
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 June 2017
Sec. Agroecology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01064
This article is part of the Research TopicHerbicide resistance in weeds: early detection, mechanisms, dispersal, new insights and management issuesView all 29 articles
Climate change endangers food security and our ability to feed the ever-increasing human population. Weeds are the most important biotic stress, reducing crop-plant productivity worldwide. Chemical control, the main approach for weed management, can be strongly affected by temperature. Previously, we have shown that temperature-dependent non-target site (NTS) resistance of Brachypodium hybridum is due to enhanced detoxification of acetyl-CoA carboxylase inhibitors. Here, we explored the transcriptional basis of this phenomenon. Plants were characterized for the transcriptional response to herbicide application, high-temperature and their combination, in an attempt to uncover the genetic basis of temperature-dependent pinoxaden resistance. Even though most of the variance among treatments was due to pinoxaden application (61%), plants were able to survive pinoxaden application only when grown under high-temperatures. Biological pathways and expression patterns of members of specific gene families, previously shown to be involved in NTS metabolic resistance to different herbicides, were examined. Cytochrome P450, glucosyl transferase and glutathione-S-transferase genes were found to be up-regulated in response to pinoxaden application under both control and high-temperature conditions. However, biological pathways related to oxidation and glucose conjugation were found to be significantly enriched only under the combination of pinoxaden application and high-temperature. Analysis of reactive oxygen species (ROS) was conducted at several time points after treatment using a probe detecting H2O2/peroxides. Comparison of ROS accumulation among treatments revealed a significant reduction in ROS quantities 24 h after pinoxaden application only under high-temperature conditions. These results may indicate significant activity of enzymatic ROS scavengers that can be correlated with the activation of herbicide-resistance mechanisms. This study shows that up-regulation of genes related to metabolic resistance is not sufficient to explain temperature-dependent pinoxaden resistance. We suggest that elevated activity of enzymatic processes at high-temperature may induce rapid and efficient pinoxaden metabolism leading to temperature-dependent herbicide resistance.
Anthropogenic greenhouse gas emissions and climate change pose risks to long-term food security due to their detrimental effects on agriculture productivity (Myers et al., 2017). To feed the 9.6 billion people expected by 2050 (FAOSTAT, 2017) a significant increase in cereal-grain yield will be needed (reviewed by Tester and Langridge, 2010). In the long-term (2030–2050), climatic changes in the Middle East are expected to affect mean temperatures by 1–2°C (Parry et al., 2007; Nelson et al., 2009). However, greater risks to food security may be posed by changes in between-year and within-year variability and the increasing frequency and severity of extreme weather events (Gornall et al., 2010; Lelieveld et al., 2016; Stott, 2016). These environmental changes will affect the development and productivity of both crops and weeds. Weed infestation has already been acknowledged as a major factor causing yield reduction in various crops such as maize (Zea mays, Soltani et al., 2016), rice (Oryza sativa, Chauhan and Johnson, 2011; Chauhan and Opena, 2012), and hazelnut (Corylus avellana, Kaya-Altop et al., 2016).
Since their introduction in 1940, herbicides are the most cost-effective and efficient practice for weed control. In recent years, this method of weed control has become less efficient due to the evolution of herbicide-resistant weeds (Heap, 2017). Herbicide resistance is a consequence of strong selection pressure imposed by repeated application of the same herbicide to a weed population. Herbicide resistance can result from modification of the target site (TS) or via other mechanisms involved in non-target site (NTS) resistance (Rubin, 1991). Mechanisms of TS resistance have been well studied; they involve structural changes at herbicide-binding sites or increased expression of target proteins. NTS resistance can be endowed through reduced absorption (Koger and Reddy, 2005), reduced translocation (Feng et al., 2004) or sequestration (Kleinman and Rubin, 2016). The underlying mechanisms involved in NTS resistance are still not thoroughly understood (Délye, 2013).
Acetyl-CoA carboxylase (ACCase) inhibitors are commonly used to control grass weeds in various crops. In most plants, two isoforms of the ACCase enzyme, heteromeric and homomeric, exist in different cell compartments (i.e., cytosol and plastids; Roesler et al., 1997). In grass species, only the homomeric form of the enzyme is present and ACCase inhibitors function by blocking this form (Sasaki and Nagano, 2004). NTS resistance to ACCase inhibitors is endowed mostly by herbicide detoxification (Kaundun, 2014). The first two crucial phases of detoxification are mediated by members of the cytochrome P450 (CYP450) enzyme family (Manabe et al., 2007; Gaines et al., 2014; Iwakami et al., 2014) and both glutathione-S-transferase (GST; Cummins et al., 1999, 2013; Skipsey et al., 2011) and glucosyl transferase (GT; Baerson et al., 2005; Gardin et al., 2015; Xu et al., 2015) play key roles in the conjugation of the herbicide. Different genes from all three families (CYP450, GST, and GT) have been found to be up-regulated in herbicide-resistant grass weed populations such as Lolium rigidum (Gaines et al., 2014; Duhoux et al., 2015), Alopecurus myosuroides (Gardin et al., 2015), and Eleusine indica (An et al., 2014; Chen et al., 2015).
Environmental conditions such as temperature can affect the retention, penetration and movement of herbicides through the plant and can also modify plants growth and development, indirectly affecting herbicide activity within the plant (e.g., Hammerton, 1967; Caseley, 1989; Rubin, 1991; Sundby et al., 1993; Robinson et al., 2015). Temperature may modify the response of plants to herbicides with different modes of action (HRAC, 2017). This phenomenon has been demonstrated in the effect of paraquat (group D) on Hordeum glaucum (Lasat et al., 1996), the effect of glyphosate (group G) on Conyza sp. (Kleinman et al., 2015), the effect of mesotrione (group F) on Amaranthus palmeri (Godar et al., 2015), and the effect of pinoxaden (group A) on Brachypodium hybridum (Matzrafi et al., 2016).
Brachypodium, a small annual grass species, native to the Mediterranean region, is a valuable model system for a variety of biological processes and genome organization in cereals (reviewed by Kellogg, 2015). In recent years, it has emerged as a powerful model plant for the study of herbicide resistance in grass weeds (e.g., Gressel et al., 1983; Matzrafi et al., 2014; Frenkel et al., 2017). Recently, we demonstrated that elevated temperatures result in increased tolerance to ACCase inhibitors in various grass weed species (Matzrafi et al., 2016). Here, we employed a system biology approach to uncover the transcriptional basis of temperature-dependent NTS resistance mechanism. We hypothesized that temperature-dependent herbicide detoxification is facilitated by enhanced enzymatic efficiency at elevated temperatures. Previously, we have identified a B. hybridum accession, presenting temperature-dependent resistant to pinoxaden (Matzrafi et al., 2016). The aims of the current study were to: (i) characterize the transcriptional differences between pinoxaden-treated and untreated plants under different temperatures, (ii) elucidate the biological processes that are associated with temperature-dependent herbicide detoxification, and (iii) examine the role of metabolism-related genes known to be involved in herbicide resistance in temperature-dependent pinoxaden resistance in B. hybridum.
Seeds of B. hybridum accession BrI-782 (temperature-dependent NTS-resistant to the ACCase inhibitor pinoxaden; Matzrafi et al., 2014) were germinated in trays filled with growth mixture (Pele-Shacham, Israel). The trays were placed in a dark, cold room (16°C) until germination. After emergence, uniform seedlings were transplanted into pots (7 cm × 7 cm × 6 cm) containing similar growth mixture and transferred to a phytotron where they were kept under natural Mediterranean growth conditions [10/16°C (night/day), 10 h of light]. Two temperature regimes were used in this study: control [10/16°C (night/day)] and high temperature [28/34°C (night/day)].
At the three-leaf stage (BBCH scale 13; Hong et al., 2011), plants were treated with either water (control) or the recommended dose of the ACCase inhibitor pinoxaden (Axial®, 50 g L-1 pinoxaden + 11.25 g L-1 cloquintocet-mexyl, EC, Syngenta, Switzerland; X = recommended dose of 30 g ai ha-1). The treatment was applied using a chain-driven sprayer delivering 300 L ha-1. One hour after treatment (HAT), plants were moved back to the phytotron and each plant was assigned to one of two temperature regimes: control or high. In each room, 10 plants (five treated with pinoxaden and five treated with water) were kept for 21 days after treatment (DAT). Survival rates were visually assessed and shoot fresh weight was measured.
Samples of fresh shoot tissue were collected from treated and untreated plants at 24 HAT (Figure 1A), immediately frozen in liquid nitrogen and stored at -80°C. RNA-seq analysis was conducted using three plants from each of the following treatments: control (C), pinoxaden application (X), high-temperature (H) and the combination of pinoxaden and high-temperature (HX; Figure 1A). Total RNA was extracted using a Plant/Fungi Total RNA Purification Kit (Norgen Biotek Corp., Canada). Total RNA was treated with TURBODNase® (RNase-Free; Ambion, Warrington, United Kingdom) to eliminate DNA contamination. RNA was quantified using a NanoDrop (ND-1000) spectrophotometer (Thermo Scientific, Wilmington, DE, United States) and RNA integrity and quality were assessed with a 2100 Bioanalyzer (Agilent Technologies Inc., Germany). Additional data concerning sample quality and other parameters are presented in Supplementary Table S1.
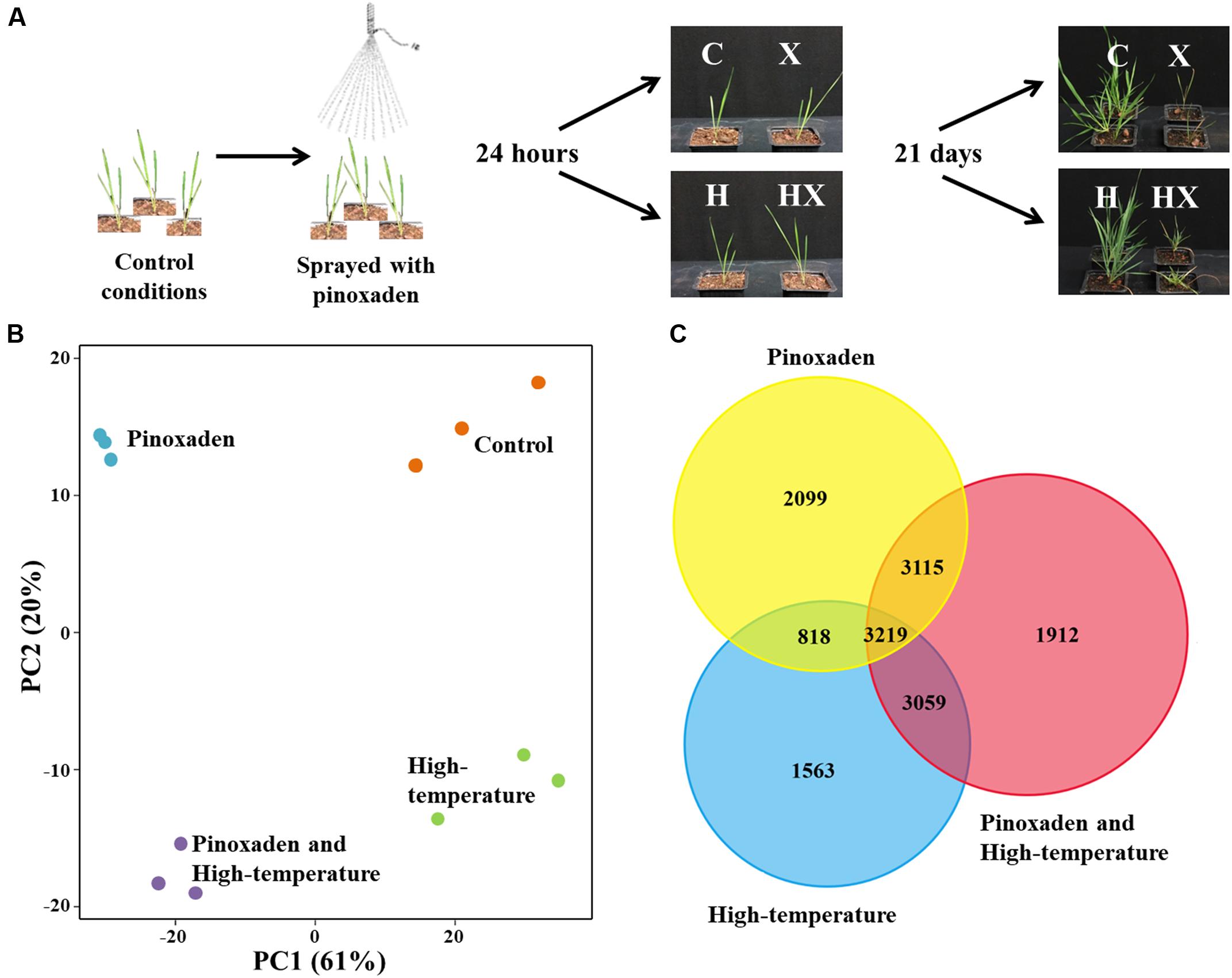
FIGURE 1. (A) Experimental design and final phenotypes of Brachypodium hybridum plants (accession BrI-782) grown under control (C, 10/16°C day/night) and high-temperature (H, 28/34°C) conditions. Plants were sprayed with pinoxaden (X) or water at the three-leaf stage; RNA-seq samples were taken 24 h after application. Plant survival rates were determined 21 days after pinoxaden application. (B) Principal-component (PC) analysis of rlog transformed gene expression data, generated by RNA-sequencing. Each treatment contains three biological repeats and is indicated by a different color. (C) Venn diagram of differentially expressed genes (DEGs) from all three treatments.
cDNA libraries were generated using a NEBNext Ultra Directional RNA Prep Kit (New England Biolabs, Ipswich, MA, United States), following the manufacturer’s instructions. After verifying their quality, libraries were indexed with six-nucleotide barcodes and sequencing was performed on the Illumina HiSeq2000 machine using multiplexing for generating 50 base-paired (bp) end reads. Sequencing was carried out at the Technion Genome Center (Haifa, Israel).
B. hybridum transcript levels were obtained from HiSeq2000 machine using a custom computational pipeline. Briefly, 50 bp end reads were trimmed and quality-filtered using Trimmomatic (Bolger et al., 2014) and then mapped to the B. distachyon genome (International Brachypodium Initiative, 2010), version 3.0 using Tophat (Kim D. et al., 2013). Tophat was run with a maximum intron length of 10,000 bases to reduce the likelihood of false positives (Walters et al., 2013). The mean percentages of overall mapped reads and multiple mapped reads across samples were 80 and 5%, respectively. Mapped reads were then checked for overlap with JGI v3.11 gene exons using htseq-count (Anders et al., 2015). The mean percentage of reads overlapping (a known feature across samples) was 90%. We then used DESeq2 (Love et al., 2014) to detect differentially expressed genes (DEGs) among samples from the different experimental treatments (high-temperature, pinoxaden and their combination) and the control. Data is presented as log2 of fold-change values (log2FC). For DEG analysis we considered genes that were significant at false discovery rate (FDR) ≤ 5%.
Differentially expressed genes were analyzed using hierarchical clustering and divided into parallel plots using the JMP (ver. 12) statistical package (SAS Institute Inc., Cary, NC, United States). Genes that were highly up-regulated under HX treatment were annotated with MapMan software (Thimm et al., 2004) using a functional data base containing 32,031 different assigned identifiers, with more than 35 general biological processes matching the B. distachyon genome (International Brachypodium Initiative, 2010). Pathway-enrichment analysis was performed with FunRich, a functional enrichment analysis software tool2 (FDR ≤ 0.05; Pathan et al., 2015), using MapMan annotation as reference data (Thimm et al., 2004).
RNA samples used for RNA-seq analysis (n = 3) as well as additional three plants that were grown and treated with the plants selected for RNA-seq analysis were used for qPCR validation (n = 6). First-strand cDNA was synthesized using qScriptTM cDNA Synthesis Kit (Quanta Biosciences Inc., United States), following the manufacturer’s instructions. qPCR was carried out using PerfeCTa® SYBR® Green FastMix® (Quanta Biosciences Inc., United States) in a PikoReal RT-PCR system (Thermo Fisher Scientific Inc., United States). Gene-specific primers were designed using Primer3 software (Supplementary Table S2). PCR mixtures included 2 μL of cDNA (diluted by four), 8 μL of SYBR Mix, and 300 nM of each primer in a final volume of 10 μL. The 2-ΔΔCT method (Livak and Schmittgen, 2001) was used for the normalization and calibration processes. Transcript values were relatively tested compared to the housekeeping gene S-adenosylmethionine decarboxylase (SamDC, BRADI2G02580; Hong et al., 2008), whose expression was not affected by the herbicide and∖or high-temperature treatment.
Plants of B. hybridum accession BrI-782 were grown under controlled conditions, as described above. At the three-leaf stage, plants were treated with the recommended dose of pinoxaden as described above. Treated and untreated plants were transferred back to controlled conditions or subjected to a high-temperature regime. The experiment was conducted three times using three replicates for each treatment. ROS measurements were taken at 2, 8, and 24 HAT. Dichlorodihydrofluorescein diacetate (H2DCF-DA) was used as a probe for H2O2/peroxides as described previously (Pena et al., 2012). A 50 μM stock of H2DCF-DA was prepared in DMSO, diluted (1:9) in a 10 mM Tris-HCl (pH 7.4) and non-ionic surfactant (Spreader L77, ADAMA-Agan, Israel) at 0.05% v/v was added. The second leaf of each plant was cut and covered with aluminum foil, to prevent light exposure for 30 min, during which time the leaf was kept at room temperature. Leaves were than washed three times with 10 mM Tris-Hcl (pH 7.4). Fluorescent stereoscope images were captured using a Nikon SMZ1500 (Nikon, Japan) zoom stereoscope, with an excitation filter of 450–490 nm and a barrier filter of 510 nm, which was used to detect H2DCFDA green fluorescence. Images were captured with a color camera (DS RI1, Nikon, Japan) operated with NIS Elements V3 software (Nikon, Japan). Exposure times were equal for all samples. Autofluorescence was not observed in unstained controls at the exposure time used. Leaf fluorescence was quantified using ImageJ software (ver. 1.63; U.S. National Institutes of Health).
Raw sequencing files of mRNA sequencing are available at the short read archive of the National Center for Biotechnology Information (https://trace.ncbi.nlm.nih.gov/Traces/sra) under accession number PRJNA360668.
As a native Mediterranean temperate grass species, B. hybridum grows during the winter season (November to April). Under control conditions (10/16°C, night/day), which mimic the Mediterranean winter, BrI-782 plants showed a higher sensitivity to pinoxaden that was manifested by lower shoot fresh weights (21%) and a lower survival rate, as compared with treated plants grown under high-temperature (Figure 1A and Supplementary Table S3). We hypothesized that the significant differences in plant response to pinoxaden under contrasting temperature regimes may be the result of temperature-dependent transcriptional modifications. In order to test our hypothesis, we used RNA-seq to detect the temperature-dependent differences in transcripts, pathways and mechanisms between pinoxaden-sensitive and pinoxaden-resistant plants.
A total of 257 million raw reads were generated from the 12 libraries (three biological replicates for each of the four treatments). After removing reads containing adaptor or ploy-N and low-quality reads, 208 million clean reads were obtained with 13.4–18.6 million reads per sample. In the absence of a fully sequenced B. hybridum genome, the annotation procedure was carried using the B. distachyon database. Our B. hybridum samples scored 80.6% mapped reads and 87.2% featured contigs (Supplementary Table S1). Principal-component analysis was used to assess the variability among biological replicates and treatments. Within each treatment, the three biological replicates showed a high degree of similarity to one another and formed an independent cluster. Among the treatments, application of herbicide either alone or in combination with high-temperature resulted in a separation of the control and high-temperature treatments. This separation explained 61% of the total experimental variance (Figure 1B).
The number of DEGs between each treatment and control conditions ranged between 11,305 for the combination of herbicide and high-temperature, 9,253 for the herbicide application, and 8,659 for the high-temperature treatment (Supplementary Table S4). A search for common DEGs among all treatments yielded 3,219 genes. The combined herbicide and high-temperature treatment was very similar to both the herbicide application and the high-temperature treatment with 3,115 and 3,059 common DEGs, respectively (Figure 1C). The similarity of the combination treatment to the other two single treatments correlates with the dual effect of both stress factors together. The combination of herbicide and high-temperature had the highest number of up-regulated DEGs (5,377), as compared to the herbicide application (4,415) and the high-temperature treatment (3,822; Supplementary Table S4).
The pattern of gene expression was analyzed using hierarchical clustering of 3,129 common DEGs across all treatments. DEGs were classified into 25 groups by parallel plots (Supplementary Table S5), which enabled us to identify specific trends in gene expression. Assuming that over-expression of DEGs can lead to enhanced herbicide detoxification (Délye, 2013), we focused on groups that included DEGs that were up-regulated mainly in the combined treatment (groups 4 and 14 in Figures 2A,B, respectively; Supplementary Table S5). Up-regulation of genes from both plots was slightly different, as plot 4 showed greater differences in the combined treatment, as compared with plot 14 (Figure 2). Enrichment analysis was conducted to identify key biological processes related to temperature-dependent pinoxaden detoxification. Both groups included genes associated with protein, RNA, stress, and transport (Figures 2A,B). Examination of specific pathways within each category showed that both groups were enriched with genes associated with oxygenase activity (P = 0.03, P < 0.01, respectively) as well as conjugation-related pathways, including genes associated with UDP-glucosyl and glucuronyl transferase (P < 0.01) and GST (P < 0.01; Figures 2A,B). Genes associated with ABC transporters and multidrug resistance, representative of the final stage in herbicide detoxification, were also significantly enriched (P = 0.05; Figure 2B).
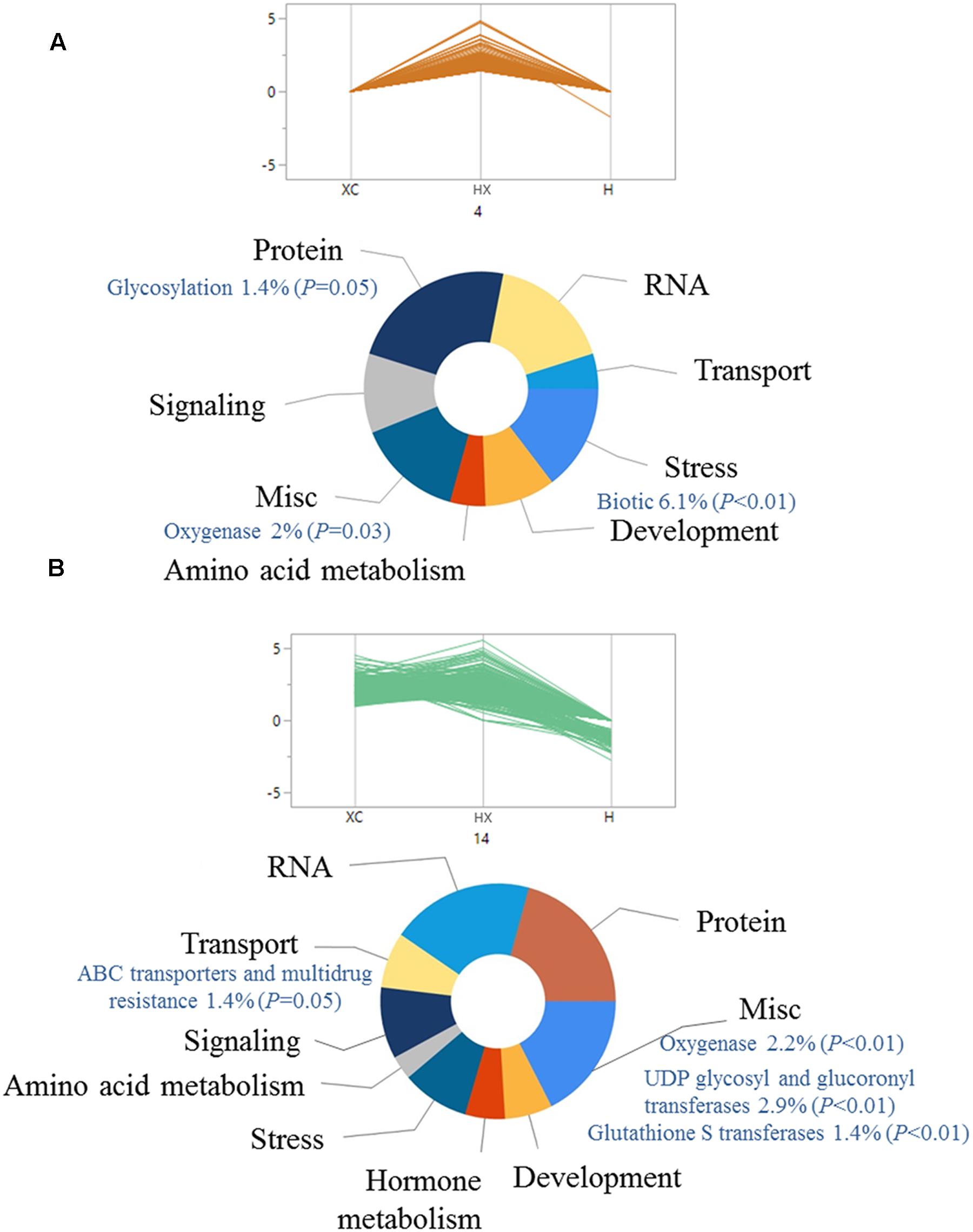
FIGURE 2. Functional classification of significantly (FDR ≤ 0.05) up-regulated genes in pinoxaden-treated plants under high-temperature regime, compared with the two other treatments (pinoxaden and high-temperature). All genes were divided into 25 groups using hierarchal clustering analysis according to the expression pattern of all three treatments. Two clusters, (A) 4 and (B) 14, were chosen as representative trends for up-regulated genes. Doughnut charts were produced based on MapMan annotations and the FunRich platform.
Herbicide metabolism is generally composed of four different phases, systematically rendering the herbicide molecule into a non-toxic product (Carvalho et al., 2009). We identified DEGs that are involved in these processes (FDR ≤ 0.05) and used qPCR to validate their expression levels under the different treatments (Supplementary Figure S1). Two DEGs, Bradi2g44160 and Bradi2g44200, were annotated as members of the CYP72A subfamily, which are associated with the first phase of herbicide metabolism. Genes of this subfamily were recently reported to be up-regulated in L. rigidum plants resistant to an ACCase inhibitor (Gaines et al., 2014). In our study, these genes were highly up-regulated (log2FC > 7.9) under both the herbicide application and the combination of herbicide and high-temperature treatments (Figure 3A and Supplementary Table S6). Other genes involved in CYP450 catalytic activity encode for NADP enzymes (Davydov, 2001), which were also found to play a role in the process of ROS quenching (Buchanan et al., 2006). NAD(P)-linked oxidoreductase (Bradi3g48197) was up-regulated with log2FC values of 3.3 and 4.6 under high-temperature and combination of herbicide and high-temperature treatments, respectively (Supplementary Table S5). Additional CYP450 genes, CYP96A10 (Bradi3g19220) and CYP79B2 (Bradi1g15695), were up-regulated only in the combination of herbicide and high-temperature, but at lower levels than the CYP72A genes (log2FC < 2.6; Figure 3A and Supplementary Table S6).

FIGURE 3. Heatmap of herbicide-associated genes among the different treatments relative to control conditions. X, pinoxaden; H, high-temperature, and HX, combination of high-temperature and pinoxaden. (A) Cytochrome P450, (B) UDP-glucosyl and glucuronyl transferase, (C) glutathione-S-transferase, and (D) peroxidase. Blue and red colors represent low and high relative expression compared with the mean value of expression across all samples, respectively. Scale is log2 of mean expression value.
With regard to the second phase of herbicide metabolism, DEGs involved in GT processes, UDP-glucosyl and glycosyl transferase (UDPGT); UDPGT88A1 (Bradi2g49067), UDPGT73B5 (Bradi2g44820), and UDPGT85A2 (Bradi3g46855) were up-regulated under herbicide application and the combination of herbicide and high-temperature (Figure 3B and Supplementary Table S7). Another major group of second-phase-related genes, GSTs, were also examined. GST DEGs (Bradi3g31777, Bradi3g61100 and others) were up-regulated in response to both herbicide treatments (herbicide application and combination of herbicide and high-temperature; Figure 3C and Supplementary Table S8). The trends observed in the RNA-seq experiment for these genes were also observed in the qPCR analysis (Supplementary Figure S1).
CYP450 activity, which produces ROS, is also regulated by the accumulation of ROS (Davydov, 2001; Narusaka et al., 2004; McIntosh et al., 2014). Peroxidases are known to act as part of ROS neutralization mechanisms (Meunier and Bernadou, 2000). A high expression of two DEGs (Bradi5g24200 and Bradi4g25660), annotated as peroxidases, was detected only under the combination of herbicide and high-temperature (log2FC > 3.5; Figure 3D and Supplementary Table S9). The multiprotein bridging factor 1C (MBF) was previously described as a key regulatory element linking ROS signaling to stress responses (Miller et al., 2008). MBF (Bradi1g37080) showed a higher expression level under the combination of herbicide and high-temperature, as compared with the other treatments (herbicide application and high-temperature treatments) with log2FC values of 8.1, 6.6 and 7.4, respectively (Supplementary Table S5). This trend can be correlated with greater activity of CYP450 enzymes, leading to more ROS neutralization, as part of the herbicide-resistance mechanism.
To learn more about the role of ROS in the herbicide-resistance mechanism, ROS staining was conducted and the patterns of ROS accumulation among the different treatments were examined. ROS quantities did not differ significantly among the different treatments at 2 and 8 HAT. However, at 24 HAT, plants subjected to the combination of herbicide and high-temperature had ROS levels that were significantly lower than those detected under control conditions (Figure 4 and Supplementary Table S10).
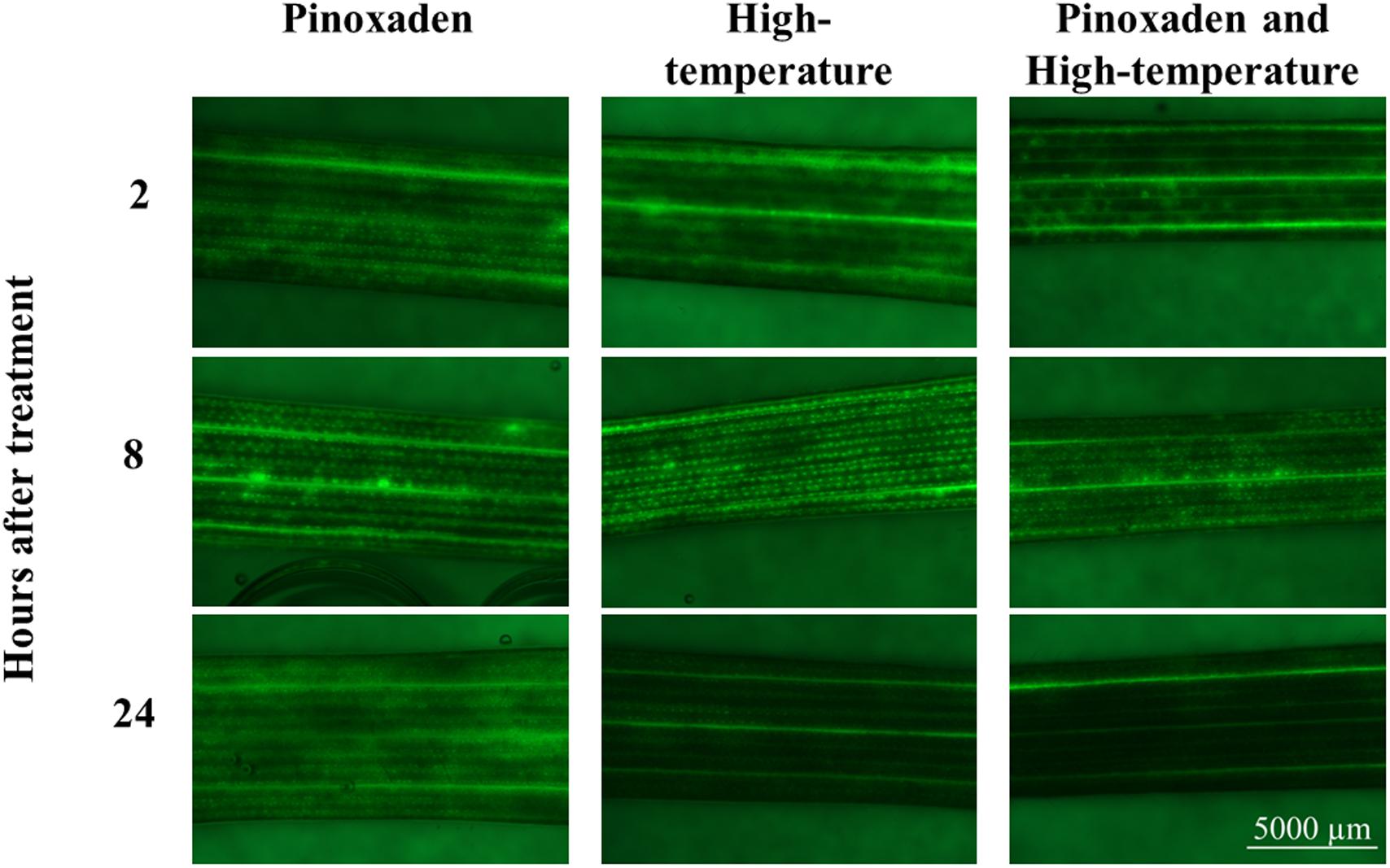
FIGURE 4. Visual presentation of the accumulation of reactive oxygen species using H2DCF-DA dye. Plants were treated with: pinoxaden, high-temperature (28/34°C) and high-temperature and pinoxaden application. Photos were taken 2, 8, and 24 h after herbicide application.
Elevated temperatures have been shown to have a direct effect on the level of NTS herbicide resistance in various weed species (Godar et al., 2015; Kleinman et al., 2015; Matzrafi et al., 2016). Here, we describe for the first time the transcriptional basis of a temperature-dependent NTS herbicide-resistance mechanism in B. hybridum and reveal the importance of temperature in herbicide-related metabolic processes. Biochemical analysis of temperature-dependent pinoxaden detoxification suggests two main stages of oxidation and glucose conjugation as key steps in the resistance mechanism (Matzrafi et al., 2016). Exploring transcriptional-enriched pathways that were differentially regulated under the combination of herbicide and high-temperature revealed up-regulation of several known herbicide defense-related genes (e.g., oxygenases, GST, GT, and ABC transporters; Figures 2A,B). The enrichment of these pathways reinforces previous findings and indicates a higher rate of pinoxaden metabolism at high-temperature. Members of the CYP72A subfamily that were previously identified as components of the mechanism of resistance to ACCase and acetolactate synthase inhibitors (Gaines et al., 2014; Saika et al., 2014) were also found to be highly expressed in pinoxaden-treated plants under both temperature regimes (Figure 3A). The abundance of genes from the CYP72A subfamily identified in different studies involving different inhibitors and weed species (Imaishi and Matumoto, 2007; Gaines et al., 2014; Iwakami et al., 2014; Saika et al., 2014) supports the assumption of their role in this mechanism of herbicide resistance. Previous studies suggested the involvement of GT activity in resistance to ACCase inhibitors (Menendez and De Prado, 1996; Brazier et al., 2002), which function mainly at the second phase of herbicide detoxification (McFadden et al., 1989; Baerson et al., 2005). Expression levels of different transcripts encoding CYP72A and UDPGT genes that were detected in our work can be associated with the formation of specific pinoxaden metabolites previously found in B. hybridum plants (Matzrafi et al., 2016). The detection of these DEGs following pinoxaden application in both temperature treatments (herbicide application and the combination of herbicide and high-temperature) suggests that the observed differences in plant response to pinoxaden are not solely due to the up-regulation of these genes. Further investigation of the role of the 2,099 DEGs that were found to be expressed only under pinoxaden treatment is needed to help to shed light on their role in herbicide response.
The catalytic activity of CYP450 enzymes includes the splitting of O2 molecules to form a hydroxylated product (Munro et al., 2013). In this process, ROS are formed and transformed into H2O (Davydov, 2001; McIntosh et al., 2014). Both NADP and peroxidases were previously suggested to play a role in CYP450 catalytic activity and the quenching of ROS (Davydov, 2001; Buchanan et al., 2006). The up-regulation of NADP and peroxidases that was unique to the combination of herbicide and high-temperature treatment can be correlated with the rapid metabolism of pinoxaden leading to herbicide resistance. The direct damage caused by ROS following the application of herbicides that function as photosystem I and II inhibitors (Fujii et al., 1990; Fuerst and Norman, 1991) is well known. In studies of other types of herbicides, this phenomenon is less familiar, but over time, as in almost any other stress response, ROS will form. The significant reduction in ROS content 24 HAT under the combination of herbicide and high-temperature, as compared with all other treatments, may indicate more efficient metabolic activity of ROS scavengers at high-temperature.
Recent studies have suggested a new approach for understanding ROS activity, as ROS have been found to act as beneficial elements in the context of many physiological functions, such as programmed cell death and cellular proliferation (Mittler, 2016). In previous studies, ROS were shown to have an effect on the expression pattern of CYP450 enzymes, inducing the expression of CYP81D8 in Arabidopsis (Desikan et al., 2001). Over-expression of additional plant defense-related genes such as GSTs [involved in antioxidant stress, lipid peroxidation (Cummins et al., 2013), detoxification of ACCase inhibitors (Edwards and Cole, 1996; Cummins et al., 1999)], and MBF, which is involved in transcriptional co-activation of ROS signaling (Miller et al., 2008), were also detected the combination of herbicide and high-temperature treatment. Herbicide metabolism processes can also be associated with several chaperoning components related to regulatory elements and activating genes. Several HSP related DEGs, mainly HSP20 (Bradi3g58590 and Bradi1g44230), were highly expressed in response to high-temperature and combination of herbicide and high-temperature (Supplementary Table S5). It can be hypothesized that this high expression is part of a general stress response (Waters, 2013), yet variation in the level of expression can suggest different activation rate of downstream resistance related components. High expression of regulatory element such as NAC related genes (Bradi1g63600 and Bradi4g44000), involved in response to biotic and abiotic stress responses (Puranik et al., 2012), was also found. Activators such as protein kinase (Bradi1g65930) and Calcium-binding (Bradi1g53830 and Bradi4g30100) related DEGs (Supplementary Table S5) were highly expressed under herbicide application. These genes can be correlated to the function of peroxidases and other enzymes related to herbicide detoxification processes (Kim Y.S. et al., 2013; Alberto et al., 2016). Our results suggest an orchestrated regulation, at the transcriptional and biochemical levels that facilitates temperature-dependent pinoxaden resistance, which is activated upon application of pinoxaden at high-temperature. These factors encourage a rapid and efficient pinoxaden detoxification that eventually results in plant survival.
Monooxygenation activity of CYP72A genes has been widely discussed in the context of herbicide resistance (Prall et al., 2016). However, only plants subjected to high-temperature survived pinoxaden treatment. Thus, the expression of CYP450 genes may not be related to their monooxygenation activity. On the other hand, temperature has been shown to play a key role in the enzymatic efficiency of CYP450 (Puntarulo and Cederbaum, 1989; Puchkaev et al., 2002). These findings help to explain the differences in plant survival between herbicide application and the combination of herbicide and high-temperature, as pinoxaden was rapidly metabolized only under high-temperature.
Our previous (Matzrafi et al., 2016) and current results on temperature-dependent herbicide resistance have emphasized the evolutionary effect of climate change on herbicide resistance and, by extension, global agriculture. We propose that temperature-dependent pinoxaden resistance may be affected more by enzymatic efficiency than by gene regulation. It is suggested that the over-expression of metabolic-related genes (CYP72A, GST and GT) is not sufficient for effective herbicide metabolism, as was previously suggested. While previous studies, quantified levels of gene expression based on comparisons between sensitive and resistant populations (i.e., different genetic background), here we used sensitive and resistant plants with the same genetic background (Accession BrI-782). It could be argued that the differences found in the current study were not detected in previous studies, due to the differences in genetic backgrounds. Thus, our strategy enabled to improve our understanding on NTS resistance mechanism under climate change. Exploring the mechanism of temperature-dependent resistance in various weed species may reveal further hub genes and up-stream regulators that affect NTS resistance.
MM designed and conducted experiments. MM and LS-M analyzed the data and drafted the paper. BR and ZP designed experiments and wrote the paper. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by the Chief Scientist of the Israeli Ministry of Agriculture. The authors would like to thank Drs. R. Hayuka and M. Sibony for their assistance with the experiments and data analysis. MM is indebted to the Teomim doctoral fellowships awards. LS-M is indebted to The Israeli President’s Scholarship for Scientific Excellence and Innovation.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01064/full#supplementary-material
Alberto, D., Serra, A.-A., Sulmon, C., Gouesbet, G., and Couée, I. (2016). Herbicide-related signaling in plants reveals novel insights for herbicide use strategies, environmental risk assessment and global change assessment challenges. Sci. Total Environ. 56, 1618–1628. doi: 10.1016/j.scitotenv.2016.06.064
An, J., Shen, X., Ma, Q., Yang, C., Liu, S., and Chen, Y. (2014). Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.). PLoS ONE 9:99940. doi:10.1371/journal.pone.0099940
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-A python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169.doi:10.1093/bioinformatics/btu638
Baerson, S. R., Sánchez-Moreiras, A., Pedrol-Bonjoch, N., Schulz, M., Kagan, I. A., Agarwal, A. K., et al. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280, 21867–21881. doi:10.1074/jbc.M500694200
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120.doi:10.1093/bioinformatics/btu170
Brazier, M., Cole, D. J., and Edwards, R. (2002). O-Glucosyltransferase activities toward phenolic natural products and xenobiotics in wheat and herbicide-resistant and herbicide-susceptible black-grass (Alopecurus myosuroides). Phytochemistry 59, 149–156. doi:10.1016/S0031-9422(01)00458-7
Buchanan, B. B., Gruissem, W., and Jones, R. (2006). Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists, 588–589.
Carvalho, S. J., De Prado, R., Nicolai, M., Ferreira, R. R., DeOliveria Figueira, A. V., and Christoffoleti, P. J. (2009). Herbicide selectivity by differential metabolism: considerations for reducing crop damages. Sci. Agric. 66, 136–142. doi:10.1590/S0103-90162009000100020
Caseley, J. C. (1989). Variation in foliar pesticide performance attributable to humidity, dew and rain effects. Asp. Appl. Biol. 21, 1215–1225.
Chauhan, B. S., and Johnson, D. E. (2011). Growth response of direct-seeded rice to oxadiazon and bispyribac-sodium in aerobic and saturated Soils. Weed Sci. 59, 119–122. doi:10.1614/WS-D-10-00075.1
Chauhan, B. S., and Opena, J. (2012). Effect of tillage systems and herbicides on weed emergence, weed growth, and grain yield in dry-seeded rice systems. Field Crop Res. 137, 56–69. doi:10.1016/j.fcr.2012.08.016
Chen, S., McElroy, J. S., Dane, F., and Peatman, E. (2015). Optimizing transcriptome assemblies for Eleusine indica leaf and seedling by combining multiple assemblies from three de novo assemblers. Plant Genome 8.doi:10.3835/plantgenome2014.10.0064
Cummins, I., Cole, D. J., and Edwards, R. (1999). A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. 18, 285–292. doi: 10.1046/j.1365-313X.1999.00452.x
Cummins, I., Wortley, D. J., Sabbadin, F., He, Z., Coxon, C. R., Straker, H. E., et al. (2013). Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. U.S.A. 110, 5812–5817. doi:10.1073/pnas.1221179110
Davydov, D. R. (2001). Microsomal monooxygenase in apoptosis: another target for cytochrome c signaling? Trends Biochem. Sci. 26, 155–160. doi:10.1016/S0968-0004(00)01749-7
Délye, C. (2013). Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 69, 176–187. doi:10.1002/ps.3318
Desikan, R., A-H-Mackerness, S., Hancock, J. T., and Neill, S. J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. doi:10.1104/pp.127.1.159
Duhoux, A., Carrère, S., Gouzy, J., and Delye, C. (2015). RNA-Seq analysis of rye-grass transcriptomic response to an herbicide inhibiting acetolactate-synthase identifies transcripts linked to non-target-site-based resistance. Plant Mol. Biol. 87, 473–487. doi:10.1007/s11103-015-0292-3
Edwards, R., and Cole, D. J. (1996). Glutathione transferases in wheat (Triticum) species with activity toward fenoxaprop-ethyl and other herbicides. Pestic. Biochem. Physiol. 54, 96–104. doi:10.1006/pest.1996.0013
FAOSTAT (2017) Food and Agriculture Organization of the United Nations-Statistics Division. Available at: http://www.fao.org/wsfs/world-summit/en
Feng, P. C. C., Tran, M., Chiu, T., Douglas Sammons, R., Heck, G. R., and Jacob, C. A. (2004). Investigations into glyphosate-resistant horseweed (Conyza canadensis): retention, uptake, translocation, and metabolism. Weed Sci. 52, 498–505. doi:10.1614/WS-03-137R
Frenkel, E., Matzrafi, M., Rubin, B., and Peleg, Z. (2017). Effects of environmental conditions on the fitness penalty in herbicide resistant Brachypodium hybridum. Front. Plant Sci. 8:94. doi:10.3389/fpls.2017.00094
Fuerst, E. P., and Norman, M. A. (1991). Interactions of herbicides with photosynthetic electron transport. Weed Sci. 39, 458–464.
Fujii, T., Yokoyama, E., Inoue, K., and Sakurai, H. (1990). The sites of electron donation of photosystem I to methyl viologen. Biochim. Biophys. Acta - Bioenerg. 1015, 41–48. doi:10.1016/0005-2728(90)90213-N
Gaines, T. A., Lorentz, L., Figge, A., Herrmann, J., Maiwald, F., Ott, M.-C., et al. (2014). RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 78, 865–876. doi:10.1111/tpj.12514
Gardin, J. A. C., Gouzy, J., Carrère, S., and Délye, C. (2015). ALOMYbase, a resource to investigate non-target-site-based resistance to herbicides inhibiting acetolactate-synthase (ALS) in the major grass weed Alopecurus myosuroides (black-grass). BMC Genomics 16:590. doi: 10.1186/s12864-015-1804-x
Godar, A. S., Varanasi, V. K., Nakka, S., Prasad, P. V. V., Thompson, C. R., and Mithila, J. (2015). Physiological and molecular mechanisms of differential sensitivity of Palmer amaranth (Amaranthus palmeri) to mesotrione at varying growth temperatures. PLoS ONE 10:0126731. doi:10.1371/journal.pone.0126731
Gornall, J., Betts, R., Burke, E., Clark, R., Camp, J., Willett, K., et al. (2010). Implications of climate change for agricultural productivity in the early twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 365, 2973–2989. doi:10.1098/rstb.2010.0158
Gressel, J., Regev, Y., Malkin, S., and Kleifeld, Y. (1983). Characterization of an s- triazine-resistant biotype of Brachypodium distachyon. Weed Sci. 31, 450–456.
Hammerton, J. L. (1967). Environmental factors and susceptibility to herbicides. Weed Sci. 15, 330–336. doi:10.2307/4041001
Heap, I. (2017). The International Survey of Herbicide-Resistant Weeds. Available at: http://www.weedscience.com
Hong, S.-Y., Park, J.-H., Cho, S.-H., Yang, M.-S., and Park, C.-M. (2011). Phenological growth stages of Brachypodium distachyon: codification and description. Weed Res. 51, 612–620. doi: 10.1111/j.1365-3180.2011.00877.x
Hong, S.-Y., Seo, P. J., Yang, M.-S., Xiang, F., and Park, C.-M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112–123. doi: 10.1186/1471-2229-8-112
HRAC (2017). Report of the Herbicide Resistance Action Committee. Available at: http://www.hracglobal.com
Imaishi, H., and Matumoto, S. (2007). Isolation and functional characterization in yeast of CYP72A18, a rice cytochrome P450 that catalyzes (ω-1)-hydroxylation of the herbicide pelargonic acid. Pestic. Biochem. Physiol. 88, 71–77.doi:10.1016/j.pestbp.2006.09.003
International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi:10.1038/nature08747
Iwakami, S., Uchino, A., Kataoka, Y., Shibaike, H., Watanabe, H., and Inamura, T. (2014). Cytochrome P450 genes induced by bispyribac-sodium treatment in a multiple-herbicide-resistant biotype of Echinochloa phyllopogon. Pest Manag. Sci. 70, 549–558. doi:10.1002/ps.3572
Kaundun, S. S. (2014). Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Sci. 70, 1405–1417. doi:10.1002/ps.3790
Kaya-Altop, E., Haghnama, K., Sariaslan, D., Phillippo, C. J., Mennan, H., and Zandstra, B. H. (2016). Long-term perennial weed control strategies: economic analyses and yield effect in hazelnut (Corylus avellana). Crop Prot. 80, 7–14. doi:10.1016/j.cropro.2015.10.022
Kellogg, E. A. (2015). Brachypodium distachyon as a genetic model system. Annu. Rev. Genet. 49, 1–20. doi:10.1146/annurev-genet-112414-055135
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, Y. S., Jung, H., Zerin, T., and Song, H. Y. (2013). Protein profiling of paraquat-exposed rat lungs following treatment with Acai (Euterpe oleracea Mart.) berry extract. Mol. Med. Rep. 7, 881–886. doi: 10.3892/mmr.2013.1259
Kleinman, Z., Ben-Ami, G., and Rubin, B. (2015). From sensitivity to resistance - factors affecting the response of Conyza spp. to glyphosate. Pest Manag. Sci. 72, 1681–1688. doi:10.1002/ps.4187
Kleinman, Z., and Rubin, B. (2016). Non-target-site glyphosate resistance in Conyza bonariensis is based on modified subcellular distribution of the herbicide. Pest Manag. Sci. 73, 246–253. doi:10.1002/ps.4293
Koger, K., and Reddy, C. (2005). Role of absorption and translocation in the mechanism of glyphosate resistance in horseweed (Conyza canadensis). Weed Sci. 53, 84–89. doi:10.1614/WS-04-102R
Lasat, M. M., DiTomaso, J. M., Hart, J. J., and Kochian, L. V. (1996). Resistance to paraquat in Hordeum glaucum is temperature dependent and not associated with enhanced apoplasmic binding. Weed Res. 36, 303–309. doi:10.1111/j.1365-3180.1996.tb01660.x
Lelieveld, J., Proestos, Y., Hadjinicolaou, P., Tanarhte, M., Tyrlis, E., and Zittis, G. (2016). Strongly increasing heat extremes in the Middle East and North Africa (MENA) in the 21st century. Clim. Change 137, 245–260. doi:10.1007/s10584-016-1665-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi:10.1186/s13059-014-0550-8
Manabe, Y., Tinker, N., Colville, A., and Miki, B. (2007). CSR1, the sole target of imidazolinone herbicide in Arabidopsis thaliana. Plant Cell Physiol. 48, 1340–1358. doi:10.1093/pcp/pcm105
Matzrafi, M., Gadri, Y., Frenkel, E., Rubin, B., and Peleg, Z. (2014). Evolution of herbicide resistance mechanisms in grass weeds. Plant Sci. 229, 43–52.doi:10.1016/j.plantsci.2014.08.013
Matzrafi, M., Seiwert, B., Reemtsma, T., Rubin, B., and Peleg, Z. (2016). Climate change increases the risk of herbicide-resistant weeds due to enhanced detoxification. Planta 244, 1217–1227. doi:10.1007/s00425-016-2577-4
McFadden, J. J., Frear, D. S., and Mansager, E. R. (1989). Aryl hydroxylation of diclofop by a cytochrome P450 dependent monooxygenase from wheat. Pestic. Biochem. Physiol. 34, 92–100. doi:10.1016/0048-3575(89)90145-4
McIntosh, J. A., Farwell, C. C., and Arnold, F. H. (2014). Expanding P450 catalytic reaction space through evolution and engineering. Curr. Opin. Chem. Biol. 19, 126–134. doi:10.1016/j.cbpa.2014.02.001
Menendez, J., and De Prado, R. (1996). Diclofop-methyl cross-resistance in a chlorotoluron-resistant biotype of Alopecurus myosuroides. Pestic. Biochem. Physiol. 56, 123–133. doi:10.1006/pest.1996.0066
Meunier, B., and Bernadou, J. (2000). Active iron-oxo and iron-peroxo species in cytochromes P450 and peroxidases; oxo-hydroxo tautomerism with water-soluble metalloporphyrins∖rmetal-oxo and metal-peroxo species in catalytic oxidations. Struct. Bond. 97, 1–35. doi:10.1007/3-540-46592-8_1
Miller, G., Shulaev, V., and Mittler, R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489. doi:10.1111/j.1399-3054.2008.01090.x
Munro, A. W., Girvan, H. M., Mason, A. E., Dunford, A. J., and Mclean, K. J. (2013). What makes a P450 tick? Trends Biochem. Sci. 38, 140–150. doi:10.1016/j.tibs.2012.11.006
Myers, S. S., Smith, M. R., Guth, S., Golden, C. D., Vaitla, B., Mueller, N. D., et al. (2017). Climate change and global food systems: potential impacts on food security and undernutrition. Annu. Rev. Public Health 38, 259–277.doi:10.1146/annurev-publhealth-031816-044356
Narusaka, Y., Narusaka, M., Seki, M., Umezawa, T., Ishida, J., Nakajima, M., et al. (2004). Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol. Biol 55, 327–342. doi:10.1007/s11103-004-0685-1
Nelson, G. C., Rosegrant, M. W., Koo, J., Robertson, R., Sulser, T., et al. (2009). Climate Change. Impact on Agriculture and Costs of Adaptation. Washington, DC: International Food Policy Research Institute.
Parry, M. L., Canziani, O. F., Palutikof, J. P., van der Linden, P. J., and Hanson, C. E. (eds) (2007). “Climate change 2007: impacts, adaptation and bulnerability,” in Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC), (Cambridge: Cambridge University Press).
Pathan, M., Keerthikumar, S., Ang, C. S., Gangoda, L., Quek, C. Y. J., Williamson, N. A., et al. (2015). FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597–2601. doi:10.1002/pmic.201400515
Pena, L. B., Barcia, R. A., Azpilicueta, C. E., Méndez, A. A. E., and Gallego, S. M. (2012). Oxidative post translational modifications of proteins related to cell cycle are involved in cadmium toxicity in wheat seedlings. Plant Sci. 196, 1–7. doi:10.1016/j.plantsci.2012.07.008
Prall, W., Hendy, O., and Thornton, L. E. (2016). Utility of a phylogenetic perspective in structural analysis of CYP72A enzymes from flowering plants. PLoS ONE 11:0163024. doi:10.1371/journal.pone.0163024
Puchkaev, A. V., Wakagi, T., and Ortiz deMontellano, P. R. (2002). CYP119 plus a Sulfolobus tokodaii strain 7 ferredoxin and 2-oxoacid: ferredoxin oxidoreductase constitute a high-temperature cytochrome P450 catalytic system. J. Am. Chem. Soc. 124, 12682–12683. doi:10.1021/ja0282036
Puntarulo, S., and Cederbaum, A. I. (1989). Temperature dependence of the microsomal oxidation of ethanol by cytochrome P450 and hydroxyl radical-dependent reactions. Arch. Biochem. Biophys. 269, 569–575. doi:10.1016/0003-9861(89)90142-2
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi:10.1016/j.tplants.2012.02.004
Robinson, M. A., Letarte, J., Cowbrough, M. J., Sikkema, P. H., and Tardif, F. J. (2015). Winter wheat (Triticum aestivum L.) response to herbicides as affected by application timing and temperature. Can. J. Plant Sci. 95, 325–333.doi:10.4141/cjps-2014-109
Roesler, K., Shintani, D., Savage, L., Boddupalli, S., and Ohlrogge, J. (1997). Targeting of the Arabidopsis homomeric acetyl-coenzyme A carboxylase to plastids of rapeseeds. Plant Physiol. 113, 75–81. doi:10.1104/pp.113.1.75
Rubin, B. (1991). “Herbicide resistance in weeds and crops, progress and prospects,” in Herbicide Resistance in Weeds and Crops, eds J. C. Caseley, G. W. Cussans, and R. K. Atkin (Oxford: Butterworth-Heinemann), 387–414. doi:10.1016/B978-0-7506-1101-5.50001-2
Saika, H., Horita, J., Taguchi-Shiobara, F., Nonaka, S., Ayako, N.-Y., Iwakami, S., et al. (2014). A novel rice cytochrome P450 gene, CYP72A31, confers tolerance to acetolactate synthase-inhibiting herbicides in rice and Arabidopsis. Plant Physiol. 166, 1232–1240. doi:10.1104/pp.113.231266
Sasaki, Y., and Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Skipsey, M., Knight, K. M., Brazier-Hicks, M., Dixon, D. P., Steel, P. G., and Edwards, R. (2011). Xenobiotic responsiveness of Arabidopsis thaliana to a chemical series derived from a herbicide safener. J. Biol. Chem. 286, 32268–32276. doi:10.1074/jbc.M111.252726
Soltani, N., Dille, A. J., Burke, I. C., Everman, W. J., VanGessel, M. J., Davis, V. M., et al. (2016). Potential corn yield losses due to weeds in North America. Weed Technol. 30, 979–984. doi:10.1614/WT-D-16-00046.1
Stott, P. (2016). How climate change affects extreme weather events. Science 352, 1517–1518. doi:10.1126/science.aaf7271
Sundby, C., Chow, W. S., and Anderson, J. M. (1993). Effects on photosystem II function, photoinhibition, and plant performance of the spontaneous mutation of serine-264 in the photosystem II reaction center D1 protein in triazine-resistant Brassica napus L. Plant Physiol. 103, 105–113. doi: 10.1104/pp.103.1.105
Tester, M., and Langridge, P. (2010). Breeding technologies to increase crop production in a changing world. Science 327, 818–822. doi:10.1126/science.1183700
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi:10.1111/j.1365-313X.2004.02016.x
Walters, B., Lum, G., Sablok, G., and Min, X. J. (2013). Genome-wide landscape of alternative splicing events in Brachypodium distachyon. DNA Res. 20, 163–171. doi:10.1093/dnares/dss041
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi:10.1093/jxb/ers355
Keywords: ACCase inhibitors, climate change, CYP450, glutathione-S-transferase, metabolic resistance, reactive oxygen species, RNA-seq, temperature-dependent response
Citation: Matzrafi M, Shaar-Moshe L, Rubin B and Peleg Z (2017) Unraveling the Transcriptional Basis of Temperature-Dependent Pinoxaden Resistance in Brachypodium hybridum. Front. Plant Sci. 8:1064. doi: 10.3389/fpls.2017.01064
Received: 05 January 2017; Accepted: 02 June 2017;
Published: 21 June 2017.
Edited by:
Demosthenis Chachalis, Benaki Phytopathological Institute, GreeceReviewed by:
Ioannis Ganopoulos, Institute of Plant Breeding and Genetic Resources-ELGO DEMETER, GreeceCopyright © 2017 Matzrafi, Shaar-Moshe, Rubin and Peleg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zvi Peleg, enZpLnBlbGVnQG1haWwuaHVqaS5hYy5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.