
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 February 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00178
 Rajkumar U. Zunjare1
Rajkumar U. Zunjare1 Firoz Hossain1*
Firoz Hossain1* Vignesh Muthusamy1
Vignesh Muthusamy1 Aanchal Baveja1
Aanchal Baveja1 Hema S. Chauhan1
Hema S. Chauhan1 Jayant S. Bhat2
Jayant S. Bhat2 Nepolean Thirunavukkarasu1
Nepolean Thirunavukkarasu1 Supradip Saha3
Supradip Saha3 Hari S. Gupta1
Hari S. Gupta1Traditional yellow maize though contains high kernel carotenoids, the concentration of provitamin A (proA) is quite low (<2 μg/g), compared to recommended level (15 μg/g). It also possesses poor endosperm protein quality due to low concentration of lysine and tryptophan. Natural variant of crtRB1 (β-carotene hydroxylase) and lcyE (lycopene-ε-cyclase) cause significant enhancement of proA concentration, while recessive allele, opaque2 (o2) enhances the level of these amino acids. Development of biofortified maize enriched in proA, lysine and tryptophan thus holds significance in alleviation of micronutrient malnutrition. In the present study, marker-assisted stacking of crtRB1, lcyE and o2 was undertaken in the genetic background of four maize hybrids (HQPM1, HQPM4, HQPM5, and HQPM7) popularly grown in India. HP704-22 and HP704-23 were used as donors, while four elite QPM parents viz., HKI161, HKI163, HKI193-1, and HKI193-2 were used as recipients. CrtRB1 showed severe segregation distortion, while lcyE segregated as per the expectation. Recovery of recurrent parent genome (RPG) among selected backcross progenies ranged from 89 to 93%. Introgressed progenies possessed high concentration of proA (7.38–13.59 μg/g), compared to 1.65–2.04 μg/g in the recurrent parents. The reconstituted hybrids showed an average of 4.5-fold increase in proA with a range of 9.25–12.88 μg/g, compared to original hybrids (2.14–2.48 μg/g). Similar plant-, ear-, and grain- characteristics of improved versions of both inbreds and hybrids were observed when evaluated with their respective original versions. Mean lysine (0.334%) and tryptophan (0.080%) of the improved hybrids were at par with the original versions (lysine: 0.340%, tryptophan: 0.083%). Improved hybrids also possessed similar grain yield potential (6,301–8,545 kg/ha) with their original versions (6,135–8,479 kg/ha) evaluated at two locations. This is the first study of staking crtRB1-, lcyE-, and o2-, favorable alleles in single genetic background. The improved inbreds can be effectively used as potential donor for independent and/or simultaneous introgression of crtRB1, lcyE, and o2 in the future breeding programme. These biofortified maize hybrids, rich in proA, lysine and tryptophan will hold great promise for nutritional security.
Micronutrient malnutrition popularly known as “hidden hunger” is a serious health problem worldwide, particularly in the under-developed and developing countries (Bouis and Saltzman, 2017). Nearly two billion people suffer from deficiency of micronutrients, while 815 million people are under-nourished (Global Nutrition Report, 2017). Among micronutrients, vitamin A plays, key role in human metabolism. This deficiency lead to visual blindness which may cause eye sight damage to millions preschool-age children. According to HarvestPlus, nearly 20 million pregnant women are vitamin A deficient, while out of which about one-third are clinically night-blind. There are about one-half of these cases occur in India with severe form of vision impairment. The deficiency of lysine and tryptophan leads to fatigue, delayed growth, loss of appetite, depression, anxiety in children (Nuss and Tanumihardjo, 2010; Jompuk et al., 2011). Moreover, unbalanced protein in the diet leads to protein energy malnutrition (PEM) that affects more than a billion people across the world (Bain et al., 2013). The adoption of quality protein maize (QPM) varieties possessing balanced protein due to higher lysine and tryptophan which has shown significant promise in solving problem of PEM across the world (Nyakurwa et al., 2017).
Cereals are rich source of energy, but lacking the required content of micronutrients (Nuss and Tanumihardjo, 2010). Genetic enhancement of micronutrient in crops through plant breeding known as “biofortification” which is a cost-effective and sustainable process, where micronutrients reach the target group in their natural form (Pfeiffer and McClafferty, 2007; Gupta et al., 2015; Neeraja et al., 2017). Maize occupies an important position in the world economy. It along with rice and wheat provides at least 30% of the food calories to more than 4.5 billion people in 94 developing countries, besides serving as a major component of animal feed (Shiferaw et al., 2011). In India, maize is the third most cereal after rice and wheat, and used as an important source of both food and feed (Yadav et al., 2015). Normal maize protein contains lower level of lysine (0.16–0.26%) and tryptophan (0.02–0.06%) which is less than half of the recommended dose specified for human nutrition (Bjarnason and Vasal, 1992; Vivek et al., 2008). Further, traditional yellow maize contains enough kernel carotenoids as compared to other cereals. However, it is predominated by non-proA fractions and contains only 0.25–2.50 μg/g of proA carotenoids which is far below the nutritional requirement (15 μg/g) for humans (Pixley et al., 2013).
Favorable alleles of lycopene ε-cyclase (lcyE) and β-carotene hydroxylase1 (crtRB1) genes causes enhancement in proA in maize (Harjes et al., 2008; Yan et al., 2010; Babu et al., 2013). The recessive opaque2 (o2) allele enhances endosperm lysine and tryptophan by almost 2-folds (Mertz et al., 1964). Marker-assisted selection (MAS) using very low expensive DNA markers helps in stacking of multiple target genes into a genetic background without progeny testing (Das et al., 2017). It also significantly reduces the breeding cycles required to reconstitute the recurrent parent genome (RPG) (Gupta et al., 2013). Further, high cost of HPLC (High Performance Liquid Chromatography) analyses for estimation of micronutrients among individuals of segregating populations could be avoided through usage of molecular markers. The successful examples of application MAS in development of nutritious maize hybrids in India have been the commercial release of “Vivek QPM9” (Gupta et al., 2013), “Pusa Vivek QPM9 Improved” (Muthusamy et al., 2014), “Pusa HM4 Improved,” “Pusa HM8 Improved,” and “Pusa HM9 Improved” (Hossain et al., 2017). Lysine and tryptophan rich QPM hybrids of late maturity so far released in the India do not contain recommended level of proA concentration. The present study was thus aimed to (i) stack favorable alleles of crtRB1, lcyE and opaque2 genes into elite inbreds/hybrids by using marker-assisted backcross breeding (MABB) and (ii) evaluate the MABB-derived –inbreds/hybrids for nutritional quality, agronomic and yield related traits.
The parental inbreds viz., HKI161, HKI163, HKI193-1, and HKI193-2 of four QPM hybrids, [HQPM1 (HKI193-1 × HKI163), HQPM4 (HKI193-2 × HKI161), HQPM5 (HKI163 × HKI161) and HQPM7 (HKI193-1 × HKI161)], were targeted for enrichment of micronutrients. The popular and commercial maize hybrids are adapted to diverse agro-ecologies of India (Table 1). Recurrent parents were crossed with donor lines and four crosses viz., cross-I (HKI161 × HP704-23), cross-II (HKI163 × HP704-22), cross-III (HKI193-1 × HP704-23), cross-IV (HKI193-2 × HP704-22) were attempted to stack crtRB1, lcyE, and o2 in the genetic background of recurrent parents. The pedigree information of the recurrent parents and donors is given in Table S1.
Backcross- and self- generations which were grown at different locations are described in Table S2, and the MABB scheme followed is represented as Figure 1. The recipients and donors showing polymorphism for gene-based markers were crossed during rainy season (July-November 2012) at IARI, New Delhi (28°089N, 77°129E, 229 MSL). Hybridity of the F1s was tested using gene-based markers, and the true F1s were backcrossed to their corresponding recurrent parent during winter season (December, 2012-April, 2013) at Winter Nursery Centre (WNC), Hyderabad (17°199N, 78°249E, 542.6 MSL). The BC1F1 progenies were grown at IARI, New Delhi during rainy season (2013), and foreground selection was carried out (Figure S1). The foreground positive plants with high recovery of RPG (RPG) and maximum phenotypic similarity were further backcrossed to the recurrent parent. The BC2F1 progenies were grown at WNC, Hyderabad during winter season (2013-14), and were subjected to foreground-, background- and phenotypic selection. The BC2F2 progenies were raised during rainy season (2014) at IARI, New Delhi. Foreground positive plants homozygous for all genes were subjected to background- and phenotypic- selection. The selected plants were subsequently self-pollinated to generate BC2F3 and BC2F4 progenies (Table S2).
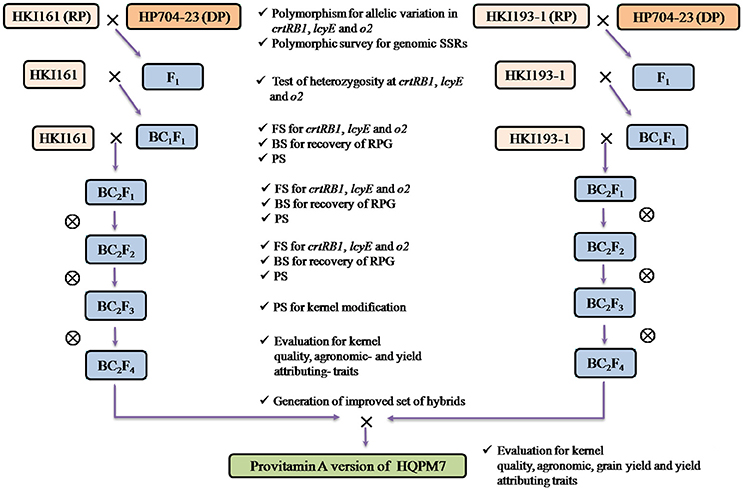
Figure 1. Marker-assisted backcross breeding scheme followed for development of provitamin A, lysine and tryptophan rich maize hybrid (e.g., HQPM7). RP, Recurrent Parent; DP, Donor Parent.
Three SSR markers based on o2 gene, viz., phi057, phi112, and umc1066 were screened to distinguish the parental lines, of which, phi057 marker revealed polymorphic pattern between recipients and donors (Gupta et al., 2013). Polymerase chain reaction (PCR) amplification for SSRs was followed as per Hossain et al. (2017). Four percent of Seakem LE agarose (Lonza, Rockland, ME USA) gel was used for electrophoretic separation of PCR products at 120 V for 3–5 h along with 100 bp DNA ladder (MBA-Fermentas). Gene-based InDel marker present in 3'TE and 5'TE region of crtRB1 and lcyE were used for foreground selection (Harjes et al., 2008; Yan et al., 2010; Figure S1; Table 2). PCR mediated amplification of crtRB1 and lcyE was performed using protocol standardized at Maize Genetics Unit, IARI (Zunjare et al., 2017a). Agarose of 1.5% concentration (Lonza, Rockland, ME USA) was used for separating the amplicon at 120 V for 2–3 h along with 100 bp DNA ladder (MBA-Fermentas). The amplified products were visualized using a gel documentation system (Alpha Innotech, California, USA) and scored for the presence and absence of designated allele.
SSRs with near uniform coverage across 10 chromosomes of maize genome were used for polymorphism survey between the respective recurrent and donor genotypes (Table 3, Table S3). The primer sequences of the SSRs were retrieved from the maize genome database (www.maizegdb.org) and were custom synthesized (Sigma Tech., USA). PCR amplification and scoring of amlicons was undertaken as per Hossain et al. (2017). The markers which were polymorphic between the recurrent and their respective donor parents were employed for recovering the RPG in individuals of BC1F1, BC2F1, and BC2F2 generations.
Selection of plant-, ear-, and grain-characteristics was performed among the individuals of each backcross- and self- generations for their similarity with their respective recurrent parents. The harvested BC2F3 seeds from the introgressed progenies were subjected to standard light box test along with the original recurrent parental seeds to measure the intensity of opaqueness (Hossain et al., 2008). The seeds with similar degree of opaqueness of the original inbreds were forwarded for further generation and the reconstitution of hybrids (Vivek et al., 2008; Gupta et al., 2013).
The selfed seeds of BC2F4 plants (two BC2F3 populations for HKI163) were utilized for biochemical analysis. The selfed ears were harvested at moisture level 12–14%, and then cleaned and dried under the shade. The equal amount of grains shelled from ears of same families was bulked together, and the samples thus drawn were stored in ambient temperature (22–26°C) for 2 months before biochemical analysis.
The extraction of β-carotene (BC) and β-cryptoxanthin (BCX) from maize seeds was carried out using procedures described by Kurilich and Juvik (1999) and Vignesh et al. (2012). Quantification of the BC and BCX was carried out with a Dionex Ultimate 3000 UHPLC System (Ultra High Performance Liquid Chromatography; Thermo Scientific, Massachusetts, USA). Samples were eluted through Carotenoid C30 column (5 μm, 4.6 × 250 mm, YMC) and detected with a diode array detector-3000 (RS). The mobile phase consisted of methanol: tert-butyl methyl ether (80:20, v/v), and the flow rate was 1 ml min−1. The standards of BC and BCX (Sigma Aldrich, USA) were used to make the regression curves and estimate the components with an absorbance at 450 nm. The proA concentration (μg/g on dry weight basis) was estimated as sum of BC plus half the BCX concentration (Babu et al., 2013).
The protocol standardized by Sarika et al. (2017) was followed to estimate lysine and tryptophan content of maize endosperm. Amino acids were estimated by using Dionex Ultimate 3000 UHPLC system (Thermo Scientific, Massachusetts, USA). The samples were eluted through AcclaimTM 120 C18 column (5 μm, 120A°, 4.6 × 150 mm, Thermo Scientific) and detected with a RS photodiode array detector (PDA) with absorbance in 265 and 280 nm wavelength, respectively for lysine and tryptophan. Final concentration of the amino acids in each sample was estimated by standard regression using external standards (AAS 18-5 ML, Sigma Aldrich).
Twelve improved progenies (BC2F3/BC2F4) along with the respective recurrent parents were evaluated during rainy season (2015) at IARI Experimental Farm, New Delhi. Two-three plants per entry were self-pollinated, and selfed grains were analyzed for proA, lysine and tryptophan. Characters viz., days to 50% male flowering (MF), days to 50% female flowering (FF), plant height (PH), ear height (EH), ear length (EL), ear width (EW), number of rows (NR), number of kernels per row (NKR), and 100-seed weight (TW) were recorded from open pollinated plants.
Selected 12 (BC2F4/BC2F5) progenies of the four improved inbreds were used to reconstitute twelve F1 hybrids during winter season (2015-16) at WNC, IIMR, Hyderabad. Three versions of the reconstituted hybrids (-A, -B, and -C) and their corresponding original hybrids were evaluated in Randomized Complete Block Design (RCBD) with two replications at two diverse maize growing zones of the country viz., IARI Experimental Farm, New Delhi in Northern India and IARI Regional Research Centre, Dharwad (15°219N, 75°059E, 750 MSL) Karnataka in Southern India during rainy season of 2016. Two to three plants in each of the hybrid entries were self-pollinated. Since, proA (Vignesh et al., 2012), lysine and tryptophan (Pixley and Bjarnason, 2002) do not vary much across locations, selfed seeds from IARI Experimental Farm, New Delhi were used for analysis of quality traits. However, morphological traits viz., MF, FF, PH, EH, EL, EW, NR, NKR, TW, and grain yield (GY) were recorded in open pollinated plants at both the locations.
The observed segregation pattern of crtRB1 and lcyE across segregating populations (BC1F1, BC2F1, and BC2F2), and o2 in BC1F1 generation of four crosses was tested for goodness of fit by χ2 analysis. The amplicons of markers used in background selection were scored as “A” for the recipient allele, “B” for the donor allele, and “H” for the heterozygous genotype. Recovery percentage of RPG was estimated using formula, RPG (%) = [A + (0.5H)/(A + B + H)] × 100 (Benchimol et al., 2005). The recovery of RPG among selected backcross-derived progenies was also established using Graphical Geno Types (GGT) version 3.0 (Van-Berloo, 1999). Graphical representation based on mean of improved proA, lysine and tryptophan was ascertained by Microsoft Excel (2013). Agronomic and biochemical data of hybrids were analyzed using Windostat 8.5 software package (Khetan, 2005).
All the four recurrent parents (HKI161, HKI163, HKI193-1, and HKI193-2) revealed unfavorable allele (C+: 296 bp), while the donors possessed favorable allele (C: 543 bp) of crtRB1 gene. Polymorphism test for lcyE revealed the presence of favorable allele (L: 650 bp) in two recurrent (HKI161 and HKI163) and two donor (HP704-22 and HP704-23) parents, while HKI193-1 and HKI193-2 possessed unfavorable allele (L+: 300 bp). All the recurrent parents possessed low proA concentration (HKI161: 2.04 μg/g, HKI163: 1.65 μg/g, HKI193-1: 1.84 μg/g, and HKI193-2: 1.74 μg/g), while donor parents possessed high proA concentration (HP704-22: 16.05 μg/g and HP704-23: 15.28 μg/g).Based on phi057, recessive allele of o2 (165 bp) was present in all QPM recurrent parents while, the donors possessed dominant allele, O2 (159 bp). The lysine (HKI161: 0.308%, HKI163: 0.347%, HKI193-1: 0.323%, and HKI193-2: 304%) and tryptophan (HKI161: 0.076%, HKI163: 0.082%, HKI193-1: 0.078%, and HKI193-2: 0.071%) content of recurrent parents was higher than their donor parents (Lysine, HP704-22: 0.176% and HP704-23: 0.192%; Tryptophan, HP704-22: 0.028% and HP704-23: 0.035%). A total of 114, 127, 133, and 124 polymorphic SSRs with polymorphism of 54.81, 61.06, 63.94, and 59.62% were observed in HKI161 × HP704-23, HKI163 × HP704-22, HKI193-1 × HP704-23, and HKI193-2 × HP704-22, respectively (Table 3, Table S3). The number of polymorphic markers in each chromosome ranged 7–17. These polymorphic markers were used for background selection for recovering the RPG in the backcross-derived populations.
The hybridity test in F1 generation has confirmed the success of crossing of parental lines. In BC1F1 and BC2F1 populations, the range of 100–120 and 106–122 plants, respectively were subjected to foreground selection of crtRB1 gene (Figure S1). The heterozygous progenies for crtRB1 were then subjected to foreground selection of lcyE. The polymorphic pattern for lcyE was observed only for viz., HKI193-1 × HP704-23 and HKI193-2 × HP704-22. The progenies (C+C/LL in cross-I and -II, and C+C/L+L in cross-III and -IV) were further subjected to foreground selection for o2 allele using phi057. The progenies with o2 allele in homozygous state were selected in BC1F1. The population size used for analysis, segregation pattern, chi-square test results are mentioned in Table 4. Subsequently, foreground positive plants were analyzed for background selection using polymorphic markers. The recovery of RPG varied from 70.47 to 80.83% across four BC1F1, while RPG varied from 83.07 to 90.60% in the four BC2F1 were observed (Table 5). Stringent phenotypic selection was also applied considering plant- architecture, ear- and grain- related traits. Foreground selection was executed among the plants in cross-I, -III and –IV, respectively to identify plants of genotype CC/LL/o2o2 (Table 3). For cross-II, two BC2F2 populations, BC2F2-I and BC2F2-II were raised in rainy season 2014 and winter season 2014-15, respectively. The RPG recovery in the selected plants ranged from 83.86 to 92.98% across four crosses (Table 4, Figure 2). Selection of morphological traits helped in deriving phenotypically similar progenies with their original versions. However, in case of HKI193-2-based progenies, CC/LL/o2o2 possessed undesirable characteristics of tip opening of ear and irregular grain arrangements, thus were not selected. Instead, progenies of genetic constitution “CC/L+L/o2o2” with desirable characteristics was selfed to develop BC2F3 population in winter season 2014-15 to recover “CC/LL/o2o2” genotypes (Table S2).The segregation pattern of crtRB1 locus showed deviation from the expected Mendelian ratio in all populations across the generations of four crosses, but lcyE segregated as per the expectation. o2 gene was also showed Mendelian monohybrid pattern of inheritance except in cross-II.
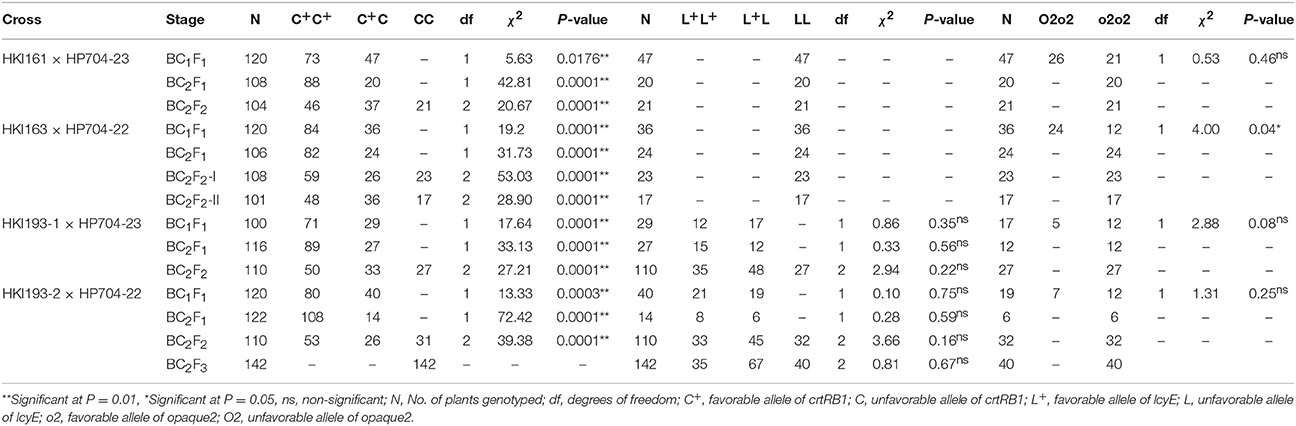
Table 4. Segregation pattern of crtRB1, lcyE and opaque2 in different backcross- and self- generations of the four crosses.
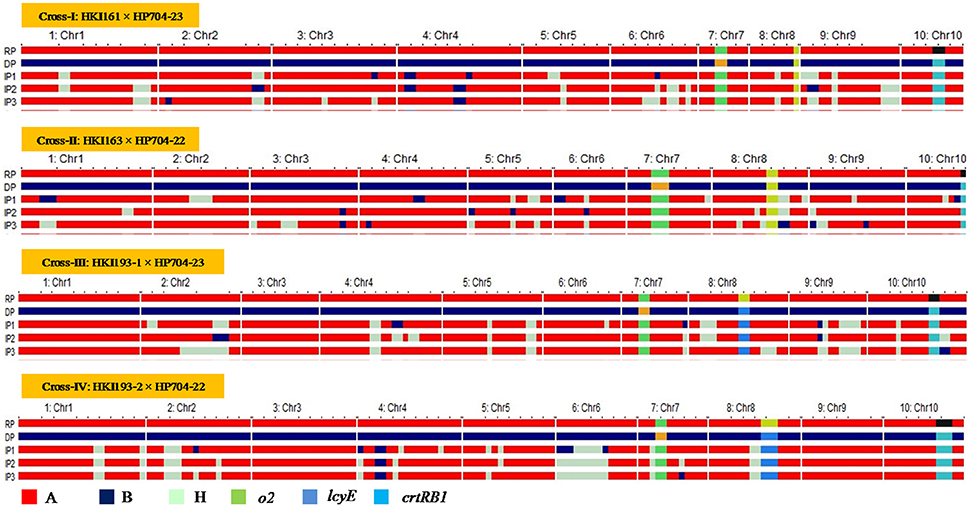
Figure 2. Graphical genotype of introgressed progenies across the four crosses. RP, Recurrent Parent; DP, Donor Parent; IP, Introgressed Progeny; Chr, Chromosome.
The carotenoid analysis of MABB-derived selected introgressed progenies of HKI161, HKI163, HKI193-1, and HKI193-2 showed a significant increase over their respective recurrent parents (Table 6). The proA concentration among improved inbreds ranged from 7.38 to 13.59 μg/g, compared to 1.65–2.04 μg/g among recurrent parents (Table 6). An average of 6-fold increase in proA was recorded among introgressed progenies. The lysine and tryptophan among the MABB-derived progenies (lysine: 0.274–0.394%, tryptophan: 0.071–0.084%) were at par with their respective parental lines (Table 6). The plant phenotypic characteristics and grain yield attributing traits of the introgressed lines were similar to their respective recurrent parents (Table 7). The opaqueness of 25–50% was recorded among introgressed progenies of HKI161 and HKI163, while 50–75% and 95–100% was observed among HKI193-1 and HKI193-2 -based introgressed lines, respectively. The degree of opaqueness is similar to proportion observed among the recurrent parents.
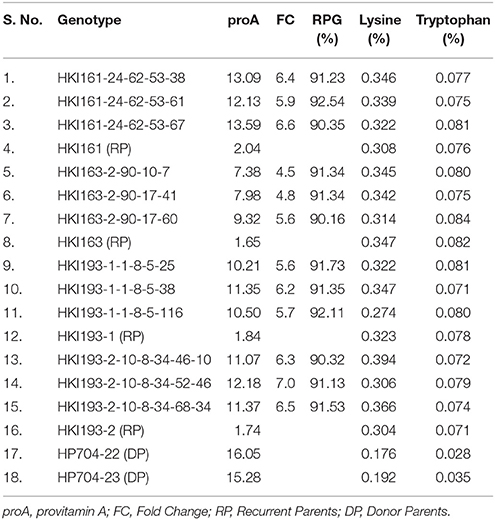
Table 6. Biochemical evaluation of selected introgressed progenies with their respective recurrent and donor parents.
The proA among reconstituted hybrids showed an average of 4.5-folds increase over their original versions. The proA of the newly derived hybrids ranged from 9.25 to 12.88 μg/g compared to 2.14–2.48 μg/g among the original hybrids (Figure 3). The mean proA in HQPM1-, HQPM4-, HQPM5-, and HQPM7-based reconstituted hybrids was 9.95, 10.47, 9.63, and 12.27 μg/g, respectively (Figure 3). The proA fold change was as high as 4.6 times in HQPM1-B over its original hybrid, while HQPM4-A, HQPM5-C, and HQPM7-B had 4.7-, 4.7-, and 5.1-fold increase in proA over their respective original hybrids. The lysine and tryptophan among the MABB-derived versions was at par with their respective original versions of hybrid (Figure 4). Among reconstituted hybrids, lysine ranged from 0.291 to 0.365%, while tryptophan varied from 0.072 to 0.085%. The improved hybrids showed high degree of resemblance for agronomic traits with their respective original hybrids across locations (Table 8 and Table S4). The grain yield and attributing traits of MABB-derived hybrids were also at par with their respective original versions (Table 8, Table S4).
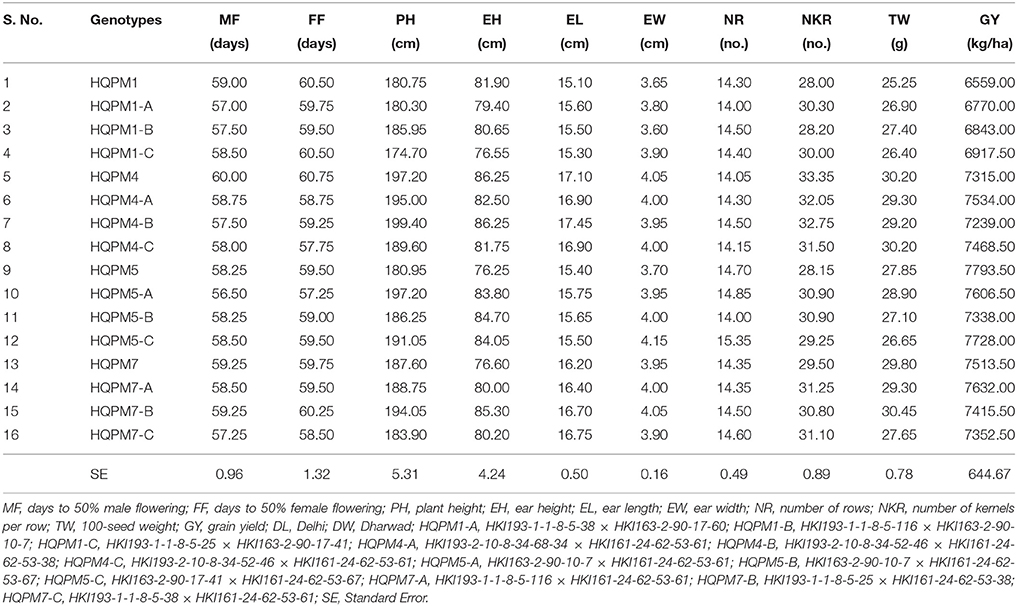
Table 8. Combined analysis of reconstituted hybrids along with original hybrids across the locations.
Normal maize endosperm contains low lysine and tryptophan, however their level is elevated by almost double in QPM genotypes due to recessive o2 present on chromosome 7 (Mertz et al., 1964; Vasal, 2000). However QPM like traditional normal maize genotypes also possesses very low proA carotenoids (Gupta et al., 2015). Animal's metabolism cannot synthesize lysine, tryptophan, and proA in their body, therefore the requirement is to be met from food sources (Pixley et al., 2013). Mutant version of crtRB1 and lcyE enhances proA level and makes maize grain more nutritious for human/ animal consumption (Harjes et al., 2008; Yan et al., 2010; Babu et al., 2013). The present study used MABB for combining the favorable alleles of crtRB1, lcyE, and o2 into four elite QPM inbreds, viz., HKI161, HKI163, HKI193-1, and HKI193-2 to develop biofortified maize hybrids rich in proA, lysine and tryptophan. In the present study, distinct variation for target traits revealed through marker polymorphism for crtRB1 (Muthusamy et al., 2014; Liu et al., 2015), lcyE, and o2 (Gupta et al., 2013; Hossain et al., 2017) was observed between recurrent and donor parents
Severe segregation distortion (SD) was realized for crtRB1 across the generations and crosses (Babu et al., 2013; Muthusamy et al., 2014; Liu et al., 2015). This SD is possibly due to the presence of gametophytic factors, mutants like defective kernel, male sterility and embryo-specific mutation (Neuffer et al., 1997; Lu et al., 2002). SD thus necessitates assaying of large population for achieving sufficient foreground positive genotypes in MABB programme. Conversely, lcyE gene did not show SD. In majority of populations, o2 also segregated as per Mendelian inheritance (Gupta et al., 2013; Hossain et al., 2017) with an exception in one population (Jompuk et al., 2011). Marker-assisted background selection using SSRs helped to achieve high recovery of RPG in just two backcross generations (Gupta et al., 2013; Muthusamy et al., 2014; Feng et al., 2015; Liu et al., 2015; Hossain et al., 2017). The introgressed progenies with high RPG showed high degree of resemblance with their corresponding recurrent parent for plant architecture and ear- and grain- characteristics (Gupta et al., 2013; Choudhary et al., 2014; Muthusamy et al., 2014). This high degree of phenotypic similarity among the reconstituted hybrids is also attributed to high RPG of the parental inbreds (Gupta et al., 2013; Muthusamy et al., 2014; Hossain et al., 2017).
Introgressed inbreds possessed 5–7-folds more proA than their respective recipient parents, while the reconstituted hybrids had 4–5-folds higher proA over their original versions. Expression analysis revealed that mutant crtRB1-transcripts was drastically reduced, leading to lesser amount of β-hydroxylase and lesser conversion of β-carotene to further components (Vallabhaneni et al., 2009; Yan et al., 2010). Similarly, reduced transcript level of lcyE-mutant produces lesser concentration of ε-cyclase compared to wild type allele, thereby shifting more lycopene flux from α-branch to β-branch of the carotenoid biosynthesis pathway (Harjes et al., 2008). The difference in expression levels of crtRB1 and lcyE genes was significant in endosperm, but not in embryos and leaves (Babu et al., 2013). The cumulative advantage of crtRB1 and lcyE for proA over individual effects has been reported in various studies (Azmach et al., 2013; Gebremeskel et al., 2017; Zunjare et al., 2017b). However, the favorable alleles of the both genes (crtRB1 and lcyE) occur in low frequency in the maize germplasm (Azmach et al., 2013; Babu et al., 2013; Muthusamy et al., 2015; Gebremeskel et al., 2017). Even in association mapping panel used by Harjes et al. (2008) and Yan et al. (2010) did not find any genotypes with favorable allele of both the genes (crtRB1 and lcyE).
The range of proA concentration was observed among both MABB-derived inbreds and hybrids, despite having the same allele of crtRB1 and lcyE. This variation is possibly due to varied interaction of crtRB1 and lcyE with the genome (Babu et al., 2013; Muthusamy et al., 2014). Also, improved progenies of four crosses revealed kernel proA concentration lower than their respective donor parents (Table 2). This suggests that other genetic loci or QTLs apart from favorable alleles of the crtRB1 and lcyE genes, contribute to the accumulation of proA (Wong et al., 2004; Chander et al., 2008; Zhou et al., 2012; Kandianis et al., 2013). Current study has achieved 70% of target level 15 μg/g proA in reconstituted hybrids (mean: 10.58 μg/g) which emphasize the need for further introgression of genetic loci like crtRB3, CCD1, and ZEP1 (Zhou et al., 2012; Suwarno et al., 2015).
The nutritional benefit of QPM with enhanced lysine and tryptophan were also conserved in the MABB-derived lines and their reconstituted hybrids. The o2 leads to reduction of zein proteins, with a concurrent increase in non-zein proteins rich in lysine and tryptophan (Ueda et al., 1992). o2 also down regulates the synthesis of lysine ketoglutarate reductase resulting in increased levels of free lysine (Kemper et al., 1999). Besides, it is also involved in regulation of various lysine-rich proteins and enzymes (Jia et al., 2013). The variation of lysine and tryptophan observed in the o2-based introgressed progenies is due to various modifier loci including opaque16 that affect regulation of amino acid biosynthesis (Wu et al., 2002; Yang et al., 2005; Pandey et al., 2015; Sarika et al., 2017). Similarly, the variation for lysine and tryptophan among o2-introgressed progenies was also observed by Gupta et al. (2013) and Hossain et al. (2017) in their MABB programmes.
The grain yield of reconstituted hybrids was also at par with the original hybrids. The similarity was due to indirect selection of loci for yield potential and various agronomic traits through background selection. Yield has not been used as the criterion of selecting the segregants, however the introgressed progenies led to the development of heterotic hybrids which were similar to the original hybrids (Gupta et al., 2013; Muthusamy et al., 2014; Hossain et al., 2017). The study thus implemented a successful demonstration of MABB augmented with stringent phenotypic selection for agro-morphological characters. The present investigation was the first report of combining favorable alleles of crtRB1, lcyE, and o2 in a single genetic background. During the year 2017, o2, and crtRB1-based “Pusa Vivek QPM9 Improved” maize hybrid with high proA, lysine and tryptophan has been released by ICAR in India (Muthusamy et al., 2014). Pusa Vivek QPM9 Improved provides an average grain yield of 5,588 and 5,916 kg/ha in Zone-I and Zone-IV, respectively (Annual Progress Report Kharif Maize, 2016). In comparison, the newly developed proA rich QPM hybrids in the present study possessed higher average grain yield (mean: 7,314 kg/ha). Moreover, these hybrids (Zone-II, III, IV, and V, Table 1) are also adapted to diverse agro-ecological zones.
The improved inbreds thus developed here can be used as donor lines for simultaneous introgression of o2, crtRB1, and lcyE in the breeding programme. Further, the improved inbreds can be crossed among them to generate different F2 populations, where from new inbreds with high proA, lysine and tryptophan can be derived using pedigree method. The nutritionally improved hybrids can be grown for cultivation for commercial usage of biofortified grains as food and feed. The significance of biofortified maize for human health has very well observed in many countries (Bouis and Saltzman, 2017). The benefit of QPM in human health and poultry birds is also well documented (Gunaratna et al., 2010; Panda et al., 2014). Biofortified orange maize was found to be as efficacious as a vitamin A supplement in children (Gannon et al., 2014). Dubey et al. (2017) using caco-2 cell model demonstrated that proA rich maize hybrids having crtRB1 allele possessed enhanced bioavailability of β-carotene. Chickens fed with biofortifid maize produced eggs rich in proA (Liu et al., 2012; Heying et al., 2014; Moreno et al., 2016; Sowa et al., 2017). A study further revealed that proA biofortified fed chickens had higher redness and yellowness and lower lightness in the meat and skin color than white maize fed chickens (Odunitan-Wayas et al., 2016). Thus, both direct consumption through foods and indirect consumption through chicken -eggs and -meats, proA rich maize contributes to nutritional security. Lividini and Fiedler (2015) demonstrated the great promise of proA rich maize for becoming a highly cost-effective strategy for reducing malnutrition. Biofortified high yielding maize hybrid rich in proA, lysine and tryptophan nutrients would be a sustainable delivery tools to overcome micronutrient malnutrition.
We report here the development of four maize hybrids using marker-assisted stacking of o2, crtRB1, and lcyE. The hybrids were evaluated at two locations and provided similar grain yield potential of the original hybrids. The inbreds with elevated lysine, tryptophan and proA concentration can be used as potential donors for development of nutrient rich maize cultivars in future breeding programmes. The biofortified maize hybrids enriched with proA, lysine and tryptophan possess great potential to simultaneously alleviate vitamin A deficiency and protein-energy malnutrition across the world.
Conduct of all experiments: RZ; Development of segregating progenies: FH and VM; Morphological characterization: FH and JB; Phenotyping for kernel quality: VM, AB and SS; Analysis on kernel modification: HC; Statistical analyses: FH and NT; Drafting of the manuscript: RZ and FH; Designing of the experiment: FH and HG.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
First author is thankful to the University Grant Commission (UGC) for Rajiv Gandhi National Fellowship (RGNF) for the doctoral programme [F1-17.1/2012-13/RGNF-2012-13-SC-MAH-28430/(SAIII/Website)]. Financial support of Department of Biotechnology (DBT) through the project entitled “Enrichment of nutritional quality in maize through molecular breeding” (SAN No. BT/PR10922/AGII/106/944/2014), and ICAR-IARI through in-house project is gratefully acknowledged. The authors thank Drs. Natalia Palacios Rojas, Raman Babu, and Boddupalli M. Prasanna (CIMMYT, Mexico and Maize HarvestPlus Programme) for providing proA enriched donor lines. We also thank the breeders of CCSHAU, Uchani center for sharing the QPM inbreds. We sincerely acknowledge Director, IIMR, Ludhiana, and Dr. Javaji C. Sekhar, In-Charge, WNC-IIMR, Hyderabad for providing the off-season nursery during the winter season. The help of Mr. Manish Kapasia for management of field activities is duly acknowledged.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00178/full#supplementary-material
Figure S1. Foreground selection for crtRB1, lcyE, and o2 in BC1F1 generation. Star indicates plants heterozygous for crtRB1/lcyE and homozygous for o2 allele. RP, Recurrent Parent; DP, Donor Parent.
Table S1. Details of genetic materials used in MABB.
Table S2. Details of populations generated under MABB.
Table S3. List of SSR markers used in background selection.
Table S4. Morphological characterization of reconstituted hybrids along with their original versions.
Annual Progress Report Kharif Maize (2016). All India Coordinated Research Project on Maize. New Delhi: Indian Institute of Maize Research, Pusa Campus.
Azmach, G., Gedil, M., Menkir, A., and Spillane, C. (2013). Marker-trait association analysis of functional gene markers for provitamin A levels across diverse tropical yellow maize inbred lines. BMC Plant Biol. 13:227. doi: 10.1186/1471-2229-13-227
Babu, R., Rojas, N. P., Gao, S., Yan, J., and Pixley, K. (2013). Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin a concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399. doi: 10.1007/s00122-012-1987-3
Bain, L. E., Awah, P. K., Geraldine, N., Kindong, N. P., Sigal, Y., Bernard, N., et al. (2013). Malnutrition in Sub - Saharan Africa: burden, causes and prospects. Pan Afr. Med. J. 15, 1–9. doi: 10.11604/pamj.2013.15.120.2535
Benchimol, L. L., de Souza, C. L., and de Souza, A. P. (2005). Microsatellite-assisted backcross selection in maize. Genet. Mol. Biol. 28, 789–797. doi: 10.1590/S1415-47572005000500022
Bjarnason, M., and Vasal, S. K. (1992). Breeding of quality protein maize. Plant Breed. Rev. 9, 181–216. doi: 10.1002/9780470650363.ch7
Bouis, H. E., and Saltzman, A. (2017). Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Sec. 12, 49–58. doi: 10.1016/j.gfs.2017.01.009
Chander, S., Guo, Y. Q., Yang, X. H., Zhang, J., Lu, X. Q., Yan, J. B., et al. (2008). Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor. Appl. Genet. 116, 223–233. doi: 10.1007/s00122-007-0661-7
Choudhary, M., Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Pandey, N., Jha, S. K., et al. (2014). Characterization of β-carotene rich MAS-derived maize inbreds possessing rare genetic variation in β-carotene hydroxylase gene. Indian J. Genet. Plant Breed. 74, 620–623. doi: 10.5958/0975-6906.2014.00900.6
Das, G., Patra, J. K., and Baek, K. H. (2017). Insight into MAS: a molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 8:985. doi: 10.3389/fpls.2017.00985
Dubey, N., Mashurabad, P. C., Hossain, F., Pullakhandam, R., Longvah, T., and Bhardwaj, D. K. (2017). β-carotene bio-accessibility from biofortified 1 maize (Zea mays L.) is related to its density and is negatively influenced by lutein and zeaxanthin. Food Funct. 9, 379–388. doi: 10.1039/c7fo01034f
Feng, F., Wang, Q., Liang, C., Yang, R., and Li, X. (2015). Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206, 513–521. doi: 10.1007/s10681-015-1519-8
Gannon, B., Kaliwile, C., Arscott, S. A., Schmaelzle, S., Chileshe, J., Kalungwana, N., et al. (2014). Biofortified orange maize is as efficacious as a vitamin a supplement in Zambian children even in the presence of high liver reserves of vitamin a: a community-based, randomized placebo-controlled trial. Am. J. Clin. Nutr. 100, 1541–1550. doi: 10.3945/ajcn.114.087379
Gebremeskel, S., Garcia-oliveira, A. L., Menkir, A., Adetimirin, V., and Gedil, M. (2017). Effectiveness of predictive markers for marker assisted selection of pro-vitamin A carotenoids in medium-late maturing maize (Zea mays L.) inbred lines. J. Cereal Sci. 79, 27–34. doi: 10.1016/j.jcs.2017.09.001
Global Nutrition Report (2017). Availble online at: https://www.globalnutritionreport.org.
Gunaratna, N. S., De Groote, H., Nestel, P., Pixley, K. V., and McCabe, G. P. (2010). A meta-analysis of community-based studies on quality protein maize. Food Policy 35, 202–210. doi: 10.1016/j.foodpol.2009.11.003
Gupta, H. S., Hossain, F., and Muthusamy, V. (2015). Biofortification of maize: an Indian perspective. Indian J. Genet. Plant Breed. 75, 1–22. doi: 10.5958/0975-6906.2015.00001.2
Gupta, H. S., Raman, B., Agrawal, P. K., Mahajan, V., Hossain, F., and Thirunavukkarasu, N. (2013). Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82. doi: 10.1111/pbr.12009
Harjes, C. E., Rocheford, T. R., Bai, L., Brutnell, T. P., Kandianis, C. B., Sowinski, S. G., et al. (2008). Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 319, 330–333. doi: 10.1126/science.1150255
Heying, E. K., Tanumihardjo, J. P., Vasic, V., Cook, M., Palacios-Rojas, N., and Tanumihardjo, S. A. (2014). Biofortified orange maize enhances β-cryptoxanthin concentrations in egg yolks of laying hens better than tangerine peel fortificant. J. Agric. Food Chem. 62, 11892–11900. doi: 10.1021/jf5037195
Hossain, F., Muthusamy, V., Pandey, N., Vishwakarma, A. K., Baveja, A., Zunjare, R. U., et al. (2017). Marker-assisted introgression of opaque2 allele for rapid conversion of elite maize hybrids into quality protein maize. J. Genet. Availble online at: http://www.ias.ac.in/public/Resources/General/jgen/JGEN-D-16-00919R2.pdf
Hossain, F., Prasanna, B. M., Kumar, R., and Singh, B. B. (2008). Genetic analysis of kernel modification in quality protein maize (QPM) genotypes. Indian J. Genet. Plant Breed. 68, 1–9.
Jia, M., Wu, H., Clay, K., Jung, R., Larkins, B. A., and Gibbon, B. C. (2013). Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2 mutant by transcriptional and proteomic analysis. BMC Plant Biol. 13:60. doi: 10.1186/1471-2229-13-60
Jompuk, C., Cheuchart, P., Jompuk, P., and Apisitwanich, S. (2011). Improved tryptophan content in maize with opaque-2 gene using marker assisted selection (MAS) in backcross and selfing generations. Kasetsart J. Nat. Sci. 45, 666–674.
Kandianis, C. B., Stevens, R., Liu, W., Palacios, N., Montgomery, K., Pixley, K., et al. (2013). Genetic architecture controlling variation in grain carotenoid composition and concentrations in two maize populations. Theor. Appl. Genet. 126, 2879–2895. doi: 10.1007/s00122-013-2179-5
Kemper, E. L., Neto, G. C., Papes, F., Moraes, K. C., Leite, A., and Arruda, P. (1999). The role of opaque2 in the control of lysine-degrading activities in developing maize endosperm. Plant Cell 11, 1981–1993. doi: 10.1105/tpc.11.10.1981
Kurilich, A. C., and Juvik, J. A. (1999). Quantification of carotenoid and tocopherol antioxidants in Zea mays. J. Agric. Food Chem. 47, 1948–1955. doi: 10.1021/jf981029d
Liu, L., Jeffers, D., Zhang, Y., Ding, M., Chen, W., Kang, M. S., et al. (2015). Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol. Breed. 35:154. doi: 10.1007/s11032-015-0349-7
Liu, Y. Q., Davis, C. R., Schmaelzle, S. T., Rocheford, T., Cook, M. E., and Tanumihardjo, S. A. (2012). β-Cryptoxanthin biofortified maize (Zea mays) increases β-cryptoxanthin concentration and enhances the color of chicken egg yolk. Poultry Sci. 91, 432–438. doi: 10.3382/ps.2011-01719
Lividini, K., and Fiedler, J. L. (2015). Assessing the promise of biofortification: a case study of high provitamin A maize in Zambia. Food Policy 54, 65–277. doi: 10.1016/j.foodpol.2015.04.007
Lu, H., Romero-Severson, J., and Bernardo, R. (2002). Chromosomal regions associated with segregation distortion in maize. Theor. Appl. Genet. 105, 622–628. doi: 10.1007/s00122-002-0970-9
Mertz, E. T., Bates, L. S., and Nelson, O. E. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280. doi: 10.1126/science.145.3629.279
Moreno, J. A., Díaz-Gómez, J., Nogareda, C., Angulo, E., Sandmann, G., Portero-Otin, M., et al. (2016).The distribution of carotenoids in hens fed on biofortified maize is influenced by feed composition, absorption, resource allocation and storage. Sci. Rep. 6:35346. doi: 10.1038/srep35346
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Choudhary, M., Saha, S., Bhat, J. S., et al. (2014). Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9:e113583. doi: 10.1371/journal.pone.0113583
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Saha, S., Shanker Gupta, H., and Gupta, H. S. (2015). Allelic variations for lycopene-ε-cyclase and β-carotene hydroxylase genes in maize inbreds and their utilization in β-carotene enrichment programme. Cogent Food Agric. 1:1033141. doi: 10.1080/23311932.2015.1033141
Neeraja, C. N., Ravindra Babu, V., Ram, S., Hossain, F., Hariprasanna, K., and Rajpurohit, B. S. (2017). Biofortification in cereals : progress and prospects. Curr. Sci. 113, 1050–1057. doi: 10.18520/cs/v113/i06/1050-1057
Neuffer, M. G., Coe, E. H., and Wessler, S. R. (1997). Mutants of Maize. New York, NY: Cold Spring Harbor Laboratory Press.
Nuss, E. T., and Tanumihardjo, S. A. (2010). Maize: a paramount staple crop in the context of global nutrition. Compr. Rev. Food Sci. Food Saf. 9, 417–436. doi: 10.1111/j.1541-4337.2010.00117.x
Nyakurwa, C. S., Gasura, E., and Mabasa, S. (2017). Potential for quality protein maize for reducing protein energy undernutrition in maize dependent sub-saharan African countries: a review. African Crop Sci. J. 25, 521–537. doi: 10.4314/acsj.v25i4.9
Odunitan-Wayas, F. A., Kolanisi, U., Chimonyo, M., and Siwela, M. (2016). Effect of provitamin A biofortified maize inclusion on quality of meat from indigenous chickens. J. Appl. Poult. Res. 25, 581–590 doi: 10.3382/japr/pfw040
Panda, A. K., Zaidi, P. H., Rama Rao, S. V., and Raju, M. V. L. N. (2014). Efficacy of quality protein maize in meeting energy and essential amino acid requirements in broiler chicken production, J. Appl. Anim. Res. 42, 133–5139, doi: 10.1080/09712119.2013.822812
Pandey, N., Hossain, F., Kumar, K., Vishwakarma, A. K., Muthusamy, V., Saha, S., et al. (2015). Molecular characterization of endosperm- and amino acids- modifications among quality protein maize inbreds. Plant Breed. 135, 47–54. doi: 10.1111/pbr.12328
Pfeiffer, W. H., and McClafferty, B. (2007). HarvestPlus: breeding crops for better nutrition. Crop Sci. 47, S89–S105. doi: 10.2135/cropsci2007.09.0020IPBS
Pixley, K., Palacios-Rojas, N., Babu, R., Mutale, R., Surles, R., and Simpungwe, E. (2013). “Biofortification of maize with provitamin A carotenoids,” in Carotenoids and Human Health, ed S. Tanumihardjo (New York, NY: Springer Science + Business Media), 271–292.
Pixley, K. V., and Bjarnason, M. S. (2002). Stability of grain yield, endosperm modification, and protein quality of hybrid and open-pollinated quality protein maize (QPM) cultivars. Crop. Sci. 42, 1882–1890. doi: 10.2135/cropsci2002.1882
Sarika, K., Hossain, F., Muthusamy, V., Baveja, A., Zunjare, R., and Goswami, R. (2017). Exploration of novel opaque16 mutation as a source for high lysine and tryptophan in maize endosperm. Indian J. Genet. 77, 59–64. doi: 10.5958/0975-6906.2017.00008.6
Shiferaw, B., Prasanna, B. M., Hellin, J., and Bänziger, M. (2011). Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327. doi: 10.1007/s12571-011-0140-5
Sowa, M., Yu, J., Palacios-Rojas, N., Goltz, S. R., Howe, J. A., Davis, C. R., et al. (2017). Retention of carotenoids in Biofortified maize flour and β-cryptoxanthin-enhanced eggs after household cooking. ACS Omega 2, 7320–7328. doi: 10.1021/acsomega.7b01202
Suwarno, W. B., Pixley, K. V., Palacios-Rojas, N., Kaeppler, S. M., and Babu, R. (2015). Genome-wide association analysis reveals new targets for carotenoid biofortification in maize. Theor. Appl. Genet. 128, 851–864. doi: 10.1007/s00122-015-2475-3
Ueda, T., Waverczak, W., Ward, K., Sher, N., Ketudat, M., Schmidt, R. J., et al. (1992). Mutations of the 22- and 27-kD zein promoters affect transactivation by the Opaque-2 protein. Plant Cell 4, 701–709. doi: 10.1105/tpc.4.6.701
Vallabhaneni, R., Gallagher, C. E., Licciardello, N., Cuttriss, A. J., Quinlan, R. F., and Wurtzel, E. T. (2009). Metabolite sorting of a germplasm collection reveals the hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol. 151, 1635–1645. doi: 10.1104/pp.109.145177
Van-Berloo (1999). GGT: software for display of graphical genotypes. J. Hered. 90, 328–329. doi: 10.1093/jhered/90.2.328
Vasal, S. K. (2000). The quality protein maize story. Food Nutr. Bull. 21, 445–450. doi: 10.1177/156482650002100420
Vignesh, M., Hossain, F., Nepolean, T., Supradip, S., Agrawal, P. K., Guleria, S. K., et al. (2012). Genetic variability for kernel beta-carotene and utilization of crtRB1 3'TE gene for biofortification in maize (Zea mays L.). Indian J Genet Plant Breed 72, 189–194.
Vivek, B. S., Krivanek, A. F., Palacios-Rojas, N., Twumasi-Afriyie, S., and Diallo, A. O. (2008). Breeding Quality Protein Maize (QPM): Protocols for Developing QPM Cultivars. Mexico: CIMMYT.
Wong, J. C., Lambert, R. J., Wurtzel, E. T., and Rocheford, T. R. (2004). QTL and candidate genes phytoene synthase and ζ-carotene desaturase associated with the accumulation of carotenoids in maize. Theor. Appl. Genet. 108, 349–359. doi: 10.1007/s00122-003-1436-4
Wu, R., Lou, X. Y., Ma, C. X., Wang, X., Larkins, B., and Casella, G. (2002). An improved genetic model generates high-resolution mapping of QTL for protein quality in maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 99, 11281–11286. doi: 10.1073/pnas.112345699
Yadav, O. P., Hossain, F., Karjagi, C. G., Kumar, B., Zaidi, P. H., Jat, S. L., et al. (2015). Genetic improvement of maize in india: retrospect and prospects. Agric. Res. 4, 325–338. doi: 10.1007/s40003-015-0180-8
Yan, J., Kandianis, C. B., Harjes, C. E., Bai, L., Kim, E.-H., Yang, X., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322–327. doi: 10.1038/ng.551
Yang, W., Zheng, Y., Zheng, W., and Feng, R. (2005). Molecular genetic mapping of a high-lysine mutant gene (opaque-16) and the double recessive effect with opaque-2 in maize. Mol. Breed. 15, 257–269. doi: 10.1007/s11032-004-5947-8
Zhou, Y., Han, Y., Li, Z., Fu, Y., Fu, Z., Xu, S., et al. (2012). ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize. J Integr Plant Biol. 54, 260–269. doi: 10.1111/j.1744-7909.2012.01106.x
Zunjare, R. U., Chhabra, R., Hossain, F., Muthusamy, V., Baveja, A., and Gupta, H. S. (2017a). Development and validation of multiplex-PCR assay for simultaneous detection of rare alleles of crtRB1 and lcyE governing higher accumulation of provitamin A in maize kernel. J. Plant Biochem. Biotechnol. 1–7. doi: 10.1007/s13562-017-0432-8
Zunjare, R. U., Hossain, F., Muthusamy, V., Baveja, A., Chauhan, H. S., Thirunavukkarasu, N., et al. (2017b). Influence of rare alleles of β-carotene hydroxylase and lycopene epsilon cyclase genes on accumulation of provitamin A carotenoids in maize kernels. Plant Breed. 136, 872–880. doi: 10.1111/pbr.12548
Keywords: provitamin A, crtRB1, lcyE, QPM, marker-assisted selection, biofortification
Citation: Zunjare RU, Hossain F, Muthusamy V, Baveja A, Chauhan HS, Bhat JS, Thirunavukkarasu N, Saha S and Gupta HS (2018) Development of Biofortified Maize Hybrids through Marker-Assisted Stacking of β-Carotene Hydroxylase, Lycopene-ε-Cyclase and Opaque2 Genes. Front. Plant Sci. 9:178. doi: 10.3389/fpls.2018.00178
Received: 17 November 2017; Accepted: 30 January 2018;
Published: 20 February 2018.
Edited by:
Rodomiro Ortiz, Swedish University of Agricultural Sciences, SwedenReviewed by:
Natalia Palacios, International Maize and Wheat Improvement Center (Mexico), MexicoCopyright © 2018 Zunjare, Hossain, Muthusamy, Baveja, Chauhan, Bhat, Thirunavukkarasu, Saha and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Firoz Hossain, ZmhfZ3BiQHlhaG9vLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.