- Ministry of Agriculture Key Laboratory of Biology and Genetic Resources of Rubber Tree and State Key Laboratory Breeding Base of Cultivation and Physiology for Tropical Crops, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China
Latex exploitation enhances latex regeneration in rubber trees. The latex exploitation-caused latex flow lasts from 10 min to a few hours, which is convenient for exploring the transcript profiling of latex metabolism-related genes at the different stages of latex flow. In the present study, the expression pattern of 62 latex metabolism-related genes involved in water transportation, carbohydrate metabolism, natural rubber biosynthesis, hormone signaling, ROS generation and scavenging, and latex coagulum across three stages of latex flow between rubber tree clones CATAS7-33-97 and CATAS8-79 were comparatively analyzed by quantitative real-time PCR. The two clones show differences in latex regeneration and have a different duration of latex flow. The results showed that the expression levels of 38 genes were significantly higher in CATAS8-79 latex than in CATAS7-33-97 during latex regeneration, while 45 genes had a notably higher expression level in CATAS8-79 latex during latex flow. Together with the activation of the MEP pathway and jasmonate pathway in CATAS8-79 latex, HbPIP1;3, HbPIP1;4, HbSUT3, HbSus3, HbHMGS1-2, HbMK should contribute to the high latex regeneration ability. The up-regulation of ethylene signaling and Hb44KD and the down-regulation of latex coagulation-related genes in CATAS8-79 latex might contribute to its longer latex flow duration. This study provides some cues for revealing the regulation of latex metabolism in rubber trees.
Introduction
Natural rubber (cis-1,4-polyisoprene) is an essential industrial substance around the world. Due to its high yield and excellent physical properties, the para rubber tree (Hevea brasiliensis) is the main source of natural rubber (Eschbach and Lacrotte, 1989). Laticifers located at the inner bark of rubber tree serve as the location of natural rubber biosynthesis. Laticifer cells have a specialized cytoplasm containing 30–50% rubber for natural rubber refinement (Chrestin et al., 1997). In natural rubber production, latex is collected by severing the laticifer rings every 2–3 days. This process is termed tapping (d’ Auzac, 1989). After tapping, 10 to a few 100 ml of latex are expelled from the severed laticifers. The latex flow is terminated by plug formation at the end of the laticifer’s wounded site after several minutes to a few hours after tapping. The duration of latex flow after tapping is one of the crucial factors that determines the rubber yield of the rubber tree. It is influenced by multiple factors such as ethylene application, temperature, latex redox homeostasis, etc. (Zhu and Zhang, 2009; Chao et al., 2016). Ethylene is a phytohormone that regulates numerous developmental as well as physiological processes in plants (Van de Poel et al., 2015). Ethrel, an ethylene releaser, is widely used to increase the latex production per tapping since it can significantly prolong the duration of latex flow (Zhu and Zhang, 2009).
It is well known that latex exploitation enhances latex regeneration in the laticifer cells within the drainage areas. Latex regeneration is a complex molecular reconstruction process, which is not only involved in de novo protein synthesis, but also in the rebuilding of the lost organelles, such as rubber particles, lutoids and ribosomes, etc. The duration of the latex flow after tapping usually lasts several minutes to a few hours. Although latex regeneration occurs at two tapping intervals, it should be initiated by the latex flow. Available data show that several rubber biosynthesis-related genes and homologues of the transcriptional complex genes significantly fluctuated during latex flow (Chao et al., 2016). Exploring the transcript profiling of latex metabolism-related genes at different stages of latex flow will provide some new cues about the regulation of latex regeneration (the early stage of latex flow) and latex flow (the late stage of latex flow).
Both the latex production and the duration of latex flow are much higher in rubber tree clone CATAS8-79 than in rubber tree clone CATAS7-33-97. CATAS8-79 originated from the offspring of a CATAS88-13 and CATAS217 cross, while rubber tree clone CATAS7-33-97 arose from the offspring of a RRIM600 and PR107 cross. The changes in the overall latex production between the two clones may be associated with the difference in latex regeneration and the duration of latex flow after tapping. In the present study, 62 genes involved in water transport regulation, carbohydrate metabolism, rubber biosynthesis, jasmonate and ethylene signaling, ROS generation and scavenging and latex coagulation were analyzed by qRT-PCR between CATAS8-79 and CATAS7-33-97 during latex flow following tapping. The results provide an initial transcript profiling of latex metabolism, which is beneficial to understand the mechanism for latex regeneration and latex flow in rubber trees.
Materials and Methods
Plant Materials
Eleven-year-old rubber tree clones, CATAS7-33-97 and CATAS8-79, with the same circumference were used in the present study. The trees were grown at the Experimental Station of the Rubber Research Institute at the Chinese Academy of Tropical Agricultural Sciences in Danzhou city, Hainan province. These trees were regularly tapped for latex collection using a half spiral pattern, every 3 days, without Ethrel stimulation (S/2, d/3). Ten trees of each clone were selected and tapped in an S/2 d/3 system. After tapping, the latex samples were collected at 1, 30, and 60 min for CATAS7-33-97, and at 1, 80, and 150 min for CATAS8-79, which respectively represented the early, middle, and late stage of the latex flow. Each of the three batches of latex samples were individually collected from ten trees of each clone, and placed on ice for determination of the rubber content in the latex or stored at -80°C for total RNA extraction (Chao et al., 2015a).
RNA Isolation and cDNA Synthesis
Total RNA was extracted using the protocol of RNAprep pure Plant Kit protocol (Tiangen, China). The concentration and quality of RNA were examined by NanoDrop 2000 (Thermo Scientific Inc., United States), and the integrity of the RNA samples was checked by 1.5% agarose gel electrophoresis. Synthesis of cDNA was performed using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Canada) following the manufacturer’s protocol.
qRT-PCR Analysis
The expression pattern of 62 latex metabolism-related genes (Hb44KD, HbAACT1-3, HbACO1-2, HbAPX1, HbCAT, HbChit, HbCMK, HbCOI1, HbCuZnSOD, HbDXR, HbDXS1-2, HbEIN2-3, HbETR1-2, HbFDPS, HbGluc, HbHDR, HbHDS, HbHevein, HbHMGR1, HbHMGS1-2, HbHRT1-2, HbIPPI1, HbJAZ2-3, HbLOX, HbMCT1-2, HbMDC1-2, HbMDS1-2, HbMnSOD, HbMK, HbMYC1, HbMYC3, HbNIN1-3, HbPDC4, HbPIP1;3-4, HbPIP2;1,3,5,7, HbPK, HbPMK, HbREF, HbRBOHA-B, HbSAMS, HbSRPP, HbSus3 and HbSUT3) reported in the previous study were used here (Tang et al., 2010; He, 2013; Xiao et al., 2014; An et al., 2015; Chao et al., 2015b; Liu et al., 2015; Long et al., 2015; Putranto et al., 2015; Deng et al., 2016; Shi et al., 2016; Makita et al., 2017). Reactions were carried out in 384-well plates as follows: 95°C for 3 min followed by 45 cycles of 95°C for 15 s, 60°C for 60 s and 72°C for 30 s, and a melting curve from 55 to 95°C, which increased by 0.5°C every 30 s. Each real-time PCR reaction was performed in triplicate. The Bio-Rad CFX384 Manager 3.0 software was used for visualizing and analyzing the data, including the quantification cycle values and the efficiency of PCR reactions. The relative expression levels of target genes were normalized with HbUBC2b (Chao et al., 2016), and displayed using a heat map. All primer pairs used in this article were list in Supplementary Table S1.
Rubber Content Determination
For rubber content determination, 100 μl of acetic acid were dropped into 1 g of fresh latex to obtain the rubber coagula. The rubber coagula were washed in water for 2 h, then dried overnight at 55°C and weighed. The experiments were repeated three times (Chao et al., 2015a).
Statistical Analysis
For multiple group comparisons, statistical analysis was performed with SPSS Statistics 17.05 using the analysis of variance (ANOVA) based on Duncan’s test. The capital letter represents P < 0.01, while the lower case letter represents P < 0.05. The same letter indicates no significant difference among groups. For two group comparisons, statistical analysis was performed with GraphPad Prism 5 based on T-test.
Results
Determination of Rubber Content in Latex during Latex Flow
The duration of latex flow in rubber tree clone CATAS7-33-97 was approximately 70 min, which was much shorter than that in rubber tree clone CATAS8-79 (more than 160 min) (Chao et al., 2016). Here, the rubber content in the latex during latex flow was determined. Changes in the rubber content of both clones occurred during the latex flow (Figure 1). A differential fluctuation pattern of the rubber content was apparent between the two clones (Figure 1). The decrease in the rubber content in the latex of CATAS7-33-97 was significant at the middle stage (30 min) (P < 0.05) and highly significant at the late stage (60 min) (P < 0.01) of latex flow. In contrast, there was no significant difference in the rubber content of the latex of CATAS8-79 between at the early stage (1 min) and the middle stage (80 min), but there was at the late stage (150 min) when the rubber content showed a highly significantly decreased (P < 0.01) and was significantly lower (P < 0.05) than the rubber content in the latex of CATAS7-33-97 at the late stage of latex flow (Figure 1).
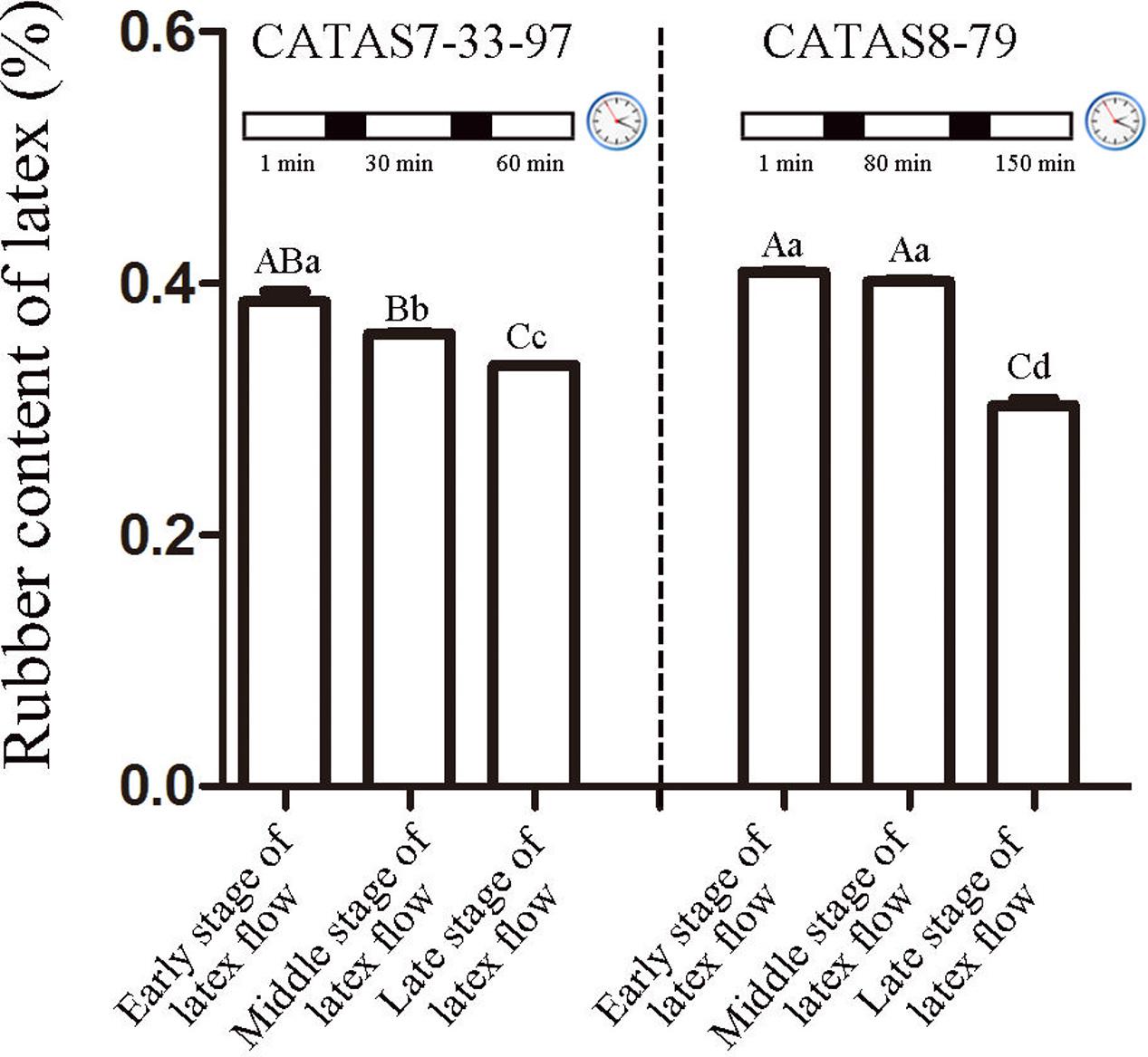
FIGURE 1. Rubber content of latex across three stages of latex flow between CATAS7-33-97 and CATAS8-79. The capital letter represents p < 0.01 while lower case represents p < 0.05. The same letter indicated no significant difference among groups.
Expression Pattern of Water Transport-Related Genes during Latex Flow
Upon tapping, aquaporin controls the water entering the laticifer cells and plays a crucial role in both latex flow and latex regeneration (Zou et al., 2015). The expression of six water transport-related genes (HbPIP1;3, HbPIP1;4, HbPIP2;1, HbPIP2;3, HbPIP2;5 and HbPIP2;7) was analyzed during the latex flow (Figure 2 and Supplementary Figure S1). Among the tested six genes, the expression of most genes had no obvious difference in the latex of CATAS7-33-97 except for HbPIP2;5 and HbPIP2;7 which were significantly up-regulated at the late stage of the latex flow. The differentially activated expression of most genes was present in the latex of CATAS8-79 during latex flow. Of these, the HbPIP1;3, HbPIP1;4 and HbPIP2;3 were significantly up-regulated at the late stage of latex flow and their expression levels were higher than that of the corresponding genes at the early stage of latex flow in CATAS7-33-97. Additionally, HbPIP2;1 was up-regulated at the middle stage and at the late stage of latex flow in both clones.
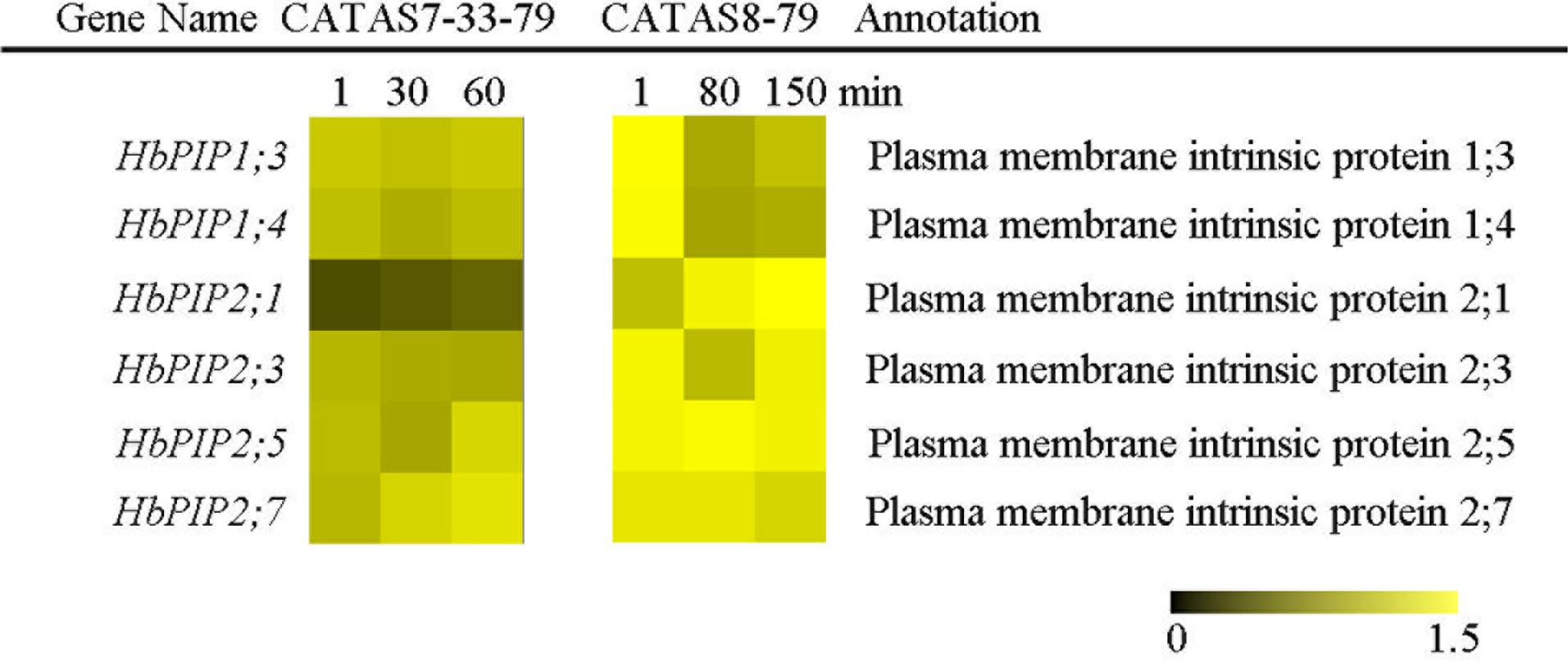
FIGURE 2. The qRT-PCR analysis of aquaporin encoding genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79. The qRT-PCR result was displayed by heatmap. Yellow showed high expression and black showed low expression. Bar represented the relative expression data (the same below).
Expression Pattern of Carbohydrate Metabolism-Related Genes during Latex Flow
In plants, carbohydrate metabolism, such as sucrose formation and degradation through glycolysis, provides both energy and a carbon skeleton for organic compound formation (Kunz et al., 2014). The expression of all the tested seven sucrose metabolism and glycolysis related genes changed in the latex of both clones during latex flow (Figure 3 and Supplementary Figure S1). In general, the expression pattern of HbNIN1, HbNIN2 and HbNIN3 was similar between the two clones. Their expression levels were high within 1 min (at the early stage of latex flow) and decreased thereafter during latex flow. The levels of HbSUT3 at the early stage and HbSus3 at the late stage in the latex of CATAS8-79 were respectively higher than those in the latex of CATAS7-33-97, although their expression patterns were similar between the two clones. In contrast to the expression pattern of HbPK, which was down-regulated in the latex of CATAS7-33-97 during latex flow, it was up-regulated in the latex of CATAS8-79 during latex flow. Similarly, the expression level of HbPDC4 was significantly higher at the late stage in the latex of CATAS8-79 than in the latex of CATAS7-33-97.
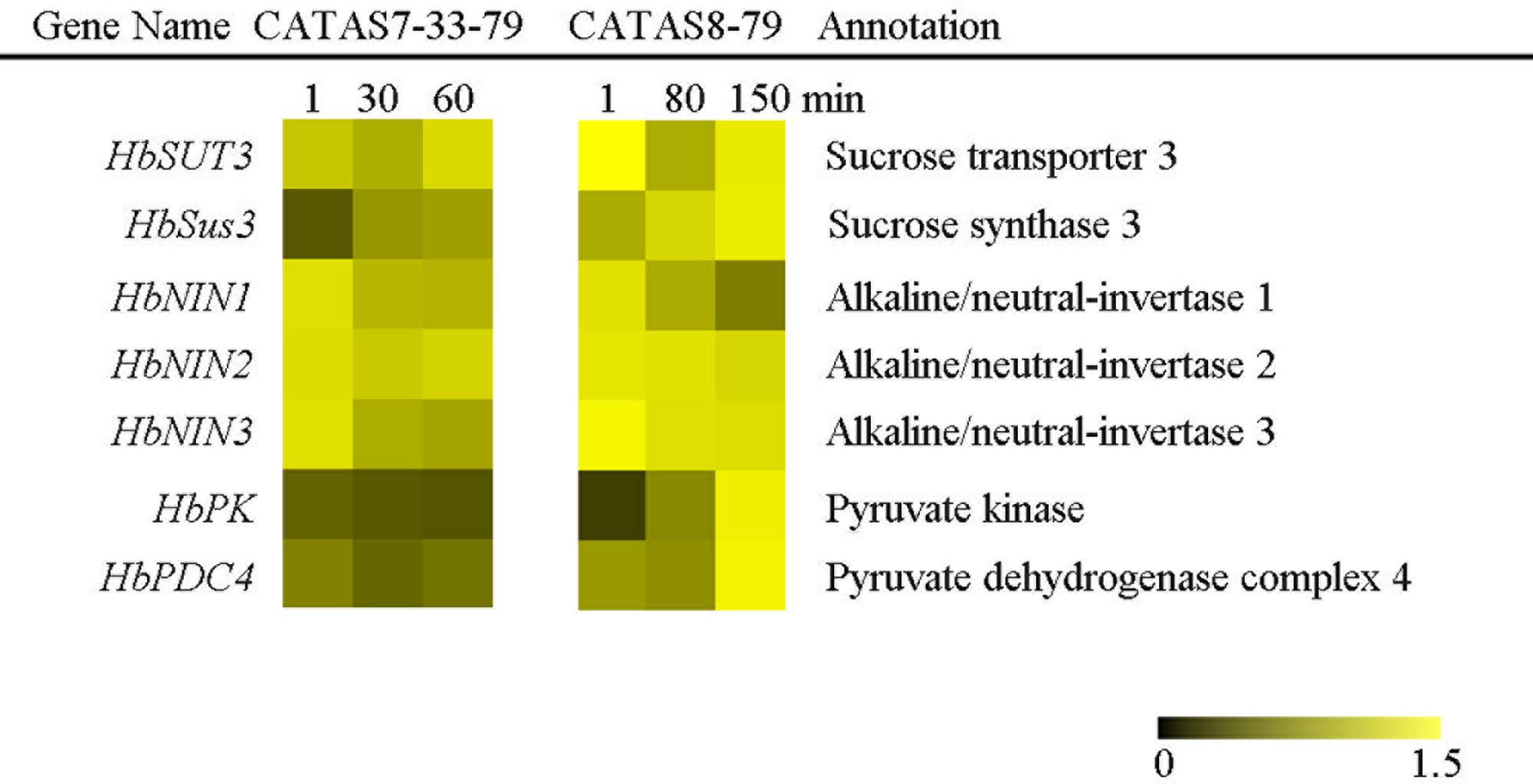
FIGURE 3. The qRT-PCR analysis of carbohydrate metabolism-related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79.
Expression Pattern of Natural Rubber Biosynthesis-Related Genes during Latex Flow
Isopentenyl pyrophosphate is the direct precursor for natural rubber (Chow et al., 2012). The expression of 26 genes related to pre-IPP and post-IPP stages of the natural rubber biosynthesis pathway were analyzed during latex flow by qRT-PCR (Figure 4 and Supplementary Figure S1). The expression of nearly all the tested genes changed during latex flow. Of these, the transcript abundance of 13 genes (HbHMGS1, HbHMGS2, HbMK, HbMDC1, HbMDC2, HbDXS1, HbDXS2, HbDXR, HbMCT2, HbCMK, HbMDS1, HbHDR and HbSRPP) at both the early and late stages in the latex of CATAS8-79 were higher than those in the latex of CATAS7-33-97. Moreover, the expression pattern of 15 genes, HbAACT1, HbHMGS1, HbHMGS2, HbHMGR1, HbMK, HbMDC1, HbMDC2, HbDXS1, HbMCT1, HbMCT2, HbHDR, HbIPPI1, HbHRT1, HbREF and HbSRPP, were generally similar between the rubber tree clone CATAS7-33-97 and CATAS8-79. The other seven genes had differential expression patterns during the latex flow between the two clones. They were HbAACT3, HbDXR, HbCMK, HbMDS1, HbMDS2, HbFDPS and HbHRT2. Of these, the expressions of HbFDPS and HbHRT2, were down-regulated in the latex of CATAS7-33-97 during latex flow while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow. Moreover, five genes were individually changed only in one clone. For CATAS8-79, the expression of HbDXR, HbMDS1 and HbMDS2 were up-regulated at the late stage of latex flow. For CATAS7-33-97, the expression of HbAACT3 was up-regulated while HbCMK was down-regulated at the late stage of latex flow.
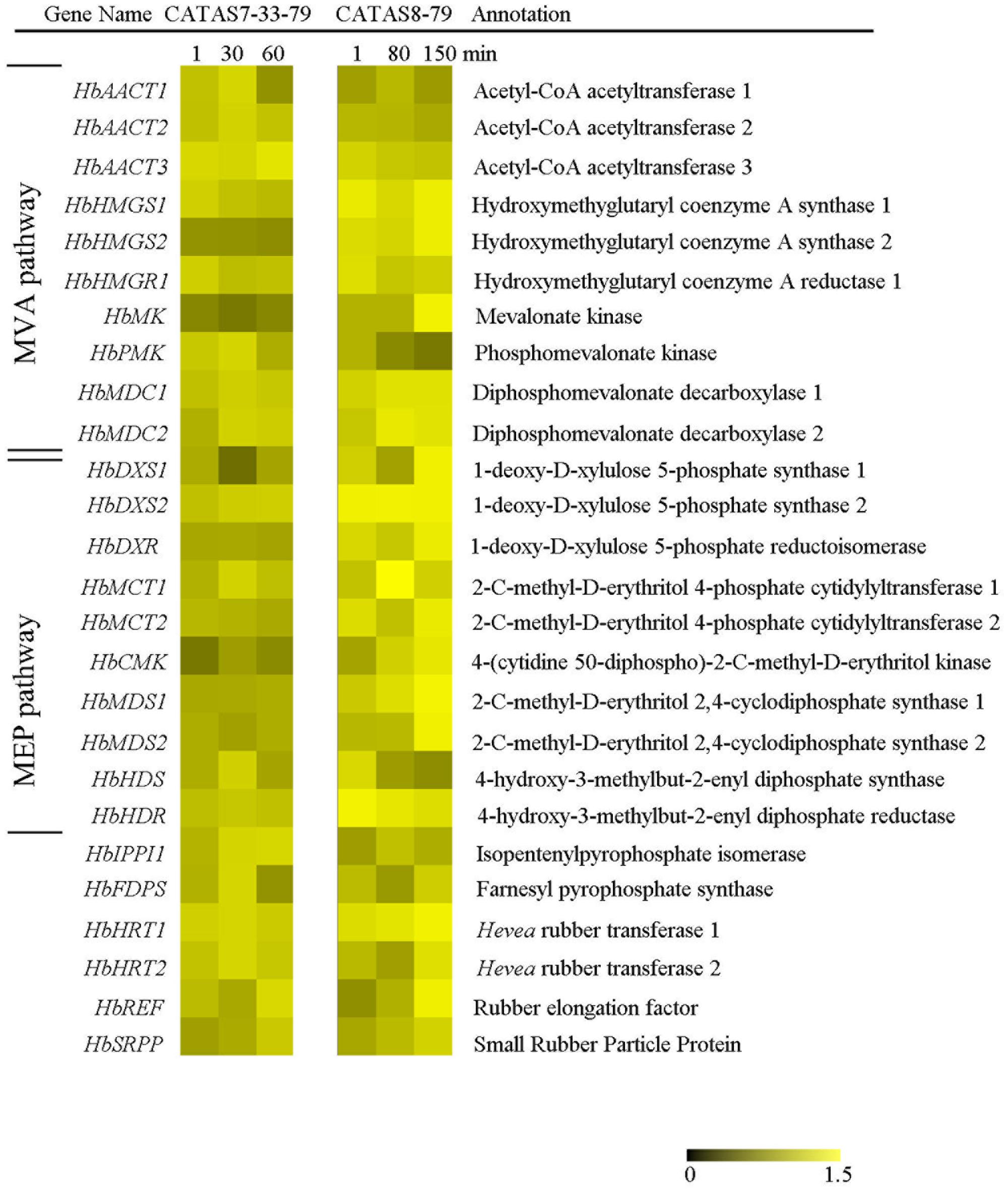
FIGURE 4. The qRT-PCR analysis of natural rubber biosynthesis metabolism-related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79.
Expression Pattern of Jasmonate and Ethylene Signaling-Related Genes during Latex Flow
It is known that phytohormones play a key role in increasing the natural rubber production (Zhu and Zhang, 2009; Zhao et al., 2011). The differential expressions of six jasmonate signaling-related genes and seven ethylene signaling-related genes during latex flow were revealed between the rubber tree clone CATAS-7-33-97 and CATAS8-79 (Figure 5 and Supplementary Figure S1). In comparison with the rubber tree clone CATAS7-33-97, all five jasmonate signaling-related genes, HbCOI1, HbJAZ2, HbJAZ3, HbMYC1 and HbMYC3, were significantly up-regulated in the latex of the rubber tree clone CATAS8-79 at the late stage of latex flow. The expression pattern of HbCOI1 and HbJAZ3 was reversed between the two clones during latex flow. They were down-regulated in the latex of CATAS7-33-97, while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow. Furthermore, HbJAZ2, HbMYC1 and HbMYC3 showed no change in the latex of CATAS7-33-97 while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow. Among the seven ethylene signaling-related genes, HbACO1, HbACO2, HbETR2 and HbEIN3 showed no change in the latex of CATAS7-33-97 while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow. Similarly, HbSAMA was down-regulated during latex flow in the latex of CATAS7-33-97 while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow.

FIGURE 5. The qRT-PCR analysis of hormone metabolism-related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79.
Expression Patterns of ROS Generation and Scavenging System Related Genes during Latex Flow
The balance of ROS generation and scavenging in laticifer cells influenced the state of latex exploitation (Zhang et al., 2017). The expression of two ROS generation-related genes (HbRBOHA and HbRBOHB) showed no change in the latex of CATAS7-33-97, while they were up-regulated in the latex of CATAS8-79 at the late stage of latex flow (Figure 6 and Supplementary Figure S1). The expression level of HbRBOHA at the early stage of latex flow in the latex of CATAS8-79 was significantly higher than that in the latex of CATAS7-33-97. Among the four ROS scavenging-related genes (HbAPX, HbCAT, HbCuZnSOD and HbMnSOD), HbAPX and HbCAT were down-regulated in the latex of CATAS7-33-97, while they showed no change (HbAPX) or were up-regulated (HbCAT) in the latex of CATAS8-79 at the late stage of latex flow. In contrast to HbMnSOD and HbCuZnSOD which were not reprogrammed by latex flow in CATAS7-33-97, the two genes were activated the latex of CATAS8-79 at the late stage of latex flow.
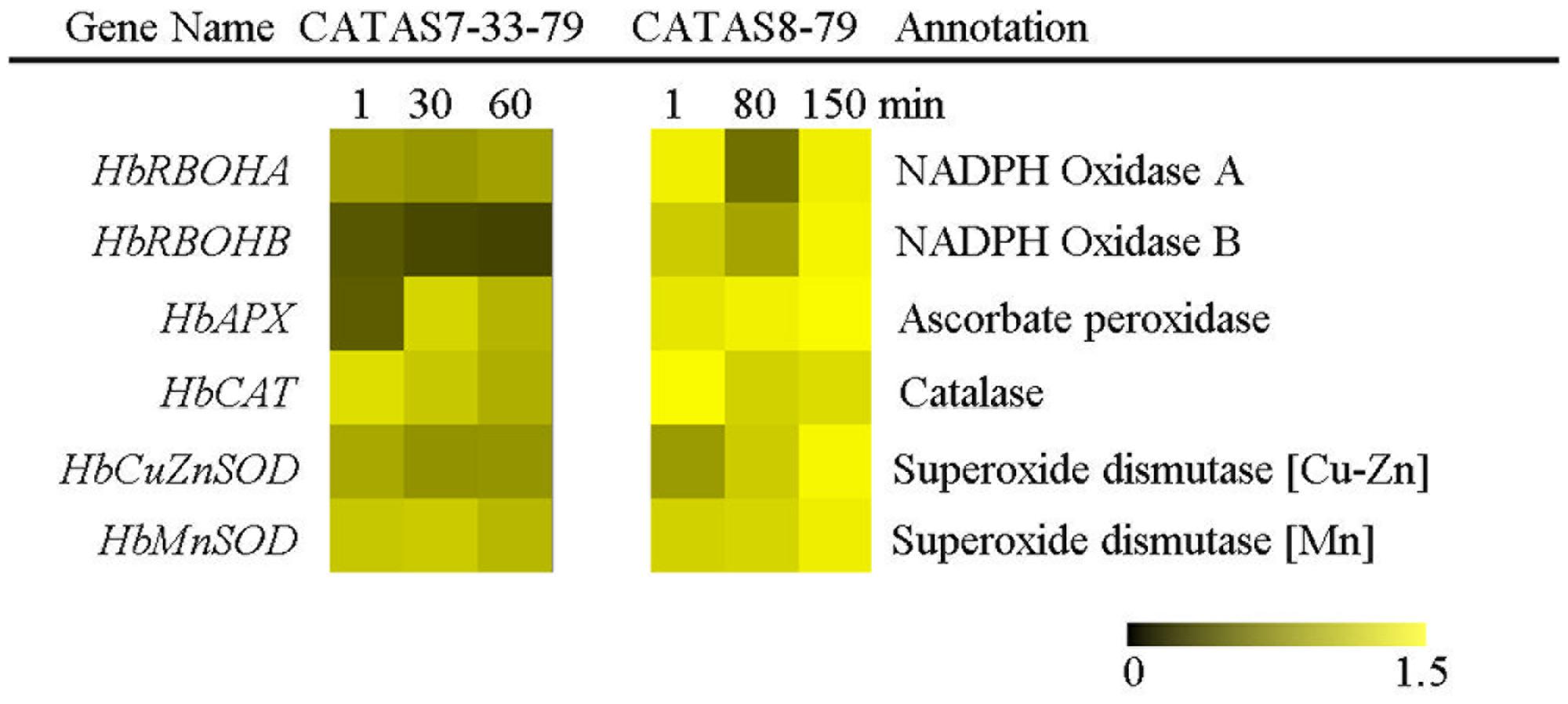
FIGURE 6. The qRT-PCR analysis of ROS generation and scavenging system related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79.
Expression Pattern of Latex Coagulation- Related Genes during Latex Flow
The expression pattern of four latex coagulation-related genes, Hb44KD, HbChit, HbGluc and HbHevein, were different between rubber tree clones CATAS7-33-97 and CATAS8-79 (Figure 7 and Supplementary Figure S1). There were slight changes in the expression levels of the four genes among the three stages of latex flow in the latex of CATAS7-33-97. By contrast, the Hb44KD gene was up-regulated, while all three genes, including HbChit, HbGluc and HbHevein, were down-regulated during latex flow in the latex of CATAS8-79.
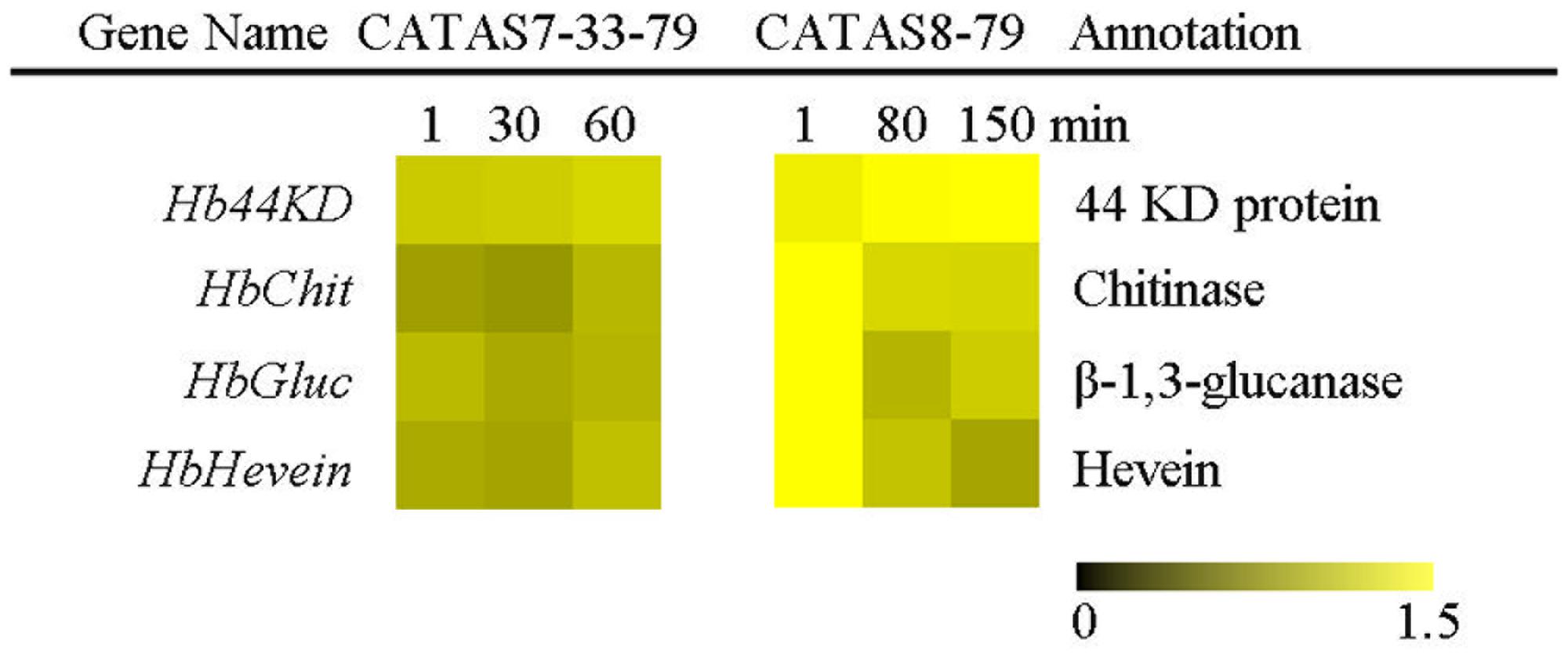
FIGURE 7. The qRT-PCR analysis of latex coagulation related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79.
Discussion
Transcription and translation are two crucial steps that determine the transfer of genetic information from genes to protein. It is known that the transcript level of a gene can largely reflect its translation state based on the central dogma. In the rubber tree, studies show the expression level of HbAPX, HbSUT3 and HbNIN2 is positively related to its protein activity both in vivo and in vitro (Tang et al., 2010; Liu et al., 2015; Chao et al., 2015b). The duration of latex flow and latex regeneration are two factors that determine the rubber yield of rubber trees (Serres et al., 1994; Chao et al., 2015a). After tapping, latex exploitation usually lasts 10 min to a few hours. However, most of the investigation on the latex metabolism focuses on latex samples after latex flow and neglect changes during latex flow (Tang et al., 2010; He, 2013; Xiao et al., 2014; An et al., 2015; Chao et al., 2015b; Liu et al., 2015; Long et al., 2015; Putranto et al., 2015; Deng et al., 2016; Shi et al., 2016; Makita et al., 2017). Considering that the expression of some candidate reference genes in the latex of the rubber tree are influenced by latex flow (Chao et al., 2016), the expression of latex metabolism-related genes should be influenced in this process. The rubber tree clones CATAS7-33-97 and CATAS8-79 have a different latex flow duration and latex regeneration (Chao et al., 2016). Accordingly, 62 latex metabolism-related genes are differentially expressed during latex flow between the two clones (Figure 8). Of these, the expression levels of 38 genes were higher in the latex of CATAS8-79 than that in the latex of CATAS7-33-97 at the early stage of the latex flow (1 min). At the late stage of the latex flow, there were 45 genes with higher expression levels in the latex of CATAS8-79 (Supplementary Figure S1). The changes in the gene expression at the late stage should be caused by the current latex flow. The expression status at the early stage should mainly be the state of latex regeneration after latex flow caused by the last tapping, though the slight influence caused by the morphological difference between two clones could not be excluded.
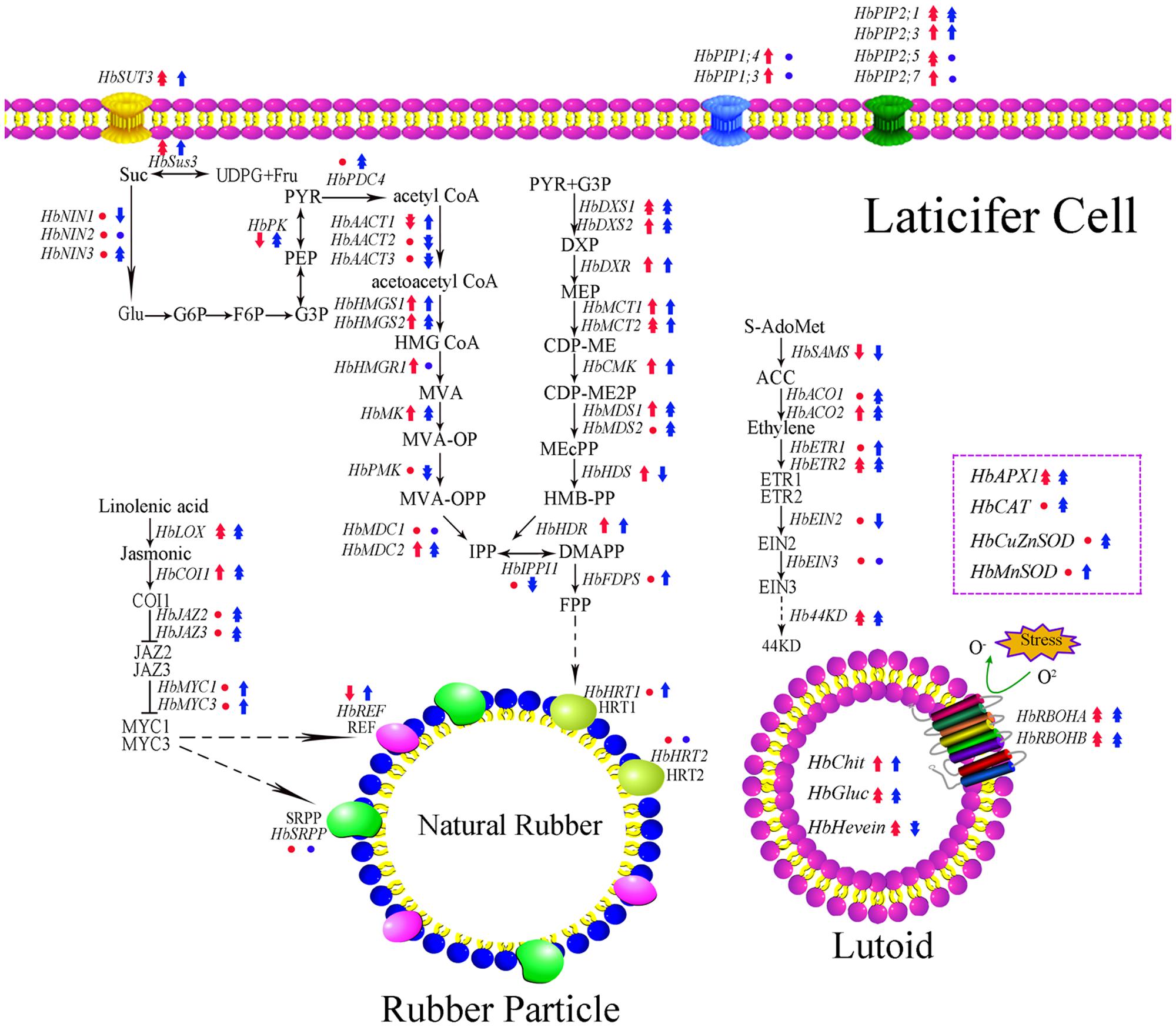
FIGURE 8. Schematic representation of expression pattern of latex metabolism-related genes in laticifer cell. The genes expression pattern in early or late stage of latex flow between two clones is obtained by CATAS7-33-97 versus CATAS8-79. Red represented early stage of latex flow while blue represented late stage of latex flow. Circle, single arrow, double arrow represented no change, 0.05, 0.01 significant difference. Up-regulation showed high expression in CATAS8-79 and down-regulation showed high expression in CATAS7-33-97.
The latex regeneration ability is a crucial factor that determines latex production. In our previous research, the duration of latex flow and latex production are notably different between CATAS8-79 and CATAS7-33-97 (Chao et al., 2016). Here, we further show the differential changes in the rubber content during latex flow between the two clones (Figure 1). The high rubber content remains unchangeable within 80 min after tapping in CATAS8-79, but significantly decreases at 30 min after tapping in CATAS7-33-97, suggesting that the rubber tree clone CATAS8-79 is more effective in latex regeneration than CATAS7-33-97. Carbohydrate metabolism, such as sucrose metabolism and glycolysis, not only supplies energy but also generates a carbon skeleton for the formation of organic compounds, including natural rubber (Tang et al., 2010). The SUT and NIN or sucrose synthase take part in sucrose transport and decomposition or synthesis, respectively (Zhang C. et al., 2016). Recently, Liu et al. (2015) cloned three invertases (HbNIN1-3) in the latex of the rubber tree and identified that HbNIN2 is the crucial isoform for latex regeneration. In the current research, only HbNIN3 was up-regulated at the late stage of latex flow in the latex of CATAS8-79 compared with that of CATAS7-33-97 (Figure 8). Moreover, the expression levels of HbSUT3 and HbSus3 at both the early and late stage of latex flow in the latex of CATAS8-79 are significantly higher than that of CATAS7-33-97 (Figure 8). The results suggest that sucrose synthesis and transport rather than decomposition might be the rate-limiting steps for latex regeneration, and HbNIN3 should play a key role in initiating the regeneration of latex after tapping. PK and pyruvate dehydrogenase complex (PDC) are major contributors to the control of glycolysis (Fleige et al., 2007; Megguer et al., 2017). In contrast to CATAS7-33-97, both HbPK and HbPCD4 are significantly activated at the late stage of latex flow in the latex of CATAS8-79 (Figure 8). This might provide much of the acetyl coenzyme-A for synthesis of IPP, the direct precursor of natural rubber, in the rubber tree clone CATAS8-79. The cytoplastic MVA pathway is the main enzymatic reaction for IPP biosynthesis (Kang et al., 2016). In the MVA pathway, HMG CoA is synthesized though 3-hydroxy-3-methylglutaryl coenzyme-A synthase (HMGS) and then reduced to MVA by 3-hydroxy-3-methylglutaryl coenzyme-A reductase (HMGR). It has long been known that HMGR is the key rate-limiting enzyme of the MVA pathway in many species, such as Homo, yeast, and Arabidopsis (Omkumar et al., 1994; Dale et al., 1995; Burg et al., 2008). In the rubber tree, four HbHMGRs have been cloned, and HbHMGR1 was previously recognized as the key member involved in natural rubber biosynthesis (Sando et al., 2008). In the present study, its expression pattern is similar between CATAS7-33-97 and CATAS8-79. In contrast, the expression levels of HbHMGS1 and HbHMGS2 at the early and late stage of latex flow in the latex of CATAS8-79 are significantly higher than those in the latex of CATAS7-33-97 (Figure 8). It seems that the biosynthesis of HMG CoA by HMGS is more important than the biosynthesis of MVA by HMGR for enhanced natural rubber biosynthesis in the rubber tree. In addition to the MVA pathway, the plastidic MEP pathway is supposed to act as an alternative pathway for IPP biosynthesis in the rubber tree (Chow et al., 2012). In the present study, most of the tested genes involved in the MEP pathway are activated while only three of the ten genes in the MVA pathway (HbHMGS1-2 and HbMK) are activated at both the early and late stages of latex flow in the latex of CATAS8-79 (Figure 8). The overall activation of the MEP pathway in CATAS8-79 may supply much more IPP for natural rubber biosynthesis. Jasmonates have important roles in the regulation of secondary metabolite biosynthesis (Zhou and Memelink, 2016). In the present research, it is of interest that in comparison with the expression pattern of CATAS7-33-97, all jasmonate biosynthesis and signaling-related genes are activated at the late stage of latex flow in the latex of CATAS8-79. This activation may contribute to enhanced rubber biosynthesis between two tappings in CATAS8-79.
The duration of latex flow is a factor that determines latex production. The duration of the latex flow of CATAS8-79 is notably longer than that of CATAS7-33-97 (Chao et al., 2016). The phloem turgor pressure is recognized as the initial power of latex exploitation after tapping. In laticifer cells, water as well as natural rubber accumulation causes a huge turgor pressure (∼10 Pa) (An et al., 2014). Aquaporins are a group of proteins that mediate the trans-membrane transport of water and other small solutes. The PIP is one subfamily of aquaporins and plays a crucial role in laticifers water transport in the rubber tree (Zou et al., 2015). The transcript levels of all the genes at the early stage of latex flow in CATAS8-79 are higher than that in CATAS7-33-97, suggesting that water entrance into laticifer cells is more active in CATAS8-79 than in CATAS7-33-97. The available data show that HbPIP2;1 is important for water metabolism (Tungngoen et al., 2009). In the present study, HbPIP1;3 and HbPIP1;4 exhibit high transcript abundance at the early stage of latex flow but low expression abundance at the late stage of latex flow in CATAS8-79 (Figure 2). By contrast, the transcript level of HbPIP2;1 is very low during latex regeneration (at the early stage of latex flow) in both CATAS7-33-97 and CATAS8-79, while they are up-regulated especially in CATAS8-79 at the late stage of latex flow. This differential expression pattern suggests that HbPIP1;3 and HbPIP1;4 play a crucial role in the process of latex regeneration while HbPIP2;1 may be active in regulating water entrance into laticifer cells during latex flow. In natural rubber production, Ethrel (an ethylene releaser) is widely used to increase rubber yield per tapping by prolonging the duration of latex flow (Zhu and Zhang, 2009). Additionally, ACO is a key enzyme that catalyzes the last step of ethylene biosynthesis, while the ETR is essential for ethylene signaling (Hall and Bleecker, 2003; Sun et al., 2017). Here, we show that the expression levels of HbACO2 and HbETR2 at both early and late stages of latex flow in the latex of CATAS8-79 are significantly higher than that of CATAS7-33-97, hinting that ethylene production and ethylene signaling are more active in the latex of CATAS8-79. The activation of ethylene signaling in CATAS8-79 may contribute to the longer duration of latex flow. It may also be related to the up-regulation of HbRBOHA and HbRBOHB at both the early and late stages of latex flow in the latex of CATAS8-79. The available data show that ethylene signaling has a synergistic role with ROS generation (Zhang H. et al., 2016; Zhang M. et al., 2016). In Arabidopsis, ERF74, an ERF, can bind to the promoter of RbohD and activate its expression (Yao et al., 2017). In rice, OsEIL1, a rice homologue of AtEIN3 and can bind to the promoters of OsRbohA and OsRbohB directly (Yang et al., 2017). The RBOH located at the surface of the lutoids is the main source of ROS, while antioxidant proteins (CAT, SOD, APX) play a key role in scavenging ROS in the latex (Zhang et al., 2017). In the current research, it is clearly shown that compared with other ROS scavenging genes (HbAPX, HbCuZnSOD and HbMnSOD), HbCAT exhibits an opposite expression pattern during latex flow in both clones (Figure 6). Compared with the high expression level of HbCAT at the early stage of latex flow, the transcript abundance of the gene is low at the late stage in both rubber tree clones. We thus deduce that the expression level of HbCAT should be up-regulated until the next tapping and the gene may play a key role in latex regeneration. Since CAT has a lower affinity for H2O2 than APX, the enzyme is effective only in the presence of a massive level of H2O2 (Anjum et al., 2016). The higher abundance of HbCAT in CATAS8-79 suggests that the clone has a high concentration of H2O2 in laticifer cells, which may be the main reason for CATAS8-79 having a higher rate of tapping panel dryness during production (Li and Liu, 2014). The activation of HbAPX, HbCuZnSOD and HbMnSOD at the late stage of latex flow in the latex of CATAS8-79 may contribute to scavenging ROS and maintaining latex exploitation. On the other hand, the plug formation at the end of the severed laticifers results in the termination of the latex flow. Since the fractured lutoid effectively causes latex coagulation, it is traditionally believed that the ethylene-caused prolongation of the latex flow duration is associated with the increased stability of lutoids (Wititsuwannakul et al., 2008). Recently, we demonstrate that the ethylene-caused prolongation of latex flow is associated with increased levels of a 44 kDa protein in the C-serum (Shi et al., 2016). The protein acts as a universal antagonist of rubber particle aggregation that is caused by proteins from the lutoids. These proteins include hevein, chitinase and glucanase. The hevein, glucanase and the combination of chitinase and glucanase are effective in aggregating rubber particles (Gidrol et al., 1994; Wititsuwannakul et al., 2008; Wang et al., 2013), which contribute to plug formation at the end of the severed laticifers. In the present study, the expression level of Hb44KD at any stage of latex flow in the CATAS8-79 latex is higher than that in CATAS7-33-97 latex. By contrast, the expression levels of the latex coagulation-related genes, HbChit, HbCluc and HbHevein, are up-regulated at the late stage of latex flow in CATAS7-33-97 latex, while there was no change or down-regulation in CATAS8-79.
Taken together, the expression levels of 62 latex metabolism related genes were monitored in two rubber tree clones with a differential latex regeneration and latex flow duration. It is speculated that the up-regulation of the antagonist Hb44KD, aquaporin HbPIP2;1, ethylene biosynthesis key gene HbACO1, and the down-regulation of the coagulation factors HbChit, HbGluc and HbHevein, are the key members that determine the duration of latex flow after tapping. While the up-regulation of the aquaporins HbPIP1;3 and HbPIP1;4, the sucrose metabolism members HbSUT3 and HbSus3, the key MVA pathway members HbHMGS1-2 and HbMK, and the activation of the MEP pathway and jasmonate pathway, are suggested to play key roles in promoting latex regeneration between two tappings. A schematic representing the molecular difference of latex metabolism between CATAS7-33-97 and CATAS8-79 is displayed (Figure 8), which provides some clues for revealing the regulation of latex metabolism and the benefits of molecular breeding in the rubber tree.
Author Contributions
JC designed and carried out the experiment of this study, and wrote the manuscript. SY and YC participated and analyzed data in the experiment. W-MT planned the study and participated in the design of the experiment. All authors have read and approved the manuscript in its final form.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by the earmarked Fund for Modern Agro-industry Technology Research System (CARS-34-GW1).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01904/full#supplementary-material
FIGURE S1 | The qRT-PCR analysis of all latex metabolism related genes across three stages of latex flow between CATAS7-33-97 and CATAS8-79. The capital letter represents p < 0.01 while lower case represents p < 0.05. The same letter indicated no significant difference among groups.
Abbreviations
AACT, Acetyl coenzyme A acetyltransferase; ACO, 1-aminocyclopropane-1-carboxylate oxidase; APX, L-ascorbate peroxidase; CAT, catalase; Chit, chitinase; CMK, 4-(cytidine 5-diphospho)-2-C-methyl-D-erythritol kinase; COI, coronatine insensitive; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; ERF, ethylene response factor; ETH, ethylene; ETR, ethylene receptor; EIN, ethylene insensitive; FDPS, farnesyl diphosphate synthase; Gluc, glucanase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HMGR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; HMGS, 3-hydroxy-3-methylglutaryl-coenzyme A synthase; HRT, Hevea rubber transferase; IPP, isopentenyl pyrophosphate; IPPI, isopentenyl pyrophosphate isomerase; JA, jasmonic acid; JAZ, jasmonate-ZIM-domain; LOX, lipoxygenase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; MDC, Diphosphomevalonate decarboxylase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; MEP, 2-C-methyl-D-erythritol 4-phosphate; MVA, mevalonate; MK, mevalonate kinase; NIN, alkaline/neutral invertase; PDC, pyruvate decarboxylase; PIP, plasma membrane intrinsic protein; PK, pyruvate kinase; PMK, Phosphomevalonate kinase; qRT-PCR, quantitative real-time PCR; RBOH, respiratory burst oxidase homolog; REF, Rubber elongation factor; ROS, reactive oxygen species; SAMS, S-adenosylmethionine synthetase; SOD, superoxide dismutase; SRPP, small rubber particle protein; Sus, sucrose synthase; SUT, sucrose transporter; UBC2b, ubiquitin-protein ligase (AtUBC2).
References
An, F., Cahill, D., Rookes, J., Lin, W., and Kong, L. (2014). Real-time measurement of phloem turgor pressure in Hevea brasiliensis with a modified cell pressure probe. Bot. Stud. 55:19. doi: 10.1186/1999-3110-55-19
An, F., Zou, Z., Cai, X., Wang, J., Rookes, J., Lin, W., et al. (2015). Regulation of HbPIP2;3, a latex-abundant water transporter, is associated with latex dilution and yield in the rubber tree (Hevea brasiliensis Muell. Arg.). PLOS ONE 10:e0125595. doi: 10.1371/journal.pone.0125595
Anjum, N. A., Sharma, P., Gill, S. S., Hasanuzzaman, M., Khan, E. A., Kachhap, K., et al. (2016). Catalase and ascorbate peroxidase-representative H2O2-detoxifying heme enzymes in plants. Environ. Sci. Pollut. Res. Int. 23, 19002–19029. doi: 10.1007/s11356-016-7309-6
Burg, J. S., Powell, D. W., Chai, R., Hughes, A. L., Link, A. J., and Espenshade, P. J. (2008). Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab. 8, 522–531. doi: 10.1016/j.cmet.2008.09.004
Chao, J., Chen, Y., Wu, S., and Tian, W. M. (2015a). Comparative transcriptome analysis of latex from rubber tree clone CATAS8-79 and PR107 reveals new cues for the regulation of latex regeneration and duration of latex flow. BMC Plant Biol. 15:104. doi: 10.1186/s12870-015-0488-3
Chao, J., Zhang, S., Chen, Y., and Tian, W. M. (2015b). Cloning, heterologous expression and characterization of ascorbate peroxidase (APX) gene in laticifer cells of rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Biochem. 97, 331–338. doi: 10.1016/j.plaphy.2015.10.023
Chao, J., Yang, S., Chen, Y., and Tian, W. M. (2016). Evaluation of reference genes for quantitative real-time PCR analysis of the gene expression in laticifers on the basis of latex flow in rubber tree (Hevea brasiliensis Muell. Arg.). Front. Plant Sci. 7:1149. doi: 10.3389/fpls.2016.01149
Chow, K. S., Mat-Isa, M. N., Bahari, A., Ghazali, A. K., Alias, H., Mohd-Zainuddin, Z., et al. (2012). Metabolic routes affecting rubber biosynthesis in Hevea brasiliensis latex. J. Exp. Bot. 63, 1863–1871. doi: 10.1093/jxb/err363
Chrestin, H., Gidrol, X., and Kush, A. (1997). Towards a latex molecular diagnostic of yield potential and the genetic engineering of the rubber tree. Euphytica 96, 77–82. doi: 10.1023/A:1002950300536
d’ Auzac, J. (1989). “Tapping systems and area of drained bark,” in Physiology of Rubber Tree Latex, eds J. d’Auzac, J. L. Jacob, and H. Chrestin (Boca Raton, FL: CRC Press), 221–232.
Dale, S., Arro, M., Becerra, B., Morrice, N. G., Boronat, A., Hardie, D. G., et al. (1995). Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at serine-577 by Brassica oleracea 3-hydroxy-3-methylglutaryl CoA reductase kinase. Eur. J. Biochem. 233, 506–513. doi: 10.1111/j.1432-1033.1995.506_2.x
Deng, X. M., Wu, S. H., Dai, X. M., and Tian, W. M. (2016). Expression analysis of MVA and MEP metabolic pathways genes in latex and suspension cells of Hevea brasiliensis. Guihaia 36, 449–455.
Eschbach, J. M., and Lacrotte, R. (1989). “Factors influencing response to hormonal yield stimulation: limits of this stimulation,” in Physiology of Rubber Tree Latex, eds J. d’Auzac, J. L. Jacob, and H. Chrestin (Boca Raton, FL: CRC Press), 321–342.
Fleige, T., Fischer, K., Ferguson, D. J., Gross, U., and Bohne, W. (2007). Carbohydrate metabolism in the Toxoplasma gondii apicoplast: localization of three glycolytic isoenzymes, the single pyruvate dehydrogenase complex, and a plastid phosphate translocator. Eukaryot. Cell 6, 984–996. doi: 10.1128/EC.00061-07
Gidrol, X., Chrestin, H., Tan, H. L., and Kush, A. (1994). Hevein, a lectin-like protein from Hevea brasiliensis (rubber tree) is involved in the coagulation of latex. J. Biol. Chem. 269, 9278–9283.
Hall, A. E., and Bleecker, A. B. (2003). Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell 15, 2032–2041. doi: 10.1105/tpc.013060
He, X. (2013). Research on the Expression of Several Members of JAZ and MYC Genes Family Correlated with Natural Rubber Yield in Hevea brasiliensis Muell. Arg. [D]. Master thesis, Hainan University, Haikou.
Kang, A., George, K. W., Wang, G., Baidoo, E., Keasling, J. D., and Lee, T. S. (2016). Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab. Eng. 34, 25–35. doi: 10.1016/j.ymben.2015.12.002
Kunz, H. H., Zamani-Nour, S., Häusler, R. E., Ludewig, K., Schroeder, J. I., Malinova, I., et al. (2014). Loss of cytosolic phosphoglucose isomerase affects carbohydrate metabolism in leaves and is essential for fertility of Arabidopsis. Plant Physiol. 166, 753–765. doi: 10.1104/pp.114.241091
Li, Y. J., and Liu, J. P. (2014). Tapping panel dryness survey of three clones of rubber tree (Hevea brasiliensis Muell. Arg.). Chin. J. Trop. Agric. 34, 58–65.
Liu, S., Lan, J., Zhou, B., Qin, Y., Zhou, Y., Xiao, X., et al. (2015). HbNIN2, a cytosolic alkaline/neutral-invertase, is responsible for sucrose catabolism in rubber-producing laticifers of Hevea brasiliensis (para rubber tree). New Phytol. 206, 709–725. doi: 10.1111/nph.13257
Long, X., He, B., Wang, C., Fang, Y., Qi, J., and Tang, C. (2015). Molecular identification and characterization of the pyruvate decarboxylase gene family associated with latex regeneration and stress response in rubber tree. Plant Physiol. Biochem. 87, 35–44. doi: 10.1016/j.plaphy.2014.12.005
Makita, Y., Ng, K. K., Veera Singham, G., Kawashima, M., Hirakawa, H., Sato, S., et al. (2017). Large-scale collection of full-length cDNA and transcriptome analysis in Hevea brasiliensis. DNA Res. 24, 159–167. doi: 10.1093/dnares/dsw056
Megguer, C. A., Fugate, K. K., Lafta, A. M., Ferrareze, J. P., Deckard, E. L., Campbell, L. G., et al. (2017). Glycolysis is dynamic and relates closely to respiration rate in stored sugarbeet roots. Front. Plant Sci. 8:861. doi: 10.3389/fpls.2017.00861
Omkumar, R. V., Darnay, B. G., and Rodwell, V. W. (1994). Modulation of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase activity by phosphorylation role of serine 871. J. Biol. Chem. 269, 6810–6814.
Putranto, R. A., Herlinawati, E., Rio, M., Leclercq, J., Piyatrakul, P., Gohet, E., et al. (2015). Involvement of ethylene in the latex metabolism and tapping panel dryness of Hevea brasiliensis. Int. J. Mol. Sci. 16, 17885–17908. doi: 10.3390/ijms160817885
Sando, T., Takaoka, C., Mukai, Y., Yamashita, A., Hattori, M., Ogasawara, N., et al. (2008). Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72, 2049–2060. doi: 10.1271/bbb.80165
Serres, E., Lacrotte, R., Prevot, J. C., Clement, A., Commere, J., and Jacob, J. L. (1994). Metabolic aspects of latex regeneration in situ for three Hevea clones. Indian J. Nat. Rubber Res. 7, 72–88.
Shi, M. J., Cai, F. G., and Tian, W. M. (2016). Ethrel-stimulated prolongation of latex flow in the rubber tree (Hevea brasiliensis Muell. Arg.): an Hev b 7-like protein acts as a universal antagonist of rubber particle aggregating factors from lutoids and C-serum. J. Biochem. 159, 209–216. doi: 10.1093/jb/mvv095
Sun, X., Li, Y., He, W., Ji, C., Xia, P., Wang, Y., et al. (2017). Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nat. Commun. 8:15758. doi: 10.1038/ncomms15758
Tang, C., Huang, D., Yang, J., Liu, S., Sakr, S., Li, H., et al. (2010). The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of Hevea brasiliensis (para rubber tree). Plant Cell Environ. 33, 1708–1720. doi: 10.1111/j.1365-3040.2010.02175.x
Tungngoen, K., Kongsawadworakul, P., Viboonjun, U., Katsuhara, M., Brunel, N., Sakr, S., et al. (2009). Involvement of HbPIP2;1 and HbTIP1;1 aquaporins in ethylene stimulation of latex yield through regulation of water exchanges between inner liber and latex cells in Hevea brasiliensis. Plant Physiol. 151, 843–856. doi: 10.1104/pp.109.140228
Van de Poel, B., Smet, D., and Van Der Straeten, D. (2015). Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. doi: 10.1104/pp.15.00724
Wang, X., Shi, M., Wang, D., Chen, Y., Cai, F., Zhang, S., et al. (2013). Comparative proteomics of primary and secondary lutoids reveals that chitinase and glucanase play a crucial combined role in rubber particle aggregation in Hevea brasiliensis. J. Proteome Res. 12, 5146–5159. doi: 10.1021/pr400378c
Wititsuwannakul, R., Pasitkul, P., Kanokwiroon, K., and Wititsuwannakul, D. (2008). A role for a Hevea latex lectin-like protein in mediating rubber particle aggregation and latex coagulation. Phytochemistry 69, 339–347. doi: 10.1016/j.phytochem.2007.08.019
Xiao, X., Tang, C., Fang, Y., Yang, M., Zhou, B., Qi, J., et al. (2014). Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 281, 291–305. doi: 10.1111/febs.12595
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Zhang, C., Zhang, H., Zhan, Z., Liu, B., Chen, Z., Liang, Y., et al. (2016). Transcriptome analysis of sucrose metabolism during bulb swelling and development in onion (Allium cepa L.). Front. Plant Sci. 7:1425. doi: 10.3389/fpls.2016.01425
Zhang, H., Li, A., Zhang, Z., Huang, Z., Lu, P., Zhang, D., et al. (2016). Ethylene response factor TERF1, regulated by ETHYLENE-INSENSITIVE3-like factors, functions in reactive oxygen species (ROS) scavenging in tobacco (Nicotiana tabacum L.). Sci. Rep. 6:29948. doi: 10.1038/srep29948
Zhang, M., Smith, J. A., Harberd, N. P., and Jiang, C. (2016). The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 91, 651–659. doi: 10.1007/s11103-016-0488-1
Zhang, Y., Leclercq, J., and Montoro, P. (2017). Reactive oxygen species in Hevea brasiliensis latex and relevance to tapping panel dryness. Tree Physiol. 37, 261–269. doi: 10.1093/treephys/tpw106
Zhao, Y., Zhou, L. M., Chen, Y. Y., Yang, S. G., and Tian, W. M. (2011). MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.). J. Plant Physiol. 168, 1649–1658. doi: 10.1016/j.jplph.2011.02.010
Zhou, M., and Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.biotechadv.2016.02.004
Zhu, J., and Zhang, Z. (2009). Ethylene stimulation of latex production in Hevea brasiliensis. Plant Signal. Behav. 4, 1072–1074. doi: 10.4161/psb.4.11.9738
Zou, Z., Gong, J., An, F., Xie, G., Wang, J., Mo, Y., et al. (2015). Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genomics 16:1001. doi: 10.1186/s12864-015-2152-6
Keywords: Hevea brasiliensis Muell. Arg., duration of latex flow, latex metabolism, gene expression, qRT-PCR
Citation: Chao J, Yang S, Chen Y and Tian W - M (2017) Transcript Profiling of Hevea brasiliensis during Latex Flow. Front. Plant Sci. 8:1904. doi: 10.3389/fpls.2017.01904
Received: 31 July 2017; Accepted: 23 October 2017;
Published: 07 November 2017.
Edited by:
Ján A. Miernyk, Agricultural Research Service (USDA), United StatesReviewed by:
Doug K. Allen, Plant Genetics Research (USDA ARS), United StatesRalph Andrew Backhaus, PharmaPacific, Inc., United States
Copyright © 2017 Chao, Yang, Chen and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Min Tian, d210aWFuQDE2My5jb20=
 Jinquan Chao
Jinquan Chao Shuguang Yang
Shuguang Yang Yueyi Chen
Yueyi Chen Wei-Min Tian
Wei-Min Tian