- 1Departamento de Genética Molecular de Plantas, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas, Universidad Autónoma de Madrid, Madrid, Spain
- 2Agricultural Biotechnology Research Center, Academia Sinica, Taipei, Taiwan
Almost half of known plant viral species rely on proteolytic cleavages as key co- and post-translational modifications throughout their infection cycle. Most of these viruses encode their own endopeptidases, proteases with high substrate specificity that internally cleave large polyprotein precursors for the release of functional sub-units. Processing of the polyprotein, however, is not an all-or-nothing process in which endopeptidases act as simple peptide cutters. On the contrary, spatial-temporal modulation of these polyprotein cleavage events is crucial for a successful viral infection. In this way, the processing of the polyprotein coordinates viral replication, assembly and movement, and has significant impact on pathogen fitness and virulence. In this mini-review, we give an overview of plant viral proteases emphasizing their importance during viral infections and the varied functionalities that result from their proteolytic activities.
Introduction
Viruses are the most abundant biological entities in the planet (Suttle, 2007). With the exception of giant viruses (Wilhelm et al., 2017), viruses share a reduced genome size and optimize a confined genetic space utilizing several strategies of alternative protein production (Firth and Brierley, 2012; Miras et al., 2017). One of these strategies commonly employed by viruses is to produce polyproteins that are further processed by proteases into smaller working units. This strategy ensures production of multiple components required for viral infection in a single molecule and at the same time saves space in the genome by using a single set of transcriptional and translational control elements. It also provides the option to yield partially processed protein products with specific activities, and to alter functionality of a particular protein in a controlled manner (Spall et al., 1997; Konvalinka et al., 2015). However, gene expression through polyproteins relies on proteases for its proper functioning and as such, these enzymes play a central role regulating infectivity and the viral cycle.
Since the discovery of tobacco mosaic virus (TMV) in the late 19th century (Zaitlin, 1998), more than 4000 viral species have been assigned and classified in a total of 131 families (ICTV, 2017; Simmonds et al., 2017). Out of these, 27 families and 9 orphan genera include plant-infecting viruses. The largest family of plant viruses is the Geminiviridae, whose members carry a single stranded DNA genome. In eukaryotes, RNA viruses account, however, for the majority of the virome diversity (Koonin et al., 2015). The plant virome is dominated by viruses with positive-stranded RNA genomes, which can be further subdivided into superfamilies based on RNA-dependent RNA-polymerase (RdRp) phylogenetic relationships: Alphavirus-like, and Picornavirus-like (Goldbach et al., 1991; Koonin, 1991; Dolja and Koonin, 2011), Potyviridae being the largest representative family of the latter class (Ivanov et al., 2014). Among plant viruses there are also pararetroviruses and viruses with negative-stranded and double-stranded RNA genomes.
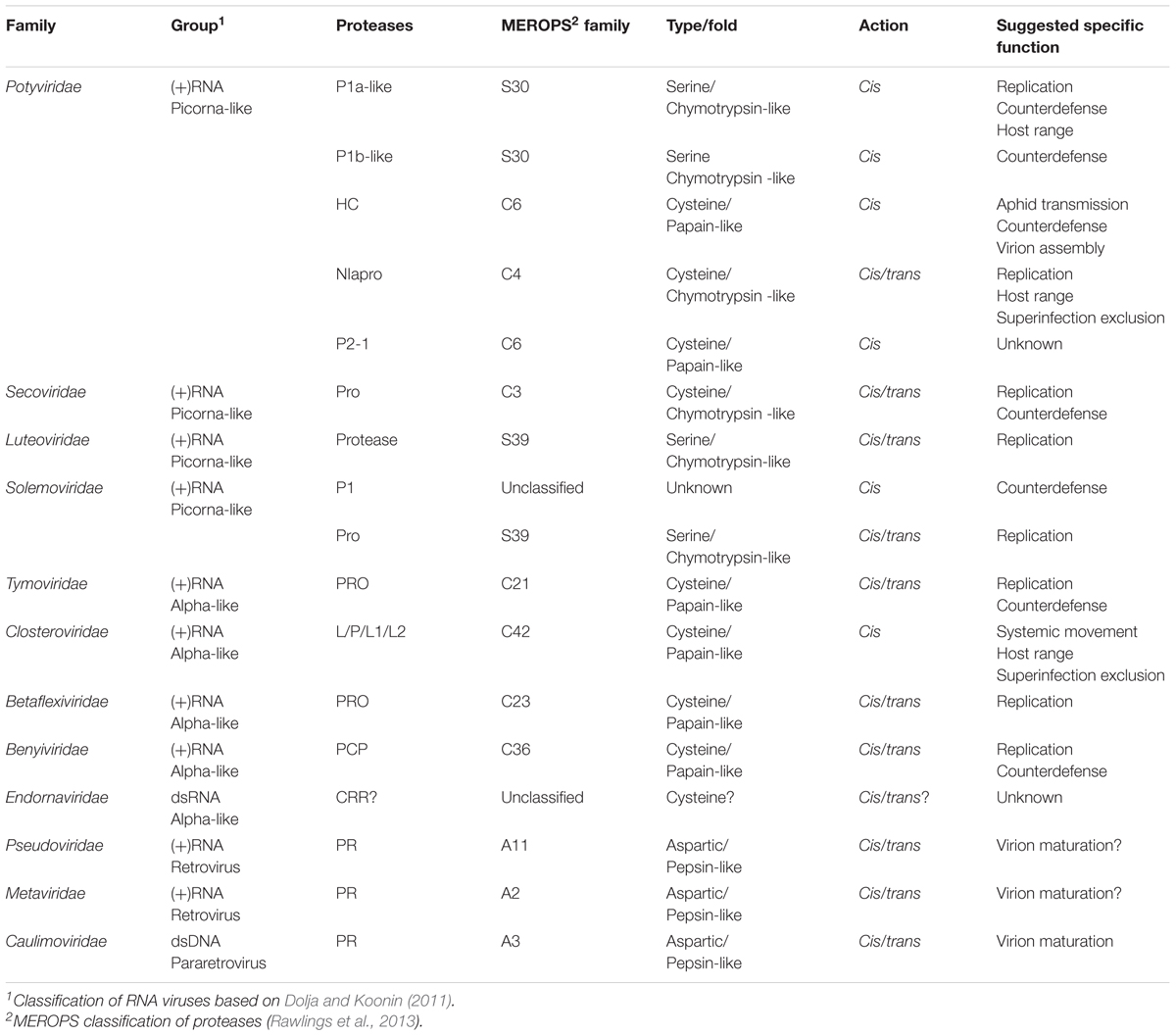
Synthesis of viral endopeptidases occurs in ∼45% of plant-infecting species (Figure 1), grouped into 12 families (Figure 2 and Table 1) (ICTV, 2017). These viruses encode three types of proteases: cysteine (67.2%), aspartic (9.0%), and serine (23.8%) proteases (Figure 1), which belong to 12 catalytic families (Table 1), according to the peptidase database MEROPS (Rawlings et al., 2013). Viral endopeptidases share certain features that make them distinct from host proteases: (i) they are smaller, (ii) they present little sequence similarity that might be restricted to active site residues, (iii) they can adapt to multiple roles, and (iv) they are very specific in their cutting requirements (Babe and Craik, 1997; Tong, 2002; Verdaguer et al., 2014). This stringent specificity of viral proteases makes them successful targets as biotechnological tools (Kim et al., 2012; Fernandez-Rodriguez and Voigt, 2016; Tran et al., 2017) and for antiviral therapies (Shamsi et al., 2016). Different drugs targeting proteases have been used effectively for treating animal viral infections (Anderson et al., 2009; Clark et al., 2013; Lv et al., 2015), and have also had moderate success in the plant world (García et al., 1993; Gutiérrez-Campos et al., 1999, 2001; Wen et al., 2004; Gholizadeh et al., 2005; Habib and Fazili, 2007; Kim et al., 2016).
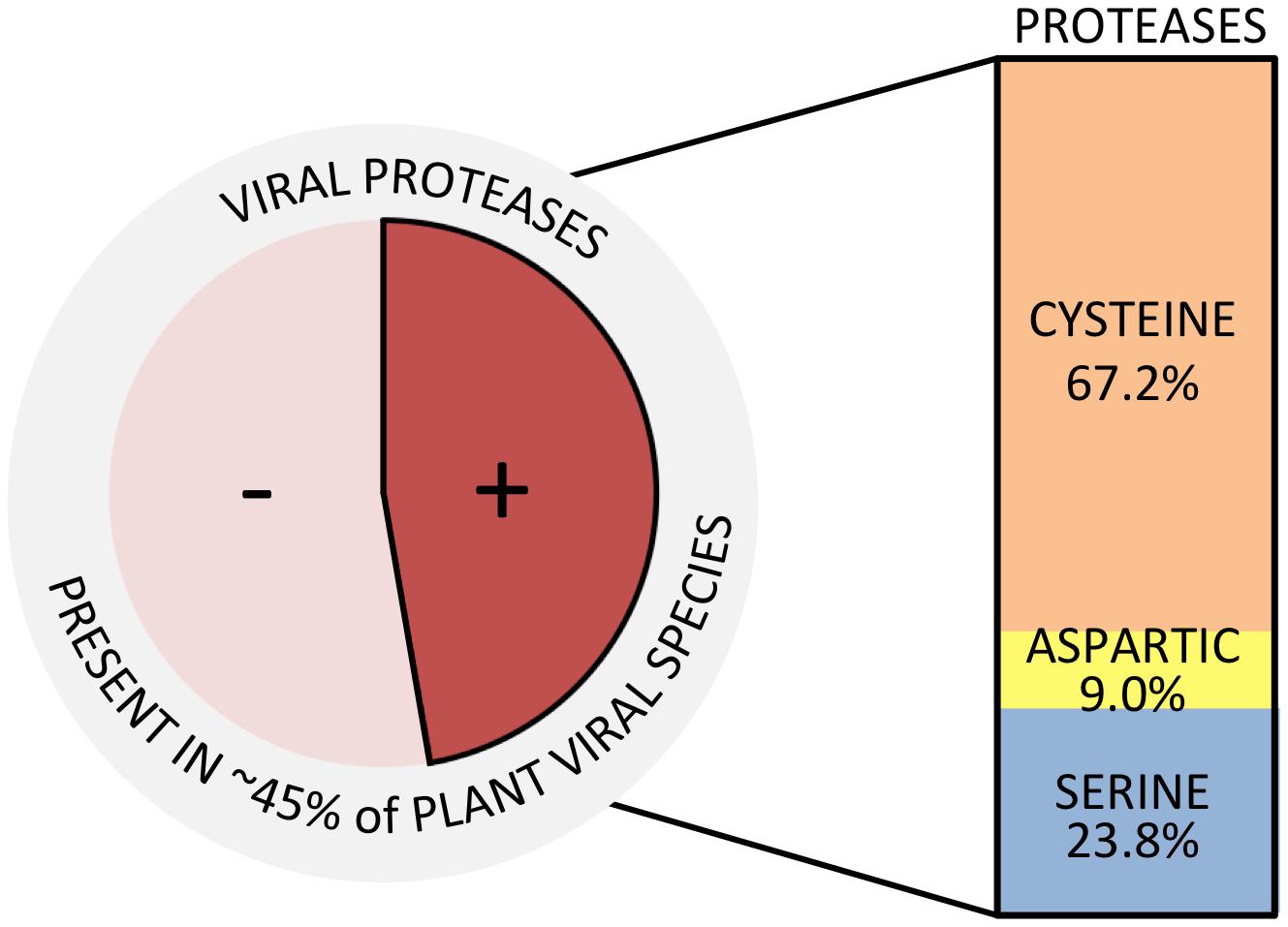
FIGURE 1. Plant viral species encoding proteases. Total number of plant viral species was based on the ICTV Master Species List (ICTV, 2017). Types of proteases and percentages were calculated counting the total number of proteases present in each of the accounted viral species.
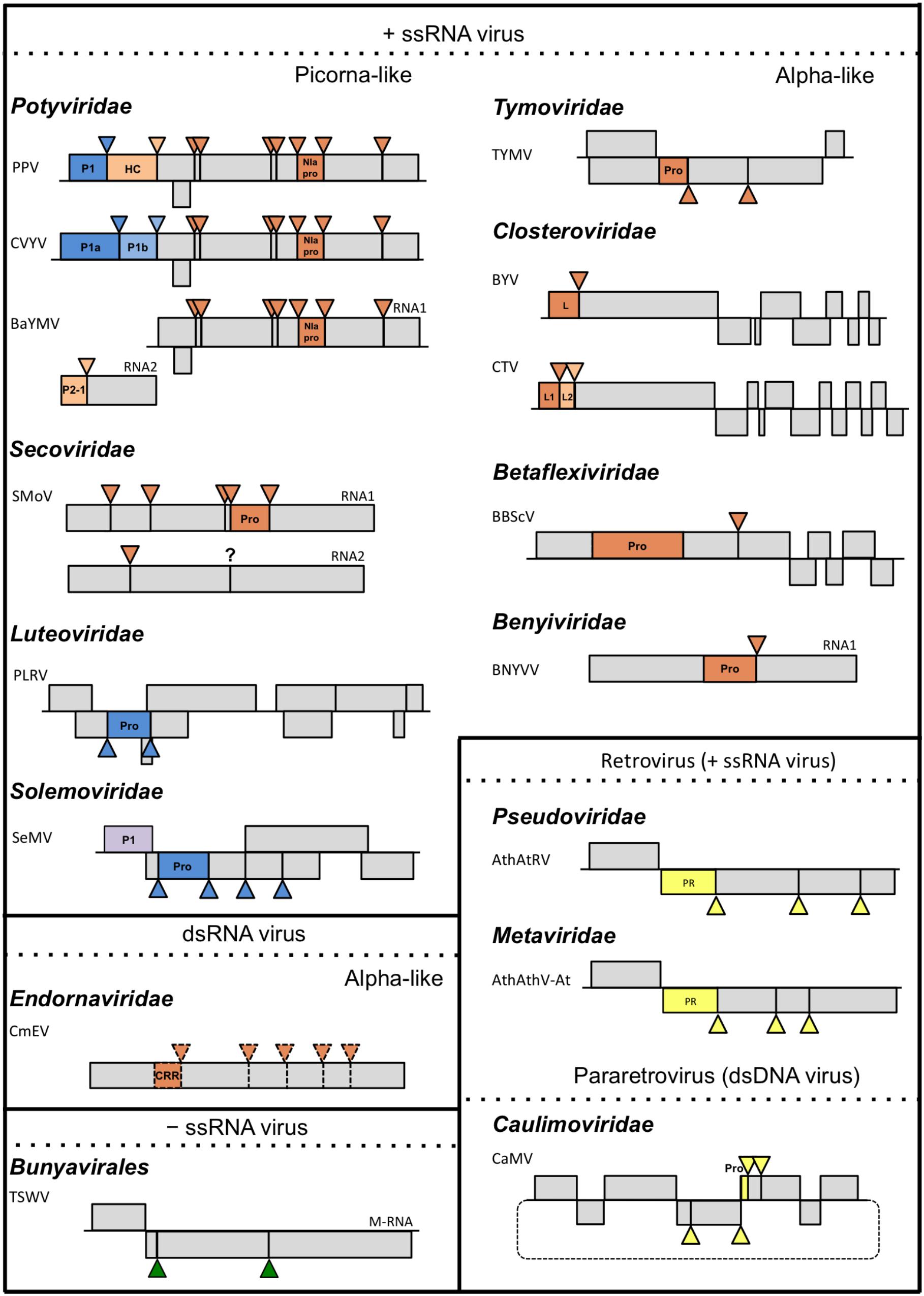
FIGURE 2. Schematic representation of plant viruses and their proteolytic cleavage sites. Triangles represent cleavage sites of endopeptidases. Colors of the triangles match the colors of the corresponding endopeptidases: orange for cysteine, blue for serine, yellow for aspartic, purple for unknown and green for plant proteases; only genomes, or sub-genomes encoding polyproteins subject to proteolytic cleavage are depicted. For each family, a representative species covering the different endopeptidases are depicted. TSWV is included as representative member of the order Bunyavirales. Scale of the genome map is maintained only within each viral species. Dotted lines used in the Endornaviridae family indicate that processing is only theoretical. Question mark indicates that the way of processing is unknown. PPV, Plum pox virus, Potyvirus; CVYV, Cucumber vein yellowing virus, Ipomovirus; BaYMV, Barley yellow mosaic virus, Bymovirus; SMoV, Strawberry mottle virus, unassigned; PLRV, Potato leafroll virus, Polerovirus; SeMV, Sesbania mosaic virus, Sobemovirus; TYMV, Turnip yellow mosaic virus, Tymovirus; BYV, Beet yellow virus, Closterovirus; CTV, Citrus tristeza virus, Closterovirus; BBScV, Blueberry scorch virus, Carlavirus; BNYVV, Beet necrotic yellow vein virus, Benyvirus; CeMV, Cucumis melo alphaendornavirus, Alphaendornavirus; TSWV, Tomato spotted wilt orthotospovirus, Orthotospovirus; AthAtRV, Arabidopsis thaliana AtRE1 virus, Pseudovirus; AthAthV-At, Arabidopsis thaliana Athila virus, Metavirus; CaMV, Cauliflower mosaic virus, Caulimovirus.
Plant viral proteases carry out multiple roles during viral infection independent of their protease activity: RNA silencing suppression, aphid transmission, systemic transport, viral accumulation, viral particle maturation, etc. (Liu et al., 2009; Csorba et al., 2015; Valli et al., 2018). As proteases, however, the primary role they play in viral infection is processing of and from viral polyproteins. But there is more to this protease activity than just acting as peptide cutters. Polyprotein processing is not an all-or-nothing process in which all products are separated at the same time with perfect efficiency. Cleavage of the polyprotein into functional units is essential for viral survival and it is a highly modulated process. Its regulation modifies the time and place of the final products as well as the possible accumulation of intermediate products, which can play distinct roles in the life cycle. In addition, processing of host proteins can also alter the function of these viral proteases. In general, plant viral proteases have been understudied when compared to their animal counterparts in terms of processing regulation and structure, probably due to the relevant role that the latter play in human health. Nonetheless, a large amount of information has been published about plant viral proteases in recent years leading us to write a review on the subject. This review will give an overview of different roles that lie behind the proteolytic activity of plant virus proteases and emphasize their relevance during viral infection.
Replication
Key to viral infection is genome replication. It takes place at specific sites in the cell, compartments termed viral factories in which multiple viral and plant factors required for replication are concentrated (Heinlein, 2015). Involvement of viral proteases in these factories has been demonstrated for some cases, but information is not always available. For animal viruses, the role that endopeptidases play in regulating replication is well-established (Sawicki and Sawicki, 1994; Racaniello, 2001; Vasiljeva et al., 2003; Yost and Marcotrigiano, 2013; Rausalu et al., 2016). Information is scarcer in the case of plant infecting viruses.
The Potyviridae is a family of positive-stranded RNA viruses that belongs to the picornavirus-like supergroup. It comprises 10 genera and presents the highest protease variety among plant viruses, coding in their genomes up to five different proteases with varied specificities [P1 (P1a- and P1b-like), HC, NIapro, P2-1 (HC-like)] (Adams et al., 2005a,b; Rodamilans et al., 2013; Revers and García, 2015). One hallmark of the picorna-like viruses, other than a conserved RdRp, is the presence of a 3C-like protease in charge of polyprotein processing. For the Potyviridae, this is NIapro. Indeed, this is the best characterized plant viral protease, functionally and structurally, which modulates replication by polyprotein processing (Carrington and Dougherty, 1987). NIapro is a chymotrypsin-like cysteine protease that acts in cis and in trans and it is involved in the generation of intermediate (such as P3-6K1, CI-6K2, and 6K2-NIa) and final products at different stages of infection. These products are implicated in the formation of the replication complex and its anchoring to, and release from, ER-derived membranes (Restrepo-Hartwig and Carrington, 1994; Riechmann et al., 1995; Schaad et al., 1997; Merits et al., 2002; Beauchemin et al., 2007; García et al., 2014; Cui and Wang, 2016).
The Secoviridae (Thompson et al., 2014), Luteoviridae (Prüfer et al., 1999; Li et al., 2000), and Solemoviridae families (Satheshkumar et al., 2004; Sõmera et al., 2015) belong to the picornavirus-like supergroup and share equivalent proteases. In the Secoviridae family, studies with the waikavirus rice tungro spherical virus (RTSV) (Thole and Hull, 1998), the nepovirus tomato ringspot virus (TomRSV) (Wang et al., 1999; Wang and Sanfaçon, 2000) and strawberry mottle virus (SMoV) (Mann et al., 2017), have characterized the viral protease (Pro) and their cleavage sites, but so far little is known about the specific involvement of these proteases in viral replication. The same is true for the Luteoviridae family. The serine protease encoded by the ORF1 of the polerovirus potato leafroll virus (PLRV) is able to act in cis and in trans and to separate the membrane anchoring portion, the protease and the genome-linked viral protein (VPg) domains; whether this is part of a regulatory mechanism for viral replication is still unknown (Li et al., 2007). Viruses of the Solemoviridae family express two versions of a polyprotein, from ORF2a and ORF2b, having different C terminus. The N-terminal common part includes a membrane anchor domain, the protease Pro, and VPg. Polyprotein 2a (P2a) C-terminal part codes for P10 and P8 proteins. Polyprotein 2ab (P2ab) codes for RdRp and is originated by ribosomal frameshift. Studies with sesbania mosaic virus (SeMV) indicate that the serine protease performs differently in P2a and in P2ab (Nair and Savithri, 2010a,b). In the first case, processing occurs at the predicted sites separating all components from the polyprotein. However, in the latter case, processing of VPg from RdRp is not fulfilled even though the protease and cleavage sequence are conserved in P2a and P2ab. This points to a regulatory process in protease activity that might have an influence in replication considering the inhibitory effect observed in vitro that VPg has over the polymerase when present at its N terminus. In addition, mutational analysis of cleavage sites indicated that all sites at P2a/P2ab are essential for viral replication, and the products are only functional when released at the site of replication (Govind et al., 2012) reinforcing the modulatory role of the protease.
Another example of plant viral protease involved in replication comes from the Tymoviridae family that belongs to the alphavirus-like supergroup. Turnip yellow mosaic virus (TYMV) encodes a papain-like cysteine protease, termed PRO (Rozanov et al., 1995; Lombardi et al., 2013). Involvement of PRO in replication comes from two different lines of evidence: (i) the processing ability of the protease to act in cis and in trans similarly to the proteases of rubiviruses and alphaviruses, which share a similar polyprotein structure (Jakubiec et al., 2004, 2007) and (ii) its deubiquitination activity (Camborde et al., 2010; Chenon et al., 2012). TYMV is the type member of the genus Tymovirus, a single positive-stranded RNA spherical virus that produces two overlapping ORFs from a single RNA. One of them encodes a polyprotein of 206 KDa that contains sequence domains of methyltransferase (MET), PRO, helicase (HEL) and RdRp. PRO was shown not only to separate RdRp from the rest of the polyprotein, but also process HEL in a secondary event. This and the ability of PRO to act in trans appear to reflect the evolutionary relationship of this virus to rubiviruses and alphaviruses, and as it occurs in these animal viruses, it is likely that temporal regulation of polyprotein processing controls the synthesis of different RNA species (negative- and positive-strands). Whether the specific cleavage observed in TYMV also shuts off the synthesis of negative-strand RNA is still unknown (Jakubiec et al., 2007). In addition to this, TYMV PRO is a functional ovarian tumor-like deubiquitylating enzyme (DUB) and this activity helps PRO to modulate viral replication by stabilizing the viral polymerase preventing degradation by the ubiquitin-proteosome system (Camborde et al., 2010; Chenon et al., 2012; Bailey-Elkin et al., 2014; Jupin et al., 2017).
The alphavirus-like supergroup does not maintain a conserved protease in all members as the picornavirus-like does. In this way, the Closteroviridae family, although sharing in ORF1a the MET, HEL organization followed by RdRp in ORF1b, does not encode a protease that acts in trans to process these products, but contains a leader proteinase(s) with autocatalytic activity (Dolja et al., 2006; Agranovsky, 2016). On the other hand, some members of the Betaflexiviridae family, do encode in ORF1 similar MET, PRO, HEL, RdRp domains as members of the Tymoviridae family do, although there is little information regarding polyprotein processing and no data regarding involvement of PRO in replication (Foster and Mills, 1992; Lawrence et al., 1995). A similar lack of information is encountered in the Benyiviridae family. Its most studied member, beet necrotic yellow vein virus (BNYVV), encodes a papain-like cysteine protease domain (PCP) (Hehn et al., 1997), and it has been hypothesized that it might act as a DUB to favor RdRp transcription (Pakdel et al., 2015), similar to the mode of action of the PCP domain of hepatitis E virus (HEV), although in the latter case, PCP acted as a DUB to counteract cellular antiviral pathways (Karpe and Lole, 2011).
But not only trans-acting proteases are involved in the regulation of replication. Recently, the leader protease P1 of the Potyvirus genus has also been assigned to this role. Work performed with plum pox virus (PPV) P1 showed that the N-terminal part of this cis-acting serine proteinase, the most variable region, acts as a negative regulator of P1 self-processing, modulating in this way potyviral replication (Pasin et al., 2014). Removal of the N-terminal part of P1, not only makes the protein co-factor independent, but also potentiates viral replication at early times of infection emphasizing the regulatory role of this protein in the potyviral life cycle. The way PPV P1 is modulating replication through host factor interactions resembles the mode of action of the NS2 protease of animal virus bovine viral diarrhea virus (BVDV) (Lackner et al., 2004, 2006). In this pestivirus, the NS2 protease modulates replication indirectly by downregulating NS2-NS3 processing. Similarly, PPV P1 modulates P1HC processing and indirectly affects viral replication.
Viral Counterdefense
Sometimes, when a protease potentiates a positive effect on replication it is not due to a specific role in this viral process, but it is the consequence of an indirect effect caused by an enhanced ability of the virus to escape plant defenses. Thus, proteases could be considered as having a counterdefense role instead of a role in viral replication. For example, PPV P1 was described as having a modulatory role in replication, but this is likely derived from the modulation of the RNA silencing suppressor HC. It can be considered that P1 is actually modulating host defense responses and that the effect observed in viral replication is just a by-product of this role. The same can be argued in the case of TYMV PRO activity as DUB, which can be viewed not in terms of modulating replication, but if RdRp degradation is considered as part of plant defense, it can be viewed as a counterdefense mechanism (Camborde et al., 2010; Chenon et al., 2012; Lombardi et al., 2013; Jupin et al., 2017).
Using DUBs as a means of protection against host defenses is something well-established in the animal viral world. Examples can be found among viruses of the order Nidovirales such as the coronavirus severe acute respiratory syndrome-related coronavirus (SARS-CoV) or the arterivirus equine arteritis virus (EAV) that use this strategy of interfering with the innate immune signaling pathway through the DUB activity of their cysteine proteases (Clementz et al., 2010; van Kasteren et al., 2013). The same is true for viruses of the order Picornavirales such as the aphtovirus foot-and-mouth disease virus (FMDV) and its Lpro leader protease (Wang et al., 2011a,b). In all these cases, however, although the counterdefense activity is well-documented, it appears that the DUB and the protease activity are not strictly interrelated. In the case of TYMV, these two activities can be uncoupled by mutations that selectively suppress the DUB activity without altering PRO (Jupin et al., 2017).
Probably, the best characterized proteases acting as viral counterdefense barriers by degrading host proteins are the ones from the Picornaviridae family (Agol and Gmyl, 2010). Thus, FMDV Lpro not only disrupts the interferon signaling pathway through its deubiquitinase activity but also cleaves eIF4G shutting off host cap-dependent translation and downregulating Type I interferons (Guarné et al., 1998; Chase and Semler, 2012; Liu et al., 2015). Moreover, FMDV produces, as the rest of the members of the Picornaviridae family, 3Cpro, a protease that is in charge of processing the different elements of the polyprotein acting in cis and in trans, and also degrades several host proteins in order to potentiate viral transcription and translation (Sun et al., 2016). In the same family, rhinoviruses and enteroviruses produce another protease termed 2Apro, which also develops these degrading functions (Seipelt et al., 1999; Chase and Semler, 2012).
Taking these activities into account it is reasonable to ask the question of whether the 3C-like proteases of plant picorna-like viruses perform similar host degrading activities to counteract plant defenses or not. In the case of NIapro, the 3C-like protease of the Potyviridae, no specific host proteins affected by its catalytic activity have been described and, only recently, a study was published describing possible interacting partners in plants (Martínez et al., 2016). However, it cannot be ruled out that NIapro might be processing more proteins than the viral ones taking into consideration its demonstrated ability to act on proteins with an engineered target sequence (Rohila et al., 2004; Cesaratto et al., 2016) or even on proteins with a naturally occurring target cleavage site, such as the amyloid-β peptide (Han et al., 2010; Kim et al., 2012). Likewise, NIapro from potato virus Y (PVY) acts as elicitor of the hypersensitive response mediated by the gene Ry in potato, and its protease activity, likely acting on a host factor, appears to be involved in this eliciting response (Mestre et al., 2000, 2003). More recent studies have described a role of potyviral NIapro in enhancing aphid transmission and suggested that this role might be related to its ability to degrade vacuolar defense proteins (Casteel et al., 2014; Bak et al., 2017).
Some newly published reports add more information to the scarce available data about activities of 3C-like proteases related with defense and counterdefense responses. The RNA silencing suppressor R78 of the waikavirus maize chlorotic dwarf virus (MCDV) is cleaved by Pro, raising the possibility that this cleavage might have some influence in R78 silencing suppression activity over the course of the infection (Stewart et al., 2017). Moreover, NIapro of the tritimovirus wheat streak mosaic virus (WSMV) contributes to prevent superinfection by related viruses, and it has been suggested that the protease activity of this protein is required for superinfection exclusion (Tatineni and French, 2016).
Virion Maturation
A good example of a viral protease directly involved in virion formation is togavirin from viruses of the genus Alphavirus. Structurally related to chymotrypsin-like serine proteases, togavirin is the actual core protein. It self-processes from the polyprotein precursor, binds viral RNA, and assembles into the capsid (Krupovic and Koonin, 2017). Apart from this versatile endopeptidase, the role of proteases in virion maturation has been well-studied for animal retroviruses such as human immunodeficiency virus (HIV), Rous sarcoma virus (RSV) or murine leukemia virus (MLV), amongst others (Konvalinka et al., 2015). In these viruses, cleavage of viral polyproteins at specific sites and in an orderly fashion is crucial for transforming the immature shell into an active infectious particle. Pseudoviridae and Metaviridae are two viral families that include plant retroviruses (Peterson-Burch and Voytas, 2002; Wright and Voytas, 2002; Eickbush and Jamburuthugoda, 2008), but there is not much information regarding the regulation of proteolytic processing. More data is available about the Caulimoviridae, the single family of plant pararetroviruses (Torruella et al., 1989). The genome of all replication-competent retroviruses consists of structural, replication and envelope proteins (gag, pol, and env) (Marmey et al., 2005). The protease (PR), an aspartate peptidase with no homology to other viral proteases, is generally included in the pol domain. Viruses of the Caulimoviridae family, the only plant viruses with dsDNA genomes, encode the gag-pol core, but unlike retroviruses, lack an integrase, which is not required because the caulimoviral DNA is not integrated in the host chromosome. The type virus of the family is cauliflower mosaic virus (CaMV), a member of the Caulimovirus genus. The capsid protein (CP) of this virus is produced as a precursor (pre-CP) with N- and C-terminal extensions. CP is involved in virion assembly, packaging of viral RNA and delivery of the genome to the nucleus. Processing of the CP extensions is thought to regulate these functions. The N-terminal extension of CP appears to be involved in keeping the pre-CP in the cytoplasm and may operate as an anchoring domain for the initiation of viral assembly, similar to what occurs to HIV viral matrix protein (Champagne et al., 2004). Virion maturation is completed by removal of the first 76 aa and about 40 aa from the C terminus by the viral aspartic proteinase (Karsies et al., 2002; Champagne et al., 2004). The fact that pre-CP is excluded from the nucleus, would assure that only mature virions, containing the genomic DNA, enter in the nucleus (Karsies et al., 2002). Studies done with another plant pararetrovirus, the badnavirus rice tungro bacilliform virus (RTBV), showed that its aspartic protease cuts independently of plant-specific host factors since it retained its proteolytic activity in baculovirus (Laco et al., 1995) and bacteria (Marmey et al., 2005). In the case of animal retroviruses, PR is expressed in an inactive monomeric form and needs to dimerize to acquire an active conformation in which each unit contributes an aspartate to the active site. Proper redox environment is likely to also play a role in PR activation (Ingr et al., 2003; Konvalinka et al., 2015). Based on active site comparison, it is anticipated that PR of Caulimoviridae also acts as dimers (Torruella et al., 1989). Its activation requirements are still pending further investigation.
Host Range Definition
Plant viruses have definite host ranges, which in some cases are very narrow. The complex network of interactions between plant and virus that needs to be established in order for the infection to progress makes it difficult for the virus to have broad host spectrum. In terms of viral proteases, the best examples of host range modulation come from the Potyviridae family (Adams et al., 2005a,b; Revers and García, 2015). Potyviruses, rymoviruses, and some ipomoviruses have P1a-like leader serine proteases whose cleavage is essential for virus infectivity (Verchot and Carrington, 1995). These proteases rely on a plant factor(s) to develop their proteolytic activity, a feature that separates them from P1b-like serine proteases in the family and whose cleavage is co-factor independent (Verchot et al., 1992; Valli et al., 2006; Rodamilans et al., 2013). The comparison of two PPV isolates, which differed in their reciprocal capacity of infecting woody and herbaceous hosts, showed the relevance of P1 among other viral proteins for host adaptation (Salvador et al., 2008a). Similarly, analyses of PPV chimeras including P1 sequences of tobacco vein mottling virus (TVMV) and of virus variants with different biological properties sorted from a single PPV isolate also pointed toward the involvement of P1 in host range definition (Salvador et al., 2008b; Maliogka et al., 2012). All these works show how relevant P1 is in terms of host spectrum characterization, but do not necessarily implicate the protease activity of P1 in this role. More direct evidence of the involvement of P1-mediated proteolytic processing in compatibility with the host comes from works performed with P1a of cucumber vein yellowing virus (CVYV) and P1 of PPV, both P1a-like proteases. In these studies, it was shown that one of the factors limiting PPV infection in Cucumis sativus was likely the incompatibility of PPV P1 with a host co-factor required for its protease activity. Either replacing P1 with P1a, supposedly compatible with a cucumber co-factor, or with a host factor-independent P1 mutant, provided PPV the ability to partially break the non-host resistance of cucumber (Carbonell et al., 2012; Shan et al., 2015, 2017).
From the same Potyviridae family, NIapro has also been described to play a role in host range determination. In the papaya ringspot virus (PRSV), a single amino acid substitution in this chymotrypsin-like protease allows a host-shift from cucurbits to papaya, although the specific involvement of the protease activity of NIapro in this effect is only a possibility (Chen et al., 2008). More direct evidence of the involvement of the protease activity of NIapro in host range determination comes from work performed with PPV (Calvo et al., 2014). This study showed that alternative adaptation to Nicotiana and Prunus hosts was determined, not by peculiarities of the NIapro sequence, but by differences in the NIapro target sequence placed between 6K1 and CI, suggesting modulation of NIapro processing at this site in a host-specific manner.
Proteolytic Activity-Unrelated Functions
The small size of the genome of plant RNA viruses forces the proteins from these viruses to acquire multiple functions. This is best exemplified by the potyviral protein HC (Valli et al., 2018). HC is a cysteine proteinase whose first identified function was to aid in aphid transmission of viral particles (de Mejia et al., 1985). However, the main function of the potyviral HC appears to be suppressing antiviral RNA silencing (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998), and an independent function of HC in the correct assembly of potyviral virions has been more recently reported (Valli et al., 2014). Interestingly, all these HC functions do not rely on its proteolytic activity, as it is also the case for the RNA silencing suppression activity of the serine proteinase P1b of the ipomovirus CVYV (Valli et al., 2008), illustrating how proteolysis-related and -unrelated roles can concur in a single viral protein. Probably also unrelated to its protease activity is the role suggested for P1 of tobacco etch virus (TEV) in stimulating viral RNA translation (Martínez and Daròs, 2014).
Viral proteinases with functions that appear unrelated to their proteolytic activity are not restricted to the family Potyviridae. The self-cleaving leader proteinases of viruses of the Closteroviridae family are a good example of this. These proteinases are involved in virus accumulation, systemic transport, host range expansion or virus superinfection exclusion, but all these roles appeared to be independent of their protease activities (Peng et al., 2001, 2003; Liu et al., 2009; Atallah et al., 2016). Contrary to what was observed for the leader proteinase of FMDV, the closterovirus proteases show no DUB activity and have not been described to be involved in further processing of host or viral proteins.
Concluding Remarks
It is well-established that viral proteases are not just proteolytic machines acting without proper modulation of time and/or space. Much effort has been put into defining what these extra roles are and characterizing the different mechanisms of action and their peculiarities. Involved in regulating replication, virion maturation, host range determination or even displaying a more active role as viral counterdefense barriers, proteases, when present, are essential in practically all aspects of the viral cycle. However, there are still many proteinases from plant viruses for which information about the integration of its enzymatic activity in the infection process is still unavailable. Viruses of the family Endornaviridae are a fine example. These viruses have been understudied probably because they do not usually cause any noticeable damage on their hosts. They have a monocistronic RNA genome that encodes a large polyprotein, but there are only hints about how this polyprotein is processed (Roossinck et al., 2011; Sabanadzovic et al., 2016). The case of P1 of the sobemovirus rice yellow mottle virus (RYMV) is another good example of a viral protease with a puzzling role (Weinheimer et al., 2010). RYMV P1, a protein with RNA silencing suppression activity, is expressed as a mature protein, rather than as part of a protein precursor; however, in experimental conditions it displays self-cleaving activity able to precisely remove engineered C-terminal extensions. Maintaining a function that seems to be superfluous raises the possibility that this protease, and by similarity other leader proteases, might have an extra unknown biological function.
We have focused this short review on the roles of virus-encoded proteinases in viral infection. However, control of gene expression by proteolytic processing of protein precursors not only relies on viral proteinases. For instance, host aspartyl proteases are in charge of the processing of the primary product of the M genomic RNA of plant viruses of the order Bunyavirales to yield two mature glycoproteins (Whitfield et al., 2005; Li et al., 2015; Shi et al., 2016). The involvement of cellular proteases in modulating plant virus infection is another exciting target for future research.
Author Contributions
BR drafted the manuscript with the collaboration of JAG. HS and FP contributed to the conception and design of the review and revised the manuscript. All the authors read, critiqued, and approved the final manuscript.
Funding
HS was supported by the China Scholarship Council; FP was supported by a post-doctoral fellowship from Academia Sinica. This work was funded by Grant No. BIO2016-80572-R and the FEDER program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Patricia Elhazaz Walsh for editorial assistance. They acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI). They would like to apologize to those colleagues whose relevant publications have not been cited because of space constraints or involuntary forgetfulness.
References
Adams, M. J., Antoniw, J. F., and Beaudoin, F. (2005a). Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6, 471–487. doi: 10.1111/j.1364-3703.2005.00296.x
Adams, M. J., Antoniw, J. F., and Fauquet, C. M. (2005b). Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. doi: 10.1007/s00705-004-0440-6
Agol, V. I., and Gmyl, A. P. (2010). Viral security proteins: counteracting host defences. Nat. Rev. Microbiol. 8, 867–878. doi: 10.1038/nrmicro2452
Agranovsky, A. A. (2016). “Closteroviruses: molecular biology, evolution and interactions with cells,” in Plant Viruses: Evolution and Management, eds R. K. Gaur, N. M. Petrov, B. L. Patil, and M. I. Stoyanova (Singapore: Springer).
Anandalakshmi, R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C., Smith, T. H., et al. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. U.S.A. 95, 13079–13084. doi: 10.1073/pnas.95.22.13079
Anderson, J., Schiffer, C., Lee, S., and Swanstrom, R. (2009). “Viral protease inhibitors,” in Antiviral Strategies, Handbook of Experimental Pharmacology, eds H. G. Kräusslich and R. Bartenschlager (Berlin: Springer), 85–110.
Atallah, O. O., Kang, S. H., El-Mohtar, C. A., Shilts, T., Bergua, M., and Folimonova, S. Y. (2016). A 5′-proximal region of the Citrus tristeza virus genome encoding two leader proteases is involved in virus superinfection exclusion. Virology 489, 108–115. doi: 10.1016/j.virol.2015.12.008
Babe, L. M., and Craik, C. S. (1997). Viral proteases: evolution of diverse structural motifs to optimize function. Cell 91, 427–430. doi: 10.1016/S0092-8674(00)80426-2
Bailey-Elkin, B. A., van Kasteren, P. B., Snijder, E. J., Kikkert, M., and Mark, B. L. (2014). Viral OTU deubiquitinases: a structural and functional comparison. PLoS Pathog. 10:e1003894. doi: 10.1371/journal.ppat.1003894
Bak, A., Cheung, A. L., Yang, C., Whitham, S. A., and Casteel, C. L. (2017). A viral protease relocalizes in the presence of the vector to promote vector performance. Nat. Commun. 8:14493. doi: 10.1038/ncomms14493
Beauchemin, C., Boutet, N., and Laliberté, J. F. (2007). Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782. doi: 10.1128/JVI.01277-06
Calvo, M., Malinowski, T., and García, J. A. (2014). Single amino acid changes in the 6K1-CI region can promote the alternative adaptation of Prunus- and Nicotiana-propagated Plum pox virus C isolates to either host. Mol. Plant Microbe Interact. 27, 136–149. doi: 10.1094/MPMI-08-13-0242-R
Camborde, L., Planchais, S., Tournier, V., Jakubiec, A., Drugeon, G., Lacassagne, E., et al. (2010). The ubiquitin-proteasome system regulates the accumulation of Turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 22, 3142–3152. doi: 10.1105/tpc.109.072090
Carbonell, A., Dujovny, G., García, J. A., and Valli, A. (2012). The Cucumber vein yellowing virus silencing suppressor P1b can functionally replace HCPro in Plum pox virus infection in a host-specific manner. Mol. Plant Microbe Interact. 25, 151–164. doi: 10.1094/MPMI-08-11-0216
Carrington, J. C., and Dougherty, W. G. (1987). Small nuclear inclusion protein encoded by a plant potyvirus genome is a protease. J. Virol. 61, 2540–2548.
Casteel, C. L., Yang, C., Nanduri, A. C., De Jong, H. N., Whitham, S. A., and Jander, G. (2014). The NIa-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid). Plant J. 77, 653–663. doi: 10.1111/tpj.12417
Cesaratto, F., Burrone, O. R., and Petris, G. (2016). Tobacco etch virus protease: a shortcut across biotechnologies. J. Biotechnol. 231, 239–249. doi: 10.1016/j.jbiotec.2016.06.012
Champagne, J., Benhamou, N., and Leclerc, D. (2004). Localization of the N-terminal domain of cauliflower mosaic virus coat protein precursor. Virology 324, 257–262. doi: 10.1016/j.virol.2004.04.014
Chase, A. J., and Semler, B. L. (2012). Viral subversion of host functions for picornavirus translation and RNA replication. Future Virol. 7, 179–191. doi: 10.2217/fvl.12.2
Chen, K. C., Chiang, C. H., Raja, J. A., Liu, F. L., Tai, C. H., and Yeh, S. D. (2008). A single amino acid of NIapro of Papaya ringspot virus determines host specificity for infection of papaya. Mol. Plant Microbe Interact. 21, 1046–1057. doi: 10.1094/MPMI-21-8-1046
Chenon, M., Camborde, L., Cheminant, S., and Jupin, I. (2012). A viral deubiquitylating enzyme targets viral RNA-dependent RNA polymerase and affects viral infectivity. EMBO J. 31, 741–753. doi: 10.1038/emboj.2011.424
Clark, V. C., Peter, J. A., and Nelson, D. R. (2013). New therapeutic strategies in HCV: second-generation protease inhibitors. Liver Int. 33(Suppl. 1), 80–84. doi: 10.1111/liv.12061
Clementz, M. A., Chen, Z., Banach, B. S., Wang, Y., Sun, L., Ratia, K., et al. (2010). Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84, 4619–4629. doi: 10.1128/JVI.02406-09
Csorba, T., Kontra, L., and Burgyan, J. (2015). Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480, 85–103. doi: 10.1016/j.virol.2015.02.028
Cui, H., and Wang, A. (2016). Plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early stage of infection. J. Virol. 90, 5119–5131. doi: 10.1128/JVI.00024-16
de Mejia, M. V., Hiebert, E., Purcifull, D. E., Thornbury, D. W., and Pirone, T. P. (1985). Identification of potyviral amorphous inclusion protein as a nonstructural, virus-specific protein related to helper component. Virology 142, 34–43. doi: 10.1016/0042-6822(85)90420-9
Dolja, V. V., and Koonin, E. V. (2011). Common origins and host-dependent diversity of plant and animal viromes. Curr. Opin. Virol. 1, 322–331. doi: 10.1016/j.coviro.2011.09.007
Dolja, V. V., Kreuze, J. F., and Valkonen, J. P. (2006). Comparative and functional genomics of closteroviruses. Virus Res. 117, 38–51. doi: 10.1016/j.virusres.2006.02.002
Eickbush, T. H., and Jamburuthugoda, V. K. (2008). The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 134, 221–234. doi: 10.1016/j.virusres.2007.12.010
Fernandez-Rodriguez, J., and Voigt, C. A. (2016). Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 44, 6493–6502. doi: 10.1093/nar/gkw537
Firth, A. E., and Brierley, I. (2012). Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409. doi: 10.1099/vir.0.042499-0
Foster, G. D., and Mills, P. R. (1992). Translation of potato virus S RNA in vitro: evidence of protein processing. Virus Genes 6, 47–52. doi: 10.1007/BF01703756
García, J. A., Cervera, M. T., Riechmann, J. L., and López-Otín, C. (1993). Inhibitory effects of human cystatin C on plum pox potyvirus proteases. Plant Mol. Biol. 22, 697–701. doi: 10.1007/BF00047410
García, J. A., Glasa, M., Cambra, M., and Candresse, T. (2014). Plum pox virus and sharka: a model potyvirus and a major disease. Mol. Plant Pathol. 15, 226–241. doi: 10.1111/mpp.12083
Gholizadeh, A., Santha, I. M., Kohnehrouz, B. B., Lodha, M. L., and Kapoor, H. C. (2005). Cystatins may confer viral resistance in plants by inhibition of a virus-induced cell death phenomenon in which cysteine proteinases are active: cloning and molecular characterization of a cDNA encoding cysteine-proteinase inhibitor (celostatin) from Celosia cristata (crested cock’s comb). Biotechnol. Appl. Biochem. 42, 197–204. doi: 10.1042/BA20050029
Goldbach, R., Le Gall, O., and Wellink, J. (1991). Alpha-like viruses in plants. Semin. Virol. 2, 19–25.
Govind, K., Mäkinen, K., and Savithri, H. S. (2012). Sesbania mosaic virus (SeMV) infectious clone: possible mechanism of 3′ and 5′ end repair and role of polyprotein processing in viral replication. PLoS One 7:e31190. doi: 10.1371/journal.pone.0031190
Guarné, A., Tormo, J., Kirchweger, R., Pfistermueller, D., Fita, I., and Skern, T. (1998). Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17, 7469–7479. doi: 10.1093/emboj/17.24.7469
Gutiérrez-Campos, R., Torres-Acosta, J. A., Pérez-Martínez, J. J., and Gómez-Lim, M. A. (2001). Pleiotropic effects in transgenic tobacco plants expressing the oryzacystatin I gene. Hortscience 36, 118–119.
Gutiérrez-Campos, R., Torres-Acosta, J. A., Saucedo-Arias, L. J., and Gomez-Lim, M. A. (1999). The use of cysteine proteinase inhibitors to engineer resistance against potyviruses in transgenic tobacco plants. Nat. Biotechnol. 17, 1223–1226.
Habib, H., and Fazili, K. M. (2007). Plant protease inhibitors: a defense strategy in plants. Biotechnol. Mol. Biol. Rev. 2, 68–85.
Han, H. E., Sellamuthu, S., Shin, B. H., Lee, Y. J., Song, S., Seo, J. S., et al. (2010). The nuclear inclusion a (NIa) protease of turnip mosaic virus (TuMV) cleaves amyloid-ß. PLoS One 5:e15645. doi: 10.1371/journal.pone.0015645
Hehn, A., Fritsch, C., Richards, K. E., Guilley, H., and Jonard, G. (1997). Evidence for in vitro and in vivo autocatalytic processing of the primary translation product of beet necrotic yellow vein virus RNA 1 by a papain-like proteinase. Arch. Virol. 142, 1051–1058. doi: 10.1007/s007050050141
Heinlein, M. (2015). Plant virus replication and movement. Virology 47, 657–671. doi: 10.1016/j.virol.2015.01.025
ICTV (2017). ICTV Master Species List for 2017. Available at: http://ictvonline.org/virusTaxonomy.asp [accessed April 5, 2018].
Ingr, M., Uhlikova, T., Strisovsky, K., Majerova, E., and Konvalinka, J. (2003). Kinetics of the dimerization of retroviral proteases: the “fireman’s grip” and dimerization. Protein Sci. 12, 2173–2182. doi: 10.1110/ps.03171903
Ivanov, K. I., Eskelin, K., Lõhmus, A., and Mäkinen, K. (2014). Molecular and cellular mechanisms underlying potyvirus infection. J. Gen. Virol. 95, 1415–1429. doi: 10.1099/vir.0.064220-0
Jakubiec, A., Drugeon, G., Camborde, L., and Jupin, I. (2007). Proteolytic processing of Turnip yellow mosaic virus replication proteins and functional impact on infectivity. J. Virol. 81, 11402–11412. doi: 10.1128/JVI.01428-07
Jakubiec, A., Notaise, J., Tournier, V., Hericourt, F., Block, M. A., Drugeon, G., et al. (2004). Assembly of Turnip yellow mosaic virus replication complexes: interaction between the proteinase and polymerase domains of the replication proteins. J. Virol. 78, 7945–7957. doi: 10.1128/JVI.78.15.7945-7957.2004
Jupin, I., Ayach, M., Jomat, L., Fieulaine, S., and Bressanelli, S. (2017). A mobile loop near the active site acts as a switch between the dual activities of a viral protease/deubiquitinase. PLoS Pathog. 13:e1006714. doi: 10.1371/journal.ppat.1006714
Karpe, Y. A., and Lole, K. S. (2011). Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J. Gen. Virol. 92, 2088–2092. doi: 10.1099/vir.0.033738-0
Karsies, A., Merkle, T., Szurek, B., Bonas, U., Hohn, T., and Leclerc, D. (2002). Regulated nuclear targeting of cauliflower mosaic virus. J. Gen. Virol. 83, 1783–1790. doi: 10.1099/0022-1317-83-7-1783
Kasschau, K. D., and Carrington, J. C. (1998). A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95, 461–470. doi: 10.1016/S0092-8674(00)81614-1
Kim, S. H., Qi, D., Ashfield, T., Helm, M., and Innes, R. W. (2016). Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351, 684–687. doi: 10.1126/science.aad3436
Kim, T. K., Han, H. E., Kim, H., Lee, J. E., Choi, D., Park, W. J., et al. (2012). Expression of the plant viral protease NIa in the brain of a mouse model of Alzheimer’s disease mitigates Aß pathology and improves cognitive function. Exp. Mol. Med. 44, 740–748. doi: 10.3858/emm.2012.44.12.082
Konvalinka, J., Krausslich, H. G., and Muller, B. (2015). Retroviral proteases and their roles in virion maturation. Virology 479–480, 403–417. doi: 10.1016/j.virol.2015.03.021
Koonin, E. V. (1991). The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72, 2197–2206. doi: 10.1099/0022-1317-72-9-2197
Koonin, E. V., Dolja, V. V., and Krupovic, M. (2015). Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479–480, 2–25. doi: 10.1016/j.virol.2015.02.039
Krupovic, M., and Koonin, E. V. (2017). Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U.S.A. 114, E2401–E2410. doi: 10.1073/pnas.1621061114
Lackner, T., Muller, A., Pankraz, A., Becher, P., Thiel, H. J., Gorbalenya, A. E., et al. (2004). Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J. Virol. 78, 10765–10775. doi: 10.1128/JVI.78.19.10765-10775.2004
Lackner, T., Thiel, H. J., and Tautz, N. (2006). Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc. Natl. Acad. Sci. U.S.A. 103, 1510–1515. doi: 10.1073/pnas.0508247103
Laco, G. S., Kent, S. B., and Beachy, R. N. (1995). Analysis of the proteolytic processing and activation of the rice tungro bacilliform virus reverse transcriptase. Virology 208, 207–214. doi: 10.1006/viro.1995.1144
Lawrence, D. M., Rozanov, M. N., and Hillman, B. I. (1995). Autocatalytic processing of the 223-kDa protein of blueberry scorch carlavirus by a papain-like proteinase. Virology 207, 127–135. doi: 10.1006/viro.1995.1058
Li, J., Feng, Z., Wu, J., Huang, Y., Lu, G., Zhu, M., et al. (2015). Structure and function analysis of nucleocapsid protein of tomato spotted wilt virus interacting with RNA using homology modeling. J. Biol. Chem. 290, 3950–3961. doi: 10.1074/jbc.M114.604678
Li, X., Halpin, C., and Ryan, M. D. (2007). A novel cleavage site within the potato leafroll virus P1 polyprotein. J. Gen. Virol. 88, 1620–1623. doi: 10.1099/vir.0.82627-0
Li, X., Ryan, M. D., and Lamb, J. W. (2000). Potato leafroll virus protein P1 contains a serine proteinase domain. J. Gen. Virol. 81, 1857–1864. doi: 10.1099/0022-1317-81-7-1857
Liu, Y., Zhu, Z., Zhang, M., and Zheng, H. (2015). Multifunctional roles of leader protein of foot-and-mouth disease viruses in suppressing host antiviral responses. Vet. Res. 46:127. doi: 10.1186/s13567-015-0273-1
Liu, Y. P., Peremyslov, V. V., Medina, V., and Dolja, V. V. (2009). Tandem leader proteases of Grapevine leafroll-associated virus-2: host-specific functions in the infection cycle. Virology 383, 291–299. doi: 10.1016/j.virol.2008.09.035
Lombardi, C., Ayach, M., Beaurepaire, L., Chenon, M., Andreani, J., Guerois, R., et al. (2013). A compact viral processing proteinase/ubiquitin hydrolase from the OTU family. PLoS Pathog. 9:e1003560. doi: 10.1371/journal.ppat.1003560
Lv, Z., Chu, Y., and Wang, Y. (2015). HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS 7, 95–104. doi: 10.2147/HIV.S79956
Maliogka, V. I., Salvador, B., Carbonell, A., Sáenz, P., San León, D., Oliveros, J. C., et al. (2012). Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Mol. Plant Pathol. 13, 877–886. doi: 10.1111/j.1364-3703.2012.00796.x
Mann, K. S., Walker, M., and Sanfaçon, H. (2017). Identification of cleavage sites recognized by the 3C-like cysteine protease within the two polyproteins of strawberry mottle virus. Front. Microbiol. 8:745. doi: 10.3389/fmicb.2017.00745
Marmey, P., Rojas-Mendoza, A., de Kochko, A., Beachy, R. N., and Fauquet, C. M. (2005). Characterization of the protease domain of rice tungro bacilliform virus responsible for the processing of the capsid protein from the polyprotein. Virol. J. 2:33. doi: 10.1186/1743-422X-2-33
Martínez, F., and Daròs, J. A. (2014). Tobacco etch virus protein P1 traffics to the nucleolus and associates with the host 60S ribosomal subunits during infection. J. Virol. 88, 10725–10737. doi: 10.1128/JVI.00928-14
Martínez, F., Rodrigo, G., Aragonés, V., Ruiz, M., Lodewijk, I., Fernández, U., et al. (2016). Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genomics 17:87. doi: 10.1186/s12864-016-2394-y
Merits, A., Rajamäki, M. L., Lindholm, P., Runeberg-Roos, P., Kekarainen, T., Puustinen, P., et al. (2002). Proteolytic processing of potyviral proteins and polyprotein processing intermediates in insect and plant cells. J. Gen. Virol. 83, 1211–1221. doi: 10.1099/0022-1317-83-5-1211
Mestre, P., Brigneti, G., and Baulcombe, D. C. (2000). An Ry-mediated resistance response in potato requires the intact active site of the NIa proteinase from potato virus Y. Plant J. 23, 653–661. doi: 10.1046/j.1365-313x.2000.00834.x
Mestre, P., Brigneti, G., Durrant, M. C., and Baulcombe, D. C. (2003). Potato virus Y NIa protease activity is not sufficient for elicitation of Ry-mediated disease resistance in potato. Plant J. 36, 755–761. doi: 10.1046/j.1365-313X.2003.01917.x
Miras, M., Miller, W. A., Truniger, V., and Aranda, M. A. (2017). Non-canonical translation in plant RNA viruses. Front. Plant Sci. 8:494. doi: 10.3389/fpls.2017.00494
Nair, S., and Savithri, H. S. (2010a). Natively unfolded nucleic acid binding P8 domain of SeMV polyprotein 2a affects the novel ATPase activity of the preceding P10 domain. FEBS Lett. 584, 571–576. doi: 10.1016/j.febslet.2009.12.003
Nair, S., and Savithri, H. S. (2010b). Processing of SeMV polyproteins revisited. Virology 396, 106–117. doi: 10.1016/j.virol.2009.09.025
Pakdel, A., Mounier, C., Klein, E., Hleibieh, K., Monsion, B., Mutterer, J., et al. (2015). On the interaction and localization of the beet necrotic yellow vein virus replicase. Virus Res. 196, 94–104. doi: 10.1016/j.virusres.2014.11.001
Pasin, F., Simón-Mateo, C., and García, J. A. (2014). The hypervariable amino-terminus of P1 protease modulates potyviral replication and host defense responses. PLoS Pathog. 10:e1003985. doi: 10.1371/journal.ppat.1003985
Peng, C. W., Napuli, A. J., and Dolja, V. V. (2003). Leader proteinase of Beet yellows virus functions in long-distance transport. J. Virol. 77, 2843–2849. doi: 10.1128/JVI.77.5.2843-2849.2003
Peng, C. W., Peremyslov, V. V., Mushegian, A. R., Dawson, W. O., and Dolja, V. V. (2001). Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75, 12153–12160. doi: 10.1128/JVI.75.24.12153-12160.2001
Peterson-Burch, B. D., and Voytas, D. F. (2002). Genes of the Pseudoviridae (Ty1/copia retrotransposons). Mol. Biol. Evol. 19, 1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008
Prüfer, D., Kawchuk, L., Monecke, M., Nowok, S., Fischer, R., and Rohde, W. (1999). Immunological analysis of potato leafroll luteovirus (PLRV) P1 expression identifies a 25 kDa RNA-binding protein derived via P1 processing. Nucleic Acids Res. 27, 421–425. doi: 10.1093/nar/27.2.421
Racaniello, V. R. (2001). “Picornaviridae: the viruses and their replication,” in Fields Virology, 4th Edn, eds D. M. Knipe and P. M. Howley (Philadelphia, PA: Lippincott-Raven Publishers), 685–722.
Rausalu, K., Utt, A., Quirin, T., Varghese, F. S., Zusinaite, E., Das, P. K., et al. (2016). Chikungunya virus infectivity, RNA replication and non-structural polyprotein processing depend on the nsP2 protease’s active site cysteine residue. Sci. Rep. 6:37124. doi: 10.1038/srep37124
Rawlings, N. D., Waller, M., Barrett, A. J., and Bateman, A. (2013). MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509. doi: 10.1093/nar/gkr987
Restrepo-Hartwig, M. A., and Carrington, J. C. (1994). The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68, 2388–2397.
Revers, F., and García, J. A. (2015). Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. doi: 10.1016/bs.aivir.2014.11.006
Riechmann, J. L., Cervera, M. T., and Garcia, J. A. (1995). Processing of the plum pox virus polyprotein at the P3-6K1 junction is not required for virus viability. J. Gen. Virol. 76, 951–956. doi: 10.1099/0022-1317-76-4-951
Rodamilans, B., Valli, A., and García, J. A. (2013). Mechanistic divergence between P1 proteases of the family Potyviridae. J. Gen. Virol. 94, 1407–1414. doi: 10.1099/vir.0.050781-0
Rohila, J. S., Chen, M., Cerny, R., and Fromm, M. E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38, 172–181. doi: 10.1111/j.1365-313X.2004.02031.x
Roossinck, M. J., Sabanadzovic, S., Okada, R., and Valverde, R. A. (2011). The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 92, 2674–2678. doi: 10.1099/vir.0.034702-0
Rozanov, M. N., Drugeon, G., and Haenni, A. L. (1995). Papain-like proteinase of turnip yellow mosaic virus: a prototype of a new viral proteinase group. Arch. Virol. 140, 273–288. doi: 10.1007/BF01309862
Sabanadzovic, S., Wintermantel, W. M., Valverde, R. A., McCreight, J. D., and Aboughanem-Sabanadzovic, N. (2016). Cucumis melo endornavirus: genome organization, host range and co-divergence with the host. Virus Res. 214, 49–58. doi: 10.1016/j.virusres.2016.01.001
Salvador, B., Delgadillo, M. O., Sáenz, P., García, J. A., and Simón-Mateo, C. (2008a). Identification of Plum pox virus pathogenicity determinants in herbaceous and woody hosts. Mol. Plant Microbe Interact. 21, 20–29. doi: 10.1094/MPMI-21-1-0020
Salvador, B., Sáenz, P., Yángüez, E., Quiot, J. B., Quiot, L., Delgadillo, M. O., et al. (2008b). Host-specific effect of P1 exchange between two potyviruses. Mol. Plant Pathol. 9, 147–155. doi: 10.1111/j.1364-3703.2007.00450.x
Satheshkumar, P. S., Lokesh, G. L., and Savithri, H. S. (2004). Polyprotein processing: cis and trans proteolytic activities of Sesbania mosaic virus serine protease. Virology 318, 429–438. doi: 10.1016/j.virol.2003.09.035
Sawicki, D. L., and Sawicki, S. G. (1994). Alphavirus positive and negative strand RNA synthesis and the role of polyproteins in formation of viral replication complexes. Arch. Virol. 9, 393–405. doi: 10.1007/978-3-7091-9326-6_39
Schaad, M. C., Jensen, P. E., and Carrington, J. C. (1997). Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16, 4049–4059. doi: 10.1093/emboj/16.13.4049
Seipelt, J., Guarné, A., Bergmann, E., James, M., Sommergruber, W., Fita, I., et al. (1999). The structures of picornaviral proteinases. Virus Res. 62, 159–168. doi: 10.1016/S0168-1702(99)00043-X
Shamsi, T. N., Parveen, R., and Fatima, S. (2016). Characterization, biomedical and agricultural applications of protease inhibitors: a review. Int. J. Biol. Macromol. 91, 1120–1133. doi: 10.1016/j.ijbiomac.2016.02.069
Shan, H., Pasin, F., Tzanetakis, I. E., Simón-Mateo, C., García, J. A., and Rodamilans, B. (2017). Truncation of a P1 leader proteinase facilitates potyvirus replication in a non-permissive host. Mol. Plant Pathol. 19, 1504–1510. doi: 10.1111/mpp.12640
Shan, H., Pasín, F., Valli, A., Castillo, C., Rajulu, C., Carbonell, A., et al. (2015). The Potyviridae P1a leader protease contributes to host range specificity. Virology 476, 264–270. doi: 10.1016/j.virol.2014.12.013
Shi, X., Botting, C. H., Li, P., Niglas, M., Brennan, B., Shirran, S. L., et al. (2016). Bunyamwera orthobunyavirus glycoprotein precursor is processed by cellular signal peptidase and signal peptide peptidase. Proc. Natl. Acad. Sci. U.S.A. 113, 8825–8830. doi: 10.1073/pnas.1603364113
Simmonds, P., Adams, M. J., Benko, M., Breitbart, M., Brister, J. R., Carstens, E. B., et al. (2017). Consensus statement: virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161–168. doi: 10.1038/nrmicro.2016.177
Sõmera, M., Sarmiento, C., and Truve, E. (2015). Overview on sobemoviruses and a proposal for the creation of the family Sobemoviridae. Viruses 7, 3076–3115. doi: 10.3390/v7062761
Spall, V. E., Shanks, M., and Lomonossoff, G. P. (1997). Polyprotein processing as a strategy for gene expression in RNA viruses. Semin. Virol. 8, 15–23. doi: 10.1006/smvy.1997.0102
Stewart, L. R., Jarugula, S., Zhao, Y., Qu, F., and Marty, D. (2017). Identification of a maize chlorotic dwarf virus silencing suppressor protein. Virology 504, 88–95. doi: 10.1016/j.virol.2016.11.017
Sun, D., Chen, S., Cheng, A., and Wang, M. (2016). Roles of the picornaviral 3C proteinase in the viral life cycle and host cells. Viruses 8:82. doi: 10.3390/v8030082
Suttle, C. A. (2007). Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. doi: 10.1038/nrmicro1750
Tatineni, S., and French, R. (2016). The coat protein and NIa protease of two Potyviridae family members independently confer superinfection exclusion. J. Virol. 90, 10886–10905. doi: 10.1128/JVI.01697-16
Thole, V., and Hull, R. (1998). Rice tungro spherical virus polyprotein processing: identification of a virus-encoded protease and mutational analysis of putative cleavage sites. Virology 247, 106–114. doi: 10.1006/viro.1998.9225
Thompson, J. R., Kamath, N., and Perry, K. L. (2014). An evolutionary analysis of the Secoviridae family of viruses. PLoS One 9:e106305. doi: 10.1371/journal.pone.0106305
Torruella, M., Gordon, K., and Hohn, T. (1989). Cauliflower mosaic virus produces an aspartic proteinase to cleave its polyproteins. EMBO J. 8, 2819–2825.
Tran, P. T., Widyasari, K., Park, J. Y., and Kim, K. H. (2017). Engineering an auto-activated R protein that is in vivo activated by a viral protease. Virology 510, 242–247. doi: 10.1016/j.virol.2017.07.020
Valli, A., Dujovny, G., and García, J. A. (2008). Protease activity, self interaction, and small interfering RNA binding of the silencing suppressor p1b from Cucumber vein yellowing ipomovirus. J. Virol. 82, 974–986. doi: 10.1128/JVI.01664-07
Valli, A., Gallo, A., Calvo, M., Pérez, J. J., and García, J. A. (2014). A novel role of the potyviral helper component proteinase contributes to enhance the yield of viral particles. J. Virol. 88, 9808–9818. doi: 10.1128/JVI.01010-14
Valli, A., Martín-Hernández, A. M., López-Moya, J. J., and García, J. A. (2006). RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J. Virol. 80, 10055–10063. doi: 10.1128/JVI.00985-06
Valli, A. A., Gallo, A., Rodamilans, B., López-Moya, J. J., and García, J. A. (2018). The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 19, 744–763. doi: 10.1111/mpp.12553
van Kasteren, P. B., Bailey-Elkin, B. A., James, T. W., Ninaber, D. K., Beugeling, C., Khajehpour, M., et al. (2013). Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc. Natl. Acad. Sci. U.S.A. 110, E838–E847. doi: 10.1073/pnas.1218464110
Vasiljeva, L., Merits, A., Golubtsov, A., Sizemskaja, V., Kaariainen, L., and Ahola, T. (2003). Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J. Biol. Chem. 278, 41636–41645. doi: 10.1074/jbc.M307481200
Verchot, J., and Carrington, J. C. (1995). Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J. Virol. 69, 1582–1590.
Verchot, J., Herndon, K. L., and Carrington, J. C. (1992). Mutational analysis of the tobacco etch potyviral 35-kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology 190, 298–306. doi: 10.1016/0042-6822(92)91216-H
Verdaguer, N., Ferrero, D., and Murthy, M. R. (2014). Viruses and viral proteins. IUCrJ 1(Pt 6), 492–504. doi: 10.1107/S205225251402003X
Wang, A., Carrier, K., Chisholm, J., Wieczorek, A., Huguenot, C., and Sanfaçon, H. (1999). Proteolytic processing of tomato ringspot nepovirus 3C-like protease precursors: definition of the domains for the VPg, protease and putative RNA-dependent RNA polymerase. J. Gen. Virol. 80, 799–809. doi: 10.1099/0022-1317-80-3-799
Wang, A., and Sanfaçon, H. (2000). Proteolytic processing at a novel cleavage site in the N-terminal region of the tomato ringspot nepovirus RNA-1-encoded polyprotein in vitro. J. Gen. Virol. 81, 2771–2781. doi: 10.1099/0022-1317-81-11-2771
Wang, D., Fang, L., Li, P., Sun, L., Fan, J., Zhang, Q., et al. (2011a). The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85, 3758–3766. doi: 10.1128/JVI.02589-10
Wang, D., Fang, L., Liu, L., Zhong, H., Chen, Q., Luo, R., et al. (2011b). Foot-and-mouth disease virus (FMDV) leader proteinase negatively regulates the porcine interferon-1 pathway. Mol. Immunol. 49, 407–412. doi: 10.1016/j.molimm.2011.09.009
Weinheimer, I., Boonrod, K., Moser, M., Zwiebel, M., Fullgrabe, M., Krczal, G., et al. (2010). Analysis of an autoproteolytic activity of rice yellow mottle virus silencing suppressor P1. Biol. Chem. 391, 271–281. doi: 10.1515/BC.2010.022
Wen, R., Zhang, S. C., Michaud, D., and Sanfaçon, H. (2004). Inhibitory effects of cystatins on proteolytic activities of the Plum pox potyvirus cysteine proteinases. Virus Res. 105, 175–182. doi: 10.1016/j.virusres.2004.05.008
Whitfield, A. E., Ullman, D. E., and German, T. L. (2005). Tomato spotted wilt virus glycoprotein G(C) is cleaved at acidic pH. Virus Res. 110, 183–186. doi: 10.1016/j.virusres.2005.01.007
Wilhelm, S. W., Bird, J. T., Bonifer, K. S., Calfee, B. C., Chen, T., Coy, S. R., et al. (2017). A student’s guide to giant viruses infecting small eukaryotes: from Acanthamoeba to Zooxanthellae. Viruses 9:E46. doi: 10.3390/v9030046
Wright, D. A., and Voytas, D. F. (2002). Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res. 12, 122–131. doi: 10.1101/gr.196001
Yost, S. A., and Marcotrigiano, J. (2013). Viral precursor polyproteins: keys of regulation from replication to maturation. Curr. Opin. Virol. 3, 137–142. doi: 10.1016/j.coviro.2013.03.009
Keywords: viral proteases, viral polyprotein, plant viruses, viral replication, virion formation, host range, defense and counterdefense
Citation: Rodamilans B, Shan H, Pasin F and García JA (2018) Plant Viral Proteases: Beyond the Role of Peptide Cutters. Front. Plant Sci. 9:666. doi: 10.3389/fpls.2018.00666
Received: 15 February 2018; Accepted: 30 April 2018;
Published: 17 May 2018.
Edited by:
Helene Sanfaçon, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Michael Rozanov, Pirogov Russian National Research Medical University, RussiaJeremy Thompson, Cornell University, United States
Valerian V. Dolja, Oregon State University, United States
Copyright © 2018 Rodamilans, Shan, Pasin and García. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernardo Rodamilans, YnJvZGFtaWxhbnNAY25iLmNzaWMuZXM=
 Bernardo Rodamilans
Bernardo Rodamilans Hongying Shan1
Hongying Shan1 Fabio Pasin
Fabio Pasin Juan Antonio García
Juan Antonio García