
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 November 2017
Sec. Plant Cell Biology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01969
Beige and Chediak Higashi (BEACH) domain-containing proteins (BDCPs) are facilitators of membrane-dependent cellular processes in eukaryotes. Mutations in BDCPs cause malfunctions of endosomal compartments in various cell types. Recently, the molecular analysis of the BDCP homolog gene SPIRRIG (SPI) has revealed a molecular function in P-bodies and the regulation of RNA stability. We therefore aimed to analyze, whether SPI has also a role in membrane-dependent processes. In this study, we show that SPI physically interacts with endosomal sorting complex required for transport associated ATPase Suppressor of K+-transport growth defect1 (SKD1) and its positive regulator, LYST Interacting Protein 5 (LIP5) and report genetic interactions between SPI and SKD1 and LIP5. We further show that the endosomal transport route of soluble proteins to the lytic vacuole is disturbed in spi lip5 double mutants but not in the single mutants. These vacuolar transport defects were suppressed by additional expression of SKD1. Our results indicate that the BEACH domain protein SPI has in addition to a role in P-bodies a function in endosomal transport routes.
Beige and Chediak Higashi (BEACH) domain-containing proteins (BDCPs) were first identified during the characterization of the human Chediak Higashi Syndrome (CHS) and the Beige phenotype in mouse (Barbosa et al., 1996; Perou et al., 1996). CHS is a lethal autosomal recessive disorder, hallmarked by complex clinical manifestations comprising bleeding diathesis, albinism, immunodeficiency, and neurodegeneration (Introne et al., 1993; Kaplan et al., 2008). At the cellular level, CHS patients show abnormally enlarged lysosomes and protein sorting and secretion defects in various cell types (Brandt et al., 1975; Zhao et al., 1994; Barbosa et al., 1996; Perou et al., 1996; Spritz, 1998; Introne et al., 1999; Ward et al., 2000). The corresponding gene was therefore termed Lysosomal Trafficking Regulator (LYST) (Barbosa et al., 1996; Nagle et al., 1996; Perou et al., 1996).
The LYST protein contains three conserved motifs at the C-terminus: a Pleckstrin-Homology Domain (PH-Domain), the BEACH domain and a WD40 repeat motif. WD40 repeats and PH-Domains are known to mediate heterotypic protein interactions and protein recruitments to membranes via phospholipid binding, respectively (Neer et al., 1994; Lemmon, 2007). The molecular function of BEACH domains is not known. BDCPs, exhibiting a protein domain organization like LYST, are present in all eukaryotic species and mutants have been isolated in various organisms. Some of the most prominent examples are Neurobeachin (Nbea) deficient mice, characterized by impaired retrieval and post-Golgi trafficking of neuronal neurotransmitter receptors (Wang et al., 2000; del Pino et al., 2011); Drosophila Blue Cheese (bchs) mutants in which Ras related in brain protein 11 (RAB11)-dependent vesicle trafficking events at the Trans-Golgi Network (TGN), Golgi-derived vesicles and recycling endosomes are disturbed (Khodosh et al., 2006); C. elegans Suppressor/Enhancer of Lin-12 Protein 2 (SEL2) mutants that exhibit protein sorting defects between the cell surface and lysosomes (de Souza et al., 2007); Dictyostelium deficient for LvsA or LvsB (Large Volume Sphere A and B), showing hyper-accumulations of the vacuolar H+-ATPase in early endo- and phagocytotic compartments (Gerald et al., 2002; Wu et al., 2004) and enhanced lysosomal enzyme secretion under starvation (Harris et al., 2002); and Arabidopsis thaliana bchD (BEACH domain protein D, also termed green fluorescent seed 12) mutants, in which protein sorting to the plant specific protein storage vacuoles (PSVs) in seeds, but not in vacuolated vegetative tissues, is compromised (Teh et al., 2015). Because of the commonalities between these phenotypes, BDCPs are considered to be involved in the regulation of endosomal sorting processes (Cullinane et al., 2013).
In plants, the SPIRRIG gene in Arabidopsis thaliana is the best-studied BDCP representative [SPI, also termed BEACH domain protein A1 (BchA1) (Teh et al., 2015)]. SPI deficient plants share all morphological phenotypes observed in Arabidopsis mutants with defects in the ARP2/3- (actin related proteins 2 and 3) and SCAR/WAVE- (suppressor of cAMP receptor from Dictyostelium/Wiskott Aldrich syndrome protein family verprolin-homologous protein) complex mediated actin filament polymerization and branching (Hulskamp et al., 1994; Szymanski et al., 1999; Saedler et al., 2009). These include twisted and wavy trichomes, reduced length of root hairs, cell attachment defects and less complex epidermal pavement cells (Saedler et al., 2009). In contrast to all other mutants of this class, spi mutant cells do not show any defect in actin cytoskeleton organization (Schwab et al., 2003). In addition, SPI is involved in salt stress response (Steffens et al., 2015). In this context the analysis of its molecular function revealed a role in post-transcriptional stabilization of mRNAs (Steffens et al., 2015). This raises the question, whether the BDCP SPI gene has a different molecular function as reported for other BDCPs or whether SPI has a dual role in post-transcriptional regulation of mRNA and endomembrane dynamics. A possible role of the Arabidopsis SPI protein in endomembrane dynamics was initially suggested by the finding that root hairs show fragmented vacuoles (Saedler et al., 2009). Plant vacuoles are thought to originate from the endoplasmic reticulum (ER) as part of the secretory pathway (Matile, 1968; Mesquita, 1969; Viotti et al., 2013; Zhang et al., 2014). As a consequence, vacuoles receive cargo molecules from both, the anterograde transport route from the ER and the retrograde trafficking pathway from the plasma membrane (Saint-Jore-Dupas et al., 2004; Scheuring et al., 2011). The latter pathway involves the evolutionarily highly conserved ESCRT (endosomal sorting complex required for transport) machinery that is essential for the recognition of ubiquitinated cargo molecules destined for the vacuolar/lysosomal decay at membranes of maturating endosomes (Hurley and Emr, 2006; Spitzer et al., 2009; Richardson et al., 2011; Scheuring et al., 2012; Gao et al., 2015). Following the deubiquitination of cargo molecules (Katsiarimpa et al., 2014), the AAA+-type ATPase SKD1 (Suppressor of K+-transport growth defect 1/also termed VPS4; Vacuolar Protein Sorting-associated protein 4) triggers the final steps of endosome maturation to multivesicular bodies (MVBs), the fission of intraluminal vesicles (ILVs) and dissociation of ESCRT components from the endosomal membrane (Babst et al., 1998; Sachse et al., 2004; Babst, 2005; Lata et al., 2008; Landsberg et al., 2009). The expression of dominant negative SKD1 causes the formation of abnormally large MVBs containing a reduced number of ILVs concomitant with a reduced vacuolar transport and secretion of intraluminal cargo (Raymond et al., 1992; Fujita et al., 2003; Haas et al., 2007; Shahriari et al., 2010b; Scheuring et al., 2012). A possible function of BDCPs in the ESCRT pathway is suggested by the finding that human LYST interacting protein 5 (LIP5) interacts with the LYST in yeast two-hybrid assays (Tchernev et al., 2002). LIP5 is a positive regulator of SKD1 in mammals, yeast, and plants (Fujita et al., 2004; Azmi et al., 2006; Haas et al., 2007). The expression of dominant negative SKD1 enhances the membrane association of LYST in cultured human cells (Fujita et al., 2004) suggesting that the interaction of LIP5 and LYST is functionally relevant.
In this study, we assessed the role of the BDCP SPI protein in the ESCRT regulatory pathway. We demonstrate that SPI physically interacts with LIP5 and SKD1 from Arabidopsis thaliana. By combining molecular and biochemical approaches, we demonstrate that SPI is involved in the regulation of SKD1 and LIP5 and the route of endomembrane trafficking.
In a first step, we examined the protein interactions of the Arabidopsis BDCP SPI with LIP5 and SKD1. As large size of the SPI cDNA renders the molecular analysis extremely difficult, we used the evolutionarily conserved C-terminal part containing the PH-BEACH-WD40 domains (called SPI-PBW hereafter, Supplementary Figure S1A) for protein-interaction assays. Yeast two-hybrid assays revealed interactions between SPI-PBW and both, LIP5 and SKD1 (Figure 1A). The interaction between SPI-PBW and SKD1 was confirmed by co-precipitations (co-Ps) of bacterially expressed Glutathione-S-Transferase (GST)/His6-fusions of SPI-PBW and His6-fusions of SKD1 (Figure 1B). His6-SKD1 did not bind to GST labeled resins, confirming the specificity of the SPI-PBW/SKD1 interaction. As the bacterial expression of His6-LIP5 failed, we confirmed the SPI/LIP5 interaction in co-immunoprecipitation (co-IP) experiments with proteins expressed in Nicotiana benthamiana (N. benthamiana) leaf cells. Here, the expression of SPI-PBW was very weak and just above the detection level. We therefore used a slightly smaller SPI fragment lacking the WD40-repeats (termed SPI-PB; Supplementary Figure S1A). SPI-PB was C-terminally fused to a HA-tag (SPI-PB-HA). LIP5 was used as N-terminal fusion with three FLAG tags (3xFLAG-LIP5). These experiments revealed a clear co-IP suggesting an interaction between SPI-PB and LIP5. 3xFLAG-LIP5 did not bind to unlabeled beads, indicating that the SPI-PB/LIP5 interaction is specific (Figure 1C). The specificity of the immunoprecipitation was confirmed, by showing that SPI-PB-HA and 3xFLAG-LIP5 did not bind to α-ProteinA beads (Supplementary Figure S1B).
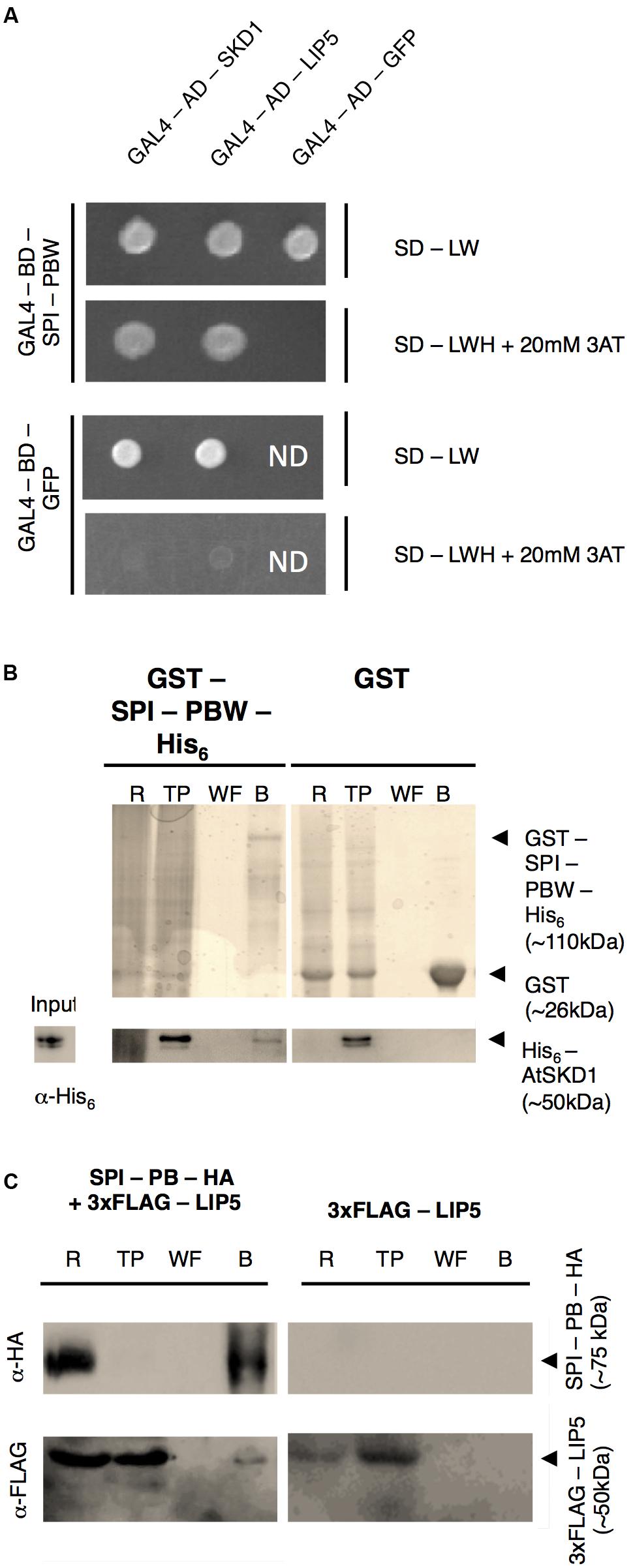
FIGURE 1. SPI interacts with SKD1 and LIP5. (A) Yeast two-hybrid interactions between SPI-PBW, SKD1, and LIP5. Top: double transformed yeast cells on dropout medium lacking leucine (-L) and tryptophan (-W). Bottom: interaction between GAL4-Binding Domain (BD) fusions of SPI-PBW, and SKD1 or LIP5, N-terminally fused to the GAL4 Activation Domain (AD), on dropout medium lacking leucine (-L), tryptophan (-W) and histidine (-H), supplemented with 20 mM 3-Aminotrizole (3AT). GAL4-AD-GFP (Green Fluorescent Protein) and GAL4-BD-GFP vectors served as negative controls. ND, not determined. (B) Co-precipitation of bacterially expressed SPI-PBW and SKD1. Top: Purifications of GST-SPI-PBW-His6 (∼110 kDa) and the negative control GST (∼26 kDa) are shown on Coomassie stained gels. Bottom: Input of purified His6-SKD1 (∼50 kDa) and co-precipitations of His6-SKD1/GST-SPI-PBW-His6 detected by α-His6 antibody staining. No co-precipitation was observed between GST and His6-SKD1. Expected protein sizes are indicated by arrowheads. R, raw extract; TP, throughput; WF, last wash fraction; B, beads fraction. (C) Co-immunoprecipitation of SPI-PB-HA and 3xFLAG-LIP5 from lysates of transfected N. benthamiana leaves. Top: Immunoprecipitation of SPI-PB-HA (∼75 kDa) was detected by α-HA antibody staining on a Western blot. Bottom: 3xFLAG-tagged LIP5 (∼50 kDa) was detected in R, TP and the B of SPI-PB-HA co-transfected leaves by α-FLAG antibody staining on Western blots. 3xFLAG-tagged LIP5 alone was not precipitated with α-HA-beads. Expected protein sizes are indicated by arrowheads. ProteinA beads were included as a negative control for all proteins tested (Supplementary Figure S1B).
To monitor the intracellular localization of SPI, LIP5 and SKD1, we transiently expressed fluorescently marked proteins. As shown before, we observed YFP-SDK1 either evenly distributed in the cytoplasm or at one or several cytoplasmic dots in transiently transformed Arabidopsis leaf epidermal cells (Supplementary Figures S2A,B) (Shahriari et al., 2010b). Quantification of YFP-SKD1 localization revealed its localization to cytoplasmic dots in about one thirds of the cells (Supplementary Figure S2C). The YFP-SKD1 dots partially co-localized with two endosomal marker proteins, the Rab5 related small GTPases ARA6 (RabF1) and ARA7 (RabF2B) (Ueda et al., 2004; Ebine et al., 2011). Sixty-two percentage of YFP-SKD1 positive dots were labeled with ARA6-mCHERRY and 47.4% were marked with mCHERRY-ARA7 (Figures 2A–D). As reported before, LIP5-YFP localized to endosomes (Brandt et al., 1975; Shim et al., 2008; van Balkom et al., 2009). 59.3% of LIP5-YFP labeled dots showed also a ARA6-mCHERRY signal and 58.2% co-localized with mCHERRY-ARA7 (Figures 2E–H). In contrast to SKD1 and LIP5, the N-terminal YFP-fusions of SPI-PBW and full length genomic SPI (genSPI) were evenly distributed in the cytoplasm (Supplementary Figures S2D,E).
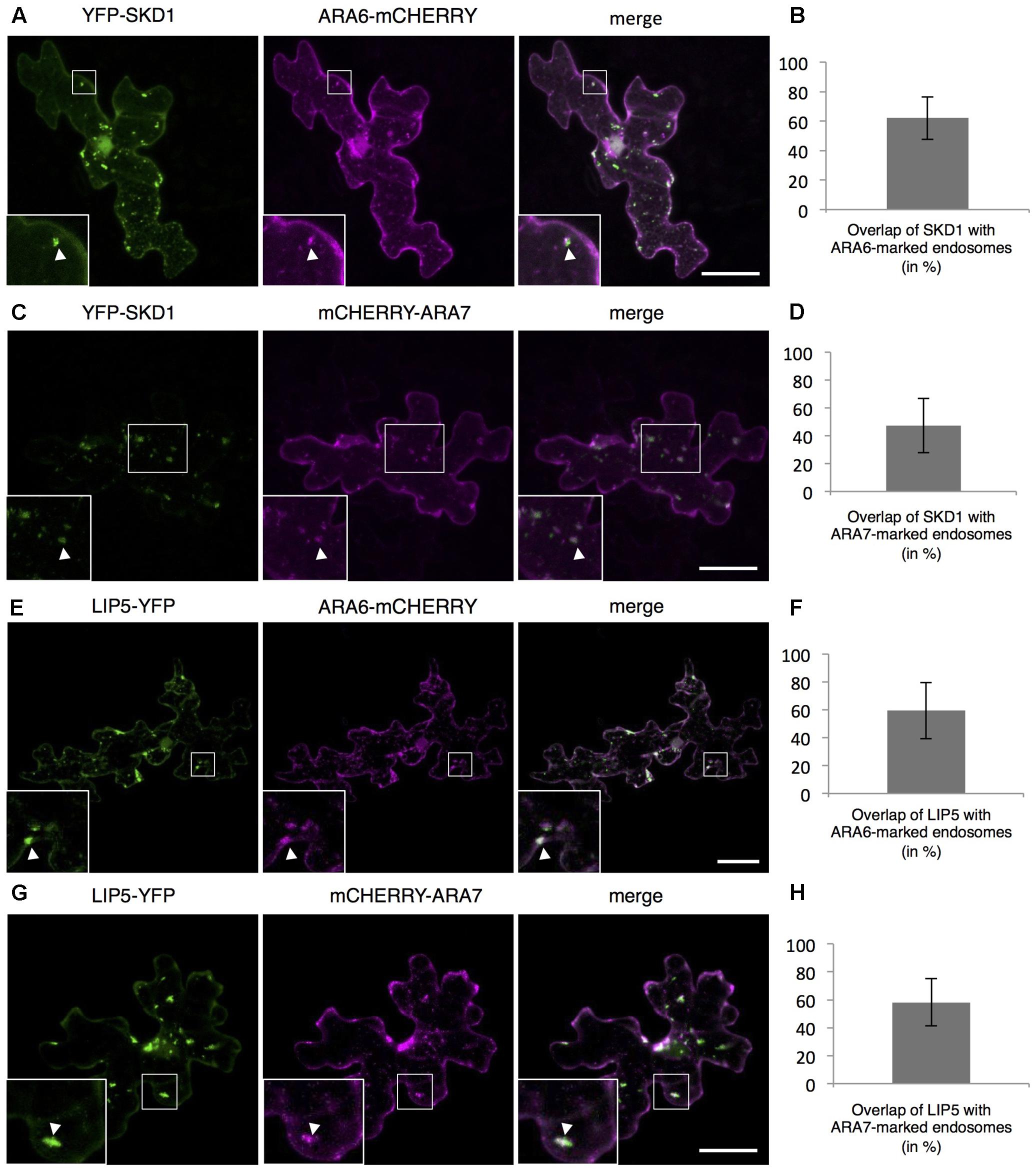
FIGURE 2. Localization of SPI, LIP5, and SKD1 in plant cells. Representative images of SKD1 or LIP5 co-transformed with endosomal marker proteins and the quantification of co-localization by Manders. (A) YFP-SKD1, ARA6-mCHERRY and their overlay. (B) Manders coefficient (in %) between YFP-SKD1 and ARA6-mCHERRY. (C) YFP-SKD1, mCHERRY-ARA7 and their overlay. (D) Manders coefficient (in %) between YFP-SKD1 and mCHERRY-ARA7. (E) LIP5-YFP, ARA6-mCHERRY and their overlay. (F) Manders coefficient (in %) between LIP5-YFP and ARA6-mCHERRY. (G) LIP5-YFP, mCHERRY-ARA7 and their overlay. (H) Manders coefficient (in %) between LIP5-YFP and mCHERRY-ARA7. Data denote the average of 20 cells and error bars the corresponding standard deviations (SDs). Scale bars represent 25 μm.
The intracellular localization of the interaction between SPI-PBW, LIP5, and SKD1 was assessed by Bimolecular fluorescence complementation (BiFC) assays in transfected N. benthamiana leaf epidermis cells. We found BiFC interactions of SPI-PBW with LIP5 and SKD1 in cytoplasmic dots. The Manders co-efficient of the SPI-PBW/SKD1 BiFC signal was 54.96% with ARA6-mCHERRY and 45.76% with mCHERRY-ARA7 (Figures 3A,B). 49.3% of the SPI-PBW/LIP5 BiFC signal co-localized with ARA6-mCHERRY and 44.6% with mCHERRY-ARA7 (Figures 3C,D). As SPI protein fusions are ubiquitously distributed in the cytoplasm we used also a cytoplasmically localized negative control. We found no interactions of SPI-PBW and LIP5 with AtMYC1 (MYC related protein1) (Pesch et al., 2013) (Supplementary Figure S2F). The functional integrity of AtMYC1 N-terminally fused to YFP-fragments was confirmed by demonstrating its BiFC interaction with TTG1 (TRANSPARENT TESTA GLABRA1) (Zimmermann et al., 2004; Zhao et al., 2012) (Supplementary Figure S2G). TTG1-YFPC was included as negative control for YFPN-SKD1 (Supplementary Figure S2F). In summary, our data suggest that the interaction between SPI-PBW and LIP5 and SKD1 can occur at endosomes.
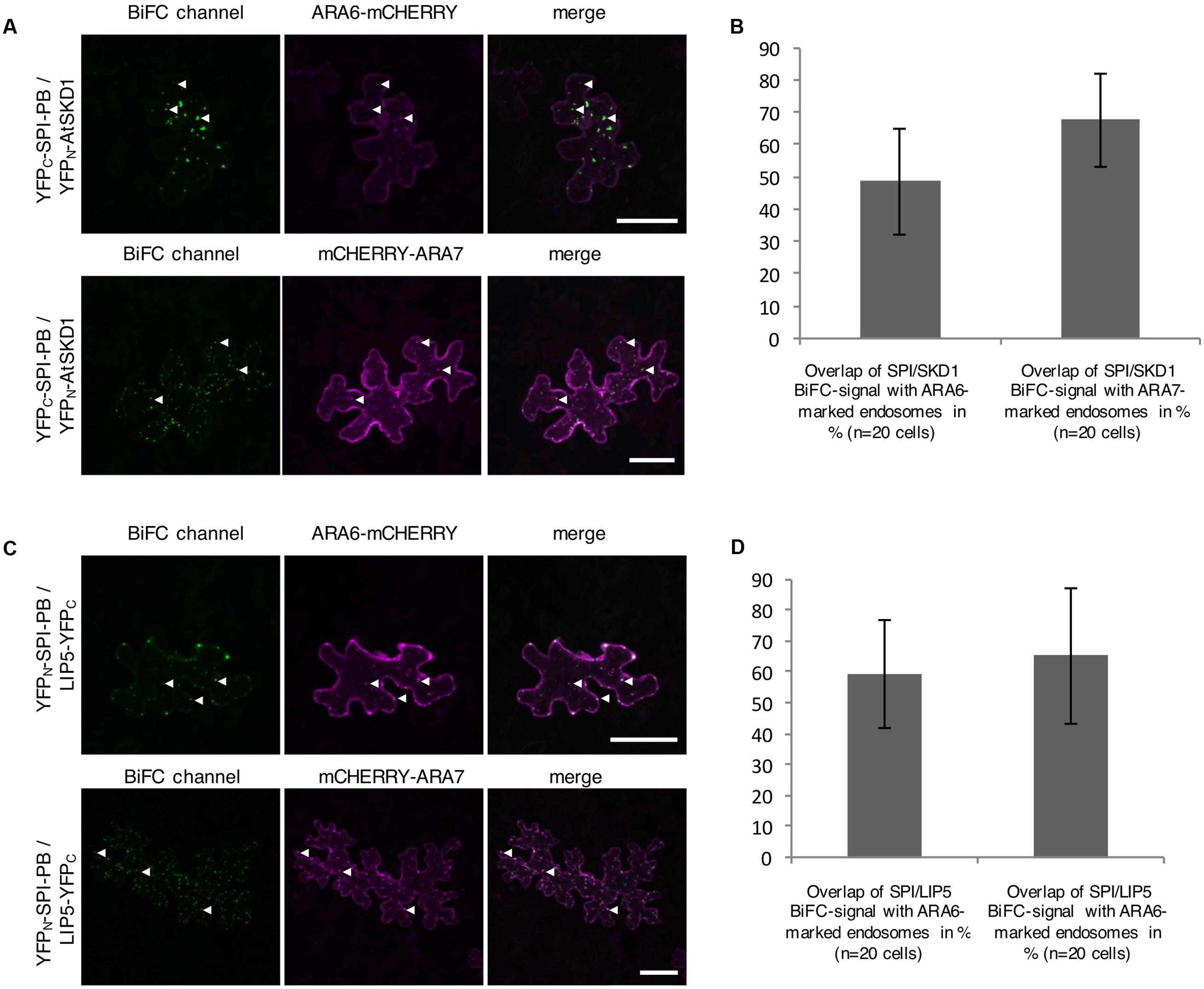
FIGURE 3. Bimolecular fluorescence complementation (BiFC) interactions of SPI-PBW on endosomes in infiltrated N. benthamiana leaf epidermis cells (related to Supplementary Figure S2). (A) BiFC interactions of YFPC-SPI-PBW and YFPN-SKD1 in cells co-expressing ARA6-mCHERRY (Upper) or mCHERRY-ARA7 (Lower). Column III shows the merged pictures of columns I and II. Scale bars represent 50 μm. (B) Overlap (in %) between SPI/SKD1 BiFC signals and ARA6- or ARA7-labeled endosomes. Data denote the average of 10 cells. Error bars represent SDs. (C) BiFC interaction of YFPN-SPI-PBW and LIP5-YFPC in cells co-expressing ARA6-mCHERRY (Upper) or mCHERRY-ARA7 (Lower). Column III shows the merged pictures of columns I and II. Scale bars represent 50 μm. (D) Overlap (in %) of SPI-PBW/LIP5 BiFC signals overlapping ARA6- and ARA7-labeled endosomes. Data denote the average of 15 and 12 cells, respectively. Error bars represent SDs.
Because SPI contains a PH domain known to mediate binding to phospholipids (Ferguson et al., 1995; Lemmon, 2007) we assessed the binding-ability of bacterially expressed GST-SPI-PBW-His6 to a selection of membrane-anchored lipids in protein-lipid overlay assays (Supplementary Figures S3A–C). GST was included as a negative control and the PH domain from PLC-d1 as a positive control (Supplementary Figure S3B). We found no binding of GST-SPI-PBW-His6 to any of the phospholipids tested, including the endosomal enriched Phosphatidylinositol-3-phosphate (PI3P) (Voigt et al., 2005; Vermeer et al., 2006) (Supplementary Figure S3C). These data suggest that the association of SPI to endosomes is not mediated by phosphoinositides.
To test the functional relevance of the SPI/LIP5/SKD1 interactions, we performed a genetic analysis. We focused on the seed coat mucilage phenotype because plants expressing dominant negative SKD1 produce seeds lacking seed mucilage completely (Shahriari et al., 2010a). Seed coat mucilage is enriched in polymeric sugars that are secreted from the outer seed coat cells. Its presence can be monitored by Ruthenium Red staining of whole seeds (Western et al., 2000). We found no difference between spi and wild-type seeds (Figures 4A,B). The mucilage layer of lip5 mutant seeds was still formed, but its area was significantly smaller than in wild-type seeds. This observation is consistent with the finding that LIP5 acts as an enhancer of SKD1 ATPase activity (Haas et al., 2007). The spi lip5 double mutant seeds exhibited a strong reduction of the mucilage layer that was significantly different to wild type as well as lip5 single mutant seeds (Figures 4A,B). Apart from this mucilage phenotype we did not note any differences to spi mutants in spi lip5 double mutant plants with respect to growth and morphological features. These data indicate that the secretion of seed coat mucilage requires the SKD1/SPI/LIP5 pathway and that SPI and LIP5 act synergistically.
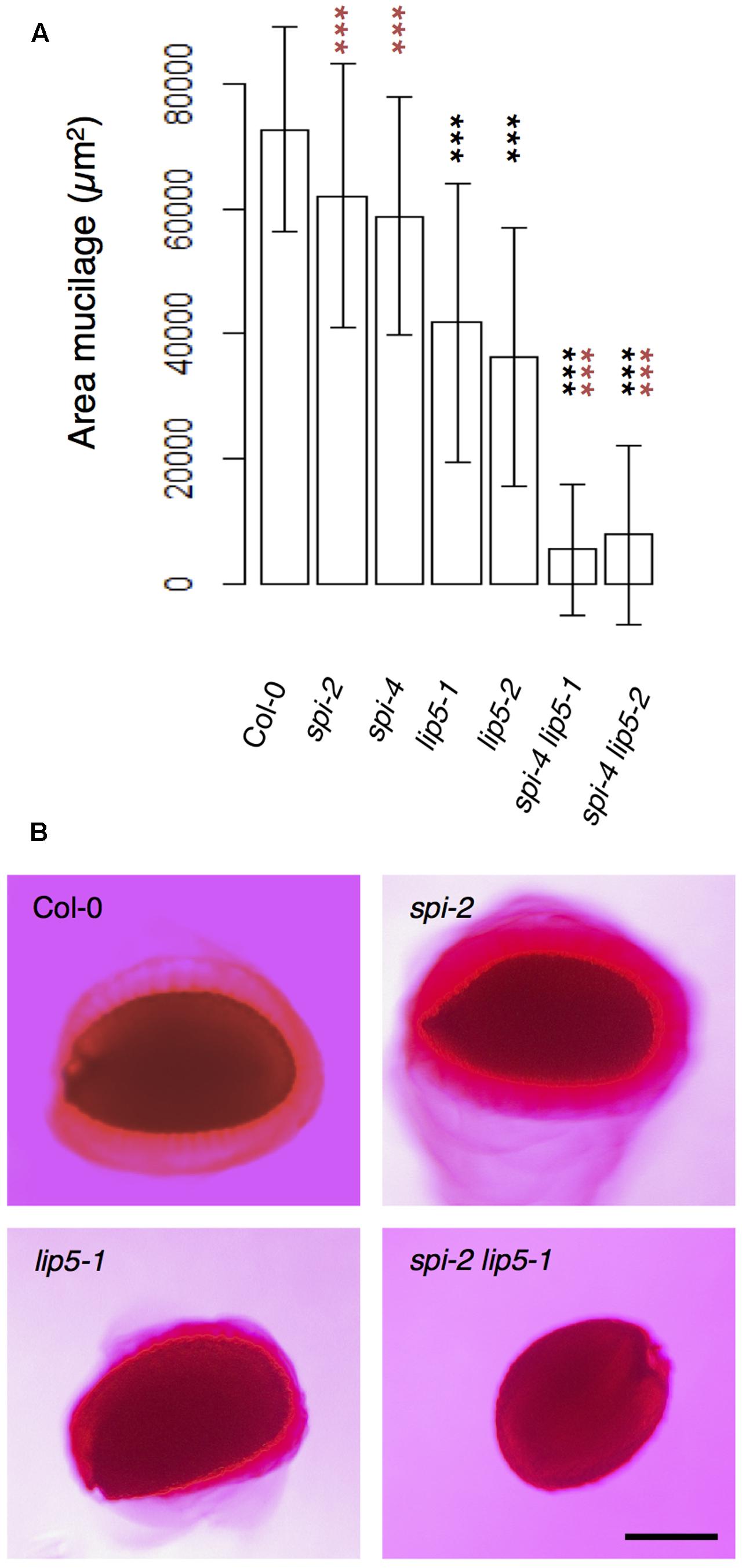
FIGURE 4. Mucilage formation in Ruthenium Red-stained seeds of spi and lip5 mutants. (A) Mucilage areas and seed sizes are presented. Data denote the average of 30 seeds. Error bars represent SDs. Black asterisks represent statistically significant changes in comparison to Col-0 and red asterisks statistically significant changes in comparison to both lip5 mutants (two-tailed Student’s t-test; ∗∗∗p < 0.001). (B) Representative images of Ruthenium Red-stained seeds. Scale bar: 200 μm.
To test whether SPI is functionally involved in the endosomal-vacuolar transport route, we examined the intracellular localization of two soluble vacuolar enzymes CPY and AALP in spi mutants (Rojo et al., 2003; Shahriari et al., 2010b). Both, CPY-CFP and AALP-mCHERRY were found in the vacuole in more than 90% of the cells. Their distribution was unaffected in both spi and lip5 single mutants. However, in spi lip5 double mutants we found a severe reduction of cells displaying vacuolar CPY and AALP (Figures 5A–D). The vacuolar sorting defect was even more obvious after controlled induction of CPY-mCHERRY expression using an EtOH-inducible vector system (Roslan et al., 2001) (Figures 5E,F and Supplementary Figure S4).
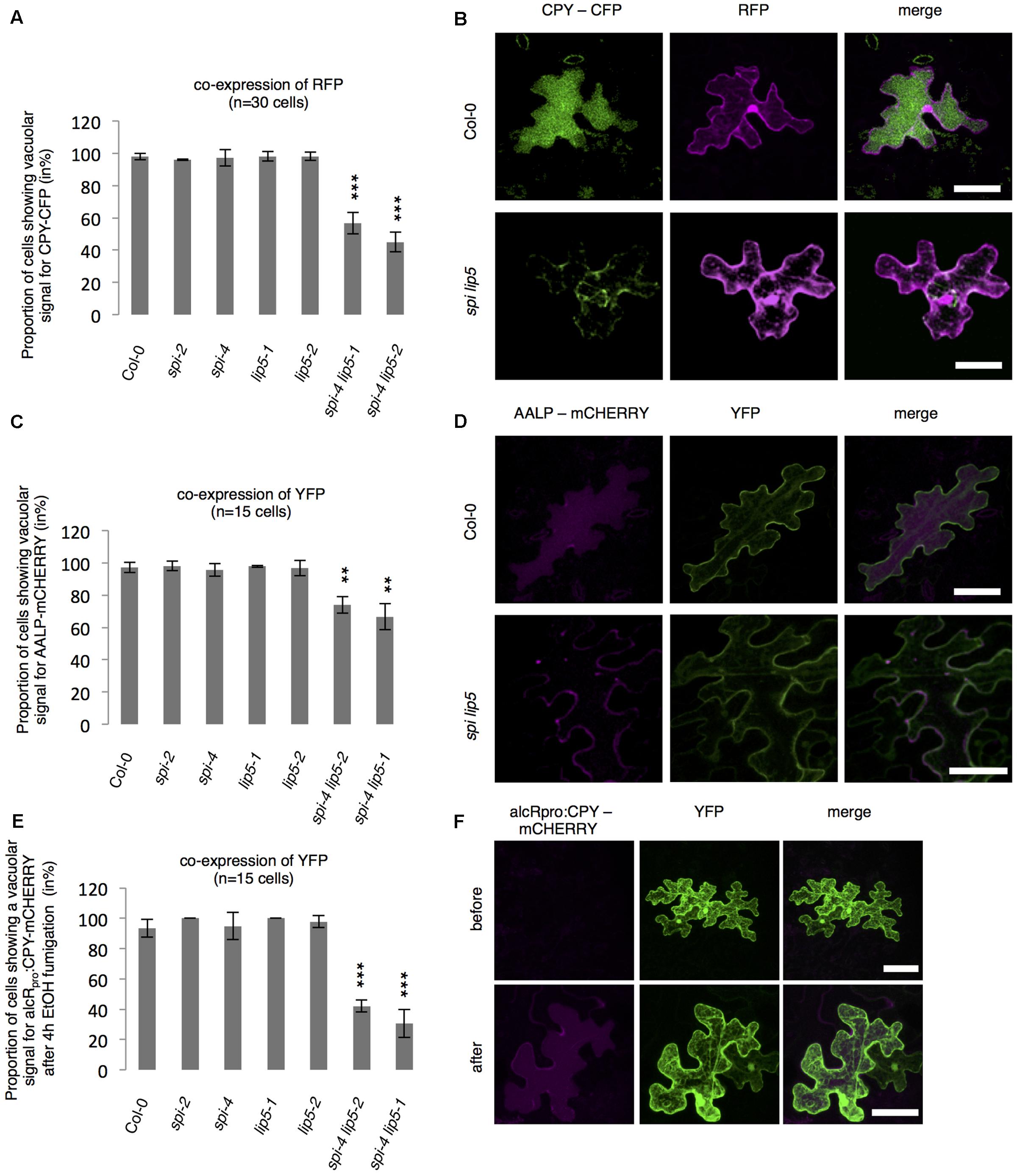
FIGURE 5. Analysis of vacuolar transport in spi and lip5 mutants. (A) Number of cells (in %) showing a vacuolar signal for CPY-CFP in cells co-expressing free RFP. Data denote the average of three biological replicates (n = 30 cells each). (B) Representative maximum projections of CPY-CFP (column I) in transfected leaf epidermis cells of Col-0 (Upper) and spi lip5 double mutants (Lower) co-expressing free RFP (column II) as transformation control. Column III presents the overlay picture of columns I and II. (C) Number of cells (in %) showing a vacuolar signal for AALP-mCHERRY in cells co-expressing free YFP. Data denote the average of three biological replicates (n = 15 cells each). (D) Representative maximum projections of AALP-mCHERRY (column I) in transfected leaf epidermis cells of Col-0 (Upper) and spi lip5 double mutants (Lower) co-expressing free YFP (column II) as transformation control. Column III presents the overlay picture of columns I and II. (E) Number of cells (in %) showing vacuolar signal for alcRpro:CPY-mCHERRY in cells co-expressing free YFP 4 h after fumigation with 2% EtOH (related to Supplementary Figure S4). Data denote the average of three biological replicates (n = 15 cells each). Error bars in A,C,E represent SDs. Asterisks represent statistically significant changes in comparison to Col-0 (two-tailed Student’s t-test; ∗∗p < 0.01, ∗∗∗p < 0.001). (F) Representative maximum projections of alcRpro:CPY-mCHERRY (column I) before (Upper) and after induction of gene expression (Lower) in transfected leaf epidermis cells of Col-0 co-expressing free YFP (column II) as transformation control. Column III presents the overlay picture of columns I and II. Scale bars: 30 μm.
We reasoned that the vacuolar sorting defect might result in an increased secretion similar as observed in plants expressing dominant negative SKD1 (Shahriari et al., 2010b). We therefore performed a secretion assay using Col-0 and spi lip5 leaf mesophyll protoplasts. We compared the amounts of CPY present in the fraction of protoplasts with those secreted to the surrounding medium after 1 h of incubation (Figure 6). As compared to wild type, we found a stronger signal of CPY in the medium of spi lip5 protoplasts in two independent experiments (one shown in Figure 6). We excluded that the CPY signals from the medium samples were caused by impurities from damaged protoplast by using α-cytoplasmic Fructose-1.6-Bisphosphatase (cF1.6BPase) as a control (Figure 6).
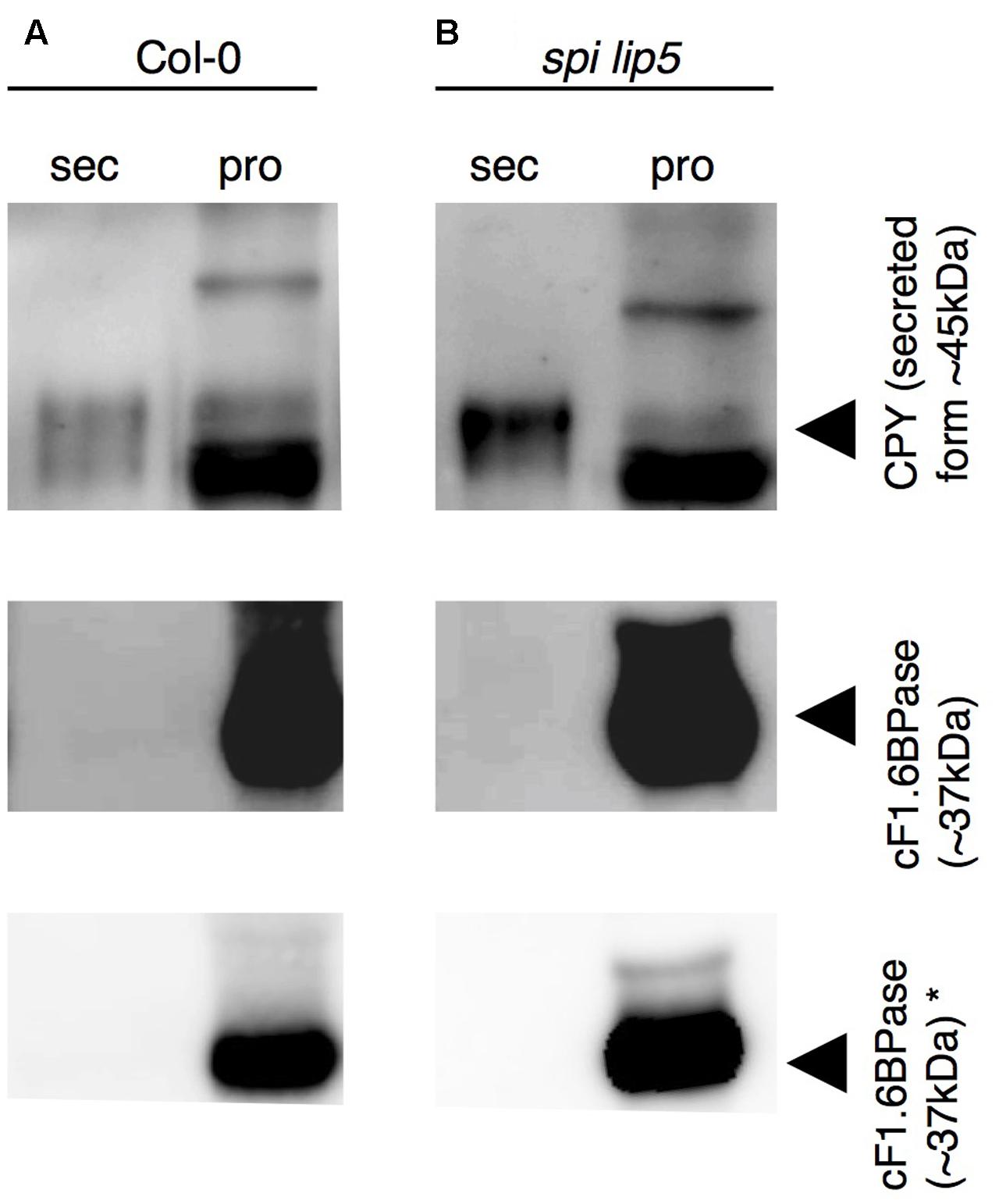
FIGURE 6. CPY is aberrantly secreted in protoplasts from spi lip5 double mutants. Detection of CPY in total protein samples from (A) Col-0 and (B) spi lip5-derived leaf mesophyll protoplasts before their incubation in secretion medium (raw), the secretion medium (sec) after 60 min of incubation and the corresponding intact protoplasts fraction (pro), on α-CPY (Rojo et al., 2003) stained Western blots. The positions of CPY precursor and intermediate forms are indicated by arrowheads. cF1.6BPase was detected by α-cF1.6BPase antibody (Agrisera antibodies) staining and used as control. Please note that the contrasts of α-CPY and α-cF1.6BPase stained Western blots have been adjusted to easily visualize any protein present in the secretory fraction. Non-manipulated α-cF1.6BPase stained Western blots are presented in the lowest row (indicated by ∗). The same pattern of CPY and cF1.6BPase was observed in a second independent experiment.
The finding that the spi lip5 double mutants share many phenotypes with the dominant negative SKD1 mutants (mucilage phenotype, vacuolar transport defects, aberrant protein secretion) suggests that they may act together in promoting SKD1 activity. In this case, one would expect that the overexpression of SKD1 can compensate the reduced activation of SKD1 in spi lip5 double mutants. We therefore tested whether overexpression of SKD1 can rescue the spi lip5 vacuolar transport phenotype. Toward this end, we assessed the localization of AALP-mCHERRY and CPY-CFP in spi lip5 double mutants co-expressing SKD1. The vacuolar transport of AALP was restored in 100% (Figures 7A,B) and of CPY-CFP in 80% of cells (Figures 7C,D). Co-expression of the late endosomal marker ARA7 did not restore the vacuolar transport of CPY-CFP (Figures 7E,F), highlighting the SKD1-specificity of the rescue. These data suggest that LIP5 and SPI regulate endosomal-vacuolar transport by acting as positive regulators of SKD1.
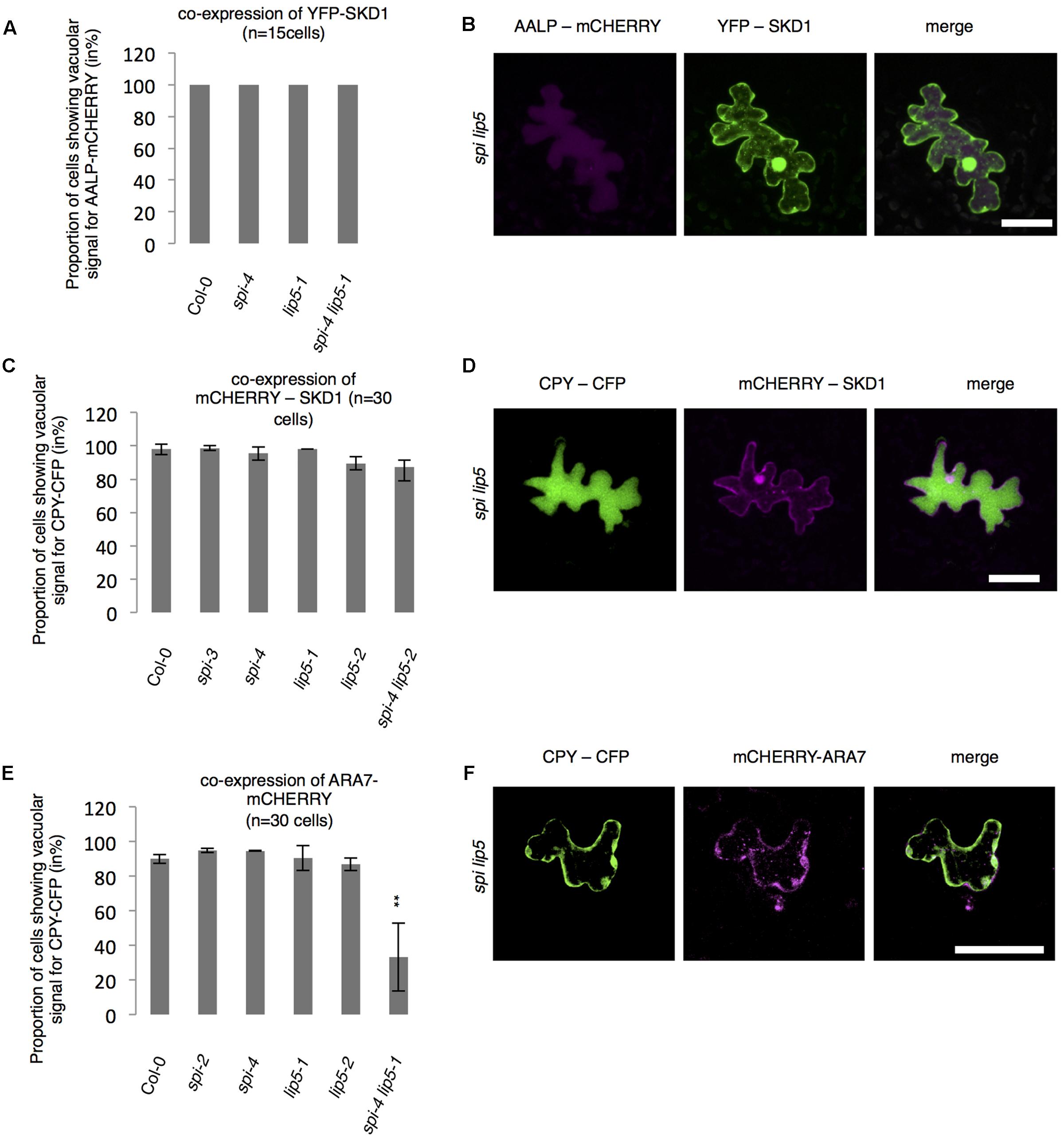
FIGURE 7. Vacuolar transport in spi lip5 mutants can be restored by SKD1. (A) Number of cells (in %) showing a vacuolar signal for AALP-mCHERRY in cells co-expressing SKD1. (B) Representative images of AALP-mCHERRY (column I) in transfected spi lip5 mutant leaf epidermis cells co-expressing SDK1-mCHERRY (column II). Column III presents the overlay-picture of columns I and II. Scale bar: 30 μm. (C) Number of cells (in %) showing a vacuolar signal for CPY-CFP in cells co-expressing SKD1. (D) Representative images of CPY-CFP (column I) in transfected spi lip5 mutant leaf epidermis cells co-expressing SDK1-mCHERRY (column II). Column III presents the overlay-picture of columns I and II. Scale bar: 30 μm. (E) Number of cells (in %) showing a vacuolar signal for CPY-CFP in cells co-expressing mCHERRY-ARA7. Data of A,C,E denote the average numbers (in %) of three biological replicates. Error bars represent SDs. Asterisks represent statistically significant changes in comparison to Col-0 (two-tailed Student’s t-test; ∗∗p < 0.01). (F) Representative image of CPY-CFP (column I) in transfected spi lip5 mutant leaf epidermis cells co-expressing mCHERRY-ARA7 (column II). Column III shows the overlay picture of columns I and II. Scale bar: 30 μm.
Initially spi has been described as a member of the class of Arabidopsis distorted mutants. In contrast to all other mutants in this class, spi displays no actin phenotype suggesting that it may regulate cell morphogenesis in a cytoskeleton-independent manner. As SPI encodes a BDCP homolog it was initially postulated that SPI may function in endomembrane dynamics. The finding, however, that SPI has a molecular function in the post-transcriptional regulation of mRNA stability raised the question, whether SPI has an additional role in membrane dynamics. Here, we provide genetic and molecular data that functionally link SPI and SKD1 and LIP5. A connection between the distorted phenotype and endosomal transport has not been revealed so far. Neither the lip5 mutant nor expression of dominant negative SKD1 has been reported to trigger a distorted phenotype. Further studies might aim to investigate if there is any direct connection between these fields of research.
The common phenotypes of BDCP mutants points to their function in the control of membrane fusion and fission events at different steps along the endomembrane system (Cullinane et al., 2013). Recently, a working model was developed that proposes BDCPs to act as scaffold proteins that regulate membrane dynamics in a binding partner-dependent manner (Cullinane et al., 2013). However, only few binding partners of BDCPs have been identified (Adam-Klages et al., 1996; Segui et al., 2001; Tchernev et al., 2002; Filimonenko et al., 2010; Hocking et al., 2010) and their contribution to the pathophysiology observed in BDCP mutants was not characterized in detail so far. A possible link between BDCPs and the LIP5/SKD1 complex had been suggested by the findings that LIP5 interacts with LYST in yeast two-hybrid assays (Tchernev et al., 2002) and that over-expression of a dominant negative SKD1 protein leads to an association of LYST to membranes (Fujita et al., 2004). Our finding that SPI directly interacts not only with LIP5 but also with SKD1 strongly suggest that a functional link between BDCPs and LIP5/SKD1 is evolutionarily conserved.
As shown for human LYST, the SPI protein is evenly distributed in the cytoplasm (Perou et al., 1997; Fujita et al., 2004). The previous finding that LYST relocates to membranes only upon co-expression of dominant negative SKD1 (E235Q) suggests that it is either recruited under these conditions, or that it is present at endosomal membranes at low levels and becomes detectable because of the formation of large endosomal class E compartments. Consistent with both scenarios, we observe SPI interactions with LIP5 and SKD1 at endosomes in BiFC experiments. As BiFC tends to stabilize protein–protein interactions (Kerppola, 2008) a recruitment of SPI to endosomes by its binding partners is conceivable.
Membrane association of other BDCPs has been shown to depend on their binding to phospholipids. The human BDCP factor associated with neutral sphingomyelinase activation (FAN) binds directly to plasma membrane associated phosphoinositides (Haubert et al., 2007). Other BDCPs such as the mammalian homologs NBEA and lipopolysaccharide-responsive and beige-like anchor protein (LRBA) do not bind to phospholipids. Crystal structure analysis of LRBA revealed an uncommon PH domain, lacking a cluster of basic residues essential to form the classical phospholipid binding fold (Gebauer et al., 2004; Haubert et al., 2007; Lemmon, 2007). SPI protein does not appear to bind to phosphoinositides suggesting that the recruitment of SPI to membranes may be mediated by binding to other proteins.
The ATPase activity of SKD1 is essential for the disassembly of the ESCRT machinery from membranes, and thereby the formation of ILVs in late endosomal compartments (Babst et al., 1998; Azmi et al., 2006; Cai et al., 2014). The depletion of SKD1 and expression of its dominant negative SKD1 versions cause the formation of aberrantly enlarged pre-vacuolar structures (class E compartments) in various species (Fujita et al., 2003; Shahriari et al., 2010b). In plants, the expression of dominant negative SKD1 causes vacuole fragmentation suggesting that the membrane transport between MVBs and the vacuole is impaired (Shahriari et al., 2010b). Although spi mutants exhibit fragmented vacuoles (Saedler et al., 2009), we found no obvious defects in endosomal trafficking. Neither the internalization of the styryl dye FM4-64 (Saedler et al., 2009) nor the vacuolar transport of soluble cargo proteins (this study) were affected in spi mutants. Thus, the cellular spi mutant phenotype differs to other class E (Fujita et al., 2003; Haas et al., 2007) or BDCP mutants (Faigle et al., 1998; Cornillon et al., 2002). Similar results were reported for lip5 mutant plants (Cai et al., 2014). Although LIP5 promotes the enzymatic activity of SKD1, lip5 mutants exhibit no global mutant phenotypes (Haas et al., 2007). However, a role of LIP5 in MVB biogenesis was reported in the context of stress responses (Wang et al., 2014, 2015). In addition, trafficking defects of PIN proteins in roots were correlated with impaired gravitropism (Buono et al., 2016). Together these data suggest that LIP5 is a regulator of SKD1 but not an absolute requirement for SKD1 activity. Our genetic studies, showing a redundant function of LIP5 and SPI suggest that the two proteins act together in the regulation of SKD1. As the additional expression of SKD1 can overcome the requirement of LIP5 and SPI it is likely that SKD1 acts genetically downstream of LIP5 and SPI. It is conceivable that SPI acts together with LIP5 as a positive regulator of SKD1. This conclusion appears to be in conflict with data obtained in human cell culture systems. Here, overexpression of SKD1 was not sufficient to restore the giant lysosome phenotype in LYST deficient fibroblasts suggesting that LYST acts downstream of SKD1-mediated membrane transport (Fujita et al., 2004). This suggests that the molecular function we describe for the BDCP SPI is mechanistically different to that of BDCPs in humans. An alternative explanation would be that a rescue of protein/membrane trafficking by additional expression of SKD1 is independent of a rescue of endosomal sizes.
Arabidopsis thaliana spi-2 (GK_205G08), spi-3 (SALK_065311), spi-4 (GK_420D09), lip5-1 (SAIL_854F08) and lip5-2 (GK_351B10) have been previously described (Haas et al., 2007; Wang et al., 2014); and were obtained from the National Arabidopsis Stock Center. Positions of T-DNA insertions were confirmed by sequencing the flanking genomic regions. Seeds were grown on soil under long day conditions (22°C; 110 μmol m-2 s-1). Ruthenium Red Staining of seed coat mucilage was performed in H2O (+0.1% Tween20) as described (Western et al., 2000). Isolation of protoplasts from Arabidopsis leaves was performed according to the Tape-Arabidopsis Sandwich method (Wu et al., 2009). Fifty microliters protoplasts were harvested and washed with 150 μl of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 2 mM MES, pH 5.7) in a 1.5 ml Eppendorf tube at 100g for 5 min. For determination of protein secretion 50 μl of washed protoplasts were incubated in 50 μl of W5 solution for 60 min at room temperature in the dark. After centrifugation (5 min, 100g), supernatant (secretion fraction) and intact protoplasts were divided and boiled in protein loading buffer for further analysis.
Coding sequences (CDS) of SPI-PBW (AT1G03060) was described before (Steffens et al., 2015). The CDS of SPI-PB and AALP (AT5G60360) were amplified from Col-0 cDNA. Primer sequences are provided in Supplementary Table S1. Entry clones of SKD1 (AT2G27600), LIP5 (AT4G26750), CPY (AT3G10410), ARA6 (AT3G54840) and ARA7 (AT4G19640) were published before (Shahriari et al., 2010b). All constructs were confirmed by sequencing. A list of GATEWAY®-vectors used in this study is provided in Supplementary Table S2.
Yeast two-hybrid assays were performed as described (Gietz et al., 1995). Interactions were analyzed on dropout media lacking leucine (L), tryptophan (W) and histidine (H) [+20 mM 3-Amino-1,2,3-Triazole (3AT)]. For biochemical assays GST-SPI-PBW-His6, GST and His6-tagged SKD1 were expressed and purified as described before (Steffens et al., 2014). Lysis of bacteria expressing GST-SPI-PBW-His6 was performed in STE-Buffer (10 mM Tris pH8.0, 150 mM NaCl, 1 mM EDTA). After centrifugation of the lysates, PBS (pH 7.5) was used for washing and elution according to (Steffens et al., 2014). GST-SPI was eluted in PBS, supplemented with 40 mM reduced L-glutathione. His6-fusions were eluted in Tris-Elution buffer (100 mM Tris pH 8.0, 150 mM NaCl) supplemented with 500 mM imidazole. For co-precipitation and protein-lipid binding assays (Echelon bioscience; performed with 1.5 μg purified protein according to the manufacturers instructions), purified proteins were concentrated using Amicons Ultra-4 Centrifugal Filters (Merck Millipore) and dialyzed (Xpress Micro Dialyzer Cartridge MD300; Scienova) against Tris-Elution Buffer. Purifications and co-precipitations were analyzed by immunoblotting (Steffens et al., 2014). HA- and 3xFLAG fusions were (co)-immunoprecipitated using antibody conjugated μ-columns according to the manufacturer’s instructions (Miltenyi; μMACSTM) from total protein lysates derived from infiltrated N. benthamiana leaves. Bound proteins were eluted and analyzed by immunoblotting, using either a monoclonal horseradish peroxidase (HRP) coupled with anti-HA antibody from rat (Roche; 1:1000 dilution) or a primary monoclonal anti-FLAG antibody from mouse (Sigma–Aldrich; 1:2000 dilution) and a secondary anti-mouse antibody from goat coupled to HRP (Sigma–Aldrich; 1:10,000).
Nicotiana benthamiana leaves were transformed as described (Yang et al., 2000). Biolistic transformation of Arabidopsis leaves was performed as described (Mathur et al., 2003) and analyzed after 12 to 16 h by Confocal Laser Scanning Microscopy (CLSM). CLSM was performed as described (Steffens et al., 2014). Expression of alcRpro-fusion proteins (pCAMPARI-vector) was induced by fumigation of transfected leaves with 2% ethanol (v/v) for 4 h (Roslan et al., 2001). Before induction of protein expression, transfected cells were identified by the presence of constantly co-expressed free YFP. To ensure comparisons between equally strong transfected cells, laser intensities and exposure times were fixed.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers: AT1G03060 (SPI), AT5G60360 (AALP), AT2G27600 (SKD1), AT4G26750 (LIP5), AT3G10410 (CPY), AT3G54840 (ARA6), and AT4G19640 (ARA7).
AS and MH designed this study. AS performed the research and analyzed the data. AS and MJ generated plasmids and established mutant lines. AS and MH wrote the paper. MH directed the project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by a DFG grant (project number 246375609) to MH. The authors would like to thank Dr. Stefanie Herberth for her support in protein-lipid binding assays; Birgit Kernebeck for excellent technical assistance; Dr. Aurelien Boisson-Dernier, Dr. Swen Schellmann and Lisa Stephan for critically reading the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01969/full#supplementary-material
Adam-Klages, S., Adam, D., Wiegmann, K., Struve, S., Kolanus, W., Schneider-Mergener, J., et al. (1996). FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell 86, 937–947. doi: 10.1016/S0092-8674(00)80169-5
Azmi, I., Davies, B., Dimaano, C., Payne, J., Eckert, D., Babst, M., et al. (2006). Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172, 705–717. doi: 10.1083/jcb.200508166
Babst, M., Wendland, B., Estepa, E. J., and Emr, S. D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993. doi: 10.1093/emboj/17.11.2982
Barbosa, M. D., Nguyen, Q. A., Tchernev, V. T., Ashley, J. A., Detter, J. C., Blaydes, S. M., et al. (1996). Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 382, 262–265. doi: 10.1038/382262a0
Brandt, E. J., Elliott, R. W., and Swank, R. T. (1975). Defective lysosomal enzyme secretion in kidneys of Chediak-Higashi (beige) mice. J. Cell Biol. 67, 774–788. doi: 10.1083/jcb.67.3.774
Buono, R. A., Paez-Valencia, J., Miller, N. D., Goodman, K., Spitzer, C., Spalding, E. P., et al. (2016). Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol. 171, 251–264. doi: 10.1104/pp.16.00240
Cai, Y., Zhuang, X., Gao, C., Wang, X., and Jiang, L. (2014). The Arabidopsis endosomal sorting complex required for transport III regulates internal vesicle formation of the prevacuolar compartment and is required for plant development. Plant Physiol. 165, 1328–1343. doi: 10.1104/pp.114.238378
Cornillon, S., Dubois, A., Bruckert, F., Lefkir, Y., Marchetti, A., Benghezal, M., et al. (2002). Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J. Cell Sci. 115, 737–744.
Cullinane, A. R., Schaffer, A. A., and Huizing, M. (2013). The BEACH is hot: a LYST of emerging roles for BEACH-domain containing proteins in human disease. Traffic 14, 749–766. doi: 10.1111/tra.12069
de Souza, N., Vallier, L. G., Fares, H., and Greenwald, I. (2007). SEL-2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development 134, 691–702. doi: 10.1242/dev.02767
del Pino, I., Paarmann, I., Karas, M., Kilimann, M. W., and Betz, H. (2011). The trafficking proteins Vacuolar Protein Sorting 35 and Neurobeachin interact with the glycine receptor beta-subunit. Biochem. Biophys. Res. Commun. 412, 435–440. doi: 10.1016/j.bbrc.2011.07.110
Ebine, K., Fujimoto, M., Okatani, Y., Nishiyama, T., Goh, T., Ito, E., et al. (2011). A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853–859. doi: 10.1038/ncb2270
Faigle, W., Raposo, G., Tenza, D., Pinet, V., Vogt, A. B., Kropshofer, H., et al. (1998). Deficient peptide loading and MHC class II endosomal sorting in a human genetic immunodeficiency disease: the Chediak-Higashi syndrome. J. Cell Biol. 141, 1121–1134. doi: 10.1083/jcb.141.5.1121
Ferguson, K. M., Lemmon, M. A., Schlessinger, J., and Sigler, P. B. (1995). Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046. doi: 10.1016/0092-8674(95)90219-8
Filimonenko, M., Isakson, P., Finley, K. D., Anderson, M., Jeong, H., Melia, T. J., et al. (2010). The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 38, 265–279. doi: 10.1016/j.molcel.2010.04.007
Fujita, H., Umezuki, Y., Imamura, K., Ishikawa, D., Uchimura, S., Nara, A., et al. (2004). Mammalian class E Vps proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J. Cell Sci. 117, 2997–3009. doi: 10.1242/jcs.01170
Fujita, H., Yamanaka, M., Imamura, K., Tanaka, Y., Nara, A., Yoshimori, T., et al. (2003). A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 116, 401–414. doi: 10.1242/jcs.00213
Gao, C., Zhuang, X., Cui, Y., Fu, X., He, Y., Zhao, Q., et al. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 112, 1886–1891. doi: 10.1073/pnas.1421271112
Gebauer, D., Li, J., Jogl, G., Shen, Y., Myszka, D. G., and Tong, L. (2004). Crystal structure of the PH-BEACH domains of human LRBA/BGL. Biochemistry 43, 14873–14880. doi: 10.1021/bi049498y
Gerald, N. J., Siano, M., and De Lozanne, A. (2002). The Dictyostelium LvsA protein is localized on the contractile vacuole and is required for osmoregulation. Traffic 3, 50–60. doi: 10.1034/j.1600-0854.2002.30107.x
Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. doi: 10.1002/yea.320110408
Haas, T. J., Sliwinski, M. K., Martinez, D. E., Preuss, M., Ebine, K., Ueda, T., et al. (2007). The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19, 1295–1312. doi: 10.1105/tpc.106.049346
Harris, E., Wang, N., Wu, W. L., Weatherford, A., De Lozanne, A., and Cardelli, J. (2002). Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol. Biol. Cell 13, 656–669. doi: 10.1091/mbc.01-09-0454
Haubert, D., Gharib, N., Rivero, F., Wiegmann, K., Hosel, M., Kronke, M., et al. (2007). PtdIns(4,5)P-restricted plasma membrane localization of FAN is involved in TNF-induced actin reorganization. EMBO J. 26, 3308–3321. doi: 10.1038/sj.emboj.7601778
Hocking, L. J., Mellis, D. J., McCabe, P. S., Helfrich, M. H., and Rogers, M. J. (2010). Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochem. Biophys. Res. Commun. 402, 543–548. doi: 10.1016/j.bbrc.2010.10.076
Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Hurley, J. H., and Emr, S. D. (2006). The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35, 277–298. doi: 10.1146/annurev.biophys.35.040405.102126
Introne, W., Boissy, R. E., and Gahl, W. A. (1999). Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome. Mol. Genet. Metab. 68, 283–303. doi: 10.1006/mgme.1999.2927
Introne, W. J., Westbroek, W., Golas, G. A., and Adams, D. (1993). “Chediak-Higashi Syndrome,” in SourceGeneReviews® [Internet], eds M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, H. C. Mefford, et al. (Seattle, WA: University of Washington), 1993–2017.
Kaplan, J., De Domenico, I., and Ward, D. M. (2008). Chediak-Higashi syndrome. Curr. Opin. Hematol. 15, 22–29. doi: 10.1097/MOH.0b013e3282f2bcce
Katsiarimpa, A., Munoz, A., Kalinowska, K., Uemura, T., Rojo, E., and Isono, E. (2014). The ESCRT-III-interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana. Plant Cell Physiol. 55, 727–736. doi: 10.1093/pcp/pcu019
Kerppola, T. K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487. doi: 10.1146/annurev.biophys.37.032807.125842
Khodosh, R., Augsburger, A., Schwarz, T. L., and Garrity, P. A. (2006). Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133, 4655–4665. doi: 10.1242/dev.02650
Landsberg, M. J., Vajjhala, P. R., Rothnagel, R., Munn, A. L., and Hankamer, B. (2009). Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure 17, 427–437. doi: 10.1016/j.str.2008.12.020
Lata, S., Schoehn, G., Jain, A., Pires, R., Piehler, J., Gottlinger, H. G., et al. (2008). Helical structures of ESCRT-III are disassembled by VPS4. Science 321, 1354–1357. doi: 10.1126/science.1161070
Lemmon, M. A. (2007). Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81–93. doi: 10.1042/BSS2007c08
Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B. P., and Hulskamp, M. (2003). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. doi: 10.1242/dev.00549
Matile, P. (1968). Lysosomes of root tip cells in corn seedlings. Planta 79, 181–196. doi: 10.1007/BF00396026
Mesquita, J. F. (1969). Electron microscope study of the origin and development of the vacuoles in root-tip cells of Lupinus albus L. J. Ultrastruct. Res. 26, 242–250. doi: 10.1016/S0022-5320(69)80004-3
Nagle, D. L., Karim, M. A., Woolf, E. A., Holmgren, L., Bork, P., Misumi, D. J., et al. (1996). Identification and mutation analysis of the complete gene for Chediak-Higashi syndrome. Nat. Genet. 14, 307–311. doi: 10.1038/ng1196-307
Neer, E. J., Schmidt, C. J., Nambudripad, R., and Smith, T. F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. doi: 10.1038/371297a0
Perou, C. M., Leslie, J. D., Green, W., Li, L., Ward, D. M., and Kaplan, J. (1997). The Beige/Chediak-Higashi syndrome gene encodes a widely expressed cytosolic protein. J. Biol. Chem. 272, 29790–29794. doi: 10.1074/jbc.272.47.29790
Perou, C. M., Moore, K. J., Nagle, D. L., Misumi, D. J., Woolf, E. A., McGrail, S. H., et al. (1996). Identification of the murine beige gene by YAC complementation and positional cloning. Nat. Genet. 13, 303–308. doi: 10.1038/ng0796-303
Pesch, M., Schultheiss, I., Digiuni, S., Uhrig, J. F., and Hulskamp, M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140, 3456–3467. doi: 10.1242/dev.094698
Raymond, C. K., Howald-Stevenson, I., Vater, C. A., and Stevens, T. H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389–1402. doi: 10.1091/mbc.3.12.1389
Richardson, L. G., Howard, A. S., Khuu, N., Gidda, S. K., McCartney, A., Morphy, B. J., et al. (2011). Protein-protein interaction network and subcellular localization of the Arabidopsis thaliana ESCRT Machinery. Front. Plant Sci. 2:20. doi: 10.3389/fpls.2011.00020
Rojo, E., Zouhar, J., Carter, C., Kovaleva, V., and Raikhel, N. V. (2003). A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 7389–7394. doi: 10.1073/pnas.1230987100
Roslan, H. A., Salter, M. G., Wood, C. D., White, M. R., Croft, K. P., Robson, F., et al. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28, 225–235. doi: 10.1046/j.1365-313X.2001.01146.x
Sachse, M., Strous, G. J., and Klumperman, J. (2004). ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J. Cell Sci. 117, 1699–1708. doi: 10.1242/jcs.00998
Saedler, R., Jakoby, M., Marin, B., Galiana-Jaime, E., and Hulskamp, M. (2009). The cell morphogenesis gene SPIRRIG in Arabidopsis encodes a WD/BEACH domain protein. Plant J. 59, 612–621. doi: 10.1111/j.1365-313X.2009.03900.x
Saint-Jore-Dupas, C., Gomord, V., and Paris, N. (2004). Protein localization in the plant Golgi apparatus and the trans-Golgi network. Cell Mol. Life Sci. 61, 159–171. doi: 10.1007/s00018-003-3354-7
Scheuring, D., Kunzl, F., Viotti, C., Yan, M. S., Jiang, L., Schellmann, S., et al. (2012). Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol. 12:164. doi: 10.1186/1471-2229-12-164
Scheuring, D., Viotti, C., Kruger, F., Kunzl, F., Sturm, S., Bubeck, J., et al. (2011). Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell 23, 3463–3481. doi: 10.1105/tpc.111.086918
Schwab, B., Mathur, J., Saedler, R., Schwarz, H., Frey, B., Scheidegger, C., et al. (2003). Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: actin controls the spatial organization of microtubules. Mol. Genet. Genomics 269, 350–360. doi: 10.1007/s00438-003-0843-1
Segui, B., Cuvillier, O., Adam-Klages, S., Garcia, V., Malagarie-Cazenave, S., Leveque, S., et al. (2001). Involvement of FAN in TNF-induced apoptosis. J. Clin. Invest. 108, 143–151. doi: 10.1172/JCI11498
Shahriari, M., Hulskamp, M., and Schellmann, S. (2010a). Seeds of Arabidopsis plants expressing dominant-negative AtSKD1 under control of the GL2 promoter show a transparent testa phenotype and a mucilage defect. Plant Signal. Behav. 5, 1308–1310. doi: 10.4161/psb.5.10.13134
Shahriari, M., Keshavaiah, C., Scheuring, D., Sabovljevic, A., Pimpl, P., Hausler, R. E., et al. (2010b). The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64, 71–85. doi: 10.1111/j.1365-313X.2010.04310.x
Shim, S., Merrill, S. A., and Hanson, P. I. (2008). Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19, 2661–2672. doi: 10.1091/mbc.E07-12-1263
Spitzer, C., Reyes, F. C., Buono, R., Sliwinski, M. K., Haas, T. J., and Otegui, M. S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21, 749–766. doi: 10.1105/tpc.108.064865
Spritz, R. A. (1998). Genetic defects in Chediak-Higashi syndrome and the beige mouse. J. Clin. Immunol. 18, 97–105. doi: 10.1023/A:1023247215374
Steffens, A., Brautigam, A., Jakoby, M., and Hulskamp, M. (2015). The BEACH domain protein SPIRRIG is essential for Arabidopsis salt stress tolerance and functions as a regulator of transcript stabilization and localization. PLOS Biol. 13:e1002188. doi: 10.1371/journal.pbio.1002188
Steffens, A., Jaegle, B., Tresch, A., Hulskamp, M., and Jakoby, M. (2014). Processing-body movement in Arabidopsis depends on an interaction between myosins and DECAPPING PROTEIN1. Plant Physiol. 164, 1879–1892. doi: 10.1104/pp.113.233031
Szymanski, D. B., Marks, M. D., and Wick, S. M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2347. doi: 10.1105/tpc.11.12.2331
Tchernev, V. T., Mansfield, T. A., Giot, L., Kumar, A. M., Nandabalan, K., Li, Y., et al. (2002). The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol. Med. 8, 56–64.
Teh, O. K., Hatsugai, N., Tamura, K., Fuji, K., Tabata, R., Yamaguchi, K., et al. (2015). BEACH-domain proteins act together in a cascade to mediate vacuolar protein trafficking and disease resistance in Arabidopsis. Mol. Plant 8, 389–398. doi: 10.1016/j.molp.2014.11.015
Ueda, T., Uemura, T., Sato, M. H., and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40, 783–789. doi: 10.1111/j.1365-313X.2004.02249.x
van Balkom, B. W., Boone, M., Hendriks, G., Kamsteeg, E. J., Robben, J. H., Stronks, H. C., et al. (2009). LIP5 interacts with aquaporin 2 and facilitates its lysosomal degradation. J. Am. Soc. Nephrol. 20, 990–1001. doi: 10.1681/ASN.2008060648
Vermeer, J. E., van Leeuwen, W., Tobena-Santamaria, R., Laxalt, A. M., Jones, D. R., Divecha, N., et al. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47, 687–700. doi: 10.1111/j.1365-313X.2006.02830.x
Viotti, C., Kruger, F., Krebs, M., Neubert, C., Fink, F., Lupanga, U., et al. (2013). The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25, 3434–3449. doi: 10.1105/tpc.113.114827
Voigt, B., Timmers, A. C., Samaj, J., Hlavacka, A., Ueda, T., Preuss, M., et al. (2005). Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur. J. Cell Biol. 84, 609–621. doi: 10.1016/j.ejcb.2004.12.029
Wang, F., Shang, Y., Fan, B., Yu, J. Q., and Chen, Z. (2014). Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLOS Pathog. 10:e1004243. doi: 10.1371/journal.ppat.1004243
Wang, F., Yang, Y., Wang, Z., Zhou, J., Fan, B., and Chen, Z. (2015). A critical role of lyst-interacting protein5, a positive regulator of multivesicular body biogenesis, in plant responses to heat and salt stresses. Plant Physiol. 169, 497–511. doi: 10.1104/pp.15.00518
Wang, X., Herberg, F. W., Laue, M. M., Wullner, C., Hu, B., Petrasch-Parwez, E., et al. (2000). Neurobeachin: A protein kinase A-anchoring, beige/Chediak-higashi protein homolog implicated in neuronal membrane traffic. J. Neurosci. 20, 8551–8565.
Ward, D. M., Griffiths, G. M., Stinchcombe, J. C., and Kaplan, J. (2000). Analysis of the lysosomal storage disease Chediak-Higashi syndrome. Traffic 1, 816–822. doi: 10.1034/j.1600-0854.2000.011102.x
Western, T. L., Skinner, D. J., and Haughn, G. W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122, 345–356. doi: 10.1104/pp.122.2.345
Wu, F. H., Shen, S. C., Lee, L. Y., Lee, S. H., Chan, M. T., and Lin, C. S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. doi: 10.1186/1746-4811-5-16
Wu, W. I., Yajnik, J., Siano, M., and De Lozanne, A. (2004). Structure-function analysis of the BEACH protein LvsA. Traffic 5, 346–355. doi: 10.1111/j.1600-0854.2004.00177.x
Yang, Y., Li, R., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi: 10.1046/j.1365-313x.2000.00760.x
Zhang, C., Hicks, G. R., and Raikhel, N. V. (2014). Plant vacuole morphology and vacuolar trafficking. Front. Plant Sci. 5:476. doi: 10.3389/fpls.2014.00476
Zhao, H., Boissy, Y. L., Abdel-Malek, Z., King, R. A., Nordlund, J. J., and Boissy, R. E. (1994). On the analysis of the pathophysiology of Chediak-Higashi syndrome. Defects expressed by cultured melanocytes. Lab. Invest. 71, 25–34.
Zhao, H., Wang, X., Zhu, D., Cui, S., Li, X., Cao, Y., et al. (2012). A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 287, k14109–14121. doi: 10.1074/jbc.M111.280735
Keywords: SPIRRIG, BEACH domain containing protein, Arabidopsis, endosomes, vacuolar transport
Citation: Steffens A, Jakoby M and Hülskamp M (2017) Physical, Functional and Genetic Interactions between the BEACH Domain Protein SPIRRIG and LIP5 and SKD1 and Its Role in Endosomal Trafficking to the Vacuole in Arabidopsis. Front. Plant Sci. 8:1969. doi: 10.3389/fpls.2017.01969
Received: 10 August 2017; Accepted: 01 November 2017;
Published: 20 November 2017.
Edited by:
Diane C. Bassham, Iowa State University, United StatesReviewed by:
Lorena Norambuena, Universidad de Chile, ChileCopyright © 2017 Steffens, Jakoby and Hülskamp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hülskamp, bWFydGluLmh1ZWxza2FtcEB1bmkta29lbG4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.