
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 April 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00560
 Min Wang1†
Min Wang1† Tao Zhang2†
Tao Zhang2† Hao Peng3†
Hao Peng3† Sheng Luo1
Sheng Luo1 Juejie Tan1
Juejie Tan1 Kaifeng Jiang2
Kaifeng Jiang2 Yueqin Heng1
Yueqin Heng1 Xin Zhang1
Xin Zhang1 Xiuping Guo1
Xiuping Guo1 Jiakui Zheng2*
Jiakui Zheng2* Zhijun Cheng1*
Zhijun Cheng1*Premature leaf senescence (PLS), which has a significant impact on yield, is caused by various underlying mechanisms. Glycosyltransferases, which function in glycosyl transfer from activated nucleotides to aglycones, are involved in diverse biological processes, but their roles in rice leaf senescence remain elusive. Here, we isolated and characterized a leaf senescence-related gene from the Premature Leaf Senescent mutant (pls2). The mutant phenotype began with leaf yellowing at tillering and resulted in PLS during the reproductive stage. Leaf senescence was associated with an increase in hydrogen peroxide (H2O2) content accompanied with pronounced decreases in net photosynthetic rate, stomatal conductance, and transpiration rate. Map-based cloning revealed that a mutation in LOC_Os03g15840 (PLS2), a putative glycosyltransferase- encoding gene, was responsible for the defective phenotype. PLS2 expression was detected in all tissues surveyed, but predominantly in leaf mesophyll cells. Subcellular localization of the PLS2 was in the endoplasmic reticulum. The pls2 mutant accumulated higher levels of sucrose together with decreased expression of sucrose metabolizing genes compared with wild type. These data suggested that the PLS2 allele is essential for normal leaf senescence and its mutation resulted in PLS.
Plant senescence is an age-dependent behavior in plant development under normal growth condition (Lee et al., 2001; Woo et al., 2001). During senescence, leaf cells undergo dramatic metabolic changes, including chlorophyll breakdown and hydrolysis of macromolecules (lipids, proteins, and nucleic acids), that results in leaf cell death (Kong et al., 2006; Schippers et al., 2015). In agriculture, delayed leaf senescence (stay green) provides opportunities to prolong photosynthetic capacity and increase crop yield (Thomas and Howarth, 2000). PLS is triggered by various external factors (such as drought, salinity, shading, or biotic stress) as well as physiological factors such as endogenous sugar content, or plant hormone levels (van Doorn, 2008; Schippers et al., 2015; Abdelrahman et al., 2017), and usually causes yield loss (Bai et al., 2015; Rao et al., 2015). Therefore, an in-depth understanding of the molecular mechanism of leaf senescence is important in delaying leaf senescence and increasing cereal crop production (Wu et al., 2012; Gregersen et al., 2013).
By now, many research advances of leaf senescence at the molecular level have been achieved through the isolation and characterization of dozens of senescence-related mutants and senescence-associated genes (SAGs) (Buchanan-Wollaston et al., 2003; Lim et al., 2007; Li et al., 2012). These SAGs are usually involved in various biological processes, such as the breakdown of chlorophyll, degradation of chloroplasts, plant hormone synthesis and signaling, and biotic and abiotic stress responses (Kong et al., 2006; Jiao et al., 2012; Li et al., 2012; Liang et al., 2014; Sakuraba et al., 2015). Several SAGs in rice have been isolated and functionally characterized. For example, the NB-domain-containing protein encoding gene, RLS1 (rapid leaf senescence 1), is involved in an autophagy-like process of chloroplast degradation (Jiao et al., 2012). The highly increased transcription level of OsABC1-2 (an Abc1 kinase family gene), a chloroplast membrane-localized kinase encoding gene, is dramatically suppressed by dark treatment and its over-expression improves plant resistance in extended periods of darkness (Gao et al., 2012). OsNAP (rice NAC-like, activated by apetala3/pistillata) exerts roles in regulating expressions of an age-dependent manner SAGs and ABA biosynthesis related genes (Liang et al., 2014). Overexpressing rice OsWRKY42 (one transcription factor of WRKY family) exhibited early leaf senescence with accumulation of hydrogen peroxide and reduced chlorophyll content (Han et al., 2014). The Stay-Green Rice (SGR) gene, encoding a chloroplast protein, is necessary for the initiation of chlorophyll breakdown (Park et al., 2007; Hörtensteiner, 2009) and it’s up-regulated expression induced leaf senescence (Ren et al., 2007; Pilkington et al., 2012). In addition, Rapid Leaf Senescence 3 (RLS3), which produces a protein with an AAAt domain, functions in delaying leaf senescence in rice (Lin et al., 2016). Mutation of DEL1 (Early Senescence Leaf 1) decreases the enzymatic activity of PEL (pectate lyase) and increases expressions of SAGs (Leng et al., 2017). Although, various kinds of genes have been studied in rice, further investigation on leaf senescence-related genes is essential in order to establish a better understanding of regulatory mechanisms of senescence.
Glycosylation, a process of glycosyltransferases (GTs, EC 2.4.x.y) catalyzing the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, is considered a single modification reaction on plant hormones, secondary metabolites, and xenobiotics by glycosidic bonds (Jones and Vogt, 2001; Lairson et al., 2008; Li et al., 2015). There are about 452 and 609 GT members in the Arabidopsis and rice genomes, respectively, and most of them have not been functionally characterized (Ko et al., 2006; Cao et al., 2008). UDP-glycosyltransferases (UGTs) utilize UDP-glucoses as donor in regulating various biological processes (Coutinho et al., 2003). Accumulating evidence suggests a critical role of UGTs in plant developmental processes and stress reactions. Reduced expression level of gene UGT71B6 in Arabidopsis induces early senescence and enhances susceptibility to the necrotrophic pathogen Alternaria brassicicola (von Saint Paul et al., 2012). UGT74E2 modulates plant architecture as well as conferring drought stress tolerance (Tognetti et al., 2010), and UGT76C2 was found to be involved in adaptation to drought stress (Li et al., 2015). UGT75D1 modulates cotyledon development and stress tolerance during seed germination (Zhang et al., 2016). Ectopic expression of UGT85A5 in tobacco and SrUGT74G1 in Arabidopsis promotes seed germination in tobacco (Sun et al., 2013) and catechin accumulation in Arabidopsis (Guleria and Yadav, 2014), respectively. Overexpression of UGT80B1 increases resistance to freezing and heat stress (Mishra et al., 2015). Ectopic expression of UGT85U1, UGT85U2, and UGT85V1 in Arabidopsis improved salt and oxidative stress tolerance (Ahrazem et al., 2015). In rice, the expression level of OsGT61-1 was significantly responsive to exogenous treatment of ABA and NaCl (Singh et al., 2010). In addition, XAX1 from GT 61, could mediate xylosyl transfer to rice xylan (Chiniquy et al., 2012). GT43 family is involved in xylan biosynthesis of rice (Lee et al., 2014). OsGT47A has a role in plant secondary cell wall thickness (Zhang B. et al., 2014).
Although several SAGs have been cloned and studied, there is no insight about the role of GT in leaf senescence. In the previous study, we described Premature Leaf Senescence mutant (pls2) in rice, mapped the PLS2 locus on chromosome 3, and postulated the LOC_Os3g15840, a glycosyltransferase encoding gene, as the candidate gene (Zhang T. et al., 2014). In the present study, we remappedPLS2 using a newly developed genetic population, and confirmed LOC_Os3g15840 as the target gene. PLS2 expression was detected in all tissue types, but predominantly in leaf mesophyll cells, with a sub-cellular localization of endoplasmic reticulum. The mutation in the PLS2 caused PLS. Compared to the wild type (WT), the pls2 mutant accumulates sucrose together with decreased expression of sucrose metabolizing related genes. The collected data suggest that PLS2 is essential for normal senescence.
Rice premature leaf senescence pls2 was obtained as a space-radiation mutant in indica var. Luhui H103. The pls2 mutant was crossed with japonica var. Nipponbare and a F2mapping population was grown in a paddy field in Beijing (39°54′N, summer season, temperate climate).
Chlorophyll content was measured according to the procedure described by Suzuki and Makino (2012). About 0.2 g of leaves were homogenized in 5 ml of a 9: 1 acetone to 0.1 M NH4OH solution and centrifuged at 3000 × g for 20 min. These supernatants were then washed three times using hexane (1:1 ration of supernatants to hexane) and the pigment content was measured by spectrophotometer at the absorption wavelengths of 663 and 645 nm (Beckman Coulter DU-800, CITY, United States). The experiment was carried out with three technical and three biological replicates, respectively.
H2O2 was detected by DAB staining as described by Thordal-Christensen et al. (1997). Fully expanded flag leaves were vacuum-infiltrated with DAB solution (1 mg of DAB dissolved in 1 ml of distilled water, pH 3.8) for 24 h at 25°C and washed in boiling ethanol (96%) for 10 min before photographing. For H2O2 quantitative measurement, H2O2 was extracted from leaves at heading according to the method described by Rao et al. (2000). MDA content was measured by the following steps: 0.5 g leaves were ground into powder, and then dipped into 0.5% TCA buffer, followed by treatment of 100°C for 30 min. After centrifuged at 3000 × g for 30 min, the supernatant (2 ml) was added to 2 ml 0.5% TBA, then subjected to 100°C for 30 min again. After the mixture was dropped to room temperature, it was centrifuged at 3000 × g for 20 min. Finally, the light absorption of the supernatant was measured at the absorption wavelength of 450-, 532-, and 600 nm, respectively. The MDA content was calculated according to the formula: (CMDA = 6.45∗[A532-A600]-0.56∗A450 [μmol/L]). The experiment of H2O2 and MDA were performed with three technical and three biological replicates.
Flag leaves of the wild type and pls2 mutant were used to measure the net photosynthetic rate, stomatal conductance, and transpiration rate from 9:00 am to 11:00 am. The detailed methods of photosynthetic parameters were to procedures (Wang et al., 2015). In order to allow flag leaves to reach steady-state photosynthesis, flag leaves were kept under each level of CO2 concentration for 5 min before these photosynthetic parameters were recorded on portable photosynthetic system (CIRAS-2, PP Systems, Hitchin, United Kingdom). The assay was carried out with three technical and biological replicates, respectively.
Leaves at heading were cut into small pieces, fixed in 2.5% glutaraldehyde in a phosphate buffer (pH 7.2), vacuum infiltrated, rinsed, and incubated overnight at 4°C in a solution of 1% OsO4. Samples were dehydrated in a series of 10, 30, 50, 70, 90, and 100% ethanol and infiltrated in epoxy resin, and embedded in Epon 812 resin. A series of 80 nm sections was cut using a Reichert OM2 ultramicrotome, stained in 2% uranylacetate and 10 mM lead citrate (pH12), before observation in a HitachiH-7650 transmission electron microscope.
DNA was extracted according to CTAB method described by Telzur et al. (1999). Eight-hundred-and twenty pls2-like individuals were sampled from the segregating F2 population for linkage analysis. All InDel markers used in this study were developed according to sequence diversity between H103 and Nipponbare, which are available at the Gramene website1. dCAPS markers were automatically designed using the web server program dCAPS Finder 2.0.2 The PLS2 locus was finally mapped to a 90-Kb region of chromosome 3 delimited by two dCAPS of C-2 and SL-1-9. The candidate gene was identified by DNA sequencing.
For overexpression of PLS2, we cloned the coding region of PLS2 into the pCAMBIA1390 vector under the maize Ubi promoter to produce the fusion vector pUbi::PLS2. The pUbi::PLS2 vector was transformed into pls2 plants by Agrobacterium-mediated transformation as described previously by Hiei and Komari (2008).
To obtain a crispr-PLS2 mutant line, we used the CRISPR-Cas9 system according to the method previously described by Miao et al. (2013). A 20 bp PLS2-specific spacer sequence was cloned into the entry vector pOs-sgRNA, followed by subcloning into a pCAS9 binary vector by means of the Gateway cloning system. The fused vector was transformed into Nipponbare as described above. The molecular markers used for vector construction are listed in Supplementary Table S1.
Total RNA was extracted according to the instructions with TRIZOL Kit (TaKaRa, Japan). First-strand cDNA was obtained from 2 μg of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Germany). qRT-PCR (20 μl reaction volume) was carried out with 0.5 μl of cDNA, 0.2 μM of primer mix, and the SYBR Premix Ex Taq Kit (TaKaRa, Japan). The endogenous rice UBQ gene (LOC_Os03g13170) and OsActin gene (LOC_Os03g50885) were used as the reference genes, respectively. The assay was carried out with three technical and biological replicates respectively. All qRT-PCR primers are listed in the Supplementary Table S1.
Assays were performed as described previously (Bradley et al., 1993). For preparation of materials, flag leaves of wild type plants at heading were fixed using an RNase-free formalin/acetic acid fixative solution, followed by a series of dehydration steps and embedded in paraffin for sectioning. To prepare the probe, we used a pair of primers, PLS2-in situ-F (5′-CGTCAGTAGCTATTGCCGAGGACTTTGA-3′) and PLS2-in situ-R (5′-GCTTGTGAGAGCTCCTCGCCTT-3′), to amplify a 335 bp unique sequence of PLS2 from a cDNA clone. The fragment was then inserted into the pGEM-T vector (Promega) for RNA transcription. Digoxigenin-labeled RNA probes were prepared using a DIG Northern Starter Kit (Roche3). Hybridization signals were visualized and photographed using a Leica DMR microscope equipped with a Micro Color charge-coupled device camera (Apogee Instruments4).
To create the integrated vector pCAMBIA1305-d35S-PLS2-GFP, the PLS2 CDS fragment was cloned into the pCAMBIA1305-GFP vector at the Bgl II site. Constructs were transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells as described previously (Batoko et al., 2000). Tobacco leaf protoplasts were obtained as described by Miao et al. (2006) to analyze transient expression of PLS2. The GFP signal was photographed by a laser scanning confocal microscope (LSCM 700; Carl Zeiss).
A 3,393 bp upstream fragment of the PLS2 gene was amplified using the primers (Supplementary Table S1) and sub-cloned into a pCAMBIA 1305:GUS vector with restriction enzyme sites EcoR I and Noc I to get a PLS2Pro: GUS construct, and then introduced into japonica var. Kitaake by Agrobacterium described above. For histochemical analysis, we used excised tissues of independent T2 transgenic plants, as reported by Jefferson (1987).
Flag leaf pieces (0.05 g) were dissolved in 1 ml of 80% ethanol, incubated at 50°C for about 30 min, and then centrifuged at 3,000 g for 15 min, repeated twice. The three supernatants were vacuum dried at 45°C, dissolved in 50°C preheated ultrapure water, and subjected to a 5 μm C18 extraction column (150 × 4.6 mm; Agilent Zorbax) for removing impurities. After that, the soluble sugar of supernatants was analyzed by High Performance Liquid Chromatography instrument (Shimadzu Company, Japan). The standard curve of sucrose was previously made including concentrations of 10 mg/mL, 5 mg/mL, 2.5 mg/mL, 1.25 mg/mL, and 0.625 mg/mL. The chromatography analysis and sample recovery steps were carried out according to the method described by Brushwood (1997). The assay was conducted with three technical and biological replicates, respectively.
The senescence symptoms of the pls2 mutant gradually developed from the tillering stage when emerging leaves slowly became yellowish or developed senescence (Figure 1). Senescent leaves had reduced chlorophyll content (Figures 1D–F). The pls2 mutant had reduced height and internode length, and low seed setting (Supplementary Figures S1A–C). Detailed analysis of grain plumpness at different positions on the panicle revealed that the mutant had a higher proportion of semi-filled grain (SFGN), and shriveled grain (SGN) compared to WT (Supplementary Figures S1D,E). 1000-grain weight was decreased slightly, but not significantly in the pls2 mutant (Supplementary Figure S1F). The results indicated that the PLS in pls2 had a detrimental effect on vegetative growth as well as grain development (Supplementary Figure S1). In addition, expressions of several SAG genes (PAO, SGR, NYC3, and Osl85) were also significantly increased in the pls2 mutant at heading stage in comparison with WT (Figure 1G).
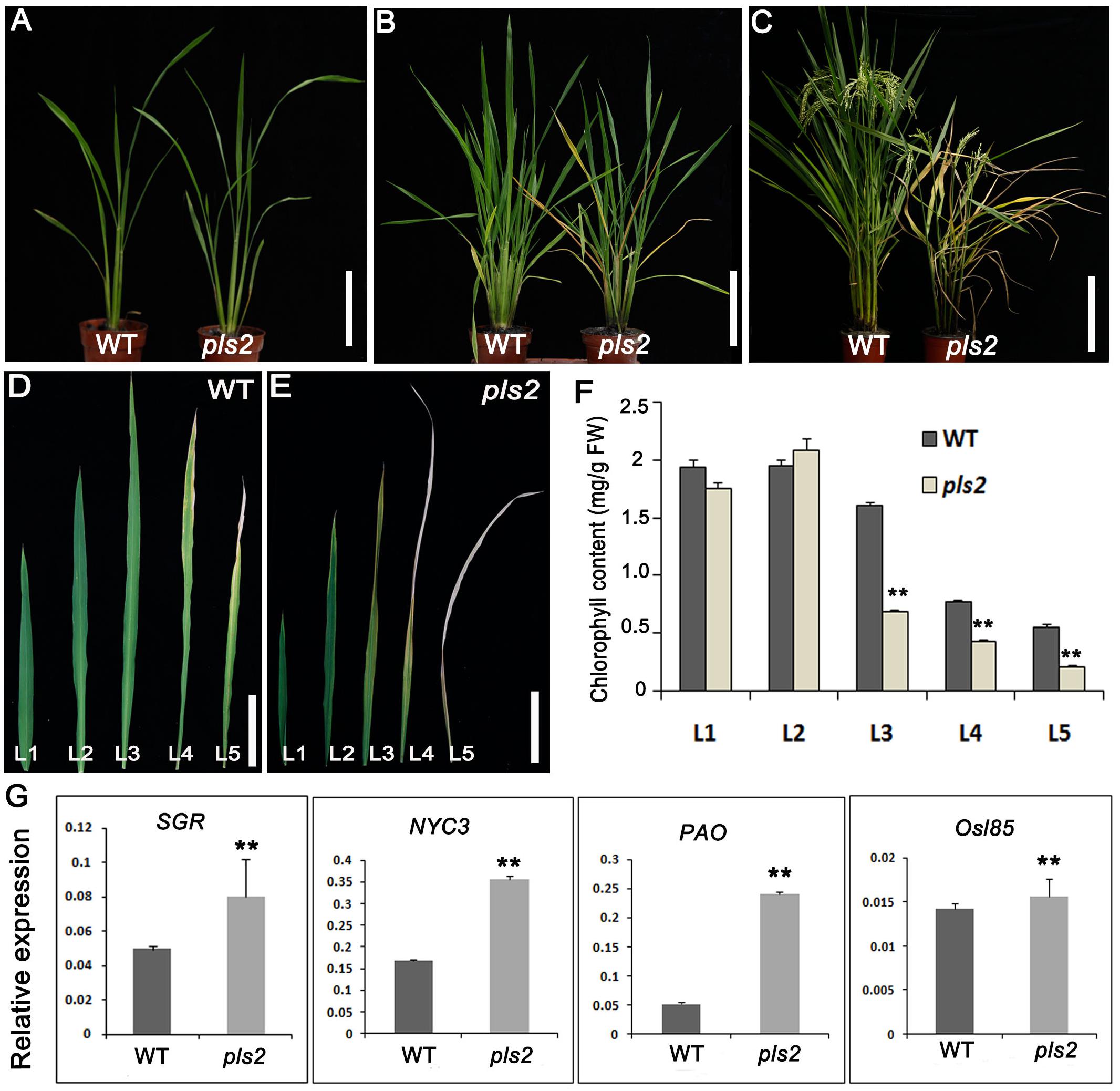
FIGURE 1. pls2 is a premature leaf senescence mutant. (A–D) Phenotypes of wild type and pls2 mutant at the seedling (A), tillering (B), and heading (C) stages. Bar in (A–C) 15 cm. (D,E) Uppermost five leaves from the main culms of WT (D) and pls2 (E). Bar in (D,E) 3 cm. L1–L5 represent leaves from the flag leaf downward. (F) Chlorophyll contents of the five uppermost leaves from WT and pls2. (G) qRT-PCR analysis of four senescence associated genes (SAGs) between WT and pls2. Data is presented as the mean ± standard deviation (n = 9). ∗∗P ≤ 0.01; Student’s t-test.
We examined H2O2 [a reactive oxygen species (ROS)] levels by DAB in both WT and pls2, and strong DAB brown straining appeared in the pls2 flag leaves (Figure 2A), consistent with quantitative results showing that the pls2 mutant exhibited excess H2O2 accumulation (Figure 2B). As the end-product of membrane lipid peroxidation caused by ROS (Draper and Hadley, 1989), the malonic dialdehyde content (MDA) was accumulated highly in the pls2 (Figure 2C). Meanwhile, leaf photosynthesis capability determined by net photosynthetic rate (Figure 2D), stomatal conductance (Figure 2E), and transpiration rate (Figure 2F) was accordingly reduced in pls2.
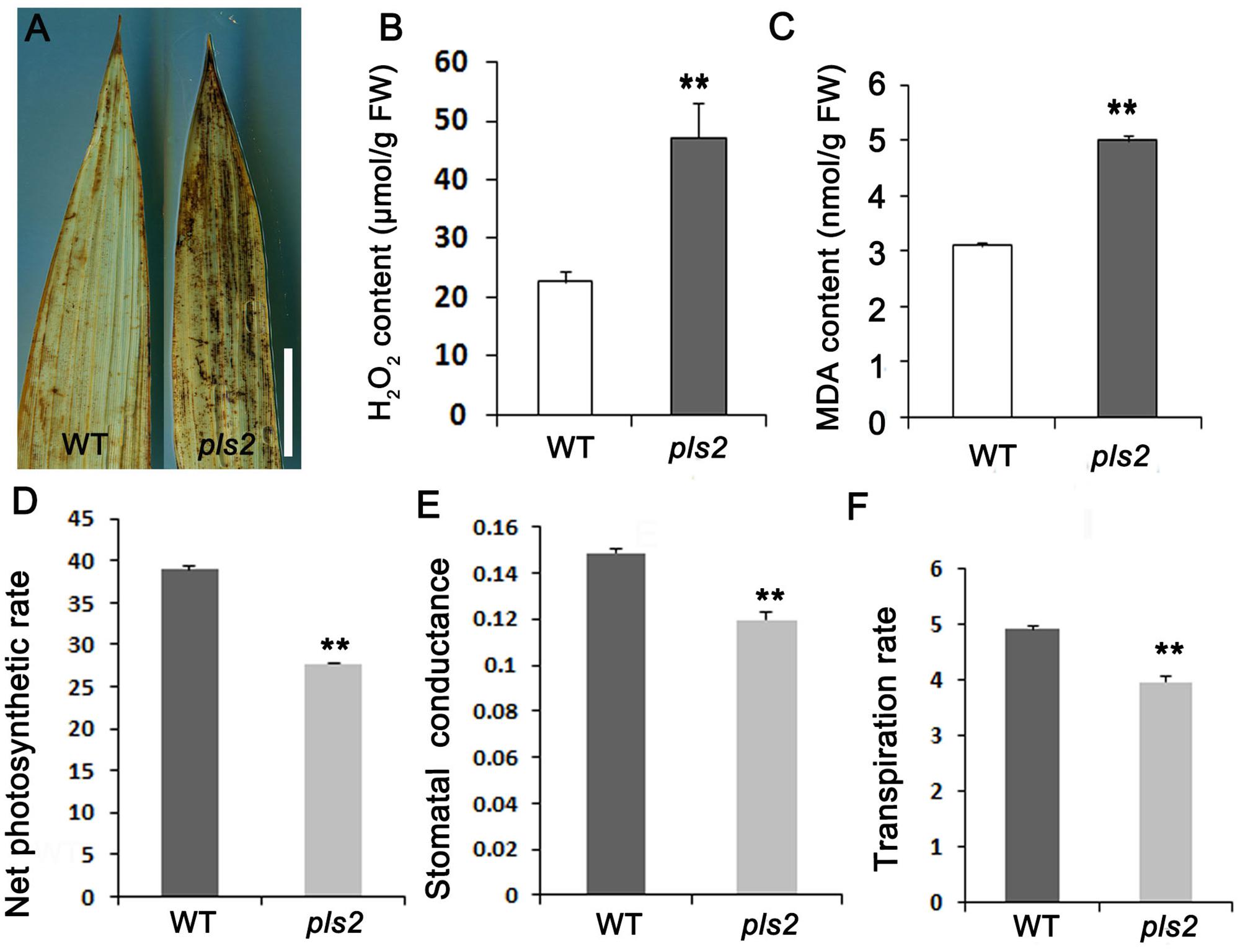
FIGURE 2. pls2 accumulated excessive ROS. (A) DAB staining of leaves of WT and pls2. Bar, 4 cm. (B,C) H2O2 (B) and MDA (C) quantitation assays at heading. (D–F) Net photosynthetic, stomatal conductance, and transpiration rates in flag leaves of WT and pls2. Data is presented as the mean ± standard deviation (n = 9). ∗0.01 ≤P ≤ 0.05; ∗∗P ≤ 0.01; Student’s t-test.
Segregation in the redeveloped F2 population of pls2 × japonica cv. Nipponbare was 2,285 plants with normal phenotype and 820 with mutant phenotype, confirming that a single recessive gene caused the senescent phenotype (χ23:1 = 0.06: P > 0.05). We previously placed PLS2 between markers of RM14704 and SL-1-5 on chromosome 3, represented by a physical distance of 84.11 Kb (Zhang T. et al., 2014). Using extra 820 F2 mutant individuals, we confirmed the interval by the newly developed marker C-2 and SL-1-9 (Figure 3A), which mapped the similar location compared with previous study. The determined 90 Kb candidate region contained 15 ORFs, and a C→T substitution in the ninth exon of gene LOC_Os03g15840 (ORF4), putatively causing an R→C amino acid alternation (Figure 3B).
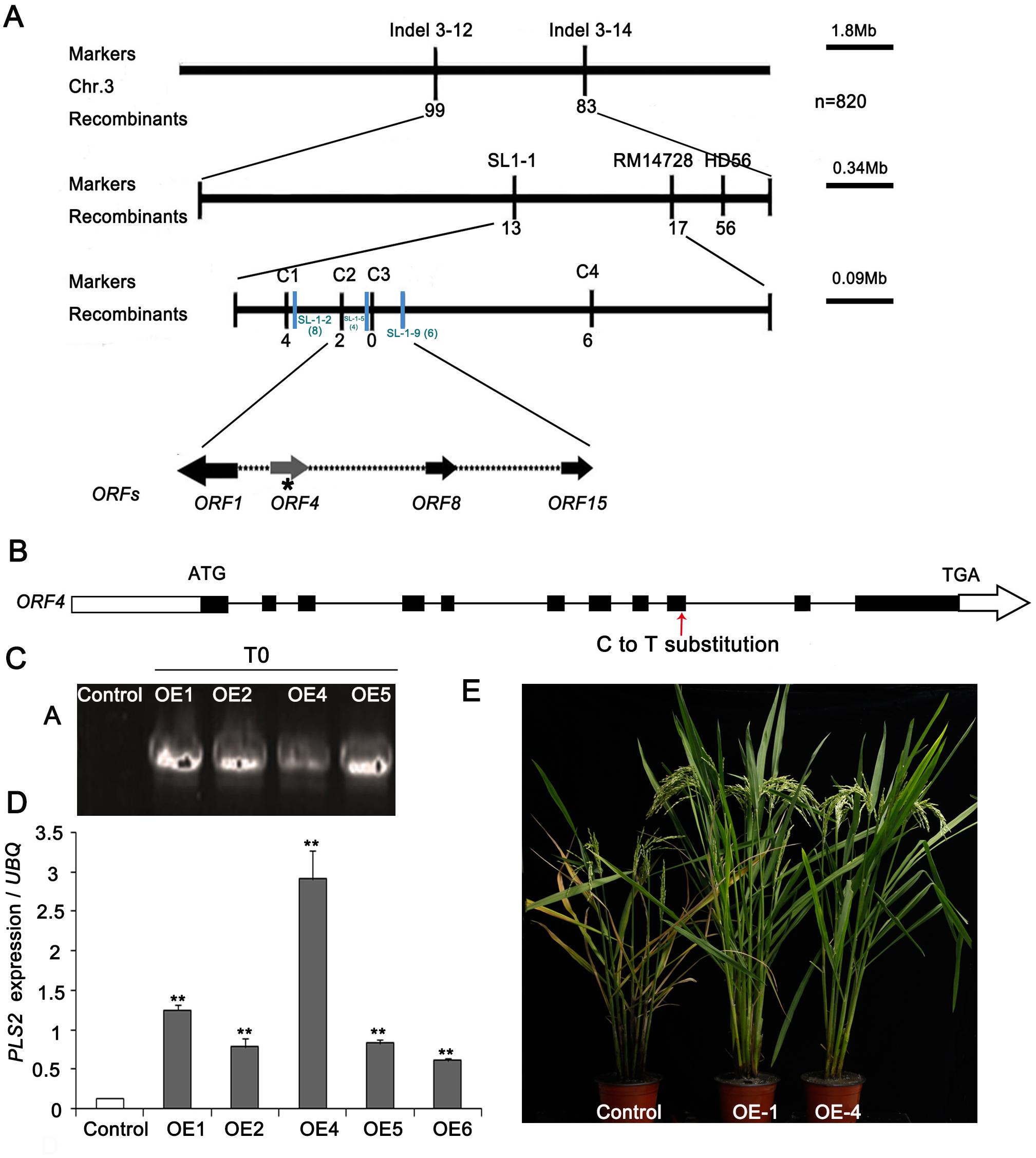
FIGURE 3. Map-based cloning of the PLS2 gene. (A) Fine mapping of PLS2 in the F2 population of pls2 × Nipponbare. (B) Schematic of the PLS2 gene structure. Black boxes indicate exons and black lines between the boxes represent introns. The red arrow indicated the position where the C→T substitution occurred. (C,D) Overexpression analyses of positive plants by qRT-PCR. (E) PLS2 over-expressing plants in pls2 background. Bar, 15 cm. ∗∗P ≤ 0.01; Student’s t-test. Data is presented as the mean ± standard deviation (n = 9).
To validate the candidate gene, we transformed the WT CDS of LOC_Os03g15840 driven by the Ubi promoter into the pls2 mutant. Four independent positive T2 transgenic plants were identified by PCR genotyping (Figure 3C) and qRT-PCR, which expressed LOC_Os03g15840 significantly higher than the control (Figure 3D) and rescued the leaf senescence phenotype of the pls2 mutant (Figure 3E), confirming that an aberrant LOC_Os03g15840 was the cause of pls2. We further verified the candidate gene by knock-out of the gene in Nipponbare background using Crispr/Cas9 technology. Finally, two Crispr-PLS2 positive transgenic plants exhibited early leaf senescent and reduced plant height (Supplementary Figure S2). These results indicated that the mutation in LOC_Os03g15840 caused the leaf senescence of the pls2, and the locus therefore was designated PLS2.
LOC_Os03g15840 encodes a glycosyltransferase containing the Glyco_trans_4_1 and Glyco_trans_4_2 domains that catalyze sugar transfer from donor to acceptor (Supplementary Figure S3A). A phylogenetic tree analysis showed that the rice PLS2 gene Os03g0265100 is highly homologous to Brachypodium sylvaticum gene XP003558286.1 and Aegilops tauschii gene EMT15626.1 (Supplementary Figure S3B).
The results of qRT-PCR showed that PLS2 was expressed in all tissues, i.e., roots, leaf sheaths, stems, young leaves, and panicles, and there was strong expression in leaves, followed by roots and stems (Figure 4A and Supplementary Figure S4). β-glucuronidase (GUS) staining was detected in various tissues, especially seedlings (Figures 4B,C), leaf sheaths (Figure 4D), leaves (Figure 4E), and panicles (Figure 4F). Observations on root sections indicated strong GUS signals in the phloem (Figure 4C). In situ hybridization (ISH) assays demonstrated that PLS2 was highly expressed in leaf mesophyll cells (Figures 4G,H). The PLS2-GFP completely merged with the ER marker HDEL-mRFP (Figure I) (Gomord et al., 1997) in epidermal protoplasts of N. benthamiana leaves, suggesting subcellular localization of the PLS2 in the endoplasmic reticulum (ER).
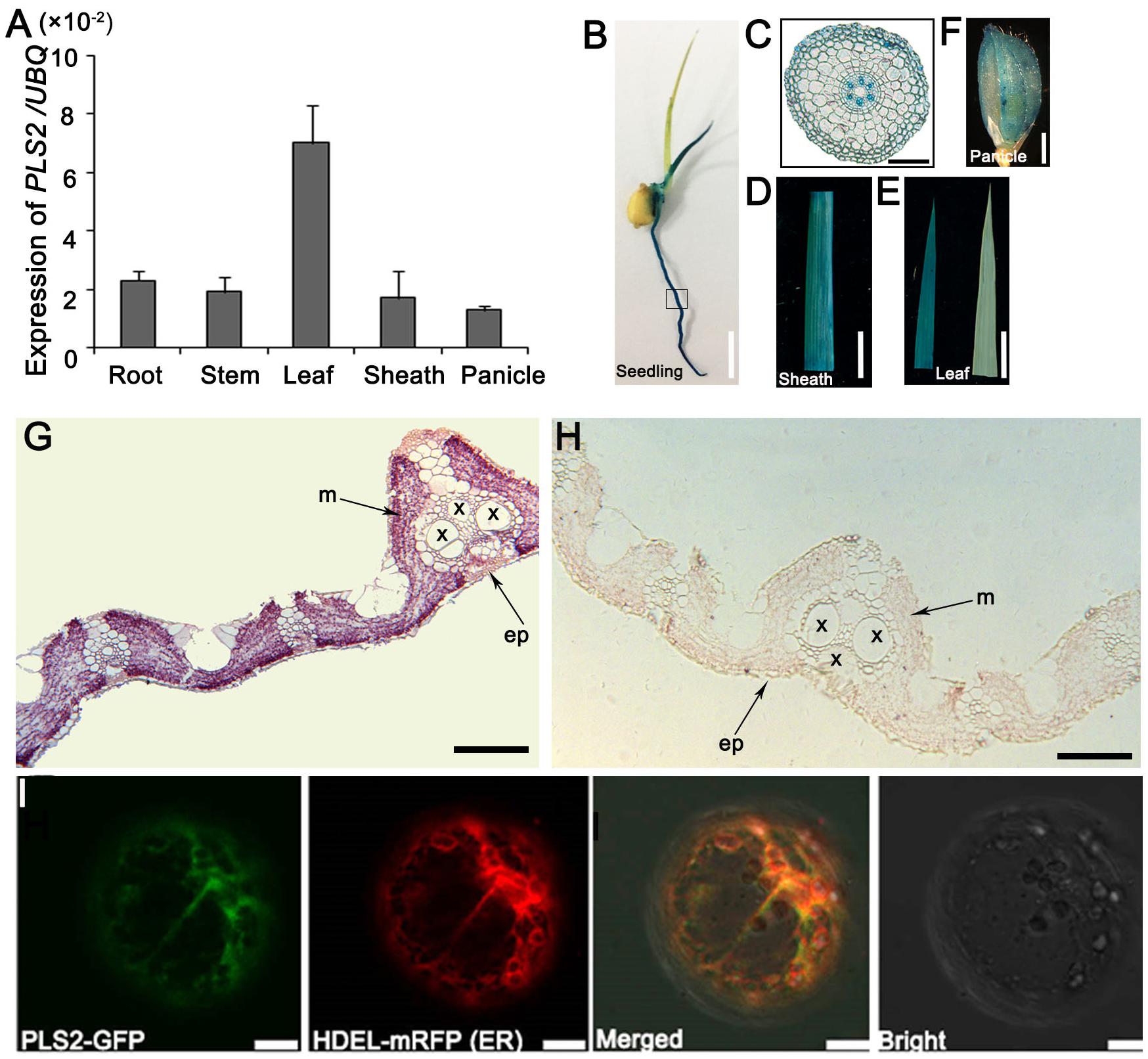
FIGURE 4. Expression and localization of PLS2. (A) Expression levels in various tissues revealed by qRT-PCR using the UBQ as the reference gene. Data is presented as the mean ± standard deviation (n = 9). (B–E) GUS expression patten of PPLS2:GUS transgenic rice plants on 5-day-old seedlings (B), root cross-scection of the box area in B (C), leaf sheath (D), leaf (E; left, PPLS2:GUS transgenic leaf; right, control). (G,H) RNA in situ hybridization analysis of PLS2. Flag leaves of wild type plants at heading were cross-sectioned and hybridized with PLS2-specific antisense (G) or sense (H) probes. x, xylem; p, phloem; ep, epidermis; m, mesophyll. (I) Co-expression of PLS2-GFP fusion protein with HDEL-mRFP (ER marker). ER, endoplasmic reticulum. Scar bars: 12 mm in (B), 100 μm in (C), 1 cm in (D,E), 3 mm in (F), 200 μm in (G,H), 10 μm in (I).
Through TEM assay, we found that cells in green sections of pls2 leaves were heteroplastidic with more starch grains (Figures 5A,B). It is well known that glycosyltransferases catalyze the transfer of activated sugars to various acceptor molecules (Li et al., 2015; Zhang et al., 2016). In order to investigate the impact of PLS2 mutation on the sugar metabolism, sucrose levels in flag leaves at grain filling stage were measured in the WT, pls2, Ubi::PLS2-OE, and CRISPR/Cas9-based knockout-plants. Results revealed that the pls2 accumulated more sucrose than WT whether in their green leaves or senesced leaves (Figure 5C). Transgenic over-expressing lines showed similar sugar levels to WT (Figure 5C). Two CRISPR knockout plants accumulated excess sucrose compared to the control (Figure 5D).
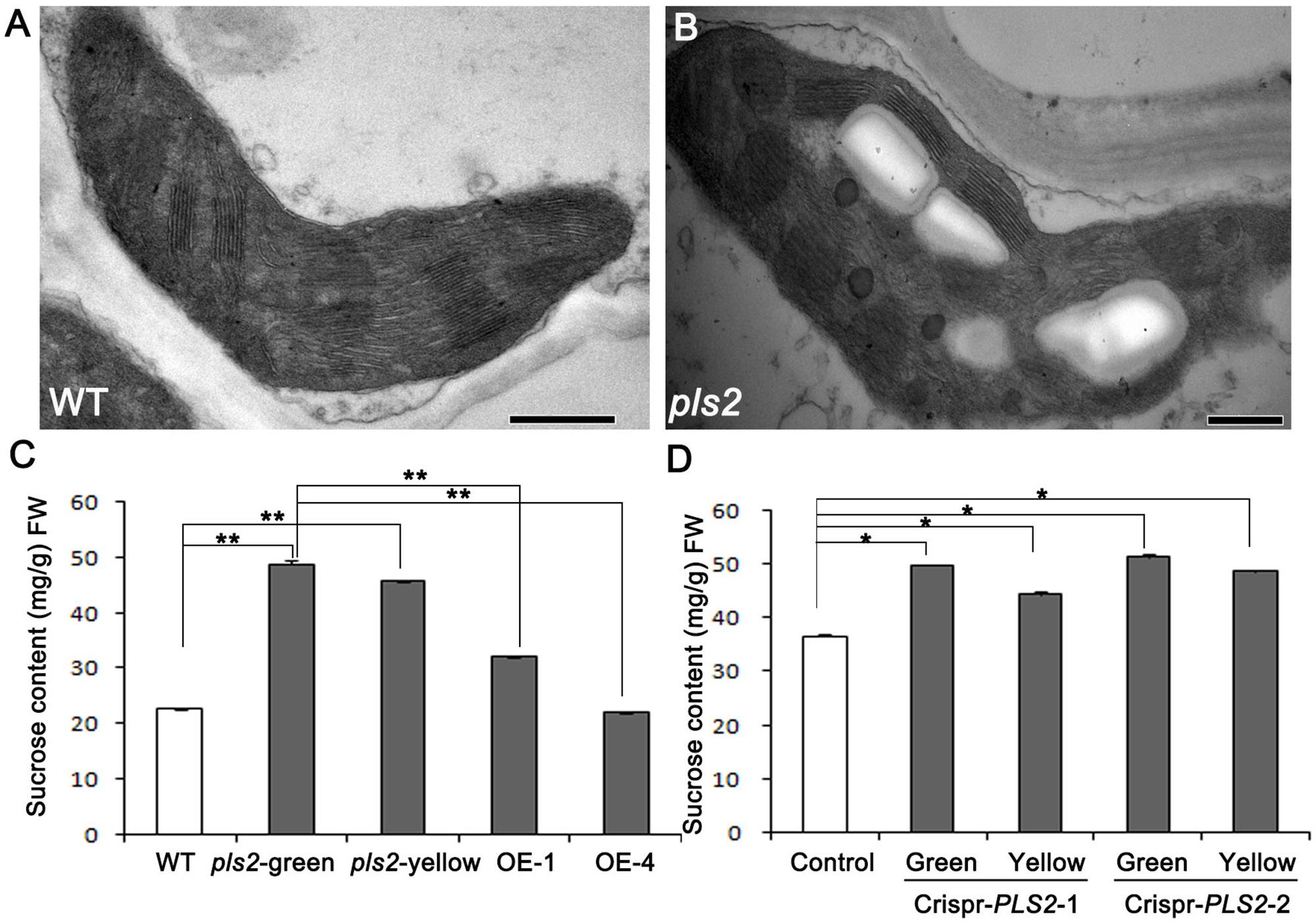
FIGURE 5. Sucrose contents of plants at heading. (A,B) Transmission electron microscopic images of flag leaf cells from wild type (A) and pls2 mutant (B). Bar 500 nm in (A,B). (C) Sucrose content in WT, pls2, and OE lines of OE1 and OE4. (D) Sucrose content in the control and two crispr plants. Data is presented as the mean ± standard deviation (n = 9). ∗0.01 ≤P ≤ 0.05; ∗∗P ≤ 0.01; student’s t-test.
Sucrose synthase (SuSy, EC 2.4.1.13) mainly mediates the sucrose metabolism by catalyzing the reversible transfer of a glucosyl moiety between fructose and a nucleoside diphosphate (NDP) (Chourey et al., 1998; Baroja-Fernández, 2012). There are at least six members (OsSUS1-6) of sucrose synthase family in rice (Tatsuro et al., 2008). We assayed the expression levels of sucrose synthase genes OsSUS1, OsSUS2, OsSUS4, and OsSUS5 (Hirose et al., 2008). qRT-PCR analysis showed that mRNA levels of all except OsSUS4 were remarkably down-regulated in pls2 (Figure 6A), suggesting that lower expression suppresses sucrose turnover and metabolism, and result in sucrose accumulation in pls2. We next detected the expression level of sugar transporters including OsSUT1 (Siahpoosh et al., 2012), OsSUT2 (Siao et al., 2011), OsSWEET4, OsSWEET5 (Zhou et al., 2014), OsSWEET11, OsSWEET14 and OsSWEET15 in WT and the pls2 mutant, respectively, and found that all mRNA levels were significantly down-regulated in pls2 (Figure 6B), suggesting that mutation in the PLS2 gene affected the sugar transport, leading to over-accumulation of starch in chloroplast of pls2.
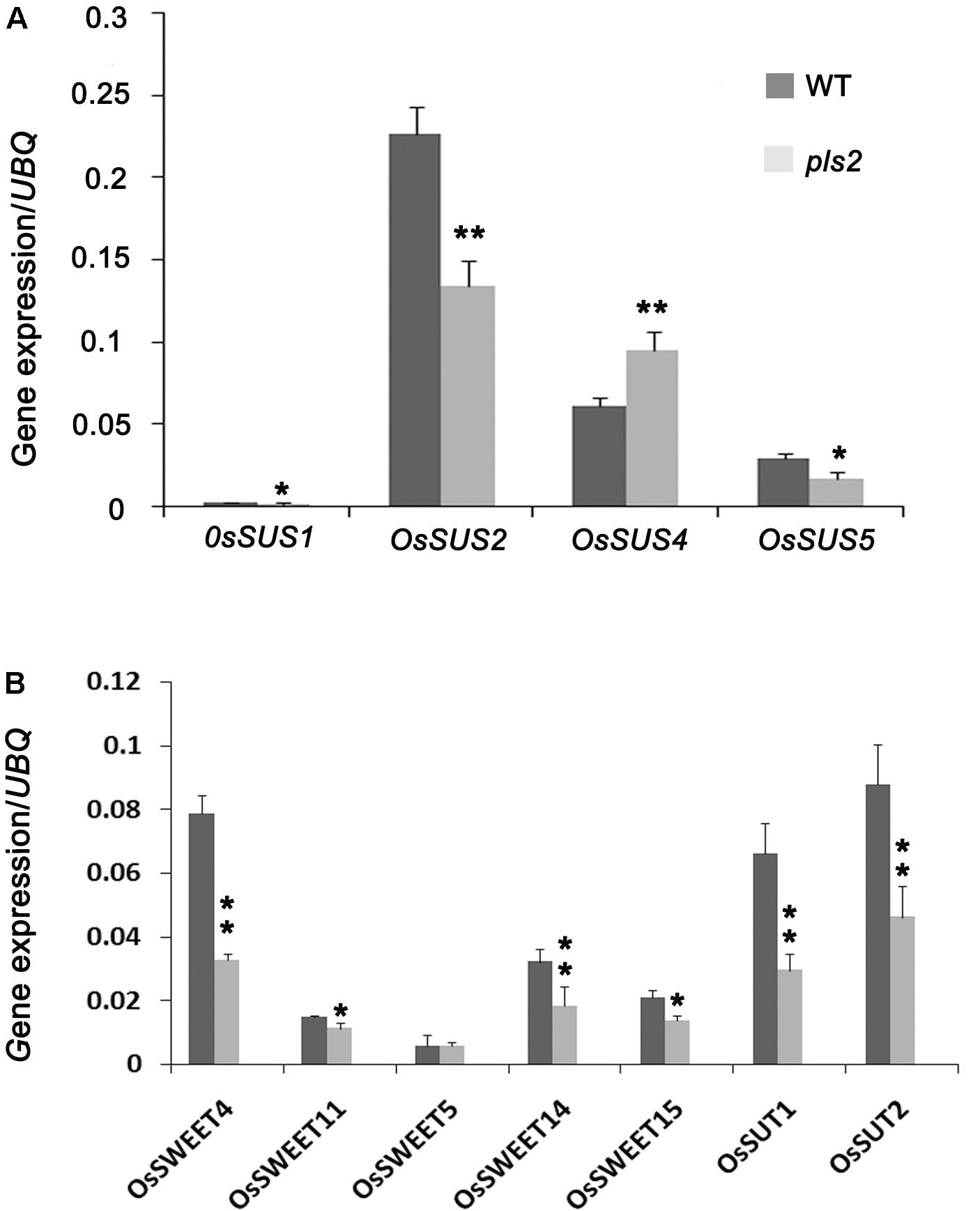
FIGURE 6. Expression analyses of genes associated with sucrose synthesis genes OsSUS1, 2, 4, 5 (A) and sucrose transportation genes OsSWEET4, 5, 11, 14 and OsSUT1, 2 (B). Data is presented as the mean ± standard deviation (n = 9). ∗0.01 ≤P ≤ 0.05, ∗∗P ≤ 0.01, Student’s t-test.
Premature leaf senescence causes detrimental plant growth and reduced crop productivity (Chen L.J. et al., 2013; Chen Y. et al., 2013; Liang et al., 2014). Previous studies on leaf senescence revealed that many genes are involved (Woo et al., 2001; Liang et al., 2014; Schippers et al., 2015) and SAGs usually up-regulated during senescence (Liang et al., 2014; Sakuraba et al., 2015; Yang et al., 2015; Zhao et al., 2015, 2016; Zhu et al., 2015). Our study found that several SAGs such as the pheophorbidea oxygenase gene PAO (Schelbert et al., 2009), chloroplast degradation related gene (SGR) (Hörtensteiner, 2009), and senescence associated genes NYC3 and Osl85 (Lee et al., 2001), were prominently up-regulated in the pls2 mutant, suggesting pls2 was undergoing a typical senescence process. However, identification of more SAGs could be helpful in elucidating the leaf senescence processes involved in normal plant development. Here, we characterized a rice pls2 mutant that displayed PLS under normal conditions and isolated a putative glycosyltransferase encoding gene LOC_Os03g15840by map-based cloning. A mutation in PLS2 was responsible for the defective phenotype. PLS2 expression was detected in all tissues surveyed, but predominantly in leaf mesophyll cells, with a sub-cellular localization of the endoplasmic reticulum. The pls2 mutant accumulated much higher levels of sucrose together with decreased expression of sucrose metabolizing related genes compared to wild type. Our results indicated that the PLS2 is essential for normal leaf senescence and its mutation resulted in the PLS.
Based on the substrate specificity of GT, all GT members can be classified into 105 GT subfamilies in Arabidopsis and 41 OsGT in rice (Ko et al., 2006; Cao et al., 2008). UDP glycosyltransferases are the most common GT enzymes that catalyze glycosylation in the plant kingdom. They transfer donor molecules to specific acceptors and participate in adversities with environmental conditions (Coutinho et al., 2003; Lairson et al., 2008). A recent study shows that excessive UGT results in programmed cell death (PCD) (Xiao et al., 2018). However, information on GT function in PLS is scarce. According to the gene annotation, PLS2 is an unidentified substrate GT and our results provide evidence that abnormal sugar metabolism leads to leaf senescence.
Reactive oxygen species are continuously produced in plants as products of aerobic metabolism (Maurino and Flügge, 2008). Excess ROS accumulation leads to oxidative damage to thylakoid membranes and other cellular components (Apel and Hirt, 2004). Previous studies showed that early leaf senescence is usually associated with excessive ROS (Han et al., 2014). In our study, one species of ROS, H2O2, was much higher in the pls2 mutant than wild type, indicating that H2O2 accumulation in pls2 might result in oxidative impair to thylakoid membranes, and finally causing PLS in the mutant. Previous studies suggested that glycosyltransferases in rice and Arabidopsis were mostly localized in cytoplasm (Dong et al., 2014; Liu et al., 2015). Our investigation with marker HDEL-mRFP (Gomord et al., 1997) indicated PLS2 was completely merged with ER in epidermal protoplasts of N. benthamiana leaves, suggesting that PLS2 perform its role in the ER.
Sugars, especially sucrose, are not only essential carbon and energy sources, but also exert regulatory roles in metabolism control, stress immunity, growth, and development (Rolland et al., 2006, a review). Previous studies showed that exogenous sucrose supplied to leaves affects sugar metabolism and inhibits photosynthesis by down-regulating Rubisco abundance activity in 4-month-old sugarcane (Lobo et al., 2015). Feeding glucose or sucrose to mesophyll protoplasts in basic maize medium decreased photosynthetic gene expression (Sheen, 1990). In addition, leaf senescence was induced by adding glucose in combination with low nitrogen levels in Arabidopsis (Pourtau et al., 2004; Wingler et al., 2004). Over-accumulation of sugar accelerated the synthesis of starch grains in chloroplast stroma, which resulted in oppressing thylakoid and inhibiting light absorption in photosynthetic membranes (?). Here, we found that PLS2 mutation caused significant sucrose accumulation and cells containing chloroplasts were heteroplastidic with starch grain accumulation in pls2, speculating that larger numbers of starch grains in chloroplasts and the descending expression of photosynthesis genes might cause the premature leaf senescence in pls2.
Sucrose is the main form of photosynthetic product and its translocation and distribution are mainly regulated by SUTs. SUTs are mainly involved in phloem loading, long-distance transportation, “library” unloading of sucrose (Kühn et al., 2003; Hackel et al., 2006), and regulating sucrose storage and distribution (Rae et al., 2005; Hackel et al., 2006). One study showed that OsSUT1 expression is inhibited by higher sucrose content and induced by drought and salt stress in excised rice tissues (Ibraheem et al., 2011). An Ossut2 mutant showed sucrose accumulation and lower sucrose output capability that finally disturbed plant development (Eom et al., 2011). When excised tobacco (Krapp et al., 1991) and barley (Parrott et al., 2005) leaves were exposed to strong sunlight more sugar accumulated and caused PLS. The pls2 mutant similarly undergoes sugar accumulation that appears to be the cause of PLS. However, due to lack of knowledge of the substrate of the glycosyltransferase encoded by PLS2 and its interacting protein, the mechanism of senescence underlying pls2 remains to be explored.
MW and ZC designed the experiment. MW, TZ, and HP performed most of the experiments. SL, JT, KJ, YH, XZ, and XG participated in some part of the study. MW wrote the paper. JZ and ZC edited the manuscript.
This research was supported by the grants from the National Transgenic Science and Technology Program (2016ZX08009003-003), and CAAS project of Y2016XT05.
All The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00560/full#supplementary-material
FIGURE S1 | Agronomic trait comparisons between the wild type (WT) and pls2 in plant height (A), and internode lengths (B), seed setting (C), three types of grain (D), their proportions (E), and 1000-grain weight (F). FGN, fully filled grain; SFGN, semi-filled grain; SGN, shriveled grain, bar, 5 mm. Data is presented as the mean ± standard deviation (n = 9). ∗0.01 ≤P ≤ 0.05; ∗∗P ≤ 0.01; Student’s t-test.
FIGURE S2 |PLS2 crispr analyses. (A) Crispr plants in Nipponbare background. Bar, 15 cm. (B) Plant height of crispr plants with the control. (C) Sequence analysis of PLS2 for Crispr lines. ∗∗P ≤ 0.01; student’s t-test. Data is presented as the mean ± standard deviation (n = 9).
FIGURE S3 | Phylogenetic analysis of PLS2 protein and homologous proteins among plant species. (A) The domains of PLS2 protein. (B) Phylogenetic and blast analyses of PLS2 among plant species.
FIGURE S4 | Expression levels in various tissues revealed by qRT-PCR using the OsAction as the reference gene. Data is presented as the mean ± standard deviation (n = 9).
TABLE S1 | Primers used in the study.
Abdelrahman, M., El-Sayed, M., Jogaiah, S., Burritt, D. J., and Tran, L. P. (2017). The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 36, 1009–1025. doi: 10.1007/s00299-017-2119-y
Ahrazem, O., Rubio-Moraga, A., Trapero-Mozos, A., Climent, M. F. L., Gómez-Cadenas, A., and Gómez-Gómez, L. (2015). Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci. 234, 60–73. doi: 10.1016/j.plantsci.2015.02.004
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bai, J. T., Zhu, X. D., Wang, Q., Zhang, J., Chen, H. Q., Dong, G. J., et al. (2015). Rice TUTOU1 encodes a suppressor of cAMP receptor-like protein that is important for actin organization and panicle development. Plant Physiol. 169, 1179–1191. doi: 10.1104/pp.15.00229
Baroja-Fernández, E., Muñoz, F. J., Li, J., Bahaji, A., Almagro, G., Montero, M., et al. (2012). Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. U.S.A. 109, 321–326. doi: 10.1073/pnas.1117099109
Batoko, H., Zheng, H. Q., Hawes, C., and Moore, I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12, 2201–2218. doi: 10.1105/tpc.12.11.2201
Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72, 85–95. doi: 10.1016/0092-8674(93)90052-R
Brushwood, D. E. (1997). “Measurement of sugar on raw cotton by HPLC, individual carbohydrate concentrations and their relationship to stickiness potential,” in Proceedings of the Beltwide Cotton Conferences, New Orleans, LA.
Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., et al. (2003). The molecular analysis of leaf senescence-a genomics approach. Plant Biotechnol. J. 1, 3–22. doi: 10.1046/j.1467-7652.2003.00004.x
Cao, P., Bartley, L. E., Jung, K., and Ronald, P. C. (2008). Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol. Plant 1, 858–877. doi: 10.1093/mp/ssn052
Chen, L. J., Wuriyanghan, H., Zhang, Y. Q., Duan, K. X., Chen, H. W., Li, Q. T., et al. (2013). An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 163, 1752–1765. doi: 10.1104/pp.113.224881
Chen, Y., Xu, Y., Luo, W., Li, W., Chen, N., Zhang, D., et al. (2013). The F-Box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol. 163, 1673–1685. doi: 10.1104/pp.113.224527
Chiniquy, D., Sharma, V., Schultink, A., Baidoo, E. E., Rautengarten, C., Cheng, K., et al. (2012). XAX1 from glycosyltransferase family 61 mediates xylosyltransfer to rice xylan. Proc. Natl. Acad. Sci. U.S.A. 109, 17117–17122. doi: 10.1073/pnas.1202079109
Chourey, P. S., Taliercio, E. W., Carlson, S. J., and Ruan, Y. L. (1998). Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 259, 88–96. doi: 10.1007/s004380050792
Coutinho, P. M., Deleury, E., Davies, G. J., and Henrissat, B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317. doi: 10.1016/S0022-2836(03)00307-3
Dong, T., Xu, Z. Y., Park, Y., Kim, D. H., Lee, Y., and Hwang, I. (2014). Abscisic acid uridine diphosphate glucosyltransferases play a crucial role in abscisic acid homeostasis in Arabidopsis. Plant Physiol. 165, 277–289. doi: 10.1104/pp.114.239210
Draper, H. H., and Hadley, M. (1989). Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 186, 421–431. doi: 10.1016/0076-6879(90)86135-I
Eom, J. S., Cho, J. I., Reinders, A., Lee, S. W., Yoo, Y., Tuan, P. Q., et al. (2011). Impaired function of the tonoplast-localized sucrose transporter in rice, ossut2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 157, 109–119. doi: 10.1104/pp.111.176982
Gao, Q., Yang, Z., Zhou, Y., Yin, Z., Qiu, J., Liang, G., et al. (2012). Characterization of an Abc1 kinase family gene OsABC1-2 conferring enhanced tolerance to dark-induced stress in rice. Gene 498, 155–163. doi: 10.1016/j.gene.2012.02.017
Gomord, V., Denmat, L. A., FitchetteLaine, A. C., SatiatJeunemaitre, B., Hawes, C., and Faye, L. (1997). The C-terminalHDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) butpromotes vacuolar targeting of proteins that escape the ER. Plant J. 11, 313–325. doi: 10.1046/j.1365-313X.1997.11020313.x
Gregersen, P. L., Culetic, A., Boschian, L., and Krupinska, K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82, 603–622. doi: 10.1007/s11103-013-0013-8
Guleria, P., and Yadav, S. K. (2014). Overexpression of a glycosyltransferase gene SrUGT74G1 from Stevia improved growth and yield of transgenic Arabidopsis by catechin accumulation. Mol. Biol. Rep. 41, 1741–1752. doi: 10.1007/s11033-014-3023-y
Hackel, A., Schauer, N., Carrari, F., Fernie, A. R., Grimm, B., and Kühn, C. (2006). Sucrose transporter lesut1 and lesut2 inhibition affects tomato fruit development in different ways. Plant J. Cell Mol. Biol. 45, 180–192. doi: 10.1111/j.1365-313X.2005.02572.x
Han, M., Kim, C., Lee, J., Lee, S., and Jeon, J. (2014). OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol. Cell 37, 532–539. doi: 10.14348/molcells.2014.0128
Hiei, Y., and Komari, T. (2008). Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834. doi: 10.1038/nprot.2008.46
Hirose, T., Scofield, G. N., and Terao, T. (2008). An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174, 534–543. doi: 10.1016/j.plantsci.2008.02.009
Hörtensteiner, S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14, 155–162. doi: 10.1016/j.tplants.2009.01.002
Ibraheem, O., Dealtry, G., Roux, S., and Bradley, G. (2011). The effect of drought and salinity on the expressional levels of sucrose transporters in rice (Oryza sativa Nipponbare) cultivar plants. Plant Omics 4, 68–74.
Jefferson, R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Jiao, B., Wang, J., Zhu, X., Zeng, L., Li, Q., and He, Z. (2012). A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant 5, 205–217. doi: 10.1093/mp/ssr081
Jones, P., and Vogt, T. (2001). Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta 213, 164–174. doi: 10.1007/s004250000492
Ko, J. H., Kim, B. G., Hur, H., Lim, Y., and Ahn, J. (2006). Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep. 25, 741–746. doi: 10.1007/s00299-006-0119-4
Kong, Z., Li, M., Yang, W., Xu, W., and Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. doi: 10.1104/pp.106.082941
Krapp, A., Quick, W., and Stitt, M. (1991). Ribulose-1, 5-biphosphate carboxylase/oxygenase, other Calvin enzymes, and chlorophyll decrease when glucose is supplied to mature spinach leaves via the transpiration stream. Planta 186, 58–69. doi: 10.1007/BF00201498
Kühn, C., Fernie, A. R., Roessner-Tunali, U., Hirner, B., and Frommer, W. B. (2003). The sucrose transporter stsut1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 131, 102–113. doi: 10.1104/pp.011676
Lairson, L. L., Henrissat, B., Davies, G. J., and Withers, S. G. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555. doi: 10.1146/annurev.biochem.76.061005.092322
Lee, C., Teng, Q., Zhong, R., Yuan, Y., and Ye, Z. H. (2014). Functional roles of rice glycosyltransferase family GT43 in xylan biosynthesis. Plant Signal. Behav. 9:e27809. doi: 10.4161/psb.27809
Lee, R. H., Wang, C. H., Huang, L. T., and Chen, S. C. (2001). Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 52, 1117–1121. doi: 10.1093/jexbot/52.358.1117
Leng, Y., Yang, Y., Ren, D., Huang, L., Dai, L., Wang, Y., et al. (2017). A rice pectate lyase-like gene is required for plant growth and leaf senescence. Plant Physiol. 174, 1151–1166. doi: 10.1104/pp.16.01625
Li, Y., Wang, B., Dong, R., and Hou, B. (2015). AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Sci. 236, 157–167. doi: 10.1016/j.plantsci.2015.04.002
Li, Z., Peng, J., Wen, X., and Guo, H. (2012). Gene network analysis and functional studies of senescence- associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 54, 526–539. doi: 10.1111/j.1744-7909.2012.01136.x
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, Y. H., Tan, L. B., Zhao, L., Sun, X. Y., and Sun, C. Q. (2016). RLS3, a protein with AAAt domain localized in chloroplast, sustains leaf longevity in rice. J. Integr. Plant Biol. 58, 971–982. doi: 10.1111/jipb.12487
Liu, Z., Yan, J., Li, D., Luo, Q., Yan, Q., Liu, Z., et al. (2015). UDP-glucosyltransferase71C5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 167, 1659–1670. doi: 10.1104/pp.15.00053
Lobo, A. K. M., de Oliveira Martins, M., Lima Neto, M. C., Machado, E. C., Ribeiro, R. V., and Silveira, J. A. G. (2015). Exogenous sucrose supply changes sugar metabolism and reduces photosynthesis of sugarcane through the down-regulation of Rubisco abundance and activity. J. Plant Physiol. 179, 113–121. doi: 10.1016/j.jplph.2015.03.007
Maurino, V. G., and Flügge, U. I. (2008). Experimental systems to assess the effects of reactive oxygen species in plant tissues. Plant Signal. Behav. 3, 923–928. doi: 10.4161/psb.7036
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Miao, Y., Yan, P. K., Kim, H., Hwang, I., and Jiang, L. (2006). Localization of green fluorescent protein fusions with the seven Arabidopsis vacuolar sorting receptors to prevacuolar compartments in tobacco BY-2 cells. Plant Physiol. 142, 945–962. doi: 10.1104/pp.106.083618
Mishra, M. K., Singh, G., Tiwari, S., Singh, R., Kumari, N., and Misra, P. (2015). Characterization of Arabidopsis sterol glycosyltransferase TTG15/UGT80B1 role during freeze and heat stress. Plant Signal. Behav. 10:e1075682. doi: 10.1080/15592324.2015.1075682
Park, S. Y., Yu, J. W., Park, J. S., Li, J. J., Yoo, S. C., Lee, N. Y., et al. (2007). The senescence-induced Stay green protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Parrott, D., Yang, L., Shama, L., and Fischer, A. M. (2005). Senescence is accelerated, and several proteases are induced by carbon “feast” conditions in barley (Hordeum vulgare L.) leaves. Planta 222, 989–1000. doi: 10.1007/s00425-005-0042-x
Pilkington, S. M., Montefiori, M., Jameson, P. E., and Allan, A. C. (2012). The control of Chlorophyll levels in maturing kiwifruit. Planta 236, 1615–1628. doi: 10.1007/s00425-012-1723-x
Pourtau, N., Marès, M., Purdy, S., Quentin, N., Ruel, A., and Wingler, A. (2004). Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219, 765–772. doi: 10.1007/s00425-004-1279-5
Rae, A. L., Perroux, J. M., and Grof, C. P. (2005). Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem: a potential role for the ShSUT1 sucrose transporter. Planta 220, 817–825. doi: 10.1007/s00425-004-1399-y
Rao, M. V., Lee, H., Creelman, R. A., Mullet, J. E., and Davis, K. R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646. doi: 10.1105/tpc.12.9.1633
Rao, Y., Yang, Y., Xu, J., Li, X., Leng, Y., Dai, L., et al. (2015). EARLY SENESCENCE1 encodes a SCAR-LIKE PROTEIN2 that affects water loss in rice. Plant Physiol. 169, 1225–1239. doi: 10.1104/pp.15.00991
Ren, G., An, K., Liao, Y., Zhou, X., Cao, Y., Zhao, H., et al. (2007). Identification of a novel chloroplast protein AtNYE1 regulating Chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144, 1429–1441. doi: 10.1104/pp.107.100172
Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Sakuraba, Y., Piao, W., Lim, J., Han, S., Kim, Y., An, G., et al. (2015). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339. doi: 10.1093/pcp/pcv144
Schelbert, S., Aubry, S., Burla, B., Agne, B., Kessler, F., Krupinska, K., et al. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll break down during leaf senescence in Arabidopsis. Plant Cell 21, 767–785. doi: 10.1105/tpc.108.064089
Schippers, J. H. M., Schmidt, R., Wagstaff, C., and Jing, H. (2015). Living to die and dying to live: the survival strategy behind leaf senescence. Plant Physiol. 169, 914–930. doi: 10.1104/pp.15.00498
Sheen, J. (1990). Metabolic repression of transcription in higher plants. Plant Cell 2, 1027–1038. doi: 10.1105/tpc.2.10.1027
Siahpoosh, M. R., Sanchez, D. H., Schlereth, A., Scofield, G. N., Furbank, R. T., van Dongen, J. T., et al. (2012). Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309. Plant Sci. 182, 101–111. doi: 10.1016/j.plantsci.2011.01.001
Siao, W., Chen, J., Hsiao, H., Chung, P., and Wang, S. (2011). Characterization of OsSUT2 expression and regulation in germinating embryos of rice seeds. Rice 4, 39–49. doi: 10.1007/s12284-011-9063-1
Singh, A., Singh, U., Mittal, D., and Grover, A. (2010). Transcript expression and regulatory characteristics of a rice glycosyltransferase OsGT61-1 gene. Plant Sci. 179, 114–122. doi: 10.1016/j.plantsci.2010.03.005
Sun, Y. G., Wang, B., Jin, S. H., Qu, X. X., Li, Y. J., and Hou, B. K. (2013). Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS One 8:e59924. doi: 10.1371/journal.pone.0059924
Suzuki, Y., and Makino, A. (2012). Availability of rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol. 160, 533–540. doi: 10.1104/pp.112.201459
Tatsuro, H., Grahamn, S., and Tomio, T. (2008). An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 174, 534–543. doi: 10.1016/j.plantsci.2008.02.009
Telzur, N., Abbo, S., Myslabodski, D., and Mizrahi, Y. (1999). Modified CTAB procedure for DNA isolation from epiphytic cacti of the genera Hylocereus and Selenicereus (Cactaceae). Plant Mol. Biol. Rep. 17, 249–254. doi: 10.1023/A:1007656315275
Thomas, H., and Howarth, C. J. (2000). Five ways to stay green. J. Exp. Bot. 51, 329–337. doi: 10.1093/jexbot/51.suppl_1.329
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tognetti, V. B., Van Aken, O., Morreel, K., Vandenbroucke, K., van de Cotte, B., De Clercq, I., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679. doi: 10.1105/tpc.109.071316
van Doorn, W. G. (2008). Is the onset of senescence in leaf cells of intact plants due to low or high sugar levels? J. Exp. Bot. 59, 1963–1972. doi: 10.1093/jxb/ern076
von Saint Paul, V., Zhang, W., Kanawati, B., Geist, B., Faus-Keßler, T., Schmitt-Kopplin, P., et al. (2012). The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23, 4124–4145. doi: 10.1105/tpc.111.088443
Wang, B., Ma, M., Lu, H., Meng, Q., Li, G., and Yang, X. (2015). Photosynthesis, sucrose metabolism, and starch accumulation in two NILs of winter wheat. Photosynth. Res. 126, 363–373. doi: 10.1007/s11120-015-0126-9
Wingler, A., Mare‘s, M., and Pourtau, N. (2004). Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol. 161, 781–789. doi: 10.1111/j.1469-8137.2004.00996.x
Woo, H. R., Chung, K. M., Park, J. H., Oh, S. A., Ahn, T., Hong, S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. doi: 10.1105/tpc.13.8.1779
Wu, X. Y., Kuai, B. K., Jia, J. Z., and Jing, H. C. (2012). Regulation of leaf senescence and crop genetic inprovement. J. Integr. Plant Biol. 54, 936–952. doi: 10.1111/jipb.12005
Xiao, G., Zhou, J., Lu, X., Huang, R., and Zhang, H. (2018). Excessive UDPG resulting from the mutation of UAP1 causes programmed cell death by triggering reactive oxygen species accumulation and caspase-like activity in rice. New Phytol. 217, 332–343. doi: 10.1111/nph.14818
Yang, J., Worley, E., and Udvardi, M. (2015). A NAP-AAO3 regulatory module promotes chlorophyll degradation via ABA biosynthesis in Arabidopsis leaves. Plant Cell 26, 4862–4874. doi: 10.1105/tpc.114.133769
Zhang, B., Zhao, T., Yu, W., Kuang, B., Yao, Y., Liu, T., et al. (2014). Functional conservation of the glycosyltransferase gene GT47A in the monocot rice. J. Plant Res. 127, 423–432. doi: 10.1007/s10265-014-0631-5
Zhang, G., Jin, S., Jiang, X., Dong, R., Li, P., Li, Y., et al. (2016). Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol. Biol. 90, 77–93. doi: 10.1007/s11103-015-0395-x
Zhang, T., Sun, Y. Y., Zheng, J. M., Cheng, Z. J., Jiang, K. F., Yang, L., et al. (2014). Genetic analysis and fine mapping of a premature leaf senescence mutant in rice (Oryza sativa L.). Acta Agron. Sin. 40, 2070–2080. doi: 10.3724/SP.J.1006.2014.02070
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Zhao, D., Derkx, A. P., Liu, D. C., Buchner, P., and Hawkesford, M. J. (2015). Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 17, 904–913. doi: 10.1111/plb.12296
Zhou, Y., Liu, L., Huang, W., Yuan, M., Zhou, F., Li, X., et al. (2014). Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One 9:e94210. doi: 10.1371/journal.pone.0094210
Keywords: glycosyltransferase, GT, leaf senescence, Oryza sativa, PLS2, sucrose
Citation: Wang M, Zhang T, Peng H, Luo S, Tan J, Jiang K, Heng Y, Zhang X, Guo X, Zheng J and Cheng Z (2018) Rice Premature Leaf Senescence 2, Encoding a Glycosyltransferase (GT), Is Involved in Leaf Senescence. Front. Plant Sci. 9:560. doi: 10.3389/fpls.2018.00560
Received: 08 February 2018; Accepted: 10 April 2018;
Published: 26 April 2018.
Edited by:
Gerrit T. S. Beemster, University of Antwerp, BelgiumReviewed by:
Henry Christopher Janse van Rensburg, KU Leuven, BelgiumCopyright © 2018 Wang, Zhang, Peng, Luo, Tan, Jiang, Heng, Zhang, Guo, Zheng and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiakui Zheng, emhlbjYxMDJAMTI2LmNvbQ== Zhijun Cheng, Y2hlbmd6aGlqdW5AY2Fhcy5jbg==
† These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.