These results confirmed that ROS accumulation in the leaf sheath of sles mutant lead to cell death and ultimately accelerated senescence.
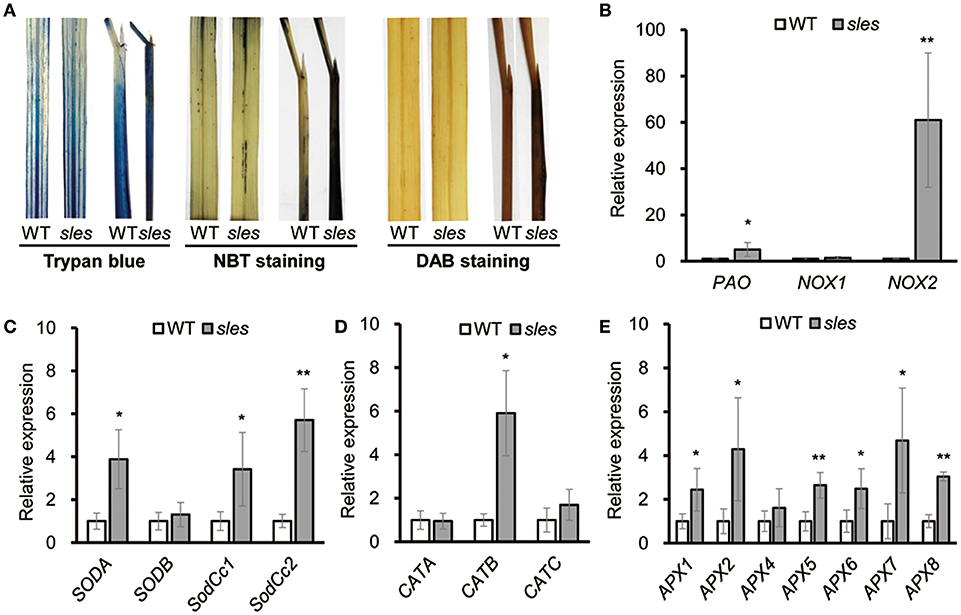
Figure 4. ROS accumulation in the sles mutant leaf sheath. (A) Trypan blue, NBT and DAB staining of penultimate leaves and leaf sheaths in wild-type and sles mutant plants after heading. (B) Expression of genes encoding ROS-generating enzymes in wild-type and sles mutant (C–E) Expression levels of ROS detoxification-related genes in wild-type and sles mutant. Real-time PCR (three biological replicates and three technical replicates) was performed with leaf sheath samples from wild-type and from areas with legion mimic spots in the sles mutant. Asterisks indicate the statistical significance level according to Student's t-test: **P < 0.01 and *P < 0.05.
NADPH oxidase (NOX) and polyamine oxidase (PAO) are major ROS sources. Expression of NOX1, NOX2, and PAO was significantly increased in the spotted region of sles mutant leaf sheath (Figure 4B). Complex antioxidant systems in diverse subcellular compartments tightly regulate the abundance of intercellular ROS. These ROS scavenging systems include major enzymes, such as superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX), that coordinately function in ROS detoxification (Mittler et al., 2004). As sles mutants exhibited enhanced ROS accumulation, we next examined gene expression of ROS scavenging genes (SODA, SODB, SodCc1, SodCc2, CATA, CATB, CATC, APX1, APX2, APX3, APX4, APX5, APX6, APX7, and APX8) (Figures 4C–E) and found that most were significantly upregulated in the sles mutant compared to wild type.
ROS contribute to accelerated transcriptional activation of PR genes, leading to production of antimicrobial secondary metabolites and localized cell death (Zurbriggen et al., 2010). As ROS accumulation was observed in the spotted region of the sles mutant leaf sheath, expression of three PR marker genes (PR1a, PR5, and PR10) associated with the defense response was examined (Figure 5A). All the PR genes were significantly upregulated in the sles mutant compared to wild type.
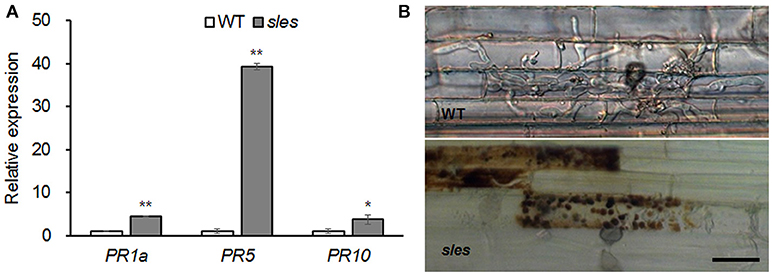
Figure 5. Blast resistance in the sles mutant leaf sheath. (A) Expression of pathogenesis-related marker genes. Real-time PCR was performed with leaf sheath samples from wild-type and from areas with legion mimic spots in the sles mutant. Asterisks indicate the statistical significance level according to Student's t-test: **P < 0.01 and *P < 0.05. (B) The excised leaf sheath from 50-day-old rice seedlings of WT and sles mutant was inoculated with conidial suspension (1 × 104 conidia/ml). Samples were harvested and observed 48 h after inoculation. Three biological replicates and three technical replicates were performed for each experiment. Bar = 25 μm.
To evaluate response of the sles mutant to rice blast, the development of infectious hyphae (IH) within the host cells was observed using an excised leaf sheath assay (Figure 5B). IH actively grew and occupied 5–7 cells neighboring the primary infected cells by 48 h after inoculation in wild type. However, IH were mostly restricted to the primary infected cell, and there was an abundant accumulation of dark brown granules along IH in sles mutant. These results indicated that sles mutant conferred significantly enhanced resistance to rice blast compared to wild type.
F1 and F2 plants from crosses between the sles mutant and M.23 were used to determine whether the phenotype was dominant or recessive, and whether the sles mutant phenotype was controlled by multiple genes or by a single gene. F1 plants exhibited the wild-type phenotype, indicating that the sles mutant phenotype was recessive. The F2 population contained 492 wild-type plants and 136 plants with the sles phenotype, fitting a 3:1 ratio [χ = 3.75 < χ = 3.84, P = 0.06]. In another population of 55 F2 individuals derived from sles mutant/Koshihikari, the phenotype of 46 plants were wild type and 9 plants were sles mutant phenotype, matching a 3:1 ratio [χ = 2.19 < χ = 3.84, P = 0.14]. This indicated that the sles phenotype was controlled by a single recessive nuclear gene.
An F2 population derived from a cross between the sles mutant and M.23 was used to map the locus responsible for the sles mutant phenotype. BSA using 60 polymorphic STS markers evenly distributed across the 12 rice chromosomes was used for preliminary genetic mapping. BSA mapped the sles locus to the interval between STS markers S07050a and S07053 (Figure 6A). Using 628 F2 individuals with newly designed STS markers between the two flanking markers, the sles locus was mapped to the interval between markers 147-1 and 147-2, an ~66 kb physical distance in Nipponbare (Figure 6A). Eight predicted candidate genes were located within the 66 kb candidate region, and the region was encompassed by BAC clone AP005101 (Supplementary Table 2).
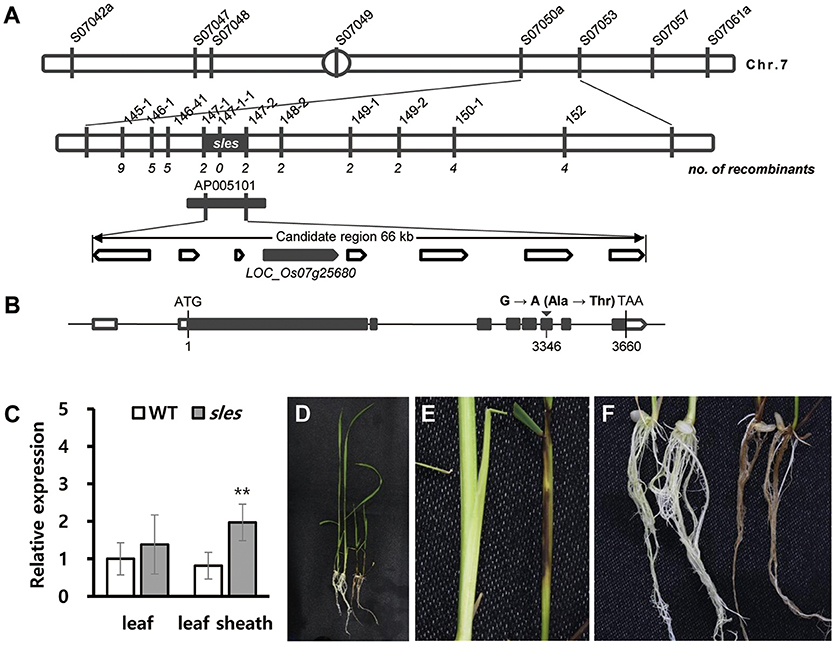
Figure 6. Fine-mapping and identification of SLES. (A) Fine-mapping of SLES. The sles locus was mapped to a 66 kb region on chromosome 7. (B) Schematic diagram of SLES. Black rectangles represent exons and the black inverted triangle represents the mutation site. (C) Expression of SLES gene on leaf and leaf sheath. (D–F) Phenotypic comparison of wild-type Dongjinbyeo (left) and the homozygous T-DNA insertion line (right). (D) Seedling. (E) Leaf sheath. (F) Root. Real-time PCR (three biological replicates and three technical replicates) was performed. Asterisks indicate the statistical significance levels according to Student's t-test: **P < 0.01 and *P < 0.05.
Sequence comparisons of candidate genes between wild type and the sles mutant revealed a single point mutation in the 6th exon of the LOC_Os07g25680 candidate gene. Guanine (G) in the wild-type gene was changed to adenine (A) in the sles mutant gene, resulting in a single amino acid change from alanine to threonine at position 3,346 (Figure 6B). For coding regions, no other DNA sequence differences were detected in any other candidate genes. To verify the SNP, dCAPS markers were designed and used to screen the F2 mapping population. Primers used for PCR are listed in Supplementary Table 1. The genotype exhibited complete co-segregation with the matching phenotypes (Supplementary Figure 1A). To examine whether the SNP was present as a natural variant in other cultivars, dCAPS analysis of eight japonica and five indica rice cultivars was performed. None of the 13 cultivars exhibited an additional restriction fragment (Supplementary Figure 1B).
To understand the possible role of SLES in premature senescence with lesion mimic spots, we examined the transcriptional level of SLES during the formation of lesion mimic spots on the leaf sheath in sles mutant. The results showed that the SLES exhibited significantly elevated expression in the leaf sheath of the sles mutant compared to wild type (Figure 6C). However, there was no remarkable difference of the expression level in the leaf between wild type and sles mutant (Figure 6C).
A T-DNA insertion line (3A-11526.R) from the Crop Biotech Institute, Department of Plant Systems Biotech, Kyung Hee University, was used to confirm that a single functional base substitution in SLES gene was responsible for the abnormal phenotype of sles mutants. This line has a T-DNA inserted into the first intron of LOC_Os07g25680 (Supplementary Figure 2A), which was confirmed by PCR analysis. Seven homozygous and five heterozygous T-DNA tagging mutants were identified (Supplementary Figure 2B). As with the sles mutant, even severer, lesion mimic spots appeared on leaf sheaths and roots in the homozygous T-DNA insertion lines, and seedling height was shorter than the wild type (the cultivar Dongjin) (Figures 6D–F). Homozygous T-DNA insertion lines exhibited weak growth vigor compared to wild type and eventually died within 4 weeks after germination. These results confirmed that the mutation in SLES was responsible for the sles mutant phenotype.
Examination of the rice genome database revealed that the coding sequence (CDS) of SLES consisted of 3,660 nucleotides over 8 exons, and encoded a putative 1,219-amino acid protein. SLES contained Phox and Bem1p (PB1) domain at the N terminus and KD at the C terminus (Figure 7A). The SLES KD contained all 11 subdomains common to known protein kinases (Hanks and Quinn, 1991; Figure 7B). Bioinformatic analysis and multiple amino acid sequence alignment of the predicted KD indicated a conserved catalytic and RAF-specific signature GTXX (W/Y) MAPE, which classified SLES as a Raf MAPKKK (Figure 7B; Rao et al., 2010). SLES homologs were identified within monocots such as Oryza brachyantha, Setaria italica, Brachypodium distachyon, Sorghum bicolor, and Zea mays with 60–92% amino acid identity. However, no clear co-orthologues were identified in eudicots. None of these predicted proteins have been characterized to date.

Figure 7. Protein sequence analysis of SLES. (A) Predicted schematic of the SLES protein. (B) Amino acid sequence alignment of the SLES kinase domain with that of other proteins indicated that SLES had a highly conserved kinase domain. Black boxes indicate identical residues and gray boxes indicate conservative substitutions. Roman numerals indicate the 11 characteristic sub-domains of protein kinases. The Raf specific motif is shown in red boxes. Asterisk indicates the position where a single amino acid change occurred in the sles mutant. Shown are: Oryza brachyantha (XP_006657649), Setaria italic (XP_004956079), Zea mays (XP_008664484), Brachypodium distachyon (XP_003560267), and Sorghum bicolor (XP_002466055).
Several rice mutants associated with lesion mimic spots that result in early senescence, such as spl5, lmes1, and lmes2, have been identified (Chen et al., 2012; Li et al., 2014; Xing et al., 2016). However, to the best of our knowledge, no LMMs in which lesion mimic spots are found on the leaf sheath have been identified in rice to date. The sles mutant identified in this study exhibited lesion mimic spots on the leaf sheath. Further analysis revealed that these lesion mimic spots were attributable to ROS accumulation.
In the sles mutant, total chlorophyll content and Chla and Chlb levels were significantly lower in the spotted region than in wild type, whereas chlorophyll levels in non-spotted regions in the sles mutant did not differ from wild type. The ratios of Chla/Chlb in the non-spotted regions and in wild type were in the 2.5–4.0 range. The Chla/Chlb ratio in the sles mutant spotted regions was significantly lower than that in wild type, indicating that Chla levels in the mutant were relatively more diminished than Chlb levels. SGR, NYC1, NYC3, and NOL play important roles in chlorophyll degradation. Overexpression of SGR and NYC3 accelerated chlorophyll degradation in developing leaves. Moreover, Chlb content was slightly lowered at the late stage of senescence in nol-1, nyc1-2, and nol-1 nyc1-2 mutants (Park et al., 2007; Sato et al., 2009; Wei et al., 2013). In the sles mutant, expression of chlorophyll degradation genes, particularly SGR, was markedly higher than in wild type. These results suggest that chlorophyll content in the sles mutant was reduced by activation of chlorophyll degradation genes resulting in senescence of the leaf sheath. Leaf senescence is mediated by a large number of genes, such as senescence transcription factors (WRKY23 and WRKY72) and SAGs (Osl2, Osl30, Osl43, Osl85, Osh36, and Osh69) (Lee et al., 2001; Zhou et al., 2013). Expression of WRKYs and SAGs, particularly Osl43 (stress response) and Osl85 (fatty acid metabolism), was significantly increased in the leaf sheath of the sles mutant compared to wild type. Moreover, a large amount of irreversible membrane damage or cell death were observed in the leaf sheath of sles mutant. Taken together, the phenotypic, physiological, biochemical and molecular observations indicate that early senescence occurs in the leaf sheath of the sles mutant.
The results outlined above showed that HR-like cell death, leading to early senescence, occurred in the sles mutant; however, the cause of the lesion mimic spots on the leaf sheath remained unclear. Substantial ROS accumulation [superoxide (O) and hydrogen peroxide (H2O2)] was detected in leaf sheath of the sles mutant. NADPH oxidase (NOX) and polyamine oxidase (PAO) are the main ROS sources (Langebartels et al., 2002). An OsSRFP1 overexpression line with enhanced levels of NOX showed high levels of ROS accumulation (Fang et al., 2015). Overexpression of AtPAO3 also resulted in increased production of ROS (Sagor et al., 2016). Expression of NOX2 and PAO was substantially elevated in the spotted leaf sheath of the sles mutant compared to wild type. These results reveal that the elevated expression of genes encoding ROS-generating enzymes may have led to ROS accumulation in spotted regions of the sles mutant leaf sheath. As a large amounts of hydrogen peroxide and superoxide anion accumulated in the spotted region of the sles mutant leaf sheath, the expression of gene encoding scavenging enzymes, especially those located in the chloroplast, such as SodCc1, SodCc2, APX5, APX6, APX7, and APX8, was significantly higher than in wild type.
ROS accumulation not only triggers senescence but also activates the expression of defense genes such as PR genes (Dangl and Jones, 2001; Zentgraf and Hemleben, 2008). Overexpression of these PR genes may enhance plant tolerance to pathogen infections. For instance, overexpression of OsPR1 in tobacco showed enhanced host tolerance to Phytophthora nicotianae, Palstonia solanacearum, and Pseudomonas syringae (Sarowar et al., 2005); overexpression of JIOsPR10 in rice enhanced tolerance to Magnaporthe oryzae (Wu et al., 2016); and overexpression of PR5 in rice enhanced tolerance to Rhizoctonia solani (Datta et al., 1999). Expression of PR genes, especially PR5, was significantly higher in the leaf sheath of the sles mutant than in wild type and showed significantly enhanced disease resistance to M. oryzae by restricting the development of infectious hyphae.
A database search and sequence analysis suggested that SLES contained a conserved protein KD and was a member of the Raf MAPKKK family. Although MAPK cascades have been identified and characterized in rice, little is known about members of the MAPKKK gene family and their functions and regulation in rice. MAPKKKs act upstream of MAPK cascade composed of three classes of enzymes: MAPKKK, MAPKK, and MAPK. Upstream signals activate MAPKKKs, which then phosphorylate MAPKKs. MAPKKs in turn activate a specific MAPK. The downstream targets of MAPKs can be transcription factors, phospholipases, or cytoskeletal proteins (Sturgill and Ray, 1986; Lin et al., 1993; Tian et al., 2017). In general, substrates of kinase are found at relatively constant level in most tissues. However, Gould et al. (1984) revealed that some kinase substrates are only expressed at high level in certain tissues. Since sles protein loses the normal function of kinase activity, we suggest that a specific protein kinase substrate regulating ROS homeostasis specifically in leaf sheath might not regularly conduct its normal function, resulting in ROS accumulation in the leaf sheath of sles mutant. However, further experiments are necessary.
Several studies reveal that MAPKKK gene family is involved in plant defense/stress responses as well as ROS homeostasis regulation. Studies of MEKK1, the MAPKKK of the flagellin cascade, revealed that MEKK1 conferred resistance to both bacterial and fungal pathogens (Asai et al., 2002). Overexpression of TaFLR (a wheat MAPKKK gene) activated PR genes, such as PR2a and PR3, and resulted in increased resistance to Fusarium graminearum (Gao et al., 2016). In the sles mutant, markedly increased ROS accumulation was observed in accordance with induced expression of genes encoding ROS generating enzymes. Moreover, pathogenesis-related genes, especially PR5, were activated and pathogen resistance was enhanced in the sles mutant compared to wild type. Taken together, we suggest that SLES might suppress production of ROS associated with pathogen defense mechanism, thus leading to HR-like cell death on the leaf sheath and prevents the further pathogen infection of the sles mutant.
In this study, the SLES gene was characterized and isolated. Further examination of SLES will facilitate a better understanding of the molecular mechanisms involved in ROS homeostasis and may also provide opportunities to improve pathogen resistance in rice. Furthermore, as SLES is a Raf MAPKKK, the sles mutant is ideal for studies of MAPK cascades.
DL performed research, analyzed data, and wrote the manuscript. GL, BK, YY, and JS performed research and analyzed data. SJ and YL phenotyped samples and analyzed data. Y-HL and SeK performed blast resistance evaluation and analyzed data. JL designed research and performed research. SuK and H-JK designed research and wrote the manuscript.
This work was supported by a Grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center number PJ013165), Rural Development Administration, Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01274/full#supplementary-material
Agrawal, G. K., Jwa, N. S., and Rakwal, R. (2000a). A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut phytohormones and protein phosphatase inhibitors. Biochem. Biophys. Res. Commun. 274, 157–165. doi: 10.1006/bbrc.2000.3114
Agrawal, G. K., Rakwal, R., and Jwa, N. S. (2000b). Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 278, 290–298. doi: 10.1006/bbrc.2000.3781
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Boch, J., Verbsky, M., Robertson, T., Larkin, J., and Kunkel, B. (1998). Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol. Plant Microbe. Interact. 11, 1196–1206. doi: 10.1094/MPMI.1998.11.12.1196
Bryngelsson, T., and Green, B. (1989). Characterization of a pathogenesis-related, thaumatin-like protein isolated from barley challenged with an incompatible race of mildew. Physiol. Mol. Plant Pathol. 35, 45–52. doi: 10.1016/0885-5765(89)90006-4
Chen, X., Hao, L., Pan, J., Zheng, X., Jiang, G., Jin, Y., et al. (2012). SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol. Breed. 30, 939–949. doi: 10.1007/s11032-011-9677-4
Chin, J., Kim, J., Jiang, W., Chu, S., Woo, M., Han, L., et al. (2007). Identification of subspecies-specific STS markers and their association with segregation distortion in rice (Oryza sativa L.). J. Crop Sci. Biotechnol. 10, 175–184.
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Datta, K., Velazhahan, R., Oliva, N., Ona, I., Mew, T., Khush, G., et al. (1999). Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor. Appl. Genet. 98, 1138–1145. doi: 10.1007/s001220051178
Dietrich, R. A., Richberg, M. H., Schmidt, R., Dean, C., and Dangl, J., L. (1997). A novel zinc finger protein is encoded by the arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. doi: 10.1016/S0092-8674(00)81911-X
Fang, H., Meng, Q., Xu, J., Tang, H., Tang, S., Zhang, H., et al. (2015). Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol. Biol. 87, 441–458. doi: 10.1007/s11103-015-0294-1
Gao, L., and Xiang, C. (2008). The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 67, 125–134. doi: 10.1007/s11103-008-9306-8
Gao, Y., Stebbing, J., Tubei, K., Tian, L., Li, X., and Xing, T. (2016). Response of TaFLR MAPKKK to wheat leaf rust and Fusarium head blight and the activation of downstream components. Trop. Plant Pathol. 41, 15–23. doi: 10.1007/s40858-015-0063-3
Gould, K. L., Cooper, J. A., and Hunter, T. (1984). The 46,000-dalton tyrosine protein kinase substrate is widespread, whereas the 36,000-dalton substrate is only expressed at high levels in certain rodent tissues. J. Cell. Biol. 98, 487–497. doi: 10.1083/jcb.98.2.487
Greenberg, J. T., Silverman, F., and Liang, H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350.
Hanks, S. K., and Quinn, A. M. (1991). Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods. Enzymol. 200, 38–62. doi: 10.1016/0076-6879(91)00126-H
Jambunathan, N., Siani, J. M., and Mcnellis, T. W., (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. doi: 10.1105/tpc.13.10.2225
Jwa, N., Agrawal, G., Rakwal, R., Park, C., and Agrawal, V. (2001). Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. 286, 973–983. doi: 10.1006/bbrc.2001.5507
Kieber, J., Rothenberg, M., Roman, G., Feldmann, K., and Ecker, J. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. doi: 10.1016/0092-8674(93)90119-B
Kim, T. W., Michniewicz, M., Bergmann, D. C., and Wang, Z. Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. doi: 10.1038/nature10794
Koga, H., Dohi, K., Nakayachi, O., and Mori, M. (2004). A novel inoculation method of Magnaporthe grisea for cytological observation of the infection process using intact leaf sheaths of rice plants. Physiol. Mol. Plant Pathol. 64, 67–72. doi: 10.1016/j.pmpp.2004.07.002
Krysan, P. J., Jester, P. J., Gottwald, J. R., and Sussman, M. R. (2002). An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14, 1109–1120. doi: 10.1105/tpc.001164
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Lam, E., Kato, N., and Lawton, M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. doi: 10.1038/35081184
Langebartels, C., Wohlgemuth, H., Kschieschan, S., Grun, S., and Sandermann, H. (2002). Oxidative burst and cell death in ozone-exposed plants. Plant Physiol. Biochem. 40, 567–575. doi: 10.1016/S0981-9428(02)01416-X
Lee, R., Wang, C., Huang, L., and Chen, S. (2001). Leaf senescence in rice plants cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 52, 1117–1121. doi: 10.1093/jexbot/52.358.1117
Li, Z., Zhang, Y., Liu, L., Liu, Q., Bi, Z., Yu, N., et al. (2014). Fine mapping of the lesion mimic and early senescence 1 (lmes1) in rice (Oryza sativa). Plant Physiol. Biochem. 80, 300–307. doi: 10.1016/j.plaphy.2014.03.031
Lim, P., Kim, H., and Nam, H. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, B., and De, W. (1999). Identification and characterization of a novel Arabidopsis mutant, svn1, exhibiting aberrant regulation of cell death. Mol. Plant Microbe. Interact. 2, 416–421.
Lin, L., Wartmann, M., Lin, A. Y., Knopf, J. L., Seth, A., and Davis, R. J. (1993). cPLA2 is phosphorylated and activated by MAP kinase. Cell 72, 269–278. doi: 10.1016/0092-8674(93)90666-E
Lorrain, S., Vailleau, F., Balagué, C., and Roby, D. (2003). Lesion mimic mutants keys for deciphering cell death and defense pathways in plants. Trends Plant Sci. 8, 263–271. doi: 10.1016/S1360-1385(03)00108-0
Mach, J. M., Castillo, A. R., Hoogstraten, R., and Greenberg, J. T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. U.S.A. 98, 771–776. doi: 10.1073/pnas.98.2.771
Mittler, R., Vanderauwera, S., Gollery, M., and Van, B. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., and Kusaba, M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Mosher, S., Moeder, W., Nishimura, N., Jikumaru, Y., Joo, S. H., Urquhart, W., et al. (2010). The Lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 152, 1901–1913. doi: 10.1104/pp.109.152603
Navabpour, S., Morris, K., Allen, R., Harrison, E., A-H-Mackerness, S., and Buchanan-Wollaston, V. (2003). Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 54, 2285–2292. doi: 10.1093/jxb/erg267
Noutoshi, Y., Kuromori, T., Wada, T., Hirayama, T., Kamiya, A., Imura, Y., et al. (2006). Loss of Necrotic Spotted Lesions 1 associates with cell death and defense responses in Arabidopsis thaliana. Plant Mol. Biol. 62, 29–42. doi: 10.1007/s11103-006-9001-6
Park, S., Yu, J., Park, J., Li, J., Yoo, S., Lee, N. Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Qiao, Y., Jiang, W., Lee, J., Park, B., Choi, M. S., Piao, R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Rao, K., Richa, T., Kumar, K., Raghuram, B., and Sinha, A. K. (2010). In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res. 17, 139–153. doi: 10.1093/dnares/dsq011
Sagor, G., Zhang, S., Kojima, S., Simm, S., Berberich, T., and Kusano, T. (2016). Reducing cytoplasmic polyamine oxidase activity in arabidopsis increases salt and drought tolerance by reducing reactive oxygen species production and increasing defense gene expression. Front. Plant Sci. 7:214. doi: 10.3389/fpls.2016.00214
Sarowar, S., Kim, Y., Kim, E., Kim, K., Hwang, B., Islam, R., et al. (2005). Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 24, 216–224. doi: 10.1007/s00299-005-0928-x
Sato, Y., Morita, R., Katsuma, S., Nishimura, M., Tanaka, A., and Kusaba, M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57, 120–131. doi: 10.1111/j.1365-313X.2008.03670.x
Sturgill, T. W., and Ray, L. B. (1986). Muscle proteins related to microtubule associated protein-2 are substrates for an insulin-stimulatable kinase. Biochem. Bioph. Res. Co. 134, 565–571. doi: 10.1016/S0006-291X(86)80457-0
Suarez-Rodriguez, M., Adams-Phillips, L., Liu, Y., Wang, H., Su, S. H., Jester, P. J., et al. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669. doi: 10.1104/pp.106.091389
Tian, X., Li, X., Zhou, W., Ren, Y., Wang, Z., Liu, Z., et al. (2017). Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol. 175, 1337–1349. doi: 10.1104/pp.17.00946
Wang, L., Pei, Z., Tian, Y., and He, C. (2005). OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol. Plant Microbe. Interact. 18, 375–384. doi: 10.1094/MPMI-18-0375
Wang, S., Lim, J., Kim, S., Cho, S., Yoo, S., Koh, H., et al. (2015). Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J. Exp. Bot. 66, 7045–7059. doi: 10.1093/jxb/erv401
Wei, Q., Cao, H., Li, Z., Kuai, B., and Ding, Y. (2013). Identification of an AtCRN1-like chloroplast protein BeCRN1 and its distinctive role in chlorophyll breakdown during leaf senescence in bamboo (Bambusa emeiensis 'Viridiflavus'). Plant Cell Tissue Organ Cult. 114, 1–10. doi: 10.1007/s11240-013-0298-y
Wu, J., Kim, S. G., Kang, K., Kim. J., Park, S., Gupta, R., et al. (2016). Overexpression of a Pathogenesis-Related Protein 10 Enhances Biotic and Abiotic Stress Tolerance in Rice. Plant Pathol. J. 32, 552–562. doi: 10.5423/PPJ.OA.06.2016.0141
Xing, Y., Du, D., Xiao, Y., Zhang, T., Chen, X., Feng, P., et al. (2016). Fine Mapping of a New Lesion Mimic and Early Senescence 2 (lmes2) Mutant in Rice. Crop Sci. 56, 1550–1560. doi: 10.2135/cropsci2015.09.0541
Yamanouchi, U., Yano, M., Lin, H., Ashikari, M., and Yamada, K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. doi: 10.1073/pnas.112209199
Zeng, L., R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Zentgraf, U., and Hemleben, V. (2008). Molecular cell biology: are reactive oxygen species regulators of leaf senescence? Prog. Bot. 69, 117–138. doi: 10.1007/978-3-540-72954-9_5
Zhou, Y., Huang, W., Liu, L., Chen, T., Zhou, F., and Lin, Y. (2013). Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13:132. doi: 10.1186/1471-2229-13-132
Keywords: lesion mimic mutant (LMM), leaf sheath, early senescence, reactive oxygen species (ROS), blast resistance, Mitogen-Activated Protein Kinase Kinase Kinase (MAPKKK)
Citation: Lee D, Lee G, Kim B, Jang S, Lee Y, Yu Y, Seo J, Kim S, Lee Y-H, Lee J, Kim S and Koh H-J (2018) Identification of a Spotted Leaf Sheath Gene Involved in Early Senescence and Defense Response in Rice. Front. Plant Sci. 9:1274. doi: 10.3389/fpls.2018.01274
Received: 17 April 2018; Accepted: 14 August 2018;
Published: 05 September 2018.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Jong-Seong Jeon, Kyung Hee University, South KoreaCopyright © 2018 Lee, Lee, Kim, Jang, Lee, Yu, Seo, Kim, Lee, Lee, Kim and Koh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hee-Jong Koh, aGVlamtvaEBzbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.