- 1Institute of Biomembranes, Bioenergetics and Molecular Biotechnologies, Consiglio Nazionale delle Ricerche, Bari, Italy
- 2Department of Biosciences, Biotechnology and Biopharmaceutics, University of Bari A. Moro, Bari, Italy
RNA editing is an important epigenetic mechanism by which genome-encoded transcripts are modified by substitutions, insertions and/or deletions. It was first discovered in kinetoplastid protozoa followed by its reporting in a wide range of organisms. In plants, RNA editing occurs mostly by cytidine (C) to uridine (U) conversion in translated regions of organelle mRNAs and tends to modify affected codons restoring evolutionary conserved aminoacid residues. RNA editing has also been described in non-protein coding regions such as group II introns and structural RNAs. Despite its impact on organellar transcriptome and proteome complexity, current primary databases still do not provide a specific field for RNA editing events. To overcome these limitations, we developed REDIdb a specialized database for RNA editing modifications in plant organelles. Hereafter we describe its third release containing more than 26,000 events in a completely novel web interface to accommodate RNA editing in its genomics, biological and evolutionary context through whole genome maps and multiple sequence alignments. REDIdb is freely available at http://srv00.recas.ba.infn.it/redidb/index.html
Introduction
RNA editing is an essential co/post transcriptional process able to expand transcriptome and proteome diversity in addition to alternative splicing. The term RNA editing was first introduced in 1986 to describe the addition and deletion of uridine nucleotides to and from mRNAs in trypanosome mitochondria (Benne et al., 1986). Since then, RNA editing events have been found in a wide range of organisms and can occur in the nucleus and cytoplasm as well as in organelles (Bowe and depamphilis, 1996). Modifications due to RNA editing comprise nucleotide substitutions and insertions or deletions that can affect both protein coding and Non-protein coding RNAs (Maier et al., 1996; Steinhauser et al., 1999).
In humans, the most prevalent type of RNA editing event is the deamination of adenosine (A) in inosine (I) in double RNA strands (dsRNAs) through the catalytic activity of the adenosine deaminase (ADAR) family of enzymes. To date, more than 4 million events have been collected and annotated in dedicated resources such as DARNED, RADAR, and REDIportal (Kiran et al., 2013; Ramaswami and Li, 2014; Picardi et al., 2017).
In plants, RNA editing occurs mostly in organelles in the form of cytidine (C) to uridine (U) conversion particularly in translated regions of mRNAs, albeit the opposite event (U-to-C substitutions) has been observed in some taxa, especially in chloroplasts RNAs (Takenaka et al., 2013). Plant RNA editing sites are recognized by specific pentatricopeptide repeat (PPR) proteins that are encoded in the nuclear genome. In flowering plants, the editosome machinery requires several additional Non-PPR protein factors, even though its molecular assembly has yet to be clarified (Sun et al., 2016).
Most of the C-to-U changes in the protein coding regions tends to modify affected codons restoring evolutionary conserved aminoacid residues (Gray, 2003). Therefore, plant RNA editing is believed to act as an additional proofreading mechanism to generate fully functional proteins. Occasionally, C-to-U modifications occur in untranslated regions, structural RNAs and intervening sequencing, affecting splicing and translation efficiency. Indeed, RNA editing changes in the domain V of plant group II introns is mandatory for the splicing process (Castandet et al., 2010).
With the advent of high-throughput sequencing technologies, many complete plant organellar genomes have been released and numerous novel RNA editing events uncovered. Nevertheless, RNA editing changes are not always correctly or completely annotated in primary databases (GenBank, ENA and DDBJ) and an appropriate field to unambiguously describe them is not provided. RNA editing modifications are often reported as misc_feature or even as simple exception notes. With the aim to overcome these limitations and create a cured catalog of plant RNA editing events, we developed the specialized REDIdb database. Its first release stored 9,964 modifications distributed over 706 different nucleotide sequences, increased to 11,897 in the following update.
After 10 years of massively parallel sequencing, we present here REDIdb 3.0, an upgraded release that annotates 26,618 RNA editing events distributed among 281 organisms and 85 complete organellar genomes.
All changes have been recovered from Genbank and literature using a semi-automated bioinformatics procedure in which each annotation has been manually checked to avoid redundancy or inconsistencies due to errors in flatfiles.
The web-interface was totally restyled and developed using the latest computational technologies in the field of database querying and managing.
Furthermore, many computational facilities have been integrated to improve the user experience and ensure continuous and future updates of the database. Indeed, REDIdb 3.0 accommodates RNA editing in its genomics, biological and evolutionary context through whole genome maps and multiple sequence alignments.
Although a variety of RNA editing databases have been released such as DARNED (Kiran et al., 2013), RADAR (Ramaswami and Li, 2014), and REDIportal (Picardi et al., 2017), REDIdb is the only one devoted to editing changes in plant organelles. Indeed, similar resources such as dbRES (He et al., 2007), RESOPS (Yura et al., 2009), ChloroplastDB (Cui et al., 2006), or GOBASE (O'Brien et al., 2009) have been dismissed or not updated.
Materials and Methods
All editing events stored in REDIdb derive from GenBank flatfiles through a semi-automated parsing algorithm implemented in custom python (2.7.13) scripts. Each flatfile is screened for RNA editing features using the SeqIO parser included in the Biopython (1.68) module (Cock et al., 2009).
All annotations have been manually checked to identify and correct potential errors, taking into account other related flatfile fields or literature. REDIdb database is organized in MySQL tables and queries are in python employing the MySQL-python (1.2.5) module, a data access library to MySQL engine. The web interface, instead, is built in BootStrap (3.3.7), while data presentation is based on DataTables, an ad hoc Javascript library (1.10.13) to efficiently show large tables in html documents. Genome rendering, available for complete organellar genomes, has been developed in pure python, mimicking OGDraw graphics (Lohse et al., 2013).
Query results are dynamically generated using the CGI (common gateway interface) technology. Multiple sequence alignments of edited cDNAs and proteins have been generated by ClustalOmega (Sievers et al., 2011) and displayed in html pages through the MSAViewer (Yachdav et al., 2016), a JavaScript component of the BioJS collection (https://biojs.net/).
The distribution of RNA editing events along functional domains and predicted protein secondary structures are shown by the feature-viewer JavaScript library (https://github.com/calipho-sib/feature-viewer) based on the powerful D3 JavaScript library for visualizing data using web standards (https://d3js.org/). Functional domains have been detected using InterPro engine (Jones et al., 2014), while protein secondary structures have been predicted using the stand-alone version of Spider2 program (Yang et al., 2017).
All the scripts to parse multiple alignments, InterPro html files and Spider2 outputs have been created in Python. Scripts used to extract RNA editing positions from Genbank flatfiles are freely available at the REDIdb help page. Additional details and supplementary scripts are available upon request.
Results
Database Content
Previous REDIdb release contained 11,897 editing events distributed over 198 organisms and 929 different nucleotide sequences. This upgraded version, instead, collects more than 26,000 editing events from 281 organisms, 85 complete organellar genomes and 3,467 sequences. REDIdb 3.0 includes 26,545 events in protein coding sequences and 73 in untranslated regions, structural RNAs and introns. The vast majority of editing changes occur in the mitochondrion, accounting for a total of 23,553 events over 2,300 sequences.
The most recurrent RNA editing modification is the C-to-U substitution, that accounts for more than 92% of all annotated events and, when located in protein coding regions, tends to modify the aminoacid coded by the edited codon. Indeed, the majority of RNA editing events affects the first and second codon position leading to aminoacid changes resulting the most conserved in the comparison with related orthologs.
Differently from the previous releases, the novel REDIdb database annotates 85 complete organellar genomes. Of these 57 are mitochondrial genomes and include 7791 events. As reported in Table 1, the most edited mitochondrial genomes are those from Liriodendron tulipifera, Nelumbo nucifera and Ginkgo biloba with 888, 847, and 717 events, respectively. Of 27 annotated chloroplast genomes, instead, the one from Anthoceros formosae comprising 564 modifications results the richest in editing events.
All REDIdb sequences including RNA editing events are identified by unique accession numbers (e.g., EDI0000.). To preserve the full compatibility with previous database versions, accession numbers linked to old entries have been maintained unchanged.
Query Form and Output Tables
REDIdb implements a modular query form (Figure 1A) allowing users to make flexible searches by selecting the organism or the intracellular location or the gene name. Regarding nucleotide sequences, users can retrieve the original sequence submitted to the primary database or the RefSeq version or both. In addition, the search can be limited to full open reading frames and include individual exons in case of interrupted genes.
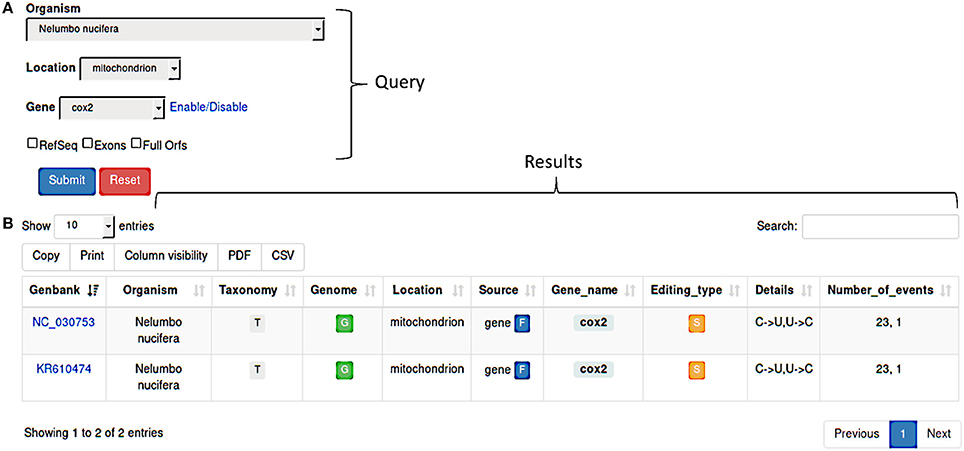
Figure 1. (A) REDIdb query form. Searches can be performed by Organism, Location, Gene or a combination of them. Additional filters (RefSeq, Exons, Full Orfs) are also available in order to refine the results. (B) Once a query has been submitted, the corresponding results are displayed in a sortable and exportable table report.
Query results are shown in a sortable and exportable summary table (Figure 1B) comprising several info such as the GenBank accession number, the organism and the link to the related taxonomy, the organelle type and the link to the complete genome (if available), the gene name and a flag indicating its partial or full nature, the editing types and details and the total number of events. Column can be selectively included in the final table and results are downloadable in pdf or csv format. The “Taxonomy” column includes a link to an interactive taxonomy chart, while the “Genome” column contains a link to the complete genome (if available in primary databases) chart in which RNA editing events are displayed in their genomics context.
Using the link in the “Gene_name” column, users can browse individual RNA editing events organized in flatfiles.
Entry Organization
RNA editing events stored in REDIdb are organized in specific flat-files comprising four main sections. The first section (Figure 2A) contains a general description of the entry including the organism name, the taxonomy (according with the NCBI Taxonomy database), the GenBank and PubMed accession numbers, the intracellular location (mitochondrion or chloroplast) and the official gene name.
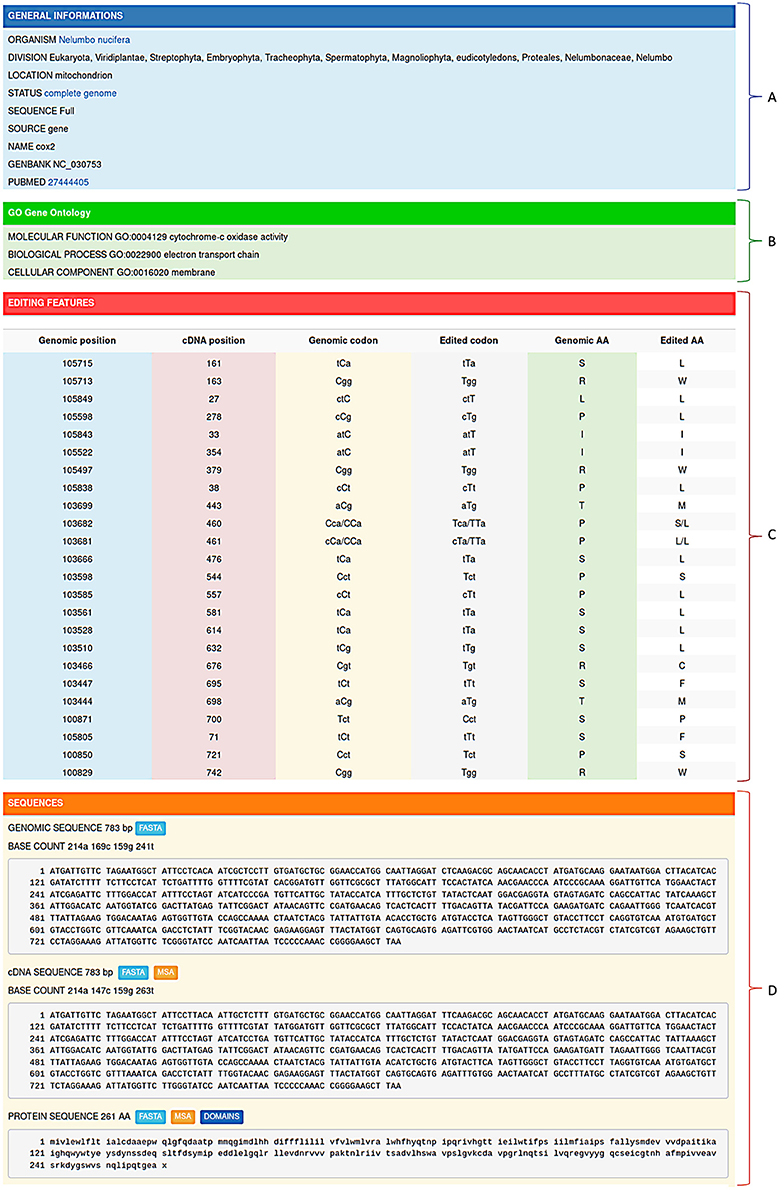
Figure 2. Editing informations stored in REDIdb are organized in specific flat-files in which it is possible to distinguish a header (A) containing the main features of the record (organism, Genbank accession, intracellular location, gene name, PubMed references, ecc.), a gene ontology box (B) describing the gene product properties, a feature table (C) with all the editing events and a sequence zone (D) with both the genomic sequence and the corresponding edited transcript/protein.
The second section (Figure 2B) is devoted to Gene Ontologies (GO), obtained by matching each protein sequence contained in REDIdb against the InterPro database (Finn et al., 2017). In the case of protein coding genes, it contains information regarding the molecular functions, the biological processes and the cellular localization of the protein product. The third section (Figure 2C) shows all the editing features that characterize the record. Here, for each editing event the position on the transcript is reported and, if the complete reference genome is available, also the genomic location. In case of editing within protein coding genes, the genomic codon, edited codon and aminoacidic change are determined and reported. Finally, the fourth section (Figure 2D) contains the genomic sequence and the corresponding edited transcript. In coding protein genes, also the edited protein is displayed. Genomic sequences as well as edited transcripts and proteins can be retrieved in Fasta format.
Graphical Visualization
Edited cDNA and protein sequences can be explored in their evolutionary context through multiple alignments of available orthologs sequences. Since plant RNA editing tends to increase the sequence conservation along the evolution, annotated RNA editing changes are marked and visualized in the multiple alignment by the MSAViewer, to give rise to conservation levels and provide valuable comparative genomics information (Figure 3A).
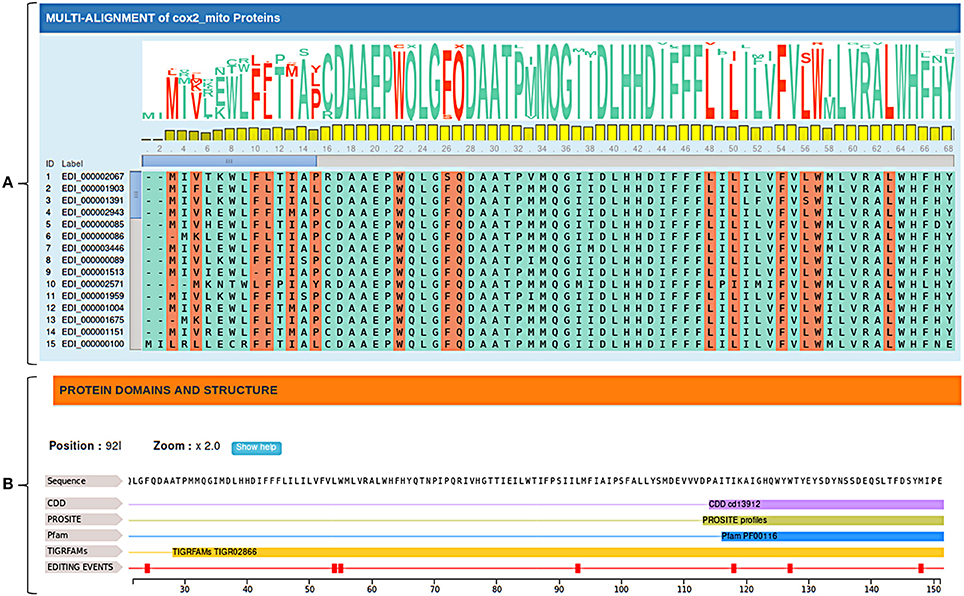
Figure 3. (A) Multiple alignments of cox2 orthologous sequences. Editing conservation across species can be easily obtained considering both the sequence logo and the bar chart relative to each position. (B) Protein domains and structure of Nelumbo nucifera's cox2 protein. Protein domains are obtained by querying multiple databases (CDD, PROSITE, Pfam ecc.).
In addition, RNA editing events are displayed along the edited sequence showing known functional domains and predicted secondary protein structures in order to better interpret the biological role of specific C-to-U or U-to-C changes (Figure 3B).
In case of complete organellar genomes, each genome is graphically rendered and edited genes can be selectively highlighted. Genome graphs are generated in SVG and include links to edited genes by mousing over. Further statistics such as the coding potential of the genome as well as the fraction of edited genes are also reported (Figure 4).
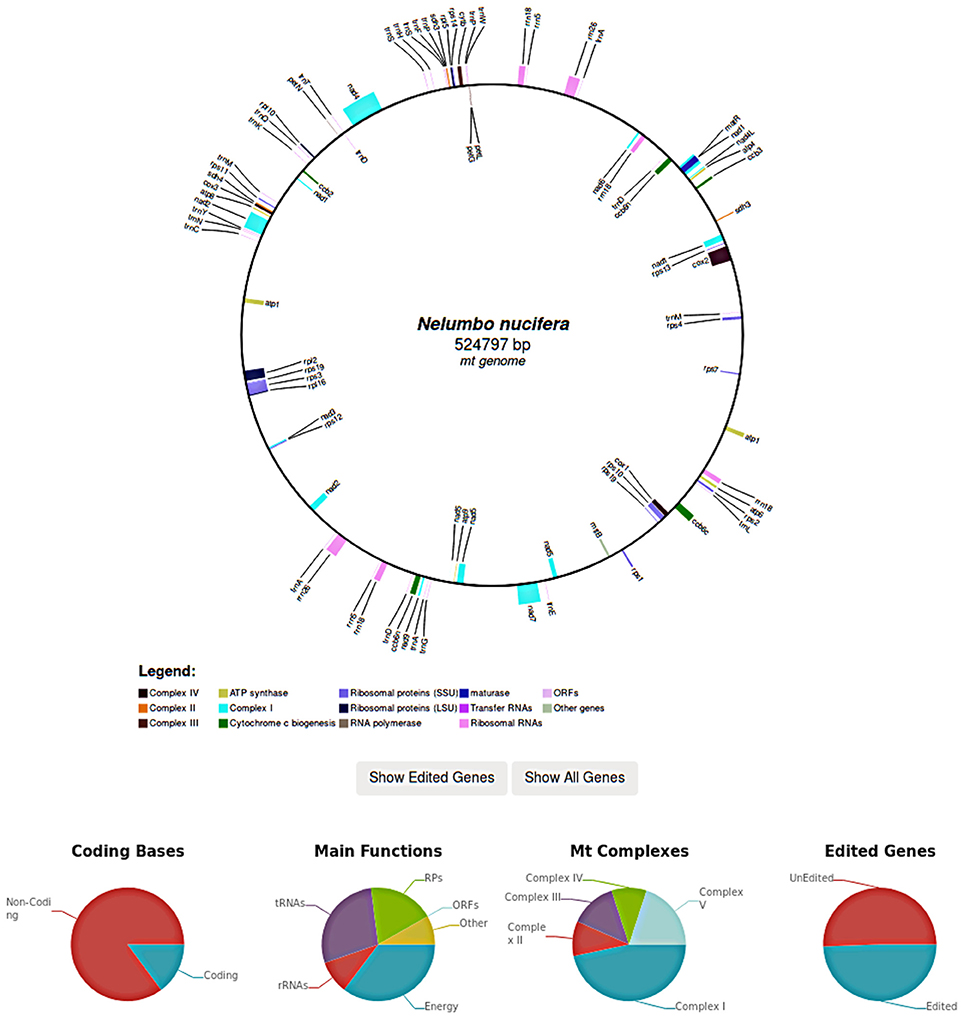
Figure 4. In REDIdb 3.0 complete genomes are graphically rendered, allowing users to visualize the reciprocal order of the edited genes. Further statistics such as the coding potential of the genome as well as the fraction of edited genes are reported in the same page.
Conclusions and Perspectives
As already mentioned, RNA editing plays an important role in transcriptome and proteome diversity. Since its first discovery in 1986 (Benne et al., 1986), a large number of events have been found in a wide range of eukaryotic organisms (Ichinose and Sugita, 2016). Only in humans more than 4 million events have been reported and dedicated resources such as DARNED, RADAR, and REDIportal have been developed to contain them into suitable specialized databases (Kiran et al., 2013; Ramaswami and Li, 2014; Picardi et al., 2017).
In the plant kingdom, RNA editing was first identified as C-to-U substitutions in mitochondrial transcripts (Hiesel et al., 1989), followed by its identification also in chloroplasts (Höch et al., 1991). In order to maintain a cured catalog of such events, we developed the specialized REDIdb database. Its third release, described here, contains three times more entries than the first version and two times more entries than the second version. To date, REDIdb is the unique bioinformatics resource collecting plant organellar RNA editing events. Indeed, similar databases such as dbRES (He et al., 2007) or RESOPS (Yura et al., 2009) have been dismissed or are no more updated. Plant RNA editing events are also annotated in CloroplastDB (Cui et al., 2006), devoted to chloroplast genomes, and GOBASE (O'Brien et al., 2009), the organelle genome database. However, such resources are not specialized for RNA editing and include potential not fixed errors due to the lack of manual curation (Picardi et al., 2011).
REDIdb 3.0 has been completely redrawn keeping in mind the simplicity as its working principle. RNA editing events are always shown in their biological context and novel graphical facilities have been added. Edited genes are now depicted in complete genome maps and RNA editing conservation can be investigated in pre-calculated multiple alignments of orthologous sequences. REDIdb 3.0 allows also the visualization of aminoacid changes induced by RNA editing in protein domains or secondary structures, providing insights into the potential functional consequences.
Next generation sequencing technologies, now arrived at their third generation, are expected to greatly increase the number of RNA editing candidates in the next future. Therefore, it will be indispensable to collect and annotate them in their biological context taking into account also the RNA editing levels.
Due to the unicity in its field, REDIdb is planned to be maintained and updated over time (as new editing sites or complete genomes are released), taking into account, as much as possible, eventual feedbacks from the users.
Author Contributions
CL conducted the bioinformatics analyses and wrote the first manuscript draft; EP and GP conceived the study and contributed to writing and revising the manuscript.
Funding
This work was supported by ELIXIR IIB (CNR).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We kindly thank TMR Regina and M. Takenaka for revising the database and fruitful suggestions, and L. Marra for technical and editorial assistance.
References
Benne, R., Van Den Burg, J., Brakenhoff, J. P. J., Sloof, P., Van Boom, J. H., and Tromp, M. C. (1986). Major transcript of the frameshifted coxll gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826. doi: 10.1016/0092-8674(86)90063-2
Bowe, L. M., and depamphilis, C. W. (1996). Effects of RNA editing and gene processing on phylogenetic reconstruction. Mol. Biol. Evol. 13, 1159–1166. doi: 10.1093/oxfordjournals.molbev.a025680
Castandet, B., Choury, D., Bégu, D., Jordana, X., and Araya, A. (2010). Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res. 38, 7112–7121. doi: 10.1093/nar/gkq591
Cock, P. J., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Cui, L., Veeraraghavan, N., Richter, A., Wall, K., Jansen, R. K., Leebens-Mack, J., et al. (2006). ChloroplastDB: the chloroplast genome database. Nucleic Acids Res. 34, D692–D696. doi: 10.1093/nar/gkj055
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017–beyond protein family and domain annotations. Nucleic. Acids. Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Gray, M. W. (2003). Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 55, 227–233. doi: 10.1080/1521654031000119425
He, T., Du, P., and Li, Y. (2007). dbRES: a web-oriented database for annotated RNA editing sites. Nucleic Acids Res. 35, D141–D144. doi: 10.1093/nar/gkl815
Hiesel, R., Wissinger, B., Schuster, W., and Brennicke, A. (1989). RNA editing in plant mitochondria. Science 246, 1632–1634. doi: 10.1126/science.2480644
Höch, B., Maier, R. M., Appel, K., Igloi, G. L., and Kossel, H. (1991). Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353, 178–180. doi: 10.1038/353178a0
Ichinose, M., and Sugita, M. (2016). RNA editing and its molecular mechanism in plant organelles. Genes 8:5. doi: 10.3390/genes8010005
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kiran, A. M., O'Mahony, J. J., Sanjeev, K., and Baranov, P. V. (2013). Darned in 2013: inclusion of model organisms and linking with Wikipedia. Nucleic Acids Res. 41, D258–D261. doi: 10.1093/nar/gks961
Lohse, M., Drechsel, O., Kahlau, S., and Bock, R. (2013). OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. doi: 10.1093/nar/gkt289
Maier, R. M., Zeltz, P., Kössel, H., Bonnard, G., Gualberto, J. M., and Grienenberger, J. M. (1996). RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 32, 343–365. doi: 10.1007/BF00039390
O'Brien, E. A., Zhang, Y., Wang, E., Marie, V., Badejoko, W., Lang, B. F., et al. (2009). GOBASE: an organelle genome database. Nucleic Acids Res. 37, D946–D950. doi: 10.1093/nar/gkn819
Picardi, E., D'erchia, A. M., Lo Giudice, C., and Pesole, G. (2017). REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45, D750–D757. doi: 10.1093/nar/gkw767
Picardi, E., Regina, T. M., Verbitskiy, D., Brennicke, A., and Quagliariello, C. (2011). REDIdb: an upgraded bioinformatics resource for organellar RNA editing sites. Mitochondrion 11, 360–365. doi: 10.1016/j.mito.2010.10.005
Ramaswami, G., and Li, J. B. (2014). RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 42, D109–D113. doi: 10.1093/nar/gkt996
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Steinhauser, S., Beckert, S., Capesius, I., Malek, O., and Knoop, V. (1999). Plant mitochondrial RNA editing. J. Mol. Evol. 48, 303–312. doi: 10.1007/PL00006473
Sun, T., Bentolila, S., and Hanson, M. R. (2016). The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 21, 962–973. doi: 10.1016/j.tplants.2016.07.005
Takenaka, M., Zehrmann, A., Verbitskiy, D., Härtel, B., and Brennicke, A. (2013). RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352. doi: 10.1146/annurev-genet-111212-133519
Yachdav, G., Wilzbach, S., Rauscher, B., Sheridan, R., Sillitoe, I., Procter, J., et al. (2016). MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics 32, 3501–3503. doi: 10.1093/bioinformatics/btw474
Yang, Y., Heffernan, R., Paliwal, K., Lyons, J., Dehzangi, A., Sharma, A., et al. (2017). SPIDER2: a package to predict secondary structure, accessible surface area, and main-chain torsional angles by deep neural networks. Methods Mol. Biol. 1484, 55–63. doi: 10.1007/978-1-4939-6406-2_6
Keywords: organellar genomes, RNA editing, plant database, mitochondria, chloroplasts
Citation: Lo Giudice C, Pesole G and Picardi E (2018) REDIdb 3.0: A Comprehensive Collection of RNA Editing Events in Plant Organellar Genomes. Front. Plant Sci. 9:482. doi: 10.3389/fpls.2018.00482
Received: 01 February 2018; Accepted: 29 March 2018;
Published: 11 April 2018.
Edited by:
Giovanni Nigita, The Ohio State University, United StatesReviewed by:
Giorgio Giurato, Università degli Studi di Salerno, ItalyShihao Shen, University of California, Los Angeles, United States
Copyright © 2018 Lo Giudice, Pesole and Picardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ernesto Picardi, ZXJuZXN0by5waWNhcmRpQHVuaWJhLml0
 Claudio Lo Giudice
Claudio Lo Giudice Graziano Pesole
Graziano Pesole Ernesto Picardi
Ernesto Picardi