- 1College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Center for Plant Aging Research, Institute for Basic Science, Daegu, South Korea
- 3Department of New Biology, Daegu Gyeongbuk Institute of Science and Technology, Daegu, South Korea
Cellular calcium acts as a second messenger and regulates diverse developmental events and stress responses. Cytosolic calcium has long been considered as an important regulator of senescence, however, the role of Ca2+ in plant senescence has remained elusive. Here we show that the Calmodulin 1 (CaM1) gene, which encodes Ca2+-binding protein calmodulin 1, positively regulates leaf senescence in Arabidopsis. Yellowing of leaves, accumulation of reactive oxygen species (ROS), and expression of the senescence-associated gene 12 (SAG12) were significantly enhanced in CaM1 overexpression plants. In contrast, abscisic acid (ABA)-triggered ROS production and stomatal closure were reduced in amiRNA-CaM1 plants. We found a positive-feedback regulation loop among three signaling components, CaM1, RPK1, and RbohF, which physically associate with each other. RPK1 positively regulates the expression of the CaM1 gene, and the CaM1 protein, in turn, up-regulates RbohF gene expression. Interestingly, the expression of CaM1 was down-regulated in rbohD, rbohF, and rbohD/F mutants. We show that CaM1 positively regulates ROS production, leaf senescence, and ABA response in Arabidopsis.
Introduction
Leaf senescence is the terminal stage of leaf development and is genetically programmed. Apparent morphological changes involved in leaf senescence include the yellowing of leaves caused by the degradation of chlorophyll, followed by reduction in photosynthesis and protein synthesis. During senescence, the metabolism and structure of leaf cells continuously change to effectively utilize plant nutrients for the developing parts of the plant, including young leaves, seeds, and fruits (Lim et al., 2007).
Calcium is a universal second messenger that exerts an allosteric effect on many enzymes and proteins in various cellular responses. In plants, calcium signaling is evoked by endogenous and environmental cues, such as drought, salt or osmotic stresses, temperature, light, and plant hormones (Dodd et al., 2010; Steinhorst and Kudla, 2014; Edel and Kudla, 2016; Ranty et al., 2016). Ca2+ ions appear to play an important role in plant senescence as well. For instance, exogenously supplied Ca2+ delays the senescence of a detached leaf (Poovaiah and Leopold, 1973), and the Ca2+ ionophore A23187 rescues MeJA-mediated leaf senescence (Chou and Kao, 1992). Moreover, Ca2+-mediated (NO production negatively regulates the expression of senescence-associated genes and production of H2O2 during the initiation of leaf senescence (Ma et al., 2010).
Calmodulin (CaM), a small Ca2+-binding protein, is one of the major calcium sensor proteins conserved in eukaryotes (Chin and Means, 2000; McCormack et al., 2005). Ca2+ binding to CaMs causes a conformational change in the protein structure, thereby modifying its interaction with various target proteins, which leads to the transduction of Ca2+ signals (Zielinski, 1998, 2002). In Arabidopsis, seven genes encode four CaM isoforms: CaM1/CaM4, CaM2/CaM3/CaM5, CaM6, and CaM7 (McCormack et al., 2005). Four amino acid substitutions differentiate CaM7 from CaM1/CaM4, and one amino acid substitutions differentiate CaM7 from CaM2/CaM3/CaM5, and CaM6. CaM7 is a transcriptional regulator that directly interacts with the promoters of several light-inducible genes, contributing to the regulation of photomorphogenesis (Kushwaha et al., 2008).
Receptor-like Protein Kinase 1 (RPK1) localizes to the plasma membrane and plays an important role in embryo development, plant growth, ABA signaling, and stress responses (Hong et al., 1997; Osakabe et al., 2005, 2010; Nodine et al., 2007; Nodine and Tax, 2008). Additionally, RPK1 positively regulates leaf aging (Lee et al., 2011). The RbohF, an NADPH oxidase, produces reactive oxidation species (ROS) in response to biotic and abiotic stresses in Arabidopsis (Torres et al., 2002; Kwak et al., 2003; Joo et al., 2005; Desikan et al., 2006; Zhang et al., 2009). The presence of EF-hand Ca2+ binding motifs on NADPH oxidases has suggested a regulatory effect of Ca2+ on the enzymatic activity (Torres et al., 1998, 2002), which was further supported by the finding that Ca2+ and phosphorylation synergistically activate RbohD NADPH oxidase (Ogasawara et al., 2008). Furthermore, it has been recently shown that RbohF is responsible for RPK1-mediated ROS production, senescence-associated gene expression, and age-induced cell death (Koo et al., 2017).
Several lines of evidence have suggested that cytosolic Ca2+ acts as a regulator of senescence in plant, but how senescence is regulated by cytosolic Ca2+ remains elusive. Here, we report that Arabidopsis CaM1 plays a positive role in RPK1-mediated leaf senescence and ABA response. Senescence phenotypes, including leaf yellowing, ROS accumulation, and expression of the SAG12 were drastically promoted in CaM1-overexpressing plants, whereas plants expressing amiRNA-CaM1 showed no visible senescence phenotype, which is likely to be due to functional redundancy in the calmodulin gene family. Expression analyses revealed that CaM1 is positively regulated by RPK1, RbohF by CaM1, and CaM1 by RbohD and RbohF, suggesting a positive-feedback loop among the three signaling components at the transcriptional level.
Results
CaM1 Gene Expression Is Associated With Aging
To identify calmodulin genes that are involved in plant senescence, in silico analysis using publicly available microarray data was conducted and showed that the expression of Arabidopsis CaM genes can be divided into two groups during leaf senescence: age-dependent increase (CaM1, CaM3, and CaM4) and age-dependent decrease (CaM2, CaM5, and CaM7) (no data was available for CaM6; Figure 1A) (Schmid et al., 2005). This implies that CaM1, CaM3, and CaM4 probably play a positive role in Arabidopsis leaf senescence. To verify the microarray data, we examined age dependent- expression pattern of the CaM genes: The fourth rosette leaves of Arabidopsis plants were collected every 3 days starting with 13-day-old plants and transcript levels of the CaM1, CaM2, and CaM4 genes were analyzed using quantitative RT-PCR (Figure 1B). We found that CaM1 and CaM4 were indeed up-regulated in the older leaves compared with the younger leaves. The expression pattern was similar to that of the leaf senescence marker, SAG12 whose expression gradually increased from 13 days after sowing in an age-dependent manner (Figure 1C) (Woo et al., 2001). By contrast, the expression of CaM2 was down-regulated (Figure 1B).
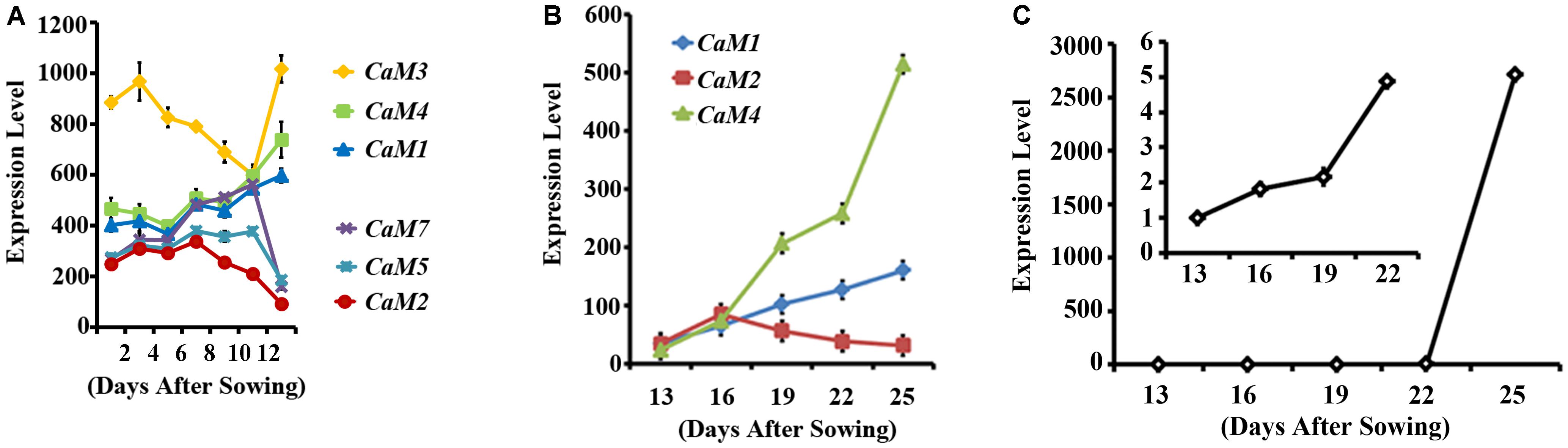
FIGURE 1. Expression profiles of CaM family members in leaves during senescence. (A) Expression of the CaM genes in rosette leaves, including leaves undergoing senescence 35 days after sowing. Data corresponding to three arrays for each data point were obtained from Genevestigator (https://genevestigator.com/gv/). (B) qRT-PCR data showing the expression of CaM1, CaM2, and CaM4 genes in the fourth rosette leaves at the indicated time points. (C) qRT-PCR analysis of SAG12 expression in the fourth rosette leaves of WT plants. The inset shows SAG12 expression until 22 days after sowing (DAS). Expression levels of genes were normalized to Actin2. Error bars represent means ± SEM of three independent experiments.
In silico analysis showed that the expression level of CaM1 was similar to that of CaM4 in most of the tissues, except for reproductive organs (Supplementary Figures S1A,B), which was confirmed by qRT-PCR analyses (Supplementary Figure S1C). CaM1 is highly expressed in pollen whereas CaM4 is highly expressed in dry seeds (Supplementary Figures S1A,B). To verify the expression pattern of CaM1, at least 10 independent pCaM1::GUS transgenic lines were subjected to GUS histochemical analysis. Data revealed that CaM1 was highly expressed in vascular bundles of cotyledons, roots, and both rosette and cauline leaves (Figures 2A–E). CaM1 was also detected in reproductive tissues, including flowers and siliques (Figures 2F–K). Dissection of stained flowers revealed that CaM1 was expressed in sepals, petals, carpels and filaments, but not in anthers (Figures 2I–K). Interestingly, CaM1 was highly expressed in guard cells (Figure 2B), suggesting a role of CaM1 in stomatal development and/or movement.
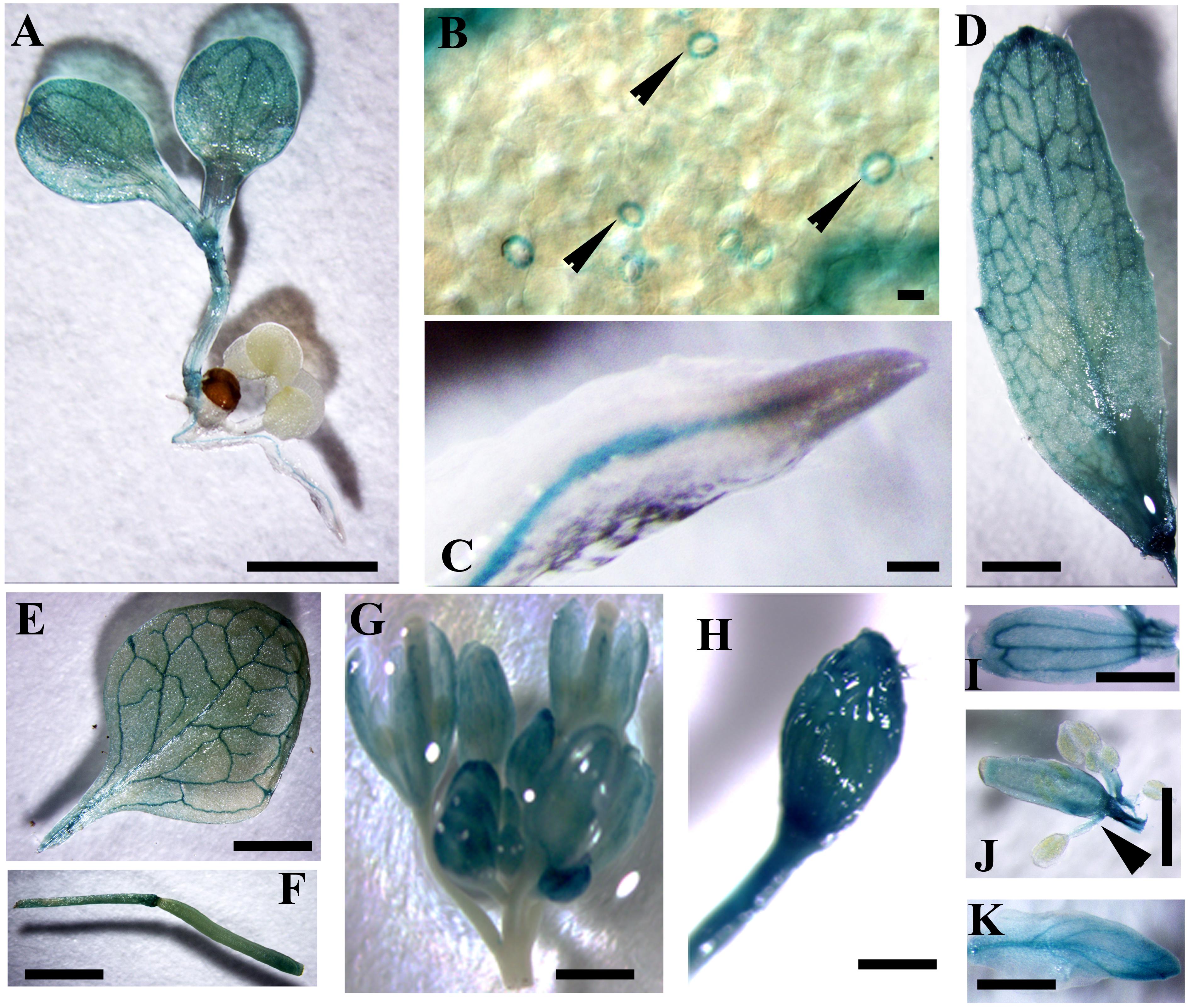
FIGURE 2. Visualization of CaM1 expression in Arabidopsis transgenic plants harboring a CaM1pro-GUS construct. Histochemical GUS staining images of seedlings (A,B), roots (C), cauline leaves (D), rosette leaves (E), silliques (F), flowers (G,H), petals (I), anthers (J), and sepals (K) are shown. Arrowheads indicate GUS staining in the guard cells of leaf (B) and in filaments (J). Scale bar, 1 cm.
Leaf Senescence Is Accelerated in CaM1-Overexpressing Plants
Previously, we have shown that CaM4 plays a role in RPK1-mediated leaf senescence in Arabidopsis (Koo et al., 2017). Thus, CaM1 was chosen to investigate its role in plant senescence. As a first step, transgenic plants expressing 35S::CaM1-GFP were generated. Western blot analyses using anti-GFP antibody showed enhanced accumulation of CaM1 in the transgenic plants (Figure 3A). Leaves of three independent 35S::CaM1-GFP transgenic lines turned yellow earlier than the WT (Figures 3B,C). No other growth abnormalities including seed size and mass were observed in the 35S::CaM1-GFP transgenic lines compared with the WT (Supplementary Figure S2). It is known that the production of ROS is induced during senescence in both animals and plants (Lim et al., 2007; Vigneron and Vousden, 2010). Thus, we monitored the accumulation of H2O2 and superoxide, respectively in the leaves of WT and 35S::CaM1-GFP transgenic lines at different developmental stages using DAB and NBT staining. We found that ROS accumulation was detected earlier in the transgenic plants than in WT (Figures 3D–F). Higher accumulation of H2O2 and superoxide was detected at 16 and 19 days, respectively, in the transgenic plant leaves after sowing (Figures 3E,F).
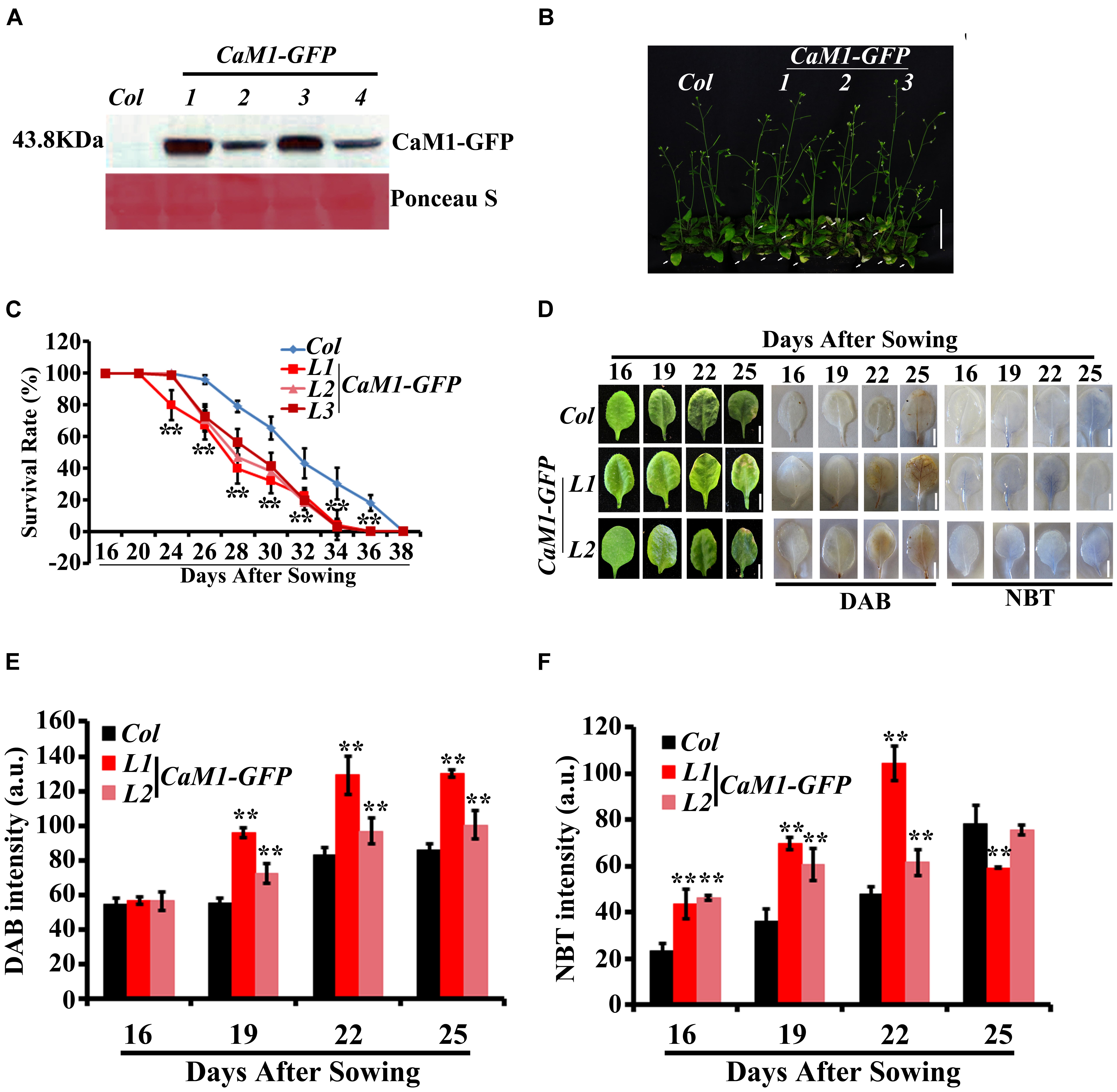
FIGURE 3. Age-dependent leaf senescence is accelerated in 35S::CaM1-GFP transgenic lines. (A) Detection of CaM1-GFP fusion protein in four independent transgenic lines using Western blot analysis with anti-GFP antibody. Approximately 10-μg protein of each sample was loaded on the gel and stained with Ponceau S. (B) Yellowing phenotypes of rosette leaves of 35S::CaM1-GFP transgenic lines. The picture was taken 31 days after sowing. Arrows indicate leaf yellowing. (C) Survival rates of WT and CaM1-GFP. Plants with third and fourth rosette leaves showing yellowing were counted. Data are from two independent experiments. For each experiment, 20 plants were analyzed. (D–F) Analysis of superoxide and H2O2 levels using NBT and DAB, respectively, in the fourth rosette leaves of WT and 35S::CaM1-GFP transgenic lines L1 and L2 at the indicated time points (D). Quantitative analysis of NBT (E) and DAB (F). Error bars represent means ± SEM of three independent experiments. Statistical analysis was performed using heteroscedastic t-test (∗p < 0.05; ∗∗p < 0.01). Scale bar, 1 cm.
To further examine the role of CaM1 in leaf senescence, we generated transgenic plants in which the expression of CaM1 was knocked down using an artificial microRNA targeting CaM1 (amiRNA-CaM1). In the amiRNA-CaM1 transgenic lines, the transcript levels of CaM2, CaM3, and CaM4 were not significantly changed, whereas CaM1 transcript level was significantly down-regulated compared with the WT (Figures 4A,B). This result implies that the amiRNA-CaM1 specifically down-regulates CaM1 transcripts, allowing us to examine cellular responses that CaM1 particularly mediates.
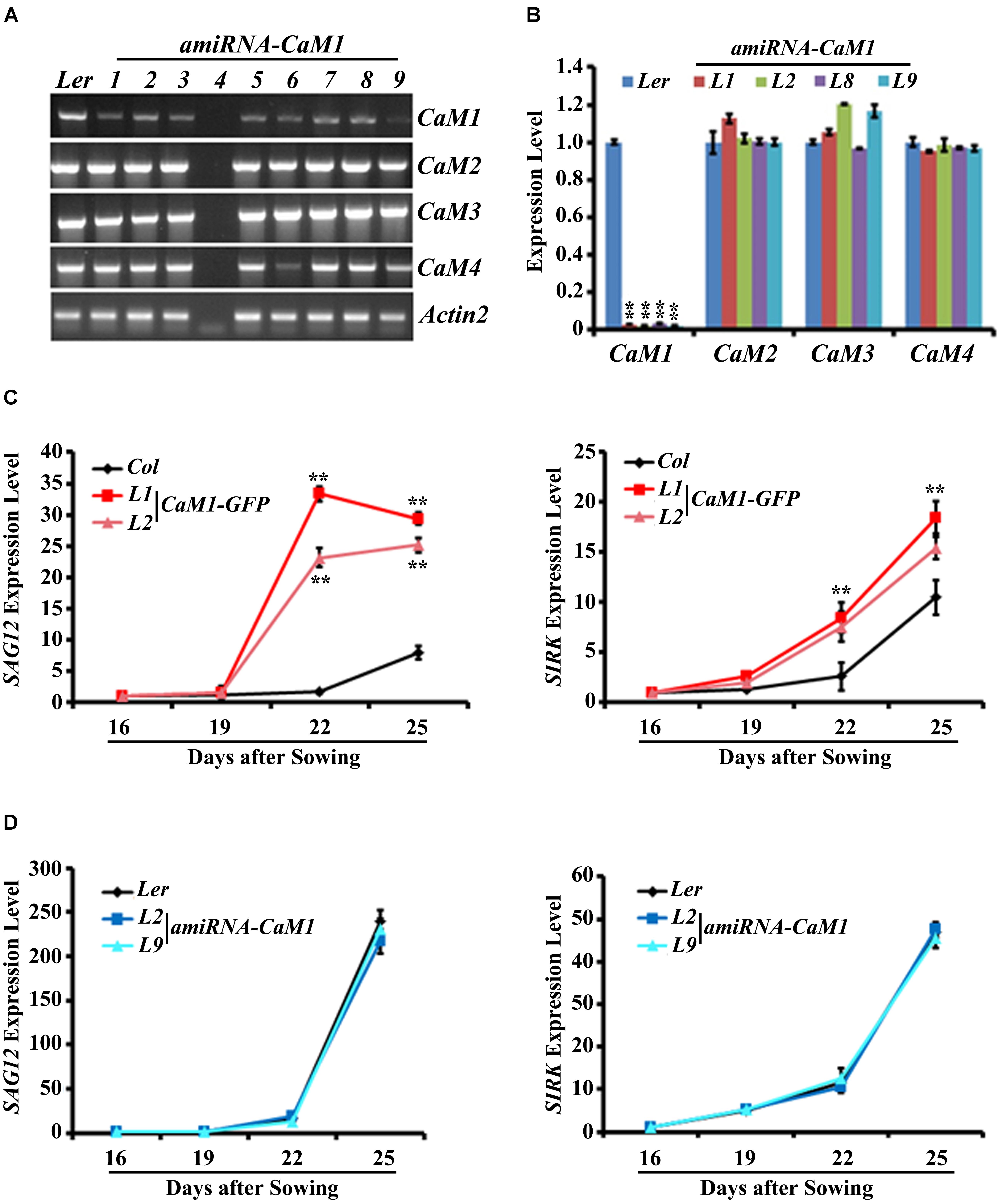
FIGURE 4. The expression of senescence marker genes is accelerated in the 35S::CaM1-GFP transgenic lines. (A,B) Expression analysis of CaM genes in T3 homozygous 35S::amiRNA-CaM1 lines by RT-PCR (A) and qRT-PCR (B). (C,D) qRT-PCR analysis of senescence-related genes in 35S::CaM1-GFP transgenic lines (C) and 35S::amiRNA-CaM1 plants (D). Expression levels of the genes were normalized to Actin2. Error bars represent means ± SEM of three independent experiments. Statistical analysis was performed using heteroscedastic t-test (∗∗p < 0.01).
qRT-PCR analyses showed that early leaf senescence phenotype of the 35S::CaM1-GFP transgenic lines was strongly correlated with the drastically enhanced expression of senescence markers SAG12, SIRK, ATG2, and ATG5 compared with the WT (Figure 4C and Supplementary Figure S3A). This suggests that CaM1 overexpression results in various senescence-associated changes, including yellowing of leaves, enhanced production of H2O2 and superoxide, and up-regulation of the senescence marker genes. In contrast, no altered senescence phenotypes were observed in the amiRNA-CaM1 transgenic lines, and the expression level of SAG12, SIRK, ATG2, and ATG5 in the amiRNA-CaM1 transgenic lines was comparable with that of the WT (Figure 4D and Supplementary Figure S3B). cam4 knockout mutants displayed no altered senescence phenotype (Supplementary Figures S3C,D). This result suggests that there is functional redundancy between CaM1 and CaM4 and that CaM1/CaM4 functions as a positive regulator of age-dependent leaf senescence.
CaM1 Positively Regulates ABA-Induced ROS Production
We found that the production of H2O2 and superoxide was increased in the 35S::CaM1-GFP transgenic plants during leaf senescence (Figure 3C). Since ABA is also known to induce ROS production (Pei et al., 2000; Kwak et al., 2003) as well as leaf senescence (Zhao et al., 2016), we examined whether CaM1 is linked to ABA-mediated ROS production. The expression of CaM1 was induced within 30 min of ABA treatment, reaching a peak at 3 h (Figure 5A). We also determined ROS levels in leaves of 3-week-old plants before and after the ABA treatment using DAB and NBT staining. The steady-state H2O2 level in 35S::CaM1-GFP plants was higher than in WT plants prior to the ABA treatment (Figure 5B). Although ABA induced H2O2 and superoxide production in both WT and the 35S::CaM1-GFP transgenic plants, we found a higher production of H2O2 and superoxide in the 35S::CaM1-GFP transgenic plants (Figure 5B). By contrast, ABA-induced ROS production was reduced in amiRNA-CaM1 transgenic lines (Figure 5C). These results imply that CaM1 positively regulates ABA-induced ROS production.
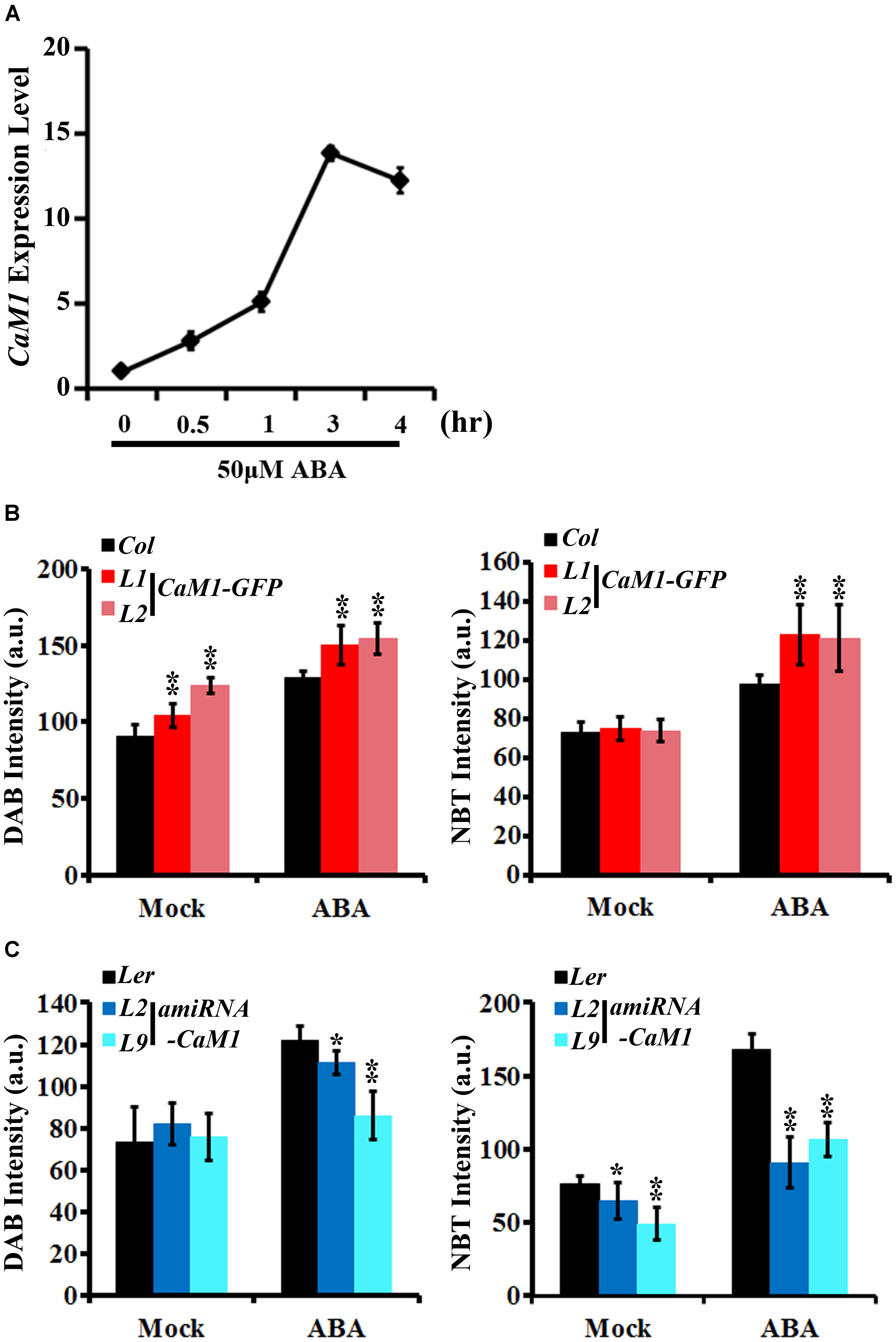
FIGURE 5. CaM1 positively regulates ABA-induced ROS production. (A) qRT-PCR analysis reveals that the expression of CaM1 is induced 0.5 h after ABA treatment (50 μM). Actin2 was used as an internal control. (B,C) Quantitative analyses of ROS staining in the fourth rosette leaves of 3-week-old plants expressing 35S::CaM1-GFP (B) and 35S::amiRNA-CaM1 (C). Leaves were treated with 100 μM ABA for 1 h and stained with DAB and NBT to visualize accumulation of H2O2 and superoxide, respectively. Leaves without ABA treatment (mock) were used as a control. Error bars represent means ± SEM of two independent experiments. Each data point represents 10 leaves. Statistical analysis was performed using heteroscedastic t-test (∗p < 0.05; ∗∗p < 0.01).
Stomatal aperture is one of the targets of ABA-mediated signaling in plants. Furthermore, ROS have been shown to act as a positive regulator of ABA signaling in guard cells (Pei et al., 2000; Kwak et al., 2003). Thus, we investigated whether CaM1 functions in guard cell ABA response. We determined ROS levels in guard cells of the fourth rosette leaves harvested from 25-day-old plants before and after ABA treatment. H2-DCFDA mainly detects H2O2 and can be easily quantitated in guard cells (Kwak et al., 2003). The steady-state ROS level in 35S::CaM1-GFP guard cells was higher than in WT guard cells (Figure 6A). Although no significant differences were detected between WT and the 35S::CaM1-GFP transgenic lines after ABA treatment (Figure 6A), it is likely to be due to the saturation of DCF fluorescence in the cells analyzed. By contrast, ABA failed to trigger ROS production in guard cells of amiRNA-CaM1 transgenic plants (Figure 6B). Overexpression of CaM1 enhanced ABA-induced stomatal closure, whereas down-regulation of CaM1 inhibited ABA-induced stomatal closure (Figures 6C,D). These results imply that CaM1 positively regulates ABA-induced ROS production in guard cells, thereby modulating stomatal movements.
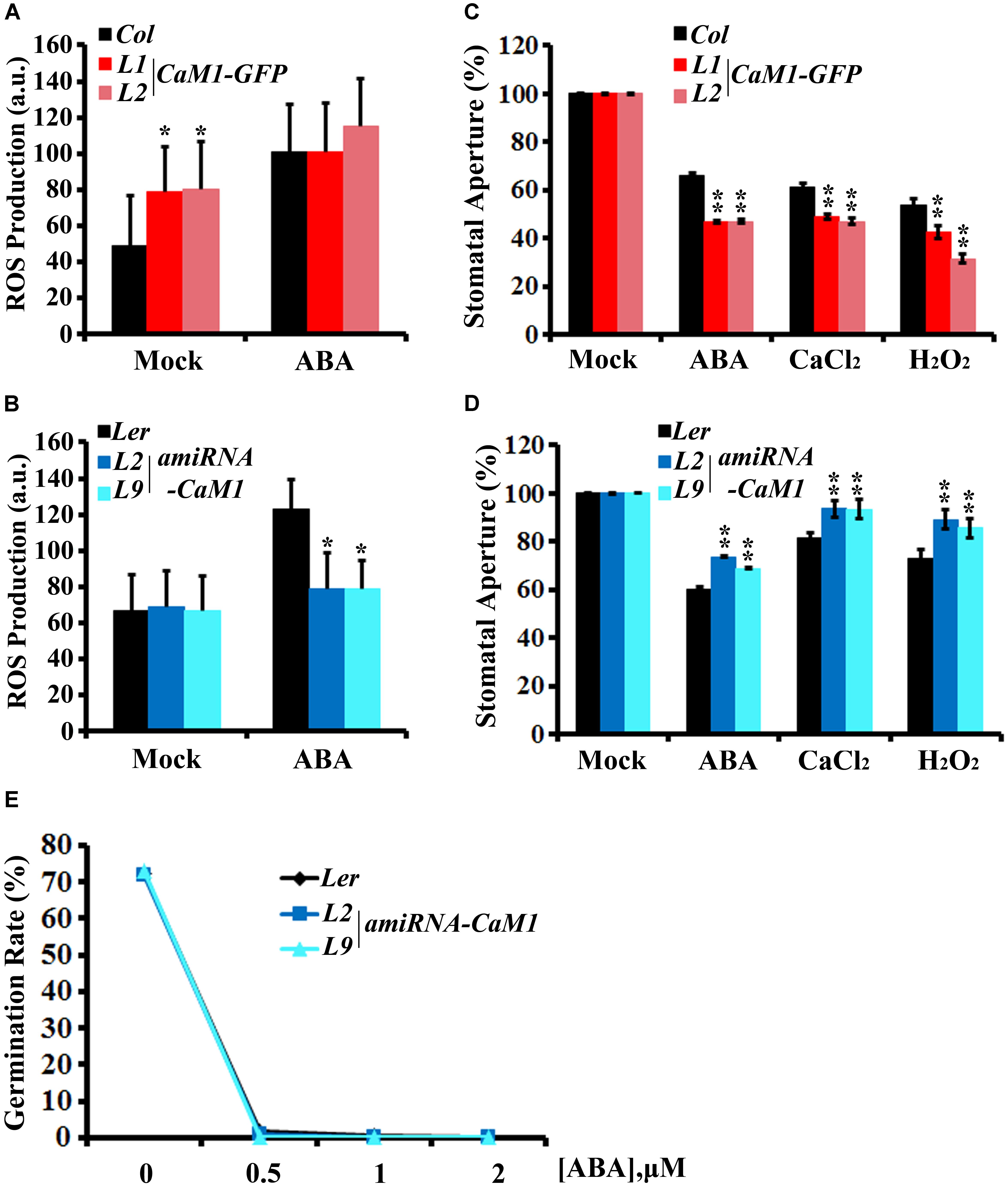
FIGURE 6. CaM1 mediates ABA-induced ROS production in guard cells and stomatal closure. (A,B) ABA-induced ROS accumulation in guard cells of 35S::CaM1-GFP plants (A) and of the 35S::amiRNA-CaM1 transgenic plants (B). ROS levels were measured 30 min after ABA treatment (50 μM) using 2′7′-dichlorofluorescein diacetate. Error bars represent means ± SEM of three independent experiments (n = 50). (C,D) ABA (5 μM)-, CaCl2 (2 mM)-, and H2O2 (100 μM)-induced stomatal closure in 35S::CaM1-GFP plants (C) and amiRNA-CaM1 plants (D) (n = 100). (E) Germination rate of WT (Ler) and transgenic amiRNA-CaM1 L2 and L9 seeds were measured after 7 days of incubation on MS medium containing 0, 0.5, 1, or 2 μM ABA without sucrose. Error bars represent means ± SEM of three independent experiments. Statistical analysis was performed using heteroscedastic t-test (∗p < 0.05; ∗∗p < 0.01).
Calcium and H2O2 are second messengers that induce stomatal closure (Pei et al., 2000; Zhang et al., 2001; Kwak et al., 2003). In the CaM1 overexpression plants, calcium- and H2O2-induced stomatal closure was enhanced (Figure 6C). In contrast, down-regulation of CaM1 reduced the calcium- and H2O2-induced stomatal closure (Figure 6D), implying that CaM1 functions in calcium- and H2O2-mediated stomatal closure. More importantly, even though there is functional redundancy between CaM1 and CaM4 in leaf senescence (Figure 4 and Supplementary Figure S3) and ABA-mediated seed germination (Figure 6E), CaM1 appears to play a unique role in the regulation of ROS production in guard cells.
Positive-Feedback Regulation Among RPK1, CaM1, and RbohF
Peptide sequence alignments of CaM proteins revealed a high level of sequence similarity (Supplementary Figure S4). Phylogenetic analysis indicated that CaM1 is closest to CaM4 (Supplementary Figure S5). Although the sequence similarity between CaM1 and CaM4 coding sequences was 89.56% at the nucleotide level, the amino acid sequences encoded by these genes were identical (Supplementary Figure S4). We have previously shown that CaM4 physically interacts with RPK1 and RbohF (Koo et al., 2017). To test whether CaM1 also acts with RPK1 and RbohF and to investigate potential regulation among RPK1, CaM1, and RbohF at the transcriptional level, we examined the expression of CaM1 in rpk1-5 null mutants and iRPK1 transgenic plants in which the expression of RPK1 is under the control of ecdysone-inducible promoter (Lee et al., 2011). In rpk1-5, the expression of CaM1 was significantly down-regulated in leaves of 25-day-old plants (Figure 7A; p < 0.01). By contrast, RPK1 expression was notably up-regulated in the iRPK1 plants when treated with methoxyfenozide, the chemical inducer of the promoter (Padidam, 2003) (Figure 7B). These results indicate that the expression of CaM1 is positively regulated by RPK1. However, no significant changes in the expression of RPK1 were detected in the 35S::CaM1-GFP or 35S::amiRNA-CaM1 transgenic lines (data not shown), suggesting that RPK1 is not transcriptionally regulated by CaM1.
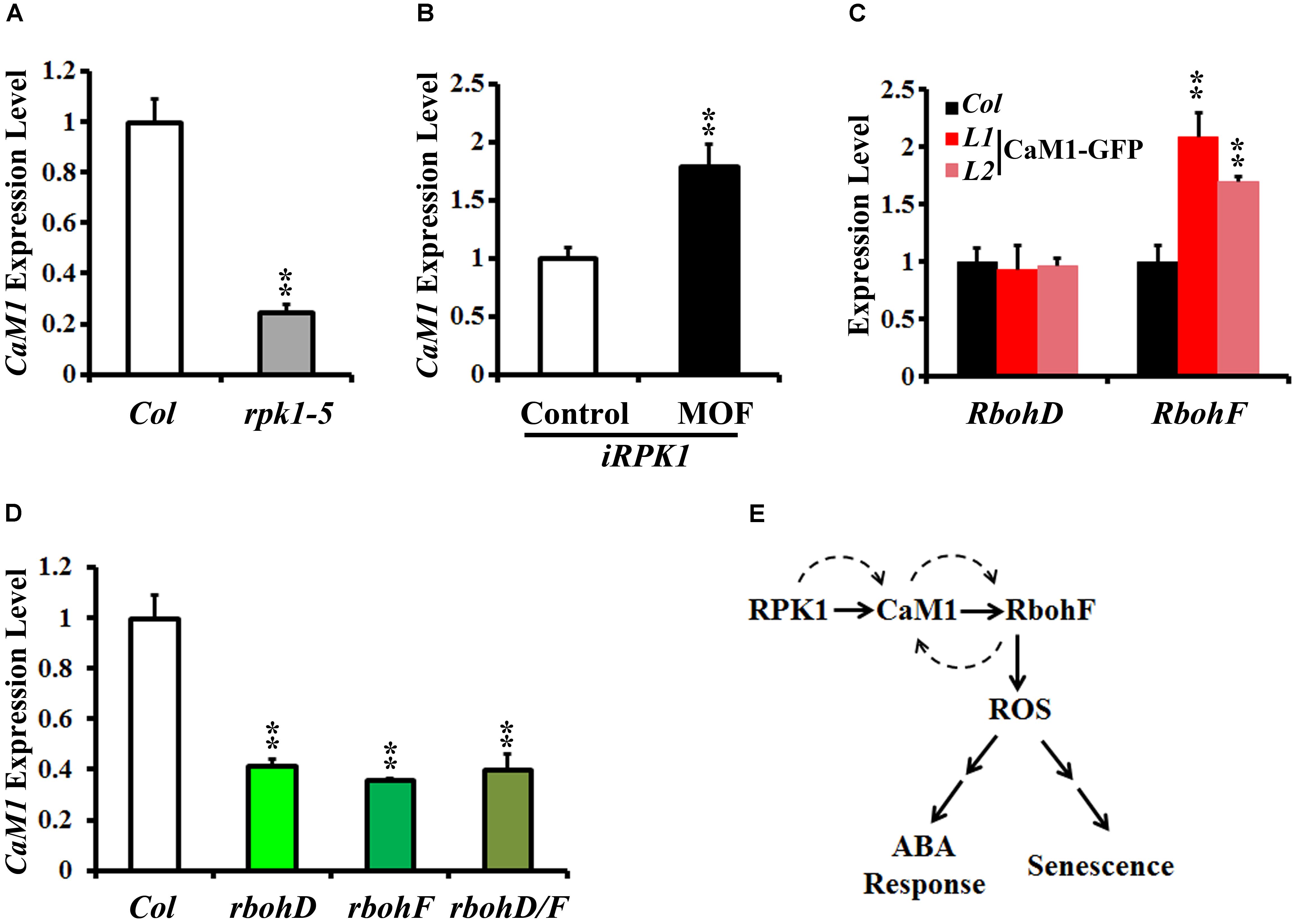
FIGURE 7. RPK1 positively regulates the expression of CaM1, which in turn activates a positive-feedback loop between CaM1 and RbohD/F. (A,B) qRT-PCR analysis of CaM1 expression in rpk1-5 mutants (A) and iRPK1 transgenic plants after methoxyfenozide (MOF) treatment (B). (C) qRT-PCR analysis of RbohD and RbohF transcript level in the 35S:CaM1-GFP transgenic plants. (D) qRT-PCR analysis of CaM1 in rbohD, rbohF, and rbohD/F mutants. Error bars represent means ± SEM of three independent experiments. (E) A working model for the regulation among RPK1, CaM1, and RbohF genes. Solid arrows indicate cellular events at the protein level, and dashed arrows indicate feed-forward and feedback regulation at the transcription level. Statistical analysis was performed using heteroscedastic t-test (∗∗p < 0.01).
In addition, we found that the expression of RbohF was up-regulated in the 25-day-old 35S::CaM1-GFP transgenic plants, whereas the expression of RbohD was not altered (Figure 7C). Interestingly, the expression of CaM1 was down-regulated in rbohD, rbohF, and rbohD/F double mutants (Figure 7D). Taken together, these data suggest a positive-feedback loop among the CaM1, RbohF, and RPK1 genes at the transcriptional level (Figure 7E).
Materials and Methods
Plant Materials and Growth Conditions
T-DNA insertional line GK-309E09 (cam4, AT1G66410) was obtained from the Arabidopsis Biological Resource Center. Seeds of wild type (WT) Arabidopsis, cam4, and 35S::CaM1-GFP (Columbia background) and 35S::amiRNA-CaM1 (Landsberg background) transgenic lines were germinated in soil. Seedlings were grown in the growth chamber at 21°C ± 1°C under 16 h light/8 h dark photoperiod. Plants at the 3–5-week-old stage were used for various experiments described below.
Dichlorofluorescein Diacetate (H2DCF-DA) Assay of Guard Cells
The production of H2O2 in guard cells was examined using 2′7′-(H2DCF-DA, Molecular Probes, Eugene, OR, United States) as described previously (Murata et al., 2001) with slight modifications. Epidermal strips were prepared from 4 to 5-week-old WT, and transgenic 35S::CaM1-GFP and 35S::amiRNA-CaM1 plants using a blender and incubated in buffer (5 mM KCl, 10 mM MES–Tris, pH 6.15) under white light (95 E m-2 s-1) for 2 h. Subsequently, H2DCF-DA (50 μM) was added to the solution containing the epidermal strips and the mixture was incubated on an orbital shaker at 70 rpm for 30 min. Finally, 50 μM ABA was added to the buffer and incubated for 10 min. Images of guard cells were taken under UV light (one 2-second UV exposure per sample) using a fluorescence microscope equipped with a digital camera (Axiovert 200, Zeiss) (Murata et al., 2001). The fluorescence intensity of guard cells was measured using Image J.
Constructs and Plant Transformation
Artificial microRNA of CaM1 (amiRNA-CaM1) was designed by WMD33. Primers, including attb1-oligo A, attb2-oligo B, CaM1-I-miR-s, CaM1-II-miR-a, CaM1-III-miR∗s, and CaM1-IV miR∗a were used to PCR amplify on pRS300 (Schwab et al., 2006). The PCR product, amiRNA-CaM1 was first cloned into the Entry vector pDONRTM/Zeo (12535-035, Invitrogen) and then into the pMDC32 vector using Gateway LR cloning (Gateway® LR Clonase® II Enzyme mix, 11791-100, Invitrogen) (Curtis and Grossniklaus, 2003). The promoter region of CaM1 was amplified by pair of primers CaM1p-attB1 and CaM1p-attB2, and the coding region of CaM1 was amplified by pair of primers CaM1-attB1 and CaM1-attB2. The PCR products were cloned into the Entry vector pDONRTM/Zeo and then into the pMDC162 vector for pCaM1::GUS and pMDC43 for 35S::CaM1-GFP (Schwab et al., 2006). These constructs were introduced into WT Arabidopsis using Agrobacterium-mediated transformation via the floral dip method (Clough and Bent, 1998). The primer sequences used for plasmid construction are provided in Table 1.
RT-PCR Analysis
Total RNA was extracted from WT and transgenic lines using the TRIzol reagent (15596-026, Invitrogen) and subsequently used for cDNA synthesis using RevertAid Reverse Transcriptase kit (#EP0441, Thermo). The primer pairs CaM1-qs and CaM2-qa, CaM2-qs and CaM1-qa, CaM4-qs/-qa, RPK1-qs/-qa, SAG12-qs/-qa, SIRK-qs/-qa, ATG2-qs/-qa, ATG5-qs/-qa, RbohD-qs/-qa, and RbohF-qs/-qa were used to examine the expression of CaM1, CaM2, CaM4, RPK1, SAG12, SIRK, ATG2, ATG5, RbohD, and RbohF, respectively, using qPCR. Actin2 amplified using the primer pair Actin-1/-2 was used as an internal positive control.
For semi-quantitative RT-PCR, equal amounts of first-strand cDNAs were used as templates for PCR amplification. The primer pairs CaM1-s/-a, CaM2-s/-a, CaM3-s/-a, and CaM4-s/-a were used to analyze the expression pattern of CaM1, CaM2, CaM3, and CaM4, respectively, in the different amiRNA-CaM1 transgenic lines. The Actin2 gene was used as an internal control. The primer sequences used for qRT and RT-PCR analysis are provided in Table 2.
Diaminobenzidine (DAB) and Nitroblue Tetrazolium (NBT) Staining
For all histochemical staining experiments, fourth rosette leaves were detached at different time points (13, 16, 19, 22, and 25 days) and treated with 100 μM ABA, 100 μM H2O2, or 2 mM CaCl2 for one hour. DAB and NBT staining were performed as described (Lee et al., 2011). The leaves were submerged in NBT solution [1 mg/mL (NBT, N5514, Sigma), 10 mM NaN3, and 10 mM potassium phosphate, pH 7.8] or DAB solution [1mg/ml 3,3′-DAB, D12384, Sigma), pH 3.8] and stained for 30 min at room temperature. Samples were then boiled in 95% ethanol for 10 min and stored in 60% glycerol. After staining, all samples were mounted on slides and photographed using a digital camera (G12, Canon). To quantify DAB and NBT staining, the stained pixels were obtained from 6 to 12 leaves per genotype using the channels function in Adobe Photoshop and their intensity was measured using the TotalLab100 program (Nonlinear Dynamics).
Stomata Movement Assay
Stomatal assays were performed as described (Desikan et al., 2002). Briefly, epidermal strips were prepared from 3- to 4-week-old rosette leaves and floated abaxial side down in 10 mM MES buffer (pH 6.15) containing 20 mM KCl for 2.5–3 h under white light (95 E m-2 s-1) to open the stomata. Following this, 100 μM H2O2 or 2 mM CaCl2 was added to the buffer and incubated for another 1–2 h to assay stomatal closure. Pictures of stomata were taken using a microscope equipped with a digital camera (Axiovert 200, Zeiss). The guard cell aperture was analyzed using Image J.
Seed Germination Assay
Dry seeds were collected and stored in a dehumidifier cabinet for at least 2 months before testing the germination capacity of seeds. Seeds were surface-sterilized using 20% commercial Clorox bleach for 15 min, and washed five times with sterile water. At least 100 sterilized seeds were plated on MS medium (MSP02-10LT, Caisson) containing 0.8% (w/v) Bacto Agar (Difco, BD) supplemented with different concentrations (0.5, 1, 2 μM) of ABA solution (A4906, Sigma-Aldrich) prepared in methanol. Seeds plated on MS media without ABA were used as control. Seeds were stratified in the dark at 4°C for 3 d and then transferred to the growth chamber maintained at 22°C with a 16 h light/8 h dark photoperiod. Germination was defined as the first sign of emergence of green cotyledons and scored daily for 7 days. The germination results were calculated based on at least three independent experiments.
Histochemical Staining
Positive transgenic plants were identified using GUS staining as described (Jefferson et al., 1987). Pictures were taken using a digital camera (G12, Canon) or microscope (STEMI SV6, Zeiss) equipped with a digital camera.
Phylogenetic and Alignment Analysis
Homolog sequences of Zea mays (gi|162463080, gi|162463001, gi|162462264, gi| 162464382, gi|226490894, gi|226496461, gi|226507438, gi|226507713, gi| 226958443, and gi|293334895), Glycine max (gi|351721559, gi|310563, gi|351721835, gi|351722047, gi|170072, gi|170076, and gi|170074), Chlamydomonas reinhardtii (gi|159490918 and gi|158280344), Acyrthosiphon pisum (gi|209870032), Triticum aestivum (gi|291246022), Brassica napus (gi|497992), B. juncea (gi|899058), B. oleracea (gi|374922809, gi|374922811, gi|374922813, and gi|374922807), Hevea brasiliensis (gi|313767030), Arabidopsis thaliana (CaM1 to CaM7), Medicago truncatula (gi|355509904 and gi|357477127), Vitis vinifera (VIT_18s0001g03880.t01) and Oryza sativa [LOC_Os05g41200.1 (OsCaM9), LOC_Os07g48780 (OsCaM1-1), LOC_Os03g20370 (OsCaM1-2), LOC_Os01g16240 (OsCaM1-3), LOC_Os11g03980 (OsCLM2) and gi|17066588] were obtained from NCBI4 or ABRC (see footnote 2). Sequence alignments were generated using Clustal Omega5. Alignments between different species were adjusted for the construction of phylogenetic tree. Neighbor-joining analyses were performed using MEGA5 (Tamura et al., 2011) with pair-wise deletion, Poisson correction set for the distance model and 1,000 bootstrap replicates.
Analysis of Seed Yield
Average seed mass was determined by weighing 1,000 dry seeds. The weight of at least five independent batches was measured. To analyze relative seed size, seed images were captured on an Epson V370 scanner with the supplied software, which were then analyzed by SmartGrain software (Tanabata et al., 2012). More than 200 seeds per plant were analyzed.
Discussion
In this report, we investigated the function of the senescence-induced gene CaM1. Overexpression of CaM1 in WT plants enhanced several aspects of age-dependent senescence, including chlorophyll content, ROS production, and expression of senescence marker genes (Figures 3, 4). These results suggest that CaM1 acts as a positive regulator of age-dependent leaf senescence. Our previous study has shown that Ca2+ induces the expression of the senescence marker gene SIRK via CaM4 (Koo et al., 2017). In this study, we found enhanced senescence phenotypes in CaM1-overexpressing plants (Figure 3). These two studies demonstrate that CaM1/CaM4 together with Ca2+ positively regulate age-dependent leaf senescence. On the contrary, calcium signaling plays a negative role in MeJA- and NO-mediated leaf senescence (Chou and Kao, 1992; Ma et al., 2010). Thus, it is plausible that other members of the CaM gene family might function in the process of leaf senescence. Differences between the expression profiles of various CaM genes and antagonistic functions of calcium signaling in leaf senescence suggest that CaM proteins fine-tune the balance of calcium signaling in leaf senescence.
CaM1 Functions in RPK1-Mediated Leaf Senescence
Receptor-like potein kinase 1 is a plasma membrane-localized receptor kinase (Osakabe et al., 2005) and plays multiple roles in various cellular signaling and developmental processes, including embryo development (Nodine et al., 2007; Nodine and Tax, 2008), plant growth, stomatal opening, stress response (Hong et al., 1997; Osakabe et al., 2005, 2010), and senescence (Lee et al., 2011; Koo et al., 2017). How RPK1 controls these diverse cellular processes may be partly explained by the proteins interacting with RPK1 in those processes. It has been suggested that calcium and CaM play a role in receptor kinase-mediated cellular processes (Oh et al., 2012). Previous studies have shown that RPK1 interacts and phosphorylates a serine residue on CaM4 (Koo et al., 2017). The expression of CaM1 and CaM4 was induced during leaf aging (Figure 1B), and knockdown of CaM1 or CaM4 did not affect leaf senescence (Figure 4 and Supplementary Figure S3). These data indicate that CaM1 and CaM4 are functionally redundant in RPK1-mediated leaf senescence. Ectopic expression of RPK1 in Arabidopsis at a young developmental stage did not result in senescence (Lee et al., 2011), and CaM1 overexpression did not result in detectable senescence phenotypes at early developmental stages (16-day-old plants; Figure 3D), suggesting that the RPK1–CaM1 module regulates senescence in conjunction with other senescence-related components.
CaM1 Positively Regulates ROS Production
Cellular ROS homeostasis and signaling are crucial parts of signaling networks in plants (Miller et al., 2009). Several lines of evidence show crosstalk between ROS and calcium signaling. For example, NADPH oxidase RHD2/RbohC produces ROS that stimulate hyperpolarization-activated Ca2+ channels, leading to the formation of a tip-focused Ca2+ gradient (Foreman et al., 2003; Takeda et al., 2008). Plasma membrane Ca2+-permeable channels in guard cells are activated by ABA via ROS (Pei et al., 2000; Murata et al., 2001). Because the Arabidopsis genome encodes 10 NADPH oxidase genes that contain the conserved EF-hand Ca2+-binding motifs in their extended N-terminal regions, a regulatory effect of Ca2+ on the activity of NADPH oxidases in plants is expected (Torres et al., 1998). Here, we present data showing a reduction in ABA-induced ROS production in amiRNA-CaM1 plants (Figure 5C) and enhanced ROS levels in 35S::CaM1-GFP plants without exogenous ABA treatment (Figure 5B), suggesting that CaM1-mediated calcium signaling promotes ABA-mediated ROS production. In addition, our data provide evidence that the expression of RbohF is positively regulated by CaM1 (Figure 7C) and show a positive-feedback regulation between RbohF and CaM1 (Figure 7D). Since RbohD also positively regulates CaM1 expression (Figure 7D), it will be of interest to test the specificity of the regulation between calmodulins and NADPH oxidases.
ROS modulate activities of target proteins or expression of genes by changing the redox state in the cell (Schmidt and Schippers, 2015; Saxena et al., 2016). A recent study has shown that the ratio between superoxide and H2O2 in roots determines the cell fate of either differentiation or proliferation, in which NADPH oxidases appear to function (Tsukagoshi et al., 2010). It would be interesting to test whether the ratio between different kinds of ROS also plays a role in age-dependent cell death in leaves. Furthermore, identification of the direct cellular targets of ROS will further shed light on our understanding of ROS-mediated plant senescence.
Diversified Functions of CaMs With High Level of Sequence Similarity
Peptide sequence alignments of CaM proteins and phylogenetic analysis revealed a high level of sequence similarity among the family members (Supplementary Figures S4, S5). Among the seven genes encoding CaM in Arabidopsis, CaM7 is the least related genes with others (Supplementary Figure S4), but only one and four amino acid substitutions differentiate CaM7 from CaM2/CaM3/CaM5 and CaM1/CaM4, respectively. One possible explanation for the conservation of Arabidopsis CaM genes is that CaMs share similar functions. Alternatively, different CaM genes might have evolved distinct expression patterns or regulatory behaviors to ensure important cellular functions. Recent evidence supports the latter. An expression analysis using the publicly available data1 shows that spatial expression of six CaM genes is differently regulated (Supplementary Figure S1), and biochemical analysis indicates functional non-redundancy in CaMs (Liao et al., 1996; Reddy et al., 1999; Kohler and Neuhaus, 2000). CaM2, CaM4, and CaM6 activates NAD kinase and phosphodiesterase with different kinetics in vitro (Liao et al., 1996; Reddy et al., 1999), and CaM2 and CaM4 bind to cyclic nucleotide-gated ion channels and a kinesin-like motor protein with different affinities (Reddy et al., 1999; Kohler and Neuhaus, 2000). These findings suggest that CaMs may exert different functions through their binding targets that may be located in different cellular compartments.
The expression analysis shows that CaM genes encoding identical proteins (CAM2/CAM3/CAM5 and CAM1/CAM4) share a similar expression pattern (Supplementary Figure S1), implying functional redundancy in the closest paralogs, as observed in the senescence phenotype of the amiRNA-CaM1 and cam4 plants (Figure 4). Nevertheless we identified a specific role for CaM1 in the ABA-mediated ROS production and stomatal closure (Figures 5, 6). Fine-tuning of gene expression in a cell type-specific or stimulus-specific manner and/or different post-transcriptional regulation between CaM1 and CaM4 could diversify their functions. Interestingly, potential miRNA target sites on CaM1 differ from them on CaM4 (Adai et al., 2005). Furthermore, various forms of splicing variants of CaM1 are distinct from those of CaM42. It would be of interest to figure out the molecular mechanism that renders the specificity of each CaM gene in plants.
Author Contributions
JK and CD conceived the study and designed the experiments. CD and IL performed the experiments and analyzed the data with YL, HN, and JK. CD, YL, HN, and JK wrote the manuscript.
Funding
This work was supported by IBS-R013-D1 to HN and IBS-R013-G2 to JK from the Institute for Basic Science, and in part by start-up funds from DGIST to JK. CD was supported by a grant from the National Researcher Support Program (No. 20100020417 to HN), the Natural Science Foundation of Hubei Province (No. 2016CFB141), and the Fundamental Research Funds for the Central Universities (Program No. 2662015BQ031).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00803/full#supplementary-material
FIGURE S1 | The expression pattern of CaM genes. (A) Expression of the CaM family genes in different tissues. Data were obtained from ePF-Browser (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Log2-transformed read counts without normalization were used in the computation. (B) Relative expression of CaM1 compared to CaM4 (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). The result indicates the expression of CaM1 and CaM4 are almost identical in most of the tissues, except CaM1 is highly expressed in pollen while CaM4 highly expressed in dry seeds. (C) qRT-PCR analysis of CaM1 and CaM4 in different plant tissue. Expression levels of the genes were normalized to Actin2. Error bars represent means ± SEM of three independent experiments.
FIGURE S2 | Overexpression of CaM1-GFP has no effect on seed size and mass. (A) Representative images of mature seeds of WT and 35S::CaM1-GFP transgenic plants. (B,C) The relative seed size (B) and weight of 1,000 seeds (C) were measured in WT and 35S::CaM1-GFP transgenic plants. Error bars represent means ± SEM (n = 5). Scale bars, 5 mm.
FIGURE S3 | Functional redundancy between CaM1 and CaM4 in age-dependent leaf senescence. (A,B) qRT-PCR analysis of senescence related genes in 35S::CaM1-GFP transgenic lines (A) and 35S::amiRNA-CaM1 plants (B). Expression levels of the genes were normalized to Actin2. Error bars represent means ± SEM of three independent experiments. (C) Senescence phenotypes of WT and cam4. (D) Survival rates of WT and cam4. Plants with third and fourth rosette leaves showing complete yellowing were counted in WT (n = 4) and cam4 (n = 8). In each experiment, 21–25 plants were analyzed. Statistical analysis was performed using heteroscedastic t-test (∗∗p < 0.01). Scale bars, 1 cm.
FIGURE S4 | Protein sequence alignment of the calmodulin family. The EF-hand motifs of CaM1 were conserved across various homologs, including Zea mays (gi|162462264), Glycine max (gi|351721559), Chlamydomonas reinhardtii (gi|167411, gi|159490918), Acyrthosiphon pisum (gi|209870032), Triticum aestivum (gi|291246022), B. napus (gi|497992), B. juncea (gi|899058), B. oleracea (gi| 374922807), Hevea brasiliensis (gi|313767030), Arabidopsis thaliana (CaM2 to CaM7), and Oryza sativa (gi|17066588). Black lines indicate EF-hand motifs. Red asterisks indicate phosphorylation sites.
FIGURE S5 | Phylogenetic analysis of the calmodulin family proteins.
Abbreviations
ABA, abscisic acid; CaM1, calmodulin 1; DAB, diaminobenzidine; H2DCF-DA, dichlorofluorescein diacetate; H2O2, hydrogen peroxide; MeJA, methyl jasmonate; NBT, nitroblue tetrazolium; NO, nitric oxide; RbohF, reactive burst oxidase homolog F; RPK1, receptor-like potein kinase 1; SAG12, senescence-associated gene 12.
Footnotes
- ^ http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi
- ^ http://www.arabidopsis.org
- ^ http://wmd3.weigelworld.org/cgi-bin/webapp.cgi
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://www.ebi.ac.uk/Tools/msa/clustalo/
References
Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V., et al. (2005). Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 15, 78–91. doi: 10.1101/gr.2908205
Chin, D., and Means, A. R. (2000). Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10, 322–328. doi: 10.1016/S0962-8924(00)01800-6
Chou, C. M., and Kao, C. H. (1992). Methyl jasmonate, calcium, and leaf senescence in rice. Plant Physiol. 99, 1693–1694. doi: 10.1104/pp.99.4.1693
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Desikan, R., Griffiths, R., Hancock, J., and Neill, S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 16314–16318. doi: 10.1073/pnas.252461999
Desikan, R., Last, K., Harrett-Williams, R., Tagliavia, C., Harter, K., Hooley, R., et al. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47, 907–916. doi: 10.1111/j.1365-313X.2006.02842.x
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Edel, K. H., and Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Foreman, J., Demidchik, V., Bothwell, J. H., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Hong, S. W., Jon, J. H., Kwak, J. M., and Nam, H. G. (1997). Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 113, 1203–1212. doi: 10.1104/pp.113.4.1203
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). Gus fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Joo, J. H., Wang, S., Chen, J. G., Jones, A. M., and Fedoroff, N. V. (2005). Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17, 957–970. doi: 10.1105/tpc.104.029603
Kohler, C., and Neuhaus, G. (2000). Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471, 133–136. doi: 10.1016/S0014-5793(00)01383-1
Koo, J. C., Lee, I. C., Dai, C., Lee, Y., Cho, H. K., Kim, Y., et al. (2017). The protein trio RPK1-CaM4-RbohF mediates transient superoxide production to trigger age-dependent cell death in Arabidopsis. Cell Rep. 21, 3373–3380. doi: 10.1016/j.celrep.2017.11.077
Kushwaha, R., Singh, A., and Chattopadhyay, S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20, 1747–1759. doi: 10.1105/tpc.107.057612
Kwak, J. M., Mori, I., Pei, Z.-M., Leonhardt, N., Torres, M. A., Dangl, J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signalin in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Lee, I. C., Hong, S. W., Whang, S. S., Lim, P. O., Nam, H. G., and Koo, J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52, 651–662. doi: 10.1093/pcp/pcr026
Liao, B., Gawienowski, M. C., and Zielinski, R. E. (1996). Differential stimulation of NAD kinase and binding of peptide substrates by wild-type and mutant plant calmodulin isoforms. Arch. Biochem. Biophys. 327, 53–60. doi: 10.1006/abbi.1996.0092
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Ma, W., Smigel, A., Walker, R. K., Moeder, W., Yoshioka, K., and Berkowitz, G. A. (2010). Leaf senescence signaling: The Ca2+-conducting Arabidopsis cyclic nucleotide gated channel2 acts through nitric oxide to repress senescence programming. Plant Physiol. 154, 733–743. doi: 10.1104/pp.110.161356
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Miller, G., Schlauch, K., Tam, R., Cortes, D., Torres, M. A., Shulaev, V., et al. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2:ra45. doi: 10.1126/scisignal.2000448
Murata, Y., Pei, Z. M., Mori, I. C., and Schroeder, J. (2001). Abscisic acid activation of plasma membrane Ca2+ channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523. doi: 10.1105/tpc.13.11.2513
Nodine, M. D., and Tax, F. E. (2008). Two receptor-like kinases required together for the establishment of Arabidopsis cotyledon primordia. Dev. Biol. 314, 161–170. doi: 10.1016/j.ydbio.2007.11.021
Nodine, M. D., Yadegari, R., and Tax, F. E. (2007). RPK1 and TOAD2 are two receptor-like kinases redundantly required for arabidopsis embryonic pattern formation. Dev. Cell 12, 943–956. doi: 10.1016/j.devcel.2007.04.003
Ogasawara, Y., Kaya, H., Hiraoka, G., Yumoto, F., Kimura, S., Kadota, Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283, 8885–8892. doi: 10.1074/jbc.M708106200
Oh, M. H., Kim, H. S., Wu, X., Clouse, S. D., Zielinski, R. E., and Huber, S. C. (2012). Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signalling. Biochem. J. 443, 515–523. doi: 10.1042/BJ20111871
Osakabe, Y., Maruyama, K., Seki, M., Satou, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17, 1105–1119. doi: 10.1105/tpc.104.027474
Osakabe, Y., Mizuno, S., Tanaka, H., Maruyama, K., Osakabe, K., Todaka, D., et al. (2010). Overproduction of the membrane-bound receptor-like protein kinase 1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 285, 9190–9201. doi: 10.1074/jbc.M109.051938
Padidam, M. (2003). Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 6, 169–177. doi: 10.1016/S1369-5266(03)00005-0
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Poovaiah, B. W., and Leopold, A. C. (1973). Deferral of leaf senescence with calcium. Plant Physiol. 52, 236–239. doi: 10.1104/pp.52.3.236
Ranty, B., Aldon, D., Cotelle, V., Galaud, J. P., Thuleau, P., and Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7:327, doi: 10.3389/fpls.2016.00327
Reddy, V. S., Safadi, F., Zielinski, R. E., and Reddy, A. S. (1999). Interaction of a kinesin-like protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274, 31727–31733. doi: 10.1074/jbc.274.44.31727
Saxena, I., Srikanth, S., and Chen, Z. (2016). Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7:570, doi: 10.3389/fpls.2016.00570
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Schmidt, R., and Schippers, J. H. (2015). ROS-mediated redox signaling during cell differentiation in plants. Biochim. Biophys. Acta. 1850, 1497–1508. doi: 10.1016/j.bbagen.2014.12.020
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Steinhorst, L., and Kudla, J. (2014). Signaling in cells and organisms - calcium holds the line. Curr. Opin. Plant Biol. 22, 14–21. doi: 10.1016/j.pbi.2014.08.003
Takeda, S., Gapper, C., Kaya, H., Bell, E., Kuchitsu, K., and Dolan, L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244. doi: 10.1126/science.1152505
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanabata, T., Shibaya, T., Hori, K., Ebana, K., and Yano, M. (2012). SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160, 1871–1880. doi: 10.1104/pp.112.205120
Torres, M. A., Dangl, J. L., and Jones, J. D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. doi: 10.1046/j.1365-313X.1998.00136.x
Tsukagoshi, H., Busch, W., and Benfey, P. N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. doi: 10.1016/j.cell.2010.10.020
Vigneron, A., and Vousden, K. H. (2010). p53, ROS and senescence in the control of aging. Aging (Albany NY) 2, 471–474. doi: 10.18632/aging.100189
Woo, H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. doi: 10.1105/tpc.13.8.1779
Zhang, X., Zhang, L., Dong, F. C., Gao, J. F., Galbraith, D. W., and Song, C. P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126, 1438–1448. doi: 10.1104/pp.126.4.1438
Zhang, Y., Zhu, H., Zhang, Q., Li, M., Yan, M., Wang, R., et al. (2009). Phospholipase dalpha1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377. doi: 10.1105/tpc.108.062992
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Zielinski, R. E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725. doi: 10.1146/annurev.arplant.49.1.697
Keywords: calmodulin 1, NADPH oxidase, reactive oxygen species, RPK1, senescence
Citation: Dai C, Lee Y, Lee IC, Nam HG and Kwak JM (2018) Calmodulin 1 Regulates Senescence and ABA Response in Arabidopsis. Front. Plant Sci. 9:803. doi: 10.3389/fpls.2018.00803
Received: 05 February 2018; Accepted: 25 May 2018;
Published: 02 July 2018.
Edited by:
Jingjuan Zhang, Murdoch University, AustraliaCopyright © 2018 Dai, Lee, Lee, Nam and Kwak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Dai, Y2RhaUBtYWlsLmh6YXUuZWR1LmNu June M. Kwak, amt3YWtAZGdpc3QuYWMua3I=
 Cheng Dai1*
Cheng Dai1* Yuree Lee
Yuree Lee Hong G. Nam
Hong G. Nam June M. Kwak
June M. Kwak
