
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 04 July 2017
Sec. Computational Genomics
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01163
G quadruplexes (G4) are higher-order DNA and RNA secondary structures formed by G-rich sequences that are built around tetrads of hydrogen-bonded guanine bases. Potential G4 quadruplex sequences have been identified in G-rich eukaryotic non-telomeric and telomeric genomic regions. Upon function, G4 formation is known to involve in chromatin remodeling, gene regulation and has been associated with genomic instability, genetic diseases and cancer progression. The natural role and biological validation of G4 structures is starting to be explored, and is of particular interest for the therapeutic interventions for human diseases. However, the existence and physiological role of G4 DNA and G4 RNA in plants species have not been much investigated yet and therefore, is of great interest for the development of improved crop varieties for sustainable agriculture. In this context, several recent studies suggests that these highly diverse G4 structures in plants can be employed to regulate expression of genes involved in several pathophysiological conditions including stress response to biotic and abiotic stresses as well as DNA damage. In the current review, we summarize the recent findings regarding the emerging functional significance of G4 structures in plants and discuss their potential value in the development of improved crop varieties.
Double helical B-DNA is the predominant nucleic acid structure of the genome. In addition, DNA may adopt various extrahelical, non B-DNA secondary confirmations depending on the nucleotide content. These secondary structures are prevalent in all living organisms and play a pivotal role in the physiology of organisms. G quadruplex or G4 DNA is one of these structures adopted by spontaneous folding of sequences containing multiple runs of guanines (Bochman et al., 2012). Structurally, G4 DNA comprises of G-quartets or G-tetrads, in which the four guanine bases are bound together via Hoogsteen hydrogen bonds in a square planar conformation (Huppert and Balasubramanian, 2005). G-quartets stack on top of each other to form an advanced nucleic acid structure, G4 DNA (Figure 1). Adding to the complexity of the G4 DNA structure, the stacks of G-quartets are connected by loops of variable sizes (1–7 nucleotides) and orientations (parallel or antiparallel) (Wang and Patel, 1993; Parkinson et al., 2002; Huppert, 2010). These secondary structures are stabilized by cations, preferably potassium ion (K+) (Largy et al., 2016). G quadruplex structures form in RNA as well as DNA and may be intermolecular or intramolecular depending on number of nucleic acid strands involved in the quadruplex formation. For identification of potential G4 forming sequences in the in genome, G quadruplex prediction algorithms, such as Quadparser (Huppert and Balasubramanian, 2007), G4 calculator (Eddy and Maizels, 2006), and Quadbase (Dhapola and Chowdhury, 2016), are easily accessible and used widely. G quadruplex forming sequences (GQFS) have been categorized into different types on the basis of the number of guanine repeats (G2- two G4 repeats, G3- three G4 repeats, G4- four G4 repeats) and the number of nucleotide in the loop (1–3 bp, 1–7 bp., etc.) (Table 1). The stability of G4 structure depends on the length of loop with prediction of increased stability with shorter loop length. For instance, G3 type GQFS are more stable with 1–3 bp than 4–7 bp loop length, similarly G2 type GQFS are more stable with loop length of 1–2 bp than 3–4 bp (Bugaut and Balasubramanian, 2008).
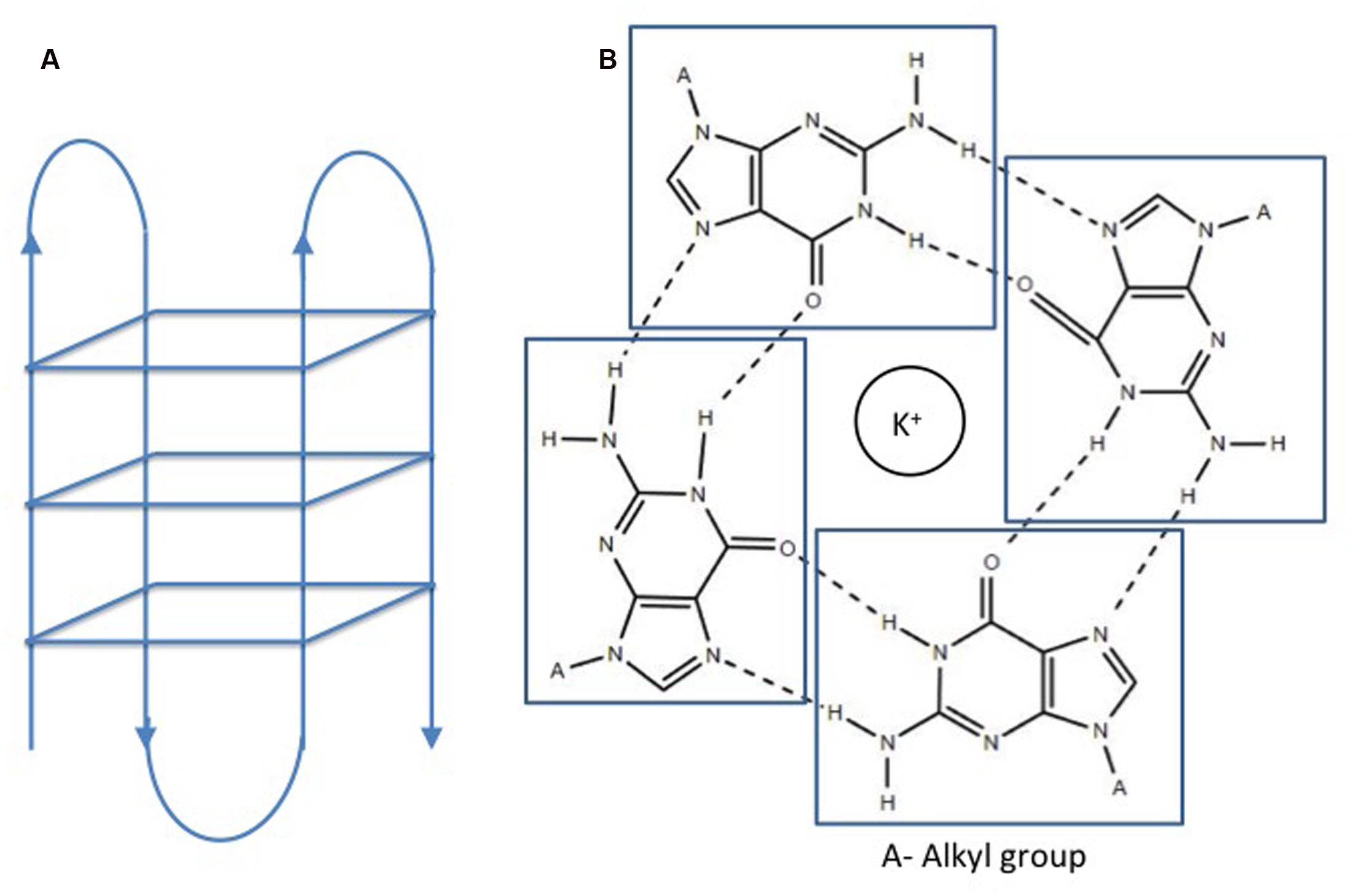
FIGURE 1. Structure of a G quadruplex. (A) Intramolecular G quadruplex. (B) G quartet formation in presence of cations (Huppert and Balasubramanian, 2005). No permission is required for the modification and reproduction of this figure under the terms of the Creative Commons CC BY license.
The study of G4 DNA has emerged as a forefront area of research because of its proposed role in several biological functions ranging from physiology to pathology. These secondary structures are found to be abundant in a wide range of eukaryotic and prokaryotic genomes. In bacteria, yeast and humans, genome-wide analyses of GQFS have revealed the non-random distribution of these secondary structures (König et al., 2010). It is evident that GQFS are particularly abundant in, but not limited to, promoters (Evans et al., 1984; Kilpatrick et al., 1986), telomeres (Blackburn, 1994), ribosomal DNA (Sun et al., 1998), untranslated region (UTR) of mRNA, micro- and mini-satellite repeats (Nakagama et al., 2006), and immunoglobulin heavy chain switch regions (Yu et al., 2003).
Formation of these secondary structures has been associated with genomic instability and thus cancer progression. The highly significant correlation between GQFS and chromosomal translocation breakpoints in cancers demonstrates the biological relevance of G4-associated genome instability in mammalian cells. Studies in the model eukaryote Saccharomyces cerevisiae showed that topoisomerase enzymes and DNA helicases actively function to suppress G4-induced genome instability (Yadav et al., 2014, 2016).
In addition, occurence and distribution of these secondary structures in protooncogenes further suggest their role in the development and progression of cancer (Huppert and Balasubramanian, 2007; Verma et al., 2008). These protooncogenes includes c-myc (Siddiqui-Jain et al., 2002), bcl-2 (Dexheimer et al., 2006), c-kit (Rankin et al., 2005; Fernando et al., 2006), c-myb (Palumbo et al., 2008), KRAS (Cogoi and Xodo, 2006; Cogoi et al., 2009) proto-oncogenes, VEGF (Sun et al., 2005), and HIF-1 (De Armond et al., 2005).
In bacteria, GQFSs are evolutionarily conserved and enriched non-randomly in the promoter region of the genes that are associated with specific functions such as transcription, secondary metabolite biosynthesis, and signal transduction, suggesting a regulatory role of G4 DNA at global level in prokaryotes as well (Rawal et al., 2006; Beaume et al., 2013). In addition, G4 DNA and RNA play a key role in recombination-mediated antigenic variation mechanism that effectively varies the amino acid sequence of the surface expressed protein pilin and, thus, evades detection by the host adaptive immune system, in the bacterial pathogen Neisseria gonorrhoeae (Cahoon and Seifert, 2009).
The emerging pattern of association of GQFS with specific genomic regions suggests a regulatory role of GQFS in biologically significant pathways (Garg et al., 2016). However, in contrast to the significant information available on the distribution and role of G quadruplex-forming sequences in humans and microbial pathogens, similar studies in plant systems has been very limited. Recent investigation showed that G4 DNA and RNA are also generally conserved across plant species. In this context, several studies on bioinformatics analysis of plant genomes have been accompanied with identification and functional characterization of these secondary structures. In plants, genome wide distribution of G quadruplexes and their association with different genomic features led to the identification of putative G4 forming sequences within gene body or promoter region of orthologs genes in monocot and dicot plant species (Garg et al., 2016). Given the significant regulatory roles ascribed to G4 DNA in multiple systems, understanding the mechanism of gene regulation through G quadruplexes in plants may provide significant information for crop improvement. Very recently, several studies have been conducted to identify GQFS in a wide variety of plant species, including many important crop plants. Here, we will review the current state of understanding of the biological pathways where a significant role of G4 DNA is implicated.
Similar to other organisms, the prevalence and distribution of GQFS in plant genomes vary according to the specific GQFS type. The G3 type GQFS were more abundant in the intergenic region, whereas, G2 type GQFS were found to be located in the genic region (Table 1). The specific association of different type of GQFS with different genomic regions suggest their vital role in various cellular processes for instance G2 GQFS may play a role in regulation of translation and transcription, while G3 GQFS are important for promoter regulation. In plant genome (Arabidopsis thaliana, Oryza sativa, Glycine max, Cypripedium arietinum), several G4 sequences were identified and confirmed to form parallel, antiparallel, intramolecular or intermolecular G4 DNA conformations in vitro by using circular dichroism (CD) spectroscopy and gel electrophoresis (Garg et al., 2016). Based on the gene ontology (GO) enrichment analyses, it have been shown that, in a variety of dicot plant species, orthologous genes harboring GQFS were involved in important biological pathways such as chromatin modification, regulating phosphorylation and intracellular signaling, auxin transport, seed development and GTPase activity. In monocot plant species, orthologous genes with GQFS are involved in biological processes such as development, ion transportation, regulation of transcription and protein folding (Garg et al., 2016).
G4 DNA forming sequences are evolutionarily conserved from bacterial to single cell eukaryotes to metazoans. Among closely related fungal species, GQFS are evolutionary conserved at the nucleotide level and associated with distinct genomic features (Capra et al., 2010). In order to assess the evolutionary conservation of G4 sequences in plant species, a genome wide analysis was conducted for monocot and dicot plant genomes. The result conclusively showed that G2 type GQFS were abundant, comprising more than 90% of GQFS found in all the plant species analyzed, while G3 type GQFS were found less frequently, comprising 5% of the total GQFS in each of the plant species (Garg et al., 2016). In addition, frequency of GQFS distribution varied between monocot (∼80–1500 GQFS/Mb) and dicot (∼10–20 GQFS/Mb), this disparity in GQFS distribution may be due to high GC content of monocot genomes (Garg et al., 2016). The evolutionary conservation of GQFSs among plant species and their association with specific genomic features as described below suggest that G4 DNAs are integral parts of plant biology and are under evolutionary constraints.
G DNAs are considered a molecular switch for gene expression in metazoan cells (Eddy and Maizels, 2006), it is imperative to study the positional relationship of GQFS in plant genomes (Figure 2). In the genomes of A. thaliana, Vitis vinifera, O. sativa and Populus trichocarpa, GQFS are frequently located near the transcribed units or genes (Mullen et al., 2010). In particular, significant GQFS enrichment was observed in the transcription start site proximal regions [TPR], which are generally conserved across plant species. This suggests that G4 motifs in plants, similar to their proposed function in mammalian systems, play a role in regulating gene expression (Andorf et al., 2014). G4 motifs are also enriched at 5′ UTR, 3′ UTR, and 5′ end of introns implicating the role of G4 quadruplex in post-transcriptional regulation of the genes (Andorf et al., 2014; Wang et al., 2015). Comparative analyses of the genomes of Oryza sativa japonica and O. sativa, widely cultivated Asian variety of rice species, showed the enrichment of GQFS in TPR region (149.57GQFS/Mb and 131.34GQFS/Mb, respectively) relative to coding regions, introns, and 5′- and 3′-UTRs. Overall, the conserved pattern of high density of GQFS at TPR across the variety of plant species suggests the role of G quadruplex in transcriptional regulation in these species. Overall, the density of GQFS among monocot species was higher than that among dicot species studied (Wang et al., 2015).
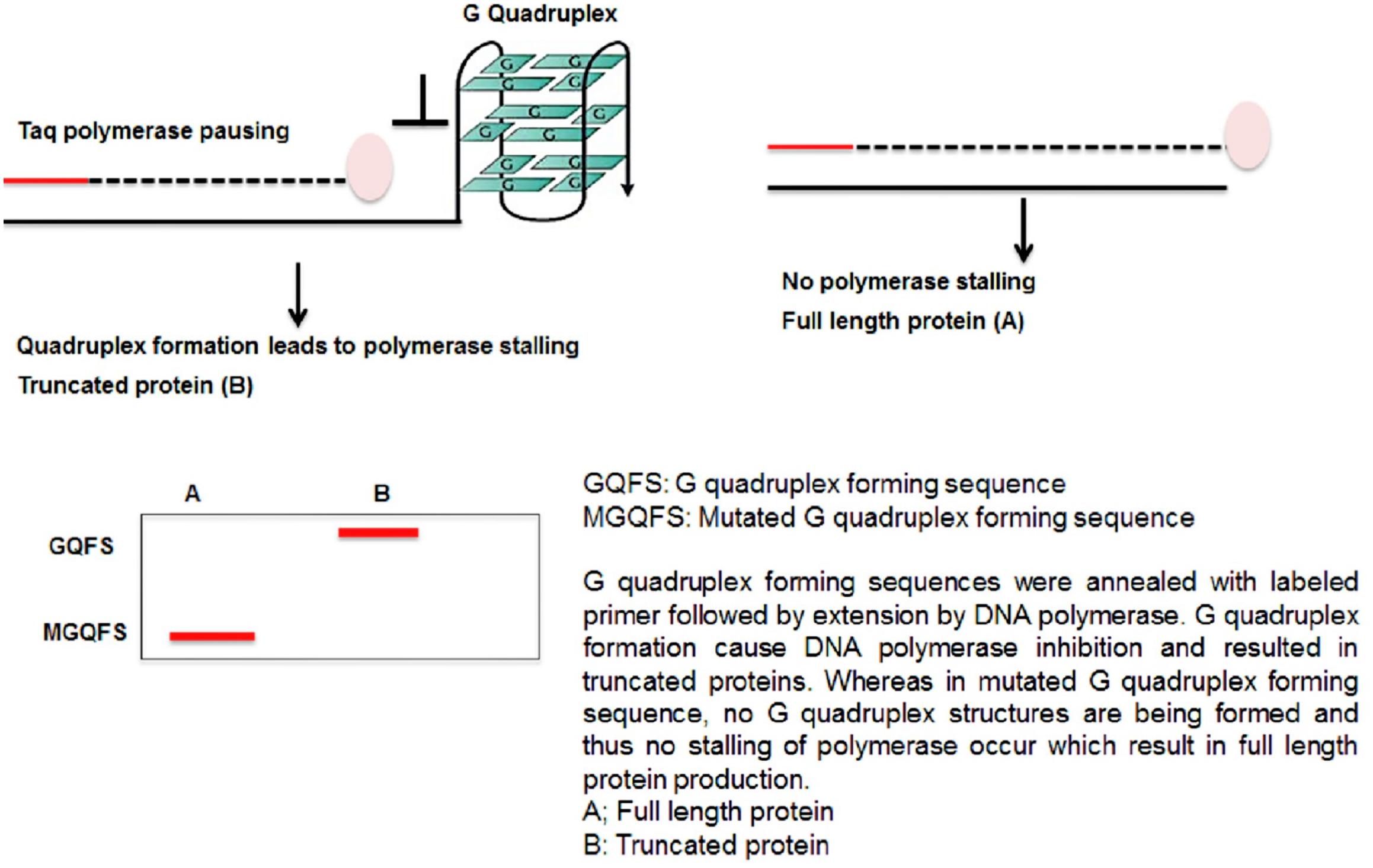
FIGURE 2. Genome wide occurance of G quadrplex forming sequences in different part of the gene (Garg et al., 2016).
G4 has also been identified in RNA in plants. For example, a combination of biophysical and biochemical assays was carried out to confirm G4 structure formation by a GQFS located in 5′ UTR of ATR mRNA in A. thaliana (Kwok et al., 2015). The ATR gene encodes a protein kinase, which is activated upon DNA damage and required for the ensuing DNA damage response of the cell including repair, cell cycle arrest and telomere maintenance. Further study into the role of the GQFS identified at the 5′ UT of the ATR mRNA showed its inhibitory effect during translation initiation. Search of ATR homologs among 31 plant species resulted in identification of 35 ATR homologs. At least one GQFS was present in the 5′ UTR of 16 (14 plant species) of 35 ATR mRNA. Whether the conserved GQFS present in the ATR mRNA also have conserved function such as the negative regulation of translation as in A. thaliana is yet to be resolved. GQFS mediated gene regulation appears to be prevalent and of functional importance in plant kingdom. Table 2 describe the genome wide distribution of GQFS in different plant species.
Several putative G quadruplex structures have been identified by bioinformatics analyses in microRNA. In human genome, there are ∼16% of pre-mi-RNA that contains putative GQFS and can adopt these secondary structures to modulate canonical stem-loop structure of mi-RNA to adopt G quadruplex structure and thus impeded dicer mediated cleavage of mi-RNA (Mirihana Arachchilage et al., 2015). In addition, the equilibrium between the G quadruplex structure and stem loop structure influence the miRNA functionality as dicer enzyme recognize canonical stem loop structure in pre mi-RNA to produce mature miRNA and thus in turn formation of G quadruplex affect miRNA maturation [The RNA Stem–Loop to G Quadruplex Equilibrium Controls Mature MicroRNA Production inside the Cell (Pandey et al., 2015)]. In silico transcriptome wide analyses have identified significant number of G quadruplex motifs in human long non-coding RNA (lncRNA). Further, biophysical methods provide the information that approximately 60% of these putative structures form stable quadruplex in vitro and a further analyses of these secondary structures would give a better insight about the functional relevance of G4 structures in cellular function (Potential G quadruplexes in the human long non-coding transcriptome (Jayaraj et al., 2012). The plant genome have not been evaluated extensively for the presence of G quadruplex in their non-coding RNAs.
During unfavorable conditions such as abiotic (environmental factors such as high salt, high or low temperature) and biotic stress (damage to plants mediated by living organisms), plants must adapt to survive. During abiotic stress such as drought, the cytosolic concentrations of cations become elevated (Leigh and Wyn Jones, 1984). Since, higher potassium (K+) level is a condition that is known to facilitate the G quadruplex formation and under high salinity conditions, the K+ ion concentration in the cell increases (Zhang and Blumwald, 2001). This elevated levels of K+ ions facilitate the formation of G quadruplex genome wide and might be involved in salinity tolerance. Differential gene regulation mediated by G4 DNA or RNA structure formation is thereby hypothesized to be a potential mechanism to cope up with drought conditions. This hypothesis recently gained support by Mullen et al. (2010) who showed that GQFS are enriched at those genes differentially regulated during drought. In this study, transcriptome study of Arabidopsis originally conducted by Matsui et al. (2008) were analyzed to conclude that 16% of all genes in A. thaliana are drought-responsive and 45% of these genes contained at least one GQFS. Many similar studies followed since, (Andorf et al., 2014) demonstrated the abundance of GQFS in hypoxia-responsive genes in maize (Bailey-Serres et al., 2012). GQFS were also found to be frequent in genes associated with energy homeostasis signaling as well as many genes associated with TOR, AMP kinase, and oxidative stress signaling pathways. Kinases in TOR pathway are also directly regulated by the level of sugar availability and play a crucial role in nutrient and energy sensing. Occurrence of GQFS in genes encoding these kinases suggests that G quadruplex plays important role in regulation, signaling, and metabolic adjustment to energy status (Xu et al., 2010; Robaglia et al., 2012; Dobrenel et al., 2013; Figure 3). In Sapium sebiferum or Chinese tallow, which is an important agricultural crop species in east Asian countries, a bioinformatic analysis predicted the enrichment of GQFS at genes in the lipid biosynthesis and stress response pathways (Yang et al., 2015). Overall, genes that are differentially regulated during various stress conditions are more likely to contain a GQFS, and formation of G quadruplex may be one of multiple adaptive mechanisms utilized by plants during environmental stresses. Understanding how the stress response pathways are regulated in agriculturally important plant species through identification and functional analyses of GQFS can facilitate development of stress-tolerant plant varieties possibly through transgenic techniques and ultimately lead to higher-yield crops (Yang et al., 2015).
G4-forming synthetic oligonucleotides impede DNA polymerase activity in vitro and produced truncated product in presence of K+ ions, which stabilizes G4 structure (Garg et al., 2016). This result leads to the postulation that replication would be obstructed by G4 DNA in vivo, causing replication fork stall and collapse and thus causes genome instability (Figure 4).
Transposable elements are a significant part of eukaryotic genomes as they contain many regulatory sequences and serve as machinery to disseminate the genes present within. Interestingly, GQFS has been found to be present in long terminal repeats (LTR) of plant-transposons and -retrotransposons and possibly effects not only transcription and translation but reverse transcription as well. (Lexa et al., 2014; Kejnovsky et al., 2015). Formation of these secondary structures causes the conformational changes in DNA and DNA can become nucleosome free when its confirmation changes and It is well known that preferred site of integration for TE is open chromatin (Huppert and Balasubramanian, 2005; Liu et al., 2009; Wong and Huppert, 2009). G quadruplex in TE inhibits transcription and formation of these structures in TE can serve as hot spot for recombination and TE serves a vehicle for spread of G4 structure in the genome. These secondary structure are not only formed inside the TE due to open chromatin confirmation but can also become the genomic targets for insertion of new TE during which changed DNA confirmations are recognized by transposase or by integrase. Proteins originating from TE retain their affinity to open configuration and prefer binding to secondary structures such as G4 DNA, i.e., RAG1 protein (Nambiar and Raghavan, 2011). Formation of G quadruplex structures in TE-derived RNA participate in many important cellular processes (Kapusta et al., 2013).
Moreover, long stretches of guanines were identified upstream and downstream of the promoter region of retrotransposons. These sequences have been confirmed to readily adopt parallel- or antiparallel-stranded G quadruplexes by CD spectroscopy (Lexa et al., 2014). Occurrence of GQFS, at these specific locations suggests their role during initiation of transcription and elongation of retrotransposon RNA. Alternatively, these GQFS might act as the check point of transcription and reverse transcription. Biological role of GQFS in transposable elements life cycle have been established, as long stretch of guanines is found in young and active LTR and lower number of guanines is found in old elements due to supress elongation of RNA strand (Lexa et al., 2014). Such enrichment of GQFS, within specific region of LTR retrotransposons propose functional role of G quadruplex in genome instability (recombination based reshuffling) of plant genomes (Lexa et al., 2014; Kejnovsky et al., 2015).
Several G quadruplex binding proteins have been identified in yeast and metazoans. Upon binding, these proteins may either help resolve the G4 structures or may enhance the stability of the secondary structures. G4 resolving proteins include DNA helicases such as BLM (RecQ family), WRN (RecQ family), and FANCJ in humans (Sun et al., 1998; Fry and Loeb, 1999; Mohaghegh et al., 2001; Cheok et al., 2005; Wu et al., 2008), Dog-1 in the nematode Caenorhabditis elegans (Youds et al., 2008), and Sgs1 (RecQ family) and Pif1 in yeast (Eddy and Maizels, 2006).
RecQ helicases, conserved from bacteria to humans, are involved in unwinding of a wide variety of DNA substrates including G quadruplexes and thus are important in maintaining genome integrity. In humans, there are five RecQ-family helicases (BLM, WRN, RTS/RECQ4) while yeast and bacteria possess only one – Sgs1 and RecQ, respectively. In case of plants, A. thaliana genome contains seven different genes that encode RecQ family helicase; RECQ1, RECQ2, RECQ3, RECQ4, RECQ4B, RECQ5, and RECQsim (Hartung et al., 2000; Hartung and Puchta, 2006). AtRecQ4A and 4B are two of the genes of this family that evolved due to recent duplication and are 70% identical on protein level to other members of this family. Role of AtRecQ4A have been suggested to be equivalent to yeast Sgs1 and human BLM. AtRecQ4B is distinct among all eukaryotic RecQ homologs as it appears to promote rather than suppress crossover recombinations (Hartung et al., 2007; Schröpfer et al., 2014). RecQsim contains a unique insertion of acidic amino acids in its helicase domain. Homologs of RecQsim of A. thaliana have been found in other plant species including rice and rape (Bagherieh-Najjar et al., 2003). The expression of A. thaliana RecQsim gene in yeast lacking Sgs1 compensates the loss of Sgs1 and rescues the hypersenstivity to the DNA damaging drug methyl methanesulfonate (MMS), indicating functional conservation between these helicases.
In yeast, other non-helicase G4 DNA binding proteins, such as the co-transcription factor Sub1, also contribute to the stability of the genomic loci containing GQFS (Lopez et al., 2017). In case of plants, very little information is available regarding potential G4-binding proteins. In maize (Zea mays), a G4 binding protein known as ZmNDPK1 have been identified in a a ligand-binding screening of a cDNA-expression library. ZmNDPK1, which is a nucleoside diphosphate kinase 1, interacts with folded GQFS-containing oligos, with low nanomolar-range affinity (Kopylov et al., 2015). Electrophoretic mobility shift assay (EMSA) using nuclear extracts from rice plants revealed proteins stably binding G4 DNA of both parallel and antiparallel conformations; the identities of these proteins, however, are still pending (Garg et al., 2016). Additionally, certain medicinal plant extracts such as theaflavin-digallate from tea and saffron carotenoids from Crocus sativus, were shown to contain non-protein, small molecule ligands with G quadruplex binding activity (Hoshyar et al., 2012; Mikutis et al., 2013; Wang et al., 2015). As shown in yeast, the interaction of GQFS with proteins with specific affinity to the non-canonical secondary structure is an important mechanism in G4-associated regulatory functions. Therefore, in order to fully understand the function of G4 in plant biology, identification and further characterization of G4 binding proteins and small molecule G4 ligands in plant species are of high priority.
Genome-wide analyses elucidated numerous GQFS in several plant species including A. thaliana, Z. mays, O. japonicum and O. sativa and others and conclusively showed the abundance of G2 type GQFS in the genic and coding regions and G3 type GQFS in the intergenic regions. Biophysical characterization of a subset of these GQFSs have also been accomplished. G quadruplex play a regulatory role during cellular responses to DNA damage and other internal and external cues such as sugar availability; metabolic and energy status; stress response etcs. In addition, G quadruplex formation can induce fluorescence activation with high selectivity and sensitivity (DasGupta et al., 2015). Current agricultural production is heavily reliant on many biotic and abiotic factors; stress conditions such as drought and soil salinity are main factors responsible for crop yield reduction. Further work into Identification and functional analyses of G quadruplexes in plants can be a particular interest as a target for biotic and abiotic stress response in plants. It is not yet clear whether the tolerance mechanisms of plants in response to various kind of stresses are directly or indirectly regulated by G4 quadruplexes. Future work is expected to focus on defining the detailed the molecular pathway(s) governed by G4 DNA in response to both biotic and abiotic stresses. Generating transgenic plants tolerant to drought and other stresses is a central hypothesis for the agricultural industry. Understanding how the structural transformation of G4 DNA assembly is regulated and in turn regulate gene expression, therefore, could have valuable implications for the development of transgenic plant varieties with higher yield.
PY and VY contributed to the conception of the review article. PY and VY drafted the work. PY, VY, H, and NK wrote the review article. PY, VY, NT, and NK revised it critically. PY, NK, and NT helped in literature search. PY, VY, H, NT, and NK gave Final approval of the version to be published.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported, in part, by a grant to PY, ECR/2015/000431 from Science and Engineering Research Board, Department of Science and Technology, Government of India; and a grant to VY Ramalingaswamy fellowship, BT/RLF/RE-ENTRY/29/2014 from Department of Biotechnology, Government of India; and grant to NK, GM116007 from National Institutes of Health, United States and AU1875 from Welch Foundation.
Andorf, C. M., Kopylov, M., Dobbs, D., Koch, K. E., Stroupe, M. E., Lawrence, C. J., et al. (2014). G-quadruplex (G4) motifs in the maize (Zea mays L.) genome are enriched at specific locations in thousands of genes coupled to energy status, hypoxia, low sugar, and nutrient deprivation. J. Genet. Genomics 41, 627–647. doi: 10.1016/j.jgg.2014.10.004
Bagherieh-Najjar, M. B., de Vries, O. M. H., Kroon, J. T. M., Wright, E. L., Elborough, K. M., Hille, J., et al. (2003). Arabidopsis RecQsim, a plant-specific member of the RecQ helicase family, can suppress the MMS hypersensitivity of the yeast sgs1 mutant. Plant Mol. Biol. 52, 273–284.
Bailey-Serres, J., Fukao, T., Gibbs, D. J., Holdsworth, M. J., Lee, S. C., Licausi, F., et al. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138. doi: 10.1016/j.tplants.2011.12.004
Beaume, N., Pathak, R., Yadav, V. K., Kota, S., Misra, H. S., Gautam, H. K., et al. (2013). Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 41, 76–89. doi: 10.1093/nar/gks1071
Blackburn, E. H. (1994). Telomeres: no end in sight. Cell 77, 621–623. doi: 10.1016/0092-8674(94)90046-9
Bochman, M. L., Paeschke, K., and Zakian, V. A. (2012). DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780. doi: 10.1038/nrg3296
Bugaut, A., and Balasubramanian, S. (2008). A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry 47, 689–697. doi: 10.1021/bi701873c
Cahoon, L. A., and Seifert, H. S. (2009). An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325, 764–767. doi: 10.1126/science.1175653
Capra, J. A., Paeschke, K., Singh, M., and Zakian, V. A. (2010). G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 6:e1000861. doi: 10.1371/journal.pcbi.1000861
Cheok, C. F., Bachrati, C. Z., Chan, K. L., Ralf, C., Wu, L., and Hickson, I. D. (2005). Roles of the Bloom’s syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 33, 1456–1459. doi: 10.1042/BST0331456
Cogoi, S., Paramasivam, M., Filichev, V., Géci, I., Pedersen, E. B., and Xodo, L. E. (2009). Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 52, 564–568. doi: 10.1021/jm800874t
Cogoi, S., and Xodo, L. E. (2006). G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549. doi: 10.1093/nar/gkl286
DasGupta, S., Shelke, S. A., Li, N., and Piccirilli, J. A. (2015). Spinach RNA aptamer detects lead(II) with high selectivity. Chem. Commun. 51, 9034–9037. doi: 10.1039/c5cc01526j
De Armond, R., Wood, S., Sun, D., Hurley, L. H., and Ebbinghaus, S. W. (2005). Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry 44, 16341–16350. doi: 10.1021/bi051618u
Dexheimer, T. S., Sun, D., and Hurley, L. H. (2006). Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 128, 5404–5415. doi: 10.1021/ja0563861
Dhapola, P., and Chowdhury, S. (2016). QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 44, W277–W283. doi: 10.1093/nar/gkw425
Dobrenel, T., Marchive, C., Azzopardi, M., Clément, G., Moreau, M., Sormani, R., et al. (2013). Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front. Plant Sci. 4:93. doi: 10.3389/fpls.2013.00093
Eddy, J., and Maizels, N. (2006). Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 34, 3887–3896. doi: 10.1093/nar/gkl529
Evans, T., Schon, E., Gora-Maslak, G., Patterson, J., and Efstratiadis, A. (1984). S1-hypersensitive sites in eukaryotic promoter regions. Nucleic Acids Res. 12, 8043–8058. doi: 10.1093/nar/12.21.8043
Fernando, H., Reszka, A. P., Huppert, J., Ladame, S., Rankin, S., Venkitaraman, A. R., et al. (2006). A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 45, 7854–7860. doi: 10.1021/bi0601510
Fry, M., and Loeb, L. A. (1999). Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 274, 12797–12802. doi: 10.1074/jbc.274.18.12797
Garg, R., Aggarwal, J., and Thakkar, B. (2016). Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. 6:28211. doi: 10.1038/srep28211
Hartung, F., Plchová, H., and Puchta, H. (2000). Molecular characterisation of RecQ homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 4275–4282.
Hartung, F., and Puchta, H. (2006). The RecQ gene family in plants. J. Plant Physiol. 163, 287–296. doi: 10.1016/j.jplph.2005.10.013
Hartung, F., Suer, S., and Puchta, H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 18836–18841. doi: 10.1073/pnas.0705998104
Hoshyar, R., Bathaie, S. Z., Kyani, A., and Mousavi, M. F. (2012). Is there any interaction between telomeric DNA structures, G-quadruplex and I-motif, with saffron active metabolites? Nucleosides Nucleotides Nucleic Acids 31, 801–812. doi: 10.1080/15257770.2012.730164
Huppert, J. L. (2010). Structure, location and interactions of G-quadruplexes. FEBS J. 277, 3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x
Huppert, J. L., and Balasubramanian, S. (2005). Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916. doi: 10.1093/nar/gki609
Huppert, J. L., and Balasubramanian, S. (2007). G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 406–413. doi: 10.1093/nar/gkl1057
Jayaraj, G. G., Pandey, S., Scaria, V., and Maiti, S. (2012). Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. 9, 81–86. doi: 10.4161/rna.9.1.18047
Kapusta, A., Kronenberg, Z., Lynch, V. J., Zhuo, X., Ramsay, L., Bourque, G., et al. (2013). Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. doi: 10.1371/journal.pgen.1003470
Kejnovsky, E., Tokan, V., and Lexa, M. (2015). Transposable elements and G-quadruplexes. Chromosome Res. 23, 615–623. doi: 10.1007/s10577-015-9491-7
Kilpatrick, M. W., Torri, A., Kang, D. S., Engler, J. A., and Wells, R. D. (1986). Unusual DNA structures in the adenovirus genome. J. Biol. Chem. 261, 11350–11354.
König, S. L. B., Evans, A. C., and Huppert, J. L. (2010). Seven essential questions on G-quadruplexes. Biomol. Concepts 1, 197–213. doi: 10.1515/bmc.2010.011
Kopylov, M., Bass, H. W., and Stroupe, M. E. (2015). The maize (Zea mays L.) nucleoside diphosphate kinase1 (ZmNDPK1) gene encodes a human NM23-H2 homologue that binds and stabilizes G-quadruplex DNA. Biochemistry 54, 1743–1757. doi: 10.1021/bi501284g
Kwok, C. K., Ding, Y., Shahid, S., Assmann, S. M., and Bevilacqua, P. C. (2015). A stable RNA G-quadruplex within the 5’-UTR of Arabidopsis thaliana ATR mRNA inhibits translation. Biochem. J. 467, 91–102. doi: 10.1042/BJ20141063
Largy, E., Mergny, J. -L., and Gabelica, V. (2016). Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 16, 203–258. doi: 10.1007/978-3-319-21756-7_7
Leigh, R. A., and Wyn Jones, R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 97, 1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x
Lexa, M., Kejnovsky, E., Steflova, P., Konvalinova, H., Vorlickova, M., and Vyskot, B. (2014). Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 42, 968–978. doi: 10.1093/nar/gkt893
Liu, S., Yeh, C. -T., Ji, T., Ying, K., Wu, H., Tang, H. M., et al. (2009). Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5:e1000733. doi: 10.1371/journal.pgen.1000733
Lopez, C. R., Singh, S., Hambarde, S., Griffin, W. C., Gao, J., Chib, S., et al. (2017). Yeast Sub1 and human PC4 are G-quadruplex binding proteins that suppress genome instability at co-transcriptionally formed G4 DNA. Nucleic Acids Res. 45, 5850–5862. doi: 10.1093/nar/gkx201
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Mikutis, G., Karaköse, H., Jaiswal, R., LeGresley, A., Islam, T., Fernandez-Lahore, M., et al. (2013). Phenolic promiscuity in the cell nucleus–epigallocatechingallate (EGCG) and theaflavin-3,3’-digallate from green and black tea bind to model cell nuclear structures including histone proteins, double stranded DNA and telomeric quadruplex DNA. Food Funct. 4, 328–337. doi: 10.1039/c2fo30159h
Mirihana Arachchilage, G., Dassanayake, A. C., and Basu, S. (2015). A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 22, 262–272. doi: 10.1016/j.chembiol.2014.12.013
Mohaghegh, P., Karow, J. K., Brosh, R. M. Jr., Bohr, V. A., and Hickson, I. D. (2001). The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849. doi: 10.1093/nar/29.13.2843
Mullen, M. A., Olson, K. J., Dallaire, P., Major, F., Assmann, S. M., and Bevilacqua, P. C. (2010). RNA G-Quadruplexes in the model plant species Arabidopsis thaliana: prevalence and possible functional roles. Nucleic Acids Res. 38, 8149–8163. doi: 10.1093/nar/gkq804
Nakagama, H., Higuchi, K., Tanaka, E., Tsuchiya, N., Nakashima, K., Katahira, M., et al. (2006). Molecular mechanisms for maintenance of G-rich short tandem repeats capable of adopting G4 DNA structures. Mutat. Res. 598, 120–131. doi: 10.1016/j.mrfmmm.2006.01.014
Nambiar, M., and Raghavan, S. C. (2011). How does DNA break during chromosomal translocations? Nucleic Acids Res. 39, 5813–5825. doi: 10.1093/nar/gkr223
Palumbo, S. L., Memmott, R. M., Uribe, D. J., Krotova-Khan, Y., Hurley, L. H., and Ebbinghaus, S. W. (2008). A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 36, 1755–1769. doi: 10.1093/nar/gkm1069
Pandey, S., Agarwala, P., Jayaraj, G. G., Gargallo, R., and Maiti, S. (2015). The RNA stem–loop to G-quadruplex equilibrium controls mature MicroRNA production inside the cell. Biochemistry 54, 7067–7078. doi: 10.1021/acs.biochem.5b00574
Parkinson, G. N., Lee, M. P. H., and Neidle, S. (2002). Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880. doi: 10.1038/nature755
Rankin, W. W., Brennan, S., Schell, E., Laviwa, J., and Rankin, S. H. (2005). The stigma of being HIV-positive in Africa. PLoS Med. 2:e247. doi: 10.1371/journal.pmed.0020247
Rawal, P., Kummarasetti, V. B. R., Ravindran, J., Kumar, N., Halder, K., Sharma, R., et al. (2006). Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 16, 644–655. doi: 10.1101/gr.4508806
Robaglia, C., Thomas, M., and Meyer, C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307. doi: 10.1016/j.pbi.2012.01.012
Schröpfer, S., Kobbe, D., Hartung, F., Knoll, A., and Puchta, H. (2014). Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 42, 1684–1697. doi: 10.1093/nar/gkt1004
Siddiqui-Jain, A., Grand, C. L., Bearss, D. J., and Hurley, L. H. (2002). Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598. doi: 10.1073/pnas.182256799
Sun, D., Guo, K., Rusche, J. J., and Hurley, L. H. (2005). Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 33, 6070–6080. doi: 10.1093/nar/gki917
Sun, H., Karow, J. K., Hickson, I. D., and Maizels, N. (1998). The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273, 27587–27592. doi: 10.1074/jbc.273.42.27587
Verma, A., Halder, K., Halder, R., Yadav, V. K., Rawal, P., Thakur, R. K., et al. (2008). Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 51, 5641–5649. doi: 10.1021/jm800448a
Wang, Y., and Patel, D. J. (1993). Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 234, 1171–1183. doi: 10.1006/jmbi.1993.1668
Wang, Y., Zhao, M., Zhang, Q., Zhu, G.-F., Li, F.-F., and Du, L.-F. (2015). Genomic distribution and possible functional roles of putative G-quadruplex motifs in two subspecies of Oryza sativa. Comput. Biol. Chem. 56, 122–130. doi: 10.1016/j.compbiolchem.2015.04.009
Wong, H. M., and Huppert, J. L. (2009). Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 5, 1713–1719. doi: 10.1039/b905848f
Wu, Y., Shin-ya, K., and Brosh, R. M. (2008). FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128. doi: 10.1128/MCB.02210-07
Xu, X. M., Lin, H., Maple, J., Björkblom, B., Alves, G., Larsen, J. P., et al. (2010). The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J. Cell Sci. 123, 1644–1651. doi: 10.1242/jcs.063222
Yadav, P., Harcy, V., Argueso, J. L., Dominska, M., Jinks-Robertson, S., and Kim, N. (2014). Topoisomerase I plays a critical role in suppressing genome instability at a highly transcribed G-quadruplex-forming sequence. PLoS Genet. 10:e1004839. doi: 10.1371/journal.pgen.1004839
Yadav, P., Owiti, N., and Kim, N. (2016). The role of topoisomerase I in suppressing genome instability associated with a highly transcribed guanine-rich sequence is not restricted to preventing RNA:DNA hybrid accumulation. Nucleic Acids Res. 44, 718–729. doi: 10.1093/nar/gkv1152
Yang, M., Wu, Y., Jin, S., Hou, J., Mao, Y., Liu, W., et al. (2015). Flower bud transcriptome analysis of Sapium sebiferum (Linn.) Roxb. and primary investigation of drought induced flowering: pathway construction and G-quadruplex prediction based on transcriptome. PLoS ONE 10:e0118479. doi: 10.1371/journal.pone.0118479
Youds, J. L., Barber, L. J., Ward, J. D., Collis, S. J., O’Neil, N. J., Boulton, S. J., et al. (2008). DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 28, 1470–1479. doi: 10.1128/MCB.01641-07
Yu, K., Chedin, F., Hsieh, C.-L., Wilson, T. E., and Lieber, M. R. (2003). R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4, 442–451. doi: 10.1038/ni919
Keywords: G quadruplex, genome stability, recombination, DNA damage and repair, transcriptional and translational regulation
Citation: Yadav V, Hemansi, Kim N, Tuteja N and Yadav P (2017) G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 8:1163. doi: 10.3389/fpls.2017.01163
Received: 28 March 2017; Accepted: 16 June 2017;
Published: 04 July 2017.
Edited by:
Manisha Goel, University of Delhi, IndiaReviewed by:
Sudhir P. Singh, Center of Innovative and Applied Bioprocessing (CIAB), IndiaCopyright © 2017 Yadav, Hemansi, Kim, Tuteja and Yadav. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Puja Yadav, cHVqYXlhZGF2QGN1aC5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.