
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 11 July 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01181
Rising temperatures during growing seasons coupled with altered precipitation rates presents a challenging task of improving crop productivity for overcoming such altered weather patterns and cater to a growing population. Light is a critical environmental factor that exerts a powerful influence on plant growth and development ranging from seed germination to flowering and fruiting. Higher plants utilize a suite of complex photoreceptor proteins to perceive surrounding red/far-red (phytochromes), blue/UV-A (cryptochromes, phototropins, ZTL/FKF1/LKP2), and UV-B light (UVR8). While genomic studies have also shown that light induces extensive reprogramming of gene expression patterns in plants, molecular genetic studies have shown that manipulation of one or more photoreceptors can result in modification of agronomically beneficial traits. Such information can assist researchers to engineer photoreceptors via genome editing technologies to alter expression or even sensitivity thresholds of native photoreceptors for targeting aspects of plant growth that can confer superior agronomic value to the engineered crops. Here we summarize the agronomically important plant growth processes influenced by photoreceptors in crop species, alongwith the functional interactions between different photoreceptors and phytohormones in regulating these responses. We also discuss the potential utility of synthetic biology approaches in photobiology for improving agronomically beneficial traits of crop plants by engineering designer photoreceptors.
“Light exerts a powerful influence on most vegetable tissues, and there can be no doubt that it generally tends to check their growth” – Charles Darwin, 1880 (The Power of Movement in Plants)
Light is a critical environmental factor that influences growth and development in plants. After seed germination in the soil, etiolated growth enables the germinated seedling to grow toward the soil surface in search of light. Upon exposure to light, the seedling undergoes photomorphogenesis characterized by de-etiolation, chlorophyll synthesis and development of chloroplast all of which enable the seedling to establish itself as an independent autotroph. Quality as well as quantity of incident light influence the developmental and growth of plants (Kami et al., 2010; Li J. et al., 2012). Research in plant photobiology has led to the discovery of several light-absorbing photoreceptor proteins that initiate plant responses to light. These include the red/far-red absorbing phytochromes (Chen and Chory, 2011), blue/UV-A absorbing cryptochromes and phototropins (Christie et al., 2015) and UV-B absorbing UVR8 (Rizzini et al., 2011). Most of these photoreceptors except for UVR8 contain more than one member, with each individual member being encoded by a different gene and sharing a high degree of similarity among the individual members of the same family. Higher plants contain multiple phytochromes (phyA to phy E), three cryptochromes (cry1, cry2, and cry3), two phototropins (phot1 and phot2), and one UVR8 photoreceptor. Land plants also contain a family of blue-light absorbing proteins referred to as ZTL/FKF1/LKP2 (ZEITLUPE/FLAVIN-BINDING KELCH REPEAT F-BOX 1/LOV KELCH PROTEIN 2) having a combination of photoreceptor and F-box protein activities within the same protein (Ito et al., 2012). Photoreceptors are widely distributed among land plants, with most of the commonly cultivated crop species reported to contain members of at least one major photoreceptor family (Kharshiing and Sinha, 2015). While most of our understanding on the roles of photoreceptors in plant development has been derived from research on Arabidopsis, there is an increasing interest in photoreceptor-mediated developmental responses in an agricultural environment (Ballaré et al., 1992; Sawers et al., 2005; Hudson, 2008; Wargent and Jordan, 2013; González et al., 2015) since several plant developmental responses involve action of one or more of these photoreceptors.
Plant growth and development involves complex signaling networks which are tightly regulated by genetic and environmental factors. These factors influence a plant’s ability to germinate, adapt, survive, and reproduce in natural conditions. In agriculture, the genetic constitution of different crop species have been altered via various crop breeding programs for enhancing their survival and productivity under different environmental conditions (Osakabe et al., 2011; Slade and Moehs, 2011; Suprasanna and Nakagawa, 2011; Ukai and Nakagawa, 2011). Regulating plant developmental processes such as biomass production, flowering, fruiting and senescence are of particular interest to agricultural scientists for enhancing productivity. Additionally, processes that regulate disease and/or pest resistance (Jeong et al., 2010), starch metabolism (Zeeman et al., 2010), fruit size and quality (Causse et al., 2006), shelf-life of fruits and tubers (Dahmani-Mardas et al., 2010) and plant productivity under high planting index (Warnasooriya and Brutnell, 2014) can also influence productivity of agricultural crops. Various reports have shown that plant photoreceptor action can influence plant growth at different developmental stages including but not limited to seed germination, plant architecture, flowering, reproduction, biomass accumulation and senescence. Plant photoreceptors and/or their signaling components are therefore attractive targets for altering productivity and yield in crop plants for future food and non-food applications (Kharshiing and Sinha, 2015). Here we discuss various photoreceptor-controlled developmental processes that can affect plant productivity and the practical implications for engineering such photoreceptors toward enhancing productivity of crop plants.
Molecular genetics approaches in plant photobiology have led to the isolation and characterization of photoreceptor genes from various species, most of which play critical roles in determining plant survival and development. Phytochromes, which are principal receptors for light in the red/far-red region of the spectrum (600–750 nm), play an essential role in regulating seed germination and seedling establishment in agriculturally important crops such as rice and tomato (Chung and Paek, 2003; Appenroth et al., 2006; Eckstein et al., 2016), while in maize, phytochromes have been implicated to contribute to the transcription of genes essential for photosynthesis (Markelz et al., 2003). Under natural or field conditions, establishment of the emergent seedling post germination is a vital developmental process that determines the ability of the plant to survive, grow, and reproduce. In photosynthesising organisms such as green plants, light is a crucial factor in determining the establishment of an emergent seedling. Maximizing photosynthesis during early development and following the formation of gaps during growth under dense canopies could therefore be critical for seedling survival. Under high plant densities, shade provided by neighboring vegetation triggers a series of developmental changes in growing plants involving several of the known photoreceptors (Ballaré and Pierik, 2017). The actions of phytochromes enables a plant to quantify shade around a seedling’s environment by detecting changes in the Red/Far-red ratios (R:FR) and trigger a series of developmental responses that is thought to provide the plant with a competitive advantage over its neighbors (Schmitt et al., 2003, Figure 1). Such responses include stimulation of elongation growth, coupled with reduced leaf development, increased apical dominance, and a reduction in branching (Franklin, 2008). While such developmental plasticity to diminished light, termed as shade avoidance responses (SAR), might increase survival percentage under limiting light conditions, it could result in compromised productivity of crop plants especially in modern intensive cropping methods with high planting densities (Weijschedé et al., 2006; Weinig et al., 2006).
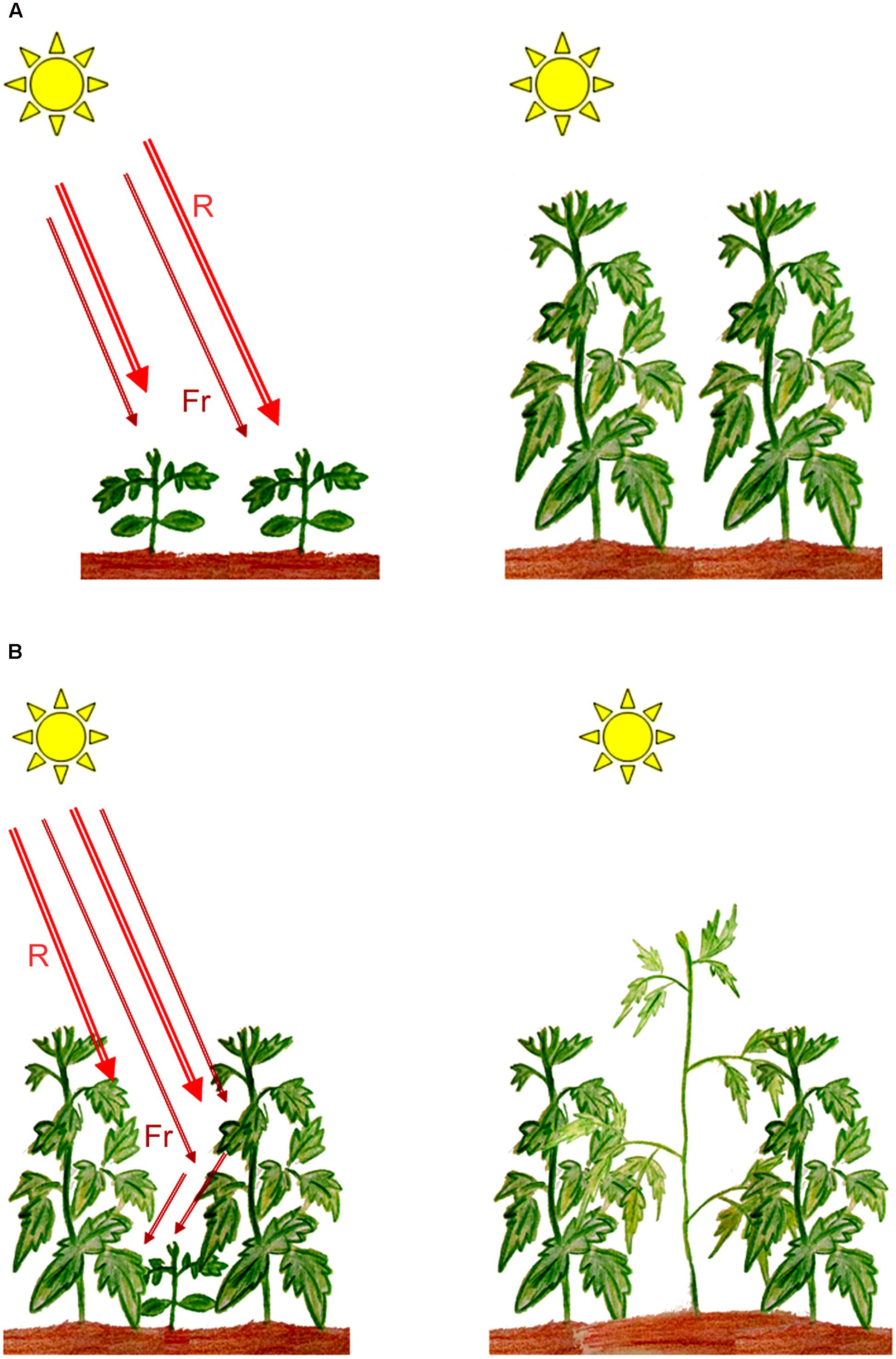
FIGURE 1. Induction of shade avoidance responses (SAR) in plants growing under shade. (A) Plants growing in full sunlight; (B) Low Red/Far-red ratios perceived by plants growing under shade resulting in SAR. R, red light; Fr, far-red light. (Artwork by Eros Kharshiing).
In modern agricultural practices, enhanced yield in food crops such as maize have been achieved by use of varieties that perform optimally at high planting densities but require higher inputs of fertilizer (Warnasooriya and Brutnell, 2014). Modulating the responses of crop plants to vegetative shade for increasing harvestable biomass under high-density planting by manipulating light signaling networks that are fundamental to shade response therefore presents itself as an attractive alternative. Mutant lines harboring lesions in PHYA and PHYB, two of the more functionally important phytochrome genes in plants, show extreme SAR (Kendrick et al., 1997; Weller et al., 2001; Franklin, 2008; Casal, 2012). The overexpression of these genes could, however, compensate for reduced available light in densely grown crops resulting in increased yield (Robson et al., 1996; Boccalandro et al., 2003; Garg et al., 2006). In Arabidopsis, Devlin et al. (2003) have identified a number of shade responsive genes that are also regulated by phyA and phyB. Recently in Arabidopsis and tomato, phytochromes have been suggested to influence plant biomass along with carbon assimilation and starch metabolism (Kharshiing and Sinha, 2016; Yang et al., 2016). Altering responses of crop plants to light spectral quality by targetting genes such as phytochromes or their signaling components can therefore influence both resource partitioning and growth patterns in crop plants, which would presumably result in higher productivity even under intensive cropping patterns (Warnasooriya and Brutnell, 2014).
Apart from phytochromes, two additional classes of photoreceptors have been reported in plants, which absorb blue/UV-A light, viz., cryptochromes and phototropins (Yu et al., 2010; Christie et al., 2015). In plants, the cryptochrome family photoreceptors participate in several plant process, ranging from photomorphogenesis to entrainment of the circadian clock (Lin, 2002; Botto et al., 2003; Millar, 2003; Giliberto et al., 2005). Developmental processes such as plant height and flowering time which are agronomically important traits are also linked to cryptochrome activity (Mockler et al., 2003; Yu and Lin, 2005; Sharma P. et al., 2014). Since the isolation of the first CRY gene from Arabidopsis, CRYs have been reported from most major crops investigated (Kharshiing and Sinha, 2015) having roles in seed germination, leaf senescence, stress responses and regulation of transcription (Lopez et al., 2012; Meng et al., 2013; Barrero et al., 2014; Facella et al., 2017). As Arabidopsis is limited as a model plant for examining the molecular networks influencing agronomically important traits (Kimura and Sinha, 2008) several workers have evaluated the role of photoreceptors in other crop models. In Brassica napus, an oilseed crop, the overexpression of CRY1 produced short-statured plants, which confer resistance to wind and water lodging (Sharma P. et al., 2014), providing such plants with an adaptive advantage in these conditions. Tomato plants overexpressing CRY2 also displayed short hypocotyl and internode length alongwith delayed flowering (Giliberto et al., 2005). Interestingly, in Arabidopsis, cryptochromes have also been reported to promote growth in a shaded environment (Pedmale et al., 2016), suggesting new a molecular target for altering shade responses in plants. Tomato transgenic lines overexpressing CRY2 overproduced anthocyanins and chlorophyll in leaves alongwith enhanced flavonoids and lycopene content in fruits. In soybean, transgenic lines with upregulated CRY2a expression show delayed senescence while senescence is accelerated in lines with downregulated CRY2a expression (Meng et al., 2013). Manipulation of CRY2 also profoundly affects the molecular pathways related to biotic/abiotic stress, photorespiration, photosynthesis, as well as secondary metabolism pathways, such as biosynthesis of phenylpropanoids, phenolic, and flavonoid/anthocyanin in tomato (Lopez et al., 2012). Tomato lines overexpressing CRY2 show major changes in the rhythmic oscillations of several genes involved in the entrainment of the endogenous clock (Facella et al., 2008) suggesting a role for the photoreceptors in the input to the tomato biological clock. Interestingly, analyses of circadian clock mutants in Arabidopsis, revealed that genes involved in carbohydrate metabolism including starch synthesis and degradation were affected in all mutant lines tested (Graf et al., 2017). This suggests that inputs to the endogenous clock is vital to the proper regulation of critical physiological and developmental responses in plants. Since cryptochromes play critical roles in the entrainment of the endogenous clock in response to light (Millar, 2003), its genetic manipulation could affect agronomically important traits including starch metabolism which would consequently impact productivity in crop plants.
Phototropins are the principal photoreceptors for blue-light phototropism in plants (Briggs and Christie, 2002; Christie et al., 2015). Besides phototropism, phototropins mediate other critical adaptive responses of plants to the surrounding light environment, which serve to enhance the photosynthetic status of the plants. The opening of stomatal pores which allow for exchange of water and carbon dioxide is redundantly controlled by phototropins (Boccalandro et al., 2012; Sharma S. et al., 2014). The movement of chloroplasts in response to different light intensities is also regulated by the phototropins. When plants are exposed to low/weak light conditions, both phot1 and phot2 induce accumulation of the chloroplasts to the upper surface of the palisade mesophyll cells of leaves to maximize photosynthetic light capture. Under strong light conditions, phot2 mediates the rearrangement of chloroplasts parallel to the direction of the light source so as to minimize photo-damage (Briggs and Christie, 2002; Kasahara et al., 2004). In field conditions, Arabidopsis mutants lacking phototropins have been shown to have reduced photosynthesis (Boccalandro et al., 2012), which could be partially due to the inability of the plants to maximally utilize photosynthetically active radiation (PAR) as the lack of phototropin-mediated adjustment of chloroplast position would inhibit optimal capture of PAR. It is also interesting to note that under laboratory conditions, phototropins promote growth in response to blue light under low light environments (Takemiya et al., 2005). While both phot1 and phot2 are involved in growth enhancement, phot1 is more sensitive than phot2 in promoting growth under low blue light, which can affect plant development in natural conditions of low light. On the other hand, phototropin mutants in Chlamydomonas have been shown to display reduced fitness under excessive light indicating a role for phototropins in photosynthetic regulation under high light conditions (Petroutsos et al., 2016). While similar findings have not yet been reported for higher plants, such reports underline the versatile roles that photoreceptors play in growth and development of plants.
Ultraviolet-B radiation (UV-B) is a key component of the radiation environment that is utilized by plants as a signal for UV acclimation and survival in sunlight (Jansen, 2002; Jenkins, 2014). The discovery of the UV-B responsive UVR8 locus in plants (Kliebenstein et al., 2002; Rizzini et al., 2011) and resultant works (reviewed in Tilbrook et al., 2013; Jenkins, 2014) has tremendously progressed our understanding of how this photoreceptor responds to UV-B at the molecular and biochemical levels. While our knowledge of UVR8 function in vivo is still at a very nascent stage, its manipulation might have potential applications in crop improvement. The vegetative phase of plant growth is characterized by key developmental processes such as cell division and elongation, directional growth and branching. Low fluence UV-B regulates several of these developmental responses in plants right from seedling to adult stages (Suesslin and Frohnmeyer, 2003; Shinkle et al., 2004; Wargent et al., 2009). In adult plants, morphogenic responses to UV-B include alterations in leaf characteristics such as leaf area, leaf thickness, leaf mass, and stomatal index (Wargent et al., 2009; Robson et al., 2015), Plants exposed to UV-B radiation also show decrease in chlorophyll content and in chla/b ratio (Lidon and Ramalho, 2011) alongwith decrease in photosynthetic efficiency (Lidon et al., 2012). Conversely, promotion of photosynthetic efficiency of Arabidopsis seedlings exposed to elevated levels of UV-B occurs via UVR8 (Davey et al., 2012). Furthermore, lettuce seedlings exposed to UV-B radiation at early stages of development also resulted in increase in productivity and biomass (Tsormpatsidis et al., 2010; Wargent et al., 2011), which could be due to the photoprotective effects of early UV-B exposure. UV-B radiation has also been reported to induce accumulation of secondary metabolites such as flavonoids (Treutter, 2005), which is reportedly controlled by UVR8 (Jenkins, 2014).
Land plants also contain a family of LOV (Light Oxygen or Voltage) photoreceptors referred to as ZEITLUPE/FLAVIN-BINDING KELCH REPEAT, F-BOX1/LOV KELCH PROTEIN2 (ZTL/FKF1/LKP2) having both photoreceptor and F-box protein activities within the same protein (Ito et al., 2012). Interestingly genes coding for this family of photoreceptors are phyllogenetically divided into two groups in dicots and monocots (Boxall et al., 2005; Taylor et al., 2010), suggesting different functions for these genes. However, the high levels of structural conservedness of ZTL and FKF1 homologs among different monocots and dicots may be suggestive of a certain level of functional conservedness of these genes across species (Taylor et al., 2010). Indeed, mutant analyses in Arabidopsis and other species have revealed that ZTL/FKF1/LPK2 regulate similar developmental pathways across different species (Somers et al., 2004; Yon et al., 2016). In Arabidopsis ZTL mutants, presence of FKF1 and LKP2 compensates for the absence of ZTL in as far as in determining circadian rhythm (Baudry et al., 2010) suggesting some level of functional redundancy between these photoreceptors even though they are reported to have distinct roles in photoperiodic flowering (Song et al., 2014). In crop plants, such as soybean GmZTL3 a homolog of Arabidopsis ZTL has also been suggested to function as a photoreceptor controlling timing of flowering (Xue et al., 2012), a critical developmental response for crop plants. For a comprehensive review on structure and functions of ZTL/FKF1/LKP2 proteins, readers can refer the works of Suetsugu and Wada (2013) and Zoltowski and Imaizumi (2014). Table 1 provides a summary of the effects of photoreceptors on agronomic traits in crops.
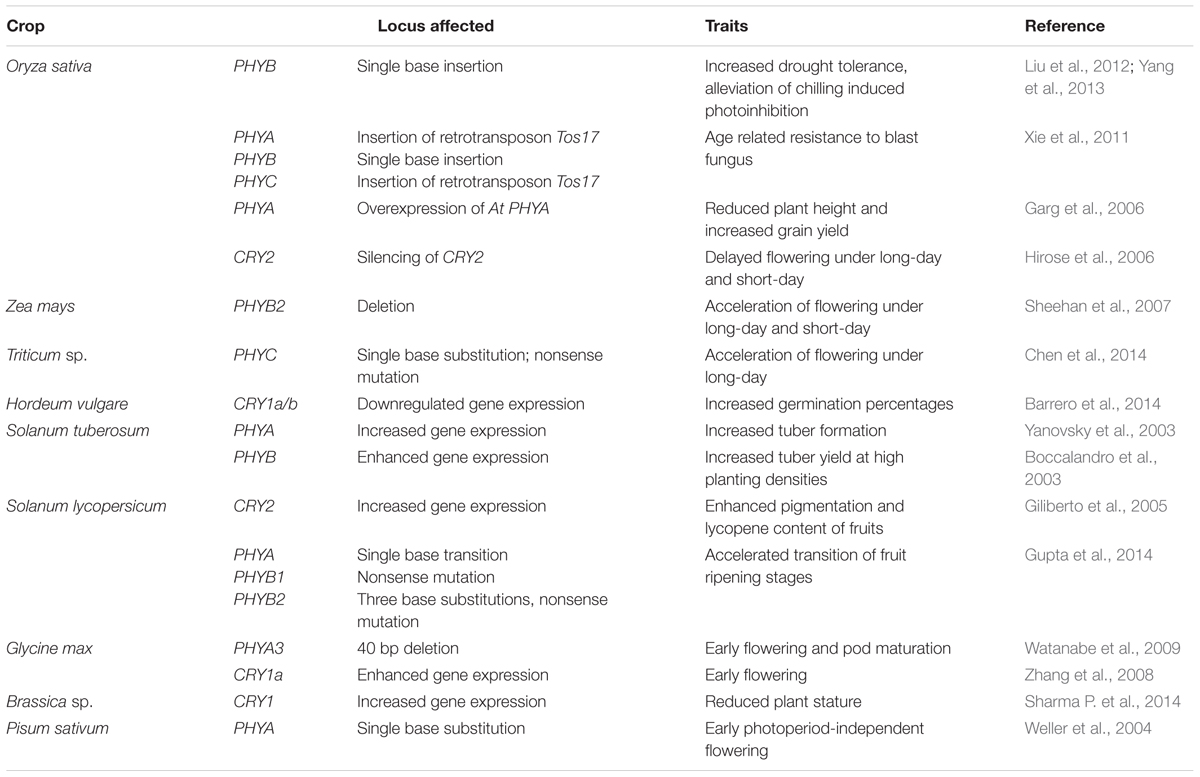
TABLE 1. Agronomic traits in few crops affected by mutations in photoreceptor genes or by altered expression of photoreceptor genes.
Since natural light is composed of different wavelengths, higher plant growing under natural conditions will invariably have activation of more than one photoreceptor at the same time. Such simultaneous activation could thus result in signaling networks where action of one photoreceptor can be affected by the activity of other photoreceptors. While some developmental responses in plants are specifically triggered by activation of a single photoreceptor, there are several instances where signaling networks downstream of light perception of more than one photoreceptor of the same or different family are co-ordinately activated to ensure proper growth and development. Such developmental processes include seedling germination (Bertram et al., 2004; Dechaine et al., 2009), photomorphogenesis (Neff and Chory, 1998; Weller et al., 2001; Kami et al., 2010), plant and leaf architecture (Takemiya et al., 2005; Kozuka et al., 2013), flowering (Más et al., 2000; Weller et al., 2001; Mockler et al., 2003; Endo et al., 2016), and fruit quality (Gupta et al., 2014; González et al., 2015) all of which eventually affect plant productivity. The availability of photomorphogenic mutant and transgenic lines in Arabidopsis, rice, tomato, and other model species have provided valuable information into the functional redundancy and interactions of the signaling pathways of different photoreceptors during plant development (Table 2). These interactions become especially relevant in the field where plants are subject to uncontrolled environmental conditions of light, temperature, moisture, etc. A detailed understanding of such functional interactions between photoreceptors would further enable researchers to utilize the information for enhancing productivity of crop plants for food and non-food applications.
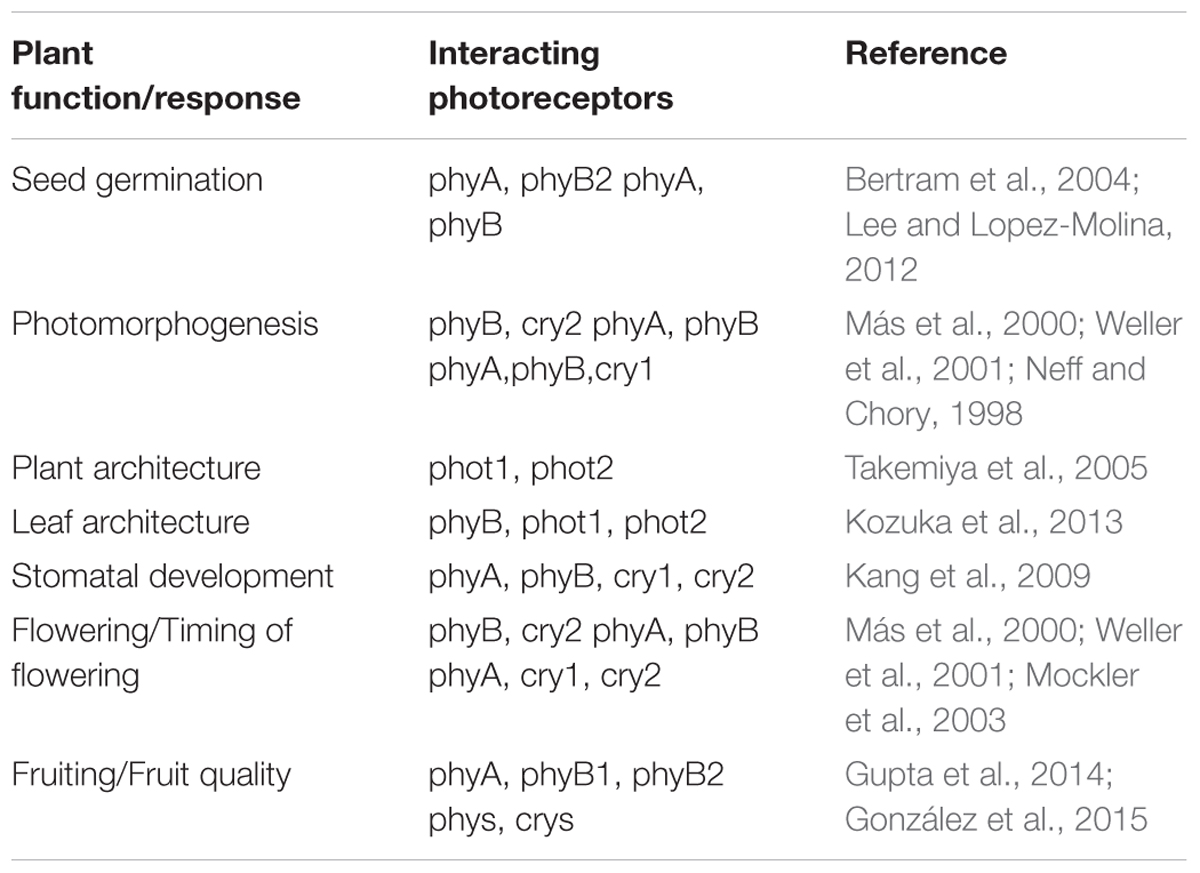
TABLE 2. Examples of plant developmental processes involving functional interaction of two or more photoreceptors.
Light is an important signal that functions as a developmental switch in germination and photomorphogenesis. Seedling emergence from soil and subsequent photomorphogenic development involve a vast array of photoreceptors which enable the establishment of emergent seedlings. Activation of these photoreceptors by light results in a range of signaling events which co-ordinate plant growth and development that direct seedling emergence from soil and establish them as autotrophs. There is growing evidence that most of these signaling cascades involve interaction of light and phytohormones signaling pathways (Wang et al., 2013). Since the initial proposal of the Cholodny–Went theory regarding the asymmetric distribution of auxin during shoot phototropism (Koepfli et al., 1938), much information has emerged on the links between light perception and hormonal regulation in plant growth responses. In plants seed germination is inhibited by abscisic acid (ABA) while gibberellic acid (GA) induces germination (Jacobsen et al., 2002; Seo et al., 2006). It is now known that light induces seed germination via the interaction of phytochromes and its partner PIF1 (PHYTOCHROME INTERACTING FACTOR 1) also known as PIF3-LIKE 5 (PIL5), which in turn regulates both ABA and GA signaling through the same downstream regulators (Bae and Choi, 2008; Seo et al., 2008; Oh et al., 2009; de Wit et al., 2016). It seems that phytochrome-mediated degradation of PIF1 is therefore a central mechanism by which light induces seed germination by altering ABA and GA metabolism in seeds. PIFs are also implicated to play a role during seedling transition from a skotomorphogenic to a photomorphogenic mode of development in light by regulating GA levels (Oh et al., 2007). In Arabidopsis, many regulators of hormone signaling including auxin and cytokinin (Cluis et al., 2004), ABA (Chen et al., 2008), ethylene (Yu et al., 2013), jasmonic acid (JA) (Prasad et al., 2012), GA (Alabadí et al., 2008), and brassinosteroid (Li and He, 2016) are reported to share a common signaling node involving the basic leucine zipper (bZIP) transcription factor ELONGATED HYPOCOTYL 5 (HY5), whose cellular accumulation involves light signaling transduction through phytochromes and cryptochromes (Osterlund et al., 2000; Wang et al., 2001).
Timing of flowering is an essential developmental process in flowering plants that is important for reproduction. In plants, the timing of flowering is determined by the length of the daylight period during a day (or photoperiod). Broadly, flowering plants are categorized into those that flower when length of daylight exceeds a particular length (called critical length) or long-day plants, those that flower when length of daylight is below the critical length or short-day plants and those that flower independent of the critical length or day-neutral plants. phyA, phyB, and cry2 were the first photoreceptors reported to be involved in regulating the timing of flowering in Arabidopsis as well as in crop plants. In rice, mutations in PHYB or PHYC cause moderate alteration in flowering while lines carrying mutations in PHYA coupled with mutations in either PHYB or PHYC show very early flowering (Takano et al., 2005). Under long-day conditions, both PHYC and PHYB-null mutants of wheat also exhibit severe delay in flowering (Chen et al., 2014; Pearce et al., 2016). A study of the transcriptomes of PHYB-null and PHYC-null mutants indicate that phyB also plays a prominent role in regulating GA, BR, auxin, ABA, and ethylene biosynthesis (Pearce et al., 2016), all of which have been implicated in the photoperiodic control of flowering (Galvão and Schmid, 2014). Of these, the regulation of endogenous GA levels may be important for light-induced flowering. Downregulation of the GA biosynthesis gene GA20OX showed delayed flowering in FR enriched environment suggesting a role for this gene in phytochrome-dependent flowering (Hisamatsu and King, 2008). While the association of light and auxin signaling is well established, the tomato pct1-2 mutant having enhanced polar transport of auxin shows delayed phototropism (Kharshiing et al., 2010a,b) as well as increased number of flowers which bloom at the same time as compared to wild type which have lesser flowers and which bloom sequentially (Al-Hammadi et al., 2003). However, the flowers of the pct1-2 mutant are male sterile because the anthers lack dehiscence. Similarly in Arabidopsis anther dehiscence is regulated by auxin synthesis (Cecchetti et al., 2008) and also involves the auxin transporters ABCB1 and ABCB19 (Cecchetti et al., 2015). The involvement of ABCB19 as a substrate target for phot1 during shoot phototropism in Arabidopsis (Christie et al., 2011) is further illustration of the overlap of photoreceptor and hormone signaling events in plants. Furthermore, while phytochromes are reported to regulate fruit development and ripening in tomato (Gupta et al., 2014), the overproduction of nitric oxide, a secondary messenger in signaling pathways for several plant hormones, in the shr mutant of tomato suppresses fruit growth and ripening (Negi et al., 2010; Bodanapu et al., 2016) implying a possible cross-talk of the two signaling pathways.
In plants, while the effect of light on photomorphogenesis and photosynthesis has been known for a long time, it is now becoming increasingly evident that light signaling is also an integral component in determining the outcome of plant-pathogen interactions (Griebel and Zeier, 2008; Ballaré et al., 2012; Demkura and Ballaré, 2012; Erb et al., 2012). Several studies have shown that phytochromes and UVR8 influence plant defense responses (reviewed in Ballaré, 2014; Mazza and Ballaré, 2015; Gommers et al., 2017) while cryptochrome 2 and phototropin 2 have been reported to mediate resistance protein-mediated plant defense against viral but not bacterial pathogens (Jeong et al., 2010). These photoreceptors have been shown to influence plant defense responses by regulating hormone signaling pathways such as that of salicylic acid (SA) and JA (Xie et al., 2011; Moreno and Ballaré, 2014). For a more detailed review on light and hormone signaling, readers can refer to Ballaré (2014), Lucas and Prat (2014); de Wit et al. (2016) and references mentioned therein. The reports mentioned here are few examples of how light and hormone metabolism seem to affect related aspects of plant development that could eventually impact productivity (Figure 2). As the mechanisms of signal integration of light and hormone signaling pathways are starting to become clearer, we will be in a better position to understand how light signaling interacts with hormone signaling to regulate traits of agronomic interest in plants.
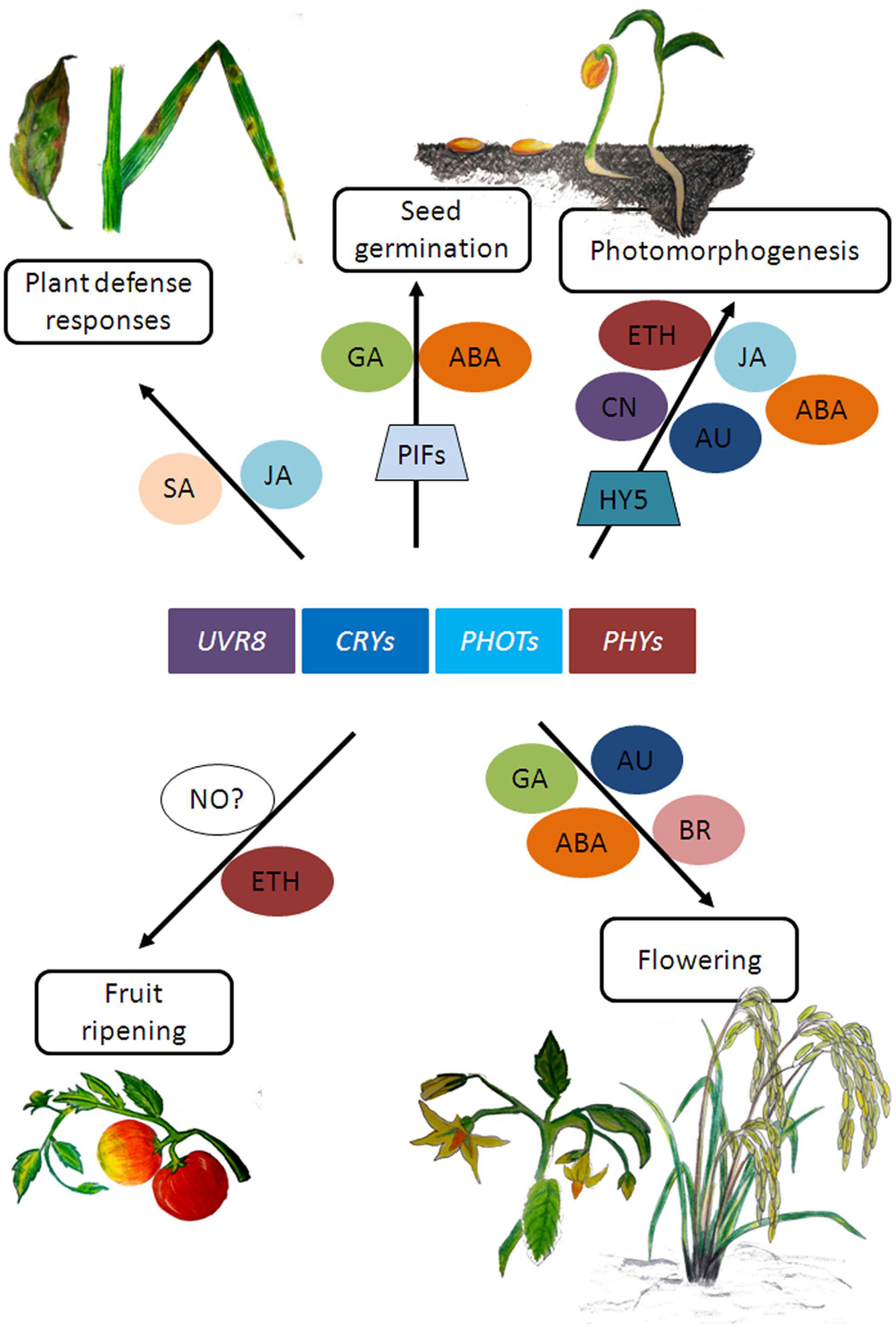
FIGURE 2. Schematic representation of few plant growth responses involving photoreceptor and hormone signaling. SA, salicylic acid; JA, jasmonic acid; GA, gibberellic acid; ABA, abscisic acid; CN, cytokinins; ETH, ethylene; NO, nitric oxide; BR, brassinosteroids; AU, auxin. (Artwork by Rimeia C. Lyngdoh and Eros Kharshiing).
The myriad developmental processes regulated by light suggest that the manipulation of genes involved in light signaling pathways could be a viable tool for crop improvement (Kharshiing and Sinha, 2015). Rapid advancement in genomics is generating new tools for editing genomes which can be utilized for modification of molecular components regulating development in plants (Belhaj et al., 2013; Song et al., 2016). Functional genomics studies, large-scale sequencing and eco-tilling strategies are rapidly identifying polymorphisms between cultivars and landraces resulting in large datasets of molecular diversity among crop plants. Such data is likely to result in the identification of natural alleles of photomorphogenic genes, as well as those that have arisen as a consequence of crop breeding. Genome projects of crops such as rice, maize, tomato, soybean, and others are increasingly generating information which can be used to effect targeted dwarfing, alter SAR, enhance yield and regulate fruiting and ripening in fruit crops by modifying photoreceptors and/or their signaling pathways. Unlike genetically modified transgenics, the upregulation or downregulation of genes in an organism does not necessarily require the introduction of foreign genes into the organism. Several reports on mutation studies also provide evidence that traits in an organism can be modified without the introduction of foreign genes into the system. Many agronomically valuable phenotypes and naturally occurring variants of crop plants have been caused by point, or only a few, mutations. During the last 70 years, more than 3200 crop varieties derived directly as mutant or their progenies have been released worldwide (Pathirana, 2011; Manova and Gruszka, 2015). Many of these mutant-derived varieties have significant economic value such as shorter growth cycle (Ahloowalia et al., 2004), semi-dwarf habit, high harvest index and drought tolerance (D’Souza et al., 2009) and disease resistance (D’Souza et al., 2009; Pathirana, 2011).
Gene targeting via site-directed mutagenesis is a technique that is commonly used in molecular biology to introduce mutations in defined site(s) on the genome. Apart from its utility in studying gene function, the versatility of these techniques also enables researchers to produce gene knockouts or point mutations in plants which can be used for molecular breeding purposes. For a more detailed summary of the various gene targeting strategies in higher plants via site-directed mutagenesis, the reader can refer to Osakabe and Osakabe (2014). While gene targeting in plant photobiology has been commonly utilized for elucidating photoreceptor function and/or signaling, such strategies have been commonly employed for improving the efficiency of reporter genes such as GFP (Cinelli et al., 2000) or iLOV (Christie et al., 2012) which is a derivative of the Light Oxygen Voltage (LOV) domains of phototropin blue-light receptors (Chapman et al., 2008). Similarly, the availability of sequence information of various plant photoreceptors coupled with information from functional genomic studies presents an ideal situation for researchers to engineer photoreceptors or even their downstream signaling partners for improving agronomically valuable traits in crop plants. These could include altering the sensitivity thresholds, photocycles or kinase activity of the different photoreceptor proteins. The emergence of large-scale screening technologies such as TILLING (McCallum et al., 2000; Henikoff et al., 2004; Kurowska et al., 2011) would also allow researchers to couple random mutagenesis with targeted-selected mutagenesis for selecting mutations in photoreceptor genes for subsequent evaluation for desirable traits without the involvement of transgenic modifications.
For many centuries, humans have been constantly modifying plants that are beneficial to them. With advancements in crop improvement practices, modification of plant characteristics that were beneficial especially in terms of yield, were favorably targeted. For many decades, traditional breeding practices along with mutation breeding programs have been instrumental in modifying agricultural traits in crop plants. However, a growing global population coupled with altered weather patterns, present multifaceted challenges to future agricultural production both for food and non-food applications. Synthetic biology has emerged as a viable technology for rapid, precise, and robust engineering of organisms for useful societal purposes (Shih et al., 2016). Synthetic biology attempts to create user-designed biological systems including plants which can display various characteristics such as responses to nutrition status, infections or to changes in environment. One defining emphasis of synthetic biology is the designed control of gene expression. Through various approaches, gene expression can be controlled at the DNA, RNA, or protein level, depending on the strategy or application. The increasing availability of resources to efficiently inactivate or replace genes in complex organisms such as plants, has led to a revolution in genome editing in the plant science community. Gene editing tools allow targeting of specific DNA sequences within the plant genome thereby enabling researchers to engineer genes and genomes with higher precision than would have been possible earlier. The various tools and technologies utilized in plant synthetic biology have been discussed in previous reviews (Bortesi and Fischer, 2015; Schaeffer and Nakata, 2015; Puchta, 2016; Weeks et al., 2016). While most of the focus of synthetic biology has been on microbes, plant synthetic biology has made rapid progress in recent years. Within the last few years, synthetic biology approaches for regulating plant responses has been successfully demonstrated in several crop species (Table 3). As gene editing techniques become more precise, the modulation of photoreceptor activity by engineering designer photoreceptors provides novel opportunities for improving productivity of crop plants. While different engineered photoreceptor systems may require different designs for optimal function (Schmidt and Cho, 2015), such photoreceptor systems can be utilized to determine how the light input affects agronomically beneficial phenotypes.

TABLE 3. Examples of agronomically beneficial traits in some crop plants through application of genome editing technologies.
Genomic studies have shown that light induces extensive reprogramming of gene expression patterns in plants (Li L. et al., 2012; Petrillo et al., 2014; Perrella and Kaiserli, 2016). The emergence of more precise and robust gene modification technologies provides researchers with exciting options to engineer photoreceptors and/or their signaling components for modulating the response of plants to light inputs. Such targeted gene editing technologies would allow for subsequent engineering of light responses for development of useful agronomic traits in both food and non-food crops. As seen in Table 1, alterations in gene function arising due to mutations or altered expression levels of photoreceptors can produce agronomically desirable traits in crops. While research on crops is gaining momentum, the rapid pace of research in Arabidopsis and other established model species is continually contributing large amounts of information on photoreceptor signaling which is yet to be translated to research in crops. However, the information gained from such studies can assist researchers to engineer photoreceptors via genome editing technologies to alter expression or even sensitivity thresholds of native photoreceptors for targeting aspects of plant development that can confer superior agronomic value to the engineered crops. While genome editing in crop plants may have perceived safety concerns (Cardi, 2016) which would influence the future integration of these crops into society (Araki and Ishii, 2015), the myriad of agronomically desirable traits that are developmentally regulated by different photoreceptors either alone or in concert with other photoreceptors and signaling pathways favorably places these genes as most suitable candidates for molecular breeding approaches for enhancing the agronomic value of domesticated lines.
EK designed and directed the study, OM and EK carried out the study, EK wrote the manuscript. All authors read and approved the manuscript.
EK is grateful to the Science and Engineering Research Board (Department of Science and Technology), New Delhi, India for supporting research in his laboratory vide grants no. SB/EMEQ-152/2014 and SR/FT/LS-82/2011.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Rimeia C. Lyngdoh, Department of Botany, St. Edmund’s College for assisting with the artwork.
Ahloowalia, B. S., Maluszynski, M., and Nichterlein, K. (2004). Global impact of mutation-derived varieties. Euphytica 135, 187–204. doi: 10.1023/B:EUPH.0000014914.85465.4f
Ainley, W. M., Sastry-Dent, L., Welter, M. E., Murray, M. G., Zeitler, B., Amora, R., et al. (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. doi: 10.1111/pbi.12107
Alabadí, D., Gallego-Bartolomé, J., Orlando, L., García Cárcel, L., Rubio, V., Martínez, C., et al. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53, 324–335. doi: 10.1111/j.1365-313X.2007.03346.x
Al-Hammadi, A. S., Sreelakshmi, Y., Negi, S., Siddiqi, I., and Sharma, R. (2003). The polycotyledon mutant of tomato shows enhanced polar auxin transport. Plant Physiol. 133, 113–125. doi: 10.1104/pp.103.025478
Appenroth, K. J., Lenk, G., Goldau, L., and Sharma, R. (2006). Tomato seed germination: regulation of different response modes by phytochrome B2 and phytochrome A. Plant Cell Environ. 29, 701–709. doi: 10.1111/j.1365-3040.2005.01455.x
Araki, M., and Ishii, T. (2015). Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 20, 145–149. doi: 10.1016/j.tplants.2015.01.010
Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59, 281–311. doi: 10.1146/annurev.arplant.59.032607.092859
Ballaré, C. L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65, 335–363. doi: 10.1146/annurev-arplant-050213-040145
Ballaré, C. L., Mazza, C. A., Austin, A. T., and Pierik, R. (2012). Canopy light and plant health. Plant Physiol. 160, 145–155. doi: 10.1104/pp.112.200733
Ballaré, C. L., and Pierik, R. (2017). The shade avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. [Epub ahead of print]. doi: 10.1111/pce.12914
Ballaré, C. L., Scopel, A. L., Sánchez, R. A., and Radosevich, S. R. (1992). Photomorphogenic processes in the agricultural environment. Photochem. Photobiol. 56, 777–788. doi: 10.1111/j.1751-1097.1992.tb02234.x
Barrero, J. M., Downie, A. B., Xu, Q., and Gubler, F. (2014). A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 26, 1094–1104. doi: 10.1105/tpc.113.121830
Baudry, A., Ito, S., Song, Y. H., Strait, A. A., Kiba, T., Lu, S., et al. (2010). F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622. doi: 10.1105/tpc.109.072843
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., and Nekrasov, V. (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39. doi: 10.1186/1746-4811-9-39
Bertram, L., Diara, C., and Lercari, B. (2004). Seed germination in tomato is controlled by physiological interactions of multiple phytochromes. Adv. Horticult. Sci. 18, 3–10.
Boccalandro, H. E., Ploschuk, E. L., Yanovsky, M. J., Sánchez, R. A., Gatz, C., and Casal, J. J. (2003). Increased phytochrome B alleviates density effects on tuber yield of field potato crops. Plant Physiol. 133, 1539–1546. doi: 10.1104/pp.103.029579
Boccalandro, H. N. E., Giordano, C. V., Ploschuk, E. L., Piccoli, P. N., Bottini, R. N., and Casal, J. J. (2012). Phototropins but not cryptochromes mediate the blue light-specific promotion of stomatal conductance, while both enhance photosynthesis and transpiration under full sunlight. Plant Physiol. 158, 1475–1484. doi: 10.1104/pp.111.187237
Bodanapu, R., Gupta, S. K., Basha, P. O., Sakthivel, K., Sadhana, Sreelakshmi, Y., and Sharma, R. (2016). Nitric oxide overproduction in tomato shr mutant shifts metabolic profiles and suppresses fruit growth and ripening. Front. Plant Sci. 7:1714. doi: 10.3389/fpls.2016.01714
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Botto, J. F., Alonso-Blanco, C., Garzarón, I., Sánchez, R. A., and Casal, J. J. (2003). the cape verde islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiol. 133, 1547–1556. doi: 10.1104/pp.103.029546
Boxall, S. F., Foster, J. M., Bohnert, H. J., Cushman, J. C., Nimmo, H. G., and Hartwell, J. (2005). Conservation and divergence of circadian clock operation in a stress-inducible crassulacean acid metabolism species reveals clock compensation against Stress. Plant Physiol. 137, 969–982. doi: 10.1104/pp.104.054577
Briggs, W. R., and Christie, J. M. (2002). Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. doi: 10.1016/S1360-1385(02)02245-8
Cardi, T. (2016). Cisgenesis and genome editing: combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 135, 139–147. doi: 10.1111/pbr.12345
Causse, M., Damidaux, R., and Rousselle, P. (2006). “Traditional and enhanced breeding for quality traits in tomato,” in Genetic Improvement of Solanaceous Crops: Tomato, Vol. 2, eds M. K. Razdan and A. Mattoo (Enfield, NH: Science Publishers), 153–193. doi: 10.1201/b10744-6
Cecchetti, V., Altamura, M. M., Falasca, G., Costantino, P., and Cardarelli, M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20, 1760–1774. doi: 10.1105/tpc.107.057570
Cecchetti, V., Brunetti, P., Napoli, N., Fattorini, L., Altamura, M. M., Costantino, P., et al. (2015). ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. Integr. J. Plant Biol. 57, 1089–1098. doi: 10.1111/jipb.12332
Chapman, S., Faulkner, C., Kaiserli, E., Garcia-Mata, C., Savenkov, E. I., Roberts, A. G., et al. (2008). The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. U.S.A. 105, 20038–20043. doi: 10.1073/pnas.0807551105
Chen, A., Li, C., Hu, W., Lau, M. Y., Lin, H., Rockwell, N. C., et al. (2014). PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. U.S.A. 111, 10037–10044. doi: 10.1073/pnas.1409795111
Chen, H., Zhang, J., Neff, M. M., Hong, S. W., Zhang, H., Deng, X., et al. (2008). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. U.S.A. 105, 4495–4500. doi: 10.1073/pnas.0710778105
Chen, M., and Chory, J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671. doi: 10.1016/j.tcb.2011.07.002
Christie, J. M., Blackwood, L., Petersen, J., and Sullivan, S. (2015). Plant flavoprotein photoreceptors. Plant Cell Physiol. 56, 401–413. doi: 10.1093/pcp/pcu196
Christie, J. M., Hitomi, K., Arvai, A. S., Hartfield, K. A., Mettlen, M., Pratt, A. J., et al. (2012). Structural tuning of the fluorescent protein iLOV for improved photostability. Biol. J. Chem. 287, 22295–22304. doi: 10.1074/jbc.M111.318881
Christie, J. M., Yang, H., Richter, G. L., Sullivan, S., Thomson, C. E., Lin, J., et al. (2011). phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9:1001076. doi: 10.1371/journal.pbio.1001076
Chung, N. J., and Paek, N. C. (2003). Photoblastism and ecophysiology of seed germination in weedy rice. Agron. J. 95, 184–190. doi: 10.2134/agronj2003.0184
Cinelli, R. A. G., Ferrari, A., Pellegrini, V., Tyagi, M., Giacca, M., and Beltram, F. (2000). The enhanced green fluorescent protein as a tool for the analysis of protein dynamics and localization: local fluorescence study at the single-molecule level. Photochem. Photobiol. 71, 771–776. doi: 10.1562/0031-8655(2000)0710771TEGFPA2.0.CO2
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14, 169–176. doi: 10.1111/pbi.12370
Cluis, C. P., Mouchel, C. F., and Hardtke, C. S. (2004). The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38, 332–347. doi: 10.1111/j.1365-313X.2004.02052.x
Dahmani-Mardas, F., Troadec, C., Boualem, A., Lévêque, S., Alsadon, A. A., Aldoss, A. A., et al. (2010). Engineering melon plants with improved fruit shelf life using the TILLING approach. PLoS ONE 5:e15776. doi: 10.1371/journal.pone.0015776.
Davey, M. P., Susanti, N. I., Wargent, J. J., Findlay, J. E., Quick, W. P., Paul, N. D., et al. (2012). The UV-B photoreceptor UVR8 promotes photosynthetic efficiency in Arabidopsis thaliana exposed to elevated levels of UV-B. Photosynth. Res. 114, 121–131. doi: 10.1007/s11120-012-9785-y
de Toledo Thomazella, D. P., Brail, Q., Dahlbeck, D., and Staskawicz, B. J. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 064824. doi: 10.1101/064824
de Wit, M., Galvão, V. C., and Fankhauser, C. (2016). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 513–537. doi: 10.1146/annurev-arplant-043015-112252
Dechaine, J. M., Gardner, G., and Weinig, C. (2009). Phytochromes differentially regulate seed germination responses to light quality and temperature cues during seed maturation. Plant Cell Environ. 32, 1297–1309. doi: 10.1111/j.1365-3040.2009.01998.x
Demkura, P. V., and Ballaré, C. L. (2012). UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol. Plant 5, 642–652. doi: 10.1093/mp/sss025
Devlin, P. F., Yanovsky, M. J., and Kay, S. A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133, 1617–1629. doi: 10.1104/pp.103.034397
D’Halluin, K., Vanderstraeten, C., Hulle, J. V., Rosolowska, J., Brande, I. V. D., Pennewaert, A., et al. (2013). Targeted molecular trait stacking in cotton through targeted double-strand break induction. Plant Biotechnol. J. 11, 933–941. doi: 10.1111/pbi.12085
Djukanovic, V., Smith, J., Lowe, K., Yang, M., Gao, H., Jones, S., et al. (2013). Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a redesigned I-CreI homing endonuclease. Plant J. 76, 888–899. doi: 10.1111/tpj.12335
D’Souza, S. F., Reddy, K. S., Badigannavar, A. M., Manjaya, J. G., and Jambhulkar, S. J. (2009). “Mutation breeding in oilseeds and grain legumes in India: accomplishments and socio-economic impact,” in Proceedings of an International Joint FAO/IAEA Symposium (Vienna: Interantional Atomic Energy Agency), 55–57.
Eckstein, A., Jagiełło-Flasiñska, D., Lewandowska, A., Hermanowicz, P., Appenroth, K. J., and Gabryś, H. (2016). Mobilization of storage materials during light-induced germination of tomato (Solanum lycopersicum) seeds. Plant Physiol. Biochem. 105, 271–281. doi: 10.1016/j.plaphy.2016.05.008
Endo, M., Araki, T., and Nagatani, A. (2016). Tissue-specific regulation of flowering by photoreceptors. Cell. Mol. Life Sci. 73, 829–839. doi: 10.1007/s00018-015-2095-8
Erb, M., Meldau, S., and Howe, G. A. (2012). Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259. doi: 10.1016/j.tplants.2012.01.003
Facella, P., Lopez, L., Carbone, F., Galbraith, D. W., Giuliano, G., and Perrotta, G. (2008). Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE 3:2798. doi: 10.1371/journal.pone.0002798
Facella, P., Carbone, F., Placido, A., and Perrotta, G. (2017). Cryptochrome 2 extensively regulates transcription of the chloroplast genome in tomato. FEBS Open Bio 7, 456–471. doi: 10.1002/2211-5463.12082
Franklin, K. A. (2008). Shade avoidance. New Phytol. 179, 930–944. doi: 10.1111/j.1469-8137.2008.02507.x
Galvão, V. C., and Schmid, M. (2014). Regulation of flowering by endogenous signals. Mol. Genet. Floral Trans. Flower Dev. 72, 63–102. doi: 10.1016/B978-0-12-417162-6.00003-1
Garg, A. K., Sawers, R. J. H., Wang, H., Kim, J., Walker, J. M., Brutnell, T. P., et al. (2006). Light-regulated overexpression of an Arabidopsis phytochrome A gene in rice alters plant architecture and increases grain yield. Planta 223, 627–636. doi: 10.1007/s00425-005-0101-3
Giliberto, L., Perrotta, G., Pallara, P., Weller, J. L., Fraser, P. D., Bramley, P. M., et al. (2005). Manipulation of the blue light photoreceptor Cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137, 199–208. doi: 10.1104/pp.104.051987
Gommers, C. M. M., Keuskamp, D. H., Buti, S., Van Veen, H., Koevoets, I. T., Reinen, E., et al. (2017). Molecular profiles of contrasting shade response strategies in wild plants: differential control of immunity and shoot elongation. Plant Cell 29, 331–334. doi: 10.1105/tpc.16.00790
González, C. V., Fanzone, M. L., Cortés, L. E., Bottini, R., Lijavetzky, D. C., Ballaré, C. L., et al. (2015). Fruit-localized photoreceptors increase phenolic compounds in berry skins of field-grown Vitis vinifera cv, L. Malbec. Phytochemistry 110, 46–57. doi: 10.1016/j.phytochem.2014.11.018
Graf, A., Coman, D., Uhrig, R. G., Walsh, S., Flis, A., Stitt, M., et al. (2017). Parallel analysis of Arabidopsis circadian clock mutants reveals different scales of transcriptome and proteome regulation. Open Biol. 7:160333. doi: 10.1098/rsob.160333
Griebel, T., and Zeier, J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147, 790–801. doi: 10.1104/pp.108.119503
Gupta, S. K., Sharma, S., Santisree, P., Kilambi, H. V., Appenroth, K., Sreelakshmi, Y., et al. (2014). Complex and shifting interactions of phytochromes regulate fruit development in tomato. Plant Cell Environ. 37, 1688–1702. doi: 10.1111/pce.12279
Haun, W., Coffman, A., Clasen, B. M., Demorest, Z. L., Lowy, A., Ray, E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201
Henikoff, S., Till, B. J., and Comai, L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. doi: 10.1104/pp.104.041061
Hirose, F., Shinomura, T., Tanabata, T., Shimada, H., and Takano, M. (2006). Involvement of rice cryptochrome in de-etiolation responses in flowering. Plant Cell Physiol. 47, 915–925. doi: 10.1093/pcp/pcj064
Hisamatsu, T., and King, R. W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. Exp. J. Bot. 59, 3821–3829. doi: 10.1093/jxb/ern232
Ito, S., Song, Y. H., and Imaizumi, T. (2012). LOV domain-containing f-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5, 47–56. doi: 10.1093/mp/sss013
Jacobsen, J. V., Pearce, D. W., Poole, A. T., Pharis, R. P., and Mander, L. N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant. 115, 428–441. doi: 10.1034/j.1399-3054.2002.1150313.x
Jansen, M. A. K. (2002). Ultraviolet-B radiation effects on plants: induction of morphogenic responses. Physiol. Plant. 116, 423–429. doi: 10.1034/j.1399-3054.2002.1160319.x
Jenkins, G. I. (2014). The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26, 21–37. doi: 10.1105/tpc.113.119446
Jeong, R., Chandra-Shekara, A. C., Barman, S. R., Navarre, D., Klessig, D. F., Kachroo, A., et al. (2010). Cryptochrome 2 and phototropin 2 regulate resistance protein-mediated viral defense by negatively regulating an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 107, 13538–13543. doi: 10.1073/pnas.1004529107
Kami, C., Lorrain, S., Hornitschek, P., Fankhauser, C., and Marja, C. P. T. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. doi: 10.1016/S0070-2153(10)91002-8
Kang, C., Lian, H., Wang, F., Huang, J., and Yang, H. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21, 2624–2641. doi: 10.1105/tpc.109.069765
Kasahara, M., Kagawa, T., Sato, Y., Kiyosue, T., and Wada, M. (2004). Phototropins mediate blue and red light-induced chloroplast movements in physcomitrella patens. Plant Physiol. 135, 1388–1397. doi: 10.1104/pp.104.042705
Kendrick, R. E., Kerckhoffs, L. H. J., Van Tuinen, A., and Koornneef, M. (1997). Photomorphogenic mutants of tomato. Plant Cell Environ. 20, 746–751. doi: 10.1046/j.1365-3040.1997.d01-109.x
Kharshiing, E., and Sinha, S. P. (2015). Plant productivity: can photoreceptors light the way? J. Plant Growth Regul. 34, 206–214. doi: 10.1007/s00344-014-9454-9
Kharshiing, E., and Sinha, S. P. (2016). Deficiency in phytochrome A alters photosynthetic activity, leaf starch metabolism and shoot biomass production in tomato. Photochem. J. Photobiol. B Biol. 165, 157–162. doi: 10.1016/j.jphotobiol.2016.10.026
Kharshiing, E. V., Kumar, G. P., Ditengou, F. A., Li, X., Palme, K., and Sharma, R. (2010a). The polycotyledon (pct1-2) mutant of tomato shows enhanced accumulation of PIN1 auxin transport facilitator protein. Plant Biol. 12, 224–228. doi: 10.1111/j.1438-8677.2009.00267.x
Kharshiing, E. V., Kumar, G. P., and Sharma, R. (2010b). PIN it on auxin: the role of PIN1 and PAT in tomato development. Plant Signal. Behav. 5, 1379–1383. doi: 10.4161/psb.5.11.13035
Kimura, S., and Sinha, N. (2008). Tomato (Solanum lycopersicum): a model fruit-bearing crop. Cold Spring Harb. Protoc. 2008:pdb.emo105.
Kliebenstein, D. J., Lim, J. E., Landry, L. G., and Last, R. L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130, 234–243. doi: 10.1104/pp.005041
Koepfli, J. B., Thimann, K. V., and Went, F. W. (1938). Phytohormones: structure and physiological activity. Biol I Chem. J. 122, 763–780.
Kozuka, T., Suetsugu, N., Wada, M., and Nagatani, A. (2013). Antagonistic regulation of leaf flattening by phytochrome B and phototropin in Arabidopsis thaliana. Plant Cell Physiol. 54, 69–79. doi: 10.1093/pcp/pcs134
Kurowska, M., Daszkowska-Golec, A., Gruszka, D., Marzec, M., Szurman, M., Szarejko, I., et al. (2011). TILLING-a shortcut in functional genomics. Appl. J. Genet. 52, 371–390. doi: 10.1007/s13353-011-0061-1
Lee, K. P., and Lopez-Molina, L. (2012). Control of seed germination in the shade. Cell Cycle 11, 4489–4490. doi: 10.4161/cc.22667
Li, J., Terzaghi, W., and Deng, X. W. (2012). Genomic basis for light control of plant development. Protein Cell 3, 106–116. doi: 10.1007/s13238-012-2016-7
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, Q., and He, J. (2016). BZR1 interacts with HY5 to mediate brassinosteroid-and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9, 113–125. doi: 10.1016/j.molp.2015.08.014
Lidon, F. C., and Ramalho, J. C. (2011). Impact of UV-B irradiation on photosynthetic performance and chloroplast membrane components in Oryza sativa L. Photochem. Photobiol. J. B Biol. 104, 457–466. doi: 10.1016/j.jphotobiol.2011.05.004
Lidon, F. J. C., Reboredo, F. H., Leitáo, A. E., Silva, M. M. A., Duarte, M. P., and Ramalho, J. C. (2012). Impact of UV-B radiation on photosynthesis-an overview. Emir. J. Food Agric. 24, 546–556. doi: 10.9755/ejfa.v24i6.546556
Liu, J., Zhang, F., Zhou, J., Chen, F., Wang, B., and Xie, X. (2012). Phytochrome B control of total leaf area and stomatal density affects drought tolerance in tomato. Plant Mol. Biol. 78, 289–300. doi: 10.1007/s11103-011-9860-3
Lopez, L., Carbone, F., Bianco, L., Giuliano, G., Facella, P., and Perrotta, G. (2012). Tomato plants overexpressing cryptochrome 2 reveal altered expression of energy and stress-related gene products in response to diurnal cues. Plant Cell Environ. 35, 994–1012. doi: 10.1111/j.1365-3040.2011.02467.x
Lucas, M., and Prat, S. (2014). PIFs get BRright: phytochrome interacting factors as integrators of light and hormonal signals. New Phytol. 202, 1126–1141. doi: 10.1111/nph.12725
Manova, V., and Gruszka, D. (2015). DNA damage and repair in plants-from models to crops. Front. Plant Sci. 6:885. doi: 10.3389/fpls.2015.00885
Markelz, N. H., Costich, D. E., and Brutnell, T. P. (2003). Photomorphogenic responses in maize seedling development. Plant Physiol. 133, 1578–1591. doi: 10.1104/pp.103.029694
Más, P., Devlin, P. F., Panda, S., and Kay, S. A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211. doi: 10.1038/35041583
Mazza, C. A., and Ballaré, C. L. (2015). Photoreceptors UVR8 and phytochrome B cooperate to optimize plant growth and defense in patchy canopies. New Phytol. 207, 4–9. doi: 10.1111/nph.13332
McCallum, C. M., Comai, L., Greene, E. A., and Henikoff, S. (2000). Targeting induced locallesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. doi: 10.1104/pp.123.2.439
Meng, Y., Li, H., Wang, Q., Liu, B., and Lin, C. (2013). Blue light-dependent interaction between Cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 25, 4405–4420. doi: 10.1105/tpc.113.116590
Millar, A. J. (2003). A suite of photoreceptors entrains the plant circadian clock. Biol. J. Rhythms 18, 217–226. doi: 10.1177/0748730403018003004
Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y., Dolan, S., et al. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 100, 2140–2145. doi: 10.1073/pnas.0437826100
Moreno, J. E., and Ballaré, C. L. (2014). Phytochrome regulation of plant immunity in vegetation canopies. J. Chem. Ecol. 40, 848–857. doi: 10.1007/s10886-014-0471-8
Neff, M. M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. doi: 10.1104/pp.118.1.27
Negi, S., Santisree, P., Kharshiing, E. V., and Sharma, R. (2010). Inhibition of the Ubiquitin-proteasome pathway alters cellular levels of nitric oxide in tomato seedlings. Mol. Plant 3, 854–869. doi: 10.1093/mp/ssq033
Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., et al. (2009). Genome-wide analysis of genes targeted by phytochrome interacting factor 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21, 403–419. doi: 10.1105/tpc.108.064691
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Osakabe, K., Saika, H., Okuzaki, A., and Toki, S. (2011). “Site-directed mutagenesis in higher plants,” in Plant Mutation Breeding and Biotechnology, eds Q. Y. Shu, B. P. Forster, and H. Nakagawa (Wallingford: CABI), 523–534.
Osakabe, Y., and Osakabe, K. (2014). Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400. doi: 10.1093/pcp/pcu170
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Pathirana, R. (2011). “Plant mutation breeding in agriculture,” in Plant Science Reviews, ed. D. Hemming (Cambridge: CABI), 107–126.
Pearce, S., Kippes, N., Chen, A., Debernardi, J. M., and Dubcovsky, J. (2016). RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 16:141. doi: 10.1186/s12870-016-0831-3
Pedmale, U. V., Huang, S. S., Zander, M., B Cole, J., Hetzel, J., Ljung, K., et al. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. doi: 10.1016/j.cell.2015.12.018
Perrella, G., and Kaiserli, E. (2016). Light behind the curtain: photoregulation of nuclear architecture and chromatin dynamics in plants. New Phytol. 212, 908–919. doi: 10.1111/nph.14269
Petrillo, E., Godoy Herz, M. A., Barta, A., Kalyna, M., and Kornblihtt, A. R. (2014). Let there be light: regulation of gene expression in plants. RNA Biol. 11, 1215–1220. doi: 10.4161/15476286.2014.972852
Petroutsos, D., Tokutsu, R., Maruyama, S., Flori, S., Greiner, A., Magneschi, L., et al. (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537, 563–566. doi: 10.1038/nature19358
Prasad, B. R. V., Kumar, S. V., Nandi, A., and Chattopadhyay, S. (2012). Functional interconnections of HY1 with MYC2 and HY5 in Arabidopsis seedling development. BMC Plant Biol. 12, 37. doi: 10.1186/1471-2229-12-37
Puchta, H. (2016). Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J. 87, 5–15. doi: 10.1111/tpj.13100
Rizzini, L., Favory, J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 Protein. Science 332, 103–106. doi: 10.1126/science.1200660
Robson, P. R. H., McCormac, A. C., Irvine, A. S., and Smith, H. (1996). Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat. Biotechnol. 14, 995–998. doi: 10.1038/nbt0896-995
Robson, T. M., Klem, K., Urban, O., and Jansen, M. A. K. (2015). Re-interpreting plant morphological responses to UV-B radiation. Plant Cell Environ. 38, 856–866. doi: 10.1111/pce.12374
Sawers, R. J. H., Sheehan, M. J., and Brutnell, T. P. (2005). Cereal phytochromes: targets of selection, targets for manipulation? Trends Plant Sci. 10, 138–143. doi: 10.1016/j.tplants.2005.01.004
Schaeffer, S. M., and Nakata, P. A. (2015). CRISPR/Cas9-mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci. 240, 130–142. doi: 10.1016/j.plantsci.2015.09.011
Schmidt, D., and Cho, Y. (2015). Natural photoreceptors and their application to synthetic biology. Trends Biotechnol. 33, 80–91. doi: 10.1016/j.tibtech.2014.10.007
Schmitt, J., Stinchcombe, J. R., Heschel, M. S., and Huber, H. (2003). The adaptive evolution of plasticity: phytochrome-mediated shade avoidance responses1. Integr. Compar. Biol. 43, 459–469. doi: 10.1093/icb/43.3.459
Seo, M., Hanada, A., Kuwahara, A., Endo, A., Okamoto, M., Yamauchi, Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366. doi: 10.1111/j.1365-313X.2006.02881.x
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2008). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69:463. doi: 10.1007/s11103-008-9429-y
Sharma, P., Chatterjee, M., Burman, N., and Khurana, J. P. (2014). Cryptochrome 1 regulates growth, and development in Brassica through alteration in the expression of genes involved in light, phytohormone, and stress signalling. Plant Cell Environ. 37, 961–977. doi: 10.1111/pce.12212
Sharma, S., Kharshiing, E., Srinivas, A., Zikihara, K., Tokutomi, S., Nagatani, A., et al. (2014). A dominant mutation in the light-oxygen, and Voltage2 domain vicinity impairs phototropin1 signaling in tomato. Plant Physiol. 164, 2030–2044. doi: 10.1104/pp.113.232306
Sheehan, M. J., Kennedy, M. L., Costich, D. E., and Brutnell, T. P. (2007). Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. Plant J. 49, 338–353. doi: 10.1111/j.1365-313X.2006.02962.x
Shih, P. M., Liang, Y., and Loqué, D. (2016). Biotechnology and synthetic biology approaches for metabolic engineering of bioenergy crops. Plant J. 87, 103–117. doi: 10.1111/tpj.13176
Shinkle, J. R., Atkins, A. K., Humphrey, E. E., Rodgers, C. W., Wheeler, S. L., and Barnes, P. W. (2004). Growth and morphological responses to different UV wavebands in cucumber (Cucumis sativum) and other dicotyledonous seedlings. Physiol. Plant. 120, 240–248. doi: 10.1111/j.0031-9317.2004.0237.x
Shukla, V. K., Doyon, Y., Miller, J. C., DeKelver, R. C., Moehle, E. A., Worden, S. E., et al. (2009). Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459, 437–441. doi: 10.1038/nature07992
Slade, A., and Moehs, C. (2011). “Uses of TILLING for crop improvement,” in Plant Mutation Breeding and Biotechnology, eds Q. Y. Shu, B. P. Forster, and H. Nakagawa (Wallingford: CABI), 359–369.
Somers, D. E., Kim, W., and Geng, R. (2004). The F-Box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16, 769–782. doi: 10.1105/tpc.016808
Song, G., Jia, M., Chen, K., Kong, X., Khattak, B., Xie, C., et al. (2016). CRISPR/Cas9: a powerful tool for crop genome editing. Crop J. 4, 75–82. doi: 10.1016/j.cj.2015.12.002
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., MacCoss, M. J., et al. (2014). Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Soyk, S., Muller, N. A., Park, S. J., Schmalenbach, I., Jiang, K., Hayama, R., J. et al. (2017). Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 49, 162–168. doi: 10.1038/ng.3733
Suesslin, C., and Frohnmeyer, H. (2003). An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J. 33, 591–601. doi: 10.1046/j.1365-313X.2003.01649.x
Suetsugu, N., and Wada, M. (2013). Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol. 54, 8–23. doi: 10.1093/pcp/pcs165
Suprasanna, P., and Nakagawa, H. (2011). “Mutation breeding of vegetatively propagated crops,” in Plant Mutation Breeding and Biotechnology, eds Q. Y. Shu, B. P. Forster, and H. Nakagawa (Wallingford: CABI), 347–358.
Takano, M., Inagaki, N., Xie, X., Yuzurihara, N., Hihara, F., Ishizuka, T., et al. (2005). Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17, 3311–3325. doi: 10.1105/tpc.105.035899
Takemiya, A., Inoue, S., Doi, M., Kinoshita, T., and Shimazaki, K. (2005). Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17, 1120–1127. doi: 10.1105/tpc.104.030049
Taylor, A., Massiah, A. J., and Thomas, B. (2010). Conservation of Arabidopsis thaliana photoperiodic flowering time genes in onion (Allium cepa L.). Plant Cell Physiol. 51, 1638–1647. doi: 10.1093/pcp/pcq120
Tilbrook, K., Arongaus, A. B., Binkert, M., Heijde, M., Yin, R., and Ulm, R. (2013). The UVR8 UV-b photoreceptor: perception, signaling and response. Arabidopsis Book 11:e0164. doi: 10.1199/tab.0164
Treutter, D. (2005). Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 7, 581–591. doi: 10.1055/s-2005-873009
Tsormpatsidis, E., Henbest, R. G. C., Battey, N. H., and Hadley, P. (2010). The influence of ultraviolet radiation on growth, photosynthesis and phenolic levels of green and red lettuce: potential for exploiting effects of ultraviolet radiation in a production system. Ann. Appl. Biol. 156, 357–366. doi: 10.1111/j.1744-7348.2010.00393.x
Ukai, N., and Nakagawa, H. (2011). “Strategies and approaches in mutant population development for mutant selection in seed propagated crops,” in Plant Mutation Breeding and Biotechnology, eds Q. Y. Shu, B. P. Forster, and H. Nakagawa (Wallingford: CABI), 209–221.
Wang, H., Ma, L., Li, J., Zhao, H., and Deng, X. W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294, 154–158. doi: 10.1126/science.1063630
Wang, Q., Zhu, Z., Ozkardesh, K., and Lin, C. (2013). Phytochromes and phytohormones: the shrinking degree of separation. Mol. Plant 6, 5–7. doi: 10.1093/mp/sss102
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wargent, J. J., Elfadly, E. M., Moore, J. P., and Paul, N. D. (2011). Increased exposure to UV-B radiation during early development leads to enhanced photoprotection and improved long-term performance in Lactuca sativa. Plant Cell Environ. 34, 1401–1413. doi: 10.1111/j.1365-3040.2011.02342.x
Wargent, J. J., Gegas, V. C., Jenkins, G. I., Doonan, J. H., and Paul, N. D. (2009). UVR8 in Arabidopsis thaliana regulates multiple aspects of cellular differentiation during leaf development in response to ultraviolet B radiation. New Phytol. 183, 315–326. doi: 10.1111/j.1469-8137.2009.02855.x
Wargent, J. J., and Jordan, B. R. (2013). From ozone depletion to agriculture: understanding the role of UV radiation in sustainable crop production. New Phytol. 197, 1058–1076. doi: 10.1111/nph.12132
Warnasooriya, S. N., and Brutnell, T. P. (2014). Enhancing the productivity of grasses under high-density planting by engineering light responses: from model systems to feedstocks. Exp. J. Bot. 65, 2825–2834. doi: 10.1093/jxb/eru221
Watanabe, S., Hideshima, R., Xia, Z., Tsubokura, Y., Sato, S., Nakamoto, Y., et al. (2009). Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–62. doi: 10.1534/genetics.108.098772
Weeks, D. P., Spalding, M. H., and Yang, B. (2016). Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 14, 483–495. doi: 10.1111/pbi.12448
Weijschedé, J., Martínková, J., De Kroon, H., and Huber, H. (2006). Shade avoidance in Trifolium repens: costs and benefits of plasticity in petiole length and leaf size. New Phytol. 172, 655–666. doi: 10.1111/j.1469-8137.2006.01885.x
Weinig, C., Johnston, J., German, Z. M., and Demink, L. M. (2006). Local and global costs of adaptive plasticity to density in Arabidopsis thaliana. Am. Nat. 167, 826–836. doi: 10.1086/503530
Weller, J. L., Batge, S. L., Smith, J. J., Kerckhoffs, L. H., Sineshchekov, V. A., Murfet, I. C., et al. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135, 2186–2195. doi: 10.1104/pp.103.036103
Weller, J. L., Beauchamp, N., Kerckhoffs, L. J., Platten, J. D., and Reid, J. B. (2001). Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 26, 283–294. doi: 10.1046/j.1365-313X.2001.01027.x
Xie, X., Xue, Y., Zhou, J., Zhang, B., Chang, H., and Takano, M. (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 4, 688–696. doi: 10.1093/mp/ssr005
Xue, Z. G., Zhang, X. M., Lei, C. F., Chen, X. J., and Fu, Y. F. (2012). Molecular cloning and functional analysis of one ZEITLUPE homolog GmZTL3 in soybean. Mol. Biol. Rep. 39, 1411–1418. doi: 10.1007/s11033-011-0875-2
Yang, D., Seaton, D. D., Krahmer, J., and Halliday, K. J. (2016). Photoreceptor effects on plant biomass, resource allocation, and metabolic state. Proc. Natl. Acad. Sci. U.S.A. 113, 7667–7672. doi: 10.1073/pnas.1601309113
Yang, J. C., Li, M., Xie, X. Z., Han, G. L., Sui, N., and Wang, B. S. (2013). Deficiency of phytochrome B alleviates chilling-induced photoinhibition in rice. Am. Bot. J. 100, 1860–1870. doi: 10.3732/ajb.1200574
Yanovsky, M. J., Izaguirre, M., Wagmaister, J. A., Gatz, C., Jackson, S. D., Thomas, B., et al. (2003). Phytochrome A resets the circadian clock and delays tuber formation under long days in potato. Plant J. 23, 223–232. doi: 10.1046/j.1365-313x.2000.00775.x
Yon, F., Joo, Y., Cortés Llorca, L., Rothe, E., Baldwin, I. T., and Kim, S. C. (2016). Silencing Nicotiana attenuata LHY and ZTL alters circadian rhythms in flowers. New Phytol. 209, 1058–1066. doi: 10.1111/nph.13681
Yu, X., and Lin, C. (2005). “Light sensing plants” in Light Regulation of Flowering Time in Arabidopsis, eds M. Wada, K. Shimazaki, and M. Iino (Berlin: Springer), 325–332.
Yu, X., Liu, H., Klejnot, J., and Lin, C. (2010). The Cryptochrome Blue Light Receptors. Arabidopsis Book 8:e0135. doi: 10.1199/tab.0135
Yu, Y., Wang, J., Zhang, B., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9:1004025. doi: 10.1371/journal.pgen.1004025
Zeeman, S. C., Kossmann, J., and Smith, A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. doi: 10.1146/annurev-arplant-042809-112301
Zhang, Q., Li, H., Li, R., Hu, R., Fan, C., Chen, F., et al. (2008). Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. U.S.A. 105, 21028–21033. doi: 10.1073/pnas.0810585105
Keywords: plant growth and development, light signaling, plant photoreceptors, photoreceptor engineering, crop productivity
Citation: Mawphlang OIL and Kharshiing EV (2017) Photoreceptor Mediated Plant Growth Responses: Implications for Photoreceptor Engineering toward Improved Performance in Crops. Front. Plant Sci. 8:1181. doi: 10.3389/fpls.2017.01181
Received: 10 May 2017; Accepted: 20 June 2017;
Published: 11 July 2017.
Edited by:
Gerrit T. S. Beemster, University of Antwerp, BelgiumReviewed by:
Roslyn Gleadow, Monash University, AustraliaCopyright © 2017 Mawphlang and Kharshiing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eros V. Kharshiing, ZXJvcy5raGFyc2hpaW5nQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.