- 1Triticeae Research Institute, Sichuan Agricultural University, Ya’an, China
- 2Crop Research Institute, Sichuan Academy of Agricultural Science, Chengdu, China
Powdery mildew, caused by the fungus Blumeria graminis f. sp. tritici, represents a yield constraint in many parts of the world. Here, the introduction of a resistance gene carried by the cereal rye cv. Qinling chromosome 6R was transferred into wheat in the form of spontaneous balanced translocation induced in plants doubly monosomic for chromosomes 6R and 6A. The translocation, along with other structural variants, was detected using in situ hybridization and genetic markers. The differential disease response of plants harboring various fragments of 6R indicated that a powdery mildew resistance gene(s) was present on both arms of rye chromosome 6R. Based on karyotyping, the short arm gene, designated Pm56, was mapped to the subtelomere region of the arm. The Robertsonian translocation 6AL⋅6RS can be exploited by wheat breeders as a novel resistance resource.
Introduction
Blumeria graminis f. sp. tritici (Bgt), the causative fungus of powdery mildew disease, can be a highly destructive pathogen of wheat. The disease is effectively controlled by host genetic resistance, but historically the pathogen has quite rapidly overcome widely deployed host resistance genes. Up to now, more than 70 formally designated powdery mildew resistance genes (Pm) have been identified in wheat (McIntosh et al., 2013; Liu et al., 2017; Wiersma et al., 2017). A significant number of these genes are derived from cultivated or wild relative species in the tertiary gene pool; species whose chromosomes do not normally pair with wheat homoeologs; for example, Thinopyrum intermedium (Pm40, Pm43, PmL962) (He et al., 2009; Luo et al., 2009; Shen et al., 2015), Th. ponticum (Pm51) (Zhang et al., 2014), Dasypyrum villosum (Pm21 and Pm55) (Chen et al., 1995; Zhang et al., 2016), Agropyron cristatum (a gene on chromosome 2P, as yet unnamed) (Li H. et al., 2016), and cereal rye (Secale cereale) (Pm7, Pm8/Pm17, and Pm20) (Heun and Friebe, 1990; Friebe et al., 1994; Hsam and Zeller, 1997; Ren et al., 2009). Of the rye-derived genes, only Pm20 is currently still effective in China (Zhou et al., 2002).
Balanced translocations, in which a wheat chromosome arm is fused to the opposite arm of a homoeologous chromosome present in a donor species belonging to the tertiary gene pool, represent a key intermediate in the process of alien introgression. Such so-called Robertsonian translocations are rather frequently induced during meiosis via the breakage-fusion of a pair of non-paired monosomes (Lukaszewski and Gustafson, 1983; Lukaszewski, 1997; Friebe et al., 2005). One of the best known of these in wheat is the 1BL⋅1RS translocation (Mettin et al., 1973; Zeller, 1973), which was used widely until recent times. A 6AL⋅6VS translocation carrying Pm21 is present in some Chinese cultivars (Chen et al., 1995; Cao et al., 2011).
The Chinese cereal rye cultivar Qinling (QL) is immune to powdery mildew. Here, as a means of introgressing its resistance into wheat, a disomic 6R(6A) substitution line was developed, which proved to be immune to powdery mildew. To induce Robertsonian translocations, double monosomic (6A + 6R) individuals were produced by crossing the substitution line with euploid, and these plants were allowed to self-pollinate. A number of cytogenetic and genotypic assays were applied to demonstrate the induction of breakage-fusion events, leading to the recognition that the 6R chromosome harbored Pm genes on both of its arms. Plants carrying a 6AL⋅6RS Robertsonian translocation proved to be immune to powdery mildew.
Materials and Methods
Plant Materials
The wheat lines used comprised cv. Kaixian-luohanmai (KL), the breeding lines D-2-3-4 and SY95-71, and the Chinese spring (CS) ph1b mutant (Sears, 1977), while the cereal rye cultivar used was cv. QL. D-2-3-4 is a selection from the cross KL × CS, and was used because its F1 hybrids with rye were known to be partially fertile (our unpublished data). SY95-71 is a highly powdery mildew susceptible breeder’s line used as a disease spreader. D-2-3-4 was crossed as female with cv. QL, and a single F2 progeny was advanced to the F3 generation. A number of the resulting F3 plants were repeatedly selfed to F11, at which point the material was analyzed both for karyotype and reaction to powdery mildew infection in the field. One of the powdery mildew resistant selections (female), which proved to be a disomic substitution of 6A by 6R [6R(6A)], was crossed to both KL (male) and CSph1b (male) to generate double monosomic (6A + 6R) plants, the selfed progeny of which were assessed for powdery mildew reaction in the greenhouse.
Molecular Marker Analysis
Genomic DNA was isolated from seedling leaves using a Plant Genomic DNA kit (Tiangen, Beijing, China) and screened with the PCR-based markers KU.825, KU.962, KU.496, and KU.824; the former two markers detect loci on 6RS proximal and distal regions, respectively, and the latter two loci on 6RL proximal and unknown regions, respectively (Qiu et al., 2016, Table 1). The chromosome-based draft genome sequence of CS (International Wheat Genome Sequencing Consortium [IWGSC], 2014) was used to develop markers specific to both 6AS and 6AL (Zeng et al., 2016): the former (6AS-LM) was based on the Traes_6AS_FA2CBD782 and the latter (6AL-LM) on the Traes_6AL_FEF586A96 sequence (Table 1). Each 20 μl PCR comprised 8 μl ddH2O, 10 μl 2× Taq PCR MasterMix (Biomed, Beijing, China), 0.5 μl 10 μM forward primer, 0.5 μl 10 μM reverse primer, and 1 μl of ∼100 ng/μl DNA template. The reactions were subjected to a denaturation of 94°C/5 min, followed by 35 cycles of 95°C/30 s, 58°C/30 s, 72°C/90 s, and were completed by an extension of 72°C/10 min. The resulting amplicons were separated by 1.5% agarose gel electrophoresis and visualized by EtBr staining.
Cytogenetic Assays
Genomic in situ hybridization (GISH) was performed as described by Hao et al. (2011). The hybridization mixture was composed of 7.5 μl 100% formamide, 1.5 μl 20× SSC, 1 μl ∼75 ng/μl labeled QL genomic DNA, 2 μl ∼4.5 μg/μl sheared CS genomic DNA, 1.5 μl 10 mg/ml sheared salmon sperm DNA, and 3 μl 5% w/v dextran sulfate. Fluorescence in situ hybridization (FISH) was performed as described by Komuro et al. (2013). The FISH probes used were oligo-pSc119.2 (Rayburn and Gill, 1985), (GAA)5 (Pedersen et al., 1996), oligo-pTa535, oligo-pTa713 (Zhao et al., 2016), oligo-CCS1 (centromere probe) (Tang et al., 2014), and telomere probe (TTTAGGG)3 (Werner et al., 1992). Oligo-pSc119.2 preferentially paints tandem repeats on B- and R-genome chromosomes. (GAA)5 preferentially paints tandem repeats on A- and B-genome chromosomes. Oligo-pTa535 preferentially paints tandem repeats on D- and A-genome chromosome. Oligo-pTa713 results in distinctive bands on multiple chromosome arms and especially useful in identifying individual chromosome arms of wheat. The probes were synthesized and either FAM- or TAMRA-labeled by TsingKe Biological Technology Company (Chengdu, China).
Assessment of Reaction to Powdery Mildew Infection
Powdery mildew reactions were observed in field-grown plants raised at Sichuan Agricultural University’s Ya’an (2010–2011) and Wenjiang (2010–2014) Experimental Stations, where natural infection is common (Liu et al., 2015). Each genotype was represented by three replicates of 20 plants grown in 2 m long rows separated from one another by 30 cm. The highly susceptible common wheat line SY95-71 was planted on both sides of each experimental row and used to increase Bgt inoculum and as the susceptible control. The host reaction was evaluated at the heading and grain filling stages by scoring on a 0–4 scale following Si et al. (1992). Powdery mildew reactions of progenies from double monosomic plants were determined in a small (15 m2 in area) greenhouse in order to a well-controlled condition. Greenhouse-based disease reactions were determined by inoculating seedlings at the three leaf stage of progenies and susceptible wheat parents held under a 16 h photoperiod, 22°C (light)/16°C (dark) regime at a relative humidity of 75%; the inoculum was a mixture of local Bgt conidiospores, which were dusted onto the leaves. Plants were inspected at 15-day intervals, and were scored using the Si et al. (1992) scale.
Results
Substitution and Addition Lines Resistant to Powdery Mildew
Of 10 D-2-3-4 × QL F1 hybrids, one produced three F2 progeny, and of these, only one was self-fertile. After selfing this individual through to the F11 generation with selection for good fertility, 42 lines were selected for karyotyping using GISH and FISH. Ten of these lines were found to contain rye chromatin: nine were 6R(6A) disomic substitutions and one was a disomic 2R addition line. All the 10 lines with rye chromatin were immune to powdery mildew in the field over the 2010–2014 seasons, whereas the other 32 lines were fully susceptible.
Detecting 6R Structural Variants in the Progeny of the 6A + 6R Double Monosomic Plants
The DS6R(6A) line was crossed with KL and CSph1b to generate double monosomic plants. The 6A + 6R double monosomic plants were fully self-fertile. In all, 69 progenies were characterized genotypically and karyotypically (Supplementary Table S1); of these, 25 were descended from three 6R(6A) × CSph1b hybrids (HM922-4, -17, and -19) and 44 from four 6R(6A) × KL hybrids (HM923-2, -4, -5, and -9). Ten progenies [two from the 6R(6A) × CSph1b cross and eight from the 6R(6A) × KL cross] lacked both the 6AS and 6AL markers, and hence were deemed to lack the entire chromosome; similarly, 18 [5 from the 6R(6A) × CSph1b cross and 13 from the 6R(6A) × KL cross] lacked the markers for both arms of 6R, so were deemed to have lost the entire 6R (Figure 1). Among the 6R(6A) × CSph1b progeny, 14 were monosomic for an intact copy of 6R and 2 were disomic, while 5 harbored either a misdivided or a translocated version of 6R (Table 2); the equivalent numbers for the 6R(6A) × KL progeny were 18 6R monosomics, 6 6R disomics, and 9 harboring a misdivided or a translocated version of 6R. The 14 misdivided or translocated versions were classified into the following six types (Figure 2A and Table 2):
(1) A telosome, either 6RS (LM52, LM74, HM151) or 6RL (LM61, LM98).
(2) An isochromosome for 6RL (HM136).
(3) A near entire chromosome 6RL⋅6RSd lacking the telomeric end of 6RS (LM99 and HM99).
(4) A chromosome 6RSpi possibly carrying an paracentric inversion (HM109, Figure 2B). All two 6RS markers had been retained (Figure 1), but the two pSc119.2 sites in the center of a normal 6RS (Figure 2A, red arrow) appeared as two weaker sites near the telomere (Figure 2A, white arrow). FISH results using the centromere probe indicated that it contained a centromere in the middle (Figure 2A, green arrow). FISH based on the telomere sequence indicated that a full telomere was only present on the arm carrying the major pSc119.2 terminal site (Figure 2A, yellow arrow).
(5) The Robertsonian translocations 6AL⋅6RS (LM47), 6AS⋅6RS (HM148, HM156), and 4AL⋅6RS (LM30).
(6) A form of 6R (6Ra) carrying two additional major pSc119.2 sites toward the end of 6RL (LM43).
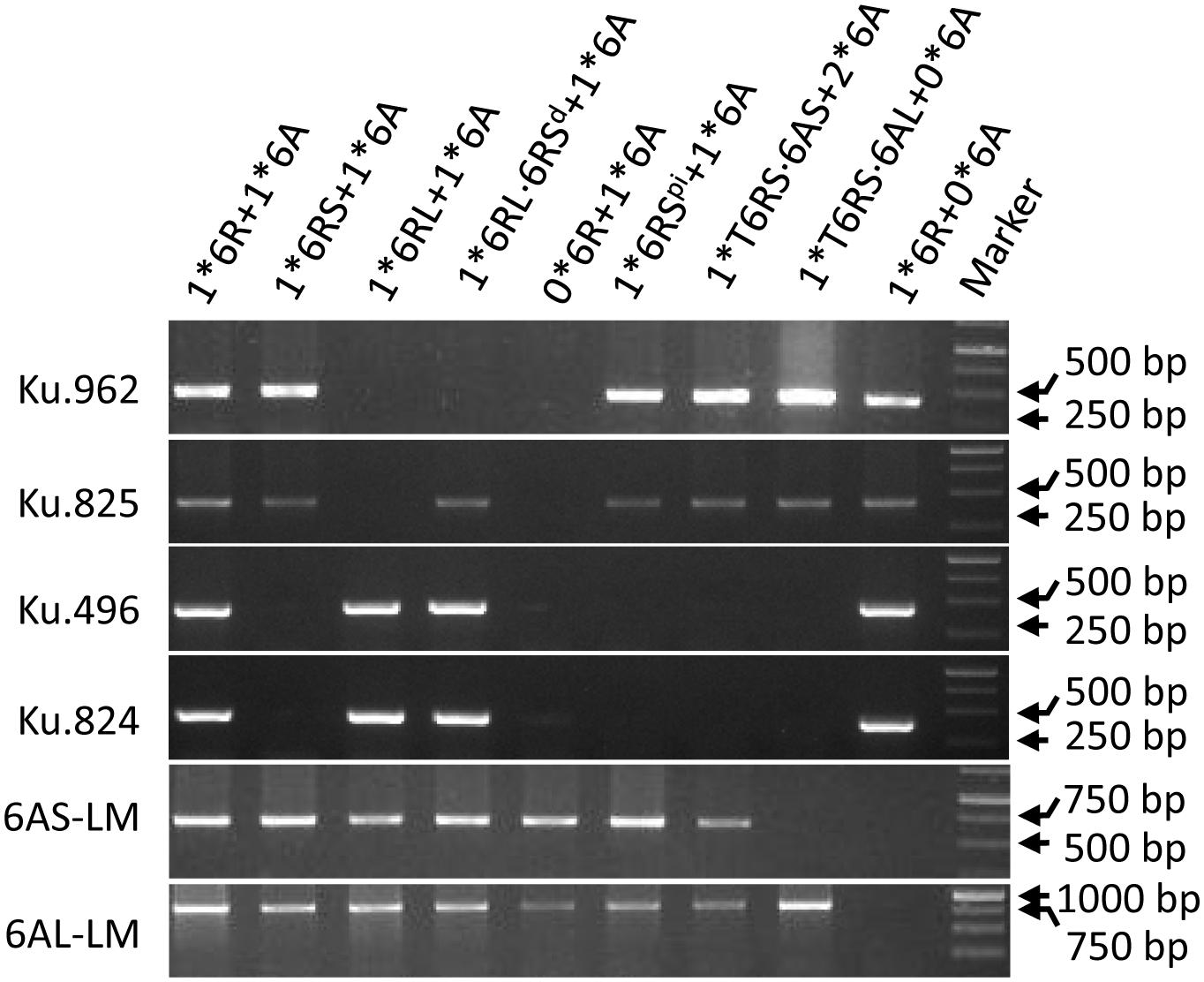
FIGURE 1. PCR profiling of (from left to right) LM51, LM52, LM61, HM99, LM57, HM109, HM148, LM47, and HM100, using assays for loci on chromosomes 6A and 6R.
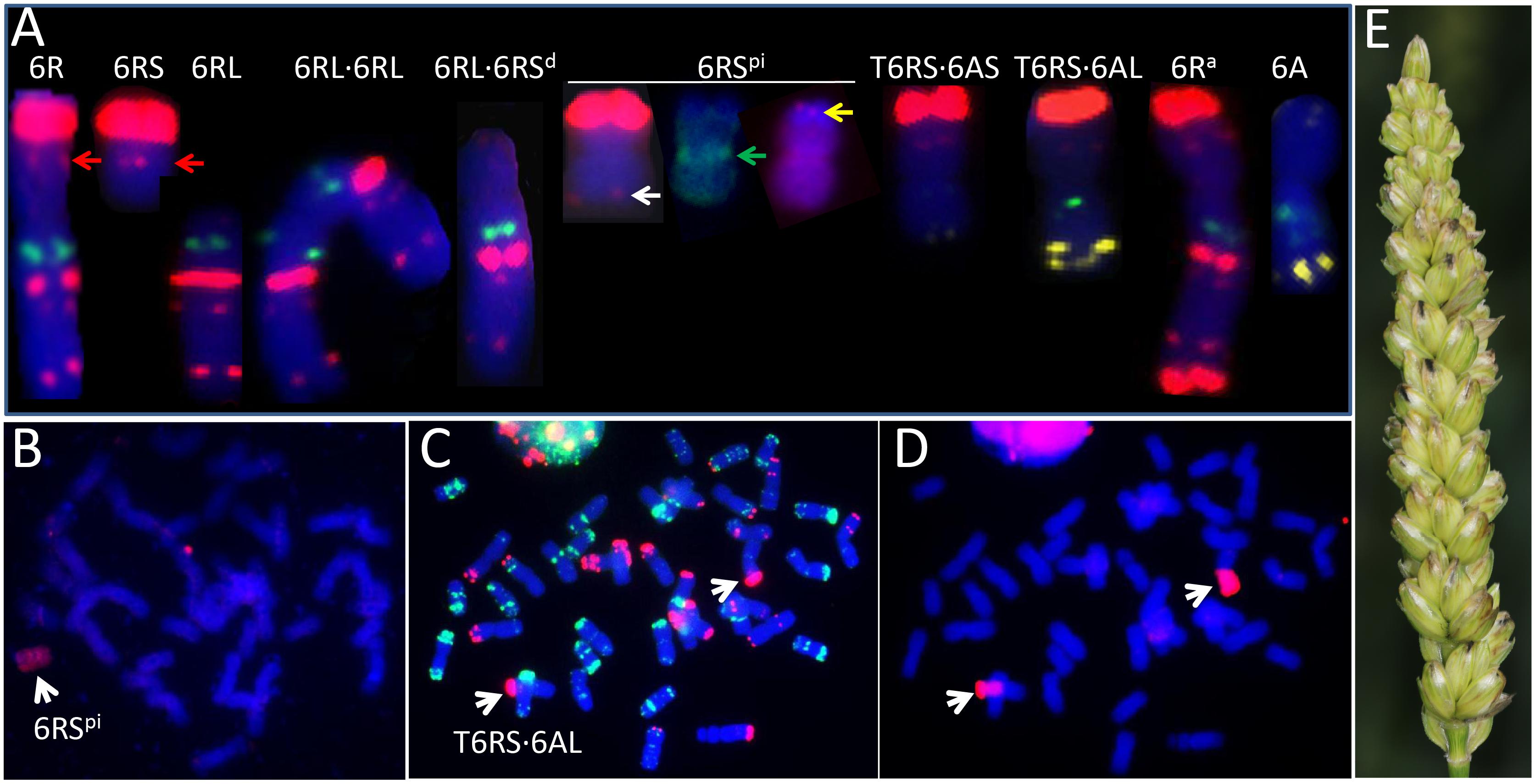
FIGURE 2. Structural variations of chromosome 6R recovered among the progeny of 6A + 6R double monosomic plants. (A) Variants recovered in the first selfing generation. pSc119.2 sites and the telomere [(TTTAGGG)3] (6RSpi, yellow arrow) are shown in red, pTa713 sites in yellow, (GAA)5 and CCS1 (6RSpi, green arrow) sites in green. The red arrow shows the pSc119.2 site in the center of a normal 6RS arm and the white arrow a minor pSc119.2 site at the terminus of the 6R paracentric inversion (6RSpi). (B) GISH karyotype of HM109, carrying the 6R paracentric inversion (6RSpi). Rye chromatin is colored red. Sequential FISH (C) and GISH (D) karyotype of LM47-6, which carries a pair of 6AL⋅6RS translocations. The red and green colored sites, respectively, are pSc119.2 and pTa535 sites (C), while the rye chromatin is colored red (D). (E) A spike of homozygous T6RS⋅6AL translocation line. Pi indicates paracentric inversion.
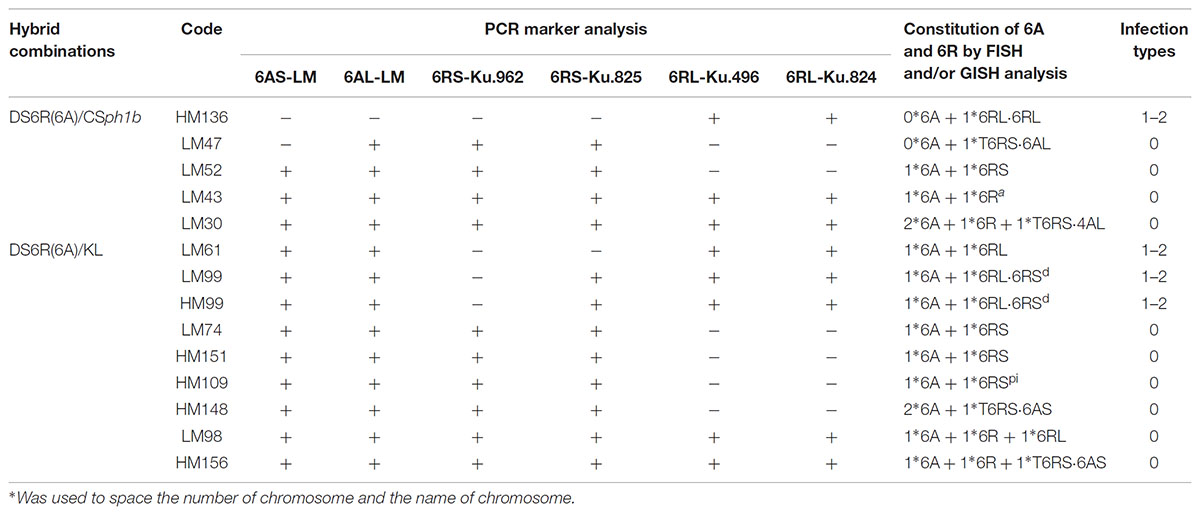
TABLE 2. First generation selfed progeny of 6A + 6R double monosomic plants carrying variant forms of 6R and their response to powdery mildew infection.
Transmission of the Modified Forms of 6R and the Occurrence of de Novo Modified Forms
The transmission of the modified forms of 6R identified in the progeny of the first selfing generation of the 6A + 6R double monosomic was quantified by an analysis of the derived second generation families (Table 3). The 6AS⋅6RS translocation present in HM148 was retained by 6 out of 13 individuals, one of which was disomic for the translocation. The near entire chromosome 6R present in HM99 was transmitted to two out of seven progeny, and the same transmission rate was observed for the 6RS telosome present in HM151. The rate of transmission of the paracentric inversion present in HM109 was only one out of eight. The 6AL⋅6RS translocation in LM47 was present in 9 out of 11 progeny, one of which carried two copies of the translocation (Figures 2C–E). Four further structural variants were identified in the second generation (Table 3). The first was a 6RL telosome found in one individual derived from HM99 (which carried the truncated form of 6R); the second was a truncated form of 6R lacking a terminal part of the short arm, derived from HM103 (a plant carrying a 6R monosome); the third was a 6RS isochromosome derived from LM47 (which harbored 6AL⋅6RS); and the fourth, a 6AL⋅6RS translocation detected among the progeny of HM166, the karyotype of which had not been determined.
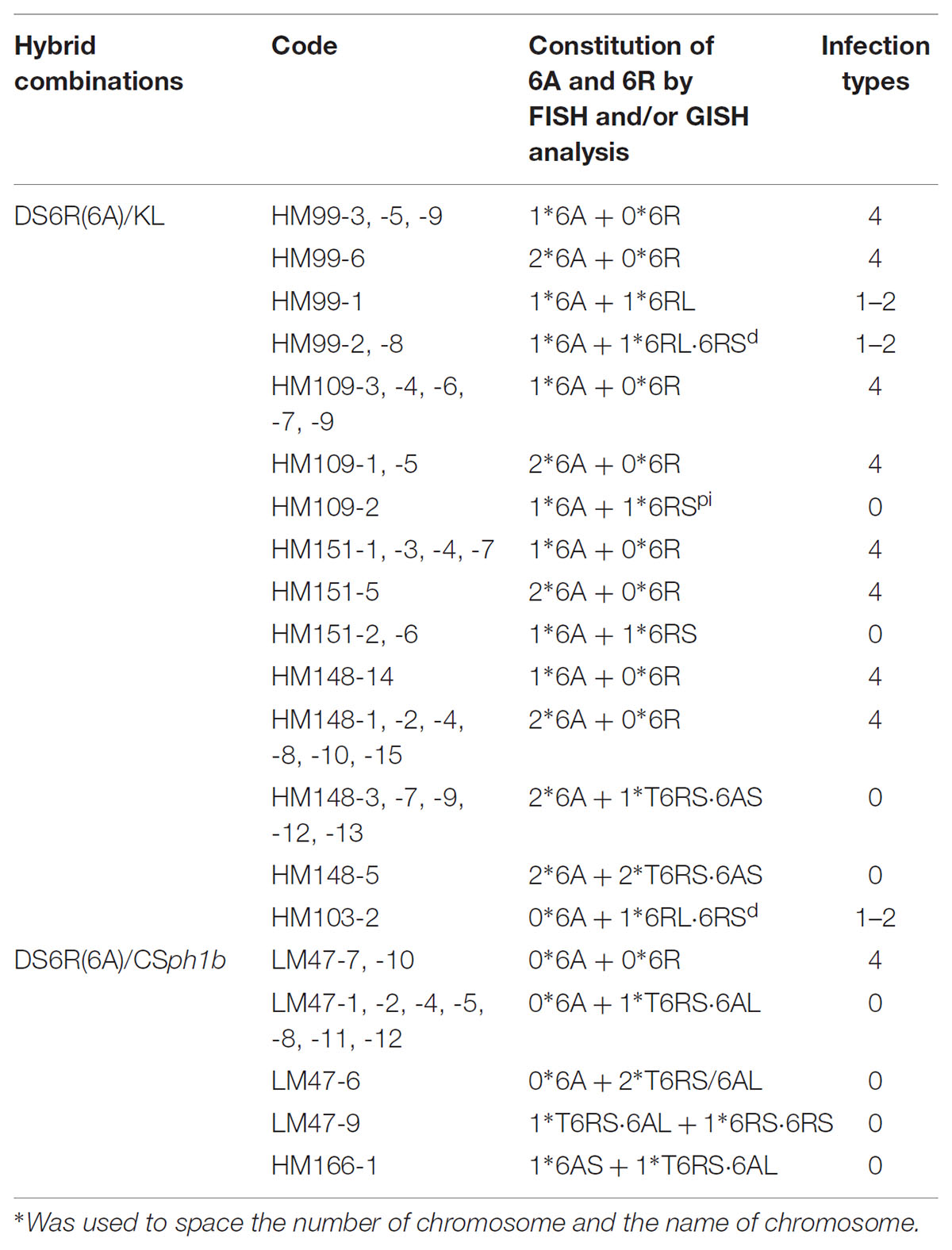
TABLE 3. The chromosome 6R constitution of the second selfing generation of 6A/6R double monosomic plants and their responses to powdery mildew infection.
Both Arms of Chromosome 6R Harbor Genes Conditioning Resistance Against Powdery Mildew
All of the first selfing generation plants harboring an entire 6R proved to be immune to powdery mildew when tested in the greenhouse, whereas all those without 6R chromatin were highly susceptible (Supplementary Table S1), as were the nine second generation plants lacking 6R (Table 3). This outcome was consistent with the field assessment, confirming that powdery mildew resistance was conferred by gene(s) mapping to chromosome 6R. When the disease response of plants derived from the 6R(6A) × CSph1b cross harboring incomplete forms of 6R (Figure 3 and Tables 2, 3) were compared, it was clear that the presence of the short arm was associated with immunity, while the presence of the long arm alone did not confer immunity, but rather only a high level of resistance. The implication was that both arms harbor a resistance gene(s), with the one(s) on the short arm being more effective than the one(s) on the long arm. A consistent conclusion was drawn from the equivalent analysis of the progeny of the 6R(6A) × KL cross: all plants which had retained the entire 6RS arm were immune, while those which retained 6RL but lacked the terminal end of 6RS were moderately to highly resistant. Since the truncated form of 6R (LM99, HM99, HM99-2, HM99-8, and HM103-2) lacked the 6RS subtelomeric region, the assumption was that the gene conferring immunity to powdery mildew lies in the truncated segment (Figure 3C). The PCR marker KU.962 specific for 6RS also failed to amplify in plants with 6RL⋅6RSd (Figure 1), indicating that the sequence is also located in this chromosome region and that this PCR-based marker will be useful in further investigation or chromosome engineering of the resistance gene. However, it is unclear for the genetic distance between marker KU.962 and the resistance gene.
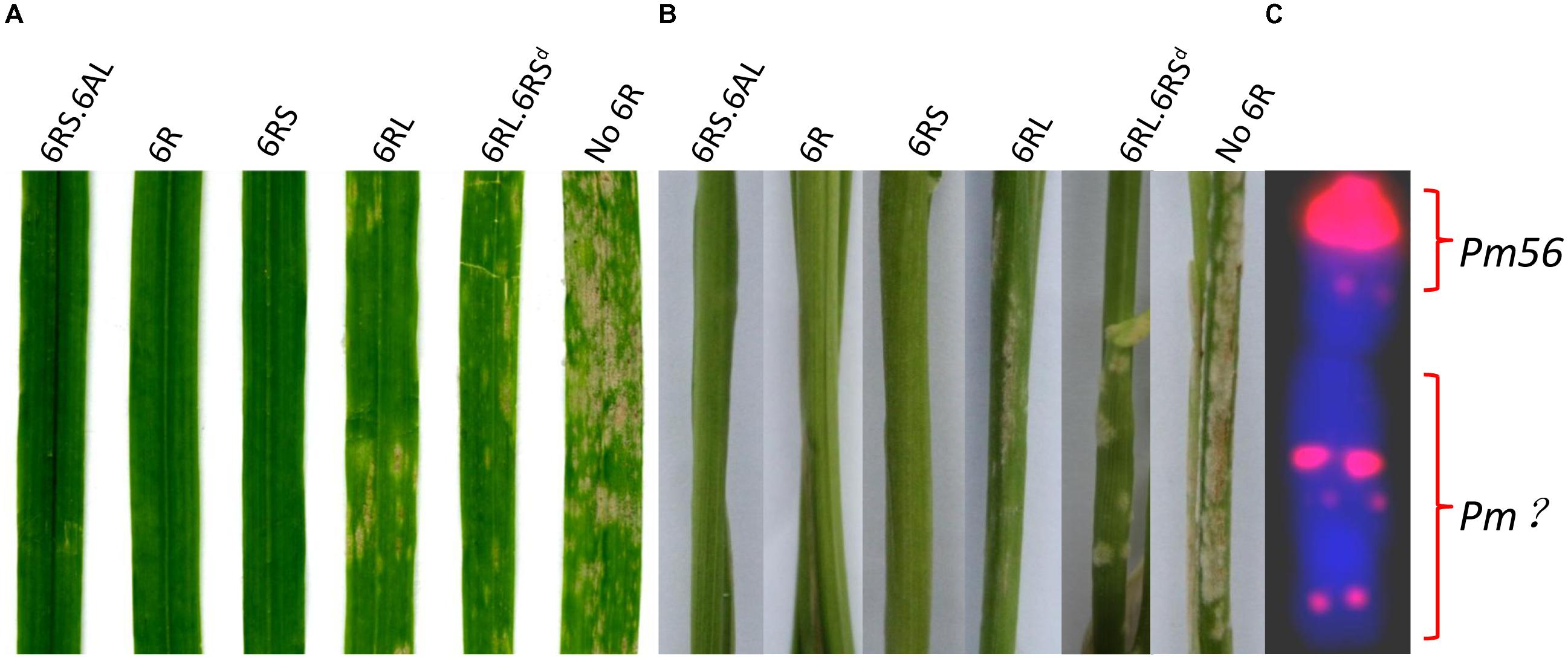
FIGURE 3. The response to powdery mildew infection on (A) the leaf and (B) the leaf sheath of plants containing various forms of chromosome 6R. (C) The location of Pm genes on cv. QL chromosome 6R. Left to right: carriers of T6AS⋅6RS, 6R, 6RS, 6RL, a truncated form of 6R lacking a subtelomeric region of 6RS (6RL⋅6RSd), and a plant lacking 6R (No 6R). The red signals on the 6R chromosome in (C) represent pSc119.2 sites.
Discussion
Powdery mildew is a damaging disease of the world wheat crops, especially in Sichuan Province, China, where it can cause very high yield losses (Liu et al., 2012). The race structure of Bgt in Sichuan is complex, with as many as 109 distinct isolates having been obtained from a set of 327 infected leaves. Of 28 known Pm genes, only Pm21 was able to mount an immune response to these isolates (Liu et al., 2015).
The present experiments have established that wheat plants carrying chromosome 6R of cereal rye cv. QL were immune to powdery mildew in Sichuan, and that resistance genes were present on both of its arms. The long arm of 6R carried by cv. Prolific is known to harbor Pm20, a gene which conditions immunity against a range of Bgt isolates (Heun and Friebe, 1990; Friebe et al., 1994), while Li M. et al. (2016) have mapped a powdery mildew resistance gene to the same arm in cv. Kustro and wheat plants carrying 6RL transferred from the rye germplasm accession PI 252003 manifest an isolate-specific response to infection (Friebe et al., 1994). Here, the presence of chromosome arm 6RS was associated with an immune reaction, although no Pm gene has previously been reported to map to this arm. Assuming that this immunity is conditioned by a single gene, the gene can be provisionally designated Pm56. Structural variation in the 6RS arm has allowed Pm56 to be localized to the terminal region of the arm, lying between the telomere and the most distal pSc119.2 site (Figure 3C). The 6RS arm is largely syntenic with the wheat homeologous group 6 short arms (Martis et al., 2013), although its terminal segment was translocated to 2R (Devos et al., 1993). Thus, of the two wheat/rye Robertsonian translocations isolated, the expectation is that 6AL⋅6RS will provide a higher level of genetic compensation than 6AS⋅6RS, so is likely to prove more useful for breeding purposes.
Wheat plants in which both a wheat and a non-wheat chromosome are present in the monosomic state have long been known to provide a source of Robertsonian translocations: some recent examples have been provided by Ardalani et al. (2016) and Rahmatov et al. (2016). Here, the use of in situ hybridization and genotyping has led to the detection of a number of novel chromosomes involving segments of chromosome 6R; notably, among 69 selfed progeny of a double monosomic for 6A and 6R, 4 carried a Robertsonian translocation, assumed to have been induced by the breakage-fusion of the two unpaired chromosomes (Lukaszewski, 1997; Friebe et al., 2005).
The 6R(6A) substitution arose following the repeated selfing of a partially fertile wheat-rye amphihaploid plant. Although such plants are generally self-sterile, a low level of fertility can be restored through the process of first division restitution (Ramanna and Jacobsen, 2003). The phenomenon has been recorded in synthetic wheat/rye amphihaploids (Zeng et al., 2014): the selfed progeny of these crosses include some partial amphiploids (Zeng et al., 2014). Repeated selfing of these individuals allied to selection for fertility favors the production of hexaploids of genomic constitution AABBRR (Hao et al., 2013). However, in both the present case and that described by Yuan et al. (2014), the final products were either euploid bread wheat or a disomic substitution line, reflecting the elimination of all or most of the rye, rather than of the D genome chromosomes. A possible cytological explanation for the low fertility of the amphihaploid and the subsequent loss of the rye complement has been suggested by Silkova et al. (2011), who observed a high frequency of equational and reductional divisions in bread wheat/rye hybrids. If equational division predominated for the wheat chromosomes and reductional division for the rye chromosomes, then gametes carrying an unreduced wheat complement along with a reduced rye one could have formed. The union of such uniparental unreduced gametes can be expected to result in the formation of a partial amphidiploids containing two sets of all or most of the wheat complement, leaving the rye chromosomes in the monosomic state. In subsequent generations, the rye chromosomes would be largely eliminated.
Data Availability
The seeds harboring homozygous 6AL⋅6RS translocation is available upon request to MH, Triticeae Research Institute, Sichuan Agricultural University, at haomingluo@foxmail.com.
Author Contributions
MH and DL conceived and designed the experiments. DL, LZ, and MH developed the substitution lines. ML, JL, CF, and YY performed the cytogenetic experiments. ML, CF, and SN molecular marker analysis. ML, JL, LZ, and ZY evaluated the powdery mildew resistance. YZ and DL supervised the study. ML, MH, and DL wrote the paper.
Funding
This study was supported by the National Key Research and Development Program (2016YFD0102000), Key Projects of Sichuan Provincial Department of Education (16ZA0028), and National Natural Science Foundation of China (NSFC) (31671689).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully appreciate the help of R.A. McIntosh, University of Sydney, with English editing and comments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01040/full#supplementary-material
References
Ardalani, S., Mirzaghaderi, G., and Badakhshan, H. (2016). A Robertsonian translocation from Thinopyrum bessarabicum into bread wheat confers high iron and zinc contents. Plant Breed. 135, 286–290. doi: 10.1111/pbr.12359
Cao, A., Xing, L., Wang, X., Yang, X., Wang, W., Sun, Y., et al. (2011). Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U.S.A. 108, 7727–7732. doi: 10.1073/pnas.1016981108
Chen, P. D., Qi, L. L., Zhou, B., Zhang, S. Z., and Liu, D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128. doi: 10.1007/BF00223930
Devos, K. M., Atkinson, M. D., Chinoy, C. N., Francis, H. A., Harcourt, R. L., Koebner, R. M. D., et al. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. doi: 10.1007/BF00225004
Friebe, B., Heun, M., Tuleen, N., Zeller, F. J., and Gill, B. S. (1994). Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 34, 621–625. doi: 10.2135/cropsci1994.0011183X003400030003x
Friebe, B., Zhang, P., Linc, G., and Gill, B. S. (2005). Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet. Genome Res. 109, 293–297. doi: 10.1159/000082412
Hao, M., Luo, J. T., Yang, M., Zhang, L. Q., Yan, Z. H., Yuan, Z. W., et al. (2011). Comparison of homoeologous chromosome pairing between hybrids of wheat genotypes Chinese spring ph1b and Kaixian-luohanmai with rye. Genome 54, 959–964 doi: 10.1139/g11-062
Hao, M., Luo, J., Zhang, L., Yuan, Z., Yang, Y., Wu, M., et al. (2013). Production of hexaploid triticale by a synthetic hexaploid wheat-rye hybrid method. Euphytica 193, 347–357. doi: 10.1007/s10681-013-0930-2
He, R., Chang, Z., Yang, Z., Yuan, Z., Zhan, H., Zhang, X., et al. (2009). Inheritance and mapping of a powdery mildew resistance gene Pm43 introgressed from Thinopyrum intermedium into wheat. Theor. Appl. Genet. 118, 1173–1180. doi: 10.1007/s00122-009-0971-z
Heun, M., and Friebe, B. (1990). Introgression of powdery mildew resistance from rye into wheat. Phytopathology 80, 242–245. doi: 10.1094/Phyto-80-242
Hsam, S. L. K., and Zeller, F. J. (1997). Evidence of allelism between genes Pm8 and Pm17 and chromosomal location of powdery mildew and leaf rust resistance genes in the common wheat cultivar ‘Amigo’. Plant Breed. 116, 119–122. doi: 10.1111/j.1439-0523.1997.tb02164.x
International Wheat Genome Sequencing Consortium [IWGSC] (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788.
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 131–137. doi: 10.1139/gen-2013-0003
Li, H., Lv, M., Song, L., Zhang, J., Gao, A., Li, L., et al. (2016). Production and identification of wheat-Agropyron cristatum 2P translocation lines. PLoS One 11:e0145928. doi: 10.1371/journal.pone.0145928
Li, M., Tang, Z., Qiu, L., Wang, Y., Tang, S., and Fu, S. (2016). Identification and physical mapping of new PCR-based markers specific for the long arm of rye (Secale cereale L.) chromosome 6. J. Genet. Genomics 43, 209–216. doi: 10.1016/j.jgg.2015.11.005
Liu, N., Gong, G., Zhang, M., Zhou, Y., Chen, Z., Yang, J., et al. (2012). Over-summering of wheat powdery mildew in sichuan province, China. Crop Prot. 34, 112–118. doi: 10.1016/j.cropro.2011.12.011
Liu, N., Liu, Z. L., Gong, G., Zhang, M., Wang, X., Zhou, Y., et al. (2015). Virulence structure of Blumeria graminis f. sp. tritici and its genetic diversity by ISSR and SRAP profiling analyses. PLoS One 10:e0130881. doi: 10.1371/journal.pone.0130881
Liu, W., Koo, D. H., Xia, Q., Li, C., Bai, F., Song, Y., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
Lukaszewski, A. J. (1997). Further manipulation by centric misdivision of the 1RS.1BL translocation in wheat. Euphytica 94, 257–261. doi: 10.1023/A:1002916323085
Lukaszewski, A. J., and Gustafson, J. P. (1983). Translocations and modifications of chromosomes in triticale × wheat hybrids. Theor. Appl. Genet. 64, 239–248. doi: 10.1007/BF00303771
Luo, P. G., Luo, H. Y., Chang, Z. J., Zhang, H. Y., Zhang, M., and Ren, Z. L. (2009). Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl. Genet. 118, 1059–1064. doi: 10.1007/s00122-009-0962-0
Martis, M. M., Zhou, R., Haseneyer, G., Schmutzer, T., Vrána, J., Kubaláková, M., et al. (2013). Reticulate evolution of the rye genome. Plant Cell 25, 3685–3698. doi: 10.1105/tpc.113.114553
McIntosh, R. A., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X. C. (2013). Catalogue of Gene Symbols for Wheat in KOMUGI-Integrated Wheat Science. Available at: http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp
Mettin, D., Bluthner, W. D., and Schlegel, G. (1973). “Additional evidence of spontaneous 1B/1R wheat-rye substitutions and translocations,” in Proceedings of the 4th International Wheat Genetetics Symposium (Columbia, MO: Agricultural Experiment Station), 179–184.
Pedersen, C., Rasmussen, S. K., and Linde-Laursen, I. (1996). Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome 39, 93–104. doi: 10.1139/g96-013
Qiu, L., Tang, Z. X., Li, M., and Fu, S. L. (2016). Development of new PCR-based markers specific for chromosome arms of rye (Secale cereale L.). Genome 59, 159–165. doi: 10.1139/gen-2015-0154
Rahmatov, M., Rouse, M. N., Nirmala, J., Danilova, T., Friebe, B., Steffenson, B. J., et al. (2016). A new 2DS⋅2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. doi: 10.1007/s00122-016-2710-6
Ramanna, M. S., and Jacobsen, E. (2003). Relevance of sexual polyploidization for crop improvement–A review. Euphytica 133, 3–8. doi: 10.1023/A:1025600824483
Rayburn, A. L., and Gill, B. S. (1985). Use of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. J. Hered. 76, 78–81. doi: 10.1093/oxfordjournals.jhered.a110049
Ren, T. H., Yang, Z. J., Yan, B. J., Zhang, H. Q., Fu, S. L., and Ren, Z. L. (2009). Development and characterization of a new 1BL.1RS translocation line with resistance to stripe rust and powdery mildew of wheat. Euphytica 169, 207–213. doi: 10.1007/s10681-009-9924-5
Sears, E. R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. doi: 10.1139/g77-063
Shen, X. K., Ma, L. X., Zhong, S. F., Liu, N., Zhang, M., Chen, W. Q., et al. (2015). Identification and genetic mapping of the putative Thinopyrum intermedium-derived dominant powdery mildew resistance gene PmL962 on wheat chromosome arm 2BS. Theor. Appl. Genet. 128, 517–528. doi: 10.1007/s00122-014-2449-x
Si, Q. M., Zhang, X. X., Duan, X. Y., Sheng, B. Q., and Zhou, Y. L. (1992). On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol. Sin. 22, 349–355.
Silkova, O. G., Shchapova, A. I., and Shumny, V. K. (2011). Patterns of meiosis in ABDR amphihaploids depend on the specific type of univalent chromosome division. Euphytica 178, 415–426. doi: 10.1007/s10681-010-0325-6
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc19. 12, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Werner, J. E., Kota, R. S., Gill, B. S., and Endo, T. R. (1992). Distribution of telomeric repeats and their role in the healing of broken chromosome ends in wheat. Genome 35, 844–848. doi: 10.1139/g92-128
Wiersma, A. T., Pulman, J. A., Brown, L. K., Cowger, C., and Olson, E. L. (2017). Identification of Pm58 from Aegilops tauschii. Theor. Appl. Genet. 130, 1123–1133. doi: 10.1007/s00122-017-2874-8
Yuan, Z., Liu, M., Ouyang, Y., Zeng, X., Hao, M., Zhang, L., et al. (2014). The detection of a de novo allele of the Glu-1Dx gene in wheat–rye hybrid offspring. Theor. Appl. Genet. 127, 2173–2182. doi: 10.1007/s00122-014-2370-3
Zeller, F. J. (1973). “1B/1R wheat-rye chromosome substitutions and translocations,” in Proceedings of the 4th International Wheat Genetics Symposium (Columbia, MO: Agricultural Experiment Station), 209–221.
Zeng, D., Luo, J., Li, Z., Chen, G., Zhang, L., Ning, S., et al. (2016). High transferability of homoeolog-specific markers between bread wheat and newly synthesized hexaploid wheat lines. PLoS One 11:e0162847. doi: 10.1371/journal.pone.0162847
Zeng, D. Y., Hao, M., Luo, J. T., Zhang, L. Q., Yuan, Z. W., Ning, S. Z., et al. (2014). Amphitelic orientation of centromeres at metaphase I is an important feature for univalent-dependent meiotic nonreduction. J. Genet. 93, 531–534. doi: 10.1007/s12041-014-0393-9
Zhang, H., Li, G. R., Zhang, X. J., Li, X., Guo, H. J., Gong, W. P., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS One 9:e113455. doi: 10.1371/journal.pone.0113455
Zhang, R., Sun, B., Chen, J., Cao, A., Xing, L., Feng, Y., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129, 1975–1984. doi: 10.1007/s00122-016-2753-8
Zhao, L., Ning, S., Yu, J., Hao, M., Zhang, L., Yuan, Z., et al. (2016). Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed. Sci. 66, 522–529. doi: 10.1270/jsbbs.16011
Keywords: Blumeria graminis, cereal rye, powdery mildew, Secale cereale, Triticum aestivum
Citation: Hao M, Liu M, Luo J, Fan C, Yi Y, Zhang L, Yuan Z, Ning S, Zheng Y and Liu D (2018) Introgression of Powdery Mildew Resistance Gene Pm56 on Rye Chromosome Arm 6RS Into Wheat. Front. Plant Sci. 9:1040. doi: 10.3389/fpls.2018.01040
Received: 13 March 2018; Accepted: 26 June 2018;
Published: 17 July 2018.
Edited by:
Thomas Miedaner, University of Hohenheim, GermanyReviewed by:
Zhang Ruiqi, Nanjing Agricultural University, ChinaFangpu Han, Institute of Genetics and Developmental Biology (CAS), China
George Fedak, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2018 Hao, Liu, Luo, Fan, Yi, Zhang, Yuan, Ning, Zheng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dengcai Liu, ZGNsaXU3QHlhaG9vLmNvbQ==
†These authors have contributed equally to this work.
 Ming Hao1†
Ming Hao1† Chaolan Fan
Chaolan Fan Youliang Zheng
Youliang Zheng Dengcai Liu
Dengcai Liu