
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 June 2018
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00797
 Bingwei Yu1,2
Bingwei Yu1,2 Shuangshuang Yan1,2
Shuangshuang Yan1,2 Huoyan Zhou1,2
Huoyan Zhou1,2 Riyue Dong1,2
Riyue Dong1,2 Jianjun Lei1,2
Jianjun Lei1,2 Changming Chen1,2
Changming Chen1,2 Bihao Cao1,2*
Bihao Cao1,2*High temperature (HT) stress affects the growth and production of cucumbers, but genetic resources with high heat tolerance are very scarce in this crop. Calmodulin (CaM) has been confirmed to be related to the regulation of HT stress resistance in plants. CsCaM3, a CaM gene, was isolated from cucumber inbred line “02-8.” Its expression was characterized in the present study. CsCaM3 transcripts differed among the organs and tissues of cucumber plants and could be induced by HTs or abscisic acid, but not by salicylic acid. CsCaM3 transcripts exhibited subcellular localization to the cytoplasm and nuclei of cells. Overexpression of CsCaM3 in cucumber plants has the potential to improve their heat tolerance and protect against oxidative damage and photosynthesis system damage by regulating the expression of HT-responsive genes in plants, including chlorophyll catabolism-related genes under HT stress. Taken together, our results provide useful insights into stress tolerance in cucumber.
Plants often endure various abiotic stresses throughout their growth and development, including low and high temperatures, salinity, and drought. High temperatures (HTs) is a serious abiotic factor that not only causes the loss of yield and diminishes quality of crops, but also leads to direct damage of crop plants, including protein denaturation and aggregation as well as oxidative injuries to membrane lipids. Accordingly, enhancing the environmental stress tolerance of crops is very important for agriculture (Varshney et al., 2012); this objective can alleviate the harm caused by HT stress and reduce associated agricultural losses.
Plant calmodulins (CaMs) comprise a multigene family that has been reported in many species. CaM, a conserved, small acidic protein, is one of the best characterized Ca2+-sensors and has four EF-hand motifs, each of which is involved in binding Ca2+. After binding to Ca2+, conformational changes in CaM lead to interactions with different target CaM-binding proteins (CaMBPs), which comprise disparate protein family members that perform a diverse range of roles in the cell (Crivici and Ikura, 1995). CaMs is a central component that modulates many cellular processes across multifarious physiological functions that regulate downstream target protein activities via Ca2+/CaM complex-mediated processes (Kim et al., 2009), including adjustment of metabolism, phytohormone signaling, ion transportation, and transcriptional regulation (Snedden and Fromm, 2001; Yang and Poovaiah, 2003; Bouché et al., 2005).
Many reports have indicated that plant CaMs play important roles in the modulation of tolerance to different stress (Kim et al., 2009), such as cold stress (Doherty et al., 2009), drought stress (Wan et al., 2012), heat stress (Li et al., 2005; Li and Gong, 2009) and so on. For example, overexpression of AtCaM3 in Arabidopsis increased heat tolerance (Zhang et al., 2009). In tobacco, CaM isoforms were confirmed to be related to regulating resistance to two fungal pathogens (Rhizoctonia solani and Pythium aphanidermatum) and one bacterial pathogen (Ralstonia solanacearum; Takabatake et al., 2007). SCaM-4 from soybean was significantly induced by the presence of a pathogen (Park et al., 2004) or NaCl stress (Park et al., 2009). The expression level of OsCaM1 in rice was enhanced by NaCl, mannitol, or wounding, and OsCaM3 was also induced by NaCl or damage (Phean-O-Pas et al., 2005). Ca2+ and CaM are also able to protect against oxidative harm under heat stress (Larkindale and Knight, 2002) and increase the heat tolerance of suspension-cultured tobacco cells (Li and Gong, 2009). Those reports clearly indicated that plant CaMs enhance stress-tolerance.
Cucumber (Cucumis sativus L.) is an important vegetable crop that is planted worldwide and originated from subtropical areas. As a thermophilic crop, the optimum temperature of cucumber growth is 25–30°C. Temperatures above 35°C can often lead to thermal damage to cucumber plants, and cucumber leaves and stems show wilt and dead tissue within a short time at temperatures of 50°C (Talanova et al., 2006). In China, cucumber is cultivated in open fields during the summer growing season, where cucumber plants usually face the combined stress of strong light and heat, or sharp temperature increases in greenhouses. Even if ventilation systems are used, it is impossible to prevent substantial heat accumulation in greenhouses. Consequently, cucumber growth is often hindered by excessive daytime temperatures. This influences normal growth, photosynthesis and fruit of cucumber plants seriously, causing premature aging, yield reduction, and diminished quality. The breeding of cucumbers for heat tolerance has been slow because of the scarcity of heat-tolerant genetic resources in the species, and in research of heat resistance in cucumber (Ding et al., 2016; Li et al., 2016a,b), in where few reports of transgenic CaM genes inserted into the cucumber genome for the purpose of increasing stress-tolerance. This study examined the overexpression of the CaM gene CsCaM3 in cucumber in order to enhance the heat tolerance of cucumber and provide the foundation for further improvements in the tolerance of cucumber plants to heat stress.
The heat-susceptible cucumber inbred line “02-8” produced by our laboratory was used for all experiments in this study. DNA sequences of the CaM gene were found in the NCBI database and Cucumber Genome Database1. A fragment of the CsCaM3 gene was amplified by PCR with the P1 forward primer (5′-CTCTAGAATGGCGGATCAGCTCACCGA-3′) and P2 reverse primer (5′-CGGATCCCCTTAGCCATCATGACCTTAAC-3′); underlined sequences show the XbaI and BamHI restriction sites.
Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The optimal tree with a summed branch length of 0.02022066 is shown (next to the branches). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are within the units of the number of amino acid substitutions per site. The analysis involved eight amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 149 positions in the final dataset.
The expression vector pBI-CsCaM3 (Supplementary Figure S1) with the CaMV35S promoter and nptII gene was constructed and transferred to the Agrobacterium tumefaciens strain EHA105 by the freeze–thaw method. The transformation of cucumber was conducted as described by Cao et al. (2011). The first filial generation of the transgenic plants (OE-1, OE-2, and OE-3) were used in all experiments conducted at the three-leaf stage. The identification of transgenic cucumber plants was by Southern and northern blots. The hybridization was conducted using a DNA labeling and detection kit (Boehringer Mannheim, Mannheim, Germany), and the dosage of DNA and total RNA were 10 μg and 12 μg, respectively. A fragment of the 35S promoter was PCR amplified using the 35S-F forward primer (5′-ATAGTGGGATTGTGCGTCA-3′) and 35S-R reverse primer (5′-GCACCTACAAATGCCATCA-3′) and was then used as a probe for Southern blotting. The resulting fragment of CsCaM3 was used as the probe for northern blotting. The heat treatment conditions for all experiments were under 43 and 25°C during the day and night, respectively.
The expression levels of CsCaM3 were analyzed at the three-leaf stage, with three biological and technical replicates. From the tissues of first top leaf, stem, and root of cucumber inbred line “02-8” seedlings were extracted total RNA with TRIzol reagents (Invitrogen, Carlsbad, CA, United States) after 43°C treatment for 12 h, and RNA was extracted from the first top leaf of seedlings after 0, 6, 12, and 24 h of temperature and hormone treatments. Solutions of 5 μM abscisic acid (ABA) and 100 μM salicylic acid (SA) were sprayed onto the leaves of seedlings under a 43°C temperature treatment. RNA was converted into cDNA with a kit (Genstar, Beijing, China). All qRT-PCR analyses were conducted by gene-specific primers (Supplementary Table S1) and a SYBR Premix Ex Taq kit (TaKaRa, Dalian, China). The RT-PCR protocol consisted of one cycle at 94°C for 3 min; 28 cycles of 94°C for 1 min, 55°C for 45 s, and 72°C for 90 s; and one cycle of 72°C for 7 min. The 2-ΔCt method (Livak and Schmittgen, 2001) was used to assess the relative gene expression.
Amplification of CsCaM3 was conducted by PCR with the sub-Cam3F forward primer (5′-ATGGCGGATCAGCTCACCGA-3′) and the sub-Cam3R reverse primer (5′-CTTGGCCATCATGACCTTCA-3′), which does not contain a stop codon. In order to control CsCaM3-GFP vectors with a CaMV 35S promoter, the PCR products were fused into the pEZS-NL-GFP vector in-frame with the sequence of green fluorescent protein (GFP). The vector was transferred into A. tumefaciens strain EHA105, and the leaf epidermis of Nicotiana benthamiana leaves were injected with the transformed EHA105 (Na et al., 2016). GFP fluorescence of the transformed plants was observed after two days in darkness using a laser scanning confocal microscope (Leica TCS SP2, Leica Microsystems, Wetzlar, Germany).
The CsCaM3 gene was selected for localizing mRNA in the leaf and root tissues of cucumbers grown under HT stress (i.e., 43°C). Digoxigenin-labeled RNA probes were hybridized to the cucumber tissues of young leaf and root as described by Ma et al. (2012). The gene fragment was labeled using the DIG RNA Labeling Kit (SP6/T7; Roche, Shanghai, China). The fragments of probes were amplified from cDNA using gene-specific primers including SP6 and T7 RNA polymerase-binding sites (CsCaM3-SP6, 5′-ATTTAGGTGACACTATAGAAGATGGCGGATCAGCTCACCG A-3′; CsCaM3-T7, 5′-TAATACGACTCACTATAGGGAGACC TTAGCCATCATGA CCTTAAC-3′).
The measurement of chlorophyll fluorescence under minimal fluorescence (Fo) was measured without actinic light. Then, the leaf was exposed to a saturation pulse to measure maximal fluorescence (Fm). The optimal quantum yield of photosystem II (PSII) (Fv/Fm, Fv = Fm -Fo) was derived from measured Fo and Fm values, where Fv was the difference between Fo and Fm. Actinic light was turned on after saturation pulse, and the fluorescent signals reached the steady state slowly with intermittent saturation pulses. Fs determined the steady-state fluorescence yield, and Fm′ was the resulting light-adapted maximum fluorescence. In this process, photochemical quenching of open PSII was measured, revealing photo-protective non-photochemical quenching (NPQ) and other heat dissipation mechanisms. qP is the fraction of open PSII reaction centers, characterized as the coefficients of photochemical fluorescence quenching (Genty et al., 1989; van Kooten and Snel, 1990). Electron transport rate (ETR) and effective quantum yield of PSII (ΦPSII) are valuable measurements in various plant stress studies. The Fv/Fm, ΦPSII, NPQ, and qP values were determined from the same individual using the following equations: ΦPSII = (Fm′ -Fs)/Fm′, NPQ = (Fm/Fm′) - 1, qP = (Fm′ -Fs)/(Fm′ -Fo), and ETR = ΦPSII × PFD (photon flux density) × 0.5. Images were obtained using a Chlorophyll fluorescence imager (IMAGING-PAM, WALZ, Germany). Total chlorophyll, chlorophyll a, and chlorophyll b contents were assessed using a spectrophotometric method (Porra et al., 1989).
The malondialdehyde (MDA) content and peroxidase (POD) activities were tested according to the method described by Velikova et al. (2000). Superoxide dismutase (SOD) activity was measured using nitro blue tetrazolium (NBT) methods (Attar et al., 2006). Catalase (CAT) activities were measured using the method described by Dong et al. (2014).
Accumulations of superoxide and hydrogen peroxide (H2O2) were visually analyzed with NBT and 3,3-diaminobenzidine (DAB) staining using the methods described by Driever et al. (2009). Measurement of reactive oxygen species (ROS) was conducted using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) staining. Leaf epidermal strips were peeled from cucumber leaves without chemical treatment. The leaf epidermal strips were floated on a solution of 50 μM DCFH-DA (Sigma Chemicals, St. Louis, MO, United States) for 30 min before treatment. ROS accumulation was observed by fluorescence microscopy using a Leica MZ FL III (excitation, 450 ± 490 nm; barrier 520 ± 560 nm).
The viability of root tissue was assessed using the fluorescein diacetate/propidium iodide (FDA/PI) staining mixture method as described by Pan et al. (2001). The tips of roots (about 2.5 cm in length) were gently incubated in a FDA/PI mixture (containing FDA 12.5 g mL-1 and PI 5.0 g mL-1) for 10 min and then washed with distilled water. Images were observed with a fluorescent microscope (Olympus BX41; Olympus Corporation, Tokyo, Japan) under blue light excitation (460–495 nm) and emission at 510 nm and analyzed using a photographic apparatus (Olympus CamediaC-5060; Olympus, Tokyo, Japan) and analySIS software (Soft Imaging Solutions GmbH, Münster, Germany).
Stomata were measured as described previously (Zhu et al., 2016). ABA content was measured as previously described (Kara et al., 1997). Enzyme-linked immunosorbent assay kits and 96-well (12 × 8) plates used in this study were purchased from the Crop Chemical Control Center of China Agricultural University. All assays and measurements were conducted in triplicate.
The NCBI database and Cucumber Genome Database (see text footnote 1) were searched to find DNA sequences of CaM genes. From these sequences, primers were designed to PCR amplify the CaM gene CsCaM3 from cucumber total cDNA, resulting in the isolation of CsCaM3, a 450-bp ORF encoding 149 amino acids. The amino acid sequence of CsCaM3 showed high similarity to CsCaM (XM_011655459.1) from cucumber, CmCaM (XM_008466393) from melon, AtCaM3 AY091301) from Arabidopsis, JrCaM7 (XM_018967153.1) from Juglans regia, StCaM (JX576246.1) from Solanum tuberosum, NtCaM7 (XM_016640697.1) from Nicotiana tabacum, BoCaM (XM_013776345) from Brassica oleracea, and SlCaM3 (NM_001321494) from Solanum lycopersicum (Figure 1).
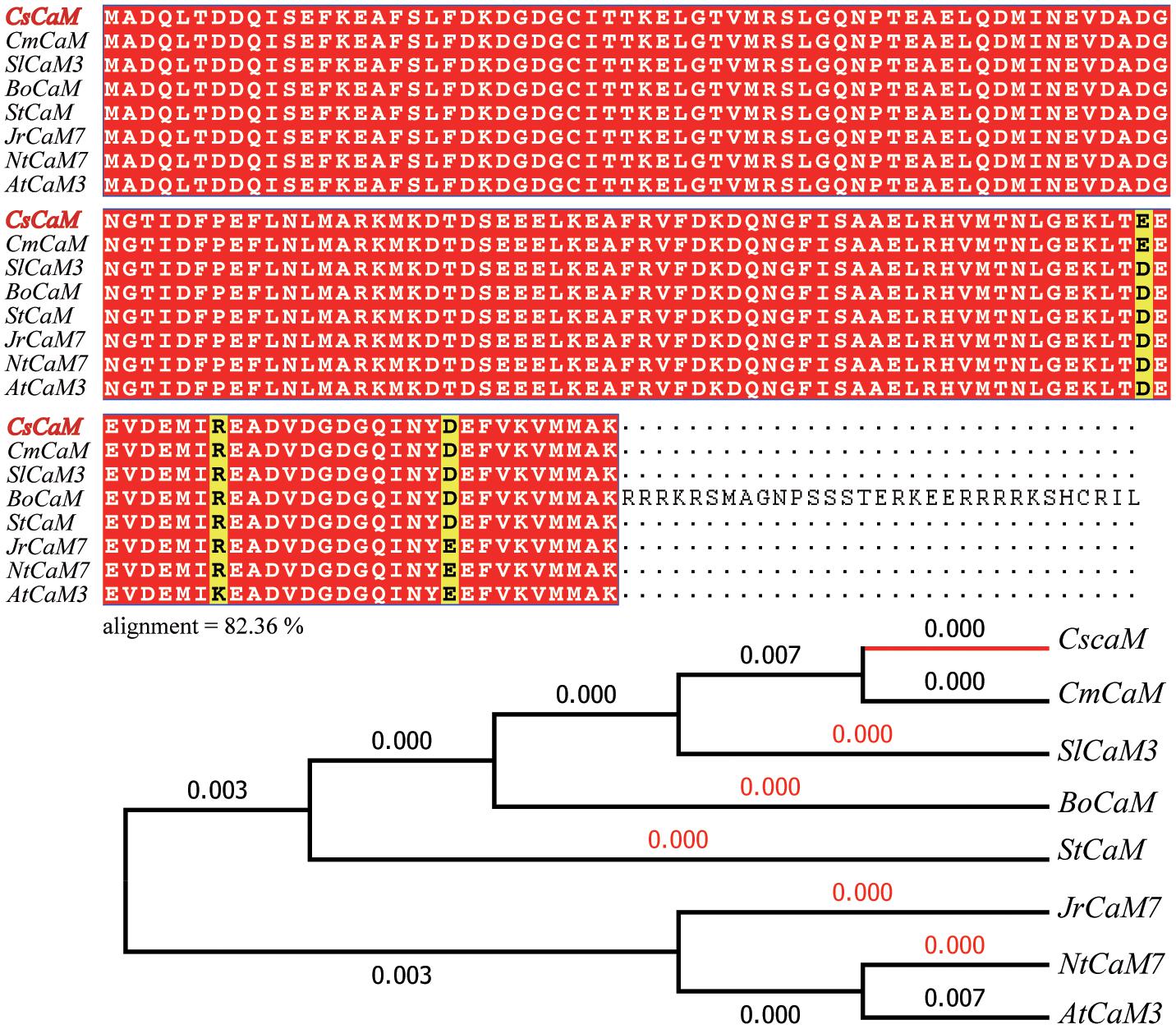
FIGURE 1. Sequence alignment and phylogenetic tree of CsCaM3. Alignment analysis of the inferred CsCaM3 protein sequence with other inferred plant calmodulin proteins: CsCaM from Cucumis sativus, CmCaM from Cucumis melo, AtCaM3 from Arabidopsis thaliana, JrCaM7 from Juglans regia, StCaM from Solanum tuberosum, NtCaM7 from Nicotiana tabacum, BoCaM from Brassica oleracea, and SlCaM3 from Solanum lycopersicum. The numbers indicate branch lengths.
The expression pattern of CsCaM3 was assessed in root, stem, and leaf tissues from cucumber inbred line “02-8.” The expression level of the gene was highest in leaves and lowest in roots (Figure 2A). ABA and HT stress treatment increased the expression of CsCaM3, but SA did not (Figures 2B–D). These results concurred with the results of situ hybridization of tissues was observed in leaf and root tissues, and the hybridization signals subjected to 43°C high-temperature stress treatment were stronger than that in the control group (Figures 2E–H). Subcellular location results indicated that CsCaM3 proteins are located in the cell membrane and nuclei (Figure 3). Taken together, CsCaM3 is likely involved in the regulation of HT stress tolerance in cucumber.
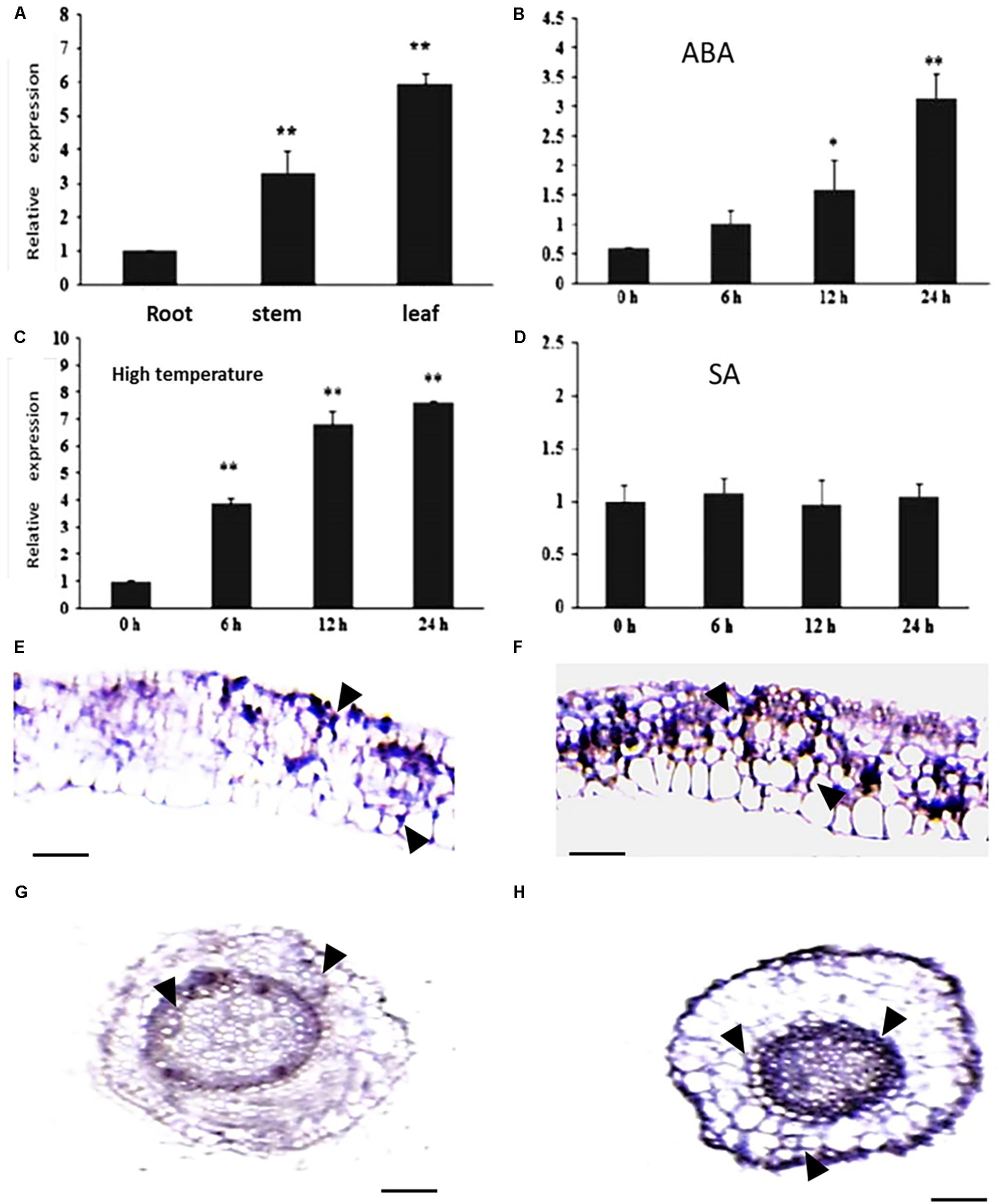
FIGURE 2. Analysis of expression characterization of CsCaM3 and in situ hybridization. (A) The expression of CsCaM3 in stem, leaf, and root tissues from cucumber inbred line “02-8.” (B) The expression of CsCaM3 in leaf tissue after 5 μM ABA treatment at different times. (C) The expression of CsCaM3 in leaf tissue after 43°C HT treatment at different times. (D) The expression of CsCaM3 in leaf tissue after 100 μM SA treatment at different times. (E,F) In situ hybridization of CsCaM3 in leaf tissue in wild-type cucumber control plants and under 43°C HT treatments for 6 h, respectively. Scale bars = 100 μm. The arrows indicate hybridization signals. (G,H) In situ hybridization of CsCaM3 in root tissue in wild-type cucumber control plants and under 43°C HT treatment for 6 h, respectively. ∗P < 0.05 and ∗∗P < 0.01 in comparisons of each time points. Scale bars = 50 μm. Each treatment was repeated three times.
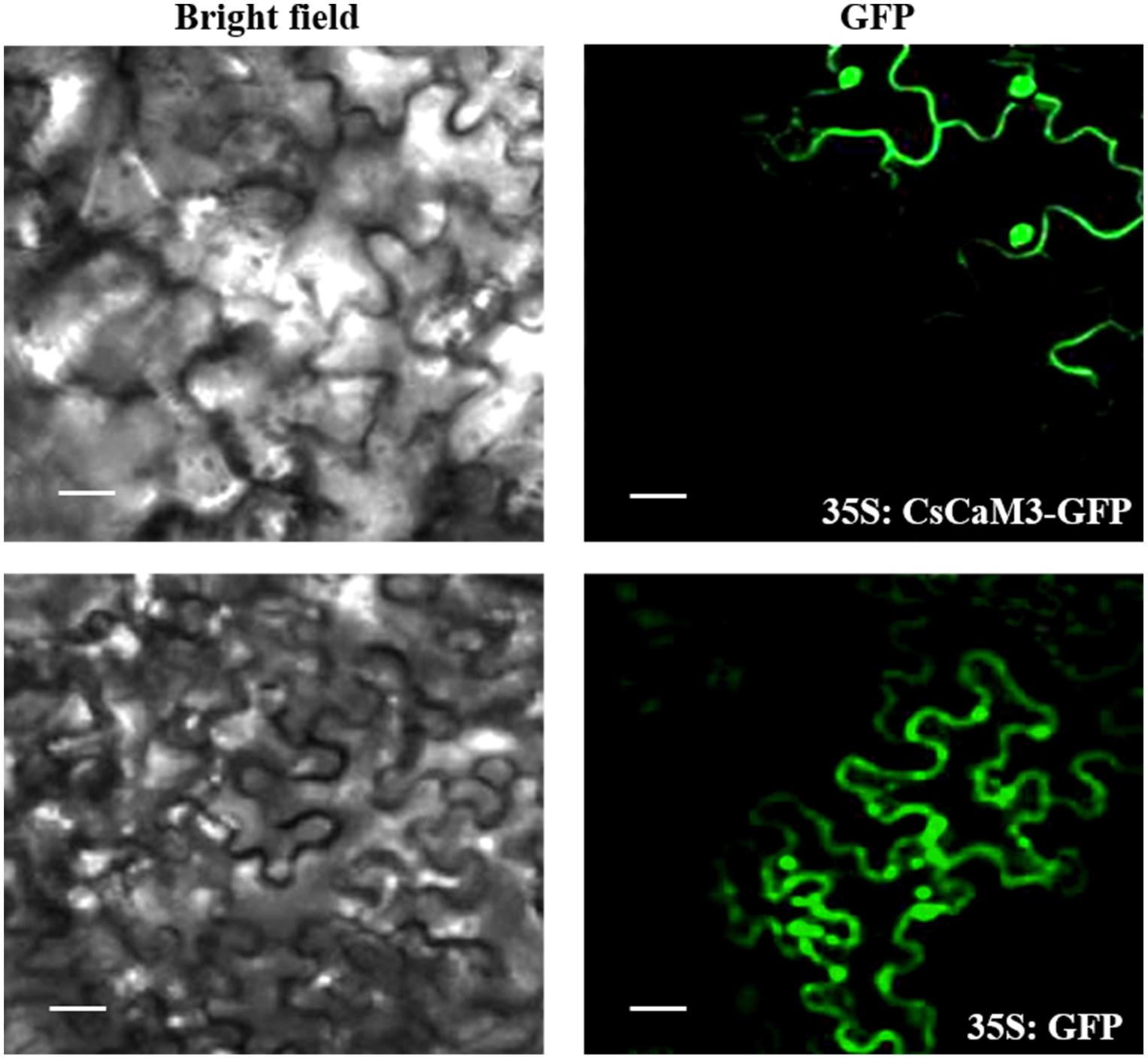
FIGURE 3. Subcellular localization of CsCaM3. The PCR products were fused with the pEZS-NL-GFP vector in-frame with the green fluorescent protein (GFP) sequence, and the CaMV 35S promoter controlled the CsCaM3-GFP vectors upstream. Subcellular distribution pattern of 35S:CsCaM3-GFP showing a nuclear and cell membrane distribution pattern at 24 h after infiltration into tobacco leaf epidermis tissues. Images of either dark field (green fluorescence) or bright field modes were acquired by fluorescence microscopy. Scale bars = 20 μm.
In order to identify the function of CsCaM3, the CsCaM3-overexpression vector (i.e., pBI-CsCaM3) was constructed, from about 1000 explants used for transformation, four transgenic plants were obtained (Supplementary Figure S2), which were identified by genomic Southern and northern blotting (Figures 4A,B). The hybridization signal of CsCaM3 in transgenic plants was determined to be stronger relative to the wild-type (WT) plants (Figure 4B). Three T1 transgenic plants (OE-1, OE-2, and OE-3) were obtained by selfing and used to assess heat-tolerance. The conditions of heat treatment were 43 and 25°C under day and night, respectively. More than 30 seedlings of each strains were used. After heat treatment for 5 days, more than 30% of WT plants were dead, but only 5% of the T1 transgenic plants were dead. After 10 days of HT treatment, all WT plants were dead, while about 60% of the T1 transgenic plants still survived (Figures 4C,E,F). The growth potential of transgenic plant radicles was stronger than that of WT plants (Figure 4D). Overexpression of CsCaM3 may thus have improved the heat tolerance of cucumber plants.
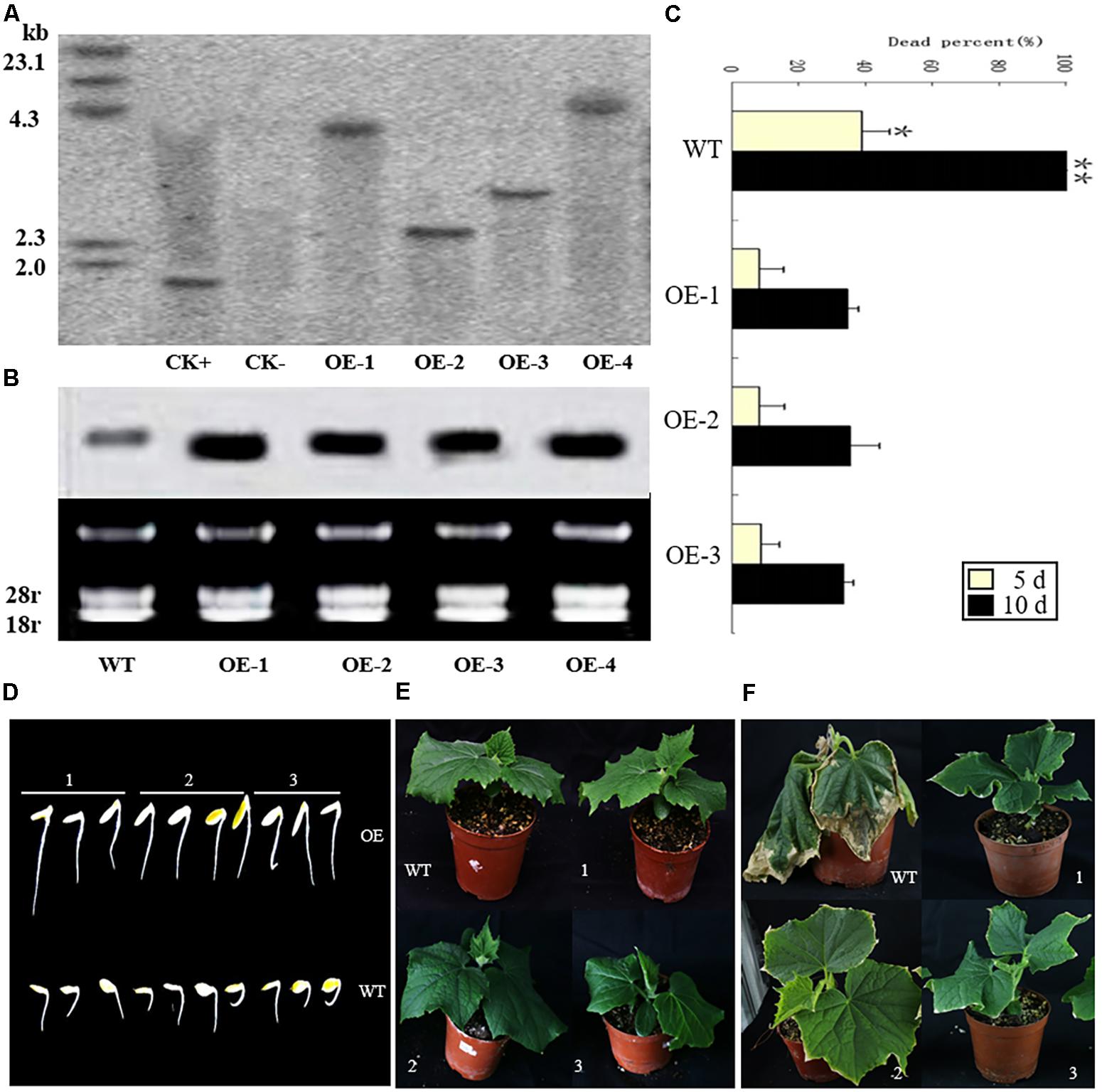
FIGURE 4. Identification of transgenic cucumber plants with Southern and northern blots. Heat-tolerance of wild-type (WT) and transgenic cucumber plants under 43 and 25°C day and night conditions, respectively. (A) Genomic DNA from transgenic Km-resistant plants was digested with HindIII, and a NPT-II gene fragment was used as a probe: CK+, the recombinant plasmid overexpressing CsCaM; CK–, the WT plants; OE-1, OE-2, OE-3, and OE-4, the transgenic plants. (B) Northern blots of the WT and transgenic plants. (C) Percentage of dead plants among the transgenic and WT plants under the HT treatment for 5 and 10 days. OE-1, OE-2, and OE-3 represent the T1 transgenic plants. There were more than 30 seedlings for each variety (WT, OE1, OE2, OE3). ∗P < 0.05 and ∗∗P < 0.01 in comparisons of WT and transgenic plants. (D) Germination of seeds from the T1 transgenic OE-1, OE-2, and OE-3 (1, 2, and 3, respectively) and WT lines under the 43°C HT treatment after 1 h on the first day. (E) The growth of T1 transgenic and WT plants under 25°C conditions. (F) Heat treatment symptoms of the T1 transgenic plants and WT plants under the HT treatments after 3 days.
In order to identify the sensitivity of transgenic cucumber plants to HT stress, the viability of root tips was assessed. The root tip viabilities of both WT and transgenic cucumber plants were decreased under the 43°C HT treatment, but there were more dead cells in WT plants relative to the transgenic plants (Figure 5A). Moreover, the heat resistance of transgenic plants was stronger than that of WT plants, and HTs affected the root viability of the plants.
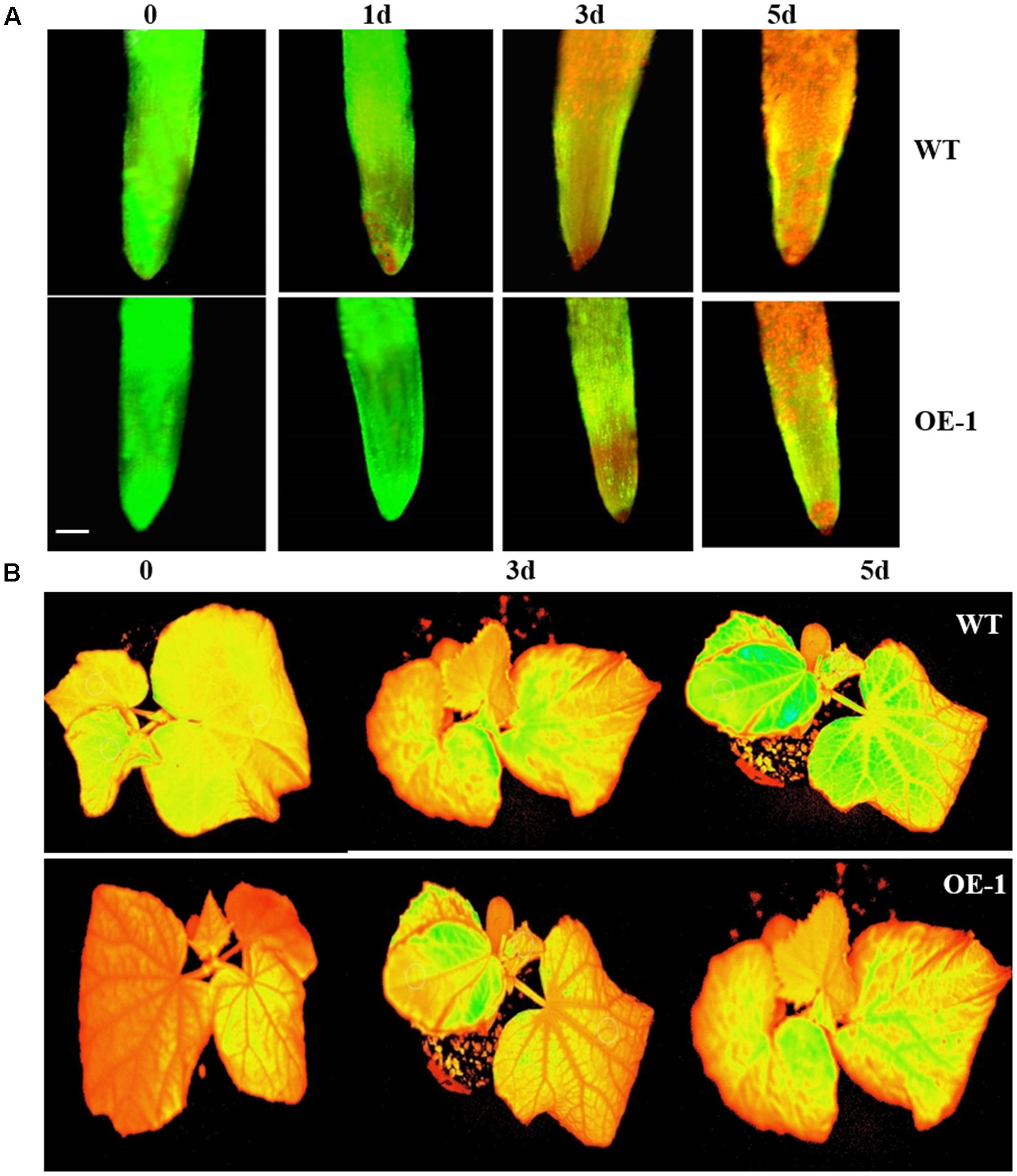
FIGURE 5. Viability of root cells and images of Ft (the instantaneous fluorescent signal) of cucumber plants under HT stress. (A) The viability of root cells of the cucumber plants under 43°C temperature treatment at different times. The yellow–green fluorescence indicates viable cells, while the red fluorescence indicates dead cells. Scale bars = 50 μm. (B) Images of Ft of cucumber plant under 43°C temperature treatment at different times. Green fluorescence indicates leaf damage.
The transgenic and WT plants were treated at 43°C for three days, after which their chlorophyll fluorescence parameters were measured. The chlorophyll fluorescence parameters of both were significantly affected, but all parameters of the WT plants were lower than those of the transgenic plants (Table 1). The damage to the photosynthesis systems of the WT plants was more serious than to that to the transgenic plants under HT stress (Figure 5B), suggesting overexpression of CsCaM3 could increase heat-resistance of cucumber plants.
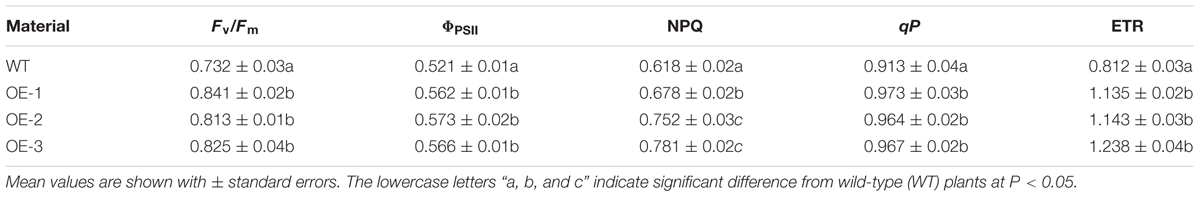
TABLE 1. Effects of HT stress on chlorophyll fluorescence parameters of transgenic and wild-type (WT) plants.
The contents of overall ROS, H2O2 and superoxide in the leaves of transgenic plants were lower than those of the WT plants, but under normal conditions, the ROS content of both the transgenic and WT did not significantly differ (Figure 6). Compared to the control plants, the chlorophyll contents of both the WT and transgenic plants were not remarkably different. However, the activities of SOD, POD, and CAT of the transgenic plants were significantly higher than those of the WT plants, and the MDA contents of WT plants were significantly higher than those of the transgenic plants (Table 2). These results again demonstrate that overexpression of CsCaM3 in cucumber could increase the activities of antioxidant enzymes and decrease MDA content and oxidative stress damage.
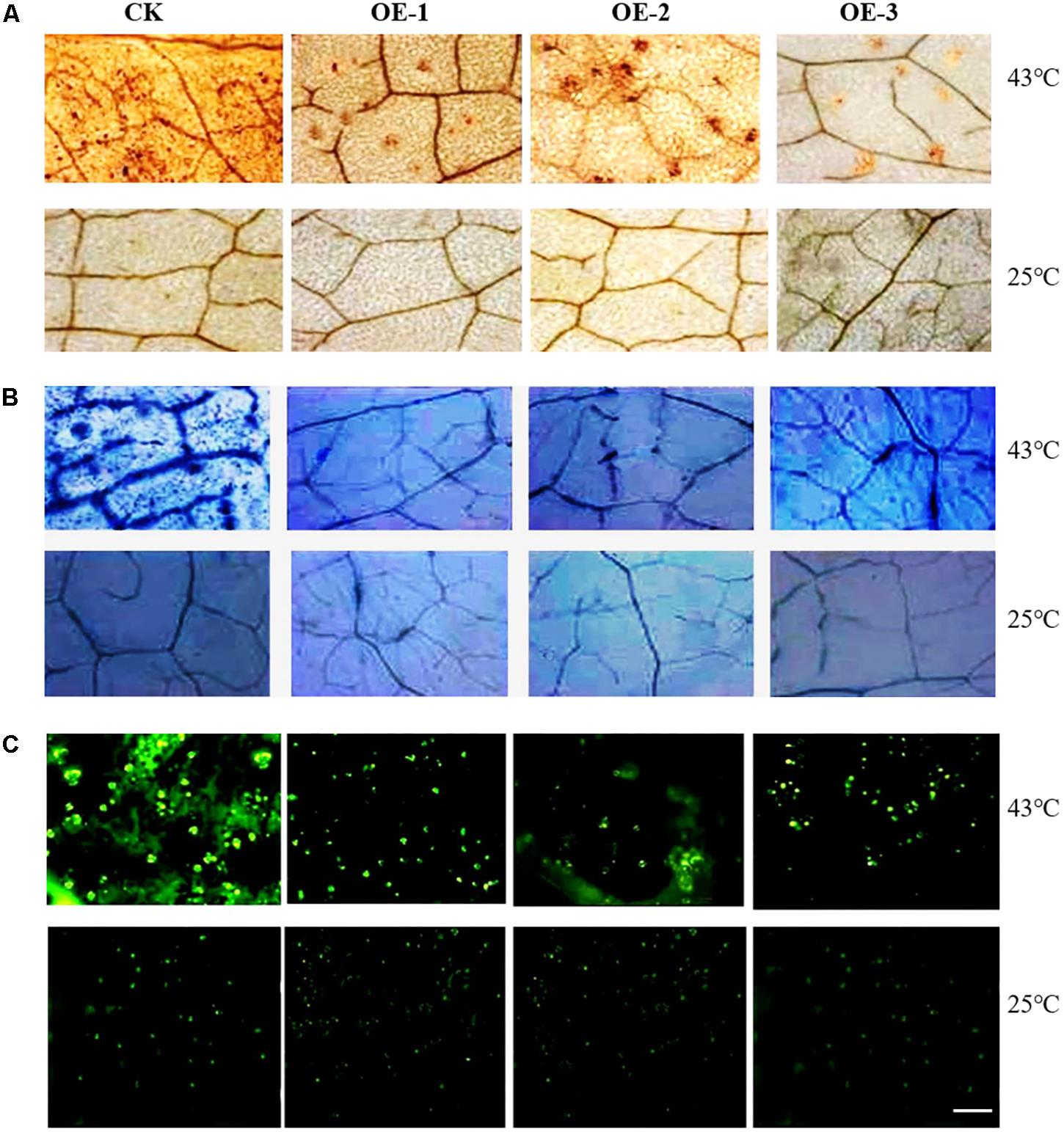
FIGURE 6. ROS detection of transgenic cucumber plant leaves under HT stress. (A) H2O2 was detected by staining with 3,3-diaminobenzidine (DAB). Brown spots indicate H2O2 accumulation in leaves. (B) Superoxide was detected by nitro blue tetrazolium (NBT) staining. Blue spots indicate the superoxide accumulation in leaves. (C) ROS was assayed with 2′,7′-dichloro dihydro fluorescein diacetate (DCFH-DA) staining, and green spots show the degree of overall ROS stress in leaves. Scale bar = 2.5 mm.

TABLE 2. Chlorophyll contents, antioxidant enzymes activities, and MDA content in transgenic plants under HT stress treatment for 3 days.
The CLH, CBR1, PAO, and RCCR genes are related to the chlorophyll catabolic pathway in plants, and their expression reflects the process of photosynthesis. Therefore, the expression levels of these four genes were measured. The transcript levels of the four genes substantially differed between the WT and transgenic plants. The expressions of CBR1, PAO, and RCCR in transgenic plants were higher than those of WT plants, but the expression of CLH in transgenic plants was lower than that of the control plants under the 43°C temperature treatment for 3 days. At the same time, the expression levels of seven genes (CsSOD, CsPOD, CsCAT, CscAPX, AOX, HSP70, and HSP90) in the transgenic plants were markedly elevated and were higher than those of the WT plants. The expression levels of CsABI1, CsABI2, and CsABI3, which are involved in the ABA catabolic pathway in plants, were significantly increased compared with the WT plants (Figure 7). These results suggest that overexpression of CsCaM could affect the transcription of genes not only in the chlorophyll pathway, but also in the ABA catabolic pathway.

FIGURE 7. Expression of genes in transgenic and WT plants under 43°C-day/25°C-night temperature stress for 3 days in WT plants and transgenic OE-1, OE-2, and OE-3 plants (1, 2, and 3, respectively). Asterisks indicate significant differences from WT at ∗P < 0.05 and ∗∗P < 0.01.
After 3 days of the 43°C temperature treatment, the ABA content of the transgenic and WT plants was assayed. The ABA content of all transgenic plants was significantly higher than that of the WT plants (Table 3). On the other hand, the opening degree of stomata of WT plants leaves was greater than that of the transgenic plants under the temperature stress treatment (Figure 8).
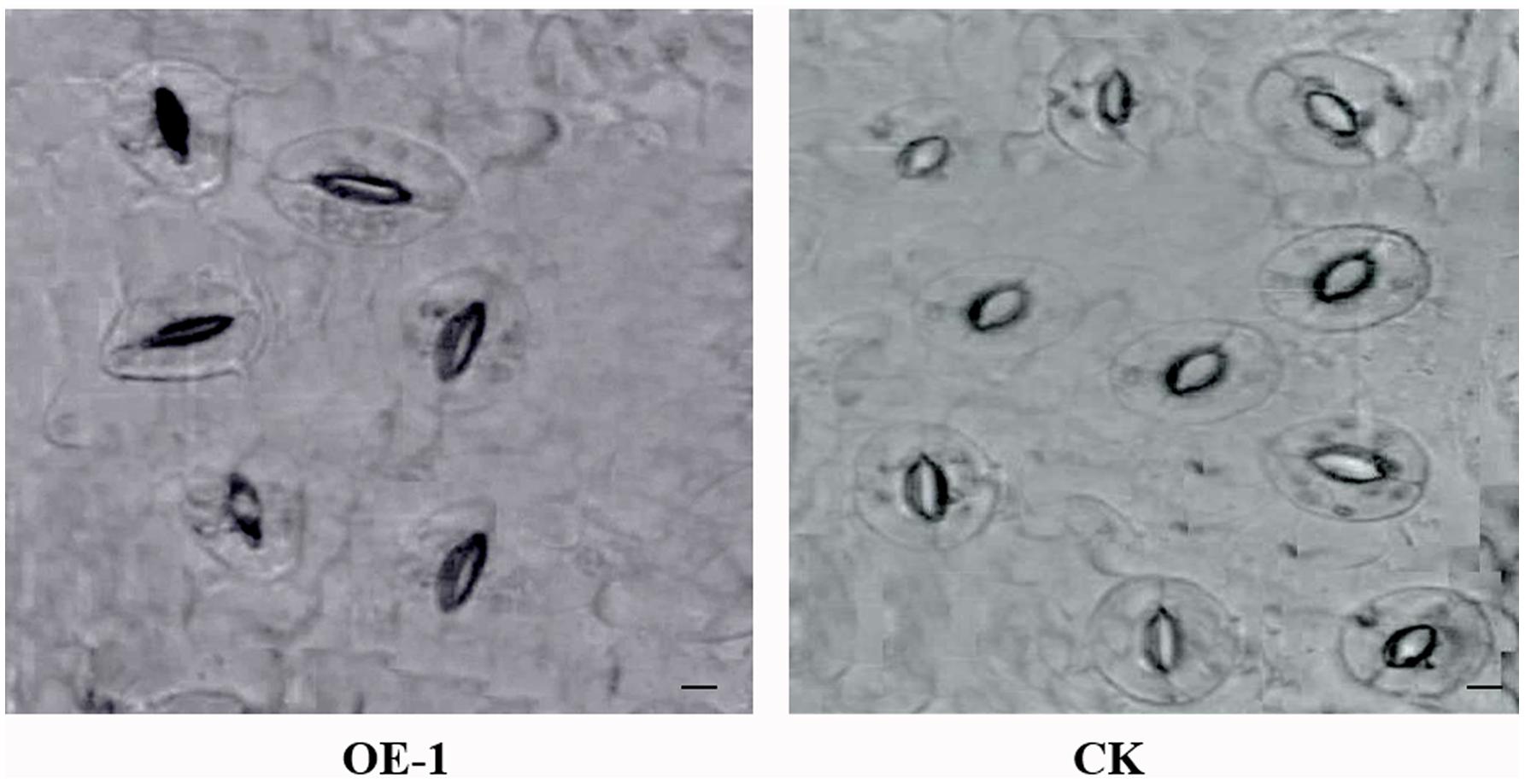
FIGURE 8. Images of stomata in transgenic and wild-type (WT) plants leaves under HT stress. OE-1 corresponds to a transgenic plant, and WT represents wild-type plants. Stomata were measured after 43°C HT treatment for 3 days. Scale bar = 20 μm.
Plants have multiple pathways that allow them to physiologically adapt to environmental stress; HTs are a major stress factor affecting plant ecology and agricultural production (Mittler and Blumwald, 2010). CaM is a ubiquitous and conserved Ca2+-binding protein that regulates a vast array of Ca2+-sensitive biological events including Ca2+ transport, long-term intracellular signaling, and transcriptional regulation (Kortvely and Gulya, 2004). CaM is also involved in stress tolerance in plants (Kim et al., 2009; Singh et al., 2017). Overexpression of CaM in plants has been reported to improve the heat tolerance of plants (Larkindale and Knight, 2002; Li and Gong, 2009; Zhang et al., 2009), but there is a lack of reports that have been published about the transformation of CaM into cucumber. In our study, CsCaM3 had high similarity to CaM genes that are related to stress-regulation in other plants and was verified to improve heat tolerance in cucumber through overexpression. Our results indicated that overexpression of CsCaM in cucumber could protect against oxidative damage and photosynthesis system damage under HT stress (Figure 4), concurring with results obtained in Arabidopsis (Larkindale and Knight, 2002) and tobacco (Li and Gong, 2009).
Subcellular localization revealed that CsCaM3 proteins were located in the membranes and nuclei of cells (Figure 3), reflecting similar results in pea (Dauwalder et al., 1986), Petunia (Rodriguez-Concepcion et al., 1999; VanDer Luit et al., 1999). CaM is also localized in peroxisomes, plastids, mitochondria, and the extracellular matrix (Jarrett et al., 1982; Roberts et al., 1983; Ma et al., 1999; Reddy et al., 2002; Yang and Poovaiah, 2002), which is consistent with the location of majority of its target proteins that in the plasma, membrane, or nucleus (Virdi et al., 2015). Target proteins affected by CsCaM are still unknown in this study and should be isolated and identified in the future.
The expression of CsCaM3 differed among plant organs and tissues (Figure 2A), and could be induced by HT stress and ABA application (Figures 2B,C), concurring with results reported by other researchers (Liu et al., 2005; Zhang et al., 2013; Munir et al., 2016). However, CaM3 transcripts in tall fescue decreased under HTs (Wang and Huang, 2017); thus, different plants may have different stress-tolerance regulation mechanisms. It was reported that calcium-activated CaM (Ca2+-CaM) was directly involved in the heat stress signal transduction pathway in wheat (Triticum aestivum) and up-regulated heat shock proteins (HSPs; Liu et al., 2003). HSPs (such as HSP18, HSP70, HSP90, and HSP101) are known to be positive regulators of heat tolerance in various plant species (Zhou and Abaraha, 2007). Some reports have shown that CaM interacts with HSP90 and HSP70, of which steady state levels are regulated through Ca2+-CaM pathways (Sun et al., 2000; Virdi et al., 2011). And in our research, expression levels of HSP70 and HSP90 in transgenic cucumber plants were significantly higher compared with WT plants (Figure 7), and the way in which CsCaM3 interacts with HSP70 and HSP90 remains to be identified in the future. The complex crosstalk between signaling molecules, HSPs, and partner proteins appears to be critical for heat stress responses (Wang and Huang, 2017).
Under adversity, plants will produce ROS, inducing oxidative stress, which damages cell membrane system. To counteract the deleterious effects of ROS, a complex antioxidative defense system evolved in plants, which includes antioxidant enzymes such as SOD, POD, CAT, and so on. Through the determination of the activity of antioxidant enzymes, the strength of stress resistance of plants can be identified (Suzuki et al., 2011; Gupta et al., 2013). H2O2 is one of the key elements in the ROS signaling pathway and was found to induce plant heat tolerance through exogenous application (Larkindale and Huang, 2005). H2O2 is also known to be involved in the early phase of heat stress and is required for effective expression of HSPs and HSFs in Arabidopsis and rice (Oryza sativa; Volkov et al., 2006; Wang et al., 2009). In our data, under HT stress, the contents of H2O2, superoxide, and the overall ROS in WT plants were higher in comparison with transgenic plants (Figure 6). In the current research, the antioxidant enzyme activities of SOD, POD, and CAT were all higher in transgenic plants, and the MDA content of transgenic plants was also lower compared with WT plants (Table 2), which may indicate increased oxidative stress tolerance in transgenic plants. At the same time, the expression levels of genes related to oxidative stress (such as CsSOD, CsPOD, CsCAT, CsAPX, and AOX) were all higher in transgenic plants (Figure 7); similar results were observed by other researchers (Kolupaev et al., 2008; Xu, 2010). For example, the expression of SPCAT in sweet potato was regulated by SPCAM (Afiyanti and Chen, 2014). However, a continuous influx of Ca2+ is required for activity associated with the production of H2O2 from NADPH, which is catalyzed by NADPH oxidase (Keller et al., 1998). The activity of NAD kinase is also adjusted through Ca2+-CaM (Harding et al., 1997), which is activated by H2O2 (Pei et al., 2000). These studies have elucidated the complex mechanisms of feedback modulation of different enzymes through Ca2+ and CaM, which may enable plants to maintain H2O2 homeostasis, despite stress-induced damage (Virdi et al., 2015). In our research, the above-mentioned genes related to heat stress all had active responses that included the overexpression of CsCaM3, but the mechanism through which overexpression of CsCaM3 induced the expression of those genes remains unclear.
Enhanced ABA content in plants can improve stress tolerance (Larkindale and Knight, 2002). In our study, the ABA content of the transgenic plants was much higher relative to WT plants (Table 3), and the expression of CsABI1, CsABI2, and CsABI3 in the ABA catabolic pathway was also upregulated under HTs (Figure 7); these genes might promote an increase in ABA content. We inferred that overexpression of CsCaM in cucumber increases the heat stress response of plants via the ABA signal pathway, but the regulation mechanism remains to be clarified. Under HT stress, the opening degree of stomata of transgenic plants leaves was lower than that of the control plants (Table 3). Because stomatal dynamic variation is a crucial ABA-regulated process, we deduced that high levels of ABA might make stomata close more quickly, as has been observed in Chinese kale (Zhu et al., 2016).
CLH is a key gene that regulates chlorophyll catabolism. The transcription of CLH in transgenic plants in the present study was significantly lower than that in WT plants under HTs (Figure 7), but the total chlorophyll, chlorophyll a, and chlorophyll b contents of the transgenic plant leaves were significantly higher than those of WT plants (Table 2). These results suggested that overexpression of CsCaM3 may downregulate the transcription of CLH to reduce chlorophyll degradation under HTs. CBR1, PAO, and RCCR are also involved in the chlorophyll catabolic pathway. Chlorophyll b reductase (CBR) is a key enzyme for the transformation of chlorophyll b into chlorophyll a. Pheophorbide a is catalytically degraded to red chl catabolites (RCCs) by pheophorbide a oxygenase (PAO), and red chlorophyll catabolite reductase (RCCR) plays a key role for the conversion of RCCs into non-fluorescent chlorophyll catabolites (NCCs; Vezitskii and Shcherbakov, 2002; Pružinská et al., 2007). In our study, under HTs, the expression levels of CBR1, PAO, and RCCR in transgenic plants were markedly higher than those in WT plants (Figure 7), which may enhance the conversion of chlorophyll b into chlorophyll a. Chlorophyll b can enter the degradation pathway, and only after being converted into chlorophyll a can it be further metabolized (Hortensteiner and Krautler, 2011). Mutations of PAO and RCCR can be further metabolized and lead to an increase in degraded chlorophyll intermediates, thus accelerating cell death (Ginsburg and Matile, 1993; Ginsburg et al., 1994). We infer that high expression levels of PAO and RCCR can decrease the accumulation of potentially phototoxic chlorophyll intermediates and thus prevent them from injuring plants. The mechanism by which CsCaM regulates the expression of these genes under HT stress is a subject for future investigations.
BC contributed to designing the experiments. BC, BY, HZ, and RD performed the experiments, collected and analyzed the data. BC, JL, and CC contributed to data interpretation and preparation of the manuscript. All authors read and approved the final manuscript.
This work was financially supported by the Programs for Science and Technology of Guangdong Province (2016A020210069, 2016B020201008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00797/full#supplementary-material
FIGURE S1 | Diagram of the pBI-CsCaM3 expression cassette transformation vector.
FIGURE S2 | The overexpression of CsCaM3 in transgenic cucumber plants. Wild-type (WT) plants are shown beside the transgenic plants (OE-1, OE-2, OE-3, and OE-4).
TABLE S1 | Primer sequences for qRT-PCR.
Afiyanti, M., and Chen, H. J. (2014). Catalase activity is modulated by calcium and calmodulin in detached mature leaves of sweet potato. J. Plant Physiol. 171, 35–47. doi: 10.1016/j.jplph.2013.10.003
Attar, F., Keyhani, E., and Keyhani, J. (2006). Comparative study of superoxide dismutase activity assays in Crocus sativus L. corms. Appl. Biochem. Microbiol. 42, 101–106. doi: 10.1134/s0003683806010169
Bouché, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005). Plant specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Cao, B., Lei, J., Chen, G., Cao, P., Liu, X., Chen, Q., et al. (2011). Testing of disease-resistance of pokeweed antiviral protein gene (PacPAP) in transgenic cucumber (Cucumis sativus). Afr. J. Biotechnol. 10, 6883–6890. doi: 10.5897/AJB10.1730
Crivici, A., and Ikura, M. (1995). Molecular and structural basis of target recognition by calmodulin. Annu. Rev. Biophys. Biomol. Struct. 24, 85–116. doi: 10.1146/annurev.biophys.24.1.85
Dauwalder, M., Roux, S. J., and Hardison, L. (1986). Distribution of calmodulin in pea seedlings: immunocytochemical localization in plumules and root apices. Planta 168, 461–470. doi: 10.1007/BF00392265
Ding, X., Jiang, Y., He, L., Zhou, Q., Yu, J., Hui, D., et al. (2016). Exogenous glutathione improves high root-zone temperature tolerance by modulating photosynthesis, antioxidant and osmolytes systems in cucumber seedlings. Sci Rep. 6, 35424. doi: 10.1038/srep35424
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, C. J., Li, L., Shang, Q. M., Liu, X. Y., and Zhang, Z. G. (2014). Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta 240, 687–700. doi: 10.1007/s00425-014-2115-1
Driever, S. M., Fryer, M. J., Mullineaux, P. M., and Baker, N. R. (2009). Imaging of reactive oxygen species in vivo. Methods Mol. Biol. 479, 109–116. doi: 10.1007/978-1-59745-289-2_7
Genty, B., Briantais, J. M., and Baker, N. R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990, 87–92. doi: 10.1016/s0304-4165(89)80016-9
Ginsburg, S., and Matile, P. (1993). Identification of catabolites of chlorophyll-porphyrin in senescent rape cotyledons. Plant Physiol. 102, 521–527. doi: 10.1104/pp.102.2.521
Ginsburg, S., Schellenberg, M., and Matile, P. (1994). Cleavage of chlorophyll-porphyrin (requirement for reduced ferredoxin and oxygen). Plant Physiol. 105, 545–554. doi: 10.1104/pp.105.2.545
Gupta, N. K., Shubhi, A., Agarwal, V. P., Nathawat, N. S., Sunita, G., and Singh, G. (2013). Effect of short-term heat stress on growth, physiology and antioxidative defence system in wheat seedlings. Acta Physiol. Plant. 35, 1837–1842. doi: 10.1016/j.ecss.2007.03.033
Harding, S. A., Oh, S. H., and Roberts, D. M. (1997). Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16, 1137–1144. doi: 10.1093/emboj/16.6.1137
Hortensteiner, S., and Krautler, B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807, 977–988. doi: 10.1016/j.bbabio.2010.12.007
Jarrett, H. W., Brown, C. J., Black, C. C., and Cormier, M. J. (1982). Evidence that calmodulin is in the chloroplast of peas and serves a regulatory role in photosynthesis. J. Biol. Chem. 257, 13795–13804.
Kara, A. N., Kotov, A. A., and Bukhov, N. G. (1997). Specific distribution of gibberellins, cytokinins, indole-3-acetic acid and abscisic acid in radish plants closely correlates with photomorpho-genetic responses to blue or red light. J. Plant Physiol. 151, 51–59. doi: 10.1016/s0176-1617(97)80035-1
Keller, T., Damude, H. G., Werner, D., Doerner, P., Dixon, R. A., and Lamb, C. (1998). A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 10, 255–266. doi: 10.2307/3870703
Kim, M. C., Chung, W. S., Yun, D. J., and Cho, M. J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2, 13–21. doi: 10.1093/mp/ssn091
Kolupaev, Y. Y., Karpets, Y. V., and Kosakivska, I. V. (2008). The importance of reactive oxygen species in the induction of plant resistance to heat stress. Gen. Appl. Plant Physiol. 34, 251–266.
Kortvely, K., and Gulya, K. (2004). Calmodulin, and various ways to regulate its activity. Life Sci. 74, 1065–1070. doi: 10.1016/j.lfs.2003.07.026
Larkindale, J., and Huang, B. R. (2005). Effects of abscisic acid, salicylic acid, ethylene and hydrogen peroxide in thermo-tolerance and recovery for creeping bentgrass. Plant Growth Regul. 47, 17–28. doi: 10.1007/s10725-005-1536-z
Larkindale, J., and Knight, M. R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128, 628–695. doi: 10.1104/pp.128.2.682
Li, H., Ahammed, G. J., Zhou, G., Xia, X., Zhou, J., Shi, K., et al. (2016a). Unraveling main limiting sites of photosynthesis under below- and above-ground heat stress in cucumber and the alleviatory role of Luffa rootstock. Front. Plant Sci. 2:746. doi: 10.3389/fpls.2016.00746
Li, H., Wang, Y., Wang, Z., Guo, X., Wang, F., Xia, X. J., et al. (2016b). Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 39, 1790–1804. doi: 10.1111/pce.12745
Li, Z. G., Du, C. K., and Gong, M. (2005). Involvement of Ca2+ and calmodulin in the regulation of H2O2-induced heat tolerance in maize seedlings. J. Plant Physiol. Mol. Biol. 31, 515–519.
Li, Z. G., and Gong, M. (2009). Involvement of calcium and calmodulin in mechanical stimulation-induced heat tolerance in tobacco (Nicotiana tabacum L.) suspension cultured cells. Plant Physiol. Commun. 45, 363–365.
Liu, H. T., Li, B., Shang, Z. L., Li, X. Z., Mu, R. L., Sun, D. Y., et al. (2003). Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiol. 132, 1186–1195. doi: 10.1104/pp.102.018564
Liu, H. T., Sun, D. Y., and Zhou, R. G. (2005). Ca2+ and AtCaM3 are involved in the expression of heat shock protein gene in Arabidopsis. Plant Cell Environ. 28, 1276–1284. doi: 10.1111/j.1365-3040.2005.01365.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., He, Y. H., Hu, Z. Y., Xu, W. T., Xia, J. X., and Guo, C. H. (2012). Characterization and expression analysis of AcSERK2, a somatic embryogenesis and stress resistance related gene in pineapple. Gene 500, 115–123. doi: 10.1016/j.gene.2012.03.013
Ma, L., Xu, X., Cui, S., and Sun, D. (1999). The presence of a heterotrimeric G protein and its role in signal transduction of extracellular calmodulin in pollen germination and tube growth. Plant Cell 11, 1351–1364. doi: 10.1105/tpc.11.7.1351
Mittler, R., and Blumwald, E. (2010). Genetic engineering for modern agriculture: challenges and perspectives. Ann. Rev. Plant Biol. 61, 443–462. doi: 10.1146/annurev-arplant-042809-112116
Munir, S., Gul Khan, M. R., Song, J., Munir, S., Zhang, Y., Ye, Z., et al. (2016). Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 102, 167–179. doi: 10.1016/j.plaphy.2016.02.020
Na, C., Shuanghua, W., Jinglong, F., Bihao, C., Jianjun, L., Changming, C., et al. (2016). Overexpression of the eggplant (Solanum melongena) NAC family transcription factor SmNAC suppresses resistance to bacterial wilt. Sci. Rep. 6:31568. doi: 10.1038/srep31568
Pan, J. W., Zhu, M. Y., and Chen, H. (2001). Aluminum-induced cell death in root-tip cells of barley. Environ. Exp. Bot. 46, 71–79. doi: 10.1016/s0098-8472(01)00083-1
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135, 2150–2161. doi: 10.1104/pp.104.041442
Park, H. C., Kim, M. L., Kang, Y. H., Jeong, J. C., Cheong, M. S., Choi, W., et al. (2009). Functional analysis of the stress-inducible soybean calmodulin isoform-4 (GmCaM-4) promoter in transgenic tobacco plants. Mol. Cells 27, 475–480. doi: 10.1007/s10059-009-0063-6
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klüsener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Phean-O-Pas, S., Punteeranurak, P., and Buaboocha, T. (2005). Calcium signaling-mediated and differential induction of calmodulin gene expression by stress in Oryza sativa L. J. Biochem. Mol. Biol. 38, 432–439. doi: 10.5483/bmbrep.2005.38.4.432
Porra, R., Thompson, W., and Kriedemann, P. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. doi: 10.1016/s0005-2728(89)80347-0
Pružinská, A., Anders, I., Aubry, S., Schenk, N., Tapernoux-Lüthi, E., and Müller, T. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19, 369–387. doi: 10.1105/tpc.106.044404
Reddy, V. S., Ali, G. S., and Reddy, A. S. (2002). Genes encoding calmodulin binding proteins in the Arabidopsis genome. J. Biol. Chem. 277, 9840–9852. doi: 10.1074/jbc.m111626200
Roberts, D. M., Zielinski, R. E., Schleicher, M., and Watterson, D. M. (1983). Analysis of suborganellar fractions from spinach and pea chloroplasts for calmodulin-binding proteins. J. Cell Biol. 97, 1644–1647. doi: 10.1083/jcb.97.5.1644
Rodriguez-Concepcion, M., Yalovsky, S., Zik, M., Fromm, H., and Gruissem, W. (1999). The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 18, 1996–2007. doi: 10.1093/emboj/18.7.1996
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Singh, S., Virdi, A. S., Jaswal, R., Chawla, M., Kapoor, S., Mohapatra, S. B., et al. (2017). A temperature-responsive gene in sorghum encodes a glycine-rich protein that interacts with calmodulin. Biochimie 137, 115–123. doi: 10.1016/j.biochi.2017.03.010
Snedden, W. A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151, 35–66. doi: 10.1046/j.1469-8137.2001.00154.x
Sun, X. T., Li, B., Zhou, G. M., Tang, W. Q., Bai, J., Sun, D. Y., et al. (2000). Binding of the maize cytosolic HSP70 to calmodulin, and identification of calmodulin-binding site in HSP70. Plant Cell Physiol. 41, 804–810. doi: 10.1093/pcp/41.6.804
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2011). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Takabatake, R., Karita, E., Seo, S., Mitsuhara, I., Kuchitsu, K., and Ohashi, Y. (2007). Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48, 414–423. doi: 10.1093/pcp/pcm011
Talanova, V. V., Topchieva, L. V., and Titov, A. F. (2006). Effect of abscisic acid on the resistance of cucumber seedlings to combined exposure to high temperature and chloride. Biol. Bull. 33, 619–622. doi: 10.1134/s1062359006060136
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
van Kooten, O., and Snel, J. F. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25, 147–150. doi: 10.1007/bf00033156
VanDer Luit, A. H., Olivari, C., Haley, A., Knight, M. R., and Trewavas, A. J. (1999). Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol. 121, 705–714. doi: 10.1104/pp.121.3.705
Varshney, R. K., Ribaut, J. M., Buckler, E. S., Tuberosa, R., Rafalski, J. A., and Langridge, P. (2012). Can genomics boost productivity of orphan crops? Nat. Biotechnol. 2012, 1172–1176. doi: 10.1038/nbt.2440
Velikova, V., Yordanov, I., and Edreva, A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants protective role of exogenous polyamines. Plant Sci. 151, 59–66. doi: 10.1016/s0168-9452(99)00197-1
Vezitskii, A. Y., and Shcherbakov, R. A. (2002). Interconversions of chlorophylls a and b synthesized from exogenous chlorophyllides a and b in etiolated and post-etiolated rye seedlings. Russ. J. Plant Physiol. 49, 605–609. doi: 10.1023/A:1020276515669
Virdi, A. S., Pareek, A., and Singh, P. (2011). Evidence for the possible involvement of calmodulin in regulation of steady state levels of Hsp90 family members (Hsp87 and Hsp85) in response to heat shock in sorghum. Plant Signal. Behav. 6, 393–399. doi: 10.4161/psb.6.3.13867
Virdi, A. S., Supreet, S., and Prabhjeet, S. (2015). Abiotic stress responses in plants: roles of calmodulin-regulated proteins. Front. Plant Sci. 6:809. doi: 10.3389/fpls.2015.00809
Volkov, R. A., Panchuk, I. I., Mullineaux, P. M., and Schoffl, F. (2006). Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol. Biol. 61, 733–746. doi: 10.1007/s11103-006-0045-4
Wan, D., Li, R., Zou, B., Zhang, X., Cong, J., Wang, R., et al. (2012). Calmodulin-binding protein CBP60g is a positive regulator of both disease resistance and drought tolerance in Arabidopsis. Plant Cell Rep. 31, 1269–1281. doi: 10.1007/s00299-012-1247-7
Wang, C., Zhang, Q., and Shou, H. X. (2009). Identification and expression analysis of OsHsfs in rice. J. Zhejiang Univ. Sci. B 10, 291–300. doi: 10.1631/jzus.b0820190
Wang, X., and Huang, B. (2017). Lipid- and calcium-signaling regulation of HsfA2c-mediated heat tolerance in tall fescue. Environ. Exp. Bot. 136, 59–67. doi: 10.1016/j.envexpbot.2017.01.008
Xu, S. (2010). Abscisic acid activates a Ca2+-calmodulin-stimulated protein kinase involved in antioxidant defense in maize leaves. Acta Biochim. Biophys. Sin. 42, 646–655. doi: 10.1093/abbs/gmq064
Yang, T., and Poovaiah, B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.m207941200
Yang, T., and Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal net work in plants. Trends Plant Sci. 8, 505–512. doi: 10.1016/j.tplants.2003.09.004
Zhang, W., Zhou, R. G., Gao, Y. J., Zheng, S. Z., Xu, P., Zhang, S. Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149, 1773–1784. doi: 10.1104/pp.108.133744
Zhang, X., Zhou, Y., Ding, L., Wu, Z., Liu, R., and Meyerowitz, E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101. doi: 10.1105/tpc.112.107854
Zhou, S., and Abaraha, A. (2007). Response to heat stress in warm season and cool season turf grass cultivars. Sci. Res. Essays 2, 95–100.
Zhu, Z., Sun, B., Xu, X., Chen, H., Zou, L., Chen, G., et al. (2016). Overexpression of AtEDT1/HDG11 in Chinese kale (Brassica oleracea var. alboglabra) enhances drought and osmotic stress tolerance. Front. Plant Sci. 7:1285. doi: 10.3389/fpls.2016.01285
Keywords: CsCaM3, calmodulin, high temperature tolerance, genes expression, cucumber
Citation: Yu B, Yan S, Zhou H, Dong R, Lei J, Chen C and Cao B (2018) Overexpression of CsCaM3 Improves High Temperature Tolerance in Cucumber. Front. Plant Sci. 9:797. doi: 10.3389/fpls.2018.00797
Received: 22 August 2017; Accepted: 24 May 2018;
Published: 12 June 2018.
Edited by:
Eric Ruelland, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Ratna Karan, University of Florida, United StatesCopyright © 2018 Yu, Yan, Zhou, Dong, Lei, Chen and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bihao Cao, Y2FvYmgwMUAxNjMuY29t; Y2FvYmgwMUBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.