- 1School of Life Sciences and Biotechnology, Dalian University of Technology, Dalian, China
- 2School of Computer Science and Technology, Dalian University of Technology, Dalian, China
MicroRNAs, a group of non-coding RNA molecules, play essential roles in a wide range of cellular processes in different molecules, cells, and organisms. In plants, microRNAs are a class of 20- to 24-nucleotides endogenous small RNAs that repress gene expression. The microRNA guide strand (miRNA) and its complementary strand (miRNA∗) both originate from the miRNA/miRNA∗ duplex. Generally, the guide strands act as post-transcriptional regulators that suppress gene expression by cleaving their target mRNA transcripts, whereas the complementary strands were thought to be degraded as ‘passenger strands.’ However, the complementary strand has been confirmed to possess significant biological functionality in recent reports. In this review, we summarized the binding characteristics of the miRNA∗ strands with ARGONAUTE proteins, their tissue-specific accumulations and their biological functions, illustrating the essential roles of miRNA∗s in biological processes and therefore providing directions for further exploration.
Introduction
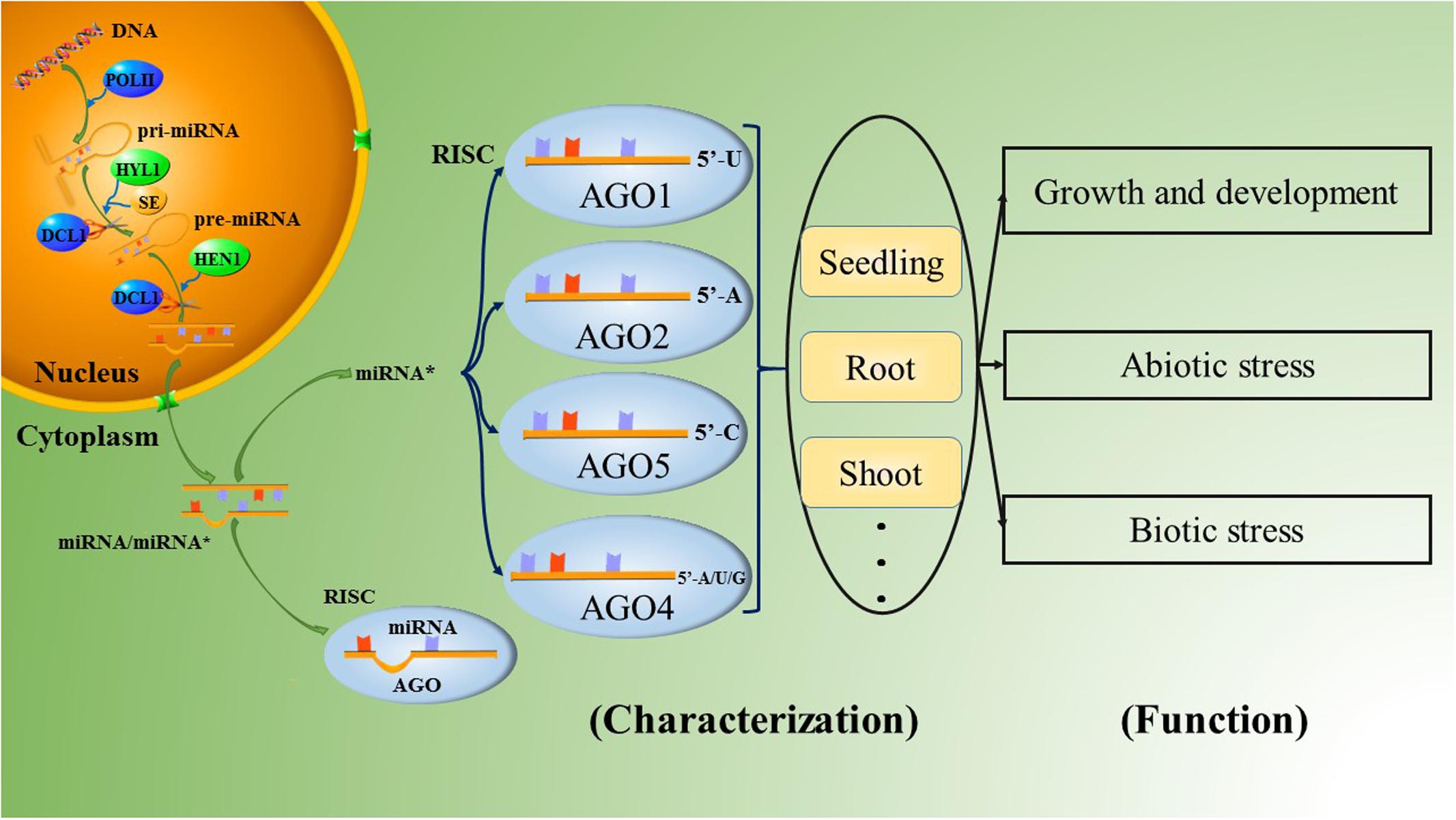
MicroRNAs are a group of non-coding RNAs that were first discovered as temporal regulators of larval differentiation in the nematode Caenorhabditis elegans (Lee et al., 1993); microRNAs play important roles in the control of diverse cellular pathways and participate in most biological processes in both plants and animals (Bartel and Bartel, 2003; Stefani and Slack, 2008). In vivo, the maturation of microRNAs requires a series of complex processes. First, endogenous genes are transcribed by RNA polymerase II (Pol II) into long primary microRNAs (pri-miRNAs) ranging from hundreds to thousands of nucleotides in length; pri-miRNAs are polyadenylated, single-stranded RNA molecules that fold into hairpin-like structures (Cai et al., 2004; Lee et al., 2004). The pri-miRNAs are then processed by the endonuclease RNase III. The species of RNase III and the mechanisms of further microRNA processing differ significantly between animals and plants (Du and Zamore, 2005). Unlike animals, plants lack Drosha homologs. Thus, after the formation of pri-miRNAs, the RNase III enzyme DICER-LIKE 1 (DCL1) regulates both the first step, which in animals involves cuts made by Drosha, and the second step, which in animals is reprocessing by Dicer, with the aid of HYPONASTIC LEAVES 1 (HYL1) and Serrate (SE); this process produces a miRNA/miRNA∗ duplex in the nucleus (Chen, 2009; Voinnet, 2009). The mature microRNA duplex consists of the active miRNA strand, called the guide strand, and the complementary miRNA∗ strand, called the passenger strand. Recently, the liberated strands have also been defined as miRNA-3ps and miRNA-5ps, according to the 5′ and 3′ arms of the hairpin precursor, after renaming by the miRBase registry (Kozomara and Griffiths-Jones, 2014; Yaish et al., 2015). The imperfect pairing of the miRNA/miRNA∗ duplex results in variable stability at the 5′ ends of the two strands. Once liberated from the duplex, the guide strands with lower thermodynamic stability at the 5′ end and high abundance are commonly loaded into specific ARGONAUTE (AGO)-associated RNA-induced silencing complexes (RISCs) and guide the RISCs to their targets. Originally, the passenger strands were thought to be degraded, since the accumulation of most passenger strands is much lower than that of the guide strands, and thus miRNA∗s were presumed to be mere by-products of the miRNA biogenesis pathway (Khvorova et al., 2003; Schwarz et al., 2003). However, an increasing number of reports have demonstrated that miRNA∗s may also act as important regulatory factors in organisms, and a large number of miRNA∗s have been confirmed to possess DCL1-dependency as similar to the miRNAs in Arabidopsis and rice (Meng et al., 2011).
The Combination Preferences of Specific Ago Proteins
In plants, miRNAs are involved in RISCs and induce the repression of gene expression in either a transcriptional or a post-transcriptional manner. One difference between animal and plant miRNAs is that the latter bind to regulatory targets with highly complementary recognition sites, a general characteristic of miRNA-guided cleavage actions (Jones-Rhoades et al., 2006; Voinnet, 2009). Because of relaxed selection pressure, miRNA∗s are generally less conserved than miRNAs and more often polymorphic in both animals and plants (Guo and Lu, 2010; Smith et al., 2015). In recent years, several reports have also indicated that miRNA∗s take part in gene regulation via incorporation into AGO-associated RISCs in both animals and plants (Okamura et al., 2008; Ghildiyal et al., 2010; Zhou et al., 2010; Zhang et al., 2011).
The AGO family in plants has undergone extensive diversification, giving rise to plant-specific AGOs (Poulsen et al., 2013), including 10 AGO proteins in Arabidopsis. Among them, AGO1 has been demonstrated to be associated with most miRNAs, forming RISCs to cleave the corresponding targets (Qi et al., 2005, 2006; Zhang et al., 2011). Furthermore, AGO1 has a preference for sequences with a uridine nucleotide at the 5′ terminus (Qi et al., 2006). Some of the other AGO proteins also have slicer activity. Takeda et al. (2008) analyzed the roles of AGO2 and AGO5 in Arabidopsis thaliana. AGO2- and AGO5-associated small RNAs were initially found to have obvious preferences for the nucleotides adenine and cytosine at their respective 5′ ends by 5′ labeling of the immunoprecipitated small RNAs. The two most abundantly cloned small RNAs were miR163-LL and miR390 in the AGO2 library and miR163-UL and miR390∗ in the AGO5 library; these molecules could form the small RNA duplexes miR163-LL/miR163-UL and miR390/miR390∗. (Note that the small RNAs derived from the lower left and the upper left of mature miR163 in pre-miR163 were named miR163-LL and miR163-UL, respectively.) Furthermore, if the 5′ nucleotides of miR163-LL and miR163-UL are exchanged, the strand of the miR163-LL/miR163-UL duplex that is preferentially incorporated into either AGO2 or AGO5 is also exchanged. This result indicates that the 5′ nucleotide plays an important role in guide strand selection in both AGO2 and AGO5 to form specific AGO-small RNA complexes in Arabidopsis. This article also provides evidence that the ‘passenger strand,’ which was previously thought to have no function, forms a RISC complex by combining with an AGO protein, functioning as an additional guide strand, as shown by miR390∗ in AGO5 (Takeda et al., 2008). The other typical species of AGO2-bound small RNA is miR393b∗, which possesses an adenine at the 5′ end and mediates disease resistance by targeting a Golgi-localized SNARE gene, MEMB12 (Zhang et al., 2011; Zhang Z. et al., 2016). Overall, Arabidopsis thaliana AGO1 (AtAGO1) prefers sequences with a uridine nucleotide at the 5′ end, such as miR170a∗, miR171a∗ and miR173∗; AtAGO2 favors those with a 5′ adenine, such as miR393b∗ and miR837-5p∗; AtAGO5 has a bias toward miRNA∗s with a 5′ cytosine, such as miR158a∗ and miR390a∗; and AtAGO4 accepts variable 5′-terminal nucleotides, especially adenine, uridine or guanine (Figure 1; Mi et al., 2008; Takeda et al., 2008; Meng et al., 2011; Wang et al., 2011; Zhang et al., 2011, 2015; Manavella et al., 2012, 2013). However, the 5′-terminal nucleotide is not the only determinant of miRNA∗ combination with AGO. For example, the abundant miRNA∗s of miR396a and miR396b in Arabidopsis are designated miR396a-3p and miR396b-3p, respectively. Both of these molecules accumulated on AGO2 with a 5′-terminal guanine instead of a 5′-terminal adenine. In addition, miR396a-3p was also found to accumulate on AGO1 and AGO4 (Jeong et al., 2013).
Characteristics of miRna∗ Accumulation
In the past, it was generally believed that the accumulation of miRNA∗s in organisms was much lower than that of miRNAs. Nevertheless, several studies have demonstrated that many miRNA∗s have abundances similar to or higher than their corresponding miRNAs in specific biological processes, in specific tissues and at specific times. For instance, miR156a∗, miR164b∗, and miR535∗ of Betula luminifera; miR82∗ of Morus notabilis in leaf tissue; miR166∗, miR159∗, and miR171∗ of Orchis italica; miR171c∗, miR369a∗, and miR2612a∗/b∗ of Medicago truncatula; and miR169h∗, miR408∗, and miR529a∗ of rice were all more abundant than their corresponding miRNAs (Devers et al., 2011; Peng et al., 2011, 2013; Aceto et al., 2014; Jia et al., 2014; Yan et al., 2014; Zhang J. et al., 2016). However, these miRNA∗s did not necessarily maintain this trend at all times. For example, rice miR1425∗ accumulation was lower than that of miR1425 at the later grain filling stage, indicating that it played an important physiological role in rice grain filling (Peng et al., 2013). Some miRNA∗s also show similar abundance to their partners, such as Morus notabilis miR166∗, miR76a-2∗, and miR82∗ in male flowers; miR172∗ and miR399c∗/j∗/k∗ of Medicago truncatula; and miR162c∗, miR162b∗, miR157a∗ and others of Brassica oleracea (Devers et al., 2011; Lukasik et al., 2013; Jia et al., 2014).
The tissue specificity of miRNA∗s is reflected in their different patterns of abundance. Some preferentially accumulate in specific tissues. In rice, miR169g∗, miR169p∗, and miR396c∗ were dominantly distributed in seedlings; miR396a∗, miR396b∗, and miR2121a∗ were accumulated in the roots; miR5159∗ and miR1423b∗ were highly accumulated in shoots; miR1428f∗ was highly expressed in the inflorescences; and miR166e∗, miR1433∗, and miR5533∗ were expressed in the panicles, indicating their potential activities in rice development (Shao et al., 2013; Hu et al., 2014). Similarly, in Arabidopsis, miR396a∗ was nearly undetectable in roots but was detected in flowers, and miR172b∗ was detected in leaves; meanwhile, miR2111a∗ and miR2111b∗ accumulated in seedlings (Meng et al., 2012; Jeong et al., 2013). Manavella et al. (2013) have confirmed miR171a∗ as important for normal development in Arabidopsis; this miRNA∗ triggers silencing of SU(VAR)3-9 HOMOLOG8 in specific tissues such as the shoot apical meristem, the tops of anther filaments, the bases of young leaves, and the stomata. Interestingly, in Medicago truncatula, miR169d∗/l∗/m∗/e.2∗ target MtBcp1, which is localized in the plasma membrane and shapes the plasma membrane to the perihyphal membrane; these species were more abundant in mycorrhizal roots than in non-mycorrhizal roots, indicating that they played a role in mycorrhizal symbiosis by causing the roots to become more conducive to the absorption of nutrition (Pumplin and Harrison, 2009; Devers et al., 2011). Furthermore, the formation of some miRNA∗s is related to the growth phases of plants, such as miR172∗ and miR390∗ in cotton. These miRNA∗s were up-regulated in seedlings but down-regulated in other growth stages, while cotton miR171∗, miR2949∗ and others were regulated in the opposite manner (Xie and Zhang, 2015). Therefore, miRNA∗s can be specifically expressed in different tissues to maintain the steady state of the organism.
Responses to Stress
With further research into miRNA∗s, more of them have been confirmed to respond to many stresses. As an example, miR399b-5p in barley, which was defined as the passenger strand, showed a much higher increase in expression under P deficiency than that of the guide strand (Hackenberg et al., 2013). An inverse result was found for miR169g∗ and miR172b∗ in tomato leaves; they were down-regulated under Pi deficient conditions compared with Pi sufficiency (Gu et al., 2010). Recent studies have indicated that nitrate significantly affects the expression of several miRNA∗s, such as miR169i∗/j∗/k∗ and miR528a∗/b∗ in maize roots (Trevisan et al., 2012). MiR169∗ possesses a large number of targets in sugarcane. Specifically, Elongation Factor 1-alpha (EF 1-α), which was encoded by the majority of miR169∗ targets, has been identified in response to abiotic stress such as water depletion. Thus, many miRNA∗s participate in regulating more than one stress response. In addition, miRNA∗s also respond to other stresses, such as gamma irradiation and high salt (Table 1).
The roles of miRNA∗s in the regulation of biological stresses (Figure 1) have mainly been studied in Arabidopsis and rice to date. Arabidopsis miR825∗, a 22-nt small RNA that can potentially initiate the production of secondary siRNAs (Zhai et al., 2011; Li et al., 2012), targets toll-interleukin-like receptor (TIR)-nucleotide binding site (NBS) and leucine-rich repeat (LRR)-type resistance (R) genes to enhance resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 infection in a manner strictly dependent on Bacillus cereus AR156 pretreatment (Niu et al., 2016). In Rice stripe virus (RSV)-infected rice plants, many miRNA∗s were also found to accumulate at higher levels than in normal plants; these miRNA∗s include the miR160∗, miR166∗, and miR396∗ families (Seo et al., 2013).
Co-Regulation
miRNA∗s are more divergent than miRNAs, but there is no doubt about their importance to biological functions (Smith et al., 2015; Jain and Das, 2016). miRNA∗s cannot only function independently in biological processes, moreover, some miRNA∗s and their corresponding miRNAs act together to regulate intracellular activity (Hsieh et al., 2009; Zhang et al., 2011; Hu et al., 2014; Niu et al., 2016; Liu and Sun, 2017). Zhang et al. (2011) explored the function of miR393/miR393∗ in Arabidopsis, showing that miR393 is loaded into AGO1 and mediates pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) by negatively regulating auxin signaling pathways, whereas miR393∗ is loaded into AGO2 and mainly regulates plant effector-triggered immunity (ETI) responses by suppressing MEMB12 and promoting the exocytosis of antimicrobial PR proteins; thus, both strands contribute to antibacterial responses in a synergistic manner (Navarro et al., 2006). Analogously, miR825 and miR825∗ were significantly down-regulated in Pst DC3000-infected Arabidopsis plants after Bacillus cereus AR156 pretreatment, and both of them were confirmed suppressing genes involved in bacterial pathogen defense (Niu et al., 2016). In addition, other experimental results have shown that miRNA and the corresponding miRNA∗ cannot only regulate a common biological pathway but also cooperate to cleave a single target gene. Luo et al. (2012) found that both miR1511 and miR1511∗ in soybean cleaved the target gene GmRPL4a, which belongs to the 60S ribosomal protein L4 family and shows a greater than 80% similarity to the RPL4A and RPL4D proteins in Arabidopsis. Therefore, they suggested that miR1511/1511∗ may function in regulating the development of soybean leaves (Luo et al., 2012).
Furthermore, because of the high complementarity between miRNA and miRNA∗ sequences, certain miRNA∗s could potentially act as anti-miRNAs to inhibit the binding of their homologous miRNAs to the target transcripts, subsequently reducing the activities of those miRNAs (Ma et al., 2015). Meanwhile, the accumulation of the miRNA∗ might regulate the mature miRNA or the precursor itself (German et al., 2008; Li et al., 2010; Meng et al., 2010). In addition, base mutations of one strand in the duplex may result in binding to a different AGO protein (Zhang and Zhang, 2012; Zhang et al., 2015). However, little research exists on the self-regulation of miRNA∗, and thus, this aspect needs to be further explored to deepen our understanding.
Conclusion
miRNA∗, the passenger strand of the miRNA/miRNA∗ duplex, was generally thought to be degraded after the formation of mature miRNAs. However, all studies of miRNA∗ have shown that the abundance of miRNA∗s and their biological functionality are not an occasional event but are universal in plant species. Based on the discoveries to date regarding the characteristics of miRNA∗ accumulation in specific tissues, immune responses to biotic and abiotic stresses, regulation of growth and development and co-regulation with mature miRNA, the miRNA∗ mediating mechanism represents a complex system of gene expression regulation. Although the binding of both miRNAs and miRNA∗s to AGO proteins has a 5′-terminal nucleotide preference, a few miRNAs and miRNA∗s still do not conform to this general rule. In addition to the 5′-terminal nucleotides, many other factors may also affect RNA binding to different AGO proteins, such as secondary structure and nucleotide sequence length. Furthermore, several miRNA∗s, such as ath-miR393b∗, osa-miR810a∗, and osa-miR1433∗, are expressed in various tissues and organs but not just in special tissue, indicating their widespread activities in plants (Shao et al., 2013). While some areas remain to be explored, the exploration of miRNA∗ characterization and function to date has further enriched our knowledge of the complex regulatory networks of microRNA systems and provided important clues for research into organism gene expression and regulation.
Author Contributions
W-wL wrote the manuscript. JC, Y-sL, and JM contributed in revising manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31471880 and 61472061).
References
Aceto, S., Sica, M., Paolo, S. D., D’Argenio, V., Cantiello, P., Salvatore, F., et al. (2014). The analysis of the inflorescence miRNome of the orchid Orchis italica reveals a DEF-Like MADS-Box gene as a new miRNA target. PLOS ONE 9:e97839. doi: 10.1371/journal.pone.0097839
Barciszewska-Pacak, M., Milanowska, K., Knop, K., Bielewicz, D., Nuc, P., Plewka, P., et al. (2015). Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 6:410. doi: 10.3389/fpls.2015.00410
Bartel, B., and Bartel, D. P. (2003). MicroRNAs: at the root of plant development? Plant Physiol. 132, 709–717. doi: 10.1104/pp.103.023630
Cai, X., Hagedorn, C. H., and Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966. doi: 10.1261/rna.7135204
Casati, P. (2013). Analysis of UV-B regulated miRNAs and their targets in maize leaves. Plant Signal. Behav. 8:e26758. doi: 10.4161/psb.26758
Chen, M., Bao, H., Wu, Q., and Wang, Y. (2015). Transcriptome-wide identification of miRNA targets under nitrogen deficiency in Populus tomentosa using degradome sequencing. Int. J. Mol. Sci. 16, 13937–13958. doi: 10.3390/ijms160613937
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/annurev.cellbio.042308.113417
Devers, E. A., Branscheid, A., May, P., and Krajinski, F. (2011). Stars and symbiosis: microRNA- and microRNA∗-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156, 1990–2010. doi: 10.1104/pp.111.172627
Du, P., Wu, J., Zhang, J., Zhao, S., Zheng, H., Gao, G., et al. (2011). Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLOS Pathog. 7:e1002176. doi: 10.1371/journal.ppat.1002176
Du, Q., Zhao, M., Gao, W., Sun, S., and Li, W.-X. (2017). MicroRNA/microRNA∗ complementarity is important for the regulation pattern of NFYA5 by miR169 under dehydration shock in Arabidopsis. Plant J. 91, 22–33. doi: 10.1111/tpj.13540
Du, T., and Zamore, P. D. (2005). MicroPrimer: the biogenesis and function of microRNA. Primer 132, 4645–4652. doi: 10.1242/dev.02070
German, M. A., Pillay, M., Jeong, D.-H., Hetawal, A., Luo, S., Janardhanan, P., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. doi: 10.1038/nbt1417
Ghildiyal, M., Xu, J., Seitz, H., Weng, Z., and Zamore, P. D. (2010). Sorting of Drosophila small silencing RNAs partitions microRNA∗ strands into the RNA interference pathway. RNA 16, 43–56. doi: 10.1261/rna.1972910
Gu, M., Xu, K., Chen, A., Zhu, Y., Tang, G., and Xu, G. (2010). Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant. 138, 226–237. doi: 10.1111/j.1399-3054.2009.01320.x
Guo, L., and Lu, Z. (2010). The fate of miRNA∗ strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLOS ONE 5:e11387. doi: 10.1371/journal.pone.0011387
Hackenberg, M., Shi, B.-J., Gustafson, P., and Langridge, P. (2013). Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. 13:214. doi: 10.1186/1471-2229-13-214
Hivrale, V., Zheng, Y., Puli, C. O. R., Jagadeeswaran, G., Gowdu, K., Kakani, V. G., et al. (2016). Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 242, 214–223. doi: 10.1016/j.plantsci.2015.07.018
Hsieh, L.-C., Lin, S.-I., Shih, A. C.-C., Chen, J.-W., Lin, W.-Y., Tseng, C.-Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151, 2120–2132. doi: 10.1104/pp.109.147280
Hu, W., Wang, T., Yue, E., Zheng, S., and Xu, J.-H. (2014). Flexible microRNA arm selection in rice. Biochem. Biophys. Res. Commun. 447, 526–530. doi: 10.1016/j.bbrc.2014.04.036
Jain, A., and Das, S. (2016). Synteny and comparative analysis of miRNA retention, conservation, and structure across Brassicaceae reveals lineage- and sub-genome-specific changes. Funct. Integr. Genomics 16, 253–268. doi: 10.1007/s10142-016-0484-1
Jeong, D.-H., Thatcher, S. R., Brown, R. S. H., Zhai, J., Park, S., Rymarquis, L. A., et al. (2013). Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiol. 162, 1225–1245. doi: 10.1104/pp.113.219873
Jia, L., Zhang, D., Qi, X., Ma, B., Xiang, Z., and He, N. (2014). Identification of the conserved and novel miRNAs in Mulberry by high-throughput sequencing. PLOS ONE 9:e104409. doi: 10.1371/journal.pone.0104409
Jones-Rhoades, M. W., Bartel, D. P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Khvorova, A., Reynolds, A., and Jayasena, S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216. doi: 10.1016/S0092-8674(03)00801-8
Kim, J. H., Go, Y. S., Kim, J. K., and Chung, B. Y. (2016). Characterization of microRNAs and their target genes associated with transcriptomic changes in gamma-irradiated Arabidopsis. Genet. Mol. Res. 15:gmr.15038386. doi: 10.4238/gmr.15038386
Kozomara, A., and Griffiths-Jones, S. (2014). MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Lee, R. C., Feinbaum, R. L., and Ambrost, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lee, Y., Kim, M., Han, J., Yeom, K.-H., Lee, S., Baek, S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060. doi: 10.1038/sj.emboj.7600385
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, Y.-F., Zheng, Y., Addo-Quaye, C., Zhang, L., Saini, A., Jagadeeswaran, G., et al. (2010). Transcriptome-wide identification of microRNA targets in rice. Plant J. 62, 742–759. doi: 10.1111/j.1365-313X.2010.04187.x
Liu, B., and Sun, G. (2017). MicroRNAs contribute to enhanced salt adaptation of the autopolyploid Hordeum bulbosum compared with its diploid ancestor. Plant J. 91, 57–69. doi: 10.1111/tpj.13546
Lukasik, A., Pietrykowska, H., Paczek, L., Szweykowska-Kulinska, Z., and Zielenkiewicz, P. (2013). High-throughput sequencing identification of novel and conserved miRNAs in the Brassica oleracea leaves. BMC Genomics 14:801. doi: 10.1186/1471-2164-14-801
Luo, Z., Jin, L., and Qiu, L. (2012). MiR1511 co-regulates with miR1511∗ to cleave the GmRPL4a gene in soybean. Chin. Sci. Bull. 57, 3804–3810. doi: 10.1007/s11434-012-5197-9
Ma, X., Tang, Z., Qin, J., and Meng, Y. (2015). The use of high-throughput sequencing methods for plant microRNA research. RNA Biol. 12, 709–719. doi: 10.1080/15476286.2015.1053686
Manavella, P. A., Koenig, D., Rubio-Somoza, I., Burbano, H. A., Becker, C., and Weigel, D. (2013). Tissue-specific silencing of Arabidopsis SU(VAR)3-9 HOMOLOG8 by miR171a∗. Plant Physiol. 161, 805–812. doi: 10.1104/pp.112.207068
Manavella, P. A., Koenig, D., and Weigel, D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U.S.A. 109, 2461–2466. doi: 10.1073/pnas.1200169109
Meng, Y., Gou, L., Chen, D., Wu, P., and Chen, M. (2010). High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J. Exp. Bot. 61, 3833–3837. doi: 10.1093/jxb/erq209
Meng, Y., Shao, C., Gou, L., Ji, Y., and Chen, M. (2011). Construction of MicroRNA- and MicroRNA∗-mediated regulatory networks in plants. RNA Biol. 8, 1124–1148. doi: 10.4161/rna.8.6.17743
Meng, Y., Shao, C., Ma, X., Wang, H., and Chen, M. (2012). Expression-based functional investigation of the organ specific microRNAs in Arabidopsis. PLOS ONE 7:e50870. doi: 10.1371/journal.pone.0050870
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Niu, D., Xia, J., Jiang, C., Qi, B., Ling, X., Lin, S., et al. (2016). Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825∗ and activating defense-related genes in Arabidopsis. J. Integr. Plant Biol. 58, 426–439. doi: 10.1111/jipb.12446
Okamura, K., Phillips, M. D., Tyler, D. M., Duan, H., Chou, Y., and Lai, E. C. (2008). The regulatory activity of microRNA∗ species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 15, 354–363. doi: 10.1038/nsmb.1409
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W.-R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Pant, B. D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J., et al. (2009). Identification of nutrient-responsive Arabidopsis and Rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541–1555. doi: 10.1104/pp.109.139139
Peng, T., Lv, Q., Zhang, J., Li, J., Du, Y., and Zhao, Q. (2011). Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa). J. Exp. Bot. 62, 4943–4954. doi: 10.1093/jxb/err205
Peng, T., Sun, H., Du, Y., Zhang, J., Li, J., Liu, Y., et al. (2013). Characterization and expression patterns of microRNAs involved in rice grain filling. PLOS ONE 8:e54148. doi: 10.1371/journal.pone.0054148
Poulsen, C., Vaucheret, H., and Brodersen, P. (2013). Lessons on RNA silencing mechanisms in plants from eukaryotic Argonaute structure. Plant Cell 25, 22–37. doi: 10.1105/tpc.112.105643
Pumplin, N., and Harrison, M. J. (2009). Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 151, 809–819. doi: 10.1104/pp.109.141879
Qi, Y., Denli, A. M., and Hannon, G. J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19, 421–428. doi: 10.1016/j.molcel.2005.06.014
Qi, Y., He, X., Wang, X.-J., Kohany, O., Jurka, J., and Hannon, G. J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012. doi: 10.1038/nature05198
Ren, Y., Sun, F., Hou, J., Chen, L., Zhang, Y., Kang, X., et al. (2015). Differential profiling analysis of miRNAs reveals a regulatory role in low N stress response of Populus. Funct. Integr. Genomics 15, 93–105. doi: 10.1007/s10142-014-0408-x
Schwarz, D. S., Hutvágner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208. doi: 10.1016/S0092-8674(03)00759-1
Seo, J.-K., Wu, J., Lii, Y., Li, Y., and Jin, H. (2013). Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 6, 617–625. doi: 10.1094/MPMI-10-12-0255-IA
Shao, C., Ma, X., Xu, X., and Meng, Y. (2013). Identification of the highly accumulated microRNA∗s in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Gene 515, 123–127. doi: 10.1016/j.gene.2012.xyb11.015
Smith, L. M., Burbano, H. A., Wang, X., Fitz, J., Wang, G., Ural-Blimke, Y., et al. (2015). Rapid divergence and high diversity of miRNAs and miRNA targets in the Camelineae. Plant J. 81, 597–610. doi: 10.1111/tpj.12754
Stefani, G., and Slack, F. J. (2008). Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230. doi: 10.1038/nrm2347
Takeda, A., Iwasaki, S., Watanabe, T., Utsumi, M., and Watanabe, Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 49, 493–500. doi: 10.1093/pcp/pcn043
Thiebaut, F., Grativol, C., Tanurdzic, M., Carnavale-Bottino, M., Vieira, T., Motta, M. R., et al. (2014). Differential sRNA regulation in leaves and roots of Sugarcane under water depletion. PLOS ONE 9:e93822. doi: 10.1371/journal.pone.0093822
Trevisan, S., Nonis, A., Begheldo, M., Manoli, A., Palme, K., Caporale, G., et al. (2012). Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ. 35, 1137–1155. doi: 10.1111/j.1365-3040.2011.02478.x
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, H., Zhang, X., Liu, J., Kiba, T., Woo, J., Ojo, T., et al. (2011). Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J. 67, 292–304. doi: 10.1111/j.1365-313X.2011.04594.x
Xie, F., and Zhang, B. (2015). MicroRNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol. J. 13, 421–434. doi: 10.1111/pbi.12295
Yaish, M. W., Sunkar, R., Zheng, Y., Ji, B., Al-Yahyai, R., and Farooq, S. A. (2015). A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front. Plant Sci. 6:946. doi: 10.3389/fpls.2015.00946
Yan, J., Zhang, H., Zheng, Y., and Ding, Y. (2014). Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta 241, 109–123. doi: 10.1007/s00425-014-2167-2
Yu, X., Wang, H., Lu, Y., de Ruiter, M., Cariaso, M., Prins, M., et al. (2012). Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 63, 1025–1038. doi: 10.1093/jxb/err337
Zhai, J., Jeong, D.-H., De Paoli, E., Park, S., Rosen, B. D., Li, Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhang, J., Huang, M., Liang, J., Pan, Y., Cheng, L., Wu, J., et al. (2016). Genome-wide mining for microRNAs and their targets in Betula luminifera using high-throughput sequencing and degradome analyses. Tree Genet. Genomes 12:99. doi: 10.1007/s11295-016-1047-2
Zhang, X., Niu, D., Carbonell, A., Wang, A., Lee, A., Tun, V., et al. (2015). ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 5:5468. doi: 10.1038/ncomms6468
Zhang, X., Zhao, H., Gao, S., Wang, W.-C., KatiyarAgarwal, S., Huang, H.-D., et al. (2011). Arabidopsis Argonaute 2 regulates innate immunity via miRNA393∗-mediated silencing of a Golgi-localized SNARE gene MEMB12. Mol. Cell 42, 356–366. doi: 10.1016/j.molcel.2011.04.010
Zhang, Z., Liu, X., Guo, X., Wang, X.-J., and Zhang, X. (2016). Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat. Plants 2, 1–7. doi: 10.1038/nplants.2016.49
Zhang, Z., and Zhang, X. (2012). Argonautes compete for miR165/166 to regulate shoot apical meristem development. Curr. Opin. Plant Biol. 15, 652–658. doi: 10.1016/j.pbi.2012.05.007
Keywords: miRNA, miRNA∗, passenger strand, ARGONAUTE protein, stress response
Citation: Liu W-w, Meng J, Cui J and Luan Y-s (2017) Characterization and Function of MicroRNA∗s in Plants. Front. Plant Sci. 8:2200. doi: 10.3389/fpls.2017.02200
Received: 13 October 2017; Accepted: 14 December 2017;
Published: 22 December 2017.
Edited by:
Elisabeth Jamet, Université Toulouse III Paul Sabatier, FranceReviewed by:
Jean-philippe Combier, Centre National de la Recherche Scientifique (CNRS), FranceShabir Hussain Wani, Michigan State University, United States
Copyright © 2017 Liu, Meng, Cui and Luan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Meng, bWVuZ2p1bkBkbHV0LmVkdS5jbg== Yu-shi Luan, eXNsdWFuQGRsdXQuZWR1LmNu
 Wei-wei Liu
Wei-wei Liu Jun Meng
Jun Meng Jun Cui
Jun Cui Yu-shi Luan
Yu-shi Luan