
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 June 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00842
 Zhennan Zhang1†
Zhennan Zhang1† Xiaoyang Ge2†
Xiaoyang Ge2† Xiaoli Luo3†
Xiaoli Luo3† Peng Wang1,2
Peng Wang1,2 Qiang Fan1
Qiang Fan1 Guang Hu1
Guang Hu1 Juanli Xiao3
Juanli Xiao3 Fuguang Li2*
Fuguang Li2* Jiahe Wu1*
Jiahe Wu1*Gossypium hirsutum is an allotetraploid species, meaning that mutants that are difficult to be generated by classical approaches due to gene redundancy. The CRISPR/Cas9 genome editing system is a robust and highly efficient tool for generating target gene mutants, by which the genes of interest may be functionally dissected and applied through genotype-to-phenotype approaches. In this study, the CRISPR/Cas9 genome editing system was developed in G. hirsutum through editing the Gh14-3-3d gene. In T0 transgenic plants, lots of insertions and deletions (indels) in Gh14-3-3d at the expected target site were detected in the allotetraploid cotton At or Dt subgenomes. The results of the PCR, T7EI digestion and sequencing analyses showed that the indels in Gh14-3-3d gene can be stably transmitted to the next generation. Additionally, the indels in the At and Dt subgenomes were segregated in the T1 transgenic plants following Mendelian law, independing on the T-DNA segregation. Two homozygous Gh14-3-3d-edited plants free of T-DNA were chosen by PCR and sequencing assays in the T1 plants, which were called transgene-clean editing plants and were designated ce1 and ce2 in the T2 lines showed higher resistance to Verticillium dahliae infestation compared to the wild-type plants. Thus, the two transgene-clean edited lines can be used as a germplasm to breed disease-resistant cotton cultivars, possibly avoiding complex and expensive safety assessments of the transgenic plants.
Cotton is an important economical crop due to its fiber and derivative production, playing crucial roles in human daily life and economical production worldwide. Cotton is planted in approximately 150 countries and is involved in the income of almost 100 million families (Guan et al., 2014; Li and Zhang, 2016). Commercial species of cotton plants are Gossypium hirsutum (>90% of world production), G. barbadense (3–4%), G. arboreum, and G. herbaceum (together, 2%). G. herbaceum and G. arboretum are diploid species with A or D genomes, respectively, while G. hirsutum and G. barbadense are allotetraploid species, consisting of two sets of subgenomes: At and Dt (Wendel, 1989; Guan et al., 2014). This polyploidization confers many excellent properties to tetraploid cotton, including fiber quality and defense (Li et al., 2015; Zhang et al., 2015). However, the complex genome of allotetraploid cotton brings many challenges for functional analyses and genetic manipulation of cotton genes, mainly due to gene functional redundancy, gene dose effect, and less phenotype of the inserting mutant (Wang Y. et al., 2014; Braatz et al., 2017). Regulation strategies of the expression level, including conventional RNAi and gene overexpression, have been used in the identification of cotton genes, for example, cotton fiber development (Walford et al., 2011; Deng et al., 2012; Tan et al., 2013; Tang et al., 2014; Wang L. et al., 2014; Li Y. et al., 2016; Wan et al., 2016) and stress responses (Min et al., 2013; Li C. et al., 2014; Li Y.B. et al., 2016; Guo et al., 2016). However, “genotype-to-phenotype" approaches are more important in identifying interesting genes. Additionally, cotton genome sequences have been published in many databases, including diploid and allotetraploid species, within recent years (Paterson et al., 2012; Wang K. et al., 2012; Li F. et al., 2014; Yuan et al., 2015; Zhang et al., 2015). Thus, novel gene or genome manipulation urgently needs to be developed to meet the demand for the rapid and precise dissection of cotton gene function.
Recently, the CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (CRISPR-associated) genome editing system was developed and has become a robust and highly effective tool for acquiring novel desired mutations in animal and plant. The RuvC-like and HNH domains of the Cas9 protein can form complexes with a synthetic sgRNA, recognizing target sequences that generate double-strand breaks (DSBs) at expected target sites (Jinek et al., 2012; Cong et al., 2013). Those breaks are quickly mended by the innate repair system via non-homologous end joining (NHEJ) and homology-directed repair (HDR). However, the NHEJ repair mechanism frequently creates small insertions and deletions (indels) at the DNA break sites. These indels can generate a frameshift mutation or disrupt important functional domains, damaging or changing the functions of the target genes (Jinek et al., 2012; Cong et al., 2013; Shan et al., 2013). Although the CRISPR/Cas9-mediated genome editing system is a new tool for gene-targeted mutagenesis, it has been successfully applied in genome editing in many plants, such as Arabidopsis (Feng et al., 2013, 2014; Fauser et al., 2014), wheat (Upadhyay et al., 2013; Wang Y. et al., 2014), tomato (Brooks et al., 2014), rice (Shan et al., 2013; Wang et al., 2016; Lu et al., 2017; Meng et al., 2017), sorghum (Jiang et al., 2013), maize (Liang et al., 2014), and oilseed rape (Lawrenson et al., 2015; Braatz et al., 2017). In addition, the Cas9 protein, when directed against multiple target sites, can induce mutations simultaneously in different (homologous) sequences, as has already been demonstrated in the tetraploid potato (Wang et al., 2015; Andersson et al., 2017), hexaploid wheat (Upadhyay et al., 2013; Wang Y. et al., 2014), and oilseed rape (Braatz et al., 2017). Very recently, application of genome editing in many important genes, excluding exogenous or endogenous marker genes, has been increasingly presented, such as simultaneously targeted mutagenesis of three homeologs of TaEDR1 to enhance powdery mildew resistance in wheat (Zhang et al., 2017), editing targeted mutagenesis of GmFt2a to delay the flowering time in soybeans (Cai et al., 2017), targeted mutagenesis of γ-aminobutyric acid synthesis genes to increase its levels in Solanum lycopersicum (Li R. et al., 2017), and targeting the mutagenesis of two BnALC homeologs to reduce seed shattering in oilseed (Braatz et al., 2017), etc. In 2017, there were several papers that documented the development of CRISPR/Cas9-mediated genome editing systems in G. hirsutum, primarily through endogenous and exogenous marker genes, including GhCLA1, DsRed2, and GFP, as well as the GhMYB25-like gene (Chen et al., 2017; Janga et al., 2017; Wang P. et al., 2017). Thus, interesting and ecological genes, especially negative regulation genes that function in defense and development, remain to be edited for improving cotton cultivars by the CRISPR/Cas9 genome editing system.
Cotton verticillium wilt, called “cotton cancer,” is a destructive disease, annually leading to 250–310 million US dollars in economic losses in China (Wang et al., 2016). The breed of disease-resistant cultivars is the best measure to prevent plants from pathogen damage by Verticillium dahliae. However, few resistant genes or germplasm resources against V. dahliae are naturally found in G. hirsutum. Thus, it is pivotal to generate novel defense genes or defense mutants using CRISPR/Cas9 targeting the negative regulator of disease-resistance, including cotton 14-3-3c/d, NINJA, and CYP82D, which had high sensitivity against V. dahliae infestation as confirmed by RNAi approaches(Gao et al., 2013; Sun et al., 2014; Wang L. et al., 2017). Among these negative defense proteins, the 14-3-3 proteins, a family of conserved regulatory molecules, are found in all eukaryotic cells, which bind to functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors (Obsil et al., 2001). In plants, the 14-3-3c/d proteins have been demonstrated to be negative regulators of BR signaling by regulating two key transcription factors, Brassinazole resistant 1 (BZR1) and Brassinosteroid insensitive 2 (Gampala et al., 2007), and regulating plant response to biotic stress (Nakashita et al., 2003; Vriet et al., 2012; Wang H. et al., 2012). Recently, four 14-3-3 proteins involved in BR signaling were identified in proteomic analysis to have decreased in abundance in cotton plants inoculated with V. dahliae (Gao et al., 2013). Silencing of 14-3-3c and 14-3-3d through the virus-induced gene silencing (VIGS) method significantly enhanced the resistance of cotton plants to V. dahliae (Gao et al., 2013). Thus, the cotton 14-3-3c/d genes can be used as candidate target genes for generating disease-resistant mutants by the CRISPR/Cas9 genome editing system.
In this study, we developed a CRISPR/Cas9-mediated genome editing system in plants with easy and convenient target sequences, by which the cotton target gene, Gh14-3-3d, were edited for generating indels at expected target sites. Lots of nucleotide insertions and deletions at the expected sites of the Gh14-3-3d target genes were generated in T0 plants induced by the CRISPR/Cas9 genome editing system. We screened 16 T1 lines and acquired two transgene-clean editing plants with homozygous indels in the tetraploid cotton At and Dt subgenomes, designated ce1 and ce2 lines in T2 had high resistance to V. dahliae compared to the wild type, which can be directly used as a germplasm to breed resistant cultivars and can be avoided to perform safety assessment of transgenic plants, a time-consuming, expensive and tedious process. This successful target gene editing will promote more studies in exploring gene functions of interest and improve agricultural traits.
Cotton cultivar CCR35 was used as the transformation receptor for Gh14-3-3d gene editing in this study. The hypocotyl of cotton seedlings were cultured in the dark from sterilized seeds in a chamber for 6 days at 30°C and were used as explants for Agrobacterium-mediated transformation (Wu et al., 2005). The regenerated plants with perfect roots were directly transplanted in pots with 1:3 vermiculite and organic matter soil, and other plants with poor root systems were grafted on the wild-type seedling, which grew in the greenhouse.
Gossypium hirsutum cv. CCR35, transgenic plants and transgenic offspring were grown in a greenhouse under 28°C and 14/10 h light/dark conditions. The T0 transgenic plants (CRISPR/Cas9 editing mutants) in the greenhouse were self-pollinated, and the seeds were finally collected. Due to the abnormal phenotypes in the T0 plants with stunted growth and fewer flowers, we acquired an 11 to 64 seeds from the 16 Gh14-3-3d-edited plants. For genetic analysis of the mutant genotypes, T1 plants were then planted under natural conditions in Yuncheng, Shanxi province, China (N35°04′, E110°58′).
Based on simplifying the assembly process of the target sequence of sgRNAs, the sequence of AtU3b-double BsaI-sgRNA (Supplementary Data S1) was synthesized, then cloned into pYLCRISPR/Cas9-N through BsaI sites (Ma et al., 2015), resulting in vector designated as pYLCRISPR/Cas9-NDB.
The sgRNA targets were predicted using the online tool Cas-Designer based on the G. hirsutum database (Bae et al., 2014). Among these target sites, a target site in exon1 with higher scores that simultaneously targeted the two copies of the Gh14-3-3d gene, Gh14-3-3d-A and Gh14-3-3d-D located in the allotetraploid cotton At and Dt subgenome, respectively, was selected. The target sequence (AGATGAAGGGAGATTACCAT) was directly inserted into pYLCRISPR/Cas9-NDB by double BsaI to obtain the Gh14-3-3d editing vector, pYLCRISPR/Cas9-NC14.
A specific Gh14-3-3d fragment was amplified by PCR and directly cloned into the tobacco rattle virus (TRV) vector pYL-156, a gift from Professor Yule Liu at Tsinghua University. The resulting vector was named pYL-156-Gh14-3-3d. The pYL-156-Gh14-3-3d, pYL-156 (empty vector) and the pYL-192 helper vector were transformed into the Agrobacterium tumefaciens GV3101 strains by electroporation.
Agrobacteria culture and inoculation solution preparation were carried out according to a previous report (Wang L. et al., 2017). Agrobacteria containing pYL-156-Gh14-3-3d or pYL-156 were equally mixed with pYL-192, and injected into fully expanded cotyledons using a needleless syringe. After 14 days, the plants injected with Agrobacteria were inoculated with V. dahliae for defense analysis.
Genomic DNA of the cotton leaves was extracted using the Plant Genome Extraction Kit (TianGen, Beijing, China) according to the instruction manual. PCR was performed to detect the T-DNA components and monitor the indels of the Gh14-3-3d genes in transgenic plants and their offspring. Cas9, Npt II, and three sequence fragments, including Npt II-nos-ter, p35s-Cas9, and sgRNA expression cassette, were analyzed by common PCR methods, whose specific primers were listed in Supplementary Table S1.
To amplify the genomic region surrounding the CRISPR target sites, two rounds of PCR reactions were performed in independent transgenic cotton plants and their offspring. In the first round of PCR, the two specific PCR fragments of Gh14-3-3d-A (1984 bp) and Gh14-3-3d-D (1957 bp) were amplified from the genomic DNA by a special forward primer located in the At or Dt subgenomes (PA or PD) and a universal reverse primer (PR). In the second round of PCR, a specific fragment of approximately 620 bp with a target site was acquired through a pair of universal primers (PF and PR) when the specific PCR fragment of the At or Dt subgenomes from the first-round PCR products was employed as a PCR template. To identify the indels of the mutants, these PCR products were directly sequenced, or cloned into the pEASY-T3 (TransGen) vector and then sequenced by Sanger method.
Fungal biomass quantification was performed through comparing the V. dahliae β-tubulin DNA levels to the cotton UB7 DNA levels after V. dahliae inoculation by qPCR according to the method described previously (Atallah et al., 2007; Wang L. et al., 2017). The relative primers were shown in Supplementary Table S1. The same experiment was carried out using three biological replicates.
The second-round PCR products of each sample mentioned above, a 620 bp fragment with target sites, were equivalently mixed with the PCR fragments amplified from the WT plant, which were used to detect the mutation with T7 Endonuclease I (Vazyme, Nanjing, China) according to the instruction manual. Final digested reaction products were analyzed with 1.5% agarose gel electrophoresis.
Total RNA of the cotton leaves and roots was isolated using the Plant Total RNA Extraction Kit (Sangon Biotech, Shanghai, China) according to the instruction manual. Two micrograms of RNA was reverse transcribed for first strand cDNA synthesized following the manufacturer’s protocol using the TransScript First-Strand cDNA Synthesis kit (TransGen).
According to the protocols of the Minimum Information for Publication of Quantitative Real Time PCR Experiments (Bustin et al., 2009), diluted cDNA was used for qPCR with SYBR green using the CFX96 TouchTM Real-Time PCR detection systems (Bio-Rad, Foster City, CA, United States). All gene expression was calculated using the dCt or ddCt algorithm. To normalize the gene expression, the UB7 gene was used as an internal standard. All the gene specific primers involved in this study were listed in Supplementary Table S1.
The defoliating isolate V991 of V. dahliae was cultured on potato dextrose agar for 3 days, and then the fungus was incubated in Czapek’s medium (sucrose, 3% w/v; NaNO3, 0.3% w/v; KCl, 0.1% w/v; KH2PO4, 0.1% w/v; MgSO4, 0.1% w/v; FeSO4, 0.0002% w/v; pH 6.0) and grown in the dark at 25°C for 5 days. The spore concentration of the fungus was adjusted to 105 conidia/ml with 5% deionized sucrose solution for inoculation. The 21-day-old seedlings were inoculated with V. dahliae or sucrose solution (mock) through the roots. The inoculated seedlings were incubated in a chamber at 25°C under a 14/10 h light/dark photoperiod.
To evaluate plant resistance to V. dahliae, the rate of disease in plants and the disease index (DI) of the plants were investigated according to the methods reported by Wang L. et al. (2017). Additionally, a fungal recovery experiment was performed 14 days after infection according to the method described previously (Wang L. et al., 2017). The same experiment was carried out using three biological replicates.
To simplify the assembly process of the target sgRNA sequences, we constructed a CRISPR/Cas9 toolkit with double BsaI for easy and convenient insertion of target sequences in plant target genes or DNA editing (Supplementary Figure S1). A previous study has shown that 14-3-3d is a negative regulator in the BR signal pathway that participates in cotton defense. In this study, the Gh14-3-3d-silenced plants by VIGS were produced to confirm its function in defense against V. dahliae. As shown in Supplementary Figure S2, the Gh14-3-3d-silenced plants showed a higher resistance compared to the wild-type plants. Thus, the 14-3-3d gene was chosen and edited for increasing cotton plant defense to V. dahliae by CRISPR/Cas9-mediated mutations. The Gh14-3-3d gene has two copies located in the tetraploid cotton At and Dt subgenomes (GenBank accession Nos. NM_001327374 and XM_016860147), which were referred to as Gh14-3-3d-A and Gh14-3-3d-D. Both share a highly similar nucleotide sequence and the same gene structure (Figure 1A). The two copies of this gene encode proteins that share 98.9% identification in amino acids and there are just three different sites among 261 amino acids (Supplementary Figure S3). We chose an excellent target site with 20-bp followed by the AGG PAM motif located in exon1 downstream of the Gh14-3-3d gene (Figure 1A), as sgRNA targeted both Gh14-3-3d-A and Gh14-3-3d-D. For designing the CRISPR/Cas9 vector, a 20-bp target sequence of Gh14-3-3d was inserted directly in the vector mentioned above by BsaI digestion and driven by AtU3b promoter (Figure 1B).
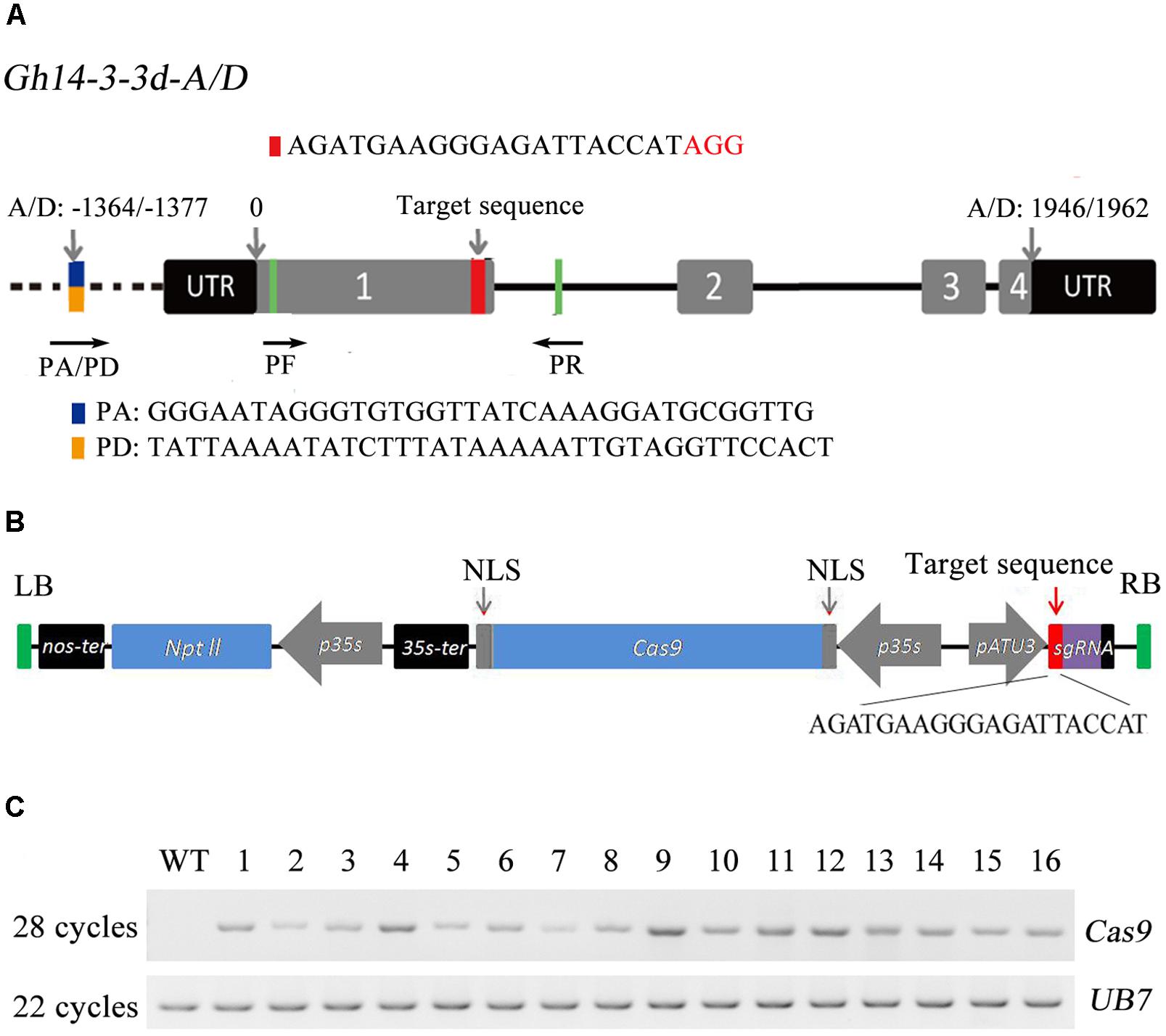
FIGURE 1. The structure of the Gh14-3-3d gene, editing vector construction and PCR analyses of the transgenic plants. (A) The structure of Gh14-3-3d-A and Gh14-3-3d-D (Gh14-3-3dA/D) and the target site of the sgRNA in exon1. The target site was highlighted in red, the difference between sequences of Gh14-3-3dA/D are labeled in blue-yellow (special forward primers, PA/PD, were designed in this site), and the universal primer sites (PF and PR) were highlighted in green. The AGG in red represents the PAM motif. The different numbers of nucleotides noted by arrows indicated different sites, start codons and stop codon. (B) The schematic of the T-DNA region of the editing vector pYLCRISPR/Cas9-NC14. The target sequence of the Gh14-3-3dA/D genes was listed under the schematic. (C) RT-PCR analysis of the Cas9 transcript level in the WT and transgenic plants. The UB7 gene was regarded as the inner control. WT, wild-type plant; lanes labeled 1 to 16 represent the 16 independent transformants.
It took 1 year to perform cotton genetic transformation, and we acquired 31 regenerating plants with kanamycin resistance that could grow in the greenhouse (Supplementary Figure S4). Among these plants, 16 plants with Cas9 gene was successfully expressed in T0 plants by RT-PCR analysis (Figure 1C). Finally, the 16 T0 plants were subject to analyses of nucleotide insertion and deletion mediated by the CRISPR/Cas9 genome editing system.
PCR products of the At and Dt subgenomes from 16 transgenic plants in T0 were acquired using two pairs of special primers (listed in Supplementary Table S1) through two cycles of PCR amplification and then were cleaved by the T7EI digestion to examine whether the Gh14-3-3d-A and Gh14-3-3d-D of these T0 plants were edited by the CRISPR/Cas9 system. Most of the PCR fragments were cleaved into two parts at the expected target site, indicating that two copies of the Gh14-3-3d gene were edited, generating indels at the expected target site (Figure 2A). Notably, if the PCR amplification of the T0 plants was obtained by a pair of common primers for the At and Dt subgenomes, they could be cleaved into lots of parts by the T7EI digestion due to sequence differences of the two copies of Gh14-3-3d (Supplementary Figure S5A). Thus, it was necessary to distinguish from different copies of polyploidy plants for employing the T7EI digestion to identify mutagenesis by the CRISPR/Cas9 genome editing system.
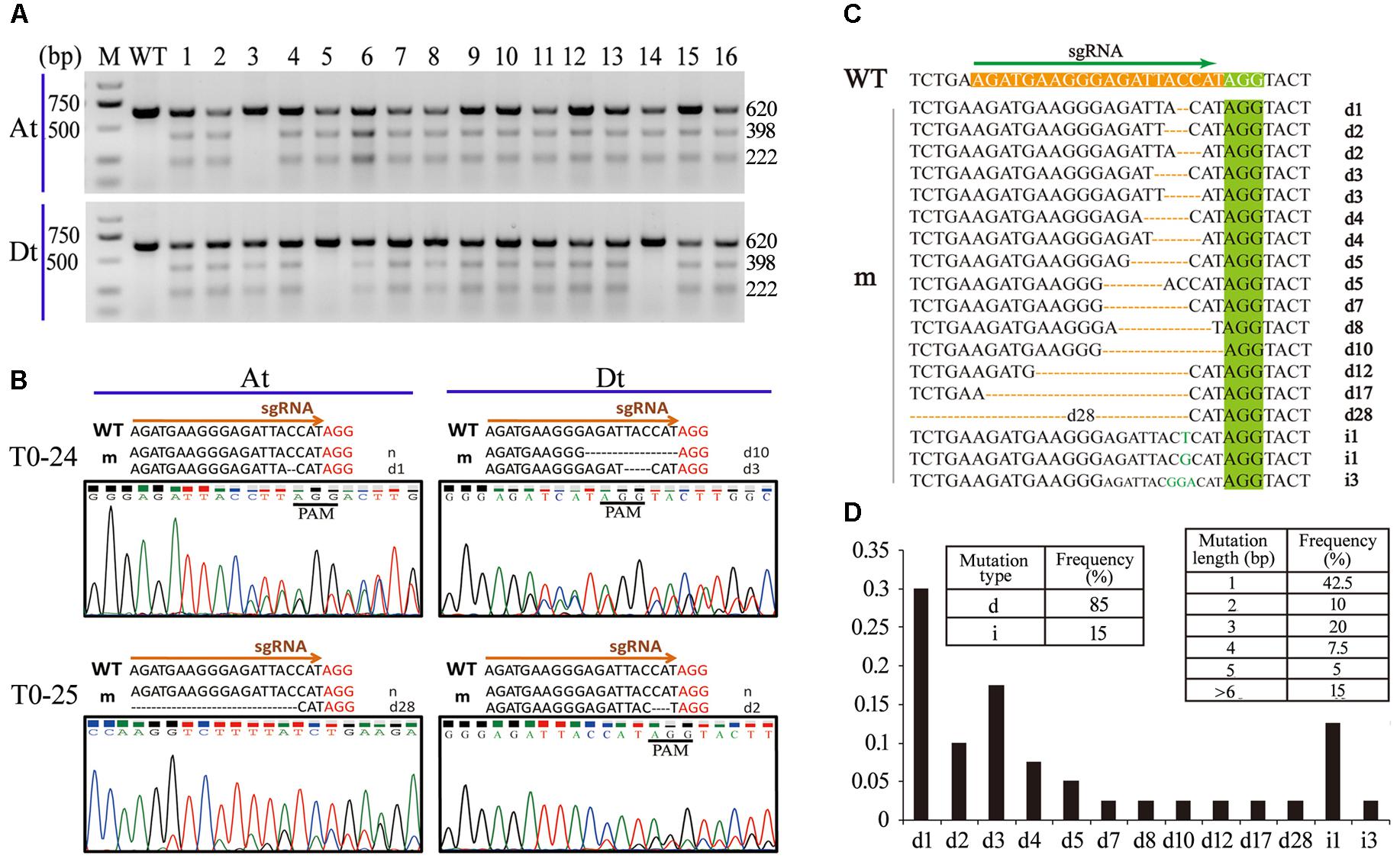
FIGURE 2. Analyses of CRISPR/Cas9-mediated mutagenesis of Gh14-3-3d in T0 plants. (A) T7EI digestion assay of the mutants at the target sites. The sizes (bp) of the digested amplifications for the Gh14-3-3d at At (Top) or Dt (Bottom) were indicated on the right side. Each amplification from the transgenic plants was equally mixed with the PCR product of the WT. M, DNA molecular marker; WT, wild-type plant; lanes labeled 1 to 16 represent the 16 independent transformants (mutants) mentioned above. (B) The indels of Gh14-3-3d at the target site of the At or Dt subgenome in T0–24 and T0–25 were illustrated based on the nucleotides and chromatograms by Sanger sequencing, respectively. The PAM sequence was shown in red. The detailed indels labeled at sequence right. d#, # of base pairs (bp) deleted from the target site; i#, # of base pair (bp) inserted at the target site, which was used thereafter. (C) Different indels of Gh14-3-3dA/D occurred at expected target site in the 16 mutants. The PAM motif was highlighted in green. The indel types are listed at the right. WT, the wild type; m, mutants. (D) The indel types and frequency in the 16 mutants. d, deletion; i, insertion.
To intensively explore the generated indels, the PCR products were directly sequenced by Sanger sequencing. Because G. hirsutum is an allotetraploid cotton species, PCR products of 16 T0 plants acquired using a common primer pair showed a notable phenomenon in the sequencing chromatogram. As shown in Supplementary Figure S5B, a typical sequencing chromatogram for these PCR fragments presented single peaks extending up to the mutation sites of the sgRNA target sequence, but immediately after the mutation sites multiple peaks often started to appear in each nucleotide position. Although Sanger sequencing chromatograms can be read by an artificial method and special program (Supplementary Figure S5B), it was unclear if these indels occurred at Gh14-3-3d-A or Gh14-3-3d-D. For distinguishing these indels from the At or Dt subgenomes, the PCR fragments amplified by two pairs of special primers were sequenced. The results showed 1–2 types of indels at the expected target site in the At or Dt subgenomes, so there were 1–4 types of indels in each plant mediated by CRISPR/Cas9 system (Table 1 and Figure 2B). For instance, T0–30 transgenic plants presented 4 types of indels, d1d3d5i3 (1 and 3 bp deletions at two alleles of At subgenome, 5 bp deletion and 3 bp insertion at two alleles of Dt subgenome), while T0–6 and T0–9 containing 1 indel, d1 at the Dt subgenome and d3 at the At subgenome, respectively. Other plants possess 2–3 types of indels at the target site. According to the genotype analysis, there were some homozygote or bi-allele indels at Gh14-3-3d-A or Gh14-3-3d-D, but only 2 plants, T0–12 and T0–30, possessed homozygote or bi-allele indels at both the At and Dt genomes (Table 1).
To precisely analyze these indels of each allele at both the At and Dt genomes, more than 150 positive colonies from PCR amplification of the edited Gh14-3-3d-A or Gh14-3-3d-D in the 16 T0 plants were randomly picked for the sequencing analysis. The result showed that 40 editing events independently occurred at the At or Dt subgenomes of 16 plants. Eighteen of these sequences showed different indels at the expected target site, suggesting that many types of genome editing events precisely occurred at the examined target gene, Gh14-3-3d (Figure 2C and Table 1). These indels were randomly presented at the target site of Gh14-3-3d-A or Gh14-3-3d-D, with 11 types of indels at both the At and Dt subgenome (Table 1). The results of these editing sequences showed that 35 of 40 editing events were nucleotide deletions and the others were insertion events, which suggested that deletions were more common than insertions in the cotton CRISPR/Cas9 genome editing system (Figure 2D). Among the 40 editing events, 12 were 1 bp nucleotide deletions, 5 exhibited 1 bp nucleotide insertions, indicating that 1 bp nucleotide indels mutants were readily generated during CRISPR/Cas9-mediated genome editing in cotton (Figure 2D). These results indicated that the sgRNA targeted Gh14-3-3d genes effectively and precisely guided Cas9-mediated genome cleavage, resulting in a highly effected target sequence mutant including nucleotide deletion and insertion. Thus, this CRISPR/Cas9 genome editing system has a high potential for producing different indel mutants on the tetraploid cotton genome for improving the cotton cultivars.
For clarifying the stabilization and genetic pattern of these indels at the At and Dt subgenomes of tetraploid cotton, the mutated sequence in editing mutants and their offsprings were analyzed. In T0, few of the seeds from the 16 Gh14-3-3d-edited plants were harvested because of the stunted growth and fewer flowers of the regenerated plants. T1 plants were then planted under natural conditions for genetic analysis of the mutant genotypes in Yuncheng, Shanxi province, China. The results of the PCR analysis suggested that the segregation ratios of the Cas9 gene mostly followed Mendelian laws in the 16 T1 lines, possibly presenting a 3:1 segregation ratio in T0–1, 6, 15, 20, 22, and 24 lines (Table 2). More importantly, the genetic patterns of the indels in the 16 T1 lines were analyzed by PCR and sequencing approaches to investigate the mutant genotype segregation of the At and Dt subgenome in the offspring. All indels generated in the T0 plants can stably transmit to T1 plants, which mostly met the genetic laws in the At and Dt subgenomes (Tables 1, 2). Interestingly, we found that there was a new indel (d3 in the At subgenome) at the target site in an offspring plant of the T0–6, indicating that Cas9 functioned in offspring plants to produce novel target gene editing (Table 2 and Supplementary Figure S6). As shown in Table 2, the Gh14-3-3d gene indels in the T1 lines showed independent segregation in T-DNA insertion, resulting in a few of mutant plants whose T-DNA were segregated and had already been eliminated. Thus, we may easily choose mutagenized plants free of T-DNA from these T1 lines.
To evaluate the function of Gh14-3-3d, the mutant plants with homozygous or bi-alleles in the tetraploid cotton At and Dt subgenomes were screened. As shown in Table 2, 10 lines with homozygous or bi-allele indels were found in the T1 segregation groups. Considering the safety of transgenic plants, the T-DNA of the Gh14-3-3d-edited plants must be ruled out, which were called transgene-clean editing target gene plants. There were 2 transgene-clean plants with homozygous Gh14-3-3d editing that were segregated from the T0–15 and T0–24 lines, whose genotypes were d2d2d1d1 and d1d1d10d10, respectively, designated ce1 and ce2 (Table 2 and Figure 3A). To confirm whether the ce1 and ce2 lines contained transgenic components, the three fragments of the T-DNA from LB to RB, including the Npt II-nos-ter (912 bp), p35s-Cas9 (1012 bp), and sgRNA expression cassettes (725 bp), were amplified by PCR. The results showed that no amplifications were present in the two transgene-clean plants and WT plants, while the three corresponding fragments as well as the Cas9 gene were amplified in the transgenic plants shown in Table 2, suggesting that the T-DNA had been ruled out in two transgene-clean plants (Figure 3B). Additionally, the result of the kanamycin-resistance assay showed that the leaf dots of the transgene-clean plants painted with 0.5% kanamycin solution became yellow 5 days later, while the transgene leaves were still green (Figure 3C). To investigate the genetic stability of the indels in ce1 and ce2, 20 offspring plants (T2) were used to carry out PCR and sequencing analyses. The results showed that the T2 plants contained the same indels as ce1 or ce2 mutagenesis (Figure 3A), showing d2d2d1d1 or d1d1d10d10 in the genotypes, respectively, suggesting that Gh14-3-3d editing genotypes were stably transmitted into the next generation at the At and Dt subgenomes, and there were no novel indels generated in the offspring plants. Altogether, these results showed that the ce1 and ce2 lines were successfully developed through the CRISPR/Cas9-targeted genome editing system.
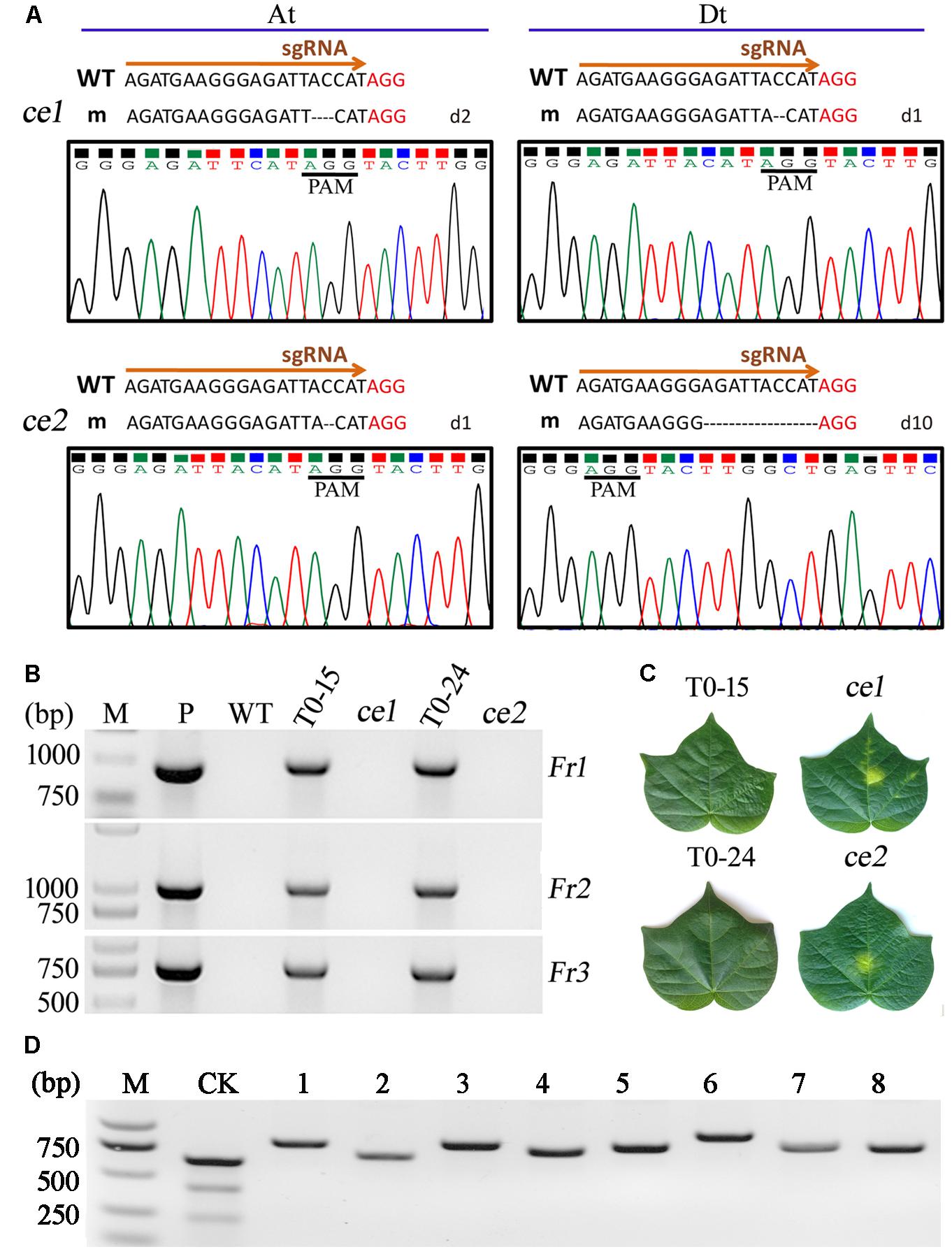
FIGURE 3. Analyses of the two transgene-clean editing lines with homozygous mutagenesis of Gh14-3-3d. (A) The two transgene-clean editing lines, ce1 and ce2, exhibited indels at the target site of the At or Dt subgenome based on the nucleotides and chromatograms by Sanger sequencing, respectively. The PAM sequence was shown in red. (B) The T-DNA free in the ce1 and ce2 plants detected by PCR analysis. M, DNA molecular marker; P, plasmid vector as a positive control; WT, wild-type plant. The ce1 and ce2 were segregated from the T0–15 and T0–24 transformants, respectively. Fr1, Fr2, and Fr3 were fragments of the Npt II-nos-ter (912 bp), p35s-Cas9 (1012 bp), and sgRNA expression cassettes (725 bp), respectively. (C) The leaf parts painted with 0.5% kanamycin solution were yellow in the ce1 and ce2 plants, while they were still green in the T0–15 and T0–24 plants. Photographs of the leaves were taken at 7 days after painting. (D) The PCR products from eight genes containing putative off-target sequences with 1–3 bp mismatches in ce1 and ce2 were not cleaved by T7EI.
To evaluate the off-target potential in ce1 and ce2 to affect other phenotypes, we analyzed the off-target effects of the putative off-target sequences obtained through blasting the cotton genome database1. These potential off-target sites contained 14 bp mismatches compared to the on-target guide sequences of Gh14-3-3d gene as shown in Supplementary Table S2. The eight putative off-target sequences with 1–3 bp mismatches located in genes were selected for further analysis. In ce1 and ce2, the PCR products from eight genes containing putative off-target sequences were not cleaved by T7EI, which was unlike the results in Figure 2A, showing the novel cleaved fragments (Figure 3D). The sequencing results of the PCR amplifications showed that there were no differences among the sequences of potential off-target sites in ce1, ce2, and WT plants, indicating that no editing was detected in these putative off-target sites (Supplementary Table S2). Additionally, the results of Sanger sequencing showed that putative off-target sequences of eight genes from the cotton gene database were the same as real sequences of the potential off-target sites by PCR in ce1, ce2, and WT plants. Those data showed that no mutations were observed in the examined putative off-target genes of ce1 and ce2 plants, possibly indicating that of the CRISPR/Cas9 genome editing toolkit has high specificity in plants.
To examine the resistance of the transgene-clean editing lines with Gh14-3-3d mutations, 21-days seedlings of ce1 and ce2 were infected by V. dahliae along with the WT plants. At 18 days after inoculation, two transgene-clean editing lines and the WT plants healthy grew under the treatment with mock, while both had increased resistance to V. dahliae with less severe defoliation and yellowing symptoms compared to the WT plants (Figure 4A). A fungal recovery assay was performed to further investigate the response of ce1 and ce2 plants to V. dahliae infestation. The result showed that obviously fewer stem sections from the transgene-clean editing plants grew mycelium compared to the WT (Figure 4B). The disease plant rate and disease index in the Gh14-3-3d-edited plants was significantly lower than those in WT (Figures 4C,D). Additionally, the fungal biomass of the leaves from the ce1 and ce2 plants at 18 days after infection obviously decreased compared to that of the WT as determined by qRT-PCR analysis, just reaching 0.19- and 0.22-fold of the WT, respectively (Figure 4E). Taken together, the data suggested that the two transgene-clean Gh14-3-3d-edited lines showed higher resistance to V. dahliae.
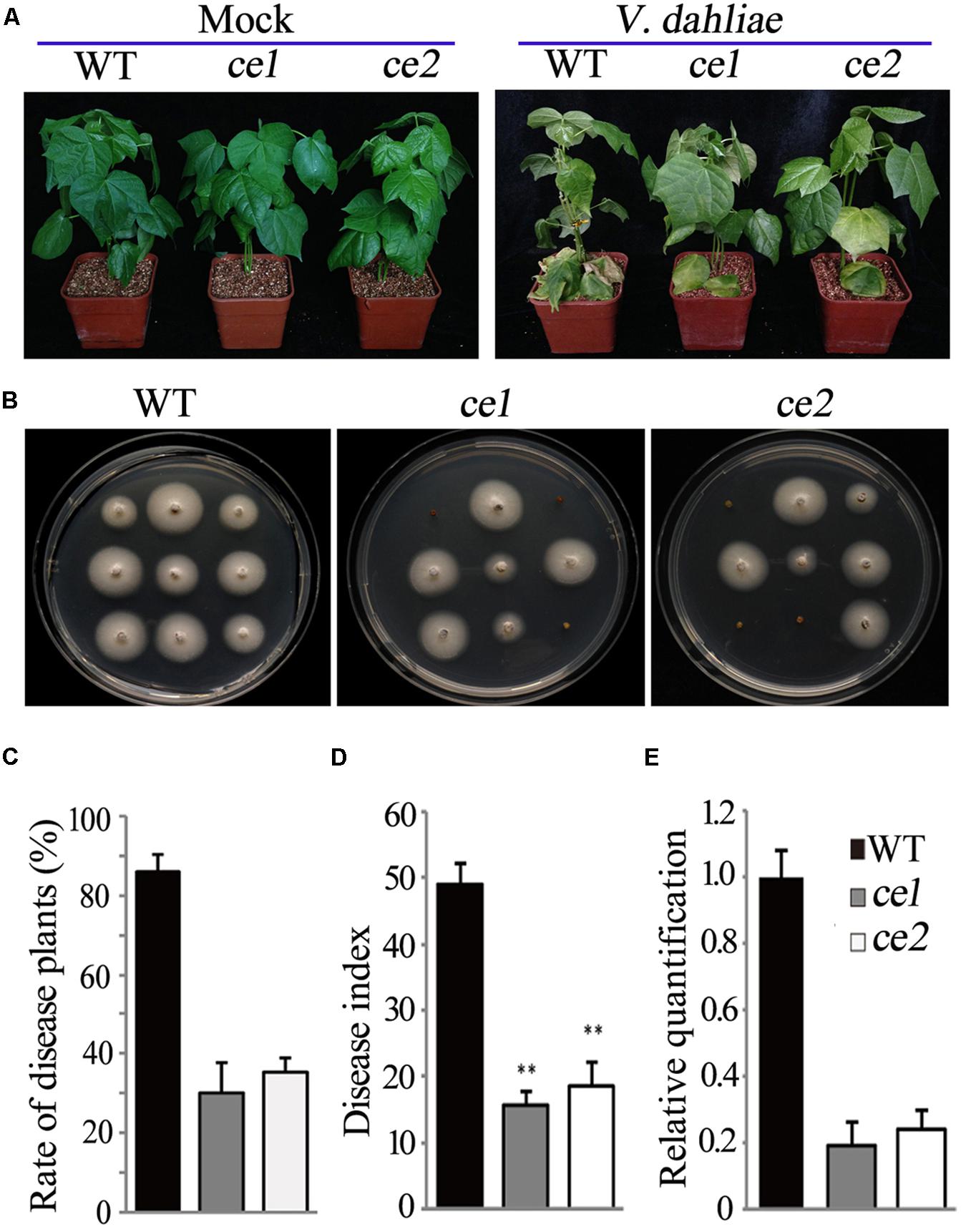
FIGURE 4. Disease symptom analysis of the ce1 and ce2 lines inoculated with V. dahliae. (A) Photographs of the representative WT, ce1 and ce2 plants taken at 18 days after inoculation with V. dahliae (105 conidia/ml) or mock. (B) Fungal recovery assay. The stem sections of the plants inoculated with V. dahliae (105 conidia/ml) 18 days later were placed on potato dextrose agar medium, and then photographs were taken 4 days after culture. (C,D) The rate of diseased plants and disease index in the WT, ce1, and ce2 plants. Error bars represent the SD of three biological replicates (n ≥ 30). Double asterisks indicate statistically significant differences compared to the control, which were analyzed using Student’s t-test (P < 0.01). (E) The relative quantification of the fungal biomass of the cotton leaves among the WT, ce1, and ce2 plants by qPCR. In comparing the V. dahliae β-tubulin gene DNA levels to the cotton UB7 DNA levels 18 days after V. dahliae inoculation, the data in the WT was normalized as “1.” Error bars represent the standard deviation of the three biological replicates.
To explore whether the enhanced resistance to V. dahliae in Gh14-3-3d-edited plants was involved in the BR signal pathway and defense-related marker genes, the expression levels of Brassinosteroid insensitive 1 (BRI1), BZR1, BIN2, PDF1.2, and PR4 were monitored by qRT-PCR analysis. The expression pattern of the three BR signal genes in the roots of ce1 and ce2 inoculated with V. dahliae showed significant differences compared to the WT, with BRI1 and BZR1 exhibiting up-regulated expression and BIN2 showing down-regulated expression (Figure 5A), suggesting that Gh14-3-d-3d participated in plant defense against V. dahliae possibly through the BR signal pathway. Most of the genes involved in JA signaling were up-regulated in cotton plants after treatment of BR, but there were no obvious changes in the transcripts of the SA signaling pathway-related genes (Gao et al., 2013). Thus, we examined the expression levels of PDF1.2 and PR4, two well-known JA-regulated defense-related marker genes; both significantly up-regulated expression in the two transgene-clean Gh14-3-3d-edited lines infected with V. dahliae, showing nearly twofold and threefold higher of the WT, respectively (Figure 5B). The result suggested that the ce1 and ce2 lines possessed higher resistance to V. dahliae, possibly by modulating BR and JA signaling gene expression.
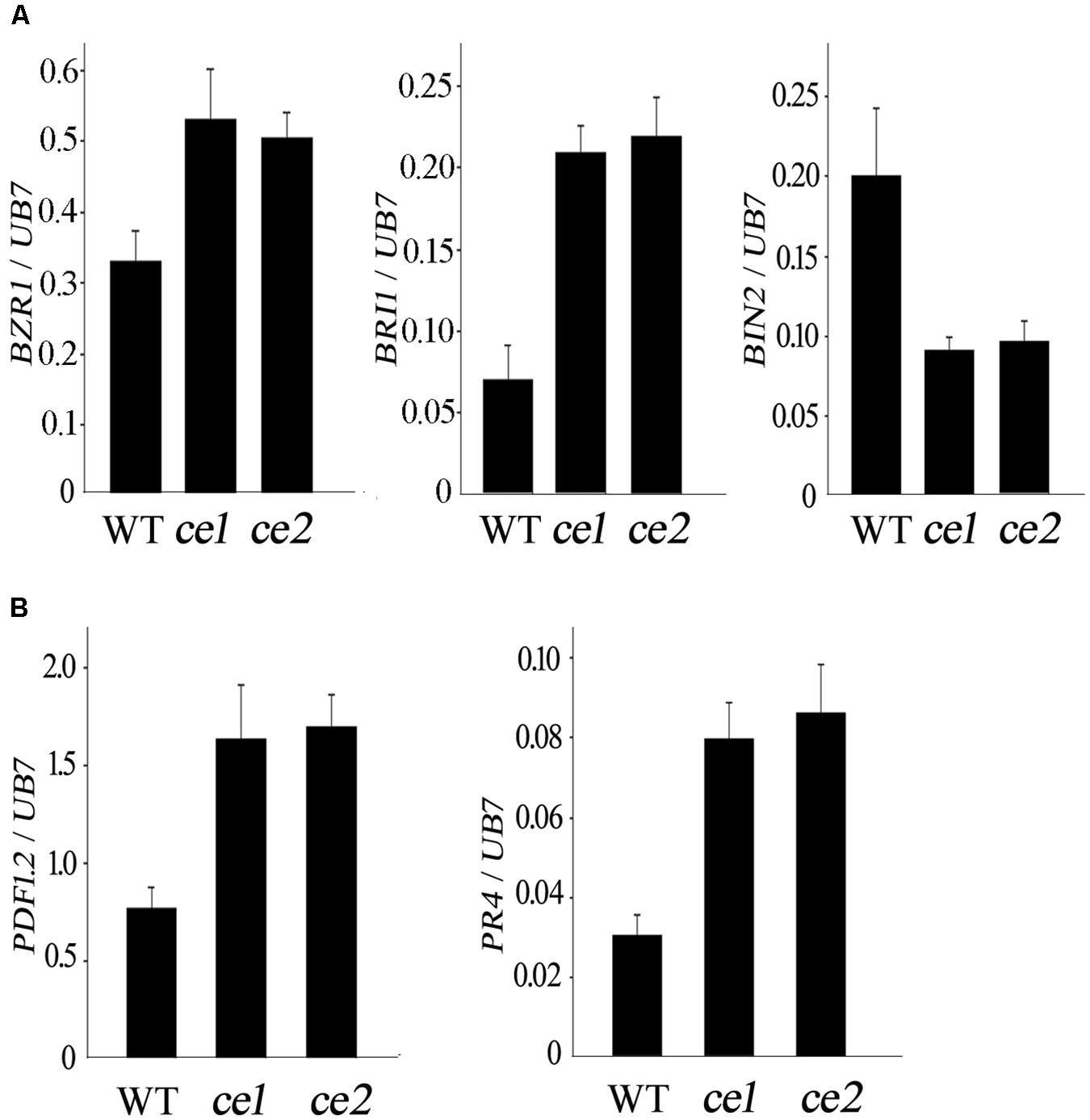
FIGURE 5. Expression patterns of the three BR signal genes (A) and two JA defense-related marker genes (B) in the WT, ce1, and ce2 plants treated with V. dahliae.
The CRISPR/Cas9 genome editing system had been developed in G. hirsutum as determined by editing endogenous and exogenous marker genes (Chen et al., 2017; Janga et al., 2017; Li C. et al., 2017; Wang P. et al., 2017). Thus, the interesting and economical genes edited by the CRISPR/Cas9 technique remained for the study of improving cotton cultivars. In this study, lots of Gh14-3-3d indels were generated in the tetraploid cotton At and Dt subgenomes mediated by our CRISPR/Cas9 genome editing toolkit. More importantly, the transgene-clean T2 lines with homozygous editing mutagenesis of Gh14-3-3d were developed, which possessed high resistance to V. dahliae infestation.
We employed the CRISPR/Cas9-targeted gene editing system to generate lots of indels of the Gh14-3-3d gene in the tetraploid cotton At and Dt subgenomes, and successfully bred the homozygous Gh14-3-3d-edited plants. Lots of plant genomes are polyploid in nature, which leads to high production and defense potential, such as allotetraploid cotton, tetraploid potato, hexaploidy wheat, and so on. In general, it is difficult for polyploid plants to generate mutants by natural mutation and artificial mutation due to the functional redundancy of multiple copies of genes (Wang Y. et al., 2014; Braatz et al., 2017; Lu et al., 2017; Meng et al., 2017). Now it is feasible to produce target gene mutants by CRISPR/Cas9 genome editing system. For example, Wang Y. et al. (2014) reported that the three copies of the TaMLO gene were simultaneously edited in a plant, resulting in increasing resistance to powdery mildew. Similar study in wheat was performed through editing three homeologs of TaEDR1 to increase the resistance to powdery mildew (Zhang et al., 2017). Three different regions of the gene encoding granule-bound starch synthase were targeted to change the starch in the tetraploid potato using the CRISPR/Cas9 technique (Andersson et al., 2017). Braatz et al. (2017) employed the CRISPR/Cas9 genome editing system to target two BnALC homeologs, resulting in reducing seed shattering in the mutant of oilseed.
Plant 14-3-3 proteins play important roles in development and defense through kinases, phosphatases and member acceptors (Obsil et al., 2001; Gao et al., 2013). In cotton, 14-3-3 proteins mainly participate in cotton fiber development (Shi et al., 2007; Zhang et al., 2010; Zhou et al., 2015) and defense (Hill et al., 1999; Yan et al., 2004; Sun et al., 2011). For instance, three Gh14-3-3 family proteins, Gh14-3-3e, Gh14-3-3h, and Gh14-3-3L, promoted fiber initiation and elongation (Zhou et al., 2015). Gao et al. (2013) showed that Gb14-3-3c and Gb14-3-3d negatively regulate cotton plant resistance to V. dahliae through participating in the BR signaling pathway. Notably, these studies on cotton 14-3-3 protein function mostly depended on regulating RNA transcript levels by RNAi and overexpression approaches. In this study, we developed two transgene-clean Gh14-3-3d-edited lines, ce1 and ce2, using the CRISPR/Cas9 genome editing system. Both mutant lines had increasing resistance against V. dahliae infestation and can be used as a germplasm to breed cotton disease-resistant cultivars. Notably, fiber length of the mutants ce1 and ce2 was comparable with the WT possibly due to Gh14-3-3d specially responding to defense, which was consistent with the result in G. barbadense (Gao et al., 2013).
In the past 20 years, application of transgenic cotton, including insect- and herbicide-resistant cultivars, has brought large economic benefits, such as 1.5 billion dollars in increased income due to the Bt cotton plant each year in China (Li and Zhang, 2016). However, before the transgenic plants grow in the open environment, a strict safety assessment must be carried out according to the policies and regulations issued by the countries or organizations. The safety assessment of the transgene crops is a time-consuming, expensive and complex process. Thus, it is a pivotal to breed transgene-clean editing plants from transgenic lines with target gene editing. Recently, the development of transgene-clean editing plants emerged in crops, including soybean, oilseed rape and wheat (Braatz et al., 2017; Cai et al., 2017; Li R. et al., 2017; Zhang et al., 2017). We developed transgene-clean cotton lines by editing Gh14-3-3d, which can highly resist V. dahliae infestation.
Lots of Gh14-3-3d indels in editing plants were identified by PCR and sequencing analyses. These indels in tetraploid cotton At and Dt subgenomes could be stably transmitted into the next generation and were segregated in T1 populations according to Mendelian laws. The two transgene-clean editing plants with homozygous mutations, ce1 and ce2, were produced in T1. The Gh14-3-3d-edited plants in T2 showed a higher resistance to V. dahliae compared to the wild-type plants. The two transgene-clean lines were directly used as germplasms to breed defense cultivars, which could be free of the safety assessments for transgenic crops.
JW and FL conceived and designed the experiments. ZZ and XL performed the experiments. PW, QF, GH, and JX constructed the gene editing vectors and data analysis. XG and JW wrote the paper. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31471544 and 31771848) and sponsored by Funds for Creative Research Groups of China (31621005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00842/full#supplementary-material
FIGURE S1 | Schematic of the T-DNA region of plant gene editing vector pYLCRISPR/Cas9-CBD. The sequence of BsaI recognition motifs were highlighted in yellow, digestion site of BsaI were shown by red lines.
FIGURE S2 | Increased resistance of the Gh14-3-3d-silenced plants to V. dahliae. (A) Gh14-3-3d expression levels in silenced plants (TRV: Gh14-3-3d) and the control (TRV: 00) were determined by qPCR. The Gh14-3-3d expression level of the control was designated 1, and the average expression level in Gh14-3-3d -silenced plants was determined from 15 independent tested plants (3 replicates and 5 plants per replicate). (B) Disease symptoms on Gb14-3-3d-silenced plants and the control at 10th day post-inoculated with V. dahliae. (C) The disease index of the Gh14-3-3d-silenced plants and the control. Error bars represent the SD of three biological replicates (n ≥ 36). Asterisk indicates statistically significant differences compared to the control using Student’s t-test (P < 0.05).
FIGURE S3 | Amino acid alignment of the two copies of Gh14-3-3d encoding proteins, Gh14-3-3d-A and Gh14-3-3d-D.
FIGURE S4 | The putative transgenic plants in T0 in greenhouse. The regenerate plantlets were directly transplanted, or grafted on receptor seedlings in pots with 1:3 vermiculite and organic matter soil.
FIGURE S5 | Two copies of Gh14-3-3d affect the analyses of CRISPR/Cas9-mediated mutagenesis in mutant plants. (A) T7EI digestion assay of the mutants at the target sites. The PCR products for Gh14-3-3d from At, Dt, and total DNA (AtDt) were digested, response fragments in size (bp) were indicated on the right side. M, DNA molecular marker; T0–16 and T0–30 were represents plants for T7EI digestion assay. (B) The indels of Gh14-3-3d at target site of AtDt genome in T0–16 and T0–30 illustrated based on nucleotides and chromatograms by Sanger sequencing, respectively. The PAM sequence was shown in red. The PCR products were acquired by genome DNA and the universal primers, digested and sequenced for analyzing Gh14-3-3d gene editing.
FIGURE S6 | A novel indel that occurred in T1. T0–6–8 with a new indel (d3) was an offspring plant segregated from T0–6 mutant.
TABLE S1 | The PCR, RT-PCR, and qPCR primers.
TABLE S2 | Mutations detected in the putative CRISPR/Cas9 off target sites in WT, ce1, and ce2 plants.
DATA S1 | The artificial synthesizing sequence of sgRNA expression cassettes in vector of pYLCRISPR/Cas9-CBD.
Andersson, M., Turesson, H., Nicolia, A., Falt, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Atallah, Z. K., Bae, J., Jansky, S. H., Rouse, D. I., and Stevenson, W. R. (2007). Multiplex real-time quantitative PCR to detect and quantify Verticillium dahliae colonization in Potato lines that differ in response to Verticillium wilt. Phytopathology 97, 865–872. doi: 10.1094/PHYTO-97-7-0865
Bae, S., Park, J., and Kim, J.-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. doi: 10.1093/bioinformatics/btu048
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., and Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different Homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. doi: 10.1104/pp.17.00426
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Chen, X., Lu, X., Shu, N., Wang, S., Wang, J., Wang, D., et al. (2017). Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci. Rep. 7:44304. doi: 10.1038/srep44304
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Deng, F., Tu, L., Tan, J., Li, Y., Nie, Y., and Zhang, X. (2012). GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 158, 890–904. doi: 10.1104/pp.111.186742
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189. doi: 10.1016/j.devcel.2007.06.009
Gao, W., Long, L., Zhu, L. F., Xu, L., Gao, W. H., Sun, L. Q., et al. (2013). Proteomic and Virus-induced Gene Silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 8, 3690–3703. doi: 10.1074/mcp.M113.031013
Guan, X., Song, Q., and Chen, Z. J. (2014). Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci. 19, 516–528. doi: 10.1016/j.tplants.2014.04.007
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
Hill, M. K., Lyon, K. J., and Lyon, B. R. (1999). Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol. Biol. 40, 289–296. doi: 10.1023/A:1006146419544
Janga, M. R., Campbell, L. M., and Rathore, K. S. (2017). CRISPR/Cas9-mediated targeted mutagenesis in upland cotton (Gossypium hirsutum L.). Plant Mol. Biol. 94, 349–360. doi: 10.1007/s11103-017-0599-3
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Ostergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16:258. doi: 10.1186/s13059-015-0826-7
Li, C., He, X., Luo, X., Xu, L., Liu, L., Min, L., et al. (2014). Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 166, 2179–2194. doi: 10.1104/pp.114.246694
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, C., Unver, T., and Zhang, B. (2017). A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in Cotton (Gossypium hirsutum L.). Sci. Rep. 7:43902. doi: 10.1038/srep43902
Li, R., Li, R., Li, X., Fu, D., Zhu, B., Tian, H., et al. (2017). Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 16, 415–427. doi: 10.1111/pbi.12781
Li, C., and Zhang, B. (2016). MicroRNAs in control of plant development. J. Cell. Physiol. 231, 303–313. doi: 10.1002/jcp.25125
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, Y., Tu, L., Pettolino, F. A., Ji, S., Hao, J., Yuan, D., et al. (2016). GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol. J. 14, 951–963. doi: 10.1111/pbi.12450
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in Cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lu, Y., Ye, X., Guo, R., Huang, J., Wang, W., Tang, J., et al. (2017). Genome-wide targeted mutagenesis in Rice using the CRISPR/Cas9 system. Mol. Plant 10, 1242–1245. doi: 10.1016/j.molp.2017.06.007
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Meng, X., Yu, H., Zhang, Y., Zhuang, F., Song, X., Gao, S., et al. (2017). Construction of a genome-wide mutant library in Rice using CRISPR/Cas9. Mol. Plant 10, 1238–1241. doi: 10.1016/j.molp.2017.06.006
Min, L., Zhu, L., Tu, L., Deng, F., Yuan, D., and Zhang, X. (2013). Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J. 75, 823–835. doi: 10.1111/tpj.12245
Nakashita, H., Yasuda, M., Nitta, T., Asami, T., Fujioka, S., Arai, Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. doi: 10.1046/j.1365-313X.2003.01675.x
Obsil, T., Ghirlando, R., Klein, D. C., Ganguly, S., and Dyda, F. (2001). Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell 105, 257–267. doi: 10.1016/S0092-8674(01)00316-6
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shi, H., Wang, X., Li, D., Tang, W., Wang, H., Xu, W., et al. (2007). Molecular characterization of Cotton 14-3-3L gene preferentially expressed during fiber elongation. J. Genet. Genomics 34, 151–159. doi: 10.1016/s1673-8527(07)60016-2
Sun, G., Xie, F., and Zhang, B. (2011). Transcriptome-wide identification and stress properties of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). Funct. Integr. Genomics 11, 627–636. doi: 10.1007/s10142-011-0242-3
Sun, L., Zhu, L., Xu, L., Yuan, D., Min, L., and Zhang, X. (2014). Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5:5372. doi: 10.1038/ncomms6372
Tan, J., Tu, L., Deng, F., Hu, H., Nie, Y., and Zhang, X. (2013). A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 162, 86–95. doi: 10.1104/pp.112.212142
Tang, W., Tu, L., Yang, X., Tan, J., Deng, F., Hao, J., et al. (2014). The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 202, 509–520. doi: 10.1111/nph.12676
Upadhyay, S. K., Kumar, J., Alok, A., and Tuli, R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3 3, 2233–2238. doi: 10.1534/g3.113.008847
Vriet, C., Russinova, E., and Reuzeau, C. (2012). Boosting crop yields with plant steroids. Plant Cell 24, 842–857. doi: 10.1105/tpc.111.094912
Walford, S. A., Wu, Y., Llewellyn, D. J., and Dennis, E. S. (2011). GhMYB25-like: a key factor in early cotton fibre development. Plant J. 65, 785–797. doi: 10.1111/j.1365-313X.2010.04464.x
Wan, Q., Guan, X., Yang, N., Wu, H., Pan, M., Liu, B., et al. (2016). Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 210, 1298–1310. doi: 10.1111/nph.13860
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-Targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, H., Nagegowda, D. A., Rawat, R., Bouvier-Nave, P., Guo, D., Bach, T. J., et al. (2012). Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol. J. 10, 31–42. doi: 10.1111/j.1467-7652.2011.00631.x
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, L., Cook, A., Patrick, J. W., Chen, X. Y., and Ruan, Y. L. (2014). Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J. 78, 686–696. doi: 10.1111/tpj.12512
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, L., Wu, S. M., Zhu, Y., Fan, Q., Zhang, Z. N., Hu, G., et al. (2017). Functional characterization of a novel jasmonate ZIM-domain interactor (NINJA) from upland cotton (Gossypium hirsutum). Plant Physiol. Biochem. 112, 152–160. doi: 10.1016/j.plaphy.2017.01.005
Wang, P., Zhang, J., Sun, L., Ma, Y., Xu, J., Liang, S., et al. (2017). High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16, 137–150. doi: 10.1111/pbi.12755
Wang, S., Zhang, S., Wang, W., Xiong, X., Meng, F., and Cui, X. (2015). Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 34, 1473–1476. doi: 10.1007/s00299-015-1816-7
Wang, Y., Liang, C., Wu, S., Zhang, X., Tang, J., Jian, G., et al. (2016). Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant 9, 1436–1439. doi: 10.1016/j.molp.2016.06.013
Wendel, J. F. (1989). New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 86, 4132–4136. doi: 10.1073/pnas.86.11.4132
Wu, J., Zhang, X., Nie, Y., and Luo, X. (2005). High-efficiency transformation of Gossypium hirsutum embryogenic calli mediated by Agrobacterium tumefaciens and regeneration of insect-resistant plants. Plant Breed. 124, 142–146. doi: 10.1111/j.1439-0523.2004.01056.x
Yan, J., He, C., Wang, J., Mao, Z., Holaday, S. A., Allen, R. D., et al. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a ”stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45, 1007–1014. doi: 10.1093/pcp/pch115
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, Z. T., Zhou, Y., Li, Y., Shao, S. Q., Li, B. Y., Shi, H. Y., et al. (2010). Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J. Exp. Bot. 61, 3331–3344. doi: 10.1093/jxb/erq155
Keywords: Gossypium hirsutum L., allotetraploid cotton, simultaneous editing, transgene-clean, CRISPR/Cas9, Verticillium dahliae
Citation: Zhang Z, Ge X, Luo X, Wang P, Fan Q, Hu G, Xiao J, Li F and Wu J (2018) Simultaneous Editing of Two Copies of Gh14-3-3d Confers Enhanced Transgene-Clean Plant Defense Against Verticillium dahliae in Allotetraploid Upland Cotton. Front. Plant Sci. 9:842. doi: 10.3389/fpls.2018.00842
Received: 31 January 2018; Accepted: 30 May 2018;
Published: 28 June 2018.
Edited by:
Junhua Peng, Center for Life Sci&Tech of China National Seed Group Co. Ltd., ChinaReviewed by:
Kaijun Zhao, Institute of Crop Science (CAS), ChinaCopyright © 2018 Zhang, Ge, Luo, Wang, Fan, Hu, Xiao, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuguang Li, YXlsaWZ1Z0AxNjMuY29t Jiahe Wu, d3VqaWFoZUBpbS5hYy5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.