- 1Institute of Pharmacy and Biochemistry, Johannes Gutenberg University Mainz, Mainz, Germany
- 2Department of Biotechnology, University of Malakand, Malakand, Pakistan
- 3Department of Biological and Environmental Sciences, University of Gothenburg, Gothenburg, Sweden
Dynamin-like proteins (DLPs) are a family of membrane-active proteins with low sequence identity. The proteins operate in different organelles in eukaryotic cells, where they trigger vesicle formation, membrane fusion, or organelle division. As discussed here, representatives of this protein family have also been identified in chloroplasts and DLPs are very common in cyanobacteria. Since cyanobacteria and chloroplasts, an organelle of bacterial origin, have similar internal membrane systems, we suggest that DLPs are involved in membrane dynamics in cyanobacteria and chloroplasts. Here, we discuss the features and activities of DLPs with a focus on their potential presence and activity in chloroplasts and cyanobacteria.
Chloroplasts and Cyanobacteria Contain Two Inner Membrane Systems
Cyanobacteria and eukaryotic chloroplasts are evolutionary deeply connected, as primordial eukaryotic organisms had engulfed cyanobacterial ancestors in an endosymbiotic event. Incorporation of the bacteria into the cell metabolism finally resulted in development of a new organelle, the chloroplast, and the first oxygenic photosynthetic eukaryotes arose (reviewed in greater detail in Hohmann-Marriott and Blankenship, 2011; Jensen and Leister, 2014). During the course of evolution, several metabolic functions were lost in this newly developed organelle, and many genes of cyanobacterial origin were transferred from the new organelle into the genome of the host eukaryote (Martin and Herrmann, 1998; Martin et al., 2002; Kleine et al., 2009). Nevertheless, the ultrastructure of cyanobacteria and chloroplasts is still very similar, and the process of oxygenic photosynthesis as well as the proteins and cofactors involved therein are largely conserved (Hohmann-Marriott and Blankenship, 2011). In both cyanobacteria and chloroplasts, the components of the photosynthetic electron transport chain are localized within a specialized and unique internal membrane system, the thylakoid membranes (TMs). While the TM system is a completely separated and enclosed membrane system in chloroplasts and cyanobacteria, the exact fine structure of TMs can differ. In chloroplasts, TMs typically form multiple membrane stacks, which are connected by unstacked TMs, called grana and stroma lamellae, respectively (Adam et al., 2011), whereas TMs are organized in a sheet-like structure in cyanobacterial cells. However, the detailed TM arrangement can vary significantly in cyanobacterial strains (Herrero and Flores, 2008).
The structure of the TM system is highly dynamic in plant chloroplasts, and the amounts as well as the subcellular organization of TMs alter in response to changing environmental conditions (Chuartzman et al., 2008; Kirchhoff et al., 2011; Herbstova et al., 2012; Iwai et al., 2015). It has been stated that the dynamics observed in plant chloroplasts can essentially only be explained by the existence of a protein machinery that controls membrane fission and fusion processes (Chuartzman et al., 2008; Kirchhoff et al., 2011; Herbstova et al., 2012), analogous to the machineries involved in fusion of other organelle membranes, such as the ER, the Golgi apparatus, and the mitochondria (e.g., reviewed by Bonifacino, 2014). In fact, membrane connections and membrane fusion/fission processes have already been discussed for a long time (Nickelsen et al., 2011; Karim and Aronsson, 2014; Rast et al., 2015), but experimental evidence still is scarce. While described in chloroplasts, TM dynamics are essentially not studied in cyanobacteria yet. In cyanobacteria, individual TM layers appear to be as interconnected as in chloroplasts, and individual TM layers fuse and form “holes,” which are discussed to be required for intracellular transport (Nevo et al., 2007; Liberton et al., 2011a,b). Thus, the structure of TMs is most likely as dynamic in cyanobacteria as in chloroplasts. However, in contrast to chloroplasts, where TMs potentially develop de novo from the chloroplast inner envelope membrane (Muehlethaler and Frey-Wyssling, 1959; Morré et al., 1991), in cyanobacteria TMs do probably not form de novo but assemble from existing structures (Barthel et al., 2013).
Dynamin(-Like) Proteins Are Involved in Membrane Remodeling Processes in Prokaryotes and Eukaryotes
Dynamin-like proteins (DLPs) [also called, dynamin-related proteins (DRPs)] are involved in diverse membrane-related processes in prokaryotic and eukaryotic cells, involving membrane fusion, membrane scission, membrane protection, and/or membrane stabilization (Figure 1). DLPs are members of a protein superfamily of GTPases. To separate DLPs from other GTPases, such as Ras-like GTPases involved in signal transduction, proliferation and survival of cells, DLPs are also entitled “large GTPases” (Praefcke and McMahon, 2004; Lu et al., 2016). The first identified member of the dynamin family, MxA and its yeast homolog Vps1 were found being involved in virus resistance, vacuolar protein sorting, fission of endosomal membranes and in endocytic events (Staeheli et al., 1986; Smaczynska-de Rooij et al., 2010; Chi et al., 2014). Simultaneously, the founder of the dynamin superfamily, the prototypical dynamin protein (Dyn), was shown to be crucial for the scission of clathrin-coated endocytic vesicles from eukaryotic plasma membranes (Shpetner and Vallee, 1989; Hinshaw and Schmid, 1995). Upon triggering membrane fission, MxA and Dyn form dimers that interacts head to tail, resulting in formation of inactive tetramers. In presence of lipids, the structure reorganizes resulting in rearrangement of this auto-inhibitory structure, and the GTP hydrolysis rate of Dyn increases from 2.6 to 105 min-1 (Song et al., 2004; Reubold et al., 2015). The mechanistic details of this GTP-driven process are described in more detail elsewhere (Praefcke and McMahon, 2004; Antonny et al., 2016). However, besides the prototypical Dyn protein, a group of related DLPs is active in/at different eukaryotic organelles. E.g., the Drosophila melanogaster DLP Atlastin and its yeast counterpart Sey1p are required for fusion of ER membranes (Hu et al., 2009; Orso et al., 2010), and the yeast protein Dnm1 and its mammalian homolog Drp1 are involved in mitochondria scission (Smirnova et al., 2001; Legesse-Miller et al., 2003; Ingerman et al., 2005; Mears et al., 2011; Ugarte-Uribe et al., 2014). Similar to Dyn, the GTPase activity of Drp1 also increases in presence of lipids (Bustillo-Zabalbeitia et al., 2014; Reubold et al., 2015). The DLPs Mitofusin and FZO mediate fusion of the mitochondrial outer membrane, whereas OPA1 and Mgm1 are involved in fusion of the mitochondrial inner membrane system (Hermann et al., 1998; Santel and Fuller, 2001; Frezza et al., 2006; Meeusen et al., 2006).
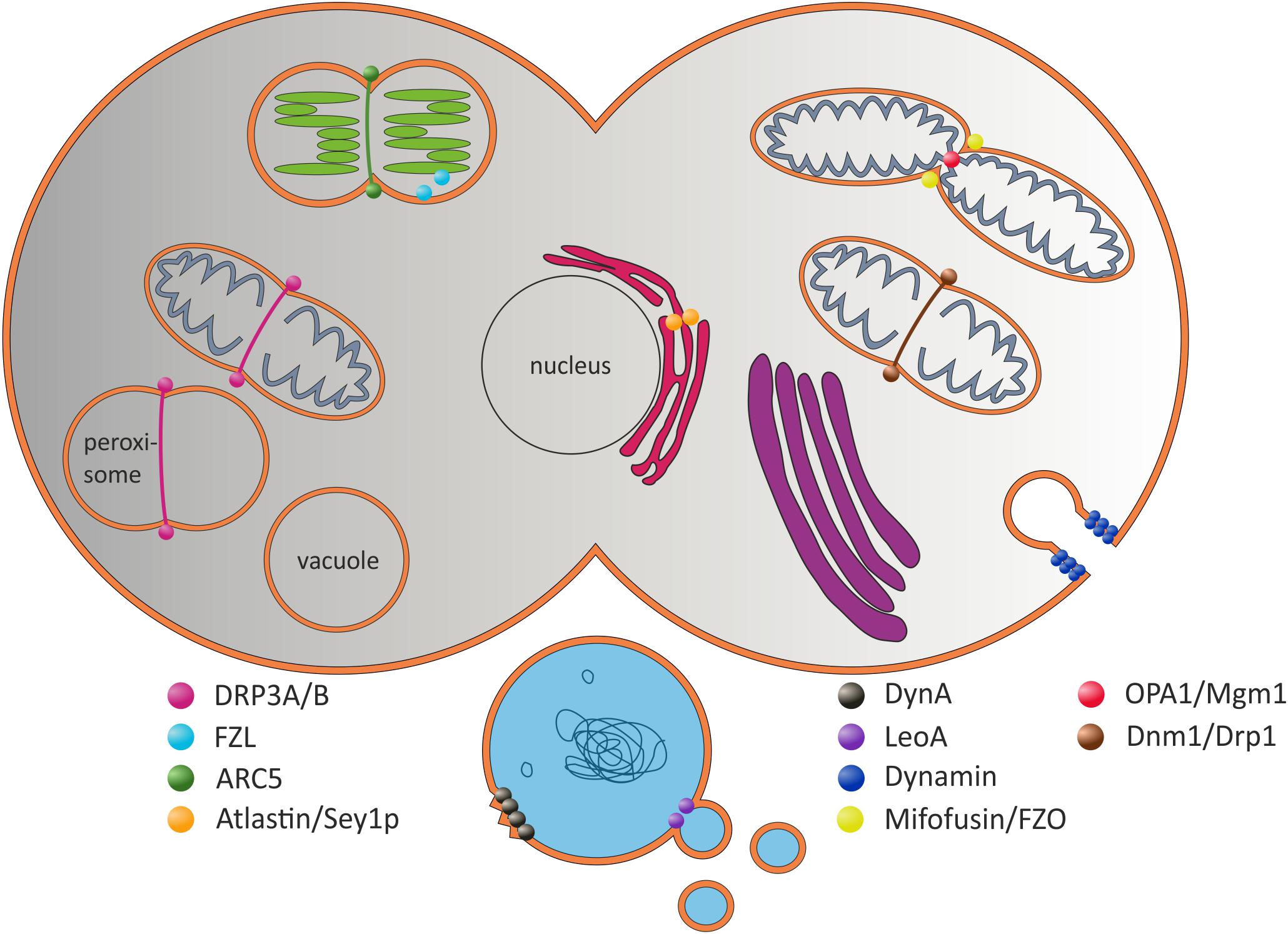
FIGURE 1. Selected (B)DLPs and their proposed in vivo functions. The prototypical Dyn protein is involved in the formation of clathrin-coated vesicles at the plasma membrane (Hinshaw and Schmid, 1995). Dnm1-like proteins are involved in mitochondrial scission. ARC5 is involved in chloroplast division (Smirnova et al., 2001; Gao et al., 2003; Mears et al., 2011). Atlastin/Sey1p are potentially involved in ER fusion, whereas Mitofusin/FZO fuse the inner and OPA1/Mgm1 the outer mitochondrial membranes (Hermann et al., 1998; Santel and Fuller, 2001; Frezza et al., 2006; Meeusen et al., 2006; Hu et al., 2009; Orso et al., 2010). The plant FZL protein is localized at the chloroplast inner envelope and the TM (Gao et al., 2006). In prokaryotic cells, DynA is proposed to be involved in membrane protection and/or membrane repair, and LeoA has been suggested being a component of a vesicle release system (Michie et al., 2014; Sawant et al., 2016).
As in yeast, Drosophila and mammals, DLPs are also present in plants and are currently best characterized in Arabidopsis thaliana. The DLP ARC5 is localized at the outer membrane of chloroplasts and is involved in chloroplast division (Gao et al., 2003), whereas FZL was found at the stromal side of the chloroplasts envelope and the TM (Gao et al., 2006). This issue makes FZL unique, because it is the only plant DLP within chloroplasts. Potential chloroplast DLPs, their subcellular localizations and potential activities are introduced and further discussed in more detail below (see section “DLPs in Arabidopsis thaliana”).
While DLPs are also predicted to exist in prokaryotes, this protein family was ignored for a long time in these organisms. About 10 years ago, the structure and (in vitro) activity of the first DLP was described, and the protein of the cyanobacterium Nostoc punctiforme was named “bacterial dynamin-like protein” (BDLP, hereafter named NosDLP) (Low and Löwe, 2006). While the exact in vivo function of this protein is still to be resolved, NosDLP behaves like other DLPs in vitro (Low et al., 2009; Bramkamp, 2012). While DLPs are rather common in cyanobacteria and some species encode multiple DLPs (as further outlined below), further representatives are not experimentally studied yet. Recently, two additional BDLPs were identified and partly analyzed. DynA of Bacillus subtilis can mediate membrane fusion in vitro (Bürmann et al., 2011), and as there is no obvious need for membrane fusion processes in B. subtilis, the in vivo activity of DynA was proposed to involve repair of disordered membranes (de Sousa Borges and Scheffers, 2016; Sawant et al., 2016). It was suggested that environmental stress (e.g., induced by phage infection or antibiotics) results in membrane pore formation, and DynA is recruited to these stressed membrane regions where it oligomerizes and fuses opposite bilayer patches in order to seal the membrane (Sawant et al., 2016). Furthermore, the Escherichia coli DLP LeoA was suggested to be involved in secretion of toxin-containing vesicles (Brown and Hardwidge, 2007; Michie et al., 2014). More recently two new BDLPs, DynA and DynB, were described to play a key role in a multiprotein cell division complex in Streptomyces venezuelae (Schlimpert et al., 2017). DynB is anchored to the cytoplasmic membrane where it interacts with DynA. Both BDLPs colocalize with the tubulin-like GTPase FtsZ and they might be involved, together with additional proteins, in the formation of sporulation septa during cell division (Schlimpert et al., 2017).
All thus far analyzed BDLPs hydrolyze GTP with much lower rates than eukaryotic DLPs (LeoA does not show any GTPase activity) and their GTP hydrolysis rates are typically not affected by lipids (Low and Löwe, 2006; Bürmann et al., 2011; Michie et al., 2014). However, being characteristic for DLPs, NosDLP homodimerizes in its GDP-bound state via its GTPase domain, and in presence of GTP and lipids, the protein self-assembles around liposomes and forms lipid tubes (Low and Löwe, 2006). The B. subtilis DynA is an internally fused protein containing two BDLP subunits, and thus, this protein works per definition as a dimer. Consequently, it shows nucleotide independent self-assembly on liposomes (Bürmann et al., 2011). As for LeoA, homo-dimerization has not been shown, suggesting that activation via heterodimerization with LeoBC is crucial (Michie et al., 2014).
Besides these few BDLPs being studied to some extent, the physiological function of DLPs is not clarified in bacteria yet. It is interesting to notice that in chloroplasts and cyanobacteria, both having similar internal membrane systems, membrane dynamics are observed (as discussed above), albeit no clear machinery is yet defined mediating membrane remodeling. Thus, it is not unlikely that BDLPs are involved in membrane dynamics in both chloroplasts and cyanobacteria.
Common Structural Elements and Domains of (B)DLPs
All (B)DLPs have a very similar domain structure (Figure 2A). Typically, a (B)DLP has a G-domain with the GTPase activity, followed by a middle (MID) domain, a region needed for membrane interaction (MI domain) and a GTPase effector domain (GED). The structure of all DLPs is dominated by α-helices, and individual helices often form helical bundle structures. The only highly conserved sequence of (B)DLPs is the G-domain, which is usually located at a (B)DLP N-terminus (Figure 2A). It harbors the GTPase domain and contains the amino acid motifs highly conserved in (B)DLPs. Like other GTPases, all (B)DLP share four motifs in the GTP binding/hydrolysis side: G1 or the P-Loop (GxxxxGKS/T), G2 or Switch I (T/S), G3 or Switch II (DxxG), and G4 (RD or N/TxxD). The G1 motif is involved in β-phosphate and Mg2+-binding. While the G2 and G3 (D) motifs are also involved in Mg2+-binding, they additionally interact with the γ-phosphate of GTP. The G4 motif is also involved in GTP binding but less conserved in (B)DLPs.
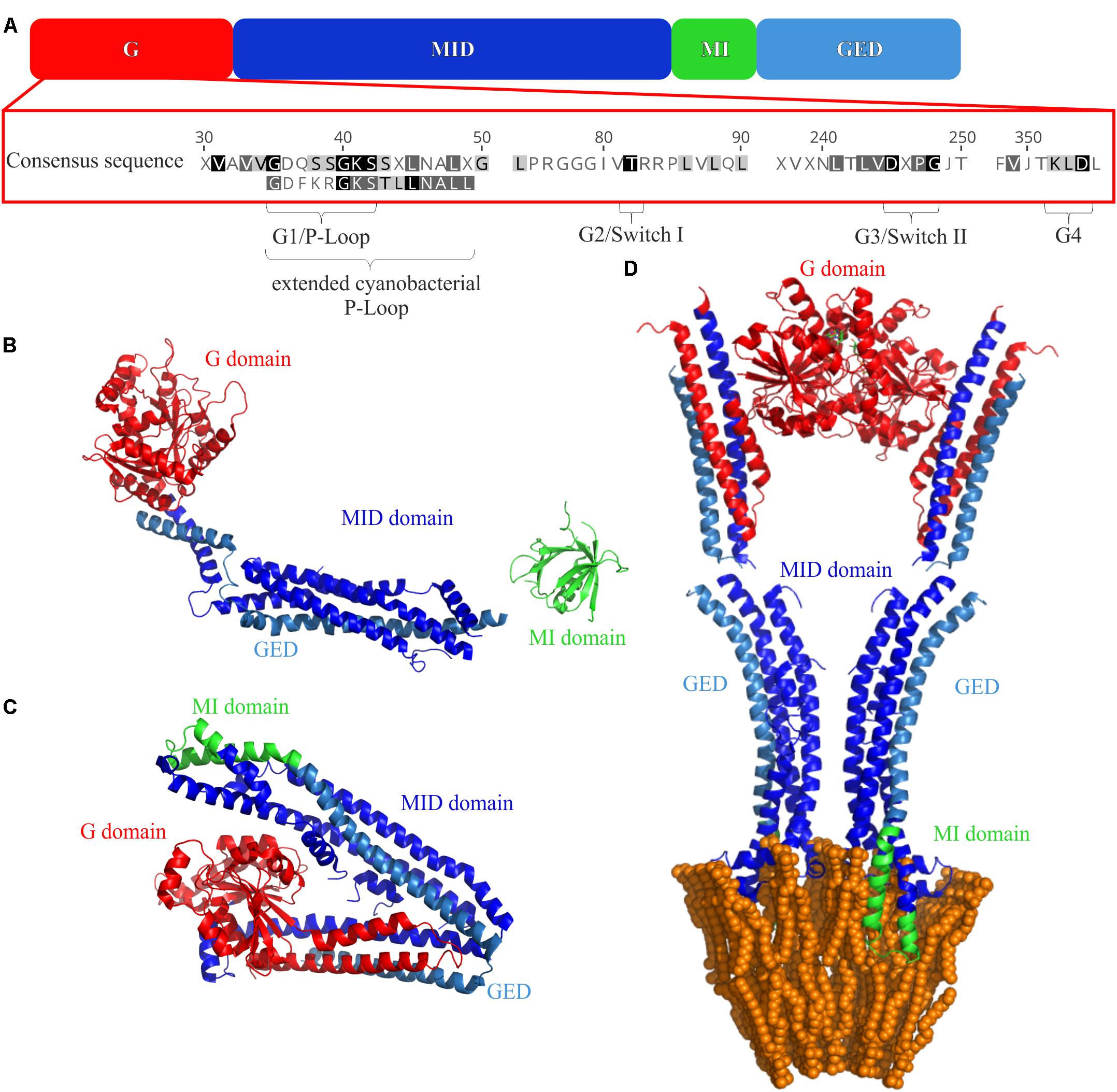
FIGURE 2. Domain structures of (B)DLPs. (A) The G domain carries the GTPase activity, the MID domain is crucial for protein oligomerization, the membrane binding MI domain and the GED are also involved in protein oligomerization. The crystal structures of (B) monomeric Dyn G397D ΔPRD (PDB 3ZVR) (Ford et al., 2011), (C) NosDLP in its GDP-bound closed monomeric form (PDB 2J69) (Low and Löwe, 2006), and (D) an open NosDLP dimer (GDP-bound) anchored in a tubulated lipid bilayer (PDB 2W6D) (Low et al., 2009). The G-domain is colored red, the MID domain in blue and the GED in light blue. The MI domains are highlighted in green.
In contrast to the G-domain, the remaining (B)DLP domains can vary strongly in their amino acid sequence, but are similar in their function. Often, these domains cannot be identified by sequence comparison and/or homology searches but solely by functional studies.
The MID domain is structurally defined by the presence of α-helical bundles (Figure 2), which are crucial for mediating oligomerization of (B)DLPs (Low and Löwe, 2006; Low et al., 2009; Gao et al., 2010; Faelber et al., 2013; Fröhlich et al., 2013; Reubold et al., 2015).
Since (B)DLPs are membrane-active proteins, they harbor one or more MI domains that interact with biological membranes, and these MI domains usually follow the MID domain. However, while membrane interaction is a key feature of DLPs and the basis for membrane remodeling, the exact mode of membrane interaction is not conserved, and different (B)DLPs interact differently with membranes. FZO/Mitofusin, OPA1/Mgm1 as well as Atlastin are anchored to membranes via transmembrane helices. In contrast, the prototypical Dyn interacts with membrane surfaces via the pleckstrin homology (PH) domain, which binds specifically to phosphatidylinositol lipids (Zheng et al., 1996). The DLP MxA harbors a disordered membrane binding loop (L4), and Dnm1 or the yeast homolog Drp1 uses the B insert loop that binds specifically to cardiolipin-enriched bilayers (Mitchell et al., 2013; Bustillo-Zabalbeitia et al., 2014). Furthermore, in EHD2, a polybasic motif was shown to mediate membrane interaction, and in GBP1, a specific Cys residue can be enzymatically isoprenylated, resulting in membrane anchoring (Nantais et al., 1996; Daumke et al., 2007; Vestal and Jeyaratnam, 2011). In case of DynA and NosDLP, the membrane interacting domain is called paddle domain (P), and it is named Tip in case of LeoA. The P/Tip domain is dominated by hydrophobic amino acids that mediate interactions with membrane surfaces (Low and Löwe, 2006; Low et al., 2009; Bürmann et al., 2011; Michie et al., 2014).
With only a few exceptions (GBP1 and Atlastin), the GTPase effector domain (GED) follows the MI domain (Figure 2). The GED is also part of a helical bundle, and in Dyn, MxA and NosDLP, the GED is additionally involved in the formation of higher-ordered structures (Schumacher and Staeheli, 1998; Chappie et al., 2009; Low et al., 2009).
Furthermore, besides the above described domains, additional domains might exist with more specialized functions.
In summary, all (B)DLPs appear to share three key features: (i) All (B)DLPs oligomerize and form higher ordered structures (Daumke and Praefcke, 2016). While in case of eukaryotic DLPs homo-oligomerization controls the GTPase activity, heterodimerization might be crucial for the activity of BDLPs (Bramkamp, 2012; Michie et al., 2014). (ii) All (B)DLPs are membrane-active and are involved in remodeling nearly every kind of cellular membrane system (reviewed in Bramkamp, 2012; Antonny et al., 2016; Daumke and Praefcke, 2016). For several DLPs it has been shown that they oligomerize in vitro and form helical structures around liposomes in the presence of a non-hydrolysable GTP analog, resulting in formation of tube-like membrane structures (Hinshaw, 2000; Low and Löwe, 2006; Bürmann et al., 2011; Mears et al., 2011; Shah et al., 2014; Ugarte-Uribe et al., 2014). (iii) (B)DLPs display a high sequence variability. Besides the G-domain, other domains can typically not be easily predicted in new classes of (B)DLPs and must be experimentally identified.
DLPs in Arabidopsis thaliana
In the model plant A. thaliana, 16 DLPs (or DRPs) are encoded. Based on their amino acid sequence and domain structure, these proteins can be grouped in six subfamilies, DRP1-DRP6 (Hong et al., 2003; Backues et al., 2010; Bednarek and Backues, 2010). When using the dynamin signature domain DYN1 (PF00350) for identification of DRPs in A. thaliana, combined with the literature and the plant subcellular localization integrative predictor (PSI1) (Liu et al., 2013), six DRPs are identified with a putative chloroplast localization. Five of these are designated DRPs in the literature; AtDRP1a/AtADL1a (At5g42080), AtDRP3a/AtADL2a (At4g33650), AtDRP3b/AtADL2b (At2g14120), AtDRP5A (At1g53140), and AtDRP5B/AtARC5 (At3g19720), whereas one is named fuzzy onion (FZO)-like protein (FZL). AtDRP1a/AtADL1a, AtDRP3a/AtADL2a, and AtDRP3b/AtADL2b all contain the GTPase (DYN1, PF00350), the dynamin MID region (DYN2, PF01031) and the GTPase-effector domain (GED, PF02212) (Miyagishima et al., 2008; Heymann and Hinshaw, 2009). The proteins AtDRP5A and AtDRP5B/AtARC5 additionally contain a pleckstrin homology domain (PH, PF00169) that binds to membrane phospholipids. In contrast, the FZL protein (At1g03160) contains solely the DYN1 signature domain.
AtDRP1a/AtADL1a is one of five proteins in the DRP1 subfamily. AtDRP1a/AtADL1a was found in TMs and was suggested to be involved in vesicle formation inside chloroplasts due to impaired chloroplast development including reduced amount of chloroplast membranes (Park et al., 1998). However, AtDRP1a/AtADL1a was also identified at the cell plate (Lauber et al., 1997). This apparent discrepancy was explained by a shortcoming of the antibody used in the study of Park et al. (1997). While the antibody was expected to recognize the GTPase domain of AtDRP1a/AtADL1a, the GTPase domains of DLPs are generally highly conserved (as discussed above), and thus the antibody could well have detected other DLPs besides AtDRP1a/AtADL1a (Kang et al., 2001). Subsequent studies have further challenged the assumption of chloroplast localization, as AtDRP1a/AtADL1a is targeted to other cellular compartments and was shown to have other roles (Kang et al., 1998, 2003; Collings et al., 2008; Konopka and Bednarek, 2008; Fujimoto et al., 2010; Yoshinari et al., 2016). The protein is targeted to the cell plate during cytokinesis (Kang et al., 1998), and in mutants lacking AtDRP1a/AtADL1a, an unusual plasma membrane accumulation is observed. This seems to inhibit efficient targeting and fusion of exocytic vesicles to the cell surface, which disturbs cell wall production (Kang et al., 2003). Moreover, the protein has also been shown to have a role in endocytic events at the plasma membrane, possibly associated with formation of clathrin-coated vesicles (Collings et al., 2008; Konopka and Bednarek, 2008; Fujimoto et al., 2010; Yoshinari et al., 2016). Thus, AtDRP1 is currently not considered to be active in chloroplasts.
AtDRP3a/AtADL2a has also been predicted to be localized in chloroplasts. A GFP-tagged version of the A. thaliana DRP3a has been shown to be chloroplast-localized in soybean and tobacco, where the N-terminal 35 amino acid residues were shown to be sufficient for chloroplast targeting (Kang et al., 1998). However, in a later study a GFP-DRP3a fusion protein was observed in mitochondria rather than in chloroplasts and was shown to be involved in mitochondrial division (Arimura et al., 2004). Moreover, AtDRP3 has also been partially targeted to peroxisomes where it has been suggested to have an essential role in peroxisome fission and replication (Lingard et al., 2008; Zhang and Hu, 2009; Mano et al., 2011). Thus, although an AtDRP3 mutant displays slow growth and a pale color at the seedling stage (Zhang and Hu, 2009), AtDRP3a less likely functions inside chloroplasts, but instead the protein localizes to mitochondria and peroxisomes.
AtDRP3b, also known as “Arabidopsis dynamin-like 2b” (ADL2b), shares 76% sequence identity with AtDRP3a/AtADL2a (Hong et al., 2003). While the protein is predicted (by the plant subcellular tool PSI) to be chloroplast-localized, is has so far not been shown to be targeted to chloroplasts. Instead, it has been identified in mitochondria and peroxisomes where it is suggested to support mitochondrial and peroxisomal division, respectively (Arimura and Tsutsumi, 2002; Fujimoto et al., 2009; Zhang and Hu, 2009). The phenotype of a mutant is similar to a AtDRP3a mutant, i.e., retarded growth and pale colored at the seedling stage (Zhang and Hu, 2009). While both, AtDRP3a/AtADL2a and AtDRP3b/AtADL2b, are suggested to be involved in mitochondrial as well as in peroxisomal division, in mitochondria the proteins have redundant functions while in peroxisomes they appear to have more distinct functions (Fujimoto et al., 2009).
Similar to AtDRP3b, the PSI tool predicts the DRP5 subfamily member AtDRP5a to be targeted to chloroplasts, although this localization has yet to be confirmed experimentally. The phenotype of mutant plants shows retarded seedling growth with no altered chloroplast (Miyagishima et al., 2008). Thus, there is currently no clear link to chloroplasts except for a putative localization prediction. Instead, AtDRP5a has been identified via GFP-tagging and immunoblot analyses in the cytosol of meristematic and meristemoid cells, and the protein is mainly found within dividing cells and suggested to be involved in cytokinesis (Miyagishima et al., 2008).
AtDRP5b is localized in both chloroplasts and peroxisomes, and mutant plants show retarded plant growth with yellowish leaves and enlarged and dumbbell-shaped chloroplasts (Gao et al., 2003; Zhang and Hu, 2010). AtDRP5b, also known as “accumulation and replication of chloroplasts 5” (ARC5), has no predicted signal sequence-mediating protein import into chloroplasts, albeit it clearly localizes to this organelle. However, AtDRP5b/ARC5 is found at the outer chloroplasts envelope membrane facing the cytosol, where it is enrolled in division ring construction at the late stage of the chloroplast division (Pyke and Leech, 1994; Gao et al., 2003; Miyagishima et al., 2006). The protein is recruited to the plastid division site by two plastid division proteins (PDV1 and PDV2), which also regulate the GTPase activity of AtDRP5b/ARC5 (Gao et al., 2013; Holtsmark et al., 2013). However, AtDRP5b is also present in peroxisomes as revealed by bimolecular fluorescence complementation and co-immunoprecipitation assays, and when the AtDRP5b gene was mutated, impaired peroxisome division and function was observed (Zhang and Hu, 2010).
The A. thaliana protein AtFZL is related to fuzzy onion (FZO) proteins that are part of the dynamin superfamily of remodeling GTPases. FZO is a protein that is located in the outer mitochondria membrane where it is involved in fusion of opposing outer mitochondrial membranes in animals and fungi (Koshiba, 2004; Meeusen, 2004). However, the A. thaliana FZL protein shows low homology both to the Mitofusin domain found in the FZO family as well as to the dynamin domain (DYN1). Despite some sequence homology and similarities with FZO in respect of existing domains and their arrangement (GTPase, coiled-coil, transmembrane helices), absence of AtFZL does not affect mitochondria morphology in A. thaliana but instead the morphology of chloroplasts (Gao et al., 2006). AtFZL is located inside the chloroplasts at the TM but also at the chloroplast inner envelope (Gao et al., 2006). It is believed to be anchored to these membranes via two transmembrane domains located within the C-terminal part of the protein, leaving the GTPase and coiled-coil domains protruding into the chloroplast stroma (Gao et al., 2006). Whether the AtFZL operates in a similar fashion as the classical FZO, i.e., whether it brings two membranes into close contact resulting in membrane fusion, is currently unknown. However, such an activity is indicated and an involvement of AtFZL in the transport of lipids between the inner envelope and the TM has been suggested, since vesicles appear to not fuse in plants lacking AtFZL (Gao et al., 2006) and since the chloroplasts show a disorganized TM morphology. Thus, AtFZL likely is a membrane-remodeling GTPase, involved in TM biogenesis and dynamics in chloroplasts (Gao et al., 2006).
Thus, out of the six predicted DRPs, evidence for chloroplast localization and function is only strong for AtDRP5b and AtFZL. However, as AtDRP5b is localized at the outer envelope membrane, AtFZL is the one remaining that could facilitate membrane remodeling, potentially involving fission and fusion of vesicles budding off from the inner envelope membrane and being targeted to the TM (Kroll et al., 2001; Wang et al., 2004; Garcia et al., 2010; Tanz et al., 2012; Armbruster et al., 2013; Karim and Aronsson, 2014; Karim et al., 2014). Thus, DRPs can be important to secure a dynamic but organized thylakoid network in chloroplasts.
DLPs Are Conserved in Cyanobacteria
The BDLP of the cyanobacterium N. punctiforme has been characterized to some extent. As typical for DLPs, the smallest structural unit of this BDLP appears to be a dimer, which can further self-assemble (Low and Löwe, 2006; Low et al., 2009). NosDLP is thought to mediate membrane fusion by inducing formation of highly curved membrane regions (Low and Löwe, 2006). After GTP-binding, NosDLP self-oligomerizes in vitro around a membrane and forces the membrane into a tube-like structure with high curvature, similar to the prototypical eukaryotic Dyn protein that is involved in vesicle fission. After GTP hydrolyzes, NosDLP is released from the lipid, and adjacent membrane regions spontaneously fuse (Low et al., 2009). A NosDLP-GFP fusion protein was found in the cell periphery in N. punctiforme as well as in rare ring-like structures at the cell septa (Low and Löwe, 2006), highlighting that the protein is interacting with membranes and is membrane-active. Nevertheless, the in vivo function of this protein is still unclear.
To obtain further information about potential cyanobacterial BDLPs (hereafter named cBDLPs to distinguish them from other bacteria DLPs), we searched the pfam database (dynamin_N family PF00350), for cyanobacterial proteins carrying a dynamin GTPase domain, the only key marker for DLPs. Based on this analysis, 279 potential cBDLPs were identified being encoded in 74 different cyanobacterial species. However, we further limited our results and removed small GTPases, which sometimes also have a predicted dynamin GTPase domain. Finally, we ended up with 121 genes that likely encode BDLPs in 56 different cyanobacterial strains (Figure 3 and Supplementary Data Sheet S1). It should be noted that due to the rather rigid search, we might have overlooked some cBDLPs. While all potential cBDLPs have a highly conserved dynamin-like GTPase domain within the N-terminal protein region, we did not limit our search to the identification of other known domains, since the sequences of these domains are typically not conserved (as discussed above). However, nearly all of the cBDLP sequences have an extended and conserved P-loop region within the G-domain: beside the common GxxxxGKS/T P-loop motif, cBDLPs have an additional L/INALL/I motif, which extends the P-loop motif to GxxxxGKS/TxL/INALL/I.
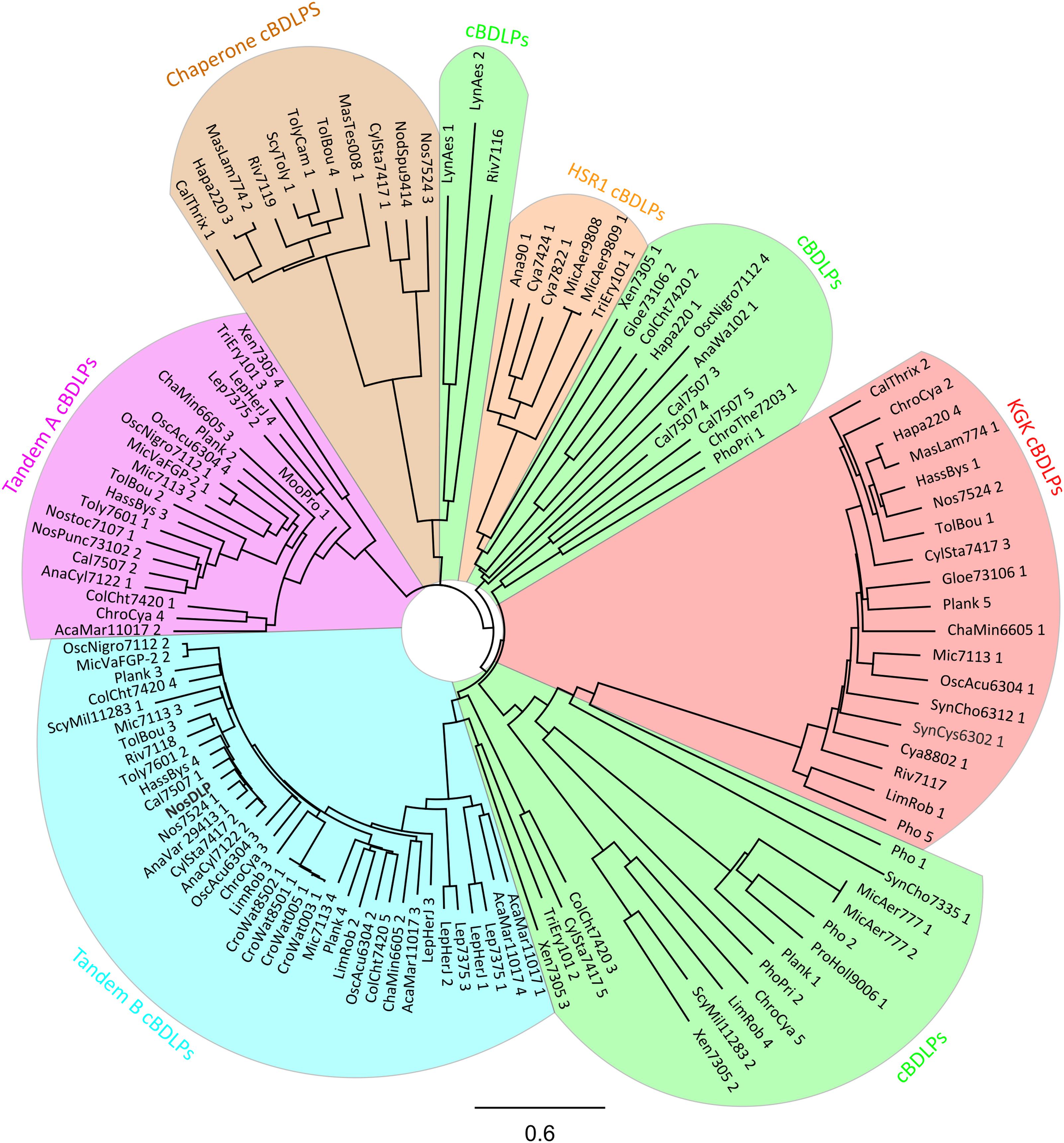
FIGURE 3. Phylogenetic tree of 121 cBDLPs encoded in 56 cyanobacteria species (including NosDLP). The DLP discussed in the text is shown in bold. The name of the potential dynamins is abbreviated. For full information about the strain, the gene name and the gene locus, view the Supplementary Data Sheet S1. The cBDLPs can be classified into six different clades, depending on their sequence and genomic environment: the KGK clade (red), the HSR1 clade (orange), and the chaperone clade (brown). Upstream of the Tandem A (pink) clade, another cBDLP (Tandem B, light blue) is encoded. cBDLPs (green) could not be classified further and thus, represents a group of diverse cBDLPs. It is worth mentioning that boundaries between the cBDLPs clade and the Tandem B or HSR1 clades, respectively, are not sharp and it might be that proteins share characteristics of both clades. Furthermore, while Xen7305 3 does not show significant sequence similarity to Tandem B members, downstream of this gene, the Tandem A protein Xen7305 4 is encoded. The here described group characteristics refer to the majority of the cBDLPs but do not represent every encoded protein. For further details, view the Supplementary Data Sheet S1. The phylogenetic tree (model: jukes-cantor, neighbor-joining) was created by a full-length multiprotein sequence alignment (Geneious global alignment, Matrix Blosum62) implemented in the software Geneious version 11.0.4 (http://www.geneious.com; Kearse et al., 2012). The cyanobacterial sequences were obtained from “cyanobase” (Nakamura et al., 1998; Fujisawa et al., 2017).
Unfortunately, none of the predicted cDLPs share a high sequence identity with the AtFZL protein in chloroplasts. However, based on a phylogenetic analysis we categorized the identified cBDLPs into six groups, where the sequences of individual cBDLPs are highly conserved within the defined clades but clearly differ in between the clades (Figure 3). Interestingly, the genetic context of some cBDLP is conserved in some clades and proteins are, e.g., part of conserved gene clusters (Supplementary Data Sheet S1).
Within the first cBDLP group, typically a KGK domain protein is encoded downstream of the cBDLP. Members of the KGK protein family (PF08872) are small cyanobacterial proteins (around 120 amino acids) that contain a KGK domain (Finn et al., 2014). Unfortunately, the precise function of this domain is enigmatic, albeit this domain potentially mediates protein–protein interactions. The second cBDLP group is termed Tandem B cBDLPs, and the only yet characterized cBLPD, the BDLP of N. punctiforme, belongs to this group (Low and Löwe, 2006). Upstream of the encoding gene, a second cBDLP is encoded (clade Tandem A), and thus, translation of the clustered genes likely results in expression of two different BDLPs. While it has been shown that NosDLP forms homodimers, its GTPase activity was not at all affected by homo-oligomerization and/or lipid binding. Therefore, it has already been suggested that hetero-oligomerization with another, different BDLP might control the activity of this BDLP in a way, as observed in the case of eukaryotic DLPs (DeVay et al., 2009; Michie et al., 2014). Based on our analyses, the Tandem A representative of N. punctiforme is a likely candidate. Noteworthy, the sequences of the Tandem A and Tandem B cBDLPs differ substantially.
In the HSR1 group of cBDLPs, a potential protein of the HSR1 protein family is encoded downstream of the cBDLP. HSR1-releated proteins are not well-characterized GTP-binding proteins that, however, have no apparent GTPase enzymatic function (Finn et al., 2017). Thus, the cyanobacterial HSR1 proteins encoded adjacent of the cBDLPs potentially have a regulatory function. In the chaperone group, conserved proteins carrying a DnaK domain are encoded downstream of the respective cBLDP. DnaK proteins belong to the group of Hsp70 chaperones (Mayer and Bukau, 2005; Young, 2010; Mayer, 2013), and thus, here the activity of a membrane remodeling DLP is linked to the activity of an Hsp70 chaperone. In fact, membrane activity of Hsp70 members has been described in recent years (Armijo et al., 2014; Mahalka et al., 2014). In humans, selected Hsp70 proteins have been suggested to, e.g., carry immunogenic peptides for antigen presentation (Haug et al., 2005), and thus, DnaK-like proteins encoded in the vicinity of a cBDLP might be involved in protein sorting during membrane remodeling processes.
The remaining cBDLPs are not significantly related to one another and could not be further categorized (named cBDLP in Figure 3).
Are (B)DLPs Involved in TM Biogenesis and Dynamics in Chloroplast and Cyanobacteria?
At least one DLP is present in chloroplasts and the activity of the FZL protein has been linked to membrane biogenesis and dynamics (Gao et al., 2006). As discussed before, DLPs are also encoded in cyanobacteria and at least the NosDLP can remodel membranes resulting in membrane fission (Low et al., 2009). However, the exact in vivo function of (B)DLPs in chloroplasts and cyanobacteria still is enigmatic. Clearly, membrane disruption would be deleterious in chloroplasts as well as in cyanobacteria, e.g., disruption of TMs would result in a breakdown of the electrochemical gradient across the TM. Thus, it is feasible to propose a membrane protective function of the DLPs and a crucial role in the repair of ruptured membrane regions, as suggested, e.g., for the BDLP of B. subtilis (Sawant et al., 2016). However, in cyanobacteria different proteins and systems are described that are suggested to be involved in membrane stabilization and membrane repair, involving small heat shock proteins and the PspA system (Torok et al., 2001; Nitta et al., 2005; Manganelli and Gennaro, 2017). Thus, the DLPs might have acquired additional functions in chloroplasts and cyanobacteria.
In recent years, evidence has accumulated indicating that membrane fusion and fission events are involved in TM development and dynamics in chloroplasts and cyanobacteria (Chuartzman et al., 2008; Kirchhoff et al., 2011; Herbstova et al., 2012; Nevo et al., 2012; Iwai et al., 2015). As spontaneous, uncontrolled membrane fusion would be deleterious to organisms, defined fusion and fission machineries likely control such remodeling processes (Chuartzman et al., 2008). However, proteins involved in membrane dynamics in chloroplast and cyanobacteria still need to be better characterized and more need to be identified. In bioinformatic analyses, several genes have been identified that code for putative chloroplast-localized proteins with homology to proteins involved in the secretory pathway, operating in the cytoplasm of eukaryotic cells, and some of these proteins are also conserved in cyanobacterial genomes (Nakai et al., 1993; Srivastava et al., 2005; Keller and Schneider, 2013; Khan et al., 2013; Paul et al., 2014). Nevertheless, potential involvement of these proteins in TM biogenesis and/or maintenance has only sparsely been shown experimentally yet, and it appears to be rather unlikely that vesicle fission and fusion is regulated identical in chloroplasts as in the secretory pathway. However, some of the identified (putative) membrane-active proteins might fulfill similar functions in chloroplasts as in the secretory pathway but work together with other, chloroplast- and cyanobacteria-specific proteins. Such chloroplast and cyanobacteria-specific proteins likely involve the recently identified Vipp1/IM30 protein (Kroll et al., 2001; Westphal et al., 2001), a protein that can fuse membranes in presence of Mg2+, at least in vitro (Hennig et al., 2015). Moreover, IM30-depleted chloroplasts and cyanobacteria have a significantly reduced TM network (Kroll et al., 2001; Fuhrmann et al., 2009). Within the secretory pathway, several small GTPases are involved in vesicle formation and fission (Hutagalung and Novick, 2011; Barlowe and Miller, 2013), and involvement of the small GTPases AR1 and CPRabA5e in TM biogenesis has been shown in A. thaliana (Garcia et al., 2010; Karim et al., 2014). Since especially dynamin-like GTPases are directly associated with membrane remodeling processes in many eukaryotic organelles, it appears possible that DLPs are also involved in membrane biogenesis and/or remodeling processes in chloroplasts and cyanobacteria. In fact, FZL has been suggested to mediate contact of two adjacent membranes in A. thaliana chloroplasts, finally resulting in membrane fusion (Gao et al., 2006). The here presented analysis clearly demonstrates that DLPs are also highly abundant in cyanobacteria. However, thus far solely the DLP of the cyanobacterium N. punctiforme has been studied to some extent. The in vitro analyses clearly indicate that the protein behaves like the classical Dyn and thus might be involved in vesicle fission in cyanobacteria. Nevertheless, we initially expected to identify a prototypical cDLP in our analysis that is conserved in all cyanobacterial species. Surprisingly, we did not identify such a candidate; rather, while most cyanobacterial genomes encode at least one cDLP, the proteins belong to different clades. Thus, the sequences of cBDLPs are highly variable. While the exact physiological function of the cyanobacterial proteins is enigmatic, it is reasonable to assume that proteins with a membrane remodeling activity will be involved in membrane dynamics in chloroplasts and cyanobacteria. The exact physiological function of the proteins, i.e., their involvement in processes such as membrane protection, membrane repair, membrane fission and/or membrane fusion, however, still needs to be established. Nevertheless, the in vivo observation of FZL being involved in TM dynamics and vesicle fusion and the in vitro observation of NosDLP behaving like the prototypical Dyn clearly indicates a crucial membrane-active role of DLPs in chloroplasts and cyanobacteria. Based on the described membrane activities of DLPs and on the need of membrane remodeling processes, in Figure 4 we summarize a potential involvement of DLPs in chloroplasts and cyanobacteria. We hope this article will stimulate future research on the involvement of this membrane-active protein family in membrane dynamics in chloroplasts and cyanobacteria.
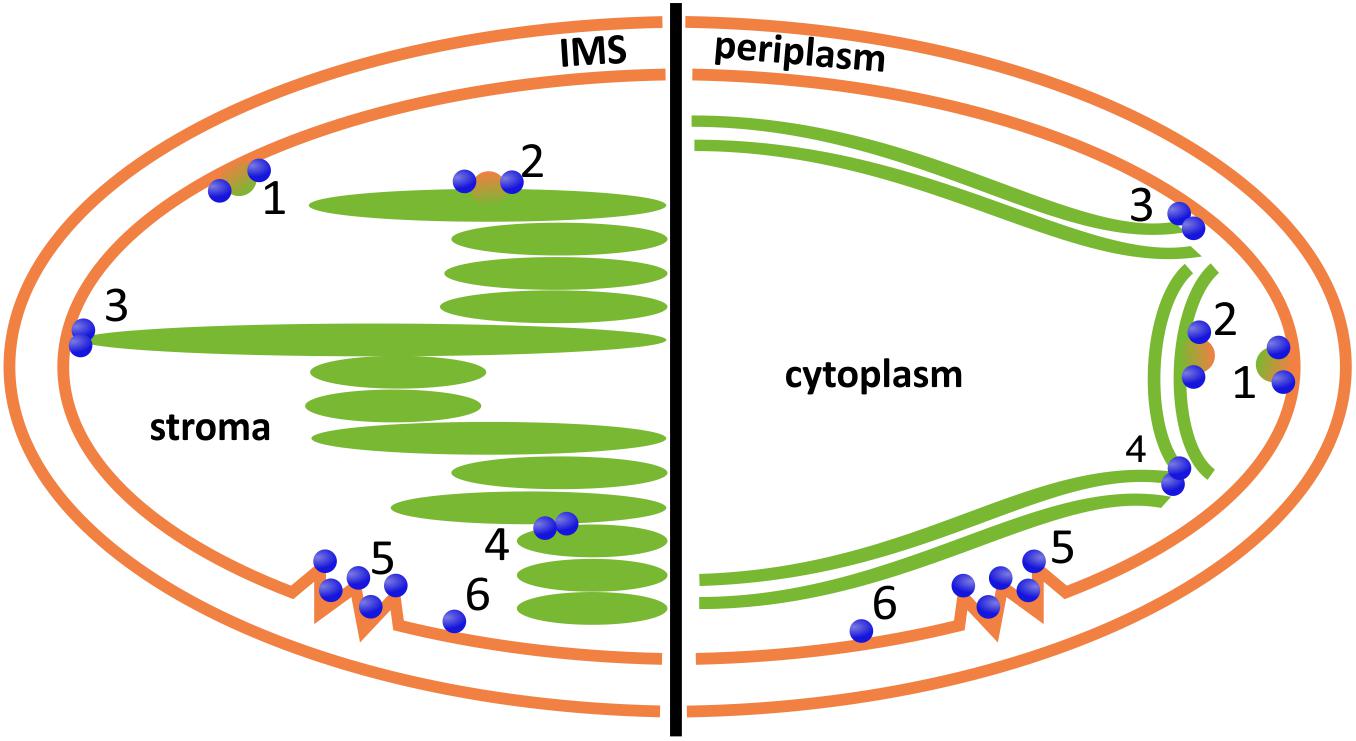
FIGURE 4. Potential involvement of DLPs in chloroplast (left) and cyanobacterial (right) inner membrane dynamics. DLPs might be involved in vesicle formation, release or fusion at the chloroplast inner envelope or the cyanobacterial cytoplasmic membrane, respectively (1), or at the TMs (2); the fusion of the inner envelope or cytoplasmic membrane, respectively, with the TM system (3); and/or the fusion/constriction of individual TM layers (4). Moreover, DLPs are potentially involved in membrane repair (5) and/or membrane protection (6). IMS, inter membrane space. Note that DLPs located outside of the chloroplast, involving AtDRP5B/ARC5, are excluded.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work and approved it for publication.
Funding
This work was funded by the Max Planck Graduate Center at the Max Planck Institutes and the University of Mainz (to RJ and DS) and by the Carl Trygger Foundation (HA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00206/full#supplementary-material
DATA SHEET S1 | Cyanobacterial species.
Footnotes
References
Adam, Z., Charuvi, D., Tsabari, O., Knopf, R. R., and Reich, Z. (2011). Biogenesis of thylakoid networks in angiosperms: knowns and unknowns. Plant Mol. Biol. 76, 221–234. doi: 10.1007/s11103-010-9693-5
Antonny, B., Burd, C., De Camilli, P., Chen, E., Daumke, O., Faelber, K., et al. (2016). Membrane fission by dynamin: what we know and what we need to know. EMBO J. 35, 2270–2284. doi: 10.15252/embj.201694613
Arimura, S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. U.S.A. 99, 5727–5731. doi: 10.1073/pnas.082663299
Arimura, S. I., Aida, G. P., Fujimoto, M., Nakazono, M., and Tsutsumi, N. (2004). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45, 236–242. doi: 10.1093/pcp/pch024
Armbruster, U., Labs, M., Pribil, M., Viola, S., Xu, W., Scharfenberg, M., et al. (2013). Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell 25, 2661–2678. doi: 10.1105/tpc.113.113118
Armijo, G., Okerblom, J., Cauvi, D. M., Lopez, V., Schlamadinger, D. E., Kim, J., et al. (2014). Interaction of heat shock protein 70 with membranes depends on the lipid environment. Cell Stress Chaperones 19, 877–886. doi: 10.1007/s12192-014-0511-x
Backues, S. K., Korasick, D. A., Heese, A., and Bednarek, S. Y. (2010). The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 22, 3218–3231. doi: 10.1105/tpc.110.077727
Barlowe, C. K., and Miller, E. A. (2013). Secretory protein biogenesis and traffic in the early secretory pathway. Genetics 193, 383–410. doi: 10.1534/genetics.112.142810
Barthel, S., Bernát, G., Seidel, T., Rupprecht, E., Kahmann, U., and Schneider, D. (2013). Thylakoid membrane maturation and PSII activation are linked in greening Synechocystis sp. PCC 6803 cells. Plant Physiol. 163, 1037–1046. doi: 10.1104/pp.113.224428
Bednarek, S. Y., and Backues, S. K. (2010). Plant dynamin-related protein families DRP1 and DRP2 in plant development. Biochem. Soc. Trans. 38, 797–806. doi: 10.1042/BST0380797
Bonifacino, J. S. (2014). Vesicular transport earns a Nobel. Trends Cell Biol. 24, 3–5. doi: 10.1016/j.tcb.2013.11.001
Bramkamp, M. (2012). Structure and function of bacterial dynamin-like proteins. Biol. Chem. 393, 1203–1214. doi: 10.1515/hsz-2012-0185
Brown, E. A., and Hardwidge, P. R. (2007). Biochemical characterization of the enterotoxigenic Escherichia coli LeoA protein. Microbiology 153, 3776–3784. doi: 10.1099/mic.0.2007/009084-0
Bürmann, F., Ebert, N., van Baarle, S., and Bramkamp, M. (2011). A bacterial dynamin-like protein mediating nucleotide-independent membrane fusion. Mol. Microbiol. 79, 1294–1304. doi: 10.1111/j.1365-2958.2011.07523.x
Bustillo-Zabalbeitia, I., Montessuit, S., Raemy, E., Basañez, G., Terrones, O., and Martinou, J. C. (2014). Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS One 9:e102738. doi: 10.1371/journal.pone.0102738
Chappie, J. S., Acharya, S., Liu, Y. W., Leonard, M., Pucadyil, T. J., and Schmid, S. L. (2009). An intramolecular signaling element that modulates dynamin function in vitro and in vivo. Mol. Biol. Cell 20, 3561–3571. doi: 10.1091/mbc.E09-04-0318
Chi, R. J., Liu, J., West, M., Wang, J., Odorizzi, G., and Burd, C. G. (2014). Fission of SNX-BAR-coated endosomal retrograde transport carriers is promoted by the dynamin-related protein Vps1. J. Cell Biol. 204, 793–806. doi: 10.1083/jcb.201309084
Chuartzman, S. G., Nevo, R., Shimoni, E., Charuvi, D., Kiss, V., Ohad, I., et al. (2008). Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell 20, 1029–1039. doi: 10.1105/tpc.107.055830
Collings, D. A., Gebbie, L. K., Howles, P. A., Hurley, U. A., Birch, R. J., Cork, A. H., et al. (2008). Arabidopsis dynamin-like protein DRP1A: a null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 59, 361–376. doi: 10.1093/jxb/erm324
Daumke, O., Lundmark, R., Vallis, Y., Martens, S., Butler, P. J. G., and McMahon, H. T. (2007). Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927. doi: 10.1038/nature06173
Daumke, O., and Praefcke, G. J. K. (2016). Invited review: mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers 105, 580–593. doi: 10.1002/bip.22855
de Sousa Borges, A., and Scheffers, D. J. (2016). Bacterial dynamin as a membrane puncture repair kit. Environ. Microbiol. 18, 2298–2301. doi: 10.1111/1462-2920.13218
DeVay, R. M., Dominguez-Ramirez, L., Lackner, L. L., Hoppins, S., Stahlberg, H., and Nunnari, J. (2009). Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803. doi: 10.1083/jcb.200906098
Faelber, K., Gao, S., Held, M., Posor, Y., Haucke, V., Noé, F., et al. (2013). Oligomerization of dynamin superfamily proteins in health and disease. Prog. Mol. Biol. Transl. Sci. 117, 411–443. doi: 10.1016/B978-0-12-386931-9.00015-5
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Ford, M. G. J., Jenni, S., and Nunnari, J. (2011). The crystal structure of dynamin. Nature 477, 561–566. doi: 10.1038/nature10441
Frezza, C., Cipolat, S., Martins de Brito, O., Micaroni, M., Beznoussenko, G. V., Rudka, T., et al. (2006). OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189. doi: 10.1016/j.cell.2006.06.025
Fröhlich, C., Grabiger, S., Schwefel, D., Faelber, K., Rosenbaum, E., Mears, J., et al. (2013). Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32, 1280–1292. doi: 10.1038/emboj.2013.74
Fuhrmann, E., Gathmann, S., Rupprecht, E., Golecki, J., and Schneider, D. (2009). Thylakoid membrane reduction affects the photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 735–744. doi: 10.1104/pp.108.132373
Fujimoto, M., Arimura, S. I., Mano, S., Kondo, M., Saito, C., Ueda, T., et al. (2009). Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58, 388–400. doi: 10.1111/j.1365-313X.2009.03786.x
Fujimoto, M., Arimura, S.-I., Ueda, T., Takanashi, H., Hayashi, Y., Nakano, A., et al. (2010). Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. U.S.A. 107, 6094–6099. doi: 10.1073/pnas.0913562107
Fujisawa, T., Narikawa, R., Maeda, S.-I., Watanabe, S., Kanesaki, Y., Kobayashi, K., et al. (2017). CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res. 45, D551–D554. doi: 10.1093/nar/gkw1131
Gao, H., Kadirjan-Kalbach, D., Froehlich, J. E., and Osteryoung, K. W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. U.S.A. 100, 4328–4333. doi: 10.1073/pnas.0530206100
Gao, H., Sage, T. L., and Osteryoung, K. W. (2006). FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc. Natl. Acad. Sci. U.S.A. 103, 6759–6764. doi: 10.1073/pnas.0507287103
Gao, S., von der Malsburg, A., Paeschke, S., Behlke, J., Haller, O., Kochs, G., et al. (2010). Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506. doi: 10.1038/nature08972
Gao, Y., Liu, H., An, C., Shi, Y., Liu, X., Yuan, W., et al. (2013). Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 75, 795–807. doi: 10.1111/tpj.12240
Garcia, C., Khan, N. Z., Nannmark, U., and Aronsson, H. (2010). The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 63, 73–85. doi: 10.1111/j.1365-313X.2010.04225.x
Haug, M., Dannecker, L., Schepp, C. P., Kwok, W. W., Wernet, D., Buckner, J. H., et al. (2005). The heat shock protein Hsp70 enhances antigen-specific proliferation of human CD4+ memory T cells. Eur. J. Immunol. 35, 3163–3172. doi: 10.1002/eji.200535050
Hennig, R., Heidrich, J., Saur, M., Schmu, L., Roeters, S. J., Hellmann, N., et al. (2015). IM30 triggers membrane fusion in cyanobacteria and chloroplasts. Nat. Commun. 6:7018. doi: 10.1038/ncomms8018
Herbstova, M., Tietz, S., Kinzel, C., Turkina, M. V., and Kirchhoff, H. (2012). Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl. Acad. Sci. U.S.A. 109, 20130–20135. doi: 10.1073/pnas.1214265109
Hermann, G. J., Thatcher, J. W., Mills, J. P., Hales, K. G., Fuller, M. T., Nunnari, J., et al. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373. doi: 10.1083/jcb.143.2.359
Herrero, A., and Flores, E. (2008). The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Poole: Caister Academic Press.
Heymann, J. A. W., and Hinshaw, J. E. (2009). Dynamins at a glance. J. Cell Sci. 122, 3427–3431. doi: 10.1242/jcs.051714
Hinshaw, J. E. (2000). Dynamin and its role in membrane fission. Annu. Rev. Cell Dev. Biol. 16, 483–519. doi: 10.1146/annurev.cellbio.16.1.483
Hinshaw, J. E., and Schmid, S. L. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. doi: 10.1038/374190a0
Hohmann-Marriott, M. F., and Blankenship, R. E. (2011). Evolution of photosynthesis. Annu. Rev. Plant Biol. 62, 515–548. doi: 10.1146/annurev-arplant-042110-103811
Holtsmark, I., Lee, S., Lunde, K. A., Auestad, K., Maple-Grødem, J., and Møller, S. G. (2013). Plastid division control: The PDV proteins regulate DRP5B dynamin activity. Plant Mol. Biol. 82, 255–266. doi: 10.1007/s11103-013-0059-7
Hong, Z., Bednarek, S. Y., Blumwald, E., Hwang, I., Jurgens, G., Menzel, D., et al. (2003). A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53, 261–265. doi: 10.1023/B:PLAN.0000007000.29697.81
Hu, J., Shibata, Y., Zhu, P. P., Voss, C., Rismanchi, N., Prinz, W. A., et al. (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. doi: 10.1016/j.cell.2009.05.025
Hutagalung, A. H., and Novick, P. J. (2011). Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149. doi: 10.1152/physrev.00059.2009
Ingerman, E., Perkins, E. M., Marino, M., Mears, J. A., McCaffery, J. M., Hinshaw, J. E., et al. (2005). Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027. doi: 10.1083/jcb.200506078
Iwai, M., Yokono, M., and Nakano, A. (2015). Visualizing structural dynamics of thylakoid membranes. Sci. Rep. 4:3768. doi: 10.1038/srep03768
Jensen, P. E., and Leister, D. (2014). Chloroplast evolution, structure and functions. F1000Prime Rep. 6:40. doi: 10.12703/P6-40
Kang, B. H., Busse, J. S., and Bednarek, S. Y. (2003). Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913. doi: 10.1105/tpc.009670
Kang, B. H., Busse, J. S., Dickey, C., Rancour, D. M., and Bednarek, S. Y. (2001). The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 126, 47–68. doi: 10.1104/pp.126.1.47
Kang, S. G., Jin, J. B., Piao, H. L., Pih, K. T., Jang, H. J., Lim, J. H., et al. (1998). Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38, 437–447. doi: 10.1023/A:1006099718761
Karim, S., Alezzawi, M., Garcia-Petit, C., Solymosi, K., Khan, N. Z., Lindquist, E., et al. (2014). A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol. Biol. 84, 675–692. doi: 10.1007/s11103-013-0161-x
Karim, S., and Aronsson, H. (2014). The puzzle of chloroplast vesicle transport - involvement of GTPases. Front. Plant Sci. 5:472. doi: 10.3389/fpls.2014.00472
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Keller, R., and Schneider, D. (2013). Homologs of the yeast Tvp38 vesicle-associated protein are conserved in chloroplasts and cyanobacteria. Front. Plant Sci. 4:467. doi: 10.3389/fpls.2013.00467
Khan, N. Z., Lindquist, E., Aronsson, H., Preisinger, C., and Fuchs, E. (2013). New putative chloroplast vesicle transport components and cargo proteins revealed using a bioinformatics approach: an Arabidopsis model. PLoS One 8:e59898. doi: 10.1371/journal.pone.0059898
Kirchhoff, H., Hall, C., Wood, M., Herbstova, M., Tsabari, O., Nevo, R., et al. (2011). Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl. Acad. Sci. U.S.A. 108, 20248–20253. doi: 10.1073/pnas.1104141109
Kleine, T., Maier, U. G., and Leister, D. (2009). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138. doi: 10.1146/annurev.arplant.043008.092119
Konopka, C. A., and Bednarek, S. Y. (2008). Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 147, 1590–1602. doi: 10.1104/pp.108.116863
Koshiba, T. (2004). Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858–862. doi: 10.1126/science.1099793
Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., Vothknecht, U. C., et al. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. U.S.A. 98, 4238–4242. doi: 10.1073/pnas.061500998
Lauber, M. H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., et al. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. doi: 10.1083/jcb.139.6.1485
Legesse-Miller, A., Massol, R. H., and Kirchhausen, T. (2003). Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell 14, 1953–1963. doi: 10.1091/mbc.E02-10-0657
Liberton, M., Austin, J. R., Berg, R. H., and Pakrasi, H. B. (2011a). Unique thylakoid membrane architecture of a unicellular N2-fixing Cyanobacterium revealed by electron tomography. Plant Physiol. 155, 1656–1666. doi: 10.1104/pp.110.165332
Liberton, M., Austin, J. R., Berg, R. H., and Pakrasi, H. B. (2011b). Insights into the complex 3-D architecture of thylakoid membranes in unicellular cyanobacterium Cyanothece sp. ATCC 51142. Plant Signal. Behav. 6, 566–569. doi: 10.4161/psb.6.4.14946
Lingard, M. J., Gidda, S. K., Bingham, S., Rothstein, S. J., Mullen, R. T., and Trelease, R. N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A Cooperate in Cell Cycle-Associated Replication of Peroxisomes. Plant Cell 20, 1567–1585. doi: 10.1105/tpc.107.057679
Liu, L., Zhang, Z., Mei, Q., and Chen, M. (2013). PSI: a comprehensive and integrative approach for accurate plant subcellular localization prediction. PLoS One 8:e75826. doi: 10.1371/journal.pone.0075826
Low, H. H., and Löwe, J. (2006). A bacterial dynamin-like protein. Nature 444, 766–769. doi: 10.1038/nature05312
Low, H. H., Sachse, C., Amos, L. A., and Löwe, J. (2009). Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342–1352. doi: 10.1016/j.cell.2009.11.003
Lu, S., Jang, H., Gu, S., Zhang, J., and Nussinov, R. (2016). Drugging ras GTPase: a comprehensive mechanistic and signaling structural view. Chem. Soc. Rev. 45, 4929–4952. doi: 10.1039/C5CS00911A
Mahalka, A. K., Kirkegaard, T., Jukola, L. T. I., Jäättelä, M., and Kinnunen, P. K. J. (2014). Human heat shock protein 70 (Hsp70) as a peripheral membrane protein. Biochim. Biophys. Acta 1838, 1344–1361. doi: 10.1016/j.bbamem.2014.01.022
Manganelli, R., and Gennaro, M. L. (2017). Protecting from envelope stress: variations on the phage-shock-protein theme. Trends Microbiol. 25, 205–216. doi: 10.1016/j.tim.2016.10.001
Mano, S., Nakamori, C., Fukao, Y., Araki, M., Matsuda, A., Kondo, M., et al. (2011). A defect of peroxisomal membrane protein 38 causes enlargement of peroxisomes. Plant Cell Physiol. 52, 2157–2172. doi: 10.1093/pcp/pcr147
Martin, W., and Herrmann, R. G. (1998). Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118, 9–17. doi: 10.1104/pp.118.1.9
Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251. doi: 10.1073/pnas.182432999
Mayer, M. P. (2013). Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514. doi: 10.1016/j.tibs.2013.08.001
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Mears, J. A., Lackner, L. L., Fang, S., Ingerman, E., Nunnari, J., and Hinshaw, J. E. (2011). Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26. doi: 10.1038/nsmb.1949
Meeusen, S. (2004). Mitochondrial fusion intermediates revealed in Vitro. Science 305, 1747–1752. doi: 10.1126/science.1100612
Meeusen, S., DeVay, R., Block, J., Cassidy-Stone, A., Wayson, S., McCaffery, J. M., et al. (2006). Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127, 383–395. doi: 10.1016/j.cell.2006.09.021
Michie, K. A., Boysen, A., Low, H. H., Møller-Jensen, J., and Löwe, J. (2014). LeoA, B and C from enterotoxigenic Escherichia coli (ETEC) are bacterial dynamins. PLoS One 9:e107211. doi: 10.1371/journal.pone.0107211
Mitchell, P. S., Emerman, M., and Malik, H. S. (2013). An evolutionary perspective on the broad antiviral specificity of MxA. Curr. Opin. Microbiol. 16, 493–499. doi: 10.1016/j.mib.2013.04.005
Miyagishima, S., Froehlich, J. E., and Osteryoung, K. W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18, 2517–2530. doi: 10.1105/tpc.106.045484
Miyagishima, S., Kuwayama, H., Urushihara, H., and Nakanishi, H. (2008). Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 15202–15207. doi: 10.1073/pnas.0802412105
Morré, D. J., Selldén, G., Sundqvist, C., and Sandelius, A. S. (1991). Stromal low temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiol. 97, 1558–1564. doi: 10.1104/pp.97.4.1558
Muehlethaler, K., and Frey-Wyssling, A. (1959). Development and structure of proplastids. J. Biophys. Biochem. Cytol. 6, 507–512.
Nakai, M., Sugita, D., Omata, T., and Endo, T. (1993). Sec-Y protein is localized in both the cytoplasmic and thylakoid membranes in the Cyanobacterium Synechococcus PCC7942. Biochem. Biophys. Res. Commun. 193, 228–234. doi: 10.1006/BBRC.1993.1613
Nakamura, Y., Kaneko, T., Hirosawa, M., Miyajima, N., and Tabata, S. (1998). CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26, 63–67. doi: 10.1093/nar/26.1.63
Nantais, D. E., Schwemmle, M., Stickney, J. T., Vestal, D. J., and Buss, J. E. (1996). Prenylation of an interferon-gamma-induced GTP-binding protein: the human guanylate binding protein, huGBP1. J. Leukoc. Biol. 60, 423–431. doi: 10.1002/jlb.60.3.423
Nevo, R., Charuvi, D., Shimoni, E., Schwarz, R., Kaplan, A., Ohad, I., et al. (2007). Thylakoid membrane perforations and connectivity enable intracellular traffic in cyanobacteria. EMBO J. 26, 1467–1473. doi: 10.1038/sj.emboj.7601594
Nevo, R., Charuvi, D., Tsabari, O., and Reich, Z. (2012). Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 70, 157–176. doi: 10.1111/j.1365-313X.2011.04876.x
Nickelsen, J., Rengstl, B., Stengel, A., Schottkowski, M., Soll, J., and Ankele, E. (2011). Biogenesis of the cyanobacterial thylakoid membrane system - an update. FEMS Microbiol. Lett. 315, 1–5. doi: 10.1111/j.1574-6968.2010.02096.x
Nitta, K., Suzuki, N., Honma, D., Kaneko, Y., and Nakamoto, H. (2005). Ultrastructural stability under high temperature or intensive light stress conferred by a small heat shock protein in cyanobacteria. FEBS Lett. 579, 1235–1242. doi: 10.1016/j.febslet.2004.12.095
Orso, G., Pendin, D., Liu, S., Tosetto, J., Moss, T. J., Faust, J. E., et al. (2010). Homotypic fusion of ER membranes requires the dynamin-like GTPase Atlastin. Nature 464, 942–942. doi: 10.1038/nature08886
Park, J. M., Cho, J. H., Kang, S. G., Jang, H. J., Pih, K. T., Piao, H. L., et al. (1998). A dynamin-like protein in Arabidopsis thaliana is involved in biogenesis of thylakoid membranes. EMBO J. 17, 859–867. doi: 10.1093/emboj/17.4.859
Park, J. M., Kang, S. G., Pih, K. T., Jang, H. J., Piao, H. L., Yoon, H. W., et al. (1997). A dynamin-like protein, ADL1, is present in membranes as a high-molecular -mass complex in Arabidopsis thaliana. Plant Physiol. 115, 763–771. doi: 10.1104/pp.115.2.763
Paul, P., Simm, S., Mirus, O., Scharf, K. D., Fragkostefanakis, S., and Schleiff, E. (2014). The complexity of vesicle transport factors in plants examined by orthology search. PLoS One 9:e97745. doi: 10.1371/journal.pone.0097745
Praefcke, G. J. K., and McMahon, H. T. (2004). The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5, k133–147. doi: 10.1038/nrm1313
Pyke, K. A., and Leech, R. M. (1994). A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 104, 201–207. doi: 10.1104/pp.110.160382
Rast, A., Heinz, S., and Nickelsen, J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847, 821–830. doi: 10.1016/j.bbabio.2015.01.007
Reubold, T. F., Faelber, K., Plattner, N., Posor, Y., Ketel, K., Curth, U., et al. (2015). Crystal structure of the dynamin tetramer. Nature 525, 404–408. doi: 10.1038/nature14880
Santel, A., and Fuller, M. T. (2001). Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867–874.
Sawant, P., Eissenberger, K., Karier, L., Mascher, T., and Bramkamp, M. (2016). A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environ. Microbiol. 18, 2705–2720. doi: 10.1111/1462-2920.13110
Schlimpert, S., Wasserstrom, S., Chandra, G., Bibb, M. J., Findlay, K. C., Flärdh, K., et al. (2017). Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl. Acad. Sci. U.S.A. 114, E6176–E6183. doi: 10.1073/pnas.1704612114
Schumacher, B., and Staeheli, P. (1998). Domains mediating intramolecular folding and oligomerization of MxA GTPase. J. Biol. Chem. 273, 28365–28370. doi: 10.1074/jbc.273.43.28365
Shah, C., Hegde, B. G., Morén, B., Behrmann, E., Mielke, T., Moenke, G., et al. (2014). Structural insights into membrane interaction and caveolar targeting of dynamin-like EHD2. Structure 22, 409–420. doi: 10.1016/j.str.2013.12.015
Shpetner, H. S., and Vallee, R. B. (1989). Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell 59, 421–432. doi: 10.1016/0092-8674(89)90027-5
Smaczynska-de Rooij, I. I., Allwood, E. G., Aghamohammadzadeh, S., Hettema, E. H., Goldberg, M. W., and Ayscough, K. R. (2010). A role for the dynamin-like protein Vps1 during endocytosis in yeast. J. Cell Sci. 123, 3496–3506. doi: 10.1242/jcs.070508
Smirnova, E., Griparic, L., Shurland, D.-L., and van der Bliek, A. M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256. doi: 10.1091/mbc.12.8.2245
Song, B. D., Leonard, M., and Schmid, S. L. (2004). Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J. Biol. Chem. 279, 40431–40436. doi: 10.1074/jbc.M407007200
Srivastava, R., Pisareva, T., and Norling, B. (2005). Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5, 4905–4916. doi: 10.1002/pmic.200500111
Staeheli, P., Haller, O., Boll, W., Lindenmann, J., and Weissmann, C. (1986). Mx protein: Constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44, 147–158. doi: 10.1016/0092-8674(86)90493-9
Tanz, S. K., Kilian, J., Johnsson, C., Apel, K., Small, I., Harter, K., et al. (2012). The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LCHB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J. 69, 743–754. doi: 10.1111/j.1365-313X.2011.04833.x
Torok, Z., Goloubinoff, P., Horvath, I., Tsvetkova, N. M., Glatz, A., Balogh, G., et al. (2001). Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc. Natl. Acad. Sci. U.S.A. 98, 3098–3103. doi: 10.1073/pnas.051619498
Ugarte-Uribe, B., Müller, H. M., Otsuki, M., Nickel, W., and García-Sáez, A. J. (2014). Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J. Biol. Chem. 289, 30645–30656. doi: 10.1074/jbc.M114.575779
Vestal, D. J., and Jeyaratnam, J. A. (2011). The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J. Interfereon Cytokine Res. 31, 89–97. doi: 10.1089/jir.2010.0102
Wang, Q., Sullivan, R. W., Kight, A., Henry, R. L., Huang, J., Jones, A. M., et al. (2004). Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604. doi: 10.1104/pp.104.049841
Westphal, S., Heins, L., Soll, J., and Vothknecht, U. C. (2001). Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl. Acad. Sci. U.S.A. 98, 4243–4248. doi: 10.1073/pnas.061501198
Yoshinari, A., Fujimoto, M., Ueda, T., Inada, N., Naito, S., and Takano, J. (2016). DRP1-dependent endocytosis is essential for polar localization and boron-induced degradation of the borate transporter BOR1 in Arabidopsis thaliana. Plant Cell Physiol. 57, 1985–2000. doi: 10.1093/pcp/pcw121
Young, J. C. (2010). Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88, 291–300. doi: 10.1139/o09-175
Zhang, X., and Hu, J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57, 146–159. doi: 10.1111/j.1365-313X.2008.03677.x
Zhang, X., and Hu, J. (2010). The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 22, 431–442. doi: 10.1105/tpc.109.071324
Keywords: dynamin, thylakoid membrane, membrane fusion, membrane biogenesis, cyanobacteria, chloroplasts
Citation: Jilly R, Khan NZ, Aronsson H and Schneider D (2018) Dynamin-Like Proteins Are Potentially Involved in Membrane Dynamics within Chloroplasts and Cyanobacteria. Front. Plant Sci. 9:206. doi: 10.3389/fpls.2018.00206
Received: 27 October 2017; Accepted: 02 February 2018;
Published: 22 February 2018.
Edited by:
Rebecca L. Roston, University of Nebraska–Lincoln, United StatesReviewed by:
William Zerges, Concordia University, CanadaWataru Sakamoto, Okayama University, Japan
Copyright © 2018 Jilly, Khan, Aronsson and Schneider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dirk Schneider, dirk.schneider@uni-mainz.de
 Ruven Jilly1
Ruven Jilly1