
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 05 June 2018
Sec. Plant Metabolism and Chemodiversity
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00761
This article is part of the Research Topic Revealing Distinct Functions of Starch Biosynthetic Enzymes View all 17 articles
Starch is the primary storage carbohydrate in most photosynthetic organisms and allows the accumulation of carbon and energy in form of an insoluble and semi-crystalline particle. In the last decades large progress, especially in the model plant Arabidopsis thaliana, was made in understanding the structure and metabolism of starch and its conjunction. The process underlying the initiation of starch granules remains obscure, although this is a fundamental process and seems to be strongly regulated, as in Arabidopsis leaves the starch granule number per chloroplast is fixed with 5-7. Several single, double, and triple mutants were reported in the last years that showed massively alterations in the starch granule number per chloroplast and allowed further insights in this important process. This mini review provides an overview of the current knowledge of processes involved in the initiation and formation of starch granules. We discuss the central role of starch synthase 4 and further proteins for starch genesis and affecting metabolic factors.
Starch is one of the most abundant biopolymers on earth. The entire process of starch biosynthesis encompasses: the generation of a suitable primer (glucosyl acceptor); the biosynthesis of a glucosyl donor (in most cases ADPglucose); the elongation of the primer by repetitive glucosyl transfer reactions via starch synthases (SS) and most probably via plastidial phosphorylase (see below); the introduction of branching points via branching enzymes and the removal of excess branching points via debranching enzymes; and the ordering of the glucans to the final physical organization. Starch granule synthesis in green algae, mosses, ferns, and higher plants occurs inside the plastids. In many other eukaryotic algae starch is formed in the cytosol, which is assumed to represent an earlier evolutionary state (Colleoni et al., 2010; Ball et al., 2011). This review will focus on the plastidial starch biosynthesis, more precisely the induction of the starch biosynthesis in Arabidopsis thaliana leaves.
To some extent, the initial steps of the starch biosynthesis, the generation of a suitable primer (a glucosyl acceptor, most likely a glucan), and the further elongation of the glucosyl acceptor by the action of glucosyl transferases is not precise to separate. The first elongation processes as well as the first branching/debranching events does not immediately result in a native starch granule rather generate a polyglucan. Furthermore, SS specifically interact with branching enzymes during the process of starch synthesis (Brust et al., 2014). But only following multiplicity of these reactions a starch granule is formed. Therefore, in this review we divide the starch synthesis in two steps. We define starch initiation in a strict sense as a process in which a primer-like structure/initiation complex is generated, that can in principle be further used for polymerization reactions. This also includes the possibility that no starch granules are detectable since the primer-like structure/initiation complex is not further synthesized. The term starch formation indicate when these potential primer-like structures/initiation complexes are used to form starch granules, and so starch granules are detectable in chloroplasts.
Starch synthases (SSs) are the enzymes responsible for elongation of amylose and amylopectin chains. These enzymes transfer glucosyl residue from ADPglucose to the non-reducing end of a preexisting glucan. It was discussed whether SSs are able to synthesize glucan chains without a glucan primer (Szydlowski et al., 2009; D’Hulst and Mérida, 2012). However, using heterologously expressed and purified SSs from Arabidopsis a primer was always required, that contained at least two glucosyl residues. Thus, maltose but not glucose was elongated by the SSs (Brust et al., 2013).
Analyses of SS mutants revealed that SSs have overlapping and in parts distinct functions during starch synthesis (Delvallé et al., 2005; Zhang et al., 2005, 2008; Roldán et al., 2007; Szydlowski et al., 2009, 2011). SS4 seems to be most important for formation of starch granules. Thus, the ss4 mutant reveals mostly one starch granule per chloroplast (Roldán et al., 2007). However, other SS isoforms may partially substitute the SS4 function and so additional loss of SS3 further reduces the starch granule number in chloroplasts (Szydlowski et al., 2009; D’Hulst and Mérida, 2012). Interestingly, the number of starch granules in ss4 is still flexible as the number shows variability in regards to growth conditions/developmental stage and enzymatic setup (see below). Thus, immature leaves of ss4 have no detectable starch granules in contrast to mature leaves (Crumpton-Taylor et al., 2013). The number of starch granules per chloroplast depends of the length of the light phase as under continuous light the starch granule number increases to up to three (Malinova et al., 2017). Furthermore, the additional lack of α-amylase 3 (Seung et al., 2016) and an increased capacity for glucan synthesis via expression of a bacterial glycogen synthase (Crumpton-Taylor et al., 2013) result in a higher number of starch granules per chloroplast. An over expression of SS4 in Arabidopsis causes an elevated in an increased starch content, although an increase in starch granule number per chloroplast was not reported (Gamez-Arjona et al., 2011).
x
All five SSs in Arabidopsis share a core sequence of approximately 60 kDa at the C-terminus responsible for the catalytic activity. The N-terminal sequences are largely different. Interestingly, SS3 and SS4 possess a large N-terminal extension. The N-terminal region of SS4 is predicted to contain two long coiled-coil domains (Leterrier et al., 2008; D’Hulst and Mérida, 2012) and was reported to enable the association with the thylakoid membrane and interaction with thylakoid- and plastoglobule-bound fibrillin proteins (Gamez-Arjona et al., 2014; Raynaud et al., 2016). Furthermore, a subchloroplastidial localization of SS4 peripheral to starch granules was shown (Gamez-Arjona et al., 2014). Expression of the N-terminal region of SS4 in ss4 did not result in alterations of the starch granule number. However, the expression of the C-terminal catalytically active region of SS4 in ss4 resulted in an increased number of starch granules per chloroplast (Lu et al., 2018). It was shown that SS4 forms dimers necessary for its enzymatic activity and dimerization is mediated by a region located between the coiled-coil domain and the glycosyltransferase domain (Raynaud et al., 2016).
Summarizing, SS4 has impact on formation of starch granules and seems to be important for the generation of the regular number of granules per chloroplast. However, the observed reduced number of starch granules by the lack of SS4 can be in parts reversed by metabolic factors (see below).
Recently, a double mutant lacking the plastidial phosphorylase (PHS1; Zeeman et al., 2004) and the disproportionating enzyme 2 (DPE2; Chia et al., 2004; Lu and Sharkey, 2004; Fettke et al., 2006) was published (Malinova et al., 2014). Both parental lines have 4-7 starch granules per chloroplast similar to wild type. Interestingly, dpe2/phs1 revealed mostly one starch granule per chloroplast when grown under 12 h light and 12 h dark conditions, similar to ss4. However, expression of SS4 was unaltered in this mutant (Malinova et al., 2014). The triple mutant dpe2/phs1/ss4 also has only one starch granule per chloroplast when grown under 12 h light and 12 h dark (Malinova et al., 2017). More interestingly, when the light/dark regime was altered ss4, dpe2/phs1, and the triple mutant plants revealed variability in the starch granule number per chloroplast. An elongated light phase and continuous light resulted in an increased number of starch granules per chloroplast in all mutants but with different quantities. In ss4 the number increased up to three granules whereas dpe2/phs1 showed with 5-7 the wild type number of starch granules per chloroplast. The dpe2/phs1/ss4 revealed an intermediate number of starch granules with 1-4 granules per chloroplast (Malinova et al., 2017). The observed increased number of starch granules in dpe2/phs1 similar to wild type was supposed to be linked to the missing dark phase and the lack of starch degradation (Malinova et al., 2014). To test whether ongoing starch degradation in dpe2/phs1 is necessary for the observed reduction of the starch granule number a triple mutant lacking the glucan, water dikinase (GWD; sex1-8; Ritte et al., 2002; Mahlow et al., 2014) in the background of dpe2/phs1 was generated. GWD is involved in the phosphorylation/dephosphorylation cycle at the starch granule surface and is the key enzyme of the initiation of starch degradation (Hejazi et al., 2012; Mahlow et al., 2016). However, sex1-8 did not reveal a reduced starch granule number per chloroplast (Mahlow et al., 2014). Similarly, dpe2/phs1/sex1-8 revealed 5-7 starch granules even when plants were grown at 12 h light and 12 h dark conditions (Malinova and Fettke, 2017). Thus, it was concluded that ongoing starch degradation is essential for the observed reduced number of starch granules in dpe2/phs1 (Malinova and Fettke, 2017).
Recently, three new starch granule bound proteins were discovered sharing high homology and building up a new protein family, PROTEIN TARGETING TO STARCH (PTST; Seung et al., 2015, 2017). All three proteins contain conserved coiled coil domains and a CBM48. While PTST1 was shown to play an essential role in amylose synthesis by localizing granule bound SSs to starch granules (Seung et al., 2015), PTST2 and PTST3 were demonstrated to be involved in alteration of the number of starch granules per chloroplast (Seung et al., 2017). In ptst2 a reduced starch granule number was observed with mostly one starch granule per chloroplast. PTST3 has partially redundant function to PTST2 but reduction of granule number was less pronounced in ptst3 compared with ptst2. Furthermore, it was shown that an overexpression of PTST2 in wild type background resulted in an increased number of small granules. In contrast, an overexpression of PTST2 in ss4 background resulted in a further reduction of the starch granule number per chloroplast. Interestingly, PTST2 was shown to interact with SS4 and longer soluble maltodextrins (Seung et al., 2017).
For ss4 as well as for ptst2 and ptst2/ptst3 an accumulation of ADPglucose was reported (Crumpton-Taylor et al., 2013; Ragel et al., 2013; Seung et al., 2017). The highest ADPglucose accumulation was observed for ss4 (with ∼100-fold increase compared to wild type). ptst2/ptst3 revealed also a high accumulation of ADPglucose (with ∼30-fold compared to wild type) that was twice of that of ptst2 (Seung et al., 2017). This accumulation of ADPglucose, the substrate of SSs, is thought to show that the SS4 acts upstream of all other SSs and that the other SSs have limited activity when SS4 is lacking. However, ss4/ss3 showed a similar high accumulation of ADPglucose as ss4 (with ∼100-fold increase compared to wild type). dpe2/phs1 and especially dpe2/phs1/ss4 also revealed mostly one starch granule per chloroplast but showed only a very moderate increase in ADPglucose content (two- and fourfold, respectively, compared to wild type). Furthermore, dpe2/ss4 and phs1/ss4, each having one starch granule per chloroplast, revealed totally different ADPglucose values (dpe2/ss4: approximately twofold, phs1/ss4: 250-fold increase compared to wild type). Under continuous light, an increase in the granule number per chloroplast was observed for ss4, dpe2/phs1, and dpe2/phs1/ss4. But these observations were not accompanied by a reduction in the ADPglucose amounts (Malinova et al., 2017). Thus, a simple direct connection of a high ADPglucose content and the lack of SS4 or a reduced granule number is not given.
Another factor influencing the starch granule number is probably the availability of soluble maltodextrins as glucan primers for the initiation of a starch granule. The generation and elongation of soluble maltodextrins is proposed to be important during granule initiation (Nakamura, 2015). It was postulated that longer soluble maltodextrins (e.g., maltodecaose), long enough to produce a helical secondary structure, are bound by PTST2. PTST2 further interacts with SS4 and initiate the synthesis of starch granules (Seung et al., 2017). Interestingly, increased levels of soluble glucans were detected in ss4 (Crumpton-Taylor et al., 2013; Malinova et al., 2017). Most of these glucans were rather shorter with a degree of polymerization (DP) less than six. In dpe2/phs1 and dpe2/phs1/ss4 a massive increase in these short glucans was observed (Malinova et al., 2014, 2017). The origin of the chloroplastidial maltodextrins necessary for initiation of starch granules is unknown so far. However, the chloroplastidial maltodextrin pool is certainly also influenced by starch degradation processes.
PHS1 could be involved in metabolism of the chloroplastidial maltodextrins and therefore in starch synthesis. Several indications for a principle involvement of PHS1 in starch synthesis were published. For Chlamydomonas reinhardtii, potato tubers, and for rice endosperm it was shown that the plastidial phosphorylase is involved in starch synthesis (Dauvillée et al., 2006; Orawetz et al., 2016) and in granule initiation (Satoh et al., 2008). Interestingly, the alteration of starch quantities and qualities in rice and potato mutants were exclusively detected when plants were grown under low temperature conditions. It was speculated that at low temperatures SSs cannot sufficiently substitute the lack of the plastidial phosphorylase (Fettke et al., 2012b; Orawetz et al., 2016). Furthermore, for endosperm starch evidences exist for a protein–protein interaction of the plastidial phosphorylase with branching enzyme and disproportionating enzyme 1 (DPE1) and production of longer soluble maltodextrins (Tetlow et al., 2004; Hwang et al., 2016; Nakamura et al., 2017). Interestingly, in Arabidopsis PHS1 interacts with SS4 (Figure 1; Qasim, 2018).
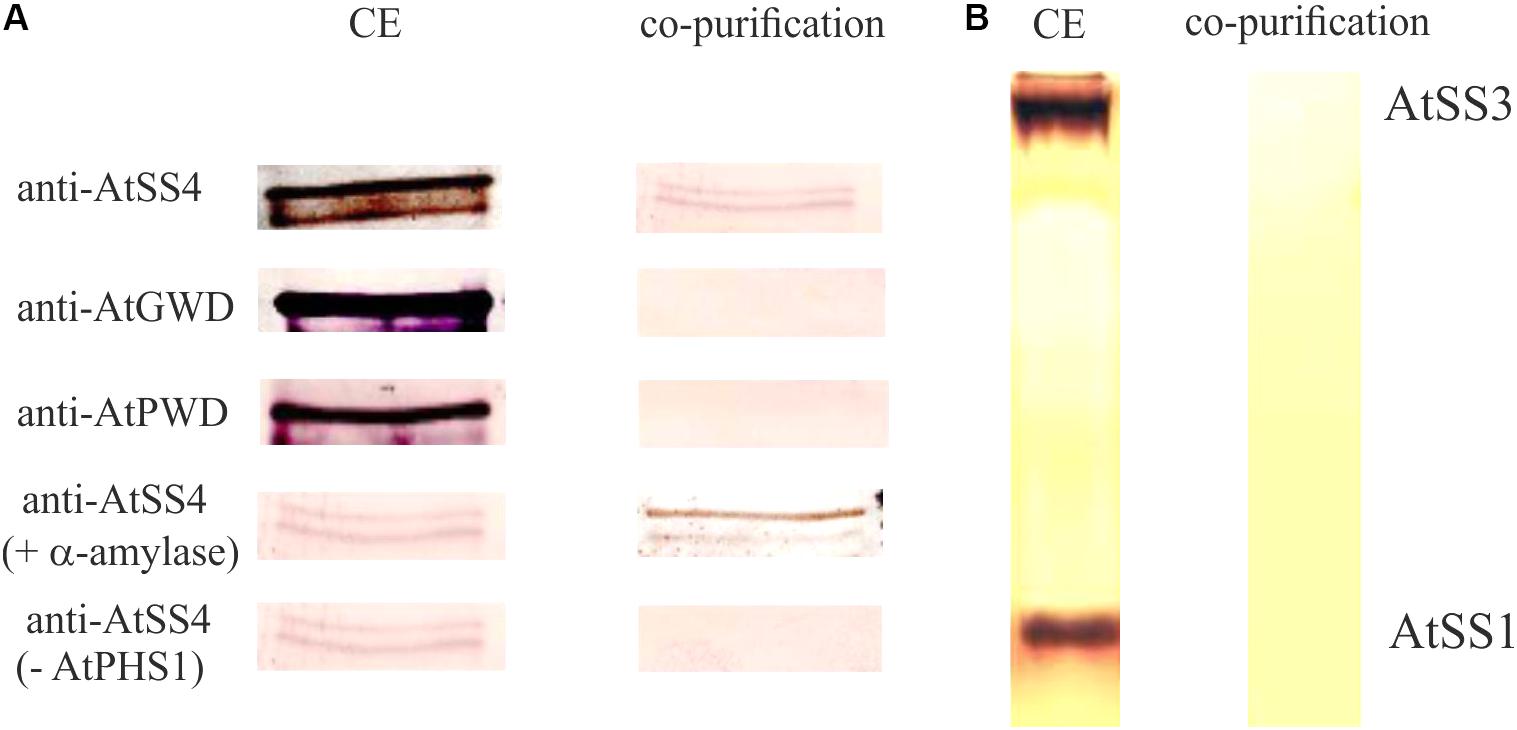
FIGURE 1. Interaction of starch synthase 4 and plastidial phosphorylase. (A) Purified C-terminal 6× His-taged recombinant plastidial phosphorylase protein from Arabidopsis thaliana (40 μg) expressed in E. coli was incubated with 200 μg protein crude extract (CE) isolated from Col-0 for 5 min at 21°C. Following purification of the recombinant plastidial phosphorylase using Ni-NTA agarose the plastidial phosphorylase containing fraction was analyzed by SDS-PAGE and the proteins were transferred to a membrane. The membrane was analyzed by antibodies specific for AtSS4, AtGWD, and AtPWD. In addition the recombinant plastidial phosphorylase and the protein crude extract were pre incubated with 20 units a-amylase for 10 min at 37° C prior incubation and purification, to exclude a potential interaction of AtPHS1 and AtSS4 via a bound glucan (AtSS4 + a-amylase). As control the experiment was performed identical but the recombinant plastidial phosphorylase was omitted (AtSS4-AtPHS1). (B) Following the purification as described in (A) the plastidial phosphorylase containing fraction was analyzed by native gel and activity staining for SSs (see also Brust et al., 2014). As control 50μg crude extract protein isolated from Col-0 was loaded on the native gel. Thus, no co-purification of plastidial phosphorylase with AtSS1 or AtSS3 was observed.
The reduced starch granule number per chloroplast observed in dpe2/phs1 is not detected in the phs1. The single mutant revealed a starch granule number similar to wild type (Malinova et al., 2014, 2017). It could be speculated that other proteins compensate the lack of PHS1 but if in addition the starch turn-over is altered an effect on starch initiation could be observed. A balance between generation and degradation of maltodextrins could be important. In this regards, it can be supposed, that an additional lack of degradation enzymes such as α-amylase 3 (AMY3; Seung et al., 2016) and DPE2 (Malinova et al., 2014, 2017) influence this balance. An increase of the starch granule number per chloroplasts was detected in the ss4/amy3 mutant, which point to a degradation of glucans in ss4 and this degradation generates primers that can be used by other SSs. In this context it can also be seen that an introduction of the glycogen synthase from Agrobacterium tumefaciens (able to initiate and elongate glucan chains using ADPglucose as substrate; Ugalde et al., 2003) in ss4 resulted in an increase in starch granule number per chloroplast by generating more of such primers (Crumpton-Taylor et al., 2013; Lu et al., 2018). In agreement with the observed binding of longer soluble maltodextrins by PTST2, it could be speculated that soluble maltodextrins longer than DP10 are involved in the starch granule initiation, as in dpe2/phs1 no limitation for maltodextrins shorter than DP10 were observed (Malinova et al., 2014). Similarly, when in the background of dpe2/phs1 the starch degradation was blocked by the additional lack of GWD, this glucan balance was again influenced and resulted in an increased number of starch granules with 5-7 granules per chloroplast similar to wild type (Malinova and Fettke, 2017). However, the additional lack of GWD in ss4 has no effect on the starch granule number (Crumpton-Taylor et al., 2013). Interestingly, phs1/mex1 double mutant lacking PHS1 and in addition the maltose exporter 1 [essential for export of the starch degradation product maltose (Niittylä et al., 2004) and thus it acts upstream to DPE2 (Fettke et al., 2012a)] in contrast to dpe2/phs1 has a similar to wild type starch granule number per chloroplast. In regards to the soluble maltodextrins both mutants are different too, thus only a small increase in soluble maltodextrins (length up to DP10) was observed in mex1/phs1 compared to wild type (Malinova et al., 2014). A further rather indirect indication of this balanced maltodextrin system is related to the observed dependency of the length of the light and dark phase on the starch granule number in ss4, dpe2/phs1, and dpe2/phs1/ss4 (Malinova et al., 2017).
Summarizing, as depicted in Figure 2, in a strict sense, the initiation of starch granules seems to be linked to maltodextrins serving as glucosyl acceptors. Increasing evidences reveal that these maltodextrin primers are influenced by the overall glucan metabolism in chloroplast. Thus, proteins such as cytosolic DPE2, PHS1, PTST2 and 3 are directly and/or indirectly involved in this process and so in starch granule initiation. Furthermore, this would also imply that the observed alteration of the starch granule number in dpe2/phs1 as well as in ptst2 and ptst3 are upstream of the action of SS4. In agreement to this dpe2/phs1 and dpe2/phs1/ss4 contained mostly one starch granule. The double mutant revealed higher variability in the starch granule number in connection with metabolic alterations than the triple mutant. Thus, generation of the glucan primer seems more variable than formation of the starch granules regulated via SS4. Unfortunately, for both ptst2 and ptst3 such metabolic data are currently missing. However, the increase of the starch granule number when PTST2 is overexpressed in wild type but not when overexpressed in the ss4 mutant is also in agreement with this assumption. Likewise, the observed increase of the starch granule number per chloroplast in ss4 by expression of the glycogen synthase is conforming.
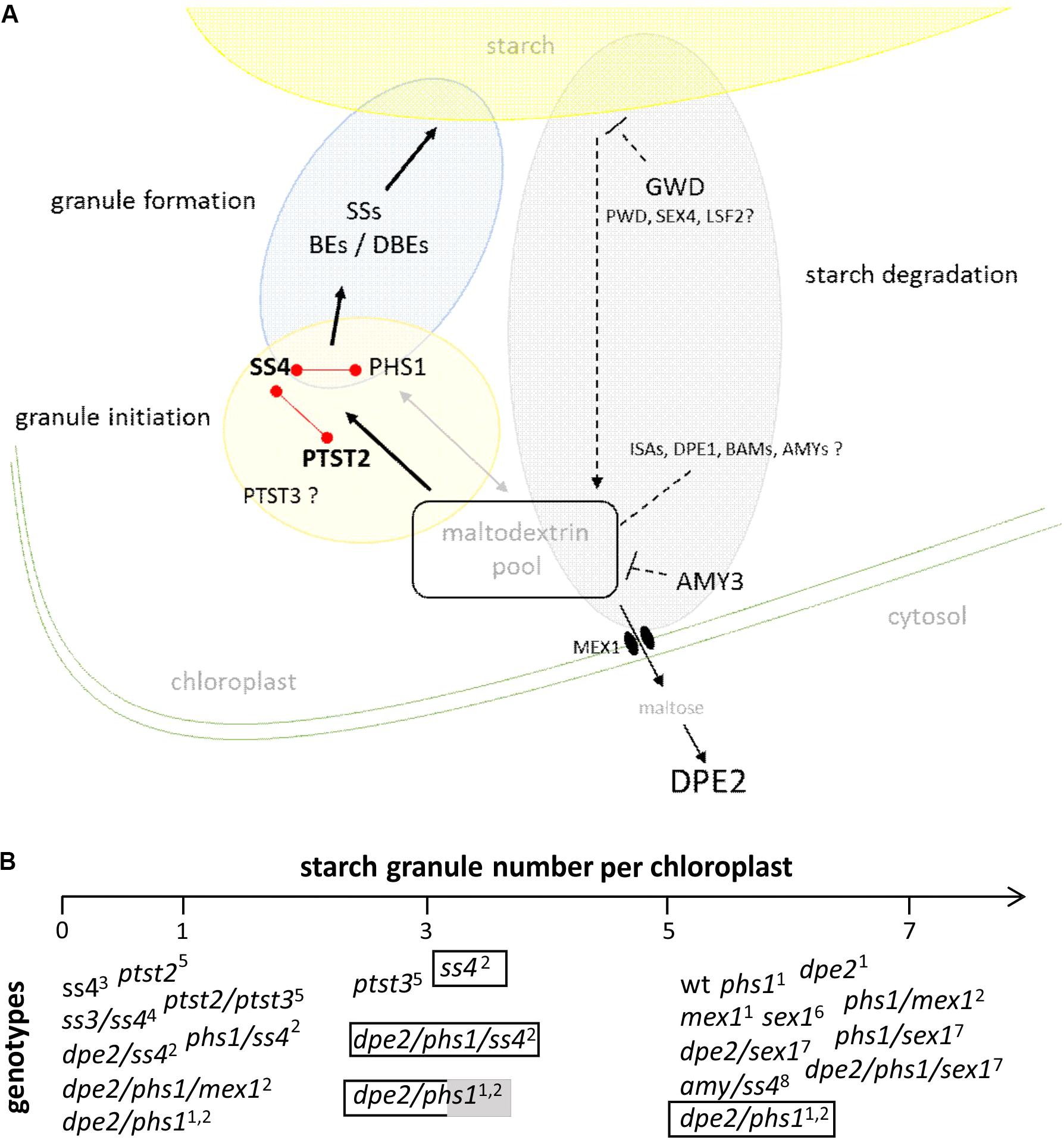
FIGURE 2. (A) Schematic presentation of starch granule synthesis. For starch granule initiation maltodextrins from the chloroplastidial maltodextrin pool are used by protein targeting to starch 2 (PTST2) and starch synthase 4 (SS4) to produce a granule initiation complex. A lack of the related enzymes massively influences the starch granule number. Enzymes related to initiation reveal protein-protein interaction as depicted by red links. The granule initiation complex is processed by SSs, branching (BEs), and debranching enzymes (DBEs) and a starch granule is formed. The chloroplastidial maltodextrin pool is affected by enzymes acting on glucans, efflux of carbohydrates to cytosol, and influx of carbohydrates via starch degradation. Thus, evidences were published for the involvement of glucan, water dikinase (GWD), disproportionating enzyme 2 (DPE2), α-amylase3 (AMY3), and plastidial phosphorylase (PHS1) during starch granule synthesis. For other enzymes acting on glucans (labeled with ?) the impact on the starch granule synthesis is lacking so far. ISAs- isoamylases; DPE1 and DPE2– disproportionating enzyme 1 and 2; BAMs – β-amylases; AMYs – α-amylases; PWD- phosphoglucan, water dikinase, SEX4 – starch excess 4, LSF2 – like starch excess4, MEX1 – maltose exporter 1 (see also Fettke et al., 2012a). Dashed arrow represents multiple enzymatic reactions. (B) Starch granule number per chloroplast in different genotypes. Plants were grown under 12 h ligth/12 h dark unless otherwise specified: transparent bar – continuous light, transparent/grey bar – 14 h light, 10 h dark.1Malinova et al., 2014; 2Malinova et al., 2017; 3Roldán et al., 2007; 4D’Hulst and Mérida, 2012; 5Seung et al., 2017; 6Mahlow et al., 2014; 7Malinova and Fettke, 2017; 8Seung et al., 2016.
Starch isolated from mutants with reduced number of starch granules per chloroplast reveal distinct alterations in the starch granule morphology. Starch granules from Arabidopsis leaves are thin and discoid. In contrast, starch from mutants lacking SS4 or DPE2/PHS1 show big and spherical starch granules (Crumpton-Taylor et al., 2013; Malinova et al., 2014, 2017).
It was reported that the N-terminal domain of SS4 influences the morphology of starch granules (Lu et al., 2018). Furthermore, the lack of PTST2 and PTST3 revealed single and spherical starch granules (Seung et al., 2017). Very interesting in this regards is again the dpe2/phs1 mutant. When one starch granule per chloroplast was observed (under 12 h light/ 12 h dark regime) the granule was large and spherical, whereas when 5-7 starch granules were monitored (under continuous light) the starch granule morphology was similar to wild type (Malinova et al., 2017). It seems that the generation of a reduced number of starch granules number per chloroplasts is connected with formation of spherical rather than thin and flattened starch granules. However, also other starch-related mutants reveal alterations in morphology albeit these are less pronounced (SEX4; Kötting et al., 2009; LSF1; Comparot-Moss et al., 2010; GWD; Mahlow et al., 2014).
In the last years we got more insights into starch granule biosynthesis. The appearance of starch granules is a rather complex mechanism influenced by various factors. According to this, it is very important to distinguish between the possibility to initiate starch and the formation of starch granules. It is quite clear that SSs, or more precisely SS4, is important in the formation of the typical number of starch granules per chloroplast in Arabidopsis (5-7, Crumpton-Taylor et al., 2012). However, the lack of SS4 is not limiting the formation of starch granules. The formation of starch granules occur independent of SS4 in several mutants. Starch isolated from these mutants analyzed so far did not reveal large inner structural alterations, although the starch morphology reveals considerable differences. Thus, following the starch initiation, the process of starch granule formation is rather similar in these mutants and point to the overlapping functions of the further SSs. Interestingly, the majority of mutants affected in the starch granule number display mostly one starch granule per chloroplast (Figure 2). The initiation of starch seems to depend on a chloroplastidial maltodextrin metabolism. At least PTST2 and PHS1 seem to be involved in this process. As this maltodextrin metabolism can be influenced by the ongoing starch turn-over and distinct enzymes of the starch metabolism (e.g., AMY3) the involvement of further proteins/enzymes is highly likely. In yeast cells, starch-like synthesis was recreated by introduction of Arabidopsis SSs without plant enzymes related to synthesis of a primer, thus the intrinsic primers of yeast seems to be sufficient for the formation of starch like polyglucans. Furthermore, the experiments again showed that SS4 can be substituted by other SSs, as starch-like polyglucans were observed not exclusively when SS4 was introduced (Pfister et al., 2016).
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
We acknowledge the support of the Deutsche Forschungsgemeinschaft FE1030/5-1 and Open Access Publishing Fund of University of Potsdam.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer NC and handling Editor declared their shared affiliation.
Ball, S., Colleoni, C., Cencil, I., Raj, I. N., and Tirtiaux, C. (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understanding the establishment of plastid endosymbiosis. J. Exp. Bot. 62, 1776–1801. doi: 10.1093/jxb/erq411
Brust, H., Lehmann, T., D’Hulst, C., and Fettke, J. (2014). Analysis of the functional interaction of Arabidopsis starch synthase and branching enzyme isoforms reveals that the cooperative action of SSI and BEs results in glucans with polymodal chain lenght distribution similar to amylopectin. PLoS One 9:e102364. doi: 10.1371/journal.pone.0102364
Brust, H., Orzechowski, S., Fettke, J., and Steup, M. (2013). Starch synthesizing reactions and paths: in vitro and in vivo studies. J. Appl. Glycosci. 60, 3–20. doi: 10.5458/jag.jag.JAG-2012_018
Chia, T., Thorneycroft, D., Chapple, A., Messerli, G., Chen, J., Zeeman, S. C., et al. (2004). A cytosolic glucosyltransferase is required for conversion of starch in Arabidopsis leaves at night. Plant J. 37, 853–863. doi: 10.1111/j.1365-313X.2003.02012.x
Colleoni, C., Linka, M., Deschamps, P., Handford, M. G., Dupree, P., Weber, A. P. M., et al. (2010). Phylogenetic and biochemical evidence supports the recruitment of an ADP-glucose translocator for the export of photosynthate during plastid endosymbiosis. Mol. Biol. Evol. 27, 2691–2701. doi: 10.1093/molbev/msq158
Comparot-Moss, S., Kötting, O., Stettler, M., Edner, C., Graf, A., Weise, S. E., et al. (2010). A putative phosphatase LSF1 is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152, 685–697. doi: 10.1104/pp.109.148981
Crumpton-Taylor, M., Grandison, S., Png, K. M. Y., Bushby, A. J., and Smith, A. M. (2012). Control of starch granule numbers in Arabidopsis chloroplasts. Plant Physiol. 158, 905–916. doi: 10.1104/pp.111.186957
Crumpton-Taylor, M., Pike, M., Lu, K. J., Hylton, C. M., Feil, R., Eicke, S., et al. (2013). Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol. 200, 1064–1075. doi: 10.1111/nph.12455
Dauvillée, D., Chochois, V., Steup, M., Haebel, S., Eckermann, N., Ritte, G., et al. (2006). Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J. 48, 274–285. doi: 10.1111/j.1365-313X.2006.02870.x
Delvallé, D., Dumez, S., Wattebled, F., Roldan, I., Planchot, V., Berbezy, P., et al. (2005). Soluble starch synthase I: a major determinant for the synthesis of amylpectin in Arabidopsis thaliana leaves. Plant J. 43, 398–412. doi: 10.1111/j.1365-313X.2005.02462.x
D’Hulst, C., and Mérida, A. (2012). “Once upon a prime: inception of the understanding of starch initiation in plants,”,” in Essential Reviews in Experimental Biol, Vol. 5, The Synthesis and Breakdown of Starch, ed. I. J. Tetlow (London: The Society for Experimental Biology), 55–76.
Fettke, J., Chia, T., Eckermann, N., Smith, A. M., and Steup, M. (2006). A transglucosidase necessary for starch degradation and maltose metabolism in leaves at night acts on cytosolic heteroglycans (SHG). Plant J. 46, 668–684. doi: 10.1111/j.1365-313X.2006.02732.x
Fettke, J., Fernie, A. R., and Steup, M. (2012a). “Transitory starch and its degradation in higher plant cells,” in Essential Reviews in Experimental Biology: Starch: Origins, Structure and Metabolism, Vol. 5, ed. I. J. Tetlow (London: SEB), 311–374.
Fettke, J., Leifels, L., Brust, H., Herbst, K., and Steup, M. (2012b). Two carbon fluxes to reserve starch in potato (Solanum tuberosum L.) tuber cells are closely interconnected but differently modulated by temperature. J. Exp. Bot. 63, 3011–3029. doi: 10.1093/jxb/ers014
Gamez-Arjona, F. M., Li, J., Raynaud, S., Baroja-Fernanddez, E., Munoz, F. J., Ovecka, M., et al. (2011). Enhancing the expression of starch synthase class IV results in increased levels of both transitory and longterm storage starch. Plant Biotechnol. J. 9, 1049–1060. doi: 10.1111/j.1467-7652.2011.00626.x
Gamez-Arjona, F. M., Raynaud, S., Ragel, P., and Merida, A. (2014). Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 80, 305–316. doi: 10.1111/tpj.12633
Hejazi, M., Fettke, J., and Steup, M. (2012). “Starch phosphorylation and dephosphorylation: the consecutive action of starch-related dikinases and phosphatases,” in Essential Reviews in Experimental Biology: Starch: Origins, Structure and Metabolism, Vol. 5, ed. I. J. Tetlow (London: SEB), 279–308.
Hwang, S. K., Koper, K., Satoh, H., and Okita, T. W. (2016). Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J. Biol. Chem. 291, 19994–20007. doi: 10.1074/jbc.M116.735449
Kötting, O., Santelia, D., Edner, C., Eicke, S., Marthaler, T., Gentry, M. S., et al. (2009). STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21, 334–346. doi: 10.1105/tpc.108.064360
Leterrier, M., Holappa, L. D., Broglie, K. E., and Beckler, D. M. (2008). Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol. 8, 98–119. doi: 10.1186/1471-2229-8-98
Lu, K. J., Pfister, B., Jenny, C., Eicke, S., and Zeeman, S. C. (2018). Distinct functions of starch synthase 4 domains in starch granule formation. Plant Physiol. 176, 566–581. doi: 10.1104/pp.17.01008
Lu, Y., and Sharkey, T. D. (2004). The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218, 466–473. doi: 10.1007/s00425-003-1127-z
Mahlow, S., Hejazi, M., Kuhnert, F., Garz, A., Brust, H., Baumann, O., et al. (2014). Phosphorylation of transitory starch by α-glucan, water dikinase during starch turnover affects the surface properties and morphology of starch granules. New Phytol. 203, 495–507. doi: 10.1111/nph.12801
Mahlow, S., Orzechowski, S., and Fettke, J. (2016). Starch phosphorylation: insights and perspectives. Cell. Mol. Life Sci. 73, 2753–2764. doi: 10.1007/s00018-016-2248-4
Malinova, I., Alseekh, S., Feil, R., Fernie, A. R., Baumann, O., Schöttler, M. A., et al. (2017). Starch synthase 4 and plastidal phosphorylase differentially affect starch granule number and morphology. Plant Physiol. 174, 73–85. doi: 10.1104/pp.16.01859
Malinova, I., and Fettke, J. (2017). Reduced starch granule number per chloroplast in the dpe2/phs1 mutant is dependent on initiation of starch degradation. PLoS One 12:e0187985. doi: 10.1371/journal.pone.0187985
Malinova, I., Mahlow, S., Alseekh, S., Orawetz, T., Fernie, A. R., Baumann, O., et al. (2014). Double knock-out mutants of Arabidopsis thaliana grown under normal conditions reveal that the plastidial phosphorylase isozyme (PHS1) participates in transitory starch metabolism. Plant Physiol. 164, 907–921. doi: 10.1104/pp.113.227843
Nakamura, Y. (2015). “Initiation process of starch biosynthesis,” in Starch: Metabolism and Structure, ed. Y. Nakamura (Tokyo: Springer), 315–332.
Nakamura, Y., Ono, M., Sawada, T., Crofts, N., Fujita, N., and Steup, M. (2017). Characterization of the functional interactions of plastidial starch phosphorylase and starch branching enzymes from rice endosperm during reserve starch biosynthesis. Plant Sci. 264, 83–95. doi: 10.1016/j.plantsci.2017.09.002
Niittylä, T., Messerli, G., Trevisan, M., Chen, J., Smith, A. M., and Zeeman, S. C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303, 87–89. doi: 10.1126/science.1091811
Orawetz, T., Malinova, I., Orzechowski, S., and Fettke, J. (2016). Reduction of the plastidial phosphorylase in potato (Solanum tuberosum L.) reveals impact on storage starch structure during growth at low temperature. Plant Physiol. Biochem. 100, 141–149. doi: 10.1016/j.plaphy.2016.01.013
Pfister, B., Sanchez-Ferrer, A., Diaz, A., Lu, K. J., Otto, C., Holler, M., et al. (2016). Recreating the synthesis of starch granules in yeast. eLife 5:e15552. doi: 10.7554/eLife.15552
Qasim, H. M. (2018). Protein Interactions During Starch Granule Initiation in Arabidopsis thaliana. Ph.D. thesis report. Potsdam: University of Potsdam.
Ragel, P., Streb, S., Feil, R., Sahrawy, M., Annunziata, M. G., Lunn, J. E., et al. (2013). Loss of starch granule initiation has a deleterious effect on the growth of Arabidopsis plants due to an accumulation of ADP-glucose. Plant Physiol. 163, 75–85. doi: 10.1104/pp.113.223420
Raynaud, S., Ragel, P., Rojas, T., and Merida, A. (2016). The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. J. Biol. Chem. 291, 10759–10771. doi: 10.1074/jbc.M115.698332
Ritte, G., Lloyd, J. R., Eckermann, N., Rottmann, A., Kossmann, J., and Steup, M. (2002). The starch-related R1 protein is an alpha-glucan water dikinase. Proc. Natl. Acad. Sci. U.S.A. 99, 7166–7171. doi: 10.1073/pnas.062053099
Roldán, I., Lucas, M. M., Delvalle, D., Planchot, V., Jimenez, S., Perez, R., et al. (2007). The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 492–504. doi: 10.1111/j.1365-313X.2006.02968.x
Satoh, H., Shibahara, K., Tokunaga, T., Nishi, A., Tasaki, M., Hwang, S. K., et al. (2008). Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20, 1833–1849. doi: 10.1105/tpc.107.054007
Seung, D., Boudet, J., Monroe, J., Schreier, T. B., David, L. C., Abt, M., et al. (2017). Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell 29, 1657–1677. doi: 10.1105/tpc.17.00222
Seung, D., Lu, K. J., Stettler, M., Streb, S., and Zeeman, S. C. (2016). Degradation of glucan primers in the absence of starch synthase 4 disrupts starch granule initiation in Arabidopsis. J. Biol. Chem. 291, 20718–20728. doi: 10.1074/jbc.M116.730648
Seung, D., Soyk, S., Coiro, M., Maier, B. A., Eicke, S., and Zeeman, S. C. (2015). PROTEIN TARGETING TO STARCH is required for localizing GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13:e1002080. doi: 10.1371/journal.pbio.1002080
Szydlowski, N., Ragel, P., Hennen-Bierwagen, T. A., Planchot, V. R., Myers, A. M., Merida, A., et al. (2011). Integrated functions among multiple starch synthases determine both amylopectin chain length and branch linkage location in Arabidopsis leaf starch. J. Exp. Bot. 62, 4547–4559. doi: 10.1093/jxb/err172
Szydlowski, N., Ragel, P., Raynaud, S., Lucas, M. M., Roldan, I., Montero, M., et al. (2009). Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21, 2443–2457. doi: 10.1105/tpc.109.066522
Tetlow, I. J., Wait, R., Lu, Z., Akkasaeng, R., Bowsher, C. G., Esposito, S., et al. (2004). Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16, 694–708. doi: 10.1105/tpc.017400
Ugalde, J. E., Parodi, A. J., and Ugalde, R. A. (2003). De novo synthesis of bacterial glycogen: Agrobacterium tumefaciens glycogen synthase is involved in glucan initiation and elongation. Proc. Natl. Acad. Sci. U.S.A 100, 10659–10663. doi: 10.1073/pnas.1534787100
Zeeman, S. C., Thorneycroft, D., Schupp, N., Chapple, A., Weck, M., Dunstan, H., et al. (2004). Plastidial α-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 135, 849–858. doi: 10.1104/pp.103.032631
Zhang, X., Myers, A. M., and James, M. G. (2005). Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol. 138, 663–674. doi: 10.1104/pp.105.060319
Keywords: starch biosynthesis, starch granule biogenesis, starch synthase, plastidial phosphorylase, malto-oligosaccharides
Citation: Malinova I, Qasim HM, Brust H and Fettke J (2018) Parameters of Starch Granule Genesis in Chloroplasts of Arabidopsis thaliana. Front. Plant Sci. 9:761. doi: 10.3389/fpls.2018.00761
Received: 20 March 2018; Accepted: 17 May 2018;
Published: 05 June 2018.
Edited by:
Yasunori Nakamura, Akita Prefectural University, JapanReviewed by:
Christophe D’Hulst, Lille University of Science and Technology, FranceCopyright © 2018 Malinova, Qasim, Brust and Fettke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joerg Fettke, ZmV0dGtlQHVuaS1wb3RzZGFtLmRl
†Present address: Irina Malinova, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany Henrike Brust, Plasma Agriculture, Leibniz-Institute for Plasma Science and Technology, Greifswald, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.