
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Plant Sci. , 18 July 2017
Sec. Plant Breeding
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01266
This article is part of the Research Topic Harnessing Crop Biodiversity and Genomics Assisted Pre-Breeding Approaches for Next Generation Climate-Smart Varieties View all 27 articles
Panicoids (subfamily: panicoideae) are a group of C4 grasses, which include agronomically important crops such as sorghum and maize, bioenergy feedstocks including sugarcane and miscanthus, nutri-cereals such as millets, and biofuel crops including switchgrass, napier grass and guinea grass. Among these, millets are known for their climate-resilient features including adaptation to a wide range of ecological conditions, less irrigational requirements, better growth and productivity in low nutrient input conditions, less reliance on synthetic fertilizers, and minimum vulnerability to environmental stresses (Kole et al., 2015). Also, millets are nutritionally superior to other major cereals as they are rich in dietary fibers, resistant starches, vitamins, essential amino acids, storage proteins and other bioactive compounds (Amadou et al., 2013). These attributes have made millets a crop of choice for cultivation in arid and semi-arid regions of the world; however, the less attempt has been made to study the climate-resilient features of millets compared to other major cereals. Among millets, foxtail millet (Setaria italica) and its wild progenitor, green foxtail (S. viridis) are extensively studied since they are considered as models for studying the traits related to C4 photosynthesis, stress biology, and bioenergy characteristics (Muthamilarasan and Prasad, 2015). The availability of genome sequence information of these two species (Bennetzen et al., 2012; Zhang et al., 2012) has unlocked the wealth of information pertaining to stress tolerance and biofuel characteristics. It has also expedited the development of large-scale genomic resources for crop improvement. On the other hand, studies on other millets are still in their infancy. The challenge to feed the ever-growing population with a healthy balanced diet and the threats faced by agricultural crops due to changing climate highlight the immediate requirement to exploit the beneficial attributes of millets. This could be utilized for the improvement of millets per se as well as other related grass species. The extensive gene-level synteny shared between the grass genomes would facilitate the transfer and introgression of useful genes, alleles and quantitative trait loci (QTL) of agronomic importance identified in millets to other major cereals. In the above context, this article advocates for initiating extensive research on millets to dissect their agronomic, nutritional as well as stress tolerance traits and develop strategies to transfer the useful traits to cultivated major cereals such as rice, wheat, maize, and sorghum.
Millets possess several morpho-physiological, molecular and biochemical characteristics which confer better tolerance to environmental stresses than major cereals. Primarily, the short life-cycle of millets assists in escaping from stress as they require 12–14 weeks to complete their life-cycle (seed to seed) whereas rice and wheat requires a maximum of 20–24 weeks. However, the prevalence of stress conditions and their consequences are circumvented by several traits such as short stature, small leaf area, thickened cell walls, and the capability to form dense root system (Li and Brutnell, 2011). Also, the C4 photosynthetic trait is highly advantageous to millets. In the C4 system, carbon dioxide (CO2) is concentrated around ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), which in turn suppresses ribulose 1,5-bisphosphate (RuBP) oxygenation and photorespiration (Aubry et al., 2011). Thus, C4 mechanism enhances the concentration of CO2 in bundle sheath, which suppresses photorespiration (around 80%) depending on the temperature and increases the in planta catalytic activity of RuBisCO (Sage et al., 2011). Since RuBisCO of C4 plants works at elevated CO2 levels, millets have enhanced photosynthetic rates at warm conditions and confers immediate water use efficiency (WUE) and nitrogen use efficiency (NUE) which are ~1.5 to 4-fold higher than C3 photosynthesis (Sage and Zhu, 2011). For instance, foxtail millet requires just 257 g of water to produce a dry biomass of 1 g, whereas maize and wheat require 470 and 510 g, respectively (Li and Brutnell, 2011). In addition to conferring WUE and NUE, C4 photosynthesis provides secondary benefit to millets including improved growth and ecological enactment in warm temperatures, enhanced flexible allocation patterns of biomass and reduced hydraulic conductivity per unit leaf area (Sage and Zhu, 2011). These attributes of millets make them next-generation crops holding the potential for research to explore the climate-resilient traits and exploit the information for the improvement of major cereals. One such effort undertaken so far is the engineering of C4 traits in rice using millet as models; however, through the realization of stress tolerance potential of millets is imperative for expedited progress in developing climate-resilient crop species.
The exceptional tolerance of millets toward diverse abiotic stresses including drought, salinity, light and heat makes them a tractable system to study their stress-responsive traits at the cellular, molecular and physiological levels. Several morpho-physiological and biochemical studies in millets have shown their stress adaptation strategies. For example, Bidinger et al. (2007) have shown that pearl millet adjusts flowering phenology according to the pattern of rainfall. Balsamo et al. (2006) observed an increase in leaf tensile strength in teff during drought, and in little millet, an increase in root length was reported by Ajithkumar and Panneerselvam (2014). Similarly, increase in biochemical activities such as enhanced levels of antioxidants, reactive oxygen species and their scavenging enzymes, enzyme activity of catalase and superoxide, and synthesis of osmolytes and other stress-related proteins has been reported in response to abiotic stresses in foxtail millet (Lata et al., 2011), little millet (Ajithkumar and Panneerselvam, 2014) and teff (Smirnoff and Colombe, 1988). van der Weerd et al. (2001) showed the dynamics of membrane permeability for water in pearl millet in comparison to maize for achieving better water status during osmotic stress. In addition, several novel genes, alleles and QTLs have been identified in millets whose functional characterization has revealed their roles in conferring stress tolerance (Table 1). Compared to other millets, foxtail millet has been studied extensively, and several genetic and genomic resources have been developed (Muthamilarasan and Prasad, 2015). Whole genome sequencing of foxtail millet and comparison of gene families among 15 sequenced plant genomes showed that 1517 genes were specific only to foxtail millet (Zhang et al., 2012). Among this, 586 genes were annotated as “response to water,” which could be playing significant roles in conferring drought and dehydration stresses, thus facilitating the adaptation of this crop to arid and semi-arid zones. The genes involved in C4 pathway namely, carbonic anhydrase (CAH), malate dehydrogenase (MDH), malic enzyme (ME), phosphoenolpyruvate carboxylase (PEPC), phosphoenolpyruvate carboxylase kinase (PPCK) and pyruvate orthophosphate dikinase (PPDK) were also identified and compared to that of sorghum, maize, rice and Brachypodium. The study showed that foxtail millet has a higher number of MDH (7 genes) and PPDK (3 genes) than other crops. Zhang et al. (2012) have also performed phylogenetic and evolutionary analysis of CAH homologs among all the five grass genomes, which showed that Ft_CA1 was highly expressed in the mesophyll, and this could be a potential candidate for studying C4 pathway in foxtail millet. Despite this progress, studies providing insights into the molecular machinery underlying stress tolerance is largely lacking in millets. In addition, knowledge on the genetic determinants of stress tolerance identified through association mapping and biparental mapping is limited. In this context, extensive phenotypic screening to observe the natural genetic variations in stress tolerance across diverse millet germplasms is greatly needed to fully harness the underlying genetic potential through conventional/molecular breeding approaches and transgenic technologies. This is required to facilitate crop improvements in millets and non-millet crops in the wake of increased desertification and salinity of the farmlands due to climate change.
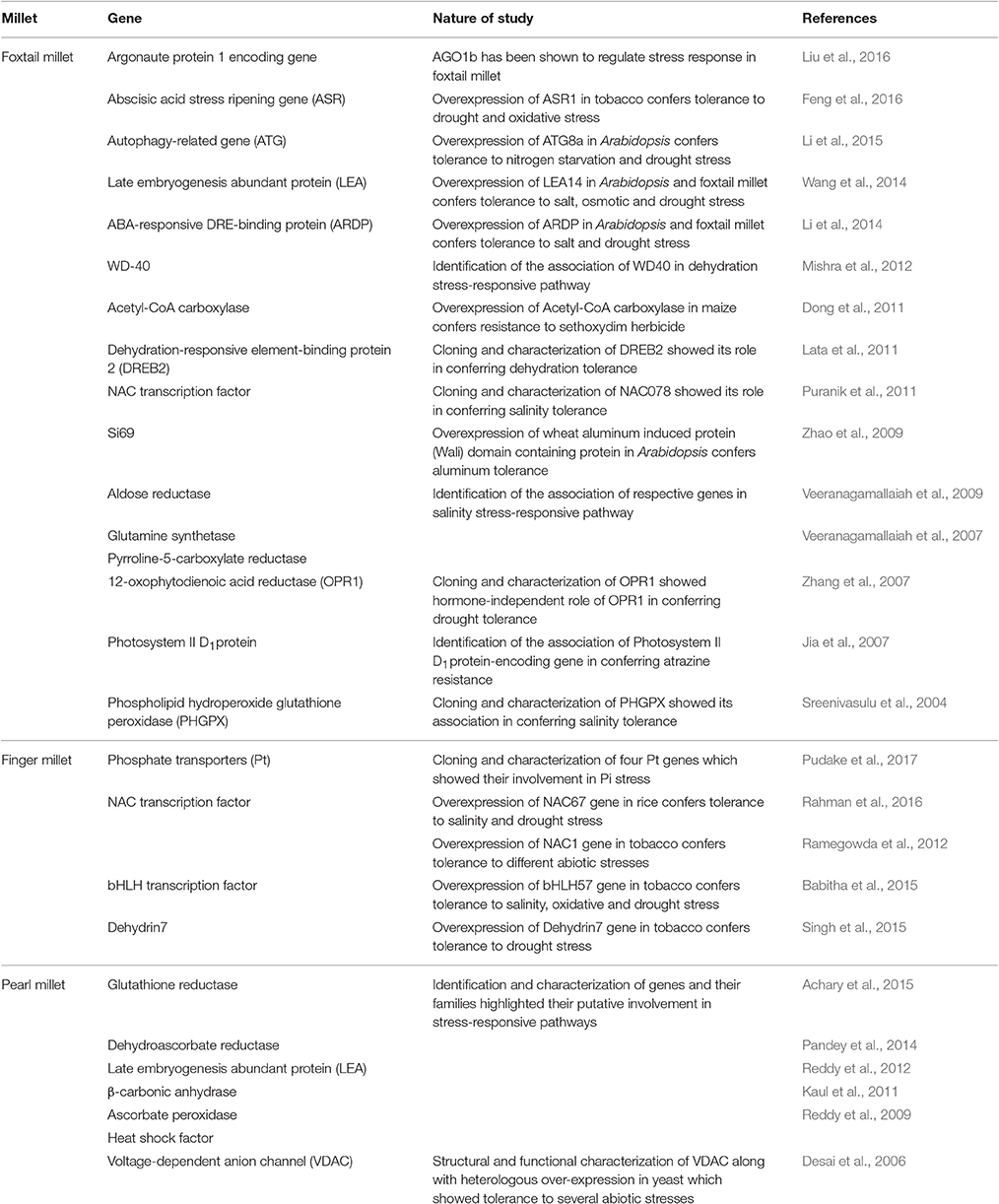
Table 1. Summary of genes identified and characterized in different millets for their roles in conferring tolerance to abiotic stresses.
Genetic and genomic resources are imperative for the improvement of any crop species, where genetic resources serve as primary input for breeding while genomic resources facilitate efficient characterization of genetic resources and their subsequent utilization in the identification of useful genes, alleles, and QTLs for crop improvement. In the case of millets, ample genetic resources are available (Goron and Raizada, 2015; Saha et al., 2016); however, the information available on genomic resources including molecular markers and physical/genetic maps are scarce as compared to major cereals. Availability of genome sequence information of foxtail millet has facilitated the development of several high-throughput genome-wide molecular markers (Jia et al., 2013; Kumari et al., 2013; Pandey et al., 2013; Muthamilarasan et al., 2014; Yadav et al., 2014, 2015; Zhang et al., 2014) and integrated marker databases (Muthamilarasan and Prasad, 2015). These resources would be inevitable for several large-scale genotyping studies including genetic diversity analysis, construction of high-density physical as well as genetic-linkage maps, and mapping of QTLs related to nutritional traits. One such example for utilization of these resources in genomics-assisted breeding (GAB) was reported by Gupta et al. (2014). In this study, association mapping of 20 yield-contributing agronomic traits among 200 foxtail millet accessions was performed, and eight microsatellite markers associated with nine different agronomic traits were identified, which contribute up to 25% of the phenotypic variation (Gupta et al., 2014). These trait-associated SSR markers will be useful for identification of genes/QTLs regulating the agronomic as well as nutritional traits, and eventually for marker-assisted genetic enhancement of foxtail millet per se and millets and cereals.
Recently, GAB has gained momentum in the arena of crop improvement as it implicates the next-generation genome analysis platforms and conventional as well as molecular breeding strategies for crop improvement. The scope for development of genomic resources for GAB in foxtail millet is relatively higher than other millets owing to the availability of genome sequence information. Moreover, the advent of next-generation genome sequencing (NGS) has enabled the development of high-throughput molecular markers in other millet crops. In finger millet, the transcriptome of high and low seed calcium genotypes was sequenced, and thousands of simple sequence repeat (SSR) containing microsatellite markers were identified (Kumar et al., 2015). Similarly, novel SSR markers were developed through sequencing the genome of allotetraploid tef (Eragrostis tef) (Cannarozzi et al., 2014). This information on the genome as well as genome-wide markers coupled high-throughput approaches including genotyping-by-sequencing (GBS) and genome-wide association mapping studies (GWAS) potentiate the discovery of novel genes/alleles or QTLs responsible for nutritional traits (Varshney et al., 2014; Muthamilarasan et al., 2016). Considering the importance of whole-genome sequence information, genome sequencing of few other millets including pearl millet and finger millet is underway.
It is realized that millets hold great promise for food security and nutrition amid ever-increasing agricultural costs, climate change and burgeoning mouths to feed worldwide. They are nutritious, possess additional health benefits, requires significantly fewer input costs for cultivation and are naturally tolerant to most biotic and abiotic stresses. These features accentuate millets as crops of choice for the world population amid growing concerns about climate change. Given the same, there is a growing need to investigate the natural genetic variation across their diverse germplasms to exploit them for crop improvement with regard to many agronomically and nutritionally important traits. With the advent of NGS technologies and high-throughput GWAS platforms, identification of candidate genes/alleles/QTLs regulating such traits is possible at a pace and precision not contemplated before, which in turn would facilitate the development of breeding lines for crop improvement. Moreover, a renewed focus on millets has important implications for the improvement of cereals and bioenergy grasses given their common ancestry from poaceae family and the presence of significant synteny between the genomes.
MP and TB conceived the idea and drafted the opinion article. MM improved the manuscript and provided revisions to the manuscript. All the authors have read and approved the final version of the manuscript.
Authors' work in millet genetics and genomics is supported by the core grant of National Institute of Plant Genome Research, New Delhi, India and Science and Engineering Research Board, Department of Science and Technology, New Delhi, India (File Number: EMR/2015/000464).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Achary, V. M., Reddy, C. S., Pandey, P., Islam, T., Kaul, T., and Reddy, M. K. (2015). Glutathione reductase a unique enzyme: molecular cloning, expression and biochemical characterization from the stress adapted C4 plant, Pennisetum glaucum (L.) R. Br. Mol. Biol. Rep. 42, 947–962. doi: 10.1007/s11033-014-3832-z
Ajithkumar, I. P., and Panneerselvam, R. (2014). ROS scavenging system, osmotic maintenance, pigment and growth status of Panicum sumatrense Roth. under drought stress. Cell Biochem. Biophys. 68, 587–595. doi: 10.1007/s12013-013-9746-x
Amadou, I., Gounga, M. E., and Le, G. W. (2013). Millets: nutritional composition, some health benefits and processing - a review. Emir. J. Food Agric. 25, 501–508. doi: 10.9755/ejfa.v25i7.12045
Aubry, S., Brown, N. J., and Hibberd, J. M. (2011). The role of proteins in C3 plants prior to their recruitment into the C4 pathway. J. Exp. Bot. 62, 3049–3059. doi: 10.1093/jxb/err012
Babitha, K. C., Vemanna, R. S., Nataraja, K. N., and Udayakumar, M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 10:e0137098. doi: 10.1371/journal.pone.0137098
Balsamo, R. A., Willigen, C. V., Bauer, A. M., and Farrant, J. (2006). Drought tolerance of selected Eragrostis species correlates with leaf tensile properties. Ann. Bot. 97, 985–991. doi: 10.1093/aob/mcl068
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Bidinger, F. R., Napolean, T., and Hash, C. T. (2007). Quantitative trait loci for grain yield in pearl millet under variable post flowering moisture conditions. Crop Sci. 47, 969–980. doi: 10.2135/cropsci2006.07.0465
Cannarozzi, G., Plaza-Wüthrich, S., Esfeld, K., Larti, S., Wilson, Y. S., Girma, D., et al. (2014). Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics 15:581. doi: 10.1186/1471-2164-15-581
Desai, M. K., Mishra, R. N., Verma, D., Nair, S., Sopory, S. K., and Reddy, M. K. (2006). Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol. Biochem. 44, 483–493. doi: 10.1016/j.plaphy.2006.08.008
Dong, Z., Zhao, H., He, J., Huai, V., Lin, H., Zheng, J., et al. (2011). Overexpression of a foxtail millet Acetyl-CoA carboxylase gene in maize increases sethoxydim resistance and oil content. Afr. J. Biotechnol. 10, 3986–3995. doi: 10.5897/AJB11.053
Feng, Z. J., Xu, Z. S., Sun, J., Li, L. C., Chen, M., Yang, G. X., et al. (2016). Investigation of the ASR family in foxtail millet and the role of ASR1 in drought/oxidative stress tolerance. Plant Cell Rep. 35, 115–128. doi: 10.1007/s00299-015-1873-y
Goron, T. L., and Raizada, M. N. (2015). Genetic diversity and genomic resources available for the small millet crops to accelerate a new green revolution. Front. Plant Sci. 6:157. doi: 10.3389/fpls.2015.00157
Gupta, S., Kumari, K., Muthamilarasan, M., Parida, S. K., and Prasad, M. (2014). Population structure and association mapping of yield contributing agronomic traits in foxtail millet. Plant Cell Rep. 33, 881–893. doi: 10.1007/s00299-014-1564-0
Jia, G., Huang, X., Zhi, H., Zhao, Y., Zhao, Q., Li, W., et al. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–961. doi: 10.1038/ng.2673
Jia, X., Yuan, J., Shi, Y., Song, Y., Wang, G., Wang, T., et al. (2007). A ser-Gly substitution in plastid-encoded photosystem II D1 protein is responsible for atrazine resistance in foxtail millet (Setaria italica). Plant Growth Regul. 52, 81–89. doi: 10.1007/s10725-007-9181-3
Kaul, T., Reddy, P. S., Mahanty, S., Thirulogachandar, V., Reddy, R. A., Kumar, B., et al. (2011). Biochemical and molecular characterization of stress-induced β-carbonic anhydrase from a C4 plant, Pennisetum glaucum. J. Plant. Physiol. 168, 601–610. doi: 10.1016/j.jplph.2010.08.007
Kole, C., Muthamilarasan, M., Henry, R., Edwards, D., Sharma, R., Abberton, M., et al. (2015). Application of genomics-assisted breeding for generation of climate resilient crops: progress and prospects. Front. Plant Sci. 6:563. doi: 10.3389/fpls.2015.00563
Kumar, A., Gaur, V. S., Goel, A., and Gupta, A. K. (2015). De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol. Biol. Rep. 33, 905–922. doi: 10.1007/s11105-014-0802-5
Kumari, K., Muthamilarasan, M., Misra, G., Gupta, S., Subramanian, A., Parida, S. K., et al. (2013). Development of eSSR-markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS ONE 8:e67742. doi: 10.1371/journal.pone.0067742
Lata, C., Bhutty, S., Bahadur, R. P., Majee, M., and Prasad, M. (2011). Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. J. Exp. Bot. 62, 3387–3401. doi: 10.1093/jxb/err016
Li, C., Yue, J., Wu, X., Xu, C., and Yu, J. (2014). ABA-responsive DRE-binding protein gene from Setaria italica, SiARDP, the target gene of SiAREB, plays a critical role under drought stress. J. Exp. Bot. 65, 5415–5427. doi: 10.1093/jxb/eru302
Li, P., and Brutnell, T. P. (2011). Setaria viridis and Setaria italica, model genetic systems for the panicoid grasses. J. Exp. Bot. 62, 3031–3037. doi: 10.1093/jxb/err096
Li, W. W., Chen, M., Zhong, L., Liu, J. M., Xu, Z. S., Li, L. C., et al. (2015). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Liu, X., Tang, S., Jia, G., Schnable, J. C., Su, H., Tang, C., et al. (2016). The C-terminal motif of SiAGO1b is required for the regulation of growth, development and stress responses in foxtail millet (Setaria italica (L.) P. Beauv). J. Exp. Bot. 67, 3237–3249. doi: 10.1093/jxb/erw135
Mishra, A. K., Puranik, S., Bahadur, R. P., and Prasad, M. (2012). The DNA-binding activity of an AP2 protein is involved in transcriptional regulation of a stress-responsive gene, SiWD40, in foxtail millet. Genomics 100, 252–263. doi: 10.1016/j.ygeno.2012.06.012
Muthamilarasan, M., Dhaka, A., Yadav, R., and Prasad, M. (2016). Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 242, 89–97. doi: 10.1016/j.plantsci.2015.08.023
Muthamilarasan, M., and Prasad, M. (2015). Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor. Appl. Genet. 128, 1–14. doi: 10.1007/s00122-014-2399-3
Muthamilarasan, M., Venkata, S. B., Pandey, G., Kumari, K., Parida, S. K., and Prasad, M. (2014). Development of 5123 intron-length polymorphic markers for large-scale genotyping applications in foxtail millet. DNA Res. 21, 41–52. doi: 10.1093/dnares/dst039
Pandey, G., Misra, G., Kumari, K., Gupta, S., Parida, S. K., Chattopadhyay, D., et al. (2013). Genome wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res. 20, 197–207. doi: 10.1093/dnares/dst002
Pandey, P., Achary, V. M., Kalasamudramu, V., Mahanty, S., Reddy, G. M., and Reddy, M. K. (2014). Molecular and biochemical characterization of dehydroascorbate reductase from a stress adapted C4 plant, pearl millet [Pennisetum glaucum (L.) R. Br]. Plant Cell Rep. 33, 435–445. doi: 10.1007/s00299-013-1544-9
Pudake, R. N., Mehta, C. M., Mohanta, T. K., Sharma, S., Varma, A., and Sharma, A. K. (2017). Expression of four phosphate transporter genes from Finger millet (Eleusine coracana L.) in response to mycorrhizal colonization and Pi stress. 3 Biotech 7:17. doi: 10.1007/s13205-017-0609-9
Puranik, S., Bahadur, R. P., Srivastava, P. S., and Prasad, M. (2011). Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.]. Mol. Biotechnol. 49, 138–150. doi: 10.1007/s12033-011-9385-7
Rahman, H., Ramanathan, V., Nallathambi, J., Duraialagaraja, S., and Muthurajan, R. (2016). Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol. 11:35. doi: 10.1186/s12896-016-0261-1
Ramegowda, V., Senthil-Kumar, M., Nataraja, K. N., Reddy, M. K., Mysore, K. S., and Udayakumar, M. (2012). Expression of a finger millet transcription factor, EcNAC1, in tobacco confers abiotic stress-tolerance. PLoS ONE 7:e40397. doi: 10.1371/journal.pone.0040397
Reddy, P. S., Reddy, G. M., Pandey, P., Chandrasekhar, K., and Reddy, M. K. (2012). Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: protection against abiotic stresses. Mol. Biol. Rep. 39, 7163–7174. doi: 10.1007/s11033-012-1548-5
Reddy, R. A., Kumar, B., Reddy, P. S., Mishra, R. N., Mahanty, S., Kaul, T., et al. (2009). Molecular cloning and characterization of genes encoding Pennisetum glaucum ascorbate peroxidase and heat-shock factor: interlinking oxidative and heat-stress responses. J. Plant Physiol. 166, 1646–1659. doi: 10.1016/j.jplph.2009.04.007
Sage, R. F., Christin, P. A., and Edwards, E. A. (2011). The lineages of C4 photosynthesis on planet Earth. J. Exp. Bot. 62, 3155–3169. doi: 10.1093/jxb/err048
Sage, R. F., and Zhu, X.-G. (2011). Exploiting the engine of C4 photosynthesis. J. Exp. Bot. 62, 2989–3000. doi: 10.1093/jxb/err179
Saha, D., Gowda, M. V. C., Arya, L., Verma, M., and Bansal, K. C. (2016). Genetic and genomic resources of small millets. Crit. Rev. Plant Sci. 35, 56–79. doi: 10.1080/07352689.2016.1147907
Singh, R. K., Singh, V. K., Raghavendrarao, S., Phanindra, M. L., Venkat Raman, K., Solanke, A. U., et al. (2015). Expression of finger millet EcDehydrin7 in transgenic tobacco confers tolerance to drought stress. Appl. Biochem. Biotechnol. 177, 207–216. doi: 10.1007/s12010-015-1738-4
Smirnoff, N., and Colombe, S. V. (1988). Drought influences the activity of enzymes of the chloroplast hydrogen-peroxide scavenging system. J. Exp. Bot. 39, 1097–1108. doi: 10.1093/jxb/39.8.1097
Sreenivasulu, N., Miranda, M., Prakash, H. S., Wobus, U., and Weschke, W. (2004). Transcriptome changes in foxtail millet genotypes at high salinity: identification and characterization of a PHGPX gene pecifically upregulated by NaCl in a salt-tolerant line. J. Plant Physiol. 161, 467–477. doi: 10.1078/0176-1617-01112
van der Weerd, L., Claessens, M. M., Ruttink, T., Vergeldt, F. J., Schaafsma, T. J., and Van As, H. (2001). Quantitative NMR microscopy of osmotic stress responses in maize and pearl millet. J. Exp. Bot. 52, 2333–2343. doi: 10.1093/jexbot/52.365.2333
Varshney, R. K., Terauchi, R., and McCouch, S. R. (2014). Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12:e1001883. doi: 10.1371/journal.pbio.1001883
Veeranagamallaiah, G., Chandraobulreddy, P., Jyothsnakumari, G., and Sudhakar, C. (2007). Glutamine synthetase expression and pyrroline-5-carboxylate reductase activity influence proline accumulation in two cultivars of foxtail millet (Setaria italica L.) with differential salt sensitivity. Environ. Exp. Bot. 60, 239–244. doi: 10.1016/j.envexpbot.2006.10.012
Veeranagamallaiah, G., Ranganayakulu, G. S., Thippeswamy, M., Sivakumar, M., Reddy, E. K., Pandurangaiah, M., et al. (2009). Aldose reductase expression contributes in sorbitol accumulation and 4-hydroxynon-2-enal detoxification in two foxtail millet (Setaria italica L.) cultivars with different salt stress tolerance. Plant Growth Regul. 59, 137–143. doi: 10.1007/s10725-009-9396-6
Wang, M., Li, P., Li, C., Pan, Y., Jiang, X., Zhu, D., et al. (2014). SiLEA14, a novel atypical LEA protein, confers abiotic stress resistance in foxtail millet. BMC Plant Biol. 14:290. doi: 10.1186/s12870-014-0290-7
Yadav, C. B., Bonthala, V. S., Muthamilarasan, M., Pandey, G., Khan, Y., and Prasad, M. (2015). Genome-wide development of transposable elements-based markers in foxtail millet and construction of an integrated database. DNA Res. 22, 79–90. doi: 10.1093/dnares/dsu039
Yadav, C. B., Muthamilarasan, M., Pandey, G., Khan, Y., and Prasad, M. (2014). Development of novel microRNA-based genetic markers in foxtail millet for genotyping applications in related grass species. Mol. Breed. 34, 2219–2224. doi: 10.1007/s11032-014-0137-9
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi: 10.1038/nbt.2195
Zhang, J. P., Liu, T. S., Zheng, J., Jin, Z., Zhu, Y., Guo, J. F., et al. (2007). Cloning and characterization of a putative 12-oxophytodienoic acid reductase cDNA induced by osmotic stress in roots of foxtail millet. DNA Seq. 18, 138–144. doi: 10.1080/10425170601060764
Zhang, S., Tang, C., Zhao, Q., Li, J., Yang, L., Qie, L., et al. (2014). Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in Foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 2:15. doi: 10.1186/1471-2164-15-78
Keywords: millets, climate-smart agriculture, foxtail millet, crop improvement, food security, nutrition security
Citation: Bandyopadhyay T, Muthamilarasan M and Prasad M (2017) Millets for Next Generation Climate-Smart Agriculture. Front. Plant Sci. 8:1266. doi: 10.3389/fpls.2017.01266
Received: 03 February 2017; Accepted: 05 July 2017;
Published: 18 July 2017.
Edited by:
Harsh Raman, NSW Department of Primary Industries, AustraliaCopyright © 2017 Bandyopadhyay, Muthamilarasan and Prasad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manoj Prasad, bWFub2pfcHJhc2FkQG5pcGdyLmFjLmlu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.