- National Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan, China
Many Xanthomonas bacteria use transcription activator-like effector (TALE) proteins to activate plant disease susceptibility (S) genes, and this activation contributes to disease. We recently reported that rice basal transcription factor IIA gamma subunit, OsTFIIAγ5, is hijacked by TALE-carrying Xanthomonas oryzae infecting the plants. However, whether TFIIAγs are also involved in TALE-carrying Xanthomonas-caused diseases in other plants is unknown. Here, molecular and genetic approaches were used to investigate the role of TFIIAγs in other plants. We found that TFIIAγs are also used by TALE-carrying Xanthomonas to cause disease in other plants. The TALEs of Xanthomonas citri pv. citri (Xcc) causing canker in citrus and Xanthomonas campestris pv. vesicatoria (Xcv) causing bacterial spot in pepper and tomato interacted with corresponding host TFIIAγs as in rice. Transcriptionally suppressing TFIIAγ led to resistance to Xcc in citrus and Xcv in pepper and tomato. The 39th residue of OsTFIIAγ5 and citrus CsTFIIAγ is vital for TALE-dependent induction of plant S genes. As mutated OsTFIIAγ5V 39E, CsTFIIAγV 39E, pepper CaTFIIAγV 39E, and tomato SlTFIIAγV 39E also did not interact with TALEs to prevent disease. These results suggest that TALE-carrying bacteria share a common mechanism for infecting plants. Using TFIIAγV 39E-type mutation could be a general strategy for improving resistance to TALE-carrying pathogens in crops.
Introduction
Many Xanthomonas bacteria use transcription activator-like effector (TALE) proteins to induce transcription of host disease susceptibility (S) genes, which can cause devastating diseases in plants (Zhang et al., 2015). The TALEs are the largest family of type III effectors in Xanthomonas, a particularly widespread phytobacterial genus consisting of almost 30 species that cause disease in more than 400 plant species, including 11 monocotyledonous and 57 dicotyledonous families (Ryan et al., 2011). Each TALE typically has a modular architecture containing an N-terminal secretion signal that guides the protein’s translocation into host cells through a type III secretion system; a central repeat domain that binds to the effector binding element (EBE) of the host gene promoter via its repeat variable di-residues (RVDs); a newly identified transcription factor binding (TFB) motif that interacts with the host basal transcription factor IIA gamma subunit (TFIIAγ); three nuclear localization signals that guide the protein’s translocation into the host nucleus; and a highly conserved C-terminal acidic activation domain that permits the TALE protein to activate transcription (Doyle et al., 2013; Zhang et al., 2015; Yuan et al., 2016).
Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc) cause bacterial blight and bacterial streak diseases in rice (Oryza sativa), respectively. A recent study revealed that the binding of Xoo and Xoc TALEs to rice EBEs via their RVDs alone cannot efficiently induce rice S gene expression to cause disease. The TFB motif of TALEs is vital for Xoo and Xoc to successfully invade plant cells (Yuan et al., 2016). Xoo and Xoc TALEs directly interact with rice TFIIAγ (OsTFIIAγ5) via their TFB motif to activate the transcription of rice S genes and enable colonization. The attenuation or absence of this interaction could eliminate or weaken the activation of rice S gene expression (Yuan et al., 2016). For example, rice recessive disease resistance gene xa5, which encodes a mutated OsTFIIAγ5 in which valine (V) is changed to glutamic acid (E) at the 39th amino acid residue (OsTFIIAγ5V 39E), confers broad-spectrum resistance to both Xoo and Xoc because most TALEs interact either weakly or not at all with OsTFIIAγ5V 39E. Partial suppression of OsTFIIAγ5 expression also results in resistance to Xoo and Xoc (Yuan et al., 2016). Further, the rice carrying OsTFIIAγ5V 39E is indistinguishable from carrying OsTFIIAγ5 in plant development and yield.
At least eight other Xanthomonas species [Xanthomonas campestris pv. musacearum, Xanthomonas axonopodis pv. manihotis, Xanthomonas citri subsp. citri (Xcc), Xanthomonas campestris pv. malvacearum, Xanthomonas campestris pv. campestris, Xanthomonas campestris pv. vesicatoria (Xcv), Xanthomonas axonopodis pv. vasculorum, and Xanthomonas translucens] also use TALEs to infect plants, including banana, cassava, citrus, cotton, brassicaceae, pepper, tomato, sugarcane, or cereal species, with severe adverse effects on crop yield and quality (Schornack et al., 2013; Boch et al., 2014). In addition to the Xanthomonas species, the plant bacterium Ralstonia solanacearum, the endosymbiont Burkholderia rhizoxinica, and unclassified marine microorganisms also carry TALEs that may also have a role in host-microbe interactions (de Lange et al., 2013, 2015; Juillerat et al., 2014).
TFIIA is a conserved transcription factor of eukaryotes, and it is involved in RNA polymerase II-dependent transcription. It contains two subunits, the large subunit TFIIAαβ and the small subunit TFIIAγ, encoded by separate genes (Orphanides et al., 1996). TFIIAγs from different species have high amino acid sequence similarity (Yuan et al., 2016). TFIIA interacts with other transcriptional regulatory proteins to form a transcription pre-initiation complex, which binds to the TATA-box of promoters and initiates gene expression (Høiby et al., 2007). However, it is unknown whether other TALE-carrying bacteria also hijack their corresponding host TFIIAγ to cause plant disease.
To investigate whether host TFIIAγs are required for TALE-carrying bacteria to infect other crops, we analyzed the interactions between TFIIAγs of citrus, pepper and tomato and TALEs from Xcc, which causes canker disease in citrus, and Xcv, which causes bacterial leaf spot disease in pepper and tomato. We also analyzed the importance of citrus, pepper, and tomato TFIIAγs in diseases caused by Xcc and Xcv. The results suggest that Xcc and Xcv TALEs also interact with plant TFIIAγs via their TFB motifs. The 39th amino acid residue of TFIIAγs is essential for the interaction between TFIIAγs and TALE TFB motifs. Suppressing the expression of citrus TFIIAγ gene significantly improves resistance to Xcc, which is associated with compromised induction of host S gene. Suppressing the expression of pepper and tomato TFIIAγ genes also improves resistance to Xcv in these plants. Therefore, our results suggest that TALE-carrying bacteria use a common mechanism to hijack host basal transcription factor for successful infection.
Materials and Methods
Plant and Bacterial Materials
Indica rice (O. sativa ssp. indica) IR24 and IRBB5 are near-isogenic lines. IR24 carries OsTFIIAγ5 gene and is susceptible to Xoo and Xoc. IRBB5, with the genetic background of IR24, carries recessive disease resistance gene xa5 encoding a mutated OsTFIIAγ5 (OsTFIIAγ5V 39E) and is resistant to Xoo and Xoc. Plants were grown during a normal rice-growing season under natural field conditions. The wild Oryza species used in this study were from our collection (Yuan et al., 2014). Sweet orange (Citrus sinensis L. Osbeck cv. Newhall) plants were grown in a greenhouse at 25 to 30°C. Pepper (Capsicum annuum L. cv. Hua 50) and tomato (Solanum lycopersicum L. cv. Ailsa Craig) plants were germinated and grown in a growth chamber under a photosynthetic photo flux density of approximately 120 μmol photons m-2 s-1 with a 16-h light/8-h dark photoperiod cycle at 25°C and a relative humidity of approximately 60%.
The Philippine Xoo strains PXO99 and PXO341 have commonly been used in studies of rice resistance to bacterial blight disease (Li et al., 2012). The Xoc strain RH3 is also frequently used in research (Yuan et al., 2016). Xcc strain X02-007 (Fu et al., 2012) and Xcv strain 23-1 (Sun et al., 1999) were also previously used in research. All the Xanthomonas strains were grown at 28°C on nutrient agar medium. When genetic manipulation of bacteria was undertaken, antibiotics were used at the following final concentrations as required: ampicillin at 100 μg ml-1, rifampicin at 75 μg ml-1, and kanamycin at 25 μg ml-1.
Transformation
For stable transformation of rice, Agrobacterium-mediated transformation was performed using calli derived from mature embryos of indica rice line IRBB5 according to previously reported methods (Ge et al., 2006). For construction of POsTFIIAγ5:OsTFIIAγ5 derivative vectors, the 2055-bp promoter, 321-bp full-length cDNA and 1229-bp terminator of OsTFIIAγ5 were amplified with primers listed in Supplementary Table 1 and inserted into vector pCAMBIA1301 in order. For construction of PUbi:OsTFIIAγ5 derivative vectors, 321-bp full-length cDNA of OsTFIIAγ5 and its derivatives were amplified with primers listed in Supplementary Table 1 and inserted into vector pU1301 (Yuan et al., 2010).
For transiently suppressing the target gene in sweet orange, Xcc-facilitated agroinfiltration was performed. In brief, a 162-bp cDNA fragment of CsTFIIAγ was amplified with primers listed in Supplementary Table 1 and inserted into vector pDS1301, and the construct was then transformed into Agrobacterium tumefaciens strain GV3101 (Yuan et al., 2010). Sweet orange young leaves were inoculated with Xcc [5 × 108 colony-forming unit (cfu)/ml] by the penetration method using a needleless syringe. Eight hours later, the same inoculated leaf areas were subjected to agroinfiltration with Agrobacterium carrying the recombinant construct (Hu et al., 2014; Jia and Wang, 2014). The Xcc-facilitated agroinfiltration plants were grown in a greenhouse at 25 to 30°C with a 16-h light/8-h dark photoperiod cycle.
For transiently suppressing target gene in pepper and tomato, virus-induced gene silencing (VIGS) was performed. In brief, the DNA segment of the target gene or green fluorescence protein (GFP) gene was inserted into the tobacco rattle virus (TRV) vector pTRV2 as described previously (Liu et al., 2002a). The recombinant vectors were introduced into GV3101. Agrobacterium culture was infiltrated into the cotyledons of germinating pepper plants or the first two true leaves of the four-leaf stage tomato plants using a 1-ml needleless syringe (Liu et al., 2002b; Chung et al., 2004). The Agrobacterium-infiltrated plants were first kept in a growth chamber at 16°C for 1 day and then grown in a chamber at 25°C with a 16-h light/8-h dark photoperiod cycle.
Protein–Protein Interaction
To study the interaction between TALE and host proteins in yeast two-hybrid assays (Yuan et al., 2010), the TFB motif of TALE genes and plant TFIIAγ genes were amplified using PCR primers listed in Supplementary Table 1. The amplified bacterial DNA segments were ligated into pGBKT7 vector, and the amplified plant DNA segments were ligated into pGADT7 Rec vector. The pGBKT7 and pGADT7 plasmids were then transformed into yeast strain AH109 (Yuan et al., 2010). The yeast clones were scribed on the synthetic defined premixes (SD) medium lacking leucine (L) and tryptophan (W) (-LW) and selective SD medium lacking L, W, histidine (H), and adenine (A) (-LWHA). The interactions of examined proteins were assessed by growth of yeast cells on selective medium and by examination of β-D-galactopyranoside (X-α-gal) activity and β-galactosidase (LacZ) activity as described previously (Yuan et al., 2010).
To study the interaction between TALE and plant proteins in planta, co-immunoprecipitation (CoIP) assays were performed (Yuan et al., 2016). The DNA segments of TFB motifs of TALEs were ligated into the pU1031-9myc vector, and the DNA segments of plant genes were ligated into the pU1301-3FLAG vector (Yuan et al., 2010). The recombinant vectors were introduced into GV3101. Agrobacterium-mediated transformation was performed by infiltrating into Nicotiana benthamiana leaves using a needleless syringe. CoIP assays were conducted using anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO, United States) and anti-myc antibody (Tiangen, Beijing, China) as described previously (Yuan et al., 2016). Each CoIP assay was repeated at least twice. The original western blotting images are provided in Supplementary Figure 13.
Site-Directed Mutation
Mutation of plant genes was performed using the GeneTailor Site-Directed Mutagenesis System (Invitrogen Life Technologies, Carlsbad, CA, United States) as described previously (Yuan et al., 2011). The mutagenic primers are listed in Supplementary Table 1.
Pathogen Inoculation
To evaluate rice bacterial blight disease, rice plants were inoculated with Xoo by the leaf-clipping method at the booting (panicle development) stage (Chen et al., 2002). Disease was scored by measuring the lesion length at 14 days after inoculation.
To evaluate rice bacterial streak disease, rice plants were inoculated with Xoc strains by the needle stab method at the tillering stage (Yuan et al., 2016). The disease was scored by measuring the lesion length at 14 days after inoculation.
To evaluate citrus bacterial canker disease, sweet orange leaves were inoculated with Xcc at a concentration of 5 × 108 cfu/ml in 10 mM MgCl2 using a needleless syringe. The bacterial growth rate in sweet orange leaves was measured by counting the cfu as described previously (Hu et al., 2014).
To evaluate bacterial spot disease of pepper and tomato, 4-week-old VIGS-treated plants were sprayed with Xcv at a concentration of 5 × 105 cfu/ml in 10 mM MgCl2. The bacterial growth rate in pepper or tomato leaves was measured by counting the cfu as described previously (Bonas et al., 1991).
Gene Expression Analysis
For gene expression analysis, 2-cm rice leaf fragments near the bacterial infection sites or citrus, pepper and tomato leaf tissues next to the infiltration sites were collected for RNA isolation. Quantitative reverse transcription-PCR (qRT-PCR) was conducted using gene-specific primers (Supplementary Table 2) as described previously (Yuan et al., 2010). The expression level of rice, pepper or tomato actin gene or citrus EF1a gene was used to standardize the RNA sample of rice, pepper, tomato or citrus, respectively. The expression level relative to that of controls was assessed. Each qRT-PCR assay was repeated at least twice with similar result, with each repetition having three replicates.
Statistical Analysis
Differences between samples were analyzed for statistical significance by using the SPSS software and the Student’s t-test (two tailed).
Results
Xcc Uses Host TFIIAγ to Cause Disease in Sweet Orange
Xcc causes citrus canker. Xcc strain X02-007 carries TALE genes, which contained TFB motifs of TALEs that showed high sequence similarity with the motifs of Xoo and Xoc TALEs (Supplementary Figure 1; Yuan et al., 2016). The TFB motifs of Xcc TALEs interacted with sweet orange TFIIAγ (CsTFIIAγ) both in yeast and in planta (Figures 1A,B). To investigate whether CsTFIIAγ is required for TALE-dependent induction of the host S gene during Xcc invasion, we suppressed CsTFIIAγ gene expression in the leaves of sweet orange by RNA interference (RNAi). The plants with suppressed expression of CsTFIIAγ appeared markedly less diseased, with less cell division and proliferation or hyperplasia after Xcc inoculation (Figures 1C–E) and significantly lower (P < 0.01) Xcc growth rate compared to control plants (Figure 1F). Citrus CsLOB1 is a S gene to Xcc (Hu et al., 2014; Li et al., 2014). Consistent with reduced disease symptoms, the CsTFIIAγ-RNAi leaves showed significantly suppressed Xcc-induced expression of CsLOB1 compared with control (Figure 1G).
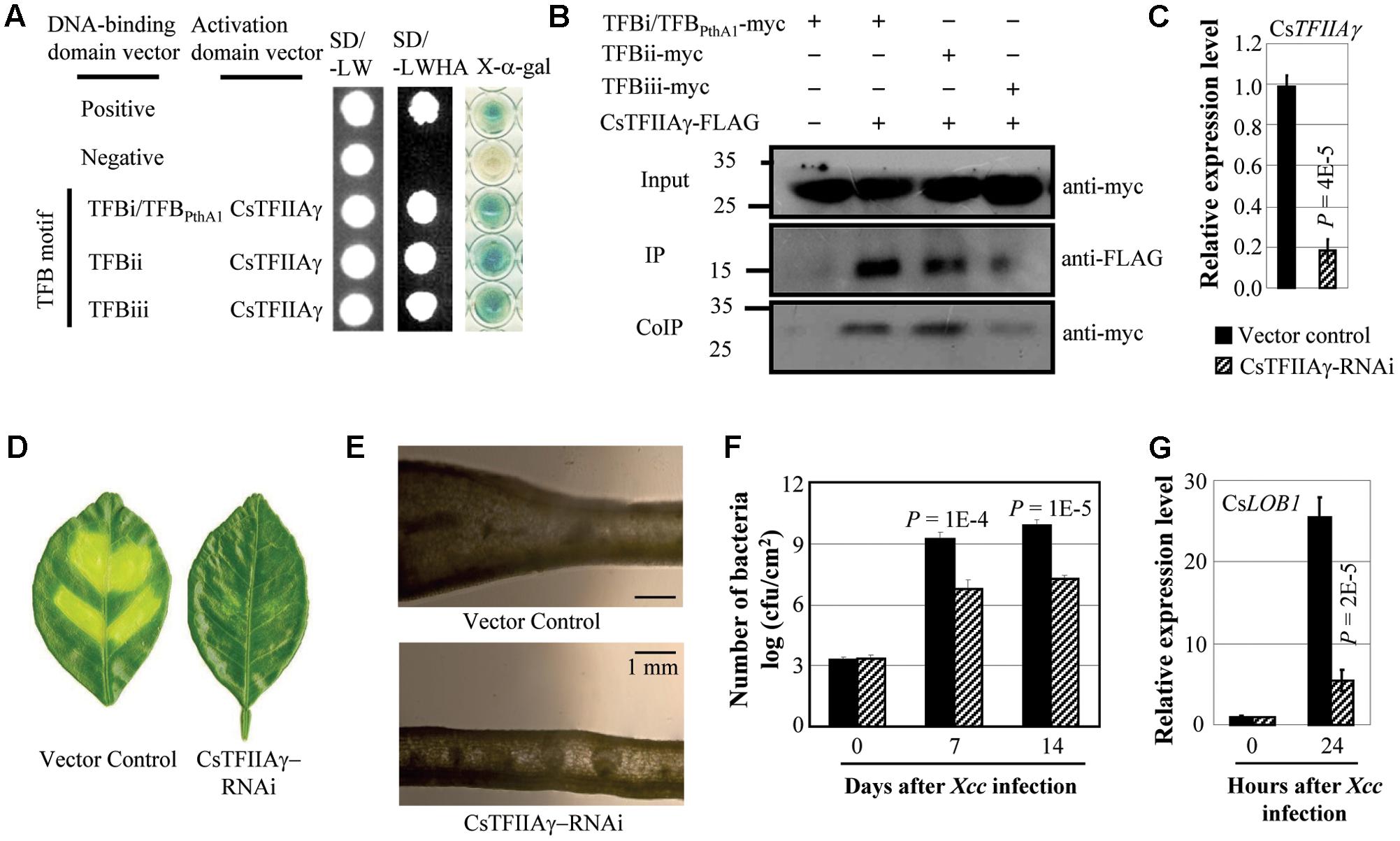
FIGURE 1. Effect of CsTFIIAγ in TALE-carrying bacterium Xcc caused infection of sweet orange. (A) The TFB motifs of all three TALEs from Xcc strain X02-007 interacted with CsTFIIAγ analyzed using yeast two-hybrid assay. Among the three TFB motifs, the sequence of TFBi is the same as the TFB motif of TALE PthA1 from sequenced Xcc strain 306. The interactions were assessed by growth of yeast cells on synthetic defined premixes (SD) medium lacking leucine (L), tryptophan (W), histidine (H), and adenine (A). (B) The myc-labeled TFBs from Xcc interacted with FLAG-labeled CsTFIIAγ in Nicotiana benthamiana leaf cells analyzed by co-immunoprecipitation (CoIP) assay. Proteins before (input) and after immunoprecipitation (IP) were detected with anti-myc and anti-FLAG antibodies. (C) Expression of CsTFIIAγ in transgenic and control plants. (D) The CsTFIIAγ-RNAi plants showed resistance to Xcc 14 days after bacteria inoculation. (E) Thin cross-section images of leaves at 14 days after inoculation with Xcc. (F) CsTFIIAγ-RNAi plants had less Xcc growth. Bar represents mean (three replicates) ± standard deviation. cfu, colony-forming unit. (G) Expression of disease susceptibility gene CsLOB1 after infection of Xcc. The corresponding P-values were determined using Student’s t-test (two tailed) comparing data from the control and suppressing plants.
Further, different citrus species, such as hongkong kumquat, grapefruit and trifoliate orange, have a TFIIAγ protein sequence identical to CsTFIIAγ in sweet orange, although the gene sequences differ for a few nucleotides (Supplementary Figure 2A). All the citrus species were similarly susceptible to Xcc (Supplementary Figure 2B). These results suggest that, like OsTFIIAγ5, CsTFIIAγ is also hijacked by TALE-carrying bacteria, with the host becoming infected via TALE-induced expression of the host S gene.
The 39th Amino Acid Residue of Plant TFIIAγs Is Essential for the Virulence of TALE-Carrying Bacteria
Rice plants carrying recessive xa5, which encodes the mutated OsTFIIAγ5V 39E, have broad-spectrum resistance to TALE-carrying bacteria and no effect on rice yield and development (Yuan et al., 2016). To analyze whether OsTFIIAγ5V 39E is the only type of mutation that confers disease resistance in rice and whether mutations similar to OsTFIIAγ5V 39E can confer disease resistance in other plant species, we first examined the sequences of OsTFIIAγ5 coding region in different Oryza species.
The Oryza genus consists of two cultivated rice species, Asian cultivated rice (O. sativa, AA genome) and African cultivated rice (O. glaberrima, AA genome), and wild (undomesticated) rice species (AA, BB, CC, BBCC, CCDD, EE, FF, GG, HHJJ, KKLL genomes) (Ge et al., 1999; Ammiraju et al., 2010). We sequenced the OsTFIIAγ5 locus in 15 different Oryza genomes, including nine accessions of AA genome (O. sativa indica, O. sativa japonica, O. rufipogon, O. nivara, O. glaberrima, O. longistaminata, O. meridionalis, O. gluaepatula, and O. barthii), one accession of BB genome (O. punctata), two accessions of CC genome (O. officinalis and O. rhizomatis), one accession of EE genome (O. australiensis), one accession of FF genome (O. brachyantha), and one accession of GG genome (O. meyeriana). Sequence alignment analysis showed that different Oryza species contained the same sequences of the OsTFIIAγ5 coding region and encoded the identical OsTFIIAγ5 protein, which is consistent with its molecular function as a basal transcription factor.
We searched the putative OsTFIIAγ5 variant from database OryzaGenome1, which contains genotype information of 463 accessions of wild rice O. rufipogon, which is evolutionally closely related to cultivated rice (Ohyanagi et al., 2016). Thirty-four single nucleotide polymorphisms (SNPs) were distributed in the OsTFIIAγ5 gene region, all within the introns and none in the exons. Thus the 463 O. rufipogon accessions encode the identical OsTFIIAγ5 protein. Moreover, we also searched the putative OsTFIIAγ5 variants from Rice SNP-Seek Database2, which contain genotype information of 3024 O. sativa varieties from the International Rice GenBank Collection Information System (IRGCIS3). There were 93 SNPs distributed in the 6260-bp region of OsTFIIAγ5 gene, and 105 rice varieties contained SNPs in the coding region causing amino acid mutation. Of these 105 rice varieties, 104 have a mutated OsTFIIAγ5 gene encoding OsTFIIAγ5V 39E, identical to the protein encoded by recessive disease resistance gene xa5. One variety has a different type of mutated OsTFIIAγ5 gene, which encodes a protein with valine changed to aspartic acid (D) at the 39th residue (OsTFIIAγ5V 39D) (Supplementary Table 3). Eighty-five of the 104 rice varieties carrying the mutated OsTFIIAγ5V 39E gene and one carrying the mutated OsTFIIAγ5V 39D gene belong to the Aus group, which is mainly from South Asia, and the other 19 carrying the mutated OsTFIIAγ5V 39E gene belong to Indica II, Indica IB, Indx and Admix groups, mainly from Southeast Asia (Huang et al., 2011). The distribution of OsTFIIAγ5V 39E resistance allele in these groups likely reflects the high disease pressure in these regions, where Xoo is a devastating disease. The results suggest that the OsTFIIAγ5 gene mutation probably appeared in cultivated rice.
Amino acid sequence alignment of TFIIAγs from different species showed that they share a high sequence similarity, with the 39th amino acid residue having three types, V, leucine (L), and E (Yuan et al., 2016). The V type occurs in plants (O. sativa, Arabidopsis thaliana, Citrus sinensis, Capsicum annuum, and Solanum lycopersicum). The L type occurs in animals (Rattus norvegicus, Homo sapiens, Danio rerio, and Drosophila melanogaster). The E type was only detected in rice OsTFIIAγ5 locus (OsTFIIAγ5V 39E), which is recessively inherited. In addition, we identified a D type in OsTFIIAγ5 locus (OsTFIIAγ5V 39D) from rice germplasm (Supplementary Table 3). To determine whether the 39th amino acid of OsTFIIAγ5 is vital for TALE-dependent induction of rice genes, we first produced four OsTFIIAγ5 derivatives at the 39th residue, substituting V with alanine (A), D, L or glutamine (Q); V, A and L are hydrophobic amino acids, and D and Q are acidic hydrophilic amino acids. The TFB motifs of TALE PthXo1 from Xoo and TALE Tal3c from Xoc interacted as strongly with mutants OsTFIIAγ5V 39A, OsTFIIAγ5V 39D, OsTFIIAγ5V 39L, and OsTFIIAγ5V 39Q as with wild-type OsTFIIAγ5 in yeast cells (Supplementary Figure 3). The PthXo1 TFB interacted weakly and Tal3c TFB did not interact with OsTFIIAγ5V 39E (E being an acidic hydrophilic amino acid) as reported previously (Yuan et al., 2016).
We then generated OsTFIIAγ5 transgenic plants with the four different amino acid substitutions (A, D, L, and Q) at the 39th residue driven by the native promoter of OsTFIIAγ5 (POsTFIIAγ5) in the xa5 (OsTFIIAγ5V 39E) resistance allele background (IRBB5). The POsTFIIAγ5:OsTFIIAγ5V 39A, POsTFIIAγ5:OsTFIIAγ5V 39D, POsTFIIAγ5:OsTFIIAγ5V 39L, and POsTFIIAγ5:OsTFIIAγ5V 39Q plants showed susceptibility to Xoo strains PXO99 and PXO341, also susceptibility to Xoc strain RH3 compared to the wild-type plant (Figure 2A). The susceptibility of these transgenic plants to Xoo was associated with more efficiently induced expression of known rice S genes (Xa13/Os8N3/SWEET11, OsTFIIAγ1, and OsTFX1) to Xoo (Figure 2B; Chu et al., 2006; Sugio et al., 2007). The susceptibility of these transgenic plants to Xoc was associated with more efficiently induced expression of the known rice S gene (OsSULTR3;6) to Xoc (Figure 2B; Cernadas et al., 2014). We also generated OsTFIIAγ5 overexpressing plants with the same mutations listed above driven by maize ubiquitin promoter (PUbi). These transgenic plants also showed increased susceptibility to Xoo and Xoc, which was associated with increased induced expression of rice susceptibility genes, compared to wild-type plants (Supplementary Figures 4, 5). These results suggest that the 39th amino acid residue of OsTFIIAγ5 is essential for TALE-dependent induction of rice S genes, and it seems that only the V to E mutation at this residue (OsTFIIAγ5V 39E) affects the interaction between OsTFIIAγ5 and TFB of TALEs.
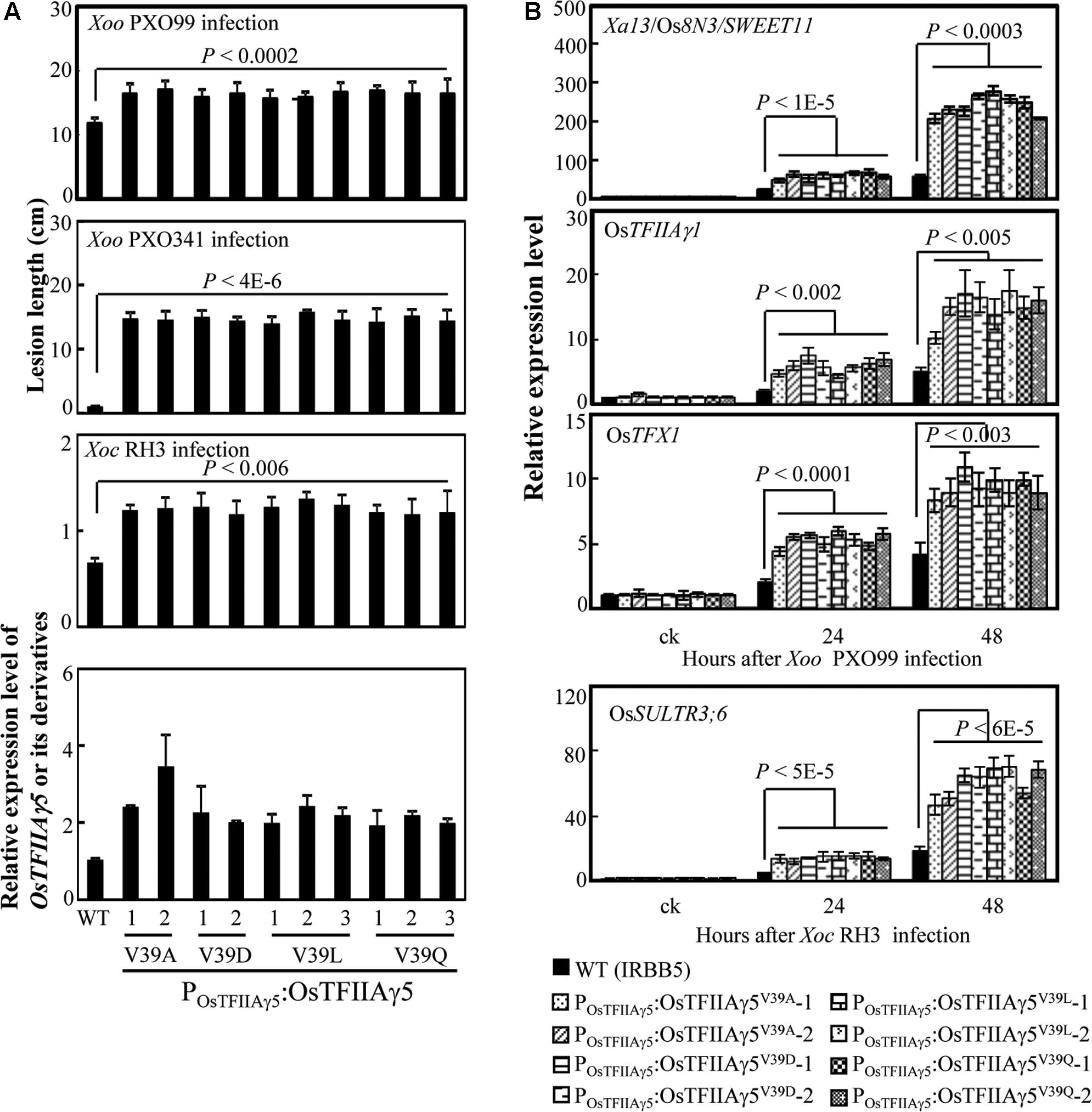
FIGURE 2. Effects of OsTFIIAγ5 mutants in rice responses to Xoo and Xoc. (A) Transgenic rice plants carrying OsTFIIAγ5 mutant were susceptible to Xoo stains PXO99 and PXO341 and Xoc strain RH3 compared to wild-type (WT) IRBB5, which carries the mutated OsTFIIAγ5V 39E. T1 plants were inoculated with Xoo at the booting stage and Xoc at the tillering stage. Data represent mean (total 20 to 25 leaves from four plants) ± standard deviation. (B) Expression of disease susceptibility genes Xa13/Os8N3/SWEET11, OsTFIIAγ1, and OsTFX1 in rice after infection of PXO99 and OsSULTR3;6 after infection of RH3. Bar represents mean (three replicates) ± standard deviation. The corresponding P-values were determined using Student’s t-test (two tailed) comparing data from the WT and transgenic plants.
The 39th amino acid residue of citrus CsTFIIAγ is also V, and CsTFIIAγ and OsTFIIAγ5 share 93/89% sequence similarity/identity (Supplementary Figure 6; Yuan et al., 2016). Furthermore, the TFB motifs of Xcc TALEs have high sequence homology with TFBs of Xoo and Xoc TALEs (Supplementary Figure 1). To investigate whether the OsTFIIAγ5V 39E type mutation in CsTFIIAγ (CsTFIIAγV 39E) could prevent TALE-carrying bacteria from infecting plants, we first examined the interactions between the TALE TFB motifs of different Xanthomonas bacteria and CsTFIIAγV 39E. The TFB motifs of Xoo, Xoc, and Xcc TALEs could not interact with CsTFIIAγV 39E (Supplementary Figures 7A, 8A), although these TFBs interacted strongly with both OsTFIIAγ5 and CsTFIIAγ in yeast cells and in planta (Figures 1, 3 and Supplementary Figure 7B). The only exception was the TFB motif of Xoo Tal7a. It interacted with CsTFIIAγV 39E (Supplementary Figure 7A), which is consistent with this TFB also interacting with OsTFIIAγ5V 39E (Yuan et al., 2016).
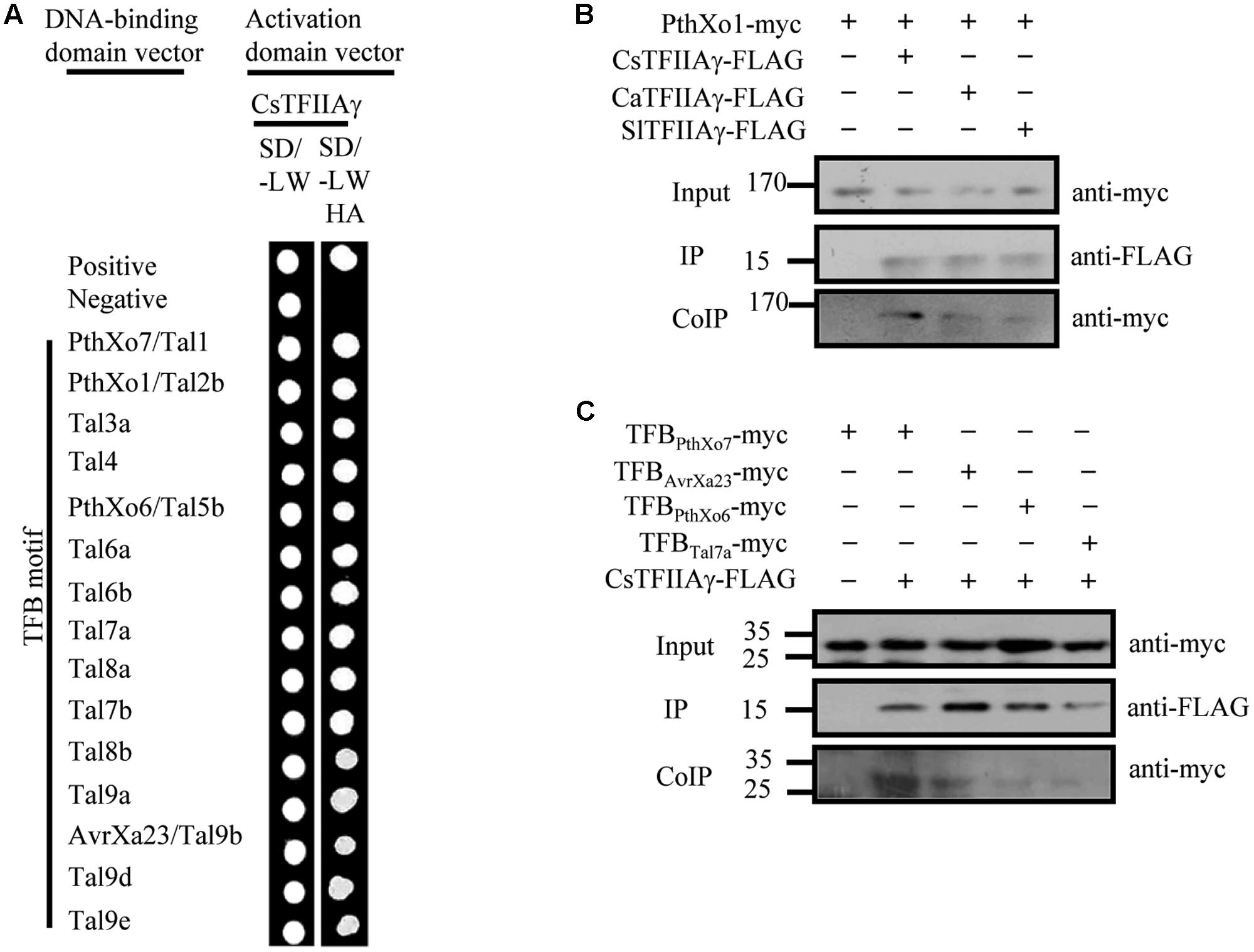
FIGURE 3. Analysis of the interactions between Xoo TALEs and plant TFIIAγs (citrus CsTFIIAγ, pepper CaTFIIAγ, and tomato SlTFIIAγ). (A) Interactions between TFB motif of Xoo TALEs and CsTFIIAγ as determined by yeast two-hybrid. (B) Interactions of the myc-labeled full-length TALE PthXo1 with FLAG-labeled plant TFIIAγs in N. benthamiana leaf cells by CoIP. (C) Interaction of the myc-labeled TFB motif of four TALEs with FLAG-labeled plant TFIIAγs in N. benthamiana leaf cells by CoIP.
Furthermore, we overexpressed CsTFIIAγ in rice IRBB5 carrying the recessive xa5 gene. The transgenic plants displayed susceptibility to Xoo strains PXO99 and PXO341 (Figure 4A). The susceptibility was associated with overexpression of CsTFIIAγ, which was further confirmed in two independent T1 families (Supplementary Figure 9). The induced expression of known S genes Xa13/Os8N3/SWEET11, OsTFIIAγ1, and OsTFX1, each of which is targeted by a different TALE, was significantly higher (P < 0.01) in transgenic plants than in wild-type plants (Figure 4B). Similarly, overexpressing CsTFIIAγ in rice increased susceptibility to Xoc, which was associated with significantly promoted Xoc-induced expression of rice S gene OsSULTR3;6 compared to wild-type plants (Supplementary Figure 10). However, the transgenic plants with overexpression of CsTFIIAγV 39E showed similar lesion length compared to wild-type plants upon inoculation with Xoo strain PXO99 (Supplementary Figure 11). These results suggest that CsTFIIAγ can also facilitate the infection of Xoo and Xoc in rice by helping TALE-induced activation of rice S genes, and the 39th amino acid residue of CsTFIIAγ is also essential for TALE-dependent induction of host S genes.
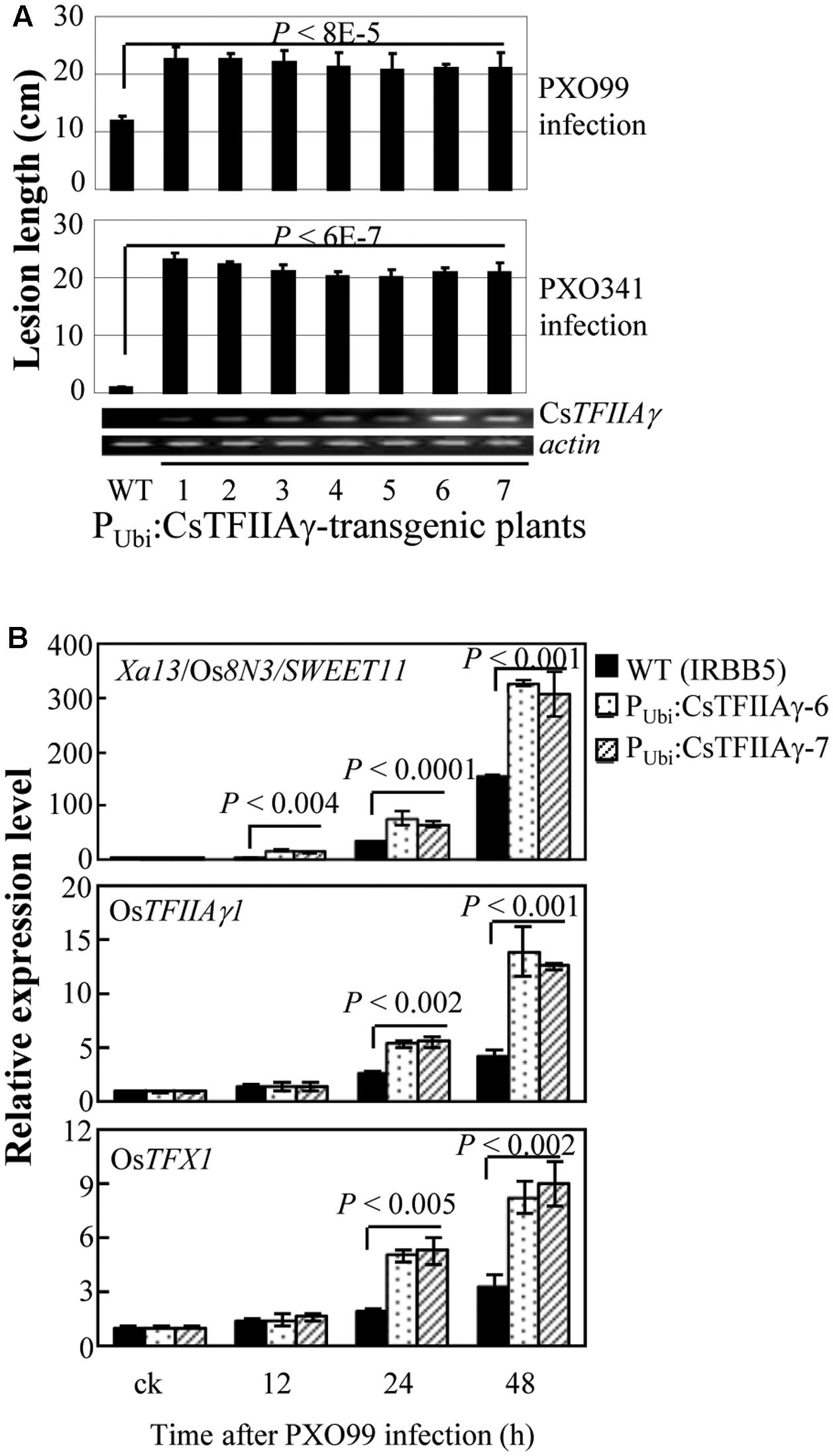
FIGURE 4. Effect of overexpressing CsTFIIAγ (PUbi:CsTFIIAγ) in rice response to Xoo. Plants were inoculated with Xoo strains PXO99 and PXO341 at the booting (panicle development) stage. (A) Transgenic rice plants (T1 generation) were susceptible to Xoo compared to WT. Data represent mean (5 to 10 replicates from one plant) ± standard deviation. RT-PCR analysis of CsTFIIAγ transcripts in transgenic plants. Actin transcripts were detected as control. (B) Expression of disease susceptibility genes Xa13/Os8N3/SWEET11, OsTFIIAγ1, and OsTFX1 in rice after infection of Xoo strain PXO99. Bar represents mean (three replicates) ± standard deviation. The corresponding P-values were determined using Student’s t-test (two tailed) comparing data from the WT and transgenic plants.
Xcv Also Uses Host TFIIAγ to Cause Disease in Pepper and Tomato
We also examined the relationship between pepper (Capsicum annuum L. cv. Hua 50) CaTFIIAγ or tomato (Solanum lycopersicum L. cv. Ailsa Craig) SlTFIIAγ and Xcv, which causes bacterial spot disease in the two crops. CaTFIIAγ and SlTFIIAγ have identical sequences, and Xcv strain 23-1 carries TALE genes (Yuan et al., 2016). The TFB motifs of Xcv 23-1 TALEs, which featured high sequence similarity with the TFB motifs of Xoo, Xoc, and Xcc TALEs (Supplementary Figure 1), interacted with CaTFIIAγ/SlTFIIAγ both in yeast and in planta (Figures 5A,B), but they did not interact with Ca/SlTFIIAγV 39E (Supplementary Figures 8B, 12). Transiently suppressing CaTFIIAγ or SlTFIIAγ by VIGS enhanced the resistance of pepper and tomato to Xcv. The pepper plants with suppressed expression of CaTFIIAγ (TRV2:CaTFIIAγ) and tomato plants with suppressed expression of SlTFIIAγ (TRV2:SlTFIIAγ) showed markedly reduced disease symptoms and significantly lower (P < 0.01) Xcv growth rates compared to corresponding control plants expressing TRV2:GFP (Figures 5C,D). These results suggest that TALE-containing Xcv causes diseases in pepper and tomato and may also need the help of host TFIIAγs.
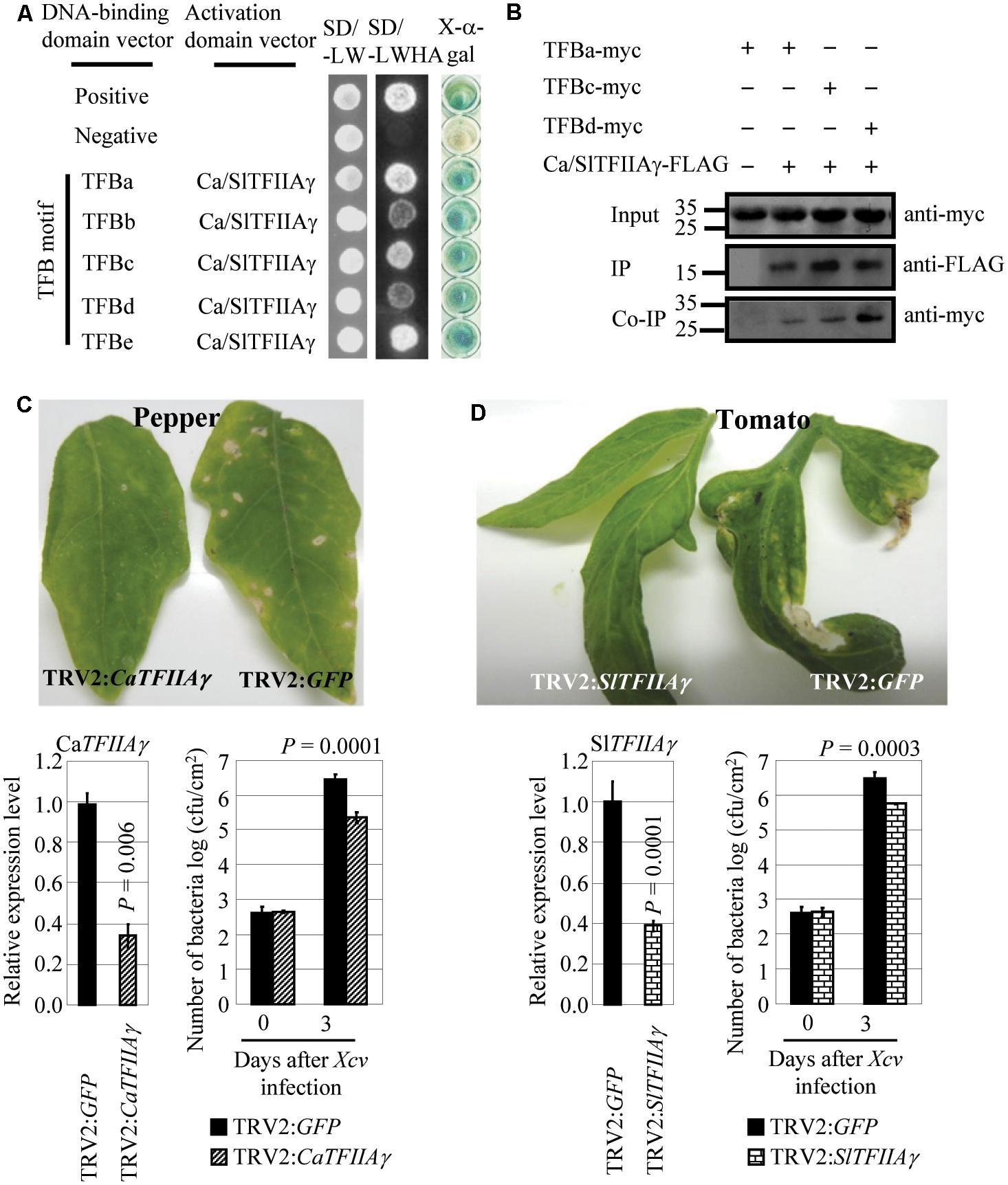
FIGURE 5. Effects of CaTFIIAγ and SlTFIIAγ in TALE-carrying bacterium Xcv-caused infection of pepper and tomato. (A) The TFB motifs of all the five TALEs from Xcv strain 23-1 interacted with CaTFIIAγ and SlTFIIAγ analyzed using yeast two-hybrid assay. (B) The myc-labeled TFB motifs of Xcv TALEs interacted with FLAG-labeled CaTFIIAγ and SlTFIIAγ in N. benthamiana leaf cells analyzed by CoIP. Proteins before (input) and after immunoprecipitation (IP) were detected with anti-myc and anti-FLAG antibodies. (C) Suppressing CaTFIIAγ enhanced pepper resistance to Xcv. Bar represents mean (three replicates) ± standard deviation. cfu, colony-forming unit. (D) Suppressing SlTFIIAγ enhanced tomato resistance to Xcv. Bar represents mean (three replicates) ± standard deviation. The corresponding P-values were determined using Student’s t-test (two tailed) comparing data from the control and suppressing plants.
Discussion
In addition to TALE-carrying Xoo and Xoc needing the help of host TFIIAγ (OsTFIIAγ5) to cause disease in rice (Yuan et al., 2016), the present results show that TALE-carrying Xcc also requires host TFIIAγ (CsTFIIAγ) for successful colonization in sweet orange. Transcriptional suppression of CsTFIIAγ can reduce disease symptoms. Xoo and Xoc employ OsTFIIAγ5 to facilitate TALE-induced expression of host S genes for infection (Yuan et al., 2016). Efficient binding of the TFB motifs of Xoo and Xoc TALEs to OsTFIIAγ5 is important for the induction of S genes. The present results suggest that Xcc likely uses the same mechanism as Xoo and Xoc to infect its host. This inference is supported by the following evidence. First, the TFBs of Xcc TALEs also interacted with CsTFIIAγ. Second, transcriptional suppression of CsTFIIAγ resulted in reduced disease of sweet orange and was associated with compromised induction of S gene CsLOB1. Last, because of the high amino acid homology among TALE TFBs of different Xanthomonas species, such as Xoo, Xoc, Xcc, and Xcv (Supplementary Figure 1), and among different TFIIAγs of different eukaryote species, such as OsTFIIAγ5, CsTFIIAγ, CaTFIIAγ, and SlTFIIAγ (Yuan et al., 2016), CsTFIIAγ could be used by Xoo and Xoc to cause disease in rice, which was associated with induced expression of rice S genes. Although high amino acid sequence similarity was present among TALE TFBs of different Xanthomonas species, there was still some dissimilarity in the motif (Supplementary Figure 1; Yuan et al., 2016). Protein residues diversity may influence the interaction strength with plant TFIIAγ, which might be supported by the relatively weak band in some of the CoIP assays.
The present results also show that Xcv TALEs can bind to pepper CaTFIIAγ and tomato SlTFIIAγ through their TFBs. Suppression of CaTFIIAγ and SlTFIIAγ reduces Xcv-caused disease symptoms. Although the present study does not identify which host gene is targeted by the TALEs of Xcv strain 23-1, the results suggest that CaTFIIAγ and SlTFIIAγ may also be hijacked by Xcv for infection. This inference is also supported by the localization characteristic of the TALE binding sites (EBEs) of host gene promoters in different plants. We analyzed 45 EBEs of known S genes and executor resistance genes that can be directly targeted by Xoo, Xoc, Xcv, Xcc, Xam, or Xg TALEs in rice, pepper, citrus, cassava, or tomato (Supplementary Table 4). These EBEs either overlapped their corresponding TATA-box or were located close to it, and some TATA-boxes are the core part of EBEs, which is consistent with computational model-based predictions that target sites of TALEs are preferentially located at most 300 bp upstream and 200 bp downstream of the transcription start site of target genes (Grau et al., 2013). TFIIAγ is a component of a transcription pre-initiation complex that binds to the TATA-box of promoters to initiate gene expression in eukaryotes (Høiby et al., 2007). The overlap or adjacent physical position of TALE-binding sites in relation to the positions of TATA-box supports the possibility that Xcv TALEs also use CaTFIIAγ and SlTFIIAγ for infection in pepper and tomato. Furthermore, the localization relationship between EBE and TATA-box also suggests that other TALE-carrying bacteria, in addition to Xoo, Xoc, Xcc, and Xcv, may also require the help of host TFIIAγ for infection.
Our previous and present results indicate that the difference in interaction strength between different TFB motifs from diverse Xanthomonas TALEs and corresponding host plant TFIIAγ with V39E mutation could be correlated with specific differences in the amino acids of the TFB motifs (Supplementary Figures 1, 3, 7, 12). The present results undoubtedly suggest that the 39th amino acid residue of OsTFIIAγ5 is important for TALE-dependent induction of host genes. The OsTFIIAγ5V 39E encoded by recessive disease resistance gene xa5 is the only type of mutation identified so far that can prevent or attenuate the infection of Xoo and Xoc in rice (Supplementary Table 3; Yuan et al., 2016). The V39E substitution of CsTFIIAγ (CsTFIIAγV 39E) also prevented its interaction with TFBs of Xcc TALEs and abolished or compromised CsTFIIAγ-facilitated infection of Xoo and Xoc in rice, suggesting that the CsTFIIAγV 39E mutation may make citrus resistance to Xcc. In addition, the TFBs of Xcv TALEs cannot interact with CaTFIIAγV 39E and SlTFIIAγV 39E, suggesting that the TFIIAγV 39E-type mutation may also be used to prevent Xcv-caused diseases in pepper and tomato. Moreover, TFBs with mutation in the 134 amino acids motif might be found with more Xcc or Xcv strains been sequenced, which can break CsTFIIAγV 39E, CaTFIIAγV 39E, and SlTFIIAγV 39E mediated resistance to Xcc or Xcv. An example is that the TFBs of pthXo1, tal7a, and tal8a of Xoo pv. PXO99 differed by 1 to 20 residues from the other 12 TFBs could interact with OsTFIIAγV 39E, and the rice variety IRBB5 carrying OsTFIIAγV 39E is susceptible to PXO99 (Yuan et al., 2016).
In summary, the previous report (Yuan et al., 2016) and the present results suggest that TALE-carrying bacteria probably use a common mechanism that is employing host TFIIAγs to cause disease in rice, citrus, pepper, and tomato. The genus Xanthomonas infects a wide variety of hosts. Because of the high amino acid sequence homology among TFIIAγs from different plant species and among the TFB motifs of TALEs from different Xanthomonas species, other TALE-carrying Xanthomonas species, which have not been examined, likely also hijack host TFIIAγs for successful infection. Since rice plant OsTFIIAγ5V 39E confers broad-spectrum resistance to Xoo and Xoc without affecting rice development and yield (Yuan et al., 2016) and the present results suggest that CsTFIIAγV 39E can also compromise bacterial infection, use of TFIIAγV 39E-type of mutation—either by exploring germplasm for natural mutants or using gene-editing technologies—may provide a general strategy for improving resistance to Xcv and other TALE-carrying pathogens in crops in addition to rice and citrus.
Additional Information
Gene sequences of CsTFIIAγ from different Citrus species have been deposited in GenBank with the following accession codes KU377725 (in Citrus sinensis Hongkong Kumquat), KU377726 (in Citrus sinensis Sweet orange), KU377727 (in Citrus sinensis Grapefruit), and KU377728 (in Citrus sinensis Trifoliate orange).
Author Contributions
MY, RH, and SH performed most of the experiments and analyzed the data; MZ and PL helped to generate transgenic rice plants, analyze protein–protein interactions, and amplify TALE; JX and XL provided biochemical and molecular analysis support and management; MY and SW designed the research, supervised the project, interpreted data, and drafted and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100600 and 2016YFD0100903), the National Natural Science Foundation of China (31371926), and the Fundamental Research Funds for the Central Universities (2662017PY014). We thank professors Gongyou Chen of Shanghai Jiao Tong University and Jihong Liu of Huazhong Agricultural University for kindly providing Xanthomonas bacteria and professors Zhibiao Ye and Hanxia Li for kindly providing pepper and tomato seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01919/full#supplementary-material
Footnotes
- ^ http://viewer.shigen.info/oryzagenome/mapview/Top.do
- ^ http://snp-seek.irri.org/
- ^ http://irgcis.irri.org:81/grc/irgcishome.html
References
Ammiraju, J. S., Fan, C., Yu, Y., Song, X., Cranston, K. A., Pontaroli, A. C., et al. (2010). Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. Plant J. 63, 430–442. doi: 10.1111/j.1365-313X.2010.04251.x
Boch, J., Bonas, U., and Lahaye, T. (2014). TAL effectors-pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015
Bonas, U., Schulte, R., Fenselau, S., Minsavage, G., Staskawicz, B., and Stall, R. (1991). Isolation of a gene-cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant Microbe Interact. 4, 81–88. doi: 10.1094/MPMI-4-081
Cernadas, R. A., Doyle, E. L., Nino-Liu, D. O., Wilkins, K. E., Bancroft, T., Wang, L., et al. (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLOS Pathog. 10:e1003972. doi: 10.1371/journal.ppat.1003972
Chen, H., Wang, S., and Zhang, Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology 92, 750–754. doi: 10.1094/PHYTO.2002.92.7.750
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Chung, E., Seong, E., Kim, Y. C., Chung, E. J., Oh, S. K., Lee, S., et al. (2004). A method of high frequency virus-induced gene silencing in chili pepper (Capsicum annuum L. Bukang). Mol. Cells 17, 377–380.
de Lange, O., Schreiber, T., Schandry, N., Radeck, J., Braun, K. H., Koszinowski, J., et al. (2013). Breaking the DNA-binding codes of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 199, 773–786. doi: 10.1111/nph.12324
de Lange, O., Wolf, C., Thiel, P., Krüger, J., Kleusch, C., Kohlbacher, O., et al. (2015). DNA-binding proteins from marine bacteria expand the known sequence diversity of TALE-like repeats. Nucleic Acids Res. 43, 10065–10080. doi: 10.1093/nar.gkv1053
Doyle, E. L., Stoddard, B. L., Voytas, D. F., and Bogdanove, A. J. (2013). TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 23, 390–398. doi: 10.1016/j.tcb.2013.04.003
Fu, X., Gong, X., Zhang, Y., Wang, Y., and Liu, J. (2012). Different transcriptional response to Xanthomonas citri subsp. citri between kumquat and sweet orange with contrasting canker tolerance. PLOS ONE 7:e41790. doi: 10.1371/journal.pone.0041790
Ge, S., Sang, T., Lu, B. R., and Hong, D. Y. (1999). Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc. Natl. Acad. Sci. U.S.A. 96, 14400–14405. doi: 10.1073/pnas.96.25.14400
Ge, X., Chu, Z., Lin, Y., and Wang, S. (2006). A tissue culture system for different germplasms of indica rice. Plant Cell Rep. 25, 392–402. doi: 10.1007/s00299-005-0100-7
Grau, J., Wolf, A., Reschke, M., Bonas, U., Posch, S., and Boch, J. (2013). Computational predictions provide insights into the biology of TAL effector target sites. PLOS Comput. Biol. 9:e1002962. doi: 10.1371/journal.pcbi.1002962
Høiby, T., Zhou, H., Mitsiou, D. J., and Stunnenberg, H. G. (2007). A facelift for the general transcription factor TFIIA. Biochim. Biophys. Acta 1769, 429–436. doi: 10.1016/j.bbaexp.2007.04.008
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Huang, X., Zhao, Y., Wei, X., Li, C., Wang, A., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Jia, H., and Wang, N. (2014). Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 33, 1993–2001. doi: 10.1007/s00299-014-1673-9
Juillerat, A., Bertonati, C., Dubois, G., Guyot, V., Thomas, S., Valton, J., et al. (2014). BurrH: a new modular DNA binding protein for genome engineering. Sci. Rep. 4:3831. doi: 10.1038/srep03831
Li, H., Li, X., Xiao, J., Wing, R. A., and Wang, S. (2012). Ortholog alleles at Xa3/Xa26 locus confer conserved race-specific resistance against Xanthomonas oryzae in rice. Mol. Plant 5, 281–290. doi: 10.1093/mp/ssr079
Li, Z., Zou, L., Ye, G., Xiong, L., Ji, Z., Zakria, M., et al. (2014). A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Mol. Plant 7, 912–915. doi: 10.1093/mp/sst176
Liu, Y., Schiff, M., and Dinesh-Kumar, S. (2002a). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. (2002b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Ohyanagi, H., Ebata, T., Huang, X., Gong, H., Fujita, M., Mochizuki, T., et al. (2016). OryzaGenome: genome diversity database of wild Oryza species. Plant Cell Physiol. 57, e1. doi: 10.1093/pcp/pcv171
Orphanides, G., Largrange, T., and Reinberg, D. (1996). The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683. doi: 10.1101/gad.10.21.2657
Ryan, R. P., Vorhölter, F. J., Potnis, N., Jones, J. B., Van Sluys, M. A., Bogdanove, A. J., et al. (2011). Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat. Rev. Microbiol. 9, 344–355. doi: 10.1038/nrmicro2558
Schornack, S., Moscou, M. J., Ward, E. R., and Horvath, D. M. (2013). Engineering plant disease resistance based on TAL effectors. Annu. Rev. Phytopathol. 51, 383–406. doi: 10.1146/annurev-phyto-082712-102255
Sugio, A., Yang, B., Zhu, T., and White, F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAγ1 and OsTFX1 during bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 104, 10720–10725. doi: 10.1073/pnas.0701742104
Sun, F., Du, Z., Jiao, Z., Zhao, T., and Cheng, B. (1999). Pathogen and race identification of bacterial spot of pepper and tomato. Acta Phytopathol. Sin. 29, 265–269. doi: 10.13926/j.cnki.apps.1999.03.014
Yuan, M., Chu, Z., Li, X., Xu, C., and Wang, S. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell 22, 3164–3176. doi: 10.1105/tpc.110.078022
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., et al. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5:e19605. doi: 10.7554/eLife.19605
Yuan, M., Zhao, J., Huang, R., Li, X., Xiao, J., and Wang, S. (2014). Rice MtN3/saliva/SWEET gene family: evolution, expression profiling, and sugar transport. J. Integr. Plant Biol. 56, 559–570. doi: 10.1111/jipb.12173
Yuan, T., Li, X., Xiao, J., and Wang, S. (2011). Characterization of Xanthomonas oryzae-responsive cis-acting element in the promoter of rice race-specific susceptibility gene Xa13. Mol. Plant 4, 300–309. doi: 10.1093/mp/ssq076
Keywords: bacterial disease, basal transcription factor, transcription activator-like effector, crop, Xanthomonas
Citation: Huang R, Hui S, Zhang M, Li P, Xiao J, Li X, Yuan M and Wang S (2017) A Conserved Basal Transcription Factor Is Required for the Function of Diverse TAL Effectors in Multiple Plant Hosts. Front. Plant Sci. 8:1919. doi: 10.3389/fpls.2017.01919
Received: 25 July 2017; Accepted: 23 October 2017;
Published: 07 November 2017.
Edited by:
Essaid Ait Barka, University of Reims Champagne-Ardenne, FranceReviewed by:
Brian H. Kvitko, University of Georgia, United StatesJan Grau, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2017 Huang, Hui, Zhang, Li, Xiao, Li, Yuan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Yuan, myuan@mail.hzau.edu.cn Shiping Wang, swang@mail.hzau.edu.cn
 Renyan Huang
Renyan Huang Shugang Hui
Shugang Hui Meng Zhang
Meng Zhang Xianghua Li
Xianghua Li Meng Yuan
Meng Yuan Shiping Wang
Shiping Wang