
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci., 11 April 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00474
Nitrate reductase (NR) is the key enzyme for nitrogen assimilation in plant cells and also works as an important enzymatic source of nitric oxide (NO), which then regulates plant growth and resistance to biotic and abiotic stresses. However, how NR activities are finely tuned to modulate these biological processes remain largely unknown. Here we present a SWISSPROT 3D analysis of different NR from plant sources indicating the possible sites of S-nitrosylation, and show some evidence of immunoblottings to S-nitrosated (SNO-) proteins. We also found that S-nitrosylation status of NR is negatively correlated with the enzyme activity. The production of NO via NR in vitro represents only 1% of its nitrate reduction activity, possibly due to NO generated through NR reaction may deactivate the enzyme by this S-nitrosylation-mediated negative-feedback regulation. NR-mediated NO generation also plays a key role in protecting plants from abiotic stresses through activating antioxidant enzymes and increasing antioxidants. Putative connections between NR S-nitrosylation and NO biosynthesis under pathogen attacks and abiotic stresses are discussed in this Perspective.
Nitrate reductase (NR) is the key enzyme for nitrogen assimilation in multiple organisms (Chamizo-Ampudia et al., 2017), which catalyzes the nitrate to nitrite reduction in plant cell cytoplasm (Kolbert et al., 2008). Nitrate, its main substrate, has been shown to be necessary for cellular-signaling and commonly distributed in multiple plant tissues (Kolbert et al., 2008).
Nitrate reductase is only active as a dimer with two subunits of approximate 100 kDa, each containing three prosthetic groups, heme b557, flavin adenine dinucleotide (FAD) and molybdenum cofactor (Campbell, 2001). The prosthetic groups are bound to separate polypeptide domains connected by flexible hinge regions. Hinge 1 is located between the heme and molybdenum domains, and hinge 2 between the FAD and heme domains (Campbell, 2001; Chamizo-Ampudia et al., 2017). Furthermore, the dimerization domain is linked to the hinge region 2 and to the molybdenum domain. They allow energetical transfer of the electron from one domain to another, starting with the electron donors NADH and NADPH toward nitrate (Campbell and Kinghorn, 1990; Chamizo-Ampudia et al., 2017).
Nitrate reductase functions as a very important enzyme for nitric oxide (NO) biosynthesis. It was established in Chlamydomonas that NR plays a key role in NO homeostasis by providing electrons from NADH or NADPH via its catalytic domain both to a truncated hemoglobin THB1 (Sanz-Luque et al., 2015), which functions in NO scavenging through the dioxygenase activity, and to a molybdo-enzyme NO-formation NR (NOFNiR) that is involved in its NO biosynthesis (Chamizo-Ampudia et al., 2016, 2017).
Phosphorylation modifications on NR modulate the NO generation rate (Rockel et al., 2002; Lillo et al., 2004). Besides phosphorylation, NR may be modified by the cysteine (Cys) S-nitrosylation on the protein surface. Here we present a SWISSPROT 3D analysis of different NR from plant sources indicating the possible sites of S-nitrosylation, and show an evidence of immunoblottings to S-nitrosated (SNO-) proteins to support this structure analysis. NO generated through NR may deactivate NR itself and down-regulate NO synthesis via this feedback loop.
Recent phytopathological studies suggested that pathogen-resistance-associated NO-synthesis influences nutrition homeostasis to favor nitrite diversions either away from infection sites or toward defensive-related metabolism (Mur et al., 2017). It is well known that NR participates in tissue development and abiotic stress adaption by regulating both the NO accumulation level and the metabolic flux of nitrate assimilation (Ziogas et al., 2013). S-nitrosylation modification on NR and its physiological significance on NO-synthesis and stress acclimation are illuminated in this Perspective.
It was well-known that for plant cells, NR is activated or inactivated by de-phosphorylation or phosphorylation modification, respectively (Lea et al., 2004). The phosphorylation site is located on a specific motif in the hinge 1 between the heme-binding domain and the molybdenum cofactor-binding domain. The phosphorylatable amino acid is Ser534 in Arabidopsis, Ser543 in spinach or Ser521 in tobacco (Bachmann et al., 1996; Su et al., 1996). Nevertheless, the post-translational regulatory mechanism is more complicated than Ser phosphorylation, because that there are also several Cysteine (Cys) residues distributed on the NR protein surface, which may be subjected to S-nitrosylation modification.
Homology models of NR protein from eight species were constructed in the SWISS-MODEL Workspace1. The molecular surface and the electrostatic potential were computed with Swiss-PdbViewer v.4.0.1 software (Biasini et al., 2014). Cys residues distributed on the protein surface were further analyzed (Figure 1). As a previous report suggested, negative or neutral electrostatic potentials on the protein surface or on the amino acid residues adjacent to the Cys residues on the protein surface are considered to be required for the S-nitrosylation reaction (Zhang et al., 2017). From Figure 1, we know that at least one Cys residue meets this requirement could be always found on the protein surface for every plant species. Therefore, S-nitrosylation on NR may occur presumably. The Cys S-nitrosylation may hamper assembly of the protein oligomer or folding of the nascent peptide chain, and therefore inhibits the enzyme activity (Lindermayr et al., 2005; Zhang et al., 2017). In other words, NO generated through NR may deactivate NR itself, which is a negative-feedback regulatory pathway and may be important for the balance between the N-mediated growth signals and the NO-mediated stress-responsive signals. We proved this hypothesis by the biotin switch assay (Figure 2). NO donor SNP (sodium nitroprusside) treatment significantly increased NR S-nitrosylation levels in both lower plants (Physcomitrella patens) and higher plants (Arabidopsis thaliana, Oryza sativa, and Zea mays). S-nitrosylation status of NR is positively correlated with the NO level, but negatively correlated with the enzyme activity (Figure 2). For example in P. patens, after the SNP treatment, the strongest NR S-nitrosylation was observed accompanying with the lowest NR activity (Figure 2).
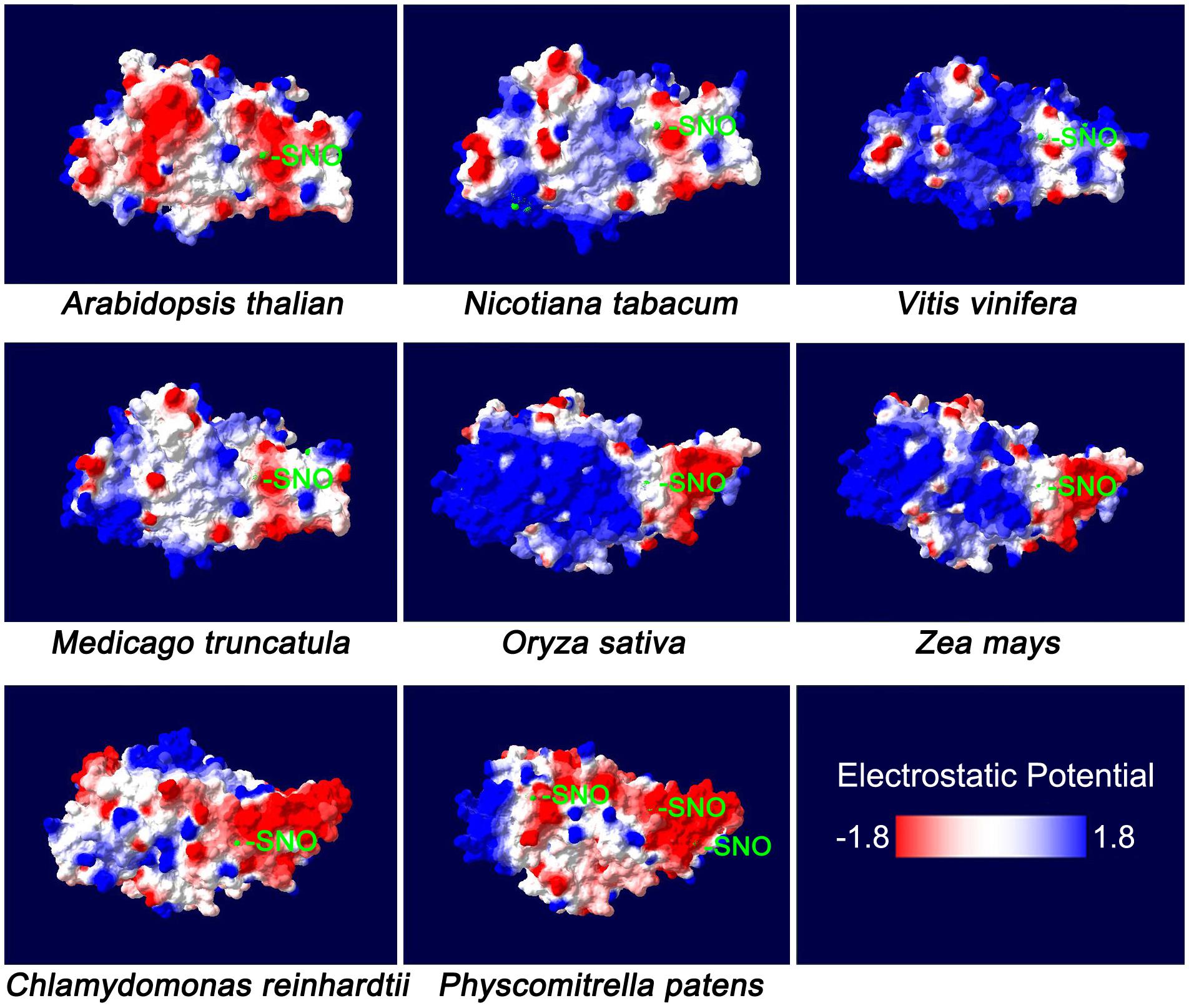
FIGURE 1. The molecular surface of plant nitrate reductase. The NR sequences of Arabidopsis thaliana (Accession ID: X13434.1), Nicotiana tabacum (XP_016462202.1), Vitis vinifera (XP_019082269.1), Medicago truncatula (XP_003614820.1), Oryza sativa Japonica Group (XP_015650300.1), Zea mays (NP_001292785.1), Chlamydomonas reinhardtii (AAF17595.1) and Physcomitrella patens (XP_001763055.1) were selected for the analysis of homology models which were constructed in the SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace/). The molecular surface and the electrostatic potential were computed with Swiss-PdbViewer v. 4.0.1 software (Biasini et al., 2014). The colors on the molecular surface indicate electrostatic potentials (red: –1.8; blue: +1.8). The cysteine (Cys) residues are marked with light-green color, some of which may be subjected to S-nitrosylation (-SNO).
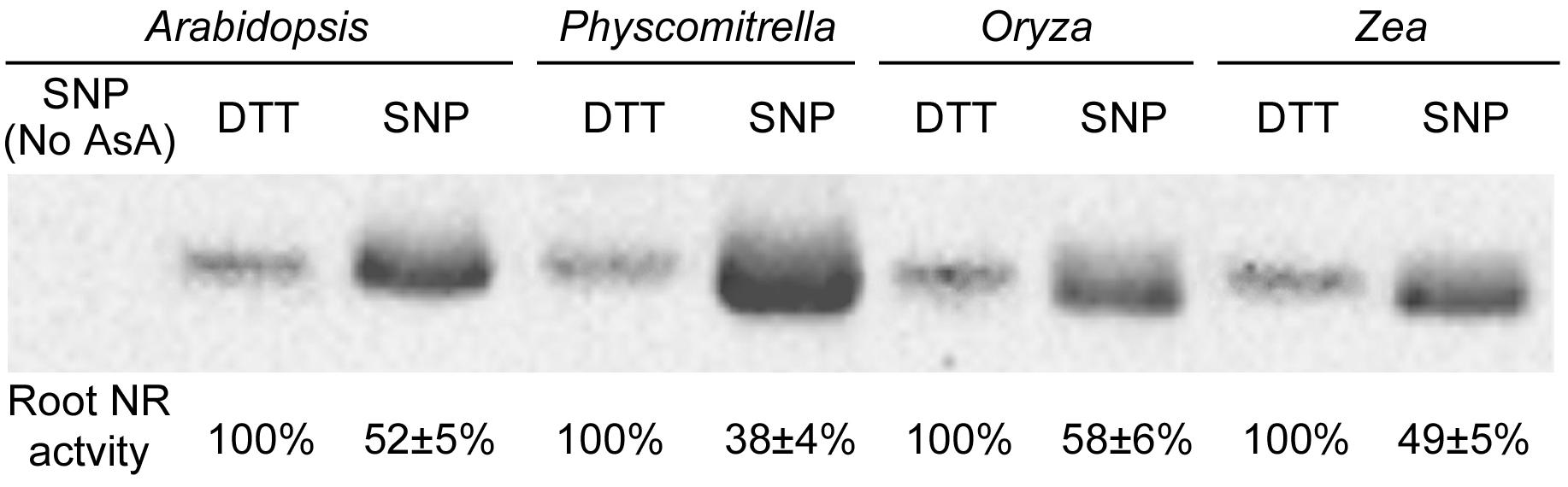
FIGURE 2. S-Nitrosylation levels and root enzyme activities of nitrate reductase from four plant species. The 14-day-old Arabidopsis thaliana, Physcomitrella patens, Oryza sativa and Zea mays seedlings treated grown with or without 0.2 mM SNP (sodium nitroprusside; a NO donor) or 2 mM DTT (dithiothreitol; a NO inhibitor) were harvested for protein extraction. Total cellular proteins were extracted with HEN buffer (250 mM Hepes-NaOH pH 7.7, 1 mM EDTA, 0.1 mM neocuproine and proteinase inhibitor cocktail). After placing on ice for 20 min, the homogenate was centrifuged at 4°C at 14,000 rpm for 5 min. The supernatant was placed into a new tube and centrifuged it at 4°C at 14,000 rpm for 15 min. About 200 μg total cellular proteins were diluted into 125 μl HEN buffer with 500 μl blocking buffer [2.5% SDS and 20 mM methylmethane thiosulphonate (MMTS) in HEN buffer]. After 60 min incubation in 50°C under continuous shaking, MMTS was removed by acetone precipitation. The protein was diluted with 100 μl HEN buffer plus 1% SDS. The labeling reaction was initiated by adding 25 μl of 400 μM sodium ascorbate and 25 μl biotin-M (Sigma-Aldrich) and incubating at 25°C 2 h on a shaker. All proteins after biotin-switch were immunoprecipitated with the biotin antibody (Abcam), and then were subjected to Western blotting with the Arabidopsis NR antibody (Agrisera: AS08 310). 1/10 of the immunoprecipitated proteins (corresponding to 20 μg total cellular proteins) were loaded into each lane (Zhang et al., 2017). Biotin-switch assay and Western blotting were repeated 3 times, and typical results were presented. Protein extracts without ascorbate treatments were used as the negative control (No AsA). The NR activity was measured by mixing 1 volume of protein extracts with 5 volumes of pre-warmed (25°C) assay buffer (100 mM HEPES-KOH, pH 7.5, 5 mM KNO3, and 0.25 mM NADH). The reaction was started by the addition of assay buffer, incubated at 25°C for 30 min, and then stopped by adding 0.1 M zinc acetate. After 15 min, the tubes were centrifuged at 13,000 ×g for 5 min. The nitrite produced was measured at 520 nm by adding 1 mL of 1% (w/v) sulfanilamide in 3 M HCl plus 1 mL of 0.02% (v/v) N-(1-naphthyl)-ethylenediamine (Hu et al., 2015). Root NR activity of each plant treated with DTT was normalized to 100%. Enzyme activities under the SNP treatment are shown as mean ± standard deviations (n = 3).
The inhibitory effect of NO on NR has been reported before. Sanz-Luque et al. (2015) showed that the cytosol truncated hemoglobin, THB1, plays a key role in NO-mediated NR deactivation. THB1 has the NO di-oxygenase activity and gets the electron from the FAD group on NR (the di-aphorase activity), which then reduces the heme group. In the presence of NO, the reduced THB1 may catalyze NO conversion into NO3- and inhibit NO3- reduction by decreasing the reducing power available to the enzyme (Sanz-Luque et al., 2015). Similar S-nitrosylation-mediated negative-feedback regulations to nitrate transporters and reductases involved in nitrate assimilation and NO production have been proposed before (Frungillo et al., 2016). Putative correlations between THB1-mediated NR deactivation and S-nitrosylation-mediated negative-feedback regulatory pathway to NR need further investigations.
It is well-known that the plants preferring nitrate show higher NR activities; in contrast, ammonium-adsorbing plants present lower NR activities, particularly in roots (Marwaha and Juliano, 1976). For instance, although rice (in favor of NH4+) nitrite reductase activities are higher in the seedlings grown in nitrate than in those grown in ammonium, its root NR activity is considerably low (Marwaha and Juliano, 1976). So its ability of using NO3- may be lower than the plants in favor of NO3-, which may be connected with different densities of NR distributed in the roots of different species (Marwaha and Juliano, 1976).
Nitric oxide synthesis in plants is a highly controversial research field (Corpas and Barroso, 2017; Santolini et al., 2017). Nitric oxide synthase (NOS) activity could be detected in fungi and some higher plants. Despite evidences of the arginine-dependent pathway for NO synthesis in higher plants, no NOS homolog has been identified in plants (Corpas and Barroso, 2017; Santolini et al., 2017). Nitric Oxide Associated 1 (NOA1) was originally reported to encode a gene of NOS (Guo et al., 2003). However, the accumulated evidence shows that NOA1 has a distinct function from the direct NO synthesis (Corpas and Barroso, 2017; Santolini et al., 2017). Whereas there are evidences for enzymatic synthesis of NO by NR, it has been suggested that NR’s main role is to provide nitrite, which in turn can be further reduced to NO in other cellular compartments (Rockel et al., 2002). The facts that NO could be generated through nitrite by NR and also by the orthologous enzymes from bacteria, fungi and green algae (Yamasaki et al., 1999; Marcos et al., 2016; Chamizo-Ampudia et al., 2017), thus indicating that this reaction may be conserved and more general than suspected. Although data supporting the relation between NO and NR production in plant cells have steadily accumulated (Hao et al., 2010; Mur et al., 2013; Lu et al., 2014; Yuan et al., 2016), important issues remain unresolved, such as the low rate of NO production by NR and the generally unclear correlation between NR activity and NO producing efficiency. In fact, the production of NO via NR in vitro represents only 1% of its nitrate reduction activity at saturating nitrite and NADH contents, and it could be inhibited by the nitrate competitively (Gupta et al., 2011). Not with standing, the cytosol nitrate level is about the mmol range, so the NO generation from nitrite would be very inefficient, unless NR was exponentially activated (Miller and Smith, 2008). One of the reasons may be that NO generated through NR deactivates NR itself by the S-nitrosylation feedback inhibitions of nitrate transporters and reductases. Therefore, we should not overlook the NOS-like pathways. Although it is a yet cryptic pathway, synthesis of NO from L-arginine intuitively goes side-by-side with the nitrate assimilatory pathway.
Nitrate assimilatory pathway involves a lot of reductases after the trans-amination process, which is responsible for the amino acid biosynthesis (Lam et al., 1996). When pathogens come into (infect) the plant tissues, they are often nutritional-starved such that fast nitrate assimilation from the hosts is necessary for the valid infection (López-Berges et al., 2010). On the other hand, plants may re-allocate their nutrients to the defensive responses or away from the infection sites (Mur et al., 2017). Exogenous applications of N fertilizer could thus change the balance between the hosts and the pathogens. The increases of N have been suggested either to enhance or to reduce plant’s resistances to biotic stresses (Robert et al., 2002; Solomon et al., 2003), reflecting the difference in defensive strategies of different plants (Mur et al., 2017). Nitrate or ammonium fertilizers affect the outcomes of pathogen-plant interactions differently. In general, nitrate feeding enhances pathogen resistance, while ammonium compromises the defensive response (Mur et al., 2017). Metabolically, NO3- enhances polyamine production, which is a well-known defense signal. While NH4+ nutrition leads to an increase in γ-aminobutyric acid, which might be a nutritional component used by some pathogens (Abrahamian et al., 2016).
Companying with the defensive N metabolism, the role of NO should also be considered. It has been suggested that both nitrate and ammonium could increase NR activity and the related NO production (Gupta et al., 2005; Planchet et al., 2005). The pathogen-induced NO accumulation influences the N homeostasis to favor nitrite diversions either away from infection sites or toward defensive-related metabolism (Mur et al., 2017). This NO accumulation is mainly derived from NR and is promoted by both Pathogen Associated Molecular Pattern (PAMP) and gene-to-gene mediated defense. NO accumulation and the correlated defense responses are nitrate dependent and are therefore largely compromised by ammonium (Mur et al., 2017). Yamamoto-Katou et al. (2006) indicated that silence of NR resulted in decreased NO contents after the elicitin treatment; these proteins are secreted by Phytophthora and Pythium species during the phytopathogenic attack. A recent report indicated that, during the compatible interaction between tobacco and Pseudomonas syringae, the mitochondrial NO production via NR-mediated mechanism increased plant resistance to pathogens (Gupta et al., 2013). Detailed biochemical study revealed that the P. syringae infection induced mitochondrial NO accumulation and nitrosylation of the photo-respiratory glycine decarboxylase complex (GDC) in mitochondria (Palmieri et al., 2010). Considering that NADH is needed for the redox balance, nitrosylation results in GDC inhibition, which could influence overall redox homeostasis and induce the cell-death (Palmieri et al., 2010).
Despite these negative arguments for the role of NR in NO production, many studies have been confirmed that NR is involved in plant development and stress responses by modulating not only the content of the signaling molecule NO but also the level of nitrite available for nitrate assimilation (Campbell, 2001). NO not only leads to the production of peroxy-nitrite and the subsequent cellular damages, but also functions as a molecule in signaling, regulating stress responses and controlling stomatal closure (Garcia-Mata and Lamattina, 2003). Recent study demonstrated that NR affects potassium nutrition status as well as NO-mediated regulation to guard cell ion channels in Arabidopsis (Chen et al., 2016). NR also drives re-partitioning of carbon flux by regulating genes encoding enzymes for chlorophyll biosynthesis and carbon fixation and metabolism in the model pennate diatom Phaeodactylum tricornutum (McCarthy et al., 2017). Therefore NR plays a key role in controlling plant nutritional stress acclimations.
Nitrate reductase is correlated with NO synthesis and NO is a stress signaling molecule. Therefore they regulate plant acclimation to adverse environments (Chamizo-Ampudia et al., 2017). For example, a recent study indicated that NR-mediated early NO generation in the shoots of hulless barley plays an irreplaceable role in protecting seedlings from copper toxicity through enhancing antioxidant enzyme activities and antioxidant levels (Hu et al., 2015). It is interesting that the early NO burst at 24 h was produced through NR, but not through the NO synthase (Hu et al., 2015).
It was also recognized that auxin shares common steps with NO in the signal transduction cascade toward the auxin-induced adventitious and lateral root formation (Lamattina et al., 2003; Correa-Aragunde et al., 2004). This increased level of NO was present only in the lateral-root initials in contrast to the primary roots where it remained at the control level. Thus, NR is involved in auxin-induced NO formation in roots (Kolbert et al., 2008) and participates in biotic and abiotic stress responses indirectly.
An interesting study showed that in a NR-null mutant, the auxin receptor AFB3 mRNA could be induced by the NO3- treatment with the maximum level at 1 h, as well as the wild-type seedlings. However, the AFB3 transcript did not decrease after that time in the Arabidopsis nr mutant under the same conditions, contrasting to the wild-type seedlings (Vidal et al., 2010). Therefore, besides auxin signaling, NR may also regulate auxin metabolism. The authors suggested that some N metabolite downstream of nitrate reduction may be involved in AFB3 gene expression regulation (Vidal et al., 2010), which requires further explorations. These results could be ascribed to the role of NR-mediated NO production. Crosstalk between NO signaling/metabolism and auxin signaling/metabolism in nutritional stress responses also needs further investigations.
Nitrate reductase is an important enzymatic source of biosynthesis of NO. While NO functions as a signaling molecule, regulating plant development and growth, resistance to abiotic and biotic stresses and metabolic repartition for optimizing nutrient utilization. Analysis to Cys residues distributed on the protein surface implies the possible S-nitrosylation (by the free radical NO) on NR protein. Understanding how NR activities are finely tuned to modulate these biological processes would require future researches.
Although NO generated through NR may deactivate NR itself by the S-nitrosylation feedback inhibition, NR-mediated NO production is a common plant’s resistance strategy against multiple biotic and abiotic stresses (Baudouin and Hancock, 2014; Simontacchi et al., 2015; Zhang et al., 2017). Besides these signaling events and developmental processes, NO induces the post-translational modification of target proteins by NO-dependent protein nitrosylation. However, a study from Ziogas et al. (2013) showed that in citrus plants, the S-nitrosylation pattern was enhanced by drought, cold or heat, but was suppressed by dark or salinity. Correspondingly, peroxynitrite (ONOO-) scavenging ability of these plants was elicited by dark, continuous-light or drought but was suppressed by the salt stress. In contrast, nitration levels were elevated by salinity and suppressed by dark or continuous-light. They suggested that the nitrosative responses of citrus plants may be differentially regulated depending on how they are stressed. Alternatively, the roles of NR-mediated NO production and its related protein nitrosylation on biotic and abiotic stress resistance may be varied depending on the stress type or the plant species, which needs further studies.
Y-FF and SY conceived the perspective, collected data, and wrote the manuscript. Z-WZ helped to write the manuscript.
This research was supported by the National Natural Science Foundation of China (31770322) and the Preeminent Youth Fund of Sichuan Province (2015JQO045).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Abrahamian, M., Ah-Fong, A. M., Davis, C., Andreeva, K., and Judelson, H. S. (2016). Gene expression and silencing studies in Phytophthora infestans reveal infection-specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Pathog. 12:e1006097. doi: 10.1371/journal.ppat.1006097
Bachmann, M., Shiraishi, N., Campbell, W. H., Yoo, B. C., Harmon, A. C., and Huber, S. C. (1996). Identification of Ser-543 as the major regultatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell 8, 505–517. doi: 10.1105/tpc.8.3.505
Baudouin, E., and Hancock, J. T. (2014). Nitric oxide signaling in plants. Front. Plant Sci. 4:553. doi: 10.3389/fpls.2013.00553
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Campbell, W. H. (2001). Structure and function of eukaryotic NAD(P)H: nitrate reductase. Cell. Mol. Life Sci. 58, 194–204. doi: 10.1007/PL00000847
Campbell, W. H., and Kinghorn, K. R. (1990). Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem. Sci. 15, 315–319. doi: 10.1016/0968-0004(90)90021-3
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas,Á., Ocaña-Calahorro, F., Mariscal, V., Carreras, A., et al. (2016). A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 39, 2097–2107. doi: 10.1111/pce.12739
Chen, Z. H., Wang, Y., Wang, J. W., Babla, M., Zhao, C., García-Mata, C., et al. (2016). Nitrate reductase mutation alters potassium nutrition as well as nitric oxide-mediated control of guard cell ion channels in Arabidopsis. New Phytol. 209, 1456–1469. doi: 10.1111/nph.13714
Corpas, F. J., and Barroso, J. B. (2017). Nitric oxide synthase-like activity in higher plants. Nitric Oxide 68, 5–6. doi: 10.1016/j.niox.2016.10.009
Correa-Aragunde, N., Graziano, M., and Lamattina, L. (2004). Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. doi: 10.1007/s00425-003-1172-7
Frungillo, L., Spoel, S. H., and Salgado, I. (2016). Control of nitrogen assimilation in plants through S-nitrosothiols. Adv. Bot. Res. 77, 55–78. doi: 10.1016/bs.abr.2015.10.011
Garcia-Mata, C., and Lamattina, L. (2003). Abscisic acid, nitric oxide and stomatal closure - is nitrate reductase one of the missing links? Trends Plant Sci. 8, 20–26.
Guo, F. Q., Okamoto, M., and Crawford, N. M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103. doi: 10.1126/science.1086770
Gupta, K. J., Brotman, Y., Segu, S., Zeier, T., Zeier, J., Persijn, S. T., et al. (2013). The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 64, 553–568. doi: 10.1093/jxb/ers348
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and van Dongen, J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. doi: 10.1016/j.tplants.2010.11.007
Gupta, K. J., Stoimenova, M., and Kaiser, W. M. (2005). In higher plants, only root mitochondria, but not leaf mitochondria reduce nitrite to NO, in vitro and in situ. J. Exp. Bot. 56, 2601–2609. doi: 10.1093/jxb/eri252
Hao, F., Zhao, S., Dong, H., Zhang, H., Sun, L., and Miao, C. (2010). Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J. Integr. Plant Biol. 52, 298–307. doi: 10.1111/j.1744-7909.2010.00920.x
Hu, Y., You, J., and Liang, X. (2015). Nitrate reductase-mediated nitric oxide production is involved in copper tolerance in shoots of hulless barley. Plant Cell Rep. 34, 367–379. doi: 10.1007/s00299-014-1715-3
Kolbert, Z., Bartha, B., and Erdei, L. (2008). Exogenous auxin-induced NO synthesis is nitrate reductase-associated in Arabidopsis thaliana root primordia. J. Plant Physiol. 165, 967–975. doi: 10.1016/j.jplph.2007.07.019
Lam, H. M., Coschigano, K. T., Oliveira, I. C., MeloOliveira, R., and Coruzzi, G. M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. doi: 10.1146/annurev.arplant.47.1.569
Lamattina, L., García-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Lea, U. S., Ten Hoopen, F., Provan, F., Kaiser, W. M., Meyer, C., and Lillo, C. (2004). Mutation of the regulatory phosphorylation site of tobacco nitrate reductase results in high nitrite excretion and NO emission from leaf and roots tissue. Planta 219, 59–65. doi: 10.1007/s00425-004-1209-6
Lillo, C., Meyer, C., Lea, U. S., Provan, F., and Oltedal, S. (2004). Mechanism and importance of post-translational regulation of nitrate reductase. J. Exp. Bot. 55, 1275–1282. doi: 10.1093/jxb/erh132
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
López-Berges, M. S., Rispail, N., Prados-Rosales, R. C., and Di Pietro, A. (2010). A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22, 2459–2475. doi: 10.1105/tpc.110.075937
Lu, S., Zhuo, C., Wang, X., and Guo, Z. (2014). Nitrate reductase (NR)-dependent NO production mediates ABA- and H2O2-induced antioxidant enzymes. Plant Physiol. Biochem. 74, 9–15. doi: 10.1016/j.plaphy.2013.10.030
Marcos, A. T., Ramos, M. S., Marcos, J. F., Carmona, L., Strauss, J., and Cánovas, D. (2016). Nitric oxide synthesis by nitrate reductase is regulated during development in Aspergillus. Mol. Microbiol. 99, 15–33. doi: 10.1111/mmi.13211
Marwaha, R. S., and Juliano, B. O. (1976). Aspects of nitrogen metabolism in the rice seedling. Plant Physiol. 57, 923–927. doi: 10.1104/pp.57.6.923
McCarthy, J. K., Smith, S. R., McCrow, J. P., Tan, M., Zheng, H., Beeri, K., et al. (2017). Nitrate reductase knockout uncouples nitrate transport from nitrate assimilation and drives repartitioning of carbon flux in a model pennate diatom. Plant Cell 29, 2047–2070. doi: 10.1105/tpc.16.00910
Miller, A. J., and Smith, S. J. (2008). Cytosolic nitrate ion homeostasis: could it have a role in sensing nitrogen status? Ann. Bot. 101, 485–489. doi: 10.1093/aob/mcm313
Mur, L. A., Mandon, J., Persijn, S., Cristescu, S. M., Moshkov, I. E., Novikova, G. V., et al. (2013). Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5, ls052. doi: 10.1093/aobpla/pls052
Mur, L. A. J., Simpson, C., Kumari, A., Gupta, A. K., and Gupta, K. J. (2017). Moving nitrogen to the centre of plant defence against pathogens. Ann. Bot. 119, 703–709. doi: 10.1093/aob/mcw179
Palmieri, M. C., Lindermayr, C., Bauwe, H., Steinhauser, C., and Durner, J. (2010). Regulation of plant glycine decarboxylase by s-nitrosylation and glutathionylation. Plant Physiol. 152, 1514–1528. doi: 10.1104/pp.109.152579
Planchet, E., Jagadis Gupta, K., Sonoda, M., and Kaiser, W. M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41, 732–743. doi: 10.1111/j.1365-313X.2005.02335.x
Robert, C., Bancal, M. O., and Lannou, C. (2002). Wheat leaf rust uredospore production and carbon and nitrogen export in relation to lesion size and density. Phytopathology 92, 762–768. doi: 10.1094/PHYTO.2002.92.7.762
Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W. M. (2002). Regulation of nitrite oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. doi: 10.1093/jexbot/53.366.103
Santolini, J., André, F., Jeandroz, S., and Wendehenne, D. (2017). Nitric oxide synthase in plants: where do we stand? Nitric Oxide 63, 30–38. doi: 10.1016/j.niox.2016.09.005
Sanz-Luque, E., Ocaña-Calahorro, F., de Montaigu, A., Chamizo-Ampudia, A., Llamas,Á., Galván, A., et al. (2015). THB1, a truncated hemoglobin, modulates nitric oxide levels and nitrate reductase activity. Plant J. 81, 467–479. doi: 10.1111/tpj.12744
Simontacchi, M., Galatro, A., Ramos-Artuso, F., and Santa-María, G. E. (2015). Plant survival in a changing environment: the role of nitric oxide in plant responses to abiotic stress. Front. Plant Sci. 6:977. doi: 10.3389/fpls.2015.00977
Solomon, P. S., Tan, K. C., and Oliver, R. P. (2003). The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 4, 203–210. doi: 10.1046/j.1364-3703.2003.00161.x
Su, W., Huber, S. C., and Crawford, N. M. (1996). Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell 8, 519–527. doi: 10.1105/tpc.8.3.519
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Yamamoto-Katou, A., Katou, S., Yoshioka, H., Doke, N., and Kawakita, K. (2006). Nitrate reductase is responsible for elicitin-induced nitric oxide production in Nicotiana benthamiana. Plant Cell Physiol. 47, 726–735. doi: 10.1093/pcp/pcj044
Yamasaki, H., Sakihama, Y., and Takahashi, S. (1999). An alternative pathway for nitric oxide production in plants: new features of an old enzyme. Trends Plant Sci. 4, 128–129. doi: 10.1016/S1360-1385(99)01393-X
Yuan, S., Zhang, Z. W., Zheng, C., Zhao, Z. Y., Wang, Y., Feng, L. Y., et al. (2016). Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc. Natl. Acad. Sci. U.S.A. 113, 7661–7666. doi: 10.1073/pnas.1602004113
Zhang, Z. W., Luo, S., Zhang, G. C., Feng, L. Y., Zheng, C., Zhou, Y. H., et al. (2017). Nitric oxide induces monosaccharide accumulation through enzyme S-nitrosylation. Plant Cell Environ. 40, 1834–1848. doi: 10.1111/pce.12989
Keywords: abiotic stress responses, nitrate reductase, NO synthesis, pathogen attacks, S-nitrosylation
Citation: Fu Y-F, Zhang Z-W and Yuan S (2018) Putative Connections Between Nitrate Reductase S-Nitrosylation and NO Synthesis Under Pathogen Attacks and Abiotic Stresses. Front. Plant Sci. 9:474. doi: 10.3389/fpls.2018.00474
Received: 10 January 2018; Accepted: 26 March 2018;
Published: 11 April 2018.
Edited by:
John Hancock, University of the West of England, Bristol, United KingdomReviewed by:
Luis A. J. Mur, Aberystwyth University, United KingdomCopyright © 2018 Fu, Zhang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Yuan, cm91bmR0cmVlMzE4QGhvdG1haWwuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.