
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 18 October 2017
Sec. Plant Metabolism and Chemodiversity
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01789
Vegetable oils rich in oleic acid are more desirable than oils rich in polyunsaturated and saturated fatty acids. The biological switch of oleic acid to linoleic acid is facilitated by fatty acid desaturase 2 enzyme that is further classified into FAD2-1, FAD2-2, FAD2-3, and FAD2-4. The genes coding these enzymes have high sequence similarity, but differ mostly in their expression patterns. The seed-type FAD2 genes had evolved independently after segregation by duplication from constitutively expressed FAD2 genes. Temperature, light and wounding effectively regulate FAD2 expression in plants. FAD2 genes are expressed differently in different tissues of the plant, and the over-expression of FAD2 modifies physiological and vegetative characteristics. The activity of FAD2 leads to an increase in the content of dienoic fatty acids, and hence increases the resistance toward cold and salt stress. The thorough study of the FAD2 gene is important for understanding the expression, regulation and mechanism that will help in improving the quality of oil and stress resistance in plants.
Vegetable oils are important renewable resources rich in fatty acids that are broadly used in industrial applications and as an important supplement in the human diet (Yang and Xu, 2007). Fatty acids and their derivatives are not only energy reserves in plant seeds, but also play key roles in plant metabolism, membrane structural components, and signaling molecule precursors that are involved in stress-response and plant development (Ohlrogge and Browse, 1995; Harwood, 1996; Weber, 2002). Fatty acids are synthesized from acetyl-CoA in the plastids, later exported into the cytosol, and finally oil is synthesized in the endoplasmic reticulum (ER) (Browse and Somerville, 1991). The 30 enzymatic reactions taking place in the stroma of plastids produce C16- and C18-carbon fatty acids, about of which 75% is unsaturated (Ohlrogge and Browse, 1995; Somerville et al., 2000). Desaturation of the membrane phospholipids takes place by desaturases of the membrane-bound ER and chloroplast. The desaturases 2 and 3 (FAD2 and FAD3) that are integral membrane proteins in the ER primarily desaturate extra chloroplast lipids (Los and Murata, 1998; Shanklin and Cahoon, 1998). The desaturation of stearic acid (C18:0) to oleic acid (C18:1) is catalyzed by stearoyl-acyl carrier protein desaturase (SAD). Further desaturation of oleic acid to linoleic acid (18:2) is catalyzed by FAD2 in the ER and FAD6 in the plastid, whereas linoleic acid desaturation to γ-linolenic acid (C18:3, n6) is catalyzed by FAD3 in the ER and FAD7/FAD8 in the plastid (Zhang et al., 2012; Bhunia et al., 2016).
Linoleic and linolenic acids are polyunsaturated fatty acids (PUFAs) that are essential for health and nutrition, as these cannot be synthesized in humans and have to be supplied through diet (Guan et al., 2012b). Despite health benefits of PUFAs, they make the edible oil more vulnerable to rancidity, decrease its flavor, and shorten its shelf life (Pandey et al., 2014). The oxidative stability and nutritional value of the edible oil are dependent upon the fatty acid content of the oil, especially of oleic and linoleic acids (Cao et al., 2013). Oleic acid was found to have higher oxidative stability than linoleic acid, resulting in the extension of its shelf life (Ge et al., 2015). Therefore, there is a high demand for premium quality oil rich in monounsaturated fatty acids and poor in PUFAs. Such oils are more desirable, both nutritionally and commercially (Khadake et al., 2009; Guan et al., 2012b). Consumption of oils rich in monounsaturated fatty acids helps to reduce cholesterol, suppresses tumor formation, and protects from inflammatory diseases (O’Byrne et al., 1997; Yamaki et al., 2005). Therefore, increasing the oleic acid content in the oil is important for the development of oilseed crops to produce stable and healthy oils (Ge et al., 2015). The desaturation of fatty acids is one of the important biochemical processes that define the quality and economic significance of the vegetable oil (Guan et al., 2012b).
In this review, we will discuss about the features and scope of the FAD2 gene. We shall also focus on the regulation, characterization and expression of the gene, and review the role of the gene in fatty acid biosynthesis, plant development, cold and salt tolerance, and also the future prospects in altering the gene for improvement of oilseed crops, and hence the quality of the oil.
Plants have numerous fatty acid desaturase enzymes, which desaturate the majority of glycerolipids present in the tissues. The desaturases are mainly soluble or membrane bound, and present in chloroplasts and the ER, respectively. These enzymes are divided into three major classes: acyl-CoA, acyl-lipid and acyl-ACP desaturases. The acyl-CoA membrane-bound desaturases associated with ER are normally found in animals, yeast and fungi. These enzymes insert unsaturated bonds into the CoA esters of the fatty acid. These acyl-CoA desaturases, for example, the Δ5, Δ6, and Δ9 acyl-CoA are the electron acceptors of the electron-transport complex that contain cytochrome b5 and NADH-dependent cytochrome b5 reductase (Mitchell and Martin, 1995). In contrast, the desaturases in the cytoplasm of plant cells require a system that consists of cytochrome b5 and a NADH:cytochrome b5 oxidoreductase (Kearns et al., 1991). The Δ-9 desaturase synthesizes oleic acid that is used for phospholipid and cholesteryl ester synthesis. Delta-6 and Δ5 desaturases are required for the synthesis of highly unsaturated fatty acids (HUFAs), which are mainly esterified into phospholipids, and helps in maintaining membrane fluidity. The role of HUFAs may be for cold tolerance in plants and fish, and cell signaling in mammals (Nakamura and Nara, 2004). The other class of integral membrane-bound desaturases is the acyl-lipid desaturases found in cyanobacterial cells and chloroplasts. They introduce double bond into the fatty acyl chain of polar glycerolipids and use ferredoxin as the electron donor (Los and Murata, 1998; Tocher et al., 1998). The structure of this enzyme is similar to acyl-CoA desaturases, which are mainly transmembrane proteins (Murata and Wada, 1995). The Δ3, Δ6, Δ9, and Δ12-acyl-lipid desaturases have been characterized and are specified by the double bond that is inserted nearest the carboxyl or the methyl terminus of the fatty acid (Reddy et al., 1993; Sakamoto et al., 1994; Murata and Wada, 1995; Sakamoto and Bryant, 1997). The soluble desaturases, for instance, acyl-ACP desaturases, present in the plastidial stroma of plants, utilize ferredoxin as the electron donor. They incorporate double bonds in fatty acids that are esterified with acyl carrier protein (ACP). The best example is the Δ9 acyl-ACP desaturase, which catalyzes desaturation of stearic to oleic acid in the stroma of chloroplasts. This desaturase enzyme predominantly converts saturated fatty acids to unsaturated ones in vegetable oils (Slabas and Fawcett, 1992). The crystallographic examination of stearoyl-ACP desaturase isolated from castor seeds (Ricinus communis) indicates that the desaturase forms a di-iron-oxo active center with the two iron atoms bound in the symmetric structure (Figure 1). The two iron atoms interact with O2 with a consensus-binding motif of [(D/E)X2H2] (Fox et al., 1993; Los and Murata, 1998; Shanklin and Cahoon, 1998). The deep channel of an extended surface may be the fatty acyl chain binding site and the substrate, stearic acid in this channel places the Δ9-carbon atom in the neighborhood of one of its iron ion (Lindqvist et al., 1996).
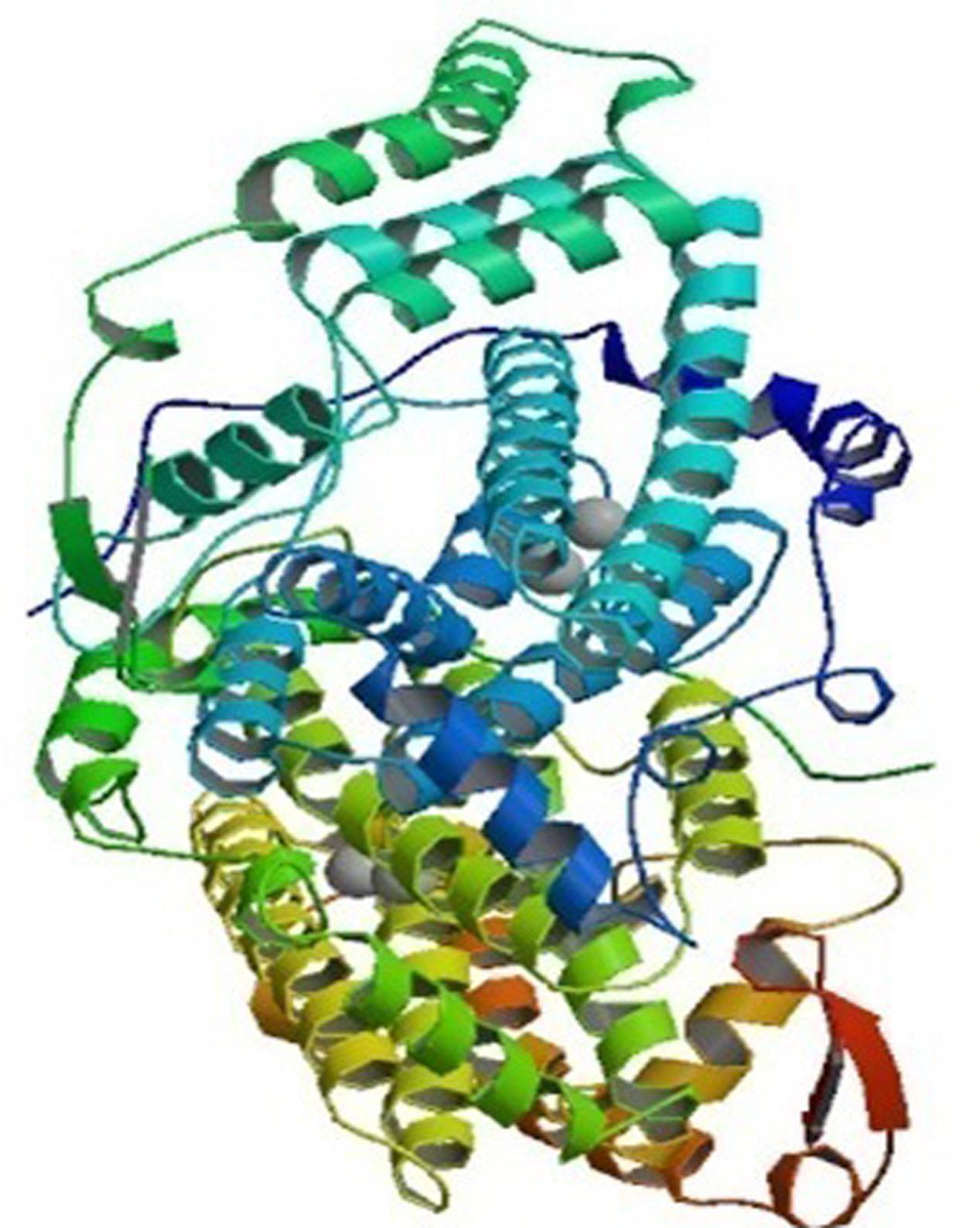
FIGURE 1. The crystal structure of stearoyl acyl protein desaturase of castor bean. The 2.40 A resolution structure shows two metal ions representing Fe-II (spheres) located at the center of the 4-helical bundle. Picture obtained from the RCSB PDB with ID 1AFR (Source: www.rcsb.org) (Lindqvist et al., 1996).
Membrane-bound desaturases, such as the Δ12- and Δ15-desaturases convert oleate into linoleate and linolenate, respectively. Since these desaturases are ER integral membrane proteins and are quite difficult to isolate and characterize by biochemical methods. The information about plant desaturases has been obtained by isolation and characterization of a series of mutants involved in lipid biosynthesis in Arabidopsis thaliana. There are four chloroplast desaturase loci: FADA, FADB, FADC and FADD, which are also called as FAD4, FAD5, FAD6, and FAD7, respectively (Ohlrogge and Browse, 1995). The microsomal oleate Δ12-desaturase or fatty acid desaturase 2 (FAD2; EC 1.3.1.35) is a hydrophobic transmembrane ER protein, acting on fatty acids, inserting a cis double bond between the C12 and C13 position of monounsaturated oleic acid, thereby producing polyunsaturated linoleic acid (Ohlrogge and Browse, 1995; Shanklin and Cahoon, 1998; Somerville et al., 2000). FAD2 is the key enzyme accountable for biosynthesis of polyunsaturated fats in non-photosynthetic tissues, for instance, roots and developing seeds of oilseed plants (Miquel and Browse, 1992; Zhang et al., 2012). The FAD2 in the ER utilizes phospholipids as substrates with NADH, NADH-cytochrome b5 reductase, and cytochrome b5 as electron donors. On the other hand, the plastidial oleate desaturase (FAD6) primarily uses glycolipids as acyl carriers, and ferredoxin reduced by ferredoxin-NAD(P) reductase as electron donors (Hernández et al., 2009). The FAD2 gene was first reported in Arabidopsis with a single copy, which is constitutively expressed (Okuley et al., 1994; Beisson et al., 2003). Further studies have identified more than one FAD2 gene in a variety of crops, such as sesame (Sesamum indicum), corn (Zea mays), canola (Brassica napus), olive (Olea europaea), soybean (Glycine max), sunflower (Helianthus annuus), and cotton (Gossypium hirsutum) (Jin et al., 2001; Pirtle et al., 2001; Kinney et al., 2002; Hernández et al., 2005; Li et al., 2007; Rolletschek et al., 2007; Kargiotidou et al., 2008). FAD2 is 1,164 bp long with an open reading frame coding for about 387 amino acids. The FAD2 gene consists of a single large intron in the 5′-untranslated region (UTR), which is evolutionarily conserved. However, the exon number may vary across the plant species, for example, Arabidopsis, castor bean, and soybean had only one exon, in contrast, Indian mustard contains two (Okuley et al., 1994; Chi et al., 2011a; Sharma and Chauhan, 2012; Suresha and Santha, 2013). The intron could be important for transcriptional regulation of FAD2 gene expression. The FAD2 gene has been classified into four types, namely, FAD2-1, FAD2-2, FAD2-3, and FAD2-4 on the basis of their site and pattern of expression. The four variations of the FAD2 gene show high sequence similarity, but show differences in their expression patterns and functions in fatty acid modification (Kongcharoensuntorn, 2001). The FAD2-1 is a seed-specific desaturase that synthesizes polyunsaturated fatty acids in young seed and developing flower buds (Liu et al., 2001). FAD2-2 is expressed at a low level from vegetative stage to maturing phase during seed development (Pirtle et al., 2001). FAD2-2 is the major gene responsible for the synthesis of linoleic acid (Hernández et al., 2009). FAD2-3 and FAD2-4 synthesize mostly polyunsaturated fatty acids almost in all the tissues. It was reported that FAD2-4 has 98% similarity with the FAD2-3 polypeptide (Zhang D. et al., 2009). Similarly, FAD6 is also ω-6 desaturase, synthesizing linoleic acid from oleic acid in plastids, unlike FAD2 in ER. The ω-3 desaturases, such as FAD3, FAD7 and FAD8 synthesize linolenic acid (C18:3) from linoleic in the ER (FAD3) and plastids (FAD7 and FAD8) (Gibson et al., 1994; Berberich et al., 1998). The FAD3 is a microsomal enzyme located in the ER facing the cytosol, whereas FAD7 and FAD8 are plastidial enzymes located in the inner membrane of the chloroplast envelope. In addition, FAD7 can also be found in the thylakoid of the chloroplasts (Andreu et al., 2007; Bhunia et al., 2016). In contrast to this, FAD4 and FAD5 produce C16:1 from C16:0 in particular for phosphatidyl glycerol and mono-galactosyldiacylglycerols (MGDG), respectively (Murphy and Piffanelli, 1998).
About 20 distinct motifs were identified in fatty acid desaturases. These motifs mainly belong to the transmembrane region of unknown complexity. Proteins of the FAD2 subfamily contain the motifs 2, 5, 6, 15, 16, and 17. The FAD2 enzyme contains 6 transmembrane domains and 8 conserved histidine residues in three clusters (HXXXH, HXXHH, and HXXHH), harboring eight iron-binding domains necessary for reduction of oxygen during desaturation, and is characteristic of all membrane-bound desaturases (Okuley et al., 1994; Shanklin et al., 1994) (Figure 2). It was found that four (valine, alanine, leucine, valine) out of the eight amino acids (threonine, histidine, valine, alanine, histidine, histidine, leucine, valine) belonged to hydrophobic residues in the third histidine cluster, which implied that these hydrophobic residues may be located in the interior of the active site of the enzyme. The top ten amino acids were all found to be leucine in the second histidine cluster (Tanhuanpaa et al., 1995; Ge et al., 2015). This structure might be one of the active sites of the FAD2 enzyme. Moreover, threonine was identified as the last three residues in the second histidine cluster and as the last four residues in the third histidine cluster, which play an important key role in the desaturation and hydroxylation of the FAD2 enzyme (Broadwater et al., 2002; Ge et al., 2015). The histidine boxes contain the catalytic center, which forms ligands to a diiron cluster, and exchange of a histidine with a different amino acid disrupts the function of desaturase (Shanklin et al., 1994; Shanklin and Cahoon, 1998; Guan et al., 2012b). The amino acid sequence of FAD2 is almost similar in most plants (Tao et al., 2006). Bioinformatics tools didn’t show any N-terminal signal peptide for probable localization of the FAD2 protein in organelles, like Golgi body, chloroplasts, and mitochondria. The C-terminal signaling motif (YKNK) allows the FAD2 protein to bind selectively to and integrate into the ER (Nielsen et al., 1997; Schell, 1998; McCartney et al., 2004; Nayeri and Yarizade, 2014).
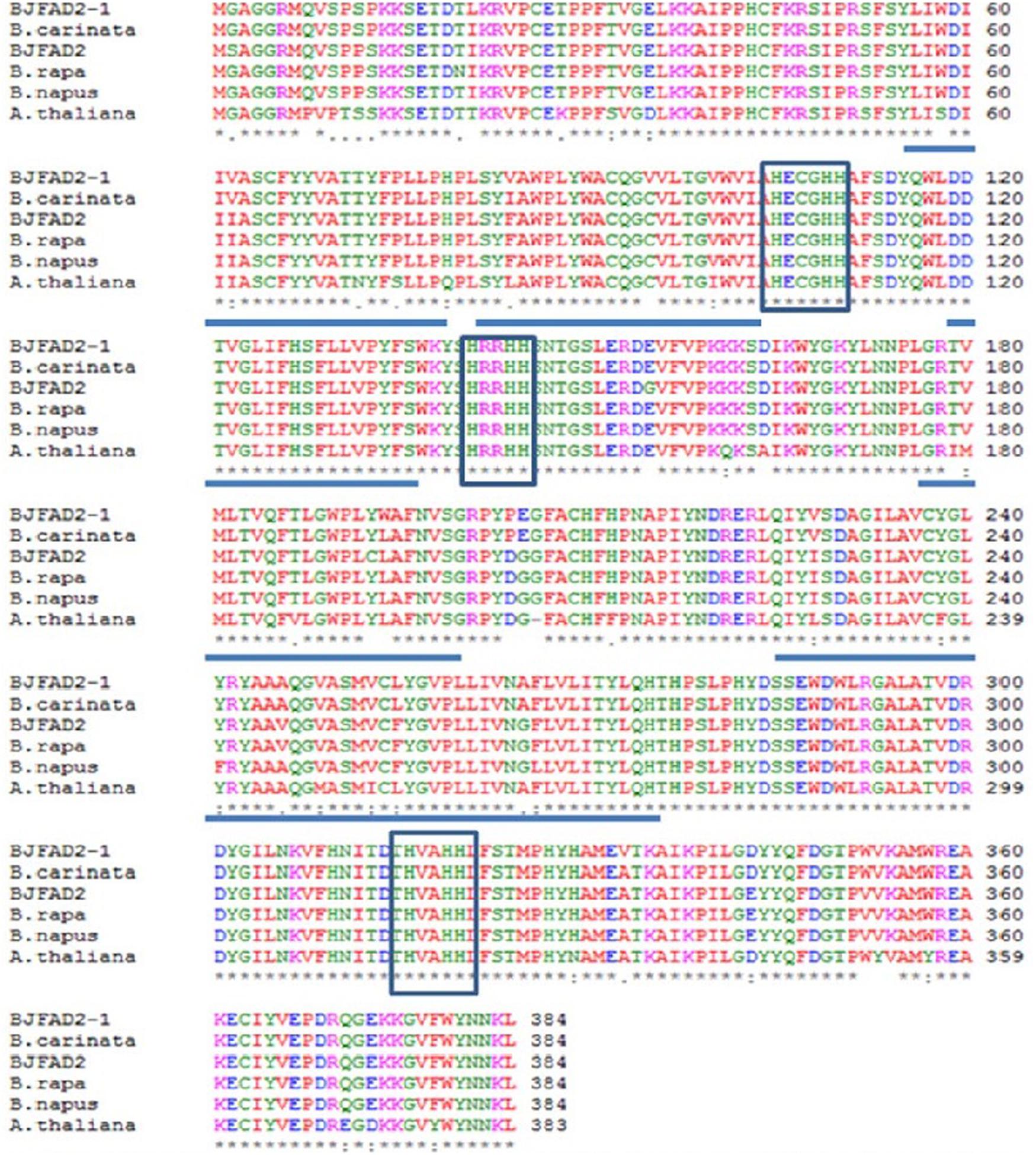
FIGURE 2. Comparative amino acid sequence alignment of FAD2 genes in Brassica juncea(X91139), B. rapa (AJ459107), B. napus (AF243045), B. carinata (AF124360) and Arabidopsis thaliana (L26296). Boxes represent three histidine motifs (HXXXHH, HXXHH and HXXHH) and underlined regions represent five hydrophobic regions (Image taken from Suresha et al., 2012) (This picture is reproduced here after taking permission from copyright holder).
As we have studied different types and subtypes of FAD genes in plants, it would be now interesting to know the evolutionary relationship of these genes in different crops. This would make us understand, how much they are diverged from other FAD genes that may help us in identifying their origin and existence.
The phylogenetic relationships of FAD2 genes were explained clearly by alignment of their coded amino acid sequences with other FAD genes of oil seed crops. A dendrogram was constructed that included all plant oleate desaturase enzymes, either plastidial (FAD6) or microsomal (FAD2). As shown in Figure 3, the coding sequences of ω-6 fatty acid desaturase in plants were classified into three main groups, namely, house-keeping FAD2, seed type FAD2 and FAD6 (Hernández et al., 2005; Guan et al., 2012a,b; Suresha et al., 2012). The huge separation between branches of FAD2 and FAD6 showed that they had diverged during early gene evolution (Reiser et al., 2000). The FAD2 seed-type genes have evolved independently after segregation by duplication from constitutive expressed FAD2 genes (Martínez-Rivas et al., 2001). This also suggests that the ancestral FAD2 gene had diverged prior to speciation, and the diverged FAD2 genes with the same function were probably found to be either seed-type or constitutive (Lee et al., 2012).
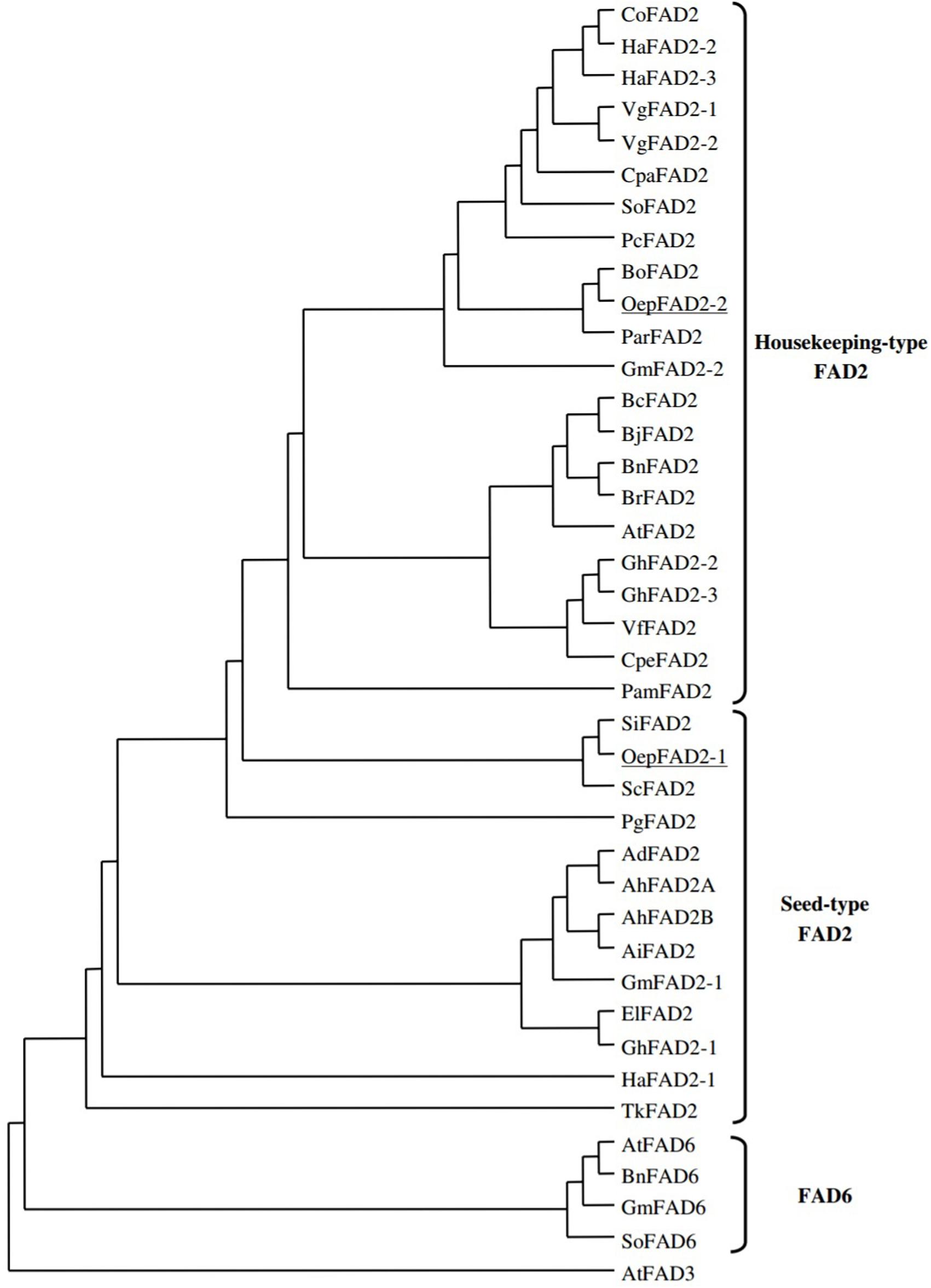
FIGURE 3. Phylogenetic analysis of FAD2 and FAD 6 enzymes. Accession numbers of the different desaturases included in the analysis are Arabidopsis thaliana (AtFAD2. L26296; AtFAD3; D17579; AtFAD6, U09503), Arachis duranensis (AdFAD2: AF272951), Arachis hypogaea (AhFAD2A, AF030319; AhFAD2B, AF272950), Arachis ipaensis (AiFAD2: AF272952), Borago officinalis (BoFAD2: AF074324), Brassica carinata (BcFAD2: AF124360), Brassica juncea (BjFAD2: X91139), Brassica napus (BnFAD2; AF243045; BnFAD6: L29214), Brassica rapa (BrFAD2: AJ459107), Calendula officinalis (CoFAD2: AF343065), Crepis palestina (CpaFAD2; Y16284), Cucurbita pepo (CpeFAD2: AY525163), Euphorbia lagascae (E1FAD2, AY486148), Glycine max (GmFAD2-l, L43920; GmFAD2-2, L43921; GmFAD6, L29215), Gossypium hirsutum (GhFAD2-l, X97016; GhFAD2-2, Y10112; GhFAD2-3, AF331163), Helianthus annuus (HaFAD2-1, AF251842; HaFAD2-2, AF251843; H3FAD2-3, AF251844), Persea americana (PamFAD2, AY057406), Petroselinum crispum (PcFAD2: U86072), Punica granatum (PgFAD2, AJ437139), Sesamum indicum (SiFAD2, AF192486), Solanum commersonii (SoFAD2; X92847), Spinacia oleracea (SoFAD2; AB094415; SoFAD6; X78311), Trichosanthes kirilowii (TkFAD2; AY188445), Vernicia fordii (VfFAD2, AF525535), Vernonia galamensis (VgFAD2-1, AF188263; VgFAD2-2, AF188264) (Picture taken from Hernández et al., 2005) (This picture is reproduced here after taking permission from copyright holder).
It would be interesting to look out on, how these divergent FAD genes regulate fatty acid desaturation in plants and carry out the specific functions. The transcriptional control of the FAD genes occurs through the promoter DNA and promoter DNA-specific regulatory proteins. The regulation has been clearly explained in sesame by Kim et al. (2006). The transcriptional control of the FAD2 gene during seed development is spatially and temporally regulated. The region from -179 to -53 in the sesame FAD2 promoter contained positive cis-elements for FAD2 gene expression. On the other side, the FAD2 promoter region from -547 to -180 region harbors negative cis-elements for the gene repression. The eight potential cis-elements in developing seeds that regulate the gene expression are (CA)2 element, E-box (CANNTG), CCAAT box, ABRE motif (ACGTGKC), G-box (CACGTG), G-box-like element (ACGT), Prolamin-box (AAAG), and RY repeat element (CATGCA) (Figure 4). The SeFAD2 gene expression revealed that abscisic acid (ABA) was responsible for the regulation during seed development. In the SeFAD2 promoter, ABA-responsive elements were found from the region of -660 to -548 and -179 to -53 (Kim et al., 2006). The FAD7 promoter contains cis-acting elements, such as BoxII (GT-1 sites) and G-box-like (CCACTTGG) motifs that are members of light-responsive promoters (Guilfoyle, 1997). Different cis-regulatory elements in FAD2 promoter are involved in the abiotic and biotic stress responses, and influence the control of gene expression, specifically in seeds (Nayeri and Yarizade, 2014).
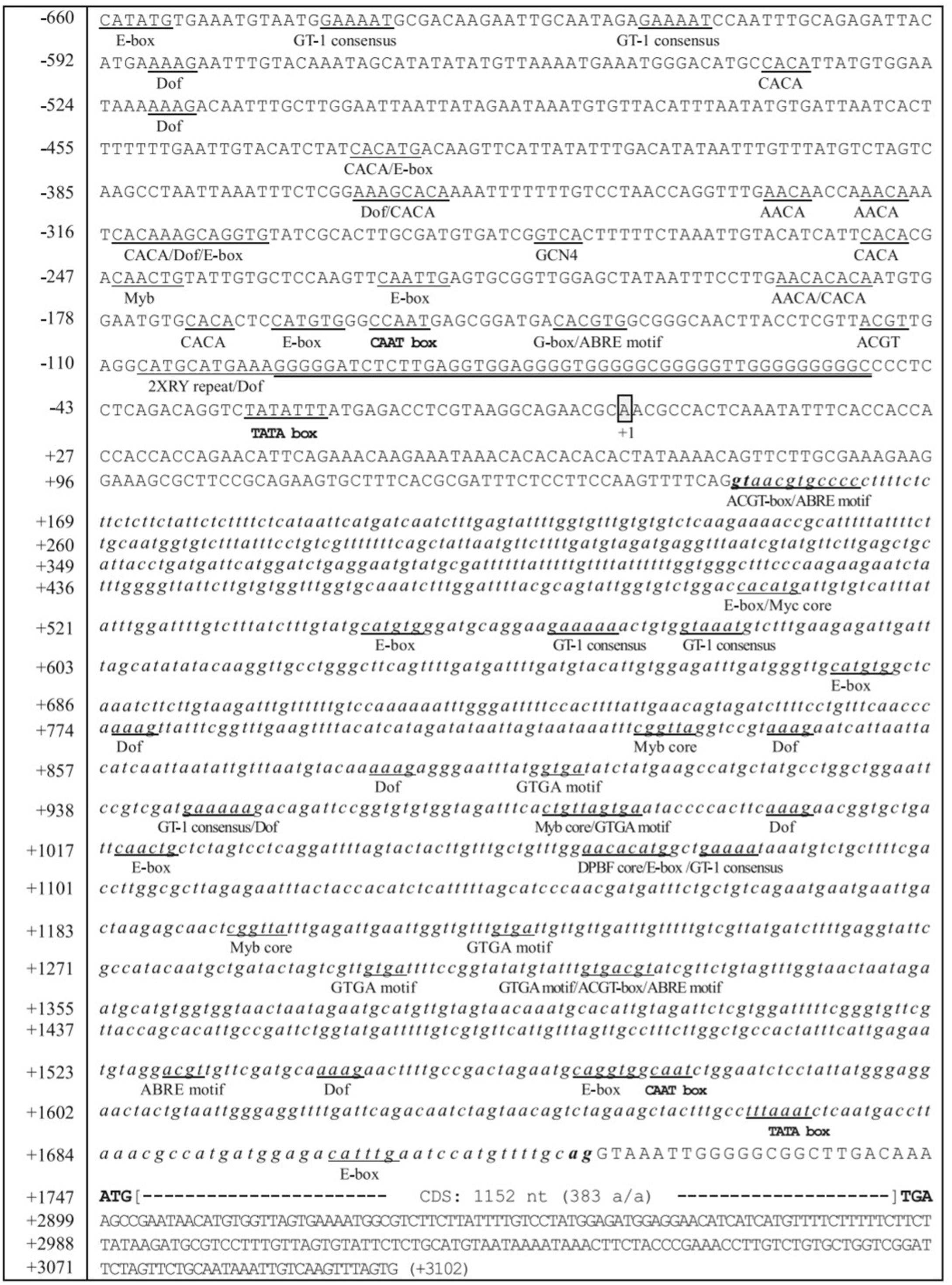
FIGURE 4. Nucleotide sequences of the 5′-flanking region, intron and regulatory elements of the Sesamum FAD2 gene. A box represents the transcription initiation site designated by the + 1 position. The numbering on the left refers to the nucleotide sequences. The sequences of the SeFAD2 intron are shown in small italic letters. The putative TATA box occurs between -25 and -31. Translational initiation (ATG) and termination codon (TGA) are in bold, and the coding region of the SeFAD2 gene (1,152 nucleotides; 383 amino acids) is shown by large brackets. Several potential regulatory cis-elements are underlined and designated with the names of each of the motifs. G-rich sequences are double-underlined with thick appearance between -49 and -96 (Image taken from Kim et al., 2006) (This picture is reproduced here after taking permission from copyright holder).
It was also reported that the regulatory mechanism based on introns was also involved in the expression of plant FAD2 genes (Kim et al., 2006). Xiao et al. (2014) reported that both promoter and intron are involved in controlling the expression of FAD2 gene in Brassica napus. The quantitative trait locus (QTL) analysis and genome-wide association studies (GWAS) using the multiparent advanced generation intercross (MAGIC) population in Arabidopsis, suggested that ω-6 desaturation can be largely controlled by cis-acting sequence variants of the FAD2 intron (Menard et al., 2017).
The regulation of FAD2 gene may be important in understanding the fatty acid composition of plant membranes and membrane fluidity in the cold tolerance of plants (Kongcharoensuntorn, 2001). Fatty acid desaturase genes in plants normally undergo different types of regulation by temperature, light, and wounding.
Temperature is a major environmental factor that regulates fatty acid desaturation in plants. The regulation of gene expression appears to vary with the species, tissue, and gene. The effect of temperature on the FAD gene expression levels was well studied in olive fruit (Hernández et al., 2009, 2011). At 15°C, a small increase of FAD6 transcript levels was observed with a temporary induction of FAD2 genes (slight for FAD2-1 and intense for FAD2-2). But when the temperature was increased from 15°C to 35°C, the expression of FAD2 genes was decreased. Similarly, an upregulation of FAD2 gene expression by cold stress was observed in avocado fruits and cotton cotyledons (Wang et al., 2004; Kargiotidou et al., 2008; Teixeira et al., 2009). On the contrary, when Arabidopsis and soybean cultures were grown under low temperatures, no such significant differences were observed in expression levels of FAD2 and FAD6 genes (Okuley et al., 1994; Heppard et al., 1996). Low temperature normally increases the content of polyunsaturated fatty acids that maintain the fluidity of membranes in cold climate (Los and Murata, 1998; Routaboul et al., 2000). The fad6 mutant of Arabidopsis had a high level of monounsaturated fatty acids and reduced levels of polyunsaturated fatty acids in membrane lipids, and mutants, such as fad5, fad6, and fad3: fad7: fad8 were found more vulnerable to photoinhibition as compared to wild-type plants after chilling treatment (Zhang et al., 2012). Recently, Botella et al. (2015) depicted the role of ALA10, a P4 type-ATPase that interacts with FAD2 gene and affects fatty acyl desaturation by downregulating the FAD2 and FAD3 activities in Arabidopsis leaves. The effect of ALA10 on leaf development was found significant, when plants were grown at chilling temperatures (10°C). These studies showed a significant up-regulation of FAD2 or FAD3 isogenes at a lower temperature, whereas some studies have also observed either down-regulation or no appreciable change.
The change in gene expression levels and protein content with temperature was observed in case of Δ9, Δ12, and Δ15 desaturases (Vega et al., 2004; Teixeira et al., 2009, 2010). It was reported that two domains in FAD2-1 were important in mediating temperature-dependent stability of FAD2-1A isoform. In addition, the N-terminus of FAD2-1 and FAD3 was shown to be important in the temperature-dependent turnover of proteins. The FAD2 and FAD3 proteins were thus less stable and hence, less abundant at high temperatures (Khuu et al., 2011). Domain-swapping and mutagenesis experiments revealed that each protein contained a degradation signal in its N-terminus, and the PEST-like sequence within this region was largely responsible for the rates of protein turnover. A PEST is a signal sequence present on a protein that confers rapid protein degradation at high temperatures, usually via the ubiquitination/26S proteasome (UPS) pathway. The E3 ligases seem to play an important role in determining the substrate specificity of proteins degraded through this pathway (Elsasser and Finley, 2005). In addition, the protein degradation requires other specific components of the ER including the Cdc48 adaptor proteins Doa1, Shp1, and Ufd2. It was thus clearly indicated that FAD2 and FAD3 protein abundance was regulated by a combination of cis-acting degradation signals and the UPS pathway, and the modulation of these protein amounts in response to temperature may represent one mechanism of homeoviscous adaptation in plants (Schrader et al., 2009; O’Quin et al., 2010).
The other way of regulating desaturase activity was by post-transcriptional mechanisms as observed in wheat roots for FAD3, FAD8 in Arabidopsis leaves, and seed-specific FAD2-1 in soybean (Horiguchi et al., 2000; Matsuda et al., 2005; Tang et al., 2005). Li et al. (2007) found that FAD2-3 increased di-unsaturated fatty acids under cold stress due to post-transcriptional/post-translational modifications of gene rather than increase in mRNA levels. Promoter analysis of FAD-2 gene showed that the response of the gene under low temperatures could be due to the interaction of various factors generating signaling pathways, which would then determine gene response and plant adaptation to the new condition. The activation of the acclimation genes induced by ABA takes place by ABA-dependent and ABA-independent pathways. The promoter analysis of the FAD2 from G. hirsutum and A. thaliana indicated that ABRE1/2 and ABRE3 elements were present in the promoters of FAD2-3 and FAD2-4 of G. hirsutum, but absent in the promoters of FAD2 from A. thaliana (Kargiotidou et al., 2008).
The glycerolipid pathway in Arabidopsis was also involved under temperature stress and was largely acting by rebalancing of the two pathways. In Arabidopsis thaliana, the prokaryotic (chloroplast) pathway was upregulated in response to low temperature that induced MGDG biosynthesis, whereas high temperature had increased di-galactosyldiacylglycerols (DGDG) biosynthesis by redirection of acyl channeling through the eukaryotic pathway (ER) (Li et al., 2015). It was reported that serine-185 of FAD2-1 enzyme was phosphorylated by calcium-dependent protein kinases (CDPKs) during seed development. The serine motif was found conserved in the amino acid sequence of many plant FAD2 enzymes. The expression studies showed that phosphorylation had downregulated enzyme activity, but there was no connection found between serine-185 phosphorylation and temperature regulation (Huang and Huber, 2001; Tang et al., 2005).
Light is another factor that affects desaturation of fatty acid in plants. A light-dependent increase of polyunsaturated fatty acids was reported in photosynthetic tissues. There was an increase in linoleic and α-linolenic acids in cotyledons of cucumber in response to light (Murphy and Stumpf, 1979), however, light had elevated only the α-linolenic acid level in oat leaves and Arabidopsis callus (Ohnishi and Yamada, 1983; Brockman et al., 1990). To confirm the effect of light on fatty acid desaturation, soybean cell cultures and olive fruits were kept under darkness, such effect decreased FAD3 and FAD8 expression levels in former and decreased FAD2 expression levels in latter, hence indicating a light-dependent transcriptional regulation of FAD genes (Collados et al., 2006; Hernández et al., 2011). It was also reported that the expression of FAD2-3 and FAD2-4 genes under cold stress was found light-dependent and was because of an indirect hormonal effect or a direct effect of light regulatory elements on the FAD2-3 and FAD2-4 promoters. It is known that light inhibits ethylene synthesis, which in turn affects auxin distribution. Thus the influence of light on gene expression under cold stress could be regulated by the ethylene/auxin gradient, which in turn, is regulated by light (Vandenbussche et al., 2003; Kargiotidou et al., 2008).
Plants use linoleic and α-linolenic acids as signaling precursor molecules for defense system against pathogen attack and wounding (Farmer, 1994). The FAD7 gene induced by wounding with the parallel increase of α-linolenic acid was reported in many plants (Teixeira et al., 2010). Further, the increase of FAD2 and Δ9 stearoyl-ACP desaturase levels was also reported in avocado fruits when infected with Colletotrichum (Wang et al., 2004), and the induction of parsley FAD2 and FAD7 genes by a fungal elicitor (Kirsch et al., 1997a,b). The FAD2 enzyme was thought to be involved in the wounding reaction by increasing the biosynthesis of linoleic and palmito-linoleic acid in the ER of olive fruit. It was reported that FAD2-1 and FAD2-2 expression levels were slightly increased on the injury, whereas FAD6 transcript levels were not altered in response to wounding. In addition, the presence of palmito-linoleic acid was also noticed in microsomal lipids, but not in plastids after wounding (Hernández et al., 2011). Such reports clearly indicate that FAD genes are also regulated by wounding and pathogen attack.
The research on FAD genes has been progressed well from the past to present. Here in this column, we will give some idea about isolation and characterization of FAD genes, especially FAD2 gene from the year 1990 to 2016 (Table 1). The techniques often used for conducting such study were reverse transcription-polymerase chain reaction (RT-PCR), rapid amplification of cDNA ends (RACE), and real-time quantitative PCR (RT-qPCR). The FAD2 gene was first cloned and identified in Arabidopsis by the T-DNA tagging method (Okuley et al., 1994). Hernández et al. (2009) studied the correlation between the expression level of FAD2 and FAD6 genes with the linoleic acid level in olive fruit. They observed an increase of linoleic acid with the increase in FAD2-2 gene expression level, and thus concluded that FAD2-2 is the main gene responsible for the linoleic acid content. To elucidate the connection of Δ12 FAD in salt and freezing tolerance, Lu et al. (2010) identified the role of FAD6 gene in low temperature and high salinity acclimatization in Antarctic microalga, Chlorella vulgaris NJ-7. The three FAD2 genes that were isolated and characterized in Camelina sativa were tissue and developmentally regulated (Kang et al., 2011). Chi et al. (2011a) identified orthologous genes from soybean encoding fatty acid desaturases. The identification of these desaturases is useful for the reconstruction of the pathways concerned in the biosynthesis of unsaturated fatty acids. Zhang et al. (2011) cloned and characterized cDNAs encoding fatty acid desaturases from lima beans, and their expression patterns were investigated in different tissues under various stress conditions, especially low temperature. Their results provided a better understanding of regulation and structure of the desaturase genes in plants. Guan et al. (2012c) used microarray technology for determining the distinct gene clusters linked with the oleic acid synthesis in rapeseed. The comparison of gene expression profiles between high and low-oleic acid genotypes grouped a large number of differentially expressed genes linked with the high-oleic acid trait. This provided a basis for further studies on the mechanisms behind oleic acid synthesis and accumulation. Lee et al. (2012) reported the cloning and characterization of two distinct FAD2 genes from Vitis labrusca, and revealed their differential expression patterns. The FAD2 genes from V. labrusca functionally complemented the FAD2 mutation in Arabidopsis. Suresha et al. (2012) isolated FAD2 gene through RT-PCR technique from Brassica juncea, and studied its tissue- and growth temperature-dependent expression. They observed that FAD2 gene is developmentally regulated with increased expression in mid-maturation stage as compared to early and late stages of seed development. The results also showed one-fold higher and three fold lower expression rate of fad2 under the lower and the higher temperature, respectively. Cao et al. (2013) isolated and characterized a FAD2 gene family from safflower by RT-qPCR. The phylogenetic analysis reported their non-allelic nature and was evolved by gene duplication. The functional divergence of the FAD2 family was discovered by heterologous and transient expression in yeast and Nicotiana benthamiana, respectively. Chen et al. (2015a) cloned FAD2 from Vernicia fordii and transformed in Rhodotorula glutinis, and observed the synergistic effect on unsaturated fatty acid metabolism. Recently, Sun et al. (2016) cloned and identified the role of microsomal FAD2 from Elaeis guineensis. They reported that FAD2 used only oleic acid as a substrate and converted it to linoleic acid. The characterization of the above FAD genes will help the researchers in the construction of the pathways involved in biosynthesis of unsaturated fatty acids, and can provide many candidate genes for the genetic engineering of stress tolerance in plants.
Genetic engineering is a recent useful technique of crop improvement that produces novel plant varieties. The genetic engineering with the aim of genetic improvement was achieved by silencing the FAD2 gene to increase oleic acid and simultaneously to reduce linoleic acid. This was reported in various plants, such as canola (Stoutjesdijk et al., 2000), cotton (Liu et al., 2000), groundnut (Yin et al., 2007), Jatropa (Utomo et al., 2015), and linseed (Chen et al., 2015b). It was reported that the plastidic desaturase activities are partly accountable for the instability of the high-oleic acid FAD2-silenced plants (Zhang et al., 2016). Marine algae were known to possess certain functional activities that help in the lipid metabolism, and hence can be useful for human and animal nutrition. Norambuena et al. (2015) conducted such type of study that assessed the potentials of two commercially available algae for including them into diet of Atlantic salmon fish. They reported increase in PUFA content in whole body of fish that displayed the importance of bioactive carotenoids found in algae. However, the observed increase of PUFA content in fish tissues was statistically significant, but from a nutritional point of view was considered minimal for consumers.
The FAD2 gene expression has provided many new challenges and opportunities for studying the gene in the enhancement of oil quality. The FAD2 gene is expressed throughout the plant kingdom and is identified by Northern blot and RT PCR techniques. The functions of both FAD2-1 and FAD2-2 in soybean were investigated by Heppard et al. (1996). They reported that the FAD2-1 gene was expressed strongly in developing seeds, whereas FAD2-2 gene was constitutively expressed in both developing seeds and vegetative tissues. It was reported that FAD2-2 was mainly responsible for the conversion of oleic to linoleic acid (Jin et al., 2001). The FAD2-1 in Olea europaea desaturates storage lipids in young seed, whereas FAD2-2 desaturates lipids in maturing seeds and mesocarp (Hernández et al., 2005). Semi-quantitative RT-PCR was used for determining the expression pattern of FAD2 gene in maize. The expression of FAD2 was found higher in immature embryos than leaves, stem, and roots (Tao et al., 2006). The FAD2-4 and FAD2-3 genes were found to be expressed in all tissues of the cotton plant, such as seeds, seedlings, roots, stems, leaves, developing flower buds, and ovule fibers. This constitutive expression pattern was found different from the FAD2-1 gene, which was limited to flower buds and seeds. However, the expression of a FAD2-2 gene was expressed in all other tissues, but hardly found in hypocotyls, roots, and stems (Zhang D. et al., 2009). The results of quantitative RT-PCR found that the transcript levels of FAD2 and FAD2-1 in Arachis hypogaea L. were elevated in seed than the other tissues. Alternatively, the transcript level of FAD2-2, FAD6, and SLD1 (sphingolipid Δ8 desaturase) were higher in leaves. These genes had different expression patterns with different biochemical functions throughout seed development and vegetative growth (Chi et al., 2011b). Suresha et al. (2012) reported that FAD2 was expressed constitutively in all the tissues of Brassica juncea and was developmentally regulated during oil biosynthesis. The expressions of FAD2-1 and FAD2-3-1 in Camelina sativa were mainly reported in flowers and seeds (Kim et al., 2014). Transcript profiling of six microsomal desaturase genes, such as SAD1, SAD2, FAD2, FAD2-2, FAD3A and FAD3B was performed by real-time PCR (Rajwade et al., 2014). SAD2 was the most highly expressed gene found throughout all stages of seed development. The expression of SAD1 was lower than SAD2 and remained constant throughout all developmental stages. Analysis of FAD2 transcript distribution in peanut found that the FAD2-1 gene had a 70-fold higher expression rate in developing seeds than the FAD2-2 gene, but the FAD2-2 gene was abundantly expressed in flowers (Wang et al., 2015). Recently, Dong et al. (2016) reported that the FAD2 gene in Cucumis sativus L. was expressed in all tissues, while all other FAD genes were found expressed mainly in the cotyledons and leaves. The story of FAD2 gene continues also hereafter its expression in order to play a key role in fatty acid synthesis, plant development, cold and salt tolerance for the plant survival.
The two major and dominant biosynthetic pathways, prokaryotic and the eukaryotic, work together in plant cells for the synthesis of glycerolipids and polyunsaturated fatty acids (Browse and Somerville, 1991). The biosynthetic pathway of fatty acids and lipids in plants is well presented in Figure 5. Many enzymes in fatty acid biosynthesis had been biochemically characterized, as well as the encoding genes from plants like Brassica napus and Arabidopsis thaliana (Slabas and Fawcett, 1992; Ohlrogge and Browse, 1995; Beisson et al., 2003; Wang et al., 2010). Fatty acids are normally synthesized by a fatty acid synthase (FAS) complex located in the plastids that uses acetyl-CoA as a precursor and malonyl-ACP as an elongator. The malonyl-thioester undergoes a series of condensation reactions with an acetyl-CoA catalyzed by a 3-ketoacyl-ACP synthase-III (KAS-III) that produces propionyl-ACP (C4:0-ACP). Subsequent condensation reactions takes place up to the formation of palmitoyl-ACP (C16:0-ACP) catalyzed by a KAS-I isoforms, and finally KAS-II elongates the C16:0-ACP to stearoyl-ACP (C18:0-ACP) (Harwood, 1996). The Δ9-stearoyl-ACP desaturase then converts most of the stearoyl-ACP to oleoyl-ACP (C18:1-ACP). Here C18:1-ACP after conversion to oleic acid (C18:1) gets desaturated to linoleic acid by the ω-6 fatty acid desaturase (FAD6) that is further desaturated to linolenic acid by the ω-3 fatty acid desaturase (FAD7/FAD8) (Shanklin and Cahoon, 1998; Hernández et al., 2011; Zhang et al., 2012).
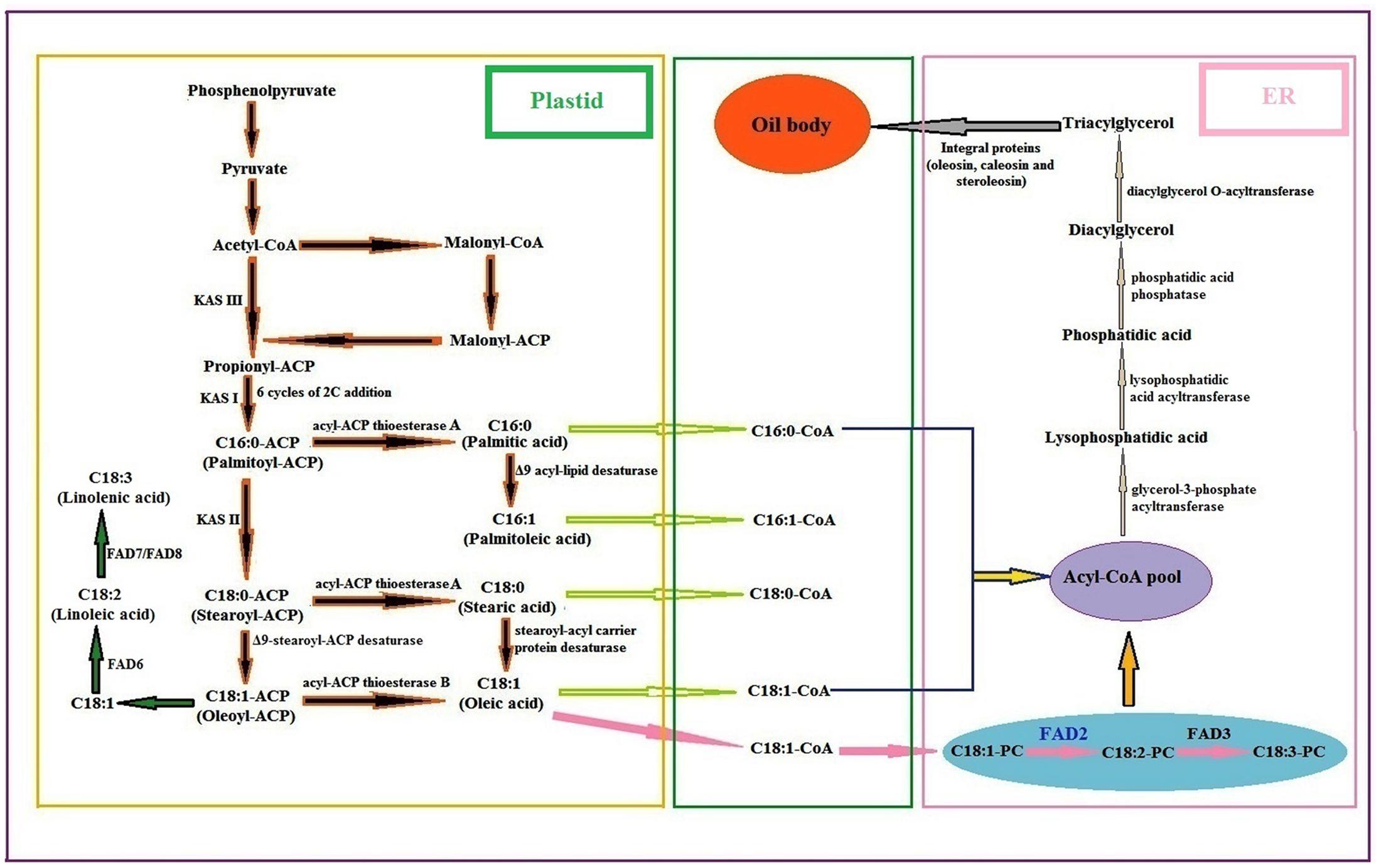
FIGURE 5. General biosynthetic pathway of fatty acids and lipids in plants [PC phosphocholine; FAD fatty acid desaturase: KAS ketoacyl-ACP synthase] (Guan et al., 2012c: Suresha et al., 2012; Bhunia et al., 2016).
After fatty acid synthesis in plastids, the fatty acyl-ACP moieties, mainly palmitoyl, stearoyl and oleoyl-ACPs are either used directly for lipid biosynthesis in the plastid or can be hydrolysed by fatty acyl-ACP thioesterases (FATA or FATB) to free fatty acids. They are later exported to ER in the form of acyl-CoA pool. The acyl-CoA pool undergoes several modifications regarding elongation, desaturation and exchange, catalyzed by a different ER membrane bound proteins, which constitute the eukaryotic pathway of lipid biosynthesis. In the ER, C18:1-CoA is incorporated into membrane phosphatidylcholine (PC) and desaturated to C18:2-PC by the ω-6 fatty acid desaturase (FAD2), and then C18:2-PC is desaturated to C18:3-PC by the ω-3 fatty acid desaturase (FAD3) (Bhunia et al., 2016). The first acylation reaction catalyzed by a glycerol-3-phosphate acyltransferase (GPAT) yields lysophosphatidic acid (LPA), which is further acylated by a lysophosphatidic acid acyltransferase (LPAAT) to the phosphatidic acid (PA). PA is then converted to 1, 2-sn-diacylglycerol (DAG) by the action of phosphatidic acid phosphatase (PAP). Finally triacylglycerol (TAG) is synthesized from DAG by 1, 2-sn-diacylglycerol acyltransferase (DGAT) followed by packaging into oil bodies with the aid of many integral proteins as reported in sesame (oleosin, caleosin, and steroleosin) (Baud and Lepiniec, 2010; Tzen, 2012).
FAD2 overexpression modifies many physiological features in transgenic seedlings, like seed germination and hypocotyl elongation, but such effects could not be observed after transformation with genes other than fatty acid desaturases (Wang et al., 2010). It was reported that fad2 mutants formed dwarf phenotypes at a temperature of 22°C in Arabidopsis, when compared with the wild type plant (Miquel et al., 1993). The change in PUFA content due to FAD2 mutation affects development in plants through the salicylic acid (SA), oxidase, abscisic acid, and jasmonic acid (JA) pathways (Martínez-Rivas et al., 2000; Kachroo A. et al., 2005; Regente et al., 2008). The cytochrome c oxidase expression was normally found inhibited in A. thaliana cells of the wild-type and mutant fad2, when exposed to a low temperature, unlike than reported in FAD3 over expression lines (Martínez-Rivas et al., 2000). Kachroo A. et al. (2005) and Kachroo P. et al. (2005) reported that stearoyl-acyl carrier desaturase of A. thaliana mutants induced JA-responsive gene PDF1.2 resistant to Botrytis cinerea pathogen, whereas Regente et al. (2008) observed major changes in the extracellular phospholipids, when sunflower seeds were treated with JA and ABA. Further, it was observed that the fad2 mutant of B. napus has variable phenotypes, regarding leaf epidermal structure and permeability (Wang et al., 2003). This shows that the FAD2 mutation is in charge for the changes in agricultural phenotypes of plants by affecting their development.
The skill of adjusting membrane fluidity by varying the unsaturated fatty acid contents is characteristic of cold-responsive plants (Upchurch, 2008). Under cold-stress conditions, FAD2 is of particular interest because of their modifying ability to increase unsaturated fatty acids (Falcone et al., 2004; Tang et al., 2005). The induction of Δ9 desaturase in A. thaliana (Kreps et al., 2002) and enhanced rate of FAD2 gene expression in cotton was observed under cold stress environment (Kargiotidou et al., 2008). The cold stress in Arabidopsis also effected the transcription of the FAD2 isogenes (FAD2.2 and FAD2.1) (Gibson et al., 1994; Matsuda et al., 2005). Similar effect was observed in the olive drupes as there was an enhancement in FAD2.2 and FAD7 expression rate, and reduction in FAD2.1 and FAD6 in the first weeks of oil biogenesis (Matteucci et al., 2011). The mRNA levels of FAD2-3 and FAD2-4 increased, when temperatures were kept below the germination temperature (Kargiotidou et al., 2008). The treatment of cold stress also increased FAD2.2 and FAD7 mRNA levels in the epi-mesocarp cells of Canino and Moraiolo (Matteucci et al., 2011). The induction of other desaturase genes with the exception of FAD8 and FAD7, which were induced under low-temperature conditions, was not reported in higher plants (Berberich et al., 1998; Falcone et al., 2004; Matsuda et al., 2005).
The Arabidopsis fad6 mutant under chilling stress accumulated palmitoleic and oleic acids, and also thylakoid number was decreased in the chloroplast. In addition, the Arabidopsis fad2 mutant has a low level of polyunsaturates in the extra chloroplast membrane lipids, and plant withering may result after long exposure to low temperatures (Miquel et al., 1993). In the Synechocystis sp., the mRNA levels of Δ6, Δ12, and ω-3 desaturases increased about 10-fold on reducing the temperature from 34°C to 22°C, in contrast, mRNA levels of Δ9-desaturase remained unchanged (Los et al., 1997). The difference in the FAD2 expression patterns observed under cold stress conditions was correlated with the rise in unsaturated fatty acids, suggesting the direct role of the FAD2 genes in membrane adaptation to cold stress.
One of the environmental stresses, such as salt induces the changes in fatty acid composition and many fatty acid desaturases participate in this event. It was reported that FAD6 expression was responsive to osmotic and salt stress (Zhang J.T. et al., 2009). Tobacco plants transgenic with the antisense FAD7 gene had low levels of polyunsaturated fatty acids, and were found to be more sensitive to drought and salt stress (Im et al., 2002). In contrast, heterologous expression of sunflower FAD2-1 or FAD2-3 in yeast had higher levels of dienoic fatty acids and thus showed increased yeast cell tolerance to salt (Rodríguez-Vargas et al., 2007). The polyunsaturated fatty acid composition in fad2 mutants reduced the mobility of membrane lipids and as a result impaired the Na+/H+ pump function localized on the plasma membrane and tonoplast, when compared with vesicles isolated from control plants. In addition, the proton translocating activity of enzymes, such as V-ATPase, V-PPase and PM-ATPase was also reported lower in fad2 mutants. It was now obvious that the role of Na+/H+ antiporters was inhibited in fad2 mutants under salt stress conditions (Zhang et al., 2012). The FAD2 gene thus played a key role in regulating and maintaining the lipid composition of intracellular membranes, biophysical characteristics, and proper functioning of membrane-attached proteins in salt stress conditions (Deuticke and Haest, 1987; Cooke and Burden, 1990; Zhang et al., 2012).
Plants have many fatty acid desaturases, most of which are located in ER and chloroplast. FAD2 is an important desaturase enzyme, accountable for most of the polyunsaturated fatty acid synthesis in oilseed crops. The regulation of FAD2 gene is important in understanding the composition of fatty acids and biosynthesis, plant development, and essential role in biotic and abiotic stresses like cold and salt tolerance. The changes in oil composition of edible seeds, especially the oleic and linoleic acid content can be modified genetically by silencing the FAD genes for a number of applications in industry, human health, and nutrition. High level of oleic acid in oil is one of the favored traits in oil engineering due to its high stability and several applications. Efforts taken for modifying plants genetically to produce pleasing unsaturated fatty acids had modest success. The identification of molecular markers for QTL can be designed for breeding high oleic varieties that could make possible the development of high or low oil content with the high oleic acid character. It is imperative to understand the mechanisms of regulation of fatty acid genes that will provide a base to modify the fatty acid compositions of membranes for the improvement of vitality and vigor of oilseed crops. Further study should also be focused on discovering other important microsomal oleate desaturase genes, and the regulation of enzymes at transcription and post-transcription levels. Effective strategies should be designed in plants to increase the accumulation of conjugated fatty acids. The approaches of metabolic engineering of oil traits can be an effective strategy for generating substantial levels of oil in crop plants.
AD has downloaded literature and written the manuscript and finally edited it. AC has also helped in writing the manuscript. PK has assisted in downloading the material for the manuscript. NA has given the idea of contents.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge UGC SAP for financial support.
Andreu, V., Collados, R., Testillano, P. S., Risueño, M. C., Picorel, R., and Alfonso, M. (2007). In situ molecular identification of the plastid ω-3 fatty acid desaturase FAD7 from soybean: evidence of thylakoid membrane localization. Plant Physiol. 145, 1336–1344. doi: 10.1104/pp.107.109637
Arondel, V., Lemieux, B., Hwang, I., Gibson, S., Goodman, H. M., and Somerville, C. R. (1992). Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258, 1353–1355. doi: 10.1126/science.1455229
Baud, S., and Lepiniec, L. (2010). Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49, 235–249. doi: 10.1016/j.plipres.2010.01.001
Beisson, F., Koo, A. J. K., Ruuska, S., Schwender, J., Pollard, M., Thelen, J. J., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132, 681–697. doi: 10.1104/pp.103.022988
Berberich, T., Harada, M., Sugawara, K., Kodama, H., Iba, K., and Kusano, T. (1998). Two maize genes encoding ω-3 fatty-acid desaturase and their differential expression to temperature. Plant Mol. Biol. 36, 297–306. doi: 10.1023/A:1005993408270
Bhunia, R. K., Kaur, R., Mrinal, K., and Maiti, M. K. (2016). Metabolic engineering of fatty acid biosynthetic pathway in sesame (Sesamum indicum L.): assembling tools to develop nutritionally desirable sesame seed oil. Phytochem. Rev. 15, 799–811. doi: 10.1007/s11101-015-9424-2
Botella, C., Sautron, E., Boudiere, L., Michaud, M., Dubots, E., Yamaryo-Botté, Y., et al. (2015). ALA10, a phospholipid flippase, controls FAD2/FAD3 desaturation of phosphatidylcholine in the ER, and affects chloroplast lipid composition in Arabidopsis thaliana. Plant Physiol. 30:01557. doi: 10.1104/pp.15.01557
Broadwater, J. A., Whittle, E., and Shanklin, J. (2002). Desaturation and hydroxylation. Residues 148 and 324 of Arabidopsis FAD2, in addition to substrate chain length, exert a major influence in partitioning of catalytic specificity. J. Biol. Chem. 277, 15613–15620. doi: 10.1074/jbc.M200231200
Brockman, J. A., Norman, H. A., and Hildebrand, D. F. (1990). Effects of temperature, light and a chemical modulator on linolenate biosynthesis in mutant and wild type Arabidopsis calli. Phytochemistry 29, 1447–1453. doi: 10.1016/0031-9422(90)80098-2
Browse, J., and Somerville, C. (1991). Glycerolipid synthesis: biochemistry and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42, 456–506. doi: 10.1146/annurev.pp.42.060191.002343
Cao, S. J., Zhou, X. R., Wood, C. C., Green, A. G., Singh, S. P., Liu, L., et al. (2013). A large and functionally diverse family of FAD2 genes in safflower (Carthamus tinctorius L.). BMC Plant Biol. 13:5. doi: 10.1186/1471-2229-13-5
Chen, Y., Cui, Q., Xu, Y., Yang, S., Gao, M., and Wang, Y. (2015a). Effects of tung oilseed FAD2 and DGAT2 genes on unsaturated fatty acid accumulation in Rhodotorula glutinis and Arabidopsis thaliana. Mol. Genet. Genom. 290, 1605–1613. doi: 10.1007/s00438-015-1011-0
Chen, Y., Zhou, X., Zhang, Z. J., Dribnenki, P., Singh, S., and Green, A. (2015b). Development of high oleic oil crop platform in flax through RNAi-mediated multiple FAD2 gene silencing. Plant Cell Rep. 34, 643–653. doi: 10.1007/s00299-015-1737-5
Chi, X., Yang, Q., Lu, Y., Wang, J., Zhang, Q., Pan, L., et al. (2011a). Genome-wide analysis of fatty acid desaturases in soybean (Glycine max). Plant Mol. Biol. Rep. 29, 769–783. doi: 10.1007/s11105-010-0284-z
Chi, X., Yang, Q., Pan, L., Chen, M., He, Y., Yang, Z., et al. (2011b). Isolation and characterization of fatty acid desaturase genes from peanut (Arachis hypogaea L.). Plant Cell Rep. 30, 1393–1404. doi: 10.1007/s00299-011-1048-4
Collados, R., Andreu, V., Picorel, R., and Alfonso, M. (2006). A light-sensitive mechanism differently regulates transcription and transcript stability of ω-3 fatty-acid desaturases (FAD3, FAD7 and FAD8) in soybean photosynthetic cell suspensions. FEBS Lett. 580, 4934–4940. doi: 10.1016/j.febslet.2006.07.087
Cooke, D. T., and Burden, R. S. (1990). Lipid modulation of plasma membrane-bound ATPases. Physiol. Plant. 78, 152–159. doi: 10.1111/j.1399-3054.1990.tb08730.x
Dong, C. J., Cao, N., Zhang, Z. G., and Shang, Q. M. (2016). Characterization of the Fatty Acid Desaturase genes in cucumber: structure, phylogeny, and expression patterns. PLOS ONE 11:e0149917. doi: 10.1371/journal.pone.0149917
Deuticke, B., and Haest, C. W. M. (1987). Lipid modulation of transport proteins in vertebrate cell membranes. Annu. Rev. Physiol. 49, 221–235. doi: 10.1146/annurev.ph.49.030187.001253
Elsasser, S., and Finley, D. (2005). Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 7, 742–749. doi: 10.1038/ncb0805-742
Falcone, D. L., Ogas, J. P., and Somerville, C. R. (2004). Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 4:17. doi: 10.1186/1471-2229-4-17
Farmer, E. E. (1994). Fatty acid signalling in plants and their associated microorganisms. Plant Mol. Biol. 26, 1423–1437. doi: 10.1007/BF00016483
Fox, B. G., Shanklin, J., Somerville, C., and Munck, E. (1993). Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc. Natl. Acad. Sci. U.S.A. 90, 2486–2490. doi: 10.1073/pnas.90.6.2486
Ge, Y., Chang, Y., Xu, W. L., Cui, C. S., and Qu, S. P. (2015). Sequence variations in the FAD2 gene in seeded pumpkins. Genet. Mol. Res. 14, 17482–17488. doi: 10.4238/2015.December.21.19
Gibson, S., Arondel, V., Iba, K., and Somerville, C. (1994). Cloning of a temperature-regulated gene encoding a chloroplast ω-3 desaturase from Arabidopsis thaliana. Plant Physiol. 106, 1615–1621. doi: 10.1104/pp.106.4.1615
Guan, L., Wang, Y., Shen, H., Hou, K., Xu, Y., and Wu, W. (2012b). Molecular cloning and expression analysis of genes encoding two microsomal oleate desaturases (FAD2) from Safflower (Carthamus tinctorius L.). Plant Mol. Biol. Rep. 30, 139–148. doi: 10.1007/s11105-011-0322-5
Guan, L., Xu, Y., Wang, Y., Chen, L., Shao, J., and Wu, W. (2012a). Isolation and characterization of a temperature-regulated microsomal oleate desaturase gene (CtFAD2-1) from Safflower (Carthamus tinctorius L.). Plant Mol. Biol. 30, 391–402. doi: 10.1007/s11105-011-0349-7
Guan, M., Li, X., and Guan, C. (2012c). Microarray analysis of differentially expressed genes between Brassica napus strains with high- and low-oleic acid contents. Plant Cell Rep. 31, 929–943. doi: 10.1007/s00299-011-1213-9
Guilfoyle, T. J. (1997). The structure of plant gene promoters. Genet. Eng. 9, 15–47. doi: 10.1007/978-1-4615-5925-2_2
Harwood, J. L. (1996). Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta 1301, 7–56. doi: 10.1016/0005-2760(95)00242-1
Heppard, E. P., Kinney, A. J., Stecca, K. L., and Miao, G. (1996). Developmental and growth temperature regulation of two different microsomal ω-6 desaturase genes in soybeans. Plant Physiol. 110, 311–319. doi: 10.1104/pp.110.1.311
Hernández, M. L., Mancha, M., and Martínez-Rivas, J. M. (2005). Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 66, 1417–1426. doi: 10.1016/j.phytochem.2005.04.004
Hernández, M. L., Padilla, M. N., Mancha, M., and Martínez-Rivas, J. M. (2009). Expression analysis identifies FAD2-2 as the olive oleate desaturase gene mainly responsible for the linoleic acid content in virgin olive oil. J. Agric. Food Chem. 57, 6199–6206. doi: 10.1021/jf900678z
Hernández, M. L., Padilla, M. N., Sicardo, M. D., Mancha, M., and Martínez-Rivas, J. M. (2011). Effect of different environmental stresses on the expression of oleate desaturase genes and fatty acid composition in olive fruit. Phytochemistry 72, 178–187. doi: 10.1016/j.phytochem.2010.11.026
Hitz, W. D., Carlson, T. J., Booth, J. R., Kinney, A. J., Stecca, K. L., and Yadav, N. S. (1994). Cloning of a higher-plant plastid ω-6 fatty acid desaturase cDNA and its expression in a cyanobacterium. Plant Physiol. 105, 635–641. doi: 10.1104/pp.105.2.635
Horiguchi, G., Fuse, T., Kawakami, N., Kodama, H., and Iba, K. (2000). Temperature dependent translational regulation of the ER ω-3 fatty-acid desaturase gene in wheat root tips. Plant J. 24, 805–813. doi: 10.1046/j.1365-313x.2000.00925.x
Huang, J. Z., and Huber, S. C. (2001). Phosphorylation of synthetic peptides by a CDPK and a plant SNF1-related protein kinase. Influence of proline and basic amino acidresidues at selected positions. Plant Cell Physiol. 42, 1079–1087. doi: 10.1093/pcp/pce137
Iba, K., Gibson, S., Nishiuchi, T., Fuso, T., Nishimura, M., Arondel, V., et al. (1993). A gene encoding a chloroplast ω-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 268, 24099–24105.
Im, Y. J., Han, O., Chung, G. C., and Cho, B. H. (2002). Antisense expression of an Arabidopsis ω-3 fatty acid desaturase gene reduces salt/drought tolerance in transgenic tobacco plants. Mol. Cells 13, 264–271.
Jin, U. H., Lee, J. W., Chung, Y. S., Lee, J. H., Yi, Y. B., Kim, Y. K., et al. (2001). Characterization and temporal expression of a ω-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci. 161, 935–941. doi: 10.1016/S0168-9452(01)00489-7
Kachroo, A., Lapchyk, L., Fukushige, H., Hildebrand, D., Klessig, D., and Kachroo, P. (2005). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15, 2952–2965. doi: 10.1105/tpc.017301
Kachroo, P., Venugopal, S. C., Navarre, D. A., Lapchyk, L., and Kachroo, A. (2005). Role of salicylic acid and fatty acid desaturation pathways in ssi2-mediated signaling. Plant Physiol. 139, 1717–1735. doi: 10.1104/pp.105.071662
Kang, J., Snapp, A. R., and Lu, C. (2011). Identification of three genes encoding microsomal oleate desaturases (FAD2) from the oilseed crop Camelina sativa. Plant Physiol. Biochem. 49, 223–229. doi: 10.1016/j.plaphy.2010.12.004
Kargiotidou, A., Deli, D., Galanopoulou, D., Tsaftaris, A., and Farmaki, T. (2008). Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 59, 2043–2056. doi: 10.1093/jxb/ern065
Kearns, E. V., Hugly, S., and Somerville, C. R. (1991). The role of cytochrome b5 in Δ12 desaturation of oleic acid by microsomes of safflower (Carthamus tinctorius L.). Arch. Biochem. Biophys. 284, 431–436. doi: 10.1016/0003-9861(91)90319-E
Khadake, R. M., Ranjekar, P. K., and Harsulkar, A. M. (2009). Cloning of a novel omega-6 desaturase from flax (Linum usitatissimum L.) and its functional analysis in Saccharomyces cerevisiae. Mol. Biotechnol. 42, 168–174. doi: 10.1007/s12033-009-9150-3
Khuu, N., Gidda, S., Shockey, J. M., Dyer, J. M., and Mullen, R. T. (2011). The N termini of Brassica and tung omega-3 fatty acid desaturases mediate proteasome-dependent protein degradation in plant cells. Plant Signal. Behav. 6, 422–425. doi: 10.4161/psb.6.3.14522
Kim, H., Go, Y. S., Kim, A. Y., Lee, S., Kim, K., Lee, G., et al. (2014). Isolation and functional analysis of three microsomal delta-12 fatty acid desaturase genes from Camelina sativa (L.) cv. CAME. Plant Biotechnol. 41, 146–158. doi: 10.5010/JPB.2014.41.3.146
Kim, M. J., Kim, H., Shin, J. S., Chung, C., Ohlrogge, J. B., and Suh, M. C. (2006). Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol. Gen. Genomics 276, 351–368. doi: 10.1007/s00438-006-0148-2
Kinney, A. J., Cahoon, E. B., and Hitz, W. D. (2002). Manipulating desaturase activities in transgenic crop plants. Biochem. Soc. Trans. 30, 1099–1103. doi: 10.1042/bst0301099
Kirsch, C., Hahlbrock, K., and Somssich, I. E. (1997a). Rapid and transient induction of a parsley microsomal Δ12 fatty acid desaturase mRNA by fungal elicitor. Plant Physiol. 115, 283–289. doi: 10.1023/A:1005726210977
Kirsch, C., Takamiya-Wik, M., Reinold, S., Hahlbrock, K., and Somssich, I. E. (1997b). Rapid, transient, and highly localized induction of plastidial ω-3 fatty acid desaturase mRNA at fungal infection sites in Petroselinum crispum. Proc. Natl. Acad. Sci. U.S.A. 94, 2079–2084. doi: 10.1104/pp.008532
Kodama, H., Akagi, H., Kusumi, K., Fujimura, T., and Iba, K. (1997). Structure, chromosomal location and expression of a rice gene encoding the microsome ω-3- fatty acid desaturase. Plant Mol. Biol. 33, 493–502. doi: 10.1016/j.gene.2012.08.032
Kongcharoensuntorn, W. (2001). Isolation and Analysis of Cotton Genomic Clones Encompassing a Fatty Acid Desaturase (FAD2) Gene. Ph.D. thesis, University of North Texas, Denton, TX, 162. doi: 10.1007/s10535-010-0006-2
Kreps, J. A., Wu, Y., Chang, H. S., Zhu, T., Wang, X., and Harper, J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. doi: 10.1016/j.jplph.2006.08.007
Lee, K., Kim, S. H., Go, Y., Jung, S. M., Roh, K. H., Kim, J., et al. (2012). Molecular cloning and functional analysis of two FAD2 genes from American grape (Vitis labrusca L.). Gene 509, 189–194. doi: 10.1016/j.gene.2012.08.032
Lei, N., Peng, S., Niu, B., Chen, J., Zhou, J., Tang, L., et al. (2010). Molecular cloning and characterization of a novel microsomal oleate desaturase gene DiFAD2 from Davidia involucrata Baill. Biol. Plant. 54, 41–46. doi: 10.1007/s13562-014-0284-4
Li, L. Y., Wang, X. L., Gai, J. Y., and Yu, D. Y. (2007). Molecular cloning and characterization of a novel microsomal oleate desaturase gene from soybean. J. Plant Physiol. 164, 1516–1526. doi: 10.2307/2657130
Li, Q., Zheng, Q., Shen, W., Cram, D., Fowler, D. B., Wei, Y., et al. (2015). Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27, 86–103. doi: 10.1105/tpc.114.134338
Lindqvist, Y., Huang, W., Schneider, G., and Shanklin, J. (1996). Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 15, 4081–4092. doi: 10.1016/S0005-2760(98)00091-5
Liu, F., Zhao, Y., Wang, X., Li, Y., and Sun, J. (2015). Isolation and functional characterization of seed-specific FAD2-1 promoter from cotton (Gossypium hirsutum L). J. Plant Biochem. Biotechnol. 24, 369–375. doi: 10.1007/s11745-009-3381-8
Liu, Q., Brubaker, C. L., Green, A. G., Marshall, D. R., Sharp, P. J., and Singh, S. P. (2001). Evolution of the FAD2-1 fatty acid desaturase 5′ UTR intron and the molecular systematics of Gossypium (Malvaceae). Am. J. Bot. 88, 92–102. doi: 10.1042/bst0280890
Liu, Q., Singh, S., and Green, A. (2000). Genetic modification of cotton seed oil using inverted-repeat gene-silencing techniques. Biochem. Soc. Trans. 28, 927–929.
Los, D. A., and Murata, N. (1998). Structure and expression of fatty acid desaturases. Biochem. Biophys. Acta 1394, 3–15.
Los, D. A., Ray, M. K., and Murata, N. (1997). Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol. Microbiol. 25, 1167–1175. doi: 10.1093/jxb/err013
Lu, Y., Chi, X., Li, Z., Yang, Q., Li, F., Liu, S., et al. (2010). Isolation and characterisation of a stress-dependent plastidial Δ12 fatty acid desaturase from the Antarctic microalga Chlorella vulgaris NJ-7. Lipids 45, 179–187. doi: 10.1007/s11745-009-3381-8
Martínez-Rivas, J. M., Garcia-Diaz, M. T., and Mancha, M. (2000). Temperature and oxygen regulation of microsomal oleate desaturase (FAD2) from sunflower. Biochem. Soc. Trans. 28, 890–892. doi: 10.1042/bst0280890
Martínez-Rivas, J. M., Sperling, P., Lühs, W., and Heinz, E. (2001). Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L.). Mol. Breed. 8, 159–168. doi: 10.1023/A:1013324329322
Matsuda, O., Sakamoto, H., Hashimoto, T., and Iba, K. (2005). A temperature-sensitive mechanism that regulates post-translational stability of a plastidial omega-3 fatty acid desaturase (FAD8) in Arabidopsis leaf tissues. J. Biol. Chem. 280, 3597–3604. doi: 10.1074/jbc.M407226200
Matteucci, M., D’Angeli, S., Errico, S., Lamanna, R., Perrotta, G., and Altamura, M. M. (2011). Cold affects the transcription of fatty acid desaturases and oil quality in the fruit of Olea europaea L. genotypes with different cold hardiness. J. Exp. Bot. 62, 3403–3420. doi: 10.1093/jxb/err013
McCartney, A. W., Dyer, J. M., Kim, P. K., Andrews, D. W., McNew, J. A., and Mullen, R. T. (2004). Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J. 37, 156–173. doi: 10.1111/j.1365-313X.2004.01949.x
Menard, G. N., Moreno, J. M., Bryant, F. N., Munoz-Azcarate, O., Kelly, A. A., Hassani-Pak, K., et al. (2017). Genome wide analysis of fatty acid desaturation and its response to temperature. Plant Physiol. 173, 1594–1605. doi: 10.1104/pp.16.01907
Miquel, M., and Browse, J. (1992). Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J. Biol. Chem. 267, 1502–1509. doi: 10.1007/s11033-014-3373-5
Miquel, M., James, D., Dooner, H., and Browse, J. (1993). Arabidopsis requires polyunsaturated lipids for low temperature survival. Proc. Natl. Acad. Sci. U.S.A. 90, 6208–6212. 10.1073/pnas.90.13.6208
Mitchell, A. G., and Martin, C. E. (1995). A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae Δ9 fatty acid desaturase. J. Biol. Chem. 270, 29766–29772. doi: 10.1074/jbc.270.50.29766
Murata, N., and Wada, H. (1995). Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochem. J. 308, 1–8. doi: 10.1042/bj3080001
Murphy, D. J., and Piffanelli, P. (1998). “Fatty acid desaturases: structure, mechanism and regulation,” in Plant Lipid Biosynthesis, ed. H. L. Harwood (Cambridge: Cambridge University Press), 95–130.
Murphy, D. J., and Stumpf, P. K. (1979). Light-dependent induction of polyunsaturated fatty acid biosynthesis in greening cucumber cotyledons. Plant Physiol. 63, 328–335. doi: 10.1104/pp.63.2.328
Nakamura, M. T., and Nara, T. Y. (2004). Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 24, 345–376. doi: 10.1146/annurev.nutr.24.121803.063211
Nayeri, F. D., and Yarizade, K. (2014). Bioinformatics study of delta-12 fatty acid desaturase 2 (FAD2) gene in oilseeds. Mol. Biol. Rep. 41, 5077–5087. doi: 10.1007/s11033-014-3373-5
Nielsen, H., Engelbrecht, J., Brunak, S., and Heijne, G. V. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. doi: 10.1093/protein/10.1.1
Norambuena, F., Hermon, K., Skrzypczyk, V., Emery, J. A., Sharon, Y., Beard, A., et al. (2015). Algae in fish feed: performances and fatty acid metabolism in juvenile Atlantic salmon. PLOS ONE 10:e0124042. doi: 10.1371/journal.pone.0124042
O’Byrne, D. J., Knauft, D. A., and Shireman, R. B. (1997). Low fat monounsaturated rich diets containing high-oleic Peanuts improve serum lipoprotein profiles. Lipids 32, 687–695. doi: 10.1007/s11745-997-0088-y
Ohlrogge, J., and Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. doi: 10.1105/tpc.7.7.957
Ohnishi, J., and Yamada, M. (1983). Glycerolipid synthesis in Avena leaves during greening of etiolated seedlings IV. Effect of light on fatty acid desaturation. Plant Cell Physiol. 24, 1553–1557. doi: 10.1093/pcp/21.8.1595
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., and Browse, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.1105/tpc.6.1.147
O’Quin, J. B., Bourassa, L., Zhang, D., Shockey, J. M., Gidda, S. K., Fosnot, S., et al. (2010). Temperature-sensitive post-translational regulation of plant omega-3 fatty-acid desaturases is mediated by the endoplasmic reticulum-associated degradation pathway. J. Biol. Chem. 285, 21781–21796. doi: 10.1074/jbc.M110.135236
Pandey, M. K., Wang, M. L., Qiao, L., Feng, S., Khera, P., Wang, H., et al. (2014). Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genetics 15:133. doi: 10.1186/s12863-014-0133-4
Pirtle, I. L., Kongcharoensuntorn, W., Nampaisansuk, M., Knesek, J. D., Chapman, K. D., and Pirtle, R. M. (2001). Molecular cloning and functional expression of the gene for a cotton Δ-12 fatty acid desaturase (FAD2). Biochim. Biophys. Acta 1522, 122–129. doi: 10.1016/S0167-4781(01)00312-8
Rajwade, A. V., Kadoo, N. Y., Borikar, S. P., Harsulkar, A. M., Ghorpade, P. B., and Gupta, V. S. (2014). Differential transcriptional activity of SAD, FAD2 and FAD3 desaturase genes in developing seeds of linseed contributes to varietal variation in α-linolenic acid content. Phytochemistry 98, 41–53. doi: 10.1016/j.phytochem.2013.12.002
Reddy, A. S., Naccio, M. L., Gross, L. M., and Thomas, T. L. (1993). Isolation of a ω-6- desaturase gene from the cyanobacterium Synechocystis sp. Strain PCC6803 by gain-of-function expression in Anabaena sp. Strain PCC 7120. Plant Mol. Biol. 22, 293–300.
Regente, M., Monzo’n, G. C., and Canal, L. (2008). Phospholipids are present in extracellular fluids of imbibing sunflower seeds and are modulated by hormonal treatments. J. Exp. Bot. 59, 553–562. doi: 10.1093/jxb/erm329
Reiser, L., Sánchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: evolutionary relationships and functions of knox homeo box genes. Plant Mol. Biol. 42, 151–166. doi: 10.1023/A:1006384122567
Rodríguez-Vargas, S., Sánchez-García, A., Martínez-Rivas, J. M., Prieto, J. A., and Randez-Gil, F. (2007). Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl. Environ. Microbiol. 73, 110–116. doi: 10.1128/AEM.01360-06
Rolletschek, H., Borisjuk, L., Sánchez-García, A., Gotor, C., Romero, L. C., Martínez-Rivas, J. M., et al. (2007). Temperature dependent endogenous oxygen concentration regulates microsomal oleate desaturase in developing sunflower seeds. J. Exp. Bot. 58, 3171–3181. doi: 10.1093/jxb/erm154
Routaboul, J. M., Fischer, S. F., and Browse, J. (2000). Trienoic fatty acids are required to maintain chloroplast function at low temperatures. Plant Physiol. 124, 1697–1705. doi: 10.1104/pp.124.4.1697
Sakamoto, T., and Bryant, D. A. (1997). Temperature-regulated mRNA accumulation and stabilization for fatty acid desaturase genes in the cyanobacterium Synechococcus sp. Strain PCC 7002. Mol. Microbiol. 23, 1281–1292.
Sakamoto, T., Wada, H., Nishida, I., Ohmori, M., and Murata, N. (1994). Δ9 Acyl-lipid desaturases of cyanobacteria. Molecular cloning and substrate specificities in terms of fatty acid sn-positions, and polar head groups. J. Biol. Chem. 269, 25576–25580.
Schell, D. J. (1998). Protein targeting to the thylakoid membrane. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 97–126. doi: 10.1146/annurev.arplant.49.1.97
Schmidt, H., and Heinz, E. (1990). Desaturation of oleoyl groups in envelope membranes from spinach chloroplast. Proc. Natl. Acad. Sci. U.S.A. 87, 9477–9480. doi: 10.1073/pnas.87.23.9477
Schrader, E. K., Harstad, K. G., and Matouschek, A. (2009). Targeting proteins for degradation. Nat. Chem. Biol. 5, 815–822. doi: 10.1038/nchembio.250
Shanklin, J., and Cahoon, E. B. (1998). Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641. doi: 10.1146/annurev.arplant.49.1.611
Shanklin, J., Whittle, E., and Fox, B. G. (1994). Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33, 12787–12794. doi: 10.1021/bi00209a009
Sharma, A., and Chauhan, R. S. (2012). In silico identification and comparative genomics of candidate genes involved in biosynthesis and accumulation of seed oil in plants. Comp. Funct. Genomics 2012:914843. doi: 10.1155/2012/914843
Slabas, A. R., and Fawcett, T. (1992). The biochemistry and molecular biology of plant lipid biosynthesis. Plant Mol. Biol. 19, 169–191. doi: 10.1007/BF00015613
Somerville, C., Browse, J., Jaworski, J. G., and Ohlrogge, J. B. (2000). “Lipids,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Rockville, MD: American Society of Plant Physiologists), 456–527.
Stoutjesdijk, P. A., Hurlestone, C., Singh, S. P., and Green, A. G. (2000). High-oleic acid Australian Brassica napus and B. juncea varieties produced by co-suppression of endogenous Δ12-desaturases. Biochem. Soc. Trans. 28, 938–940. doi: 10.1042/bst0280938
Sun, R., Gao, L., Yu, X., Zheng, Y., Li, D., and Wang, X. (2016). Identification of a Δ12 fatty acid desaturase from oil palm (Elaeis guineensis Jacq.) involved in the biosynthesis of linoleic acid by heterologous expression in Saccharomyces cerevisiae. Gene 591, 21–26. doi: 10.1016/j.gene.2016.06.039
Suresha, G. S., Rai, R. D., and Santha, I. M. (2012). Molecular cloning, expression analysis and growth temperature dependent regulation of a novel oleate desaturase gene (fad2) homologue from Brassica juncea. Austr. J. Crop Sci. 6, 296–308.
Suresha, G. S., and Santha, I. M. (2013). Molecular cloning and in silico analysis of novel oleate desaturase gene homologues from Brassica juncea through sub-genomic library approach. Plant Omics 6, 55–64. doi: 10.5402/2012/173954
Tang, G. Q., Novitzky, W. P., Griffin, C. H., Huber, S. C., and Dewey, R. E. (2005). Oleate desaturase enzymes of soybean: evidence of regulation through differential stability and phosphorylation. Plant J. 44, 433–446. doi: 10.1111/j.1365-313X.2005.02535.x
Tanhuanpaa, P. K., Vilkki, J. P., and Vilkki, H. J. (1995). Identification of an RAPD marker for palmitic-acid concentration in the seed oil of spring turnip rape (Brassica rapa ssp. oleifera). Theor. Appl. Genet. 91, 477–480. doi: 10.1007/BF00222976
Tao, F., Zhu, S. W., Fan, J., and Cheng, B. J. (2006). Cloning and sequence analysis of maize FAD2 gene. J. Plant Physiol. Mol. Biol. 32, 649–656.
Teixeira, M. C., Carvalho, I. S., and Brodelius, M. (2010). ω-3 fatty acid desaturase genes isolated from purslane (Portulaca oleracea L.): expression in different tissues and response to cold and wound stress. J. Agric. Food Chem. 58, 1870–1877. doi: 10.1021/jf902684v
Teixeira, M. C., Coelho, N., Olsson, M. E., Brodelius, P. E., Carvalho, I. S., and Brodelius, M. (2009). Molecular cloning and expression analysis of three omega-6 desaturase genes from purslane (Portulaca oleracea L.). Biotechnol. Lett. 31, 1089–1101. doi: 10.1007/s10529-009-9956-x
Tocher, D. R., Leaver, M. J., and Hodgson, P. A. (1998). Recent advances in the biochemistry and molecular biology of fatty acyl desaturases. Prog. Lipid Res. 37, 73–117. doi: 10.1016/S0163-7827(98)00005-8
Tzen, J. T. C. (2012). Integral proeins in plant oil bodies. ISRN Bot. 2012:173954. doi: 10.5402/2012/173954
Upchurch, R. G. (2008). Fatty acid unsaturation, mobilization, and regulation the response of plants to stress. Biotechnol. Lett. 30, 967–977. doi: 10.1007/s10529-008-9639-z
Utomo, C., Subroto, A. P., Darmawan, C., Setyobudi, R. H., Nugroho, Y. A., and Liwang, T. (2015). Construction for Δ12 fatty acid desaturase (FAD2) silencing to improve oil quality of Jatropha curcas Linn. Energy Proc. 65, 36–41. doi: 10.1016/j.egypro.2015.01.027
Vandenbussche, F., Vriezen, W. H., Smalle, J., Laarhoven, L. J., Harren, F. J., and Van Der Straeten, D. (2003). Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 133, 517–527. doi: 10.1104/pp.103.022665
Vega, S. E., del-Rio, A. H., Bamberg, J. B., and Palta, J. P. (2004). Evidence for the up-regulation of stearoyl-ACP (Δ9) desaturase gene expression during cold acclimation. Am. J. Potato Res. 81, 125–135. doi: 10.1007/BF02853610
Wang, J., Chen, W., and Zen, L. (2003). Effects of Δ12 fatty acid desaturase gene (fad2) mutation on epidermal construct and permeability of Brassica napus leaf (in Chinese). J. Plant Physiol. Mol. Biol. 29, 192–198.
Wang, M., Liu, M., Li, D., Wu, J., Li, X., and Yang, Y. (2010). Overexpression of FAD2 promotes seed germination and hypocotyl elongation in Brassica napus. Plant Cell Tiss. Organ. Cult. 102, 205–211. doi: 10.1007/s11240-010-9723-7
Wang, X., Beno-Moualem, D., Kobiler, I., Leiking-Frenkel, A., Lichter, A., and Prusky, D. (2004). Expression of Δ12 fatty acid desaturase during the induced accumulation of the antifungal diene in avocado fruits. Mol. Plant Pathol. 5, 575–585. doi: 10.1111/j.1364-3703.2004.00249.x
Wang, Y., Zhang, X., Zhao, Y., Prakash, C. S., He, G., and Yin, D. (2015). Insights into the novel members of the FAD2 gene family involved in high-oleate fluxes in peanut. Genome 58, 375–383. doi: 10.1139/gen-2015-0008
Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. doi: 10.1016/S1360-1385(02)02250-1
Xiao, G., Zhang, Z. Q., Yin, C. F., Liu, R. Y., Wu, X. M., Tan, T. L., et al. (2014). Characterization of the promoter and 5′-UTR intron of oleic acid desaturase (FAD2) gene in Brassica napus. Gene 545, 45–55. doi: 10.1016/j.gene.2014.05.008
Yadav, N. S., Wierzbicki, A., Aegerter, M., Caster, C. S., Pérez-Grau, L., Kinney, A. J., et al. (1993). Cloning of higher plant ω-3 fatty acid desaturases. Plant Physiol. 103, 467–476. doi: 10.1104/pp.103.2.467
Yamaki, T., Nagamine, I., Fukumoto, K., Yano, T., Miyahara, M., and Sakurai, H. (2005). High oleic peanut oil modulates promotion stage in lung tumorigenesis of mice treated with methyl nitrosourea. Food Sci. Technol. Res. 11, 231–235. doi: 10.3136/fstr.11.231
Yang, M. F., and Xu, Y. N. (2007). Oleate accumulation, induced by silencing of microsomal omega-6 desaturase, declines with leaf expansion in transgenic tobacco. J. Plant Physiol. 164, 23–30. doi: 10.1016/j.jplph.2005.11.002
Yin, D., Deng, S., Zhan, K., and Cui, D. (2007). High oleic oils produced by Hp RNA-mediated gene silencing of oleate desaturase. Plant Mol. Biol. Rep. 25, 154–163. doi: 10.1007/s11105-007-0017-0
Zhang, D., Pirtle, I. L., Park, S. J., Nampaisansuk, M., Neogi, P., Wanjie, S. W., et al. (2009). Identification and expression of a new delta-12 fatty acid desaturase (FAD2-4) gene in upland cotton and its functional expression in yeast and Arabidopsis thaliana plants. Plant Physiol. Biochem. 47, 462–471. doi: 10.1016/j.plaphy.2008.12.024
Zhang, J., Liu, H., Sun, J., Li, B., Zhu, Q., Chen, S., et al. (2012). Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLOS ONE 7:e30355. doi: 10.1371/journal.pone.0030355
Zhang, J. T., Zhu, J. Q., Zhu, Q., Liu, H., Gao, X. S., and Zhang, H. X. (2009). Fatty acid desaturase-6 (FAD6) is required for plant salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 390, 469–474. doi: 10.1016/j.bbrc.2009.09.095
Zhang, L., Lu, H., Liu, C., Xue, F., Yang, J., Ma, L., et al. (2016). Lipid desaturation in prokaryotic pathway abates the high-oleic phenotype of FAD2-silenced tobacco at a lower temperature. J. Plant Biochem. Biotechnol. 25, 375–381. doi: 10.1007/s13562-016-0349-7
Keywords: FAD2 gene, fatty acid desaturase, fatty acids, cold and salt stress, plant development
Citation: Dar AA, Choudhury AR, Kancharla PK and Arumugam N (2017) The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 8:1789. doi: 10.3389/fpls.2017.01789
Received: 07 June 2017; Accepted: 02 October 2017;
Published: 18 October 2017.
Edited by:
Jens Rohloff, Norwegian University of Science and Technology, NorwayReviewed by:
Thomas J. Bach, University of Strasbourg, FranceCopyright © 2017 Dar, Choudhury, Kancharla and Arumugam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aejaz A. Dar, YWVqYXpkYnQucGJpb3RAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.