- 1Agriculture Division of DowDuPont™, Corteva Agriscience™, Johnston, IA, United States
- 2Center of Genomics and Bioinformatics, Academy of Sciences of the Republic of Uzbekistan, Republic of Uzbekistan, Tashkent, Uzbekistan
Global food demand is expected to nearly double by 2050 due to an increase in the world's population. The Green Revolution has played a key role in the past century by increasing agricultural productivity worldwide, however, limited availability and continued depletion of natural resources such as arable land and water will continue to pose a serious challenge for global food security in the coming decades. High yielding varieties with proven tolerance to biotic and abiotic stresses, superior nutritional profiles, and the ability to adapt to the changing environment are needed for continued agricultural sustainability. The narrow genetic base of modern cultivars is becoming a major bottleneck for crop improvement efforts and, therefore, the use of crop wild relatives (CWRs) is a promising approach to enhance genetic diversity of cultivated crops. This article provides a review of the efforts to date on the exploration of CWRs as a source of tolerance to multiple biotic and abiotic stresses in four global crops of importance; maize, rice, cotton, and soybean. In addition to the overview of the repertoire and geographical spread of CWRs in each of the respective crops, we have provided a comprehensive discussion on the morphological and/or genetic basis of the traits along with some examples, when available, of the research in the transfer of traits from CWRs to cultivated varieties. The emergence of modern molecular and genomic technologies has not only accelerated the pace of dissecting the genetics underlying the traits found in CWRs, but also enabled rapid and efficient trait transfer and genome manipulation. The potential and promise of these technologies has also been highlighted in this review.
Introduction
Understanding the origins of crop plants and their relationships to wild relatives has been a major focus for plant biologists for many years. This knowledge continues to be of great importance in dissecting the process of crop domestication and the ability to leverage wild relatives for crop improvement. Germplasm characterization studies and breeding programs over decades have shown that cultivated plants, in general, have a relatively lower level of tolerance to biotic and abiotic stresses when compared to crop wild relatives (CWRs). One-dimensional selection for increased yield has been hypothesized to result in metabolic resource allocation toward accelerated growth and reproduction and away from plant's tolerance to biotic and abiotic factors (Rosenthal and Dirzo, 1997). Alternatively, plant breeders and population geneticists believe that artificial selection for very small percentage of genes has created breeding bottlenecks that has drastically reduced genetic variation of modern crops and led to the loss of genes derived from CWRs (Hufford et al., 2012). Although tolerance genes have traditionally been considered as negatively correlated with yield (Strauss et al., 2002; Wise, 2007), it has been recently reported that breeding for multiple plant defense traits can be achieved without compromising crop yield (Kaplan et al., 2009).
CWRs offer a diverse array of traits with the potential to decrease the amount of yield loss as a result of biotic and abiotic stresses and pest damage. These CWR- derived resistant traits could be brought into susceptible modern crops through conventional breeding (if there is a sexual compatibility), transgenesis, or other emerging technologies. Introgression of traits of interest from a CWR to a sexually compatible conventional line through traditional breeding could add complications due to the substantial amount of linkage drag resulting from the CWR. However, marker-assisted backcrossing has proven to be a technique capable of rapidly eliminating linkage drag with a minimum number of generations (Peng et al., 2014; Vishwakarma et al., 2014). If biological constraints are creating barriers to transfer desirable loci from CWRs to crops, then transgenesis can be a method of choice. Theoretically, transgenesis is attractive because it eliminates linkage drag, however, practically, it is a very cumbersome and expensive process. Transgenesis also depends on the complexity of the donor genome, availability of an efficient transformation system for the recipient crop, and, last but not least, the final product will have to undergo a lengthy, expensive, and complicated deregulation process.
The use of CWRs in crop improvement has been extensively reviewed by Hajjar and Hodgkin (2007), Prescott-Allen and Prescott-Allen (2013), and Yumurtaci (2015). Hajjar and Hodgkin (2007) and Prescott-Allen and Prescott-Allen (2013) reviews have covered research activities prior to 2005, whereas Yumurtaci (2015) has focused on studies published between 1997 and 2014 covering wheat, barley, maize, and oat. The purpose of this review was to summarize information related to economically important biotic and abiotic traits characterized in CWRs of two monocot crops (maize and rice) and two dicot crops (soybean and cotton).
Maize
Maize (Zea mays L., ssp. mays) is one of the most important crops in the world cultivated primarily for use in animal feed and biofuel. Teosinte (Z. mays ssp. parviglumis Iltis & Doebley) and Tripsacum are two CWRs that have been extensively characterized as donors of economically important traits that could be used for improvement of maize (Table 1). It has taken nearly a century to confirm that Balsas teosinte (Z. mays ssp. parviglumis Iltis & Doebley) is a progenitor of maize (Matsuoka et al., 2002). Teosinte is a wild grass natively grown in Mexico and some Central American countries including Nicaragua (Iltis and Benz, 2000), Guatemala (Wilkes, 1977), and Honduras (Standley, 2015); refer to Figure 1 for geographical representation. Genus Zea comprises annual species Zea luxurians, diploid perennial Z. diploperennis, tertraploid perennial Z. perennis, and polytypic annual species Z. mays, which in turn includes four subspecies; ssp. mays (maize), ssp. mexicana, ssp. parviglumis, and ssp. huehuetanangensis (Fukunaga et al., 2005). Tripsacum has been considered closely related to Zea due to morphological similarities including the highly specialized cupulate fruitcase, and the ability to cross with Zea and produce viable but generally infertile hybrids (Galinat, 1961). The genus Tripsacum comprises nine species of warm-season, perennial grasses that are native to the area starting in southern Canada (North America) and extending as far south as Chile (South America) (Doebley, 1983; Eubanks, 2006; Figure 1). One species of Tripsacum that has been broadly used to generate intergeneric hybrids with maize is T. dactyloides, or Eastern gamagrass (De Wet et al., 1981).
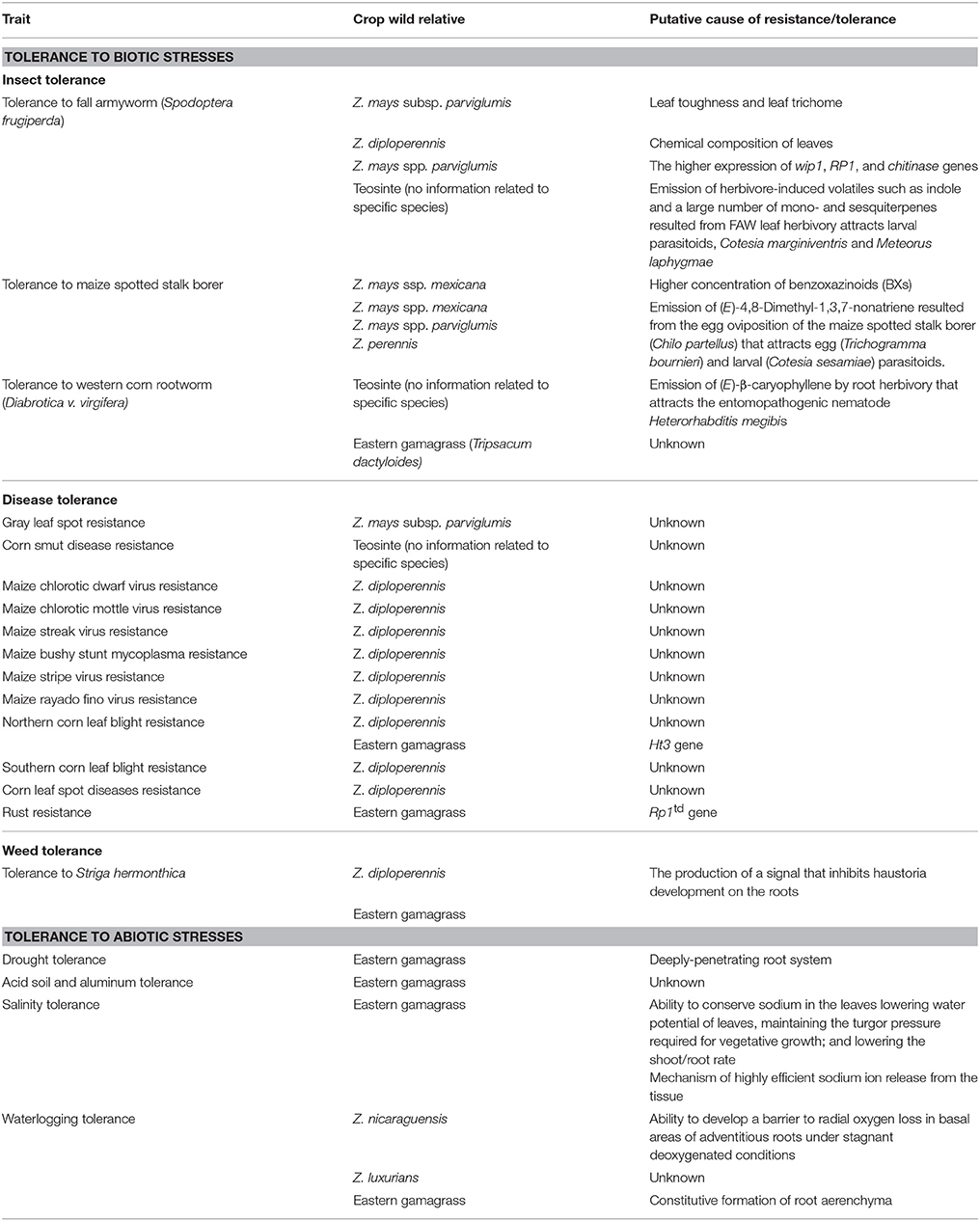
Table 1. Summary of biotic and abiotic stress tolerance traits of wild relatives of maize (Zea mays subsp. mays).
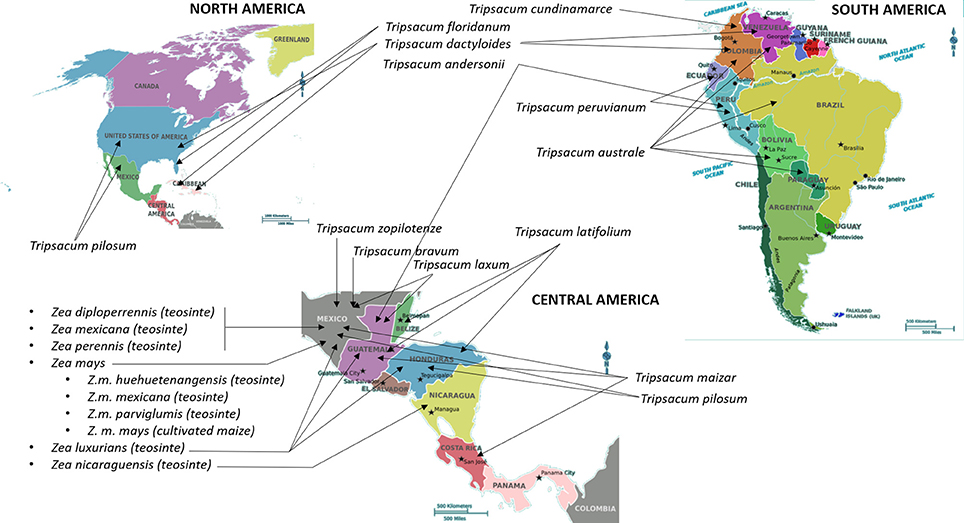
Figure 1. Centers of origin and primary geographical distribution of cultivated maize and its wild relatives. Information on the distribution and centers of origin was gathered from Wilkes (1977), Doebley (1983), Iltis and Benz (2000), and Standley (2015). International Maize and Wheat Improvement Center (CIMMYT) (https://www.cimmyt.org/) is a major maize “living catalog of genetic diversity comprising over 28,000 unique seed collections of maize,” including teosinte species. In addition to CIMMYT, seeds of maize, teosinte, and Tripsacum species are stored and could be obtained from the Germplasm Resources Information Network in Beltsville, USA (https://npgsweb.ars-grin.gov/gringlobal/search.aspx) and N. I. Vavilov Research Institute of Plant Industry in Saint Petersburg, Russia (http://www.vir.nw.ru/).
Tolerance to Biotic Stresses
Insect Tolerance
The leaves of Balsas teosinte possess certain physical barriers that are believed to make them less desirable for herbivory when compared to cultivated maize. For instance, leaf toughness was reported as a possible reason why fall armyworm (FAW) (Spodoptera frugiperda) and leafhopper (Dalbulus maidis) prefer cultivated maize for oviposition in order to provide a better food source for freshly hatched larvae (Takahashi et al., 2012; Bellota et al., 2013; Dávila-Flores et al., 2013; Bernal et al., 2015). Another physical barrier against the parasite invasion is the density of the leaf trichome. This is particularly effective in early leaf development where it is reported to play a significant role in preventing damage by FAW in teosinte species as compared to maize lines (Moya-Raygoza, 2016). Pubescence, measured by trichome density, did not significantly decline through domestication transitions as was expected. Research has also demonstrated that trichome density poses very little barrier against D. maidis oviposition (Dávila-Flores et al., 2013).
In addition to physical barriers, chemical composition of teosinte leaves in particular, toxic secondary metabolites, could result in tolerance to herbivore insect consumption (Howe and Jander, 2008). It was shown by Farias-Rivera et al. that leaf extracts and residual fiber from Z. diploperennis affected the growth of FAW resulting in diminished length and width of pupae and an increased cumulative mortality rate (Farias-Rivera et al., 2003). The increased expression of wip1, RP1, and chitinase genes is believed to result in a greater tolerance of Balsas teosinte to FAW (Szczepaniec et al., 2013). Teosinte accessions of Z. mays ssp. mexicana that demonstrated resistance to maize spotted stalk borer (MSSB) produced a higher concentration of benzoxazinoids (BXs) compared to susceptible maize lines. This was hypothesized to be the underlying molecular basis of teosinte resistance to MSSB (Niazi et al., 2014). Benzoxazinoids have also been reported to be involved in resistance of maize to lepidopterans, in particular, European corn borer (Niemeyer, 2009). In contrast, Maag and colleagues rejected the hypothesis that higher concentrations of BXs and maysin make teosinte more insect tolerant than cultivated maize varieties (Maag et al., 2015). Researchers suggested that teosinte tolerance to herbivore insects is more likely controlled by the other defense traits than BXs and maysin.
In maize and teosinte, herbivory induces the emission of volatile compounds (VOCs) that attract beneficial insects which in turn attack pests (McMullen et al., 2009; Lange et al., 2014; Tamiru et al., 2015). The emission of VOCs is not completely a domesticated trait in maize as the crop also uses VOC as a weapon to fight pests such as FAW and leafhoppers by attracting predators and parasitoids (Hoballah et al., 2004). However, teosinte was reported to attract quantitatively more beneficial insects than cultivated maize (Lange et al., 2014). Depending on which organ of maize or teosinte is damaged by herbivory, the spectrum of the beneficial insect attracted changes. Foliage damage releases a blend of VOCs which consists of indole and a large number of mono- and sesquiterpenes, which are strongly expressed after herbivory by Lepidoptera (Köllner et al., 2004). These VOCs attract parasitic wasps such as Cotesia marginiventris and Meteorus laphygmae which have a tendency to attack FAW (Hoballah et al., 2004). However, the latter parasitic wasp “learned” to recognize these VOCs to detect their source of food (Carroll et al., 2006). Teosinte was also reported to release the VOC (E)-4,8-dimethyl-1,3,7-nonatriene as a response to the spotted stalk borer oviposition that attracted both egg (Trichogramma bourneiri) and larval (Cotesia sesamiae) parasitoids (Mutyambai et al., 2015). It was also reported that maize and teosinte roots release VOCs in response to herbivory damage by Coleoptera. In particular, they release a (E)-β-caryophyllene in a response to damage by D. v. virgifera, which attracts the entomopathogenic nematode Heterorhabditis megibis (Rasmann et al., 2005). Interestingly, North American maize lines do not emit a (E)-β-caryophyllene, whereas European lines do so in response to D. v. virgifera attack (Rasmann et al., 2005). Teosinte, particularly Z. diploperennis, seems to attract more root damaging insects, including western corn rootworm (WCR) (Diabrotica virgifera virgifera) than maize. However, due to larger amount of root biomass in teosinte it is still more tolerant to insect root damage than maize (reviewed by Lange et al., 2014). In contrast to teosinte, eastern gamagrass has been observed to be resistant to WCR and the resistance has been reported to have a quantitative nature (Prischmann et al., 2009). Eastern gamagrass was also identified as a donor of resistance to the maize weevil, Sitophilus zeamais Motschulsky, one of the major pests of maize worldwide (Throne and Eubanks, 2015).
Disease Tolerance
The genetics of resistance of Balsas teosinte to the fungal disease, gray leaf spot (GLS), was studied by Lennon and colleagues using classical quantitative trait loci (QTL) mapping and employing a population of near isogenic lines (Lennon et al., 2016). Using these approaches Lennon et al was able to identify the teosinte-derived GLS resistance QTL located in bin 4.07. Teosinte was also reported to manifest resistance to corn smut disease (Chavan and Smith, 2014). Using genomic in situ hybridization technology, Wei et al. (2001) identified the chromosomal locations of the Z. diploperennis-derived segments in maize × Z. diploperennis crosses that carried loci resistant to fungal diseases including northern corn leaf blight, southern corn leaf blight, and corn leaf spot diseases. In addition, several other species of teosinte such as Z. mays ssp. mexicana have been shown to be a donor of downy mildew and Fusarium resistance (reviewed by Maazou et al., 2017). Perennial teosinte, Z. diploperennis, has demonstrated immunity to many viral and micoplasmal diseases, including maize chlorotic dwarf virus, maize chlorotic mottle virus, maize streak virus, maize bushy stunt mycoplasma, maize stripe and rayado fino viruses (Nault and Findley, 1982). Eastern gamagrass can be utilized as a donor of a novel rust resistance gene Rp1td. This gene was successfully introgressed into a susceptible maize background and proved capable of controlling resistance to rust in the cooler regions of the tropics (Bergquist, 1981). The northern corn leaf blight resistance gene, Ht3, is another disease resistance gene that has been successfully introgressed from eastern gamagrass into maize (Hooker, 1981).
Weed Tolerance
Striga hermonthica is very devastating root parasite of maize in Africa. The parasitism of S. hermonthica results from the arrays of signal exchanges between the host (maize or sorghum) and the weed. It has been reported that the host plant will produce specific substances, called strigolactones that serve as a seed germination signal for S. hermonthica (Awad et al., 2006). In maize, only qualitative resistance to S. hermonthica has been reported (Gethi and Smith, 2004; Amusan et al., 2008). Although neither maize nor its wild relatives show 100% resistance to S. hermonthica, Z. diploperennis has demonstrated higher level of tolerance to the parasite compared to a majority of maize cultivars (Kling et al., 2000; Yallou et al., 2009). The notable manifestations of Z. diploperennis' tolerance to the parasite are (1) resisting the attachment of nearby germinating S. hermonthica to its roots, and (2) restricting the penetration of the vascular system of the host by the parasitic seedlings, which would eventually result in host death (Lane et al., 1997; Rich and Ejeta, 2008). Gurney and colleagues demonstrated that eastern gamagrass also possesses tolerance to S. hermonthica through the production of a signal that inhibits haustoria development on its roots (Gurney et al., 2003). This finding of resistance of eastern gamagrass to this parasite plant was determined to be quantitatively inherited.
Tolerance to Abiotic Stresses
Drought Tolerance
If a maize plant is subjected to drought stress within a 2–10 week window before anthesis, yield reductions can be substantial because this is a very critical period for ear development (reviewed in Eubanks, 2006). Eastern gamagrass has been known for its ability to tolerate drought stress due to its morphologic and metabolic characteristics. Roots of Tripsacum can penetrate deeply into soil regardless of quality (Clark et al., 1998; Gilker et al., 2002; Gitz et al., 2013). This plant also demonstrates enhanced water use efficiency and photosynthesis during drought stress (Coyne and Bradford, 1985). Although drought tolerance is a complex trait, Eubanks has demonstrated that it can be successfully introgressed into a cultivated maize background (Eubanks, 2006).
Acid Soil and Aluminum Tolerance
Soil acidification is a process that occurs naturally due to decomposition of organic matter and air pollution-caused acid rain. Excessive use of nitrogen-containing fertilizers in agriculture can also contribute to an elevated soil acidity. Acidic soil negatively impacts crops by increasing bioavailability of toxic metal ions, including aluminum. Aluminum toxicity in maize is observed at a pH > 5.5 (Von Uexküll and Mutert, 1995) and results in the inhibition of root growth (reviewed in Aggarwal et al., 2015). Eastern gamagrass has been reported as tolerant to aluminum toxicity and efforts to introgress loci controlling tolerance to aluminum toxicity into maize background have been studied (Eubanks, 2006). Although results were promising, no further reports have been published on developing aluminum tolerant maize cultivars. The mechanism of aluminum tolerance in eastern gamagrass remains unknown.
Salinity Tolerance
Soil salinity negatively affects crops by causing a hyperionic and hyperosmotic stress environment that slows down plant growth and significantly decreases yield (Shrivastava and Kumar, 2015). Eastern gamagrass has shown tolerance to high salt concentrations in soil associated with the ability to conserve sodium in its leaves lowering the water potential and therefore, providing the turgor pressure necessary for vegetative growth (Pesqueira et al., 2003, 2006). Additional benefits of eastern gamagrass include lowering the shoot/root rate which is a favorable aspect for plant water balance. Another study attributed salinity tolerance of eastern gamagrass to the ability of highly efficient sodium ion release from the tissue (Shavrukov and Sokolov, 2015).
Waterlogging Tolerance
Z. nicaraguensis grows in the coastal plains of Nicaragua and is known for higher waterlogging tolerance than any other species of Zea (Iltis and Benz, 2000). Mano and his colleagues identified the genetic components underlying three major traits that makes Z. nicaraguensis waterlogging tolerant: (1) formation of constitutive aerenchyma in drained soil; (2) the ability to grow adventitious roots at the soil surface in waterlogged conditions; and (3) tolerance to reduced soil toxins (Mano et al., 2007, 2009; Mano and Omori, 2008). However, in recent research they did not find a significant correlation between the ability to form constitutive aerenchyma and waterlogging tolerance and, therefore, indicated the presence of other flood tolerance-related traits (Mano and Omori, 2013). Recently Abiko and colleagues concluded that Z. nicaraguensis has an ability to develop a barrier to radial oxygen loss (ROL) in basal areas of adventitious roots under stagnant deoxygenated conditions (Abiko et al., 2012). ROL drives a higher internal oxygen supply to the root tip allowing it to grow in waterlogged soil. Another species of teosinte, Z. luxurians, was declared a source of a putative waterlogging tolerance due to the nodal root angle (Omori and Mano, 2007). Eastern gamagrass exhibits waterlogging tolerance due to the constitutive formation of root aerenchyma (Hardin, 1994; Comis, 1997; Ray et al., 1998), the tissue that allows oxygen to penetrate to the distal regions of the roots (Mujeeb-Kazi and Jewell, 1985; Gitz et al., 2013). Attempts to introgress waterlogging tolerance into maize through conventional breeding have not been successful due to the complexity of the trait inheritance (Ray et al., 1999).
Rice
Rice (Oryza sativa L.) is the world's most important staple food crop contributing to the majority of dietary intake for nearly half of the global population. Biotic and abiotic stresses have seriously affected rice production in recent times due to changing climatic conditions and non-durability of resistance incorporated into cultivars (Normile, 2008). Genetic variability for resistance/tolerance to these stresses is limited in the germplasm of cultivated rice, however, wild species of Oryza are considered rich sources of unexplored genes for these traits (Jena, 2010; Sanchez et al., 2013; Shakiba and Eizenga, 2014). Identification and transfer of these genes from wild species to modern cultivars is, therefore, an attractive option.
The genus Oryza consists of 24 species of which two, Oryza sativa L. and Oryza glaberrima S., are cultivated and the remaining are wild relatives distributed around the world (Sanchez et al., 2013; Shakiba and Eizenga, 2014); refer to Figure 2 for geographical resprentations of these locations. Based on taxonomy, isozyme, and DNA marker analysis, and characterization of genome sequences, the species of the genus Oryza were assembled into several groups. These groups include O. sativa, O. ridleyi, O. granulata, and O. officinalis. The species belonging to the O. sativa group include the cultivated species, O. sativa L. and O. glaberrima S., which are diploids and possess the AA genome type (2n = 24). Species belonging to groups farther away from O. sativa are diploids or tetraploids and contain other genome types (Sanchez et al., 2013; Shakiba and Eizenga, 2014).
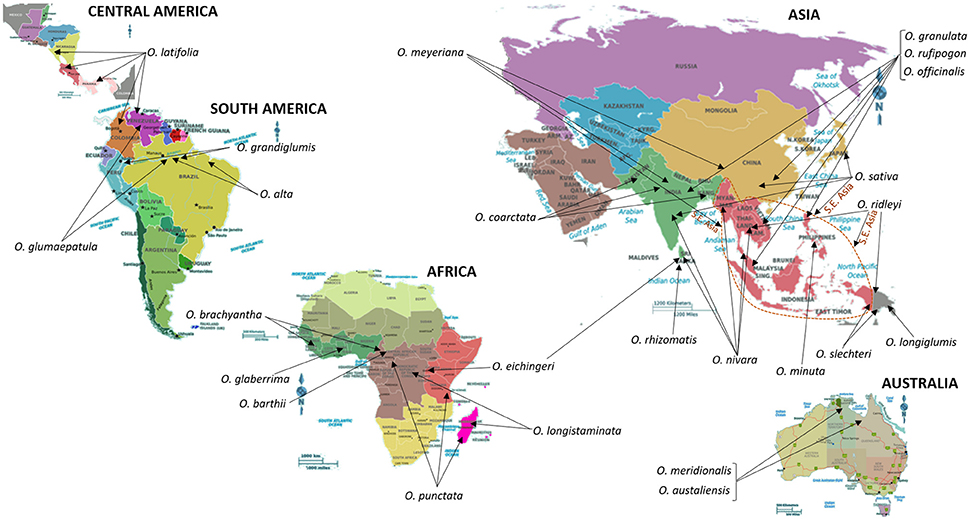
Figure 2. Centers of origin and/or primary sites of diversity and distribution of wild relatives of Rice. South East Asia (S.E. Asia), a key region for the origin and distribution of several wild relatives of Rice, is indicated by a dotted line. Information on the distribution and centers of origin was gathered from Shakiba and Eizenga (2014), and references therein. “The International Rice Genebank, maintained by International Rice Research Institute, holds more than 124,000 rice accessions that include modern and traditional varieties and wild relatives of rice. It is the biggest collection of rice genetic diversity in the world. Countries from all over the world have sent their rice samples to IRRI for safe keeping as well as for sharing” (http://irri.org/). In addition to IRRI, the Germplasm Resources Information Network in Beltsville, USA (https://npgsweb.ars-grin.gov/gringlobal/search.aspx) also stores and distributes genetic material of cultivated rice and its wild relatives.
Early breeding efforts for introgressing genes of interest from wild relatives into cultivated species of rice were intentionally focused on wild species with the AA genome because F1 hybrids derived from these crosses display regular chromosome pairing and recombination. In contrast, hybrids between cultivated and wild species that possess alternative genomes (for example, AA genome species crossed with non-AA genome species) are typically more difficult to generate due to incompatibility issues resulting in sub-optimal crossability and non-viable embryos (Brar and Khush, 1997; McCouch et al., 2007). However, embryo rescue can be used to produce male-sterile interspecific F1 hybrids for utilization in a backcrossing program resulting in fertile diploid plants (Jena, 2010). Several tools can be used to enable chromosomal assignment, dissection of traits in wild species, and gene transfer from wild to cultivated species including: monosomic alien addition lines, disomic introgression lines, chromosome segment substitution lines, and backcross inbred lines (Ali et al., 2010; Jena, 2010).
Tolerance to Biotic Stresses
Insect Tolerance
The brown planthopper (BPH), Nilaparvata lugens Stål, is a migratory insect that has become the most devastating pest of rice. In addition to causing severe plant damage resulting in significant production losses, BPH also transmits two disease causing viruses, rice grassy stunt virus and rice ragged stunt virus. Although control of BPH using pesticides such as imidacloprid is effective, host-plant resistance is the preferred method. Host-plant resistance has additional benefits including reducing production costs and the risk of possible environmental contamination when compared to chemical control of BPH (Tanaka et al., 2000; Hu et al., 2016).
Several studies indicate that wild species are an important source of planthopper resistance genes (Jena, 2010; Fujita et al., 2013; Hu et al., 2016; Sarao et al., 2016) (Table 2). Studies on the exploration of resistance genes to date have identified a total of 30 Bph resistance genes from indica and wild species of rice (Fujita et al., 2013; Hu et al., 2016; Ling and Weilin, 2016). Out of the 30 Bph resistance genes more than a dozen are from wild relatives. As of today, four resistance genes have been cloned including, Bph14 and bph29, which are from O. officinalis and O. rufipogon, respectively. Parallel to gene identification and cloning efforts, introgressions of BPH resistance into cultivated/elite lines has also been performed (Table 2; Ram et al., 2010; Sarao et al., 2016).
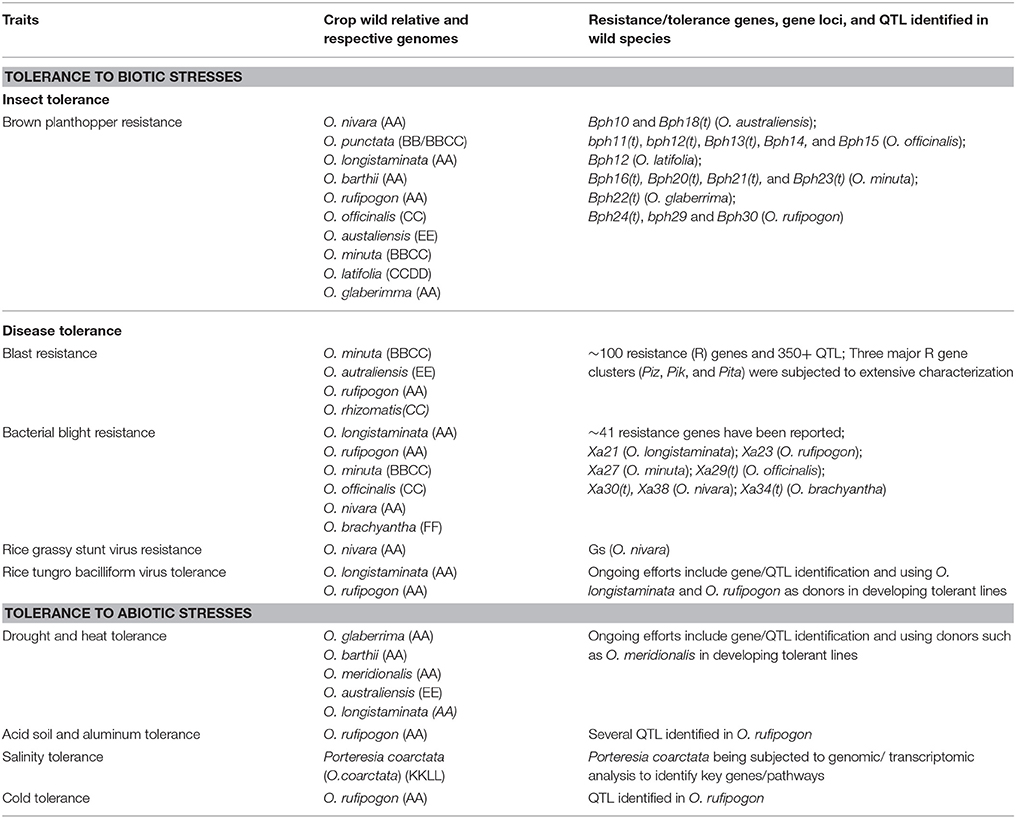
Table 2. Summary of biotic and abiotic stress tolerance traits of wild relatives of rice (Oryza sativa).
Disease Tolerance
Rice blast is considered the most serious and economically important disease caused by a fungal pathogen in rice crop. The causative agent is Magnaporthe oryzae (M. oryzae). Although M. oryzae infects other grasses, the primary host is rice. Since the late nineteenth century when rice blast was first observed in the United States it has been reported in 85 rice-growing countries around the globe and caused severe economic damage in several of these countries (Wang et al., 2014). Since host plant resistance is the preferred mode of control efforts in rice blast research has focused on identifying resistance genes and QTLs in wild species. To date, more than 100 resistance genes (called R genes) and over 350 QTL regions have been identified (Wang et al., 2014; Ashkani et al., 2015; Vasudevan et al., 2015). Out of all of the R genes identified, 96% were found in japonica and indica cultivars, and the remaining 4% in wild relatives (Wang et al., 2014). Studies on R gene mapping revealed their presence in all rice chromosomes except chromosome 3. Three major R gene clusters, Piz, Pik, and Pita, were identified and mapped to chromosomes 6, 11, and 12, respectively. These clusters have been the focus of extensive characterization research over the years and apart from a few exceptions, most R genes cloned have been found to encode nucleotide binding site-leucine-rich repeat (NBS-LRR) proteins, similar to other plant R genes known to date (Wang et al., 2014). Wild species including, O. minuta, O. autraliensis, O. rufipogon, and O. rhizomatis, have been found to harbor resistance genes to rice blast. These genes have been introgressed into susceptible lines and proven effective against rice blast disease (Table 2; Sharma et al., 2012; Wang et al., 2014; Ashkani et al., 2015). Although many varieties were developed containing a single resistance gene resistance it was broken down in 3–5 years of release due to the emergence of new races of rice blast. Therefore, it is important to develop varieties containing broad spectrum and/or durable by stacking different R genes with overlapping resistance spectra (Sharma et al., 2012). This requires the availability of a pool of R genes to utilize. Since a very small portion (4%) of the R genes identified to date are from wild species, exploring them further is important.
Bacterial blight, caused by the pathogen Xanthomonas oryzae pv. oryzae (Xoo), is endemic to most of the rice growing regions in Asia and West Africa and has recorded yield losses as high as 75% in India, Indonesia, and the Philippines (Shakiba and Eizenga, 2014). Extensive research efforts to date have resulted in the identification of 41 reported resistance genes, of which eight have been characterized (Ellur et al., 2016). Table 2 lists some of the wild relatives and the blight resistance genes identified in their genomes. A widely used resistance gene, Xa21, was isolated using a positional cloning strategy, and found to encode a receptor kinase-like protein (Song et al., 1995). It was one of the first genes tagged with DNA markers and transferred to many rice cultivars using marker-assisted breeding, however, in China, new strains of X. oryzae pv. oryzae broke the resistance conferred by Xa21 highlighting the need to explore more novel genes with durable resistance. Out of all known resistance genes Xa23, a single, completely dominant gene identified from O. rufipogon, has recorded efficacy at all growth stages of rice, possesses broad spectrum resistance, and was acknowledged as the most promising gene highly resistant to all the 20 strains of Xoo (Zhang et al., 2001; Zhang and Xie, 2014). A novel resistance gene, Xa38, has been identified recently in O. nivara and its deployment in varieties is expected to counter the breakdown of resistance to Xoo (Ellur et al., 2016).
More than 20 viruses infect rice and a majority of them use insects as vectors for their transmission. Two virus-associated diseases that cause significant damage to rice are: rice grassy stunt disease; caused by rice grassy stunt virus (RGSV), and rice tungro disease; caused by mixed infection of rice tungro bacilliform virus (RTBV) and rice tungro spherical virus (RTSV). RGSV uses rice brown planthopper as its vector for transmission. Infection results in stunted plants that produce very few to no panicles with deformed grains. Screening of thousands of accessions of cultivated and wild species led to the identification of O. nivara (AA-genome) as the only source of resistance due to a single dominant gene, Gs. The transfer of RGSV resistance from O. nivara into cultivated O. sativa represented the first successful introduction of a useful, agronomic gene from a wild to cultivated species of rice (Khush et al., 1977). Rice tungro disease is another economically important viral disease in Southern and Southeastern Asia and is transmitted by green leafhopper Nephotettix virescens. Of the two Rice tungro disease-causing viruses, RTBV and RTSV, RTBV is the main cause of symptoms. Cultivated rice germplasm has limited variability for resistance to RTBV. Tolerance to RTBV has been identified in two wild species, O. longistaminata and O. rufipogon, and many tungro-tolerant lines have been developed utilizing them as donors (Khush et al., 2004; Table 2).
Tolerance to Abiotic Stresses
Drought and Heat Tolerance
Drought is one of the main environmental stressors that reduces agricultural productivity in rice. Exploratory studies have been performed to better understand the effects of drought using backcross inbred lines from a cross between an O. sativa line, WAB56-104, and an O. glaberrima line, CG14 (Ndjiondjop et al., 2010). The findings indicated that drought reduces grain yield and adversely affects yield stability by affecting several morphological traits, including delay of flowering time and plant maturity. Although several QTLs for drought tolerance have been identified in O. sativa and the underlying genes cloned, wild relatives are considered to harbor stronger/novel alternatives and, therefore, several promising species are being investigated (Ndjiondjop et al., 2010; Menguer et al., 2017; Table 2). For example, although low yielding, O. glaberrima, has been found to be an excellent source of tolerance for drought. Screening several accessions of three wild species, O. barthii, O. australiensis, and O. meridionalis, has shown variability for two key traits; plant height and tillering ability. These species, therefore, can be used as donors in breeding programs aimed at developing tolerance to drought and heat (Ndjiondjop et al., 2010; Sanchez et al., 2013). While the exploration of candidate wild relatives continues, some donors have already been used in crossing programs to develop varieties tolerant to heat (Sanchez et al., 2013).
Acid Soil and Aluminum Tolerance
Aluminum toxicity is of utmost concern when rice is grown in acidic soils since it adversely effects root development, water and nutrient uptake, and growth resulting yield loss. Tolerance to aluminum is a quantitative trait with high variability among various rice species and therefore, the identification and introgression of an associated QTL into cultivated varieties is a very promising option. IRGC106424, an accession of O. rufipogon, was originally identified growing in acidic soils in Vietnam and has since proven to be a valuable resource for imparting aluminum tolerance to rice cultivars (Sanchez et al., 2013; Table 2). The evaluation of recombinant inbred lines derived from indica and O. rufipogon crosses by Nguyen et al. led to the identification of several QTLs for stress-associated root length, including a major QTL on chromosome 3 (Nguyen et al., 2003). An acid sulfate tolerant rice variety, AS996, has been developed by introgressing tolerance from O. rufipogon into the IR64 background (Sanchez et al., 2013). Studies on understanding the physiological and genetic basis of aluminum tolerance in O. rufipogon have indicated an association with cell division and altered photosynthesis (Cao et al., 2011).
Salinity Tolerance
While high soil salinity is a serious problem for most of the major agricultural crops, it is especially important for rice which is one of the most salt-sensitive crops. High salinity is reported to reduce seed germination, decrease growth, and survival of seedlings, damage chloroplast structure, reduce photosynthesis and negatively impact seed set and grain yield. Porteresia coarctata (Oryza coarctata), an Asian halophyte and wild relative of rice, occurring in coastal environments shows high salinity and submergence tolerance (Zhang and Xie, 2014; Table 2). Transcriptome sequence of P. coarctata suggested salinity and submergence tolerance in this species is due to substantial transcriptional reprogramming (Garg et al., 2014). Such efforts pave the way for dissecting the mechanism(s) of tolerance and identifying key genes for salinity tolerance in rice. Ultimately, resistance could be transferred to O. sativa through bridge crossing or leveraging molecular techniques.
Cold Tolerance
Cold tolerance is a concern for rice cultivation and productivity since stress induced by low temperatures can adversely affect germination, growth, and pollen development (Andaya and Tai, 2007). Initial efforts focused on identifying variability for cold tolerance in indica and japonica subspecies, which are better adapted to tropical and temperate climates. Large scale screening of indica and japonica cultivars demonstrated that japonica rice possesses relatively high tolerance to low temperatures at seedling, vegetative and reproductive stages and, therefore, efforts focused on identification of underlying QTL(s) on progeny derived from japonica and indica crosses (Lou et al., 2007; Zeng et al., 2009; Cruz et al., 2013). In addition, the wild relative O. rufipogon was also found to harbor a QTL for cold tolerance and, therefore, an ideal donor for breeding programs (Koseki et al., 2010; Table 2). QTL analysis of progeny from crosses between O. rufipogon (cold tolerant) and indica (cold sensitive) or japonica cultivars yielded a major QTL explaining 40% of the phenotypic variation (Koseki et al., 2010). This identified cold tolerant QTL was later fine mapped.
Cotton
Cotton (Gossypium spp.) is the most commonly grown natural fiber and oil seed crop throughout the world. Understanding the evolutionary history of cotton is as equally important as that of developing new cultivars. The cotton genus (Gossypium) includes ~50 species, including five allotetraploids, and other diploid species distributed across Africa, Australia, Central and South America, the Galapagos Islands, Hawaii, the Indian sub-continent, and Arabia (Fryxell, 1992; Wendel and Cronn, 2003). Figures 3A,B show the native locations of these cotton species across the world. Gossypium species express wide morphological differences from herbivorous perennials to small trees with enormous diversity in vegetative and reproductive features (Wendel and Cronn, 2003). Most of the world's textile fiber is derived from four species of the genus Gossypium, including two New World tetraploid species, G. hirsutum L. and G. barbadense L (Figure 3A) and two Old World Asian-African diploids G. arboreum L., G. herbaceum L. (Figure 3B). Diploid Gossypium species diverged into multiple genomes designated A–G, and K, based on meiotic chromosomal pairing behavior (Wendel and Cronn, 2003). The new world tetraploid species arose ~1–2 million years ago resulting from hybridization of A-genome diploid species, G. herbaceum and G. arboreum (2n = 2x = 26), with “D-genome” diploid species, G. raimondii Ulbrich and G. gossipioides L. (2n = 2x = 26) (Wendel, 1989; Wendel and Cronn, 2003). Currently, a diverse Gossypium germplasm containing wild species, race and seed stocks is conserved through national germplasm banks worldwide. The inventory of the World Cotton Germplasm Resources shows there are ~53,000–63,946 world cotton accessions preserved in major cotton-growing countries (Campbell et al., 2010; Abdurakhmonov, 2014, 2016). The wealth of the diversity available among wild and cultivated cotton species is a valuable resource for cotton breeders to hasten selection of traits of agronomic importance. Although the transfer of beneficial traits from wild diploid species into cultivated tetraploid cottons is a challenging task, many researchers have been successful through the development of interspecific hybrids involving donor and recipient genotype. These interspecific hybrids have been developed through bridge crosses or other means, including finding genetic recombinants between donor and recipient genotype chromosomes using cytogenetics and other techniques (Robinson et al., 2004a; Konan et al., 2007; Table 3).
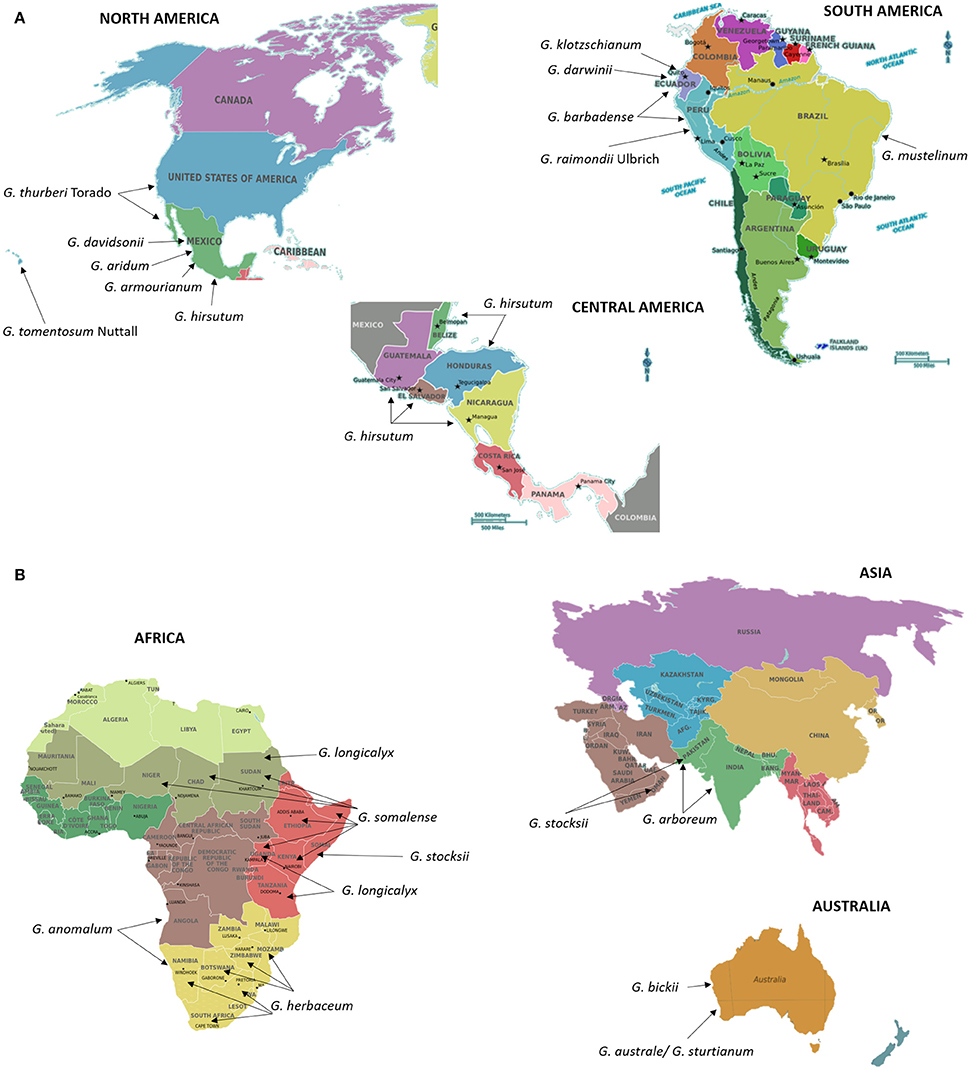
Figure 3. (AB) Centers of origin or primary geographic distribution sites of cultivated cotton species G. hirsutum L. (A), G. barbadense L (A), G. arboreum L. (B), and G. herbaceum L. (B), and their wild relatives (A,B). Information on the distribution and centers of origin was gathered from Fryxell (1992) and Wendel and Cronn (2003). Seeds of cotton and its wild relatives are stored and could be obtained from the Germplasm Resources Information Network in Beltsville, USA (https://npgsweb.ars-grin.gov/gringlobal/search.aspx).
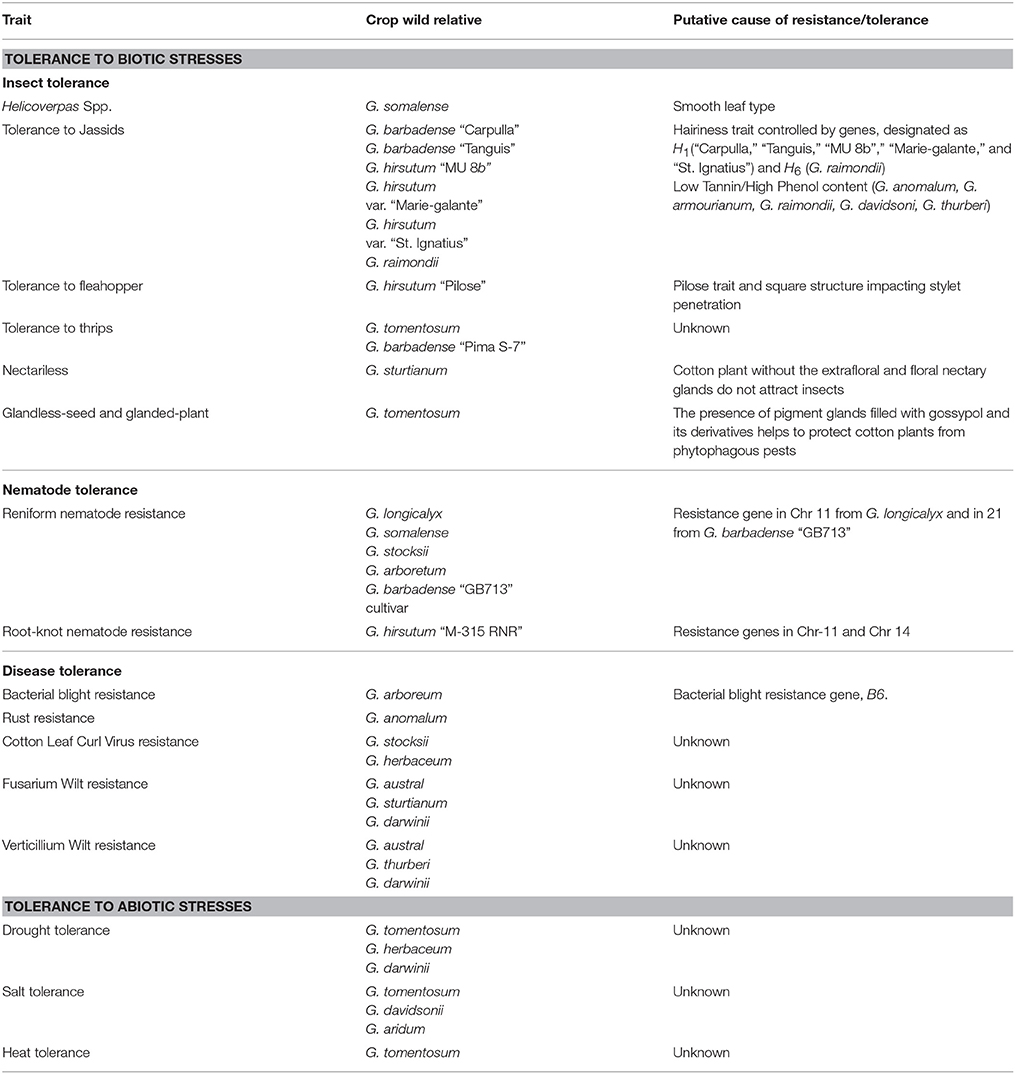
Table 3. Summary of biotic and abiotic stress tolerance traits of wild relatives of cultivated cotton species.
Tolerance to Biotic Stresses
Insect Tolerance
Cotton attracts many pests and host tolerance can be improved through morphological and biochemical traits including hairiness, okra leaf shape, nectariless, and polyphenol compounds which may confer broad-spectrum insect tolerance. Cotton displays various densities of trichomes (pubescence), on leaves and stem. Depending on the density of trichomes, pubescence phenotypes are rated as smooth (no trichomes), hirsute (moderate density), and pilose (high density) and most of the modern cotton cultivars belong to smooth category (Wright et al., 1999). Though the smooth leaf trait is related to reduced oviposition by the Heliothis spp. (Hassan et al., 1990), and is a desirable feature to minimize the heavy reliance on pesticide usage, it may increase susceptibility to other pests such as jassids. However, several explorations of jassid resistance have demonstrated that hairs of reasonable length and density presented on the underside of a cotton leaf can impart immunity to jassids. A basic, partially dominant, hairiness gene, designated H1, is shown to be responsible for the resistance of two G. barbadense perennial types, Carpulla and Tanguis, a commercial type from India, MU 8b (G. hirsutum), and the “St Ignatius” variety (G. hirsutum var. marie-galante) from British Guiana (Zafar et al., 2009). Another hairiness contributing gene (H6) was successfully transferred from G. raimondii to the G. hirsutum race punctatum (Saunders, 1965). Cotton plants with high phenolic content showed low incidence of jassids. Diploid species such as G. anomalum (B1), G. armourianum (D2), G. raimondii (D5), G. davidsoni (D3), and G. thurberi (D1) with low tannin and high phenol contents are genotypes less susceptible to jassids (Shinde et al., 2014).
Cotton fleahopper, Pseudatomoscelis seriatus, is a piercing–sucking pest of cotton that feeds preferentially on developing flower buds, called squares. It is observed that square structure along with the reproductive tissue morphology affect the stylet penetration of fleahoppers and thus contribute to resistance in cotton genotypes derived from the crosses between fleahopper resistant “Pilose” and other and high-yielding susceptible lines (McLoud et al., 2016).
Thrips are one of the most damaging early growing season insects and can cause yield losses up to 1% in spite of one insecticide application. Among the five allotetraploid cotton species, G. tomentosum with the Pilose trait was the most resistant, followed by G. mustelinum, G. barbadense, and G. darwinii, and G. hirsutum was the most susceptible (Zhang et al., 2013). Zhang et al. (2013) were able to transfer thrips resistance from Pima S-7, a G. barbadense accession, into Upland cotton which is ideal because host resistance is the most attractive strategy for the control of thrip damage.
Upland cotton has leaf, extrafloral, and floral nectary glands which secrete nectar that attracts many insects. In contrast, G. tomentosum Nuttall, a wild cotton tetraploid species native to Hawaii, does not contain leaf or extrafloral nectaries. Through interspecific crosses between G. hirsutum and G. tomentosum, the absence of leaf and extrafloral nectaries (nectariless trait) was transferred to Upland cotton (Meyer and Meyer, 1961). The presence of a plant alkaloid, gossypol, and its derivatives above 0.02–0.04% (WHO/FAO recommended limits) in cotton seed oil and meal limits its usage as food and feed. However, gossypol and its derivatives in pigment glands protect cotton plants from phytophagous insects. Thus, a desirable cotton plant would have glandless seeds on a glanded plant to protect cotton plants from phytophagous pests, as well as, increase the value of the seed derived products (Zhu et al., 2005). Two tri-specific hybrids were created using either G. thurberi Torado (2n = 2x = 26, D1 genome) or G. raimondii Ulbrich (2n = 2x = 26, D5 genome) as bridge species to introgress the “glandless-seed and glanded-plant” trait from G. sturtianum Willis (2n = 2x = 26, C1 genome) into the upland cotton G. hirsutum L. Further crosses of these tri-specific hybrids by G. hirsutum produced the first backcross progenies (BCl) to serve as donors of these traits for other elite cotton lines (Bi et al., 1998). A similar introgression effort was attempted from G. bickii into upland cotton by crossing the amphidiploid F1 (G. arboreum × G. bickii) as female with various glanded accessions of G. hirsutum as male parents (Zhu et al., 2005).
Nematode Tolerance
In recent years, reniform nematode (Rotylenchulus reniformis Linford and Oliveira), has been causing significant economic damage to cotton industry with losses exceeding $100 M annually (Blasingame, 2005). Germplasm mining into the collection of 2,000 primitive G. hirsutum accessions for sources of reniform nematode resistance (Robinson and Percival, 1997) resulted in the identification of only six accessions with moderate resistance and highly resistant cultivars were non-existent (Robinson et al., 2004b). Even though highly resistant sources were previously identified in wild diploid species, including G. longicalyx J.B. Hutch. & B.J.S. Lee; G. somalense (Gürke) J.B. Hutch.; G. stocksii Mast.; G. arboreum L.; and tetraploid species of G. barbadense L. (Yik and Birchfield, 1984), genetic incompatibility, ploidy, climbing growth habit, photoperiodism, and agronomic issues were major challenges for introgressing the resistance trait from these alien genomes. Through the development of three-species hybrids named HLA [(G. hirsutum × G. longicalyx)2 × G. armourianum] and HHL [(G. hirsutum × G. herbaceum)2 × G. longicalyx] at USDA-ARS, Texas in the United States (Robinson et al., 2004a) and HTL [(G. hirsutum × G. thurberi)2 × G. longicalyx] at Gembloux Agricultural University, Belgium (Konan et al., 2007), researchers were successful in identifying donor plants which were fertile and had reniform nematode resistance. In another effort to simultaneously provide host tolerance for two major nematode pests in cotton (reniform and root-knot nematodes), Bell et al. (2015), were successful in introgressing reniform nematode resistance from GB 713, a photoperiodic G. barbadense race stock accession, into M-315 RNR, a root-knot nematode resistance accession. Lonren-1, Lonren-2, and Barbren were the agronomically improved Upland cotton germplasm releases by USDA-ARS, College Station, that have reniform nematode resistance derived from G. longicalyx (Bell et al., 2014) and G. barbadense (Bell et al., 2015), respectively. Such resistant sources add significant value for breeding programs to improve their germplasm.
Disease Tolerance
Bacterial blight, caused by Xanthomonas axonopodis pv. malvacearum, is a key disease in many parts of the world. Gossypium taxa harboring A-genome were shown to possess near-immunity for this pathogen and, therefore, these sources are valuable for introgression into cultivated tetraploids. Bacterial blight resistance gene, B6, found in G. arboreum was successfully transferred into G. barbadense (Zafar et al., 2009). Cotton rust resistance genes were successfully transferred from G. anomalum into G. hirsutum through interspecific hybridization, polyploidy induction and continuous screening for resistance in back-cross populations (Blank and Leathers, 1963).
Cotton leaf curl virus (CLCuV) is the wide spread and most damaging disease in northern India and Pakistan and is capable of causing yield loss up to 90%. Since none of the existing G. hirsutum varieties have recorded resistance to CLCuV, wild relatives have been explored. G. arboreum, was used as a donor of CLCuV resistance to transfer it to G. hirsutum using conventional hybridization and backcrossing. In another effort, resistance to CLCuV from G. stocksii was successfully introgressed into “MNH-786,” a G. hirsutum cultivar, through interspecific hybridization (Nazeer et al., 2014).
G. australe, a wild G-genome species possesses resistance to Fusarium wilt and Verticillium wilt diseases apart from other economically important traits such as resistance to aphids and mites. G. herbaceum, a cultivated diploid species (A-genome), has resistance to leaf curl virus along with other favorable traits including tolerance to sucking pests and drought tolerance (Liu Q. et al., 2015). Recently, a novel synthetic allotetraploid (A1A1G1G1) was developed from two diploid species and capable to lay the foundation for transferring favorable alleles into Upland cotton (Liu Q. et al., 2015). It was reported that throughout Australia no Fusarium spores were isolated from G. sturtianum stems suggesting that this species might possess Fusarium resistance (Wang et al., 2004), whereas G. bickii showed higher affinity with the pathogen (McFadden et al., 2004). G. thurberi is another wild species containing high Verticillium wilt tolerance which was supported by research using two-dimensional electrophoresis (2-DE) and tandem time-of-flight mass spectrometry (MALDI-TOF-MS) to identify 57 different proteins in G. thurberi infested with Verticillium dahliae (Zhao et al., 2012).
Tolerance to Abiotic Stresses
G. tomentosum, a wild cotton species, is host to many unique agronomic traits, including drought tolerance, salt tolerance, heat tolerance, nectarilessness, insect-pest resistance and lint color. G. darwinii, another wild allotetraploid species, with AD5 genome has many useful traits, including drought tolerance, fiber fineness, Fusarium wilt, and Verticillium wilt resistance (Liu F. et al., 2015). Soil salinity adversely affects crop growth along with cotton yield and fiber quality. Salt tolerance can improve plant emergency and ensure adequate uniformity stand in cotton. Through an interspecific cross between G. tomentosum and G. hirsutum, Oluoch et al. (2016), identified eight QTL regions contributing to salt tolerance and Zheng et al. (2016) identified multiple QTLs contributing toward drought tolerance. By applying RNA-Seq technology, Zhang F. et al. (2016) identified differentially expressed genes from G. davidsonii, a superior salt tolerant diploid cotton species. Similarly, Fan et al. (2015) detected 109 WRKY genes from transcriptome analyses in a salt-tolerant wild cotton species G. aridum to investigate the roles of these genes in cotton salt tolerance. From another salt-tolerant wild cotton species, G. klotzschianum, genes regulating salt tolerance mechanisms such as hormone biosynthesis and signal transduction, reactive oxygen species, and salt overly sensitive signal transduction related genes were elucidated though RNA-seq experiments from a time controlled NaCl (300 mM) experiments at 0, 3, 12, and 48 h intervals (Wei et al., 2017).
Soybean
Soybean (Glycine max [L.] Merr.) is an annual legume crop with major economic significance, contributing to more than half of the global oilseed production (Wilson, 2008). Domestication of cultivated soybean is thought to have happened in China ~5,000 years ago from wild soybean (Glycine soja Sieb. & Zucc.) (Boerma and Specht, 2004). Soybean introduction to the Korean peninsula and then Japan is estimated to have occured about 2,000 years ago, whereas, the crop first appeared in North America in 1765. Finally, soybean introduction to Central and South Americas happened during the first half of the last century (Wilson, 2008). Soybean has undergone natural phenotypic changes in plant development, flowering time, seed size and color, seed shattering, and dormancy during the process of domestication. Understanding the process of domestication and crop improvement at a genetic level is crucial for soybean breeding and crop improvement (Zhou et al., 2015). Genetic diversity of soybean has eroded due to domestication and breeding for yield to meet food demand. The genus Glycine consists of two subgenera, Soja and Glycine. Subgenus Soja includes G. max and its annual wild relative G. soja. G. soja has been distributed across broad geographical ranges and has a high genetic diversity due to the ability to adapt to a wide range of ecological conditions (Qiu et al., 2013; Sherman-Broyles et al., 2014). These geographical locations can be found for soybean in Figure 4. Because of the adaptability of G. soja it has become an important source of novel genes and alleles for soybean improvement necessary to meet the requirements of a rapidly growing world population and environmental changes (Table 4). The subgenus Glycine includes several perennial species such as G. canescens, G. clandestina, G.latifolia, G. microphylla, G. tabacina, G. tomentella, G. falcate, G. argyrea, G. carvata, and G. hirticaulis, which grow primarily in Australia (Brown et al., 1989; Sherman-Broyles et al., 2014; Zhou et al., 2015; Figure 4).
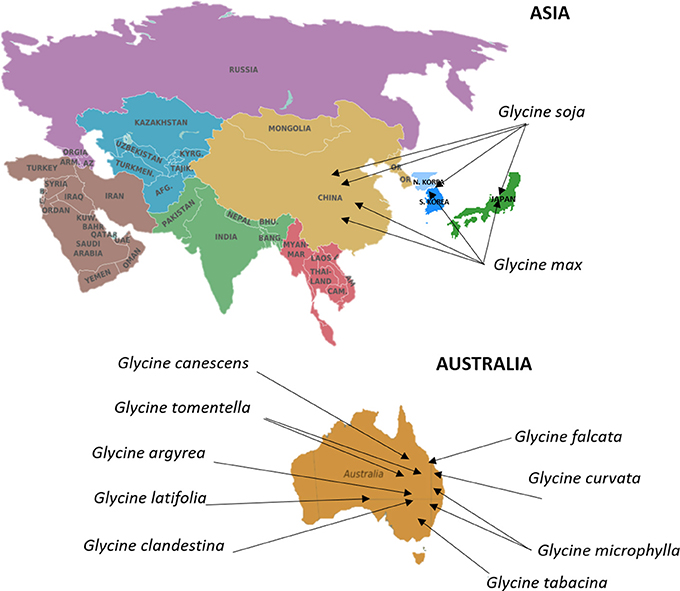
Figure 4. Centers of origin and primary geographical distribution of cultivated soybean and its wild relatives. Information on the distribution and centers of origin was gathered from Brown et al. (1989), Sherman-Broyles et al. (2014), Zhou et al. (2015). The Germplasm Resources Information Network in Beltsville, USA (https://npgsweb.ars-grin.gov/gringlobal/search.aspx) stores large global collection of soybean cultivars. Commonwealth Scientific and Industrial Research Organization in Canberra, Australia, encompasses a major collection of wild Glycine species.
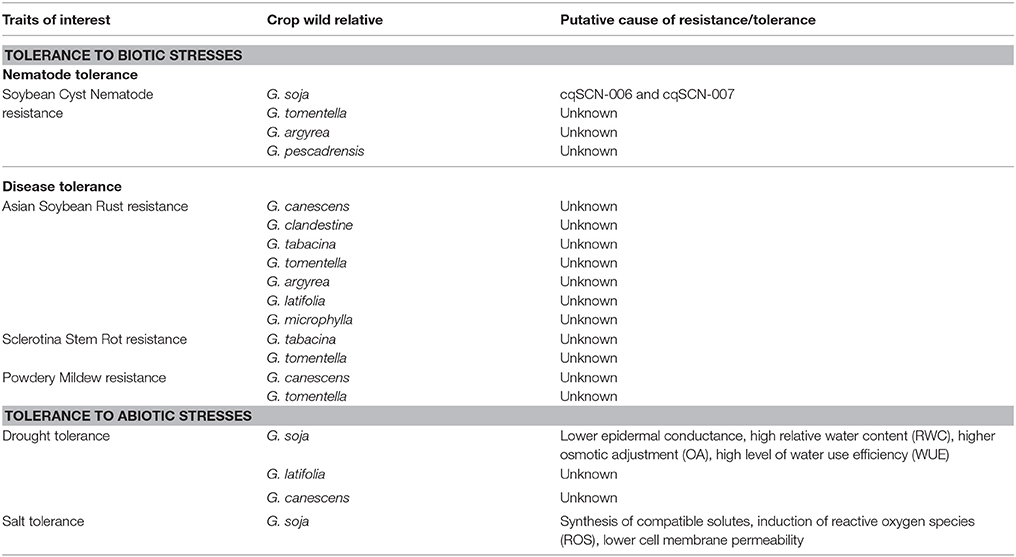
Table 4. Summary of biotic and abiotic stress tolerance traits of wild relatives of cultivated soybean (Glycine max).
Tolerance to Biotic Stresses
Nematode Tolerance
Soybean cyst nematode (SCN) (Heterodera glycines) is one of the major pathogens that affects soy. Concibido et al reviewed research efforts toward the identification of new sources of SCN-resistance both in cultivated soybean and G. soja as of 2004 (Concibido et al., 2004). Several new reports on the discovery of novel SCN-resistance QTL(s) in G. soja have been published based on traditional genetic linkage mapping or linkage disequilibrium based association mapping (Winter et al., 2007; Vuong et al., 2015; Zhang H. et al., 2016; Yu and Diers, 2017). Stacking of three SCN resistance alleles, rhg1-a, rhg1-b, and Rhg4, from the cultivated G. max with two SCN-resistance QTLs, cqSCN-006, and cqSCN-007, derived from G. soja, resulted in more durable resistance to the disease (Kim et al., 2011). Among 223 G. tomentella plant introductions tested, 86 accessions showed resistance to three H. glycines (HG) types, HG0, HG2, and HG1.2.3, 69 accessions showed resistance to two HG types and 22 showed resistance to one HG type. Of the other 12 perennial Glycine species evaluated, all accessions of G. argyrea and G. pescadrensis were resistant to all three HG types (Wen et al., 2017). So far no SCN resistance genes derived from soybean's CWR have been cloned.
Disease Tolerance
Numerous diseases caused by bacteria, fungi, nematodes, oomycetes, and viruses have played a significant role in soybean production resulting in tremendous yield losses among susceptible varieties. Compared to the first report published in 1975 (Sinclair and Backman, 1993), the fifth edition of the Compendium of Soybean Diseases and Pests shows the dramatic increase in the number of diseases affecting soybean (Hartman et al., 2015) which is believed to have resulted from the accelerated growth in production and expansion of the crop to new geographies (Hartman et al., 2011). Molecular mechanisms of soybean disease resistance have been extensively studied. The majority of soybean disease resistance (R) genes cloned to date have encoded proteins with two conserved motifs, including nucleotide binding sites (NBS) and leucine rich repeats (LRR). NBS-LRR genes confer resistance to pathogens through a gene-for-gene interaction mechanism (Innes et al., 2008).
Asian soybean rust (ASR) is a major fungal disease caused by Phakopsora pachyrhizi and has the potential to cause significant soybean yield loss. Perennial Glycine species have demonstrated resistance to this pathogen. Screening of 294 accessions from 17 Glycine species identified ASR resistance sources within G. canescens, G. clandestine, G. tabacina, G. argyrea, G. latifolia, G. microphylla, and G. tomentella (Hartman et al., 1992). Other research studies toward the identifying new sources of ASR resistance have been reviewed by Langenbach et al. (2016).
Sclerotinia stem rot, commonly known as white mold, is caused by the pathogen Sclerotinia sclerotiorum. This pathogen is one of most important diseases in North and South America and ranked second to SCN yield reductions within the USA. Several accessions of G. tabacina and G. tomentella showed partial resistance to S. sclerotiorum (Hartman et al., 2000). SCN resistance loci were also identified in G. lalifolia (Chang, 2015). Several accessions of G. canescens and G. tomentella showed resistance to powdery mildew disease and sudden death syndrome (Mignucci and Chamberlain, 1978; Hartman et al., 2000).
Tolerance to Abiotic Stresses
Drought Tolerance
Genotypic variation for epidermal conductance, relative water content and osmotic adjustment was assessed in 58 G. max genotypes, G. soja, and nine genotypes from six different perennial wild soybean. Results suggested that accessions of G. latifolia and G. canescens are more likely to be drought tolerant than G. max and G. soja due to their lowered epidermal conductance, relatively higher water content, and higher osmotic adjustment (James et al., 2008). PI407155, a genotype of G. soja, demonstrated a higher level of water use efficiency compared to “Essex” genotype of G. max (Chen et al., 2006).
Salinity Tolerance
CWRs of soybean have shown a notable level of tolerance to soil salinity. It has been reported that G. max and G. soja have reciprocal mechanisms of tolerance to soil salinity: G. soja accessions showed a higher leaf tolerance to Cl− toxicity than G. max but were more susceptible to Na+ accumulation (Luo et al., 2005). Tolerance to salinity in G. soja was believed to be due to the release of sodium ions which subsequently reduced the accumulation at toxic concentrations in plant organs. At the biochemical level, it is believed that induction of plant hormones, reactive oxygen species, cell membrane modifications, and synthesis of compatible solutes control the observed salt tolerance of G. soja (Lu et al., 2008). A single dominant gene derived from the wild soybean accession PI483463 was reported to control salt tolerance (Lee et al., 2009). The gene GsWRKY20, which encodes a WRKY-type transcription factor, was isolated from G. soja and was capable of increasing salt and drought tolerance of susceptible alfalfa after transformation. Relatively lower membrane permeability and lower malondialdehyde content were also observed in the transgenic alfalfa, as well as, a higher accumulation of free proline and soluble sugars compared with wild-type plants under high-salinity and water-deficit conditions (Tang et al., 2014). Over-expression of G. soja gene, GsJAZ2, in Arabidopsis resulted in enhanced plant tolerance to salt and alkali stress (Zhu et al., 2012).
Emerging Technologies for the Identification, Characterization, and Transfer of Traits from CWRs
Although wild relatives of crops are an important source of genetic diversity, their genepool has not been sufficiently explored (Li Y. H. et al., 2014). These wild relatives can increase the adaptive capacity of agricultural systems around the world by offering new allelic variations that are required to address disease pressures, farming practices, market demands, and climatic conditions. However, the process of introducing genetic diversity from wild species into cultivars requires a significant amount of time, resources, and human capacity (Dempewolf et al., 2017). Molecular markers and genotyping platforms can enhance our ability to rapidly dissect useful agronomic traits found in wild relatives and transfer these traits to elite germplasm through QTL introgression, marker assisted selection, and other genomic approaches. The recent developments of 50 K maize, 50 K soybean, 50 K rice, and 63 K cotton SNP arrays will further aid in the detection of useful variation from wild relatives through QTL mapping, genome wide association studies, and genomic selection studies. These studies could also be used for positive selection of traits of interest and negative selection of wild genomic segments causing agronomic penalties in interspecific hybridization and pre-breeding programs (Ganal et al., 2011; Song et al., 2013; Hulse-Kemp et al., 2015; Singh et al., 2015).
Wide availability of cost-effective next generation sequencing technologies have created opportunities to generate whole genome sequence for several crop species and their wild relatives. These genome resources enhance our ability to mine economically important traits from wild relatives through structural, functional and comparative genomics approaches. Availability of reference genome sequences for maize (Schnable et al., 2009; Lai et al., 2010), rice (Matsumoto et al., 2005; Kawahara et al., 2013), soybean (Haun et al., 2011), cotton diploid progenitors (Paterson et al., 2012; Wang et al., 2012; Li F. et al., 2014), cultivated Upland cotton (Li et al., 2015; Zhang et al., 2015), and Pima cotton (Yuan et al., 2015) has further created opportunities to generate draft assemblies for wild relatives enhancing our ability to mine economically important traits in these gene pools using robust bioinformatics tools (Sripathi et al., 2016). The generation of genomic resources, such as those listed above, from the wild relatives of major crops would allow for characterization of their genome contents and structural variations to understand the evolution, as well as, enable large scale comparisons of wild relatives to their cultivated counterparts to realize the benefits of a Pan-genome for a crop species. The Pan-genome would constitute a common sets of genes available at the species level along with unique gene sets available within a subset of individuals, including wild relatives. Such meta-analyses would enable genome wide comparison of gene pools across the genetic stocks and enhance our understanding of evolutionary history of domesticated crop species, genetic erosion during the domestication process, and shed light on avenues for recovering useful genetic variation lost during domestication from these wild relatives. Pan-genome studies on wild relatives of maize have elucidated the underlying genetic basis of two traits important for fitness and adaptation: timing of the juvenile-to-adult vegetative and vegetative-to-reproductive developmental transitions (Hirsch et al., 2014). In soybean, Pan-genome studies have helped identify soybean lineage-specific copy number variants allowing for positive selection for biotic resistance, seed composition, flowering and maturity time, organ size, and final biomass (Li Y.H. et al., 2014). Exploitation of novel genes and alleles in wild relatives through pan-genome studies would prove highly valuable in cotton to overcome the diminishing genetic gains in yield, fiber quality, biotic, and abiotic stress tolerance caused by narrow gene pools in cultivated cotton species.
Genomic technologies are providing a holistic perspective of gene structure, organization, and regulation in the genome, as well as, their role in biological pathways; and such information greatly accelerates crop breeding (Huang et al., 2016). The functional roles of genes and alleles are being explored through transcription and translational genomic studies to identify genes impacting the crop productivity positively. Such genes have been candidates for testing the cross-transferability of phenotype to other crops and organisms through transformation experiments. In cotton, one of the most highly accepted transgenic crops across the globe, transformation has proved highly challenging. However, recent genome editing technologies and novel plant breeding techniques are promising to allow for manipulation of gene sequences in crops. Sequence-specific nucleases such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), or clustered, regularly interspaced short palindromic repeat–associated endonucleases (CRISPR/Cas) are typically introduced through transformation and create double stranded breaks at the target locus due to their sequence specificity and can also add or delete nucleotides at that site. Transgenes and other off-target mutations can be further removed through self- or back-crossing (Huang et al., 2016). Knowledge of the novel genes associated with valuable traits from wild relatives can be sourced to make changes through targeted mutagenesis in the genes of cultivated crops to introduce a new phenotype or variation to improve crop performance. Genome editing of bacterial leaf blight disease susceptible S genes in rice such as the SWEET14 promoter and SWEET-type S genes using TALENs (Li et al., 2012) and CRISPR-Cas9 (Zhou et al., 2014) systems have resulted in resistant genotypes. Van de Wiel et al. have thoroughly reviewed deployment of these genome editing technologies on candidate genes controlling disease resistance, product quality, allergens, male fertility, yield, and other traits (van de Wiel et al., 2017). In order to take advantage of genome editing tools for crop improvement, apart from the knowledge of large effect gene variants from wild relatives, development of highly efficient transformation systems is necessary.
Conclusion
Diverse biotic and abiotic stresses, climate change, and rapidly increasing demand for food production are posing unprecedented challenges for global agriculture. Development of superior cultivars by harnessing diverse sources of variation and modern molecular and genomic tools have the potential to improve food production process. While plant domestication over the millennia has contributed to development of numerous cultivars in crop plants, those efforts primarily relied on yield improvement, edibility, and very few other traits. Such a strong selection has led to the creation of genetic bottlenecks and thereby resulted in the reduction of genetic variation. In contrast, CWRs in their natural environments have constantly been challenged and maintained higher levels of genetic diversity. Leveraging the untapped genetic diversity available in CWRs for improvement of crops is an attractive option for improving crops. The use of modern technologies for identifying and dissecting the molecular, genetic, and genomic bases of traits in CWRs can accelerate this process. The review presented here has discussed the wealth of traits in CWRs of four important crops and the efforts that have gone in this area of research toward harnessing this valuable resource for crop improvement.
Author Contributions
JM, SK, SG, and RB have substantially contributed to the conception, design, and writing of this review paper. KP has contributed to the critical review of entire article and improved the quality and uniformity of the language. IA has contributed to the expansion and critical review of cotton section.
Conflict of Interest Statement
JM, SG, RB, KP, and SK were employed by company Dow AgroSciences.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Wes Marchione, Rajat Aggarwal, Phil Poirier, and Kathryn Clayton of Corteva Agriscience™, Agriculture Division of DowDuPont™, for the review and final approval of the content of the manuscript for external release.
References
Abdurakhmonov, I. Y. (ed.). (2016). “Introductory chapter: introduction to cotton research highlights,” in Cotton Research (London: IntechOpen), 3–15.
Abiko, T., Kotula, L., Shiono, K., Malik, A. I., Colmer, T. D., and Nakazono, M. (2012). Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea nicaraguensis contribute to its waterlogging tolerance as compared with maize (Zea mays ssp. mays). Plant Cell Environ. 35, 1618–1630. doi: 10.1111/j.1365-3040.2012.02513.x
Aggarwal, A., Ezaki, B., Munjal, A., and Tripathi, B. N. (2015). “Physiology and biochemistry of aluminum toxicity and tolerance in crops,” in Stress Responses in Plants, eds B. N. Tripathi and M. Müller (Basel: Springer International Publishing Switzerland), 35–57.
Ali, M. L., Sanchez, P. L., Yu, S.- B., Lorieux, M., and Eizenga, G. C. (2010). Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3, 218–234. doi: 10.1007/s12284-010-9058-3
Amusan, I. O., Rich, P. J., Menkir, A., Housley, T., and Ejeta, G. (2008). Resistance to Striga hermonthica in a maize inbred line derived from Zea diploperennis. New Phytol. 178, 157–166. doi: 10.1111/j.1469-8137.2007.02355.x
Andaya, V. C., and Tai, T. H. (2007). Fine mapping of the qCTS4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol. Breed. 20, 349–358. doi: 10.1007/s11032-007-9096-8
Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Miah, G., Sahebi, M., Azizi, P., et al. (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 6:886. doi: 10.3389/fpls.2015.00886
Awad, A. A., Sato, D., Kusumoto, D., Kamioka, H., Takeuchi, Y., and Yoneyama, K. (2006). Characterization of strigolactones, germination stimulants for the root parasitic plants Striga and Orobanche, produced by maize, millet and sorghum. Plant Growth Regul. 48, 221–227. doi: 10.1007/s10725-006-0009-3
Bell, A. A., Forest Robinson, A., Quintana, J., Dighe, N. D., Menz, M. A., Stelly, D. M., et al. (2014). Registration of LONREN-1 and LONREN-2 germplasm lines of upland cotton resistant to reniform nematode. J. Plant Registr. 8, 187–190. doi: 10.3198/jpr2013.11.0069crg
Bell, A. A., Robinson, A., Quintana, J., Duke, S. E., Starr, J. L., Stelly, D. M., et al. (2015). Registration of BARBREN-713 germplasm line of upland cotton resistant to reniform and root-knot nematodes. J. Plant Registr. 9, 89–93. doi: 10.3198/jpr2014.04.0021crg
Bellota, E., Medina, R. F., and Bernal, J. S. (2013). Physical leaf defenses–altered by Zea life-history evolution, domestication, and breeding–mediate oviposition preference of a specialist leafhopper. Entomol. Exp. Appl. 149, 185–195. doi: 10.1111/eea.12122
Bergquist, R. (1981). Transfer from Tripsacum dactyloides to corn of a major gene locus conditioning resistance to Puccinia sorghi. Phytopathology 71, 518–520. doi: 10.1094/Phyto-71-518
Bernal, J. S., Melancon, J. E., and Zhu-Salzman, K. (2015). Clear advantages for fall armyworm larvae from feeding on maize relative to its ancestor Balsas teosinte may not be reflected in their mother's host choice. Entomol. Exp. Appl. 155, 206–217. doi: 10.1111/eea.12299
Bi, I. V., Baudoin, J. P., and Mergeai, G. (1998). Cytogenetics of the ‘glandless-seed and glanded-plant’ trait from Gossypium sturtianum Willis introgressed into upland cotton (Gossypium hirsutum L.). Plant Breed. 117, 235–241.
Blank, L. M., and Leathers, C. R. (1963). Environmental and other factors influencing development of southwestern cotton rust (Puccinia stakmanii). Phytopathology 53, 921–928.
Blasingame, D. (2005). “2005 cotton disease loss estimate,” in Proceedings Beltwide Cotton Conferences (Cordova) 155–157.
Boerma, H. R., and Specht, J. E. (2004). Soybeans: Improvement, Production and Uses. Madison: American Society of Agronomy.
Brar, D. S., and Khush, G. (1997). Alien introgression in rice. Plant Mol. Biol. 35, 35–47. doi: 10.1023/A:1005825519998
Brown, A. H. D., Frankel, O. H., Marshall, D. R., and Williams, J. T. (eds.). (1989). The Use of Plant Genetic Resources. Cambridge University Press.
Campbell, B., Saha, S., Percy, R., Frelichowski, J., Jenkins, J. N., Park, W., et al. (2010). Status of the global cotton germplasm resources. Crop Sci. 50, 1161–1179. doi: 10.2135/cropsci2009.09.0551
Cao, Y., Lou, Y., Han, Y., Shi, J., Wang, Y., Wang, W., et al. (2011). Al toxicity leads to enhanced cell division and changed photosynthesis in Oryza rufipogon L. Mol. Biol. Rep. 38, 4839–4846. doi: 10.1007/s11033-010-0618-9
Carroll, M. J., Schmelz, E. A., Meagher, R. L., and Teal, P. E. (2006). Attraction of Spodoptera frugiperda larvae to volatiles from herbivore-damaged maize seedlings. J. Chem. Ecol. 32, 1911–1924. doi: 10.1007/s10886-006-9117-9
Chang, S. (2015). Identification of Loci for Resistance to Sclerotinia Stem Rot (white mold) in Accessions of Perennial Relative of Soybean (Glycine latifolia). Dissertation, University of Illinois at Urbana-Champaign, Urbana-Champaign, IL.
Chavan, S., and Smith, S. M. (2014). A rapid and efficient method for assessing pathogenicity of Ustilago maydis on maize and teosinte lines. J. Vis. Exp. 83:e50712. doi: 10.3791/50712
Chen, Y., Chen, P., and de los Reyes, B. G. (2006). Differential responses of the cultivated and wild species of soybean to dehydration stress. Crop Sci. 46, 2041–2046. doi: 10.2135/cropsci2005.12.0466
Clark, R., Alberts, E., Zobel, R., Sinclair, T., Miller, M., Kemper, W., et al. (1998). “Eastern gamagrass (Tripsacumdactyloides) root penetration into and chemical properties of claypan soils,” in Root Demographics and Their Efficiencies in Sustainable Agriculture, Grasslands and Forest Ecosystems, ed E. J. Box Jr. (Dordrecht: Kluwer Academic Publishers), 191–211.
Concibido, V. C., Diers, B. W., and Arelli, P. R. (2004). A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 44, 1121–1131. doi: 10.2135/cropsci2004.1121
Coyne, P. I., and Bradford, J. A. (1985). Comparison of leaf gas exchange and water-use efficiency in two eastern gamagrass accessions. Crop Sci. 25, 65–75. doi: 10.2135/cropsci1985.0011183X002500010018x
Cruz, R. P., Sperotto, R. A., Cargnelutti, D., Adamski, J. M., FreitasTerra, T., and Fett, J. P. (2013). Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur. 2, 96–119. doi: 10.1002/fes3.25
Dávila-Flores, A. M., DeWitt, T. J., and Bernal, J. S. (2013). Facilitated by nature and agriculture: performance of a specialist herbivore improves with host-plant life history evolution, domestication, and breeding. Oecologia 173, 1425–1437. doi: 10.1007/s00442-013-2728-2
Dempewolf, H., Baute, G., Anderson, J., Kilian, B., Smith, C., and Guarino, L. (2017). Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. doi: 10.2135/cropsci2016.10.0885
De Wet, J., Timothy, D., Hilu, K., and Fletcher, G. (1981). Systematics of South American Tripsacum (Gramineae). Am. J. Bot. 68, 269–276. doi: 10.1002/j.1537-2197.1981.tb12387.x
Doebley, J. F. (1983). “The taxonomy and evolution of Tripsacum and teosinte, the closest relatives of maize,” in International Maize Virus Disease Colloquium and Workshop, 2-6 Aug 1982 (Wooster, OH: Ohio Agricultural Research and Development Center).
Ellur, R. K., Khanna, A., Bhowmick, P. K., Vinod, K., Nagarajan, M., Mondal, K. K., et al. (2016). Marker-aided incorporation of Xa38, a novel bacterial blight resistance gene, in PB1121 and comparison of its resistance spectrum with xa13+ Xa21. Sci. Rep. 6:29188. doi: 10.1038/srep29188
Eubanks, M. (2006). A genetic bridge to utilize Tripsacum germplasm in maize improvement. Maydica 51, 315–327.
Fan, X., Guo, Q., Xu, P., Gong, Y., Shu, H., Yang, Y., et al. (2015). Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS ONE 10:e0126148. doi: 10.1371/journal.pone.0126148
Farias-Rivera, L. A., Hernandez-Mendoza, J. L., Molina-Ochoa, J., and Pescador-Rubio, A. (2003). Effect of leaf extracts of teosinte, Zea diploperennis L., and a mexican maize variety, criollo'uruapeño', on the growth and survival of the fall armyworm (Lepidoptera: Noctuidae). Florida Entomol. 86, 239–243. doi: 10.1653/0015-4040(2003)086[0239:EOLEOT]2.0.CO;2
Fryxell, P. A. (1992). A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea 2, 108–165.
Fujita, D., Kohli, A., and Horgan, F. G. (2013). Rice resistance to planthoppers and leafhoppers. Crit. Rev. Plant Sci. 32, 162–191. doi: 10.1080/07352689.2012.735986
Fukunaga, K., Hill, J., Vigouroux, Y., Matsuoka, Y., Sanchez, J., Liu, K., et al. (2005). Genetic diversity and population structure of teosinte. Genetics 169, 2241–2254. doi: 10.1534/genetics.104.031393
Galinat, W. (1961). Tripsacum floridanum crosses readily with corn. Maize Genet. Coop. Newsl 35, 38–39.
Ganal, M. W., Durstewitz, G., Polley, A., Bérard, A., Buckler, E. S., Charcosset, A., et al. (2011). A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6:e28334. doi: 10.1371/journal.pone.0028334
Garg, R., Verma, M., Agrawal, S., Shankar, R., Majee, M., and Jain, M. (2014). Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 21, 69–84. doi: 10.1093/dnares/dst042
Gethi, J. G., and Smith, M. E. (2004). Genetic responses of single crosses of maize to (Del.) Benth. and (L.) Kuntze. Crop Sci. 44, 2068–2077. doi: 10.2135/cropsci2004.2068
Gilker, R. E., Weil, R. R., Krizek, D. T., and Momen, B. (2002). Eastern gamagrass root penetration in adverse subsoil conditions. Soil Sci. Soc. Am. J. 66, 931–938. doi: 10.2136/sssaj2002.9310
Gitz, D. C., Baker, J. T., Stout, J. E., Brauer, D. K., Lascano, R. J., and Velten, J. P. (2013). Suitability of eastern gamagrass for in situ precipitation catchment forage production in playas. Agron. J. 105, 907–914. doi: 10.2134/agronj2012.0358
Gurney, A., Grimanelli, D., Kanampiu, F., Hoisington, D., Scholes, J., and Press, M. (2003). Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytol. 160, 557–568. doi: 10.1046/j.1469-8137.2003.00904.x
Hajjar, R., and Hodgkin, T. (2007). The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica 156, 1–13. doi: 10.1007/s10681-007-9363-0
Hartman, G., Gardner, M., Hymowitz, T., and Naidoo, G. (2000). Evaluation of perennial species for resistance to soybean fungal pathogens that cause sclerotinia stem rot and sudden death syndrome. Crop Sci. 40, 545–549. doi: 10.2135/cropsci2000.402545x
Hartman, G. L., Rupe, J. C., Sikora, E., Domier, L. L., Davis, J. A., and Steffey, K. L. (2015). Compendium of Soybean Diseases and Pests. St. Paul, MN: American Phytopathological Society.
Hartman, G. L., Wang, T. C., and Hymowitz, T. (1992). Sources of resistance to soybean rust in perennial Glycine species. Plant Dis. 76, 396–399. doi: 10.1094/PD-76-0396
Hartman, G. L., West, E. D., and Herman, T. K. (2011). Crops that feed the World 2. soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 3, 5–17. doi: 10.1007/s12571-010-0108-x
Hassan, S., Wilson, L., and Blood, P. (1990). Oviposition by Heliothis armigera and H. punctigera (Lepidoptera: Noctuidae) on okra leaf and smooth-leaf cotton. Environ. Entomol. 19, 710–716. doi: 10.1093/ee/19.3.710
Haun, W. J., Hyten, D. L., Xu, W. W., Gerhardt, D. J., Albert, T. J., Richmond, T., et al. (2011). The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 155, 645–655. doi: 10.1104/pp.110.166736
Hirsch, C. N., Foerster, J. M., Johnson, J. M., Sekhon, R. S., Muttoni, G., Vaillancourt, B., et al. (2014). Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26, 121–135. doi: 10.1105/tpc.113.119982
Hoballah, M. E., Degen, T., Bergvinson, D., Savidan, A., Tamò, C., and Turlings, T. C. (2004). Occurrence and direct control potential of parasitoids and predators of the fall armyworm (Lepidoptera: Noctuidae) on maize in the subtropical lowlands of Mexico. Agric. For. Entomol. 6, 83–88. doi: 10.1111/j.1461-9555.2004.00207.x
Hooker, A. (1981). Resistance to Helminthosporium turcicum from Tripsacum floridanum incorporated into corn. Maize Genet. Coop. Newsl. 55, 87–88.
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Hu, J., Xiao, C., and He, Y. (2016). Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice 9, 1–12. doi: 10.1186/s12284-016-0099-0
Huang, S., Weigel, D., Beachy, R. N., and Li, J. (2016). A proposed regulatory framework for genome-edited crops. Nat. Genet. 48, 109–111. doi: 10.1038/ng.3484
Hufford, M. B., Xu, X., Van Heerwaarden, J., Pyhäjärvi, T., Chia, J.-M., Cartwright, R. A., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44, 808–811. doi: 10.1038/ng.2309
Hulse-Kemp, A. M., Lemm, J., Plieske, J., Ashrafi, H., Buyyarapu, R., Fang, D. D., et al. (2015). Development of a 63K SNP array for cotton and high-density mapping of intra-and inter-specific populations of Gossypium spp. G3 5, 1187–1209. doi: 10.1534/g3.115.018416
Iltis, H. H., and Benz, B. F. (2000). Zea nicaraguensis (Poaceae), a new teosinte from Pacific coastal Nicaragua. Novon 10, 382–390. doi: 10.2307/3392992
Innes, R. W., Ameline-Torregrosa, C., Ashfield, T., Cannon, E., Cannon, S. B., Chacko, B., et al. (2008). Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148, 1740–1759. doi: 10.1104/pp.108.127902
James, R. A., von Caemmerer, S., Condon, A. T., Zwart, A. B., and Munns, R. (2008). Genetic variation in tolerance to the osmotic stress componentof salinity stress in durum wheat. Funct. Plant Biol. 35, 111–123. doi: 10.1071/FP07234
Jena, K. K. (2010). The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breed. Sci. 60, 518–523. doi: 10.1270/jsbbs.60.518
Kaplan, I., Dively, G. P., and Denno, R. F. (2009). The costs of anti-herbivore defense traits in agricultural crop plants: a case study involving leafhoppers and trichomes. Ecol. Appl. 19, 864–872. doi: 10.1890/07-1566.1
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Khush, G., Angeles, E., Virak, P., and Brar, D. (2004). Breeding rice for resistance to tungro virus at IRRI. SABRAO J. Breed. Genet. 36, 101–106. Available online at: http://eprints.icrisat.ac.in/id/eprint/2899
Khush, G., Ling, K., Aquino, R., and Aguiero, V. (1977). Breeding for resistance to grassy stunt in rice. Plant Breed. Papers Int. Congr. Soc. Adv. Breed. Res. Asia Ocean. 1, 3–9.
Kim, M., Hyten, D. L., Niblack, T. L., and Diers, B. W. (2011). Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci. 51, 934–943. doi: 10.2135/cropsci2010.08.0459
Kling, J., Fajemisin, J., Badu-Apraku, B., Diallo, A., Menkir, A., and Melake-Berhan, A. (2000). “Striga resistance breeding in maize. Breeding for Striga resistance in cereals. Proceedings of a Workshop; Ibadan, Nigeria; 18-20 Aug 1999,” in Breeding for Striga Resistance in Cereals. Proceedings of a Workshop; Ibadan, Nigeria; 18-20 Aug 1999, eds A. Haussmann, B. I. G. Hess, D. E. Koyama, M. L. Grivet, L. Rattunde, H. W. Geiger, and H. H. A. Weikersheim (Weikersheim: Margraf Verlag), 376.
Köllner, T. G., Schnee, C., Gershenzon, J., and Degenhardt, J. (2004). The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distributions. Phytochemistry 65, 1895–1902. doi: 10.1016/j.phytochem.2004.05.021
Konan, O. N., D'Hont, A., Baudoin, J. P., and Mergeai, G. (2007). Cytogenetics of a new trispecies hybrid in cotton: [(Gossypium hirsutum L. × G. thurberi Tod.)2 × G. longicalyx Hutch. & Lee]. Plant Breed. 126, 176–181. doi: 10.1111/j.1439-0523.2007.01325.x
Koseki, M., Kitazawa, N., Yonebayashi, S., Maehara, Y., Wang, Z.-X., and Minobe, Y. (2010). Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol. Genet. Genomics 284, 45–54. doi: 10.1007/s00438-010-0548-1
Lai, J., Li, R., Xu, X., Jin, W., Xu, M., Zhao, H., et al. (2010). Genome-wide patterns of genetic variation among elite maize inbred lines. Nat. Genet. 42, 1027–1030. doi: 10.1038/ng.684
Lane, J., Child, D., Moore, T., Arnold, G., and Bailey, J. (1997). Phenotypic characterisation of resistance in Zea diploperennis to Striga hermonthica. Maydica 42, 45–51.
Lange, E. S., Balmer, D., Mauch-Mani, B., and Turlings, T. C. (2014). Insect and pathogen attack and resistance in maize and its wild ancestors, the teosintes. New Phytol. 204, 329–341. doi: 10.1111/nph.13005
Langenbach, C., Campe, R., Beyer, S. F., Mueller, A. N., and Conrath, U. (2016). Fighting asian soybean rust. Front. Plant Sci. 7:797. doi: 10.3389/fpls.2016.00797
Lee, J. D., Shannon, J. G., Vuong, T. D., and Nguyen, H. T. (2009). Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Hered. 100, 798–801. doi: 10.1093/jhered/esp027
Lennon, J. R., Krakowsky, M., Goodman, M., Flint-Garcia, S., and Balint-Kurti, P. J. (2016). Identification of alleles conferring resistance to gray leaf spot in maize derived from its wild progenitor species teosinte. Crop Sci. 56, 209–218. doi: 10.2135/cropsci2014.07.0468
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi: 10.1038/nbt.2979
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Ling, Y., and Weilin, Z. (2016). Genetic and biochemical mechanisms of rice resistance to planthopper. Plant Cell Rep. 35, 1559–1572. doi: 10.1007/s00299-016-1962-6
Liu, F., Zhou, Z. L., Wang, C. Y., Wang, Y. H., Cai, X. Y., Wang, X.-X., et al. (2015). Collinearity analysis of allotetraploid Gossypium tomentosum and Gossypium darwinii. Genet. Mol. Res. 15, 1–12. doi: 10.1101/031104
Liu, Q., Chen, Y., Chen, Y., Wang, Y., Chen, J., Zhang, T., et al. (2015). A New Synthetic Allotetraploid (A1A1G2G2) between Gossypium herbaceum and G. australe: bridging for simultaneously transferring favorable genes from these two diploid species into upland cotton. PLoS ONE 10:e0123209. doi: 10.1371/journal.pone.0123209
Lou, Q., Chen, L., Sun, Z., Xing, Y., Li, J., Xu, X., et al. (2007). A major QTL associated with cold tolerance at seedling stage in rice (Oryza sativa L.). Euphytica 158, 87–94. doi: 10.1007/s10681-007-9431-5
Lu, W., Bennett, B. D., and Rabinowitz, J. D. (2008). Analytical strategies for LC–MS-based targeted metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871, 236–242. doi: 10.1016/j.jchromb.2008.04.031
Luo, Q., Yu, B., and Liu, Y. (2005). Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J. Plant Physiol. 162, 1003–1012. doi: 10.1016/j.jplph.2004.11.008
Maag, D., Erb, M., Bernal, J. S., Wolfender, J. L., Turlings, T. C., and Glauser, G. (2015). Maize domestication and anti-herbivore defences: leaf-specific dynamics during early ontogeny of maize and its wild ancestors. PLoS ONE 10:e0135722. doi: 10.1371/journal.pone.0135722
Maazou, A.-R. S., Qiu, J., Mu, J., and Liu, Z. (2017). Utilization of wild relatives for maize (Zea mays L.) improvement. Afr. J. Plant Sci. 11, 105–113. doi: 10.5897/AJPS2017.1521
Mano, Y., and Omori, F. (2008). Verification of QTL controlling root aerenchyma formation in a maize* teosinte “Zea nicaraguensis” advanced backcross population. Breed. Sci. 58, 217–223. doi: 10.1270/jsbbs.58.217
Mano, Y., and Omori, F. (2013). Flooding tolerance in interspecific introgression lines containing chromosome segments from teosinte (Zea nicaraguensis) in maize (Zea mays subsp. mays). Ann. Bot. 112, 1125–1139. doi: 10.1093/aob/mct160
Mano, Y., Omori, F., Loaisiga, C. H., and Bird, R. M. (2009). QTL mapping of above-ground adventitious roots during flooding in maize x teosinte “Zea nicaraguensis” backcross population. Plant Root 3, 3–9. doi: 10.3117/plantroot.3.3
Mano, Y., Omori, F., Takamizo, T., Kindiger, B., Bird, R. M., Loaisiga, C., et al. (2007). QTL mapping of root aerenchyma formation in seedlings of a maize × rare teosinte “Zea nicaraguensis” cross. Plant Soil 295, 103–113. doi: 10.1007/s11104-007-9266-9
Matsumoto, T., Wu, J., Kanamori, H., and Katayose, Y. (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Matsuoka, Y., Vigouroux, Y., Goodman, M. M., Sanchez, J., Buckler, E., and Doebley, J. (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084. doi: 10.1073/pnas.052125199
McCouch, S. R., Sweeney, M., Li, J., Jiang, H., Thomson, M., Septiningsih, E., et al. (2007). Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica 154, 317–339. doi: 10.1007/s10681-006-9210-8
McFadden, H., Beasley, D., and Brubaker, C. L. (2004). Assessment of Gossypium sturtianum and G. australe as potential sources of Fusarium wilt resistance to cotton. Euphytica 138, 61–72. doi: 10.1023/B:EUPH.0000047076.38747.81
McLoud, L. A., Hague, S., Knutson, A., Smith, C. W., and Brewer, M. (2016). Cotton square morphology offers new insights into host plant resistance to cotton fleahopper (Hemiptera: Miridae) in Upland cotton. J. Econ. Entomol. 109, 392–398. doi: 10.1093/jee/tov275
McMullen, M. D., Frey, M., and Degenhardt, J. (2009). “Genetics and biochemistry of insect resistance in maize,” in Handbook of Maize: Its Biology, eds J. L. Bennetzen and S. C. Hake (New York, NY: Springer), 271–289.
Menguer, P. K., Sperotto, R. A., and Ricachenevsky, F. K. (2017). A walk on the wild side: oryza species as source for rice abiotic stress tolerance. Genet. Mol. Biol. 40(Suppl. 1), 238–252. doi: 10.1590/1678-4685-gmb-2016-0093
Meyer, J. R., and Meyer, V. G. (1961). Origin and inheritance of nectariless cotton. Crop Sci. 1, 167–169. doi: 10.2135/cropsci1961.0011183X000100030004x
Mignucci, J. S., and Chamberlain, D. (1978). Interactions of Microsphaera diffusa with soybeans and other legumes. Phytopathology 68, 169–173. doi: 10.1094/Phyto-68-169
Moya-Raygoza, G. (2016). Early development of leaf trichomes is associated with decreased damage in teosinte, compared with maize, by Spodoptera frugiperda (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 109, 737–743. doi: 10.1093/aesa/saw049
Mujeeb-Kazi, A., and Jewell, D. (1985). “CIMMYT's wide cross program for wheat and maize improvement,” in Biotechnology in International Agricultural Research, Proceedings of the Intercenter Seminar on International Agricultural Research Centers and Biotechnology 23–27 April 1984 (Manila: International Rice Research Institute), 219–226.
Mutyambai, D. M., Bruce, T. J., Midega, C. A., Woodcock, C. M., Caulfield, J. C., Van Den Berg, J., et al. (2015). Responses of parasitoids to volatiles induced by Chilo partellus oviposition on teosinte, a wild ancestor of maize. J. Chem. Ecol. 41, 323–329. doi: 10.1007/s10886-015-0570-1
Nault, L., and Findley, W. (1982). Zea diploperennis: a primitive relative offers new traits to improve corn. Desert Plants 3, 203–205.
Nazeer, W., Ahmad, S., Mahmood, K., Tipu, A., Mahmood, A., and Zhou, B. (2014). Introgression of genes for cotton leaf curl virus resistance and increased fiber strength from Gossypium stocksii into upland cotton (Gossypium hirsutum). Genet. Mol. Res. 13, 1133–1143. doi: 10.4238/2014.February.21.2
Ndjiondjop, M. N., Manneh, B., Cissoko, M., Drame, N. K., Kakai, R. G., Bocco, R., et al. (2010). Drought resistance in an interspecific backcross population of rice (Oryza spp.) derived from the cross WAB56-104 (O. sativa) × CG14 (O. glaberrima). Plant Sci. 179, 364–373. doi: 10.1016/j.plantsci.2010.06.006
Nguyen, B. D., Brar, D. S., Bui, B. C., Nguyen, T. V., Pham, L. N., and Nguyen, H. T. (2003). Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor. Appl. Genet. 106, 583–593. doi: 10.1007/s00122-002-1072-4
Niazi, I. A. K., Rafique, A., Rauf, S., da Silva, J. A. T., and Afzal, M. (2014). Simultaneous selection for stem borer resistance and forage related traits in maize (Zea mays ssp. mays L.) × teosinte (Zea mays ssp. mexicana L.) derived populations. Crop Protect. 57, 27–34. doi: 10.1016/j.cropro.2013.10.026
Niemeyer, H. M. (2009). Hydroxamic acids derived from 2-hydroxy-2 H-1, 4-benzoxazin-3 (4 H)-one: key defense chemicals of cereals. J. Agric. Food Chem. 57, 1677–1696. doi: 10.1021/jf8034034
Normile, D. (2008). Reinventing rice to feed the world. Science 321, 330–333. doi: 10.1126/science.321.5887.330
Oluoch, G., Zheng, J., Wang, X., Khan, M. K. R., Zhou, Z., Cai, X., et al. (2016). QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum. Euphytica 209, 223–235. doi: 10.1007/s10681-016-1674-6
Omori, F., and Mano, Y. (2007). QTL mapping of root angle in F2 populations from maize ‘B73’ × teosinte ‘Zea luxurians’. Plant Root 1, 57–65. doi: 10.3117/plantroot.1.57
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Peng, T., Sun, X., and Mumm, R. H. (2014). Optimized breeding strategies for multiple trait integration: I. Minimizing linkage drag in single event introgression. Mol. Breed. 33, 89–104. doi: 10.1007/s11032-013-9936-7
Pesqueira, J., García, M. D., Staltari, S., and Molina, M. d. C. (2006). NaCl effects in Zea mays L. × Tripsacum dactyloides (L.) L. hybrid calli and plants. Electr. J. Biotechnol. 9, 286–290. doi: 10.2225/vol9-issue3-fulltext-29
Pesqueira, J., Garcia, M., and Molina, M. C. (2003). NaCl tolerance in maize (Zea mays ssp. mays) x Tripsacum dactyloides L. hybrid calli in regenerated plants. Spanish J. Agric. Res. 1, 59–63. doi: 10.5424/sjar/2003012-21
Prescott-Allen, R., and Prescott-Allen, C. (2013). Genes From the Wild: Using Wild Genetic Resources for Food and Raw Materials. Sterling, TX: Routledge.
Prischmann, D., Dashiell, K., Schneider, D., and Eubanks, M. (2009). Evaluating Tripsacum-introgressed maize germplasm after infestation with western corn rootworms (Coleoptera: Chrysomelidae). J. Appl. Entomol. 133, 10–20. doi: 10.1111/j.1439-0418.2008.01311.x
Qiu, L.-J., Xing, L.-L., Guo, Y., Wang, J., Jackson, S. A., and Chang, R.-Z. (2013). A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol. Biol. 83, 41–50. doi: 10.1007/s11103-013-0076-6
Ram, T., Deen, R., Gautam, S., Ramesh, K., Rao, Y., and Brar, D. (2010). Identification of new genes for brown planthopper resistance in rice introgressed from O. glaberrima and O. minuta. Rice Genet. Newsl. 25, 67–69. doi: 10.1186/s12284-017-0178-x
Rasmann, S., Köllner, T. G., Degenhardt, J., Hiltpold, I., Toepfer, S., Kuhlmann, U., et al. (2005). Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434, 732–737. doi: 10.1038/nature03451
Ray, J., Kindiger, B., Dewald, C., and Sinclair, T. (1998). Preliminary survey of root aerenchyma in Tripsacum. Maydica 43, 49–54.
Ray, J., Sinclair, T., and Kindiger, B. (1999). Introgressing root aerenchyma into maize [Zea mays L.]. Maydica 44, 113–117.
Rich, P. J., and Ejeta, G. (2008). Towards effective resistance to Striga in African maize. Plant Signal. Behav. 3, 618–621. doi: 10.4161/psb.3.9.5750
Robinson, A., Bell, A., Dinghe, N., and Stelly, D. (2004a). “Status report on introgression of reniform nematode resistance from Gossypium longicalyx,” in National Cotton Council Beltwide Cotton Conference (San Antonio, TX).
Robinson, A., Bridges, A., and Percival, A. (2004b). New sources of resistance to the reniform (Rotylenchulus reniformis) and root-knot (Meloidogyne incognita) nematode in upland (Gossypium hirsutum L.) and sea island (G. barbadense L.) cotton. J. Cotton Sci. 8, 191–197. Available online at: http://www.cotton.org/journal/2004-08/3/upload/jcs08-191.pdf
Robinson, A., and Percival, A. (1997). Resistance to Meloidogyne incognita race 3 and Rotylenchulus reniformis in wild accessions of Gossypium hirsutum and G. barbadense from Mexico. J. Nematol. 29:746.
Rosenthal, J. P., and Dirzo, R. (1997). Effects of life history, domestication and agronomic selection on plant defence against insects: evidence from maizes and wild relatives. Evol. Ecol. 11, 337–355. doi: 10.1023/A:1018420504439
Sanchez, P. L., Wing, R. A., and Brar, D. S. (2013). “The wild relative of rice: genomes and genomics,” in Genetics and Genomics of Rice, eds Q. Zhang and R. Wing (Berlin: Springer Science+Business Media, LLC), 9–25.
Sarao, P. S., Sahi, G. K., Neelam, K., Mangat, G. S., Patra, B. C., and Singh, K. (2016). Donors for resistance to brown planthopper Nilaparvata lugens (stål) from wild rice species. Rice Sci. 23, 219–224. doi: 10.1016/j.rsci.2016.06.005
Saunders, J. (1965). Genetics of hairiness transferred from Gossypium raimondii to G. hirsutum. Euphytica 14, 276–282. doi: 10.1007/BF00149511
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. doi: 10.1126/science.1178534
Shakiba, E., and Eizenga, G. C. (2014). “Unraveling the secrets of rice wild species,” in Rice-Germplasm, Genetics and Improvement, ed J. Bao (London: IntechOpen), 1–58.
Sharma, T., Rai, A., Gupta, S., Vijayan, J., Devanna, B., and Ray, S. (2012). Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52. doi: 10.1007/s40003-011-0003-5
Shavrukov, Y., and Sokolov, V. (2015). Maize-Gamagrass interspecific hybrid, Zea mays x Tripsacum dactyloides, shows better salinity tolerance and higher Na+ exclusion than maize and sorghum. Int. J. Latest Res. Sci. Technol. 4, 128–133. Available online at: https://www.mnkpublication.com/journal/ijlrst/Article.php?paper_id=10475
Sherman-Broyles, S., Bombarely, A., Powell, A. F., Doyle, J. L., Egan, A. N., Coate, J. E., et al. (2014). The wild side of a major crop: soybean's perennial cousins from Down Under. Am. J. Bot. 101, 1651–1665. doi: 10.3732/ajb.1400121
Shinde, B., Gurve, S., Gonde, A., and Hole, U. (2014). Studies on resistance of cotton genotypes against jassids (Amrasca biguttula biguttula Ishida). BIOINFOLET-A Q. J. Life Sci. 11, 758–762. doi: 10.20546/ijcmas.2017.604.312
Shrivastava, P., and Kumar, R. (2015). Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Sinclair, J. B., and Backman, P. (1993). Compendium of Soybean Diseases. St. Paul, MN: American Phytopathological Society.
Singh, N., Jayaswal, P. K., Panda, K., Mandal, P., Kumar, V., Singh, B., et al. (2015). Single-copy gene based 50 K SNP chip for genetic studies and molecular breeding in rice. Sci. Rep. 5:11600. doi: 10.1038/srep11600
Song, Q., Hyten, D. L., Jia, G., Quigley, C. V., Fickus, E. W., Nelson, R. L., et al. (2013). Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 8:e54985. doi: 10.1371/journal.pone.0054985
Song, W. Y., Wang, G. L., Chen, L. L., and Kim, H. S. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804. doi: 10.1126/science.270.5243.1804
Sripathi, V. R., Buyyarapu, R., Kumpatla, S. P., Williams, A. J., Nyaku, S. T., Tilahun, Y., et al. (2016). Bioinformatics tools and genomic resources available in understanding the structure and function of gossypium. Bioinformatics 231, 233–258. doi: 10.5772/64325
Strauss, S. Y., Rudgers, J. A., Lau, J. A., and Irwin, R. E. (2002). Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 17, 278–285. doi: 10.1016/S0169-5347(02)02483-7
Szczepaniec, A., Widney, S. E., Bernal, J. S., and Eubanks, M. D. (2013). Higher expression of induced defenses in teosintes (Zea spp.) is correlated with greater resistance to fall armyworm, Spodoptera frugiperda. Entomol. Exp. Appl. 146, 242–251. doi: 10.1111/eea.12014
Takahashi, C. G., Kalns, L. L., and Bernal, J. S. (2012). Plant defense against fall armyworm in micro-sympatric maize (Zea mays ssp. mays) and Balsas teosinte (Zea mays ssp. parviglumis). Entomol. Exp. Appl. 145, 191–200. doi: 10.1111/eea.12004
Tamiru, A., Khan, Z. R., and Bruce, T. J. (2015). New directions for improving crop resistance to insects by breeding for egg induced defence. Curr. Opin. Insect Sci. 9, 51–55. doi: 10.1016/j.cois.2015.02.011
Tanaka, K., Endo, S., and Kazano, H. (2000). Toxicity of insecticides to predators of rice planthoppers: spiders, the mirid bug and the dryinid wasp. Appl. Entomol. Zool. 35, 177–187. doi: 10.1303/aez.2000.177
Tang, L., Cai, H., Zhai, H., Luo, X., Wang, Z., Cui, L., et al. (2014). Overexpression of Glycine soja WRKY20 enhances both drought and salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Cell Tissue Organ Cult. 118, 77–86. doi: 10.1007/s11240-014-0463-y
Throne, J. E., and Eubanks, M. W. (2015). Resistance of Tripsacorn-introgressed maize lines to Sitophilus zeamais. J. Stored Prod. Res. 64, 62–64. doi: 10.1016/j.jspr.2015.08.006
van de Wiel, C. C. M., Schaart, J. G., Lotz, L. A. P., and Smulders, M. J. M. (2017). New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol. Rep. 11, 1–8. doi: 10.1007/s11816-017-0425-z
Vasudevan, K., Gruissem, W., and Bhullar, N. K. (2015). Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 5:15678. doi: 10.1038/srep15678
Vishwakarma, M. K., Mishra, V., Gupta, P., Yadav, P., Kumar, H., and Joshi, A. K. (2014). Introgression of the high grain protein gene Gpc-B1 in an elite wheat variety of Indo-Gangetic Plains through marker assisted backcross breeding. Curr. Plant Biol. 1, 60–67. doi: 10.1016/j.cpb.2014.09.003
Von Uexküll, H., and Mutert, E. (1995). “Global extent, development and economic impact of acid soils,” in Plant-Soil Interactions at Low pH: Principles and Management, eds R. A. Date, N. J. Grundon, G. E. Rayment and M. E. Probert (Dordrecht: Kluwer Academic Publishers), 5–19.
Vuong, T. D., Sonah, H., Meinhardt, C., Deshmukh, R., Kadam, S., Nelson, R., et al. (2015). Genetic architecture of cyst nematode resistance revealed by genome-wide association study in soybean. BMC Genomics 16:593. doi: 10.1186/s12864-015-1811-y
Wang, B., Brubaker, C. L., and Burdon, J. J. (2004). Fusarium species and Fusarium wilt pathogens associated with native Gossypium populations in Australia. Mycol. Res. 108, 35–44. doi: 10.1017/S0953756203008803
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, X., Lee, S., Wang, J., Ma, J., Bianco, T., and Jia, Y. (2014). “Current advances on genetic resistance to rice blast disease,” in Rice—Germplasm, Genetics and Improvement, eds W. Yan and J. Bao (Rijeka: InTech), 195–217.
Wei, W.-H., Qin, R., Song, Y.-C., Guo, L.-Q., and Gu, M.-G. (2001). Comparative analyses of disease resistant and nonresistant lines from maize × Zea diploperennis by GISH. Bot. Bull. Acad. Sin. 42, 109–114. Available online at: https://ejournal.sinica.edu.tw/bbas/content/2001/2/bot422-04.html
Wei, Y., Xu, Y., Lu, P., Wang, X., Li, Z., Cai, X., et al. (2017). Salt stress responsiveness of a wild cotton species (Gossypium klotzschianum) based on transcriptomic analysis. PLoS ONE 12:e0178313. doi: 10.1371/journal.pone.0178313
Wen, L., Yuan, C., Herman, T., and Hartman, G. (2017). Accessions of perennial glycine species with resistance to multiple types of soybean cyst nematode (Heterodera glycines). Plant Dis. 101, 1201–1206. doi: 10.1094/PDIS-10-16-1472-RE
Wendel, J. F. (1989). New World tetraploid cottons contain old world cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 86, 4132–4136. doi: 10.1073/pnas.86.11.4132
Wendel, J. F., and Cronn, R. C. (2003). Polyploidy and the evolutionary histroy of cotton. Adv. Agron. 78, 139–186. doi: 10.1016/S0065-2113(02)78004-8
Wilkes, H. (1977). Hybridization of maize and teosinte, in Mexico and Guatemala and the improvement of maize. Econ. Bot. 254–293. doi: 10.1007/BF02866877
Wilson, R. F. (2008). “Soybean: market driven research needs,” in Genetics and Genomics of Soybean, ed G. Stacey (Berlin: Springer Science+Business Media, LLC), 3–15.
Winter, S. M., Shelp, B. J., Anderson, T. R., Welacky, T. W., and Rajcan, I. (2007). QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI464925B. Theor. Appl. Genet. 114, 461–472. doi: 10.1007/s00122-006-0446-4
Wise, M. J. (2007). Evolutionary ecology of resistance to herbivory: an investigation of potential genetic constraints in the multiple-herbivore community of Solanum carolinense. New Phytol. 175, 773–784. doi: 10.1111/j.1469-8137.2007.02143.x
Wright, R. J., Thaxton, P. M., El-Zik, K. M., and Paterson, A. H. (1999). Molecular mapping of genes affecting pubescence of cotton. J. Hered. 90, 215–219. doi: 10.1093/jhered/90.1.215
Yallou, C., Menkir, A., Adetimirin, V., and Kling, J. (2009). Combining ability of maize inbred lines containing genes from Zea diploperennis for resistance to Striga hermonthica (Del.) Benth. Plant Breed. 128, 143–148. doi: 10.1111/j.1439-0523.2008.01583.x
Yik, C. P., and Birchfield, W. (1984). Resistant germplasm in Gossypium species and related plants to Rotylenchulus reniformis. J. Nematol. 16, 146.
Yu, N., and Diers, B. W. (2017). Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 213:54. doi: 10.1007/s10681-016-1791-2
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Yumurtaci, A. (2015). Utilization of wild relatives of wheat, barley, maize and oat in developing abiotic and biotic stress tolerant new varieties. Emir. J. Food Agric. 27, 1–23. doi: 10.9755/ejfa.v27i1.17852
Zafar, Y., Asif, M., Ashraf, M., Riaz, S., Zafar, S., Wahid, A., et al. (2009). Development of genetic linkage map of leaf hairiness in Gossypium hirsutum (cotton) using molecular markers. Pak. J. Bot. 41, 1627–1635. Available online at: www.pakbs.org/pjbot/PDFs/41(4)/PJB41(4)2009.pdf
Zeng, Y., Yang, S., Cui, H., Yang, X., Xu, L., Du, J., et al. (2009). QTLs of cold tolerance-related traits at the booting stage for NIL-RILs in rice revealed by SSR. Genes Genomics 31, 143–154. doi: 10.1007/BF03191147
Zhang, F., and Xie, J. (2014). “Genes and QTLs resistant to biotic and abiotic stresses from wild rice and their applications in cultivar improvements,” in Rice-Germplasm, Genetics and Improvement, ed J. Bao (London: IntechOpen), 59–78.
Zhang, F., Zhu, G., Du, L., Shang, X., Cheng, C., Yang, B., et al. (2016). Genetic regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species, Gossypium davidsonii. Sci. Rep. 6:20582. doi: 10.1038/srep20582
Zhang, H., Li, C., Davis, E. L., Wang, J., Griffin, J. D., Kofsky, J., et al. (2016). Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG Type 2.5. 7 in wild soybean (Glycine soja). Front. Plant Sci. 7:1214. doi: 10.3389/fpls.2016.01214
Zhang, J., Fang, H., Zhou, H., Hughs, S., and Jones, D. C. (2013). Inheritance and transfer of thrips resistance from Pima cotton to Upland cotton. J. Cotton Sci. 17, 163–169. Available online at: http://www.cotton.org/journal/2013-17/3/loader.cfm?csModule=security/getfile&pageid=147394
Zhang, Q., Wang, C., Zhao, K., Zhao, Y., Caslana, V., Zhu, X., et al. (2001). The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet. Newslett. 18, 71–72. Available online at: http://archive.gramene.org/newsletters/rice_genetics/rgn18/c31.html
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhao, F. A., Fang, W., Xie, D., Zhao, Y., Tang, Z., Li, W., et al. (2012). Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae. Plant Sci. 185–186, 176–184. doi: 10.1016/j.plantsci.2011.10.007
Zheng, J., Oluoch, G., Riaz, K. M., Wang, X., Cai, X., Zhou, Z., et al. (2016). Mapping QTLs for drought tolerance in an F2: 3 population from an inter-specific cross between Gossypium tomentosum and Gossypium hirsutum. Genet. Mol. Res. 15, 1–14. doi: 10.4238/gmr.15038477
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H., and Yang, B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914. doi: 10.1093/nar/gku806
Zhou, Z., Jiang, Y., Wang, Z., Gou, Z., Lyu, J., Li, W., et al. (2015). Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 33, 408–414. doi: 10.1038/nbt.3096
Zhu, D., Cai, H., Luo, X., Bai, X., Deyholos, M. K., Chen, Q., et al. (2012). Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 426, 273–279. doi: 10.1016/j.bbrc.2012.08.086
Keywords: crop wild relatives (CWRs), maize, rice, cotton, soybean, tolerance to biotic stress, tolerance to abiotic stress
Citation: Mammadov J, Buyyarapu R, Guttikonda SK, Parliament K, Abdurakhmonov IY and Kumpatla SP (2018) Wild Relatives of Maize, Rice, Cotton, and Soybean: Treasure Troves for Tolerance to Biotic and Abiotic Stresses. Front. Plant Sci. 9:886. doi: 10.3389/fpls.2018.00886
Received: 18 September 2017; Accepted: 07 June 2018;
Published: 28 June 2018.
Edited by:
Alma Balestrazzi, University of Pavia, ItalyReviewed by:
Charu Lata, National Botanical Research Institute (CSIR), IndiaShabir Hussain Wani, Sher-e-Kashmir University of Agricultural Sciences and Technology, India
Nacira Belen Muñoz, National Institute of Agricultural Technology (INTA), Argentina
Copyright © 2018 Mammadov, Buyyarapu, Guttikonda, Parliament, Abdurakhmonov and Kumpatla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jafar Mammadov, amFtYW1tYWRvdkBkb3cuY29t
 Jafar Mammadov
Jafar Mammadov Ramesh Buyyarapu
Ramesh Buyyarapu Satish K. Guttikonda
Satish K. Guttikonda Kelly Parliament
Kelly Parliament Ibrokhim Y. Abdurakhmonov
Ibrokhim Y. Abdurakhmonov Siva P. Kumpatla
Siva P. Kumpatla