- 1School of Environmental and Life Sciences, University of Newcastle, Callaghan, NSW, Australia
- 2School of Mathematical and Physical Sciences, University of Newcastle, Callaghan, NSW, Australia
Transfer cells (TCs) support high rates of membrane transport of nutrients conferred by a plasma membrane area amplified by lining a wall labyrinth comprised of an uniform wall layer (UWL) upon which intricate wall ingrowth (WI) papillae are deposited. A signal cascade of auxin, ethylene, extracellular hydrogen peroxide (H2O2) and cytosolic Ca2+ regulates wall labyrinth assembly. To identify gene cohorts regulated by each signal, a RNA- sequencing study was undertaken using Vicia faba cotyledons. When cotyledons are placed in culture, their adaxial epidermal cells spontaneously undergo trans-differentiation to epidermal TCs (ETCs). Expressed genes encoding proteins central to wall labyrinth formation (signaling, intracellular organization, cell wall) and TC function of nutrient transport were assembled. Transcriptional profiles identified 9,742 annotated ETC-specific differentially expressed genes (DEGs; Log2fold change > 1; FDR p ≤ 0.05) of which 1,371 belonged to signaling (50%), intracellular organization (27%), cell wall (15%) and nutrient transporters (9%) functional categories. Expression levels of 941 ETC-specific DEGs were found to be sensitive to the known signals regulating ETC trans-differentiation. Significantly, signals acting alone, or in various combinations, impacted similar numbers of ETC-specific DEGs across the four functional gene categories. Amongst the signals acting alone, H2O2 exerted most influence affecting expression levels of 56% of the ETC-specific DEGs followed by Ca2+ (21%), auxin (18%) and ethylene (5%). The dominance by H2O2 was evident across all functional categories, but became more attenuated once trans-differentiation transitioned into WI papillae formation. Amongst the eleven signal combinations, H2O2/Ca2+ elicited the greatest impact across all functional categories accounting for 20% of the ETC-specific DEG cohort. The relative influence of the other signals acting alone, or in various combinations, varied across the four functional categories and two phases of wall labyrinth construction. These transcriptome data provide a powerful information platform from which to examine signal transduction pathways and how these regulate expression of genes encoding proteins engaged in intracellular organization, cell wall construction and nutrient transport.
Introduction
Transfer cells (TCs) differentiate at cellular sites supporting high rates of apo-/symplasmic exchange of nutrients. These sites can include interfaces between soil/root, maternal/filial tissues of developing seeds and biotroph/plant host as well as loading/unloading regions of phloem and xylem (Offler et al., 2003). The TC capacity for apo-/symplasmic exchange of nutrients is conferred by development of a wall labyrinth that forms a scaffold supporting an amplified (10- to 20-fold) plasma membrane area enriched in transporters. As a result, TCs can contribute to plant productivity and crop yield by enhancing overall (e.g., soil/root interface – Schmidt and Bartels, 1996) or directing preferential (e.g., developing seeds – Andriunas et al., 2013) nutrient flows to nourish whole plant or selective organ development respectively. Equally, TCs can compromise plant productivity and crop yield in those circumstances where the invading pathogen (e.g., root knot nematodes – Cabrera et al., 2014) has hijacked the developmental program responsible for constructing the TC phenotype to support their nutritional requirements. Thus, in the quest to improve crop yield and crop protection, there are compelling imperatives to discover the cell and molecular mechanisms underpinning TC development.
A major impediment to discovering cell and molecular mechanisms underpinning TC development is that, in most circumstances, TCs are embedded within tissue matrices and occur in low numbers (Offler et al., 2003). An experimental system that overcomes this technical impasse is one in which 90% of the adaxial epidermal cell population of developing Vicia faba cotyledons spontaneously undergo quasi-synchronous trans-differentiation to a TC phenotype upon excision and transfer to a culture medium (Offler et al., 1997; Wardini et al., 2007). The phenotype of the adaxial epidermal TC (ETC), structurally and functionally mirrors that of their abaxial ETC counterparts (Farley et al., 2000; Talbot et al., 2001). Their location on the cotyledon surface in large numbers (thousands) and ready isolation in epidermal peels has opened up opportunities for replicated cell (both live and preserved) and molecular biology investigations of their development (Andriunas et al., 2013). A complex of sequential signals comprising auxin (Dibley et al., 2009), ethylene (Zhou et al., 2010) negatively regulated by glucose through a hexokinase pathway (Andriunas et al., 2011), extracellular H2O2 (Andriunas et al., 2012; Xia et al., 2012) and cytosolic Ca2+ (Ca2+; Zhang et al., 2015a) participate in regulating construction of the wall labyrinth comprised of an UWL upon which WI papillae form (Andriunas et al., 2013). The phasic temporal development of the UWL followed by that of WI papillae has permitted identification of the ETC-specific transcriptomes associated with each of these phases of wall labyrinth construction (Zhang et al., 2015d; Arun-Chinnappa and McCurdy, 2016). Comparable temporal studies have been undertaken on developing basal endosperm TCs of barley (Thiel, 2014) and maize (Zhan et al., 2015) grains as well as on nematode-induced giant cells formed in roots (Cabrera et al., 2014, 2016). Collectively this work has identified TC-specific genes encoding signal cascades (Yuan et al., 2016) and downstream machinery responsible for constructing the TC wall labyrinth and membrane transporters that confer TC function (Cabrera et al., 2014; Thiel, 2014; Zhan et al., 2015; Zhang et al., 2015d). A significant outstanding question is to discover the cohorts of downstream genes regulated by each of the various signaling cascades.
As a first step in filling this knowledge gap described above, we designed a RNA-seq experiment to identify gene cohorts controlled by each signal currently known to participate in regulating ETC trans-differentiation (i.e., auxin, ethylene, H2O2 and Ca2+) of cultured V. faba cotyledons. To this end, cotyledons were cultured on media in the absence (control) and presence (treated) of pharmacological blockers of each signal. Transcriptome comparisons between ETC and underlying SPC, obtained from control cotyledons, identified the assemblage of DEGs specifically expressed in the trans-differentiating ETCs. The analysis was further refined by identifying those ETC-specific DEGs that were located in functional categories (Mapman and KEGG) considered to contribute to wall labyrinth construction (i.e., signaling, intracellular organization and cell wall) and to TC function in nutrient transport. Numbers of ETC-specific DEGs in these TC functional categories regulated by one or more signals were deduced from determining whether their transcript levels were significantly altered in cotyledons cultured on each of the pharmacological blockers. Overall, the findings demonstrated that signals acting alone exerted control over the greater number of ETC-specific DEGS during UWL construction while various signal combinations assumed dominance during the subsequent developmental phase of WI papillae formation. H2O2, acting alone or in combination with other signals, regulated expression of the greatest number of ETC-specific DEGs during both phases of wall labyrinth construction and within each of the four gene functional categories examined. Numbers of ETC-specific DEGs influenced by the remaining signals in descending order, were Ca2+, auxin and ethylene with H2O2/Ca2+ being the strongest signal combination.
Materials and Methods
Plant Growth Conditions, Cotyledon Culture and Collection of Tissue Samples for Sequencing
Vicia faba L. (cv. Fiord) plants were raised under controlled environmental conditions (Dibley et al., 2009). Cotyledons were freshly harvested or cultured for specified times on liquid MS medium containing ± specified pharmacological agents to block signal action (see Experimental Design for choice of culture time and agents). Each pharmacological agent was applied at a concentration that did not impact cell viability as verified by staining tissue sections of cultured cotyledons with 0.1% (w/v) tetrazolium blue for 20 min. At harvest, cotyledons were immediately fixed in 75% ethanol and 25% acetic acid for 1 h at 4 °C. Peels of the trans-differentiating adaxial ETCs were collected from replicate batches of fixed cotyledons exposed to each pharmacological treatment and pooled (ca 16 mg fresh weight of epidermal peels/replicate batch), immediately snap-frozen in liquid nitrogen and stored at -80 °C until used for RNA extraction (Dibley et al., 2009).
RNA Isolation, cDNA-Library Construction and Illumina Sequencing
Total RNA was extracted using Qiagen (USA) RNeasy plant mini kits. Contaminating genomic DNA was removed using DNase I. Total RNA quality was verified by determining the integrity of the 25S and 18S RNA with an Agilent 2100 Bioanalyzer (Agilent, United States; Zhang et al., 2015d).
cDNA libraries were prepared from poly-A mRNA isolated from 1 μg of total RNA of each biological replicate using a TruSeq® RNA V2 sample preparation kit (Illumina, United States) according to the manufacturer’s instructions. cDNA quality was evaluated by determining size and purity using an Agilent 2100 Bioanalyzer. cDNA fragments, ranging from 100 to 700 bp, were selected by agarose gel purification and indexed with unique nucleic acid identifiers (Illumina TruSeq V2 index sequence). The indexed cDNA libraries were diluted to an average concentration of 10 nM and pooled in equal volumes (10 μL of each library) to generate the final mixed cDNA pool for sequencing. The pool was then sequenced using 100 bp pair-end sequencing strategy on an Illumina HiSeq 2000 platform (Australian Genome Research Facility, Melbourne, VIC, Australia).
Filtering, Mapping and Annotation of Sequenced Reads
Using Illumina CASAVA pipeline version 1.8.2, raw reads were trimmed with adaptor filtering and a read length cut-off of 50%. Thereafter, filtered reads with over 20% of their nucleotides having a Q score < 20 (probability of sequencing error > 0.01) or their sequences having an N reading over 5% were removed. Filtered reads were mapped to the previously constructed transcriptome library of trans-differentiating adaxial ETCs of V. faba cotyledons (Zhang et al., 2015d) using SOAPallgner/SOAP2 (BGI, China) to obtain abundance of transcripts, normalized as RPKM.
All mapped unigenes were re-annotated using the Mercator pipeline1 (Lohse et al., 2014) against the following publically available databases: The Arabidopsis Information Resource (TAIR10) proteins (Swarbreck et al., 2010), SwissProt/Uniprot plant proteins2, CDD (Marchler-Bauer et al., 2007), COG3, JGI Chlamy release 4 Augustus models (CHLAMY; Nordberg et al., 2014), TIGR5 rice proteins (ORYZA; Kawahara et al., 2013) and InterProScan (Zdobnov and Apweiler, 2001). A KEGG4 enzymatic pathway annotation was also performed. In all annotations, an e-value threshold of 1e-5 was used as a cut-off for meaningful annotation.
Differentially Expressed Gene Analysis and Other Statistical Tests
Any outlier replicate RPKM values within each culture time/treatment were identified (Grubbs, 1969) and removed. Only genes exhibiting mean RPKM values > 1 were retained for further analysis. DEGs between culture times of control cotyledons were identified in the trans-differentiating ETCs using limmaR (Ritchie et al., 2015) and in SPCs employing edgeR (McCarthy et al., 2012; rationale outlined in the Experimental Design section). Genes were considered differentially expressed if their Log2-fold change was greater than 1 with a FDR of p ≤ 0.05 (Benjamini and Hochberg, 1995).
Impacts of blocking inductive signals on TC-specific DEGs were assessed by comparing RPKM values for control against those for each signal using lmFit function in limmaR to conduct independent sample t-tests, with a two-tailed probability value threshold of 0.05 and no FDR restriction.
Quantitative RT-PCR Validation
Aliquots from the RNA samples sent for Illumina sequencing were converted to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen, United States). Primers were designed using Primer 3 plus (Whitehead Institute for Biomedical Research, United States) and synthesized by Sigma–Aldrich Australia (see Supplementary Table S1 for primer sequences). For each qRT-PCR reaction, a 15 μL mix containing 7.5 μL SYBR Green master mix (Qiagen, United States), 0.375 μL of forward and reverse primers (10 μM), 1.75 μL of nuclease free H2O and 5 μL cDNA was set up. The following PCR cycle was used: 95°C for 5 min, 95°C for 15 s, 60°C for 20 s, 72°C for 30 s; steps 2–4 were repeated 50 times. High-resolution melting curves (72–95°C) following the final PCR cycle checked the specificity of the PCR products. For each cDNA sample, technical duplicates for each biological replicate (n = 3) were tested.
Four housekeeping gene candidates, elongation factor 2-α (VfEF2α), NADH dehydrogenase subunit 4 (VfNADHD4), 60S ribosomal protein subunit L2 (Vf60SL2), and multi-domain cyclophilin type peptidyl-prolyl cis-trans isomerase G (VfPPlaseG) were selected based on their stable expression in adaxial epidermal cells during TC trans-differentiation (Supplementary Figure S1). These candidates were further assessed (see Supplementary Table S1 for primer sequences) using GeNorm analysis of real-time PCR data derived from cDNA of epidermal peels that collectively yielded a reliable reference M value of <0.15 (Vandesompele et al., 2002). Relative expression levels of each unigene were determined using the ΔΔct method.
Scanning Electron Microscopy
Protoplasts of adaxial epidermal peels (Dibley et al., 2009) were removed by washing in 2% (v/v) NaClO for 3 h at room temperature with hourly changes followed by three 10-min washes in dH2O to remove the bleach. Peels then were placed in small steel cages and dehydrated at 4°C through a 10% step-graded ethanol/dH2O series, changed at 30-min intervals from 10% up to 100% ethanol. After holding in 100% ethanol at least overnight, peels were critical-point dried with liquid CO2 in a critical-point drier (Balzers Union, Liechtenstein). The dried peels were orientated outer face down on sticky carbon tabs to reveal the cytoplasmic face of their outer periclinal cell walls. Samples were sputter coated with gold to a thickness of 20 nm in a sputter-coating unit (SPI Suppliers, United States) and viewed at 15 kV with a Philips XL30 scanning electron microscope.
Results and Discussion
Experimental Design
Selection of Cotyledon Culture Times and Pharmacological Agents for RNA-Sequencing
Cotyledons were cultured for specified times to capture transcriptional activities during the two phases of wall labyrinth construction. Freshly harvested cotyledons were used as a baseline transcriptome of the adaxial epidermal cells before their induction to undergo trans-differentiation to a TC-morphology (Figure 1). A 3-h culture time was chosen as representative of the transcriptome during UWL formation with >40% of the trans-differentiating ETCs exhibiting an UWL (Zhang et al., 2015d) while WI papillae formation was evident in <10% of cells (Wardini et al., 2007). Following 12 h of cotyledon culture, UWL formation has ceased (Zhang et al., 2015d) while WI papillae construction continues in over 75% of cells (Wardini et al., 2007). Thus 12-h cultured cotyledons offer a transcriptome representative of trans-differentiating ETCs supporting WI papillae construction alone (Figure 1).
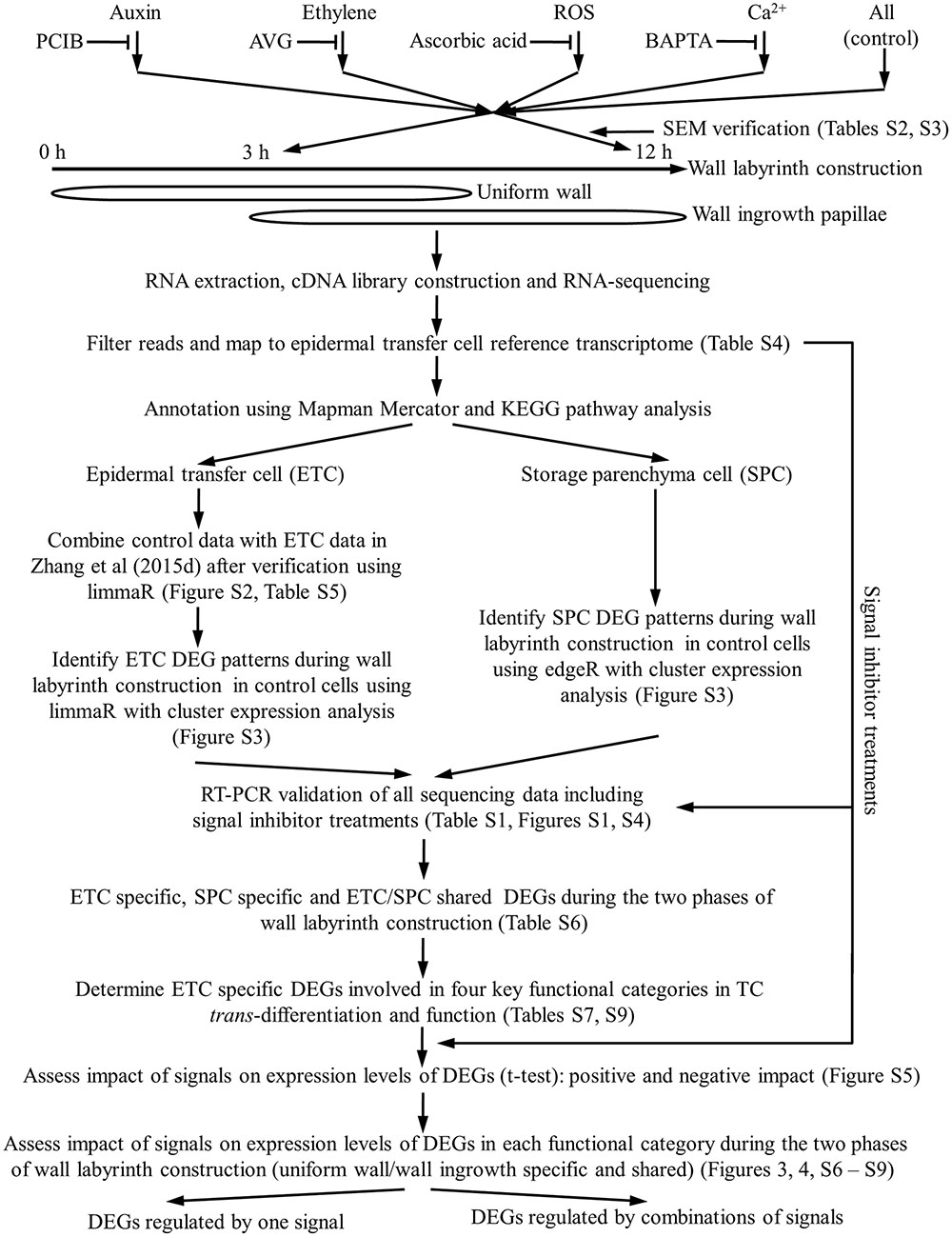
FIGURE 1. Schematic flow chart illustrating the experimental design used to investigate signal regulation of the transcriptional network underpinning TC trans-differentiation and function.
The four signals (auxin, ethylene, H2O2 and Ca2+) identified to date as responsible for regulating trans-differentiation to a TC-morphology, were blocked using pharmacological agents known to be effective in the V. faba cotyledon culture system (Figure 1 and, for more details, see Supplementary Table S2). Cotyledons were either freshly harvested, or cultured in the absence/presence of the signal blockers (Supplementary Table S2), for 4 h at 4°C to allow the blocker to enter the cotyledons prior to their epidermal cells being rapidly induced to undergo trans-differentiation following transfer to 26°C for a further 3 or 12 h of culture (see for example, Zhang et al., 2015b). To verify that each set of cultured cotyledons was trans-differentiating and responding to the pharmacological blockades, percentages of ETCs forming WI papillae of a sub-set of cotyledons at 12 h of culture were scored along with ETC viability (Supplementary Table S3). Only those biological replicate batches of cotyledons that complied with previously reported responses of WI papillae formation to the pharmacological agents (Supplementary Table S3) were processed for RNA-seq analysis to determine their transcriptomes (Figure 1). That the pharmacological agents specifically block ETC trans-differentiation has been demonstrated previously whereby an exogenous supply of each signal fully restored WI papillae formation in the presence of the respective pharmacological blockade (see Supplementary Table S3).
Preliminary Analysis of RNA-Seq Reads and Strategy to Identify TC-Specific Differentially Expressed Genes
Fifty-nine to 71 million clean reads were generated for each biological replicate (Supplementary Table S4; Zhang et al., 2015d). Unigene transcript abundance was determined for only uniquely mapped reads (58% of total mapped reads) normalized as RPKM values. All unigenes then had their biochemical functions annotated using Mapman Mercator and KEGG pathway analyses.
To increase the statistical power of the control RNA-seq RPKM data set, we evaluated whether these data could be combined with that reported in Zhang et al. (2015d). Multidimensional scaling plots of these two data sets using limmaR (Ritchie et al., 2015) showed tight coupling of gene expression profiles between the two data sets at each time point (Supplementary Figure S2). Further analysis using limmaR DEG analysis showed that between these two data sets, at each time point, >99% of genes shared identical expression levels (FDR corrected p > 0.05; Supplementary Table S5). For transcripts fulfilling this criterion, their RPKM values from the two data sets were combined increasing replicate numbers to six. Those not satisfying this criterion were excluded from further analysis (Figure 1).
Temporal changes of control RPKM values of unigenes expressed in ETCs were assessed using limmaR (for details see Materials and Methods). Nine temporal expression profile patterns were identified for up- and down-regulated DEGs (Supplementary Figure S3). These patterns were aggregated into three broad temporal expression profiles of UWL specific (0–3 h), WI papillae specific (3 – 12 h) and shared between the two phases of wall labyrinth construction. In each temporal expression profile, DEGs that were up- or down-regulated are presented separately (Figure 1 and Table 1). To identify ETC-specific DEGs in these profiles, a similar analysis of temporal gene expression was undertaken for the SPCs. To ensure a conservative identification of ETC-specific DEGs, the rigor of limmaR analysis applied to the epidermal cell data set was relaxed somewhat using edgeR (McCarthy et al., 2012) to identify DEGs in the SPCs (Figure 1 and Supplementary Figure S3). Comparison of the temporal expression profiles for the two cell types in each of the three broad temporal expression profiles, identified DEGs that were ETC-specific, SPC specific and shared between the two cell types (Figure 1, Table 1, and Supplementary Table S6).
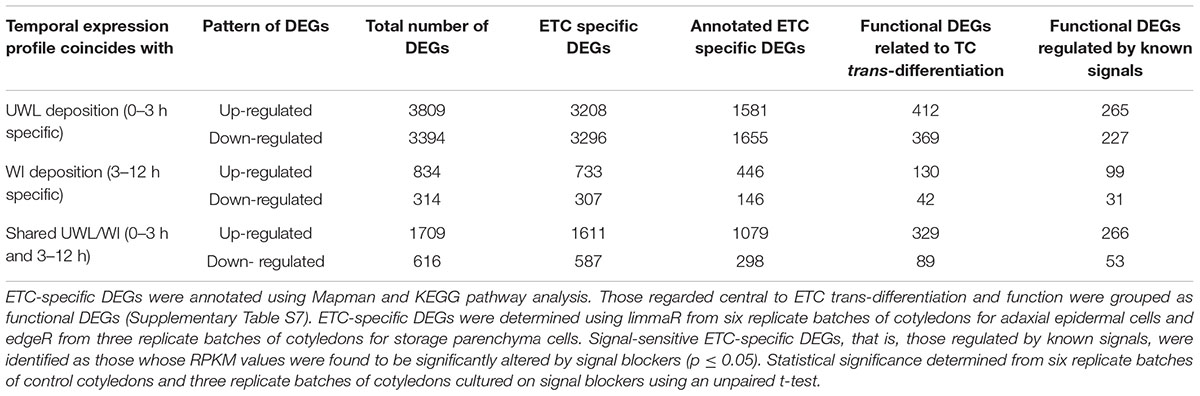
TABLE 1. Global identification of DEGs in trans-differentiating ETCs expressed at the two phases of wall labyrinth construction.
RT-qPCR Validation of RNA-Seq Expression Data
Gene expression profiles obtained using RNA-seq were validated by RT-PCR (Figure 1). Transcripts of 14 genes encoding proteins linked with the four identified signals were selected. Of these, nine were UWL-specific (six up-regulated, three down-regulated) and five WI papillae specific (three up-regulated, two down-regulated). RT-qPCR estimates of gene expression were normalized as Log2 fold change against their 0 h expression levels and plotted against the corresponding RNA-seq data. When fitted to a linear regression analysis the two estimates of gene expression were highly correlated indicating that the DEG estimates were reliable (Supplementary Figure S4).
DEGs in Key Functional Categories Central to TC Trans-Differentiation and Nutrient Transport
Genes encoding proteins participating in signaling, intracellular organization and cell wall construction are considered central to TC trans-differentiation while nutrient transporters are core to TC function (Figure 1). Genes were assigned to these categories based on biochemical and enzymatic functions as annotated by Mapman Mercator and KEGG (Supplementary Table S7). Mapman and KEGG annotations have been effectively used to investigate genes encoding proteins contributing to mycorrhizal associations (Calabrese et al., 2017) and root hair growth (Damiani et al., 2016).
Inclusion of Mapman and KEGG categories into each functional category was informed by previously reported transcriptome analyses of developing ETCs (Zhang et al., 2015d). Thus, the signaling category was allocated genes encoding proteins for biosynthesis, action and metabolism of hormones, respiratory burst oxidases, superoxide dismutases and peroxidases as well as Ca2+ transporters and proteins participating in Ca2+ signaling pathways (Andriunas et al., 2013; Thiel, 2014; Zhang et al., 2015a). Map kinases were added on the basis that these act as messengers for H2O2 and Ca2+ signaling in plant cells (e.g., Wurzinger et al., 2011; Meng and Zhang, 2013). The intracellular organization category was ascribed genes encoding proteins contributing to microdomain formation in plasma membranes (Thiel, 2014), cytoskeleton and vesicle trafficking (Thiel, 2014; Zhang et al., 2015b, 2017). The cell wall category captured genes encoding proteins involved in the biosynthesis, modification and degradation of cell walls (Thiel, 2014; Zhang et al., 2015d). The central role TCs play in nutrient transport (Offler et al., 2003) justified a category covering genes encoding nutrient transporters (Figure 1).
Strategies to Assess Impact of Signaling Molecules on Expression of ETC-Specific DEGs
How key the signals, auxin, ethylene, H2O2 and Ca2+, impacted gene expression during wall labyrinth construction was assessed by performing RNA-seq on ETCs isolated from cotyledons cultured in the presence of their pharmacological inhibitors (Figure 1 and Supplementary Tables S2, S3). The analysis was confined to control ETC-specific DEGs in the four functional gene categories (Supplementary Table S7) across each temporal expression profile (UWL-, WI papillae-specific and shared across both phases of wall labyrinth construction –Figure 1). Impacts of pharmacological removal of each signal were detected by determining whether the agent caused a statistically significant change of the control RPKM value of each DEG using an unpaired t-test. These tests were carried out on 3-h RPKM data for the UWL-specific DEGs and on 12-h RPKM data for WI papillae-specific and UWL/WI papillae shared DEGs (Supplementary Figure S5).
Overall Spatiotemporal Expression Profiles of ETC DEGs
RNA-seq analysis detected 33,423 and 27,437 expressed unigenes in the epidermal/ETCs and SPCs of cultured V. faba cotyledons respectively (Supplementary Table S8). As reported by Zhang et al. (2015d), numbers of unigenes expressed at the two phases of wall labyrinth construction in trans-differentiating ETCs were temporally stable (Supplementary Table S8). A similar temporal profile was detected in nematode induced giant cells of rice roots (Ji et al., 2013). The 12–16% higher number of unigenes detected in the epidermal and ETCs compared to SPCs (Supplementary Table S8) reflects the fact that the reference library used to map expressed genes was generated from peels of epidermal cells and developing ETCs (Zhang et al., 2015d).
The total number of DEGs detected in the ETCs and underlying SPCs during TC trans-differentiation were 10,676 and 3,772 respectively (Supplementary Table S6). Thus 30% of the unigenes expressed in the ETCs underwent significant change compared to 14% in the SPCs (cf. Supplementary Table S8). For the ETCs, the proportion of DEGs was higher than the 22% recorded for nematode-induced giant cells of rice roots across two phases of their wall labyrinth development (Ji et al., 2013). Comparable DEG percentages have been recorded for other single cell transcriptomes including those of tip growing pollen tubes (Zhao et al., 2016) and root hairs (Hey et al., 2017).
Consistent with a rapid shift in cell fate, 67% of ETC DEGs were detected during the first 3 h of trans-differentiation to a TC-morphology with comparable numbers of up- and down-regulated DEGs. During the later stages of trans-differentiation, the ratio of up- to down-regulated DEGs increased two to three-fold (Table 1). Such a developmental change in the ratio of up- to down-regulated DEGs also occurs during trans-differentiation of cucumber fruit epidermal cells into trichomes (Chen et al., 2014).
A 2.8-fold higher DEG number in the ETCs compared to SPCs is consistent with the former undergoing trans-differentiation from an epidermal to ETC fate while the SPCs remain committed to a SPC fate. This is further reflected by the differential in ratios between up- and down-regulated DEGs for the two cell types with a 2.3-fold higher ratio exhibited by the SPCs. This differential is most pronounced during the first 3 h of ETC induction. Here the ratio is 1.1 and 5.1 for the ETCs and SPCs respectively indicating a large cohort of epidermal cell fate genes were switched off as the epidermal cells transition to an ETC fate. Thereafter, the ratios between the cell types fell into the range of two to three (Supplementary Table S6). Similar ratios of up- and down-regulated genes were detected in nematode-induced TC giant cells of tomato roots at 7 days post infection (Portillo et al., 2013). In contrast, nematode-induced TC giant cells of rice roots at the stage of WI papillae formation, strongly favored up-regulated genes (Ji et al., 2013). The observed differences are suggested to be more apparent than real. They likely reflect the rapid (h) temporal changes in gene expression found for ETCs. For example, transcript levels of ethylene and ROS biosynthetic genes undergo marked changes within the first 3 h of cotyledon culture (Zhou et al., 2010; Andriunas et al., 2012).
Of the total DEGs expressed in the trans-differentiating ETCs, a high proportion (91%) representing 9,742 DEGS, were found to be ETC specific (Table 1). This indicates that ETC-centric differences in quantitative gene expression profiles between ETCs and SPCs contribute significantly to driving ETC trans-differentiation (Supplementary Table S6). Of the 579 DEGs shared between the ETCs and SPCs, up-regulated DEGs across 0 – 3 h or 3 – 12 h exhibited the higher proportion (12 and 16% respectively) compared to 3 and 2% for down-regulated DEGs across these phases of wall labyrinth construction (Supplementary Table S6).
Profiling ETC-Specific DEGS Within Each TC Functional Category
The TC-related functional categories of expressed genes chosen (Supplementary Table S7) represent 26% of the annotated ETC-specific DEG population with this proportion evenly represented by up- and down-regulated DEGs across both phases of wall labyrinth construction (Table 1). This proportion is somewhat higher than that found for ETC-specific expression of unigenes allocated to these functional categories (i.e., 17% – Zhang et al., 2015d). However, consistent with total DEG numbers, those found to be ETC-specific (i.e., 1,371; Table 1) were 3.8-fold greater than those of ETC-specific transcripts (365) reported in Zhang et al. (2015d). This finding further highlights the role DEGs play in regulating developmental pathways underpinning trans-differentiation to a TC phenotype.
The principal categories accounting for the remaining 74% of ETC-specific DEGs were general transcriptional regulation (21%, Mapman bins ‘RNA,’ ‘DNA’), general protein biosynthesis and modification (19%, Mapman bin ‘protein’), miscellaneous cell function and development (13%, Mapman bins ‘misc,’ ‘cell,’ ‘development’), general metabolism (12%, Mapman bins ‘major CHO metabolism,’ ‘minor CHO metabolism,’ ‘lipid metabolism,’ ‘amino acid metabolism’ and ‘secondary metabolism’) and stress response (9%, Mapman bin ‘stress’; for more details, see Supplementary Table S9).
Transcript numbers of ETC-specific DEGs detected within each functional category were in descending order signaling, intracellular organization, cell wall and nutrient transporters. These respectively accounted for 50, 27, 15, and 9% of the total ETC-specific DEG numbers in the four functional TC categories (Figure 2A). Up- and down-regulated DEG numbers also reflected the broad trend of DEG allocation between functional categories (Figures 2B,C). This finding indicated a substantive relative investment into DEGs encoding proteins participating in signaling pathways to orchestrate trans-differentiation to a TC phenotype. A similar proportionate investment into signaling was noted for expression of ETC-specific genes (Zhang et al., 2015d). However, in contrast to the disproportionate allocation of ETC-specific DEGs across the remaining functional categories, comparable portions of transcript numbers of ETC-specific genes were detected in groups encoding proteins involved in intracellular organization, cell wall and nutrient transporters (Zhang et al., 2015d). This suggests that ETC-specific DEGs play a greater role in the transcriptional regulation of genes in these functional categories compared to those encoding proteins with a signaling function.
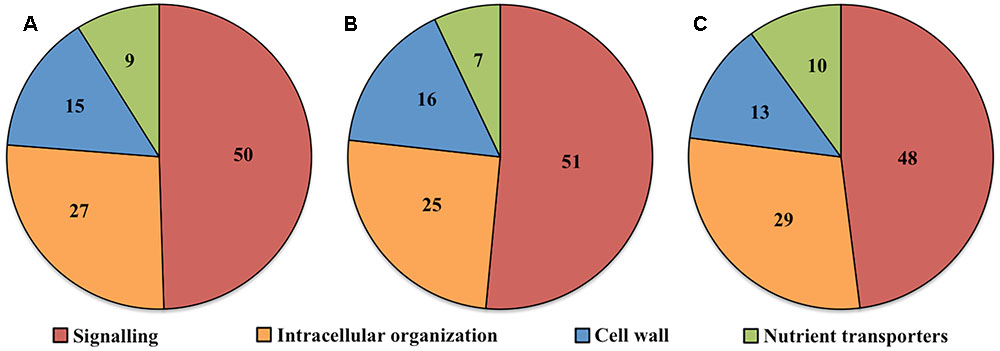
FIGURE 2. Pie charts showing the proportion (%) of (A) total, (B) up-regulated and (C) down-regulated ETC-specific DEGs in each key TC functional category (signaling, intracellular organization, cell wall, nutrient transporters) during wall labyrinth construction. DEGs in each functional category were classified based on their annotation in Mapman Mercator and KEGG functional analysis (see Supplementary Table S7).
Ratios of DEGs encoding proteins involved in signaling compared to cell wall building and nutrient transporters, 3.5X and 5.5X respectively (Table 2), were similar to those reported for trans-differentiating root cells penetrated by hyphae of mycorrhizal fungi (Calabrese et al., 2017). In contrast, investment into expression of genes encoding intracellular organization proteins was relatively higher in the ETCs, highlighting the complexity of building their wall labyrinth. A significant decrease of 67% in up-regulated DEGs involved in signaling specific for UWL versus WI papillae formation fits with the rapid change in cell fate during the former phase of ETC development (Table 2). However, an 80% decrease in up-regulated DEGs encoding intracellular organization proteins is more difficult to reconcile with a perceived greater complexity in constructing WI papillae compared to depositing the UWL (Table 2). A possible explanation could be that a significant portion of genes in this functional category are transcribed during UWL formation and are translated or post-translationally activated during WI papillae formation (e.g., see Zhang et al., 2017).
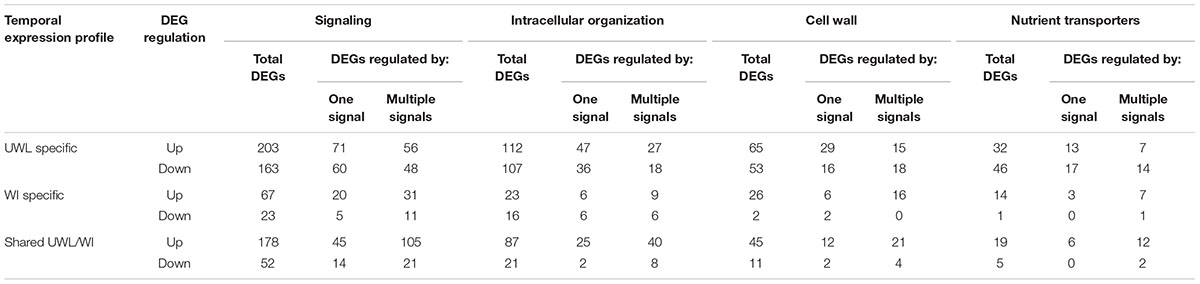
TABLE 2. Numbers of ETC-specific DEGs, within each TC functional category, responsive to one or a combination of ETC regulatory signals during the two phases of wall labyrinth construction (p ≤ 0.05).
Global Evaluation of ETC-Specific DEGs Regulated by Signals within each Functional TC Category
Expression levels of DEGs regulated by one or more of the four known signals, auxin, ethylene, H2O2 (Andriunas et al., 2013) and Ca2+ (Zhang et al., 2015a), accounted for 69% of the ETC-specific DEGs associated with the four functional gene categories linked with ETC development and membrane transport of nutrients (Table 1). The greater proportions of ETC-specific DEGs acted upon by the known signals were those up- and down-regulated specifically during WI papillae formation and for those up-regulated across both phases of wall labyrinth formation (Table 1).
Signaling molecules, yet to be confirmed, control expression levels of the remaining 31% of the ETC-specific functional categories of DEGs (Table 1). Additional signaling candidates that are expressed during ETC development are ABA and gibberellins (Zhang et al., 2015d). Other signaling candidates suggested to regulate wall labyrinth construction in other cell systems include jasmonic acid shown to influence phloem parenchyma TC development in Arabidopsis leaves (Amiard et al., 2007) and cytokinins driving basal endosperm TC development in maize caryopses (Yuan et al., 2016). These signaling candidates are being explored in another study.
To explore the relationship between the cohort size of ETC-specific DEGs regulated by one or various combinations of the signals known to orchestrate wall labyrinth assembly, Venn diagrams were constructed for the number of ETC-specific DEGs linked with the four TC functional categories for each temporal transcript profile (Figure 3 and Supplementary Figures S6–S9). Overall, the influence of the signals acting alone (48%) or in combination (52%) on expression levels of ETC-specific DEGs was shared equally across the four functional TC categories (Figure 4 and Table 2). However, there was a shift in dominance as wall labyrinth construction proceeded from DEGs being regulated by one signal during UWL formation (60%) to combinations of signals exerting an influence during WI papillae assembly (58%; Table 2). The structural complexity of assembling WI papillae extending vertically into the cytoplasm from loci on the UWL is consistent with a need for an increased sophistication of the signaling network regulating such a complex form of cell wall construction. Consistent with this interpretation, the cell wall functional category was impacted the most by signal combinations (72%; Table 2).
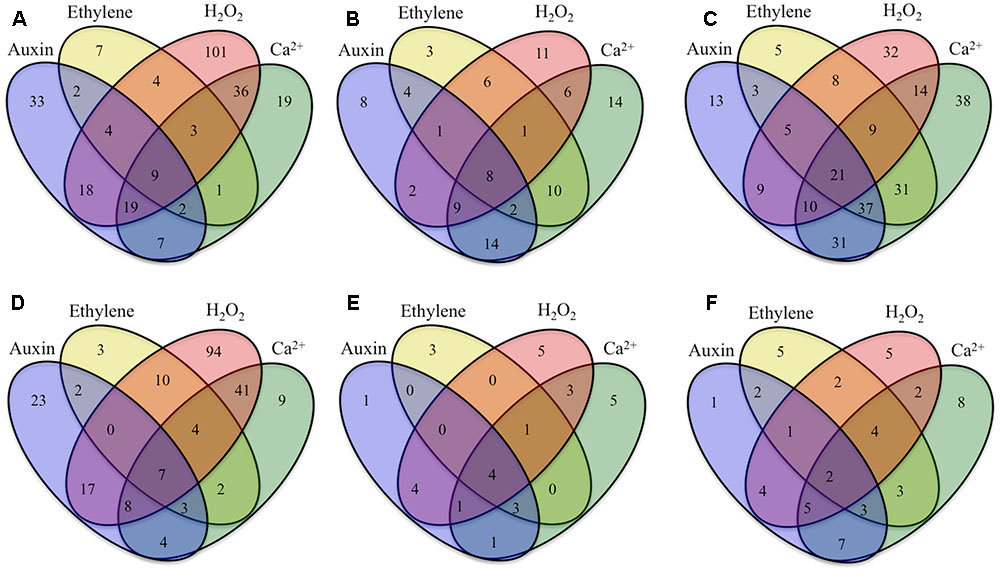
FIGURE 3. Venn diagrams showing numbers of ETC-specific DEGs in key TC functional categories (signaling, intracellular organization, cell wall, nutrient transporters), sensitive to signaling molecules known to regulate ETC trans-differentiation (auxin, ethylene, H2O2 and Ca2+) that were (A–C) up-regulated or (D–F) down-regulated specifically during construction of (A,D) UWL, (B,E) WI papillae, or (C,F) shared between UWL and WI papillae construction phases. Signal-sensitive ETC-specific DEGs were identified as those whose RPKM values were found to be significantly altered by signal blockers (p ≤ 0.05). Statistical significance was determined from six replicate batches of control cotyledons and three replicate batches of cotyledons cultured on signaling blockers using an unpaired t-test.
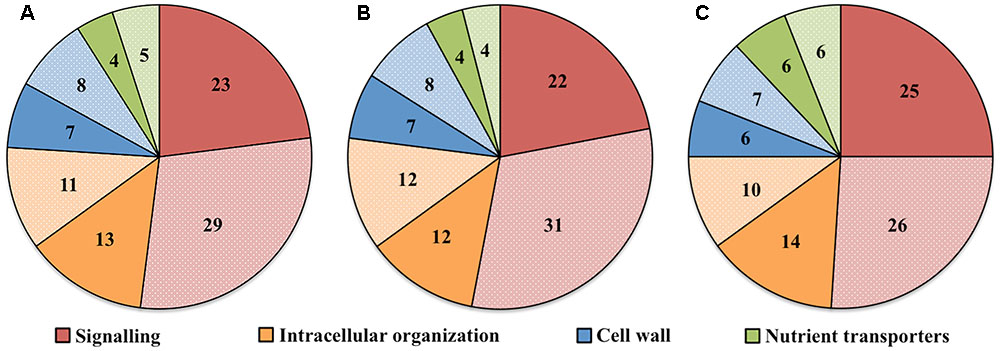
FIGURE 4. Pie charts showing the proportion (%) of (A) total number, (B) up-regulated or (C) down-regulated ETC-specific DEGs in key TC functional categories (signaling, intracellular organization, cell wall, nutrient transporters) whose change in expression was regulated by one (dark color fill) or multiple (light color fill) signaling molecules (auxin, ethylene, H2O2 and Ca2+) known to regulate ETC trans-differentiation during wall labyrinth construction.
Cohorts of ETC-Specific TC Functional DEGs Regulated by Signals Acting Alone
For those ETC-specific DEGs, within the specified TC functional categories, responsive to one of the four signals (Figure 4 and Table 2), the data demonstrated that, across wall labyrinth formation, the descending order of regulated DEG numbers was H2O2 (56%), Ca2+ (21%), auxin (18%) and ethylene (5%; Figure 3). Domination by H2O2 applied to up- and down-regulated DEGs in all functional TC categories (Table 3; Supplementary Figures S6–S9) and was most pronounced during UWL formation (Figures 3A,D). This observation is consistent with the central role played by H2O2 in initiating and directing cell wall deposition to the outer planar cell wall domain of developing ETCs to construct the wall labyrinth (Andriunas et al., 2012; Xia et al., 2012). The significant role played by H2O2 in regulating the transcriptome during ETC trans-differentiation contrasts with auxin and ethylene functioning as master transcriptional regulatory molecules in root hairs (Velasquez et al., 2016) and trichomes (Zhao et al., 2015a,b). Never-the-less, in other systems ROS plays a core regulatory role in initiating substantive transcriptional cascades for stress response and developmental pathways (Willems et al., 2016). In the context of the present study, H2O2 has been shown to exert a significant regulatory influence over expression of genes encoding cellulose biosynthesis in elongating root hairs (Nestler et al., 2014) and of plant hormonal and signal transduction pathways, as well as cell wall modifying enzymes during H2O2-induced adventitious rooting (Li et al., 2017). Roles for ROS in controlling expression of genes encoding proteins forming structural elements, or controlling the dynamics, of the plant cytoskeleton as well as membrane transporters of nutrients are less well known. The influence of H2O2 in regulating ETC-specific DEGs decreased as ETC development transitioned from UWL to WI papillae formation to a shared role with Ca2+ (Figure 3) in the signaling, intracellular organization and nutrient transport functional categories (Supplementary Figures S6, S7, S9). In contrast, Ca2+ controlled two-fold more DEGs in the cell wall functional category (Supplementary Figure S8).
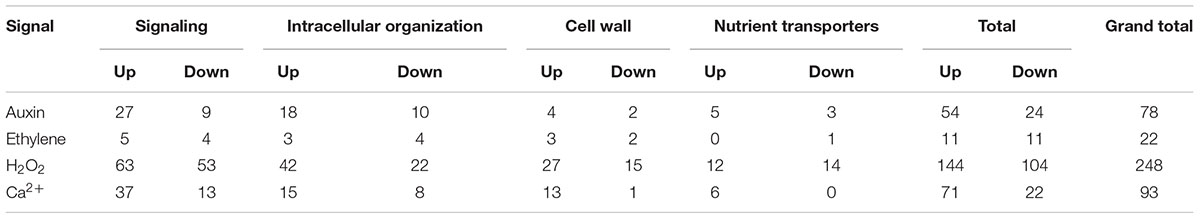
TABLE 3. Numbers of ETC-specific DEGs, within each TC functional category, responsive to each ETC regulatory signal across wall labyrinth construction (p ≤ 0.05).
Auxin and Ca2+ were the second ranked influential signals acting alone to regulate expression of ETC-specific DEGs (Table 2). Across ETC development Ca2+ exerted greater control over the signaling and cell wall TC functional categories while auxin regulated a slightly higher number of ETC-specific DEGs encoding proteins involved in intracellular organization and nutrient transporter categories (Table 3). In addition to the well described Ca2+ signaling pathways regulating plant development (Niu and Liao, 2016), a recent study has documented expression of genes encoding cell wall metabolizing enzymes positively regulated by Ca2+ along with similar impacts on genes encoding nutrient transporters (Wang et al., 2015). Similarly, as reported for other plant systems, auxin regulates expression of genes encoding proteins contributing to intracellular organization and to membrane transport of nutrients (e.g., Dai et al., 2009; Omelyanchuk et al., 2017). During UWL formation, two-fold more ETC-specific DEGs were regulated by auxin compared to Ca2+ (Figures 3A,D); an effect evident across all TC functional categories (Supplementary Figures S6–S9). Comparable to the role played by auxin in root hair initiation (Velasquez et al., 2016), this gene expression profile is consistent with auxin being positioned as an upstream component in the signaling cascade regulating ETC trans-differentiation (Zhou et al., 2010). The relative contributions of auxin and Ca2+ reversed once WI papillae formation commenced (Figures 3B,E; Supplementary Figures S6–S9). Consistent with WI papillae assembly being Ca2+-regulated (Zhang et al., 2015a), Ca2+ impacted 75% of the WI-specific DEGs in the cell wall functional category with the remainder being comprised of ethylene-sensitive DEGs (Supplementary Figure S8).
Ethylene acting alone is the least influential signal affecting transcription of ETC-specific DEGs across all functional categories. This feature is most pronounced for genes encoding nutrient transporters where only one down-regulated DEG specifically expressed during UWL formation was found to be ethylene sensitive (Table 3 and Supplementary Figure S9). However, ethylene exerted a four-fold greater influence than auxin on expression of genes in the cell wall functional category as the wall labyrinth transitioned from UWL to WI papillae construction (Supplementary Figure S8) indicating a possible ethylene regulation of WI papillae deposition (Andriunas et al., 2012). A similar conclusion can be reached for auxin since ETC-specific DEGs, sensitive to auxin, were detected during this phase of wall labyrinth construction in all TC functional categories (Figures 3C,F and Supplementary Figures S6–S9). We tested this claim by blocking auxin and ethylene signaling following cessation of UWL deposition by culturing cotyledons on media containing pharmacological inhibitors of both hormones. The auxin block halted on-going development of WI papillae while that for ethylene elicited a partial inhibition (Supplementary Table S10). The latter response is consistent with ethylene regulating expression of Ca2+-permeable channels responsible for forming plumes of elevated [Ca2+]cyt defining loci for WI papillae formation (Zhang et al., 2015c). The greater influence of auxin over ethylene on WI-specific DEG expression (Supplementary Figures S6–S9) and on WI papillae formation (Supplementary Table S10) is not readily reconciled with the proposed linear signaling cascade regulating wall labyrinth assembly with ethylene positioned downstream of auxin (Zhou et al., 2010). Together, the observations reported above suggest that auxin likely acts at more than one point in the signaling cascade regulating wall labyrinth development.
ETC-Specific DEGs within Each Functional TC Category Regulated by Signal Combinations
All 11 combinations of the four signals affected expression levels of a cohort of ETC-specific DEGs encoding transcripts that fall into the four specified functional categories across wall labyrinth formation (Figure 3). The exceptions to this generalization were an absence of up- and down-regulated DEGs sensitive to the combination of auxin/ethylene/H2O2 and of down-regulated DEGs sensitive to auxin/ethylene, auxin/ethylene/Ca2+, auxin/ethylene/H2O2/Ca2+, ethylene/Ca2+ and ethylene/H2O2/Ca2+ in some of the functional categories (Table 4). Of the eleven signal combinations, seven exerted predominant control accounting for 84% of DEGs regulated by all possible signal combinations. The standout was the H2O2/Ca2+ combination affecting expression levels of approximately two-fold more ETC-specific DEGs than each of the remaining six combinations impacting a range of 47–63 ETC-specific DEGs per combination (Table 4). Significantly, either or both H2O2 and Ca2+ were present in all the latter signal combinations with Ca2+ acting on a greater portion of up-regulated ETC-specific DEGs compared to H2O2 (Table 4). Overall these findings highlight the central role played by H2O2 and Ca2+ signals in regulating gene expression of proteins orchestrating wall labyrinth formation and nutrient transporters (Andriunas et al., 2012; Zhang et al., 2015a). Cross talk between H2O2 and Ca2+ signaling has been identified in a number of plant developmental programs (Niu and Liao, 2016) including polarized tip growth of pollen tubes and root hairs (Mangano et al., 2016). The remaining four signal combinations, which affected the least number of ETC-specific DEGS (11–30), had ethylene as one of the partners (Table 4). However, Ca2+ offset the negative influence of ethylene except when H2O2 was present in the absence of auxin.
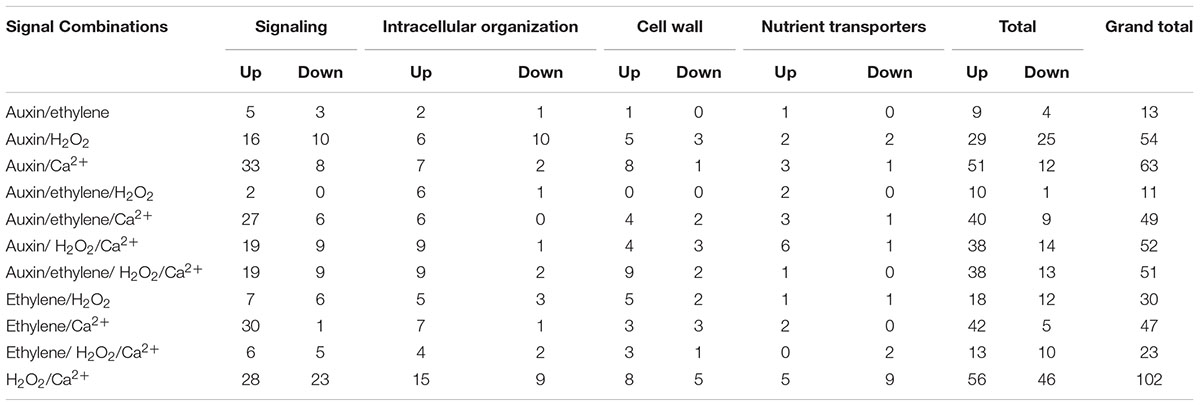
TABLE 4. Numbers of ETC-specific DEGs, within each TC functional category, responsive to multiple combinations of ETC regulatory signals across wall labyrinth construction (p ≤ 0.05).
The global impacts of the various signal combinations described above were manifested in the expression responses of the signaling functional category of genes (Table 4) that contained the largest number of ETC-specific DEGs regulated by signal combinations (Table 2). Except for the dominance displayed by the H2O2/Ca2+ combination, the proportions of ETC-specific DEG numbers responding to the various signal combinations departed from the overall pattern and were functional category centric (Table 4). For instance, in the intracellular organization function category, auxin/H2O2 elicited the second strongest response and auxin/ethylene the least, with remaining signal combinations affecting a narrow range (6–10) of ETC-specific DEG numbers. In contrast, for the cell wall functional category, comparable responses were shared across all combinations except for auxin/ethylene and auxin/ethylene/H2O2 that affected proportionally lower numbers of ETC-specific DEGs (Table 4).
Conclusion
An experimental design employing culture-induced trans-differentiating ETCs of V. faba cotyledons, exposed to media in the presence or absence of pharmacological blockers of signals known to regulate ETC trans-differentiation, identified an assemblage of ETC-specific DEGs within functional categories of genes encoding proteins considered responsible for wall labyrinth construction and TC function of nutrient transport. Significant alterations in transcript levels of these ETC-specific DEGs to pharmacological blockers of the known signals revealed that H2O2 acting alone, and in combination with Ca2+, controlled expression of the largest cohort (37% of the total) in the four TC functional gene categories across the two phases of wall labyrinth construction. Collectively these data offer a powerful resource to elucidate signal transduction pathways regulating expression of genes encoding proteins involved in intracellular organization and cell wall construction to assemble the wall labyrinth and transporters responsible for nutrient transport.
Data Deposition
The cDNA sequence datasets of raw reads and assembled reference transcriptome library (Zhang et al., 2015d) supporting this article are available in the repository of the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) with the ENA accession number: PRJEB8906. Lists of ETC-specific DEGs in each functional category and the impact of pharmacological blockers of known signals on their expression are provided in Supplementary Table S11.
Author Contributions
JP and CO conceived and designed the research project. H-MZ, SW, and XX performed the experiments and compiled the data sets. H-MZ assisted by KC and JP analyzed the data. H-MZ wrote the first draft of the manuscript that was revised by JP and reviewed by CO, KC, SW, XX, and H-MZ.
Funding
This work was supported by the Australian Research Council-Discovery Project scheme (DP130101396) to JP and CO. SW and XX acknowledge the support of ARC/UON RHD scholarships.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Joseph Enright ensured a continuous supply of healthy experimental plant material. The EM/X-ray unit of the University of Newcastle provided support for the scanning and transmission electron microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02021/full#supplementary-material
Abbreviations
AA, ascorbic acid; AVG, aminoethoxyvinylglycine; BAPTA, 1, 2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; CDD, Conserved Domain Database; COG, Clusters of Orthologous Groups; DEG, differentially expressed gene; ETC, epidermal transfer cell; FDR, false discovery rate; H2O2, hydrogen peroxide; KEGG, Kyoto Encyclopedia of Genes and Genomes; MS, Murashige and Skoog; PCIB, p-chlorophenoxyisobutyric acid; RNA-seq, RNA-sequencing; ROS, reactive oxygen species; RPKM, reads per kilo base per million reads; SPC, storage parenchyma cell; TC, transfer cell; UWL, uniform wall layer; WI, wall ingrowth.
Footnotes
- ^http://mapman.gabipd.org/web/guest/mercator
- ^http://www.uniprot.org/
- ^https://www.ncbi.nlm.nih.gov/COG/
- ^http://www.genome.jp/kegg/
References
Amiard, V., Demmig-Adams, B., Mueh, K. E., Turgeon, R., Combs, A. F., and Adams, W. W. III (2007). Role of light and jasmonic acid signaling in regulating foliar phloem cell wall ingrowth development. New Phytol. 173, 722–731. doi: 10.1111/j.1469-8137.2006.01954.x
Andriunas, F. A., Zhang, H. M., Weber, H., McCurdy, D. W., Offler, C. E., and Patrick, J. W. (2011). Glucose and ethylene signalling pathways converge to regulate trans- differentiation of epidermal transfer cells in Vicia narbonensis cotyledons. Plant J. 68, 987–998. doi: 10.1111/j.1365-313X.2011.04749.x
Andriunas, F. A., Zhang, H. M., Xia, X., Offler, C. E., McCurdy, D. W., and Patrick, J. W. (2012). Reactive oxygen species form part of a regulatory pathway initiating trans-differentiation of epidermal transfer cells in Vicia faba cotyledons. J. Exp. Bot. 63, 3617–3629. doi: 10.1093/jxb/ers029
Andriunas, F. A., Zhang, H. M., Xia, X., Patrick, J. W., and Offler, C. E. (2013). Intersection of transfer cells with phloem biology - broad evolutionary trends, function, and induction. Front. Plant Sci. 4:221. doi: 10.3389/fpls.2013.00221
Arun-Chinnappa, K. S., and McCurdy, D. W. (2016). Identification of candidate transcriptional regulators of epidermal transfer cell development in Vicia faba cotyledons. Front. Plant Sci. 7:717. doi: 10.3389/fpls.2016.00717
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
Cabrera, J., Barcala, M., Fenoll, C., and Escobar, C. (2014). Transcriptomic signatures of transfer cells in early developing nematode feeding cells of Arabidopsis focused on auxin and ethylene signaling. Front. Plant Sci. 5:107. doi: 10.3389/fpls.2014.00107
Cabrera, J., Barcala, M., Garcia, A., Rio-Machin, A., Medina, C., Jaubert-Possamai, S., et al. (2016). Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for MiR390 and its TAS3-derived tasiRNAs. New Phytol. 209, 1625–1640. doi: 10.1111/nph.13735
Calabrese, S., Kohler, A., Niehl, A., Veneault-Fourrey, C., Boller, T., and Courty, P. E. (2017). Transcriptome analysis of the Populus trichocarpa-Rhizophagus irregularis mycorrhizal symbiosis: regulation of plant and fungal transportomes under nitrogen starvation. Plant Cell Physiol. 58, 1003–1017. doi: 10.1093/pcp/pcx044
Chen, C., Liu, M., Jiang, L., Liu, X., Zhao, J., Yan, S., et al. (2014). Transcriptome profiling reveals roles of meristem regulators and polarity genes during fruit trichome development in cucumber (Cucumis sativus L.). J. Exp. Bot. 65, 4943–4958. doi: 10.1093/jxb/eru258
Dai, X., Su, Y. R., Wei, W. X., Wu, J. S., and Fan, Y. K. (2009). Effects of top excision on the potassium accumulation and expression of potassium channel genes in tobacco. J. Exp. Bot. 60, 279–289. doi: 10.1093/jxb/ern285
Damiani, I., Drain, A., Guichard, M., Balzergue, S., Boscari, A., Boyer, J. C., et al. (2016). Nod factor effects on root hair-specific transcriptome of Medicago truncatula: focus on plasma membrane transport systems and reactive oxygen species networks. Front. Plant Sci. 7:794. doi: 10.3389/fpls.2016.00794
Dibley, S. J., Zhou, Y., Andriunas, F. A., Talbot, M. J., Offler, C. E., Patrick, J. W., et al. (2009). Early gene expression programs accompanying trans-differentiation of epidermal cells of Vicia faba cotyledons into transfer cells. New Phytol. 182, 863–877. doi: 10.1111/j.1469-8137.2009.02822.x
Farley, S. J., Patrick, J. W., and Offler, C. E. (2000). Functional transfer cells differentiate in cultured cotyledons of Vicia faba L. seeds. Protoplasma 214, 102–117. doi: 10.1007/BF02524267
Grubbs, F. (1969). Procedures for detecting outlying observations in samples. Technometrics 11, 1–21. doi: 10.1080/00401706.1969.10490657
Hey, S., Baldauf, J., Opitz, N., Lithio, A., Pasha, A., Provart, N., et al. (2017). Complexity and specificity of the maize (Zea mays L.) root hair transcriptome. J. Exp. Bot. 68, 2175–2185. doi: 10.1093/jxb/erx104
Ji, H., Gheysen, G., Denil, S., Lindsey, K., Topping, J. F., Nahar, K., et al. (2013). Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 64, 3885–3898. doi: 10.1093/jxb/ert219
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Li, S.-W., Leng, Y., and Shi, R.-F. (2017). Transcriptome profiling provides molecular insights into hydrogen peroxide-induced adventitious rooting in mung bean seedlings. BMC Genomics 18:188. doi: 10.1186/s12864-017-3576-y
Lohse, M., Nagel, A., Herter, T., May, P., Schroda, M., Zrenner, R., et al. (2014). Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37, 1250–1258. doi: 10.1111/pce.12231
Mangano, S., Juárez, S. P., and Estevez, J. M. (2016). ROS regulation of polar growth in plant cells. Plant Physiol. 171, 1593–1605. doi: 10.1104/pp.16.00191
Marchler-Bauer, A., Anderson, J. B., Derbyshire, M. K., DeWeese-Scott, C., Gonzales, N. R., Gwadz, M., et al. (2007). CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35, D237–D240. doi: 10.1093/nar/gkl951
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Nestler, J., Liu, S., Wen, T.-J., Paschold, A., Marcon, C., Tang, H. M., et al. (2014). Roothairles5, which functions in maize (Zea mays L) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J. 79, 729–740. doi: 10.1111/tpj.12578
Niu, L., and Liao, W. (2016). Hydrogen peroxide signaling in plant development and abiotic responses: crosstalk with nitric oxide and calcium. Front. Plant Sci. 7:230. doi: 10.3389/fpls.2016.00230
Nordberg, H., Cantor, M., Dusheyko, S., Hua, S., Poliakov, A., Shabalov, I., et al. (2014). The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. doi: 10.1093/nar/gkt1069
Offler, C. E., Liet, E., and Sutton, E. G. (1997). Transfer cell induction in cotyledons of Vicia faba L. Protoplasma 200, 51–64. doi: 10.1007/BF01280734
Offler, C. E., McCurdy, D. W., Patrick, J. W., and Talbot, M. J. (2003). Transfer cells: cells specialized for a special purpose. Annu. Rev. Plant Biol. 54, 431–454. doi: 10.1146/annurev.arplant.54.031902.134812
Omelyanchuk, N. A., Wiebe, D. S., Novikova, D. D., Levitsky, V. G., Kllimova, N., Gorelova, V., et al. (2017). Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci. Rep. 7, 2489. doi: 10.1038/s41598-017-02476-8
Portillo, M., Cabrera, J., Lindsey, K., Topping, J., Andrés, M. F., and Emiliozzi, M. (2013). Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 197, 1276–1290. doi: 10.1111/nph.12121
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Schmidt, W., and Bartels, M. (1996). Formation of root epidermal transfer cells in Plantago. Plant Physiol. 110, 217–225. doi: 10.1104/pp.110.1.217
Swarbreck, D., Wilks, C., Lamesch, P., Berardini, T. Z., Garcia-Hernandez, M., Foerster, H., et al. (2010). The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 36, D1009–D1014. doi: 10.1093/nur/skm965
Talbot, M. J., Franceschi, V. R., McCurdy, D. W., and Offler, C. E. (2001). Wall ingrowth architecture in epidermal transfer cells of Vicia faba cotyledons. Protoplasma 215, 191–203. doi: 10.1007/BF01280314
Thiel, J. (2014). Development of endosperm transfer cells in barley. Front. Plant Sci. 5:108. doi: 10.3389/fpls.2014.00108
Vandesompele, J., De Paepe, A., and Speleman, F. (2002). Elimination of primer-dimer artifacts and genomic co-amplification using a two-step SYBR green 1 real-time PCR. Ann. Biochem. 303, 95–98. doi: 10.1006/abio.2001.5564
Velasquez, S. M., Barbez, E., Kleine-Vehn, J., and Estevez, J. M. (2016). Auxin and cellular elongation. Plant Physiol. 170, 1206–1215. doi: 10.1104/pp.15.01863
Wang, J., Tergel, T., Chen, J., Yang, J., Kang, Y., and Qi, Z. (2015). Arabidopsis transcriptional response to extracellular Ca2+ depletion involves a transient rise in cytosolic Ca2+. J. Integr. Plant Biol. 57, 138–150. doi: 10.1111/jipb.12218
Wardini, T., Wang, X. D., Offler, C. E., and Patrick, J. W. (2007). Induction of wall ingrowths of transfer cells occurs rapidly and depends upon gene expression in cotyledons of developing Vicia faba seeds. Protoplasma 231, 15–23. doi: 10.1007/s00709-007-0244-0
Willems, P., Mhamdi, A., Stael, S., Storme, V., Kerchev, P., Noctor, K. G., et al. (2016). The ROS wheel: refining ROS transcriptional footprints. Plant Physiol. 171, 1720–1733. doi: 10.1104/pp.16.00420
Wurzinger, B., Mair, A., Pfister, B., and Teige, M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6, 8–12. doi: 10.4161/psb.6.1.14012
Xia, X., Zhang, H.-M., Andriunas, F. A., Offler, C. E., and Patrick, J. W. (2012). Extracellular hydrogen peroxide, produced through a respiratory burst/superoxide dismutase pathway, directs ingrowth wall formation in epidermal cells of Vicia faba cotyledons. Plant Signal. Behav. 7, 1125–1128. doi: 10.4161/psb.21320
Yuan, J., Bateman, P., and Gutierrez-Marcos, J. (2016). Genetic and epigenetic control of transfer cell development in plants. J. Genet. Genomics 43, 533–539. doi: 10.1016/j.jgg.2016.08.002
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848. doi: 10.1093/bioinformatics/17.9.847
Zhan, J., Thakare, D., Ma, C., Lloyd, A., Nixon, N. M., Arakaki, A. M., et al. (2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27, 513–531. doi: 10.1105/tpc.114.135657
Zhang, H.-M., Colyvas, K., Patrick, J. W., and Offler, C. E. (2017). A Ca2+-dependent remodelled actin network directs vesicle trafficking to build wall ingrowth papillae in transfer cells. J. Exp. Bot. 68, 4749–4764. doi: 10.1093/jxb/erx315
Zhang, H.-M., Imtiaz, M. S., Laver, D. R., McCurdy, D. W., Offler, C. E., van Helden, D. F., et al. (2015a). Polarized and persistent Ca2+ plumes define loci for formation of wall ingrowth papillae in transfer cells. J. Exp. Bot. 66, 1179–1190. doi: 10.1093/jxb/eru460
Zhang, H.-M., Talbot, M. J., McCurdy, D. W., Patrick, J. W., and Offler, C. E. (2015b). Calcium-dependent depletion zones in the cortical microtubule array coincide with sites of, but do not regulate, wall ingrowth papillae deposition in epidermal transfer cells. J. Exp. Bot. 66, 6021–6033. doi: 10.1093/jxb/erv317
Zhang, H.-M., van Helden, D. F., McCurdy, D. W., Offler, C. E., and Patrick, J. W. (2015c). Plasma membrane Ca2+-permeable channels are differentially regulated by ethylene and hydrogen peroxide to generate persistent plumes of elevated cytosolic Ca2+ during transfer trans-differentiation. Plant Cell Physiol. 56, 1711–1720. doi: 10.1093/pcp/pcv100
Zhang, H.-M., Wheeler, S., Xia, X., Radchuk, R., Weber, H., Offler, C. E., et al. (2015d). Differential transcriptional networks associated with key phases of ingrowth wall construction in trans-differentiating epidermal transfer cells of Vicia faba cotyledons. BMC Plant Biol. 15:103. doi: 10.1186/s12870-015-0486-5
Zhao, J. L., Pan, J. S., Guan, Y., Nie, J. T., Yang, J. J., Qu, M. L., et al. (2015a). Transcriptome analysis in Cucumis sativus identifies genes involved in multicellular trichome development. Genomics 105, 296–303. doi: 10.1016/j.ygeno.2015.01.010
Zhao, J. L., Wang, Y. L., Yao, D. Q., Zhu, W. Y., Chen, L., He, H. L., et al. (2015b). Transcriptome profiling of trichome-less reveals genes associated with multicellular trichome development in Cucumis sativus. Mol. Genet. Genomics 290, 2007–2018. doi: 10.1007/s00438-015-1057-z
Zhao, L. J., Yuan, H. M., Guo, W. D., and Yang, C. P. (2016). Digital gene expression analysis of Populus simonii × P. nigra pollen germination and tube growth. Front. Plant Sci. 7:825. doi: 10.3389/fpls.2016.00825
Keywords: RNA-sequencing, signaling, cell wall, transfer cells, wall ingrowth papillae, wall labyrinth
Citation: Zhang H-M, Wheeler SL, Xia X, Colyvas K, Offler CE and Patrick JW (2017) Transcript Profiling Identifies Gene Cohorts Controlled by Each Signal Regulating Trans-Differentiation of Epidermal Cells of Vicia faba Cotyledons to a Transfer Cell Phenotype. Front. Plant Sci. 8:2021. doi: 10.3389/fpls.2017.02021
Received: 12 September 2017; Accepted: 14 November 2017;
Published: 28 November 2017.
Edited by:
Rujin Chen, Noble Research Institute, LLC, United StatesReviewed by:
Jing Yuan, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology (CAS), ChinaFrantisek Baluska, University of Bonn, Germany
Copyright © 2017 Zhang, Wheeler, Xia, Colyvas, Offler and Patrick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John W. Patrick, am9obi5wYXRyaWNrQG5ld2Nhc3RsZS5lZHUuYXU= Christina E. Offler, dGluYS5vZmZsZXJAbmV3Y2FzdGxlLmVkdS5hdQ==
 Hui-Ming Zhang
Hui-Ming Zhang Simon L. Wheeler
Simon L. Wheeler Xue Xia
Xue Xia Kim Colyvas2
Kim Colyvas2 Christina E. Offler
Christina E. Offler John W. Patrick
John W. Patrick