- 1National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 2University of Chinese Academy of Sciences, Shanghai, China
- 3Key Laboratory of Insect Developmental and Evolutionary Biology, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
- 4Ministry of Agriculture Key Laboratory of Urban Agriculture (South), Plant Biotechnology Research Center, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
The panicle architecture of rice is an important characteristic that influences reproductive success and yield. It is largely determined by the number and length of the primary and secondary branches. The number of panicle branches is defined by the inflorescence meristem state between determinacy and indeterminacy; for example, the maize ramosa2 (ra2) mutant has more branches in its tassel through loss of spikelet determinacy. Some genes and factors influencing the number of primary and secondary branches have been studied, but little is known about the molecular mechanism underlying pedicel development, which also influences panicle architecture. We report here that rice OsRAMOSA2 (OsRA2) gene modifies panicle architecture through regulating pedicel length. Ectopic expression of OsRA2 resulted in a shortened pedicel while inhibition of OsRA2 through RNA interference produced elongated pedicel. In addition, OsRA2 influenced seed morphology. The OsRA2 protein localized to the nucleus and showed transcriptional activation in yeast; in accordance with its function in pedicel development, OsRA2 mRNA was enriched in the anlagen of axillary meristems, such as primary and secondary branch meristems and the spikelet meristems of young panicles. This indicates a conserved role of OsRA2 for shaping the initial steps of inflorescence architecture. Genetic analysis revealed that OsRA2 may control panicle architecture using the same pathway as that of the axillary meristem gene LAX1 (LAX PANICLE1). Moreover, OsRA2 acted downstream of RCN2 in regulating pedicel and branch lengths, but upstream of RCN2 for control of the number of secondary branches, indicating that branch number and length development in the panicle were respectively regulated using parallel pathway. Functional conservation between OsRA2 and AtLOB, and the conservation and diversification of RA2 in maize and rice are also discussed.
Introduction
Rice (Oryza sativa L.) provides a staple food for more than half of the world’s population and a model plant for the molecular study of cereal crops. Panicle architecture is one of the main features influencing reproductive success and contributes directly to grain yield (Sakamoto and Matsuoka, 2008; Li et al., 2011), so has been a major target of cereal crop domestication.
The panicle architecture of cereal plants is mainly determined by the number and length of primary branches (PBs) and secondary branches (SBs). These are largely established by iterations of branching that are governed by spatiotemporal developmental decisions in inflorescence, branch and spikelet meristems between the maintenance of determinacy and indeterminacy (Kellogg et al., 2013; Teo et al., 2014; Zhang and Yuan, 2014). Arabidopsis genes LEAFY (LFY), APETALA1 (AP1), CAULIFLOWER (CAL), FRUITFULL (FUL) and Antirrhinum genes FLORICAULA (FLO) and SQUAMOSA (SQUA) promote the formation of terminal floral and determinacy (Coen et al., 1990; Huijser et al., 1992; Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995; Ferrandiz et al., 2000), while TERMINAL FLOWER1 (TFL1) in Arabidopsis and CENTRORADIALIS (CEN) in Antirrhinum promote an indeterminate inflorescence (Bradley et al., 1996, 1997). Accordingly, RCN1 and RCN2, rice homologs of TFL/CEN, lead to an indeterminate inflorescence and a more branched panicle when ectopically expressed (Nakagawa et al., 2002). These indeterminacy promoting and inhibiting factors also interact with each other, for instance, CEN interacts with FLO in regulating inflorescence architecture (Bradley et al., 1996, 1997). However, it is not known if OsRA2 interact with other genes in panicle development?
The maize ra2 mutant has an increased number of tassel branches through loss of spikelet determinacy (Bortiri et al., 2006). The RA2 protein, a member of the Lateral Organ Boundaries Domain (LBD) protein family, is a specific regulator of plant organ development, and has been shown to function not only in various developmental processes in the leaf, root, inflorescence, microspore, and pollen, but also in plant regeneration, photomorphogenesis, and plant defense (Shuai et al., 2002; Xu et al., 2016). Barley HvRA2 plays conserved role in determining spikelet determinacy and controlling lateral spikelet fertility (Koppolu et al., 2013).
Branch meristems in the panicle are axillary meristems (AMs) that determine the complexity of the whole plant architecture (Tabuchi et al., 2011). LAX1 is a key regulator of AM initiation and maintenance in rice (Komatsu K. et al., 2003; Oikawa and Kyozuka, 2009), MONOCULM1 (MOC1) regulates AM formation and when mutated leads to fewer branches in the panicle (Li et al., 2003), while LAX2 is expressed in all the AMs that give rise to PBs, SBs and spikelets, and physically interacts with LAX1, the lax1lax2 double mutant develops almost no branches in the panicle (Tabuchi et al., 2011). FRIZZY PANICLE (FZP) restrains overgrowth of AM and results in excessive ramification of rachis-branches when mutated (Komatsu et al., 2001; Komatsu M. et al., 2003). MOC1 and LAX1, and some microRNAs (miRNAs) are involved in the consistent development of panicle architecture and tillering (Wang L. et al., 2015), but mostly, panicle development is distinctly regulated (Liang et al., 2014). It is not known if any genetic crosstalk exists between AM development and meristem determinacy during panicle development.
Phytohormones are also thought to influence panicle architecture (Mcsteen, 2009), with up-regulation of OsCKX2 (CYTOKININ OXIDASE) in the cytokinin pathway resulting in an increased number of PBs and SBs (Ashikari et al., 2005; Han et al., 2014). Moreover, post-transcriptional regulation by miRNAs and their targets was found to regulate panicle branching (Wang L. et al., 2015), as demonstrated by the loss of function of maize miR172 which caused a loss of spikelet determinacy and an excessive number of branches (Chuck et al., 2007). OsSPL14 negatively modulates the transition to spikelet meristem (Rhoades et al., 2002; Jiao et al., 2010; Miura et al., 2010; Luo et al., 2012; Wang et al., 2017).
The pedicel length is another key contributor to the diversity of inflorescence architecture. Although a few genes and factors regulating the number of PBs and SBs have been identified, little is known about the regulation of their length. SP1 was reported to negatively regulate the panicle length (Li et al., 2009). And even less is known about the control of pedicel development, with only two associated genes in Arabidopsis and one in tomato reported (Yamaguchi and Komeda, 2013; Wang D. et al., 2015).
Because of the complexity of panicle constitution and development, its molecular mechanism and regulation network is poorly understood. Therefore, the present study investigated the function of OsRA2 in rice. OsRA2 was found to regulate seed morphology and pedicel development in the panicle, and its expression profile corresponded well with its function in these aspects. Preliminary genetic analysis revealed that OsRA2 might control rice panicle architecture using the same pathway as that of LAX1. Moreover, OsRA2 acted downstream of RCN2 in regulating pedicel and branch lengths, but upstream of RCN2 for control of the number of SBs, indicating that branch number and length development in the panicle were respectively regulated using parallel pathway.
Materials and Methods
All experimental protocols and plant materials were approved by Shanghai Institutes for Biological Science, Chinese Academy of Sciences.
Plant Materials
Wild type rice ZH11 (Oryza sativa L. subsp. japonica cv. Zhonghua No.11), the lax1, A989 mutants, and all the other transgenic plants used in this study were grown in a paddy field or in pots in a greenhouse under standard growth conditions.
Phylogenetic Analysis
Homolog sequences of RA2 from Schizachyrium sanguineum (SsRA2), Sorghum bicolor (SbRA2), Andropogon hallii (AhRA2), Cymbopogon flexuosus (CfRA2), Andropterum stolzii (AsRA2), Phacelurus digitatus (PdRA2), Zea mays (ZmRA2), Chrysopogon gryllus (CgRA2), Hordeum vulgare (HvRA2), Oryza sativa (OsRA2), Loudetia sp. MCE (LsRA2), AtASL4 and AtLBD25 were obtained from Uniprot1 (Supplementary Table S1), homologous sequences of AtASL4 and AtLBD25 were obtained from the National Centre for Biotechnology Information2. Multiple sequence alignments of proteins were conducted using ClustalX Version 1.83 (Thompson et al., 1997). A phylogenetic tree of the sequences was reconstructed by the neighbor-joining (NJ) method by MEGA3 (Kumar et al., 2004).
Plasmids and Constructs
To generate OsRA2 RNAi plasmid pdsRNAiOsRA2, a gene-specific fragment was amplified by KOD-plus DNA polymerase (TOYOBO) using primers OsRA2-RNAiF/OsRA2-RNAiR and cloned into pCAMBIA1301RNAi vector. To obtain the pUbi::OsRA2 plasmid, full-length cDNA of OsRA2 was amplified using primers OsRA2-BamHIA/OsRA2-KpnI and cloned into pCAMBIA1301UbiNos vector using BamHI and KpnI.
Genetic Transformation of Rice
The embryogenic calli induced from immature ZH11 seeds were transformed by Agrobacterium tumefaciens infection and transgenic plants were regenerated (Hiei et al., 1994). Primer pairs Ubi 90+/OsRA2-KpnI and ocs-160/OsRA2-RNAiF were used to check pUbi::OsRA2 and pdsRNAiOsRA2 transgenic plants respectively.
Quantitative Real-Time (qRT-PCR) PCR Assays
For qRT-PCR assays, total RNAs were extracted from various tissues, panicles and seeds at different developmental stages using TRIzol reagent (Invitrogen), and reverse transcribed using oligo(dT) primer and ReverAce (TOYOBO). cDNA was synthesized from 2 μg of total RNA treated with DnaseI (TOYOBO) and used as templates for RT-PCR or qRT-PCR. Rice actin gene was used for normalization. Single and double asterisks represent significant difference determined by the Student’s t-test at ∗P < 0.05 and ∗∗P < 0.01 respectively.
For OsRA2 expression analysis, the roots, leaves, sheaths, culms, shoot apical meristem (SAM) were collected from around 30-day-old plants; panicles meristems less than 1 cm were collected under microscope, and flowers were collected from heading stage.
In Situ Hybridization Analysis
In situ hybridization was performed according to previous description (Coen et al., 1990). Young panicles at different developmental stages were fixed in 4% paraformaldehyde PBS solution overnight at 4°C, dehydrated through a concentration grade of ethanol, cleared through a dimethylbenzene series, then infiltrated through a series of paraffin (Sigma–Aldrich) melted at 60°C, and finally embedded in 100% paraffin. The longitudinal sections of young panicle were sectioned into 7 μm strips and mounted on RNase-free glass slides (Sigma). Gene-specific fragments of OsRA2 and OSH1 were amplified using primer pairs OsRA2-insitu F/OsRA2-insitu R and OSH1F/OSH1R respectively, and cloned into pBSK(-) vector, then linearized and used as templates for digoxigenin-labeled sense and antisense RNA probes, which were transcribed in vitro using a DIG RNA labeling kit (Promega).
Measurement of Grain Traits
Fifty grains from each sample were used for measurement of grain traits, including grain length (GL), grain width (GW) and grain thickness (GT). Length-to-width ratio (LWR) was obtained by dividing GL by GW. The 1,000-grain weight was got by measuring 100 grains in 10 biological repeats. Data was shown as mean ± SD.
Anatomical Analysis
Pedicels of ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants were cut transversely and fixed in 50% FAA at 4°C overnight after vacuuming. After dehydration in several concentrations of ethanol, samples were embedded in epoxide resin and cut into 2–3 μm slices and spread at 42°C on a hot platform overnight, stained using 0.5% toluidine Blue O and sealed for observation under the microscope (Wang et al., 2010). For the thickness measurement, at least 10 pedicels were made into histological sections and measured under microscope.
Trans-Activation Assay
Full-length cDNA of OsRA2 was amplified and cloned in-frame with the LexA DNA-binding domain using primers Osa_yRA2EcoRI and Osa_ yRA2BamHI in PEG202 (His3, 2 μm, Ampr, ADH constitutive promoter, LexA DNA-binding domain) (DupLEX-ATM system) (OriGene Technologies). The pEG202-OsRA2 and pEG202 plasmids were introduced into yeast strain EGY48 (MATtrp1his3ura3leu2::6lexAop-LEU2) harboring the reporter plasmid pSH18-34 (URA3, 2 mm, Ampr, LexAops-lacZ). Yeast clones were grown on SD medium without histidine or uracil in the presence of X-gal (5-bromo-4-chloro-3-indolyl-β-galactopyranoside) for 2 days at 30°C.
Sub-cellular Localization of OsRA2
Full length cDNA of OsRA2 was cloned in frame with GFP into p1301-GFP using primers OsRA2-BamHIB/OsRA2-PstI and genetically transformed into ZH11. Root apices from the transgenic plants were observed through a confocal laser scanning microscopy (OLYMPUS FV1000).
Primer sequences used in this study were listed in Supplementary Table S2.
Results
Identification of OsRA2 Gene As Homologous to Maize RA2
A search of the public databases3,4 ,5 revealed that only one gene, OsRA2, of the rice genome was homologues to maize RA2. OsRA2 contains four exons and three introns, with an open reading frame (ORF) in the third exon encoding a protein of 251 amino acids (Figure 1A). The deduced OsRA2 protein is composed of a characteristic N-terminal LOB domain and a variable C-terminal RA2 domain. The LOB domain is characteristic of the LBD family proteins (Bortiri et al., 2006), and in OsRA2 consists of a C block that functions in DNA-binding, a leucine zipper coiled coil motif for protein dimerization, and a Gly-Ala-Ser block (Figure 1B). The C block is represented by a CX2CX6CX3C motif containing four conserved cysteine (C) residues and other non-conserved residues (X), and the leucine-zipper-like motif includes five hydrophobic amino acids separated by six variable amino acid residues (Shuai et al., 2002; Matsumura et al., 2009; Majer and Hochholdinger, 2011). Phylogenetic alignment of protein sequences revealed that RA2 is highly conserved among monocotyledons, while some diversification has occurred between grass RA2 proteins and Arabidopsis LBD proteins (Figures 1B,C).
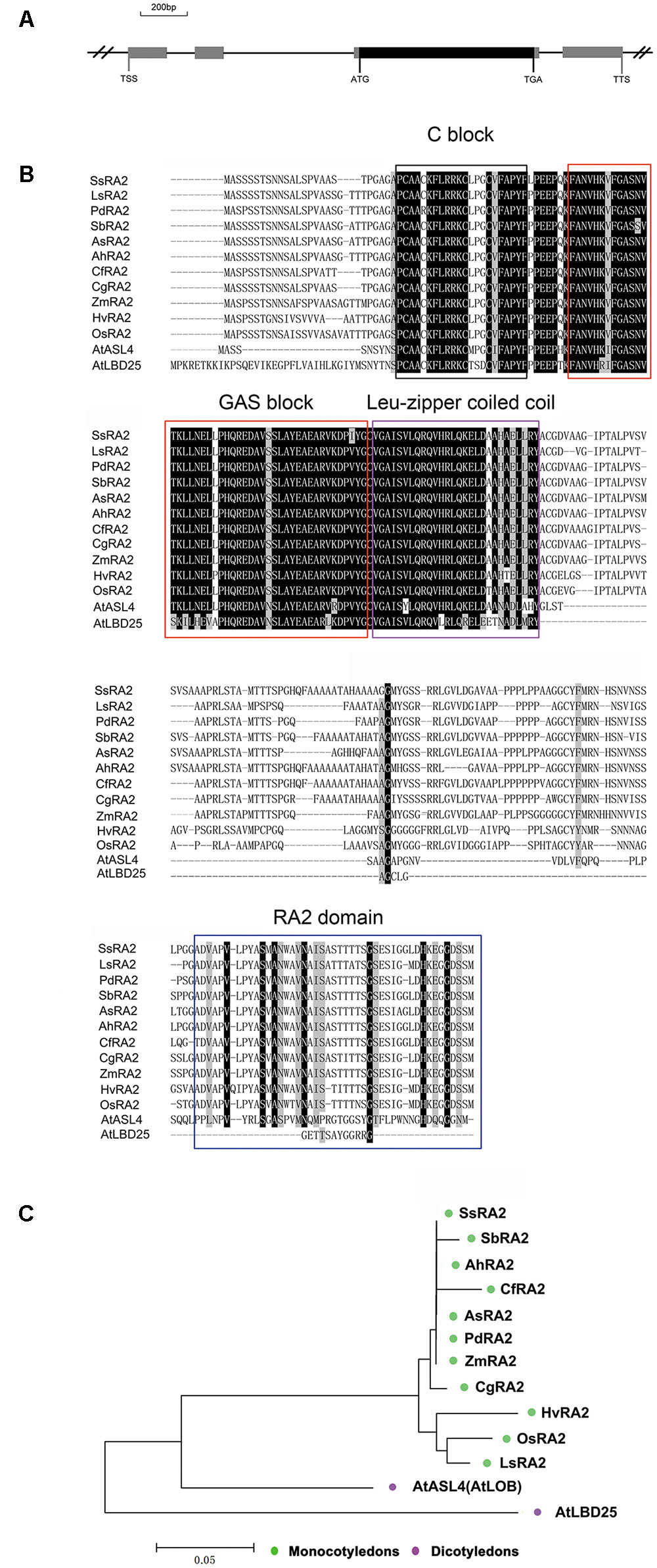
FIGURE 1. Analysis of OsRA2 and comparison with other grass RA2 proteins and Arabidopsis LBD proteins. (A) Schematic of OsRA2. Black box represents the protein-coding region and gray ones represent exons. (B) Alignment of the deduced full-length amino acid sequences of SsRA2, SbRA2, AhRA2, CfRA2, AsRA2, PdRA2, ZmRA2, CgRA2, HvRA2, OsRA2, LsRA2, AtASL4 and AtLBD25. The respective species are listed in the Materials and methods. Protein domains are framed in black for C-block, red for GAS-block, purple for Leu-zipper coiled coil and blue for the RA2 domain. (C) Phylogenetic tree of grass RA2 proteins and Arabidopsis LBD proteins.
OsRA2 Regulated Pedicel Development and Other Panicle Characteristics in Rice
To investigate the function of OsRA2, we carried out genetic analysis, using gene-specific doubled–stranded RNA interference (dsRNAi) and ectopic expression technologies.
We obtained a total of 27 T0 generation plants from the dsRNAiOsRA2 transformation, of which 24 showed elongated pedicels (Figures 2A–C). Successful down-regulation of OsRA2 in these lines was verified (Supplementary Figure S1). The pedicels of the panicles in dsRNAiOsRA2 plants were longer than those in wild type (WT) ZH11 (>1.4 cm vs. 1 cm; Figures 2B,C,F), but there was little change in the number and length of the PBs and SBs (Supplementary Figures S2A–D).
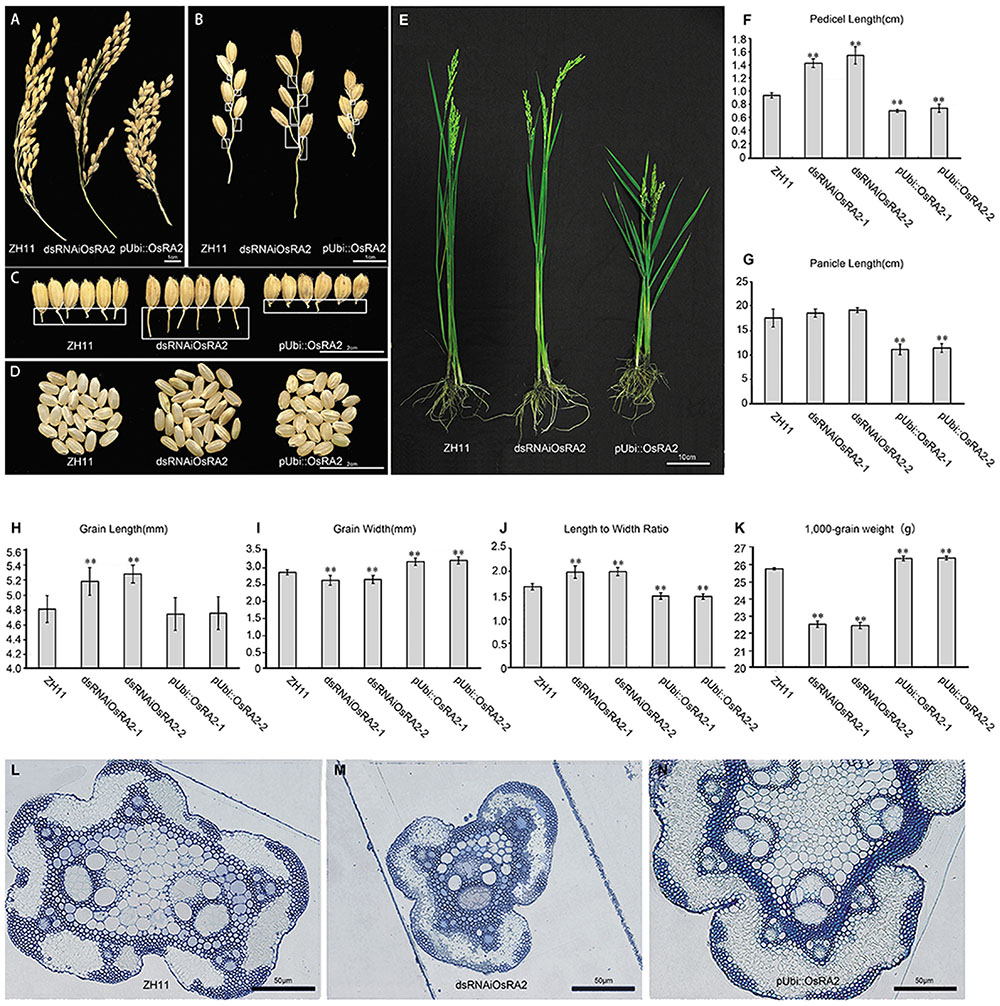
FIGURE 2. Phenotypes of OsRA2 transgenic plants. (A–C) Phenotypes of ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants with respect to panicles, PBs and pedicels, respectively. White rectangles in (B) indicate the position of pedicels in (C). (D) The seed phenotype of ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants. (E) Morphology of ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants. (F,G) Statistical analysis of pedicel and panicle length among ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants. (H–K) Statistical analysis of grain traits among ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants. Values in (F–K) are means ± SE. n = 30 panicles in (F–K) and 10 replicates in (K). (L–N) Cross sections of the pedicel of ZH11, dsRNAiOsRA2 and pUbi::OsRA2 plants. Student’s t-test at ∗P < 0.05 and ∗∗P < 0.01.
By contrast, pUbi::OsRA2 plants in which OsRA2 was over expressed showed a compact panicle and shortened pedicel (Figure 2A and Supplementary Figure S1), with an average pedicel length of 0.8 cm (Figure 2F), which was obviously shorter than WT (Figures 2B,C,F). Furthermore, the PBs, SBs, and the entire panicle were shortened (Figure 2G and Supplementary Figures S2A,B). There was little change in the number of the PBs (Supplementary Figure S2C), but the number of SBs decreased (Supplementary Figure S2D), such that the entire panicle was shortened and condensed (Figure 2A).
Based on the phenotype of dsRNAiOsRA2 and pUbi::OsRA2 transgenic plants, OsRA2 appears to regulate the pedicel length in rice.
OsRA2 Influenced the Longitudinal Elongation of the Entire Plant
Besides controlling pedicel length, OsRA2 also regulated the thickness of the pedicel, which was decreased in dsRNAiOsRA2 plants and increased in pUbi::OsRA2 plants (Figure 2C and Supplementary Figure S3); the thicknesses of the branches and the culms in their panicles were altered accordingly (Figures 2A,B). We further examined the transverse structure of the pedicels, and identified fewer vascular bundles in dsRNAiOsRA2 plants (Figure 2M), and more in pUbi::OsRA2 plants (Figure 2N), as compared with ZH11 (Figure 2L). So that, dsRNAiOsRA2 plants had thinner pedicels, while pUbi::OsRA2 plants had thicker ones.
OsRA2 also influenced the shape of the grains in transgenic plants. In dsRNAiOsRA2 plants, the grains were elongated (Figures 2D,H), but less wide (Figure 2I), so that the GL to width ratio was increased (Figure 2J). In pUbi::OsRA2 plants, the GL did not change (Figure 2H), but the GW increased (Figure 2I), so that the length to width ratio was decreased (Figure 2J). As a result, the 1,000-grain weight of dsRNAiOsRA2 plants decreased, while that of pUbi::OsRA2 plants increased (Figure 2K).
At the whole plant level, the height of pUbi::OsRA2 plants decreased, while that of dsRNAiOsRA2 plants remained unchanged (Figure 2E and Supplementary Figure S2E).
Taken together, OsRA2 appears to regulate pedicel development and seed development, and its over-expression also influences plant height in rice.
The OsRA2 Expression Pattern Conformed to Its Function in Panicle Development
Gene expression is tightly linked to its function. To detect OsRA2 expression, we carried out quantitative real-time RT-PCR (qRT-PCR), semi-quantitative RT-PCR and in situ hybridization analyses. OsRA2 was shown to mainly express in young panicles and in shoot apical meristems (SAMs) at the vegetative stage. Weaker expression was also detected in the culms and flowers, and basal expression was revealed in the roots, leaves, and sheaths (Figure 3A). Based on our observation that OsRA2 regulates pedicel development, and that OsRA2 over-expression results in shortened PBs, SBs, and panicles, we next examined the expression of OsRA2 at different stages of panicle development (indicated by panicle length). Expression of OsRA2 gradually increased and then decreased during panicle development, with the highest transcription level observed in 0.2–0.6 mm and 0.6–0.9 mm panicle meristems (Figure 3B). The 0.2–0.6 mm panicle meristem corresponds to the stage of PBs formation and development, while the 0.6–0.9 mm panicle meristem corresponds to the formation and development of SBs, tertiary branches and pedicels. This suggests that levels of OsRA2 expression are in accordance with its function in pedicel development and panicle development.
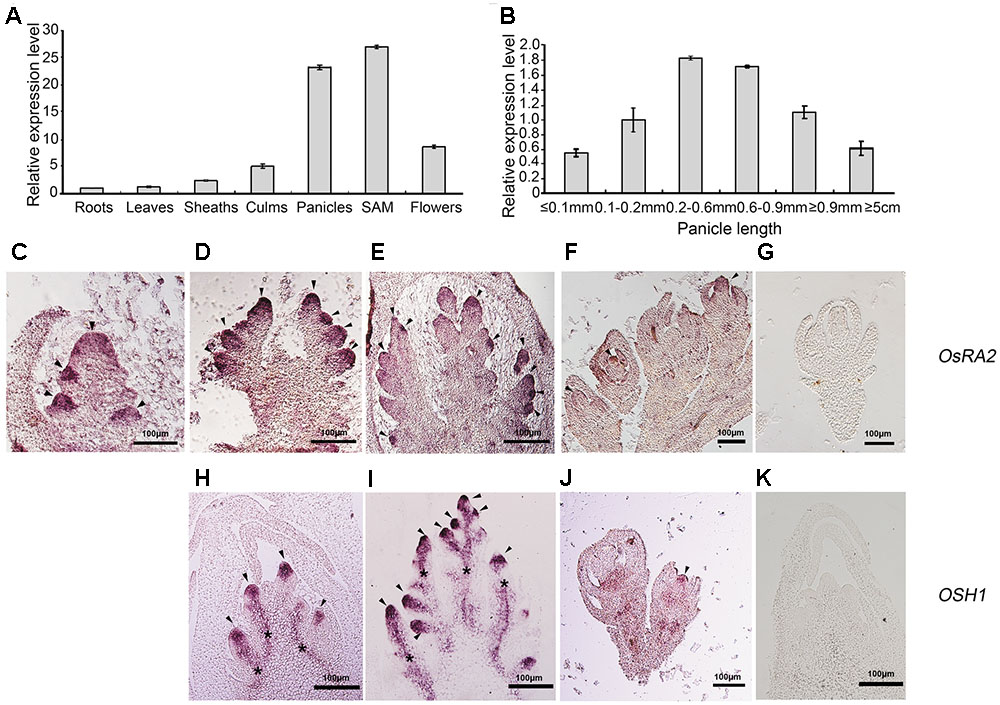
FIGURE 3. Expression profile of OsRA2. (A) OsRA2 expression in various plant organs. (B) OsRA2 expression in various stages of panicle development. (C–G) mRNA in situ hybridization of OsRA2. (C) OsRA2 expression in the primordia of the PBs (arrowheads). (D) OsRA2 expression in the primordia of the SBs (arrowheads). (E) OsRA2 expression in the primordia of the spikelet meristem (arrowheads). (F) OsRA2 expression in the developed spikelets (arrowhead). (G) Sense probe of OsRA2 hybridization. (H–K) mRNA in situ hybridization of OSH1. (H) OSH1 expression in the primordia of the PBs (arrowheads). (I) OSH1 expression in the primordia of the SBs (arrowheads). (J) OsRA2 expression in the developed spikelets (arrowheads). (K) Sense probe of OSH1 hybridization.
We further used in situ hybridization to locate OsRA2 mRNA in the young panicles. OsRA2 mRNA was highly enriched in the anlagen of PB meristems (Figure 3C), SB meristems (Figure 3D), spikelet meristems (Figure 3E), and floral organ meristems (Figure 3F) as compared with the sense probe (Figure 3G). ORYZA SATIVA HOMEOBOX1 (OSH1), an ortholog of KNOTTED1 (KN1) in rice, is highly expressed in meristematic cells and is required for meristem maintenance (Sentoku et al., 1999; Oikawa and Kyozuka, 2009; Tsuda et al., 2011). Similarly, OSH1 transcripts were detected in the anlagen of PB meristems (Figure 3H), SB meristems (Figure 3I), and floral organ meristems (Figure 3J), as compared with the sense probe (Figure 3K), indicating the meristematic character of the region enriched in OsRA2 mRNA transcripts.
Characterization of the OsRA2 Protein
Most LBD proteins are transcription factors (Husbands et al., 2007). OsRA2 was shown to have a putative nuclear localization signal in its N-terminal LOB domain (Figure 1B)6, indicating that it is also likely to be a transcription factor.
To study the cellular localization of OsRA2, we constructed transgenic plants containing OsRA2 fused in-frame to Green Fluorescent Protein (GFP) at the C-terminal. The fluorescence signal from the fusion protein was observed in the nuclei of p35S::GFP-OsRA2 root tips (Figure 4A); but was ubiquitously distributed in the cytoplasm of control plants carrying an empty GFP vector (p35S::GFP) (Figure 4A), suggesting that OsRA2 is a nuclear-localized protein. Examination of the phenotype of transgenic plants suggested that OsRA2 functions normally when fused with GFP, because p35S::GFP-OsRA2 plants showed a dwarf phenotype similar to pUbi::OsRA2 plants (Supplementary Figure S4A), and the panicles of p35S::GFP-OsRA2 plants were shortened (Supplementary Figure S4B), similar to pUbi::OsRA2 plants.
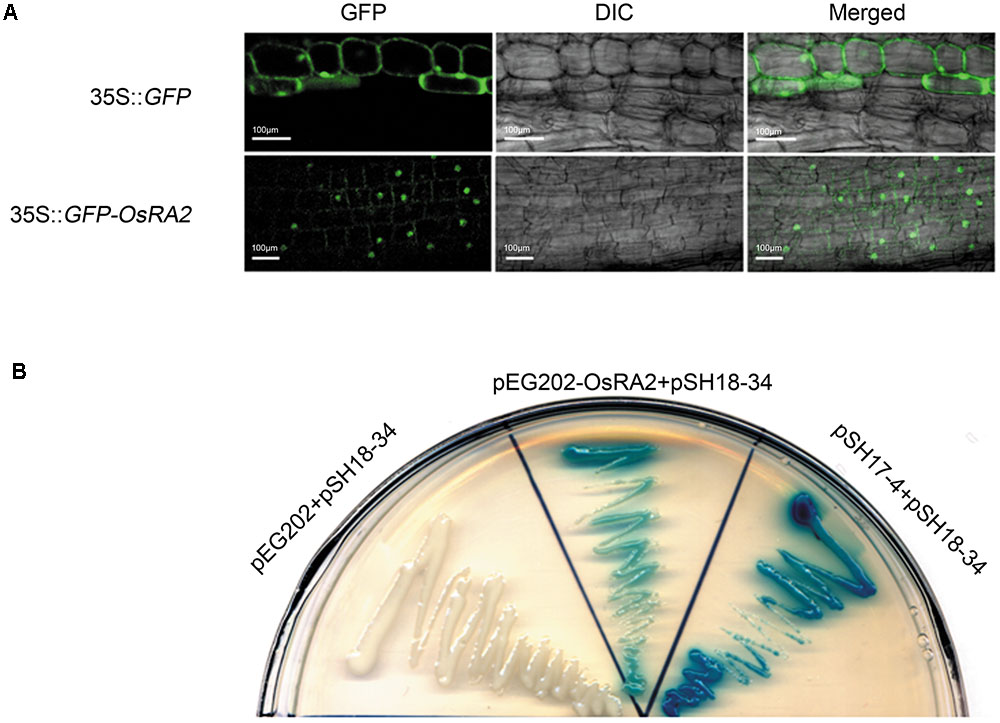
FIGURE 4. Characterization of the OsRA2 protein. (A) Sub-cellular localization of the GFP signal in the 35S::GFP-OsRA2 and the 35S::GFP transgenic plants. (B) OsRA2 showing trans-activating activity in the yeast two-hybrid system. Yeast cells transformed with OsRA2 and reporter pSH18-34 turned blue similar to the positive controls pSH17-4 and pSH18-34.
We next detected if OsRA2 contained transcriptional activity. We used two plasmids (effector pEG202 and reporter pSH18-34) selected from a DupLEX-ATM yeast two-hybrid system (Zhu et al., 2003). Full-length OsRA2 cDNA was fused in-frame with LexA protein and transformed into recipient yeast containing LacZ, whose promoter elements can be recognized by LexA. Yeast clones transformed with OsRA2-lexA showed a blue color similar to the positive control when supplemented with the β-galactosidase substrate X-gal, indicating that the OsRA2 protein has trans-activation activity (Figure 4B).
Together, these experiments strongly indicate that OsRA2 protein is a transcription factor.
Possible Cross-Talk between OsRA2 and Other Panicle Development Related Genes
To explore the pathway by which OsRA2 regulates pedicel development, we detected the expression of panicle-regulating genes in dsRNAiOsRA2 and pUbi::OsRA2 plants (Supplementary Figure S5). Among them, LAX1, MOC1 and the RCNs are involved in meristem initiation and development (Nakagawa et al., 2002; Komatsu K. et al., 2003; Li et al., 2003), SP1 regulates the panicle length (Li et al., 2009), DEP1 regulates number of branches and panicle length (Huang et al., 2009), and OSH1 is a meristem marker gene (Tsuda et al., 2011). LAX1 was shown to be up-regulated in pUbi::OsRA2 plants but down-regulated in dsRNAiOsRA2 plants (Figure 5A). lax1 mutants contain longer pedicel because of defects in initiation or maintenance of the lateral and terminal spikelets (Komatsu K. et al., 2003; Tabuchi et al., 2011), so we wondered if LAX1 is transcriptionally regulated by OsRA2. However,we found no OsRA2 binding motifs in the LAX1 promoter7. We also performed a yeast two-hybrid assay to check an interaction between OsRA2 and LAX1, but negative result turned out. Therefore, it remains unclear if any direct regulation or interaction between OsRA2 and LAX1exists.
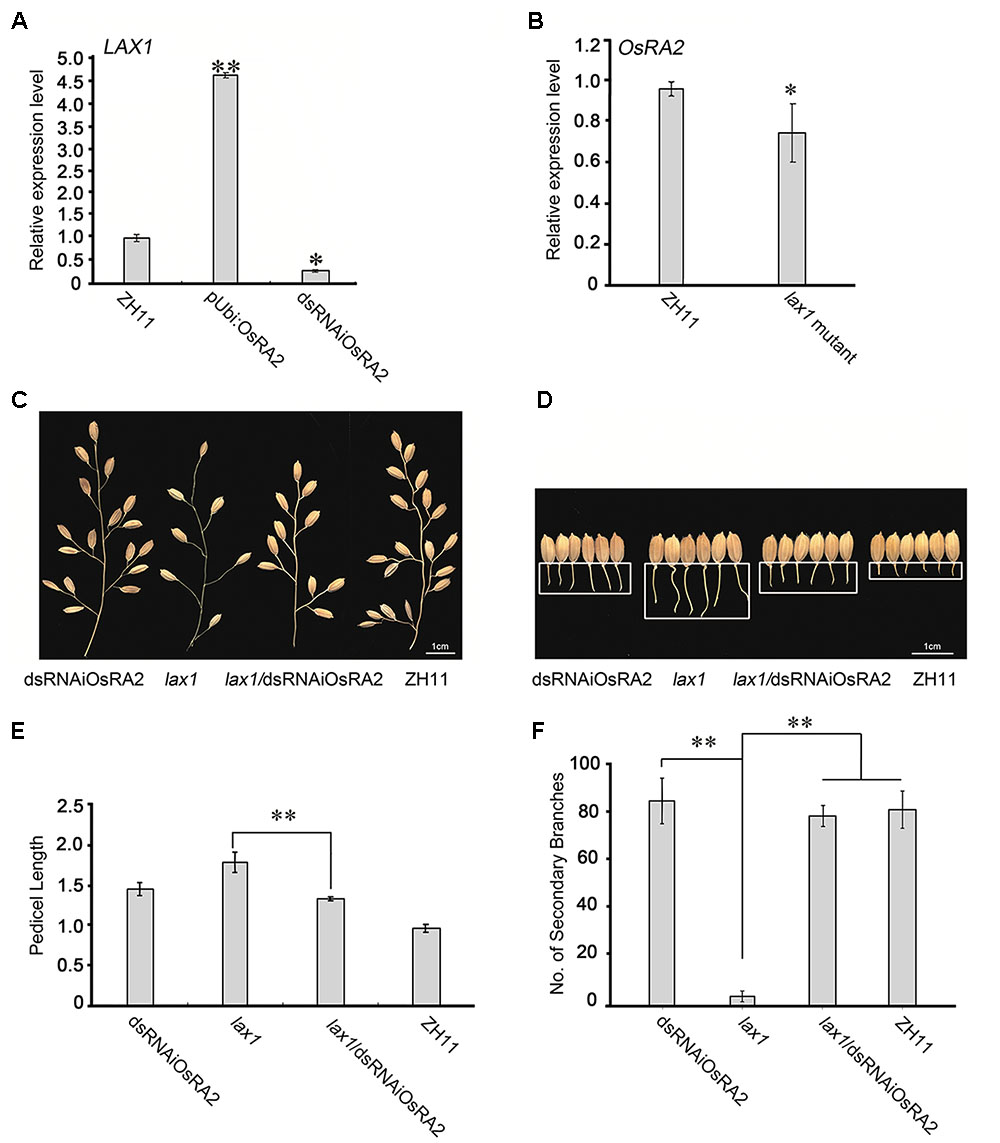
FIGURE 5. Possible relationships between OsRA2 and other genes. (A) qRT-PCR analyses of LAX1 in the ∼2 cm young panicles of pUbi::OsRA2 and dsRNAiOsRA2 plants. (B) qRT-PCR analyses of OsRA2 in the lax1 mutant. (C,D) PBs and pedicels of the dsRNAiOsRA2 plant, lax1 mutant, lax1/dsRNAiOsRA2 plant, and ZH11. (E,F) Statistical analysis of the pedicel length and number of SBs in dsRNAiOsRA2 plants, lax1 mutants, lax1/dsRNAiOsRA2 plants and ZH11. Values are means ± SE n = 15 panicles. Single and double asterisks represent significant difference determined by the Student’s t-test at ∗P < 0.05 and ∗∗P < 0.01 respectively.
OsRA2 expression was down-regulated in the lax1 mutant (Figure 5B), so to investigate the genetic relationship between OsRA2 and LAX1, we crossed the lax1 mutant with dsRNAiOsRA2 plants. And the number of SBs in the hybrid increased as compared with that in the lax1 mutant (Figures 5C,F), suggesting that LAX1 functions upstream of OsRA2 in determining the number of SBs. F2 lax1/dsRNAiOsRA2 plants showed longer pedicels compared with WT, which were more similar to those of dsRNAiOsRA2 plants (Figures 5D,E). Thus, LAX1 might also function upstream of OsRA2 in regulating pedicel length.
The over expression of RCN2 showed indeterminacy with increased PBs and SBs (Ikeda-Kawakatsu et al., 2012). To study the possible genetic relation between RCN2 and OsRA2, we crossed the pUbi::OsRA2 plants to the T-DNA insertion mutant of RCN2 in our lab (A989, Figure 6A), in the F2 generation, the characteristics of pUbi::OsRA2 plants and those of A989 were effectively additive, with the hybrid showing the shortened branches and panicles similar to those of pUbi::OsRA2 plants (Figures 6A,B,D,E), and an increased number of SBs similar to A989 (Figures 6A,C). This indicates that OsRA2 acts downstream of RCN2 in terms of pedicel and branch lengths, but upstream of RCN2 in terms of the number of SBs.
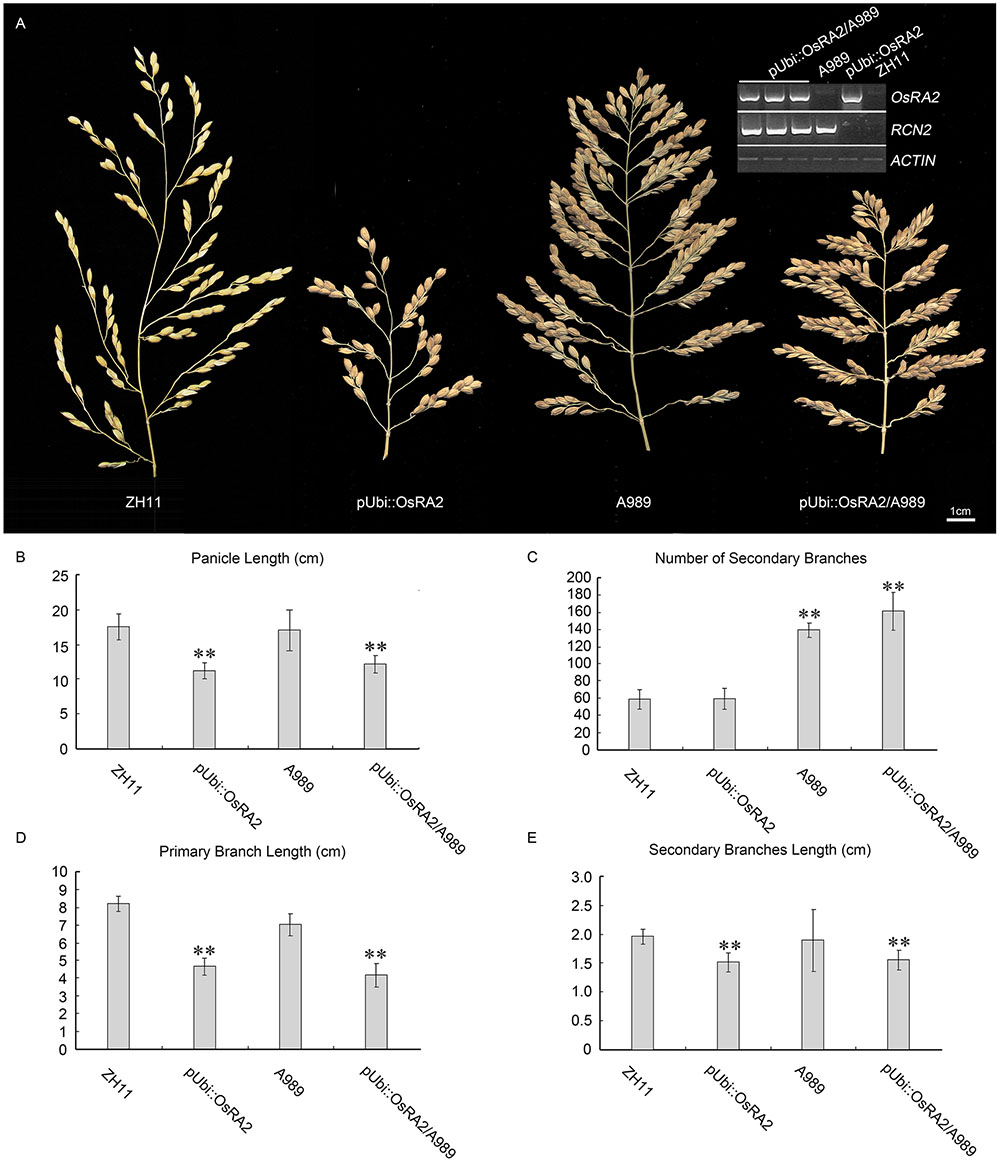
FIGURE 6. Phenotypic analysis of pUbi::OsRA2 plants and the A989 mutant. (A) Panicle characteristic of the pUbi::OsRA2 plant and the A989 mutant and their hybrid. Expression levels of RCN2 and OsRA2 in the hybrids are shown in the gel image (top right, inset). (B–E) Respective panicle characters of the pUbi::OsRA2 plant, A989 mutant and their hybrid. Student’s t-test at ∗∗P < 0.01.
Discussion
OsRA2 encodes a plant specific LBD protein. Previous studies have suggested that the function and expression pattern of RA2 genes are evolutionarily conserved among grasses (Bortiri et al., 2006). OsRA2 mRNA can be detected from the P3 stage (corresponding to the stage of PB meristems) and in the anlagen of PB meristems and spikelet meristems (Bortiri et al., 2006; Oikawa and Kyozuka, 2009). In this study, we provide further evidence that the sequences of the RA2 protein are highly conserved among several monocotyledons and dicotyledons (Figures 1B,C). We also demonstrate that OsRA2 has a novel expression profile and function in seed morphology compared with RA2 in maize. OsRA2 mRNA expression is highly enriched in AMs such as PB and SB meristems and spikelet meristems, which is in accordance with its function in regulating pedicel length and the shortened PBs and SBs in pUbi::OsRA2 plants. OsRA2 is also expressed in the floral organ meristem (Figure 3F), which is in accordance with its function in seed morphology.
Protein sequence conservation among species implies a conserved function. Many LBD proteins are characterized by their expression in the boundary between SAM and lateral organs (Shuai et al., 2002; Bortiri et al., 2006; Koppolu et al., 2013), usually boundary formation is associated with reduced cell division (Wang et al., 2016), so the reduced plant height (Figure 2E) and shortened pedicels (Figures 2B,C) of pUbi::OsRA2 plants could be explained by the functional conservation of LBD proteins, similar to the stunt phenotype of AtLOB ectopic expression in Arabidopsis (Shuai et al., 2002).
Maize exhibits two types of reproductive inflorescences, the male tassel and the female ear. Pairs of staminate flowers develop within the tassel while spikelet pairs with two pistillate flowers initiate in the ear (Liu et al., 2009; Thompson and Hake, 2009; Yuan et al., 2009; Zhang and Yuan, 2014). In both maize and barley, RA2 genes determine the spikelet determinacy and the ra2 mutant shows increased number of branches in the reproductive organs (Bortiri et al., 2006; Koppolu et al., 2013). However, in the present study, down regulation of OsRA2 in dsRNAiOsRA2 plants showed distinct pedicel elongation, but not increased branches. Therefore, OsRA2 does not seem to influence determinacy in rice, which differs from the associations in maize and barley. Although some spikelet pairs of maize ra2 mutants became single spikelets in the tassel, and some showed longer pedicels, the function of ra2 in regulating pedicel length is less obvious than OsRA2 in rice. RA2 predicts the position of branch anlagen in maize inflorescence (Bortiri et al., 2006); similarly, OsRA2 functions within rice PB and SB anlagens (Figures 3C–E). Therefore, expression profile conservation indicates that RA2 is critical for shaping the initial steps of inflorescence architecture in both maize and rice.
To date, although a few genes associated with the panicle architecture have been studied, little is known about the underlying mechanism of their functional cross-talks. In the present study, genetic analysis suggested that LAX1 functions upstream of OsRA2 in regulating the number of SBs (Figures 5C,F), and may also do so in regulating pedicel length (Figures 5D,E), although this result might have been influenced by the expression level of OsRA2 in RNAi plants. The expression of LAX1 was also influenced by OsRA2 expression (Figure 5A), indicating the possible existence of feed-back regulation between OsRA2 and LAX1. The fact that higher OsRA2 expression led to an increased number of vascular bundles suggests that it might benefit vascular development. The cross between A989 (gain of function of RCN2) and OsRA2 over expressing lines resulted in plants with more branches and shorter branch lengths, indicating that OsRA2 acts downstream of RCN2 in regulating branch lengths, while upstream of RCN2 in regulating the number of SBs. This provides evidence for the respective regulation of the number and length of branches in the panicle by two parallel pathways.
Author Contributions
Experimental design: XM and ZS; Experiments: HL, ZD, LL, and JW; Data analysis: JW and ZS; Manuscript preparation: HL, ZD, ZS, and XM; Supervision, funding and reagents: ZS and XM.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0100600), Ministry of Science and Technology of China (2012AA10A302-2), the National Special Program on Research and Commercialization of Transgenic Plant (2014ZX08009-003-003), Scholarship Foundation from Shanghai Institutes for Biological Sciences (2007KIP206) and the SA-SIBS 2009 Young Faculty Award.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We would like to thank Xiaoyan Gao from Shanghai Institutes for Biological Sciences for his help in Anatomical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01538/full#supplementary-material
Footnotes
- ^http://www.uniprot.org/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://rice.plantbiology.msu.edu/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://www.uniprot.org/
- ^http://www.uniprot.org/
- ^http://www.dna.affrc.go.jp/PLACE/signalscan.html
References
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18, 574–585. doi: 10.1105/tpc.105.039032
Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S., and Coen, E. (1996). Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797. doi: 10.1038/379791a0
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. doi: 10.1126/science.275.5296.80
Chuck, G., Meeley, R., Irish, E., Sakai, H., and Hake, S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521. doi: 10.1038/ng.2007.20
Coen, E. S., Romero, J. M., Doyle, S., Elliott, R., Murphy, G., and Carpenter, R. (1990). floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell 63, 1311–1322. doi: 10.1016/0092-8674(90)90426-F
Ferrandiz, C., Gu, Q., Martienssen, R., and Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Han, Y., Yang, H., and Jiao, Y. (2014). Regulation of inflorescence architecture by cytokinins. Front. Plant Sci. 5:669. doi: 10.3389/fpls.2014.00669
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Huang, X., Qian, Q., Liu, Z., Sun, H., He, S., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Huijser, P., Klein, J., Lonnig, W. E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11, 1239–1249.
Husbands, A., Bell, E. M., Shuai, B., Smith, H. M., and Springer, P. S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671. doi: 10.1093/nar/gkm775
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J., and Nagato, Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kellogg, E. A., Camara, P. E., Rudall, P. J., Ladd, P., Malcomber, S. T., Whipple, C. J., et al. (2013). Early inflorescence development in the grasses (Poaceae). Front. Plant Sci. 4:250. doi: 10.3389/fpls.2013.00250
Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., et al. (2003). LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. U.S.A. 100, 11765–11770. doi: 10.1073/pnas.1932414100
Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K., and Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. doi: 10.1242/dev.00564
Komatsu, M., Maekawa, M., Shimamoto, K., and Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. doi: 10.1006/dbio.2000.9988
Koppolu, R., Anwar, N., Sakuma, S., Tagiri, A., Lundqvist, U., Pourkheirandish, M., et al. (2013). Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. U.S.A. 110, 13198–13203. doi: 10.1073/pnas.1221950110
Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5, 150–163. doi: 10.1093/bib/5.2.150
Li, M., Tang, D., Wang, K., Wu, X., Lu, L., Yu, H., et al. (2011). Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol. J. 9, 1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x
Li, S., Qian, Q., Fu, Z., Zeng, D., Meng, X., Kyozuka, J., et al. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58, 592–605. doi: 10.1111/j.1365-313X.2009.03799.x
Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., et al. (2003). Control of tillering in rice. Nature 422, 618–621. doi: 10.1038/nature01518
Liang, W. H., Shang, F., Lin, Q. T., Lou, C., and Zhang, J. (2014). Tillering and panicle branching genes in rice. Gene 537, 1–5. doi: 10.1016/j.gene.2013.11.058
Liu, C., Thong, Z., and Yu, H. (2009). Coming into bloom: the specification of floral meristems. Development 136, 3379–3391. doi: 10.1242/dev.033076
Luo, L., Li, W., Miura, K., Ashikari, M., and Kyozuka, J. (2012). Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 53, 1793–1801. doi: 10.1093/pcp/pcs122
Majer, C., and Hochholdinger, F. (2011). Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. doi: 10.1016/j.tplants.2010.09.009
Mandel, M. A., and Yanofsky, M. F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. doi: 10.1038/377522a0
Matsumura, Y., Iwakawa, H., Machida, Y., and Machida, C. (2009). Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 58, 525–537. doi: 10.1111/j.1365-313X.2009.03797.x
Mcsteen, P. (2009). Hormonal regulation of branching in grasses. Plant Physiol. 149, 46–55. doi: 10.1104/pp.108.129056
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Nakagawa, M., Shimamoto, K., and Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313X.2002.01255.x
Oikawa, T., and Kyozuka, J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21, 1095–1108. doi: 10.1105/tpc.108.065425
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Sakamoto, T., and Matsuoka, M. (2008). Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11, 209–214. doi: 10.1016/j.pbi.2008.01.009
Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H., and Matsuoka, M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11, 1651–1664. doi: 10.1105/tpc.11.9.1651
Shuai, B., Reynaga-Pena, C. G., and Springer, P. S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Tabuchi, H., Zhang, Y., Hattori, S., Omae, M., Shimizu-Sato, S., Oikawa, T., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765
Teo, Z. W., Song, S., Wang, Y. Q., Liu, J., and Yu, H. (2014). New insights into the regulation of inflorescence architecture. Trends Plant Sci. 19, 158–165. doi: 10.1016/j.tplants.2013.11.001
Thompson, B. E., and Hake, S. (2009). Translational biology: from Arabidopsis flowers to grass inflorescence architecture. Plant Physiol. 149, 38–45. doi: 10.1104/pp.108.129619
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tsuda, K., Ito, Y., Sato, Y., and Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
Wang, D., Chen, X., Zhang, Z., Liu, D., Song, G., Kong, X., et al. (2015). A MADS-box gene NtSVP regulates pedicel elongation by directly suppressing a KNAT1-like KNOX gene NtBPL in tobacco (Nicotiana tabacum L.). J. Exp. Bot. 66, 6233–6244. doi: 10.1093/jxb/erv332
Wang, L., Sun, S., Jin, J., Fu, D., Yang, X., Weng, X., et al. (2015). Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 15504–15509. doi: 10.1073/pnas.1521949112
Wang, J., Gao, X., Li, L., Shi, X., Zhang, J., and Shi, Z. (2010). Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J. Exp. Bot. 61, 1885–1895. doi: 10.1093/jxb/erp378
Wang, J., Yu, H., Xiong, G., Lu, Z., Jiao, Y., Meng, X., et al. (2017). Tissue-specific ubiquitination by IPA1 INTERACTING PROTEIN 1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29, 697–707. doi: 10.1105/tpc.16.00879
Wang, Q., Hasson, A., Rossmann, S., and Theres, K. (2016). Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol. 209, 485–498. doi: 10.1111/nph.13641
Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377, 495–500. doi: 10.1038/377495a0
Xu, C., Luo, F., and Hochholdinger, F. (2016). LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. doi: 10.1016/j.tplants.2015.10.010
Yamaguchi, N., and Komeda, Y. (2013). The role of CORYMBOSA1/BIG and auxin in the growth of Arabidopsis pedicel and internode. Plant Sci. 209, 64–74. doi: 10.1016/j.plantsci.2013.04.009
Yuan, Z., Gao, S., Xue, D. W., Luo, D., Li, L. T., Ding, S. Y., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149, 235–244. doi: 10.1104/pp.108.128231
Zhang, D., and Yuan, Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65, 553–578. doi: 10.1146/annurev-arplant-050213-040104
Keywords: LBD protein, RA2 gene, panicle architecture, pedicel, transcriptional factors
Citation: Lu H, Dai Z, Li L, Wang J, Miao X and Shi Z (2017) OsRAMOSA2 Shapes Panicle Architecture through Regulating Pedicel Length. Front. Plant Sci. 8:1538. doi: 10.3389/fpls.2017.01538
Received: 11 April 2017; Accepted: 22 August 2017;
Published: 12 September 2017.
Edited by:
Patricia Springer, University of California, Riverside, United StatesReviewed by:
Yanhai Yin, Iowa State University, United StatesBeth Thompson, East Carolina University, United States
Copyright © 2017 Lu, Dai, Li, Wang, Miao and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenying Shi, enlzaGlAc2licy5hYy5jbg==
†These authors have contributed equally to this work.
 Huan Lu1,2†
Huan Lu1,2† Ling Li
Ling Li Zhenying Shi
Zhenying Shi