
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 September 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01525
This article is part of the Research Topic Hormonal Control of Important Agronomic Traits View all 31 articles
The phytohormone jasmonates (JAs) regulate various defense responses and diverse developmental processes including stamen development and fertility. Previous studies showed that JA induces CORONATINE INSENSITIVE 1-mediated degradation of JA ZIM-domain (JAZ) proteins, and activates the MYB transcription factors (such as MYB21 and MYB24) to regulate stamen development. In this study, we further uncover the mechanism underlying how MYB24 interacts with JAZs to control JA-regulated stamen development. We show that N-terminus of MYB21/24 interacts with 10 out of 12 JAZ proteins while both N-terminus and C-terminus of MYB24 are involved in dimerization of MYB21 and MYB24. Interestingly, male sterility of the JA-deficient mutant opr3 can be rescued by suitable level of the MYB24 overexpression but not by excessive high level of MYB24. Surprisingly, overexpression of MYB24NT, but not MYB24CT, could cause male sterility. These results provide new insights on MYB factors in JA-regulated stamen development.
Jasmonates (JAs), a class of lipid-derived phytohormones (Browse, 2009; Wasternack and Song, 2017), are crucial players in various aspects of plant developmental processes (Huang et al., 2017), including root growth (Fernandez-Calvo et al., 2011; Lischweski et al., 2015), stamen development (Mandaokar et al., 2006; Mandaokar and Browse, 2009; Song et al., 2011; Reeves et al., 2012; Song et al., 2013b), flowering (Zhai et al., 2015), trichome initiation (Qi et al., 2011), and leaf senescence (Qi et al., 2015b); they also mediate plant abiotic stress tolerance and defenses against herbivores and necrotrophic pathogens (Howe and Jander, 2008; Campos et al., 2014; Kazan, 2015; Goossens et al., 2016).
In response to developmental signals or environmental cue-triggered JA biosynthesis, the JA receptor CORONATINE INSENSITIVE 1 (COI1) (Xie et al., 1998; Yan et al., 2009) perceives bioactive molecules of JA (Fonseca et al., 2009; Yan et al., 2016) to recruit JA ZIM-domain (JAZ) proteins for ubiquitination and subsequent degradation via the 26S-proteasome (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007), thereby de-repressing JAZ-inhibited transcription factors, such as MYC2/3/4/5 (Cheng et al., 2011; Fernandez-Calvo et al., 2011; Niu et al., 2011; Song et al., 2014a; Figueroa and Browse, 2015; Qi et al., 2015a; Gimenez-Ibanez et al., 2017; Major et al., 2017), MYB21/24 (Song et al., 2011), IIId bHLH factors (Nakata et al., 2013; Nakata and Ohme-Takagi, 2013; Song et al., 2013a), and TTG1/bHLH/MYB complexes (Qi et al., 2011) to modulate distinct JA responses.
Jasmonate-deficient mutants (e.g., aos and opr3), the JA receptor mutant coi1-1, and JAZ dominant-negative transgenic plants (JAZ1Δ3A) are all male sterile with defects in filament elongation, anther dehiscence, and pollen maturation (Xie et al., 1998; Stintzi and Browse, 2000; Park et al., 2002; Thines et al., 2007). The R2R3–MYB transcription factors MYB21 and MYB24 associate with IIIe bHLH factors (MYC2, MYC3, MYC4, and MYC5) to form MYB–MYC complexes, and interact with JAZs to mediate late stamen development (Qi et al., 2015a).
We previously showed that JAZ1/8/11 interact with MYB21/MYB24 in yeast and plants (Song et al., 2011). In this study, we further showed that MYB21 and MYB24 interact with most JAZs via their N-terminal R2R3 domains, and both the N-terminus and C-terminus of MYB24 mediate the dimeric interactions of MYB21 and MYB24. Proper overexpression of MYB24 partially restores male fertility of opr3. Overexpression of N-terminus of MYB24, but not C-terminus, causes male sterility in wild-type. Furthermore, young flower buds from myb21 myb24 myb57 accumulate more jasmonic acid than that of wild-type.
The Arabidopsis mutants opr3 (Stintzi and Browse, 2000) and myb21 myb24 myb57 (Cheng et al., 2009) were described previously. Arabidopsis thaliana seeds were disinfected, germinated on Murashige and Skoog (MS) medium, stored at 4°C for 3 days, and transferred to a growth room for another 7 days before being transferred to soil under a 16 h (22–24°C)/8 h (17–19°C) light/dark photoperiod. Nicotiana benthamiana seeds were sown in soil and grown under a 16 h (26°C)/8 h (22°C) light/dark photoperiod.
The full lengths of coding regions of MYB21, MYB24, 12 JAZs, and truncated domains of MYB24 were individually fused with the activation domain (AD) in pB42AD, or the DNA binding domain (BD) in pLexA. The primer pairs used for vector construction are listed in Supplementary Table 1. Yeast transformation and protein–protein interaction assays were performed according to the Matchmaker LexA Two-Hybrid System (Clontech) as described previously (Song et al., 2011).
JAZ5, MYB21, MYB24, MYB24NT, and MYB24CT were individually inserted into pCAMBIA-nLUC or pCAMBIA-cLUC for fusion with the N-terminal half of LUC (nLUC) or C-terminal half of LUC (cLUC). The primers used to construct the LCI vector are listed in Supplementary Table 1. Agrobacterium tumefaciens cells containing the indicated plasmids were co-infiltrated into N. benthamiana leaves and LUC activity was detected as described previously (Song et al., 2011).
To obtain transgenic plants overexpressing MYB24, the coding sequence of MYB24 was amplified and cloned using XbaI and SacI into pCAMBIA1301 under the control of the CaMV 35S promoter. The construct was transformed into OPR3/opr3 heterozygous plants using the Agrobacterium-mediated floral dip method. The primers used for vector construction are presented in Supplementary Table 1. pCAMBIA-MYB24NT-nLUC and pCAMBIA-MYB24CT-nLUC were transformed into Arabidopsis Col-0 wild-type to generate MYB24NT- and MYB24CT-overexpressing plants.
Young flower buds of plants were collected for total RNA extraction and subsequent reverse transcription. Quantitative real-time PCR was performed with RealMasterMix (SYBR Green I; Takara; Bio Inc., Otsu, Japan) using the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States), and ACTIN8 was used as an internal control. The primers used are listed in Supplementary Table 1.
For flower phenotype analysis, flowers of each genotype at stage 13 were photographed under a microscope. Pollen germination assay was conducted as described previously (Song et al., 2011). Pollen grains were germinated on pollen germination medium [1 mM MgSO4, 5 mM CaCl2, 5 mM KCl, 10% (w/v) sucrose, 0.01% boric acid, and 1.5% agar at pH 7.5], incubated for 10 h at 22°C in the dark, and observed under a microscope.
Five hundred milligrams of young flower buds before floral stage 13 from 5-week-old wild-type and the myb21 myb24 myb57 mutant was harvested, and jasmonic acid was extracted and quantified as described previously using a liquid chromatography–tandem mass spectrometry system (Cheng et al., 2009).
The Arabidopsis Genome Initiative numbers for genes mentioned in this article are as follows: JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ4 (At1g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10 (At5g13220), JAZ11 (At3g43440), JAZ12 (At5g20900), MYB21 (At3g27810), MYB24 (At5g40350), MYB57 (At3g01530), OPR3 (AT2G06050), and ACTIN8 (At1g49240).
MYB21 and MYB24 were divided into MYB21NT and MYB24NT harboring the R2R3 DNA BD, and MYB21CT and MYB24CT containing the C-terminal NYWG/SM/VDDI/LWS/P transcriptional activation motif (Figures 1A,B). Binding domain-fused MYB21CT and MYB24CT exhibited auto-activation, while MYB21NT and MYB24NT did not. Binding domain-fused MYB21NT and MYB24NT were used to detect interactions with AD-fused JAZ proteins. The results (Figure 1C) showed that MYB21NT and MYB24NT interact with JAZ1, JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ8, JAZ10, JAZ11, and JAZ12, but not with JAZ7 or JAZ9, demonstrating that MYB21 and MYB24 interact with multiple JAZs through their N-terminus.
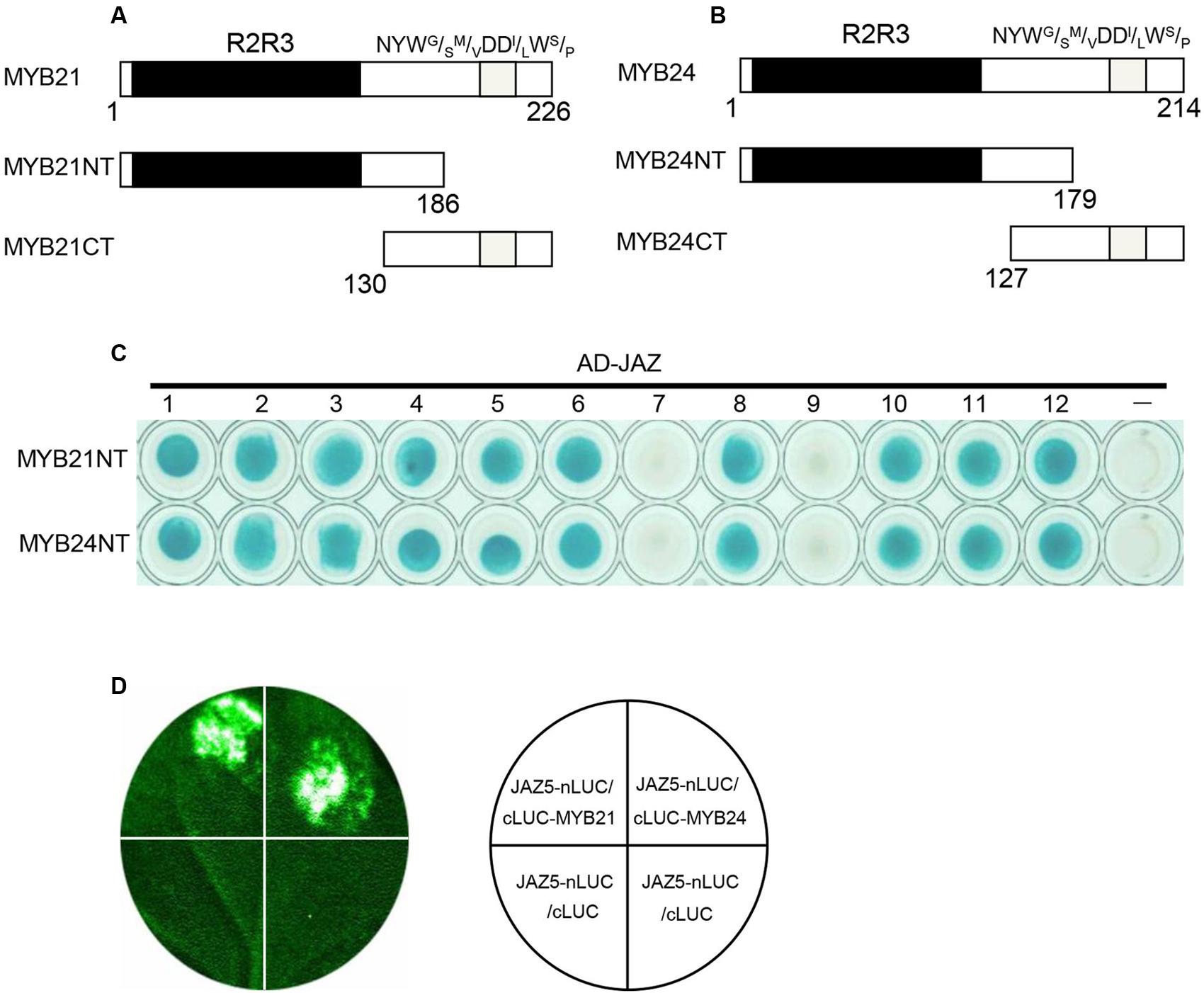
FIGURE 1. MYB21 and MYB24 interact with JAZ proteins via their N-terminus. (A,B) Schematic diagram of MYB21 (A) and MYB24 (B) with the R2R3 domain (black) and NYWG/SM/VDDl/LWS/P motif (gray). (C) Y2H assays to show interactions between MYB21NT/MYB24NT and JAZs. MYB21NT and MYB24NT were fused with DNA binding domain (BD) in pLexA, and JAZs were fused with activation domain (AD) in pB42AD, respectively. (D) JAZ5 interacts with MYB21 and MYB24 in LCI assay. JAZ5 was fused with N-terminal fragment of LUC (nLUC), while MYB21/MYB24 were fused with C-terminal fragment of LUC (cLUC), respectively. LUC signals were detected 60 h after co-infiltration of the indicated constructs into N. benthamiana leaves.
We next performed a firefly LCI assay to test the interactions of JAZ5 with MYB21/24 in plant. JAZ5 was fused with nLUC, while MYB21 and MYB24 were fused with cLUC. The results showed that co-infiltration of JAZ5-nLUC/cLUC-MYB21 or JAZ5-nLUC/cLUC-MYB24 in N. benthamiana leaves resulted in strong LUC signals while the negative control did not (Figure 1D), suggesting that MYB21 and MYB24 interact with JAZ5 in plant.
We next examined whether MYB24 overexpression could escape from inhibition by multiple JAZs to rescue stamen development and fertility in the JA-deficient mutant opr3. As shown in Figure 2A, opr3 exhibited unelongated filaments, indehiscent anthers, and inviable pollen grains at floral stage 13. MYB24 expression decreased in young opr3 flower buds (Figure 2D). MYB24 overexpression in opr3 (opr3 MYB24OE37, with fivefold to sixfold of wild-type level) restored filament elongation, anther dehiscence, and pollen viability (Figure 2A). Further, the opr3 MYB24OE37 plants were able to set seeds (Figures 2B,C). These results suggest that overexpression of MYB24 could partially restore stamen development and fertility in opr3.
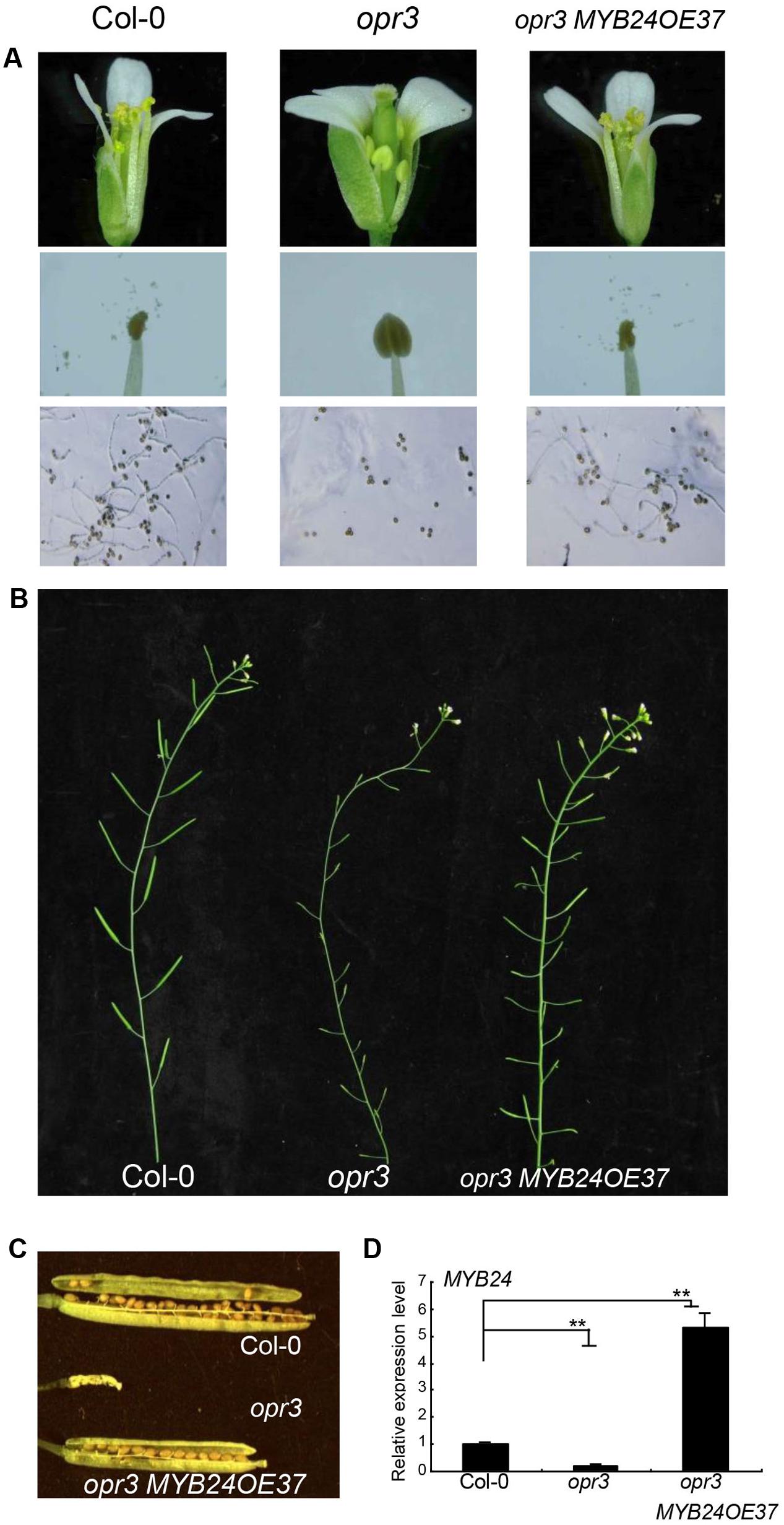
FIGURE 2. Overexpression of MYB24 partially restores the stamen development and fertility of opr3. (A–C) Comparison of flowers, stamens, and pollen germination (A) at floral stage 13, main inflorescence (B), and seed setting (C) in Col-0, opr3, and opr3 MYB24OE37. MYB24 overexpression partially restored the fertility of opr3 (A–C). (D) Quantitative real-time PCR analysis of MYB24 expression in young flower buds of Col-0, opr3, and opr3 MYB24OE37. ACTIN8 was used as the internal control. Data are means (±SE) of three biological replicates. Asterisks represent Student’s t-test significance between pairs indicated with brackets (∗∗p < 0.01).
Previous studies showed that strong MYB24 overexpression inhibits stamen development (Yang et al., 2007; Song et al., 2011). We therefore tested whether excess expression of MYB24 could restore fertility in opr3. As shown in Figure 3, transgenic plants that expressed excessive amounts of MYB24 (∼75-fold of the wild-type level) were male sterile. Further, transgenic opr3 plants that expressed excessive amounts of MYB24 (∼70-fold of the wild-type level) were still male sterile, suggesting that excess expression of MYB24 could not restore stamen development and fertility in opr3 plants.
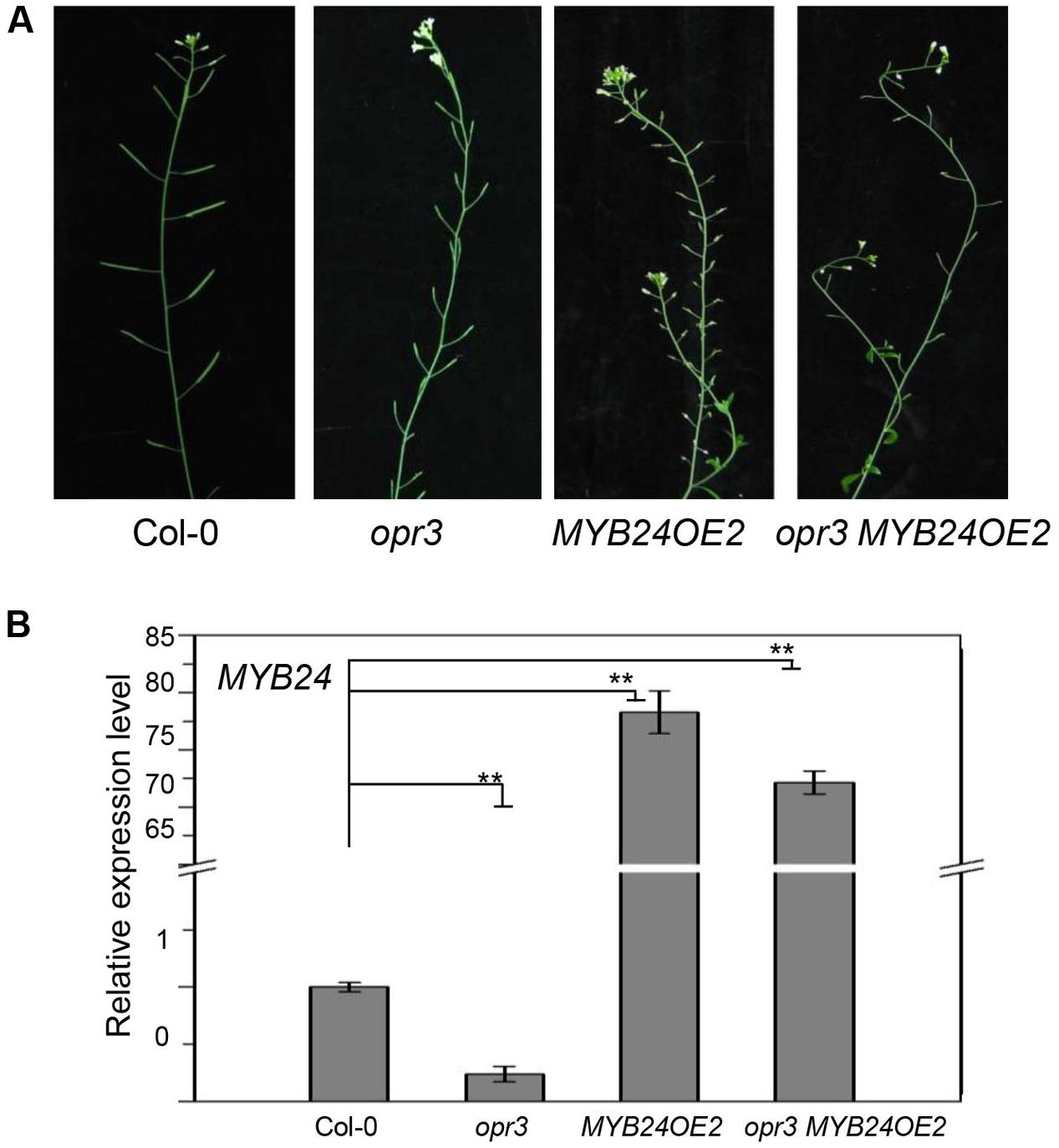
FIGURE 3. Excess expression of MYB24 fails to rescue the fertility of opr3. (A) Main inflorescences of Col-0, opr3, MYB24OE2, and opr3 MYB24OE2. (B) Quantitative real-time PCR analysis of MYB24 expression in young flower buds. ACTIN8 was used as the internal control. Data are means (±SE) of three biological replicates. Asterisks represent Student’s t-test significance between pairs indicated with brackets (∗∗p < 0.01).
We next investigated the dimeric interactions of MYB21 and MYB24 in detail. As shown in Figure 4A, Y2H analysis showed that BD-fused MYB24NT interacted strongly with AD-fused MYB24NT, and weakly with MYB24CT. Further, LCI assay exhibited that the co-expression of MYB24NT-nLUC/cLUC-MYB21, MYB24CT-nLUC/cLUC-MYB21, MYB24NT-nLUC/cLUC-MYB24, and MYB24CT-nLUC/cLUC-MYB24 resulted in strong LUC activity, while the negative controls did not (Figures 4B,C). These results demonstrate that both N-terminus and C-terminus of MYB24 are involved in dimeric interactions of MYB21 and MYB24.
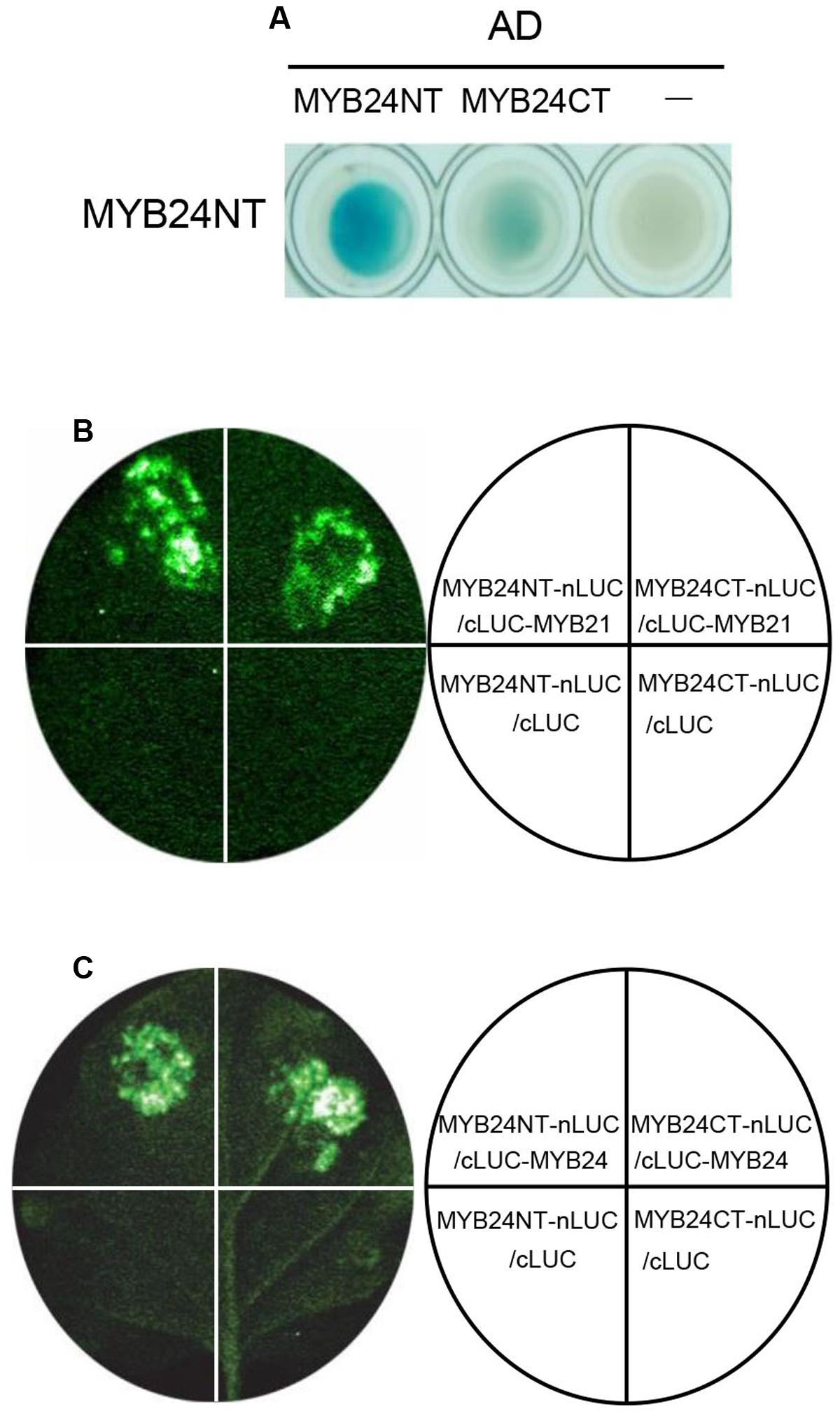
FIGURE 4. MYB24NT and MYB24CT mediate dimeric interactions. (A) Y2H assays to show the interactions of BD-fused MYB24NT with AD-fused MYB24NT and MYB24CT. (B,C) LCI assays show that both MYB24NT and MYB24CT interact with MYB21 (B) and MYB24 (C). MYB24NT and MYB24CT were fused with nLUC, while MYB21/MYB24 were fused with cLUC, respectively. LUC signals were collected 60 h after co-infiltration.
We next examined whether the overexpression of MYB24NT and MYB24CT could dominantly repress stamen development and male fertility. MYB24NT overexpression (∼20–40-fold of the wild-type level) inhibited stamen development, including filament elongation, anther dehiscence, and male fertility (Figures 5A,B and Supplementary Figure 1A), while all MYB24CT transgenic lines showed no obvious influence on male fertility (Figure 5C and Supplementary Figure 1B). These data indicate that overexpression of N-terminus of MYB24 could dominantly repress stamen development and fertility.
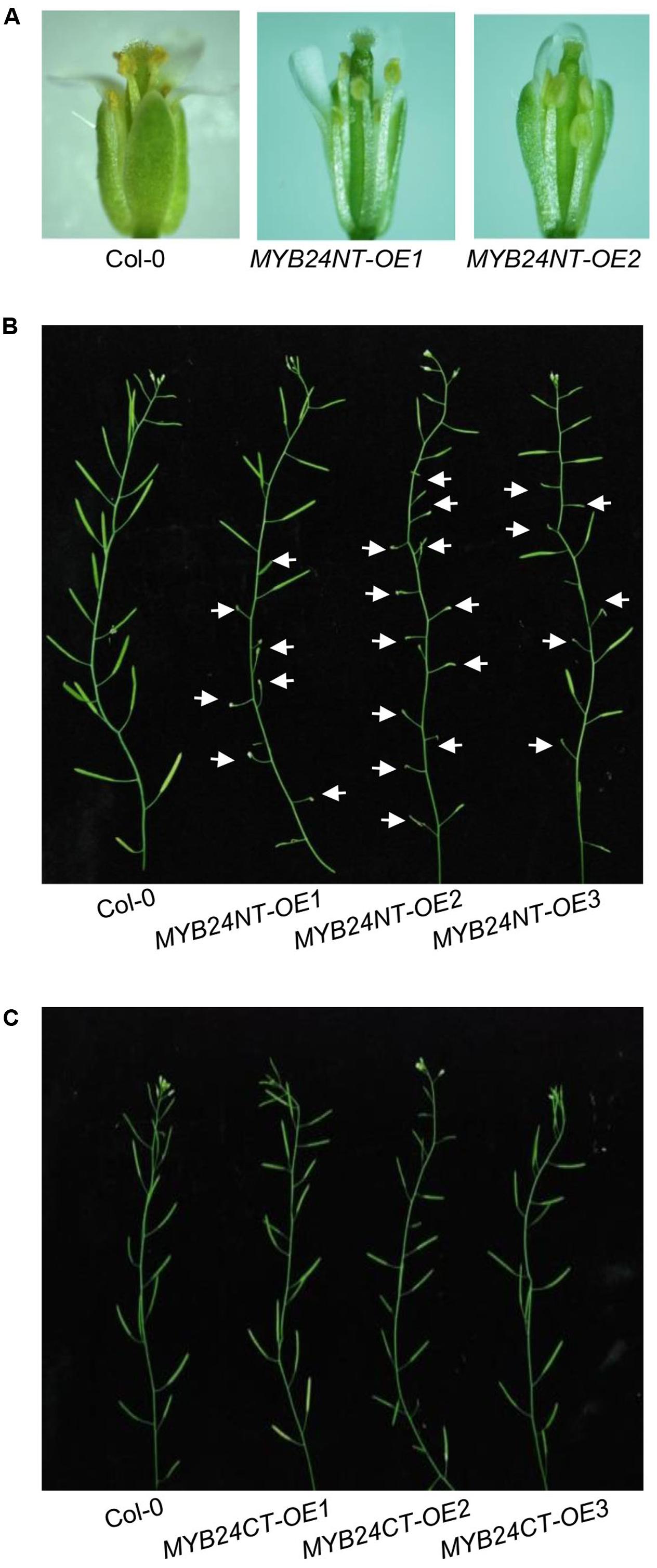
FIGURE 5. Excess expression of MYB24NT causes male sterility. (A) Comparison of flowers at stage 13 from Col-0 and two MYB24NT overexpression lines (MYB24NT-OE1 and MYB24NT-OE2). (B) Main inflorescences of Col-0, MYB24NT-OE1, MYB24NT-OE2, and MYB24NT-OE3. White arrows indicate sterile siliques. (C) Main inflorescences of Col-0, MYB24CT-OE1, MYB24CT-OE2, and MYB24CT-OE3.
MYB21, MYB24, and MYB57 are all responsive to JA (Figure 1; Mandaokar et al., 2006; Cheng et al., 2009; Mandaokar and Browse, 2009). We thus tested whether MYB21, MYB24, and MYB57 in turn regulate JA biosynthesis in young flower buds (before floral stage 13). The jasmonic acid content of young myb21 myb24 myb57 mutant flower buds was approximately twofold of the wild-type level (Figure 6), suggesting that MYB21, MYB24, and MYB57 negatively regulate JA biosynthesis as part of a negative feedback loop.
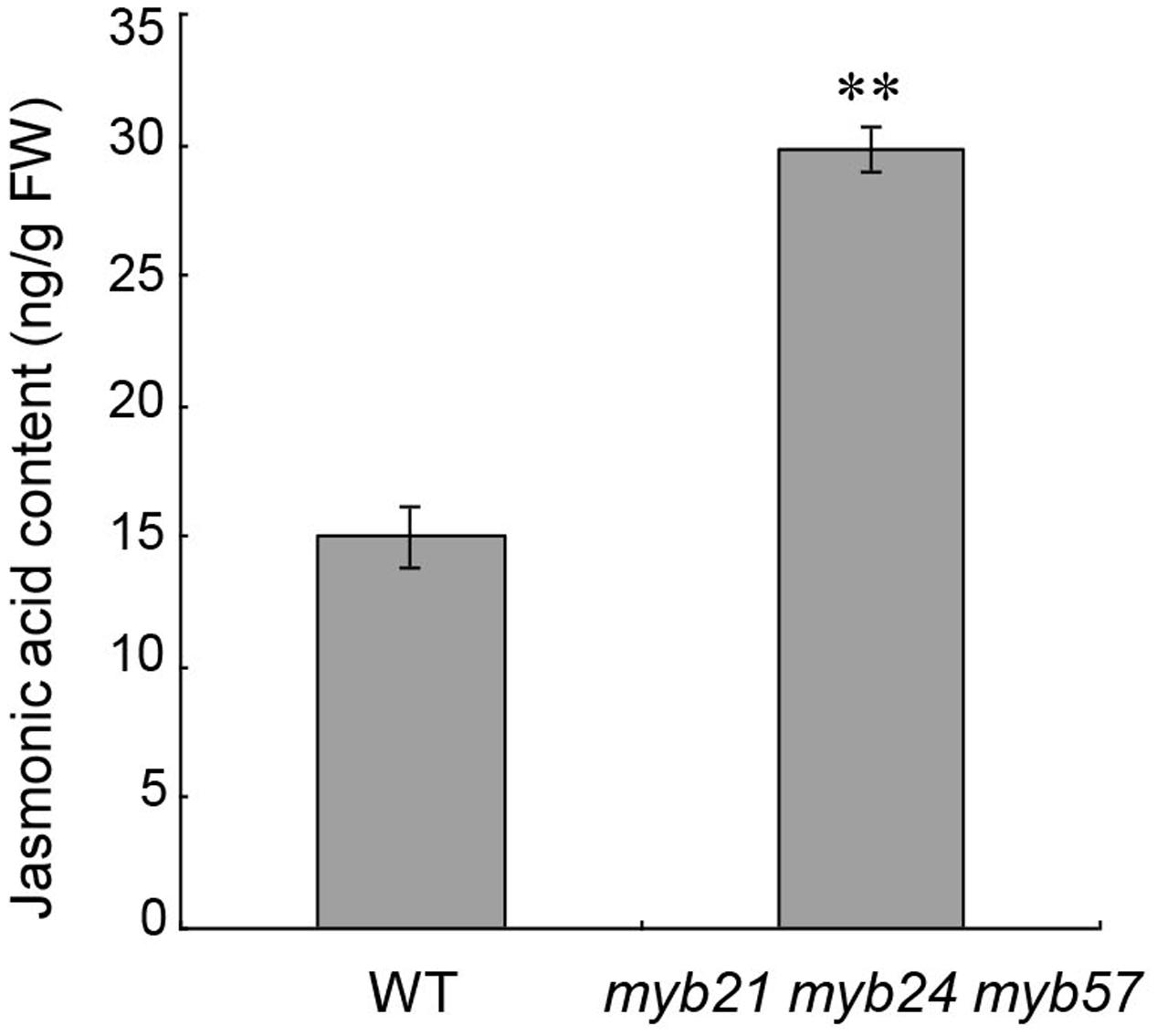
FIGURE 6. Jasmonic acid contents of young flower buds from wild-type and myb21 myb24 myb57. Jasmonic acid contents of young flower buds before floral stage 13 from wild-type and myb21 myb24 myb57 plants. Data are means (±SE) of three biological replicates. Asterisks represent Student’s t-test significance compared with Col-0 wild-type (∗∗p < 0.01).
Jasmonate ZIM-domain proteins serve as repressors that target specific transcription factors and control their downstream pathways to modulate distinct JA responses for coordinated regulation of development, growth, and defense (Song et al., 2014b; Chini et al., 2016; Gimenez-Ibanez et al., 2017; Major et al., 2017). Our previous study showed that JAZ1, JAZ8, and JAZ11 interact with MYB21 and MYB24 (Song et al., 2011). In this study, we further showed that MYB21 and MYB24 act through N-terminus to interact with 10 out of 12 JAZs (Figure 1), suggesting that most JAZs may act through interfering the DNA binding function of MYB21/24 to attenuate their function in regulating stamen development. Excess expression of MYB24 is unable to restore the stamen development of opr3, whereas suitable overexpression of MYB24 could recover stamen development and male fertility (Figures 2, 3). Exploring the downstream pathways of MYB24 would help to understand that the suitable MYB24 expression level is essential for proper stamen development.
Both the N-terminal DNA BD and C-terminal transcriptional activation motif mediate dimerization of MYB21 and MYB24 (Figure 4). Determination of crystal structure of MYB21/24 will help to further elucidate the interaction. Interestingly, overexpression of N-terminus of MYB24, but not the C-terminus of MYB24, attenuates stamen development and male fertility (Figure 5). MYB24NT-OE2 with the highest expression level of MYB24NT confers the most severe male sterility (Figures 5A,B and Supplementary Figure 1A), suggesting that the expression level of MYB24NT is correlated with male sterility. It remains to study whether MYB24NT affects the dimeric interaction of MYBs to affect stamen development.
We also found that young flower buds of myb21 myb24 myb57 accumulated more jasmonic acid (Figure 6), suggesting that MYBs negatively regulate JA biosynthesis to attenuate JA-induced expression of MYBs and to elaborately regulate stamen development, and that the restored fertility in opr3 (Stintzi and Browse, 2000; Chehab et al., 2011) by suitable MYB24 overexpression (Figure 2) is not due to recovery of JA biosynthesis. It would be useful for understanding the MYB21/24 module in stamen development if the links between MYB21/24 and JA biosynthetic genes are elucidated.
DX and SS designed the study; HH, HG, BL, TQ, JT, LX, and SS performed the experiments; HH, HG, BL, TQ, JT, LX, and SS analyzed the data; and HH, HG, and SS wrote the manuscript.
This work was financially supported by the National Natural Science Foundation of China (31670315 and 31570372), the Ministry of Science and Technology of China (2016YFA0500501), and Beijing Nova Program (Z171100001117037).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. John Browse for providing the opr3 mutant and Dr. Jianmin Zhou for providing the vectors used in the LCI assay.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01525/full#supplementary-material
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Campos, M. L., Kang, J. H., and Howe, G. A. (2014). Jasmonate-triggered plant immunity. J. Chem. Ecol. 40, 657–675. doi: 10.1007/s10886-014-0468-3
Chehab, E. W., Kim, S., Savchenko, T., Kliebenstein, D., Dehesh, K., and Braam, J. (2011). Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol. 156, 770–778. doi: 10.1104/pp.111.174169
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., et al. (2011). The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288. doi: 10.1093/mp/ssq073
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Figueroa, P., and Browse, J. (2015). Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 81, 849–860. doi: 10.1111/tpj.12776
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Gimenez-Ibanez, S., Boter, M., Ortigosa, A., Garcia-Casado, G., Chini, A., Lewsey, M. G., et al. (2017). JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 213, 1378–1392. doi: 10.1111/nph.14354
Goossens, J., Fernandez-Calvo, P., Schweizer, F., and Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. doi: 10.1007/s11103-016-0480-9
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Huang, H., Liu, B., Liu, L., and Song, S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68, 1349–1359. doi: 10.1093/jxb/erw495
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Lischweski, S., Muchow, A., Guthorl, D., and Hause, B. (2015). Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 15:229. doi: 10.1186/s12870-015-0615-1
Major, I. T., Yoshida, Y., Campos, M. L., Kapali, G., Xin, X. F., Sugimoto, K., et al. (2017). Regulation of growth-defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 215, 1533–1547. doi: 10.1111/nph.14638
Mandaokar, A., and Browse, J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149, 851–862. doi: 10.1104/pp.108.132597
Mandaokar, A., Thines, B., Shin, B., Lange, B. M., Choi, G., Koo, Y. J., et al. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46, 984–1008. doi: 10.1111/j.1365-313X.2006.02756.x
Nakata, M., Mitsuda, N., Herde, M., Koo, A. J., Moreno, J. E., Suzuki, K., et al. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell 25, 1641–1656. doi: 10.1105/tpc.113.111112
Nakata, M., and Ohme-Takagi, M. (2013). Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal. Behav. 8:e26473. doi: 10.4161/psb.26473
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Park, J. H., Halitschke, R., Kim, H. B., Baldwin, I. T., Feldmann, K. A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. doi: 10.1046/j.1365-313X.2002.01328.x
Qi, T., Huang, H., Song, S., and Xie, D. (2015a). Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27, 1620–1633. doi: 10.1105/tpc.15.00116
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., et al. (2015b). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649. doi: 10.1105/tpc.15.00110
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Reeves, P. H., Ellis, C. M., Ploense, S. E., Wu, M. F., Yadav, V., Tholl, D., et al. (2012). A regulatory network for coordinated flower maturation. PLoS Genet. 8:e1002506. doi: 10.1371/journal.pgen.1002506
Song, S., Huang, H., Gao, H., Wang, J., Wu, D., Liu, X., et al. (2014a). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26, 263–279. doi: 10.1105/tpc.113.120394
Song, S., Qi, T., Fan, M., Zhang, X., Gao, H., Huang, H., et al. (2013a). The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 9:e1003653. doi: 10.1371/journal.pgen.1003653
Song, S., Qi, T., Huang, H., and Xie, D. (2013b). Regulation of stamen development by coordinated actions of jasmonate, auxin and gibberellin in Arabidopsis. Mol. Plant 6, 1065–1073. doi: 10.1093/mp/sst054
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Song, S., Qi, T., Wasternack, C., and Xie, D. (2014b). Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 21, 112–119. doi: 10.1016/j.pbi.2014.07.005
Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630. doi: 10.1073/pnas.190264497
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Wasternack, C., and Song, S. (2017). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yan, J., Li, S., Gu, M., Yao, R., Li, Y., Chen, J., et al. (2016). Endogenous bioactive jasmonate is composed of a set of (+)-7-iso-JA-amino acid conjugates. Plant Physiol. 172, 2154–2164. doi: 10.1104/pp.16.00906
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Yan, Y. X., Stolz, S., Chetelat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Yang, X. Y., Li, J. G., Pei, M., Gu, H., Chen, Z. L., and Qu, L. J. (2007). Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Rep. 26, 219–228. doi: 10.1007/s00299-006-0229-z
Keywords: fertility, interaction, JAZs, MYB24, OPR3, stamen
Citation: Huang H, Gao H, Liu B, Qi T, Tong J, Xiao L, Xie D and Song S (2017) Arabidopsis MYB24 Regulates Jasmonate-Mediated Stamen Development. Front. Plant Sci. 8:1525. doi: 10.3389/fpls.2017.01525
Received: 30 June 2017; Accepted: 21 August 2017;
Published: 05 September 2017.
Edited by:
Chi-Kuang Wen, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Yang Do Choi, Seoul National University, South KoreaCopyright © 2017 Huang, Gao, Liu, Qi, Tong, Xiao, Xie and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daoxin Xie, ZGFveGlubGFiQHRzaW5naHVhLmVkdS5jbg== Susheng Song, c29uZ3NzbGFiQDE2My5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.