- 1Oilseed Crops Research Centre, Plant Breeding and Acclimatization Institute-National Research Institute, Poznań, Poland
- 2School of Biological Sciences, The University of Western Australia, Crawley, WA, Australia
The world-wide demand for additional protein sources for human nutrition and animal feed keeps rising due to rapidly growing world population. Oilseed rape is a second important oil producing crop and the by-product of the oil production is a protein rich meal. The protein in rapeseed meal finds its application in animal feed and various industrial purposes, but its improvement is of great interest, especially for non-ruminants and poultry feed. To be able to manipulate the quality and quantity of seed protein in oilseed rape, understanding genetic architecture of seed storage protein (SSPs) synthesis and accumulation in this crop species is of great interest. For this, application of modern molecular breeding tools such as whole genome sequencing, genotyping, association mapping, and genome editing methods implemented in oilseed rape seed protein improvement would be of great interest. This review examines current knowledge and opportunities to manipulate of SSPs in oilseed rape to improve its quality, quantity and digestibility.
Introduction
It has been estimated that the world global food demand will more than double by 2050, mainly due to the rapidly growing world population, increasing urbanization and changes in food preferences. Protein has been identified as the most limiting macronutrient, and sufficient protein quantity and quality will be required as the global food demand increases (FAO, 20131). Oilseed rape (Brassica napus, canola) is the second largest oil producing crop in the world after soybean, and the by-product of the oil production is a protein rich meal or cake (OECD-FAO, 20152; Oil World, 20173). At present canola meal is primarily used in animal feed, but it can have application as a high quality food protein source, as well as in bio products such as formation of films, gels, foams, emulsions (Wanasundara et al., 2016).
The seeds of B. napus contain around 45% oil and 25% seed storage proteins (SSPs), which are predominantly composed of cruciferin (60%), napin (20%) and other minor proteins such as oleosins and lipid transfer proteins (Gehrig et al., 1996). When compared to soybean and legumes, the protein of rapeseed contains an excellent balance of essential amino acids, including high levels of sulfur containing amino acids (cysteine, methionine in napin), which are very desirable for human consumption, and a slightly limited amount of lysine. However, the introduction of modern varieties of oilseed rape with low glucosinolate (GLS) content lowered the amount of the desired napin in seeds. The pathways of amino acid and glucosinolate biosynthesis share common enzymes, therefore perturbation of glucosinolate in double low varieties could affect the level of napin (Field et al., 2004; Nesi et al., 2008). The level of napin and cruciferin in seeds also affects the functionality of canola protein products, including solubility, emulsifying ability and heat induced gel formation. Both cruciferin and napin are soluble above pH 5.5, whereas napin is also soluble at a lower pH 3-4. Napin exhibits poorer emulsifying abilities, therefore cruciferin rich oilseed rape proteins show higher emulsion capacities. Cruciferin has also stronger tendency to form heat-induced gels when compared to napin (Wanasundara et al., 2016). Various industrial purposes require different level of cruciferin and napin in the seed proteins of B. napus.
Current goals in oilseed rape meal improvement is to make it a useful protein source for non-ruminants, especially for poultry nutrition (Wanasundara et al., 2016). For this it is necessary to reduce the most limiting anti nutritional factors, such as glucosinolates, phytic acid, fiber, sinapates and non-starch polysaccharides (NSP). Sinapates have a negative effect on the smell of the eggs therefore their use is limited for laying hens. Phenolic compounds, such as sinapates, flavonoids and lignins are present in the maternally derived seed coat tissue and play an important role in pigmentation and protection against various environmental injuries (Qu et al., 2016). In Arabidopsis several genes regulating phenolic compounds named TRANSPARENT TESTA (TT) were identified, and the seeds of the mutants defective in flavonoids accumulation are pale yellow to pale brown (Johnson et al., 2002). TT8 was also found to regulate activity of the four master regulator genes and inhibits seed fatty acid accumulation (Chen et al., 2014). In oilseed rape yellow seeded genotypes derived through breeding are deprived of tannins, contain lower level of fiber, higher level of protein content but their digestibility is not improved (Swiech et al., 2016). The yellow seeded genotypes are advantageous to incorporate into genomic studies in order to identify genetic variation underlying the important trait of interest. Current challenges would also include providing consistent quality of protein meal on the market. There is also a great interest to improve oilseed rape meal for human consumption. It would mean elevating the content of proteins in the seeds and manipulating the ratio of napins/cruciferins in the composition of the proteins within B. napus seeds. As oilseed rape is mainly used for animal feed, the knowledge of its nutritional value for humans is quite limited. The digestibility of oilseed rape protein was found to be similar to that of soy protein (Bos et al., 2007). The analysis of oilseed rape protein products (Isolexx) showed a low level of anti nutritional factors which make them good candidates for a novel food ingredient (EFSA NDA Panel [EFSA Panel on Dietetic Products and Nutrition and Allergies], 2013). The studies of oilseed rape peptide mixtures have proved their role in lowering the blood pressure of hypertensive mice and showed their antioxidant, antidiabetic, anorexigenic, anticancer, antiviral, hypercholesterolemic and bile acid binding activities (Wanasundara, 2011; Aachary and Thiyam, 2012; Alashi et al., 2013).
In this review we will look into current knowledge in genetic regulation of biosynthesis and accumulation of SSPs in the model species Arabidopsis, Brassicas and other crop species including maize, beans, pea, broad beans and Vicia narbonensis. We will also summarize recent advances in genomic studies related to seed protein content in B. napus and show the prospects to improve the quality and quantity of protein in this crop species for animal feed, human consumption and industrial purposes.
Seed Storage Proteins of B. napus
The SSPs, together with oil and starch, form seed storage reserves which accumulate in the matured embryo of B. napus seed and supply energy for seed germination, growth, as well as provide energy for humans and animals. The analysis of spatial and temporal napin, cruciferin and oleosin accumulation revealed that their storage during early and middle stages of seed development was mostly restricted to the radicle of the embryo. By the late stage of seed growth the accumulation of SSPs ceased. The pattern of SSPs accumulation corresponded with that of lipids which indicates that the two compete for substrate, energy and space (Borisjuk et al., 2013).
The identified in Arabidopsis cruciferin encoding genes were named CRUCIFERINA1, CRUCIFERINA2, CRU3/CRUCIFERIN C, and CRUCIFERIN B and 2S precursors (2S1–2S5) (Gruis et al., 2002, 2004). Genes encoding cruciferin precursors have been also identified in B. napus and comprise of a five-member gene family (Breen and Crouch, 1992) (Table 1). Napin in B. napus is also encoded by multi-gene families (Josefsson et al., 1987; Scofield and Crouch, 1987; Baszczynski and Fallis, 1990; Sjodahl et al., 1991) and in order to manipulate the composition of SSPs in oilseed rape to a more desirable level of napin, genetic engineering studies have been performed in this crop species (Altenbach et al., 1992; Hannoufa et al., 2014). Introduction of the antisense gene for cruciferin resulted in increased levels of cysteine, lysine and methionine essential amino acids in the SSPs of the B. napus seeds. Downregulation of napins in oilseed rape resulted in increased level of cruciferins and lower amounts of cysteine and lysine (Guerche et al., 1990; Kohnomurase et al., 1994, 1995). Despite the genetic engineering studies, genomic studies are desired to identify key genes regulating napin content in the SSPs of oilseed rape in order to introduce that genetic variation into the modern, double low varieties of oilseed rape.
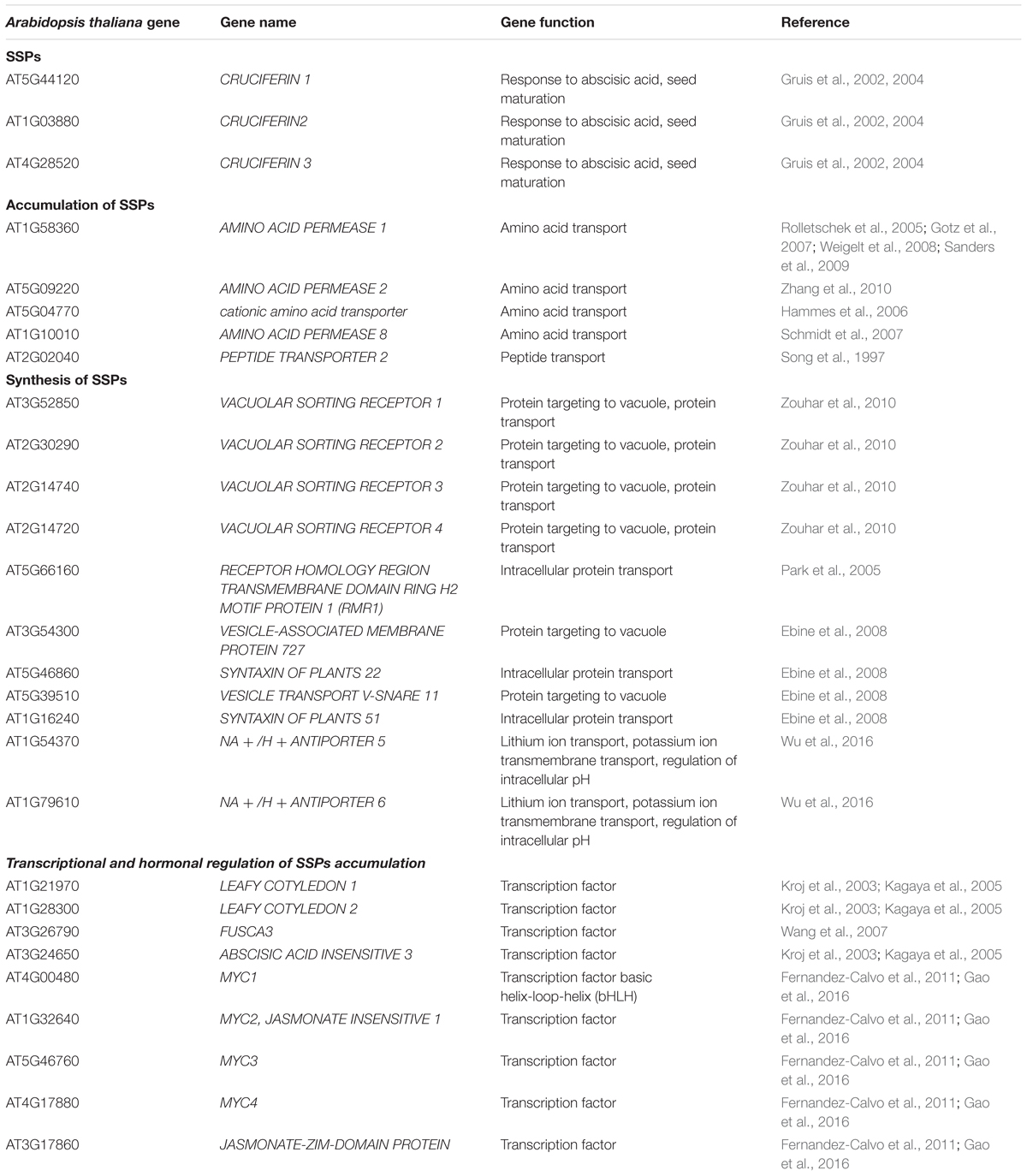
TABLE 1. List of genes identified in previous studies, known to be involved in accumulation, synthesis and transcriptional regulation of SSPs.
Genetic Regulation of Synthesis and Accumulation of SSPs During Seed Growth and Development
The majority of genes responsible for regulation of SSPs synthesis and accumulation in seeds have been identified in genetic studies using model species Arabidopsis thaliana. High collinearity between A. thaliana and B. napus genomes is a great advantage therefore the identified genes are great candidates to study in B. napus (Figure 1 and Table 1).
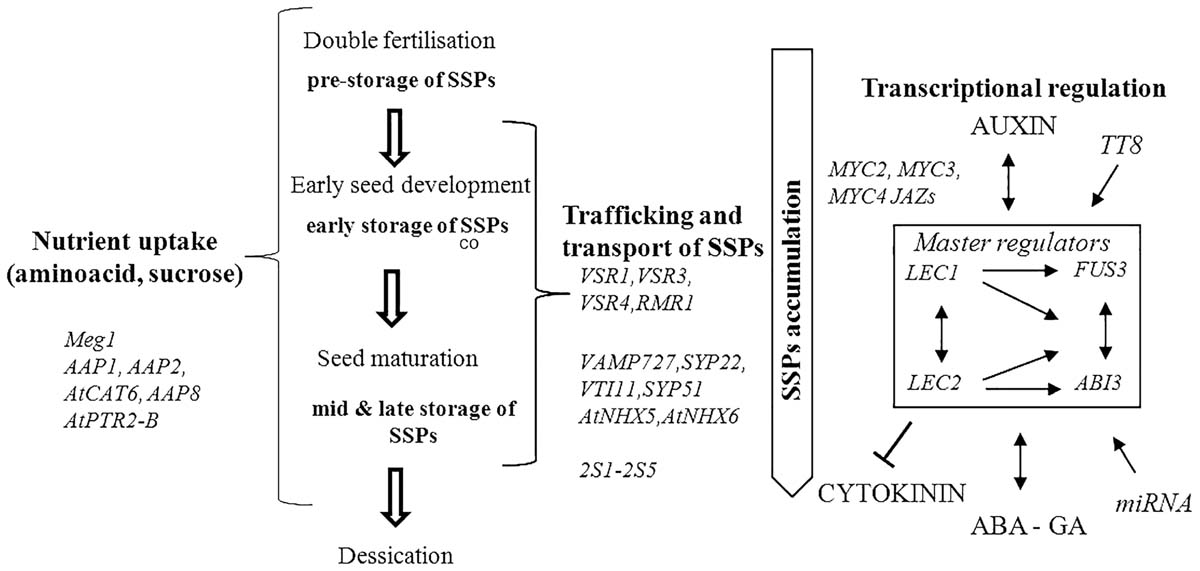
FIGURE 1. Summary of genetic and transcriptional regulation of seed storage proteins (SSPs). Accumulation of SSPs occurs from early to late storage stage of seed development; en, endosperm. The genes regulating SSPs in seed consist of genes identified during nutrient uptake, trafficking and transport of SSPs. SSPs accumulation is also orchestrated by the regulatory network of transcription factors (master regulators), hormones, miRNA and other genes during seed development. The interactions of genes are depicted with arrows and T-bar.
Synthesis of SSPs
Seed storage proteins are synthesized in the endoplasmic reticulum (ER) and transported to protein storage vacuoles (PSV) where they are primarily accumulated. The trafficking of the mature SSPs are transported in the Golgi dependent and independent manner (Hohl et al., 1996; Hara-Nishimura et al., 1998). Several vacuolar sorting receptors (VSR) genes including VSR1, VSR3, VSR4 and RMR1 have been identified in this process in Arabidopsis (Park et al., 2005; Zouhar et al., 2010). VAMP727, SYP22, VTI11, and SYP51 compose the SNARE complex which mediates fusion between PVC and vacuole (Ebine et al., 2008). AtNHX5 and AtNHX6 regulate localization of the SNARE complex and thus its function in protein transport (Wu et al., 2016). The genes are listed in Table 1 and Figure 1.
Accumulation of SSPs
Accumulation of seed storage reserves is highly coupled with seed growth and development (Figure 1). Seed growth is initiated with a process of double fertilization, when one sperm cell fertilizes an egg cell to generate a diploid embryo and a second sperm cell fuses with the central cell giving rise to the triploid endosperm. Embryo and endosperm are surrounded by maternal integuments, future seed coat. In the early stages of development, organic nutrients in the form of sucrose, amino acids and potassium are imported from the maternal tissue to the seeds via vascular tissue within the funiculus which terminates in the chalazal part of the seed coat. Many genes involved in the transport of sugar, amino acids, lipids and hormonal regulation were found to be specifically expressed in this part of the seed, which indicates its importance in seed filling (Millar et al., 2015). Released nutrients are mainly accumulated in the endosperm, which acts as a nourishment to the developing embryo. In maize, genes specifically expressed in the region of endosperm which surrounds the embryo (ESR genes) were found to regulate nutrient transfer to the embryo (Bonello et al., 2000, 2002). Meg1 genes identified in the endosperm of maize seeds ensure maternal nutrient uptake, sucrose partitioning and seed biomass yield in maize (Costa et al., 2012). In the endospermic seeds, such as Arabidopsis and Brassica, the nutrients are also thought to be released from the seed coat by as yet unknown transport proteins. In Vicia faba, Pisum sativum, Vicia narbonensis, and A. thaliana Amino acid permase AAP1 mediates uptake of amino acids by the embryos and thus regulates SSP accumulation and seed yield (Rolletschek et al., 2005; Gotz et al., 2007; Weigelt et al., 2008; Sanders et al., 2009).
In Arabidopsis another amino acid permase AAP2 and cationic amino acid transporter AtCAT6 were also shown to play a role in supplying amino acid and higher SSP accumulation in seeds (Hammes et al., 2006; Zhang et al., 2010). Amino acid permases AAP8 was identified with a role in the uptake of amino acids into the endosperm during early embryogenesis (Schmidt et al., 2007). Peptide transport gene, AtPTR2-B was shown to play a role in general plant nutrition and seed development (Song et al., 1997). The translocation of amino-N and loading process of amino acids in the phloem sap of leaves affect protein content in the seeds of the Brassica species (Lohaus and Moellers, 2000; Tilsner et al., 2005). At maturity the endosperm of brassicas remains as a one cell layer and together with the cells of embryo are packed full of PSV and oil bodies (Herman and Larkins, 1999). The orthologous genes identified to play a role in SSP accumulation in here mentioned crop species and model Arabidopsis have not been investigated in oilseed rape. These genes are excellent candidate genes to investigate in this crop in seed protein improvement programs. The genes are listed in Table 1.
Transcriptional and Hormonal Regulation of SSPs Accumulation
Transcriptional regulation of the accumulation of SSPs in seeds occurs during the maturation phase of the seed. In Arabidopsis four ‘master regulator’ transcription factors (TFs) LEC1, LEC2, FUS3, and ABI3 specifically expressed in seeds are known to play a role in seed maturation, accumulation of SSPs and controlling expression of many other regulators of metabolic pathways (Parcy et al., 1994; Verdier and Thompson, 2008). LEC1 controls the expression of the SSP genes in a hierarchical manner, which involves ABI3 and FUS3 (Kroj et al., 2003; Kagaya et al., 2005). In Arabidopsis, FUS3 was found to be associated with the induction of genes encoding SSPs: napin and cruciferin as well as enzymes involved in FA biosynthesis (Wang et al., 2007). The seeds of the loss of function mutant of B. napus, BnFus3 showed decreased level of total seed oil content while seed protein content was increased. BnFus3 mutant seeds showed lower expression level of genes involved in sucrose photo-assimilation and glycolysis, which might have affected the biosynthesis of both protein and oil and can explain their altered level in the seeds (Elahi et al., 2015). Overexpression of BnLEC1 in oilseed rape enhanced seed oil production but did not affect the level of seed protein content (Tan et al., 2011). Other identified TFs which act in the accumulation of SSPs include basic helix-loop-helix (bHLH) MYC2, MYC3, and MYC4 which interact with JASMONATE ZIM-DOMAIN (JAZs) (Fernandez-Calvo et al., 2011; Gao et al., 2016). Recent studies allowed identification of miRNAs which regulate expression of TFs during seed growth and maturation (Gupta et al., 2017). In beans, a TF ROM1 was found to modulate transcription from lectin and storage protein genes (Chern et al., 1996).
Regulation of seed storage accumulation is coupled with complex interplay of hormones with TFs (Verdier and Thompson, 2008). Abscisic acid (ABA) is key regulator of gene expression in the maturing seeds (Delisle and Crouch, 1989) and interacts with the four master regulator genes (Kagaya et al., 2005). In B. napus, the level of SSPs (cruciferin and napin) is higher when induced by abscisic acid (Delisle and Crouch, 1989). Protein phosphatase ABI1 which regulates abscisic acid responses in plants was found to regulate the expression of napin genes in B. napus (Rask et al., 1998; Ezcurra et al., 2000). LEC2 interplays with auxin and induces auxin responsive genes and downregulates cytokinin (Stone et al., 2008). LEC1 also interacts with auxin (Casson and Lindsey, 2006) whereas FUS3 acts as a negative regulator of gibberellic acid biosynthesis. The genes known to be involved in transcriptional regulation of SSPs are listed in Table 1.
Genomic Studies in Seed Storage Protein in B. napus
Genetic mapping is an effective way to dissect the genetic mechanisms regulating important agronomic traits at the whole genome level. B. napus L. has a complex tetraploid genome AACC, 2n = 38 as it originated from spontaneous hybridization between two diploid species: B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18). Since oil is of a major economic interest in this crop species, the majority of QTL genetic mapping studies in B. napus so far have been performed to identify markers for seed oil content and only a few of such studies were conducted to unravel the genetic regulation of seed protein content in this crop. The results of those studies have shown that QTL for seed oil and protein content are closely linked and that there is a negative correlation between protein and oil content (Grami et al., 1977; Gül et al., 2003). These findings are not surprising as both protein and oil compete for the same basic substrates in the biochemical pathway and therefore must be partly controlled by the same genes. To unravel this problem, Zhao et al. (2006) applied a conditional mapping approach which allowed them to analyze the influence of genetic variation on protein content in oilseed rape seeds independently from oil content. The conditional QTL mapping allowed detection of five QTL for protein synthesis on separate linkage groups from oil content localized on chromosome A07, A09, C01, C08, C09 (Table 2) (Zhao et al., 2006). Another QTL genetic mapping study allowed identification of major QTL for protein content in oilseed rape seeds on chromosome A07 (Wurschum et al., 2012). Three QTL for napin content was detected on linkage groups A2, C6, C9 and two QTL for cruciferin were detected in linkage group A2 and C19 (Schatzki et al., 2014). Due to low mapping resolution and lack of marker sequence information, integration of these QTL onto the physical map and identification of underlying candidate genes was impossible.
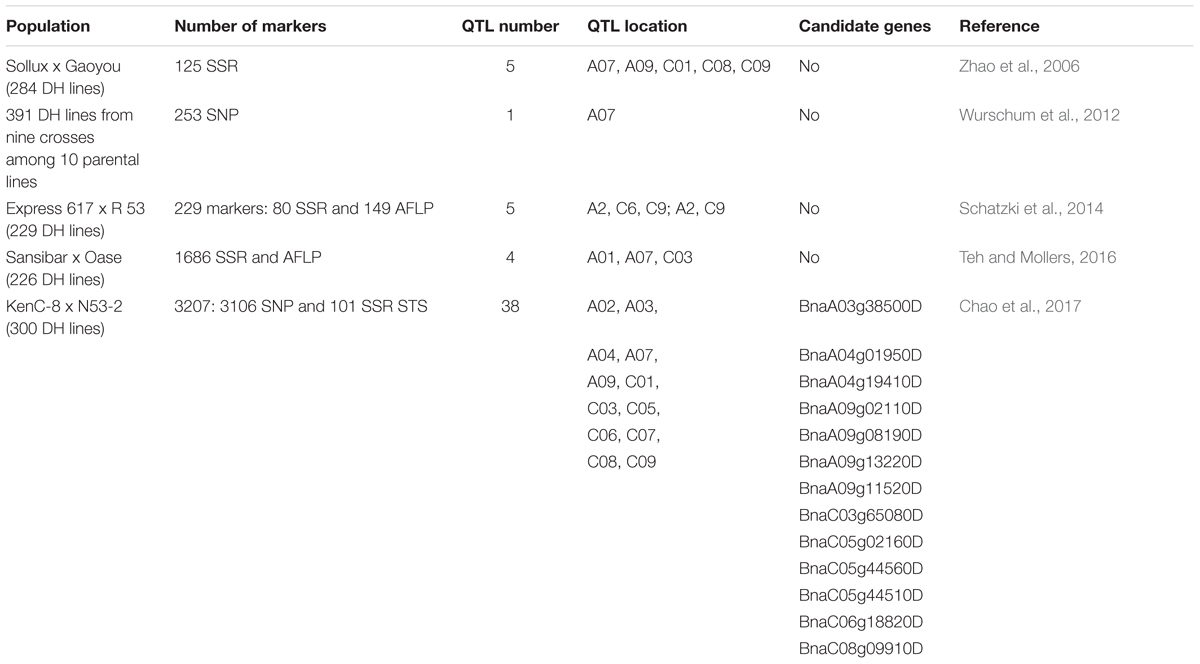
TABLE 2. Summary of genetic mapping studies and list of the identified QTL for protein content in B. napus seeds.
The recent release of the oilseed rape reference genome (Darmour cultivar) (Chalhoub et al., 2014) and high resolution sequencing technologies (Edwards and Batley, 2010; Edwards et al., 2013) allow more accurate genetic mapping studies which lead to identification of potential candidate genes regulating the trait of interest. The high collinearity between B. napus and Arabidopsis is a great advantage as a lot of genes which affect biosynthesis and accumulation of SSPs in seeds have been already identified in this model plant and other crop species. Orthologous genes mapped in genetic mapping studies on protein content in B. napus seeds would be of interest. Teh and Mollers (2016) identified four minor QTL for protein content of defatted meal on chromosomes A01, A07 and C03 (Table 2). The identified QTL had the largest effect but no candidate genes were elucidated in this study. Interestingly, the most interesting hotspot of QTL were located on chromosome A01 which were linked to palmitic, oleic and linoleic fatty acids, oil content and protein content. The variation in this region led to decrease of palmitic, linoleic fatty acids and protein content in the defatted meal which led to increase of oil content in the seed (Teh and Mollers, 2016). Recently, the application of the 60K SNP Infinium Array allowed identification of 3700 single nucleotide polymorphisms (SNPs) in 38 QTL regions for seed protein content in B. napus (Chao et al., 2017). The potential candidate genes identified in this study encode seed storage 2S, caleosin, oleosin, and cruciferin. To date, this is the most accurate genetic mapping of protein content in B. napus which allowed identification of candidate genes involved in regulation of protein content in seeds of oilseed rape (Table 2).
Summary and Future Prospects
As the world population increases, the demand of protein for animal feed and human consumption in the world will also be rising. Oilseed rape meal is a great source of plant protein but still requires improvement of its quality and quantity. As the quantity of seed protein is highly interconnected with oil content, therefore enhanced protein content should not affect the level of the primary desired seed oil. It is of great interest to introduce desirable traits present in the yellow seeded genotypes (less fiber, higher protein content), improve the level of lysine, reduce anti nutritional factors and engineer the oleosins to produce valuable proteins. Future prospects of improvement of oilseed rape as a protein crop would be integration of recent advances in genomic studies and bioinformatics methods to identify key genes regulating quantity of seed protein and its amino acids composition to implement them as molecular markers into marker assisted breeding. Recent advances in genome sequencing technology, knowledge of the oilseed rape reference genome together with the latest bioinformatics methods (Edwards et al., 2013; Glaubitz et al., 2014; Bayer et al., 2015) allow identification of high resolution sequence based markers which are very useful in genome wide association studies (GWAS). GWAS studies have already been proved to be a very useful tool in genetic dissection of agronomically important traits in B. napus (Pasam et al., 2012; Gacek et al., 2017). Implementation of the identified markers in GWAS or QTL studies into breeding practice includes validation of the genetic variant linked to a trait of interest with available molecular methods. One of such methods includes targeting of the identified genes with latest genome editing methods such as CRISPR/Cas9 technology (Bortesi and Fischer, 2015), which are great future prospects for breeding. The validated marker can only then become a functional genetic marker. Standard PCR based genotyping methods are then used to screen the pre-breeding material for identification of the desired genetic variant which allows selection of the most promising genotypes for further breeding programs. This can shorten the length of the breeding cycle of a new variety by a few years. For this reason integration of latest bioinformatics, sequencing technology methods and genetic mapping studies should be applied in breeding programs of seed protein quantity and quality in oilseed rape.
Author Contributions
KG developed the idea and wrote the manuscript. IB-B and JB edited the manuscript and contributed ideas and suggestions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank for the financial support The National Centre for Research and Development (NCBiR), Poland. This review was performed within project CORNET/22/2/2017 (ProRapeSeed), NCBiR.
Footnotes
- ^ http://www.fao.org/docrep/007/y5019e/y5019e03.htm
- ^ http://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Oilcrops/Documents/Food_outlook_oilseeds/FO_May_2015.pdf
- ^ https://www.oilworld.biz/
References
Aachary, A. A., and Thiyam, U. (2012). A pursuit of the functional nutritional and bioactive properties of canola proteins and peptides. Crit. Rev. Food Sci. Nutr. 52, 965–979. doi: 10.1080/10408398.2010.516033
Alashi, A. M., Blanchard, C. L., Mailer, R. J., and Agboola, S. O. (2013). Technological and bioactive functionalities of canola meal proteins and hydrolysates. Food Rev. Int. 29, 231–260. doi: 10.1080/87559129.2013.790046
Altenbach, S. B., Kuo, C. C., Staraci, L. C., Pearson, K. W., Wainwright, C., Georgescu, A., et al. (1992). Accumulation of a BRAZIL nut albumin in seeds of transgenic canola results in enhanced levels of seed protein methionine. Plant Mol. Biol. 18, 235–245. doi: 10.1007/BF00034952
Baszczynski, C. L., and Fallis, L. (1990). Isolation and nucleotide-sequence of a genomic clone encoding a new Brassica napus napin gene. Plant Mol. Biol. 14, 633–635. doi: 10.1007/BF00027511
Bayer, P. E., Ruperao, P., Mason, A. S., Stiller, J., Chan, C.-K., Hayashi, S., et al. (2015). High-resolution skim genotyping by sequencing reveals the distribution of crossovers and gene conversions in Cicer arietinum and Brassica napus. Theor. Appl. Genet. 128, 1039–1047. doi: 10.1007/s00122-015-2488-y
Bonello, J. F., Opsahl-Ferstad, H. G., Perez, P., Dumas, C., and Rogowsky, P. M. (2000). Esr genes show different levels of expression in the same region of maize endosperm. Gene 246, 219–227. doi: 10.1016/S0378-1119(00)00088-3
Bonello, J. F., Sevilla-Lecoq, S., Berne, A., Risueno, M. C., Dumas, C., and Rogowsky, P. M. (2002). Esr proteins are secreted by the cells of the embryo surrounding region. J. Exp. Bot. 53, 1559–1568. doi: 10.1093/jxb/erf010
Borisjuk, L., Neuberger, T., Schwender, J., Heinzel, N., Sunderhaus, S., Fuchs, J., et al. (2013). Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell 25, 1625–1640. doi: 10.1105/tpc.113.111740
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Bos, C., Airinei, G., Mariotti, F., Benamouzig, R., Berot, S., Evrard, J., et al. (2007). The poor digestibility of rapeseed protein is balanced by its very high metabolic utilization in humans. J. Nutr. 137, 594–600. doi: 10.1093/jn/137.3.594
Breen, J. P., and Crouch, M. L. (1992). Molecular analysis of a cruciferin storage protein gene family of Brassica napus. Plant Mol. Biol. 19, 1049–1055. doi: 10.1007/BF00040536
Casson, S. A., and Lindsey, K. (2006). The turnip mutant of arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 142, 526–541. doi: 10.1104/pp.106.080895
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chao, H. B., Wang, H., Wang, X. D., Guo, L. X., Gu, J. W., Zhao, W. G., et al. (2017). Genetic dissection of seed oil and protein content and identification of networks associated with oil content in Brassica napus. Sci. Rep. 7:46295. doi: 10.1038/srep46295
Chen, M., Xuan, L., Wang, Z., Zhou, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 165, 905–916. doi: 10.1104/pp.114.235507
Chern, M. S., Eiben, H. G., and Bustos, M. M. (1996). The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos. Plant J. 10, 135–148. doi: 10.1046/j.1365-313X.1996.10010135.x
Costa, L. M., Yuan, J., Rouster, J., Paul, W., Dickinson, H., and Gutierrez-Marcos, J. F. (2012). Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 22, 160–165. doi: 10.1016/j.cub.2011.11.059
Delisle, A. J., and Crouch, M. L. (1989). Seed storage protein transcription and messenger-RNA levels in Brassica napus during development and in response to exogenous abscisic-acid. Plant Physiol. 91, 617–623. doi: 10.1104/pp.91.2.617
Ebine, K., Okatani, Y., Uemura, T., Goh, T., Shoda, K., Niihama, M., et al. (2008). A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20, 3006–3021. doi: 10.1105/tpc.107.057711
Edwards, D., and Batley, J. (2010). Plant genome sequencing: applications for crop improvement. Plant Biotechnol. J. 8, 2–9. doi: 10.1111/j.1467-7652.2009.00459.x
Edwards, D., Batley, J., and Snowdon, R. J. (2013). Accessing complex crop genomes with next-generation sequencing. Theor. Appl. Genet. 126, 1–11. doi: 10.1007/s00122-012-1964-x
EFSA NDA Panel [EFSA Panel on Dietetic Products and Nutrition and Allergies] (2013). Scientific opinion on the safety of “rapeseed protein isolate” as a novel food ingredient. EFSA J. 11:3420. doi: 10.2903/j.efsa.2013.3420
Elahi, N., Duncan, R. W., and Stasolla, C. (2015). Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol. Biochem. 96, 222–230. doi: 10.1016/j.plaphy.2015.08.002
Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerstrom, M., and Rask, L. (2000). Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 24, 57–66. doi: 10.1046/j.1365-313x.2000.00857.x
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Field, B., Cardon, G., Traka, M., Botterman, J., Vancanneyt, G., and Mithen, R. (2004). Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol. 135, 828–839. doi: 10.1104/pp.104.039347
Gacek, K., Bayer, P. E., Bartkowiak-Broda, I., Szala, L., Bocianowski, J., Edwards, D., et al. (2017). Genome-wide association study of genetic control of seed fatty acid biosynthesis in Brassica napus. Front. Plant Sci. 7:2062. doi: 10.3389/fpls.2016.02062
Gao, C. H., Qi, S. H., Liu, K. G., Li, D., Jin, C. Y., Li, Z. W., et al. (2016). MYC2, MYC3, and MYC4 function redundantly in seed storage protein accumulation in Arabidopsis. Plant Physiol. Biochem. 108, 63–70. doi: 10.1016/j.plaphy.2016.07.004
Gehrig, P. M., Krzyzaniak, A., Barciszewski, J., and Biemann, K. (1996). Mass spectrometric amino acid sequencing of a mixture of seed storage proteins (napin) from Brassica napus, products of a multigene family. Proc. Natl. Acad. Sci. U.S.A. 93, 3647–3652. doi: 10.1073/pnas.93.8.3647
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9:e90346. doi: 10.1371/journal.pone.0090346
Gotz, K. P., Staroske, N., Radchuk, R., Emery, R. J. N., Wutzke, K. D., Herzog, H., et al. (2007). Uptake and allocation of carbon and nitrogen in Vicia narbonensis plants with increased seed sink strength achieved by seed-specific expression of an amino acid permease. J. Exp. Bot. 58, 3183–3195. doi: 10.1093/jxb/erm164
Grami, B., Baker, R. J., and Stefansson, B. R. (1977). Genetics of protein and oil content in summer rape - heritability, number of effective factors, and correlations. Can. J. Plant Sci. 57, 937–943. doi: 10.4141/cjps77-134
Gruis, D., Selinger, D. A., Curran, J. M., and Jung, R. (2002). Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 14, 2863–2882. doi: 10.1105/tpc.005009
Gruis, D. F., Schulze, J., and Jung, R. (2004). Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16, 270–290. doi: 10.1105/tpc.016378
Guerche, P., Dealmeida, E. R. P., Schwarztein, M. A., Gander, E., Krebbers, E., and Pelletier, G. (1990). Expression of the 2S albumin from bertholletia-excelsa in Brassica napus. Mol. Gen. Genet. 221, 306–314. doi: 10.1007/BF00259393
Gül, M., Becker, H. C., and Ecke, W. (2003). “QTL mapping and analysis of QTL x nitrogen interactions for protein and oil contents in Brassica napus L.,” in Proceedings of the 11th International Rapeseed Congress, Copenhagen, 91–99.
Gupta, M., Bhaskar, P. B., Sriram, S., and Wang, P. H. (2017). Integration of omics approaches to understand oil/protein content during seed development in oilseed crops. Plant Cell Rep. 36, 637–652. doi: 10.1007/s00299-016-2064-1
Hammes, U. Z., Nielsen, E., Honaas, L. A., Taylor, C. G., and Schachtman, D. P. (2006). AtCAT6, a sink-tissue-localized transporter for essential amino acids in Arabidopsis. Plant J. 48, 414–426. doi: 10.1111/j.1365-313X.2006.02880.x
Hannoufa, A., Pillai, B. V. S., and Chellamma, S. (2014). Genetic enhancement of Brassica napus seed quality. Transgenic Res. 23, 39–52. doi: 10.1007/s11248-013-9742-3
Hara-Nishimura, I., Shimada, T., Hatano, K., Takeuchi, Y., and Nishimura, M. (1998). Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10, 825–836. doi: 10.1105/tpc.10.5.825
Herman, E. M., and Larkins, B. A. (1999). Protein storage bodies and vacuoles. Plant Cell 11, 601–613. doi: 10.1105/tpc.11.4.601
Hohl, I., Robinson, D. G., Chrispeels, M. J., and Hinz, G. (1996). Transport of storage proteins to the vacuole is mediated by vesicles without a clathrin coat. J. Cell Sci. 109, 2539–2550.
Johnson, C. S., Kolevski, B., and Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404
Josefsson, L. G., Lenman, M., Ericson, M. L., and Rask, L. (1987). Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J. Biol. Chem. 262, 12196–12201.
Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., and Hattori, T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406. doi: 10.1093/pcp/pci048
Kohnomurase, J., Murase, M., Ichikawa, H., and Imamura, J. (1994). Effects of an antisense napin gene on seed storage compounds in transgenic Brassica napus seeds. Plant Mol. Biol. 26, 1115–1124. doi: 10.1007/BF00040693
Kohnomurase, J., Murase, M., Ichikawa, H., and Imamura, J. (1995). Improvement in the quality of seed storage protein by transformation of Brassica napus with an antisense gene for cruciferin. Theor. Appl. Genet. 91, 627–631. doi: 10.1007/BF00223289
Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130, 6065–6073. doi: 10.1242/dev.00814
Lohaus, G., and Moellers, C. (2000). Phloem transport of amino acids in two Brassica napus L. genotypes and one B-carinata genotype in relation to their seed protein content. Planta 211, 833–840. doi: 10.1007/s004250000349
Millar, J. L., Khan, D., Becker, M. G., Chan, A., Dufresne, A., Sumner, M., et al. (2015). Chalazal seed coat development in Brassica napus. Plant Sci. 241, 45–54. doi: 10.1016/j.plantsci.2015.09.019
Nesi, N., Delourme, R., Bregeon, M., Falentin, C., and Renard, M. (2008). Genetic and molecular approaches to improve nutritional value of Brassica napus L. seed. Comptes Rendus Biologies 331, 763–771. doi: 10.1016/j.crvi.2008.07.018
Parcy, F., Valon, C., Raynal, M., Gaubiercomella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene-expression programs during arabidopsis seed development - roles of the abi3 locus and of endogenous abscisic-acid. Plant Cell 6, 1567–1582. doi: 10.1105/tpc.6.11.1567
Park, M., Lee, D., Lee, G. J., and Hwang, I. (2005). AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170, 757–767. doi: 10.1083/jcb.200504112
Pasam, R. K., Sharma, R., Malosetti, M., van Eeuwijk, F. A., Haseneyer, G., Kilian, B., et al. (2012). Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 12:16. doi: 10.1186/1471-2229-12-16
Qu, C., Zhao, H., Fu, F., Wang, Z., Zhang, K., Zhou, Y., et al. (2016). Genome-wide survey of flavonoid biosynthesis genes and gene expression analysis between black- and yellow-seeded Brassica napus. Front. Plant Sci. 7:1755. doi: 10.3389/fpls.2016.01755
Rask, L., Ellerstrom, M., Ezcurra, I., Stalberg, K., and Wycliffe, P. (1998). Seed-specific regulation of the napin promoter in Brassica napus. J. Plant Physiol. 152, 595–599. doi: 10.1016/S0176-1617(98)80017-5
Rolletschek, H., Hosein, F., Miranda, M., Heim, U., Gotz, K. P., Schlereth, A., et al. (2005). Ectopic expression of an amino acid transporter (VfAAP1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiol. 137, 1236–1249. doi: 10.1104/pp.104.056523
Sanders, A., Collier, R., Trethewy, A., Gould, G., Sieker, R., and Tegeder, M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59, 540–552. doi: 10.1111/j.1365-313X.2009.03890.x
Schatzki, J., Ecke, W., Becker, H. C., and Mollers, C. (2014). Mapping of QTL for the seed storage proteins cruciferin and napin in a winter oilseed rape doubled haploid population and their inheritance in relation to other seed traits. Theor. Appl. Genet. 127, 1213–1222. doi: 10.1007/s00122-014-2292-0
Schmidt, R., Stransky, H., and Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226, 805–813. doi: 10.1007/s00425-007-0527-x
Scofield, S. R., and Crouch, M. L. (1987). Nucleotide sequence of a member of the napin storage protein family from Brassica napus. J. Biol. Chem. 262, 12202–12208.
Sjodahl, S., Rodin, J., and Rask, L. (1991). Characterization of the 12S globulin complex of Brassica napus - evolutionary relationship to other 11-12S storage globulins. Eur. J. Biochem. 196, 617–621. doi: 10.1111/j.1432-1033.1991.tb15857.x
Song, W., Koh, S., Czako, M., Marton, L., Drenkard, E., Becker, J. M., et al. (1997). Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol. 114, 927–935. doi: 10.1104/pp.114.3.927
Stone, S. L., Braybrook, S. A., Paula, S. L., Kwong, L. W., Meuser, J., Pelletier, J., et al. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3151–3156. doi: 10.1073/pnas.0712364105
Swiech, E., Raj, S., Pastuszewska, B., Taciak, M., Bartkowiak-Broda, I., and Smulikowska, S. (2016). Nutritional value of yellow-seeded winter rapeseed cakes for growing pigs. Agric. Food Sci. 25, 99–110.
Tan, H. L., Yang, X. H., Zhang, F. X., Zheng, X., Qu, C. M., Mu, J. Y., et al. (2011). Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 156, 1577–1588. doi: 10.1104/pp.111.175000
Teh, L., and Mollers, C. (2016). Genetic variation and inheritance of phytosterol and oil content in a doubled haploid population derived from the winter oilseed rape Sansibar x Oase cross. Theor. Appl. Genet. 129, 181–199. doi: 10.1007/s00122-015-2621-y
Tilsner, J., Kassner, N., Struck, C., and Lohaus, G. (2005). Amino acid contents and transport in oilseed rape (Brassica napus L.) under different nitrogen conditions. Planta 221, 328–338. doi: 10.1007/s00425-004-1446-8
Verdier, J., and Thompson, R. D. (2008). Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 49, 1263–1271. doi: 10.1093/pcp/pcn116
Wanasundara, J. P. D. (2011). Proteins of Brassicaceae oilseeds and their potential as a plant protein source. Crit. Rev. Food Sci. Nutr. 51, 635–677. doi: 10.1080/10408391003749942
Wanasundara, J. P. D., McIntosh, T. C., Perera, S. P., Withana-Gamage, T. S., and Mitra, P. (2016). Canola/rapeseed protein-functionality and nutrition. OCL 23:15. doi: 10.1051/ocl/2016028
Wang, H. Y., Guo, J. H., Lambert, K. N., and Lin, Y. (2007). Developmental control of Arabidopsis seed oil biosynthesis. Planta 226, 773–783. doi: 10.1007/s00425-007-0524-0
Weigelt, K., Kuster, H., Radchuk, R., Muller, M., Weichert, H., Fait, A., et al. (2008). Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. Plant J. 55, 909–926. doi: 10.1111/j.1365-313X.2008.03560.x
Wu, X., Ebine, K., Ueda, T., and Qiu, Q. S. (2016). AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLoS One 11:e0151658. doi: 10.1371/journal.pone.0151658
Wurschum, T., Liu, W. X., Maurer, H. P., Abel, S., and Reif, J. C. (2012). Dissecting the genetic architecture of agronomic traits in multiple segregating populations in rapeseed (Brassica napus L.). Theor. Appl. Genet. 124, 153–161. doi: 10.1007/s00122-011-1694-5
Zhang, L., Tan, Q., Lee, R., Trethewy, A., Lee, Y.-H., and Tegeder, M. (2010). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22, 3603–3620. doi: 10.1105/tpc.110.073833
Zhao, J. Y., Becker, H. C., Zhang, D., Zhang, Y. F., and Ecke, W. (2006). Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor. Appl. Genet. 113, 33–38. doi: 10.1007/s00122-006-0267-5
Keywords: Brassica napus, oilseed rape, seed protein, seed storage protein, genetic regulation
Citation: Gacek K, Bartkowiak-Broda I and Batley J (2018) Genetic and Molecular Regulation of Seed Storage Proteins (SSPs) to Improve Protein Nutritional Value of Oilseed Rape (Brassica napus L.) Seeds. Front. Plant Sci. 9:890. doi: 10.3389/fpls.2018.00890
Received: 28 February 2018; Accepted: 07 June 2018;
Published: 02 July 2018.
Edited by:
Maoteng Li, Huazhong University of Science and Technology, ChinaReviewed by:
Yan Long, Chinese Academy of Agricultural Sciences, ChinaLiezhao Liu, Southwest University, China
Xiao-Li Tan, Jiangsu University, China
Copyright © 2018 Gacek, Bartkowiak-Broda and Batley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jacqueline Batley, SmFjcXVlbGluZS5iYXRsZXlAdXdhLmVkdS5hdQ==
 Katarzyna Gacek
Katarzyna Gacek Iwona Bartkowiak-Broda1
Iwona Bartkowiak-Broda1 Jacqueline Batley
Jacqueline Batley