- Laboratory of Molecular Biology, Department of Plant Sciences, Wageningen University & Research, Wageningen, Netherlands
Parasponia represents five fast-growing tropical tree species in the Cannabaceae and is the only plant lineage besides legumes that can establish nitrogen-fixing nodules with rhizobium. Comparative analyses between legumes and Parasponia allows identification of conserved genetic networks controlling this symbiosis. However, such studies are hampered due to the absence of powerful reverse genetic tools for Parasponia. Here, we present a fast and efficient protocol for Agrobacterium tumefaciens-mediated transformation and CRISPR/Cas9 mutagenesis of Parasponia andersonii. Using this protocol, knockout mutants are obtained within 3 months. Due to efficient micro-propagation, bi-allelic mutants can be studied in the T0 generation, allowing phenotypic evaluation within 6 months after transformation. We mutated four genes – PanHK4, PanEIN2, PanNSP1, and PanNSP2 – that control cytokinin, ethylene, or strigolactone hormonal networks and that in legumes commit essential symbiotic functions. Knockout mutants in Panhk4 and Panein2 displayed developmental phenotypes, namely reduced procambium activity in Panhk4 and disturbed sex differentiation in Panein2 mutants. The symbiotic phenotypes of Panhk4 and Panein2 mutant lines differ from those in legumes. In contrast, PanNSP1 and PanNSP2 are essential for nodule formation, a phenotype similar as reported for legumes. This indicates a conserved role for these GRAS-type transcriptional regulators in rhizobium symbiosis, illustrating the value of Parasponia trees as a research model for reverse genetic studies.
Introduction
Parasponia are tropical tree species belonging to the Cannabis family (Cannabaceae) and are known as the only non-legume plants that can establish a nitrogen-fixing endosymbiosis with rhizobium (Clason, 1936; Trinick, 1973; Akkermans et al., 1978). The Parasponia genus consists of five species indigenous to the Malay Archipelago and Papua New Guinea, where they grow on the slopes of volcanic mountains (Clason, 1936; Soepadmo, 1974; Becking, 1992). Parasponia spp. are typical fast-growing pioneer plants, capable of covering nitrogen-poor eroded soils in a relatively short time span (Becking, 1992). Under suitable greenhouse conditions, young Parasponia trees can grow at speeds exceeding 45 centimeters per month, and fix up to 850 kg N ha-1 year-1 in association with rhizobium (Trinick, 1980, 1981; Trinick and Hadobas, 1989). As Parasponia is the only non-legume that can establish rhizobium symbiosis, it may represent a valuable model to study the core genetic networks underlying this symbiosis (Geurts et al., 2012, 2016; Behm et al., 2014).
Like legumes, Parasponia develops specialized root nodular organs to host the rhizobium partner. Nodules provide the rhizobium bacteria with suitable environmental conditions to convert atmospheric nitrogen into ammonium. The Cannabaceae and legume family (Fabaceae) diverged about a 100 million years ago (Wang et al., 2009), underlining that the rhizobium symbiosis in legumes and Parasponia evolved largely independent (Li et al., 2015). This is reflected in the distinct nodule-types found in both lineages (Behm et al., 2014). Legume nodules possess a large central zone of infected cells, which is surrounded by peripheral vascular bundles. In contrast, Parasponia nodules have a central vascular bundle and infected cells in the peripheral zone, giving these nodules a lateral root-like appearance. Nevertheless, initial comparative studies revealed that both symbioses are founded on conserved signaling networks. In legumes as well as Parasponia, root nodule formation is induced upon recognition of rhizobial secreted lipo-chitooligosaccharide (LCO) signals (Marvel et al., 1987; Op den Camp et al., 2011; Granqvist et al., 2015). Research on model legumes, like Medicago truncatula and Lotus japonicus, showed that the perception of these symbiotic signals requires a signaling cascade that has been co-opted from the much older endomycorrhizal symbiosis (Geurts et al., 2012; Oldroyd, 2013). In legumes, activation of the LCO signaling network results in a massive transcriptional reprogramming, requiring among others the GRAS-type transcriptional regulators NODULATION SIGNALLING PATHWAY 1 (NSP1) and NSP2 and the cytokinin receptor MtCRE1/LjLHK1 (Kaló et al., 2005; Smit et al., 2005; Gonzalez-Rizzo et al., 2006; Heckmann et al., 2006; Murray et al., 2007; Tirichine et al., 2007; Plet et al., 2011). Subsequent nodule formation is tightly controlled by regulatory feedback loops, including negative regulation by ethylene signaling (Penmetsa et al., 2008; Miyata et al., 2013; van Zeijl et al., 2015b).
A reference quality genome sequence for Parasponia andersonii and draft genome sequences of two additional Parasponia species have been generated (van Velzen et al., 2017). Mining these genomes uncovered ∼1,800 putative symbiosis genes, of which 100s are close homologs of legume symbiosis genes (van Velzen et al., 2017). Initial reverse genetic studies in P. andersonii, using a transient Agrobacterium rhizogenes-based root transformation system, revealed that at least two genes – NOD FACTOR PERCEPTION 1 (PanNFP1) and CALCIUM AND CALMODULIN-DEPENDENT PROTEIN KINASE (PanCCaMK) – commit conserved functions in the Parasponia and legume LCO signaling pathways (Op den Camp et al., 2011). We argue that a more comprehensive comparative analysis between legumes and Parasponia will allow identification of conserved genetic networks that are essential to establish symbiosis with rhizobium. However, to use Parasponia as an effective research model – alongside the legume models M. truncatula and L. japonicus – efficient transformation and genome editing tools are required.
Here, we exploit an efficient in vitro micro-propagation system available for P. andersonii to establish stable transformation and CRISPR/Cas9-mediated mutagenesis for this species (Davey et al., 1993; Webster et al., 1995; Cao et al., 2012). We show that using Agrobacterium tumefaciens-mediated transformation, stable transgenic lines of P. andersonii can be obtained in ∼3–4 months. Additionally, we show that P. andersonii is amenable to targeted mutagenesis using the CRISPR/Cas9 system. As ∼40% of the resulting T0 lines harbor bi-allelic mutations, these can be phenotyped upon in vitro propagation. As proof of concept, we mutated four genes in P. andersonii that in legumes control hormonal pathways as well as commit symbiotic functions. These include: the GRAS-type transcriptional regulators NSP1 and NSP2 that are essential for nodule organogenesis (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006) and control strigolactone biosynthesis by mediating DWARF27 (D27) expression (Liu et al., 2011; van Zeijl et al., 2015a); the cytokinin receptor HISTIDINE KINASE 4 (HK4) that in legumes is essential for nodule organogenesis (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Plet et al., 2011); and the ethylene signaling hub ETHYLENE INSENSITIVE 2 (EIN2) that is a negative regulator of nodulation in legumes (Penmetsa and Cook, 1997; Penmetsa et al., 2008; Miyata et al., 2013).
Materials and Methods
Plant Materials and Growth Conditions
All experiments were conducted using P. andersonii WU1 or offspring thereof (Op den Camp et al., 2011; van Velzen et al., 2017). P. andersonii trees were grown in a conditioned greenhouse at 28°C, 85% humidity and a 16/8 h day/night regime. For in vitro culturing, P. andersonii was grown in an Elbanton growth cabinet at 28°C, 16/8 h day/night. Growth of young P. andersonii plantlets for nodulation assays or qRT-PCR analysis was performed in 1 L crystal-clear polypropelene containers equipped with a gas exchange filter (OS140BOX, Duchefa Biochemie, Netherlands). Pots were half-filled with agraperlite (Maasmond-Westland, Netherlands) and watered with modified EKM medium [3 mM MES (C6H13NO4) pH 6.6, 2.08 mM MgSO4, 0.88 mM KH2PO4, 2.07 mM K2HPO4, 1.45 mM CaCl2, 0.70 mM Na2SO4, 0.375 mM NH4NO3, 15 μM Fe-citrate, 6.6 μM MnSO4, 1.5 μM ZnSO4, 1.6 μM CuSO4, 4 μM H3BO3, 4.1 μM Na2MoO4] (Becking, 1983) and placed in a climate room set at 28°C, 16/8 h day/night. For nodulation assays, EKM medium was inoculated with Mesorhizobium plurifarium BOR2 (OD600 = 0.025) (van Velzen et al., 2017).
Vectors and Constructs
For CRISPR/Cas9-mediated mutagenesis, binary transformation constructs were created using Golden Gate assembly (Engler et al., 2009). For an overview of all Golden Gate clones used in this study, see Supplementary Table 1. sgRNAs were designed based on the principles described in Doench et al. (2014) and PCR amplified using specific forward primers and a universal reverse primer (Supplementary Table 2), using Addgene plasmid # 46966 as template (Nekrasov et al., 2013). These were cloned behind the AtU6p small RNA promoter and inserted behind the neomycin phosphotransferease II gene (NPTII) and an Arabidopsis thaliana codon-optimized variant of Cas9 (Fauser et al., 2014) fused to an N-terminal nuclear localization signal and driven by the 35S promoter (Supplementary Table 1). As negative control, a binary vector was created containing only the NPTII- and NLS-Cas9-encoding sequences (Supplementary Table 1). To setup P. andersonii stable transformation, vector pKGWFS7-RR was used (Karimi et al., 2002).
Phylogenetic Reconstruction
Protein sequences of Glycine max (Wm82.a2.v1) (Schmutz et al., 2010), M. truncatula (Mt4.0v1) (Young et al., 2011; Tang et al., 2014) and Populus trichocarpa (v3.0) (Tuskan et al., 2006) were obtained through Phytozome 101. Protein sequences of P. andersonii (PanWU01x14_asm01_ann01) and Trema orientalis (TorRG33x02_asm01_ann01) were obtained from www.parasponia.org (van Velzen et al., 2017). These sequences were mined using sequences from A. thaliana (TAIR102) (Lamesch et al., 2012) and M. truncatula. Protein sequences were aligned using MAFFT v7.017 (Katoh et al., 2002) implemented in Geneious 8.1.9 (Biomatters, New Zealand), using default parameter settings. Approximately-maximum-likelihood phylogenetic trees were constructed using FastTree (Price et al., 2009) implemented in Geneious 8.1.9. Mid-point rooting was applied for better tree visualization using FigTree v1.4.23.
Plant Transformation
Stable transformation of P. andersonii was performed using A. tumefaciens strain AGL1 (Lazo et al., 1991). A. tumefaciens was grown for 2 days on agar-solidified LB medium containing appropriate antibiotics. For each P. andersonii transformation, two Petri dishes (Ø 9 cm) of A. tumefaciens were used. Bacteria were scraped from plate and resuspended in 25 ml of infiltration medium [SH10 (Supplementary Table 3), 20 mg/l acetosyringone (Sigma, United States), 0.001% (v/v) Silwet L-774]. P. andersonii tissue explants used for transformation were harvested from mature trees grown under greenhouse conditions and sterilized in 2% commercial bleach for 15 min. Tissue explants were cut at both ends inside the A. tumefaciens suspension, creating fresh wound surfaces, and kept inside the suspension for about 20 min. Subsequently, excess liquid was removed from tissue explants using sterilized filter paper and explants were placed on co-cultivation medium [Root-inducing medium (Supplementary Table 3), 20 mg/l acetosyringone (Sigma, United States)]. Plates were incubated for 2 days at 21°C in darkness. After 2 days, tissue explants were washed three times using SH10 (Supplementary Table 3) and subsequently dried using filter paper. Tissue explants were placed on root-inducing medium containing 50 mg/l kanamycin and 300 mg/l cefotaxime and incubated at 28°C, 16/8 h day/night. Nine days after transformation, tissue explants were transferred to propagation medium (Supplementary Table 3) containing 50 mg/l kanamycin and 300 mg/l cefotaxime. Plates were refreshed every other week. When regenerative calli reached ∼2 mm in size they were separated from tissue explants to stimulate shoot formation. A single shoot was selected per tissue explant. These shoots were propagated on propagation medium (Supplementary Table 3), as previously described (Cao et al., 2012). Rooted plantlets were generated by placing individual shoots on root-inducing medium (Supplementary Table 3) (Cao et al., 2012).
Characterization of Transgenic Lines
For T-DNA copy number estimates based on qPCR analysis, genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Germany). qPCR was set up in a 10 μl reaction system with 2x iQ SYBR Green Super-mix (Bio-Rad, United States) and 5 ng template DNA. The experimental setup and procedure were executed on a CFX Connect optical cycler, according to the manufacturer’s protocol (Bio-Rad, United States). T-DNA copy number was estimated using two primer pairs amplifying part of the T-DNA and two primer pairs amplifying single copy P. andersonii genes (PanAGT1 and PanWU01x14_asm01_ann01_338920) that were selected based on a study by Duarte et al. (2010). Primer sequences are listed in Supplementary Table 2. Data analysis was performed using CFX Manager 3.0 software (Bio-Rad, United States). For T-DNA copy number estimates based on Southern blotting, genomic DNA was separately digested with XbaI, HindIII, and EcoRI. Blots were hybridized with a 516 bp α-32P-labeled probe corresponding to part of the NPTII gene that was amplified using primers nptII_Fw and nptII_Rv listed in Supplementary Table 2.
Genotyping of transgenic lines was performed using the Phire Plant Direct PCR Kit (Thermo Scientific, United States) and gene specific primers listed in Supplementary Table 2. Ploidy estimates of transgenic lines were determined by FACS as described by van Velzen et al. (2017).
To determine ethylene sensitivity of Panein2 mutants, tips of young branches of 4 months-old trees were covered with 1 L plastic bags and injected with 1 ml of pure ethylene gas. After 3 days, bags were removed and leaf abscission examined. Total number of leaves on treated branches varied from 6 to 18.
Microtome Sectioning
Stem cross-sections were made from the primary stem, 5 cm below the apical meristem, of 2 month-old trees. Shoot tissue was fixed in 5% glutaraldehyde and embedded in Technovit 7100 (Heraeus-Kulzer, Germany), according to the manufacturer’s protocol. Semi-thin (7 μm) sections were cut using a microtome (Reichert-Jung, Leica Microsystems, Netherlands) and stained with 0.05% Toluidine Blue O. Images were taken using a Leica DM5500B microscope equipped with a DFC425C camera (Leica Microsystems, Germany). Average procambium cell number was quantified by averaging the number of cells within 25–40 cell files for each of the biological replicates.
Nodule tissue fixation and embedding was performed as previously described (Fedorova et al., 1999). Semi-thin (0.6 μm) sections were cut using a Leica Ultracut microtome (Leica Microsystems, Germany) and photographed as described above.
RNA Isolation and qRT-PCR Analysis
RNA was isolated from snap-frozen root tips (∼2–3 cm) as described by van Velzen et al. (2017). cDNA was prepared from 1 μg of total RNA using the i-script cDNA synthesis kit (Bio-Rad, United States), following the manufacturer’s instructions. RT-qPCR was set up as described above. Normalization was performed based on two stably expressed reference genes [UNKNOWN 2 (PanUNK2) and ELONGATION FACTOR 1α (PanEF1α)], chosen based on previous study (Czechowski et al., 2005; Bansal et al., 2015). All primer sequences are listed in Supplementary Table 2.
Statistical Analysis
Statistical differences were determined based on one-way ANOVA and Tukey post hoc tests. Statistical analyses were performed using IBM SPSS Statistics 23.0 (IBM, United States).
Results
Agrobacterium tumefaciens-Mediated Transformation of Parasponia
To establish a protocol for stable transformation of P. andersonii, we first determined the most optimal conditions for regeneration of non-transgenic tissue. We compared regeneration efficiencies of nine tissue explant types in combination with 11 different media, including the propagation and root-inducing media previously used for P. andersonii (Supplementary Tables 4, 5) (Op den Camp et al., 2011; Cao et al., 2012). This revealed that young stem pieces and petioles placed on original propagation medium regenerate plantlets most efficiently (Supplementary Table 4). Next, we questioned whether stem pieces and petioles could be transformed efficiently using A. tumefaciens. To this end, we used A. tumefaciens AGL1 carrying a binary transformation vector containing in its T-DNA the kanamycin resistance gene NPTII and the red fluorescent protein DsRED1. Co-cultivation of A. tumefaciens and P. andersonii stem or petiole explants was conducted in darkness for 2 days at 21°C to promote T-DNA transfer (Cao et al., 2012). Afterward, tissue explants were placed on selective medium and incubated at 28°C in the light. These latter conditions are most favorable for P. andersonii regeneration (Cao et al., 2012). From day 8 onwards, DsRED1-fluorescent cells could be observed near the wound surface indicating a successful transfer of the T-DNA.
Recent research on A. thaliana showed that acquisition of pluripotency requires activation of a root developmental program (Kareem et al., 2015). We tested whether an initial culturing period on root-inducing medium further improves the transformation efficiency of P. andersonii. This showed to be the case (Supplementary Table 6). About half of the explants formed regenerative calli at 4 weeks after co-cultivation (Figure 1A). When 2 mm in size, transgenic calli were separated from tissue explants, which stimulated shoot formation (Figures 1B,C). Two to three months after the start of transformation, a single shoot was selected from each explant to ensure that the transgenic lines represent independent transformation events. These shoots can be genotyped and vegetatively propagated (Supplementary Figure 1). The latter allows clonal multiplication of individual transgenic lines in a period of ∼4–6 weeks, which means that phenotyping assays could be initiated at ∼4 months after the start of transformation.
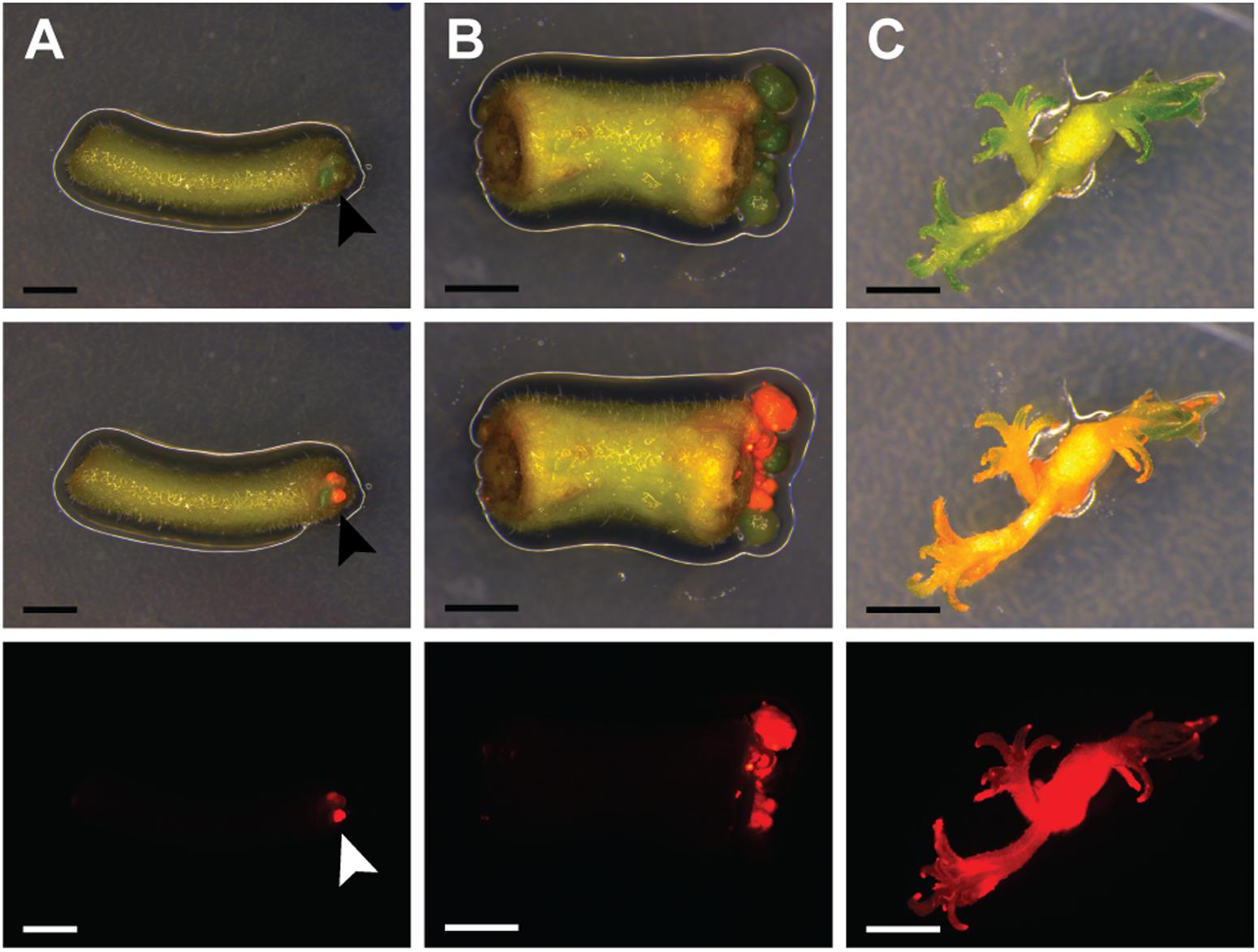
FIGURE 1. Parasponia andersonii stem and petiole explants can be efficiently transformed using Agrobacterium tumefaciens. (A) Petiole explant at 4 weeks after transformation using A. tumefaciens. Arrowheads indicate transgenic micro-calli. (B) Stem explant at 5 weeks after transformation using A. tumefaciens. (C) Small transgenic shoots at 10 weeks after transformation. Explants were incubated on root-inducing medium for 9 days, prior to transfer to propagation medium. DsRED fluorescence indicates transgenic tissue. Scale bars are equal to 2.5 mm. Shown from top to bottom are bright-field images, overlays of bright-field and DsRED fluorescence and DsRED fluorescence images.
To characterize the resulting transgenic P. andersonii lines at the molecular level, we selected – based on red fluorescence – 20 independent transformants for further analyses. PCR reactions using primers amplifying a sequence near the right T-DNA border indicated complete T-DNA integration in 19 out of 20 lines (Supplementary Table 7). To determine whether the transformation procedure might affect ploidy level of the regenerated transgenic lines, we estimated genome size based on flow cytometry. This showed no effect of the transformation procedure on the genome size of transgenic lines (Supplementary Table 7). To estimate the number of T-DNA integrations, we used quantitative RT-PCR (qRT-PCR) as well as Southern blotting. This showed an overall low T-DNA copy number, varying between one and three integrations per line (Supplementary Table 7). We selected three transgenic lines with a single T-DNA integration to examine T-DNA stability. In greenhouse-grown trees as well as in vitro propagated material, DsRED1 fluorescence could still be observed at 6–12 months after transgenic lines were selected (Supplementary Figures 1, 2). This indicates that trans-genes remain stably integrated into the P. andersonii genome and actively transcribed, even after multiple rounds of vegetative propagation. Taken together, the protocol described above allows generating A. tumefaciens-transformed P. andersonii plantlets within 3 months, which can be phenotyped upon vegetative propagation.
Parasponia Is Amenable to CRISPR/Cas9-Mediated Mutagenesis
To test whether CRISPR/Cas9 could be used for targeted mutagenesis in P. andersonii, we aimed at mutating the P. andersonii putative orthologs of EIN2, MtCRE1/LjLHK1, NSP1, and NSP2. These genes were selected, because they control legume root nodule formation as well as commit essential non-symbiotic functions in hormone homeostasis. Putative orthologs of all four genes were previously identified from the P. andersonii genome and named PanEIN2, PanHK4, PanNSP1, and PanNSP2, respectively (van Velzen et al., 2017). Phylogenetic reconstruction based on protein sequences confirmed that these represent the most likely orthologs of legume symbiotic genes (Supplementary Figures 3–6). To mutate PanEIN2, PanHK4, PanNSP1 and PanNSP2, three single guide RNAs (sgRNAs) targeting PanHK4 and PanNSP2 and single sgRNAs targeting PanEIN2 and PanNSP1 were placed under an A. thaliana AtU6 small RNA promoter (Nekrasov et al., 2013). These were cloned into a binary transformation vector containing the NPTII kanamycin resistance gene as well as a Cas9-encoding sequence fused to an N-terminal nuclear-localization signal and driven by the CaMV 35S promoter (Engler et al., 2014; Fauser et al., 2014). The resulting constructs as well a control construct containing only the NPTII- and Cas9-encoding sequences were transformed to P. andersonii using the method described above. For all constructs, transgenic shoots were obtained, although in case of the construct targeting PanHK4 regeneration took considerably longer (up to 6 months). Genotyping of regenerated shoots showed that >85% contained the Cas9 gene, indicating successful transformation. Potential mutations at any of the target sites were identified through PCR amplification and subsequent sequencing of the PCR product. This revealed mutations at the target site in about half of the transgenic shoots examined, of which the majority were bi-allelic (Table 1). Most mutations represent small insertions and deletions but also larger deletions and inversions were identified, some of which occur in between two target sites (Supplementary Figures 7–10). In case of PanHK4, most mutants contained small in-frame deletions of 3 or 6 bp that most likely do not disrupt protein function. In fact, only two bi-allelic knockout mutants could be identified (Supplementary Figure 8). For the remaining three constructs, multiple bi-allelic knockout mutants were identified of which three individuals were selected for further studies (for an overview of mutant alleles see Supplementary Figures 7–10).
For phenotypic evaluation, P. andersonii T0 transgenic lines are propagated vegetatively. Therefore, we first evaluated whether any of the mutant lines might be chimeric. To this end, tissue samples were taken from at least three different positions and genotyped for the corresponding target mutation. For each of the mutant lines, except Pannsp2–9, the same mutations were retrieved, suggesting that genome editing occurred soon after T-DNA integration. In case of Pannsp2–9, chimeric mutations were detected at the first of three target sites (Supplementary Figure 10C). However, the nature of the mutations at the second and third target site prevent that gene function could be restored in this line. Therefore, all 11 mutants are suitable for phenotypic evaluation. This proves that CRISPR/Cas9 can be used to efficiently mutagenize P. andersonii in the T0 generation.
Non-symbiotic Phenotypes in Parasponia ein2, hk4, nsp1, and nsp2 Mutant Lines
To characterize the resulting Panein2, Panhk4, Pannsp1 and Pannsp2 mutant lines, we studied their non-symbiotic phenotypes. PanEIN2 putatively encodes a central component of the ethylene signaling pathway and therefore Panein2 mutants are expected to be ethylene insensitive. One phenotype triggered in response to ethylene treatment is abscission of leaves and flowers, as shown in amongst others common bean (Phaseolus vulgaris), cotton (Gossypium hirsutum), and citrus (Citrus clementina) (Jackson and Osborne, 1970; Brown, 1997; Agustí et al., 2009). We exploited this phenotype to assess ethylene sensitivity of Panein2 mutants. To this end, the tips of young shoot branches of greenhouse grown trees were exposed to ethylene gas. Within 3 days, ethylene triggered abscission of ∼65% of treated leaves on wild-type (WT) P. andersonii as well as control transgenic lines (Figure 2). In contrast, leaf abscission was not observed on Panein2 mutant trees (Figure 2). This demonstrates that Panein2 mutants are indeed ethylene insensitive.
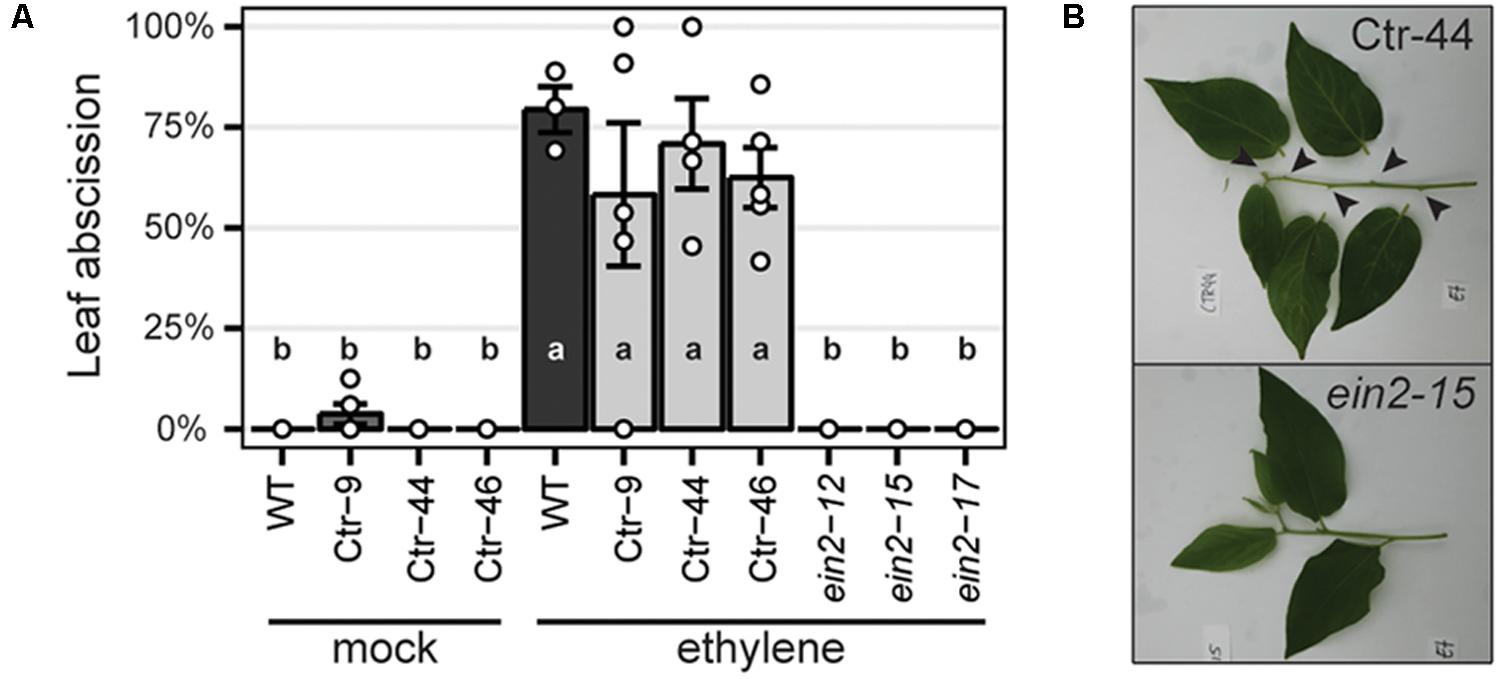
FIGURE 2. Panein2 mutants are insensitive to ethylene treatment. (A) Percentage of abscised leaves after mock or ethylene treatment. Data represent means of 3–5 biological replicates ± SEM. Dots represent measurement values of biological repeats. Different letters indicate statistical significance (p < 0.05) as determined by ANOVA in combination with Tukey post hoc test. (B) Representative images showing abscission of leaves on a transgenic control line (Ctr-44), but not on a Panein2 mutant. Abscission points are indicated by arrowheads.
Inspection of Panein2 mutant trees revealed an additional non-symbiotic phenotype. These trees form bisexual flowers containing both male and female reproductive organs (Figures 3A–C). In contrast, WT P. andersonii trees form unisexual flowers that contain either stamens or carpels (Becking, 1992) (Figures 3D,E). This suggests that ethylene is involved in the regulation of Parasponia sex type.
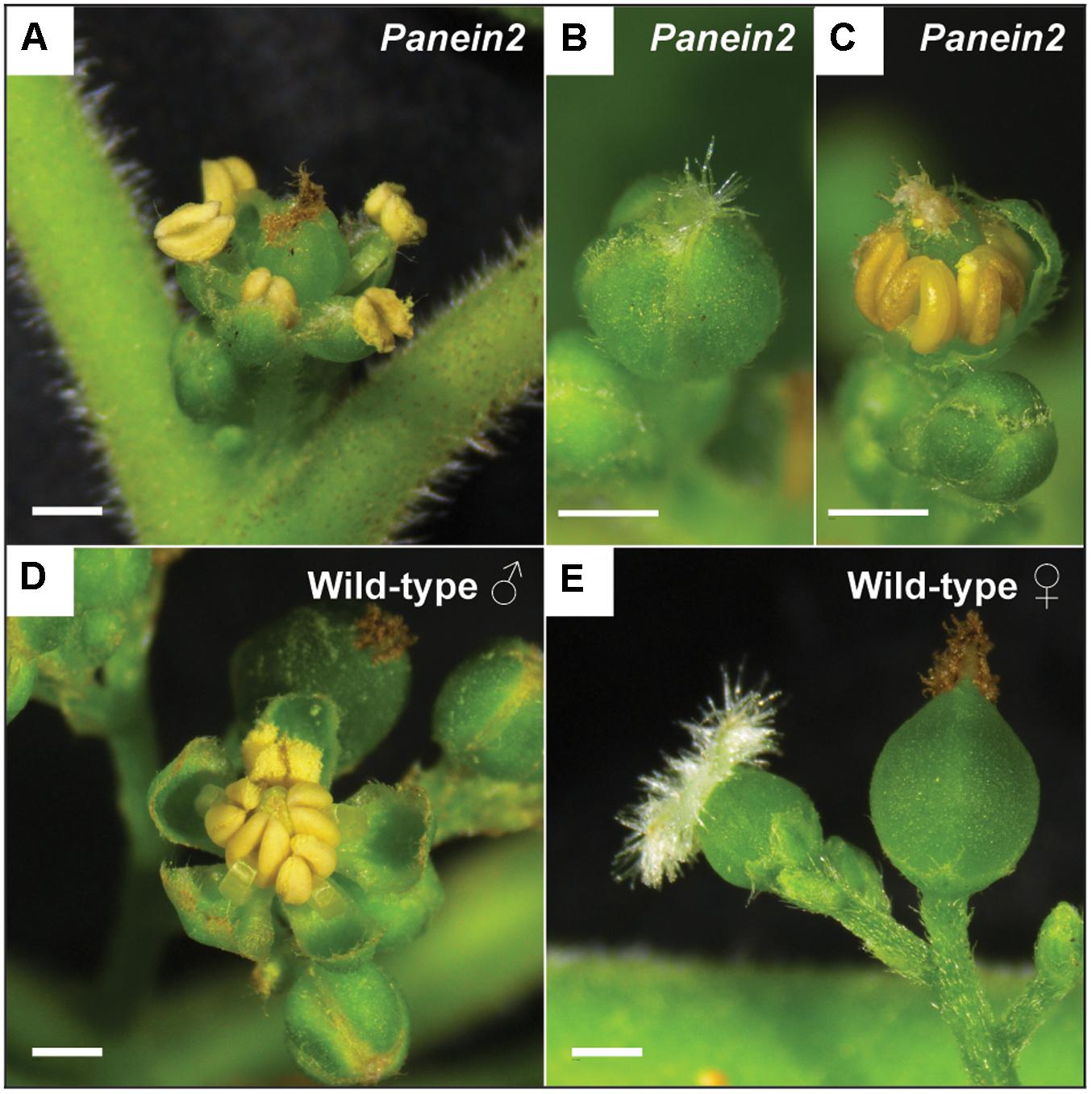
FIGURE 3. Panein2 mutants form bisexual flowers. (A) Mature Panein2 flower. Note the presence of both stamen and a carpel inside Panein2 flowers. (B) Immature Panein2 flower. Note the presence of stigmata, indicating presence of a carpel inside the flower. (C) Immature Panein2 flower of which sepals have been removed to show the presence of stamen. (D) Mature wild-type (WT) male flower. (E) Mature WT female flowers. Left: young female flower. Right: older female flower. Scale bars are equal to 1 mm.
Cytokinins are important regulators of cambial activity, as shown in A. thaliana and poplar (Populus tremula x tremuloides) (Matsumoto-Kitano et al., 2008; Nieminen et al., 2008; Bhalerao and Fischer, 2017). To determine whether reduced cytokinin sensitivity in Panhk4 mutant lines affects the activity of the procambium, we sectioned young primary stems, 5 cm below the apical meristem. This showed that procambium activity is reduced in Panhk4 mutant lines compared to transgenic controls (Figure 4). Therefore, we conclude that PanHK4-mediated cytokinin signaling is required for regulation of P. andersonii secondary growth.
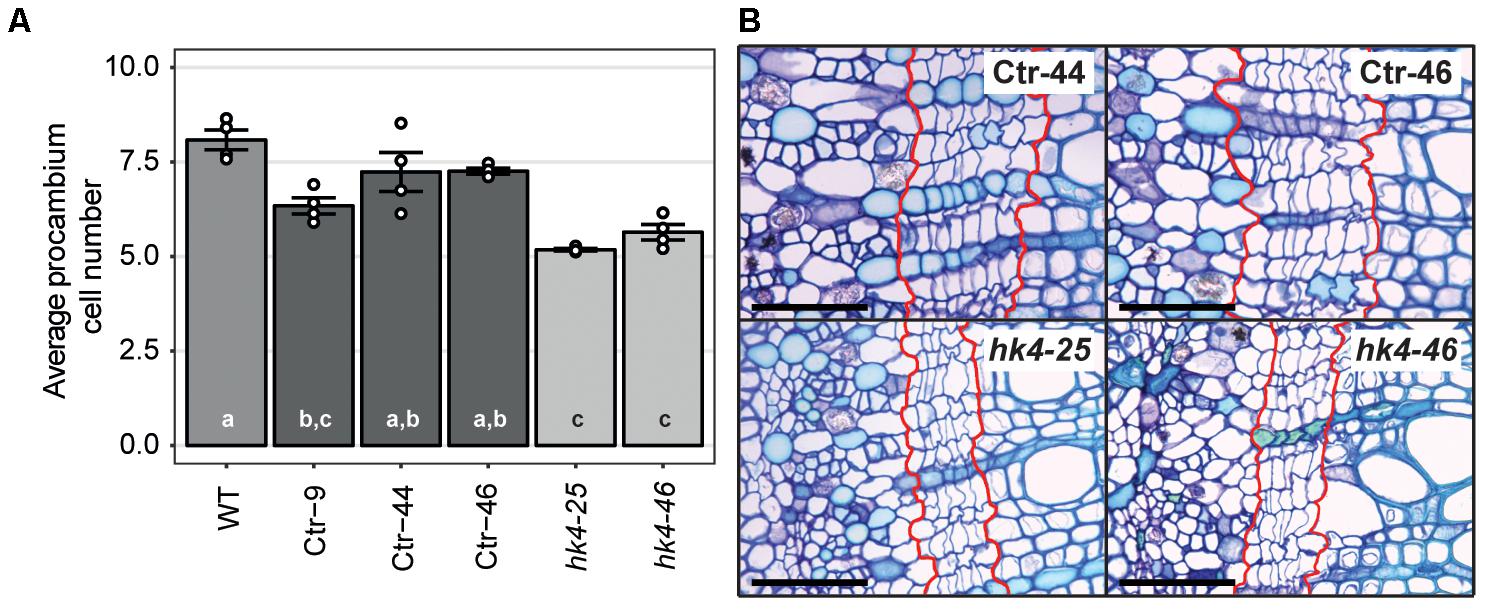
FIGURE 4. Panhk4 mutants display reduced procambium activity. (A) Average number of procambium cells in WT, transgenic control (Ctr) and Panhk4 mutant stems. Data represent means of four biological replicates ± SEM. Dots represent measurement values of biological repeats. Different letters indicate statistical significance (p < 0.05) as determined by ANOVA in combination with Tukey post hoc test. (B) Representative images of stem cross-sections of control transgenic (Ctr) and Panhk4 mutant lines. The location of the procambium is indicated by red lines. Scale bars represent 50 μm.
Expression studies in M. truncatula previously identified a set of genes downregulated in roots of Mtnsp1 and Mtnsp2 mutants (Liu et al., 2011). Among these are DWARF27 (MtD27; Medtr1g471050) and MORE AXILLARY BRANCHING 1 (MtMAX1; Medtr3g104560) that are putatively involved in strigolactone biosynthesis (Liu et al., 2011; Cardoso et al., 2014; Zhang et al., 2014; van Zeijl et al., 2015a). We identified putative P. andersonii orthologs of these genes (Supplementary Figures 11, 12) and compared their expression levels in young root segments of three Pannsp1, Pannsp2 and control plants by qRT-PCR. This showed that expression of PanD27 and PanMAX1 is reduced in roots of Pannsp1 and Pannsp2 mutant lines (Figure 5). We noted that Pannsp1 mutant lines differ in the level of PanD27 and PanMAX1 expression. Both genes have an intermediate expression level in Pannsp1–6 and Pannsp1–13, compared to Pannsp1–39 and Pannsp2 mutants (Figure 5). The three Pannsp1 mutant lines differ from each other in the type of mutations that were created. Pannsp1–6 and Pannsp1–13 contain a 1 bp insertion and 5 bp deletion close to the 5′-end of the coding region, respectively. These mutations are immediately followed by a second in-frame ATG that in WT PanNSP1 encodes a methionine at position 16. In contrast, Pannsp1–39 contains a large 232 bp deletion that removes this in-frame ATG (see Supplementary Figure 9). Together, this suggests that Pannsp1–6 and Pannsp1–13 might represent weak alleles. Overall, these data suggest that regulation of D27 and MAX1 expression by NSP1 and NSP2 is conserved between M. truncatula and P. andersonii.
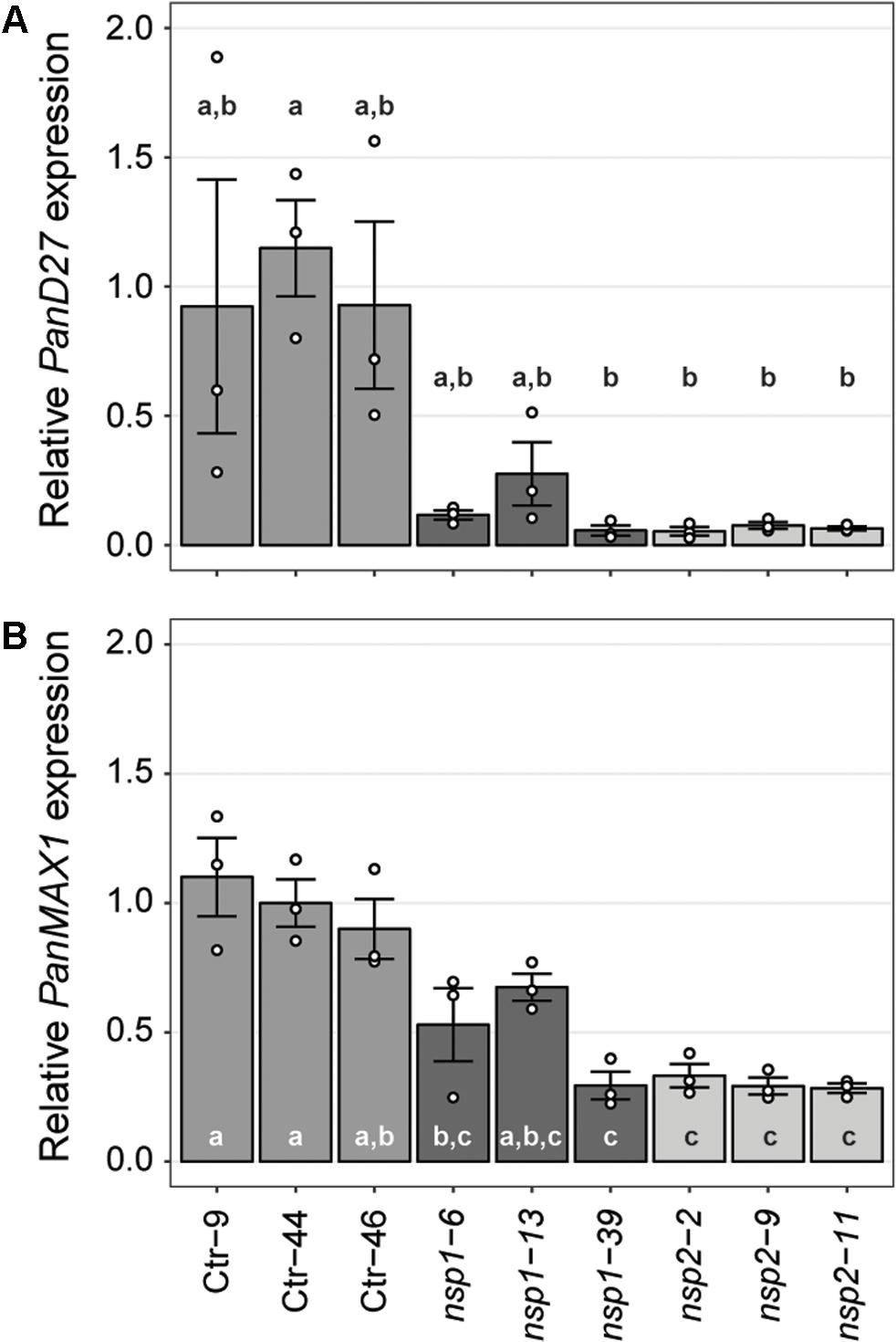
FIGURE 5. Expression of PanD27 and PanMAX1 is reduced in Pannsp1 and Pannsp2 mutant roots. Relative expression of PanD27 (A) and PanMAX1 (B) in roots of transgenic control (Ctr) and Pannsp1 and Pannsp2 mutant lines. Data represent means of three biological replicates ± SEM. Dots represent measurement values of biological repeats. Different letters indicate statistical significance (p < 0.05) as determined by ANOVA in combination with Tukey post hoc test.
Taken together, we showed that EIN2, HK4, NSP1, and NSP2 in P. andersonii commit non-symbiotic functions in hormonal homeostasis. These functions are in line with what is described for other plant species, suggesting that the generated P. andersonii lines represent true mutants.
Nodulation Phenotypes of Parasponia Panein2 and panhk4 Mutants Differ From Their Legume Counterparts
To determine whether PanEIN2, PanHK4, PanNSP1, and PanNSP2 perform similar functions during nodule formation as their legume orthologs, P. andersonii mutant plantlets were inoculated with Mesorhizobium plurifarium BOR2 (van Velzen et al., 2017). Nodulation phenotypes were examined 1 month after inoculation.
The strong Pannsp1–39 mutant allele and all three Pannsp2 mutant lines are unable to form root nodules (Figure 6A and Supplementary Figure 13). This is similar as described for M. truncatula, L. japonicus, and Pisum sativum nsp1 and nsp2 mutants (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Shtark et al., 2016). In contrast, the weak Pannsp1 alleles Pannsp1–6 and Pannsp1–13 could be nodulated similar as WT or control transgenic plants (Figure 6A and Supplementary Figure 13), suggesting that residual PanNSP1 activity is sufficient to support root nodule formation. Overall, these data show that NSP1 and NSP2 functioning is essential for root nodule formation in Parasponia.
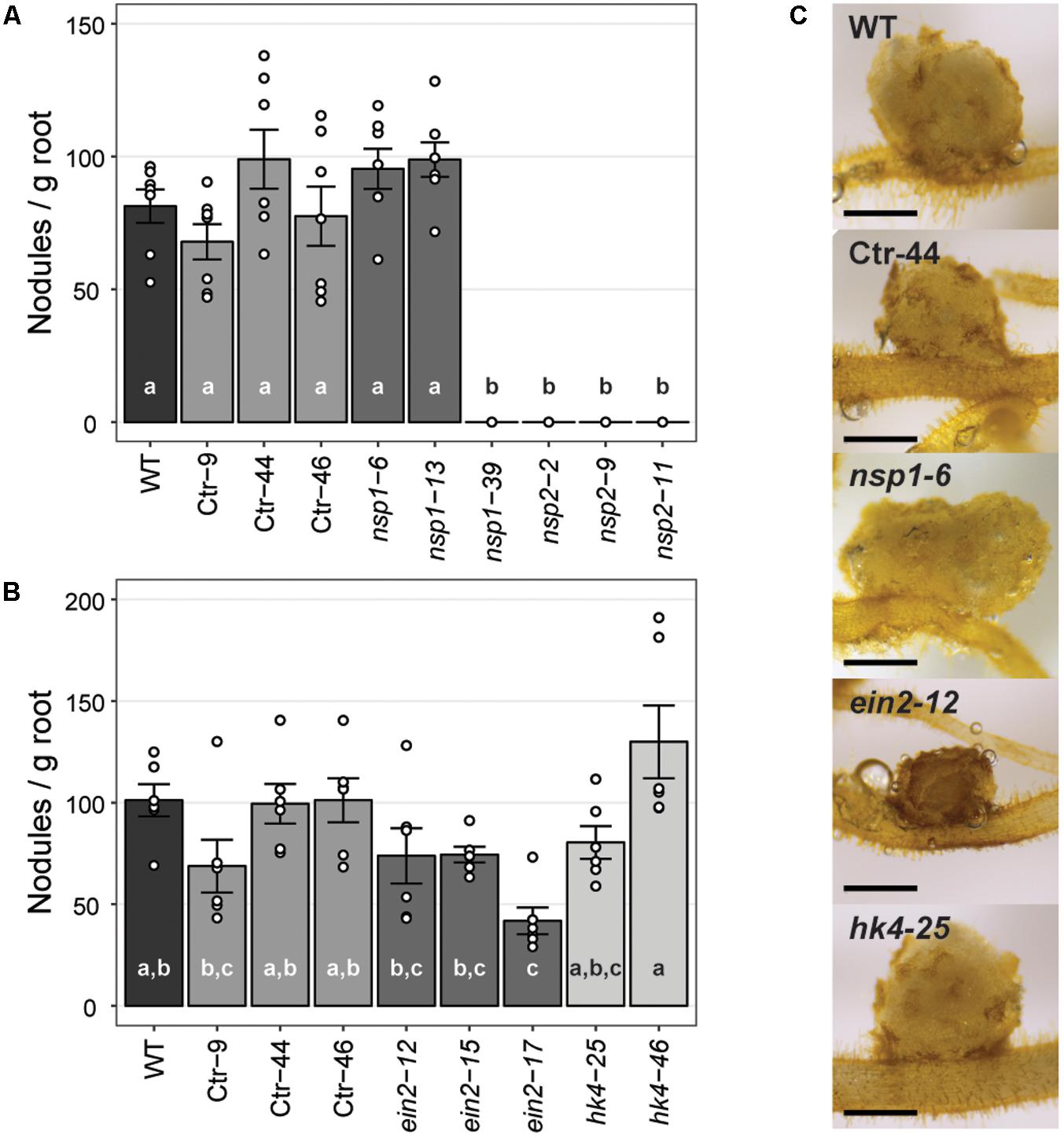
FIGURE 6. Nodule formation on P. andersonii CRISPR/Cas9 mutant lines. (A) Nodule number per gram root fresh weight on WT, transgenic control (Ctr) and Pannsp1 and Pannsp2 mutant lines. Nodule number was determined at 1 month after inoculation with Mesorhizobium plurifarium BOR2. (B) Nodule number per gram root fresh weight on WT, transgenic control (Ctr) and Panein2 and Panhk4 mutant lines. Nodule number was determined at 1 month after inoculation with Mesorhizobium plurifarium BOR2. (C) Representative images of 1 month-old nodules. Note the dark color of Panein2 nodules. Scale bars are equal to 0.5 mm. Data in (A,B) represent means of 5–7 biological replicates ± SEM. Dots represent measurement values of biological repeats. Different letters indicate statistical significance (p < 0.05) as determined by ANOVA in combination with Tukey post hoc test. Data on nodule number and root weight are shown in Supplementary Figure 13.
Analysis of the nodulation phenotype of P. andersonii Panhk4 mutants showed that PanHK4 is not required for root nodule formation. Both Panhk4 mutant lines formed a similar amount of nodules as WT and transgenic controls (Figure 6B and Supplementary Figure 13). This is different from the corresponding legume mutants – M. truncatula Mtcre1 and L. japonicus Ljlhk1 – that are generally not forming root nodules (Murray et al., 2007; Plet et al., 2011).
The phenotype of P. andersonii Panein2 mutants also differs from that of legume mutants. M. truncatula ein2 mutants – as well as L. japonicus plants in which both EIN2-encoding genes have been silenced – form more nodules than WT, which are clustered in distinct zones along the root (Penmetsa and Cook, 1997; Miyata et al., 2013). Panein2 mutants do not form such nodule clusters and nodule number is not higher than WT (Figure 6B and Supplementary Figure 13). However, nodules formed on Panein2 mutant plants are smaller and dark colored when compared to nodules of control plants (Figure 6C). This suggests impaired nodule development in P. andersonii ein2 mutants.
To determine the cytoarchitecture of Panein2, Panhk4, and Pannsp1–6/Pannsp1–13 mutant nodules, we sectioned ∼10 nodules for each mutant line and studied these by light microscopy. Wild-type P. andersonii nodules harbor an apical meristem, followed by several cell layers that contain infection threads (Figure 7A) (Op den Camp et al., 2012). Below this infection zone, 2–3 cell layers are present that display vacuolar fragmentation and increase in size compared to non-infected cells (Figure 7B). These cells are followed by cells that are filled with fixation threads (Figures 7B,C). The general cytoarchitecture of Panhk4 and Pannsp1–6/Pannsp1–13 mutant nodules does not differ from that of WT or transgenic control nodules (Figures 7A,D,E), suggesting that these are functional. In contrast, in Panein2 mutant nodules intracellular infection is hampered (Figures 7F–I). Most (>75%) Panein2 mutant nodules harbor only infection threads as well as large apoplastic colonies (Figure 7I). Some mutant nodules, harbor cells that contain fixation threads. However, even in the best nodules, fixation thread formation is severely delayed and many cells in the fixation zone still show vacuolar fragmentation (Figure 7I). This shows that ethylene signaling is required for efficient fixation thread formation in P. andersonii nodules.
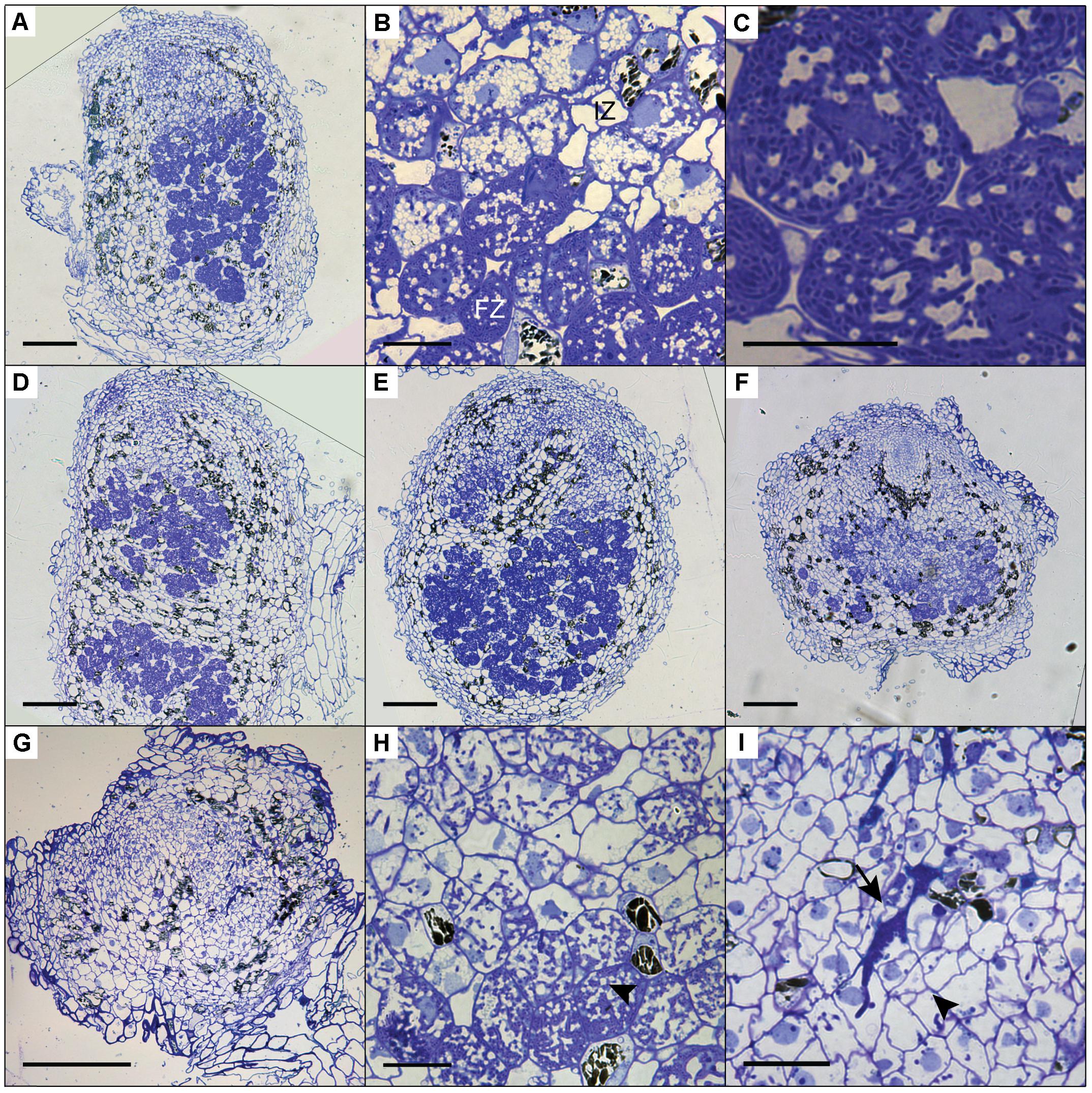
FIGURE 7. Cytoarchitecture of CRISPR/Cas9 mutant nodules. (A) Longitudinal nodule sections of 1 month-old nodule formed on transgenic control line Ctr-9. (B) Zoom in on cells in the infection (IZ) and fixation zone (FZ) of the transgenic control nodule shown in (A). Note the presence of small fragmented vacuoles in infected cells in the infection zone. (C) Zoom in on a cell in the fixation zone of a transgenic control nodule showing the presence of fixation threads. (D) Longitudinal nodule sections of 1 month-old nodule formed on Pannsp1–13. (E) Longitudinal nodule sections of 1 month-old nodule formed on Panhk4–25. (F) Longitudinal nodule sections of 1 month-old nodule formed on Panein2–15. (G) Longitudinal nodule sections of 1 month-old nodule formed on Panein2–17. (H) Zoom in on cells in the basal part of the Panein2–15 nodule shown in (F). Indicated by an arrowhead are cells containing fixation threads. (I) Zoom in of the Panein2–17 nodule shown in (G). Indicated by an arrowhead are infection threads. Indicated by an arrow are large apoplastic colonies. Scale bars in (A,D–G) are equal to 150 and 25 μm in (B,C,H,I).
Taken together, these data reveal symbiotic mutant phenotypes for nsp1, nsp2 and ein2, whereas no effect on nodule formation was found by knocking out hk4 in P. andersonii. Interestingly, we uncovered a novel role for the ethylene signaling component EIN2 in intracellular infection of P. andersonii nodules.
Discussion
Comparative studies between legumes and the Cannabaceae tree Parasponia can provide insights into ‘core’ genetic networks underlying rhizobium symbiosis (van Velzen et al., 2017). To facilitate such studies, we aimed to establish a reverse genetics platform for P. andersonii based on CRISPR/Cas9 genome editing. We show that using A. tumefaciens transformation, P. andersonii stable transgenic lines can be obtained in 3–4 months. In combination with CRISPR/Cas9 mutagenesis, this allows efficient generation of bi-allelic knockout mutants. As a proof-of-concept, we mutated four genes that commit essential symbiotic functions in legumes as well as control different hormonal networks. Characterization of the resulting lines revealed both symbiotic as well as non-symbiotic mutant phenotypes. Therefore, we conclude that stable A. tumefaciens-mediated transformation in combination with CRISPR/Cas9 genome editing can be efficiently used for reverse genetic analysis in P. andersonii.
Plant transformation efficiency is the main bottleneck in plant genome editing (Altpeter et al., 2016; Ledford, 2016). Especially regeneration of an entire transgenic plant out of a single transformed cell remains difficult for most plant species. We took advantage of an efficient micro-propagation system available for Parasponia spp. to establish a protocol for stable transformation (Davey et al., 1993; Webster et al., 1995; Cao et al., 2012). About 8–12 weeks after cocultivation with A. tumefaciens, ∼50% of explants develop transgenic shoots. This relatively high efficiency is, in part, obtained through an initial 9-day culturing period on root-inducing medium, before incubation on standard propagation medium. This adaptation in the protocol was inspired by a recent study that showed that regeneration of plant cells consists of two distinctive steps (Kareem et al., 2015). Regenerative competence is established through activation of a root developmental program, followed by activation of shoot promoting factors that are required to complete shoot regeneration (Kareem et al., 2015). The latter explains why transfer to propagation medium is required to regenerate P. andersonii transgenic shoots. However, this promoting effect of rooting medium on regeneration of transgenic shoots might differ between different explant types, as noted for P. andersonii stems and petioles (Supplementary Table 3).
An advantage of the Parasponia system is that T0 transgenic knockout mutants can be clonally propagated through in vitro micro-propagation (Davey et al., 1993; Webster et al., 1995; Cao et al., 2012). This allows a large number of rooted plantlets to be generated in a relatively short time span. As a result, phenotypic characterization can be initiated already at 4 months after the start of the transformation. However, a disadvantage of clonal propagation in combination with CRISPR/Cas9 mutagenesis is the possibility of obtaining chimeric mutants. Among the mutant lines we created, we identified one line (out of 11) that was chimeric for one out of three CRISPR target sites (Supplementary Figure 10C). Most mutant lines were genetically homogeneous, suggesting that mutations are induced soon after T-DNA integration. This is consistent with results in poplar, which also revealed a low percentage of chimeric mutants (Fan et al., 2015). Since chimeras are observed occasionally, thorough genotypic analysis will be required when phenotyping is performed in the T0 generation. Besides vegetative propagation, Parasponia trees can also be propagated generatively. Under suitable greenhouse conditions, Parasponia trees flower within ∼6–9 months and are self-compatible (Becking, 1992). However, Parasponia trees can be monoecious or diecious and female flowers are wind pollinated (Soepadmo, 1974). This complicates selfing of trees and the production of pure seed badges. Additionally, Parasponia trees are fast growing and occupy a substantial amount of space in a tropical greenhouse (28°C, ∼100% relative humidity), making generative propagation of multiple mutant lines logistically somewhat challenging. An alternative to generative propagation is in vitro maintenance of transgenic lines. Additionally, the fast and efficient transformation procedure presented here will allow recreation of a particular mutant in less than 6 months.
Among the mutants we created, Panhk4 and Panein2 showed symbiotic phenotypes that differ from corresponding legume mutants. P. andersonii Panhk4 mutants form nodules with a WT cytoarchitecture, indicating that these nodules are most likely functional. Analysis of stem cross-sections showed that Panhk4 mutants possess a reduced procambial activity. Similar phenotypes are observed in homologous mutants in A. thaliana (Mahonen et al., 2006a,b). Procambium activity is slightly reduced in the orthologs receptor mutant arabidopsis histidine kinase 4 (ahk4), whereas it is completely abolished in the ahk2 ahk3 ahk4 triple mutant (Mahonen et al., 2006a,b). The comparable phenotypes in cambium activity upon mutating histidine kinases suggest that PanHK4 encodes a functional cytokinin receptor. M. truncatula and L. japonicus mutants in the cytokinin receptors orthologs to PanHK4 are characterized as nodulation deficient (Murray et al., 2007; Plet et al., 2011). However, these mutants occasionally form nodules (Plet et al., 2011; Held et al., 2014; Boivin et al., 2016). This suggests redundant functioning of additional cytokinin receptors in both legume species. The P. andersonii genome also encodes two additional cytokinin receptors: PanHK2 and PanHK3 (van Velzen et al., 2017) (Supplementary Figure 4). Therefore, redundant functioning of one of these receptors cannot be excluded. In legumes, cell divisions associated with nodule development are initiated in the root cortex in response to epidermal perception of rhizobial signals (Timmers et al., 1999; Xiao et al., 2014). Cytokinin appears important for activation of this cortical organogenesis program (Vernie et al., 2015; Gamas et al., 2017). In Parasponia, cell divisions associated with nodule development are first observed in the epidermis, the cell layer that is in direct contact with the rhizobium bacteria (Lancelle and Torrey, 1984; Geurts et al., 2016). This difference in mitotically-responding tissues could create different dependencies on cytokinin signaling between legumes and Parasponia. However, whether this explains the absence of a symbiotic phenotype of Panhk4 mutants requires further experimentation.
Panein2 mutants are ethylene insensitive, as indicated by the absence of leaf abscission following ethylene treatment. Additionally, we noticed a disturbed sex differentiation in Panein2 flowers. Functioning of ethylene in flower sex differentiation is known in cucurbit species, like cucumber (Cucumis sativus) and melon (Cucumis melo) (Rudich et al., 1972; Yin and Quinn, 1995; Tanurdzic and Banks, 2004). Molecular genetic studies revealed that flower bud-specific expression of ACC synthase (ACS) genes, which are essential for biosynthesis of the ethylene precursor ACC, inhibits stamen development (Boualem et al., 2008, 2015). In line with these findings in cucurbits, we hypothesize that EIN2-mediated ethylene signaling commits a similar function in sex differentiation in Parasponia species.
In symbiotic context, EIN2 knockout mutations result in different phenotypes between Parasponia and legumes. In legumes, ethylene negatively regulates rhizobial infection and root nodule formation (Penmetsa and Cook, 1997; Penmetsa et al., 2008; Miyata et al., 2013). This is illustrated by the phenotype of the M. truncatula ein2 mutant (named sickle) that forms extensive epidermal infection threads and clusters of small nodules (Penmetsa and Cook, 1997; Xiao et al., 2014). P. andersonii ein2 mutants also form smaller nodules than WT. However, in contrast to the Mtein2 mutant, these nodules are regularly spaced on the root system. This suggests that in Parasponia ethylene signaling is not involved in regulating nodule number. Additionally, also the infection phenotype of Panein2 mutants differs from that in legumes. In M. truncatula and L. japonicus, interference with ethylene signaling increases the number of epidermal infection threads but does not affect intracellular colonization of nodule cells (Penmetsa and Cook, 1997; Nukui et al., 2004; Lohar et al., 2009). In contrast, in P. andersonii Panein2 mutants, intracellular colonization is hampered. Inside nodules, large apoplastic colonies are observed and fixation thread formation is severely reduced or even absent. This suggests that in Parasponia a functional ethylene signaling pathway is required for efficient intracellular infection of nodule cells.
Mutagenesis of the NSP2 ortholog of P. andersonii indicated a conserved symbiotic role for this GRAS-type transcriptional regulator. In legumes, NSP2 works in concert with NSP1 to control root nodule formation (Hirsch et al., 2009). Mutagenesis of the NSP1 ortholog of P. andersonii resulted in contrasting nodulation phenotypes. Two mutant lines, Pannsp1–6 and Pannsp1–13, form nodules with a WT cytoarchitecture, whereas mutant line Pannsp1–39 is unable to form nodules (Figures 6, 7). However, all three mutants are affected in transcriptional regulation of strigolactone biosynthesis genes PanD27 and PanMAX1 (Figure 5). The three Pannsp1 mutant lines differ from each other in the type of mutations that were created. Pannsp1–6 and Pannsp1–13 contain small deletions that are immediately followed by a second in-frame ATG that in WT PanNSP1 encodes a methionine at position 16. In contrast, Pannsp1–39 contains a larger deletion that removes this in-frame ATG (see Supplementary Figure 9). Several reports have shown that alternative start codons are occasionally used to initiate transcription (Chabregas et al., 2003; Thatcher et al., 2007; Bazykin and Kochetov, 2011). Therefore, Pannsp1–6 and Pannsp1–13 most probably represent weak alleles that still possess residual PanNSP1 function. Such residual levels of PanNSP1 are affecting the expression of strigolactone biosynthesis genes, but are still sufficient to allow nodule formation. Therefore, we argue that the P. andersonii Pannsp1–39 line carries a knockout mutation, indicating that in P. andersonii both NSP1 and NSP2 are essential for rhizobium root nodule formation.
Taken together, we showed that P. andersonii can be efficiently transformed using A. tumefaciens and is amenable to targeted mutagenesis using CRISPR/Cas9. This protocol takes only marginally more time than the transient A. rhizogenes transformation system that is generally used to study root nodule formation (e.g., Boisson-Dernier et al., 2001; Kumagai and Kouchi, 2003; Limpens et al., 2004; Op den Camp et al., 2011; Cao et al., 2012) but has several advantages. One of these is the absence of the A. rhizogenes root inducing locus (rol) that interferes with hormone homeostasis (Nilsson and Olsson, 1997). The protocol we developed will allow studies on P. andersonii symbiosis genes to determine to what extent legumes and Parasponia use a similar mechanism to establish a nitrogen-fixing symbiosis with rhizobium.
Data Availability Statement
All datasets analyzed for this study are included in the manuscript and the supplementary files. Gene identifiers for all P. andersonii genes used in this study can be found in Supplementary Table 8. Sequences can be downloaded from www.parasponia.org.
Author Contributions
Conceptualization, AvZ and RG; Methodology, AvZ, MH, SL, and WK; Investigation, AvZ, TW, MSK, LR, FB, MH, SL, EF, and WK; Formal analysis, AvZ, TW, and EF; Visualization, AvZ; Writing – original draft, AvZ; Writing – review and editing, AvZ and RG; Funding acquisition, TB and RG; Supervision, RG.
Funding
This work was supported by NWO-VICI (865.13.001) to RG, NWO-VENI (863.15.010) to WK, European Research Council (ERC-2011-AdG294790) to TB, and China Scholarship Council (201303250067) to FB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors like to thank Renze Heidstra for help with FACS analysis and Michiel Lammers and Renze Heidstra for useful tips regarding CRISPR/Cas9 strategy. Ethylene gas was kindly provided by Arjen van de Peppel and Julian Verdonk. Golden Gate parts and cloning vectors were kindly provided by Mark Youles, Sophien Kamoun, and Sylvestre Marillonnet through the Addgene database. The work described here has not been previously published, except in the form of a chapter in the publicly defended Ph.D. thesis of AvZ (van Zeijl, 2017). The publication of this thesis has occurred in accordance with the policy of Wageningen University & Research, Wageningen, Netherlands.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00284/full#supplementary-material
Footnotes
- ^ http://phytozome.jgi.doe.gov/
- ^ www.arabidopsis.org
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ www.arabidopsis.com
References
Agustí, J., Merelo, P., Cercós, M., Tadeo, F. R., and Talón, M. (2009). Comparative transcriptional survey between laser-microdissected cells from laminar abscission zone and petiolar cortical tissue during ethylene-promoted abscission in citrus leaves. BMC Plant Biol. 9:127. doi: 10.1186/1471-2229-9-127
Akkermans, A. D. L., Abdulkadir, S., and Trinick, M. J. (1978). Nitrogen-fixing root nodules in Ulmaceae. Nature 274:190. doi: 10.1038/274190c0
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520. doi: 10.1105/tpc.16.00196
Bansal, R., Mittapelly, P., Cassone, B. J., Mamidala, P., Redinbaugh, M. G., and Michel, A. (2015). Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS One 10:e0134890. doi: 10.1371/journal.pone.0134890
Bazykin, G. A., and Kochetov, A. V. (2011). Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 39, 567–577. doi: 10.1093/nar/gkq806
Becking, J. H. (1983). The Parasponia parviflora-rhizobium symbiosis-isotopic nitrogen-fixation, hydrogen evolution and nitrogen-fixation efficiency, and oxygen relations. Plant Soil 75, 343–360. doi: 10.1007/BF02369970
Becking, J. H. (1992). “The Rhizobium symbiosis of the nonlegume Parasponia,” in Biological Nitrogen Fixation, eds G. S. Stacey, H. J. Evans, and R. H. Burris (New York, NY: Routledge), 497–559.
Behm, J. E., Geurts, R., and Kiers, E. T. (2014). Parasponia: a novel system for studying mutualism stability. Trends Plant Sci. 19, 757–763. doi: 10.1016/j.tplants.2014.08.007
Bhalerao, R. P., and Fischer, U. (2017). Environmental and hormonal control of cambial stem cell dynamics. J. Exp. Bot. 68, 79–87. doi: 10.1093/jxb/erw466
Boisson-Dernier, A., Chabaud, M., Garcia, F., Bécard, G., Rosenberg, C., and Barker, D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700. doi: 10.1094/MPMI.2001.14.6.695
Boivin, S., Kazmierczak, T., Brault, M., Wen, J. Q., Gamas, P., Mysore, K. S., et al. (2016). Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 39, 2198–2209. doi: 10.1111/pce.12779
Boualem, A., Fergany, M., Fernandez, R., Troadec, C., Martin, A., Morin, H., et al. (2008). A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321, 836–838. doi: 10.1126/science.1159023
Boualem, A., Troadec, C., Camps, C., Lemhemdi, A., Morin, H., Sari, M. A., et al. (2015). A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350, 688–691. doi: 10.1126/science.aac8370
Brown, K. M. (1997). Ethylene and abscission. Physiol. Plant. 100, 567–576. doi: 10.1111/j.1399-3054.1997.tb03062.x
Cao, Q., Op den Camp, R., Seifi Kalhor, M., Bisseling, T., and Geurts, R. (2012). Efficiency of Agrobacterium rhizogenes–mediated root transformation of Parasponia and Trema is temperature dependent. Plant Growth Regul. 68, 459–465. doi: 10.1007/s10725-012-9734-y
Cardoso, C., Zhang, Y. X., Jamil, M., Hepworth, J., Charnikhova, T., Dimkpa, S. O. N., et al. (2014). Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc. Natl. Acad. Sci. U.S.A. 111, 2379–2384. doi: 10.1073/pnas.1317360111
Chabregas, S. M., Luche, D. D., Van Sluys, M. A., Menck, C. F. M., and Silva-Filho, M. C. (2003). Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J. Cell Sci. 116, 285–291. doi: 10.1242/jcs.00228
Clason, E. W. (1936). The vegetation of the upper-Badak region of mount Kelut (east java). Bull. Jard. Bot. Buitenz. III 13, 509–518. doi: 10.1073/pnas.1714977115
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Davey, M. R., Webster, G., Manders, G., Ringrose, F. L., Power, J. B., and Cocking, E. C. (1993). Effective nodulation of micro-propagated shoots of the non-legume Parasponia andersonii by Bradyrhizobium. J. Exp. Bot. 44, 863–867. doi: 10.1093/jxb/44.5.863
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Duarte, J. M., Wall, P. K., Edger, P. P., Landherr, L. L., Ma, H., Pires, J. C., et al. (2010). Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol. 10:61. doi: 10.1186/1471-2148-10-61
Engler, C., Gruetzner, R., Kandzia, R., and Marillonnet, S. (2009). Golden Gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553. doi: 10.1371/journal.pone.0005553
Engler, C., Youles, M., Gruetzner, R., Ehnert, T. M., Werner, S., Jones, J. D. G., et al. (2014). A Golden Gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843. doi: 10.1021/sb4001504
Fan, D., Liu, T. T., Li, C. F., Jiao, B., Li, S., Hou, Y. S., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Fedorova, E., Thomson, R., Whitehead, L. F., Maudoux, O., Udvardi, M. K., and Day, D. A. (1999). Localization of H+-ATPases in soybean root nodules. Planta 209, 25–32. doi: 10.1007/s004250050603
Gamas, P., Brault, M., Jardinaud, M.-F., and Frugier, F. (2017). Cytokinins in symbiotic nodulation: when, where, what for? Trends Plant Sci. 22, 792–802. doi: 10.1016/j.tplants.2017.06.012
Geurts, R., Lillo, A., and Bisseling, T. (2012). Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr. Opin. Plant Biol. 15, 438–443. doi: 10.1016/j.pbi.2012.04.004
Geurts, R., Xiao, T. T., and Reinhold-Hurek, B. (2016). What does it take to evolve a nitrogen-fixing endosymbiosis? Trends Plant Sci. 21, 199–208. doi: 10.1016/j.tplants.2016.01.012
Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18, 2680–2693. doi: 10.1105/tpc.106.043778
Granqvist, E., Sun, J., Op den Camp, R., Pujic, P., Hill, L., Normand, P., et al. (2015). Bacterial-induced calcium oscillations are common to nitrogen-fixing associations of nodulating legumes and non-legumes. New Phytol. 207, 551–558. doi: 10.1111/nph.13464
Heckmann, A. B., Lombardo, F., Miwa, H., Perry, J. A., Bunnewell, S., Parniske, M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. doi: 10.1104/pp.106.089508
Held, M., Hou, H., Miri, M., Huynh, C., Ross, L., Hossain, M. S., et al. (2014). Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26, 678–694. doi: 10.1105/tpc.113.119362
Hirsch, S., Kim, J., Muñoz, A., Heckmann, A. B., Downie, J. A., and Oldroyd, G. E. D. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. doi: 10.1105/tpc.108.064501
Jackson, M. B., and Osborne, D. J. (1970). Ethylene, the natural regulator of leaf abscission. Nature 225, 1019–1022. doi: 10.1038/2251019a0
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kareem, A., Durgaprasad, K., Sugimoto, K., Du, Y., Pulianmackal, A. J., Trivedi, Z. B., et al. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25, 1017–1030. doi: 10.1016/j.cub.2015.02.022
Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kumagai, H., and Kouchi, H. (2003). Gene silencing by expression of hairpin RNA in Lotus japonicus roots and root nodules. Mol. Plant Microbe Interact. 16, 663–668. doi: 10.1094/MPMI.2003.16.8.663
Lamesch, P., Berardini, T. Z., Li, D. H., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Lancelle, S. A., and Torrey, J. G. (1984). Early development of Rhizobium-induced root nodules of Parasponia rigida. I. Infection and early nodule initiation. Protoplasma 123, 26–37. doi: 10.1007/BF01283179
Lazo, G. R., Stein, P. A., and Ludwig, R. A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. doi: 10.1038/nbt1091-963
Li, H.-L., Wang, W., Mortimer, P. E., Li, R.-Q., Li, D.-Z., Hyde, K. D., et al. (2015). Large-scale phylogenetic analyses reveal multiple gains of actinorhizal nitrogen-fixing symbioses in angiosperms associated with climate change. Sci. Rep. 5:14023. doi: 10.1038/srep14023
Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., et al. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55, 983–992. doi: 10.1093/jxb/erh122
Liu, W., Kohlen, W., Lillo, A., Op den Camp, R., Ivanov, S., Hartog, M., et al. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23, 3853–3865. doi: 10.1105/tpc.111.089771
Lohar, D., Stiller, J., Kam, J., Stacey, G., and Gresshoff, P. M. (2009). Ethylene insensitivity conferred by a mutated Arabidopsis ethylene receptor gene alters nodulation in transgenic Lotus japonicus. Ann. Bot. 104, 277–285. doi: 10.1093/aob/mcp132
Mahonen, A. P., Bishopp, A., Higuchi, M., Nieminen, K. M., Kinoshita, K., Tormakangas, K., et al. (2006a). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311, 94–98.
Mahonen, A. P., Higuchi, M., Tormakangas, K., Miyawaki, K., Pischke, M. S., Sussman, M. R., et al. (2006b). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 16, 1116–1122.
Marvel, D. J., Torrey, J. G., and Ausubel, F. M. (1987). Rhizobium symbiotic genes required for nodulation of legume and nonlegume hosts. Proc. Natl. Acad. Sci. U.S.A. 84, 1319–1323. doi: 10.1073/pnas.84.5.1319
Matsumoto-Kitano, M., Kusumoto, T., Tarkowski, P., Kinoshita-Tsujimura, K., Václavíková, K., Miyawaki, K., et al. (2008). Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. U.S.A. 105, 20027–20031. doi: 10.1073/pnas.0805619105
Miyata, K., Kawaguchi, M., and Nakagawa, T. (2013). Two distinct EIN2 genes cooperatively regulate ethylene signaling in Lotus japonicus. Plant Cell Physiol. 54, 1469–1477. doi: 10.1093/pcp/pct095
Murray, J. D., Karas, B. J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315, 101–104. doi: 10.1126/science.1132514
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D. G., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nieminen, K., Immanen, J., Laxell, M., Kauppinen, L., Tarkowski, P., Dolezal, K., et al. (2008). Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. U.S.A. 105, 20032–20037. doi: 10.1073/pnas.0805617106
Nilsson, O., and Olsson, O. (1997). Getting to the root: the role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol. Plant. 100, 463–473. doi: 10.1111/j.1399-3054.1997.tb03050.x
Nukui, N., Ezura, H., and Minamisawa, K. (2004). Transgenic Lotus japonicus with an ethylene receptor gene Cm-ERS1/H70A enhances formation of infection threads and nodule primordia. Plant Cell Physiol. 45, 427–435. doi: 10.1093/pcp/pch046
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Op, den Camp, R. H. M., Polone, E., Fedorova, E., Roelofsen, W., Squartini, A., et al. (2012). Nonlegume Parasponia andersonii deploys a broad Rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Mol. Plant Microbe Interact. 25, 954–963. doi: 10.1094/MPMI-11-11-0304
Op den Camp, R., Streng, A., De Mita, S., Cao, Q., Polone, E., Liu, W., et al. (2011). LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331, 909–912. doi: 10.1126/science.1198181
Penmetsa, R. V., and Cook, D. R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530. doi: 10.1126/science.275.5299.527
Penmetsa, V. R., Uribe, P., Anderson, J., Lichtenzveig, J., Gish, J.-C., Nam, Y. W., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. doi: 10.1111/j.1365-313X.2008.03531.x
Plet, J., Wasson, A., Ariel, F., Le Signor, C., Baker, D., Mathesius, U., et al. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65, 622–633. doi: 10.1111/j.1365-313X.2010.04447.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rudich, J., Kedar, N., and Halevy, A. H. (1972). Ethylene evolution from cucumber plants as related to sex expression. Plant Physiol. 49, 998–999. doi: 10.1104/pp.49.6.998
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J. X., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shtark, O. Y., Sulima, A. S., Zhernakov, A. I., Kliukova, M. S., Fedorina, J. V., Pinaev, A. G., et al. (2016). Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 68, 129–144. doi: 10.1007/s13199-016-0382-2
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Tang, H. B., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A. N., Zhou, S. G., et al. (2014). An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. doi: 10.1186/1471-2164-15-312
Tanurdzic, M., and Banks, J. A. (2004). Sex-determining mechanisms in land plants. Plant Cell 16, S61–S71. doi: 10.1105/tpc.016667
Thatcher, L. F., Carrie, C., Andersson, C. R., Sivasithamparam, K., Whelan, J., and Singh, K. B. (2007). Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. J. Biol. Chem. 282, 28915–28928. doi: 10.1074/jbc.M702207200
Timmers, A. C. J., Auriac, M.-C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628.
Tirichine, L., Sandal, N., Madsen, L. H., Radutoiu, S., Albrektsen, A. S., Sato, S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. doi: 10.1126/science.1132397
Trinick, M. J. (1973). Symbiosis between Rhizobium and the non-legume. Trema aspera. Nature 244, 459–460. doi: 10.1038/244459a0
Trinick, M. J. (1980). Growth of Parasponia in agar tube culture and symbiotic effectiveness of isolates from Parasponia spp. New Phytol. 85, 37–45. doi: 10.1111/j.1469-8137.1980.tb04446.x
Trinick, M. J. (1981). “The effective rhizobium symbiosis with the non-legume Parasponia andersonii,” in Current Perspectives in Nitrogen Fixation, eds A. H. Gibson and W. E. Newton (Canberra: Australian Academy of Sciences), 480.
Trinick, M. J., and Hadobas, P. A. (1989). “Biology of the Parasponia-Bradyrhizobium symbiosis,” in Nitrogen Fixation with Non-Legumes, eds J. K. Ladha and K. A. Malik (Netherlands: Springer), 25–33. doi: 10.1007/978-94-009-0889-5_3
Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
van Velzen, R., Holmer, R., Bu, F., Rutten, L., van Zeijl, A., Liu, W., et al. (2017). Parallel loss of symbiosis genes in relatives of nitrogen-fixing non-legume Parasponia. bioRxiv 169706. doi: 10.1101/169706
van Zeijl, A. (2017). Dissecting Hormonal Pathways in Nitrogen-Fixing Rhizobium Symbioses. Ph.D. thesis, Wageningen University: Wageningen, doi: 10.18174/419674
van Zeijl, A., Liu, W., Xiao, T. T., Kohlen, W., Yang, W.-C., Bisseling, T., et al. (2015a). The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol. 15:260. doi: 10.1186/s12870-015-0651-x
van Zeijl, A., Op den Camp, R. H. M., Deinum, E. E., Charnikhova, T., Franssen, H., Op den Camp, H. J. M., et al. (2015b). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8, 1213–1226. doi: 10.1016/j.molp.2015.03.010
Vernie, T., Kim, J., Frances, L., Ding, Y., Sun, J., Guan, D., et al. (2015). The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27, 3410–3424. doi: 10.1105/tpc.15.00461
Wang, H., Moore, M. J., Soltis, P. S., Bell, C. D., Brockington, S. F., Alexandre, R., et al. (2009). Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. U.S.A. 106, 3853–3858. doi: 10.1073/pnas.0813376106
Webster, G., Poulton, P. R., Cocking, E. C., and Davey, M. R. (1995). The nodulation of micro-propagated plants of Parasponia andersonii by tropical legume rhizobia. J. Exp. Bot. 46, 1131–1137. doi: 10.1093/jxb/46.9.1131
Xiao, T. T., Schilderink, S., Moling, S., Deinum, E. E., Kondorosi, E., Franssen, H., et al. (2014). Fate map of Medicago truncatula root nodules. Development 141, 3517–3528. doi: 10.1242/dev.110775
Yin, T. J., and Quinn, J. A. (1995). Tests of a mechanistic model of one hormone regulating both sexes in Cucumis sativus (Cucurbitaceae). Am. J. Bot. 82, 1537–1546. doi: 10.1002/j.1537-2197.1995.tb13856.x
Young, N. D., Debelle, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Keywords: Parasponia andersonii, rhizobium, nodule, symbiosis, CRISPR/Cas9, stable transformation
Citation: van Zeijl A, Wardhani TAK, Seifi Kalhor M, Rutten L, Bu F, Hartog M, Linders S, Fedorova EE, Bisseling T, Kohlen W and Geurts R (2018) CRISPR/Cas9-Mediated Mutagenesis of Four Putative Symbiosis Genes of the Tropical Tree Parasponia andersonii Reveals Novel Phenotypes. Front. Plant Sci. 9:284. doi: 10.3389/fpls.2018.00284
Received: 05 January 2018; Accepted: 19 February 2018;
Published: 06 March 2018.
Edited by:
Jeanne Marie Harris, University of Vermont, United StatesReviewed by:
Jean-Francois Arrighi, Institut de Recherche pour le Développement (IRD), FranceValérie Hocher, Institut de Recherche pour le Développement (IRD), France
Copyright © 2018 van Zeijl, Wardhani, Seifi Kalhor, Rutten, Bu, Hartog, Linders, Fedorova, Bisseling, Kohlen and Geurts. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rene Geurts, cmVuZS5nZXVydHNAd3VyLm5s
†Present address: Elena E. Fedorova, K.A. Timiryazev Institute of Plant Physiology, Russian Academy of Sciences, Moscow, Russia
 Arjan van Zeijl
Arjan van Zeijl Titis A. K. Wardhani
Titis A. K. Wardhani Maryam Seifi Kalhor
Maryam Seifi Kalhor Luuk Rutten
Luuk Rutten Fengjiao Bu
Fengjiao Bu Sidney Linders
Sidney Linders Elena E. Fedorova
Elena E. Fedorova Ton Bisseling
Ton Bisseling Wouter Kohlen
Wouter Kohlen Rene Geurts
Rene Geurts