- State Key Laboratory of Agricultural Microbiology, College of Life Sciences and Technology, Huazhong Agricultural University, Wuhan, China
Symbiosis receptor kinase (SYMRK) is a cell membrane-localized protein kinase containing extracellular malectin-like domain (MLD) and leucine-rich repeat (LRR) domains, which is critically required for both root nodule symbiosis (RNS) and arbuscular mycorrhizal symbiosis (AMS). SYMRK is widely distributed in the genomes of different plant species; however, the contribution of different domains of SYMRK and its homologs from other plant species to RNS is largely unclear. In this study, SYMRK and its homologs from three typical plant species including Medicago truncatula (for both RNS and AMS), Oryza sativa (for AMS but not RNS), and Arabidopsis thaliana (for neither RNS or AMS) were investigated using domain swap approach in response to rhizobia in Lotus japonicus. Full-length SYMRK from rice and Medicago but not from Arabidopsis could complement Lotus symrk-409 mutant plants to contribute RNS. The chimeric protein with the extracellular domain (ED) of LjSYMRK and cytoplasmic domains (CD) of SYMRK from both Medicago and rice but not Arabidopsis could contribute to RNS in Lotus, suggesting that the CD of SYMRK is required for symbiotic signaling. The chimeric receptors containing the CD of LjSYMRK (SYMRKCD) and the EDs of MtDMI2 (MtDMI2ED), OsSYMRK (OsSYMRKED), AtSYMRK (AtSYMRKED), NFR1 (NFR1ED), and NFR5 (NFR5ED) could complement Lotus symrk-409 mutant plants to develop nodules. However, MtDMI2 could partially complement Lotus symrk-409 mutants to form both effective nodules and ineffective bumps, which is similar to the complementation results from MtDMI2ED-LjSYMRKCD and LjSYMRKGDLC in Lotus symrk-409 mutants, suggesting that ED of SYMRK has a very fine-tune regulation for RNS in Lotus. The deletion of either MLD or LRR on SYMRKGDLC (a mutant version of SYMRK with GDPC motif replaced by GDLC) could contribute to RNS when overexpressed in Lotus symrk-409 mutants, suggesting that MLD and LRR domains might work together to be involved in symbiotic signaling and the LRR domain might play a negative role in LjSYMRKGDLC-mediated RNS. By mutagenizing the conserved amino acids on LRR domain, five serine residues were found to be required for the function of LjSYMRKGDLC in RNS. These finding precisely refine the molecular mechanisms of SYMRK function in symbiotic signaling in L. japonicus.
Introduction
In barren soils, leguminous plants form symbiosis with rhizobia leading to the development of a new organ called a nodule, where rhizobia reside and subsequently reduce nitrogen into ammonium for the plant host in exchange for nutrients. In comparison to the root nodule symbiosis (RNS), which is mostly restricted in legumes, the arbuscular mycorrhizal symbiosis (AMS) is widespread in most landed plants, with a few exceptions (for example cruciferous plants). It is estimated that AMS originated about 400 million years ago, while RNS originated about 60 million years ago (Remy et al., 1994; Young et al., 2011), consistent with the generally accepted theory that RNS evolved from AMS and both of them evolved from plant–pathogens interaction (Parniske, 2008; Markmann and Parniske, 2009).
For almost all the symbiosis between legumes and rhizobia, the initiation of compatible interaction begins with a molecular dialog between two partners (Oldroyd, 2013). The flavonoids secreted by plant hosts induce the biosynthesis and secretion of lipo-chitooligosaccharide called Nod factor (NF) by rhizobia (Fisher and Long, 1992). In the root hairs, two LysM receptor-like kinases, for example, NFR1/NFR5 in Lotus, or LYK3/NFP in Medicago, could recognize the NF to initiate the symbiotic signaling leading to the formation of root nodules for rhizobial colonization (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Broghammer et al., 2012; Moling et al., 2014). Recently, another LysM-RLK (exopolysaccharide receptor 3, EPR3) was shown to recognize rhizobia EPS, indicating that plants could recognize at least two symbiotic signals to mediate symbiosis (Kawaharada et al., 2015, 2017). A leucine-rich repeat (LRR) receptor-like kinase, Lotus SYMRK or Medicago Does Not make Infections (DMI2), plays a key role in symbiotic signaling acting as a downstream component of Nod Factor Receptors (Ané et al., 2002; Stracke et al., 2002; Miwa et al., 2006). All these receptor-like kinases are required for calcium spiking and expression of symbiosis-relate genes via a common symbiosis pathway (CSP) (Oldroyd and Downie, 2008; Kouchi et al., 2010). Similar to RNS, LjNFR1/MtLYK3 also participates in AMS with a possible function to recognize Myc Factor produced by arbuscular mycorrhizal fungi (Zhang et al., 2015). The Myc and Nod factors initiate a common signaling pathway (CSP) with overlapped signaling components identified (Oldroyd, 2013; Genre and Russo, 2016). Symbiosis receptor kinase (SYMRK) is widespread in most plant species such as leguminous and cereal plants. Furthermore, in the Arabidopsis who could not have the capability of either RNS or AMS has two SYMRK homologs, i.e., AT1G67720.1 and AT2G37050.1 (Shiu and Bleecker, 2001).
In Lotus, knock-out of SYMRK completely abolished the ability to form ITs or produce nodules with rhizobia. However, in the symrk-14 mutant plants that harbor a point mutation at GDPC motif linking malectin and LRR domains, epidermal responses including infection thread formation were significantly reduced, while the formation of nodule primordia and cortical infection were slightly changed (Kosuta et al., 2011), suggesting that epidermal response and cortical program might be differentially regulated during symbiotic interaction with rhizobia. Since GDPC motif was shown to be required for the cleavage of malectin-like domain (MLD) from SYMRK protein (Antolín-Llovera et al., 2014), malectin and LRR might have a fine-tune regulation mechanism in mediating symbiotic response. The truncated version SYMRKΔMLD (lacking the MLD) was shown to be able to interact with NFR5 and transfer the symbiotic signal to downstream targets (Antolín-Llovera et al., 2014). In Lotus, overexpression of the full-length SYMRK could produce spontaneous nodules in the absence of rhizobia (Ried et al., 2014). Similarly, overexpression of the kinase domain of Arachis hypogaea SYMRK (AhSYMRKKD) could also induce spontaneous nodule formation in the absence of rhizobia (Saha et al., 2014), suggesting that the kinase domain of SYMRK might have a dominant positive role in nodule organogenesis. However, the autophosphorylation of SYMRK seems not to be essential to mediate symbiotic signaling, since mutation at an autophosphorylation site (Tyr-670) in AhSYMRK could partially complement symrk mutants by producing infection threads and uninfected nodule primordia (Samaddar et al., 2013; Paul et al., 2014; Saha et al., 2016). In addition, expression of OsSYMRK under the control of the SYMRK promoter could only complement symrk mutants to produce AMS but not RNS in Lotus (Markmann et al., 2008). All these results not only indicated the critical role of SYMRK protein in RNS, but also suggested that SYMRK uncouples the rhizobia infection and nodule formation. However, how the extracellular domains (EDs) of SYMRK is involved in RNS is largely unknown.
In this study, we demonstrate that MtDMI2 (SYMRK homolog protein in Medicago) and OsSYMRK but not AtSYMRK could complement Lotus SYMRK−/− plants. In addition, multiple chimeric SYMRK proteins were developed to analyze the function of different domains of SYMRK in RNS. The data refine the molecular mechanisms of each domain of SYMRK in RNS in Lotus.
Results
Homology Analysis of SYMRK in Legumes and Non-legumes
SYMRK is critically required for both root nodule symbiosis (RNS) and AMS. To study its function involved in RNS, SYMRK homologs were identified based on sequence similarity with Lotus SYMRK using BLASTp as a search engine at the National Center for Biotechnology Information database (NCBI), and 18 SYMRK homologs were identified. A phylogenetic tree based on various SYMRK proteins was made using the MEGA 5.1 software (Figure 1). In the phylogenetic tree, SYMRK proteins and homologs fall into three different clades. Clade I contains SYMRK proteins from plants that could have both RNS and AMS; clade II contains SYMKR from plants having AMS but not RNS, while proteins from Cruciferae plants that could not possess either AMS or RNS fallen into clade III.
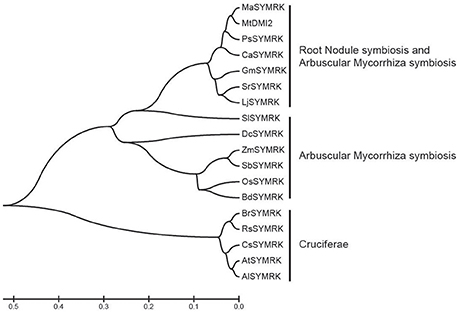
Figure 1. Phylogenetic tree of SYMRK proteins in different species. The Phylogenetic tree was constructed using MEGA 5.1 software with 1,000 bootstrap replications. Abbreviation of each species names were used to represent the SYMRK protein cloned from this species. Ma, Melilotus albus; Mt, M. truncatula; Ps, Pisum sativum; Ca, Cicer arietinum; Gm, Glycine max; Sr, Sesbania rostrata; Lj, L. japonicus; Sl, Solanum lycopersicum; Dc, Dendrobium catenatum; Zm, Zea mays; Sb, Sorghum bicolor; Os, Oryza sativa; Bd, Brachypodium distachyon; Br, Brassica rapa; Rs, Raphanus sativus; Cs, Camelina sativa; At, Arabidopsis thaliana; Al, Arabidopsis lyrata.
Both OsSYMRK and MtDMI2 but not AtSYMRK Could Complement symrk-409 Mutant in Lotus
To study the function of SYMRK involved in RNS in Lotus, an LORE1-insertion symrk mutant were identified from the Centre for Carbohydrate Recognition and Signaling (CARB, http://users-mb.au.dk/pmgrp/) (Fukai et al., 2012; Urbanski et al., 2012; Małolepszy et al., 2016). The line 30010361 (symrk-409) contains an LORE1-insertion between exon 4 and intron 4, leading to the early termination of translation of SYMRK (Figures S1A–C). In response to rhizobia treatment, infection threads and nodules were not detected in the symrk-409 mutant plants when compared with Lotus B-129 Gifu 7- and 21-day post-inoculated (DPI) with rhizobia (Figures S1D–G), indicating that the symrk-409 is another knock-out mutant that could be used for future study.
To analyze the symbiotic function of various SYMRK proteins, one SYMRK protein from each clade-based phylogenetic tree, respectively, was chosen for study, i.e., MtDMI2 from Medicago that could have both RNS and AMS, OsSYMRK from rice that could only have AMS, and AtSYMRK from Arabidopsis that could have neither RNS or AMS.
To test the function of each SYMRK as indicated above, SYMRK proteins including LjSYMRK, MtDMI2, OsSYMRK, and AtSYMRK and control vector were transgenically expressed in the symrk-409 mutant plants under the control of the Ljubiquitin promoter using hairy root transformation. The Ljubiquitin promoter could induce a strong expression of multiple genes in previous publications (Maekawa et al., 2009; Yoro et al., 2014; Wang et al., 2016). Compared with no nodules observed in the control transformation (Figures 2A1–5), overexpression of LjSYMRK or OsSYMRK or MtDMI2 but not AtSYMRK-induced nodule formation 21 DPI with rhizobia in symrk-409 mutant plants (Figure 2A). There was no significant difference in the number of nodules per plant root transformed with either LjSYMRK or MtDMI2 or OsSYMRK 21 DPI with rhizobia (Figure 2B). In comparison to wild type and LjSYMRK-transgenic Lotus nodules, decreased nitrogenase activities from transgenic nodules with expression of either MtDMI2 or OsSYMRK were measured. While no nitrogenase activity was detected in roots transformed with either vector control or AtSYMRK (Figure 2C). In consistence with the low levels of nitrogenase activity from MtDMI2-transgenic nodules, about 30% ineffective bumps with no rhizobia infection were observed (Figure 2D). However, no ineffective bumps were observed in OsSYMRK-transgenic Lotus symrk-409 mutant roots. These data indicate that both MtDMI2 and OsSYMRK but not AtSYMRK could complement symrk-409 mutant to form nodules.
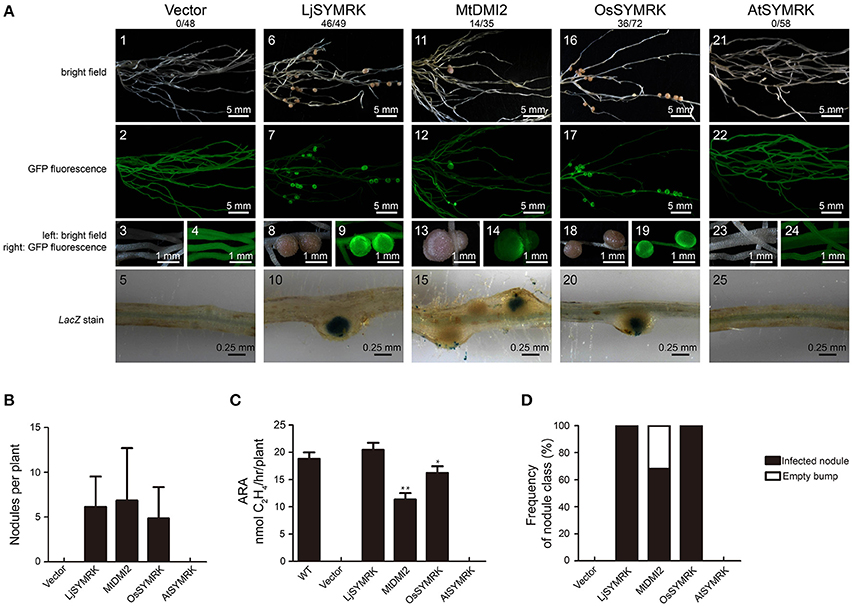
Figure 2. Complementation of Lotus symrk-409 mutants with LjSYMRK, MtDMI2, OsSYMRK, and AtSYMRK. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed with empty control (A1–5), LjSYMRK (A6–10), MtDMI2 (A11–15), OsSYMRK (A16–20), and AtSYMRK (A21–25). Digital numbers on top of (A) indicate the number of nodulated plants out of the positive transgens. (B) Numbers of nodules and bumps generated on different transgenic roots 21 DPI with rhizobia. (C) Nitrogenase activity of root nodules 21 DPI with rhizobia determined using acetylene reduction method. *P < 0.05, **P < 0.01 indicate significant differences (t-test). (D) Frequency of nodules and bumps produced on the transgenic roots.
Since both Lotus and Medicago belong to leguminous plants, we examined whether LjSYMRK could complement MtDMI2-knock-out mutant plants dmi2-1 (or named TR25). Both LjSYMRK and MtDMI2 and control vector were introduced into dmi2-1 using hairy root transformation. In comparison with no infection threads and nodules observed on the dmi2-1 mutant transformed with empty vector (Figures S2A1–5,B), a significant number of nodules and infected nodule primordium was detected on roots transformed with either MtDMI2 or LjSYMRK (Figures S2A6–15,2B), which is consistent with nitrogenase activity measured in these two transgenic nodules (Figure S2C). However, about 20% nodule cases formed on dmi2-1 mutant plants expressing LjSYMRK are ineffective since no rhizobia were detected using lacZ staining (Figure S2D), suggesting that MtDMI2 and LjSYMRK have overlapping function to complement each other but with exceptional roles in mediating symbiosis in their own hosts.
The CD of SYMRK Is Essential but the ED Is Important in RNS in Lotus
To study the function of each domain of SYMRK from different plant species, a variety of constructs was made using domain swaps and functionally tested in symrk-409 mutant plants using hairy root transformation (Figure S3). To study the function of the cytoplasmic domain (CD) of SYMRK, the CD of LjSYMRK replaced with CDs of MtDMI2, OsSYMRK, and AtSYMRK (Figure S3). In symrk-409, roots transformed with LjSYMRKED-MtDMI2CD and LjSYMRKED-OsSYMRKCD formed normal infected nodule primordia and effective nodules (Figure 3A1–10,B–D) with similar nitrogenase activities measured (Figure 3C). However, LjSYMRKED-AtSYMRKCD could not complement symrk-409 to form infection threads and nodules (Figures 3A11–15). These data suggest that the CD of SYMRK plays an essential role in RNS.
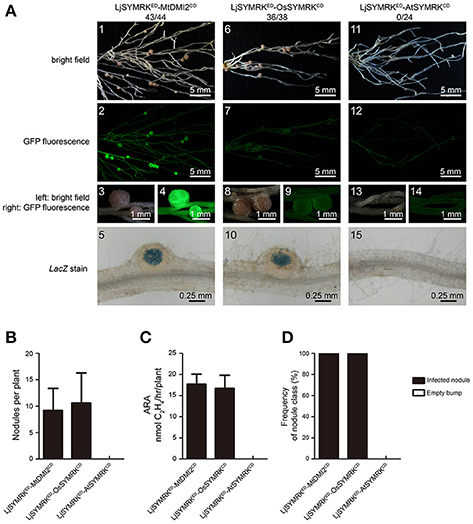
Figure 3. Complementation of Lotus symrk-409 mutants with LjSYMRKED-MtDMI2CD, LjSYMRKED-OsSYMRKCD, and LjSYMRKED-AtSYMRKCD. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed LjSYMRKED-MtDMI2CD (A1–5), LjSYMRKED-OsSYMRKCD (A6–10), and LjSYMRKED-AtSYMRKCD (A11–15). Digital numbers on top of panel A indicate the number of nodulated plants out of total positive trasngens. (B) Numbers of nodules produced per transgenic roots 21 DPI with rhizobia. (C) Nitrogenase activities of root nodules 21 DPI with rhizobia determined using acetylene reduction method. (D) Frequency of nodules and bumps generated on the transgenic roots.
To analyze the symbiotic function of the ED of SYMRK, the ED of LjSYMRK was replaced with the ED of MtDMI2, OsSYMRK, and AtSYMRK to obtain the chimeric protein MtDMI2ED-LjSYMRKCD, OsSYMRKED-LjSYMRKCD, and AtSYMRKED-LjSYMRKCD, respectively (Figure S3). After transgenically expressed in symrk-409 roots, the function of these three chimeric proteins in nodulation were analyzed. All these three chimeric proteins could confer significant number of nodules formed on symrk-409 roots 21 DPI with rhizobia (Figures 4A,B). However, MtDMI2ED-LjSYMRKCD-transgenic plants produced 30% bumps with no LacZ activity detected (Figure 4D) and lower nitrogenase activity than the other two transgenic nodules (Figure 4C). The ED of LjSYMRK was also replaced with ED of LjNFR1 or LjNFR5 to generate LjNFR1ED-LjSYMRKCD and LjNFR5ED-LjSYMRKCD, respectively, and transgenically expressed these chimeric proteins in symrk-409 mutant plants. Surprisingly, normal nodules numbers and significantly high nitrogenase activities were measured in the symrk-409 mutant transformed with either LjNFR1ED-LjSYMRKCD or LjNFR5ED-LjSYMRKCD (Figure S4). These data suggest that ED of SYMRK is important but might not be necessary while the CD of LjSYMRK is essential for RNS in Lotus.
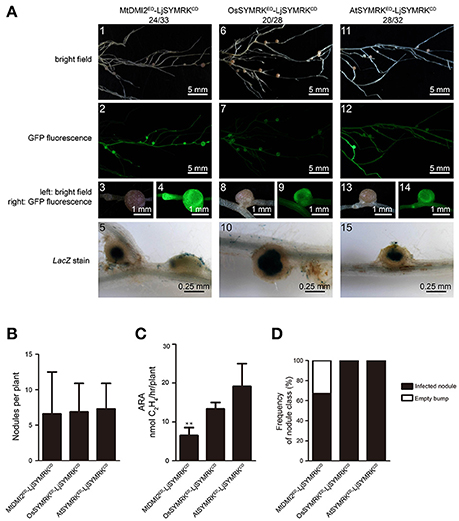
Figure 4. Complementation of Lotus symrk-409 mutants with MtDMI2ED-LjSYMRKCD, OsSYMRKED-LjSYMRKCD, and AtSYMRKED-LjSYMRKCD. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed MtDMI2ED-LjSYMRKCD (A1–5), OsSYMRKED-LjSYMRKCD (A6–10), and AtSYMRED-LjSYMRKCD (A11–15). Digital numbers on panel A indicate the number of nodulated plants per total number of GFP-positive plants. (B) Numbers of nodules produced per transgenic roots 21 DPI with rhizobia. (C) Nitrogenase activities of transgenic nodules 21 DPI with rhizobia determined using acetylene reduction method. **P < 0.01 (t-test). (D) Frequency of nodules and bumps generated on the transgenic roots.
MLD and LRR Domain Have a Fine-Tune Regulation on SYMRK-Mediated Symbiotic Response
Complementation with either MtDMI2 or MtDMI2ED-LjSYMRKCD in symrk-409 plants could produce both effective nodules and ineffective bumps. To test which domain is required for its function in negative regulate rhizobia infection, we made four truncated versions of LjSYMRK (Figure S3), i.e., LjSYMRKΔED (LjSYMRK without ED), LjSYMRKΔMLD (LjSYMRK without MLD), LjSYMRKΔLRR (LjSYMRK without LRR domain and “GDPC” motif), and LjSYMRKGDLC (Proline replaced with leucine). LjSYMRK deleted with MLD, or LRR, or ED could rescue the defects on nodulation phenotype in symrk-409 mutant plants (Figures 5A5–20). The transgenic plants with overexpression of LjSYMRKΔMLD, or LjSYMRKΔLRR, or LjSYMRKΔED formed a significant number of nodules with high nitrogenase activities measured (Figures 5B–D), suggesting that the EDs of SYMRK might not be necessary to be involved in symbiosis. However, LjSYMRKGDLC (a mutation at the conserved residues “GDPC” motif) whose MLD was not detected to be cleaved during symbiosis could not completely complement symrk-409 mutant plants with only ineffective bumps developed (Figures 5A1–5,B–D). These data suggest that MLD or LRR domain alone is dispensable but might work together to reduce rhizobial infection in mutant plants expressing LjSYMRKGDLC.
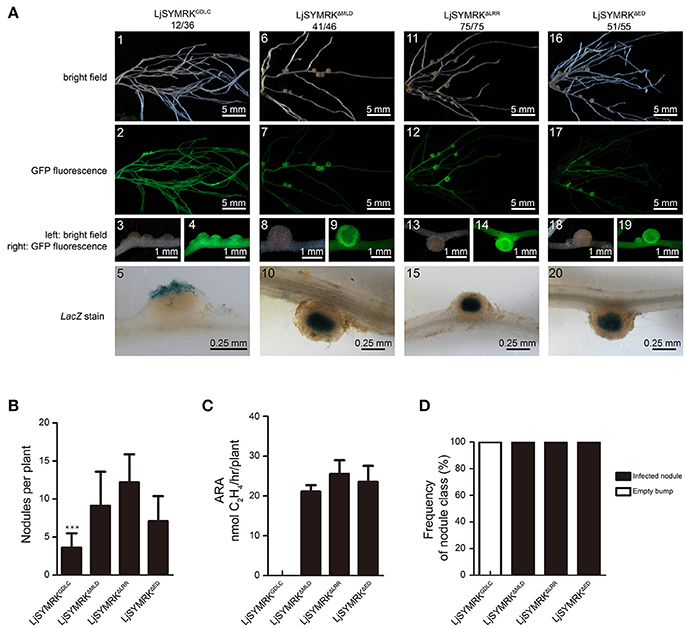
Figure 5. Complementation of Lotus symrk-409 mutants with LjSYMRKGDLC, LjSYMRKΔMLD, LjSYMRKΔLRR, and LjSYMRKΔED. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed LjSYMRKGDLC (A1–5), LjSYMRKΔMLD (A6–10), LjSYMRKΔLRR (A11–15), and LjSYMRKΔED (A16–20). Digital numbers on panel A indicate the number of nodulated plants per total positive roots. (B) Numbers of nodules and bumps produced on the transgenic roots 21 DPI with rhizobia. ***P < 0.01 (t-test). (C) Nitrogenase activities of transgenic nodules 21 DPI with rhizobia determined using acetylene reduction method. (D) Frequency of nodules and bumps generated on the transgenic roots.
The LRR Domain Plays a Crucial Role in LjSYMRKGDLC-Mediated RNS
A previous conclusion indicated that mutation at the GDPC motif that connects MLD and LRR disrupts the cleavage of MLD from SYMRK leading to the block of RNS in Lotus. We sought to examine whether LRR domain is required for its function in symbiosis. Chimeric constructs containing LjSYMRKGDLC with LRR domains of MtDMI2, OsSYMRK, and AtSYMRK were made to generate LjSYMRKGDLC-MtLRR, LjSYMRKGDLC-OsLRR, and LjSYMRKGDLC-AtLRR, respectively (Figure S3), and functionally tested in symrk-409 mutant plants using hairy root transformation approach. Transgenic roots overexpressing either LjSYMRKGDLC-OsLRR or LjSYMRKGDLC-AtLRR but not LjSYMRKGDLC-MtLRR could generate effective nodules with high nitrogenase activity per root tested (Figure 6). These findings implicate that the LRR domain is critical among SYMRK homologous proteins in the RNS.
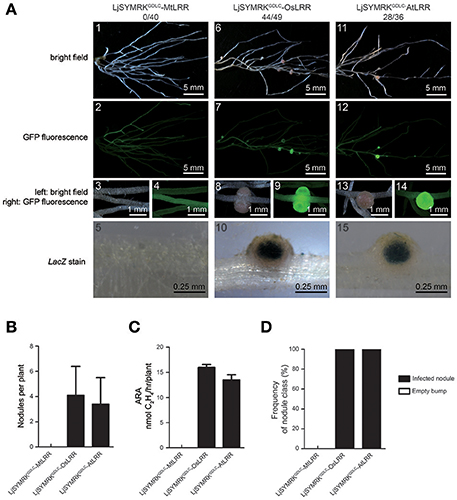
Figure 6. Complementation of Lotus symrk-409 mutants with LjSYMRKGDLC-MtLRR, LjSYMRKGDLC-OsLRR, and LjSYMRKGDLC-AtLRR. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed LjSYMRKGDLC-MtLRR (A1–5), LjSYMRKGDLC-OsLRR (A6–10), and LjSYMRKGDLC-AtLRR (A11–15). Digital numbers on (A) indicate the number of nodulated plants per total positive roots. (B) Numbers of nodules and bumps produced on the transgenic roots 21 DPI with rhizobia. (C) Nitrogenase activities of transgenic nodules 21 DPI with rhizobia determined using acetylene reduction method. (D) Frequency of nodules and bumps generated on the transgenic roots.
In order to verify the importance of LRR domains, the alignment of amino acid sequence of LRR domains of SYMRK from different plant species were made (Figure S5) and nine conserved residues were chosen for point mutation into alanine based on the construct containing LjSYMRKGDLC. All these variants were transgenically overexpressed in symrk-409 mutant plants and nodulation phenotype was assayed 21 DPI with rhizobia. Transgenic roots expressing LjSYMRKGDLC with mutations at H437, P448, Y460, and E469, respectively, only developed ineffective nodules with no nitrogenase activity detected (Figure 7), which is similar to the phenotype on LjSYMRKGDLC-expressing transgenic roots. However, mutations at five serine residues (S422, S450, S451, S455, and S470) in LjSYMRKGDLC induced the formation of nodules in symrk-409 mutant plants, some of which are rhizobia-infected nodules with measurable nitrogenase activity (Figure 7), indicating that these five serine residues are critical in SYMRKGDLC.
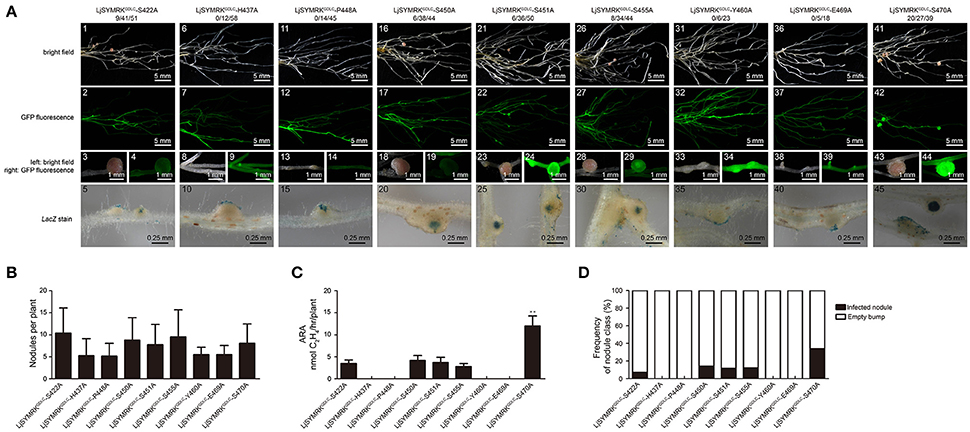
Figure 7. Complementation of Lotus symrk-409 mutants with constructs of point mutations on the LRR domain of LjSYMRKGDLC. (A) Representative micrographs of different transgenic nodules. Positive transgenic roots identified using GFP signals were inoculated with M. loti strain NZP2235 containing a LacZ report gene. Nodules and bumps were stained with X-gal. Images showed nodules produced on symrk-409 roots transgenically expressed LjSYMRKGDLC-S422A (A1–5), LjSYMRKGDLC-H437A (A6–10), LjSYMRKGDLC-P448A (A11–15), LjSYMRKGDLC-S450A (A16–20), LjSYMRKGDLC-S451A (A21–25), LjSYMRKGDLC-S455A (A26–30), LjSYMRKGDLC-Y460A (A31–35), LjSYMRKGDLC-E469A (A36–40), and LjSYMRKGDLC-S470A (A41–45). Digital numbers on (A) indicate the number of nodulated plants per total positive roots. (B) Numbers of nodules and bumps produced on the transgenic roots 21 DPI with rhizobia. (C) Nitrogenase activities of transgenic nodules 21 DPI with rhizobia determined using acetylene reduction method. **P < 0.01 (t-test). (D) Frequency of nodules and bumps generated on the transgenic roots.
Discussion
SYMRK protein plays a crucial role in root nodule symbiosis in leguminous plants in response to compatible rhizobia. In this study, domain swap approach was used to study the function of different domains of SYMRK involved in RNS in Lotus. The broad goal of this study was to refine the molecular function of different domains of SYMRK in the contribution to root nodule symbiosis using domain swap approach in Lotus japonicus.
SYMRK is a key component in the symbiotic signaling pathway, which is essential for both RNS and AMS. In leguminous plants, such as in Medicago, MtDMI2 is required for both RNS and AMS, while in rice, OsSYMRK was only shown to be required for AMS and AtSYMRK was shown not to be involved in symbiosis in Arabidopsis. To test the roles of different domains of SYMRK in RNS, several chimeric SYMRK protein-encoding genes were constructed under the control of the Ljubiquitin promoter and multiple individual transgenic roots were used to test their functions in Lotus. The diverse function of SYMRK homologous proteins was further confirmed with the finding that MtDMI2 and OsSYMRK but not AtSYMRK could complement Lotus SYMRK−/− mutant to mediate RNS. The function of SYMRK proteins were tested in Lotus SYMRK−/− plants in response to rhizobia. The partial complementation by MtDMI2 by forming some ineffective nodules suggests the slight different function of SYMRK between Lotus and Medicago might exist. Since Medicago forms indeterminate nodules while Lotus forms determinate nodules with rhizobia, it is possible that SYMRK-mediated RNS have a little difference within these two plant species. Domain swap approach by replacing the CD of LjSYMRK with CDs from other SYMRK homologs confirmed that AtSYMRK is not functional in Lotus to mediate RNS. While the ED of LjSYMRK seems to be not so important since replacing LjSYMRKED with ED from other SYMRK proteins or even from NFR1 and NFR5 is still functional to mediate RNS in Lotus. The critical function of SYMRKCD in symbiosis was also reported that spontaneous nodule formation was observed in Medicago and Lotus when the kinase domain of AhSYMRK and the full length LjSYMRK were overexpressed, respectively (Ried et al., 2014; Saha et al., 2014). Chimeric protein AtSYMRKED-LjSYMRKCD but not full length of AtSYMRK could complement Lotus SYMRK1−/− mutants indicates that the kinase domain of SYMRK is critical to induce nodule initiation even regardless of its ED or without rhizobia treatment when overexpressed in plants.
The ED of SYMRK contains two important domains, i.e., MLD and LRR linked by a GDPC motif. The well-established model about SYMRK reveals that cleavage of MLD is required for its association with NFR5 to mediate symbiotic signaling transduction in Lotus (Antolín-Llovera et al., 2014). It seems that the presence of MLD suppresses SYMRK-mediated symbiotic signaling since Lotus symrk-14 mutants having a point mutation at the linker (“GDPC” motif was mutated into “GDLC”) between MLD and LRR showed strong inhibition on bacterial infection but slight suppression on the development of nodules (Ried et al., 2014). The abnormal function of SYMRK-14 (or SYMRKGDLC) in RNS was exaggerated with only ineffective nodules formed in Lotus when SYMRKGDLC was overexpressed. However, overexpression of SYMRKGDLC with deletion of either MLD or LRR domain only produced effective nodules, suggesting that both MLD and LRR domain of SYMRK might play a negative rule but might work together in SYMRKGDLC-mediated nodule organogenesis in Lotus. The same phenotype with effective nodule formation was also observed in Lotus when LjSYMRKGDLC with its LRR replaced by the LRR domains from OsSYMRK or AtSYMRK but not from MtDMI2 was overexpressed, suggesting that LRR domains from OsSYMRK or AtSYMRK might be not functional in mediating RNS in Lotus. The negative role of LRR domain in SYMRKGDLC-mediated response is further confirmed by point mutations at five conserved serine residues, leaving us a possibility that the negative function of LRR might be regulated by an unknown phosphorylation.
The ED of SYMRK seems to have a fine-tune regulation on the activation of symbiotic signaling in Lotus. Replacement ED with EDs from OsSYMRK or AtSYMRK proteins or even with NFR1 and NFR5 could contribute symbiotic response when overexpressed in Lotus plants. Since these proteins located far from LjSYMRK based on the phylogenetic tree, it is possible that these chimeric proteins with nonfunctional EDs still keep some function as LjSYMRKΔED to induce symbiotic response. However, the chimeric protein MtDMI2ED-LjSYMRKCD could not completely complement Lotus SYMRK−/− plants, indicating that the functional difference between Lotus and Medicago possibly due to that different nodule types formed on these two plants. The abovementioned hypothesis is further confirmed by the finding that LjSYMRKGDLC-MtLRR but not LjSYMRKGDLC-AtLRR or LjSYMRKGDLC-OsLRR is not functional to mediate RNS in Lotus. The whole ED from LjSYMRK might be nonfunctional when LRR domain was replaced with LRR from AtSYMRK or OsSYMRK. However, since the LRR from MtDMI2 might have difference to mediate RNS, the MLD of LjSYMRK seems not to match with the LRR from MtDMI2 which finally made the chimeric protein LjSYMRKGDLC-MtLRR lose its function in RNS. The data indicated that MLD and LRR must have a fine-tune regulation on the activation of symbiotic response.
In conclusion, the intracellular domain of SYMRK is critical while the ED of SYMRK plays a relatively minor role in mediating RNS in Lotus. GDPC motif that connects MLD and LRR domain required for MLD cleavage in response to RNS is responsible for nodule organogenesis. Both MLD and LRR domains might have a fine-tune regulation of SYMRK-mediated nodule organogenesis in Lotus. This progress in defining the molecular functions of SYMRK in RNS. However, identification of the direct targets of SYMRK and crystal structure of SYMRK are the future topics for understanding the molecular mechanisms mediated by SYMRK in NRS.
Materials and Methods
Homology Analysis
SYMRK homologous proteins were identified based on the sequence similarity to Lotus SYMRK with the protein blast suite at the National Center for Biotechnology Information database (https://blast.ncbi.nlm.nih.gov) used. The phylogenetic tree was built by MEGA 5.1 software (Tamura et al., 2011).
Plant Growth and Rhizobia Inoculation
Lotus seeds (B-129 Gifu and symrk mutant) and hair root transgenic seedlings were grown in vermiculite and perlite mixed (2:1 volume ratio) supplied a 1/2 B&D medium and placed in a growth chamber set at 22°C for 16-h-light/8-h-dark cycle. Plants were inoculated with Mesorhizobium loti strain NZP2235 expressing beta-galactosidase (LacZ) after 5 days. Rhizobia grown in liquid TY medium adding tetracycline at OD600 of 1.0. They were then pelleted and resuspended in 1/2 B&D medium containing 0.5 mM KNO3 at OD600 of 0.02.
Medicago hair root transgenic seedlings were grown in vermiculite and perlite mixed (2:1 volume ratio) supplied a 1/2 FM medium and placed in a growth chamber set at 22°C for 16-h-light/8-h-dark cycle. Plants were inoculated with Sinorhizobium meliloti strain Sm2011 expressing LacZ after 5 days. Rhizobia grown in liquid TY medium adding tetracycline at OD600 of 1.0. They were then pelleted and resuspended in 1/2 FM medium containing 0.5 mM KNO3 at OD600 of 0.02.
Lotus symrk Mutant Identification
Genomic DNA was extracted from putative mutant plants and used for PCR amplification of 2 min soak at 95°C, followed by 35 cycles of 30 s at 94°C, 30 s at 58°C, and 50 s at 72°C, followed by 5 min soak at 72°C. Primers for genotyping of symrk-409 insertion alleles are provided in Table S1, which were performed as described previously (Fukai et al., 2012; Urbanski et al., 2012).
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from roots at 7 DPI using the EASYspin Plant RNA Kit (Aidlab, China). Primescript RT Reagent Kit (TaKaRa, Japan) was used to remove gDNA of RNA samples and reverse transcribe RNA into cDNA. Quantitative real-time PCR was performed on an ABI ViiATM7 Real-Time PCR System (ABI, USA) using One-Step SYBR PrimeScript RT-PCR kit II (Takara, Japan). Lotus ATP synthase (Genbank ID: AW719841) and ubiquitin (Genbank ID: AW720576) were used as reference genes, which are stably expressed in all plant tissues. Primers for qRT-PCR are listed in Table S1.
Plasmid Construction for Mutant Complementation
The pUB-GFP vector was digested by restriction endonucleases XbaI and StuI, then linked to a fragment or multi-fragments by Gibson assembly (Gibson et al., 2009). Primers for PCR are listed in Table S1.
Lotus Hairy Root Transformation
The pUB-GFP vectors carrying a variety of chimeric SYMRK constructs were transformed into symrk-409 seedlings using Agrobacterium rhizogenes LBA1334 as described by Wang et al. (2013). The GFP marker was used for selection of transgenic hairy roots by a fluorescence stereo microscope (Nikon SMZ18, Japan).
Medicago Hairy Root Transformation
Transgenic root on dmi2-1 seedlings were induced using A. rhizogenes MSU440 as described by Wang et al. (2016). Positive transgenic hairy roots were identified by GFP marker using a fluorescence stereo microscope (Nikon SMZ18, Japan).
X-Gal Staining
Plants were inoculated with rhizobia carrying a lacZ reporter gene. At 7 or 21 DPI, roots were vacuum infiltrated for 5 min in fixative solution (1.25% Glutaraldehyde dissolved in 0.1 M potassium phosphate buffer pH 7.4), and placed at room temperature for 40 min, washed twice 10 min in 0.1 M potassium phosphate buffer. Roots were vacuum infiltrated for 5 min with X-Gal staining solution [0.1 M phosphate buffer, 6.25 mM K4Fe(CN)6, 6.25 mM K3Fe(CN)6, 0.75% X-Gal in DMF] and kept overnight at 28°C in the dark. Roots were washed twice in 0.1 M potassium phosphate buffer for 5 min, then rinsed twice in ddH2O for 5 min. Stained roots were examined using a fluorescence stereo microscope (Nikon SMZ18, Japan) and a fluorescence microscope (LEICA DM2500, Germany).
Detection of Nitrogenase Activity
Acetylene reduction activity (ARA) was used to detect nitrogenase activity of root nodules at 21 DPI. Four nodulated seedlings were transferred into test tubes with 2 ml acetylene added for additional growth at 28°C for 2 h. At least three repeats of each experiment were analyzed. Acetylene was surveyed using a GC-4000A gas chromatograph (Dongxi Company, China).
Accession Numbers
Sequence data from this work can be found under the following GenBank accession numbers: AAV88623.1 for SrSYMRK, XP_004512550.1 for CaSYMRK, CAD22013.1 for MaSYMRK, CAD10812.1 for PsSYMRK, XP_003517193.1 for GmSYMRK, CAD10811.1 for MtSYMRK, AAM67418.1 for LjSYMRK, NP_001234869.1 for SlSYMRK, NP_001105860.1 for ZmSYMRK, XP_015646949.1 for OsSYMRK, XP_014752741.1 for BdSYMRK, XP_002460874.1 for SbSYMRK, XP_020677592.1 for DcSYMRK, NP_564904.1 for AtSYMRK, XP_020890981.1 for AlSYMRK, XP_010511724.1 for CsSYMRK, XP_009105317.1 for BrSYMRK, and XP_018445268.1 for RsSYMRK.
Author Contributions
HL, YC, and ZZ designed the experiments and analyzed the results. HL, MC, TZ, and LD performed the experiments and analyzed the data. HL, YC, and ZZ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Centre for Carbohydrate Recognition and Signaling (CARB) for providing LORE1 symrk-409 (30010361) mutant seeds. We thank Dr. Giles E. D. Oldroyd (John Innes Centre, UK) for kindly providing seeds of dmi2-1 mutant seeds. We thank Dr. Fang Xie (Shanghai Institute of Plant Physiology & Ecology) for providing S. meliloti strain Sm2011 expressing lacZ. This work was supported by the National Key R&D Program of China (2016YF0100700), the National Natural Science Foundation of China (31670240), and the State Key Laboratory of Agricultural Microbiology (AMLKF201503 and AMLKF201608).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00697/full#supplementary-material
References
Ané, J. M., Lévy, J., Thoquet, P., Kulikova, O., de Billy, F., Penmetsa, V., et al. (2002). Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant Microbe Interact. 15, 1108–1118. doi: 10.1094/MPMI.2002.15.11.1108
Antolín-Llovera, M., Ried, M. K., and Parniske, M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod Factor Receptor 5. Curr. Biol. 24, 422–427. doi: 10.1016/j.cub.2013.12.053
Arrighi, J. F., Barre, A., Ben Amor, B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Broghammer, A., Krusell, L., Blaise, M., Sauer, J., Sullivan, J. T., Maolanon, N., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109, 13859–13864. doi: 10.1073/pnas.1205171109
Fisher, R. F., and Long, S. R. (1992). Rhizobium–plant signal exchange. Nature 357, 655–660. doi: 10.1038/357655a0
Fukai, E., Soyano, T., Umehara, Y., Nakayama, S., Hirakawa, H., Tabata, S., et al. (2012). Establishment of a Lotus japonicus gene tagging population using the exon-targeting endogenous retrotransposon LORE1. Plant J. 69, 720–730. doi: 10.1111/j.1365-313X.2011.04826.x
Genre, A., and Russo, G. (2016). Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 7:96. doi: 10.3389/fpls.2016.00096
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A. III., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Kawaharada, Y., Kelly, S., Nielsen, M. W., Hjuler, C. T., Gysel, K., Muszynski, A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312. doi: 10.1038/nature14611
Kawaharada, Y., Nielsen, M. W., Kelly, S., James, E. K., Andersen, K. R., Rasmussen, S. R., et al. (2017). Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat. Commun. 8:14534. doi: 10.1038/ncomms14534
Kosuta, S., Held, M., Hossain, M. S., Morieri, G., MacGillivary, A., Johansen, C., et al. (2011). Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 67, 929–940. doi: 10.1111/j.1365-313X.2011.04645.x
Kouchi, H., Imaizumi-Anraku, H., Hayashi, M., Hakoyama, T., Nakagawa, T., Umehara, Y., et al. (2010). How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51, 1381–1397. doi: 10.1093/pcp/pcq107
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. doi: 10.1126/science.1090074
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Maekawa, T., Maekawa-Yoshikawa, M., Takeda, N., Imaizumi-Anraku, H., Murooka, Y., and Hayashi, M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194. doi: 10.1111/j.1365-313X.2008.03774.x
Małolepszy, A., Mun, T., Sandal, N., Gupta, V., Dubin, M., Urbanski, D., et al. (2016). The LORE1 insertion mutant resource. Plant J. 88, 306–317. doi: 10.1111/tpj.13243
Markmann, K., Giczey, G., and Parniske, M. (2008). Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 6:e68. doi: 10.1371/journal.pbio.0060068
Markmann, K., and Parniske, M. (2009). Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci. 14, 77–86. doi: 10.1016/j.tplants.2008.11.009
Miwa, H., Sun, J., Oldroyd, G. E. D., and Downie, J. A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19, 914–923. doi: 10.1094/MPMI-19-0914
Moling, S., Pietraszewska-Bogiel, A., Postma, M., Fedorova, E., Hink, M. A., Limpens, E., et al. (2014). Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26, 4188–4199. doi: 10.1105/tpc.114.129502
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E., and Downie, J. M. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Paul, A., Samaddar, S., Bhattacharya, A., Banerjee, A., Das, A., Chakrabarti, S., et al. (2014). Gatekeeper tyrosine phosphorylation is autoinhibitory for Symbiosis Receptor Kinase. FEBS Lett. 588, 2881–2889. doi: 10.1016/j.febslet.2014.06.056
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Remy, W., Taylor, T. N., Hass, H., and Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U.S.A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Ried, M. K., Antolín-Llovera, M., and Parniske, M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife 3:e03891. doi: 10.7554/eLife.03891
Saha, S., Dutta, A., Bhattacharya, A., and DasGupta, M. (2014). Intracellular catalytic domain of symbiosis receptor kinase hyperactivates spontaneous nodulation in absence of rhizobia. Plant Physiol. 166, 1699–U1826. doi: 10.1104/pp.114.250084
Saha, S., Paul, A., Herring, L., Dutta, A., Bhattacharya, A., Samaddar, S., et al. (2016). Gatekeeper tyrosine phosphorylation of SYMRK is essential for synchronizing the epidermal and cortical responses in root nodule symbiosis. Plant Physiol. 171, 71–81. doi: 10.1104/pp.15.01962
Samaddar, S., Dutta, A., Sinharoy, S., Paul, A., Bhattacharya, A., Saha, S., et al. (2013). Autophosphorylation of gatekeeper tyrosine by symbiosis receptor kinase. FEBS Lett. 587, 2972–2979. doi: 10.1016/j.febslet.2013.07.050
Shiu, S.-H., and Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. doi: 10.1038/nature00841
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Urbanski, D. F., Małolepszy, A., Stougaard, J., and Andersen, S. U. (2012). Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. Plant J. 69, 731–741. doi: 10.1111/j.1365-313X.2011.04827.x
Wang, C., Yu, H., Luo, L., Duan, L., Cai, L., He, X., et al. (2016). NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol. 212, 176–191. doi: 10.1111/nph.14017
Wang, C., Zhu, H., Jin, L., Chen, T., Wang, L., Kang, H., et al. (2013). Splice variants of the SIP1 transcripts play a role in nodule organogenesis in Lotus japonicus. Plant Mol. Biol. 82, 97–111. doi: 10.1007/s11103-013-0042-3
Yoro, E., Suzaki, T., Toyokura, K., Miyazawa, H., Fukaki, H., and Kawaguchi, M. (2014). A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 165, 747–758. doi: 10.1104/pp.113.233379
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Keywords: Lotus japonicus, root nodule symbiosis, symbiosis receptor kinase, symbiotic signaling, domain swap, nitrogenase activity
Citation: Li H, Chen M, Duan L, Zhang T, Cao Y and Zhang Z (2018) Domain Swap Approach Reveals the Critical Roles of Different Domains of SYMRK in Root Nodule Symbiosis in Lotus japonicus. Front. Plant Sci. 9:697. doi: 10.3389/fpls.2018.00697
Received: 01 February 2018; Accepted: 07 May 2018;
Published: 05 June 2018.
Edited by:
Jeanne Marie Harris, University of Vermont, United StatesReviewed by:
Yusuke Saijo, Nara Institute of Science and Technology (NAIST), JapanFang Xie, Shanghai Institutes for Biological Sciences (CAS), China
Copyright © 2018 Li, Chen, Duan, Zhang, Cao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongming Zhang, em16aGFuZ0BtYWlsLmh6YXUuZWR1LmNu
 Hao Li
Hao Li Mengxiao Chen
Mengxiao Chen Liujian Duan
Liujian Duan Yangrong Cao
Yangrong Cao Zhongming Zhang
Zhongming Zhang