
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 July 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01221
This article is part of the Research Topic Plant Pathogenic Ralstonia spp. From the Field to the Lab and Back Again: mechanisms of pathogen virulence and host resistance, population biology, community ecology and strategies for bacterial wilt disease management. View all 34 articles
The current bacterial wilt infestation level in the potato fields in the Peruvian Andes was investigated by collecting stem samples from wilted plants and detecting Ralstonia solanacearum. In total 39 farmers’ fields located in the central and northern Peru between the altitudes 2111 and 3742 m above sea level were sampled. R. solanacearum was detected in 19 fields, and in 153 out of the 358 samples analyzed. Phylogenetic analysis using the partial sequence of the endoglucanase gene on strains collected in Peru between 1966 and 2016 from potato, pepper, tomato, plantain or soil, divided the strains in phylotypes I, IIA, and IIB. The Phylotype IIB isolates formed seven sequevar groups including the previously identified sequevars 1, 2, 3, 4, and 25. In addition to this, three new sequevars of phylotype IIB were identified. Phylotype IIA isolates from Peru clustered together with reference strains previously assigned to sequevars 5, 39, 41, and 50, and additionally one new sequevar was identified. The Phylotype I strain was similar to the sequevar 18. Most of the Peruvian R. solanacearum isolates were IIB-1 strains. In the old collection sampled between 1966 and 2013, 72% were IIB-1 and in the new collection at 2016 no other strains were found. The pathogenicity of 25 isolates representing the IIA and IIB sequevar groups was tested on potato, tomato, eggplant and tobacco. All were highly aggressive on potato, but differed in pathogenicity on the other hosts, especially on tobacco. All IIA strains caused latent infection on tobacco and some strains also caused wilting, while IIB strains caused only few latent infections on this species. In conclusion, high molecular diversity was found among the R. solanacearum strains in Peru. Most of the variability was found in areas that are no longer used for potato cultivation and thus these strains do not pose a real threat for potato production in the country. Compared to the previous data from the 1990s, the incidence of bacterial wilt has decreased in Peru. The epidemics are likely caused by infected seed tubers carrying the clonal brown rot strain IIB-1.
Potato is an important food security crop with great potential for poverty alleviation and combating malnutrition in the developing world (FAO, 2009; Devaux et al., 2014). However, significant yield gaps have been observed in the small-holder’s fields caused by several constraints such as poor seed quality, soil fertility and plant pathogens (Gildemacher et al., 2011). One of the most notorious diseases affecting potato production in the developing world is bacterial wilt, caused by Ralstonia solanacearum. It is part of a diverse species complex (Fegan and Prior, 2005) affecting many plant species (Kelman, 1953; Hayward, 1994), including potato, tobacco, tomato, banana, peanut, and ginger (Elphinstone, 2005). The bacterium is difficult to control because it survives in water and in residues of infected plants in the soil. (Graham et al., 1979; Pradhanang et al., 2000; Caruso et al., 2005). Distribution of asymptomatic infected plant material has resulted in establishment of this pathogen in numerous ecoregions around the world (Hayward, 1991).
Recently, based on the combination of genomic and proteomic methods the taxonomy of the R. solanacearum species complex was revised to consist of three species: R. pseudosolanacearum, R. solanacearum, and R. syzygii (Safni et al., 2014; Prior et al., 2016). The species division is congruent with the previous Phylotype classification based on the sequence analysis of the ITS region and further subdivision by the DNA sequence of the endoglucanase gene (egl) (Fegan and Prior, 2005) so that the Phylotype II strains belong to the species R. solanacearum, Phylotype IV strains to R. syzygii and Phylotypes I and III to R. pseudosolanacearum. Before the use of DNA based analysis R. solanacearum was subdivided into five races based on host range (Buddenhagen et al., 1962; Pegg and Moffett, 1971; He et al., 1983; Hayward, 1991) and to five biovars, based on the capacity to use disaccharides or oxidize several hexose alcohols (Hayward, 1964; He et al., 1983; Hayward et al., 1989; French et al., 1995). At present the most common characterization method for Ralstonia populations involves partial sequencing of the egl gene coding for the Endoglucanase enzyme and assigning the strains into sequevars based on phylogenetic analysis (Fegan and Prior, 2005). Endoglucanase has an important role in pathogenicity of Ralstonia solanacearum being necessary for efficient invasion of plant roots and colonization of stems (Roberts et al., 1988; Denny et al., 1990).
Potato bacterial wilt is caused mostly by race 3/biovar 2A, which is now classified as phylotype IIB/ sequevar 1 (IIB-1) (Janse, 1996; Perez et al., 2008; Cellier and Prior, 2010; Siri et al., 2011). which probably originated from the Brazilian Amazon basin and can have co-evolved with many plant species (Wicker et al., 2012; Santiago et al., 2016). This potato infecting taxon has spread world-wide and the reference sequences from all over the world are highly homologous suggesting clonal propagation and a relatively recent international dissemination. In addition to the dominating IIB-1 strain, also other phylogenetically distinct strains have been found infecting potato, such as IIB-3, IIB-25, IIA-7 and IIA-35 (Cellier and Prior, 2010; Hong et al., 2012; Santiago et al., 2016).
In Peru, bacterial wilt was first recognized in banana and plantain (Revilla, 1967) and in tomato along the Amazon river (French and Sequeira, 1968, 1970). In potato, it was first discovered in the highlands of Ancash, Cajamarca, and La Libertad (Herrera and French, 1970) and in central Peru in San Ramon (Torres et al., 1985). Surprisingly it was also found in potato planted on virgin soils in the Amazon basin in the Northern part of the country in Yurimaguas suggesting that it was endemic to the area (Martin et al., 1981). Peruvian R. solanacearum isolates have been extensively characterized for phenotypic markers, such as biovar, virulence and pathogenicity using several hosts. These studies identified that the biovars 1, 2 and 3 exist in Peru and all can infect potato, however, biovar 3 is less pathogenic on potato (Martin et al., 1981). Biovar 2 was further divided in two different phenotypes: 2T (first known as phenotype A) and 2A (phenotype B), which had different virulence patterns and host ranges (French et al., 1993; Marin and El-Nashar, 1993). In addition, biovar 2A is much more aggressive than biovars 1 and 2T on potato (Gutarra et al., 2014). Thus, it has been known that the R. solanacearum population infecting potato in Peru is variable, but a detailed molecular characterization of the isolates has not been done. Similarly, the current infestation level of the potato fields in the main productions areas is unknown.
The objective of this study was to analyze the occurrence, genetic diversity and pathogenicity of R. solanacearum bacterium population that currently affects farmers’ fields in Peru and compare that to the diversity found in the old collection dating from 1960 to 2013. In this paper, we characterize the phylotype, endoglucanase (egl) sequevar and biovar of over 200 isolates to identify patterns of geographical distribution and pathogenicity on potato, tomato, eggplant and tobacco.
The sampling locations at 2016 were selected based on the previous reports on the presence of potato brown rot in the area and consulting the local farmers and local phytosanitary officials. The samples were collected at the beginning of the potato flowering period, when tuber symptoms are rarely observed, thus we focused on collecting stem samples. Plants with wilting symptoms were uprooted in the field and a fragment of one principal stem cut at the stem base (ca. 10 cm above the ground) was collected from each plant. The scissors were disinfected between the cuttings of each sample by dipping in a 2% solution of NaOCl for 10 s. The stem piece of each sample was wrapped in towel paper and put inside a paper bag. In the laboratory, the stem pieces were washed individually with tap water and then allowed to dry on clean paper towel. From each of the 10-cm piece, a section of approximately 5 cm in length (eliminating 2–3 cm at each side) was cut using horticultural scissors. The scissors were flame sterilized between each sample. Each sample was dipped in a solution of 0.5% NaOCl contained in a plastic bag for 1 min; this solution was then discarded and a stem section was rinsed in the same plastic bag first with sterile distilled water, then with 96% alcohol, and finally with sterile distilled water (at least twice). From each of the 5-cm stem pieces cut previously, a 1–2-cm long fragment was cut using horticultural scissors. The stem fragments obtained from each sample was again placed in a new plastic bag and crushed with a hammer. Then 3 ml of sterile citrate extraction buffer (0.1 M citric acid, 0.1 M sodium citrate, pH 5.6) was added and the sample homogenized.
For isolation of bacteria 20 μl of stem extract was plated on modified Kelman’s medium (MKM) (French et al., 1995), diluted by streaking and plates were incubated at 30°C for 2 days. Single colonies matching the description of that of R. solanacearum (irregular shape, fluidal, and entirely white or with a pink center) were selected and sub cultured on MKM without TZC (2, 3, 5 triphenyl tetrazolium chloride) at 30°C for two days. The bacteria colonies were transferred in screw cap test tubes containing 10 ml sterile water and maintained at 15°C. To verify that the cultures were pathogenic, all strains were inoculated in susceptible potato plants of the variety Desiree by injecting 20 μl of bacterial cell suspension (108 cells ml-1) into the axil of the third leaf from the apical meristem and re-isolating the bacteria within a week from a wilted plant on MKM.
The pathogenicity test was performed using tomato (Rio Grande), potato (Canchan INIA), eggplant (Vivian) and tobacco (Samsun) plants. Seeds of tobacco, tomato and eggplant, and potato tubers were sown in plastic trays (30 cm × 40 cm) containing Promix Bx substrate. Approximately two weeks later when the plants were 10-15 cm long and had adequate roots, they were transplanted in 3.5 inch-diameter pots. 50 ml solution of fertilizer (N–P–K: 6-7-22) was applied at the time of planting. Plants were irrigated daily, except for 1 day prior to inoculation.
Bacterial suspensions were prepared by culturing strains for 48-h. at 30°C on MKM without TZC. The cells were harvested in sterile distilled water and the bacterial suspensions were measured using a spectrophotometer to determine concentrations.
For inoculation, 2 weeks after transplanting, the plant roots were damaged with a scalpel and immediately 40 ml of the bacterial suspension was poured into the soil (the final concentration of each isolate was of 5 × 107 cells/g soil). A set of six plants was inoculated with each strain and placed in separated trays to prevent cross contamination. Plants were maintained until one month after inoculation, during which temperatures ranged from 16.9 to 28.5°C. The wilt incidence was recorded twice a week after the first wilting symptoms began to appear. Stems of those plants exhibiting no wilt were also analyzed, to detect latent infections by isolating the bacteria on MKM (French et al., 1995).
For DNA extraction, the bacteria were streaked on MKM without TZC and the plates incubated at 30°C for 2 days. Then one colony was suspended in 100 μl of sterile NFW, boiled for 10 min and kept at -20°C prior to use.
The taxonomic identity of R. solanacearum was verified with primers 759/760 (Opina et al., 1997). PCR amplification was performed in a total volume of 15 μl containing 1X of PCR Buffer, 2.5 mM MgCl2, 0.2 μM each dNTP, 0.2 μM of each primer, 0.3 U of GoTaqG2 Flexi DNA polymerase (PROMEGA) and 1 μl DNA template. Amplification was performed in an Applied Biosystem Veriti thermocycler as follows: an initial denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, extension at 72°C for 17 s, and a final extension step at 72°C for 5 min. PCR products (10 μl) were analyzed by electrophoresis through 1% (w/v) agarose gels with 0.01 μl /ml GelRedTM 10,000X (Biotium) and photographed under UV light in a The ChemidocTM MP Photodocumentation System (BIO-RAD). Fragments were compared with a 1 Kb Plus marker ladder. A positive identification was based on the presence of a 282 bp amplicon.
Biovars of the strains were determined following the protocol described by Hayward (1964) and Hayward et al. (1989) using the basal medium (NH4H2PO4 1.0 g, KCl 0.2 g, MgSO4.7H2O 0.2 g, Difco bacto peptone 1.0 g, Agar 3.0 g, and Bromothymol blue 0.03 g per liter) containing 1% of each type of sugar (lactose, maltose, mannitol and sorbitol, D (+) trehalose or 1% of L (+) tartrate). An inoculum suspension (O.D. + 0.05 at 600 nm) of each strain was prepared in sterile distilled water from 2-day-old cultures. For the determination of biovars 40 μl of bacterial suspension was added in the microtiter plate containing 170 μl of the basal medium and for the phenotypes differentiation (2A and 2T) 200 μl of the suspension was added to 3 ml the basal medium without agar contained in test tubes. The sealed tubes and plates were incubated at 30°C for 6 days and the color reaction was visually evaluated.
Phylotype was determined by multiplex PCR using a set of phylotype-specific primers Nmult:21:1F, Nmult:21:2F, Nmult:22:InF, Nmult:23:AF, and Nmult21:RR (Fegan and Prior, 2005). Amplification was carried out in a total volume of 15 μl containing 1 X PCR buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 0.2 μM of each primer, 0.3 U of GoTaq G2 Flexi DNA polymerase (PROMEGA) and 1 μl DNA template. Amplifications were performed in an Applied Biosystem Veriti thermocycler as follows: an initial denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 59°C for 30 s, extension at 72°C for 23 s, and a final extension step at 72°C for 5 min. PCR products (10 μl) were analyzed by electrophoresis through 1% (w/v) agarose gels with 0.01 μl /ml GelRedTM 10,000X (Biotium) and photographed under UV light in The ChemidocTM MP Photodocumentation System (BIO-RAD). DNA template from strains CIP-277 (phylotype I), CIP-435 (phylotype II), and CIP-358 (phylotype III) were used as positive amplification controls. The size of the amplified fragments was estimated by comparison with a 1 Kb Plus marker ladder.
PCR amplification of a 750-bp region of the egl gene was done using the primer pair Endo-F (5-ATGCATGCCGCTGGTCGCCGC-3) and Endo-R (5-GCGTTGCCCGGCACGAACACC-3) (Fegan et al., 1998; Fegan and Prior, 2005). Amplification was carried out in a total volume of 75 μl containing PCR buffer, 1.25 mM MgCl2, and 0.2 mM of each dNTP, 0.2 μM of each primer, 1.5 U of GoTaq G2 Flexi DNA polymerase (PROMEGA) and 3 μl DNA template. PCR was performed using an Applied Biosystem Veriti thermocycler. Amplifications were performed with an initial denaturation step at 95°C for 2 min, followed by 28 cycles of denaturation at 95°C for 30 s, annealing at 70°C for 30 s, extension at 72°C for 50 s, and a final extension step at 72°C for 5 min. PCR products (5 μl) were checked by electrophoresis through 1% (w/v) agarose gels with 0.01 μl /ml GelRedTM 10,000X (Biotium) and photographed under UV light in The ChemidocTM MP Photodocumentation System (BIO-RAD). The size of the amplified fragments was estimated by comparison with a 1 Kb Plus marker ladder. Then PCR products were purified and sequenced by Macrogen services (Kumchun-ku, Seoul, Korea) using Endo-F and Endo-R primers. The raw sequences were assembled and aligned together with reference sequences of R. solanacearum strains obtained from NCBI sequence database. The DNA sequences of the isolates were assigned accession numbers MF461731 – MF461866 in the NCBI sequence database. The phylogenetic analysis was done based on partial sequences using MEGA7 (Kumar et al., 2016), phylogenetic trees were constructed from the genetic distance data by the Neighbor-Joining (NJ) and Maximum Likelihood (ML) method using the algorithm of Jukes–Cantor with 1000 bootstrap resampling of the data to test the tree topologies.
Samples were collected from the central and northern departments of Peru, Piura, Amazonas, Ancash, Cajamarca, and Huanuco, based on consultation with the local phytosanitary organization SENASA and agricultural innovation center INIA. The specialists identified the locations of potato fields or other Solanaceous crops with symptoms similar to bacterial wilt infection. The approximate field size varied from 0.1 to 3 ha, destined either for own consumption or for selling and the seed was either self-produced or commercially purchased. Infected samples came from both types of fields, confirming that although diagnostic tests for R. solanacearum are available in Peru (Priou et al., 1999, 2010) formal disease diagnostics is not common practice before seed trading. The most popular cultivars were Yungay and Amarilis, followed by Unica, Perricholi, Capiro, Chaucha Amarilla, and Huayro Blanco (Table 1).
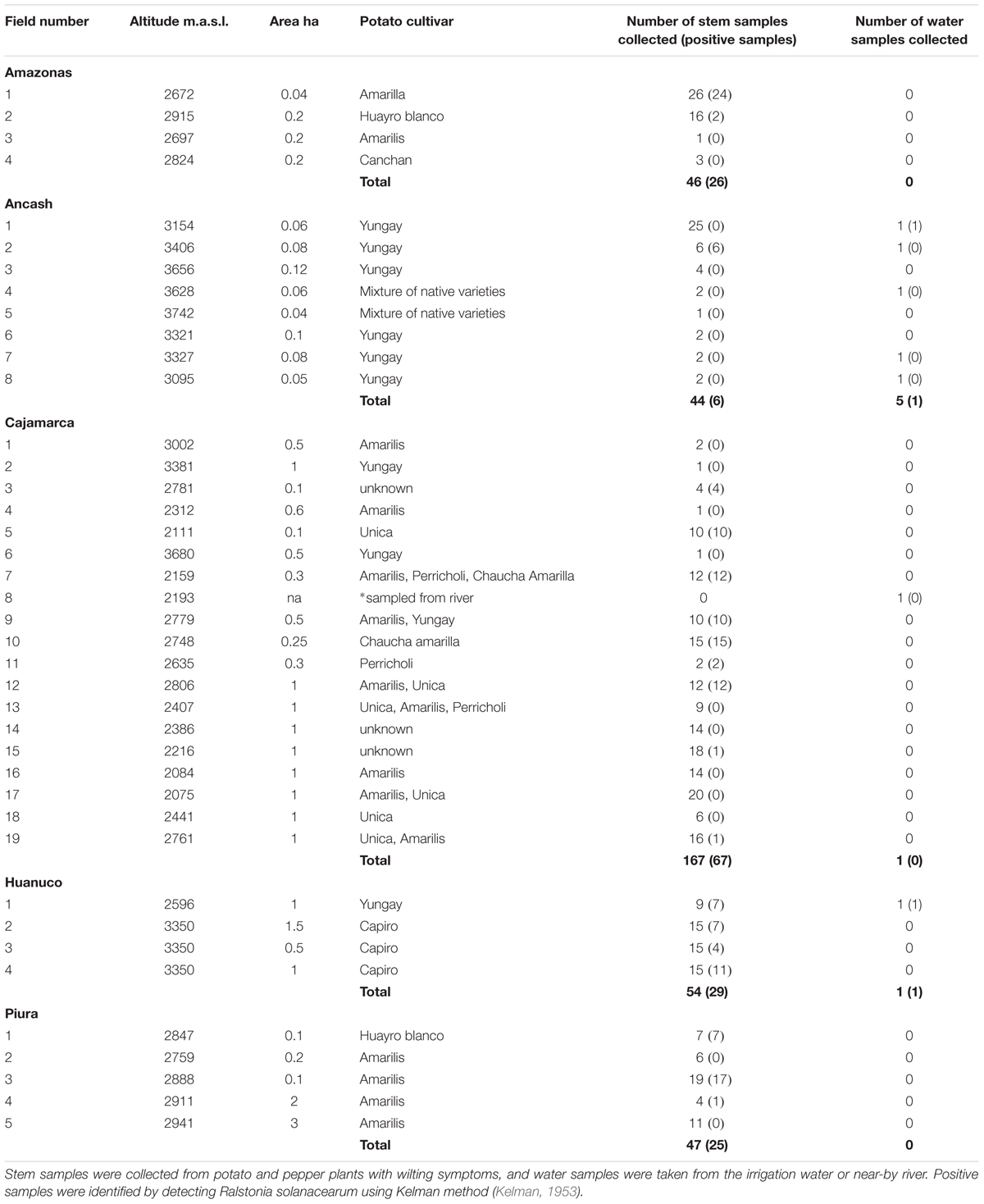
TABLE 1. Details of the fields or water ways sampled in the departments of Peru during 2016, and number of infected samples detected.
The sampling focus was in potato crop, but if the farmers had other crops with wilting symptoms those were also collected. In total samples were collected from 38 potato fields and one pepper field. Of these 19, including the pepper field, were found infected with R. solanacearum (Table 1). The highest proportion of infections was found in Huánuco, where all fields with wilted samples had infected plants. In Ancash, R. solanacearum was found in the water sample collected from the irrigation canal of one of the fields, but nevertheless the wilted samples from this field were not infected. Only one of the fields sampled in Ancash was infected. Two separate prospections were made in Cajamarca, the first one in March and the second one in October 2016 and out of the 18 fields sampled 8 were found with infected plants. In Piura four of the five fields we visited had infected samples. In Amazonas, we visited four fields and two of them had infected samples. In many cases the wilting observed was apparently caused by other pathogens since of the 358 wilted samples collected, only 43% were found to contain R. solanacearum. The altitude of the sampling sites ranged from 2111 to 3742 m.a.s.l. and R. solanacearum was found in samples up to the altitude of 3406 m.a.s.l (Table 1).
The 265 R. solanacearum strains from Peru characterized in this study have been collected in Peru between the years 1966 and 2016 mostly from potato, but also from pepper, tomato, plantain, or soil (Supplementary Table S1). Based on the partial sequence of the endoglucanase gene, all new isolates collected at 2016 and all but one old isolate, were Phylotype II. One single isolate collected from tomato at the 1960’s turned out to be phylotype I.
Phylogenetic analysis divided the Peruvian R. solanacearum isolates into three main clusters following the Phylotype classification (Figure 1 and Table 2). The Phylotype IIB isolates formed seven sequevar groups including the previously identified sequevars 1, 2, 3, and 4 (Poussier et al., 2000; Wicker et al., 2007; Cellier and Prior, 2010). CIP-184, CIP-172, CIP-162, and CIP-10 share similarity to sequevar 25 (UW477, Poussier et al., 2000), but the grouping does not receive strong bootstrap support. Nevertheless, these isolates are here considered belonging to sequevar 25 (Figure 1 and Table 2). In fact, CIP10 has been sequenced before and is classified as sequevar 25 (Poussier et al., 2000). Some Peruvian isolates that group together with the reference sequevars previously assigned as Phylotype IIB strains IIB-57, IIB-51, IIB-28, IIB-26, IIB-27, IIB-55, IIB-56, IIB-54 (Poussier et al., 2000; Wicker et al., 2007; Cellier and Prior, 2010; Santiago et al., 2016) cluster in sub-groups supported by high bootstrap values in the phylogenetic analysis and are thus considered as new sequevars (Figure 1). Isolates CIP-61 and CIP-63 form a new sequevar of their own (IIB-new2), while CIP-312, CIP-313, CIP-314 belong to another new sequevar (IIB-new3). Phylotype IIA isolates from Peru clustered together with reference strains previously assigned to sequevars IIA-5, IIA-39, IIA-41, and IIA-50 (Wicker et al., 2007: Toukam et al., 2009; Cellier and Prior, 2010). Isolates CIP-301, CIP-303 and CIP-307 are classified as a new sequevar (IIA-new) since they are separated from the other previously identified IIA reference sequevars IIA-38, IIA-24, IIA-36, IIA-6, IIA-35, IIA-52 (Wicker et al., 2007, 2012; Toukam et al., 2009; Cellier and Prior, 2010; Lebeau et al., 2011; Hong et al., 2012) included in the phylogenetic analysis (Figure 1). The single Phylotype I isolate, CIP-072, was found in the banks of Amazon river in Loreto and had been determined as a biovar 3 (Martin et al., 1981). It shares similarity to sequevar 18 strain GM1000 isolated from tomato in French Quiana (Cellier and Prior, 2010).
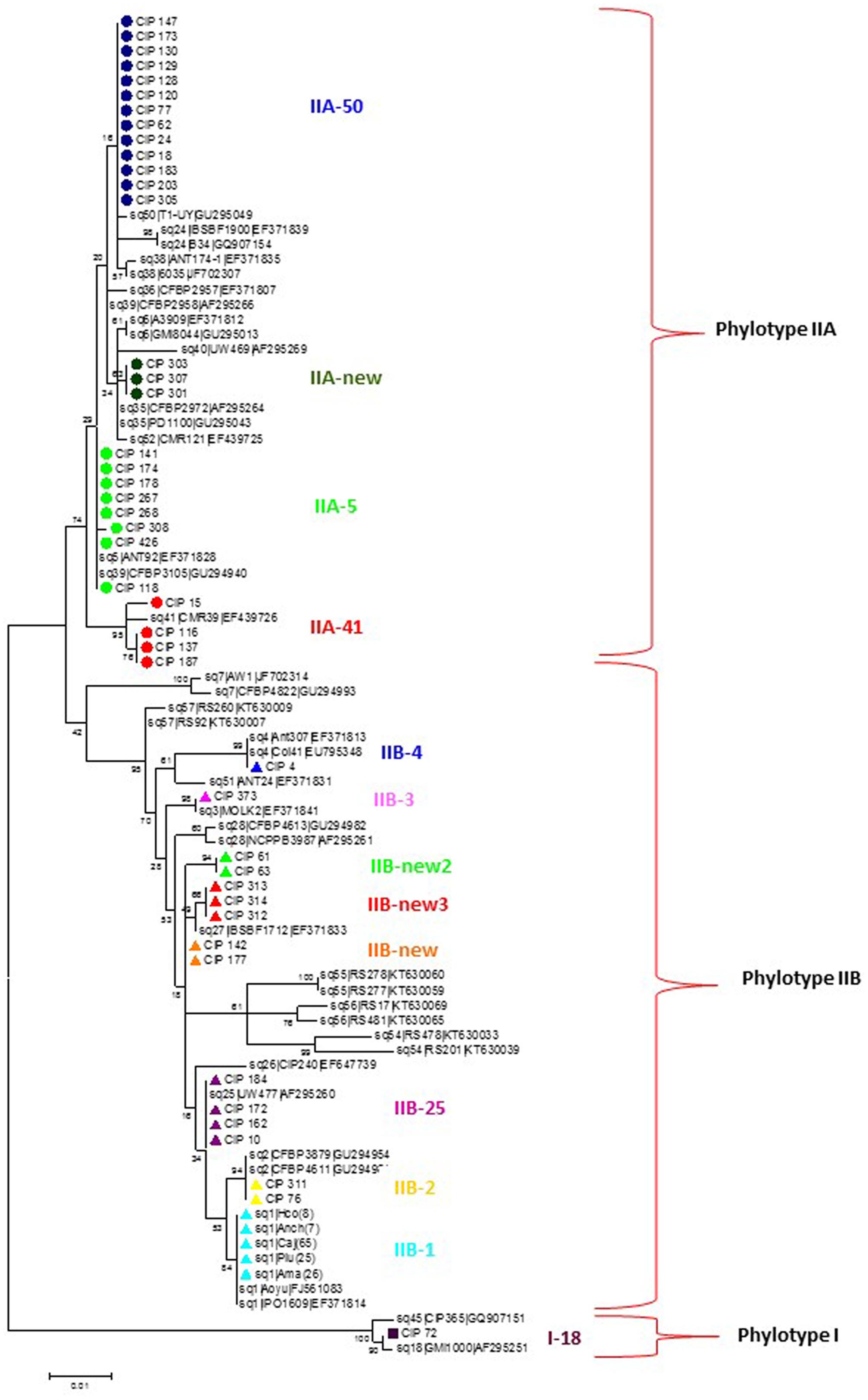
FIGURE 1. Molecular Phylogenetic analysis of the partial endoglucanase gene sequences (622 nt) of 90 Ralstonia solanacearum isolates from Peru and reference isolate sequences obtained from NCBI sequence database. The phylogeny was constructed using the Maximum Likelihood method. The tree with the highest log likelihood (–2108.4692) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Circles, triangles and squares indicate sequences of phylotypes IIA, IIB and I, respectively, determined in this study, colors indicate different sequevars. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
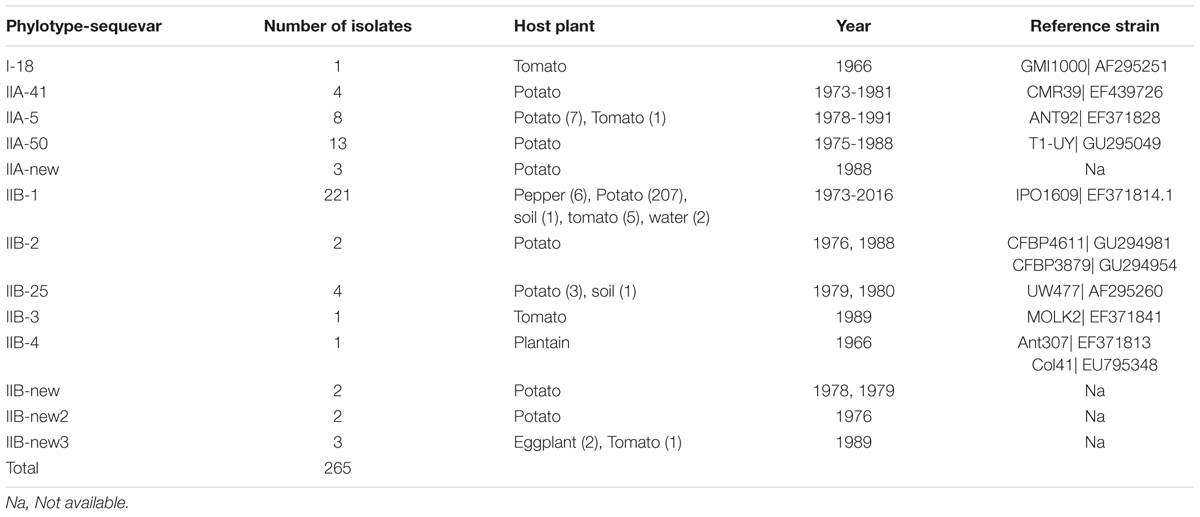
TABLE 2. Endoglucanase (egl) sequevars identified among the sample of 265 R. solanacearum isolates collected from different hosts, soil, or water in Peru 1966–2016.
As expected, most of the Peruvian R. solanacearum isolates were IIB-1 strains. In the old collection sampled between 1966 and 2013, 72% (92/121) were IIB-1 and in the new collection at 2016 no other strains were found. Since all isolates analyzed at 2016 had identical egl sequences only five were left in the final phylogenetic tree to represent each of the Peruvian departments Huanuco (Hco), Ancash (Anch), Cajamarca (Caj), Piura (Piu), and Amazonas (Ama) that were sampled. Looking at the geographical differentiation in terms of sequevar diversity, departments of Junin with seven and Loreto with five different sequevar groups, contained the greatest diversity (Figure 2). Unfortunately, we were unable to obtain samples from these locations at 2016 and thus cannot confirm the level of current diversity at these locations.
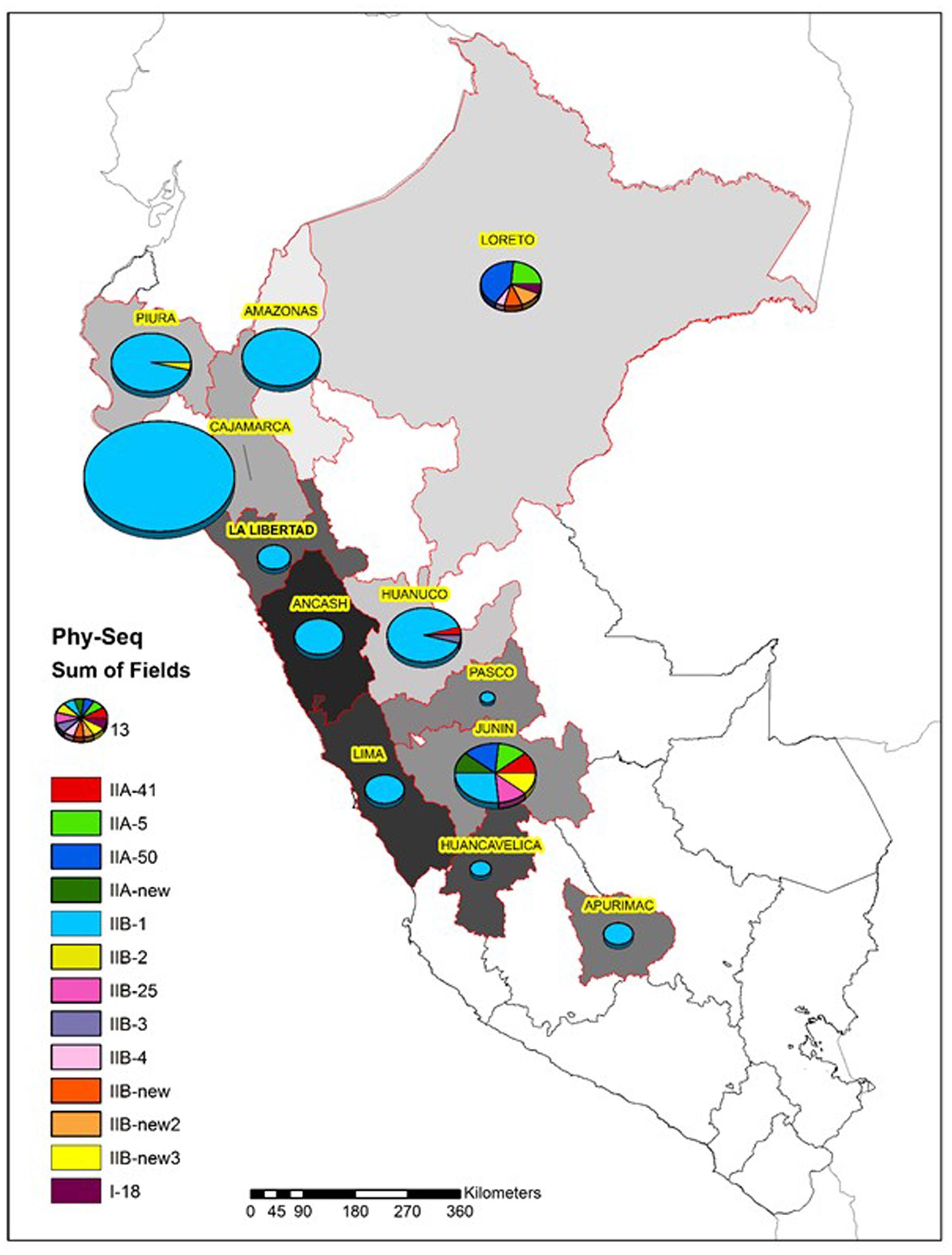
FIGURE 2. Diversity of the R. solanacearum isolates collected in Peru 1966–2016 based on the phylotype and the partial sequence of the endoglucanase gene (egl) (Fegan and Prior, 2005). The size of the pie is proportional to the number of isolates analyzed from each Peruvian department.
All Peruvian Phylotype IIA isolates had the biovar 1 phenotype while the Phylotype IIB isolates were more variable in this respect consisting of isolates belonging to biovar 2A, 2T or 1 phenotypes (Table 3).
We selected 25 isolates representing the different egl sequevar groups (Figure 1) for pathogenicity tests using pepper, potato, tomato and tobacco. Most of these isolates originate from potato, but two are from tomato, one from eggplant and one from plantain (Figure 3). All isolates, except for the one originating from plantain, were pathogenic on at least one of the tested plants. The plantain isolate may have lost its pathogenicity during storage, but we cannot be sure since we did not attempt to inoculate its original host. Potato was susceptible to all the other isolates, resulting in wilting symptoms in 50–100% of the plants. Almost all isolates caused latent infection in tomato and eggplant, but wilting was observed much less frequently. Tobacco was least affected by the isolates tested as most of the isolates were nonpathogenic to this plant. Only two isolates belonging the phylotype IIA (CIP-426 and CIP-130) caused wilting in all hosts tested. The phylotype IIA seems more adapted to infect tobacco than the phylotype IIB strains as nearly all IIA isolates caused latent infection in this plant. IIB-1 and IIB-2 isolates were very aggressive on potato, tomato and eggplant, but all other IIB sequevars, except for IIB-4, caused wilting only on potato. Thus, these other IIB sequevars may have different pathogenicity mechanisms as they do not cause wilting in tomato but result in only latent infection.
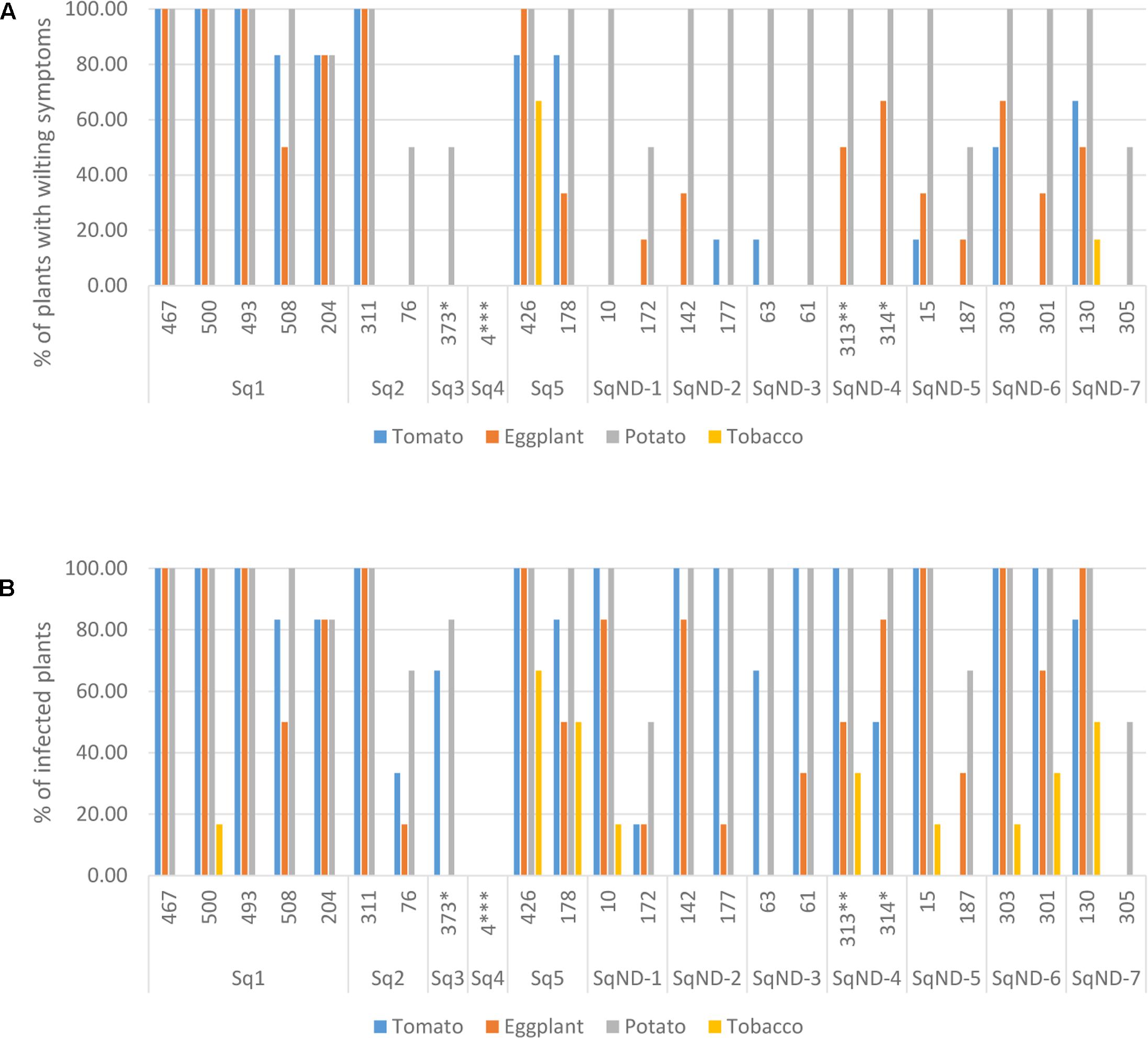
FIGURE 3. Pathogenicity of 25 R. solanacearum isolates on tomato, eggplant, potato, and tobacco measured as proportion of wilted plants (A) and proportion of latent infections by MKM test (B). The ID number and the egl sequevar group of each isolate are indicated on the x-axis. ∗isolated from tomato, ∗∗isolated from eggplant, ∗∗∗isolated from plantain.
This study describes the current bacterial wilt infestation level in the potato fields in the Peruvian Andes and is the first extensive molecular characterization of the bacterial wilt pathogen R. solanacearum in Peru. To investigate the occurrence of wilting due to this pathogen and to unravel its genetic diversity we embarked on several prospections that included the main potato growing areas in the central and northern parts of the country in the slopes of the Andes mountains. We sampled in total 39 farmers’ fields located between the latitudes of S05°09′53.0″ and S10°16′19.1″ and altitudes 2111–3742 m.a.s.l. where wilting symptoms had been reported by farmers or phytosanitary officials. Symptoms in the fields included wilting, in some cases of one branch in the hill only, a symptom that is very typical for R. solanacearum infection. In some cases, after removing the plants from the soil, the underground part of the stem was found damaged, either by insects or other pathogens. While symptomatic plants were found in almost all the fields visited, the frequency of R. solanacearum detected on potato was relatively low in the areas sampled during 2016. Although fields were pre-selected based on reported wilting symptoms, in less than 50% of the fields we found plants infected by R. solanacearum suggesting that other pathogens are in many cases responsible for the wilting symptoms observed. This means that the presence of the R. solanacearum in the field cannot be reliably determined only based on symptoms, but needs to be verified by a detection method such as NCM ELISA or PCR using specific primers. We used the Kelman’s cultivation method (Kelman, 1953) to detect the bacterium, which should be sensitive enough to enable the detection also of latent infections since it has been successfully used to confirm positive NCM ELISA tests from un-symptomatic samples (Priou et al., 1999, 2000, 2010). Taking a representative tuber sample after the harvest is recommended to catch the symptomless latent infections. However, since our prospections were made during the flowering period, we focused on sampling only stems. The previous prospections were made in the 1990s when seed lots from symptomless potato crops from Peru and Bolivia were tested for latent infection by R. solanacearum (Priou et al., 1999, 2010). In that study, tuber lots and stems from sixteen fields in Cajamarca and La Libertad departments exhibiting either no symptoms or low incidence of wilted plants (0.1–3%) were sampled and analyzed for presence of R. solanacearum by NCM ELISA test. All seed lots from the fields with low incidence of bacterial wilt were found positive for R. solanacearum, and roughly 50% of the seed lots coming from the apparently healthy fields were also positive (Priou et al., 1999). Cajamarca was also sampled in our study, where we collected wilted samples from 19 fields and found R. solanacearum only from nine of them suggesting that at least in Cajamarca the overall incidence of brown rot is now much less than it used to be. Some of this difference could be explained by the different detection methods and sampling units (stem vs tuber) used between the two studies, but most likely the reduction has taken place because of the intensive farmer training conducted by SENASA and CIP at 2003–2005 in the integrated management of bacterial wilt in potato (Anonymous, 2005). While this is a positive development, the disease is far from being eradicated and if the use of uncertified seed in potato production remains common practice, brown rot will also continue to be present. Only extremely strict hygiene measures in seed production and separating commercial seed from the ware potatoes can lead to a successful eradication of this bacterium (French, 1994; Janse, 2012). Most of the fields sampled were rather small plots (less than 1ha) and the smallest ones were used for family’s own consumption rather than selling in the market, which may affect the choice of tuber seed source and other cultivation methods. Our research confirms again that the bacterial wilt pathogen can survive in high altitudes as we found infected samples from the fields located above 3000 m.a.s.l. This has been reported before in the Andes of Peru and Bolivia (French, 1986; Castillo and Plata, 2016). The higher altitudes in the Andes are often used as potato seed production areas because these generally have lower disease incidence. The ability of brown rot to survive in the high altitudes means that there are less and less areas available for clean seed production by farmer’s conventional means.
We analyzed the genetic diversity and phylogeny of the R. solanacearum isolates collected in Peru from potato, tomato, pepper, plantain, soil, and water between 1966 and 2016. As expected all isolates, with one exception, belonged to the phylotype II supporting the association between phylotype and geographic region of origin (South America). The departments and areas visited at 2016 included many of those that were also represented in the historical collection and for all these the same IIB-1 strain was dominating. The Andes is thought to be the origin of the R. solanacearum phylotype IIB sequevars 1 and 2 (previously race3 biovar 2) causing potato brown rot (Champoiseau et al., 2009). This strain has also spread through the tropical highlands and subtropical warm-temperate areas throughout the world, and is considered a quarantine pathogen in Europe (Janse, 2012) and a select agent in the United States under the Agricultural Bioterrorism Protection Act (Hawks, 2002). The IIB-1 strain is considered cold-tolerant as it can cause disease in cool conditions, but has a relatively narrow host range (Milling et al., 2009; Cellier and Prior, 2010). In our pathogenicity tests the IIB-1 and IIB-2 strains, the biovar 2T strains IIB-25, IIB-new and all IIA strains were highly aggressive on potato and causing latent infection also in tomato and eggplant. There was a clear difference in pathogenicity on tobacco between the two phylotypes IIA and IIB. All IIA strains were causing at least latent infection on tobacco and the strains IIA-5 and IIA-50 also caused wilting of this species, while IIB strains caused only few latent infections. IIB-3 isolate CIP-373 originating from tomato caused wilting in potato but in tomato only led to latent infection, while IIB-4 isolate CIP-4 originating from plantain was found nonvirulent. Previous research has shown that IIB-3 and IIB-4 strains are generally virulent to susceptible potato and tomato varieties (Cellier and Prior, 2010). The reduced virulence of some isolates could be attributed to the long time they have spent in the storage.
Evidently IIB-1 is currently the dominating R. solanacearum strain infecting potato in Peru and it is not known whether the other phylogenetically distinct strains still exist. Two isolates collected in Piura and Cajamarca at 1982 and 1976, respectively, grouped together with IIB-2 reference strains (Cellier and Prior, 2010) forming a phylogenetically separated group from the IIB-1 strains (Figure 1). This separation has been noted before for potato and plantain infecting strains from Colombia by microarrays (Cellier et al., 2012). The IIB-25, IIB-new, IIA-5, IIA-50, IIA-41, IIA-new, and I-18 were originally discovered in lowland areas where potato is normally not cultivated, because of the too warm climate for this crop. Research in 1970s and 1980s was conducted in these locations to study potato heat tolerance and the R. solanacearum strains were assumed endemic to the area (Martin et al., 1981). Since potato seed production usually takes place at higher altitudes with cooler climates the cold tolerant IIB-1 strain may have a better fitness and thus better chance for survival than the low land strains. The finding that the strains IIA-50 and IIA-41 that were also collected in Cajamarca and Huanuco, respectively, in the early 1980’s were no longer discovered in these areas in 2016 supports this idea.
The greatest diversity was discovered in the department of Junin in the warm jungle areas of San Ramon and in the department of Loreto at Yurimaguas, where some new sequevars of phylotype II and one isolate of phylotype I were found. Considering that these strains are probably endemic the high level of diversity encountered is expected and similar variability has been reported before in Brazil, especially for phylotype IIB by Santiago et al. (2016) who reported four new sequevars for this phylotype some of which were found on potato. Here we report additional three new sequevars of phylotype IIB that are phylogenetically distinct from any of those described before. The next prospections in Peru should focus on these diversity hotspots to check for the current occurrence of these strains. The old biovar 3 strain CIP-072 collected by French and Sequeira (1970) in the Amazon basin turned out to be phylotype I sequevar 18, which has been also found in other warmer climates in the Americas, such as Brazil (Santiago et al., 2016), Guatemala (Sanchez Perez et al., 2008), French Guiana (Deberdt et al., 2014), and Trinidad y Tobago (Ramsubhag et al., 2012). The I-18 strain has mostly been associated with tomato
High molecular diversity was found among the historical collection of R. solanacearum strains sampled in Peru. However, current epidemics are caused by a clonal brown rot strain IIB-1. Nearly all isolates were highly aggressive on potato causing severe wilting and were also virulent on tomato, eggplant and tobacco to variable degrees. The most variable strains in the historical collection were found in areas that are no longer used for potato cultivation and thus these strains do not pose a real threat in the current landscape of potato production in the country. More discriminant molecular analysis using multilocus markers are under way to identify sources of contamination and patterns of distribution within the clonal dominating strain IIB-1.
LG: Collected samples from the field, conducted and supervised the laboratory work, conducted and supervised the pathogenicity assays, drafted and revised the manuscript. JH: Collected samples from the field, conducted laboratory work and pathogenicity assays, conducted sequence analysis, drafted the manuscript. EF: Collected samples from the field, conducted laboratory work and sequence analysis, revised the manuscript. JK: Conceived funding for the project, designed the work, supervised and conducted analysis of sequence data, did data interpretation, revised the manuscript. HL-K: Conceived funding for the project, designed the work, collected samples from the field, supervised and conducted analysis of sequence data, did data interpretation, drafted and revised the manuscript.
Research was conducted within the project PNIA-IE-10460-2015.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank our colleagues at CIP: Willmer Pérez, Soledad Gamboa, Myriam Izarra, Mirian Yanina and Eva Huaman for help in sampling and laboratory work, our colleagues at SENASA: Alicia Villar, Ronald Llique, Luis Rubio, Augusto Ruiz and Arbel Gamonal for the identification of the fields and Liz Garcia at INIA for the help in sampling. Special thanks also to Léonardo Espinoza for the identification of the fields and sampling in Huanuco.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01221/full#supplementary-material
Buddenhagen, I., Sequeira, L., and Kelman, A. (1962). Designation of races in Pseudomonas solanacearum. Phytopathology 52, 726.
Caruso, P., Palomo, J. L., Bertolini, E., Álvarez, B., López, M. M., and Biosca, E. G. (2005). Seasonal variation of Ralstonia solanacearum biovar 2 populations in a Spanish river: recovery of stressed cells at low temperatures. Appl. Environ. Microbiol. 71, 140–148. doi: 10.1128/AEM.71.1.140-148.2005
Castillo, J. A., and Plata, G. (2016). The expansion of brown rot disease throughout Bolivia: possible role of climate change. Can. J. Microbiol. 62, 442–448. doi: 10.1139/cjm-2015-0665
Cellier, G., and Prior, P. (2010). Deciphering phenotypic diversity of Ralstonia solanacearum strains pathogenic to potato. Phytopathology 100, 1250–1261. doi: 10.1094/PHYTO-02-10-0059
Cellier, G., Remenant, B., Chiroleu, F., Lefeuvre, P., and Prior, P. (2012). Phylogeny and population structure of brown Rot- and Moko disease-causing strains of Ralstonia solanacearum phylotype II. Appl. Environ. Microbiol. 78, 2367–2375. doi: 10.1128/AEM.06123-11
Champoiseau, P. G., Jones, J. B., and Allen, C. (2009). Ralstonia solanacearum race 3 biovar 2 causes tropical losses and temperate anxieties. Plant Health Prog. 10, 1–10. doi: 10.1094/PHP-2009-0313-01-RV
Deberdt, P., Guyot, J., Coranson-Beaudu, R., Launay, J., Noreskal, M., Rivière, P., et al. (2014). Diversity of Ralstonia solanacearum in French Guiana expands knowledge of the ‘emerging ecotype’. Phytopathology 104, 586–596. doi: 10.1094/PHYTO-09-13-0264-R
Denny, T. P., Carney, B. F., and Schell, M. A. (1990). Inactivation of multiple virulence genes reduces the ability of Pseudomonas solanacearum to cause wilt symptoms. Mol. Plant Microbe Interact. 3, 293–300. doi: 10.1094/MPMI-3-293
Devaux, A., Kromann, P., and Ortiz, O. (2014). Potatoes for sustainable global food security. Potato Res. 57, 185–199. doi: 10.1007/s11540-014-9265-1
Elphinstone, J. G. (2005). “The current bacterial wilt situation: global overview,” in Bacterial Wilt Diseases and the Ralstonia solanacearum Species Complex, eds C. Allen, P. Prior, and A. C. Hayward (St. Paul, MN: American Phytopathological Society), 9–28.
FAO (2009). International Year of the Potato 2008: New Light on a Hidden Treasure. Available at: http://www.fao.org/potato-2008/en/events/book.html
Fegan, M., and Prior, P. (2005). “How complex is the Ralstonia solanacearum species complex,” in Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex, eds C. Allen, P. Prior, and A. C. Hayward (Madison, WI: APS Press), 449–462.
Fegan, M., Taghavi, M., Sly, L. I., and Hayward, A. C. (1998). “Phylogeny, diversity and molecular diagnostics of Ralstonia solanacearum,” in Bacterial Wilt Disease: Molecular and Ecological Aspects, eds P. Prior, C. Allen, and J. Elphinstone (New York: Springer-Verlag), 19–33.
French, E. R. (1986). “Interaction between strains of Pseudomonas solanacearum, its hosts and the environment,” in Proceedings of the ACIR Proceedings No. 13 Bacterial Wilt Disease in Asia and the South Pacific, ed. G. J. Persley (Canberra, ACT: Australian Centre for International Agricultural Research), 99–104.
French, E. R. (1994). “Strategies for integrated control of bacterial wilt of potatoes,” in Bacterial Wilt: The Disease and its Causative Agent Pseudomonas solanacearum, eds A. C. Hayward and G. L. Hartman (Wallingford: CAB International), 199–207.
French, E. R., Aley, P., Torres, E., and Nydegger, U. (1993). “Diversity of Pseudomonas solanacearum in Perú and Brasil,” in Proceedings of the International Conference Held at Kaohsiung, Taiwan, 28-31 October 1992: Bacterial Wilt ACIAR Proceedings No. 45, eds G. L. Hartman and A. C. Hayward (Canberra, ACT: Australian Centre for International Agricultural Research), 70–77.
French, E. R., Gutarra, L., Aley, P., and Elphinstone, J. (1995). Culture media for Pseudomonas solanacearum: isolation, identification and maintenance. Fitopatologia 30, 126–130.
French, E. R., and Sequeira, L. (1968). Marchitez bacteriana o moko del plátano en el Perú. Fitopatología 3, 27–38.
French, E. R., and Sequeira, L. (1970). Strains of Pseudomonas solanacearum from Central and South America, a comparative study. Phytopathology 60, 506–512. doi: 10.1094/Phyto-60-506
Gildemacher, P., Schulte-Geldermann, E., Borus, D., Demo, P., Kinyae, P., Mundia, P., et al. (2011). Seed potato quality improvement through positive selection by smallholder farmers in Kenya. Potato Res. 54, 253–266. doi: 10.1007/s11540-011-9190-5
Graham, J., Jones, D. A., and Llyd, A. B. (1979). Survival of Pseudomonas solanacearum race 3 in plant debris and in latently infected potato tubers. Phytopathology 69, 1100–1103. doi: 10.1094/Phyto-69-1100
Gutarra, L., Kreuze, J., Lindqvist-Kreuze, H., and De Mendiburu, F. (2014). Variation of resistance to different strains of Ralstonia solanacearum in highland tropics adapted potato genotypes. Am. J. Potato Res. 92, 258–265. doi: 10.1007/s12230-014-9426-4
Hawks, B. (2002). Agricultural Bioterrorism Protection Act of 2002; Listing of Biological Agents and Toxins and Requirements and Procedures for Notification of Possession. Available at: http://www.fas.org/sgp/news/2002/08/fr081202.html
Hayward, A. C. (1964). Characteristics of Pseudomonas solanacearum. J. Appl. Bacteriol. 27, 265–277. doi: 10.1111/j.1365-2672.1964.tb04912.x
Hayward, A. C. (1991). Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87. doi: 10.1146/annurev.py.29.090191.000433
Hayward, A. C. (1994). “The hosts of Pseudomonas solanacearum,” in Bacterial Wilt: The Disease and its Causative Agent, Pseudomonas solanacearum, eds A. C. Hayward and G. L. Hartman (Wallingford: CAB International), 9–24.
Hayward, A. C., El-Nashaar, H. M., De Lindo, L., and Nyddegger, U. (1989). “The use of microtiter plates in the phenotypic characterization of phytopathogenic pseudomonads,” in Proceedings of the 7th International Conference on Plant Pathogenic Bacteria, Budapest, 593–598.
He, Y. L., Sequeira, L., and Kelman, A. (1983). Characteristics of strains of Pseudomonas solanacearum from China. Plant Dis. 67, 1357–1361. doi: 10.1094/PD-67-1357
Herrera, I. A., and French, E. (1970). Strains de Pseudomonas solanacearum aisladas de papa en el Perú. Invest. Agropecuarias 1, 47–52.
Hong, J. C., Norman, D. J., Reed, D. L., Momol, M. T., and Jones, J. B. (2012). Diversity among Ralstonia solanacearum strains isolated from the southeastern United States. Phytopathology 102, 924–936. doi: 10.1094/PHYTO-12-11-0342
Janse, J. D. (1996). Potato brown rot in western Europe - history, present occurrence and some remarks on possible origin, epidemiology and control strategies. Bull. OEPP 26, 679–695. doi: 10.1111/j.1365-2338.1996.tb01512.x
Janse, J. D. (2012). Review on brown rot (Ralstonia solanacearum race 3, biovar3, phylotype IIb) epidemiology and control in the Netherlands since 1995: a success story of integrated pest management. J. Plant Pathol. 94, 257–272.
Kelman, A. (1953). The Bacterial Wilt Caused by Pseudomonas solanacearum. A Literature Review and Bibliography. Raleigh, NC: North Carolina State College.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lebeau, A., Daunay, M. C., Frary, A., Palloix, A., Wang, J. F., Dintinger, J., et al. (2011). Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101, 154–165. doi: 10.1094/PHYTO-02-10-0048
Marin, J. E., and El-Nashar, H. M. (1993). “Pathogenicity of the new phenotypes of Pseudomonas solanacearum from Perú,” in Proceedings of the International Conference Held at Kaohsiung, Taiwan, 28-31 October 1992: Bacterial Wilt ACIAR Proceedings No. 45, eds G. L. Hartman and A. C. Hayward (Canberra, ACT: Australian Centre for International Agricultural Research), 78–84.
Martin, C., French, E. R., and Nydegger, U. (1981). Bacterial wilt of potato in the Amazon Basin. Plant Dis. 65, 246–248. doi: 10.1094/PD-65-246
Milling, A., Meng, F., Denny, T. P., and Allen, C. (2009). Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99, 1127–1134. doi: 10.1094/PHYTO-99-10-1127
Opina, N., Tavner, F., Holloway, G., Wang, J. F., Li, T. H., Maghirang, R., et al. (1997). A novel method for development of species and strain-specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formerly Pseudomonas solanacearum). Asia Pac. J. Mol. Biol. Biotechnol. 5, 19–30.
Pegg, K., and Moffett, M. (1971). Host range of the ginger strain of Pseudomonas solanacearum in Queensland. Aust. J. Exp. Agric. Anim. Husb. 11, 696–698. doi: 10.1071/EA9710696
Perez, A. S., Mejia, L., Fegan, M., and Allen, C. (2008). Diversity and distribution of Ralstonia solanacearum strains in Guatemala and rare occurrence of tomato fruit infection. Plant Pathol. 57, 320–331. doi: 10.1111/j.1365-3059.2007.01769.x
Poussier, S., Prior, P., Luisetti, J., Hayward, C., and Fegan, M. (2000). Partial sequencing of the hrpB and endoglucanase genes confirms and expands the known diversity within the Ralstonia solanacearum species complex. Syst. Appl. Microbiol. 23, 479–486. doi: 10.1016/S0723-2020(00)80021-1
Pradhanang, P. M., Elphinstone, J. G., and Fox, R. T. V. (2000). Identification of crop and weed hosts of Ralstonia solanacearum biovar 2 in the hill of Nepal. Plant Pathol. 49, 403–413. doi: 10.1046/j.1365-3059.2000.00480.x
Prior, P., Ailloud, F., Dalsing, B. L., Remenant, B., Sanchez, B., and Allen, C. (2016). Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17:90. doi: 10.1186/s12864-016-2413-z
Priou, S., Gutarra, L., and Aley, P. (1999). Highly sensitive detection of Ralstonia solanacearum in latently infected potato tubers by post-enrichment ELISA on nitrocellulose membrane. OEPP/EPPO Bull. 29, 117–125. doi: 10.1111/j.1365-2338.1999.tb00805.x
Priou, S., Gutarra, L., De Mendiburu, F., and Llique, R. (2010). Detection of Ralstonia solanacearum (Biovar 2A) in stems of symptomless plants before harvest of the potato crop using post-enrichment DAS-ELISA. Plant Pathol. 59, 59–67. doi: 10.1111/j.1365-3059.2009.02155
Priou, S., Torres, S., Villar, A., Martinez, J., Barrea, O., Gutarra, L., et al. (2000). “Optimization of simple size for the detection of latent infection by Ralstonia solanacearum in potato seed tubers in the highlands of Peru and Bolivia,” in Scientist and Farmer – Partners in Research for the 21st Century, Program Report 1999-2000, (Lima: International Potato Center), 143–153.
Ramsubhag, A., Lawrence, D., Cassie, D., Fraser, R., Umaharan, P., Prior, P., et al. (2012). Wide genetic diversity of Ralstonia solanacearum strains affecting tomato in Trinidad, West Indies. Plant Pathol. 61, 844–857. doi: 10.1111/j.1365-3059.2011.02572.x
Roberts, D. P., Denny, T. P., and Schell, M. A. (1988). Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451. doi: 10.1128/jb.170.4.1445-1451.1988
Safni, I., Cleenwerck, I., De Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Sanchez Perez, A., Meija, L., Fegan, M., and Allen, C. (2008). Diversity and distribution of Ralstonia solanacearum strains in Guatemala and rare occurrence of tomato fruit infection. Plant Pathol. 57, 320–331. doi: 10.1111/j.1365-3059.2007.01769.x
Santiago, T., Lopes, C., Caetano-Anollés, G., and Mizubuti, E. (2016). Phylotype and sequevar variability of Ralstonia solanacearum in Brazil, an ancient centre of diversity of the pathogen. Plant Pathol. 66, 383–392. doi: 10.1111/ppa.12586
Siri, M. I., Sanabria, A., and Pianzzola, M. J. (2011). Genetic diversity and aggressiveness of Ralstonia solanacearum strains causing bacterial wilt of potato in Uruguay. Plant Dis. 95, 1292–1301. doi: 10.1094/PDIS-09-10-0626
Torres, E. M., French, E. R., and Martin, C. (1985). Differential reaction of potato clones to Pseudomonas solanacearum at two sites in Peru. Fitopatologia 20, 1–6.
Toukam, G. M. S., Cellier, G., Wicker, E., Guilbaud, C., Kahane, R., Allen, C., et al. (2009). Broad diversity of Ralstonia solanacearum strains in Cameroon. Plant Dis. 93, 1123–1130. doi: 10.1094/PHYTO-11-11-0300
Wicker, E., Grassart, L., Coranson-Beaudu, R., Mian, D., Guilbaud, C., Fegan, M., et al. (2007). Ralstonia solanacearum strains from Martinique (French west indies) exhibiting a new pathogenic potential. Appl. Environ. Microbiol. 73, 6790–6801. doi: 10.1128/AEM.00841-07
Keywords: bacterial wilt, brown rot, potato, Ralstonia solanacearum, phylotype, sequevar, pathogenicity
Citation: Gutarra L, Herrera J, Fernandez E, Kreuze J and Lindqvist-Kreuze H (2017) Diversity, Pathogenicity, and Current Occurrence of Bacterial Wilt Bacterium Ralstonia solanacearum in Peru. Front. Plant Sci. 8:1221. doi: 10.3389/fpls.2017.01221
Received: 31 March 2017; Accepted: 28 June 2017;
Published: 18 July 2017.
Edited by:
Boshou Liao, Oil Crops Research Institute (CAAS), ChinaReviewed by:
Lucy N. Moleleki, University of Pretoria, South AfricaCopyright © 2017 Gutarra, Herrera, Fernandez, Kreuze and Lindqvist-Kreuze. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannele Lindqvist-Kreuze, aC5saW5kcXZpc3Qta3JldXplQGNnaWFyLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.