
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 February 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00094
This article is part of the Research Topic Crop Breeding for Drought Resistance View all 16 articles
 Liyu Huang1,2†
Liyu Huang1,2† Yinxiao Wang1†
Yinxiao Wang1† Wensheng Wang1
Wensheng Wang1 Xiuqin Zhao1
Xiuqin Zhao1 Qiao Qin1
Qiao Qin1 Fan Sun1
Fan Sun1 Fengyi Hu2
Fengyi Hu2 Yan Zhao3
Yan Zhao3 Zichao Li3
Zichao Li3 Binying Fu1,4*
Binying Fu1,4* Zhikang Li1,4*
Zhikang Li1,4*HIGHLIGHTS
• Overexpressing and RNA interfering OsDRAP1 transgenic rice plants exhibited significantly improved and reduced drought tolerance, but accompanied with negative effects on development and yield.
The dehydration responsive element binding (DREBs) genes are important transcription factors which play a crucial role in plant abiotic stress tolerances. In this study, we functionally characterized a DREB2-like gene, OsDRAP1 conferring drought tolerance (DT) in rice. OsDRAP1, containing many cis-elements in its promoter region, was expressed in all organs (mainly expressed in vascular tissues) of rice, and induced by a variety of environmental stresses and plant hormones. Overexpressing OsDRAP1 transgenic plants exhibited significantly improved DT; while OsDRAP1 RNA interfering plants exhibited significantly reduced DT which also accompanied with significant negative effects on development and yield. Overexpression of OsDRAP1 has a positive impact on maintaining water balance, redox homeostasis and vascular development in transgenic rice plants under drought stress. OsDRAP1 interacted with many genes/proteins and could activate many downstream DT related genes, including important transcription factors such as OsCBSX3 to response drought stress, indicating the OsDRAP1-mediated pathways for DT involve complex genes networks. All these results provide a basis for further complete understanding of the OsDRAP1 mediated gene networks and their related phenotypic effects.
Rice (Oryza sativa. L) is the staple food for half of the world's population. Rice production normally requires large amounts of water. Water availability and drought are becoming major constraints for rice cultivation because of greatly fluctuated and generally reduced rainfall in the majority of rice growing areas worldwide resulting from the global climate change. Rice production was dramatically affected by severe drought occurred every year in rainfed rice-growing areas worldwide (Pandey, 2009; Lenka et al., 2011). To ensure food security, it is imperative to develop drought tolerant high-yielding rice varieties.
DT is a complex trait controlled by many genes and shows significant genotype by environment interaction (Babu, 2010). Genetically, many QTLs affecting DT in rice has been identified (Li et al., 2005; Xu and Vision, 2005; Kumar et al., 2014), but few of them have been cloned and functionally characterized (Uga et al., 2013; Li et al., 2015). At the transcriptomic level, it is well known that drought can induce differential expression of large numbers of genes in rice (Wang D. et al., 2011). Of these drought responsive genes, several families of transcription factor (TF) such as AP2/ERFs, NAC, bZIPs and MYBs were found to play an important role in drought responses via regulating large numbers of downstream genes and pathways through complex transcriptional networks (Hadiarto and Tran, 2011; Todaka et al., 2015). Many TF families typically each contains many member genes, but only a portion of a TF family are functionally involved in plant responses to abiotic stresses (Joshi et al., 2016). Genes of the AP2/ERF family are characterized by a conserved AP2/ERF DNA binding domain (Riechmann and Meyerowitz, 1998; Sakuma et al., 2002). This TF family is further classified as four major subfamilies including AP2 (Apetala), RAV (related to ABI3/VP1), DREB (dehydration responsive element-binding protein), and ERF (ethylene responsive factor). There are 163 AP2/ERF genes in the rice genome (including 57 DREB members), and 70 of them were differentially regulated under different abiotic stresses (Sharoni et al., 2011). Most DREB member genes have been found to manipulate downstream stress-responsive genes by binding their drought responsive element (DRE) and GCC-box cis-element (Liu et al., 1998; Dubouzet et al., 2003). The DREB TFs could be further divided into DREB1 and DREB2, which were reportedly involved in two separate signal transduction pathways in plants in response to low temperature and dehydration stresses, respectively (Lata and Prasad, 2011). Four DREB1 genes (OsDREB1A, OsDREB1B, OsDREB1C, and OsDREB1D) were firstly identified in rice. Overexpression of OsDREB1A could enhance abiotic stress tolerance in Arabidopsis (Dubouzet et al., 2003) OsDREB1F is highly induced by drought stress and exogenous ABA application and overexpression of OsDREB1F is reportedly to lead to enhanced tolerance to salt, drought, and low temperature in both rice and Arabidopsis (Wang et al., 2008). In addition, higher expression of OsDREB1G in transgenic rice plants could significantly improve their tolerance to water deficit stress (Chen et al., 2008). Similarly, DREB2-type TFs are involved in a conserved regulatory mechanism in several crop plants in response to drought, salinity, and heat stresses (Lata and Prasad, 2011; Mizoi et al., 2012). There are five DREB2 genes (OsDREB2A, OsDREB2B, OsDREB2C, OsDREB2E, and OsABI4) in the rice genome (Matsukura et al., 2010; Srivastav et al., 2010). OsDREB2A expression in rice was evidently induced by water deficit and exogenous ABA application, which could result in improved DT (Cui et al., 2011). The transcript of OsDREB2B has both functional and non-functional forms. The former was markedly increased during stress conditions and was able to enhance DT by drought-induced alternative splicing of its pre-mRNA (Matsukura et al., 2010). All these results indicated that OsDREB2s also play important roles in plant DT.
In our breeding program, we developed a DT pyramiding line-DK151 from an F2 population of a cross between two IR64 introgression lines (Wang W. S. et al., 2011). In the following transcriptomic experiment using DK151 and its background parent, IR64, we found that large numbers of genes were specifically up-regulated in DK151 when compared with IR64 (Wang D. et al., 2011). Of these drought up-regulated genes in DK151, we identified a new rice DREB2-like TF gene, OsDRAP1 (Drought Responsive AP2/EREBP gene) located in an introgressed segment on chromosome 8 of DK151 and whose expression was greatly up-regulated in DK151 by drought, suggesting its potential role in rice DT. Here, we report the functional characterization of OsDRAP1 and evidence for its important role in improving rice DT.
Seeds of three rice genotypes, including Nipponbare, its transgenic plants overexpressing coding region of OsDRAP1 and RNAi knock-down transgenic plants generated by Agrobacterium-mediated transformation, were grown under the controlled conditions in the growth chamber. For stress and hormones treatments, 2-week-old rice plants were transferred from the basal nutrient solution to nutrient solution containing 20% PEG (PEG6000), 150 mM NaCl, 100 μM ABA, 100 μM methyl jasmonate (MeJA) or 20 mM H2O2 and exposed to low temperature (4°C). Leaf tissues were then harvested as the indicated times and stored at −80°C for further analysis.
The transgenic rice plants and their background parent, Nipponbare, were evaluated for their DT performances in a pot experiment in the greenhouse of Chinese Academy of Agricultural Sciences (Beijing, China). Three healthy seedlings were transplanted equidistantly into a strip pot (50 cm in length, 15 cm in width and 15 cm in height) filled with 2 kg of sterilized field soil. Seedlings of each genotype were planted in six pots. The pots were maintained under controlled conditions with 14 h daylight at 28°C and a 10 h dark period at 25°C. In the stress treatment, drought stress was initiated at seedling and tillering stages by withholding watering at precisely determined time intervals when the soil water content reached 15, 10, and 7.5% at 1, 3, and 5 days after the drought treatment, respectively, as measured by soil moisture meters (TZS-W, Zhejiang Top Instrument Co. Ltd).
Total RNA was extracted from rice tissues of the three genotypes using TRIzol Reagent (Invitrogen, USA). cDNA synthesis was performed using EasyScript First-strand cDNA Synthesis SuperMix (TransGene, Beijing, China) according to the manufacturer's instructions. qRT-PCR analysis was carried out using TaKaRa SYBR premix Ex Taq™ according to the manufacturer's instructions. The relative expression of each gene was calculated according to the method of 2−ΔΔCt (Livak and Schmittgen, 2001). Primers used for qRT-PCR were subsequently tested in a dissociation curve analysis and verified for the absence of nonspecific amplification. All analyses were performed with three biological replicates.
The coding region of OsDRAP1, was amplified from the rice cDNA by PCR using KpnI and BamHI linker primers. The resulting OsDRAP1 fragment was inserted into the KpnI and BamHI site of pCUbi1390 (Peng et al., 2009), generating Ubipro::OsDRAP1. 275-bp OsDRAP1 specific sequence was amplified from the rice cDNA and inserted into RNAi vector pH7GWIWG2 (II) (Karimi et al., 2005) using the Gateway technology in end-to-end orientations by using an intron as spacer. 1.8 kb-fragments of the OsDRAP1 promoter were amplified from rice genomic DNA (Nipponbare) by PCR and inserted into the vector pMDC162 (Curtis and Grossniklaus, 2003), respectively using the Gateway technology, generating OsDRAP1-pro::GUS. All the vectors were introduced into Agrobacterium tumefaciens strain EHA105, and then transferred into Nipponbare plants via Agrobacterium-mediated transformation following the protocol described by Duan (Duan et al., 2012).
To determine the localization of OsDRAP1 in rice plant tissues, the transverse sections (100 μm thick) of the OsDRAP1-pro1::GUS transgenic seedlings and roots with GUS stained were fixed for over 12 h in 2.5% glutaraldehyde buffered with 0.2 M phosphate buffer (pH 7.2). The sections were treated as described by Takemoto et al. (2002). For histological analyses, freshly collected leaves at the tillering stage were fixed immediately in FAA (3.7% formaldehyde, 5% acetic acid, 50% absolute ethanol). Plant tissues were then dehydrated, embedded, sliced, and pre-treated. Transverse section slices were ultimately stained with Haematoxylin and eosin (HE) and viewed and photographed on a microscope (DM-LS2, Leica, http://www.leica.com/).
Relative Water Content (RWC) was calculated using the following formula: RWC (%) = [(FM−DM)/(TM−DM)] × 100, where FM, DM, and TM were the fresh, dry, and turgid masses of the leaves weighed, respectively. Monitoring the fresh weight loss at the indicated time points (per hour) measured the WLRs (water loss rates) of the detached leaves (You et al., 2013). Relative electrolyte leakage (REL) or solute leakage from the sampled rice leaves were evaluated using the method of Arora et al. (1998). Percent injury arising from each treatment was calculated from the conductivity data using the equation: % injury = [(% L(t)-% L(c))/(100-% L(c))] × 100), where % L (t) and % L(c) are percent conductivity for treated and control samples, respectively. Antioxidant enzyme activity such as catalase (CAT), was determined following previously reported methods (Bonnecarrère et al., 2011). Proline and MDA concentrations of the sampled leaves were measured according to the protocol of Shukla et al. (2012).
The open reading frames (ORFs) of OsDRAP1 were inserted into pMDC43 as C-terminal fusions to the green fluorescent protein (GFP) reporter gene driven by the CaMV 35S promoter (Curtis and Grossniklaus, 2003). For transient expression, the GFP-OsDRAP1 fusion vector constructs were transformed into Nicotiana benthamiana protoplasts. Protoplast isolation from tobacco leaf tissues and PEG-mediated transformation were performed according to Bart et al. (2006). Cells were incubated at 28°C in the dark overnight. The resulting green fluorescence of protoplasts expressing GFP-OsDRAP1 was observed using a confocal laser scanning microscope (LSM700, Zeiss, Jena, Germany). The 35S::GFP construct was used as a control. AHL22 was used as a nucleus marker protein (Xiao et al., 2009).
For GUS staining analysis, sample tissues or whole seedlings were submerged in the GUS staining buffer (containing 2 mM 5-bromo-4-chloro-3-indolyl glucuronide, 0.1 M sodium phosphate buffer [pH 7.0], 0.1% [v/v] Triton X-100, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, and 10 mM EDTA), vacuum infiltrated for 10 min, and then incubated overnight at 37°C. The staining buffer was removed before the samples were cleared with 95% (v/v) ethanol and then observed using a stereoscope (LEICA, 10447157, Germany).
OsDRAP1 in transgenic plants was examined for the presence of one of its activation domains using a yeast assay system as described (Liu et al., 2013). A series of coding region fragments of OsDRAP1 were amplified by PCR using NcoI and PstI linker primers. The resultant PCR products were digested with NcoI and PstI and then cloned into the NcoI and PstI sites of pGBKT7, generating BD-OsDRAP1, BD-OsDRAP1ΔA, BD-OsDRAP1ΔAB, BD-OsDRAP1ΔD, BD-OsDRAP1ΔCD, BD-OsDRAP1ΔAD. An empty vector (BD) was used as a negative control. The constructs and the negative control BD were transformed into AH109, respectively according to the protocol of the manufacturer in the Leu-medium. About 2 days, the positive transformants verified by PCR were dropped on Leu- and Leu-His-Ade- medium, respectively. The transcriptional activation activities were evaluated according to their growth status.
A previously described dual-luc method using Nicotiana benthamiana plants (Liu et al., 2008) was used for transient transcription assays. The effector plasmids, pMDC43-OsDRAP1 and pMDC43-OsDRAP1ΔD were constructed as described above. The reporter plasmid, pGreen-GCC/DRE-LUC, encodes two luciferases, the firefly luciferase controlled by the recombinant GCC box promoter or DRE cis-element, and the Renilla (REN) luciferase controlled by the constitutive 35S promoter. The recombinant GCC box and DRE cis-element promoters contain two wild-type GCC box (ATAAGAGCCGCCACTCATAAGAGCCGCCACT) and DRE cis-element (ATACTACCGACATGAGATACTACCGACATGAG), respectively. Their mutant GCC box (mGCC, ATAAGATCCTCCACTCATAAGATCCTCCACT) and DRE cis-element (mDRE, ATACTACTGATATGAGATACTACTGATATGAG) fused to the minimum 35S promoter, were PCR-amplified from the 35S template and cloned into the ApaI and SacII sites of the vector pGreen-0800-LUC, then transformed into Agrobacterium (strain EHA105) containing the helper plasmid, pSoup-P19, that also encodes a repressor of co-suppression. The Agrobacterium strain containing both the reporter pGreen-GCC/DRE-LUC and helper pSoup-P19 was used alone or mixed with the Agrobacterium strain containing the effector plasmids pMDC43-OsDRAP1 or pMDC43-OsDRAP1ΔD.
The reporter strain was either incubated alone or incubated as a mixture with the effector strain (at the reporter: effector ratio of 1:9 or 2:8). Agrobacteria suspension in a 10 ml syringe was carefully press-infiltrated manually onto healthy leaves of Nicotiana benthamiana. Plants were left for 3 days after infiltration. Leaf samples were collected for the dual-luc assay using commercial Dual-LUC reaction (DLR) reagents, according to the manufacturer's instruction (Promega). After measurement of the firefly luciferase activity, 40 ul of the Stop and Glow buffer (Promega) was added to the reaction to quench the firefly luciferase and to initiate the REN luciferase (REN) reaction.
The protein interaction analysis was performed using Matchmaker Two-Hybrid System 3 (Clontech, http://www.clontech.com/). A bait gene is expressed as a fusion to the GAL4 DNA-binding domain (DNA-BD), while cDNA is expressed as a fusion to the GAL4 activation domain-AD (Fields and Song, 1989; Chien et al., 1991). Construct pGBKT7-OsDRAP1ΔD above was used as a bait protein. An AD fusion library of rice leaves under drought condition (pGADT7-library) was constructed in Oebiotech (Shanghai). pGBKT7-OsDRAP1ΔD and pGADT7-library plasmids were co-transformed into AH109, and then were spread on SD/–Ade/–His/–Leu/–Trp/X-α-gal plates. These plates were incubated at 30°C until colonies appear. To identify the gene responsible for a positive two-hybrid interaction, we rescue the gene by PCR colony screening. After that, we sequenced the cDNA inserts and blast the sequences in NCBI databases. Finally, we retest the interaction by cotransformation into the AH109. In the same way, cotransformation mixtures were spread on SD/–Ade/–His/–Leu/–Trp/X-α-gal plates. These plates were incubated at 30°C until colonies appear.
Bimolecular fluorescence complementation (BiFC) assay analyses were performed as previously described (Sparkes et al., 2006). Complementary DNAs of OsDRAP1 and OsCBSX3 were cloned into BiFC vectors pnYFP-X and pcCFP-X, respectively. Pair of constructs was co-transformed into the leaves of 3-week-old tobacco (Nicotiana benthamiana) by A. tumefaciens infiltration as the above protocol in dual-LUC assays. Cells co-transformed with pnYFP-OsDRAP1/pcCFP-GUS, pnYFP-GUS/pcCFP-OsCBSX3, were used as negative controls. AHL22-mRFP was used as a nuclear localization marker protein (Xiao et al., 2009).
Recombinant constructs GFP-OsCBSX3 and Myc-OsDRAP1 were introduced into tobacco leaves as the above protocol in dual-LUC or BiFC assays, respectively, and protein extracts were prepared as described by Sawa et al. (2007). The protein extracts were precipitated with anti-Flag agarose beads (Abmart, http://www.ab-mart.com/) or anti-Myc agarose beads (CMC Scientific, http://www.cmcscientific.com) overnight. Then proteins bound to beads were resolved by SDS-PAGE and detected with Western blot using anti-GFP antibody (Sigma), and anti-Myc antibody (MBL, http://www.mblintl.com/).
Three top leaves for each sample (two replicates for each sample) were harvested for each genotype under 1 day and 3 days of drought stress and control conditions. The RNA-seq sequencing and assembly were performed by Beijing CapitalBio Corporation as described in previous study (Huang et al., 2014). The number of mapped clean reads for each gene was counted and normalized into the reads per kilo base per million value (Mortazavi et al., 2008); Cuffdiff (Trapnell et al., 2009) was then used to identify DEGs. Finally, genes with a p ≤ 0.001 were designated as significantly differential expressed between each pair of samples. Gene function annotations were performed based on the Rice Genome Annotation Project version 7 (Kawahara et al., 2013). AgriGO was used to perform GO enrichment analysis (Du et al., 2010). The raw RNA-seq data are available in the Genome Sequence Archive in BIG Data Center (Beijing Institute of Genomics, Chinese Academy of Sciences) under the accession number PRJCA000683.
In our previous transcriptomic analyses (Wang D. et al., 2011), we identified a set of AP2/EREBP TF genes that were greatly up-regulated in leaves and panicles in drought tolerant pyramiding line DK151 under drought stress. One of these AP2/EREBP TF genes, designated as OsDRAP1 (LOC_Os08g31580), was selected for further function confirmation in this study.
The OsDRAP1 gene sequence has a full-length of 843 bp without any intron and encodes a polypeptide of 280 amino acids, consisting of an AP2 domain (103–166 aa) and four putative unknown domains (A: 25–38 aa, B: 82–100 aa, C: 190–201 aa, D: 210–239 aa; Figure 1C). The phylogenetic analysis indicated that OsDRAP1 was closely related to members of the DREB2 subfamily in rice and wheat (Figure S1; Sharoni et al., 2011). OsDRAP1 contains several stress- or hormone-responsive cis-elements in its promoter region (2.0 Kb upstream of the start codon), including 5 MYB binding sites (MBS), 6 ABA-responsive elements (ABRE), 2 gibberellin-responsive elements (GARE), and a CGTCA-motif for the MeJA-responsiveness (Table 1), implying that the expression of OsDRAP1 could be regulated by phytohormones. Our results from cytological analyses (Figures 1A,B) indicated that OsDRAP1 was localized in the nucleus and functioned as a nuclear protein, with its transactivation activity located primarily in region D (207-280 aa) (Figures 1C,D).
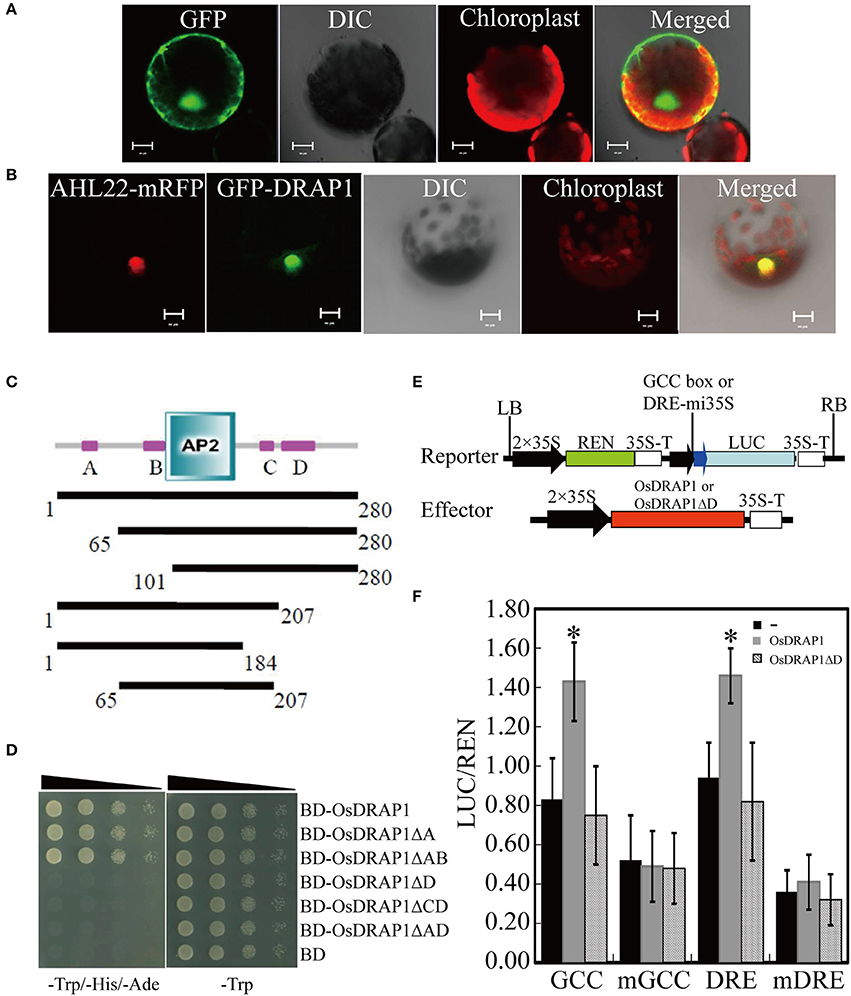
Figure 1. OsDRAP1 is a typical transcription activator. (A) GFP extensively localized in both nucleus andcytoplasm in the N. benthamiana protoplast; (B) GFP-OsDRAP1 positively localized in the nucleus, in which AHL22 used as the nucleus marker protein, DIC indicating its differential interference contrast transmission, chlorophyll fluorescence in red and the merged image, with the scale bars in 5 μm. (C) The structure of the OsDRAP1 protein and analysis of its transcription activating domains in yeast; Domains of the OsDRAP1 protein and different lengths of bait proteins with different domain deletions; A–D indicate four unknown domains. (D) Transcription activating domain identification of OsDRAP1 in yeast in which Yeast suspension was diluted to 10−1, 10−2, 10−3 times and dripped to the SD (-Trp/-His/-Ade) and SD (-Trp) medium, with pGKBT7 (BD) used as the negative control. (E) The schematic diagram of the reporters and effectors; (F) The transcription activation assay of OsDRAP1 and OsDRAP1ΔD in N. benthamiana, with “-,” OsDRAP1 and OsDRAP1ΔD indicating no effector control, full length and D domain deletion of the OsDRAP1 proteins, respectively, with the asterisks indicating significant differences at p < 0.05 in t-tests (vs. no effector control, n > 10).
To determine if OsDRAP1 is involved in the activation of multiple stress signaling pathways by interacting with GCC box or DRE element (Hao et al., 1998; Cheng et al., 2013), we performed a dual luciferase reporter assay by constructing reporter vectors containing GCC box mi35S-LUC, DRE mi35S-LUC, mutated GCC- and DRE-LUC (mGCC box mi35S-LUC, mDRE mi35S-LUC), and effector vectors containing 35S::OsDRAP1 and 35S::OsDRAP1ΔD. The results (Figures 1E,F) showed that the full length of OsDRAP1 could significantly enhance the LUC activity in the reporters of the GCC box mi35S-LUC and DRE mi35S-LUC, but had no effect on the two mutated reporters. The effector containing the truncated OsDRAP1 with deletion of region D (35S::OsDRAP1ΔD) could not activate all reporters. All these results indicated that OsDRAP1 was able to activate the expression of downstream genes by binding cis-elements of the GCC box and DRE.
Because OsDRAP1 was differentially expressed under drought stress in a spatial-temporal manner (Wang D. et al., 2011), we examined its expression patterns in different tissues at different developmental stages using qRT-PCR. The results showed that OsDRAP1 was highly expressed in the mature leaves and roots, but had relatively lower expression in young panicles, leaf sheaths and internodes (Figure 2A). The expression of OsDRAP1 was induced by PEG-simulated drought, high salt, low temperature, H2O2, ABA (Figure 3). Generally, OsDRAP1 showed similarly high expression in response to drought, salt and cold with the peak expression at 1 h after the treatments and maintaining a relatively high level of expression afterwards. Of the three plant hormones, OsDRAP1 responded much more strongly to ABA and H2O2 than to JA, though it showed the highest expression at 1 or 3 h after the treatments and decline afterwards in the H2O2 and ABA treatments. All these results indicated that the OsDRAP1 expression is induced by abiotic stresses, primarily by drought and salt, in a more H2O2 and/or ABA dependent manner.
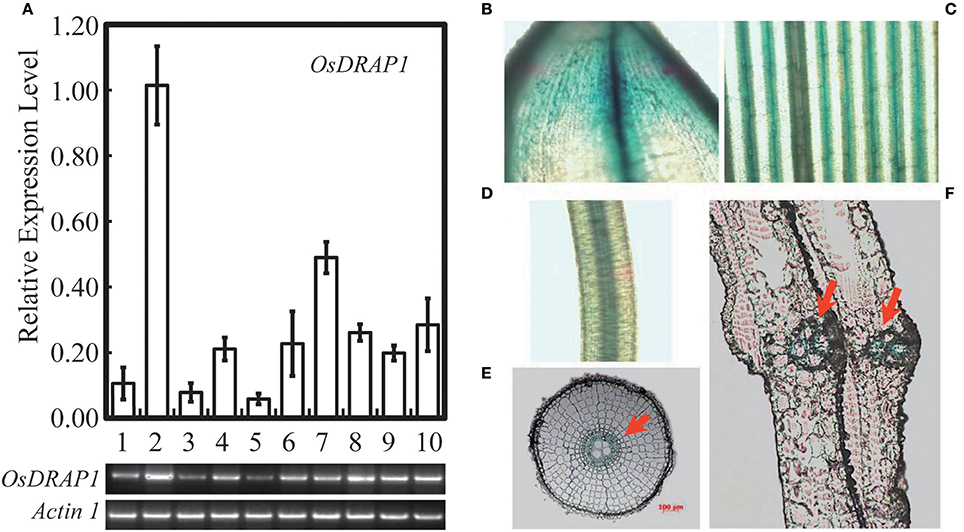
Figure 2. Expression model of OsDRAP1 in different tissues of rice, in which (A) the transcript levels were determined by qRT-PCR and semi RT-PCR, OsDRAP1 transcript levels in young panicles (1), matured leaves (2), leaf sheaths (3), nodes (4), internodes (5), stem bases (6), matured roots (7), young leaves (8), young roots (9) and callus (10) with Actin1 used as the reference gene. The OsDRAP1 expression in different tissues of the OsDRAP1-Pro::GUS transgenic rice plants by GUS staining analysis, in the shell (B), leaf blade (C), root (D), root cross section (E) and sheath cross section (F) with red arrows indicating the vascular bundles (the scale bars are in 100 μm).
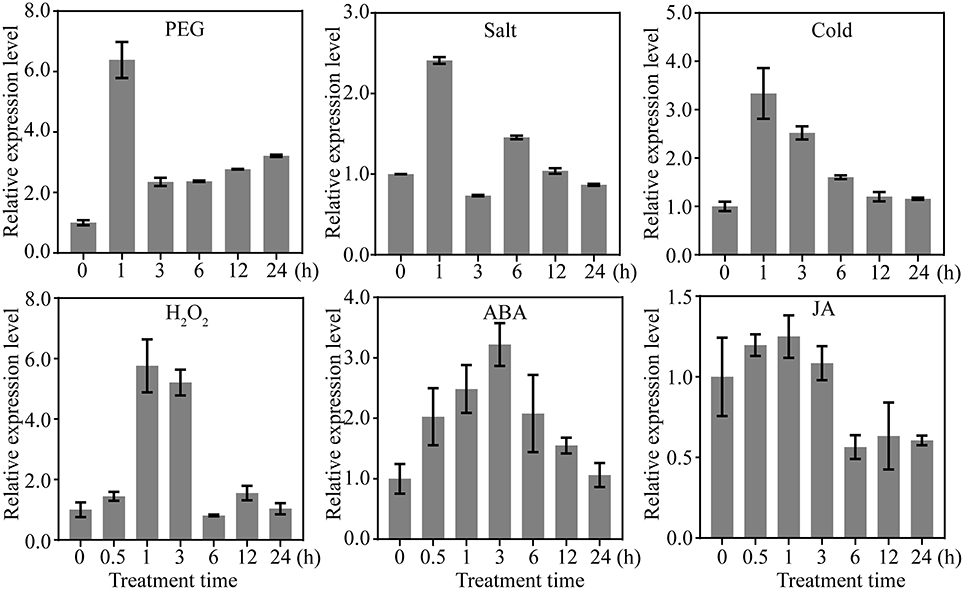
Figure 3. Expression profiles of OsDRAP1 in response to various abiotic stresses and exogenous hormones; in which 14-day-old seedlings of Nipponbare treated with PEG (20%), salt (150 mM), cold (4 C°), 100 mμ MeJA, 100 μM ABA and 20 mM H2O2, with Actin 1 used as the endogenous control.
To determine the biological function(s) of OsDRAP1 in response to abiotic stresses, we constructed overexpressing and RNAi knockdown vectors including UbiPro::OsDRAP1 and OsDRAP1-RNAi (see details in Materials and Methods). Thirteen OsDRAP1-overexpressing lines (OE-1 to 13) and four RNAi lines (RNAi-1 to 4) were obtained. The qRT-PCR results showed that the expression of OsDRAP1 was up-regulated in OE lines, but down-regulated in RNAi lines to various extents when compared with WT under the normal growth conditions (Figure S2). The seedlings of the OE lines and WT plants showed no evident differences before the stress treatments and 5 d after drought (Figures 4A–F), but all transgenic OE lines exhibited improved DT compared to WT with significantly higher seedling survival rates 7 days after rewatering (Figure 4G). As expected, the two RNAi lines were more sensitive to drought stress with significantly decreased survival rates compared to WT (Figures 4H,I). These results showed that the expression of OsDRAP1 was positively involved in DT. Based on these results, the transgenic lines, OE-7 and RNAi-2 were selected for further characterizing the functionality of OsDRAP1 in the following experiments.
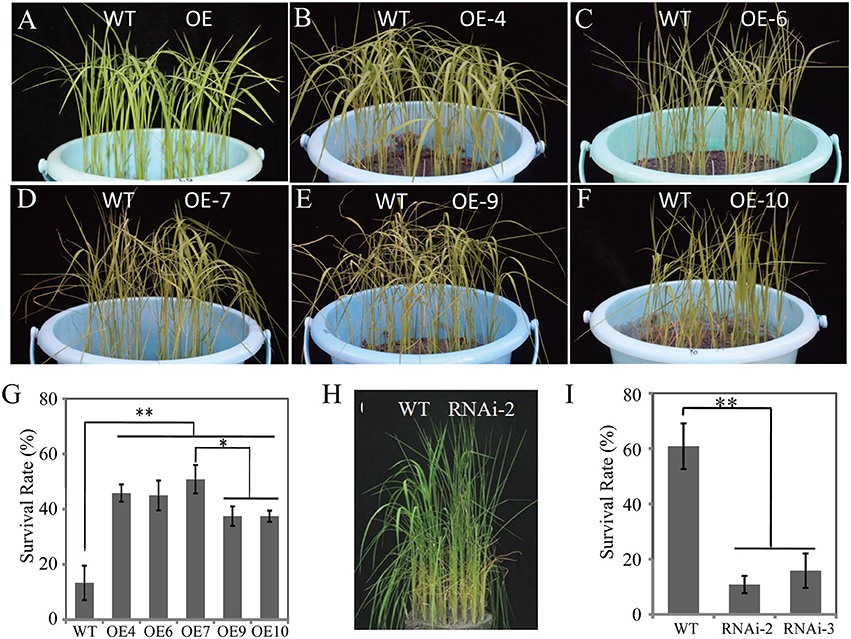
Figure 4. The seedling phenotypes of OsDRAP1 OE and RNAi lines at the seedling stage under the drought stress treatments: before drought (A) and 5 days after drought (B–F,H), and the seedling survival rates (G,I) evaluated 7 days after re-watering, in which the error bars indicate SD based on data of 3 replicates and the asterisks (* or **) indicate the significant differences at p < 0.05 or p < 0.01 in t-tests when compared with WT (n > 100).
Figure 5 shows the results on the evaluation of the OsDRAP1 overexpression line (OE-7), RNAi line (RNAi-2) and WT at the tillering stage for phenotypic and physiological trait performances under 3 and 5 days drought stress. There were no significant differences among three lines under the normal growth conditions. However, the WT and RNAi-2 plants started to show leaf rolling and wilting 3 d after drought stress, while the OE-7 was more viable than RNAi-2 and WT 5 days after drought stress and showed much better recovery 5 days after rewatering from the drought stress damage when the RNAi-2 plant was still wilted (Figure 5D). Furthermore, the OE-7 showed the lowest water loss, highest relative leaf water content, the lowest relative electrolyte leakage, and highest activities of ROS-scavenging enzymes during drought stress, while the opposite was true for the RNAi-2 plant (Figures 5E–H). All these results strongly suggested that OsDRAP1 overexpression improved drought tolerance in rice by maintaining higher leaf water content and enhancing activities of ROS-scavenging enzymes. However, at the maturity, the OE-7 and RNAi-2 plants showed significantly reduced height by 6 cm and 16 cm when compared with the WT plant (Figure S3). In addition, when compared to the WT plant, the OE-7 plant showed significantly reduced spikelet fertility while the RNAi-2 plant showed significantly reduced panicle numbers/plant (Figures S3C,E). As a result, both the OE-7 and RNAi-2 plants had lower grain yield/plant than the WT plant (Figure S3F). All these results indicated that overexpression or repression of OsDRAP1 had negative effects on the development and grain yield of rice, though the latter's effect was more severe.
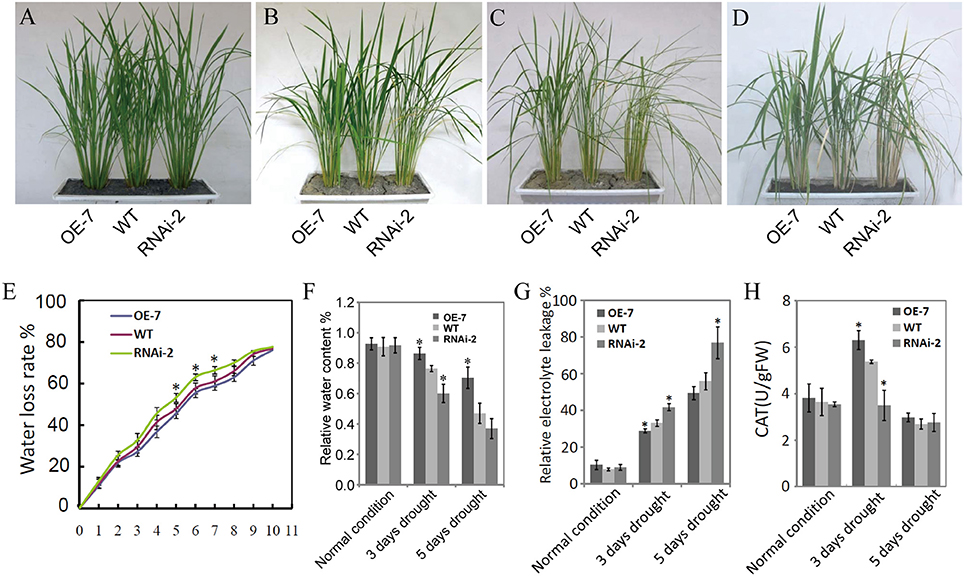
Figure 5. Phenotypic and physiological trait performances of the OsDRAP1 overexpression line (OE-7), RNAi line (RNAi-2) and wild type under drought stress; The seedling growth phenotypes of OE-7, RNAi-2 and WT under the normal irrigation (A), 3d and 5d drought stress (B,C), and 5d after re-watering (D); (E) The assay of water loss rates of OE-7, WT and RNAi-2; (F) The relative leaf water content of OE-7, WT and RNAi-3 under normal control, 3d and 5d drought stress; (G) Detection of relative electrolyte leakage of OE-7, WT and RNAi-2 under control, 3d and 5d drought stress conditions; (H) Detection of activities of ROS-scavenging enzymes (CAT) in three rice genotypes subjected to drought stress, with each column representing mean ± SD (three replicates) and * indicating a significant difference at p < 0.05 vs. WT based on the Dunnett's multiple comparison tests in ANOVA.
GUS staining results of the OsDRAP1-Pro::GUS transgenic plants showed that OsDRAP1 was mainly expressed in rice vascular tissues, especially in the vascular bundle of glume, leaf vein, pericycle and sheaths (Figures 2B–F). Electron microscope scanning of the stem cross sections revealed significant differences in cell number and diameter of vascular bundle between of the RNAi-2 and WT or OE-7 rice plants. Specifically, the RNAi-2 plant had fewer vascular cells and smaller diameter of vascular bundle than the OE-7 and WT plants (Figure 6), suggesting that OsDRAP1 may play an important role in vascular development.
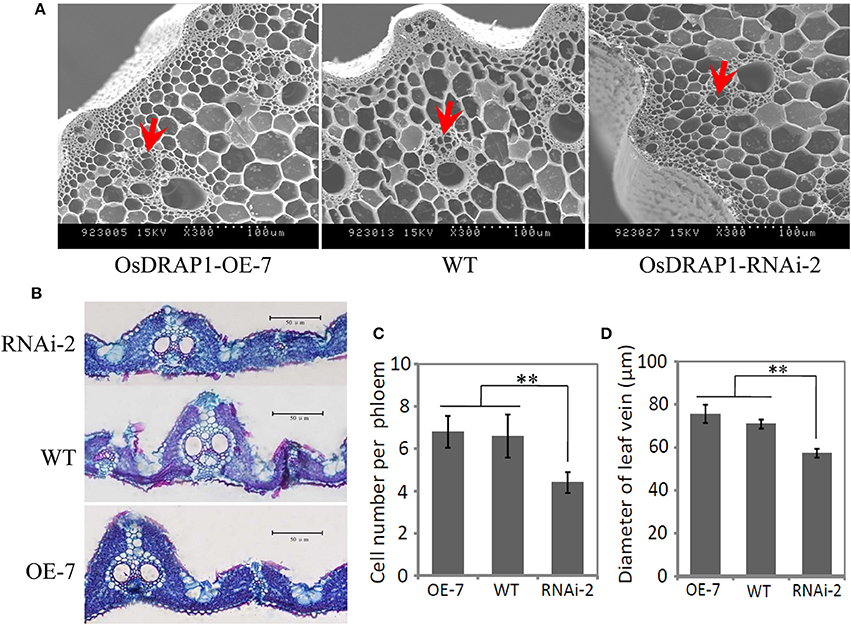
Figure 6. The cellular observation of the vascular tissues in one overexpression line (OE-7), one RNAi line (RNAi-2) and non-transgenic wild type (WT) for OsDRAP1, (A); and the cross section of the stem and leaf blade (B) at the tillering stage. Red arrows indicate the vascular bundles (phloem), with scale bars in 100 μm; (C) The mean cell number per phloem in the vascular bundle of the OE-7, WT and RNAi-2 plants; and (D) The diameter of leaf veins of the OE-7, WT and RNAi-2 plants, with the asterisks (**) indicating significant differences at p < 0.01 in t-tests when compared with WT (n > 10).
To gain insights into genes and pathways for DT regulated by OsDRAP1, we performed transcriptome sequencing to screen the differentially expressed genes in OE-7 compared to WT plants under normal growth and drought conditions, respectively. Table S1 shows 42 and 46 genes that were up- and down-regulated in OE-7, respectively, when compared to WT under the normal growth conditions. GO analysis revealed that the majority of these genes were functionally related to “response to stress” and “metabolic process.” After analyzing the cis-elements in the promoters of the 42 up-regulated genes in OE-7 using PLACE, we found that many of them have the binding sites of DREB TFs, GCC box and DRE cis-elements (Table S1), including GCCCOREC (S000430), DRECOREZMRAB17 (S000402) and DRECRTCOREAT (S000418), indicating that these genes were most likely directly regulated by OsDRAP1 and function in pathway(s) contributing in the basal DT of rice.
Under drought stress, there were 85 and 53 genes up- and down-regulated in OE-7 as compared to WT (Table S2). Most of the up-regulated genes in OE-7 were functionally categorized in metabolic processes (19), response to stress (14) and transcription regulation (6). Several TFs (OsEATB, OsERF5, OsMYB4, and OsWRKY89) were evidently induced in OE-7 by drought stress. In addition, a gene encoding cellulose synthase-like family A (CSLA11) reportedly involved in cell wall biogenesis (Wang et al., 2010), was remarkably up-regulated in OE-7. Meanwhile, a set of genes related to the JA signaling pathway such as OsJAZ8 (Yamada et al., 2012), OsJAZ13 (Singh et al., 2015), Jasmonate-zim-domain protein 1, and Jasmonate O-methyltransferase were simultaneously induced in OE-7 under drought, indicating that JA was also involved in the OsDRAP1 mediated regulating pathways of DT.
To identify proteins that directly interact with OsDRAP1, we performed yeast two-hybrid (Y2H) screening, which identified 30 interacting proteins from 45 positive clones screened according to their α-galactosidase activity (Table S3). Genes encoding these interacting proteins were predominant in function categories of response to stress (9), metabolic process (8) and nucleotide binding (7). These genes included several previously reported genes involved in response to abiotic stresses, such as OsRZFP34 for stomata opening, OsAPX2 and OsCATC involved in redox homeostasis under abiotic stress, OsRACK1A in ABA and H2O2 signaling pathways and OsCBSX3 involved in biotic stress tolerance (Lin et al., 2012; Zhang et al., 2013, 2014; Hsu et al., 2014; Mou et al., 2015). Results from bimolecular fluorescence complementation and co-immunoprecipitation assays revealed the direct interaction between A CBS (cystathionine beta-synthase) domain containing protein (OsCBSX3) and OsDRAP1 in the nucleus (Figure 7), indicating that OsCBSX3 is involved in the OsDRAP1-regulated pathway in rice.
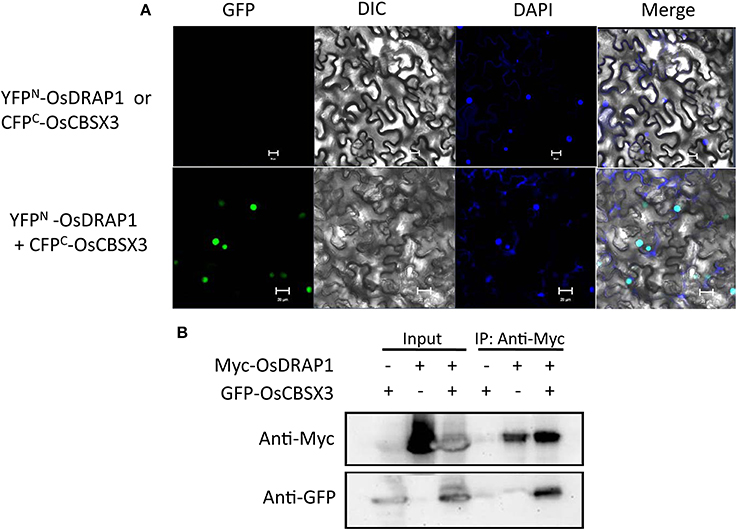
Figure 7. Evidence of the interaction of OsDRAP1 with OsCBSX3. (A) the BiFC assay showing the interaction of OsDRAP1 with OsCBSX3 in the nuclei of N. benthamiana leaf cells with YFPN-OsDRAP1 or CFPC-OsCBSX3 co-transformed withCFPC-GUS orYFPN-GUSserved as the negative control and scale bars in 10 μm; (B) the Coimmunoprecipitation assays of OsDRAP1 and OsCBSX3 with GFP-OsCBSX3 or Myc-OsDRAP1 used as the negative control.
In plants, many DREB1 genes are known to be responsive to abiotic stresses and have been characterized, but only a few DREB2 genes have been functionally elucidated (Lata and Prasad, 2011). In this study, we have demonstrated that OsDRAP1 in rice encodes a DREB2-like protein and functioned, as one of the DREB proteins in the AP2/ERF sub-family and plays an important role involved in several signal transduction pathways in response to different abiotic stresses, including drought, salt, cold and H2O2 stresses. OsDRAP1 was previously characterized as a drought stress-induced AP2/EREBP gene (Wang D. et al., 2011). In this study, OsDRAP1 was proved as a nuclear-localized protein and possessed transactivation activity which involved in ABA and JA transcriptional regulation, and OsDRAP1 could activate the transcription of downstream genes by binding GCC box and DRE elements. The unique spatial expression pattern of OsDRAP1 in vascular tissues and its effects causing reduced cell number and diameter of vascular bundle in RNAi plants suggest that OsDRAP1 is involved in vascular bundle development. Although the vascular tissue is responsible the transport of water and nutrients in plants (Campbell and Turner, 2017), how vascular tissue responds to drought stress remains to be elucidated at the molecular level.
The presence of many different functional motifs in the promoter region of OsDRAP1 suggests that OsDRAP1 functions by mediating many downstream genes and pathways. Indeed, we were able to detect many genes functioning in response to stress and metabolic process were differentially expressed in the OsDRAP1 OE lines, and most of these genes have GCC box and DRE elements in their promoters regulated by OsDRAP1 (Cheng et al., 2013). Those genes also included a few TF genes such as OsEATB, OsERF5, OsMYB4 and OsWRKY89 that were previously reported to be involved in plant development or abiotic stress tolerance (Vannini et al., 2007; Wang et al., 2007; Zhou et al., 2010; Qi et al., 2011). Clearly, the OsDRAP1 mediated DT genetic pathways underlying abiotic stress tolerances are involved in complex synergistic relationships with these TFs. Other important DT genes downstream of OsDRAP1 included CSLA11 that encodes a cellulose synthase-like family A protein. The CSLA gene family is reported to be involved in cell wall biogenesis (Wang et al., 2010; Liepman and Cavalier, 2012) and increased expression of CSLA11 which might explain the strong effect of OsDRAP1 on vascular development. We noted that several other genes the JA signaling pathway were strongly activated by OsDRAP1 in the OE line, including OsJAZ8, OsJAZ13, Jasmonate-zim-domain protein 1 (LOC_Os03g08330) and Jasmonate O-methyltransferase (LOC_Os06g21820), implying that JA was involved in coordinate regulation of downstream genes in the OsDRAP1 mediated drought stress responses, as reported previously (Ahmad et al., 2016). Other genes interacting with OsDRAP1 included OsCBSX3/OsBi1 (LOC_Os02g57280) and a CBS (cystathionine beta-synthase) domain containing protein (CDCPs). OsCBSX3/OsBi1 was previously reported to be induced in rice by herbivore feeding and water deficit stress (Wang et al., 2004) and have a positive regulating role in rice resistance to M. oryzae in JA-mediated signaling pathways (Mou et al., 2015). CDCPs were reported to play an important role in plant responses to abiotic stresses by regulating many enzymes and maintaining the intracellular redox balance (Kushwaha et al., 2009; Yoo et al., 2011; Singh et al., 2012). OsCBSX3 might interact with OsDRAP1 and function positively in DT by maintaining the redox homeostasis under drought stress, however, the detailed molecular mechanisms how OsCBSX3 is involved in the OsDRAP1-mediated DT genetic pathways remains to be elucidated.
Moreover, our results indicated that like many other plant DREB proteins, overexpression of OsDRAP1 enhanced rice tolerance to drought and salinity, while knockdown of OsDRAP1 lead to more sensitivity to drought in addition to significant negative effects on rice development and grain yield, as reported previously in overexpression experiments of other plant DREB genes (Kasuga et al., 1999, 2004; Ito et al., 2006). Clearly, this kind of fitness costs was much more severe in the OsDRAP1-knockdown plants than its constitutive overexpression-plants. This suggests it may be difficult to improve rice DT or salt tolerance by transgenic overexpression of OsDRAP1. Therefore, efficient utilization of OsDRAP1 for improving rice tolerance to drought and salt requires more complete understanding of the OsDRAP1 mediated gene networks and pathways and their related phenotypic effects.
OsDRAP1 is a DREB2-like TF gene which affects DT by maintaining water balance and redox homeostasis in rice under water deficit conditions. OsDRAP1 appears to be involved in the regulation of the development of the vascular tissues. Constitutive overexpression of OsDRAP1 could improve drought stress tolerance by affecting downstream genes with GCC and DRE cis-elements and synergistically regulating several TF genes including OsERF5, OsMYB4, and OsWRKY89 and those related to JA pathways (Figure 8). The interaction protein OsCBSX3 could participate in the OsDRAP1-mediated regulation network in drought stress tolerance in rice.
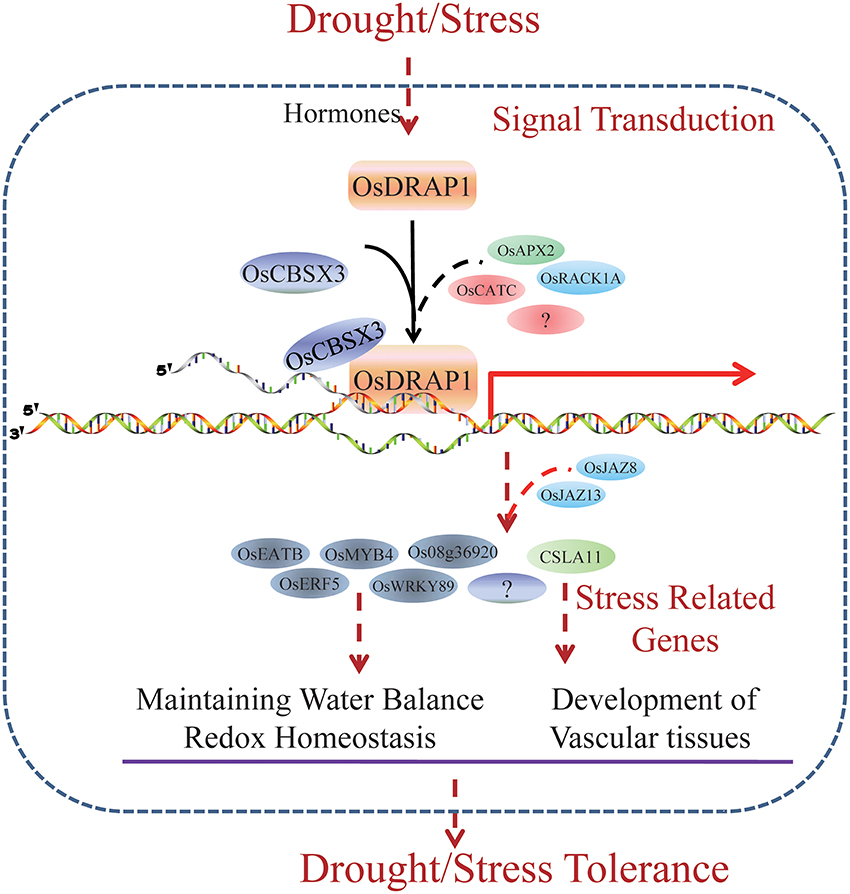
Figure 8. A model for OsDRAP1-Mediated Stress Tolerance. Through a set of plant hormones pathways, the rice feels the stress signal and induce the expression of OsDRAP1. Interacting with other stress related proteins, including OsCBSX3, OsRACK1A, OsRZFP34, OsAPX2, and OsCATC, OsDRAP1 activate stress responsive genes which regulate water balance, redox homeostasis and vascular development directly or indirectly. Several stress responsive TFs (OsEATB, OsERF5, OsMYB4, and OsWRKY89) and a gene encoding cellulose synthase-like family A (CSLA11) induced by OsDRAP1 complex are likely to mediated by JA signaling pathway (OsJAZ8, OsJAZ13, Jasmonate-zim-domain protein 1, and Jasmonate O-methyltransferase). OsDRAP1 mediate stress tolerance especially DT via its regulation of these stress responding genes.
LH and YW generated the transgenic materials, designed and performed the experiments, WW and ZcL analyzed RNA-Seq data, FS and QQ provided assistance on vector construction, XZ, FH, and YZ provided assistance on the drought phenotyping experiment, BF and ZL designed the experiments and wrote the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31571631 and 31501291), the Shenzhen Peacock Plan (20130415095710361), the CAAS Innovative Team Award, and the Bill & Melinda Gates Foundation (OPP1130530).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00094/full#supplementary-material
Figure S1. The phylogenetic analysis of the DREB proteins in plants. The maximum parsimony tree of the DREB proteins from various plants (A) and rice (B), based on the sequence database in UniProt Knowledgebase (UniProtKB): Arabidopsis thaliana, At; Oryza sativa, Os; Zea mays, Zm; Phyllostachysedulis, Pe; Helianthus annuus, Ha; Oryzabrachyantha, Ob; Aegilopsbiuncialis, Ab; Nicotianabenthamiana, Nb; Poapratensis, Pp; Triticumaestivum, Ta; Glycine max, Gm; Solanumlycopersicum, Sl; Festucaarundinacea, Fa.
Figure S2. Expression analysis of OsDRAP1 in the transgenic rice plants. With red and green columns indicating OsDRAP1 overexpression (OsDRAP1-OE-1~13) and OsDRAP1 RNAi (OsDRAP1-RNAi-1~4) in transgenic lines, respectively, the error bars indicating SD based on data of 3 replicates, Actin 1 used as the endogenous control and Arrows indicating the two lines (OE-7 and RNAi-2) that were used for further functional analysis.
Figure S3. The negative effect on the plants overexpressing and knock-down of OsDRAP1. (A) The growth of OsDRAP1-OE-7, WT and OsDRAP1-RNAi-2 plants. Investigation of plant height (B), number of tillering (C), root performance (D), fertility (E), and grain yield (F) of the transgenic lines. The asterisk indicated the significant difference in t-test (p < 0.05 vs. WT, n > 10).
Table S1. List of the differentially expressed gene in the OsDRAP1 overexpression line compared with WT under normal growth conditions.
Table S2. List of the differentially expressed gene in OsDRAP1 overexpression line compared with WT under drought stress conditions.
Table S3. List of interacting proteins of OsDRAP1 identified by yeast two-hybridization screening.
Ahmad, P., Rasool, S., Gul, A., Sheikh, S. A., Akram, N. A., Ashraf, M., et al. (2016). Jasmonates: Multifunctional Roles in Stress Tolerance. Front. Plant Sci. 7:813. doi: 10.3389/fpls.2016.00813
Arora, R., Pitchay, D. S., and Bearce, B. C. (1998). Water-stress-induced heat tolerance in geranium leaf tissues: a possible linkage through stress proteins? Plant Physiol. 103, 24–34. doi: 10.1034/j.1399-3054.1998.1030104.x
Babu, R. C. (2010). Breeding for drought resistance in rice: an integrated view from physiology to genomics. Electron. J. Plant Breed. 1, 1133–1141.
Bart, R., Chern, M., Park, C. J., Bartley, L., and Ronald, P. C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2:13. doi: 10.1186/1746-4811-2-13
Bonnecarrère, V., Borsani, O., Díaz, P., Capdevielle, F., Blanco, P., and Monza, J. (2011). Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci. 180, 726–732. doi: 10.1016/j.plantsci.2011.01.023
Campbell, L., and Turner, S. (2017). Regulation of vascular cell division. J. Exp. Bot. 68, 27–43. doi: 10.1093/jxb/erw448
Chen, J. Q., Meng, X. P., Zhang, Y., Xia, M., and Wang, X. P. (2008). Over- expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 30, 2191–2198. doi: 10.1007/s10529-008-9811-5
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Chien, C. T., Bartel, P. L., Sternglanz, R., and Fields, S. (1991). The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Nati. Acad. Sci. U.S.A. 88, 9578–9582. doi: 10.1073/pnas.88.21.9578
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Curtis, M. D., and Grossniklaus, U. (2003). A Gateway TM cloning vector set for high-throughput functional analysis of genes in plants. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, 64–70. doi: 10.1093/nar/gkq310
Duan, Y., Zhai, C., Li, H., Li, J., Mei, W., Gui, H., et al. (2012). An efficient and high-throughput protocol for Agrobacterium-mediated transformation based on phosphomannoseisomerase positive selection in Japonica rice (Oryza sativa L.). Plant Cell Rep. 31, 1611–1624. doi: 10.1007/s00299-012-1275-3
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245–246. doi: 10.1038/340245a0
Hadiarto, T., and Tran, L. S. (2011). Progress studies of drought-responsive genes in rice. Plant Cell Rep. 30, 297–310. doi: 10.1007/s00299-010-0956-z
Hao, D., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273, 26857–26861. doi: 10.1074/jbc.273.41.26857
Hsu, K. H., Liu, C. C., Wu, S. J., Kuo, Y. Y., Lu, C. A., Wu, C. R., et al. (2014). Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 86, 125–137. doi: 10.1007/s11103-014-0217-6
Huang, L., Zhang, F., Zhang, F., Wang, W., Zhou, Y., Fu, B., et al. (2014). Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics 15:1026. doi: 10.1186/1471-2164-15-1026
Ito, Y., Katsura, K., and Maruyama, K. (2006). Functional analysis of rice DREB1/CBF type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning and expression of tagged fluorescent protein in plant cells. Trends Plant Sci. 10, 103–105. doi: 10.1016/j.tplants.2005.01.008
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of Arabidopsis DREB1A gene and stress inducible rd29Apromoter improved drought and low temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Kumar, A., Dixit, S., Ram, T., Yadaw, R. B., Mishra, K. K., and Mandal, N. P. (2014). Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J. Exp. Bot. 65, 6265–6278. doi: 10.1093/jxb/eru363
Kushwaha, H. R., Singh, A. K., Sopory, S. K., Singla-Pareek, S. L., and Pareek, A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 10:200. doi: 10.1186/1471-2164-10-200
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lenka, S. K., Katiyar, A., Chinnusamy, V., and Bansal, K. C. (2011). Comparative analysis of drought-responsive transcriptome in indica rice genotypes with contrasting drought tolerance. Plant Biotechnol. J. 9, 315–327. doi: 10.1111/j.1467-7652.2010.00560.x
Li, J., Han, Y., Liu, L., Chen, Y., Du, Y., Zhang, J., et al. (2015). qRT9, a quantitative trait locus controlling root thickness and root length in upland rice. J. Exp. Bot. 66, 2723–2732. doi: 10.1093/jxb/erv076
Li, Z. K., Fu, B. Y., Gao, Y. M., Xu, J. L., Ali, J., Lafitte, H. R., et al. (2005). Genome-wide introgression lines and their use in genetic and molecular dissection of complex phenotypes in rice (Oryza sativa L.). Plant Mol. Biol. 59, 33–52. doi: 10.1007/s11103-005-8519-3
Liepman, A. H., and Cavalier, D. M. (2012). The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front. Plant Sci. 3:109. doi: 10.3389/fpls.2012.00109
Lin, A., Wang, Y., Tang, J., Xue, P., Li, C., Liu, L., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. doi: 10.1104/pp.111.184531
Liu, C. W., Fukumoto, T., Matsumoto, T., Gena, P., Frascaria, D., Kaneko, T., et al. (2013). Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 63, 151–158. doi: 10.1016/j.plaphy.2012.11.018
Liu, H. T., Yu, X. H., Li, K. W., Klejnot, J., Yang, H. Y., Lisiero, D., et al. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. doi: 10.1126/science.1163927
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−DeltaDeltaC(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mou, S., Shi, L., Lin, W., Liu, Y., Shen, L., Guan, D., et al. (2015). Over-expression of rice CBS domain containing protein, OsCBSX3, confers rice resistance to Magnaporthe oryzae inoculation. Int. J. Mol. Sci. 16, 15903–15917. doi: 10.3390/ijms160715903
Pandey, S. (2009). Economic costs of drought and rice farmers' coping mechanisms. Int. Rice Res. Notes 1, 5–11. doi: 10.3860/irrn.v32i1.1078
Peng, H., ·Zhang, Q., ·Li, Y. D., Lei, C. L., Zhai, Y., Sun, X. H., et al. (2009). A putative leucine-rich repeat receptor kinase, OsBRR1, is involved in rice blast resistance. Planta 230, 377–385. doi: 10.1007/s00425-009-0951-1
Qi, W., Sun, F., Wang, Q., Chen, M., Huang, Y., Feng, Y., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646.
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem.Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Sharoni, A. M., Nuruzzaman, M., Satoh, K., Shimizu, T., Kondoh, H., Sasaya, T., et al. (2011). Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 52, 344–360. doi: 10.1093/pcp/pcq196
Shukla, N., Awasthi, R. P., Rawat, L., and Kumar, J. (2012). Biochemical and physiological responses of rice (Oryza sativa L.) as influenced by Trichodermaharzianum under drought stress. Plant Physiol. Biochem. 54, 78–88. doi: 10.1016/j.plaphy.2012.02.001
Singh, A. K., Kumar, R., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2012). Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 52, 205–216. doi: 10.1007/s12033-011-9487-2
Singh, A. P., Pandey, B. K., Deveshwar, P., Narnoliya, L., Parida, S. K., and Giri, J. (2015). JAZ repressors: potential involvement in nutrients deficiency response in rice and chickpea. Front. Plant Sci. 6:975. doi: 10.3389/fpls.2015.00975
Sparkes, I. A., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Srivastav, A., Mehta, S., Lindlof, A., and Bhargava, S. (2010). Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal. Behav. 5, 775–784. doi: 10.4161/psb.5.7.11769
Takemoto, Y., Coughlan, S. J., Okita, T. W., Satoh, H., Ogawa, M., and Kumamaru, T. (2002). The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128, 1212–1222. doi: 10.1104/pp.010624
Todaka, D., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6:84. doi: 10.3389/fpls.2015.00084
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Vannini, C., Campa, M., Iriti, M., Genga, A., Faoro, F., Carravieri, S., et al. (2007). Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci. 173, 231–239. doi: 10.1016/j.plantsci.2007.05.007
Wang, D., Pan, Y., Zhao, X., Zhu, L., Fu, B., and Li, Z. (2011). Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics. 12:149. doi: 10.1186/1471-2164-12-149
Wang, H., Hao, J., Chen, X., Hao, Z., Wang, X., Lou, U., et al. (2007). Overexpression of rice WRKY89 enhances ultravioletB tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wang, L., Guo, K., Li, Y., Tu, Y., Hu, H., Wang, B., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Wang, Q., Guan, Y., Wu, Y., Chen, H., Chen, F., and Chu, C. (2008). Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 67, 589–602. doi: 10.1007/s11103-008-9340-6
Wang, W. S., Pan, Y. J., Zhao, X. Q., Dwivedi, D., Zhu, L. H., Ali, J., et al. (2011). Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 62, 1951–1960. doi: 10.1093/jxb/erq391
Wang, X., Ren, X., Zhu, L., and He, G. (2004). OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Sci. 166, 1581–1588. doi: 10.1016/j.plantsci.2004.02.011
Xiao, C., Chen, F., Yu, X., Lin, C., and Fu, Y. F. (2009). Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 71, 39–50. doi: 10.1007/s11103-009-9507-9
Xu, Z., and Vision, T. J. (2005). Improving quantitative trait loci mapping resolution in experimental crosses by the use of genotypically selected samples. Genetics 170, 401–408. doi: 10.1534/genetics.104.033746
Yamada, S., Kano, A., Tamaoki, D., Miyamoto, A., Shishido, H., Miyoshi, S., et al. (2012). Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 53, 2060–2072. doi: 10.1093/pcp/pcs145
Yoo, K. S., Ok, S. H., Jeong, B. C., Jung, K. W., Cui, M. H., Hyoung, S., et al. (2011). Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 23, 3577–3594. doi: 10.1105/tpc.111.089847
You, J., Zong, W., Li, X. K., Ning, J., Hu, H. H., Li, X. H., et al. (2013). The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 64, 569–583. doi: 10.1093/jxb/ers349
Zhang, D., Chen, L., Li, D., Lv, B., Chen, Y., Chen, J., et al. (2014). OsRACK1 is involved in abscisic acid- and H2O2- mediated signaling to regulate seed germination in rice (Oryza sativa, L.). PLoS ONE 9:e97120. doi: 10.1371/journal.pone.0097120
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8:e57472. doi: 10.1371/journal.pone.0057472
Zhou, Y. L., Xu, M. R., Zhao, M. F., Xie, X. W., Zhu, L. H., Fu, B. Y., et al. (2010). Genome-wide gene responses in a transgenic rice line carrying the maize resistance gene Rxo1 to the rice bacterial streak pathogen, Xanthomonas oryzae pv. oryzicola. BMC Genomics 11:78. doi: 10.1186/1471-2164-11-78
Keywords: DREB, drought tolerance, OsDRAP1, overexpression, rice, transcription factor
Citation: Huang L, Wang Y, Wang W, Zhao X, Qin Q, Sun F, Hu F, Zhao Y, Li Z, Fu B and Li Z (2018) Characterization of Transcription Factor Gene OsDRAP1 Conferring Drought Tolerance in Rice. Front. Plant Sci. 9:94. doi: 10.3389/fpls.2018.00094
Received: 11 December 2017; Accepted: 17 January 2018;
Published: 01 February 2018.
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Lizhong Xiong, Huazhong Agricultural University, ChinaCopyright © 2018 Huang, Wang, Wang, Zhao, Qin, Sun, Hu, Zhao, Li, Fu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binying Fu, ZnViaW55aW5nQGNhYXMuY24=
Zhikang Li, bGl6aGlrYW5nQGNhYXMuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.