- 1State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
- 2Section of Cell and Developmental Biology, Division of Biological Sciences, University of California, San Diego, La Jolla, CA, United States
Nitrate is a key nutrient that affects maize growth and yield, and much has yet to be learned about nitrate regulatory genes and mechanisms in maize. Here, we identified nine ZmNLP genes in maize and analyzed the functions of two ZmNLP members in nitrate signaling. qPCR results revealed a broad pattern of expression for ZmNLP genes in different stages and organs with the highest levels of transcript expression of ZmNLP6 and ZmNLP8. When ZmNLP6 and ZmNLP8 were overexpressed in the Arabidopsis nitrate regulatory gene mutant nlp7-4, nitrate assimilation and induction of nitrate-responsive genes in the transgenic plants were recovered to WT levels, indicating that ZmNLP6 and ZmNLP8 can replace the essential roles of the master nitrate regulatory gene AtNLP7 in nitrate signaling and metabolism. ZmNLP6 and ZmNLP8 are localized in the nucleus and can bind candidate nitrate-responsive cis-elements in vitro. The biomass and yield of transgenic Arabidopsis lines overexpressing ZmNLP6 and ZmNLP8 showed significant increase compared with WT and nlp7-4 mutant line in low nitrate conditions. Thus, ZmNLP6 and ZmNLP8 regulate nitrate signaling in transgenic Arabidopsis plants and may be potential candidates for improving nitrogen use efficiency of maize.
Introduction
Nitrogen is one of the most critical macronutrients for plant growth, development, and production (Crawford and Forde, 2002; Kusano et al., 2011). Most plants that grow in aerobic soils, such as maize and wheat absorb nitrogen mainly in the form of nitrate. In agriculture, nitrogen fertilizer has been widely used to maintain the high yield of crops. Because of low NUE, nitrate cannot be completely absorbed by plants and therefore a large part is lost to the environment, resulting in serious environmental pollution. Improving NUE is key to solving these problems. However, the current progress in NUE is limited by the fact that genes and mechanisms involved in nitrate regulation in plants are still poorly understood. Thus, identification and characterization of nitrate regulatory genes and underlining mechanisms is of great importance, especially in crop plants, for developing sustainable agriculture.
The uptake and transport of nitrate in plants have been found to be achieved by NRT1, NRT2, CLC, and SLAH gene families (Krapp et al., 2014). After importation into the cells, nitrate will be reduced then assimilated into amino acids by the action of NR, NiR and GS/glutamate synthase (GOGAT). Nitrate serves not only as a nutrient, but also as a signal. Previous reports on nitrate regulatory genes mainly focused on components involved in regulating root architecture and the PNR in Arabidopsis (Alvarez et al., 2012; Forde, 2014; Medici and Krouk, 2014). AtANR1, an Arabidopsis MADS-box gene, was the first identified nitrate regulator to function in lateral root branching in response to nitrate (Zhang and Forde, 1998). Later, the nitrate transporter AtNRT1.1 was found to work upstream of AtANR1 in regulating lateral root development on nitrate-rich patches (Remans et al., 2006). A unique nitrogen-responsive module AtmiR393/AFB3 controls the growth of both primary and lateral root in response to external and internal nitrogen availability in Arabidopsis (Vidal et al., 2010). Recently, AtTCP20 was characterized to redirect root growth to nitrate-rich regions by a systemic signaling pathway and to support root meristem growth under nitrate starvation with the interaction of AtNLP7 (Guan et al., 2014, 2017). In addition, CLE, a peptide generated in the root cells, was identified to interact with AtCLV1 to control the development of lateral root in nitrogen-deficient environment (Araya et al., 2014). Another peptide CEP induced by nitrogen starvation in roots is translocated to shoots to bind CEP receptor to regulate the lateral root foraging (Tabata et al., 2014). AtABI2, a member of protein phosphatase 2C family, dephosphorylates AtCIPK23 and AtCBL1 to modulate nitrate uptake and lateral root development in a AtNRT1.1-dependent manner in Arabidopsis (Liu et al., 2015).
During the last several years, a few of the regulatory genes involved in PNR have also been identified in Arabidopsis. AtNRT1.1 (NPF6.3), which was previously identified as a dual-affinity nitrate transporter, has been found to be a nitrate sensor (transceptor) to regulate nitrate signaling (Ho et al., 2009; Wang et al., 2009; Gojon et al., 2011). The AtNRT1.1 can be phosphorylated by AtCIPK23 to respond to different nitrate concentrations (Ho et al., 2009). Furthermore, AtNRG2 has been isolated by a forward genetics approach and functions as an essential nitrate signaling component (Xu et al., 2016). It regulates and works upstream of AtNRT1.1. AtLBD37/38/39 repressed the expression of genes involved in nitrate uptake and assimilation to respond to nitrate status (Rubin et al., 2009). AtSPL9, AtNAC4, AtTGA1, and AtTGA4 have been identified by using system approaches to function in regulating nitrate response (Krouk et al., 2010; Alvarez et al., 2014; Vidal et al., 2014). AtCPK10/30/32, Ca2+-depended protein kinases, were recently identified by a nitrate-sensitized and targeted functional genomic screen and play key roles in nitrate signaling (Liu K.H. et al., 2017).
Notably, AtNLP7 has been demonstrated as a central component in regulating nitrate response in Arabidopsis. Nitrate induction of NIA1, NIA2, NRT2.1, and NRT2.2 were all severely reduced in nlp7 mutants (Castaings et al., 2009). The subcellular localization of AtNLP7 is regulated by nitrate: it accumulates in the nucleus when nitrate is present (Marchive et al., 2013). The nuclear retention is controlled by the phosphorylation of the conserved serine205 by AtCPK10/30/32 in the N-terminal region of AtNLP7 (Liu K.H. et al., 2017). In addition, other members of the AtNLP family can bind the NRE and modulate nitrate responsive genes (Konishi and Yanagisawa, 2013).
Maize is an essential food and cash crop in many parts of the world. Much of the world’s nitrogen fertilizer is used to support the growth and high yield of maize. However, little is known about the regulation of nitrate assimilation in maize at the molecular and genetic levels. It has been reported that the expression levels of maize nitrate transporters ZmNRT2.1 and ZmNRT2.2 exhibit two distinct peaks in different growth stages under low nitrate conditions (Garnett et al., 2013). On the other hand, the expression of ZmNRT2.1 and ZmNRT2.2 in the roots of seedlings and adult plants are inhibited by local low nitrate while enhanced by local high nitrate (Yu et al., 2014). Additionally, most ZmNRT2 family members are responsive to nitrogen treatments (Garnett et al., 2015). GS gene ZmGln1-3 is localized in mesophyll cells and plays an important role in grain production (Martin et al., 2006). However, there have been no nitrate regulatory genes characterized in maize. In this study, we identified all nine ZmNLP genes in maize and analyzed their expression profiles. Genetic and molecular analyses suggest that ZmNLP6 and ZmNLP8 play essential roles in nitrate signaling and assimilation. Both genes are localized in the nucleus. Y1H assay revealed that ZmNLP6 and ZmNLP8 could bind NRE of ZmNRT1.2 and ZmNiR2 directly in vitro. Furthermore, constitutively overexpressing ZmNLP6 and ZmNLP8 in Arabidopsis can promote growth and yield under low nitrate conditions, implicating their potential in improving maize NUE.
Materials and Methods
The Characterization of the NLP Genes in Plants
To character the NLP members, the protein sequences of NLP family in Arabidopsis and rice were used as query sequences to perform BLASTp in 34 species in plant genomics resource1. The BLASTp were used with an e-value cutoff set to 1e-003. The isolated protein sequences were examined using the domain analysis program Hidden Markov Model (HMM) of SMART2 and Pfam3 with the default cutoff parameters (Bateman et al., 2002; Letunic et al., 2012). We obtained the sequences with the Pfam number PF02042.10 and PF00564.19, which contained a typical RWP-RK and PB1 domains, from the maize genome sequences using a Perl-based script.
Phylogenetic Analysis, Gene and Protein Structure of the NLP Members
The full length NLP protein sequence of Arabidopsis, maize, sorghum, and rice were used for constructing phylogenetic tree with the neighbor-joining (NJ) method of the MEGA7 program4 using the p-distance and complete default option parameters. The reliability of the obtained trees was tested using a bootstrapping method with 1000 replicates. The images of the phylogenetic trees were drawn using MEGA7. The genomic and CDS of NLP members were downloaded from the plantGDB database5. The gene structures of NLP genes were generated using the GSDS6. The position of GAF-Like, RWP-RK and PB1 domain was indicated from the Pfam results.
Sequence Alignment, Chromosomal Location and Characterization of NLP Members in Maize
The amino acid sequences of ZmNLPs were aligned using the ClustalX program with BLOSUM30 as the protein-weight matrix. The chromosomal location was retrieved from the maize genome data7, and the genes were mapped to the chromosomes using MapDraw program. The isoelectric points and molecular weights of these proteins were obtained with the help of the proteomics and sequence analysis tools on the ExPASy Proteomics Server8.
The conserved motif was discovered by Motif discovery in MEME9 using 248 NLP proteins in 34 species. The logos were created and downloaded by Multiple Em for Motif Elicitation.
Plant Growth and Treatments
Maize B73 inbred seeds were grown in the fields every year (2014–2016) and a part of 0–20 cm underground roots and ear leaves(whole) were collected at the stage vegetative 3 (V3), vegetative 5 (V5), vegetative 9 (V9), vegetative 13 (V13), reproductive 1 (R1), and vegetative 18 (V18) (Sekhon et al., 2011). These are important stages for the growth and development of maize. A part of 0–20 cm underground roots, ear leaf (whole), ear stem (whole), ear sheath (whole), tassels (whole), and grains in reproductive 1 stage (15 days after pollination) were collected for testing the gene expression every year. Maize plants used for testing the induction of nitrate responsive genes were grown on matrix watered with B5 solution to 3-leaf stage, and then the endosperm were removed from the seedlings. The uniform seedlings were transferred to hydroponics system (Liu W. et al., 2017) with 2.5 mM ammonium succinate for 2 days, followed by the treatments with 0.2 mM or 10 mM KNO3/KCl for 2 h. The roots were collected for testing the expression of ZmNLP genes. Maize seeds were grown on matrix watered with different nitrate concentrations (0, 0.2, 2.5, 5, and 10 mM KNO3) for 2 weeks and then the shoots and roots were collected for detecting the expression of ZmNLP genes.
Arabidopsis plants used for testing the YFP fluorescence intensity were grown on plates with nitrate medium (10 mM KNO3) for 4 days and observed under a fluorescence microscope (Nikon Eclipse Ti-s). The fluorescence of roots was quantified using Image J. For Arabidopsis plants used for qPCR analysis of nitrate responsive genes, seedlings were grown in aseptic hydroponics (initial medium with 2.5 mM ammonium succinate) as previously described (Xu et al., 2016) for 7 days and then treated with 10 mM KNO3 or KCl for 2 h. RNA was extracted from the roots for testing the expression of nitrate responsive genes.
qPCR
Total RNA was isolated using a total RNA miniprep kit (CWBIO). Template cDNA samples were prepared using the EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TRAN). qPCR reaction was prepared with UltraSYBR Mixture (With Rox I) followed by performing qPCR using QuantStudioTM 6 (Thermo Fisher Scientific). ZmActin1 (J01238), ZmUBCE (GRMZM2G027378) (Manoli et al., 2012), AtTUB2 (At5g62690), and AtActin8 (At1g49240) were used as the internal reference genes.
Subcellular Localization Assay
The coding regions of ZmNLP6 and ZmNLP8 containing the stop codon were cloned into pMDC43 (Invitrogen). The pMDC43-GFP-ZmNLP6 and pMDC43-GFP-ZmNLP8 were transiently transformed into Nicotiana benthamiana epidermal cells and Arabidopsis mesophyll protoplasts. The GFP fluorescence was observed using a confocal microscope (Leica TCS SP5II).
Yeast One-Hybrid (Y1H) Assay
Yeast one-hybrid (Y1H) assay was performed using the system as described previously (Lin et al., 2007). The full length of cDNA for ZmNLP6 and ZmNLP8 were ligated into the pJG4 vector and the putative NRE from the 5′ and 3′ flanking sequences of nitrate uptake and assimilation genes in maize were ligated into the pLacZ-2U vector. The constructed vectors were co-transferred into yeast EGY48 competent cells and grew the cells on SD/-Trp/-Ura medium for 4 days, and then positive clones were transferred into a new plate of SD/-Trp/-Ura with X-gal.
Molecular Constructs and Transactivation Activity Assay
The nlp7-4 mutant was isolated from EMS-treated seeds of transgenic lines containing a nitrate-regulated promoter fused to a yellow fluorescent protein (NRP::YFP) (Wang et al., 2009). ZmNLP6 and ZmNLP8 were amplified from the B73 inbred line by PCR. The constructs p35S::ZmNLP6 and p35S::ZmNLP8 in binary vector pSuper1300 were transformed into the nlp7-4 mutant (Xu et al., 2016) using the Agrobacterium-mediated floral dip method. Homozygous transgenic lines were isolated and two of these lines for each ZmNLP gene were selected for further investigation. All primers were listed in Supplementary Table S1.
Nitrate Metabolite Measurement
For nitrate content detection, seedlings of WT, nlp7-4, and transgenic lines were grown in 1/2 MS medium (pH = 5.7, 10 mM KNO3 and 10 mM NH4NO3) for 7 days and then collected 0.05–0.1 g of the whole seedlings. The samples were milled into powder using a RETCH MM400 and then added into l mL ddH2O followed by boiling at 100 °C for 30 min. The cooled sample was centrifuged at 13000 rpm for 10 min and 400 μL supernatant was collected to the flow cell. The nitrate content was detected by hydrazine reduction method using AutoAnalyzer 3 continuous flow analytical system (SEAL Analytical).
Nitrate reductase activity was detected by sulfanilamide colorimetric method as described previously (Ferrario-Mery et al., 1998). Amino acid content in tissue was determined using ninhydrin colorimetric analysis as described previously (Rosen, 1957).
Growth and Yield Measurement
For root architecture, WT, nlp7-4, ZmNLP6 and ZmNLP8 transgenic lines grown vertically on agar plates containing 0.2, 2.5, and 5 mM KNO3, respectively, for 10 days. The primary root length was measured by Image J and the lateral root number was counted under a dissecting microscope (Optec). For biomass measurement, seedlings were grown on KNO3 solid medium 0.2, 2.5, and 5 mM KNO3, respectively, for 10 days and fresh weight was measured or seedlings were dried under 65°C for 3 days to obtain the dry weight.
For yield measurement, seedlings were grown in pots with matrix at 22°C, 16 h light/8 h dark photoperiod in growth room and watered with 0.2 mM or 5 mM KNO3. The seeds were collected when plants were mature and dried at 30°C for 2 weeks.
Results
NLP Gene Family in Maize
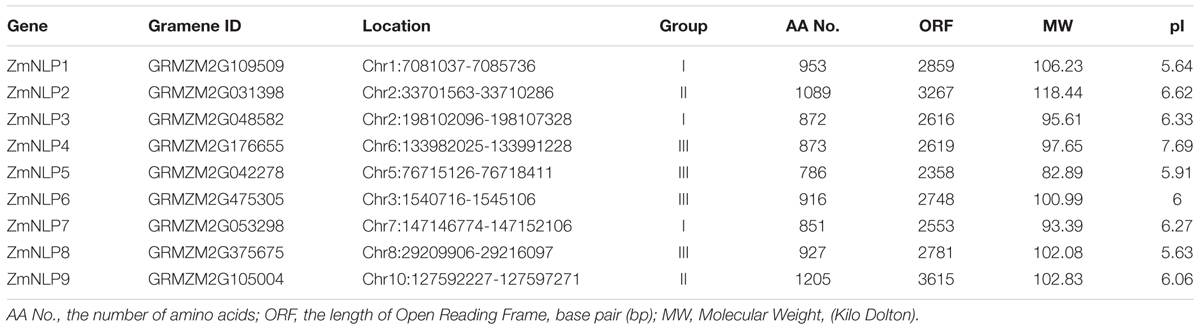
To find the NLP homologous genes in maize, BLASTp of 34 species using NLP family amino acid sequences of Arabidopsis and rice as queries were performed from the plant genomics resource10. The search results were further verified by HMM of SMART and Pfam tools, and the proteins containing RWP-RK and PB1 domains were considered as NLP homologs. By using this strategy, 248 proteins were defined as NLP homologs in 34 species (Supplementary Figure S1). NLP proteins exist in all land plants we searched. Nine NLP members were found in the maize genome (Table 1). We named these nine members, ZmNLP1 to 9, mainly following their chromosomal locations except GRMZM2G475305 as ZmNLP6 to be consistent with nomenclatures proposed in the previous study (Burdo et al., 2014) and GRMZM2G176655 on chromosome 6 alternately named ZmNLP4. The genes of the ZmNLP family are distributed over eight of the 10 maize chromosomes (except Chr.4 and Chr.9) (Supplementary Figure S2). Pairwise comparison of the ZmNLP proteins revealed levels of identity ranging from 16.67% between ZmNLP5 and ZmNLP7 to 66.08% between ZmNLP6 and ZmNLP8 (Supplementary Figure S3). Three sister pairs (ZmNLP3/7, ZmNLP2/9, and ZmNLP6/8) of paralogs with high identity (>60%) and strong bootstrap support (>90%) (Figure 1A) were found, implicating a paralogous pattern of ZmNLP divergence by gene duplication in maize. The alignment results of the nine full length ZmNLP amino acid sequences were shown in Supplementary Figure S4. Two conserved domains, RWP-RK and PB1, were found to be separated by an area of low similarity in the C-termini of ZmNLPs.
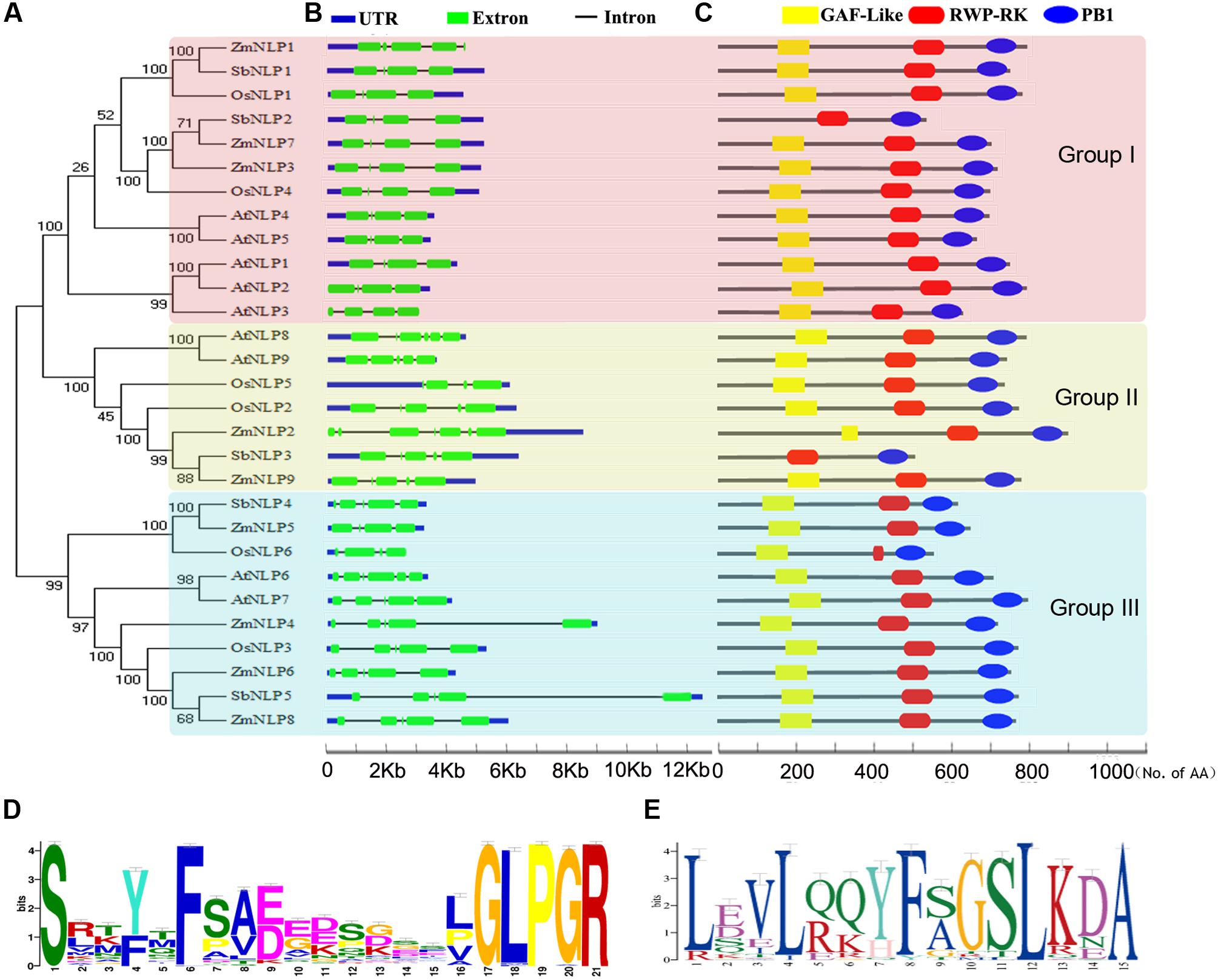
FIGURE 1. Phylogenetic analysis, schematic diagram for gene and protein structures, and conserved motifs of NLP proteins in Arabidopsis, maize, sorghum, and rice. (A) The phylogenetic analysis. The phylogenetic tree was constructed based on a complete protein sequence alignment of the NLPs by the neighbor-joining method using MEGA7 software. (B) The schematic diagram for exon/intron structure of the NLP family genes in Arabidopsis, maize, sorghum, and rice. The exon, intron, and UTR are represented by the green box, black line, and blue box, respectively. The scale bar represents 12 kb. (C) The schematic diagram for protein structure. The black lines indicate the peptide chain. The yellow squares, red cylinders, and blue ellipses represent the GAF-like, RWP-RK, and PB1 domains, respectively. The scale bar represents 100 amino acids. (D,E) The conserved motifs of NLP proteins in Arabidopsis, maize, sorghum, and rice. (D) The GAF motif logo; (E) The GSL motif logo.
To analyze the phylogenetic relationship of NLP proteins, the unrooted phylogenetic tree was generated based on the full length amino acid sequences of NLP proteins from Arabidopsis (9), maize (9), sorghum (5), and rice (6) using MEGA7.0. As shown in Figure 1A, the NLPs were divided into 3 groups (Group I, II, and III with different colors). In each group, the NLP proteins from maize, sorghum, and rice were closely clustered. Exon/intron structures of these NLPs, shown in Figure 1B, revealed an obvious difference among groups: a module with four exons and three introns was found in most group I members with the smallest size in the second exon. The group II members mainly contain five exons and four introns, and two smaller exons separate three bigger ones. A module of five exons and four introns also found in group III members, but the third exon is the smallest one. Thus, the length and position of introns make great contributions to the diversity of gene structure in each group. For the ZmNLP proteins, the RWP-RK and PB1 domains were found to be well conserved in all tested members, and GAF-like domain were also found in the N-termini of ZmNLPs (Figure 1C). In addition, the results revealed two novel motifs, GAF motif and GSL motif, in the NLP family by using MEME analysis (Figures 1D,E and Supplementary Figure S4). The GAF motif (SX4FX10GLPGR), which is located in the N-termini of NLPs, is the main part of the GAF-Like domain and critical for nitrate signaling transduction in Arabidopsis (Wang et al., 2009; Liu K.H. et al., 2017). GSL motif (LX2LX3FXGSLKD) exists just in front of the RWP-PK domain and with conserved GSL residues as a core. The GSL motif was well conserved in 236 NLP proteins in 34 species, but not found in the RKD family proteins, so that it can be regarded as a marker for distinguishing the NLP proteins from RKD proteins.
Expression Profiles of ZmNLP Genes
To better understand the function of each gene in the ZmNLP family, we examined their temporal and spatial expressions in maize B73 inbred lines by using qPCR. The results showed that different ZmNLP genes exhibited various expression patterns (Figures 2A,B). Among these genes, ZmNLP6 and ZmNLP8 showed significant higher expression levels in almost all tested stages and tissues, while the expression levels of ZmNLP1 and ZmNLP7 were much lower. ZmNLP4 and ZmNL9 exhibited the lowest expression levels in diverse organs and stages. Remarkably, the expression of ZmNLP6 and ZmNLP8 was the highest in the vegetative 13 (V13) stage in leaves and reproductive 1 (R1) stage in roots. V13 is the stage of tasseling and R1 is grouting stage, both of which are essential for maize production, suggesting that ZmNLP6 and ZmNLP8 may play outsized roles in these stages critical to maximizing maize yield. The expression of ZmNLP6 and ZmNLP8 was higher in roots, tassels, and grains, while ZmNLP2 and ZmNLP3 was higher in leaves than in other organs (Figure 2B). The expression of ZmNLP1 was relatively uniform, while ZmNLP7 was more highly expressed in both roots and tassels. The expression of ZmNLP5 was relatively low, mainly expressed in leaves, tassels, and grains. We also used the second housekeeping gene ZmUBCE for testing the expression of these genes and similar results were found (Supplementary Figure S5). These results suggest that ZmNLP6 and ZmNLP8 may play more important roles in different developmental stages and tissues compared with the other members of ZmNLPs in maize.
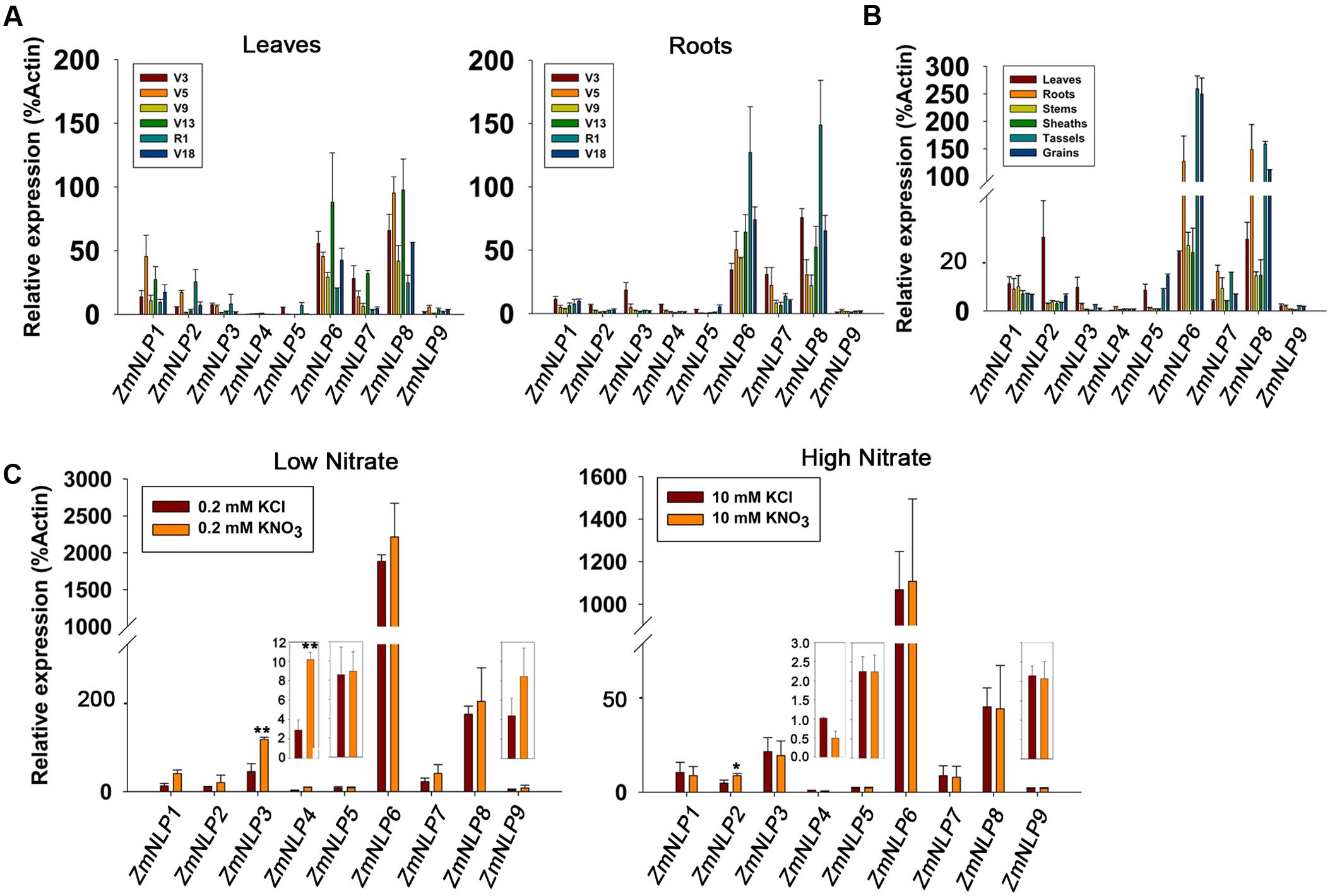
FIGURE 2. The temporal, spatial, and induction expression patterns of ZmNLP genes. (A) The temporal expression pattern of ZmNLPs in leaves and roots. The expanded leaves and roots were sampled at different developmental stages including vegetative 3 (V3), vegetative 5 (V5), vegetative 9 (V9), vegetative 13 (V13), reproductive 1 (R1), and vegetative 18 (V18) stages. The relative expression levels of each gene in leaves and roots were normalized to the percentage of ZmActin1 gene. Error bars represent SD of three biological replicates. (B) The spatial expression pattern of ZmNLP genes. The expression of ZmNLP genes in leaves, roots, stems, sheaths, tassels, and grains was detected by qPCR. The relative expression levels of each gene in different tissues were normalized to the percentage of ZmActin1 gene. Error bars represent SD of three biological replicates. (C) The relative expression of ZmNLP genes after low and high nitrate treatments. The 2-week-old seedlings grown under normal conditions were transferred into the medium with 2.5 mM ammonium succinate for 2 days, and then treated with 0.2 or 10 mM KNO3 for 2 h. KCl treatments were used as a control. The roots were collected for testing the expression of ZmNLP genes. The relative expression levels of each gene were normalized to the percentage of ZmActin1 gene. Error bars represent SD of four biological replicates, (∗∗P < 0.01, ∗P < 0.05, u-test).
To test if ZmNLPs are responsive to different nitrate conditions, we investigated the expression of all ZmNLP genes in roots. Seedlings were grown on 1/2MS medium (10 mM KNO3 and 10 mM NH4NO3) for 2 weeks, and then transferred into 2.5 mM ammonium succinate for 2 days followed by treatments with 0.2 or 10 mM KNO3 for 2 h. As shown in the Figure 2C, ZmNLP3 and ZmNLP4 were induced by 0.2 mM KNO3 in roots. After 10 mM KNO3 treatment, the expression of most ZmNLPs was not induced except ZmNLP2 (Figure 2C and Supplementary Figure S5). Thus, most ZmNLP genes were not induced by nitrate.
To test the expression patterns of ZmNLP genes under various nitrate conditions, 2-week-old seedlings were grown under 0, 0.2, 2.5, 5, and 10 mM KNO3 conditions and RNAs were extracted from the shoots and roots for qPCR analysis. The results showed that the expression levels of the ZmNLP4, ZmNLP6, and ZmNP8 were generally increased with the higher concentrations of nitrate in shoots, while the highest peak was observed at 2.5 mM nitrate concentration for ZmNLP6 and ZmNLP8 in the roots (Figure 3). However, the expression of ZmNLP3 and ZmNLP7 generally decreased in both shoots and roots with the increase of nitrate concentrations. For ZmNLP1, the expression was stable in shoots while decreasing with increased nitrate concentrations (Figure 3). These data indicate that different ZmNLPs exhibit various expression profiles. The expression of ZmNLP6 and ZmNLP8 in shoots was the highest and increased with increasing nitrate concentrations, indicating that both genes are the strongest candidates for playing important roles in regulating nitrate metabolism in maize. The second housekeeping gene ZmUBCE were used for testing the expression of these genes and the results were similar to those obtained by ZmActin1 (Supplementary Figure S6). Therefore, we focused on ZmNLP6 and ZmNLP8 in the following studies.
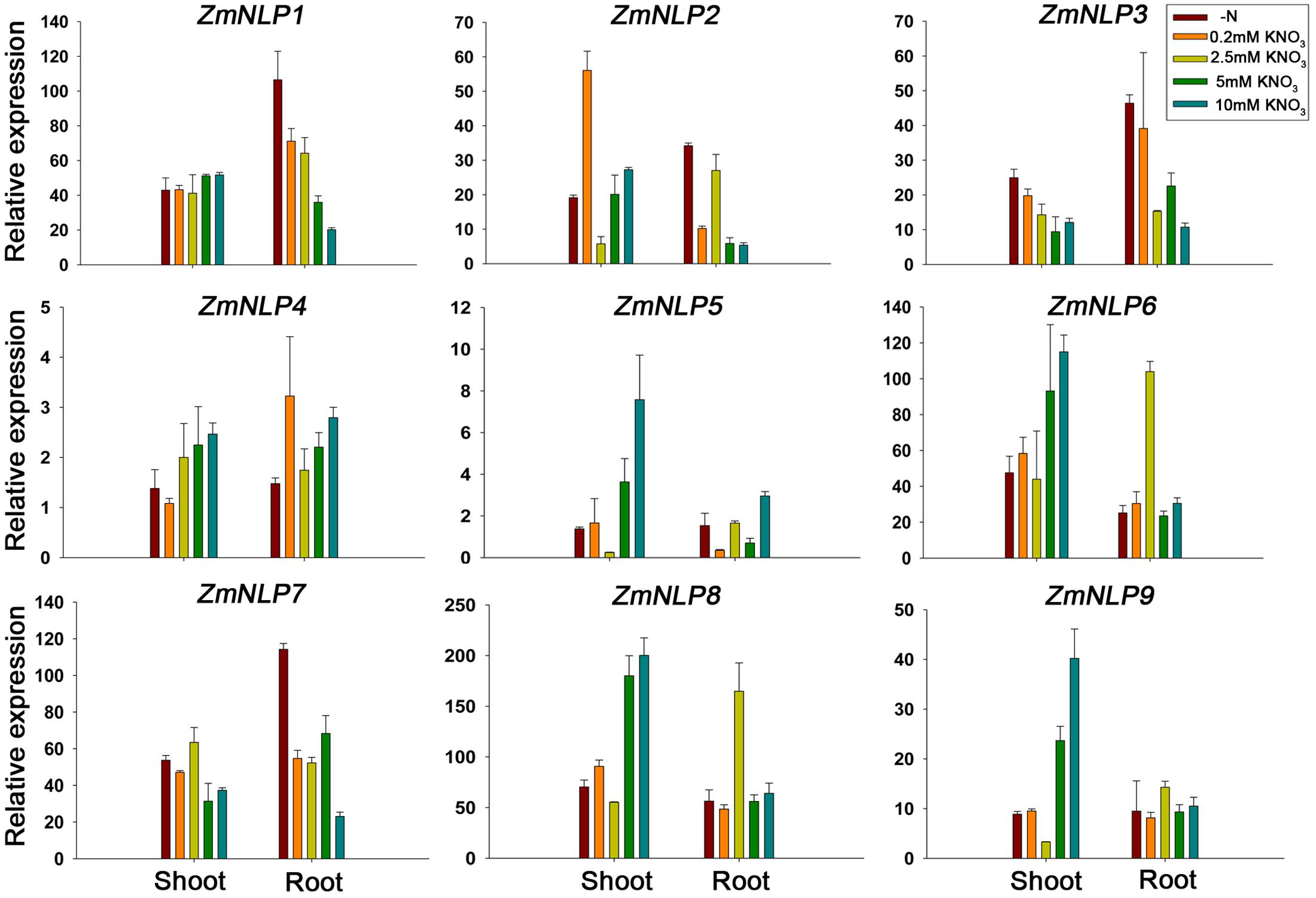
FIGURE 3. Expression patterns of ZmNLP genes under different nitrate conditions. The maize seedlings were grown on matrix for 2 weeks and watered with media containing 0, 0.2, 2.5, 5, or 10 mM KNO3. The shoots and roots were collected for detecting the expression levels of each ZmNLP. The relative expression levels of each gene in shoots and roots were normalized to the percentage of ZmActin1 gene. Error bars represent SD of three biological replicates.
Cloning and Functional Identification of ZmNLP Genes in Nitrate Signaling
AtNLP7 has been reported as a key regulator in nitrate signaling (Marchive et al., 2013). The Arabidopsis mutant nlp7-4 with a mutation (C to T) at the position 62, converting Gln to a stop codon, was identified by a forward genetic screen (Xu et al., 2016). The nlp7-4 plants contain NRP::YFP (a nitrate-regulated promoter fused to a yellow fluorescent protein) construct and are defective in nitrate regulation of nitrate responsive genes. To investigate if ZmNLP6 and ZmNLP8 are involved in nitrate signaling, we cloned them from the cDNAs of B73 inbred line and overexpressed them in the mutant nlp7-4, respectively, using a 35S promoter. Transgenic plants overexpressing the ZmNLP6 and ZmNLP8 were generated and two independent homozygous lines (ZmNLP6/nlp7-4-1 and ZmNLP6/nlp7-4-12 for ZmNLP6 transgenic lines, ZmNLP8/nlp7-4-8 and ZmNLP8/nlp7-4-16 for ZmNLP8 transgenic lines) were selected for following investigation (Figure 4A). Nitrate-responsive expression was monitored using a construct containing the YFP fused to a nitrate-responsive synthetic promoter (NRP) (Wang et al., 2009).
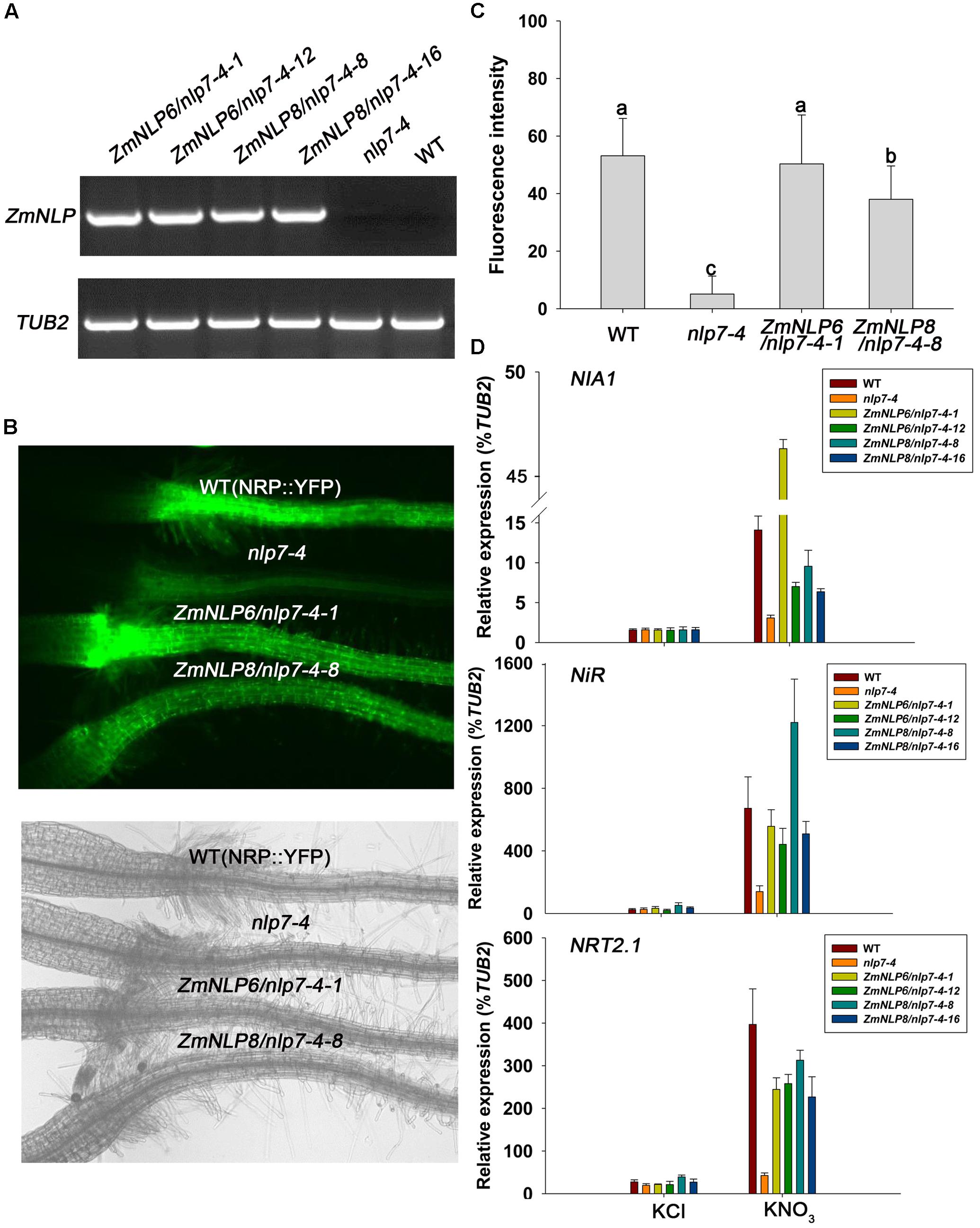
FIGURE 4. ZmNLP6 and ZmNLP8 modulate nitrate signaling. (A) The transcript expression of ZmNLP6 and ZmNLP8 in WT, nlp7-4, and transgenic lines by RT-PCR. Arabidopsis plants were grown on 1/2 MS medium (pH = 5.7) for 7 days and then the whole seedlings were collected for RNA extraction. The primers qZmNLP6-F, qZmNLP8-F, and 1300RT-R were used for confirming WT and the transgenic lines. TUB2 was used as a reference gene. (B) Root fluorescence phenotypes of ZmNLP6 and ZmNLP8 transgenic lines. The seedlings were grown on 10 mM KNO3 medium for 4 days. Fluorescence and visible light images were captured with a fluorescent microscope. (C) Quantification of root fluorescence of WT, nlp7-4, and transgenic lines. The plants were grown on the same conditions as (B). Error bars represent SD (n = 60). Different letters indicate statistically significant difference (P < 0.05, t-test). (D) The expression of nitrate responsive genes in WT, nlp7-4, and ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines after nitrate treatment. The seedlings grown on medium with 2.5 mM ammonium succinate for 7 days were treated with 10 mM KNO3 for 2 h. 10 mM KCl treatment was used as a control. The roots were collected for testing the expression of nitrate responsive genes. The relative expression levels of each gene were normalized to the percentage of TUB2 gene. Error bars represent SD of five biological replicates.
The transgenic lines were grown on nitrate medium and YFP fluorescence of these lines was found to be partially or completely restored to WT levels (Figures 4B,C), indicating that the ZmNLP6 and ZmNLP8 may function in nitrate signaling. Furthermore, we investigated the expression of endogenous nitrate-responsive genes including NIA1, NiR, and NRT2.1 and found that the induction levels of all three genes were recovered in ZmNLP6 and ZmNLP8 transgenic lines after nitrate treatments (Figure 4D). The expressions of these genes were also normalized to the percentage of second housekeeping gene AtActin8 and similar results were found (Supplementary Figure S7). These results suggest that ZmNLP6 and ZmNLP8 can complement the Atnlp7 mutant and thus play important roles in nitrate signaling.
Subcellular Localization of ZmNLP6 and ZmNLP8
To investigate the subcellular localization of ZmNLP6 and ZmNLP8 proteins, the full length CDS of the two genes driven by 35S promoter were fused with GFP in N-terminal. The 35S::GFP-ZmNLP6 and 35S::GFP-ZmNLP8 constructs were transiently transformed into Nicotiana benthamiana epidermal cells and Arabidopsis mesophyll protoplasts, the results showed that both ZmNLP6 and ZmNLP8 were mainly localized in the nucleus and slightly in cytosol (Figures 5A,B). As AtNLP7 has been reported to be regulated by nitrate via a nuclear retention mechanism (Marchive et al., 2013), we next tested the subcellular localization of ZmNLP6 and ZmNLP8 proteins under nitrate starvation and nitrate re-addition conditions. Both ZmNLP6 and ZmNLP8 were found to be localized in cytosol after nitrate starvation (Figures 5C,D) while in the nucleus when nitrate was resupplied (Figures 5E,F). Thus, ZmNLP6 and ZmNLP8 are mainly localized in the nucleus and slightly in cytosol in the presence of nitrate while both proteins are localized in cytosol when nitrate is absent.
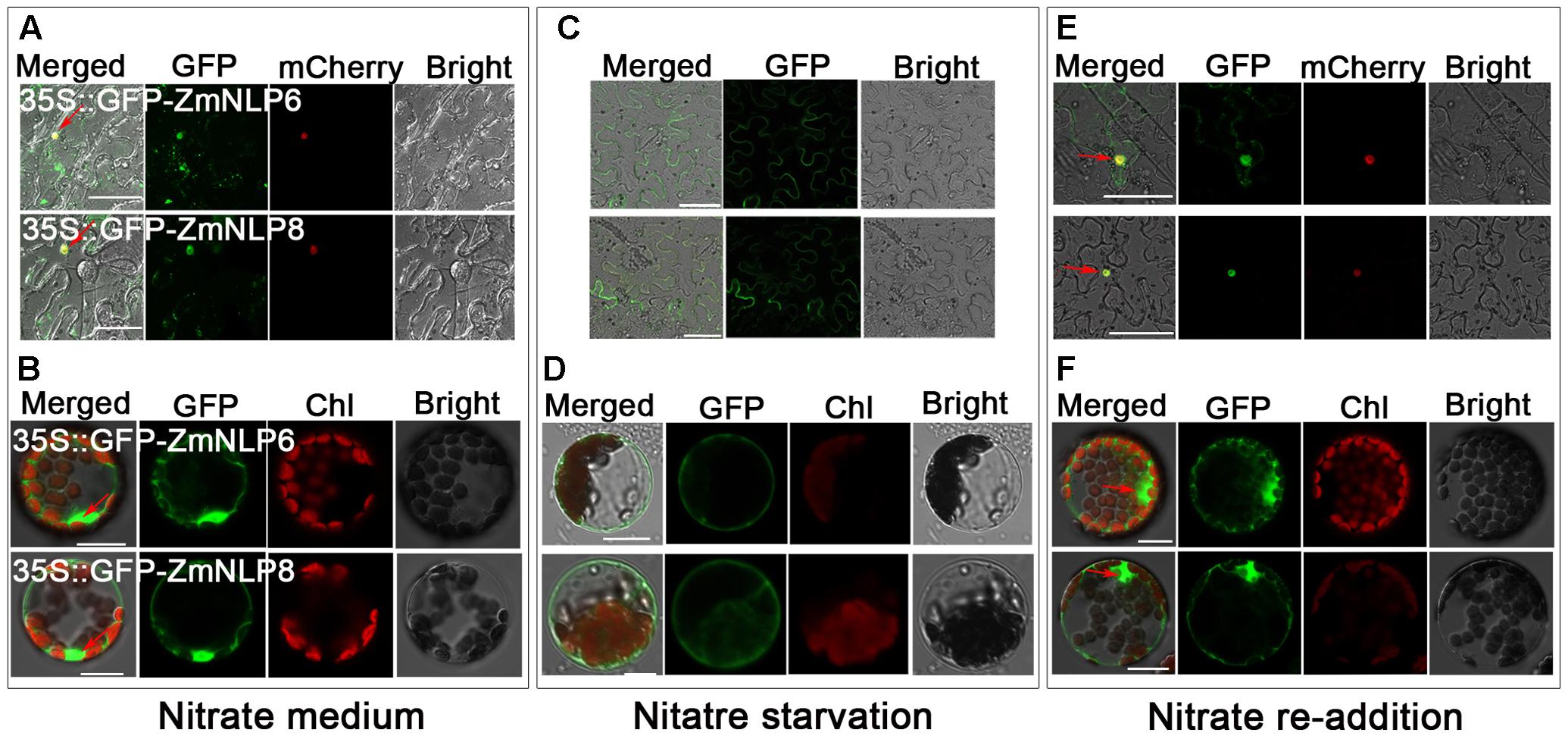
FIGURE 5. ZmNLP6 and ZmNLP8 are localized in the nucleus and are regulated by nitrate. (A,C,E) The subcellular localization of ZmNLP6 and ZmNLP8 in Nicotiana benthamiana epidermal cells. Nicotiana benthamiana seedlings were grown on 5 mM KNO3 for 5 weeks (A) and then treated with 2.5 mM ammonium succinate for 4 days (C) followed with 5 mM KNO3 for 12 h (E). The constructs pMDC43-ZmNLP6 and pMDC43-ZmNLP8 were transiently co-transformed into Nicotiana benthamiana epidermal cells with the mCherry construct, respectively. Red mCherry fused with H2B native promoter and H2B protein was used as a marker for nucleus localization. The scale bar represents 50 μm. GFP fluorescence in the nucleus was observed and marked by red arrows. (B,D,F) The subcellular localization of ZmNLP6 and ZmNLP8 in Arabidopsis mesophyll protoplasts. The Arabidopsis WT seedlings were grown on 5 mM KNO3 for 4 weeks (B) and then treated with 2.5 mM ammonium succinate for 6 days (D) followed with 5 mM KNO3 for 12 h (F). The constructs pMDC43-ZmNLP6 and pMDC43-ZmNLP8 were transiently into Arabidopsis mesophyll protoplasts. The scale bar represents 20 μm. GFP fluorescence in the nucleus was observed and marked by red arrows.
ZmNLP6 and ZmNLP8 Regulate Nitrate Assimilation When Overexpressed in Arabidopsis
Previous studies have shown that nitrate assimilation is impaired and the nitrate content is increased in nlp7 mutants (Castaings et al., 2009). To test if ZmNLP6 and ZmNLP8 affect nitrate assimilation, we measured nitrate content in transgenic lines and found that the increased nitrate content in nlp7-4 was recovered to WT levels (Figure 6A). To investigate if this recovery is associated with nitrate reduction, we examined the NR activity and found that this activity was restored in the transgenic lines (Figure 6B). Furthermore, the deficiency of amino acid in nlp7-4 mutant was completely rescued when ZmNLP6 and ZmNLP8 were overexpressed in nlp7-4 mutant (Figure 6C). Taken together, these data indicate that ZmNLP6 and ZmNLP8 are involved in regulating nitrate assimilation when overexpressed in Arabidopsis.
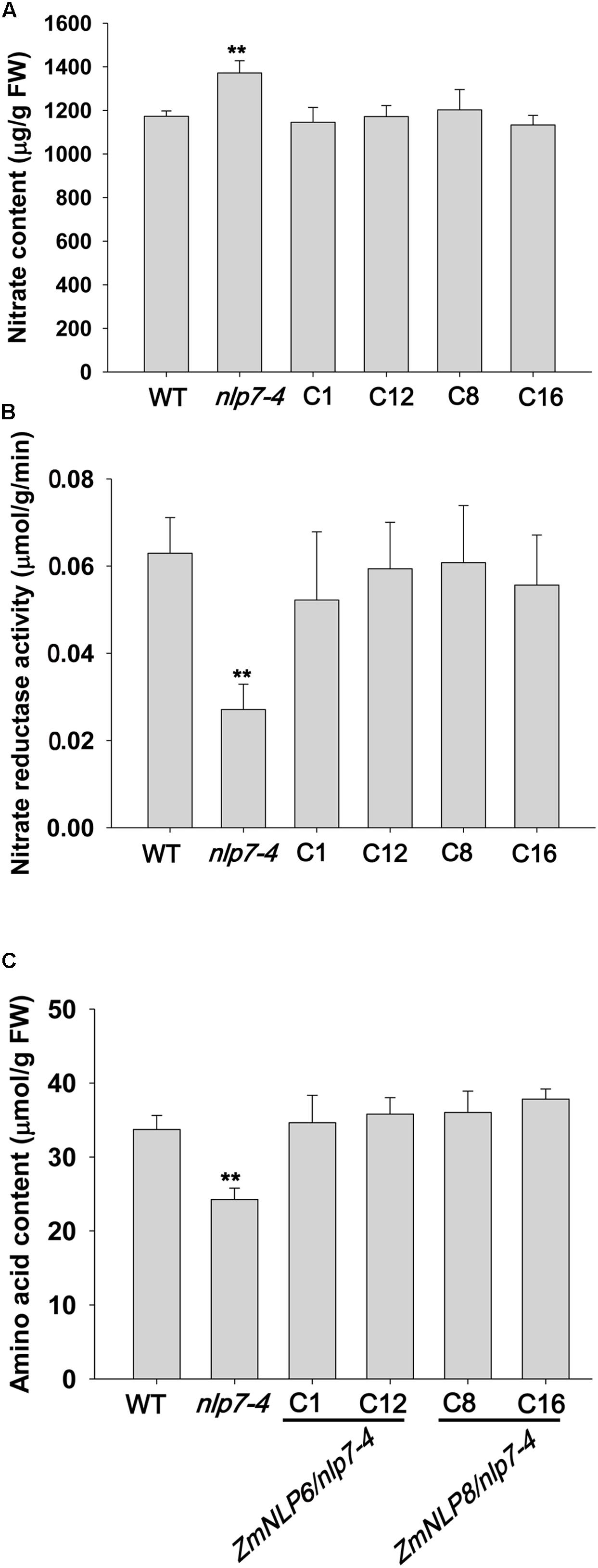
FIGURE 6. ZmNLP6 and ZmNLP8 affect nitrate accumulation and assimilation. (A) Nitrate content in seedlings. (B) Nitrate reductase activity in seedlings. (C) Amino acid content in seedlings. Seedlings of WT, nlp7-4, and ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines were grown on 1/2 MS medium (pH = 5.7) for 7 days and collected for testing nitrate content, nitrate reductase activity, and amino acid content. Error bars represent SD of six biological replicates (∗∗P < 0.01, u-test).
In order to further explore the underling mechanism whereby ZmNLP6 and ZmNLP8 affect nitrate content, we examined nitrate accumulation and the expression of several genes involved in nitrate assimilation. Plants were grown on 2.5 mM ammonium succinate for 7 days and then treated with 5 mM KNO3 for 0, 0.25, 0.5, 1, 2, and 4 h in the presence of 2.5 mM ammonium succinate. The nitrate content in whole seedlings was investigated and the results showed that no difference was found in nitrate accumulation among the WT, nlp7-4, and transgenic lines (Supplementary Figure S8). However, the expression of Gln1.1, Gln1.3, NIA2, and NiR in transgenic lines was recovered to the levels even higher than in WT (Supplementary Figure S9). These findings suggest that ZmNLP6 and ZmNLP8 can regulate the nitrate assimilation more strongly than nitrate accumulation.
ZmNLP6 and ZmNLP8 Can Bind NRE of ZmNRT1.2 and ZmNiR2 Directly In Vitro
It has been found that Arabidopsis NLP proteins can bind nitrate regulatory elements (NREs) to regulate the nitrate responsive genes (Konishi and Yanagisawa, 2013). To investigate whether ZmNLP6 and ZmNLP8 can bind NREs in maize, Y1H assay was performed. Firstly, we searched for NREs with a module (tGACcCTTN10AAGagtcc) from the 5′ and 3′ franking sequences of nitrate uptake and assimilation genes. Nine putative NREs were obtained as shown in Supplementary Table S2. Then, the candidate NREs were used for testing the binding activity of ZmNLP6 and ZmNLP8. The results showed that ZmNLP6 and ZmNLP8 could bind the NRE-like motifs of ZmNRT1.2 (3′UTR, with position 7299–7326) and ZmNiR2 (promoter, -149-124), respectively (Figures 7A–C). But no binding activity was found between ZmNLP6/ZmNLP8 and other NREs (Supplemental Figure S10). These data suggest that the ZmNLP6 and ZmNLP8 proteins can bind putative NREs similar to what was reported for Arabidopsis.
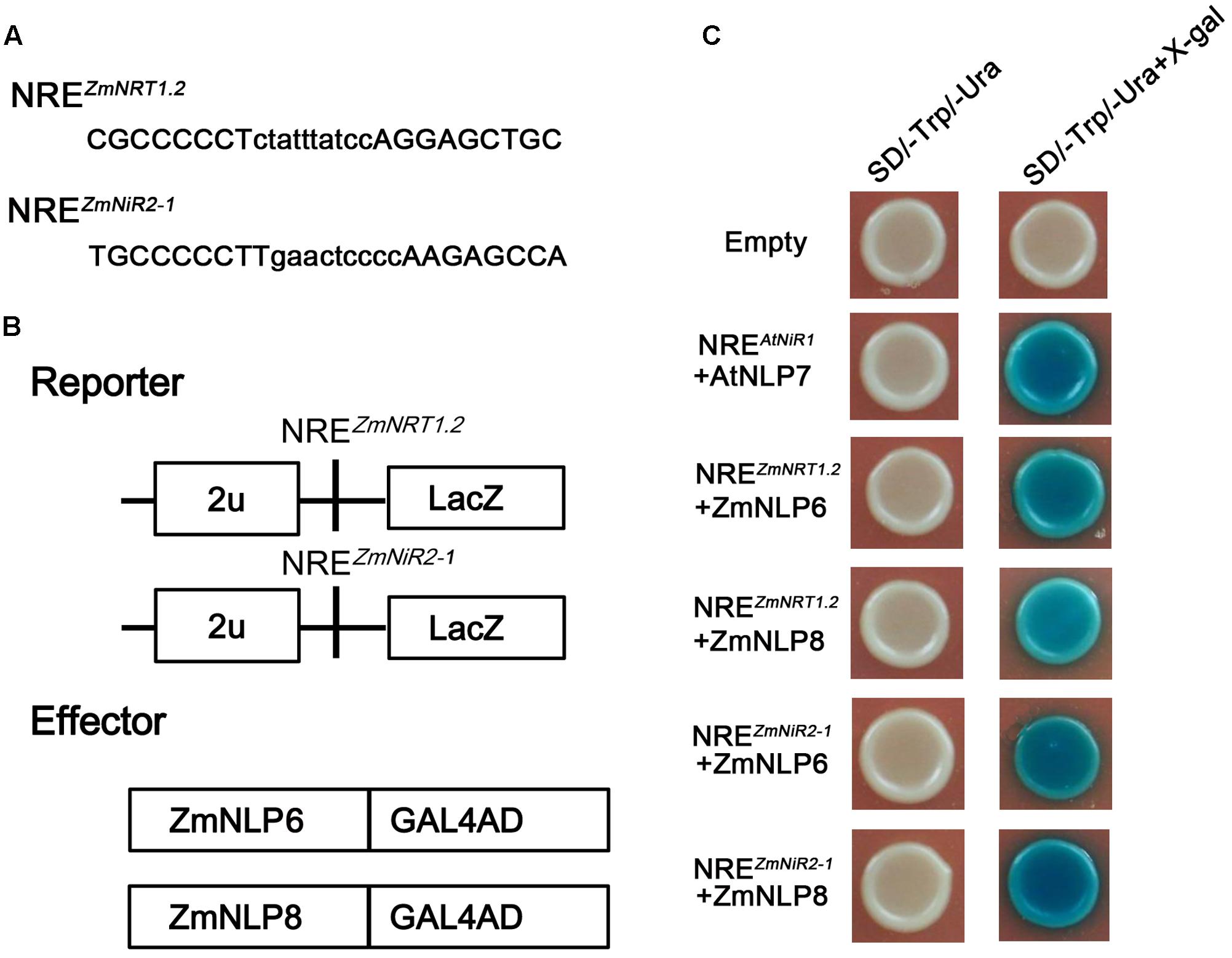
FIGURE 7. ZmNLP6 and ZmNLP8 can bind NRE of ZmNRT1.2 and ZmNiR2 in vitro. (A) NRE sequences and (B) the reporters and effectors used in Y1H assay. (C) Y1H assay. LexA-ZmNLP6 and LexA-ZmNLP8 fusion proteins activate the expression of LacZ reporter gene driven by the NRE of ZmNRT2.1 and ZmNiR2, respectively, in yeast. The empty vector was used as a negative control and the NREAtNiR1+AtNLP7 was used as a positive control.
Overexpression of ZmNLP6 and ZmNLP8 Can Alter Root Architecture and Improve Nitrogen Use Efficiency (NUE)
Low NUE in agriculture system is a global problem and therefore we assessed the potential of both genes to affect root architecture and NUE of in Arabidopsis. We first examined the primary root length and lateral root number in plants grown vertically on the media with different nitrate concentrations (0.2, 2.5, and 5 mM KNO3). The results showed that the length of primary roots and number of lateral roots were higher in ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines than in WT and nlp7-4 mutant under these three nitrate conditions (Figures 8A–C). To determine whether ZmNLP6 and ZmNLP8 can enhance NUE in plants, we investigated the biomass of WT, nlp7-4, and ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines under different nitrate concentrations. The results showed that the transgenic seedlings grew bigger than WT and nlp7-4 and the biomass of the whole seedlings increased by 15, 35, and 40% more than WT under 0.2, 2.5, and 5 mM KNO3 conditions (Figures 8D–F), indicating that the ZmNLP6 and ZmNLP8 can rescue the deficient growth phenotype of nlp7-4 mutant and promote plant growth under both low and high nitrate conditions.
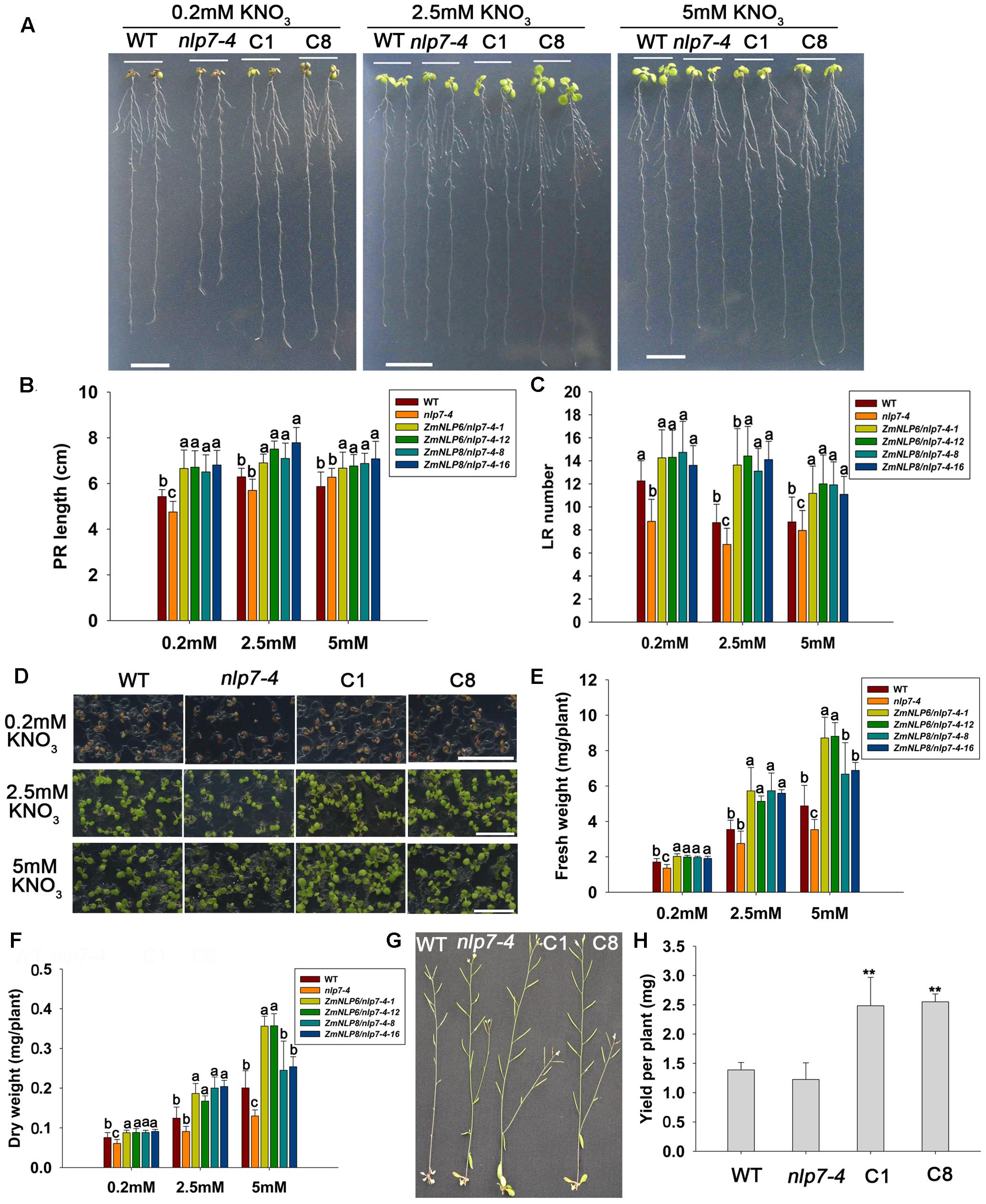
FIGURE 8. ZmNLP6 and ZmNLP8 can improve plant biomass under both high and low nitrate conditions and promote the seed yield. (A) Seedlings grown vertically on the media with 0.2, 2.5, and 5 mM KNO3 for 10 days, respectively. The scale bar represents 1 cm. C1 represents ZmNLP6/nlp7-4-1 transgenic line and C8 represents ZmNLP8/nlp7-4-8 transgenic line. (B) Primary root length and (C) lateral root number of the plants. Error bars represent SD of four biological replicates and each replicate contains 15 plants. Different letters indicate statistically significant difference (P < 0.05, u-test). (D) Seedlings grown horizontally on the media with 0.2, 2.5, and 5 mM KNO3 for 10 days, respectively. The scale bar represents 1 cm. (E) Fresh weigh and (F) dry weight of the plants. Error bars represent SD of four biological replicates and each replicate contains 40 plants. Different letters indicate statistically significant difference (P < 0.05, t-test). (G) WT, nlp7-4, and transgenic plants grown in vermiculite watered with 0.2 mM KNO3. (H) The yield per plant in WT, nlp7-4, and transgenic lines. Error bars represent SD of five biological replicates and each replicate contains 9 plants (∗∗P < 0.01, u-test).
Seed yield is an important trait for agricultural production and also for assessing the NUE of plants. Thus, we investigated the seed yield of ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines grown under both high and low nitrate conditions. The results showed that the yield per plant was higher in ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines by 44 and 45%, respectively, than in WT under low nitrate conditions (Figures 8G,H). However, no significant difference was found between the transgenic lines and WT when grown under high nitrate conditions (Supplementary Figure S11). These data suggest that ZmNLP6 and ZmNLP8 may improve plant NUE under low nitrate conditions.
Discussion
It has been reported that Arabidopsis NLP genes are involved in nitrate regulation, but the functions of maize NLP genes remain unknown. As maize is one of the main crops of the world, identifying the genes associated with nitrate signaling and deciphering the corresponding gene networks are of great importance for improving NUE and reducing environmental pollution. In this study, we identified nine ZmNLP genes containing RWP-RK and PB1 domains by genome-wide analysis in maize. The RWP-RK superfamily includes NLP and RKD families, both of which contain RWP-RK domain (Schauser et al., 2005). NLP family is conserved in the land plants we searched, especially in maize (9 members), sorghum (5 members), rice (6 members), and Arabidopsis (9 members) (Chardin et al., 2014). This family can be divided into three subfamilies and each subfamily shows different gene structure characteristics from each other (Figure 1A). A previous study also reported a similar subfamily division in Arabidopsis, rice, and Lotus japonicus (Schauser et al., 2005). We found two novel NLP motifs among these searched 34 species: GAF motif (SX4FX10GLPGR) and GSL motif (LX2LX3 FXGSLKD) (Figures 1D,E). The GAF motif exists in the N-terminus of NLP protein, a region involved in receiving the nitrate signal (Konishi and Yanagisawa, 2013). The most conserved signature structure in GAF motif is the first serine, the fifth phenylalanine, and GLPGR. The effects of serine and proline in GAF motif for nitrate signaling transduction have been reported (Wang et al., 2009; Liu K.H. et al., 2017). The GSL motif, located in the front of the RWP-RK domain, is conserved only in the NLP proteins but not in RDK proteins.
The expression profiles of ZmNLP genes showed that ZmNLP6 and ZmNLP8 exhibited the highest expression levels among the whole gene family, especially in R1 in roots and V13 in leaves (Figure 2A). As roots of R1 and the leaves of V13 are important for absorbing and remobilizing nitrate to pool organs and critical for yield of maize (Hirel et al., 2001), ZmNLP6 and ZmNLP8 may be important for nutrient uptake and translocation. The nitrate induction was tested for all ZmNLP genes and the results showed that only the expression of ZmNLP3 and ZmNLP4 were induced by low nitrate, and induced poorly or not at all by high nitrate (Figure 2C), implying that a post-transcriptional regulation response to nitrate may exist. Under different nitrate conditions, ZmNLP1, ZmNLP2, ZmNLP3, and ZmNLP7 exhibited relatively higher expression in roots under nitrogen starvation and they mainly belongs to ZmNLP Group I, implying that the function of Group I may be involved in nitrogen starvation (Figure 3). Moreover, ZmNLP6 and ZmNLP8 showed the highest expression levels under higher concentrations of nitrate (5 and 10 mM) in shoots while the highest levels on 2.5 mM nitrate condition in roots (Figure 3) and may participated in nitrate absorb and allocation. The expression patterns of ZmNLPs implicate that functional redundancy of ZmNLP family members in nitrate regulation may exist, and these NLP genes may play important roles in nitrate regulation in different stages and organs under various nitrate conditions.
In Arabidopsis, several nitrate regulatory genes have been identified and these genes can modulate genes involved in nitrate transport, assimilation, and response. But so far, no nitrate regulators have been reported in maize. We investigated the function of maize NLP genes and found that overexpression of ZmNLP6 and ZmNLP8 in Arabidopsis nlp7-4 mutant could recover the YFP fluorescence from the NRP-YFP transgene product and the induction of nitrate responsive genes to WT levels (Figure 4), indicating that ZmNLP6 and ZmNLP8 can restore the nitrate signaling in nlp7-4 mutant. Previous studies have reported that NRE works as an important nitrate-responsive cis-acting element and can be bound by NLPs in Arabidopsis (Konishi and Yanagisawa, 2013). Our Y1H results showed that ZmNLP6 and ZmNLP8 proteins can bind potential NREs of ZmNRT1.2 and ZmNiR2 in vitro (Figure 7), suggesting a direct regulation of ZmNRT1.2 and ZmNiR2 by ZmNLP6 and ZmNLP8 may exist in maize. In Arabidopsis, NLP7 has a profound influence on nitrate responsive genes at the transcriptional level and some target genes of NLP7 are regulated by binding to their NREs (Konishi and Yanagisawa, 2013; Guan et al., 2017; Sato et al., 2017). Thus, the ZmNLPs may bind NRE to regulate nitrate signaling in maize similar to that in Arabidopsis, and this regulation mechanism may be conserved in monocots and dicots. The subcellular localization of ZmNLP6 and ZmNLP8 is regulated by nitrate (Figure 5), similar to a mechanism controlling AtNLP7 localization in Arabidopsis. Our physiological and molecular analyses further revealed that the nitrate reduction process could be recovered in the ZmNLP6/nlp7-4 and ZmNLP8/nlp7-4 transgenic lines, indicating that both ZmNLP6 and ZmNLP8 can modulate nitrate assimilation when constitutively overexpressed in Arabidopsis. It has been reported that overexpression AtNLP7 in Arabidopsis can increase fresh weight and modify root architecture (including the longer primary root and increased lateral roots) under low and high nitrate conditions. In our study, overexpression of ZmNLP6 and ZmNLP8 in nlp7-4 mutant can also enhance the biomass and root development. However, the nitrate content, amino acid content, and NR activity were increased in AtNLP7 overexpression lines while restored to WT levels in ZmNLP transgenic lines. In addition, we found an increase in seed yield in ZmNLP6 and ZmNLP8 transgenic Arabidopsis lines under low nitrate conditions. It remains to be further investigated if the seed yield can be increased in AtNLP7 overexpression lines. Results shown in this study suggest the function of the group III NLPs in Arabidopsis and maize may be partially conserved in nitrate regulation.
Improving NUE of crops is of great importance for sustainable agriculture. Several nitrate-related genes have been implicating in improving NUE. OsDEP1, encoding a highly cysteine (Cys)-rich G protein γ subunit, has been reported to increase rice harvest index and grain yield under moderate levels of nitrogen fertilization (Sun et al., 2014). OsNRT1.1B-indica variation has been identified to enhance the ability of nitrate uptake and root-to-shoot transport to improve NUE in rice (Hu et al., 2015). In addition, overexpression of OsNRT2.3b can improve grain yield and NUE by increasing the capacity of pH-buffering and uptake of N, Fe, and P in rice (Fan et al., 2016). In Arabidopsis, overexpression of AtNLP7 can improve plant growth under both nitrogen-limiting and -sufficient conditions (Yu et al., 2016). In this paper, our results showed that ZmNLP6 and ZmNLP8 could promote plant growth under both low and high nitrate conditions, and increase seed yield under low nitrate conditions (Figure 8). Therefore, both ZmNLP6 and ZmNLP8 genes may be of great potential in improving NUE of maize. It would be also interesting to assess the role of other NLP members in promoting NUE of maize in the near future.
Author Contributions
YW, the corresponding author of the manuscript, designed the research and analysis of data for the work with the help of NC. YW, HC, and SQ: agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. HC and SQ, designed the work, drafted the work or revised it critically for important intellectual content; Final approval of the version to be published. MS, ZL, and YY revised it critically for important intellectual content.
Funding
This work was supported by The National Key R&D Program of China [grant number 2016YFD0100701 to YW], Funds of Shandong “Double Tops” Program [grant number SYL2017YSTD01 to YW], and Natural Science Foundation of Shandong [grant number BS2014NY003 to SQ].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
CDS, coding sequence; GFP, green fluorescent protein; GS, glutamine synthetase; NiR, nitrite reductase; NR, nitrate reductase; NRE, nitrate-responsive cis-element; NUE, nitrogen use efficiency; PNR, primary nitrate response; WT, wide type; Y1H, Yeast one-hybrid; YFP, yellow fluorescent protein.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01703/full#supplementary-material
Footnotes
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://smart.emblheidelberg.de/
- ^ http://pfam.xfam.org/
- ^ http://www.megasoftware.net/
- ^ http://www.plantgdb.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://www.maizegdb.org/
- ^ http://expasy.org/
- ^ http://meme-suite.org/tools/meme
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
References
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-Lopez, O., Tamayo, K. P., et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 80, 1–13. doi: 10.1111/tpj.12618
Alvarez, J. M., Vidal, E. A., and Gutierrez, R. A. (2012). Integration of local and systemic signaling pathways for plant N responses. Curr. Opin. Plant Biol. 15, 185–191. doi: 10.1016/j.pbi.2012.03.009
Araya, T., Miyamoto, M., Wibowo, J., Suzuki, A., Kojima, S., Tsuchiya, Y. N., et al. (2014). CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 2029–2034. doi: 10.1073/pnas.1319953111
Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., et al. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. doi: 10.1093/nar/30.1.276
Burdo, B., Gray, J., Goetting-Minesky, M. P., Wittler, B., Hunt, M., Li, T., et al. (2014). The Maize TFome–development of a transcription factor open reading frame collection for functional genomics. Plant J. 80, 356–366. doi: 10.1111/tpj.12623
Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435. doi: 10.1111/j.1365-313X.2008.03695.x
Chardin, C., Girin, T., Roudier, F., Meyer, C., and Krapp, A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587. doi: 10.1093/jxb/eru261
Crawford, N. M., and Forde, B. G. (2002). Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book 1, e0011. doi: 10.1199/tab.0011
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U.S.A. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Ferrario-Mery, S., Valadier, M. H., and Foyer, C. H. (1998). Overexpression of nitrate reductase in tobacco delays drought-induced decreases in nitrate reductase activity and mRNA. Plant Physiol. 117, 293–302. doi: 10.1104/pp.117.1.293
Forde, B. G. (2014). Nitrogen signalling pathways shaping root system architecture: an update. Curr. Opin. Plant Biol. 21, 30–36. doi: 10.1016/j.pbi.2014.06.004
Garnett, T., Conn, V., Plett, D., Conn, S., Zanghellini, J., Mackenzie, N., et al. (2013). The response of the maize nitrate transport system to nitrogen demand and supply across the lifecycle. New Phytol. 198, 82–94. doi: 10.1111/nph.12166
Garnett, T., Plett, D., Conn, V., Conn, S., Rabie, H., Rafalski, J. A., et al. (2015). Variation for N uptake system in maize: genotypic response to N supply. Front. Plant Sci. 6:936. doi: 10.3389/fpls.2015.00936
Gojon, A., Krouk, G., Perrine-Walker, F., and Laugier, E. (2011). Nitrate transceptor(s) in plants. J. Exp. Bot. 62, 2299–2308. doi: 10.1093/jxb/erq419
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Hirel, B., Bertin, P., Quillere, I., Bourdoncle, W., Attagnant, C., Dellay, C., et al. (2001). Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 125, 1258–1270. doi: 10.1104/pp.125.3.1258
Ho, C. H., Lin, S. H., Hu, H. C., and Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Konishi, M., and Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4, 1617. doi: 10.1038/ncomms2621
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A. S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Krouk, G., Mirowski, P., LeCun, Y., Shasha, D. E., and Coruzzi, G. M. (2010). Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 11:R123. doi: 10.1186/gb-2010-11-12-r123
Kusano, M., Fukushima, A., Redestig, H., and Saito, K. (2011). Metabolomic approaches toward understanding nitrogen metabolism in plants. J. Exp. Bot. 62, 1439–1453. doi: 10.1093/jxb/erq417
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Lin, R., Ding, L., Casola, C., Ripoll, D. R., Feschotte, C., and Wang, H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305. doi: 10.1126/science.1146281
Liu, K. H., Niu, Y., Konishi, M., Wu, Y., Du, H., Sun Chung, H., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316. doi: 10.1038/nature22077
Liu, W., Sun, Q., Wang, K., Du, Q., and Li, W. X. (2017). Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol. 214, 734–744. doi: 10.1111/nph.14396
Liu, Q., Chen, X., Wu, K., and Fu, X. (2015). Nitrogen signaling and use efficiency in plants: what’s new? Curr. Opin. Plant Biol. 27, 192–198. doi: 10.1016/j.pbi.2015.08.002
Manoli, A., Sturaro, A., Trevisan, S., Quaggiotti, S., and Nonis, A. (2012). Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 169, 807–815. doi: 10.1016/j.jplph.2012.01.019
Marchive, C., Roudier, F., Castaings, L., Brehaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4, 1713. doi: 10.1038/ncomms2650
Martin, A., Lee, J., Kichey, T., Gerentes, D., Zivy, M., Tatout, C., et al. (2006). Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18, 3252–3274. doi: 10.1105/tpc.106.042689
Medici, A., and Krouk, G. (2014). The primary nitrate response: a multifaceted signalling pathway. J. Exp. Bot. 65, 5567–5576. doi: 10.1093/jxb/eru245
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. U.S.A. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Rosen, H. (1957). A modified ninhydrin colorimetric analysis for amino acids. Arch. Biochem. Biophys. 67, 10–15. doi: 10.1016/0003-9861(57)90241-2
Rubin, G., Tohge, T., Matsuda, F., Saito, K., and Scheible, W. R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21, 3567–3584. doi: 10.1105/tpc.109.067041
Sato, T., Maekawa, S., Konishi, M., Yoshioka, N., Sasaki, Y., Maeda, H., et al. (2017). Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochem. Biophys. Res. Commun. 483, 380–386. doi: 10.1016/j.bbrc.2016.12.135
Schauser, L., Wieloch, W., and Stougaard, J. (2005). Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60, 229–237. doi: 10.1007/s00239-004-0144-2
Sekhon, R. S., Lin, H., Childs, K. L., Hansey, C. N., Buell, C. R., de Leon, N., et al. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563. doi: 10.1111/j.1365-313X.2011.04527.x
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Tabata, R., Sumida, K., Yoshii, T., Ohyama, K., Shinohara, H., and Matsubayashi, Y. (2014). Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. doi: 10.1126/science.1257800
Vidal, E. A., Alvarez, J. M., and Gutierrez, R. A. (2014). Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal. Behav. 9:e28501.
Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 4477–4482. doi: 10.1073/pnas.0909571107
Wang, R., Xing, X., Wang, Y., Tran, A., and Crawford, N. M. (2009). A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 151, 472–478. doi: 10.1104/pp.109.140434
Xu, N., Wang, R., Zhao, L., Zhang, C., Li, Z., Lei, Z., et al. (2016). The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 28, 485–504. doi: 10.1105/tpc.15.00567
Yu, L. H., Wu, J., Tang, H., Yuan, Y., Wang, S. M., Wang, Y. P., et al. (2016). Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6:27795. doi: 10.1038/srep27795
Yu, P., Li, X., Yuan, L., and Li, C. (2014). A novel morphological response of maize (Zea mays) adult roots to heterogeneous nitrate supply revealed by a split-root experiment. Physiol. Plant. 150, 133–144. doi: 10.1111/ppl.12075
Keywords: Maize, nitrate assimilation, nitrate regulatory gene, nitrate signaling, ZmNLP6 and ZmNLP8
Citation: Cao H, Qi S, Sun M, Li Z, Yang Y, Crawford NM and Wang Y (2017) Overexpression of the Maize ZmNLP6 and ZmNLP8 Can Complement the Arabidopsis Nitrate Regulatory Mutant nlp7 by Restoring Nitrate Signaling and Assimilation. Front. Plant Sci. 8:1703. doi: 10.3389/fpls.2017.01703
Received: 18 July 2017; Accepted: 19 September 2017;
Published: 05 October 2017.
Edited by:
Hideki Takahashi, Michigan State University, United StatesReviewed by:
Thomas Girin, INRA Centre Versailles-Grignon, FranceEiji Nambara, University of Toronto, Canada
Copyright © 2017 Cao, Qi, Sun, Li, Yang, Crawford and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Wang, d2FuZ3lvbmdAc2RhdS5lZHUuY24=
†These authors have contributed equally to this work.
 Huairong Cao
Huairong Cao Shengdong Qi
Shengdong Qi Mengwei Sun
Mengwei Sun Zehui Li
Zehui Li Yi Yang
Yi Yang Nigel M. Crawford
Nigel M. Crawford Yong Wang
Yong Wang