- 1School of Food Science and Nutrition Engineering, China Agricultural University, Beijing, China
- 2School of Chemical Engineering and Technology, Tianjin University, Tianjin, China
Salt stress affects the plant quality, which affects the productivity of plants and the quality of water storage. In a recent study, we conducted the Technique for Order Preference by Similarity to Ideal Solution (TOPSIS) analysis and RNA-Seq, bioinformatics study methods, and detection of the key genes with qRT-PCR. Our findings suggested that the optimum salt treatment conditions are 200 mM and 19d for the identification of salt tolerance in tomato. Based on the RNA-Seq, we found 17 amino acid metabolic and 17 carbohydrate metabolic pathways enriched in the biological metabolism during the response to salt stress in tomato. We found 7 amino acid metabolic and 6 carbohydrate metabolic pathways that were significantly enriched in the adaption to salt stress. Moreover, we screened 17 and 19 key genes in 7 amino acid metabolic and 6 carbohydrate metabolic pathways respectively. We chose some of the key genes for verifying by qRT-PCR. The results showed that the expression of these genes was the same as that of RNA-seq. We found that these significant pathways and vital genes occupy an important roles in a whole process of adaptation to salt stress. These results provide valuable information, improve the ability to resist pressure, and improve the quality of the plant.
Introduction
Tomato is an important and nutritious vegetable crop worldwide. However, cultivated tomatoes are susceptible under a wide range of environmental pressures (Zhu et al., 2014; Zushi et al., 2014; Sasidharan and Voesenek, 2015). High salinity, for example, can have harmful effects, such as reducing germination, inhibiting growth and reducing fruit production (Xie et al., 2000; Cuartero et al., 2006).
Salt stress can adversely affect the growth and productivity of plants. In the course of growth and development, plants have developed many physiological and biochemical mechanisms to adapt to the stress of the environment (Zhu et al., 2014), and a large number of physiological and metabolic reactions are produced in plants to accommodate this change (Yamaguchi-Shinozaki and Shinozaki, 2006; Hirayama and Shinozaki, 2010; Lata and Prasad, 2011; Tang et al., 2012). By studying the patterns of gene expression in different stress conditions, the adaptive mechanism of different stresses was analyzed (Zhu, 2002).
Salt stress has been observed that amino acids and carbohydrates increase greatly (Fougère et al., 1991). Haitao et al. (2013) demonstrated that arginase expression modulates abiotic stress tolerance in Arabidopsis. It was suggested by Tattini et al. (1996) that in olive leaves, mannitol and glucose play an active role in the osmotic adaptation of plants to salinity. These studies clearly demonstrate that the protein metabolism and carbohydrate metabolism play very important roles in the stress response. We can more fully understand gene function by studying the plant transcriptome. The RNA sequence is a new, efficient and rapid way to conduct transcriptional research and explore the mechanisms that exist in cells (Gal, 1980; Sultan et al., 2008; Trapnell et al., 2010). Despite progress in understanding how protein metabolism and carbohydrate metabolism participate in the adaptation to salt stress, the key genes and signaling pathways involved in these processes remain unclear.
Therefore, we use the RNA-seq screen for obvious pathways and key genes and protein metabolism and carbohydrate metabolism based on the Web gene ontology (GO) (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (http://www.funnet.ws/). TOPSIS (the Technique for Order Preference by Similarity to Ideal Solution) also test and determine the salt concentration and processing time, to provide a base for other researchers. Our aim is to explore the mechanism of protein metabolism regulation and carbohydrate metabolism regulation during the salt stress response in tomato and to provide valuable information to enhance the ability to resist pressure, and improve the quality of the plant. The results of the study can also be used as a reference for the selection of candidate genes in tomato for further functional characterization.
Materials and Methods
Plant Materials and Growth Conditions
Tomato (Solanum lycopersicum) seeds included the Micro-Tom cultivar. A total of 120 seeds of uniform size were surface-sterilized in 70% (v/v) ethanol for 15 s, followed by 4% (w/v) sodium hypochlorite for 15 min, and then use sterile distilled water rinse several times (Cavalcanti et al., 2004). The seeds were germinated at 24°C on wet filter paper and under darkness. When a 0.3 cm-radicle protrusion appeared, the seeds were laid on an agar-solidified MS medium (Murashige and Skoog, 1962) in triplicate, without or supplemented with 50–250 mM sodium chloride (NaCl). They were then moved to an incubator at 24°C in a dark 16 h/light 8 h system. Additionally, the plant height, leaf blade number and antioxidant enzyme activities were measured and analyzed after 7 d, 11 d, 15 d, 19 d, and 23 d treatment. The POD, SOD, and CAT activities were determined according to previous work (Cavalcanti et al., 2004).
Comprehensive Evaluation of Salt Treatment Conditions
The Technique for Order Preference by Similarity to Ideal Solution (TOPSIS) was provided by Hwang and Yoon (1981). The detailed steps are listed below:
Step 1: Create a ranking decision matrix:
where xij is the rating of alternative Ai (m) with respect to the criterion Cj(n) evaluated.
Step2: Calculate the normalized decision matrix rij:
Step 3: A weighted standardized decision matrix is constructed by multiplying the standardized decision matrix times the associated weights. The weighted standardized value vij is calculated in the following way:
Where wi is the weight of the jth criterion.
Step 4: Identify positive and negative ideal solutions:
Where I is associated with the benefit criteria and J is associated with the cost criteria.
Step 5: Use Measure two Euclidean distances for both the positive and the negative.
where dj+ and dj− represent the distance from Aj which is expressed separately from the ideal solution of positive and negative.
Step 6: Calculate the relative closeness of the ideal solution and compare the Rj value to the alternatives.
where Rj represents the relative closeness.
All treatments have three replication operations presented as the mean ± the standard deviation of each experiment. The correlations were estimated using Pearson's correlation coefficient in the IBM SPSS Statistics 20 (SPSS, Chicago, Illinois, USA) software package. We chose the tomato material to be used for RNA-Seq according to the results of the correlation analysis and the TOPSIS analysis.
RNA-Seq Analysis
For building the RNA library based on mRNA-seq Illumina company, the tomatos were sequenced using HiSeq 2000 from Shanghai OE Biotech. Co., Ltd. Each sample use 50 ng. The results were compared with the database, each of which was anotated for a follow-up experiment. The standard for RNA is RIN ≥ 7, 28S/18S>0.7. Both the control and salt treated samples had three repetitions, respectively.
Quantitative Real-Time PCR (qRT-PCR) Verification
Extract the total RNA use the RNeasy extraction tool. Then reverse-transcribed it into cDNA for the next analysis. Based on the equipment instructions, we used the SYBR fluorescent reagent and the 7900 system to conduct real-time PCR, and analyzed data using Microsoft Excel with the 2−ΔΔCt relative quantitative method.
Results
Comprehensive Evaluations of the Salt Concentration and Treatment Time
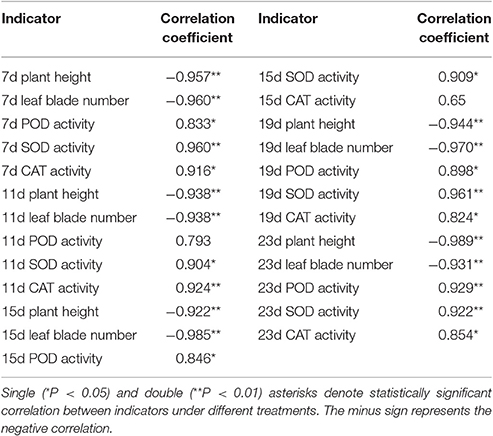
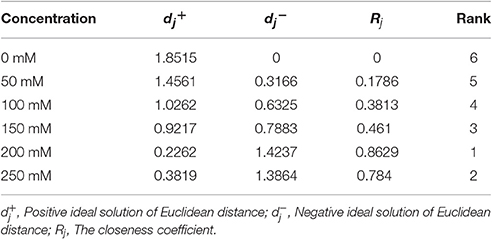
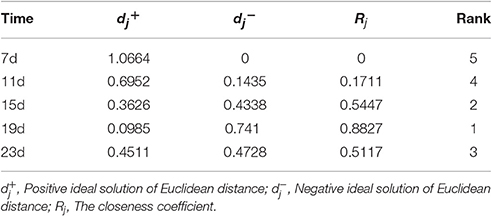
Based on a correlation analysis (Table 1), all of the indicators are strongly correlated with the salt concentration, suggesting effective indicators that represent the salt tolerance of tomato. The TOPSIS analysis revealed the rank of Rj considering all indicator values for different salt concentrations during the response to salt stress (Table 2); 200 mM was the optimum NaCl concentration for the identification of salt tolerance. Furthermore, the TOPSIS analysis examined the rank of Rj considering of the indicator values for different salt treatment times under the 200 mM NaCl treatment (Table 3) and indicated that 19 d is the optimum salt treatment time for the identification of salt tolerance. Accordingly, tomatoes treated without or with 200 mM NaCl for 19 d were chosen for further RNA-Seq analysis.
The Differential Expression Analysis and Go Classification of Expressed Genes
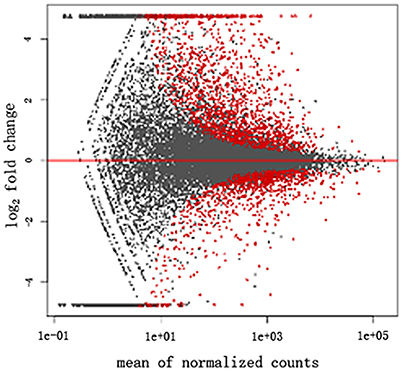
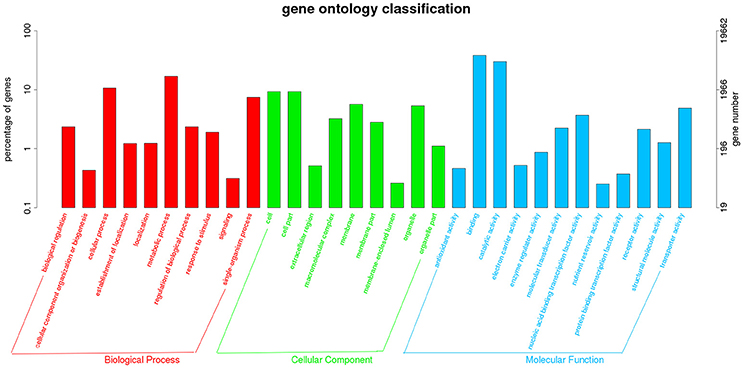
The transcriptome data were analyzed with RNA-Seq technology. Based on RNA-seq, we did the variance analysis and the differential expression gene was selected according to the standard of P < 0.05. The false discovery rate (FDR) was set to 0.001 to determine the threshold of the P-value for multiple tests. The absolute value of |log2Ratio| ≥ 1 was used to determine the difference between the gene expression transcription group and the database. The 3,792 different genes were discovered during the salt stress response, including 1,498 up-regulated genes and 2,294 down-regulated genes (Figure 1). Gene function, annotation, and classification were researched by GO analysis. The 2,776 genes were founded to participated in the metabolic processes (Figure 2).
Pathway Enrichment Analysis of Differentially Displayed Genes in the Amino Acid Metabolic Pathway and Carbohydrate Metabolic Pathway
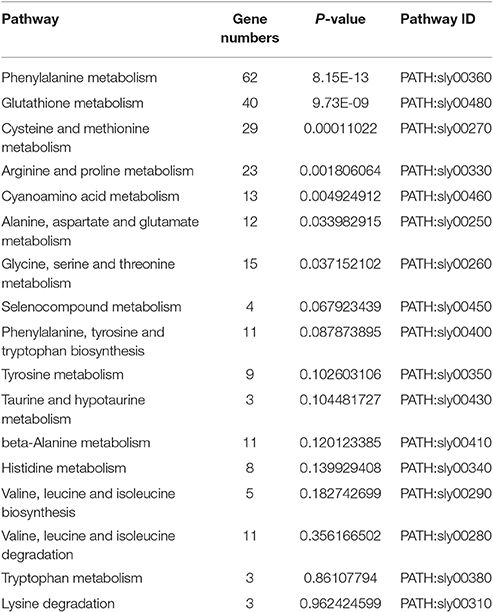
The KEGG database analysis revealed 45 biological metabolic pathways involved in the response to salt stress, including 17 essential amino acid metabolic pathways. A total of 262 genes that participated in amino acid metabolism were found to be enriched in the transcriptome. In these amino acid metabolic pathways, there are seven distinct pathways (P < 0.05): phenylalanine metabolism; glutathione metabolism; cysteine and methionine metabolism; arginine and proline metabolism; cyanoamino acid metabolism; alanine, aspartate and glutamate metabolism; and glycine, serine and threonine metabolism. The remaining 10 metabolic pathways exhibited no significant difference (Table 4, Figure 3A).
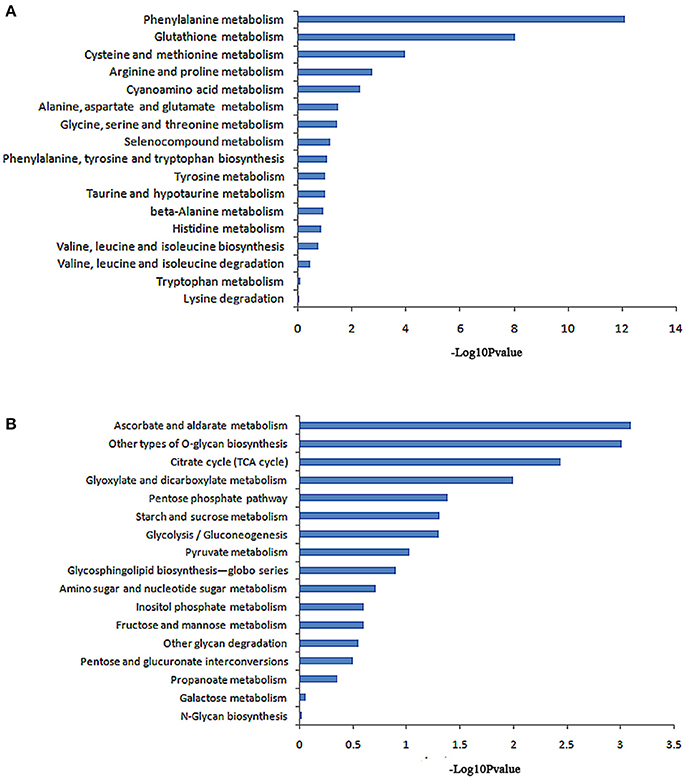
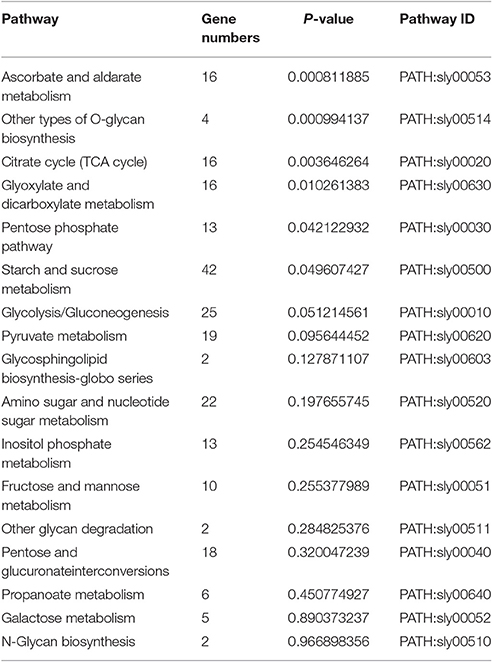
Figure 3. Pathway enrichment analysis of differentially displayed genes. (A) Amino acid metabolic pathway, (B) Carbohydrate metabolic pathway.
A total of 231 genes that participated in carbohydrate metabolism were found to be enriched in the transcriptome. KEGG database analysis revealed 17 carbohydrate metabolism pathways. In these pathways, six greatly different pathways were detected (P < 0.05): an ascorbate and aldarate metabolic pathway; other types of O-glycan biosynthesis; a citrate cycle (TCA cycle); glyoxylate and dicarboxylate metabolism; a pentose phosphate pathway; and starch and sucrose metabolism. The remaining 11 metabolic pathways showed no significant difference (Table 5, Figure 3B).
Screening of Key Regulatory Genes Participating in Amino Acid Metabolism and Carbohydrate Metabolism during the Response to Salt Stress
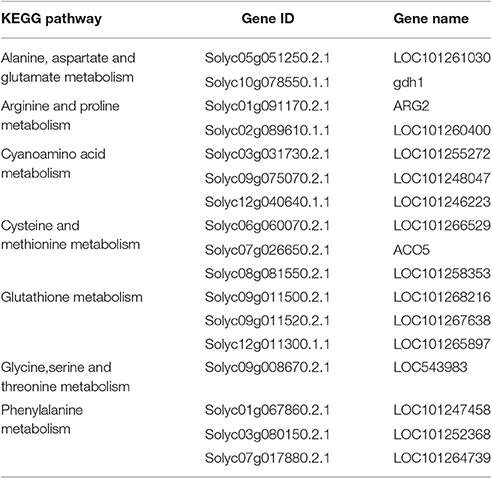
In addition to P < 0.05, we further screened differentially expressed genes with the two new standards to ensure the availability of screening results: fragments per kb per million reads (fpkm)>0.05 and |log2Fold Change| ≥ 1. Based on these standards, we screened 17 genes in seven amino acid metabolic pathways (Table 6) and 19 genes in six carbohydrate metabolic pathways (Table 7). To forecast the expression profiles of the genes during the tomato salt tolerance process, we conducted a cluster analysis using a heat map (Figure 4). As shown in Figure 4A, Solyc05g051250.2.1 and Solyc02g089610.1.1 exhibited an increase in expression during the response to salt stress. The opposite expression patterns were observed in the15 other genes in the amino acid metabolic pathway. As shown in Figure 4B, Solyc06g062430.2.1, Solyc09g007270.2.1, Solyc07g055840.2.1, Solyc05g051250.2.1, Solyc10g007600.2.1, Solyc01g110360.2.1, and Solyc07g064180.2.1 exhibited an increase in expression during the salt stress response. Opposite expression patterns were observed for 12 other genes in the carbohydrate metabolic pathway.
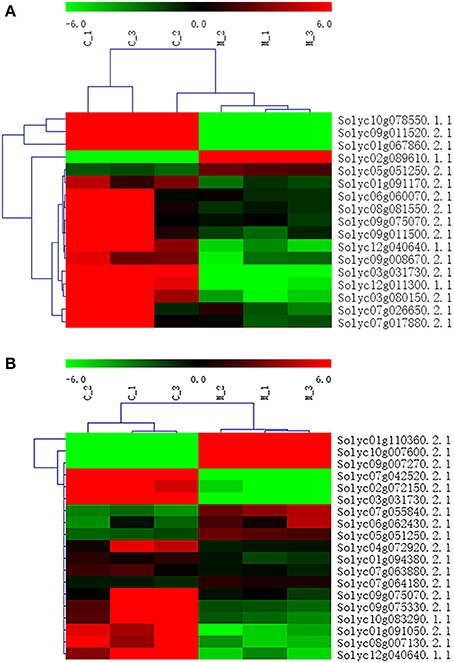
Figure 4. Heat map representation of differentially displayed genes. (A) Amino acid metabolic pathway, (B) Carbohydrate metabolic pathway.
qRT-PCR Verification
Some key genes were chosen in Tables 6, 7. We analyed their expression levels in tomatoes treated without NaCl as a control and in tomatoes treated with 20 mM NaCl (Figure 5). According to the results, Solyc05g051250.2.1 was up-regulated, which is involved in Alanine, aspartate and glutamate metabolism, by contrast, Solyc01g091170.2.1 (ARG2) involved in Arginine and proline metabolism, Solyc09g075070.2.1 involved in Cyanoamino acid metabolism, Solyc06g060070.2.1 and Solyc07g026650.2.1 (ACO5) involved in Cysteine and methionine metabolism, Solyc12g011300.1.1 involved in Glutathione metabolism, Solyc09g008670.2.1 involved in Glycine,serine and threonine metabolism and Solyc01g067860.2.1 involved in Phenylalanine metabolism were down-regulated (Figure 5A). Meanwhile, Solyc06g062430.2.1 involved in Ascorbate and alarate metabolism, Solyc07g055840.2.1 involved in Citrate cycle (TCA cycle), Solyc05g051250.2.1 involved in Glyoxylate and dicarboxylate metabolism and Solyc01g110360.2.1 involved in Pentose phosphate pathway were up-regulated, by contrast, Solyc01g094380.2.1 involved in Other types of O-glycan biosynthesis, Solyc09g075330.2.1 and Solyc10g083290.1.1 (Wiv-1) involved in Starch and sucrose metabolism were down-regulated (Figure 5B). The expression of these genes is mainly the same as the RNA-seq results.
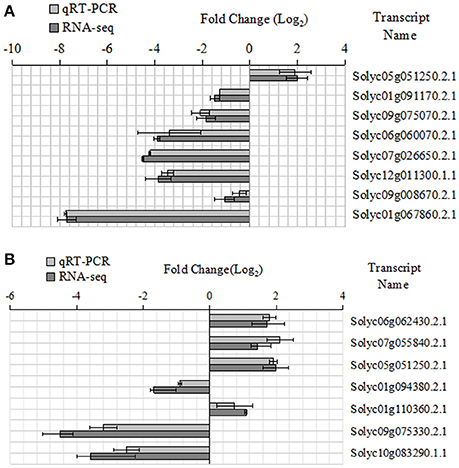
Figure 5. Expression patterns of the different genes between the qRT-PCR and RNA-seq. (A) Amino acid metabolic pathway, (B) Carbohydrate metabolic pathway.
Discussion
The adaptation of plant cells in salt stress is closely relative to various metabolic processes. It has been suggested that proline metabolism (Khedr et al., 2003), ascorbic acid (ASA) metabolism (Zushi et al., 2014), and other pathways play important roles during the salt stress response. In these metabolic processes, amino acid and carbohydrates are the main players in various metabolic and regulatory pathways. They are responsible for biological cell adaptation. Therefore, amino acid metabolism and carbohydrate metabolism are crucial to the salt stress response. In our study, we found that in the adaptation process, the differentially expressed genes were significantly enriched in 7 amino acid metabolic pathways and in 6 carbohydrate metabolic pathways.
Many studies have found that amino acid metabolism is closely related to abiotic stress tolerance (Fougère et al., 1991; Zörb et al., 2004; Haitao et al., 2013). We identified 7 significant amino acid metabolic pathways and verified 8 key genes. In our study, the ARG2 gene encoding arginase 2 (Solyc01g091170.2.1), involved in arginine and proline metabolism, was significantly enriched. Arginine is an vital amino acid to transport and storage the nitrogen and occupy a precursor to synthesis other amino acids or polyamines (Flores et al., 2008; Brauc et al., 2012). The arginase 2 genes variations associate with steroid response, which probably plays an important role in asthma development, severity and progression (Vonk et al., 2010). The AtARG2-knockout Arabidopsis could enhance environmental stress tolerance compared with the wild type, and increased tolerance was suggested by changes in physiological parameters, containing electrolyte leakage, water loss, porosity and surial rate (Haitao et al., 2013). However, in the past, most attention has focused on the function of proline which occupy a compatible osmolyte (Yancey et al., 1982) and osmoprotectant (Serrano and Gaxiola, 1994), It is much less concerned with the further role of stress tolerance. Our study suggested that proline metabolic pathways exhibit significant differences during the salt stress response (P < 0.05). Consistently, Khedr et al. (2003) reported that proline induces the expression of salt-stress-responsive proteins, which may increase the adaptation of Pancratium maritimum L. to response salt stress. The gene encoding beta-glucosidase 11-like (Solyc09g075070.2.1) was down-regulated in our study, which is involved in Cyanoamino acid metabolism. The role of beta-glucosidase is the hydrolysis of terminal, non-reducing beta-D-glucose residues with the increase of beta-D-glucose. Compared to the control group, the enzyme increased in the corn roots and buds (Zörb et al., 2004). Structure of acid beta-glucosidase with pharmacological chaperone provides insight into Gaucher disease (Raquel et al., 2007). López-Berenguer et al. (2008) demonstrated that cysteine and methionine decreased significantly in broccoli after salt application. Furthermore, the expression of the ACO5 gene encoding the 1-aminocyclopropane-1-carboxylate oxidase (Solyc07g026650.2.1), which is involved in cysteine and methionine metabolism, was observed to be down-regulated throughout the salt stress response. An increase in ethylene biosynthesis of arabidopsis is releated to the transcript of ACO5 under waterlogging stress (López-Berenguer et al., 2008; Sasidharan and Voesenek, 2015). ACO5 osmoregulation in cotton could probably response to water stress (Tschaplinski and Blake, 1989). ACO5 is the specific target gene of the NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF], and cup-shaped cotyledon [CUC2]) transcription factors), and many studies have found that NAC is involved in the plant abiotic stressresponse (Xie et al., 2000; Nuruzzaman et al., 2010; Rauf et al., 2013). Hernández et al. (2000) found that the activity of glutathione reductase (GR) accumulated in the salt-tolerant pea compared with the salt-sensitive cultivar after long-term NaCl treatment, indicating that glutathione metabolism is participated in the adaption to salt stress. In our study, the gene encoding a probable glutathione S-transferase (Solyc12g011300.1.1) involved in glutathione metabolism was significantly down-regulated. Cho et al. (2001) reported that Glutathione S-Transferase Mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. Roxas et al. (1997) found that hyperexpression glutathione s-transferase enhanced the growth of genetically modified tobacco seedlings suffering stress. Moreover, the gene encoding peroxidase 24 (Solyc01g067860.2.1), which is involved in phenylalanine metabolism, was observed to be significantly down-regulated. Huang et al. (2009) reported that a rice peroxidase 24 precursor was down-regulated and could exhibit peroxidase activity ofscavenging H2O2 under salt treatment. The total peroxidase activity improved under salt stress, and elevation activity depend on NaCl concentration (Sreenivasulu et al., 1999). SE-glutathione peroxidase play the important role for cell survival against oxidative stress (Carine et al., 1994).
Many studies have shown that carbohydrate metabolism occupies a vital function in abiotic stress tolerance (Fougère et al., 1991; Zörb et al., 2004; Haitao et al., 2013). We identified 6 significant carbohydrate metabolic pathways and verified 7 key genes. Downton (1977) found that NaCl-stressed grapevine leaves included decreased sucrose and starch but improved the reducing sugars levels. In this study, Wiv-1 (Solyc10g083290.1.1), encoding the acid invertase involved in starch and sucrose metabolism, was down-regulated during the response to salt stress. Consistently, Dubey and Singh (1999) reported that acid invertase activity decreased on the shoots of NaCl-tolerant rice but increased on NaCl-sensitive rice. It is suggested that the increasion of sugars and other compatible solutes, contributes to osmotic adjustment under salt stress. In addition, Kim et al. (2000) reported that a maize vacuolar invertase is induced by water stress. In our study, the gene encoding fructose-bisphosphate aldolase 1 (Solyc01g110360.2.1), which is participated in the pentose phosphate pathway, was significantly up-regulated. Chaves et al. (2009) also demonstrated that fructose-bisphosphate aldolase was differently affected by salt stress. Lu et al. (2012) reported that fructose 1,6-bisphosphate aldolase genes in Arabidopsis response to abiotic stresses. The gene encoding citrate synthase 3 (Solyc07g055840.2.1) was up-regulated in our study, which is participated in the Citrate cycle (TCA cycle) pathway. Citrate synthase activity is an indicator of metabolic potential in mitochondria and should increase if there is a net increase in the volume of mitochondria in the tissue. However, there was no difference in citrate synthase activity among freshwater- and seawater-acclimated fish (Marshall et al., 1999). The gene encoding myo-inositol oxygenase 1 (Solyc06g062430.2.1) was up-regulated in our study, which is participated in the Ascorbate and alarate metabolism. The vital function of myo-inositol is showed to be relative to osmotic balance and transport the Na + from roots to shoots (Nelson and Bohnert, 1999). The activity of inositol and inositol oxygenase has been found to catalyze the oxidation of the d-glucuronate free inositol (Lorence et al., 2004). Cotsaftis et al. (2011) reported that myo-inositol oxygenase was down-regulated in salt-tolerant rice.
The gene encoding glutamine synthetase-like (Solyc05g051250.2.1) was up-regulated in our study and is involved in both amino acid and carbohydrate metabolism. Glutamine synthetase is beneficial to nitrogen assimilation and has been founded in prokaryotes and eukaryotes (Doskočilová et al., 2011). Viégas and Silveira (1999) found that salinity imposed sensitivity on nitrogen assimilation. The higher the sensitivity imposed, the more severe the salt-injurious affects on plant development. Meanwhile, uptake and its assimilation were suggested to limit the nitrogen assimilation under salt stress (Silveira et al., 2001). Furthermore, genes involved in starch and sucrose metabolism (Solyc09g075330.2.1), cysteine and methionine metabolism (Solyc06g060070.2.1), glycine, serine and threonine metabolism (Solyc09g008670.2.1) and other types of o-glycan biosynthesis (Solyc01g094380.2.1) were observed to exhibit significant expression after salt stress, indicating that they were involved in the response to salt stress, but their impact on stress adaptation has not yet been reported. Therefore, these specific regulatory mechanisms need to be explored in future.
In conclusion, 200 mM and 19d are the optimum salt treatment conditions for the identification of salt tolerance in tomato. Based on RNA-Seq, we analyzed the metabolic pathways and identified some of the relevant critical genes participating in amino acid metabolism and carbohydrate metabolism during the response to salt stress in tomato. These metabolic pathways and key genes play vital roles to response salt stress, which provides valuable information to enhance the ability to resist pressure, improve the quality of the plant and lay a solid foundation for future research.
Author Contributions
XK planned the research. ZZ, CM, and ZS conducted experiments and data acquisition. ZZ wrote the manuscript closely with all authors. XK and CM modified the manuscript. All the authors were involved in many discussion and revised the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant Number 31671899 and Grant Number 31470091).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to Dr. Daqi Fu (School of Food Science and Nutrition Engineering, China Agricultural University) for the tomato (cv Ailsa Craig) seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01231/full#supplementary-material
References
Brauc, S., De, V. E., Claeys, M., Geuns, J. M., Höfte, M., and Angenon, G. (2012). Overexpression of arginase in Arabidopsis thaliana influences defence responses against botrytis cinerea. Plant Biol. 14(Suppl. 1), 39–45. doi: 10.1111/j.1438-8677.2011.00520.x
Carine, M., Martine, R., Olivier, T., and Jose, R. (1994). Importance of SE-glutathione peroxidase, catalase, and CU/ZN-SOD for cell survival against oxidative stress. Free Radic. Biol. Med. 17, 235–248. doi: 10.1016/0891-5849(94)90079-5
Cavalcanti, F. R., Martins-Miranda, A. S., and Viégas, R. A. (2004). Superoxide dismutase, catalase and peroxidase activities do not confer protection against oxidative damage in salt-stressed cowpea leaves. New Phytol. 163, 563–571. doi: 10.1111/j.1469-8137.2004.01139.x
Chaves, M. M., Flexas, J., and Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. doi: 10.1093/aob/mcn125
Cho, S., Lee, Y., Park, H., Ryoo, K., Kang, K., Park, J. M., et al. (2001). Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1 the journal of biological Chemistry 276, 12749–12755. doi: 10.1074/jbc.M005561200
Cotsaftis, O., Plett, D., Johnson, A. A. T., Walia, H., Wilson, C., Ismail, A. M., et al. (2011). Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol. Plant. 4, 25–41. doi: 10.1093/mp/ssq056
Cuartero, J., Bolarín, M. C., Asíns, M. J., and Moreno, V. (2006). Increasing salt tolerance in the tomato. J. Exp. Bot. 57, 1045–1058. doi: 10.1093/jxb/erj102
Doskočilová, A., Plíhal, O., Volc, J., Chumová, J., Kourová, H., and Halada, P. (2011). A nodulin/glutamine synthetase-like fusion protein is implicated in the regulation of root morphogenesis and in signalling triggered by flagellin. Planta 234, 459–476. doi: 10.1007/s00425-011-1419-7
Downton, W. J. S. (1977). Photosynthesis in salt-stressed grapevines. Austral. J. Plant Physiol. 4, 183–192. doi: 10.1071/PP9770183
Dubey, R. S., and Singh, A. K. (1999). Salinity induces accumulation of soluble sugars and alters the activity of sugar metabolising enzymes in rice plants. Biol. Plant. 42, 233–239. doi: 10.1023/A:1002160618700
Flores, T., Todd, C. D., Tovarmendez, A., Dhanoa, P. K., Correaaragunde, N., Hoyos, M. E., et al. (2008). Arginase-negative mutants of Arabidopsis exhibit increased nitric oxide signaling in root development. Plant Physiol. 147, 1936–1946. doi: 10.1104/pp.108.121459
Fougère, F., Rudulier, D. L., and Streeter, J. G. (1991). Effects of salt stress on amino acid, organic acid, and carbohydrate composition of roots, bacteroids, and cytosol of alfalfa (Medicago sativa L.). Plant Physiol. 96, 1228–1236. doi: 10.1104/pp.96.4.1228
Gal, T. (1980). Multiple Objective Decision Making - Methods and Applications: A State-of-the Art Survey, Vol. 375. New York, NY: Springer-Verlag, Inc. 1–531.
Haitao, S., Tiantian, Y., Fangfang, C., Zhangmin, C., Yanping, W., Pingfang, Y., et al. (2013). Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 64, 1367–1379. doi: 10.1093/jxb/ers400
Hernández, J. A., Jiménez, A., Mullineaux, P., and Sevilia, F. (2000). Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 23, 853–862. doi: 10.1046/j.1365-3040.2000.00602.x
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Huang, X. Y., Chao, D. Y., Gao, J. P., Zhu, M. Z., Min, S., and Lin, H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. doi: 10.1101/gad.1812409
Hwang, C. L., and Yoon, K. (1981). Multiple Attribute Decision Making: Methods and Applications, a State of the Art Survey. New York, NY: Springer-Verlag.
Khedr, A. H. A., Abbas, M. A., Wahid, A. A. A., Quick, W. P., and Abogadallah, G. M. (2003). Proline induces the expression of salt-stress-responsive proteins and may improve the adaptation of Pancratiummaritimum L. to salt-stress. J. Exp. Bot. 54, 2553–2562. doi: 10.1093/jxb/erg277
Kim, J. Y., Mahe, A., Brangeon, J., and Prioul, J. L. (2000). A maize vacuolar invertase, IVR2, is induced by water stress. Organ/tissue specificity and diurnal modulation of expression. Plant Physiol. 124, 171–184. doi: 10.1104/pp.124.1.71
Lata, C., and Prasad, M. (2011). Role of drebs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
López-Berenguer, C., Martínez-Ballesta, M. C., García-Viguera, C., and Carvajal, M. (2008). Leaf water balance mediated by aquaporins under salt stress and associated glucosinolate synthesis in broccoli. Plant Sci. 174, 321–328. doi: 10.1016/j.plantsci.2007.11.012
Lorence, A., Chevone, B. I., Mendes, P., and Nessler, C. L. (2004). Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 134, 1200–1205. doi: 10.1104/pp.103.033936
Lu, W., Tang, X. L., Huo, Y. Q., Xu, R., Qi, S. D., Huang, J. G., et al. (2012). Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 503, 65–74. doi: 10.1016/j.gene.2012.04.042
Marshall, W. S., Emberley, T. R., Singer, T. D., Bryson, S. E., and Mccormick, S. D. (1999). Time course of salinity adaptation in a strongly euryhaline estuarine teleost, fundulus heteroclitus: a multivariable approach. J. Exp. Biol. 202, 1535–1544.
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nelson, D. E., and Bohnert, H. J. (1999). Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol. 119, 165–172. doi: 10.1104/pp.119.1.165
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Raquel, L., Brandon, A. W., Pedro, H., Allan, C., Corey, W., Richie, K., et al. (2007). Structure of acid [beta]-glucosidase with pharmacological chaperone provides insight into Gaucher disease. Nat. Chem. Biol. 3, 101–107. doi: 10.1038/nchembio850
Rauf, M., Arif, M., Fisahn, J., Xue, G. P., Balazadeh, S., and Muellerroeber, B. (2013). NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 25, 4941–4955. doi: 10.1105/tpc.113.117861
Roxas, V. P. Jr., Allen, E. R., and Allen, R. D. (1997). Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15, 988–991. doi: 10.1038/nbt1097-988
Sasidharan, R., and Voesenek, L. A. C. J. (2015). Ethylene-mediated acclimations to flooding stress. Plant Physiol. 169, 3–12. doi: 10.1104/pp.15.00387
Serrano, R., and Gaxiola, R. (1994). Microbial models and salt stress tolerance in plants. Crit. Rev. Plant Sci. 13, 121–138. doi: 10.1080/07352689409701911
Silveira, J. A. G., Melo, A. R. B., Viégas, R. A., and Oliveira, J. T. A. (2001). Salinity-induced effects on nitrogen assimilation related to growth in cowpea plants. Environ. Exp. Bot. 46, 171–179. doi: 10.1016/S0098-8472(01)00095-8
Sreenivasulu, N., Ramanjulu, S., Ramachandra-Kini, K., Prakash, H. S., Shekar-Shetty, H., Savithri, H. S., et al. (1999). Total peroxidase activity and peroxidase isoforms as modified by salt stress in two cultivars of fox-tail millet with differential salt tolerance. Plant Sci. 141, 1–9. doi: 10.1016/S0168-9452(98)00204-0
Sultan, M., Schulz, M. H., Richard, H., Magen, A., Klingenhoff, A., Scherf, M., et al. (2008). A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321, 956–960. doi: 10.1126/science.1160342
Tang, N., Zhang, H., Li, X., Xiao, J., and Xiong, L. (2012). Constitutive activation of transcription factor osbzip46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Tattini, M., Gucci, R., Romani, A., Baldi, A., and Everard, J. D. (1996). Changes in non-structural carbohydrates in olive (Olea europaea) leaves during root zone salinity stress. Physiol. Plant. 98, 117–124. doi: 10.1111/j.1399-3054.1996.tb00682.x
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tschaplinski, T. J., and Blake, T. J. (1989). Water-stress tolerance and late-season organic solute accumulation in hybrid poplar. Can. J. Bot. 67, 1681–1688. doi: 10.1139/b89-212
Viégas, R. A., and Silveira, J. D. (1999). Ammonia assimilation and proline accumulation in young cashew plants during long term exposure to NaCl-salinity. Revistabrasileira de fisiología vegetal. 11, 153–159.
Vonk, J. M., Postma, D. S., Maarsingh, H., Bruinenberg, M., Koppelman, G. H., and Meurs, H. (2010). Arginase 1 and arginase 2 variations associate with asthma, asthma severity and 2 agonist and steroid response. Pharmacogenet. Genomics 20, 179–186. doi: 10.1097/FPC.0b013e328336c7fd
Xie, Q., Frugis, G., Colgan, D., and Chua, N.-H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Develop. 14, 3024–3036. doi: 10.1101/gad.852200
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D., and Somero, G. N. (1982). Living with water stress: evolution of osmolyte systems. Science 217, 1214–1222. doi: 10.1126/science.7112124
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, M., Chen, G., Zhang, J., Zhang, Y., Xie, Q., Zhao, Z., et al. (2014). The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (Solanum lycopersicum). Plant Cell Rep. 33, 1851–1863. doi: 10.1007/s00299-014-1662-z
Zörb, C., Schmitt, S., Neeb, A., Karl, S., Linder, M., and Schubert, S. (2004). The biochemical reaction of maize (Zea mays L.) to salt stress is characterized by a mitigation of symptoms and not by a specific adaptation. Plant Sci. 167, 91–100. doi: 10.1016/j.plantsci.2004.03.004
Keywords: RNA-seq, tomato, salt stress, amino acid metabolism, carbohydrate metabolism, TOPSIS
Citation: Zhang Z, Mao C, Shi Z and Kou X (2017) The Amino Acid Metabolic and Carbohydrate Metabolic Pathway Play Important Roles during Salt-Stress Response in Tomato. Front. Plant Sci. 8:1231. doi: 10.3389/fpls.2017.01231
Received: 07 May 2017; Accepted: 29 June 2017;
Published: 17 July 2017.
Edited by:
Junhua Peng, Center for Life Sci & Tech of China National Seed Group Co., Ltd., ChinaReviewed by:
Liang Chen, University of Chinese Academy of Sciences (UCAS), ChinaXian Li, Zhejiang University, China
Copyright © 2017 Zhang, Mao, Shi and Kou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohong Kou, a291eGlhb2hvbmdAdGp1LmVkdS5jbg==
 Zhi Zhang1
Zhi Zhang1 Xiaohong Kou
Xiaohong Kou