
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 January 2018
Sec. Agroecology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00029
This article is part of the Research TopicHerbicide resistance in weeds: early detection, mechanisms, dispersal, new insights and management issuesView all 29 articles
 Anita Küpper1
Anita Küpper1 Harish K. Manmathan2
Harish K. Manmathan2 Darci Giacomini3
Darci Giacomini3 Eric L. Patterson1
Eric L. Patterson1 William B. McCloskey4
William B. McCloskey4 Todd A. Gaines1*
Todd A. Gaines1*Palmer amaranth (Amaranthus palmeri) is a major weed in United States cotton and soybean production systems. Originally native to the Southwest, the species has spread throughout the country. In 2004 a population of A. palmeri was identified with resistance to glyphosate, a herbicide heavily relied on in modern no-tillage and transgenic glyphosate-resistant (GR) crop systems. This project aims to determine the degree of genetic relatedness among eight different populations of GR and glyphosate-susceptible (GS) A. palmeri from various geographic regions in the United States by analyzing patterns of phylogeography and diversity to ascertain whether resistance evolved independently or spread from outside to an Arizona locality (AZ-R). Shikimic acid accumulation and EPSPS genomic copy assays confirmed resistance or susceptibility. With a set of 1,351 single nucleotide polymorphisms (SNPs), discovered by genotyping-by-sequencing (GBS), UPGMA phylogenetic analysis, principal component analysis, Bayesian model-based clustering, and pairwise comparisons of genetic distances were conducted. A GR population from Tennessee and two GS populations from Georgia and Arizona were identified as genetically distinct while the remaining GS populations from Kansas, Arizona, and Nebraska clustered together with two GR populations from Arizona and Georgia. Within the latter group, AZ-R was most closely related to the GS populations from Kansas and Arizona followed by the GR population from Georgia. GR populations from Georgia and Tennessee were genetically distinct from each other. No isolation by distance was detected and A. palmeri was revealed to be a species with high genetic diversity. The data suggest the following two possible scenarios: either glyphosate resistance was introduced to the Arizona locality from the east, or resistance evolved independently in Arizona. Glyphosate resistance in the Georgia and Tennessee localities most likely evolved separately. Thus, modern farmers need to continue to diversify weed management practices and prevent seed dispersal to mitigate herbicide resistance evolution in A. palmeri.
Since the introduction of transgenic soybean, corn, and cotton in the mid-1990s, herbicide-resistant varieties of these crops have largely replaced conventional varieties in the United States (Coupe and Capel, 2016). In 1996, glyphosate-resistant (GR) (Roundup Ready) crops were commercialized and as a result global glyphosate usage rose by about 15-fold (Benbrook, 2016), dominating the current herbicide market (Duke, 2017). The widespread reliance on glyphosate to the exclusion of all other weed control methods has resulted in high selection pressure and the evolution of GR weeds, including Palmer amaranth (Amaranthus palmeri S. Wats.) (Culpepper et al., 2006), which is now a major threat to many U.S. food production systems (Beckie, 2011).
Amaranthus palmeri is a dioecious, annual species with prolific seed production, pollen-mediated gene flow due to obligate outcrossing, and high genetic variability (Franssen et al., 2001; Sellers et al., 2003; Ward et al., 2013). As a member of the Amaranthaceae family, A. palmeri is native to the southwestern United States and northwestern Mexico, having first been documented in Sonora, California, Arizona, New Mexico, and Texas in the late 19th century. During the early 20th century, the species started to spread east and northeast, probably because of human mediated seed dispersal (Sauer, 1957; Ward et al., 2013). In recent years, A. palmeri has expanded its distribution as far north as Ontario, Canada and as far east as Massachusetts, United States (Kartescz, 2014). The species made its first occurrence on the annual listing of most troublesome weeds in South Carolina in 1989 (Webster and Coble, 1997). By 2009 the weed was ranked the most troublesome weed in cotton in the Southern United States (Webster and Nichols, 2012; Ward et al., 2013).
Resistance to glyphosate in A. palmeri was first reported from a GR cotton field in Georgia in 2004. Shortly after, another case was reported from North Carolina in 2005 (Culpepper et al., 2006, 2008). As of 2017, GR A. palmeri was found in 27 U.S. states, Argentina, and Brazil (Scott et al., 2007; Norsworthy et al., 2008; Steckel et al., 2008; Berger et al., 2016; Heap, 2017; Küpper et al., 2017). The primary mechanism of glyphosate resistance in A. palmeri has been identified as the amplification of the gene encoding the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) which produces increased EPSPS transcription and protein activity (Gaines et al., 2010). The same glyphosate resistance mechanism has independently evolved in six other species (Salas et al., 2012; Jugulam et al., 2014; Lorentz et al., 2014; Chatham et al., 2015; Chen et al., 2015; Wiersma et al., 2015; Malone et al., 2016; Ngo et al., 2017). EPSPS gene amplification has also transferred via pollen-mediated inter-specific hybridization from A. palmeri to A. spinosus (Nandula et al., 2014).
Evolutionary models have identified that herbicide resistance dynamics are largely influenced by gene flow, seed immigration, and fitness cost (Maxwell et al., 1990). Further factors include mutation rate, the mode of inheritance, dominance of the resistance trait, seed bank turnover rate, herbicide chemistry and persistence, as well as herbicide usage patterns (Georghiou and Taylor, 1986; Jasieniuk et al., 1996; Neve, 2008). For instance, glyphosate used prior to crop emergence is predicted to have a low risk of resistance evolution while post-emergence use increases the risk, and reliance on glyphosate exclusively increases the risk even further (Neve, 2008). A simulation model for A. palmeri predicted that five applications of glyphosate each year with no other herbicides would result in resistance evolving in 74% of the simulated populations (Neve et al., 2011).
Amaranthus palmeri management is complicated by the fact that this species evolved resistance to five different modes of action (Chahal et al., 2015; Heap, 2017; Nakka et al., 2017; Schwartz-Lazaro et al., 2017), the lack of discovery of new modes of action for the past three decades, and the high cost of bringing new herbicides to the market (Duke, 2012). The overuse of and sole reliance on glyphosate and the resulting evolution of resistant weeds exhausted the lifespan of a once-in-a-century herbicide (Duke and Powles, 2008) and threatens current crop production practices by diminishing available weed management options further. Therefore, knowledge about the origin and geographical pathways of glyphosate resistance in A. palmeri, one of the most problematic GR weeds in the United States, is crucial to avoid repeating the same mistakes made with glyphosate with other modes of action that are still successful at controlling weeds in the field.
This study focuses on a GR population identified in a no-till cotton-wheat double crop system near Phoenix, Arizona (AZ), United States. Glyphosate was used as the sole weed management technique for the cotton portion of the production cycle for more than 10 year before glyphosate resistance was first suspected in 2012, 8 years after the first report in the species. The objective was to determine whether GR A. palmeri immigrated to the AZ locality from an outside location via seed or pollen-mediated gene flow, or if resistance evolved at or nearby the location in AZ independently via parallel evolution. To answer this question, single nucleotide polymorphisms (SNPs) generated by genotyping-by-sequencing (GBS) to identify numerous sequence differences at presumably random parts of the genome (Brumfield et al., 2003), were used. The GR population from AZ and seven other populations from different locations in the United States were investigated for their degree of genetic relatedness to identify patterns of phylogeography and variation on an intraspecific level.
Twelve A. palmeri individuals (six males and six females) from each of eight different locations in the United States were used for the analyses (Table 1), except for AZ-S2 for which only eleven individuals were used to leave a blank on the plate. Locations AZ-S1, AZ-S2, KS-S, GA-S and NE-S were verified as glyphosate-susceptible (GS) and locations AZ-R (Molin et al., 2017b), GA-R (Culpepper et al., 2006), and TN-R (Steckel et al., 2008) were verified as GR. The populations were collected between 2004 and 2012, except for AZ-S2 which was maintained by the USDA-ARS Germplasm Resource Information Network (accession number: Ames 5370) since its collection in 1981 and serves as an outgroup to prevent ascertainment bias (Wakeley et al., 2001; Akey et al., 2003). AZ-S1 was collected about 240 km southeast of Buckeye, AZ (AZ-R) where no agronomic crop production has occurred since the 1960s to provide a recently collected Arizona-native GS population that is fairly sympatric with AZ-R.
For DNA extraction, young leaf tissue was collected, immediately frozen in liquid nitrogen, and stored at -80°C. For GR samples only individuals that survived 800 g a.e. ha-1 glyphosate (Roundup WeatherMAX, Monsanto) were used. DNA extraction was performed following a modified cetyltrimethylammonium bromide (CTAB) extraction protocol (Doyle, 1991; Küpper et al., 2017) and quantified on a NanoDrop spectrophotometer (Thermo Scientific) followed by normalization. Gel electrophoresis and enzyme digestion with HindIII (Thermo Scientific) were performed on all or 10% of the samples, respectively, to confirm DNA quality and normalization.
To confirm glyphosate resistance and susceptibility for the individuals used for GBS, an in vivo shikimate accumulation assay with excised leaf tissue (Shaner et al., 2005) was conducted. Additionally, EPSPS gene copy number was determined for all samples. Four-mm leaf disks from each individual were exposed to glyphosate at 0, 100, 500, and 1000 μm glyphosate for 16 h. Shikimate accumulation was measured on a spectrophotometer (Synergy 2 Multi-Mode Reader, BioTek). A shikimate standard curve was used to calculate the ng shikimate μl-1 accumulation above the background level. Each biological sample was run in three technical replicates for each dose.
For EPSPS gene copy number determination, DNA concentrations were adjusted to 5 ng μl-1 and primer sets (ALSF2 and ALSR2, EPSF1 and EPSR8) and qPCR conditions were used as previously described (Gaines et al., 2010). Quantitative PCR was performed using SYBR green master-mix (BioRad) on a CFX ConnectTM Real-Time PCR Detection System (BioRad). EPSPS gene copy number relative to ALS was determined using the 2ΔCT method where ΔCT = CT(ALS) - CT(EPSPS). Each biological sample was run in three technical replicates.
Greenhouse dose response studies were conducted to confirm pyrithiobac-sodium [acetolactate synthase (ALS inhibitor)] resistance in AZ-R with AZ-S1 as a susceptible control. The experiments took place at the University of Arizona Campus Agricultural Center in Tucson, AZ, United States. Seeds were planted in artificial soil mix in 10 cm pots and after emergence seedlings were thinned, fertilized, and irrigated as needed. ALS-inhibitor treatments included 0, 0.0001, 0.0005, 0.001, 0.002, 0.005, 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, and 1 kg a.i. ha-1 pyrithiobac-sodium (Staple LX, DuPont) with 0.25% v/v non-ionic surfactant (Activator 90, Loveland Products). Plants were sprayed at the six-leaf stage using a CO2 pressurized backpack sprayer equipped with a three nozzle (TeeJet XR8001VS) boom delivering a carrier volume of 112 L ha-1 at 172 kPa at 4 km h-1. The experimental design was random with five replications per dose. Above-ground biomass was harvested 27 days after treatment (DAT), dried at 60°C and dry weight was measured.
The ALS gene was sequenced from three individuals each of the AZ-R, GA-R, and TN-R populations using the same DNA used for the EPSPS copy number test and SNP calling. ALS gene sequencing was conducted as previously described (Küpper et al., 2017).
After DNA extraction, GBS and bi-allelic SNP calling was conducted by the Biotechnology Resource Center at Cornell University, Ithaca, NY, United States (Elshire et al., 2011). A total of 95 samples (eleven samples for AZ-S2 and twelve samples for the remaining populations) were digested with ApeKI, individually barcoded, run on an Illumina HiSeq2500 single-end 100 bp sequencing lane, and later trimmed to 64 bp for analysis. The GBS UNEAK pipeline in TASSEL v. 3.0.173 (Bradbury et al., 2007; Lu et al., 2013; Glaubitz et al., 2014) was used for de novo clustering of the sequences. The resulting SNP calls were then filtered for depth and missing values at any given locus with VCFtools v. 0.1.11 (Danecek et al., 2011) after which 4,566 filtered SNPs remained. Through further pruning, 70.4% of the filtered SNPs were excluded due to percentage of missing data points (>5%), minor allele frequency (MAF) values lower than 0.05, or more than 80% loci with more than one allele, leaving 1,351 SNPs which were informative (Supplementary Information Figure 2). Except where indicated, all analyses were performed on the panel of 1,351 SNPs.
EPSPS gene copies in GR A. palmeri individuals are randomly dispersed throughout the whole genome (Gaines et al., 2010). They can be found embedded in a complex array of repetitive elements and putative helitron sequences referred to as the ‘EPSPS cassette’ (Molin et al., 2017a). Because SNPs are called genome-wide, an overrepresentation of called SNPs within these sequences could potentially lead to clustering of GR individuals regardless of their actual genetic relatedness. To avoid such bias, the sequences flanking the 1,351 SNPs were aligned to the A. palmeri 1,044 bp EPSPS sequence (Gaines et al., 2010) and the 297,445 bp A. palmeri EPSPS cassette (Molin et al., 2017a). The 1,351 SNP sequences were also aligned to the chloroplast genome of spinach (Spinacia oleracea) and the mitochondrial genome of sugar beet (Beta vulgaris) to identify SNPs specific to the cytoplasmic regions.
The putative population genetic structure was explored using the model-based Bayesian analysis implemented in STRUCTURE v2.3.4 (Pritchard et al., 2000). The number of sub-populations K in the dataset was determined by the averaged likelihood at each K [In Pr (X|K) or In (Kn)] and the variance between replicates was determined by running a continuous series of K = 1–15 to determine the optimal number of populations present within the 95 individuals. The analysis was carried out using a burn-in of 30,000 iterations and a run length of 100,000 Markov Chain Monte Carlo (MCMC) replications in ten independent runs. Prior knowledge about the number of populations was not included. The optimum number of clusters was predicted following the ad hoc statistic ΔK (Evanno et al., 2005) using Structure Harvester v0.6.94 (Earl, 2012). For the final K analysis a burn-in of 30,000 with a run length of 500,000 MCMC replications and 20 independent runs were used. To be conservative, the analyses were run assuming admixture and correlated allele frequencies (Porras-Hurtado et al., 2013). The Greedy algorithm by CLUMPP v1.1.2 (Jakobsson and Rosenberg, 2007) was used to obtain the individual and cluster membership coefficient matrices over the 20 runs which were then plotted using distruct 1.1 (Rosenberg, 2004).
The following information and tests were calculated in R v3.4.1. The number of alleles (Na) and allelic richness (AR) per population were calculated using the package ‘PopGenReport.’ Observed (HO) and expected heterozygosity (HE) were calculated with ‘adegenet’ (Jombart and Ahmed, 2011; Adamack and Gruber, 2014). The inbreeding coefficient (FIS) was calculated following the formula 1 - (Ho/HE). Principal component analysis (PCA) was conducted using ‘SNPRelate’ and ‘gdsfmt’ (Zheng et al., 2012). Calculations for Nei’s distance (DST) (Nei, 1972) and pairwise fixation index (FST) among populations were performed with 1,000 bootstrap replications using ‘StAMPP’ (Pembleton et al., 2013). The analysis of molecular variance (AMOVA) (10,000 permutations) and the Mantel test (10,000 permutations) for isolation by distance analysis were performed using ‘poppr’ (Kamvar et al., 2014) and ‘adegenet’ (Dray and Dufour, 2007), respectively. The phylogenetic analysis was based on the UPGMA clustering method using the Hasegawa-Kishino-Yano (HKY) genetic distance model in the software Geneious v10.0.6.
Glyphosate-susceptible A. palmeri populations showed higher shikimate accumulation (11.8–146.3 ng μl-1 at 500 μm glyphosate) than GR populations (0–3.8 ng μl-1) (Figure 1A) while GR populations showed higher genomic EPSPS copy number (individuals measured from 25- to 250-fold) than GS populations (individuals measured from onefold to twofold) (mean EPSPS copy number shown in Figure 1B). Thus, the mechanism of glyphosate resistance was determined to be EPSPS gene duplication in all the sampled GR populations (Gaines et al., 2010). The average copy numbers for the GR populations were within a similar range (Figure 1B). The 500 μm glyphosate concentration was a clear discriminating dose between GR and GS individuals.
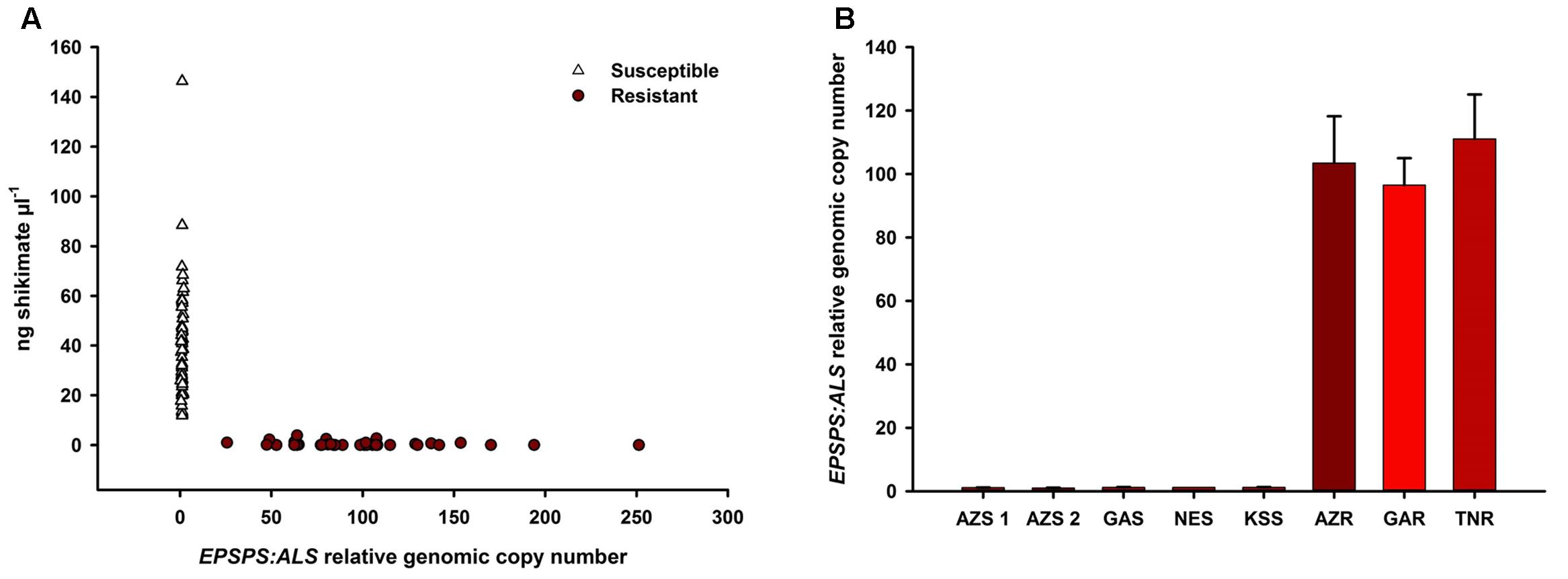
FIGURE 1. Correlation of shikimate accumulation and EPSPS genomic copy number in all individuals of each of the glyphosate-resistant and -susceptible A. palmeri populations. Shikimate accumulation was measured after incubation in 500 μm glyphosate in an in vivo leaf disk assay Increase in genomic copy number of EPSPS is relative to ALS as measured using qPCR on genomic DNA (A). EPSPS genomic copy number by population (B).
Resistance to the ALS-inhibitors, commonly used in cotton, was suspected in AZ-R as well, thus a dose response with pyrithiobac-sodium and sequencing of the ALS gene was conducted. The ED50 values for dry weight (pyrithiobac-sodium dose causing 50% reduction in dry weight) were 6.9 and 1.3 g a.i. ha-1 for AZ-R and AZ-S1, respectively (P = 0.027) (Supplementary Information Figure 1).
Sequencing the ALS gene in three GR individuals each from AZ-R, GA-R and TN-R, showed that one TN and one AZ plant were heterozygous for a mutation from TGG → TTG resulting in an amino acid change from tryptophan to leucine at position 574 (W574L). A different AZ plant was heterozygous for a AGC → AAC mutation resulting in a change from serine to asparagine at position 653 (S653N). No individual carried both mutations within the same allele. The remaining individuals tested showed no mutations at these positions (Supplementary Information Table 1). Both mutations have been reported before in A. palmeri from Mississippi, United States, and Brazil (Molin et al., 2016; Küpper et al., 2017) while only S653N was reported from GA (Berger et al., 2016). The mutation at W574L is known to confer resistance to triazolopyrimidines, sulfonylureas, imidazolinones, and pyrimidinylthio-benzoates (including pyrithiobac-sodium), whereas the S653N mutation confers resistance to imidazolinones and the pyrimidinylthio-benzoates only (McNaughton et al., 2005; Whaley et al., 2006; Patzoldt and Tranel, 2007; Laplante et al., 2009; Yu et al., 2012). Both mutations are known to be inherited as a dominant trait (Tranel and Wright, 2002; Powles and Yu, 2010). It is suspected that a non-target site mechanism conferring resistance to ALS inhibitors exists (Küpper et al., 2017) and such a mechanism may also be present in AZ-R ALS-resistant individuals that lack target-site ALS mutations.
The EPSPS gene was found to have five potential cutting sites for the enzyme ApeKI used in this GBS study, while the entire EPSPS cassette has 289 potential cutting sites. No SNPs were called within the EPSPS gene and only one SNP was called from within the EPSPS cassette which was removed from further analysis. Thus, the mechanism of glyphosate resistance (repetitive EPSPS gene copies) is not expected to influence the analysis of genetic relatedness in this case.
The 1,351 loci used for this study had an average percentage of missing data of 1.07% and an average MAF of 0.159. A high degree of polymorphism (MAF ≥ 0.30) was found in 14.41% of the dataset. The proportion of MAF < 0.1 was 45.37%. One AZ-S2 individual was removed from all future analysis because it was an extreme outlier. The observed number of alleles within a population ranged from 2,017 (AZ-S2) to 2,395 (KS-S), with an average of 2,217. Levels of heterogeneity were compared among populations to examine genetic variability within populations. Allelic richness (AR) ranged from 1.445 (AZ-S2) to 1.654 (KS-S) with an average of 1.560. The observed (HO) and expected heterozygosity (HE) values ranged from 0.161 (AZ-S1) to 0.219 (TN-R) and from 0.163 (AZ-S2) to 0.211 (KS-S/GA-R), respectively, with an average of 0.193. Low values for HO indicate small effective population sizes or population bottlenecks. The HO values in most populations were less than the HE values (Supplementary Information Figure 3), with the exception of GA-S, TN-R and AZ-S2. The inbreeding coefficient (FIS) for these three populations was negative. AZ-R was the population with the highest FIS value (0.121) (Table 2).
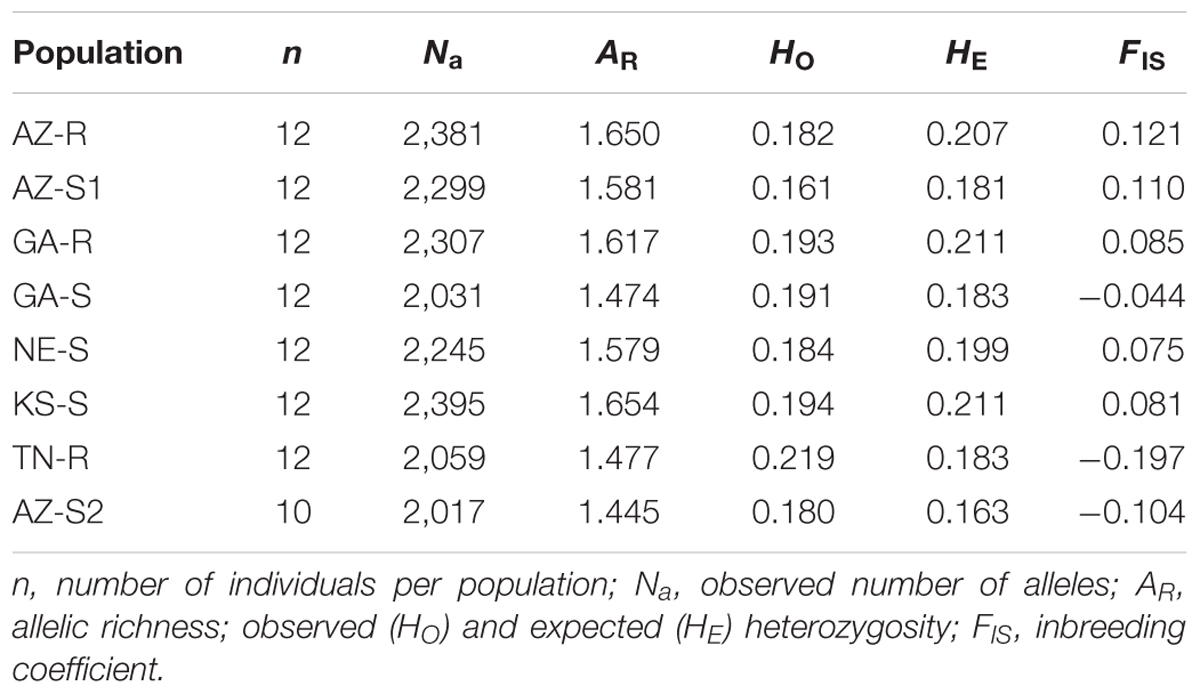
TABLE 2. Population information and genetic variability estimates based on 1,351 SNP loci in eight populations of A. palmeri.
The consensus tree separated GA-S, TN-R, NE-S, and AZ-S2 with over 86% certainty with GAS, TN-R, and AZ-S2 being the most divergent populations. AZ-S1, AZ-R, and KS-S clustered together. Except for KS-S and AZ-R, all individuals clustered within their sampling location (Figure 2), The long branch lengths for the individuals indicate high within-individual genetic variability.
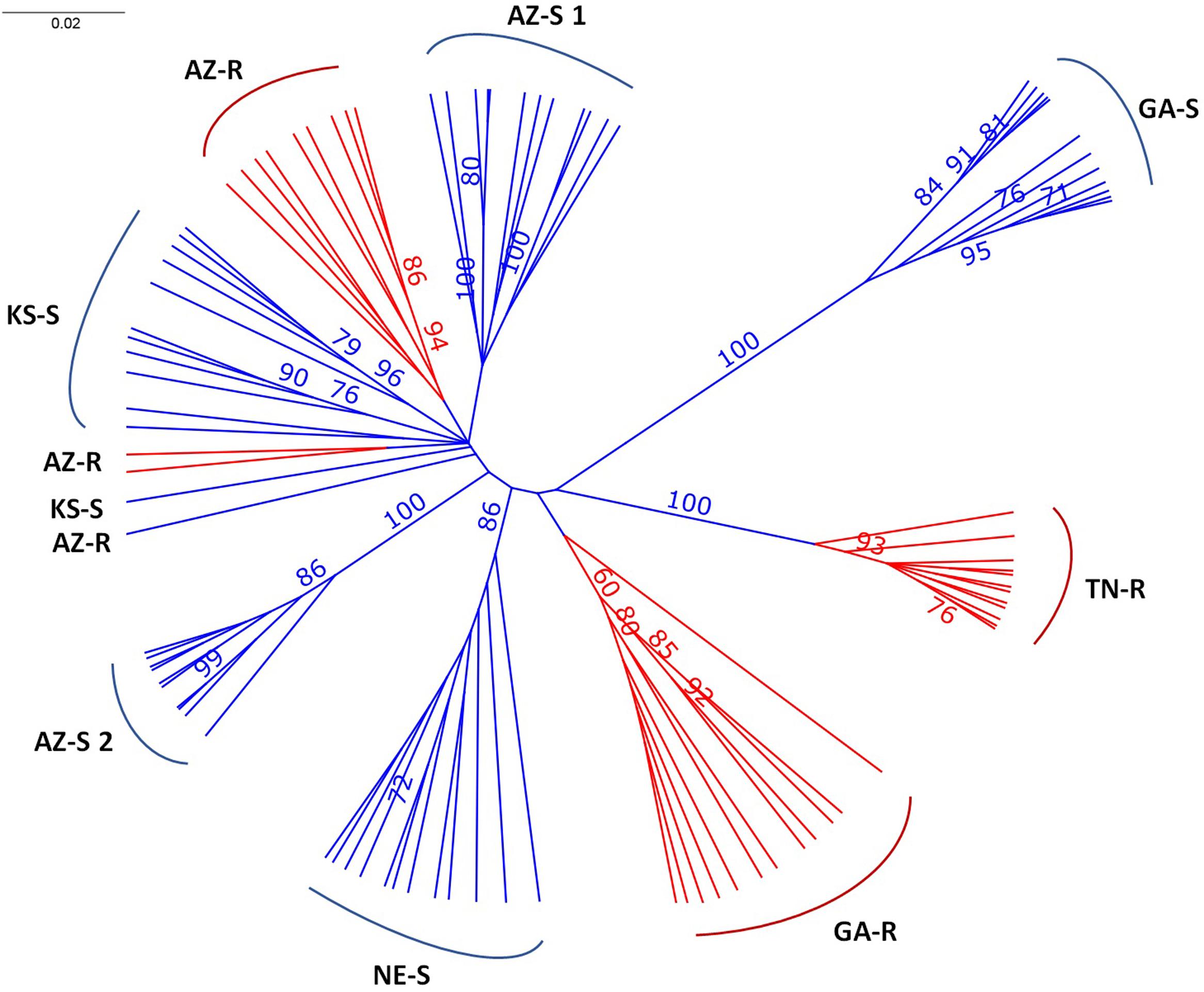
FIGURE 2. Unrooted UPGMA consensus tree after 1,000 bootstrap replications depicting the relationships of A. palmeri individuals from eight populations. Bootstrap values > 70% at nodes are indicated.
To confirm this clustering, a similar pattern of differentiation among populations was constructed using PCA which is used to bring out strong patterns in the dataset based on their variance. The first two principal component (PC) axes cumulatively accounted for 16.69% of the total variation. PCA showed that all individuals clustered according to their collection site. Three distinct outgroups (GA-S, TN-R, and AZ-S2) emerged while the remaining individuals from the other five populations clustered into one group. The first dimension (PC 1) accounted for 8.91% of the variation and roughly separated GR from GS individuals (Figure 3A). After removing GA-S, TN-R, and AZ-S2, AZ-R did not separate from the cluster with KS-S and AZ-S1, while GA-R and NE-S clustered distinctively according to PC 2, supporting the UPGMA consensus tree. PC 1 in the second PCA on the subset of populations accounted for 6.87% of the variation in the dataset and again roughly separated GR from GS individuals. Individuals from the same population occupied different areas of the cluster, which indicates a population substructure (Figure 3B).
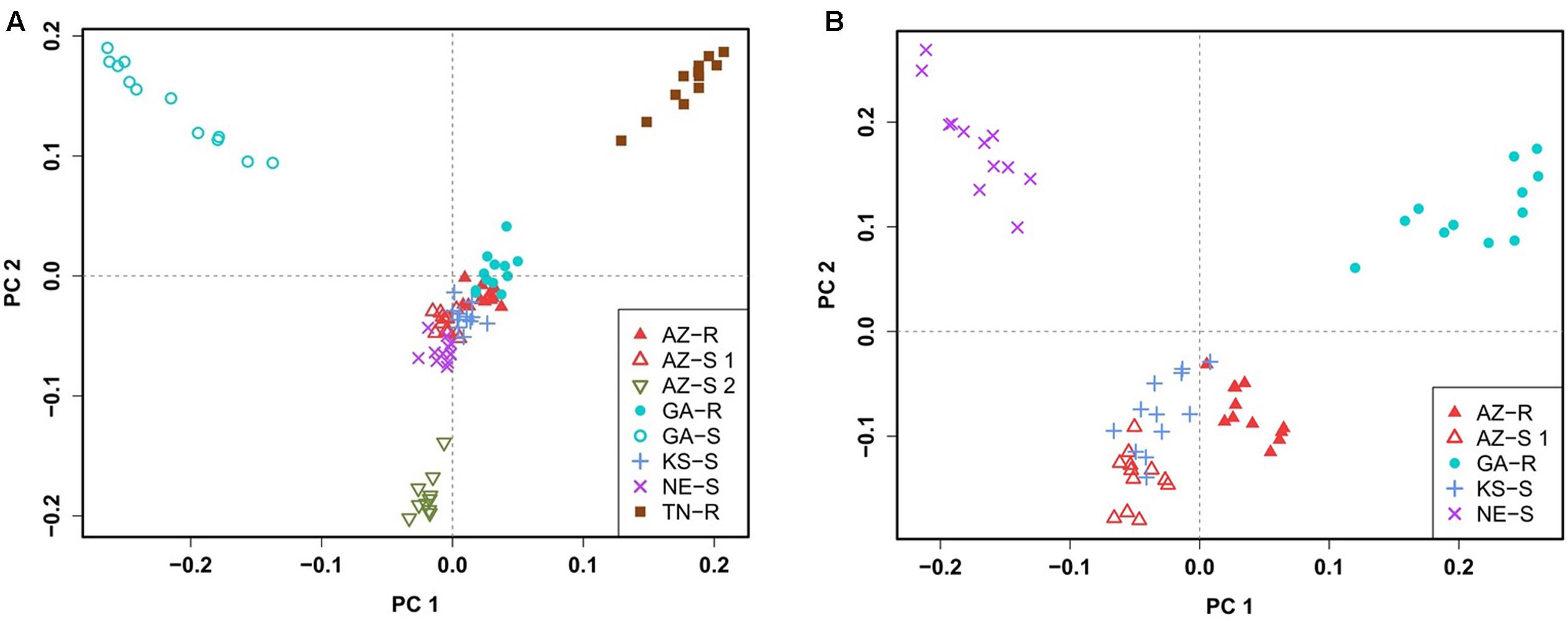
FIGURE 3. Clustering of A. palmeri populations based on principal component analysis (PCA) using the filtered and pruned whole dataset of 1,351 SNPs. Analysis was done on all eight populations (A) and on a subset of populations removing the three outlier groups AZ-S2, GA-S, and TN-R (B). Each point represents an individual colored according to the collection site. Glyphosate-resistant individuals are marked by filled symbols and susceptible individuals are marked by empty symbols. Individuals from the same U.S. state have the same symbols.
Model based clustering was used to assign individuals to sub-populations based on allele frequency differences. Initially, the putative number of populations (K) in the dataset required to explain the total sum of genetic variation observed was determined. Evanno’s test (Evanno et al., 2005) on the whole dataset of 1,351 SNPs indicated that the K distribution was bimodal and that the most informative numbers of subpopulations were four and six with K = 6 being most probable (Supplementary Information Figure 4). At K = 4, consistent with the previous findings, sub-population structure analysis revealed that individuals from GA-S, TN-R, and AZ-S2 appeared distinct from the other populations. The same analysis also showed that individuals from AZ-R and GA-R shared a proportion of their alleles with TN-R while AZ-S1 shared a small proportion with AZ-S2. At K = 6, AZ-R, KS-S, and AZ-S1 showed the highest membership coefficient for a shared cluster (beige) while the remaining populations contained unique alleles. This is supported by a high number of shared alleles among these three populations at K = 8 where KS-S displayed a high degree of admixture with AZ-R and AZ-S1. Although less than with KS-S and AZ-S1, AZ-R still shared alleles with GA-R while AZ-S1 and GA-R shared none (Figure 4). Investigating the dataset without the three outgroups GA-S, TN-R, and AZ-S2 at K = 5 supports that AZ-R shares alleles with KS-S, AZ-S1, and GA-R and very few with NE-S (Supplementary Information Figure 5).
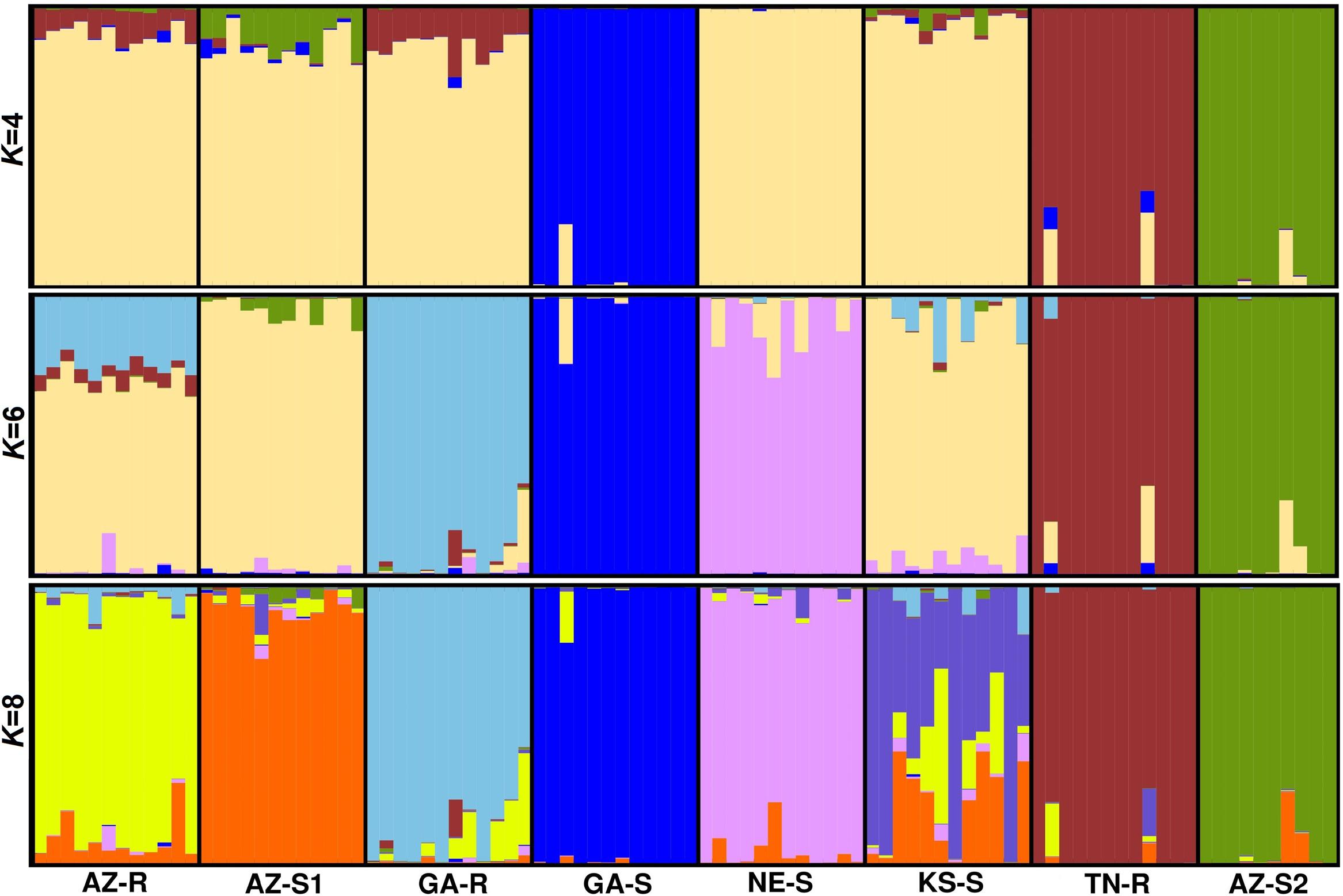
FIGURE 4. Population structure analysis with K = 4, K = 6, and K = 8 based on 1,351 SNPs of eight A. palmeri populations. Each individual is represented by a vertical bar that is divided by K colored segments representing the likelihood of a membership to each cluster.
As expected, very high genetic distances (DST) (Nei, 1972) and FST values were found for the three outgroups (TN-R, AZ-S2, and GA-S) while the genetic distance was lower among AZ-R, AZ-S1, and KS-S. Thus, AZ-R was most closely related to AZ-S1 (FST = 0.052, DST = 0.026) and KS-S (FST = 0.049, DST = 0.028) and most distantly related to the three outgroups GA-S (FST = 0.201, DST = 0.079), TN-R (FST = 0.176, DST = 0.067), and AZ-S2 (FST = 0.179, DST = 0.067). This was further visualized by a heatmap in Supplementary Information Figure 6. The bootstrap analysis of FST values indicated that all populations were significantly different from each other, except for AZ-R and KS-S, where only 5% of the genetic differences between populations were attributable to their geographic origin (Table 3).
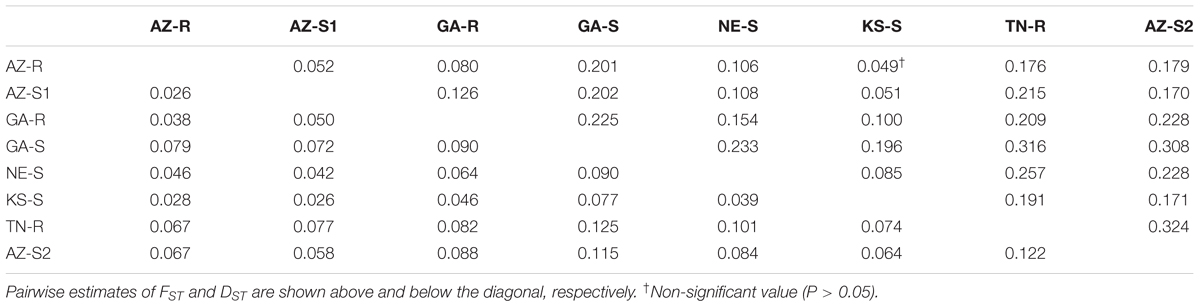
TABLE 3. Pairwise estimates of FST and Nei’s standard genetic distance (DST) between eight A. palmeri populations.
An AMOVA revealed that 17.78% (P < 0.001) of the total genetic variation was among populations, 4.87% was among individuals within a population (P < 0.01) and the remaining 77.35% (P < 0.001) of the genetic variation was within individuals (Supplementary Information Figure 7). Population differentiation exists at all levels but the variation within individuals was the largest. The high genetic variation within individuals suggests a lack of population structure, even though FST values up to 0.324 (Table 3) indicate that genetic differentiation between populations was high.
The geographical distance between any two populations ranged from about 60 to 1,930 km. The Mantel test revealed that no pattern of isolation by distance was evident between genetic and geographic distance (R2 = 0.006, P = 0.259). The observed correlation of 0.076 further suggests that the two distances are not associated (Supplementary Information Figure 8).
Forty-two SNPs specific to the chloroplast genome and fifty-four SNPs specific to the mitochondrial genome were identified. PCA with chloroplast SNPs identified GA-S and TN-R as distinct groups (Figure 5A). Structure analysis with the identified sub-populations of K = 4 and K = 5 (Supplementary Information Figure 9A) supported this observation. AZ-S2, however, shared considerably more alleles with NE-S, AZ-S1, KS-S, and GA-R than previously observed when including the loci from the nuclear and mitochondrial genomes (Figure 6A). At K = 8 AZ-R was closest related to AZ-S1 (FST = 0.058, DST = 0.279) (Supplementary Information Table 2).
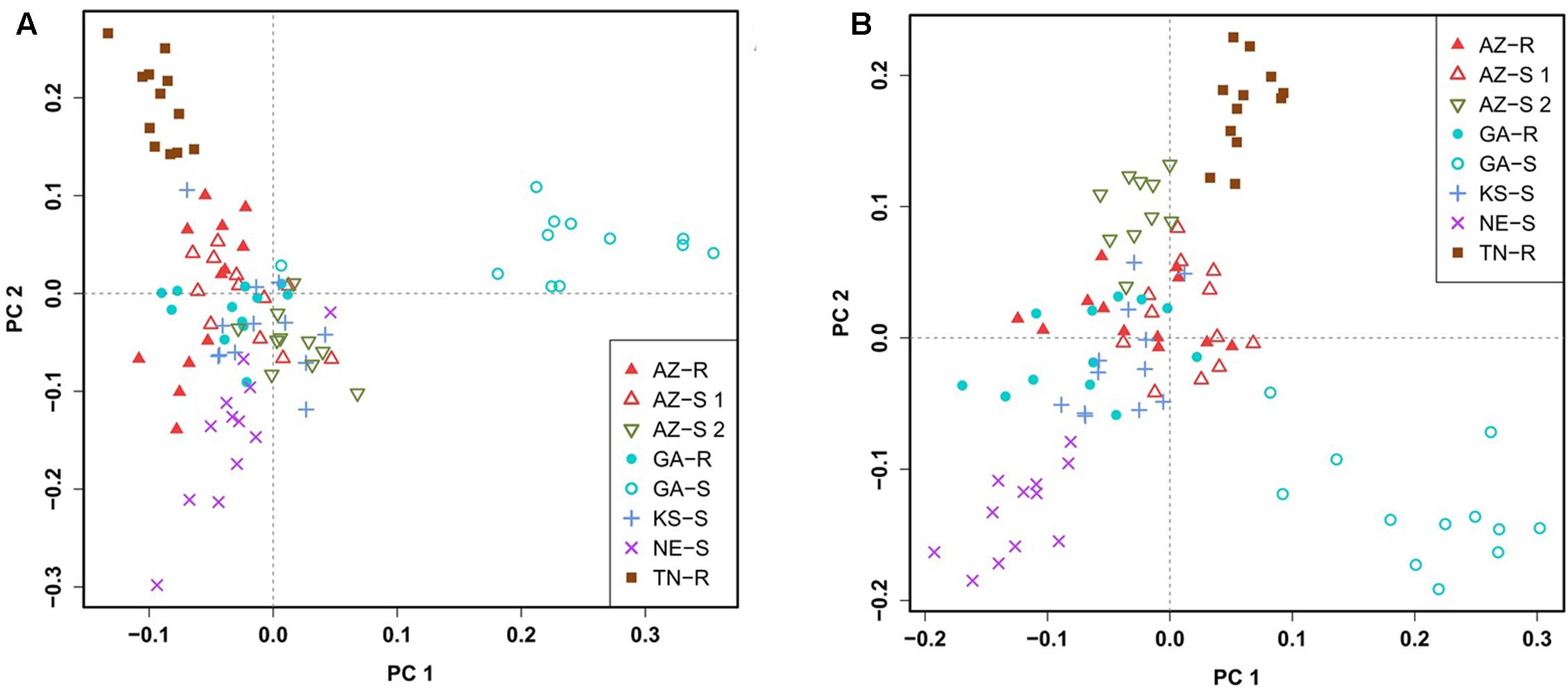
FIGURE 5. Clustering of A. palmeri populations based on principal component analysis (PCA) for SNPs found in all eight populations in the chloroplast genome (A) and the mitochondrial genome (B) using the filtered and pruned whole dataset. Each point represents an individual colored according to the collection site. Glyphosate-resistant individuals are marked by filled symbols and susceptible individuals are marked by empty symbols. Individuals from the same U.S. state have the same symbols.
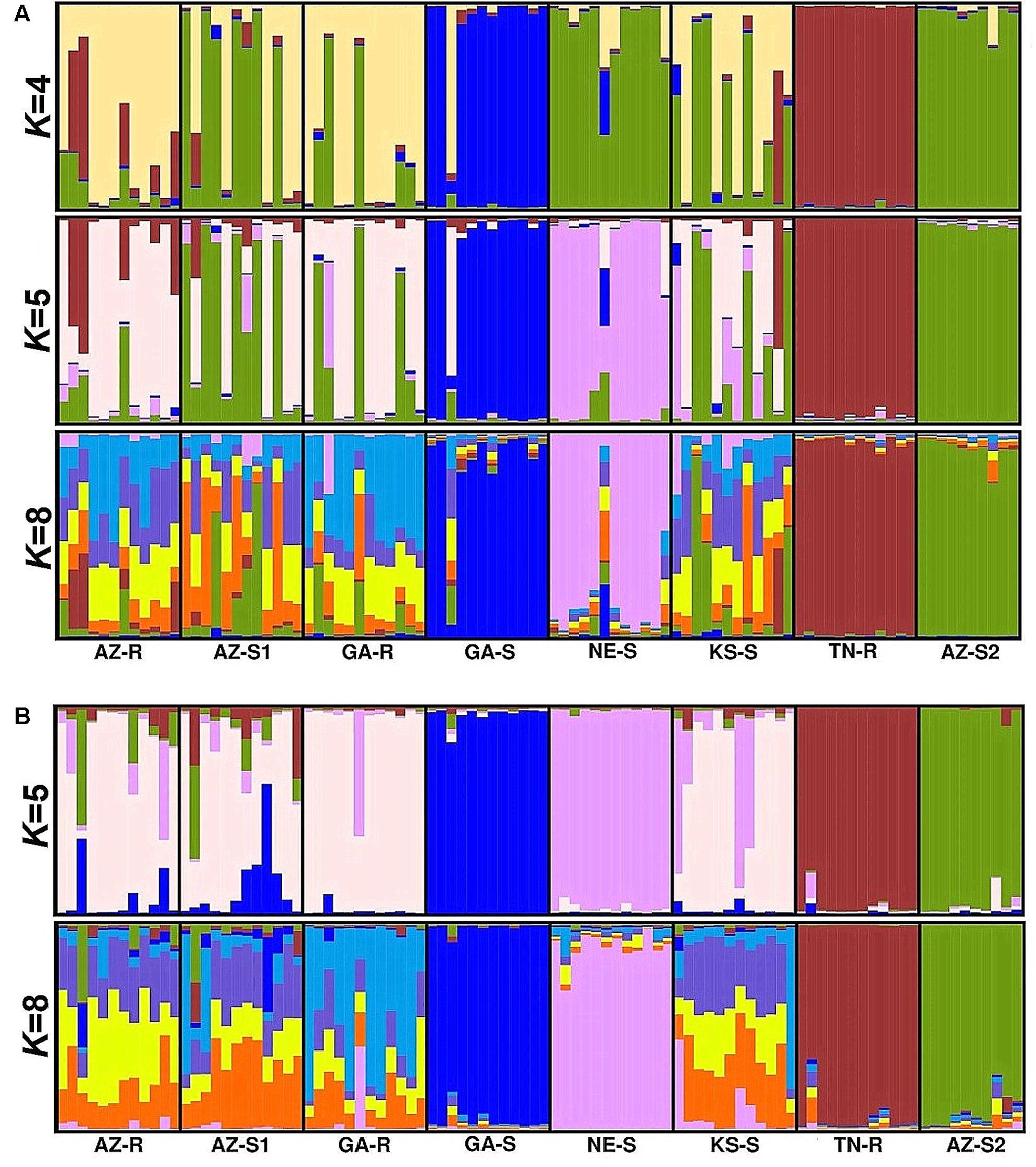
FIGURE 6. Population structure analysis with K = 4, K = 5, and K = 8 based on SNPs from the chloroplast genome for eight A. palmeri populations (A) and K = 5, and K = 8 based on SNPs from the mitochondrial genome for eight A. palmeri populations (B). Each individual is represented by a vertical bar that is divided by K colored segments representing the likelihood of a membership to each cluster.
Consistent with the analysis with all 1,351 SNPs, mitochondrial SNPs identified GA-S, TN-R, NE-S, and also AZ-S2 as distinct groups (Figure 5B), while the remaining populations AZ-R, AZ-S1, GA-R, and KS-S clustered together (Figure 6B) leaving K = 5 identifiable clusters among the eight populations (Supplementary Information Figure 9B). At K = 8, AZ-R was closest related to KS-S (FST = 0.053, DST = 0.23) (Supplementary Information Table 3).
Previous population genetics studies investigating the phylogeographic structure of pesticide resistant organisms reveal either a single origin (Raymond and Callaghan, 1991; Linda and Alan, 1997; Daborn et al., 2002) or, more frequently, redundant independent, parallel evolution events shaped by variations in selection pressure (Cavan et al., 1998; Anstead et al., 2005; Menchari et al., 2006; Chen et al., 2007; Pinto et al., 2007; Délye et al., 2010). As an example, it was found that glyphosate resistance in horseweed (Conyza canadensis) from California had multiple independent origins within the Central Valley and evolved many years before its first detection. From there it spread, possibly due to increased selection by the herbicide (Okada et al., 2013). The resistance mechanism(s) for the C. canadensis populations used in this study were unknown but most likely involved reduced translocation (Wang et al., 2014) and vacuolar sequestration (Ge et al., 2010). Similarly, investigations into the frequency of target site mutations in the EPSPS gene of GR Italian ryegrass (Lolium perenne L. ssp. multiflorum) populations (Karn and Jasieniuk, 2017) as well as simple sequence repeats (SSR) genotyping of GR common morning glory (Ipomoea purpurea) (Kuester et al., 2015), and GR Johnsongrass (Sorghum halepense L. Pers) (Fernández et al., 2013) found multiple evolutionary origins for glyphosate resistance.
Two previous studies have examined population genetics in A. palmeri with glyphosate resistance due to EPSPS gene amplification. An investigation using four genomic loci as markers of GR A. palmeri from several sampling sites within North Carolina suggested that adaptation to glyphosate application took place in parallel. The authors based this conclusion on the fact that four out of five identified population clusters were statistically associated with increased glyphosate resistance (Beard et al., 2014). In contrast, sequencing of selected regions of the 287 kb EPSPS cassette in GR populations from geographically distant locations within the United States showed strong homology between sequences and the authors concluded that the conserved nature of the cassette indicated that glyphosate resistance via amplification evolved once from a point source and then rapidly spread across the United States (Molin et al., 2017b).
Information on the factors that influence the evolutionary origin, demographic history, and geographical pathways of glyphosate resistance in A. palmeri is crucial for the formulation of successful strategies to delay and manage herbicide resistance. The aim of this study was to investigate population structure and genetic differentiation among eight geographically distant A. palmeri populations to assess if glyphosate resistance evolved in the southeastern United States and migrated to the southwestern United States, or if it evolved independently in AZ as a result of local management practices. Glyphosate resistance and susceptibility were determined by EPSPS copy number and shikimate assay test in all sampled individuals confirming that the resistance mechanism was EPSPS gene amplification. EPSPS genomic copy number was similar among the resistant populations; thus, spread of glyphosate resistance from a single origin is possible. GBS was used to identify numerous genome-wide sequence differences used as putative neutral markers due to its fast and simple application, cost-effectiveness and high resolution (Brumfield et al., 2003; Morin et al., 2004; Deschamps et al., 2012; Narum et al., 2013). The technique is widely applicable for studying non-model organisms, such as weeds, because the consensus of read clusters around the sequence site becomes the reference sequence and therefore a complete reference genome sequence is not required (Baxter et al., 2011; Elshire et al., 2011; Reitzel et al., 2013). For this study, 1,351 SNPs were used that remained after filtering.
Genetic diversity for each of the A. palmeri populations was estimated by the number of alleles, allelic richness, observed and expected heterozygosity, as well as inbreeding coefficient. The varying levels of heterozygosity found can most likely be attributed to differing collection dates and subsequent seed increase events which may have caused inbreeding depression. In particular AZ-S2, collected in 1981, is expected to have undergone severe inbreeding.
UPGMA phylogenetic tree analysis, PCA, Bayesian model-based clustering, and pairwise comparisons of genetic distances were used to determine the genetic relationship among the eight different A. palmeri populations and yielded congruent results. GA-S, TN-R, and AZ-S2 were genetically distinct while the remaining populations AZ-R, KS-S, AZ-S1, GA-R, and NE-S clustered together more closely. AZ-R was most closely related to KS-S, followed by AZ-S1, with GA-R being the next most similar population to AZ-R.
Cytoplasmic genomes are maternally inherited and do not undergo recombination. Thus, they permit a more conserved examination of intraspecific phylogeography in plants. They further have the potential to allow for higher differentiation (Petit et al., 2005). Chloroplast and mitochondrial SNPs were evaluated separately because they might support different phylogenies (Washburn et al., 2015; Zhu et al., 2016), since mitochondrial genomes have lower nucleotide sequence variation than chloroplast genomes (Wolfe et al., 1987). Analyses with SNPs in cytoplasmic genomes supported GA-S and TN-R to be genetically distinct. Chloroplast SNPs, however, placed AZ-S2 closer to the remaining populations than NE-S, consistent with the geographical distribution. AZ-R was closest to AZ-S1 based on chloroplast SNPs while mitochondrial SNPs placed the population closest to KS-S. Sequencing of the ALS gene revealed that two out of three AZ-R individuals carried a W574L and S653N mutation each, showing high diversity of the ALS sequence within the population. Since only the W574L mutation was found in one out of three individuals from TN-R, while GA-R and GA-S individuals had none (Küpper et al., 2017), mutations in the ALS gene do not support clustering of AZ-R and GA-R.
According to the observed population genetic structure, two scenarios are possible for AZ-R: Either glyphosate resistance evolved independently in AZ or GR A. palmeri from GA migrated west via KS to AZ, against the species expansion direction (Supplementary Information Figure 10). The small amount of shared sequences with GA-R and the moderate amount of shared sequences with KS-S individuals support such an introduction route, as does the chronological order of reports of glyphosate resistance (Georgia: 2004, Kansas: 2011, Arizona: 2012). AZ-R individuals shared alleles with AZ-S1 which could be attributed to crossing events with the native, GS population since resistance is likely to have been reported some time after the introduction event (Cavan et al., 1998). The high degree of unique sequence in AZ-R suggests that the exact origin of the population could not be identified. It can, however, be predicted that AZ-R individuals were not introduced from around the sampling location in TN.
Interestingly, TN-R and GA-R did not share any alleles and seemed unrelated in all analyses. Such strong population differentiation and monophyly can stem from a past divergence event and subsequent adaptation to environmental conditions through intraspecies convergent evolution (Ralph and Coop, 2015) or isolation due to limited dispersal and low connectivity (Reitzel et al., 2013). Further, agricultural practices might have strongly modified weed communities and disturbed genetic equilibrium (Menchari et al., 2007). TN and GA/NC coastal regions are geographically separated by the Appalachian mountain range and have dissimilar cropping systems with one primarily focusing on soybean and corn production and the other on cotton. As resistance to glyphosate was reported within a time frame of 2 years in these states, it is very possible that the populations represent independent glyphosate resistance origins. GA-R and GA-S, however, were genetically distinct from each other in all analyses even though collected from about 115 km apart and without any major geographical obstacles in the way. If glyphosate resistance evolved at the GA-R location, a more panmictic structure would have been expected (Chauvel and Gasquez, 1994). Such differences could be attributed to locally differing conditions, a high degree of natural spatial genetic diversity within the species, or the possibility that glyphosate resistance did not originate in GA. It is also possible that continuous selection with glyphosate created a genetic bottleneck and subsequent inbreeding of resistant individuals.
Amaranthus palmeri is a species with high genetic variability which makes it a challenge to draw a definite conclusion about an introduction event without very specific sampling. This study has shown that genetic relatedness does not decrease with distance. Hence, if GS individuals collected from within a 50 km radius can have the high level of genetic differentiation observed in this study (e.g., AZ-S1 and AZ-S2), it may be difficult to identify a parallel adaptation event. The nativity of A. palmeri to the southwestern United States and adaptation to local and heterogenous environments (Clements et al., 2004) as well as the species’ obligate outcrossing nature are drivers for heterozygosity. Genetic diversity, in turn, increases the likelihood of resistance to evolve, as does high selection pressure due to frequent usage of glyphosate which has been the case in all areas of GR A. palmeri collection sites. Future research should incorporate a more extensive collection of GR A. palmeri populations, always coupled with at least one geographically close GS population. Furthermore, all seed should be collected by the exact same sampling technique to increase the precision and accuracy with which questions of genetic relatedness and geographic migration patterns can be answered.
A major management question for growers is how much of the resistance issue results from previous selection intensity from management practices in their own fields, and how much results from gene flow from neighboring fields. Although this study was not able to definitively determine whether AZ-R evolved independently or if glyphosate resistance migrated to AZ, the recent geographical expansion of A. palmeri to the upper United States Midwest (Kartescz, 2014), Argentina (Berger et al., 2016), and Brazil (Küpper et al., 2017) shows that migration via seed movement is an important factor for A. palmeri. Long-distance seed dispersal is possible through irrigation and rainfall events (Norsworthy et al., 2014), buying and selling of used harvest equipment, custom harvesting crews moving around the country (Schwartz et al., 2016), contaminated crop seed and feed, as well as transportation through migrating wildlife such as ducks and geese (Farmer et al., 2017). Aside from harvest equipment hygiene requirements, it is difficult to prevent such seed dispersal. Early detection and rapid response approaches already used in invasive species management (Westbrooks, 2004) and disease outbreaks (Fasina et al., 2014) could be useful to adopt for herbicide resistance management. Delaying resistance evolution and prolonging the utility of remaining effective modes of actions for which resistance is not yet widespread, such as synthetic auxins, glutamine synthetase-, and phytoene desaturase (PDS)-inhibitors, is critical for future A. palmeri management.
Conceived and designed the experiments: AK, HM, WM, and TG. Performed the experiments: AK, WM, and DG. Analyzed the data: AK and EP. Contributed materials: WM and TG. Drafted the manuscript and figures: AK. All authors contributed to the revision of the final manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the National Institute of Food and Agriculture (NIFA) or the United States Department of Agriculture (USDA).
This work was partially financially supported by the USDA National Institute of Food and Agriculture, Hatch project COL00719 to the Colorado State University Agricultural Experiment Station. The authors acknowledge the Colorado State University Libraries Open Access Research and Scholarship Fund for supporting open access publication of the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MJG and handling Editor declared their shared affiliation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00029/full#supplementary-material
Adamack, A. T., and Gruber, B. (2014). PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol. Evol. 5, 384–387. doi: 10.1111/2041-210X.12158
Akey, J. M., Zhang, K., Xiong, M., and Jin, L. (2003). The effect of single nucleotide polymorphism identification strategies on estimates of linkage disequilibrium. Mol. Biol. Evol. 20, 232–242. doi: 10.1093/molbev/msg032
Anstead, J. A., Williamson, M. S., and Denholm, I. (2005). Evidence for multiple origins of identical insecticide resistance mutations in the aphid Myzus persicae. Insect Biochem. Mol. Biol. 35, 249–256. doi: 10.1016/j.ibmb.2004.12.004
Baxter, S. W., Davey, J. W., Johnston, J. S., Shelton, A. M., Heckel, D. G., Jiggins, C. D., et al. (2011). Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLOS ONE 6:e19315. doi: 10.1371/journal.pone.0019315
Beard, K. E., Lay, K. S., Burton, J. D., Burgos, N., and Lawton-Rauh, A. L. (2014). Can Very Rapid Adaptation Arise without Ancestral Variation? Insight from the Molecular Evolution of Herbicide Resistance in Genus Amaranthus. Doctoral dissertation, Clemson University, Clemson, SC.
Beckie, H. J. (2011). Herbicide-resistant weed management: focus on glyphosate. Pest Manag. Sci. 67, 1037–1048. doi: 10.1002/ps.2195
Benbrook, C. M. (2016). Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 28:3. doi: 10.1186/s12302-016-0070-0
Berger, S., Madeira, P. T., Ferrell, J., Gettys, L., Morichetti, S., Cantero, J. J., et al. (2016). Palmer amaranth (Amaranthus palmeri) identification and documentation of ALS-resistance in Argentina. Weed Sci. 64, 312–320. doi: 10.1614/WS-D-15-00125.1
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Brumfield, R. T., Beerli, P., Nickerson, D. A., and Edwards, S. V. (2003). The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol. 18, 249–256. doi: 10.1016/s0169-5347(03)00018-1
Cavan, G., Biss, P., and Moss, S. (1998). Localized origins of herbicide resistance in Alopecurus myosuroides. Weed Res. 38, 239–245. doi: 10.1046/j.1365-3180.1998.00093.x
Chahal, P. S., Aulakh, J. S., Jugulam, M., and Jhala, A. J. (2015). Herbicide-resistant Palmer Amaranth (Amaranthus palmeri S. Wats.) in the United States—Mechanisms of Resistance, Impact, and Management. Rijeka: InTechOpen.
Chatham, L. A., Wu, C., Riggins, C. W., Hager, A. G., Young, B. G., Roskamp, G. K., et al. (2015). EPSPS gene amplification is present in the majority of glyphosate-resistant Illinois waterhemp (Amaranthus tuberculatus) populations. Weed Technol. 29, 48–55. doi: 10.1614/WT-D-14-00064.1
Chauvel, B., and Gasquez, J. (1994). Relationships between genetic polymorphism and herbicide resistance within Alopecurus myosuroides Huds. Heredity 72, 336–344. doi: 10.1038/hdy.1994.50
Chen, J., Huang, H., Zhang, C., Wei, S., Huang, Z., Chen, J., et al. (2015). Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242, 859–868. doi: 10.1007/s00425-015-2324-2
Chen, W.-J., Delmotte, F., Cervera, S. R., Douence, L., Greif, C., and Corio-Costet, M.-F. (2007). At least two origins of fungicide resistance in grapevine downy mildew populations. Appl. Environ. Microbiol. 73, 5162–5172. doi: 10.1128/AEM.00507-07
Clements, D. R., DiTommaso, A., Jordan, N., Booth, B. D., Cardina, J., Doohan, D., et al. (2004). Adaptability of plants invading North American cropland. Agric. Ecosyst. Environ. 104, 379–398. doi: 10.1016/j.agee.2004.03.003
Coupe, R. H., and Capel, P. D. (2016). Trends in pesticide use on soybean, corn and cotton since the introduction of major genetically modified crops in the United States. Pest Manag. Sci. 72, 1013–1022. doi: 10.1002/ps.4082
Culpepper, A., Whitaker, J., MacRae, A., and York, A. (2008). Distribution of glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in Georgia and North Carolina during 2005 and 2006. J. Cotton Sci. 12, 306–310.
Culpepper, A. S., Grey, T. L., Vencill, W. K., Kichler, J. M., Webster, T. M., Brown, S. M., et al. (2006). Glyphosate-resistant Palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci. 54, 620–626. doi: 10.1021/jf104719k
Daborn, P., Yen, J., Bogwitz, M., Le Goff, G., Feil, E., Jeffers, S., et al. (2002). A single P450 allele associated with insecticide resistance in Drosophila. Science 297, 2253–2256. doi: 10.1126/science.1074170
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Délye, C., Michel, S., Bérard, A., Chauvel, B., Brunel, D., Guillemin, J. P., et al. (2010). Geographical variation in resistance to acetyl-coenzyme A carboxylase-inhibiting herbicides across the range of the arable weed Alopecurus myosuroides (black-grass). New Phytol. 186, 1005–1017. doi: 10.1111/j.1469-8137.2010.03233.x
Deschamps, S., Llaca, V., and May, G. D. (2012). Genotyping-by-sequencing in plants. Biology 1, 460–483. doi: 10.3390/biology1030460
Dray, S., and Dufour, A.-B. (2007). The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20. doi: 10.18637/jss.v022.i04
Duke, S. O. (2012). Why have no new herbicide modes of action appeared in recent years? Pest Manag. Sci. 68, 505–512. doi: 10.1002/ps.2333
Duke, S. O. (2017). The history and current status of glyphosate. Pest Manag. Sci. doi: 10.1002/ps.4652 [Epub ahead of print].
Duke, S. O., and Powles, S. B. (2008). Glyphosate: a once-in-a-century herbicide. Pest Manag. Sci. 64, 319–325. doi: 10.1002/ps.1518
Earl, D. A. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLOS ONE 6:e19379. doi: 10.1371/journal.pone.0019379
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Farmer, J. A., Webb, E. B., Pierce, R. A., and Bradley, K. W. (2017). Evaluating the potential for weed seed dispersal based on waterfowl consumption and seed viability. Pest Manag. Sci. 73, 2592–2603. doi: 10.1002/ps.4710
Fasina, F. O., Shittu, A., Lazarus, D., Tomori, O., Simonsen, L., Viboud, C., et al. (2014). Transmission dynamics and control of Ebola virus disease outbreak in Nigeria, July to September 2014. Euro Surveill. 19:20920. doi: 10.2807/1560-7917.ES2014.19.40.20920
Fernández, L., Haro, L. A., Distefano, A. J., Carolina Martínez, M., Lía, V., Papa, J. C., et al. (2013). Population genetics structure of glyphosate-resistant Johnsongrass (Sorghum halepense L. Pers) does not support a single origin of the resistance. Ecol. Evol. 3, 3388–3400. doi: 10.1002/ece3.671
Franssen, A. S., Skinner, D. Z., Al-Khatib, K., Horak, M. J., and Kulakow, P. A. (2001). Interspecific hybridization and gene flow of ALS resistance in Amaranthus species. Weed Sci. 49, 598–606. doi: 10.1614/0043-1745(2001)049[0598:IHAGFO]2.0.CO;2
Gaines, T. A., Zhang, W., Wang, D., Bukun, B., Chisholm, S. T., Shaner, D. L., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. U.S.A. 107, 1029–1034. doi: 10.1073/pnas.0906649107
Ge, X., d’Avignon, D. A., Ackerman, J. J., and Sammons, R. D. (2010). Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manag. Sci. 66, 345–348. doi: 10.1002/ps.1911
Georghiou, G. P., and Taylor, C. E. (1986). Factors Influencing the Evolution of Resistance. Washington, DC: National Academy Press.
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLOS ONE 9:e90346. doi: 10.1371/journal.pone.0090346
Heap, I. (2017). The International Survey of Herbicide Resistant Weeds. Available at: http://www.weedscience.org [accessed July 12, 2017].
Jakobsson, M., and Rosenberg, N. A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Jasieniuk, M., Brûlé-Babel, A. L., and Morrison, I. N. (1996). The evolution and genetics of herbicide resistance in weeds. Weed Sci. 44, 176–193.
Jombart, T., and Ahmed, I. (2011). adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. doi: 10.1093/bioinformatics/btr521
Jugulam, M., Niehues, K., Godar, A. S., Koo, D.-H., Danilova, T., Friebe, B., et al. (2014). Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia. Plant Physiol. 166, 1200–1207. doi: 10.1104/pp.114.242826
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281
Karn, E., and Jasieniuk, M. (2017). Nucleotide diversity at site 106 of EPSPS in Lolium perenne L. ssp. multiflorum from California indicates multiple evolutionary origins of herbicide resistance. Front. Plant Sci. 8:777. doi: 10.3389/fpls.2017.00777
Kartescz, J. T. (2014). The Biota of North America Program (BONAP). Chapel Hill, NC: Taxonomic Data Center.
Kuester, A., Chang, S. M., and Baucom, R. S. (2015). The geographic mosaic of herbicide resistance evolution in the common morning glory, Ipomoea purpurea: evidence for resistance hotspots and low genetic differentiation across the landscape. Evol. Appl. 8, 821–833. doi: 10.1111/eva.12290
Küpper, A., Borgato, E. A., Patterson, E. L., Netto, A. G., Nicolai, M., Carvalho, S. J. D., et al. (2017). Multiple resistance to glyphosate and acetolactate synthase inhibitors in Palmer amaranth (Amaranthus palmeri) identified in Brazil. Weed Sci. 65, 317–326. doi: 10.1017/wsc.2017.1
Laplante, J., Rajcan, I., and Tardif, F. J. (2009). Multiple allelic forms of acetohydroxyacid synthase are responsible for herbicide resistance in Setaria viridis. Theor. Appl. Genet. 119, 577–585. doi: 10.1007/s00122-009-1067-5
Linda, M., and Alan, L. (1997). Structure and organization of amplicons containing the E4 esterase genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer). Biochem. J. 322, 867–871. doi: 10.1042/bj3220867
Lorentz, L., Gaines, T. A., Nissen, S. J., Westra, P., Strek, H. J., Dehne, H. W., et al. (2014). Characterization of glyphosate resistance in Amaranthus tuberculatus populations. J. Agric. Food Chem. 62, 8134–8142. doi: 10.1021/jf501040x
Lu, F., Lipka, A. E., Glaubitz, J., Elshire, R., Cherney, J. H., Casler, M. D., et al. (2013). Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLOS Genet. 9:e1003215. doi: 10.1371/journal.pgen.1003215
Malone, J. M., Morran, S., Shirley, N., Boutsalis, P., and Preston, C. (2016). EPSPS gene amplification in glyphosate-resistant Bromus diandrus. Pest Manag. Sci. 72, 81–88. doi: 10.1002/ps.4019
Maxwell, B. D., Roush, M. L., and Radosevich, S. R. (1990). Predicting the evolution and dynamics of herbicide resistance in weed populations. Weed Technol. 4, 2–13.
McNaughton, K. E., Letarte, J., Lee, E. A., and Tardif, F. J. (2005). Mutations in ALS confer herbicide resistance in redroot pigweed (Amaranthus retroflexus) and Powell amaranth (Amaranthus powellii). Weed Sci. 53, 17–22. doi: 10.1614/WS-04-109
Menchari, Y., Camilleri, C., Michel, S., Brunel, D., Dessaint, F., Le Corre, V., et al. (2006). Weed response to herbicides: regional-scale distribution of herbicide resistance alleles in the grass weed Alopecurus myosuroides. New Phytol. 171, 861–874. doi: 10.1111/j.1469-8137.2006.01788.x
Menchari, Y., Délye, C., and Le Corre, V. (2007). Genetic variation and population structure in black-grass (Alopecurus myosuroides Huds.), a successful, herbicide-resistant, annual grass weed of winter cereal fields. Mol. Ecol. 16, 3161–3172. doi: 10.1111/j.1365-294X.2007.03390.x
Molin, W. T., Nandula, V. K., Wright, A. A., and Bond, J. A. (2016). Transfer and expression of ALS inhibitor resistance from Palmer Amaranth (Amaranthus palmeri) to an A. spinosus × A. palmeri hybrid. Weed Sci. 64, 240–247. doi: 10.1614/WS-D-15-00172.1
Molin, W. T., Wright, A. A., Lawton-Rauh, A., and Saski, C. A. (2017a). The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: a repetitive path to resistance. BMC Genomics 18:91. doi: 10.1186/s12864-12016-13336-12864
Molin, W. T., Wright, A. A., VanGessel, M. J., McCloskey, W. B., Jugulam, M., and Hoagland, R. E. (2017b). Survey of the genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri from geographically distant populations in the United States. Pest Manag. Sci. doi: 10.1002/ps.4659 [Epub ahead of print].
Morin, P. A., Luikart, G., Wayne, R. K., and Palsboll, P. (2004). SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 19, 208–216. doi: 10.1016/j.tree.2004.01.009
Nakka, S., Godar, A. S., Wani, P. S., Thompson, C. R., Peterson, D. E., Roelofs, J., et al. (2017). Physiological and molecular characterization of hydroxyphenylpyruvate dioxygenase (HPPD)-inhibitor resistance in Palmer amaranth (Amaranthus palmeri S. Wats.). Front. Plant Sci. 8:555. doi: 10.3389/fpls.2017.00555
Nandula, V. K., Wright, A. A., Bond, J. A., Ray, J. D., Eubank, T. W., and Molin, W. T. (2014). EPSPS amplification in glyphosate-resistant spiny amaranth (Amaranthus spinosus): a case of gene transfer via interspecific hybridization from glyphosate-resistant Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 70, 1902–1909. doi: 10.1002/ps.3754
Narum, S. R., Buerkle, C. A., Davey, J. W., Miller, M. R., and Hohenlohe, P. A. (2013). Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22, 2841–2847. doi: 10.1111/mec.12350
Neve, P. (2008). Simulation modelling to understand the evolution and management of glyphosate resistance in weeds. Pest Manag. Sci. 64, 392–401. doi: 10.1002/ps.1495
Neve, P., Norsworthy, J. K., Smith, K. L., and Zelaya, I. A. (2011). Modelling evolution and management of glyphosate resistance in Amaranthus palmeri. Weed Res. 51, 99–112. doi: 10.1111/j.1365-3180.2010.00838.x
Ngo, T. D., Malone, J. M., Boutsalis, P., Gill, G., and Preston, C. (2017). EPSPS gene amplification conferring resistance to glyphosate in windmill grass (Chloris truncata) in Australia. Pest Manag. Sci. doi: 10.1002/ps.4573 [Epub ahead of print].
Norsworthy, J. K., Griffith, G., Griffin, T., Bagavathiannan, M., and Gbur, E. E. (2014). In-field movement of glyphosate-resistant Palmer amaranth (Amaranthus palmeri) and its impact on cotton lint yield: evidence supporting a zero-threshold strategy. Weed Sci. 62, 237–249. doi: 10.1614/WS-D-13-00145.1
Norsworthy, J. K., Griffith, G. M., Scott, R. C., Smith, K. L., and Oliver, L. R. (2008). Confirmation and control of glyphosate-resistant Palmer amaranth (Amaranthus palmeri) in Arkansas. Weed Technol. 22, 108–113. doi: 10.1614/WT-07-128.1
Okada, M., Hanson, B. D., Hembree, K. J., Peng, Y., Shrestha, A., Stewart, C. N., et al. (2013). Evolution and spread of glyphosate resistance in Conyza canadensis in California. Evol. Appl. 6, 761–777. doi: 10.1111/eva.12061
Patzoldt, W. L., and Tranel, P. J. (2007). Multiple ALS mutations confer herbicide resistance in waterhemp (Amaranthus tuberculatus). Weed Sci. 55, 421–428. doi: 10.1614/WS-06-213.1
Pembleton, L. W., Cogan, N. O., and Forster, J. W. (2013). StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 13, 946–952. doi: 10.1111/1755-0998.12129
Petit, R. J., Duminil, J., Fineschi, S., Hampe, A., Salvini, D., and Vendramin, G. G. (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. doi: 10.1111/j.1365-294X.2004.02410.x
Pinto, J., Lynd, A., Vicente, J. L., Santolamazza, F., Randle, N. P., Gentile, G., et al. (2007). Multiple origins of knockdown resistance mutations in the Afrotropical mosquito vector Anopheles gambiae. PLOS ONE 2:e1243. doi: 10.1371/journal.pone.0001243
Porras-Hurtado, L., Ruiz, Y., Santos, C., Phillips, C., Carracedo,Á., and Lareu, M. V. (2013). An overview of STRUCTURE: applications, parameter settings, and supporting software. Front. Genet. 4:98. doi: 10.3389/fgene.2013.00098
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Ralph, P. L., and Coop, G. (2015). Convergent evolution during local adaptation to patchy landscapes. PLOS Genet. 11:e1005630. doi: 10.1371/journal.pgen.1005630
Raymond, M., and Callaghan, A. (1991). Worldwide migration of amplified insecticide resistance genes in mosquitoes. Nature 350, 151–153. doi: 10.1038/350151a0
Reitzel, A. M., Herrera, S., Layden, M. J., Martindale, M. Q., and Shank, T. M. (2013). Going where traditional markers have not gone before: utility of and promise for RAD sequencing in marine invertebrate phylogeography and population genomics. Mol. Ecol. 22, 2953–2970. doi: 10.1111/mec.12228
Rosenberg, N. A. (2004). DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Resour. 4, 137–138. doi: 10.1046/j.1471-8286.2003.00566.x
Salas, R. A., Dayan, F. E., Pan, Z., Watson, S. B., Dickson, J. W., Scott, R. C., et al. (2012). EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag. Sci. 68, 1223–1230. doi: 10.1002/ps.3342
Sauer, J. (1957). Recent migration and evolution of the dioecious amaranths. Evolution 11, 11–31. doi: 10.1111/j.1558-5646.1957.tb02872.x
Schwartz, L. M., Norsworthy, J. K., Young, B. G., Bradley, K. W., Kruger, G. R., Davis, V. M., et al. (2016). Tall waterhemp (Amaranthus tuberculatus) and palmer amaranth (Amaranthus palmeri) seed production and retention at soybean maturity. Weed Technol. 30, 284–290. doi: 10.1614/WT-D-15-00130.1
Schwartz-Lazaro, L. M., Norsworthy, J. K., Scott, R. C., and Barber, L. T. (2017). Resistance of two Arkansas Palmer amaranth populations to multiple herbicide sites of action. Crop Prot. 96, 158–163. doi: 10.1371/journal.pone.0148748
Scott, R., Steckel, L., Smith, K., Mueller, S., Oliver, L., and Norsworthy, J. (2007). “Glyphosate-resistant Palmer amaranth in Tennessee and Arkansas,” in Proceedings of the Southern Weed Science Society, Las Cruces, NM, 226.
Sellers, B. A., Smeda, R. J., Johnson, W. G., Kendig, J. A., and Ellersieck, M. R. (2003). Comparative growth of six Amaranthus species in Missouri. Weed Sci. 51, 329–333. doi: 10.1614/0043-1745(2003)051[0329:CGOSAS]2.0.CO;2
Shaner, D. L., Nadler-Hassar, T., Henry, W. B., and Koger, C. H. (2005). A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 53, 769–774. doi: 10.1614/WS-05-009R.1
Steckel, L. E., Main, C. L., Ellis, A. T., and Mueller, T. C. (2008). Palmer amaranth (Amaranthus palmeri) in Tennessee has low level glyphosate resistance. Weed Technol. 22, 119–123. doi: 10.1614/WT-07-061.1
Tranel, P. J., and Wright, T. R. (2002). Resistance of weeds to ALS-inhibiting herbicides: what have we learned? Weed Sci. 50, 700–712. doi: 10.1614/0043-1745(2002)050[0700:RROWTA]2.0.CO;2
Wakeley, J., Nielsen, R., Liu-Cordero, S. N., and Ardlie, K. (2001). The discovery of single-nucleotide polymorphisms—and inferences about human demographic history. Am. J. Hum. Genet. 69, 1332–1347. doi: 10.1086/324521
Wang, W., Xia, H., Yang, X., Xu, T., Si, H. J., Cai, X. X., et al. (2014). A novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) synthase transgene for glyphosate resistance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide. New Phytol. 202, 679–688. doi: 10.1111/nph.12428
Ward, S. M., Webster, T. M., and Steckel, L. E. (2013). Palmer amaranth (Amaranthus palmeri): a review. Weed Technol. 27, 12–27. doi: 10.1614/WT-D-12-00113.1
Washburn, J. D., Schnable, J. C., Davidse, G., and Pires, J. C. (2015). Phylogeny and photosynthesis of the grass tribe Paniceae. Am. J. Bot. 102, 1493–1505. doi: 10.3732/ajb.1500222
Webster, T. M., and Coble, H. D. (1997). Changes in the weed species composition of the southern United States: 1974 to 1995. Weed Technol. 11, 308–317.
Webster, T. M., and Nichols, R. L. (2012). Changes in the prevalence of weed species in the major agronomic crops of the Southern United States: 1994/1995 to 2008/2009. Weed Sci. 60, 145–157. doi: 10.1614/WS-D-11-00092.1
Westbrooks, R. G. (2004). New approaches for early detection and rapid response to invasive plants in the United States. Weed Technol. 18, 1468–1471. doi: 10.1614/0890-037X (2004)018[1468:NAFEDA]2.0.CO;2
Whaley, C. M., Wilson, H. P., and Westwood, J. H. (2006). ALS resistance in several smooth pigweed (Amaranthus hybridus) biotypes. Weed Sci. 54, 828–832. doi: 10.1614/WS-05-040R.1
Wiersma, A. T., Gaines, T. A., Preston, C., Hamilton, J. P., Giacomini, D., Buell, C. R., et al. (2015). Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia. Planta 241, 463–474. doi: 10.1007/s00425-014-2197-9
Wolfe, K. H., Li, W.-H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Yu, Q., Han, H., Li, M., Purba, E., Walsh, M., and Powles, S. (2012). Resistance evaluation for herbicide resistance–endowing acetolactate synthase (ALS) gene mutations using Raphanus raphanistrum populations homozygous for specific ALS mutations. Weed Res. 52, 178–186. doi: 10.1111/j.1365-3180.2012.00902.x
Zheng, X., Levine, D., Shen, J., Gogarten, S. M., Laurie, C., and Weir, B. S. (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. doi: 10.1093/bioinformatics/bts606
Zhu, Q., Gao, P., Liu, S., Amanullah, S., and Luan, F. (2016). Comparative analysis of single nucleotide polymorphisms in the nuclear, chloroplast, and mitochondrial genomes in identification of phylogenetic association among seven melon (Cucumis melo L.) cultivars. Breed. Sci. 66, 711–719. doi: 10.1270/jsbbs.16066
Keywords: Palmer amaranth, population genetics, glyphosate, herbicide resistance, genetic relatedness, SNP molecular markers, phylogeography
Citation: Küpper A, Manmathan HK, Giacomini D, Patterson EL, McCloskey WB and Gaines TA (2018) Population Genetic Structure in Glyphosate-Resistant and -Susceptible Palmer Amaranth (Amaranthus palmeri) Populations Using Genotyping-by-sequencing (GBS). Front. Plant Sci. 9:29. doi: 10.3389/fpls.2018.00029
Received: 09 October 2017; Accepted: 09 January 2018;
Published: 25 January 2018.
Edited by:
Rafael De Prado, Universidad de Córdoba, SpainReviewed by:
Shabir Hussain Wani, Michigan State University, United StatesCopyright © 2018 Küpper, Manmathan, Giacomini, Patterson, McCloskey and Gaines. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Todd A. Gaines, dG9kZC5nYWluZXNAY29sb3N0YXRlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.