
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 05 October 2017
Sec. Plant Breeding
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01730
 Zhongshan He1,2†
Zhongshan He1,2† Jing Zeng1,2†
Jing Zeng1,2† Yun Ren1,2
Yun Ren1,2 Dan Chen1,2
Dan Chen1,2 Wenjie Li1,2
Wenjie Li1,2 Fengyan Gao1,2
Fengyan Gao1,2 Ye Cao1,2
Ye Cao1,2 Tao Luo1,2
Tao Luo1,2 Guoqiang Yuan1,2
Guoqiang Yuan1,2 Xianghong Wu1,2
Xianghong Wu1,2 Yueyang Liang1,2
Yueyang Liang1,2 Qiming Deng1,2
Qiming Deng1,2 Shiquan Wang1,2
Shiquan Wang1,2 Aiping Zheng1,2
Aiping Zheng1,2 Jun Zhu1,2
Jun Zhu1,2 Huainian Liu1,2
Huainian Liu1,2 Lingxia Wang1,2
Lingxia Wang1,2 Ping Li1,2,3*
Ping Li1,2,3* Shuangcheng Li1,2*
Shuangcheng Li1,2*Growth-regulating factor (GRF) interacting factors (GIFs) are involved in several developmental processes in Arabidopsis. We previously showed that upregulation of OsGIF1 expression improves rice grain size. However, whether OsGIF1 is involved in other developmental processes remains unclear. Here, we report pleiotropic effects of OsGIF1 on rice organ size regulation. Overexpression and functional knock-out via a CRISPR/Cas9 strategy revealed that OsGIF1 not only positively regulates the sizes of rice leaf, stem, and grain but also influences rice reproduction. Expression profiles based on both qRT-PCR and GUS (β-glucuronidase) histochemical staining suggested that OsGIF1 is differentially expressed across various rice tissues, consistent with its roles in regulating the development of multiple rice organs. Additionally, we found that OsGIF1-GFP localized preferentially in the nucleus, which supports its proposed role as a transcriptional cofactor. Further histological analysis suggested that OsGIF1 affected rice organ size possibly by regulating cell size. Our results suggest that OsGIF1 plays important roles in vegetative and reproductive developmental processes, with important implications for rice breeding.
Growth-regulating factor (GRF) interacting factor (GIF) was first reported as the major partner of the plant-specific transcription factor GRF (Kim and Kende, 2004; Horiguchi et al., 2005), which has been implicated in stem and leaf development (van der Knaap et al., 2000; Kim et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005; Omidbakhshfard et al., 2015). The amino acid (AA) sequences of GIFs are homologous to those of the human synovial translocation family protein SYT (Thaete et al., 1999; Kim and Kende, 2004; Horiguchi et al., 2005; de Bruijn and Geurts van Kessel, 2006; Kim and Tsukaya, 2015), which is also known as SS18. Both proteins lack the DNA binding domain (DBD) and act as transcriptional co-activators by interacting with human SWI/SNF ATPases (Brett et al., 1997; dos Santos et al., 1997; Thaete et al., 1999; Kato et al., 2002; Aizawa et al., 2004).
The SYT N-terminal homology (SNH) domain of a GIF is necessary because of its direct interaction with the QLQ domain of GRF in Arabidopsis and rice (Kim and Kende, 2004; Horiguchi et al., 2005; Liu et al., 2014; Duan et al., 2015; Li et al., 2016). Although GIF proteins do not have DBDs and NLSs, AtGIF1/AN3 protein is preferentially localized in the nucleus (Kim and Kende, 2004). Additionally, several pairs of GRF and GIF in Arabidopsis and rice have been demonstrated to work as complexes in the nucleus (Liang et al., 2014; Liu et al., 2014; Kim and Tsukaya, 2015). Multiple reports have tested the transactivation activities of AtGIF1/AN3 (Kim and Kende, 2004; Liu et al., 2014; Li et al., 2016). Three copies of GIFs have been annotated in the Arabidopsis, rice, and maize genomes, while nine, 12 and 17 GRF members have been found in Arabidopsis, rice and maize, respectively (Choi et al., 2004; Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015). The interacting partnership between GRF and GIF, which is required for several developmental processes, has been confirmed in nearly all Arabidopsis members and has also been seen in rice (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015).
Growth-regulating factor interacting factor is involved in several vegetative and reproductive developmental processes in Arabidopsis (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015). In earlier studies, GIF was reported to be required in controlling cell proliferation during leaf development by interacting with GRF (Kim and Kende, 2004; Horiguchi et al., 2005). Interestingly, although the AN3/GIF1 transcripts are not detectable in leaf epidermal cells, the AN3/GIF1 protein can move into epidermal cells after being synthesized within mesophyll cells and helps to control epidermal cell proliferation (Kawade et al., 2013). The “compensation effect” phenomenon was also found in the an3/gif1 mutant and further investigation demonstrated that the an3-dependent compensation was a non-cell autonomous process (Kim and Kende, 2004; Horiguchi et al., 2005; Kawade et al., 2010). GIF also works in adaxial/abaxial (Ad-Ab) patterning by the interaction of GIF1 with ASYMMETRIC LEAVES 2, a nuclear protein important for leaf Ad-Ab patterning (Iwakawa et al., 2002, 2007; Xu et al., 2003; Horiguchi et al., 2011). Moreover, GIF contributes to establishment of cotyledon identity by repressing the expression of an embryonic apical fate determination gene PLETHORA1, by cooperating with HAN, a GATA-type transcription factor (Kanei et al., 2012). In addition, GIF plays an important role in the determination of carpel number and male and female reproductive development in Arabidopsis and in husk/lemma development in rice, suggesting a role of OsGIF in floral organ determination and development (Lee et al., 2014; Liang et al., 2014; Liu et al., 2014; Duan et al., 2015; Li et al., 2016; Meng et al., 2016b). Interactions with numerous other proteins involved in chromatin remodeling processes, such as ATPases of the SWI/SNF family, have also been presented (Debernardi et al., 2014; Vercruyssen et al., 2014; Nelissen et al., 2015). Very recently, the AN3/GIF1-YODA cascade has been implicated in anthocyanin accumulation (Meng et al., 2016a), water-use efficiency and drought tolerance (Meng and Yao, 2015) in Arabidopsis. However, the function of rice GIF members remains ambiguous.
Plant organ size is a complex trait and is determined majorly by the process of cell proliferation and cell expansion. Significant progresses in dissecting of genetic factors that control plant organ size have been achieved in Arabidopsis. Several pathways, such as plant hormones, ubiquitination degredations, cytochrome P450 pathway and microRNAs, are implicated in this developmental process. Among which, ERBB-3 BINDING PROTEIN 1 (Gingras et al., 2001), AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (Hu et al., 2003) and AINTEGUMENTA (Elliott et al., 1996) were auxin related positive regulators for cell proliferation; The A-type and B-type ARABIDOPSIS RESPONSE REGULATORS could transmit the CK signal to the downstream gene for the promotion of cell proliferation (Dello Ioio et al., 2007). The BIN2 and BES1/BZR1 genes were key factors in the BR signaling pathway and positively regulate cell expansion (Halliday, 2004). While the GIBBERELLIN INSENSITIVE (Ubeda-Tomas et al., 2009) and REPERSSOR OF GA1-3 (Silverstone et al., 2001) genes in GA signaling negatively regulated cell expansion. Besides, BIG BROTHER (Disch et al., 2006), an ubiquitin protein ligase, and DA1 (Li et al., 2008), an ubiquitin receptor, were both negatively regulators for cell proliferation, while KLUH (Anastasiou et al., 2007) and EOD3 (Fang et al., 2012), both encoded cytochrome P450 monooxygenases, positively regulated cell proliferation. Moreover, miR396 was reported to negatively regulate cell proliferation by target degradation of several GRF members in Arabidopsis (Rodriguez et al., 2010), and the OsmiR396-GRF4-GIF1 regulatory module was demonstrated affecting rice grain size by influencing the cell size via simultaneously regulating the BR and GA pathways (Che et al., 2015; Duan et al., 2015; Li et al., 2016).
We previously found that OsGIF1 interacts directly with OsGRF4 and its upregulation improves rice grain size (Li et al., 2016). Thus, OsGIF1 plays a role in regulating rice grain size. However, further analysis found that OsGIF1 expression was not restricted to the spikelet, which might suggest that OsGIF1 is involved in other developmental process. Here, we report the pleiotropic effects of OsGIF1 on rice organ size regulation by analyzing the overexpression and functional knock-out (KO) of OsGIF1 in rice. OsGIF1 not only positively regulated the sizes of rice leaf, stem, and grain but also affected the rice reproductive process. The results suggest that OsGIF1 plays an important role in vegetative and reproductive developmental processes, which has implications for rice breeding.
Three KO lines and three overexpression lines were used in this study. The three overexpression lines, in which the OsGIF1 (LOC_Os03g52320) was driven by the 2x35S promoter (Mao et al., 2005), were obtained by self-pollinating the T0 plants described in our previous report (Li et al., 2016). The japonica variety Nipponbare (Nipp) was used as control. All plants were planted in the experimental field at the Rice Research Institute, Sichuan Agricultural University, Wenjiang. Phenotypic data were collected at the maturing stage. The data were analyzed using Excel (Microsoft) for mean values and standard errors of mean (SEM). Statistical significance was assessed by conducting Student’s t-test.
The primer sequences for molecular cloning and constructions are listed in Supplementary Table 1. We verified OsGIF1 function by generating two gRNA constructs, in which the gRNA was driven by the rice U6 promoter, and the plant-optimized Cas9 was driven by the UBI promoter (Miao et al., 2013). These constructs were introduced into the WT (Nipp) (Hiei et al., 1997). Then, the transgenic plants were subjected to PCR and sequencing analysis to determine the occurrence of mutations. To verify the association between the mutation in OsGIF1 and the mutant phenotype, we performed segregation analysis in some populations generated by back-crossing these mutants with WT and confirmed the co-segregation of the mutant phenotype and mutations. To evaluate off-target effects, four putative off-target sites were identified by similar sequence searches within the rice genome (Supplementary Table 2). These sites were then sequenced for mutation analysis.
The grain length, width, and 1,000-grain weight were measured by an automatic seed-size analyzing system (SC-G, Wanshen, Hangzhou). Other traits were investigated using conventional methods at maturing stage. An environmental scanning electronic microscope (QUANTA 450, Nikon) was employed to observe the outer surface of the leaf, stem internode, and outer glume. For histological analysis, samples of the leaf and stem internode were placed in the FAA solution for 12 h at 4°C, dehydrated in a graded ethanol series, followed by substitution using 3-methylbutyl acetate (Li et al., 2016). The samples were dissected and observed under a microscope (80I, Nikon) for determinations of cell number and size.
To localize the transcripts of OsGIF1 in rice tissue, a OsGIF1pro::GUS construct in which expression of the GUS gene was driven by native promoters was generated. Then, the OsGIF1pro::GUS construct was introduced into the Agrobacterium tumefaciens strain EHA105 to transform WT plants. Histochemical GUS staining was performed in the transgenic plants as described previously (Li et al., 2013).
Total RNAs were isolated from various rice tissues at different developmental stages using the TriPure Isolation reagent (Roche). The cDNAs were then reverse-transcribed using the Transcriptor First-Strand cDNA Synthesis kit (Roche). qRT-PCR was conducted in a total volume of 10 μl, with 0.3 μl of the reverse-transcribed product, 0.08 mM gene-specific primers, and 5.0 ml of Sso Advanced TM SYBR Green Supermix (Bio-Rad) using a Bio-Rad CFX96 Real-Time PCR System according to the manufacturer’s instructions. Data were analyzed by the relative quantification method (Livak and Schmittgen, 2001). The rice Actin gene was used as internal control. Each measurement was determined for at least two biological samples using three replicates for each sample.
The full-length cDNA of OsGIF1 was cloned into the pA7-GFP vector to generate the 2x35S::OsGIF1-GFP cassette. This plasmid was then introduced into rice protoplast cells for transient expression (Li et al., 2013). GFP signals were visualized using a confocal scanning microscope (Nikon A1, Kanagawa, Japan) 24–48 h after transformation.
In order to investigate the function of OsGIF1, plasmids containing CRISPR-Cas9 guide RNAs (gRNAs) against two target sites within the first exon of OsGIF1 were made (Figure 1B). The plasmids were then transformed into the wild-type (WT) variety of Nipponbare (Nipp), and approximately 30 transgenic plants were obtained for each plasmid. Sequencing of PCR-amplified OsGIF1 genomic DNA from transgenic plants showed five types of homozygous mutations within the target sites: one in target site 1 (an A insertion) and four in target site 2 (3-base deletions, 18-base deletions, 10-base deletions, and an T insertion) (Figure 1B). Although no biallelic plants were obtained, several heterozygous transgenic plants of the five mutations were found, and no phenotypes were observed.

FIGURE 1. Knock-out (KO) of OsGIF1 by a CRISPR/Cas9 strategy. (A) Comparisons of the OsGIF1 KO plants, scale bar: 10 cm; (B) Identification of mutation by direct sequencing of target sites in transgenic plants.
Interestingly, the phenotype of the homozygous osgif1 mutant differed from its mutation type. Slight effects were observed on plant height in the 3-/18-base-deletion plants. By contrast, the A/T insertion plants had severe effects on plant height, leaf development, and panicle construction. Additionally, the 10-base-deletion plants had moderate impact on these traits, compared with plants bearing the other two mutations (Figures 1A,B). These phenotypic differences are to some extent in accordance with the effects of each mutation on the protein sequence of OsGIF1. The 3-/18-base deletions only deleted 1 or 6 AAs without changing the coding frame of OsGIF1, while the A/T insertion and 10-base deletions resulted in evidently truncated OsGIF1 by introducing premature stop codons (Supplementary Figure 1). To elucidate whether these differences in phenotype were also linked with off-target effects of the gRNAs, we sequenced several potential off-target sites and found no off-target editing for both gRNAs (Supplementary Table 2). Further linkage analysis of a F2 populations (28 plants in total) generated by the crosses of the T408-6 mutants and the WT found that all the plants with homozygotic mutations were showed in the mutant phenotype (6 plants), whereas other plants carrying no (10 plants) or heterozygous mutations (12 plants) were all normal in phenotype, confirming the association between the mutation and the mutant phenotype. Thus, the phenotype was directly caused by OsGIF1 mutation, and the phenotypic diversity of the different mutation types might have resulted from varying degrees loss of OsGIF1 function.
The most noticeable phenotype in the OsGIF1 KO plants was reduction of plant height. Three types of mutant phenotypes were found (Figure 1A). Thus, we selected three representative mutant plants, namely, T408-14, T408-6, and T409-17 to analyze the phenotype and function of OsGIF1. The plant height of all the KO plants decreased highly significantly compared with that of the WT (Figures 2A,B). Consistent with decrease in height, most of the mutant panicles became smaller, and the number of grains per panicle in the mutants was reduced (Figures 2C,D). We also examined the stems and internodes of mutant plants, since rice plant height is primarily determined by internode length of the stem. The results showed that almost all mutant stems and their internodes were considerably shortened, compared with their WT counterparts (Figures 2A,E). These results demonstrated that functional loss of OsGIF1 led to reduced plant height of rice by shortening the internode length of stems.
OsGIF1 KO plants also shows phenotypes related to leaf development (Figure 2F). Leaf lengths and widths of all the mutant plants were significantly reduced compared with those of the WT (Figures 2G,H). Besides, these mutants also showed different degrees of leaf rolling. The leaf rolling indices of plants with the most severe mutations, namely T408-6 and T409-17, were sharply increased, with almost all leaves rolled compared with those of the WT (Figures 2F,I). These results indicated that OsGIF1 affected rice leaf development by regulating both leaf size and leaf rolling.
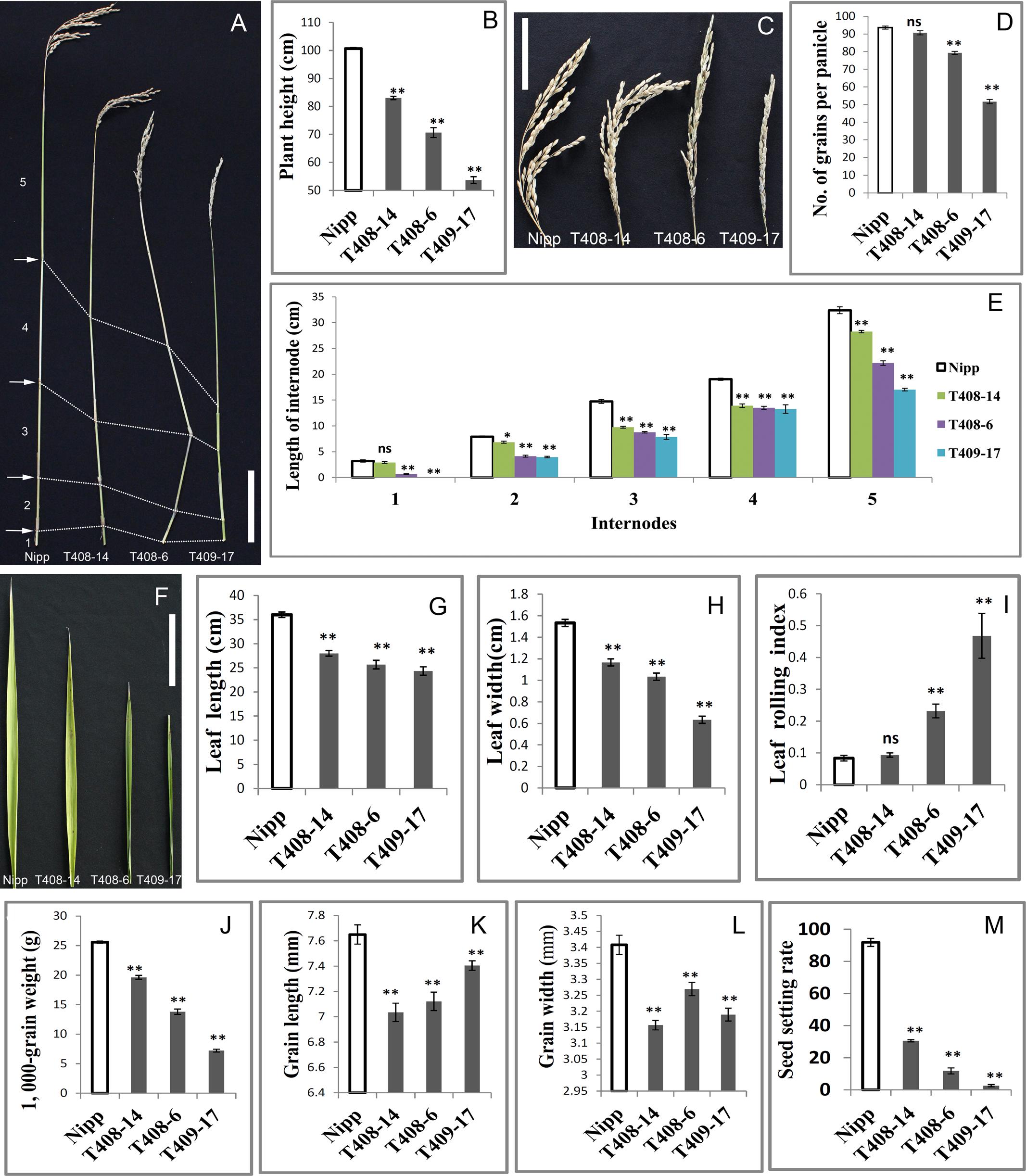
FIGURE 2. Pleiotropic effects of OsGIF1 KO on rice development. Comparisons of several important traits between the OsGIF1 KO and control plants. (A) Stem; (B) plant height; (C) panicle; (D) number of grains per panicle; (E) internode length; (F) leaf; (G) flag leaf length; (H) flag leaf width; (I) leaf rolling index; leaf rolling index = (Lw – Ln)/Lw; Lw, distance of leaf blade margins at unfolding state; Ln, distance of leaf blade margins at natural state; (J) 1000-grain weight; (K) grain length; (L) grain width; and (M) seed setting rate. Scale bars: 10 cm. Values are all shown as means ± standard error of the mean (SEM) (n = 3). Asterisks indicate significant differences between the WT and KO plants as determined by Student’s t-test: ns, not significant; ∗∗p ≤ 0.01; ∗p ≤ 0.05.
Rice is an important crop as a source of carbohydrate for more than half of the world population. Therefore, we investigated whether OsGIF1 affects the grain yield of rice. We found that grain weights of all mutant plants were significantly decreased compared with that of the WT (Figure 2J and Supplementary Figure 2). The reduced grain weights were further observed to be mainly caused by significant decreases in grain length and grain width of the OsGIF1 KO plants (Figures 2K,L). This indicated that OsGIF1 also functioned in rice seed development, which is consistent with our previous findings (Li et al., 2016).
Reductions in the panicle seed setting rate were also observed in the OsGIF1 KO plants (Figure 2M), suggesting a role of OsGIF1 in regulating the rice reproduction process. The degree of reduction was consistent with the mutant type. In the most severely mutated plant, T409-17, the seed setting rate was extremely low, and the plant became almost completely sterile (Figure 2M). The floral structures in T409-17 at the heading date manifested the following series of abnormalities in several whirls of floral organs: tightly wrapped spikelets, shorter, twisted, or ectopic paleas/lemmas, decreased stamens, increased pistils, and white-colored sterile anthers (Supplementary Figure 2). These results indicated that serious KO of OsGIF1 led to various floral organ abnormalities and apparent reductions in the seed setting rate, suggesting an important role of OsGIF1 in the determination of rice floral organs.
We performed detailed investigations of multiple organs of several representative overexpression plants designated as T381 1-3, T395 1-6, and T382 1-1. First, qRT-PCR was performed to confirm the overexpression of OsGIF1 within these representative plants (Figure 3C). The plant heights of the overexpressing plants were slightly or sometimes significantly increased (Figures 3A,D). The transgenic plants also exhibited larger panicles than the WT (Figure 3B) although the number of grains per main panicle did not differ significantly between the transgenic and WT plants (Figures 3E,L). Consistent with our previous work (Li et al., 2016), overexpression of OsGIF1 in these plants also significantly increased grain size and weight by synchronously increasing grain length and width (Figures 3B,F–H). Additionally, overexpression of OsGIF1 apparently affected leaf development, because all of these plants exhibited highly significantly increased leaf length and width (Figures 3I–K). These results confirmed that overexpression of OsGIF1 increased the size of multiple rice organs, indicating a positive role of OsGIF1 in regulating rice organ size.
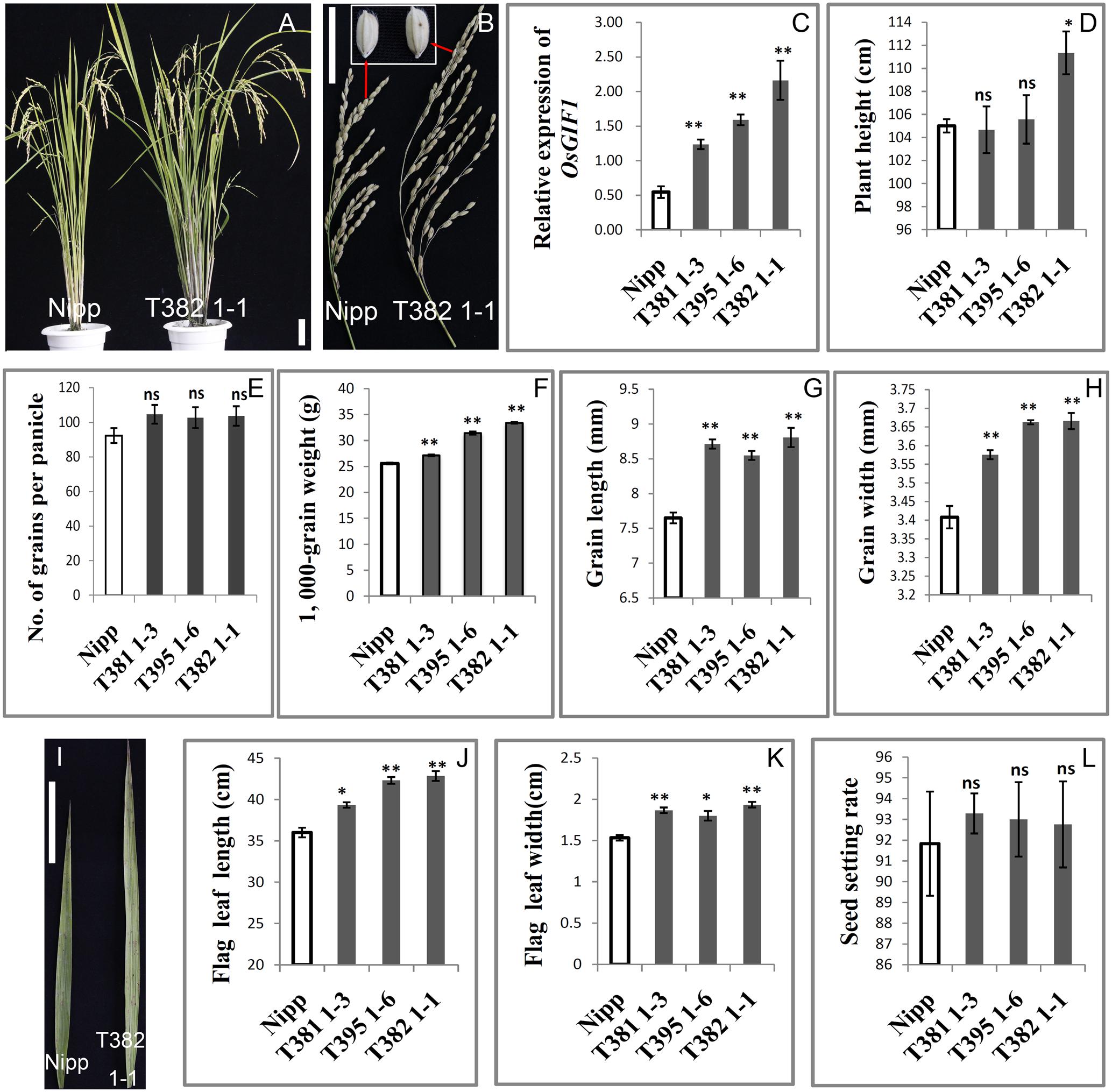
FIGURE 3. Increase in the size of multiple rice organs caused by overexpression of OsGIF1. Comparisons of several important traits between the OsGIF1 overexpression and control plants. (A) Plant phenotype; (B) panicle and grain; (C) relative expression level of OsGIF1; (D) plant height; (E) number of grains per panicle; (F) 1000-grain weight; (G) grain length; (H) grain width; (I) leaf; (J) flag leaf length; (K) flag leaf width; and (L) seed setting rate. Scale bars: 10 cm. Values are all shown as mean ± SEM (n = 3). Asterisks indicate significant differences between the WT and overexpression plants as determined by Student’s t-test: ns, not significant; ∗∗p ≤ 0.01; ∗p ≤ 0.05.
To investigate the cellular effect of OsGIF1, we first performed paraffin sectioning of stem internodes of these plants. Longitudinal histological sectioning analysis showed a clear trend toward reduced cell volume in the KO plants, especially in the severe KO plants (Figure 4A). Overall, these results are consistent with the degree of organ size alternation in these plants (Figure 1A). To further address this, we then counted cell numbers per unit area and found significant increases in T408-14 (+19.92%), T408-6 (+47.39%), and T409-17 (+94.8%) when compared to WT (Figure 4D), suggesting that cell sizes of the KO plants were altered. Direct cell measurement showed that the cell perimeters were significantly reduced in T408-14 (-7.39%), T408-6 (-16.6%) and T409-17 (-30.77%) (Figure 4E). Transverse sectioning of these internodes (Figure 4B) further confirmed the significantly increased cell numbers per unit area (+28.86% in T408-6 and +75.03% in T409-17) (Figure 4F) and reduced cell size (-18.57% in T408-6 and -27.23% in T409-17) (Figure 4G) in the stems of severe mutants. Consistent with the result of stems, transverse section of flag leaves (Figure 4C) also showed significantly reduced cell size in T408-14 (-12.93%), T408-6 (-16.33%), and T409-17 (-23.06%) (Figure 4H). Since grain size is influenced by spikelet hull, we next performed scanning electron microscopy (SEM) analysis of the spikelet hulls of the overexpression and KO plants. Results showed that T382 1-1 exhibited significantly enlarged cell volume compared to WT, while T409-17 showed opposite effect of reduced outer glume cells (Supplementary Figure 3). Cellular measurement further showed that, compared with those in the WT, the length and width of epidermal cells of the outer glumes increased by 43.8 and 15.22% in T382 1-1, and decreased by 16.64 and 12.18% in T409-17, respectively (Supplementary Figure 3). Taken together, these results suggest that OsGIF1 enhances the size of multiple important rice organs predominantly by promoting cell enlargement.
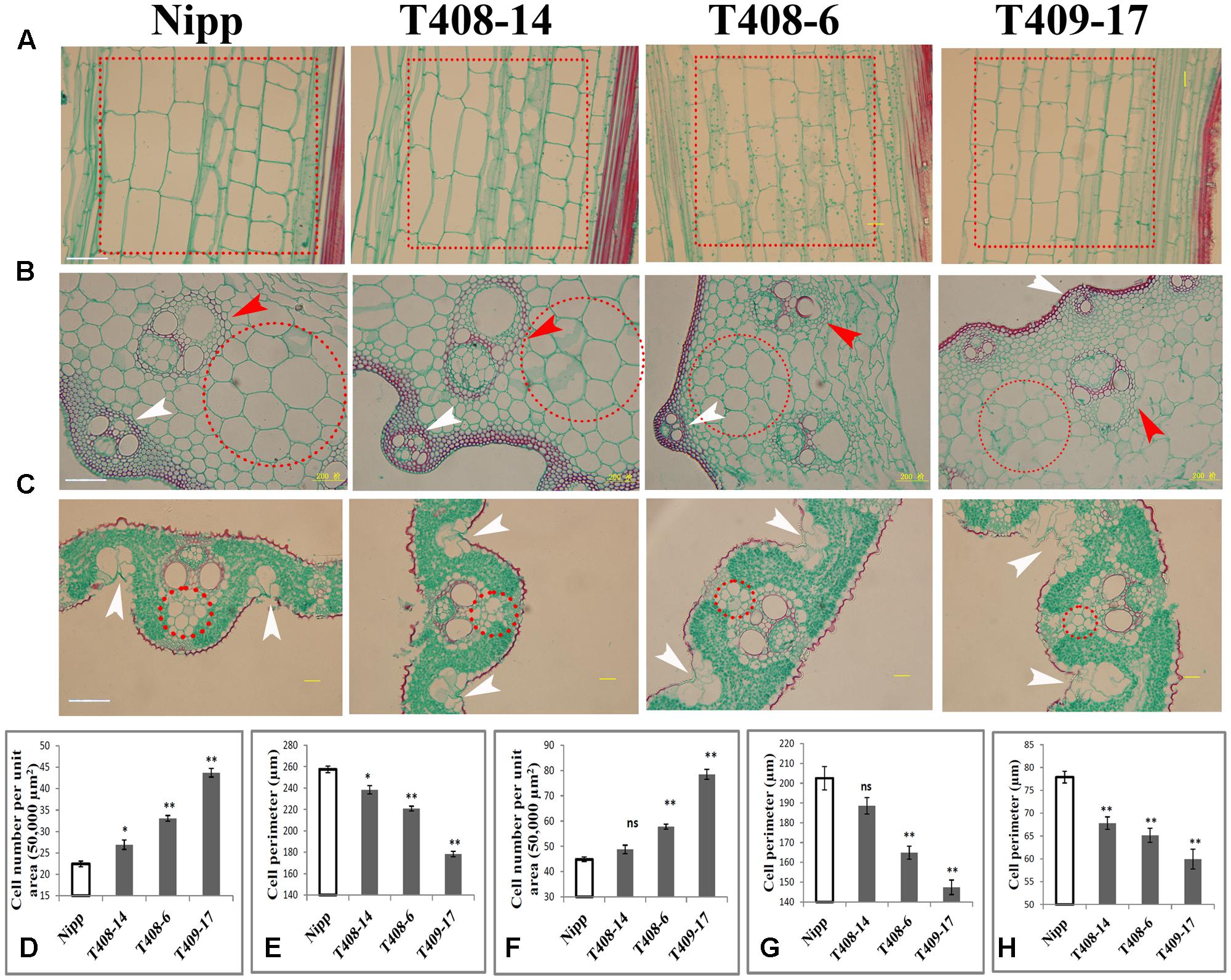
FIGURE 4. OsGIF1 regulation of cell size to control rice organ size. (A–C) Paraffin section analysis of the stems and leaves of KO plants. (A) Longitudinal section of stem internode; dotted boxes in red indicate the major cell types selected for cell counting and measuring. (B) Cross section of stem internode. Red arrow heads indicate inner vascular bundles, and white arrow heads indicate outer vascular bundles. Dotted circles in red indicate the major cell types selected for cell counting and measuring. (C) Cross section of leaves. White arrow heads indicate bulliform cells. Dotted circles in red indicate the major cell types selected for cell counting and measurement. The first internode from the bottom and the middle region of the flag leaf of all plants were collected for observation. Scale bars (50 μm) are shown in Nipp. (D–H) Cell number and size determinations. (D) Cell number per unit area in A; (E), cell size in A; (F), cell number per unit area in B; (G), cell size in B; (H), cell size in C. For cell numbers (per unit area) counting, a total area of 50,000 μm2 for each sample was investigated. For cell volume determination, 10 representative cells within the dotted circles/boxes regions were selected for cell size measuring. Values are all shown as mean ± SEM (n = 3). Asterisks indicate significant differences between the WT and KO plants as determined by Student’s t-test: ns, not significant; ∗∗p ≤ 0.01; ∗p ≤ 0.05.
We also observed other morphological abnormalities except for cell size alternation in these plants. Transverse section of internodes revealed that layers of sclerenchyma cells of the stem vascular bundle and the number and size of the stem vascular bundle were apparently decreased in the KO plants (Figure 4B). Development of the stem vascular bundle was also affected, with incomplete development of outer vascular bundle occurring in the most serious KO plants (Figure 4B). Besides, SEM analysis of flag leaf revealed that the morphologies and organization patterns of cells were also altered in T382 1-1 and T409-17 (Supplementary Figure 4). Stoma guard cells are typically arranged in the longitudinal direction and separated by a longitudinal line of dumbbell silicon cells in the WT. However, two longitudinal lines of dumbbell silicon cells and several incompletely developed lines of stoma guard cells were observed both in T382 1-1 and T409-17. Several cells from these regions were morphologically abnormal compared to the WT. Interestingly, the number of silicon papillae on the leaf stoma guard cells seemed to have increased both in the overexpressing and KO plants (Supplementary Figure 4). Moreover, compared to the WT, organization patterns of stem exterior cells were slightly changed both in overexpressing and KO plants. However, abnormal development of stoma guard cells was especially apparent in T409-17, with significantly fewer stoma guard cells in the stem (Supplementary Figure 4), which is consistent with the findings of a recent study in which a role of AN3/GIF1 in modulating stomatal density was reported (Meng and Yao, 2015). Notably, the number of bulliform cells in the leaves of the serious KO plants was increased (Figure 4C), which abolished the balance of the Ad-Ab patterning and consequently led to rolled leaves. Thus, our findings reveal a cellular mechanism of the leaf-rolling phenotype in the most severe KO plants (Figures 2F,I), consistent with the function of SEMI-ROLLED LEAF1, a gene that modulates rice leaf rolling by regulating the formation of bulliform cells (Xiang et al., 2012). These results suggest that OsGIF1 might also affect other cellular processes during rice organ or tissue development.
We generated a construct in which the GUS gene was driven by the approximately 1.8 kb OsGIF1 promoter to examine the temporal and spatial expression patterns of OsGIF1. Histochemical staining suggested that OsGIF1 was differentially expressed in various rice tissues. Expression was relatively weak in the root (Figure 5A) and mature glume (Figure 5H) but was strong in the internode (Figure 5B), node (Figure 5D), the developing spikelet (Figures 5E–G), and especially in the developing anther (Figures 5E–G). Results of qRT-PCR assays further confirmed this result by showing that OsGIF1 was highly expressed in developing stems but moderately in the root, young panicle, and flower (Figure 5I). Overall, the expression pattern of OsGIF1 is consistent with its function. However, both GUS staining and qRT-PCR analysis indicated extremely low expression of OsGIF1 in leaves approaching maturity (Figures 5C,I). Taken together with our results that OsGIF1 is involved in leaf development (Figures 2F–I, 3I–K), these findings point toward a role for OsGIF1 in the early development of rice leaf.
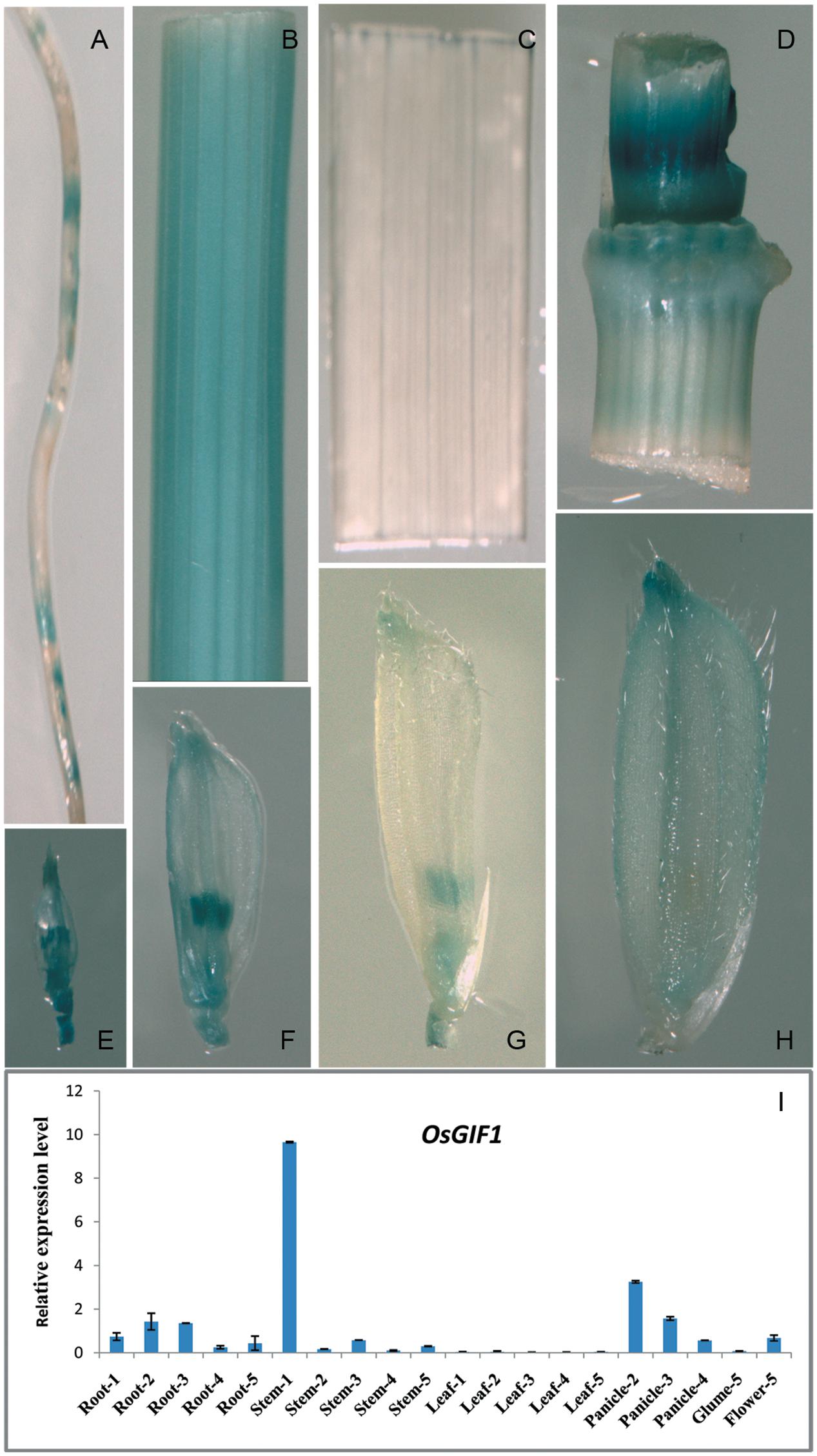
FIGURE 5. Expression profile of OsGIF1. (A–H) GUS staining of various tissues of OsGIF1pro::GUS transgenic plants. (A) Root; (B) internode; (C) leaf; (D) stem node; (E–H) Spikelets of plants with panicle lengths of 2 (E), 5 (F), 10 (G), and 15 cm (H), respectively. (I) Relative expression levels of OsGIF1 in various tissues of WT plants. (1), (2), (3), (4), and (5) indicate plants with panicle lengths of 0, 2, 5, 10, and 15 cm, respectively. Values are all shown as mean ± SEM (n = 3).
In order to investigate the subcellular localization of OsGIF1, we constructed an OsGIF1-GFP (green fluorescent protein) fusion construct with its expression driven by the CaMV 35S promoter. Transient expression in the rice protoplast cells showed that OsGIF1-GFP localized preferentially in the nucleus and was weakly expressed in the cytoplasm (Figure 6). This result is consistent with the idea that GIF proteins function as transcriptional co-activators by forming complexes with GRF transcription factors.
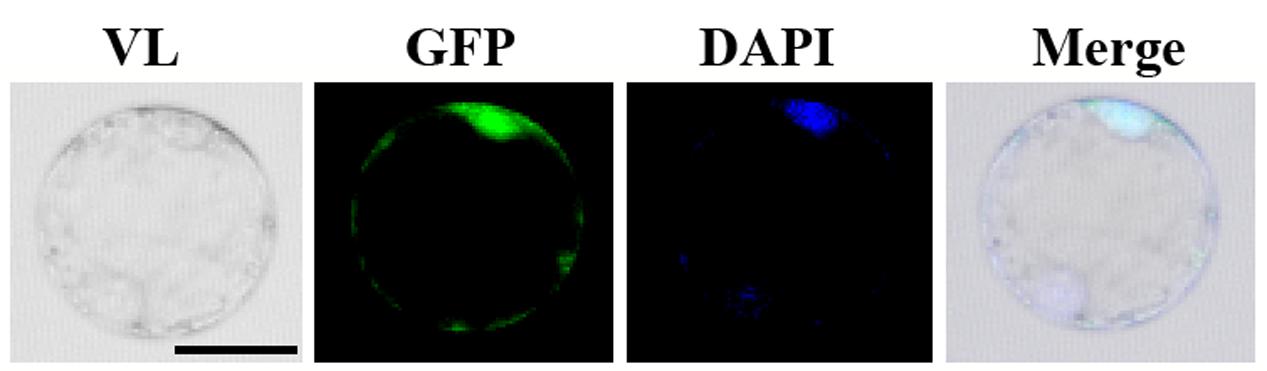
FIGURE 6. Subcellular localization of OsGIF1. Confocal images of rice protoplast cells after 48 h of infection. The OsGIF1 protein (visualized based on GFP signal) was mainly localized in the nucleus in rice cells transfected with the 2x35S::OsGIF1-GFP construct. Scale bar: 25 μm.
Many lines of evidence indicate that GIF genes are involved in regulating the vegetative growth of multiple organs (Iwakawa et al., 2002, 2007; Xu et al., 2003; Kim and Kende, 2004; Horiguchi et al., 2005, 2011; Kanei et al., 2012; Debernardi et al., 2014; Vercruyssen et al., 2014; Kim and Tsukaya, 2015; Nelissen et al., 2015; Omidbakhshfard et al., 2015), reproductive development (Lee et al., 2014; Liang et al., 2014; Meng et al., 2016b), and stress tolerance (Meng and Yao, 2015) in Arabidopsis plants. In the present study, KO of OsGIF1 led to the following pleiotropic defects in rice development: reduction of plant height caused by significantly shortened internodes; decreased leaf size due to reduction in leaf length and width; and small seed size owing to reduction in grain length and width. The functions of OsGIF1 were further elucidated by results of our overexpression studies. Rolled leaves and various kinds of floral organ abnormalities were also observed in the most severe KO of OsGIF1. Our results as a whole are in agreement with those reported previously in Arabidopsis and suggest various roles of GIF1/OsGIF1 during plant organ development.
The most important function of GIF1/OsGIF1 appears to be the determination of the size of different organs. However, the cellular effect of OsGIF1 in rice may differ from its counterpart GIF1 in Arabidopsis. Organ sizes of plants have been reported to be enlarged by two major mechanisms, namely, by promoting cell expansion or increasing cell proliferation. The majority of reports in Arabidopsis indicate that GIF1 influences organ size by promoting cell proliferation (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015). In the present study, we demonstrated that OsGIF1 affects leaf size, length of the stem internodes, and seed size possibly by promoting cell expansion. Recently, a major QTL, namely GLW2/GL2/GS2, the allelic mutation of OsGRF4, was reported to positively regulate grain weight and size in rice by several independent groups (Che et al., 2015; Duan et al., 2015; Hu et al., 2015; Li et al., 2016). Further investigations demonstrated that OsGRF4 directly interacts with OsGIF1 to positively regulate seed size in rice mainly by promoting cell expansion (Che et al., 2015; Duan et al., 2015; Li et al., 2016). Although, the detailed cellular mechanism underlying this function of OsGRF4 remains unknown, the results are consistent with our findings that OsGIF1 might be involved in organ size regulation via mechanisms controlling cell expansion. The difference in GIF1 functions between rice and Arabidopsis is very interesting but remains elusive due to a fundamental lack of understanding of functions of the rice GIF proteins and their possible partners. How and when this difference occurs should be clarified in future investigations.
However, our results also suggest a role of OsGIF1 in regulating cell proliferation of some tissues, such as the leaf bulliform cells and silicon papillae of stoma guard cells in rice. Recent works in rice also suggest that the cell proliferation pathway contributes to grain size regulation, but is far less important than cell expansion (Che et al., 2015; Duan et al., 2015; Li et al., 2016). These results might also suggest that both cellular pathways work simultaneously in rice. Another difference in GIF1 function between rice and Arabidopsis is that, although GIFI is involved in floral organ development, and despite differences in their mutant phenotypes, only the Arabidopsis gif1 gif2 gif3 triple mutant, but not the single or double mutants showed obvious phenotypes in reproductive organs (Lee et al., 2014). By contrast, in rice, severe KO of OsGIF1 single gene produced several reproductive organ abnormalities. These results suggest real functional differences between OsGIF1 and GIF.
Interestingly, Che et al. (2015) demonstrated that OsGRF4/GL2 functions by activating brassinosteroid response, and simultaneously, the elevated BR response stimulates cell elongation by promoting gibberellin (GA) biosynthesis in rice seedlings. Consistently, in the present study, we showed that OsGIF1 affects the size of leaf, stem internodes, and seed majorly by promoting cell expansion. Considering that GIFs have been widely proved to function in complexes with GRFs in Arabidopsis and rice (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015), it is reasonable that the similar biochemical pathways, like the BR and GA pathways, may also influence the leaf/stem cell size in rice. However, several other direct evidences are necessarily needed to apply the OsmiR396-GRF4-GIF1 regulatory module and its related biochemical mechanism to other organs.
Sequencing of OsGIF1 KO plants revealed five types of homozygous mutations within the target sites. The phenotype of the homozygous osgif1 mutant differed from its mutation type as follows: 3-/18-base deletion (resulted in 1 or 6 AA deletions) plants showed slight effects; A/T insertion (resulting in premature TGA stop codon at the 173rd AA) plants exhibited the most severe phenotypes; and the 10-base-deletion plants (resulting in premature TGA stop codon at 101st AA) have moderate effects. The range of mutant phenotypes was not caused by off-target effects of the gRNAs and might have resulted instead from varying degrees of loss of OsGIF1 function. The phenotypic difference between the 3-/18-base-deletion and A/T insertion plants suggest that the 101–226 AAs polypeptide segment of OsGIF1 has an important function in regulating rice organ size. However, phenotypic differences between 10-base-deletion and A/T insertion plants suggest that the new 101–173 AAs polypeptide segment produced by A/T insertion frameshift mutation might have antagonistic effects against the residual function of the truncated OsGIF1 protein (1–101 AAs). Although, the major functions of GIF are being gradually elucidated in plants, further efforts are necessary to gain a deeper understanding of the detailed molecular function of each polypeptide fragment of OsGIF1.
SL and PL designed the experiments and directed the project; JiZ and ZH performed the cloning and functional analysis and collected almost all of the data; YR and FG performed the expression analysis and tissue localization and cell localization; DC, WL, and YC performed genetic transformations; TL, GY, and XW performed the phenotypic characterization of the mutant and transgenic plants; SW, HL, LW, and QD carried out the field experiments and investigations; YL, JuZ, and AZ constructed all the vectors; SL analyzed the data and wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (91435102, 31570004, and 31471474), The Sichuan Provincial Funding for Distinguished Young Scholars (2015JQ0048), and the Open Research Fund of the State Key Laboratory of Hybrid Rice (Hunan Hybrid Rice Research Center, 2016KF10).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01730/full#supplementary-material
Aizawa, H., Hu, S. C., Bobb, K., Balakrishnan, K., Ince, G., Gurevich, I., et al. (2004). Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303, 197–202. doi: 10.1126/science.1089845
Anastasiou, E., Kenz, S., Gerstung, M., MacLean, D., Timmer, J., Fleck, C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856. doi: 10.1016/j.devcel.2007.10.001
Brett, D., Whitehouse, S., Antonson, P., Shipley, J., Cooper, C., and Goodwin, G. (1997). The SYT protein involved in the t(X;18) synovial sarcoma translocation is a transcriptional activator localised in nuclear bodies. Hum. Mol. Genet. 6, 1559–1564. doi: 10.1093/hmg/6.9.1559
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2:15195. doi: 10.1038/nplants.2015.195
Choi, D., Kim, J. H., and Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
de Bruijn, D. R., and Geurts van Kessel, A. (2006). Common origin of the human synovial sarcoma associated SS18 and SS18L1 gene loci. Cytogenet. Genome Res. 112, 222–226. doi: 10.1159/000089874
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79, 413–426. doi: 10.1111/tpj.12567
Dello Ioio, R., Linhares, F. S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., et al. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678–682. doi: 10.1016/j.cub.2007.02.047
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
dos Santos, N. R., de Bruijn, D. R., Balemans, M., Janssen, B., Gartner, F., Lopes, J. M., et al. (1997). Nuclear localization of SYT, SSX and the synovial sarcoma-associated SYT-SSX fusion proteins. Hum. Mol. Genet. 6, 1549–1558. doi: 10.1093/hmg/6.9.1549
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2:15203. doi: 10.1038/nplants.2015.203
Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q., Gerentes, D., et al. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. doi: 10.1105/tpc.8.2.155
Fang, W., Wang, Z., Cui, R., Li, J., and Li, Y. (2012). Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 70, 929–939. doi: 10.1111/j.1365-313X.2012.04907.x
Gingras, A. C., Raught, B., Gygi, S. P., Niedzwiecka, A., Miron, M., Burley, S. K., et al. (2001). Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864. doi: 10.1101/gad.912401
Halliday, K. J. (2004). Plant hormones: the interplay of brassinosteroids and auxin. Curr. Biol. 14, R1008–R1010. doi: 10.1016/j.cub.2004.11.025
Hiei, Y., Komari, T., and Kubo, T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218. doi: 10.1023/A:1005847615493
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Horiguchi, G., Nakayama, H., Ishikawa, N., Kubo, M., Demura, T., Fukuda, H., et al. (2011). ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 52, 112–124. doi: 10.1093/pcp/pcq178
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Hu, Y., Xie, Q., and Chua, N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. doi: 10.1105/tpc.013557
Iwakawa, H., Iwasaki, M., Kojima, S., Ueno, Y., Soma, T., Tanaka, H., et al. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51, 173–184. doi: 10.1111/j.1365-313X.2007.03132.x
Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., et al. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. doi: 10.1093/pcp/pcf077
Kanei, M., Horiguchi, G., and Tsukaya, H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139, 2436–2446. doi: 10.1242/dev.081547
Kato, H., Tjernberg, A., Zhang, W., Krutchinsky, A. N., An, W., Takeuchi, T., et al. (2002). SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J. Biol. Chem. 277, 5498–5505. doi: 10.1074/jbc.M108702200
Kawade, K., Horiguchi, G., and Tsukaya, H. (2010). Non-cell-autonomously coordinated organ size regulation in leaf development. Development 137, 4221–4227. doi: 10.1242/dev.057117
Kawade, K., Horiguchi, G., Usami, T., Hirai, M. Y., and Tsukaya, H. (2013). ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr. Biol. 23, 788–792. doi: 10.1016/j.cub.2013.03.044
Kim, J., and Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Lee, B. H., Wynn, A. N., Franks, R. G., Hwang, Y. S., Lim, J., and Kim, J. H. (2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386, 12–24. doi: 10.1016/j.ydbio.2013.12.009
Li, S., Gao, F., Xie, K., Zeng, X., Cao, Y., Zeng, J., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14, 2134–2146. doi: 10.1111/pbi.12569
Li, S., Li, W., Huang, B., Cao, X., Zhou, X., Ye, S., et al. (2013). Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat. Commun. 4:2793. doi: 10.1038/ncomms3793
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Liang, G., He, H., Li, Y., Wang, F., and Yu, D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Liu, H., Guo, S., Xu, Y., Li, C., Zhang, Z., Zhang, D., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mao, J., Zhang, Y. C., Sang, Y., Li, Q. H., and Yang, H. Q. (2005). From the cover: a role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. U.S.A. 102, 12270–12275. doi: 10.1073/pnas.0501011102
Meng, L. S., Li, Y. Q., Liu, M. Q., and Jiang, J. H. (2016a). The Arabidopsis ANGUSTIFOLIA3-YODA gene cascade induces anthocyanin accumulation by regulating sucrose levels. Front. Plant Sci. 7:1728. doi: 10.3389/fpls.2016.01728
Meng, L. S., Wang, Y. B., Loake, G. J., and Jiang, J. H. (2016b). Seed embryo development is regulated via an AN3-MINI3 gene cascade. Front. Plant Sci. 7:1645. doi: 10.3389/fpls.2016.01645
Meng, L. S., and Yao, S. Q. (2015). Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnol. J. 13, 893–902. doi: 10.1111/pbi.12324
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Nelissen, H., Eeckhout, D., Demuynck, K., Persiau, G., Walton, A., van Bel, M., et al. (2015). Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27, 1605–1619. doi: 10.1105/tpc.15.00269
Omidbakhshfard, M. A., Proost, S., Fujikura, U., and Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Silverstone, A. L., Jung, H. S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T. P. (2001). Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1566. doi: 10.1105/tpc.13.7.1555
Thaete, C., Brett, D., Monaghan, P., Whitehouse, S., Rennie, G., Rayner, E., et al. (1999). Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum. Mol. Genet. 8, 585–591. doi: 10.1093/hmg/8.4.585
Ubeda-Tomas, S., Federici, F., Casimiro, I., Beemster, G. T., Bhalerao, R., Swarup, R., et al. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194–1199. doi: 10.1016/j.cub.2009.06.023
van der Knaap, E., Kim, J. H., and Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S. K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Xiang, J. J., Zhang, G. H., Qian, Q., and Xue, H. W. (2012). Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. doi: 10.1104/pp.112.199968
Keywords: rice, OsGIF1, organ size, cell size, knock-out
Citation: He Z, Zeng J, Ren Y, Chen D, Li W, Gao F, Cao Y, Luo T, Yuan G, Wu X, Liang Y, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P and Li S (2017) OsGIF1 Positively Regulates the Sizes of Stems, Leaves, and Grains in Rice. Front. Plant Sci. 8:1730. doi: 10.3389/fpls.2017.01730
Received: 25 April 2017; Accepted: 21 September 2017;
Published: 05 October 2017.
Edited by:
Chengdao Li, Murdoch University, AustraliaReviewed by:
Yongzhong Xing, Huazhong Agricultural University, ChinaCopyright © 2017 He, Zeng, Ren, Chen, Li, Gao, Cao, Luo, Yuan, Wu, Liang, Deng, Wang, Zheng, Zhu, Liu, Wang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Li, bGlwaW5nNjU3NUAxNjMuY29t Shuangcheng Li, bGlzYzkyNjEwNUAxNjMuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.