- School of Life and Environmental Sciences, Sydney Institute of Agriculture, The University of Sydney, Brownlow Hill, NSW, Australia
The nitrate transporter 1/peptide transporter (NPF) family represents a growing list of putative nitrate permeable transport proteins expressed within multiple cell types and tissues across a diverse range of plant species. Their designation as nitrate permeable and/or selective transporters is slowly being defined as more genes are characterized and their functional activities tested both in planta and in vitro. The most notable of the NPF family has been the Arabidopsis thaliana homolog, AtNPF6.3, previously known as AtNRT1.1 or CHL1. AtNPF6.3 has traditionally been characterized as a dual-affinity nitrate transporter contributing to root nitrate uptake in Arabidopsis. It has also been identified as a nitrate sensor which regulates the expression of high-affinity nitrate transport proteins NRT2s and lateral root development as a part of the primary nitrate response in plants. The sensor function of AtNPF6.3 has also been attributed to its auxin transport activity. Other homologs of AtNPF6.3 are now being described highlighting the variability in their functional capabilities (alternative substrates and kinetics) linking to structural aspects of the proteins. This review focusses on NPF6.3-like transport proteins and the knowledge that has been gained since their initial discovery over two decades ago. The review will investigate from a structural point of view how NPF6.3-like proteins may transport nitrate as well as other ions and what can be learned from structural uniqueness about predicted activities in plants.
Introduction
Arabidopsis NPF6.3, also known as CHL1 and NRT1.1, is undoubtedly the most well studied plant NRT in the NPF family. Since NPF6.3s discovery in 1993 (Tsay et al., 1993), numerous studies have examined its in planta function and transport mechanism (Figures 1, 2). From these studies, NPF6.3 is a versatile transport protein in its substrate specificity, proposed physiological function and its location across cell types and organs. In roots, NPF6.3 has been suggested to participate directly in both high-affinity and low-affinity nitrate transport (Huang et al., 1996; Wang et al., 1998; Liu et al., 1999). Its activity across concentration gradients has been linked to the regulation of nitrate transport and assimilation genes as part of the primary nitrate response in plant roots resulting in root growth toward nitrate-rich soils (Remans et al., 2006; Ho et al., 2009). In leaves, NPF6.3 is expressed in guard cells and linked to stomata function and water use efficiency (Guo et al., 2003). NPF6.3 also transports auxin, a process negatively regulated by nitrate. The interaction between auxin and nitrate is linked to nitrate sensing and regulation of nitrate-dependent root development (Remans et al., 2006; Krouk et al., 2010; Bouguyon et al., 2015). NPF6.3 activity has also been detected in nascent organs and considered important for the development and growth of roots, stems, leaves and flower buds (Guo et al., 2001).
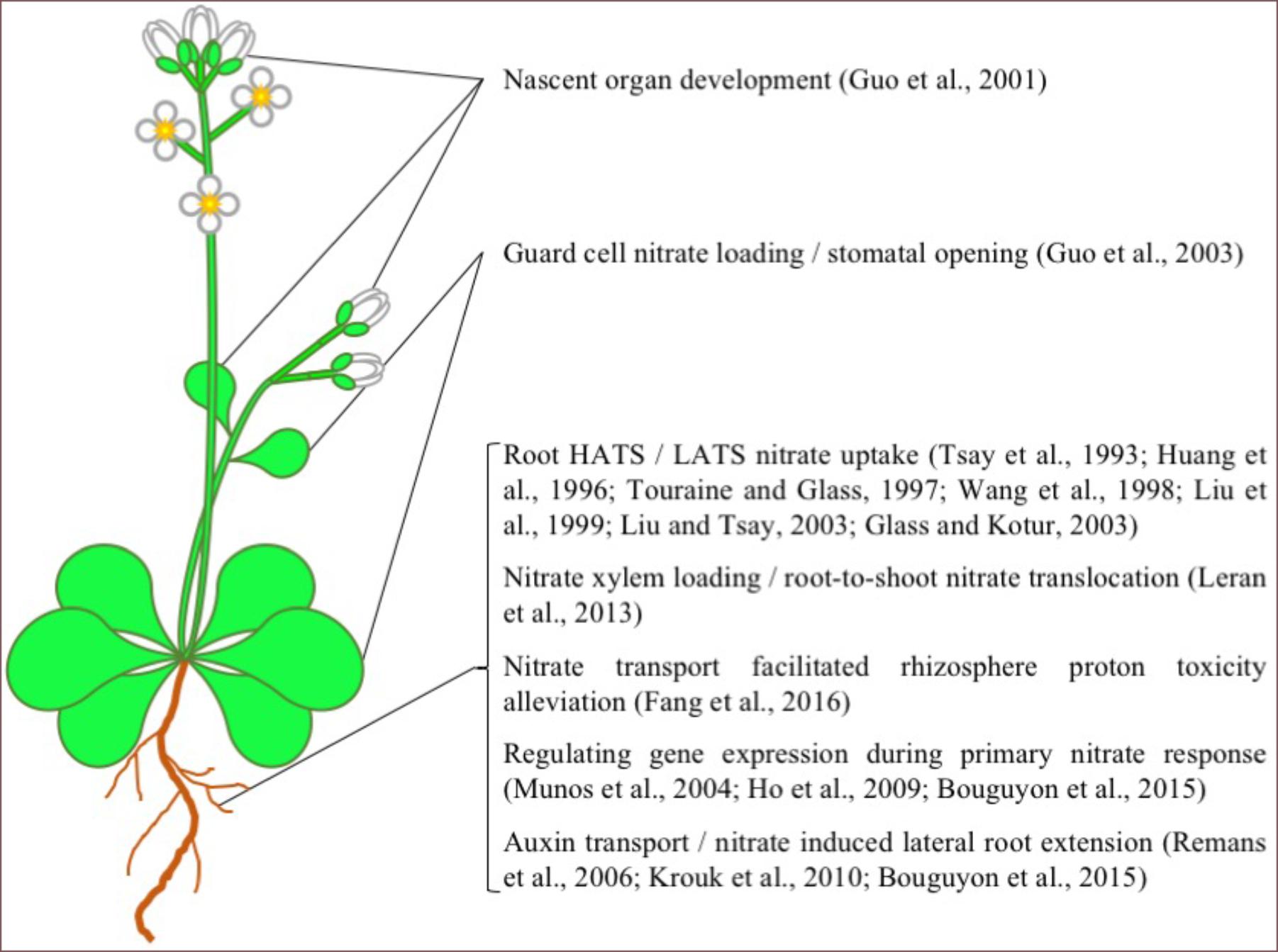
FIGURE 1. AtNPF6.3 Function in Arabidopsis. A list of the physiological functions of AtNPF6.3 in Arabidopsis. Picture source: http://laoblogger.com/arabidopsis-clipart.html.
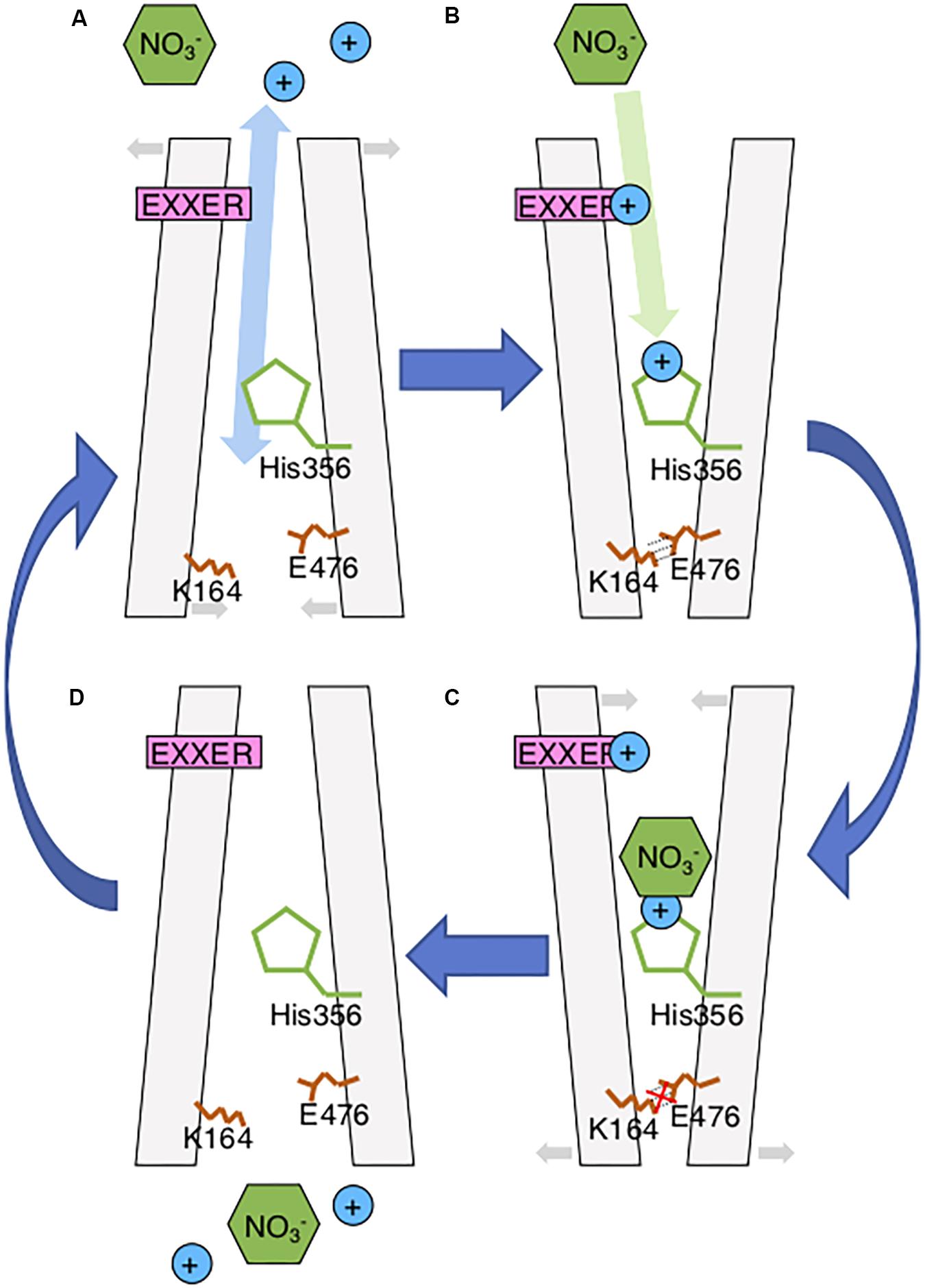
FIGURE 2. Updated AtNPF6.3 Model. (A) During the inward-facing stage, extracellular protons may enter and bind to the EXXER motif and His356 through a possible water molecule network observed in bacterial NPFs initiating the conformational rearrangement toward outward-facing stage; (B) While the salt bridge consists of K164 and E476 holding the outward-facing conformation, nitrate molecules enter the protein and bind to the protonated His356; (C) Nitrate-binding the His356 and disrupting the salt bridge. This causes a rearrangement of the protein toward the inward-facing stage; (D) Nitrate and protons are released through the intracellular gate of the protein.
The structure of NPF6.3 was recently described (Parker and Newstead, 2014; Sun et al., 2014). In both models, it was concluded that NPF6.3 transports nitrate using an “alternating access” mechanism similar to bacterial homologs, where substrate transport is facilitated by conformational rearrangement between outward-facing, occluded and inward-facing stages (Figure 2) (Solcan et al., 2012). The transport cycle begins with the protonation of the substrate binding residue, H356, followed by nitrate binding to the protonated His356 through a charge–charge interaction when NPF6.3 is in the outward-facing stage. However, a recent study of a bacterial NPF/POT/PTR homolog (PepTXc) suggested that proton and substrate binding of NPF members could involve two independent events occurring during the inward-facing and outward-facing stages, respectively (Parker et al., 2017). In its inward-facing structure, a water molecule network is observed in the central cavity extending from the substrate binding site to the ExxER motif. This could facilitate extracellular water-bonded protons entering and binding to the proton binding sites to initiate conformation change even when the extracellular gate is closed (Figure 2). The conformational change of NPF6.3 during nitrate transport was confirmed using FRET analysis of a fluorescent NPF6.3 sensor positioned between an acceptor and a donor fluorophore in the presence or absence of nitrate (Ho and Frommer, 2014). This FRET signal change was nitrate-specific and cannot be affected by other ions or dipeptides.
The two independent structures explain the nitrate transport mechanism of NPF6.3. They also confirm the 2:1 stoichiometry of proton/nitrate symport observed in Arabidopsis roots from physiological and electrophysiological studies (Meharg and Blatt, 1995; Miller and Smith, 1996). However, there are still questions around NPF6.3, such as its dual-affinity transport activity, auxin transport and its link to nitrate signaling. In this review, these questions are discussed with emphasis on the recent developments in NPF6.3 and its homologs.
His356 is the Key Structural Element for Nitrate Transport
AtNPF6.3 shares conserved structural elements (an ExxER motif and an intercellular gate salt bridge) with bacterial NPF homologs. However, the His356 substrate binding residue is unique to a select number of plant NPFs (Parker and Newstead, 2014; Sun et al., 2014; Hu et al., 2015; Wen et al., 2017). His356 replaces and consolidates the function of the peptide binding/specificity pocket and the proton-binding residues located on TM7 of bacterial NPFs (Newstead, 2017). The nitrate binding activity of His356 was demonstrated by mutagenesis studies where H356A abolished nitrate transport activity of AtNPF6.3 (Parker and Newstead, 2014; Sun et al., 2014). The nitrate selective function has recently been observed in an AtNPF6.3 maize homolog, ZmNPF6.4, where replacing Tyr370 to His (equivalent to AtNPF6.3:His356) altered substrate selectivity from chloride to nitrate (Wen et al., 2017). Apart from AtNPF6.3, only two other published NPFs contain an equivalent His residue, ZmNPF6.6 and OsNPF6.5 (Hu et al., 2015; Wen et al., 2017). Interestingly, while sharing a nitrate selectivity residue, both NPFs facilitate nitrate transport at low concentrations (0.25 mM), suggesting the His residue is also important for high-affinity nitrate transport.
Chloride Transport in NPF6s
Our group recently characterized the chloride transport activity in AtNPF6.3, and its His-containing homolog, ZmNPF6.6 (Wen et al., 2017). AtNPF6.3 and ZmNPF6.6 transport chloride when nitrate is absent, following a linear un-saturable pattern. According to Ho and Frommer (2014), chloride does not change the FRET signal of the AtNPF6.3 fluorescent sensor. Therefore AtNPF6.3 must maintain its conformation without going through an “alternative access” cycle when transporting chloride. Additionally, chloride transport in AtNPF6.3 is unlikely to involve a substrate-transporter binding episode as no chloride molecule was trapped in the AtNPF6.3 structure during crystallization even though chloride was present (Sun et al., 2014). Given the strong inhibition of nitrate on chloride transport in both AtNPF6.3 and ZmNPF6.6, we suggest nitrate occupation of the substrate binding pocket must block chloride transport (Wen et al., 2017). The chloride transport properties of AtNPF6.3 and ZmNPF6.6 indicate a channel-like activity similar to activities observed in protein families, including LacY, AKT, KUP, and CLC (Conde et al., 2010). Nevertheless, both proteins must overcome an inherent internal chloride concentration gradient (∼24–62 mM) to permit chloride uptake into Xenopus oocytes (Weber, 1999; Bröer, 2010). This suggests that some level of facilitated chloride transport may occur in AtNPF6.3 and ZmNPF6.6. The chloride permeability of these NPFs may provide plants a physiological opportunity for chloride uptake (Wege et al., 2017). Correspondingly, nitrate selectivity could act to inhibit excessive chloride uptake to avoid chloride toxicity (Wen et al., 2017). In addition, given the similar effect of nitrate on auxin transport in AtNPF6.3, the auxin transport activity may share a similar channel-like mechanism with that for chloride (Krouk et al., 2010). Further structural evidence will be required to confirm if this is the case.
Chloride transport has been reported in another NPF member, ZmNPF6.4, although with distinct differences (Wen et al., 2017). Unlike AtNPF6.3 and ZmNPF6.6, ZmNPF6.4 transports chloride selectively over nitrate. We believe the high-affinity chloride transport must also be facilitated by a substrate-binding residue located in the central pore of ZmNPF6.4. This mechanism would function in a similar manner to the His residue in AtNPF6.3 and ZmNPF6.6 but instead of nitrate, it binds chloride. Alternatively, the selective and binding residues could be two different structural elements, similar to those reported in bacterial NPFs. A ZmNPF6.4 protein structure will help identify the chloride transport mechanism as would a fluorescent sensor (Ho and Frommer, 2014) designed against ZmNPF6.4 to validate chloride induced conformational rearrangement during transport.
Dual-Affinity in Vitro?
From a biophysical perspective, all evidence supporting dual-affinity nitrate transport activity of AtNPF6.3 (and MtNPF6.8 discussed below) are from concentration gradient nitrate flux experiments using the Xenopus laevis oocytes. In these experiments, nitrate concentrations were defined as high-affinity (below 0.25 mM) and low-affinity (above 0.25 mM), based on HATS and LATS nitrate uptake rates from plant studies (Glass, 2009; Wang et al., 2012). Rates of nitrate uptake were fitted using two separate Michaelis-Menten curves, resulting in two Km values for high- and low-affinity activities, respectively (Liu et al., 1999; Morère-Le Paven et al., 2011). We believe this method may not be appropriate for separating the two affinity ranges using a value (0.25 mM) derived from physiological studies. If the dual-affinity model is true, to observe a bi-phasic flux curve, there must be a proportion of NPF6 in the high-affinity mode and the rest in the low-affinity mode. Then, at any given concentration, nitrate uptake should be the sum of the nitrate transported through transporters in the high- and the low-affinity modes, described using a double Michaelis-Menten equation:
Where “Y” is the total nitrate uptake amount, “X” is the nitrate concentration, “a%” is the percentage of the high-affinity mode transporter, VmaxHA, VmaxLA, KmHA, and KmLA are the Michaelis-Menten constants for two action modes. However, when tested against published data of AtNPF6.3 and MtNPF6.8 (Liu et al., 1999; Morère-Le Paven et al., 2011; Sun et al., 2014), the curve fits were found to be poor (Figure 3). Instead, applying a normal Michaelis-Menten equation provided the best fit to the data, suggesting AtNPF6.3 and MtNPF6.8 are most-likely single-phasic nitrate transporters when expressed in oocytes. Further, kinetic studies of AtNPF6.3 by Martin et al. (2008) in yeast and our group (Wen et al., 2017) in Xenopus also support the single-phasic model.
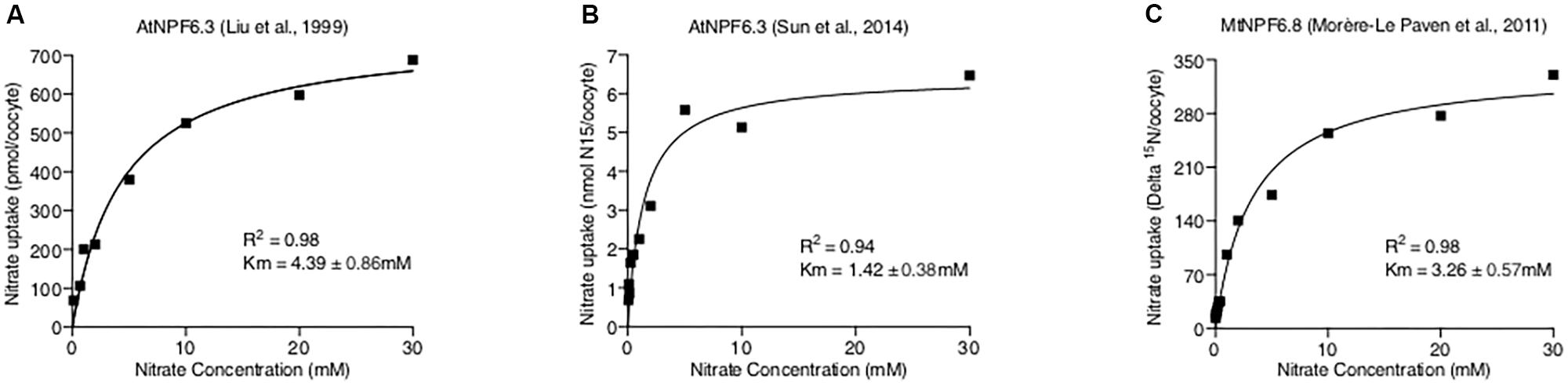
FIGURE 3. Curve Fitting NPF6.3 Comparisons. Published NPF6 dual-affinity nitrate uptake data (A. Liu et al., 1999; B. Sun et al., 2014; C. Morère-Le Paven et al., 2011) were directly measured from original figures and fitted with both Michaelis-Menten and double Michaelis-Menten equations using Prism 7 software. The detailed curve fitting results are listed in the tables below figures of each data set with the preferred model.
Dual-Affinity in Planta?
The dual-affinity nitrate transport activity of NPF6.3 as well as its net contribution to in planta nitrate uptake remains a controversial topic. As the activity of NPF6.3 was recently reviewed by Glass and Kotur (2013), we will only comment on a few key points about its potential activity and role. As discussed previously, experimental evidence of a dual-affinity (HATS and LATS) nitrate transport activity has been documented when NPF6.3 is expressed in Xenopus laevis oocytes and indirectly using the NPF6.3 knockout mutant, chl1-5 (Liu et al., 1999; Liu and Tsay, 2003). In Arabidopsis, the evidence of dual-affinity activity linked to NPF6.3 remains limited as most nitrate transport measurements from low to high concentrations fail to see the distinct saturation kinetics required to support a biphasic transport system (Huang et al., 1996; Touraine and Glass, 1997; Glass and Kotur, 2013). Furthermore, evidence of a HATS activity and a corresponding nitrate uptake was missing in both the nrt3.1 mutant (Okamoto et al., 2006) and the nrt2.1 ×nrt2.2 double mutant (Li et al., 2007), which in theory both should still express NPF6.3, and if active would display a HATS activity. Liu and Tsay (2003) do show a loss of potential HATS activity in chl1-5 relative to a wildtype control. Unfortunately, the evidence that suggests nitrate uptake across the HATS range is in fact hyperbolic in the WT control but not in the chl1-5 mutant is compromised by the limited range of external nitrate concentrations tested, the extended period of which nitrate influx was measured and the prior growth exposure to NH4+ which is known to influence the LATS activity in chl1-5 (inhibiting transport when grown on NH4NO3, but not KNO3 (see Touraine and Glass, 1997). This lack of a strong relationship with nitrate uptake in planta suggests NPF6.3 may not have a direct role in the process, but rather a minor or targeted contribution to the overall transport of nitrate in plants.
Given the proposed nitrate sensing function of NPF6.3, a loss of activity could alter the expression of other genes responsible for nitrate uptake and assimilation, a process which may influence the nitrate uptake pathways in the chl1-5 mutant as external nitrogen provision varies (Ho et al., 2009; Bouguyon et al., 2015). The sensor function of NPF6.3 may also help to explain the uncertainty on why chl1-5 can tolerate chlorate even though it still able to absorb chlorate (Touraine and Glass, 1997). In the absence of NPF6.3, a disruption in nitrate-linked signaling could influence nitrate reductase activity that may alleviate toxic concentrations of chlorite being generated through the reduction of chlorate by nitrate reductase.
The nitrate uptake contribution of NPF6s has also been evaluated in other species. In dwarf maize Gaspe, the gene activity of ZmNPF6.4 and ZmNPF6.6 were found to be very low relative to ZmNRT2.1 and ZmNRT2.2 where transcript levels were 200–300 × higher and subsequently presumed the primary nitrate uptake mechanism (Garnett et al., 2013). Although the transcriptional responses of NPF6.3-like proteins in Gaspe lines suggest a limited role to either HATS or LATS activities, their functional role still needs to be resolved. Recently, we have demonstrated that ZmNPF6.6 is a significant contributor to the nitrate response, where transcript abundance is linked to increased nitrate uptake in crown roots of the maize inbred F44 relative to the common inbred B73 (Dechorgnat et al., 2018). These data demonstrate that ZmNPF6.6 may in fact be acquired for specific purposes and able to contribute to net plant nitrate uptake.
Thr101
If we consider AtNPF6.3 as only a single-phasic nitrate transporters, then the proposed role of Thr101 as a substrate affinity switch needs to be reconsidered. Liu and Tsay (2003) suggested that when T101 is not phosphorylated (or mimicked by T101A mutation), AtNPF6.3 exhibits a low-affinity transport activity, and vice versa. In a AtNPF6.3 maize homolog ZmNPF6.6, we failed to observe similar effects of T104 mutations (equivalent to AtNPF6.3:T101) on nitrate transport (Wen et al., 2017), where both mutations kept similar kinetics with the transporter, suggesting the nitrate binding mechanism in ZmNPF6.6 was not affected. This is consistent with Parker and Newstead (2014) who show the KD value for nitrate binding of AtNPF6.3 is not changed by the T101 mutation. However, the phosphorylation of this highly conserved Thr residue seems to have different effects to different NPF members and/or when transporting different substrates. For example, the high-affinity chloride transporter ZmNPF6.4 was converted into a low-affinity transporter by equivalent Thr mutations (Wen et al., 2017). Another example is that the dephosphorylated AtNPF6.3 loses its auxin permeability (Bouguyon et al., 2015). So far, the Thr residue has only been proposed to have an effect on the structural conformation of AtNPF6.3, either by distorting the packing of N-terminal TM bundles or disrupting protein dimerization (Parker and Newstead, 2014; Sun et al., 2014). As for how it affects the transport activity of NPF proteins this will require further study.
NPF6 Center Loop
Another unique feature of plant NPFs is the intercellular loop linking the two six-helical bundles between TM6 and TM7. The structure of this 84-amino acid center loop in AtNPF6.3 has only been partially solved since most of the loop is disordered in crystal structures. This center loop contains three conserved positively charged residues, Arg264-Arg266-Lys267. Parker and Newstead (2014) have suggested these residues could play a role in stabilizing the protein from the intercellular side. Indeed, Ho and Frommer (2014) confirmed the importance of these structural elements where nitrate transport activity is inhibited in a R264A-R266A-K267A mutant. Sun et al. (2014) suggested that the stable amphipathic alpha-helix in the N-terminal region (including Arg264-Arg266-Lys267) of the center loop could be a potential protein-docking site. In rice, differential nitrate sensing patterns exist through natural variation in the center loop region of an AtNPF6.3 rice homolog, OsNPF6.5 (Hu et al., 2015). The authors identified that variants with the OsNPF6.5:Met327 substitution rather than those with OsNPF6.5:Thr327, influenced the primary nitrate response (nitrate uptake, root-to-shoot nitrate transport) and upregulation of the nitrate reductase genes OsNIA1 and OsNIA2. Collectively, the activity of the loop region suggests a post-translational regulatory site of NPF6s and possibly an insight into the nitrate sensing mechanism?
Bi-Directional Nitrate Transport
Previous studies suggest nitrate transport activity of AtNPF6.3 may be partially bi-directional. Leran et al. (2013) reported that AtNPF6.3 facilitated nitrate efflux in Xenopus oocytes, an activity later confirmed by Wen et al. (2017). These observations would suggest that the “alternating access” transport cycle of AtNPF6.3-like proteins can be reversed. Unfortunately, there is no structural evidence to support this suggestion which would require further investigation. It is interesting that NPF6.3-like proteins display both influx and efflux activities when plants already express a dedicated nitrate efflux system such as AtNPF2.7 (AtNAXT1) (Segonzac et al., 2007). Apart from structural confirmation, in planta estimates of bidirectional transport needs to be verified.
Tonoplast Localized NPF6
Recently a tonoplast localized NPF6, OsNPF6.3 was characterized in rice, where overexpression enhanced both yield and nitrogen use-efficiency (Wang et al., 2018). An osnpf6.3 knock-out mutant was able to phenocopy many of the chl1-5 features while also exhibiting a strong repression of nitrogen assimilation associated genes. These observations would suggest OsNPF6.3 may also act as a nitrate sensor? It is already known using the uptake/sensing decoupling mutant, chl1-9, that the sensor function of AtNPF6.3 does not require plasma membrane targeting of the protein (Bouguyon et al., 2015). Further research is required to confirm whether tonoplast localized sensors are involved in the OsNPF6.3 nitrate signal transduction pathway and why a tonoplast location is important.
Other NPFs
His356 is an important structural element for nitrate transport of AtNPF6.3. However, most of the NPF nitrate transporters that have been characterized do not have an equivalent His residue, or other charged residues that can bind nitrate electrostatically in the equivalent position of His356. Nitrate is then probably transported using a similar channel-like mechanism to that of the chloride transport activity in AtNPF6.3. If this occurs, then it is reasonable to question what is the correct substrate that could induce the alternative access cycle of these NPFs. It is important to stress that without examination, it is premature to conclude a NPF as a nitrate transporter. For example, AtNPF4.6 (also known as AtNRT1.2) was first characterized by Huang et al. (1999) as a constitutive LATS nitrate transporter with a low-affinity Km of ∼5.9 mM. However, a much higher transport affinity of AtNPF4.6 toward ABA (Km ∼5 μM) was reported by Kanno et al. (2012) and nitrate dose not compete with ABA transport regardless of concentration. These results suggest ABA is probably the primary substrate of AtNPF4.6, not nitrate. Physiological studies also do not agree with the nitrate transport activity of AtNPF4.6 in vivo as (1) nitrate has the same seed germination induction effect in both atnpf4.6 mutants and wild type plants and (2) atnpf4.6 mutant did not exhibit the ABA-hypersensitive effect, which would expect to happen if AtNPF4.6 was a nitrate transporters (Kanno et al., 2012, 2013). Reduced stomatal closing and associated leaf/stem surface temperature decreases were observed in atnpf4.6 knockoff mutants, but also in overexpression mutants. Accordingly, Kanno et al. (2012) suggested a physiological role of AtNPF4.6 as an ABA pool size regulator during the biosynthesis of ABA in vascular parenchyma cells. Similarly, the role of another ABA permeable nitrate transporters, MtNPF6.8, may require further validation as it may have a nitrate sensor function regulating root architecture through its ABA transport activity (Morère-Le Paven et al., 2011; Pellizzaro et al., 2014).
Unlike the nitrate binding His residue, the structural element, ExxER, is a highly conserved motif across the NPF family (Solcan et al., 2012). It has been suggested the ExxER is an essential structural component of the proton-coupling substrate import activity in NPFs (Jorgensen et al., 2015). However, some characterized NPF nitrate transporters do not contain the ExxER motif or the nitrate binding His residue. For example, AtNPF7.3 (AtNRT1.5) lacks both elements although it was originally characterized as a bidirectional nitrate transporters expressed in root pericycle cells involved in root-to-shoot nitrate translocation (Lin et al., 2008). Recently, a second atnpf7.3 mutant (lks2) was identified based on a low potassium leaf chlorosis phenotype (Li et al., 2017). The transport activity of NPF7.3 was redefined as a proton-coupled H+/K+ antiporter using the NMT/MIFE technique. Therefore, NPF7.3 may have a role in potassium translocation from root to shoot, although root-to-shoot relocation of nitrate was also reduced in the atnpf7.3 mutant. The detailed mechanism behind this nitrate phenotype, or whether it is related to the transporter activity of NPF7.3 remains unclear. Similarly, the transporter function and physiological role of AtNPF7.2 (AtNRT1.8), which also does not have the ExxER motif or a nitrate binding His residue requires further study (Li et al., 2010).
Perspectives
With each NPF characterization, the breadth and complexity of the transport family continues to expand. This is evident in the range of transported substrates linked to individual NPF proteins as well as their cellular localization and relevance to plant growth. What is evident is the difficulty in prescribing in planta function from sequence homology and current technological measurements. The recent crystallization studies examining Arabidopsis NPF6.3 identified new insights on its structural state and functional activities. New crystal structures of plant NPF proteins trapped in different conformations (outward-facing, occluded, and inward-facing) will be required to define the substrate transport cycle for each protein. When combined with existing biochemical, electrophysiological and physiological approaches the improved knowledge base will help describe the variability of new and existing NPF proteins. For NPFs, the reliance on the traditional TEVC system needs to accommodate alternative technologies including chemical flux analysis, a greater use of MIFE to evaluate the cross-membrane movement of particular cations or anions in real time (Shabala et al., 2013) and a greater use of the liposome system to describe transport activities relative to protein content (Allen and Cullis, 2013). In summary, a cross-disciplinary approach is required for plant NPF transport research to provide the necessary level of detail regarding function the community expects.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by an Australian Research Council Linkage Grant awarded to BK, S.D. Tyerman, A. Raflaski, and K. Dhugga (LP110200878).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ABA, abscisic acid; HATS, high-affinity transport system; LATS, low-affinity transport system; NPF, nitrate transporter 1/peptide transporter; NRT, nitrate transporter; POT, proton-dependent oligopeptide transporter; PTR, peptide transporter; TM, transmembrane domain.
References
Allen, T. M., and Cullis, P. R. (2013). Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 65, 36–48. doi: 10.1016/j.addr.2012.09.037
Bouguyon, E., Brun, F., Meynard, D., Kubeš, M., Pervent, M., Leran, S., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 1:15015. doi: 10.1038/nplants.2015.15
Bröer, S. (2010). “Xenopus laevis Oocytes,” in Membrane Transporters in Drug Discovery and Development: Methods and Protocols, ed. Q. Yan (Totowa, NJ: Humana Press), 295–310. doi: 10.1007/978-1-60761-700-6_16
Conde, A., Diallinas, G., Chaumont, F., Chaves, M., and Geros, H. (2010). Transporters, channels, or simple diffusion? Dogmas, atypical roles and complexity in transport systems. Int. J. Biochem. Cell Biol. 42, 857–868. doi: 10.1016/j.biocel.2009.12.012
Dechorgnat, J., Francis, K. L., Dhugga, K. S., Rafalski, J. A., Tyerman, S. D., and Kaiser, B. N. (2018). Root ideotype influences nitrogen transport and assimilation in maize. Front. Plant Sci. 9:531. doi: 10.3389/fpls.2018.00531
Garnett, T., Conn, V., Plett, D., Conn, S., Zanghellini, J., Mackenzie, N., et al. (2013). The response of the maize nitrate transport system to nitrogen demand and supply across the lifecycle. New Phytol. 198, 82–94. doi: 10.1111/nph.12166
Glass, A. D., and Kotur, Z. (2013). A reevaluation of the role of Arabidopsis NRT1.1 in high-affinity nitrate transport. Plant Physiol. 163, 1103–1106. doi: 10.1104/pp.113.229161
Guo, F., Young, J., and Crawford, N. M. (2003). The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in arabidopsis. Plant Cell 15, 107–117. doi: 10.1105/tpc.006312
Guo, F.-Q., Wang, R., Chen, M., and Crawford, N. M. (2001). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) Is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13, 1761–1777. doi: 10.1105/tpc.13.8.1761
Ho, C. H., and Frommer, W. B. (2014). Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. eLife 3:e01917. doi: 10.7554/eLife.01917
Ho, C.-H., Lin, S.-H., Hu, H.-C., and Tsay, Y.-F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hu, B., Wang, W., Ou, S. J., Tang, J. Y., Li, H., Che, R. H., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, N., Chiang, C., Crawford, N. M., and Tsay, Y. (1996). CHL1 encodes a component of the low-affinity nitrate uptake system in arabidopsis and shows cell type-specific expression in roots. Plant Cell 8, 2183–2191. doi: 10.1105/tpc.8.12.2183
Huang, N. C., Liu, K. H., Lo, H. J., and Tsay, Y. F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Jorgensen, M. E., Olsen, C. E., Geiger, D., Mirza, O., Halkier, B. A., and Nour-Eldin, H. H. (2015). A functional EXXEK motif is essential for proton coupling and active glucosinolate transport by NPF2.11. Plant Cell Physiol. 56, 2340–2350. doi: 10.1093/pcp/pcv145
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. doi: 10.1073/pnas.1203567109
Kanno, Y., Kamiya, Y., and Seo, M. (2013). Nitrate does not compete with abscisic acid as a substrate of AtNPF4.6/NRT1.2/AIT1 in Arabidopsis. Plant Signal. Behav. 8:e26624. doi: 10.4161/psb.26624
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Leran, S., Munos, S., Brachet, C., Tillard, P., Gojon, A., and Lacombe, B. (2013). Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Mol. Plant 6, 1984–1987. doi: 10.1093/mp/sst068
Li, H., Yu, M., Du, X. Q., Wang, Z. F., Wu, W. H., Quintero, F. J., et al. (2017). NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ Antiporter for K+ Loading into the Xylem in Arabidopsis. Plant Cell 29, 2016–2026. doi: 10.1105/tpc.16.00972
Li, J.-Y., Fu, Y.-L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., and Glass, A. D. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Lin, S.-H., Kuo, H.-F., Canivenc, G., Lin, C.-S., Lepetit, M., Hsu, P.-K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Liu, K. H., Huang, C. Y., and Tsay, Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, K. H., and Tsay, Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Martin, Y., Navarro, F. J., and Siverio, J. M. (2008). Functional characterization of the Arabidopsis thaliana nitrate transporter CHL1 in the yeast Hansenula polymorpha. Plant Mol. Biol. 68, 215–224. doi: 10.1007/s11103-008-9363-z
Meharg, A. A., and Blatt, M. R. (1995). No3- transport across the plasma-membrane of Arabidopsis-thaliana root hairs - kinetic control by Ph and membrane voltage. J. Membr. Biol. 145, 49–66. doi: 10.1007/BF00233306
Miller, A. J., and Smith, S. J. (1996). Nitrate transport and compartmentation in cereal root cells. J. Exp. Bot. 47, 843–854. doi: 10.1093/jxb/47.7.843
Morère-Le Paven, M.-C., Frugier, F., Legros, C., Limami, A. M., Viau, L., Hamon, A., et al. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 62, 5595–5605. doi: 10.1093/jxb/err243
Newstead, S. (2017). Recent advances in understanding proton coupled peptide transport via the POT family. Curr. Opin. Struct. Biol. 45, 17–24. doi: 10.1016/j.sbi.2016.10.018
Okamoto, M., Kumar, A., Li, W. B., Wang, Y., Siddiqi, M. Y., Crawford, N. M., et al. (2006). High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 140, 1036–1046. doi: 10.1104/pp.105.074385
Parker, J. L., Li, C. H., Brinth, A., Wang, Z., Vogeley, L., Solcan, N., et al. (2017). Proton movement and coupling in the POT family of peptide transporters. Proc. Natl. Acad. Sci. U.S.A. 114, 13182–13187. doi: 10.1073/pnas.1710727114
Parker, J. L., and Newstead, S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. doi: 10.1038/nature13116
Pellizzaro, A., Clochard, T., Cukier, C., Bourdin, C., Juchaux, M., Montrichard, F., et al. (2014). The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol. 166, 2152–2165. doi: 10.1104/pp.114.250811
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U.S.A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Segonzac, C., Boyer, J.-C., Ipotesi, E., Szponarski, W., Tillard, P., Touraine, B., et al. (2007). Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19, 3760–3777. doi: 10.1105/tpc.106.048173
Shabala, S., Shabala, L., Bose, J., Cuin, T., and Newman, I. (2013). Ion flux measurements using the MIFE technique. Methods Mol. Biol. 953, 171–183. doi: 10.1007/978-1-62703-152-3_11
Solcan, N., Kwok, J., Fowler, P. W., Cameron, A. D., Drew, D., Iwata, S., et al. (2012). Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 31, 3411–3421. doi: 10.1038/emboj.2012.157
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., and Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Touraine, B., and Glass, A. D. M. (1997). NO3- and ClO3 - fluxes in the chl1-5 mutant of Arabidopsis thaliana: Does the CHL1-5 gene encode a low-affinity NO3 - transporter? Plant Physiol. 114, 137–144. doi: 10.1104/pp.114.1.137
Tsay, Y.-F., Schroeder, J. I., Feldmann, K. A., and Crawford, N. M. (1993). The herbicide sensitivity gene CHL1 of arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. doi: 10.1016/0092-8674(93)90399-b
Wang, R. C., Liu, D., and Crawford, N. M. (1998). The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. U.S.A. 95, 15134–15139. doi: 10.1073/pnas.95.25.15134
Wang, W., Hu, B., Yuan, D. Y., Liu, Y. Q., Che, R. H., Hu, Y. C., et al. (2018). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30, 638–651. doi: 10.1105/tpc.17.00809
Wang, Y.-Y., Hsu, P.-K., and Tsay, Y.-F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17, 624–667. doi: 10.1016/j.tplants.2012.08.007
Weber, W. (1999). Ion currents of Xenopus laevis oocytes: state of the art. Biochim. Biophys. Acta 1421, 213–233. doi: 10.1016/S0005-2736(99)00135-2
Wege, S., Gilliham, M., and Henderson, S. W. (2017). Chloride: not simply a ‘cheap osmoticum’, but a beneficial plant macronutrient. J. Exp. Bot. 68, 3057–3069. doi: 10.1093/jxb/erx050
Keywords: NPF6.3, NRT1.1, dual-affinity, nitrate transport, Arabidopsis
Citation: Wen Z and Kaiser BN (2018) Unraveling the Functional Role of NPF6 Transporters. Front. Plant Sci. 9:973. doi: 10.3389/fpls.2018.00973
Received: 31 March 2018; Accepted: 15 June 2018;
Published: 10 July 2018.
Edited by:
Sebastien Thomine, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Lixing Yuan, China Agricultural University, ChinaEmilio Fernandez, Universidad de Córdoba, Spain
Copyright © 2018 Wen and Kaiser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengyu Wen, emhlbmd5dS53ZW5Ac3lkbmV5LmVkdS5hdQ== Brent N. Kaiser, YnJlbnQua2Fpc2VyQHN5ZG5leS5lZHUuYXU=
 Zhengyu Wen
Zhengyu Wen Brent N. Kaiser
Brent N. Kaiser