- 1Laboratory of Cell and Molecular Biology, Institute of Vegetable Science, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Ministry of Agriculture – Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Hangzhou, China
- 3Department of Horticulture, College of Agriculture and Food Science, Zhejiang A & F University, Hangzhou, China
- 4Department of Biology, The Pennsylvania State University, University Park, Pennsylvania, PA, United States
- 5Center for Lignocellulose Structure and Formation, The Pennsylvania State University, University Park, Pennsylvania, PA, United States
In plants, the construction, differentiation, maturation, and degradation of the cell wall are essential for development. Pectins, which are major constituents of primary cell walls in eudicots, function in multiple developmental processes through their synthesis, modification, and degradation. Several pectin modifying enzymes regulate pectin degradation via different modes of action. Polygalacturonases (PGs), which function in the last step of pectin degradation, are a crucial class of pectin-modifying enzymes. Based on differences in their hydrolyzing activities, PGs can be divided into three main types: exo-PGs, endo-PGs, and rhamno-PGs. Their functions were initially investigated based on the expression patterns of PG genes and measurements of total PG activity in organs. In most plant species, PGs are encoded by a large, multigene family. However, due to the lack of genome sequencing data in early studies, the number of identified PG genes was initially limited. Little was initially known about the evolution and expression patterns of PG family members in different species. Furthermore, the functions of PGs in cell dynamics and developmental processes, as well as the regulatory pathways that govern these functions, are far from fully understood. In this review, we focus on how recent studies have begun to fill in these blanks. On the basis of identified PG family members in multiple species, we review their structural characteristics, classification, and molecular evolution in terms of plant phylogenetics. We also highlight the diverse expression patterns and biological functions of PGs during various developmental processes, as well as their mechanisms of action in cell dynamic processes. How PG functions are potentially regulated by hormones, transcription factors, environmental factors, pH and Ca2+ is discussed, indicating directions for future research into PG function and regulation.
Introduction
The cell walls of plants are unique extracellular structures, the construction, differentiation, maturation, and degradation of which lay the foundations for tissue differentiation, organ patterning and developmental transitions. Their structural variability in different plant species not only reflects the phylogenetic diversity of plants, but is also associated with the complexity of plant development and plant resistance to biotic and abiotic stresses (Somerville et al., 2004; Cosgrove, 2005; Braybrook and Joensson, 2016). In primary cell walls, 90% of the non-water mass is composed of cellulose, hemicelluloses, and pectins (Albersheim et al., 1996). Pectins, which are acidic polysaccharides that surround cellulose and hemicelluloses in a matrix, control wall porosity, wall hydration, and intercellular adhesion (Daher and Braybrook, 2015; Anderson, 2016). They can be classified into homogalacturonan (HG), rhamnogalacturonan-I, rhamnogalacturonan-II, and xylogalacturonan domains (Sénéchal et al., 2014). As major constituents of primary cell walls, the pollen intine, pollen tube walls, and the middle lamella (Aouali et al., 2001; Dardelle et al., 2010), pectins and their synthesis and degradation influence tissue elongation, pollen development, fruit ripening, and organ abscission (Willats et al., 2001; Sénéchal et al., 2014). The metabolism of pectins in the cell wall is regulated by different classes of pectin-modifying enzymes (Willats et al., 2001; Sénéchal et al., 2014). The polygalacturonases (PGs), which comprise one of these classes, catalyze the hydrolysis of pectins and are involved in numerous developmental processes. Based on differences in hydrolyzing activity, PGs can be divided into three main types: exo-PGs, endo-PGs, and rhamno-PGs. Therefore, illuminating the functions of PGs is of great importance in understanding the dynamics of cell walls during plant growth and strengthening breeding strategies for improving the productivity of crop varieties.
Early studies succeeded in cloning PG genes from species like maize (Zea mays) (Allen and Lonsdale, 1993), tobacco (Nicotiana tabacum) (Tebbutt et al., 1994), peach (Prunus persica) (Lester et al., 1996), and tomato (Solanum lycopersicum) (Kalaitzis et al., 1997). Total PG activity was measured in various organs of these species, indicating the functions of PGs in fruit ripening, pollen development, and organ abscission, in line with the expression patterns of these genes. Hadfield and Bennett (1998) reviewed this initial progress and proposed the first classification system for PGs (Hadfield et al., 1998). However, without genome data, little was known about the composition, classification, and evolution of the PG family. Although the sequences of some PG genes were reported, only a few of these were characterized in terms of their expression patterns and functions. At the turn of the 21st century, it remained unclear how pectin hydrolysis by PGs alters cell wall structure, modulates cell growth, and regulates organ growth.
In the last 18 years, the expression patterns and putative functions of hundreds of PG genes have been delineated. Some regulatory factors for PGs at both the transcriptional and post-translational levels have also been reported. Innovative research methods and a flood of genome sequencing data have both contributed to the systematic identification and phylogenetic analysis of PG families in numerous plant species. This review highlights the latest advances in the structural analysis and classification of PG families, the molecular evolution of PG genes in the context of plant evolution, and the expression, functions, and regulators of PGs during different developmental processes in plants. It also explores the basis for a better understanding of how cell wall dynamics influence cell and plant growth.
Structural Characteristics and Classification of Polygalacturonases
Structural Characteristics of Polygalacturonases
Polygalacturonases belong to glycoside hydrolase family 28 and contain at least one GH28 (Pfam00295) domain (Markovič and Janeček, 2001; Kim et al., 2006). This domain is replaced by a Pectate Lyase 3 domain (Pfam12708) in Arabidopsis (Arabidopsis thaliana) QRT3 and its homologs in other species (Rhee et al., 2003; Yu et al., 2014). The encoded protein of Arabidopsis QRT3 exhibits PG activity, as demonstrated by heterologous expression in Saccharomyces cerevisiae (Rhee et al., 2003). Most PG genes (91.2% of PG genes in Arabidopsis and 87.9% of PG genes in Chinese cabbage (Brassica campestris, syn. Brassica rapa) are predicted to encode a signal peptide upstream of the GH28 domain. Since signal peptides typically function in guiding proteins through secretory pathways that end with exocytosis (Babu et al., 2013), their presence implies that most PGs should be located in the apoplast. This hypothesis has been supported in recent studies of PG localization (Irshad et al., 2008; Xiao et al., 2014, 2017; Rui et al., 2017). In addition to the apoplast, one PG with a signal peptide has been localized in Golgi bodies and vesicles in vitro, presumably as part of its secretory journey (Nakashima et al., 2004).
In plant and fungal PGs, there are four commonly conserved, functional domain motifs known as SPNTDG (motif I), GDDC (motif II), CGPGHGISIGSLG (motif III), and RIK (motif IV), although domain III shows lower conservation and is missing in rhamno-PGs (Park et al., 2008). Typically, a PG gene encodes at least one of these domains (Torki et al., 2000). Within these four motifs, NTD, DD, GHG, and RIK amino acid segments are conserved active-site residues in plant, fungal, bacterial, and insect PGs, and have been demonstrated to be essential for fungal PG activity (Bussink et al., 1991; Markovič and Janeček, 2001). Within motifs I and II, the aspartic acid (D) residues in NTD and DD are components of the catalytic site (Rexovabenkova, 1990). In motif III, the histidine (H) residue acts in catalysis (Rao et al., 1996; Markovič and Janeček, 2001). Finally, the positively charged motif IV is thought to form interactions with the carboxylate groups of pectate substrates (Bussink et al., 1991).
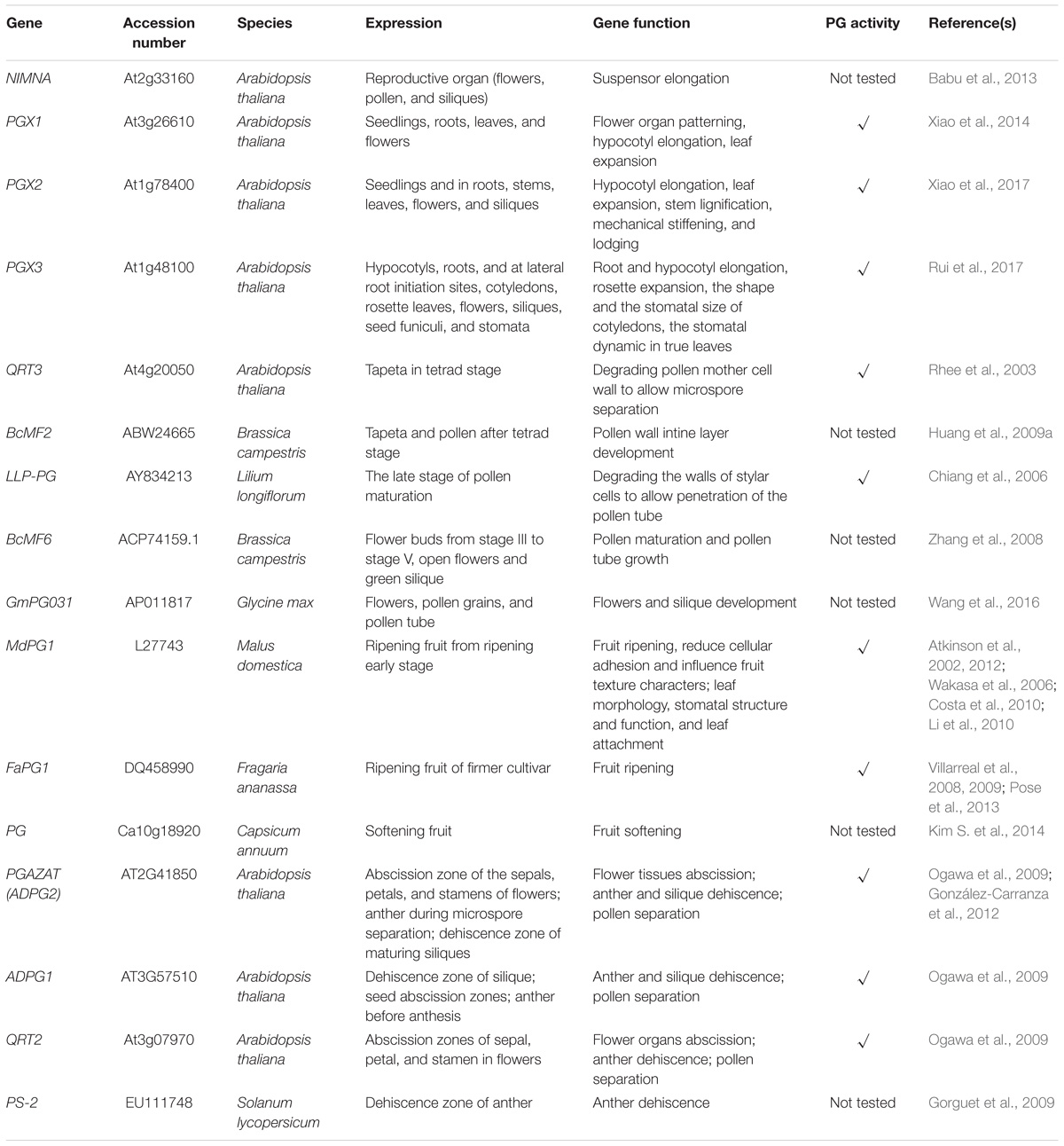
Crystal structures of PGs can reveal the mechanisms by which they select substrates or act in catalysis. Although crystal structures of plant PGs are not yet published, we can infer their modes of action by inspecting the reported structures of fungal and bacterial enzymes due to the high conservation of functional domains between plant, fungal, and bacterial PGs. A typical PG displays 10 complete turns of β-structure, which is formed by four parallel sheets extending along the longitudinal axis. This structure can be used to distinguish PGs from polysaccharide lyases, which have three-sheet topology (Kluskens et al., 2005; Abbott and Boraston, 2007). Endo-PGs and exo-PGs show significant differences in crystal structures, indicating their different substrates and modes of action (Figure 1A). The active site of a typical endo-PG is a tunnel-like substrate-binding cleft lying between two loop regions. Hence, endo-PGs can potentially bind polysaccharides in either direction and produce oligosaccharides with varying degrees of polymerization (Jenkins et al., 1998). The adjacent loop regions help identify substrates and guide them to the active site (Pickersgill et al., 1998). For example, in Achaetomium sp. Xz8 endo-PG, the Asn94 residue of the T3 loop binds to substrates in the active site cleft by forming a hydrogen bond, which ensures correct positioning of substrates (Tu et al., 2015). In contrast, exo-PGs have a closed-pocket active site that only binds to the non-reducing ends of pectins due to inserted stretches of amino acids (Abbott and Boraston, 2007). Rhamno-PGs (RGs), which hydrolyze GalA-rhamnose bonds of rhamnogalacturonan-I, can be further divided into exo-RGs and endo-RGs (Mutter et al., 1998; Choi et al., 2004; Damak et al., 2015). However, tertiary structures of exo-RGs remain unreported. A predicted structure of an endo-RG has only been modeled in Aspergillus aculeatus (Choi et al., 2004). Compared with endo-PGs, endo-RGs are also predicted to have a tunnel-like active-site with two open ends. Differences in loop structure are likely to provide endo-RGs with more space in the active site to bind more complex substrates. The most significant difference between the structures of endo-PGs and endo-RGs is that endo-RGs have long tails of 19 and 45 residues at the N-terminus and C-terminus, respectively, whereas endo-PGs lack these tails (Choi et al., 2004). As a result of their unique structures, exo-PGs can only remove galacturonic acid residues from the non-reducing ends of HG chains; endo-PGs catalyze the random hydrolytic cleavage of α-1,4 glycosidic bonds in HG chains; and rhamno-PGs catalyze the hydrolytic cleavage of α-1,2 glycosidic bonds randomly within or from the non-reducing ends of rhamnogalacturonan-I main chains (Figure 1A) (Markovič and Janeček, 2001; Park et al., 2010).
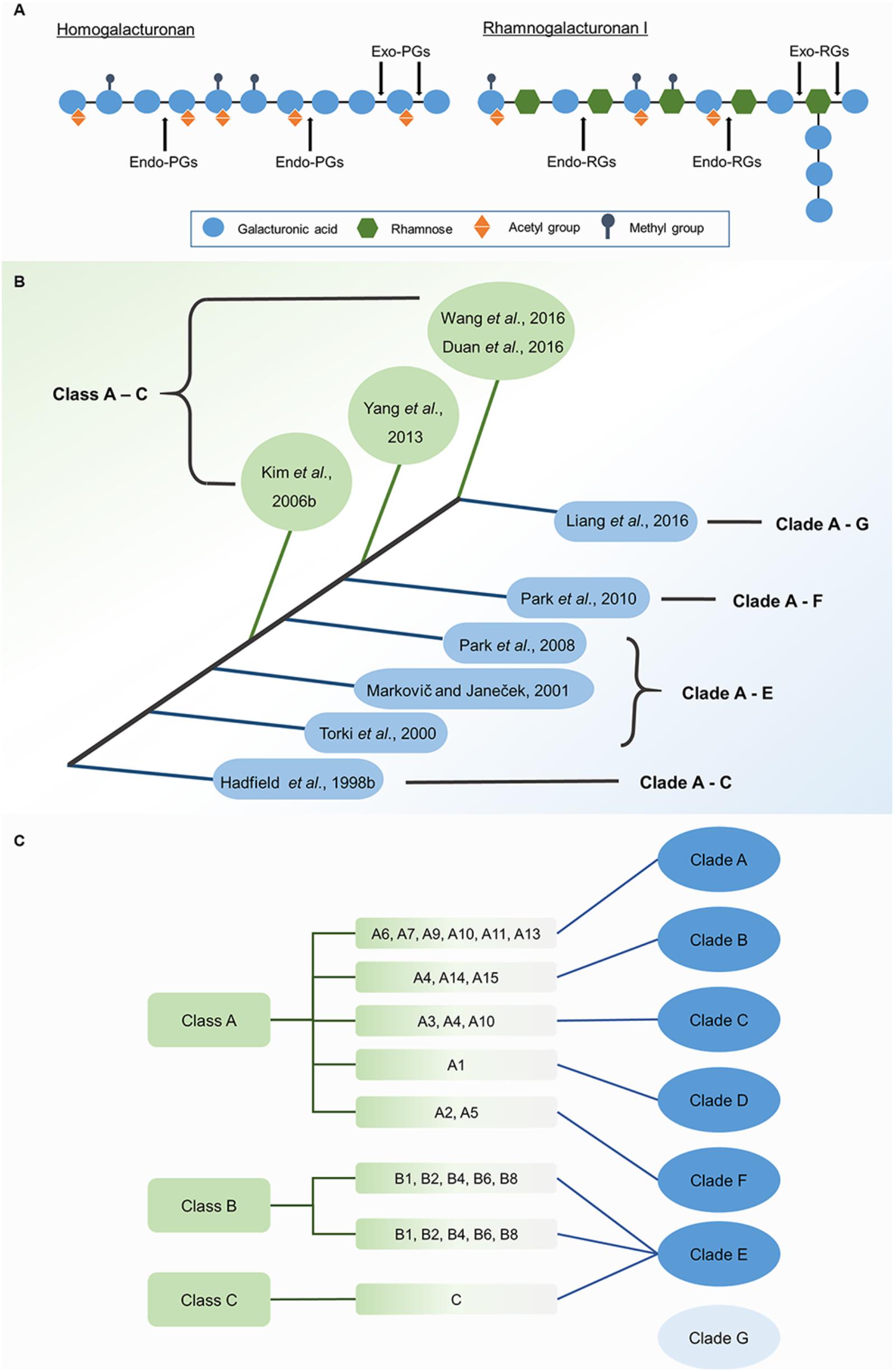
FIGURE 1. Modes of action of PGs of different types (A) and classification of PG genes (B). (A) Different types of PGs differ in the selection of substrates and the action site of pectins main chain. (B) Based on the system first proposed by Kim, PG genes were all grouped into three classes (with green background). Based on the system first proposed by Hadfield, PG genes were divided into three to seven clades in different studies (with blue background). (C) A comparison of Kim’s system (with green background) and Liang’s system (with blue background) in the classification of Arabidopsis PG gene family.
Classification and Molecular Evolution of Polygalacturonases
As mentioned above, PGs can be divided into exo-PGs, endo-PGs and rhamno-PGs, based on their modes of hydrolysis. PGs of these different types emerged at different times during plant evolution. Rhamno-PGs, which are regarded as the earliest type, appear in both algae and land plants, and endo-PGs exist across land plants, whereas exo-PGs only appear in angiosperms (Park et al., 2010). The PG family in land plants had five common ancestral genes rather than a single one (McCarthy et al., 2014), which might be explained by this early divergence of rhamno-PGs and endo-PGs.
Using bioinformatics, PG genes can be grouped by their phylogenetic relationships. Two main classification systems have been proposed (Figure 1B) for analyzing these relationships according to amino acid sequence. The first system was put forward by Hadfield et al. (1998), who grouped three PG genes from melon (Cucumis melo) and 17 homologous genes from other plants and fungi into Clades A–C. In agreement with Hadfield’s system, Torki et al. (2000), Markovič and Janeček (2001), and Park et al. (2008) grouped PG genes into five clades: Clades A–E. Clades A, B, and C were found to have invariant conserved residues (Gly264 and Phe294 in Clade A, Asn104 in Clade B, and Lys176 in Clade C), revealing distinct structural characteristics in different clades. However, Clades D and E lack exclusively invariant residues (Markovič and Janeček, 2001). With the accumulation of genomic data for numerous species, PG classification entered into a new phase in which cluster analysis of all PG gene family members for a species could be performed. Park et al. (2010) divided 225 genes into six clades, which contain eight PG gene subfamilies from algae to angiosperms. Later, Liang et al. (2015) classified PG genes from five species ranging from algae to eudicots into seven clades, as supported by the classification of 557 PG genes from five grass and five eudicot species (Liang et al., 2016). Arabidopsis QRT3 and its homologous genes in core angiosperms are grouped into the new clade (Clade G). The second classification system was proposed by Kim et al. (2006) and divided 125 PG genes from Arabidopsis and rice (Oryza sativa) into three classes, A–C. The subsequent studies that used this system also grouped 75 PG genes from poplar (Populus trichocarpa) and 100 PG genes from Chinese cabbage into three classes (Yang et al., 2013; Duan et al., 2016).
Each classification system has advantages and shortcomings for analyzing molecular evolution. The first system is more suitable for analyzing the emergence of PG genes over time and the compositional changes in PG families. PG genes in different clades emerged at different times (Figure 2). Based on this system, PG genes in Clade E exist from algae to angiosperms, those in Clades A and B appear in land plants, and the genes in Clades C, D, F, and G only appear in flowering plants (Park et al., 2010). Clade emergence shows a pattern that is consistent with species evolution (Figure 2). The dominant clade in non-vascular plants is Clade E, which diversifies into Clades B, D, and E in monocots. The proportions of Clades B and E decrease in eudicots [except in soybean (Glycine max)], and Clades C, D and F are instead predominant. Further analysis shows that Clade D is the principal clade in Cruciferae, Cucurbitaceae, and Solanaceae, whereas the most-represented clades in poplar and soybean are Clades C and E, respectively. The second system also reveals different appearance times for different PG classes. However, PG genes within the same class also emerged at different times in this system. For example, the two major subgroups of Class A, A1, and A2, exist in angiosperms and non-vascular plants, respectively (Duan et al., 2016), and the proportion of Class C is lower than Classes A and B in species from moss (Physcomitrella patens) to vascular plants. The representation of Class A outpaces Class B in moss and vascular plants, but the converse is true in lycophytes (Selaginella moellendorffii). The various expansion rates of PG classes were likely driven by different selection pressures on those classes, accounting for their different proportionalities across taxa (Kim et al., 2006; Yang et al., 2013; Wang et al., 2016).
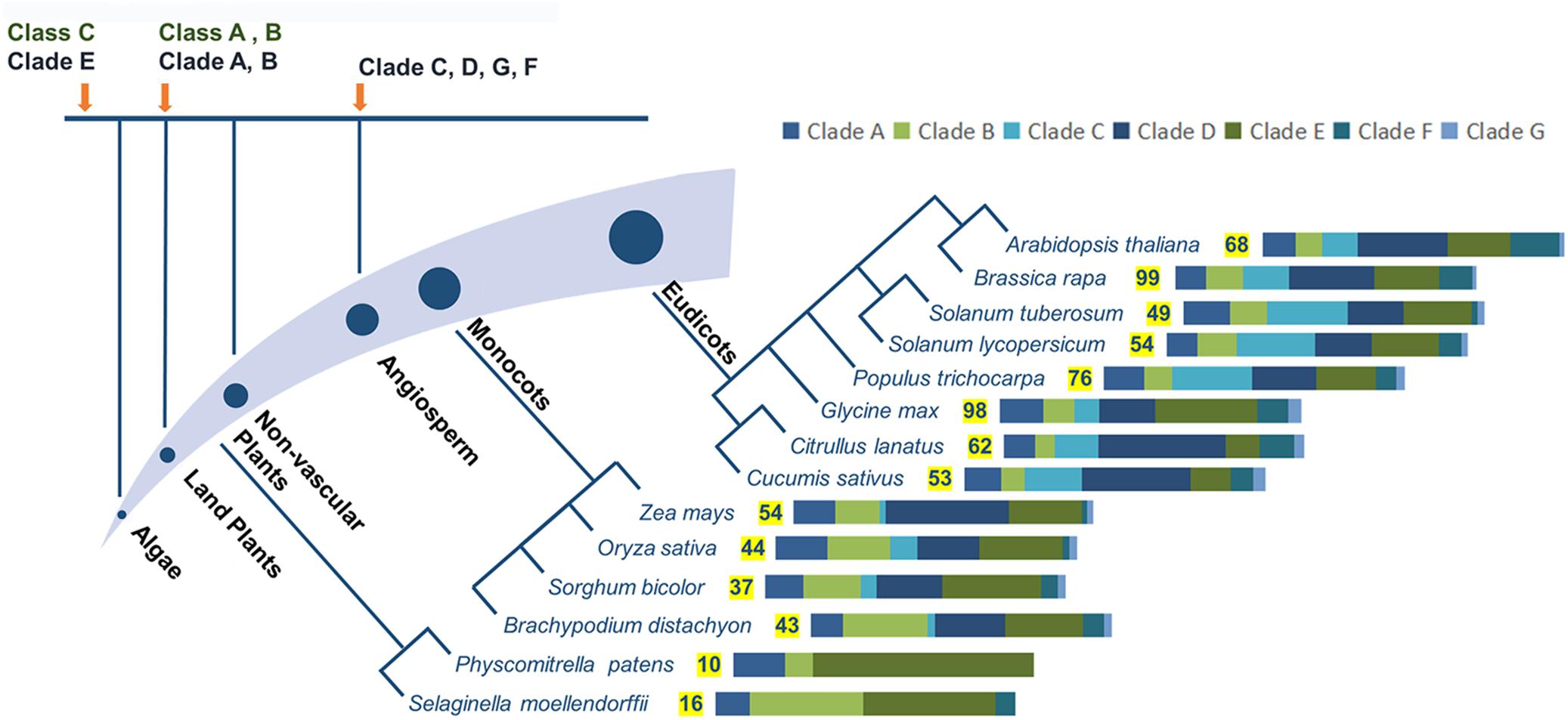
FIGURE 2. Evolutionary analysis of PG genes, numbers of family members, and proportions of different clades in representing species. Different clades or classes emerged at different periods during evolution, with appearance times indicated by orange arrows. PG family members are shown for different species of land plants, with significant changes in the proportions of different clades. Different bar lengths represent different proportions of clades. The numbers of family members in representative species are from reported studies (Tyler et al., 2010; McCarthy et al., 2014; Sénéchal et al., 2014; Yu et al., 2014; Liang et al., 2015, 2016) with some modifications.
The first classification system has the advantage of analyzing large numbers of PG genes, which contributes to a comprehensive understanding of the phylogenetic relationships between genes across multiple species. Neighbor-joining and maximum parsimony algorithms were mainly used in this system, enabling concise and fast calculations to analyze large quantities of sequence data. However, the increased amount of data might lead to lower confidence in the existence of a new clade and reduce statistical accuracy. Based on the first system, the number of PG family members shows an increasing trend from non-vascular plants to monocots to eudicots. Gene duplication and loss are both likely to be contributors to these differences in gene number. For example, more old and recent duplications exist in Clade B in grasses, likely explaining the larger numbers of genes in this family in grasses than in eudicots (Liang et al., 2016). The difference in wall composition between grasses and eudicots, wherein pectins are less abundant in grasses than in eudicots, is another likely explanation for the different scales of PG families (McCarthy et al., 2014; Liang et al., 2016). In comparison, the second system built high-value phylogenetic trees, mainly by applying maximum likelihood algorithms. Although 212 PG genes from five species were successfully classified in the second system (Yang et al., 2013), these time-consuming calculations are inadequate for analyzing complex phylogenetic relationships among PG genes across a large number of species.
Instead, the second system proposed by Kim has the potential to reveal orthologous relationships and ancestral PGs between two species. In total, 21, 39, and 54 common ancestral PG genes were found between Arabidopsis and rice, poplar, and Chinese cabbage, respectively (Kim et al., 2006; Yang et al., 2013; Duan et al., 2016). The difference in ancestral gene number can be explained by the phylogenetic distance between these species. Genes clustered in the same group are orthologs. Therefore, the possible biological functions of candidate PG genes from a given species can be inferred by referring to the function of orthologous genes from other species, if this information is available.
Expression, Functions, and Mechanisms of Action of Polygalacturonases
Expression Patterns of Polygalacturonase Genes
Numerous PG genes have been identified in various species. Here, we summarize PG genes with defined accession numbers and expression patterns as analyzed by qRT-PCR and/or promoter activity (Table 1 and Supplementary Table 1). According to their expression patterns, these genes are divided into five groups: ubiquitously expressed in multiple organs, specifically expressed in flowers and pollen, expressed during fruit ripening, expressed at sites of organ abscission and dehiscence, and expressed in other organs. Expression patterns among different clades/classes and PG genes with close phylogenetic relationships are discussed below.
Polygalacturonase Genes Among Different Clades Are Conserved and Diversified in Expression
Previous studies revealed that the expression patterns of PG genes are conserved in the same clade, but diversified between clades. PG genes in Clades A and B are expressed in fruits and abscission and dehiscence zones, and genes in Clade C are expressed in flowers (Hadfield et al., 1998). This pattern is supported by the expression of most identified genes. PG genes such as PS-2, ADPG1, ADPG2, RDPG1, and SDPG (Table 1 and Supplementary Table 1D) are expressed during organ abscission and dehiscence, and belong to Clade B with the conserved motif FGAKGDG (Rodriguez-Llorente et al., 2004); PG genes such as PcPG1, VvPG1, VvPG2 and MAPG3, which are expressed during fruit ripening, belong to Clade B, and sPG belongs to Clade A (Supplementary Table 1C). PG genes such as BcMF2, BcMF6, BcMF9, BcMF16, BcMF24, and PGA4 (Table 1 and Supplementary Table 1B), are expressed in flowers and pollen and belong to Clade C. However, FaPG1, which belongs to Clade C and has a cysteine residue conserved in pollen-specific genes, is expressed in the middle-late stage of fruit ripening and is involved in flesh softening (Villarreal et al., 2008).
Genes from the same PG clade can also sometimes show distinct expression patterns. Hadfield’s hypothesis that orthologous genes are expressed similarly has proven to be partly incorrect. Expression patterns of PG genes are conserved in Clades D and E, with expression in inflorescences and ubiquitous expression in multiple organs, respectively (Yu et al., 2014; Liang et al., 2015, 2016). These results illustrate that these PG genes play crucial roles during multiple developmental processes. Members of Clades C and F, which are mainly expressed in reproductive organs in grasses and eudicots, are also sometimes expressed during root and seed development. A few eudicot PG genes of Clade C show expression in roots and root nodules. In some members of Clade F, expression can be detected in embryos and developing seeds as well. However, expression patterns are diversified in other clades. Members of Clades A, B, and G are either expressed specifically in different organs or widely across various organs. Some Clade G members, for example Arabidopsis QRT3 and its homologs, are expressed either in one specific organ (such as seeds, stems, flowers, or siliques) or in multiple organs (Liang et al., 2016). Cucumber (Cucumis sativus) PG genes in Clade A are related to fruit development. However, these PG genes are expressed ubiquitously instead of showing fruit-specific expression (Yu et al., 2014).
Based on Kim’s system, Classes B and C have particular expression patterns in moss, lycophytes, rice, Arabidopsis, and poplar. However, members of Class A, which contains PG genes from lycophytes to angiosperms, are expressed with variable patterns (Kim et al., 2006; Yang et al., 2013). The requirement of angiosperm PG genes to function in the flower, which is the unique organ of angiosperms, might partly explain the diverse expression patterns of Class A genes. This diversity also likely relies on lower selective pressure compared with other classes (Yang et al., 2013). Specific expression patterns for genes within Class A, the largest class, can be found in its subgroups (Kim et al., 2006; Duan et al., 2016). To explore the relationship between these two systems of PG classification in a model species, we analyzed the Arabidopsis PG family using Kim’s system (Kim et al., 2006) and Liang’s system (Liang et al., 2015). Interestingly, almost all PG genes within the same subgroup of Kim’s system are grouped into a specific clade of Liang’s system, and different classes are composed of certain clade(s) (Figure 1C). This relationship can explain the specific expression of Classes B and C, which only contain PG genes from the deeply conserved Clade E (Figure 2). The complex composition of Class A accounts for its various expression patterns. Hence, the classification system of Hadfield has some advantages in connecting expression patterns to possible functions.
It is noteworthy that the conservation of expression patterns for PG genes is relative. PG genes expressed in the same general organs can still differ in specific expression patterns when they are studied in more detail. For example, PcPG1 and PcPG3 are both expressed in pear (Pyrus communis) fruit, but are expressed at different stages of fruit ripening and function separately in affecting fruit ripening and flesh texture, respectively (Sekine et al., 2006). Promoter activity analysis of ADPG2 and its three closest homologs reveal that three of these genes are specifically expressed in different organs, and that another one had no detectable expression, even though ADPG2 and its three closest homologs are all grouped into Clade B (González-Carranza et al., 2007). Expression specificity is also supported by the expression patterns of PG genes in other species. For example, 16 fruit-specific PG genes in cucumber can be divided into three subgroups, with highest expression occurring for each subgroup at 3 days after pollination, 6–9 days after pollination, or 27 days after pollination, respectively (Yu et al., 2014). In Arabidopsis, 47 abscission-related PG genes have nine significantly different expression patterns during five stages of floral organ abscission (Kim and Patterson, 2006). The diversity of expression patterns in the same organ implies that these genes play different roles in the development of a single organ, but this idea requires further research to be validated.
Duplicated Polygalacturonase Genes Show Divergent Expression
Duplicated PG genes, which are highly similar in sequence, are more likely to have different expression patterns. These gene pairs result from duplication events such as whole genome duplication (WGD) and tandem duplication (TD) during plant evolution. Two tandemly duplicated PG genes, PpendoPGM and PpendoPGF, promote the softening of peach flesh, but only the latter is involved in stone adhesion (Gu et al., 2016). MaPG1 and MaPG2, of which the cDNA sequences are 98% similar, are expressed in different organs: the former is expressed in roots, stem, leaves, and flowers, whereas the latter is expressed in the late stage of fruit ripening (Mbéguié-A-Mbéguié et al., 2009). PG paralog pairs generated by TD are more likely to be expressed differently than pairs generated by WGD. For example, based on microarray and RNA-seq-based data, paralogous PG pairs with different expression patterns constitute 71.4%, 75%, and 75% of pairs resulting from TD in rice, poplar, and cucumber (Kim et al., 2006; Yang et al., 2013; Yu et al., 2014), whereas PG copies with diversified expression patterns cover 56% and 67.8% of PG copies caused by WGD in poplar and soybean, respectively (Yang et al., 2013; Wang et al., 2016). Expression diversity of duplicated genes is also demonstrated by variable expression between paralogs in Arabidopsis and Chinese cabbage, which underwent WGD twice and tree times, respectively (Liang et al., 2015). The difference in expression of duplicated PG copies highlights the diversity of their functions and exemplifies the duplicate retention mechanisms known as neofunctionalization and subfunctionalization (Innan and Kondrashov, 2010).
Functions of Polygalacturonases in Pectin Degradation, Cell Dynamics, and Plant Development
Polygalacturonases and Pectin Degradation
Homogalacturonan, which is the most abundant pectin domain, has been extensively studied in terms of its synthesis, modification, and degradation. HG is synthesized in the Golgi apparatus, and its polysaccharide chain is extended by galacturonic acid residues being added to the non-reducing end. Under the action of pectin methyltransferases (PMTs) and acetyltransferases (PATs), HG can be modified with methyl-ester or acetyl groups. Newly synthesized HG polymers are apportioned into vesicles and transported to the plasma membrane, and finally are secreted into the apoplast for incorporation into the cell wall (Ridley et al., 2001; Sénéchal et al., 2014).
In the wall, HG can be metabolized by the action of HG modifying-enzymes (HGMEs), and participates in the regulation of plant development and responses to external stimuli. PGs, pectin methylesterases (PMEs), pectin acetylesterases (PAEs), and pectin/pectate lyase-like proteins (PLLs, including pectin lyases and pectate lyases) all are HGMEs. Their coding genes have distinct expression patterns (Sénéchal et al., 2014), and they modify pectin in different ways. PMEs and PAEs remove modifications from the HG backbone, whereas PGs and PLLs cleave or depolymerize HG. More specifically, PMEs control the degree of methylesterification of HG by removing methyl-ester groups, resulting in negatively charged galacturonic acid residues (Jolie et al., 2010). PAEs hydrolyze O-acetyl groups and produce linear HG (Gou et al., 2012). PGs hydrolyze the α-1,4 glycosidic bonds of demethylesterified HG, releasing oligogalacturonides (OGs) or galacturonic acid monomers (Markovič and Janeček, 2001). In contrast to PGs, pectin lyases and pectate lyases can cleave the α-1,4 glycosidic bonds of methylesterified and demethylesterified HG, respectively, by β-elimination (Mayans et al., 1997; Herron et al., 2000).
Models of HG remodeling have been proposed that take distinct cell wall microenvironments into consideration (Sénéchal et al., 2014; Hocq et al., 2017). In an acidic cell wall context, HG that is randomly demethylesterified by acidic PMEs can be a substrate for PGs. Basic PMEs, which can be easily trapped by free carboxyl groups in this circumstance, would inefficiently remove methyl-ester groups from HG. In a slightly alkaline context, H+ diffuse into the cytoplasm by producing IAAH, due to in the absence of auxin (IAA-). As a result, the cell wall can be high in the concentrations of ionic cations. Under this condition, basic PMEs play the main role in HG de-methylesterification, whereas acidic PMEs will be charged and unable to bind HG. Continuous stretches of demethylesterified HG can crosslink via Ca2+ to form stable “egg-box” structures, which restrict cell wall loosening (Figure 3). Other regions of HG can be hydrolyzed by PGs, releasing OGs. HG has been proposed to go through a cycle of crosslinking, modification, cell wall loosening, and cell growth (Sénéchal et al., 2014; Boyer, 2016; Hocq et al., 2017).
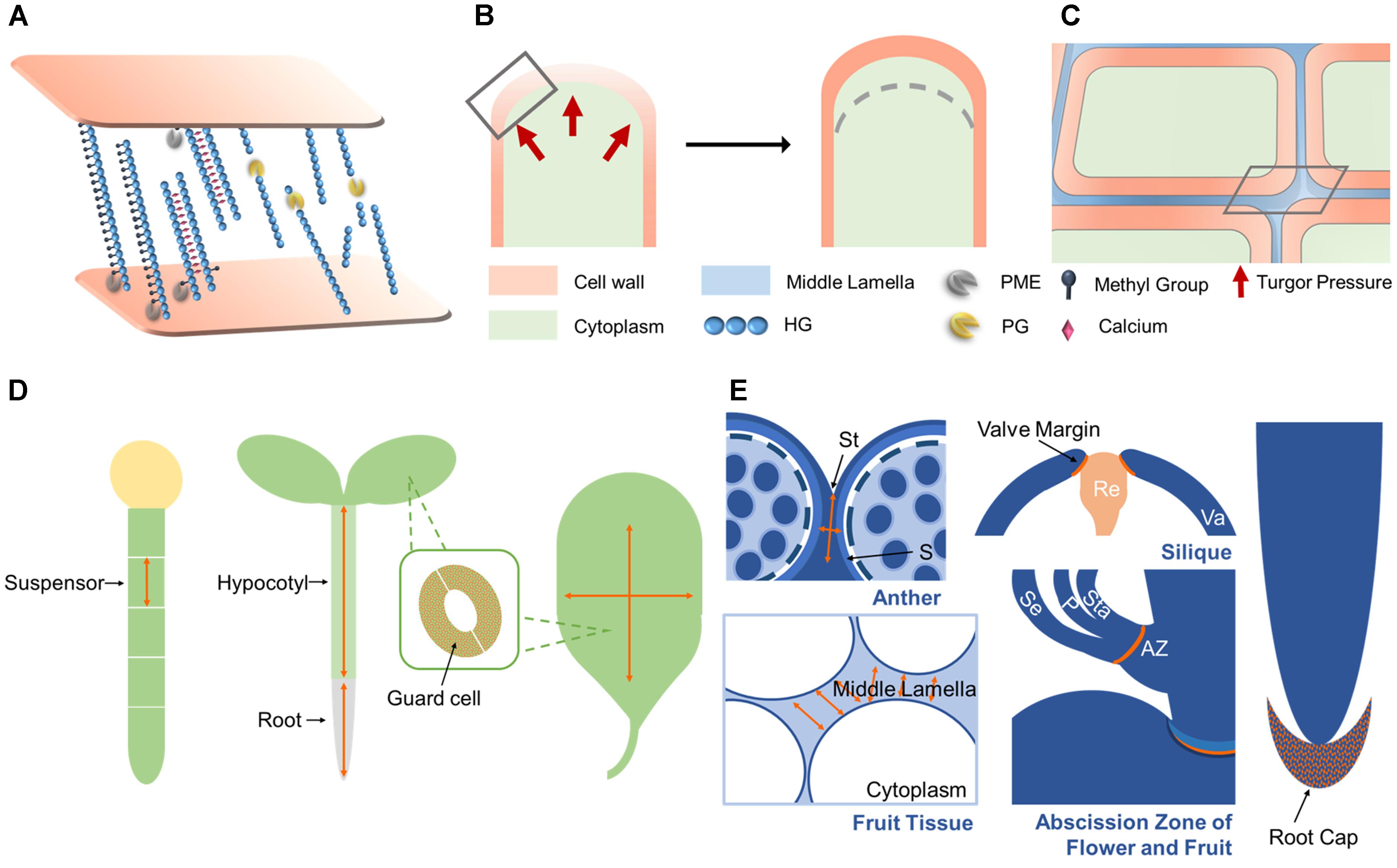
FIGURE 3. PG functions and mechanisms of action. (A) PG genes are involved in suspensor, root, and hypocotyl elongation, leaf expansion, and stomatal dynamics by functioning in cell expansion. (B) PG genes function in root call detachment, anther and silique dehiscence, organ abscission, and fruit softening by functioning in cell separation. (C) During cell expansion, the cell wall loosens due to the hydrolysis of pectins by PGs. Under intracellular turgor pressure, the cell wall expands outward and then cell elongated. (D) During cell separation, PGs function in the degradation of pectins within the middle lamella. As a result, adjacent cells are reduced in adhesion, then finally separate from each other. (E) The subcellular actions in (C,D) marked by gray rectangle are shown here. In the cell wall, de-esterified HG produced by PMEs can be crosslinked by calcium, stiffening the wall, or cleaved by PGs, resulting in wall loosening. In the middle lamella, PGs hydrolyze de-esterified HG in the same way as in the cell wall. Activity sites are noted in orange and the expansion direction is indicated by orange arrows in (A,B). A lighter shade of cell wall and middle lamella means looser wall integrity in (C–E). St, stomium; S, septum; Va, valve; Re, replum; Se, sepal; P, petal; Sta, stamen; AZ, abscission zone.
Polygalacturonase Mechanisms of Action in Cell Dynamics
Cell proliferation, expansion, and separation form the foundation of plant morphogenesis and development. During cell expansion and separation, pectin is degraded in the primary cell wall and middle lamella, decreasing cell wall stiffness and increasing wall fluidity. The identified roles of PGs in plant development reveal that they can function in both cell separation and expansion. Here, we summarize the possible mechanisms of action of PGs in cell dynamics.
Separation between cells in the abscission zone (AZ) results in organ abscission and dehiscence. After receiving the abscission signal in the AZ, the cell wall and middle lamella begin to detach via the action of cell wall degrading enzymes (Figures 3A,C). Cells separate along the middle lamella, allowing for organ separation and dehiscence (Kim et al., 2015). After organ detachment, cells in AZ can elongate, implying that cell elongation is also involved in organ abscission (Patterson and Bleecker, 2004). AZ-expressed PG genes function in root cap detachment, anther and fruit dehiscence, leaf shedding, and fruit maturation, highlighting their central roles in cell separation (Figure 3E). Crosslinks between adjacent cells are formed under the control of Ca2+, and cell adhesion is maintained by HG methylesterification (Daher and Braybrook, 2015). During cell separation, PMEs remove the methyl-ester groups from HG to provide substrates for PGs. PGs can then cleave HG backbones and reduce the HG-mediated cell adhesion that is maintained by Ca2+. Finally, cells detach from each other (Figures 3A,C) (Daher and Braybrook, 2015). This model is supported by the possible role of MdPG1 during leaf shedding. In MdPG1 overexpression lines, more abundant low-esterified pectins are found in AZs with increased PG activity in leaves (Atkinson et al., 2002). Noticeably, tetrad separation is a special case of tissue separation. Instead of cell separation, the release of microspores results from the degradation of the primary pollen mother wall. PGs like QRT3, which are secreted from tapetum cells during early microspore stage, are responsible for micropore separation by degrading this cell wall (Francis et al., 2006).
Cell expansion is required by numerous developmental processes such as suspensor elongation, hypocotyl elongation, stomatal opening and closure, and leaf expansion (Figure 3D). Therefore, PG genes like POLYGALACTURONASE INVOLVED IN EXPANSION (PGX) genes, which affect the elongation and expansion of several organs, might have their mechanism of action explained as facilitating cell expansion (Xiao et al., 2014, 2017; Rui et al., 2017). Lower demethylesterified HG level, higher PG activity and smaller HG size in PGX3 overexpression lines provide further evidence for the functions of PGs in regulating stomata dynamics and leaf expansion (Rui et al., 2017). The aforementioned fact that cells in the AZ elongate after organ shedding (Patterson and Bleecker, 2004) suggests that abscission-related PGs might influence both cell separation and cell expansion. Cell expansion can be regulated by intracellular turgor pressure and mechanical interactions between cell wall components. The direction of cell expansion is influenced by both the orientation of cellulose microfibrils (Cosgrove, 2005; Palin and Geitmann, 2012) and the modification of pectins (Peaucelle et al., 2015). Turgor pressure, in combination with increased cell wall extensibility, is thought to be the driving force for cell expansion (Figure 3B). Pectins affect cell expansion by modulating the mechanical properties of cell walls (Peaucelle et al., 2011; Kozioł et al., 2017). By effecting pectin degradation, PGs are hypothesized to cause a decrease in cell wall stiffness (Figures 3A,B), which is supported by the identified role of PGX2 in regulating the mechanical properties of stems (Xiao et al., 2017). PGs might also function in cell expansion by modulating the directionality of expansion. A mutant Arabidopsis line resistant to the drug cobtorin can suppress the cell-swelling phenotype induced by cobtorin, due to the overexpression of a PG gene in this line. Exogenously supplied PGs can also rescue this phenotype in Arabidopsis, restoring the deposition of cellulose microfibrils in parallel with cortical microtubules with the help of PME (Yoneda et al., 2010). Exactly how PGs might influence expansion direction requires further study.
It is noteworthy that PGs might participate in cell and tissue patterning. PME5, which works upstream of PGs, plays a key role in controlling phyllotactic patterning (Peaucelle et al., 2008). Changes in the degree of methylesterification of pectins can enhance cell wall loosening, which underlies organ initiation (Peaucelle et al., 2011). One PME inhibitor protein, PMEI3, can suppress morphogenesis in inflorescence meristems (Peaucelle et al., 2008). The deformed pollen grains that result from abnormal intine formation in BcMF2, BcMF9 antisense lines indicate that PGs might also function in determining cell morphology during pollen development. However, whether PGs act in organ patterning through regulating cell wall mechanics directly or by changing wall integrity signaling remains unknown.
Biological Functions of Polygalacturonase Genes in Plant Development and Fruit Ripening
The functions of PG genes, which have been identified by mutants or transgenic lines (Table 1) or have in some cases been inferred from their expression patterns (Supplementary Table 1), reveal their significant roles in both vegetative and reproductive development in plants. Ubiquitously expressed PG genes have been identified as functioning in multiple processes during development, which might be important for plant survival. For instance, the expression of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION1 (PGX1) is detected in seedlings, roots, leaves, and flowers (Xiao et al., 2014). Overexpression of this PG leads to significant elongation of etiolated hypocotyls and enhanced rosette leaf expansion. Its functions in cell and tissue expansion are supported by the opposite phenotypes in a pgx1 knockout mutant. In flower development, abnormal angles between flower primordia and extra petals are found in both overexpression lines and mutants, which indicates that this PG might function in floral organ patterning (Xiao et al., 2014). Overexpressing Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 (PGX2) leads to faster flowering and enhanced stem lignification with enhanced tensile stiffness but lower bending stiffness, as well as enhanced etiolated hypocotyl length and rosette leaf area (Xiao et al., 2017). PGX2 represents the first PG gene that has been found to regulate stem lignification. Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION3 (PGX3) also functions in rosette expansion and root elongation (Rui et al., 2017). Overexpressing this PG gene also causes larger stomatal pores in cotyledons, whereas its knockout mutant has the opposite phenotype. Without influencing stomatal dimensions in true leaves, PGX3 asymmetrically affects mature stomatal opening and closure (Rui et al., 2017). Interestingly, despite their common functions in rosette leaf expansion, PGX1, PGX2, and PGX3 are not closely related in the phylogeny of Arabidopsis PGs (McCarthy et al., 2014).
How PG genes influence pollen development and fertilization has been a recent research priority. The expression of PG genes in tapeta, pollen grains, stigmas and pollinated pistils implies their role in tapetum degradation, pollen maturation, pollen tube growth, and pollination, as evinced by their functional characterization (Table 1). Suppressing the expression of QRT3 in Arabidopsis and BcMF6 in Chinese cabbage interferes with microspore separation after the tetrad stage (Rhee et al., 2003; Zhang et al., 2008). In addition, the latter mutant has smaller floral organs and a lower pollen germination rate caused by the disruption of microspore maturation (Zhang et al., 2008). RNA antisense lines of Chinese cabbage BcMF2 and BcMF9 show disturbed development of the pollen wall intine layer and of the pollen tube wall that is the continuation of the intine layer (Huang et al., 2009a,b). The downregulation of BcMF2 causes pollen deformity and balloon-like swelling in the pollen tube tip, along with premature tapetum formation (Huang et al., 2009a). Downregulating BcMF9 influences pollen wall exine layer formation as well (Huang et al., 2009b). When a soybean PG is heterologously overexpressed in Arabidopsis, inflorescence mortality is over 50%, and siliques and seeds significantly decrease in number (Wang et al., 2016). NIMNA, which plays a role in early embryo cells and suspensor elongation, is the only Arabidopsis PG gene that has been identified as functioning in embryo development (Babu et al., 2013).
Early studies examined the relationship between PGs and fruit development and mainly focused on the activity of PG proteins and their responses to exogenous stimuli. In recent studies, fruit-related PG gene functions have gradually attracted more attention, especially in species with edible fruit. The suppression of FaPG1 leads to significant increase in strawberry (Fragaria ananassa) fruit firmness (Pose et al., 2013). A similar change is also found in PG1 suppression lines of apple (Malus × domestica), with an enhancement of cell adhesion (Atkinson et al., 2012). Despite MdPG1 showing fruit-specific expression, its overexpression lines display various novel phenotypes in vegetative growth, such as the silvery colored leaves, earlier leaf shedding, and abnormal stomatal structure (Atkinson et al., 2002). Its function in organ abscission is also supported by heterologous expression studies in Arabidopsis. Overexpressing MdPG1 in Arabidopsis can drive early silique dehiscence, whereas the siliques of MdPG1 antisense lines do not open normally, possibly due to the downregulation of endogenous PGs (Li et al., 2013). In non-softening fruit species like pepper (Capsicum annuum), PG genes perform crucial functions as well. A point mutation in one pepper PG gene (CA10g18920) in wild type downregulates its expression by generating a premature stop code in its 3′ splicer accepted site. This results in lower water-soluble pectin levels compared with a soft flesh mutant, preventing aberrant fruit softening (Kim S. et al., 2014).
Abscission and dehiscence help to remove old, impaired and infected tissues as well as release leaves, floral organs, pollen or seeds, and are of great value in maintaining normal growth and reproduction. PG genes have been found to play a pivotal role in this process (Supplementary Table 1D). Mutation of tomato PS-2 in intron recognition splice sites leads to non-dehiscent anthers and functional sterility (Gorguet et al., 2009). Similarly, an Arabidopsis adpg1 adpg2 qrt2 triple mutant is delayed in anther dehiscence (Ogawa et al., 2009). This phenotype is not found in double mutants or single mutants, indicating that anther dehiscence is co-regulated by these three PG genes. However, non-dehiscent siliques are found in single mutants of adpg1, adpg2 and more prevalently in an adpg1 adpg2 double mutant. ADPG1 and ADPG2 potentially function in silique dehiscence by hydrolyzing only a small amount of pectin (Ogawa et al., 2009).
Polygalacturonase genes also participate in biotic and abiotic stress responses in plants. These findings have been nicely summarized by Sénéchal et al. (2014). When treating strawberry fruit with resistance inducers like chitosan, benzothiadiazole or a mixture of calcium and organic acids, the expression of PG genes changes significantly (Landi et al., 2014). In the same study, when stimulated by abiotic stresses, the expression of PG genes is up-regulated, which might elicit responses to environmental stresses (Liu et al., 2014).
Regulatory Networks of Polygalacturonase Genes and Encoded Proteins
Regulation of Polygalacturonase Gene Expression
The expression patterns of PG genes are regulated by various factors such as hormones, transcription factors, and environmental factors. These factors can function separately or be combined to form regulatory networks. Based on reported gene regulation in processes like lateral root initiation, root cap detachment, fruit ripening, and organ abscission, we summarize the regulatory network of PG genes in cell separation in Figure 4.
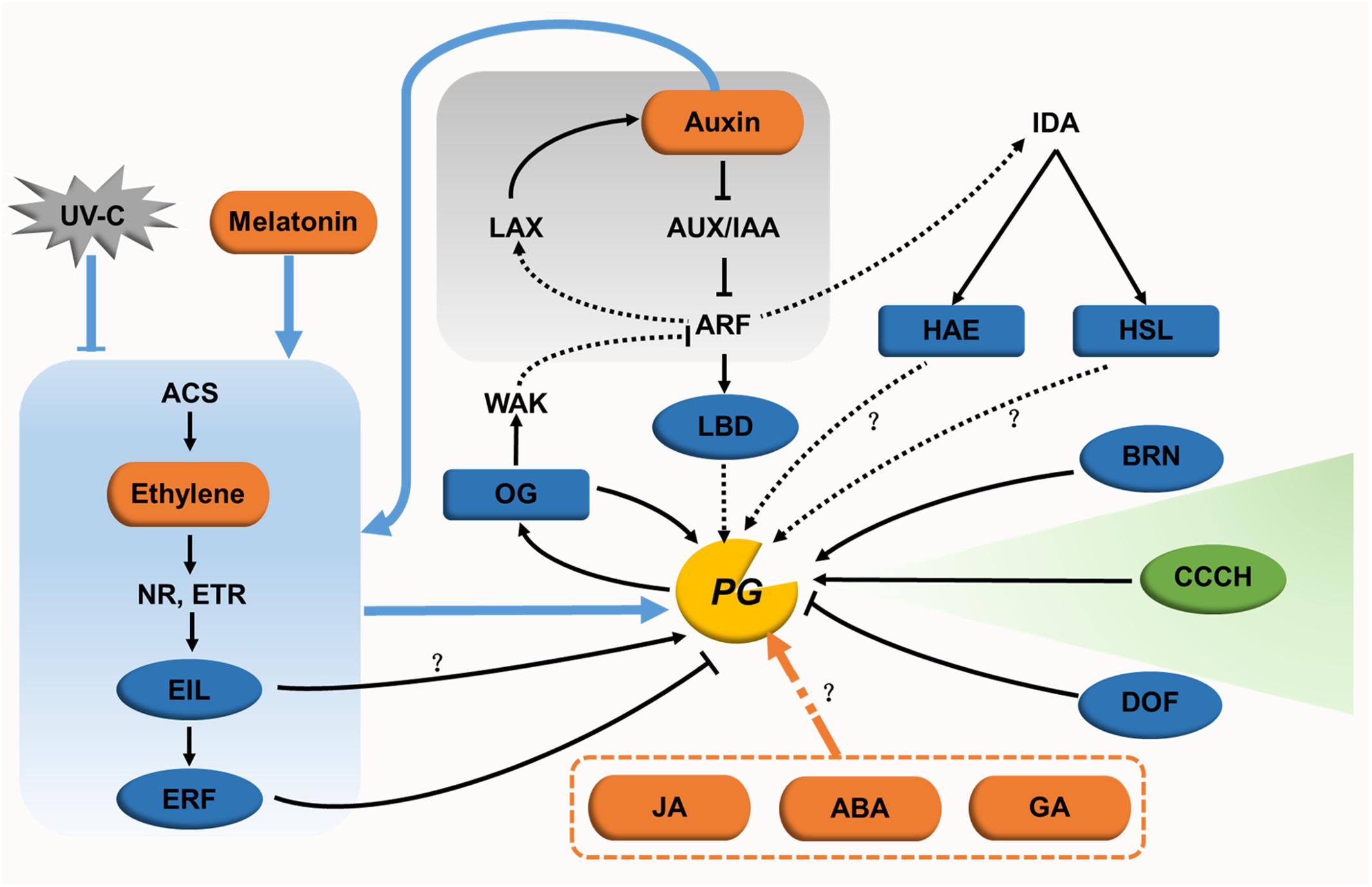
FIGURE 4. Regulatory networks for PGs in cell expansion and separation. In cell expansion, only one regulator (green oval) has been identified. The regulatory network for this cell dynamic processes remain unclear. In cell separation, hormones (orange ovals) and their signaling pathways (with gray or blue background), transcription factors (blue ovals), other endogenous factors (blue rectangles), and the exogenous factor UV-C (gray polygon) form a genetic regulatory network for PGs. The direct or indirect action of some regulators needs to be further studied (black dotted lines). The regulatory pathways of JA, ABA, and GA in relation to PGs also need to be characterized further.
Lateral root initiation requires the primordium to emerge successively through the endodermal layer, cortical layer, and epidermal layer. PGs likely act to separate cells in the cortical and epidermal layers, contributing to the elongation of the primordium (Swarup et al., 2008; Kumpf et al., 2013). During this stage, a regulatory network (Figure 4) is formed centering on the auxin signal pathway and connecting with transcription factors, receptor-like kinases and OGs. Auxin influx carrier LIKE-AUXIN3 (LAX3) and its downstream auxin-inducing transcription factor LATERAL ORGAN BOUNDARIES DOMAIN18 (LBD18) indirectly increases the expression of AtPG10 (Swarup et al., 2008; Kumpf et al., 2013; Lee et al., 2015). INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) is the key regulator of cell separation, the expression of which is induced by AUXIN RESPONSE FACTOR7. After receiving a signal from IDA, the receptor-like kinase HAESA promotes the expression of ADPG2 in lateral root development (Kumpf et al., 2013). However, whether HAESA regulates ADPG2 directly has not been reported. OGs, the product of pectin degradation by PGs, might in turn control PG gene expression (Moscatiello et al., 2006). Exogenous OGs inhibit lateral root formation by suppressing the expression of auxin synthesis- and signaling-associated genes, such as IAA5 and SAUR16 (Savatin et al., 2011). Taking these results together, the interaction between auxin and PG-induced OGs is assumed to form a feedback regulation network (Ferrari et al., 2013). In this model, PG expression is induced by auxin, and the resulting PG proteins then release OGs in the apoplast. The accumulation of OGs in turn perturbs auxin responses. In root cap detachment, the transcription factor BEARSKIN1 directly binds to the ROOT CAP POLYGALACTURONASE (RCPG) promoter at a region containing a NAC-BINDING ELEMENT motif. The activation of RCPG accelerates cell detachment at the root cap (Kamiya et al., 2016).
In the process of fruit ripening, ethylene synthesis and signaling are crucial for regulating PG expression. Upon treatment with exogenous ethylene in the full bloom stage, expression of apple PG1 is upregulated in the pericarp, leading to fruit softening (Tacken et al., 2010). The same expression pattern is found for the ethylene synthesis gene ACC OXIDASE1, which indicates that ethylene might regulate PG1 expression. This idea was confirmed by the finding that EIN-LIKE3 transactivates the expression of PG1 in transient assays (Tacken et al., 2010). Furthermore, ETHYLENE RESPONSE FACTOR9 (ERF9) in papaya (Carica papaya), which functions downstream of EIN3-LIKE, represses the expression of CpPG5 in fruit ripening by specifically binding to its promoter at a GCC-BOX motif. Its repressive role relies on the ERF-ASSOCIATED AMPHIPHILIC REPRESSION motif within ERF9 (Fu et al., 2016). Auxin functions in this process in concert with ethylene. With naphthalene acetic acid (NAA) treatment, the effect of an ethylene repressor is enhanced in downregulating the expression of ethylene-independent MdPG2 in the cortex and abscission layer, and ethylene-dependent MdPG1 in the abscission layer as well. However, NAA conversely upregulates the expression of MdPG1 in the cortex, which results in the increased accumulation of ethylene in this layer (Li and Yuan, 2008). Apart from auxin, UV-C and melatonin also interact with ethylene in regulating PG genes. Treating postharvest tomato fruits with UV-C decreases the synthesis of ethylene, and suppresses the expression of PG genes together with other cell wall modifying genes during fruit softening (Bu et al., 2013). These data reveal that the regulatory mode of UV-C may be through the action of ethylene. Melatonin influences ethylene signaling in postharvest tomato ripening by promoting the expression of ethylene related genes (such as NR, ETR4, EILs, and ERF2), causing the upregulation of PG genes (Sun et al., 2015). ABA and GA also take part in this regulatory network. ABA treated tomato is downregulated in the expression of SlPG and other genes encoding wall-modifying enzymes, showing a delay in fruit softening (Sun et al., 2012). However, strawberry fruit treated with ABA showed slightly increased FaPG1 expression without influencing its protein activity (Villarreal et al., 2009). GA affects flower blooming and fruit maturity in grapes (Vitis labrusca × V. vinifera) with two 230-fold upregulated PG genes (Cheng et al., 2015). However, the regulatory pathways for these two hormones need to be studied further.
During organ abscission, jasmonic acid (JA), transcription factors, and receptor-like kinases are involved in regulating PG expression. Microarray transcriptome data of a JA mutant treated with JA show that the expression of ADPG1, ADPG2, and QRT2 are upregulated 10-fold upon JA treatment (Mandaokar et al., 2006). GUS activity of the QRT2 promoter is detected in the anther of a JA mutant treated with methyl jasmonate (Ogawa et al., 2009). Taken together with the delayed dehiscence of anthers and siliques in the adpg1 adpg2 qrt2 mutant, the non-dehiscent anther of the JA mutant may be caused by decreased PG gene expression (Ogawa et al., 2009). Additionally, the transcription of ADPG2 in the AZ might be regulated directly via the binding of the transcription factor FOR DNA BINDING WITH ONE FINGER4.7 to the AAAG cis-regulatory element of its promoter (Wei et al., 2010). During lateral root emergence, both HAESA and HAESA-LIKE2 act downstream of IDA. These two kinases promote the expression of ADPG2 in floral organ AZ via regulatory mechanisms that are currently unclear (González-Carranza et al., 2012).
Surprisingly little is known about the regulation of PG expression in cell expansion. Only a plant-specific tandem CCCH zinc-finger C3H14 in Arabidopsis has been demonstrated to affect cell elongation by binding to the AU-RICH ELEMENT motif of the ADPG1 promoter and activating its transcription (Kim W.C. et al., 2014). The roles of hormones, other transcription factors and environmental factors in regulating PG-mediated cell expansion need to be studied further.
Regulation of Polygalacturonase Biochemical Activities
External factors, other proteins, and the apoplastic microenvironment can all act as regulators of PG biochemical activities. PG activities can be controlled by temperature, pressure, exogenous polyamine and expansin (Crelier et al., 2001; Fachin et al., 2002; Martinez-Tellez et al., 2002; Karakurt and Huber, 2003; Yuan et al., 2011; Nardi et al., 2013). UV-C, which was mentioned above as a transcriptional inhibitor, delays tomato fruit softening by negatively affecting PG activities (Barka et al., 2000). Hormones can also influence PG activities. During strawberry fruit ripening, exogenous NAA and an inhibitor of ethylene perception (1-MCP) decrease the expression of FaPG1 and the activity of FaPG1, resulting in the delay of ripening. However, this phenotype is not necessarily caused by the difference in gene expression, because treatment with GA3 reduces the activity of FaPG1 without significantly changing gene expression (Villarreal et al., 2009).
The wall microenvironment influences PG activities mainly through cations and pH. Ca2+ deficiency in potato leaves leads to a dramatic increase in PG enzyme activity (Seling et al., 2000), and the inhibitory role of Ca2+ for PG activities was also supported by treating grape berries with 1 mM Ca2+ (Cabanne and Doneche, 2001). In mango (Mangifera indica) fruit, cations like Li+, Mg2+, and a high substrate (polygalacturonic acid) concentration suppress PG activities, whereas Fe3+ activates PG activities (Prasanna et al., 2006). PGs show high activity within physiological pH ranges. Two PG isoforms isolated from avocado (Persea americana) fruit mesocarp show highest activity when the pH is 6.0 (Wakabayashi and Huber, 2001). The influence of pH on PG activity can be altered by intercellular cations. The optimal pH for PG activity in tomato is 4–4.5 with NaCl, and changes to 5–6 with KCl (Chun and Huber, 1998). pH and Ca2+ are also likely to play complex roles in pectin degradation by influencing the action of PMEs and the formation of PG substrates.
Posttranslational modifications and interacting proteins can also influence PG activities. Polygalacturonase inhibiting proteins (PGIPs) can specifically bind and inhibit PG activities. Studies on the PGIP–PG interaction have been well summarized before (Kalunke et al., 2015). The β subunit of a tomato fruit polygalacturonase inhibits the interaction between PG and pectins (Zheng et al., 1992). To figure out whether a posttranslational modification works in affecting PG activities, phenylalanine residues within the FxxY sequence in a β subunit were modified to α, β-didehydro-Phe (Sergeant et al., 2017). This modification promoted βPG binding to pectins. It also contributed to the interaction of βPG with the catalytic subunit of PG (Sergeant et al., 2017). In addition, an 8-kD non-specific lipid transfer protein, ACTIVATOR, which is contained in a PG1 multiprotein complex, might change the form of associated PG into a heat-stable and active one (Tomassen et al., 2007). N-terminal prosequences have been hypothesized to control PG activity through influencing protein folding or protein inactivation (Hadfield and Bennett, 1998; Torki et al., 2000), but in at least some cases, the N-terminal prosequence instead controls the secretion of PG to the cell wall, as demonstrated in Pichia pastoris and Arabidopsis (Dal et al., 2001; Babu et al., 2013). To explore the posttranslational modifications of PG activity, an advanced approach for producing custom-folded proteins is required for further studies.
Conclusion
Major advances in our knowledge of the expression patterns, biological functions, and mechanisms of action of PGs have deepened our understanding of their roles in vegetative growth, pollen development, fruit ripening, and organ abscission. Although PGs are not the only pectin hydrolases, these studies provide convincing evidence for their unique importance in plant development. Based on genome data and current classification methods, we have a much clearer knowledge of PG structural characteristics, evolution, and expression patterns. However, shortcomings exist for each classification system, which calls for a better system that combines convenient calculations of phylogenetic relationships with high confidence values when dealing with PG families at large scales. PG genes with diverse expression patterns in the same tissues need to have their biological functions pinpointed for a better understanding of their individual activities and retention mechanisms. To illuminate the ways in which PGs function in developmental processes, a comprehensive regulatory network needs to be built based on well-studied pathways that regulate PG expression and activity. As a wider suite of dyes and antibodies become available for observing pectin dynamics (Mravec et al., 2014, 2017; Pattathil et al., 2015; Anderson, 2016; Rydahl et al., 2018; Voiniciuc et al., 2018), we can dive into the mechanisms of PG action in cell wall dynamics. These and other studies can help us gain insights into the role of pectins in plant cell wall construction and dynamics as well as their relationships with other cell wall components.
Author Contributions
YgY, YjY, and YL wrote the manuscript. CA and JC revised the paper. All the authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31471877 and 31501769) and the Natural Science Foundation of Zhejiang Province (Grant No. LQ16C150005). The contributions of CA to this work were supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the United States Department of Energy, Office of Science, Basic Energy Sciences (Award No. DE-SC0001090).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01208/full#supplementary-material
References
Abbott, D. W., and Boraston, A. B. (2007). The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 368, 1215–1222. doi: 10.1016/j.jmb.2007.02.083
Albersheim, P., Darvill, A. G., O’Neill, M. A., Schols, H. A., and Voragen, A. G. J. (1996). An hypothesis: the same six polysaccharides are components of the primary cell walls of all higher plants. Biotechnol. Prog. 14, 47–55. doi: 10.1016/S0921-0423(96)80245-0
Allen, R. L., and Lonsdale, D. M. (1993). Molecular characterization of one of the maize polygalacturonase gene family members which are expressed during late pollen development. Plant J. 3, 261–271. doi: 10.1111/j.1365-313X.1993.tb00177.x
Anderson, C. T. (2016). We be Jammin’: an update on pectin biosynthesis, trafficking and dynamics. J. Exp. Bot. 67, 495–502. doi: 10.1093/jxb/erv501
Aouali, N., Laporte, P., and Clement, C. (2001). Pectin secretion and distribution in the anther during pollen development in Lilium. Planta 213, 71–79. doi: 10.1007/s004250000469
Atkinson, R. G., Schroder, R., Hallett, I. C., Cohen, D., and MacRae, E. A. (2002). Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 129, 122–133. doi: 10.1104/pp.010986
Atkinson, R. G., Sutherland, P. W., Johnston, S. L., Gunaseelan, K., Hallett, I. C., Mitra, D., et al. (2012). Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 12:129. doi: 10.1186/1471-2229-12-129
Babu, Y., Musielak, T., Henschen, A., and Bayer, M. (2013). Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiol. 162, 1448–1458. doi: 10.1104/pp.113.217166
Barka, E. A., Kalantari, S., Makhlouf, J., and Arul, J. (2000). Impact of UV-C irradiation on the cell wall-degrading enzymes during ripening of tomato (Lycopersicon esculentum L.) fruit. J. Agric. Food Chem. 48, 667–671. doi: 10.1021/jf9906174
Boyer, J. S. (2016). Enzyme-less growth in Chara and terrestrial plants. Front. Plant Sci. 7:866. doi: 10.3389/fpls.2016.00866
Braybrook, S. A., and Joensson, H. (2016). Shifting foundations: the mechanical cell wall and development. Curr. Opin. Plant Biol. 29, 115–120. doi: 10.1016/j.pbi.2015.12.009
Bu, J., Yu, Y., Aisikaer, G., and Ying, T. (2013). Postharvest UV-C irradiation inhibits the production of ethylene and the activity of cell wall-degrading enzymes during softening of tomato (Lycopersicon esculentum L.) fruit. Postharvest Biol. Technol. 86, 337–345. doi: 10.1016/j.postharvbio.2013.07.026
Bussink, H. J. D., Buxton, F. P., and Visser, J. (1991). Expression and sequence comparison of the Aspergillus niger and Aspergillus tubingensis genes encoding polygalacturonase-II. Curr. Genet. 19, 467–474. doi: 10.1007/BF00312738
Cabanne, C., and Doneche, B. (2001). Changes in polygalacturonase activity and calcium content during ripening of grape berries. Am. J. Enol. Vitic. 52, 331–335.
Cheng, C., Jiao, C., Singer, S. D., Gao, M., Xu, X., Zhou, Y., et al. (2015). Gibberellin-induced changes in the transcriptome of grapevine (Vitis labrusca x V-vinifera) cv. Kyoho flowers. BMC Genomics 16:128. doi: 10.1186/s12864-015-1324-8
Chiang, J. Y., Shu, S. W., Ko, C. W., and Wang, C. S. (2006). Biochemical characterization of a pollen-specific cDNA encoding polygalacturonase in Lilium longiflorum. Plant Sci. 170, 433–440. doi: 10.1016/j.plantsci.2005.09.010
Choi, J. K., Lee, B. H., Chae, C. H., and Shin, W. (2004). Computer modeling of the rhamnogalacturonase - ”Hairy” pectin complex. Proteins 55, 22–33. doi: 10.1002/prot.10434
Chun, J. P., and Huber, D. J. (1998). Polygalacturonase-mediated solubilization and depolymerization of pectic polymers in tomato fruit cell walls - Regulation by pH and ionic conditions. Plant Physiol. 117, 1293–1299. doi: 10.1104/pp.117.4.1293
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell. Bio. 6, 850–861. doi: 10.1038/nrm1746
Costa, F., Peace, C. P., Stella, S., Serra, S., Musacchi, S., Bazzani, M., et al. (2010). QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus x domestica Borkh.). J. Exp. Bot. 61, 3029–3039. doi: 10.1093/jxb/erq130
Crelier, S., Robert, M. C., Claude, J., and Juillerat, M. A. (2001). Tomato (Lycopersicon esculentum) pectin methylesterase and polygalacturonase behaviors regarding heat- and pressure-induced inactivation. J. Agric. Food Chem. 49, 5566–5575. doi: 10.1021/jf010202u
Daher, F. B., and Braybrook, S. A. (2015). How to let go: pectin and plant cell adhesion. Front. Plant Sci. 6:523. doi: 10.3389/fpls.2015.00523
Dal, F. D., Child, R., Svendsen, I., and Ulvskov, P. (2001). The cleavable N-terminal domain of plant endopolygalacturonases from clade B may be involved in a regulated secretion mechanism. J. Biol. Chem. 276, 35297–35304. doi: 10.1074/jbc.M102136200
Damak, N., Abdeljalil, S., Taeib, N. H., and Gargouri, A. (2015). Cloning and genomic organization of a rhamnogalacturonase gene from locally isolated strain of Aspergillus niger. Appl. Biochem. Biotechnol. 176, 2314–2327. doi: 10.1007/s12010-015-1720-1
Dardelle, F., Lehner, A., Ramdani, Y., Bardor, M., Lerouge, P., Driouich, A., et al. (2010). Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol. 153, 1563–1576. doi: 10.1104/pp.110.158881
Duan, W., Huang, Z., Song, X., Liu, T., Liu, H., Hou, X., et al. (2016). Comprehensive analysis of the polygalacturonase and pectin methylesterase genes in Brassica rapa shed light on their different evolutionary patterns. Sci. Rep. 6:25107. doi: 10.1038/srep25107
Fachin, D., Van Loey, A., VanLoeyIndrawati, A., Ludikhuyze, L., and Hendrickx, M. (2002). Thermal and high-pressure inactivation of tomato polygalacturonase: a kinetic study. J. Food Sci. 67, 1610–1615. doi: 10.1111/j.1365-2621.2002.tb08692.x
Ferrari, S., Savatin, D. V., Sicilia, F., Gramegna, G., Cervone, F., and De Lorenzo, G. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49. doi: 10.3389/fpls.2013.00049
Francis, K. E., Lam, S. Y., and Copenhaver, G. P. (2006). Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142, 1004–1013. doi: 10.1104/pp.106.085274
Fu, C. C., Han, Y. C., Qi, X. Y., Shan, W., Chen, J. Y., Lu, W. J., et al. (2016). Papaya CpERF9 acts as a transcriptional repressor of cell-wall-modifying genes CpPME1/2 and CpPG5 involved in fruit ripening. Plant Cell Rep. 35, 2341–2352. doi: 10.1007/s00299-016-2038-3
González-Carranza, Z. H., Elliott, K. A., and Roberts, J. A. (2007). Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58, 3719–3730. doi: 10.1093/jxb/erm222
González-Carranza, Z. H., Shahid, A. A., Zhang, L., Liu, Y., Ninsuwan, U., and Roberts, J. A. (2012). A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol. 160, 1342–1356. doi: 10.1104/pp.112.205955
Gorguet, B., Schipper, D., van Lammeren, A., Visser, R. G. F., and van Heusden, A. W. (2009). ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor. Appl. Genet. 118, 1199–1209. doi: 10.1007/s00122-009-0974-9
Gou, J. Y., Miller, L. M., Hou, G., Yu, X. H., Chen, X. Y., and Liu, C. J. (2012). Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 24, 50–65. doi: 10.1105/tpc.111.092411
Gu, C., Wang, L., Wang, W., Zhou, H., Ma, B., Zheng, H., et al. (2016). Copy number variation of a gene cluster encoding endopolygalacturonase mediates flesh texture and stone adhesion in peach. J. Exp. Bot. 67, 1993–2005. doi: 10.1093/jxb/erw021
Hadfield, K. A., and Bennett, A. B. (1998). Polygalacturonases: many genes in search of a function. Plant Physiol. 117, 337–343. doi: 10.1104/pp.117.2.337
Hadfield, K. A., Rose, J. K. C., Yaver, D. S., Berka, R. M., and Bennett, A. B. (1998). Polygalacturonase gene expression in ripe melon fruit supports a role for polygalacturonase in ripening-associated pectin disassembly. Plant Physiol. 117, 363–373. doi: 10.1104/pp.117.2.363
Herron, S. R., Benen, J. A. E., Scavetta, R. D., Visser, J., and Jurnak, F. (2000). Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 97, 8762–8769. doi: 10.1073/pnas.97.16.8762
Hocq, L., Pelloux, J., and Lefebvre, V. (2017). Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 22, 20–29. doi: 10.1016/j.tplants.2016.10.009
Huang, L., Cao, J., Zhang, A., Ye, Y., Zhang, Y., and Liu, T. (2009a). The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J. Exp. Bot. 60, 301–313. doi: 10.1093/jxb/ern295
Huang, L., Ye, Y., Zhang, Y., Zhang, A., Liu, T., and Cao, J. (2009b). BcMF9, a novel polygalacturonase gene, is required for both Brassica campestris intine and exine formation. Ann. Bot. 104, 1339–1351. doi: 10.1093/aob/mcp244
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11, 97–108. doi: 10.1038/nrg2689
Irshad, M., Canut, H., Borderies, G., Pont-Lezica, R., and Jamet, E. (2008). A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol. 8:94. doi: 10.1186/1471-2229-8-94
Jenkins, J., Mayans, O., and Pickersgill, R. (1998). Structure and evolution of parallel beta-helix proteins. J. Struct. Biol. 122, 236–246. doi: 10.1006/jsbi.1998.3985
Jolie, R. P., Duvetter, T., Van Loey, A. M., and Hendrickx, M. E. (2010). Pectin methylesterase and its proteinaceous inhibitor: a review. Carbohyd. Res. 345, 2583–2595. doi: 10.1016/j.carres.2010.10.002
Kalaitzis, P., Solomos, T., and Tucker, M. L. (1997). Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 113, 1303–1308. doi: 10.1104/pp.113.4.1303
Kalunke, R. M., Tundo, S., Benedetti, M., Cervone, F., De Lorenzo, G., and D’Ovidio, R. (2015). An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 6:146. doi: 10.3389/fpls.2015.00146
Kamiya, M., Higashio, S. Y., Isomoto, A., Kim, J. M., Seki, M., Miyashima, S., et al. (2016). Control of root cap maturation and cell detachment by BEARSKIN transcription factors in Arabidopsis. Development 143, 4063–4072. doi: 10.1242/dev.142331
Karakurt, Y., and Huber, D. J. (2003). Activities of several membrane and cell-wall hydrolases, ethylene biosynthetic enzymes, and cell wall polyuronide degradation during low-temperature storage of intact and fresh-cut papaya (Carica papaya) fruit. Postharvest Biol. Technol. 28, 219–229. doi: 10.1016/S0925-5214(02)00177-1
Kim, J., and Patterson, S. E. (2006). Expression divergence and functional redundancy of polygalacturonases in floral organ abscission. Plant Signal. Behav. 1, 281–283. doi: 10.4161/psb.1.6.3541
Kim, J., Shiu, S. H., Thoma, S., Li, W. H., and Patterson, S. E. (2006). Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 7:R87. doi: 10.1186/gb-2006-7-9-r87
Kim, J., Sundaresan, S., Philosoph-Hadas, S., Yang, R., Meir, S., and Tucker, M. L. (2015). Examination of the abscission-associated transcriptomes for soybean, tomato, and Arabidopsis highlights the conserved biosynthesis of an extensible extracellular matrix and boundary layer. Front. Plant Sci. 6:1109. doi: 10.3389/fpls.2015.01109
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kim, W. C., Kim, J. Y., Ko, J. H., Kang, H., Kim, J., and Han, K. H. (2014). AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mRNAs in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. Plant J. 80, 772–784. doi: 10.1111/tpj.12667
Kluskens, L. D., van Alebeek, G., Walther, J., Voragen, A. G. J., de Vos, W. M., and van der Oost, J. (2005). Characterization and mode of action of an exopolygalacturonase from the hyperthermophilic bacterium Thermotoga maritima. FEBS J. 272, 5464–5473. doi: 10.1111/j.1742-4658.2005.04935.x
Kozioł, A., Cybulska, J., Pieczywek, P. M., and Zdunek, A. (2017). Changes of pectin nanostructure and cell wall stiffness induced in vitro by pectinase. Carbohyd. Polym. 161, 197–207. doi: 10.1016/j.carbpol.2017.01.014
Kumpf, R. P., Shi, C. L., Larrieu, A., Sto, I. M., Butenko, M. A., Peret, B., et al. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. U.S.A. 110, 5235–5240. doi: 10.1073/pnas.1210835110
Landi, L., Feliziani, E., and Romanazzi, G. (2014). Expression of defense genes in strawberry fruits treated with different resistance inducers. J. Agric. Food Chem. 62, 3047–3056. doi: 10.1021/jf404423x
Lee, H. W., Cho, C., and Kim, J. (2015). Lateral organ boundaries Domain16 and 18 Act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 168, 1792–U1177. doi: 10.1104/pp.15.00578
Lester, D. R., Sherman, W. B., and Atwell, B. J. (1996). Endopolygalacturonase and the Melting flesh (M) locus in peach. J. Am. Soc. Hortic. Sci. 121, 231–235.
Li, J., and Yuan, R. (2008). NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. J. Plant Growth Regul. 27, 283–295. doi: 10.1007/s00344-008-9055-6
Li, J., Zhu, H., and Yuan, R. (2010). Profiling the expression of genes related to ethylene biosynthesis, ethylene perception, and cell wall degradation during fruit abscission and fruit ripening in apple. J. Am. Soc. Hortic. Sci. 135, 391–401.
Li, M., Zhang, Y., Zhang, Z., Ji, X., Zhang, R., Liu, D., et al. (2013). Hypersensitive ethylene signaling and ZMdPG1 expression dead to fruit softening and dehiscence. PLoS One 8:e58745. doi: 10.1371/journal.pone.0058745
Liang, Y., Yu, Y., Cui, J., Lyu, M., Xu, L., and Cao, J. (2016). A comparative analysis of the evolution, expression, and cis-regulatory element of polygalacturonase genes in grasses and dicots. Funct. Integr. Genomics 16, 641–656. doi: 10.1007/s10142-016-0503-2
Liang, Y., Yu, Y., Shen, X., Dong, H., Lyu, M., Xu, L., et al. (2015). Dissecting the complex molecular evolution and expression of polygalacturonase gene family in Brassica rapa ssp chinensis. Plant Mol. Biol. 89, 629–646. doi: 10.1007/s11103-015-0390-2
Liu, H., Ma, Y., Chen, N., Guo, S., Liu, H., Guo, X., et al. (2014). Overexpression of stress-inducible OsBURP16, the beta subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 37, 1144–1158. doi: 10.1111/pce.12223
Mandaokar, A., Thines, B., Shin, B., Lange, B. M., Choi, G., Koo, Y. J., et al. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46, 984–1008. doi: 10.1111/j.1365-313X.2006.02756.x
Markovič, O., and Janeček, S. (2001). Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng. 14, 615–631. doi: 10.1093/protein/14.9.615
Martinez-Tellez, M. A., Ramos-Clamont, M. G., Gardea, A. A., and Vargas-Arispuro, I. (2002). Effect of infiltrated polyamines on polygalacturonase activity and chilling injury responses in zucchini squash (Cucurbita pepo L.). Biochem. Biophys. Res. Commun. 295, 98–101. doi: 10.1016/S0006-291X(02)00631-9
Mayans, O., Scott, M., Connerton, I., Gravesen, T., Benen, J., Visser, J., et al. (1997). Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure 5, 677–689. doi: 10.1016/S0969-2126(97)00222-0
Mbéguié-A-Mbéguié, D., Hubert, O., Baurens, F. C., Matsumoto, T., Chillet, M., Fils-Lycaon, B., et al. (2009). Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. J. Exp. Bot. 60, 2021–2034. doi: 10.1093/jxb/erp079
McCarthy, T. W., Der, J. P., Honaas, L. A., dePamphilis, C. W., and Anderson, C. T. (2014). Phylogenetic analysis of pectin-related gene families in Physcomitrella patens and nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol. 14:79. doi: 10.1186/1471-2229-14-79
Moscatiello, R., Mariani, P., Sanders, D., and Maathuis, F. J. M. (2006). Transcriptional analysis of calcium-dependent and calcium-independent signalling pathways induced by oligogalacturonides. J. Exp. Bot. 57, 2847–2865. doi: 10.1093/jxb/erl043
Mravec, J., Kračun, S. K., Rydahl, M. G., Westereng, B., Miart, F., Clausen, M. H., et al. (2014). Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development 141, 4841–4850. doi: 10.1242/dev.113365
Mravec, J., Kracun, S. K., Rydahl, M. G., Westereng, B., Pontiggia, D., De Lorenzo, G., et al. (2017). An oligogalacturonide-derived molecular probe demonstrates the dynamics of calcium-mediated pectin complexation in cell walls of tip-growing structures. Plant J. 91, 534–546. doi: 10.1111/tpj.13574
Mutter, M., Beldman, G., Pitson, S. M., Schols, H. A., and Voragen, A. G. J. (1998). Rhamnogalacturonan alpha-D-galactopyranosyluronohydrolase - An enzyme that specifically removes the terminal nonreducing galacturonosyl residue in rhamnogalacturonan regions of pectin. Plant Physiol. 117, 153–163. doi: 10.1104/pp.117.1.153
Nakashima, J., Endo, S., and Fukuda, H. (2004). Immunocytochemical localization of polygalacturonase during tracheary element differentiation in Zinnia elegans. Planta 218, 729–739. doi: 10.1007/s00425-003-1167-4
Nardi, C., Escudero, C., Villarreal, N., Martinez, G., and Marcos Civello, P. (2013). The carbohydrate-binding module of Fragaria x ananassa expansin 2 (CBM-FaExp2) binds to cell wall polysaccharides and decreases cell wall enzyme activities ”in vitro”. J. Plant Res. 126, 151–159. doi: 10.1007/s10265-012-0504-8
Ogawa, M., Kay, P., Wilson, S., and Swain, S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233. doi: 10.1105/tpc.108.063768
Palin, R., and Geitmann, A. (2012). The role of pectin in plant morphogenesis. Biosystems 109, 397–402. doi: 10.1016/j.biosystems.2012.04.006
Park, K. C., Kwon, S. J., and Kim, N. S. (2010). Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genomics 32, 570–577. doi: 10.1007/s13258-010-0076-8
Park, K. C., Kwon, S. J., Kim, P. H., Bureau, T., and Kim, N. S. (2008). Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 51, 30–40. doi: 10.1139/G07-093
Pattathil, S., Hahn, M. G., Dale, B. E., and Chundawat, S. P. S. (2015). Insights into plant cell wall structure, architecture, and integrity using glycome profiling of native and AFEX (TM)-pre-treated biomass. J. Exp. Bot. 66, 4279–4294. doi: 10.1093/jxb/erv107
Patterson, S. E., and Bleecker, A. B. (2004). Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 134, 194–203. doi: 10.1104/pp.103.028027
Peaucelle, A., Braybrook, S. A., Le Guillou, L., Bron, E., Kuhlemeier, C., and Hoefte, H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726. doi: 10.1016/j.cub.2011.08.057
Peaucelle, A., Louvet, R., Johansen, J. N., Hoefte, H., Laufs, P., Pelloux, J., et al. (2008). Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr. Biol. 18, 1943–1948. doi: 10.1016/j.cub.2008.10.065
Peaucelle, A., Wightman, R., and Hoefte, H. (2015). The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr. Biol. 25, 1746–1752. doi: 10.1016/j.cub.2015.06.032
Pickersgill, R., Smith, D., Worboys, K., and Jenkins, J. (1998). Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J. Biol. Chem. 273, 24660–24664.
Pose, S., Paniagua, C., Cifuentes, M., Blanco-Portales, R., Quesada, M. A., and Mercado, J. A. (2013). Insights into the effects of polygalacturonase FaPG1 gene silencing on pectin matrix disassembly, enhanced tissue integrity, and firmness in ripe strawberry fruits. J. Exp. Bot. 64, 3803–3815. doi: 10.1093/jxb/ert210
Prasanna, V., Prabha, T. N., and Tharanathan, R. N. (2006). Multiple forms of polygalacturonase from mango (Mangifera indica L. cv Alphonso) fruit. Food Chem. 95, 30–36. doi: 10.1016/j.foodchem.2004.12.014
Rao, M. N., Kembhavi, A. A., and Pant, A. (1996). Implication of tryptophan and histidine in the active site of endo-polygalacturonase from Aspergillus ustus: elucidation of the reaction mechanism. Biochim. Biophys. Acta 1296, 167–173.
Rexovabenkova, L. (1990). Evidence for the role of carboxyl groups in activity of endopolygalacturonase of Aspergillus niger chemical modification by carbodiimide reagent. Collect. Czechoslov. Chem. Commun. 55, 1389–1395. doi: 10.1135/cccc19901389
Rhee, S. Y., Osborne, E., Poindexter, P. D., and Somerville, C. R. (2003). Microspore separation in the quartet3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133, 1170–1180. doi: 10.1104/pp.103.028266
Ridley, B. L., O’Neill, M. A., and Mohnen, D. A. (2001). Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57,929–967. doi: 10.1016/S0031-9422(01)00113-3
Rodriguez-Llorente, I. D., Pérez-Hormaeche, J., El Mounadi, K., Dary, M., Caviedes, M. A., Cosson, V., et al. (2004). From pollen tubes to infection threads: recruitment of Medicago floral pectic genes for symbiosis. Plant J. 39, 587–598. doi: 10.1111/j.1365-313X.2004.02155.x
Rui, Y., Xiao, C., Yi, H., Kandemir, B., Wang, J. Z., Puri, V. M., et al. (2017). POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 29, 2413–2432. doi: 10.1105/tpc.17.00568
Rydahl, M. G., Hansen, A. R., Kracun, S. K., and Mravec, J. (2018). Report on the current inventory of the toolbox for plant cell wall analysis: proteinaceous and small molecular probes. Front. Plant Sci. 9:581. doi: 10.3389/fpls.2018.00581
Savatin, D. V., Ferrari, S., Sicilia, F., and De Lorenzo, G. (2011). Oligogalacturonide-auxin antagonism does not require posttranscriptional gene silencing or stabilization of auxin response repressors in Arabidopsis. Plant Physiol. 157, 1163–1174. doi: 10.1104/pp.111.184663
Sekine, D., Munemura, I., Gao, M., Mitsuhashi, W., Toyomasu, T., and Murayama, H. (2006). Cloning of cDNAs encoding cell-wall hydrolases from pear (Pyrus communis) fruit and their involvement in fruit softening and development of melting texture. Physiol. Plant. 126, 163–174. doi: 10.1111/j.1399-3054.2006.00583.x
Seling, S., Wissemeier, A. H., Cambier, P., and Van Cutsem, P. (2000). Calcium deficiency in potato (Solanum tuberosum ssp tuberosum) leaves and its effects on the pectic composition of the apoplastic fluid. Physiol. Plant. 109, 44–50. doi: 10.1034/j.1399-3054.2000.100107.x
Sénéchal, F., Wattier, C., Rusterucci, C., and Pelloux, J. (2014). Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J. Exp. Bot. 65, 5125–5160. doi: 10.1093/jxb/eru272
Sergeant, K., Printz, B., Gutsch, A., Behr, M., Renaut, J., and Hausman, J. F. (2017). Didehydrophenylalanine, an abundant modification in the beta subunit of plant polygalacturonases. PLoS One 12:e0171990. doi: 10.1371/journal.pone.0171990
Somerville, C., Bauer, S., Brininstool, G., Facette, M., Hamann, T., Milne, J., et al. (2004). Toward a systems approach to understanding plant-cell walls. Science 306, 2206–2211. doi: 10.1126/science.1102765
Sun, L., Sun, Y., Zhang, M., Wang, L., Ren, J., Cui, M., et al. (2012). Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 158, 283–298. doi: 10.1104/pp.111.186866
Sun, Q., Zhang, N., Wang, J., Zhang, H., Li, D., Shi, J., et al. (2015). Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J. Exp. Bot. 66, 657–668. doi: 10.1093/jxb/eru332
Swarup, K., Benkova, E., Swarup, R., Casimiro, I., Peret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Tacken, E., Ireland, H., Gunaseelan, K., Karunairetnam, S., Wang, D., Schultz, K., et al. (2010). The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol. 153, 294–305. doi: 10.1104/pp.109.151092
Tebbutt, S. J., Rogers, H. J., and Lonsdale, D. M. (1994). Characterization of a tobacco gene encoding a pollen-specific polygalacturonase. Plant Mol. Biol. 25, 283–297. doi: 10.1007/BF00023244
Tomassen, M. M. M., Barrett, D. M., van der Valk, H. C. P. M., and Woltering, E. J. (2007). Isolation and characterization of a tomato non-specific lipid transfer protein involved in polygalacturonase-mediated pectin degradation. J. Exp. Bot. 58, 1151–1160. doi: 10.1093/jxb/erl288
Torki, M., Mandaron, P., Mache, R., and Falconet, D. (2000). Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 242, 427–436. doi: 10.1016/S0378-1119(99)00497-7
Tu, T., Meng, K., Luo, H., Turunen, O., Zhang, L., Cheng, Y., et al. (2015). New insights into the role of T3 loop in determining catalytic efficiency of GH28 endo-polygalacturonases. PLoS One 10:e0135413. doi: 10.1371/journal.pone.0135413
Tyler, L., Bragg, J. N., Wu, J., Yang, X., Tuskan, G. A., and Vogel, J. P. (2010). Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC Genomics 11:600. doi: 10.1186/1471-2164-11-600
Villarreal, N. M., Martinez, G. A., and Civello, P. M. (2009). Influence of plant growth regulators on polygalacturonase expression in strawberry fruit. Plant Sci. 176, 749–757. doi: 10.1016/j.plantsci.2009.02.019
Villarreal, N. M., Rosli, H. G., Martinez, G. A., and Civello, M. (2008). Polygalacturonase activity and expression of related genes during ripening of strawberry cultivars with contrasting fruit firmness. Postharvest Biol. Technol. 47, 141–150. doi: 10.1016/j.postharvbio.2007.06.011
Voiniciuc, C., Pauly, M., and Usadel, B. (2018). Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 176, 2590–2600. doi: 10.1104/pp.17.01776
Wakabayashi, K., and Huber, D. J. (2001). Purification and catalytic properties of polygalacturonase isoforms from ripe avocado (Persea americana) fruit mesocarp. Physiol. Plant. 113, 210–216. doi: 10.1034/j.1399-3054.2001.1130208.x
Wakasa, Y., Kudo, H., Ishikawa, R., Akada, S., Senda, M., Niizeki, M., et al. (2006). Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol. Technol. 39, 193–198. doi: 10.1016/j.postharvbio.2005.10.005
Wang, F., Sun, X., Shi, X., Zhai, H., Tian, C., Kong, F., et al. (2016). A global analysis of the polygalacturonase gene family in soybean (Glycine max). PLoS One 11:e0163012. doi: 10.1371/journal.pone.0163012
Wei, P. C., Tan, F., Gao, X. Q., Zhang, X. Q., Wang, G. Q., Xu, H., et al. (2010). Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiol. 153, 1031–1045. doi: 10.1104/pp.110.153247
Willats, W. G. T., McCartney, L., Mackie, W., and Knox, J. P. (2001). Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 47, 9–27. doi: 10.1007/978-94-010-0668-2_2
Xiao, C., Barnes, W. J., Zamil, M. S., Yi, H., Puri, V. M., and Anderson, C. T. (2017). Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. Plant J. 89, 1159–1173. doi: 10.1111/tpj.13453
Xiao, C., Somerville, C., and Anderson, C. T. (2014). POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell Elongation and flower development in Arabidopsis. Plant Cell 26, 1018–1035. doi: 10.1105/tpc.114.123968
Yang, Z. L., Liu, H. J., Wang, X. R., and Zeng, Q. Y. (2013). Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 197, 1353–1365. doi: 10.1007/s11103-015-0390-2
Yoneda, A., Ito, T., Higaki, T., Kutsuna, N., Saito, T., Ishimizu, T., et al. (2010). Cobtorin target analysis reveals that pectin functions in the deposition of cellulose microfibrils in parallel with cortical microtubules. Plant J. 64, 657–667. doi: 10.1111/j.1365-313X.2010.04356.x
Yu, Y., Liang, Y., Lv, M., Wu, J., Lu, G., and Cao, J. (2014). Genome-wide identification and characterization of polygalacturonase genes in Cucumis sativus and Citrullus lanatus. Plant Physiol. Biochem. 74, 263–275. doi: 10.1016/j.plaphy.2013.11.022
Yuan, P., Meng, K., Huang, H., Shi, P., Luo, H., Yang, P., et al. (2011). A novel acidic and low-temperature-active endo-polygalacturonase from Penicillium sp CGMCC 1669 with potential for application in apple juice clarification. Food Chem. 129, 1369–1375. doi: 10.1016/j.foodchem.2011.05.065
Zhang, Q., Huang, L., Liu, T., Yu, X., and Cao, J. (2008). Functional analysis of a pollen-expressed polygalacturonase gene BcMF6 in Chinese cabbage (Brassica campestris L. ssp chinensis Makino). Plant Cell Rep. 27, 1207–1215. doi: 10.1007/s00299-008-0541-x
Keywords: polygalacturonase, pectin modification, cell wall, classification, expression pattern, function
Citation: Yang Y, Yu Y, Liang Y, Anderson CT and Cao J (2018) A Profusion of Molecular Scissors for Pectins: Classification, Expression, and Functions of Plant Polygalacturonases. Front. Plant Sci. 9:1208. doi: 10.3389/fpls.2018.01208
Received: 27 February 2018; Accepted: 27 July 2018;
Published: 14 August 2018.
Edited by:
Dan Szymanski, Purdue University, United StatesReviewed by:
Olga A. Zabotina, Iowa State University, United StatesKim Johnson, La Trobe University, Australia
Copyright © 2018 Yang, Yu, Liang, Anderson and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiashu Cao, anNoY2FvQHpqdS5lZHUuY24=
 Yang Yang1,2
Yang Yang1,2 Charles T. Anderson
Charles T. Anderson Jiashu Cao
Jiashu Cao