- 1Faculty of Applied Biological Sciences, Gifu University, Gifu, Japan
- 2Faculty of Life Science and Bioinformatics, Assam University, Silchar, India
Aluminum (Al) rhizotoxicity is one of the major environmental stresses that decrease global food production. Clarifying the molecular mechanisms underlying Al tolerance may contribute to the breeding of Al-tolerant crops. Recent studies identified various Al-tolerance genes. The expression of these genes is inducible by Al. Studies of the major Arabidopsis thaliana Al-tolerance gene, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 (AtALMT1), which encodes an Al-activated malate transporter, revealed that the Al-inducible expression is regulated by a SENSITIVE TO PROTON RHIXOTOXICITY 1 (STOP1) zinc-finger transcription factor. This system, which involves STOP1 and organic acid transporters, is conserved in diverse plant species. The expression of AtALMT1 is also upregulated by several phytohormones and hydrogen peroxide, suggesting there is crosstalk among the signals involved in the transcriptional regulation of AtALMT1. Additionally, phytohormones and reactive oxygen species (ROS) activate various transcriptional responses, including the expression of genes related to increased Al tolerance or the suppression of root growth under Al stress conditions. For example, Al suppressed root growth due to abnormal accumulation of auxin and cytokinin. It activates transcription of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 and other phytohormone responsive genes in distal transition zone, which causes suppression of root elongation. On the other hand, overexpression of Al inducible genes for ROS-detoxifying enzymes such as GLUTATHIONE–S-TRANSFERASE, PEROXIDASE, SUPEROXIDE DISMUTASE enhances Al resistance in several plant species. We herein summarize the complex transcriptional regulation of an Al-inducible genes affected by STOP1, phytohormones, and ROS.
Introduction
The insoluble oxidized form of aluminum (Al) in soil clay becomes soluble in acidic soil solutions at pH < 5.5 (Kinraide and Parker, 1989). Among the various Al ion species, Al3+ is the most toxic form. This ion can exist in naturally acidic soils, and is toxic at sub-micromolar concentrations (Kinraide et al., 1985; Kinraide, 2003). Crop growth is severely suppressed in Al-solubilizing soils. This is primarily because of the root growth inhibition due to Al rhizotoxicity (i.e., cytotoxic and genotoxic effects of Al3+), which restricts root tip cell elongation and division (Nezames et al., 2012). Improving crop tolerance to Al rhizotoxicity is one of the most important targets for increasing crop production in regions with acidic soil, which are dominant in the sub-tropical and tropical regions of many highly populated developing countries. Recent studies revealed that the transcriptional regulation of stress-tolerance genes is important for Al tolerance (Kochian et al., 2015).
Differences in the expression levels of Al-tolerance genes influence the extent of Al tolerance in several important crops such as wheat (Sasaki et al., 2004), rice (Yamaji et al., 2009), and sorghum (Magalhaes et al., 2007). Additionally, an analysis of expression level polymorphisms among Arabidopsis thaliana accessions identified several novel genes that regulate Al tolerance (Kusunoki et al., 2017). In contrast, the identification and characterization of transcription factors and regulatory proteins (e.g., protein kinases) revealed that the transcription of Al-tolerance genes is likely regulated by a very complex mechanism. This mechanism involves repressors and activators, co-regulation with other Al-tolerance genes (e.g., Delhaize et al., 2012; Tokizawa et al., 2015), and crosstalk with mechanisms controlling other stress responses. A more comprehensive characterization of these complex regulatory mechanisms may be useful for accelerating the breeding of Al-tolerant crops.
The different types of Al-tolerance mechanisms in various crops involve the exclusion of Al, an internal tolerance mechanism, and recovery from Al-induced damages (Taylor, 1987, 1988, 1991; Kochian, 1995). The excretion of Al-detoxifying ligands, such as organic acids (OAs) and Pi, to the apoplast or rhizosphere is the most common feature of Al-exclusion mechanisms in several crop plants. The OA types differ among plant species (Ma et al., 1998; Zheng et al., 1998; Wenzl et al., 2001; Kobayashi et al., 2005). The internal tolerance mechanisms involve the sequestration of Al into vacuoles and the detoxification of Al by chelation. Meanwhile, recovery from Al-induced damages is mediated by the detoxification of the reactive oxygen species (ROS) produced following exposures to excessive amounts of Al. By combining these mechanisms with the transcriptional regulation of Al-tolerance genes, plants can protect the most sensitive part of the root apex from Al rhizotoxicity. The expression of genes encoding OA transporters is inducible by Al. These genes include ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 (ALMT1) and members of the multidrug and toxic compound extrusion (MATE) citrate transporter gene family in A. thaliana (Sawaki et al., 2009) and tobacco (Nicotiana tabacum; Ohyama et al., 2013). A previous study of A. thaliana detected a typically complex regulation of the Al-inducible expression of AtALMT1, which encodes a protein that mediates protein phosphorylation/dephosphorylation processes (Kobayashi et al., 2007). Other studies concluded that AtALMT1 expression is regulated by transcription factors (Ding et al., 2013; Tokizawa et al., 2015) and phytohormone-signaling networks (e.g., jasmonate and ethylene; Kobayashi et al., 2013). The expression of AtALMT1 is tightly regulated by the STOP1 (sensitive to proton rhizotoxicity 1; Iuchi et al., 2007) zinc-finger transcription factor, and is co-regulated with several other Al- and proton-tolerance genes (Sawaki et al., 2009). This complex regulation may account for the pleiotropic roles of ALMT1 (Koyama et al., 2015).
Reactive oxygen species can cause irreversible damage to growing tissues, in part because of Al-induced metabolic changes. This damage induces the transcription of genes encoding ROS-detoxifying enzymes (e.g., glutathione S-transferase, peroxidase, alternative oxidase, and malate dehydrogenase). The overexpression of these genes usually confers Al tolerance, which suggests that plants activate Al-tolerance mechanisms to recover from Al-induced ROS damages. Additionally, recent developments in next-generation sequencing technologies have resulted in the identification of several novel Al-inducible genes that influence the level of Al tolerance (Kusunoki et al., 2017). Overall, we speculate that transcriptional regulation is critical for mechanisms mediating the Al tolerance of crops (Figure 1). We herein summarize our current understanding of the transcriptional regulation of Al-tolerance genes and its relevance to the breeding of Al-tolerant varieties.
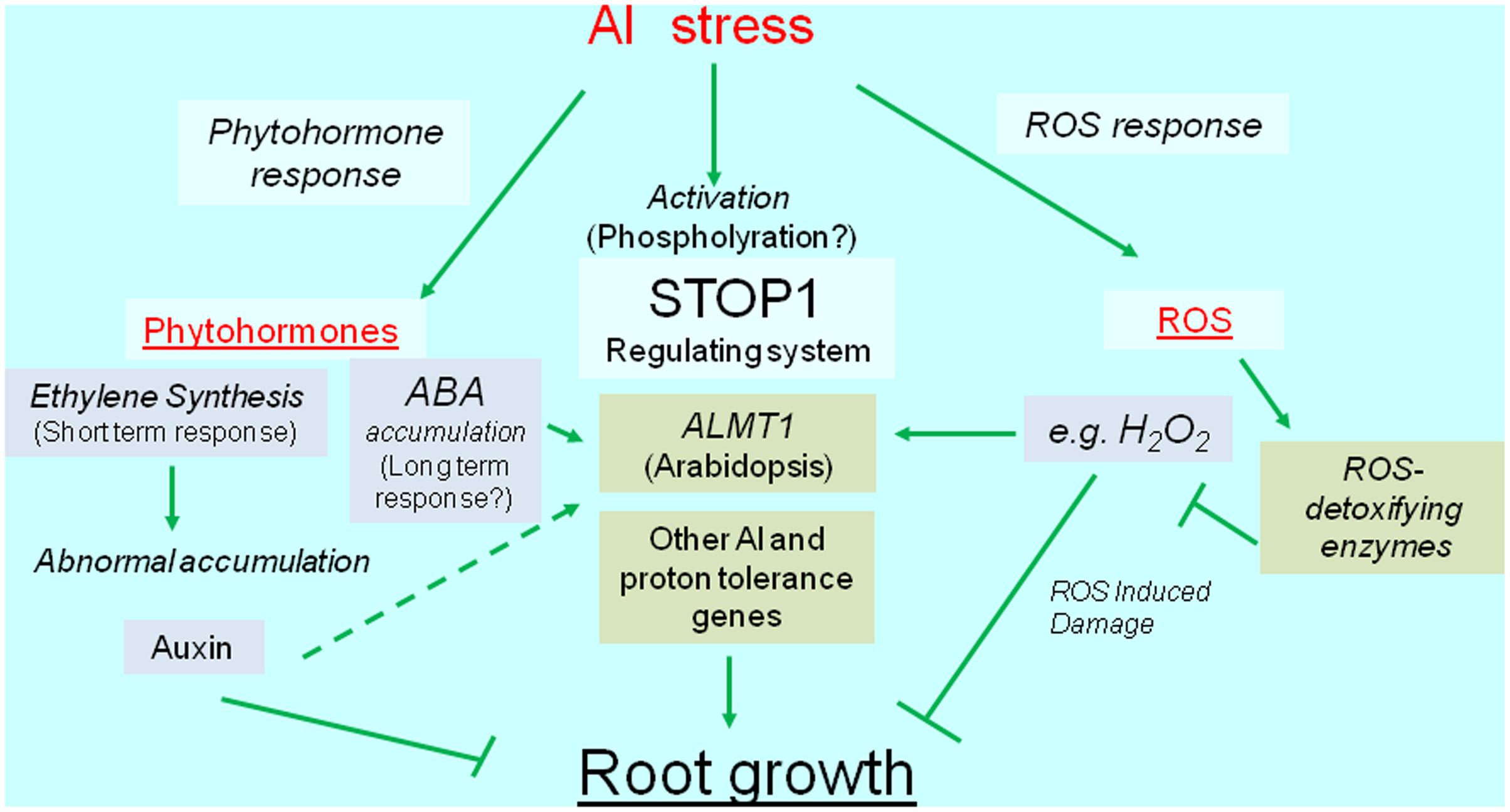
FIGURE 1. Diagrammatic representation of transcriptional regulation of Al tolerance genes. Al stress activates transcription of Al tolerance genes (green box), which are regulated the transcription via phytohormone, SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1), and reactive oxygen species (ROS) mediated pathways. The major Al tolerance gene, such as ALUMINUM ACTIVATED MALATE TRANSPORTER1 (ALMT1) -type malate transporter, is regulated the transcription by STOP1-like protein, but it’s likely activated by phytohormone(s) and ROS. ROS induces various genes including ROS detoxifying enzymes.
Transcriptional Regulation of Al-Tolerance Genes by the Stop1 Protein
The SENSITIVE TO PROTON RHIZOTOXICITY (STOP1) gene was discovered based on the positional cloning of an A. thaliana mutant with short roots in an acidic medium (Iuchi et al., 2007). The mutant was also hypersensitive to Al because of the suppressed expression of AtALMT1 (Iuchi et al., 2007). In A. thaliana, the Al-inducible expression of AtALMT1 is critical for Al tolerance (Hoekenga et al., 2006), while the expression of AtALMT1 is completely suppressed in the stop1 mutant (Iuchi et al., 2007). STOP1 contains four zinc-finger domains, suggesting it is a critical transcription factor regulating the expression of ALMT1 and proton-tolerance genes. Additionally, AtALMT1 is co-regulated with genes for proton tolerance under the control of STOP1, which is a protein that is essential for AtALMT1 transcription. Functional orthologs of AtSTOP1 are key regulators of Al-tolerance genes in various plant species. In this section, we describe our current understanding of the STOP1-like protein, including its effects on Al-inducible expression of Al-tolerance genes.
STOP1-Regulated Genes in Arabidopsis thaliana
A systems biology study uncovered the co-regulation of multiple Al- and proton-tolerance genes by AtSTOP1 in A. thaliana, including ALUMINUM SENSITIVE 3 (ALS3; possibly encodes UDP glucose transporter, homolog of rice STAR2) (i.e., Al tolerance) and various genes affecting ion homeostasis [e.g., CBL-INTERACTING PROTEIN KINASE 23 (CIPK23), which phosphorylates ARABIDOPSIS POTASSIUM TRANSPORTER1; Xu et al., 2006], pH-regulated metabolic activities (e.g., GABA-shunt and biochemical pH stat pathways), and cell wall stabilization (i.e., proton tolerance) (Sawaki et al., 2009). Additionally, Al-inducible expression of MULTIDRUG AND TOXIC COMPOUND EXTRUSION (AtMATE; encoding a citrate transporter) is regulated by STOP1 (Liu et al., 2009). These results indicate that STOP1 is a key regulatory transcription factor for Al and proton tolerance.
The Al- and proton-tolerance genes are differentially regulated by STOP1. In A. thaliana, STOP2 is a unique ortholog of STOP1, with a shorter C-terminus. It rescues the proton tolerance in the stop1 mutant by activating the expression of several proton-tolerance genes (e.g., CIPK23 encoding regulator of K+ and NO3- transporters, polygalacturonase inhibitor protein 1, stabilizing pectin at low pH, and others; Kobayashi et al., 2014). Although STOP2 expression is regulated by STOP1, it does not recover Al tolerance in the stop1 mutant because of the very limited recovery of AtALMT1 and ALS3 expression. This suggests that STOP2 only enhances the proton-tolerance mechanism controlled by STOP1. Additionally, an in planta complementation of STOP1-like proteins usually fails to recover Al tolerance in the A. thaliana stop1 mutant, while it confers proton tolerance (e.g., Ohyama et al., 2013). These observations suggest that the transcriptional activation of major Al-tolerance genes by STOP1 (e.g., AtALMT1 and ALS3) requires additional mechanisms (e.g., co-activators or post-translational mechanisms), which are sensitive to the STOP1 protein structure (Figure 2).
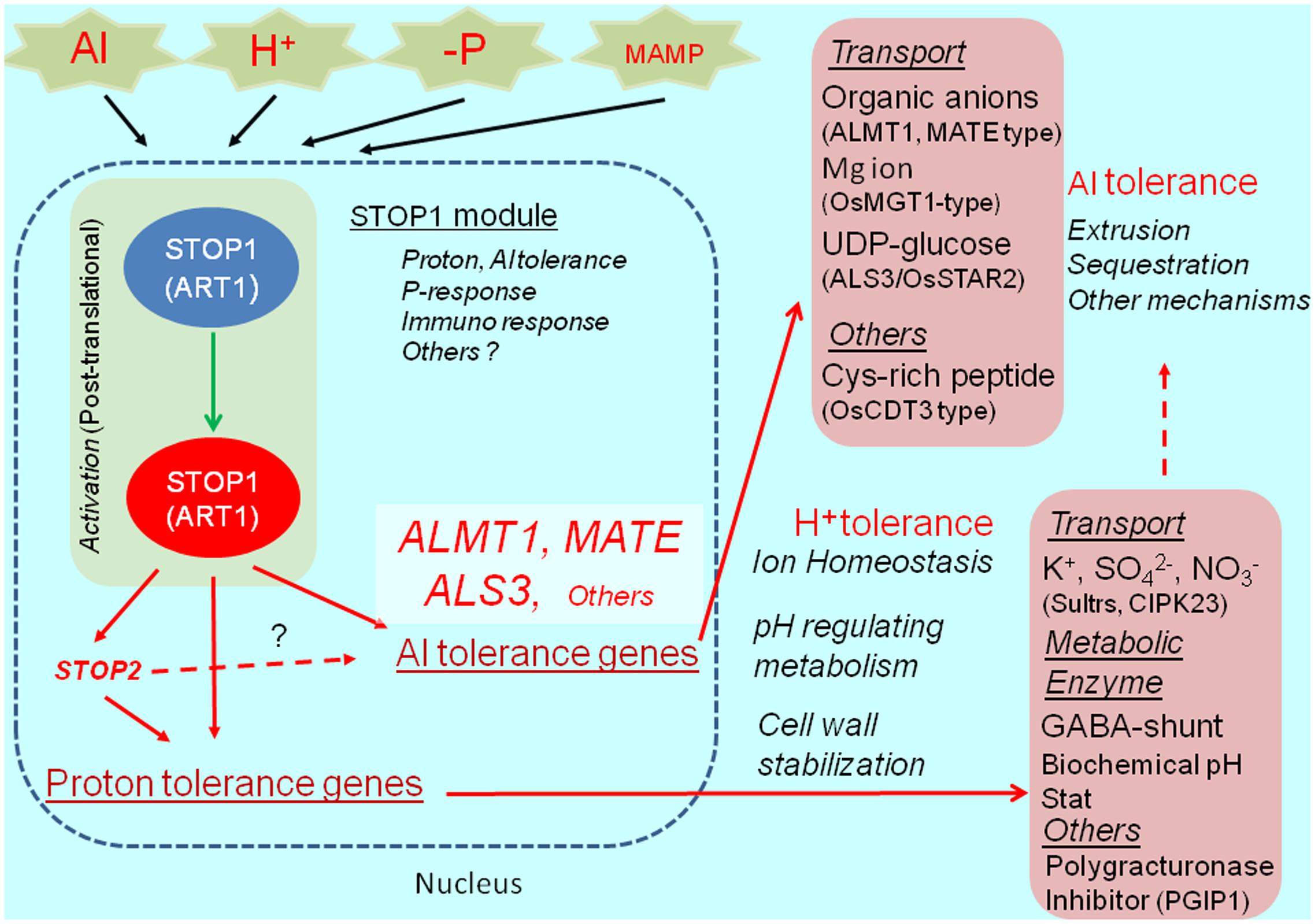
FIGURE 2. STOP1 module and ALMT1 regulation. The SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1) module, which is consisted of STOP1-like protein (in Arabidopsis, STOP1; in rice ALUMINUM RESISTANCE TRANSCRIPTION FACTOR 1), OA transporters ALUMINUM ACTIVATED MALATE TRANSPORTER1 (ALMT1), malate transporter; MULTIDRUG AND TOXIC COMPOUNDS EXTRUSION (MATE); citrate transporter, and ALUMINUM SENSITIVE 3 (ALS3), are conserved in wide range of plant species. The module includes various Al tolerance genes and possibly H+ tolerance genes (pink box), which play critical role in Al detoxification, ion homeostasis (transport and metabolisms), cell wall stabilization and other roles for tolerance. In Arabidopsis, STOP1 regulates STOP2 expression, while STOP2 only regulates expression of H+ tolerance genes. STOP1-regulating gene, such as ALMT1, is activated by various environmental stimuli (Al, H+ -P and MAMP, Microbe Associated Molecular Pattern).
Conservation of the STOP1-Regulated System in Various Plant Species
The ALUMINUM-RESISTANCE TRANSCRIPTION FACTOR1 (OsART1) gene is a rice ortholog of AtSTOP1 that was identified during the positional cloning of an Al-sensitive mutant (Yamaji et al., 2009). The art1 mutant exhibits repressed expression of STAR2 [(rice homolog of AtALS3 (Larsen et al., 2005; Yamaji et al., 2009)] and Oryza sativa FERRIC REDUCTASE DEFECTIVE 4 (OsFRDL4) (rice homolog of AtMATE; Yamaji et al., 2009). Additionally, a magnesium transporter (Chen et al., 2012) and a plasma-membrane–localized cysteine-rich peptide CDT3 (Xia et al., 2013; see also in ROS section) are critical for Al resistance in rice, and the expression of the corresponding genes is regulated by the ART1 transcription factor. These observations strongly suggest that a STOP1-like protein regulates the expression of multiple Al-tolerance genes in various plant species. In fact, functional orthologs of STOP1 (hereafter called STOP1-like proteins) have been identified in a wide range of plant species, including dicotyledons and monocotyledons, tree and grass species, Brassicaceae species, legumes, and mosses (Ohyama et al., 2013; Sawaki et al., 2014; Fan et al., 2015; Wang et al., 2017). A Physcomitrella patens subsp. patens knock-out of STOP1 is reportedly more sensitive to Al than the wild-type (WT) plants. These findings indicate that STOP1-like protein(s), and regulating core Al tolerance gens (STOP1-module), are conserved among land plant species.
The suppression of NtSTOP1 expression (NtSTOP1-KD) based on RNA interference represses the Al tolerance of tobacco plants (N. tabacum) because the Al-inducible expression of NtMATE1 is inhibited (Ohyama et al., 2013). The NtMATE1 expression level in NtSTOP1-KD plants is more than 100-fold lower than in WT plants. Additionally, Al accumulates in the root tips of the NtSTOP1-KD plants (see Figure 4A in Ohyama et al., 2013). This complete suppression of NtMATE expression is very similar to the lack of AtAMLT1 expression in the stop1 A. thaliana mutant. In contrast, the Al-inducible expression of NtALS3 is regulated by NtSTOP1 (Ohyama et al., 2013). Thus, the STOP1-like protein tightly regulates the genes encoding OA transporters and ALS3 orthologs, with diverse transporters likely being regulated in different plant species. The Al-inducible transcription of genes encoding OA-transporters involves a STOP1-binding process. A yeast one-hybrid assay revealed that VuSTOP1 binds to the promoter of VuMATE1 to regulate expression (Fan et al., 2015). Gel-shift assays involving OsART1 confirmed that GGN(T/g/a/C)V(C/A/g)S(C/G) (simply, GGNVS-consensus) is the canonical OsART1-binding sequence in the promoters of 29 of the 31 genes regulated by ART1 (e.g., OsFRDL4) (Tsutsui et al., 2011). In contrast, the AtALMT1 promoter carries a STOP1-binding site that is longer (about 15 bp) than the corresponding rice sequence. This binding site is suitable for the four zinc-finger domains of the STOP1-like protein (Tokizawa et al., 2015). Several studies concluded that the number of STOP1-binding sites in the promoter influences the OA transporter gene expression-level differences among cultivars (Chen et al., 2013). A Holcus lanatus accession adapted to an acid plot was observed to carry several GGN(T/g/a/C)V(C/A/g)S(C/G) sequences in the ALMT1 promoter (Chen et al., 2013), while wheat near-isogenic line ET8, which exhibits upregulated TaALMT1 expression, carries three sets of the STOP1-binding site identified in the Al-sensitive near-isogenic line ES8 (Tokizawa et al., 2015) (Figure 2).
Complexity of the STOP1-Regulated Expression of OA Transporter Genes, and the Associated Pleiotropic Functions
Recent studies have confirmed that the mechanism underlying the STOP1 regulation of the expression of OA transporter genes is complex because several other transcription factors are involved. The Al-inducible AtALMT1 expression is associated with the upregulated expression of calmodulin-binding transcription activator 2 (Tokizawa et al., 2015) and the downregulated expression of AtWRKY46, which encodes a repressor of AtALMT1 expression (Ding et al., 2013). abscisic acid (ABA) induce the AtALMT1 expression (see the below; Phytohormone section), while the promoter deletion analysis identified that differential regulation of transcription by ABA and Al in short term (6 h, Kobayashi et al., 2013). On the other hand, AtWRKY46 reported as repressor of AtALMT1 (Ding et al., 2013) and ABA repress the expression of AtWRKY46 (Ding et al., 2015). Furthermore, analyses of the AtALMT1 promoter (i.e., bioinformatics based promoter-GUS reporter assay) identified multiple cis-elements responsible for the short- and long-term Al-inducible expression (Tokizawa et al., 2015). The cbl1 mutant exposed to Al stress conditions reportedly exhibits inhibited root growth, decreased malate secretion, and increased accumulation of Al in the root tips (Ligaba-Osena et al., 2017). This suggests that the ARABIDOPSIS THALIANA CALCINEURIN B-LIKE PROTEIN 1 (AtCBL1) gene product helps activate STOP1 in A. thaliana. Because CBL1 activates several regulatory protein kinases, it may also be involved in the protein phosphorylation that activates AtALMT1 expression (Kobayashi et al., 2007). However, as described above, the in planta complementation of STOP1-like proteins in the A. thaliana stop1 mutant often fail to induce AtALMT1 transcription, while activating the expression of proton-tolerance genes. This also applies to the functional ortholog, NtSTOP1, which can activate NtMATE expression in tobacco (Ohyama et al., 2013). The reasons for these observations may be related to the differential structures of the N- and C-termini, which are specifically targeted for post-translational modifications or for interactions with a co-activator of AtALMT1 expression.
Although the relevant regulatory mechanisms have not been fully characterized, STOP1 and ALMT1 have pleiotropic roles related to adaptations to other stressors. Rudrappa et al. (2008) reported that infections to the aerial parts of A. thaliana plants by pathogenic bacteria upregulate AtALMT1 expression and malate excretion in the roots. The excreted malate recruits beneficial rhizobacteria that stimulate the A. thaliana immune system. The molecular mechanism underlying the long-distance signaling from the shoots to the roots has not been elucidated. However, AtALMT1 expression is induced in roots treated with the FLG22 peptide (i.e., conserved peptide in the bacterial flagella), which likely involves the FLG22 receptor FLAGELLIN-SENSITIVE 2, a type of MAMP (microbe associated molecular pattern) (Kobayashi et al., 2013). Additionally, a recent study determined that STOP1 and ALMT1 trigger a malate-exudate–dependent Fe relocation in the root apical meristem, which is essential for the reprogramming of root growth under low-Pi conditions (Balzergue et al., 2017; Macias et al., 2017). The fact that AtALMT1 expression is dependent on STOP1 binding to the promoter suggests that STOP1 is a HUB molecule of these multiple responses. Al induces malate excretion via STOP1/ALMT1 activities, with pleiotropic consequences for stress tolerance. Further research is required to uncover the molecular mechanisms related to the activation of STOP1.
Phytohormones Involved in Al-Stress Responses and their Role in the Transcription of Al- and Proton-Tolerance Genes
Most phytohormones are important for root development and elongation (see reviews; Benkova and Hejatko, 2009; Jung and McCouch, 2013). Additionally, phytohormones help mediate biotic and abiotic stress responses (e.g., by activating the transcription of various genes). Many studies have revealed that several phytohormones affect the Al-induced inhibition of root growth. The abnormal accumulation of phytohormones disrupts normal root growth. Additionally, the accumulated phytohormones modify the transcriptome, and affect the activation of some Al-tolerance genes. In this section, we describe the relationships between phytohormones and plant responses to Al stress by categorizing the phytohormones according to whether they negatively or positively affect root growth under Al stress conditions (Figure 3).
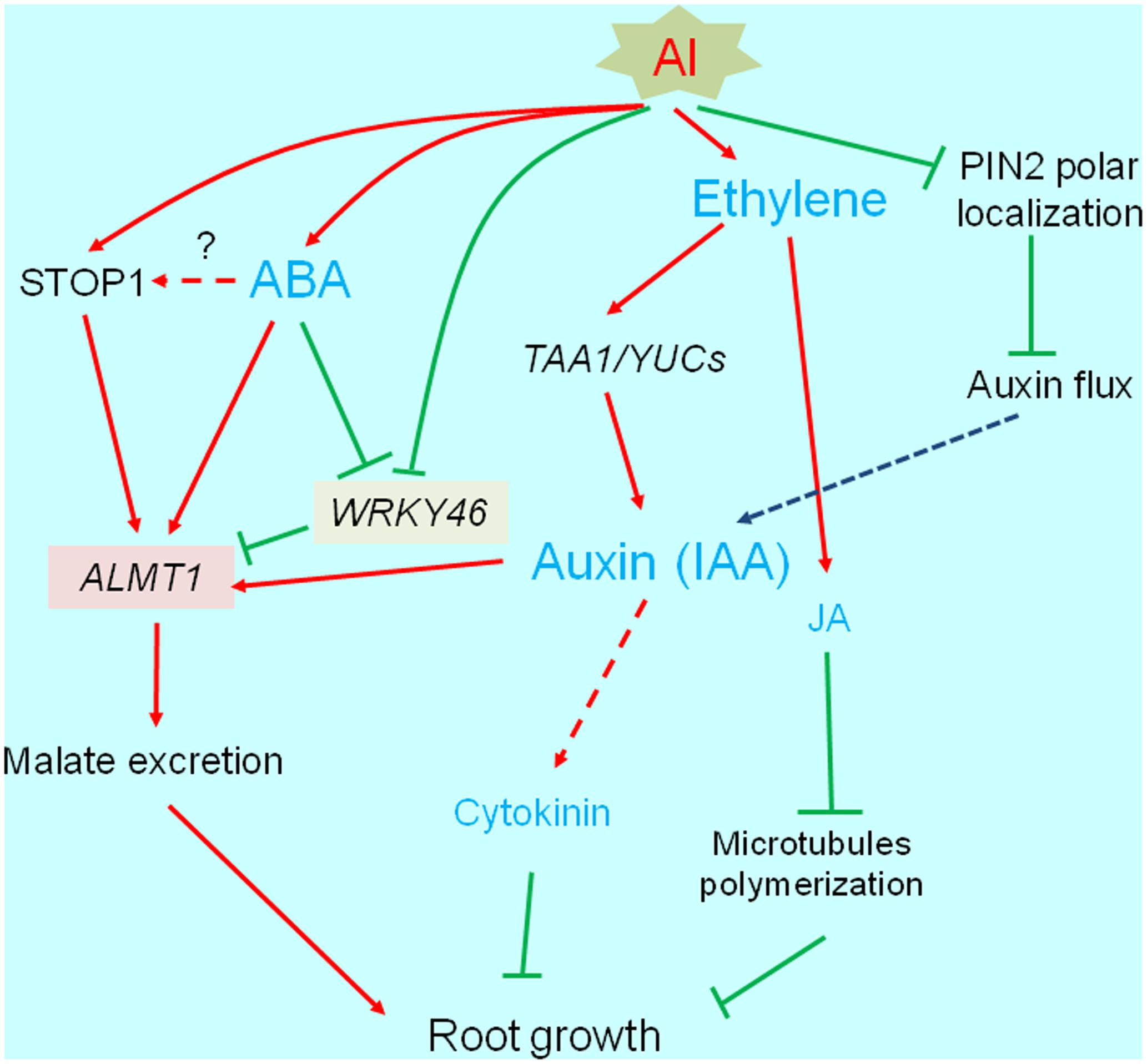
FIGURE 3. Transcriptional regulation of phytohormone responsive root growth under Al stress. Al induces root growth inhibition, which is associated with increased ethylene synthesis followed by accumulation of auxin (IAA) and jasmonic acid (JA). These events occur within short-term range (3 h), which is associated with PIN-FORMED (PIN2). Finally, IAA and JA activated pathways are likely blocking root growth. IAA and abscisic acid activate ALUMINUM ACTIVATED MALATE TRANSPORTER1 (ALMT1) expression, which may increase Al tolerance.
Negative Effects of Phytohormones on Root Growth under Al Stress Conditions and the Consequences for Transcriptional Regulation
Auxin is a key regulatory phytohormone for root development and elongation. The auxin gradient in the root apex is formed by a series of auxin polar transporters such as the PIN-FORMED (PIN) family proteins (Blilou et al., 2005; Leyser, 2005; Baluska et al., 2010; Takatsuka and Umeda, 2014). The root apex auxin gradient along with the high and low auxin concentrations in the meristem and elongation zone, respectively, are essential for continuous root growth. Normal root growth is disrupted by Al stress conditions, especially in the distal transition zone (DST) (i.e., 1–3 mm behind the root tip, corresponding to the transition from cell division to cell elongation; Kollmeier et al., 2000). Moreover, Al interferes with the plasma membrane localization of PIN2, which regulates the auxin flux from the root tip to the elongation zone (Shen et al., 2008). These results imply that the disruption of PIN-mediated auxin flow is one of the causes of inhibited root growth due to Al toxicity. This possibility is supported in A. thaliana and rice by the observed changes in the Al-sensitivity of mutants as well as the phenotypes of transgenic plants overexpressing the genes encoding certain PIN proteins (Sun et al., 2010; Wu et al., 2014, 2015). Yang et al. (2014) characterized the mechanisms regulating the indole-3-acetic acid (IAA)-mediated inhibition of root growth under Al stress conditions. They determined that the Al-induced accumulation of IAA in the DST of A. thaliana plants is caused by TAA1-mediated auxin biosynthesis, which is concomitant with activation of transcription of other genes for IAA synthesizing proteins (e.g., YUCCA; Liu et al., 2016). The inhibition of root elongation by accumulating IAA occurs simultaneously with the increased production of ethylene in response to Al stress. This increase in ethylene contents suppresses the expression of cell-wall modification genes mediated by auxin-response factors 10 and 16 (Yang et al., 2014). Ethylene is a key regulator of auxin biosynthesis and basipetal auxin transport in the root apex (Ruzicka et al., 2007; Stepanova et al., 2007; Swarup et al., 2007; Muday et al., 2012). Thus, it can inhibit root growth in Al-stressed plants. In fact, Al induces rapid and considerable increases in ethylene levels in A. thaliana (Sun et al., 2010), Lotus japonicus (Sun et al., 2007), and Phaseolus vulgaris (Massot et al., 2002). This increase is due to the upregulated expression of genes encoding ethylene biosynthesis enzymes, including ACC-synthase and ACC-oxidase (Sun et al., 2010). An in planta GFP reporter assay involving the auxin-responsive promoter (DR5) confirmed that Al-inducible IAA accumulation can be suppressed by the ethylene synthesis inhibitor aminoethoxyvinylglycine (Yang et al., 2014). This inhibition is induced after a 1.5-h exposure to Al, which follows a very quick Al inhibition (initiated at 5 min), possibly via the inhibition of cell-wall loosening resulting from the binding of Al (Kopittke et al., 2015). Yang et al. (2017a,b) also demonstrated that Al-induced upregulation of ethylene synthesis suppresses root growth by mediating the jasmonic acid (JA)-responsive and cytokinin-responsive pathways. The accumulation of JA-isoleucine (i.e., active form of JA) in the root tips is induced by Al, but is suppressed by aminoethoxyvinylglycine. The accumulated JA-isoleucine downregulates the expression of microtubule-associated genes, resulting in inhibited root growth (Yang et al., 2014).
Indole-3-acetic acid can activate the transcription of Al-tolerance genes. For example, the application of exogenous IAA induces considerable increases in AtALMT1 expression levels, as well as slight increases in AtMATE expression levels (Kobayashi et al., 2013). Additionally, an acidic external environment (i.e., pH approximately 4.5) leads to transcriptome-level changes in A. thaliana roots that resemble the transcriptomic changes induced by short-term auxin treatments. Because most Al-inducible genes are also inducible by acidic conditions (Sawaki et al., 2009), the cross-talk between auxin and low pH/Al responses may be important for the regulation of Al- and proton-tolerance genes.
Positive Effects of ABA on Root Growth under Al Stress Conditions and the Consequences for Transcriptional Regulation
Previous studies have examined the endogenous accumulation of ABA in response to Al in buckwheat (Reyna-Llorens et al., 2015), soybean (Shen et al., 2004), and barley (Kasai et al., 1993). The application of exogenous ABA can activate the release of OAs from the roots of some plants (Ma et al., 2001; Shen et al., 2004). Hou et al. (2010) reported that Al-induced soybean root growth inhibition is alleviated in plants treated with exogenous ABA. This alleviation is suppressed by the addition of an ABA biosynthesis inhibitor (e.g., furidone). These results imply that ABA modulates Al-tolerance mechanisms, possibly through the transcriptional regulation of Al-tolerance genes. In fact, AtALMT1 and ALS3 expression levels are upregulated by ABA (Kobayashi et al., 2013). Additionally, the AtALMT1 promoter differentially regulates Al and ABA responses (Kobayashi et al., 2013). Furthermore, ABA activates the release of malate from roots. These results indicate that ABA can activate ALMT1 expression as well as malate transport activity, suggesting that Al-induced ABA accumulation induces OA transporter gene expression and activates the resulting protein (Figure 3). These processes may have important functions in the Al-tolerance mechanism of plants.
A comparative A. thaliana microarray revealed that the expression of several ABA-responsive genes (e.g., DREB1A and DREB1A-regulated genes) is induced by Al treatments (Sawaki et al., 2016). Although the consequences of the upregulated expression of DREB1A-regulated genes have not been studied in terms of Al tolerance, they may include increased drought tolerance that can overcome the effects of Al-induced root growth inhibition.
Reactive Oxygen Species-Mediated Transcriptional Regulation of Al-Tolerance Genes Under Al Stress Conditions
Exposure to Al stress conditions alters the cellular ROS levels in different root regions. ROS, including hydrogen peroxide (H2O2), superoxide (O2-), and hydroxyl radicals (•OH), adversely affect root cells (e.g., apoptosis and damages to various molecules) and influence signaling pathways for essential processes such as growth and stress adaptations (e.g., activation of Al-tolerance gene expression). In this section, we describe ROS production under Al stress conditions as well as the transcriptional regulation of plant ROS responses to mitigate ROS-induced damages (Figure 4).
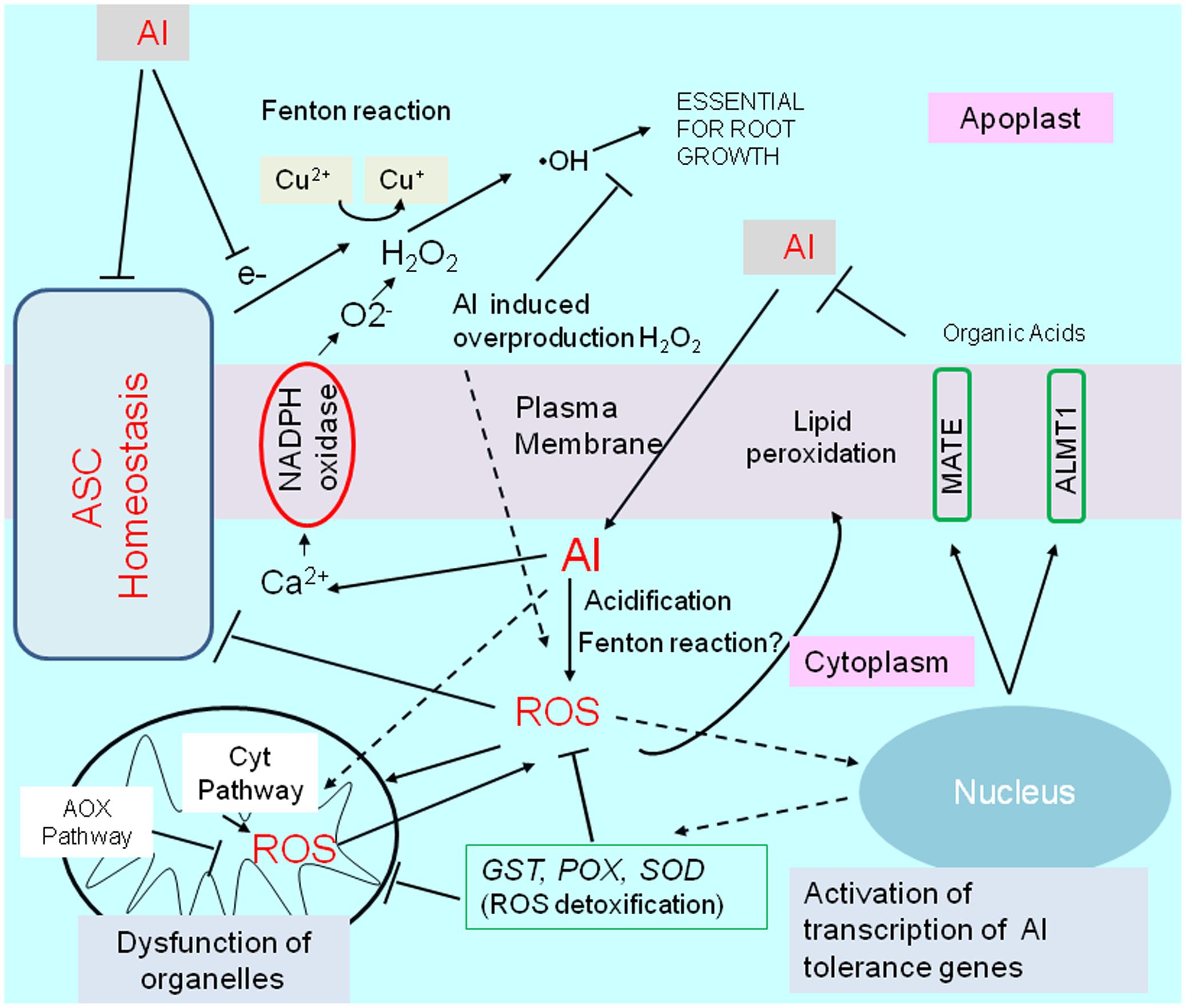
FIGURE 4. Schematic representation of aluminum-induced reactive oxygen species (ROS) stress, and activation of tolerance mechanisms by ROS. Aluminum disturbs an essential fenton reaction in the cell wall for root growth, but is induced ROS production by (1) increases cytosolic Ca2+ concentrations, which triggers activation of NADPH-oxidase, (2) inducing fenton reaction in cytosol, (3) cytosol acidification, and by other mechanisms. Disturbance of ascorbate (ASC) homeostasis enhance abnormal fenton reaction in apoplast, and ROS production in the cytoplasm. Aluminum (or by generated ROS) shifts mitochondrial respiration to an alternative pathway (AOX pathway) from Cyt pathway (cytochrome pathway), which is likely important to protect mitochondrial function. ROS activates transcription of several genes for ROS detoxifying enzymes such as GLUTATHIONE–S-TRANSFERASE (GST); PEROXIDASE (POX); SUPEROXIDE DISMUTASE (SOD), which can confer Al resistance in some plant species by overexpression. ROS, in particular, H2O2 is known as a inducer of an Al tolerance genes such as ALUMINUM ACTIVATED MALATE TRANSPORTER1 (ALMT1) and MULTIDRUG AND TOXIC COMPOUNDS EXTRUSION (MATE) in Arabidopsis.
Reactive Oxygen Species Accumulation and Toxicity under Al Stress Conditions
The enhanced ROS production induced by Al stress conditions is mediated by several mechanisms. In the apoplast, activated plasma membrane NADPH oxidase is the primary source of ROS in Al-stressed plants. The Al transiently increases the cytoplasmic Ca2+ concentration (Bhuja et al., 2004), which activates the plasma membrane NADPH oxidase, leading to the production of O2- (Sagi and Fluhr, 2001) and H2O2 in the apoplast. Under normal conditions, the Cu-mediated Fenton reaction is critical for generating ⋅OH and loosening the cell wall, which is essential for cell elongation, because it can cleave the sugar–sugar bond in cell wall polysaccharides. Normal Fenton reaction activity is regulated by the coupling of Cu2+ ↔ Cu+ of the cell-wall–localizing blue-copper–binding proteins and ascorbate (ASC) ↔ monodehydroascorbate (MDHA). However, Al can activate the Fenton reaction by coupling with other metals, including Cu, leading to the excessive formation of the •OH radical (Mujika et al., 2011; Ruipérez et al., 2012). In contrast, a shortage of ASC in the apoplast, which may result from the excessive conversion of ASC to oxalate (Al-responsive gene; Hamel et al., 1998), leads to the accumulation of H2O2. In fact, Al treatments generate diverse ROS in the apoplast (Maltais and Houde, 2002), while the maintenance of high levels of ascorbate is characteristic of Al tolerance in tobacco and rice plants (Devi et al., 2003; Guo et al., 2005). A comparison of wheat transcriptomes revealed that Al-tolerant varieties exhibited considerably upregulated expression of genes encoding proteins belonging to such a system, including enzymes involved in ASC-homeostasis and cell-wall–loosening proteins associated with the Fenton reaction (Houde and Diallo, 2008).
Plants treated with Al usually accumulate ROS in the symplast (e.g., accumulation of H2O2 in Al-sensitive varieties; Kobayashi et al., 2005), leading to the peroxidation of lipids in the plasma membrane and the production of dysfunctional organelles (Yamamoto et al., 2002). In tobacco, Al stress is associated with swollen/dysfunctional mitochondria, fragmented vacuoles, and pre-apoptotic nuclear structures (Yamamoto et al., 2002; Panda et al., 2008), which may ultimately induce the mitochondrial pathway to initiate programmed cell death (Huang et al., 2014). The manner in which Al induces the production of excess ROS in the cytoplasm (symplast) is complex. The H2O2 accumulating in the apoplast due to NADPH oxidase or the Fenton reaction may be introduced to the cytosol (Bienert et al., 2006). Additionally, Al can quickly cross the plasma membrane (Taylor et al., 2000) and activate the Fenton reaction in the cytoplasm. These mechanisms increase the cytosolic ROS concentration. However, ROS production in the cytosol and mitochondria may also be enhanced by the acidification of the cytosol by Al (Moseyko and Feldman, 2001). This acidification disrupts the redox metabolic activities in the cytosol by inactivating -SH residues (pH < 7) and destabilizing NAD+. It also enhances ROS toxicity in the cytoplasm, and inhibits the production of excess ROS in mitochondria. Disrupting the redox molecules (e.g., -SH, NAD+) in the cytosol interferes with the regulation of NAD(P)H/NAD(P)+ contents in other cellular components, including mitochondria. Moreover, the generation of toxic O2- in mitochondria is enhanced by a relatively low ATP demand or a high NADH/NAD+ ratio (Murphy, 2009). Under the crisis, tolerant cultivars synthesize cysteine-rich proteins to reduce ROS production (Hamel et al., 1998), while one of such proteins had been identified as an Al tolerance gene regulated the transcription by STOP1-like protein (ART1) in rice (Xia et al., 2013).
Inducible Expression of ROS-Mediated Al-Tolerance Genes
Aluminum treatments upregulate the expression of various genes that help plants survive the effects of ROS stress/damages. A transcriptome analysis confirmed that Al induces the expression of several genes to decrease ROS production, detoxify ROS, and stimulate the recovery from ROS-induced damages (Richards et al., 1998; Kumari et al., 2008; Chowra et al., 2017). Most of these genes are responsive to diverse biotic and abiotic stresses because their expression is induced by ROS. However, some of these ROS-related genes have an active role in Al-tolerance mechanisms.
The ectopic expression of several genes confers Al tolerance to A. thaliana and several crops (Inostroza-Blancheteau et al., 2012). This suggests that genes encoding ROS-scavenging proteins may be useful for breeding transgenic crops that are tolerant to Al stress conditions. For example, transgenic A. thaliana plants overexpressing three glutathione S-transferase genes and two peroxidase genes from tobacco, all of which are inducible by Al, were observed to be tolerant to Al stress conditions (Ezaki et al., 2000). Additionally, the ectopic expression of wheat WMnSOD1 confers Al tolerance to transgenic mustard plants (Basu et al., 2001). Meanwhile, Panda et al. (2013) reported that the overexpression of alternative oxidase enhances Al resistance. The shift in the regular electron transfer reaction of mitochondria (Cyt pathway) to the alternative oxidase pathway decreases ROS production under stress conditions. These results indicate that Al-inducible ROS-mediated genes help protect plants from Al-induced ROS damages.
The metabolic engineering of redox metabolic activities is another potential approach for improving Al tolerance in terms of ROS damages. As described above, the metabolism of ascorbate and tissue ascorbate levels affect ROS production in the apoplast. Yin et al. (2010) demonstrated that the overexpression of a dehydroascorbate reductase gene increases the ascorbate levels in tobacco and enhances Al tolerance. This may explain the results of the wheat transcriptome comparisons that indicated genes related to the metabolism of ascorbate were more highly expressed in Al-tolerant plants than in WT plants (Houde and Diallo, 2008). Manipulating non-enzymatic antioxidant defense molecules also improved Al tolerance, while downregulating polyamine synthesis resulted in Al sensitivity (Nezames et al., 2012). Another study concluded that the application of exogenous polyamine improved Al tolerance in saffron plants (Chen et al., 2008). This is likely because polyamines have protective roles against ROS (Alcazar et al., 2010). In contrast, the overexpression of a gene encoding an MDH exhibiting a unique kinetic property conferred Al tolerance to alfalfa plants (Tesfaye et al., 2001). This MDH converted oxaloacetate to malate, meaning its function is to convert NADH to NAD+. The overexpression of another gene encoding a plastid-localized MDH with a similar kinetic property also conferred Al tolerance to transgenic A. thaliana plants (Li et al., 2016). The transgenic lines exhibited an enhanced reducing capacity for 2,3,5-triphenyl tetrazolium chloride, indicating that increased respiration may improve Al tolerance. Finally, it is important to note that Al-induced ROS production can also activate the transcription of several Al-tolerance genes. For example, H2O2 induces the transcription of AtALMT1 and AtMATE in A. thaliana (Kobayashi et al., 2013). Furthermore, SbMATE (i.e., citrate transporter in sorghum) expression is upregulated by the accumulation of ROS, which serves as an indicator of Al exposure (Sivaguru et al., 2013).
Concluding Remarks and Future Interests
Recent studies clarified the complex transcriptional regulation of Al-tolerance genes (Table 1). The expression of major Al-tolerance genes, such as ALMT1 and MATE, is regulated by STOP1-type zinc-finger proteins. The STOP1-regulated system likely also affects proton tolerance, plant immunity, and root development under P-starvation conditions. The transcription of major Al-tolerance genes, including ALMT, is also regulated by phytohormones and ROS. These signal inducers regulate Al tolerance via the transcriptional regulation of diverse genes. However, recent studies also have shown that abnormal accumulation of phytohormones (e.g., IAA) is involved in the Al-induced suppression of root growth, which is concomitant with transcriptional regulation of various genes. These findings clearly indicate there is crosstalk between the transcription of Al-tolerance genes and various stress response mechanisms. Future studies should characterize the molecular mechanisms underlying this crosstalk. Additionally, the mechanisms regulating specific responses to Al will need to be elucidated. Combination of genome-wide approaches such as genome-wide association study and expression level polymorphism analysis, and its integration with genome-wide functional genomics may be useful to elucidate true nature of complex transcriptomic regulation of Al tolerance.
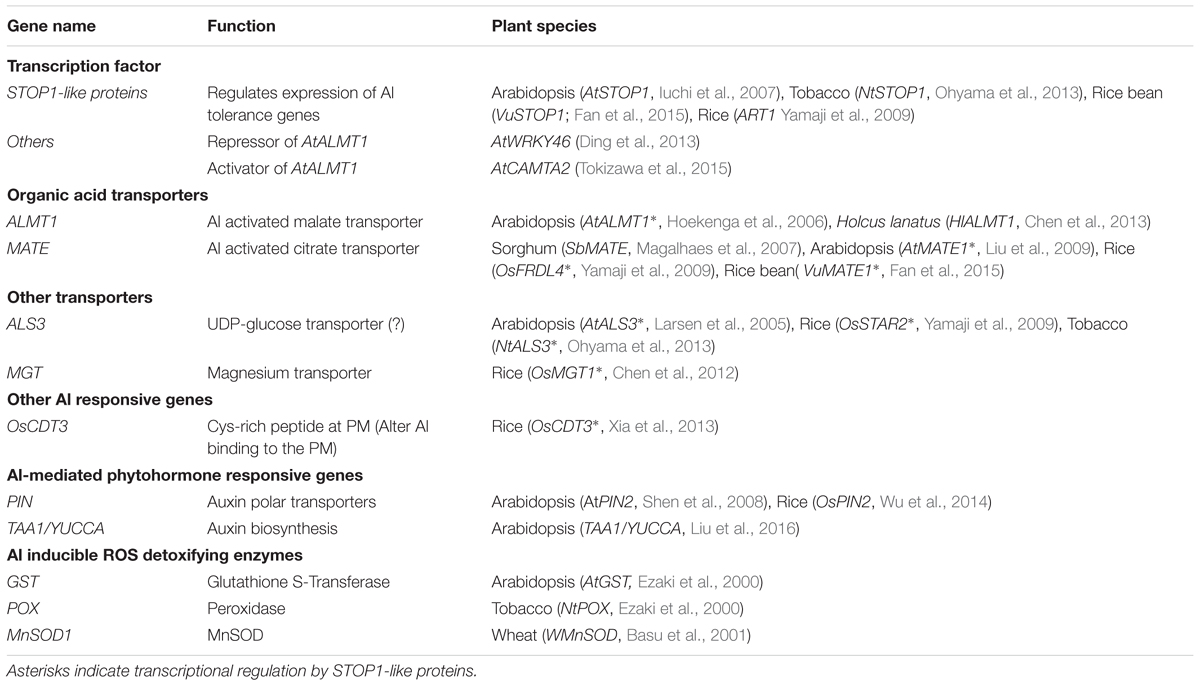
TABLE 1. Summary of transcriptionally regulated Al tolerance genes, and their regulatory genes mediating Al, phytohormone and reactive oxygen species signaling.
Author Contributions
AD wrote the article, revised the text based on feedback from the co-authors, and prepared the illustrations. AS and MT helped prepare the figures. SP provided editorial suggestions regarding the article. HK and YK conceptualized the overall structure of the review article and critically edited it. All authors have read and approved the final draft.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15H04468).
References
Alcazar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. doi: 10.1007/s00425-010-1130-0
Baluska, F., Mancuso, S., Volkmann, D., and Barlow, P. W. (2010). Root apex transition zone: a signalling-response nexus in the root. Trends Plant Sci. 15, 402–408. doi: 10.1016/j.tplants.2010.04.007
Balzergue, C., Dartevelle, T., Godon, C., Laugier, E., Meisrimler, C., Teulon, J. M., et al. (2017). Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commun. 8:15300. doi: 10.1038/ncomms15300
Basu, U., Good, A. G., and Taylor, G. J. (2001). Transgenic Brassica napus plants overexpressing aluminum-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminum. Plant Cell Environ. 24, 1269–1278. doi: 10.1046/j.0016-8025.2001.00783.x
Benkova, E., and Hejatko, J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69, 383–396. doi: 10.1007/s11103-008-9393-6
Bhuja, P., McLachlan, K., Stephens, J., and Taylor, G. J. (2004). Accumulation of 1,3-β-D-glucans, in response to aluminum and cytosolic calcium in Triticum aestivum. Plant Cell Physiol. 45, 543–549. doi: 10.1093/pcp/pch068
Bienert, G. P., Schjoerring, J. K., and Jahn, T. P. (2006). Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 1758, 994–1003. doi: 10.1016/j.bbamem.2006.02.015
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Chen, W., Xu, C., Zhao, B., Wang, X., and Wang, Y. (2008). Improved Al tolerance of saffron (Crocus sativus L.) by exogenous polyamines. Acta Physiol. Plant. 30, 121–127. doi: 10.1007/s11738-007-0100-z
Chen, Z. C., Yamaji, N., Motoyama, R., Nagamura, Y., and Ma, J. F. (2012). Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 159, 1624–1633. doi: 10.1104/pp.112.199778
Chen, Z. C., Yokosho, K., Kashino, M., Zhao, F. J., Yamaji, N., and Ma, J. F. (2013). Adaptation to acidic soil is achieved by increased numbers of cis-acting elements regulating ALMT1 expression in Holcus lanatus. Plant J. 76, 10–23. doi: 10.1111/tpj.12266
Chowra, U., Yanase, E., Koyama, H., and Panda, S. K. (2017). Aluminium-induced excessive ROS causes cellular damage and metabolic shifts in black gram Vigna mungo (L.) Hepper. Protoplasma 254, 293–302. doi: 10.1007/s00709-016-0943-5
Delhaize, E., Ma, J. F., and Ryan, P. R. (2012). Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 17, 341–348. doi: 10.1016/j.tplants.2012.02.008
Devi, S. R., Yamamoto, Y., and Matsumoto, H. (2003). An intracellular mechanism of aluminum tolerance associated with high antioxidant status in cultured tobacco cells. J. Inorg. Biochem. 97, 59–68. doi: 10.1016/S0162-0134(03)00182-X
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., and Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmatic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., and Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced ma- late secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Ezaki, B., Gardner, R. C., Ezaki, Y., and Matsumoto, H. (2000). Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 122, 657–665. doi: 10.1104/pp.122.3.657
Fan, W., Lou, H. Q., Gong, Y. L., Liu, M. Y., Cao, M. J., Liu, Y., et al. (2015). Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 208, 456–468. doi: 10.1111/nph.13456
Guo, Z., Tan, H., Zhu, Z., Lu, S., and Zhou, B. (2005). Effect of intermediates on ascorbic acid and oxalate biosynthesis of rice and in relation to its stress resistance. Plant Physiol. Biochem. 43, 955–962. doi: 10.1016/j.plaphy.2005.08.007
Hamel, F., Breton, C., and Houde, M. (1998). Isolation and characterization of wheat aluminum-regulated genes: possible involvement of aluminum as a pathogenesis response elicitor. Planta 205, 531–538. doi: 10.1007/s004250050352
Hoekenga, O. A., Maron, L. G., Piñeros, M. A., Cançado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1,which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 9738–9743. doi: 10.1073/pnas.0602868103
Hou, N., You, J., Pang, J., Xu, M., Chen, G., and Yang, Z. (2010). The accumulation and transport of abscisic acid in soybean (Glycine max L.) under aluminum stress. Plant Soil 330, 127–137. doi: 10.1007/s11104-009-0184-x
Houde, M., and Diallo, A. O. (2008). Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics 9:400. doi: 10.1186/1471-2164-9-400
Huang, W., Yang, X., Yao, S., LwinOo, T., He, H., Wang, A., et al. (2014). Reactive oxygen species burst induced by aluminum stress triggers mitochondria-dependent programmed cell death in peanut root tip cells. Plant Physiol. Biochem. 82, 76–84. doi: 10.1016/j.plaphy.2014.03.037
Inostroza-Blancheteau, C., Rengel, Z., Alberdi, M., De La Luz Mora, M., Aquea, F., Arce-Johnson, P., et al. (2012). Molecular and physiological strategies to increase aluminum resistance in plants. Mol. Biol. Rep. 39, 2069–2079. doi: 10.1007/s11033-011-0954-4
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Kobayashi, Y., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and co- regulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. U.S.A 104, 9900–9905. doi: 10.1073/pnas.0700117104
Jung, J. K., and McCouch, S. (2013). Getting to the roots of it: genetic and hormonal control of root architecture. Front. Plant Sci. 4:186. doi: 10.3389/fpls.2013.00186
Kasai, M., Sasaki, M., Tanakamaru, S., Yamamoto, Y., and Matsumoto, H. (1993). Possible involvement of abscisic acid in increases in activities of two vacuolar H+- pumps in barley roots under aluminum stress. Plant Cell Physiol. 34, 1335–1338.
Kinraide, T., and Parker, D. (1989). Assessing the phytotoxicity of mononuclear hydroxy- aluminum. Plant Cell Environ. 12, 478–487. doi: 10.1111/j.1365-3040.1989.tb02120.x
Kinraide, T. B. (2003). Toxicity factors in acidic forest soils, attempts to evaluate separately the toxic effects of excessive Al3+ and H+ and insufficient Ca2+ and Mg2+ upon root elongation. Eur. J. Soil Sci. 54, 323–333. doi: 10.1046/j.1365-2389.2003.00538.x
Kinraide, T. V., Arnold, R. C., and Baligar, V. C. (1985). A Rapid assay for aluminium phytoticity at submicromolar concentrations. Physiol. Plant 65, 245–250. doi: 10.1111/j.1399-3054.1985.tb02390.x
Kobayashi, Y., Furuta, Y., Ohno, T., Hara, T., and Koyama, H. (2005). Quantitative trait loci controlling aluminium tolerance in two accessions of Arabidopsis thaliana (Landsberg erecta and Cape Verde Islands). Plant Cell Environ. 28, 1516–1524. doi: 10.1111/j.1365-3040.2005.01388.x
Kobayashi, Y., Hoekenga, O. A., Itoh, H., Nakashima, M., Saito, S., Shaff, J. E., et al. (2007). Characteriza- tion of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145, 843–852. doi: 10.1104/pp.107.102335
Kobayashi, Y., Kobayashi, Y., Sugimoto, M., Lakshmanan, V., Iuchi, S., Kobayashi, M., et al. (2013). Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol. 162, 732–740. doi: 10.1104/pp.113.218065
Kobayashi, Y., Ohyama, Y., Kobayashi, Y., Ito, H., Iuchi, S., Fujita, M., et al. (2014). STOP2 activates transcription of several genes for Al- and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol. Plant 7, 311–322. doi: 10.1093/mp/sst116
Kochian, L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. 46, 237–260. doi: 10.1146/annurev.pp.46.060195.001321
Kochian, L. V., Piñeros, M. A., Liu, J., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 1–28. doi: 10.1146/annurev-arplant-043014-114822
Kollmeier, M., Felle, H. H., and Horst, W. J. (2000). Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. Is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 122, 945–956.
Kopittke, P. M., Moore, K. L., Lombi, E., Gianoncelli, A., Ferguson, B. J., Blamey, F. P., et al. (2015). Identification of the primary lesion of toxic aluminum in plant roots. Plant Physiol. 167, 1402–1411. doi: 10.1104/pp.114.253229
Koyama, H., Kobayashi, Y., Panda, S. K., and Taylor, G. J. (2015). “The molecular physiology and regulation of aluminum resistance in higher plants,” in Aluminum Stress Adaptation in Plants, Signaling and Communication in Plants, Vol. 24, eds S. K. Panda and F. Baluska (Cham: Springer). doi: 10.1007/978-3-319-19968-9_9
Kumari, M., Taylor, G. J., and Deyholos, M. K. (2008). Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol. Genet. Genomics. 279, 339–357. doi: 10.1007/s00438-007-0316-z
Kusunoki, K., Nakano, Y., Tanaka, K., Sakata, Y., Koyama, H., and Kobayashi, Y. (2017). Transcriptomic variation among six Arabidopsis thaliana accessions identified several novel genes controlling aluminium tolerance. Plant Cell Environ. 40, 249–263. doi: 10.1111/pce.12866
Larsen, P. B., Geisler, M. J. B., Jones, C. A., Williams, K. M., and Cancel, J. D. (2005). ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 41, 353–363. doi: 10.1111/j.1365-313X.2004.02306.x
Leyser, O. (2005). Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell 121, 819–822.
Li, Q.-F., Zhao, J., Zhang, J., Dai, Z.-H., and Zhang, L. G. (2016). Ectopic expression of the Chinese cabbage malate dehydrogenase gene promotes growth and aluminum resistance in Arabidopsis. Front. Plant Sci. 7:1180. doi: 10.3389/fpls.2016.01180
Ligaba-Osena, A., Fei, Z., Liu, J., Xu, Y., Shaff, J., Lee, S. C., et al. (2017). Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis. New Phytol. 214, 830–841. doi: 10.1111/nph.14420
Liu, G., Gao, S., Tian, H., Wu, W., Robert, H. S., and Ding, Z. (2016). Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet 12:e1006360. doi: 10.1371/journal.pgen.1006360
Liu, J., Jurandir, V., Magalhaes, J. V., Shaff, J., and Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Ma, J. F., Hiradate, S., and Matsumoto, H. (1998). High aluminum resistance in buckwheat. II. Oxalic acid detoxifies aluminum internally. Plant Physiol. 117, 753–759. doi: 10.1104/pp.117.3.753
Ma, J. F., Zhang, W., and Zhao, Z. (2001). “Regulatory mechanisms of Al-induced secretion of organic acids anions- Involvement of ABA in the Al-induced secretion of oxalate in buckwheat,” in Plant Nutrition-Food Security and Sustainability of Agro-Ecosystems, eds W. J. Hortst, H. Flessa, and B. Sattelmacher (Dordrecht: Kluwer Academic).
Macias, J. M., Rivera, J. O. O., Alanís, G. D., Villalobos, L. Y., Aburto, A. O., González, J. R., et al. (2017). Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc. Natl. Acad. Sci. U.S.A 114, 3563–3572. doi: 10.1073/pnas.1701952114
Magalhaes, J. V., Liu, J., Guimaraes, C. T., Lana, U. G., Alves, V. M., Wang, Y. H., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161. doi: 10.1038/ng2074
Maltais, K., and Houde, M. (2002). A new biochemical marker for aluminium tolerance in plants. Physiol. Plant. 115, 81–86. doi: 10.1034/j.1399-3054.2002.1150109.x
Massot, N., Nicander, B., Barcelo, J., Poschenrieder, C., and Tillberg, E. (2002). A rapid increase in cytokinin levels and enhanced ethylene evolution precede Al3+-induced inhibition of root growth in bean seedlings (Phaseolus vulgaris L.). Plant Growth Regul. 37, 105–112. doi: 10.1023/A:1020511007138
Moseyko, N., and Feldman, L. J. (2001). Expression of pH-sensitive green fluorescent protein in Arabidopsis thaliana. Plant Cell Environ. 24, 557–563. doi: 10.1046/j.1365-3040.2001.00703.x
Muday, G. K., Rahman, A., and Binder, B. M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195. doi: 10.1016/j.tplants.2012.02.001
Mujika, J., Ruipérez, F., Infante, I., Ugalde, J., Exley, C., and Lopez, X. (2011). Pro-oxidant activity ofaluminum: stabilization of the aluminum superoxide radical ion. J. Phys. Chem. A 115, 6717–6723. doi: 10.1021/jp203290b
Murphy, M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13. doi: 10.1042/BJ20081386
Nezames, C. D., Ochoa, V., and Larsen, P. B. (2012). Mutational loss of Arabidopsis SLOW WALKER2 results in reduced endogenous spermine concomitant with increased aluminum sensitivity. Funct. Plant Biol. 40, 67–78. doi: 10.1071/FP12234
Ohyama, Y., Ito, H., Kobayashi, Y., Ikka, T., Morita, A., Kobayashi, M., et al. (2013). Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162, 1937–1946. doi: 10.1104/pp.113.218958
Panda, S. K., Sahoo, L., Katsuhara, M., and Matsumoto, H. (2013). Overexpression of alternative oxidase gene confers aluminum tolerance by altering the respiratory capacity and the response to oxidative stress in tobacco cells. Mol. Biotechnol. 54, 551–563. doi: 10.1007/s12033-012-9595-7
Panda, S. K., Yamamoto, Y., Kondo, H., and Matsumoto, H. (2008). Mitochondrial alterations related to programmed cell death in tobacco cells under aluminium stress. Compt. Rend. Biol. 331, 597–610. doi: 10.1016/j.crvi.2008.04.008
Reyna-Llorens, I., Corrales, I., Poschenrieder, C., Barcelo, J., and Cruz, O. R. (2015). Both aluminum and ABA induce the expression of an ABC-like transporter gene (FeALS3) in the Al-tolerant species Fagopyrumesculentum. Environ. Exp. Bot. 111, 74–82. doi: 10.1016/j.envexpbot.2014.11.005
Richards, K. D., Schott, E. J., Sharma, Y. K., Davis, K. R., and Gardner, R. C. (1998). Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 116, 409–418. doi: 10.1104/pp.116.1.409
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Ruipérez, F., Mujika, J., Ugalde, J., Exley, C., and Lopez, X. (2012). Pro-oxidant activity of aluminum: promoting the Fenton reaction by reducing Fe (III) to Fe (II). J. Inorg. Biochem. 117, 118–123. doi: 10.1016/j.jinorgbio.2012.09.008
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Sagi, M., and Fluhr, R. (2001). Superoxide production by plant homologues of the gp91phox NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126, 1281–1290. doi: 10.1104/pp.126.3.1281
Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313X.2003.01991.x
Sawaki, K., Sawaki, Y., Zhao, C. R., Kobayashi, Y., and Koyama, H. (2016). Specific transcriptomic response in the shoots of Arabidopsis thaliana after exposure to Al rhizotoxicity?: - potential gene expression biomarkers for evaluating Al toxicity in soils. Plant Soil 409, 131–142. doi: 10.1007/s11104-016-2960-8
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that pro- tect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. doi: 10.1104/pp.108.134700
Sawaki, Y., Kobayashia, Y., Kihara-Doic, T., Nishikuboc, N., Kawazuc, T., Kobayashid, M., et al. (2014). Identification of a STOP1-like protein in Eucalyptus that regulates transcription of Al tolerance genes. Plant Sci. 223, 8–15. doi: 10.1016/j.plantsci.2014.02.011
Shen, H., Hou, N., Schlicht, M., Wan, Y., Mancuso, S., and Baluska, F. (2008). Aluminium toxicity targets PIN2 in Arabidopsis root apices: effects on PIN2 endocytosis, vesicular recycling, and polar auxin transport. Chin. Sci. Bull. 53, 2480–2487. doi: 10.1007/s11434-008-0332-3
Shen, H., Ligaba, A., Yamaguchi, M., Osawa, H., Shibata, K., Yan, X., et al. (2004). Effect of K-252a and abscisic acid on the efflux of citrate from soybean roots. J. Exp. Bot. 55, 663–671. doi: 10.1093/jxb/erh058
Sivaguru, M., Liu, J., and Kochian, L. V. (2013). Targeted expression of SbMATE in the root distal transition zone is responsible for sorghum aluminum resistance. Plant J. 76, 297–307. doi: 10.1111/tpj.12290
Stepanova, A. N., Yun, J., Likhacheva, A. V., and Alonso, J. M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185. doi: 10.1105/tpc.107.052068
Sun, P., Tian, Q. Y., Chen, J., and Zhang, W. H. (2010). Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 61, 347–356. doi: 10.1093/jxb/erp306
Sun, P., Tian, Q.-Y., Zhao, M.-G., Dai, X.-Y., Huang, J.-H., Li, L.-H., et al. (2007). Aluminum-induced ethylene production is associated with inhibition of root elongation in Lotus japonicus L. Plant Cell Physiol. 48, 1229–1235. doi: 10.1093/pcp/pcm077
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Takatsuka, H., and Umeda, M. (2014). Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 65, 2633–2643. doi: 10.1093/jxb/ert485
Taylor, G. J. (1987). Exclusion of metals from the symplasm: a possible mechanism of metal tolerance in higher plants. J. Plant Nutr. 10, 1213–1222. doi: 10.1080/01904168709363649
Taylor, G. J. (1988). “The physiology of aluminum phytotoxicity,” in Aluminum and Its Role in Biology, Metal Ions in Biological Systems, Vol. 24, ed. H. Sigel (New York, NY: Marcel Dekker), 123–163.
Taylor, G. J. (1991). Current views of the aluminum stress response: the physiological basis of tolerance. Curr. Top Plant Biochem. Physiol. 10, 57–93.
Taylor, G. J., McDonald-Stephens, J. L., Hunter, D. B., Bertsch, P. M., Elmore, D., Rengel, Z., et al. (2000). Direct measurement of aluminum uptake and distribution in single cells of Chara corallina. Plant Physiol. 123, 987–996. doi: 10.1104/pp.123.3.987
Tesfaye, M., Temple, S. J., Allan, D. L., Vance, C. P., and Samac, D. A. (2001). Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol. 127, 1836–1844. doi: 10.1007/s11738-010-0522-x
Tokizawa, M., Kobayashi, Y., Saito, T., Kobayashi, M., Iuchi, S., Nomoto, M., et al. (2015). Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 167, 991–1003. doi: 10.1104/pp.114.256552
Tsutsui, T., Yamaji, N., and Ma, J. (2011). Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 156, 925–931. doi: 10.1104/pp.111.175802
Wang, J., Hou, Q., Li, P., Yang, L., Sun, X., Benedito, V. A., et al. (2017). Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula. Plant J. 90, 79–95. doi: 10.1111/tpj.13471
Wenzl, P., Patino, G. M., Chaves, A. L., Mayer, J. E., and Rao, I. M. (2001). The high level of aluminum resistance in signalgrass is not associated with known mechanisms of external aluminum detoxification in root apices. Plant Physiol. 125, 1473–1484. doi: 10.1104/pp.125.3.1473
Wu, D., Shen, H., Yokawa, K., and Baluska, F. (2014). Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 65, 5305–5315. doi: 10.1093/jxb/eru292
Wu, D., Shen, H., Yokawa, K., and Baluška, F. (2015). Overexpressing OsPIN2 enhances aluminium internalization by elevating vesicular trafficking in rice root apex. J. Exp. Bot. 66, 6791–6801. doi: 10.1093/jxb/erv385
Xia, J., Yamaji, N., and Ma, J. F. (2013). A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 76, 345–355. doi: 10.1111/tpj.12296
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yamaji, N., Huang, C. F., Nagao, S., Yano, M., Sato, Y., Nagamura, Y., et al. (2009). Azinc finger transcription factor ART1 regulates multiple genes impli- cated in aluminum tolerance in rice. Plant Cell 21, 3339–3349. doi: 10.1105/tpc.109.070771
Yamamoto, Y., Kobayashi, Y., Devi, S. R., Rikiishi, S., and Matsumoto, H. (2002). Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 128, 63–72. doi: 10.1104/pp.010417
Yang, Z. B., Geng, X., He, C., Zhang, F., Wang, R., Horst, W. J., et al. (2014). TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26, 2889–2904. doi: 10.1105/tpc.114.127993
Yang, Z. B., He, C., Ma, Y., Herde, M., and Ding, Z. (2017a). Jasmonic acid enhances Al-induced root growth inhibition. Plant Physiol. 173, 1420–1433. doi: 10.1104/pp.16.01756
Yang, Z. B., Liu, G., Liu, J., Zhang, B., Meng, W., Müller, B., et al. (2017b). Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 18, 1213–1230. doi: 10.15252/embr.201643806
Yin, L. N., Wang, S. W., Eltayeb, A. E., Uddin, M. I., Yamamoto, Y., Tsuji, W., et al. (2010). Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231, 609–621. doi: 10.1007/s00425-009-1075-3
Keywords: Al, ALMT1, phytohormone, ROS, STOP1
Citation: Daspute AA, Sadhukhan A, Tokizawa M, Kobayashi Y, Panda SK and Koyama H (2017) Transcriptional Regulation of Aluminum-Tolerance Genes in Higher Plants: Clarifying the Underlying Molecular Mechanisms. Front. Plant Sci. 8:1358. doi: 10.3389/fpls.2017.01358
Received: 25 May 2017; Accepted: 20 July 2017;
Published: 08 August 2017.
Edited by:
Rudra Deo Tripathi, National Botanical Research Institute (CSIR), IndiaReviewed by:
Sudhakar Srivastava, Banaras Hindu University, IndiaFrantisek Baluska, University of Bonn, Germany
Meetu Gupta, Jamia Millia Islamia, India
Copyright © 2017 Daspute, Sadhukhan, Tokizawa, Kobayashi, Panda and Koyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroyuki Koyama, koyama@gifu-u.ac.jp
 Abhijit A. Daspute1
Abhijit A. Daspute1