- 1School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2National Engineering Laboratory of Crop Stress Resistance Breeding, Anhui Agricultural University, Hefei, China
- 3College of Agronomy, Anhui Agricultural University, Hefei, China
Nucleotide-binding site-leucine-rich repeat (NBS-LRR) domain proteins are immune sensors and play critical roles in plant disease resistance. In this study, we cloned and characterized a novel NBS-LRR gene ZmNBS25 in maize. We found that ZmNBS25 could response to pathogen inoculation and salicylic acid (SA) treatment in maize, and transient overexpression of ZmNBS25 induced a hypersensitive response in tobacco. High-performance liquid chromatography (HPLC) analysis showed that, compared to control plants, ZmNBS25 overexpression (ZmNBS25-OE) in Arabidopsis and rice resulted in higher SA levels. By triggering the expression of certain defense-responsive genes, ZmNBS25-OE enhanced the resistance of Arabidopsis and rice to Pseudomonas syringae pv. tomato DC3000 and sheath blight disease, respectively. Moreover, we found little change of grain size and 1000-grain weight between ZmNBS25-OE rice lines and controls. Together, our results suggest that ZmNBS25 can function as a disease resistance gene across different species, being a valuable candidate for engineering resistance in breeding programs.
Introduction
Plants have evolved multiple defense strategies against pathogen infections (Thomma et al., 2011; Henry et al., 2013) and have different immune systems that are highly effective against most microbial pathogens (Jones and Dangl, 2006; Chen and Ronald, 2011; Dangl et al., 2013). The first defense system recognizes conserved pathogen-associated molecular patterns (PAMPs) and is called PAMP-triggered immunity. This first line of defense kills many pathogens, while the other systems suppress or bypass infection (Jones and Dangl, 2006; Zhang and Zhou, 2010; Chen and Ronald, 2011). The second defense system recognizes specific pathogen effectors, such as the avirulence protein (Avr), which is produced by pathogens in an attempt to suppress host defenses and cause disease. This process is called effector-triggered immunity (ETI) (Nurnberger et al., 2004) and relies on the specific recognition of pathogen effectors by disease resistance (R) proteins. The rapid defense reaction initiated by ETI, i.e., the hypersensitive response (HR) (Dangl et al., 1996; Greenberg, 1997), prevents the spread of microbial pathogens and infection in plants (Zvereva and Pooggin, 2012). It ultimately leads to systemic acquired resistance (SAR), which confers long-lasting protection against a broad spectrum of microorganisms (Chen and Ronald, 2011; Molinari, 2016).
R proteins can act as receptors that directly or indirectly recognize the Avr and form R-Avr complexes to activate various resistance responses (Liu et al., 2007; Cesari et al., 2013; Sohn et al., 2014). Most reported plant R genes belong to the nucleotide-binding site-leucine-rich repeat (NBS-LRR) gene family (Jones and Dangl, 2006) and comprise C-terminal leucine-rich repeat (LRR) and central nucleotide-binding site (NBS) domains. In most cases, a Toll/interleukin-1 receptor (TIR) domain homology region or coiled-coil (CC) domain is located at their N-terminal, and these genes are called TIR-NBS-LRR and CC-NBS-LRR genes, respectively (Takken and Goverse, 2012). Plant NBS-LRR genes interact with pathogen effector proteins to activate signal transduction pathways involved in innate immunity while TIR and CC domains specifically recognize R-Avr complexes and initiate downstream defense signaling (Burkhard et al., 2001; Liu et al., 2007). The LRR domain may determine resistance specificity and be primarily responsible for the recognition of R-Avr complexes (Jones and Dangl, 2006; Liu et al., 2007).
The functions of NBS-LRR genes have been studied in several species. For example, ZmRXO1 isolated from maize (Zea mays L.) is involved in transformed rice (Oryza sativa L.) resistance against Xanthomonas oryzae pv. oryzicola, which causes rice blast, revealing the feasibility of non-host R gene transfer between crops (Zhao et al., 2005). AhRRS5, a novel NBS-LRR resistance gene from peanut (Arachis hypogaea L.), was up-regulated in response to Ralstonia solanacearum, and its transient overexpression in Nicotiana benthamiana leaves induces HR (Zhang et al., 2017). Overexpression of AhRRS5 in tobacco (N. tabacum L.) significantly enhanced its resistance to R. solanacearum (Zhang et al., 2017). Heterologous overexpression of the Prunus sogdiana Vassilcz NBS-LRR gene PsoRPM2 in tobacco enhanced its resistance to root-knot nematode (Meloidogyne incognita) infection (Zhu et al., 2017), and overexpression of the grapevine Vitis amurensis Rupr. TIR-NBS-LRR gene VaRGA1 in tobacco enhanced its resistance to Phytophthora parasitica (Li et al., 2016). In addition, the expression of NBS-LRR genes is always correlated with pathogen infection or salicylic acid (SA) treatment. For instance, the Arabidopsis RPP2 gene, which includes the RPP2A and RPP2B isoforms, confers downy mildew resistance and is required for SA-dependent ETI (Sinapidou et al., 2004; Bonardi et al., 2011).
Maize is an important crop grown worldwide and it is susceptible to many diseases that can significantly lower its yield. For example, southern leaf blight (SLB) caused by the filamentous ascomycete Bipolaris maydis is widespread and has high infectivity in maize (Shurtleff, 1980). Host plants are infected by asexual spores (conidia) of B. maydis through wind and rain (Sumita et al., 2017). It is estimated that, in 1970, an SLB epidemic caused 15% drop in total maize production and a loss of one billion dollars (Ullstrup, 1972). Therefore, a better understanding of the NBS-LRR gene in maize might enable predicting and identifying genes that play important roles in heterologous systems and such knowledge might be used to greatly improve the stability of agricultural system. In our previous study, we identified 109 NBS-encoding genes based on the complete genome sequence of maize and found that some of these genes responded to B. maydis infection (Cheng et al., 2012). The phylogenetic analysis of the 109 NBS-encoding gene sequences revealed that some maize NBS-encoding genes shared high similarity to NBS-encoding genes with known functions. ZmNBS25 was included in this set of NBS-encoding genes and clustered with Arabidopsis thaliana L. AtRPM1 in the phylogenetic tree (Cheng et al., 2012). However, the response of ZmNBS25 to B. maydis or SA and its functions are yet to be elucidated. In the present study, we isolated the NBS-LRR gene ZmNBS25 from maize and investigated its disease resistance functions in Arabidopsis and rice. Our results indicate that ZmNBS25 plays important roles and can be functional across different species against diverse pathogens. Thus, it might be a valuable candidate for engineering pathogen resistance in breeding programs.
Materials and Methods
Maize Materials and Treatments
Maize CMT030 plants were grown in a greenhouse at 28°C under a 16 h light/8 h dark cycle. The fungal strain was grown at 28°C in potato dextrose agar (PDA) medium for 7 days. Healthy maize seedlings grown for 14 days (3-leaf stage) were treated with a B. maydis spores suspension (105 mL−1) in sterile deionized water. The B. maydis spore suspension was sprayed onto maize leaves, and these were covered with plastic film for 24 h to maintain moisture in the treated areas. For SA treatment, 1 mM SA (Sangon Biotech Co., Ltd., Shanghai, China) was sprayed onto maize seedlings not subject to B. maydis infection. The leaves were harvested for RNA extraction at 0, 12, 24, 48, and 60 h post B. maydis inoculation, and at 0, 1, 6, 12, and 24 h post SA treatment. Untreated plants were harvested at the same time points and used as controls. Six maize plants were collected per treatment at each time point. Three biological replicates were used for all treatments.
Bioinformatics Analysis
The GSDS website (Gene Structure Display Server1) was used to analyze ZmNBS25 gene structure (Zhang et al., 2013). Alignments between ZmNBS25 and other functional R proteins were performed in MEGA6 (Koichiro et al., 2013), and a phylogenetic tree was constructed by the neighbor-joining method based on whole protein sequences and considering 1,000 bootstrap replicates (Liu et al., 2016). Spatio-temporal expression of ZmNBS25 during maize development was investigated using the microarray data of B73 maize from PLEXdb (Sekhon et al., 2011; Dash et al., 2012). A heat map of the spatio-temporal expression of ZmNBS25 was generated in R/Bioconductor2. Promoters of ZmNBS25 were analyzed by RSAT3 (Medina-Rivera et al., 2015).
Full-Length cDNA Cloning
Total RNA extracted from the leaves of CMT030 seedlings was used to synthesize first-strand cDNA. The full-length cDNA products of ZmNBS25 were obtained using PrimeSTAR Max DNA Polymerase (TaKaRa Bio Inc., Kusatsu, Shiga, Japan) and forward (5′-ATGGCAGAAGCTGTGGTGTT-3′) and reverse (5′-CTATATGCGCAACTCCAGACC-3′) primers, under 35 cycles of 98°C for 10 s, 55°C for 15 s, and 72°C for 15 s. The products were then cloned and sequenced.
Vector Construction and Cell Death Assays in N. benthamiana
The full-length coding sequence of ZmNBS25 without a termination codon was inserted into the vector pCAMBIA1301 to generate the 35S::ZmNBS25 vector. The 35S::ZmNBS25 and pCAMBIA1301 (control) vectors were transformed into the Agrobacterium tumefaciens strain GV3101. About 100 μL of A. tumefaciens suspensions carrying 35S::ZmNBS25 or control constructs were injected into 4-week N. benthamiana leaves using a small syringe as previously described (Cesari et al., 2014; Zhang et al., 2017). Staining with 1 g L−1 3′-Diaminobenzidine (DAB; Sigma-Aldrich, St. Louis, MO, United States) and 1 g L−1 lactophenol-trypan blue (Sangon Biotech Co., Ltd., Shanghai, China) was performed as previously described (Hwang and Hwang, 2011; Zhang et al., 2017). N. benthamiana leaves were treated overnight with DAB and the stained leaves were cleared with 95% ethanol. For trypan blue staining, leaves were boiled in the trypan blue solution for 5 min and destained overnight in 2.5 g mL−1 chloral hydrate. The destained leaves were observed under a microscope (Leica DM5000 B, Leica Microsystems Ltd., Heerbrugg, Switzerland). Three biological replicates were used for the experiment.
Measurement of Electrolyte Leakage
Electrolyte leakage was determined according to Hwang and Hwang (2011). N. benthamiana leaves were infiltrated with A. tumefaciens GV3101 carrying 35S::ZmNBS25 or control constructs to determine electrolyte leakage. Thirty N. benthamiana leaf disks approximately 1 cm in diameter were punched out with a cork borer, washed in 20 mL double distilled water for 30 min, and then transferred to fresh double distilled water (20 mL). Ion leakage was measured at 0 and 48 h after infiltration. Conductance was measured with a conductivity meter (DDS-11A) and the electrical conductivities of the media were expressed as μS cm−1. Three biological replicates were set for the experiment.
Rice Transformation
The rice cultivar Zhonghua 11 was selected for ZmNBS25 transformation. Embryogenic calli were obtained using 14-day-old immature embryos. Active embryogenic calli were harvested for transformation with A. tumefaciens GV3101 containing the 35S::ZmNBS25 constructs, which was performed as previously described (Datta et al., 2000). Briefly, embryogenic calli were co-cultivated for 2 days with A. tumefaciens GV3101 in N6 medium (Sigma-Aldrich) containing 200 μM acetosyringone (Sangon Biotech Co., Ltd.). They were then thoroughly washed with sterile water and transferred to fresh medium containing 250 mg L−1 cefotaxime and 50 mg L−1 hygromycin (both Sangon Biotech Co., Ltd.). After several selection cycles, the embryogenic calli were used to propagate plants in regeneration medium. The transgenic plants were transplanted to pots filled with soil. Independent transgenic lines expressing 35S::ZmNBS25 were identified by Southern blot analysis as previously described (Jia et al., 2011; Zhao et al., 2014). Genomic DNA was extracted from the leaves of T0 transgenic lines using CTAB method (cetyl trimethyl ammonium bromide) (Clarke, 2009), and digested by the restriction enzyme EcoRV (TaKaRa Bio Inc., Kusatsu, Shiga, Japan) overnight at 37°C. A fragment of the hygromycin gene, labeled with digoxigenin by a PCR DIG Probe Synthesis Kit (Roche Molecular Diagnostics, Pleasanton, CA, United States) was used as the hybridization probe.
Arabidopsis Transformation
Arabidopsis plants were transformed with the floral dip method (Clough and Bent, 1998) using A. tumefaciens GV3101 carrying the 35S::ZmNBS25 construct or the control vector. Transformed Arabidopsis seeds were sown onto Murashige and Skoog (MS; Sigma-Aldrich) agar plates containing 20 mg mL−1 hygromycin (Roche Molecular Diagnostics) to screen for positive transformants, which were then transplanted for further growth. Genomic DNA was extracted from the leaves of transgenic Arabidopsis lines using the CTAB (cetyl trimethyl ammonium bromide) method (Clarke, 2009). The DNA extracted from each transgenic Arabidopsis plant was used as template to determine positive transgene integration by PCR. The 2× Taq Master Mix (Dye Plus; Vazyme Biotech Co., Ltd., Nanjing City, PRC) was used for PCR amplification under the following profile: 95°C for 10 s, 32 cycles of 55°C for 30 s, and 72°C for 30 s. Seeds from positive transformants were harvested, subjected to hygromycin screening, and the process was repeated until only seeds capable of growing in hygromycin medium remained. The homozygous T3 generation was selected for subsequent experiments.
Measurement of SA
In Arabidopsis and rice, SA was extracted and quantified according to a previously described method (Meuwly and Metraux, 1993; Cho et al., 2013). Briefly, leaves were flash-frozen in liquid nitrogen and ground to a very fine powder. The SA extracted from 0.5 g Arabidopsis leaf powder was determined by high-performance liquid chromatography (HPLC) (Agilent 1100 series with a C18 column, Agilent Technologies, Santa Clara, CA, United States), using SA from Dikma (Beijing, China) as the internal standard. The SA extracted from 0.3 g rice leaf powder was also determined by HPLC (Agilent 1200 series with a C18 column, Agilent Technologies) using SA from Sigma-Aldrich as the internal standard.
Arabidopsis Pathogen Inoculation and Disease Index
Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) was grown for 2 days at 28°C on King’s B (KB) medium (tryptone 20 g L−1, glycerol 10 mg L−1, K2HPO4 1.5 g L−1, MgSO4 1.5 g L−1, and agar 15 g L−1). After this period, a single Pst DC3000 colony was used to inoculate 5 mL KB medium and left to proliferate for 1 day at 28°C. One milliliter of the culture was used to inoculate 100 mL KB and left to grow for 1 day. The bacterial suspension was centrifuged at 3,000 rpm for 15 min and the pellet was homogenized in a resuspension solution of 0.01% Silwet L-77 and 10 mM MgSO4. The final optical density at 600 nm (OD600) was adjusted to 1.0 and Arabidopsis presenting 14 to 18 rosette leaves were sprayed with the bacterial suspension [105 colony forming units (CFU) mL−1] and control were sprayed with resuspension solution without Pst DC3000 (mock). Plants were then covered with plastic film for 3 days to retain moisture in the treated area. Evaluation of the disease index (DI) was performed 7 days post inoculation (dpi) as follows: DI (%) = [Σ(ni × vi)/(V × N)/100], where vi = disease rating; ni = number of plants with that disease rating; V = highest disease rating; and N = total number of observed plants (Niu et al., 2011). Three biological replicates were set and each replicate contained nine Arabidopsis plants. Cell death was measured by Evans blue staining as previously described (Xie et al., 2017).
Rice Pathogen Inoculation
To evaluate the resistance of transgenic rice to disease, rice overexpressing ZmNBS25 (ZmNBS25-OE) and control rice (EV) were inoculated with Rhizoctonia solani at the seedling stage using the leaf sheath inoculation method. Rhizoctonia solani cultured on PDA plates was cut into equal size pieces and then inoculated in rice leaf sheath (ZmNBS25-OE transgenic and control plant). Plants were then covered with plastic film to maintain moisture. Disease phenotype was obtained at 14 days post R. solani inoculation.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was isolated with TRIzol (Thermo Fisher Scientific, Waltham, MA, United States) from 6 maize plants, 12 Arabidopsis plants, and 6 rice plants subject to each treatment including maize treatment with SA, B. maydis and control, Arabidopsis treatment with Pst. DC3000 and control, rice treatment with Rhizoctonia solani and control. Three biological repeats were performed for each treatment. DNase (TaKaRa Bio Inc., Kusatsu, Shiga, Japan) was used to eliminate genomic DNA contamination. A reverse transcription kit (Roche Molecular Systems, Inc., Pleasanton, CA, United States) was used to synthesize first-strand cDNA from 1 μg total RNA from each sample. Quantitative real-time polymerase chain reactions (qRT-PCR) were run on an Applied Biosystems 7300 system (Applied Biosystems, Foster City, CA, United States) using the primers listed in Supplementary Table S1. The relative expression level of genes investigated in this study was calculated with the formula 2−ΔΔCt, ΔΔCt = (CTgene - CTactin)treat - (CTgene - CTactin)control (Zhang et al., 2017).
Statistical Analyses
The data were analyzed by Student’s t-test to evaluate differences between control and treated samples. Statistical significance was set at ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Each assay contained three independent replicates.
Results
Characterization of ZmNBS25
Our previous study has identified 109 NBS encoding genes in maize (Cheng et al., 2012), including ZmNBS25. The protein encoded by this gene clustered with the Arabidopsis disease resistance protein AtRPM1 (Cheng et al., 2012). The full-length cDNA sequence of ZmNBS25 (GRMZM2G050959) is 3077 bp long, containing a 2736 bp coding sequence (Supplementary Figure S1). The phylogenetic analysis, based on whole protein sequences, showed that ZmNBS25 clustered with HvSL8 of Hordeum vulgare, ZmMRPR1 of Z. mays, SiRPM1-like of Setaria italica, and OsYR5 of O. sativa (Figure 1A and Supplementary Table S2). Whole-protein sequence identities between ZmNBS25 and HvSL8, ZmMRPR1, SiRPM1-like, OsYR5 were 45.84, 45.54, 47.46, and 46.92%, respectively. Protein sequence motif analysis showed that ZmNBS25 has conserved NBS motifs, including P-Loop, GLPL, kinase-2, MHD motif, and a C-terminal LRR domain (Bertioli et al., 2003; Cheng et al., 2012; Cesari et al., 2014) (Figure 1B).

FIGURE 1. Comparison of ZmNBS25 with other disease resistance proteins. (A) Phylogenetic analysis of ZmNBS25 and other plant resistance proteins. Bootstrap values (1,000 replicates) are shown as percentages at the branch nodes. Bar = 0.2. GenBank accession numbers and plant sources are: AtRPM1 (CAA61131.1) from Arabidopsis thaliana, HvSL8 (AJ507098) and HvMla1 (AAG37356.1) from Hordeum vulgare, OsXA1 (BAA25068.1), OsRGA5 (AGM61351.1), and OsYR5 (AF456245.1) from Oryza sativa, SbPc-A (ACB72454.1) from Sorghum bicolor, SiRPM1-like (XM_004978992.1) from Setaria italica, TaLr10 (ADM65840.1) from Triticum dicoccoides, ZmMRPR1 (NM_001112339.1), ZmRp1 (AAP81261.1), ZmRXO1 (AAX31149.1), and ZmRGA4 (NP_001147651.1) from Zea mays. (B) Conserved domain comparisons between the amino acid sequences of ZmNBS25 and other resistance proteins. Black, red, and blue shading represent 100%, ≥75%, and ≥50% amino acid sequence similarity, respectively.
Expression patterns of ZmNBS25 in various maize organs and different developmental stages were investigated by using the microarray data. We found that ZmNBS25 expressed highly in root and stem, but relatively low in other tissues, such as leaf and cob (Figure 2A). We further examined the spatial expression patterns of ZmNBS25 in seven tissues (root, stem, leaf, tassel, silk, husk, and cob) of maize by qRT-PCR and found that, similar to the microarray analysis, ZmNBS25 expression was high in root and stem, and low in leaf and cob (Figure 2B).
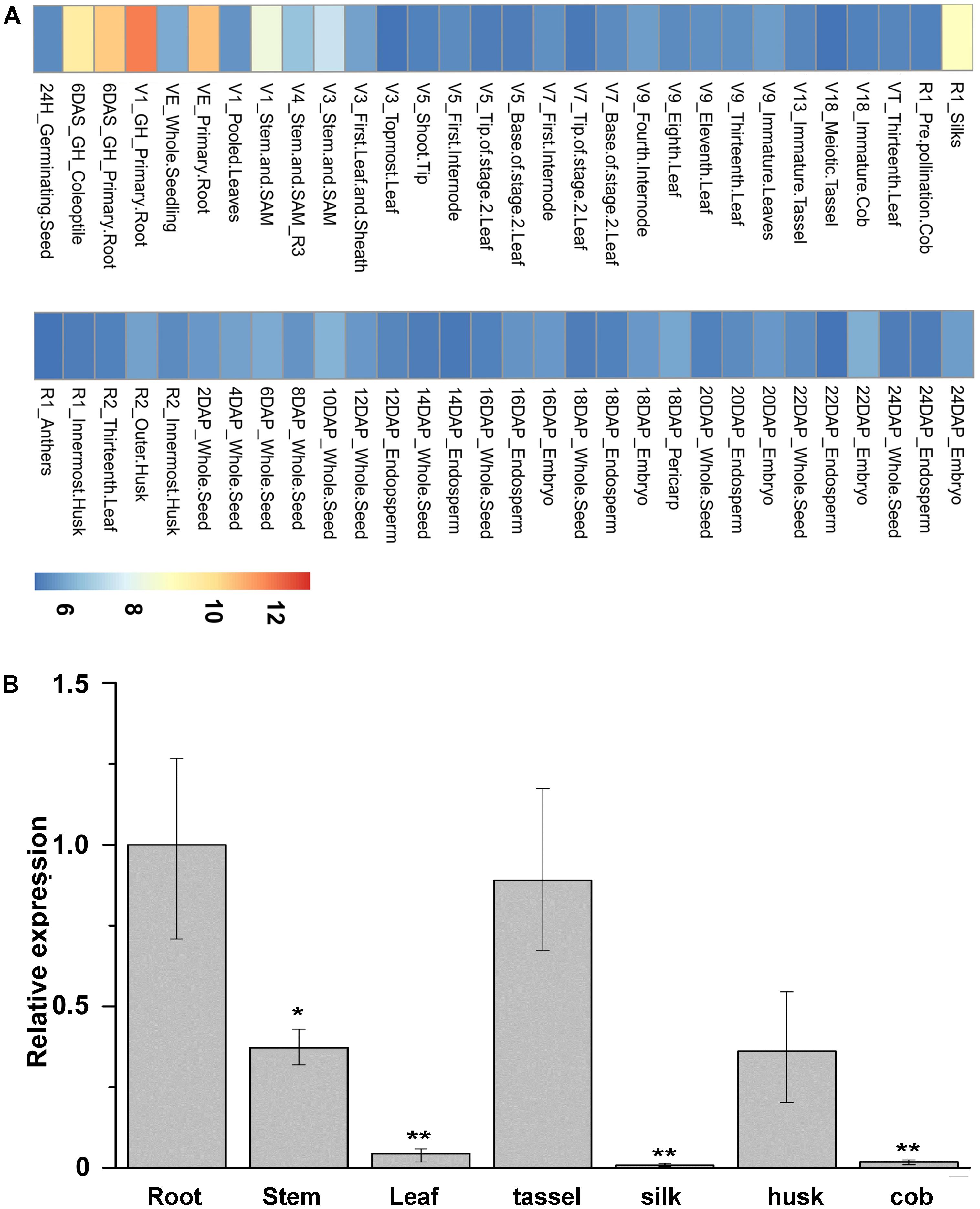
FIGURE 2. Spatial expression pattern of ZmNBS25 in maize. (A) Heat map of ZmNBS25 gene expression in maize. High, medium, and low expression levels are represented in red, yellow, and blue in the map, respectively. V, vegetative growth stage; R, reproductive growth stage; DAS, days after sowing, and DAP, days after pollination. (B) Expression pattern of ZmNBS25 in different maize CMT030 tissues. Data represent mean relative expression values ± standard deviation from three independent experiments. The expression level in root was used as the control and assigned the value of 1. Asterisks indicate statistically significant differences between root and other tissues by Student’s t-test (∗P< 0.05, ∗∗P< 0.01).
ZmNBS25 Is Induced in Leaf Upon B. maydis Infection and SA Treatment
To test whether ZmNBS25 is responsible for disease resistance in maize, we sprayed either a spore suspension of B. maydis or SA on leaves to mimic the natural disease environment. After inoculating maize CMT030 with B. maydis, the expression level of ZmNBS25 was first reduced at 12 h post inoculation (hpi), followed by a significantly increased at 24, 48, and 60 hpi (Figure 3A). On the other hand, after SA treatment, the expression level of ZmNBS25 was consistently increased from 1 to 24 hpi when compared to control samples (Figure 3B).
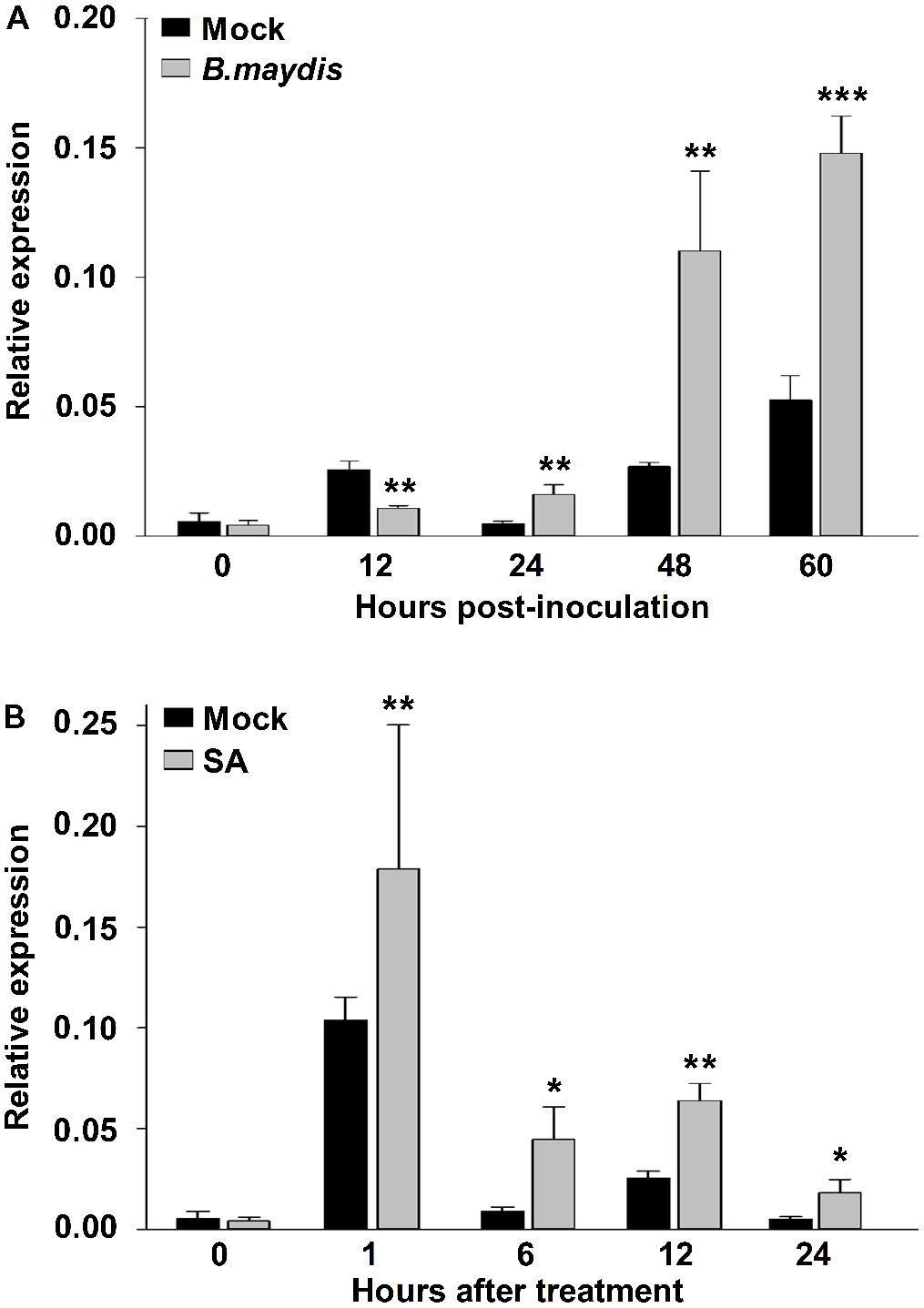
FIGURE 3. Gene expression analysis of ZmNBS25 in maize leaf upon Bipolaris maydis inoculation and salicylic acid (SA) treatment. (A) Expression levels of ZmNBS25 upon B. maydis inoculation. Maize seedlings at the 3-leaf stage were infected with B. maydis, and leaves were harvested at 0, 12, 24, 48, and 60 h post inoculation. (B) Expression levels of ZmNBS25 treated with SA. Three-leaf stage maize seedlings received SA and leaves were harvested at 0, 1, 6, 12, and 24 h after SA treatment. Data were normalized using the transcript level of ZmActin and Zmtubulin, and relative expression levels of ZmNBS25 at different time points are shown as folds of the level of ZmActin. Data represent mean relative expression values ± standard deviation from three independent experiments. Asterisks indicate statistically significant differences between treated and untreated maize by Student’s t-test (∗P< 0.05, ∗∗P< 0.01, ∗∗∗P< 0.001).
Transient Overexpression of ZmNBS25 in N. benthamiana Leaves Induces HR
To determine whether ZmNBS25 is involved in disease resistance in other crops, we cloned ZmNBS25 from maize CMT030 and generated an overexpression vector of ZmNBS25 using the pCAMBIA1301 vector with a CaMV35S promoter. After transforming the plasmid in A. tumefaciens GV3101, this was infiltrated into N. benthamiana leaves to verify whether transient ZmNBS25 overexpression could cause HR cell death. Large amounts of H2O2 accumulation were observed in N. benthamiana leaves after ZmNBS25 transient overexpression for 48 h by DAB staining (Figure 4A and Supplementary Figure S2). Trypan blue staining and electrical conductivity are two well established indicators of electrolyte leakage and correlate with the severity of visual damage (Zhang et al., 2016). As shown in Figure 4B, ion conductivity significantly increased in plants overexpressing ZmNBS25 compared with plants expressing pCAMBIA1301. Furthermore, trypan blue staining was darker in the leaves of ZmNBS25-OE plants than in the leaves of pCAMBIA1301 transgenic plants (Figure 4A and Supplementary Figure S2). These results suggested that the transient overexpression of ZmNBS25 in tobacco leaves induced HR and H2O2 accumulation, acting as a defense response to stress.
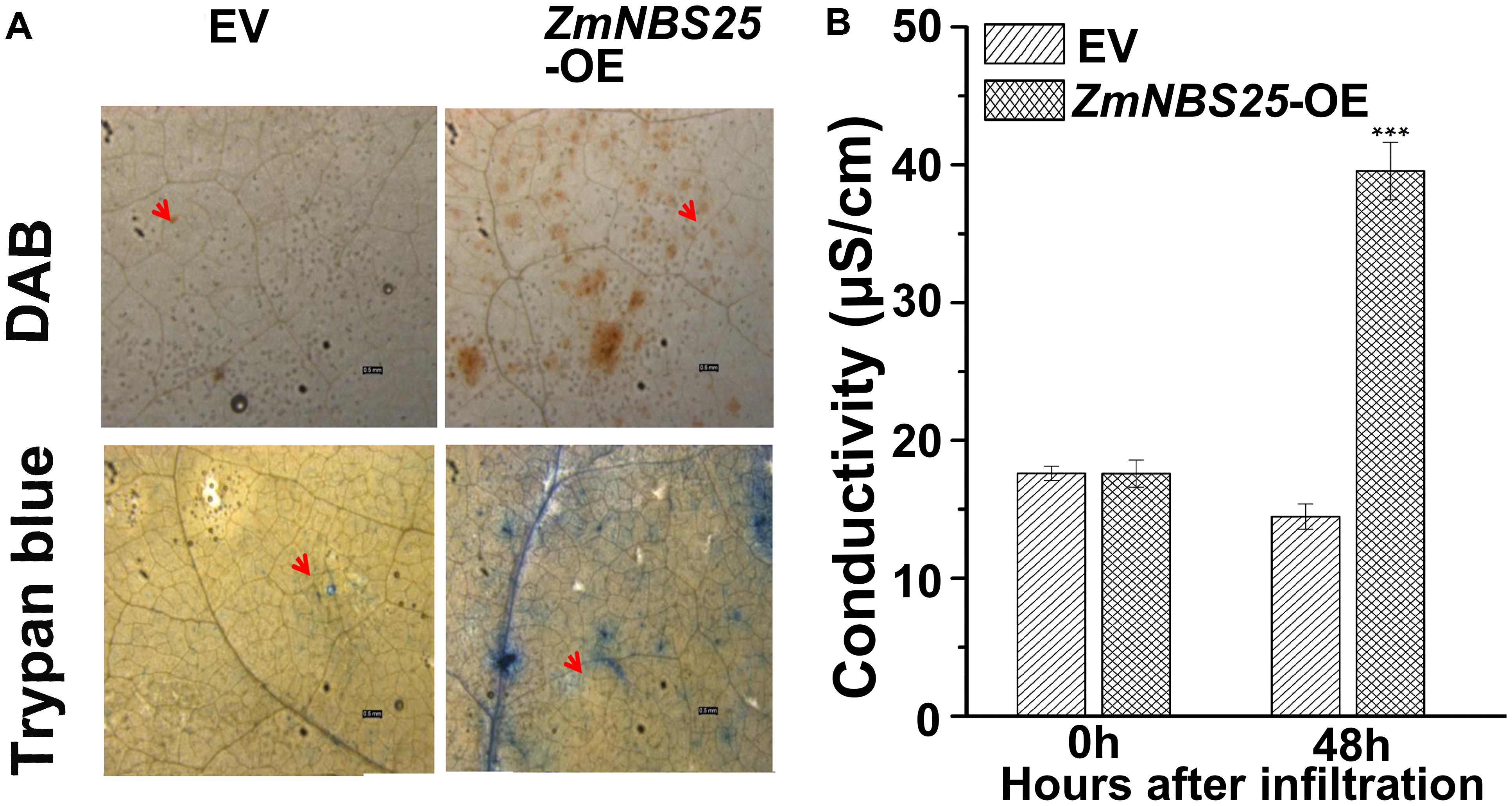
FIGURE 4. Effect of transient ZmNBS25 expression on immunity induction in Nicotiana benthamiana. (A) DAB staining (above) and trypan blue staining (below) of N. benthamiana leaves that transiently express 35S::ZmNBS25 and pCAMBIA1301. (B) Electrolyte leakage of N. benthamiana leaves infiltrated with either 35S::ZmNBS25 or pCAMBIA1301. Bars = 0.5 mm. The arrow indicates the infiltration spot. Data represent mean relative expression values ± standard deviation from three independent experiments. Asterisks indicate statistically significant differences between pCAMBIA1301 and 35S::ZmNBS25 tobacco by Student’s t-test (∗∗∗P< 0.001).
ZmNBS25 Overexpression in Arabidopsis Enhances Resistance to Pst DC3000
To investigate whether ZmNBS25 has a universal role in disease resistance, we cloned ZmNBS25 into a pCAMBIA1301 binary vector and generated ZmNBS25-OE transgenic Arabidopsis plants (Supplementary Figure S3). As shown in the HPLC chromatogram, total SA levels were drastically increased in ZmNBS25-OE plants (Peak area of OE1 was 460.4 g−1 and peak area of OE2 was 486.2 g−1) compared with non-transgenic Col-0 plants (Peak area was 75.5 g−1) without any treatment (Figure 5A). We then examined the effect of ZmNBS25 overexpression after Pst DC3000 inoculation. Two transgenic Arabidopsis lines and Col-0 were sprayed with a virulent Pst DC3000 suspension (105 CFU mL−1). Seven days after inoculation, the ZmNBS25-OE lines showed significant resistance to Pst DC3000 (Figures 5B–D). Evans blue staining of infected Arabidopsis plants showed that cell death was more evident in Col-0 plants than in ZmNBS25-OE plants (Figure 5B). The disease severity of ZmNBS25-OE lines was 23.66% on average, while that of Col-0 plants was 45.58% (Figure 5C). The density of Pst DC3000 on ZmNBS25-OE plants was significantly lower than in Col-0 plants at 7 dpi (Figure 5D). Moreover, the density of Pst DC3000 grown on the ZmNBS25-OE1 plants were significant lower than that on the ZmNBS25-OE2 plants at 7 dpi (P = 0.0114) (Figure 5D). This might be caused by the higher ZmNBS25 expression in ZmNBS25-OE1 Arabidopsis plants than in ZmNBS25-OE2 plants (Supplementary Figure S3). These results strongly suggest that ZmNBS25 plays important roles in disease resistance.
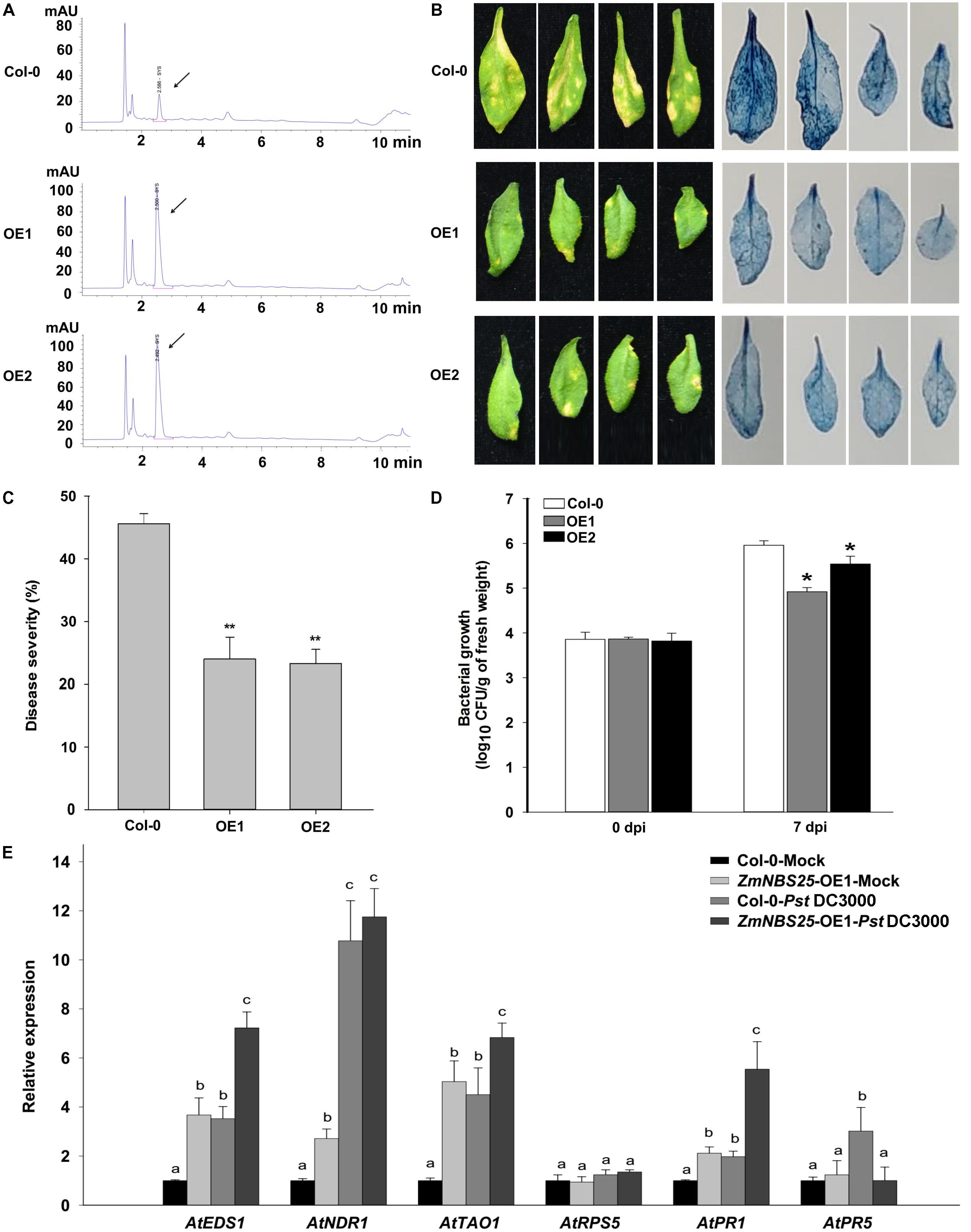
FIGURE 5. Enhanced resistance of ZmNBS25-OE transgenic Arabidopsis plants to Pst DC3000. (A) Chromatogram of SA extracted from ZmNBS25-OE and Col-0 plants without pathogen inoculation. mAU indicates peak height. The arrow indicates the SA peak. (B) Symptoms and Evans blue staining of ZmNBS25-OE and Col-0 leaves after 7 days of Pst DC3000 infection. Blue areas represent cell death. (C) Disease index of ZmNBS25-OE and Col-0 plants upon Pst DC3000 infection. (D) Pst DC3000 density on the infected leaves of ZmNBS25-OE and Col-0 plants. Leaf samples were collected at 0 and 7 days post inoculation (dpi). CFU represents colony-forming unit. (E) Expression levels of defense related genes in ZmNBS25-OE and Col-0 plants with or without Pst DC3000 infection. OE1 and OE2 represent two different transgenic lines of ZmNBS25-overexpressed Arabidopsis. (C,D) Asterisks indicate statistically significant differences between ZmNBS25-OE lines and Col-0 plants by Student’s t-test (∗P< 0.05, ∗∗P< 0.01). (E) Data were normalized using the transcript level of AtActin and AtUbiquitin. The expression levels of genes in Col-0 plants without Pst DC3000 infection were used as controls and assigned the value of 1. Different letters above the columns indicate significant differences at P< 0.05 level among Col-0-Mock, ZmNBS25-OE1-Mock, Col-0-Pst DC3000, and ZmNBS25-OE1-Pst DC3000. Data represent mean ± standard deviation from three independent experiments.
To examine if the enhanced resistance to Pst DC3000 is related to changes in defense responsive genes, we measured the expression levels of some defense related genes in Col-0 and ZmNBS25-OE plants upon Pst DC3000 infection. Particularly, we examined the relative expression levels of genes involved in pathogen resistance (PR) and on the SA-dependent defense signaling pathway, including AtEDS1 (Enhanced Disease Susceptibility 1) (Falk et al., 1999), AtNDR1 (Non-race Specific Disease Resistance 1) (Day et al., 2006), AtTAO1 (Target of AvrB Operation) (Eitas et al., 2008), AtRPS5 (Resistance to P. syringae protein 5) (Warren et al., 1998), AtPR1, and AtPR5 in Col-0 and ZmNBS25-OE plants. Except for AtRPS5 and AtPR5, defense genes showed higher expression levels in ZmNBS25-OE plants than in Col-0 plants without Pst DC3000 infection. On the other hand, the expression levels of AtEDS1, AtNDR1, AtTAO1, AtPR1, and AtPR5 in Col-0 plants were significantly induced by Pst DC3000 (Figure 5E and Supplementary Tables S3, S4). Similar inductions were observed in Pst DC3000-infected ZmNBS25-OE plants compared with control ZmNBS25-OE plants, except AtPR5. When compared with Pst DC3000-infected Col-0 plants, the expression levels of AtEDS1, AtTAO1, and AtPR1 were significantly increased in the Pst DC3000-infected ZmNBS25-OE plants (Figure 5E and Supplementary Tables S3, S4). These results indicate that ZmNBS25 overexpression enhanced disease resistance of transgenic Arabidopsis against Pst DC3000.
Transgenic Overexpression of ZmNBS25 in Rice Confers Disease Resistance Without Affecting Grain Yield
To determine whether ZmNBS25 overexpression confers disease resistance in other heterologous plant systems, we generated ZmNBS25-OE transgenic rice lines. Southern blot analysis showed three separate transfer (T)-DNA insertions events with only one copy each (1, 5, and 7; Supplementary Figure S4); thus, we selected two independent transgenic lines, L5 and L7, for subsequent experiments. Plant resistant against biotrophic pathogens is usually regulated by the SA-dependent pathway (Felton and Korth, 2000), so we first measured the SA content in ZmNBS25-OE and control plants. HPLC chromatograms showed that ZmNBS25-OE lines accumulated more SA (peak area 309.489 g−1) than control transgenic plants (peak area 74.749 g−1) without pathogen inoculation (Figure 6A). To further determine the disease resistance capacity of ZmNBS25 overexpression, we inoculated R. solani, which can cause rice sheath blight, at rice leaf sheath. ZmNBS25-OE transgenic rice lines developed fewer and smaller disease lesions than control transgenic plants at 14 dpi (Figure 6B). We found that two typical defense related genes, OsPAL06 and OsPXa5, were significantly upregulated in ZmNBS25-OE lines after R. solani infection (Figure 6C).
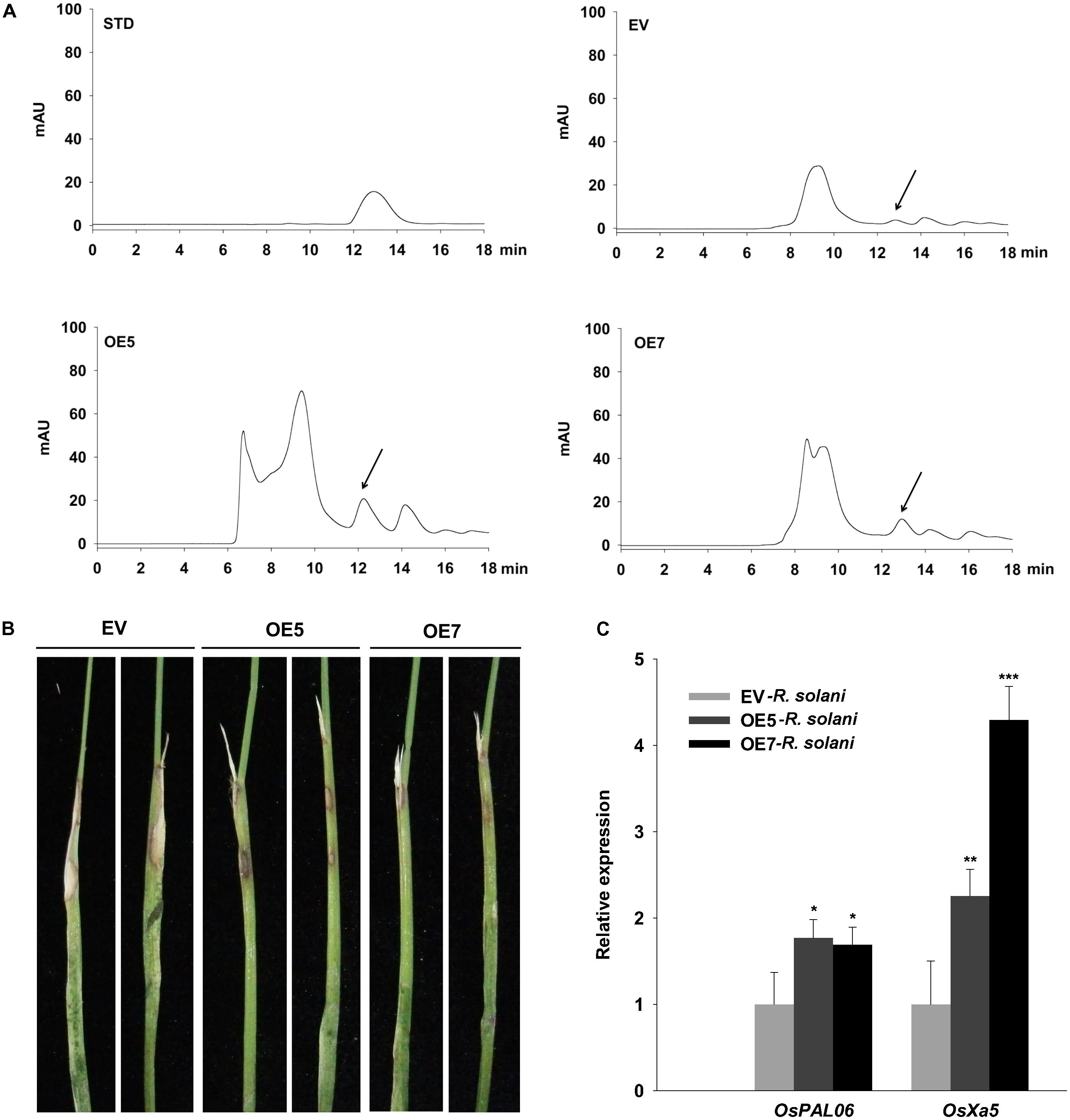
FIGURE 6. Functional analysis of the ZmNBS25 gene in rice. (A) Chromatogram of SA extracted from ZmNBS25-OE and control (EV) rice plants without pathogen inoculation. STD represents standard SA; mAU indicates peak height. The arrow indicates the SA peak. (B) Resistance of ZmNBS25-OE plants to rice sheath blight. (C) Expression levels of defense related genes in ZmNBS25-OE and control plants inoculated with Ralstonia solani. Data were normalized using the transcript level of Ostubulin and 18srRNA. OE5 and OE7 represent two different transgenic lines of ZmNBS25-overexpressed rice. The expression levels of genes in R. solani-infected EV plants were used as controls and assigned the value of 1. Asterisks indicate statistically significant differences between treated and untreated maize by Student’s t-test (∗P< 0.05, ∗∗P< 0.01, ∗∗∗P< 0.001). Data represent mean relative expression values ± standard deviation from three independent experiments.
To investigate the effect of overexpressing the ZmNBS25 gene on yield-related traits, we compared seeds’ size and 1000-grain weight between wild type (WT) and ZmNBS25-OE rice lines (Supplementary Figure S5). No phenotypic differences, including seed length and width, were observed between WT and ZmNBS25-OE rice lines (Supplementary Figures S5A,C,D). Except for ZmNBS25-OE line L7, no significant reductions in 1000-grain weight were observed in ZmNBS25-OE lines (L1, L5) (Supplementary Figure S5B). In addition, we observed similar results in Arabidopsis, where seeds showed no phenotypic differences between WT and ZmNBS25-OE lines (Supplementary Figure S6).
Discussion
ZmNBS25 Is a Novel Maize NBS-LRR Resistance Gene
Nucleotide-binding site-leucine-rich repeat proteins play important roles in pathogen recognition and defense response signal transduction (Ameline-Torregrosa et al., 2008; Gao et al., 2010). Some NBS-LRRs that confer resistance to microbial pathogens and certain environmental stressors have been cloned from higher plants (Liu et al., 2007). In the present study, we cloned and systematically characterized a novel NBS-LRR-encoding gene, ZmNBS25, in maize. We found that ZmNBS25 could be induced by B. maydis inoculation and SA treatment. Overexpression of ZmNBS25 provides enhanced disease resistance in transgenic rice and Arabidopsis. This is similar to the function of ZmRXO1, which is a maize NBS-LRR gene involved in resistance to diverse pathogens, including rice blast (Zhao et al., 2004, 2005). Our phylogenetic analysis based on whole-protein sequences also showed that ZmNBS25 is closely related to ZmRXO1. In addition, ZmNBS25 has typical NBS-ARC, P-Loop, GLPL, kinase-2, MHD, and other conserved motifs similar to S-L8, MRPR1, RPM1-like, and YR5 proteins, suggesting that ZmNBS25 can participate in maize pathogen interactions or defense responses.
ZmNBS25 Is Involved in Defense Responses to Biotic Stresses
Our results indicated that ZmNBS25 was expressed at relatively low levels in uninfected maize leaves; however, it was significantly upregulated at 24 h post B. maydis inoculation, indicating that ZmNBS25 might be associated with B. maydis resistance in maize. Similar expression patterns have been observed in other plants NBS-LRR genes, such as: AhRRS5, a R. solanacearum resistance gene in peanut (Zhang et al., 2017); Xa1, a bacterial resistance gene from rice (Yoshimura et al., 1998); and SacMi, a M. incognita resistance gene (Zhou et al., 2018). In addition, the expression level of ZmNBS25 was significantly upregulated by SA treatment. SA is a well-known signaling molecule involved in defense against stress (Divi et al., 2010). It accumulates in plant tissues challenged by pathogen infections, and it can induce SAR and increase the expression of certain PR genes, which generally increase resistance to a wide range of diseases (Gaffney et al., 1993; Yang et al., 2013; Novakova et al., 2014). The expression of disease resistance genes such as RCY1 and NPR1 in Arabidopsis partially depends upon SA signaling (Cao et al., 1997; Takken and Joosten, 2000). Given the similar expression pattern of ZmNBS25 upon SA treatment and B. maydis induction, the enhanced defense response to B. maydis in ZmNBS25-OE lines is likely associated with SA signaling. Moreover, we also found that binding elements such as WBOXATNPR1, WRKY71OS, BIHD1OS, and GCCCORE could participate in plant disease resistance and were enriched in the ZmNBS25 promoter (Supplementary Figure S7). For example, WBOXATNPR1 recognizes the WRKY DNA binding proteins induced by SA (Eulgem et al., 2000), and WRKY71OS interacts with certain WRKY family members that play vital roles in plant disease resistance (Eulgem et al., 2000; Shimono et al., 2012). Together, these results suggest that ZmNBS25 plays a vital role in the defense response to diseases.
ZmNBS25 Confers Disease Resistance in Heterologous Plant Systems
Activation of NBS-LRR proteins triggers programmed cell death and reactive oxygen species (ROS) production in plants (Andersson et al., 2006; Qi and Innes, 2013). Indeed, we found that ZmNBS25 might positively regulate SA-dependent cell death and disease resistance during pathogen infection. Transient overexpression of ZmNBS25 in N. benthamiana induced a HR and caused cell death and H2O2 accumulation without pathogenic infection. Therefore, ZmNBS25 could be involved in ROS signaling pathways for disease resistance. An R gene that can be integrated into a heterologous cereal crop will significantly improve disease resistance strategies for that species. An earlier study on the ZmRXO1 gene in rice showed that it is possible to transfer a NBS-LRR-type R gene to a distantly related cereal species (Zhao et al., 2005). Studies have demonstrated that SA is involved in defense responses mediated by plant NBS-LRR R proteins (Roberts et al., 2013). For instance, the expression of ADR1-L2D848V in Arabidopsis caused increased disease resistance and constitutively high SA levels (Roberts et al., 2013). In our study, ZmNBS25 overexpression in transgenic Arabidopsis and rice produced more SA than in control plants and enhanced transgenic plants resistance to disease, indicating that ZmNBS25 might participate in disease responses by inducing SA accumulation.
ZmNBS25-Mediated Pathogen Resistance Is Concomitant With the Upregulation of Defense-Related Genes
Without Pst DC3000 infection, the expression of defense genes AtEDS1, AtNDR1, AtTAO1, and AtPR1 was higher in ZmNBS25-OE plants than in Col-0 plants, indicating that ZmNBS25 overexpression affected the expression of defense-related genes and resulted in enhanced resistance to disease. In Pst DC3000 infected ZmNBS25-OE transgenic Arabidopsis plants, disease severity was reduced, and ZmNBS25-OE also affected the expression of some defense genes after Pst DC3000 inoculation as defense genes were significantly upregulated in ZmNBS25-OE plants after Pst DC3000 infection. Moreover, ZmNBS25-OE rice plants showed similar changes of defense-related genes after R. solani infection. These data indicated that ZmNBS25 overexpression could promote the activation of defense response in Arabidopsis and rice plants infected with Pst DC3000 and R. solani, respectively. Taken together, our results suggest that ZmNBS25 can function as a positive regulator to prime defense response upon pathogen infection.
Outlook
The results in our current study strongly suggest that ZmNBS25 can function as a disease resistance gene across different species, being a valuable candidate for engineering resistance in breeding programs. Given the common difficulties of gene editing in maize, a cost-effective and high-efficient transformation method is desired in the future to further evaluate the disease resistance function of ZmNBS25 in maize by manipulating its gene expression.
Author Contributions
YX, FL, SZ and XL conceived the project. YX carried out the experiments and FL performed the statistical analysis. YX, FL, SZ and XL wrote the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (No. 2016YFD0300300), National Natural Science Foundation of China (No. 31201217), and Graduate Innovation Fund of Anhui Agricultural University (No. 2018yjs-40).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Shanshan Xie for providing the P. syringae pv. tomato DC3000 strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01033/full#supplementary-material
FIGURE S1 | Gene structure of ZmNBS25.
FIGURE S2 | Trypan blue and DAB staining of N. benthamiana leaves transiently expressing 35S::ZmNBS25 and pCAMBIA1301. Bar = 0.5 mm. The arrow indicates the infiltration spot.
FIGURE S3 | Relative expression levels of ZmNBS25 in ZmNBS25-OE1 and ZmNBS25-OE2 transgenic Arabidopsis lines.
FIGURE S4 | Southern blot analysis of ZmNBS25-OE transgenic rice lines.
FIGURE S5 | Phenotypes of grain yields of wild type (WT) and the ZmNBS25-OE rice lines. (A) Grain size comparison between WT and ZmNBS25-OE seeds.(B) 1000-grain weights of WT and ZmNBS25-OE seeds. (C) Grain lengths of WT and ZmNBS25-OE seeds. (D) Grain widths of WT and ZmNBS25-OE seeds. Scale bars = 5mm. Student’s t-tests were performed between WT and ZmNBS25-OE rice lines (∗∗∗P < 0.001).
FIGURE S6 | Seed phenotypes of WT and ZmNBS25-OE Arabidopsis lines.
FIGURE S7 | Analysis of regulatory elements in the predicted promoter of ZmNBS25. The predicted promoter sequence was 2 kb upstream the ZmNBS25 gene.
TABLE S1 | Primers used for qRT-PCR in this study.
TABLE S2 | Disease resistance proteins in various plant species.
TABLE S3 | The cycle threshold (Ct) of defense-related genes in Arabidopsis from qRT-PCR program for Figure 5E.
TABLE S4 | Statistical analyses of genes relative expression levels for Figure 5E.
Footnotes
References
Ameline-Torregrosa, C., Wang, B. B., O’bleness, M. S., Deshpande, S., Zhu, H., Roe, B., et al. (2008). Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 146, 5–21. doi: 10.1104/pp.107.104588
Andersson, M. X., Kourtchenko, O., Dangl, J. L., Mackey, D., and Ellerström, M. (2006). Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J. 47, 947–959. doi: 10.1111/j.1365-313X.2006.02844.x
Bertioli, D. J., Leal-Bertioli, S. C., Lion, M. B., Santos, V. L., Pappas, G. Jr., Cannon, S. B., et al. (2003). A large scale analysis of resistance gene homologues in Arachis. Mol. Genet. Genomics 270, 34–45. doi: 10.1007/s00438-003-0893-4
Bonardi, V., Tang, S. J., Stallmann, A., Roberts, M., Cherkis, K., and Dangl, J. L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 16463–16468. doi: 10.1073/pnas.1113726108
Burkhard, P., Stetefeld, J., and Strelkov, S. V. (2001). Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88. doi: 10.1016/S0962-8924(00)01898-5
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Cesari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. doi: 10.15252/embj.201487923
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae Effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Chen, X., and Ronald, P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16, 451–459. doi: 10.1016/j.tplants.2011.04.003
Cheng, Y., Li, X., Jiang, H., Ma, W., Miao, W., Yamada, T., et al. (2012). Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J. 279, 2431–2443. doi: 10.1111/j.1742-4658.2012.08621.x
Cho, K., Han, O., Tamogami, S., Shibato, J., Kubo, A., Agrawal, G. K., et al. (2013). Quantification of jasmonic and salicylic acids in rice seedling leaves. Methods Mol. Biol. 956, 185–200. doi: 10.1007/978-1-62703-194-3_13
Clarke, J. D. (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009:dbrot5177. doi: 10.1101/pdb.prot5177
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dangl, J. L., Dietrich, R. A., and Richberg, M. H. (1996). Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8, 1793–1807. doi: 10.1105/tpc.8.10.1793
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Dash, S., Van Hemert, J., Hong, L., Wise, R. P., and Dickerson, J. A. (2012). PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Res. 40, D1194–D1201. doi: 10.1093/nar/gkr938
Datta, K., Koukolíkova-Nicolá, Z., Baisakh, N., Oliva, N., and Datta, S. K. (2000). Agrobacterium-mediated engineering for sheath blight resistance of indica rice cultivars from different ecosystems. Theor. Appl. Genet. 100, 832–839. doi: 10.1007/s001220051359
Day, B., Dahlbeck, D., and Staskawicz, B. J. (2006). NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18, 2782–2791. doi: 10.1105/tpc.106.044693
Divi, U. K., Rahman, T., and Krishna, P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10:151. doi: 10.1186/1471-2229-10-151
Eitas, T. K., Nimchuk, Z. L., and Dangl, J. L. (2008). Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. U.S. A. 105, 6475–6480. doi: 10.1073/pnas.0802157105
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Falk, A., Feys, B. J., Frost, L. N., Jones, J. D., Daniels, M. J., and Parker, J. E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. U.S. A. 96, 3292. doi: 10.1073/pnas.96.6.3292
Felton, G. W., and Korth, K. L. (2000). Trade-offs between pathogen and herbivore resistance. Curr. Opin. Plant Biol. 3, 309–314. doi: 10.1016/S1369-5266(00)00086-8
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Gao, Y., Xu, Z., Jiao, F., Yu, H., Xiao, B., Li, Y., et al. (2010). Cloning, structural features, and expression analysis of resistance gene analogs in tobacco. Mol. Biol. Rep. 37, 345–354. doi: 10.1007/s11033-009-9749-2
Greenberg, J. T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. doi: 10.1146/annurev.arplant.48.1.525
Henry, E., Yadeta, K. A., and Coaker, G. (2013). Recognition of bacterial plant pathogens: local, systemic and transgene rational immunity. New Phytol. 199, 908–915. doi: 10.1111/nph.12214
Hwang, I. S., and Hwang, B. K. (2011). The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 155, 447–463. doi: 10.1104/pp.110.164848
Jia, H. F., Ren, H. Y., Gu, M., Zhao, J. N., Sun, S. B., Zhang, X., et al. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175. doi: 10.1104/pp.111.175240
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Koichiro, T., Glen, S., Daniel, P., Alan, F., and Sudhir, K. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Li, X., Zhang, Y., Yin, L., and Lu, J. (2016). Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma 254, 1–13. doi: 10.1007/s00709-016-1005-8
Liu, F., Xu, Y. J., Jiang, H. H., Jiang, C. S., Du, Y. B., Gong, C., et al. (2016). Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 17:930. doi: 10.3390/ijms17060930
Liu, J., Liu, X., Dai, L., and Wang, G. (2007). Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genomics 34, 765–776. doi: 10.1016/S1673-8527(07)60087-3
Medina-Rivera, A., Defrance, M., Sand, O., Herrmann, C., Castro-Mondragon, J. A., Delerce, J., et al. (2015). RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 43, W50–W56. doi: 10.1093/nar/gkv362
Meuwly, P., and Metraux, J. P. (1993). Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 214, 500–505. doi: 10.1006/abio.1993.1529
Molinari, S. (2016). Systemic acquired resistance activation in solanaceous crops as a management strategy against root-knot nematodes. Pest Manag. Sci. 72, 888–896. doi: 10.1002/ps.4063
Niu, D. D., Liu, H. X., Jiang, C. H., Wang, Y. P., Wang, Q. Y., Jin, H. L., et al. (2011). The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol. Plant Microbe Interact. 24, 533–542. doi: 10.1094/MPMI-09-10-0213
Novakova, M., Sasek, V., Dobrev, P. I., Valentova, O., and Burketova, L. (2014). Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum - reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 80, 308–317. doi: 10.1016/j.plaphy.2014.04.019
Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Qi, D., and Innes, R. W. (2013). Recent advances in plant NLR structure, function, localization, and signaling. Front. Immunol. 4:348. doi: 10.3389/fimmu.2013.00348
Roberts, M., Tang, S., Stallmann, A., Dangl, J. L., and Bonardi, V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 9:e1003465. doi: 10.1371/journal.pgen.1003465
Sekhon, R. S., Lin, H. N., Childs, K. L., Hansey, C. N., Buell, C. R., De Leon, N., et al. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563. doi: 10.1111/j.1365-313X.2011.04527.x
Shimono, M., Koga, H., Akagi, A., Hayashi, N., Goto, S., Sawada, M., et al. (2012). Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94. doi: 10.1111/j.1364-3703.2011.00732.x
Sinapidou, E., Williams, K., Nott, L., Bahkt, S., Tor, M., Crute, I., et al. (2004). Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38, 898–909. doi: 10.1111/j.1365-313X.2004.02099.x
Sohn, K. H., Segonzac, C., Rallapalli, G., Sarris, P. F., Woo, J. Y., Williams, S. J., et al. (2014). The nuclear immune receptor RPS4 is required for RRS1 (SLH1)-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10:e1004655. doi: 10.1371/journal.pgen.1004655
Sumita, T., Izumitsu, K., and Tanaka, C. (2017). Characterization of the autophagy-related gene BmATG8 in Bipolaris maydis. Fungal Biol. 121, 785–797. doi: 10.1016/j.funbio.2017.05.008
Takken, F. L. W., and Goverse, A. (2012). How to build a pathogen detector: structural basis of NB-LRR function. Curr. Opin. Plant Biol. 15, 375–384. doi: 10.1016/j.pbi.2012.05.001
Takken, F. L. W., and Joosten, M. H. A. J. (2000). Plant resistance genes: their structure, function and evolution. Eur. J. Plant Pathol. 106, 699–713. doi: 10.1023/A:1026571130477
Thomma, B. P., Nurnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Ullstrup, A. J. (1972). The impacts of the southern corn leaf blight epidemics of 1970-1971. Annu. Rev. Phytopathol. 10, 37–50. doi: 10.1146/annurev.py.10.090172.000345
Warren, R. F., Henk, A., Mowery, P., Holub, E., and Innes, R. W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452. doi: 10.1105/tpc.10.9.1439
Xie, S., Jiang, H., Ding, T., Xu, Q., Chai, W., and Cheng, B. (2017). Bacillus amyloliquefaciens FZB42 repressed plant miR846 to induce systemic resistance via jasmonic acid-dependent signaling pathway. Mol. Plant Pathol. 246, 1215–1231.
Yang, D. L., Yang, Y. N., and He, Z. H. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6, 675–685. doi: 10.1093/mp/sst056
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S. A. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Zhang, C., Chen, H., Cai, T., Deng, Y., Zhuang, R., Zhang, N., et al. (2017). Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 15, 39–55. doi: 10.1111/pbi.12589
Zhang, C., Zhang, H., Zhao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2013). Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 32, 1543–1555. doi: 10.1007/s00299-013-1466-6
Zhang, H., Teng, W., Liang, J., Liu, X., Zhang, H., Zhang, Z., et al. (2016). MADS1, a novel MADS-box protein, is involved in the response of Nicotiana benthamiana to bacterial harpin (Xoo). J. Exp. Bot. 67, 131–141. doi: 10.1093/jxb/erv448
Zhang, J., and Zhou, J. M. (2010). Plant immunity triggered by microbial molecular signatures. Mol. Plant 3, 783–793. doi: 10.1093/mp/ssq035
Zhao, B., Ardales, E. Y., Raymundo, A., Bai, J., Trick, H. N., Leach, J. E., et al. (2004). The avrRxo1 gene from the rice pathogen Xanthomonas oryzae pv. oryzicola confers a nonhost defense reaction on maize with resistance gene Rxo1. Mol. Plant Microbe Interact. 17, 771–779. doi: 10.1094/MPMI.2004.17.7.771
Zhao, B., Lin, X., Poland, J., Trick, H., Leach, J., and Hulbert, S. (2005). A maize resistance gene functions against bacterial streak disease in rice. Proc. Natl. Acad. Sci. U.S.A. 102, 15383–15388. doi: 10.1073/pnas.0503023102
Zhao, Y., Ma, Q., Jin, X. L., Peng, X. J., Liu, J. Y., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Zhou, X., Liu, J., Bao, S., Yang, Y., and Zhuang, Y. (2018). Molecular cloning and characterization of a wild eggplant Solanum aculeatissimum NBS-LRR gene, involved in plant resistance to Meloidogyne incognita. Int. J. Mol. Sci. 19:E583. doi: 10.3390/ijms19020583
Zhu, X., Xiao, K., Cui, H., and Hu, J. (2017). Overexpression of the Prunus sogdiana NBS-LRR subgroup gene PsoRPM2 promotes resistance to the root-knot nematode Meloidogyne incognita in tobacco. Front. Microbiol. 8:2113. doi: 10.3389/fmicb.2017.02113
Keywords: disease resistance, NBS-LRR, maize, rice, Arabidopsis
Citation: Xu Y, Liu F, Zhu S and Li X (2018) The Maize NBS-LRR Gene ZmNBS25 Enhances Disease Resistance in Rice and Arabidopsis. Front. Plant Sci. 9:1033. doi: 10.3389/fpls.2018.01033
Received: 04 December 2017; Accepted: 25 June 2018;
Published: 17 July 2018.
Edited by:
Dirk Albert Balmer, Syngenta, SwitzerlandReviewed by:
Eduard Venter, University of Johannesburg, South AfricaFei Gao, University of Missouri, United States
Copyright © 2018 Xu, Liu, Zhu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyu Li, bGl4aWFveXVAYWhhdS5lZHUuY24=; bGl4aWFveXVhaGF1QDE2My5jb20=
 Yunjian Xu
Yunjian Xu Fang Liu
Fang Liu Suwen Zhu1,2
Suwen Zhu1,2