- 1State Key Laboratory for Crop Genetics and Germplasm Enhancement, Jiangsu Plant Gene Engineering Research Center, Nanjing Agricultural University, Nanjing, China
- 2National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
The chloroplast is a self-independent organelle and contains its own transcription and translation systems. The establishment of genetic systems is vital for normal plant growth and development. We isolated a rice zebra leaf 16 (zl16) mutant derived from rice cultivar 9311. The zl16 mutant showed chlorotic abnormalities in the transverse sectors of the young leaves of seedlings. The use of transmission electron microscopy (TEM) demonstrated that dramatic defects occurred in variegated zl16 leaves during the early development of a chloroplast. Map-based cloning revealed that ZL16 encodes a β-hydroxyacyl-ACP dehydratase (HAD) involved in de novo fatty acid synthesis. Compared with the wild type, a missense mutation (Arg164Trp) in the zl16 mutant was identified, which significantly reduced enzymatic activity and altered the three-dimensional modeling structure of the putative protein. ZL16 was ubiquitously expressed in various plant organs, with a pronounced level in the young leaf. A subcellular localization experiment indicated that ZL16 was targeted in the chloroplast. Furthermore, we analyzed the expression of some nuclear genes involved in chloroplast development, and found they were altered in the zl16 mutant. RNA-Seq analysis indicated that some genes related to cell membrane constituents were downregulated in the mutant. An in vivo metabolic assay revealed that the total fatty acid content in the mutant was significantly decreased relative to the wild type. Our results indicate that HAD is essential for the development of chloroplasts by regulating the synthesis of fatty acids in rice.
Introduction
A chloroplast is a plant organelle that originated from an endosymbiotic event between a photosynthetic cyanobacteria and its eukaryotic host (Moreira et al., 2000; Kusumi et al., 2004). Its main function is to conduct photosynthesis and convert the energy from solar irradiation into chemical energy. Thus, the normal formation and development of chloroplasts is vital to sustain crop yield. Many studies have been conducted to uncover the molecular mechanisms of chloroplast development (Barkan et al., 1995; de Souza et al., 2016).
In addition to being a photosynthetic organelle, the plastid is also a major synthetic site for fatty acids and many amino acids in plants. In plant plastids, the de novo biosynthesis of fatty acids is mainly catalyzed by several conserved enzymes, including acetyl-CoA carboxylase (ACCase), malonyl-CoA:ACP transacylase (MAT), β-ketoacyl- ACP synthase (KAS), β-ketoacyl-ACP reductase (KAR), HAD, and enoyl-ACP reductase (ENR) (Gardner et al., 2002).
HAD conducts the third step in the fatty acid synthesis in a plant fatty acid synthase complex (González-Thuillier et al., 2016). Previous studies of Escherichia coli determined that HAD possesses two isozymes, viz., β-hydroxydecanoyl-ACP dehydratase/isomerase (FabA) and HAD (FabZ) (Mohan et al., 1994; Heath and Rock, 1996). FabA produces unsaturated fatty acids and is only found in gram-negative bacteria, while FabZ allows the synthesis of saturated as well as unsaturated fatty acids and functions ubiquitously in both gram-positive and negative bacteria. Unlike the dehydratase/isomerase bifunctional enzyme activity of FabA, FabZ only has a dehydration function. Additionally, FabZ is more active than FabA for catalyzing short-chain primer substrates (Mohan et al., 1994; Pillai et al., 2003; Sharma et al., 2003). Because bacteria and plants share a low sequence similarity, sequence-based searching for plant HAD with E. coli. HAD homologs was difficult until the recent characterization of a mitochondrial HAD (mtHAD) in Arabidopsis. RNA interference (RNAi) mutants, with reduced mtHAD expression, exhibit traits such as a significantly reduced size of aerial organs, photorespiratory deficiency, altered chloroplastic starch granule morphology, and reduced catalytic activity of lipoylated enzymes (Guan et al., 2017). Despite the clarification of the various HADs in the fatty acid biosynthesis of E. coli and Arabidopsis, the roles of HAD proteins in rice (Oryza sativa), a staple crop for over half of the world’s population and a monocot model plant, remain uncertain.
In this study, we identified a rice chlorophyll deficient mutant (zebra leaf 16, zl16) that develops zebra leaves with reduced chlorophyll contents and abnormal chloroplast microstructures during the early leaf developmental stage. Map-based cloning showed that ZL16 encodes a FabA-like HAD, targeting the chloroplast. An expression experiment revealed that the zl16 mutation was likely involved in an alteration of the transcriptional levels of many of the nuclear- and plastid-encoded genes related to chloroplast development. Our results indicated that fatty acid synthesis is closely related to chloroplast development in rice.
Materials and Methods
Plant Materials and Growth Conditions
The zl16 mutant was selected from an ethyl methyl sulfonate (EMS)-induced mutant pool of indica c.v. 93-11. F2 populations derived from a crossing between zl16 and 02428 or Wuyunjing 7 were used for genetic analysis. All plants were grown in a paddy field at Nanjing, China (32°N latitude) unless otherwise stated. For a shading treatment, plants were germinated and kept in a growth chamber (ATC40, Conviron, MB, Canada) under a 12:12 h light (440 μmol m-2 s-1)/darkness cycle at temperatures of 30°C (day)/20°C (night). For temperature treatments, the chamber containing plants was set at 12:12 h light/darkness cycle at a constant temperature of 20°C or 30°C. The genetic materials generated in this study will be provided in a timely manner for non-profit use.
Quantitative Analysis of Chlorophyll Content
According to a previous study (Lichtenthaler, 1987), pigments from 8 and 12 d seedlings of the 93-11 and zl16 mutants were extracted. Equal weights of fresh leaves (approximately 30 mg fresh weight) were cut into small pieces, and then immersed in 5 ml of 95% ethanol in darkness, with intermittent shocking. After a 48-h treatment, the absorbance of the supernatants was measured by a spectrophotometer (SpectraMax M3, Molecular Devices, San Jose, CA, United States) at 470, 649, and 665 nm, respectively.
Transmission Electron Microscopy (TEM) Analysis
To observe the structure of chloroplasts, seeds were soaked in tap water for 2 days and then germinated in a growth chamber. The temperature was set for 16 h at 30°C in the light (photon flux 440 μmol m-2 s) and 8 h at 20°C in the dark. The second leaves from 8 and 12-day-old seedlings were harvested. The samples were processed and sectioned according to a method used previously (Wang et al., 2017), and the sections were observed by TEM (H7650, Hitachi, Tokyo, Japan).
Fine-Mapping and Cloning of zl16 Locus
An F2 population derived from a cross between zl16 (in the indica background) and 02428 (in the japonica background) was used for the map-based cloning of zl16. Seedlings with the typical mutated phenotype were collected for DNA extraction. Initially, 10 individuals were used for preliminary linkage analysis. Then 944 additional individuals were used to delimit the zl16 locus to a region flanked by the markers VF3 and VF4. Within this region, the gene divergences between 93-11 and zl16 were examined by sequencing genomic DNA. Simple sequence repeat (SSR), insert-deletion (In-Del), and derived cleaved amplified polymorphic sequences (dCAPS) markers were developed to confirm the ZL16 locus. The sequences of the newly developed primers are shown in Supplementary Table S1. The polymerase chain reaction (PCR) procedure was the same as that used in a previous study (Wang et al., 2017).
Genetic Complementation and Artificial Suppression of ZL16
For the complementation of the zl16 mutant, a full-length ZL16 cDNA fragment including a 2.4-kb upstream sequence from the start codon of a genomic fragment were amplified with the HUQ and HUC prime pairs (Supplementary Table S2) using KOD FX polymerase (Toboyo, Osaka, Japan). Then the correct PCR products were cloned into the binary vector pCUbi1390 with restricted enzyme sites KpnI/BamHI. The resultant recombinant construct pCUbi1390-ZL16 was introduced into Agrobacterium tumefaciens EHA105. The zl16 mutant calli was infected using the same method as described previously (Hiei et al., 1994).
A 20 bp fragment targeted to ZL16 was designed using the CRISPR-P web tool1. The targeted fragment was inserted into the CRISPR/Cas9 vector pCAMBIA1305.1 carrying the CaMV35S promoter. The sequenced construct was introduced into cv. Nipponbare as described previously (Hiei et al., 1994). Positive T1 plant lines were obtained by sequencing the targeted gene.
Heterologous Expression of Recombinant ZL16 Proteins in E. coli
The cDNA sequences from wild-type and mutant genes of ZL16 were amplified using the primer pairs pGEX-4T-2F and pGEX-4T-2R (Supplementary Table S2), respectively. The PCR products, containing each gene, missing the 56-amino-acid signal peptide, were inserted into the pGEX-4T-2 vector. The constructions pGEX:: ZL16 and pGEX:: zl16 were transformed into a competent E. coli BL21(DE3) strain (Genescript, Nanjing, China). Heterologous expression of the genes was induced at OD 0.6 with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). The cells were collected and lysed by sonication on ice. The protein was extracted from the resulting supernatant and purified by a glutathione S-transferase (GST) protein purification kit (ProfiniaTM, Bio-Rad, Hercules, CA, United States). HAD activities were assessed with a spectrophotometric method as described previously (Sharma et al., 2003; González-Thuillier et al., 2016). The substrate crotonoyl-CoA was purchased from Merck (Kenilworth, NJ, United States; catalog No. 28007). Protein concentration was determined by a BCA Protein Assay kit (Thermo Fisher Scientific, Waltham, MA, United States). SDS polyacrylamide gel electrophoresis (PAGE) was used to determine the purity of recombinant proteins.
Subcellular Localization of ZL16 Protein
The online tools TargetP2 and ChloroP1.13 were used to predict the transit peptide (Emanuelsson et al., 2007). To validate the subcellular localization of ZL16 protein, the coding sequence of ZL16 (1-648, excluding the stop codon) was amplified using the primer pair Zl16- green fluorescent protein (GFP) (Supplementary Table S2). ZL16 cDNA was inserted into the vector pET30a and then cloned into the transient expression vector pAN580 with the CaMV35S promoter to generate a chimeric gene at the N-terminus of the GFP as described previously (Hu et al., 2012). The constructs were then transformed to rice protoplasts as described previously (Yoo et al., 2007). Protoplast extraction was achieved using a method adopted in a previous study (Zhang et al., 2011). GFP fluorescence signals and autofluorescence of chlorophylls were analyzed with a confocal laser scanning microscope (LSM780, Carl Zeiss, Oberkochen, Germany). Empty vector pAN580 was used as the control.
Quantitative Reverse Transcription-PCR (qRT-PCR) Analysis
QRT-PCR was conducted to investigate the expression of ZL16, and chlorophyll synthesis- and plastid development-related genes. Total RNA extraction from rice seedlings, cDNA preparation, and PCR analysis were performed as previously described (Zhou et al., 2012; Liu and Charng, 2013). The primers for the qRT-PCR are listed in Supplementary Table S3. The ubiquitin gene (LOC_Os03g13170) served as the internal control. The associated gene expressions were normalized to that of ubiquitin. For each sample, three technical replicates on three biological replicates were conducted.
Phylogenetic Analysis
A ZL16 peptide sequence, with 216 amino acids, was obtained from Gramene4. A homologous analysis of ZL16 was conducted at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/).The peptide sequences were used to construct a phylogenetic tree using the MEGA version 5.0 software (http://www.megasoftware.net/; Peng et al., 2014) by the bootstrap method with 1,000 replicates.
Modeling of the Three-Dimensional (3D) Structures
The 3D structures of zl16 and its wild type were modeled with the protein data bank (PDB) server5 and PDB KING viewer (Berman et al., 2000). The structural data of the Plasmodium falciparum HAD sequence (accession number UniProtKB Q965D7, Kostrewa et al., 2005; Maity et al., 2011) was used as a template for both rice sequences.
RNA-Seq Library Preparation and Sequencing
Leaf samples from 8-day-old plant seedlings were ground immediately in liquid nitrogen and RNA was isolated using TriZol (Invitrogen) and cleaned with DNase1 using a Total RNA kit (Sigma-Aldrich). An Agilent 2100 BioAnalyzer was used to assess the quality of the resulting RNA. Library construction, PCR amplification and sequencing were conducted at Berry Hekang Biotech Ltd. (Beijing, China). Each RNA sample for sequencing library was repeated at least three times, and sequenced on the Illumina HiSeqTM 2500 platform, which produced an average read length of 100 bp. The mapping of sequencing reads onto the rice genome (The MSU Rice Genome Annotation Project Data base version 7.0) was conducted using Bowtie software (bowtie2-2.2.9) with the default parameters (Langmead and Salzberg, 2012). A gene ontology (GO) functional classification for each sequence was determined using the Blast2go program (Conesa et al., 2005). Gene expression between the wild type and mutant was considered significantly different if the absolute value of | log2 (fold change) | was ≥1 and q ≤ 0.05.
Measurement of the Fatty Acid Content
Fatty acids were extracted from rice seedlings and analyzed by the gas chromatography–mass spectrometry (GC-MS) of methyl ester derivatives as described previously (Bligh and Dyer, 1959). Lipids were extracted and separated by thin-layer chromatography (TLC). Each individual lipid was isolated and treated with methanolic HCl. Then the fatty acid methyl esters (FAMEs) were analyzed by GC-MS (TRACETM 1310, Thermo Fisher Scientific) with a 30-m (length) × 0.25-mm (inner diameter) × 0.25-μm (gel thickness) TG-5MS column. The parameters were set as following: sample size, 1 μL; injection temperature, 250°C; column temperature, 260°C; split ratio, 10:1; and carrier gas flow velocity, 0.5 mL/min. The fatty acids were detected by comparing retention times with FAME standards (Sigma, Aldrich, St Louis, MO, United States). Most tests were conducted at Qingdao Kechuang Quality Analysis Ltd. (Shandong, China).
Statistical Analysis
Student’s t-tests were performed with MicrosoftTM Excel 2007. For each experiment, the number of biological replicates (n) is shown in the corresponding figure legend. P < 0.05 indicated a statistically significant difference and P < 0.01 indicated a very significant difference.
Results
Isolation and Phenotypic Description of the zl16 Mutant
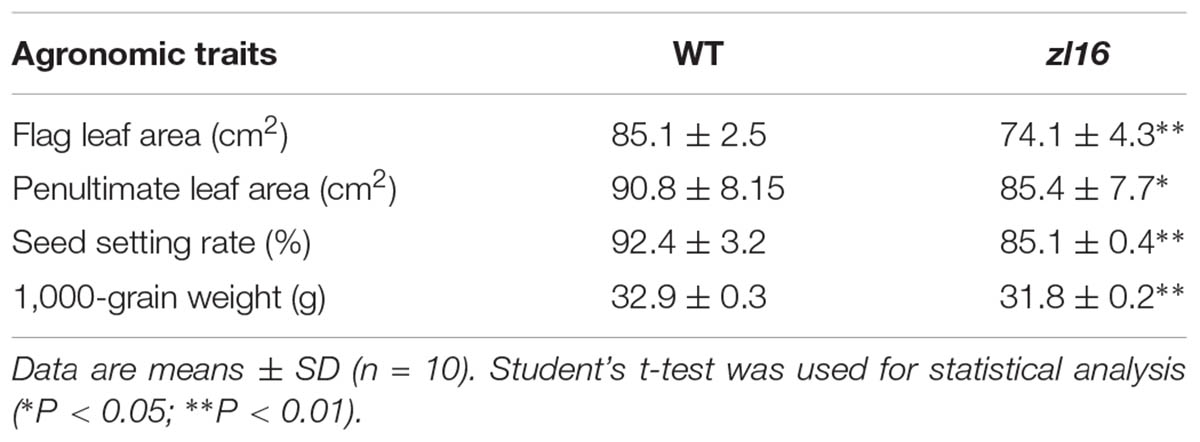
The zl16 mutant was derived from the indica variety 93-11 mutagenized by EMS. Under high light intensity conditions (paddy field with a maximum light intensity of 3,000 μmol photons m-2 s), the zl16 seedling displayed a transverse zebra leaf symptom during early growth (Figure 1A). As the leaf developed, the defective phenotype was gradually restored and disappeared after the fifth-leaf stage. Previously, a mutant (zebra2) was reported, which developed a zebra leaf phenotype under diurnal light–dark cycles in a natural field, whereas this was suppressed under shaded conditions (Chai et al., 2011; Han et al., 2012). Thus, zl16 plants were grown in a growth chamber with an average of 30% of the light intensity of field conditions. Compared to those grown in a natural paddy field, the zl16 seedlings exhibited much weaker lesions or stripes, and under growth chamber conditions, all seedlings displayed the light-green phenotype, with the most obvious symptoms observed at the second-leaf stage (Figures 1B–D). After the fifth-leaf stage, all leaves were as green as wild-type plants. When the plants initiated flowering, no leaf-color difference was found between the wild type and zl16 mutant. Additionally, the mature zl16 mutant plants showed a significant reduction in traits including flag and penultimate leaf areas, seed setting rate, and 1000 grain weight (Table 1), compared with the wild type (t-test, n = 10). Consistent with these observations, the zl16 mutant leaves had a much lower chlorophyll and carotenoid contents than the wild-type seedlings at the earlier stages of leaf development (Figure 1E), but gradually recovered to wild-type levels in the later stages of development (Figure 1F).
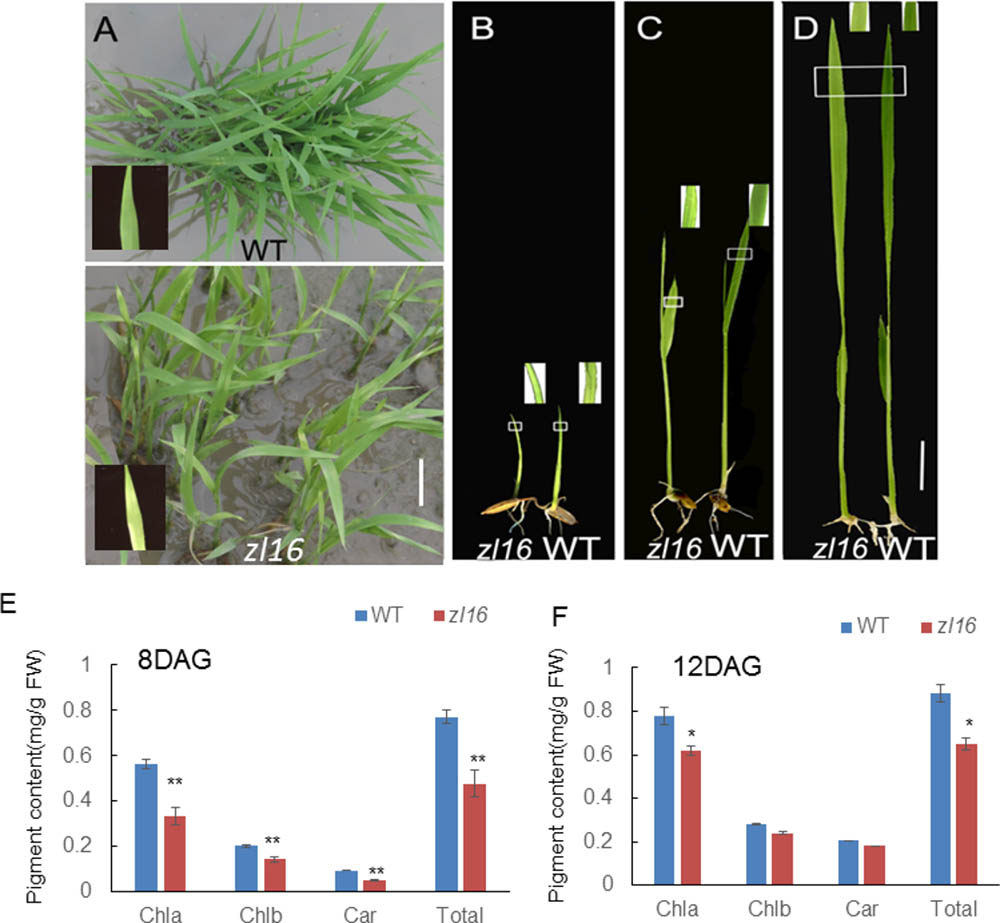
FIGURE 1. Phenotypic characteristics of zl16 mutant. (A) Phenotypic comparison between the wild type (WT) and zl16 mutant seedlings in the paddy field after 20 days of germination. The inset means enlarged detail of leaf blade. (B–D)The WT and zl16 mutant seedlings in a growth chamber at the 5th- (B), 8th (C), and 12th-day after germination, respectively. Bars = 2 cm. The inset indicates enlargement of leaf sector in box. (E–F) Measurement of pigment contents in WT and zl16 mutant seedlings at the 8th (E) and 12th- day (F) after germination (DAG), respectively. Chla, Chlorophyll a; Chlb, Chlorophyll b; Car, total carotenoids; FW, fresh weight. Bar denotes standard error (SD, n = 6). Asterisks indicate the significance levels according to Student’s t-test: ∗P < 0.05, ∗∗P < 0.01.
To investigate the effect of different temperatures on the zl16 phenotype, we treated zl16 and wild type seedlings at two temperatures (i.e., high temperature, 30°C; low temperature, 20°C). We found that, despite much slower growth at the low temperature, the zl16 mutant displayed a consistent leaf phenotype under different temperature conditions (Supplementary Figure S1). These results indicate that zl16 is a temperature insensitive mutant.
Chloroplast Ultrastructure of the zl16 Mutant
To determine whether the color deficiency in the zl16 mutant was related to ultrastructural changes in chloroplasts, we compared the chloroplast features from wild-type and zl16 mutant leaves using TEM. As shown in Figures 2A–C, 8-day wild-type chloroplasts had well developed lamellar structures and were equipped with normal thylakoid membranes and stacked grana. In contrast, the chloroplasts in the white striped regions from 8-day mutant seedlings were obviously shriveled and lacked an organized lamellar structure (Figures 2D–F). As the seedlings developed, the chloroplast structural damage in the 12-day mutant leaves was alleviated to some extent and the chloroplast was almost restored to its wild-type status (Figures 2G–I).
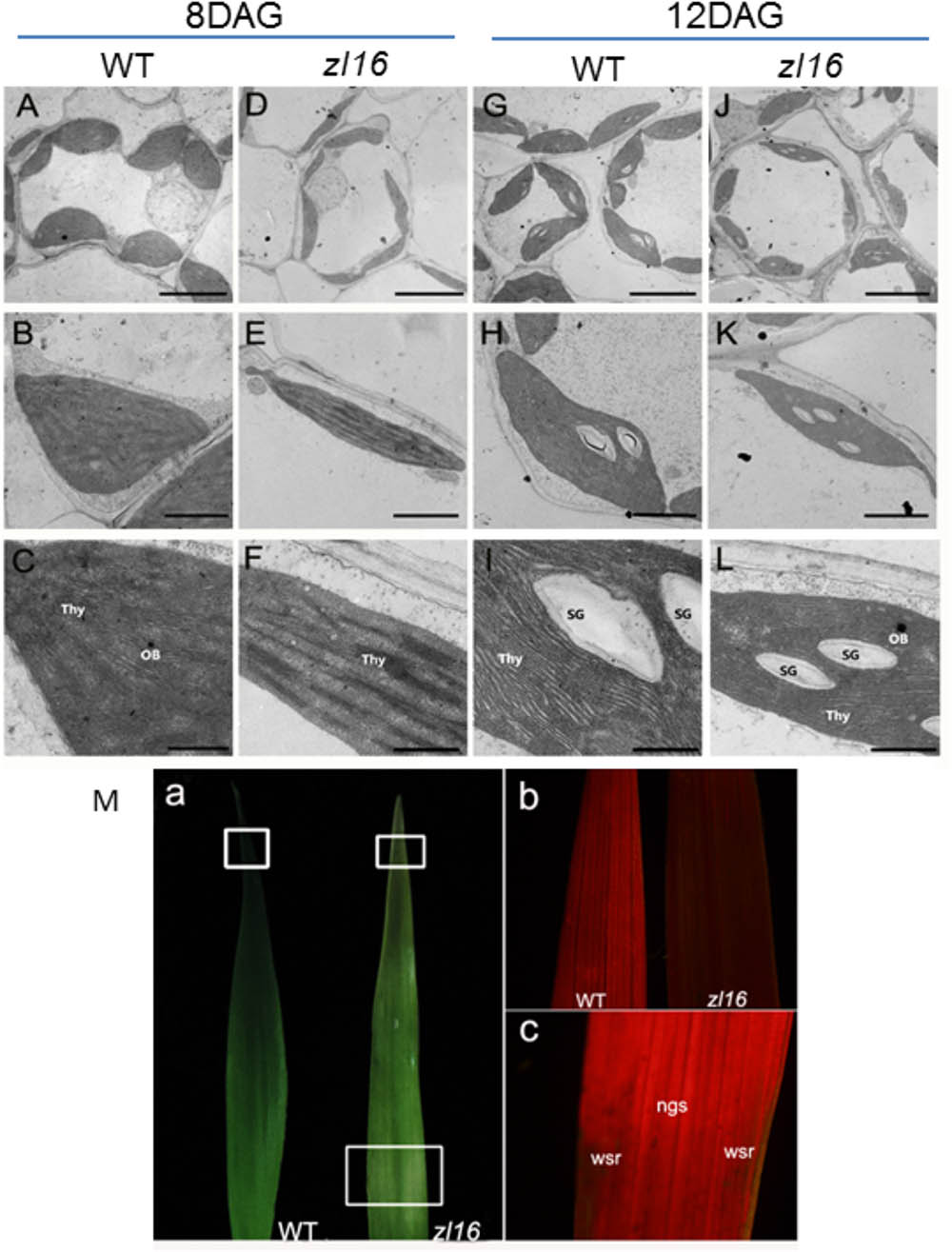
FIGURE 2. Ultrastructure of the chloroplast and chlorophyll autofluorescene observations of wild type (WT) and zl16 mutant. (A–L) Transmission electron micrographs (TEM) images of WT (A–C and G–I) and zl16 (D–F and J–L) chloroplasts. Seedlings were grown for 8 and 12 days after germination, respectively. OB, osmiphilic body; SG, starch granule; Thy, thylakoid lamellar. Bars: 2 μm (A,D,G,J), 1 μm (B,E,H,K), and 0.5 μm (C,F,I,L). (M) Observation of autofluorescence of leaves. Panel a, observation of leaves under natural light conditions. The boxes in the upper and lower parts denote the regions that were enlarged in panel b and c, respectively. Panel b, autofluorescene of leaf tips from wild-type and zl16 seedlings under UV light, respectively, at 8-d after germination. Panel c, autofluorescene from the white striped regions (wsr) and normal green sectors (ngs) in the zl16 mutant seedlings under UV light, respectively.
A previous study showed that chlorophyll autofluorescence is an indicator of normal chloroplast development (Kitajima and Butler, 1975). We observed chlorophyll autofluorescence in wild type and zl16 mutant seedlings under UV light (Figure 2M panels a–c). After 8 days of germination, chlorophyll autofluorescence at the leaf tip in the zl16 mutant was obviously weaker than its counterpart in the wild type (Figure 2M, panel b). In the same way, the autofluorescence from the white striped regions was significantly lower than that in the normal green sectors of zl16 leaves (Figure 2M, panel c). Collectively, the results indicated that chloroplast development was blocked in the zl16 mutant.
Isolation and Confirmation of the Rice zl16 Gene
For a genetic analysis of the zl16 locus, two F2 populations were produced from zl16/ Wuyunjing 7 and zl16/02428. All heterozygous F1 seedlings had the wild-type phenotype and the F2 segregation pattern fitted a 3:1 normal green to zebra leaf ratio under a x2 test (Supplementary Table S4). These data indicate that the zebra leaf in the zl16 mutant is controlled by a single recessive nuclear gene.
Genetic mapping of the zl16 gene was conducted using the F2 population from the zl16/02428 cross. Initially the zl16 locus was mapped to a region between the markers In8-3 and RM8243 on the short arm of chromosome 8 based on ten typical zebra leaf F2 individuals (Figure 3A). Then 94 additional zebra-leaf-like individuals were used to narrow the zl16 lous into a 2.8 Mb distance between the markers L1 and L3. With the newly developed SSR and In-Del markers (Supplementary Table S1), as well as other 944 F2 homozygous mutant-like plants, we finally delimited the zl16 locus to a 58.2 kb region between In-Del markers VF3 and VF4. Within this interval, eight open reading frames (ORFs) were predicted with the program RiceVarMap v2.06 (Figure 3B, Supplementary Table S5).
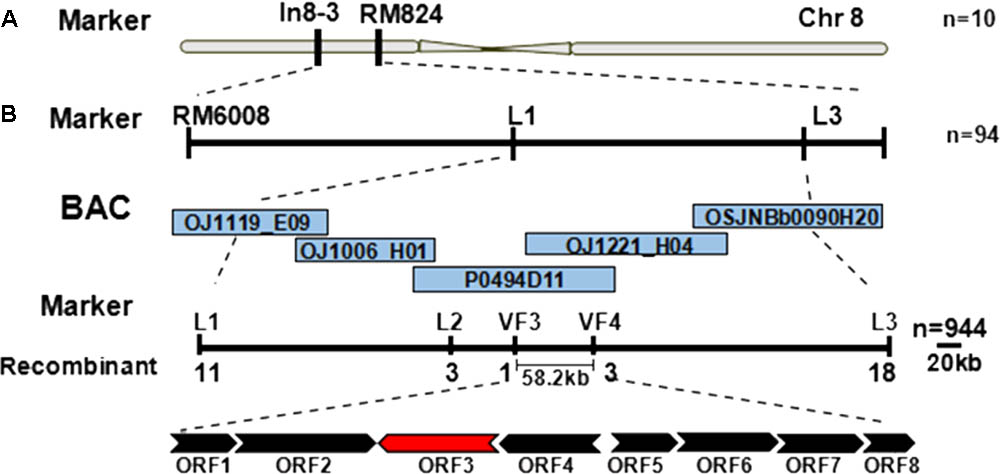
FIGURE 3. Map-based cloning of the zl16 gene. (A) Rough mapping of the zl16 locus with SSR and In-Del markers. n, the number of zebra leaf-like plants for mapping in the F2 population from zl16/02428 cross. Chr, Chromosome. (B) Fine-mapping of the zl16 locus. BACs denote bacterial artificial chromosomes of rice whole genome. ORF, open reading frame; the most likely candidate gene is indicated in red. The arrows indicate transcriptional direction.
To determine the mutation site, all eight ORFs were separately sequenced using genome DNA. Sequence alignments with wild-type genes revealed that an A-to-T transition in LOC_Os08g12840 was present in the fourth exon of the third ORF in the zl16 mutant (Figures 4A,B). This resulted in a substitution of Arg by Trp at the + 164 position of the encoding protein (Figure 4C). No difference was detected in the other seven ORFs.
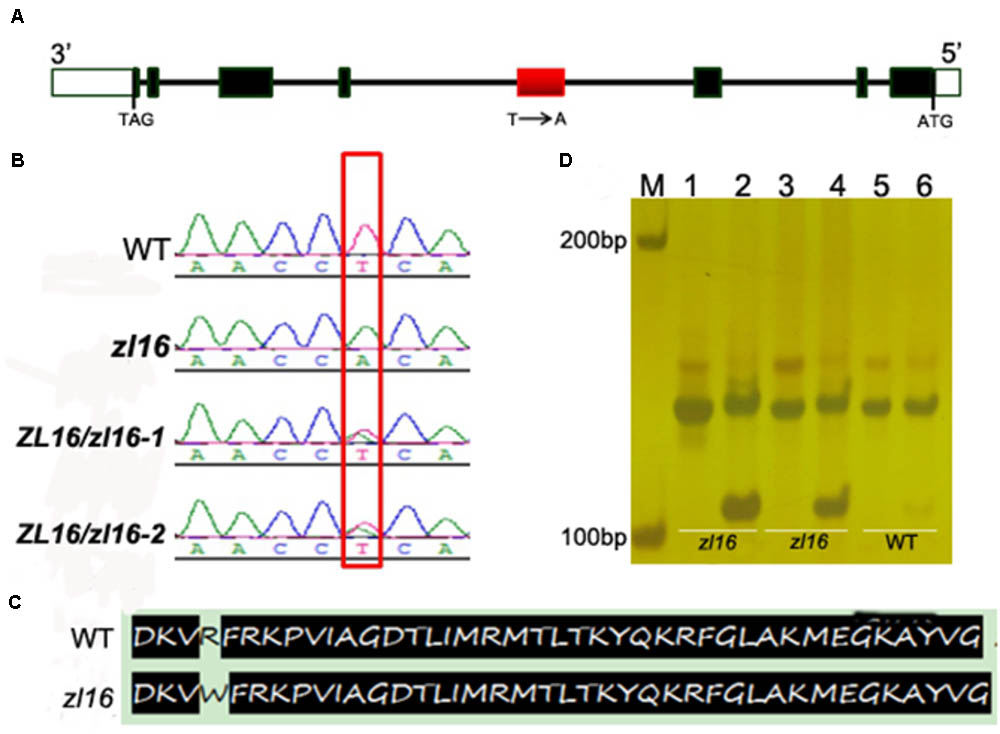
FIGURE 4. Identification of the mutation site and structure of zl16. (A) The structure of ZL16 gene. ATG and TAG represent the start and stop codons, respectively. Black box indicates the exon and the line denotes intron. The region where base substitution happens is shown in red box. (B) cDNA sequence comparison between the wild type, homozygous zl16 mutant (zl16/ zl16), and two heterozygous (ZL16/ zl16-1 and 2) genotypes. The point mutation was shown in box. (C) Comparison of translated protein sequences in the wild type (WT) and zl16 mutant. (D) Identification of the base substitution between wild type and zl16 with a dCAPs marker. Lane M means the molecular weight marker; Lanes 1, 3, and 5 indicate PCR products without AluI digest; Lanes 2, 4, and 6, PCR products with AluI digest. Lanes 1–4, DNA samples from the two homozygotes of the zl16 mutant. Lanes 5–6, DNA samples from the wild-type cultivar, 9311.
To confirm the point mutation of LOC_Os08g12840 in the zl16 mutant, we designed a dCAPS marker and used it to genotype the wild type, heterozygous zl16 plant, and homozygous zl16 mutant. AluI could cut the homozygous zl16 allele but could not digest the wild-type allele (Figure 4D). We then used the dCAPS marker to detect zebra seedlings in the F2 population and found that all the recessive recombinants surrounding the zl16 locus were co-segregated with this marker. Thus, LOC_Os08g12840 (ZL16) was thought to be a candidate gene for the zl16 phenotype.
Complementation tests were conducted to examine whether the missense mutation led to the zebra phenotype. The seedlings regenerated from the zl16 mutant calli that expressed the wild-type ZL16 version restored the normal leaf color (Figure 5A) and chloroplast microstructure (Figure 5B), which was similar to the wild type. Homozygous lines of the transgenic plants with a CRISPR event were also isolated for analysis. Phenotyping results indicated that a knockouting of the ZL16 gene by CRISPR-Cas9 demonstrated a chlorophyll deficient phenotype under a japonica rice ‘Nipponbare’ background (Supplementary Figure S2). Hence LOC_Os08g12840 corresponds to the ZL16 gene.
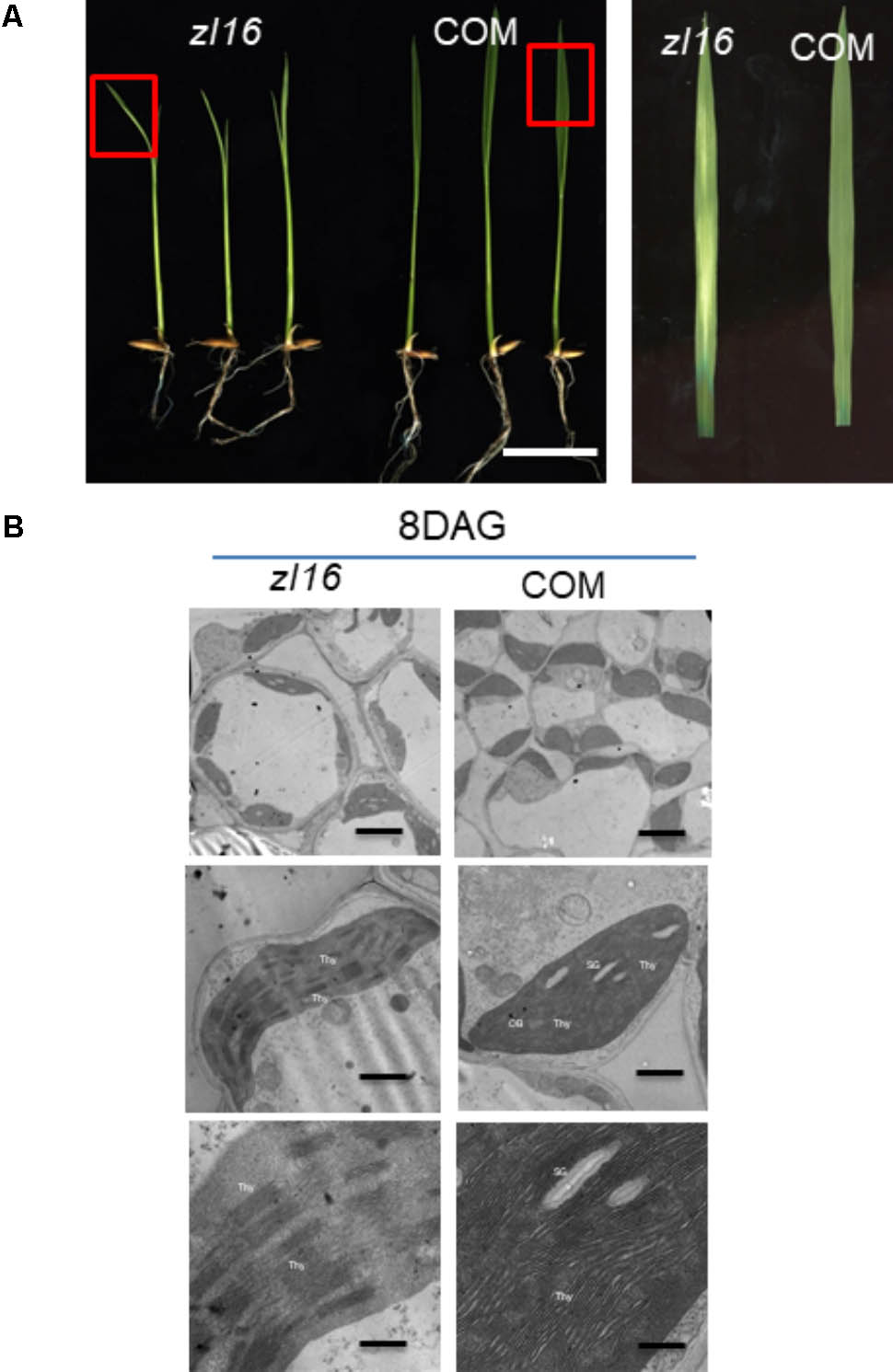
FIGURE 5. Functional confirmation of ZL16 gene. (A) Genetic complementation. The functional wild-type (WT) ZL16 was inserted in the zl16 mutant genome and zl16 recover to normal green color. “COM” indicates three representative positive complemented plants generated by callus transformation. Bar = 2cm. The boxes in the left panel denote the regions that are enlarged in the right panel. (B). Transmission electron micrographs (TEM) images of zl16 (left panel) and complemented (COM) plant (right panel) chloroplasts. Seedlings were grown for 8 days after germination. OB, osmiphilic body; SG, starch granule; Thy, thylakoid lamellar. Bars: 2 μm (top row), 1 μm (middle row), and 0.5 μm (bottom row).
Features of ZL16
The whole amino acid sequence from ZL16 was used for a BLASTP search of the NCBI website7. We found that the ZL16 gene had only a single copy in the rice genome, consisting of eight exons (Figure 4A) and encoding a polypeptide with 216 amino acid residues (Figure 6A). The ZL16 protein shared a high sequence similarity with its homologs in algae, monocotyledon, and dicotyledon (Figure 6B), suggesting its evolutionary sequence is highly conserved in plants. Many of these homologs were annotated as HAD; thus, we named ZL16 as OsHAD1 and another ZL16 homolog on chromosome 5, which was identified by BLAST analysis, as OsHAD2. OsHAD1 and OsHAD2 shared a 45.9% identity in amino acid sequences and little reference information is available for both OsHAD members in plants.
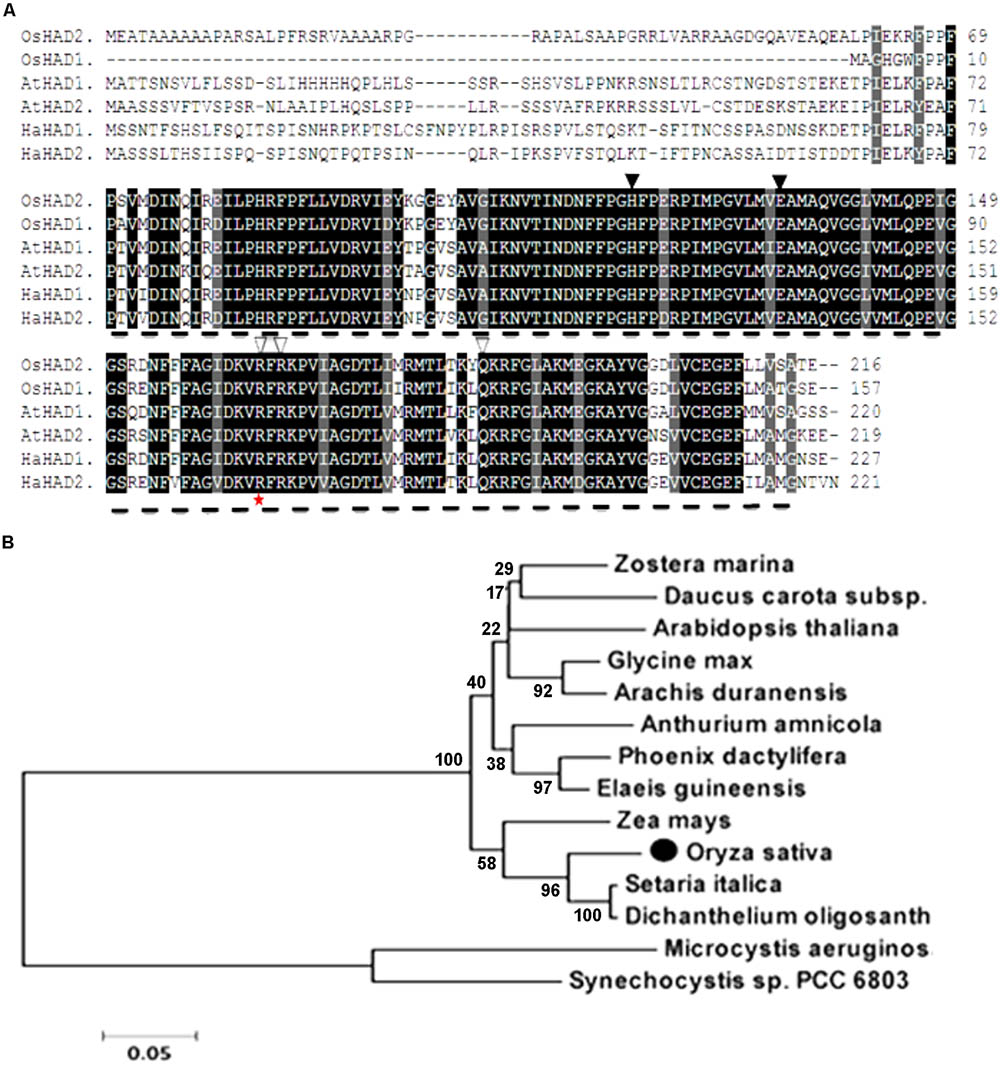
FIGURE 6. Alignment of the predicted amino acid sequence and phylogeny of ZL16. (A) Amino acid sequence alignment of ZL16 (OsHAD1). Dash line denotes FabA-like domain. Identical residues are highlighted as black shadings, highly conserved residues as dark gray shadings and weakly conserved residues as light gray shadings. Black triangles indicate the active sites, and white triangles are the residues participating in the ACP recognition and binding sites. The red star indicates the mutant site in the zl16 mutant. Os, Oryza sativa; At, Arabidopsis thaliana; Ha, Helianthus annuus (sunflower). (B) Phylogenetic analysis of ZL16 proteins (black solid dot). A maximum likelihood phylogenetic tree was constructed based on the amino acid sequences using the MAGA5.0 software.
ZL16 contains a FabA-like domain and belongs to the hot-dog protein superfamily. There are two critical areas in the FabA-like domain. One is considered to be a catalytic activity center and includes two conserved residues (His119 and Glu133), while the other is considered to be involved in ACP recognition and binding, and harbors three critical residues (Arg164, Arg166, and Glu181) (González-Thuillier et al., 2016). The point mutation in the zl16 mutant (Arg164Trp) was found to reside at the first conserved residue in the ACP recognition and binding sites (Figure 6A). Based on the 3D structure model, the mutation in zl16 led to the enlargement of a curve angle near the + 164 site in the tertiary structure compared to the wild type (Supplementary Figure S3), which might compromise HAD recognition and its binding ability to its substrates. To further verify the hypothesis, we successfully expressed the wild-type and mutant zl16 coding proteins in E. coli with the expected molecular weight sizes (Figure 7A). A comparison of the enzyme activity experiments revealed that purified mutant zl16 recombinant protein had a significantly lower activity than its wild-type counterpart, which amounted to only 17.1% of the wild-type level (Figure 7B). The near loss of HAD enzyme activity in the mutant was consistent with the recessive genetic feature of the zl16 mutation.
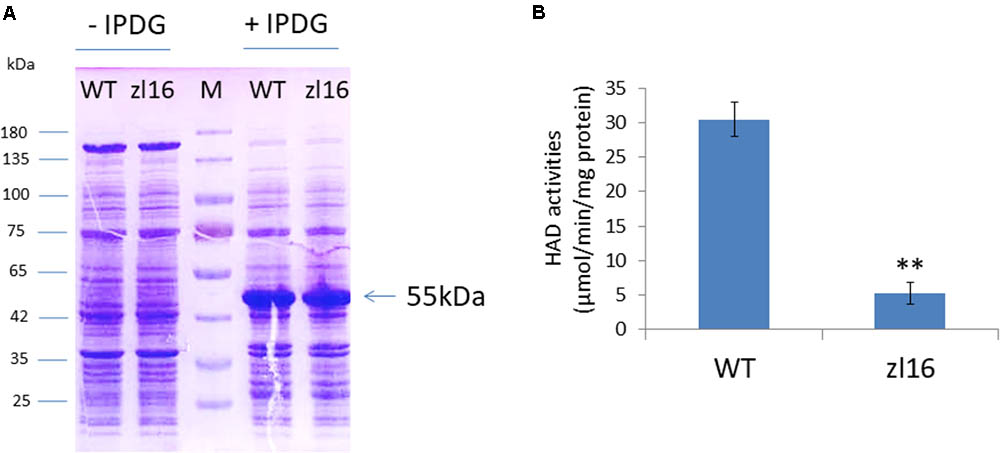
FIGURE 7. Heterologous expression of recombinant ZL16 proteins in E. coli. (A) Expression of wild-type (WT) and zl16 recombinant proteins in E. coli. –/+ denote before and after adding 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) for protein induction. M means molecular weight marker. Arrow indicates the target protein from ZL16 or zl16 (29 kDa) fusing with glutathione S-transferase (26 kDa). (B) Comparison of β-hydroxyacyl-ACP dehydratase (HAD) enzyme activities between the wild type (WT) and zl16 after purification of the recombinant proteins. Data are means ± SD of six replicates. ∗∗P < 0.01.
QRT-PCR revealed that ZL16 transcripts were detected in all tissues examined including seed, root, sheath, stem, mature leaf, and young leaf, with a relatively preferential expression in the young leaf (Figure 8A). During seedling growth, OsHAD1 reached the highest expression level in 8-day-old seedlings, and then decreased as the plant developed. In contrast, OsHAD2 showed low expression during early seedling growth, but there was a constant increase during leaf development. OsHAD2 had a higher or equal expression than OsHAD1 in all tissues other than young leaves.
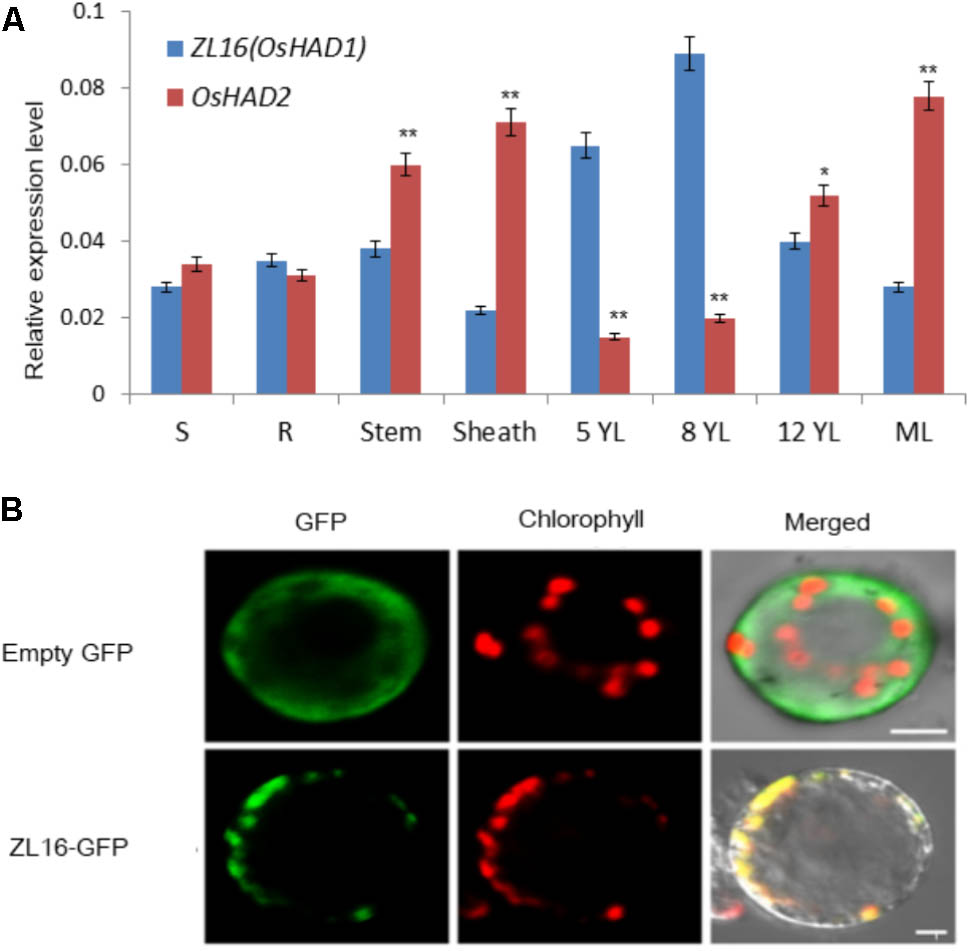
FIGURE 8. Expression pattern and subcellular localization of ZL16. (A) Expression analysis of ZL16(OsHAD1) and OsHAD2 genes in rice various tissues. S, seed; R, young root; ML, mature leaf during flowering; 5, 8 and 12 YL, young top leaves from 5-, 8- and 12-day-old seedlings after germination. Ubiquitin gene was used as an internal control. Data are means ± SD of three biological replicates. For t-test, OsHAD2 values were compared with expression level of ZL16(OsHAD1). ∗P < 0.05; ∗∗P < 0.01. (B) Transient expression of ZL16–GFP in rice protoplasts. Empty GFP indicates vector construct from GFP alone for subcellular localization, ZL16-GFP indicates construct from fused ZL16 and GFP for subcellular localization. GFP, GFP signal of ZL16 fusions; Chlorophyll, chlorophyll autofluorescence; Merged, merged images of GFP and chlorophyll autofluorescence. Bar = 10 um
Both online tools, ChloroP and TargetP, predicted ZL16 harbors a chloroplast transit peptide at its N-terminus, suggesting ZL16 is localized to chloroplasts. To verify this, we generated the full-length ZL16 cDNA (not including the stop codon) fusion with GFP. In rice protoplasts, confocal microscopy showed that the green fluorescence of GFP-ZL16 was entirely co-localized with the autofluorescence signal of chlorophyll in the chloroplasts, while the empty GFP protein was mainly located in the cytosol (Figure 8B). These results indicate that ZL16 is located within the chloroplast, which is a cellular compartment where fatty acids are mainly synthesized in plants (Ohlrogge and Jaworski, 1997).
Expression Analysis of Other Related Transcripts
The zl16 mutant had a zebra leaf lesion and produced less chlorophyll than its wild type (Figure 1), and therefore it was presumed that the mutant might have transcriptional defects in chlorophyll synthesis. We performed a qRT-PCR to analyze the expression of chlorophyll synthesis-related genes. In 23 tested genes, 11 were down-regulated: HEMA (encodes glutamyl-tRNA reductase), HEMB (encodes porphobilinogen synthase), CHLH (encodes Mg-chelatase subunit H), CRD [encodes Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase], PORA (encodes NADPH:protochlorophyllide oxidoreductase A), CAO (encodes chlorophyll a oxygenase 1), HO (encodes heme oxidase 1), GUN4 (encodes GENOMES UNCOUPLED 4), PIF3, PIF4, and PIF7 (encode phytochrome-interacting factors 3, 4, and 7, respectively). Of these genes, PORA was the most down-regulated (Figure 9A). These results indicate that chlorophyll biosynthesis might be impaired in the zl16 mutant.
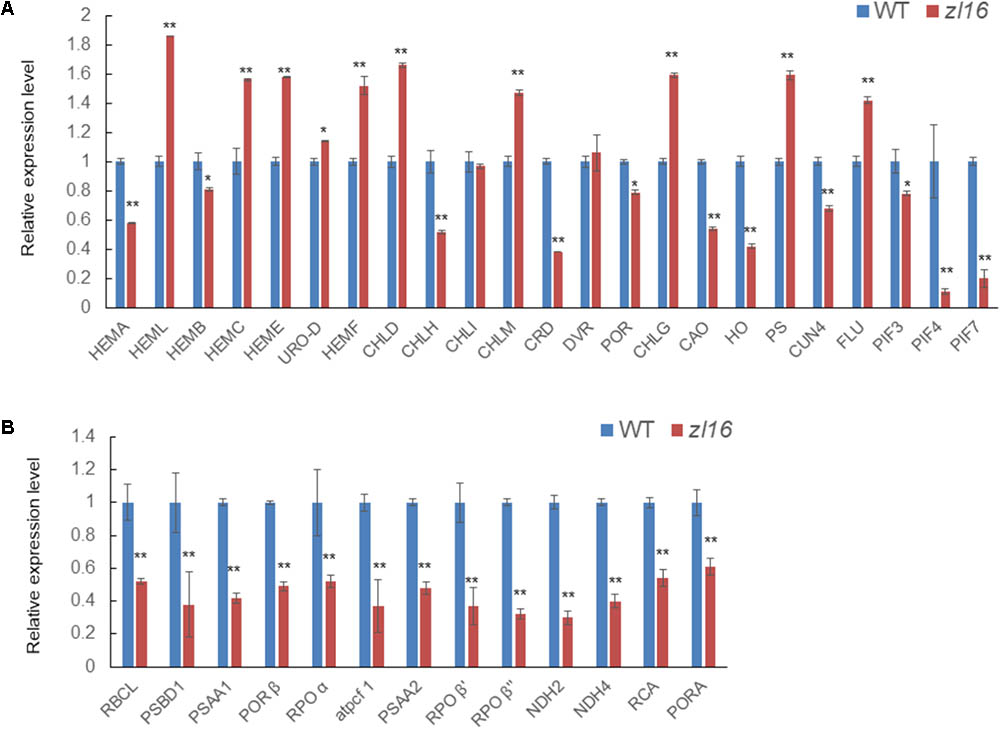
FIGURE 9. Expression analysis of genes associated with chlorophyll biosynthesis and plastid development in the wild type (WT) and zl16. (A) Relative expression of genes involved in chlorophyll biosynthesis. (B) Relative expression of genes involved in plastid development. Experiments were normalized using the Ubiquitin gene as an internal control. Data are means ± SD of three replicates.
In the TEM observation, chloroplast development of the mutant was found to be abnormal relative to the wild type (Figure 2). Previous studies have revealed that impaired plastid development could trigger plastid retrograde signaling to regulate nuclear-encoded gene expression (Chi et al., 2013), and we speculated that the transcription levels of some relevant genes in the mutant might be altered. As expected, the qRT-PCR revealed that 13 detected genes participating in plastid development were all significantly down-regulated in 8-day-old zl16 seedlings (Figure 9B), suggesting an important role for ZL16 in the development of rice plastids.
To obtain a comprehensive view of the transcriptome affected by the zl16 mutation, we conducted an RNA-Seq analysis to detect the differential genes between the zl16 mutant and its wild type. Under stringent conditions (q < 0.001; |log2 (fold change)| > 3), a total of 195 differential genes encoded by the nuclear genome were identified, among which 124 were up-regulated and 71 down-regulated. The GO analysis of these down-regulated genes revealed that, apart from chlorophyll biosynthesis and plastid development, most of them were involved in the biosynthesis of cell constituents, especially for membrane systems (Supplementary Figure S4).
Determination of the Fatty Acid Content
To further investigate the function of ZL16, we measured the kinds and contents of various fatty acids in wild type and mutant seedlings at 8 days after germination. TLC analysis revealed in the mutant the contents of galactolipids [monogalactosyl-diacylglycerol (MGDG) and digalactosyl-diacylglycerol (DGDG)] were reduced by 25 and 21%, respectively, compared to wild type. In contrast, a slight increase (ranging from 6∼10%) in the abundance of other membrane lipids such as phosphatidylcholine (PC), phosphatidylglycerol (PG), and phosphatidylethanolamine (PE) were observed, while the levels of sulfoquinovosyldiacylglycerol (SQDG) and phosphatidylinositol (PI) were not changed significantly in the mutant (Figure 10A). Because the two galactolipids (MGDG and DGDG) and SQDG are main components of thylakoid membrane lipids (Kobayashi, 2016); this result suggests that the mutant had a disturbed chloroplast membrane lipid composition relative to wild type plants.
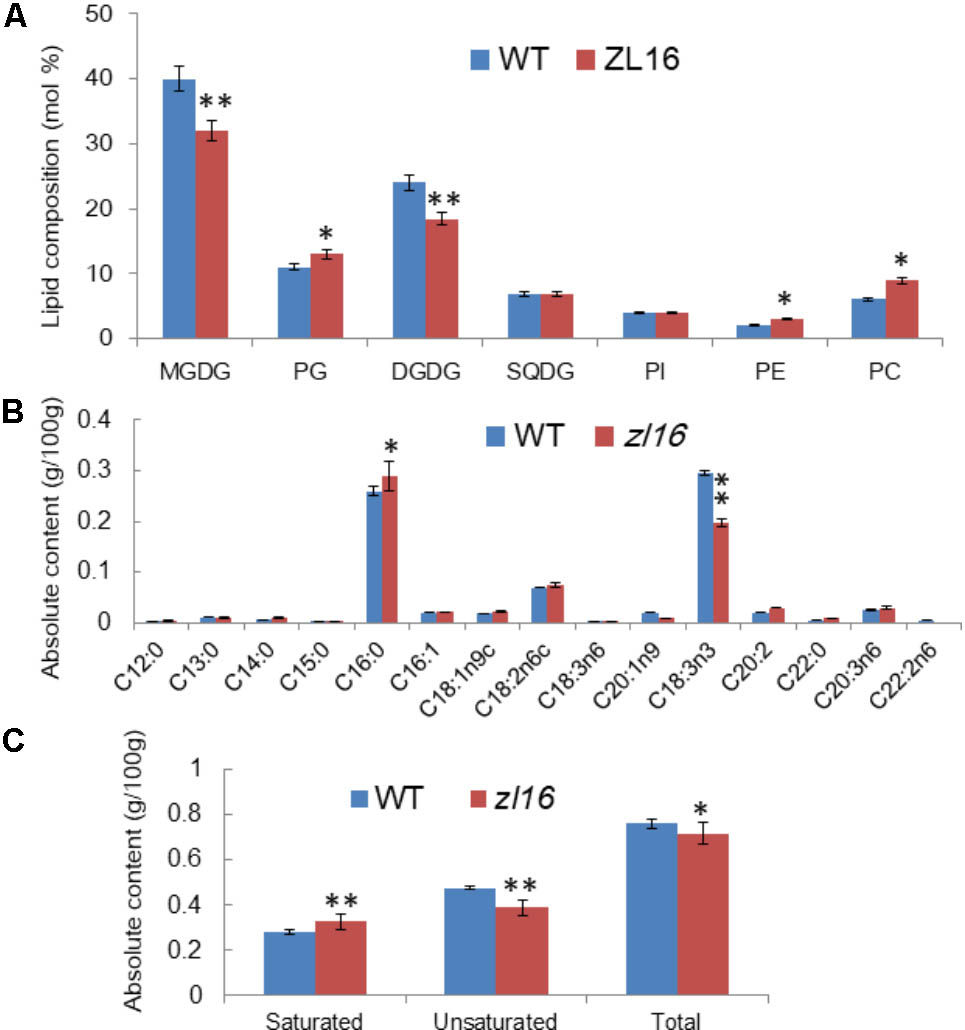
FIGURE 10. Determination of fatty acids in wild-type (WT) and zl16 seedlings. (A) Comparison of polar lipid classes (percentage of total polar glycerolipids analyzed) in leaves of the wild type and zl16 mutant. The bars indicate means ± SD of three measurements. MGDG, monogalactosyl-diacylglycerol; DGDG, digalactosyl-diacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; SQDG, sulfoquinovosyldiacylglycerol. (B)The content of each fatty acid in wild-type and zl16 mutant seedlings. (C) Comparison of saturated and unsaturated fatty acids in wild-type and zl16 mutant seedlings. n = 6. Asterisks indicate the significance levels according to Student’s t-test: ∗P < 0.05, ∗∗P < 0.01.
A total of 13 different fatty acids were identified in the wild type seedlings, among which four (C13: 0, C14: 0, C16: 0, and C22: 0) were saturated fatty acids, and the remainder were unsaturated fatty acids. In zl16 seedlings, a total of 14 different fatty acids were identified. Compared with the wild type, two more saturated fatty acids (C12: 0 and C15: 0) were present, but an unsaturated fatty acid (C22: 2n6) was missing in the mutant. Remarkably, in contrast to the wild type where the most abundant fatty acids was octadecenoic acid (C18: 3n3, an unsaturated fatty acid), palmitic acid (C16: 0, a saturated fatty acid) was predominant in the mutant (Figure 10B). A previous study showed that C18: 3n3 is a major component of MGDG in the chloroplast membrane (Xu et al., 2010). Thus, the significant reduction of C18: 3n3 in the mutant was consistent with the impaired chloroplasts. Overall, the total fatty acid content of the mutant was reduced relative to the wild type. Moreover, we found that the saturated fatty acid content was significantly higher and the unsaturated fatty acid was significantly lower than in the wild type (Figure 10C).
Discussion
Many chlorophyll- and chloroplast-related mutations that change leaf coloration and/or seedling viability have been identified and designated as virescent, stripe, albino, chlorina, zebra, and yellow variegated based on visual phenotypes. Among them, some (such as virescent1, 2, and 3) have been reported as temperature-conditional (Iba et al., 1991; Kusumi et al., 1997; Sugimoto et al., 2004), while others (such as LCM6, 7, and 8) are thermo-insensitive (Wu et al., 1999) in terms of their different responses to temperature. Zebra mutants have alternating green and white (or yellow) stripes on leaf blades and had been reported in many monocotyledonous crops, including rice, maize, sorghum, and pearl millet (Coe et al., 1987; Werner and Burton, 1991; Oki et al., 1997; He et al., 2000; Kusumi et al., 2000). At least 15 non-allelic “zebra” mutants (zebra1-15) have been identified in rice, and the causative genes had been mapped on different chromosomes8. Two of them, ZEBRA-NECROSIS and ZEBRA2, which encode a thylakoid-bound protein (Li et al., 2010) and carotenoid isomerase (Chai et al., 2011; Han et al., 2012), respectively, were cloned. In addition to temperature, the appearance of the zebra phenotype also depends on light and the seedling growth stage (He et al., 2000; Kusumi et al., 2000). In this study, the rice zebra leaf mutant zl16 was identified. Under field conditions with high light intensity, zl16 exhibited its characteristic zebra phenotype with reduced chlorophyll content, while mutant plants grown in a growth chamber under a low light intensity displayed a much less severe phenotype (Figure 1). Moreover, we found that the temperature treatment did not enhance the symptom in the zl16 mutant (Supplementary Figure S1). These results indicate that the zl16 phenotype is light-dependent rather than temperature-conditional.
Map-based cloning indicated that ZL16 encodes a HAD, which is one of four enzymes (KAS, KAR, HAD, and ENR) in the carbon chain elongation cycle of de novo fatty acid synthesis. Fatty acids and their derivatives can not only function as an important component of membrane lipids that are vital for cell growth and development, but also play an essential role as signaling molecules (Weber, 2002; Kachroo et al., 2003, 2004). In Arabidopsis, several mutants have been reported in the different steps of the fatty acid synthesis. Downregulation of KAS expression in a kas1 mutant due to a T-DNA insertion in the 5′-UTR of KAS1 causes chlorotic and curly leaves, decreased fertility, and significantly suppressed chloroplast division in rosette leaves (Wu and Xue, 2010). A mod1 mutant, with a point mutation in the sixth exon of ENR, produces reduced ENR activity, variegated leaves, abnormal chloroplast structure, and reduced fertility (Mou et al., 2000). Under UV light, the mesophyll cells in mod1 yellowish leaves irradiated a weak red fluorescence (Mou et al., 2000). Similarly, here we found a single nucleotide substitution in a conservative region of HAD that led to the reduction of both enzyme activities and total fatty acids, altered the expression of photosynthesis-related genes, and resulted in a defective development of the chloroplast during early leaf development in rice. Intriguingly, apart from leaf color, in the zl16 mutant there was also a significant reduction in traits such as the size of mature flag and penultimate leaves, seed setting rate, and 1,000-grain weight (Table 1), which are highly consistent with the Arabidopsis mod1 mutant (Mou et al., 2000). These results indicate that a lipid supply is essential to plant growth and development including the chloroplast structure in plants. There is an abundance of thylakoid membranes in chloroplasts; thus, sufficient provision of lipids is required for normal chloroplast formation. Defects in the synthesis of fatty acids can compromise chloroplast membrane assembly and development (Fan et al., 2015), in accordance with the observation of significantly reduced MGDG and DGDG in this study. It is worth noting that MGDG and DGDG account for about 50 and 25% of total thylakoid lipids, respectively, in plants (Kobayashi, 2016). The lack of the two galactolipids in the zl16 mutant confirms the close relationship between polar lipid supply and chloroplast development. Additionally, our RNA-Seq results indicated that many genes associated with the biosynthesis of cell membrane systems were downregulated in the zl16 mutant, which further supports the notion that a lipid supply is essential for ensuring chloroplast structure.
Chloroplast development was blocked in zl16 leaves at early growth stages; however, it recovered to some extent in terms of chloroplast size in the later developmental stages relative to the wild type. This is coincident with leaf color phenotypic changes in the zl16 mutant. There are two possible reasons for the restoration of chloroplasts. First, in the course of ZL16 (OsHAD1) deficiency, another OsHAD1 paralogous member (OsHAD2) might replenish the HAD activity in plastids. We found that ZL16 is predominantly expressed in young leaves, whereas OsHAD2 was mainly present in mature leaves based on qRT-PCR data, indicating that ZL16 have a function in fatty acid synthesis primarily in the early stages, while OsHAD2 is more important in the later stages of plant development. Another explanation for the presence of normal chloroplasts is that the mitochondrial fatty acid synthesis system may compensate for the defective plastidial fatty acid synthesis. In a plant cell, de novo fatty acid synthesis can occur in both plastids and mitochondria (James et al., 1995). Similarly to plastids, mitochondria contain all the enzymes associated with de novo fatty acid synthesis and can function independently to form long-chain acyl-ACP (Wada et al., 1997; Gueguen et al., 2000; Focke et al., 2003). The central function of mitochondrial fatty acid synthesis is to maintain lipoic homeostasis, which is important for supporting photorespiration (Ewald et al., 2007; Guan et al., 2015; Guan and Nikolau, 2016). Recently Guan et al. (2017) showed that in both Arabidopsis mthad and mtkas mutants, the thylakoid membrane assembly is altered relative to their wild types, and is accompanied by the appearance of many small starch granules in the chloroplasts. The deformed chloroplasts are similar to those in the zl16 mutant here. Given the functional conservation of HAD for chloroplast development between dicot and monocot plants, we speculate that in the absence of rice plastidial HAD activity (i.e., OsHAD1/ZL16), mitochondrial FA synthesis, e.g., from OsHAD2, may partially complement this gap in fatty acid chain elongation.
In summary, this study found that ZL16, encoding a HAD in rice, plays a vital role in the synthesis of fatty acids and sets a foundation for connecting the relations between lipid and chloroplast development in rice leaves. This information will be useful for gene engineering to improve photosynthetic ability in rice. Further studies should be conducted to obtain comprehensive lipidomic data, particularly focusing on chloroplast lipids, and so enable a mechanistic understanding of the findings.
Author Contributions
LL, JW, and ZL conceived the research. JW supervised the whole project. ZL, LL, ZW, HG, JY, MH, YZ, ZZ, SL, LC, XL, and YT performed the experiments. JW and YW provided the mutant materials. YW, SL, LC, XL, YT, SZ, and JL involve technical assistance. ZL analyzed the data and drafted the manuscript. LL analyzed the data and rewrote the manuscript completely.
Funding
Funds were from The National Key Research and Development Program of China (2016YFD0101801 and 2017YFD0100400), Jiangsu Science and Technology Development Program (BE2017368), the Agricultural Science and Technology Innovation Fund project of Jiangsu Province (CX(16)1029), and the National Natural Science Foundation of China Grant (31571629).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the supports by Key Laboratory of Biology, Genetics and Breeding of Japonica Rice in Mid-lower Yangtze River, Ministry of Agriculture, P.R. China, and Jiangsu Collaborative Innovation Center for Modern Crop Production.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00782/full#supplementary-material
Abbreviations
ACP, acyl carrier protein; DAF, days after flowering; DAG, days after germination; ENR, enoyl-ACP reductase; FAS, fatty acid synthase, HAD, β-Hydroxyacyl-ACP dehydratase.
Footnotes
- ^http://crispr.hzau.edu.cn/CRISPR2/
- ^http://www.cbs.dtu.dk/services/TargetP/
- ^http://www.cbs.dtu.dk/services/ChloroP/
- ^http://www.gramene.org/
- ^http://www.rcsb.org/pdb/
- ^http://ricevarmap.ncpgr.cn/cgi-bin/gb2/gbrowse/ricevarmap2/
- ^https://www.ncbi.nlm.nih.gov/
- ^http://www.shigen.nig.ac.jp/rice/oryzabase/
References
Barkan, A., Voelker, R., Mendel-Hartvig, J., Johnson, D., and Walker, M. (1995). Genetic analysis of chloroplast-biogenesis in higher plants. Physiol. Plant. 93, 163–170. doi: 10.1034/j.1399-3054.1995.930123.x
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The protein data bank. Nucleic Acids Res. 28, 235–242. doi: 10.1093/nar/28.1.235
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Chai, C. L., Fang, J., Liu, Y., Tong, H. N., Gong, Y. Q., Wang, Y. Q., et al. (2011). ZEBRA2, encoding a carotenoid isomerase, is involved in photo protection in rice. Plant Mol. Biol. 75, 211–221. doi: 10.1007/s11103-010-9719-z
Chi, W., Sun, X., and Zhang, L. (2013). Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 64, 559–582. doi: 10.1146/annurev-arplant-050312-120147
Coe, E. H. Jr., Hoisington, D. A., and Neuffer, M. G. (1987). “Linkage map of corn (maize) (Zea mays L.),” in Genetic Maps, 1987: A Compilation of Linkage and Restriction Maps of Genetically Studied Organisms, Vol. 4, ed. S. J. O’Brien (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory), 685–707.
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
de Souza, A., Wang, J. Z., and Dehesh, K. (2016). Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 58–108. doi: 10.1146/annurev-arplant-042916-041007
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Ewald, R., Kolukisaoglu, U., Bauwe, U., Mikkat, S., and Bauwe, H. (2007). Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol. 145, 41–48. doi: 10.1104/pp.107.104000
Fan, J., Zhai, Z., Yan, C., and Xu, C. (2015). Arabidopsis TRIGALACTOS YLDIACYLGLYCEROL5 interacts with TGD1, TGD2, and TGD4 to facilitate lipid transfer from the endoplasmic reticulum to plastids. Plant Cell 27, 2941–2955. doi: 10.1105/tpc.15.00394
Focke, M., Gieringer, E., Schwan, S., Jänsch, L., Binder, S., and Braun, H. P. (2003). Fatty acid biosynthesis in mitochondria of grasses: malonyl-coenzyme A is generated by a mitochondrial-localized acetyl-coenzyme A carboxylase. Plant Physiol. 133, 875–884. doi: 10.1104/pp.103.027375
Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., et al. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511. doi: 10.1038/nature01097
González-Thuillier, I., Venegas-Calerón, M., Sánchez, R., Garcés, R., von Wettstein-Knowles, P., and Martínez-Force, E. (2016). Sunflower (Helianthus annuus) fatty acid synthase complex: β-hydroxyacyl-[acyl carrier protein] dehydratase genes. Planta 243, 397–410. doi: 10.1007/s00425-015-2410-5
Guan, X., Chen, H., Abramson, A., Man, H., Wu, J., Yu, O., et al. (2015). A phosphopantetheinyl transferase that is essential for mitochondrial fatty acid biosynthesis. Plant J. 84, 718–732. doi: 10.1111/tpj.13034
Guan, X., and Nikolau, B. J. (2016). AAE13 encodes a dual-localized malonyl-CoA synthetase that is crucial for mitochondrial fatty acid biosynthesis. Plant J. 85, 581–593. doi: 10.1111/tpj.13130
Guan, X., Okazaki, Y., Lithio, A., Li, L., Zhao, X., Jin, H., et al. (2017). Discovery and characterization of the 3-hydroxyacyl-acp dehydratase component of the plant mitochondrial fatty acid synthase system. Plant Physiol. 173, 2010–2028. doi: 10.1104/pp.16.01732
Gueguen, V., Macherel, D., Jaquinod, M., Douce, R., and Bourguignon, J. (2000). Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275, 5016–5025. doi: 10.1074/jbc.275.7.5016
Han, S. H., Sakuraba, Y., Koh, H. J., and Paek, N. C. (2012). Leaf variegation in the rice zebra2 mutant is caused by photoperiodic accumulation of tetra-Cis-lycopene and singlet oxygen. Mol. Cells 33, 87–97. doi: 10.1007/s10059-012-2218-0
He, R., Ding, Y., Yu, J., and Gao, W. (2000). The changes of chlorophyll content and several enzyme activities in zebra-leaf rice. J. Wuhan Univ. 46, 761–765.
Heath, R. J., and Rock, C. O. (1996). Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271, 27795–27801. doi: 10.1074/jbc.271.44.27795
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Hu, C., Lin, S. Y., Chi, W. T., and Charng, Y. Y. (2012). Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 158, 747–758. doi: 10.1104/pp.111.187674
Iba, K., Takamiya, K. I., Toh, Y., Satoh, H., and Nishimura, M. (1991). Formation of functionally active chloroplasts is determined at a limited stage of leaf development in virescent mutants of rice. Dev. Genet. 12, 342–348. doi: 10.1002/dvg.1020120503
James, D. W. Jr., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H. K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309
Kachroo, A., Lapchyk, L., Fukushige, H., Hildebrand, D., Klessig, D., and Kachroo, P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15, 2952–2965. doi: 10.1105/tpc.017301
Kachroo, A., Venugopal, S. C., Lapchyk, L., Falcone, D., Hildebrand, D., and Kachroo, P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157. doi: 10.1073/pnas.0401315101
Kitajima, M., and Butler, W. L. (1975). Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim. Biophys. Acta 376, 105–115. doi: 10.1016/0005-2728(75)90209-1
Kobayashi, K. (2016). Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 129, 565–580. doi: 10.1007/s10265-016-0827-y
Kostrewa, D., Winkler, F. K., Folkers, G., Scapozza, L., and Perozzo, R. (2005). The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci. 14, 1570–1580. doi: 10.1110/ps.051373005
Kusumi, K., Komori, H., Satoh, H., and Iba, K. (2000). Characterization of a zebra mutant of rice with increased susceptibility to light stress. Plant Cell Physiol. 41, 158–164. doi: 10.1093/pcp/41.2.158
Kusumi, K., Mizutani, A., Nishimura, M., and Iba, K. (1997). A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J. 12, 1241–1250. doi: 10.1046/j.1365-313x.1997.12061241.x
Kusumi, K., Yara, A., Mitsui, N., Tozawa, Y., and Iba, K. (2004). Characterization of a rice nuclear-encoded plastid RNA polymerase gene OsRpoTp. Plant Cell Physiol. 45, 1194–1201. doi: 10.1093/pcp/pch133
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, J., Pandeya, D., Nath, K., Zulfugarov, I. S., Yoo, S. C., and Zhang, H. T. (2010). ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J. 62, 713–725. doi: 10.1111/j.1365-313X.2010.04183.x
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.4236/oalib.1101097
Liu, H. C., and Charng, Y. Y. (2013). Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 163, 276–290. doi: 10.1104/pp.113.221168
Maity, K., Venkata, B. S., Kapoor, N., Surolia, N., Surolia, A., and Suguna, K. (2011). Structural basis for the functional and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum. J. Struct. Biol. 176, 238–249. doi: 10.1016/j.jsb.2011.07.018
Mohan, S., Kelly, T. M., Eveland, S. S., Raetz, C. R., and Anderson, M. S. (1994). An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydratase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269, 32896–32903.
Moreira, D., Le Guyader, H., and Philippe, H. (2000). The origin of red algae and the evolution of chloroplasts. Nature 405, 69–72. doi: 10.1007/978-1-4615-2818-0_7
Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–418. doi: 10.2307/3870945
Ohlrogge, J. B., and Jaworski, J. G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 109–136. doi: 10.1146/annurev.arplant.48.1.109
Oki, S., Gu, X., Kofoid, K. D., and Liang, G. H. (1997). A light-intensity sensitive chlorophyll mutant in sorghum. Hereditas 126, 239–245. doi: 10.1111/j.1601-5223.1997.00239.x
Peng, C., Wang, Y., Liu, F., Ren, Y., Zhou, K., Lv, J., et al. (2014). FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77, 917–930. doi: 10.1111/tpj.12444
Pillai, S., Rajagopal, C., Kapoor, M., Kumar, G., Gupta, A., and Surolia, N. (2003). Functional characterization of beta-ketoacyl-ACP reductase (FabG) from Plasmodium falciparum. Biochem. Biophys. Res. Commun. 303, 387–392. doi: 10.1016/S0006-291X(03)00321-8
Sharma, S. K., Kapoor, M., Ramya, T. N., Kumar, S., Kumar, G., Modak, R., et al. (2003). Identification, characterization, and inhibition of Plasmodium falciparum betahydroxyacyl-acyl carrier protein dehydratase (FabZ). J. Biol. Chem. 278, 45661–45671. doi: 10.1074/jbc.M304283200
Sugimoto, H., Kusumi, K., Tozawa, Y., Yazaki, J., Kishimoto, N., Kikuchi, S., et al. (2004). The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant Cell Physiol. 45, 985–996. doi: 10.1093/pcp/pch111
Wada, H., Shintani, D., and Ohlrogge, J. (1997). Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. U.S.A. 94, 1591–1596. doi: 10.1073/pnas.94.4.1591
Wang, Y., Ren, Y., Zhou, K., Liu, L., Wang, J., Xu, Y., et al. (2017). WHITE STRIPE LEAF4 encodes a novel p-type PPR protein required for chloroplast biogenesis during early leaf development. Front. Plant Sci. 8:1116. doi: 10.3389/fpls.2017.01116
Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. doi: 10.1016/S1360-1385(02)02250-1
Werner, B. K., and Burton, G. W. (1991). Dominant zebra mutant in pearl millet. J. Hered. 82, 68–69. doi: 10.1093/jhered/82.1.68
Wu, D., Shu, Q., Xia, Y., Jing, X., and Liu, G. (1999). Analysis of variation in thermo-sensitive genic male sterile leaf color mutant lines induced from Indica rice (Oryza sativa L.) by 60Co irradiation. Acta Agron. Sin. 25, 64–69.
Wu, G., and Xue, H. (2010). Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell 22, 3726–3744. doi: 10.1105/tpc.110.075564
Xu, C., Moellering, E. R., Muthan, B., Fan, J., and Benning, C. (2010). Lipid transport mediated by Arabidopsis TGD proteins is unidirectional from the endoplasmic reticulum to the plastid. Plant Cell Physiol. 51, 1019–1028. doi: 10.1093/pcp/pcq053
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30–43. doi: 10.1186/1746-4811-7-30
Keywords: ZEBRA LEAF 16, β-hydroxyacyl-ACP dehydratase, chloroplast development, fatty acid, Oryza sativa L.
Citation: Liu Z, Wang Z, Gu H, You J, Hu M, Zhang Y, Zhu Z, Wang Y, Liu S, Chen L, Liu X, Tian Y, Zhou S, Jiang L, Liu L and Wan J (2018) Identification and Phenotypic Characterization of ZEBRA LEAF16 Encoding a β-Hydroxyacyl-ACP Dehydratase in Rice. Front. Plant Sci. 9:782. doi: 10.3389/fpls.2018.00782
Received: 04 January 2018; Accepted: 23 May 2018;
Published: 12 June 2018.
Edited by:
Ján A. Miernyk, Agricultural Research Service (USDA), United StatesReviewed by:
Edgar B. Cahoon, University of Nebraska–Lincoln, United StatesBasil J. Nikolau, Iowa State University, United States
Copyright © 2018 Liu, Wang, Gu, You, Hu, Zhang, Zhu, Wang, Liu, Chen, Liu, Tian, Zhou, Jiang, Liu and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linglong Liu, bGl1bGluZ2xvbmdAbmphdS5lZHUuY24=
†These authors have contributed equally to this work.
 Ziwen Liu1†
Ziwen Liu1† Linglong Liu
Linglong Liu Jianmin Wan
Jianmin Wan