
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 February 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00146
Ribosome-inactivating proteins (RIPs) are toxic N-glycosidases that depurinate eukaryotic and prokaryotic rRNAs, thereby arresting protein synthesis during translation. RIPs are widely found in various plant species and within different tissues. It is demonstrated in vitro and in transgenic plants that RIPs have been connected to defense by antifungal, antibacterial, antiviral, and insecticidal activities. However, the mechanism of these effects is still not completely clear. There are a number of reviews of RIPs. However, there are no reviews on the biological functions of RIPs in defense against pathogens and insect pests. Therefore, in this report, we focused on the effect of RIPs from plants in defense against pathogens and insect pest attacks. First, we summarize the three different types of RIPs based on their physical properties. RIPs are generally distributed in plants. Then, we discuss the distribution of RIPs that are found in various plant species and in fungi, bacteria, algae, and animals. Various RIPs have shown unique bioactive properties including antibacterial, antifungal, antiviral, and insecticidal activity. Finally, we divided the discussion into the biological roles of RIPs in defense against bacteria, fungi, viruses, and insects. This review is focused on the role of plant RIPs in defense against bacteria, fungi, viruses, and insect attacks. The role of plant RIPs in defense against pathogens and insects is being comprehended currently. Future study utilizing transgenic technology approaches to study the mechanisms of RIPs will undoubtedly generate a better comprehending of the role of plant RIPs in defense against pathogens and insects. Discovering additional crosstalk mechanisms between RIPs and phytohormones or reactive oxygen species (ROS) against pathogen and insect infections will be a significant subject in the field of biotic stress study. These studies are helpful in revealing significance of genetic control that can be beneficial to engineer crops tolerance to biotic stress.
Agricultural crops often suffer from fungi, viruses, and bacteria attacks which negatively cause the survival, biomass production and yield and quality of produces throughout the world (Culbreath et al., 2003; Rodoni, 2009). We have known that plants own some specific metabolic pathways to synthesize lots of valuable proteins, and these proteins can be used for prevention and treatment of diseases (Calixto, 2000). Such as, ribosome-inactivating proteins (RIPs) from plants have been suggested to confer disease resistance (Huang et al., 2008). The RIPs could catalytically inactivate eukaryotic ribosomes which inhibit protein synthesis at translation (de Virgilio et al., 2010).
Various RIPs have been proved to exhibit different of antimicrobial activities, for instance antitumor, antibacterial activity, antifungal activity, and broad-spectrum antiviral activity (Mock et al., 1996; Wang and Tumer, 2000; Stirpe, 2004; Puri et al., 2009; Bian et al., 2010). In agriculture, It is demonstrated in vitro and in transgenic plants that RIPs have been connected to defense by antifungal, antibacterial, antiviral and insecticidal activities (Stevens et al., 1981; Wang and Tumer, 2000; Choudhary N.L. et al., 2008; Akkouh et al., 2015). These studies have conferred a wide knowledge base for comprehending the medicinal and biochemical properties of RIPs. Nevertheless, the biological functions of plant RIPs are rarely investigated. Lots of reviews on RIPs have been reported (Nielsen and Boston, 2001; Girbés et al., 2004; Hartley and Lord, 2004b; Stirpe, 2004;Van Damme et al., 2008; de Virgilio et al., 2010; Lord and Hartley, 2010; Ng et al., 2010; Stirpe and Lappi, 2014; Schrot et al., 2015). For example, Bolognesi et al. (2016) provided a historical overview of the biological role of RIPs. However, there are rarely reviews on the biological functions of RIPs in defense against attacks by pathogens (bacteria, fungi, and viruses) and insects. Therefore, we focus on recent study advances, distribution, physiological roles of RIPs and their roles in defense against pathogen and insect attacks.
RIPs from plants have been divided into three main types based on their physical properties, including type I, type II, and type III (Olsnes and Pihl, 1973a; Lord et al., 1994; Peumans et al., 2001; de Virgilio et al., 2010).
Type I RIPs are the most widely distributed RIPs, and they are composed of a single polypeptide domain protein of about 30 kDa with N-glycosidase activity (Barbieri et al., 1993; Hartley and Lord, 2004a,b; Stirpe, 2004; de Virgilio et al., 2010) (Figure 1A). Even though type I RIPs share some of highly conserved active site cleft residues, however, the overall sequence and posttranslational modifications of them are significantly different (Monzingo and Robertus, 1992; Barbieri et al., 1993; Mlsna et al., 1993; Husain et al., 1994). The first type I RIP, pokeweed antiviral protein (PAP), was identified from American pokeweed (Dallal and Irvin, 1978). Subsequently, various type I RIPs have been identified from many different of plant species and a few bacteria species. A large number of isolated type I RIPs are composed of Cucurbitaceae, Euphorbiaceae, and Fabaceae. For example, trichosanthin (TCS), luffin α, luffin β, Mirabilis antiviral protein (MAP), camphorin, barley (Hordeum vulgare) translation inhibitor and saporin (from soapwort, S. officinalis) are type I RIPs. Meng et al. (2014) combined ion-exchange and gel filtration chromatography methods to isolate two RIPs, α-MMC and an anti-HIV protein (MAP30) from bitter melon. The α-MMC and MAP30 are also type I RIPs with molecular weights of 28.585 and 29.094 kDa, respectively.
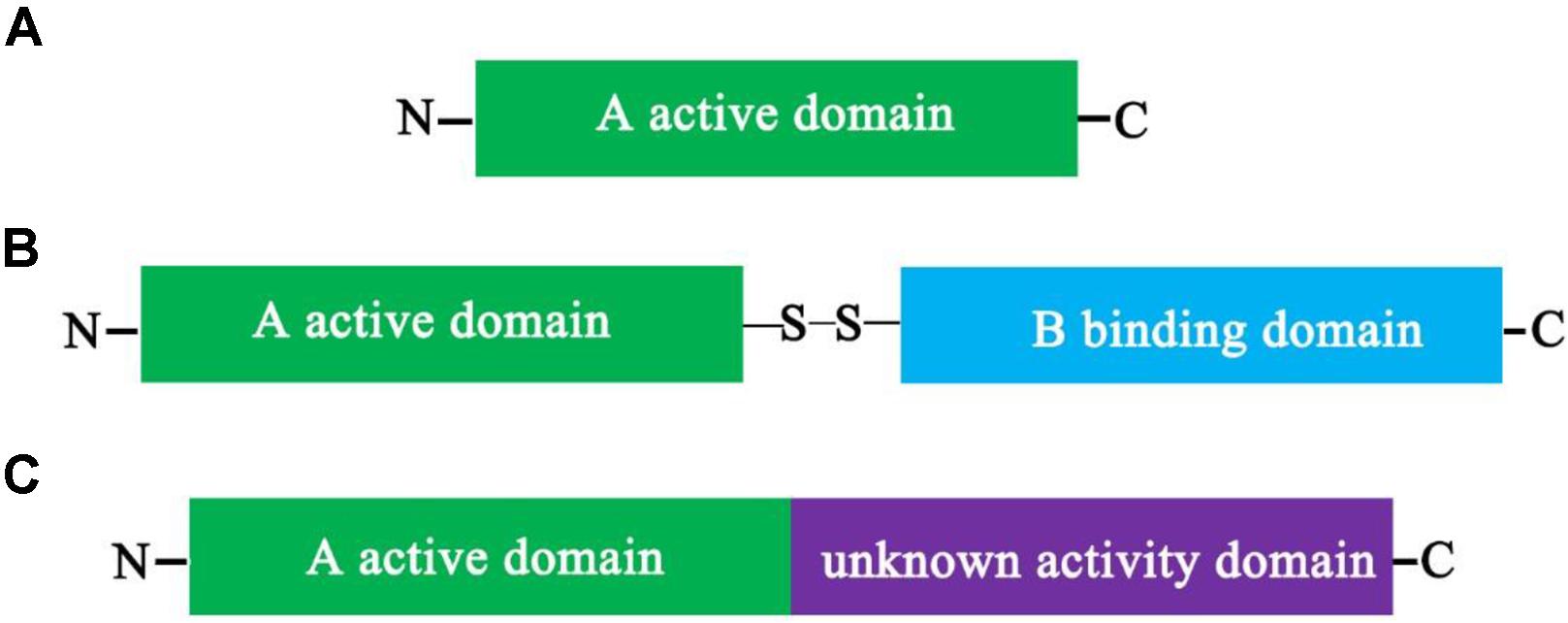
FIGURE 1. Schematic depiction of the structure of plant ribosome-inactivating proteins (RIPs). Schematic representation of the mature forms of Type I (A), Type II, (B) and Type III (C) plant RIPs. Comparison between the mature forms of a Type I (RIP) (A), such as α-MMC, composed only of a catalytically active A domain, and that of a Type II RIP (B), such as ricin, in which the active domain is linked to a lectin-binding B domain by a disulfide bond, and that of a Type III RIP (C), such as barley JIP60, in which the active domain is fused to an extra domain with an unknown function. Once the extra domain with the unknown function is removed, the processed active protein is similar in charge and enzymatic activity to type I RIPs.
In addition, there are “small type I RIPs” with a low molecular weight compared with the typical type I RIPs. These RIPs are single domain proteins with N-glycosidase activity. The N-terminal sequences of these small proteins enrich glutamine and arginine residues. Therefore, they are also considered as arginine/glutamate-rich polypeptides (AGRPs). These AGRPs manifest translation-inhibiting activity. Interestingly, so far, all low molecular weight type I RIPs are identified from Cucurbitaceae family plants. For example, luffacylin, luffangulin, and luffin P1 were isolated from the seeds of Luffa cylindrical, and the molecular weights of luffacylin, luffangulin, and luffin P1 are only 7.8, 5.6, and 5.226 kDa, respectively (Parkash et al., 2002a; Wang and Ng, 2002; Li et al., 2003b). Various low molecular weight type I RIPs were isolated and purified from Cucurbitaceae plants, such as α-benincasin and β-benincasin with molecular weights of 12 kDa, which have been isolated and purified from the seeds of Benincasa hispida (Ng et al., 2003). In addition, S-trichokirin, trichokirin S1, and trichosanthrip isolated and purified from the seeds of the Trichosanthes kirilowii were also low molecular weight type I RIPs. Their molecular weights are only 8, 11.426, and 10.964 kDa, respectively (Tai et al., 2000; Li et al., 2003a; Shu et al., 2009). In addition, Charantin and γ-momorcharin were purified from the seeds of Momordica charantia and belong to small type I RIPs, with molecular weights of 9.7 and 11.5 kDa, respectively (Pu et al., 1996; Parkash et al., 2002b).
Type II RIPs are two-domain polypeptide proteins that include an enzymatically active domain (A domain) of approximately 30 kDa, which is structurally alike to type I RIPs, and a slightly bigger binding domain (B domain) of about 35 kDa that possesses lectin properties (Figure 1B). Between the A domain and the B domain is linked by a disulfide bond (Olsnes and Pihl, 1973b; Stirpe et al., 1978). The B-lectin domain possesses sugar-binding properties. For example, it may combine to galactosyl moieties of glycoproteins and/or glycolipids that are discoverd on the surface of eukaryotic cells and mediates retrograde transport of the A-domain by the secretory pathways into the cytosol following by inhibition of protein synthesis (Sandvig and van Deurs, 1994; Steeves et al., 1999). In addition, type I RIPs have a low toxicity due to the lack of the B domain. However, type II RIPs have been broadly divided into two different of groups, nontoxic and toxic based on their cytotoxicity. For example, some type II RIPs, such as modeccin, viscumin, volkensin, abrin, and ricin are highly toxic; in contrast, others type II RIPs, such as iris lectin, cinnamomin, nigrin, and ebulin are nontoxic. The reasons for the differences in toxicities are not completely understood. Type II RIPs were only identified in plants, result in the hypothesis that the connection of RIP and lectin domains happened once in the flowering plant lineage.
Type III RIPs contain an N-terminal domain which is correlative to the A domain of RIPs and fused to an unknown functional C-terminal domain (Figure 1C). Type III RIPs, like barley JIP60 and maize b-32 are synthesized as single-domain proenzymes (Walsh et al., 1991; Chaudhry et al., 1994). RIP1 or b-32 was illustrated as two-domain type I RIP, nevertheless JIP60 was described as a true type III RIP (Nielsen and Boston, 2001; Van Damme et al., 2008). The function of the C-terminal domains (extra domains) of the type III RIP is unknown. When they are removed from the type III RIPs, the active protein is alike in charge and enzymatic activity to type I RIPs (Walsh et al., 1991; Hey et al., 1995; Krawetz and Boston, 2000). For maize, the C-terminal domains (extra domains) are unlikely protective features to prevent self-inactivation of maize ribosomes, because ribosomes from plants are resistant to active RIP and maize proRIP (Hey et al., 1995; Krawetz and Boston, 2000). These RIPs have been characterized only from barley and maize, and they are much less prevalent compared with type I or type II RIPs (Walsh et al., 1991; Bass et al., 1992; Reinbothe et al., 1994).
Different RIPs have been found in various plant species covering approximately 17 families and in bacteria, fungi and algae (Girbés et al., 2004; Stirpe, 2004). Additionally, RIP-type activity is also reported in some animal tissues (Barbieri et al., 2001). Lots of RIPs were found in a small group of plant species, such as Caryophyllaceae, Sambucaceae, Euphorbiaceae, Cucurbitaceae, Poaceae, Phytolaccaceae, and Rosaceae (Girbés et al., 2004; Domashevskiy and Goss, 2015; Shang et al., 2016). For example, a type I RIP named TCS was purified from tubers of T. kirilowii Maxim. A novel RIP, designated Trichosanthrip, was purified from mature seeds of T. kirilowii. Trichosanthrip strongly inhibited cell-free protein synthesis (Shu et al., 2009). Furthermore, RIPs have been proven to locate in various tissues, including leaves, seeds, roots, and tubers (Shu et al., 2009). Some type II RIPs termed Shiga toxins could be produced by bacteria. For example, certain strains of Escherichia coli produced type II RIPs Shiga toxin type 1 (Stx1) and Shiga toxin type 2 (Stx2), and their enzymatic activity is similar to their plant analogs (Brown et al., 1980; Sandvig and van Deurs, 2002; Bergan et al., 2012; Melton-Celsa, 2014; Russo et al., 2014). Some studies indicate that RIPs have also been found in different of fungi species, namely, Volvariella volvacea, Flammulina velutipes, Hypsizigus marmoreus, and Lyophyllum shimeji (Yao et al., 1998; Lam and Ng, 2001a,b; Wang and Ng, 2001). In addition, one RIP has been found in algae (Laminaria japonica A) (Liu et al., 2002). In addition, studies suggest that some RIP genes are present in the genome of two mosquitoes (Lapadula et al., 2013, 2017). Overall, RIPs are enzymes, which widely distribute in nature and play significant undefined biological functions.
Various RIPs have been shown some unique bioactive properties, including antibacterial (Table 1), antifungal and antiviral (Stirpe and Battelli, 2006; Shu et al., 2009). Two type I RIPs isolated from Mirabilis expansa roots were active against soilborne bacterial species at microgram levels. In bioassays, the such antibacterial activity was first demonstrated from a plant RIP (Vivanco et al., 1999). The two RIPs ME1 and ME2 exhibited antibacterial activity against Pseudomonas syringae, Agrobacterium tumefaciens and Agrobacterium radiobacter (Vivanco et al., 1999). A tobacco RIP (TRIP) was isolated and purified from Nicotiana tabacum leaves (Sharma et al., 2004). TRIP showed strong N-glycosidase activity (Sharma et al., 2004). Purified TRIP showed strong antibacterial activity against Pseudomonas solanacearum, Erwinia amylovora, Shigella asonei, Salmonella typhimurium, and Rhizobium leguminosarum (Sharma et al., 2004). TRIP at a 50 μg mL-1 concentration strongly inhibited P. solanacearum. Furthermore, a type I RIP was purified from Cucurbita moschata, and it was referred to as C. moschata RIP (Barbieri et al., 2006). And it shows antibacterial, superoxide dismutase (SOD) and antifungal activities. It inhibited the growth of two bacterial strains of E. amylovora and P. solanacearum by 70 and 50%, respectively (Barbieri et al., 2006). Interestingly, α-MMC was successfully expressed in the E. coli Rosetta (DE3) and purified by nickel–nitrilotriacetic acid affinity chromatography (Wang et al., 2012). It was active and exhibited antibacterial activity against Pseudomonas aeruginosa (Wang et al., 2012). In addition, type I RIP Balsamin isolated from Momordica balsamina exhibited broad-spectrum antibacterial activity against Staphylococcus aureus, Salmonella enterica, Staphylococcus epidermidis, and E. coli (Kaur et al., 2012; Ajji et al., 2016). A type I RIP from M. balsamina (MbRIP-1) showed significant antibacterial activity by measuring the radius of suppression from the border of paper disks (Kushwaha et al., 2012). A RIP of M. jalapa leaves at concentrations of 0.3–2.5 mg mL-1 exhibited antibacterial activity against Propionibacterium acnes and Staphylococcus epidermidis (Rumiyati et al., 2014). Ricin exhibits the highest activity to mammalian and yeast ribosomes, but presents low activity on E. coli and plant ribosomes, however, PAP depurinates ribosomes from yeasts, bacteria, plants, and animals (Barbieri et al., 1993). Furthermore, ricin is active on naked E. coli 23S rRNA, whereas is not active against the intact E. coli ribosomes (Tang et al., 2001). In a word, RIPs play significant roles in defense against bacteria (Table 1).
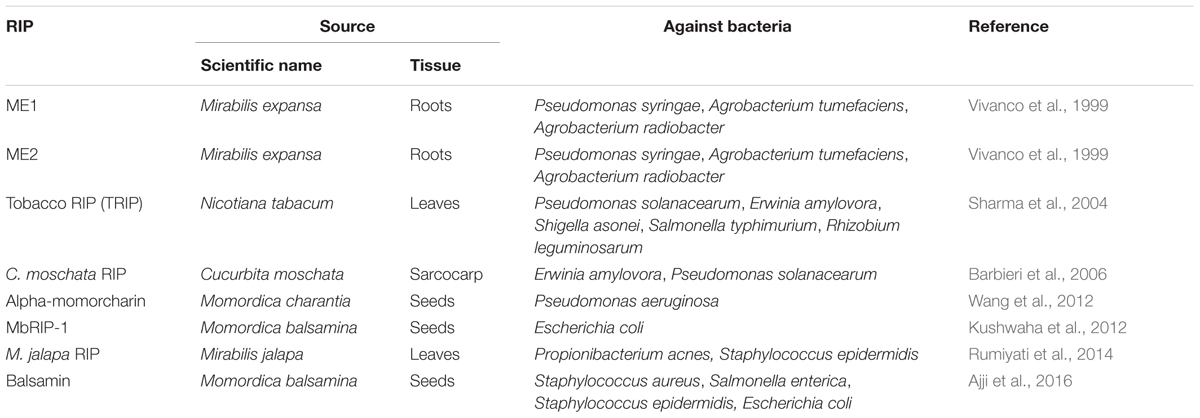
TABLE 1. The role of different source ribosome-inactivating proteins (RIPs) in defense against bacteria.
Ribosome-inactivating proteins possess extensive interest on account of their potential applications as plant defense agents against viruses and fungi. Luffacylin is a low molecular weight RIP with molecular weights of 7.8 kDa, and the N-terminal sequences of luffacylin enrich glutamate and arginine residues. Furthermore, its chromatographic behavior is alike to charantin. It demonstrates antifungal activity against Fusarium oxysporum and Mycosphaerella arachidicola in vivo (Parkash et al., 2002a). Two 11 kDa have been proteins are isolated from winter melon (Benincasa hispida) seeds (Ng et al., 2003). They exhibited antifungal activity toward Coprinus comatus and Physalospora piricola (Ng et al., 2003). The isolated TRIP was showed to inhibit several fungi pathogens, including Trichoderma reesei, Cytospora canker, Fusarium oxysporum, and Cochliobolus heterostrophus (Sharma et al., 2004). The study suggested that TRIP was most active on T. reesei among all of the fungi tested. A strong mycelial growth inhibition with 50 μg mL-1 TRIP was showed in the time course assay with T. reesei (Sharma et al., 2004). C. moschata RIP suppressed the growth of Phytophthora infestans by more than 60% (Barbieri et al., 2006). A RIP with a molecular weight of approximately 20 kDa was isolated from the seeds of the bottle melon (Lagenaria siceraria), and it exhibits N-glycosidase and antifungal activities (Wang and Ng, 2000). Two type I RIPs from M. expansa roots were found to inhibit both nonpathogenic and pathogenic fungi, Fusarium and Trichoderma species (Vivanco et al., 1999). Interestingly, the differential sensitivity was found in some cases fungal species from the same genus. For instance, Pythium irregulare was found to be sensitive; however, P. ultimum was found to be resistant. The type I barley RIP was showed to inhibit the growth of fungal strain of Trichoderma reesei on solid media (Roberts and Stewart, 1979). However, barley RIP was minimal to inhibit the growth of T. reesei in liquid media but inhibition of growth increased when chitinase was added (Leah et al., 1991). The small RIP luffacylin and Hairy melon RIP also show antifungal activity (Wong et al., 2010). Recently, recombinant DNA technology was utilized to obtain a large number of recombinant proteins (Kushwaha et al., 2012; Zhu et al., 2012). For example, the active and soluble recombinant α-MMC was obtained from the E. coli prokaryotic expression system (Wang et al., 2012). Protein activity assay suggested that α-MMC had both DNA-nuclease activity and N-glycosidase activity (Wang et al., 2012). Furthermore, the results indicated that the recombinant α-MMC showed a strong mycelial growth inhibition of the filamentous fungi including Fusarium oxysporum and Fusarium solani, with IC50 values of 4.15 and 6.23 μM, respectively (Wang et al., 2012). Further studies suggest that α-MMC isolated from seeds of Momordica charantia exhibited antifungal activity (Zhu et al., 2013). The antifungal activity of MbRIP-1 was determined using a radial growth inhibition assay (Kushwaha et al., 2012). The results indicated MbRIP-1 showed significant antifungal activity against Aspergillus niger (Kushwaha et al., 2012). Recently, RIPs from Phytolacca dioica, dioicin 2 and PD-L4 inhibited the growth of the fungus Penicillium digitatum (Iglesias et al., 2016). Novel type I RIPs isolated from oil palm (Elaeis guineensis) showed inhibition on Ganoderma boninense mycelial growth (Sargolzaei et al., 2016). Therefore, various RIPs exhibited significant antifungal activity (Table 2).
The fungal bioassay offers the advantage of testing the function of a single protein. In addition, some tests can be done with the same concentrations of the target protein equivalent to those found in plant tissues (Dowd et al., 1998). Whereas the employ of a purified protein produces an artificial condition. The actual quantity of RIP would be released from cells during fungal-plant interaction; therefore, it could only be approximated. Transgenic technologies can well solve this question by overexpression of RIPs in transgenic plants. More scientists found that overexpression of RIPs in transgenic plants exhibit enhanced resistance to pathogens attack (Table 3). For example, expressing the barley RIP in Nicotiana tabacum plants enhanced resistance against the soil-borne pathogen Rhizoctonia solani (Logemann et al., 1992; Jach et al., 1995). Co-expressing the barely transgenes β-1,3-glucanase (GLU)/chitinase (CHI) or CHI/RIP in tobacco plants increased significantly resistance against Rhizoctonia solani infection (Jach et al., 1995). Expressing the maize proRIP in tobacco plants enhanced resistance against the fungus R. solani (Maddaloni et al., 1997). A type I RIP TCS was able to inhibit rice fungus blast disease in transgenic rice (Yuan et al., 2002). A new kind of RIP (curcin 2) isolated from Jatropha curcas leaves, can be induced by different kinds of stress (Huang et al., 2008). Expression of curcin 2 in transgenic tobacco plants clearly demonstrated antifungal activity against R. solani (Huang et al., 2008). Co-expression of rice basic chitinase (RCH10) and modified maize RIP (MOD1) in rice plants enhanced resistance against R. solani (Kim et al., 2003). The barley antifungal gene chitinase and RIP were transformed into Blackgram [Vigna mungo (L.) Hepper]. The transgenic plants exhibited improved resistance to Corynespora leaf spot fungal disease (Chopra and Saini, 2014). Recently, expressing the RIP α-MMC confers enhanced resistance to rice blast fungus in rice (Qian et al., 2014). Overexpression of the PhRIP I in transgenic potato plants resulted in antifungal activity against Botrytis cinerea and R. solani (Gonzales-Salazar et al., 2017). Therefore, further work is necessary to screen the high germination rate and strong resistance to rice blast fungus in transgenic rice for applications in agriculture. From the standpoint of agricultural biotechnology applications, ectopic expression studies with transgenic plants is valuable. However, discovering that RIPs can reduce susceptibility to damage by different of pathogens does not eliminate more important roles for these proteins. To discover such roles, a comparative analysis of plants involving the alteration of RIP function by mutation, insertional inactivation, or co-suppression will be carried out.
It is believed that the antifungal proteins, namely, RIP, thaumatin-like proteins, chitinases, proteinase inhibitors, endoproteinases, plant defensins, glucanases, peroxidases, and immunophilin, utilize protective activity against fungal and bacterial invasions (Park et al., 2009). Antifungal proteins can disrupt the cell wall structure or function and suppress the synthesis of the fungal cell wall (Morais et al., 2010). Some antifungal proteins can interact with the potential fungal intracellular targets and plasma membrane, leading to changes in the cell death or membrane potential (Morais et al., 2010). Wang et al. (2012) investigated the antifungal activity and mechanism of α-MMC toward F. solani. The results of the DAPI and PI uptakes assay indicated that the apoptotic responses can be induced and the permeability of fungal membranes can be changed by α-MMC (Wang et al., 2012). The ultrastructural changes, such as cells with irregular budding sites, other aspect in the cytoplasm, the loss of integrity and rigidity of cell walls suggested that the antifungal mechanism of α-MMC was a complex process (Wang et al., 2012). Recently, the antifungal activity of the RIP BE27 isolated from sugar beet was investigated against the green mold Penicillium digitatum (Citores et al., 2016). They found that RIP BE27 can inhibit the growth of the fungus Penicillium digitatum because that it can enter the fungal cytoplasm and kill fungal cells (Citores et al., 2016).
The existence of antiviral activities of RIPs are known for about 75 years. RIPs emerged as potent antiviral agents against lots of animal, plant and human viruses. This review summarizes work related to the role of RIPs in defense against plant viruses (Tables 4, 5). PAP is a RIP that is purified from the extracts of plant leaves of pokeweed (Obrig et al., 1973). It was first shown to reduce the infectivity of TMV by inhibiting protein synthesis (Duggar and Armstrong, 1925; Dallal and Irvin, 1978). Further studies suggested that the exogenous application of PAP appears to enhance the systemic resistance of N. benthamiana to TMV infection (Zhu et al., 2016). Application of PAP I also causes concentration-dependent inhibition of zucchini yellow mosaic virus (ZYMV) infection on squash plants (Sipahioğlu et al., 2017). Dianthins isolated from the leaves of Dianthus caryophyllus mixed with TMV strongly decreased the amount of local lesions on plant leaves of Nicotiana glutinosa (Stirpe et al., 1981). The MAP isolated from M. jalapa showed highly potent activity against the mechanical transmission of viruses, including cucumber mosaic virus (CMV), TMV, potato Y virus, cucumber green mottle mosaic virus and turnip mosaic potyvirus (TuMV) (Kubo et al., 1990). The infection of artichoke mottled crinkle virus (AMCV) can be inhibited by the new single-chain RIPs from the seeds of Basella rubra and from the leaves of Bougainvillea spectabilis (Bolognesi et al., 1997). In addition, the translation progress of pokeweed mosaic virus (PMV) and brome mosaic virus (BMV) can be inhibited by an antiviral protein isolated from plant leaves of Celosia cristata (Baranwal et al., 2002). A 27 kDa RIP purified from Amaranthus tricolor leaves exhibited antiviral activity against the sunn-hemp rosette virus (SRV) (Roy et al., 2006). Choudhary N. et al. (2008) purified a recombinant RIP named BBAP1, which showed N-glycosidase activity, and emerged strong resistance against TMV. A RIP from Bougainvillea xbuttiana leaves was also successfully expressed in E. coli. And it demonstrated a high level of resistance against SRV (Choudhary N.L. et al., 2008). In addition, the activity of RIPs against capped and uncapped viral RNAs were examined. Vivanco and Tumer (2003) found that PAP, ME1, and saporin can depurinate the capped TMV and BMV RNAs. However, the uncapped luciferase RNA are not depurinated by them. Taken together, various RIPs purified from different plant species exhibited significant antiviral activity (Table 4).
Biotechnological methods have been used to show a significant role of RIPs in plant defense against viral infections (Table 5). Different RIPs, including PAP and TCS have been successfully overexpressed in different of transgenic plant species, leading to improved resistance against multiple plant viruses (Lodge et al., 1993; Taylor et al., 1994; Lam et al., 1996; Moon et al., 1997). A gene encoding a RIP, named P. insularis antiviral protein 2 (PIP2) was isolated from P. insularis (Song et al., 2000). They also found that the expression of the PIP2 gene is enhanced in leaves after treatment with phytohormones, such as abscisic acid (ABA) and jasmonic acid (JA). Furthermore, the PIP2 protein exhibited antiviral activity against TMV. The results indicate that the antiviral activity of PAP was not due to the depurination of host ribosomes. A mutant PAP, PAPn, did not bind ribosomes efficiently. PAPn did not depurinate ribosomes and was non-toxic when expressed in transgenic tobacco plants (Zoubenko et al., 2000). However, overexpression of PAPn in tobacco plants increased resistance to the potato virus X (PVX) infection (Zoubenko et al., 2000). They presented evidence that a novel SA-independent, stress-associated signal transduction pathway activated in PAPn-expressing plants play a significant role in pathogen resistance against virus infection (Zoubenko et al., 2000). Transgenic plants expressing the C-terminal deletion mutant PAP were resistant to PVX infection. However, the depurination of rRNA was not found in these plants (Tumer et al., 1997). In addition, Hudak et al. (2000) showed that PAP inhibited the translation of BMV and PVX RNAs without depurinating ribosomes. Overall, overexpression of PAP and non-toxic PAP mutants increased broad-spectrum resistance to viral infection in transgenic plants. Di and Tumer (2015) have summarized the mechanism of PAP-induced disease resistance. Ruan et al. (2007) suggested that overexpressing Cassin, a new gene of RIP isolated from Cassia occidentalis in tobacco plants exhibited different levels of resistance to TMV. The expression of curcin 2 in transgenic tobacco plants clearly demonstrated antiviral activity against TMV (Huang et al., 2008). The expression of a type I RIP TCS enhanced resistance to CMV and TMV infection (Krishna et al., 2002). The symptoms of systemic infection were weakened and delayed in the transgenic plants overexpressing TCS. Most of the evidence suggest that overexpression of the type I RIPs increases the plant’s resistance against various plant viruses in transgenic plants. In order to confirm whether overexpression of type II RIPs also enhance the plant’s resistance against viruses infection, Chen et al. (2002) transformed the Sambucus nigra type II RIP SNA-I’ into tobacco plants. Overexpression of SNA-I’ strongly enhanced the plant’s resistance against TMV infection in transgenic tobacco plants (Chen et al., 2002). These results suggest that type II RIPs are also involved in plant defense response against viral infection.
A cDNA clone, encoding RIP was isolated from the post-flowering stage of C. cristata leaves (Begam et al., 2006). The recombinant protein was successfully expressed in E. coli and showed antiviral activity against TMV and SRV (Begam et al., 2006). Similarly, a full-length cDNA encoding RIP from the leaves of B. xbuttiana was isolated, and this 35.5 kDa RIP was also successfully expressed in E. coli (Choudhary N.L. et al., 2008). The 35.5-kDa RIP can inhibit the local lesion formation of SRV infecting Cyamopsis tetragonoloba leaves, which indicates that it exhibited antiviral activity toward SRV (Choudhary N.L. et al., 2008). In addition, the antiviral potential of RIPs for plant viruses and its potential function in systemic resistance were also studied. The Clerodendrum aculeatum-systemic resistance inducing (CA-SRI) protein, which has RIP activity, plays a key role in inducing strong systemic resistance in susceptible plants against various plant viruses (Verma et al., 1996; Kumar et al., 1997). Further studies indicate that CAP-34, a protein from C. aculeatum induced systemic antiviral resistance against the papaya ringspot virus (PRSV) infection in Carica papaya (Srivastava et al., 2009). Likewise, the CIP-29 protein isolated from Clerodendrum inerme exhibited ribosome-inactivating properties. Compared with other known RIPs, CIP-29 can induce systemic resistance against virus infection in susceptible plants (Prasad et al., 1995; Olivieri et al., 1996). Further study suggest that application of CIP-29 in C. tetragonoloba plants can induce two virus inhibitory proteins, named CT-VIA-32 and CT-VIA-62 (Prasad et al., 2014). These two virus inhibitory proteins can resist virus infection. Systemic resistance in N. glutinosa and Cyamopsis tetragonoloba was induced by an antiviral protein from B. xbuttiana leaves against TMV and SRV (Narwal et al., 2001). Iglesias et al. (2005) found that sugar beet RIPs named beetins were systemically induced by AMCV infection. BDP-30, a glycoprotein isolated from Boerhavia diffusa displays homology with RIPs. It also induced systemic resistance in tobacco against TMV (Srivastava et al., 2015). Zhu et al. (2013) demonstrated that α-MMC had broad-spectrum antiviral activity against phytopathogenic viruses, including ChiVMV, CMV, TMV, and TuMV. The results demonstrated that α-MMC inhibited the accumulation of viral RNA and protein. Furthermore, application of α-MMC can increase the expression of NPR1, PR1, and PR2 in tobacco plants, which strongly indicates that the plant systemic resistance can be activated by α-MMC against multiple viruses (Zhu et al., 2013). Furthermore, application of α-MMC in M. charantia led to a significant increase of JA indicated that the anti-viral activities of α-MMC in M. charantia may be accomplished through the JA related signaling pathway (Yang et al., 2016). Therefore, it may be a candidate for the development of virus resistant crop plants, for instance cotton, tobacco, rice, tea, and clover. Although RIPs exhibited antiviral activity against various plant viruses, viruses also have evolved a viral strategy to evade host cell defenses and possible anti-cytotoxic activity against RIPs. The interactions between the turnip mosaic virus genome linked protein (VPg) and PAP was investigated (Domashevskiy et al., 2012). PAP interacts strongly with VPg in a mixed type competition. These findings correlate with the inhibition of PAP enzymatic activity by VPg in wheat germ lysate. Furthermore, depurination inhibition by VPg also indicates the use of VPg against the cytotoxic activity of RIPs and the inhibition of their biological potency (Domashevskiy et al., 2012).
A large number of type I and type II RIPs have been shown to have antiviral effect; however, the antiviral mechanism of RIPs is still not completely clear (Wang and Tumer, 2000). The antiviral effect of RIPs is supposed on the basis of their enzymatic activity and selective compartmentalization (Roberts and Lord, 1981; Ready et al., 1986; Carzaniga et al., 1994; Tarantini et al., 2010). RIPs may work on ribosomes of infected plant cells, thereby inhibiting the synthesis of viral protein (Parikh and Tumer, 2004). When plants are infected by plant viruses, RIPs can be released rapidly from their intracellular compartments. Therefore, viral replication can be prevented at an early stage by inhibiting the cell protein synthesis and leading to autonomous cell death (Wang and Tumer, 2000; Nielsen and Boston, 2001). However this antiviral mechanism of RIPs may have a direct interaction of RIP with nucleic acid of viruses. Vandenbussche et al. (2004) considered that RIP may have a direct effect on the nucleic acid of viruses. PAP-I, PAP-II, and PAP-III can cause a concentration-dependent depurination of genomic TMV RNA (Chen et al., 1993), BMV (Picard et al., 2005), and tobacco etch virus (TEV) RNA (Domashevskiy et al., 2012). The antiviral mechanism of RIPs may change among different RIPs and different viruses.
RIPs can enhance plant resistance to insect pests (Stirpe, 2013) (Table 6). Lots of studies have suggested that RIPs possess the insecticidal activity upon different insects such as Lepidoptera (Dowd et al., 1998, 2003, 2006; Zhou et al., 2000; Wei et al., 2004), Coleoptera (Gatehouse et al., 1990; Kumar et al., 1993) and Diptera (Shahidi-Noghabi et al., 2008). The artificial diets experiments were used to study the insecticidal activity of RIPs. For example, an artificial diet supplemented with different concentrations of a type-II RIP from Sambucus nigra, decreased the fecundity and survival of Acyrthosiphon pisum (Shahidi-Noghabi et al., 2008). Furthermore, the feeding of Myzus nicotianae with leaves from transgenic tobacco plants overexpressing SNA-I retarded the development and decreased the fecundity and adult survival (Shahidi-Noghabi et al., 2008). In addition, an artificial diet supplemented with different type-I RIPs reduced the fecundity and survival of Anticarsia gemmatalis Hübner and Spodoptera frugiperda (Bertholdo-Vargas et al., 2009). Recent studies have suggested that type-I and type-II RIPs from apple (Malus domestica Borkh) have strong aphicidal activity (Hamshou et al., 2016). The feeding of pea aphids (Myzus nicotianae Blackman) on an artificial diet supplemented with the purified recombinant proteins for type-I RIPs and type-II RIPs from apple reduced the nymphal survival of aphid (Hamshou et al., 2016). The studies also indicated that the feeding of green peach aphids (Myzus persicae Sulzer) on leaves of different transgenic tobacco plants lines overexpressing type-I or type-II RIPs significantly reduced nymphal survival of this important hemipteran pest. In addition, Overexpression of a maize RIP in tobacco plants enhanced resistance against Helicoverpa zea (Dowd et al., 2003). Furthermore, Maize leaves expressing higher levels of maize ribosome-inactivating protein (MRIP) and wheat germ agglutinin (WGA) were more resistant to feeding by Spodoptera frugiperda and corn earworms (Helicoverpa zea) (Dowd et al., 2012). Overexpression of type 1 or type 2 RIPs from apple in tobacco plants enhanced resistance against agronomically important insect pest, Spodoptera exigua (Hamshou et al., 2017). The mechanism of insecticidal activity of RIPs is still not completely clear. Several studies suggested that apoptosis can be induced by RIPs (Narayanan et al., 2005; Sikriwal and Batra, 2010; Das et al., 2012). The feeding of A. pisum on an artificial diet supplemented with SNA-I induced apoptosis in the midgut of A. pisum through the activation of caspase-3 (Shahidi-Noghabi et al., 2010).
As shown in Figure 2, the proposed model for the role of RIPs in defense against pathogens showed that various RIPs have shown unique bioactive properties, including antibacterial, antifungal, antiviral, and insecticidal activity (Stirpe and Battelli, 2006; Shu et al., 2009; Akkouh et al., 2015). A large number of studies suggest that the expression of RIPs could be activated by some factors, such as viral infection, phytohormones, senescence, development, and environmental stress (Reinbothe et al., 1994; Girbés et al., 1996; Stirpe et al., 1996; Rippmann et al., 1997; Wang et al., 2016). Expressing α-MMC by transgenic technologies in rice confers enhanced resistance to rice blast fungus (Qian et al., 2014). Future research employing transgenic technology approaches to study the mechanisms of RIPs will undoubtedly lead to a better comprehending of the effect of plant RIPs in defense against pathogens and insects (Figure 2). A deeper insight into antiviral mechanisms should also be carried out (Figure 2). Phytohormones salicylic acid (SA), JA, and ethylene (ET) have been proved to participate in the systemic resistance responses of plants against pathogen infections (van Wees et al., 2000). These phytohormones signaling pathways could impact each other via a complex signaling network (Koornneef and Pieterse, 2008). Therefore, how RIPs participate in the systemic resistance responses of plants or whether RIPs can regulate phytohormone signaling pathways (SA, JA, and ET) against pathogen infections needs further investigation (Figure 2). Previous studies suggest that PAP may enhance plant systemic resistance against TMV infection by regulation the reactive oxygen species (ROS) levels (Zhu et al., 2016). As shown in Figure 2, the proposed model for the role of RIPs in defense against pathogens and insects indicated that discovering additional crosstalk mechanisms between RIPs and phytohormones or ROS against pathogen and insect infections will be a significant subject in the field of biotic stress study.
FZ wrote the manuscript. Y-KZ, Z-LJ, and X-RC collected the data and the references. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Prof. Wong Sek Man (Department of Biological Sciences, National University of Singapore) for review of this manuscript. This work was supported by the National Natural Science Foundation of China (31500209 and 31671971), Natural Science Foundation of the Higher Education Institutions of Jiangsu Province of China (15KJB210007), Natural Science Foundation of Yangzhou City (China) (YZ2016121), and earmarked fund for Modern Agro-industry Technology Research System (CARS-31-2-02).
Ajji, P. K., Walder, K., and Puri, M. (2016). Functional analysis of a type-I ribosome inactivating protein balsamin from Momordica balsamina with anti-microbial and DNase activity. Plant Foods Hum. Nutr. 71, 265–271. doi: 10.1007/s11130-016-0555-4
Akkouh, O., Ng, T. B., Cheung, R. C., Wong, J. H., Pan, W., Ng, C. C., et al. (2015). Biological activities of ribosome-inactivating proteins and their possible applications as antimicrobial, anticancer, and anti-pest agents and in neuroscience research. Appl. Microbiol. Biotechnol. 99, 9847–9863. doi: 10.1007/s00253-015-6941-2
Baranwal, V. K., Tumer, N. E., and Kapoor, H. C. (2002). Depurination of ribosomal RNA and inhibition of viral RNA translation by an antiviral protein of Celosia cristata. Indian J. Exp. Biol. 40, 1195–1197.
Barbieri, L., Battelli, M. G., and Stripe, F. (1993). Ribosome inactivating protein from plants. Biochim. Biophys. Acta 1154, 237–282. doi: 10.1016/0304-4157(93)90002-6
Barbieri, L., Polito, L., Bolognesi, A., Ciani, M., Pelosi, E., Farini, V., et al. (2006). Ribosome-inactivating proteins in edible plants and purification and characterization of a new ribosome-inactivating protein from Cucurbita moschata. Biochim. Biophys. Acta 1760, 783–792. doi: 10.1016/j.bbagen.2006.01.002
Barbieri, L., Valbonesi, P., Bondioli, M., Alvarez, M. L., Dal-Monte, P., Landini, M. P., et al. (2001). Adenine glycosylase activity in mammalian tissues: an equivalent of ribosome-inactivating proteins. FEBS Lett. 505, 196–197. doi: 10.1016/S0014-5793(01)02789-2
Bass, H. W., Webster, C., Gr, O. B., Roberts, J. K., and Boston, R. S. (1992). A maize ribosome-inactivating protein is controlled by the transcriptional activator opaque-2. Plant Cell 4, 225–234. doi: 10.1105/tpc.4.2.225
Begam, M., Kumar, S., Roy, S., Campanella, J. J., and Kapoor, H. C. (2006). Molecular cloning and functional identification of a ribosome-inactivating/antiviral protein from leaves of post-flowering stage of Celosia cristata and its expression in E. coli. Phytochemistry 67, 2441–2449. doi: 10.1016/j.phytochem.2006.08.015
Bergan, J., Dyve Lingelem, A. B., Simm, R., Skotland, T., and Sandvig, K. (2012). Shiga toxins. Toxicon 60, 1085–1107. doi: 10.1016/j.toxicon.2012.07.016
Bertholdo-Vargas, L. R., Martins, J. N., Bordin, D., Salvador, M., Schafer, A. E., Barros, N. M., et al. (2009). Type 1 ribosome inactivating proteins entomotoxic, oxidative and genotoxic action on Anticarsia gemmatalis (Hübner) and Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). J. Insect Physiol. 55, 51–58. doi: 10.1016/j.jinsphys.2008.10.004
Bian, X. X., Shen, F. B., Chen, Y. W., Wang, B. N., Deng, M., and Meng, Y. F. (2010). PEGylation of alpha-momorcharin: synthesis and characterization of novel anti-tumor conjugates with therapeutic potential. Biotechnol. Lett. 32, 883–890. doi: 10.1007/s10529-010-0242-8
Bolognesi, A., Bortolotti, M., Maiello, S., Battelli, M. G., and Polito, L. (2016). Ribosome-inactivating proteins from plants: a historical overview. Molecules 21:1627. doi: 10.3390/molecules21121627
Bolognesi, A., Polito, L., Olivieri, F., Valbonesi, P., Barbieri, L., Battelli, M. G., et al. (1997). New ribosome-inactivating proteins with polynucleotide: adenosine glycosidase and antiviral activities from Basella rubra L. and Bougainvillea spectabilis Willd. Planta 203, 422–429. doi: 10.1007/s004250050209
Brown, J. E., Ussery, M. A., Leppla, S. H., and Rothman, S. W. (1980). Inhibition of protein synthesis by shiga toxin: activation of the toxin and inhibition of peptide elongation. FEBS Lett. 117, 84–88. doi: 10.1016/0014-5793(80)80918-5
Calixto, J. B. (2000). Efficacy safety quality control marketing and regulatory guidelines for herbal medicines (phytotherapeutic agents). Braz. J. Med. Biol. Res. 33, 179–189. doi: 10.1590/S0100-879X2000000200004
Carzaniga, R., Sinclair, L., Fordham-Skeleton, A. P., Harris, N., and Croy, R. R. D. (1994). Cellular and subcellular distribution of saporins, type I ribosome-inactivating proteins, in soapwort (Saponaria officinalis L.). Planta 194, 461–470. doi: 10.1007/BF00714457
Chaudhry, B., Mueller, U. F., Cameron-Mills, V., Gough, S., and Simpson, D. (1994). The barley 60 kDa jasmonate-induced protein (JIP60) is a novel ribosome inactivating protein. Plant J. 6, 815–824. doi: 10.1046/j.1365-313X.1994.6060815.x
Chen, Y., Peumans, W. J., and Van-Damme, E. J. M. (2002). The Sambucus nigra type-2 ribosome-inactivating protein SNA-1 exhibits in planta antiviral activity in transgenic tobacco. FEBS Lett. 516, 27–30. doi: 10.1016/S0014-5793(02)02455-9
Chen, Z., Antoniw, J. F., and White, R. F. (1993). A possible mechanism for the antiviral activity of pokeweed antiviral protein. Physiol. Mol. Plant Pathol. 42, 249–258. doi: 10.1006/pmpp.1993.1023
Chopra, R., and Saini, R. (2014). Transformation of blackgram (Vigna mungo (L.) Hepper) by barley chitinase and ribosome-inactivating protein genes towards improving resistance to corynespora leaf spot fungal disease. Appl. Microbiol. Biotechnol. 174, 2791–2800. doi: 10.1007/s12010-014-1226-2
Choudhary, N., Kapoor, H. C., and Lodha, M. L. (2008). Cloning and expression of antiviral/ribosome-inactivating protein from Bougainvillea xbuttiana. J. Biosci. 33, 91–101. doi: 10.1007/s12038-008-0025-8
Choudhary, N. L., Yadav, O. P., and Lodha, M. L. (2008). Ribonuclease, deoxyribonuclease, and antiviral activity of Escherichia coli-expressed Bougainvillea xbuttiana antiviral protein. Biochemistry 73, 273–277. doi: 10.1134/S000629790803005X
Citores, L., Iglesias, R., Gay, C., and Ferreras, J. M. (2016). Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet (Beta vulgaris L.) against the green mould Penicillium digitatum. Mol. Plant Pathol. 17, 261–271. doi: 10.1111/mpp.12278
Culbreath, A. K., Todd, J. W., and Brown, S. L. (2003). Epidemiology and management of tomato spotted wilt in peanut. Annu. Rev. Phytopathol. 41, 53–75. doi: 10.1146/annurev.phyto.41.052002.095522
Dallal, J. A., and Irvin, J. D. (1978). Enzymatic inactivation of eukaryotic ribosomes by the pokeweed antiviral protein. FEBS Lett. 89, 257–259. doi: 10.1016/0014-5793(78)80230-0
Das, M. K., Sharma, R. S., and Mishra, V. (2012). Induction of apoptosis by ribosome inactivating proteins. Importance of N-glycosidase activity. Appl. Biochem. Biotechnol. 166, 1552–1561. doi: 10.1007/s12010-012-9550-x
de Virgilio, M., Lombardi, A., Caliandro, R., and Fabbrini, M. S. (2010). Ribosome inactivating proteins: from plant defense to tumor attack. Toxins 2, 2699–2737. doi: 10.3390/toxins2112699
Di, R., and Tumer, N. E. (2015). Pokeweed antiviral protein: its cytotoxicity mechanisms and applications in plant disease resistance. Toxins 7, 755–772. doi: 10.3390/toxins7030755
Domashevskiy, A. V., and Goss, D. J. (2015). Pokeweed antiviral protein, a ribosome inactivating protein: activity, inhibition and prospects. Toxins 7, 274–298. doi: 10.3390/toxins7020274
Domashevskiy, A. V., Miyoshi, H., and Goss, D. J. (2012). Inhibition of pokeweed antiviral protein (PAP) by turnip mosaic virus genome-linked protein (VPg). J. Biol. Chem. 287, 29729–29738. doi: 10.1074/jbc.M112.367581
Dowd, P. F., Johnson, E. T., and Price, N. P. (2012). Enhanced pest resistance of maize leaves expressing monocot crop plant-derived ribosome-inactivating protein and agglutinin. J. Agric. Food Chem. 60, 10768–10775. doi: 10.1021/jf3041337
Dowd, P. F., Mehta, A. D., and Boston, R. S. (1998). Relative toxicity of the maize endosperm ribosome-inactivating protein to insects. J. Agric. Food Chem. 46, 3775–3779. doi: 10.1021/jf980334w
Dowd, P. F., Rober, A. H., Pinkerton, T. S., Johnson, E. T., Lagrimini, L. M., and Boston, R. S. (2006). Relative activity of a tobacco hybrid expressing high levels of a tobacco anionic peroxidase and maize ribosome-inactivating protein against Helicoverpa zea and Lasioderma serricorne. J. Agric. Food Chem. 54, 2629–2634. doi: 10.1021/jf058180p
Dowd, P. F., Zuo, W. N., Gillikin, J. W., Johnson, E. T., and Boston, R. S. (2003). Enhanced resistance to Helicoverpa zea in tobacco expressing an activated form of maize ribosome-inactivating protein. J. Agric. Food Chem. 51, 3568–3574. doi: 10.1021/jf0211433
Duggar, B. M., and Armstrong, J. K. (1925). The effect of treating virus of tobacco mosaic with juice of various plants. Ann. Mol. Bot. Gard. 12, 359–365. doi: 10.2307/2394061
Gatehouse, A. M. R., Barbieri, L., Stirpe, F., and Croy, R. R. D. (1990). Effects of ribosome inactivating proteins on insect development differences between Lepidoptera and Coleoptera. Entomol. Exp. Appl. 54, 43–51. doi: 10.1111/j.1570-7458.1990.tb01310.x
Girbés, T., de-Torre, C., Iglesias, R., Ferreras, J. M., and Mendéz, E. (1996). RIP for viruses. Nature 379, 777–778. doi: 10.1038/379777b0
Girbés, T., Ferreras, J. M., Arias, F. J., and Stirpe, F. (2004). Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria. Mini Rev. Med. Chem. 4, 461–476. doi: 10.2174/1389557043403891
Gonzales-Salazar, R., Cecere, B., Ruocco, M., Rao, R., and Corrado, G. (2017). A comparison between constitutive and inducible transgenic expression of the PhRIP I gene for broad-spectrum resistance against phytopathogens in potato. Biotechnol. Lett. 39, 1049–1058. doi: 10.1007/s10529-017-2335-0
Hamshou, M., Shang, C., De Zaeytijd, J., Van Damme, E. J. M., and Smagghe, G. (2017). Expression of ribosome-inactivating proteins from apple in tobacco plants results in enhanced resistance to Spodoptera exigua. J. Asia Pac. Entomol. 20, 1–5. doi: 10.1016/j.aspen.2016.09.009
Hamshou, M., Shang, C., Smagghe, G., and Van-Damme, E. J. M. (2016). Ribosome-inactivating proteins from apple have strong aphicidal activity in artificial diet and in planta. Crop Prot. 87, 19–24. doi: 10.1016/j.cropro.2016.04.013
Hartley, M. R., and Lord, J. M. (2004a). Cytotoxic ribosome-inactivating lectins from plants. Biochim. Biophys. Acta 1701, 1–14. doi: 10.1016/j.bbapap.2004.06.004
Hartley, M. R., and Lord, J. M. (2004b). Genetics of ribosome-inactivating proteins. Mini Rev. Med. Chem. 4, 487–492.
Hey, T. D., Hartley, M., and Walsh, T. A. (1995). Maize ribosome-inactivating protein (b-32). Homologs in related species, effects on maize ribosomes, and modulation of activity by pro-peptide deletions. Plant Physiol. 107, 1323–1332. doi: 10.1104/pp.107.4.1323
Huang, M. X., Hou, P., Wei, Q., Xu, Y., and Chen, F. (2008). A ribosome-inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regul. 54, 115–123. doi: 10.1007/s10725-007-9234-7
Hudak, K. A., Wang, P., and Tumer, N. E. (2000). A novel mechanism for inhibition of translation by pokeweed antiviral protein: depurination of the capped RNA template. RNA 6, 369–380. doi: 10.1017/S1355838200991337
Husain, J., Tickle, I. J., and Wood, S. P. (1994). Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia. FEBS Lett. 342, 154–158. doi: 10.1016/0014-5793(94)80491-5
Iglesias, R., Citores, L., Ragucci, S., Russo, R., Di Maro, A., and Ferreras, J. M. (2016). Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L. Biochim. Biophys. Acta 1860, 1256–1264. doi: 10.1016/j.bbagen.2016.03.011
Iglesias, R., Péreze, Y., de-Torre, C., Ferreras, J. M., Antolßn, P., Jiménez, P., et al. (2005). Molecular characterization and systemic induction of single-chain ribosome-inactivating protein (RIPs) in sugar beet (Beta vulgaris) leaves. J. Exp. Bot. 56, 1675–1684. doi: 10.1093/jxb/eri164
Jach, G., Görnhardt, B., Mundy, J., Logemann, J., Pinsdorf, E., Leah, R., et al. (1995). Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 8, 97–109. doi: 10.1046/j.1365-313X.1995.08010097.x
Kaur, I., Yadav, S. K., Hariprasad, G., Gupta, R. C., Srinivasan, A., Batra, J. K., et al. (2012). Balsamin, a novel ribosome-inactivating protein from the seeds of Balsam apple Momordica balsamina. Amino Acids 43, 973–981. doi: 10.1007/s00726-011-1162-1
Kim, J. K., Jang, I. C., Wu, R., Zuo, W. N., Boston, R. S., Lee, Y. H., et al. (2003). Co-expression of a modified maize ribosome-inactivating protein and a rice basic chitinase gene in transgenic rice plants confers enhanced resistance to sheath blight. Transgenic Res. 12, 475–484. doi: 10.1023/A:1024276127001
Koornneef, A., and Pieterse, C. M. J. (2008). Cross talk in defense signaling. Plant Physiol. 146, 839–844. doi: 10.1104/pp.107.112029
Krawetz, J. E., and Boston, R. S. (2000). Substrate specificity of a maize ribosome-inactivating protein differs across diverse taxa. Eur. J. Biochem. 267, 1966–1974. doi: 10.1046/j.1432-1327.2000.01200.x
Krishna, R., McDonald, K. A., Dandekar, A. M., Jackman, A. P., and Falk, B. (2002). Expression of recombinant trichosanthin, a ribosome-inactivating protein, in transgenic tobacco. J. Biotechnol. 97, 69–88. doi: 10.1016/S0168-1656(02)00058-5
Kubo, S., Ikeda, T., Imaizumi, S., Takanami, Y., and Mikami, Y. (1990). A potent plant virus inhibitor found in Mirabilis jalapa L. Ann. Phytopathol. Soc. Japan 56, 481–487. doi: 10.3186/jjphytopath.56.481
Kumar, D., Verma, H. N., Tuteja, N., and Tewari, K. K. (1997). Cloning and characterization of a gene encoding an antiviral protein from Clerodendrum aculeatum L. Plant Mol. Biol. 33, 745–751. doi: 10.1023/A:1005716103632
Kumar, M. A., Timm, D. E., Neet, K. E., Owen, W. G., Peumans, W. J., and Rao, A. G. (1993). Characterization of the lectin from the bulbs of Eranthis hyemalis (winter aconite) as an inhibitor of protein synthesis. J. Biol. Chem. 268, 25176–25183.
Kushwaha, G. S., Pandey, N., Sinha, M., Singh, S. B., Kaur, P., Sharma, S., et al. (2012). Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states. Biochim. Biophys. Acta 1824, 679–691. doi: 10.1016/j.bbapap.2012.02.005
Lam, S. K., and Ng, T. B. (2001a). First simultaneous isolation of a ribosome inactivating protein and an antifungal protein from a mushroom (Lyophyllum shimeji) together with evidence for synergism of their antifungal effects. Arch. Biochem. Biophys. 393, 271–280.
Lam, S. K., and Ng, T. B. (2001b). Hypsin, a novel thermostable ribosome-inactivating protein with antifungal and antiproliferative activities from fruiting bodies of the edible mushroom Hypsizigus marmoreus. Biochem. Biophys. Res. Commun. 285, 1071–1075.
Lam, Y. H., Wong, Y. S., Wang, B., Wong, R. N. S., Yeung, H. M., Shaw, P. C., et al. (1996). Use of trichosanthin to reduce infection by turnip mosaic virus. Plant Sci. 114, 111–117. doi: 10.1016/0168-9452(95)04310-1
Lapadula, W. J., Marcet, P. L., Mascotti, M. L., Sanchez-Puerta, M. V., and Juri Ayub, M. (2017). Metazoan ribosome inactivating protein encoding genes acquired by horizontal gene transfer. Sci. Rep. 7:1863. doi: 10.1038/s41598-017-01859-1
Lapadula, W. J., Sánchez-Puerta, M. V., and Juri-Ayub, M. (2013). Revising the taxonomic distribution, origin and evolution of ribosome inactivating protein genes. PLOS ONE 8:e72825. doi: 10.1371/journal.pone.0072825
Leah, R., Tommerup, H., Svendsen, I., and Mundy, J. (1991). Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 266, 1564–1573.
Li, F., Yang, X. X., Hu, W. G., Xia, H. C., Li, Z., and Zhang, Z. C. (2003a). Purification and characterization of trichokirin-s1, a novel ribosome-inactivating peptide from seeds of Trichosanthes kirilowii. Acta Biochem. Biophys. Sin. 35, 841–846.
Li, F., Yang, X. X., Xia, H. C., Zeng, R., Hu, W. G., Li, Z., et al. (2003b). Purification and characterization of Luffin p1, a ribosome-inactivating peptide from the seeds of Luffa cylindrica. Peptides 24, 799–805.
Liu, R. S., Yang, J. H., and Liu, W. Y. (2002). Isolation and enzymatic characterization of lamjapin, the first ribosome-inactivating protein from cryptogamic algal plant (Laminaria japonica A). Eur. J. Biochem. 269, 4746–4752. doi: 10.1046/j.1432-1033.2002.03165.x
Lodge, J. K., Kaniewski, W. K., and Tumer, N. E. (1993). Broad-spectrum virus resistance in transgenic plants expressing pokeweed antiviral protein. Proc. Natl. Acad. Sci. U.S.A. 90, 7089–7093. doi: 10.1073/pnas.90.15.7089
Logemann, J., Jach, G., Tommerup, H., Mundy, J., and Schell, J. (1992). Expression of a barley ribosome-inactivating protein leads to increased fungal protection in transgenic tobacco plants. Nat. Biotechnol. 10, 305–308. doi: 10.1038/nbt0392-305
Lord, J. M., and Hartley, M. R. (eds). (2010). Toxic Plant Proteins: Plant Cell Monographs, Vol. 18. Berlin: Springer.
Lord, J. M., Roberts, L. M., and Robertus, J. D. (1994). Ricin: structure, mode of action, and some current applications. FASEB J. 8, 201–208. doi: 10.1096/fasebj.8.2.8119491
Maddaloni, M., Forlani, F., Balmas, V., Donini, G., Stasse, L., Corazza, L., et al. (1997). Tolerance to the fungal pathogen Rhizoctonia solani AG4 of transgenic tobacco expressing the maize ribosome inactivating protein b-32. Transgenic Res. 6, 393–402. doi: 10.1023/A:1018435417740
Melton-Celsa, A. R. (2014). Shiga toxin (Stx) classification, structure, and function. Microbiol. Spectr. 9, 1–13. doi: 10.1128/microbiolspec.EHEC-0024-2013
Meng, Y., Lin, S., Liu, S. F., Fan, X., Li, G. R., and Meng, Y. F. (2014). A novel method for simultaneous production of two ribosome-inactivating proteins, a-MMC and MAP30, from Momordica charantia L. PLOS ONE 9:e101998. doi: 10.1371/journal.pone.0101998
Mlsna, D., Monzingo, A. F., Katzin, B. J., Ernst, S., and Robertus, J. D. (1993). Structure of recombinant ricin A chain at 2.3 Å. Protein Sci. 2, 429–435. doi: 10.1002/pro.5560020315
Mock, J. W., Ng, T. B., Wong, R. N., Yao, Q. Z., Yeung, H. W., and Fong, W. P. (1996). Demonstration of ribonuclease activity in the plant ribosome inactivating proteins alpha- and beta-momorcharins. Life Sci. 59, 1853–1859. doi: 10.1016/S0024-3205(96)00532-2
Monzingo, A. F., and Robertus, J. D. (1992). X-ray analysis of substrate analogs in the ricin A-chain active site. J. Mol. Biol. 227, 1136–1145. doi: 10.1016/0022-2836(92)90526-P
Moon, Y. H., Song, S. K., Choi, K. W., and Lee, J. S. (1997). Expression of a cDNA encoding Phytolacca insularis antiviral protein confers virus resistance of transgenic potato plants. Mol. Cells 7, 807–815.
Morais, J. K. S., Gomes, V. M., Oliveira, J. T. A., Santos, I. S., Da-Cunha, M., Oliveira, H. D., et al. (2010). Soybean toxin (SBTX), a protein from soybeans that inhibits the life cycle of plant and human pathogenic fungi. J. Agric. Food Chem. 58, 10356–10363. doi: 10.1021/jf101688k
Narayanan, S., Surendranath, K., Bora, N., Surolia, A., and Anjali, A. K. (2005). Ribosome inactivating proteins and apoptosis. FEBS Lett. 579, 1324–1331. doi: 10.1016/j.febslet.2005.01.038
Narwal, S., Balasubrahmanyam, A., Sadhna, P., Kapoor, H. C., and Lodha, M. L. A. (2001). Systemic resistance inducing antiviral protein with N-glycosidase activity from Bougainvillea xbuttiana leaves. Indian J. Exp. Biol. 39, 600–603.
Ng, T. B., Parkash, A., and Tso, W. W. (2003). Purification and characterization of alpha- and beta-benincasins, arginine/glutamate-rich peptides with translation-inhibiting activity from wax gourd seeds. Peptides 24, 11–16. doi: 10.1016/S0196-9781(02)00271-1
Ng, T. B., Wong, J. H., and Wang, H. (2010). Recent progress in research on ribosome inactivating proteins. Curr. Protein Pept. Sci. 11, 37–53. doi: 10.2174/138920310790274662
Nielsen, K., and Boston, R. S. (2001). Ribosome-inactivating proteins: a plant perspective. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 785–816. doi: 10.1146/annurev.arplant.52.1.785
Obrig, T. G., Irvin, J. D., and Hardesty, B. (1973). The effect of an antiviral peptide on the ribosomal reactions of the peptide elongation enzymes, EF-I and EF-II. Arch. Biochem. Biophys. 155, 278–289. doi: 10.1016/0003-9861(73)90116-1
Olivieri, F., Prasad, V., Valbonesi, P., Srivastava, S., Ghosal-Chowdhury, P., Barbieri, L., et al. (1996). A systemic antiviral resistance inducing protein isolated from Clerodendrum inerme Gaertn. is a polynucleotide: adenosine glycosidase (ribosome-inactivating protein). FEBS Lett. 396, 132–134. doi: 10.1016/0014-5793(96)01089-7
Olsnes, S., and Pihl, A. (1973a). Different biological properties of the two constituent peptide chains of ricin, a toxic protein inhibiting protein synthesis. Biochemistry 12, 3121–3126. doi: 10.1021/bi00740a028
Olsnes, S., and Pihl, A. (1973b). Isolation and properties of abrin: a toxic protein inhibiting protein synthesis. Evidence for different biological functions of its two constituent-peptide chains. Eur. J. Biochem. 35, 179–185. doi: 10.1111/j.1432-1033.1973.tb02823.x
Parikh, B. A., and Tumer, N. E. (2004). Antiviral activity of ribosome inactivating proteins in medicine. Mini Rev. Med. Chem. 4, 523–543. doi: 10.2174/1389557043403800
Park, S. C., Kim, J. Y., Lee, J. K., Hwang, I., Cheong, H., and Nah, J. W. (2009). Antifungal mechanism of a novel antifungal protein from pumpkin rinds against various fungal pathogens. J. Agric. Food Chem. 57, 9299–9304. doi: 10.1021/jf902005g
Parkash, A., Ng, T. B., and Tso, W. W. (2002a). Isolation and characterization of luffacylin, a ribosome inactivating peptide with anti-fungal activity from sponge gourd (Luffa cylindrica) seeds. Peptides 23, 1019–1024.
Parkash, A., Ng, T. B., and Tso, W. W. (2002b). Purification and characterization of charantin, a napin-like ribosome-inactivating peptide from bitter gourd (Momordica charantia) seeds. J. Pept. Res. 59, 197–202.
Peumans, W. J., Hao, Q., and Van-Damme, E. J. (2001). Ribosome-inactivating proteins from plants: more than N-glycosidases? FASEB J. 15, 1493–1506.
Picard, D., Kao, C. C., and Hudak, K. A. (2005). Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 280, 20069–20075. doi: 10.1074/jbc.M413452200
Prasad, V., Mishra, S. K., Sivastava, S., and Srivastava, A. (2014). A virus inhibitory protein isolated from Cyamopsis tetragonoloba (L.) Taub. upon induction of systemic antiviral resistance shares partial amino acid sequence homology with a lectin. Plant Cell Rep. 33, 1467–1478. doi: 10.1007/s00299-014-1630-7
Prasad, V., Srivastava, S., and Varsha-Verma, H. N. (1995). Two basic proteins isolated from Clerodendrum inerme Gaertn. are inducers of systemic antiviral resistance in susceptible plants. Plant Sci. 110, 73–82. doi: 10.1016/0168-9452(95)04192-W
Pu, Z., Lu, B. Y., Liu, W. Y., and Jin, S. W. (1996). Characterization of the enzymatic mechanism of gamma-momorcharin, a novel ribosome-inactivating protein with lower molecular weight of 11,500 purified from the seeds of bitter gourd (Momordica charantia). Biochem. Biophys. Res. Commun. 229, 287–294. doi: 10.1006/bbrc.1996.1794
Puri, M., Kaur, I., Kanwar, R. K., Gupta, R. C., Chauhan, A., and Kanwar, J. R. (2009). Ribosome inactivating proteins (RIPs) from Momordica charantia for antiviral therapy. Curr. Mol. Med. 9, 1080–1094. doi: 10.2174/156652409789839071
Qian, Q., Huang, L., Yi, R., Wang, S. Z., and Ding, Y. (2014). Enhanced resistance to blast fungus in rice (Oryza sativa L.) by expressing the ribosome-inactivating protein alpha-momorcharin. Plant Sci. 217–218, 1–7. doi: 10.1016/j.plantsci.2013.11.012
Ready, M., Brown, D. T., and Robertus, J. D. (1986). Extracellular localization of pokeweed antiviral protein. Proc. Natl. Acad. Sci. U.S.A. 83, 5053–5056. doi: 10.1073/pnas.83.14.5053
Reinbothe, S., Reinbothe, C., Lehmann, J., Becker, W., Apel, K., and Parthier, B. (1994). JIP60, a methyl jasmonate-induced ribosome-inactivating protein involved in plant stress reactions. Proc. Natl. Acad. Sci. U.S.A. 91, 7012–7016. doi: 10.1073/pnas.91.15.7012
Rippmann, J. F., Michalowski, C. B., Nelson, D. E., and Bohnert, H. J. (1997). Induction of a ribosome-inactivating protein upon environmental stress. Plant Mol. Biol. 35, 701–709. doi: 10.1023/A:1005871023944
Roberts, L. M., and Lord, J. M. (1981). The synthesis of Ricinus communis agglutinin-cotranslational and posttranslational modification of agglutinin polypeptides. Eur. J. Biochem. 119, 31–41. doi: 10.1111/j.1432-1033.1981.tb05573.x
Roberts, W., and Stewart, T. S. (1979). Purification and properties of a translational inhibitor from wheat germ. Biochemistry 18, 2615–2621. doi: 10.1021/bi00579a028
Rodoni, B. (2009). The role of plant biosecurity in preventing and controlling emerging plant virus disease epidemics. Virus Res. 141, 150–157. doi: 10.1016/j.virusres.2008.11.019
Roy, S., Sadhana, P., Begum, M., Kumar, S., Lodha, M. L., and Kapoor, H. C. (2006). Purification, characterization and cloning of antiviral/ribosome inactivating protein from Amaranthus tricolor leaves. Phytochemistry 67, 1865–1873. doi: 10.1016/j.phytochem.2006.06.011
Ruan, X. L., Liu, L. F., and Li, H. P. (2007). Transgenic tobacco plants with ribosome inactivating protein gene cassin from Cassia occidentalis and their resistance to tobacco mosaic virus. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33, 517–523.
Rumiyati, N. A. W., Sismindari-Lukitaningsih, E., and Yuliati, T. (2014). Potential of ribosome-inactivating proteins (RIPs) of Mirabilis jalapa L. as an antiacne: effect on proliferation of cultured sebocyte cells and its antibacterial activities against Propionibacterium acnes and Staphylococcus epidermidis. Int. J. Pharm. Chem. 4, 130–133.
Russo, L. M., Melton-Celsa, A. R., Smith, M. J., and O’Brien, A. D. (2014). Comparisons of native shiga toxins (stxs) type 1 and 2 with chimeric toxins indicate that the source of the binding subunit dictates degree of toxicity. PLOS ONE 9:e93463. doi: 10.1371/journal.pone.0093463
Sandvig, K., and van Deurs, B. (1994). Endocytosis and intracellular sorting of ricin and shiga toxin. FEBS Lett. 346, 99–102. doi: 10.1016/0014-5793(94)00281-9
Sandvig, K., and van Deurs, B. (2002). Transport of protein toxins into cells: pathways used by ricin, cholera toxin and Shiga toxin. FEBS Lett. 529, 49–53. doi: 10.1016/S0014-5793(02)03182-4
Sargolzaei, M., Ho, C. L., and Wong, M. Y. (2016). Characterization of novel type I ribosome-inactivating proteins isolated from oil palm (Elaeis guineensis) inoculated with Ganoderma boninense, the causal agent of basal stem rot. Physiol. Mol. Plant Pathol. 94, 53–61. doi: 10.1016/j.pmpp.2016.04.002
Schrot, J., Weng, A., and Melzig, M. F. (2015). Ribosome-inactivating and related proteins. Toxins 7, 1556–1615. doi: 10.3390/toxins7051556
Shahidi-Noghabi, S., Van-Damme, E. J. M., Mahdian, K., and Smagghe, G. (2010). Entomotoxic action of Sambucus nigra agglutinin I in Acyrthosiphon pisumaphids and Spodoptera exigua caterpillars through caspase-3 like dependent apoptosis. Arch. Insect Biochem. Physiol. 75, 207–220. doi: 10.1002/arch.20387
Shahidi-Noghabi, S., Van-Damme, E. J. M., and Smagghe, G. (2008). Carbohydrate-binding activity of the type-2 ribosome-inactivating protein SNA-I from elderberry (Sambucus nigra) is a determining factor for its insecticidal activity. Phytochemistry 69, 2972–2978. doi: 10.1016/j.phytochem.2008.09.012
Shang, C., Rougé, P., and Van Damme, E. J. M. (2016). Ribosome inactivating proteins from rosacea. Molecules 21:1105. doi: 10.3390/molecules21081105
Sharma, N., Park, S. W., Vepachedu, R., Barbieri, L., Ciani, M., Stirpe, F., et al. (2004). Isolation and characterization of an RIP (ribosome-inactivating protein)-like protein from tobacco with dual enzymatic activity. Plant Physiol. 134, 171–181. doi: 10.1104/pp.103.030205
Shu, S. H., Xie, G. Z., Guo, X. L., and Wang, M. (2009). Purification and characterization of a novel ribosome-inactivating protein from seeds of Trichosanthes kirilowii maxim. Protein Expr. Purif. 67, 120–125. doi: 10.1016/j.pep.2009.03.004
Sikriwal, D., and Batra, J. K. (2010). “Ribosome inactivating proteins and apoptosis,” in Plant Cell Monographs: Toxic Plant Proteins, eds J. M. Lord and M. R. Hartley (Berlin: Springer Verlag), 107–132.
Sipahioğlu, H. M., Kaya,İ., Usta, M.,Ünal, M., Özcan, D., Özer, M., et al. (2017). Pokeweed (Phytolacca americana L.) antiviral protein inhibits Zucchini yellow mosaic virus infection in a dose-dependent manner in squash plants. Turk. J. Agric. For. 41, 256–262. doi: 10.3906/tar-1612-30
Song, S. K., Choi, Y., Moon, Y. H., Kim, S. G., Choi, Y. D., and Lee, J. S. (2000). Systemic induction of a Phytolacca insularis antiviral protein gene by mechanical wounding, jasmonic acid, and abscisic acid. Plant Mol. Biol. 43, 439–450. doi: 10.1023/A:1006444322626
Srivastava, A., Trivedi, S., Krishna, S. K., Verma, H. N., and Prasad, V. (2009). Suppression of papaya ringspot virus infection in Carica papaya with CAP-34, a systemic antiviral resistance inducing protein from Clerodendrum aculeatum. Eur. J. Plant Pathol. 123, 241–246. doi: 10.1007/s10658-008-9358-2
Srivastava, S., Verma, H. N., Srivastava, A., and Prasad, V. (2015). BDP-30, a systemic resistance inducer from Boerhaavia diffusa L., suppresses TMV infection, and displays homology with ribosome-inactivating proteins. J. Biosci. 40, 125–135. doi: 10.1007/s12038-014-9494-0
Steeves, R. M., Denton, M. E., Barnard, F. C., Henry, A., and Lambert, J. M. (1999). Identification of three oligosaccharide binding sites in ricin. Biochemistry 38, 11677–11685. doi: 10.1021/bi990493o
Stevens, W. A., Spurdon, C., Onyon, L. J., and Stirpe, F. (1981). Effect of inhibitors of protein synthesis from plants on tobacco mosaic virus infection. Experientia 37, 257–259. doi: 10.1007/BF01991642
Stirpe, F. (2004). Ribosome-inactivating proteins. Toxicon 44, 371–383. doi: 10.1016/j.toxicon.2004.05.004
Stirpe, F. (2013). Ribosome-inactivating proteins: from Toxins to useful proteins. Toxicon 67, 12–16. doi: 10.1016/j.toxicon.2013.02.005
Stirpe, F., Barbieri, L., Gorini, P., Valbonesi, P., Bolognesi, A., and Polito, L. (1996). Activities associated with the presence of ribosome-inactivating proteins increase in senescent and stressed leaves. FEBS Lett. 382, 309–312. doi: 10.1016/0014-5793(96)00188-3
Stirpe, F., and Battelli, M. G. (2006). Ribosome-inactivating proteins: progress and problems. Cell Mol. Life Sci. 63, 1850–1866. doi: 10.1007/s00018-006-6078-7
Stirpe, F., Gasperi-Campani, A., Barbieri, L., Lorenzoni, E., and Montanaro, L. (1978). Inhibition of protein synthesis, by modeccin, the toxin of Modecca digitata. FEBS Lett. 85, 65–67. doi: 10.1016/0014-5793(78)81249-6
Stirpe, F., and Lappi, D. A. (eds). (2014). Ribosome-Inactivating Proteins Ricin, and Related Proteins. Oxford: Wiley Blackwell.
Stirpe, F., Williams, D. G., Onyon, L. J., and Legg, R. F. (1981). Dianthins, ribosome damaging proteins with antiviral properties from Dianthus caryophyllus L. (carnation). Biochem. J. 195, 399–405. doi: 10.1042/bj1950399
Tai, N. W., Li, F., Li, Z., Zhuang, D. H., and Zhang, Z. C. (2000). Purification and partial characterization of s-trichokirin, a new small ribosome-inactivating protein, from seeds of Trichosanthes kirilowii. Acta Biochim. Biophys. Sin. 32, 495–498.
Tang, S., Xie, L., Hou, F., Liu, W. Y., and Ruan, K. (2001). Non-specific deadenylation and deguanylation of naked RNA catalyzed by ricin under acidic condition. Biochim. Biophys. Acta 1519, 192–198. doi: 10.1016/S0167-4781(01)00236-6
Tarantini, A., Pittaluga, E., Marcozzi, G., Testone, G., Rodrigues-Pousada, R. A., Giannino, D., et al. (2010). Differential expression of saporin genes upon wounding, ABA treatment and leaf development. Physiol. Plant. 140, 141–152. doi: 10.1111/j.1399-3054.2010.01388.x
Taylor, S., Massiah, A., Lomonossoff, G., Roberts, L. M., Lord, J. M., and Hartley, M. (1994). Correlation between the activities of five ribosome-inactivating proteins in depurination of tobacco ribosomes and inhibition of tobacco mosaic virus infection. Plant J. 5, 827–835. doi: 10.1046/j.1365-313X.1994.5060827.x
Tumer, N. E., Hwang, D. J., and Bonness, A. M. (1997). C-terminal deletion mutant of pokeweed antiviral protein inhibits viral infection but does not depurinate host ribosomes. Proc. Natl. Acad. Sci. U.S.A. 94, 3866–3871. doi: 10.1073/pnas.94.8.3866
Van Damme, E. J., Hao, Q., Barre, A., Vandenbussche, F., Desmyter, S., Rougé, P., et al. (2008). Ribosome-inactivating proteins: a family of proteins that do more than inactivate ribosomes. Crit. Rev. Plant Sci. 20, 395–465. doi: 10.1080/07352689.2001.10131826
van Wees, S. C. M., De-Swart, E. A. M., Van-Pelt, J. A., Van-Loon, L. C., and Pieterse, C. M. J. (2000). Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 97, 8711–8716. doi: 10.1073/pnas.130425197
Vandenbussche, F., Desmyter, S., Ciani, M., Proost, P., Peumans, W. J., and Van-Damme, E. J. M. (2004). Analysis of the in planta antiviral activity of elderberry ribosome-inactivating proteins. Eur. J. Biochem. 271, 1508–1515. doi: 10.1111/j.1432-1033.2004.04059.x
Verma, H. N., Srivastava, S., and Varsha-Kumar, D. (1996). Induction of systemic resistance in plants against viruses by a basic protein from Clerodendrum aculeatum leaves. Phytopathology 86, 485–492. doi: 10.1094/Phyto-86-485
Vivanco, J. M., Savary, B. J., and Flores, H. E. (1999). Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the Andean crop Mirabilis expansa. Plant Physiol. 119, 1447–1456. doi: 10.1104/pp.119.4.1447
Vivanco, J. M., and Tumer, N. E. (2003). Translation inhibition of capped and uncapped viral RNAs mediated by ribosome-inactivating proteins. Phytopathology 93, 588–595. doi: 10.1094/PHYTO.2003.93.5.588
Walsh, T. A., Morgan, A. E., and Hey, T. D. (1991). Characterization and molecular cloning of a proenzyme form of a ribosome-inactivating protein from maize. Novel mechanism of proenzyme activation by proteolytic removal of a 2.8-kiloDalton internal peptide segment. J. Biol. Chem. 266, 23422–23427.
Wang, H., and Ng, T. B. (2000). Lagenin, a novel ribosome-inactivating protein with ribonucleolytic activity from bottle gourd (Lagenaria siceraria) seeds. Life Sci. 67, 2631–2638. doi: 10.1016/S0024-3205(00)00846-8
Wang, H., and Ng, T. B. (2001). Isolation and characterization of velutin, a novel low-molecular-weight ribosome-inactivating protein from winter mushroom (Flammulina velutipes) fruiting bodies. Life Sci. 68, 2151–2158. doi: 10.1016/S0024-3205(01)01023-2
Wang, H., and Ng, T. B. (2002). Luffangulin, a novel ribosome inactivating peptide from ridge gourd (Luffa acutangula) seeds. Life Sci. 70, 899–906. doi: 10.1016/S0024-3205(01)01466-7
Wang, P., and Tumer, N. E. (2000). Virus resistance mediated by ribosome inactivating proteins. Adv. Virus Res. 55, 325–356. doi: 10.1016/S0065-3527(00)55007-6
Wang, S., Zhang, H., Zheng, Y., Li, Z., Xiang, F., Ding, Y., et al. (2016). Environmental factors and phytohormones enhancing expression of α-momorcharin gene in Momordica charantia. Biologia 71, 155–160. doi: 10.1515/biolog-2016-0018
Wang, S., Zhang, Y., Liu, H., He, Y., Yan, J., Wu, Z., et al. (2012). Molecular cloning and functional analysis of a recombinant ribosome-inactivating protein (alpha-momorcharin) from Momordica charantia. Appl. Microbiol. Biotechnol. 96, 939–950. doi: 10.1007/s00253-012-3886-6
Wei, G. Q., Liu, R. S., Wang, Q., and Liu, W. Y. (2004). Toxicity of two type-2 ribosome-inactivating proteins (cinnamomin and ricin) to domestic silkworm larvae. Arch. Insect Biochem. Physiol. 57, 160–165. doi: 10.1002/arch.20024
Wong, J. H., Ng, T. B., Cheung, R. C. F., Ye, X. J., Wang, H. X., Lam, S. K., et al. (2010). Proteins with antifungal properties and other medicinal applications from plants and mushrooms. Appl. Microbiol. Biotechnol. 87, 1–15. doi: 10.1007/s00253-010-2690-4
Yang, T., Meng, Y., Chen, L. J., Lin, H. H., and Xi, D. H. (2016). The roles of alpha-momorcharin and jasmonic acid in modulating the response of Momordica charantia to Cucumber mosaic virus. Front. Microbiol. 7:1796. doi: 10.3389/fmicb.2016.01796
Yao, Q. Z., Yu, M. M., Ooi, L. S., Ng, T. B., Chang, S. T., Sun, S. S., et al. (1998). Isolation and characterization of a type 1 ribosome-inactivating protein from fruiting bodies of the edible mushroom (Volvariella volvacea). J. Agric. Food Chem. 46, 788–792. doi: 10.1021/jf970551h
Yuan, H., Ming, X., Wang, L., Hu, P., An, C., and Chen, Z. (2002). Expression of a gene encoding trichosanthin in transgenic rice plants enhances resistance to fungus blast disease. Plant Cell Rep. 20, 992–998. doi: 10.1007/s00299-002-0437-0
Zhou, X., Li, X. D., Yuan, J. Z., Tang, Z. H., and Liu, W. Y. (2000). Toxicity of cinnamomin: a new type-2 ribosome-inactivating protein to bollworm and mosquito. Insect Biochem. Mol. Biol. 30, 259–264. doi: 10.1016/S0965-1748(99)00126-5
Zhu, F., Xu, M. Y., Wang, S. D., Jia, S. D., Zhang, P., Lin, H. H., et al. (2012). Prokaryotic expression of pathogenesis related protein 1 gene from Nicotiana benthamiana: antifungal activity and preparation of its polyclonal antibody. Biotechnol. Lett. 34, 919–924. doi: 10.1007/s10529-012-0851-5
Zhu, F., Yuan, S., Zhang, Z. W., Qian, K., Feng, J. G., and Yang, Y. Z. (2016). Pokeweed antiviral protein (PAP) increases plant systemic resistance to Tobacco mosaic virus infection in Nicotiana benthamiana. Eur. J. Plant Pathol. 146, 541–549. doi: 10.1007/s10658-016-0938-2
Zhu, F., Zhang, P., Meng, Y. F., Xu, F., Zhang, D. W., Cheng, J., et al. (2013). Alpha-momorcharin, a RIP produced by bitter melon, enhances defense response in tobacco plants against diverse plant viruses and shows antifungal activity in vitro. Planta 237, 77–88. doi: 10.1007/s00425-012-1746-3
Keywords: antibacterial, antifungal, antiviral, defense, ribosome-inactivating proteins, systemic resistance
Citation: Zhu F, Zhou Y-K, Ji Z-L and Chen X-R (2018) The Plant Ribosome-Inactivating Proteins Play Important Roles in Defense against Pathogens and Insect Pest Attacks. Front. Plant Sci. 9:146. doi: 10.3389/fpls.2018.00146
Received: 02 November 2017; Accepted: 25 January 2018;
Published: 09 February 2018.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Victoria Pastor, Jaume I University, SpainCopyright © 2018 Zhu, Zhou, Ji and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Zhu, emh1ZmVuZ0B5enUuZWR1LmNu; emh1ZmVuZzEzNjFAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.